Abstract
β2-Glycoprotein I (β2GPI) is an abundant plasma protein displaying phospholipid-binding properties. Because it binds phospholipids, it is a target of antiphospholipid antibodies (aPLs) in antiphospholipid syndrome (APS), a life-threatening autoimmune thrombotic disease. Indeed, aPLs prefer membrane-bound β2GPI to that in solution. β2GPI exists in two almost equally populated redox states: oxidized, in which all the disulfide bonds are formed, and reduced, in which one or more disulfide bonds are broken. Furthermore, β2GPI can adopt multiple conformations (i.e. J-elongated, S-twisted, and O-circular). While strong evidence indicates that the J-form is the structure bound to aPLs, which conformation exists and predominates in solution remains controversial, and so is the conformational pathway leading to the bound state. Here, we report that human recombinant β2GPI purified under native conditions is oxidized. Moreover, under physiological pH and salt concentrations, this oxidized form adopts a J-elongated, flexible conformation, not circular or twisted, in which the N-terminal domain I (DI) and the C-terminal domain V (DV) are exposed to the solvent. Consistent with this model, binding kinetics and mutagenesis experiments revealed that in solution the J-form interacts with negatively charged liposomes and with MBB2, a monoclonal anti-DI antibody that recapitulates most of the features of pathogenic aPLs. We conclude that the preferential binding of aPLs to phospholipid-bound β2GPI arises from the ability of its preexisting J-form to accumulate on the membranes, thereby offering an ideal environment for aPL binding. We propose that targeting the J-form of β2GPI provides a strategy to block pathogenic aPLs in APS.
Keywords: beta-2 glycoprotein I, single-molecule biophysics, X-ray crystallography, structure-function, autoimmunity, thrombosis, antiphospholipid syndrome, complement system, coagulation, lipid–protein interaction, structural biology, autoimmune disease, protein–protein interaction
β2-Glycoprotein I (β2GPI) is a 50-kDa multidomain glycoprotein that circulates in the plasma at a concentration of ∼0.2 mg/ml (1, 2) (Fig. 1A). It consists of 326 amino acids organized into five domains (DI–V) connected by four short linkers, Lnk1 (residues 61–64), Lnk2 (residues 119–122), Lnk3 (residues 182–185), and Lnk4 (residues 242–244) (3). Domains I–IV are canonical complement control protein (CCP) domains, each containing 4 cysteine residues typically forming 2 disulfide bonds (4, 5). CCP domains are found in regulators of complement activation, such as factor H (FH) (6), complement receptor 1 (CR1), membrane-cofactor protein (MCP), and decay-accelerating factor (DAF) (5). In contrast, DV is aberrant, consisting of one extra pair of cysteines and a 19-residue hydrophobic loop that is responsible for anchoring the protein to negatively charged phospholipids (7, 8) (Fig. 1A). Importantly, β2GPI exists in two interconvertible biochemical variants, oxidized (54%) and reduced (46%), depending on the integrity of the disulfide bonds (9, 10). In the oxidized form, 11 disulfide bonds are formed. In the reduced form, the disulfide bonds C288–C326 in DV and C32–C60 in DI are individually or simultaneously broken.
Figure 1.
Structure of human β2GPI and current mechanism of antigen-antibody recognition for pathogenic anti-DI antibodies in APS. A, color-coded domain structure of human β2GPI (pβ2GPI). β2GPI consists of 326 amino acids organized into five domains (DI–V) connected by four short linkers, Lnk1 (residues 61–64), Lnk2 (residues 119–122), Lnk3 (residues 182–185), and Lnk4 (residues 242–244) (3). Domains I–IV are canonical complement control protein (CCP) domains, each containing two disulfide bonds. In contrast, DV is aberrant, consisting of one extra disulfide bond and a 19-residue hydrophobic loop that is responsible for anchoring the protein to negatively charged phospholipids (7, 8). Similar to other CCP-containing proteins, β2GPI is also heavily glycosylated, bearing four N-linked and one O-linked glycosylations located at positions T130, N143, N164, N174, and N234 that account for ∼20% of the total protein mass. The position of the O- and N-linked glycosylations is shown as triangles and stars, respectively. B, based on previous studies, β2GPI is believed to adopt an O-circular (29, 31), an S-twisted (32), and a J-elongated conformation (33, 34). The J-open conformation results upon interaction of DV with the phospholipids exposing the cryptic epitope R39-R43 (purple) to the solvent. The O-circular form features an intramolecular interaction between DI (blue) and DV (yellow), with amino acids K19, R39, and R43 in DI potentially making contact with K305 and K317 in DV. The S-twisted conformation features a rotation of the DI/DII module, brokered by Lnk2, relative to the rest of the protein, resulting in the blockade of the immunogenic region R39-R43 by the N-linked glycosylation (orange line).
Forgotten for many years since its discovery in 1961 (1), β2GPI gained popularity in the fields of hematology and rheumatology in 1990, when it was recognized by two independent studies as the dominant antigen of antiphospholipid antibodies (aPLs) in antiphospholipid syndrome (APS) (11–13), a life-threatening blood-clotting disorder characterized by vascular thrombosis and pregnancy morbidity (10, 14). In this context, β2GPI is heavily studied. Indeed, autoantibodies against β2GPI (anti-β2GPI) are frequently found in young patients with a history of thrombosis (15, 16); they are often associated with lupus anticoagulant, a laboratory test that indicates predisposition for blood clots (17); they induce (18) and potentiate thrombus formation in vivo (14, 18–20) and cause pregnancy complications resulting in fetal loss (21). Thus, a deeper understanding of the structural determinants of antigen-antibody recognition is likely to accelerate the development of new diagnostics and therapeutics for APS patients.
An important feature of all aPLs, and especially highly pathogenic aPLs recognizing the epitope R39-R43 in the N-terminal domain I of β2GPI, referred to here as anti-DI antibodies (22–25), is that their detection requires proper immobilization of the antigen onto negatively charged surfaces or lipid membranes (26–28), raising the question of whether the epitopes recognized by aPLs are cryptic in the circulating form of β2GPI. In support of this viewpoint, structural studies have documented that β2GPI can adopt alternative O-circular (29–31), S-twisted (32), and J-elongated conformations (29–31, 33, 34) featuring different exposures of DI and DV to the solvent (Fig. 1B) and have led to the proposition of a model whereby the O-circular form of β2GPI, which was captured by negative-stain EM (29) and atomic force microscopy (AFM) (31) using β2GPI purified from plasma using mild conditions, is the most abundant (>90%) protein conformation of β2GPI under physiological conditions, which is immunologically inert and incapable of reacting with aPLs (Fig. 1B, left). In contrast, the J-elongated form, originally described in 1999 by X-ray crystallography using β2GPI purified from plasma using the harsh oxidizing agent perchloric acid (33, 34) and, more recently, observed by EM (29) and AFM (31) using “native” β2GPI subjected to high salt (i.e. 1.15 M NaCl) and high pH (11.5) or in complex with the mAb 3B7 (29), bacterial lipopolysaccharide (LPS) labeled with gold nanoparticles (30), and protein H of Streptococcus pyogenes (35), is believed to be the immunogenic conformation of the protein that interacts with aPLs, which appears when β2GPI binds to the membranes (Fig. 1B, right). Since the S-twisted conformation of the protein, inferred by small-angle X-ray scattering (SAXS) (32), was not detected by EM, AFM, or X-ray crystallography, this model also predicts that the S-twisted form represents a transient, unreactive intermediate state that the protein populates while transitioning between the J and O forms (Fig. 1B, central).
Although very popular in the field and highly endorsed by the scientific community, there are two important caveats for this commonly accepted model. First, the three structures have been obtained using different protein preparations and experimental techniques. Second, the O-circular form has never been documented in solution. Thus, while consensus exists regarding the fact the J-form is the structure of β2GPI bound to aPLs (29) and perhaps to the membranes (30), unresolved issues remain: 1) which conformation exists and predominates in solution, and 2) what is the conformational pathway leading to the bound state.
Encouraged by our recent results with prothrombin (36–39), the second most common antigen of aPLs in APS, this work was initiated to study the solution structure of β2GPI and provide new insights into the mechanism of autoantibody recognition. Our results, based on X-ray crystallography, single-molecule FRET (smFRET), SAXS, binding kinetics, and mutational studies, were unexpected. In contrast to what was previously thought and widely accepted in the field (2, 29) (Fig. 1B), we found that, in the free form, human recombinant oxidized β2GPI adopts the J-elongated conformation, not O-circular or S-twisted, in which DI is exposed to the solvent and, therefore, is available for autoantibody recognition. Based on this new evidence and previous findings (28, 40, 41), an alternative mechanism to explain how negatively charged phospholipids enhance the affinity toward aPLs without requiring opening of the protein structure or relocation of the glycosylations away from DI is proposed, and its implications for our understanding of APS is discussed here.
Results
Expression, purification, and functional characterization of human recombinant beta-2 glycoprotein I
To get a better grasp of the structural architecture of β2GPI under conditions relevant to physiology, we set out to perform structural and biophysical studies of fully glycosylated human recombinant β2GPI (hrβ2GPI). Two versions of the proteins were successfully expressed and purified under native conditions at high yield and purity. The first version, called LT-β2GPI, contained a long multifunctional cleavable tag at the N terminus, located right before the natural N-terminal sequence 1GRTC4 (Fig. 2A and Fig. S1). The tag was then cleaved with enterokinase to generate the intact, mature protein (hrβ2GPI). Removal of the tag was confirmed by N-terminal sequencing (Fig. 2B). The second version, called ST-β2GPI, contains a shorter, noncleavable purification tag at the N terminus that, based on our previous work (37), is expected not to affect the conformational properties of the protein (Fig. 2B and Fig. S1). ST-β2GPI was made to eliminate the enterokinase cleavage step, which was very laborious and not as efficient as expected. The presence of the short tag was confirmed by N-terminal sequencing and accounted for the different electrophoretic mobilities observed between recombinant and plasma purified protein before and after enzymatic removal of the N-glycosylations (Fig. 2B).
