Abstract
Infections by many bacterial pathogens rely on their ability to degrade host glycans by producing glycoside hydrolases (GHs). Here, we discovered a conserved multifunctional GH, SsGalNagA, containing a unique combination of two family 32 carbohydrate-binding modules (CBM), a GH16 domain and a GH20 domain, in the zoonotic pathogen Streptococcus suis 05ZYH33. Enzymatic assays revealed that the SsCBM-GH16 domain displays endo-(β1,4)-galactosidase activity specifically toward the host-derived αGal epitope Gal(α1,3)Gal(β1,4)Glc(NAc)-R, whereas the SsGH20 domain has a wide spectrum of exo-β-N-acetylhexosaminidase activities, including exo-(β1,3)-N-acetylglucosaminidase activity, and employs this activity to act in tandem with SsCBM-GH16 on the αGal-epitope glycan. Further, we found that the CBM32 domain adjacent to the SsGH16 domain is indispensable for SsGH16 catalytic activity. Surface plasmon resonance experiments uncovered that both CBM32 domains specifically bind to αGal-epitope glycan, and together they had a KD of 3.5 mm toward a pentasaccharide αGal-epitope glycan. Cell-binding and αGal epitope removal assays revealed that SsGalNagA efficiently binds to both swine erythrocytes and tracheal epithelial cells and removes the αGal epitope from these cells, suggesting that SsGalNagA functions in nutrient acquisition or alters host signaling in S. suis. Both binding and removal activities were blocked by an αGal-epitope glycan. SsGalNagA is the first enzyme reported to sequentially act on a glycan containing the αGal epitope. These findings shed detailed light on the evolution of GHs and an important host-pathogen interaction.
Keywords: Streptococcus suis; glycoside hydrolase (GH); multifunctional enzyme; family 32 carbohydrate-binding module (CBM32); evolution; αGal epitope; swine tracheal epithelial cells (STEC); endo-(β1,4)-galactosidase; β-acetylhexosaminidase; GalNagA; Streptococcus; substrate specificity; multifunctional enzyme; carbohydrate-binding protein; carbohydrate processing; enzyme catalysis; enzyme kinetics; host–pathogen interaction; erythrocyte
The glycosylation of macromolecules, such as proteins and lipids, to form glycoconjugates is one of the most abundant modifications of biomolecules in nature (1). In mammals, the glycoconjugates displayed on the cell surface form the complex carbohydrate-rich glycocalyx that surrounds cells and play key roles in important cellular functions, such as cell-cell interactions and cell signaling (2–4). The N-glycosylation of Asn residues in proteins is the most frequent form of protein glycosylation (4, 5). Correspondingly, many commensal and pathogenic microbes have developed mechanisms to depolymerize N-glycans to take advantage of the carbohydrate-rich environment in the host and successfully colonize and/or infect the host (6–9). Recently, a model was developed for N-glycan depolymerization in the human pathogen Streptococcus pneumoniae. In this model, an exo-α-sialidase (NanA) and an exo-(β1,4)-galactosidase (BgaA), together with an exo-β-N-acetylglucosaminidase (StrH), all of which are anchored to the cell surface of S. pneumoniae, were shown to have sequential activity on complex N-glycans, thereby contributing to nutrient acquisition and host immune response suppression (10, 11).
The αGal-epitope glycan is a unique glycan produced on glycolipids and glycoproteins, containing a special terminal αGal-epitope structure in the form of Gal(α1,3)Gal(β1,4)GlcNAc-R (12, 13). It was first reported as a ceramide pentahexoside, Gal(α1,3)Gal(β1,4)GlcNAc(β1,3)Gal(β1,4)Glc-Cer, from rabbit erythrocytes (14). Subsequent reports demonstrated that rabbit erythrocytes contain a range of glycolipids of various lengths that terminate with Gal(α1,3)Gal(β1,4)GlcNAc-R, ranging from pentasaccharide to multibranched 35- and 40-sugar-containing glycolipids, with five sugars per branch (15–17). The αGal-epitope glycan is ubiquitously present in nonprimate mammals, but the amount varies in different organs, cells, and species (12, 18). It has been detected on glycolipids in kidney and thymus tissues from sheep, pigs, cows, and rabbits, as well as the pig respiratory epithelium and vascular endothelium, but not in kidney tissue from rats and brain tissues from any of the above animals (19, 20). The αGal-epitope glycan has also been revealed on glycoproteins, such as mouse and bovine laminin, fibrinogen, Igs, and thyroglobulins, from various species (12).
The αGal-epitope glycan is absent from humans. Instead, humans naturally generate a large amount of anti-αGal epitope antibody, accounting for ∼1% of total Igs (21, 22) and causing hyperacute rejection of pig xenograft organs in humans (23). Therefore, the αGal epitope-containing glycans have mainly been investigated in the context of xenotransplantation (20). Under these circumstances, endo–β-gal C (EndoGalC), characterized from Clostridium perfringens, and its potential application in removing the αGal epitope for xenotransplantation in humans were studied (24, 25). Recently, the host-derived αGal epitope was identified as a receptor for bovine norovirus Newbury2, revealing an expanded role of the αGal-epitope glycan in host–pathogen interactions (26). However, its potential as a target for bacterial pathogens has not been elucidated (27).
Streptococcus suis is a major swine bacterial pathogen that is responsible for severe economic losses in the porcine industry and represents a significant threat to human health (28, 29). The natural habitat of S. suis is the upper respiratory tract of pigs. Healthy pigs asymptomatically colonized with S. suis form a reservoir for this disease and play a major role in its epidemiology (29). The successful colonization of S. suis in upper respiratory tracts suggested that this microorganism can utilize the glycans enriched in these niches. Therefore, in this study, we identified a multifunctional glycoside hydrolase, SsGalNagA, from S. suis. This enzyme possesses a unique combination of activities to interact with and cleave the αGal epitope, which has been proven to be abundantly present on swine tracheal epithelial cells, potentially playing roles in upper respiratory tract colonization of S. suis. The results presented here represent the first report of the roles of the αGal epitope in the interactions of host and bacterial pathogens.
Results
S. suis does not have the same complex N-glycan degradation enzymatic system as S. pneumoniae
Previously, a complex N-glycan degradation enzymatic system, containing NanA, BgaA, and StrH, was revealed in S. pneumoniae (10, 11). To examine whether this system was present in a related bacterium, the zoonotic streptococcus S. suis, homologs for these three proteins were searched among S. suis genomes that were sequenced. However, no homolog was found for NanA and BgaA, whereas one homologous protein was found for StrH, in S. suis strain 05ZYH33, which was annotated as an N-acetyl-β-hexosaminidase (NCBI accession no. ABP89596; gene locus tag SSU05_0630). We named this protein SsGalNagA on the basis of the observations that we report in this study, representing an enzyme with both galactosidase and N-acetylglucosaminidase activity. The absence of NanA and BgaA homologs indicated that S. suis does not have the same complex N-glycan degradation enzymatic systems as S. pneumoniae.
SsGalNagA contains a unique and novel combination of GH16 and GH20 catalytic domains that is conserved in S. suis
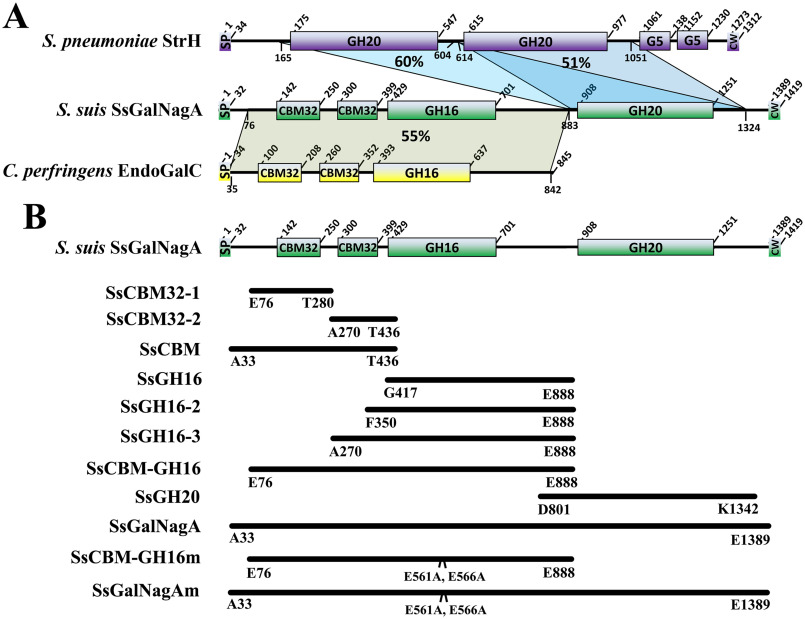
As shown in Fig. 1A, the 1419-residue SsGalNagA is composed of six domains: sequentially, a 32-residue N-terminal signal peptide (SP), a tandem repeat of two family 32 carbohydrate-binding modules (CBM32), two tandem glycosidase hydrolase (GH) catalytic domains, namely, family GH16 and GH20 domains, followed by a C-terminal canonical LPxTG motif (CW) (30). The presence of the LPxTG motif suggested that SsGalNagA is also a cell wall-anchored protein, similar to NanA, BgaA, and StrH in S. pneumoniae.
Figure 1.
Domain arrangement, alignment, and cloning strategy. A, domain arrangement and alignment of S. suis SsGalNagA (green), S. pneumoniae StrH (purple), and C. perfringens EndoGalC (yellow). The residue positions for each domain are labeled. The abbreviated domains are the following: SP, signal peptide; CW, cell wall-anchoring domain with a canonical LPxTG motif. The sequence coverage and identity are shown as shading with the percent identity. The residue ranges for the aligned domains are also labeled. B, the cloning strategy for full-length SsGalNagA, truncated proteins, and protein mutants used in this study. The start and end residues for each clone are labeled.
Unlike StrH, which contains two tandem GH20 catalytic domains (GH20A and GH20B), SsGalNagA contains two different GH catalytic domains, GH16 and GH20 (Fig. 1A), designated SsGH16 and SsGH20, respectively. StrH matched domain SsGH20 only via its two GH20 domains (Fig. 1A). Residues 76–883 of SsGalNagA, containing the two CBM32 domains and the SsGH16 domain, designated SsCBM-GH16, showed the highest similarities (55–59%) to GH16 glycosylhydrolases annotated in several bacterial genomes and to full-length EndoGalC from C. perfringens (55%) (Fig. 1A). Among these proteins, only the latter has been characterized, where it shows endo-β-galactosidase activity that catalyzes the release of Gal(α1,3)Gal from the αGal-epitope glycan (24). According to the newly developed subfamily classification of GH16, EndoGalC and its above homologs, as well as SsGH16, belong to GH16 subfamily 8, i.e. GH16_8. At present, EndoGalC is the only characterized enzyme in this subfamily (31, 32).
No homologous proteins possessing both GH16 and GH20 catalytic domains have been revealed from a species other than S. suis, indicating that the combination of GH16 and GH20 catalytic domains in one protein is unique for SsGalNagA and novel for GH. To assess whether the combination of GH16 and GH20 is conserved in S. suis, the 43 completely sequenced S. suis genomes were searched using full-length SsGalNagA as a probe. The search revealed that full-length SsGalNagA is highly conserved in S. suis from different serotypes, with 99–100% full-length coverage and 82–100% identities on the amino acid sequence level (Table S1). In summary, SsGalNagA is a multidomain protein containing a unique combination of GH16 and GH20 catalytic domains. This GH domain combination is novel for GH and conserved in S. suis.
SsGH20 has exo-(β1,2)-N-acetylglucosaminidase activity
To explore the biological function of the unique domain combination of SsGalNagA, full-length SsGalNagA and its domain truncations were cloned as summarized in Fig. 1B. The proteins of the domain variant were expressed and purified (Fig. S1).
