Abstract
SHC adaptor protein (SHCA) and lipoma-preferred partner (LPP) mediate transforming growth factor β (TGFβ)-induced breast cancer cell migration and invasion. Reduced expression of either protein diminishes breast cancer lung metastasis, but the reason for this effect is unclear. Here, using total internal reflection fluorescence (TIRF) microscopy, we found that TGFβ enhanced the assembly and disassembly rates of paxillin-containing adhesions in an SHCA-dependent manner through the phosphorylation of the specific SHCA tyrosine residues Tyr-239, Tyr-240, and Tyr-313. Using a BioID proximity labeling approach, we show that SHCA exists in a complex with a variety of actin cytoskeletal proteins, including paxillin and LPP. Consistent with a functional interaction between SHCA and LPP, TGFβ-induced LPP localization to cellular adhesions depended on SHCA. Once localized to the adhesions, LPP was required for TGFβ-induced increases in cell migration and adhesion dynamics. Mutations that impaired LPP localization to adhesions (mLIM1) or impeded interactions with the actin cytoskeleton via α-actinin (ΔABD) abrogated migratory responses to TGFβ. Live-cell TIRF microscopy revealed that SHCA clustering at the cell membrane preceded LPP recruitment. We therefore hypothesize that, in the presence of TGFβ, SHCA promotes the formation of small, dynamic adhesions by acting as a nucleator of focal complex formation. Finally, we defined a previously unknown function for SHCA in the formation of invadopodia, a process that also required LPP. Our results reveal that SHCA controls the formation and function of adhesions and invadopodia, two key cellular structures required for breast cancer metastasis.
Keywords: cell migration, focal adhesion, cell invasion, invadopodia, microscopy, adhesion dynamics, cancer, lipoma-preferred partner (LPP), SHC adaptor protein (SHCA), transforming growth factor beta (TGFβ)
Cellular migration and invasion are fundamental processes that are required for metastasis. Some cancer cells employ a mesenchymal mode of cell migration, which is highly dependent on adhesions that link the actin cytoskeleton to the extracellular matrix (ECM).11 Cellular adhesions consist of a complex network of transmembrane integrin heterodimers and cytoplasmic proteins that form a plaque containing hundreds of components with a multitude of potential protein-protein interactions (1–3). Together, these protein interactions mediate cellular signaling and allow the cell to generate traction forces that are important for controlling cell migration. Equally important to the metastatic cascade are invadopodia, which are cellular structures that mediate cancer cell invasion through barriers otherwise impenetrable to migratory cells. Invadopodia are F-actin–rich and proteolytically active cell protrusions capable of degrading ECM components and breaching epithelial and endothelial basement membranes (4). These structures share many protein components with adhesions, such as cortactin (Cttn), but can be distinguished by the presence of Tks5 (4–6). Together, adhesions and invadopodia are two fundamental structures that enable cancer cells to escape from the primary tumor and establish distant metastases (7–12). Consequently, up-regulation and/or increased signaling of proteins that enhance adhesion and invadopodia formation are often observed in invasive and metastatic cancer cells (13–17).
Previous work by our group and others showed that ErbB2 and transforming growth factor β (TGFβ) signaling pathways cooperate to enhance the metastatic ability of breast cancer cells (18–23). SHC adapter protein (SHCA) is a critical downstream integrator of these pathways and is essential for breast tumor growth, migration, invasion, and metastasis (24–32). SHCA is part of a Src-homology/collagen (SHC) family of proteins that also includes SHCB, SHCC, and SHCD. Alternative translational initiation and RNA splicing result in the expression of three different SHCA isoforms: p46, p52, and p66 (33). All breast cancer cells express p46/52 SHCA isoforms, whereas p66SHCA is more highly expressed in breast cancers with mesenchymal features (34). Recently, p66SHCA has been shown to be a context-dependent promoter of breast cancer metastasis (35). Loss of p66SHCA expression results in slower adhesion dynamics, reduced cell migration rates, and diminished lung metastasis (35). SHCA harbors an N-terminal phosphotyrosine-binding domain (PTB), a central collagen homology domain (CH1) with three key tyrosine residues (Tyr-239/Tyr-240/Tyr-313) and a C-terminal SH2 domain (33). Our previous studies revealed that the PTB domain, but not the SH2 domain, is required for TGFβ-induced migration and invasion of ErbB2+ breast cancer cells (32). Moreover, phosphorylation of tyrosine residues was required for migratory and invasive phenotypes in vitro and breast cancer lung metastasis in vivo (32).
More recently, we have characterized lipoma-preferred partner (LPP) as an important regulator of breast cancer cell migration, invasion, and metastasis (36, 37). LPP is a member of the zyxin family of LIM proteins and is known to promote mesenchymal migration (38). LPP contains three LIM domains and a proline-rich N-terminal region, which allow it to localize to adhesions and interact with numerous proteins (39). Whereas loss of SHCA negatively impacts breast tumor initiation and growth (24), LPP is dispensable for primary tumor growth (37). However, loss of LPP recapitulates the migratory and invasive defects seen in SHCA-depleted cells. Namely, breast cancer cells with diminished LPP expression do not exhibit increased migration and invasion in response to TGFβ stimulation (36). The ability of LPP to localize to adhesions via its LIM domains and interact with α-actinin is required for the pro-migratory and pro-invasive functions of LPP (36). Src-mediated phosphorylation of LPP, while dispensable for cell migration, is required for invadopodia formation and efficient breast cancer lung metastasis (37).
In the current study, we show for the first time that SHCA acts as a nucleator of focal complex formation by promoting the formation of small, dynamic adhesions in response to TGFβ. We suggest that SHCA serves as a molecular scaffold to facilitate the recruitment of actin cytoskeletal and adhesion proteins, including paxillin and LPP. Indeed, TGFβ enhances adhesion targeting of paxillin and LPP, which permits faster assembly and disassembly of these structures. TGFβ-induced migration and adhesion dynamics require LPP localization to adhesions and interaction with the actin cytoskeleton. Furthermore, we show that tyrosine phosphorylation of SHCA is required for TGFβ-induced adhesion dynamics. We also implicate SHCA as an important regulator of invadopodia formation, which requires phosphorylation of tyrosine residues within the CH1 domain. The requirement of SHCA for efficient invadopodia formation is reminiscent of the role of LPP in the formation of these structures (37). Taken together, we delineate essential roles for SHCA and LPP as critical mediators of adhesion fate and invadopodia formation.
Results
Cooperation between TGFβ and ErbB2 signaling pathways promotes single-cell migration
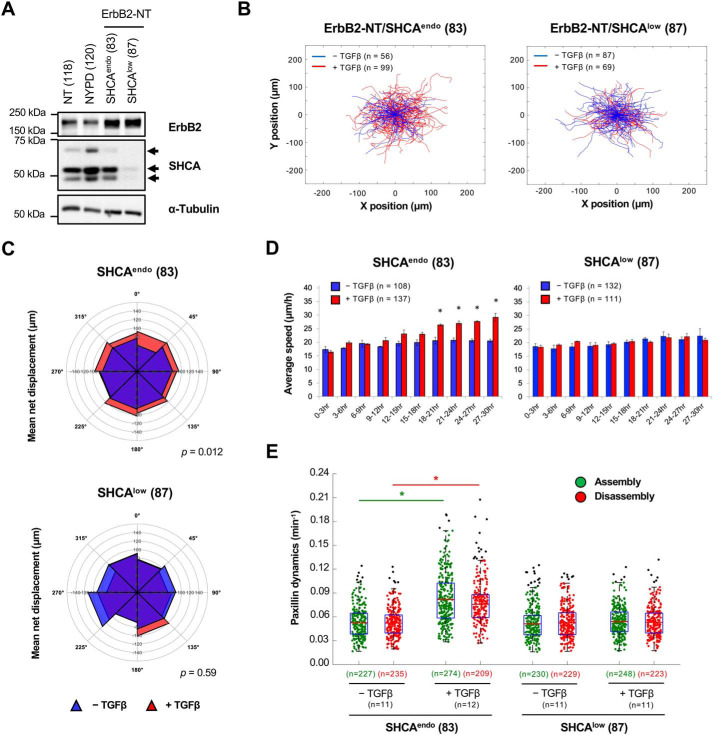
NMuMG cells expressing activated ErbB2 spontaneously metastasize to the lung from the primary tumor (31). We have previously observed that cells with constitutively active ErbB2 (ErbB2-NT) exhibit increased movement through porous membranes in response to TGFβ (31, 36). In contrast, NMuMG cells expressing a variant of ErbB2 that lacks five important tyrosine phosphorylation sites (ErbB2-NYPD) fail to exhibit this phenotype (31). Whereas transwell assays are useful for defining gross cellular phenotypes at a population level, they fail to provide insight into the mechanisms of migration and invasion at the single-cell level. To gain a more in-depth understanding of TGFβ-induced migration of ErbB2+ breast cancer cells, we employed live-cell time-lapse microscopy. Using this approach, we could readily assess the behavior, mean net displacement, and speed of individual breast cancer cells.
Rose plots of breast cancer cells treated with TGFβ demonstrated that ErbB2-NT cells stimulated with TGFβ migrated further and faster than untreated cells, a response that was not observed with ErbB2-NYPD breast cancer cells (Fig. 1 (A–C) and Movie S1). Population-based analysis of single cells showed that ErbB2-NT cells began to increase in speed after 9 h of TGFβ treatment and achieved a maximal speed of ∼33 μm h−1 (Fig. 1C). In contrast, ErbB2-NYPD–expressing cells continued to migrate at ∼21 μm h−1 even after 27 h of TGFβ stimulation (Fig. 1C). These results were confirmed with an independent set of mammary tumor explants generated from mice injected with ErbB2-NT– or ErbB2-NYPD–expressing breast cancer cells (Fig. S1).
Figure 1.
TGFβ stimulates the migration of ErbB2-NT–expressing breast cancer cells. A, live-cell migration tracks for each condition are shown 18 h after treatment with or without TGFβ (2 ng ml−1). Each line represents the migration path of a single cell over 6 h. The starting point of each cell was superimposed on the origin (0, 0). Numbers in parentheses refer to explant cell lines. B, cell displacements from A were pooled into 45° segments based on their angle of trajectory and averaged to determine mean net displacement. C, average migration speed over time was calculated by determining the mean distance traveled between each imaging time point by all cells in A. The data for each population were then averaged into 3-h segments. Data represent mean ± S.E. (error bars) from five (NT) or six (NYPD) independent experiments. (*, p < 0.05, two-tailed Student's t test). D, adhesions in protrusive cell regions were tracked over time using TIRF Spectral Diskovery System coupled to a Leica microscope equipped with a Plan-Apochromat ×63/1.47 NA oil immersion DIC objective lens and EM-CCD camera (1 pixel = 0.155 μm). Cells were transfected with mCherry-paxillin, a marker of cellular adhesions, 48 h prior to imaging and left untreated or treated with TGFβ for 24 h. Average assembly (green) or disassembly (red) rates were determined from changes in mean fluorescence intensity. Data represent individual assembly and disassembly events from three independent experiments. Coloured n values refer to the number of events while black n values refer to the number of cells. The top and bottom lines of the box indicate the third and first quartile, respectively, whereas the heavy central line indicates the mean. The whiskers extend up to 1.5 times the interquartile range. Black dots represent outliers. *, p < 0.0001 calculated from the cell averages for assembly and disassembly rates; two-tailed Student's t test.
TGFβ is known to induce an epithelial-to-mesenchymal transition (EMT) in ErbB2+ breast cancer cells (40). Cells engaging a mesenchymal mode of migration depend on integrin-mediated adhesion dynamics (41–43). Therefore, we sought to investigate the assembly and disassembly rates of adhesions using fluorescently labeled paxillin, a bona fide marker of adhesions (44, 45). TGFβ significantly increased the assembly and disassembly rates of mCherry-paxillin containing adhesions in protrusive cell regions of ErbB2-NT, but not ErbB2-NYPD, expressing breast cancer cells (Fig. 1D). In particular, a larger proportion of rapid events was observed in ErbB2-NT–expressing cells (Movie S2). The observed rapid adhesion dynamics in ErbB2-NT cells following TGFβ stimulation correlated well with the TGFβ-induced increase in migration speeds exhibited by these cells.
Signaling through the SHCA adapter protein is required for TGFβ-induced migration
SHCA is recruited to tyrosine-phosphorylated residues (Tyr-1226/Tyr-1227) in the cytoplasmic tail of the ErbB2 receptor, and this association is required for the development of aggressively growing mammary tumors (46). Indeed, SHCA has been shown to be important for primary tumor growth and metastasis of ErbB2+ breast cancer cells (26, 29, 32). Therefore, we applied time-lapse microscopy to assess the impact of SHCA loss on cell migration. NMuMG ErbB2-NT cells expressing shRNAs against the 3′-UTR of mouse SHCA mRNA (SHCAlow) or luciferase-targeting shRNAs (SHCAendo) were previously generated in our laboratory (32). Immunoblot analysis demonstrated that cells from ErbB2-NT/SHCAlow explants expressed significantly reduced levels of SHCA (p46, p52, and p66 isoforms) when compared with parental controls (Fig. 2A). Rose plots revealed that ErbB2-NT/SHCAendo cells exhibited an increase in cell migration following TGFβ treatment, whereas ErbB2-NT/SHCAlow cells did not (Fig. 2B). Treatment with TGFβ also significantly increased the mean net displacement of cells expressing endogenous levels of SHCA (SHCAendo), which was not observed in cells with low SHCA expression (SHCAlow) (Fig. 2C). This effect was due to an increase in migration speed, as ErbB2-NT breast cancer cells expressing endogenous levels of SHCA (SHCAendo) exhibited significantly faster speeds after 18 h of TGFβ treatment (Fig. 2D) rather than a change in directional persistence (data not shown). Results from an independent set of ErbB2-NT/SHCAendo and ErbB2-NT/SHCAlow mammary tumor explants supported these findings (Fig. S2).
Figure 2.
The SHCA adapter protein is required for TGFβ-induced migration. A, total cell lysates showing ErbB2 and SHCA levels in ErbB2-NT (explant 118), ErbB2-NYPD (explant 120), ErbB2-NT/SHCAendo (explant 83), and ErbB2-NT/SHCAlow (explant 87) populations. α-Tubulin was used as a loading control. B, ErbB2-NT/SHCAendo (explant 83) and ErbB2-NT/SHCAlow (explant 87) cells were seeded onto fibronectin-coated glass bottom dishes and allowed to migrate in the absence or presence of TGFβ (2 ng ml−1). Live-cell migration tracks for each condition are shown 18 h after treatment. Each line represents the migration path of a single cell over 6 h. The starting point of each cell was superimposed on the origin (0, 0). C, cell displacements from B were pooled into 45° segments based on their angle of trajectory and averaged to determine mean net displacement. D, average migration speed over time was calculated by determining the mean distance traveled between each imaging time point by all cells in B. The data for each population was then averaged into 3-h segments. Data represent mean ± S.E. (error bars) from three (ErbB2-NT/SHCAendo) or four (ErbB2-NT/SHCAlow) independent experiments. (*, p < 0.05, two-tailed Student's t test). E, adhesions in protrusive cell regions were tracked over time using TIRF microscopy. Cells were transfected with mCherry-paxillin 48 h prior to imaging and left untreated or treated with TGFβ for 24 h. Average assembly (green) or disassembly (red) rates were determined from changes in mean fluorescence intensity. Data represent individual assembly and disassembly events from three (ErbB2-NT/SHCAendo) or four (ErbB2-NT/SHCAlow) independent experiments. Coloured n values refer to the number of events while black n values refer to the number of cells. The top and bottom lines of the box indicate the third and first quartile, respectively, whereas the heavy central line indicates the mean. The whiskers extend up to 1.5 times the interquartile range. Black dots represent outliers. *, p < 0.0001 calculated from the cell averages for assembly and disassembly rates; two-tailed Student's t test.
