Abstract
Alternative splicing plays a pivotal role in modulating the function of eukaryotic proteins. In the inner ear, many genes undergo alternative splicing, and errors in this process lead to hearing loss. Cadherin 23 (CDH23) forms part of the so-called tip links, which are indispensable for mechanoelectrical transduction (MET) in the hair cells. Cdh23 gene contains 69 exons, and exon 68 is subjected to alternative splicing. Exon 68 of the Cdh23 gene is spliced into its mRNA only in a few cell types including hair cells. The mechanism responsible for the alternative splicing of Cdh23 exon 68 remains elusive. In the present work, we performed a cell-based screening to look for splicing factors that regulate the splicing of Cdh23 exon 68. RBM24 and RBM38 were identified to enhance the inclusion of Cdh23 exon 68. The splicing of Cdh23 exon 68 is affected in Rbm24 knockdown or knockout cells. Moreover, we also found that PTBP1 inhibits the inclusion of Cdh23 exon 68. Taken together, we show here that alternative splicing of Cdh23 exon 68 is regulated by RBM24, RBM38, and PTBP1.
1. Introduction
In eukaryotic cells, the exons of precursor mRNAs (pre-mRNAs) are spliced together by a large protein complex named spliceosome to give rise to mature mRNAs [1]. Many genes undergo alternative splicing, which results in distinct mature mRNAs through splicing different exons together [2]. In this way, alternative splicing helps to produce structurally and functionally similar but not identical protein isoforms, hence contributing to proteomic diversity [3]. Tissue-specific alternative splicing also contributes to tissue specificity. Alternative splicing is regulated by specific RNA sequences and related RNA-binding proteins [2]. The RNA sequences that regulate alternative splicing are divided into different classes according to their position and function, namely, exonic splicing enhancers (ESEs), exonic splicing silencers (ESSs), intronic splicing enhancers (ISEs), and intronic splicing silencers (ISSs), which are bound by different splicing regulators [2]. Tissue-specific alternative splicing is thought to be regulated mainly by differentially expressed splicing regulators [4, 5].
In the inner ear, many important genes have been shown to undergo alternative splicing, and dysregulation of this process causes syndromic or nonsyndromic hearing loss [6]. Cadherin 23 (CDH23) is an atypical cadherin, forming part of the so-called tip links that play a pivotal role in mechanoelectrical transduction (MET) in the hair cells [7–10]. Mouse Cdh23 gene contains 69 exons, and exon 68 is subjected to alternative splicing [11]. Interestingly, the inclusion of Cdh23 exon 68 is only detected in the inner ear so far [8, 12, 13]. Cdh23 exon 68 is 105 base pair (bp) long, which encodes 35 amino acids (aa) in the cytoplasmic domain of CDH23. The exon 68-encoded peptide shows no homology to any known proteins and might affect the conformation as well as protein-protein interactions of CDH23 [12–16]. At present, the mechanism responsible for the inner ear-specific alternative splicing of Cdh23 exon 68 is still unknown.
In the present work, we performed a cell-based screening in order to identify the splicing factors that regulate the alternative splicing of Cdh23 exon 68. The screening identified RBM24 and RBM38 that enhance the inclusion of Cdh23 exon 68. Moreover, we also identified PTBP1 that inhibits the inclusion of Cdh23 exon 68 through bioinformatics analysis. Our data suggest that RBM24, RBM38, and PTBP1 together regulate the alternative splicing of Cdh23 exon 68.
2. Materials and Methods
2.1. Reverse Transcription-Polymerase Chain Reaction (RT-PCR)
Total RNA from different mouse tissues or cultured cells was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. Briefly, 1 μg RNA was used for reverse transcription (RT) using PrimeScript RT Reagent Kit with gDNA Eraser (Takara, RR047A). Polymerase chain reaction (PCR) was then performed using the cDNA as template with the following primers: mouse Esrp1 forward 5′-CAATATTGCCAAGGGTGGCG-3′, reverse 5′-CTCATGCGCACGATGACTTG-3′; mouse Rbm24 forward 5′-GAGACATCGAGGAAGCGGTG-3′, reverse 5′-TCTGGATAAGGGCTGGGTGA-3′; mouse Rbm38 forward 5′-ATGGCAGATCGGGCAGC-3′, reverse 5′-AGCCCGTCTGTAAGCTCCTA-3′; mouse Ptbp1 forward 5′-AGTGCGCATTACACTGTCCA-3′, reverse 5′-CTTGAGGTCGTCCTCTGACA-3′; mouse Ptbp2 forward 5′-GCAGAAGAGGATCTGCGAAC-3′, reverse 5′-CATCTTCATCTCCCGTGCTT-3′; mouse β-actin forward 5′-CGTTGACATCCGTAAAGACC-3′, reverse 5′-AACAGTCCGCCTAGAAGCAC-3′; mouse Cdh23 forward 5′-GACAACATCGCCAAGCTG-3′, reverse (for minigene) 5′-GGCCAGCAGTGTGCC-3′; reverse (for endogenous Cdh23) 5′-GCAAGCTGTTGAGATCAGTGG-3′; and rat Cdh23 forward 5′-TGGAACTTTTGGACGTGAGC-3′, reverse 5′-GCTTGTGTCGGAAACGGAGG-3′. For different PCR reactions, 23-40 cycles were used and the annealing temperatures were adjusted between 56 and 64°C to obtain the optimal sensitivity and specificity.
2.2. DNA Constructs
The genomic DNA ranging from exon 67 to exon 69 of mouse Cdh23 gene was PCR amplified and inserted into pmCherry-N1 vector to construct Cdh23 minigene reporter. Mutation or deletion was introduced into the minigene through overlapping PCR-mediated site-directed mutagenesis. The cDNAs encoding various mouse splicing factors were inserted into pEGFP-C2 or pEGFP-N2 to express GFP-fusion proteins.
