Graphical abstract
Keywords: Non-synonymous single nucleotide variants, Protein stability, Protein function, Computational tools and databases, Machine learning, Performance bias, Mutations
Abstract
Protein stability predictions are becoming essential in medicine to develop novel immunotherapeutic agents and for drug discovery. Despite the large number of computational approaches for predicting the protein stability upon mutation, there are still critical unsolved problems: 1) the limited number of thermodynamic measurements for proteins provided by current databases; 2) the large intrinsic variability of ΔΔG values due to different experimental conditions; 3) biases in the development of predictive methods caused by ignoring the anti-symmetry of ΔΔG values between mutant and native protein forms; 4) over-optimistic prediction performance, due to sequence similarity between proteins used in training and test datasets. Here, we review these issues, highlighting new challenges required to improve current tools and to achieve more reliable predictions. In addition, we provide a perspective of how these methods will be beneficial for designing novel precision medicine approaches for several genetic disorders caused by mutations, such as cancer and neurodegenerative diseases.
1. Introduction
Protein structure is determined by the interactions among the amino acids and with the environment, resulting in a stable 3D structure. In the folding process, a stable 3D conformation of the protein corresponds to a minimum of the Gibbs free energy (ΔG) of the protein-environment system. ΔG of folding includes both the entropic contributions (hydrophobic effects and protein configurations) and the interaction energies within the protein (like Van der Waals interactions, hydrogen and electrostatic bonding, etc.). In many cases, the functional cycle of the protein is accomplished by switching among a few stable 3D conformations, implying the existence of several local free energy minima experienced by the same protein and the need of keeping the energy barriers among them relatively low. Non-synonymous DNA variations, which alter the amino acid sequence, may change protein function by either increasing or decreasing protein stability, which may prevent the conformational changes required for the protein to function [1]. Mutations occurring at the surface of a protein domain are often neutral. Yet, they may affect the binding affinities for other proteins or the inter-domain compactness in multi-domain proteins. Mutations occurring in the core may alter the stability in the domain fold [2]. Indeed, protein stability changes have been shown to constitute one of the major underlying molecular mechanisms in several mutation-induced diseases [3] and might prove to be an even more frequent cause of function loss and disease than previously thought. Understanding how specific mutations in a patient affect protein stability or interactions can identify possible drug resistance/sensitivity in that patient, allowing better therapeutic approaches. In addition, such knowledge is important for improving the protein design through site-directed or random mutagenesis, leading to promising new approaches for precision medicine.
The effects of non-synonymous variants on protein stability are quantified in terms of the Gibbs free energy of unfolding (ΔG). ΔG is a nonlinear function of several factors, like temperature, pH values, concentrations of salt, organic solvents, urea and other chemical agents. From the experimental point of view, the measure of interest is the difference of the unfolding free energy between the mutated and wild-type proteins (ΔΔGu). Thus ΔΔGu = ΔGumutant- ΔGuwild-type, which is the difference of the free energy of unfolding between the mutated and wild-type proteins (Fig. 1, central box). The sign of ΔΔG indicates if the variation decreases (ΔΔGu < 0) or increases (ΔΔGu > 0) the protein stability. However, the opposite convention is also possible (ΔΔGf = ΔGfmutant − ΔGfwild-type). Thus, it is crucial to specify which definition is adopted. In this review, we will use the symbol ΔΔG to refer to ΔΔGu.
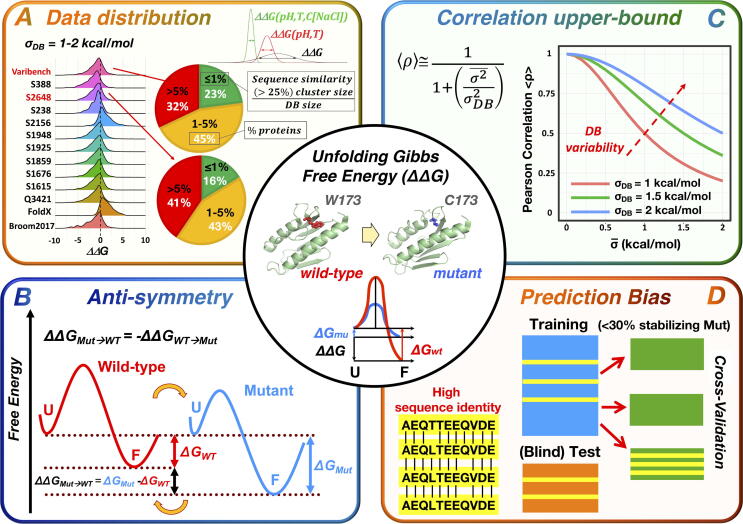
Fig. 1.
Summary of the main issues affecting the prediction of stability changes upon mutation. A) Distribution of ΔΔG values from current databases (DB), characterized by σDB < 2 kcal/mol, and intrinsic variability of the available ΔΔG values, which are obtained under different experimental conditions (upper-right corner of panel A). The pie-charts represent the distribution of proteins clustered by sequence similarity for the two largest and most widely used datasets (S2648 and VariBench). Clusters were obtained using blastclust (www.ncbi.nlm.nih.gov/Web/Newsltr/Spring04/blastlab.html), using a threshold cut-off of 25% of identity. The proportion of proteins belonging to clusters with a size representing <= 1%, 1–5% and > 5% of the size of the entire DB are highlighted in the pie-charts in green, orange and red, respectively. The smaller the cluster size, the lower the probability of choosing proteins with sequence identity or high sequence similarity. Only a small fraction of the datasets can be randomly selected (green slices). B) Anti-symmetric properties of the folding process. U = unfolded; F = folded. C) Low upper-bound for correlation estimates due to low variability of ΔΔG values in current databases. D) Biased datasets, due to unbalance between destabilizing and stabilizing (< 30%) variants and to the presence of proteins with high sequence identity, which generate inappropriate training/testing sets leading to over-fitting issues during the learning process (e.g yellow-green set in cross validation and the blind test set in panel D). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Protein stability has been traditionally measured with circular dichroism, differential scanning calorimetry, fluorescence spectroscopy using thermal and denaturant unfolding methods [4]. Although experimental approaches to measure differences between mutant and wild-type proteins are accurate, they are demanding in terms of time and costs. Therefore, several computational methods have been developed for assessing and predicting how protein stability is affected by single-point mutations [5]. Most of them aim at either classifying the effect (stabilizing/destabilizing/neutral) or quantifying the ΔΔG values.
Several tools for predicting pathogenic variants have been developed and reviewed [6], [7], [8], [9], [10]. In this work, we focus on the methods and issues related to the robust prediction of ΔΔG values, providing a comprehensive evaluation of both the limits of current approaches and some possible solutions to achieve better predictions by minimizing the bias in the performance assessment. Recent critical reviews were already provided by Pucci et al. [11] and Fang [12]. Here we will extend those reviews by introducing some further crucial points, discussing: 1) the unbalance and intrinsic variability of the thermodynamic data; 2) the failure of taking into account the anti-symmetric property of the folding process (ΔΔGu = −ΔΔGf ); 3) the best practices for providing a fair assessment of prediction performance. Beyond discussing these critical points, an additional aim of the present review is to provide a perspective in using these prediction approaches for direct biomedical applications. Such tools could help clinicians in selecting the most appropriate treatments and creating novel therapeutic strategies based on a more thorough genetic understanding of the patient's disease.
2. Stability data and benchmark sets
As pointed out by Pucci and Rooman [13], current experimental and computational techniques provide only a partial picture of protein folding and adaptation processes, making the comparison between different prediction approaches a non-trivial task. Furthermore, despite the large number of benchmark datasets provided by VariBench and VariSNP used for variation interpretation [14], only a few sources are available for protein stability studies. The main repository to retrieve protein stability information used to be ProTherm [15], which provided a collection of thermodynamic measures including Gibbs free energy change, enthalpy change, heat capacity change, transition temperature etc. for both wild-type and mutant proteins. The last version (5.0) contained ~ 17,000 entries from 771 proteins. Reported thermodynamic measurements for wild-type proteins, single, double and multiple mutants were 7014, 8202, 1277 and 620, respectively. However, this database is not available anymore, and it is known to present several inconsistencies [14], including missing thermodynamic values and ΔΔG values with opposite sign. Data need to be filtered and manually cleaned for collecting reliable datasets. For these reasons, a large number of curated benchmark datasets was derived from ProTherm. The most used ones are reported in Table 1. Despite the several considered subsets, there is an urgent need for new experimental data, particularly for stabilizing mutations: they account for <30% of the datasets reported above; the worst case is represented by S388, with only 11% stabilizing mutations. Indeed, the available datasets are still limited compared to the thousands of three-dimensional (3D) protein structures available in the Protein Data Bank (PDB) [34], which in total includes ~ 167,000 proteins (as of July 2020). A major limitation of the currently available data is the intrinsic variability characterizing the ΔΔG values. Two experimental ΔΔG measurements of the same variant may disagree if not carried out under the same conditions and by the same experimental technique. Therefore, methods have to deal with ΔΔG values that have different uncertainties associated with their measurements (see Fig. 1A) and multiple experimental ΔΔG values for the same mutation. As an example, in Keeler et al. [35] the variation of Histidine 180 to Alanine in the human prolactin measured at 25 °C, but different pH, corresponds to a ΔΔG = 1.39 kcal/mol at pH = 5.8 and ΔΔG = -0.04 at pH = 7.8. In Ferguson and Shaw [36], the variation of Leucine 3 to Serine in the calcium-binding protein S100B, measured under two different starting conditions and techniques but at the same temperature (25 °C) and pH (7.2), yielded two different ΔΔG: 1.91 and −2.77 kcal/mol. In addition, some works used all the ΔΔG values for training/testing, while other analyses focused on selected subsets of ΔΔG values obtained under specific experimental conditions (e.g. S388) or took their average or weighted average (e.g. S2156 and S2648). Considering the overall ΔΔG value distributions for the most used databases, the observed variability is high (Fig. 1A); e.g. ΔΔG standard deviation is 2.06 kcal/mol in ProTherm, 1.91 kcal/mol in Varibench [31], 1.47 kcal/mol in S2648 [25]. Actually, a meaningful comparison of different methods should be based on the same datasets.