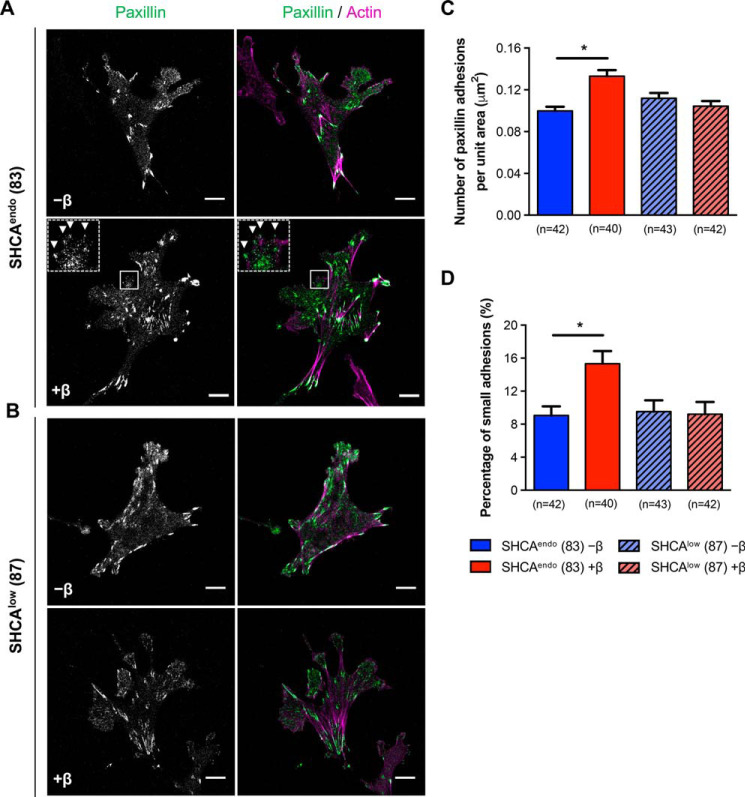
Next, we investigated adhesion dynamics in protrusive cell regions of SHCAendo- and SHCAlow-expressing cells. We found that TGFβ enhanced the assembly and disassembly rates of mCherry-paxillin–containing adhesions in cells expressing endogenous levels of SHCA, but not in ErbB2-NT/SHCAlow cells (Fig. 2E). Moreover, TGFβ increased the formation of paxillin-bearing adhesions in ErbB2-NT/SHCAendo cells (Fig. 3, A–C). These newly formed adhesions appeared to be smaller in size (Fig. 3D), which is consistent with the idea that smaller adhesions are more dynamic (47). In contrast, TGFβ did not affect the number and size distribution of adhesions in ErbB2-NT/SHCAlow cells (Fig. 3, B–D). Taken together, our results suggest that SHCA is required for the formation of small, dynamic adhesions in response to TGFβ stimulation.
Figure 3.
SHCA is required for the formation of small, dynamic adhesions in breast cancer cells exposed to TGFβ. A and B, ErbB2-NT/SHCAendo (explant 83) and ErbB2-NT/SHCAlow (explant 87) cells were infected with mCherry-paxillin, seeded onto fibronectin-coated glass bottom dishes, and cultured in the absence or presence of TGFβ (2 ng ml−1) for 24 h. Cells were then fixed with 4% PFA and stained with phalloidin (F-actin). Images were taken on a Zeiss confocal laser-scanning microscope (CLSM) using a Plan-Apochromat ×63/1.4 NA oil immersion DIC objective lens (1 pixel = 0.132 μm). Arrowheads highlight examples of small adhesions formed after treatment with TGFβ. Scale bar, 10 μm. C, images were imported into Imaris to determine the average number of cellular adhesions for the indicated conditions. Data were normalized by dividing the number of adhesions in each cell by the total cell area. Cell area was determined by drawing a contour around each cell. D, cellular adhesions from C were subsequently analyzed for size. An intensity threshold was used to classify small adhesions. Adhesions with a mean intensity less than 35 (arbitrary units) were considered to be small. The number of small adhesions in each cell was then divided by total adhesions identified. Data represent mean ± S.E. (error bars) from three independent experiments. (*, p < 0.003; Mann–Whitney U test).
Phosphorylation of SHCA is required for TGFβ-induced migration
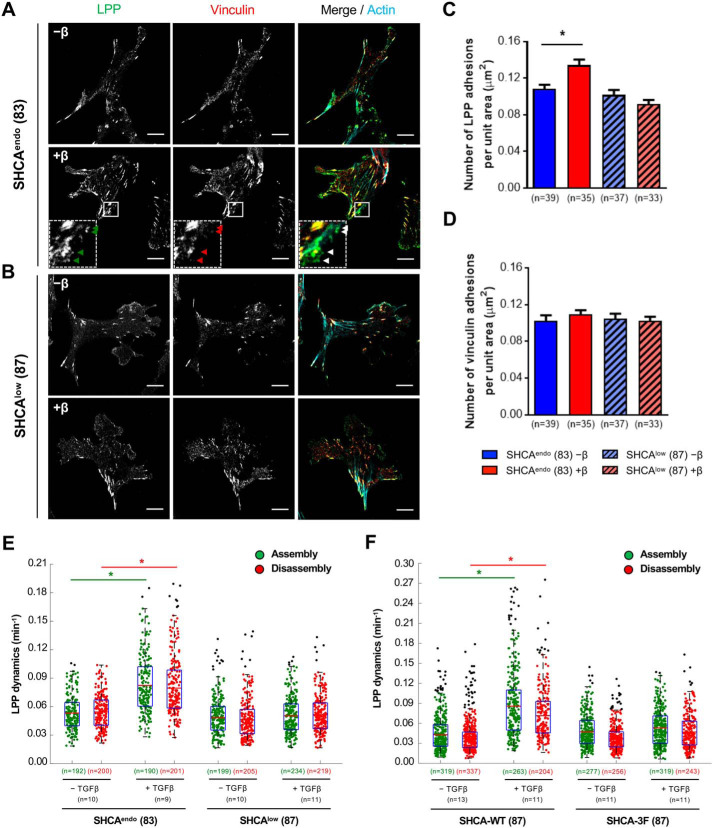
The CH1 domain of SHCA contains three phosphotyrosine residues (Tyr-239/Tyr-240/Tyr-313) that are critical for breast cancer metastasis (32). Given the importance of SHCA signaling in breast cancer progression, we sought to investigate single-cell migration of ErbB2-NT/SHCAlow cells rescued with WT p46/52SHCA or various SHCA mutants that lacked specific tyrosine phosphorylation sites (Shc313F, Y313F; Shc2F, Y239F/Y240F; Shc3F, Y239F/Y240F/Y313F) (Fig. 4A). Immunoblot analysis revealed that WT and mutant SHCA alleles were expressed in ErbB2-NT/SHCAlow breast cancer cells at comparable levels (Fig. 4B). As expected, TGFβ stimulated the migration of cells rescued with SHCA-WT in a manner similar to ErbB2-NT/SHCAendo cells (Fig. 4C). In particular, the mean net displacement and average speed of cells were increased 18 h after TGFβ treatment (Fig. 4, D and E). Additionally, TGFβ promoted an increase in adhesion assembly and disassembly rates in SHCA-WT cells (Fig. 4F). In contrast, SHCA-3F cells failed to respond to TGFβ. Indeed, the mean net displacement, migration speeds, and adhesion dynamics of SHCA-3F–expressing ErbB2-NT/SHCAlow cells was unaffected by TGFβ stimulation (Fig. 4, C–F). Expression of SHCA-313F or SHCA-2F was sufficient to restore responsiveness to TGFβ, as measured by mean net displacement and cell speed (Fig. S3). We confirmed these results by re-expressing SHCA-WT, SHCA-313F, SHCA-2F, and SHCA-3F in cells from an independent ErbB2-NT/SHCAlow explant (Fig. S4). Altogether, these results revealed that SHCA controls TGFβ-induced migration of ErbB2-overexpressing breast cancer cells by regulating adhesion dynamics, which requires pY239/pY240 or pY313-dependent SHCA signaling.
Figure 4.
Loss of SHCA phosphorylation abrogates TGFβ-induced increase in migration speed of ErbB2-expressing breast cancer cells. A, schematic diagram of SHCA showing the domain structure and location of three tyrosine residues (Y) that were mutated to phenylalanine (F). B, immunoblot analysis of whole-cell lysates showing SHCA levels (anti-SHCA or anti-FLAG antibodies) in the indicated cell populations. α-Tubulin was used as a loading control. C, ErbB2-NT/SHCA-WT (explant 87) and ErbB2-NT/SHCA-3F (explant 87) cells were seeded onto fibronectin-coated glass bottom dishes and allowed to migrate in the absence or presence of TGFβ (2 ng ml−1). Live-cell migration tracks for each condition are shown 18 h after treatment. Each line represents the migration path of a single cell over 6 h. The starting point of each cell was superimposed on the origin (0, 0). D, cell displacements from C were pooled into 45° segments based on their angle of trajectory and averaged to determine mean final displacement. E, average migration speed over time was calculated by determining the mean distance traveled between each imaging time point by all cells in C. The data for each population were then averaged into 3-h segments. Data represent mean ± S.E. (error bars) from four independent experiments. *, p < 0.05, two-tailed Student's t test. F, adhesions in protrusive cell regions were tracked over time using TIRF microscopy. Cells were transfected with mCherry-paxillin 48 h prior to imaging and left untreated or treated with TGFβ for 24 h. Average assembly (green) or disassembly (red) rates were determined from changes in mean fluorescence intensity. Data represent individual assembly and disassembly events from three independent experiments. Coloured n values refer to the number of events while black n values refer to the number of cells. The top and bottom lines of the box indicate the third and first quartile, respectively, whereas the heavy central line indicates the mean. The whiskers extend up to 1.5 times the interquartile range. Black dots represent outliers. *, p < 0.0001 calculated from the cell averages for assembly and disassembly rates; two-tailed Student's t test.
SHCA regulates LPP recruitment and retention in cellular adhesions
Our data demonstrate a role for SHCA in regulating adhesion dynamics in rapidly migrating cells. To outline the molecular mechanisms by which SHCA impacts cellular adhesions, we sought to delineate its potential protein partners in signaling complexes using BioID proximity labeling. To achieve this, we generated cells expressing WT SHCA fused to a mutant BirA biotin ligase that covalently adds biotin to adjacent proteins (Myc-SHCA-WT-BirA). Biotinylated proteins were pulled down from cell lysates and identified by MS, using Myc-BirA as a control. A biological triplicate was performed, and nonspecific interactions were eliminated using the significance analysis of interactome (SAINT) algorithm (48, 49). Strikingly, several adhesion and actin cytoskeletal proteins were identified as proximal to SHCA, including arpin, paxillin (Pxn), talin (Tln), and LPP (Table 1 and Table S1). LPP is a member of the zyxin family of LIM proteins (50). It is able to localize to adhesions and interact with LIM and SH3 protein (LASP), palladin, protein phosphatase 2A (PP2A), scrib, supervillin, and vasodilator-stimulated phosphoprotein (VASP) (39). We have previously identified LPP as a promoter of efficient breast cancer lung metastasis (37). Therefore, we decided to investigate whether SHCA regulates the function of LPP in response to TGFβ.
Table 1.
BioID analysis to identify novel SHCA-interacting proteins
| Prey | Spectral countsa | SAINT scoreb | -Fold changec | Function |
|---|---|---|---|---|
| Cellular adhesion proteins (SHCA interactors) | ||||
| Actr2 | 11.3 ± 2.3 | 0.96 | 4.3 | ATP-binding component of Arp2/3 complex |
| Arhgef10l | 9.3 ± 2.5 | 1 | 93.3 | RhoGEF |
| Arhgef5 | 5.3 ± 3.5 | 0.98 | 53.3 | RhoGEF |
| Arhgef7 | 3.3 ± 1.2 | 0.98 | 33.3 | RhoGEF |
| Arpin | 7.3 ± 2.9 | 1 | 73.3 | Regulates actin polymerization |
| Cd2ap | 24 ± 4 | 1 | 10.3 | Scaffold protein, binds actin |
| Coro1b | 37.3 ± 11.7 | 1 | 12.4 | Actin-binding protein |
| Crk | 5.7 ± 2.9 | 1 | 56.7 | Adaptor protein, cell adhesion, migration |
| Cttn | 25 ± 4.6 | 1 | 18.8 | Focal adhesion protein, cytoskeletal organization |
| Eps8l2 | 25.3 ± 7.5 | 1 | 253.3 | Enhances SOS activity, cytoskeletal organization |
| Lims1 | 4.7 ± 0.6 | 1 | 46.7 | Integrin signaling, focal adhesion protein |
| Lpp | 9.7 ± 5.5 | 0.98 | 9.7 | Focal adhesion protein, adhesion dynamics |
| Pak2 | 8.3 ± 2.3 | 1 | 25 | Ser/Thr kinase, cytoskeletal organization |
| Parva | 3.3 ± 0.6 | 0.96 | 10 | Focal adhesion protein, binds actin |
| Pdlim5 | 20.3 ± 7 | 1 | 15.3 | Cytoskeleton-associated protein, migration |
| Pxn | 2.7 ± 1.2 | 0.96 | 26.7 | Focal adhesion protein, adhesion dynamics |
| Sept10 | 8.3 ± 2.5 | 1 | 12.5 | GTP-binding protein, cytoskeletal organization |
| Sept11 | 27.7 ± 0.6 | 1 | 4.6 | GTP-binding protein, cytoskeletal organization |
| Sept2 | 33 ± 3.6 | 0.97 | 3.1 | GTP-binding protein, cytoskeletal organization |
| Sept6 | 11 ± 2.6 | 1 | 16.5 | GTP-binding protein, cytoskeletal organization |
| Sept7 | 36 ± 3 | 1 | 6.8 | GTP-binding protein, cytoskeletal organization |
| Tln1 | 37 ± 19.5 | 1 | 37 | Focal adhesion protein, binds actin |
| Known SHCA-interacting proteins | ||||
| Ptpn12 | 10.7 ± 2.9 | 1 | 32 | Protein-tyrosine phosphatase |
| Grb2 | 6.3 ± 3.2 | 1 | 63.3 | Adaptor protein; activates ERK and AKT pathways |
| Shcbp1 | 8.3 ± 1.5 | 1 | 83.3 | SHCA-binding protein |
a Average of three biological replicates.
b Cutoff: SAINT score >0.9 (from a total of 113 interacting proteins).
c -Fold enrichment relative to Myc-BirA alone.
We first validated that the Myc-SHCA-WT-BirA construct biotinylates LPP using an immunoblotting-based method that is orthogonal to BioID (Fig. S5). A BirA-only vector was included as a negative control. Next, we introduced mCherry-LPP into ErbB2-NT/SHCAendo and ErbB2-NT/SHCAlow cells to assess LPP localization in the absence or presence of TGFβ (Fig. 5, A and B). TGFβ enhanced LPP targeting to adhesions in ErbB2-NT/SHCAendo cells, which was not observed in ErbB2-NT/SHCAlow breast cancer cells (Fig. 5C). In addition to paxillin and LPP, we also investigated the number of vinculin-bearing adhesions found in ErbB2-NT/SHCAendo and ErbB2-NT/SHCAlow cells treated with TGFβ. Vinculin stabilizes and strengthens adhesions and is therefore an important indicator of more mature adhesions (51, 52). The number of vinculin-bearing mature adhesions did not change with TGFβ treatment (Fig. 5D), further supporting the notion that TGFβ promotes the formation of more nascent adhesions. Based on these observations, we performed time-lapse imaging to determine the assembly and disassembly rates of cellular adhesions containing LPP. ErbB2-NT/SHCAendo cells treated with TGFβ exhibited increased dynamics of LPP-containing adhesions in protrusive cell regions, whereas ErbB2-NT/SHCAlow cells did not (Fig. 5E). Expression of SHCA-WT in ErbB2-NT/SHCAlow cells restored TGFβ-induced increases in LPP dynamics; however, expression of the SHCA-3F phosphomutant was not sufficient to rescue this phenotype (Fig. 5F). Altogether, our data suggest that SHCA controls paxillin and LPP localization to dynamic adhesions in response to TGFβ.
Figure 5.
LPP targeting to adhesions in response to TGFβ requires SHCA. A and B, ErbB2-NT/SHCAendo (explant 83) and ErbB2-NT/SHCAlow (explant 87) cells were infected with mCherry-LPP, seeded onto fibronectin-coated glass bottom dishes, and cultured in the absence or presence of TGFβ (2 ng ml−1) for 24 h. Cells were then fixed with 4% PFA and stained with antibodies against vinculin and phalloidin (F-actin). Images were taken on a Zeiss CLSM using a Plan-Apochromat ×63/1.4 numerical aperture oil immersion DIC objective lens (1 pixel = 0.132 μm). The arrowheads highlight examples of small adhesions formed after treatment with TGFβ. Scale bar, 10 μm. C and D, images were imported into Imaris to determine the average number of LPP and vinculin-bearing adhesions over the whole cell for each condition. Data were normalized by dividing the number of adhesions in each cell by the total cell area. Cell area was determined by drawing a contour around each cell. Data represent mean ± S.E. (error bars) from three independent experiments. (*, p = 0.001, Mann–Whitney U test). E and F, adhesions in protrusive cell regions were tracked over time using TIRF microscopy. Average assembly (green) or disassembly (red) rates were determined from changes in mean fluorescence intensity after 24 h with or without TGFβ treatment. Data represent individual assembly and disassembly events from three independent experiments. Coloured n values refer to the number of events while black n values refer to the number of cells. The top and bottom lines of the box indicate the third and first quartile, respectively, whereas the heavy central line indicates the mean. The whiskers extend up to 1.5 times the interquartile range. Black dots represent outliers. *, p < 0.001 calculated from the cell averages for assembly and disassembly rates, two-tailed Student's t test.
