Abstract
PCORnet, the Patient-Centered Outcomes Research Network, is comprised of health systems and health plans that transform electronic health records (EHRs) and claims data to a common data model (CDM) to facilitate real-world clinical research. Because patients receive health care in multiple care delivery settings, linking health records across systems and health plan claims would provide a more comprehensive and accurate picture of health care for patients. The current study expanded on a PCORnet Antibiotics and Childhood Growth (ABX) study to (1) identify and implement a privacy-preserving patient linkage solution among a clinical data research network and a health plan network within the ABX Study, and (2) assess overlap in prescribed and dispensed antibiotics and additional data gained from claims among the linked patients. This manuscript describes the linkage process and resulting overlap analysis. The authors identified 549 patients from the EHR record study cohort who had claims records with the health plan. Sixty percent (n = 329) of patients had consistent antibiotic exposure data across the 2 sources, indicating antibiotic exposure (44.3%) or nonexposure (15.7%). Among total antibiotic prescribing records, 43.1% had a matched claims record for dispensing within 60 days. Among antibiotic dispense records 26.5% were not associated with a prescribing record in the linked health systems. These findings showcase the feasibility of linking health plan claims data to PCORnet CDM in a privacy-preserving manner while also demonstrating continued gaps in data that may occur. The study highlights the importance of combining multiple health data sources for comprehensive clinical research.
Keywords: patient data linkage, electronic health records, childhood antibiotics
Introduction
Patients, their families, and clinicians face a wide range of complex and often confusing choices when it comes to health and health care concerns. They need trustworthy information to make the best health care decisions for themselves and their families. The Patient-Centered Outcomes Research Institute (PCORI), a nonprofit, nongovernmental organization, was authorized by Congress in 2010 with a mandate to improve the quality and relevance of evidence available to help patients, caregivers, clinicians, employers, insurers, and policy makers make better-informed health decisions. To facilitate efficient, large-scale research to support its mission, a major initiative of PCORI and the Patient-Centered Research Foundation was the development of PCORnet, the Patient-Centered Outcomes Research Network.1 PCORnet is now comprised of 9 clinical research networks (CRNs) and 2 health plan networks (HPNs) that transform existing electronic health records (EHRs) and claims data to the PCORnet common data model (CDM).2 PCORnet has data from more than 65 million patients with an encounter in the past 5 years, and the infrastructure offers the opportunity to conduct retrospective and observational studies designed to answer critical real-world research questions.
An early PCORnet demonstration project—designed to answer important patient-centered, comparative effectiveness research questions and demonstrate PCORnet capabilities—was Short- and Long-term Effects of Antibiotics on Childhood Growth (ABX).3 This study explored the association between antibiotic use at <24 months of age and weight outcomes in later childhood across diverse multi-institutional networks using the PCORnet CDM. Study data included prescriptions recorded in the EHR, heights, weights, diagnostic codes, and demographics. The ABX study employed an observational design and tested the technical, operational, and governance aspects of PCORnet’s Distributed Research Network (DRN) as well as the ability to standardize the capture of EHR data with some claims and pharmacy dispensing data, available in the CDM. A key finding from the study was that children had a slightly higher weight at 5 years of age if they were exposed to antibiotics at <2 years of age, compared to children who received no antibiotics.4
Although access to a large network of standardized health record data, such as PCORnet, presents opportunities for significant research, challenges concerning data completeness exist because patients receive health care in multiple care delivery settings within a region; this includes provider encounters, prescription fills, and laboratory work. Integrating health records across systems and health plan claims would provide a more comprehensive and accurate picture of health care for patients than relying on a single data source.
For example, in the ABX study, most of the data available from PCORnet came from EHRs. When examining the use of antibiotics, a critical adjunct to prescribing data is pharmacy dispensing data and insurance claims for prescriptions. Children can receive antibiotics from urgent care or retail clinics, community-based primary care practices, or other health care institutions that are outside of the data network. Insurance claims and pharmacy dispensing data can help to fill in these gaps by providing additional records of medications that were given but not available in PCORnet. Linking patient data across health care sources requires a balance between data availability and privacy, which is not an easy problem to solve.