Figure 2.

Functional characterization of human recombinant β2GPI. A, color-coded domain structure of the recombinant variants used in this work (i.e. LT-β2GPI, hrβ2GPI, and ST-β2GPI), highlighting the position and chemical composition of the N-terminal tag. The long tag (LT, yellow) is composed of three parts: (i) a calcium-dependent epitope for the mAb HPC4 (EDQVDPRLIDGK); (ii) a site-specific biotinylation sequence (AviTag); and (iii) a conventional enterokinase recognition site (DDDDK). Two flexible linkers (i.e. GGGS) were introduced to separate the three functional units of the tag to avoid the formation of secondary structure and ensure exposure of the tag to solvent. Removal of the LT with enterokinase generates hrβ2GPI. The short-tag version of β2GPI contains only the HPC4 purification tag (purple). B, SDS-PAGE analysis of the recombinant proteins (sample 1, LT-β2GPI; sample 2, hrβ2GPI; sample 3, ST-β2GPI) and plasma-purified β2GPI (sample 4) before (left) and after (right) the removal of the glycosylations. Protein deglycosylation mix II, from NEB, was used to remove O-linked and N-linked glycosylations under denaturing conditions. Chemical identity was verified by N-terminal sequencing, and results are the following: band 1, EDQVD; band 2, GRT; band 3, EDQVD; band 4, GRTC. C, representative sensograms of the interaction between LT-β2GPI and liposomes (PC:PS, 80:20) monitored by SPR. Liposomes were immobilized on an L1 chip and soluble β2GPI (0–2 μm) was used in the fluid phase. D, dose-dependent curves quantifying the interaction of LT-β2GPI (red circles), hrβ2GPI (blue circles), ST-β2GPI (green circles), and pβ2GPI (gray circles) with liposomes monitored by SPR. Affinity values (Kd) are 0.19 ± 0.05 μm for LT-β2GPI, 0.33 ± 0.08 μm for hrβ2GPI, 0.22 ± 0.08 μm for ST-β2GPI, and 0.32 ± 0.09 μm for pβ2GPI. No significant binding was observed with liposomes made entirely of PC (light gray circles). Each experiment was repeated at least three times, using two distinct batches of proteins. E, reactivity of immobilized LT-β2GPI (red bars), hrβ2GPI (blue bars), ST-β2GPI (green bars), and pβ2GPI (gray bars) against IgG anti-β2GPI antibodies followed by ELISA. Comparisons between 2 groups were performed using a two-sample t test. Results were considered significant at p < 0.05 (*).
To evaluate the functional integrity of the recombinant proteins, LT-β2GPI, hrβ2GPI, and ST-β2GPI were tested in several biochemical assays. β2GPI purified from human plasma using the perchloric acid method (pβ2GPI) was used as a control. Using surface plasmon resonance (SPR), a technique that allows us to measure association (on) and dissociation (off) rate constants in real time, we found that all variants interacted avidly with liposomes containing negatively charged phospholipids, such as phosphatidylserine, yet they failed to interact with phospholipids made entirely of phosphatidylcholine (Fig. 2, C and D). Importantly, the values of the affinity constants were similar for all the constructs and consistent with published data (28), and so was the inhibitory effect of physiological concentrations of calcium chloride (data not shown). These results document structural integrity of the hydrophobic loop in DV and also prove that the phospholipid binding activity of β2GPI is not perturbed by the presence or removal of the purification tags.
In addition to properly interacting with phospholipids, the recombinant proteins were also successfully recognized in ELISAs by aPLs isolated from four triple-positive APS patients, which contain anti-DI antibodies (25, 36, 42) (Fig. 2E). In this case, however, LT-β2GPI exhibited significantly lower values of optical density at 450 nm than the other variants and plasma-purified protein, suggesting that the presence of the long tag masks some epitopes or, more likely, changes the preferential orientation of the molecule that is adsorbed onto the plastic surface. Taken together, these studies validate recombinantly made β2GPI as a proxy for plasma-purified β2GPI. They also demonstrate that, under physiological conditions, β2GPI is primed for phospholipid binding.
X-ray crystal structure of human recombinant beta-2 glycoprotein I
In 1999, two identical X-ray crystal structures of β2GPI were simultaneously solved by two independent groups (33, 34). β2GPI used in these studies was purified from human plasma using the perchloric acid method and crystallized at resolutions of 2.87 Å (1CIZ) and 2.7 Å (1QUB). Since then, no other crystal structure has been deposited in the Protein Data Bank (PDB).
To investigate the structural properties of the recombinant proteins, crystallization experiments were performed for all the protein constructs. While it was not possible to crystallize LT-β2GPI, we solved the X-ray crystal structures of hrβ2GPI (Fig. 3A) and ST-β2GPI (Fig. 3B) at 2.6- and 3.0-Å resolution, respectively. We also solved the X-ray crystal pβ2GPI at 2.4 Å as a control (Fig. 3C). Similar to what was previously reported by Bouma et al. (34) and Schwarzenbacher et al. (33), diffraction quality crystals were obtained after 2 weeks at 4 °C using ammonium sulfate as a precipitating agent. The crystals belong to the orthorhombic space group C2221 (Table 1). Notably, ST-β2GPI, for which extra electron density was observed at the N terminus (Fig. 3D), crystallized under similar conditions and in the same space group as hrβ2GPI and pβ2GPI, confirming minimal structural perturbation introduced by the artificial tag.
Figure 3.

X-ray crystal structures of human recombinant β2GPI. X-ray crystal structures of hrβ2GPI (blue) (A), ST-β2GPI (green) (B), and pβ2GPI (gray) (C) solved at 2.6-, 3.0-, and 2.4-Å resolution. All three structures document similar elongated conformations of the protein spanning ∼140 Å in length. Disulfide bonds are highlighted in yellow. D, zoom-in of three extra residues (DGK) belonging to the N-terminal tag preceding the natural sequence of β2GPI (1GRTC4) exclusively found in the structure of ST-β2GPI, as expected. The electron density 2F0-Fc map is countered at 1.0σ. E, structural architecture of the epitope R39-R43 in DI highlighting the position and interactions of R43 (magenta stick) with the nearby residues R39, G41, and T57. Hydrogen bonds between the guanidinium group of R43 and neighboring residues are shown in black. Of note, the conformation of this segment is not involved in crystal contacts and is conserved in all the available crystal structures of β2GPI solved thus far, despite the high salt concentrations in which the crystals grow, suggesting that this is a genuine structural feature of β2-GPI.
Table 1.
Crystallographic data for the structures of human beta-2 glycoprotein I
| Parameter | Value(s) for: |
||
|---|---|---|---|
| pβ2GPI | hrβ2GPI | ST-β2GPI | |
| Buffer/salt | 100 mm HEPES, pH 7.5/1.5 m AmSO4, 20 mm CaCl2, 2% glycerol | 100 mm MES, pH 6.0/1.6 m AmSO4 | 100 mm HEPES, pH 7.5/1.5 m AmSO4, 20 mm CaCl2, 2% glycerol |
| PDB entry | 6V06 | 6V08 | 6V09 |
| Data collection | |||
| Wavelength (Å) | 1.54 | 1.033 | 1.54 |
| Space group | C2221 | C2221 | C2221 |
| Unit cell dimensions (Å) | a = 160.8, b = 166.9, c = 114.0 | a = 159.3, b = 173.2, c = 115.2 | a = 160.2, b = 171.2, c = 113.4 |
| Molecules/asymmetric unit | 1 | 1 | 1 |
| Resolution range (Å) | 40–2.4 | 40–2.6 | 40–3.0 |
| No. of observations | 513,296 | 363,697 | 158,386 |
| No. of unique observations | 59,497 | 48,503 | 31,543 |
| Completeness (%) | 98.8 (97.1) | 97.1 (77.1) | 98.8 (97.7) |
| Rsym (%) | 7.9 (50.6) | 14.3 (64.7) | 9.5 (46.1) |
| I/σ(I) | 21.1 (2.4) | 10.0 (1.5) | 13.7 (2.4) |
| Refinement | |||
| Resolution (Å) | 40–2.4 | 40–2.6 | 40–3.0 |
| Rcryst, Rfree | 0.201, 0.236 | 0.200, 0.232 | 0.223, 0.246 |
| No. of reflections (working/test) | 56,556/2935 | 45,862/2599 | 29,924/1611 |
| No. of protein atoms | 2540 | 2510 | 2517 |
| No. of solvent molecules | 431 | 377 | 15 |
| RMSDa bond lengths (Å) | 0.013 | 0.010 | 0.011 |
| RMSD angles (˚) | 2.0 | 2.0 | 1.7 |
| RMSD ΔB (Å2) (mm/ms/ssb) | 5.12/5.28/6.93 | 4.54/5.13/5.85 | 3.97/3.58/4.38 |
| Protein (Å2) | 63.8 | 68.5 | 71.0 |
| Solvent (Å2) | 64.4 | 62.8 | 49.6 |
| Ramachandran plot: | |||
| Most favored (%) | 98.9 | 100.0 | 99.6 |
| Generously allowed (%) | 1.1 | 0.0 | 0.4 |
| Disallowed (%) | 0.0 | 0.0 | 0.0 |
aRMSD from ideal bond lengths and angles and RMSD in B-factors of bonded atoms.
bmm, main chain–main chain; ms, main chain–side chain; ss, side chain–side chain.
