Abstract
Alterations in ROS metabolism and redox signaling are often observed in cancer cells and play a significant role in tumor development and drug resistance. However, the mechanisms by which redox alterations impact cellular sensitivity to anticancer drugs remain elusive. Here we have identified the mitochondrial isoform of thioredoxin reductase 3 (mtTXNRD3), through RT-PCR microarray screen, as a key molecule that confers drug resistance to sorafenib and other clinical anticancer agents. High expression of mtTXNRD3 is detected in drug-resistant leukemia and hepatocellular carcinoma cells associated with significant metabolic alterations manifested by low mitochondrial respiration and high glycolysis. Mechanistically, high mtTXNRD3 activity keeps the mitochondrial thioredoxin2 (Trx2) in a reduced stage that in turn stabilizes several key survival molecules including HK2, Bcl-XL, Bcl-2, and MCL-1, leading to increased cell survival and drug resistance. Pharmacological inhibition of thioredoxin reductase by auranofin effectively overcomes such drug resistance in vitro and in vivo, suggesting that targeting this redox mechanism may be a feasible strategy to treat drug-resistant cancer.
Keywords: Drug resistance, Sorafenib, TXNRD3, Redox modulation, Mitochondria, Auranofin
1. Introduction
Development of drug resistance in tumor cells is a major challenge in clinical treatment of cancer [1]. This is particularly true in case of tyrosine kinases inhibitors used in targeted therapy, where mutations of the target genes and altered expression of the molecules involved in the relevant pathways often occur after a period of drug treatment [2,3]. The development of second- and third-generations of tyrosine kinase inhibitors is an effective ways to meet the challenge of drug resistance due to target mutations, while proper drug combination is a logical way to overcome drug resistance due to alterations of pathways that functionally compensate or alleviate the inhibition of the primary target. However, the success of the later strategy largely depends on the elucidation of the compensatory pathways for the non-mutational drug resistant mechanisms, which seems to account for a large portion of the drug resistant cases observed in clinic [[4], [5], [6]]. The mechanisms of non-mutational drug resistance to tyrosine kinase inhibitors are complex, and likely vary depending on the nature of the drugs and the pathways impacted. Although it is generally assumed that an up-regulation of pathways that compensate the inhibited targets and/or elevated expression of cell survival molecules are possible mechanisms [2,5,7,8], the detail molecular basis for non-mutational drug resistance for many tyrosine kinase inhibitors remain unclear. This situation hinders the development of effective drug combination strategies to overcome drug resistance in targeted therapy.
Sorafenib is multi-kinase inhibitor currently used in the clinical treatment of advanced liver cancer, advanced kidney cancer, and acute myelocytic leukemia (AML) with FLT3-IDT mutations [[9], [10], [11]]. Although this targeted agent provides a new therapeutic option for treatment of cancers in advanced stage, the clinical outcomes are far from satisfactory and drug resistance often occurs [12,13]. Although certain molecules/pathways such as PI3K/Akt and microRNA-122 have been implicated to be involved in the sorafenib resistance [14,15], the exact underlying mechanisms responsible for sorafenib resistance largely remain unclear. Our previous study showed that alterations of metabolism, especially changes in mitochondrial functions, contributed to resistance to multiple chemotherapeutic drugs [16,17]. Since altered metabolism is a major feature of cancer cells associated with increased cell survival and drug resistance [18], we postulated that metabolic alterations might play a significant role in the development of resistance to tyrosine kinase inhibitors, many of which directly or indirectly affect metabolic pathways.
In this study, we developed a set of PCR microarrays containing primers for 168 genes involving various metabolic pathways and mitochondrial molecules as a tool to identify potential metabolic genes that might affect cell survival and drug sensitivity. Using two pairs of cancer cells with different sensitivity to sorafinib, we unexpectedly identified a redox metabolic enzyme mtTXNRD3, a mitochondrial isoform of thioredoxin reductase 3, as a key molecule that promotes sorafinib resistance through stabilization of the mitochondrial-associated pro-survival molecules. The possibility of use mtTXNRD3 as a new therapeutic target to overcome drug resistance was also tested in vitro and in vivo.
2. Materials and methods
2.1. Reagents and antibodies
Sorafenib (Selleck Chemicals, Houston, TX, USA), auranofin (MCE, Houston, TX, USA), and Gefitinib (Selleck Chemicals, Houston, TX, USA) were dissolved in DMSO at a stock concentration of 10 mM, Erlotinib (Selleck Chemicals, Houston, TX, USA) was dissolved in DMSO at a stock concentration of 5 mM and stored in aliquots at −20 °C. MTS assay Kit (CellTiter 96 Aqueous One Solution reagent) was purchased from Promega Corporation (Madison, WI, USA). Annexin V-FITC/PI apoptosis Detection Kit was purchased from Keygen Company (Nanjing, China). Mito-SOX were purchased from Invitrogen (Carlsbad, CA, USA). Anti-TXNRD2 (ab16841), anti-Trx2 (ab185544), anti-MCL-1 (ab32087), and anti-TXNRD3 (ab134034) antibodies were purchased from Abcam (Cambridge, MA, USA); Anti-HSP60 (#4870S), anti-TXNRD1 (#15140s), anti-α-tubulin (#2144), anti-β-actin (#4970), anti-Bcl-2 (#3498s), and anti-Bcl-xL (#2762s) antibodies were from Cell Signaling Technologies (Danvers, MA, USA).
2.2. Metabolic qRT-PCR array
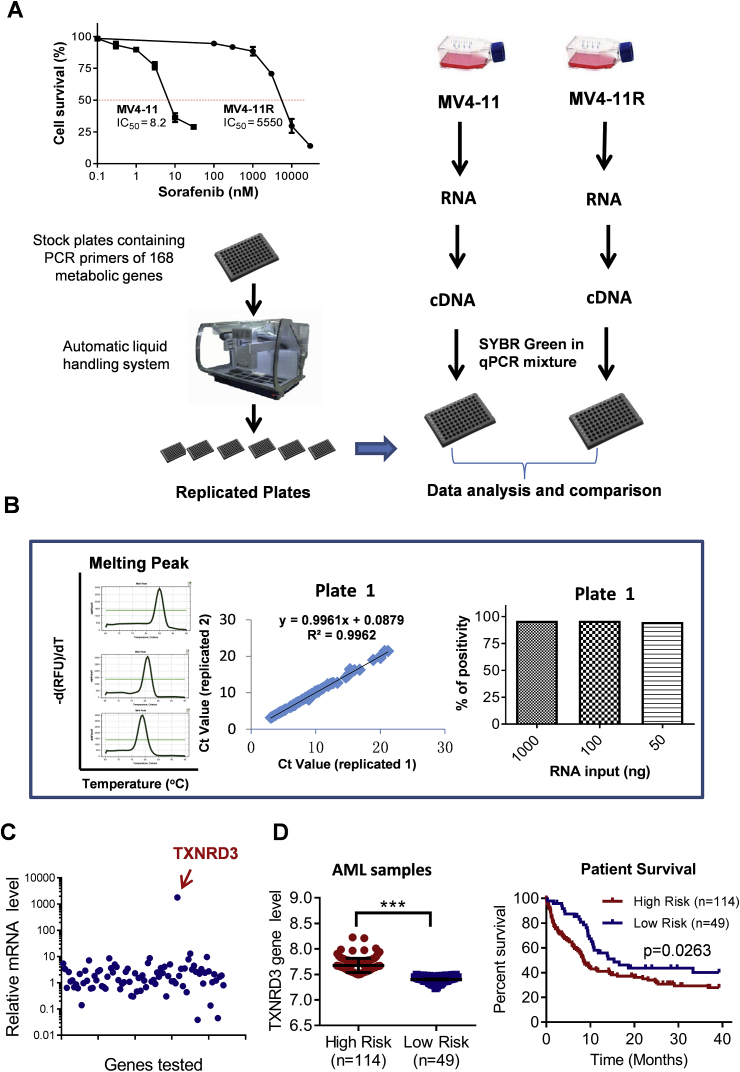
Based on their roles in various metabolic pathways, 168 genes were selected for assembling a set of metabolic qRT-PCR arrays. The oligo sequences of the primers were designed using the Primers 3 software (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) or were obtained from Primer Bank (https://pga.mgh.harvard.edu/primerbank/index.html). To ensure a uniform RT-PCR amplification conditions, primers were designed to achieve an amplicon size of 80–230 bp, a GC content of within the range of 35–65%, and a melting temperature (Tm) of 59–62 °C. The primers (10 μM, 250 μL) were loaded onto their designated wells in the 96-well micro-plates as original stock plates, which were then replicated to make assay plates using a HTS liquid handling station (Aurora Versa1100, Canada). The working concentration of primer was 20 pmole/well/gene. The assay plates were stored at −20 °C until used. For qRT-PCR analysis, the cDNA templates were mixed with the PCR master mixture (SYBR Premix Ex Taq II kit from TaKaRa Bio, Tokyo, Japan), and then run in a Bio-Rad CFX96 PCR machine (Bio-Rad, Hercules, CA, USA) using the following cycling program: 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 55 °C for 30 s, and then 72 °C 30 s. The performance of the metabolic qRT-PCR array was evaluated for specificity, reproducibility and sensitivity. Primer specificity was evaluated using dissociation (melting) curve analysis. The high quality of input primers (Supplementary Table 2) was indicated by single-band (peak) product without primer dimers or other secondary products. Reproducibility was tested by comparing two replicate sets of raw threshold data obtained in two separate operations. The degree of correlation with R2 > 0.99 was considered highly reproducible. Assay sensitivity was evaluated by using series dilutions of total RNA input (50–1000 ng) and monitoring the percentage of detectable RT-PCR gene products. Data analysis was based on the ΔΔCT method. The differential expression of the 168 metabolic genes was revealed by analyzing the ratios of the respective gene expression between the drug-resistant MV4-11R cells and the drug-sensitive MV4-11 cells. Data analysis was automated using an Excel-based data analysis tool. Fig. 1A shows the design and steps of the qRT-PCR analysis.
Fig. 1.