Table 1.
The most used datasets and subsets derived from ProTherm Database.
| Dataset | Total Variants (Proteins) | Stabilizing Variants (Proteins) | Destabilizing Variants (Proteins) | Additional Details |
|---|---|---|---|---|
| Broom2017 [16] | 605 (58) | 147 (37) | 458 (54) | Unique Variants/Background Variants |
| Cao Test [17] | 276 (37) | 79 (21) | 197 (35) | Replicated Variants |
| Cao Train [17] | 5,444 (2 0 4) | 1,233 (1 5 0) | 4,211 (1 8 5) | Replicated Variants |
| Fold-X [18] | 964 (38) | 110 (25) | 854 (36) | Unique Variants |
| Myoglobin [19] | 134 (1) | 36 (1) | 98 (1) | One Protein |
| p53 [20] | 42 (1) | 11 (1) | 31 (1) | One Protein |
| PTmul [21] | 914 (90) | 310 (57) | 604 (68) | Unique Variants/Multiple Variants |
| Q3421 [22] | 3,421 (1 4 8) | 763 (1 1 4) | 2,658 (1 3 1) | Unique Variants/Averaged ΔΔG |
| S1615 [23] | 1,615 (41) | 449 (35) | 1166 (35) | Unique Variants |
| S1676 [24] | 1676 (95) | 453 (53) | 1,223 (62) | Unique Variants/Averaged ΔΔG |
| S1859 [25] | 1,859 (64) | 583 (48) | 1,276 (55) | Replicated Variants/Averaged ΔΔG |
| S1925 [20] | 1925 (55) | 582 (42) | 1,343 (48) | Replicated Variants |
| S1948 [26] | 1,948 (58) | 592 (45) | 1,356 (50) | Replicated Variants |
| S2156 [27] | 2,156 (84) | 472 (61) | 1,684 (68) | Unique Variants/Averaged ΔΔG |
| S238 [28] | 238 (25) | 45 (16) | 193 (20) | Unique Variants/Subset of S1948 |
| S2648 [28] | 2,648 (1 3 1) | 602 (96) | 2,046 (1 1 8) | Unique Variants/Averaged ΔΔG |
| S3366[29] | 3,366 (1 3 0) | 836 (1 0 3) | 2,530 (1 1 0) | Unique/Single and Multiple Variants |
| S350 [28] | 350 (67) | 90 (35) | 260 (57) | Unique Variants/Subset of S2648 |
| S388 [23] | 388 (17) | 48 (12) | 340 (15) | Unique Variants/Physiological Conditions |
| S3568 [30] | 3,568 (1 5 4) | 947 (1 1 0) | 2,621 (1 3 8) | Replicated Variants |
| S630 [30] | 630 (39) | 467 (26) | 163 (32) | Replicated Variants |
| Ssym* [11] | 342 (15) | 90 (10) | 251 (13) | Unique/Symmetric Variants |
| VariBench [31] | 1,564 (89) | 436 (70) | 1,128 (78) | Unique Variants |
| VariBench3D[31] | 1,423(79) | 382 (60) | 1,041 (68) | Variants with available structures from [31] |
Unique Variants: Only one ΔΔG value for each variation. Replicated Variants: Multiple data for the same variant are included. Averaged ΔΔG: Multiple ΔΔG values for the same variant are replaced with their average. Multiple Variant: The dataset includes variation data for protein with multiple-site variants. Background Variants: The initial protein used as a reference for calculating the ΔGs is different from the wild-type. Physiological Conditions: Temperature 20–40 °C and pH: 6–8. *Reported data only for direct variants.
A possible solution to generate new ΔΔG data might be the application of molecular dynamics (MD) simulations. MD is a powerful tool that enables the investigation of conformational changes in proteins [32], [33], [34], [35]. However, to use this approach, an extensive number of simulations under different conditions (e.g. with different force fields) would be necessary, given the high number of experimental variations with a ΔΔG lower than single hydrogen-bond energy. This requires a super-accurate force field and MD tuning. Nonetheless, with the increasing computational power and force field accuracy, MD simulations might play a relevant role in this field in the future, at least as data-augmentation tools.
3. Computational tools for predicting protein stability change
Existing prediction algorithms use one or a combination of the following features, characterizing protein stability:
-
-
Structural-based features: residue contact networks, residue/atom distances, protein geometry, etc.
-
-
Sequence-based features: based on conserved sequences and amino acid positions. They can provide the impact on protein viability but no information at a functional level.
-
-
Energy-based features: energy of unfolding of the target protein as the sum of various energies such as Van der Waals interactions, solvation energy, extra-stabilizing free energy, etc.
-
-
Molecular-based features: solvent accessible surface area of the interface, hydrophobic and hydrophilic area. Most of these features are derived by the structure-based ones; however, some of them, such as hydrophobicity or molecular weight, are related to the chemistry of the residue, and can be described without the structure information.
Most of the current methods use a mixture of all the above-described features, although some of them focus only on a specific subset. The first predictive approaches for protein stability have been developed through the application of force fields based on physical free energy functions derived from molecular mechanics [32], which are often combined with molecular dynamics or Monte Carlo simulations [33], [34], [35]. A force field is a collection of bonded and non-bonded interaction terms that are related by a set of equations, which can be used to estimate the potential energy of a molecular system. However, these approaches are computationally intensive, and this limits their practical application to small sets of protein mutants. For this reason, problem-specific physical-based methods have been introduced, such as methods based on empirical energy functions [18], [28], [36], which apply the Maxwell-Boltzmann statistics to estimate the propensities of interaction between atoms from a set of known protein structures. These functions are also known as scoring functions, empirical potentials, knowledge-based potentials, or statistical potentials. In contrast to the force fields approaches, empirical potentials are based on geometrical descriptors, reporting information from experimental data of known protein structures. The advantage of these models, based on principles from statistical physics, is that they provide a trade-off between computational cost and accuracy of the free energy function. Among the most popular approaches, FoldX [18] uses van der Waals and electrostatic energies with additional hydrogen bond and solvation contributions. Model parameters are fitted to a given set of experimental data and the resulting model is used to predict new mutation-induced folding free energy changes. To model mutations, FoldX uses a rotamer approach, allowing conformational changes of sidechains and keeping the backbone fixed.
Another group of classical linear ΔΔG predictors are based on Molecular Mechanics/Poisson − Boltzmann Surface Area (MM/PBSA) or Molecular Mechanics/Generalized Born Surface Area (MM/GBSA) methods [37], [38]. These methods typically use ensembles of conformations from molecular dynamics simulations, combining the molecular mechanics energies with solvation energies. Among the most recent of these approaches, SAAFEC [39] combines a MM/PBSA component and a set of knowledge-based terms of biophysical characteristics into a multiple linear regression model. The goal is not only to accurately predict ΔΔG values, but also to characterize the structural changes induced by mutations and the physical nature of the predicted folding free energy changes. In general, these MM/PBSA‐based methods demonstrate a rather good prediction performance at a reasonable computational cost.
Data-driven computational tools based on both simple regression or machine-learning approaches like Support Vector Machine (SVM), Random Forest (RF) and Artificial Neural Network (ANN) methods have also been explored to predict ΔΔG. Most known and recent approaches are reported in Table 2, together with the tools mentioned above. These methods are based on a first training phase using examples of proteins and their mutants, for which the ΔΔGs have been experimentally measured. Machine learning approaches do not require a full understanding of the principles underlying the target function, since these are modeled during the learning process. This aspect increases the flexibility of these approaches in building new features, revealing unrecognized patterns, relationships and dependencies not considered by knowledge-based models. By construction, these methods can combine all kinds of computable features. In addition, these approaches are much less time-consuming compared to the previous techniques since, once a model is built from the data, the prediction is immediate. Most of these methods use statistical potentials of environmental propensities, substitution frequencies and correlations of adjacent residues found experimentally in protein structure, enabling non-energy-like terms to be incorporated into the scoring function. However, these estimations are highly dependent on the availability of large and diverse experimental training data, increasing the risks of over-fitting issues, and their results might not be easily interpreted in physical terms.
Table 2.
