Abstract
Technical advances in genome sequencing and the implementation of next-generation sequencing (NGS) in clinical oncology have paved the way for individualizing cancer patient therapy based on molecular profiles. When and how to use NGS testing in the clinic is at present an unsolved issue, although new research results provide evidence favoring this approach in some types of advanced cancer. Clinical research is evolving rapidly, from basket and umbrella trials to adaptative design precision oncology clinical studies, and genomic and molecular data often displace the classical clinical validation procedures of biomarkers. In this context, physicians must be aware of the clinical evidence behind these new biomarkers and NGS tests available, in order to use them in the right moment, and with a critical point of view. This review will present the status of currently available targeted drugs that can be effective based on actionable molecular alterations, and the NGS tests that are currently available, offering a practical guide for the application of Clinical Precision Oncology in the real world routine practice.
Keywords: Precision oncology, Personalised medicine, Next generation sequencing, Cancer Genomics, Cancer Therapy, Targeted therapy
1. Precision oncology at a glance
Precision Oncology is the form of medicine which uses cancer treatments that are targeted to individual patients on the basis of genetic, biomarker, phenotypic, or psychosocial characteristics that distinguish a given cancer patient from other patients with similar clinical presentations [1]. While this is not an entirely new approach in oncology, it takes advantage of recent advances in genome sequencing and the growing availability of clinical data, and also offers an unprecedented opportunity to make personalized precision patient care a clinical reality [2].
1.1. From the TCGA project to targeted therapy
The Cancer Genome Atlas (TCGA) project was undertaken in 2005 to map the human cancer genome [3,4], The TCGA project, a joint effort of the National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI), has identified new oncogenic point mutations, fusions and variants that have therapeutic impact, and impact the clinical course cancer patients.
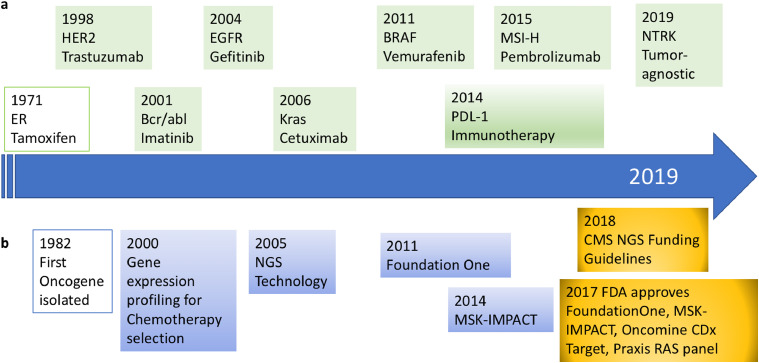
This growing data about cancer opened up the first steps towards precision oncology with the aim of ensuring that cancer patients get the right treatment at the right dose at the right time, with minimum ill consequences and maximum efficacy [5]. While an early precedent of precision oncology was the use of tamoxifen in estrogen receptor-positive breast cancer [6], the first precision cancer medicines approved specifically against a molecular target were trastuzumab (for HER2+ breast cancer in 1998) and imatinib (for Bcr/abl-positive chronic myelogenous leukemia in 2001). In the ensuing 20 years, the number of actionable alterations that have a corresponding targeted therapy has been growing steadily, and these include both single genes (such as BRAF or ALK), or composite genetic signatures (such as mismatch repair or homologous recombination deficiency). A timeline of the most remarkable precision oncology highlights is shown in Fig. 1.
Fig. 1.
Timeline showing the main clinical precision oncology highlights. a) Therapeutic landmarks and their molecular targets (in green). b) Most relevant diagnostic technologies (in blue) and regulatory landmarks (in yellow).
1.2. Actionable molecular alterations and biology-guided signatures
Molecular testing has become useful in clinical practice to detect actionable genomic alterations for both diagnostic and therapeutic purposes. As a general rule, molecular testing (like most medical testing) should be ordered when results may impact clinical management.
At the end of 2019, the number of anticancer therapies targeted against a molecular alteration was 64, and the number of targetable molecular alterations was 24. It is very relevant that in 19 of these, the detection of the alteration was required in order to effectively indicate a particular prescription (Table 1).
Table 1.
FDA approved targeted therapies in solid malignancies.†.
| GENE TARGET | GENOMIC ALTERATION | MALIGNANCY | THERAPEUTIC AGENTS |
|---|---|---|---|
| EML4-ALK* | Rearrangement | Lung Cancer | Crizotinib, Alectinib, Ceritinib, Brigatinib, Lorlatinib |
| BRAF* | Mutation | Melanoma | Vemurafenib, Dabrafenib, Trametinib, Cobimetinib, Encorafenib, Binimetinib |
| Mutation | Anaplastic thyroid cancer, lung cancer | Dabrafenib, trametinib | |
| BRCA1/2* | Mutation | Ovarian Cancer, Prostate Cancer | Olaparib, Niraparib, Talazoparib, Rucaparib |
| Mutation | Triple negative breast cancer | Olaparib | |
| CKIT* | Mutation | GIST, Mastocytosis | Imatinib, Sunitinib, Regorafenib |
| EGFR* | Mutation | Lung Cancer | Erlotinib, Gefitinib, Afatinib, Osimertinib, Dacomitinib |
| EGFR* | Expression | Colon | Cetuximab, Panitumumab |
| Lung Cancers | Necitumumab | ||
| HER2* | Amplification, overexpression | Breast Cancer | Trastuzumab, Lapatinib, Pertuzumab, Ado-trastuzumab emtansine, Neratinib |
| Amplification, overexpression | Gastric Cancer | Trastuzumab | |
| FGFR3, FGFR2* | Mutation | Bladder cancer | Erdafitinib |
| Homologous Recombination Deficiency (HRD)* | Composite | Ovarian cancer | Niraparib |
| C-KIT* | Mutation, expression | GIST | Imatinib, sunitinib, regorafenib |
| Mismatch Repair (MMR)* | Expression, mutation | Tumor-agnostic, MSI-H Cancers | Pembrolizumab |
| Expression, mutation | Colorectal MSI-H Cancers | Nivolumab | |
| NTRK* | Fusion | Tumor-agnostic, NTRK+ cancers | Entrectinib, larotrectinib |
| PDGFRA* | Mutation | GIST, Sarcoma | Imatinib, Sunitinib, Olaratumab |
| COL1A1-PDGFΒ | Rearrangement | Dermatofibrosarcoma protuberans | Imatinib |
| PDL-1* | Expression | Lung, triple negative breast, urothelial, cervical cancer | Pembrolizumab, atezolizumab |
| PI3K* | Mutation | Breast Cancer | Alpelisib |
| SMO and PTCH1 | Mutation | Basal Cell Carcinoma | Vismodegib, Sonidegib |
| K-RAS* | Mutation | Colon cancer | Cetuximab, panitumumab (in RAS-non mutated) |
| RET* | Mutation | Thyroid Cancer | Vandetanib, Cabozantinib, Lenvatinib |
| ROS-1* | Rearrangement | Lung cancer | Crizotinib, Entrectinib |
| VEGF/VEGFR | Expression | Kidney, Colon, Lung, Gastric, Cervix, Ovarian Cancers | Bevacizumab, Ramucirumab, Regorafenib, Ziv-aflibercept, Axitinib, Pazopanib, Sunitinib, Sorafenib |
| CDK4/6 | Amplification | Breast Cancer | Palbociclib, Ribociclib, Abemaciclib |
| mTOR | Mutation | Breast, Renal, Brain Cancers | Everolimus, Temsirolimus |
| Estrogen Receptor* | Expression | Breast Cancer | Tamoxifen, fulvestrant, anastrozole, letrozole, exemestane, everolimus, palbociclib, ribociclib, abemaciclib |
| Androgen Receptor | Expression | Prostate Cancer | Abiraterone, Enzalutamide, Apalutamide, Darolutamide |
in 2019(modified from[63]).