Consistent with previous structural data (33, 34), all three independently solved X-ray crystal structures depicted β2GPI featuring an elongated conformation spanning ∼140 Å in length, from the N to the C terminus. The first three domains, DI–DIII, are aligned along the vertical axis of the molecule, whereas DIV and DV bend, forcing the molecule to adopt a characteristic J-shaped elongated form resembling a hockey stick. DI and DV are located >100 Å apart, and both of them are exposed to the solvent. Interestingly, the side chain of residue R43, which is part of the epitope recognized by anti-DI antibodies, is partly exposed to the solvent as it forms a hydrogen bond network with residues R39, G41, and T57 (Fig. 3E).
β2GPI is known to circulate in alternative chemically different species, oxidized and reduced (9, 10, 43). Oxidized β2GPI contains 11 disulfide bonds, whereas reduced β2GPI contains 8 disulfide bonds and four free thiols resulting from the rupture of the disulfide bonds C32-C60 in DI and C288-C326 in DV. In our crystal structures, all 22 cysteine residues were engaged in 11 disulfide bridges (Fig. 3, A–C), indicating that the structure of human recombinant β2GPI purified by immunoaffinity and size exclusion chromatography represents the oxidized form of β2GPI.
Given the similar experimental conditions in which the crystals grew, it was not surprising to observe that, overall, the three structures were superimposable (RMSD of 0.709, 0.516, and 0.630 Å for 6V06 versus 6V08, 6V06 versus 6V09, and 6V08 versus 6V09, respectively) and also very similar to the published ones (RMSD of ≤0.810 Å) (33, 34) (Fig. 4A). There were, however, a few notable differences. First, the phospholipid binding loop in DV (residues 308–319) adopted alternative conformations (RMSD of 3.122 Å), documenting flexibility and exposure to the solvent (Fig. 4A). This is in agreement with spectroscopic data indicating that W316 is exposed to the solvent (44) and that plasmin cleaves β2GPI at position K317-T318 (45). Second, significant extra electron density appeared after molecular replacement, especially in the two datasets at a higher resolution (i.e. 2.4 and 2.6 Å) (Fig. 4B), which we attributed to the N-linked glycosylations (Fig. 4, C and D). Guided by MS analyses (46), we modeled the following sugar sequences: Gal2GlcNAc2Man3GlcNAc2 at N143, Gal2GlcNAc2Man3GlcNAc2 at N164, GlcNAc2 at N174, and Gal3GlcNAc2 at N234. The presence of a putative O-linked glycosylation at T130 could not be confirmed because of weak density, even in the datasets at higher resolution.
Figure 4.
Location and structural role of the N-linked glycosylations. A, superposition of five X-ray crystal structures of β2GPI highlights structural similarities yet diversity of the phospholipid binding loop in DV (residues 308–319). B, extra electron density detected in the structure of pβ2GPI solved at 2.4-Å resolution attributed to the N-linked glycosylations. The domains of β2GPI are color coded as shown in Fig. 1. The presence of a putative O-linked glycosylation at T130 could not be confirmed because of weak density. Side (C) and top (D) views of the N-glycosylations surrounding DIII are shown. The electron density 2F0-Fc map is countered at 0.8σ.
smFRET studies of human recombinant beta-2 glycoprotein I
The high salt concentration used in the crystallization buffers may destabilize hydrogen bonds and favor hydrophobic interactions, forcing the protein to assume a nonnative conformation. To test whether the J-elongated form exists in solution and determine its relative abundance compared with other forms, we applied smFRET to β2GPI (38, 39, 47) (Fig. 5A). By recording the energy that is transferred from an excited fluorophore (donor) to a second fluorophore with spectral overlap (acceptor), smFRET measures distances on a nanometer scale, allowing us to interrogate protein structures and protein conformational changes one molecule at a time. This overcomes averaging effects seen in traditional bulk experiments.
Figure 5.

smFRET measurements of β2GPI in solution. A, confocal microscope used to collect smFRET data of fluorescently labeled β2GPI showing the ps-pulsed laser box, the 60× objective, the observation chamber (confocal volume), the location of the pinhole, and two SPAD detectors. B, structure-based design of the FRET constructs S13C/S112C, S13C/S312C, S112C/S312C, and S190C/S312C used in this study. Ser residues mutated to Cys for conjugation with the thiol-reactive dyes AF555 and AF647 used in smFRET measurements are indicated by red spheres. C, FRET couples are listed with their respective domains. Cα-Cα distances were obtained from the crystal structure of hrβ2GPI. D, after labeling and gel filtration, selective incorporation of the fluorescence dyes was verified by loading the proteins (samples 2–5) alongside β2GPI WT (sample 1) into a gradient 4–12% polyacrylamide gel in the presence of SDS and visualized by Coomassie brilliant blue R-250 (black and white) or fluorescence intensity by exciting donor at 532 nm (red panel) and acceptor at 640 nm (blue panel). The uncut gel is shown in Fig. S2C. smFRET histograms of the mutants S13C/S312C, S112C/S312C, S13C/S112C, and S190C/S312C were labeled with AF555/647 measured in Tris 20 mm, pH 7.4, 145 mm NaCl, 0.1% Tween 20 for 1 h at room temperature at a concentration of 100 pm. Populations were fit to a single Gaussian distribution (black lines). FRET efficiency values and calculated distances are indicated. The theoretical shot noise peak highlighting conformational heterogeneity for the FRET pair 190/312 is shown as a dotted line. F, effect of sodium chloride monitored by smFRET. smFRET experiments for the mutant 13/312 (100 pm) were performed at 25, 145, and 1000 mm NaCl in 20 mm Tris, pH 7.4, 145 mm NaCl. G, effect of pH monitored by smFRET. smFRET experiments for the mutant 13/312 (100 pm) were performed at pH 4.0 (20 mm Na acetate, pH 4.0, 150 mm NaCl), pH 7.4 (20 mm Tris, pH 7.4, 145 mm NaCl), and pH 10.0 (20 mm glycine, pH 10.0, 145 mm NaCl).
Guided by our structures, we generated four FRET pairs in the ST background, S13C/S112C, S13C/S312C, S112C/S312C, and S190C/S312C (Fig. 5, B and C), by replacing the natural serine residues with isosteric cysteines and then reacting those newly engineered cysteines with Alexa Fluor 555 (AF555)-maleimide as the FRET donor and Alexa Fluor 647 (AF647)-maleimide as the FRET acceptor. Residue 13 is located in DI, residue 112 is located in DII, residue 190 is located in DIV, and residue 312 is located in DV. Labeling occurred only at the engineered sites, as no fluorescence was observed for WT β2GPI (Fig. 5C). This result is consistent with our structural data and provides additional support to our conclusion that human recombinant β2GPI secreted in the media and purified for our structural studies does not contain free thiols. Given the Förster radius, R0 = 50 Å, of the 555/647 FRET couple, the crystal structure predicts no FRET for the mutants S112C/S312C and S13C/S312C. In contrast, high FRET and low but measurable FRET values are expected for the FRET pair S13C/S112C and S190C/S312C, respectively. This is because residues 13 and 112 are located ∼24 Å apart while the Cα-Cα distance between residues 190 and 312 is ∼46 Å. Remarkably, the experimental results were fully consistent with our structure-based calculations, validating our previous structural work and proving that the elongated conformation predominates (>90%) under physiological pH and salt concentrations (Fig. 5E). Specifically, probes located at positions C13, C312, C112, and C312 reported a negligible FRET signal, whereas probes attached to the S13C/S112C and S190C/S312C mutants reported high (EFRET = 0.92) and low (EFRET = 0.26) FRET values, respectively. Interestingly, the construct 190/312 displayed a FRET distribution wider than the theoretical distribution predicted by shot noise (48), documenting the existence of multiple conformations at equilibrium mediated by the flexibility of Lnk4.
We also tested the effect of sodium chloride (0.025 versus 0.145 versus 1 m, Fig. 5F) and pH (4.0 versus 7.4 versus 10.0, Fig. 5G). In contrast to previous findings in which low pH and low concentrations of sodium chloride favored circularization of the protein (29), no significant FRET differences were observed, suggesting that variation of [NaCl] and pH produces conformational changes, such as further elongation of the protein structure or rotations of the domains, that could not be detected by our FRET pairs.
SAXS studies of human recombinant beta-2 glycoprotein I
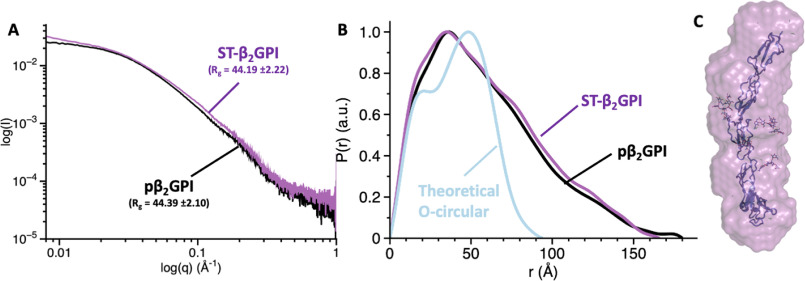
To rule out potential artifacts arising from the replacement of natural amino acids with cysteine and incorporation of fluorescent dyes, we collected SAXS data for ST-β2GPI and pβ2GPI under physiological pH and salt concentrations (Fig. 6). SAXS is a biophysical method that is particularly useful to assess the overall shape of biological macromolecules in solution (49), i.e. linear versus globular, and, similar to smFRET, therefore is ideal to detect large conformational changes in β2GPI. The radius of gyration (Rg) for ST-β2GPI and β2GPI purified from plasma was 44.19 ± 2.22 and 44.39 ± 2.10, respectively (Fig. 6A). These values and the maximum linear dimension (Dmax) obtained by computing the pair distance distribution function, P(r), shown in Fig. 6B (i.e. ∼160 Å), agree very well with the values calculated using the J-elongated structure captured by X-ray crystallography (i.e. Rg = 45.22 and Dmax ∼140 Å, Fig. 3A) and explain the lack of FRET for the FRET couples 13/312 and 112/312 (Fig. 5E), confirming that β2GPI is extended in solution, not circular. Whether the S- or J-form predominates in solution cannot be concluded from these studies due to the low resolution of SAXS. However, it is worth noting that the molecular envelope calculated ab initio from the SAXS data of ST-β2GPI returned a linear and not a twisted envelope, as documented before (32), in which the structure of ST-β2GPI fits nicely (Fig. 6C) without having to rotate or bend the domains.