Identification of TXNRD3 in Sorafenib-resistant cells by metabolic qRT-PCR array. (A) Experimental procedures to prepare metabolic qRT-PCR array for comparison of gene expression profiles of sorafenib-sensitive cells (MV4-11) and sorafenib–resistant cells (MV4-11R). Cell viability was measured by MTS assay. (B) Characterization of the metabolic qRT-PCR array showing primer specificity (product melting curves, left panel), reproducibility in replicate experiments (middle panel), and detection sensitivity over a range of 50–1000 ng RNA input (right panel). (C) Identification of TXNRD3 as a highly expressed gene in sorafenib-resistant cells (MV4-11R), revealed by metabolic qRT-PCR array analysis. (D) Expression of TXNRD3 in leukemia samples from high-risk and low-risk AML patients and their association with overall survival based on analysis of GEO dataset (GSE12417). Data represent mean ± SD; ***, p < 0.001.
2.3. Cell culture
MV4-11, SK-Hep1, Hep3B cells were obtained from the American Type Culture Collection (ATCC, USA). Cells were maintained in RPMI1640 or DMEM (GIBCO, Life Technologies, NY, USA) supplemented with 10% FBS (Biological Industries, State Israel) in a 37 °C humidified incubator with 5% CO2. Sorafenib-resistant cell line MV4-11R was maintained in RPMI 1640 medium containing 500 nM sorafenib as described previously [19]. Sorafenib-resistant hepatocellular carcinoma cells (Hep3B-R) were established by culturing Hep3B cells with increasing concentrations (up to 5 μM) of sorafenib for 2–3 months in complete DMEM medium. The cells were assayed for mycoplasma by the PCR (Thermo Fisher Scientific, Waltham, MA, USA), and all tests were negative.
2.4. Western blot analysis
Cells or isolated mitochondria were homogenized in protein lysis buffer, and debris was removed by centrifugation at 12,000 g for 10 min at 4 °C. The protein concentrations in all samples were quantified by using the Pierce™ BCA Protein assay kit (Thermo scientific, Waltham, MA, USA). Protein lysates were analyzed by standard SDS/PAGE and transferred to a PVDF membrane. Protein bands of interest were revealed by blotting with the respective antibodies, using a rabbit anti-TXNRD1 antibody (1:1000 dilution), anti-TXNRD2 antibody (1:1000 dilution), anti-TXNRD3 antibody (1:600 dilution). The density of the immune-reactive bands was analyzed using Image J.
2.5. Forced overexpression of TXNRD3
TXNRD3 plasmid containing the coding sequence of TXNRD3 with a Sec/Cys conversion, and the empty vector control were purchased from GenePharma (Shanghai, China). The detail information of the recombinant plasmid is shown in Supplementary Table 3. For transient transfection, 5 × 105 SK-Hep1 cells were seeded into 6-well plates and transiently transfected with 5 μg of TXNRD3 plasmids or empty vector using Lipofectamine 3000 transfection reagent (Invitrogen, USA). Stable TXNRD3 over-expressing cell colonies were selected in cell culture medium containing 600 μg/mL G418 (Selleck, Houston, TX, USA).
2.6. Cell proliferation and cell viability assay
Cell proliferation was measured using MTS assay (CellTiter 96 AQueous One Solution, Promega) as previously described [19]. Cell viability was calculated by the following formula: cell viability (%)=(average absorbance of treated group-average absorbance of blank)/(average absorbance of untreated group-average absorbance of blank) × 100%. IC50 values were calculated using the GraphPad Prism 5 software.
2.7. Isolation of mitochondria
Mitochondria were isolated using Qproteome mitochondria isolation kit using the procedures recommended by the manufacturer (QIAGEN, Duesseldorf, Germany).
2.8. TrxR activity assay
TrxR activity was measured using a thioredoxin reductase activity colorimetric assay kit according to the procedures recommended by the manufacturer (BioVision, Milpitas, CA, USA). Briefly, cell lysates were prepared using the assay buffer (containing 1 μl/ml protease inhibitor Cocktail). After centrifugation at 10,000×g for 15 min at 4 °C, the supernatant was collected and the TrxR activity was measured in the reaction buffer (a total 40 μl Reaction Mix: 30 μl Assay Buffer; 8 μl DTNB Solution; 2 μl NADPH). The specific TrxR activity (U/mg protein) was calculated by measuring the change in OD at 412 nm during a 20-min incubation in dark. The TrxR activity was further normalized with the quantity of protein in the samples quantified using the PierceTM BCA Protein assay kit (Thermo scientific, Waltham, MA, USA).
2.9. Clonogenic survival assay
For liver cancer cells, 5 × 103 cells per well were seeded onto a 6-well culture plate and cultured in DMEM medium with or without 5 μM sorafenib. After two weeks of incubation, the samples were washed once with PBS, fixed with methanol (5 min), and stained with crystal violet. The blue-stained colonies were imaged and counted. All analyses were performed in triplicate.
2.10. Measurement of oxygen consumption rate and extracellular acidification rate
Oxygen consumption rates (OCR) and extracellular acidification rates (ECAR) were measured using an XF24 extracellular analyzer (Seahorse Bioscience, USA) according to the manufacturer recommended procedures. Liver cancer cells in the exponential growth phase were seeded in triplicate at a density of 2.5 × 105 cells/well onto a 24-well cell culture microplate overnight. For MV-411R cells, a 24-well cell culture microplate was coated with Cell and Tissue Adhesive Corning® Cell-Tak™ (Corning Incorporated, NY, USA) to allow adhesion of the leukemia cells. The plate with seeded cells was centrifuged at 1500 rpm for 5 min (accel rate:4, brake rate:0). After calibration of the analyzer, sequential compound injections of oligomycin (1 μM), carbonyl-cyanide 4-(trifluoromethoxy) phenylhydrazone (FCCP, 1 μM), antimycin A & rotenone (0.5 μM) were applied to measure oxygen consumption rate (OCR). To test glycolytic activity, sequential compound injections of glucose (10 mM), oligomycin (1 μM) and 2-DG (50 mM) were applied to measure extracellular acidification rate (ECAR). The OCR and ECAR values were normalized by protein levels in the samples quantified using the PierceTM BCA Protein assay kit (Thermo scientific, Waltham, MA, USA).
2.11. Measurement of ROS generation
Cancer cells were incubated with serum-free medium with addition of 2 μM of Mito-SOX for 20 min at 37 °C. Following the staining, the cells were washed with 4 °C PBS twice, and then collected and analyzed with a FACSCalibur flow cytometer (Becton, Dickinson and Company, Franklin Lakes, NJ, USA). Data were analyzed using CellQuest Pro software.
2.12. Apoptosis assay
Cell death of MV4-11 and MV4-11R cells induced by sorafenib or auranofin was analyzed with flow cytometry using annexin V/PI assays according to the manufacturer's instructions. The cells were finally subjected to flow cytometric and results were analyzed using CellQuest Pro software.
2.13. Silencing of gene expression by siRNA
Small interfering RNAs (siRNA), specifically targeting TXNRD3 and negative control were purchased from Ribobio (Guangzhou, China), genOFFTM st-h-TXNRD3-1: GGAGAAGATTGGTGTCAAA; st-h-TXNRD3-2: GTGGTGATCTTCAGCAAGA. To silence the expression of mtTXNRD3, MV4-11R cells were transfected with siRNA by electroporation, using the Neon transfection system (Invitrogen, CA, USA) according to manufacturer's instructions. In brief, cells for each transfection were washed with PBS, resuspended in 100 μl of resuspension buffer (R-buffer) at a final density of 1.0 × 107 cells/mL and mixed with 10 μl (20 μM) of TXNRD3 siRNA in a sterile eppendorf tube. The cells-siRNA mixture was subjected to two pulses with pulse width 20 ms at 1400 V. After 48 or 72 h, the cells were analyzed by qRT-PCR or Western Blot.
2.14. Analysis of thioredoxin-2 redox status
The redox status of Trx2 was measured as previously described [20]. In brief, cells were washed with ice-cold PBS and collected by acid precipitation using ice-cold trichloric acetic acid (10%) for 30 min at 4 °C. Samples were centrifuged at 13000×g for 10 min and resuspended in ice-cold acetone (100%) and incubated for 30 min at 4 °C. Acetone was removed from tube by centrifugation at 12000g for 10 min, and the pellet was resuspended in lysis/derivatization buffer (50 mM Tris-HCl, pH 8, 0.1% SDS, and 15 mM AMS) by sonication, and incubated for 3 h at room temperature. Non-reducing SDS polyacrylamide (15%) gel electrophoresis was performed to separate the oxidized and reduced Trx2. Western blotting was performed using a rabbit anti-Trx2 polyclonal antibody (1:1000 dilution) and goat anti-rabbit secondary antibody (1:10000 dilution).
2.15. Tumor xenograft and animal study
Female Balb/c nude mice aged 5–6 weeks were obtained from Beijing Vital River Laboratory Animal Technology Co. Ltd (Beijing, China). All experimental procedures using these mice were carried out in accordance with a research protocol approved by the Institutional Laboratory Animal Care and Use Committee of Sun Yat-sen University Cancer Center. Each mouse was inoculated s.c. in the dorsal flank with 2 × 106 SK-Hep1OE or SK-Hep1VC cells suspended in 0.1 mL of serum-free medium. When tumors reached about 50 mm3, mice received auranofin (10 mg/kg.d, i.p., for 22 d) or sorafenib tosylate (10 mg/kg.d, p.o., for 22 d) or vehicle. To examine the antitumor effect of auranofin in combination with sorafenib, mice received a single agent or auranofin (10 mg/kg) + sorafenib tosylate (10 mg/kg) or vehicle orally (p.o.) once daily. Tumors were measured every 2–3 days, and their volumes were calculated using the following standard formula: width2 × length × 0.52 [21]. At the end of the experiment, animals were euthanized, and the tumors were harvested and subjected to histological examination.
To evaluate the effect of sorafenib on leukemia cells in vivo, 6–8 week old female NOD-SCID mice were pretreated with cyclophosphamide (150 mg/kg i.p., once a day for 2 days). After 24 h, each mouse was intravenously injected via the tail vein with 3.5 × 106 MV4-11R cells harboring GFP. Disease progression was monitored by quantifying the GFP-expressing cells in the peripheral blood by flow cytometry. Drug treatment started 11 days after mice were inoculated with GFP-expressing leukemia cells. At the end of the experiment, mice were sacrificed and leukemia cells in the blood samples were analyzed using flow cytometry to quantify GFP-positive cells.
