Abstract
Stink bugs of the superfamilies Coreoidea and Lygaeoidea establish gut symbioses with environmentally acquired bacteria of the genus Burkholderia sensu lato. In the genus Burkholderia, the stink bug-associated strains form a monophyletic clade, named stink bug-associated beneficial and environmental (SBE) clade (or Caballeronia). Recently, we revealed that members of the family Largidae of the superfamily Pyrrhocoroidea are associated with Burkholderia but not specifically with the SBE Burkholderia; largid bugs harbor symbionts that belong to a clade of plant-associated group of Burkholderia, called plant-associated beneficial and environmental (PBE) clade (or Paraburkholderia). To understand the genomic features of Burkholderia symbionts of stink bugs, we isolated two symbiotic Burkholderia strains from a bordered plant bug Physopellta gutta (Pyrrhocoroidea: Largidae) and determined their complete genomes. The genome sizes of the insect-associated PBE (iPBE) are 9.5 Mb and 11.2 Mb, both of which are larger than the genomes of the SBE Burkholderia symbionts. A whole-genome comparison between two iPBE symbionts and three SBE symbionts highlighted that all previously reported symbiosis factors are shared and that 282 genes are specifically conserved in the five stink bug symbionts, over one-third of which have unknown function. Among the symbiont-specific genes, about 40 genes formed a cluster in all five symbionts; this suggests a “symbiotic island” in the genome of stink bug-associated Burkholderia.
Keywords: insect-microbe interaction, gut symbiosis, stink bugs, bordered plant bugs, Burkholderia, environmental acquisition, evolution, comparative genomics
1. Introduction
Symbioses with microorganisms are ubiquitously found in various animals and plants in nature. Insects are the most diverse animal group in the terrestrial ecosystems and consist of over one million described species, of which almost half are associated with mutualistic microorganisms [1,2,3,4]. These microbial symbionts play important metabolic roles and provide adaptive traits such as heat tolerance and pathogen resistance that have allowed their host insects to expand habitats and become diverse [5,6,7]. Although knowledge of the biological importance of symbionts has increased, information on how beneficial symbionts evolved from environmental microorganisms are still scarce. Most characterized insect symbionts are vertically transmitted from mother to offspring and have highly adapted to host in vivo environments, which is often coupled with symbiont genome reduction [8,9]. The longer evolutionary history of most of these characterized symbiotic associations has made it difficult to reveal the evolutionary transition from non-symbionts to symbionts [9]. There are few insect systems suitable for addressing this evolutionary issue, in which both the symbionts and their closely related free-living strains are available as in vitro cultures with corresponding whole-genome sequence data.
Several phytophagous stink bugs have developed crypt-bearing symbiotic organs in their midgut that densely harbor gut symbiotic bacteria [3,10,11,12,13]. The bean bug Riptortus pedestris (superfamily Coreoidea; family Alydidae), which is the most intensively and comprehensively investigated species among stink bugs, hosts a culturable symbiont, Burkholderia insecticola (class Betaproteobacteria) in the lumen of their midgut crypts [11,14,15]. Unlike many other insects that vertically transmit their symbionts, the Burkholderia symbiont of the bean bug is not essential to their hosts and is not vertically transmitted. Instead, hatchlings of the bean bug are initially free from Burkholderia symbionts that are acquired later in life from the ambient soil environment [16]. Sorting the appropriate partner from enormously diverse soil microbiota is achieved in two steps: (i) physical selection at the narrow entrance of the crypt-bearing symbiotic organ, called the constricted region, and (ii) a subsequent microbial competition within the lumen of the symbiotic organ [17,18,19,20]. The Burkholderia symbiont has beneficial effects on the host’s fitness, although it is not essential. For example, infected stink bugs grow larger and lay a higher number of eggs than non-infected ones [16,18]. Moreover, some Burkholderia symbiont strains can degrade substances including insecticides; such insecticide-degrading Burkholderia symbiont thus confers insecticide resistance to their hosts [21,22].
The genus Burkholderia sensu lato consists of over 100 species and had been phylogenetically grouped into three main clades. Briefly, the three clades were as follows: the “Burkholderia cepacia complex and Burkholderia pseudomallei (BCC&P)” clade included many animal and plant pathogenic species; the “plant-associated beneficial and environmental (PBE)” clade included environmental species, plant growth-promoting rhizobacteria, and leguminous nodule symbionts; and the “stink bug-associated beneficial and environmental (SBE)” clade included environmental species and midgut symbionts of various stink bugs [11,23,24,25]. The symbiont of the bean bug, B. insecticola, belongs to the SBE clade. As the efforts for better taxonomic classification of this diverse bacterial genus have been continued, the genus Burkholderia has been subdivided into six nominated genera based on their phylogenetic relationships [12,26,27,28,29,30]. The BCC&P, PBE, and SBE clades currently correspond to Burkholderia sensu stricto, Paraburkholderia, and Caballeronia, respectively. The remaining three genera are Robbsia, Mycetohabitans, and Trinickia. Considering the ongoing taxonomic reclassification, in this manuscript we refer to the six subdivided genera as the genus Burkholderia. Given the close phylogenetic relationships between symbiotic and non-symbiotic environmental species and the unique, free-living life stage of the SBE Burkholderia, this stink bug-Burkholderia symbiosis provides a valuable opportunity for tracking the evolutionary transition of a free-living environmental bacterium into an insect gut symbiont.
Previous studies, including our own, have revealed that the stink bug-Burkholderia symbiosis with environmental symbiont acquisition originated at the common ancestor of the stink bug superfamilies Coreoidea, Lygaeoidea, and Pyrrhocoroidea [31,32,33,34]. However, the evolution of this interesting symbiotic system was found to be more complicated than we expected before; recent studies have shown that the symbiotic Burkholderia in the family Largidae of the Pyrrhocoroidea is phylogenetically distinct from that of the other two superfamilies [31,33]. While the symbionts of the Coreoidea and Lygaeoidea are members of the SBE clade, those of largid bugs are members of the PBE clade, wherein the largid bug-associated Burkholderia, as well as closely related species isolated from rhizosphere, form a subclade named the “insect-associated PBE (iPBE)” clade [31].
Two strains of the SBE Burkholderia have been isolated from the bean bug R. pedestris and one SBE strain from the seed bug Togo hemipterus (superfamily Lygaeoidea; family Rhyparochromidae); we also have sequenced their complete genomes [35,36,37]. However, available isolated strains of the largid bug-associated iPBE Burkholderia and their genomic information were lacking due to difficulties in culture by the simple plating of gut contents of the symbiotic organs. In this study, we succeeded in isolating iPBE Burkholderia symbionts from the midgut crypts of the bordered plant bug Physopelta gutta (superfamily Pyrrhocoroidea; family Largidae; Figure 1A) by applying the organ culture method [38]; we then determined the complete genome sequences of the two isolates. Furthermore, we performed a comparative analysis of the newly sequenced iPBE symbiont genomes with those of other Burkholderia species including the SBE Burkholderia symbionts, highlighting the genes and functions commonly conserved among stink bug-associated iPBE and SBE Burkholderia symbionts.
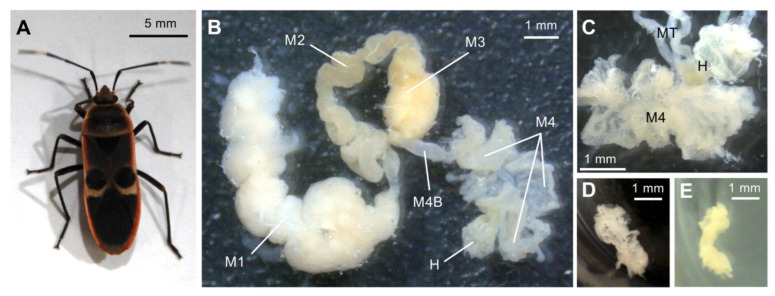
Figure 1.
The symbiont-harboring midgut crypts of Physopelta gutta. (A) An adult female P. gutta. (B) The dissected midgut of an adult female P. gutta. The Burkholderia symbiont is specifically localized in the 4th section of the crypt-bearing midgut [31]. (C) An enlarged image of the M4 section. (D,E) Pre-culture of the midgut crypts in yeast-glucose medium. The midgut crypts before pre-culture (D) and after two days of pre-culture (E). After pre-culture, the crypts became cloudy due to the bacterial growth inside the crypts. Bacterial growth was also observed outside the crypts. M1, midgut 1st section; M2, midgut 2nd section; M3, midgut 3rd section; M4, midgut 4th section; M4B, M4 bulb; MT, malpighian tubule; H, hindgut.
2. Materials and Methods
2.1. Insect Samples
Samplings of P. gutta were conducted in Okinawa prefecture, Japan in June 2016 and July 2019 (Table 1). The insects were fed with sunflower and peanut seeds and distilled water containing 0.05% ascorbic acid until examination. Burkholderia symbionts were isolated from the gut of two adult females in 2016, and of three adult females and three adult males in 2019.
Table 1.
