Abstract
Background
Human cytomegalovirus (HCMV) causes asymptomatic infections, but also causes congenital infections when women were infected with HCMV during pregnancy, and life-threatening diseases in immunocompromised patients. To better understand the mechanism of the neutralization activity against HCMV, the association of HCMV NT antibody titers was assessed with the antibody titers against each glycoprotein complex (gc) of HCMV.
Methods
Sera collected from 78 healthy adult volunteers were used. HCMV Merlin strain and HCMV clinical isolate strain 1612 were used in the NT assay with the plaque reduction assay, in which both the MRC-5 fibroblasts cells and the RPE-1 epithelial cells were used. Glycoprotein complex of gB, gH/gL complexes (gH/gL/gO and gH/gL/UL128–131A [PC]) and gM/gN were selected as target glycoproteins. 293FT cells expressed with gB, gM/gN, gH/gL/gO, or PC, were prepared and used for the measurement of the antibody titers against each gc in an indirect immunofluorescence assay (IIFA). The correlation between the IIFA titers to each gc and the HCMV-NT titers was evaluated.
Results
There were no significant correlations between gB-specific IIFA titers and the HCMV-NT titers in epithelial cells or between gM/gN complex-specific IIFA titers and the HCMV-NT titers. On the other hand, there was a statistically significant positive correlation between the IIFA titers to gH/gL complexes and HCMV-NT titers.
Conclusions
The data suggest that the gH/gL complexes might be the major target to induce NT activity against HCMV.
Keywords: Human cytomegalovirus, HCMV, Glycoprotein complex, Neutralization, Antibody
Introduction
Human cytomegalovirus (HCMV) is a member of the betaherpesvirinae sub-family of Herpesviridae, and is a cause of congenital transplacental infection of the fetus. Children with congenital HCMV infection have significant morbidity and mortality with symptoms that include permanent neurological defects, such as sensorineural deafness, developmental delay, dysopia, and epilepsy. HCMV also causes severe and sometimes lethal diseases in immunocompromised patients [1, 2].
HCMV glycoprotein (gP) complexes (gcs) expressed in the virion envelope function in the process of cell attachment and entry, and they are predominant targets of the virus-neutralization (NT) antibodies of HCMV. Among 23 gPs that are encoded in the HCMV genome, 9 gPs form 4 gcs: gB, gM/gN, gH/gL/gO and pentamer complex; gH/gL/pUL128/pUL130/pUL131A (PC), and all of these gcs are reported to have the potential to induce HCMV-NT antibodies [3–6].
Before the early 2000s, gB had been considered the main antigen in the development of the HCMV vaccine [7]. At that time, HCMV-NT activities were measured using fibroblast cells and the highly passaged laboratory HCMV strain, which lacked the expression of PC [8–11]. PC has recently been shown to be important for efficient entry of HCMV to epithelial cells (ECs), endothelial cells, and monocytes [12, 13], while the gH/gL/gO is required for the infection of both fibroblast cells and ECs [14]. Furthermore, it was reported that PC-specific antibodies might be a major component of HCMV-NT antibodies [6, 15–18].
For the development of an HCMV vaccine, the strong induction of NT activity against HCMV is desired. HCMV has many intricate evasion strategies against humoral immunity; these impede the development of an HCMV vaccine [19]. On the other hand, it is believed that potent HCMV-NT activity is required to protect a fetus from transplacental HCMV infection [20, 21]. Therefore, understanding the mechanism of HCMV NT antibody induction is necessary. We aimed to identify the antigen that actually induced the highest neutralizing activity by assessment for the correlation of anti-HCMV-NT antibody titers of healthy adult sera with the antibody titers against each of the following 4 gc antigens, gB, gM/gN, gH/gL/gO and PC.
Materials and methods
Cells
Fibroblast MRC-5 cells (American Type Culture Collection [ATCC® CCL-171]) were used. The MRC-5 cells were grown in Eagle’s minimum essential medium (MEM) supplemented with 10% heat-inactivated fetal bovine serum (Gibco, Carlsbad, CA), L-glutamine and sodium bicarbonate and 1% penicillin-streptomycin (MEM-10FBS). The retinal pigment epithelial cell line ARPE-19 (ATCC® CRL-2302™) and hTERT (human telomerase reverse transcriptase)-immortalized RPE-1 (ATCC® CRL-4000™), both of which were epithelial cell lines, were cultured in Dulbecco’s modified Eagle medium (DMEM)/F-12 (1:1), including L-glutamine and 2.438 g/L sodium bicarbonate (Gibco) supplemented with 10% heat-inactivated fetal bovine serum (Gibco or Hyclone, GE Healthcare UK Ltd., UK) (DMEM-10FBS). The culture medium for RPE-1 was additionally supplemented with hygromycin B at a concentration of 0.01 mg/mL. 293FT cells (Thermo Fisher Scientific, Waltham, MA) were grown in DMEM (Wako, Odawara, JAPAN) supplemented with 5% fetal bovine serum (Gibco) (DMEM-5FBS). The 293FT cells were cultured in collagen-coated plates (TOYOBO, Osaka, JAPAN).
Viruses
HCMV strain Merlin (ME, ATCC® VR-1590™) was used as the source for constructing plasmids and for NT antibody analyses in MRC-5 cells. For the measurement of NT titers in ECs, HCMV clinical strain 1612, which was isolated in our laboratory from the urine of a 2-month-old baby with symptomatic HCMV disease was used. The HCMV 1612 strain was propagated in MRC-5 cells with three passages followed by five passages in ARPE-19 cells. In ARPE-19 cells infected with the 1612 strain, clear CPE was confirmed at 1 week after passage. The ME strain was mass-cultured following two passages in MRC-5 cells. The ME strain- or 1612 strain-infected cells were harvested at full CPE and suspended in FBS-free medium before being stored at − 80 °C. The stored cells were repeated freezing and thawing twice, and then the infectious dose in the supernatant was determined by counting CPE using MRC-5 cells. The titers of ME strain and 1612 strain measured using MRC-5 cells were 3.6 × 104 plaque forming unit (PFU)/mL and 3.5 × 105 PFU/mL, respectively. HCMV 1612 strain was used to measure the NT antibody titers in ECs, because the strain possessed the capacity to infect ECs.