Figure 6.
Elongated conformation of β2GPI revealed by SAXS. Scattering profiles (A) and pair distribution functions (B) for pβ2GPI (black) and ST-β2GPI (magenta) collected at 2 mg/ml under physiological conditions (20 mm Tris, pH 7.4, 145 mm NaCl). The calculated values of the radius of gyration, Rg, are very similar for pβ2GPI and ST-β2GPI. The blue curve in panel B, which is significantly different from the experimental scattering profiles, represents the theoretical pair distribution function for a hypothetical circular conformation. C, ab initio envelope calculated from scattering profiles for ST-β2GPI (magenta). Note that the linear arrangement of domains in the structure 6V09 is consistent with the elongated SAXS envelope.
Autoantibody binding studies
In addition to demonstrating that oxidized β2GPI adopts an elongated conformation in solution, our structural analysis predicts that this form may be primed for autoantibody binding, especially anti-DI antibodies. To test this hypothesis, we took advantage of the reactivity of MBB2 (21), a newly developed recombinant mAb raised against DI that, upon complement fixation, recapitulates, in vivo, most of the clinical characteristics assigned to pathogenic aPLs. The binding of β2GPI to MBB2 was monitored using SPR (Fig. 7).
Figure 7.

Exposure of DI revealed by MBB2 binding and mutagenesis studies. A, interaction of MBB2 and LT-β2GPI monitored by SPR. MBB2 was immobilized onto a CM5 chip using NHS/EDC chemistry to a final density of 6000 RU. A solution of LT-β2GPI (0–20 μm) in Tris 20 mm, pH 7.4, 145 mm NaCl, 0.01% Tween 20 was injected at 30 µl/min for 60 s to observe binding, followed by 60 s of dissociation in running buffer. B, dose-dependent curves quantifying the interaction of LT-β2GPI (red circles), hrβ2GPI (blue circles), ST-β2GPI (green circles), and pβ2GPI (gray circles) with MBB2 monitored by SPR. Affinity values (Kd) are 2.2 ± 0.5 μm for LT-β2GPI, 2.1 ± 0.6 μm for hrβ2GPI, 1.8 ± 0.5 μm for ST-β2GPI, and 1.4 ± 0.6 μm for pβ2GPI. Each experiment was repeated at least three times, using two distinct batches of proteins. C, effect of sodium chloride. SPR binding experiments were performed at 15, 75, 150, and 300 mm NaCl. Analysis of the slope of the linear fit of the association constant versus [Na+] reveals a strong dependence of Na+ and the presence of at least 2.0 ionic contacts (salt bridges) in the complex (59, 60). D, electrostatic potential displaying positive (blue) and negative (red) clusters with a −2.0 to 2.0 intensity scale. The N-glycosylations are shown as magenta sticks. Locations of residues R39, R43, K44, and 120–122 were targeted by site-directed mutagenesis. E, SPR analysis of β2GPI WT and mutant R39A/R43A/RK44A reveals that residues R39 and R43 are critical for MBB2 binding. F, SPR analysis of β2GPI WT (black circles), DI (green circles), β2GPIΔ(120-122) (magenta circles), and deglycosylated β2GPI (blue circles). Removal of DII–DV, perturbation of Lnk2, and removal of the glycosylations do not affect the binding affinity of MBB2 toward β2GPI. This indicates that 1) DI is primed for autoantibody recognition in solution, 2) the exposure of residues R39 and R43 in DI is independent of the conformation of Lnk2, and 3) Lnk2 is not an epitope of MBB2. Given the different molecular weights of the constructs, the binding curves are reported as normalized fraction bound versus analyte to facilitate comparison. Each experiment was repeated at least three times, using two distinct batches of proteins.
To retain the native conformation of β2GPI in solution, we immobilized MBB2 to the chip's surface and injected β2GPI in the fluid phase. Binding between MBB2 and β2GPI should occur only if DI is exposed to the solvent. This experimental setup is different from previously reported interaction data between MBB2 and β2GPI in which β2GPI was covalently immobilized on a dextran-based chip and the antibody was used in the fluid phase to mimic binding of MBB2 to β2GPI bound to negatively charged phospholipids (21). The results of the experiments shown in Fig. 7A demonstrate that MBB2 interacts with β2GPI in solution with a modest but measurable affinity, characterized by a dissociation constant, Kd, of 2.2 ± 0.2 μm (Fig. 7B). The interaction was characterized by very fast on and off rates, suggesting that electrostatic interactions dominate the binding interface. This was demonstrated by systematic experiments in which we varied the concentration of sodium chloride in the running buffer from 300 to 15 mm. As expected, the affinity constant (Ka) of MBB2 for β2GPI increased ∼700-fold at low salt concentrations, from 4.4 × 104 m−1 at 300 mm NaCl to 3.2 × 107 m−1 at 15 mm NaCl (Fig. 7C).
Interestingly, the Kd value determined for MBB2 at 145 mm NaCl is similar to the Kd value obtained by Dienava-Verdoold et al. for patient-derived monoclonal antibodies targeting DI (50). Furthermore, the affinity of MBB2 for soluble β2GPI is 200-fold weaker than the affinity previously determined for MBB2 toward immobilized β2GPI (i.e. Kd of 11 nm) (21), as expected for pathogenic aPLs. To test whether MBB2 binds to the epitope R39-R43 in DI that is targeted by pathogenic aPLs in APS patients (22), positively charged residues R39, R43, and K44 were mutated to the neutral amino acid alanine. Remarkably, a significant ∼30-fold reduction of the affinity under physiological conditions was detected for the triple mutant (Fig. 7, D and E). This result is consistent with the electrostatic nature of the interaction and demonstrates that these three positively charged amino acids are part of the binding epitope.
The ability of MBB2 to interact with residues R39-K44 in DI provides the opportunity to further and independently test the model in Fig. 1B envisioning the epitope R39-R43 to be cryptic, either buried by DV in the O-circular form (29) or shielded by the N-linked glycosylations in the S-twisted form (32). To this goal, we measured the affinity of three new constructs, isolated DI (residues 1–60), β2GPI deletion Lnk2 (β2GPI Δ120-122), and recombinantly deglycosylated β2GPI (degβ2GPI; T130S/N143Q/N164Q/N174Q/N234Q) toward immobilized MBB2, which were designed to force the exposure of DI to the solvent and, in principle, should have higher affinity for MBB2. Our binding data shown in Fig. 7F indicate that all three constructs interact with immobilized MBB2 with micromolar affinity comparable to that of full-length β2GPI WT. This conclusively rules out a significant contribution of the neighboring domains and the glycosylations in shielding the R39-R43 epitope when the protein is free in solution and further validates the conclusions derived from our structural studies.
Discussion
In recent years, our structural understanding of multidomain clotting and complement factors, such as prothrombin (37, 39), plasminogen (51), FXII (52), ADAMTS-13 (53), factor H (6), and properdin (54), has evolved, and so have the technologies capable of capturing protein conformational changes in solution. Owing to its flexibility, the solution structure of β2GPI has remained controversial, and so is the mechanism through which pathogenic anti-DI antibodies recognize β2GPI in APS patients, thereby promoting thrombosis. To fill this gap in knowledge, this study was initiated to provide a rigorous structural and functional assessment of plasma-purified and human recombinant β2GPI under conditions relevant to physiology, using traditional (X-ray crystallography, SAXS, and mutagenesis studies) and state-of-the-art (smFRET) technologies. To our knowledge, this is the first application of smFRET to a CCP-containing protein.
A first major conclusion emerging from this study is that human β2GPI expressed in mammalian cells and purified under native conditions adopts an elongated, flexible conformation in which DV and DI are exposed to the solvent. Importantly, this conformation has 11 disulfide bonds (Fig. 3) and does not react with thiol-specific maleimide dyes (Fig. 5B), thereby representing the oxidized form of β2GPI, which accounts for 54% of the protein in human plasma (9). In the free form, under physiological pH and salt concentrations, we propose that β2GPI is primed for phospholipid binding and autoantibody recognition. As such, these findings contrast with current structural and mechanistic models envisioning β2GPI primarily (∼90%) adopting an O-circular conformation in human plasma and formation of the J-elongated form only after binding to the membranes (Fig. 1B).
A second major conclusion of this study is that recombinantly made β2GPI protein is structurally and functionally identical to β2GPI purified from plasma using the perchloric acid method. Hence, in contrast to what was previously thought (29), the elongated conformation of β2GPI is not an artifact caused by the harsh purification methods or crystallization conditions but a genuine conformation of the protein in solution. Whether the treatment of β2GPI with perchloric acid introduces additional chemical modifications to specific amino acids (e.g. oxidation and deamidation) remains a possibility that cannot be ruled out by our current studies.
In addition to settling previous controversies in the field, the recognition that the elongated form of oxidized β2GPI preexists and, according to our smFRET experiments (Fig. 5E) and data from Ioannou et al. (9), predominates in human plasma bears important implications in our understanding of the APS pathology. It also provides new ideas for the development of APS-focused diagnostics and therapeutics.