Insect samples and isolated Burkholderia symbionts.
| Host Physopelta gutta | Symbiotic Burkholderia | |||||
|---|---|---|---|---|---|---|
| Sex 1 | Sampling Date | Sampling Location (in Okinawa, Japan) |
Dissection Date | ID | Acc. No. 2 | The Closest Species (% Identity) 3 |
| F | 28 June 2016 | Sesoko island | 1 July 2016 | PGU16 | LC547415 | Burkholderia caribensis MWAP64 (99.3) |
| F | 29 June 2016 | Kunigami | 1 July 2016 | — 4 | — 4 | — 4 |
| F | 4 July 2019 | Nago | 18 July 2019 | F1 | LC547416 | Burkholderia sabiae Br3407 (99.0) |
| F | 4 July 2019 | Nago | 18 July 2019 | F2 | LC547417 | B. caribensis MWAP64 (99.3) |
| F | 4 July 2019 | Nago | 18 July 2019 | — 4 | — 4 | — 4 |
| M | 4 July 2019 | Nago | 18 July 2019 | M1 | LC547418 | B. sabiae Br3407 (99.3) |
| M | 4 July 2019 | Nago | 18 July 2019 | PGU19 | LC547419 | B. caribensis MWAP64 (99.4) |
| M | 4 July 2019 | Nago | 18 July 2019 | — 4 | — 4 | — 4 |
1 F, Female; M, Male. 2 Accession number of sequences of 16S rRNA gene is shown. 3 The result of the EzBioCloud [39] is shown. 4 Burkholderia symbiont was not isolated.
2.2. Isolation of Symbiotic Bacteria from the Midgut Crypts
Wild-captured adult insects were dissected in phosphate-buffered saline (PBS: 137 mM NaCl, 2.7 mM KCl, 8.1 mM Na2HPO4, 1.5 mM KH2PO4 [pH 7.4]) with micro-scissors and micro-tweezers under a dissection microscope (S9 D, Leica Microsystems, Wetzlar, Germany); whole midguts were photographed with a digital microscope camera (MC170 HD, Leica Microsystems). To isolate Burkholderia symbionts from the midgut of the bordered plant bug, the organ culture method originally developed by Xu et al. [38] was employed with a simple modification. The crypt-bearing midgut 4th section (Figure 1B), where the symbionts are specifically harbored, was isolated and transferred to a tissue-culture test plate (24-well; TPP Techno Plastic Products AG, Trasadingen, Switzerland). After washing three times with PBS and two times with yeast-glucose (YG) medium (0.5% yeast extract, 0.4% glucose, 0.1% NaCl), the symbiotic organ was pre-cultured in YG medium instead of Grace’s insect cell culture medium, which was applied in the original method [38], at 27 °C for 2–3 days. Subsequently, the pre-cultured symbionts were retrieved from the symbiotic organ by pipetting, streaked on YG-agar plate, and incubated at 27 °C for 2 days. After taxonomic identification by sequencing the 16S rRNA gene, new isolates of the Burkholderia symbionts were cultured overnight in YG medium at 28 °C and stocks of them were stored in 20% glycerol at −80 °C.
2.3. Sequencing of Bacterial 16S rRNA Gene
At least six isolates for each insect sample were randomly selected and subjected to diagnostic PCR for the specific detection of the PBE Burkholderia as described in our previous study [31]. Then, positive isolates were subjected to direct PCR with TaKaRa Ex Taq Hot Start Version (TaKaRa Bio, Kusatsu, Japan) for taxonomic identification by sequencing the 16S rRNA gene. All the primers used in this study are listed in Table S1. The PCR reaction mixture was prepared according to the manufacturer’s instructions. The PCR conditions in the order of initial denaturation, denaturation, annealing, elongation, and final elongation were as follows: 94 °C for 2 min; 35 cycles of 98 °C for 10 s, 52 °C for 30 s, and 72 °C for 2 min; and 72 °C for 3 min. PCR products were cleaned up with Exonuclease I (Escherichia coli) (New England Biolabs, Ipswich, MA, USA) and Alkaline Phosphatase (Shrimp) (TaKaRa Bio). Sanger sequencing was performed by Macrogen Japan (Kyoto, Japan). Assembly of the Sanger-sequenced reads was performed as previously described [31]. The 16S rRNA gene sequences of the isolates were then queried for homology with blastn of the BLAST+ 2.5.0 [40] against the NCBI 16S rRNA sequence database to confirm whether these isolates are of Burkholderia or not. To identify the closest Burkholderia species, the sequences were subsequently analyzed with EzBioCloud [39].
2.4. Genome Sequencing, Assembly, Finishing, and Annotation
The two isolates of the iPBE symbionts, PGU16 and PGU19, were subjected to whole-genome sequencing. Genomic DNA from an overnight culture in YG medium at 28 °C was extracted using the Wizard® Genomic DNA Purification Kit (Promega, Madison, WI, USA) according to the manufacturer’s instructions. Preparation of a 20-kb library with DNA Template Prep Kit 3.0 (Pacific Biosciences of California, Menlo Park, CA, USA), PacBio RSII sequencing with the P6-C4 chemistry (Pacific Biosciences of California), and de novo assembly with HGAP v3.0 [41] were performed by Macrogen (Seoul, South Korea). To close a gap in one of the replicons (Chromosome 2) of PGU16, PCR amplification with KOD -Plus- Ver.2 (Toyobo, Osaka, Japan) and Sanger sequencing were performed with specifically designed primers (Table S1). The PCR reaction mixture was prepared according to the manufacturer’s instructions, and the PCR conditions were as follows: 94 °C for 2 min; 35 cycles of 98 °C for 10 s, 60 °C for 10 s, and 68 °C for 1.5 min; and 68 °C for 3 min. The PCR products were purified with the Wizard® SV Gel and PCR Clean-Up System (Promega) and then sequenced in both directions. The Sanger-sequenced reads were manually inspected and used for circularization. The assignment of each replicon of the circularized genomes to either chromosome or plasmid was based on the comparison at protein level with the genome of Burkholderia caribensis MWAP64 [42].
Finished genomes were annotated by using DFAST v1.2.4 [43] with default settings. The protein sequences from each genome were also queried for homology with blastp against the Clusters of Orthologous Groups of proteins (COGs) database with a threshold E-value of 10−3 and were functionally categorized [44]. Circular genomes were visualized with Circos v0.69-8 [45].
2.5. Phylogenetic Analyses
To determine the phylogenetic positions of the newly isolated Burkholderia symbionts, phylogenetic analyses with the maximum likelihood (ML) method were conducted based on the 16S rRNA gene and genome sequences. The 16S rRNA gene sequences of the isolates and uncultured Burkholderia derived from the midgut crypts of largid stink bugs [31,33], as well as those of representative Burkholderia species, were aligned by using SINA (the SILVA Incremental Aligner) v1.2.11 [46]. For genome-based phylogeny, genome sequences of the representatives Burkholderia were downloaded from GenBank (Table S2). Ninety-two up-to-date bacterial core genes (UBCGs) were extracted from the data by using the UBCG pipeline [47]. After translation into protein sequences, 70 UBCGs that were found in all the analyzed Burkholderia (Table S3) were aligned independently with L-INS-i of MAFFT v7.455 [48].
Gap-including and ambiguous sites were removed from both types of multiple alignments. Phylogenetic relationships were reconstructed with RAxML v8.2.12 [49]. The GTR + Γ model was applied for 16S rRNA gene phylogeny. The WAG + Γ model was applied for genome-based phylogeny and the parameters for each gene partition were calculated independently. The bootstrap values of 1000 replicates for all internal branches were calculated with a rapid bootstrapping algorithm [50].
2.6. Comparative Genomics
To calculate the values of pairwise digital DNA-DNA hybridization (dDDH) between the isolates and the iPBE Burkholderia species, i.e., Burkholderia phymatum, B. caribensis, Burkholderia hospita, Burkholderia terrae and Burkholderia steynii, the genome sequences were analyzed with GGDC 2.1 [51]. To perform comparative genomic analysis, protein sequences of the related Burkholderia genomes were downloaded from GenBank (Table S2). The orthologous gene clustering of the five Burkholderia symbionts of stink bugs listed in Table 2, Burkholderia phytofirmans PsJN (PBE), and B. pseudomallei K96243 (BCC&P) were performed with OrthoFinder v2.3.11 [52] with the options “-S blast -M msa -T raxml”. The COG functional categories of the orthogroups were assigned based on the functional categories of genes of PGU19 and B. insecticola RPE64, which were included in the orthogroup. The genes involved in the virulence in B. pseudomallei K96243 [53,54], in symbiotic association with plants in B. phytofirmans PsJN [55,56], and in symbiosis with stink bugs in B. insecticola RPE64 [17,57,58,59,60,61,62,63,64,65,66] were searched in the resultant orthogroups and their orthologs were identified.
Table 2.
Statistics of the complete genomes of Burkholderia symbionts of stink bugs.
| Burkholderia sp. PGU16 | Burkholderia sp. PGU19 | B. insecticola RPE64 | Burkholderia cordobensis RPE67 | Burkholderia sp. THE68 | |
|---|---|---|---|---|---|
| Clade | iPBE | iPBE | SBE | SBE | SBE |
| Host species | Physopelta gutta | Physopelta gutta | Riptortus pedestris | Riptortus pedestris | Togo hepipterus |
| (Pyrrhocoroidea: Largidae) | (Pyrrhocoroidea: Largidae) | (Coreoidea: Alydidae) | (Coreoidea: Alydidae) | (Lygaeoidea: Rhyparochromidae) | |
| Genome size (Mb) | 9.47 | 11.25 | 6.96 | 8.69 | 7.98 |
| GC content (%) | 62.4 | 61.7 | 63.2 | 63.4 | 63.1 |
| Chromosome | 2 | 2 | 3 | 3 | 4 |
| Plasmid | 3 | 3 | 2 | 3 | 2 |
| CDSs 1 | 8498 | 10,280 | 6349 | 8156 | 7278 |
| Coding ratio (%) 1 | 85.0 | 81.8 | 87.7 | 87.8 | 87.9 |
| Average protein length (aa) 1 | 315.8 | 298.2 | 320.5 | 311.8 | 321.2 |
| rRNAs 1 | 12 | 14 | 10 | 15 | 16 |
| tRNAs 1 | 71 | 70 | 69 | 73 | 68 |
| Reference | this study | this study | [15,35] | [36] | [37] |
1 Statistics calculated with DFAST [43] are shown.
2.7. Data Availability
The nucleotide sequences of 16S rRNA gene determined in this study have been deposited in DDBJ/ENA/GenBank under the accession no. LC547415 to LC547419 (see Table 1). The genome sequences of PGU16 and PGU19 have been deposited in DDBJ/ENA/GenBank under the accession no. AP023174 to AP023178 and AP023179 to AP023183, respectively. The raw sequence reads have been deposited in the DDBJ Sequence Read Archive under the accession no. DRA010199.
