FYCO1 plays an important role in the kinesin-dependent microtubule plus-end-directed transport of autophagosomes. Here, the crystal structure of the RUN domain of FYCO1 has been determined.
Keywords: X-ray crystallography, autophagy, FYCO1, RUN domain, small GTPase binding
Abstract
FYCO1 is a multidomain adaptor protein that plays an important role in autophagy by mediating the kinesin-dependent microtubule plus-end-directed transport of autophagosomes. FYCO1 contains a RUN domain, which is hypothesized to function as a specific effector for members of the Ras superfamily of small GTPases, but its role has not been well characterized and its interaction partner(s) have not been identified. Here, the crystal structure of the FYCO1 RUN domain was determined at 1.3 Å resolution. The overall structure of the FYCO1 RUN domain was similar to those of previously reported RUN domains. Detailed structural comparisons with other RUN domains and docking studies suggested a possible interaction interface of the FYCO1 RUN domain with small GTPases of the Ras superfamily.
1. Introduction
Autophagy is an intracellular degradation process that is found in all eukaryotes from yeast to humans. During this process, unnecessary or damaged cytoplasmic components are delivered to the lysosome for degradation (Klionsky & Emr, 2000 ▸; Mizushima, 2007 ▸; Mizushima & Komatsu, 2011 ▸; Mackeh et al., 2013 ▸). Since autophagy is essential for the maintenance of intracellular homeostasis, its dysregulation is involved in various human diseases such as cancer, neurodegenerative diseases, infectious diseases and metabolic diseases (Mizushima & Komatsu, 2011 ▸; Jiang & Mizushima, 2014 ▸). In macroautophagy (hereafter referred to as autophagy), unique double-membrane vesicles called autophagosomes containing the target proteins or organelles are transported along microtubules to fuse with lysosomes and mature into autolysosomes, where the targets of autophagy are degraded (Klionsky & Emr, 2000 ▸; Mizushima, 2007 ▸; Mizushima & Komatsu, 2011 ▸; Mackeh et al., 2013 ▸). In this process, FYVE and coiled-coil domain-containing protein 1 (FYCO1), which has been identified as an autophagy adaptor protein, mediates microtubule plus-end-directed autophagosome transport by simultaneously interacting with kinesin motor proteins and autophagosomal membrane components such as Rab7, microtubule-associated protein 1 light chain 3 (LC3) and phosphatidylinositol 3-phosphate (PI3P) (Pankiv et al., 2010 ▸; Mackeh et al., 2013 ▸)
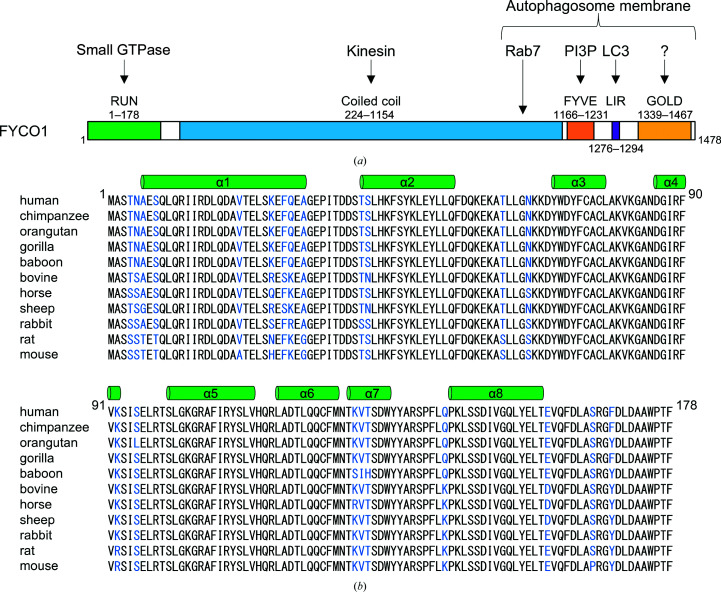
FYCO1 has an N-terminal RUN domain (an acronym for RPIP8, UNC-14 and NESCA), a central coiled-coil region, a C-terminal FYVE domain (an acronym for Fab1, YOTB/ZK632.12, Vac1 and EEA1), an LC3-interacting region (LIR) motif and a Golgi dynamics (GOLD) domain (Fig. 1 ▸ a). The central coiled-coil region, which is required for homodimerization of FYCO1, interacts with kinesin and Rab7. The FYVE domain and LIR motif are responsible for anchoring FYCO1 to the autophagosome membrane by interaction with PI3P and LC3, respectively (Gaullier et al., 1998 ▸; Birgisdottir et al., 2013 ▸). The GOLD domain is also thought to be involved in membrane anchoring (Anantharaman & Aravind, 2002 ▸). However, the function of the N-terminal RUN domain in autophagosome transport has not previously been defined. RUN domains are found in proteins which are involved in signalling pathways of small GTPases of the Ras superfamily. Moreover, specific interactions between the RUN domain and small GTPases have been reported (Callebaut et al., 2001 ▸): for example, between RPIP8 (Rap2-interacting protein 8) and Rap2 (Kukimoto-Niino et al., 2006 ▸), between Rab6IP1 (Rab6-interacting protein 1) and Rab6 (Recacha et al., 2009 ▸), between RUSC2 (RUN and SH3 domain-containing protein 2) and Rab35 (Fukuda et al., 2011 ▸) etc. Therefore, the RUN domain is thought to be a specific effector of small GTPases, but in the case of many of RUN domains their specific partners and their roles need to be determined.