Regarding the mechanism of anti-DI antibody recognition, our structural and binding data indicate that the opening of the protein structure and relocation of DI away from the glycosylations are neither necessary nor sufficient to explain how β2GPI becomes a better antigen for anti-DI antibodies upon binding to the membranes. They instead strongly suggest that, in agreement with previous oligomerization models (39, 48, 49), binding of the preexisting elongated conformation of β2GPI to the membranes gives rise to an ideal surface in which β2GPI has a sufficiently high density and adopts a favorable orientation that promotes bivalent binding. Indeed, recent theoretical calculations (55) and binding studies (56) elegantly demonstrated that the energetic gain associated with limiting antigen diffusivity is quite substantial, and such a gain is expected to be even higher for low-affinity antibodies, such as aPLs. In this context, rotation or bending of the CCP domains relative to the plane of the membrane, such as those documented here, by the FRET couple 190/312 may be important for proper packing of β2GPI onto the lipids. A contribution of local conformational changes, such as exposure of R43 upon binding to the lipids, is also possible, yet, considering the intrinsic low affinity of aPLs for their targets, the modest effect caused by mutations in isolated DI (51), and the key role of bivalency documented before (49, 52), the energetic contribution of this process is expected to be minor.
One of the strongest arguments in favor of the O-circular/J-elongated conformational model is that immunocomplexes are rarely found in patients' plasma (57, 58). If β2GPI were primed for autoantibody binding, then immunocomplexes should be seen more often in APS patients. An alternative view for this phenomenon, which is consistent with the preexistence of the J-elongated form in solution, comes from our SPR binding kinetics and analysis of the literature. Figure 7A shows that, under physiological pH and salt concentrations, the interaction of MBB2 and β2GPI is characterized by a low affinity (Kd of 2.2 μm) and fast on/off rates, indicating that, in solution, the immunogenic complexes form rapidly but also dissociate very rapidly. According to previously published data (20), the plasma level of anti-β2GPI antibodies in thrombotic APS patients represents 0.5% or less of the total IgG pool, which corresponds to molar concentrations of ∼0.1 μm. Given the micromolar affinity of MBB2 for its target, this translates into <90% complex formation at equilibrium. Hence, the immunocomplexes are extremely difficult to detect and even more challenging to purify because of their thermodynamic instability and low concentration of autoantibodies with respect to their targets. In this context, a possible role of the negatively charged surfaces, as suggested by our SPR binding experiments performed at low concentrations of NaCl (Fig. S3), could be to stimulate the binding of anti-DI antibodies to β2GPI by slowing down the dissociation rate, resulting in complex stabilization. Such a mechanism is fully consistent with recent observations (35) and previous data (49) and might be conserved among other aPLs.
An important and relatively unknown aspect of the pathogenic APS is how aPLs potentiate thrombus formation (10, 14, 61–64). The discovery that the J-elongated form dominates in solution provides new insights into this mechanism. Since binding between aPLs and oxidized β2GPI can occur in solution, our studies support the hypothesis that stimulation of circulating and endothelial cells requires clustering of the antigen onto the membranes, which leads to aPL-dependent complement activation (18) and/or aPL-induced receptor dimerization (63–65).
Regarding the development of new diagnostics and therapeutics, if the main role of the membranes is to increase the local concentration of the elongated form, we speculate that immobilization of human recombinant β2GPI produced in this work at the desired density and with a defined orientation should provide a novel, efficient, robust, and cost-effective method to detect anti-β2GPI antibodies. On the other hand, blocking the binding of the elongated form of β2GPI to cell receptors and phospholipids should limit the pathogenic effects of anti-DI antibodies. This approach could complement current strategies aimed at competing with anti-DI antibodies in solution (13, 57,58, 61), as it would theoretically block other potentially pathogenic aPLs, in addition to those targeting DI. Consistent with this premise, antibodies against DV found in APS patients do not induce thrombosis but are protective instead (66), and a novel dimeric molecule, A1-A1, protects mice from aPL-induced thrombosis by interfering with ApoER2 and phospholipid binding (63).
In conclusion, our study provides clear-cut evidence that the monomeric oxidized form of β2GPI adopts a J-elongated conformation under physiological pH and salt concentrations, not O-circular or S-twisted. Whether the reduced form of β2GPI adopts alternative structures in solution remains a possibility. However, answering this question is challenging and will require rigorous structural, biophysical, and mutagenesis studies.
Experimental procedures
Protein production and purification
β2GPI WT and mutants were expressed in BHK (LT-β2GPI) and HEK293 (ST-β2GPI) mammalian cells and purified to homogeneity by immunopurification and heparin and size exclusion chromatography after swapping the signal peptide of β2GPI with the one of the coagulation factor X to boost expression and adding a furin specific recognition motif RRKR for quantitative posttranslational processing. The purity and chemical identity of each fragment were verified by SDS-PAGE and N-terminal sequencing. Domain I (1–60) was chemically synthesized and refolded as done before (67). Plasma-derived β2GPI (pβ2GPI) was purified using the perchloric acid method, as described previously (67). MBB2 was produced as described before (21). Liposomes composed of phosphatidylcholine (PC) or phosphatidylcholine and phosphatidylserine (PS) in a 4:1 molar ratio (PC:PS) were prepared by extrusion using 100-nm polycarbonate membranes (Avanti Polar Lipids, Alabaster, Alabama), kept a 4 °C, and used within 7 days. ELISAs were performed as described before (25, 36, 67). Protein concentrations were determined by reading at 280 nm with molar extinction coefficients adjusted according to the amino acid sequence. All other chemicals were purchased from Sigma-Aldrich.
SPR experiments
Binding affinities for liposomes were measured as done before (36), using an L1 sensor chip in which liposomes were immobilized at 1600 response units (RU). Titrations were performed by injecting increasing concentrations (0–2 μm) of β2GPI and its variants in running buffer (20 mm Tris, pH 7.4, 150 mm NaCl, 0.1%, w/w, BSA) at a flow rate of 25 μl/min at 25 °C. Binding affinities for MBB2 were measured using a CM5 sensor chip in which MBB2 was immobilized at 6000 RU using NHS/EDC chemistry. Titrations were performed by injecting increasing concentrations (0–20 μm) of β2GPI and its variants in running buffer (20 mm Tris, pH 7.4, 25–300 mm NaCl, 0.01%, w/w, Tween 20) at a flow rate of 25 μl/min at 25 °C. All experiments were carried out on a BIAcore-S200 instrument (GE-Healthcare). The dissociation constants (Kd) were obtained as a fitting parameter by plotting the value of the RU at the steady state for each concentration using the BIAevaluation software and Origin Pro 2015.
X-ray studies
Crystallization of human recombinant (hrβ2GPI and ST-β2GPI) and pβ2GPI was achieved at 4 °C by the vapor diffusion technique, using the Art Robbins Instruments PhoenixTM liquid-handling robot with 10 mg/ml protein (0.3 µl) mixed with an equal volume of reservoir solution. Optimization of crystal growth was achieved by the hanging drop vapor diffusion method, mixing 3 µl of protein (10 mg/ml) with equal volumes of reservoir solution at 4 °C. After 7–10 days at 4 °C, crystals were frozen with 25% glycerol from original mother liquid. X-ray diffraction data were collected at 100° K with a home source (Rigaku 1.2-kW MMX007 generator with VHF optics) Rigaku Raxis IV++ detector for pβ2GPI and ST-β2GPI and with detector Pilatus of beamline IDD23 at the Advanced Photon Source, Argonne, IL, for hrβ2GPI. Data sets were indexed, integrated, and scaled with the HKL2000 software package (68). All structures were solved by molecular replacement using PHASER from the CCP4 suite (46) and the structure of pβ2GPI (PDB entry 1C1Z) as the starting model. Refinement and electron density generation were performed with REFMAC5 from the CCP4 package. 5% of the reflections were randomly selected as a test set for cross-validation for four structures. Model building and analysis of the structures were carried out using COOT (69). Ramachandran plots were calculated using PROCHECK. Statistics for data collection and refinement are summarized in Table 1.
Protein labeling for single-molecule detection
Selective labeling of the unpaired Cys residues with AF555–C2-maleimide as the donor and AF647–C2-maleimide as the acceptor was achieved as described recently for prothrombin, with minor modifications (37, 38). Briefly, β2GPI (8–10 μm) was used in the presence of 350 mm NaCl, 20 mm Tris, pH 7.4, at room temperature for 1 h in the dark and in the presence of DTT at a molar ratio ([−SH]:[DTT]) of 1:1.6. Constructs were then labeled with an equal molar mixture of the thiol-reactive dyes, using AF555–C2-maleimide as the donor and AF647–C2-maleimide as the acceptor (Thermo Fisher Scientific, MA). The labeling reaction was carried out for 3 h at room temperature in the dark. The monomeric protein free of unreacted dyes was purified on an analytical Superdex 200 column (GE Healthcare, PA), and the efficiency of derivatization was assessed by UV-Vis measurements.
