SUMMARY
Lipid droplets (LDs) store lipids for energy and are central to cellular lipid homeostasis. The mechanisms coordinating lipid storage in LDs with cellular metabolism are unclear but relevant to obesity-related diseases. Here we utilized genome-wide screening to identify genes that modulate lipid storage in macrophages, a cell type involved in metabolic diseases. Among ~550 identified screen hits is MLX, a basic helix-loop-helix/leucine-zipper transcription factor that regulates metabolic processes. We show that MLX and glucose-sensing family members MLXIP/MondoA and MLXIPL/ChREBP bind LDs via C-terminal amphipathic helices. When LDs accumulate in cells, these transcription factors bind to LDs, reducing their availability for transcriptional activity and attenuating the response to glucose. Conversely, the absence of LDs results in hyperactivation of MLX target genes. Our findings uncover a paradigm for a lipid storage response, in which binding of MLX transcription factors to LD surfaces adjusts the expression of metabolic genes to lipid storage levels.
Keywords: ChREBP, glucose, lipid droplets, metabolism, MLX, MLXIP, MLXIPL, MondoA, MondoB, transcription factor
Graphical Abstract
eTOC Blurb
Mejhert et al. identify genetic determinants of lipid storage in macrophages by integrating a global genome perturbation screen with lipid droplet proteomics. Among hits that localize to lipid droplets is the transcription factor MLX. By binding to lipid droplets, MLX coordinates lipid storage with metabolic gene expression regulation.
INTRODUCTION
Lipids are central to the physiology of life, as building blocks of cell membranes, signaling molecules, and reservoirs of metabolic energy. Control of lipid metabolism occurs at many levels, ranging from transcriptional regulation of enzymes to post-transcriptional modulation of their activities. To buffer fluctuations in the need and availability of lipids, cells evolved storage organelles for them, called lipid droplets (LDs) (Henne et al., 2018; Olzmann and Carvalho, 2018; Walther et al., 2017). LDs are dynamic organelles that change their numbers and size, depending on the metabolic state of the cell with excess lipid stored and mobilized as needed.
How the availability of lipid stores in LDs is integrated with the regulation of cellular energy metabolism is mostly unknown. This is an important question, however, not just for cellular metabolism, but because overwhelming the capacity of cells to store lipids is a root-cause for many metabolic diseases, including cardiovascular and liver disease (Krahmer et al., 2013). For example, storage of lipids in LDs serves to sequester potentially toxic lipid intermediates, thereby protecting cells from the consequences of lipotoxicity, which include endoplasmic reticulum stress, inflammation, and cell death (Chitraju et al., 2017; Listenberger et al., 2003). This function includes the temporary storage of excess cholesterol as cholesterol esters (CEs), as found in macrophage foam cells (Fazio et al., 2001), or of fatty acids and fatty acid derivatives, such as diacylglycerol, as triacylglycerol (TG) in hepatocytes (Villanueva et al., 2009). LDs are thus integral to the cellular mechanisms that maintain lipid homeostasis.
Among cell types, macrophages appear to be crucial for the development of diseases caused by lipid overload. In atherosclerosis, macrophages in the arterial intima take up cholesterol-rich lipoproteins and temporarily store and detoxify cholesterol as CEs in LDs, thereby becoming foam cells, before free cholesterol can be removed via transfer to high-density lipoproteins (Moore et al., 2013; Tabas and Bornfeldt, 2016). Overwhelming the capacity of macrophages to recycle cholesterol is thought to be a critical step in development of vulnerable atherosclerotic plaques (Accad et al., 2000; Fazio et al., 2001). Similarly, overwhelming macrophages in adipose tissue or liver with fatty acids contributes to development of pathologies, such as insulin resistance and non-alcoholic fatty liver disease (Lee et al., 2018; Rosen and Spiegelman, 2014). Consistent with a critical role of macrophage LDs in disease development, increasing the capacity to detoxify fatty acids by overexpressing the TG-synthesis enzyme DGAT1 in macrophages protects mice from diet-induced insulin resistance (Koliwad et al., 2010).