2.16. Database analysis
The Gene Expression Omnibus (GEO) database was used to analyze the potential correlation between TXNRD3 expression in AML patients and their survival. The pretreatment cohort of AML patients in the GEO database was analyzed. The data set (GSE12417) comprised of 115 cases with high TXNRD3 expression and 48 cases with low TXNRD3 expression from Metzeler et al. [22] were analyzed. The hazard ratio was estimated by fitting a CoxPH (Surv (time, status) model using the risk group as the covariate.
2.17. Statistical analysis
The data were expressed as means ± SD. The statistical difference between two groups of data was evaluated by Student's t-test or two-way ANOVA, and the difference among more than two groups was evaluated by one-way ANOVA (Prism GraphPad 7.0, La Jolla, CA, USA). A p value of less than 0.05 was considered statistically significant.
3. Results
3.1. Identification of TXNRD3 in sorafinib-resistant cells by RT- PCR array analysis
We have recently established a sorafenib-resistant cell line (MV4-11R) by step-wise exposure of AML leukemia cells (MV4-11) to increasing concentrations of sorafinib [19]. As shown in Fig. 1A, MV4-11R cells were highly resistant to sorafenib with an IC50 value of 5550 nM (MTS assay), approximately 670-fold more resistant than the parental MV4-11 cells. Since the sorafenib-resistant cells exhibited metabolic alterations [19], we developed a metabolic RT-PCR array in an attempt to identify potential metabolic gene(s) that might be responsible for the observed phenotype. The RT-PCR array contained 168 genes involved in various metabolic pathways and ROS/redox signaling (Supplementary Table 1). As shown in Fig. 1A, RNA was isolated from MV4-11 cells and the sorafenib-resistant MV4-11R cells. Quantitative RT-PCR was used to compare the expression of the 168 genes on the metabolic array. The results showed that this qRT-PCR analysis was both sensitive and reproducible, required only 50 ng total RNA input for reliable detection with a high degree of linear correlation (R2 > 0.99) between replicate experiments (Fig. 1B and Supplementary Fig. S1). Comparison of the gene expression in the two cell lines showed that thioredoxin reductase 3 (TXNRD3) was highly up-regulated in the sorafenib-resistant cells with an increase of over 1000 folds (Fig. 1C). Importantly, analysis of public database revealed that high-risk acute myeloid leukemia (AML) patients had high expression of TXNRD3, and exhibited worse overall survival compared to the low-risk AML patients who had better survival and lower TXNRD3 expression (p = 0.0263, Fig. 1D). These data suggest that TXNRD3 might play a major role in affecting the clinical outcome of the leukemia patients.
3.2. The elevated TXNRD3 in sorafenib-resistant cells is located in the mitochondria
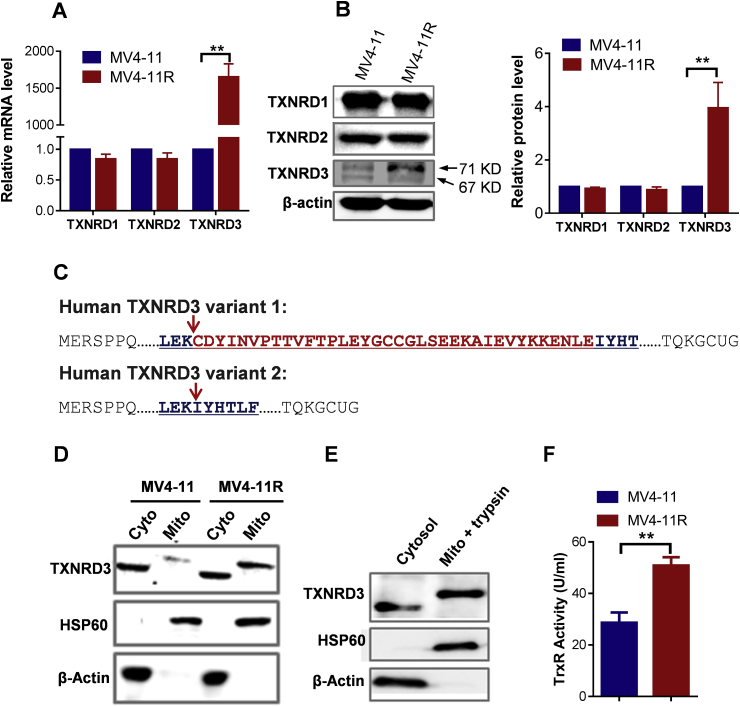
The observation that TXNRD3 mRNA expression was highly up-regulated in sorafenib-resistant cells prompted us to compare the expression of other TrxR isoforms, since it is known that there are at least three isoforms of thioredoxin reductases [23]. As shown in Fig. 2A, the expression of TXNRD3 mRNA, but not that of the TXNRD1 or TXNRD2 isoforms, significantly increased in the drug-resistant cells. This specific increase of TXNRD3 expression was also observed at the protein level (Fig. 2B). Western blot analysis using an antibody against TXNRD3 revealed two protein bands located approximately at 71 KD and 67 KD (Fig. 2B), indicating a simultaneous presence of two possible splicing variants. A search of the NCBI database revealed that there are indeed two possible TXNRD3 splicing variants with predicted molecular weights of 70.7 KD and 66.6 KD, respectively (www.ncbi.nlm.nih.gov/protein/NP443115.1; www.ncbi.nlm.nih.gov/protein/NP001166984.1). As shown in Fig. 2C, the difference between the 70.7KD and 66.6 KD TXNRD3 variants is a deletion of 36 amino acids (indicated in red color). Interestingly, we found that only the higher molecular weight variant (70.7 KD) exhibited an increase in the sorafenib-resistant cells, whereas the 66.6 kD variant remained unchanged (Fig. 2B). Further subcellular fractionation experiments showed that the 70.7KD variant was located in the mitochondrial fraction, while the 66.6 KD protein was in the cytosol (Fig. 2D). Trypsin digestion of isolated mitochondria did not cause any degradation of the 70.7kD protein (Fig. 2E), indicating that this protein variant was located within the mitochondria and was protected by the mitochondrial membranes from trypsin digestion. Thus, we designated this TXNRD3 variant as mitochondrial thioredoxin reductase 3 (mtTXNRD3). Analysis of thioredoxin reductase activity in the cell extracts of MV4-11 and MV4-11R cells showed that the enzyme activity was significantly increased in the sorafenib-resistant MV4-11R cells (Fig. 2F), suggesting that mtTXNRD3 contributed substantially to the total TrxR activity in the drug-resistant cells.
Fig. 2.
The upregulated TXNRD3 in sorafenib-resistant cells was located in the mitochondria. (A) Comparison of mRNA expression levels of TXNRD1, TXNRD2, TXNRD3 in sorafenib-sensitive (MV4-11) and -resistant (MV4-11R) cells, measured by RT-qPCR. (B) Comparison of protein expression levels of TXNRD1, TXNRD2, TXNRD3 in sorafenib-sensitive (MV4-11) and -resistant (MV4-11R) cells, measured by western blotting analyses using the respective antibodies. (C) Comparison of amino acid sequences of two possible TXNRD3 splicing variants. (D) Mitochondrial and cytosolic proteins were prepared and analyzed by immune blotting analyses using the indicated antibodies. β-actin and HSP60 were used as the loading controls for cytosolic and mitochondrial proteins, respectively. “Mito” and “Cyto” indicate mitochondrial and cytosolic, respectively. (E) Isolated mitochondria were incubated with trypsin for 2 min at 37 °C, and then resuspended in protein lysis buffer and determined by immunoblotting analyses using the indicated antibodies. (F) The TrxR activity in MV4-11R cells compared with the parental MV4-11 cells. Data represent mean ± SD; **, p < 0.01, n = 3.
Since sorafenib is currently used for clinical treatment of certain leukemia (AML) as well as liver cancer, we next tested if mtTXNRD3 might also increase in liver cancer cells that became resistant to sorafenib. To this end, we exposed liver cancer cells (Hep3B) to increasing concentrations of sorafenib in a stepwise manner over months to generate sorafenib-resistant cell population (Hep3B-R). As shown in Supplementary Figs. S2A–E, Hep3B-R cells were substantially more resistant to sorafenib treatment compared to the parental cells, as measured by flow cytometry analysis of apoptosis (Supplementary Fig. 2A), MTS assay for cell proliferation (Supplementary Fig. S2B) and colony formation assay for long-term cell replication (Supplementary Figs. S2C–E). Western blot analysis showed that the expression of mtTXNRD3 protein was significantly up-regulated in sorafenib-resistant cells (Supplementary Fig. S2F), consistent with the observation in the drug-resistant AML cells (MV4-11R). Analysis of TrxR activity also confirmed the increase of the enzyme activity in Hep3B-R cells (Supplementary Fig. S2G). Interestingly, analysis of TXNRD3 expression in human hepatocellular carcinoma samples using Kaplan-Meier Plotter Database as described by Menyhart et al. [24] revealed that high expression of TXNRD3 appeared to be associated with shorter relapse-free survival (Supplementary Fig. S3).
3.3. Over-expression of mtTXNRD3 induces metabolic shift toward glycolysis and promotes sorafenib resistance
To further evaluate the biological consequence of mtTXNRD3 high expression, we first transfected liver cancer SK-Hep1 cells, which expressed low level of endogenous mtTXNRD3 protein, with mtTXNRD3 expression vector in comparison with an empty vector control. As shown in Fig. 3A–C, the forced overexpression of mtTXNRD3 (SK-Hep1OE) moderately increased the enzyme activity, and caused a moderate decrease of cell proliferation. Importantly, the mtTXNRD3 overexpressing cells exhibited significant resistance to sorafenib measured by MTS assay (Fig. 3D). The drug-resistant phenotype of mtTXNRD3-overexpressing cells was also observed in colony formation assay (Fig. 3E and F), confirming the role of mtTXNRD3 in promoting sorafenib resistance.
Fig. 3.