A selection of the most popular methods to predict the functional effects of missense variants in terms of protein stability, i.e. prediction of ΔΔG values.
| Method | 3D/1D features required | Algorithm | Basic idea and advantages | Anti-Symmetry | Multiple mutations | URL |
|---|---|---|---|---|---|---|
| FoldX [18] | 3D | linear regression | Based on empirical physical-based energies | NO | YES | http://foldxsuite.crg.eu/ |
| MUpro [51] | 3D and 1D | ANN and SVMs | It predicts from sequence | NO | NO | http://mupro.proteomics.ics.uci.edu/ |
| CUPSAT [36] | 3D | combined statistical potentials | It provides information about the site of mutation and protein structural features | NO | NO | http://cupsat.tu-bs.de/ |
| I-Mutant(3.0/2.0) [26], [40] | 3D or 1D | SVM | It classifies the predictions into three classes: neutral, large increase or decrease | NO | NO | http://gpcr2.biocomp.unibo.it/cgi/predictors/I-Mutant3.0/I-Mutant3.0.cgi |
| iPTREE-STAB [25] | 1D | Decision trees and adaptive boosting | It also provides numerical stability values | NO | NO | http://bioinformatics.myweb.hinet.net/iptree.htm |
| AUTO-MUTE (2.0) [69] | 3D | Random Forest | It considers spatial perturbations or 3D neighbors of the modified residue | NO | NO | http://binf.gmu.edu/automute/ |
| Prethermut [29] | 3D | SVM + Random Forest | It predicts single- and multi-site mutations | NO | YES | http://www.mobioinfor.cn/prethermut/download.htm |
| POPMUSIC(3.1/2.1) [28] | 3D | ANN + Statistical potential | Linear combination of statistical potentials whose coefficients depend on the solvent accessibility of the modified residue | NO | NO | http://babylone.ulb.ac.be/popmusic |
| Pro-Maya [52] | 3D and 1D | Random Forest regression | It uses available data on mutations occurring in the same position and in other positions | NO | NO | http://bental.tau.ac.il/ProMaya |
| PROTS-RF [51] | 3D and 1D | Random Forest | Derived from a non-redundant representative collection of thousands of thermophilic and mesophilic protein structures | YES | YES | Unavailable |
| iStable(2.0) [30], [53], | 3D or 1D | meta-predictor | It uses either sequence or structure information | NO | NO | http://predictor.nchu.edu.tw/iStable/ |
| NeEMO [54] | 3D | ANN regression | Based on residue-residue interaction networks | NO | NO | http://protein.bio.unipd.it/neemo/help.html |
| DUET [41] | 3D | meta-predictor (SVM regression) | It integrates mCSM and SDM in a consensus prediction | NO | NO | http://biosig.unimelb.edu.au/duet/stability |
| mCSM [20] | 3D | Gaussian process regression and random forest | It translates the distance patterns between atoms into graph-based signatures providing complementary data to potential energy-based approaches | NO | NO | http://biosig.unimelb.edu.au/mcsm/ |
| EASE-MM [24] | 1D | SVM | It combines 5 SVM models and makes the final prediction from a consensus of 2 models selected based on the predicted secondary structure and accessible surface area of the mutated residue | NO | NO | http://sparks-lab.org/server/ease |
| INPS(3D) [43], [55] | 3D or 1D | SVM regression | It takes into account evolutionary information from sequence | YES | NO | http://inps.biocomp.unibo.it |
| STRUM [22] | 3D and 1D | Gradient boosting regression trees | It combines sequence profiles with low-resolution 3D models constructed by iterative threading assembly refinement simulations | NO | NO | https://zhanglab.ccmb.med.umich.edu/STRUM/ |
| ELASPIC [56] | 3D and 1D | Stochastic Gradient Boosting of Decision Trees | It predicts the mutation effects on protein folding using homology modeling | NO | YES | http://elaspic.kimlab.org/ |
| SAAFEC [39] | 3D | Molecular Mechanics Poisson-Boltzmann | It represents the non-polar solvation energy through a linear relation to the solvent accessible surface area | NO | NO | http://compbio.clemson.edu/SAAFEC |
| MAESTRO(web) [57] | 3D | Multi-agent prediction (ANN + SVM + linear regression) | It provides confidence estimations and multiple-site predictions | NO | YES | https://biwww.che.sbg.ac.at/maestro/web |
| SDM [46] | 3D and 1D | environment-specific substitution tables (ESSTs) | It uses conformationally constrained environment-specific substitution tables from 2,054 protein family sequence and structure alignments | YES | NO | http://marid.bioc.cam.ac.uk/sdm2 |
| TML-MP [58] | 3D and 1D | Gradient boosting | Topology-based predictor using persistent homology to reduce the geometric complexity and the number of degrees of freedom of proteins | NO | NO | http://weilab.math.msu.edu/TML/TML-MP/ |
| ThreeFoil [16] | 3D and 1D | meta-predictor using 11 tools | It assigns weight to any tool's prediction proportionally to its performance on similar types of mutations | YES | NO | http://meieringlab.uwaterloo.ca/stabilitypredict/ |
| DynaMut [59] | 3D | meta-predictor based on Normal Mode Analysis | It allows the study of harmonic motions in a system, providing insights into its dynamics and accessible conformations | YES | NO | http://biosig.unimelb.edu.au/dynamut/. |
| DDGun [21] | 3D or 1D | Linear combination of features | Untrained method introducing anti-symmetric features based on evolutionary information; it predicts also for multiple site variations | YES | YES | http://folding.biofold.org/ddgun/ |
| DeepDDG [17] | 3D and 1D | ANN | It is based on a deep neural network architecture trained on > 5,700 manually curated experimental data | YES | NO | http://protein.org.cn/ddg.html |
| ProTstab [60] | 1D | Gradient boosting of regression trees | It exploits a cellular stability based on limited proteolysis and mass spectrometry | NO | NO | http://structure.bmc.lu.se/ProTstab/index/ |
According to the type of data considered, there might be some restrictions. Structure-based approaches (e.g. methods using 3D structure in Table 2) cannot be used if the 3-dimensional structures of the proteins are unavailable. This limitation can be resolved by using sequence-based prediction tools (referred as 1D in Table 2), which use the amino acid sequence of proteins along with different machine-learning methods to predict changes in the folding free energy. These methods have the advantage of being applicable when the three-dimensional (3D) structure is not available. Methods using the 3D structure of the wild-type protein, e.g. I-Mutant [23], [26], [40], PoPMuSiC [28] or DUET [41], turned out to perform better than purely sequence-based approaches in general [42]. Interestingly, the recent sequence-based method INPS [43] was shown to complement and improve structure-based approaches. However, so far, no known polynomial-time and polynomial-space algorithms are available to solve the full-optimization of protein geometry since it is a combinatorial NP-hard problem [44]. Therefore, every tool implements heuristics and approximations. When no experimental structure is available, predicted structures can be used as models [45]. However, evaluations of the current methods have been systematically performed on the basis of the experimental structures and of the modeling procedure used for generating the predicted structures [16].
3.1. ΔΔG Anti-symmetry
An important property considered only by few predictors is the ΔΔG anti-symmetry, which represents a physical principle at the basis of any thermodynamic transformation (Fig. 1B). Given a chemical (or conformational) transformation from A to B, the relative concentrations of A and B (i.e. [A] and [B]) at equilibrium (and at constant temperature and pressure) are described by the following Gibbs free energy difference:
This relation is anti-symmetric in A and B, since the inverse transformation (from B to A) is described by:
This general principle, which can be also applied to the protein folding/unfolding process, leads to the basic anti-symmetry of the free energy change upon residue variation. In the case of a protein variation, if the unfolded and folded states of a protein X are defined as U(X) and F(X), two protein sequences p and q that differ by few aminoacids will be characterized by the following anti-symmetric equation:
Machine learning approaches have started to consider reverse variants with inverse ΔΔG signs in the training step. Referring to Table 2, only the following methods consider this aspect: SDM [46], DDGun(3D) [21], ThreeFoil [43], PROTS-RF [47], INPS [43] and DeepDDG [17]. In particular, PROTS-RF was the first approach to assess its performance by taking into account also reverse mutations, while DDGun(3D) implemented anti-symmetric scoring functions used in the prediction model, i.e. evolutionary, hydrophobicity and residue contact scores.
The simplest way to measure the anti-symmetry bias is to compute the Pearson correlation between the predictions on a variation set against the predictions on the corresponding reverse variation set. Correlation values shifted away from −1 indicate the extent of the anti-symmetry bias (Table 3). To fulfill the anti-symmetry constraint, Usmanova et al. [48] showed that the methods should generate the lowest energy structure of the mutant starting from the native one and vice versa, which may be hard, considering internal specific features of the programs. FoldX, for instance, keeps fixed the backbone and side-chain of all residues but neighbors upon mutation. As a result, the modeled structure of the mutant (B) is expected to be less stable than the native structure (A) by some value δAB and vice versa. Thus, the average mutation bias (<δ > ) can be calculated as <δ> = <(δAB + δBA)/2> = <(ΔΔGAB + ΔΔGBA)/2>. This bias does not depend on the experimental determination of the free energy of the protein structure nor on the definition of wild-type and mutant proteins.
Table 3.