Patient data linkage and associated challenges
Options for linking patient data across multiple care settings exist. A unique national patient-level identifier could be the answer to consolidate data on patients, as has been adopted in other countries, but it is still not an option in the United States because of concerns about lack of privacy, identity theft, and lack of access to care.5 Because there is no single common patient identifier, linking a patient across settings must be performed using a combination of each patient’s personally identifiable information (PII) such as social security numbers, dates of birth, names, and addresses to ensure accurate matching. Linking patients across health care data sets is very limited because of privacy and security regulations. Besides strict Health Insurance Portability and Accountability Act (HIPAA) guidelines, there are business concerns in health systems for sharing patient identifiers among care-providing entities. Secure Privacy Preserving Record Linkage (PPRL; also known as Anonymous Record Linkage) offers the best option for linking individual-level patient records across multiple institutions without the need to expose patient-level identifiers that could create privacy risks.6–8
Key considerations for PPRL include the need for:
Identification of data sources with adequate patient overlap to complete the analysis
Identification of a linkage method acceptable to participating sites
Ability to navigate contractual and governance requirements with efficiency and site cooperation
An inability to comply with any of the above considerations can lead to failure of a linkage project.
As an extension of the ABX study, the study team sought to navigate the challenges of linkage and provide a supplementary analysis to the study. To accomplish this goal, HUMnet HPN was charged with identifying a CRN linkage partner—the Research Action for Health Network (REACHnet)—and implementing a patient data linkage process. REACHnet CRN is a partnership between the Louisiana Public Health Institute, Ochsner Health System, Partnership for Achieving Total Health, Louisiana State University, Pennington Biomedical Research Center, Tulane University, Baylor Scott & White Health, and University Medical Center New Orleans. REACHnet contains EHR data from more than 6 million patients across Louisiana and Texas. HUMnet HPN is a partnership between Humana and Medical Outcomes Management and offers a research and data infrastructure resource that uses the 23-million member HUMnet administrative claims database transformed into the PCORnet CDM. HUMnet represents one of the first collaborations of a national-level health plan with PCORnet health care systems.
The study objective was to use a PPRL method to link HUMnet health plan claims to REACHnet EHR data and provide additional longitudinal data for the ABX study.9–11 The analysis was set to (1) describe the population of patients with data in HUMnet and REACHnet, and (2) identify dispensed antibiotics captured by health plan claims but otherwise not captured in the EHR from the PCORnet CDM prescribing table. The overarching goal was to demonstrate what additional data may be captured from PPRL and contribute to a more complete health record for research uses.
Methods
Linkage site selection
HUMnet initially estimated their overlapping member populations with CRN populations involved in the PCORnet ABX study simply by comparing the zip codes in which HUMnet patients lived and CRNs operated. A subsequent, more accurate overlap analysis required the use of National Provider Identifier numbers from the CRNs. Three CRNs emerged as likely partners based on a reasonably large overlapping population.
Next, HUMnet evaluated the technical method of linking data and discovered that the 3 identified CRNs used different linkage methods. Two of the CRNs used similar versions of the same robust linkage solution while the third CRN’s linkage solution required a different set of software dependencies that were technically harder to overcome. Based on multiple considerations, including ease of scalability and future research, familiarity, and the potential for developing a solution that could be disseminated widely across PCORnet, HUMnet identified REACHnet CRN as their partner for this project.
Anonymous record linkage
Privacy-preserving methods, such as anonymous linkage strategies through the creation of an anonymous hash identifier with a secret key, have been used as HIPAA-compliant means to identify cross-site patient overlap. Two PCORnet CRNs including REACHnet have successfully implemented the hashing tool provided by Datavant (Datavant, San Francisco, CA [formerly Health Data Link Inc.]) to identify overlapping patients across health data sources in their network.12
Hashing is an established technique to de-identify data by applying hash functions to individuals’ records to enable record linkage and disambiguation. However, use of functions alone is subject to dictionary or “rainbow table” attacks. Datavant’s approach addresses these shortcomings using the following:
The hashing software uses a runtime key and combination of multiple process source input (ie, PII) elements to generate individual hashes to secure against potential dictionary attacks.