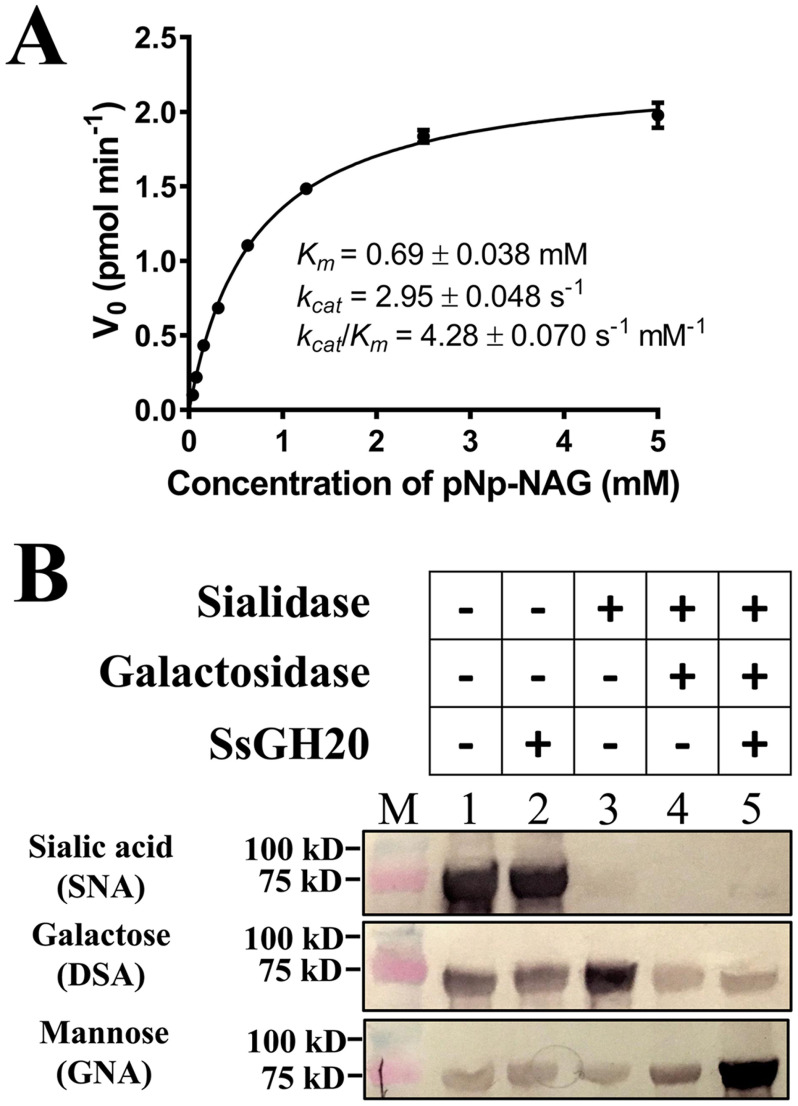
As SsGH20 showed high similarity to GH20A and GH20B of StrH, it was hypothesized that SsGH20 had the same activities as GH20A and GH20B, with catalytic activity toward 4-nitrophenyl N-acetyl-β-d-glucosaminide (pNp-NAG), and, further, would release (β1,2)-linked GlcNAc (GlcNAc) from the α1,3 or α1,6 arm of complex N-linked glycans. We first measured the activity of SsGH20 toward pNp-NAG. Purified SsGH20 demonstrated hydrolysis of pNp-NAG with a Km of 0.69 mm, a kcat of 2.95 s−1, and a kcat/Km of 4.28 s−1 mm−1 (Fig. 2A).
Figure 2.
Activities of SsGH20. A, kinetics of the SsGH20-catalyzed cleavage of pNp-NAG. B, deglycosylation of the complex N-glycan by SsGH20 in the absence (lane 2) and presence (lane 5) of sialidase and galactosidase, using complex N-glycosylated transferrin as the substrate. Transferrin (lane 1) and transferrin incubated with sialidase (lane 3) or with both sialidase and galactosidase (lane 4) were used as controls. Terminal sialic acid (Sia), galactose (Gal), and mannose (Man) of the complex N-glycan on transferrin were detected by lectins SNA, DSA, and GNA, respectively, using the DIG glycan differentiation kit (Sigma).
To determine the activity of SsGH20 on (β1,2)-linked GlcNAc from the α1,3 or α1,6 arm of a complex biantennary N-linked glycan, the deglycosylation of transferrin, which possesses a typical complex N-linked glycan (33), was performed. As shown in Fig. 2B, SsGH20 was unable to remove terminal (α2,6)-linked sialic acid (Sia) or expose additional mannose (Man) from complex N-linked glycan (Fig. 2B, lane 2), indicating that it could not release (β1,2)-linked GlcNAc through an endo-type activity. However, after sequential removal of terminal Sia and the second galactose (Gal) by sialidase and galactosidase, SsGH20 catalyzed the release of the exposed terminal (β1,2)-linked GlcNAc through an exo-type activity, exposing much more terminal Man recognized by the lectin Galanthus nivalis agglutinin (GNA) (Fig. 2B, lane 5). The results confirmed that SsGH20 has exo-(β1,2)-acetylglucosaminidase activity, as GH20A and GH20B of intact StrH do.
SsCBM-GH16 acts on the αGal epitope on erythrocytes from pigs, rabbits, and cows but not on the human blood type B epitope
SsCBM-GH16 showed high similarity to EndoGalC (Fig. 1A), which cleaves the (β1,4)-linkage between Gal(β1,4)GlcNAc in the αGal-epitope glycan to release Gal(α1,3)Gal. To detect whether SsCBM-GH16 showed the same activity as EndoGalC, based on the facts that the αGal epitope is abundant on erythrocytes of nonprimate mammals and human serum contains a large amount of anti-αGal epitope antibodies, a simple hemagglutination test was developed in this study. As shown in Fig. 3A, erythrocytes from pigs, cows, and rabbits agglutinated after the addition of human serum (Fig. 3A, lane 2) confirming the existence of αGal epitopes on these erythrocytes. In contrast, mouse erythrocytes did not show clear hemagglutination after the addition of human serum, indicating there was little or no αGal epitope on mouse erythrocytes.
Figure 3.
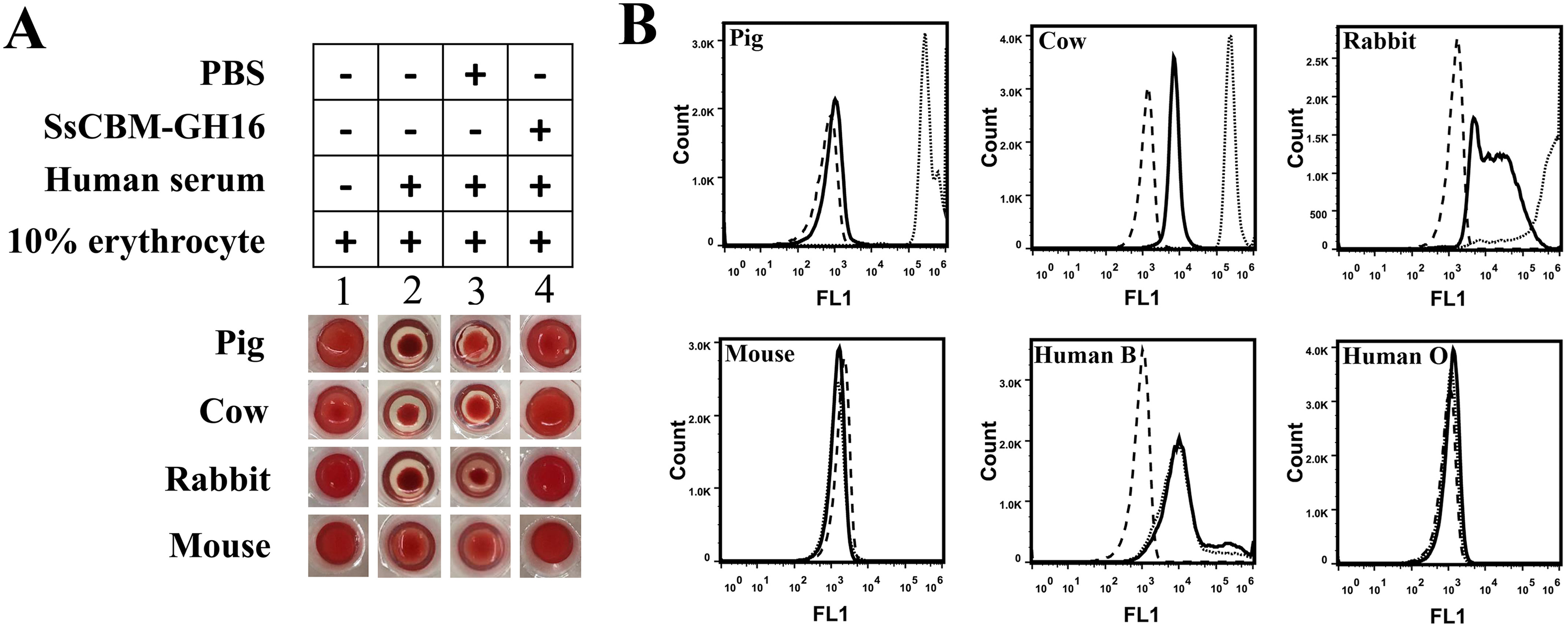
Removal of the αGal epitope from different erythrocytes by SsCBM-GH16. A, hemagglutination assay for detection of αGal epitope removal by SsCBM-GH16 from pig, cow, rabbit, and mouse erythrocytes. Ten percent erythrocyte solutions directly mixed with PBS–BSA (lane 1) or human serum (lane 2) served as hemagglutination negative and positive controls, respectively. B, FCA to detect the removal of the terminal αGal epitope by SsCBM-GH16 from different erythrocytes. The FITC signal of erythrocytes only (dashed line), erythrocytes incubated with FITC–IBS-B4 (dotted line), and SsCBM-GH16-treated erythrocytes incubated with FITC–IBS-B4 (solid line) were detected by FCA using a 488-nm laser.
These results were extended using flow cytometric analysis (FCA) with detection by FITC-conjugated Bandeiraea simplicifolia isolectin B4 (FITC–BSI-B4), which specifically binds to the terminal αGal residue. The results showed that the αGal epitope was indeed present on the erythrocytes from pigs, cows, and rabbits but absent from the erythrocytes from mice (dotted line in Fig. 3B). Further, after treatment with SsCBM-GH16, no additional hemagglutination was observed for these same erythrocytes after the addition of human serum (Fig. 3A, lane 4). FCA additionally showed that the αGal epitopes were removed by SsCBM-GH16 (Fig. 3B, solid line).
However, different degrees of activity have been observed by FCA. The removal of αGal epitopes from pig erythrocytes was more complete than that for cow and rabbit erythrocytes. Considering that Sia-containing αGal epitopes have been detected on bovine erythrocytes (18), the residual signal likely originates from FITC–BSI-B4 binding to this type of αGal epitope, which was not removed by SsCBM-GH16. Furthermore, compared with the human blood type O epitopes on erythrocytes, human blood type B epitopes were also recognized by BSI-B4 (Fig. 3B, dotted line) but not by SsCBM-GH16 (Fig. 3B, solid line). This suggested that SsCBM-GH16 could not catalyze cleavage of the fucosylated (α1,3)-galactobiose from human blood type B epitopes.
SsCBM-GH16 cleaves the (β1,4)-linkage in αGal epitopes, releasing Gal(α1,3)Gal
To further determine the exact cleavage site of SsCBM-GH16 in an αGal epitope, the Galili antigen pentaose (Gap), with the structure Gal(α1,3)Gal(β1,4)GlcNAc(β1,3)Gal(β1,4)Glc (Fig. 4A), was used as the substrate, and the hydrolytic products were detected by TLC and further identified by ultraperformance liquid chromatography–high-resolution MS (UPLC-HRMS). As shown in Fig. 4, Gap was present at the pentasaccharide position on TLC plates (Fig. 4B, lane 1) and showed a molecular mass of 869 Da (Fig. 4C), corresponding to that of intact Gap. After hydrolysis by SsCBM-GH16, disaccharides and trisaccharides were released (Fig. 4B, lane 2). The molecular masses of these products were determined as 342 Da and 545 Da by UPLC-HRMS analysis (Fig. 4D), corresponding to that of Gal(α1,3)Gal and GlcNAc(β1,3)Gal(β1,4)Glc, respectively. This clearly indicated that SsCBM-GH16 acts very similarly to EndoGalC, cleaving the (β1,4)-linkage between Gal(β1,4)GlcNAc in the αGal-epitope glycan, releasing Gal(α1,3)Gal (Fig. 4A).
Figure 4.