Forced overexpression of mtTXNRD3 led to sorafenib resistance and a metabolic shift toward low OXPHOS and high glycolysis. (A) Forced overexpression of mtTXNRD3 in liver cancer cell line (SK-Hep1) by stable transfection. Immunoblot analysis was performed to detect the TXNRD3 protein expression in SK-Hep1 cells (OE, over expression; VC, vector control). “Mito” and “Cyto” indicate mitochondrial and cytosolic, respectively. (B) Relative levels (% of control) of the TrxR enzyme activity in SK-Hep1OE cells compared with the SK-Hep1VC cells. (C) Proliferation in SK-Hep1OE and SK-Hep1VC cells. Equal number (3 × 103) of the indicated cells were started in cell culture, and MTS analysis was used to monitor cell growth every 24 h for up to 96 h. (D) Comparison of sensitivity of SK-Hep1OE and SK-Hep1VC cells to sorafenib. The indicated cells were treated with various concentrations of sorafenib for 72 h and cell viability was measured by MTS assay. (E, F) Effect of sorafenib on clonogenic formation in SK-Hep1 cells with or without overexpression of TXNRD3. Cells were treated with sorafenib as indicated. After two weeks of incubation, colonies were stained and imaged using an inverted microscope with a camera, and colony numbers were then counted. (G-J) Impact of mtTXNRD3 expression on oxygen consumption rate (OCR) and extracellular acidification rate (ECAR). OCR (G and I) and ECAR (H and J) of the indicated cells were monitored using the Seahorse XF24 analyzer. Data represent mean ± SD; **, p < 0.01; n = 3.
Since mtTXNRD3 is physically located in the mitochondrial, we then tested if its over expression might affect mitochondrial function. Analysis of cellular oxygen consumption rate (OCR), a key indicator of mitochondrial respiratory activity, showed that overexpression of mtTXNRD3 in SK-Hep1 cells led to a down-regulation of OCR (Fig. 3G) and an increase of extracellular acidification rate (ECAR, Fig. 3H), an indicator of lactate production from glycolytic metabolism. These data together suggest that overexpression of mtTXNRD3 could induce a metabolic shift from oxidative phosphorylation toward more active glycolysis. Consistently, the sorafenib-induced drug resistant Hep3B-R cells, which expressed a high level of mtTXNRD3, also exhibited a decrease in OCR (Fig. 3I) and an increase in ECAR (Fig. 3J), confirming a significant role of mtTXNRD3 in causing metabolic shift toward the “Warburg phenotype”.
A key function of thioredoxin reductase is to convert the oxidized thioredoxin to the reduced form, and thus functions as an antioxidant molecule. We tested the impact of mtTXNRD3 on cellular ROS level, and showed that overexpression of TXNRD3 could decrease the cellular ROS in both MV4-11 cells and SK-Hep1 cells, whereas a knockdown of mtTXNRD3 in SK-Hep1OE cells induced a significant increase in ROS (Supplementary Fig. S4). Since many cancer cells intrinsically have higher levels of ROS compared with normal cells, they are under oxidative stress and are more sensitive to further ROS stress [25,26]. Thus, the high expression of mtTXNRD3 might alleviate ROS stress in cancer cells and thus promote their survival.
3.4. Silencing of mtTXNRD3 expression restores cellular sensitivity to sorafenib
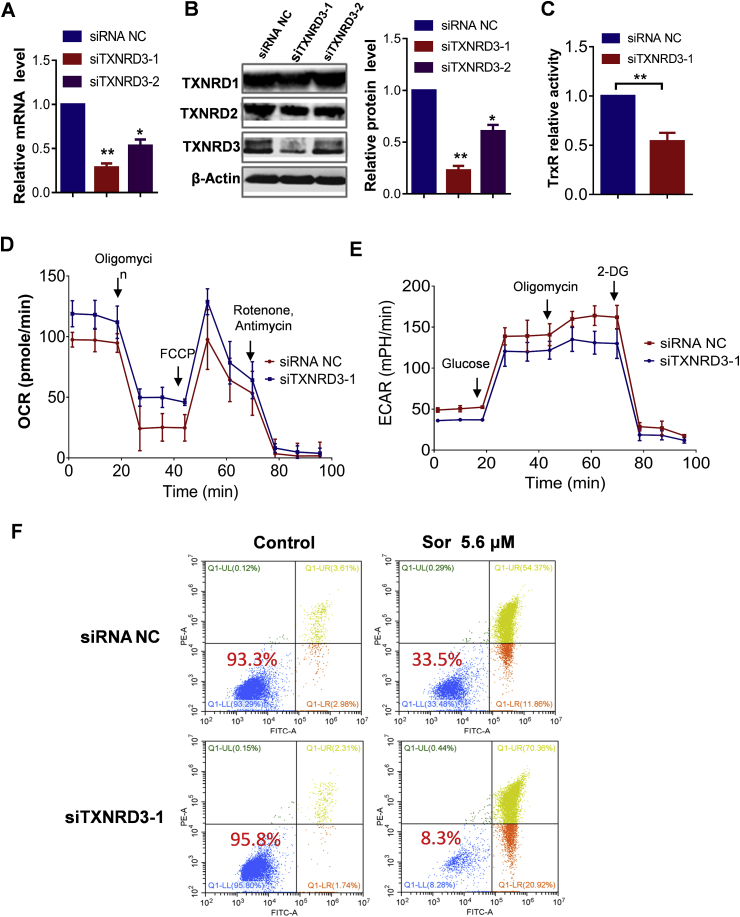
To functionally validate the role of TXNRD3 in sorafenib-resistant cells, we used siRNA to silence the expression of mtTXNRD3 in MV4-11R cells. Among the siRNAs tested, siTXNRD3-1 (siRNA #1) was most effective in suppressing mtTXNRD3 expression with 80% silencing at 48 h (Fig. 4A and B). Thus, siTXNRD3-1 was chosen for use in the subsequent experiments. We further showed that knocking down of mtTXNRD3 expression by siTXNRD3-1 was specific, and did not affect the expression of TXNRD1 and TXNRD2 at mRNA and protein levels (Fig. 4B; Supplementary Fig. S5). Analysis of TrxR activity showed that silencing of TXNRD3 by siTXNRD3-1 led to a significant decrease in the enzyme activity (Fig. 4C). We then measured OCR and ECAR to evaluate the effect of TXNRD3KD on the function of mitochondria and glycolysis. In MV4-11R cells, a knockdown of TXNRD3 expression by siTXNRD3-1 led to an increase of OCR and a decrease of ECAR (Fig. 4D and E).
Fig. 4.
Silencing of mtTXNRD3 expression in sorafenib-resistant cells restored drug sensitivity. (A)–(B): MV4-11R cells were treated with siRNAs targeting different sites of TXNRD3 (siTXNRD3) for 48–72h, cells were then collected for mRNA (48 h) and protein (72 h) isolation. TXNRD3 mRNA (A) was quantified using RT-qPCR; protein expression (B) was monitored and quantified using immunoblot analysis. NC: negative control. (C) Comparison of the relative TrxR enzyme activity in siTXNRD3 NC or siTXNRD3-1 treated MV4-11R cells. (D)–(E) Comparison of oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) in siTXNRD3 NC or siTXNRD3-1 treated MV4-11R cells. Cellular OCR (D) and ECAR (E) of the indicated cells were monitored using the Seahorse XF24 analyzer. (F) TXNRD3 knockdown by siRNA increased cellular sensitivity to sorafenib in MV4-11R cells. Cell viability was assessed by annexin-V/PI assay. The value in each panel indicates the % of survival cells. Data represent mean ± SD; *, p < 0.05; **, p < 0.01; n = 3.
We reasoned that if mtTXNRD3 played an important role in sorafenib resistance, silencing of its expression in sorafenib-resistant cells would restore their sensitivity to this compound. Indeed, when mtTXNRD3 in MV4-11R cells was knocked down by siTXNRD3-1, their apoptotic response to sorafenib was significantly increased with only 8.3% viable cells. In contrast, MV4-11R cells treated with control vector and the same concentration of sorafenib showed 33.5% viable cells (Fig. 4F).
3.5. Overexpression of mtTXNRD3 leads to accumulation of cell survival molecules
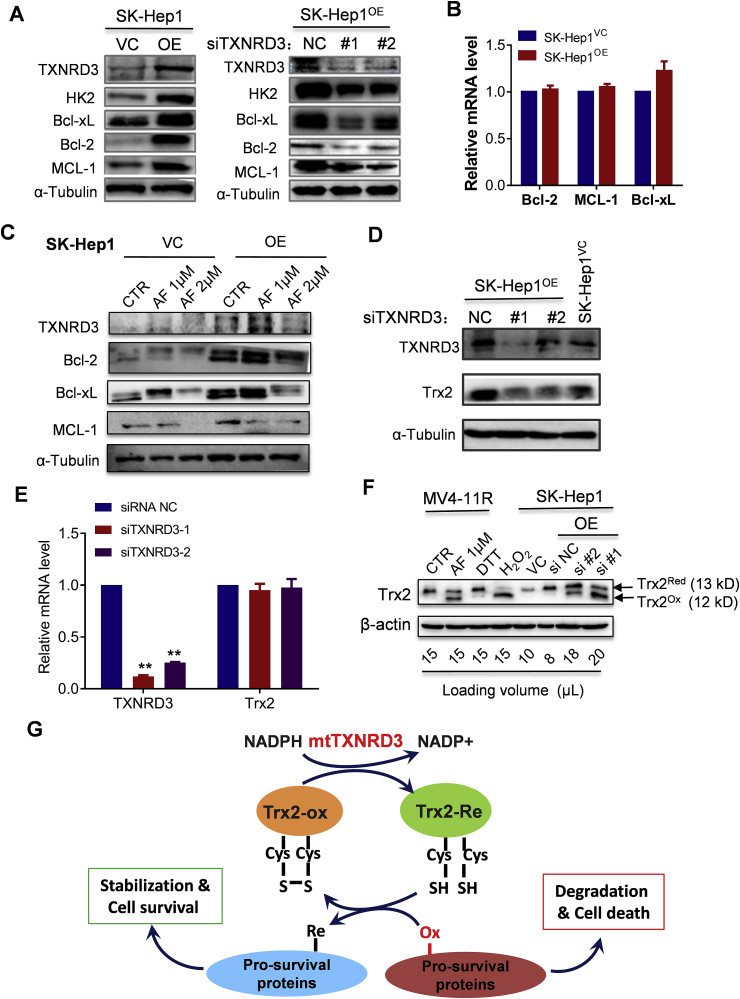
To investigate the potential mechanisms by which mtTXNRD3 induces resistance to sorafenib, we tested if mtTXNRD3 could affect the levels of key molecules involved in cell survival and drug resistance. Western blot analysis revealed that overexpression of mtTXNRD3 in SK-Hep1 cells caused a substantial increase of several mitochondrial-associated survival molecules including hexokinase-2 (HK2), Bcl-xL, Bcl-2, and MCL-1 (Fig. 5A, left panel). Consistently, a knockdown of mtTXNRD3 led to a decrease of these survival molecules (Fig. 5A, right panel). RT-qPCR analysis showed that overexpression of mtTXNRD3 did not cause any changes in mRNA expression of Bcl-2, MCL-1, or Bcl-xL (Fig. 5B), suggesting that mtTXNRD3 likely affected these survival molecules at the protein level. Consistently, treatment of SK-Hep1 cells with auranofin (AF), a known inhibitor of TrxR, led to a decrease in protein levels of the Bcl-2 family members (Fig. 5C).