Only authorized users are able to run the hashing software and each event is logged for security. Upon authentication and an institution-specific runtime key distribution, hashing software terminates the external connection to PII data.
The current study used REACHnet’s Global Patient Identifier (GPID) solution, powered by Datavant, which relies on advanced hashing and matching algorithms to provide a secure and efficient process for deduplication and data linkage without requiring the sharing of individually identifiable information. Through the hashing and matching process, a GPID is assigned to each patient and is associated with that patient’s records across all REACHnet partner-specific data marts. The GPID is then utilized to link these disparate records for the creation of cohort- and study-specific data sets. The 2 primary use cases for the GPID solution include (1) deduplication and linkage of REACHnet clinical data from their health system partners, and (2) longitudinal data linkage with care components that occur outside of that health system (eg, regional claims partners, public health department, state registries).
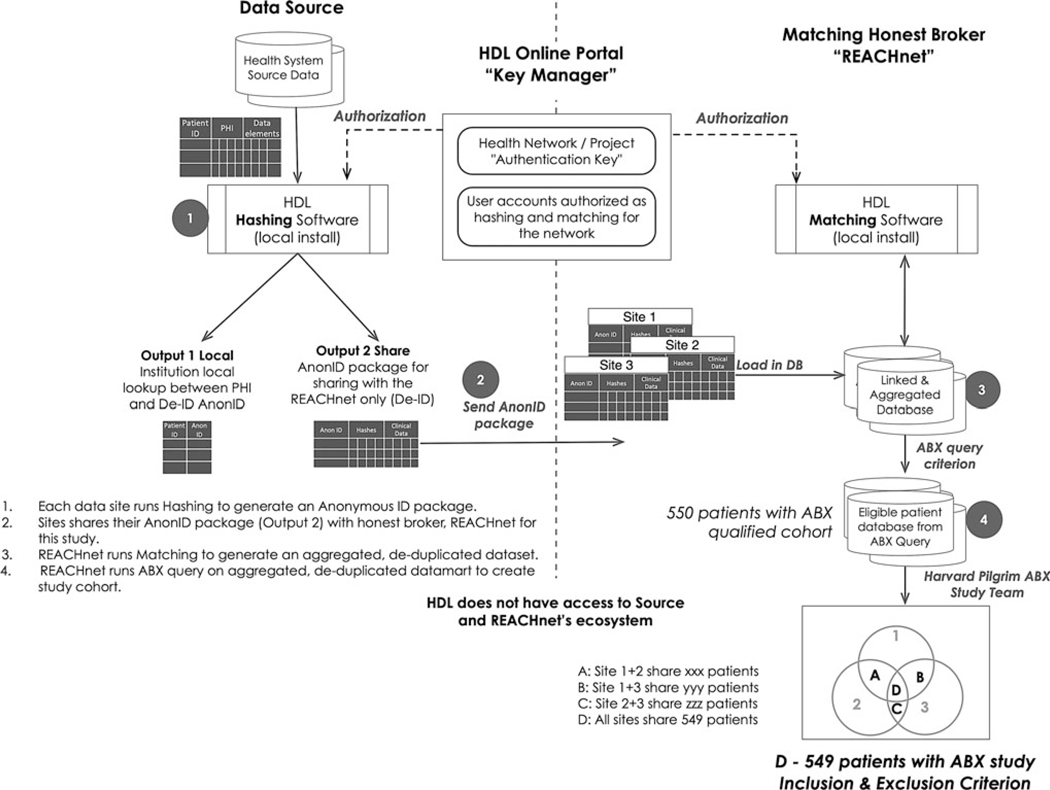
The GPID solution is composed of a hashing tool (secure agent) and a matching tool. The hashing tool is deployed behind the data source’s firewall and uses PII inputs to create hash bundles produced from a combination of attributes. The Matching Tool is deployed within REACHnet’s information technology (IT) ecosystem (the data aggregator site serving as an “honest broker”) and uses patient hashes provided by data owners. A schematic representation of the process is detailed in Figure 1. Datavant’s hashing tool uses the National Security Agency Secure Hash Algorithm 2–512 in combination with a runtime key provided by a third party (Datavant web portal) that is not part of REACHnet’s ecosystem. Datavant’s hashing tool leaves a lookup table at each data source for reidentification of records whose values may need to be updated later in the process (eg, to improve quality at the source).