2.3. Splicing Factor Screening
COS7 cells were transfected with the Cdh23 minigene reporter and vectors expressing different splicing factors using LipoMax (Sudgen) in 24-well plates. Forty-eight hours after transfection, cells were washed with PBS and fixed with 4% paraformaldehyde (PFA) in PBS for 15 minutes. Florescence was detected using a confocal microscope (LSM 700, Zeiss). ImageJ software was used to quantify the intensity of fluorescence.
2.4. RNA Interference and Quantitative Real-Time PCR (qPCR)
Chemically synthesized siRNAs were obtained from Sigma-Aldrich, and their sequences are as follows: siRNA-1, 5′-GCAAUAUGUAGCUUGAAUUdTdT-3′; siRNA-2, 5′-CUUAAGGCCUAUAGAACUUdTdT-3′; siRNA-3, 5′-CCCAAAGAGCCUGAGUAAAdTdT-3′; and control siRNA, 5′-UUCUCCGAACGUGUCACGUTT-3′. COS7 cells were transfected with siRNAs using jetPRIME transfection reagent (Polyplus) in 12-well plates. Twenty-four hours after transfection, the Cdh23 minigene reporter was transfected into the same cells using LipoMax transfection reagents. Cells were cultured for another twenty-four hours, and RNA was extracted and reversed transcribed as described above. qPCR was then performed with SYBR Premix Ex Taq (Takara, RR420A) and the QuantGene 9600 real-time system (Bioer). Sequences of primers used in the qPCR reactions are as follows: monkey Rbm24 forward 5′-AGATCGAGGAGGCGGTGGTC-3′, reverse 5′-TCTTTGTATAAGGGCTGGATGAAG-3′; mouse Cdh23(+68) forward 5′-AACTCTTTGCACAACGGATG-3′, reverse 5′-ATGGGTGGCTTGTGTCGG-3′; and monkey β-actin forward 5′-CGTGGACATCCGCAAAGACC-3′, reverse 5′-CACAGTCCGCCTAGAAGCA-3′.
2.5. RNA Immunoprecipitation (RIP)
RIP analysis was carried out according to the standard procedure with modifications [17]. Briefly, COS7 cells were transfected with expression vectors of Myc-tagged splicing factors as well as the Cdh23 minigene. Forty-eight hours after transfection, cells were lysed in lysis buffer containing 100 mM KCl, 5 mM MgCl2, 10 mM HEPES-NaOH, 0.5% NP-40, 1 mM DTT, 200 units/ml RNase inhibitor (Takara, 2313A), and EDTA-free protease inhibitor cocktail (Sigma, S8830). After centrifugation at 4°C, the supernatant was collected and incubated with immobilized anti-Myc antibody (Sigma-Aldrich, Cat. No. E6654) at 4°C for 3 hours. The immunoprecipitated RNA was then used as template for RT-PCR and qPCR analysis. Primers used are as follows: mouse Cdh23 pre-mRNA forward 5′-CAGCAGCCTAAGTGGGAAG-3′ and reverse 5 ′-AGCTCCCGAGGCTACCTC-3 ′.
2.6. Western Blot
COS7 cells were transfected with expression vectors in 6-well plates. Forty-eight hours after transfection, cells were washed with PBS and lysed in ice-cold lysis buffer containing 150 mM NaCl, 50 mM Tris at pH 7.5, 1% Triton X-100, and 1 mM PMSF. After centrifugation at 4°C, the supernatant was collected and separated by 10% polyacrylamide gel electrophoresis (PAGE), then transferred to PVDF membrane. The membrane was blocked in 5% nonfat milk for an hour at room temperature, then incubated with anti-GFP antibody (ABclonal, AE012) at 4°C overnight, followed by incubation with HRP-conjugated secondary antibody (Bio-Rad, 170-6516) for an hour at room temperature. The signals were detected with the ECL system (Cell Signaling Technology).
2.7. Animals
All animal experiments were approved by the Ethics Committee of Shandong University School of Life Sciences and conducted accordingly. Construction of Atoh1-cre; Rbm24flox/flox mice is described elsewhere.
2.8. Statistical Analysis
Each experiment was repeated at least three times. Student's t-test was used to determine the statistical significance, and p < 0.05 was considered statistically significant. For alternative splicing events, data were shown as means ± SEM.
3. Results
3.1. Screening Splicing Factors That Regulate the Alternative Splicing of Cdh23 Exon 68
To identify splicing factors that regulate the inner ear-specific alternative splicing of Cdh23 exon 68, we started with testing the known splicing factors that are expressed in the inner ear. Splicing factors SRRM4, SFSWAP, and ESRP1 play important roles in the inner ear, albeit none of them has been shown to regulate the alternative splicing of Cdh23 exon 68 [18–20]. We previously showed that deafness-related protein ILDR1 and its homolog ILDR2 regulate alternative splicing through binding to splicing factors TRA2A, TRA2B, and SRSF1 [21]. We also found that RNA-binding protein RBM24 is specifically expressed in the hair cells [22]. The expression of these splicing factors in the inner ear is confirmed by performing RT-PCR (Figure 1(a), data not shown). These splicing factors are used as the candidates for the following screening.
Figure 1.