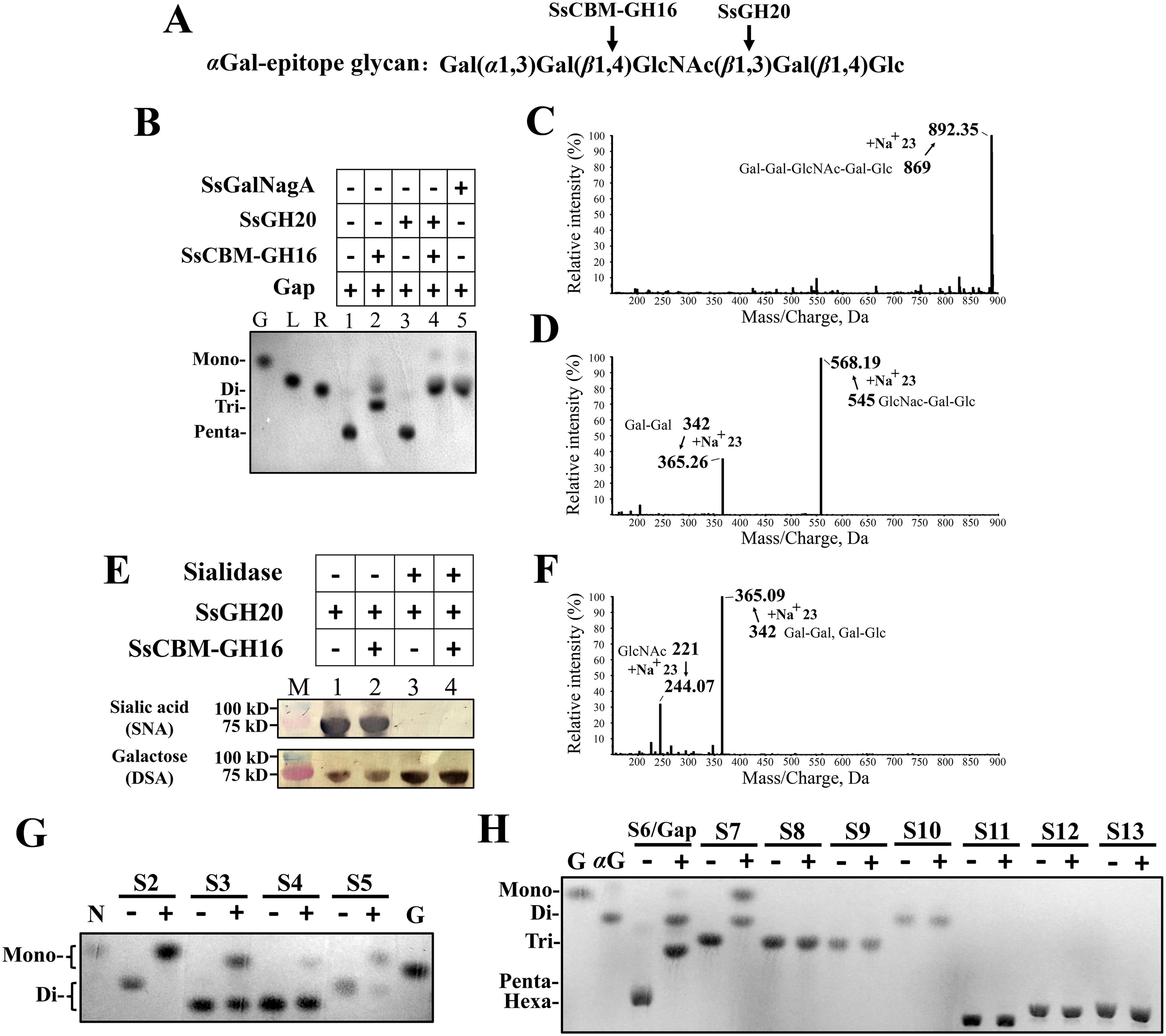
Activity assays for synergy and specificity of SsCBM-GH16 and SsGH20. A, the structure of the αGal-epitope glycan, Gap. The cleavage sites for SsCBM-GH16 and SsGH20 are indicated by arrows. B, TLC analysis of the products from Gap hydrolysis. Gal (G), lactose (L), and raffinose (R) were used to indicate the chromatographic mobilities of monosaccharide, disaccharide, and trisaccharide, respectively. C, D, and F, UPLC-HRMS analysis of the products from Gap hydrolyzed by no protein (C), SsCBM-GH16 (D), and SsCBM-GH16 plus SsGH20 (F). E, deglycosylation of the complex N-glycan on transferrin by SsCBM-GH16 and SsGH20 without (lanes 1 and 2) and with (lanes 3 and 4) sialidase. Terminal sialic acid (Sia) and galactose (Gal) of the complex N-glycan on transferrin were detected by lectins SNA and DSA, respectively, using the DIG glycan differentiation kit (Sigma). G and H, TLC analysis of the products from the indicated oligosaccharides hydrolyzed by SsGH20 (G) or SsCBM-GH16 (H). S2–S13 oligosaccharides and their structural compositions are listed in Table 1. −, without protein; +, with protein. N, GlcNAc; αG, Gal(α1-3)Gal.
SsCBM-GH16 does not synergistically act with SsGH20 on complex N-glycans
We considered a rationale for S. suis combining SsCBM-GH16 and SsGH20 in one protein. The activity of SsGH20 on complex N-glycans requires removal of the terminal Sia and the downstream Gal. S. pneumoniae achieved this by sequential cleavage of (α2,6)-linked Sia and (β1,4)-linked Gal by NanA and BgaA, respectively. S. suis does not possess the same complex N-glycan degradation system as S. pneumoniae (vide supra), because homologs of NanA and BgaA are absent from S. suis. Considering that SsCBM-GH16 shows endo-type activity on (β1,4)-linked Gal in Gal(α1,3)Gal(β1,4)GlcNAc, except that a different disaccharide Gal(α1,3)Gal is released, we considered whether SsCBM-GH16 could act on the (β1,4)-linked Gal in canonical complex N-glycans through an endo-type action to directly trim the disaccharide, Sia(α2,6)Gal, and subsequently form the substrate for SsGH20. However, incubation of transferrin with SsCBM-GH16 and SsGH20 did not remove additional terminal Sia from transferrin (Fig. 4E, lane 2) compared to that from transferrin incubated with SsGH20 alone, which does not remove terminal Sia (Fig. 4E, lane 1). Therefore, SsCBM-GH16 did not act on the (β1,4)-linked Gal in complex N-glycans through an endo-type activity. Similar assays were performed as above, except that sialidase was added. Here, SsCBM-GH16 and SsGH20 did not remove any more Gal (Fig. 4E, lane 4) than SsGH20 alone (Fig. 4E, lane 3). This result additionally indicated that SsCBM-GH16 did not act on the (β1,4)-linked Gal in complex N-glycans through exo-type action. Thus, SsCBM-GH16 does not synergistically act on complex N-glycans with SsGH20 via its (β1,4)-galactosidase activity.
SsGH20 shows exo-(β1,3)-acetylglucosaminidase activity, which synergistically acts on the αGal-epitope glycan with SsCBM-GH16
Whereas the endo-(β1,4)-galactosidase activity of SsCBM-GH16 does not provide synergy with the activity of SsGH20 on complex N-glycans, we next focused on their synergistic actions on the αGal-epitope glycan. As SsCBM-GH16 cleaved the Gal(β1,4)GlcNAc linkage in the αGal-epitope glycan and exposed a terminal (β1,3)-linked GlcNAc, we considered whether SsGH20, which was shown to cleave (β1,2)-linked GlcNAc, could act on this (β1,3)-GlcNAc linkage, the activity of which has not been revealed for StrH. To test this concept, SsGH20 was incubated with Gap in the absence and presence of SsCBM-GH16. As shown in Fig. 4B, lane 3, in the absence of SsCBM-GH16, SsGH20 alone does not act on Gap. However, in the presence of SsCBM-GH16, SsGH20 could further hydrolyze Gap (Fig. 4B, lane 4). Compared with the products released by hydrolysis by SsCBM-GH16 alone (Fig. 4B, lane 2), the disappearance of trisaccharide, the increased levels of disaccharide, and the appearance of monosaccharides, after synergistic action of SsCBM-GH16 and SsGH20, strongly indicated that the trisaccharide GlcNAc(β1,3)Gal(β1,4)Glc produced by SsCBM-GH16 was further cleaved by SsGH20, resulting in the monosaccharide, GlcNAc, and the disaccharide, Gal(β1,4)Glc. This was confirmed by UPLC-HRMS analysis, which revealed that the hydrolysis products showed molecular masses of 244 Da and 365 Da (Fig. 4F), corresponding to that of GlcNAc and Gal(α1,3)Gal/Gal(β1,4)Glc, respectively. These results clearly revealed that SsGH20 acts on the terminal (β1,3)-linked GlcNAc. Thus, in addition to exo-(β1,2)-acetylglucosaminidase activity, SsGH20 also showed exo-(β1,3)-acetylglucosaminidase activity, and, with this activity, was able to synergistically act on the αGal-epitope glycan with SsCBM-GH16.
To further confirm that this synergistic action is combined in one protein, the full length of SsGalNagA (Fig. 1B and Fig. S1) was tested with Gap as the substrate. Not surprisingly, full-length SsGalNagA hydrolyzed Gap to GlcNAc and Gal(α1,3)Gal/Gal(β1,4)Glc in the same manner as the combination of the individual domains, SsCBM-GH16 and SsGH20 (Fig. 4B, lane 5).
SsGH20 has a broad substrate spectrum, whereas SsCBM-GH16 specifically targets αGal-epitope glycan
We show above that SsGH20 has both exo-(β1,2)- and exo-(β1,3)-acetylglucosaminidase activities, indirectly or directly, whereas SsCBM-GH16 displays endo-β-galactosidase activity releasing terminal (β1,4)-linked Gal(α1,3)Gal from αGal-epitope glycans. To better explore the potential substrates of SsGalNagA, activities of SsGH20 and SsCBM-GH16 were screened on a variety of biologically relevant sugars using TLC. The results are shown in Fig. 4G and H and summarized in Table 1.
Table 1.
Activities of SsGH20 and SsCBM-GH16 on different oligosaccharides
| Substrate no. | Oligosaccharide composition | Activitya |
|---|---|---|
| SsGH20 | ||
| S1b | GalNAc(β1,3)Gal(β1,4)Glc | +++ |
| S2 | GlcNAc(β1,2)Man | +++ |
| S3 | GalNAc(β1,3)Gal | ++ |
| S4 | GalNAc(β1,4)Gal | + |
| S5 | GlcNAc(β1,4)GlcNAc | ++ |
| SsCBM-GH16 | ||
| S6/Gap | Gal(α1,3)Gal(β1,4)GlcNAc(β1,3)Gal(β1,4)Glc | +++ |
| S7 | Gal(α1,3)Gal(β1,4)Glc | +++ |
| S8 | Gal(α1,3)Gal(β1,4)Gal | − |
| S9 | Gal(α1,4)Gal(β1,4)GlcNAc | − |
| S10 | Gal(β1,4)GlcNAc | − |
| S11 | Gal(β1,4)GlcNAc(β1,3)Gal(β1,4)GlcNAc(β1,3)Gal(β1,4)Glc | − |
| S12 | GalNAc(α1,3)(Fucα1,2)Gal(β1,4)GlcNAc(β1,3)Gal(β1,4)Glc | − |
| S13 | Gal(α1,3)(Fucα1,2)Gal(β1,4)GlcNAc(β1,3)Gal(β1,4)Glc | − |
aActivities were rated according to the degree of hydrolysis.
bThe structure and activity of substrate S1 were inferred from results in Fig. 4, B and F.
For SsGH20 (Fig. 4G and Table 1), we show that SsGH20 efficiently acted on terminal (β1,2)-linked GlcNAc and completely hydrolyzed GlcNAc(β1,2)Man (S2). Additionally, cleavage of terminal (β1,4)-linked GlcNAc was observed but with lower efficiency, because SsGH20 did not completely hydrolyze GlcNAc(β1,4)GlcNAc (S5). In addition to GlcNAc, SsGH20 also recognized terminal β-linked N- acetylgalactosamine (GalNAc). It was able to catalyze the cleavage of terminal (β1,3)- and (β1,4)-linked GalNAc from GalNAc(β1,3)Gal (S3) and GalNAc(β1,4)Gal (S4), respectively. However, as indicated by the incomplete hydrolysis, SsGH20 was less active on terminal β-linked GalNAcs than on their GlcNAc counterparts.
For SsCBM-GH16, strict specificity for the terminal (β1,4)-linked Gal(α1,3)Gal and the following sugar residue at subsite +1 was observed. As shown in Fig. 4H and Table 1, SsCBM-GH16 only cleaved the terminal (β1,4)-linked Gal(α1,3)Gal (S6/Gap and S7) but not the terminal (β1,4)-linked Gal(α1,4)Gal (S9) or Gal (S10 and S11). These results indicated that SsCBM-GH16 has strict specificity for the terminal Gal(α1,3)Gal and does not perform exo-type activity. Furthermore, SsCBM-GH16 exhibited no activity on blood group B antigen (S13), which shares the highest structural similarity with αGal-epitope glycan, except for a fucosyl substitution on the Gal located at subsite −1, indicating that the −1 subsite of the enzyme does not tolerate a fucosylated Gal residue. This is consistent with the above result that SsCBM-GH16 could not remove the terminal αGal from human blood type B erythrocytes (Fig. 3B). On the other hand, for the sugar residue located at subsite +1, SsCBM-GH16 recognized either a GlcNAc (S6/Gap) or a Glc (S7) but not a Gal (S9), implying that SsCBM-GH16 also has high specificity for the sugar residue at this position. With this high specificity, it is not surprising that SsCBM-GH16 did not cleave human blood type A antigen (S12).