FIG. 1.
Anonymous record linkage and data curation process flow for building linked ABX cohort used in the study. ABX, Antibiotics and Childhood Growth; HDL, Health Data Link, Inc.; REACHnet, Research Action for Health Network.
The hash bundles created at the data sources are sent to the matching tool in the REACHnet IT ecosystem, effectively creating a de-identified and deduplicated unique patient identifier referred to as the GPID. The GPID is assigned to each patient and associated with her/his hash bundle. Calls to the matching tool through the REACHnet IT ecosystem utilize Datavant’s matching algorithm to find hashes that match across organizations. Once a match is found, the matching patient’s GPID is returned, resulting in the index needed by the network to not only link records across sites, but also to allow for reidentification by the data contributor when necessary. Steps 1 through 3 in Figure 1 demonstrate the workflow of hashing and matching for REACHnet. Reidentification is not possible at the REACHnet IT ecosystem level as the hashes are considered de-identified and lookup tables are stored locally at the data source. This process has already been validated by an expert determination as a HIPAA-compliant de-identified method (Privacy Rule Section 164.514(b)(1)).
Data curation
REACHnet worked with HUMnet to perform cross-network linkage using the Datavant/GPID method. This analysis used PCORnet CDM version 3.0. The study team extracted antibiotic prescription records from the CDM prescribing table at REACHnet between 2010 and September 2018 at the time of the data extraction. Prior to proceeding with the linkage, REACHnet identified their patients that met the inclusion criteria for the PCORnet ABX study, including children who had at least 1 same-day height and weight in each of 3 age periods: 0 to <12 months of age, 12 to <30 months, and ≤24 months. The team used these criteria to ensure that children were being followed in the health system, such that prescription records were more likely to be captured than if the child only had 1 appointment in the system, for example. The overlap for the 2nd and 3rd age groups allowed children to be included even if they received their 2-year well-child visit after their 2nd birthday; a child could qualify for inclusion in the cohort with only 2 height and weight measures if the second was between 24 and 30 months. Children included in the cohort were eligible to be linked to HUMnet data.
REACHnet received patient hashes from HUMnet and their participating sites and used the GPID to identify linked patients in their ABX study cohort. REACHnet and HUMnet patients were linked if they had a record in both databases at any time during the study period, rather than based on insurance enrollment during the time of the antibiotic prescription records. REACHnet then merged HUMnet’s dispensing data to the REACHnet ABX study data set on the GPID resulting in the linked data set. Step 4 and final Venn diagram in Figure 1 highlight data curation steps for this study. To prepare for the analysis query, the study team assigned antibiotics specifications (antibiotic spectrum and therapeutic class) to each prescribing (REACHnet) and dispensing (HUMnet) record. For efficiency in assessing record linkage, the study team deduplicated same-day records with the same antibiotic spectrum (broad or narrow) and grouped antibiotic records that were within 10 days of each other into a 10-day episode separately for prescribing and dispensing records.
Data analysis
The study team descriptively analyzed the number of patients by sex and race across the entire ABX study, the REACHnet ABX study cohort, and the REACHnet–HUMnet linked patient cohort. The team then described antibiotics use among ABX patients with health plan linkage by counting the total number of patients who had (1) true positive: both prescribing records in REACHnet data and dispensing records in HUMnet data (yPyD), (2) true negative: no prescribing nor dispensing records (nPnD), (3) false positive: had prescribing but no dispensing records (yPnD), and (4) false negative: had dispensing but no prescribing record (nPyD). (Dispensing records were used as the gold standard, or true condition). Next, the team compared 10-day prescribing and dispensing episodes by performing bidirectional matching and presented the proportion of episodes with and without corresponding matching. Antibiotics episodes linked together consecutive prescriptions to the same patient over a short period, which the team assumed was likely part of the same illness episode. The matching variable used included patient ID and 60 lag days between records to capture medications that might have been dispensed well after the prescription was written.