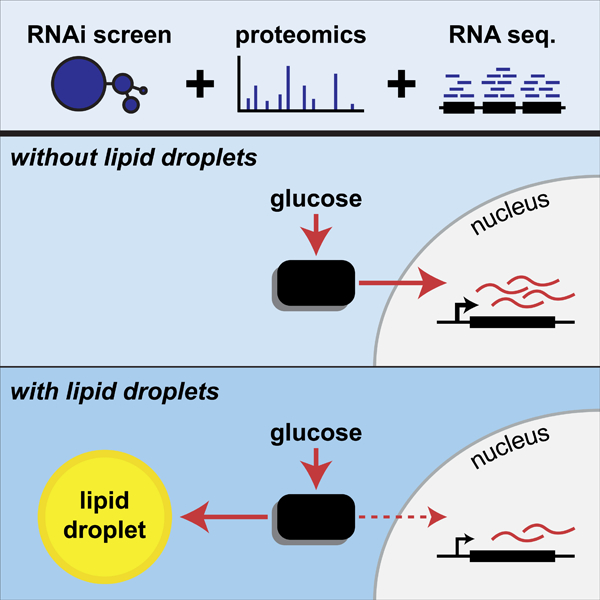
To unravel the molecular processes of cellular adaptation and dysfunction due to excess lipids, we interrogated LD formation in human macrophages that were cultured in the presence of excess lipids provided by lipoproteins. We utilized a systems-type approach to analyze the response, including studies of gene expression, subcellular proteomes, and genetic perturbations via RNA interference (RNAi) under lipid-storage conditions. Of the ~550 genes we identified that influence lipid storage, we unexpectedly found that the MLX transcription factors, MLX (Max-like protein X), MLXIP (MLX interacting protein, also known as MondoA) and MLXIPL (MLX interacting protein like, also known as ChREBP), which are key regulators of multiple metabolic adaptations to glucose, localize to LDs and are required for normal LD biology. Our mechanistic studies reveal an unexpected paradigm, in which binding of these transcription factors to the LD surface modulates MLX-regulated gene expression, adjusting the expression of metabolic genes in response to lipid storage. Thus, in addition to discovering genetic determinants of lipid storage, our screen uncovers a novel mechanism connecting LD binding of transcription factors with metabolic transcriptional regulation.
RESULTS
Systematic Classification of Regulators of Lipid Storage in Human Macrophages
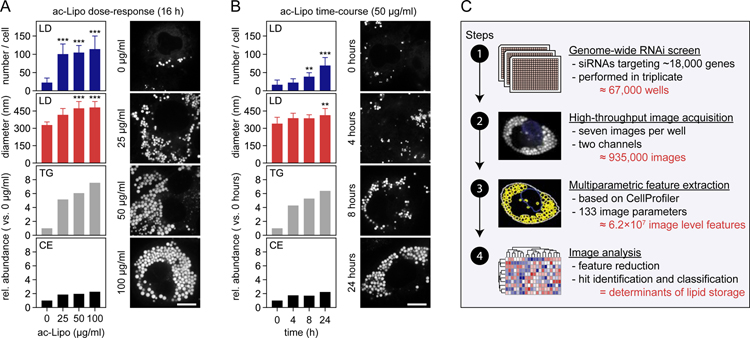
We sought to comprehensively identify the determinants of lipid storage in human cells through a combination of systematic approaches. We selected human macrophages for these studies because they store a variety of lipids, including CEs and TGs, and because they are central to the pathogenesis of metabolic diseases, including atherosclerosis where they form foam cells, a cell type which is characterized by increased lipid storage and a pro-inflammatory phenotype (Moore et al., 2013). In agreement with this, human THP-1 macrophages formed many LDs containing TGs and CEs when incubated with acetylated apolipoprotein B-containing lipoproteins (ac-Lipo) that contained both neutral lipids (Figure 1A–B).
Figure 1. Systematic Identification of Regulators of Lipid Storage in Human Macrophages.