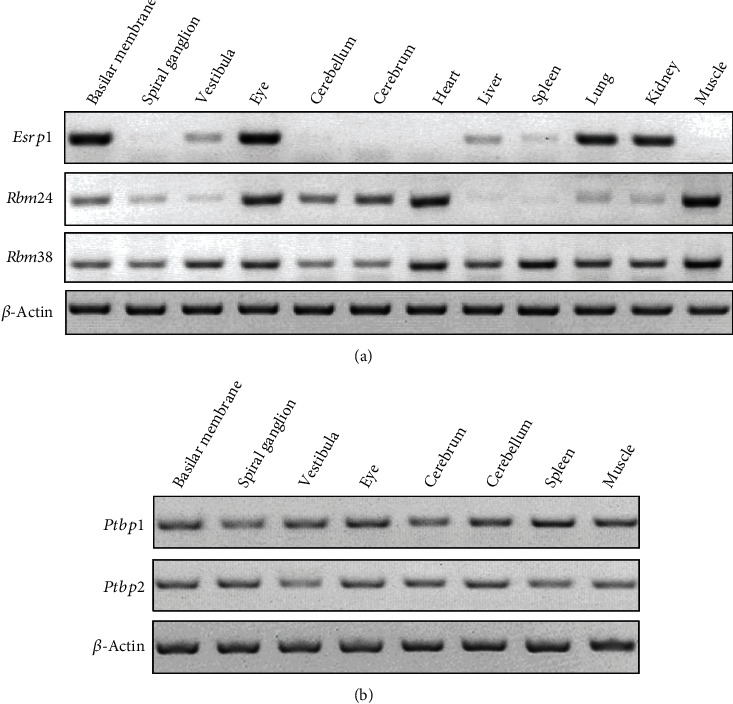
Expression of various splicing regulators in different mouse tissues. RT-PCR analysis was performed to examine the expression of splicing regulators in different tissues of P10 C57BL/6 mice. (a) Expression of Esrp1, Rbm24, and Rbm38 was examined by RT-PCR. (b) Expression of Ptbp1 and Ptbp2 was examined by RT-PCR. β-Actin was used as internal control.
A Cdh23 minigene reporter was constructed by fusing mouse Cdh23 genomic sequence ranging from exon 67 to exon 69 in frame with a mCherry-encoding sequence whose expression is driven by a CMV promoter (Figure 2(a)). To facilitate the screening, we introduced a single base pair insertion in exon 68 and a single base pair deletion in exon 69. When exon 68 is included in the mature mRNA, the insertion and deletion cancel each other out and a mCherry fusion protein is expressed. However, when exon 68 is not included in the mature mRNA, the single base pair deletion in exon 69 will cause a frameshift and mCherry fusion protein will not be expressed (Figure 2(a)). This minigene was then used as the reporter to screen splicing factors that regulate the alternative splicing of Cdh23 exon 68.
Figure 2.
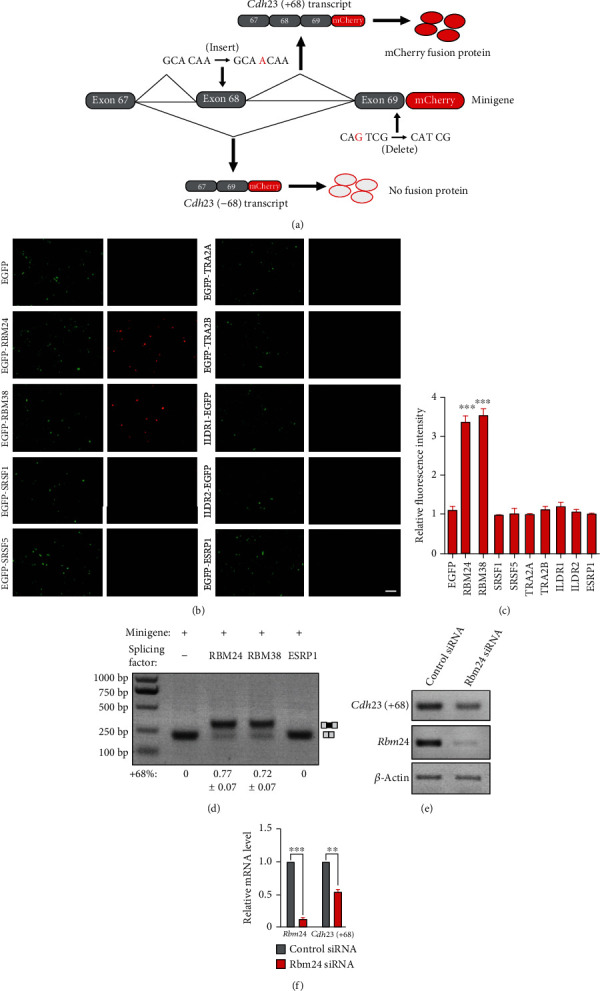
Screening splicing factors that regulate Cdh23 exon68 inclusion. (a) Schematic drawing of the screening strategy. The Cdh23 minigene reporter was constructed by fusing mouse Cdh23 genomic sequence ranging from exon 67 to exon 69 in frame with a mCherry-encoding sequence. A single base pair insertion in exon 68 and a single base pair deletion in exon 69 introduced in the minigene are indicated in red. When exon 68 is included in the mature mRNA, a mCherry fusion protein is expressed. In contrast, when exon 68 is not included in the mature mRNA, the single base pair deletion in exon 69 will cause a frameshift and mCherry fusion protein will not be expressed. (b) The Cdh23 minigene and various splicing factors were overexpressed in COS7 cells, and the resultant fluorescence was examined using a confocal microscope. Scale bar, 200 μm. (c) The relative mCherry fluorescence intensity from (b) was analyzed using ImageJ software. (d) The Cdh23 minigene and various splicing factors were overexpressed in COS7 cells, and RT-PCR was performed to examine the inclusion of exon 68. (e, f) The Cdh23 minigene and either Rbm24 or control siRNAs were transfected into COS7 cells, and the expression level of Rbm24 and Cdh23(+68) was examined by performing RT-PCR (e) and qPCR (f). β-Actin was used as internal control. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
COS7 cells were transfected with the Cdh23 minigene reporter and vectors that express different candidate splicing factors fused with GFP. Among all the splicing factors tested, RBM24 and its homolog RBM38 result in robust mCherry fluorescence (Figure 2(b)). Quantification of the red fluorescence using ImageJ shows significant increases in RBM24- or RBM38-exressing cells, but not in cells expressing other splicing factors (Figure 2(c)). This result suggests that RBM24 and RBM38 are able to promote Cdh23 exon 68 inclusion.