LPP is an important component of adhesions that interacts with α-actinin to mediate TGFβ-enhanced migration
Previous studies show that LPP is an important regulator of mesenchymal cell migration (53, 54). Given that SHCA regulates LPP recruitment to adhesions in response to TGFβ, we sought to determine whether LPP also regulates TGFβ-enhanced migration and adhesion dynamics. ErbB2-NT cells with endogenous levels of SHCA were infected with shRNAs against LPP (LPPKD) or LucA (LPPendo) (Fig. 6A and Fig. S6A). LPP knockdown was also confirmed by immunofluorescence staining and linescan analysis of paxillin-bearing adhesions (Fig. S6, B–D). Cells with endogenous levels of SHCA and LPP (LPPendo) migrated further and faster in response to TGFβ treatment (Fig. 6, B–D). Cells with LPP knockdown (LPPKD), however, did not exhibit increased migration despite remaining responsive to TGFβ-induced signaling (pSmad/Smad) (Fig. 6B and Fig. S6A). Furthermore, TGFβ did not affect assembly and disassembly rates of paxillin-bearing adhesions in LPPKD cells (Fig. 6E). Together, these data suggest that both SHCA and LPP are required for enhancing adhesion dynamics in TGFβ-stimulated cells.
Figure 6.
LPP is required for TGFβ-induced migration and adhesion dynamics. A, immunoblot analysis of whole cell lysates showing LPP levels in the indicated cell populations. α-Tubulin was used as a loading control. B, ErbB2-NT/LPPendo and ErbB2-NT/LPPKD cells were seeded onto fibronectin-coated glass bottom dishes and allowed to migrate in the absence or presence of TGFβ (2 ng ml−1). Live-cell migration tracks for each condition are shown 18 h after treatment. Each line represents the migration path of a single cell over 6 h. The starting point of each cell was superimposed on the origin (0, 0). C, cell displacements from B were pooled into 45° segments based on their angle of trajectory and averaged to determine mean final displacement. D, average migration speed over time was calculated by determining the mean distance traveled between each imaging time point by all cells in B. The data for each population were then averaged into 3-h segments. Data represent mean ± S.E. (error bars) from four independent experiments. *, p < 0.05, two-tailed Student's t test. E, adhesions in protrusive cell regions were tracked over time using TIRF microscopy. Cells were transfected with mCherry-paxillin 48 h prior to imaging and left untreated or treated with TGFβ for 24 h. Average assembly (green) or disassembly (red) rates were determined from changes in mean fluorescence intensity. Data represent individual assembly and disassembly events from three independent experiments. Coloured n values refer to the number of events while black n values refer to the number of cells. The top and bottom lines of the box indicate the third and first quartile, respectively, whereas the heavy central line indicates the mean. The whiskers extend up to 1.5 times the interquartile range. Black dots represent outliers. *, p < 0.0001 calculated from the cell averages for assembly and disassembly rates; two-tailed Student's t test.
LPP has three LIM domains, which are necessary for localization to adhesions. LPP also contains an α-actinin–binding domain (ABD), which allows it to interact with the actin cytoskeleton. Previous studies by our group show that LPP must localize to adhesions and interact with the actin cytoskeleton to promote metastasis (37). Therefore, we rescued LPPKD cells with one of three fluorescently tagged LPP constructs: WT LPP (EGFP-LPP-WT), a mutant that cannot localize to adhesions (EGFP-LPP-mLIM1), or a mutant that cannot interact with α-actinin (EGFP-LPP-ΔABD) (Fig. S7, A and B). Immunofluorescence staining and linescan analysis showed co-localization between EGFP-LPP-WT and paxillin in adhesions (Fig. S7, C–E). LPP-ΔABD also co-localized with paxillin at adhesions (Fig. S7, C–E). LPP-mLIM1, however, failed to localize to adhesions despite being expressed at levels comparable with endogenous LPP (Fig. S7, C–E). Migration and adhesion dynamics of LPP-WT, LPP-mLIM1, and LPP-ΔABD cells were then analyzed using live-cell microscopy. Re-expression of WT LPP in LPPKD cells successfully rescued the migratory phenotype observed in LPPendo cells (Fig. S8). Expression of either LPP-mLIM1 and LPP-ΔABD, however, was not able to rescue TGFβ-enhanced cell migration and adhesion dynamics (Fig. S8). Thus, LPP must localize to adhesions and interact with the actin cytoskeleton to facilitate faster cell migration and adhesion dynamics in response to TGFβ treatment.
SHCA localizes to adhesions to regulate focal complex formation
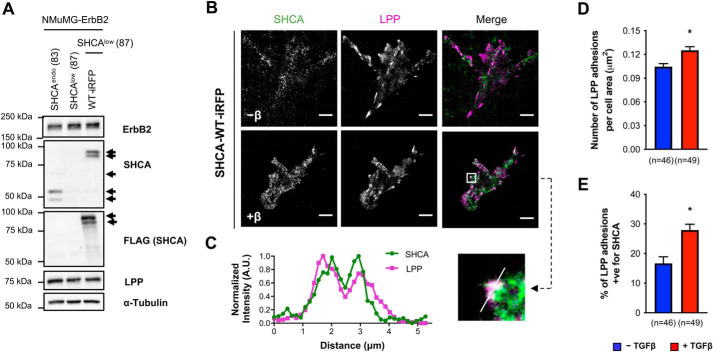
Given that SHCA interacts with several adhesion proteins, we explored the possibility that SHCA may also localize to adhesions. Using a far-red fluorescent protein (miRFP670) (55), we generated a WT SHCA construct (SHCA-WT-iRFP) and performed TIRF microscopy to limit fluorescent illumination to the first 80 nm of the cell. ErbB2-NT/SHCAlow cells were infected with SHCA-WT-iRFP or SHCA-3F-iRFP (Fig. 7A and Fig. S9A) and assessed for their responsiveness to TGFβ. The migratory phenotype was successfully rescued with SHCA-WT-iRFP but not the SHCA-3F-iRFP mutant (Fig. S9, B–D). SHCA-WT-iRFP cells were then infected with mCherry-LPP and fixed to assess the cellular localization of these adapter proteins. TIRF microscopy revealed more SHCA clusters at the plasma membrane after TGFβ treatment (Fig. 7B). SHCA-positive signals co-localized with mCherry-LPP, suggesting that SHCA is targeted to adhesions upon stimulation (Fig. 7C). Image quantification revealed that the number of LPP-bearing adhesions increased upon TGFβ treatment, as well as the percentage of LPP-containing adhesions positive for SHCA (Fig. 7, D and E). Taken together, these results suggest that SHCA facilitates the recruitment of LPP into cellular adhesions in response to TGFβ treatment.
Figure 7.
SHCA is found in adhesions and co-localizes with LPP upon TGFβ treatment. A, immunoblot analyses of whole-cell lysates showing SHCA (anti-SHCA or anti-FLAG antibodies) and LPP levels in ErbB2-NT/SHCAendo (explant 83), ErbB2-NT/SHCAlow (explant 87), and ErbB2-NT/SHCAlow/SHCA-WT-iRFP (explant 87) cells. α-Tubulin was used as a loading control. B, NMuMG ErbB2 cells expressing SHCA-WT-iRFP were infected with mCherry-LPP, seeded onto fibronectin-coated glass bottom dishes, and cultured in the absence or presence of TGFβ (2 ng ml−1) for 24 h. Cells were then fixed with 4% PFA and imaged using TIRF microscopy. Scale bar, 10 μm. C, linescan analysis of SHCA and LPP from the boxed region in B. D and E, images were imported into Imaris to determine the average number of LPP-bearing adhesions and percentage co-localization with SHCA over the whole cell. Data were normalized by dividing the number of adhesions in each cell by the total cell area. Cell area was determined by drawing a contour around each cell. Adhesions were considered positive for SHCA if the mean intensity of the SHCA channel within the adhesion exceeded the diffuse cytoplasmic signal intensity. Data represent mean ± S.E. (error bars) from three independent experiments. *, p < 0.0012; Mann–Whitney U test.
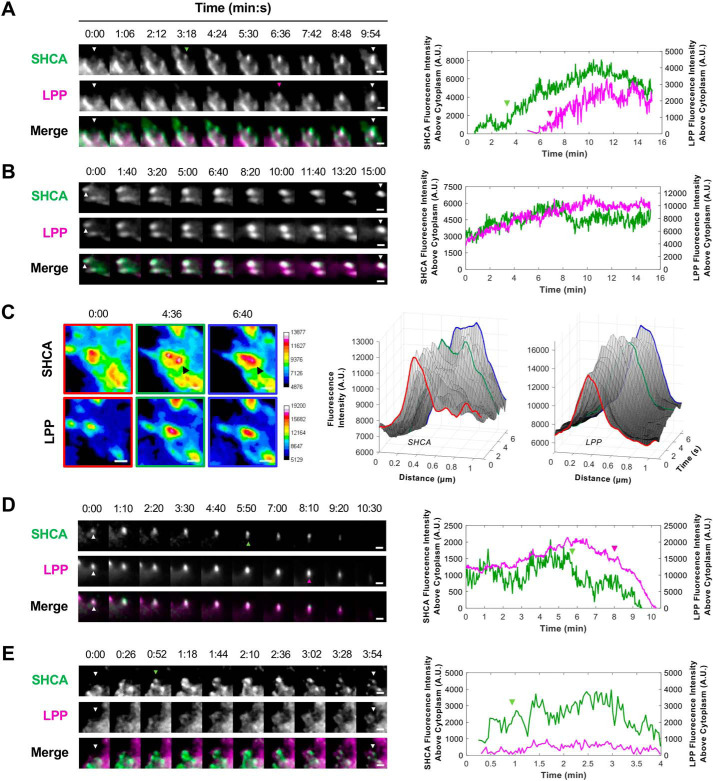
Adhesion assembly is a highly regulated process that begins with integrin binding to ECM ligands, such as fibronectin, vitronectin, and laminin. Ligand binding induces a conformational change that unmasks the short cytoplasmic tail of integrins, enabling the recruitment of scaffold and signaling proteins (56). TGFβ can regulate the expression of integrin subunits (57–59) and cause clustering of ErbB2 and integrins at the cell membrane (22). Considering that SHCA regulates the recruitment of LPP, we wondered whether SHCA directs adhesion formation. We performed semi-continuous time-lapse imaging of SHCA-WT-iRFP and EGFP-LPP to obtain high temporal resolution of adhesion dynamics (2-s intervals). SHCA appeared to assemble at the site of an adhesion prior to LPP recruitment (Fig. 8A and Movie S3). Once an adhesion formed and began to increase in size, the rate of SHCA recruitment decreased and eventually stabilized (Fig. 8B and Movie S4). Occasionally, a second nucleation point or “treadmilling” of SHCA was observed at the growing end of an adhesion (Fig. 8C). Interestingly, SHCA fluorescence intensity at some adhesions decreased prior to loss of LPP signal (Fig. 8D and Movie S5). Finally, there were instances where SHCA appeared to nucleate an adhesion but LPP was not recruited (Fig. 8E; Movie S6). In these cases, the adhesion often disassembled and was characterized by a short lifespan. These adhesions likely coincided with nascent adhesion formation and disassembly. Collectively, these four examples demonstrate that SHCA is an early component of adhesions and is required for the recruitment of LPP.
Figure 8.
SHCA localization to adhesions precedes LPP recruitment. ErbB2-NT/SHCAlow (explant 87) cells expressing SHCA-WT-iRFP and EGFP-LPP were seeded onto fibronectin-coated glass bottom dishes and imaged with TIRF microscopy. A, time-lapse montage of a newly forming adhesion (see Movie S3). White arrowheads indicate the site of adhesion nucleation; green arrowheads indicate the appearance of SHCA; and magenta arrowheads indicate the appearance of LPP. Intensity traces for SHCA and LPP are depicted in the adjacent graph. B, time-lapse montage of a maturing adhesion (see Movie S4). White arrowheads indicate the adhesion of interest. Intensity traces for SHCA and LPP are depicted in the adjacent graph. C, pseudo-colored montage (16-color) of SHCA and LPP in a mature adhesion. The black arrowhead points to a second nucleation point in the SHCA channel. Linescan analysis was performed every 20 s to show the formation of a second SHCA nucleation point at the growing end of the adhesion. Fluorescence intensity of SHCA and LPP are plotted as a function of distance and time. D, time-lapse montage of a disassembling adhesion (see Movie S5). White arrowheads indicate the adhesion of interest; green arrowheads indicate the start of SHCA disassembly; and magenta arrowheads indicate the start of LPP disassembly. Intensity traces for SHCA and LPP are depicted in the adjacent graph. E, time-lapse montage of a short-lived adhesion (see Movie S6). White arrowheads indicate the site of adhesion nucleation, and green arrowheads indicate the appearance of SHCA. Intensity traces for SHCA and LPP are depicted in the adjacent graph. Scale bars, 2 μm.
MAPK and PI3K activation are required for LPP localization to adhesions
In addition to the engagement of Smad proteins, TGFβ is known to modulate mitogen-activated protein kinase (MAPK) and phosphatidylinositol 3-kinase (PI3K)/AKT pathways (60–62). Tyrosine phosphorylation of SHCA by the TGFβ receptor complex causes the recruitment of a Grb2/son of sevenless (SOS)/Ras complex, which subsequently triggers activation of the Ras-MAPK pathway (63). Similarly, TGFβ stimulation causes the association of type I TGFβ receptor with p85, the regulatory subunit of PI3K, which mediates AKT activation and leads to the phosphorylation of S6 kinase 1 (64, 65). MAPK and PI3K pathways are also known to regulate cell migration (66–70). Therefore, LPP recruitment to adhesions was analyzed in the absence or presence of trametinib (a MEK inhibitor) or pictilisib (a PI3K inhibitor) to determine whether these signaling pathways play a role in regulating LPP recruitment to adhesions in response to TGFβ stimulation.
NMuMG cells expressing constitutively active ErbB2 (NT118) exhibited increased ERK1/2 phosphorylation (Thr-202/Tyr-204) levels following 24 h of TGFβ stimulation; however, S6 phosphorylation (Ser-240/244) levels did not increase significantly (Fig. S10, A and B). As expected, TGFβ promoted the formation of small, dynamic adhesions in control cells (Fig. S10, C (top panels) and D–F). Pretreatment with trametinib effectively eliminated ERK1/2 phosphorylation in both TGFβ-stimulated and unstimulated cells (Fig. S10, A and B). In line with previous observations (71), ERK inhibition resulted in the formation of large adhesions (Fig. S10, C (middle panels), E, and F). Cells pretreated with trametinib also exhibited large stress fibers at the trailing edge and a robust lamellipodia (Fig. S10C, middle panels). However, TGFβ stimulation did not affect the number or size of LPP adhesions compared with unstimulated trametinib-treated cells (Fig. S10, D–F). On the other hand, pretreatment with pictilisib reduced S6 phosphorylation (Fig. S10, A and B). Cells pretreated with pictilisib also exhibited large adhesions (Fig. S10, C (bottom panels) and D–F). TGFβ stimulation did not affect the number of LPP adhesions; however, the size of LPP adhesions increased significantly (Fig. S10, D–F). Together, these results suggest that both MAPK and PI3K signaling are required for LPP recruitment to small, dynamic adhesions in response to TGFβ. Additionally, the increased size of LPP-containing adhesions in pictilisib-treated cells stimulated with TGFβ suggests that PI3K signaling may be important for the regulation of adhesion disassembly.
SHCA is a mediator of invadopodia formation
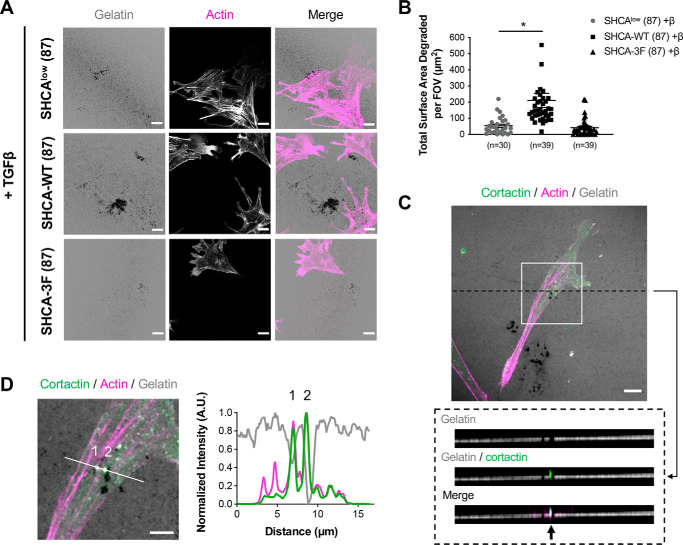
Invadopodia structures are unique mechanosensory structures that are central to the metastatic process (72). We have previously shown that LPP is required for TGFβ-induced invadopodia formation (37). Because SHCA regulates LPP recruitment to adhesions in response to TGFβ, we next investigated the effect of SHCA knockdown on invadopodia formation. ErbB2-NT/SHCAlow cells were plated onto fluorescently labeled gelatin for 24 h in the absence or presence of TGFβ. Cells were then fixed and stained with phalloidin (F-actin) and assessed for gelatin degradation (as indicated by the loss of fluorescence signal). Coverslips without cells were imaged as a control to ensure uniform gelatin coating (Fig. S11A). Our data revealed that shRNA-mediated knockdown of SHCA impaired the ability of cells to form invadopodia in response to TGFβ (Fig. 9, A and B). Of note, NMuMG ErbB2-NT cells did not degrade ECM in the absence of TGFβ stimulation (Fig. S11B). Exogenous expression of WT SHCA restored TGFβ-induced gelatin degradation; however, ErbB2-NT/SHCAlow cells expressing the SHCA-3F mutant failed to respond to TGFβ (Fig. 9, A and B). These results indicate that phosphorylation of Tyr-239/Tyr-240/Tyr-313 within SHCA is required for efficient invadopodia formation. To ensure that gelatin degradation was coincident with invadopodia formation, cells were stained for Cttn, a well-characterized marker of invadopodia (6, 11) (Fig. 9C). Linescan analysis showed multiple sites of actin and cortactin co-localization at sites of reduced gelatin fluorescence (Fig. 9D). ErbB2-NT/SHCAlow cells expressing fluorescently tagged SHCA constructs (SHCA-WT-iRFP and SHCA-3F-iRFP) demonstrated a similar phenotype, with cells expressing SHCA-WT-iRFP exhibiting significantly greater TGFβ-induced gelatin degradation relative to SHCA-3F-iRFP–expressing cells (Fig. S11, C–F). Thus, our results indicate that tyrosine phosphorylation of SHCA is required for TGFβ-induced invadopodia formation.