Performance evaluation reported for each method, highlighting possible biases.
| Method | Validation | Training data | Test Data | Correlation (ρ) | Anti-symmetry (ρ_dir-inv) | Sequence Identity/homologs |
|---|---|---|---|---|---|---|
| FoldX [18] | none | S339 | S625 | 0.82 | Biased* (−0.38) | not declared |
| MUpro [51] | 20‐fold CV and LOO | S1615 | S1615, S388 | 0.13–0.76 | Biased* (−0.02) | Homologs removed from S1615 (SR1496, SR1135, SR1023, SR1539) |
| CUPSAT [36] | 3/4/5-fold CV | S1538, S1603 | S1538, S1603 | 0.55–0.78 | Biased* (−0.54) | not declared |
| I-Mutant(3.0/2.0) [26], [40] | 10/20/30-fold CV | S1948 | S1948 | 0.62–0.71 | Biased* (0.02) | not declared |
| iPTREE-STAB [25] | 4/10/20-fold CV | S1859 | S1859 | 0.7 | not evaluated | not declared |
| AUTO-MUTE (2.0) [69] | 20‐fold CV | Subsets of S1948, S1615, S388,S1791, S1396, S2204 | Subsets of S1948, S1615, S388,S1791, S1396 | 0.74–0.79 | Biased* (−0.06) | not declared |
| Prethermut [29] | 10-fold CV | S3366, S2156 | S3366, S2156 | 0.67–0.72 | not evaluated | not declared |
| POPMUSIC(3.1/2.1) [28] | 5-fold CV | S2648 | S2648 | 0.63–0.79 | Biased* (−0.29); unbiased version POPMUSICsym (−0.77) | not declared |
| Pro-Maya [52] | 5/10-fold CV and LOO | S2648, S2156 | S2648, S2156 | 0.59–0.8 | not evaluated | <30% identity and keeping information on the mutation site |
| PROTS-RF [51] | 5/10-fold CV | S2156 | S2156 + D180, D140 (27 and 19 proteins) | 0.62–0.86 | Unbiased | <30% identity in CV |
| iStable(2.0) [30], [53] | 5-fold CV v2.0: 10-fold CV | S2648, S1948 v2.0: S3528 | M1311, M1820 (from S2648, S1948, no redundancies) v2.0: S630 | 0.85 v2.0: 0.67–0.71 | Biased* (−0.05) v2.0: not evaluated | Meta predictor combining several predictors and using the same protein variant to train the combined model |
| NeEMO [54] | 10-fold CV | S2399 (113 proteins) | IM_631 (from S2399) | 0.5–0.79 | Biased* (0.09) | Evaluated but not used in CV |
| DUET [41] | S350, p53 | S2648 | p53,S350 | 0.71–0.82 | Biased* (−0.21) | not declared |
| mCSM [20] | 5/10/20-fold CV | S2648, S1925 | S350 | 0.51–0.82 | Biased* (−0.26) | 5-fold cross-validation separating by protein (Pearson = 0.51) |
| EASE-MM [24] | 10-fold CV and blind | 1676 mutations from S1948 | S543, S236 | 0.51–0.59 | not evaluated | max 25% sequence identity between folds and train/test sets |
| INPS(3D) [43], [55] | 5-fold CV | S2648, p53 | S2648, p53 | 0.53–0.71 | Unbiased for sequence-only* (−0.99 1D, −0.86 3D) | 5-fold cross-validation separating by protein as in mCSM |
| STRUM [22] | 5-fold CV | Q3421 | S2648, S350, Q306 (subset of S2648) | 0.4–0.8 | Biased* (0.34) | Q306 as test, with sequence identity < 60% |
| ELASPIC [56] | 20‐fold CV and LOO | S3463 (159 proteins) | S2636 (134 proteins), S2104 (79 proteins) | 0.77 | not evaluated | 90% sequence identity redundancy reduction |
| SAAFEC [39] | 5-fold CV | 983 mutations from Protherm (42 proteins) | 983 mutations from Protherm (42 proteins) | 0.61 | not evaluated | not declared |
| MAESTRO(web) [57] | 5/10/20-fold CV | S2648, S1925 (from S1948), S1765, S2244 | S2648, S350 | 0.63–0.76 | Biased* (−0.34) | Lowest correlation (0.63) when separating by protein in 5-fold CV as in mCSM |
| SDM(2) [46] | None | None | S2648, S350, p53, S140 | 0.52–0.63 | Biased* (−0.75) | not declared |
| TML-MP [58] | 5-fold cross validation | S2648 S350, M233 | 2648 S350, M233 | 0.54–0.82 | not evaluated | not declared |
| ThreeFoil [16] | 2-fold CV with 1000 reshuffling | Broom2017 | Broom2017 | 0.73 | not evaluated | not declared |
| DynaMut [59] | 10-fold CV and blind TS | S2648 | S351 | 0.58–0,70 | Biased | Homology included, use in input Duet, trained on the same S2640 |
| DDGun [21] | S2648, Ssym, p53, Myoglobin | Untrained | S2648, Ssym, p53, Myoglobin | 0.45–0.71 (0.54–0.68) | Unbiased* (−0.99) | not applicable |
| DeepDDG [17] | Cao Train | Cao Test | 0.56 | not evaluated | Homologs (>25%) removed from test set | |
| ProTstab [60] | 10-fold CV | Varibench | Varibench | 0.79 | not evaluated | not declared |
Possible biases can be due to anti-symmetry and lack of checking for sequence identity on training/test data. CV = cross-validation, LOO = leave-one-out, TS = test-set, MCC = Matthews Correlation Coefficient, ρ = correlation, ρ_dir-inv = correlation between direct and inverse variation. *Evaluated on Ssym database [11].
3.2. Multiple-site variations
Another challenge is the prediction of protein stability in the occurrence of multiple mutations. Multiple-point mutations are common variations of the protein sequence that may be needed in protein engineering when a single-point mutation is not enough to yield the desired stability change. Dealing with multiple-site variations adds another level of complexity beyond the prediction of the effect of a single variant on protein stability, since it requires the learning of many types of combinatorial effects (compensatory, additive, following linear or nonlinear combinations, threshold effects, etc.) [49]. Moreover, experimental ΔΔG data for multiple-point mutations are less abundant than for single-point ones. This shortage of data, together with difficulties arising from the combinatorial nature of the problem, make the effects of multiple-point mutations hard to predict. For these reasons, most of the methods only handle single-point mutations.
Generally speaking, it is expected that substitutions that are closer (in sequence or structure) interact more than those that are farther away from each other. In this latter case, predicting the ΔΔG values due to each substitution independently and then adding the predicted values should provide a good approximation of the multiple-point mutation ΔΔG [50]. This additive approach is most naturally employed by linear methods like FoldX [18] and DDGun [21]. However, it can be trivially used to extend all single-point methods to multiple-point mutations. Another approach, implemented by PROTS-RF [47] and Prethermut [29], calculates the average input features across multiple substitutions. However, averaging may not be appropriate in case of cooperative effects.
4. Best practice and pitfalls in prediction assessment
4.1. A prediction accuracy upper bound
As shown above (see Fig. 1A), given a set of experimental ΔΔG values for a specific variation, two samples can be considered as having similar ΔΔG values if they are within the experimental error (uncertainty). However, since a certain variability characterizes each dataset, a question to address is: given the standard deviation of the dataset () and the measure uncertainty (σ), is there an upper bound to the prediction performance? Starting from a set of measured ΔΔG protein variations, the best predictor can be estimated according to its similarity to another set of experimentally-determined ΔΔG values. The assumption is that no computational method can be better than a set of similar experiments. To derive an upper bound of the Pearson correlation as a function of the noise and distribution of the ΔΔG data, the idea is that, starting from a narrow dataset distribution with a variance having the same order of magnitude of the experimental uncertainty, the theoretical upper bounds can be lower than expected. Some studies provided a theoretical estimation of the upper bound of the Pearson correlation as a function of the average uncertainty of the data (σ) and the standard deviation of the dataset ( see Fig. 1C) [61], [62]. As an example, the popular datasets S2648 and VariBench have a σDB < 2 kcal/mol, leading to an upper bound for the Pearson correlation coefficient of ~ 0.8 and a lower bound for the root mean square error between experimental and predicted ΔΔG values of ~ 1 kcal/mol. This implies that the methods using these data for estimating their performance and reporting Pearson correlation above 0.8 are likely to be affected by over-fitting biases in the training step.
4.2. The burden of sequence identity
The reliability of machine learning approaches depends on the size and quality of the training dataset. Therefore, it is essential to use high-quality experimental observations with high consistency when training and testing these methods. Table 3 reports the main features characterizing the performance evaluations of each method listed in Table 2. In particular, the size and balance of training data must be considered carefully. Datasets with a few hundred or thousand cases, as those mostly used, might be too small to identify useful descriptors in the learning process. In addition, low heterogeneity within training datasets might lead to prediction tools not able to generalize. As a consequence, the weights assigned to the descriptors identified by these predictive models can be biased by the over-representation of some partial descriptors in the training data, sometimes ignoring general predictive descriptors. A common error made by the current methods is not taking into account similarities between training and test sets, leading to over-fitting issues (see Fig. 1D).
Evolution has selected proteins that are well-suited to maintain certain specific functions. Although evolutionary pressure constrains protein structure, sequence-level variations can be high, with very different sequences having a similar structure. Two proteins with different sequences but evolutionary or functionally-related are called homologs. Quantifying these evolutionary relationships is very important for preventing undesired information leakage between data splits. However, we mainly rely on sequence identity, which measures the percentage of exact amino acid matches between aligned subsequences of proteins. For example, filtering at a 25% sequence identity threshold means that no pairs of proteins in the training and test sets have >25% exact amino acid matches. Unfortunately, not all the current approaches have considered sequence identity in dividing training and test sets (Table 3), which means measuring the performance of the method using test sets not containing proteins similar (or identical) to those of the training set. One of the most representative examples explaining the similarity issue between training and testing is shown in Pires et al. [20]. The Pearson correlation of the method mCSM ranges between 0.7 and 0.8 when a random selection of the variations is used. However, the correlation drops down to the range 0.54–0.51 when the cross-validation is performed using ‘per site-’ and ‘per protein-’ clustering, respectively. Different protein positions tend to be more or less sensitive to mutations independently of the residue substitution (Pro-Maya [52]). Thus, using the same site (or same protein) in training and testing leads to a wrong and over-optimistic performance. Furthermore, a method trained under these conditions tends not to generalize well. For these reasons, a simple random split of the datasets should be avoided.