Figure 1.
Domain structure and amino-acid sequence of FYCO1. (a) Schematic representation of the domain structure of human FYCO1. The RUN (RPIP8, UNC-14 and NESCA) domain, coiled-coil region, FYVE (Fab1, YOTB/ZK632.12, Vac1 and EEA1) domain, LIR (LC3-interacting region) motif and GOLD (Golgi dynamics) domain are shown in green, blue, orange, purple and yellow, respectively. (b) Sequence alignment of FYCO1 RUN domains from mammals. The secondary-structural elements are indicated above the alignments. Nonconserved residues are highlighted in blue.
To understand the structural features of the FYCO1 RUN domain, we determined its crystal structure. The overall structure of the FYCO1 RUN domain was similar to those of other RUN domains, but with some structural differences. In addition, detailed structural analyses and docking studies suggested a possible interaction interface of the FYCO1 RUN domain with small GTPases of the Ras superfamily.
2. Materials and methods
2.1. Preparation of the recombinant protein
The gene encoding the human FYCO1 RUN domain (residues 1–178) was inserted into the pGEX6P-1 expression vector (GE Healthcare) between the BamHI and EcoRI restriction sites. Escherichia coli BL21(DE3)pLysS cells were transformed with the vector and cultured at 37°C to a suitable cell density (OD600 of 0.7–0.8). Protein expression was then induced by the addition of 0.5 mM isopropyl β-d-1-thiogalactopyranoside and incubation was continued for 14 h at 18°C. The cells were collected by centrifugation and lysed by sonication in phosphate-buffered saline (PBS) with 2 mM dithiothreitol (DTT). The protein was purified from the clarified lysate using a Glutathione Sepharose 4B column (GE Healthcare; eluted with 20 mM reduced glutathione in PBS buffer) followed by GST-tag cleavage with 1:100(w:w) PreScission protease for 3 h at 4°C. Further purification was performed using a HiTrap Q (GE Healthcare) anion-exchange column (eluted with a 50–1000 mM NaCl gradient in 10 mM Tris–HCl pH 8.0) and a Superdex 200 (GE Healthcare) gel-filtration column (10 mM Tris–HCl pH 8.0, 150 mM NaCl). The protein was concentrated to approximately 11 mg ml−1 in 10 mM Tris–HCl pH 8.0, 150 mM NaCl buffer.
2.2. Crystallization, data collection and structure determination
Crystals of the FYCO1 RUN domain were grown at 4°C by the sitting-drop vapour-diffusion method by diluting the protein solution (11 mg ml−1) with an equal volume of reservoir solution [20%(w/v) PEG 3350, 180 mM triammonium citrate]. Mercury-derivatized FYCO1 RUN domain crystals were prepared by soaking the crystals in a solution consisting of 5 mM p-chloromercuriphenylsulfonic acid (PCMBS) in mother liquor [25%(w/v) PEG 3350, 180 mM triammonium citrate, 150 mM NaCl, 10 mM Tris–HCl pH 8.0] for two days.
X-ray diffraction data were collected on beamline PF-AR NE3A at the Photon Factory, Ibaraki, Japan under cryogenic conditions at 100 K. Crystals were equilibrated in a cryoprotectant solution consisting of mother liquor supplemented with 25% glycerol prior to flash-cooling. X-ray diffraction data were processed with HKL-2000 (Otwinowski & Minor, 1997 ▸).