3.2. RBM24 and RBM38 Enhance the Inclusion of Cdh23 Exon 68
RT-PCR was then performed to confirm the screening result. The Cdh23 minigene was expressed in COS7 cells together with RBM24, RBM38, ESRP1, or an empty vector. RT-PCR reveals that exon 68 inclusion is not detectable in mature Cdh23 mRNA in control cells transfected with an empty vector or ESRP1. However, exon 68 inclusion is significantly enhanced by either RBM24 or RBM38 overexpression (Figure 2(d)).
Small interfering RNAs (siRNAs) were then used to knock down the expression of endogenous Rbm24 in COS7 cells. All three siRNAs tested inhibit Rbm24 expression efficiently (Figures 2(e) and 2(f), data not shown). Without overexpressing the splicing enhancers, exon 68 inclusion from Cdh23 minigene is too weak to be detected with regular RT-PCR (Figure 2(d)), possibly because of the relatively low expression level of endogenous splicing enhancers in COS7 cells. We then used primers specific for Cdh23(+68) isoform to examine the inclusion of exon 68. RT-PCR and qPCR results show that Cdh23(+68) expression level is significantly decreased by Rbm24 knockdown (Figures 2(e) and 2(f)). Taken together, both overexpression and knockdown experiments confirm that RBM24/RBM38 could regulate the alternative splicing of Cdh23 exon 68.
RBM24 and RBM38 have been shown to regulate alternative splicing through binding to a GUGUG motif in the ISEs [23, 24]. There are two potential GTGTG motifs located in intron 68 of the Cdh23 gene. Motif 1 (TGTGTG) is located 30 bp downstream of Cdh23 exon 68, while motif 2 (GTGTGGT) is located 630 bp downstream of exon 68 (Figure 3(a)). We examined whether these motifs are involved in the alternative splicing of Cdh23 exon 68 through deleting either of them in the minigene. Both fluorescence assay and RT-PCR results show that exon 68 inclusion enhancement by RBM24/RBM38 is completely abolished by the deletion of motif 1 but not motif 2 (Figures 3(b) and 3(c)), indicating that motif 1 is required for RBM24/RBM38-regulated inclusion of Cdh23 exon 68.
Figure 3.
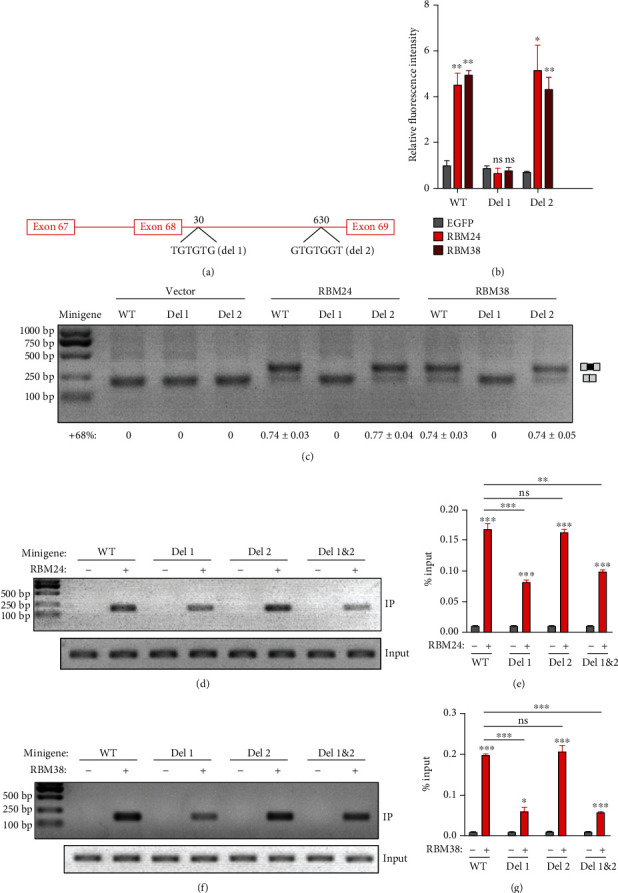
An ISE is required for RBM24-/RBM38-mediated Cdh23 exon 68 inclusion. (a) Schematic drawing of the locations of the two potential RBM24/RBM38-binding ISEs. (b) Wild-type Cdh23 minigene or mutants with deletion of the potential RBM24/RBM38-binding ISE were expressed together with RBM24 or RBM38 in COS7 cells. The mCherry fluorescence was examined using a confocal microscope and analyzed using ImageJ software. (c) Wild-type Cdh23 minigene or mutants with deletion of the potential RBM24/RBM38-binding ISE was expressed together with RBM24 or RBM38 in COS7 cells, and the inclusion of exon 68 in the mature Cdh23 mRNA was examined by performing RT-PCR. (d–g) Wild-type Cdh23 minigene or mutants with deletion of the potential RBM24/RBM38-binding ISE were expressed together with RBM24 or RBM38 in COS7 cells. Binding of RBM24/RBM38 with Cdh23 pre-mRNA was examined by performing native RIP followed by RT-PCR (d, f) or qPCR (e, g). Error bars represent mean ± SEM from triplicate experiments. ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001.
RNA immunoprecipitation (RIP) was then performed to examine the physical interaction of RBM24/RBM38 with Cdh23 pre-mRNA. RT-PCR results show that Cdh23 pre-mRNA is readily coimmunoprecipitated with RBM24 or RBM38, which is significantly reduced by deletion of motif 1 but not motif 2 (Figures 3(d) and 3(f)). Similar results were obtained by performing qPCR (Figures 3(e) and 3(g)). Taken together, our present data suggest that RNA-binding proteins RBM24 and RBM38 enhance the inclusion of Cdh23 exon 68 through binding to an ISE motif (UGUGUG).