Required for prescription.
Furthermore, TRK fusions and microsatellite instability have both been validated as histology-agnostic biomarkers for FDA approval of larotrectinib and entrectinib, and pembrolizumab, respectively. These markers are detected in some but not all NGS platforms, emphasizing the need for clinicians to know the differences between platforms and to keep in mind what is likely to be detected in different tumor types when initiating NGS testing. The European Society for Medical Oncology (ESMO) has developed a Scale of Clinical Actionability for molecular Targets (ESCAT) that defines six levels of clinical evidence for molecular targets according to the implications for patient management [7]. The number of patients eligible for a genome-driven therapy has been estimated to be 5% in 2006 and 8.33% in 2018. The estimated clinical benefit has increased from 0.7% in 2006 to 4.9% in 2018 [8].
Multigene panels allow grouping patterns of mutations into mutational signatures [9]. The most relevant currently are homologous recombination deficiency (HRD), TMB, and MSI. Defective DNA repair pathways (like mutations in BRCA1/2) cause HRD, and it has been shown that somatic substitution, insertion/deletion and rearrangement patterns are also associated with HRD. Thus, a model called “HRDetect” has been developed, which identifies HRDtumours. These tumours may also be sensitive to PARP inhibition. Using NGS, it is also possible to quantify the TMB, which might be associated with response to immunotherapy. Also, pembrolizumab has been approved for patients with tumours (of any histology or organ of origin) which are MSI-high. Yet some of these markers need a complete clinical validation in clinical cancer management [10].
2. Next generation sequencing platforms
Multigene sequencing avoids performing multiple sequential single tests with the advantages of sparing tissue samples, avoiding delays for patients and being able to direct the patient to the most appropriate clinical trial [11].
Targeted panels rely on amplicon-based or hybridization capture-based NGS, which show consistent results in detecting single nucleotide variations and insertions or deletions in a range of clinical applications. These can identify druggable alterations that show value in some (but not all) malignancies.
2.1. Approved NGS tests
The two broad molecular profiling NGS tests approved in 2017 by the FDA that interrogate a higher number of genes at once are the FoundationOne CDx (F1CDx) test, and the MSK-IMPACT test. There are also several other NGS tests that are approved by the FDA that target a specific gene or set of genes, such as Oncomine Dx Target Test for lung cancer (in 2017), Illumina Extended RAS Panel for colon cancer (in 2017), or Foundation Focus CDx BRCA LOH (in 2018).
Other NGS tests are in development. Caris MI Transcriptome CDx is a next-generation sequencing-based in vitro diagnostic test that uses RNA isolated from formalin-fixed paraffin embedded (FFPE) tumor tissue to detect structural rearrangements. In 2019, it has received Breakthrough Device designation for detection of FGFR gene fusions in solid tumors. In 2019, the FDA also granted Breakthrough Device Designation for Illumina's pan-cancer assay, TruSight Oncology 500, which utilizes DNA and RNA from tumor samples to identify small DNA variants, fusions, and splice variants, as well as tumor mutational burden (TMB) and microsatellite instability (MSI).
The characteristics of a selection of NGS platforms are displayed in Table 2. The progressive cost reduction of NGS platforms suggests that, in a near future, testing with a 300-gene panel may have a similar cost to that of testing 5 or 6 alterations individually [12]. The information generated, although not always usable immediately, may prove of extreme value in future evaluations.
Table 2.
A selection of Next Generation Sequencing Platforms and other genetic companion devices.
| PLATFORM | GENES ASSESSED | FDA APPROVAL | MUTATIONS |
|---|---|---|---|
| FoundationOne CDX (Foundation Medicine) | 324 | Yes | Copy number alterations, gene fusions, MSI, TMB, PDL-1 (IHC) |
| MSK IMPACT (Integrated Mutation Profiling Of Actionable Cancer Targets) (Memorial Sloan Kettering) | 468 | Yes | Somatic single nucleotide variants, insertions, deletions, and microsatellite instability |
| Oncomine Dx Target Test (Thermofisher) | 46 | Yes | DNA single-nucleotide variants (SNVs) and deletions in 35 genes, and RNA sequence variantions from 21 genes (Non-small cell lung cancer) |
| Caris Mollecular Intelligence CDX (Caris Life Sciences) | 592 | Partial | DNA: copy number alterations, MSI, TMB RNA: gene fusions, mRNA variants |
| Oncomine Comprehensive Assay (Thermofisher) | 161 | – | DNA sequencing: copy number alterations, gene fusions |
| Trusight Oncology 500 (Illumina) | 523 | – | DNA + RNA assay for assessment of small variants, TMB, MSI, splice variants, and fusions |
| FoundationOne Liquid | 70 | – | Plasma: DNA sequencing: copy number alterations, specific gene fusions for lung malignancies, MSI |
| Guandant360 (Guardant) | 76 | – | Plasma: DNA sequencing: copy number alterations, 6 gene fusions |
| GENETIC COMPANION DEVICES | |||
| Praxis Extended RAS Panel (Illumina) | 2 | Yes | K-ras and N-ras (colorectal cancer) |
| Therascreen KRAS RGQ PCR Kit (Qiagen) | 1 | Yes | K-ras (colorectal cancer) |
| BRACANALYSIS CDX (Myriad Genetic Laboratories) | 2 | Yes | BRCA1, BRCA2 (Ovarian and Breast cancers) |
| FoundationFocus CDX BRCA Assay (FoundationOne) | 2 | Yes | BRCA1, BRCA2 (Ovarian cancer) |
| Therascreen EGFR RGQ PCR KIT (Qiagen) | 1 | Yes | EGFR (Non-small cell lung cancer) |
| COBAS EGFR Mutation Test V2 (Roche Molecular Systems) | 1 | Yes | EGFR (Non-small cell lung cancer) |
| THXID BRAF Kit (Biomérieux) | 1 | Yes | BRAF (Melanoma) |
| COBAS 4800 BRAF V600 Mutation Test (Roche Molecular Systems) | 1 | Yes | BRAF (Melanoma) |
| Therascreen FGFR RGQ RT-PCR Kit (Qiagen) | 1 | Yes | FGFR (Urothelial cancer) |
| Therascreen PIK3CA RGQ PCR Kit (Qiagen) | 1 | Yes | PIK3CA, tissue and plasma (breast cancer) |
| Myriad MYCHOICE® CDX (Myriad Genetic Laboratories) | Combined asay | Yes | Loss of heterozygosity (LOH), telomeric-allelic imbalance (TAI), large-scale state transitions (LST) (ovarian cancer) |
In Europe, the assessment of genetic tests has traditionally been regulated mostly at the national level [13]. The European Network for Health Technology Assessment has started to centrally perform health technology assessments [14], although EUnetHTA has not yet performed evaluations of cancer-related next generation sequencing testing.
2.2. Understanding NGS limitations
There are several limitations for the detection of gene alterations in NGS. Discrepancies may be related to tumor heterogeneity, either static (in the tumor tissue) or dynamic (in different time points of tumor biopsy-plasma sampling or of plasma sampling), and different sequencing techniques. A recent analysis of concordance among tumor, normal, and replicate plasma samples along with orthogonal ctDNA assays revealed that discordance was a result of technical variations and, to a lesser extent, biologic factors such as clonal hematopoiesis of indeterminate potential and tumor heterogeneity [15].