3. Results
3.1. Isolation of Burkholderia Symbiont from the Midgut Crypts of a Largid bug
As reported in our previous study [31], the midgut of P. gutta consisted of four morphologically distinct sections, the midgut 1st section (M1) to the crypt-bearing midgut 4th section (M4), and that its Burkholderia symbionts are specifically localized inside tubular-type crypts of M4 (Figure 1B,C). Although the crypts were translucent before pre-culture, the M4 crypts became swollen and their color changed into cream after one- to three-day pre-culture (depending on the samples) in YG medium at 27 °C (Figure 1D,E). By streak-plating the content of the pre-cultured M4 crypts, bacterial colonies were successfully cultured on YG-agar plates. Among the eight pre-cultured crypt samples, colonies were observed from only five samples.
Isolated colonies from the five samples were identified as Burkholderia by diagnostic PCR and by querying their 16S rRNA gene sequences against nucleotide databases through blastn (Table 1). According to our previous study based on culture-independent approach, where symbiotic microbiota of largid species showed some level of diversity within the individuals [31], we expected that several genetically distinguishable strains would be isolated from a single insect sample. Therefore, at least six randomly selected colonies from each insect sample were sequenced. However, no sequence diversity between colonies derived from the same insect sample was observed. This loss of diversity might have been due to the pre-culture step. The described closely related species from the five isolates were B. caribensis or Burkholderia sabiae; this is consistent with our previous study based on culture-independent methods [31], although it remains unclear how much individuals in an insect population possess these isolates. The sequences of the isolates PGU16 and F2 were identical even though the host individuals were sampled with a 3-year interval (Table 1). The lowest sequence identity between the five isolates was 98.2% (between PGU19 and F1).
3.2. Phylogenetic Affiliation of the Isolated Symbionts
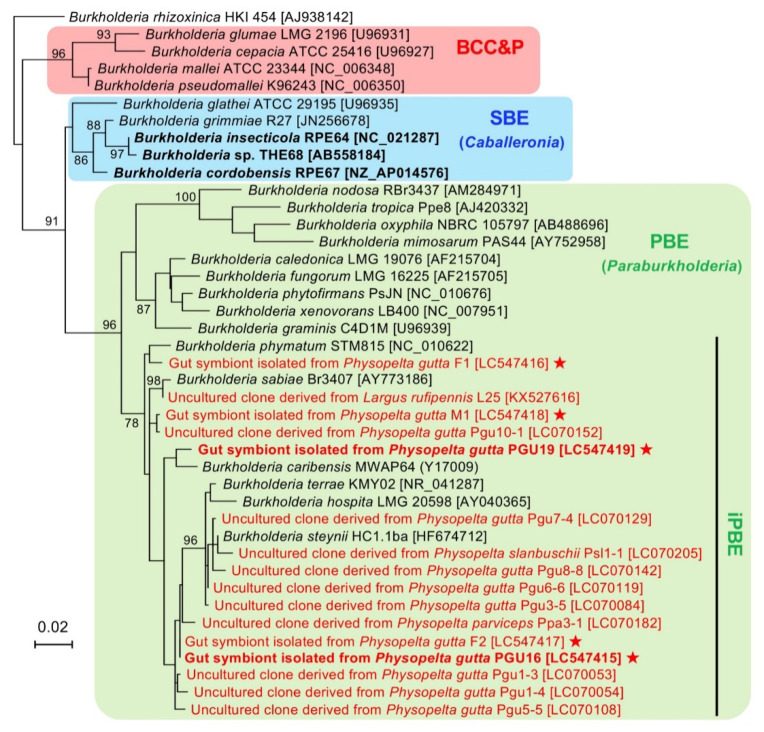
To determine the phylogenetic position of the isolated iPBE Burkholderia from P. gutta, a 16S rRNA gene-based phylogenetic analysis was performed. Figure 2 shows the ML tree of the five isolates from the largid bug and related Burkholderia species/clones based on 16S rRNA gene. All the isolates were classified under the iPBE clade, together with six species, i.e., B. caribensis, B. hospita, B. phymatum, B. sabiae, B. steynii and B. terrae, and uncultured clones identified from largid species in previous studies [31,33]. The congruence of the result with that of the culture-independent approach [31] confirmed that the five isolates were certainly gut symbiotic Burkholderia of the bordered plant bug P. gutta.
Figure 2.
Molecular phylogeny of the Burkholderia symbionts isolated from P. gutta based on 16S rRNA gene. The tree shows an ML phylogeny of the five isolates and related species/clones of the genus Burkholderia sensu lato. The multiple sequence alignment of 1320 nucleotide sites of the bacterial 16S rRNA gene was analyzed. Accession numbers in the DDBJ/EMBL/GenBank DNA database are shown in square brackets. The isolates and uncultured clones of symbiotic Burkholderia derived from largid stink bugs are shown in red. Stars indicate the isolates reported in this study. Bold indicates the Burkholderia symbionts of stink bugs with complete genome sequences. Bootstrap support values higher than 70% are shown on the internal branches. BCC&P, Burkholderia cepacia complex and Burkholderia pseudomallei clade; SBE, stink bug-associated beneficial and environmental clade; PBE, plant-associated beneficial and environmental clade; iPBE, insect-associated PBE clade.
3.3. Complete Genomes of the Isolated Symbionts
The two isolates, PGU16 and PGU19, between which the sequence identity of their 16S rRNA gene sequences was 98.7%, were then subjected to whole-genome sequencing with the PacBio RS II sequencer. The reasons why these two isolates were chosen are as follows: PGU16 is the first isolate from P. gutta; among the four isolates obtained in 2019, PGU19 shows the lowest 16S rRNA gene sequence identity against PGU16. De novo assembly of generated long-sequence-reads resulted in four circular and one non-circular replicons for PGU16 and five circular ones for PGU19. After closing a gap in the replicon of PGU16 by Sanger-sequencing, we obtained two complete genomes of the Burkholderia symbionts.
Based on the synteny between the PGU strains and their closely related species B. caribensis, which have two chromosomes and two plasmids, we assigned the five replicons to chromosomes or plasmids: the longest three replicons of each PGU strain corresponded to chromosomes 1 and 2, and plasmid 1 of B. caribensis, respectively; the remaining two replicons, which did not correspond to plasmid 2 of B. caribensis, were assigned as plasmids.
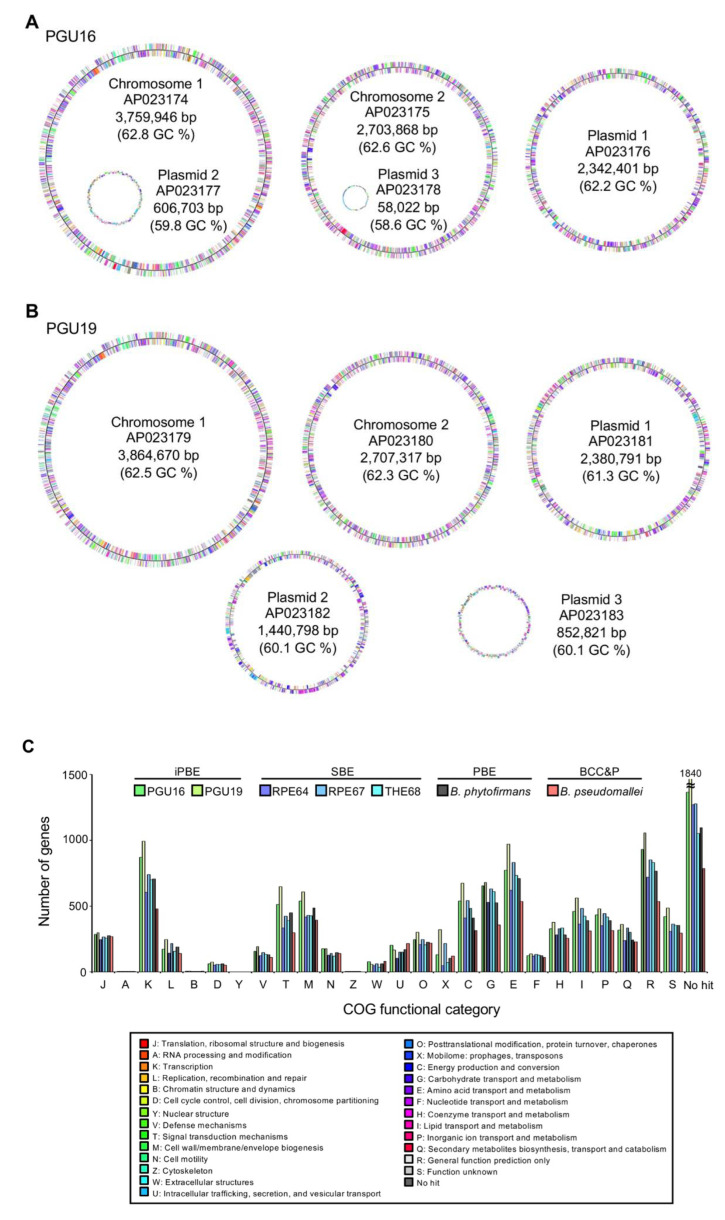
Both genomes consist of two circular chromosomes and three circular plasmids. The genome of PGU16 and PGU19 are 9,470,940 bp (141 × genome coverage) and 11,246,397 bp (86 × genome coverage) long and contain 8498 and 10,280 protein-coding genes, respectively (Figure 3A,B). Since the genome size of B. insecticola RPE64 isolated from the bean bug is 6.96 Mb [15,35], the iPBE symbionts have 1.36- and 1.62-times larger genome than RPE64. The statistics of these genomes and those of the SBE Burkholderia symbionts of other stink bugs are summarized in Table 2. The comparison of COG functional categories between the new isolates and other Burkholderia is shown in Figure 3C.