Single-molecule FRET measurements
FRET measurements of freely diffusing single molecules were performed with a MicroTime 200 confocal microscope (PicoQuant, Berlin, Germany) as detailed elsewhere (37, 38). Briefly, as shown in Fig. 5A, experiments were carried out with pulsed interleaved excitation, which reports the status of both donor and acceptor fluorophores by sorting molecules on the basis of relative donor:acceptor stoichiometry and apparent FRET efficiency. The donor and acceptor dyes were excited with a ps-pulsed diode laser at 532 and 638 nm, respectively. To achieve pulsed interleaved excitation (70), the 532-nm laser was electronically delayed 25 ns relative to the 638-nm laser (48, 71). A dual-band dichroic mirror reflecting 532 nm and 638 nm guided the light to a high-numerical-aperture apochromatic objective (60×, numeric aperture of 1.2, water immersion, Olympus) that focused the light to a confocal volume of 1.0 fl for excitation at 532 nm and detection at 575 nm. Fluorescence from excited molecules was collected with the same objective and focused onto a 50-μm-diameter pinhole. The donor and acceptor emissions were separated via a dichroic long-pass filter with a dividing edge at 620 nm. Suited bandpass filters were inserted to eliminate the respective excitation wavelength and minimize spectral cross-talk. The fluorescence was detected with two avalanche photodiodes using time-correlated single-photon counting with the TimeHarp 200 board. Data were stored in the time-tagged, time-resolved mode. Measurements were performed 25 μm deep in the solution with a total acquisition time of 1 h and repeated fresh up to four times on each protein sample (50–100 pm). Signals from single molecules were observed as bursts of fluorescence. Bursts with more than 40 counts were searched with the APBS algorithm, while the integration time was set to 0.5 ms (72). Appropriate correction for direct excitation of the acceptor at the donor excitation wavelength, leakage of the donor in the acceptor channel, and the instrumental γ factor were calculated using a mixture of dsDNA models with known FRET efficiency (E) and stoichiometry (S) labeled with dyes AF555 and AF647 (73). Only molecules with a stoichiometry in the range of 0.25–0.75 were considered in the final analysis, and their distribution was fit to Gaussian curves using Origin 2015 (OriginLab Corporation, Northampton, MA). Data recording was performed using SymphoTime software 6.4 (PicoQuant, Berlin). Data analysis was carried out with PAM (74).
Small-angle X-ray scattering measurements
SAXS data were collected at beamline 12-ID-B of the Advanced Photon Source at Argonne National Laboratory (Argonne, IL) on ST-β2GPI and pβ2GPI at concentrations of 0.5, 1, 2, and 5 mg/ml. The Rg was determined using the Guinier approximation in the low q region (qRg < 1.3), and its linearity served as an initial assessment of data and sample quality. Maximum particle dimension, Dmax, and distance distribution function, P(r), were calculated using GNOM. The low-resolution envelopes were produced using both GASBOR (75) (q up to 0.8 Å−1) and DAMMIN (76) (q up to 0.3 Å−1) by directly fitting the reciprocal space-scattering profile. Twenty models were generated for every calculation and then aligned and averaged using DAMAVER (76). The results of GASBOR and DAMMIN were very similar, but only GASBOR results are reported here. Structural figures were prepared using Pymol.
Data availability
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (accession codes 6V06 for pβ2GPI at 2.4 Å, 6V08 for hrβ2GPI at 2.6 Å, and 6V09 for ST-β2GPI at 3.0 Å).
Supplementary Material
This article contains supporting information.
Author contributions—E. A. R. software; E. A. R., Z. C., X. Z., and N. P. formal analysis; E. A. R., W. P., Z. C., and N. P. validation; E. A. R., W. P., M. C., Z. C., X. Z., V. P., V. D. F., R. K. A., P. M., F. T., and N. P. investigation; E. A. R., Z. C., and N. P. methodology; E. A. R., M. C., V. P., V. D. F., K. R. M., F. T., and N. P. writing-review and editing; N. P. conceptualization; N. P. data curation; N. P. supervision; N. P. funding acquisition; N. P. visualization; N. P. writing-original draft; N. P. project administration.
Funding and additional information—This work was supported in part by the CaRiPaRo Foundation Excellence Research Grant 2018 BPiTA-52012 (V. D. F.), the National Institutes of Health Research Grant HL150146 (N. P.), and President's Research Fund, Saint Louis University (N. P.). Z. C. was supported in part by the National Institutes of Health Research Grants HL049413, HL139554, and HL147821 to E. D. C. This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under contract no. DE-AC02-06CH11357. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- β2GPI
- β2-Glycoprotein I
- aPLs
- antiphospholipid antibodies
- APS
- antiphospholipid syndrome
- AFM
- atomic force microscopy
- SAXS
- small-angle X-ray scattering
- smFRET
- single-molecule FRET
- SPR
- surface plasmon resonance
- RMSD
- root mean square deviations
- AF555
- Alexa Fluor 555
- Rg
- radius of gyration
- Dmax
- maximum linear dimension
- RU
- response units
- CCP
- complement control protein
- PC
- phosphatidylcholine
- PS
- phosphatidylserine.
References
- 1. Schultze H. E., Heide K., and Haupt H. (1961) Uber ein bischer unb ekanntesniedermolekularis b2-globulins des human serums. Naturwissens-chaften 48, 719–719 10.1007/BF00620967 [DOI] [Google Scholar]
- 2. de Groot P. G., and Meijers J. C. (2011) Beta(2) glycoprotein I: evolution, structure and function. J. Thromb. Haemost. 9, 1275–1284 10.1111/j.1538-7836.2011.04327.x [DOI] [PubMed] [Google Scholar]
- 3. Lozier J., Takahashi N., and Putnam F. W. (1984) Complete amino acid sequence of human plasma beta 2-glycoprotein I. Proc. Natl. Acad. Sci. U S A 81, 3640–3644 10.1073/pnas.81.12.3640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lehtinen M. J., Meri S., and Jokiranta T. S. (2004) Interdomain contact regions and angles between adjacent short consensus repeat domains. J. Mol. Biol. 344, 1385–1396 10.1016/j.jmb.2004.10.017 [DOI] [PubMed] [Google Scholar]
- 5. Forneris F., Wu J., Xue X., Ricklin D., Lin Z., Sfyroera G., Tzekou A., Volokhina E., Granneman J. C., Hauhart R., Bertram P., Liszewski M. K., Atkinson J. P., Lambris J. D., and Gros P. (2016) Regulators of complement activity mediate inhibitory mechanisms through a common C3b-binding mode. EMBO J. 35, 1133–1149 10.15252/embj.201593673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Osborne A. J., Nan R., Miller A., Bhatt J. S., Gor J., and Perkins S. J. (2018) Two distinct conformations of factor H regulate discrete complement-binding functions in the fluid phase and at cell surfaces. J. Biol. Chem. 293, 17166–17187 10.1074/jbc.RA118.004767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hunt J. E., Simpson R. J., and Krilis S. A. (1993) Identification of a region of beta 2-glycoprotein I critical for lipid binding and anti-cardiolipin antibody cofactor activity. Proc. Natl. Acad. Sci. U S A 90, 2141–2145 10.1073/pnas.90.6.2141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hunt J., and Krilis S. (1994) The fifth domain of beta 2-glycoprotein I contains a phospholipid binding site (Cys281-Cys288) and a region recognized by anticardiolipin antibodies. J. Immunol. 152, 653–659 [PubMed] [Google Scholar]
- 9. Ioannou Y., Zhang J. Y., Qi M., Gao L., Qi J. C., Yu D. M., Lau H., Sturgess A. D., Vlachoyiannopoulos P. G., Moutsopoulos H. M., Rahman A., Pericleous C., Atsumi T., Koike T., Heritier S., et al. (2011) Novel assays of thrombogenic pathogenicity in the antiphospholipid syndrome based on the detection of molecular oxidative modification of the major autoantigen beta2-glycoprotein I. Arthritis Rheum. 63, 2774–2782 10.1002/art.30383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Giannakopoulos B., and Krilis S. A. (2013) The pathogenesis of the antiphospholipid syndrome. N. Engl. J. Med. 368, 1033–1044 10.1056/NEJMra1112830 [DOI] [PubMed] [Google Scholar]
- 11. Bevers E. M., and Galli M. (1990) Beta 2-glycoprotein I for binding of anticardiolipin antibodies to cardiolipin. Lancet 336, 952–953 10.1016/0140-6736(90)92330-k [DOI] [PubMed] [Google Scholar]
- 12. Galli M., Comfurius P., Maassen C., Hemker H. C., de Baets M. H., van Breda-Vriesman P. J., Barbui T., Zwaal R. F., and Bevers E. M. (1990) Anticardiolipin antibodies (ACA) directed not to cardiolipin but to a plasma protein cofactor. Lancet 335, 1544–1547 10.1016/0140-6736(90)91374-J [DOI] [PubMed] [Google Scholar]
- 13. McNeil H. P., Simpson R. J., Chesterman C. N., and Krilis S. A. (1990) Anti-phospholipid antibodies are directed against a complex antigen that includes a lipid-binding inhibitor of coagulation: beta 2-glycoprotein I (apolipoprotein H). Proc. Natl. Acad. Sci. U S A 87, 4120–4124 10.1073/pnas.87.11.4120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Meroni P. L., Borghi M. O., Raschi E., and Tedesco F. (2011) Pathogenesis of antiphospholipid syndrome: understanding the antibodies. Nat. Rev. Rheumatol. 7, 330–339 10.1038/nrrheum.2011.52 [DOI] [PubMed] [Google Scholar]