In summary, SsGH20 has a broad spectrum of exo-β-N-acetylhexosaminidase activities, with a preference for (β1,2)- and (β1,3)-linked GlcNAc over (β1,4)-linked GlcNAc and β-linked GalNAc counterparts. On the contrary, SsCBM-GH16 displays strict specificity toward αGal-epitope glycans, with some small flexibility of the sugar residue located at the subsite +1 of the enzyme, which can accommodate either a GlcNAc or a Glc but not a Gal.
The SsCBM32-2 domain is indispensable for SsGH16 catalytic activity
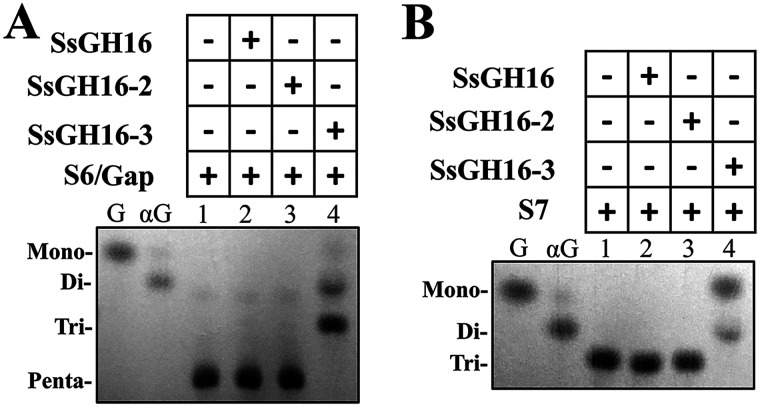
SsCBM-GH16 and its homolog contain both the tandem CBM32 domains (designated SsCBM32-1 and SsCBM32-2; Fig. 1B) and the GH16 domain. In addition, it was reported that more than 75% of enzymes from subfamily GH16_8 were linked to CBM32 (32). Therefore, we examined the contributions of the CBM32 domains to the catalytic activity of SsGH16. Three truncated proteins, viz., SsGH16, SsGH16-2, and SsGH16-3, were prepared, as shown in Fig. 1B and Fig. S1. Their activities on both of the SsCBM-GH16 substrates, S6/Gap and Gal(α1,3)Gal(β1,4)Glc (S7), were examined. As shown in Fig. 5, A and B, neither SsGH16, with no CBM32 domain, nor SsGH16-2, with a partial domain of SsCBM32-2, was active on the substrates. However, SsGH16-3, which contains the SsGH16 domain and the complete SsCBM32-2 domain, recapitulated the activities on both substrates. These findings indicated that SsCBM32-2, the CBM32 domain adjacent to SsGH16, is indispensable for the catalytic activity of the SsGH16 catalytic domain.
Figure 5.
Hydrolysis of oligosaccharides S6/Gap and S7 by SsGH16 truncations. A and B, TLC analysis of the hydrolytic products from Gal(α1,3)Gal(β1,4)GlcNAc(β1,3)Gal(β1,4)Glc (S6/Gap) (A) and Gal(α1,3)Gal(β1,4)Glc (S7) (B) by SsGH16 (lane 2), SsGH16-2 (lane 3), and SsGH16-3 (lane 4). G, Gal; αG, Gal(α1,3)Gal.
Both SsCBM32-1 and SsCBM32-2 bind αGal-epitope glycan with high specificity
The two CBM32 domains were annotated by dbCAN-seq (34). The CBMs from family 32 have been demonstrated to bind Gal, GlcNAc, Gal(β1,4)GlcNAc, and GlcNAc(α1,4)Gal (35, 36). To determine the binding specificity of these two CBM32 domains, three SsCBM truncations, viz., SsCBM32-1, SsCBM32-2, and SsCBM, were obtained (Fig. 1B andFig. S1). Their binding activities toward various biologically relevant oligosaccharides were examined by surface plasmon resonance (SPR). As shown in Fig. 6, A–C, similar binding patterns were observed for all three SsCBM truncations. With either one, or both, of the CBM32 domains, SsCBM32-1, SsCBM32-2, and SsCBM only showed significant binding activity toward the pentasaccharide αGal-epitope glycan, S6/Gap. For other oligosaccharides tested, weak or no binding signals were detected.
Figure 6.
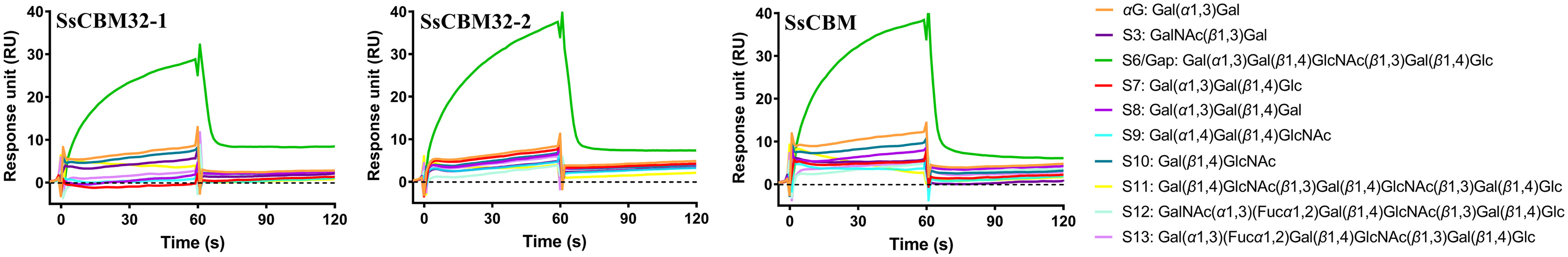
Binding of oligosaccharides to SsCBM truncations. A, B, and C, SPR sensorgrams for the indicated oligosaccharides binding to SsCBM32-1 (A), SsCBM32-2 (B), and SsCBM (C). SsCBM32-1, SsCBM32-2, and SsCBM were immobilized on CM5 chips with immobilization levels of 5781 RU, 4143 RU, and 3869 RU, respectively. The indicated oligosaccharides were passed through the control and sample flow cells. The sensorgrams were generated by subtraction of the background from the total binding.
No significant binding was observed for disaccharides Gal(α1,3)Gal (αG), GalNAc(β1,3)Gal (S3), and Gal(β1,4)GlcNAc (S10), all of which are fragments of the pentasaccharide S6/Gap, suggesting that the strong binding of S6/Gap involves more than two sugar residues. For trisaccharide Gal(α1,4)Gal(β1,4)GlcNAc (S9) and hexasaccharide Gal(β1,4)GlcNAc(β1,3)Gal(β1,4)GlcNAc(β1,3)Gal(β1,4)Glc (S11), lack of binding indicates that the terminal Gal(α1,3)Gal disaccharide structure of S6/Gap is important for the strong binding. This was further demonstrated by the inability of the proteins to bind human blood type B epitope (S13), which specified that a fucosyl substitution on the second Gal of S6/Gap impeded binding. On the other hand, none of the three proteins showed apparent binding to Gal(α1,3)Gal(β1,4)Gal (S8) and, notably, to Gal(α1,3)Gal(β1,4)Glc (S7), which is an active substrate of SsCBM-GH16, suggesting that the GlcNAc at the third position in S6/Gap is strictly specific for strong binding.
In summary, SsCBM32-1, SsCBM32-2, and SsCBM bind αGal-epitope glycan with high specificity, and the terminal Gal(α1,3)Gal(β1,4)GlcNAc trisaccharide structure of αGal-epitope glycan plays an important role in the specific binding. With this specificity, it is unsurprising that the proteins did not bind human blood type A epitope (S12).
The binding kinetics of SsCBM to the pentasaccharide Gap was measured by SPR and are the following: kon, 0.6746 ± 0.024 M−1 s−1; koff, (2.334 ± 0.008) × 10−3 s−1. The two CBM32 together interact with Gap with a KD of 3.46 mm.
SsGalNagA binds and releases αGal epitopes on swine erythrocytes
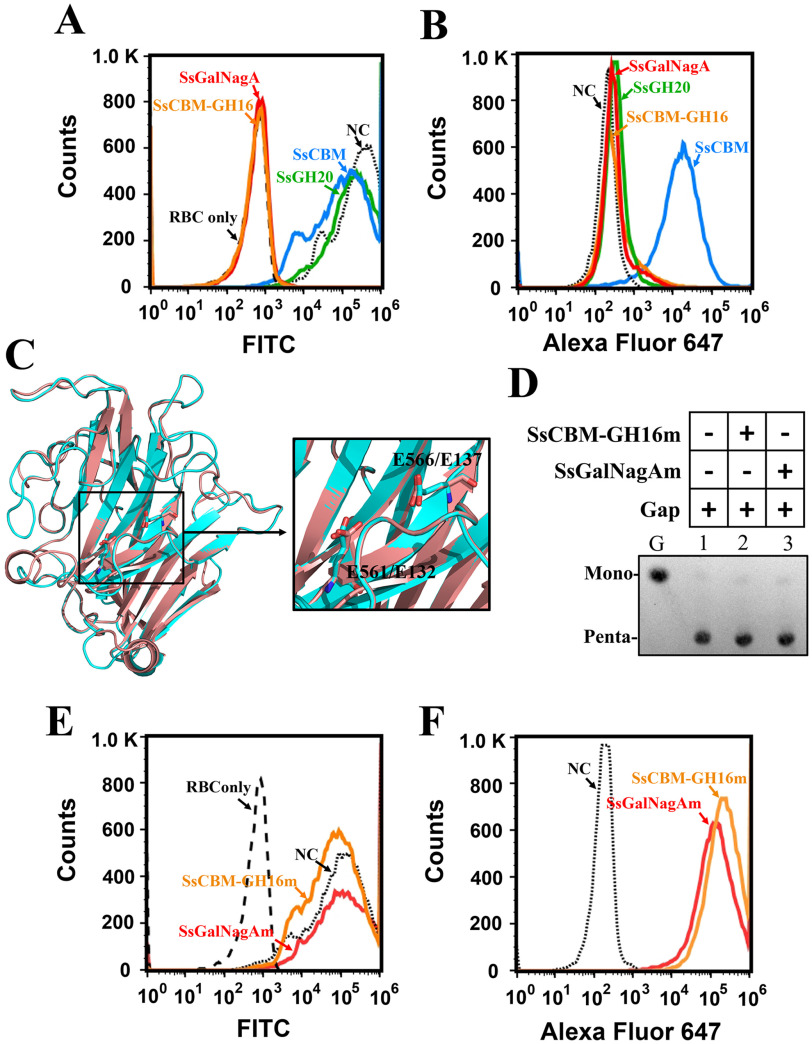
Further elucidation of the function of SsGalNagA in the removal of αGal epitopes on erythrocytes in the natural host of S. suis will assist in understanding the mechanism of diseases, such as septicemia, caused by S. suis. The αGal epitope removal and binding activities of the full-length SsGalNagA and domain variants were detected by FCA, using FITC–BSI-B4 and corresponding antibodies, respectively. As expected, neither SsCBM nor the exo-acting SsGH20 shows αGal epitope removal activity, whereas both SsCBM-GH16 and full-length SsGalNagA efficiently removed the αGal epitope from swine erythrocytes (Fig. 7A). Binding assays revealed that SsGH20 did not interact with swine erythrocytes, which was not unexpected. Surprisingly, among SsCBM, SsCBM-GH16, and SsGalNagA, all of which contain the SsCBM domain and are predicted to bind to the αGal epitope, only SsCBM binds to swine erythrocytes. Neither SsCBM-GH16 nor SsGalNagA showed any such binding activity (Fig. 7B).
Figure 7.
The removal and binding of the terminal αGal epitope on swine erythrocytes by SsCBM, SsCBM-GH16, and full-length SsGalNagA. A, FCA detection of αGal epitope removal from swine erythrocytes. The erythrocyte solution (10%) was incubated with PBS–BSA (NC, negative control) or the indicated protein. The presence of the terminal αGal epitope was detected by FITC–BSI-B4 using FCA. The erythrocyte solution was detected and used as a control (Red blood cell only (RBC only)), B, FCA detection of protein binding to swine erythrocytes. The erythrocyte suspension (10%) was incubated with PBS–BSA (NC) or the indicated protein. The bound proteins were detected by Alexa Fluor 647 using FCA. C, superpositioning of structures of SsGH16 (cyan) and GH16 laminarinase from Thermotoga maritima MSB8 (pink; PDB entry 3b01). The structure of SsGH16 was modeled by SWISS-MODEL using 3b01 as the template. The catalytic residues, viz., E561 and E566 in SsGH16 and E132 and E137 in 3b01, are shown as stick models. The image on the right is an enlargement of the area framed on the left. D, TLC analysis of the hydrolysis of Gap by SsCBM-GH16m and SsGalNagAm. E, FCA detection of αGal epitope removal from swine erythrocytes by SsCBM-GH16m and SsGalNagAm. F, FCA detection of SsCBM-GH16m and SsGalNagAm binding to swine erythrocytes.
