Abstract
Plant arabinogalactan proteins (AGPs) are a diverse group of cell surface– and wall–associated glycoproteins. Functionally important AGP glycans are synthesized in the Golgi apparatus, but the relationships among their glycosylation levels, processing, and functionalities are poorly understood. Here, we report the identification and functional characterization of two Golgi-localized exo-β-1,3-galactosidases from the glycosyl hydrolase 43 (GH43) family in Arabidopsis thaliana. GH43 loss-of-function mutants exhibited root cell expansion defects in sugar-containing growth media. This root phenotype was associated with an increase in the extent of AGP cell wall association, as demonstrated by Yariv phenylglycoside dye quantification and comprehensive microarray polymer profiling of sequentially extracted cell walls. Characterization of recombinant GH43 variants revealed that the exo-β-1,3-galactosidase activity of GH43 enzymes is hindered by β-1,6 branches on β-1,3-galactans. In line with this steric hindrance, the recombinant GH43 variants did not release galactose from cell wall–extracted glycoproteins or AGP-rich gum arabic. These results indicate that the lack of exo-β-1,3-galactosidase activity alters cell wall extensibility in roots, a phenotype that could be explained by the involvement of galactosidases in AGP glycan biosynthesis.
Keywords: glycoside hydrolase 43; Golgi; glycosylation; arabinogalactan proteins; Arabidopsis; roots; cell expansion; plant cell wall; exo-β13-galactosidase; comprehensive microarray polymer profiling (CoMPP) microarray; cell wall; Arabidopsis thaliana; glycoside hydrolase; glycosidase; glycoprotein biosynthesis; arabinogalactan protein; exo-β1,3-galactosidase
Plant cell growth is dictated by turgor-driven extension of the primary cell wall. Oriented cellulose biosynthesis and localized wall deposition create dynamic cell wall mechanics that enable turgor-driven anisotropic cell growth (1). Studies on the molecular interactions in the primary cell wall matrix have led to models in which covalent and noncovalent interactions between matrix components facilitate anisotropic cell expansion (2). The primary cell wall polysaccharide matrix contains cellulose, hemicellulose, and pectin as well as enzymes and structural proteins (3–6). The cell wall-associated glycoproteins include arabinogalactan proteins (AGPs), which are proposed to have diverse roles during cell expansion, including as structural components, modifiers of the extracellular matrix, and ligands for cell surface receptors (7, 8).
Glycans account for 90–98% of the molecular weight of AGPs, and are thought to be critical for their functionality (9). Characterization of the enzymes responsible for AGP glycosylation can elucidate AGP functions and functional redundancies (7, 10). AGP glycosylation is initiated by proline hydroxylation in the endoplasmic reticulum and is then advanced in the Golgi apparatus by specialized glycosyl transferases (GTs) (6, 11). The glycosyl transferase family 31 (GT31) is responsible for the synthesis of the hydroxyproline galactose and the β-1,3-galactan backbone of AGPs (12–15). Defects in β-1,3-galactan biosynthesis are associated with various developmental phenotypes including defects in cell expansion (13–15). The side chains, which branch off from the β-1,3-galactan backbone via β-1,6 linkages, are synthesized by GALT29A and/or GT31 family enzymes (16, 17). Further side chain residues are attached by other GTs: GT77 enzymes add α-1,3/1,5-arabinose units (18), GT14 attaches β-1,6-glucuronic acid units (17, 19), and GT37 adds α-1,2-fucose units (20, 21). These GTs acting on AGPs are expected to generate a glycan structure consisting of a β-1,3-galactan backbone with β-1,6-galactan branches containing diverse other sugars.
Despite the advances in identifying GTs involved in AGP glycosylation, the in vivo structural diversity of mature arabinogalactans is huge and largely uncharacterised. The glycan structure is also likely to vary between AGPs, tissues, and even cell types, as indicated by differences in extractability, expression during specific growth stages, and the expression of the glycosylation machinery (7, 22). NMR analysis of the glycans attached to the synthetic AGP motif repeats expressed in tobacco Bright Yellow-2 cells revealed relatively short galactan backbone with a mixture of β-1,6 and β-1,3 links (23, 24). The side chains were also short and contained a mixture of galactose, arabinose, glucuronic acid, and rhamnose. How well these structures reflect native structures is unclear. Larger glycans were found in radish roots, wheat flowers, and Arabidopsis leaves based on mass spectrometric analysis of enzymatically released AGP glycans (25–27). The evidence from these structural glycan studies and characterizations of GTs active on AGPs suggest a β-1,3-galactan backbone of varying length with β-1,6 side chains containing mainly galactan and arabinose residues with some additional sugar residues also found in pectin.
We hypothesize that hydrolytic enzymes acting on AGP glycans may also be involved in the synthesis of the arabinogalactan chains or their modification in the cell wall matrix. There is precedent for apoplastic post-deposition modification of xyloglucan by hydrolases (28, 29), but secretory pathway modification as known from N-glycan biosynthesis should also be considered. Based on the description of hydrolase enzymes in the carbohydrate-active enzyme database (CAZy), glycoside hydrolase family 43 (GH43) was identified as a potential group with AGP glycan hydrolyzing activity (16, 30, 31). The GH43 family is conserved across prokaryotes and eukaryotes; it currently contains 15,620 GH43 enzymes, making it one of the largest known hydrolase families. The family is divided into α-l-arabinofuranosidase, β-d-xylosidase, α-l-arabinanase, and β-d-galactosidase groups (30). Several GH43 enzymes from prokaryotes and eukaryotes have recently been characterized because of their potential in biomass degradation and other biotechnological applications (32, 33). Based on their diverse identified enzymatic activities and amino acid motifs, the GH43s have been further subdivided into 37 subclades (30). However, no GH43 family enzyme from plants has yet been characterized. The Arabidopsis thaliana genome contains two genes encoding amino acid sequences with similarity to GH43 enzymes belonging to GH43 subfamily 24, named GH43A and GH43B (30). Here, we characterize these GH43 enzymes and describe their role in cell expansion and root growth in A. thaliana.
Results
GH43 null mutants are defective in root cell expansion
The A. thaliana genome encodes two putative glycosyl hydrolase 43 family enzymes according to the latest version of the Arabidopsis Information Resource database (TAIR; RRID:SCR_004618). Based on publicly available expression data, both genes are expressed in the cell elongation zone above the root meristem. To investigate the functional role of these GH43 enzymes, we first obtained mutants carrying exon T-DNA insertions in GH43A and GH43B (Fig. 1A). Initial analysis of the single gh43 mutants and the double gh43a-1/gh43b-1 mutant (gh43null henceforth) revealed no obvious visual phenotypes in seedlings (Fig. 1B).
Figure 1.
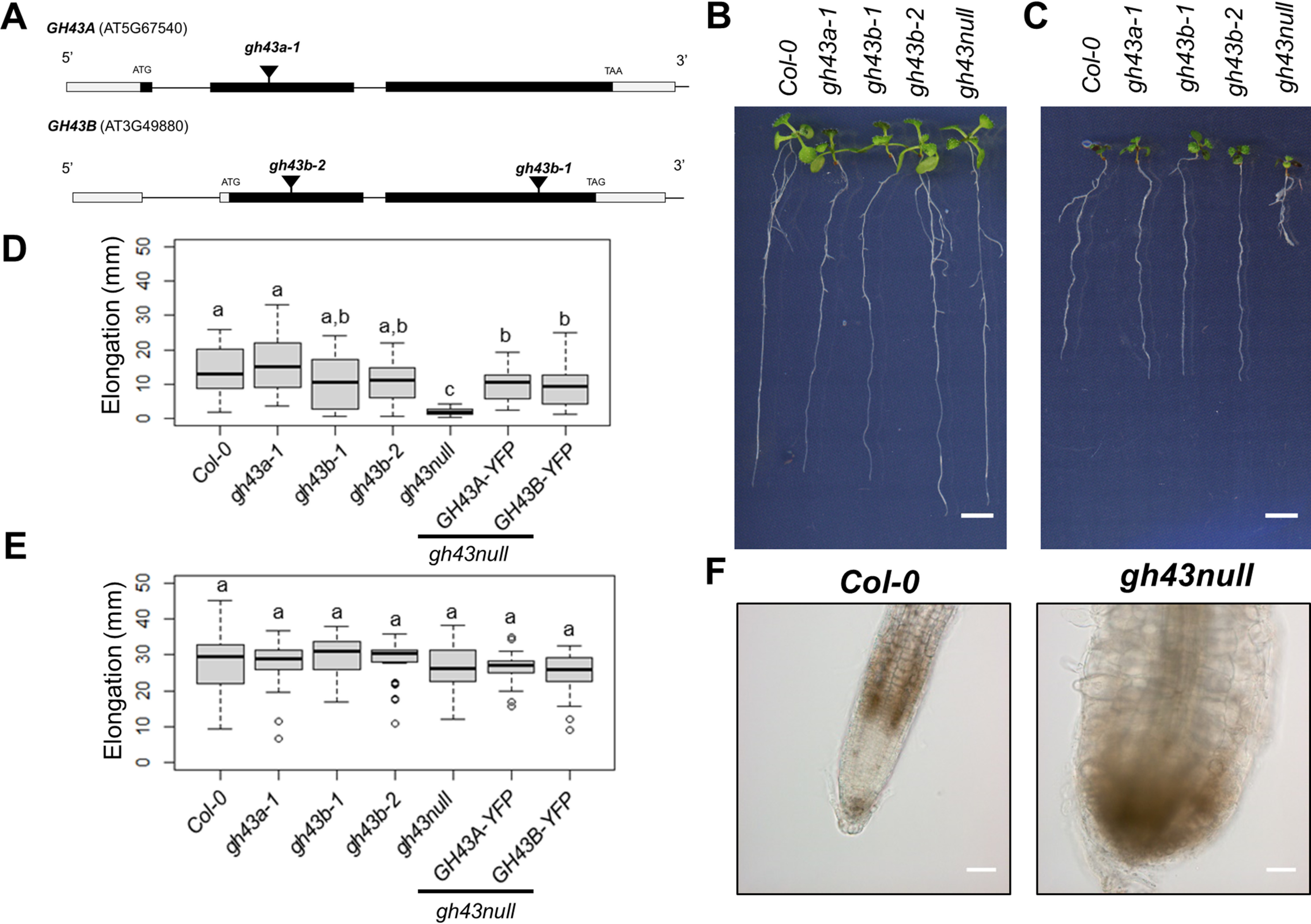
Characterization of the gh43 Arabidopsis T-DNA insertion lines. A, schematic diagram of the GH43A and GH43B gene structure and the T-DNA insertion sites. The white and black boxes indicate untranslated and translated regions, respectively. B, Col-0 and gh43 seedlings grown on nutrient media without sugar for 4 days and then moved to media without sugar for 6 days. Scale bar = 5 mm. C, Col-0 and gh43 seedlings grown on nutrient media without sugar for 4 days and then moved to media with 4.5% glucose for 6 days. Scale bar = 5 mm. D, root elongation of Col-0 and gh43 seedlings grown on nutrient media without sugar for 4 days and then 6 days on media with 4.5% glucose. The box plot's dark horizontal lines represent the median, the two gray boxes the 25th and 75th percentile, the whiskers the 1.5 interquartile limits, and the dots the outliers (n = 25–28 biological replicates). Means not sharing a common letter are significantly different at p < 0.05, as determined by Tukey's test after one-way ANOVA. E, root elongation of Col-0 and gh43 seedlings grown on nutrient media without sugar for 4 days and then 6 days on media without glucose. The box plot's dark horizontal lines represent the median, the two gray boxes the 25th and 75th percentile, the whiskers the 1.5 interquartile limits, and the dots the outliers (n = 25–28 biological replicates). Means not sharing a common letter are significantly different at p < 0.05, as determined by Tukey's test after one-way ANOVA. F, light microscopy images of Col-0 and gh43null root tip after seedlings were grown 4 days on nutrient media followed by 6 days on 4.5% glucose media. Scale bar = 100 μm.