Fig. 5.
Overexpression of mtTXNRD3 led to accumulation of cell survival molecules. (A) Comparison of protein expression of survival molecules in liver cancer cells with different TXNRD3 status. Protein extracts were prepared from SK-Hep1 cells with TXNRD3OE or TXNRD3KD as indicated, and immunoblot analysis was performed using the indicated antibodies. (B) RT-qPCR was performed to measure the mRNA level of Bcl-2, Bcl-xL and MCL-1 for indicated cell lines. (C) Effect of auranofin on expression of TXNRD3 and Bcl-2 family members. SK-Hep1OE or SK-Hep1VC cells were treated with various concentrations of auranofin for 24 h, and immunoblot analysis was performed with the indicated antibodies. (D) Effect of TXNRD3OE or TXNRD3KD on mitochondrial Trx2 protein level in SK-Hep1 cells. Immunoblot analysis was performed with the indicated antibodies. (E) RT-qPCR was performed to measure the mRNA level of TXNRD3 and Trx2 after knockdown of mtTXNRD3 in SK-Hep1OE cells. (F) Analysis of Trx2 redox status by immunoblot analysis in MV4-11R and SK-Hep1 cells under the indicated conditions. (G) Schematic illustration of the molecular mechanism by which mtTXNRD3 regulation cell survival and drug sensitivity. Data represent mean ± SD; **, p < 0.01, n = 3.
The thioredoxin reductases (TXNRD1, TXNRD2, and TXNRD3) and various thioredoxins are key components of the cellular redox systems important for antioxidant defense and redox regulation. Since thioredoxin-2 (Trx2) is mitochondria-specific member of the thioredoxin family that affects the redox status of many mitochondria-associated proteins [27], we postulated that the overexpression of mtTXNRD3 might keep Trx2 in its reduced form, which could in turn promote the stability of multiple survival molecules by altering their redox states. To test this possibility, we first examined the impact of mtTXNRD3 on Trx2 protein, and showed that mtTXNRD3KD resulted in a decrease of Trx2 protein level (Fig. 5D) without any change in mRNA level (Fig. 5E). Analysis of Trx2 protein by redox-western blotting showed that the genetic knockdown of mtTXNRD3 by siRNA or chemical inhibition of the thioredoxin reductase enzyme activity by AF induced a major shift of Trx2 from reduced state to oxidized state (Fig. 5F). Since it is the reduced form of Trx2 that provides reducing power to maintain the redox homeostasis of other proteins, the shifting of mitochondrial Trx2 to oxidized form would render other mitochondrial-associated proteins in an oxidized/unstable state. Conversely, a high expression of mtTXNRD3 led to stabilization of pro-survival proteins such as HK2, Bcl-2, Bcl-xL, and MCL-1, thus promoting cell survival and drug resistance (Fig. 5G).
To test if the overexpression of mtTXNRD3 could also confer resistance to other tyrosine kinase inhibitors, we compared three pairs of cells with high or low mtTXNRD3 expression (MV4-11 vs MV4-11R cells; SK-Hep1VC vs SK-Hep1OE cells; Hep3B vs Hep3B-R cells) for their sensitivity to gefitinib and erlotinib. The results showed that the three cell lines with high mtTXNRD3 expression (MV4-11R, SK-Hep1OE, Hep3B-R) were significantly less sensitive to the tyrosine kinase inhibitors compared with their counterparts with low mtTXNRD3 expression (Supplementary Figs. S6–S7). We also compared MV4-11 cells with MV4-11R cells and SK-Hep1VC cells with SK-Hep1OE cells for their sensitivity to doxorubicin and ara-C and found that the two cell lines with high mtTXNRD3 expression (MV4-11R and SK-Hep1OE) were more resistant to the chemotherapeutic agents compared to their respective control cells (MV4-11 and SK-Hep1VC), with approximately 3–5 folds increase in IC50 values (Table 1, Supplementary Fig. S8).
Table 1.
Comparison of chemo sensitivity in sorafenib-sensitive and sorafenib-resistant cells with different mtTXNRD3 expression levels.
| Compound | Cell Lines & IC50 (Mean ± SD) | p Value | |
|---|---|---|---|
|
Doxorubicin (μM) |
MV4-11 IC50 = 0.54 ± 0.11 | MV4-11R IC50 = 2.57 ± 0.16 |
0.0004 |
| SK-Hep1VC IC50 = 0.99 ± 0.06 |
SK-Hep1OE IC50 = 3.42 ± 0.30 |
0.0014 |
|
| Ara-C (μM) | MV4-11 IC50 = 0.86 ± 0.12 |
MV4-11R IC50 = 3.35 ± 0.18 |
0.0003 |
| SK-Hep1VC IC50 = 0.56 ± 0.06 |
SK-Hep1OE IC50 = 1.48 ± 0.18 |
0.0091 | |
The data were the results of three separate experiments. Each experiment was done in triplicate. The statistical significance of the difference between sorafenib-sensitive (low mtTXNRD3 expression) and sorafenib-resistant (high mtTXNRD3 expression) cells was analyzed using Student t-test.
3.6. Effective killing of sorafenib-resistant cells by auranofin in vitro and in vivo
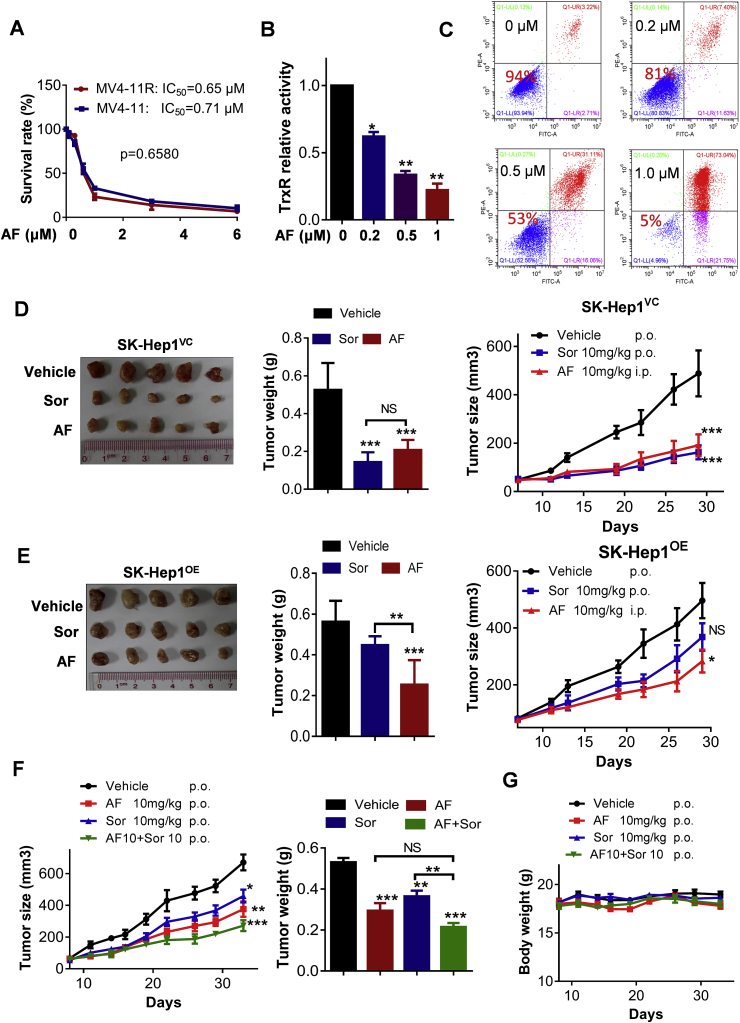
Considering the role of mtTXNRD3 in promoting cell survival and drug resistance, we hypothesized that inhibition of mtTXNRD3 could be an effective strategy to overcome cancer resistance to sorafenib. We first compared the sensitivity of MV4-11 cells and MV4-11R cells to AF, an anti-rheumatic drug known to inhibit TrxR. The results showed that MV4-11 and MV4-11R cells (670-fold more resistant to sorafenib) were equally sensitive to AF, as measured by MTS assay (Fig. 6A). Similarly, the liver cancer cells (SK-Hep1OE) that over-express mtTXNRD3 and the control SK-Hep1VC cells were also equally sensitive to AF (Supplementary Fig. S9). Biochemical analysis showed that AF inhibited TrxR activity in a dose-dependent manner (Fig. 6B). The dose-dependent cytotoxic effect of AF in sorafenib-resistant cells MV4-11R was also confirmed using another assay by flow cytometry analysis of apoptotic cell death (Fig. 6C).
Fig. 6.
Overcoming sorafenib resistance by auranofin in vitro and in vivo. (A) MV4-11/MV4-11R were treated with various concentrations of auranofin (AF) for 72 h and then subjected to MTS assay, n = 3. (B) MV4-11R cells were treated with various concentrations of AF, and the relative enzyme activity of TrxR was measured; n = 3. (C) MV4-11R cells were treated with the indicated concentration of AF for 48 h and subjected to annexin-V/PI assay. The values within each panel indicate % of survival cells. (D) In vivo therapeutic activity of Sorafenib (Sor) or AF in mice bearing SK-Hep1VC human liver cancer xenografts. Tumor images (left panel), tumor weight (middle panel), and tumor growth curves (right panel) were shown, n = 5 mice per group. (E) In vivo therapeutic activity of sorafenib (Sor) or AF in mice bearing SK-Hep1OE human liver cancer xenografts. Tumor images (left panel), tumor weight (middle panel), and tumor growth curves (right panel) were shown, n = 5 mice per group. (F) In vivo therapeutic activity of sorafenib (Sor) in combination with AF in mice bearing SK-Hep1OE human liver cancer xenografts. The tumor growth curves (left panel), tumor weight (right panel), and (G) mouse body weight are shown, n = 7 mice per group. Data represent mean ± SD; *, p < 0.05; **, p < 0.01; ***, p < 0.001; NS, not statistically significant.