In some cases in which the primary biopsy was performed years before the development of advanced disease, the tissue may not be in good conservation state for NGS test. It has been suggested that DNA extracted from FFPE cancer tissue samples of surgical specimens older than 7 years are not adequate for NGS analysis [16]. In these cases, fresh biopsies should be done if possible or plasma NGS should be considered.
Perhaps the most relevant and still not fully acknowledged limitation for NGS testing is the existence of cancer-associated mutations in normal tissue [17]. Mutations frequently seen in cancer, commonly referred to as ‘driver mutations’ (for example, TP53) are also frequently mutated in normal tissues and other benign conditions that rarely progress to malignancy. Mutations in normal tissues frequently cluster in functionally relevant portions of the cancer associated gene, such as DNA-binding domains or regions involved in protein-protein interactions, in a pattern nearly identical to the distribution of mutations seen in tumor sequencing data. This has been considered a normal phenotype of aging, and may be related to an age-related decreased efficacy of the normal tumor suppression mechanisms such as contact inhibition, senescence or immune surveillance [25].
There is an urgent need to coordinate and homogenize NGS testings and NGS results reports, in order to perform observational Real World studies that collect the experience of a very large number of patient results (that record patient characteristics, stages, and whether there is a therapeutic intervention), and to establish the real utility of these profilings based on the ESMO and the FDA criteria.
3. Precision medicine clinical trials: a new paradigm for research
Precision Medicine strategies have been studied with several clinical trial designs, although only a limited number are controlled and randomised. Many of these trials are included in the categories of umbrella trials or basket trials [18]. “Basket” trials enroll patients of different histologies that harbor the same genomic alteration; all patients receive the same drug targeting such alteration, while “umbrella” trials enroll patients of the same histology and stratify them by different genomic alterations, administering a different drug to each alteration/patient. These trials are powered to assess the efficacy of a specific drug in a molecularly- and histologically-defined subgroup of patients, allowing to run parallel phase II trials. Another major precision medicine trial category mixes multiple tumor types, multiple molecular targets and multiple drugs. Given the usually high number of molecularly- and histologically-defined subgroups of patients in these studies, they are most often not powered to assess the efficacy of the drugs in each subgroup of patients, although the efficiency of the treatment algorithm that has been used to allocate drugs to patients (“algorithm-testing studies”) [15,19]. Several different types of algorithm-testing precision medicine studies have been reported to date, and some of them have provided evidence that molecularly targeted therapies can be given in a histology-agnostic way, based only on molecular profiling.
3.1. Impact of precision clinical trials in last decades
The French SHIVA trial was the first randomised algorithm-testing precision medicine study. SHIVA01 compared molecularly-targeted therapy based on tumor molecular profile versus treatment at physician's choice in patients with diverse types of metastatic cancers that had failed standard of care treatment [20]. This controlled, phase II trial included patients for whom a molecular alteration was identified within one of three molecular pathways (hormone receptor, PI3K/AKT/mTOR, RAF/MEK), which could be matched to one of ten regimens including 11 available molecularly targeted agents (erlotinib, lapatinib plus trastuzumab, sorafenib, imatinib, dasatinib, vemurafenib, everolimus, abiraterone, letrozole, tamoxifen). 195 (26%) patients were randomized, 99 to the experimental group and 96 to the control group. Median progression-free survival was 2.3 months in the experimental group and 2.0 months in the control group (p = 0.41). The primary endpoint of the trial was not met, with no statistical difference in PFS between the two treatment arms. These results show that the specific treatment algorithm used in SHIVA01 involving targeted therapies allocation according to the presence of some genetic alterations was not able to improve patients’ outcome when they received purely empirical treatment. The authors concluded that off-label routine use of molecularly targeted agents should be discouraged, although enrolment in clinical trials should be encouraged to assess predictive biomarkers of efficacy.
The Molecular Screening for Cancer Treatment Optimization (MOSCATO 01) study was a non-randomised prospective trial that evaluated the clinical benefit of high-throughput genomic analyses in different types of advanced cancer (n = 1035). NGS testing was performed on a fresh biopsy. A total of 199 patients were finally treated with a targeted therapy, and objective responses were observed in 22 patients (11%) [21]. The authors observed that 63 patients obtained a PFS ratio (PFS2/PFS1) above the predefined threshold of 1.3, and concluded that the trial was positive because tumor sequencing improved outcome in one-third of those patients with advanced cancers that received targeted therapy.
The MyPathway study is a non-randomised multiple basket study in patients with refractory solid tumors harboring molecular alterations in HER2, EGFR, B-RAF or the Hedgehog pathway (n = 251). Therapies used in this Roche/Genentech sponsored study were trastuzumab/pertuzumab, erlotinib, vemurafenib and vismodegib, respectively. Objective responses were observed, most notably in the HER2+ colorectal cohort (38%) and the BRAF V600 lung cancer cohort (43%) [22]. The authors concluded that the targeted therapy regimens produced meaningful responses in several refractory solid tumor types not currently labeled for these agents.
The Novartis Signature Program is a series of 8 phase 2, agent-specific basket protocols (buparlisib, dovitinib, binimetinib, encorafenib, sonidegib, BGJ398, ceritinib, or ribociclib) in cancer patients with an actionable mutation (n = 595). The Signature Program uses a modified Bayesian adaptive design with a hierarchical model, and allowed enrollment based on local testing of archival or fresh tissue. Frequent genetic alterations were observed in PIK3CA, RAS, p16, and PTEN. Overall, 30 partial or complete responses were observed with 6 of the compounds studied, in 16 tumor types [23]. The authors concluded that the Program was a successful approach, because it led to rapid signal finding, reduced patient exposure to toxicity, and substantially shortened trial start-up times .
The American Society of Clinical Oncology (ASCO) sponsored the Targeted Agent and Profiling Utilization Registry (TAPUR) Study. TAPUR is an ongoing non-randomized phase II, open label that aims to define signals of drug activity of FDA-approved targeted anticancer drugs prescribed for treatment of patients with advanced cancer that have a potentially actionable genomic alteration. There are currently 113 TAPUR Study sites and nearly 1400 participants who have received study therapy [24]. TAPUR evaluates 8 drug or drug combinations from several manufacturers in a total of 44 cohorts. Currently, 15 cohorts have been closed, of which five had positive results, [25], [26], [27], [28], [29], five had negative results, and five have results pending (Table 3). Twenty-nine cohorts have been expanded. Objective responses in the positive cohorts were pertuzumab + trastuzumab in ERBB2 amplified or overexpressed colorectal cancer, 25%; vemurafenib + cobimetinib in BRAF V600E/D/K/ mutated colorectal cancer, 29%; pembrolizumab in metastatic breast cancer with high mutational burden, 21%; pembrolizumab in metastatic colorectal cancer with high mutational burden, 11%; and palbociclib in non-small-cell lung cancer with CDKN2A alterations, 3.6%. The most recent results of the TAPUR study indicate that this approach is valid in order to identify new signals of activity with the use of targeted drugs in off-label molecularly-guided indications.
Table 4.