The initial phases were obtained by the single isomorphous replacement with anomalous scattering method using phenix.autosol (Adams et al., 2002 ▸). The structure of the native crystal was solved by molecular replacement using MOLREP (Vagin & Teplyakov, 2010 ▸). The models were subjected to iterative cycles of manual model building using Coot (Emsley et al., 2010 ▸) and restrained refinement using REFMAC (Murshudov et al., 2011 ▸) (Table 1 ▸). The quality of the refined model was evaluated with MolProbity (Chen et al., 2010 ▸). Although the R and R free values of 0.212 and 0.240, respectively, were very high for a resolution of 1.3 Å (possibly owing to detector saturation of the low-angle reflections), the quality of the structural model was good enough to justify the conclusions in this study. The structural figures were prepared with Cuemol (http://www.cuemol.org). The coordinate and structure-factor data for the FYCO1 RUN domain have been deposited in the Protein Data Bank (PDB) as entry 7bqi.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Native | Mercury derivative | |
|---|---|---|
| Data collection | ||
| X-ray source | PF-AR NE3A | PF-AR NE3A |
| Wavelength (Å) | 1.0000 | 1.0085 |
| Space group | P41212 | P41212 |
| a, b, c (Å) | 48.1, 48.1, 142.7 | 48.3, 48.3, 142.8 |
| Resolution (Å) | 1.3 (1.32–1.30) | 1.6 (1.63–1.60) |
| R merge (%) | 9.2 (54.0) | 10.1 (97.4) |
| 〈I/σ(I)〉 | 41.6 (4.5) | 32.3 (2.7) |
| Completeness (%) | 90.7 (91.3) | 99.7 (100.0) |
| Multiplicity | 7.2 (7.2) | 7.0 (6.9) |
| Refinement | ||
| Resolution (Å) | 28.67–1.30 | |
| No. of reflections | 36472 | |
| R work/R free (%) | 21.2/24.0 | |
| No. of atoms | ||
| Protein | 1474 | |
| Water | 129 | |
| B factors (Å2) | ||
| Protein | 18.5 | |
| Water | 26.9 | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.014 | |
| Bond angles (°) | 1.91 | |
| Ramachandran plot | ||
| Favoured (%) | 99.3 | |
| Allowed (%) | 0.7 | |
| Outliers (%) | 0.0 | |
2.3. Docking calculation
Docking calculations were performed by the ZDOCK server (version 3.0.2; Pierce et al., 2011 ▸). For docking calculations, one molecule of the FYCO1 RUN domain and of a GTP-bound small GTPase, including H-Ras (PDB entry 1ctq; Scheidig et al., 1999 ▸), K-Ras (PDB entry 6god; Cruz-Migoni et al., 2019 ▸), Rab6 (PDB entry 1yzq; Eathiraj et al., 2005 ▸), Rab7 (PDB entry 1t91; Wu et al., 2005 ▸), RhoA (PDB entry 1a2b; Ihara et al., 1998 ▸), Rac1 (PDB entry 1mh1; Hirshberg et al., 1997 ▸), Rap2 (PDB entry 2rap; Cherfils et al., 1997 ▸), Arf6 (PDB entry 2j5x; Pasqualato et al., 2001 ▸) and ARL6 (PDB entry 2h57; Structural Genomics Consortium, unpublished work), were used as the receptor and ligand, respectively. Additional parameters such as binding or nonbinding site residues were not set.
3. Results
3.1. Structure determination and overall structure of the FYCO1 RUN domain
The RUN domain is thought to be a specific effector of small GTPases because RUN domains are found in proteins linked to the signalling pathway of small GTPases, and specific interactions between RUN domains and small GTPases have been reported (Callebaut et al., 2001 ▸; Fukuda et al., 2011 ▸; Lin et al., 2019 ▸; Recacha et al., 2009 ▸). Although FYCO1 has a RUN domain in its N-terminus (Fig. 1 ▸ a), its function and interaction partner are unknown. An amino-acid sequence alignment of the FYCO1 RUN domains from 11 mammals shows that the RUN domains of FYCO1 are highly conserved (Fig. 1 ▸ b): 156 of 178 N-terminal residues (88%) are conserved among the 11 species, suggesting that the RUN domain of FYCO1 may have an important role.