Serum sampling (subject selection) and ethical considerations
Seventy-eight healthy volunteers were recruited. The age of the volunteers ranged from 20 to 60 years (Table 1). All serum samples were first tested for HCMV IgG antibody positivity against total HCMV proteins with a commercially available enzyme-linked immunosorbent assay (ELISA) kit (DENKA SEIKEN, Tokyo, Japan), and the HCMV-IgG ELISA titers were determined according to the manufacturer’s instructions. The HCMV-IgG-positive sera were then further tested for HCMV-antibody titers by an indirect immunofluorescence assay (IIFA) and an HCMV-NT assay, as described below. The serum samples were heat-inactivated at 56 °C for 30 min before testing antibody titers with any assays.
Table 1.
The HCMV-IgG antibody positive rate, as determined by HCMV-IgG ELISA
| Age | HCMV-IgG positivity [No. of positive/No. of tested (%)] | ||
|---|---|---|---|
| Female | Male | Total | |
| 20–29 | 1/3 (33.3%) | 2/4 (50%) | 3/7 (42.9%) |
| 30–39 | 10/15 (66.7%) | 5/14 (33.3%) | 15/29 (51.7%) |
| 40–49 | 8/11 (72.7%) | 11/15 (73.3%) | 19/26 (73%) |
| 50–59 | 3/6 (50%) | 7/9 (77.8%) | 10/15 (66.7%) |
| 60–69 | 1/1 (100%) | 0/0 | 1/1 (100%) |
| Total | 23/42 (54.7%) | 25/36 (69.4%) | 48/78 (61.5%) |
HCMV neutralization assay
HCMV-NT titers of each serum were assessed with a conventional plaque reduction assay. In brief, serum samples were serially diluted with maintenance medium, MEM-2FBS or DMEM-2FBS. Sixty μL of the diluted serum sample and an equal volume of virus solution containing 60 plaque forming units (PFUs) of HCMV were mixed and incubated in a U-bottomed 96-well plate (Greiner Bio-One JAPAN, Tokyo, Japan) at 37 °C for 1 h. One hundred microliters of the mixture was then added to monolayers of ECs (RPE-1) or fibroblast (MRC-5) cells. The cell sheets were prepared by seeding cells to 15,000 cells per well in 96-well plates (CORNING, Corning, NY) on a day before performing the neutralization process. HCMV ME was used for the HCMV-NT assay in MRC-5 cells, while HCMV 1612 was used for the HCMV-NT assays in RPE-1 cells and MRC-5 cells. Fixation, staining with crystal violet, formalin and methanol treatment, and washing of the HCMV ME-MRC-5 plates were carried out at 4 days post inoculation, while the processing and measuring of NT antibody titers against HCMV 1612 in RPE-1 cells and MRC-5 cells were fixed at 2 days post inoculation. After staining, washing, and drying, CPE consisting of agglomerated cells were observed by stereomicroscopy. The 50% virus-NT titers (NT50) were defined as the reciprocal of the highest dilution level, at which the plaque number became less than half of the control. Each test was run in triplicate.
HCMV glycoprotein genome amplification with polymerase chain reaction for plasmid construction
Each open reading frame (ORF) of the gP of HCMV ME was amplified by polymerase chain reaction (PCR) using primer sets that were designed with reference to the ME sequence (GenBank Accession no; AY446894.2.). All primers and oligonucleotides were purchased from Eurofins Genomics (Tokyo, Japan). The 30-μL reaction was composed of 15 μL Q5 High-Fidelity 2X Master Mix (New England Biolabs, Ipswich, MA), 0.5 μM of each primer, and template DNA. PCR amplicon bands were isolated from 10% agarose electrophoresis gel and were purified using the FastGene Gel/PCR Extraction Kit (NIPPON Genetics, Tokyo, JAPAN). Purified DNA was quantitated using a NanoDrop 2000c Spectrophotometer (Thermo Fisher Scientific).
Plasmid construction
First, the ORF of each gP of HCMV ME was amplified from cDNA, which was derived from RNA purified from the HCMV-ME-infecting MRC-5 cells by two step conventional reverse transcription. The synthetic DNA oligonucleotide of ME-UL128 wt (G > A in UL128 was fixed) was purchased from Integrated DNA Technologies (Coralville, IA). Each of the ORF genes was cloned into the cloning site of the modified pHEK293 ULTRA Expression vector II (Takara Bio Inc., Shiga, Japan), which was used for the recombinant protein expression in mammalian cells as a form of fusion protein with a designated tag at the carboxy-terminal (Fig. 1). The insertion of the gene of interest was carried out using Fusion™ HD (Takara Bio Inc) according to the manufacturer’s instructions.
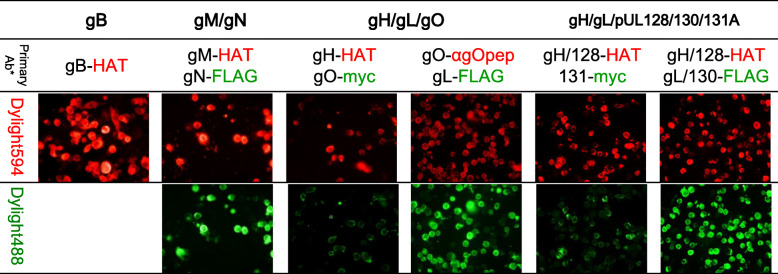
Fig. 1.
Confirmation of the expression of each membrane glycoprotein complex in 293FT cells transfected with each expression vector complex with an indirect immunofluorescence assay (IIFA) using antibodies to the tags fused with each membrane glycoprotein complex. The 293FT cells transfected with each designated plasmid or the combination of the plasmids were washed with phosphate buffered saline (PBS) (−), spotted on glass slides, and fixed and permeabilized with a methanol and acetone mixture. Glycoproteins that were expressed on each IIFA plate were described on the first line. Tag antibodies used as the primary antibodies are rabbit anti- HAT-IgG, mouse anti-FLAG, mouse myc-IgG, and rabbit anti-gO peptide antibody. The expression of Tag was distinguished by a secondary antibody (anti-rabbit IgG Dylight594 or anti-mouse IgG Dylight488). The tags fused with each gc were HAT (−gB, −gH, −gM, −pUL128), FLAG (−gL, −gN, −pUL130) and c-myc (−gO, −pUL131A)
Sanger DNA sequencing
The nucleotide sequence was determined using an ABI Prism 3130 Avant Genetic Analyzer (Applied Biosystems, Foster City, CA). The sequences were aligned to the reference using DNA Dynamo (Blue-Tractor Software, North Wales, UK). The nucleotide sequence of the constructed plasmids was confirmed to be the original sequence by Sanger DNA sequencing.