The problem of similarities among training and testing sets is more pronounced when meta-predictors are used. In the last years new stacked methods, combining multiple different predictors, have been developed, e.g. DUET [41], iStable(2.0) [30], [53], Dynamut [59] and ThreeFoil [16]. Meta-predictors offer the main advantage that even a simple majority voting approach over several methods yields better results than any individual method, each characterized by its strengths and weaknesses. However, the performance reported by these methods might be misleading. For example, ThreeFoil combines predictions from 11 freely available tools, each originally trained on different datasets, all derived from ProTherm. The meta-predictor was built such that the weight given to the prediction from any particular tool was based on that tool's performance against similar types of mutations from a training set of 605 mutations. Matthews Correlation Coefficient (MCC) values were determined through cross-validation and a dataset of 605 mutations was split into halves, with one half used to determine MCC values as weights and the other half used to test the overall performance, repeating the procedure 1000 times. However, 60% of the proteins used for this dataset were used by some of the predictors during their training step, introducing the similarity issues between training and test sets described above. Similarly, Dynamut combines the effects of mutations on protein stability and dynamics calculated by Bio3D [63], ENCoM [82] and DUET to generate an optimized and more robust predictor. DUET, in turn, combines two approaches, SDM and mCSM. These approaches were trained on S2648, which was the same dataset used by Dynamut for training the meta-predictor, making cross-validation, and for extracting a “blind” set of 351 variations for the performance evaluation. In iStable 2.0 the authors introduced two datasets for training and testing [30]. In the test set of 630 variations, 442 were derived from the same proteins used in the training sets. The two datasets contain repeated mutations, 703 and 77 for training and test sets, respectively. In terms of protein sequence similarity, only three proteins (corresponding to 81 mutations) can be safely used in S630 to test the methods. Therefore, there is an urgent need of reviewing these methods by testing their predictive models with data that have no sequence similarity (trained on similar proteins) to those of the training set.
4.3. Unbalanced experimental datasets and anti-symmetry
Unbalanced training datasets with large differences in the number of cases representing specific categories might also lead to erroneous estimations (see Fig. 1D). As an example, a training dataset with 80% of destabilizing mutations would allow the predictor to erroneously classify most mutations as destabilizing. This can hamper both classification (ΔΔG sign) and regression (ΔΔG value). Given this unbalance, the most recommended measures of accuracy in the context of classification are the MCC and the harmonic mean between precision and recall (F1) [64].
Another important feature described above, which has a great impact on the algorithm performance, is represented by the anti-symmetric properties of the free energy changes. Recently, different studies addressed this problem as bias in most predictors [11], [12], [48], [61], [65], and specifically designed datasets including both the variations and the corresponding inverses (such as A->B and B->A on the same protein) have been introduced: Ssym [11], Usmanova-DB [48] and Fang-DB [12]. The results show that only INPS and PopMusic [11] were sufficiently robust to be defined compliant with the anti-symmetric properties [61], [65]. The Pearson correlation between the predictions on a variation set and the predictions on the corresponding inverse variation set (ρ_dir-inv) can measure the method bias. In Table 3 we report the anti-symmetry bias of different methods (when measured). A perfect method should show a Pearson correlation equal to −1.
4.4. Developing a new method
Most of the predictors were trained using random splits of the selected datasets (Table 1). However, as mentioned above, this procedure is not correct since there is a huge level of redundancy in the datasets. Considering the pie-charts represented in Fig. 1A, only few protein sequences show no significant sequence similarity (<25%) with other proteins of the same database (only 15 and 5 proteins in S2648 and Varibench, respectively). The percentage of proteins clustered into large groups (i.e. clusters including > 5% of the proteins present in the entire database) with high sequence similarity is significantly high, involving 32% and 41% of the proteins annotated in S2648 and Varibench, respectively. This redundancy characterizes all the available datasets. Thus, given the difficulty of comparing methods and assessing the performance on fair bases, here we provide sequence-identity-reduced datasets (see Supplementary Materials). We clustered the variants according to the sequence identity of the protein sequences they belong to, using a threshold cut-off of 25% identity. The variants of S2648, VariBench3D, sSym, P53 and Myoglobin datasets are grouped in a way that it is possible to perform 10-fold cross-validation on S2648 and, using the models trained in the different folds, to predict the variations of the other datasets excluding sequence similarity issues. With our clustering, new methods can be developed and fairly compared. We adopted a circular cross-validation procedure (with either 10 or 5 sets): 8 (or 3) folds at a time are kept for the training while the other 2 are considered for validation and test, respectively. By rotating along all the folds, it is possible to use the whole dataset both for the validation and for the test (see Supplementary). It is compulsory that the tuning of the hyper-parameters or method selections should be done by analyzing the performance computed on the validation sets, without “peeping” at those obtained on the test sets, otherwise the whole procedure would be spoiled.
5. The test case of CAGI experiments
The Critical Assessment of Genome Interpretation (CAGI) is a community experiment aimed at fairly assessing the computational methods for genome interpretation [66]. In the latest edition (CAGI 5) data providers measured unfolding free energy of a set of variants with far-UV circular dichroism and intrinsic fluorescence spectra on Frataxin (FXN), a highly-conserved protein fundamental for the cellular iron homeostasis in both prokaryotes and eukaryotes. These measurements were used to calculate the change in unfolding free energy between the variant and wild-type proteins at zero concentration of denaturant (ΔΔGH2O). The experimental dataset [67], including eight amino acid substitutions, was used to evaluate the performance of the web-available tools for predicting the value of ΔΔGH2O associated with the variants and to classify them as destabilizing or not-destabilizing [68]. Eight performance measures were applied to test the methods, five for assessing the predictions in regression (Pearson and Spearman Correlation Coefficients, Kendall-Tau Coefficient, Root Mean Square and Mean Absolute Errors) and three for the classification performance, i.e. Accuracy, Matthews Correlation Coefficient and Area Under the Receiving Operator Characteristic Curve (AUC). A prediction threshold equal to −1 kcal/mol was set to discriminate between destabilizing (ΔΔG < −1 kcal/mol) and not-destabilizing (neutral) variants (ΔΔG−1 kcal/mol) [69]. According to this classification, 5 variants destabilize the protein structure and 3 variants are non-destabilizing. These protein variations were submitted to the web server of each method for the assessment of its predictions. The results confirmed that ELASPIC and INPS3D, as well as DDGun and PopMusic that were optimized for the anti-symmetric property, are among the best methods in predicting the ΔΔG associated with the variants included in the challenge (see Table 4). Although the FXN dataset includes only 8 variants, it represents a first attempt to validate the performance of ΔΔG prediction tools using a blind test set.
Table 4.
Performance of state-of-the-art methods on 8 variants of the CAGI FXN challenge.
| Method | rP | rS | Kτ | RMSE | MAE | Q2 | MCC | AUC |
|---|---|---|---|---|---|---|---|---|
| DDGun [21] | 0.90 | 0.69 | 0.57 | 2.15 | 1.93 | 0.70 | 0.45 | 0.80 |
| ELASPIC [56] | 0.82 | 0.69 | 0.50 | 2.38 | 1.71 | 0.80 | 0.60 | 0.93 |
| SAAFEC [39] | 0.74 | 0.83 | 0.71 | 2.71 | 2.08 | 0.83 | 0.75 | 0.93 |
| PopMusic [28] | 0.83 | 0.54 | 0.40 | 2.68 | 2.00 | 0.90 | 0.78 | 0.80 |
| EASE-MM (1D) [24] | 0.81 | 0.64 | 0.64 | 2.88 | 2.22 | 0.70 | 0.45 | 0.80 |
| INPS (3D) [55] | 0.76 | 0.60 | 0.43 | 3.18 | 2.36 | 0.80 | 0.60 | 0.87 |
| NeEMO [54] | 0.83 | 0.62 | 0.50 | 3.39 | 2.44 | 0.70 | 0.45 | 0.87 |
| INPS (1D) [43] | 0.70 | 0.60 | 0.43 | 3.35 | 2.52 | 0.80 | 0.60 | 0.87 |
| MAESTRO [57] | 0.72 | 0.60 | 0.43 | 3.40 | 2.48 | 0.70 | 0.45 | 0.67 |
| iStable [30], [53] | 0.82 | 0.55 | 0.43 | 3.57 | 2.73 | 0.70 | 0.45 | 0.80 |
| DUET [41] | 0.53 | 0.55 | 0.36 | 3.34 | 2.46 | 0.70 | 0.45 | 0.73 |
| DeepDDG [17] | 0.73 | 0.38 | 0.29 | 3.10 | 2.43 | 0.53 | 0.07 | 0.67 |
| I-Mutant2 (1D) [26], [40] | 0.69 | 0.36 | 0.21 | 3.52 | 2.68 | 0.80 | 0.60 | 0.67 |
| SDM [46] | 0.54 | 0.52 | 0.36 | 3.49 | 2.73 | 0.63 | 0.26 | 0.67 |
| mCSM [20] | 0.54 | 0.43 | 0.21 | 3.30 | 2.49 | 0.37 | −0.26 | 0.60 |
| I-Mutant2 (3D) [26], [40] | 0.75 | 0.04 | −0.05 | 3.60 | 2.55 | 0.63 | 0.35 | 0.58 |
| AUTOMUTE (TR) [69] | 0.33 | 0.26 | 0.14 | 3.64 | 2.67 | 0.53 | 0.07 | 0.73 |
| DynaMut [59] | 0.37 | 0.29 | 0.14 | 3.94 | 2.89 | 0.60 | 0.29 | 0.67 |
| MUpro (1D) [51] | 0.14 | −0.10 | −0.07 | 3.89 | 2.80 | 0.70 | 0.45 | 0.53 |
| CUPSAT (DN) [36] | 0.20 | −0.12 | −0.14 | 4.98 | 3.97 | 0.50 | 0.00 | 0.20 |
Methods are ranked using 8 measures of performance. rP and rS: Pearson and Spearman Correlation Coefficients, respectively. Kτ: Kendall-Tau Coefficient. RMSE and MAE: Root Mean Square and Mean Absolute Errors. Q2: Accuracy: MCC: Matthews Correlation Coefficient. AUC: Area Under the Receiving Operator Characteristic (ROC) Curve. A binary classification threshold between destabilizing and neutral variants has been set to −1.0 kcal/mol. Measures of performance were defined as in Baldi et al. [69]. DN: Denaturant. TR: Tree Regression. 1D and 3D: sequence-based and structure-based predictions, respectively. ΔΔGs were calculated at the experimental conditions of pH = 8 and T = 20C.