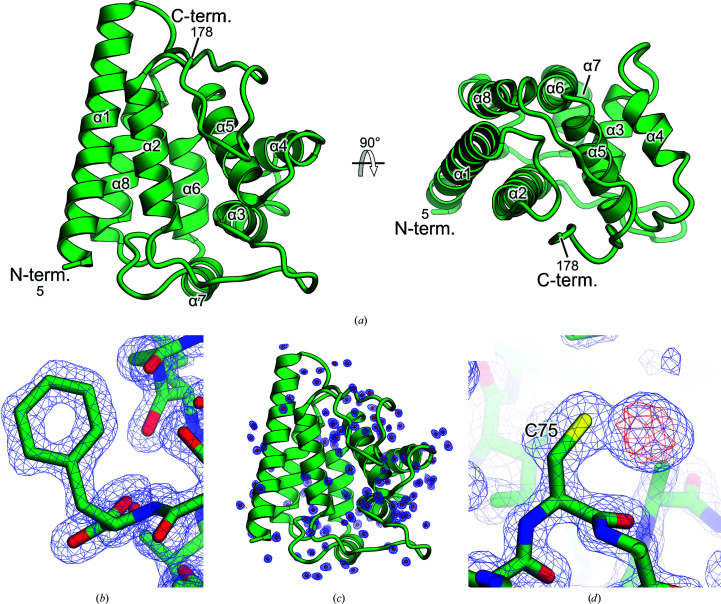
To better understand the structural features of the FYCO1 RUN domain, we crystallized the human FYCO1 RUN domain (residues 1–178) and determined the structure at 1.3 Å resolution (Fig. 2 ▸ a, Table 1 ▸). The crystal belonged to space group P41212, with unit-cell parameters a = 48.1, b = 48.1, c = 142.7 Å. The initial phases were determined by the single isomorphous replacement with anomalous scattering method using mercury-derivatized crystals (Table 1 ▸). The asymmetric unit of the crystals contained one molecule of the RUN domain (residues 5–178). The RUN domain of FYCO1 adopts a compact globular shape composed of eight α-helices (α1–α8), with dimensions of approximately 30 × 40 × 50 Å (Figs. 1 ▸ b and 2 ▸ a). Helices α1, α5 and α8 are arranged in a parallel orientation, and helices α2 and α6 are arranged in an antiparallel orientation, while helices α3, α4 and α7 are arranged in a vertical orientation. Since the structure was determined at the high resolution of 1.3 Å, the centres of the aromatic residues and numerous water molecules were observed in the electron-density map (Figs. 2 ▸ b and 2 ▸ c). In the anomalous difference Fourier map of the mercury-derivatized crystal, a single clear density corresponding to the Hg atom coordinated to Cys75 was observed (Fig. 2 ▸ d).
Figure 2.
Crystal structure of the FYCO1 RUN domain. (a) Overall structure of the FYCO1 RUN domain. The N- and C-termini and the structural elements are labelled. (b) Electron-density map of the phenylalanine residues in the native crystal. The 2F o − F c difference electron-density map is contoured at the 1.5σ level with a blue mesh. The hole in the aromatic ring is clearly visible in the density map. (c) Electron-density map of water molecules. The 2F o − F c difference electron-density map is contoured at the 1.0σ level with a blue mesh. (d) Electron density of the mercury-derivatized crystal at 1.6 Å resolution. The 2F o − F c difference electron-density map is contoured at the 1.5σ level with a blue mesh. The anomalous difference Fourier map is contoured at the 5.0σ level with a red mesh.