Considering the efficient αGal-epitope cleavage activities of SsCBM-GH16 and SsGalNagA (Fig. 4B and 7A), we predicted that the cleavage of the αGal epitope would attenuate the binding target of SsCBM. To address this issue, mutations at the catalytic sites of SsGH16 were performed. We hypothesized, by structural simulations and alignments with GH16 laminarinase from Thermotoga maritima MSB8 (PDB entry 3b01) (Fig. 7C), that E561 and E566 are essential for the activity of SsGH16. Mutations of both E561 and E566 to alanines in SsCBM-GH16 and SsGalNagA were performed, resulting in the variants SsCBM-GH16m and SsGalNagAm, respectively (Fig. 1B and Fig. S1). The loss of catalytic activities of SsCBM-GH16m and SsGalNagAm were confirmed both by Gap hydrolysis (Fig. 7D, lanes 2 and 3) and αGal epitope removal from swine erythrocytes (Fig. 7E). Finally, as predicted, abundant binding of SsCBM-GH16m and SsGalNagAm to swine erythrocytes was observed after loss of the catalytic activity (Fig. 7F).
In summary, SsGalNagA binds to the αGal epitope on swine erythrocytes through SsCBM, after which the αGal epitope is cleaved by SsGH16, followed by β(1,3)-GlcNAc cleavage by SsGH20. The steps also occur with Gap as a substrate.
SsGalNagA binds to STEC and removes αGal epitopes
The natural niche of S. suis is the upper respiratory tract of swine. Swine tracheal epithelial cells (STEC), the main cell type at this location, provide the first contact point for colonization of S. suis. To assess whether SsGalNagA plays a role in the interaction of STEC and S. suis, the αGal epitope on STEC was analyzed by confocal laser scanning microscopy (CLSM) using FITC–BSI-B4. As shown in Fig. 8A, αGal epitopes were detected on STEC by FITC–BSI-B4. Incubation of STEC with either SsGalNagA or SsCBM-GH16 significantly reduced the amount of αGal epitopes on STEC (Fig. 8A). The reduction of αGal epitopes was significantly dependent on the concentration of SsGalNagA added (Fig. 8B), clearly indicating that the reduction of αGal epitopes was due to the action of SsGalNagA. After inactivation of the catalytic activity of GH16, neither SsGalNagAm nor SsCBM-GH16m catalyzed the cleavage of the αGal epitope from STEC (Fig. 8, A and C), further confirming that the removal of the αGal epitope from STEC was attributable to the catalytic activity of SsGalNagA.
Figure 8.
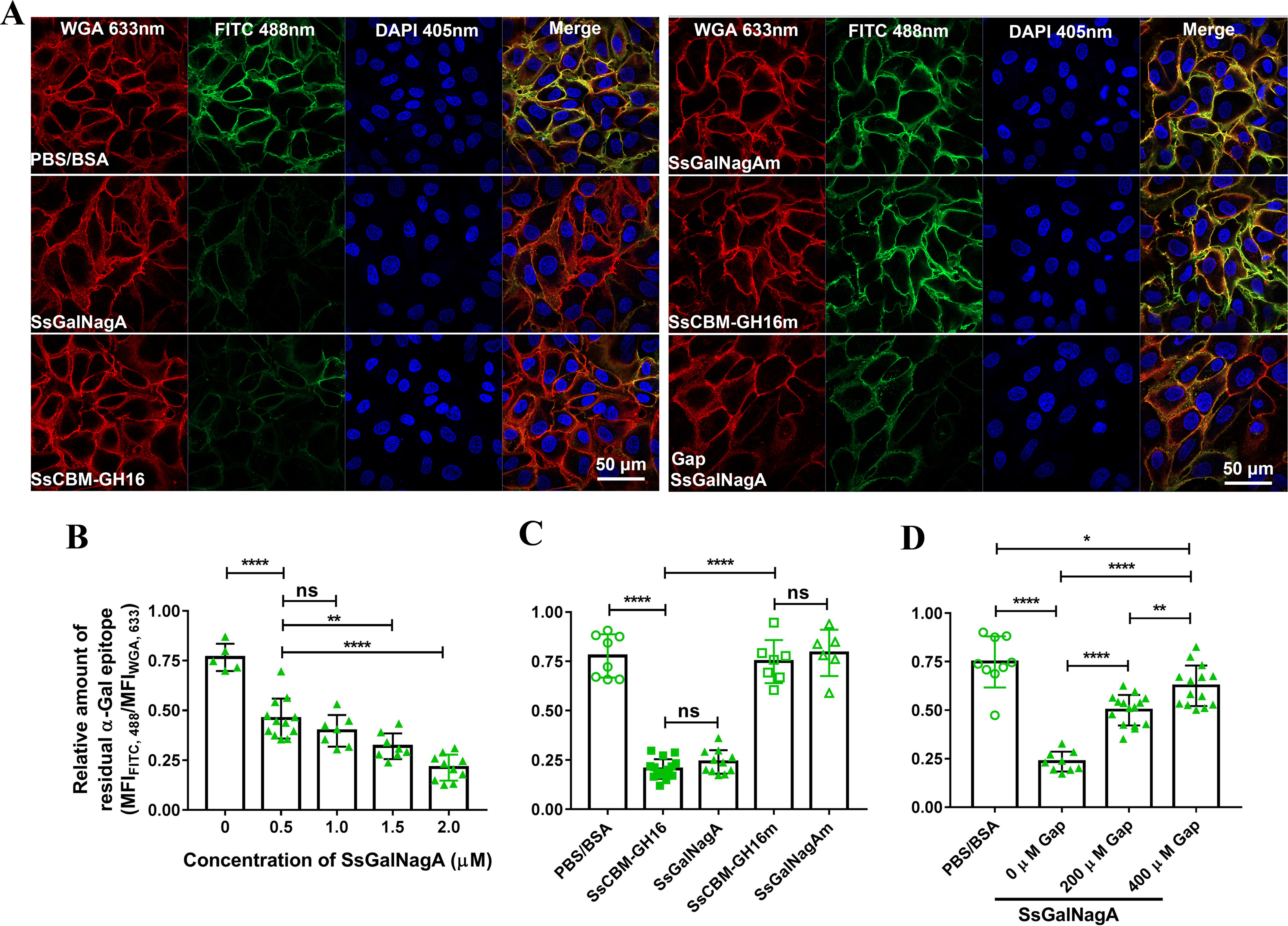
Removal of the terminal αGal epitope on STEC by SsGalNagA and its variants. A, representative CLSM images for STEC after treatment of PBS–BSA (control), the indicated protein, or the indicated protein plus Gap. The total cell membrane, residual αGal epitope, and DNA were observed with lasers at wavelengths of 633 nm (WGA), 488 nm (FITC), and 405 nm (DAPI). The removal of terminal αGal epitope was assessed by the decrease of residual αGal epitope (FITC, 488 nm) compared with the control. Images were prepared by Zen Black software (Zeiss) and organized by Photoshop. B, C, and D, calculated relative amounts of residual αGal epitope from CLSM images. The relative amount of residual αGal epitope was calculated by MFIFITC,488/MFIWGA,633 to indicate the amount of residual αGal epitope per unit of cell membrane. B, relative amounts of residual αGal epitope after treatment by the indicated concentrations of SsGalNagA. C, relative amounts of residual αGal epitope after treatment by SsGalNagA or its variants. D, relative amounts of residual αGal epitope after treatment by SsGalNagA in the presence of the indicated concentrations of Gap. Statistical significance was tested by nonpaired t test using GraphPad. Each symbol represents one MFIFITC,488/MFIWGA,633 value calculated from one image. ns, not significant.
Binding of SsGalNagA and SsCBM-GH16 was also investigated by CLSM. As shown in Fig. 9, A and B, similar to swine erythrocytes, little binding was detected for catalytically active SsCBM-GH16 and SsGalNagA, whereas high levels of binding were detected for the inactive mutants, SsCBM-GH16m and SsGalNagAm. The results indicated that, similar to the action of αGal epitopes on erythrocytes, SsGalNagA binds to the αGal epitope and removes this terminal αGal from STEC.
Figure 9.
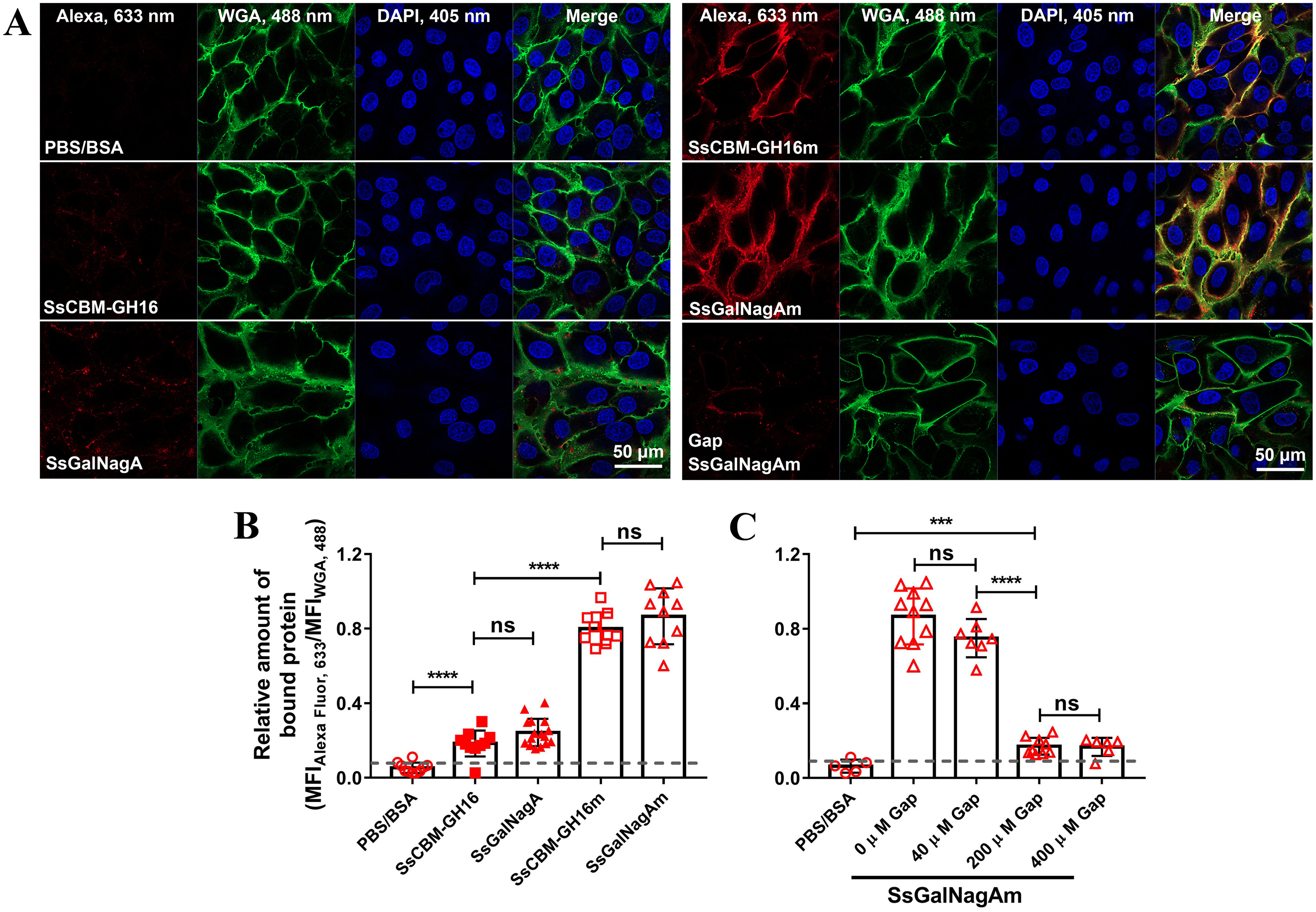
Binding of SsGalNagA and its variants to STEC. A, representative CLSM images for STEC binding by PBS–BSA (control), SsGalNagA, and its variants. The bound protein, total cell membrane, and DNA contents were observed with lasers at wavelengths of 633 nm (AlexaFluor 647), 488 nm (WGA), and 405 nm (DAPI), respectively. Images were prepared by Zen Black software (Zeiss) and organized by Photoshop. B and C, calculated relative amounts of bound protein from CLSM images. The relative amounts of bound protein were calculated by MFIAlexa,633/MFIWGA,488 to indicate the levels of bound protein per unit of cell membrane. B, relative amounts of bound SsGalNagA and its variants to STEC. C, relative amounts of SsGalNagAm bound to STEC in the presence of the indicated concentrations of Gap. The dashed lines indicate the blank signals. The statistical significance was tested by nonpaired t test using GraphPad. Each symbol represents one MFIAlexa,633/MFIWGA,488 value calculated from one image.