The sequence of the UL128L (UL128–131A locus) in strain 1612 was determined by Sanger sequencing, in which the PCR product was amplified using primers UL128L-F (GCGTATTTCGGACAAACACACA) and UL128L-R (CGCATGTTGCAGACTGAGAAAGA) [22]. It was confirmed that there were no mutations in the UL128L gene of the HCMV 1612.
Antigen preparations for the indirect immunofluorescence assay
293FT cells were transfected with pHEK293-gB for the expression of recombinant gB. The same cells were also co-transfected with pHEK-gM and pHEK-gN, with pHEK-gH, pHEK-gL, and pHEK-gO, and with pHEK-gH, pHEK-gL, pUL128, pUL130, and pUL131A for the expression of gM/gN, gH/gL/gO, and PC, respectively, using pHEK293 Enhancer Vector (Takara Bio Inc.) and HuGENE HD (Promega).
The 293FT cells transfected with each designated plasmid or the combination of the plasmids were washed with phosphate buffered saline (PBS) (−), spotted on glass slides (Matsunami Glass IND., Ltd., Osaka, Japan), and fixed with a methanol and acetone mixture mixed at a ratio of 1:1.
The expression of gc was confirmed by IIFA by detecting the respective tags. The antibodies, which were used for the detection of histidine affinity tag (HAT)-fusion protein, c-myc-fusion protein, and FLAG-tag fusion protein, were rabbit anti-HAT-tag polyclonal antibody (GenScript, Piscataway, NJ), mouse MYC-monoclonal antibody (Aviva Systems Biology, San Diego, CA), and anti-FLAG M2 monoclonal antibody (Sigma-Aldrich Japan, Tokyo, Japan), respectively. The secondary antibodies were Alexa Fluor DyLight 488-conjugated goat anti-mouse IgG H + L antibody or DyLight 594-conjugated goat anti-rabbit IgG H + L antibody (Invitrogen). Anti-gO peptide rabbit antibodies (peptide sequence: KLKRKQALVKEQPQKKNKKS [23]) were produced by Eurofins Genomics (Tokyo, Japan).
Detection of antibodies to each gc in indirect immunofluorescence assay
To measure each gc-specific IIFA titer in sera, the serum samples were two-fold serially diluted with PBS and added onto the glass slides. After incubation at 37 °C for 1 h, the cells were washed with PBS 3 times, and were then reacted with fluorescein isothiocyanate (FITC)-conjugated goat anti-human IgG H + L (Invitrogen, Carlsbad, CA). The antibody titer was defined as the reciprocal of the highest dilution level, at which the specific fluorescent signal was detected. The cells transfected with plasmid vectors without each gp insert were used as negative control. Two samples of CMV-IgG-negative sera were tested negative for antibody detection in IIFA. Since there was a concern about subjectivity with regard to the measurement of specific antibody titers in IIFA, the FITC-specific signal was observed by two experts to ensure consistency. To avoid detecting artifacts as much as possible, the signals in the fixed cells were observed at a lower magnification first, and they were again observed at a higher magnification. Clearly positive signals, which were different from those of the negative control, were determined positive.
Statistical analysis
Statistical analyses were performed using the Stata15 software program (STATA Corporation, College Station, TX). Non-parametric analyses of the correlations were performed using Spearman’s test. P values of < 0.05 were considered to indicate statistical significance.
Next generation sequencing
Amino-acid sequence homology of HCMV ME and HCMV 1612 was confirmed following base sequence determination using a next generation sequencer (NGS). Genomic DNA of HCMV 1612 was extracted from ARPE-19 cells infected with HCMV 1612 using a QIAmp DNA Mini Kit (QIAGEN, Hilden, Germany) after repeated freeze-thaw cycle treatment 2 times. The sequencing libraries were then prepared by an Ion Xpress Plus Fragment Library Kit (Thermo Fisher Scientific) in accordance with the manufacturer’s instructions. The library concentration was quantified using the Ion Library TaqMan Quantification Kit (Thermo Fisher Scientific). An emulsion PCR was performed on the library, which was adjusted to a 50-pM concentration, pooled in equimolar amounts, and mixed with capture beads on the Ion Chef System (Thermo Fisher Scientific) supplemented with the Ion Torrent Personal Genome Machine (PGM) template 200 kit (Thermo Fisher Scientific). The template libraries were sequenced with the Ion Torrent PGM using the Ion 314 Chip Kit v2 (Thermo Fisher Scientific) and the Ion Torrent PGM Sequencing 200 Kit v2 (Thermo Fisher Scientific), according to the manufacturer’s instructions. The resulting FASTQ format files were imported into CLC Genomics Workbench 9.0.1 (QIAGEN) for a homology analysis. Each gP sequence of 1612 strains were registered in GenBank with the accession numbers LC425070-LC425078.
Results
Confirmation of the gc expression for the detection of antibodies to IIFA antigen
The expression of all 4 types of gc was confirmed by IIFA through the detection of each of the fused tags (Fig. 1). The tag fused with gB, gH, gM, and pUL128 was HAT, while that with gL, gN, pUL130 was FLAG (−) and that with gO and pUL131A was c-myc. The tags assigned to the gp comprising each complex were differentiated by using two different fluorescent labels of the secondary antibodies. It was confirmed that the expression of each gp except for gO with Western blotting (data not shown). The expression of gO, which was fused with several different tags at difference position (C-terminus or N-terminus) was examined with WB and IIFA using anti-gO peptide antibodies or the anti-designated tag antibodies repeatedly, but no signals for gO expression were detected.
HCMV IgG prevalence among volunteers
The HCMV ELISA-IgG prevalence rate determined in the ELISA is shown in Table 1. The overall positive rate was 61.5%. After whole population was divided into two groups, one under 39 years of age and the other over 40 years of age. The prevalence of HCMV-IgG antibody positive rate among those aged equal to or over 40 was significantly higher among those aged less than 40 (p = 0.02, Student’s t-test). There was no significant difference in the prevalence of IgG between men and women.