Results
Linked population
The main ABX study included a total of 681,739 patients from 36 health care institutions, with 8451 patients from REACHnet. Of those patients, 549 matched patients were identified in REACHnet and HUMnet. The linked population had a similar sex distribution as the main ABX study population; however, the distribution of race was different, with a higher proportion of white patients in the linked population (Table 1).
Table 1.
Demographics of Antibiotics and Childhood Growth Linkage Study Population
| Main ABX study | REACHnet | REACHnet/HUMnet linkage | |
|---|---|---|---|
| Total patients | 681,739 | 8451 | 549 |
| Sex | |||
| Male | 356,875 (52.3%) | 151,364 (52.7%) | 295 (53.7%) |
| Female | 324,864 (47.7%) | 103,219 (47.3%) | 254 (46.3%) |
| Race | |||
| White | 363,759 (53.4%) | 5315 (62.9%) | 442 (80.5%) |
| Black or African American | 170,007 (24.9%) | 2699 (31.9%) | 91 (16.6%) |
| Asian | 28,356 (4.2%) | 227 (2.7%) | 10 (1.8%) |
| Other/unknown | 119,545 (17.5%) | 210 (2.5%) | 6 (1.0%) |
ABX, Antibiotics and Childhood Growth; REACHnet, Research Action for Health Network.
Previous studies and prior REACHnet assessments have demonstrated that the GPID methodology produces accurate results.11 A rapid “false-positive” check was performed for the linked population between REACHnet and HUMnet as an additional check. Because of the large population size, an in-network manual check for duplicates (<1% of total population) was completed within the HUMnet population. A subsegment (3520 out of total 12,528) of duplicates identified by GPID were reviewed by HUMnet staff, who found those records having identical PII attributes, providing confidence of a ~zero false-positive rate for this study. The subsegment comprised those members who had multiple enrollments (ie, discontinued enrollment in different years) in HUMnet insurance plans.
Comparison of antibiotic use by patients and prescribing records
At the patient level, for antibiotics, there was a 44.3% true positive (yPyD) rate and a 15.7% true-negative (nPnD) rate (binary, yes/no) (Table 2). In other words, 60% of patients were accurately classified as being exposed or not exposed to antibiotics considering both sources of data. Approximately one third of patients had false positives (yPnD), such that they received prescriptions but had no evidence of dispensed antibiotics; this was expected in large part because there was only one source of dispensing data, and patients could have had medications covered by different insurers over the course of time (Table 2). More importantly, ~4% of the linked population were in the false-negative category with no record of the medication episode in the EHR (nPyD), meaning their exposure would have been missed without inclusion of the claims data (Table 2).
Table 2.
Antibiotic Use* Among Linked Population
| REACHnet (N = 549) | |
|---|---|
| yPyD | 243 (44.3%) |
| nPnD | 86 (15.7%) |
| yPnD | 199 (36.2%) |
| nPyD | 21 (3.8%) |
Antibiotic use per patient based on prescribing and dispensing record.
REACHnet, Research Action for Health Network; yPyD, the patient had both prescribing and dispensing records; nPnD, the patient had no prescribing nor dispensing record; yPnD, the patient had prescribing but no dispensing records; nPyD, the patient had dispensing but no prescribing records.