- 15. Levine J. S., Branch D. W., and Rauch J. (2002) The antiphospholipid syndrome. N. Engl. J. Med. 346, 752–763 10.1056/NEJMra002974 [DOI] [PubMed] [Google Scholar]
- 16. Koike T., and Matsuura E. (1996) Anticardiolipin antibodies and beta 2-glycoprotein I. Lupus 5, 156–157 10.1177/096120339600500213 [DOI] [PubMed] [Google Scholar]
- 17. Oosting J. D., Derksen R. H., Entjes H. T., Bouma B. N., and de Groot P. G. (1992) Lupus anticoagulant activity is frequently dependent on the presence of beta 2-glycoprotein I. Thromb. Haemost. 67, 499–502 10.1055/s-0038-1648480 [DOI] [PubMed] [Google Scholar]
- 18. Fischetti F., Durigutto P., Pellis V., Debeus A., Macor P., Bulla R., Bossi F., Ziller F., Sblattero D., Meroni P., and Tedesco F. (2005) Thrombus formation induced by antibodies to beta2-glycoprotein I is complement dependent and requires a priming factor. Blood 106, 2340–2346 10.1182/blood-2005-03-1319 [DOI] [PubMed] [Google Scholar]
- 19. Pierangeli S. S., and Harris E. N. (1996) In vivo models of thrombosis for the antiphospholipid syndrome. Lupus 5, 451–455 10.1177/096120339600500524 [DOI] [PubMed] [Google Scholar]
- 20. Arad A., Proulle V., Furie R. A., Furie B. C., and Furie B. (2011) Beta(2) glycoprotein-1 autoantibodies from patients with antiphospholipid syndrome are sufficient to potentiate arterial thrombus formation in a mouse model. Blood 117, 3453–3459 10.1182/blood-2010-08-300715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Agostinis C., Durigutto P., Sblattero D., Borghi M. O., Grossi C., Guida F., Bulla R., Macor P., Pregnolato F., Meroni P. L., and Tedesco F. (2014) A non-complement-fixing antibody to beta2 glycoprotein I as a novel therapy for antiphospholipid syndrome. Blood 123, 3478–3487 10.1182/blood-2013-11-537704 [DOI] [PubMed] [Google Scholar]
- 22. de Laat B., Derksen R. H., Urbanus R. T., and de Groot P. G. (2005) IgG antibodies that recognize epitope Gly40-Arg43 in domain I of beta 2-glycoprotein I cause LAC, and their presence correlates strongly with thrombosis. Blood 105, 1540–1545 10.1182/blood-2004-09-3387 [DOI] [PubMed] [Google Scholar]
- 23. Iverson G. M., Reddel S., Victoria E. J., Cockerill K. A., Wang Y. X., Marti-Renom M. A., Sali A., Marquis D. M., Krilis S. A., and Linnik M. D. (2002) Use of single point mutations in domain I of beta 2-glycoprotein I to determine fine antigenic specificity of antiphospholipid autoantibodies. J. Immunol. 169, 7097–7103 10.4049/jimmunol.169.12.7097 [DOI] [PubMed] [Google Scholar]
- 24. Iverson G. M., Victoria E. J., and Marquis D. M. (1998) Anti-beta2 glycoprotein I (beta2GPI) autoantibodies recognize an epitope on the first domain of beta2GPI. Proc. Natl. Acad. Sci. U S A 95, 15542–15546 10.1073/pnas.95.26.15542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Banzato A., Pozzi N., Frasson R., De Filippis V., Ruffatti A., Bison E., Padayattil S. J., Denas G., and Pengo V. (2011) Antibodies to Domain I of beta(2)Glycoprotein I are in close relation to patients risk categories in Antiphospholipid Syndrome (APS). Thromb. Res. 128, 583–586 10.1016/j.thromres.2011.04.021 [DOI] [PubMed] [Google Scholar]
- 26. de Laat B., Derksen R. H., van Lummel M., Pennings M. T., and de Groot P. G. (2006) Pathogenic anti-beta2-glycoprotein I antibodies recognize domain I of beta2-glycoprotein I only after a conformational change. Blood 107, 1916–1924 10.1182/blood-2005-05-1943 [DOI] [PubMed] [Google Scholar]
- 27. de Laat B., van Berkel M., Urbanus R. T., Siregar B., de Groot P. G., Gebbink M. F., and Maas C. (2011) Immune responses against domain I of beta(2)-glycoprotein I are driven by conformational changes: domain I of beta(2)-glycoprotein I harbors a cryptic immunogenic epitope. Arthritis Rheum. 63, 3960–3968 10.1002/art.30633 [DOI] [PubMed] [Google Scholar]
- 28. Bevers E. M., Zwaal R. F., and Willems G. M. (2004) The effect of phospholipids on the formation of immune complexes between autoantibodies and beta2-glycoprotein I or prothrombin. Clin. Immunol. 112, 150–160 10.1016/j.clim.2004.02.009 [DOI] [PubMed] [Google Scholar]
- 29. Agar C., van Os G. M., Morgelin M., Sprenger R. R., Marquart J. A., Urbanus R. T., Derksen R. H., Meijers J. C., and de Groot P. G. (2010) Beta2-glycoprotein I can exist in 2 conformations: implications for our understanding of the antiphospholipid syndrome. Blood 116, 1336–1343 10.1182/blood-2009-12-260976 [DOI] [PubMed] [Google Scholar]
- 30. Agar C., de Groot P. G., Morgelin M., Monk S. D., van Os G., Levels J. H., de Laat B., Urbanus R. T., Herwald H., van der Poll T., and Meijers J. C. (2011) Beta(2) glycoprotein I: a novel component of innate immunity. Blood 117, 6939–6947 10.1182/blood-2010-12-325951 [DOI] [PubMed] [Google Scholar]
- 31. Buchholz I., Nestler P., Koppen S., and Delcea M. (2018) Lysine residues control the conformational dynamics of beta 2-glycoprotein I. Phys. Chem. Chem. Phys. 20, 26819–26829 10.1039/c8cp03234c [DOI] [PubMed] [Google Scholar]
- 32. Hammel M., Kriechbaum M., Gries A., Kostner G. M., Laggner P., and Prassl R. (2002) Solution structure of human and bovine beta(2)-glycoprotein I revealed by small-angle X-ray scattering. J. Mol. Biol. 321, 85–97 10.1016/S0022-2836(02)00621-6 [DOI] [PubMed] [Google Scholar]
- 33. Schwarzenbacher R., Zeth K., Diederichs K., Gries A., Kostner G. M., Laggner P., and Prassl R. (1999) Crystal structure of human beta2-glycoprotein I: implications for phospholipid binding and the antiphospholipid syndrome. EMBO J. 18, 6228–6239 10.1093/emboj/18.22.6228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bouma B., de Groot P. G., van den Elsen J. M., Ravelli R. B., Schouten A., Simmelink M. J., Derksen R. H., Kroon J., and Gros P. (1999) Adhesion mechanism of human beta(2)-glycoprotein I to phospholipids based on its crystal structure. EMBO J. 18, 5166–5174 10.1093/emboj/18.19.5166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van Os G. M. A., Meijers J. C. M., Agar Ç., Seron M. V., Marquart J. A., Åkesson P., Urbanus R. T., Derksen R. H. W. M., Herwald H., Mörgelin M., and D E Groot P. G. (2011) Induction of anti-beta2-glycoprotein I autoantibodies in mice by protein H of Streptococcus pyogenes. J. Thromb. Haemost. 9, 2447–2456 10.1111/j.1538-7836.2011.04532.x [DOI] [PubMed] [Google Scholar]
- 36. Chinnaraj M., Planer W., Pengo V., and Pozzi N. (2019) Discovery and characterization of 2 novel subpopulations of aPS/PT antibodies in patients at high risk of thrombosis. Blood Adv. 3, 1738–1749 10.1182/bloodadvances.2019030932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chinnaraj M., Chen Z., Pelc L. A., Grese Z., Bystranowska D., Di Cera E., and Pozzi N. (2018) Structure of prothrombin in the closed form reveals new details on the mechanism of activation. Sci. Rep. 8, 2945 10.1038/s41598-018-21304-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chinnaraj M., Planer W., and Pozzi N. (2018) Structure of coagulation factor II: molecular mechanism of thrombin generation and development of next-generation anticoagulants. Front. Med. 5, 281 10.3389/fmed.2018.00281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Pozzi N., Bystranowska D., Zuo X., and Di Cera E. (2016) Structural architecture of prothrombin in solution revealed by single molecule spectroscopy. J. Biol. Chem. 291, 18107–18116 10.1074/jbc.M116.738310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sheng Y., Kandiah D. A., and Krilis S. A. (1998) Anti-beta 2-glycoprotein I autoantibodies from patients with the “antiphospholipid” syndrome bind to beta 2-glycoprotein I with low affinity: dimerization of beta 2-glycoprotein I induces a significant increase in anti-beta 2-glycoprotein I antibody affinity. J. Immunol. 161, 2038–2043 [PubMed] [Google Scholar]
- 41. Lutters B. C., Derksen R. H., Tekelenburg W. L., Lenting P. J., Arnout J., and de Groot P. G. (2003) Dimers of beta 2-glycoprotein I increase platelet deposition to collagen via interaction with phospholipids and the apolipoprotein E receptor 2. J. Biol. Chem. 278, 33831–33838 10.1074/jbc.M212655200 [DOI] [PubMed] [Google Scholar]
- 42. Pengo V., Del Ross T., Ruffatti A., Bison E., Cattini M. G., Pontara E., Testa S., Legnani C., Pozzi N., Peterle D., Acquasaliente L., De Filippis V., and Denas G. (2018) Lupus anticoagulant identifies two distinct groups of patients with different antibody patterns. Thromb. Res. 172, 172–178 10.1016/j.thromres.2018.11.003 [DOI] [PubMed] [Google Scholar]
- 43. Passam F. H., Rahgozar S., Qi M., Raftery M. J., Wong J. W., Tanaka K., Ioannou Y., Zhang J. Y., Gemmell R., Qi J. C., Giannakopoulos B., Hughes W. E., Hogg P. J., and Krilis S. A. (2010) Beta 2 glycoprotein I is a substrate of thiol oxidoreductases. Blood 116, 1995–1997 10.1182/blood-2010-02-271494 [DOI] [PubMed] [Google Scholar]
- 44. Hammel M., Schwarzenbacher R., Gries A., Kostner G. M., Laggner P., and Prassl R. (2001) Mechanism of the interaction of beta(2)-glycoprotein I with negatively charged phospholipid membranes. Biochemistry 40, 14173–14181 10.1021/bi0114372 [DOI] [PubMed] [Google Scholar]