Correlation between the HCMV ELISA-IgG titers and the HCMV-NT antibody titers
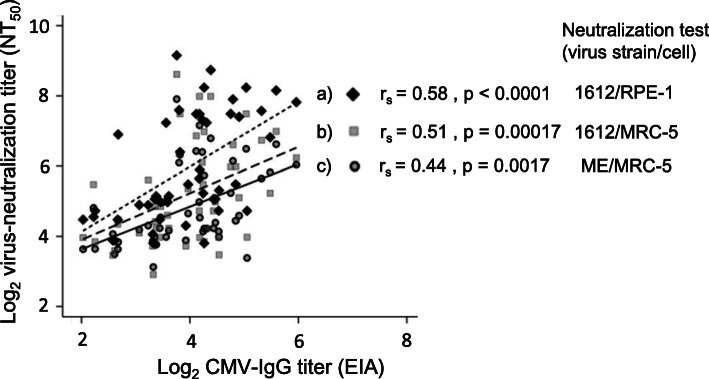
Correlation between the ELISA HCMV-IgG titers and the NT50 titers was assessed using Spearman’s test. The correlation coefficient “rs” were 0.58, 0.51 and 0.44 in NT assay using HCMV 1612/RPE-1 cells, HCMV 1612/MRC-5 cells, and HCMV ME/MRC-5 cells combinations, respectively. Thus, there was a relatively moderate statistically significant correlation between the HCMV ELISA-IgG titers and the HCMV-NT titers, regardless of the combinations between cells and virus types (Fig. 2). All P-value were less than 0.01.
Fig. 2.
Correlation between the quantitative ELISA HCMV-IgG titers and the HCMV-NT titers (NT50). The correlations were analyzed using Spearman’s test. The strength of the correlation was expressed using the correlation coefficient “rs”. P values of < 0.05 were considered to indicate statistical significance. The rs and p values were 0.58 (a; closed diamond), 0.51 (b; square) and 0.44 (c; closed circle) for HCMV 1612/cell RPE-1 (a), HCMV 1612/cell MRC-5 (b), and HCMV ME/cell MRC-5 (c) combinations, respectively
Relevance of using HCMV strain 1612
The amino acid sequence homology of gB, gH, gL, gO, gM, gN, pUL130, and pUL131A between HCMV1612 and HCMV ME was 99.4, 99.2, 98.9, 99.6, 100, 99.3, 98.6, and 100%, respectively. The amino acid sequence homology of pUL128 between strains 1612 and ME wild-type (Merlin BAC in which UL128 is repaired [21, 24]) was 99.4%. In addition to the amino acid homology, we also evaluated the phenotypic homology of strain ME with strain 1612 (i.e., the NT antibody titers to each strain determined in MRC-5 cells were compared). There was a positive correlation between the NT50 determined in fibroblasts against HCMV ME and those against HCMV 1612 (Fig. 3), indicating the validity of using the clinical isolate HCMV 1612.
Fig. 3.
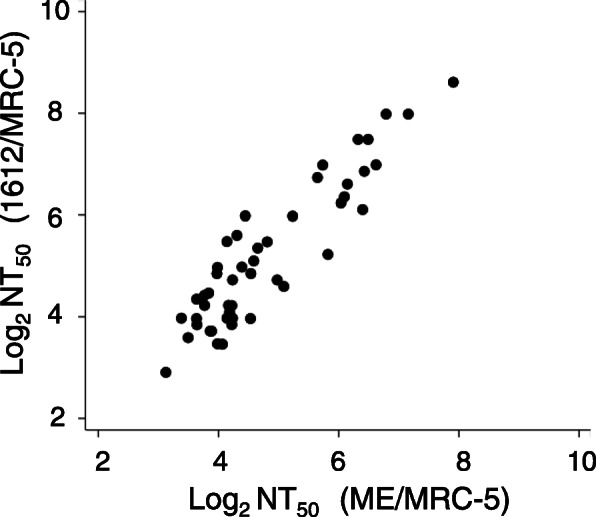
The correlation between the NT antibody titers of 78 participants determined using HCMV ME in MRC-5 fibroblast cells and those determined using HCMV 1612 in MRC-5 fibroblast cells. A strong correlation was observed with an rs value of 0.85 (p < 0.05)
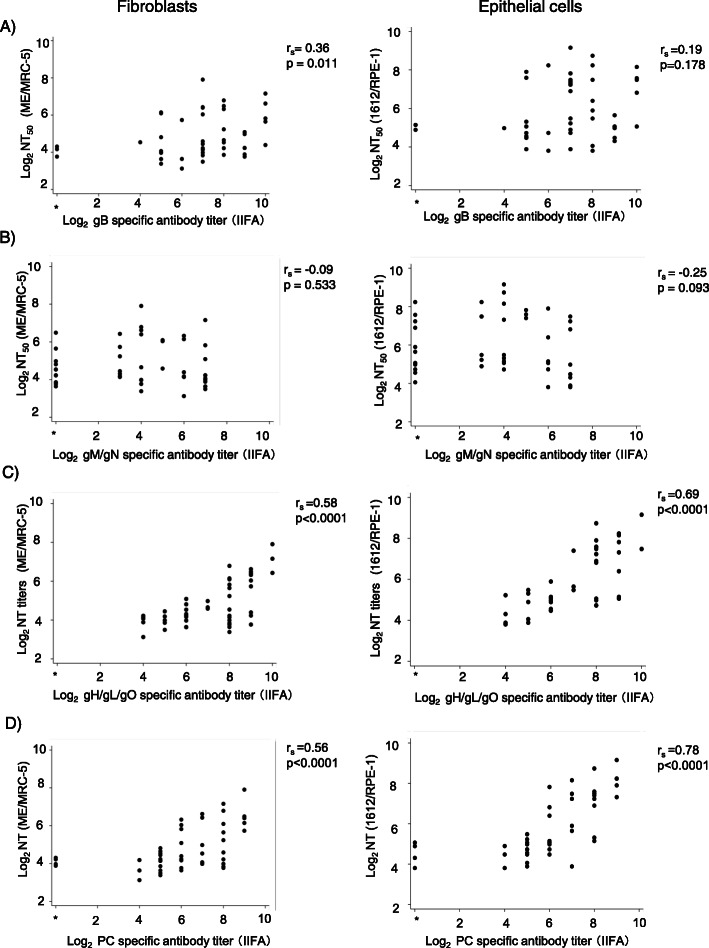
Correlation between NT50 and gc specific antibody titers determined with IIFA
The correlation between the HCMV-NT50 and each gc-specific IIFA antibody titers is shown in Fig. 4. gB-specific IIFA antibody titers showed a statistically significant positive correlation with the HCMV-NT50 titers to both HCMV ME and 1612 determined in fibroblast cells (Spearman R = 0.36, p = 0.011), but not with the HCMV-NT50 to 1612 strain determined in ECs (R = 0.19, p = 0.17). Anti-gM/gN IIFA antibody titers did not show any correlation with the HCMV-NT50 in fibroblasts or ECs (rs = − 0.09 and p = 0.533 in fibroblast cells; rs = − 0.25 and p = 0.092 in ECs) (Fig. 4b). In contrast, gH/gL complex-specific IIFA titers had a statistically significant correlation with HCMV-NT50 titers (Fig. 4c and d), as a correlation analysis, gave Spearman’s R values of 0.58, 0.69, 0.56, and 0.78 for gHLO/NT (Fibroblasts), gHLO/NT (ECs), PC/NT (Fibroblasts), and PC/NT (ECs), respectively; all p values were < 0.05.The highest correlation was demonstrated between the HCMV-NT50s determined in ECs and PC-specific IIFA titers (R = 0.78, p < 0.0001, Fig. 4d, right panel). Both gH/gL/gO-specific IIFA antibody titers and PC-specific IIFA titers showed a higher correlation with the HCMV-NT50s determined in ECs in comparison to those determined in fibroblasts. There was also a statistically significant positive correlation between the gH/gL/gO-specific IIFA titers and the PC-specific IIFA titers (R = 0.5967, p < 0.001).