The competitive effects of Gap on the removal of the αGal epitope from STEC and binding to the αGal epitope on STEC by SsGalNagA were examined. As shown in Fig. 8, A and D, significantly more residual αGal epitopes were revealed on STEC with the addition of 200 μm and 400 μm of Gap than that without the addition of Gap. These results indicate that the competition by Gap with the αGal epitope on STEC decreased the cleavage of the αGal epitope on STEC by SsGalNagA. Correspondingly, the binding of SsGalNagA to the αGal epitope on STEC was greatly attenuated upon the addition of Gap in a concentration-dependent manner (Fig. 9, A and C). This demonstrated that Gap competed with the αGal epitope for binding of SsGalNagA to STEC.
Discussion
In this study, a unique and conserved multifunctional SsGalNagA has been discovered and characterized from the bacterial pathogen S. suis. Its singular combination of GH16 and GH20 catalytic domains sheds new light on the evolution of GH. The sugar components and linkages in host glycans are complicated in structure, and their degradation requires the combined activities of multiple GHs with different activities. Commensal and pathogenic bacteria have evolved different strategies to degrade the complex host glycans. One common strategy found in many bacteria is the formation of polysaccharide utilization loci or the carbohydrate-processing locus, which encodes an array of glycoside hydrolases (GH) required to degrade complex carbohydrates at a single genomic locus (37, 38). Another strategy is to locate the required GHs at the same cellular location, although they are encoded by different gene loci. A representative example is the complex N-glycan degradation enzymes of S. pneumoniae, viz., NanA, BgaA, and StrH. All of these enzymes are equipped with the LPxTG motif, recognized by sortase A; consequently, all are likely anchored on the cell surface (11). Revealing the roles of S. suis SsGalNagA in this study represents a rare evolutionary strategy for GH by combining different GH catalytic domains needed into a single protein that targets different linkages in one glycan. Because the capacity of one protein is limited, this strategy may have developed for the glycans that contain relatively simple linkages.
In this study, a dependence of activity on the adjacent CBM32 domain has been revealed for a GH16 enzyme. This is unexpected, as GH16 enzymes are known to adopt a well-defined β-sandwich fold and have not been reported to rely on an additional ancillary module. However, this is not totally unexpected. First, until now, the activity and specificity toward αGal-epitope glycans were only reported for the members of the GH16_8 subfamily, indicative of a special substrate recognition pattern for this subfamily. Second, most of the enzymes from this subfamily are linked to CBM32, suggesting that an important role is performed by this CBM for the enzymes in this subfamily. Third, although rare, CBM has been reported to contribute to a conformational change and subsequent active-site formation for GH (35, 36). As SsCBM32-2 specifically binds the αGal epitope, the catalytic activity of SsGH16 seemingly depends on the binding ability of SsCBM32-2. However, SsCBM32-2 was also indispensable for the hydrolysis of Gal(α1,3)Gal(β1,4)Glc (Fig. 5B), to which it did not show significant binding activity (Fig. 6). This suggested that the indispensability is not due to the binding ability of SsCBM32-2 but because of its contribution to another process, e.g. conformational change, substrate recognition, or active-site formation. Nevertheless, more evidence from activity and structural studies on SsGalNagA and its GH16_8 homologs are needed to further address this question.
Before now, the potential of the host-derived αGal epitope as a target for bacterial pathogens has not been revealed (27). In this study, it was shown that SsCBM-GH16 of SsGalNagA specifically binds and acts on αGal-epitope glycans. Whereas SsGH20 has a broad substrate spectrum and potentially acts on complex N-glycans, the lack of exo-sialidase and exo-(β1,4)-galactosidase partners in S. suis suggests that this is unlikely. Thus, the putative surface protein, SsGalNagA, from S. suis, specifically and sequentially cleaved terminal sugar residues from the αGal-epitope glycan. Furthermore, αGal epitopes are present on the surface of STEC, the first contact point for the colonization of S. suis, and can be removed by SsGalNagA. Based on these results, we propose that SsGalNagA from S. suis targets the αGal-epitope glycans on STEC in the pig upper respiratory tract, through which it acquires carbohydrate nutrients for the bacterium by degradation of αGal-epitope glycan, thereby facilitating the colonization of S. suis at this niche. Further, signal transduction functions have been reported for ceramide-based glycolipids (39), and the αGal epitope-depleted glycan structure has been identified as a regulator of transforming growth factor β receptor (TβR) (2). Therefore, SsGalNagA may also play a role in host cell signaling by altering the structure of this potential glycan regulator.
In conclusion, a multifunctional SsGalNagA was identified from S. suis 05ZYH33. This protein contains a unique GH16 and GH20 catalytic domain combination, enabling it to specifically and sequentially act on host-derived αGal-epitope glycans. SsGalNagA efficiently binds to swine erythrocytes and STEC and removes αGal epitopes, indicating that it plays roles in host colonization and host cell signaling in S. suis.
Experimental procedures
Reagents
α(2,3,6,8,9)-neuraminidase A (sialidase) and (β1,4)-galactosidase S were purchased from New England Biolabs (NEB). The oligosaccharides used in this study were purchased from Elicityl (France), Dextra (UK), or Sigma (USA) and are listed in Table S2. FITC-conjugated isolectin B4 from Bandeiraea simplicifolia (Griffonia simplicifolia) (BSI-B4) and 4′,6-diamidino-2-phenylindole (DAPI) were obtained from Sigma. Alexa Fluor 488-WGA (wheat germ agglutinin), Alexa Fluor 633-WGA, and Alexa Fluor 647-goat-anti-rabbit IgG were purchased from Thermo Fisher.
Bacterial strains, plasmids, cells, and growth conditions
Streptococcus suis strain 05ZYH33, serotype 2, which caused severe human infection in China in 2005 (40), was used to clone cDNAs of ssGalNagA and its truncation fragments. The S. suis strain was cultured in Todd-Hewitt broth (BD Bacto) supplemented with 1% yeast extract (BD Bacto) (THY) at 37 °C and 5% CO2. Escherichia coli strains DH5α (Invitrogen) and BL21(DE3) and plasmid pET22b (Novagen) were used for gene cloning and protein expression. The E. coli strains were grown at 37 °C in Luria-Bertani medium, and 100 μg/ml ampicillin was added for transformant selection. Fresh normal erythrocytes were obtained by drawing heparinized blood from healthy human and other animal donors. Swine tracheal epithelial cells (STEC) were obtained from Shanghai Fusheng (China) and routinely maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. The cells were passaged at confluence after dispersal with 0.25% trypsin-EDTA (Gibco) (41).
Sequence and structure analysis
Homologs of NanA (NCBI accession no. NP_359129), BgaA (NCBI accession no. NC_003098), and StrH (NCBI accession no. NP_357651) from S. pneumoniae strain R6 were identified from the genome of S. suis 05ZYH33 using the protein Basic Local Alignment Search Tool (BLAST) (42). The domain architecture was searched by CD-Search (43), and the annotation for each domain was obtained from the dbCAN-seq database (34). Sequences were aligned with ClustalX (44). Signal peptide predictions were performed using SignalP 5.0 (RRID:SCR_015644).
Structural neighbors were searched against the Protein Data Bank (PDB) using SWISS-MODEL (45). Three-dimensional molecular visualizations, structure superimpositions, and figure preparations were performed with PyMOL (46).
Cloning, protein expression, and purification
Gene fragments encoding full-length SsGalNagA and its truncated proteins, shown in Fig. 1B, were amplified from the genomic DNA of S. suis 05ZYH33 by PCR with the primers listed in Table S3. The PCR products were cloned into an NdeI- and XhoI-linearized pET22b vector by homologous recombination using the Trelief SoSoo cloning kit Ver.2 (TsingKe, Beijing, China). The constructions were verified by DNA sequencing and transformed into E. coli BL21(DE3) to express C-terminally His-tagged SsGalNagA and domains. His-tagged proteins were expressed and purified as described previously (47) (Fig. S1). The purified protein was finally maintained in 50 mm sodium phosphate, pH 7.4. The purity of proteins was assessed with 12% (w/v) SDS-PAGE. Protein concentrations were measured by the absorbance at 280 nm using the calculated extinction coefficients (RRID:SCR_018087) listed in Table S4. The final solutions of purified protein were stored in aliquots at −80 °C for later use.
Site-directed mutagenesis
These steps were performed using the QuikChange site-directed mutagenesis kit (Stratagene) with the plasmids encoding the WT ssCBM-GH16 or ssGalNagA gene as the template. The mutant proteins were expressed and purified similarly to the WT protein.
β-N-acetylglucosaminidase activity and kinetics
The β-N-acetylglucosaminidase activity and kinetic parameters of SsGH20 were measured using the β-N-acetylglucosaminidase assay kit (Sigma). In summary, the activity was detected using 4-nitrophenyl N-acetyl-β-d-glucosaminide (pNp-NAG; Sigma) as the substrate compared with a standard curve of 4-nitrophenol. All of the assays were performed at 37 °C in 0.09 m citrate buffer, pH 4.7, containing 64 nm SsGH20. For kinetic assays, substrate concentrations from 0.04 mm to 5 mm were used. The increase in absorption at 405 nm was monitored using an Enspire multimode plate reader (PerkinElmer). Kinetic parameters (Vmax and Km) were extracted from these data by nonlinear fitting to the Michaelis-Menten equation using GraphPad Prism software.
Deglycosylation of complex N-glycan
Deglycosylation of a typical biantennary N-glycan was performed using N-glycosylated transferrin as the substrate (DIG glycan differentiation kit; Sigma). SsGH16, SsCBM-GH16, or SsGH20 (each added at 4 μm) was added to 1.5 μg of transferrin in deglycosylation buffer (50 mm sodium acetate, pH 5.5, 5 mm CaCl2) to a final volume of 10 μl and incubated at 37 °C for 12 h with the addition of protease inhibitor cocktail (PIC; Roche). Controls were performed using deglycosylation buffer alone with transferrin. Where specified, 10 units of the following commercial enzymes were added: α(2,3,6,8,9)-neuraminidase A (sialidase) and (β1,4)-galactosidase S (NEB). The reactions were terminated by adding gel-loading buffer. To investigate the deglycosylation efficacy, samples were subjected to electrophoresis on an SDS-PAGE gel (10%), transferred onto PVDF membranes (Millipore), and detected using Sambucus nigra agglutinin (SNA), Datura stramonium agglutinin (DSA), or Galanthus nivalis agglutinin (GNA) lectin from the DIG glycan differentiation kit. SNA recognizes terminal sialic acid (Sia) α(2,6)-linked to Gal; thus, it is suitable for identifying terminal Sia in complex N-glycans. DSA recognizes Gal(β1,4)GlcNAc in complex N-glycans after removal of terminal Sia. GNA recognizes terminal mannose (Man)-linked (α1,2)-, (α1,3)-, or (α1,6)-Man after removal of terminal Sia, Gal, and GlcNAc.
Hemagglutination assay
Hemagglutination assays were carried out using fresh normal pig, cow, rabbit, and mouse erythrocytes with addition of normal human serum (NHS). Fresh normal erythrocytes were obtained by drawing heparinized blood from healthy human and other animal donors and then washed 3× in PBS containing 2% BSA (PBS–BSA). Erythrocyte solutions (10%) were prepared in PBS–BSA. NHS was isolated from healthy human whole blood. Blood was drawn into a gel vacuum tube, allowed to clot at 37 °C for 0.5 h, and then centrifuged at 1000 × g for 20 min at 4 °C. The serum layer was collected, aliquoted, and frozen at −80 °C until use. PBS–BSA (5 μl) or protein (5 μm) was added to 50 μl of 10% erythrocyte suspension and incubated at 37 °C for 1 h. The erythrocytes were collected by centrifugation and washed twice with PBS–BSA and resuspended in 50 μl of PBS–BSA. Finally, 50 μl of 50% NHS was added and mixed. The solution was transferred to a round-bottom 96-well dish to observe the hemagglutination.
Removal of the αGal epitopes from erythrocytes
Removal of the αGal epitope from the erythrocytes prepared as described above was carried out, and the residual αGal epitope was labeled with FITC–BSI-B4 and detected using FCA. Erythrocyte suspensions (100 μl) were incubated with 10 μl of PBS–BSA or protein (40 μm) for 2 h. The erythrocytes were collected and washed, resuspended in 400 μl PBS–BSA containing 1 μg/ml of FITC–BSI-B4, and incubated in the dark for 2 h. The erythrocytes were collected, washed, and then resuspended in PBS–BSA. The terminal αGal epitope on the erythrocytes was assessed by FITC detection by FCA using a 488-nm laser. All incubations were performed at room temperature with gentle rotation with PIC added.