Precision Oncology NGS studies with novel designs published in 2019.
| Trial name (ref) | Design Innovation | N | Objective response, number (%) |
|---|---|---|---|
| WINTHER (30) | Fresh biopsy: DNA + RNA testing | 253 | 12 (4.7%) |
| DRUP (31) | Fresh biopsy: WGS; off-label use | 215 | 33 (15.3%) |
| I-PREDICT (32) | Genetic + Immunotherapy markers (MSI, PDL1 by IHC); Drug combinations | 149 | 17 (11%) |
| TARGET (33) | ctDNA testing | 100 | 4 (4.0%) |
Table 3.
TAPUR study cohorts that have been closed as of February 20, 2020* (adapted).
| Treatment | Cancer | Variant | Findings |
|---|---|---|---|
| Pertuzumab + Trastuzumab | Colorectal | ERBB2 amplification or overexpression | Positive |
| Colorectal | ERBB2/ERBB3 mutation | Pending | |
| Vemurafenib + Cobimetinib | Colorectal | BRAF V600E/D/K/R mutation | Positive |
| Pembrolizumab | Metastatic breast | High tumor mutational burden | Positive[25] |
| Colorectal | High tumor mutational burden | Positive | |
| Palbociclib | Non-small-cell lung | CDKN2A alterations | Positive[24] |
| Pancreatic | CDKN2A loss or mutation | Negative | |
| Gallbladder and bile ducts | CDKN2A loss or mutation | Negative | |
| Sunitinib | Colorectal | FLT-3 mutation or amplification | Negative |
| Cetuximab | Breast | KRAS, NRAS, BRAF wild type | Negative |
| Bronchus and lung | KRAS, NRAS, BRAF wild type | Negative | |
| Ovarian | KRAS, NRAS, BRAF wild type | Pending | |
| Olaparib | Colorectal | ATM mutation or deletion | Pending |
| Prostate | BRCA1/BRCA2 mutation | Pending | |
| Pancreatic | BRCA1/BRCA2 mutation | Pending |
Accessed on March 23, 2020.
Finally, some histology-specific umbrella studies assigned targeted therapy randomly based on their molecular profile. One of these is the BATTLE-2 program, a targeted therapy study performed in previously treated patients with advanced refractory NSCLC (n = 334). Activity was modest, yielding no new predictive markers and not warranting further exploration [30].
3.2. Precision clinical trials in 2019
In 2019, four precision oncology studies that use genetic NGS testing to guide cancer therapy in patients with advanced malignancies have been published. Each one of these studies has an innovative, original approach (Table 5)[31], [32], [33], [34].
The WINTHER study, from the Worldwide Innovative Network, performed fresh biopsy-derived DNA sequencing or RNA expression in tumor and normal tissues in 253 patients [30]. Oncogenic driver mutations/amplifications/translocations were detected through Next Generation Sequencing (NGS) performed by Foundation Medicine (arm A), and in patients negative for oncogene events, genome based relevant information was obtained through functional genomics (micro arrays and gene expression profiling) performed by Institut Gustave Roussy (arm B). 107 patients (69 in arm A and 38 in arm B) were evaluable for therapy. Objective response was observed in 12 patients (9 in arm A and 3 in arm B). The study identified that few previous therapies, good performance status and a high matching score correlated with improved clinical outcome. Although the WINTHER trial did not meet its primary end point related to a PFS2/PFS1 ratio of >1.5, there were some innovative advances, that included performing RNA testing, the presence of a clinical management committee that recommended therapies, and also that the trial design navigated patients to a clinical trial or to on-label-approved or off-label-approved drugs.
The Drug Rediscovery protocol (DRUP) is an adaptive trial that aims to identify signals of activity in cohorts of patients with defined tumor types and molecular variants, who are being treated with anticancer drugs outside of their approved label. In this Dutch trial, patients with all types of metastatic cancer were offered the opportunity to undergo a fresh tumor biopsy for whole-genome sequencing (WGS) before starting systemic anticancer treatment. In a total of 215 patients, 76 cohorts were initiated. Objective response rate was not reported, although clinical benefit (CB) was observed across all types of treatment, comprising immunotherapy (n = 79 patients, CB rate of 38%), treatment with small-molecule inhibitors (including PARP inhibitors) (n = 81 patients, CB rate of 36%) and with monoclonal antibodies (n = 55 patients, CB rate of 27%). The study identified a successful cohort of 30 cases with microsatellite unstable tumors who received nivolumab and had a response rate of 40%, and also an unsuccessful cohort of patients with colorectal cancer with low TMB, that had only a marginal benefit with immunotherapy.
The Investigation of Profile-Related Evidence Determining Individualized Cancer Therapy (I-PREDICT) study evaluated tissue genomic profiling using Foundation Medicine and, if possible, PD-L1 IHC, tumor mutational burden, MSI status, and the NGS of blood-derived circulating tumor DNA (ctDNA). Of the 83 treated patients, 73 were administered a personalized, precision therapy consisting of ≥1 molecularly ‘matched’ treatments; since no two molecular profiles were identical, most treatment regimens were not exactly alike. The authors achieved a matching rate of 49% (73 of 149 patients), a number considerably high. The high matching rate was based on using expanded NGS testing, and also on timely discussions of the Molecular Tumor Board, which occurred immediately on receipt of molecular results.
The British TARGET study was designed to use circulating tumor DNA (ctDNA) to identify actionable alterations in patients and direct them to clinical trials [35]. They used a 641-gene panel ctDNA NGS, with which data was generated successfully for 99% of patients, compared to tumor tissue DNA analysis in 95%. For the first 100 TARGET patients, ctDNA data showed good concordance (74.5%) with matched tumor and results were available within a clinically acceptable timeframe for Molecular Tumour Board (MTB) review. When applying a 2.5% Variant Allele Frequency (VAF) threshold, actionable mutations were identified in 41/100 patients and 11 of these patients received a matched therapy. In 4 of these, an objective response was observed. A limitation was the lack of available trials or the declining performance status that did not allow any form of treatment.
In the near future, new forms of analysis that include proteomic and epigenetic assessments will increase the number of relevant actionable molecular alterations, and possibly improve the results [36].
Currently, there are additional clinical trials ongoing and nation-wide projects, applying multigene sequencing to a number of different tumor types. Two randomised histology-agnostic studies that will end in 2020 are the IMPACT II study from MD Anderson (that has enrolled 391 cases) and the M-PACT study from the NCI (that expects to enroll 700 cases) [37]. Other ongoing examples are the non-randomised “Adjuvant Lung Cancer Enrichment Marker Identification and Sequencing Trial” (ALCHEMIST) from the NCI that expects to enroll 8300 patients, the randomised Lung Cancer Master Protocol, or Lung-MAP (SWOG S1400) that plans to enroll 10,000 patients, and the randomised SAFIR-02 (Lung) and SAFIR-02 (Breast) that plan to enroll 650 and 1460 patients, respectively.
4. When should a NGS test be considered clinically in 2020
Some authors have expressed a conservative approach towards generalized NGS testing, and have suggested that currently NGS may be mainly useful in controlled research environments or clinical trials, where off-label administration of expensive drugs is restricted to prospective patient registry cohorts [38]. Considering that only a minority of patients with cancer derive a clear benefit for matched treatments, they support that NGS for precision oncology based on emerging biomarkers remain an investigational strategy [39,40].
In contrast, other authors advocate that routine upfront NGS testing should be used for all patients with metastatic cancer with limited standard of care options [41]. In addition, the use of multiplatform technologies seems to identify a higher number of potential targets than conventional consecutive molecular testing and this might subsequently translate in a higher probability to detect an effective matching drug [42]. Furthermore, NGS testing may identify "hypermutations" or DNA damage repair signatures that can predict a response to immunotherapy (TMB, MSI), that otherwise would not be detected.