Fig. 4.
The correlation of each anti-gc specific IIFA titer with HCMV-NT50 titers. The HCMV strain and cell types used in the neutralization test were HCMV ME/MRC-5 fibroblasts (left panels of a, b, c, and d), and HCMV 1612/RPE-1 epithelial cells (right panels of a, b, c, and d). Each correlation coefficient rs and p-value is indicated in the figure. “Negative” in IIFA means a result of < 8 (1 × 23). All samples were HCMV-NT50-positive (> 8) and showed a positive CMV-IgG titer (EIA). *The correlation coefficient was calculated regarding IIFA-negative as 1, that is indicated as 1 × 20
Discussion
The HCMV IgG seroprevalence rate of subjects (approximately 60%) in the present study is compatible with previously reported data in Japan [25, 26]. The HCMV-NT50 titers showed a weak to moderate correlation with EIA IgG-antibody titers in either virus–cell combination (Fig. 2), as reported previously [27–29]. When pregnant women are infected with HCMV as the primary infection, the IgG antibody, as determined by the EIA, become positive. The levels are parallel to the NT-antibody titers, and the antibody level peaked at approximately 3 months after infection, and are maintained for approximately 1 year [18]. The present study demonstrated a moderate correlation between NT-antibody titers and EIA-HCMV IgG titers.
This study confirmed the expression of each gP, with the exception of gO, by Western blotting (data not shown). The single expression of gO was not observed by any methods (Fig. 2). The gH/gL complex is stabilized as a complex form by binding with gO or UL128, UL130 and UL131A; thus, tags and peptide antigens were considered to be exposed after conformational changing of the expressed protein [30, 31].
In the last 20 years, it has been recognized that gB might be the main target with which NT antibodies react [7]. Originally, the development of gB-based vaccines has been driven by the observation that adsorption treatment of human sera with using recombinant gB diminishes HCMV entry to fibroblast cells through neutralizing activity by approximately 50% [4, 32–34]. It was also reported that gB-specific antibody titers were correlated with NT50 titers [35]. This evidence was obtained from experiments in which the serum NT activity was explored using fibroblasts (mainly, human embryo lung fibroblast cells) and highly-passaged laboratory HCMV strains, such as the HCMV Town strain. As HCMV-NT50 titers differ greatly depending on the type of cells and HCMV strains used, we measured the HCMV-NT50 titers using 3 types of HCMV-cells combination as described above. It was revealed that there was a positive correlation between gB-specific IIFA titers and the HCMV-NT50 titers when the HCMV-NT50 was determined in MRC-5 fibroblast cells but not in RPE-1 ECs (Fig. 4a), suggesting that the correlation is dependent on the virus strain and cells used for measuring HCMV-NT antibodies. In addition, it was revealed that antigenic target sites, with which NT antibodies react, were more heavily glycosylated than those that elicit non-NT antibodies, suggesting that HCMV-gB shields NT epitopes by taking advantage of glycosylation [36]. It has also been shown that the partial effect of the gB subunit vaccine is independent on NT antibody induction [37, 38], and that the vast majority (> 90%) of gB-specific antibodies secreted from B-cell clones do not have NT activity against HCMV [39]. Furthermore, adsorption studies with recombinant gB in sera from naturally infected humans suggest that anti-gB antibodies do not contribute significantly to HCMV EC entry through neutralizing responses [15, 17]. These reports support the results obtained in the present study that anti-gB specific IIFA titers in sera showed little correlation with the NT antibody titers determined in the HCMV 1612/RPE-1 ECs combination. However, it should be note that neutralizing assays were conducted in the absence of complement. Given that at least two neutralizing epitopes in gB have been shown to be fully dependent on complement, that neutralizing activities induced by gB-subunit vaccines often exhibit profound complement dependence, and that fibroblast neutralizing activity of human convalescent sera was 2 to 4 fold enhanced by the presence of complement [40, 41]. It would be important to note that the contribution made by gB-specific antibodies to fibroblast neutralizing activities may have been under-represented because endogenous complement in the sera was heat inactivated and exogenous complement was not added after heat inactivation. Consequently, the correlation between gB antibodies and neutralizing activities might be stronger if neutralizing assays were conducted in the presence of complement. It was reported that the structural and antigenic analyses showed that the postfusion gB trimer presented complement-independent neutralizing sites [42]. Thus, modifications on the gB trimer surface that reduce exposure to non-neutralizing sites, such as avoidance of glycosylation, may result in an improvement of HCMV vaccine development.
Although the function of gM/gN is largely unknown, there are a few previous reports that indicate gM/gN can elicit a virus-neutralizing antibody response [5, 43]. However, no correlation was found between anti-gM/gN IIFA antibody titers and NT50 titers (Fig. 4b). It was reported that immune evasion from NT antibodies became possible by the glycosylation of gN [44]. The result that the IIFA gM/gN-IgG titers were not significantly correlated with the HCMV-NT50 titers in this study might be due to the glycosylation of gN. Because the gM/gN is the glycoprotein complex that is the most abundantly expressed on viral particles [45], glycosylation of gM/gN could reduce the NT effect by NT antibodies, as in the case of gB [36]. Further studies are needed to reveal the mechanism of host immune evasion by gM/gN, if gM/gN could induce the immune evasion. Furthermore, it should be clarified whether gM/gN complex has an ability to induce HCMV neutralizing antibodies.