6. Challenges in the applications of protein stability predictions in biomedicine and precision medicine
Sequence variations and structural changes that cause disease were first linked in 1949 for sickle-cell anemia [70], [71], which is an autosomal recessive disorder caused by the amino acid substitution E6V in the β-chain of human hemoglobin [72], i.e., a replacement of glutamic acid (E) by a valine (V) at position 6. Currently, the Human Gene Mutation Database (HGMD [73]), Online Mendelian Inheritance in Man (OMIM [74]), the Catalogue of Somatic Mutations in Cancer (COSMIC) [75], and other resources collect thousands of such single amino acid variants causative of, or associated with, diseases, in addition to many other types of sequence variations. However, the number of pathogenic variants in those databases represents only a small fraction of the potential number of pathogenic mutations in the world-wide human population.
Engineering stability-changing mutations are opening promising perspectives for several approaches in precision medicine. Analyses linking the effects of a mutation on a protein thermodynamic stability with pathogenicity indicate that loss of stability could be a main driver of inherited diseases. Thus, an improved understanding of the complex relationships among protein sequence, structure, folding and stability could provide new possibilities for diagnosis and even novel treatments specific for the patients. Casadio et al. [76] made the first effort in assessing the relationships among stability changes in proteins and their involvement in human diseases at large-scale by analyzing the Human Proteome [2]. By estimating for each single amino acid polymorphism the probability of being disease-related, they showed that protein stability changes can also be disease-associated at the proteome level. The probability indexes were statistically derived from a dataset of 17,170 single amino acid variations in 5,305 proteins retrieved from data available at UniProtKB (release 2010_04), dbSNP (build 132), OMIM and ProTherm [15]. SDM2, mCSM and DUET have been used to evaluate the effects of stability changes in different applications: human cancer-related genes in the COSMIC database [77], inhibition of inosine-5′-monophosphate dehydrogenase, isoniazid and rifampicin resistance in Mycobacterium Tuberculosis [78], phosphodiesterase somatic mutations implicated in cancer and retinitis pigmentosa [79], protein presenilin 2 linked to familial Alzheimer’s disease [80].
A challenging task for medical applications is predicting how mutations in proteins alter their ability to function, distinguishing mutations that are “drivers” of disease or drug resistance from “passengers” that are neutral, or even selectively advantageous for the organism. This task requires understanding the impact of missense mutations in protein function, and mapping genetic variations to 3D protein structures [81]. In particular, mutations on protein surfaces in the protein–protein interfaces were associated with larger global effects than mutations occurring elsewhere in protein surfaces [2], [81]. Protein stability may directly be related to functional activity, and changes in stability or incorrect folding could be the major effects of pathogenic missense mutations. In most cases, missense mutations are deleterious due to the decrease in stability of the corresponding protein [82]. In other cases, missense mutations may cause diseases by enhancing the stability of the corresponding protein [83]. The mutant kinase EGFR can be taken as an example of both kinds of disease-inducing changes, at the same time illustrating emblematically how complex the path from the detection of mutation-induced free energy changes to precision medicine might be. Kinase activation commonly involves a rotation and shift of the αC-helix between these states, a conformational change that is governed by an allosteric switch. Driver mutations can control that switch. Oncogenic driver mutations act by either stabilizing the active conformation, or disrupting interactions that stabilize the inactive one. 41% of the EGFR mutations in lung cancer present the oncogenic Leu858 driver mutation [84], which stabilizes the αC-helix in the active conformation. By contrast, the T790M mutation in EGFR stabilizes the hydrophobic R-spine, which destabilizes the inactive conformation. The outcome is similar: the mutations allosterically switch the preferred states towards a constitutively activated kinase. In brief, driver mutations can adopt one of three mechanisms: destabilize the inactive state, stabilize the active state, or both. In terms of free energy, those effects can be seen as changes in the relative depths of minima, with the outcome of favoring or disfavoring the asymmetric or symmetric dimer conformation [85]. This kind of knowledge clearly impacts on the choice of treatment for lung cancer patients. For instance, Gefitinib is an EGFR inhibitor used to treat lung cancer, but, as expected by the reasons described above, it improves the outcome only in a subset of patients bearing very specific genetic changes.
Another research field that might benefit from future applications of engineering stability-changing mutations is represented by the immunogenomics studies. In the last years, immunotherapies have shown high rates of success as treatments for several types of cancer. Recent approaches employ the adoptive transfer of autologous T cells from a patient, genetically modified either with an engineered Chimeric Antigen Receptor (CAR) or with an engineered T Cell Receptor (TCR), for recognition of the appropriate peptides in complex with the Major Histocompatibility Complex class I (MHC-I).
However, finding suitable candidates (for CAR-T and TCR-T cell therapy) is very costly and time-consuming. For this reason, computational methods for predicting the immunogenicity of peptide-MHC-I complexes are being developed.
In particular, the role of protein-complex stability has been recognized as crucial for the immunogenicity prediction of the peptide-MHC-I complexes (for example NetMHCstab [86] and NetMHCstabpan [87]). These methods are expected to speed up therapy implementation for each patient significantly. The intrinsic effects of fold stability on immunogenicity have wide potential applications also in the vaccine field. The first examples of stability-optimized vaccines have been demonstrated [88], and further major advances on protein design for novel therapies within the next years are expected.
7. Summary and outlook
The prediction of protein stability changes upon variation is essential for protein design and precision medicine. The current methods, although far from being perfect, achieved sufficient levels of performance to complement experimental studies [68]. However, several issues need to be addressed to enhance such levels of performance, in particular:
-
1.
increasing the quality and the size of the current datasets, adding more carefully curated experimental data;
-
2.
building methods that are intrinsically anti-symmetric (ΔΔG(A->B) = -ΔΔG(B->A));
-
3.
for machine-learning approaches, it is essential that the model training has to be performed using low levels of sequence identity between learning and testing sets. To facilitate the scientific community in this task, we provide similarity-free cross-validation folds for the most relevant datasets (Supplementary materials).
If these indications are taken into account, the next generation of predictors can achieve more reliable and more consistent performances.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have influenced the work reported in this paper.
Acknowledgements
We thank the Italian Ministry for Education, University and Research for the PRIN 2017 201744NR8S “Integrative tools for defining the molecular basis of the diseases” and for the programme “Dipartimenti di Eccellenza 20182022D15D18000410001”.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2020.07.011.
Contributor Information
Emidio Capriotti, Email: emidio.capriotti@unibo.it.