3.3. PTBP1 Inhibits the Inclusion of Cdh23 Exon 68
Next, we want to know how ubiquitously expressed RBM24/RBM38 drives tissue-specific alternative splicing of Cdh23 exon 68. It has been suggested that most alternative-spliced exons are under combinatorial regulation by both splicing enhancers and repressors [25]. Therefore, RBM24-/RBM38-mediated inclusion of exon 68 in some tissues might be counteracted by yet unknown splicing repressor(s). Bioinformatics analysis suggests a potential ESS motif TCTT in Cdh23 exon 68, which might be recognized by splicing suppressor PTBP1 (Figure 4(a)) [26]. Consistently, when overexpressed in COS7 cells, PTBP1 inhibits RBM24- or RBM38-mediated inclusion of exon 68, albeit the inhibitory effect is not very robust (Figure 4(b)). We then mutated the weak splice donor site located at the exon 68/intron 68 boundary into an artificial strong donor site, which leads to constitutive exon 68 inclusion (Figure 4(c)). When Cdh23 minigene with this donor site mutation is used, more robust inhibition of exon 68 inclusion by PTBP1 is observed (Figure 4(c)). PTBP2 is a homolog of PTBP1 that recognizes similar binding motifs [26], and RT-PCR results reveal that both Ptbp1 and Ptbp2 transcripts are expressed in the inner ear (Figure 1(b)). However, our present data suggest that PTBP2 inhibits Cdh23 exon 68 inclusion to a much lesser extent if any compared with PTBP1 (Figure 4(c)).
Figure 4.
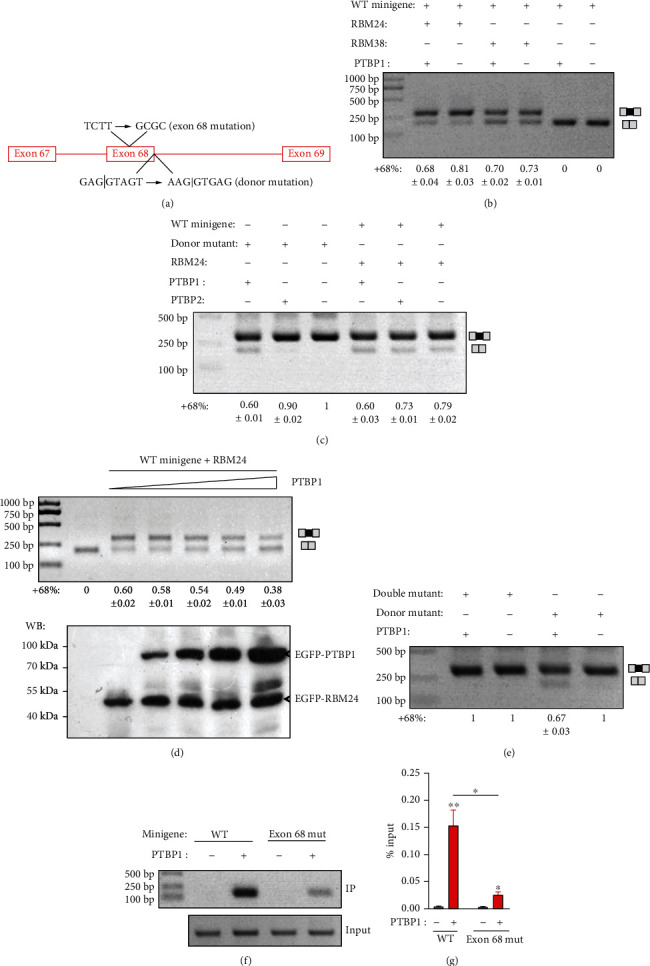
An ESS is required for PTBP1-mediated inhibition of Cdh23 exon 68 inclusion. (a) Schematic drawing of the location of the potential PTBP1-binding ESS and the donor splicing site of intron 68. (b) Cdh23 minigene was expressed in COS7 cells together with the splicing regulators as indicated, and the inclusion of Cdh23 exon 68 was examined by performing RT-PCR. (c) Wild-type Cdh23 minigene or minigene with donor site mutation was expressed in COS7 cells together with the splicing regulators as indicated, and the inclusion of Cdh23 exon 68 was examined by performing RT-PCR. (d) Cdh23 minigene was expressed in COS7 cells together with constant amount of RBM24 and various amount of PTBP1, and the inclusion of Cdh23 exon 68 was examined by performing RT-PCR. The expression level of RBM24 and PTBP1 was confirmed by performing western blot. (e) Cdh23 minigene with donor site mutation or donor site/ESS double mutations was expressed in COS7 cells together with PTBP1, and the inclusion of Cdh23 exon 68 was examined by performing RT-PCR. (f, g) Wild-type Cdh23 minigene or mutant with deletion of the potential PTBP1-binding ESS was expressed in COS7 cells with or without PTBP1. Binding of PTBP1 with Cdh23 pre-mRNA was examined by performing native RIP followed by RT-PCR (f) or qPCR (g). Error bars represent mean ± SEM from triplicate experiments. ∗p < 0.05; ∗∗p < 0.01.
To mimic the various PTBP1 expression levels in different tissues, we overexpressed PTBP1 in COS7 cells in an increasing gradient. The results show that RBM24-mediated exon 68 inclusion is inhibited by PTBP1 in a dosage-dependent manner (Figure 4(d)). Lastly, we mutated the potential PTBP1-binding site TCTT in exon 68 to GCGC in the presence of the donor site mutation. The results show that the TCTT to GCGC mutation abolishes the inhibitory effect of PTBP1 on exon 68 inclusion (Figure 4(e)). Consistently, the TCTT to GCGC mutation also significantly reduces the binding of Cdh23 pre-mRNA to PTBP1 revealed by RIP experiment (Figures 4(f) and 4(g)). Taken together, our present data suggest that PTBP1 inhibits Cdh23 exon 68 inclusion through binding to an ESS motif (UCUU).
3.4. RBM24 Regulates the Alternative Splicing of Endogenous Cdh23 Exon 68
The results discussed above are all obtained using the artificially constructed Cdh23 minigene. We then used the inner ear cell line HEI-OC-1 to investigate the alternative splicing of endogenous Cdh23 mRNA. RT-PCR results show that Cdh23(-68) is the main splicing isoform expressed in HEI-OC-1 (Figure 5(a)). When RBM24 or RBM38 is overexpressed, Cdh23(+68) expression level is significantly increased, suggesting that RBM24 and RBM38 could enhance endogenous Cdh23 exon68 inclusion (Figure 5(a)).