3.2. Structural comparison of RUN domains
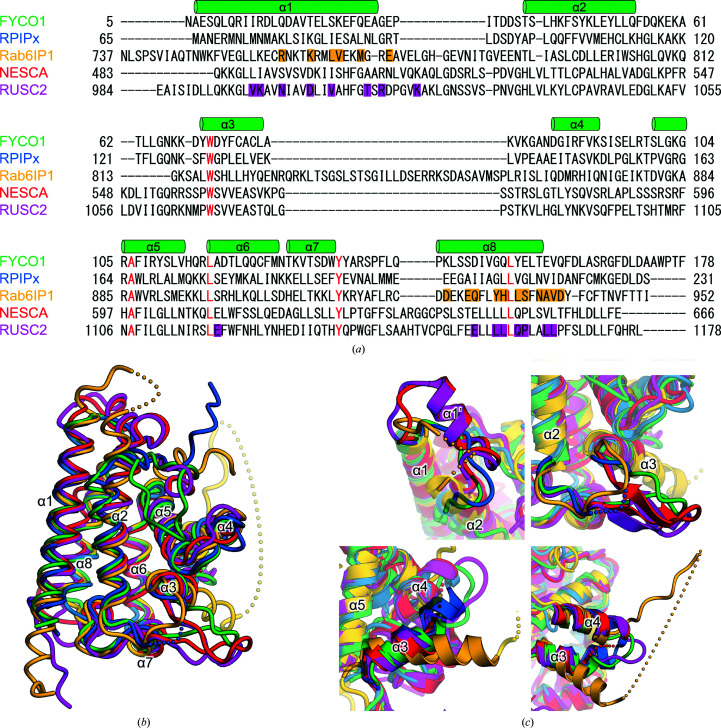
There are four RUN domains for which crystal structures have been reported: the RUN domain of Rap2-interacting protein x (RPIPx), which is a specific effector of Rap2 (Kukimoto-Niino et al., 2006 ▸), the RUN1 and subsequent PLAT domains of Rab6-interacting protein 1 (Rab6IP1) in complex with Rab6 (Recacha et al., 2009 ▸), the RUN domain of a new signalling adapter protein in the NGF pathway containing an SH3 domain at the carboxyl-terminus (NESCA; Sun et al., 2012 ▸) and the RUN domain of RUN and SH3 domain-containing protein 2 (RUSC2) in complex with Rab35 (Lin et al., 2019 ▸). The sequence identities among the RUN domains of FYCO1, RPIPx and Rab6IP1 were 13–21%; only NESCA and RUSC2 had a high sequence identity of 51%. The five residues corresponding to Trp71, Ala106, Leu117, Tyr135 and Leu154 of FYCO1 were strictly conserved (Fig. 3 ▸ a). These residues are also highly conserved in other RUN domains, suggesting their important role in the folding of the RUN domain (Callebaut et al., 2001 ▸). Next, we superimposed the structural models of the five RUN domains (Fig. 3 ▸ b). The overall structures of these RUN domains were similar, with root-mean-square deviation (r.m.s.d.) values of 1.8, 1.7, 2.2 and 2.0 Å between FYCO1 RUN and RPIPx (PDB entry 2dwg), Rab6IP1 (PDB entry 3cwz), NESCA (PDB entry 4giw) and RUSC2 (PDB entry 6if2), respectively. Several structural deviations were observed among the five RUN domains. NESCA and RUSC2 had an additional α-helix (α1′) inserted between helices α1 and α2, while the corresponding regions of FYCO1 and RPIPx were shorter than those of NESCA and RUSC2, and that of Rab6IP1 was disordered in the structure (Figs. 3 ▸ a and 3 ▸ c). The region between α2 and α3 of Rab6IP1 was slightly shorter than those of the other four RUN domains, and this region formed β-hairpin structures in NESCA and RUSC2 (Figs. 3 ▸ a and 3 ▸ c). The α3 helices of RPIPx and Rab6IP1 were rotated approximately 15° and −20°, respectively, relative to those of FYCO1, NESCA and RUSC2 (Fig. 3 ▸ c). The loop region between α3 and α4 of Rab6IP1 was about 25 residues longer than those of other RUN domains and was mostly disordered (Figs. 3 ▸ a and 3 ▸ c).
Figure 3.
Sequence and structural comparisons of RUN domains. (a) Sequence alignment of RUN domains. Secondary-structural elements are indicated above the alignments. The conserved residues are highlighted in red. The residues of Rab6IP1 that interact with Rab6 and those of RUSC2 that interact with Rab35 are highlighted in yellow and purple, respectively. (b) Superposition of RUN domains. The RUN domains of FYCO1, RPIPx (Kukimoto-Niino et al., 2006 ▸), Rab6IP1 (Recacha et al., 2009 ▸), NESCA (Sun et al., 2012 ▸) and RUSC2 (Lin et al., 2019 ▸) are shown in green, blue, yellow, red and purple, respectively. (c) Structural differences among the five RUN domains. Secondary-structural elements are indicated.