Based on the promising anticancer activity of AF against sorafenib-resistant cancer cells in-vitro, we next evaluated the in vivo therapeutic effect of AF using a subcutaneous xenograft model of SK-Hep1 cells with mtTXNRD3 overexpression (SK-Hep1OE) or the vector-control (SK-Hep1VC) cells inoculated in immune-deficient mice. As expected, sorafenib (10 mg/kg.day, p.o.) exhibited significant therapeutic effect in the drug-sensitive tumor model (SK-Hep1VC, Fig. 6D), but did not show a significant activity against the drug-resistant tumor model (SK-Hep1OE, Fig. 6E). In contrast, AF was effective in both tumor models, suggesting that this drug could potentially be used to treat liver cancers that become resistant to sorafenib.
To test if sorafenib in combination with AF could increase the therapeutic activity in sorafenib-resistant tumors, mice bearing SK-Hep1OE xenografts were treated with AF (10 mg/kg/d, p.o.), sorafenib (10 mg/kg/d, p.o.), or their combination. As shown in Fig. 6F and G, sorafenib as a single drug alone exhibited only modest activity against SK-Hep1OE tumor in vivo. Addition of AF significantly enhanced the in vivo the therapeutic activity in both animal models, although the antitumor activity of AF alone was modest (Fig. 6F). This drug combination was well tolerated in mice without significant body weight loss (Fig. 6G).
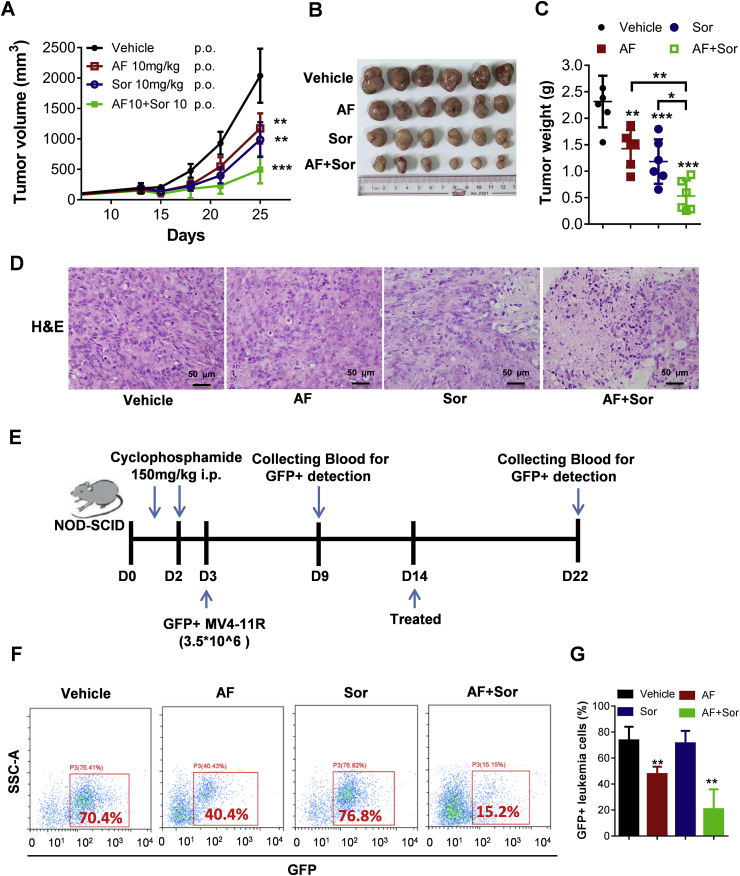
We also use mice bearing subcutaneously inoculated sorafenib-resistant MV4-11R AML cells with TXNRD3 overexpression as another animal model to further evaluate the in vivo therapeutic activity of sorafenib + AF combination. As shown in Fig. 7A–C, sorafenib or AF each as a single drug alone showed only modest activity against MV4-11R leukemia in vivo, whereas the combination of sorafenib and AF significantly enhanced the therapeutic activity in vivo. Morphologic evaluation of the Hematoxylin/eosin (H&E)-stained tumor tissues revealed that the combination of sorafenib and AF caused a structural disruption of the tumor tissue, with many cells exhibiting shrinkage and nuclear condensation indicative of apoptosis (Fig. 7D). Such morphological changes were less obvious in the samples from mice treated with single drug (sorafenib or AF alone).
Fig. 7.
Effect of sorafenib and auranofin combination on the growth of sorafinib-resistant AML cells in vivo. (A)–(C): The sorafinib-resistant MV4-11R leukemia cells were inoculated subcutaneously into Balb/C nude mice, which were treated with sorafenib (Sor), auranofin (AF), or their combination. The subcutaneous tumor growth was measured twice a week for up to 25 days (A); Upon termination of the experiment, tumors were isolated, photographed (B), and weighed (C), n = 6 mice per group. (D) Tumor tissues were fixed, sliced, and stained with H&E for pathological analysis. Scale bar: 50 μm. (E) Schematic illustration of in vivo evaluation of the impact of sorafenib and auranofin on AML leukemia burden. The sorafinib-resistant MV4-11R leukemia cells harboring green fluorescent protein (GFP)-expressing vector (MV4-11R-GFP + cells) were injected into the tail vein of NOD-SCID mice pretreated with cyclophosphamide to enable leukemia engraft (3.5 × 106 cells per mouse). The schedules of drug treatment and blood sampling for analyses of GFP + cells were indicated by the blue arrows. (F) Flow cytometry analysis of GFP + leukemia cells in the peripheral blood from mice sacrificed at the end of experiment (day 22). (G) Flow cytometry quantitation of GFP + leukemia cells in the peripheral blood from mice sacrificed on day 22, n = 6 mice per group. Data represent mean ± SD; *, p < 0.05; **, p < 0.01; ***, p < 0.001. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
We also tested the efficacy of sorafenib in combination with AF in the MV4-11R AML model, where the GFP-expressing MV4-11R leukemia cells (Supplementary Fig. S10A) were inoculated into the tail vein of NOD-SCID mice pre-conditioned with cyclophosphamide (Fig. 7E). In this model, the GFP-expressing MV4-11R cells were disseminated systemically through blood circulation and engrafted in the bone marrow, identified by flow cytometric analysis of GFP-positive cells (Supplementary Fig. S10B). Drug treatment started on day 14 after MV4-11R inoculation, and the drug impact on the leukemia burden was evaluated by analyzing the percent of GFP-positive cells in the blood circulation. As shown in Fig. 7F and G, treatment with sorafenib in combination with AF for 8 days could significantly reduce leukemia cells in the blood, whereas sorafenib or AF alone exhibited weak or no impact (Fig. 7F and G). These data together demonstrated that sorafenib + AF might be a novel drug combination that could be effective against sorafinib-resistant cancer in vivo.
4. Discussion
Using microarray analysis to compare the gene expression profiles in sorafenib-sensitive and sorafenib-resistant cells, we have identified the mitochondrial isoform of thioredoxin reductase-3 (mtTXNRD3) that was highly expressed in the drug resistant cells. Functional study further confirmed that the up-regulation of mtTXNRD3 was responsible for the drug-resistant phenotype. This conclusion is supported by several lines of evidences: (1) In a pair of leukemia cells (MV4-11 & MV4-11R), mtTXNRD3 was highly expression in the sorafenib-resistant MV4-11R cells; (2) Over-expression of mtTXNRD3 in SK-Hep1 cells led to a decrease of their sensitivity to sorafenib; (3) A TXNRD3KD expression resulted in an increase of sensitivity to sorafenib; (4) The thioredoxin reductase inhibitor auranofin was able to overcome the sorafenib-resistant phenotype in vitro and in vivo. Although it is clear that high expression of mtTXNRD3 causes drug resistance to sorafenib in cancer cells, a key question is how mtTXNRD3 could confer resistance to sorafenib?
Previous studies have suggested that the activity of Trx/TrxR system is associated with cancer cell growth and anti-apoptosis process, and that this system is up-regulated in several types of cancers, including malignant mesothelioma [28,29]. Kahlos et al. suggested that the up-regulation of thioredoxin and TrxR might contribute to drug resistance in malignant cells [30]. Higher TrxR activity was thought to enhance cellular antioxidant capacity to detoxify excessive ROS and might also promote therapeutic resistance [31], although there was no report suggesting a role of mitochondrial TrxR in this process. There are three TrxR isoenzymes. TXNRD1 and TXNRD2 are located outside and inside the mitochondria, respectively [32], while the localization of TXNRD3, which has two splicing variants, has not been previously well defined. Our study showed that the expression of TXNRD3, but not TXNRD1 or TXNRD2, was correlated with sorafenib-resistance. Importantly, we found that the 70.7 kD variant of TXNRD3 was located in the mitochondria and was up-regulated in sorafenib-resistant cells, whereas the 66.6 kD variant appeared in the cytosol and seemed to express at similar levels in sorafenib-sensitive and resistant cells. It is unclear at the present time if overexpression of mtTXNRD3 could also be found in cells resistant to other tyrosine kinase inhibitors or it is only sorafenib-specific. Based on our observations that leukemia cells and liver cancer cells with overexpression of mtTXNRD3 were significantly less sensitive to erlotinib and gefitinib (Figs. S6–S7), it is reasonable to speculate that mtTXNRD3 would likely be overexpressed in cancer cells resistant to other tyrosine kinase inhibitors. This possibility requires further study in the future.