Figure 3.
Complete genomes of the isolated iPBE Burkholderia symbionts. Circular visualization of the genomes of Burkholderia sp. strain PGU16 (A) and Burkholderia sp. strain PGU19 (B). Colors indicate the Clusters of Orthologous Group (COG) functional categories of the genes on the genomes. (C) The number of genes classified into each COG functional category in the five symbionts of stink bugs, Burkholderia phytofirmans PsJN and B. pseudomallei K96243. A gene may be classified into two or more categories; therefore, the sum of numbers here might be different from the total number of genes in a genome. The description of each COG category is shown in the bottom. “No hit” indicates no homologous sequence was detected in the COG database with blastp.
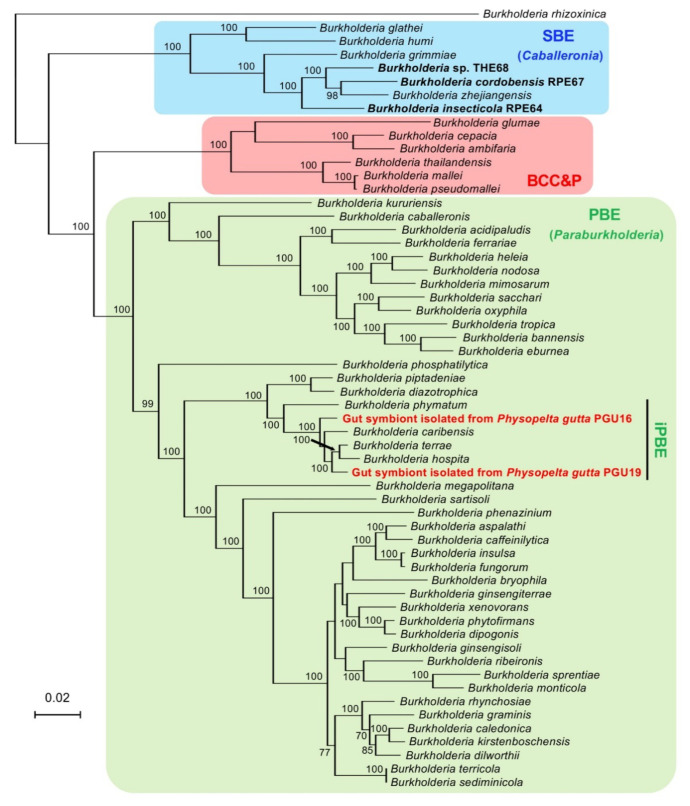
The genome-based molecular phylogeny shown in Figure 4 supported that the iPBE clade is a monophyletic subclade within the PBE clade and includes the two isolates from the bordered plant bug. The dDDH value between the isolates PGU16 and PGU19 was 47.7%. The highest dDDH value between the isolates and the described iPBE Burkholderia species with complete genomes was 57.7% (PGU19 vs. B. terrae). These values are below the proposed criterion [67], indicating that these isolates are of different species and different from any other described species of the Burkholderia iPBE clade.
Figure 4.
Genome-based molecular phylogeny of the Burkholderia symbionts isolated from P. gutta. The tree shows an ML phylogeny of the two isolates and related species of the genus Burkholderia sensu lato. The multiple sequence alignment of 22,112 amino acids of the 70 UBCGs was analyzed. Accession numbers in the DDBJ/EMBL/GenBank DNA database are shown in Table S2. Isolates of the symbiotic Burkholderia derived from the largid stink bug are shown in red. Bold indicates the Burkholderia symbionts of stink bugs. Bootstrap support values higher than 70% are shown on the internal branches. BCC&P, B. cepacia complex and B. pseudomallei clade; SBE, stink bug-associated beneficial and environmental clade; PBE, plant-associated beneficial and environmental clade; iPBE, insect-associated PBE clade.
3.4. Virulence and Symbiosis Factors Conserved in the iPBE and SBE Burkholderia Symbionts
To characterize the genomes of the stink bug symbionts that belong to the iPBE clade and/or the SBE clade of Burkholderia, comparative genomic analysis was performed. The seven complete genomes, including the five symbionts of stink bugs listed in Table 2, were compared and their orthologous genes were clustered into orthogroups. The other two genomes were B. pseudomallei K96243 and B. phytofirmans PsJN which were chosen as well-characterized representatives of BCC&P and PBE clades, respectively. In total, 53,870 protein sequences from seven genomes were clustered into 12,556 orthogroups that includes 3795 singleton genes without any orthologous gene in the analyzed data set.
We first searched whether the five symbiont genomes carry the representative virulence or symbiosis factors previously reported in pathogenic species of BCC&P, such as B. pseudomallei; plant growth-promoting rhizobacteria (PGPR) of PBE, such as B. phytofirmans; and bean bug-associated SBE, such as B. insecticola [17,53,54,55,56,57,58,59,60,61,62,63,64,65,66] (Table 3).
Table 3.
Presence/absence of the virulence and symbiosis factors in seven Burkholderia genomes.
| iPBE | SBE | BCC&P | PBE | |||||
|---|---|---|---|---|---|---|---|---|
| Functions 1 | Factors 2 | Burkholderia sp. PGU16 | Burkholderia sp. PGU19 | B. insecticola RPE64 | B. cordobensis RPE67 | Burkholderia sp. THE68 | B. pseudomallei K96243 | B. phytofirmans PsJN |
| Flagella | VPS | + | + | + | + | + | + | + |
| Chemotaxis | VPS | + | + | + | + | + | + | + |
| T3SS 3 | VP | + | + | - | - | - | + | - |
| T6SS 3 | VP | + | + | + | + | + | + | + |
| Capsular polysaccharide synthesis and export cluster | V | - | - | - | - | - | + | - |
| LPS biosynthetic cluster 3 | V | + | + | + | + | + | + | + |
| Phospholipase C | V | + | + | + | + | + | + | + |
| Metalloprotease A | V | - | - | - | - | - | + | - |
| MucD Ser protease | V | + | + | + | + | + | + | + |
| Type 1 fimbriae | V | + | + | + | + | + | + | + |
| Type 4 pili | VP | + | - | + | + | + | + | + |
| Tad pili 3 | V | + | + | + | + | + | + | + |
| T4SS 3 | VP | + | - | - | - | + | - | + |
| Siderophore biosynthesis | P | + | - | + | + | + | + | + |
| ACC deaminase 3 | P | + | + | + | + | + | + | + |
| AHL-based quorum sensing 3 | VP | + | + | - | - | - | + | + |
| 3-OH-PAME-based quorum sensing 3 | P | - | - | - | - | - | - | - |
| PHA biosynthesis 3 | S | + | + | + | + | + | + | + |
| Purine biosynthesis | S | + | + | + | + | + | + | + |
| Allantoin and urea metabolic pathway | S | + | + | + | + | + | + | + |
1 The following symbiotic genes reported in the stink bug-Burkholderia symbiosis were conserved in all the Burkholderia genomes and were omitted: uppP, amiC, wbxA, waaF, waaC. 2 V, Virulence; P, Plant growth promotion; S, Symbiosis with the bean bug. 3 T3SS, Type three secretion system; T6SS, Type six secretion system; LPS, Lipopolysaccharide; Tad, Tight adherence; T4SS, Type four secretion system; ACC, 1-aminocyclopropane-1-carboxylic acid; AHL, N-acyl homoserine lactone; and 3-OH-PAME, 3-hydroxypalmitate methyl ester; PHA, Polyhydroxyalkanoates.
The five symbionts consistently possessed the following factors that are commonly important in both pathogenesis and symbiosis: flagella, chemotaxis, lipopolysaccharide (LPS) biosynthetic cluster, phospholipase C, MucD Ser protease, 1-aminocyclopropane-1-carboxylic acid (ACC) deaminase, and all the symbiosis factors reported in B. insecticola. Type six secretion system (T6SS), type one fimbriae, and tight adherence (Tad) pili were also commonly found in all the five symbionts; however, the symbiont genomes possessed clusters of more than one for these factors and they did not show one-to-one orthologous correspondence. On the other hand, the capsular polysaccharide synthesis and export cluster and 3-hydroxypalmitate methyl ester (3-OH-PAME)-based quorum sensing system were missing in the five symbionts. Notably, although the two iPBE symbionts possessed type three secretion system (T3SS) and N-acyl homoserine lactone (AHL)-based quorum sensing system, the three SBE symbionts did not. The genes related to nodulation and nitrogen fixation, nodA and nifH, of B. phymatum STM 815 were not found in PGU16, PGU19, and the SBE symbionts.