Figure 9.
SHCA promotes breast cancer cell-mediated gelatin degradation. A, ErbB2-NT/SHCAlow, ErbB2-NT/SHCAlow/SHCA-WT and ErbB2-NT/SHCAlow/SHCA-3F (explant 87) cells were pre-treated with TGFβ (2 ng ml−1) for 24 h and seeded onto fluorescently-labelled gelatin. Cells were allowed to invade for 24 h before fixing with 4% PFA and staining with antibodies against cortactin and phalloidin (F-actin). Images were taken on a Zeiss CLSM using a Plan-Apochromat ×63/1.4 NA oil immersion DIC objective lens (1 pixel = 0.132 μm). Scale bar, 15 μm. B, images were imported into Imaris to determine the total surface area degraded per field of view (FOV). Data represent mean ± S.E. from three independent experiments. Individual data points are depicted with symbols. *, p < 0.0001; Mann–Whitney U test. C, Z-stack acquisition was performed over a depth of 2.1 μm at 0.3-μm intervals. An orthogonal view (x-z plane, dotted black box) is presented to highlight an area of gelatin degradation where cortactin and actin are co-localized (black arrow). Scale bar, 10 μm. D, linescan analysis of zoomed region from C showing a slice view (x-y plane, solid white line) of cortactin and actin co-localization at sites of gelatin degradation. Intensity traces for cortactin, actin, and gelatin are shown in the adjacent graph. Scale bar, 5 μm.
Discussion
In this study, we establish SHCA (p46/52 isoforms) as a critical mediator of cell migration and invasion in ErbB2-overexpressing breast cancer cells. SHCA nucleates focal complex formation in the presence of TGFβ by enhancing the recruitment of key adhesion proteins, such as paxillin and LPP (Fig. 10). Beyond its function within cell adhesions, we further delineate a novel role for SHCA in TGFβ-induced invadopodia formation. In line with our observations that SHCA phosphorylation at Tyr-239/Tyr-240/Tyr-313 mediates adhesion dynamics, SHCA phosphorylation is also required for these invasive structures to form.
Figure 10.
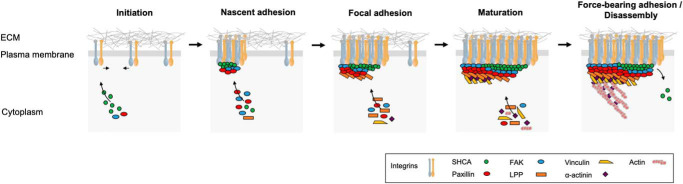
SHCA promotes the formation of small, dynamic adhesions in the presence of TGFβ by acting as a nucleator of focal complex formation. A schematic diagram illustrates the proposed role of SHCA during the different stages of adhesion formation. During initiation, TGFβ causes clustering of ErbB2 and integrins at the cell membrane. During nascent adhesion formation, SHCA associates with the cytoplasmic regions of several integrins both directly and indirectly. FAK and paxillin are also recruited. During focal adhesion formation, SHCA interacts with a variety of intracellular adapter/signaling molecules (including LPP) and enhances their recruitment to adhesions. During mature adhesion formation, SHCA exhibits treadmilling as more integrins are engaged. This promotes the recruitment of additional components to the growing end of an adhesion. In force-bearing adhesions, adhesion binding to actin cables establishes a link between the ECM and the cellular cytoskeleton for the generation of traction forces. During disassembly, SHCA begins to leave before other components.
Cells that undergo an EMT adopt a mesenchymal mode of migration, which relies on adhesions to generate traction forces for movement (73, 74). For cells to migrate, nascent adhesions must form, grow into focal contacts, couple to the actin cytoskeleton, and then disassemble as the cell moves forward, a process that generally occurs on the order of minutes (75, 76). Consequently, changes in adhesion dynamics dramatically affect the migration speed of cells (77). Our data demonstrate that TGFβ stimulates the formation of small, dynamic adhesions, which allow breast cancer cells to move at greater speeds. This is evidenced by a selective increase in the number of nascent paxillin-containing adhesions in response to TGFβ, with no change in more stable vinculin-containing mature adhesions. p46/52SHCA is required for this phenotype, as shRNA-mediated knockdown impairs the ability of NMuMG ErbB2-NT cells to enhance assembly and disassembly rates in response to TGFβ. Mutating the cytoplasmic tail of ErbB2 such that p46/52SHCA can no longer bind (ErbB2-NYPD) also prevents TGFβ-induced migration. Accordingly, lung metastatic burden is significantly reduced when SHCA signaling is disrupted (31, 32). These data are consistent with findings in triple-negative breast cancer cells, where p66SHCA is required for efficient cell migration and lung metastasis (35). Loss of p66SHCA expression results in the formation of large, elongated adhesions that exhibit slower assembly and disassembly rates, whereas exogenous expression of p66SHCA induces an EMT in ErbB2+ luminal breast cancers (34, 35).
TGFβ has been previously shown to induce the association of ErbB2 with the actin cytoskeleton (21, 22). In addition, TGFβ stimulation induces the expression of various α and β integrin subunits, including α1, α2, α3, α5, αv, β1, and β3 (57, 59, 78–80). Of these integrin subunits, collagens are bound by α1β1 and α2β1; laminin is bound by α1β1, α2β1 and αvβ3; and vitronectin is bound by αvβ1 and αvβ3 (81). Fibronectin is also a ligand for many of these integrin receptors, including α5β1, which is widely regarded as the major fibronectin receptor (81). Immunoprecipitation experiments show that p52SHCA can bind α5β1, α1β1, and αvβ3, but not α2β1, α3β1, or α6β1 (82). Binding to integrin β3 is facilitated by the PTB domain (83). SHCA can also interact with the cytoplasmic domain of integrin β4 through its SH2 or PTB domains upon phosphorylation of Tyr-1440 or Tyr-1526, respectively (84). Here, we demonstrate that SHCA localizes to integrin-dependent adhesions using a fluorescently tagged construct and time-lapse TIRF microscopy. Our BioID results show that SHCA interacts with a variety of adhesion and actin cytoskeletal proteins, such as arpin, crk, LPP, paxillin, and talin. The SHCA-BirA construct also labeled known interacting partners, including Grb2, Shc SH2 domain-binding protein 1 (Shcbp1), and protein-tyrosine phosphatase nonreceptor type 12 (Ptpn12) (85–87). Cell migration experiments were performed on fibronectin as it has been implicated in the development of multiple cancers (88–90). In fact, human breast cancer cells that overexpress α5β1 integrins show a 3-fold increase in cell invasiveness compared with ɑ5β1-depleted cells due to the generation of greater contractile forces (91). Given that SHCA regulates cell migration, invasion, and proliferation on collagen (92), laminin (93, 94), vitronectin (92), and fibronectin (31), it would be interesting to investigate adhesion dynamics and SHCA localization in response to different ECM components.
Our imaging results demonstrate that TGFβ induces clustering of SHCA in what appear to be nascent adhesions prior to LPP recruitment. In addition to nucleation, SHCA exhibits a treadmilling effect that may allow components to be recruited to the growing end of an adhesion. Three-dimensional superresolution microscopy reveals that integrins and actin filaments are separated by a core region that spans ∼40 nm (2). This region can be stratified into three spatial and function compartments: an integrin signaling layer, a force transduction layer, and an actin regulatory layer. Paxillin and focal adhesion kinase (FAK) appear to be key players in the membrane-proximal integrin signaling layer; vinculin is observed in the broader central zone responsible for force transduction; and zyxin localizes to the uppermost region that constitutes the actin regulatory layer (2). The ability of SHCA to engage with integrins and facilitate LPP localization to adhesions in the presence of TGFβ is necessary for enhanced cell migration. Moreover, LPP must interact with the actin cytoskeleton through an α-actinin–binding domain (ABD) to mediate adhesion dynamics. Based on the results presented here, it is conceivable that SHCA is localized to the signaling layer, whereas LPP is a constituent of the actin regulatory layer. Immunofluorescence analysis of motile cells shows that early nascent adhesions are largely composed of integrins, talin, and paxillin (95, 96). Mass spectrometry of the adhesion machinery shows that paxillin is also one of the first components to dissociate during nocodazole-induced adhesion disassembly, with ∼12% abundance remaining after 15 min (97). This is in contrast to vinculin, which is still an abundant constituent of adhesions (∼73%) over this time frame (97). In comparison, LPP dynamics appear to fall between these two adhesion components (∼43%) (97). It is interesting to note that p46/52SHCA does not appear in proteomics analyses of adhesions (97, 98). However, human endothelial cells treated with fibronectin-bound paramagnetic microbeads show that all three splice isoforms of SHCA (p46, p52, and p66) are found in adhesions (99). These results are in line with phosphoproteomic analyses that detect SHCA within isolated adhesion complexes and reveal SHCA phosphorylation on serine 139 (100). Importantly, our data also show that p46/52SHCA isoforms localize to adhesions following TGFβ treatment, which occurs prior to LPP localization. Given the fact that SHCA directly interacts with FAK (99), and FAK is a component of early adhesions (101, 102) that can localize prior to paxillin recruitment (103) and dissociate at roughly the same rate (97), it is conceivable that SHCA localizes to adhesions during the early stages of formation. It would be particularly interesting to generate a superresolution map of SHCA and other adhesion components in ErbB2-overexpressing breast cancer cells and investigate whether SHCA forms multiple nucleation sites along the growing end of an adhesion. Such an approach would provide clarity on the precise relationship between SHCA and LPP within adhesions.
Our data suggest that both MAPK and PI3K signaling pathways are required for LPP recruitment to small, dynamic adhesions in response to TGFβ stimulation. However, the observation that trametinib-treated cells are phenotypically different from pictilisib-treated cells suggests that distinct signaling pathways engaged downstream of SHCA differentially impact adhesion dynamics. In particular, trametinib causes LPP localization to stress fibers with no further increases in adhesion size and number following TGFβ stimulation. In contrast, pictilisib does not impact the pattern of LPP localization or result in the formation of robust lamellipodia. The size increase of LPP-containing adhesions following PI3K inhibition suggests that it plays a role in regulating adhesion disassembly. This interpretation is consistent with previous observations that knockout of FAK, which is upstream of PI3K (104, 105), specifically affects adhesion disassembly, but not adhesion assembly, rates (106).
LPP is a known regulator of migration in mesenchymal cells that has recently been characterized as a critical mediator of ErbB2+ breast cancer metastasis (37). LPP is also an important component of invadopodia (37). Invadopodia are mechanosensory structures (72, 107) that allow tumor cells to penetrate the basement membrane and move through dense environments comprised of highly cross-linked ECM fibers (108). Increasing evidence suggests that invadopodia directly contribute to poor overall survival (13). Consequently, many studies have attempted to elucidate key regulators of invadopodia formation and target precursor formation. Current models propose three stages of invadopodia formation: precursor core initiation, precursor stabilization, and maturation (109). Similar to adhesion assembly, SHCA may be involved in invadopodia formation. In the present study, we provide the first evidence that phosphotyrosine-dependent SHCA signaling is required for efficient invadopodia formation in breast cancer cells in response to TGFβ treatment. Interestingly, the results of our BioID screen show that SHCA is proximal to Cttn, a core invadopodia component. Therefore, the finding that SHCA knockdown prevents efficient TGFβ-induced invadopodia formation may be due to a loss of LPP recruitment to these structures, a loss of interaction with Cttn or both.
Several proteins (including LPP and Cttn (110)) have been identified in both adhesions and invadopodia, suggesting that these structures are intimately linked. Both cellular structures are connected to the actin cytoskeleton, albeit in a different fashion (5). The actin cytoskeleton couples to adhesions in a tangential orientation to generate traction forces, whereas invadopodia require perpendicular alignment of filaments, with respect to the underlying ECM, to create protrusive forces (111, 112). Whether invadopodia are discrete structures or derived from adhesions is still debated (113). Similar to adhesions, invadopodia are frequently surrounded by a multimeric protein complex consisting of integrins and integrin-associated proteins, such as paxillin, talin, and vinculin (114). These adhesion rings may help anchor invadopodia and promote their maturation. Given that SHCA regulates the recruitment of adhesion components in response to TGFβ, it is possible that SHCA may affect the formation of invadopodia anchors. Thus, the mechanisms through which SHCA regulates invadopodia formation awaits further investigation.
Experimental procedures
Cell culture
Normal murine mammary gland (NMuMG) cells were obtained from the American Type Culture Collection (ATCC; catalogue no. CRL-1636) and grown in high-glucose (4.5 g liter−1) Dulbecco's modified Eagle's medium (DMEM; catalogue no. 319-005-CL, Wisent Bioproducts) supplemented with 5% fetal bovine serum (FBS; catalogue no. 10082-147, Thermo Fisher Scientific), 10 μg ml−1 insulin (catalogue no. 511-016-CM, Wisent Bioproducts), 1 mm l-glutamine (catalogue no. 609-065-CM, Wisent Bioproducts), 1% penicillin-streptomycin (catalogue no. 450-201-EL, Wisent Bioproducts), and 0.2% amphotericin B (catalogue no. 450-105-QL, Wisent Bioproducts).
NMuMG cells (NT 118/119 and NYPD 120/121) were previously infected with a pMSCV-puro viral vector to express the rat orthologue of ErbB2 with an activating transmembrane point mutation V664E (31). Cells were cultured as described above with the addition of 2 μg ml−1 puromycin (catalogue no. ant-pr-1, InvivoGen) antibiotic selection to maintain ErbB2 expression. NMuMG cells (SHCAendo 83/84 and SHCAlow 87/92) were also previously infected with a pMSCV-hygro viral vector to express constitutively active ErbB2 (ErbB2-NT) and a pMSCV-puro viral vector containing an shRNA against endogenous SHCA (SHCAlow) or an shRNA against luciferase (SHCAendo) (32). Cells were maintained in 0.8 mg ml−1 hygromycin B (catalogue no. 450–141-XL, Wisent Bioproducts) to maintain ErbB2 expression and 2 μg ml−1 puromycin to maintain SHCA knockdown.
To generate individual explants (NT 118/119, NYPD 120/121, SHCAendo 83/84, and SHCAlow 87/92), NMuMG cells were injected into the mammary fat pad of athymic mice and subsequently derived from primary tumors (31, 32). Explants 87 and 92 (SHCAlow) were infected with a pMSCV-blast viral vector harboring a variety of SHCA mutants, including WT SHCA (SHCA-WT), SHCA-313F, SHCA-2F, and SHCA-3F. Finally, explant 87 (SHCAlow) was infected with WT SHCA fused to a far-red fluorescent protein (SHCA-WT-iRFP). Cells were maintained in 5 μg ml−1 blasticidin (catalogue no. ant-bl-1, InvivoGen) to maintain SHCA re-expression.
In parallel, NMuMG cells were previously infected with an shRNA against endogenous LPP (LPPKD) or an shRNA against luciferase (LPPendo) (36). Cells were then infected with constitutively active ErbB2 (36). Cells were cultured in 2 μg ml−1 puromycin to maintain LPP knockdown and 0.8 mg ml−1 hygromycin B to maintain ErbB2 expression. Finally, NMuMG-ErbB2 cells with LPP knockdown were infected with a pMSCV-blast vector containing EGFP-LPP-WT, EGFP-LPP-mLIM1, or EGFP-LPP-ΔABD (37). Cells were cultured in 5 μg ml−1 blasticidin.