(A–B) Induction of lipid storage in macrophages by acetylated apolipoprotein B-containing lipoproteins. Macrophages were incubated in the absence/presence of ac-Lipo (0–100 µg/mL) for 0–24 hours followed by assessment of LD formation using BODIPY staining and quantification of lipid composition by TLC. Results from one representative experiment are shown for TLC analyses and quantifications of LDs were based on multiple cells (n=8–19). Results were evaluated using one-way non-parametric ANOVA (Kruskal-Wallis followed by Dunn’s multiple comparisons test). Scale bar, 10 µm.
(C) Overview of the experimental and computational design of the study. Based on four major steps, regulators of lipid storage in macrophages were identified.
Abbreviations: ac-Lipo, acetylated apolipoprotein B-containing lipoprotein; CE, cholesterol ester; TG, triacylglycerol.
To systematically identify determinants of lipid storage in this system, we adapted it for a genome-wide perturbation screen by developing an imaging strategy to measure LDs in cells and by optimizing each step of a screening pipeline, including differentiation of monocytes into macrophages, transfection efficiency, and induction of lipid storage (Figure S1A–C). We then tested our platform by depleting transcripts from more than a dozen genes implicated in lipid storage by RNAi and measuring the effects on LDs with BODIPY staining and confocal microscopy, followed by multiparametric feature extraction. These analyses confirmed that our pipeline could detect and distinguish LD phenotypes with great precision, as shown for instance for knockdown of BSCL2 (encoding seipin), which yields cells with large LDs (Figure S1D–E) (Fei et al., 2008; Szymanski et al., 2007; Wang et al., 2016).
Having developed a robust platform (summarized in Figure 1C), we systematically screened the genome for determinants of lipid storage. Specifically, we screened ~18,000 genes by using pools of four siRNA duplexes per gene (in triplicate) and confirmed our results with independent replicate screens, thereby generating nearly one million images for analysis. From these images, we extracted 133 parameters, calculated robust z-scores for each of them, and determined their reproducibility and redundancy with other extracted parameters. Through such analyses, we generated a final set of 21 high-confidence image parameters, which together described five dimensions of lipid storage: the number, size, shape, intensity, and dispersion of LDs (Figure S2A–C). Utilizing these most informative parameters, we identified 558 hits with altered LDs (Table S1). To validate the results of the screen, we independently re-screened roughly 10% of these hits (targeting 51 genes) with four different siRNAs and found excellent reproducibility (Figure S2D).
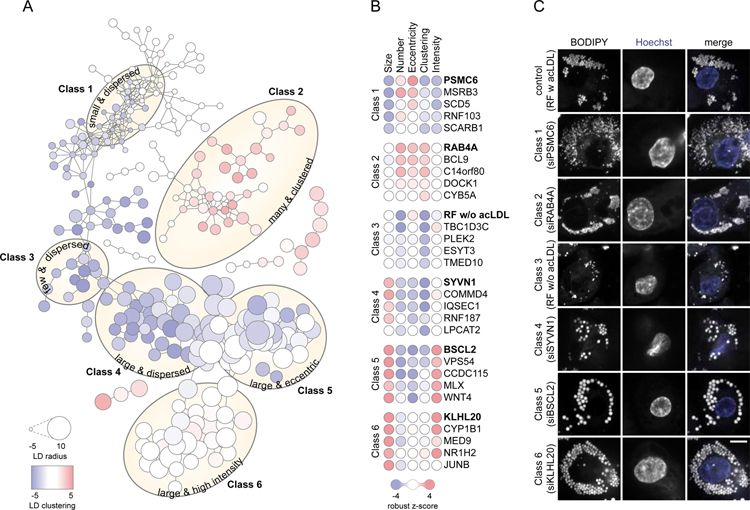
To begin analyzing the results of the screen, we categorized the hits into six major phenotypic classes, based on similarity scores (Figure 2A–C, Data S1). Class 1 screen hits were characterized by small and dispersed LDs and were enriched in subunits of the proteasome. Class 2 hits exhibited many and clustered LDs, and among these hits were two open-reading frames, C14orf80 and C22orf31, with uncharacterized functions. Class 3 hits clustered with controls that were not incubated with ac-Lipo and were characterized by few and dispersed LDs. This class included proteins involved in vesicular (e.g., TMED10) and non-vesicular (e.g., ESYT3) transport. Classes 4, 5, and 6 each contained hits with large LDs. These classes were separated from each other due to differences in the localization of LDs within the cell (e.g., Class 4 were large and dispersed), the shape of LDs (Class 5 were large and eccentric), or BODIPY-staining intensity of LDs (Class 6 with large and high intensity). These three classes included genes encoding proteins of diverse function, including for instance transcription factors [e.g., MLX (Class 5) and NR1H2 (Class 6)], E3 ligases [e.g., SYVN1 (Class 4) and KLHL20 (Class 6)], and lipid-modifying enzymes [e.g., LPCAT2 (Class 4) and CYP1B1 (Class 6)]. Collectively, our screen yielded many previously unknown genes that modify lipid storage in LDs and provides a comprehensive set for human macrophages.