Of all the combinations of cells and gc that were examined in this study for the correlation between NT50 titers and specific IIFA antibody titers against gc, the strongest correlation was found between anti-PC IIFA antibody titers and NT50 titers measured in ECs (Fig. 4d). Furthermore, the anti-gH/gL/gO IIFA titers were also significantly correlated with the HCMV-NT50 titers (Fig. 4c). These glycoprotein complexes seem to be the most important antigen in the induction of NT antibodies, as reported previously [17, 30, 46–49]. In addition, there are multiple neutralizing epitopes in gH/gL/gO. Those that are defined only by gH/gL sequences are common in both gH/gL/gO and the pentameric complex and for the most part block both fibroblast and epithelial cell entry, while those that are defined by UL128/UL130/UL131A sequences only block epithelial cell entry. There are epitopes on gH/gL within PC that probably associated with HCMV entry to fibroblast cells. Thus, the presence of the common epitopes is likely to have a major impact on the positive and significant correlation of both anti-PC and anti-gH/gL/gO IIFA antibody titers with HCMV-NT50 titer (Fig. 4c and d) [31].
One limitation of our study is that we used fixed and permeabilized cells for IIFA. Thus, as serum antibodies would have access to the viral proteins expressed not only on the virion surface but also inside the cells, suggesting the IIFA positive signals might be due to the antibodies that reacted with individual gc, misfolded gc, or partial degradation products of gc. In contrast, IIFAs based on non-permeabilized cells might reveal more robust correlations with neutralizing activities, as prior studies suggest that cell surface expression of both gH/gL and gM/gN complexes is restricted to conformationally native complexes [30, 50–53]. Thus, the antibodies, which react with these complexes in their native and quaternary conformations, would be selectively detected with IIFA, in which non-permeabilized cells were used for IIFA antigens. Therefore, further studies are needed to eliminate IIFA signals due to the antibodies react with non-native epitopes, which presumably do not exhibit neutralizing activity.
To the best our knowledge, because there have not been reported similar studies that included gO in the gH/gL complex or gM/gN. the results are well worth reporting. The findings will be of particular importance in the vaccine and immunotherapeutics development against HCMV infections.
Conclusion
Both the anti-PC antibody titers and the anti-gH/gL/gO antibody titers determined in the IIFA were highly correlated with the HCMV-NT50 titers determined in both HCMV ME-fibroblasts and HCMV 1612-epithelial cell combinations. By measuring the antibody titers to gH/gL/gO or PC in IIFA, it is possible to estimate the level of the HCMV-NT50 titer. The data obtained in this study suggest that the induction of a strong immune response to gH/gL complexes is important for the development of HCMV vaccines, which might have an ability to reduce the risk of congenital HCMV infection.
Acknowledgements
We wish to thank Ms. Yoshiko Fukui and Ms. Mihoko Tsuda for their timely help in cell culturing, propagating the HCMV clinical isolate, and accounting management.
Abbreviations
- DMEM
Dulbecco’s modified Eagle medium
- EC
Epithelial cells
- ELISA
Enzyme-linked immunosorbent assay
- FITC
Fluorescein isothiocyanate
- gc
Glycoprotein complex
- gP
Glycoprotein
- HCMV
Human cytomegalovirus
- IIFA
Indirect immunofluorescence assay
- NGS
Next generation sequencer
- NT
Neutralization
- NT50
Fifty percent virus-NT titers
- ORF
Open reading frame
- PC
Pentamer complex
- PCR
Polymerase chain reaction
- PFU
Plaque forming unit
- PGM
Personal Genome Machine
Authors’ contributions
M Shibamura performed the majority of the experiments and wrote the manuscript. M Saijo, AO, and MM planned and guided the research and supported to write this manuscript. TY, TI, SY, PHAN, HF, SH, and SF supported performing the research under discussion on this research. All authors read and revised the manuscript and approved the final manuscript.
Funding
This work was supported by grants from The Japan Foundation for Pediatric Research and a Grant-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology (Grant Numbers 15 K09675 and 18 K07894).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was carried out under the approval of the Medical Research Ethics Committee of the National Institute of Infectious Diseases (NIID) for the use of human subjects (Approval number: 797). Informed consent was obtained from all volunteer blood donors.
Consent for publication
Not applicable.
Competing interests
We have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Britt W. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr Top Microbiol Immunol. 2008;325:417–470. doi: 10.1007/978-3-540-77349-8_23. [DOI] [PubMed] [Google Scholar]
- 2.Boppana SB, Fowler KB, Pass RF, Rivera LB, Bradford RD, Lakeman FD, et al. Congenital cytomegalovirus infection: association between virus burden in infancy and hearing loss. J Pediatr. 2005;146:817–823. doi: 10.1016/j.jpeds.2005.01.059. [DOI] [PubMed] [Google Scholar]
- 3.Edward S, Mocarski J, Shenk T, Griffiths PD, Pass RF. Fields Virology. In: Knipe DM, Howley PM, editors. Chapter 62 Cytomegaloviruses. 6. Philadelphia: Walters Kluwer; 2013. pp. 1960–2014. [Google Scholar]
- 4.Marshall GS, Rabalais GP, Stout GG, Waldeyer SL. Antibodies to recombinant-derived glycoprotein B after natural human cytomegalovirus infection correlate with neutralizing activity. J Infect Dis. 1992;165:381–384. doi: 10.1093/infdis/165.2.381. [DOI] [PubMed] [Google Scholar]
- 5.Shimamura M, Mach M, Britt WJ. Human cytomegalovirus infection elicits a glycoprotein M (gM)/gN-specific virus-neutralizing antibody response. J Virol. 2006;80:4591–4600. doi: 10.1128/JVI.80.9.4591-4600.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Macagno A, Bernasconi NL, Vanzetta F, Dander E, Sarasini A, Revello MG, et al. Isolation of human monoclonal antibodies that potently neutralize human cytomegalovirus infection by targeting different epitopes on the gH/gL/UL128-131A complex. J Virol. 2010;84:1005–1013. doi: 10.1128/JVI.01809-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Plotkin S. The history of vaccination against cytomegalovirus. Med Microbiol Immunol. 2015;204(3):247–254. doi: 10.1007/s00430-015-0388-z. [DOI] [PubMed] [Google Scholar]