Hydrolysis of oligosaccharides and detection of the hydrolytic products
The hydrolysis of oligosaccharides was performed using the oligosaccharides listed in Table 1. The hydrolytic products were analyzed by TLC and UPLC-HRMS. Procedurally, in a 10-μl reaction, 2.5 mm the indicated oligosaccharide was incubated with 4 μm protein in 50 mm phosphate buffer, pH 7.4, at 37 °C for 1 h. After incubation, 2 μl of samples was spotted onto a GF254 silica gel TLC plate (Haiyang, China). The plate was developed in n-butanol–acetic acid–H2O (2:1:1), and sugar spots were visualized by spraying with acetone–diphenylamine–aniline–phosphoric acid reagent (100 ml–2 g–2 ml–10 ml) and heating. For MS, the resulting hydrolytic products from Gap were characterized by UPLC-HRMS. Chromatography was performed on a Shimidzu LC30A UPLC system. The mobile phase was run at 0.3 ml/min using 0.1% formic acid in H2O and CH3CN. MS was performed on a Triple TOF 4600 (AB SCIEX) operated in high-resolution mode in positive polarity.
SPR
Surface plasmon resonance (SPR) was carried out with a BIAcore T200 Biosensor (GE Healthcare) to measure the binding specificities of individual SsCBM32-1 and SsCBM32-2 domains and SsCBM, containing both SsCBM32 domains, toward different oligosaccharides. The proteins were immobilized onto a CM5 sensor chip (GE Healthcare) using the amine coupling method. For immobilization, 40 μg/ml of SsCBM32-1, SsCBM32-2, or SsCBM in 10 mm NaOAc, pH 4.0, was used to attain 5781 response units (RU), 4143 RU, and 3869 RU, respectively. For each protein, a similarly treated flow cell, except that protein was not injected, was used as a control surface for the subtraction of the nonspecific binding. Binding was measured for each protein and the indicated oligosaccharides in HBS-P buffer (10 mm HEPES, 150 mm NaCl, 0.005% P20, pH 7.4). One millimolar each oligosaccharide was used. The oligosaccharide solutions were injected at a flow rate of 50 μl/min for 60 s. Postinjection phase dissociation was monitored in the running buffer for 60 s. The surface was regenerated between injections using 10 mm glycine-HCl, pH 2.0, at a flow rate of 50 μl/min for 30 s. The background was subtracted by passing the oligosaccharide solution over the control chip surface.
To measure the binding kinetic for SsCBM and Gap, the same immobilization strategy was used for SsCBM, except that 10,000 RU was reached. Concentrations from 31 μm to 500 μm Gap were used. Gap was injected at a flow rate of 50 μl/min with association and dissociation times of 120 and 180 s, respectively. The kinetic constants were calculated from the sensorgrams by nonlinear fitting of the association and dissociation curves according to a 1:1 Langmuir binding model using BIAevaluation software (GE Healthcare).
Protein binding to swine erythrocytes
Protein binding to erythrocytes was performed with fresh normal erythrocytes, and the binding proteins were detected by indirect immunofluorescence using FCA. Protein or PBS–BSA (10 μl) was added to 150 μl of 10% swine erythrocyte suspension and incubated for 1 h. The erythrocytes then were collected by centrifugation, washed with PBS–BSA, and incubated with corresponding rabbit polyclonal antibody. Anti-SsCBM was used for SsCBM, SsCBM-GH16, SsGalNagA, SsCBM-GH16m, and SsGalNagAm, and anti-SsGH20 was used for SsGH20. Next, erythrocytes were collected and washed with PBS–BSA, resuspended in 1 ml PBS–BSA with Alexa Fluor 647-goat-anti-rabbit IgG, and incubated in the dark for 1 h. The erythrocytes were then collected, washed, and resuspended in PBS–BSA. All incubations were performed at room temperature with gentle rotation with PIC added. The extent of the binding of proteins to erythrocytes was detected by the fluorescence signal of Alexa Fluor 647 on erythrocytes using FCA.
FCA
FCA was performed with a CytomicsTM FC500 cytometer (Beckman) using 488-nm and 633-nm lasers for the detection of FITC and Alexa Fluor 647, respectively. The cells were fixed with 4% paraformaldehyde, washed, and resuspended in PBS–BSA to yield a cell concentration of 1 × 106 cells/ml. Acquisition and analysis were performed by gating on side scatter and fluorescence. Cells in suspension were analyzed at a flow rate of 10 μl/min, and 50,000 events were recorded for analysis. Each histogram was analyzed using FlowJo_V10 software (BD Biosciences).
Removal of the αGal epitope from STEC
The removal of the terminal αGal epitope from STEC was detected by FITC–BSI-B4 using confocal laser scanning microscopy (CLSM). STEC were plated at 5 × 105 cells/disc in 15-mm glass-bottom cell culture dishes 2 days prior to the assay. On the day of the assay, STEC were washed twice with Hank's balanced salt solution (HBSS), followed by incubation for 30 min with PBS–BSA (control) or the indicated concentrations of proteins in DMEM in a humidified 37 °C, 5% CO2, incubator. After washing with HBSS, the cells were fixed with 4% paraformaldehyde, followed by sequential incubation with 1 μg/ml FITC–BSI-B4 for 2 h, 5 μg/ml Alexa Fluor 633-WGA for 10 min, and 2 μg/ml DAPI for 5 min. The incubations all were performed in HBSS at room temperature in the dark, with addition of PIC. The cells were then washed twice with HBSS between additions, and fluorescent cells were observed by CLSM. The fluorescence was observed by a 640-nm laser for Alexa Fluor 633, for analysis of the cell membrane, and a 405-nm laser for DAPI, for cellular DNA. The residual αGal epitopes were indicated by fluorescence observed by a 488-nm laser (for FITC).
To quantify the residual αGal epitope, CLSM images were analyzed by ZEN Lite microscope software (Zeiss) to obtain the mean fluorescence intensity (MFI). To correct the errors introduced to MFI by different amounts of cell membrane, the MFI of Alexa Fluor 633-WGA was used to normalize cell counts. The resulting MFIFITC,488/MFIWGA,633 was designated the relative residual αGal epitope level and used to compare the number of residual αGal epitopes per unit of cell membrane. The removal of the αGal epitope from STEC was evaluated by reduction of the relative number of residual αGal epitopes compared with controls. The statistical significance was tested by the nonpaired t test using GraphPad.
To assess the competitive effects of free αGal epitope on its removal from STEC, the experiment was carried out as above, except that the indicated concentrations of Gap were added.
Protein binding to STEC
The binding of proteins to STEC was performed by immunofluorescence using CLSM. STEC was prepared as above 2 days prior to the assay. The cell suspensions were prepared and fixed as above. Incubation of the cells proceeded with anti-SsCBM, Alexa Fluor 647-goat anti-rabbit IgG, Alexa Fluor 488-WGA, and DAPI for 5 min in HBSS. The incubations all were performed at room temperature in the dark, with the addition of PIC. The cells were washed twice with HBSS between additions. Protein binding to the cells was observed by fluorescence with a 640-nm laser as in the previous section.
To quantify the protein bound, CLSM images were analyzed by the ZEN Lite microscope software (Zeiss) to obtain the MFI. Similarly, MFIAlexa, 633/MFIWGA,488 was designated the relative amount of bound protein and used to compare the protein bound per unit of cell membrane. Protein binding levels were evaluated by comparing the relative amount of bound protein to the control. Statistical significance was tested by nonpaired t test using GraphPad.
To test the effect of free αGal epitope on binding, the experiment was carried out as above, except that prior to incubation with STEC, protein was incubated with the indicated concentrations of Gap for 10 min.
CLSM
CLSM was conducted with an LSM800 confocal laser scanning microscope with an Airyscan (Zeiss) microscope equipped with a 63× oil objective and 405-nm, 488-nm, and 640-nm lasers. Identical parameters were set for all the images taken. Image preparation and analysis were performed using the ZEN Lite microscope software (Zeiss).
Data availability
All data are contained within the manuscript.
Supplementary Material
Acknowledgments
We thank Prof. Rui Zhou, Hongjie Fan, and Dr. Zhe Ma for helpful discussions. We also thank our colleagues Dr. Peixin Liu and Tao Yang for their assistance in equipment manipulation.
This article contains supporting information.
Author contributions—P. C. and Y. Z. data curation; P. C., R. L., M. H., J. Z., D. W., G. D., F. X., Z. C., and Y. Z. software; P. C., G. L., and Y. Z. formal analysis; P. C., R. L., M. H., J. Z., D. W., G. D., F. X., Z. C., and Y. Z. methodology; F. J. C., S. L., and Y. Z. writing-review and editing; G. L., S. L., and Y. Z. supervision; Y. Z. funding acquisition; Y. Z. validation; Y. Z. writing-original draft; Y. Z. project administration.
Funding and additional information—This research was supported by grants from the National Key R&D Program of China (2017YFD0500203 to Y. Z.), the National Natural Science Foundation of China (31772757 to Y. Z.; 31873016 to F. X.; 31672575 to G. L.), and the Natural Science Foundation of Heilongjiang Province of China (C2017075 to Y. Z.; C2017078 to G. L.).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- GH
- glycoside hydrolase
- Gap
- Galili antigen pentaose
- pNp-NAG
- 4-nitrophenyl N-acetyl-β-D-glucosaminide
- SNA
- Sambucus nigra agglutinin
- DSA
- Datura stramonium agglutinin
- GNA
- Galanthus nivalis agglutinin
- BSI-B4
- Bandeiraea simplicifolia isolectin B4
- WGA
- wheat germ agglutinin
- STEC
- swine tracheal epithelial cells
- Gal
- galactose
- Glc
- Glucose
- GlcNAc
- N-acetylglucosamine
- GalNAc
- N-acetylgalactosamine
- Sia
- sialic acid
- Man
- mannose
- CBM
- carbohydrate-binding modules
- DAPI
- 4′,6-diamidino-2-phenylindole
- FCA
- flow cytometric analysis
- UPLC-HRMS
- ultraperformance liquid chromatography–high-resolution MS
- NHS
- normal human serum
- MFI
- mean fluorescence intensity
- PIC
- protease inhibitor cocktail
- SPR
- surface plasmon resonance
- HBSS
- Hanks' balanced salt solution.