Piero Fariselli, Email: piero.fariselli@unito.it.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Stefl S., Nishi H., Petukh M., Panchenko A.R., Alexov E. Molecular mechanisms of disease-causing missense mutations. J Mol Biol. 2013;425:3919–3936. doi: 10.1016/j.jmb.2013.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martelli P.L., Fariselli P., Savojardo C., Babbi G., Aggazio F., Casadio R. Large scale analysis of protein stability in OMIM disease related human protein variants. BMC Genomics. 2016;17(Suppl 2):397. doi: 10.1186/s12864-016-2726-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hartl F.U. Protein Misfolding Diseases. 2017;86:21–26. doi: 10.1146/annurev-biochem-061516-044518. [DOI] [PubMed] [Google Scholar]
- 4.Ó’Fágáin C. Protein stability: enhancement and measurement. Methods Mol Biol. 2017;1485:101–129. doi: 10.1007/978-1-4939-6412-3_7. [DOI] [PubMed] [Google Scholar]
- 5.Compiani M., Capriotti E. Computational and theoretical methods for protein folding. Biochemistry. 2013;52:8601–8624. doi: 10.1021/bi4001529. [DOI] [PubMed] [Google Scholar]
- 6.Capriotti E., Nehrt N.L., Kann M.G., Bromberg Y. Bioinformatics for personal genome interpretation. Brief Bioinform. 2012;13:495–512. doi: 10.1093/bib/bbr070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tian R., Basu M.K., Capriotti E. Computational methods and resources for the interpretation of genomic variants in cancer. BMC Genomics. 2015;16(Suppl 8):S7. doi: 10.1186/1471-2164-16-S8-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Capriotti E., Ozturk K., Carter H. Integrating molecular networks with genetic variant interpretation for precision medicine. Wiley Interdiscip Rev Syst Biol Med. 2019;11 doi: 10.1002/wsbm.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hassan M.S., Shaalan A.A., Dessouky M.I., Abdelnaiem A.E., ElHefnawi M. A review study: computational techniques for expecting the impact of non-synonymous single nucleotide variants in human diseases. Gene. 2019;680:20–33. doi: 10.1016/j.gene.2018.09.028. [DOI] [PubMed] [Google Scholar]
- 10.Zhou Y., Fujikura K., Mkrtchian S., Lauschke V.M. Computational methods for the pharmacogenetic interpretation of next generation sequencing data. Front Pharmacol. 2018;9:1437. doi: 10.3389/fphar.2018.01437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pucci F., Bernaerts K.V., Kwasigroch J.M., Rooman M. Quantification of biases in predictions of protein stability changes upon mutations. Bioinformatics. 2018;34:3659–3665. doi: 10.1093/bioinformatics/bty348. [DOI] [PubMed] [Google Scholar]
- 12.Fang J. A critical review of five machine learning-based algorithms for predicting protein stability changes upon mutation. Brief Bioinform. 2019 doi: 10.1093/bib/bbz071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pucci F., Rooman M. Stability curve prediction of homologous proteins using temperature-dependent statistical potentials. PLoS Comput Biol. 2014;10 doi: 10.1371/journal.pcbi.1003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sarkar A., Yang Y., Vihinen M. Variation benchmark datasets: update, criteria, quality and applications. Database (Oxford) 2020;2020 doi: 10.1093/database/baz117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar M.D.S., Bava K.A., Gromiha M.M., Prabakaran P., Kitajima K. ProTherm and ProNIT: thermodynamic databases for proteins and protein-nucleic acid interactions. Nucleic Acids Res. 2006;34:D204–D206. doi: 10.1093/nar/gkj103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broom A., Jacobi Z., Trainor K., Meiering E.M. Computational tools help improve protein stability but with a solubility tradeoff. J Biol Chem. 2017;292:14349–14361. doi: 10.1074/jbc.M117.784165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cao H., Wang J., He L., Qi Y., Zhang J.Z. DeepDDG: predicting the stability change of protein point mutations using neural networks. J Chem Inf Model. 2019;59:1508–1514. doi: 10.1021/acs.jcim.8b00697. [DOI] [PubMed] [Google Scholar]
- 18.Guerois R., Nielsen J.E., Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 19.Kepp K.P. Towards a “Golden Standard” for computing globin stability: Stability and structure sensitivity of myoglobin mutants. Biochim Biophys Acta, Gene Regul Mech. 2015;1854:1239–1248. doi: 10.1016/j.bbapap.2015.06.002. [DOI] [PubMed] [Google Scholar]
- 20.Pires D.E.V., Ascher D.B., Blundell T.L. mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics. 2014;30:335–342. doi: 10.1093/bioinformatics/btt691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Montanucci L., Capriotti E., Frank Y., Ben-Tal N., Fariselli P. DDGun: an untrained method for the prediction of protein stability changes upon single and multiple point variations. BMC Bioinf. 2019;20:335. doi: 10.1186/s12859-019-2923-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Quan L., Lv Q., Zhang Y. STRUM: structure-based prediction of protein stability changes upon single-point mutation. Bioinformatics. 2016;32:2936–2946. doi: 10.1093/bioinformatics/btw361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Capriotti E., Fariselli P., Casadio R. A neural-network-based method for predicting protein stability changes upon single point mutations. Bioinformatics. 2004;20(Suppl 1):i63–i68. doi: 10.1093/bioinformatics/bth928. [DOI] [PubMed] [Google Scholar]
- 24.Folkman L., Stantic B., Sattar A., Zhou Y. EASE-MM: sequence-based prediction of mutation-induced stability changes with feature-based multiple models. J Mol Biol. 2016;428:1394–1405. doi: 10.1016/j.jmb.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 25.Huang L.-T., Gromiha M.M., Ho S.-Y. iPTREE-STAB: interpretable decision tree based method for predicting protein stability changes upon mutations. Bioinformatics. 2007;23:1292–1293. doi: 10.1093/bioinformatics/btm100. [DOI] [PubMed] [Google Scholar]
- 26.Capriotti E., Fariselli P., Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33 doi: 10.1093/nar/gki375. W306–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Potapov V., Cohen M., Schreiber G. Assessing computational methods for predicting protein stability upon mutation: good on average but not in the details. Protein Eng Des Sel. 2009;22:553–560. doi: 10.1093/protein/gzp030. [DOI] [PubMed] [Google Scholar]
- 28.Dehouck Y., Kwasigroch J.M., Gilis D., Rooman M. PoPMuSiC 2.1: a web server for the estimation of protein stability changes upon mutation and sequence optimality. BMC Bioinf. 2011;12:151. doi: 10.1186/1471-2105-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tian J., Wu N., Chu X., Fan Y. Predicting changes in protein thermostability brought about by single- or multi-site mutations. BMC Bioinf. 2010;11:370. doi: 10.1186/1471-2105-11-370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen C.W., Lin M.H., Liao C.C., Chang H.P., Chu Y.W. iStable 2.0: Predicting protein thermal stability changes by integrating various characteristic modules. Comput Struct. Biotechnol J. 2020;18:622–630. doi: 10.1016/j.csbj.2020.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasidharan Nair P., Vihinen M. VariBench: a benchmark database for variations. Hum Mutat. 2013;34:42–49. doi: 10.1002/humu.22204. [DOI] [PubMed] [Google Scholar]
- 32.Lazaridis T., Karplus M. Effective energy functions for protein structure prediction. Curr Opin Struct Biol. 2000;10:139–145. doi: 10.1016/s0959-440x(00)00063-4. [DOI] [PubMed] [Google Scholar]
- 33.Duan Y., Wang L., Kollman P.A. The early stage of folding of villin headpiece subdomain observed in a 200-nanosecond fully solvated molecular dynamics simulation. Proc Natl Acad Sci U S A. 1998;95:9897–9902. doi: 10.1073/pnas.95.17.9897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kollman P.A., Massova I., Reyes C., Kuhn B., Huo S. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. 2000;33:889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 35.Pitera J.W., Kollman P.A. Exhaustive mutagenesis in silico: multicoordinate free energy calculations on proteins and peptides. Proteins. 2000;41:385–397. [PubMed] [Google Scholar]
- 36.Parthiban V., Gromiha M.M., Schomburg D. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. 2006;34:W239–W242. doi: 10.1093/nar/gkl190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petukh M., Li M., Alexov E. Predicting binding free energy change caused by point mutations with knowledge-modified MM/PBSA method. PLoS Comput Biol. 2015;11 doi: 10.1371/journal.pcbi.1004276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang E., Sun H., Wang J., Wang Z., Liu H. End-point binding free energy calculation with MM/PBSA and MM/GBSA: strategies and applications in drug design. Chem Rev. 2019;119:9478–9508. doi: 10.1021/acs.chemrev.9b00055. [DOI] [PubMed] [Google Scholar]
- 39.Getov I., Petukh M., Alexov E. SAAFEC: predicting the effect of single point mutations on protein folding free energy using a knowledge-modified MM/PBSA approach. Int J Mol Sci. 2016;17:512. doi: 10.3390/ijms17040512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Capriotti E., Fariselli P., Rossi I., Casadio R. A three-state prediction of single point mutations on protein stability changes. BMC Bioinf. 2008;9(Suppl 2):S6. doi: 10.1186/1471-2105-9-S2-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pires D.E.V., Ascher D.B., Blundell T.L. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 2014;42:W314–W319. doi: 10.1093/nar/gku411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Khan S., Vihinen M. Performance of protein stability predictors. Hum Mutat. 2010;31:675–684. doi: 10.1002/humu.21242. [DOI] [PubMed] [Google Scholar]
- 43.Fariselli P., Martelli P.L., Savojardo C., Casadio R. INPS: predicting the impact of non-synonymous variations on protein stability from sequence. Bioinformatics. 2015;31:2816–2821. doi: 10.1093/bioinformatics/btv291. [DOI] [PubMed] [Google Scholar]
- 44.Pierce N.A., Winfree E. Protein design is NP-hard. Protein Eng. 2002;15:779–782. doi: 10.1093/protein/15.10.779. [DOI] [PubMed] [Google Scholar]
- 45.Hurst J.M., McMillan L.E.M., Porter C.T., Allen J., Fakorede A., Martin A.C.R. The SAAPdb web resource: a large-scale structural analysis of mutant proteins. Hum Mutat. 2009;30:616–624. doi: 10.1002/humu.20898. [DOI] [PubMed] [Google Scholar]