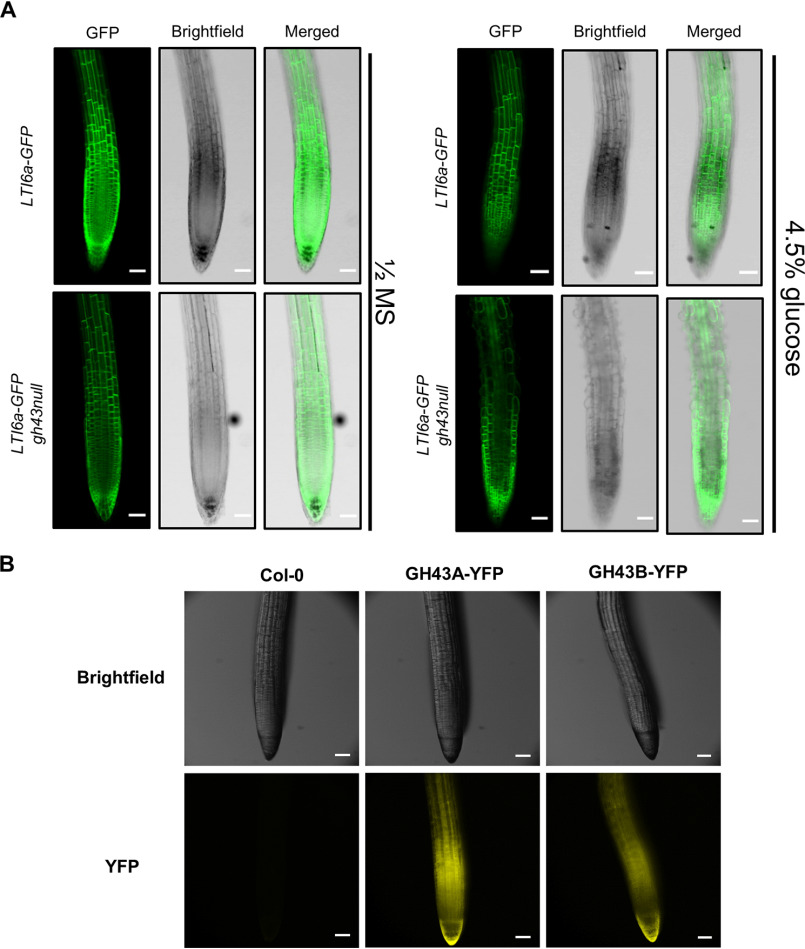
Several classic cell wall mutants in Arabidopsis show enhanced or conditional root growth defects on medium containing 4.5% exogenous sugar (34, 35). We therefore grew the gh43 mutants on nutrient media without sugar for 4 days and then moved them to media containing 4.5% glucose for 6 days. During the 6 days on glucose media, the gh43a, gh43b, and WT roots elongated at similar rates, but in gh43null root growth was severely inhibited (Fig. 1, C and D). No change in growth rate was observed in controls moved to media without sugar (Fig. 1E). On sugar-containing media, the gh43null root epidermal cells exhibited clear swelling and loss of anisotropic growth (Fig. 1F). A time course experiment comparing WT and gh43null lines expressing the plasma membrane marker LTi6a-GFP revealed that the swelling was detectable in the cell elongation zone already after 6 h and became obvious after 10 h (Fig. 2A and Fig. S1). This cell expansion defect is not observed on media containing 4.5% sorbitol or 100 mm NaCl, showing that the phenotype cannot be caused by an osmotic effect alone or salt stress (Fig. S2). To confirm the causal gene defect responsible for the sugar-inducible loss of anisotropic growth, we performed complementation experiments by introducing either GH43A-YFP or GH43B-YFP under the control of their native promoters into gh43null background. Both constructs rescued the root growth phenotype on sugar, albeit GH43B-YFP only partially (Fig. 1D). The GH43B-YFP construct contained a 1-kb promoter sequence, whereas in the GH43A-YFP the promoter length was 1.9 kb, possibly explaining the difference in complementation efficiency. The GH43-YFP constructs may not fully replicate the native GH43 expression, but in line with the root swelling phenotype a strong YFP signal was observed in the elongation zone of the GH43-YFP expressing roots (Fig. 2B). Based on these results we conclude that the GH43 proteins are functionally redundant in the elongation zone of young roots. We also observed a clear GH43-YFP signal in the root cap cells suggesting a function for GH43s also in these cells (Fig. 2B).
Figure 2.
The initiation of the sugar-induced root swelling in gh43null. A, confocal laser scanning microscope images of Col-0 and gh43null roots stably expressing the LTI6a-GFP plasma membrane marker. Images were obtained 10 h after moving 4-day–old seedlings to nutrient media without (left) and with 4.5% glucose (right). Scale bars = 100 μm. B, fluorescent stereomicroscope images of native promoter GH43-YFP signal in roots. Seedlings were grown on nutrient media without sugar for 4 days. Scale bars = 100 μm.
GH43A and GH43B are Golgi-localized exo-β1,3-galactosidases
The pGH43A:GH43A-YFP and pGH43B:GH43B-YFP lines were used to investigate the subcellular location of the GH43s. In the expanding root cells of 4-day–old seedlings, both GH43A-YFP and GH43B-YFP appeared to localize to the Golgi apparatus (Fig. 3A). To confirm the Golgi localization, the lines were crossed with lines carrying the cis-Golgi marker SYP32-mCherry or the trans-Golgi network marker SYP43-mCherry (Fig. 3A) (36, 37). Both GH43 proteins co-localized with the Golgi markers. However, the overlap with SYP32-mCherry was higher than with SYP43-mCherry, indicating an enrichment on the incoming transport vesicle side of the Golgi apparatus (Fig. 3, B and C). In further support of the Golgi localization, both Arabidopsis GH43s were identified in Golgi-enriched cell extracts in global proteomics experiments (38, 39).
Figure 3.

Co-localization of GH43A-YFP and GH43B-YFP with mCherry Golgi markers. A, confocal laser scanning microscope images of root epidermis cells in Arabidopsis seedlings stably expressing pGH43A::GH43A-YFP or pGH43B::GH43B-YFP and the cis-Golgi marker SYP32-mCherry or the trans-Golgi network marker SYP43-mCherry. Images were captured with a Zeiss LSM880 confocal microscope. Scale bars = 5 μm (1 μm for zoomed images). B, degree of co-localization of pGH43A::GH43A-YFP with SYP32-mCherry or SYP43-mCherry in the root elongation zone epidermis. Images were captured from three independent GH43A-YFP lines (L1–L3) and five seedlings per line. The values represent the degree of Pearson's correlation between the YFP and RFP channel analyzed with ImageJ. ***, p < 0.001 (unpaired t test, n = 5). C, degree of co-localization of pGH43B::GH43B-YFP with SYP32-mCherry or SYP43-mCherry in the root elongation zone epidermis. Images were captured from three independent GH43A-YFP lines (L1–L3) and five seedlings per line. The values represent the degree of Pearson's correlation between the YFP and RFP channel analyzed with ImageJ. ***, p < 0.001 (unpaired t test, n = 5).