3.3. Possible interaction surface for a small GTPase in the FYCO1 RUN domain
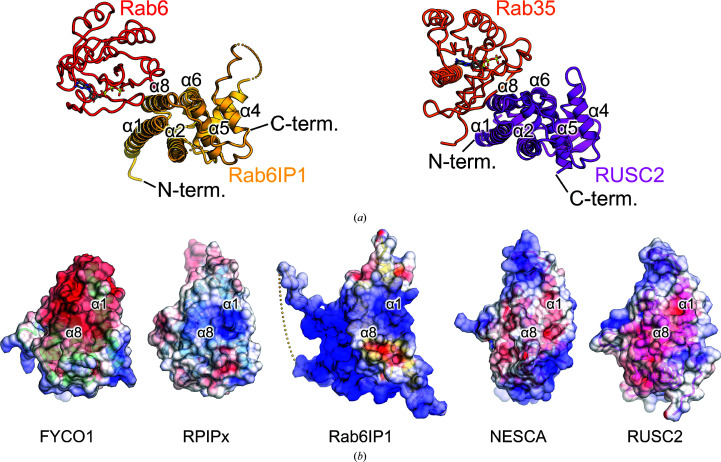
RUN domains are generally accepted to interact and function with small GTPases of the Ras superfamily (Callebaut et al., 2001 ▸; Fukuda et al., 2011 ▸). The available structures of RUN domain–Ras superfamily protein complexes are the Rab6IP1–Rab6 structure and the RUSC2–Rab35 structure (Recacha et al., 2009 ▸; Lin et al., 2019 ▸). Both Rab6IP1 and RUSC2 utilize the α1 and α8 helices for interaction with the Rab protein, although the orientations of the Rab proteins with respect to the two RUN domains were different (Figs. 3 ▸ a and 4 ▸ a). This interaction may occur in other RUN domains, including the FYCO1 RUN domain. However, most of the residues involved in the interactions with the small GTPases in the Rab6IP1–Rab6 and RUSC2–Rab35 structures are not conserved among the five RUN domains (Fig. 3 ▸ a). In addition, the distributions of electrostatic surface potential in these RUN domains were also not conserved (Fig. 4 ▸ b). A docking model of NESCA RUN and H-Ras has been proposed in which NESCA interacts with the small GTPase via the region between α2 and α3 and the loop between α4 and α5, in a different manner to that in Rab6IP1 or RUSC2 (Sun et al., 2012 ▸; Recacha et al., 2009 ▸; Lin et al., 2019 ▸). From these observations, it is possible that the RUN domain of FYCO1 utilizes a different region to interact with a small GTPase.
Figure 4.
Interaction surface of the RUN–small GTPase complex. (a) Crystal structures of the Rab6IP1–Rab6 and RUSC2–Rab35 complexes (Recacha et al., 2009 ▸; Lin et al., 2019 ▸). Rab6IP1, Rab6, RUSC2 and Rab35 are shown in yellow, red, purple and orange, respectively. (b) Electrostatic surface potentials of the five RUN domains. The electrostatic surface potential is coloured from −8 kT e−1 (red) to +8 kT e−1 (blue).
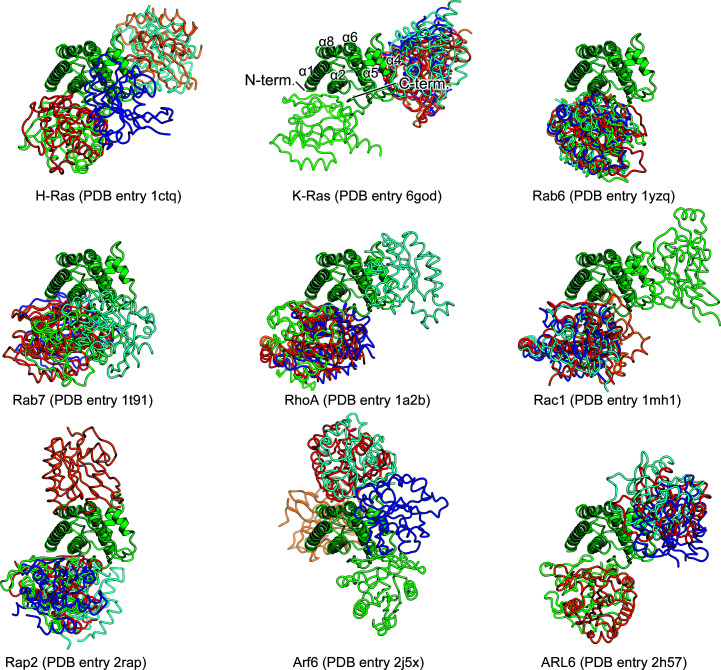
We performed docking studies to determine the potential interface between the FYCO1 RUN domain and a small GTPase using ZDOCK version 3.0.2 (Pierce et al., 2011 ▸). Since a specific small GTPase for the FYCO1 RUN domain has not been identified, we used nine crystal structures of small GTPases of the Ras superfamily in their GTP-bound forms as input models for the docking calculations, including H-Ras (PDB entry 1ctq; Scheidig et al., 1999 ▸), K-Ras (PDB entry 6god; Cruz-Migoni et al., 2019 ▸), Rab6 (PDB entry 1yzq; Eathiraj et al., 2005 ▸), Rab7 (PDB entry 1t91; Wu et al., 2005 ▸), RhoA (PDB entry 1a2b; Ihara et al., 1998 ▸), Rac1 (PDB entry 1mh1; Hirshberg et al., 1997 ▸), Rap2 (PDB entry 2rap; Cherfils et al., 1997 ▸), Arf6 (PDB entry 2j5x; Pasqualato et al., 2001 ▸) and ARL6 (PDB entry 2h57; Structural Genomics Consortium, unpublished work). We found that more than half of the predicted docking models, regardless of the small GTPase subfamily, contained the same interaction site on the FYCO1 RUN domain formed by the α1 and α2 helices and the C-terminal region after the α8 helix (Fig. 5 ▸). Although the orientations of the small GTPases were not defined, the interfaces calculated could be representative of the interaction between the FYCO1 RUN domain and small GTPases.