- 46.Pandurangan A.P., Ochoa-Montaño B., Ascher D.B., Blundell T.L. SDM: a server for predicting effects of mutations on protein stability. Nucleic Acids Res. 2017;45:W229–W235. doi: 10.1093/nar/gkx439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li Y., Fang J. PROTS-RF: a robust model for predicting mutation-induced protein stability changes. PLoS ONE. 2012;7 doi: 10.1371/journal.pone.0047247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Usmanova D.R., Bogatyreva N.S., Ariño Bernad J., Eremina A.A., Gorshkova A.A. Self-consistency test reveals systematic bias in programs for prediction change of stability upon mutation. Bioinformatics. 2018;34:3653–3658. doi: 10.1093/bioinformatics/bty340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Montanucci L., Fariselli P., Martelli P.L., Casadio R. Predicting protein thermostability changes from sequence upon multiple mutations. Bioinformatics. 2008;24:i190–i195. doi: 10.1093/bioinformatics/btn166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Huang L.T., Gromiha M.M. Reliable prediction of protein thermostability change upon double mutation from amino acid sequence. Bioinformatics. 2009;25:2181–2187. doi: 10.1093/bioinformatics/btp370. [DOI] [PubMed] [Google Scholar]
- 51.Cheng J., Randall A., Baldi P. Prediction of protein stability changes for single-site mutations using support vector machines. Proteins. 2006;62:1125–1132. doi: 10.1002/prot.20810. [DOI] [PubMed] [Google Scholar]
- 52.Wainreb G., Wolf L., Ashkenazy H., Dehouck Y., Ben-Tal N. Protein stability: a single recorded mutation aids in predicting the effects of other mutations in the same amino acid site. Bioinformatics. 2011;27:3286–3292. doi: 10.1093/bioinformatics/btr576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen C.W., Lin J., Chu Y.W. iStable: off-the-shelf predictor integration for predicting protein stability changes. BMC Bioinf. 2013;14(Suppl 2):S5. doi: 10.1186/1471-2105-14-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Giollo M., Martin A.J.M., Walsh I., Ferrari C., Tosatto S.C.E. NeEMO: a method using residue interaction networks to improve prediction of protein stability upon mutation. BMC Genomics. 2014;15(Suppl 4):S7. doi: 10.1186/1471-2164-15-S4-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Savojardo C., Fariselli P., Martelli P.L., Casadio R. INPS-MD: a web server to predict stability of protein variants from sequence and structure. Bioinformatics. 2016;32:2542–2544. doi: 10.1093/bioinformatics/btw192. [DOI] [PubMed] [Google Scholar]
- 56.Witvliet D.K., Strokach A., Giraldo-Forero A.F., Teyra J., Colak R., Kim P.M. ELASPIC web-server: proteome-wide structure-based prediction of mutation effects on protein stability and binding affinity. Bioinformatics. 2016;32:1589–1591. doi: 10.1093/bioinformatics/btw031. [DOI] [PubMed] [Google Scholar]
- 57.Laimer J., Hofer H., Fritz M., Wegenkittl S., Lackner P. MAESTRO–multi agent stability prediction upon point mutations. BMC Bioinf. 2015;16:116. doi: 10.1186/s12859-015-0548-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cang Z., Wei G.W. Analysis and prediction of protein folding energy changes upon mutation by element specific persistent homology. Bioinformatics. 2017;33:3549–3557. doi: 10.1093/bioinformatics/btx460. [DOI] [PubMed] [Google Scholar]
- 59.Rodrigues C.H., Pires D.E., Ascher D.B. DynaMut: predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46:W350–W355. doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Y., Ding X., Zhu G., Niroula A., Lv Q., Vihinen M. ProTstab – predictor for cellular protein stability. BMC Genomics. 2019;20:804. doi: 10.1186/s12864-019-6138-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Montanucci L., Savojardo C., Martelli P.L., Casadio R., Fariselli P. On the biases in predictions of protein stability changes upon variations: the INPS test case. Bioinformatics. 2019;35:2525–2527. doi: 10.1093/bioinformatics/bty979. [DOI] [PubMed] [Google Scholar]
- 62.Benevenuta S., Fariselli P. On the upper bounds of the real-valued predictions. Bioinform Biol Insights. 2019;13 doi: 10.1177/1177932219871263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Grant B.J., Rodrigues A.P.C., ElSawy K.M., McCammon J.A., Caves L.S.D. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 64.Chicco D., Jurman G. The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genomics. 2020;21:6. doi: 10.1186/s12864-019-6413-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Savojardo C., Martelli P.L., Casadio R., Fariselli P. On the critical review of five machine learning-based algorithms for predicting protein stability changes upon mutation. Brief Bioinform. 2019 doi: 10.1093/bib/bbz168. [DOI] [PubMed] [Google Scholar]
- 66.Andreoletti G., Pal L.R., Moult J., Brenner S.E. Reports from the fifth edition of CAGI: the critical assessment of genome interpretation. Hum Mutat. 2019;40(9):1197–1201. doi: 10.1002/humu.v40.910.1002/humu.23876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Petrosino M., Pasquo A., Novak L., Toto A., Gianni S. Characterization of human frataxin missense variants in cancer tissues. Hum Mutat. 2019;40:1400–1413. doi: 10.1002/humu.23789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Savojardo C., Petrosino M., Babbi G., Bovo S., Corbi-Verge C. Evaluating the predictions of the protein stability change upon single amino acid substitutions for the FXN CAGI5 challenge. Hum Mutat. 2019;40:1392–1399. doi: 10.1002/humu.23843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baldi P., Brunak S., Chauvin Y., Andersen C.A., Nielsen H. Assessing the accuracy of prediction algorithms for classification: an overview. Bioinformatics. 2000;16:412–424. doi: 10.1093/bioinformatics/16.5.412. [DOI] [PubMed] [Google Scholar]
- 70.Pauling L., Itano H.A. Sickle cell anemia a molecular disease. Science. 1949;110:543–548. doi: 10.1126/science.110.2865.543. [DOI] [PubMed] [Google Scholar]
- 71.Perutz M.F., Mitchison J.M. State of haemoglobin in sickle-cell anaemia. Nature. 1950;166:677–679. doi: 10.1038/166677a0. [DOI] [PubMed] [Google Scholar]
- 72.Hunt J.A., Ingram V.M. Allelomorphism and the chemical differences of the human haemoglobins A, S and C. Nature. 1958;181:1062–1063. doi: 10.1038/1811062a0. [DOI] [PubMed] [Google Scholar]
- 73.Stenson P.D., Mort M., Ball E.V., Shaw K., Phillips A., Cooper D.N. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Amberger J.S., Bocchini C.A., Schiettecatte F., Scott A.F., Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tate J.G., Bamford S., Jubb H.C., Sondka Z., Beare D.M. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47(D1):D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Casadio R., Vassura M., Tiwari S., Fariselli P., Luigi M.P. Correlating disease-related mutations to their effect on protein stability: a large-scale analysis of the human proteome. Hum Mutat. 2011;32:1161–1170. doi: 10.1002/humu.21555. [DOI] [PubMed] [Google Scholar]
- 77.Malhotra S., Alsulami A.F., Heiyun Y., Ochoa B.M., Jubb H. Understanding the impacts of missense mutations on structures and functions of human cancer-related genes: a preliminary computational analysis of the COSMIC Cancer Gene Census. PLoS ONE. 2019;14 doi: 10.1371/journal.pone.0219935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Singh V., Pacitto A., Donini S., Ferraris D.M., Boros S. Synthesis and Structure-Activity relationship of 1-(5-isoquinolinesulfonyl)piperazine analogues as inhibitors of Mycobacterium tuberculosis IMPDH. Eur J Med Chem. 2019;174:309–329. doi: 10.1016/j.ejmech.2019.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Munir A., Kumar N., Ramalingam S.B., Tamilzhalagan S., Shanmugam S.K. Identification and characterization of genetic determinants of isoniazid and rifampicin resistance in mycobacterium tuberculosis in Southern India. Sci Rep. 2019;9:10283. doi: 10.1038/s41598-019-46756-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Maryam A., Vedithi S.C., Khalid R.R., Alsulami A.F., Torres P.H.M. The molecular organization of human cGMP specific phosphodiesterase 6 (PDE6): structural implications of somatic mutations in cancer and retinitis pigmentosa. Comput Struct Biotechnol J. 2019;17:378–389. doi: 10.1016/j.csbj.2019.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Glusman G., Rose P.W., Prlić A., Dougherty J., Duarte J.M. Mapping genetic variations to three-dimensional protein structures to enhance variant interpretation: a proposed framework. Genome Med. 2017;9:113. doi: 10.1186/s13073-017-0509-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hashimoto K., Rogozin I.B., Panchenko A.R. Oncogenic potential is related to activating effect of cancer single and double somatic mutations in receptor tyrosine kinases. Hum Mutat. 2012;33:1566–1575. doi: 10.1002/humu.22145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chiang C.H., Grauffel C., Wu L.S., Kuo P.H., Doudeva L.G. Structural analysis of disease-related TDP-43 D169G mutation: linking enhanced stability and caspase cleavage efficiency to protein accumulation. Sci Rep. 2016;6:21581. doi: 10.1038/srep21581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Shan Y., Eastwood M.P., Zhang X., Kim E.T., Arkhipov A. Oncogenic mutations counteract intrinsic disorder in the EGFR kinase and promote receptor dimerization. Cell. 2012;149:860–870. doi: 10.1016/j.cell.2012.02.063. [DOI] [PubMed] [Google Scholar]
- 85.Nussinov R., Jang H., Tsai C.J., Cheng F. Review: precision medicine and driver mutations: Computational methods, functional assays and conformational principles for interpreting cancer drivers. PLoS Comput Biol. 2019;15 doi: 10.1371/journal.pcbi.1006658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Jørgensen K.W., Rasmussen M., Buus S., Nielsen M. NetMHCstab – predicting stability of peptide-MHC-I complexes; impacts for cytotoxic T lymphocyte epitope discovery. Immunology. 2014;141:18–26. doi: 10.1111/imm.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Rasmussen M., Fenoy E., Harndahl M., Kristensen A.B., Nielsen I.K. Pan-specific prediction of peptide-MHC class I complex stability, a correlate of T cell immunogenicity. J Immunol. 1950;2016(197):1517–1524. doi: 10.4049/jimmunol.1600582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Scheiblhofer S., Laimer J., Machado Y., Weiss R., Thalhamer J. Influence of protein fold stability on immunogenicity and its implications for vaccine design. Expert Rev Vaccines. 2017;16:479–489. doi: 10.1080/14760584.2017.1306441. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.