- 8.McVoy MA. Cytomegalovirus vaccines. Clin Infect Dis. 2013;57(Suppl 4):S196–S199. doi: 10.1093/cid/cit587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schleiss MR. Cytomegalovirus vaccine development. Curr Top Microbiol Immunol. 2008;325:361–382. doi: 10.1007/978-3-540-77349-8_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rieder F, Steininger C. Cytomegalovirus vaccine: phase II clinical trial results. Clin Microbiol Infect. 2014;20(Suppl 5):S95–102. doi: 10.1111/1469-0691.12449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fu TM, An Z, Wang D. Progress on pursuit of human cytomegalovirus vaccines for prevention of congenital infection and disease. Vaccine. 2014;32:2525–2533. doi: 10.1016/j.vaccine.2014.03.057. [DOI] [PubMed] [Google Scholar]
- 12.Hahn G, Revello MG, Patrone M, Percivalle E, Campanini G, Sarasini A, et al. Human cytomegalovirus UL131-128 genes are indispensable for virus growth in endothelial cells and virus transfer to leukocytes. J Virol. 2004;78:10023–10033. doi: 10.1128/JVI.78.18.10023-10033.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang D, Shenk T. Human cytomegalovirus UL131 open reading frame is required for epithelial cell tropism. J Virol. 2005;79:10330–10338. doi: 10.1128/JVI.79.16.10330-10338.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang D, Shenk T. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc Natl Acad Sci U S A. 2005;102:18153–18158. doi: 10.1073/pnas.0509201102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui X, Meza BP, Adler SP, McVoy MA. Cytomegalovirus vaccines fail to induce epithelial entry neutralizing antibodies comparable to natural infection. Vaccine. 2008;26:5760–5766. doi: 10.1016/j.vaccine.2008.07.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerna G, Sarasini A, Patrone M. Percivalle E1, Fiorina L, Campanini G, et al. human cytomegalovirus serum neutralizing antibodies block virus infection of endothelial/epithelial cells, but not fibroblasts, early during primary infection. J Gen Virol. 2008;89:853–865. doi: 10.1099/vir.0.83523-0. [DOI] [PubMed] [Google Scholar]
- 17.Fouts AE, Chan P, Stephan JP, Vandlen R, Feierbach B. Antibodies against the gH/gL/UL128/UL130/UL131 complex comprise the majority of the anti-cytomegalovirus (anti-CMV) neutralizing antibody response in CMV hyperimmune globulin. J Virol. 2012;86:7444–7447. doi: 10.1128/JVI.00467-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lilleri D, Kabanova A, Revello MG, Percivalle E, Sarasini A, Genini E, et al. Fetal human cytomegalovirus transmission correlates with delayed maternal antibodies to gH/gL/pUL128-130-131 complex during primary infection. PLoS One. 2013;8:e59863. doi: 10.1371/journal.pone.0059863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardner TJ, Tortorella D. Virion glycoprotein-mediated immune evasion by human Cytomegalovirus: a sticky virus makes a slick getaway. Microbiol Mol Biol Rev. 2016;80:663–677. doi: 10.1128/MMBR.00018-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pereira L, Petitt M, Tabata T. Cytomegalovirus infection and antibody protection of the developing placenta. Clin Infect Dis. 2013;57(Suppl 4):S174–S177. doi: 10.1093/cid/cit583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Maidji E, McDonagh S, Genbacev O, Tabata T, Pereira L. Maternal antibodies enhance or prevent cytomegalovirus infection in the placenta by neonatal fc receptor-mediated transcytosis. Am J Pathol. 2006;168:1210–1226. doi: 10.2353/ajpath.2006.050482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murrell I, Tomasec P, Wilkie GS, Dargan DJ, Davison AJ, Stanton RJ. Impact of sequence variation in the UL128 locus on production of human cytomegalovirus in fibroblast and epithelial cells. J Virol. 2013;87:10489–10500. doi: 10.1128/JVI.01546-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou M, Yu Q, Wechsler A, Ryckman BJ. Comparative analysis of gO isoforms reveals that strains of human cytomegalovirus differ in the ratio of gH/gL/gO and gH/gL/UL128-131 in the virion envelope. J Virol. 2013;87:9680–9690. doi: 10.1128/JVI.01167-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanton RJ, Baluchova K, Dargan DJ, Cunningham C, Sheehy O, Seirafian S, et al. Reconstruction of the complete human cytomegalovirus genome in a BAC reveals RL13 to be a potent inhibitor of replication. J Clin Invest. 2010;120:3191–3208. doi: 10.1172/JCI42955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taniguchi K, Watanabe N, Sato A, Jwa SC, Suzuki T, Yamanobe Y, et al. Changes in cytomegalovirus seroprevalence in pregnant Japanese women-a 10-year single center study. J Clin Virol. 2014;59:192–194. doi: 10.1016/j.jcv.2013.12.013. [DOI] [PubMed] [Google Scholar]
- 26.Shigemi D, Yamaguchi S, Otsuka T, Kamoi S, Takeshita T. Seroprevalence of cytomegalovirus IgG antibodies among pregnant women in Japan from 2009-2014. Am J Infect Control. 2015;43:1218–1221. doi: 10.1016/j.ajic.2015.06.026. [DOI] [PubMed] [Google Scholar]
- 27.Schmitz H, Essuman S. Comparison of the neutralizing and ELISA antibody titres to human cytomegalovirus (HCMV) in human sera and in gamma globulin preparations. J Med Virol. 1986;20:177–182. doi: 10.1002/jmv.1890200209. [DOI] [PubMed] [Google Scholar]
- 28.Wang D, Li F, Freed DC, Finnefrock AC, Tang A, Grimes SN, et al. Quantitative analysis of neutralizing antibody response to human cytomegalovirus in natural infection. Vaccine. 2011;29:9075–9080. doi: 10.1016/j.vaccine.2011.09.056. [DOI] [PubMed] [Google Scholar]
- 29.Leogrande G, Merchionne F, Lazzarotto T, Landini MP. Large-scale testing of human serum to determine cytomegalovirus neutralising antibody. J Inf Secur. 1992;24:289–299. doi: 10.1016/s0163-4453(05)80034-9. [DOI] [PubMed] [Google Scholar]