3.5. Identification of the Genes Specifically Conserved in the iPBE and SBE Burkholderia Symbionts
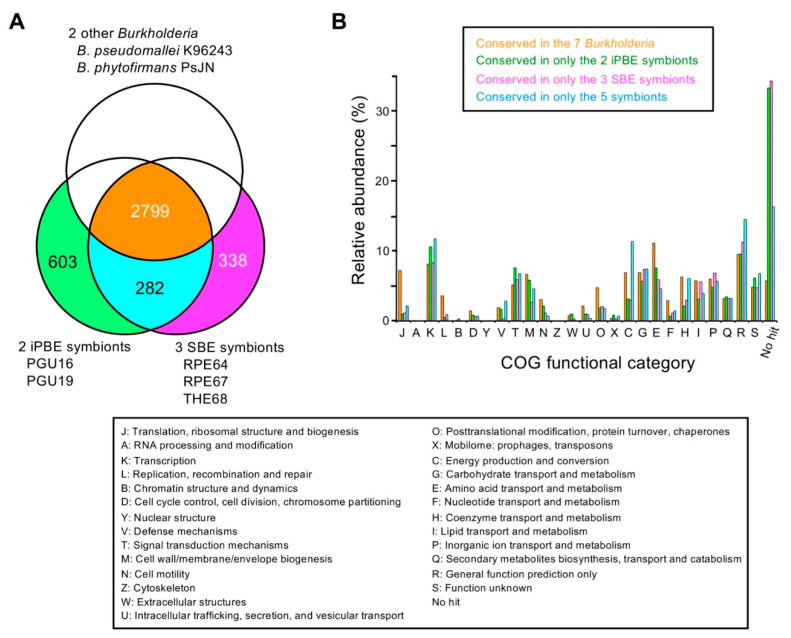
To distinguish between the specific genes conserved among the iPBE and SBE symbionts, we then classified the 12,556 orthogroups according to their presence in genomes. The analysis showed that the seven Burkholderia symbionts shared 2799 orthogroups and that 282 orthogroups were conserved in the five insect symbionts but were absent in the other two species (Figure 5A). The 282 orthogroups conserved in the five insect symbionts are listed in Table S4. The number of orthogroups conserved in only the iPBE and SBE symbionts were 603 and 338, respectively (Figure 5A).
Figure 5.
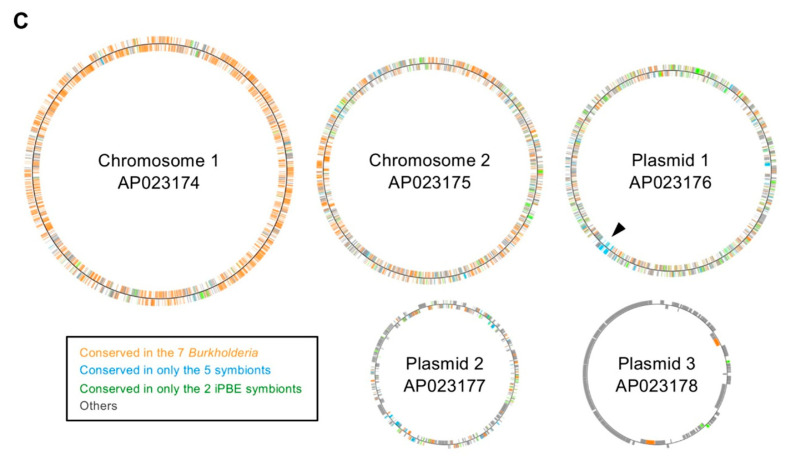
Comparative genomics for highlighting conserved genes among the five Burkholderia symbionts of stink bugs. (A) Venn diagram for the comparison between the five symbionts and the other two Burkholderia. The number of orthogroups is shown. One orthogroup may include two or more genes from a genome. (B) Relative abundance of the conserved orthogroups classified into each COG functional category. Since one orthogroup may be classified into two or more categories, the sum of relative abundance can be over 1. The description of each COG category is shown below the panels A and B. (C). The distribution of the conserved genes on the genome of the iPBE symbiont PGU16. The arrowhead indicates the region that contains the cluster of conserved genes in the five symbionts.
Figure 5B shows the comparison of COG functional categories between the above four classes of orthogroups, i.e., conserved in the seven species, conserved in only the five insect symbionts, conserved in only the two iPBE symbionts, and conserved in only the three SBE symbionts. Almost half of the orthogroups conserved in only the iPBE and SBE symbionts were functionally unknown. Figure 5C shows the distribution of the genes classified into the four classes on the genome of the iPBE symbiont PGU16. The chromosome mainly harbors genes conserved in all the seven genomes, as expected. On the other hand, most of the genes conserved in only the five symbionts and the iPBE symbionts were located on the plasmids. Notably, on plasmid 1, the region that harbors the genes conserved in the five insect symbionts was found. The conserved gene region on plasmid 1 consisted of about 40 genes (PPGU16_69460–PPGU16_69920) and interestingly, this 40-gene cluster was also conserved on the genomes of the other four insect symbionts (Table S4 and Figure S1), which is similar to the “symbiosis island” reported in rhizobia [68,69]. This gene cluster contained coenzyme pyrroloquinoline-quinone (PQQ) synthase and ribulose 1,5-bisphosphate carboxylase/oxygenase (Rubisco) (Table S4). Moreover, a partial duplicate of this conserved region was found only on plasmid 2 of the iPBE symbiont PGU19 (Table S4 and Figure S1).
4. Discussion
In this study, we first isolated Burkholderia symbionts from the largid stink bug, P. gutta, which belong to the iPBE clade of the genus Burkholderia. We subsequently determined the complete genomes of two isolated symbionts of P. gutta and then compared them with the whole genomes of other three SBE symbionts. Studies on an SBE symbiont, B. insecticola RPE64, of the bean bug have revealed that several symbiosis factors, such as genes related to motility and cell wall synthesis, are important for colonization in the midgut crypts [17,57,58,59,60,61,62,63,64,65,66]. The presence of all these symbiosis factors was confirmed in five stink bug-associated Burkholderia symbionts, which include two iPBE symbionts (Table 3). This result suggests that: (i) the gut luminal environments are fundamentally similar between the largid bug and the bean bug; and thus (ii) the iPBE symbionts can colonize the gut of largid bug upon the similar mechanism reported in B. insecticola [11,20]. Our recent comparative transcriptome analysis between the cultured and the gut-colonizing B. insecticola suggests that the symbiont plays a role in recycling host’s nitrogen wastes, such as allantoin and urea, and providing the host bean bug with essential nutrients [58]. Considering that the iPBE symbionts carry the pathways for allantoin and urea metabolisms, nitrogen recycling may be a metabolic function of the symbionts in largid bugs. It should be noted that the previously-reported symbiosis factors were also conserved in the non-symbiont Burkholderia species, B. pseudomallei and B. phytofirmans (Table 3). Considering that chinch bugs, Blissus insularis and Cavelerius saccharivorus (superfamily Lygaeoidea; family Blissidae), are associated with all BCC&P, PBE, and SBE Burkholderia [25,70] and that the genus Pandoraea, the sister group of Burkholderia sensu lato, can be beneficially associated with the bean bug under the laboratory condition [18], key symbiosis factors for association with stink bugs might be originated at the common ancestor of Burkholderia sensu lato or earlier.
Our comparative analysis also identified 282 orthogroups that were specifically conserved in five insect symbionts (Figure 5A and Table S4). These specifically conserved genes, of which more than one third are functionally unknown (Figure 5B), may include important genes that are essential for environmental adaptation inside insect guts, although the 282 orthogroups do not contain any of the symbiosis factors reported in the bean bug symbiosis to date. Interestingly, a gene mapping analysis revealed a conserved gene region that consists of about 40 genes in a plasmid and a second chromosome of the symbionts (Figure 5C and Figure S1). This conserved region, despite its relatively small size, is reminiscent of the “symbiosis island” or “symbiotic plasmid” of the nodule-forming rhizobia symbionts in leguminous plants [68,69]. Although we have identified only 527 genes of B. insecticola RPE64 (~8% in the whole genome) that are upregulated in the midgut crypts of the bean bug compared to those cultured conditions [58], more than half of the genes of this “symbiont-conserved gene cluster” were among these genes (Table S4). Furthermore, the conserved region contains functionally interesting genes: PQQ-dependent dehydrogenase and a PQQ biosynthesis cluster that consists of four genes. In Drosophila, the PQQ-dependent alcohol dehydrogenase activity of a commensal gut bacterium, Acetobacter pomorum, has been experimentally demonstrated; it modulates insulin/insulin-like growth factor signaling in the host to regulate metabolic homeostasis of the insect host. Moreover, gene-deletion mutants of the PQQ biosynthesis and PQQ-dependent alcohol dehydrogenase caused retarded growth and smaller body size [71]. Considering the genomic conservation in the five insect symbionts and higher expression under in vivo conditions, it is plausible that these PQQ genes play a similar function in the stink bug-Burkholderia symbiosis. It would be of great interest to test this possibility in the bean bug model system and the symbiosis between largid bug and iPBE symbionts.