A key question is how an over-expression of the 70.7 kD variant of TXNRD3 could render leukemia and liver cancer cells resistant to sorafenib? Since mtTXNRD3 is located in the mitochondria, which play a central role in energy metabolism as well as in regulation of the intrinsic apoptotic pathway, it is thus possible that overexpression of mtTXNRD3 might affect mitochondrial metabolism and alter mitochondrial-mediated apoptosis, thus reducing cellular sensitivity to sorafenib. Our data indeed showed that overexpression of mtTXNRD3 caused a significant decrease in mitochondrial respiration and an increase of glycolysis, as evident by a decrease in oxygen consumption rate and an increase in lactate production. The increased glycolytic activity was associated with increased expression of HK2, a key glycolytic enzyme that is known to suppress apoptosis and promote cellular resistance to chemotherapy [33]. Thus, enhancing the protein level of HK2 seems to be a possible mechanism contributing to mtTXNRD3-induced drug resistance.
Another possible mechanism by which mtTXNRD3 induces drug resistance is by increasing anti-apoptotic proteins including Bcl-2, Bcl-xL, and MCL-1. These proteins are members of the Bcl-2 family. They are associated with mitochondrial outer membranes, and play a major role in suppressing the intrinsic or mitochondria-initiated apoptosis. The cellular apoptotic response to drug treatment is in part determined by the Bcl-2 protein family [[34], [35], [36]]. In the National Cancer Institute (NCI) panel of 60 diverse cancer cell lines, the expression level of Bcl-xL is strongly correlated with drug resistance [37]. Thus, the ability of mtTXNRD3 to enhance the protein levels of Bcl-2, Bcl-xL, and MCL-1 likely contributes the sorafenib-resistant phenotype observed in leukemia and liver cancer cells.
One major issue is how the overexpression of mtTXNRD3 could lead to an increase in multiple proteins including HK2, BCL-2, Bcl-xL, and MCL-1? It is known that thioredoxins (Trx) are small (12-kDa) thiol-disulfide proteins with oxidoreductase activity critical for cellular redox regulation [38]. The oxidized form of Trx is reversibly reduced by thioredoxin reductase using NADPH as a reducing agent. Two isoforms of Trx, Trx1 and Trx2, are present in mammalian cells. Trx2 is found in mitochondria and has a protective function against mitochondrial oxidative stress [39,40]. Thus, a possible explanation is that the high level of mtTXNRD3 would be able to maintain the mitochondrial Trx2 in its reduced form, which plays a key role in repairing oxidized proteins and thus enhancing their stability. Our findings that overexpression of mtTrxT3 in SK-Hep1 cells kept Trx2 in the reduced stage associated with increased protein levels of Bcl-2, Bcl-xL, and MCL-1 are consistent this mechanism. The sorafenib-resistant cells with high expression of mtTXNRD3 also exhibited cross-resistance to other tyrosine kinase inhibitors (gefitinib and erlotinib) and certain chemotherapeutic drugs (doxorubicin and ara-C). These results were consistent with the observation that high expression of mtTXNRD3 led to stabilization of pro-survival proteins HK2 and Bcl-2 family members that promote cell survival and drug resistance.
Auranofin (AF) is a gold-containing drug used in the clinical treatment of rheumatoid arthritis. This compound has been previously reported to inhibit thioredoxin reductase and induce ROS production [28,[41], [42], [43]]. The other research also showed that AF activated caspase-3 and -8 in a concentration-dependent manner and could decrease the levels of mitochondrial anti-apoptotic factors including Bcl-2 and Bcl-xL [44]. Since we have found that elevated expression of mtTXNRD3 caused resistance to sorafenib associated with increased levels of Bcl-2 family members, thus it is logical to use AF to inhibit mtTXNRD3 and overcome drug resistance. Interestingly, a recent study by Lee et al. showed that liver cancer cells depend on antioxidants for maintaining redox homeostasis, and that inhibition of thioredoxin reductase by AF effectively suppressed the liver cancer in vivo [45].
In conclusion, our study showed that mitochondrial TXNRD3 was highly expressed in drug-resistant cancer cells and was associated with poor clinical outcome. The elevated mitochondrial TXNRD3 resulted in metabolic alterations and stabilization of important survival molecules such as Bcl-2 family members, leading to drug resistant. This study also demonstrated that it was possible to use auranofin to inhibit TrxR and thus overcome mtTXNRD3-induced drug resistance in vitro and in vivo. Since auranofin is a clinical drug for treatment of rheumatoid arthritis, it may be possible to reposition this drug for treatment of drug-resistant cancer due to mtTXNRD3 over-expression.
Author contributions
Conception and design: XX Liu and P Huang. Acquisition of data: XX Liu, Y Han, WH Lu, YY Zhang, WY Jiang and Y Luo. Analysis and interpretation of data: XX Liu, Y Han, X You, J Yang. Study supervision: SJ Wen, YM Hu and P Huang. Drafting of the manuscript: XX Liu and P Huang.
Data availability
All data supporting the findings of this study are available with the article, or from the corresponding author upon reasonable request.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The study was supported in part by grants from National Key R&D Program of China (2018YFC0910203), the National Natural Science Foundation of China (No. 81430060), and the Basic Research Funding from Sun Yat-sen University (No.19ykyjs71).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2020.101652.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Ning N., Yu Y., Wu M., Zhang R., Zhang T., Zhu C., Huang L., Yun C.H., Benes C.H., Zhang J., Deng X., Chen Q., Ren R. A novel microtubule inhibitor overcomes multidrug resistance in tumors. Cancer Res. 2018;78:5949–5957. doi: 10.1158/0008-5472.CAN-18-0455. [DOI] [PubMed] [Google Scholar]
- 2.Toyooka S., Kiura K., Mitsudomi T. EGFR mutation and response of lung cancer to gefitinib. N. Engl. J. Med. 2005;352:2136. doi: 10.1056/NEJM200505193522019. author reply 2136. [DOI] [PubMed] [Google Scholar]
- 3.Hata A.N., Niederst M.J., Archibald H.L., Gomez-Caraballo M., Siddiqui F.M., Mulvey H.E., Maruvka Y.E., Ji F., Bhang H.E. Tumor cells can follow distinct evolutionary paths to become resistant to epidermal growth factor receptor inhibition. Nat. Med. 2016;22:262–269. doi: 10.1038/nm.4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agarwal S., Zerillo C., Kolmakova J., Christensen J.G., Harris L.N., Rimm D.L., Digiovanna M.P., Stern D.F. Association of constitutively activated hepatocyte growth factor receptor (Met) with resistance to a dual EGFR/Her2 inhibitor in non-small-cell lung cancer cells. Br. J. Canc. 2009;100:941–949. doi: 10.1038/sj.bjc.6604937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Piloto O., Wright M., Brown P., Kim K.T., Levis M., Small D. Prolonged exposure to FLT3 inhibitors leads to resistance via activation of parallel signaling pathways. Blood. 2007;109:1643–1652. doi: 10.1182/blood-2006-05-023804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jun H.J., Acquaviva J., Chi D., Lessard J., Zhu H., Woolfenden S., Bronson R.T., Pfannl R., White F., Housman D.E., Iyer L., Whittaker C.A., Boskovitz A., Raval A., Charest A. Acquired MET expression confers resistance to EGFR inhibition in a mouse model of glioblastoma multiforme. Oncogene. 2012;31:3039–3050. doi: 10.1038/onc.2011.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gillis N.K., McLeod H.L. The pharmacogenomics of drug resistance to protein kinase inhibitors. Drug Resist. Updates : Rev. Comment. Antimicrob. Anticanc. Chemother. 2016;28:28–42. doi: 10.1016/j.drup.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korfi K., Mandal A., Furney S.J., Wiseman D., Somervaille T.C., Marais R. A personalised medicine approach for ponatinib-resistant chronic myeloid leukaemia. Ann. Oncol. : Off. J. Euro. Soc. Med. Oncol. 2015;26:1180–1187. doi: 10.1093/annonc/mdv110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Llovet J.M., Ricci S., Mazzaferro V., Hilgard P., Gane E., Blanc J.F., de Oliveira A.C., Santoro A., Raoul J.L., Forner A., Schwartz M., Porta C., Zeuzem S., Bolondi L., Greten T.F., Galle P.R., Seitz J.F., Borbath I., Haussinger D., Giannaris T., Shan M., Moscovici M., Voliotis D., Bruix J. Sorafenib in advanced hepatocellular carcinoma. N. Engl. J. Med. 2008;359:378–390. doi: 10.1056/NEJMoa0708857. [DOI] [PubMed] [Google Scholar]
- 10.Escudier B., Eisen T., Stadler W.M., Szczylik C., Oudard S., Siebels M., Negrier S., Chevreau C., Solska E., Desai A.A., Rolland F., Demkow T., Hutson T.E., Gore M., Freeman S., Schwartz B., Shan M., Simantov R., Bukowski R.M. Sorafenib in advanced clear-cell renal-cell carcinoma. N. Engl. J. Med. 2007;356:125–134. doi: 10.1056/NEJMoa060655. [DOI] [PubMed] [Google Scholar]
- 11.Man C.H., Fung T.K., Ho C., Han H.H., Chow H.C., Ma A.C., Choi W.W., Lok S., Cheung A.M., Eaves C., Kwong Y.L., Leung A.Y. Sorafenib treatment of FLT3-ITD(+) acute myeloid leukemia: favorable initial outcome and mechanisms of subsequent nonresponsiveness associated with the emergence of a D835 mutation. Blood. 2012;119:5133–5143. doi: 10.1182/blood-2011-06-363960. [DOI] [PubMed] [Google Scholar]