MMTV/polyoma virus middle T antigen–expressing mammary tumor cells (MT864) were obtained from the laboratory of Dr. William J. Muller. Cells were cultured in high-glucose DMEM supplemented with 5% FBS, mammary epithelial growth supplement (3 ng ml−1 EGF (catalogue no. PHG0311, Invitrogen), 0.5 μg ml−1 hydrocortisone (catalogue no. 511-002-UG, Wisent Bioproducts), 5 μg ml−1 insulin, 0.4% (v/v) bovine pituitary extract (catalogue no. 002-011-IL, Wisent Bioproducts), 1% penicillin-streptomycin, and 50 μg ml−1 gentamycin (catalogue no. 450-135-XL, Wisent Bioproducts). WT SHCA tagged with BirA (Myc-SHCA-WT-BirA) and BirA-only vector were expressed in MMTV middle T antigen mammary epithelial cells. Cells were maintained in 8 μg ml−1 blasticidin (catalogue no. 400-190-EM, Wisent Bioproducts) antibiotic selection to maintain SHCA re-expression.
Retroviral production was performed using the Retro-X universal packaging system (catalogue no. 631530, Clontech) according to the manufacturer's protocol. Cells were then incubated with Polybrene (10 μg ml−1) and virus-containing medium for 48 h to allow for infection. Mycoplasma screening was routinely performed using the MycoAlert mycoplasma detection kit (catalogue no. LT07-318, Lonza).
DNA constructs
pMSCV-puro-ErbB2, pMSCV-hygro-ErbB2, pMSCV-puro-SHCA (shRNA), pMSCV-puro-LucA (shRNA), pMSCV-blast-SHCA, pMSCV-puro-LPP (shRNA), and pMSCV-EGFP-LPP constructs were previously generated in the laboratory of P. M. S (31, 32, 37).
FLAG-tagged SHCA was subcloned into pcDNA3.1 mycBioID (catalogue no. 35700, Addgene) using the following primers and NotI and EcoRI restriction enzymes: 5′-CACGAGCGGCCGCAAAACAAGCTGAGT-3′ and 5′-GCCGGAATTCGAATTTCACTTGTCATCGTC-3′. BirA-SHCA-FLAG was then subcloned into the pQCXIB expression vector (catalogue no. 22266, Addgene) using the following primers and AgeI and EcoRI restriction enzymes: 5′-AGCTGGCAC CGGTAGCCACCATGGAACAAAAACTC-3′ (a gift from Dr. Marc Fabian) and 5′-GCCGGAATTCGAATTTCACTTGTCATCGTC-3′.
To create SHCA-iRFP, SHCA was PCR-amplified from the previously generated constructs (pMSCV-SHCA-WT and pMSCV-SHCA-3F) with the following primers: 5′-CCCTTGAACCTCCTCGTTCGACC-3′ and 5′-TAGGTACCGCCTTGTCATCGTCATCCT-3′. XhoI and KpnI restriction enzymes were then used to insert the amplified product into pCMV-miRFP670 (55). Finally, 5′-CGCAAATGGGCGGTAGGCGTG-3′ and 5′-TATAGAATTCTTAGCTCTCAAGCGCGG-3′ primers with EcoRI and BglII restriction sites were used to shuttle SHCA-iRFP into pMSCV-blast.
To create cells with stable expression of mCherry-paxillin, pSL301 (Invitrogen) and pmCherryPaxillin (catalogue no. 50526, Addgene) were digested with AgeI and XbaI restriction enzymes. The pSL301-mCherryPaxillin intermediate was then digested with HpaI and EcoRI restriction enzymes to shuttle mCherry-paxillin into pMSCV-blast.
To create cells with stable expression of mCherry-LPP, LPP was PCR-amplified from previously generated pMSCV-EGFP-LPP-WT construct with the following primers: 5′-ATTGCGGCCGCGATGTCTCACCCATCTTGG-3′ and 5′-GAGACGTGCTACTTCCATTTGTC-3′. NotI and EcoRI restriction enzymes were then used to replace paxillin with the amplified LPP product in pMSCV-blast-mCherryPaxillin.
Cell migration assays
NMuMG ErbB2 cells were seeded onto μ-slide 8-well plates (catalogue no. 80821, ibidi) coated with 5 μg cm−2 fibronectin (catalogue no. F-0895, Sigma–Aldrich) diluted in 1× PBS. Cells were allowed to adhere and grow under exponential conditions for at least 12 h prior to experimentation. Recombinant human TGFβ1 (2 ng ml−1; catalogue no. 100-21, PeproTech) was applied to the cells directly before imaging (t = 0). Images were acquired on a Zeiss AxioObserver fully automated inverted microscope equipped with a Plan-Neofluar ×10/0.3 NA Ph1 objective lens, Axiocam 506 CCD camera (Carl Zeiss, Jena, Germany) and Chamlide TC-L-Z003 stage top environmental control incubator (Live-cell Instrument, Seoul, South Korea). Cells were imaged every 10 min in phase-contrast for a total of 30 h.
Analysis of cell migration
Cells were manually tracked in MetaXpress analysis software (Molecular Devices, Sunnyvale, CA). x,y position data for each cell track was then exported to MATLAB (version 8.6, release R2015b, The MathWorks, Natick, MA). Rose plots of cell migration were created by superimposing the starting position of each track on the origin (0, 0). Windrose plots were generated by determining the angle and mean net displacement of each track. The mean net displacement for each 45° segment was then plotted. The average speed was calculated by determining the mean distance traveled between each time point over the imaging interval. Speeds were then averaged into 3-h segments. The data shown represent the mean ± S.E. for all cells analyzed from at least three independent experiments.
Imaging adhesion turnover
NMuMG ErbB2 cells were seeded onto 35-mm coverglass bottom cell culture dishes (catalogue no. FD35-100, World Precision Instruments) coated with fibronectin and transfected with 1 μg of pmCherry paxillin using Effectene reagent (catalogue no. 301425, Qiagen). Medium was changed 18–24 h after transfection, and cells were allowed to recover for an additional 24 h in the absence or presence of TGFβ. NMuMG ErbB2 cells stably expressing mCherry-LPP were also seeded onto 35-mm dishes to analyze the dynamics of LPP. Cells with stable expression of mCherry-LPP were cultured in 5 μg ml−1 blasticidin to maintain expression. Images were acquired every 30 s for 25 min on a total internal reflection fluorescence (TIRF)-Spinning Disk Spectral Diskovery System (Spectral Applied Research, Richmond Hill, Ontario, Canada) coupled to a DMI6000B Leica microscope equipped with a Plan-Apochromat ×63/1.47 NA oil immersion DIC objective lens (Leica, Wetzler, Germany), ImagEM X2 EM-CCD camera (Hamamatsu Photonics K.K., Hamamatsu City, Japan), and Chamlide CU-501 top-stage incubator system (Live Cell Instrument, Seoul, South Korea). An EM-CCD camera was chosen at the cost of resolution (1 pixel = 0.155 μm) because of increased sensitivity. A 561-nm laser with an ET 620/60-nm filter cube was used to visualize mCherry-paxillin or mCherry-LPP (where indicated). The camera exposure time was set to 500 ms with an EM gain of 255 and read speed of 22 MHz. A TIRF prism was used to limit fluorescence excitation to a depth of 100 nm.
Calculating adhesion dynamics
Images collected with the TIRF microscope were processed in Imaris (version 8.3.1; Bitplane AG, Zurich, Switzerland) using the Surfaces function. A protruding edge of each cell was manually selected using the region of interest (ROI) tool. Surface detail was smoothed and set to 0.300 μm with a local background subtraction of 0.300 μm. Adhesions were then masked by a manual refinement of the autothreshold feature. Finally, adhesions were tracked over time using an autoregressive algorithm with a maximum distance of 2 μm and maximum gap size of three time points. Surfaces smaller than 5 pixels were removed by filtering.
Mean intensity data for each adhesion tracked in Imaris were exported to Excel for further analysis. Curves were visually inspected for assembly and disassembly events. A string of five or more points upward was interpreted as assembly, whereas five or more points downward was interpreted as disassembly. A log-linear fitting method was then used to determine the rate for each event. Fits with an r2 value greater than 0.7 were considered acceptable. Finally, assembly and disassembly rates were pooled together into a box plot for each condition. The mean for each condition is denoted by a red line. The data shown represent the results from at least three independent experiments.
To verify the results of our semi-automatic analysis, a custom algorithm was created in MATLAB (35, 115). Here, a spline curve was first fitted to each intensity trace to identify segments of assembly and disassembly. The difference in intensity between each time point was calculated, and changes greater than 15% were considered significant. A log-linear fitting method was then used to determine the rate for each event, as described above. Our automatic algorithm generated the same results as the semi-automatic analysis.
Immunoblotting
Cells were cultured to 80% confluence and lysed in ice-cold TNE lysis buffer, as described previously (31). Where indicated, cells were cultured in the presence of TGFβ for 48 h. Total cell lysates (20 μg) were resolved by 6–12% SDS-PAGE. Proteins were transferred onto polyvinylidene difluoride membranes (catalogue no. IPVH00010, Millipore), and membranes were blocked in 5% fat-free milk for 1 h. Membranes were incubated with the following antibodies overnight at 4 °C: ErbB2 (1:4000; catalogue no. sc-284, Santa Cruz Biotechnology, Inc.), SHCA (1:4000; catalogue no. 610082, BD Transduction Laboratories), FLAG (1:4000; catalogue no. F1804, Sigma–Aldrich), LPP (1:4000; catalogue no. sc-101434, Santa Cruz Biotechnology), paxillin (1:4000; catalogue no. ab23510, Abcam), Smad2/3 (1:1000; catalogue no. 3102, Cell Signaling), pSmad2 (Ser-465/467) (1:1000; catalogue no. 3101, Cell Signaling), Myc (1:3000; catalogue no. TAG003, BioShop), α-tubulin (1:20,000; catalogue no. T9026, Sigma–Aldrich), and glyceraldehyde-3-phosphate dehydrogenase (1:10,000; catalogue no. MAB374, Millipore). The appropriate horseradish peroxidase–conjugated secondary antibodies (1:10,000; Jackson Immuno Research Laboratories) were added to the membranes for 1 h. Finally, the membranes were visualized using SuperSignalTM West Pico PLUS Chemiluminescent Substrate (catalogue no. 34578, Thermo Fisher Scientific).
For MAPK and PI3K inhibitor experiments, cells were pretreated with 500 nm trametinib (catalogue no. T-8123, LC Laboratories) or 500 nm pictilisib (catalogue no. 11600-10, Cayman Chemical) for 12 h. TGFβ was then added to the medium, and cells were cultured for an additional 24 h. Proteins were transferred onto polyvinylidene difluoride membranes (catalogue no. IPFL00010, Millipore), and membranes were blocked with Intercept blocking buffer (catalogue no. 927-60001, LI-COR) for 1 h. Membranes were incubated with the following antibodies overnight at 4 °C: ERK1/2 (1:1000; catalogue no. 9107S, Cell Signaling), pErk1/2 (Thr-202/Tyr-204) (1:1000; catalogue no. 9101S, Cell Signaling), S6 (1:500; catalogue no. 2317S, Cell Signaling), and pS6 (Ser-240/244) (1:500; catalogue no. 5364S, Cell Signaling). IR dyes 680RD donkey anti-mouse (1:10,000; catalogue no. 926-68072, LI-COR) and 800CW donkey anti-rabbit (1:10,000; catalogue no. 926-32213, LI-COR) were added to the membranes for 1 h. Finally, the membranes were visualized with the Odyssey Imager (LI-COR, Lincoln, NE). Quantification was performed with ImageJ (National Institutes of Health, Bethesda, MD). Phosphorylated protein levels were normalized by their respective total protein levels. Fold change for each experiment was determined relative to unstimulated cells. The data shown represent the mean ± S.E. for three independent experiments.
Immunofluorescence
Cells were plated onto 35-mm coverglass bottom dishes coated with fibronectin and allowed to settle overnight. Cells were then cultured in the absence or presence of TGFβ for 24 h before being fixed with 4% paraformaldehyde (PFA) for 10 min at 37 °C. Cells were permeabilized with 0.3% Triton X-100 for 10 min, rinsed with 100 mm glycine in PBS, and blocked with 10% FBS in PBS for 1 h. Antibodies against LPP (1:500; catalogue no. sc-27312, Santa Cruz Biotechnology) and vinculin (1:1500; catalogue no. V9131, Sigma–Aldrich) were applied overnight at 4 °C (where indicated). Alexa Fluor 488 (1:1000; catalogue no. A11055, Invitrogen) and Alexa Fluor 546 (1:1000; catalogue no. A10036, Invitrogen) dye-conjugated secondary antibodies were then applied for 1 h at room temperature to visualize LPP and vinculin, respectively. Alexa Fluor 488 phalloidin (1:500; catalogue no. A12379, Invitrogen) was used to visualize F-actin, and 4′,6-diamidino-2-phenylindole (1:20,000; catalogue no. D1306, Thermo Fisher Scientific) was used to visualize nuclei (where indicated). Cells were kept in 1× PBS with 0.05% sodium azide (catalogue no. SAZ001, BioShop).
For MAPK and PI3K inhibitor experiments, cells were seeded onto coverglass bottom dishes and pretreated with 500 nm trametinib or 500 nm pictilisib for 12 h. TGFβ was then added to the medium, and cells were cultured for an additional 24 h. Antibodies against LPP (1:1000) and paxillin (1:1000) were applied overnight at 4 °C. Alexa Fluor 546 (1:1000) and Alexa Fluor 647 (1:1000; catalogue no. A21447, Invitrogen) dye-conjugated secondary antibodies were then applied for 1 h at room temperature to visualize paxillin and LPP, respectively. Alexa Fluor 488 phalloidin (1:1000) was used to visualize F-actin. Cells were kept in 1× PBS.
Images were acquired on a Zeiss LSM 710 confocal microscope with a Plan-Apochromat ×63/1.4 NA oil immersion DIC objective lens. For Figs. 3 and 5, the following parameters were used: 1 Airy unit, 900 master gain, 1 digital gain, 3.15-μs pixel dwell time, and 4-line averaging. An Ar ion laser tuned to 488 nm (1% power; 488/594-nm beam splitter; 493–556-nm detection) was used to visualize actin, a HeNe Green 543-nm laser (8% power; 488/543/633-nm beam splitter; 548–586 nm detection) was used to visualize vinculin, and a HeNe 594-nm laser (8% power; 458/514/594-nm beam splitter; 599–638-nm detection) was used to visualize mCherry-paxillin or mCherry-LPP. Images were acquired sequentially and saved as 8-bit with 1024 × 1024 pixels. For EGFP-LPP-WT, -mLIM1, and -ΔABD localization (Fig. S7), the following parameters were used: 1 Airy unit, 875 (EGFP-LPP) and 900 (mCherry-paxillin) master gain, 1 digital gain, 0.79-μs pixel dwell time, and 16-line averaging. An Ar ion laser tuned to 488 nm (2–2.4% power; 488-nm beam splitter; 467–560-nm detection) was used to visualize EGFP-LPP constructs, and a 561-nm DPSS laser (3–4% power; 458/561-nm beam splitter; 575–692-nm detection) was used to visualize mCherry-paxillin. Images were saved as 12-bit with 1024 × 1024 pixels. For MAPK and PI3K inhibitor experiments (Fig. S10), the following parameters were used: 1 Airy unit, 850 master gain, 1 digital gain, 6.3-μs pixel dwell time, and 4-line averaging. The 488-nm laser (3.5% power; 488/594-nm beam splitter; 493–556-nm detection) was used to visualize actin, the 543-nm laser (8% power; 458/543-nm beam splitter; 548–643-nm detection) was used to visualize paxillin, and a HeNe Red 633-nm laser (5% power; 488/543/633-nm beam splitter; 638–755-nm detection) was used to visualize LPP. Images were saved as 8-bit with 512 × 512 pixels.
Images were also acquired on a Zeiss LSM 780 confocal microscope with a Plan-Apochromat ×63/1.4 NA oil immersion objective lens. For Fig. S6, the following parameters were used: 1 Airy unit, 800 master gain, 1 digital gain, 6.3-μs pixel dwell time, and 4-line averaging. A 405-nm blue diode laser (1% power; 405-nm beam splitter; 410–514-nm detection) was used to visualize 4′,6-diamidino-2-phenylindole, an Ar ion laser tuned to 488 nm (2% power; 488-nm beam splitter; 490–606-nm detection) was used to visualize LPP, and a 561-nm DPSS laser (2% power; 458/561-nm beam splitter; 578–696-nm detection) was used to visualize mCherry-paxillin. Images were saved as 8-bit with 1024 × 1024 pixels.
Determining the number and size of adhesions per cell area
Immunofluorescence images were imported into Imaris for analysis using the Surfaces function. Surface detail was smoothed and set to 0.200 μm with a local background subtraction of 0.200 μm. All cellular adhesions (vinculin, paxillin, and/or LPP signals) were masked by a manual refinement of the autothreshold feature and splitting touching objects with a seed point diameter of 0.500 μm. Surfaces smaller than 10 pixels were removed by filtering. A contour was then manually drawn around the cell to determine the total surface area in contact with the dish. Finally, the number of adhesions for each cell was divided by the total cell area. An intensity threshold was used to classify small adhesions. Adhesions with a mean intensity less than 35 (arbitrary units) were considered to be small. The number of small adhesions in each cell was then divided by the total number of adhesions identified. The data shown represent the mean ± S.E. for all cells analyzed from three independent experiments.