Figure 5.
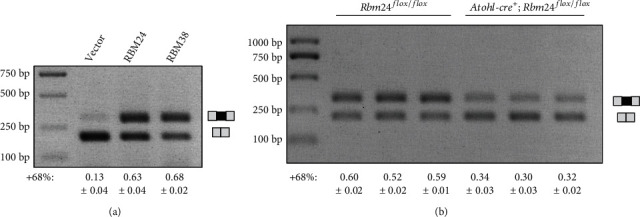
RBM24 regulates the alternative splicing of endogenous Cdh23 pre-mRNA. (a) RBM24 or RBM38 was overexpressed in HEI-OC-1 cells, and exon 68 inclusion of endogenous Cdh23 mRNA was examined by performing RT-PCR. (b) RT-PCR was performed to examine Cdh23 exon 68 inclusion in the inner ear of P2 Atoh1-cre; Rbm24flox/flox (n = 3) or Rbm24flox/flox mice (n = 3).
Lastly, RBM24-mediated alternative splicing of Cdh23 exon 68 in the inner ear was examined using Rbm24 knockout mice. Rbm24 conventional knockout mice die between embryonic day 12.5 (E12.5) and E14.5 because of cardiac problems [24]. In the present work, we used Rbm24 conditional knockout (cko) mice that disrupt Rbm24 gene expression in the hair cells. To do so, Rbm24loxp/loxp mice are crossed with Atoh1-cre mice that express Cre recombinase in the developing cochlear hair cells from E14.5 [27]. RT-PCR results show that Cdh23 exon 68 inclusion is significantly decreased in the inner ear of Rbm24 cko mice compared with that of control mice (Figure 5(b)), suggesting that RBM24 is required for Cdh23 exon 68 inclusion in vivo.
4. Discussion
Cdh23 exon 68 is subjected to alternative splicing, and Cdh23(+68) is preferentially expressed in the inner ear [8, 11–13]. The underlying mechanism for this inner ear-specific alternative splicing of Cdh23 exon 68 remained unknown. In the present work, we show that the alternative splicing of Cdh23 exon 68 is regulated by RBM24, RBM38, and PTBP1. RBM24 and RBM38 promote the inclusion of Cdh23 exon 68, whereas PTBP1 inhibits it.
Several lines of evidences support our conclusion. First, overexpression of RBM24 or RBM38 enhances exon 68 inclusion of the Cdh23 minigene, while overexpression of PTBP1 inhibits it. Second, mutations of the potential binding sites of RBM24, RBM38, or PTBP1 in the Cdh23 minigene affect exon 68 inclusion. Third, RIP experiments show that RBM24, RBM38, and PTBP1 bind Cdh23 pre-mRNA, which is affected by mutations of their potential binding sites. Fourth, knockdown of Rbm24 expression with siRNAs decreases exon 68 inclusion of the Cdh23 minigene. Fifth, overexpression of RBM24 or RBM38 promotes exon 68 inclusion of endogenous Cdh23 in HEI-OC-1 cells. Last, inclusion of Cdh23 exon 68 is reduced in the inner ear of Rbm24 cko mice.
There are two potential RBM24/RBM38-binding ISEs downstream of exon 68, and our data suggest that mutation of site 1 but not site 2 affects the alternative splicing of Cdh23 exon 68. Alignments of genomic sequences flanking Cdh23 exon 68 among different mammals show that site 1 is evolutionally conserved while site 2 is not (data not shown), consistent with an important role of site 1 in mediating Cdh23 exon 68 inclusion. There is also a potential PTBP1-binding ESS located in Cdh23 exon 68, and mutation of this site affects the inhibition of Cdh23 exon 68 inclusion by PTBP1. Interestingly, the PTBP1 homolog PTBP2 shows a much weaker inhibitory effect on Cdh23 exon 68 inclusion, albeit it is expressed in the inner ear and recognizes similar binding motifs.
A question that remained is how relatively ubiquitously expressed RBM24, RBM38, and PTBP1 regulate inner ear-specific alternative splicing of Cdh23 exon 68. Alternative splicing has been proposed to be regulated by the combinatorial action of splicing enhancers and repressors [25]. Transcriptome RNAseq reveals high expression level of Rbm24 and low expression level of Rbm38 and Ptbp1 in the auditory and vestibular hair cells, which is consistent with the inner ear-specific Cdh23 exon 68 splicing [28]. Additionally, there might be other splicing factors that regulate the alternative splicing of Cdh23 exon 68 but are not included in our screening in the present work. In line with this, Cdh23 exon 68 inclusion is decreased but still present in the inner ear of Rbm24 cko mice. qPCR reveals that Rbm38 expression remains on the similar low level in the inner ear of Rbm24 cko mice (data not shown), suggesting that splicing enhancer other than RBM24 and RBM38 might be responsible for the remnant inclusion of Cdh23 exon 68 in the Rbm24 cko mice.
At present, only a few splicing factors have been identified to play important roles in the inner ear, including SRRM4, SFSWAP, and ESRP1 [18–20]. Our present work adds RBM24 to this growing list. RBM24 is an RNA-binding protein that contains an RNA-recognition motif (RRM) and an alanine-rich low-complexity region. It has been shown that RBM24 regulates pre-mRNA alternative splicing as well as mRNA stability and translation [24, 29–38]. RBM24 is a major regulator of muscle-specific alternative splicing, and global Rbm24 inactivation affects cardiac development that eventually leads to embryonic death [24]. In the mouse inner ear, RBM24 expression is specifically detected in the hair cells, but its physiological role in the inner ear remains unknown [22, 39]. Here, we show that RBM24 regulates the alternative splicing of Cdh23; hence, it might play an important role in the development and/or function of hair cells. Meanwhile, immunostaining shows that RBM24 is also localized in the hair cell stereocilia, suggesting that it might also play some roles in the organization and/or function of this F-actin-based cell protrusion [22]. Super-resolution microscopy [40–42] and conditional knockout mice will certainly help to learn more about the role of RBM24 in the inner ear.