- 12.Won J.K., Yu S.J., Hwang C.Y., Cho S.H., Park S.M., Kim K., Choi W.M., Cho H., Cho E.J., Lee J.H., Lee K.B., Kim Y.J., Suh K.S., Jang J.J., Kim C.Y., Yoon J.H., Cho K.H. Protein disulfide isomerase inhibition synergistically enhances the efficacy of sorafenib for hepatocellular carcinoma. Hepatology. 2017;66:855–868. doi: 10.1002/hep.29237. [DOI] [PubMed] [Google Scholar]
- 13.Chen H.A., Kuo T.C., Tseng C.F., Ma J.T., Yang S.T., Yen C.J., Yang C.Y., Sung S.Y., Su J.L. Angiopoietin-like protein 1 antagonizes MET receptor activity to repress sorafenib resistance and cancer stemness in hepatocellular carcinoma. Hepatology. 2016;64:1637–1651. doi: 10.1002/hep.28773. [DOI] [PubMed] [Google Scholar]
- 14.Zhai B., Sun X.Y. Mechanisms of resistance to sorafenib and the corresponding strategies in hepatocellular carcinoma. World J. Hepatol. 2013;5:345–352. doi: 10.4254/wjh.v5.i7.345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu Y., Huang J., Ma L., Shan J., Shen J., Yang Z., Liu L., Luo Y., Yao C., Qian C. MicroRNA-122 confers sorafenib resistance to hepatocellular carcinoma cells by targeting IGF-1R to regulate RAS/RAF/ERK signaling pathways. Canc. Lett. 2016;371:171–181. doi: 10.1016/j.canlet.2015.11.034. [DOI] [PubMed] [Google Scholar]
- 16.Xu R.H., Pelicano H., Zhou Y., Carew J.S., Feng L., Bhalla K.N., Keating M.J., Huang P. Inhibition of glycolysis in cancer cells: a novel strategy to overcome drug resistance associated with mitochondrial respiratory defect and hypoxia. Canc. Res. 2005;65:613–621. [PubMed] [Google Scholar]
- 17.Zhou Y., Zhou Y., Shingu T., Feng L., Chen Z., Ogasawara M., Keating M.J., Kondo S., Huang P. Metabolic alterations in highly tumorigenic glioblastoma cells: preference for hypoxia and high dependency on glycolysis. J. Biol. Chem. 2011;286:32843–32853. doi: 10.1074/jbc.M111.260935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 19.Huang A., Ju H.Q., Liu K., Zhan G., Liu D., Wen S., Garcia-Manero G., Huang P., Hu Y. Metabolic alterations and drug sensitivity of tyrosine kinase inhibitor resistant leukemia cells with a FLT3/ITD mutation. Canc. Lett. 2016;377:149–157. doi: 10.1016/j.canlet.2016.04.040. [DOI] [PubMed] [Google Scholar]
- 20.Go Y.M., Jones D.P. 2009. Thioredoxin Redox Western Analysis, Current Protocols in Toxicology, Chapter 17. Unit17.12. [DOI] [PubMed] [Google Scholar]
- 21.Liu X., Wang W., Samarsky D., Liu L., Xu Q., Zhang W., Zhu G., Wu P., Zuo X., Deng H., Zhang J., Wu Z., Chen X., Zhao L., Qiu Z., Zhang Z., Zeng Q., Yang W., Zhang B., Ji A. Tumor-targeted in vivo gene silencing via systemic delivery of cRGD-conjugated siRNA. Nucleic Acids Res. 2014;42:11805–11817. doi: 10.1093/nar/gku831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Metzeler K.H., Hummel M., Bloomfield C.D., Spiekermann K., Braess J., Sauerland M.C., Heinecke A., Radmacher M., Marcucci G., Whitman S.P., Maharry K., Paschka P., Larson R.A., Berdel W.E., Buchner T., Wormann B., Mansmann U., Hiddemann W., Bohlander S.K., Buske C. An 86-probe-set gene-expression signature predicts survival in cytogenetically normal acute myeloid leukemia. Blood. 2008;112:4193–4201. doi: 10.1182/blood-2008-02-134411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Arner E.S. Focus on mammalian thioredoxin reductases--important selenoproteins with versatile functions. Biochim. Biophys. Acta. 2009;1790:495–526. doi: 10.1016/j.bbagen.2009.01.014. [DOI] [PubMed] [Google Scholar]
- 24.Menyhárt O., Nagy A., Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. R. Soc. Open Sci. 2018;5:181006. doi: 10.1098/rsos.181006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trachootham D., Zhang H., Zhang W., Feng L., Du M., Zhou Y., Chen Z., Pelicano H., Plunkett W., Wierda W.G., Keating M.J., Huang P. Effective elimination of fludarabine-resistant CLL cells by PEITC through a redox-mediated mechanism. Blood. 2008;112:1912–1922. doi: 10.1182/blood-2008-04-149815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng X., Holenya P., Can S., Alborzinia H., Rubbiani R., Ott I., Wolfl S. A TrxR inhibiting gold(I) NHC complex induces apoptosis through ASK1-p38-MAPK signaling in pancreatic cancer cells. Mol. Canc. 2014;13:221. doi: 10.1186/1476-4598-13-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rackham O., Shearwood A.M., Thyer R., McNamara E., Davies S.M., Callus B.A., Miranda-Vizuete A., Berners-Price S.J., Cheng Q., Arnér E.S., Filipovska A. Substrate and inhibitor specificities differ between human cytosolic and mitochondrial thioredoxin reductases: implications for development of specific inhibitors. Free Radi. Biol. Med. 2011;50:689–699. doi: 10.1016/j.freeradbiomed.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 28.Zhao F., Yan J., Deng S., Lan L., He F., Kuang B., Zeng H. A thioredoxin reductase inhibitor induces growth inhibition and apoptosis in five cultured human carcinoma cell lines. Canc. Lett. 2006;236:46–53. doi: 10.1016/j.canlet.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 29.Nilsonne G., Sun X., Nystrom C., Rundlof A.K., Potamitou Fernandes A., Bjornstedt M., Dobra K. Selenite induces apoptosis in sarcomatoid malignant mesothelioma cells through oxidative stress. Free Radi. Biol. Med. 2006;41:874–885. doi: 10.1016/j.freeradbiomed.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 30.Kahlos K., Soini Y., Saily M., Koistinen P., Kakko S., Paakko P., Holmgren A., Kinnula V.L. Up-regulation of thioredoxin and thioredoxin reductase in human malignant pleural mesothelioma. Int. J. Canc. 2001;95:198–204. doi: 10.1002/1097-0215(20010520)95:3<198::aid-ijc1034>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 31.Maiti A.K. Genetic determinants of oxidative stress-mediated sensitization of drug-resistant cancer cells. Int. J. Canc. 2012;130:1–9. doi: 10.1002/ijc.26306. [DOI] [PubMed] [Google Scholar]
- 32.Mustacich D., Powis G. Thioredoxin reductase. Biochem. J. 2000;346(Pt 1):1–8. [PMC free article] [PubMed] [Google Scholar]
- 33.Galluzzi L., Kepp O., Vander Heiden M.G., Kroemer G. Metabolic targets for cancer therapy, Nature reviews. Drug Discov. 2013;12:829–846. doi: 10.1038/nrd4145. [DOI] [PubMed] [Google Scholar]
- 34.Adams J.M., Cory S. The Bcl-2 apoptotic switch in cancer development and therapy. Oncogene. 2007;26:1324–1337. doi: 10.1038/sj.onc.1210220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozopas K.M., Yang T., Buchan H.L., Zhou P., Craig R.W. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc. Natl. Acad. Sci. U.S.A. 1993;90:3516–3520. doi: 10.1073/pnas.90.8.3516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng E.H., Wei M.C., Weiler S., Flavell R.A., Mak T.W., Lindsten T., Korsmeyer S.J. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol. Cell. 2001;8:705–711. doi: 10.1016/s1097-2765(01)00320-3. [DOI] [PubMed] [Google Scholar]
- 37.Amundson S.A., Myers T.G., Scudiero D., Kitada S., Reed J.C., Fornace A.J., Jr. An informatics approach identifying markers of chemosensitivity in human cancer cell lines. Canc. Res. 2000;60:6101–6110. [PubMed] [Google Scholar]
- 38.Chen Y., Cai J., Jones D.P. Mitochondrial thioredoxin in regulation of oxidant-induced cell death. FEBS Lett. 2006;580:6596–6602. doi: 10.1016/j.febslet.2006.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen Y., Cai J., Murphy T.J., Jones D.P. Overexpressed human mitochondrial thioredoxin confers resistance to oxidant-induced apoptosis in human osteosarcoma cells. J. Biol. Chem. 2002;277:33242–33248. doi: 10.1074/jbc.M202026200. [DOI] [PubMed] [Google Scholar]
- 40.Hansen J.M., Zhang H., Jones D.P. Mitochondrial thioredoxin-2 has a key role in determining tumor necrosis factor-alpha-induced reactive oxygen species generation, NF-kappaB activation, and apoptosis. Toxicol. Sci. : Off. J. Soc. Toxicol. 2006;91:643–650. doi: 10.1093/toxsci/kfj175. [DOI] [PubMed] [Google Scholar]
- 41.Huang H., Liao Y., Liu N., Hua X., Cai J., Yang C., Long H., Zhao C., Chen X., Lan X., Zang D., Wu J., Li X., Shi X., Wang X., Liu J. Two clinical drugs deubiquitinase inhibitor auranofin and aldehyde dehydrogenase inhibitor disulfiram trigger synergistic anti-tumor effects in vitro and in vivo. Oncotarget. 2016;7:2796–2808. doi: 10.18632/oncotarget.6425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gandin V., Fernandes A.P., Rigobello M.P., Dani B., Sorrentino F., Tisato F., Bjornstedt M., Bindoli A., Sturaro A., Rella R., Marzano C. Cancer cell death induced by phosphine gold(I) compounds targeting thioredoxin reductase. Biochem. Pharmacol. 2010;79:90–101. doi: 10.1016/j.bcp.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 43.Sze J.H., Raninga P.V., Nakamura K., Casey M., Khanna K.K., Berners-Price S.J., Di Trapani G., Tonissen K.F. Anticancer activity of a Gold(I) phosphine thioredoxin reductase inhibitor in multiple myeloma. Redox Biol. 2020;28:101310. doi: 10.1016/j.redox.2019.101310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Park N., Chun Y.J. Auranofin promotes mitochondrial apoptosis by inducing annexin A5 expression and translocation in human prostate cancer cells. J. Toxicol. Environ. Health Part A. 2014;77:1467–1476. doi: 10.1080/15287394.2014.955834. [DOI] [PubMed] [Google Scholar]
- 45.Lee D., Xu I.M., Chiu D.K., Leibold J., Tse A.P., Bao M.H., Yuen V.W., Chan C.Y., Lai R.K., Chin D.W., Chan D.F., Cheung T.T., Chok S.H., Wong C.M. Induction of oxidative stress through inhibition of thioredoxin reductase 1 is an effective therapeutic approach for hepatocellular carcinoma. Hepatology. 2019;69:1768–1786. doi: 10.1002/hep.30467. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available with the article, or from the corresponding author upon reasonable request.