For MAPK and PI3K inhibitor experiments, surface detail and local background subtraction were uniformly changed to 0.300 μm to accommodate for the larger adhesions present in some conditions. Surfaces smaller than 2 pixels were removed by filtering. Measurements of adhesion size were obtained directly from masked adhesions. The data shown represent the mean ± S.E. for all cells analyzed from two independent experiments.
BioID assay and streptavidin pulldown
MMTV/MT cells were starved overnight and subsequently treated with 50 μm biotin-supplemented 10% FBS/DMEM growth medium for 24 h. Two 15-cm plates of cells at 70% confluence were used per pulldown sample. The next day, cells were trypsinized, washed with PBS, and centrifuged at 250 × g for 5 min. Cells collected from each 15-cm plate were lysed with 700 μl of RIPA lysis buffer (1% Triton X-100, 50 mm Tris-HCl, pH 7.5, 150 mm NaCl, 1 mm EDTA, 1 mm EGTA, 0.1% SDS) supplemented with protease inhibitor mixture, 50 units of benzonase, 0.5% sodium deoxycholate for 1 h on ice. Cells were vortexed every 15 min for 10 s. Lysates were sonicated at 50% amplitude two times for 10 s on ice. Lysates were centrifuged at maximum speed for 30 min at 4 °C. Pierce Avidin-agarose beads (30-μl bead volume per pulldown; catalogue no. 20219, Thermo Fisher Scientific) were used on supernatants from the centrifuged lysates (6% by volume of the supernatant was used as input control). Samples were rotated end-over-end for 3 h at 4 °C. Afterward, beads were centrifuged at 3000 rpm for 1 min at 4 °C and washed with RIPA buffer three times. For immunoblots, both input and pulldown samples were incubated with protein loading dye for 10 min at 95 °C. For elution for MS, following the wash with RIPA buffer, samples were washed twice with LIGHT buffer (50 mm Tris-HCl, pH 7.5, using HPLC grade water) and eluted with 150 μl of 50 mm H3PO4 (pH 1.5–2) on ice for 10 min. Supernatants were collected. This was repeated two more times, and all three elutions were pooled and kept in −80 °C until MS analysis. Three technical replicates per condition were used for MS analysis.
Mass spectrometry and analysis of BioID data
Each BioID experiment was performed in triplicate, on different days, and using cells from successive passages. To distinguish background from bona fide protein associations, Myc-BirA (no biotin) and Myc-SHCA-WT-BirA (no biotin) were used as controls to perform SAINT analyses (48, 116).
Peptide identification was carried out on an Orbitrap Fusion Tribrid mass spectrometer (Thermo Fisher Scientific) equipped with a nanoelectrospray ion source (Thermo Fisher Scientific) and coupled to an UltiMate 3000 nanoRSLC (Thermo Fisher Scientific). Data-dependent acquisition of mass spectra was performed using Xcalibur (version 3.0.63; Thermo Fisher Scientific). Full scan mass spectra (350–1800 m/z) were acquired in the orbitrap at a resolution of 120,000 using a maximum injection time of 50 ms and automatic gain control target of 4e5. The quadrupole analyzer allowed for the isolation of selected ions in a window of 1.6 m/z and fragmentation by higher-energy collision–induced dissociation with 35% energy. The resulting fragments were detected by the linear ion trap at a rapid scan rate. Dynamic exclusion of previously fragmented peptides was set for a period of 20 s and a tolerance of 10 ppm. All MS/MS peak lists were generated using Thermo Proteome Discoverer (version 1.4.0.288; Thermo Fisher Scientific) with default parameters. MGF sample files were analyzed using Mascot (version 2.5.1; Matrix Science, London, UK). The software was programmed to search against the Uniprot Mus musculus database (November 2014, 84,646 entries), assuming trypsin digestion. Searches were carried out with a fragment ion mass tolerance of 0.60 Da and a parent ion tolerance of 10 ppm. Carbamidomethyl of cysteine was specified as a fixed modification. Variable modifications were deamidation of asparagine and glutamine, oxidation of methionine, and phosphorylation of serine, threonine, and tyrosine. Two miscleavages were allowed.
Validation of the MS/MS-based peptide and protein identifications was carried out using Scaffold software (version 4.7.5; Proteome Software Inc., Portland, OR). Criteria for protein identifications were fixed at greater than 99% probability to achieve an FDR less than 1% and required at least one peptide to be identified. The Protein Prophet algorithm was used to assign peptide and protein probabilities (117, 118). For proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone, the principle of parsimony was applied. After being exported from Scaffold, spectral counts were formatted according to SAINT algorithm guidelines (48, 49). For each prey, the SAINT express statistical analyses were carried out by using the maximal spectral count value from Myc-BirA (no biotin) or Myc-SHCA-WT-BirA (no biotin) as controls. Proteins with a SAINT score ≥0.9 were deemed genuine interactors.
Co-localization of SHCA and LPP
NMuMG ErbB2 cells expressing SHCA-WT-iRFP and mCherry-LPP were plated onto 35-mm coverglass bottom dishes coated with fibronectin and allowed to settle overnight. Cells were cultured in the absence or presence of TGFβ for 24 h before being fixed with 4% PFA for 10 min at 37 °C. Images of SHCA and LPP were acquired sequentially on the TIRF microscope described above. A 561-nm laser with an ET 620/60-nm filter cube was used to visualize mCherry-LPP, whereas a 642-nm laser with an ET 700/75-nm filter cube was used to visualize SHCA-WT-iRFP. The camera exposure time was set to 1 s with an EM gain of 255 and read speed of 22 MHz. The TIRF prism was set to limit fluorescence excitation to a depth of 80 nm.
Images were imported into Imaris for analysis using the Surfaces function. Surface detail was smoothed and set to 0.200 μm with a local background subtraction of 0.200 μm. The LPP signal was used to mask cellular adhesions; manual refinement of the autothreshold feature and splitting touching objects with a seed point diameter of 0.500 μm was used. A contour was then manually drawn around the cell to determine the total surface area in contact with the dish. The percentage of LPP adhesions positive for SHCA was determined by analyzing the mean intensity of SHCA within each mask. Adhesions with a mean intensity greater than diffuse cytoplasmic signal were considered positive for SHCA. The data shown represent the mean ± S.E. for all cells analyzed from three independent experiments. Linescan analysis was performed in ImageJ to show overlap of SHCA and LPP signals in fixed cells.
Rapid imaging of adhesion dynamics
NMuMG ErbB2 cells expressing SHCA-WT-iRFP and EGFP-LPP were plated onto 35-mm coverglass bottom dishes coated with fibronectin and allowed to settle overnight. Cells were cultured in the presence of TGFβ for 24 h prior to imaging. Images of SHCA and LPP were acquired sequentially on the TIRF microscope described above. A 488-nm laser with an ET 525/50 nm filter cube was used to visualize EGFP-LPP, whereas a 642-nm laser with an ET 700/75-nm filter cube was used to visualize SHCA-WT-iRFP. The camera exposure time was set to 350 ms with an EM gain of 255 and read speed of 22 MHz. Images were acquired semi-continuously with the imaging interval set to 2 s. This time delay was chosen to give the filter wheel sufficient time to switch between filters. The exact time of acquisition for each image was recorded. The TIRF prism was set to limit fluorescence excitation to a depth of 80 nm.
Image stacks were analyzed in Imaris, as described in the previous section. Given that SHCA appears at sites of adhesion before LPP, the SHCA signal was used to mask adhesions in these sets of experiments. The mean intensities of SHCA and LPP within an adhesion were then plotted against time. Fluorescence intensity signals were adjusted for local cytoplasmic intensity. Time series data were adjusted for the delay between channels. Image stacks were also analyzed in ImageJ. Linescan analysis (ImageJ) was used to show treadmilling of SHCA, or the formation of a second nucleation point, in mature adhesions. Note that EGFP-LPP was chosen for these sets of experiments because EGFP is brighter than mCherry and there is no spectral overlap with iRFP.
Gelatin degradation assays
Degradation assays were performed on fluorescently conjugated gelatin-coated coverslips. Briefly, 35-mm glass-bottom dishes were coated with a mix of 0.1 mg ml−1 poly-d-lysine (catalogue no. P6407, Sigma–Aldrich) and 5 μg cm−2 fibronectin in 1× PBS for 20 min, followed by incubation with 0.4% glutaraldehyde for 10 min. Oregon Green 488–conjugated gelatin (catalogue no. G13186, Invitrogen) was diluted 1:20 with 0.1% unconjugated gelatin (catalogue no. 07903, Stem Cell Technologies) and used to coat dishes for 10 min at 37 °C. Coverslips were then incubated with 10 mg ml−1 sodium borohydride for 2 min, followed by 70% ethanol for 20 min. Three washes with 1× PBS were performed between each step. DMEM (37 °C) was added to the coverslips 1 h before cell plating.
Cells were pretreated with TGFβ for 24 h in a cell culture dish and counted before plating. Cells seeded onto gelatin dishes (32,000 cells) were allowed to invade for 24 h before fixing with 4% PFA. Cells were then permeabilized with 0.2% Triton X-100, rinsed with 100 mm glycine in PBS, and blocked with 10% FBS in PBS. Antibodies against cortactin (1:1000; catalogue no. 05-180, EMD Millipore) and Atto 647N phalloidin for F-actin (1:2000; catalogue no. 65906, Sigma–Aldrich) were used. Alexa Fluor 546 dye-conjugated secondary antibody (1:1000) was used to visualize cortactin. Cells were kept in 1× PBS with 0.05% sodium azide.
Images were acquired on a Zeiss LSM 710 confocal microscope with a Plan-Apochromat ×63/1.4 NA oil immersion DIC objective lens with the following parameters: 1 Airy unit, 800–900 master gain, 1 digital gain, 3.15-μs pixel dwell time, 4-line averaging. An Ar ion laser tuned to 488 nm (0.5–2.5% power; 488/594-nm beam splitter; 493–549-nm detection) was used to visualize gelatin, a HeNe Green 543-nm laser (4–5% power; 458/543-nm beam splitter; 566–599-nm detection) was used to visualize cortactin, and a HeNe Red 633-nm laser (1–4% power; 488/543/633-nm beam splitter; 638–759-nm detection) was used to visualize actin. Z-stacks were acquired at 0.26–0.30-μm steps over 2.1–2.4 μm. Images of gelatin alone were captured with a ×20/0.8 NA Plan-Apochromat objective lens. Images were saved as 12-bit with 1024 × 1024 pixels.
Quantification of gelatin degradation was performed in Imaris using the Surfaces function. The lookup table was inverted to make areas of degradation appear bright. Surface detail was smoothed and set to 0.500 μm with a local background subtraction of 0.250 μm. Areas of degradation were then masked by a manual refinement of the autothreshold feature. Finally, orthogonal views were generated with the section viewer. Linescan analysis was performed in ImageJ. The data shown represent the mean ± S.E. for all cells analyzed from three independent experiments.
Statistical analyses
Statistical significance values (p values) were obtained by performing an unpaired two-tailed Student's t test or Mann–Whitney U test. Data are presented as mean ± S.E. (where indicated). For adhesion dynamics, the top and bottom lines of box plots indicate the third and first quartile, respectively, whereas the heavy central lines indicate the mean. The whiskers extend up to 1.5 times the interquartile range. Black dots represent outliers.
Data availability
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (119) partner repository with the data set identifier PXD018265 and 10.6019/PXD018265.
Author contributions
A. K., E. V., R. A., and K. J. data curation; A. K. software; A. K., E. V., J. S., R. A., and K. J. formal analysis; A. K. and J. S. validation; A. K., E. V., and R. A. investigation; A. K., E. V., J. U.-S., P. M. S., and C. M. B. visualization; A. K., E. V., R. A., M. G. A., K. J., and C. M. B. methodology; A. K., E. V., P. M. S., and C. M. B. writing-original draft; A. K., E. V., P. M. S., and C. M. B. writing-review and editing; M. G. A., J. U.-S., P. M. S., and C. M. B. resources; N. B., J. U.-S., P. M. S., and C. M. B. supervision; G. T. preliminary data.
Acknowledgments
We thank Dr. William J. Muller (McGill University) for providing MMTV middle T antigen–expressing breast cancer cells. We also thank Dr. Marc Fabian (Lady Davis Institute) for providing the AgeI-Myc primer used to create Myc-SHCA-WT-BirA. Finally, we thank members of the Ursini-Siegel, Siegel, and Brown laboratories for thoughtful discussions about the manuscript. Imaging experiments were performed at the McGill Life Sciences Complex Advanced BioImaging Facility (ABIF) and Cell Imaging and Analysis Network (CIAN).
This work was supported by Natural Sciences and Engineering Research Council of Canada (NSERC) Grants 493616-16 and 386084-12, Canadian Institutes of Health Research (CIHR) Grant CPG-146475, Canadian Cancer Society Research Institute (CCSRI) Grant 702060, and Canadian Cancer Society (CCS) i2I Grant 705838 (to J. U.-S., P. M. S., and C. M. B.). The authors declare that they have no conflicts of interest with the contents of this article.
This article was selected as one of our Editors' Picks.
This article contains Table S1, Figs. S1–S11, and Movies S1–S6.
- ECM
- extracellular matrix
- ABD
- α-actinin–binding domain
- CH
- collagen homology domain
- Cttn
- cortactin
- EMT
- epithelial-to-mesenchymal transition
- ErbB2
- avian erythroblastosis oncogene B
- ERK
- extracellular signal–related kinase
- FAK
- focal adhesion kinase
- iRFP670
- far-red fluorescent protein
- LPP
- lipoma-preferred partner
- LPPendo
- cells expressing luciferase targeting shRNA
- LPPKD
- cells expressing LPP targeting shRNA
- LPP-mLIM1
- LPP construct with mutation in the first LIM domain
- LPP-ΔABD
- LPP construct with a deletion in the α-actinin–binding domain
- MAPK
- mitogen-activated protein kinase
- NMuMG
- normal murine mammary gland
- NT
- constitutively active ErbB2 receptor (V664E point mutation)
- NYPD
- constitutively active ErbB2 receptor with five autophosphorylation sites (Tyr-1028, Tyr-1144, Tyr-1201, Tyr-1226/Tyr-1227, and Tyr-1253) mutated to phenylalanine
- PI3K
- phosphoinositide 3-kinase
- PTB
- phosphotyrosine-binding domain
- SAINT
- significance analysis of interactome
- SH2
- Src homology 2 domain
- SHCA
- Src homology and collagen A
- SHCAendo
- cells expressing luciferase targeting shRNA
- SHCAlow
- cells expressing SHCA targeting shRNA
- Src
- avian sarcoma (Schmidt–Ruppin A-2) viral oncogene
- TGFβ
- transforming growth factor β
- TIRF
- total internal reflection fluorescence
- Tks5
- tyrosine kinase substrate with five SH3 domains
- 313F
- SHCA construct with tyrosine 313 mutated to phenylalanine
- 2F
- SHCA construct with tyrosine residues 239 and 240 mutated to phenylalanine
- 3F
- SHCA construct with tyrosine residues 239, 240, and 313 mutated to phenylalanine
- MMTV
- murine mammary tumor virus
- MEK
- mitogen-activated protein kinase/extracellular signal-regulated kinase kinase
- DMEM
- Dulbecco's modified Eagle's medium
- FBS
- fetal bovine serum
- NA
- numeric aperture
- DIC
- differential interference contrast
- RIPA
- radioimmunoprecipitation assay
- PFA
- paraformaldehyde
- EGFP
- enhanced green fluorescent protein
- CLSM
- confocal laser-scanning microscope.