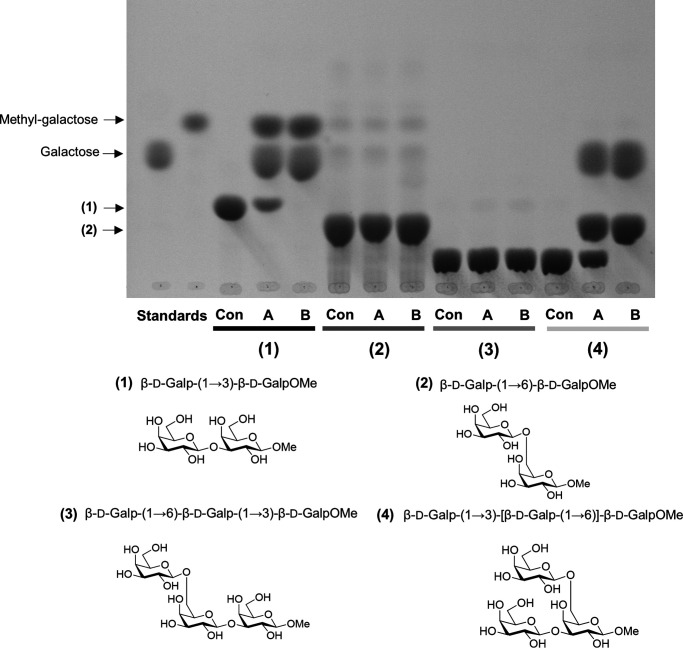
The predicted hydrolase domains of the two Arabidopsis GH43 proteins exhibit over 90% similarity at the amino acid sequence level, suggesting similar enzyme activity (Fig. S3). Sequence similarity also indicates that these proteins belong to the GH43 subfamily of enzymes with β-1,3-galactosidase activity (30, 31, 40–42). To test for this activity, we expressed GH43A and GH43B in Escherichia coli and purified the recombinant proteins using nickel ion affinity chromatography (Fig. S4A). The activity of the recombinant GH43s was assayed against various of β-1,3-galactan di- and trisaccharides (Fig. 4). Both GH43 proteins hydrolyzed β-d-Galp-(1→3)-β-d-GalpOMe, but not β-d-Galp-(1→6)-β-d-GalpOMe. To confirm that the observed β-1,3-galactosidase activity was because of the GH43 enzymes, we generated GH43 enzymes with mutations in the predicted active site. The mutation sites were selected based on sequence similarity with the active site of a Clostridium thermocellum GH43 protein whose structure was solved by crystallography (Fig. S4B) (42). Of the mutated versions, the GH43BE224Q had minor activity toward β-d-Galp-(1→3)-β-d-GalpOMe, whereas GH43BE224Q,E338Q and GH43B337-339DEL were inactive toward β-d-Galp-(1→3)-β-d-GalpOMe (Fig. S4C). These assays confirmed that the Arabidopsis GH43s are β-1,3-galactosidases and support the conservation of active sites between bacteria and plants.
Figure 4.
TLC analysis of sugars released by recombinant Arabidopsis GH43 activity. Hydrolytic activity of heterologous GH43A and GH43B against (1) methyl β-d-galactopyranosyl-(1→3)-β-d-galactopyranoside, (2) methyl β-d-galactopyranosyl-(1→6)-β-d-galactopyranoside, (3) methyl β-d-galactopyranosyl-(1→6)-β-d-galactopyranosyl-(1→3)-β-d-galactopyranoside, and (4) methyl β-d-galactopyranosyl-(1→3)-[β-d-galactopyranosyl-(1→6)]-β-d-galactopyranoside. The digestion products were separated with TLC. Con, substrate in buffer without enzyme; A,= GH43A; B, GH43B.
β-1,3-Galactan in plant cell walls is mainly found on AGPs (43). Therefore, to investigate the presence of possible GH43 substrates in Arabidopsis cell walls, we sequentially extracted cell wall glycoproteins from Arabidopsis leaves using 0.2 m CaCl2, 50 mm CDTA, 0.5 m NaCO3, and 4 m NaOH, and analyzed the extracts by TLC before and after treatment with the GH43 proteins (Fig. S5A). Interestingly, the recombinant GH43s did not release galactose from any of the cell wall fractions, suggesting that the β-1,3-linkages were inaccessible. The recombinant GH43s also failed to release galactose from gum arabic, which is rich in AGPs (Fig. S5B) (44). Even partial deglycosylation of the AGPs was insufficient to allow the GH43 proteins access to the β-1,3-linkages (Fig. S5B). These observations suggested that the β-1,3-linkages of mature AGPs may be protected against GH43 activity.
To investigate the enzymatic mechanism of the Arabidopsis GH43 proteins in more depth, we synthesized β-1,3-galactan oligosaccharides with a β-1,6-galactose branch on the reducing and nonreducing ends of β-d-Galp-(1→3)-β-d-GalpOMe. The GH43s were unable to hydrolyze β-d-Galp-(1→3)-β-d-GalpOMe when the nonreducing end bore a β-1,6-galactose unit, but did hydrolyze it when the substitution was at the reducing end (Fig. 4). Steric hindrance due to the presence of side chains at the nonreducing end of the β-1,3-galactan thus protects against hydrolysis catalyzed by GH43 proteins. We also tested the ability of GH43s to hydrolyze β-1,3- or β-1,4-glucans, but observed no activity toward these polymers (Fig. S5C). These localization and recombinant enzyme assays established the two Arabidopsis GH43s as Golgi exo-β-1,3-galactosidases with quite strict substrate specificity.
gh43null mutants have altered cell wall structure
WT and gh43 null seedlings had similar contents of cellulose, hemicellulosic, and pectic sugars (Fig. S6, Table S1). The gh43null root swelling thus cannot be explained by changes in the levels of the main cell wall polymers, although it is possible that more localized cell type-specific defects were masked by the analysis of total tissue extracts. Based on the GH43 proteins' Golgi localization, exo-β1,3-galactosidase activity and inactivity toward AGP-containing plant extracts, we hypothesised that they are involved in the synthesis of the arabinogalactans in the Golgi. It is plausible that their native targets occur only found in the Golgi and represent a small fraction of the total β-1,3-galactan pool. This hypothesis is consistent with the inactivity of the GH43 proteins toward β-1,3-galactans substituted with a β-1,6 galactan at the nonreducing end, and suggests that GH43s may be involved in the processing of the AGPs during biosynthesis.
We hypothesized that defects in arabinogalactan synthesis in the gh43null background would cause changes in the structure and/or extractability of AGPs. To investigate this possibility, we first quantified the levels of cell wall-bound AGPs (isolated by removing the CaCl2-soluble fraction followed by AIR1 treatment) and AGPs soluble in 0.18 m CaCl2 in WT and gh43null seedlings using β-Yariv. Yariv phenylglycosides selectively bind to the β-1,3-galactans of AGPs, enabling their spectrophotometric quantification (45). The fraction of cell wall-bound AGPs in gh43null was higher than in the WT (Fig. 5A), whereas no statistically significant difference was observed for the soluble fraction (Fig. 5B). We thus hypothesized that structural differences in the AGP glycans of the gh43null mutant may cause some AGPs to bind more tightly to the cell wall.
Figure 5.
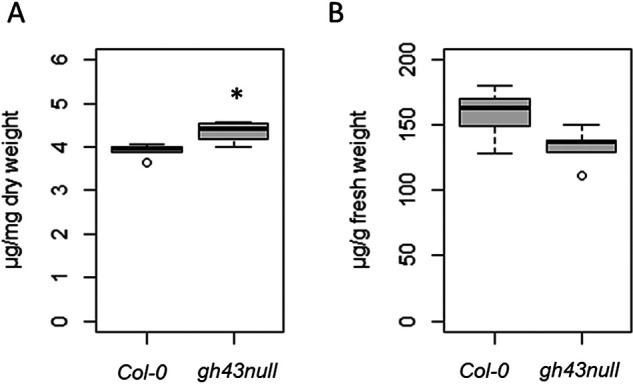
Spectrophotometric quantification of cell wall bound and soluble arabinogalactan proteins with β-Yariv. A, β-Yariv binding to WT (Col-0) and gh43null cell walls from 7-day–old seedlings after removal of CaCl2- and AIR1-soluble fractions. The amount of cell wall-bound β-Yariv was quantified against a gum arabic standard. The box plot's dark horizontal lines represent the median, the two gray boxes the 25th and 75th percentile, the whiskers the 1.5 interquartile limits, and the dots the outliers. *, p < 0.05 (unpaired t test, n = 5 biological replicate pools of seedlings). B, β-Yariv binding to the CaCl2-soluble fraction from WT (Col-0) and gh43null 7-day–old seedlings. Amount of bound β-Yariv was quantified against a gum arabic standard. The box plot's dark horizontal lines represent the median, the two gray boxes the 25th and 75th percentile, the whiskers the 1.5 interquartile limits, and the dots the outliers (n = 5 biological replicate pools of seedlings).
AGPs bound to cell walls may affect matrix properties and conceivably also cell expansion. To test whether this could explain the root swelling phenotype, we sequentially extracted cell wall fractions from 7-day–old WT and gh43null seedlings using 180 mm CaCl2, 50 mm CDTA, and 4 m NaOH, 26.5 mm NaBH4. An equal amount of each fraction was analyzed on a comprehensive microarray polymer profiling (CoMPP) microarray designed to determine different sugar epitopes in complex plant extracts (46). Similar polymer profiling has also previously been employed to demonstrate cell wall matrix separability features (47, 48). The CoMPP microarrays were probed with monoclonal antibodies specific for pectin, xylan, xyloglucan, mannan, crystalline cellulose, extension, and AGP epitopes (Table S2). For epitopes released with 180 mm CaCl2, the JIM13, LM20, and RU2 antibody signals of gh43null were significantly weaker than those of the WT (Table 1). Pectin antibodies LM20 and RU2 bind partially methyl esterified homogalacturonan and the rhamnogalacturonan-I (RG-I) backbone, respectively (49, 50), whereas JIM13 recognize an AGP epitope (51). For epitopes released with 50 mm CDTA, the LM5, LM15, and LM18 signals of the gh43null were significantly stronger than those of the WT (Table 1). Pectin epitope antibody LM5 binds to β1,4-galactan, LM18 to partially methyl esterified homogalacturonan, and LM15 to xyloglucan (49, 52, 53). The cell wall fraction exhibiting the most pronounced differences between gh43null and WT was extracted with 4 m NaOH, 26.5 mm NaBH4 released epitopes (Table 1). In accordance with the β-Yariv assay results (Fig. 5A), the signals from AGP antibodies JIM13, LM2, LM14, and MAC207 were all significantly more intense in gh43null (Table 1) (51, 54, 55). An increased signal intensity in gh43null was also observed for the pectin (galactan) antibody LM5, the xyloglucan-binding LM25, and the CBM3a crystalline cellulose and JIM20 extensin antibodies (49, 56, 57). These CoMPP results established that the defect caused by the loss of GH43 β-1,3-galactosidase activity causes changes in the abundance of cell wall-associated AGPs.
Table 1.
CoMMP of sequentially extracted cell walls from 7-day-old Col-0 and gh43null seedlings
Cell wall fractions extracted with 180 mm CaCl2, 50 mm CDTA, and 4 m NaOH, 26.5 mm NaBH4. The values represent the signal intensity of the fluorescent secondary antibody. Values are mean ± S.E. Gray cells mark significant differences: *, p < 0.05; **, p < 0.01; ***, p <0.001 (Student's t test, n = 5 biological replicates with pooled seedlings per replicate).
| CaCl2 soluble | CDTA | NaOH | |||||
|---|---|---|---|---|---|---|---|
| Antibody | Col-0 | gh43null | Col-0 | gh43null | Col-0 | gh43null | Antibody binds |
| JIM5 | 0 | 0 | 42 ± 1.1 | 43 ± 1.7 | 0 | 0 | Homogalacturonan |
| JIM7 | 60 ± 2.0 | 54 ± 3.2 | 91 ± 1.8 | 92 ± 3.5 | 0 | 0 | Homogalacturonan |
| LM5 | 21 ± 0.4 | 20 ± 1.9 | 10 ± 0.6 | 13 ± 0.5* | 19 ± 1.9 | 26 ± 0.8* | (1,4)-β-d-Gal4 |
| LM6 | 13 ± 1.3 | 11 ± 1.5 | 16 ± 1.0 | 17 ± 0.4 | 28 ± 1.2 | 31 ± 0.6 | (1,5)-α-l-Araf5 |
| LM18 | 0 | 0 | 31 ± 0.9 | 34 ± 1.0* | 0 | 0 | Homogalacturonan |
| LM19 | 0 | 0 | 47 ± 0.5 | 46 ± 0.5 | 61 ± 5.1 | 60 ± 2.1 | Homogalacturonan |
| LM20 | 35 ± 1.1 | 29 ± 2.0* | 90 ± 2.0 | 87 ± 2.0 | 0 | 0 | Homogalacturonan |
| RU1 | 0 | 0 | 46 ± 3.9 | 47 ± 1.5 | 43 ± 2.8 | 42 ± 1.0 | Backbone of RG-I |
| RU2 | 10 ± 1.2 | 4 ± 1.6* | 84 ± 6.1 | 88 ± 2.1 | 64 ± 2.5 | 69 ± 2.3 | Backbone of RG-I |
| LM15 | 0 | 0 | 3 ± 1.7 | 8 ± 0.7* | 73 ± 3.6 | 78 ± 3.0 | XXXG-motif in xyloglucan |
| LM25 | 31 ± 0.6 | 30 ± 3.1 | 31 ± 2.0 | 34 ± 1.5 | 62 ± 1.5 | 71 ± 2.6* | XLLG, XXLG, XXXG in xyloglucan |
| LM10 | 0 | 0 | 0 | 0 | 27 ± 1.4 | 25 ± 1.4 | (1, 4)-β-d-xylan |
| LM11 | 0 | 0 | 0 | 0 | 35 ± 1.7 | 35 ± 2.6 | (1, 4)-β-d-xylan |
| LM28 | 8 ± 0.8 | 8 ± 2.3 | 0 | 0 | 58 ± 3.9 | 51 ± 1.3 | Glucuronosyl-containing epitope in heteroxylan |
| LM23 | 0 | 0 | 0 | 0 | 7 ± 1.9 | 9 ± 1.7 | Nonacetylated xylosyl residues |
| BS400-4 | 0 | 0 | 0 | 0 | 28 ± 1.5 | 32 ± 1.9 | (1,4)-β-d-Mannan/galactomannan |
| CBM 3a | 0 | 0 | 17 ± 5.8 | 24 ± 1.5 | 38 ± 1.4 | 44 ± 1.7* | Crystalline cellulose |
| LM1 | 12 ± 0.6 | 10 ± 1.0 | 0 | 0 | 0 | 0 | Extensin |
| LM3 | 33 ± 1.6 | 28 ± 2.8 | 17 ± 0.7 | 18 ± 0.6 | 25 ± 2.3 | 30 ± 0.6 | Extensin |
| JIM11 | 16 ± 2.4 | 10 ± 2.8 | 13 ± 0.6 | 13 ± 0.6 | 22 ± 2.4 | 28 ± 0.5 | Extensin |
| JIM12 | 35 ± 1.8 | 32 ± 2.4 | 12 ± 0.4 | 10 ± 0.6 | 16 ± 2.0 | 20 ± 1.6 | Extensin |
| JIM20 | 37 ± 2.2 | 31 ± 2.8 | 8 ± 0.8 | 10 ± 0.6 | 20 ± 2.2 | 26 ± 0.4* | Extensin |
| LM2 | 13 ± 1.2 | 8 ± 2.2 | 0 | 0 | 10 ± 0.9 | 15 ± 1.2* | GlcA terminally attached to 1,6-linked galactan |
| LM14 | 41 ± 1.9 | 39 ± 2.2 | 16 ± 1.1 | 17 ± 0.5 | 22 ± 1.1 | 29 ± 0.6*** | AGP |
| JIM13 | 16 ± 1.0 | 12 ± 0.9* | 6 ± 0.2 | 6 ± 0.3 | 8 ± 0.5 | 12 ± 1.0* | AGP |
| JIM16 | 32 ± 1.4 | 28 ± 1.8 | 0 | 0 | 0 | 0 | β-1,3-Linked galactan backbone substituted with a single β-1,6-linked Gal residue |
| MAC 207 | 35 ± 1.6 | 34 ± 1.7 | 12 ± 1.0 | 13 ± 0.8 | 11 ± 0.7 | 15 ± 0.9** | AGP |
Discussion
β1,3-Galactan in plant cell walls is found primarily on AGPs (43). We identified and characterized the enzymatic activity of two Golgi-localized GH43 exo-β1,3-galactosidases and discovered that their activity influences the levels of cell wall-associated AGPs. Functional characterization of plant GH43 enzymes has not previously been reported. The active site of the prokaryotic GH43 from C. thermocellum and the Arabidopsis GH43s is conserved (Fig. S4). Despite this catalytic similarity, the Arabidopsis GH43s are involved in cell wall biosynthesis, whereas their prokaryotic and fungal counterparts are involved in plant cell wall degradation (30, 31, 40–42). A role for GH43 activity during root growth was revealed by experiments using nutrient media supplemented with 4.5% glucose. On such media, root growth is modestly reduced in single gh43b mutants and dramatically reduced in the gh43null double mutant (Fig. 1). Nutrient media with 4.5% sugar was also used in a classical genetic screen for conditional root growth mutants of Arabidopsis (34, 35). Several mutants identified in this way were subsequently shown to be defective in cell wall biosynthesis genes including the glycosylphosphatidylinositol-anchored plasma membrane protein COBRA (58), the chitinase-like protein CTL1 (59), and the cellulose synthase interacting 1 (CSI1) (60). The root cell expansion defects of gh43null become noticeable in the elongation zone within 6 h of transferring seedlings to sugar media (Fig. S1). Quantification of cell wall-associated AGPs with β-Yariv (Fig. 5), and the CoMPP analysis of cell wall fractions from untreated seedlings (Table 1) confirmed that the gh43null cell walls differ from WT already before the sugar treatment. Thus, a pre-existing cell wall defect is likely to underlie the sugar induced swelling.
Investigation of the gh43null cell wall defect revealed a connection to AGPs. We observed AGP epitopes in all sequentially extracted cell wall fractions (Table 1). The increase in the levels of AGPs extracted from the gh43null with 4 m NaOH (i.e. AGPs not released by 180 mm CaCl2 or 50 mm CDTA) suggests that GH43s play a role in adjusting the cell wall affinity of AGPs. The CoMPP microarray analysis of cell wall fractions also revealed subtle differences in the abundance of pectin and xyloglucan epitopes in the gh43null mutant (Table 1). We hypothesize that these changes reflect modifications of the cell wall matrix resulting from initial changes in the AGPs. Based on the inactivity of GH43s toward β-1,3-galactans with β-1,6 branching (Fig. 4), and the lack of galactose release from extracted cell wall glycoproteins or AGP-rich gum arabic (Fig. S5), we propose that the Arabidopsis GH43s are required for β-1,3-galactan processing during AGP maturation in the Golgi. It is conceivable that GH43s regulate the length of the β-1,3-galactan backbone (and thus the number of side chains that can attach to it), and/or trim erroneously synthesized β-1,3-galactans in an AGP quality control process, and/or modify the backbone length to make it accessible to other enzymes. The Golgi-associated α-mannosidase I, which is involved in structuring N-glycosylation, has been demonstrated to perform proteoglycan trimming of the latter sort in Arabidopsis (61). A similar AGP glycan structuring role could be envisaged for GH43s. Inherent to these models is that the native GH43 target(s) would only exist transiently in the Golgi. Both GH43A-YFP and GH43B-YFP were enriched on the cis-Golgi side receiving transport vesicles from the endoplasmic reticulum (Fig. 3), suggesting that GH43s are involved in an early glycan modification process. This would be in line with the models of sequential maturation of plant cell wall glycans and glycoproteins during transit through the Golgi (62, 63).
Accumulating evidence from several plant species supports the existence of cell wall matrix-bound AGPs, primarily associated with pectin (summarised by Tan et al. (48)). Pectin can be subdivided into homogalacturonan (HG), xylogalacturonan (XGA), apiogalacturonan, rhamnogalacturonan type I (RG-I), and rhamnogalacturonan type II (RG-II) (3). Of these, HG and RG-I were the most abundant in Arabidopsis primary walls, accounting for ∼65% and 20–35% of the total pectin content, respectively (64). HG is a linear α-1,4-linked galacturonic acid that is partially methyl esterified and can be O-acetylated. RG-I consists of a α-1,4-d-galacturonic acid-α-1,2-l-rhamnose backbone with several side chains (3). The RG-I side chains include galactans and arabinans as well as arabinogalactan type I (AG-I), and arabinogalactan type II (AG-II) polysaccharide chains (65). The AG-II structure resembles that of AGP glycans and it may be that some of the RG-I–attached oligosaccharides actually derive from AGP glycans. In Arabidopsis, the AGP ARABINOXYLAN PECTIN ARABINOGALACTAN PROTEIN1 (APAP1) binds covalently to RG-I/HG and was shown to increase the extractability of pectin and hemicellulosic immunoreactive epitopes, suggesting that it acts as a structural cross-linker in plant cell walls for this AGP (48). Whether the covalent bond between APAP1 and RG-I is a result of transglycosylation taking place in the apoplast or synthesizes as such in the Golgi is unknown. Interestingly, the CoMPP microarray results for the gh43null mutant revealed decreased homogalacturonan (LM20) and RGI-backbone (RU2) signals in the CaCl2-soluble fraction, and increased β-1,4-galactan (LM5) and homogalacturonan (LM18) signal in the CDTA fraction and β-1,4-galactan (LM5) in the NaOH fraction (Table 1). Together, these results show that gh43null exhibits altered pectin extractability compared with the WT, and suggest that APAP1 and/or similar cell wall-bound AGPs could be targets of GH43 activity.
Pectin in expanding cells has been proposed to function as a mechanical tether between cellulose microfibrils (66), and as a lubricant of microfibril movement during cell expansion (67). Experimental support for direct interaction between cellulose and pectin in primary cell walls of Arabidopsis was provided by the comparison of never dried, dehydrated, and rehydrated cell walls using multidimensional solid state NMR spectroscopy (68, 69). The NMR spectra showed cross-peaks between cellulose and pectin indicating that some of the pectic sugars come into subnanometer contact with cellulose microfibrils. Based on these pectin models, AGP and pectin interactions could be expected to influence primary cell wall extensibility by restricting cellulose microfibril movement and/or cell wall matrix extension. This concept was first introduced in an early cell wall model proposed in the 1970s, which depicted covalent connections between pectin and structural proteins (70). However, evidence of the role of structural cell wall proteins has been slow to emerge and therefore many current cell wall models omit them. Currently AGPs are generally not considered load-bearing in cell walls, but rather as chaperones or scaffolds facilitating cell wall polymer deposition. Cell wall-bound AGPs may, for example, play an important role as matrix proteins preventing cellulose microfibril coalescence, simultaneously hindering other cell wall polymers from binding to cellulose. In this scenario, more tightly cell wall-bound AGPs could impair wall stiffening during cell expansion, and thus contribute to the gh43null root swelling phenotype. This interpretation would imply that the distribution, measured as extractability, of other polymers in the wall would also change and this seems indeed to be the case in the gh43null mutant. However, it is worth noting that the loss of exo-β1,3-galactosidase activity may affect other glycosylation processes either directly or indirectly, and further work is required to establish the primary in vivo targets of GH43s and the cause of the cell expansion defects.
Experimental Procedures
Plant material and growth conditions
gh43a-1 (GABI_062D83), gh43b-1 (Salk_002830), and gh43b-2 (SALK_087519) were ordered from NASC and genotyped (Table S3). The T-DNA insert locations were determined by sequencing (Fig. 1A). Mutant and WT plants were grown on soil at 22 °C with a photoperiod of 16-h light and 8-h dark at 65% relative humidity.
Plant vector construction and transformation
The genomic sequences of GH43A and -B were amplified using the primers in Table S3. The amplified pieces were cloned in the pDONR207 plasmid via Gibson assembly and recombined into pHGY (71). Both constructs were transformed into gh43null. Stably transgenic plants were generated by Agrobacterium tumefaciens–mediated transformation. Transgenic seedlings were selected based on hypocotyl elongation on 0.9% agar ½ MS plates supplemented with 30 μg/ml of hygromycin. Homozygous lines were generated based on hygromycin segregation and detection of the YFP signal by confocal microscopy.
Root elongation/complementation experiments
Seeds were surface sterilized with 70% ethanol and grown on ½ MS agar plates (0.9% agar, 5 mm MES, pH 5.7) for 4 days. After this period, the seedlings were moved to ½ MS plates, ½ MS plates containing either 4.5% glucose or 4.5% sorbitol, or 100 mm NaCl plates. The seedlings were then allowed to grow for 6 days. The difference in root elongation from day 0 on the new plate to day 6 was quantified using ImageJ (RRID:SCR_003070).
GH43 protein localization and fluorescent root imaging
The lines pGH43A::GH43A-YFP and pGH43B::GH43B-YFP co-expressing the marker lines SYP32-mCherry or SYP43-mCherry were generated by plant crossing (36, 37). For observation, seedlings were grown vertically on plates containing ½ MS media with 0.9% plant agar for 5 days and then observed under a Zeiss LSM880 confocal laser scanning microscope. A Zeiss LSM880 confocal laser scanning microscope with an Airyscan detector and a LD LCI Plan-Apochromat ×40/1.2 Imm AutoCorr DIC M27 water immersion objective was used. The co-localization analysis was performed with the PSC Colocalization Plugin from ImageJ (72).
The GH43-YFP root signal distribution was imaged with a Leica M205 FA stereomicroscope with fluorescence around a ×200 magnification. Seedlings were grown on ½ MS media for 4 days and imaged on the growth plate.
The LTI6a-GFP line was crossed in the gh43null mutant background and homozygous lines were generated (73). The seedlings were grown for 4 days on ½ MS plates and then moved to ½ MS plates or ½ MS plates with 4.5% glucose. The roots were imaged with a Zeiss LSM880 confocal microscope.
Heterologous expression and purification of the GH43 proteins
The Arabidopsis GH43 coding sequences were amplified from 121-1401 bp (AT5G67540.1, GH43A) and 115-1401 bp (AT3G49880, GH43B) to avoid the predicted membrane domain. The proteins were expressed in RosettaTM (DE3) E. coli cells using a pET24d His10SUMO vector. The crude protein extract was purified by nickel-nitrilotriacetic acid, then the His10SUMO tag was cleaved and the GH43 protein isolated by nickel-nitrilotriacetic acid chromatograph.
Synthesis of methyl-β-d-galactopyranoside substrates 1--4
Chemical synthesis of methyl 3-O-β-d-galactopyranosyl-β-d-galactopyranoside, substrate 1 (74), methyl 6-O-β-d-galactopyranosyl-β-d-galactopyranoside, substrate 2 (75), and methyl 3,6-di-O-(β-d-galactopyranosyl)-β-d-galactopyranoside, substrate 4 (76), have been reported in literature. However, methyl β-d-galactopyranosyl-(1→6)-β-d-galactopyranosyl-(1→3)-β-d-galactopyranoside, substrate 3, has not previously been reported. The strategy for chemical synthesis of substrates 1--4 is outlined in Fig. S7.
GH43 protein activity assays
The enzymes' buffer was changed to 10 mm MOPS (pH 7) using the PD Spintrap™ G-25 (GE Healthcare, 28-9180-04) protocol. To determine the activity toward the β-galactan substrates and β-glucan substrates, 1 µg of GH43A and 4 µg of GH43B were incubated with 100 µg of substrate (15 µl total volume), overnight at 30 °C while shaking. For tests using the inactive enzymes, 1 µg was loaded.
Gum arabic (200 mg/ml) was hydrolyzed in 0.1 m TFA hydrolyzed at 80 °C for 45 min (loading 10 µl on a TLC plate). The TFA was then removed using a SpeedVac by reducing the reaction mixture's volume to around half of its original value. 30 mg of the resulting digestion product was further hydrolyzed with an enzyme mixture containing 1 µl of β-galactanase (E-BGLAN), 0.5 µl of β-glucuronosidase (E-BGLAEC), 18 µl of α-fucosidase (E-FUCTM), and 20 µl of α-arabinofuranosidase (E-ABFAN) from Megazyme in a 50 mm sodium acetate buffer (pH 5) at 37 °C for 4 h (again, 10 µl was loaded on a TLC plate). The enzymes were then inactivated at 95 °C for 15 min, after which their buffer was changed to 10 mm MOPS (pH 7) using a PD MidiTrap G-10 column to 10 mm MOPS (pH 7) to remove the sodium acetate and digestion products. The total sample was incubated with 2.5 µg of GH43A or 10 µg of GH43B overnight while shaking at 30 °C, then concentrated with a SpeedVac to ∼10 µl and loaded onto a TLC plate.
Sequentially extracted cell wall material (10 mg/100 µl of MOPS, pH 7) was incubated with 2 µg of GH43A or 7.5 µg of GH43B at 30 °C overnight while shaking. The samples were then concentrated with a SpeedVac to ∼10 µl and loaded onto a TLC plate.
The products were loaded on a TLC Silica Gel 60 F254 TLC plate membrane and developed with 4:1:1 (1-butanol:acetetic acid:H2O). The membranes were visualized with 5% sulfuric acid and 0.5% thymol in 96% ethanol at 105 °C.
AGP quantification with β-Yariv
Soluble AGP purification and quantification was performed according to Lamport (77). For quantification of cell wall-associated AGPs, the cell wall pellet left after removing CaCl2-soluble AGPs was flash frozen in liquid nitrogen and then lyophilized. The resulting material was ball milled, AIR1-treated (see below), and lyophilized. The lyophilized material was then incubated with β-Yariv for 4 h at room temperature, and the β-Yariv absorbance was spectrophotometrically quantified according to Lamport (77).
Sequential extraction and CoMPP
Seeds were surface sterilized with 70% ethanol and grown on ½ MS agar plates (0.9% agar, 5 mm MES, pH 5.7) for 7 days. The seedlings were then harvested, flash frozen, and ground in liquid nitrogen with a mortar and pestle. CaCl2-soluble glycoproteins were extracted with 0.18 m CaCl2 (2 ml/g fresh weight) for 2 h at room temperature and spun down at 4000 × g for 10 min. The pellet containing the cell wall material was freeze dried, ball milled, and AIR1-treated (see below) before further processing. The supernatant was precipitated with 4 volumes of ethanol at 4 °C for 16 h and spun down at 2000 × g for 2 min. The pellet was further solubilized in 45 mm CaCl2 (2 ml/g fresh weight from starting material) and freeze dried. The soluble glycoproteins and cell wall material were then analyzed by CoMPP (46). CoMPP is a semiquantitative method that relies on the spotting onto microarrays of soluble glycoproteins or cell wall biopolymers solubilized by sequential extraction. Samples are spotted in serial dilutions onto the microarrays, which are then blocked using skim milk, probed with carbohydrate-binding modules (CBMs) or monoclonal antibodies specific to glycan epitopes, and then visualized using enzyme-conjugated secondary antibodies. Spot intensities are quantified from 256-step gray scale scans of the microarrays and data are presented on a relative scale with 100 assigned to the most intense spot. CoMPP fractions extracted with 50 mm CDTA and 4 m NaOH, 26.5 mm NaBH4 fractions were dialyzed against demineralized water using Visking® 12-14 kDa dialysis tubing (45 mm) prior to determination of the monosaccharide composition.
Wet chemical analysis of cell walls
The 7-day–old seedlings were harvested, flash frozen, and lyophilized. The dried material was ball milled, and the alcohol-insoluble residue (AIR1) was removed by incubating the material first for 30 min in 80% ethanol, and then for 30 min in 70% ethanol at 95 °C, and finally in chloroform:methanol (1:1) for 5 min at room temperature before washing with acetone. This sequence of incubations and washes constitutes the AIR1 treatment mentioned above. Starch (AIR2) was then removed according to Ref. 78. The mono sugar composition was measured according to Ref. 79. The crystalline cellulose content was quantified according to Updegraff (80).
Data availability
All data are contained within the manuscript.
Supplementary Material
Acknowledgments
We thank Mikael Lindberg from the Umeå University Protein Expertise Platform (PEP), for help with expressing the recombinant Arabidopsis GH43s. We thank Dr. Junko Takahashi-Schmidt and the UPSC Biopolymer Analytical Platform for help with the cell wall monosugar analysis.
This article contains supporting information.
Author contributions—P. N., M. S. M., B. J., P. U., and T. N. formal analysis; P. N., B. L. P., M. S. M., B. J., P. U., and T. N. investigation; P. N., B. L. P., M. S. M., and P. U. methodology; P. N., P. U., and T. N. writing-original draft; B. L. P., M. S. M., B. J., P. U., and T. N. writing-review and editing; P. U. and T. N. conceptualization; P. U. and T. N. supervision; T. N. funding acquisition; T. N. project administration.
Funding and additional information—This work was supported by The Swedish Foundation for Strategic Research (Value Tree), Bio4Energy (Swedish Programme for Renewable Energy), the UPSC Centre for Forest Biotechnology funded by VINNOVA, and the Swedish Research Council for Sustainable Development (Formas).
Conflict of interest—The authors declare that they have no conflicts of interest with the contents of this article.
- AGP
- arabinogalactan protein
- GT
- glycosyl transferase
- CDTA
- cyclohexanediaminetetraacetic acid
- CoMPP
- comprehensive microarray polymer profiling
- HG
- homogalacturonan
- XGA
- xylogalacturonan
- RG-I
- rhamnogalacturonan type I
- RG-II
- rhamnogalacturonan type II
- AG-I
- arabinogalactan type I
- APAP1
- ARABINOXYLAN PECTIN ARABINOGALACTAN PROTEIN 1
- CBM
- carbohydrate-binding module
- ANOVA
- analysis of variance
- YFP
- yellow fluorescent protein.
References
- 1. Cosgrove D. J. (2005) Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6, 850–861 10.1038/nrm1746 [DOI] [PubMed] [Google Scholar]
- 2. Bashline L., Lei L., Li S. D., and Gu Y. (2014) Cell wall, cytoskeleton, and cell expansion in higher plants. Mol. Plant 7, 586–600 10.1093/mp/ssu018 [DOI] [PubMed] [Google Scholar]
- 3. Harholt J., Suttangkakul A., and Vibe Scheller H. (2010) Biosynthesis of pectin. Plant Physiol. 153, 384–395 10.1104/pp.110.156588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scheller H. V., and Ulvskov P. (2010) Hemicelluloses. Annu. Rev. Plant Biol. 61, 263–289 10.1146/annurev-arplant-042809-112315 [DOI] [PubMed] [Google Scholar]
- 5. Ellis M., Egelund J., Schultz C. J., and Bacic A. (2010) Arabinogalactan-proteins: key regulators at the cell surface? Plant Physiol. 153, 403–419 10.1104/pp.110.156000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rose J. K. C., and Lee S. J. (2010) Straying off the highway: trafficking of secreted plant proteins and complexity in the plant cell wall proteome. Plant Physiol. 153, 433–436 10.1104/pp.110.154872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Showalter A. M., Keppler B., Lichtenberg J., Gu D., and Welch L. R. (2010) A bioinformatics approach to the identification, classification, and analysis of hydroxyproline-rich glycoproteins. Plant Physiol. 153, 485–513 10.1104/pp.110.156554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xue H., Veit C., Abas L., Tryfona T., Maresch D., Ricardi M. M., Estevez J. M., Strasser R., and Seifert G. J. (2017) Arabidopsis thaliana FLA 4 functions as a glycan‐stabilized soluble factor via its carboxy‐proximal Fasciclin 1 domain. Plant J. 91, 613–630 10.1111/tpj.13591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Showalter A. M. (1993) Structure and function of plant cell wall proteins. Plant Cell 5, 9–23 10.2307/3869424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Showalter A. M., and Basu D. (2016) Extensin and arabinogalactan-protein biosynthesis: glycosyltransferases, research challenges, and biosensors. Front. Plant Sci. 7, 814 10.3389/fpls.2016.00814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hijazi M., Velasquez S. M., Jamet E., Estevez J. M., and Albenne C. (2014) An update on post-translational modifications of hydroxyproline-rich glycoproteins: toward a model highlighting their contribution to plant cell wall architecture. Front Plant Sci. 5, 395 10.3389/fpls.2014.00395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Basu D., Tian L., Wang W., Bobbs S., Herock H., Travers A., and Showalter A. M. (2015) A small multigene hydroxyproline-O-galactosyltransferase family functions in arabinogalactan-protein glycosylation, growth and development in Arabidopsis. BMC Plant Biol 15, 295 10.1186/s12870-015-0670-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Basu D., Wang W., Ma S., DeBrosse T., Poirier E., Emch K., Soukup E., Tian L., and Showalter A. M. (2015) Two hydroxyproline galactosyltransferases, GALT5 and GALT2, function in arabinogalactan-protein glycosylation, growth and development in Arabidopsis. PLoS ONE 10, e0125624 10.1371/journal.pone.0125624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ogawa-Ohnishi M., and Matsubayashi Y. (2015) Identification of three potent hydroxyproline O-galactosyltransferases in Arabidopsis. Plant J. 81, 736–746 10.1111/tpj.12764 [DOI] [PubMed] [Google Scholar]
- 15. Suzuki T., Narciso J. O., Zeng W., van de Meene A., Yasutomi M., Takemura S., Lampugnani E. R., Doblin M. S., Bacic A., and Ishiguro S. (2017) KNS4/UPEX1: a type II arabinogalactan β-(1,3)-galactosyltransferase required for pollen exine development. Plant Physiol. 173, 183–205 10.1104/pp.16.01385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Geshi N., Johansen J. N., Dilokpimol A., Rolland A., Belcram K., Verger S., Kotake T., Tsumuraya Y., Kaneko S., Tryfona T., Dupree P., Scheller H. V., Höfte H., and Mouille G. (2013) A galactosyltransferase acting on arabinogalactan protein glycans is essential for embryo development in Arabidopsis. Plant J. 76, 128–137 10.1111/tpj.12281 [DOI] [PubMed] [Google Scholar]
- 17. Dilokpimol A., Poulsen C. P., Vereb G., Kaneko S., Schulz A., and Geshi N. (2014) Galactosyltransferases from Arabidopsis thaliana in the biosynthesis of type II arabinogalactan: molecular interaction enhances enzyme activity. BMC Plant Biol. 14, 90 10.1186/1471-2229-14-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gille S., Sharma V., Baidoo E. E. K., Keasling J. D., Scheller H. V., and Pauly M. (2013) Arabinosylation of a Yariv-precipitable cell wall polymer impacts plant growth as exemplified by the Arabidopsis glycosyltransferase mutant ray1. Mol. Plant 6, 1369–1372 10.1093/mp/sst029 [DOI] [PubMed] [Google Scholar]
- 19. Knoch E., Dilokpimol A., Tryfona T., Poulsen C. P., Xiong G. Y., Harholt J., Petersen B. L., Ulvskov P., Hadi M. Z., Kotake T., Tsumuraya Y., Pauly M., Dupree P., and Geshi N. (2013) A β-glucuronosyltransferase from Arabidopsis thaliana involved in biosynthesis of type II arabinogalactan has a role in cell elongation during seedling growth. Plant J. 76, 1016–1029 10.1111/tpj.12353 [DOI] [PubMed] [Google Scholar]
- 20. Tryfona T., Theys T. E., Wagner T., Stott K., Keegstra K., and Dupree P. (2014) Characterisation of FUT4 and FUT6 α-(1 → 2)-fucosyltransferases reveals that absence of root arabinogalactan fucosylation increases Arabidopsis root growth salt sensitivity. PLoS ONE 9, e93291 10.1371/journal.pone.0093291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wu Y., Williams M., Bernard S., Driouich A., Showalter A. M., and Faik A. (2010) Functional identification of two nonredundant Arabidopsis α(1,2)fucosyltransferases specific to arabinogalactan proteins. J. Biol. Chem. 285, 13638–13645 10.1074/jbc.M110.102715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pennell R. I., Janniche L., Kjellbom P., Scofield G. N., Peart J. M., and Roberts K. (1991) Developmental regulation of a plasma-membrane arabinogalactan protein epitope in oilseed rape flowers. Plant Cell 3, 1317–1326 10.2307/3869311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tan L., Qiu F., Lamport D. T. A., and Kieliszewski M. J. (2004) Structure of a hydroxyproline (Hyp)-arabinogalactan polysaccharide from repetitive Ala-Hyp expressed in transgenic Nicotiana tabacum. J. Biol. Chem. 279, 13156–13165 10.1074/jbc.M311864200 [DOI] [PubMed] [Google Scholar]
- 24. Tan L., Varnai P., Lamport D. T., Yuan C., Xu J., Qiu F., and Kieliszewski M. J. (2010) Plant O-hydroxyproline arabinogalactans are composed of repeating trigalactosyl subunits with short bifurcated side chains. J. Biol. Chem. 285, 24575–24583 10.1074/jbc.M109.100149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Haque M., Kotake T., and Tsumuraya Y. (2005) Mode of action of β-glucuronidase from Aspergillus niger on the sugar chains of arabinogalactan-protein. Biosci. Biotechnol. Biochem. 69, 2170–2177 10.1271/bbb.69.2170 [DOI] [PubMed] [Google Scholar]
- 26. Tryfona T., Liang H. C., Kotake T., Kaneko S., Marsh J., Ichinose H., Lovegrove A., Tsumuraya Y., Shewry P. R., Stephens E., and Dupree P. (2010) Carbohydrate structural analysis of wheat flour arabinogalactan protein. Carbohydr. Res. 345, 2648–2656 10.1016/j.carres.2010.09.018 [DOI] [PubMed] [Google Scholar]
- 27. Tryfona T., Liang H. C., Kotake T., Tsumuraya Y., Stephens E., and Dupree P. (2012) Structural characterization of Arabidopsis leaf arabinogalactan polysaccharides. Plant Physiol. 160, 653–666 10.1104/pp.112.202309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Günl M., Neumetzler L., Kraemer F., de Souza A., Schultink A., Pena M., York W. S., and Pauly M. (2011) AXY8 encodes an α-fucosidase, underscoring the importance of apoplastic metabolism on the fine structure of Arabidopsis cell wall polysaccharides. Plant Cell 23, 4025–4040 10.1105/tpc.111.089193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Günl M., and Pauly M. (2011) AXY3 encodes a α-xylosidase that impacts the structure and accessibility of the hemicellulose xyloglucan in Arabidopsis plant cell walls. Planta 233, 707–719 10.1007/s00425-010-1330-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mewis K., Lenfant N., Lombard V., and Henrissat B. (2016) Dividing the large glycoside hydrolase family 43 into subfamilies: a motivation for detailed enzyme characterization. Appl. Environ. Microbiol. 82, 1686–1692 10.1128/AEM.03453-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kotake T., Kitazawa K., Takata R., Okabe K., Ichinose H., Kaneko S., and Tsumuraya Y. (2009) Molecular cloning and expression in Pichia pastoris of a Irpex lacteus exo-β-(1→3)-galactanase gene. Biosci. Biotechnol. Biochem. 73, 2303–2309 10.1271/bbb.90433 [DOI] [PubMed] [Google Scholar]
- 32. Jordan D. B., Wagschal K., Grigorescu A. A., and Braker J. D. (2013) Highly active β-xylosidases of glycoside hydrolase family 43 operating on natural and artificial substrates. Appl. Microbiol. Biotechnol. 97, 4415–4428 10.1007/s00253-012-4475-4 [DOI] [PubMed] [Google Scholar]
- 33. McCleary B. V., McKie V. A., Draga A., Rooney E., Mangan D., and Larkin J. (2015) Hydrolysis of wheat flour arabinoxylan, acid-debranched wheat flour arabinoxylan and arabino-xylo-oligosaccharides by β-xylanase, α-l-arabinofuranosidase and β-xylosidase. Carbohydr. Res. 407, 79–96 10.1016/j.carres.2015.01.017 [DOI] [PubMed] [Google Scholar]
- 34. Hauser M. T., Morikami A., and Benfey P. N. (1995) Conditional root expansion mutants of Arabidopsis. Development 121, 1237–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Benfey P. N., Linstead P. J., Roberts K., Schiefelbein J. W., Hauser M. T., and Aeschbacher R. A. (1993) Root development in Arabidopsis: four mutants with dramatically altered root morphogenesis. Development 119, 57–70 [DOI] [PubMed] [Google Scholar]
- 36. Uemura T., Ueda T., Ohniwa R. L., Nakano A., Takeyasu K., and Sato M. H. (2004) Systematic analysis of SNARE molecules in Arabidopsis: dissection of the post-Golgi network in plant cells. Cell Struct. Funct. 29, 49–65 10.1247/csf.29.49 [DOI] [PubMed] [Google Scholar]
- 37. Geldner N., Dénervaud-Tendon V., Hyman D. L., Mayer U., Stierhof Y. D., and Chory J. (2009) Rapid, combinatorial analysis of membrane compartments in intact plants with a multicolor marker set. Plant J. 59, 169–178 10.1111/j.1365-313X.2009.03851.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Parsons H. T., Stevens T. J., McFarlane H. E., Vidal-Melgosa S., Griss J., Lawrence N., Butler R., Sousa M. M. L., Salemi M., Willats W. G. T., Petzold C. J., Heazlewood J. L., and Lilley K. S. (2019) Separating Golgi proteins from cis to trans reveals underlying properties of cisternal localization. Plant Cell 31, 2010–2034 10.1105/tpc.19.00081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Parsons H. T., Christiansen K., Knierim B., Carroll A., Ito J., Batth T. S., Smith-Moritz A. M., Morrison S., McInerney P., Hadi M. Z., Auer M., Mukhopadhyay A., Petzold C. J., Scheller H. V., Loque D., et al. (2012) Isolation and proteomic characterization of the Arabidopsis Golgi defines functional and novel components involved in plant cell wall biosynthesis. Plant Physiol. 159, 12–26 10.1104/pp.111.193151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ichinose H., Kuno A., Kotake T., Yoshida M., Sakka K., Hirabayashi J., Tsumuraya Y., and Kaneko S. (2006) Characterization of an exo-β-1,3-galactanase from Clostridium thermocellum. Appl. Environ. Microbiol. 72, 3515–3523 10.1128/AEM.72.5.3515-3523.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ichinose H., Yoshida M., Kotake T., Kuno A., Igarashi K., Tsumuraya Y., Samejima M., Hirabayashi J., Kobayashi H., and Kaneko S. (2005) An exo-β-1,3-galactanase having a novel β-1,3-galactan-binding module from Phanerochaete chrysosporium. J. Biol. Chem. 280, 25820–25829 10.1074/jbc.M501024200 [DOI] [PubMed] [Google Scholar]
- 42. Jiang D., Fan J., Wang X., Zhao Y., Huang B., Liu J., and Zhang X. C. (2012) Crystal structure of 1,3Gal43A, an exo-β-1,3-galactanase from Clostridium thermocellum. J. Struct. Biol. 180, 447–457 10.1016/j.jsb.2012.08.005 [DOI] [PubMed] [Google Scholar]
- 43. Du H., Clarke A. E., and Bacic A. (1996) Arabinogalactan-proteins: a class of extracellular matrix proteoglycans involved in plant growth and development. Trends Cell Biol. 6, 411–414 10.1016/S0962-8924(96)20036-4 [DOI] [PubMed] [Google Scholar]
- 44. Akiyama Y., Eda S., and Kato K. (1984) Gum arabic is a kind of arabinogalactan protein. Agr. Biol. Chem. Tokyo 48, 235–237 10.1271/bbb1961.48.235 [DOI] [Google Scholar]
- 45. Kitazawa K., Tryfona T., Yoshimi Y., Hayashi Y., Kawauchi S., Antonov L., Tanaka H., Takahashi T., Kaneko S., Dupree P., Tsumuraya Y., and Kotake T. (2013) β-Galactosyl Yariv reagent binds to the β-1,3-galactan of arabinogalactan proteins. Plant Physiol. 161, 1117–1126 10.1104/pp.112.211722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Moller I. E., Pettolino F. A., Hart C., Lampugnani E. R., Willats W. G. T., and Bacic A. (2012) Glycan profiling of plant cell wall polymers using microarrays. J. Vis. Exp. 70 10.3791/4238 10.3791/4238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Harholt J., Sorensen I., Fangel J., Roberts A., Willats W. G. T., Scheller H. V., Petersen B. L., Banks A., and Ulvskov P. (2012) The glycosyltransferase repertoire of the spikemoss Selaginella moellendorffii and a comparative study of its cell wall. PLoS ONE 7, e35846 10.1371/journal.pone.0035846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tan L., Eberhard S., Pattathil S., Warder C., Glushka J., Yuan C., Hao Z., Zhu X., Avci U., Miller J. S., Baldwin D., Pham C., Orlando R., Darvill A., Hahn M. G., et al. (2013) An Arabidopsis cell wall proteoglycan consists of pectin and arabinoxylan covalently linked to an arabinogalactan protein. Plant Cell 25, 270–287 10.1105/tpc.112.107334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Verhertbruggen Y., Marcus S. E., Haeger A., Ordaz-Ortiz J. J., and Knox J. P. (2009) An extended set of monoclonal antibodies to pectic homogalacturonan. Carbohydr. Res. 344, 1858–1862 10.1016/j.carres.2008.11.010 [DOI] [PubMed] [Google Scholar]
- 50. Ralet M. C., Tranquet O., Poulain D., Moise A., and Guillon F. (2010) Monoclonal antibodies to rhamnogalacturonan I backbone. Planta 231, 1373–1383 10.1007/s00425-010-1116-y [DOI] [PubMed] [Google Scholar]
- 51. Yates E. A., Valdor J. F., Haslam S. M., Morris H. R., Dell A., Mackie W., and Knox J. P. (1996) Characterization of carbohydrate structural features recognized by anti-arabinogalactan-protein monoclonal antibodies. Glycobiology 6, 131–139 10.1093/glycob/6.2.131 [DOI] [PubMed] [Google Scholar]
- 52. Jones L., Seymour G. B., and Knox J. P. (1997) Localization of pectic galactan in tomato cell walls using a monoclonal antibody specific to (1→4)-β-d-galactan. Plant Physiol. 113, 1405–1412 10.1104/pp.113.4.1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marcus S. E., Verhertbruggen Y., Hervé C., Ordaz-Ortiz J. J., Farkas V., Pedersen H. L., Willats W. G., and Knox J. P. (2008) Pectic homogalacturonan masks abundant sets of xyloglucan epitopes in plant cell walls. BMC Plant Biol. 8, 60 10.1186/1471-2229-8-60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ruprecht C., Bartetzko M. P., Senf D., Dallabernadina P., Boos I., Andersen M. C. F., Kotake T., Knox J. P., Hahn M. G., Clausen M. H., and Pfrengle F. (2017) A synthetic glycan microarray enables epitope mapping of plant cell wall glycan-directed antibodies. Plant Physiol. 175, 1094–1104 10.1104/pp.17.00737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Smallwood M., Yates E. A., Willats W. G. T., Martin H., and Knox J. P. (1996) Immunochemical comparison of membrane-associated and secreted arabinogalactan-proteins in rice and carrot. Planta 198, 452–459 10.1007/BF00620063 [DOI] [Google Scholar]
- 56. Smallwood M., Beven A., Donovan N., Neill S. J., Peart J., Roberts K., and Knox J. P. (1994) Localization of cell-wall proteins in relation to the developmental anatomy of the carrot root apex. Plant J. 5, 237–246 10.1046/j.1365-313X.1994.05020237.x [DOI] [Google Scholar]
- 57. Hernandez-Gomez M. C., Rydahl M. G., Rogowski A., Morland C., Cartmell A., Crouch L., Labourel A., Fontes C. M., Willats W. G., Gilbert H. J., and Knox J. P. (2015) Recognition of xyloglucan by the crystalline cellulose-binding site of a family 3a carbohydrate-binding module. FEBS Lett. 589, 2297–2303 10.1016/j.febslet.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Schindelman G., Morikami A., Jung J., Baskin T. I., Carpita N. C., Derbyshire P., McCann M. C., and Benfey P. N. (2001) COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 15, 1115–1127 10.1101/gad.879101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Sanchez-Rodriguez C., Bauer S., Hématy K., Saxe F., Ibáñez A. B., Vodermaier V., Konlechner C., Sampathkumar A., Ruggeberg M., Aichinger E., Neumetzler L., Burgert I., Somerville C., Hauser M. T., and Persson S. (2012) Chitinase-like1/pom-pom1 and its homolog CTL2 are glucan-interacting proteins important for cellulose biosynthesis in Arabidopsis. Plant Cell 24, 589–607 10.1105/tpc.111.094672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Gu Y., Kaplinsky N., Bringmann M., Cobb A., Carroll A., Sampathkumar A., Baskin T. I., Persson S., and Somerville C. R. (2010) Identification of a cellulose synthase-associated protein required for cellulose biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 107, 12866–12871 10.1073/pnas.1007092107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Liebminger E., Huttner S., Vavra U., Fischl R., Schoberer J., Grass J., Blaukopf C., Seifert G. J., Altmann F., Mach L., and Strasser R. (2009) Class I α-mannosidases are required for N-glycan processing and root development in Arabidopsis thaliana. Plant Cell 21, 3850–3867 10.1105/tpc.109.072363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Oikawa A., Lund C. H., Sakuragi Y., and Scheller H. V. (2013) Golgi-localized enzyme complexes for plant cell wall biosynthesis. Trends Plant Sci. 18, 49–58 10.1016/j.tplants.2012.07.002 [DOI] [PubMed] [Google Scholar]
- 63. Nguema-Ona E., Vicre-Gibouin M., Gotte M., Plancot B., Lerouge P., Bardor M., and Driouich A. (2014) Cell wall O-glycoproteins and N-glycoproteins: aspects of biosynthesis and function. Front. Plant Sci. 5, 499 10.3389/fpls.2014.00499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mohnen D. (2008) Pectin structure and biosynthesis. Curr. Opin. Plant Biol. 11, 266–277 10.1016/j.pbi.2008.03.006 [DOI] [PubMed] [Google Scholar]
- 65. Yapo B. M. (2011) Rhamnogalacturonan-I: a structurally puzzling and functionally versatile polysaccharide from plant cell walls and mucilages. Polymer Rev. 51, 391–413 10.1080/15583724.2011.615962 [DOI] [Google Scholar]
- 66. Höfte H., Peaucelle A., and Braybrook S. (2012) Cell wall mechanics and growth control in plants: the role of pectins revisited. Front. Plant Sci. 3, 121 10.3389/fpls.2012.00121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cosgrove D. J. (2014) Re-constructing our models of cellulose and primary cell wall assembly. Curr. Opin. Plant Biol. 22, 122–131 10.1016/j.pbi.2014.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang T., Park Y. B., Cosgrove D. J., and Hong M. (2015) Cellulose-pectin spatial contacts are inherent to never-dried Arabidopsis primary cell walls: evidence from solid-state nuclear magnetic resonance. Plant Physiol. 168, 871–884 10.1104/pp.15.00665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Wang T., Zabotina O., and Hong M. (2012) Pectin-cellulose interactions in the Arabidopsis primary cell wall from two-dimensional magic-angle-spinning solid-state nuclear magnetic resonance. Biochemistry 51, 9846–9856 10.1021/bi3015532 [DOI] [PubMed] [Google Scholar]
- 70. Keegstra K., Talmadge K. W., Bauer W., and Albersheim P. (1973) The structure of plant cell walls: III. a model of the walls of suspension-cultured sycamore cells based on the interconnections of the macromolecular components. Plant Physiol. 51, 188–197 10.1104/pp.51.1.188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Kubo M., Udagawa M., Nishikubo N., Horiguchi G., Yamaguchi M., Ito J., Mimura T., Fukuda H., and Demura T. (2005) Transcription switches for protoxylem and metaxylem vessel formation. Genes Dev. 19, 1855–1860 10.1101/gad.1331305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. French A. P., Mills S., Swarup R., Bennett M. J., and Pridmore T. P. (2008) Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat. Protoc. 3, 619–628 10.1038/nprot.2008.31 [DOI] [PubMed] [Google Scholar]
- 73. Grebe M., Xu J., Mobius W., Ueda T., Nakano A., Geuze H. J., Rook M. B., and Scheres B. (2003) Arabidopsis sterol endocytosis involves actin-mediated trafficking via ARA6-positive early endosomes. Curr. Biol. 13, 1378–1387 10.1016/S0960-9822(03)00538-4 [DOI] [PubMed] [Google Scholar]
- 74. Kovac P., Glaudemans C. P. J., and Taylor R. B. (1985) An efficient, unambiguous synthesis of methyl 3-O-β-d-galactopyranosyl-β-d-galactopyranoside: further studies on the specificity of antigalactopyranan monoclonal antibodies. Carbohydr. Res. 142, 158–164 10.1016/s0008-6215(00)90746-8 [DOI] [PubMed] [Google Scholar]
- 75. Kovác P., Sokoloski E. A., and Glaudemans C. P. (1984) Synthesis and characterization of methyl 6-O-α- and -β-d-galactopyranosyl-β-d-galactopyranoside. Carbohydr. Res. 128, 101–109 10.1016/0008-6215(84)85088-0 [DOI] [PubMed] [Google Scholar]
- 76. Kaji E., Nishino T., Ishige K., Ohya Y., and Shirai Y. (2010) Regioselective glycosylation of fully unprotected methyl hexopyranosides by means of transient masking of hydroxy groups with arylboronic acids. Tetrahedron Lett. 51, 1570–1573 10.1016/j.tetlet.2010.01.048 [DOI] [Google Scholar]
- 77. Lamport D. T. (2013) Preparation of arabinogalactan glycoproteins from plant tissue. Bioprotocol 3, e918 10.21769/BioProtoc.918 [DOI] [Google Scholar]
- 78. Rende U., Wang W., Gandla M. L., Jönsson L. J., and Niittyla T. (2017) Cytosolic invertase contributes to the supply of substrate for cellulose biosynthesis in developing wood. New Phytol. 214, 796–807 10.1111/nph.14392 [DOI] [PubMed] [Google Scholar]
- 79. Latha Gandla M., Derba-Maceluch M., Liu X., Gerber L., Master E. R., Mellerowicz E. J., and Jönsson L. J. (2015) Expression of a fungal glucuronoyl esterase in Populus: effects on wood properties and saccharification efficiency. Phytochemistry 112, 210–220 10.1016/j.phytochem.2014.06.002 [DOI] [PubMed] [Google Scholar]
- 80. Updegraff D. M. (1969) Semimicro determination of cellulose in biological materials. Anal. Biochem. 32, 420–424 10.1016/S0003-2697(69)80009-6 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are contained within the manuscript.