From the difference between the iPBE and SBE symbionts, it should be noted that T3SS and AHL-based quorum sensing are found only in the iPBE symbionts (Table 3). In the genome of the notorious human pathogen B. pseudomallei K96243, three T3SS clusters have been known; one of them plays a role in virulence, whereas the function of other two clusters in virulence has been less characterized [53,72]. Although the biological functions of the two T3SS clusters remain unclear, it is notable that the T3SS cluster found in the iPBE symbionts is orthologous to one of these less characterized T3SS. Although a previous comparative genomic study by Angus et al. [56] reported that PBE Burkholderia generally lack T3SS clusters, a recent genomic study revealed that T3SS clusters that are orthologous to the less characterized one are conserved in the iPBE species, B. caribensis, B. terrae, and B. hospita [73]. Apart from Burkholderia, T3SS plays a pivotal function in diverse symbiotic systems, such as the legume-Rhizobium and weevil-Sodalis symbioses [74,75]. Probably, the presence of the T3SS cluster is a genomic signature of the iPBE Burkholderia and plays a specific role in the symbiosis. Quorum sensing is also known as an important factor in various symbiotic systems, including the squid-Vibrio symbiosis and root nodule symbiosis of leguminous plants [76,77,78]. Therefore, these factors possibly play important roles also in the largid bug-Burkholderia gut symbiosis. If we are to set-up an experimental system for the bordered plant bug symbiosis, these factors, i.e., T3SS and quorum sensing, would be the first candidates for functional analyses.
The iPBE clade was highly supported in the genome-based phylogeny (Figure 4) and includes not only the stink bug symbionts isolated in this study but also plant-associated species such as B. caribensis and B. phymatum [79,80,81]. This result suggests that other members of this clade are also associated with largid bugs. Such a polygamous nature of the iPBE Burkholderia might be underpinned by their remarkably large genome. Burkholderia species generally possess larger genomes, with an average of 7–8 Mb, than other bacteria [12,82]. Meanwhile, iPBE species possess even larger genomes than other Burkholderia species: the genome size of B. terrae DSM 17804T, B. hospita DSM 17164T, B. caribensis MWAP64T, and B. phymatum STM815T are 10.1 Mb, 11.5 Mb, 9.0 Mb, and 8.7 Mb, respectively [42,73,83]. This is also true for the PGU strains (Table 2). As mentioned in previous studies, larger genomes of Burkholderia may be advantageous for their adaptability [23,84]. At this stage, however, it is unknown whether the plant-associated iPBE species can be also associated with the largid bugs; a comprehensive inoculation test as shown in the bean bug R. pedestris should be performed [18]. It should also be tested whether the iPBE symbionts of largid bugs can be a symbiont of plants, although the PGU strains lack nodA and nifH genes that would have enabled them to induce nodules in leguminous plants.
Due to the advantage that the Burkholderia symbiont can be genetically manipulated, the bean bug-Burkholderia symbiosis has been widely recognized as one of the promising models for experimentally elucidating host-symbiont molecular interactions [11,12,20]. As several species of the iPBE clade Burkholderia can be genetically manipulated [85,86], the genetic manipulation of the iPBE symbionts of largid bug (i.e., the PGU strains) is probably applicable as well. As we start to propagate the bordered plant bug in laboratories, we would be able to experimentally examine the functions of the candidate genes in the largid bug-Burkholderia symbiosis in the future. The new symbiotic system, as well as the newly isolated strains with complete genome, would be a useful resource for revealing the complex evolution of insect-microbe symbiosis maintained by environmental symbiont acquisition.
Acknowledgments
The authors would like to thank Yu Matsuura for his help with insect sampling; and Jun Fukushima, Yoichiro Shimura, and Makoto Abe for granting us permission to use facilities. Computations were partially performed on the NIG supercomputer at Research Organization of Information and Systems (ROIS), National Institute of Genetics (NIG), Japan.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/7/744/s1, Table S1: Primers used in this study, Table S2: Burkholderia genomes analyzed in this study, Table S3: The 70 UBCGs used for genome-based phylogenetic analysis, Table S4: Orthogroups specifically conserved in five Burkholderia symbionts, Figure S1: The distribution of the conserved genes on the genomes of the iPBE symbiont PGU19 (A) and the SBE symbiont B. insecticola RPE64 (B).
Author Contributions
K.T. designed the study, performed the experiments, analyzed and interpreted the data, and drafted the manuscript. Y.K. designed the study, interpreted the data, and edited the manuscript. All authors have read and agree to the published version of the manuscript.
Funding
This study was supported by JSPS KAKENHI grant no. 19K15850 (K.T.) and no. 20H03303 (Y.K. and K.T.).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Buchner P. Endosymbiosis of Animals With Plant Microorganisms. Interscience Publishers; New York, NY, USA: 1965. [Google Scholar]
- 2.Bourtzis K., Miller T.A. In: Insect Symbiosis. Bourtzis K., Miller T.A., editors. CRC Press; Boca Raton, FL, USA: 2003. [Google Scholar]
- 3.Engel P., Moran N.A. The gut microbiota of insects—Diversity in structure and function. FEMS Microbiol. Rev. 2013;37:699–735. doi: 10.1111/1574-6976.12025. [DOI] [PubMed] [Google Scholar]
- 4.Douglas A.E. Multiorganismal insects: Diversity and function of resident microorganisms. Annu. Rev. Entomol. 2015;60:17–34. doi: 10.1146/annurev-ento-010814-020822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Janson E.M., Stireman J.O., Singer M.S., Abbot P. Phytophagous insect–microbe mutualisms and adaptive evolutionary diversification. Evolution. 2008;62:997–1012. doi: 10.1111/j.1558-5646.2008.00348.x. [DOI] [PubMed] [Google Scholar]
- 6.Brucker R.M., Bordenstein S.R. Speciation by symbiosis. Trends Ecol. Evol. 2012;27:443–451. doi: 10.1016/j.tree.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Sudakaran S., Kost C., Kaltenpoth M. Symbiont acquisition and replacement as a source of ecological innovation. Trends Microbiol. 2017;25:375–390. doi: 10.1016/j.tim.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 8.Moran N.A. Accelerated evolution and Muller’s rachet in endosymbiotic bacteria. Proc. Natl. Acad. Sci. USA. 1996;93:2873–2878. doi: 10.1073/pnas.93.7.2873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moran N.A., McCutcheon J.P., Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu. Rev. Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 10.Kikuchi Y., Hosokawa T., Fukatsu T. An ancient but promiscuous host-symbiont association between Burkholderia gut symbionts and their heteropteran hosts. ISME J. 2011;5:446–460. doi: 10.1038/ismej.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Takeshita K., Kikuchi Y. Riptortus pedestris and Burkholderia symbiont: An ideal model system for insect–microbe symbiotic associations. Res. Microbiol. 2017;168:175–187. doi: 10.1016/j.resmic.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 12.Kaltenpoth M., Flórez L.V. Versatile and dynamic symbioses between insects and Burkholderia bacteria. Annu. Rev. Entomol. 2020;65:145–170. doi: 10.1146/annurev-ento-011019-025025. [DOI] [PubMed] [Google Scholar]
- 13.Ohbayashi T., Itoh H., Lachat J., Kikuchi Y., Mergaert P. Burkholderia gut symbionts associated with European and Japanese populations of the dock bug Coreus marginatus (Coreoidea: Coreidae) Microbes Environ. 2019;34:219–222. doi: 10.1264/jsme2.ME19011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kikuchi Y., Meng X.Y., Fukatsu T. Gut symbiotic bacteria of the genus Burkholderia in the broad-headed bugs Riptortus clavatus and Leptocorisa chinensis (Heteroptera: Alydidae) Appl. Environ. Microbiol. 2005;71:4035–4043. doi: 10.1128/AEM.71.7.4035-4043.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeshita K., Tamaki H., Ohbayashi T., Meng X.Y., Sone T., Mitani Y., Peeters C., Kikuchi Y., Vandamme P. Burkholderia insecticola sp. nov., a gut symbiotic bacterium of the bean bug Riptortus pedestris. Int. J. Syst. Evol. Microbiol. 2018;68:2370–2374. doi: 10.1099/ijsem.0.002848. [DOI] [PubMed] [Google Scholar]
- 16.Kikuchi Y., Hosokawa T., Fukatsu T. Insect-microbe mutualism without vertical transmission: A stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl. Environ. Microbiol. 2007;73:4308–4316. doi: 10.1128/AEM.00067-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ohbayashi T., Takeshita K., Kitagawa W., Nikoh N., Koga R., Meng X.Y., Tago K., Hori T., Hayatsu M., Asano K., et al. Insect’s intestinal organ for symbiont sorting. Proc. Natl. Acad. Sci. USA. 2015;112:E5179–E5188. doi: 10.1073/pnas.1511454112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Itoh H., Jang S., Takeshita K., Ohbayashi T., Ohnishi N., Meng X.Y., Mitani Y., Kikuchi Y. Host–symbiont specificity determined by microbe–microbe competition in an insect gut. Proc. Natl. Acad. Sci. USA. 2019;116:22673–22682. doi: 10.1073/pnas.1912397116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kikuchi Y., Ohbayashi T., Jang S., Mergaert P. Burkholderia insecticola triggers midgut closure in the bean bug Riptortus pedestris to prevent secondary bacterial infections of midgut crypts. ISME J. 2020;14:1627–1638. doi: 10.1038/s41396-020-0633-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohbayashi T., Mergaert P., Kikuchi Y. Host-symbiont specificity in insects: Underpinning mechanisms and evolution. Adv. Insect Physiol. 2020 doi: 10.1016/bs.aiip.2020.03.002. [DOI] [Google Scholar]