4.1. Multi-testing
For certain diseases in which a first-line decision depends on multiple molecular markers, such as advanced non-small cell lung cancer, the use of a NGS panel at diagnosis is becoming increasingly attractive given the growing number of actionable gene alterations and the ability to obtain at the same time all the therapy-oriented information [42]. The fact that the specimens most commonly available for advanced lung cancer have a low tumor cell content (patients are commonly diagnosed only by small biopsies) is very relevant in this setting [43]. In one study of 1402 NSCLC samples, the large majority of tissue samples submitted for clinical testing were small (10% FNA, 70% core needle biopsies) [44], and the NGS test used provided a high success rate for reporting 5 or more biomarkers on core needle biopsies, and 5 or more on FNAs.
There are other tumor types where multi-testing is relevant, such as colorectal carcinoma (RAS, BRAF) or melanoma (BRAF, KIT), although rarely NGS testing is currently performed.
4.2. Infrequent molecular alterations and exceptional responders
An additional reason to favor NGS panels in lung cancer is that infrequent molecular alterations occurring in other cancers can also be present in a small proportion of NSCLCs, such as NTRK gene rearrangements that are amenable to transversal or tumor-agnostic treatment with NTRK TKIs[49] (in this case RNA-based NGS is preferred), or microsatellite instability, and NGS contributes to the opportunity of testing for these additional and rare biomarkers. The informed integration of genomic markers is therefore becoming increasingly crucial.
In a recent study, a decision analytic model showed that NGS was cost-effective over sequential single-gene testing modalities for newly diagnosed advanced NSCLC in the USA, without introducing delays in patient management [45]. However, the same model could give opposite results if applied in patients with NSCLC from a different region, such as Asian population, where the incidence of EGFR mutation is higher than in Caucasians (50% vs 11%) [40], and a different study of precision medicine treatment was not cost-effective as fourth-line treatment for metastatic lung adenocarcinoma [46].
Furthermore, next-generation sequencing may be of great value in identifying what has been defined as "exceptional responders" to anticancer drugs, or patients with extreme phenotypes [47]. A systematic search of the medical literature identified 180 cases of exceptional response, and the most common class of drug therapy used was targeted therapies [48]. Of the publications that reported duration of response to a previous drug given in the unresectable setting, 49% demonstrated a progression-free survival ratio of exceptional response to prior line of 1.3 or greater. Programs such as the NCI's Exceptional responders initiative[49] will perform NGS testing to characterize molecularly these cases and accumulate learning for the future of precision oncology [50].
4.3. Rare cancers
Rare cancers are those found in a small number of patients, and often standard second-line therapy is not established. Due to their rarity, these malignancies are often not studied in conventional phase III clinical trials that establish the value of newer therapies. Some examples are biliary tract cancers, sarcomas, mesothelioma and cancer of unknown primary.
In biliary tract cancers, cisplatin-gemcitabine combination chemotherapy is the reference first-line treatment regimen, but there is no standard second-line therapy. Mutation profiling has highlighted the genomic differences between the intra, extrahepatic cholangiocarcinoma, and gallbladder cancer [51]. There is a series of 75 cholangiocarcinoma patients in whom NGS-based testing was performed. There were significant differences in gene expression between intrahepatic and extrahepatic cholangiocarcinomas. IDH1 and DNA repair gene alterations occurred more frequently in intrahepatic cholangiocarcinomas, while ERBB2 gene alterations occurred in the extrahepatic group. BAP1 and FGFR gene pathway alterations occurred in both types of cholangiocarcinomas. Clinical benefit was noted with EGFR, FGFR, C-met, B-RAF and MEK inhibitors [52]. Very interestingly, NGS screening of intrahepatic cholangiocarcinomas identified gene fusions of NTRK, and responses have been observed with larotrectinib [53]. An estimated 15% of gallbladder cancers have Her2/neu amplification and could be targeted with antiHER2 therapies, and an estimated 10–15% of cholangiocarcinomas have DNA repair mutations and might be candidates for immune therapies. The MOSCATO trial analysed separately the 43 cases with advanced biliary tract cancer, and managed to administer molecular targeted agents in 18 cases, of which six had an objective response [54].
Sarcomas are a heterogeneous group of rare malignancies, with more than 50 subtypes recognized, and the majority of mutations detected by NGS are not drivers and do not translate into clinical benefit for patients [55]. However, for a patient with few treatment options, a clinical trial based on NGS-derived data may offer the treatment chance of research drugs.
At present, no therapies are approved in the second-line setting following progression of mesotheliomas after a platinum compound combined with pemetrexed. The mutational and transcriptomics landscape of mesothelioma has been published over the past several years, and highlights a limited number of loss-of-function tumor suppressor actionable mutations, most frequently in CDKN2A, BRCA1-associated protein-1 (BAP1), neurofibromin 2 (NF2), LATS2 and TP53 [56]. These gene alterations have been confirmed in small series of patients that were tested with NGS [57].
A recent review of 10 published studies using NGS on patients with Cancers of Unknown Primary (CUP) shows that mutations with potential therapeutic relevance were identified in 30%−85% of patients [58]. It is interesting that results from the AACR Project GENIE show that CUP were within the top 10% most highly mutated samples [59].
4.4. Clinical trial networks
An additional use of NGS and precision medicine is to direct patients to a particular clinical trial. This of course needs an organized structure of Clinical trial networks and new clinical trial designs. Currently, there is an unprecedented evolution in the design of early-stage cancer clinical trials, such as rapid phase 1 dose-escalation trials followed by remarkably large expansion cohorts, or new trials, as adaptive studies with basket and umbrella designs aimed at optimizing the biomarker–drug co-development process [60].
4.5. Clinical judgement-oriented testing
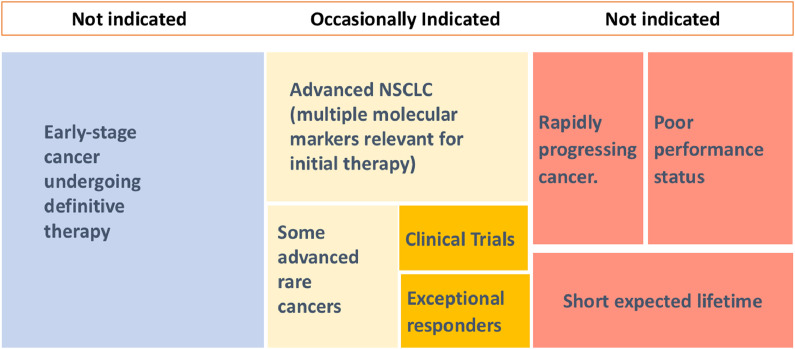
Clinical judgement must always precede molecular testing in cancer patients, since a NGS test is not indicated in a considerable number of cases. In general, a NGS test should not be ordered when the genomic test result will most likely not have an impact in cancer clinical management. For example, early-stage patients undergoing definitive treatment do not tipically require somatic gene panels. They will not have actionable alterations provided by NGS beyond what can be ascertained from standard evaluation (ER, PR, HER2 in the case of breast cancer) [44]. In another example, those cases with a rapidly progressing cancer, poor performance status, or with an expected lifetime of less than 3 months should not, as a rule, be molecularly profiled. These patients should most often be candidates for palliative therapy. (Fig. 2) Clinical judgement should also consider the frequency of targetable alterations in a specific population (e.g. advanced lung cancer in an elderly non-Asian smoker).