References
- 1. Zaidel-Bar R., Itzkovitz S., Ma'ayan A., Iyengar R., and Geiger B. (2007) Functional atlas of the integrin adhesome. Nat. Cell Biol. 9, 858–867 10.1038/ncb0807-858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kanchanawong P., Shtengel G., Pasapera A. M., Ramko E. B., Davidson M. W., Hess H. F., and Waterman C. M. (2010) Nanoscale architecture of integrin-based cell adhesions. Nature 468, 580–584 10.1038/nature09621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Winograd-Katz S. E., Fässler R., Geiger B., and Legate K. R. (2014) The integrin adhesome: from genes and proteins to human disease. Nat. Rev. Mol. Cell Biol. 15, 273–288 10.1038/nrm3769 [DOI] [PubMed] [Google Scholar]
- 4. Murphy D. A., and Courtneidge S. A. (2011) The “ins” and “outs” of podosomes and invadopodia: characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12, 413–426 10.1038/nrm3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Albiges-Rizo C., Destaing O., Fourcade B., Planus E., and Block M. R. (2009) Actin machinery and mechanosensitivity in invadopodia, podosomes and focal adhesions. J. Cell Sci. 122, 3037–3049 10.1242/jcs.052704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Beaty B. T., and Condeelis J. (2014) Digging a little deeper: the stages of invadopodium formation and maturation. Eur. J. Cell Biol. 93, 438–444 10.1016/j.ejcb.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Seals D. F., Azucena E. F. Jr, Pass I., Tesfay L., Gordon R., Woodrow M., Resau J. H., and Courtneidge S. A. (2005) The adaptor protein Tks5/Fish is required for podosome formation and function, and for the protease-driven invasion of cancer cells. Cancer Cell 7, 155–165 10.1016/j.ccr.2005.01.006 [DOI] [PubMed] [Google Scholar]
- 8. Courtneidge S. A., Azucena E. F., Pass I., Seals D. F., and Tesfay L. (2005) The SRC substrate Tks5, podosomes (invadopodia), and cancer cell invasion. Cold Spring Harb. Symp. Quant. Biol. 70, 167–171 10.1101/sqb.2005.70.014 [DOI] [PubMed] [Google Scholar]
- 9. Stylli S. S., Stacey T. T., Verhagen A. M., Xu S. S., Pass I., Courtneidge S. A., and Lock P. (2009) Nck adaptor proteins link Tks5 to invadopodia actin regulation and ECM degradation. J. Cell Sci. 122, 2727–2740 10.1242/jcs.046680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bowden E. T., Barth M., Thomas D., Glazer R. I., and Mueller S. C. (1999) An invasion-related complex of cortactin, paxillin and PKCmu associates with invadopodia at sites of extracellular matrix degradation. Oncogene 18, 4440–4449 10.1038/sj.onc.1202827 [DOI] [PubMed] [Google Scholar]
- 11. Clark E. S., Whigham A. S., Yarbrough W. G., and Weaver A. M. (2007) Cortactin is an essential regulator of matrix metalloproteinase secretion and extracellular matrix degradation in invadopodia. Cancer Res. 67, 4227–4235 10.1158/0008-5472.CAN-06-3928 [DOI] [PubMed] [Google Scholar]
- 12. Provenzano P. P., and Keely P. J. (2009) The role of focal adhesion kinase in tumor initiation and progression. Cell Adh. Migr. 3, 347–350 10.4161/cam.3.4.9458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blouw B., Patel M., Iizuka S., Abdullah C., You W. K., Huang X., Li J. L., Diaz B., Stallcup W. B., and Courtneidge S. A. (2015) The invadopodia scaffold protein Tks5 is required for the growth of human breast cancer cells in vitro and in vivo. PLoS ONE 10, e0121003 10.1371/journal.pone.0121003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yamaguchi H., and Condeelis J. (2007) Regulation of the actin cytoskeleton in cancer cell migration and invasion. Biochim. Biophys. Acta 1773, 642–652 10.1016/j.bbamcr.2006.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wendt M. K., and Schiemann W. P. (2009) Therapeutic targeting of the focal adhesion complex prevents oncogenic TGF-β signaling and metastasis. Breast Cancer Res. 11, R68 10.1186/bcr2360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sulzmaier F. J., Jean C., and Schlaepfer D. D. (2014) FAK in cancer: mechanistic findings and clinical applications. Nat. Rev. Cancer 14, 598–610 10.1038/nrc3792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sakamoto S., McCann R. O., Dhir R., and Kyprianou N. (2010) Talin1 promotes tumor invasion and metastasis via focal adhesion signaling and anoikis resistance. Cancer Res. 70, 1885–1895 10.1158/0008-5472.CAN-09-2833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ueda Y., Wang S., Dumont N., Yi J. Y., Koh Y., and Arteaga C. L. (2004) Overexpression of HER2 (erbB2) in human breast epithelial cells unmasks transforming growth factor β-induced cell motility. J. Biol. Chem. 279, 24505–24513 10.1074/jbc.M400081200 [DOI] [PubMed] [Google Scholar]
- 19. Seton-Rogers S. E., Lu Y., Hines L. M., Koundinya M., LaBaer J., Muthuswamy S. K., and Brugge J. S. (2004) Cooperation of the ErbB2 receptor and transforming growth factor beta in induction of migration and invasion in mammary epithelial cells. Proc. Natl. Acad. Sci. U.S.A. 101, 1257–1262 10.1073/pnas.0308090100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rexer B. N., and Arteaga C. L. (2012) Intrinsic and acquired resistance to HER2-targeted therapies in HER2 gene-amplified breast cancer: mechanisms and clinical implications. Crit. Rev. Oncog. 17, 1–16 10.1615/CritRevOncog.v17.i1.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang S. E., Shin I., Wu F. Y., Friedman D. B., and Arteaga C. L. (2006) HER2/Neu (ErbB2) signaling to Rac1-Pak1 is temporally and spatially modulated by transforming growth factor β. Cancer Res. 66, 9591–9600 10.1158/0008-5472.CAN-06-2071 [DOI] [PubMed] [Google Scholar]
- 22. Wang S. E., Xiang B., Zent R., Quaranta V., Pozzi A., and Arteaga C. L. (2009) Transforming growth factor β induces clustering of HER2 and integrins by activating Src-focal adhesion kinase and receptor association to the cytoskeleton. Cancer Res. 69, 475–482 10.1158/0008-5472.CAN-08-2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Siegel P. M., Shu W., Cardiff R. D., Muller W. J., and Massagué J. (2003) Transforming growth factor beta signaling impairs Neu-induced mammary tumorigenesis while promoting pulmonary metastasis. Proc. Natl. Acad. Sci. U.S.A. 100, 8430–8435 10.1073/pnas.0932636100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ursini-Siegel J., Hardy W. R., Zuo D., Lam S. H., Sanguin-Gendreau V., Cardiff R. D., Pawson T., and Muller W. J. (2008) ShcA signalling is essential for tumour progression in mouse models of human breast cancer. EMBO J. 27, 910–920 10.1038/emboj.2008.22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ursini-Siegel J., Cory S., Zuo D., Hardy W. R., Rexhepaj E., Lam S., Schade B., Jirstrom K., Bjur E., Piccirillo C. A., Denardo D., Coussens L. M., Brennan D. J., Gallagher W. M., Park M., et al. (2010) Receptor tyrosine kinase signaling favors a protumorigenic state in breast cancer cells by inhibiting the adaptive immune response. Cancer Res. 70, 7776–7787 10.1158/0008-5472.CAN-10-2229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ursini-Siegel J., Hardy W. R., Zheng Y., Ling C., Zuo D., Zhang C., Podmore L., Pawson T., and Muller W. J. (2012) The ShcA SH2 domain engages a 14-3-3/PI3′K signaling complex and promotes breast cancer cell survival. Oncogene 31, 5038–5044 10.1038/onc.2012.4 [DOI] [PubMed] [Google Scholar]
- 27. Ahn R., Sabourin V., Ha J. R., Cory S., Maric G., Im Y. K., Hardy W. R., Zhao H., Park M., Hallett M., Siegel P. M., Pawson T., and Ursini-Siegel J. (2013) The ShcA PTB domain functions as a biological sensor of phosphotyrosine signaling during breast cancer progression. Cancer Res. 73, 4521–4532 10.1158/0008-5472.CAN-12-4178 [DOI] [PubMed] [Google Scholar]
- 28. Im Y. K., La Selva R., Gandin V., Ha J. R., Sabourin V., Sonenberg N., Pawson T., Topisirovic I., and Ursini-Siegel J. (2015) The ShcA adaptor activates AKT signaling to potentiate breast tumor angiogenesis by stimulating VEGF mRNA translation in a 4E-BP-dependent manner. Oncogene 34, 1729–1735 10.1038/onc.2014.110 [DOI] [PubMed] [Google Scholar]
- 29. Ha J. R., Ahn R., Smith H. W., Sabourin V., Hébert S., Cepeda Cañedo E., Im Y. K., Kleinman C. L., Muller W. J., and Ursini-Siegel J. (2018) Integration of distinct ShcA signaling complexes promotes breast tumor growth and tyrosine kinase inhibitor resistance. Mol. Cancer Res. 16, 894–908 10.1158/1541-7786.MCR-17-0623 [DOI] [PubMed] [Google Scholar]
- 30. Wright K. D., Miller B. S., El-Meanawy S., Tsaih S. W., Banerjee A., Geurts A. M., Sheinin Y., Sun Y., Kalyanaraman B., Rui H., Flister M. J., and Sorokin A. (2019) The p52 isoform of SHC1 is a key driver of breast cancer initiation. Breast Cancer Res. 21, 74 10.1186/s13058-019-1155-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Northey J. J., Chmielecki J., Ngan E., Russo C., Annis M. G., Muller W. J., and Siegel P. M. (2008) Signaling through ShcA is required for transforming growth factor β- and Neu/ErbB-2-induced breast cancer cell motility and invasion. Mol. Cell Biol. 28, 3162–3176 10.1128/MCB.01734-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Northey J. J., Dong Z., Ngan E., Kaplan A., Hardy W. R., Pawson T., and Siegel P. M. (2013) Distinct phosphotyrosine-dependent functions of the ShcA adaptor protein are required for transforming growth factor beta (TGFβ)-induced breast cancer cell migration, invasion, and metastasis. J. Biol. Chem. 288, 5210–5222 10.1074/jbc.M112.424804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ursini-Siegel J., and Muller W. J. (2008) The ShcA adaptor protein is a critical regulator of breast cancer progression. Cell Cycle 7, 1936–1943 10.4161/cc.7.13.6205 [DOI] [PubMed] [Google Scholar]
- 34. Hudson J., Ha J. R., Sabourin V., Ahn R., La Selva R., Livingstone J., Podmore L., Knight J., Forrest L., Beauchemin N., Hallett M., Park M., and Ursini-Siegel J. (2014) p66ShcA promotes breast cancer plasticity by inducing an epithelial-to-mesenchymal transition. Mol. Cell Biol. 34, 3689–3701 10.1128/MCB.00341-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lewis K., Kiepas A., Hudson J., Senecal J., Ha J. R., Voorand E., Annis M. G., Sabourin V., Ahn R., La Selva R., Tabariès S., Hsu B. E., Siegel M. J., Dankner M., Canedo E. C., et al. (2020) p66ShcA functions as a contextual promoter of breast cancer metastasis. Breast Cancer Res. 22, 7 10.1186/s13058-020-1245-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ngan E., Northey J. J., Brown C. M., Ursini-Siegel J., and Siegel P. M. (2013) A complex containing LPP and α-actinin mediates TGFβ-induced migration and invasion of ErbB2-expressing breast cancer cells. J. Cell Sci. 126, 1981–1991 10.1242/jcs.118315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ngan E., Stoletov K., Smith H. W., Common J., Muller W. J., Lewis J. D., and Siegel P. M. (2017) LPP is a Src substrate required for invadopodia formation and efficient breast cancer lung metastasis. Nat. Commun. 8, 15059 10.1038/ncomms15059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Grunewald T. G., Pasedag S. M., and Butt E. (2009) Cell adhesion and transcriptional activity: defining the role of the novel protooncogene LPP. Transl. Oncol. 2, 107–116 10.1593/tlo.09112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ngan E., Kiepas A., Brown C. M., and Siegel P. M. (2018) Emerging roles for LPP in metastatic cancer progression. J. Cell Commun. Signal. 12, 143–156 10.1007/s12079-017-0415-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Muraoka R. S., Koh Y., Roebuck L. R., Sanders M. E., Brantley-Sieders D., Gorska A. E., Moses H. L., and Arteaga C. L. (2003) Increased malignancy of Neu-induced mammary tumors overexpressing active transforming growth factor β1. Mol. Cell Biol. 23, 8691–8703 10.1128/MCB.23.23.8691-8703.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lauffenburger D. A., and Horwitz A. F. (1996) Cell migration: a physically integrated molecular process. Cell 84, 359–369 10.1016/S0092-8674(00)81280-5 [DOI] [PubMed] [Google Scholar]
- 42. Friedl P., and Wolf K. (2003) Tumour-cell invasion and migration: diversity and escape mechanisms. Nat. Rev. Cancer 3, 362–374 10.1038/nrc1075 [DOI] [PubMed] [Google Scholar]
- 43. Friedl P., Sahai E., Weiss S., and Yamada K. M. (2012) New dimensions in cell migration. Nat. Rev. Mol. Cell Biol. 13, 743–747 10.1038/nrm3459 [DOI] [PubMed] [Google Scholar]
- 44. Glenney J. R. Jr., and Zokas L. (1989) Novel tyrosine kinase substrates from Rous sarcoma virus-transformed cells are present in the membrane skeleton. J. Cell Biol. 108, 2401–2408 10.1083/jcb.108.6.2401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Turner C. E., Glenney J. R. Jr., and Burridge K. (1990) Paxillin: a new vinculin-binding protein present in focal adhesions. J. Cell Biol. 111, 1059–1068 10.1083/jcb.111.3.1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dankort D., Maslikowski B., Warner N., Kanno N., Kim H., Wang Z., Moran M. F., Oshima R. G., Cardiff R. D., and Muller W. J. (2001) Grb2 and Shc adapter proteins play distinct roles in Neu (ErbB-2)-induced mammary tumorigenesis: implications for human breast cancer. Mol. Cell Biol. 21, 1540–1551 10.1128/MCB.21.5.1540-1551.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zaidel-Bar R., Milo R., Kam Z., and Geiger B. (2007) A paxillin tyrosine phosphorylation switch regulates the assembly and form of cell-matrix adhesions. J. Cell Sci. 120, 137–148 10.1242/jcs.03314 [DOI] [PubMed] [Google Scholar]
- 48. Choi H., Larsen B., Lin Z. Y., Breitkreutz A., Mellacheruvu D., Fermin D., Qin Z. S., Tyers M., Gingras A. C., and Nesvizhskii A. I. (2011) SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat. Methods 8, 70–73 10.1038/nmeth.1541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Teo G., Liu G., Zhang J., Nesvizhskii A. I., Gingras A. C., and Choi H. (2014) SAINTexpress: improvements and additional features in Significance Analysis of INTeractome software. J. Proteomics 100, 37–43 10.1016/j.jprot.2013.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Petit M. M., Fradelizi J., Golsteyn R. M., Ayoubi T. A., Menichi B., Louvard D., Van de Ven W. J., and Friederich E. (2000) LPP, an actin cytoskeleton protein related to zyxin, harbors a nuclear export signal and transcriptional activation capacity. Mol. Biol. Cell 11, 117–129 10.1091/mbc.11.1.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dumbauld D. W., Shin H., Gallant N. D., Michael K. E., Radhakrishna H., and García A. J. (2010) Contractility modulates cell adhesion strengthening through focal adhesion kinase and assembly of vinculin-containing focal adhesions. J. Cell Physiol. 223, 746–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Plotnikov S. V., Pasapera A. M., Sabass B., and Waterman C. M. (2012) Force fluctuations within focal adhesions mediate ECM-rigidity sensing to guide directed cell migration. Cell 151, 1513–1527 10.1016/j.cell.2012.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gorenne I., Jin L., Yoshida T., Sanders J. M., Sarembock I. J., Owens G. K., Somlyo A. P., and Somlyo A. V. (2006) LPP expression during in vitro smooth muscle differentiation and stent-induced vascular injury. Circ. Res. 98, 378–385 10.1161/01.RES.0000202802.34727.fd [DOI] [PubMed] [Google Scholar]
- 54. Jin L., Kern M. J., Otey C. A., Wamhoff B. R., and Somlyo A. V. (2007) Angiotensin II, focal adhesion kinase, and PRX1 enhance smooth muscle expression of lipoma preferred partner and its newly identified binding partner palladin to promote cell migration. Circ. Res. 100, 817–825 10.1161/01.RES.0000261351.54147.de [DOI] [PubMed] [Google Scholar]
- 55. Shcherbakova D. M., Baloban M., Emelyanov A. V., Brenowitz M., Guo P., and Verkhusha V. V. (2016) Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging. Nat. Commun. 7, 12405 10.1038/ncomms12405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Humphries J. D., Byron A., and Humphries M. J. (2006) Integrin ligands at a glance. J. Cell Sci. 119, 3901–3903 10.1242/jcs.03098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Pechkovsky D. V., Scaffidi A. K., Hackett T. L., Ballard J., Shaheen F., Thompson P. J., Thannickal V. J., and Knight D. A. (2008) Transforming growth factor β1 induces αvβ3 integrin expression in human lung fibroblasts via a β3 integrin-, c-Src-, and p38 MAPK-dependent pathway. J. Biol. Chem. 283, 12898–12908 10.1074/jbc.M708226200 [DOI] [PubMed] [Google Scholar]