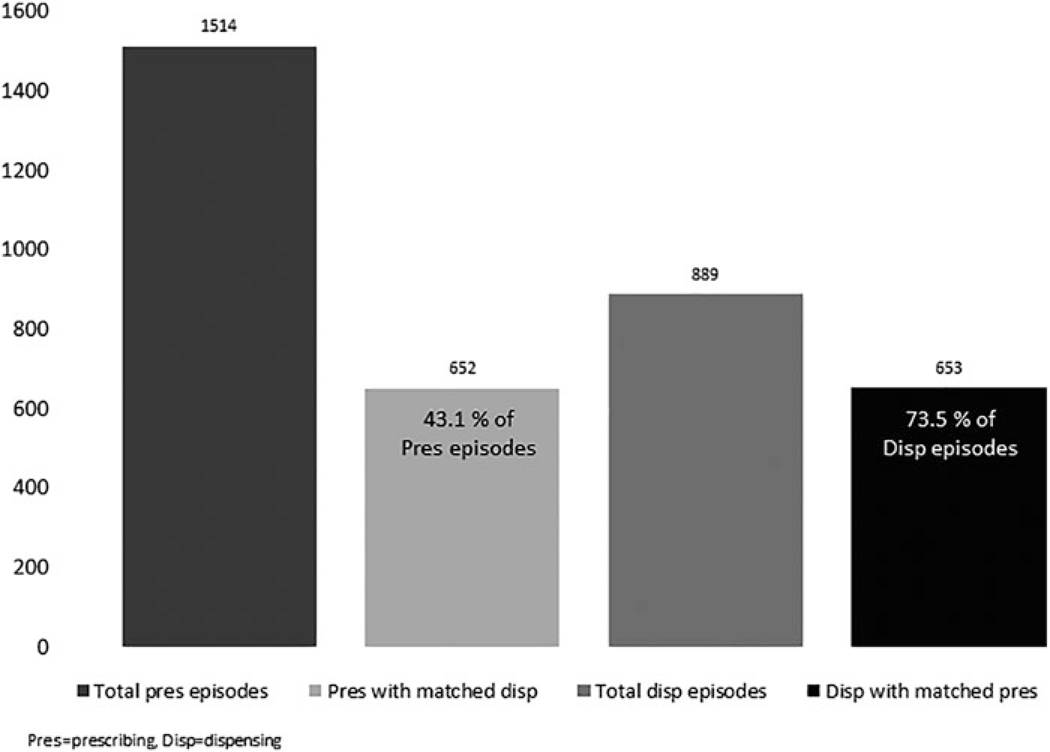
Examining the prescription records, rather than individuals, this study found ~43% (652/1514) of prescription episodes had matching dispensing records 60 days after being prescribed, with the median (interquartile range [IQR]) gap day between matched records being 0 (0, 0) day (Figure 2). The total number of linked patients who had equal numbers of prescribing and dispensing records was 167 (30.4%). For the dispensing records, 73.5% (653/889) of the dispensing episodes had matching prescribing records 60 days prior to the dispensing date, with the median (IQR) gap day between matched records being 0 (0, 0) day (Figure 2).
FIG. 2.
Number of matched antibiotic prescribing (REACHnet) and dispensing (HUMnet) episodes (0 < 2 years). REACHnet, Research Action for Health Network.
Discussion
PCORnet’s standardization of health records from across health systems and plans provides immense opportunity for real-world clinical research, but the ability to link patients across sources is essential for a comprehensive understanding of the health care process. The current study was conducted to identify and implement a PPRL solution across EHR and claims data sources and to demonstrate linkage utility by examining patient concordance in prescribed and dispensed records as part of a larger childhood antibiotic study (ABX study). The study team implemented a linkage solution, identified key requirements for success, and shared findings demonstrating the utility of cross-network patient linkage for clinical research.
For a patient data linkage project, the team identified the need for data sources with adequate patient overlap, agreement on linkage method, and ability to efficiently navigate contractual and governance requirements as key factors for successfully implementation. The partnership between REACHnet CRN and HUMnet HRN stood out as a key model for successful engagement. With future research and reducing contractual burden in mind, REACHnet and HUMnet developed a single broad contractual agreement that covers data exchange, honest broker, and software deployment. Study-specific use cases are then nested within the master agreement as addenda using an agreed upon template. Thus, although individual research uses of data may require internal approval, the infrastructure and data sharing parameters are already contractually agreed upon. The master agreement process can be lengthy, but the reduction in time and effort needed for subsequent projects is greatly reduced. The team intends to leverage the master agreement approach when engaging additional partners/sites for future linkage studies. Ultimately, the master agreement approach is expected to increase scalability by having multiple networks on the same governance and technology leading to increased sample sizes.