- 21.Kikuchi Y., Hayatsu M., Hosokawa T., Nagayama A., Tago K., Fukatsu T. Symbiont-mediated insecticide resistance. Proc. Natl. Acad. Sci. USA. 2012;109:8618–8622. doi: 10.1073/pnas.1200231109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itoh H., Tago K., Hayatsu M., Kikuchi Y. Detoxifying symbiosis: Microbe-mediated detoxification of phytotoxins and pesticides in insects. Nat. Prod. Rep. 2018;35:434–454. doi: 10.1039/C7NP00051K. [DOI] [PubMed] [Google Scholar]
- 23.Mahenthiralingam E., Urban T.A., Goldberg J.B. The multifarious, multireplicon Burkholderia cepacia complex. Nat. Rev. Microbiol. 2005;3:144–156. doi: 10.1038/nrmicro1085. [DOI] [PubMed] [Google Scholar]
- 24.Suárez-Moreno Z.R., Caballero-Mellado J., Coutinho B.G., Mendonça-Previato L., James E.K., Venturi V. Common features of environmental and potentially beneficial plant-associated Burkholderia. Microb. Ecol. 2012;63:249–266. doi: 10.1007/s00248-011-9929-1. [DOI] [PubMed] [Google Scholar]
- 25.Itoh H., Aita M., Nagayama A., Meng X.Y., Kamagata Y., Navarro R., Hori T., Ohgiya S., Kikuchi Y. Evidence of environmental and vertical transmission of Burkholderia symbionts in the oriental chinch bug, Cavelerius saccharivorus (Heteroptera: Blissidae) Appl. Environ. Microbiol. 2014;80:5974–5983. doi: 10.1128/AEM.01087-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yabuuchi E., Kosako Y., Oyaizu H., Yano I., Hotta H., Hashimoto Y., Ezaki T., Arakawa M. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas homology group II to the new genus, with the type species Burkholderia cepacia (Palleroni and Holmes 1981) comb. nov. Microbiol. Immunol. 1992;36:1251–1275. doi: 10.1111/j.1348-0421.1992.tb02129.x. [DOI] [PubMed] [Google Scholar]
- 27.Sawana A., Adeolu M., Gupta R.S. Molecular signatures and phylogenomic analysis of the genus Burkholderia: Proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring env. Front. Genet. 2014;5:429. doi: 10.3389/fgene.2014.00429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dobritsa A.P., Samadpour M. Transfer of eleven species of the genus Burkholderia to the genus Paraburkholderia and proposal of Caballeronia gen. nov. to accommodate twelve species of the genera Burkholderia and Paraburkholderia. Int. J. Syst. Evol. Microbiol. 2016;66:2836–2846. doi: 10.1099/ijsem.0.001065. [DOI] [PubMed] [Google Scholar]
- 29.Lopes-Santos L., Castro D.B.A., Ferreira-Tonin M., Corrêa D.B.A., Weir B.S., Park D., Ottoboni L.M.M., Neto J.R., Destéfano S.A.L. Reassessment of the taxonomic position of Burkholderia andropogonis and description of Robbsia andropogonis gen. nov., comb. nov. Antonie Van Leeuwenhoek. 2017;110:727–736. doi: 10.1007/s10482-017-0842-6. [DOI] [PubMed] [Google Scholar]
- 30.Estrada-de los Santos P., Palmer M., Chávez-Ramírez B., Beukes C., Steenkamp E., Briscoe L., Khan N., Maluk M., Lafos M., Humm E., et al. Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): Implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes. 2018;9:389. doi: 10.3390/genes9080389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeshita K., Matsuura Y., Itoh H., Navarro R., Hori T., Sone T., Kamagata Y., Mergaert P., Kikuchi Y. Burkholderia of plant-beneficial group are symbiotically associated with bordered plant bugs (Heteroptera: Pyrrhocoroidea: Largidae) Microbes Environ. 2015;30:321–329. doi: 10.1264/jsme2.ME15153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sudakaran S., Retz F., Kikuchi Y., Kost C., Kaltenpoth M. Evolutionary transition in symbiotic syndromes enabled diversification of phytophagous insects on an imbalanced diet. ISME J. 2015;9:2587–2604. doi: 10.1038/ismej.2015.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gordon E.R.L., McFrederick Q., Weirauch C. Phylogenetic evidence for ancient and persistent environmental symbiont reacquisition in Largidae (Hemiptera: Heteroptera) Appl. Environ. Microbiol. 2016;82:7123–7133. doi: 10.1128/AEM.02114-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kuechler S.M., Fukatsu T., Matsuura Y. Repeated evolution of bacteriocytes in lygaeoid stinkbugs. Environ. Microbiol. 2019;21:4378–4394. doi: 10.1111/1462-2920.14804. [DOI] [PubMed] [Google Scholar]
- 35.Shibata T.F., Maeda T., Nikoh N., Yamaguchi K., Oshima K., Hattori M., Nishiyama T., Hasebe M., Fukatsu T., Kikuchi Y., et al. Complete genome sequence of Burkholderia sp. strain RPE64, bacterial symbiont of the bean bug Riptortus pedestris. Genome Announc. 2013;1:e00441-13. doi: 10.1128/genomeA.00441-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takeshita K., Shibata T.F., Nikoh N., Nishiyama T., Hasebe M., Fukatsu T., Shigenobu S., Kikuchi Y. Whole-genome sequence of Burkholderia sp. strain RPE67, a bacterial gut symbiont of the bean bug Riptortus pedestris. Genome Announc. 2014;2:e00556-14. doi: 10.1128/genomeA.00556-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takeshita K., Jang S., Kikuchi Y. Complete genome sequence of Burkholderia sp. strain THE68, a bacterial symbiont isolated from midgut crypts of the seed bug Togo hemipterus. Microbiol. Resour. Announc. 2020;9:e00041-20. doi: 10.1128/MRA.00041-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu Y., Buss E.A., Boucias D.G. Culturing and characterization of gut symbiont Burkholderia spp. from the southern chinch bug, Blissus insularis (Hemiptera: Blissidae) Appl. Environ. Microbiol. 2016;82:3319–3330. doi: 10.1128/AEM.00367-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon S.H., Ha S.M., Kwon S., Lim J., Kim Y., Seo H., Chun J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017;67:1613–1617. doi: 10.1099/ijsem.0.001755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chin C.S., Alexander D.H., Marks P., Klammer A.A., Drake J., Heiner C., Clum A., Copeland A., Huddleston J., Eichler E.E., et al. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 42.Pan Y., Kong K.F., Tsang J.S.H. Complete genome sequence of the exopolysaccharide-producing Burkholderia caribensis type strain MWAP64. Genome Announc. 2016;4:e01636-15. doi: 10.1128/genomeA.01636-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanizawa Y., Fujisawa T., Nakamura Y. DFAST: A flexible prokaryotic genome annotation pipeline for faster genome publication. Bioinformatics. 2018;34:1037–1039. doi: 10.1093/bioinformatics/btx713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Galperin M.Y., Makarova K.S., Wolf Y.I., Koonin E.V. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2015;43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krzywinski M., Schein J., Birol I., Connors J., Gascoyne R., Horsman D., Jones S.J., Marra M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pruesse E., Peplies J., Glöckner F.O. SINA: Accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics. 2012;28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Na S.I., Kim Y.O., Yoon S.H., Ha S.M., Baek I., Chun J. UBCG: Up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J. Microbiol. 2018;56:281–285. doi: 10.1007/s12275-018-8014-6. [DOI] [PubMed] [Google Scholar]
- 48.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Stamatakis A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stamatakis A., Hoover P., Rougemont J. A rapid bootstrap algorithm for the RAxML web servers. Syst. Biol. 2008;57:758–771. doi: 10.1080/10635150802429642. [DOI] [PubMed] [Google Scholar]