- 58. Mamuya F. A., and Duncan M. K. (2012) aV integrins and TGF-β-induced EMT: a circle of regulation. J. Cell Mol. Med. 16, 445–455 10.1111/j.1582-4934.2011.01419.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Heino J., and Massagué J. (1989) Transforming growth factor-β switches the pattern of integrins expressed in MG-63 human osteosarcoma cells and causes a selective loss of cell adhesion to laminin. J. Biol. Chem. 264, 21806–21811 [PubMed] [Google Scholar]
- 60. Zhang L., Zhou F., and ten Dijke P. (2013) Signaling interplay between transforming growth factor-β receptor and PI3K/AKT pathways in cancer. Trends Biochem. Sci. 38, 612–620 10.1016/j.tibs.2013.10.001 [DOI] [PubMed] [Google Scholar]
- 61. Chapnick D. A., Warner L., Bernet J., Rao T., and Liu X. (2011) Partners in crime: the TGFβ and MAPK pathways in cancer progression. Cell Biosci. 1, 42 10.1186/2045-3701-1-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zhang Y. E. (2009) Non-Smad pathways in TGF-β signaling. Cell Res. 19, 128–139 10.1038/cr.2008.328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lee M. K., Pardoux C., Hall M. C., Lee P. S., Warburton D., Qing J., Smith S. M., and Derynck R. (2007) TGF-β activates Erk MAP kinase signalling through direct phosphorylation of ShcA. EMBO J. 26, 3957–3967 10.1038/sj.emboj.7601818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lamouille S., and Derynck R. (2007) Cell size and invasion in TGF-β-induced epithelial to mesenchymal transition is regulated by activation of the mTOR pathway. J. Cell Biol. 178, 437–451 10.1083/jcb.200611146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Yi J. Y., Shin I., and Arteaga C. L. (2005) Type I transforming growth factor β receptor binds to and activates phosphatidylinositol 3-kinase. J. Biol. Chem. 280, 10870–10876 10.1074/jbc.M413223200 [DOI] [PubMed] [Google Scholar]
- 66. Huang C., Jacobson K., and Schaller M. D. (2004) MAP kinases and cell migration. J. Cell Sci. 117, 4619–4628 10.1242/jcs.01481 [DOI] [PubMed] [Google Scholar]
- 67. Huang C., Rajfur Z., Borchers C., Schaller M. D., and Jacobson K. (2003) JNK phosphorylates paxillin and regulates cell migration. Nature 424, 219–223 10.1038/nature01745 [DOI] [PubMed] [Google Scholar]
- 68. Chin Y. R., and Toker A. (2009) Function of Akt/PKB signaling to cell motility, invasion and the tumor stroma in cancer. Cell. Signal. 21, 470–476 10.1016/j.cellsig.2008.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Grille S. J., Bellacosa A., Upson J., Klein-Szanto A. J., van Roy F., Lee-Kwon W., Donowitz M., Tsichlis P. N., and Larue L. (2003) The protein kinase Akt induces epithelial mesenchymal transition and promotes enhanced motility and invasiveness of squamous cell carcinoma lines. Cancer Res. 63, 2172–2178 [PubMed] [Google Scholar]
- 70. Enomoto A., Murakami H., Asai N., Morone N., Watanabe T., Kawai K., Murakumo Y., Usukura J., Kaibuchi K., and Takahashi M. (2005) Akt/PKB regulates actin organization and cell motility via Girdin/APE. Dev. Cell 9, 389–402 10.1016/j.devcel.2005.08.001 [DOI] [PubMed] [Google Scholar]
- 71. Mendoza M. C., Er E. E., Zhang W., Ballif B. A., Elliott H. L., Danuser G., and Blenis J. (2011) ERK-MAPK drives lamellipodia protrusion by activating the WAVE2 regulatory complex. Mol. Cell 41, 661–671 10.1016/j.molcel.2011.02.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Alexander N. R., Branch K. M., Parekh A., Clark E. S., Iwueke I. C., Guelcher S. A., and Weaver A. M. (2008) Extracellular matrix rigidity promotes invadopodia activity. Curr. Biol. 18, 1295–1299 10.1016/j.cub.2008.07.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Savagner P. (2001) Leaving the neighborhood: molecular mechanisms involved during epithelial-mesenchymal transition. Bioessays 23, 912–923 10.1002/bies.1132 [DOI] [PubMed] [Google Scholar]
- 74. Condeelis J., and Segall J. E. (2003) Intravital imaging of cell movement in tumours. Nat. Rev. Cancer 3, 921–930 10.1038/nrc1231 [DOI] [PubMed] [Google Scholar]
- 75. Nayal A., Webb D. J., Brown C. M., Schaefer E. M., Vicente-Manzanares M., and Horwitz A. R. (2006) Paxillin phosphorylation at Ser273 localizes a GIT1-PIX-PAK complex and regulates adhesion and protrusion dynamics. J. Cell Biol. 173, 587–589 10.1083/jcb.200509075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Berginski M. E., Vitriol E. A., Hahn K. M., and Gomez S. M. (2011) High-resolution quantification of focal adhesion spatiotemporal dynamics in living cells. PLoS ONE 6, e22025 10.1371/journal.pone.0022025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Huttenlocher A., and Horwitz A. R. (2011) Integrins in cell migration. Cold Spring Harb. Perspect. Biol. 3, a005074 10.1101/cshperspect.a005074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Heino J., Ignotz R. A., Hemler M. E., Crouse C., and Massagué J. (1989) Regulation of cell adhesion receptors by transforming growth factor-β. Concomitant regulation of integrins that share a common β1 subunit. J. Biol. Chem. 264, 380–388 [PubMed] [Google Scholar]
- 79. Honda E., Yoshida K., and Munakata H. (2010) Transforming growth factor-β upregulates the expression of integrin and related proteins in MRC-5 human myofibroblasts. Tohoku J. Exp. Med. 220, 319–327 10.1620/tjem.220.319 [DOI] [PubMed] [Google Scholar]
- 80. Yeh Y. C., Wei W. C., Wang Y. K., Lin S. C., Sung J. M., and Tang M. J. (2010) Transforming growth factor-β1 induces Smad3-dependent β1 integrin gene expression in epithelial-to-mesenchymal transition during chronic tubulointerstitial fibrosis. Am. J. Pathol. 177, 1743–1754 10.2353/ajpath.2010.091183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Plow E. F., Haas T. A., Zhang L., Loftus J., and Smith J. W. (2000) Ligand binding to integrins. J. Biol. Chem. 275, 21785–21788 10.1074/jbc.R000003200 [DOI] [PubMed] [Google Scholar]
- 82. Wary K. K., Mainiero F., Isakoff S. J., Marcantonio E. E., and Giancotti F. G. (1996) The adaptor protein Shc couples a class of integrins to the control of cell cycle progression. Cell 87, 733–743 10.1016/S0092-8674(00)81392-6 [DOI] [PubMed] [Google Scholar]
- 83. Deshmukh L., Gorbatyuk V., and Vinogradova O. (2010) Integrin β3 phosphorylation dictates its complex with the Shc phosphotyrosine-binding (PTB) domain. J. Biol. Chem. 285, 34875–34884 10.1074/jbc.M110.159087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Dans M., Gagnoux-Palacios L., Blaikie P., Klein S., Mariotti A., and Giancotti F. G. (2001) Tyrosine phosphorylation of the β4 integrin cytoplasmic domain mediates Shc signaling to extracellular signal-regulated kinase and antagonizes formation of hemidesmosomes. J. Biol. Chem. 276, 1494–1502 10.1074/jbc.M008663200 [DOI] [PubMed] [Google Scholar]
- 85. Rozakis-Adcock M., McGlade J., Mbamalu G., Pelicci G., Daly R., Li W., Batzer A., Thomas S., Brugge J., Pelicci P. G., Schlessinger J., and Pawson T. (1992) Association of the Shc and Grb2/Sem5 SH2-containing proteins is implicated in activation of the Ras pathway by tyrosine kinases. Nature 360, 689–692 10.1038/360689a0 [DOI] [PubMed] [Google Scholar]
- 86. Habib T., Herrera R., and Decker S. J. (1994) Activators of protein kinase C stimulate association of Shc and the PEST tyrosine phosphatase. J. Biol. Chem. 269, 25243–25246 [PubMed] [Google Scholar]
- 87. Schmandt R., Liu S. K., and McGlade C. J. (1999) Cloning and characterization of mPAL, a novel Shc SH2 domain-binding protein expressed in proliferating cells. Oncogene 18, 1867–1879 10.1038/sj.onc.1202507 [DOI] [PubMed] [Google Scholar]
- 88. Waalkes S., Atschekzei F., Kramer M. W., Hennenlotter J., Vetter G., Becker J. U., Stenzl A., Merseburger A. S., Schrader A. J., Kuczyk M. A., and Serth J. (2010) Fibronectin 1 mRNA expression correlates with advanced disease in renal cancer. BMC Cancer 10, 503 10.1186/1471-2407-10-503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Jia D., Yan M., Wang X., Hao X., Liang L., Liu L., Kong H., He X., Li J., and Yao M. (2010) Development of a highly metastatic model that reveals a crucial role of fibronectin in lung cancer cell migration and invasion. BMC Cancer 10, 364 10.1186/1471-2407-10-364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Ioachim E., Charchanti A., Briasoulis E., Karavasilis V., Tsanou H., Arvanitis D. L., Agnantis N. J., and Pavlidis N. (2002) Immunohistochemical expression of extracellular matrix components tenascin, fibronectin, collagen type IV and laminin in breast cancer: their prognostic value and role in tumour invasion and progression. Eur. J. Cancer 38, 2362–2370 10.1016/S0959-8049(02)00210-1 [DOI] [PubMed] [Google Scholar]
- 91. Mierke C. T., Frey B., Fellner M., Herrmann M., and Fabry B. (2011) Integrin α5β1 facilitates cancer cell invasion through enhanced contractile forces. J. Cell Sci. 124, 369–383 10.1242/jcs.071985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Collins L. R., Ricketts W. A., Yeh L., and Cheresh D. (1999) Bifurcation of cell migratory and proliferative signaling by the adaptor protein Shc. J. Cell Biol. 147, 1561–1568 10.1083/jcb.147.7.1561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Mainiero F., Murgia C., Wary K. K., Curatola A. M., Pepe A., Blumemberg M., Westwick J. K., Der C. J., and Giancotti F. G. (1997) The coupling of α6β4 integrin to Ras-MAP kinase pathways mediated by Shc controls keratinocyte proliferation. EMBO J. 16, 2365–2375 10.1093/emboj/16.9.2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Shaw L. M., Rabinovitz I., Wang H. H., Toker A., and Mercurio A. M. (1997) Activation of phosphoinositide 3-OH kinase by the α6β4 integrin promotes carcinoma invasion. Cell 91, 949–960 10.1016/S0092-8674(00)80486-9 [DOI] [PubMed] [Google Scholar]
- 95. Zaidel-Bar R., Ballestrem C., Kam Z., and Geiger B. (2003) Early molecular events in the assembly of matrix adhesions at the leading edge of migrating cells. J. Cell Sci. 116, 4605–4613 10.1242/jcs.00792 [DOI] [PubMed] [Google Scholar]
- 96. Laukaitis C. M., Webb D. J., Donais K., and Horwitz A. F. (2001) Differential dynamics of α5 integrin, paxillin, and α-actinin during formation and disassembly of adhesions in migrating cells. J. Cell Biol. 153, 1427–1440 10.1083/jcb.153.7.1427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Horton E. R., Byron A., Askari J. A., Ng D. H. J., Millon-Frémillon A., Robertson J., Koper E. J., Paul N. R., Warwood S., Knight D., Humphries J. D., and Humphries M. J. (2015) Definition of a consensus integrin adhesome and its dynamics during adhesion complex assembly and disassembly. Nat. Cell Biol. 17, 1577–1587 10.1038/ncb3257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Kuo J. C., Han X., Hsiao C. T., Yates J. R. 3rd, Waterman C. M. (2011) Analysis of the myosin-II-responsive focal adhesion proteome reveals a role for β-Pix in negative regulation of focal adhesion maturation. Nat. Cell Biol. 13, 383–393 10.1038/ncb2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Wu R. F., Liao C., Fu G., Hayenga H. N., Yang K., Ma Z., Liu Z., and Terada L. S. (2016) p66(Shc) Couples mechanical signals to RhoA through focal adhesion kinase-dependent recruitment of p115-RhoGEF and GEF-H1. Mol. Cell Biol. 36, 2824–2837 10.1128/MCB.00194-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Robertson J., Jacquemet G., Byron A., Jones M. C., Warwood S., Selley J. N., Knight D., Humphries J. D., and Humphries M. J. (2015) Defining the phospho-adhesome through the phosphoproteomic analysis of integrin signalling. Nat. Commun. 6, 6265 10.1038/ncomms7265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Lawson C., Lim S. T., Uryu S., Chen X. L., Calderwood D. A., and Schlaepfer D. D. (2012) FAK promotes recruitment of talin to nascent adhesions to control cell motility. J. Cell Biol. 196, 223–232 10.1083/jcb.201108078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Lawson C., and Schlaepfer D. D. (2012) Integrin adhesions: who's on first? What's on second? Connections between FAK and talin. Cell Adh. Migr. 6, 302–306 10.4161/cam.20488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Hu Y. L., Lu S., Szeto K. W., Sun J., Wang Y., Lasheras J. C., and Chien S. (2014) FAK and paxillin dynamics at focal adhesions in the protrusions of migrating cells. Sci. Rep. 4, 6024 10.1038/srep06024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Xia H., Nho R. S., Kahm J., Kleidon J., and Henke C. A. (2004) Focal adhesion kinase is upstream of phosphatidylinositol 3-kinase/Akt in regulating fibroblast survival in response to contraction of type I collagen matrices via a β1 integrin viability signaling pathway. J. Biol. Chem. 279, 33024–33034 10.1074/jbc.M313265200 [DOI] [PubMed] [Google Scholar]
- 105. Kim N. G., and Gumbiner B. M. (2015) Adhesion to fibronectin regulates Hippo signaling via the FAK-Src-PI3K pathway. J. Cell Biol. 210, 503–515 10.1083/jcb.201501025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Schober M., Raghavan S., Nikolova M., Polak L., Pasolli H. A., Beggs H. E., Reichardt L. F., and Fuchs E. (2007) Focal adhesion kinase modulates tension signaling to control actin and focal adhesion dynamics. J. Cell Biol. 176, 667–680 10.1083/jcb.200608010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Parekh A., and Weaver A. M. (2016) Regulation of invadopodia by mechanical signaling. Exp. Cell Res. 343, 89–95 10.1016/j.yexcr.2015.10.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Schoumacher M., Goldman R. D., Louvard D., and Vignjevic D. M. (2010) Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J. Cell Biol. 189, 541–556 10.1083/jcb.200909113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Eddy R. J., Weidmann M. D., Sharma V. P., and Condeelis J. S. (2017) Tumor cell invadopodia: invasive protrusions that orchestrate metastasis. Trends Cell Biol. 27, 595–607 10.1016/j.tcb.2017.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Tomar A., Lawson C., Ghassemian M., and Schlaepfer D. D. (2012) Cortactin as a target for FAK in the regulation of focal adhesion dynamics. PLoS ONE 7, e44041 10.1371/journal.pone.0044041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Artym V. V., Zhang Y., Seillier-Moiseiwitsch F., Yamada K. M., and Mueller S. C. (2006) Dynamic interactions of cortactin and membrane type 1 matrix metalloproteinase at invadopodia: defining the stages of invadopodia formation and function. Cancer Res. 66, 3034–3043 10.1158/0008-5472.CAN-05-2177 [DOI] [PubMed] [Google Scholar]
- 112. Weaver A. M. (2006) Invadopodia: specialized cell structures for cancer invasion. Clin. Exp. Metastasis 23, 97–105 10.1007/s10585-006-9014-1 [DOI] [PubMed] [Google Scholar]
- 113. Buccione R., Caldieri G., and Ayala I. (2009) Invadopodia: specialized tumor cell structures for the focal degradation of the extracellular matrix. Cancer Metastasis Rev. 28, 137–149 10.1007/s10555-008-9176-1 [DOI] [PubMed] [Google Scholar]
- 114. Branch K. M., Hoshino D., and Weaver A. M. (2012) Adhesion rings surround invadopodia and promote maturation. Biol. Open 1, 711–722 10.1242/bio.20121867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Kiepas A., Voorand E., Mubaid F., Siegel P. M., and Brown C. M. (2020) Optimizing live-cell fluorescence imaging conditions to minimize phototoxicity. J. Cell Sci. 133, jcs242834 10.1242/jcs.242834 [DOI] [PubMed] [Google Scholar]
- 116. Choi H., Liu G., Mellacheruvu D., Tyers M., Gingras A. C., and Nesvizhskii A. I. (2012) Analyzing protein-protein interactions from affinity purification-mass spectrometry data with SAINT. Curr. Protoc. Bioinformatics, Chapter 8, Unit 8.15 10.1002/0471250953.bi0815s39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Keller A., Nesvizhskii A. I., Kolker E., and Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 10.1021/ac025747h [DOI] [PubMed] [Google Scholar]
- 118. Nesvizhskii A. I., Keller A., Kolker E., and Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- 119. Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., Pérez E., Uszkoreit J., Pfeuffer J., Sachsenberg T., Yilmaz S., et al. (2019) The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47, D442–D450 10.1093/nar/gky1106 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (119) partner repository with the data set identifier PXD018265 and 10.6019/PXD018265.