- 30.Ciferri C, Chandramouli S, Leitner A, Donnarumma D, Cianfrocco MA, Gerrein R, et al. Antigenic characterization of the HCMV gH/gL/gO and pentamer cell entry complexes reveals binding sites for potently neutralizing human antibodies. PLoS Pathog. 2015;11:e1005230. doi: 10.1371/journal.ppat.1005230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ciferri C, Chandramouli S, Donnarumma D, Nikitin PA, Cianfrocco MA, Gerrein R, et al. Structural and biochemical studies of HCMV gH/gL/gO and Pentamer reveal mutually exclusive cell entry complexes. Proc Natl Acad Sci U S A. 2015;112:1767–1772. doi: 10.1073/pnas.1424818112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Britt WJ, Vugler L, Butfiloski EJ, Stephens EB. Cell surface expression of human cytomegalovirus (HCMV) gp55-116 (gB): use of HCMV-recombinant vaccinia virus-infected cells in analysis of the human neutralizing antibody response. J Virol. 1990;64:1079–1085. doi: 10.1128/jvi.64.3.1079-1085.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang JB, Adler SP, Hempfling S, Burke RL, Duliège AM, Starr SE, et al. Mucosal antibodies to human cytomegalovirus glycoprotein B occur following both natural infection and immunization with human cytomegalovirus vaccines. J Infect Dis. 1996;174:387–392. doi: 10.1093/infdis/174.2.387. [DOI] [PubMed] [Google Scholar]
- 34.Gönczöl E, deTaisne C, Hirka G, Berencsi K, Lin WC, Paoletti E, et al. High expression of human cytomegalovirus (HCMV)-gB protein in cells infected with a vaccinia-gB recombinant: the importance of the gB protein in HCMV immunity. Vaccine. 1991;9:631–637. doi: 10.1016/0264-410x(91)90187-b. [DOI] [PubMed] [Google Scholar]
- 35.Abai AM, Smith LR, Wloch MK. Novel microneutralization assay for HCMV using automated data collection and analysis. J Immunol Methods. 2007;322:82–93. doi: 10.1016/j.jim.2007.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Burke HG, Heldwein EE. Crystal structure of the human cytomegalovirus glycoprotein B. PLoS Pathog. 2015;11:e1005227. doi: 10.1371/journal.ppat.1005227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nelson CS, Huffman T, Jenks JA, de la Rosa EC, Xie G, Vandergrift N. HCMV glycoprotein B subunit vaccine efficacy mediated by nonneutralizing antibody effector functions. Proc Natl Acad Sci U S A. 2018;115(24):6267–6272. doi: 10.1073/pnas.1800177115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Baraniak I, Kropff B, Ambrose L, McIntosh M, McLean GR, Pichon S. Protection from cytomegalovirus viremia following glycoprotein B vaccination is not dependent on neutralizing antibodies. Proc Natl Acad Sci U S A. 2018;115(24):6273–6278. doi: 10.1073/pnas.1800224115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pötzsch S, Spindler N, Wiegers AK, Fisch T, Rücker P, Sticht H, et al. B cell repertoire analysis identifies new antigenic domains on glycoprotein B of human cytomegalovirus which are target of neutralizing antibodies. PLoS Pathog. 2011;7:e1002172. doi: 10.1371/journal.ppat.1002172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li F, Freed DC, Tang A, Rustandi RR, Troutman MC, Espeseth AS, et al. Complement enhances in vitro neutralizing potency of antibodies to human cytomegalovirus glycoprotein B (gB) and immune sera induced by gB/MF59 vaccination. NPJ Vaccines. 2017;2:36. doi: 10.1038/s41541-017-0038-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cui X, Cao Z, Wang S, Lee RB, Wang X, Murata H, et al. Novel trimeric human cytomegalovirus glycoprotein B elicits a high-titer neutralizing antibody response. Vaccine. 2018;36:5580–5590. doi: 10.1016/j.vaccine.2018.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chandramouli S, Ciferri C, Nikitin PA, et al. Structure of HCMV glycoprotein B in the postfusion conformation bound to a neutralizing human antibody. Nat Commun. 2015;6:8176. doi: 10.1038/ncomms9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shen S, Wang S, Britt WJ, Lu S. DNA vaccines expressing glycoprotein complex II antigens gM and gN elicited neutralizing antibodies against multiple human cytomegalovirus (HCMV) isolates. Vaccine. 2007;25:3319–3327. doi: 10.1016/j.vaccine.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 44.Kropff B, Burkhardt C, Schott J, Nentwich J, Fisch T, Britt W, et al. Glycoprotein N of human cytomegalovirus protects the virus from neutralizing antibodies. PLoS Pathog. 2012;8:e1002999. doi: 10.1371/journal.ppat.1002999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Varnum SM, Streblow DN, Monroe ME, Smith P, Auberry KJ, Pasa-Tolic L, et al. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J Virol. 2004;78:10960–10966. doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fouts AE, Comps-Agrar L, Stengel KF, Ellerman D, Schoeffler AJ, Warming S, et al. Mechanism for neutralizing activity by the anti-CMV gH/gL monoclonal antibody MSL-109. Proc Natl Acad Sci U S A. 2014;111:8209–8214. doi: 10.1073/pnas.1404653111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wille PT, Knoche AJ, Nelson JA, Jarvis MA, Johnson DC. A human cytomegalovirus gO-null mutant fails to incorporate gH/gL into the virion envelope and is unable to enter fibroblasts and epithelial and endothelial cells. J Virol. 2010;84:2585–2596. doi: 10.1128/JVI.02249-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Genini E, Percivalle E, Sarasini A, Revello MG, Baldanti F, Gerna G. Serum antibody response to the gH/gL/pUL128-131 five-protein complex of human cytomegalovirus (HCMV) in primary and reactivated HCMV infections. J Clin Virol. 2011;52:113–118. doi: 10.1016/j.jcv.2011.06.018. [DOI] [PubMed] [Google Scholar]
- 49.Saccoccio FM, Sauer AL, Cui X, Armstrong AE, Habib e-SE, Johnson DC, et al. Peptides from cytomegalovirus UL130 and UL131 proteins induce high titer antibodies that block viral entry into mucosal epithelial cells. Vaccine. 2011;29:2705–2711. doi: 10.1016/j.vaccine.2011.01.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wussow F, Chiuppesi F, Martinez J, Campo J, Johnson E, Flechsig C, et al. Human cytomegalovirus vaccine based on the envelope gH/gL pentamer complex. PLoS Pathog. 2014;10:e1004524. doi: 10.1371/journal.ppat.1004524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mach M, Kropff B, Dal Monte P, Britt W. Complex formation by human cytomegalovirus glycoproteins M (gpUL100) and N (gpUL73) J Virol. 2000;74:11881–11892. doi: 10.1128/jvi.74.24.11881-11892.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mach M, Kropff B, Kryzaniak M, Britt W. Complex formation by glycoproteins M and N of human cytomegalovirus: structural and functional aspects. J Virol. 2005;79:2160–2170. doi: 10.1128/JVI.79.4.2160-2170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ryckman BJ, Rainish BL, Chase MC, Borton JA, Nelson JA, Jarvis MA, Johnson DC. Characterization of the human cytomegalovirus gH/gL/UL128-131 complex that mediates entry into epithelial and endothelial cells. J Virol. 2008;82:60–70. doi: 10.1128/JVI.01910-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.