Acknowledgments
This work was supported by grants from the National Key Basic Research Program of China (2018YFC1003600), the National Natural Science Foundation of China (81771001), Shandong Provincial Key Laboratory of Animal Cell and Developmental Biology (SPKLACDB-2019000), and the Fundamental Research Funds of Shandong University (2018JC025).
Data Availability
The data supporting the findings of this study are available within the article.
Conflicts of Interest
The authors declare that there are no competing interests regarding the publication of this paper.
References
- 1.Wahl M. C., Will C. L., Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136(4):701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Chen M., Manley J. L. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nature Reviews. Molecular Cell Biology. 2009;10(11):741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nilsen T. W., Graveley B. R. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463(7280):457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grabowski P. J., Black D. L. Alternative RNA splicing in the nervous system. Progress in Neurobiology. 2001;65(3):289–308. doi: 10.1016/S0301-0082(01)00007-7. [DOI] [PubMed] [Google Scholar]
- 5.Black D. L. Mechanisms of alternative pre-messenger RNA splicing. Annual Review of Biochemistry. 2003;72(1):291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 6.Wang Y., Liu Y., Nie H., Ma X., Xu Z. Alternative splicing of inner-ear-expressed genes. Frontiers in Medicine. 2016;10(3):250–257. doi: 10.1007/s11684-016-0454-y. [DOI] [PubMed] [Google Scholar]
- 7.Assad J. A., Shepherd G. M. G., Corey D. P. Tip-link integrity and mechanical transduction in vertebrate hair cells. Neuron. 1991;7(6):985–994. doi: 10.1016/0896-6273(91)90343-X. [DOI] [PubMed] [Google Scholar]
- 8.Siemens J., Lillo C., Dumont R. A., et al. Cadherin 23 is a component of the tip link in hair-cell stereocilia. Nature. 2004;428(6986):950–955. doi: 10.1038/nature02483. [DOI] [PubMed] [Google Scholar]
- 9.Söllner C., the Tübingen 2000 Screen Consortium, Rauch G.-J., et al. Mutations in cadherin 23 affect tip links in zebrafish sensory hair cells. Nature. 2004;428(6986):955–959. doi: 10.1038/nature02484. [DOI] [PubMed] [Google Scholar]
- 10.Kazmierczak P., Sakaguchi H., Tokita J., et al. Cadherin 23 and protocadherin 15 interact to form tip-link filaments in sensory hair cells. Nature. 2007;449(7158):87–91. doi: 10.1038/nature06091. [DOI] [PubMed] [Google Scholar]
- 11.Di Palma F., Pellegrino R., Noben-Trauth K. Genomic structure, alternative splice forms and normal and mutant alleles of cadherin 23 (Cdh23) Gene. 2001;281(1-2):31–41. doi: 10.1016/S0378-1119(01)00761-2. [DOI] [PubMed] [Google Scholar]
- 12.Siemens J., Kazmierczak P., Reynolds A., Sticker M., Littlewood-Evans A., Muller U. The Usher syndrome proteins cadherin 23 and harmonin form a complex by means of PDZ-domain interactions. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(23):14946–14951. doi: 10.1073/pnas.232579599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Z., Peng A. W., Oshima K., Heller S. MAGI-1, a candidate stereociliary scaffolding protein, associates with the tip-link component cadherin 23. The Journal of Neuroscience. 2008;28(44):11269–11276. doi: 10.1523/JNEUROSCI.3833-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yonezawa S., Hanai A., Mutoh N., Moriyama A., Kageyama T. Redox-dependent structural ambivalence of the cytoplasmic domain in the inner ear-specific cadherin 23 isoform. Biochemical and Biophysical Research Communications. 2008;366(1):92–97. doi: 10.1016/j.bbrc.2007.11.102. [DOI] [PubMed] [Google Scholar]
- 15.Xu Z., Oshima K., Heller S. PIST regulates the intracellular trafficking and plasma membrane expression of cadherin 23. BMC Cell Biology. 2010;11(1):p. 80. doi: 10.1186/1471-2121-11-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu L., Pan L., Zhang C., Zhang M. Large protein assemblies formed by multivalent interactions between cadherin23 and harmonin suggest a stable anchorage structure at the tip link of stereocilia. The Journal of Biological Chemistry. 2012;287(40):33460–33471. doi: 10.1074/jbc.M112.378505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gagliardi M., Matarazzo M. R. RIP: RNA immunoprecipitation. Methods in Molecular Biology. 2016;1480:73–86. doi: 10.1007/978-1-4939-6380-5_7. [DOI] [PubMed] [Google Scholar]
- 18.Nakano Y., Jahan I., Bonde G., et al. A mutation in the Srrm4 gene causes alternative splicing defects and deafness in the Bronx waltzer mouse. PLoS Genetics. 2012;8(10, article e1002966) doi: 10.1371/journal.pgen.1002966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moayedi Y., Basch M. L., Pacheco N. L., et al. The candidate splicing factor Sfswap regulates growth and patterning of inner ear sensory organs. PLoS Genetics. 2014;10(1, article e1004055) doi: 10.1371/journal.pgen.1004055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rohacek A. M., Bebee T. W., Tilton R. K., et al. ESRP1 mutations cause hearing loss due to defects in alternative splicing that disrupt cochlear development. Developmental Cell. 2017;43(3):318–331.e5. doi: 10.1016/j.devcel.2017.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y., Nie H., Liu C., et al. Angulin proteins ILDR1 and ILDR2 regulate alternative pre-mRNA splicing through binding to splicing factors TRA2A, TRA2B, or SRSF1. Scientific Reports. 2017;7(1):p. 7466. doi: 10.1038/s41598-017-07530-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grifone R., Saquet A., Xu Z., Shi D. L. Expression patterns of Rbm24 in lens, nasal epithelium, and inner ear during mouse embryonic development. Developmental Dynamics. 2018;247(10):1160–1169. doi: 10.1002/dvdy.24666. [DOI] [PubMed] [Google Scholar]
- 23.Ray D., Kazan H., Cook K. B., et al. A compendium of RNA-binding motifs for decoding gene regulation. Nature. 2013;499(7457):172–177. doi: 10.1038/nature12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J., Hung L. H., Licht T., et al. RBM24 is a major regulator of muscle-specific alternative splicing. Developmental Cell. 2014;31(1):87–99. doi: 10.1016/j.devcel.2014.08.025. [DOI] [PubMed] [Google Scholar]
- 25.Smith C. W. J., Valcárcel J. Alternative pre-mRNA splicing: the logic of combinatorial control. Trends in Biochemical Sciences. 2000;25(8):381–388. doi: 10.1016/S0968-0004(00)01604-2. [DOI] [PubMed] [Google Scholar]
- 26.Boutz P. L., Stoilov P., Li Q., et al. A post-transcriptional regulatory switch in polypyrimidine tract-binding proteins reprograms alternative splicing in developing neurons. Genes & Development. 2007;21(13):1636–1652. doi: 10.1101/gad.1558107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang H., Xie X., Deng M., Chen X., Gan L. Generation and characterization of Atoh1-Cre knock-in mouse line. Genesis. 2010;48(6):407–413. doi: 10.1002/dvg.20633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scheffer D. I., Shen J., Corey D. P., Chen Z. Y. Gene expression by mouse inner ear hair cells during development. The Journal of Neuroscience. 2015;35(16):6366–6380. doi: 10.1523/JNEUROSCI.5126-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jiang Y., Zhang M., Qian Y., Xu E., Zhang J., Chen X. Rbm24, an RNA-binding protein and a target of p53, regulates p21 expression via mRNA stability. The Journal of Biological Chemistry. 2014;289(6):3164–3175. doi: 10.1074/jbc.M113.524413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu E., Zhang J., Zhang M., Jiang Y., Cho S. J., Chen X. RNA-binding protein RBM24 regulates p63 expression via mRNA stability. Molecular Cancer Research. 2014;12(3):359–369. doi: 10.1158/1541-7786.MCR-13-0526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ito J., Iijima M., Yoshimoto N., Niimi T., Kuroda S.'., Maturana A. D. RBM20 and RBM24 cooperatively promote the expression of short enh splice variants. FEBS Letters. 2016;590(14):2262–2274. doi: 10.1002/1873-3468.12251. [DOI] [PubMed] [Google Scholar]
- 32.Zhang T., Lin Y., Liu J., et al. Rbm24 regulates alternative splicing switch in embryonic stem cell cardiac lineage differentiation. Stem Cells. 2016;34(7):1776–1789. doi: 10.1002/stem.2366. [DOI] [PubMed] [Google Scholar]
- 33.Lin Y., Tan K. T., Liu J., Kong X., Huang Z., Xu X. Q. Global profiling of Rbm24 bound RNAs uncovers a multi-tasking RNA binding protein. The International Journal of Biochemistry & Cell Biology. 2018;94:10–21. doi: 10.1016/j.biocel.2017.11.002. [DOI] [PubMed] [Google Scholar]
- 34.Liu J., Kong X., Zhang M., Yang X., Xu X. RNA binding protein 24 deletion disrupts global alternative splicing and causes dilated cardiomyopathy. Protein & Cell. 2019;10(6):405–416. doi: 10.1007/s13238-018-0578-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van den Hoogenhof M. M. G., van der Made I., de Groot N. E., et al. AAV9-mediated Rbm24 overexpression induces fibrosis in the mouse heart. Scientific Reports. 2018;8(1):p. 11696. doi: 10.1038/s41598-018-29552-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang M., Zhang Y., Xu E., et al. Rbm24, a target of p53, is necessary for proper expression of p53 and heart development. Cell Death and Differentiation. 2018;25(6):1118–1130. doi: 10.1038/s41418-017-0029-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shao M., Lu T., Zhang C., Zhang Y. Z., Kong S. H., Shi D. L. Rbm24 controls poly(A) tail length and translation efficiency of crystallin mRNAs in the lens via cytoplasmic polyadenylation. Proceedings of the National Academy of Sciences of the United States of America. 2020;117(13):7245–7254. doi: 10.1073/pnas.1917922117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Dash S., Brastrom L. K., Patel S. D., Scott C. A., Slusarski D. C., Lachke S. A. The master transcription factor SOX2, mutated in anophthalmia/microphthalmia, is post-transcriptionally regulated by the conserved RNA-binding protein RBM24 in vertebrate eye development. Human Molecular Genetics. 2020;29(4):591–604. doi: 10.1093/hmg/ddz278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai T., Jen H. I., Kang H., Klisch T. J., Zoghbi H. Y., Groves A. K. Characterization of the transcriptome of nascent hair cells and identification of direct targets of the Atoh1 transcription factor. The Journal of Neuroscience. 2015;35(14):5870–5883. doi: 10.1523/JNEUROSCI.5083-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Y., Qi J., Chen X., et al. Critical role of spectrin in hearing development and deafness. Science Advances. 2019;5(4, article eaav7803) doi: 10.1126/sciadv.aav7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Qi J., Liu Y., Chu C., et al. A cytoskeleton structure revealed by super-resolution fluorescence imaging in inner ear hair cells. Cell Discovery. 2019;5(1):p. 12. doi: 10.1038/s41421-018-0076-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qi J., Zhang L., Tan F., et al. Espin distribution as revealed by super-resolution microscopy of stereocilia. American Journal of Translational Research. 2020;12(1):130–141. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data supporting the findings of this study are available within the article.