Fig. 2.
When to order a NGS Test in 2020. NGS is occasionally indicated in advanced cancers when there are several actionable targets (such as NSCLC), in some selected rare cancers, when there are clinical trials guided by molecular testing available, and in those cases with an exceptional response to a molecularly guided therapy. NGS is not indicated in early stage cancer, especially when definitive therapy will be delivered, or in patients with very advanced cancers that show rapid progression and have poor performance status or short expected lifetime.
4.6. Outstanding questions
Only in a number of practices single-gene sequencing has been routinely replaced with NGS sequencing and, therefore, the frequency of NGS testing is markedly regionally different.
Future issues that will need to be addressed in the near future will include establishing who should provide funding for NGS testing beyond government of insurance restrictions, improving the reproducibility of tests, improving the proportion of tests that are able to produce a meaningful result, and improving the algorithms for prediction of therapeutic benefit.
Conclusions
NGS-enabled precision oncology is adding a new layer of complexity to daily clinical decision making [61]. We have shown that NGS is most appropriate for patients who have an advanced cancer in which there are several commonly identified molecular targets, particularly when they are useful at the initial therapy. Additionally, NGS testing can be helpful in navigating patients to clinical trials, and can offer individual options in patients with rare cancers. For other cancers, NGS may be currently less appropriate because either the probability of identifying a targetable mutation is low [62], the cancer is in early stages with recognized and effective forms of standard treatment, or the patient has an irreversible disease with very short life-expectancy. As with any other laboratory test, doctors and patients must be sure before ordering an NGS test that its result will have an impact of the therapeutic plan. In any case, standard single-gene molecular testing must always be performed when indicated, since important therapeutic targets might be potentially missed if no molecular analyses were performed.
Clinical trials are showing that NGS testing can have an impact in the response rate and progression-free survival of patients, and can therefore be a very useful strategy leading to new molecularly-targeted treatment indications. Key factors responsible for improved results in precision-oriented clinical research, include refining the molecular pathways studied, developing molecular testing that integrates standarised genomic tests with transcriptomic analysis and immunohistochemistry, selecting more active targeted agents, designing combinations of targeted agents -also with other forms of therapy, and providing early treatment recommendations with available Molecular Multidisciplinary Boards. Interdisciplinary discussion are very important to help with the interpretation of unclear molecular results that are oftentimes seen with NGS testing.
Important unsolved issues that will need to be addressed in the future include deciding which is the best tissue to perform NGS (primary tumor vs metastasis, tumor DNA vs circulating tumor DNA), when is the right moment to test (at first diagnosis of advanced disease or when the disease is refractory), and whether there are NGS clinical trial designs that allow for the use of control groups. Finally, using a complete informed consent before NGS testing and communicating NGS reports to patients are two very important aspects of the procedure that have raised ethical concerns, and that must be always addressed by the practicing oncologists when ordering a NGS test.
Search strategy and selection criteria
We identified references through PubMed with the search terms “cancer AND NGS,” “cancer AND next generation sequencing,” “cancer AND genomics,” for articles published to March 30, 2020. The final reference list was generated on the basis of originality and relevance to the broad scope of this Review.
Funding
This Review was funded in part by research funds from projects PIE15/00068 and PI17/01865 (Instituto de Salud Carlos III) awarded to RC, projects JR17/00007 and PI17/008 (Instituto de Salud Carlos III), awarded to NR-L, PI15/01491 and PI19/00549 (Instituto de Salud Carlos III) awarded to AA, projects SAF2017–82886-R (Ministerio de Economía y Competitividad), INDISNET-S2011/BMD-2332 (Fundación Ramón Areces), and HR17-00016 ("La Caixa" Foundation) awarded to FS-M, and projects PI16/00354 (Instituto de Salud Carlos III) and B2017/BMD-3733 from the Consejería de Educación, Juventud y Deporte, Comunidad de Madrid, awarded to MQ-F. The manuscript is part of the activities of the endowed Chair of Personalised Precision Oncology, Universidad Autónoma de Madrid (UAM-Fundación Instituto Roche).
Authors contributions
RC, RM, NR-L, FS-M and MQ-F contributed to the study design. RC, RM, NR-L, and AA contributed to data collection and data analysis. RC, NR-L and MQ-F contributed to the interpretation of the data. RC designed and drew the figures. All authors contributed to the writing of this Review.
Declaration of Competing Interests
RC has received speaker's honoraria from BMS, AstraZeneca, Lilly, Roche, Pfizer, MSD, Janssen and Novartis, honoraria for participation as consultant or in Advisory Boards from Lilly, MSD, Roche and Servier, institutional research funding from BMS, MSD, Roche, Pfizer, AstraZeneca and Astellas, and has participated in academic activities funded by Fundacion Instituto Roche. RM has received speakers honoraria and honoraria from Advisory Boards from Merck, Roche, and Amgen, and has participated in academic activities funded by Fundacion Instituto Roche. NR-L has received speaker's honoraria from Bayer, Pfizer, Astellas Pharma, Sanofi, Janssen-Cilag, Roche, PharmaMar, and institutional research funding from Janssen-Cilag, Bayer, Astellas Pharma, Sanofi. MQ-F has received research funding from AstraZeneca and Bayer, and institutional research funding from MEI Pharma, Boehringer-Ingelheim, Bayer and Novartis. All other authors declare no competing interests.
References
- 1.Jameson J.L., Longo D.L. Precision medicine–personalized, problematic, and promising. N Engl J Med. 2015 Jun 4;372(23):2229–2234. doi: 10.1056/NEJMsb1503104. [DOI] [PubMed] [Google Scholar]
- 2.Hodson R. Precision medicine. Nature. 2016 Sep 8;537(7619):S49. doi: 10.1038/537S49a. [DOI] [PubMed] [Google Scholar]
- 3.Wheeler D.A., Wang L. From human genome to cancer genome: the first decade. Genome Res. 2013 Jul;23(7):1054–1062. doi: 10.1101/gr.157602.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Green E.D., Watson J.D., Collins F.S. Human Genome Project: twenty-five years of big biology. Nature. 2015 Oct 1;526(7571):29–31. doi: 10.1038/526029a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mirnezami R., Nicholson J., Darzi A. Preparing for precision medicine. N Engl J Med. 2012 Feb 9;366(6):489–491. doi: 10.1056/NEJMp1114866. [DOI] [PubMed] [Google Scholar]
- 6.Jordan V.C, Fourteenth Gaddum Memorial Lecture A current view of tamoxifen for the treatment and prevention of breast cancer. Br J Pharmacol. 1993 Oct;110(2):507–517. doi: 10.1111/j.1476-5381.1993.tb13840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mateo J., Chakravarty D., Dienstmann R. A framework to rank genomic alterations as targets for cancer precision medicine: the ESMO scale for clinical actionability of molecular targets (ESCAT) Ann Oncol. 2018;29(9):1895–1902. doi: 10.1093/annonc/mdy263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marquart J., Chen E.Y., Prasad V. Estimation of the percentage of us patients with cancer who benefit from genome-driven oncology. JAMA Oncol. 2018 Aug 1;4(8):1093–1098. doi: 10.1001/jamaoncol.2018.1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.BrandãoM, Sotiriou C. Multigene sequencing in breast cancer: ESMO biomarker factsheet. (15 Jan 2019) https://oncologypro.esmo.org/Education-Library/Factsheets-on-Biomarkers/Multigene-Sequencing-in-Breast-Cancer). Accessed on November 17, 2019.