- 51.Meier-Kolthoff J.P., Auch A.F., Klenk H.P., Göker M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics. 2013;14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emms D.M., Kelly S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20:238. doi: 10.1186/s13059-019-1832-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holden M.T.G., Titball R.W., Peacock S.J., Cerdeño-Tárraga A.M., Atkins T., Crossman L.C., Pitt T., Churcher C., Mungall K., Bentley S.D., et al. Genomic plasticity of the causative agent of melioidosis, Burkholderia pseudomallei. Proc. Natl. Acad. Sci. USA. 2004;101:14240–14245. doi: 10.1073/pnas.0403302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Burtnick M.N., Brett P.J., Harding S.V., Ngugi S.A., Ribot W.J., Chantratita N., Scorpio A., Milne T.S., Dean R.E., Fritz D.L., et al. The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect. Immun. 2011;79:1512–1525. doi: 10.1128/IAI.01218-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mitter B., Petric A., Shin M.W., Chain P.S.G., Hauberg-Lotte L., Reinhold-Hurek B., Nowak J., Sessitsch A. Comparative genome analysis of Burkholderia phytofirmans PsJN reveals a wide spectrum of endophytic lifestyles based on interaction strategies with host plants. Front. Plant. Sci. 2013;4:120. doi: 10.3389/fpls.2013.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Angus A.A., Agapakis C.M., Fong S., Yerrapragada S., Estrada-de Los Santos P., Yang P., Song N., Kano S., Caballero-Mellado J., De Faria S.M., et al. Plant-associated symbiotic Burkholderia species lack hallmark strategies required in mammalian pathogenesis. PLoS ONE. 2014;9:e83779. doi: 10.1371/journal.pone.0083779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kim J.K., Lee H.J., Kikuchi Y., Kitagawa W., Nikoh N., Fukatsu T., Lee B.L. Bacterial cell wall synthesis gene uppP is required for Burkholderia colonization of the stinkbug gut. Appl. Environ. Microbiol. 2013;79:4879–4886. doi: 10.1128/AEM.01269-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ohbayashi T., Futahashi R., Terashima M., Barrière Q., Lamouche F., Takeshita K., Meng X.Y., Mitani Y., Sone T., Shigenobu S., et al. Comparative cytology, physiology and transcriptomics of Burkholderia insecticola in symbiosis with the bean bug Riptortus pedestris and in culture. ISME J. 2019;13:1469–1483. doi: 10.1038/s41396-019-0361-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kim J.K., Won Y.J., Nikoh N., Nakayama H., Han S.H., Kikuchi Y., Rhee Y.H., Park H.Y., Kwon J.Y., Kurokawa K., et al. Polyester synthesis genes associated with stress resistance are involved in an insect-bacterium symbiosis. Proc. Natl. Acad. Sci. USA. 2013;110:E2381–E2389. doi: 10.1073/pnas.1303228110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim J.K., Kwon J.Y., Kim S.K., Han S.H., Won Y.J., Lee J.H., Kim C.H., Fukatsu T., Lee B.L. Purine biosynthesis, biofilm formation, and persistence of an insect-microbe gut symbiosis. Appl. Environ. Microbiol. 2014;80:4374–4382. doi: 10.1128/AEM.00739-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim J.K., Jang H.A., Won Y.J., Kikuchi Y., Han S.H., Kim C.H., Nikoh N., Fukatsu T., Lee B.L. Purine biosynthesis-deficient Burkholderia mutants are incapable of symbiotic accommodation in the stinkbug. ISME J. 2014;8:552–563. doi: 10.1038/ismej.2013.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee J.B., Byeon J.H., Jang H.A., Kim J.K., Yoo J.W., Kikuchi Y., Lee B.L. Bacterial cell motility of Burkholderia gut symbiont is required to colonize the insect gut. FEBS Lett. 2015;589:2784–2790. doi: 10.1016/j.febslet.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 63.Kim J.K., Park H.Y., Lee B.L. The symbiotic role of O-antigen of Burkholderia symbiont in association with host Riptortus pedestris. Dev. Comp. Immunol. 2016;60:202–208. doi: 10.1016/j.dci.2016.02.009. [DOI] [PubMed] [Google Scholar]
- 64.Jang S., Jang H.A., Lee J., Kim J.U., Lee S.A., Park K.-E., Kim B.H., Jo Y.H., Lee B.L. PhaR, a negative regulator of PhaP, modulates the colonization of a Burkholderia gut symbiont in the midgut of the host insect, Riptortus pedestris. Appl. Environ. Microbiol. 2017;83:e00459-17. doi: 10.1128/AEM.00459-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim J.K., Jang H.A., Kim M.S., Cho J.H., Lee J., Di Lorenzo F., Sturiale L., Silipo A., Molinaro A., Lee B.L. The lipopolysaccharide core oligosaccharide of Burkholderia plays a critical role in maintaining a proper gut symbiosis with the bean bug Riptortus pedestris. J. Biol. Chem. 2017;292:19226–19237. doi: 10.1074/jbc.M117.813832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kinosita Y., Kikuchi Y., Mikami N., Nakane D., Nishizaka T. Unforeseen swimming and gliding mode of an insect gut symbiont, Burkholderia sp. RPE64, with wrapping of the flagella around its cell body. ISME J. 2018;12:838–848. doi: 10.1038/s41396-017-0010-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chun J., Oren A., Ventosa A., Christensen H., Arahal D.R., da Costa M.S., Rooney A.P., Yi H., Xu X.-W., De Meyer S., et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int. J. Syst. Evol. Microbiol. 2018;68:461–466. doi: 10.1099/ijsem.0.002516. [DOI] [PubMed] [Google Scholar]
- 68.Freiberg C., Fellay R., Bairoch A., Broughton W.J., Rosenthal A., Perret X. Molecular basis of symbiosis between Rhizobium and legumes. Nature. 1997;387:394–401. doi: 10.1038/387394a0. [DOI] [PubMed] [Google Scholar]
- 69.Sullivan J.T., Ronson C.W. Evolution of rhizobia by acquisition of a 500-kb symbiosis island that integrates into a phe-tRNA gene. Proc. Natl. Acad. Sci. USA. 1998;95:5145–5149. doi: 10.1073/pnas.95.9.5145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Boucias D.G., Garcia-Maruniak A., Cherry R., Lu H., Maruniak J.E., Lietze V.U. Detection and characterization of bacterial symbionts in the Heteropteran, Blissus insularis. FEMS Microbiol. Ecol. 2012;82:629–641. doi: 10.1111/j.1574-6941.2012.01433.x. [DOI] [PubMed] [Google Scholar]
- 71.Shin S.C., Kim S.H., You H., Kim B., Kim A.C., Lee K.A., Yoon J.H., Ryu J.H., Lee W.J. Drosophila microbiome modulates host developmental and metabolic homeostasis via insulin signaling. Science. 2011;334:670–674. doi: 10.1126/science.1212782. [DOI] [PubMed] [Google Scholar]
- 72.Vander Broek C.W., Stevens J.M. Type III secretion in the melioidosis pathogen Burkholderia pseudomallei. Front. Cell. Infect. Microbiol. 2017;7:255. doi: 10.3389/fcimb.2017.00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pratama A.A., Jiménez D.J., Chen Q., Bunk B., Spröer C., Overmann J., van Elsas J.D. Delineation of a subgroup of the genus Paraburkholderia, including P. terrae DSM 17804T, P. hospita DSM 17164T and four soil-isolated fungiphiles, reveals remarkable genomic and ecological features—Proposal for the definition of a P. hospita species cluster. Genome Biol. Evol. 2020;12:325–344. doi: 10.1093/gbe/evaa031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Deakin W.J., Broughton W.J. Symbiotic use of pathogenic strategies: Rhizobial protein secretion systems. Nat. Rev. Microbiol. 2009;7:312–320. doi: 10.1038/nrmicro2091. [DOI] [PubMed] [Google Scholar]
- 75.Dale C., Plague G.R., Wang B., Ochman H., Moran N.A. Type III secretion systems and the evolution of mutualistic endosymbiosis. Proc. Natl. Acad. Sci. USA. 2002;99:12397–12402. doi: 10.1073/pnas.182213299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ruby E.G. Lessons from a cooperative, bacterial-animal association: The Vibrio fischeri–Euprymna scolopes light organ symbiosis. Annu. Rev. Microbiol. 1996;50:591–624. doi: 10.1146/annurev.micro.50.1.591. [DOI] [PubMed] [Google Scholar]
- 77.Calatrava-Morales N., McIntosh M., Soto M. Regulation mediated by N-acyl homoserine lactone quorum sensing signals in the Rhizobium-Legume symbiosis. Genes. 2018;9:263. doi: 10.3390/genes9050263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Girard L., Blanchet E., Stien D., Baudart J., Suzuki M., Lami R. Evidence of a large diversity of N-acyl-homoserine lactones in symbiotic Vibrio fischeri strains associated with the squid Euprymna scolopes. Microbes Environ. 2019;34:99–103. doi: 10.1264/jsme2.ME18145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen W.M., Moulin L., Bontemps C., Vandamme P., Béna G., Boivin-Masson C. Legume symbiotic nitrogen fixation by β-proteobacteria is widespread in nature. J. Bacteriol. 2003;185:7266–7272. doi: 10.1128/JB.185.24.7266-7272.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Moulin L., Munive A., Dreyfus B., Boivin-Masson C. Nodulation of legumes by members of the β-subclass of proteobacteria. Nature. 2001;411:948–950. doi: 10.1038/35082070. [DOI] [PubMed] [Google Scholar]
- 81.Chen W.M., de Farja S.M., Chou J.H., James E.K., Elliott G.N., Sprent J.I., Bontemps C., Young J.P.W., Vandamme P. Burkholderia sabiae sp. nov., isolated from root nodules of Mimosa caesalpiniifolia. Int. J. Syst. Evol. Microbiol. 2008;58:2174–2179. doi: 10.1099/ijs.0.65816-0. [DOI] [PubMed] [Google Scholar]
- 82.Mannaa M., Park I., Seo Y.-S. Genomic features and insights into the taxonomy, virulence, and benevolence of plant-associated Burkholderia species. Int. J. Mol. Sci. 2018;20:121. doi: 10.3390/ijms20010121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moulin L., Klonowska A., Caroline B., Booth K., Vriezen J.A.C., Melkonian R., James E.K., Young J.P.W., Bena G., Hauser L., et al. Complete genome sequence of Burkholderia phymatum STM815T, a broad host range and efficient nitrogen-fixing symbiont of Mimosa species. Stand. Genomic Sci. 2014;9:763–774. doi: 10.4056/sigs.4861021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Martínez-Aguilar L., Díaz R., Peña-Cabriales J.J., Estrada-de Los Santos P., Dunn M.F., Caballero-Mellado J. Multichromosomal genome structure and confirmation of diazotrophy in novel plant-associated Burkholderia species. Appl. Environ. Microbiol. 2008;74:4574–4579. doi: 10.1128/AEM.00201-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Elliott G.N., Chen W.M., Chou J.H., Wang H.C., Sheu S.Y., Perin L., Reis V.M., Moulin L., Simon M.F., Bontemps C., et al. Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. N. Phytol. 2007;173:168–180. doi: 10.1111/j.1469-8137.2006.01894.x. [DOI] [PubMed] [Google Scholar]
- 86.Stopnisek N., Zühlke D., Carlier A., Barberán A., Fierer N., Becher D., Riedel K., Eberl L., Weisskopf L. Molecular mechanisms underlying the close association between soil Burkholderia and fungi. ISME J. 2016;10:253–264. doi: 10.1038/ismej.2015.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.