Present study clinical findings revealed that for research involving EHR records, linking to claims provides a valuable supplement and highlights some considerations. The descriptive analysis of the linked population found a higher proportion of white patients compared to the EHR-based cohort. The percentage of black patients dropped by close to half from 32% in the EHR cohort to about 17% in the linked cohort. Black and other ethnic and racial minorities tend to have lower rates of private and employer-based health care coverage, which was represented in the analysis, and higher rates of government-sponsored coverage and uninsurance.13 Including linkage with public plans, such as Medicaid, should be considered in future studies to reduce this disparity.
This analysis of antibiotic concordance between EHR and claims data showed that 60% of linked patients had consistent records for prescribed and dispensed antibiotics (true positive or negative). Thus, use of one of the data sources would have provided accurate information on antibiotic exposure for a little more than half of patients. The primary reason for this was the lack of comprehensive dispensing data related to incomplete coverage over the prescribing period and use of a single dispensing source.
Among the 442 patients with a prescribing record in the EHR, 55% had a dispensing record in the health plan, and among 1514 total prescribing records, 43% had a matched dispensing record. These matches demonstrate the ability to identify corresponding records, but do not reflect the prescription filling rate because the study team was not able to ascertain whether patients had full coverage throughout the entire duration for which EHR data were available. Patients needed only 1 record present in the claims data to support linkage, and patients may have had multiple and variable insurance coverage over time. Other reasons for lacking dispensing records could be out-of-pocket payment for prescriptions, which would be missing from claims records, or nonadherence; though based on prior work, the team suspects that patients fail to fill prescriptions relatively rarely (around 10%).14
The findings from this study demonstrate the importance of including multiple data sources to gain a more complete picture of a patient’s health and heath care and to reduce bias of single-source studies. The study highlights the benefit of including EHR and claims data for research involving prescribing, though other non-EHR data sources such as laboratory tests and radiology that are often outsourced are other key areas for linkage. The study team also developed a governance strategy leveraging a master agreement that increases efficiency of future research and scalability when partnering with new sites.
Limitations/challenges
This study was an initial linkage analysis among 2 networks that were able to agree on a linkage solution and successfully negotiate contractual and governance needs. For a more complete picture of a patient’s health care journey, linkage among a wider set of health care sources is needed, which will require an ability to navigate the technical and contractual challenges of patient linkage across multiple institutions. This would allow for more complete records. This was particularly notable in the present study, which had a relatively low matching rate from prescription to dispensing. The source of the discordance between prescribing and dispensing records is not fully understood, but key factors are use of a single dispensing source and lack of information on gaps of non-enrollment. Because of business and privacy concerns, detailed claims information on gaps in care or coverage availability by data type were not shared during the linkage process. More privacy-protecting methods are in progress to overcome this limitation, which could allow for linkage based on enrollment period and with multiple health plan providers in the future. In addition, a model for linkage across PCORnet, which will scale this model to numerous health systems and several health plans, is in progress. Lastly, this study had limited scope on outcomes given the focus on technical and governance components of linkage, including only on prescribed and dispensed antibiotics. It will be important to identify other health events (other than medications) that especially benefit from health care and claims linkage for clinical research.
Conclusion
Routine health care continues to generate increasing amounts of EHR data, both in breadth and depth. Linking sources of clinical and claims data such as patient EHRs, pharmacy claims, insurance claims, electronic patient-reported outcomes, and registries for clinical and observational research can provide insights into improving the delivery of cost-effective health care and improving patient outcomes. These data can assess the value and effectiveness of pharmaceuticals, medical devices, and the ever-growing and increasingly expensive biotechnology products, and impact future development and reimbursement. Close collaboration between those organizations, companies, and industry that possess the data requires vision, cooperation, sharing of data, up-front strategizing using novel data technologies, and testing and validating data models. As this study demonstrates, this collaboration can help facilitate capture of information on care provided to patients, rather than only those components that are available in 1 institution that has data on a patient.