- 10.Addeo A., Banna G.L., Weiss G.J. Tumor mutation burden-from hopes to doubts. JAMA Oncol. 2019;5(7):934–935. doi: 10.1001/jamaoncol.2019.0626. [DOI] [PubMed] [Google Scholar]
- 11.Le Tourneau C., Kama M., Bièche I. Multigene sequencing for treatment selection: ESMO biomarker factsheet (17 Jul 2017) (https://oncologypro.esmo.org/education-library/factsheets-on-biomarkers/multigene-sequencing-for-treatment-selection) accessed November 17, 2019.
- 12.Legras A., Barritault M., Tallet A. Validity of targeted next-generation sequencing in routine care for identifying clinically relevant molecular profiles in non-small-cell lung cancer: results of a 2-year experience on 1343 samples. J Mol Diagn. 2018;20(4):550–564. doi: 10.1016/j.jmoldx.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 13.Kalokairinou L., Howard H.C., Slokenberga S., Fisher E., Flatscher-Thöni M., Hartlev M., van Hellemondt R., Juškevičius J., Kapelenska-Pregowska J., Kováč P., Lovrečić L., Nys H., de Paor A., Phillips A., Prudil L., Rial-Sebbag E., Romeo Casabona C.M., Sándor J., Schuster A., Soini S., Søvig K.H., Stoffel D., Titma T., Trokanas T., Borry P. Legislation of direct-to-consumer genetic testing in Europe: a fragmented regulatory landscape. J Commun Genet. 2018 Apr;9(2):117–132. doi: 10.1007/s12687-017-0344-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.https://www.corehta.info/ReviewResults.aspx?p=113&pg=1. (31 Jan 2013)
- 15.Stetson D., Ahmed A., Xu X., et al. Orthogonal comparison of four plasma NGS tests with tumor suggests technical factors are a major source of assay discordance. JCO Precis Oncol 2019, vol 3 - published online March 14, 2019. [DOI] [PubMed]
- 16.Nagahashi M., Shimada Y., Ichikawa H. Formalin-fixed paraffin-embedded sample conditions for deep next generation sequencing. J Surg Res. 2017;220:125–132. doi: 10.1016/j.jss.2017.06.077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kennedy S.R., Zhang Y., Risques R.A. Cancer-associated mutations but no cancer: insights into the early steps of carcinogenesis and implications for early cancer detection. Trends Cancer. 2019 Sep;5(9):531–540. doi: 10.1016/j.trecan.2019.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.West H. Novel precision medicine trial designs: umbrellas and baskets. JAMA Oncol. 2017;3(3):423. doi: 10.1001/jamaoncol.2016.5299. [DOI] [PubMed] [Google Scholar]
- 19.Le Tourneau C., Kamal M., Tsimberidou A.M. Treatment algorithms based on tumor molecular profiling: the essence of precision medicine trials. J Natl Cancer Inst. 2015;108(4) doi: 10.1093/jnci/djv362. djv362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Le Tourneau C., Delord J.P., Gonçalves A. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol. 2015 Oct;16(13):1324–1334. doi: 10.1016/S1470-2045(15)00188-6. [DOI] [PubMed] [Google Scholar]
- 21.Massard C., Michiels S., Ferté C. High-throughput genomics and clinical outcome in hard-to-treat advanced cancers: results of the MOSCATO 01 trial. Cancer Discov. 2017;7(6):586–595. doi: 10.1158/2159-8290.CD-16-1396. [DOI] [PubMed] [Google Scholar]
- 22.Hainsworth J.D., Meric-Bernstam F., Swanton C. Targeted therapy for advanced solid tumors on the basis of molecular profiles: results from mypathway, an open-label, phase IIa multiple basket study. J Clin Oncol. 2018;36(6):536–542. doi: 10.1200/JCO.2017.75.3780. [DOI] [PubMed] [Google Scholar]
- 23.Slosberg E.D., Kang B.P., Peguero J. Signature program: a platform of basket trials. Oncotarget. 2018 Apr 20;9(30):21383–21395. doi: 10.18632/oncotarget.25109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.tapur.org/news (20 Feb 2020), accessed on March 23, 2020.
- 25.Gupta R., Garrett-Mayer E., Halabi S., et al. Pertuzumab + trastuzumab in patients with colorectal cancer with ERBB2 amplification or overexpression: results from the targeted agent and profiling utilization registry (TAPUR) study. 2020 ASCO GI Cancers Symposium in San Francisco, CA - January 25, 2020.
- 26.Klute K.A., Garrett-Mayer E., Halabi S., et al. Cobimetinib + vemurafenib in patients with colorectal cancer with BRAF V600E mutations: results from the targeted agent and profiling utilization registry (TAPUR) study. 2020 ASCO GI Cancers Symposium in San Francisco, CA - January 25, 2020.
- 27.Alva Ajjai Shivaram, K Pam, Mangat Elizabeth Garrett-Mayer. Pembrolizumab (P) in patients (pts) with metastatic breast cancer (MBC) with high tumor mutational burden (HTMB): results from the targeted agent and profiling utilization registry (TAPUR) study. J Clin Oncol. May 20, 2019;37(15_suppl) doi: 10.1200/JCO.20.02923. 1014-1014. [DOI] [PubMed] [Google Scholar]
- 28.Meiri E., Garrett-Mayer E., Halabi S., et al. Pembrolizumab in patients with metastatic colorectal cancer with high tumor mutational burden (HTMB): results from the targeted agent and profiling utilization registry (TAPUR) study. 2020 ASCO GI Cancers Symposium in San Francisco, CA - January 25, 2020.