References
- 1. Reitsma S., Slaaf D. W., Vink H., van Zandvoort M. A., and Oude Egbrink M. G. (2007) The endothelial glycocalyx: composition, functions, and visualization. Pflugers Arch. 454, 345–359 10.1007/s00424-007-0212-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Watanabe S., Misawa M., Matsuzaki T., Sakurai T., Muramatsu T., and Sato M. (2011) A novel glycosylation signal regulates transforming growth factor beta receptors as evidenced by endo-beta-galactosidase C expression in rodent cells. Glycobiology 21, 482–492 10.1093/glycob/cwq186 [DOI] [PubMed] [Google Scholar]
- 3. Khoury G. A., Baliban R. C., and Floudas C. A. (2011) Proteome-wide post-translational modification statistics: frequency analysis and curation of the swiss-prot database. Sci. Rep. 1, 90 10.1038/srep00090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Helenius A., and Aebi M. (2001) Intracellular functions of N-linked glycans. Science 291, 2364–2369 10.1126/science.291.5512.2364 [DOI] [PubMed] [Google Scholar]
- 5. Ritchie G. E., Moffatt B. E., Sim R. B., Morgan B. P., Dwek R. A., and Rudd P. M. (2002) Glycosylation and the complement system. Chem. Rev. 102, 305–320 10.1021/cr990294a [DOI] [PubMed] [Google Scholar]
- 6. Cao Y. L., Rocha E. R., and Smith C. J. (2014) Efficient utilization of complex N-linked glycans is a selective advantage for Bacteroides fragilis in extraintestinal infections. Proc. Natl. Acad. Sci. U S A 111, 12901–12906 10.1073/pnas.1407344111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hobbs J. K., Pluvinage B., and Boraston A. B. (2018) Glycan-metabolizing enzymes in microbe-host interactions: the Streptococcus pneumoniae paradigm. FEBS Lett. 592, 3865–3897 10.1002/1873-3468.13045 [DOI] [PubMed] [Google Scholar]
- 8. Byers H. L., Tarelli E., Homer K. A., and Beighton D. (1999) Sequential deglycosylation and utilization of the N-linked, complex-type glycans of human alpha1-acid glycoprotein mediates growth of Streptococcus oralis. Glycobiology 9, 469–479 10.1093/glycob/9.5.469 [DOI] [PubMed] [Google Scholar]
- 9. Manfredi P., Renzi F., Mally M., Sauteur L., Schmaler M., Moes S., Jeno P., and Cornelis G. R. (2011) The genome and surface proteome of Capnocytophaga canimorsus reveal a key role of glycan foraging systems in host glycoproteins deglycosylation. Mol. Microbiol. 81, 1050–1060 10.1111/j.1365-2958.2011.07750.x [DOI] [PubMed] [Google Scholar]
- 10. Dalia A. B., Standish A. J., and Weiser J. N. (2010) Three surface exoglycosidases from Streptococcus pneumoniae, NanA, BgaA, and StrH, promote resistance to opsonophagocytic killing by human neutrophils. Infect. Immun. 78, 2108–2116 10.1128/IAI.01125-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Robb M., Hobbs J. K., Woodiga S. A., Shapiro-Ward S., Suits M. D., McGregor N., Brumer H., Yesilkaya H., King S. J., and Boraston A. B. (2017) Molecular characterization of N-glycan degradation and transport in Streptococcus pneumoniae and its contribution to virulence. PLoS Pathog. 13, e1006090 10.1371/journal.ppat.1006090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Macher B. A., and Galili U. (2008) The Galalpha1,3Galbeta1,4GlcNAc-R (alpha-Gal) epitope: a carbohydrate of unique evolution and clinical relevance. Biochim. Biophys. Acta 1780, 75–88 10.1016/j.bbagen.2007.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Huai G., Qi P., Yang H., and Wang Y. (2016) Characteristics of alpha-Gal epitope, anti-Gal antibody, alpha1,3 galactosyltransferase and its clinical exploitation (review). Int. J. Mol. Med. 37, 11–20 10.3892/ijmm.2015.2397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Stellner K., Saito H., and Hakomori S. I. (1973) Determination of aminosugar linkages in glycolipids by methylation. Aminosugar linkages of ceramide pentasaccharides of rabbit erythrocytes and of Forssman antigen. Arch. Biochem. Biophys. 155, 464–472 10.1016/0003-9861(73)90138-0 [DOI] [PubMed] [Google Scholar]
- 15. Hanfland P., Kordowicz M., Peter-Katalinic J., Egge H., Dabrowski J., and Dabrowski U. (1988) Structure elucidation of blood group B-like and I-active ceramide eicosa- and pentacosasaccharides from rabbit erythrocyte membranes by combined gas chromatography-mass spectrometry; electron-impact and fast-atom-bombardment mass spectrometry; and two-dimensional correlated, relayed-coherence transfer, and nuclear Overhauser effect 500-MHz 1H-n.m.r. spectroscopy. Carbohydr. Res. 178, 1–21 10.1016/0008-6215(88)80098-3 [DOI] [PubMed] [Google Scholar]
- 16. Egge H., Kordowicz M., Peter-Katalinic J., and Hanfland P. (1985) Immunochemistry of I/i-active oligo- and polyglycosylceramides from rabbit erythrocyte membranes. Characterization of linear, di-, and triantennary neolactoglycosphingolipids. J. Biol. Chem. 260, 4927–4935 [PubMed] [Google Scholar]
- 17. Dabrowski U., Hanfland P., Egge H., Kuhn S., and Dabrowski J. (1984) Immunochemistry of I/i-active oligo- and polyglycosylceramides from rabbit erythrocyte membranes. Determination of branching patterns of a ceramide pentadecasaccharide by 1H nuclear magnetic resonance. J. Biol. Chem. 259, 7648–7651 [PubMed] [Google Scholar]
- 18. Watanabe K., Hakomori S. I., Childs R. A., and Feizi T. (1979) Characterization of a blood group I-active ganglioside. Structural requirements for I and I specificities. J. Biol. Chem. 254, 3221–3228 [PubMed] [Google Scholar]
- 19. Hendricks S. P., He P., Stults C. L., and Macher B. A. (1990) Regulation of the expression of Gal alpha 1-3Gal beta 1-4GlcNAc glycosphingolipids in kidney. J. Biol. Chem. 265, 17621–17626 [PubMed] [Google Scholar]
- 20. Oriol R., Ye Y., Koren E., and Cooper D. K. (1993) Carbohydrate antigens of pig tissues reacting with human natural antibodies as potential targets for hyperacute vascular rejection in pig-to-man organ xenotransplantation. Transplantation 56, 1433–1442 10.1097/00007890-199312000-00031 [DOI] [PubMed] [Google Scholar]
- 21. Galili U., Rachmilewitz E. A., Peleg A., and Flechner I. (1984) A unique natural human IgG antibody with anti-alpha-galactosyl specificity. J. Exp. Med. 160, 1519–1531 10.1084/jem.160.5.1519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Galili U., Macher B. A., Buehler J., and Shohet S. B. (1985) Human natural anti-alpha-galactosyl IgG. II. The specific recognition of alpha (1-3)-linked galactose residues. J. Exp. Med. 162, 573–582 10.1084/jem.162.2.573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Galili U. (2005) The alpha-gal epitope and the anti-Gal antibody in xenotransplantation and in cancer immunotherapy. Immunol. Cell Biol. 83, 674–686 10.1111/j.1440-1711.2005.01366.x [DOI] [PubMed] [Google Scholar]
- 24. Fushuku N., Muramatsu H., Uezono M. M., and Muramatsu T. (1987) A new endo-beta-galactosidase releasing Gal alpha 1-3Gal from carbohydrate moieties of glycoproteins and from a glycolipid. J. Biol. Chem. 262, 10086–10092 [PubMed] [Google Scholar]
- 25. Ogawa H., Muramatsu H., Kobayashi T., Morozumi K., Yokoyama I., Kurosawa N., Nakao A., and Muramatsu T. (2000) Molecular cloning of endo-beta-galactosidase C and its application in removing alpha-galactosyl xenoantigen from blood vessels in the pig kidney. J. Biol. Chem. 275, 19368–19374 10.1074/jbc.M001888200 [DOI] [PubMed] [Google Scholar]
- 26. Zakhour M., Ruvoen-Clouet N., Charpilienne A., Langpap B., Poncet D., Peters T., Bovin N., and Le Pendu J. (2009) The alpha Gal epitope of the histo-blood group antigen family is a ligand for bovine norovirus Newbury2 expected to prevent cross-species transmission. PLoS Pathog. 5, e1000504 10.1371/journal.ppat.1000504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Poole J., Day C. J., von Itzstein M., Paton J. C., and Jennings M. P. (2018) Glycointeractions in bacterial pathogenesis. Nat. Rev. Microbiol. 16, 440–452 10.1038/s41579-018-0007-2 [DOI] [PubMed] [Google Scholar]
- 28. Feng Y. J., Zhang H. M., Wu Z. W., Wang S. H., Cao M., Hu D., and Wang C. J. (2014) Streptococcus suis infection. An emerging/reemerging challenge of bacterial infectious diseases? Virulence 5, 477–497 10.4161/viru.28595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fittipaldi N., Segura M., Grenier D., and Gottschalk M. (2012) Virulence factors involved in the pathogenesis of the infection caused by the swine pathogen and zoonotic agent Streptococcus suis. Future Microbiol. 7, 259–279 10.2217/fmb.11.149 [DOI] [PubMed] [Google Scholar]
- 30. Mazmanian S. K., Liu G., Ton-That H., and Schneewind O. (1999) Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 285, 760–763 10.1126/science.285.5428.760 [DOI] [PubMed] [Google Scholar]
- 31. Lombard V., Golaconda Ramulu H., Drula E., Coutinho P. M., and Henrissat B. (2014) The carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 10.1093/nar/gkt1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Viborg A. H., Terrapon N., Lombard V., Michel G., Czjzek M., Henrissat B., and Brumer H. (2019) A subfamily roadmap of the evolutionarily diverse glycoside hydrolase family 16 (GH16). J. Biol. Chem. 294, 15973–15986 10.1074/jbc.RA119.010619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Finne J., and Krusius T. (1979) Structural features of the carbohydrate units of plasma glycoproteins. Eur. J. Biochem. 102, 583–588 10.1111/j.1432-1033.1979.tb04275.x [DOI] [PubMed] [Google Scholar]
- 34. Huang L., Zhang H., Wu P., Entwistle S., Li X., Yohe T., Yi H., Yang Z., and Yin Y. (2018) dbCAN-seq: a database of carbohydrate-active enzyme (CAZyme) sequence and annotation. Nucleic Acids Res. 46, D516–D521 10.1093/nar/gkx894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lammerts van Bueren A., Ficko-Blean E., Pluvinage B., Hehemann J. H., Higgins M. A., Deng L., Ogunniyi A. D., Stroeher U. H., El Warry N., Burke R. D., Czjzek M., Paton J. C., Vocadlo D. J., and Boraston A. B. (2011) The conformation and function of a multimodular glycogen-degrading pneumococcal virulence factor. Structure 19, 640–651 10.1016/j.str.2011.03.001 [DOI] [PubMed] [Google Scholar]
- 36. Ficko-Blean E., and Boraston A. B. (2012) Insights into the recognition of the human glycome by microbial carbohydrate-binding modules. Curr. Opin. Struct. Biol. 22, 570–577 10.1016/j.sbi.2012.07.009 [DOI] [PubMed] [Google Scholar]
- 37. Cuskin F., Lowe E. C., Temple M. J., Zhu Y. P., Cameron E. A., Pudlo N. A., Porter N. T., Urs K., Thompson A. J., Cartmell A., Rogowski A., Hamilton B. S., Chen R., Tolbert T. J., Piens K., et al. (2015) Human gut Bacteroidetes can utilize yeast mannan through a selfish mechanism. Nature 517, 165–186 10.1038/nature13995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Terrapon N., Lombard V., Drula E., Lapebie P., Al-Masaudi S., Gilbert H. J., and Henrissat B. (2018) PULDB: the expanded database of polysaccharide utilization loci. Nucleic Acids Res. 46, D677–D683 10.1093/nar/gkx1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Russo D., Parashuraman S., and D'Angelo G. (2016) Glycosphingolipid-protein interaction in signal transduction. Int. J. Mol. Sci. 17, 1732 10.3390/ijms17101732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chen C., Tang J. Q., Dong W., Wang C. J., Feng Y. J., Wang J., Zheng F., Pan X. Z., Liu D., Li M., Song Y. J., Zhu X. X., Sun H. B., Feng T., Guo Z. B., et al. (2007) A glimpse of Streptococcal toxic shock syndrome from comparative genomics of S. suis 2 Chinese isolates. PLoS ONE 2, e315 10.1371/journal.pone.0000315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yu Y. F., Wang H. G., Wang J., Feng Z. X., Wu M., Liu B. B., Xin J. Q., Xiong Q. Y., Liu M. J., and Shao G. Q. (2018) Elongation factor thermo unstable (EF-Tu) moonlights as an adhesin on the surface of Mycoplasma hyopneumoniae by binding to fibronectin. Front. Microbiol. 9, 974 10.3389/fmicb.2018.00974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Johnson M., Zaretskaya I., Raytselis Y., Merezhuk Y., McGinnis S., and Madden T. L. (2008) NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–W9 10.1093/nar/gkn201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Marchler-Bauer A., Lu S., Anderson J. B., Chitsaz F., Derbyshire M. K., DeWeese-Scott C., Fong J. H., Geer L. Y., Geer R. C., Gonzales N. R., Gwadz M., Hurwitz D. I., Jackson J. D., Ke Z., Lanczycki C. J., et al. (2011) CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 39, D225–D229 10.1093/nar/gkq1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Larkin M. A., Blackshields G., Brown N. P., Chenna R., McGettigan P. A., McWilliam H., Valentin F., Wallace I. M., Wilm A., Lopez R., Thompson J. D., Gibson T. J., and Higgins D. G. (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- 45. Waterhouse A., Bertoni M., Bienert S., Studer G., Tauriello G., Gumienny R., Heer F. T., de Beer T. A. P., Rempfer C., Bordoli L., Lepore R., and Schwede T. (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 46, W296–W303 10.1093/nar/gky427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Alexander N., Woetzel N., and Meiler J. (2011) bcl::Cluster: a method for clustering biological molecules coupled with visualization in the Pymol Molecular Graphics System. IEEE Int. Conf. Comput. Adv. Bio. Med. Sci. 2011, 13–18 10.1109/ICCABS.2011.5729867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang Y., Liang Z., Hsueh H. T., Ploplis V. A., and Castellino F. J. (2012) Characterization of streptokinases from group A streptococci reveals a strong functional relationship that supports the coinheritance of plasminogen-binding M protein and cluster 2b streptokinase. J. Biol. Chem. 287, 42093–42103 10.1074/jbc.M112.417808 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript.