Abstract
Purpose
Bacterial metabolism regulators offer a novel productive strategy in the eradication of antibiotic refractory bacteria, particularly bacterial persisters. However, the potential of amino acids in the fight against Gram-negative bacterial persisters has not been fully explored. The aim of this study is to investigate the potentiation of amino acids to antibiotics in combating Gram-negative bacterial persisters and to reveal the underlying mechanisms of action.
Methods
Bactericidal activity of antibiotics in the absence or presence of amino acids was evaluated through detecting the reduction of bacterial CFUs. The ratio of NAD+/NADH in E. coli B2 persisters was determined using assay kit with WST-8. Bacterial respiration and ROS production were measured by the reduction of iodonitrotetrazolium chloride and fluorescent probe 2′,7′-dichlorodihydrofluorescein diacetate, respectively.
Results
In this study, we found that cysteine possesses excellent synergistic bactericidal activity with ciprofloxacin against multiple Gram-negative bacterial persisters. Furthermore, the potentiation of cysteine was evaluated in exponential and stationary-phase E. coli ATCC 25922 and E. coli B2. Interestingly, cysteine significantly improves three bactericidal antibiotics killing against stationary-phase bacteria, but not exponential-phase bacteria, implying that the effect of cysteine correlates with the metabolic state of bacteria. Mechanistic studies revealed that cysteine accelerates the bacterial TCA cycle and promotes bacterial respiration and ROS production. These metabolic regulation effects of cysteine re-sensitive bacterial persisters to antibiotic killing.
Conclusion
Collectively, our study highlights the synergistic bactericidal activity of bacterial metabolism regulators such as cysteine with commonly used antibiotics against Gram-negative bacterial persisters.
Keywords: amino acids, bacterial persisters, bactericidal antibiotics, cysteine, gram-negative bacteria
Introduction
Discovery and application of antibiotics are recognized as the cornerstones of modern medicine.1 However, the emergence and prevalence of antibiotic resistance and antibiotic tolerance threaten this therapeutic pipeline in treatment of bacterial infections.2 Different from antibiotic resistance, antibiotic-tolerant bacteria are genetically susceptible but phenotypically resistant to antibiotic treatment.3 In particular, the formation of non-growing subpopulations such as bacterial persisters is one of the important antibiotic tolerance phenotypes, which acts as an underappreciated role in chronic or recurrent infections.4 It has been proposed that some physiological and environmental cues such as starvation, acid stress, quorum sensing, biofilm, and indole signaling could promote persister cells formation.5 Although the accurate molecular mechanisms of persisters formation are still a matter of debate, it has been suggested that these persister cells commonly have low metabolic activity, including decreased tricarboxylic acid (TCA) cycle and proton motive force.6
Up to now, several strategies have been proposed to combat antibiotic resistant pathogens, including discovery of novel antibiotics,7,8 antibiotic adjuvants,9,11 and other alternatives.12 For example, a novel antibiotic termed teixobactin,13 discovered from uncultured bacteria, was found to effectively kill resistant Gram-positive bacteria by inhibiting cell wall synthesis. Recently, a new antimicrobial agent named darobactin14 from a screen of Photorhabdus isolates was discovered, which is active against multiple resistant Gram-negative pathogens. In addition, the discovery of novel antibiotic adjuvants such as Aspergillomarasmine A restores meropenem activity against metallo-β-lactamase-producing pathogens.15 Despite these ongoing effects, these strategies are not applicable for bacterial persisters. It is imperative to develop feasible approaches to tackle bacterial persisters.
Accordingly, the metabolic state of bacteria has been found to closely correlate with their susceptibility to antibiotics.5 Specifically, it has been demonstrated that antibiotic efficacy could be improved through altering bacterial metabolism.16,17 Therefore, we hypothesized that bacterial metabolism regulators may provide a novel strategy in the eradication of antibiotic refractory bacteria. In this study, we investigated the potential of 20 natural amino acids in restoring antibiotic activity against Gram-negative bacterial persisters. Importantly, we found that cysteine could effectively potentiate bactericidal antibiotics activity against Gram-negative bacterial persisters through promoting bacterial respiration and ROS production.
Materials and Methods
Bacterial Strains and Chemical Reagents
The bacterial strains used in this study are listed in Supplementary Table S1. Unless specifically noted, bacteria were grown in Mueller-Hinton broth (MHB) at 37°C at 200 rpm. To obtain different growth phase bacteria, overnight E. coli were diluted 1/1,000 into 5 mL fresh MHB, and grown to exponential-phase (4 hours) or stationary-phase (8 hours). To obtain bacterial persisters, Escherichia coli B2, Escherichia coli ATCC 25922, Salmonella enteritidis ATCC 13076, Acinetobacter baumannii ATCC 19609, and Pseudomonas aeruginosa PA14 were grown to late-stationary phase for 16 hours at 37°C and then treated with ciprofloxacin (10-fold MIC) for 4 hours to kill non-persistent cells, according to a previous study.18 Antibiotics were obtained from China Institute of Veterinary Drug Control and other chemical reagents were purchased from TCI chemicals (Japan).
Minimum Inhibitory Concentration (MIC) Measurements
MICs of different antibiotics were determined using the two-fold serially microdilution method according to the CLSI 2018 guidelines.19 Then, the samples were incubated at 37°C for 18 hours and the MIC values were interpreted as the lowest concentrations of antibiotics with no visible growth of bacteria.
Bactericidal Activity of Antibiotics in the Presence of Amino Acids
To evaluate the effect of different amino acids on the eradication of E. coli B2 persisters, surviving cells after treatment with ciprofloxacin (10-fold MIC) for 4 hours were pelleted and resuspended in M9 minimal medium. Then, persister cells (about 108 CFUs/mL) were treated with ciprofloxacin (10-fold MIC) and 20 amino acids alone (10 mM) or their combination. After 4 hours of treatment, 50 μL samples were removed, serially diluted, and spot-plated onto MH agar plates to determine colony-forming units (CFUs)/mL, and the percentage of surviving cells at 4 hours divided by initial CFUs/mL were calculated. In addition, the synergistic bactericidal activity of cysteine and ciprofloxacin in another four Gram-negative bacterial persisters was evaluated as described above. Meanwhile, the effect of cysteine on three antibiotics (ampicillin, kanamycin, and ciprofloxacin) killing against exponential and stationary-phase E. coli ATCC 25922 and E. coli B2 was determined by measuring the CFUs reduction of E. coli by antibiotic with or without cysteine.
NAD+/NADH Determination
E. coli B2 persisters were collected and diluted into OD600=0.5 in M9. Then, different concentrations of ciprofloxacin (0, 1, 5, and 10-fold MIC) were added with or without cysteine (10 mM). After 4 hours incubation, cell pellets were washed with 0.01 M PBS (pH=7.2) and re-suspended with 200 μL precooled extraction buffer (Beyotime, China). The lysate was centrifuged at 12,000 g for 10 minutes at 4°C, and supernatant was measured using an NAD+/NADH Assay Kit with WST-8 (Beyotime, China).
Bacterial Respiration and ROS Assay
Effect of cysteine on the bacterial respiration was monitored by reduction of iodonitrotetrazolium chloride (INT).20 Briefly, E. coli B2 persisters were diluted into OD600=0.5 in M9, and cysteine (10 mM) was added. Subsequently, 1 mM INT and 0.6 mM NADH were added as substrate and the solution was incubated for 1 hour in the dark. INT reduction was stopped by addition of 5% trichloroacetic acid. Insoluble formazan was centrifuged for 5 minutes at 13,000 g, extracted with 1 mL ethanol. Absorbance of the supernatant at 485 nm was measured during 60 minutes.
2′,7′-dichlorodihydrofluorescein diacetate21 (DCFH-DA, 10 μM) were added into E. coli B2 persisters suspension. After being incubated at 37°C for 30 minutes, 190 μL of probe-labeled bacterial cells were added to a 96-well plate and 10 μL of ciprofloxacin (0, 1, 5, and 10-fold MIC) without or with cysteine (10 mM) were added. After incubation at 37°C for 1 hour, fluorescence units were immediately measured with the excitation wavelength at 488 nm and emission wavelength at 525 nm using an Infinite M200 Microplate reader (Tecan, Switzerland).
Results and Discussion
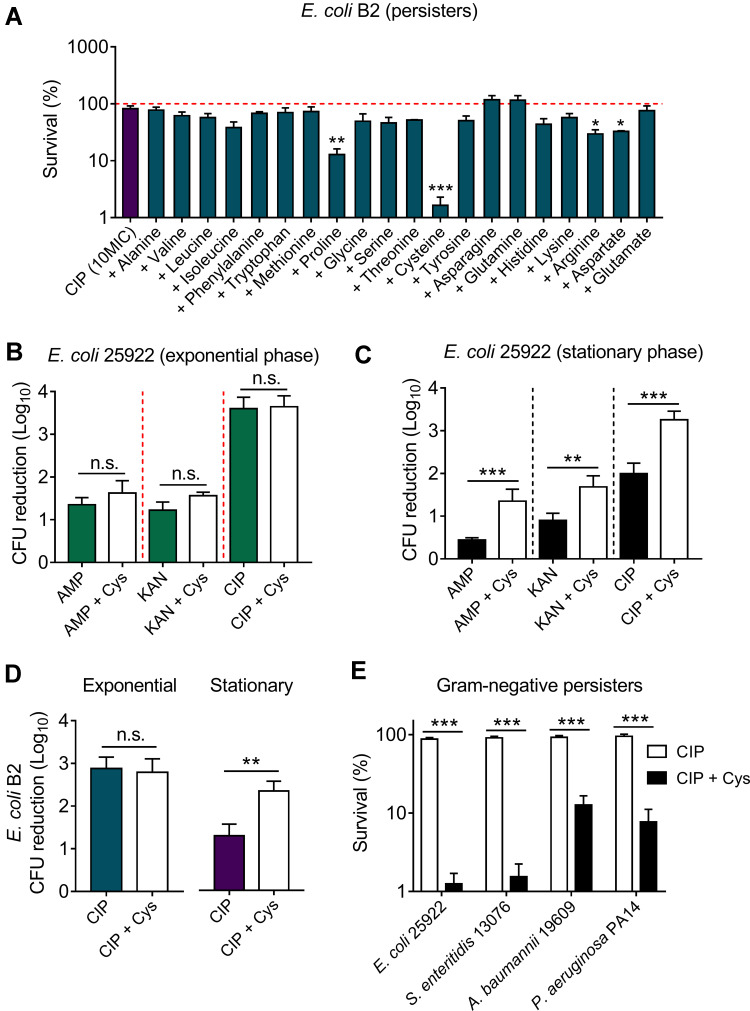
We used a multidrug-resistant (MDR) isolate E. coli B2 as an indicative strain, which is resistant to almost all commonly used antibiotics expect tigecycline.22,24 To explore the effect of 20 amino acids on ciprofloxacin killing against bacterial persisters, E. coli B2 persisters were treated with ciprofloxacin (10-fold MIC) and 20 amino acids alone or their combinations. After 4 hours treatment, the percentage of surviving E. coli B2 cells divided by initial bacteria were calculated. We found that above 85% cells survived after exposing ciprofloxacin alone (Figure 1A, first column) or 20 amino acids alone (Supplementary Figure S1), indicating that ciprofloxacin or amino acids monotreatment displayed weak bactericidal activity against E. coli B2 persisters. In combinational treatment groups (Figure 1A), the results showed that the vast majority of amino acids such as alanine and valine have no remarkable effect on ciprofloxacin killing. By contrast, two amino acids involve asparagine and glutamine modestly impaired ciprofloxacin activity and promoted bacterial replicate. Only four amino acids including proline (non-polar amino acid, pI=6.30), cysteine (polar amino acid, pI=5.02), arginine (cationic amino acid, pI=10.76), and aspartate (anionic amino acid, pI=2.97) showed a significant synergistic bactericidal activity with ciprofloxacin in the elimination of E. coli B2 persisters, implying that the potentiation potency of amino acids is independent of their physicochemical properties. Among these four amino acids, cysteine showed the best potentiation with an above 99% eradication rate of E. coli B2 persisters.
Figure 1.
Cysteine potentiates ciprofloxacin killing against stationary-phase bacteria and persisters, but not exponential-phase bacteria. (A) Effect of 20 L-amino acids (10 mM) on ciprofloxacin killing against E. coli B2 persisters. The initial persisters counts (CFUs/mL) were defined as 100% (red line). (B and C) Addition of cysteine (10 mM) has no effect on three bactericidal antibiotics activity in exponential phase (4 hours) antibiotic-sensitive E. coli ATCC 25922, whereas it significantly potentiates antibiotic activity against stationary phase bacteria (8 hours). (D) Cysteine (10 mM) improves ciprofloxacin activity against multi-drug resistant E. coli B2 in the stationary phase, but not in the exponential phase. (E) Cysteine improves ciprofloxacin killing against multiple Gram-negative bacterial persisters. All data were obtained in three independent experiments and are shown as mean±SD. n.s., not significant, *P<0.05, **P<0.01, ***P<0.001, determined by unpaired t-test.
Abbreviations: Cys, cysteine; AMP, ampicillin; KAN, kanamycin; CIP, ciprofloxacin.
Having shown the potentiation of cysteine in the fight against E. coli B2 persisters, we next evaluated the synergistic bactericidal activity of cysteine with three antibiotics from different classes and modes of action against non-persisters, including exponential-phase and stationary-phase bacteria of E. coli ATCC 25922 or E. coli B2. Intriguingly, we found that the addition of cysteine could not enhance the sterilization effect of ampicillin (cell wall), kanamycin (protein), and ciprofloxacin (DNA synthesis) against exponential-phase E. coli ATCC 25922 (Figure 1B). However, a significant bacterial CFUs reduction after treatment with the combination of antibiotics with cysteine was found in stationary-phase E. coli 25922 (Figure 1C). For MDR clinical strain E. coli B2, a similar synergistic effect in stationary-phase bacteria but not in exponential-phase bacteria were also observed (Figure 1D). In addition, we found that three antibiotics exhibited better bactericidal effects against exponential-phase bacteria than stationary-phase bacteria. This observation was consistent with the previous notion that stationary-phase bacteria are harder to treat, because most of the stationary-phase bacteria are in a low-growth state due to the limitation of nutrients and the increase of toxic metabolites.25 Similarly, bacterial persisters are commonly characterized by quiescent and slow-growing bacteria.26 We next evaluated the potentiation activity of cysteine with ciprofloxacin in other notorious Gram-negative bacterial persisters, including E. coli 25922, S. enteritidis 13076, A. baumannii 19609, and P. aeruginosa PA14. As expected, remarkably reduced bacterial persisters in combinational treatment were observed (Figure 1E), indicating the potentiation potency of cysteine in improving antibiotics activity against Gram-negative bacterial persisters. Taking these results together, we concluded that the potentiation of cysteine with antibiotics is efficacious for inactive bacteria such as stationary-phase pathogens and bacterial persisters, but ineffective for those bacteria that are already active, such as exponential-phase bacteria.
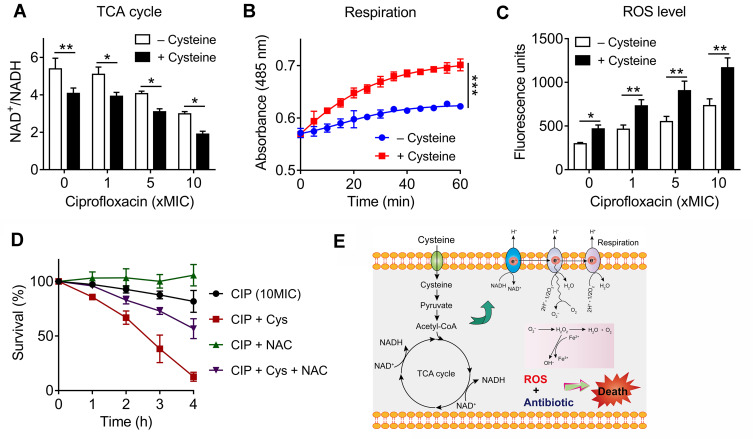
Previous studies have shown that drug-drug interactions may contribute to the improve specific drugs activity.27 To evaluate whether there is a drug-drug interaction between cysteine and antibiotics, the MICs of antibiotics in the absence and presence of cysteine (10 mM) against E. coli ATCC25922 and E. coli B2 were compared. However, we did not observe a decreased MIC values in the presence of cysteine, suggesting no direct drug-drug interaction between cysteine and antibiotics (Supplementary Table S2). Given the potentiation of cysteine is highly dependent on the metabolic states of bacteria, but independent of antibiotic types and their modes of action, we hypothesized that cysteine may specifically enhance bacterial metabolism and thereby restore antibiotics activity. In addition, cysteine could be metabolized to pyruvate and further converted into acetyl-coenzyme A,28 which is the major upstream input for the TCA cycle. Thus, we first investigated whether the addition of cysteine affects the TCA cycle, which is recognized as the hub of bacterial substance and energy metabolism.29 In this progress, NAD is reduced to NADH by a series of redox reactions, thus the production of NADH and the ratio of NAD+/NADH is an important index to characterize TCA cycle. Thus, we monitored the NAD+/NADH ratio of E. coli B2 persisters after treatment with increasing concentrations of ciprofloxacin alone or in combination with cysteine. We found that cysteine drastically decreased the bacteria NAD+/NADH ratio with or without ciprofloxacin (Figure 2A), indicating that cysteine could accelerate the bacterial TCA cycle. In agreement with our observations, a previous report has also showed that carbon sources such as fumarate could potentiate tobramycin activity by stimulating the TCA cycle.30 In contrast, inactivation of the TCA cycle promotes persister cells formation in multiple pathogens, including S. aureus and E. coli.31
Figure 2.
Exogenous cysteine promotes bacterial respiration and production of reactive oxygen species (ROS). (A) Exogenous cysteine (10 mM) decreases the ratio of NAD+/NADH in the absence or presence of ciprofloxacin. (B) Enhanced bacterial respiration by exogenous cysteine (10 mM) during 60 minutes, determined by iodonitrotetrazolium chloride, a reduction-sensitive dye. (C) Addition of cysteine significantly triggers the production of ROS with or without ciprofloxacin, probed by 2′,7′-dichlorodihydrofluorescein diacetate (DCFH-DA). (D) ROS scavenger NAC counteracts the potentiation activity of cysteine with ciprofloxacin against E. coli B2 persisters. NAC, N-acetylcysteine. (E) Scheme of potentiation mechanisms of cysteine. Cysteine might be metabolized to pyruvate and further entry into the TCA cycle, then promotes the NADH production and bacterial respiration, triggers ROS production (partly from oxidation of cysteine), and thereby improves bactericidal lethality against bacterial persisters. All data in (A–D) were obtained in three independent experiments and areshown as mean±SD. *P<0.05, **P<0.01, ***P<0.001, determined by unpaired t-test.
To explore whether the increased NADH from the TCA cycle would promote bacterial respiration, we used a reduction-sensitive dye named iodonitrotetrazolium chloride to monitor E. coli B2 persisters respiration in the absence or presence of cysteine. Consistently, a significant enhanced bacterial respiration by cysteine during 60 minutes was observed (Figure 2B). It has been demonstrated that enhanced respiration prevents drug tolerance in Mycobacterium tuberculosis and ultimately leads to mycobacterial cell death by isoniazid.32 Increased bacterial respiration is always accompanied with generation of reactive oxygen species (ROS). In addition, previous studies have demonstrated that cysteine can be oxidized and accompanied by the generation of ROS due to the presence of its thiol group.33,35 Thus, we reasoned that the addition of cystine may induce a large amount of ROS production through two different means. To test this, we next determined the ROS production of E. coli B2 persisters after treatment with ciprofloxacin and cysteine alone or their combination. Consistent with our hypothesis, cysteine alone triggered more generation of ROS than four other amino acids (Supplementary Figure S2). Most importantly, the combination of cysteine and ciprofloxacin drastically enhanced ROS production compared with ciprofloxacin alone (Figure 2C). Despite some controversies, it has been widely acknowledged that ROS mediated killing is more or less important for bactericidal antibiotics.36,37 To explore whether the enhanced ROS by cysteine play a critical role in its potentiation activity, we performed a time-killing curve of E. coli B2 persisters in the presence of ciprofloxacin and in combination with cysteine and/or N-acetylcysteine (NAC; ROS scavenger). We found that the addition of NAC not only abolished the weak bactericidal activity of ciprofloxacin but also counteracted the potentiation of cysteine to ciprofloxacin (Figure 2D), demonstrating that ROS is indispensable in the synergistic activity of cysteine with bactericidal antibiotics against Gram-negative bacterial persisters.
Conclusion
In conclusion, we reveal the important values of amino acids such as cysteine in assisting bactericidal antibiotics to eliminate bacterial persisters. Specifically, cysteine effectively potentiates multiple bactericidal antibiotics killing in the fight against various Gram-negative bacterial persisters through stimulating bacterial respiration and triggering the production of ROS, and converting persister cells to metabolically active cells (Figure 2E). These findings suggest that bacterial metabolism regulators may be an important pipeline to treat the growing infections caused by bacterial persisters in the clinical setting. Further investigations are still required to provide a better understanding of their molecular mechanisms and verify the efficacy of this combinational treatment in vivo.
Acknowledgments
This work was supported by the National Key Research and Development Program of China (2018YFA0903400), National Natural Science Foundation of China (31872526), Natural Science Foundation of Jiangsu Province of China (BK20190893), China Postdoctoral Science Foundation funded project (2019M651984), a project funded by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD) and Lift Engineering of Young Talents of Jiangsu Association for Science and Technology.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325(5944):1089–1093. doi: 10.1126/science.1176667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Povolo VR, Ackermann M. Disseminating antibiotic resistance during treatment. Science. 2019;364(6442):737–738. doi: 10.1126/science.aax6620 [DOI] [PubMed] [Google Scholar]
- 3.Fridman O, Goldberg A, Ronin I, et al. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature. 2014;513(7518):418–421. doi: 10.1038/nature13469 [DOI] [PubMed] [Google Scholar]
- 4.Fisher RA, Gollan B, Helaine S. Persistent bacterial infections and persister cells. Nat Rev Microbiol. 2017;15(8):453. doi: 10.1038/nrmicro.2017.42 [DOI] [PubMed] [Google Scholar]
- 5.Stokes JM, Lopatkin AJ, Lobritz MA, et al. Bacterial metabolism and antibiotic efficacy. Cell Metab. 2019;30:251–259. doi: 10.1016/j.cmet.2019.06.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crabbé A, Jensen PØ, Bjarnsholt T, et al. Antimicrobial tolerance and metabolic adaptations in microbial biofilms. Trends Microbiol. 2019;27(10):850–863. doi: 10.1016/j.tim.2019.05.003 [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Ding S, Shen J, et al. Nonribosomal antibacterial peptides that target multidrug-resistant bacteria. Nat Prod Rep. 2019;36:573–592. doi: 10.1039/C8NP00031J [DOI] [PubMed] [Google Scholar]
- 8.Brown ED, Wright GD. Antibacterial drug discovery in the resistance era. Nature. 2016;529(7586):336–343. doi: 10.1038/nature17042 [DOI] [PubMed] [Google Scholar]
- 9.Liu Y, Li R, Xiao X, et al. Antibiotic adjuvants: an alternative approach to overcome multi-drug resistant Gram-negative bacteria. Crit Rev Microbiol. 2019;45(3):301–314. doi: 10.1080/1040841X.2019.1599813 [DOI] [PubMed] [Google Scholar]
- 10.Liu Y, Li R, Xiao X, et al. Molecules that inhibit bacterial resistance enzymes. Molecules. 2019;24:43. doi: 10.3390/molecules24010043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wright GD. Antibiotic adjuvants: rescuing antibiotics from resistance. Trends Microbiol. 2016;24(11):862–871. doi: 10.1016/j.tim.2016.06.009 [DOI] [PubMed] [Google Scholar]
- 12.Czaplewski L, Bax R, Clokie M, et al. Alternatives to antibiotics—a pipeline portfolio review. Lancet Infect Dis. 2016;16(2):239–251. doi: 10.1016/S1473-3099(15)00466-1 [DOI] [PubMed] [Google Scholar]
- 13.Ling LL, Schneider T, Peoples AJ, et al. A new antibiotic kills pathogens without detectable resistance. Nature. 2015;517(7535):455–459. doi: 10.1038/nature14098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imai Y, Meyer KJ, Iinishi A, et al. A new antibiotic selectively kills Gram-negative pathogens. Nature. 2019;576(7787):459–464. doi: 10.1038/s41586-019-1791-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.King AM, Reid-Yu SA, Wang WL, et al. Aspergillomarasmine A overcomes metallo-beta-lactamase antibiotic resistance. Nature. 2014;510(7506):503–506. doi: 10.1038/nature13445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, Li R, Xiao X, et al. Bacterial metabolism-inspired molecules to modulate antibiotic efficacy. J Antimicrob Chemother. 2019;74:3409–3417. doi: 10.1093/jac/dkz230 [DOI] [PubMed] [Google Scholar]
- 17.Yang JH, Wright SN, Hamblin M, et al. A white-box machine learning approach for revealing antibiotic mechanisms of action. Cell. 2019;177:1649–1661 e1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morones-Ramirez JR, Winkler JA, Spina CS, et al. Silver enhances antibiotic activity against Gram-negative bacteria. Sci Transl Med. 2013;5:190ra181. doi: 10.1126/scitranslmed.3006276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing. CLSI; 2018. [Google Scholar]
- 20.Wenzel M, Chiriac AI, Otto A, et al. Small cationic antimicrobial peptides delocalize peripheral membrane proteins. Proc Natl Acad Sci USA. 2014;111(14):E1409–E1418. doi: 10.1073/pnas.1319900111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen X, Zhong Z, Xu Z, et al. 2′, 7′-Dichlorodihydrofluorescein as a fluorescent probe for reactive oxygen species measurement: forty years of application and controversy. Free Radical Res. 2010;44:587–604. doi: 10.3109/10715761003709802 [DOI] [PubMed] [Google Scholar]
- 22.Liu Y, Yang K, Jia Y, et al. Repurposing peptidomimetic as potential inhibitor of New Delhi metallo-beta-lactamases in Gram-negative bacteria. ACS Infect Dis. 2019;5:2061–2066. doi: 10.1021/acsinfecdis.9b00364 [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Jia Y, Yang K, et al. Metformin restores tetracyclines susceptibility against multidrug resistant bacteria. Adv Sci. 2020;7(12):1902227. doi: 10.1002/advs.201902227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Song M, Liu Y, Huang X, et al. A broad-spectrum antibiotic adjuvant reverses multidrug-resistant Gram-negative pathogens. Nat Microbiol. 2020. doi: 10.1038/s41564-020-0723-z [DOI] [PubMed] [Google Scholar]
- 25.Navarro Llorens JM, Tormo A, Martínez-García E. Stationary phase in Gram-negative bacteria. FEMS Microbiol Rev. 2010;34(4):476–495. doi: 10.1111/j.1574-6976.2010.00213.x [DOI] [PubMed] [Google Scholar]
- 26.Maisonneuve E, Gerdes K. Molecular mechanisms underlying bacterial persisters. Cell. 2014;157:539–548. doi: 10.1016/j.cell.2014.02.050 [DOI] [PubMed] [Google Scholar]
- 27.Tyers M, Wright GD. Drug combinations: a strategy to extend the life of antibiotics in the 21st century. Nat Rev Microbiol. 2019;17(3):141–155. doi: 10.1038/s41579-018-0141-x [DOI] [PubMed] [Google Scholar]
- 28.Pietrocola F, Galluzzi L, Bravo-San Pedro JM, et al. Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 2015;21:805–821. doi: 10.1016/j.cmet.2015.05.014 [DOI] [PubMed] [Google Scholar]
- 29.Huynen MA, Dandekar T, Bork P. Variation and evolution of the citric-acid cycle: a genomic perspective. Trends Microbiol. 1999;7(7):281–291. doi: 10.1016/S0966-842X(99)01539-5 [DOI] [PubMed] [Google Scholar]
- 30.Meylan S, Porter CBM, Yang JH, et al. Carbon sources tune antibiotic susceptibility in Pseudomonas aeruginosa via tricarboxylic acid cycle control. Cell Chem Biol. 2017;24:195–206. doi: 10.1016/j.chembiol.2016.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Y, Bojer MS, George SE, et al. Inactivation of TCA cycle enhances Staphylococcus aureus persister cell formation in stationary phase. Sci Rep. 2018;8:1–13. doi: 10.1038/s41598-017-17765-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vilchèze C, Hartman T, Weinrick B, et al. Enhanced respiration prevents drug tolerance and drug resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci USA. 2017;114(17):4495–4500. doi: 10.1073/pnas.1704376114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van der Reest J, Lilla S, Zheng L, et al. Proteome-wide analysis of cysteine oxidation reveals metabolic sensitivity to redox stress. Nat Commun. 2018;9:1581. doi: 10.1038/s41467-018-04003-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ezraty B, Gennaris A, Barras F, et al. Oxidative stress, protein damage and repair in bacteria. Nat Rev Microbiol. 2017;15(7):385–396. doi: 10.1038/nrmicro.2017.26 [DOI] [PubMed] [Google Scholar]
- 35.Park S, Imlay JA. High levels of intracellular cysteine promote oxidative DNA damage by driving the Fenton reaction. J Bacteriol. 2003;185:1942–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Acker HV, Coenye T. The role of reactive oxygen species in antibiotic-mediated killing of bacteria. Trends Microbiol. 2017;25(6):456–466. doi: 10.1016/j.tim.2016.12.008 [DOI] [PubMed] [Google Scholar]
- 37.Hong Y, Li L, Luan G, et al. Contribution of reactive oxygen species to thymineless death in Escherichia coli. Nat Microbiol. 2017;2(12):1667. doi: 10.1038/s41564-017-0037-y [DOI] [PMC free article] [PubMed] [Google Scholar]