- 29.Ahn Eugene R, Pam K, Mangat Elizabeth Garrett-Mayer. Palbociclib (P) in patients (pts) with non-small cell lung cancer (NSCLC) with CDKN2A alterations: results from the targeted agent and profiling utilization registry (TAPUR) study. J Clin Oncol. May 20, 2019;37(15_suppl) 9041-9041. [Google Scholar]
- 30.Papadimitrakopoulou V., Lee J.J., Wistuba I.I. The BATTLE-2 study: a biomarker-integrated targeted therapy study in previously treated patients with advanced non-small-cell lung cancer. J Clin Oncol. 2016;34(30):3638–3647. doi: 10.1200/JCO.2015.66.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodon J., Soria J.C., Berger R. Genomic and transcriptomic profiling expands precision cancer medicine: the WINTHER trial. Nat Med. 2019 May;25(5):751–758. doi: 10.1038/s41591-019-0424-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Velden D.L., Hoes L.R., van der Wijngaart H. The Drug Rediscovery protocol facilitates the expanded use of existing anticancer drugs. Nature. 2019 Oct;574(7776):127–131. doi: 10.1038/s41586-019-1600-x. [DOI] [PubMed] [Google Scholar]
- 33.Sicklick J.K., Kato S., Okamura R. Molecular profiling of cancer patients enables personalized combination therapy: the I-PREDICT study. Nat Med. 2019 May;25(5):744–750. doi: 10.1038/s41591-019-0407-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rothwell D.G., Ayub M., Cook N. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat Med. 2019 May;25(5):738–743. doi: 10.1038/s41591-019-0380-z. [DOI] [PubMed] [Google Scholar]
- 35.Rothwell D.G., Ayub M., Cook N. Utility of ctDNA to support patient selection for early phase clinical trials: the TARGET study. Nat Med. 2019;25(5):738–743. doi: 10.1038/s41591-019-0380-z. [DOI] [PubMed] [Google Scholar]
- 36.Le Tourneau C., Borcoman E., Kamal M. Molecular profiling in precision medicine oncology. Nat Med. 2019 May;25(5):711–712. doi: 10.1038/s41591-019-0442-2. [DOI] [PubMed] [Google Scholar]
- 37.Coyne G.O., Takebe N., Chen A.P. Defining precision: the precision medicine initiative trials NCI-MPACT and NCI-MATCH. Curr Probl Cancer. 2017;41(3):182–193. doi: 10.1016/j.currproblcancer.2017.02.001. [DOI] [PubMed] [Google Scholar]
- 38.Prasad V., De Jesús K., Mailankody S. The high price of anticancer drugs: origins, implications, barriers, solutions. Nat Rev Clin Oncol. 2017;14(6):381–390. doi: 10.1038/nrclinonc.2017.31. [DOI] [PubMed] [Google Scholar]
- 39.Remon J., Dienstmann R. Precision oncology: separating the wheat from the chaff. ESMO Open. 2018;3(6) doi: 10.1136/esmoopen-2018-000446. Published 2018 Oct 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swanton C., Soria J.C., Bardelli A. Consensus on precision medicine for metastatic cancers: a report from the MAP conference. Ann Oncol. 2016;27(8):1443–1448. doi: 10.1093/annonc/mdw192. [DOI] [PubMed] [Google Scholar]
- 41.McKenzie A.J., H Dilks H., Jones S.F., Burris H., 3rd Should next-generation sequencing tests be performed on all cancer patients? Expert Rev Mol Diagn. 2019;19(2):89–93. doi: 10.1080/14737159.2019.1564043. [DOI] [PubMed] [Google Scholar]
- 42.Schwartzberg L., Kim E.S., Liu D., Schrag D. Precision Oncology: who, How, What, When, and When Not? Am Soc Clin Oncol Educ Book. 2017;37:160–169. doi: 10.1200/EDBK_174176. [DOI] [PubMed] [Google Scholar]
- 43.Garrido P., Conde E., de Castro J. Updated guidelines for predictive biomarker testing in advanced non-small-cell lung cancer: a national consensus of the spanish society of pathology and the Spanish society of medical oncology [published online ahead of print, 2019 Oct 9] Clin Transl Oncol. 2019 doi: 10.1007/s12094-019-02218-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yu T.M., Morrison C., Gold E.J. Multiple biomarker testing tissue consumption and completion rates with single-gene tests and investigational use of oncomine Dx target test for advanced non-small-cell lung cancer: a single-center analysis. Clin Lung Cancer. 2019;20(1):20–29.e8. doi: 10.1016/j.cllc.2018.08.010. [DOI] [PubMed] [Google Scholar]
- 45.Pennell N.A., Mutebi A., Zhou Z.-.Y. Economic impact of next generation sequencing vs sequential single-gene testing modalities to detect genomic alterations in metastatic non-small cell lung cancer using a decision analytic model. J Clinical Oncol. 2018;36(15_suppl):9031. doi: 10.1200/JCO.2018.36.15_suppl.9031. 9031-9031. [DOI] [PubMed] [Google Scholar]
- 46.Doble B., John T., Thomas D., Fellowes A. Cost-effectiveness of precision medicine in the fourth-line treatment of metastatic lung adenocarcinoma: an early decision analytic model of multiplex targeted sequencing. Lung Cancer. 2017;107:22–35. doi: 10.1016/j.lungcan.2016.05.024. [DOI] [PubMed] [Google Scholar]
- 47.Pérez-Gracia J.L., Gúrpide A., Ruiz-Ilundain M.G. Selection of extreme phenotypes: the role of clinical observation in translational research. Clin Transl Oncol. 2010;12(3):174–180. doi: 10.1007/s12094-010-0487-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nishikawa G., Luo J., Prasad V. A comprehensive review of exceptional responders to anticancer drugs in the biomedical literature. Eur J Cancer. 2018;101:143–151. doi: 10.1016/j.ejca.2018.06.010. [DOI] [PubMed] [Google Scholar]
- 49.Mullard A. Learning from exceptional drug responders. Nat Rev Drug Discov. 2014;13(6):401–402. doi: 10.1038/nrd4338. [DOI] [PubMed] [Google Scholar]
- 50.Jonsson P., Taylor B.S. Transforming biomarker development with exceptional responders. Trends Cancer. 2018;4(1):3–6. doi: 10.1016/j.trecan.2017.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jain A., Kwong L.N., Javle M. Genomic profiling of biliary tract cancers and implications for clinical practice. Curr Treat Options Oncol. 2016;17(11):58. doi: 10.1007/s11864-016-0432-2. [DOI] [PubMed] [Google Scholar]
- 52.Putra J., de Abreu F.B., Peterson J.D. Molecular profiling of intrahepatic and extrahepatic cholangiocarcinoma using next generation sequencing. Exp Mol Pathol. 2015;99(2):240–244. doi: 10.1016/j.yexmp.2015.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cocco E., Scaltriti M., Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol. 2018;15(12):731–747. doi: 10.1038/s41571-018-0113-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verlingue L., Malka D., Allorant A. Precision medicine for patients with advanced biliary tract cancers: an effective strategy within the prospective MOSCATO-01 trial. Eur J Cancer. 2017 Dec;87:122–130. doi: 10.1016/j.ejca.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 55.Pestana R.C., Groisberg R., Roszik J., Subbiah V. Precision oncology in sarcomas: divide and conquer. JCO Precis Oncol. 2019;3:1–16. doi: 10.1200/PO.18.00247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hinz T.K., Heasley L.E. Translating mesothelioma molecular genomics and dependencies into precision oncology-based therapies [published online ahead of print, 2019 Sep 20] Semin Cancer Biol. 2019 doi: 10.1016/j.semcancer.2019.09.014. S1044-579X(19)30288-3. [DOI] [PubMed] [Google Scholar]
- 57.Ugurluer G., Chang K., Gamez M.E. Genome-based mutational analysis by next generation sequencing in patients with malignant pleural and peritoneal mesothelioma. Anticancer Res. 2016;36(5):2331–2338. [PubMed] [Google Scholar]
- 58.Binder C., Matthes K.L., Korol D. Cancer of unknown primary-Epidemiological trends and relevance of comprehensive genomic profiling. Cancer Med. 2018;7(9):4814–4824. doi: 10.1002/cam4.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.AACR Project GENIE Consortium AACR project GENIE: powering precision medicine through an international consortium. Cancer Discov. 2017 Aug;7(8):818–831. doi: 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Garralda E., Dienstmann R., Piris-Giménez A. New clinical trial designs in the era of precision medicine. Mol Oncol. 2019;13(3):549–557. doi: 10.1002/1878-0261.12465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horak P., Fröhling S., Glimm H. Integrating next-generation sequencing into clinical oncology: strategies, promises and pitfalls. ESMO Open. 2016;1(5) doi: 10.1136/esmoopen-2016-000094. Published 2016 Nov 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ewalt M.D., West H., Aisner D.L. Next generation sequencing-testing multiple genetic markers at once. JAMA Oncol. 2019;5(7):1076. doi: 10.1001/jamaoncol.2019.0453. [DOI] [PubMed] [Google Scholar]
- 63.Chen H.Z., Bonneville R., Roychowdhury S. Implementing precision cancer medicine in the genomic era. Semin Cancer Biol. 2019;55:16‐27. doi: 10.1016/j.semcancer.2018.05.009. [DOI] [PubMed] [Google Scholar]