- 45. Ohkura N., Hagihara Y., Yoshimura T., Goto Y., and Kato H. (1998) Plasmin can reduce the function of human beta2 glycoprotein I by cleaving domain V into a nicked form. Blood 91, 4173–4179 10.1182/blood.V91.11.4173.411k35_4173_4179 [DOI] [PubMed] [Google Scholar]
- 46. Dodson E. J., Winn M., and Ralph A. (1997) Collaborative Computational Project, number 4: providing programs for protein crystallography. Methods Enzymol. 277, 620–633 10.1016/s0076-6879(97)77034-4 [DOI] [PubMed] [Google Scholar]
- 47. Lerner E., Cordes T., Ingargiola A., Alhadid Y., Chung S., Michalet X., and Weiss S. (2018) Toward dynamic structural biology: two decades of single-molecule Forster resonance energy transfer. Science 359, eaan1133 10.1126/science.aan1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gopich I. V., and Szabo A. (2007) Single-molecule FRET with diffusion and conformational dynamics. J. Phys. Chem. B 111, 12925–12932 10.1021/jp075255e [DOI] [PubMed] [Google Scholar]
- 49. Kikhney A. G., and Svergun D. I. (2015) A practical guide to small angle X-ray scattering (SAXS) of flexible and intrinsically disordered proteins. FEBS Lett. 589, 2570–2577 10.1016/j.febslet.2015.08.027 [DOI] [PubMed] [Google Scholar]
- 50. Dienava-Verdoold I., Boon-Spijker M. G., de Groot P. G., Brinkman H. J., Voorberg J., Mertens K., Derksen R. H., and de Laat B. (2011) Patient-derived monoclonal antibodies directed towards beta2 glycoprotein-1 display lupus anticoagulant activity. J. Thromb. Haemost. 9, 738–747 10.1111/j.1538-7836.2011.04212.x [DOI] [PubMed] [Google Scholar]
- 51. Law R. H., Caradoc-Davies T., Cowieson N., Horvath A. J., Quek A. J., Encarnacao J. A., Steer D., Cowan A., Zhang Q., Lu B. G., Pike R. N., Smith A. I., Coughlin P. B., and Whisstock J. C. (2012) The X-ray crystal structure of full-length human plasminogen. Cell Rep. 1, 185–190 10.1016/j.celrep.2012.02.012 [DOI] [PubMed] [Google Scholar]
- 52. Ivanov I., Matafonov A., Sun M. F., Cheng Q., Dickeson S. K., Verhamme I. M., Emsley J., and Gailani D. (2017) Proteolytic properties of single-chain factor XII: a mechanism for triggering contact activation. Blood 129, 1527–1537 10.1182/blood-2016-10-744110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Muia J., Zhu J., Gupta G., Haberichter S. L., Friedman K. D., Feys H. B., Deforche L., Vanhoorelbeke K., Westfield L. A., Roth R., Tolia N. H., Heuser J. E., and Sadler J. E. (2014) Allosteric activation of ADAMTS13 by von Willebrand factor. Proc. Natl. Acad. Sci. U S A 111, 18584–18589 10.1073/pnas.1413282112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Pedersen D. V., Roumenina L., Jensen R. K., Gadeberg T. A., Marinozzi C., Picard C., Rybkine T., Thiel S., Sorensen U. B., Stover C., Fremeaux-Bacchi V., and Andersen G. R. (2017) Functional and structural insight into properdin control of complement alternative pathway amplification. EMBO J. 36, 1084–1099 10.15252/embj.201696173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. De Michele C., De Los Rios P., Foffi G., and Piazza F. (2016) Simulation and theory of antibody binding to crowded antigen-covered surfaces. PLoS Comput. Biol. 12, e1004752 10.1371/journal.pcbi.1004752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hadzhieva M., Pashov A. D., Kaveri S., Lacroix-Desmazes S., Mouquet H., and Dimitrov J. D. (2017) Impact of antigen density on the binding mechanism of IgG antibodies. Sci. Rep. 7, 3767 10.1038/s41598-017-03942-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Banzato A., Frasson R., Acquasaliente L., Bison E., Bracco A., Denas G., Cuffaro S., Hoxha A., Ruffatti A., Iliceto S., De Filippis V., and Pengo V. (2012) Circulating beta2 glycoprotein I-IgG anti-beta2 glycoprotein I immunocomplexes in patients with definite antiphospholipid syndrome. Lupus 21, 784–786 10.1177/0961203312440347 [DOI] [PubMed] [Google Scholar]
- 58. Biasiolo A., Rampazzo P., Brocco T., Barbero F., Rosato A., and Pengo V. (1999) [Anti-beta2 glycoprotein I-beta2 glycoprotein I] immune complexes in patients with antiphospholipid syndrome and other autoimmune diseases. Lupus 8, 121–126 10.1191/096120399678847506 [DOI] [PubMed] [Google Scholar]
- 59. Kolyada A., De Biasio A., and Beglova N. (2013) Identification of the binding site for fondaparinux on Beta2-glycoprotein I. Biochim. Biophys. Acta 1834, 2080–2088 10.1016/j.bbapap.2013.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Acquasaliente L., Peterle D., Tescari S., Pozzi N., Pengo V., and De Filippis V. (2016) Molecular mapping of alpha-thrombin (alphaT)/beta2-glycoprotein I (beta2GpI) interaction reveals how beta2GpI affects alphaT functions. Biochem. J. 473, 4629–4650 10.1042/BCJ20160603 [DOI] [PubMed] [Google Scholar]
- 61. Pierangeli S. S., Vega-Ostertag M. E., Raschi E., Liu X., Romay-Penabad Z., De Micheli V., Galli M., Moia M., Tincani A., Borghi M. O., Nguyen-Oghalai T., and Meroni P. L. (2007) Toll-like receptor and antiphospholipid mediated thrombosis: in vivo studies. Ann. Rheum. Dis. 66, 1327–1333 10.1136/ard.2006.065037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lee C. J., De Biasio A., and Beglova N. (2010) Mode of interaction between beta2GPI and lipoprotein receptors suggests mutually exclusive binding of beta2GPI to the receptors and anionic phospholipids. Structure 18, 366–376 10.1016/j.str.2009.12.013 [DOI] [PubMed] [Google Scholar]
- 63. Zhang J., and McCrae K. R. (2005) Annexin A2 mediates endothelial cell activation by antiphospholipid/anti-beta2 glycoprotein I antibodies. Blood 105, 1964–1969 10.1182/blood-2004-05-1708 [DOI] [PubMed] [Google Scholar]
- 64. Allen K. L., Fonseca F. V., Betapudi V., Willard B., Zhang J., and McCrae K. R. (2012) A novel pathway for human endothelial cell activation by antiphospholipid/anti-beta2 glycoprotein I antibodies. Blood 119, 884–893 10.1182/blood-2011-03-344671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Kolyada A., Barrios D. A., and Beglova N. (2017) Dimerized domain V of beta2-glycoprotein I is sufficient to upregulate procoagulant activity in PMA-treated U937 monocytes and require intact residues in two phospholipid-binding loops. Antibodies 6, 8 10.3390/antib6020008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Durigutto P., Grossi C., Borghi M. O., Macor P., Pregnolato F., Raschi E., Myers M. P., de Groot P. G., Meroni P. L., and Tedesco F. (2019) New insight into antiphospholipid syndrome: antibodies to beta2glycoprotein I-domain 5 fail to induce thrombi in rats. Haematologica 104, 819–826 10.3324/haematol.2018.198119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pozzi N., Banzato A., Bettin S., Bison E., Pengo V., and De Filippis V. (2010) Chemical synthesis and characterization of wild-type and biotinylated N-terminal domain 1-64 of beta2-glycoprotein I. Protein Sci. 19, 1065–1078 10.1002/pro.387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Otwinowski Z., and Minor W. (1997) Processing of x-ray diffraction data collected by oscillation methods. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 69. Emsley P., and Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 10.1107/S0907444904019158 [DOI] [PubMed] [Google Scholar]
- 70. Muller B. K., Zaychikov E., Brauchle C., and Lamb D. C. (2005) Pulsed interleaved excitation. Biophys. J. 89, 3508–3522 10.1529/biophysj.105.064766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kapanidis A. N., Lee N. K., Laurence T. A., Doose S., Margeat E., and Weiss S. (2004) Fluorescence-aided molecule sorting: analysis of structure and interactions by alternating-laser excitation of single molecules. Proc. Natl. Acad. Sci. U S A 101, 8936–8941 10.1073/pnas.0401690101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Nir E., Michalet X., Hamadani K. M., Laurence T. A., Neuhauser D., Kovchegov Y., and Weiss S. (2006) Shot-noise limited single-molecule FRET histograms: comparison between theory and experiments. J. Phys. Chem. B 110, 22103–22124 10.1021/jp063483n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Lee N. K., Kapanidis A. N., Wang Y., Michalet X., Mukhopadhyay J., Ebright R. H., and Weiss S. (2005) Accurate FRET measurements within single diffusing biomolecules using alternating-laser excitation. Biophys. J. 88, 2939–2953 10.1529/biophysj.104.054114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Schrimpf W., Barth A., Hendrix J., and Lamb D. C. (2018) PAM: a framework for integrated analysis of imaging, single-molecule, and ensemble fluorescence data. Biophys. J. 114, 1518–1528 10.1016/j.bpj.2018.02.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Svergun D. I., Petoukhov M. V., and Koch M. H. (2001) Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 80, 2946–2953 10.1016/S0006-3495(01)76260-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Svergun D. I. (1999) Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 10.1016/S0006-3495(99)77443-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (accession codes 6V06 for pβ2GPI at 2.4 Å, 6V08 for hrβ2GPI at 2.6 Å, and 6V09 for ST-β2GPI at 3.0 Å).