Figure 5.
Docking models of the FYCO1 RUN domain and small GTPases. The docking models were calculated by ZDOCK (Pierce et al., 2011 ▸). The FYCO1 RUN domain is shown as a ribbon model in green. The top five models of small GTPases from each calculation are shown as tube models in red, orange, light green, cyan and blue. The PDB entries used as input models for small GTPases are shown under the models.
4. Discussion
In this study, we determined the crystal structure of the human FYCO1 RUN domain at 1.3 Å resolution. The overall structure was very similar to those previously determined for RUN domains despite low sequence homology. Our detailed structural analyses, including docking studies, suggest a possible interaction interface with small GTPases in the FYCO1 RUN domain. Moreover, it has been suggested that not only small GTPases but also other functionally related proteins interact with RUN domains. For example, the RUN domain of NESCA interacts with TrkA as well as with the small GTPase H-Ras (Sun et al., 2012 ▸). This could also be the case for FYCO1. In addition, FYCO1 binds kinesin and autophagosome membrane-localized molecules such as Rab7, LC3 and PI3P. Currently, structural information about FYCO1 is limited to the RUN domain in this study and the LIR motif–LC3 complex (Olsvik et al., 2015 ▸; Cheng et al., 2016 ▸; Sakurai et al., 2017 ▸). Additional structural analyses of FYCO1 in complexes with Rab7, kinesin and PI3P would provide further insight into the function of FYCO1 and autophagosome transport mediated by FYCO1.
Supplementary Material
PDB reference: FYCO1 RUN domain, 7bqi
Acknowledgments
We would like to thank Dr Taisuke Tomita for providing the cDNA of human FYCO1. We also thank the beamline staff members at the Photon Factory for their assistance with data collection. The authors declare no competing financial interests. Author contributions are as follows. SS and UO designed the experiments. SS prepared the recombinant proteins and performed the structural analyses. UO and TS supervised the project. SS, TS and UO wrote the paper.
Funding Statement
This work was funded by Ministry of Education, Culture, Sports, Science and Technology grants 26711002 and 19H00976. Core Research for Evolutional Science and Technology grant . Japan Science and Technology Agency grant . Mochida Memorial Foundation for Medical and Pharmaceutical Research grant . Daiichi Sankyo Foundation of Life Science grant . Uehara Memorial Foundation grant . Naito Foundation grant .