Acknowledgments
The authors would like to acknowledge the following people: Daniel Hsia, MD for review of the manuscript and contribution as the REACHnet ABX Study Lead, Maryan Zirkle, MD, MS, MA for her encouragement and support throughout the development and writing the manuscript, and Elizabeth Crull, MPH for analytic work on the data linkage for this study.
Funding Information
This work was supported through the Patient-Centered Outcomes Research Institute (PCORI) Program Award (OBS-1505-30699). REACHnet is supported by PCORI (CDRN-1306-04864).
Footnotes
Author Disclosure Statement
The authors declare that there are no conflicts of interest.
References
- 1.Fleurence RL, Curtis LH, Califf RM, Platt R, Selby JV, Brown JS. Launching PCORnet, a national patient-centered clinical research network. J Am Med Inform Assoc 2014; 21:578–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.National Patient-Centered Clinical Research Network (PCORnet). PCORnet Common Data Model Specification. https://pcornet.org/data-driven-common-model Accessed August 26, 2019.
- 3.Block JP, Baile LC, Gillman MW, et al. PCORnet antibiotics and childhood growth study: process for cohort creation and cohort description. Acad Pediatr 2018;18:569–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Block JP, Bailey LC, Gillman MW, et al. Early antibiotic exposure and weight outcomes in young children. Pediatrics 2018;142:e20180290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sood HS, Bates DW, Halamka JD, Sheikh A. Has the time come for a unique patient identifier for the U.S.? NEJM Catalyst. Published February 21, 2018. https://catalyst.nejm.org/time-unique-patient-identifiers-us/ Accessed May 10, 2019. [Google Scholar]
- 6.Atiemo K, Skaro A, Maddur H, et al. Mortality risk factors among patients with cirrhosis and a low model for end-stage liver disease sodium score (≤15): an analysis of liver transplant allocation policy using aggregated electronic health record data. Am J Transplant 2017;17:2410–2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jackson KL, Goel S, Kho AN, Keswani RN. Distance from hospital impacts adverse event detection after outpatient endoscopy. Gastrointest Endosc 2017;85:380–386. [DOI] [PubMed] [Google Scholar]
- 8.Mays JA, Jackson KL, Derby TA, et al. An evaluation of recurrent diabetic ketoacidosis, fragmentation of care, and mortality across Chicago, Illinois. Diabetes Care 2016;39: 1671–1676. [DOI] [PubMed] [Google Scholar]
- 9.Randall SM, Ferrante AM, Boyd JH, Bauer JK, Semmens JB. Privacy-preserving record linkage on large real world datasets. J Biomed Inform 2014;50:205–212. [DOI] [PubMed] [Google Scholar]
- 10.Quantin C, Bouzelat H, Allaert FAA, Benhamiche AM, Faivre J, Dusserre L. How to ensure data security of an epidemiological follow-up: quality assessment of an anonymous record linkage procedure. Int J Med Inform 1998;49:117–122. [DOI] [PubMed] [Google Scholar]
- 11.Vatsalan D, Christen P, Verykios VS. A taxonomy of privacy-preserving record linkage techniques. Inf Syst 2013; 38:946–969. [Google Scholar]
- 12.Kho AN, Cashy JP, Jackson KL, et al. Design and implementation of a privacy preserving electronic health record linkage tool in Chicago. J Am Med Inform Assoc 2015;22:1072–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berchick ER, Hood E, Barnett JC. Health Insurance Coverage in the United States: 2017. Report No. P60–264. Washington, DC: US Government Printing Office, 2018. [Google Scholar]
- 14.Lin PI, Daley MF, Boone-Heinonen J, et al. Comparing prescribing and dispensing data of the PCORnet common data model within PCORnet Antibiotics and Childhood Growth Study. EGEMs 2019;7:11. [DOI] [PMC free article] [PubMed] [Google Scholar]