References
- Adams, P. D., Grosse-Kunstleve, R. W., Hung, L.-W., Ioerger, T. R., McCoy, A. J., Moriarty, N. W., Read, R. J., Sacchettini, J. C., Sauter, N. K. & Terwilliger, T. C. (2002). Acta Cryst. D58, 1948–1954. [DOI] [PubMed]
- Anantharaman, V. & Aravind, L. (2002). Genome Biol. 3, research0023. [DOI] [PMC free article] [PubMed]
- Birgisdottir, Å. B., Lamark, T. & Johansen, T. (2013). J. Cell Sci. 126, 3237–3247. [DOI] [PubMed]
- Callebaut, I., de Gunzburg, J., Goud, B. & Mornon, J.-P. (2001). Trends Biochem. Sci. 26, 79–83. [DOI] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Cheng, X., Wang, Y., Gong, Y., Li, F., Guo, Y., Hu, S., Liu, J. & Pan, L. (2016). Autophagy, 12, 1330–1339. [DOI] [PMC free article] [PubMed]
- Cherfils, J., Ménétrey, J., Le Bras, G., Janoueix-Lerosey, I., de Gunzburg, J., Garel, J.-R. & Auzat, I. (1997). EMBO J. 16, 5582–5591. [DOI] [PMC free article] [PubMed]
- Cruz-Migoni, A., Canning, P., Quevedo, C. E., Bataille, C. J. R., Bery, N., Miller, A., Russell, A. J., Phillips, S. E. V., Carr, S. B. & Rabbitts, T. H. (2019). Proc. Natl Acad. Sci. USA, 116, 2545–2550. [DOI] [PMC free article] [PubMed]
- Eathiraj, S., Pan, X., Ritacco, C. & Lambright, D. G. (2005). Nature, 436, 415–419. [DOI] [PMC free article] [PubMed]
- Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. (2010). Acta Cryst. D66, 486–501. [DOI] [PMC free article] [PubMed]
- Fukuda, M., Kobayashi, H., Ishibashi, K. & Ohbayashi, N. (2011). Cell Struct. Funct. 36, 155–170. [DOI] [PubMed]
- Gaullier, J.-M., Simonsen, A., D’Arrigo, A., Bremnes, B., Stenmark, H. & Aasland, R. (1998). Nature, 394, 432–433. [DOI] [PubMed]
- Hirshberg, M., Stockley, R. W., Dodson, G. & Webb, M. R. (1997). Nat. Struct. Mol. Biol. 4, 147–152. [DOI] [PubMed]
- Ihara, K., Muraguchi, S., Kato, M., Shimizu, T., Shirakawa, M., Kuroda, S., Kaibuchi, K. & Hakoshima, T. (1998). J. Biol. Chem. 273, 9656–9666. [DOI] [PubMed]
- Jiang, P. & Mizushima, N. (2014). Cell Res. 24, 69–79. [DOI] [PMC free article] [PubMed]
- Klionsky, D. J. & Emr, S. D. (2000). Science, 290, 1717–1721. [DOI] [PMC free article] [PubMed]
- Kukimoto-Niino, M., Takagi, T., Akasaka, R., Murayama, K., Uchikubo-Kamo, T., Terada, T., Inoue, M., Watanabe, S., Tanaka, A., Hayashizaki, Y., Kigawa, T., Shirouzu, M. & Yokoyama, S. (2006). J. Biol. Chem. 281, 31843–31853. [DOI] [PubMed]
- Lin, L., Shi, Y., Wang, M., Wang, C., Zhu, J. & Zhang, R. (2019). Structure, 27, 729–740. [DOI] [PubMed]
- Mackeh, R., Perdiz, D., Lorin, S., Codogno, P. & Poüs, C. (2013). J. Cell Sci. 126, 1071–1080. [DOI] [PubMed]
- Mizushima, N. (2007). Genes Dev. 21, 2861–2873. [DOI] [PubMed]
- Mizushima, N. & Komatsu, M. (2011). Cell, 147, 728–741. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Olsvik, H. L., Lamark, T., Takagi, K., Larsen, K. B., Evjen, G., Øvervatn, A., Mizushima, T. & Johansen, T. (2015). J. Biol. Chem. 290, 29361–29374. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Pankiv, S., Alemu, E. A., Brech, A., Bruun, J. A., Lamark, T., Overvatn, A., Bjørkøy, G. & Johansen, T. (2010). J. Cell Biol. 188, 253–269. [DOI] [PMC free article] [PubMed]
- Pasqualato, S., Ménétrey, J., Franco, M. & Cherfils, J. (2001). EMBO Rep. 2, 234–238. [DOI] [PMC free article] [PubMed]
- Pierce, B. G., Hourai, Y. & Weng, Z. (2011). PLoS One, 6, e24657. [DOI] [PMC free article] [PubMed]
- Recacha, R., Boulet, A., Jollivet, F., Monier, S., Houdusse, A., Goud, B. & Khan, A. R. (2009). Structure, 17, 21–30. [DOI] [PubMed]
- Sakurai, S., Tomita, T., Shimizu, T. & Ohto, U. (2017). Acta Cryst. F73, 130–137. [DOI] [PMC free article] [PubMed]
- Scheidig, A. J., Burmester, C. & Goody, R. S. (1999). Structure, 7, 1311–1324. [DOI] [PubMed]
- Sun, Q., Han, C., Liu, L., Wang, Y., Deng, H., Bai, L. & Jiang, T. (2012). Protein Cell, 3, 609–617. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Wu, M., Wang, T., Loh, E., Hong, W. & Song, H. (2005). EMBO J. 24, 1491–1501. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: FYCO1 RUN domain, 7bqi