Figure 2. Genetic Determinants of Lipid Storage Belong to Six Major Classes.
(A) RNAi screen hits cluster into six major classes. Based on pair-wise similarities derived from Spearman’s rank correlation, RNAi screen hits were interconnected by edges into major classes as indicated by yellow ellipses. Each node represents a hit and its size and color are proportional to the robust z-score of the hit for LD radius and LD clustering, respectively. Data are also available as an interactive tree view file, see Data S1.
(B–C) Representative hits for classes 1–6. (B) For each class, five hits are visualized as nodes where the color of each circle is proportional to the robust z-score of the hit. (C) Confocal images displaying LDs (stained by BODIPY) and nuclei (stained by Hoechst) of a representative hit for each class. Scale bar, 5 µm.
Identification of LD Proteins in Human THP-1 Macrophages
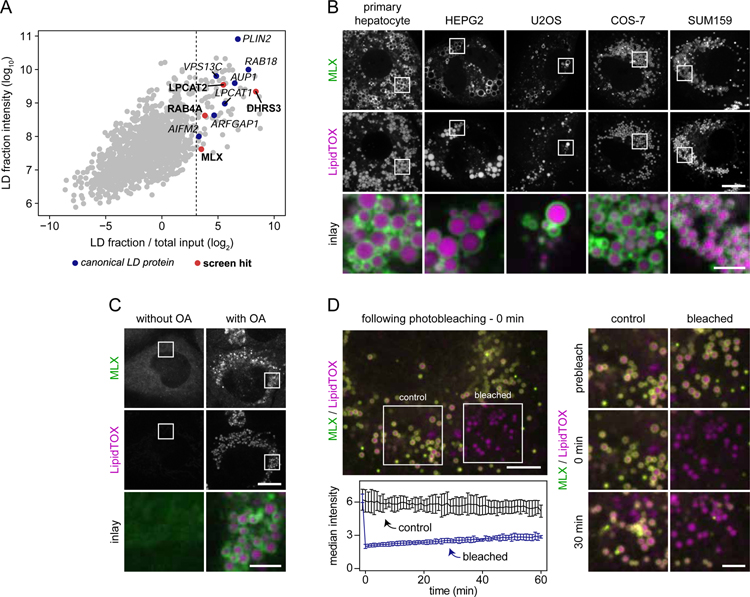
To identify genes from our screen that are directly involved in LD biology, we determined which of the hits encode proteins that are enriched at LDs. We purified LD fractions from THP-1 cells through a series of centrifugation steps, including density-gradient centrifugation. Western blot analyses of isolated fractions confirmed the separation of LDs from other major cellular organelles (Figure S3A). To determine proteins that were enriched in the LD fraction, we analyzed the total proteome and the LD-enriched fraction, using high resolution mass-spectrometry-based proteomics. Figure 3A shows the enrichment of proteins plotted against their abundance in the LD fraction. Proteins that were abundant and strongly enriched in the LD fraction included many well-established LD proteins, including PLIN2 (also known as ADRP, (Brasaemle et al., 1997), RAB18 (Martin et al., 2005; Ozeki et al., 2005), and LPCAT1 (Moessinger et al., 2011), validating our approach. By setting a threshold of 99% confidence for LD enrichment, we identified 75 proteins as candidate bona fide LD proteins (Table S2).
Figure 3. Transcription Factor MLX Is an LD Protein.
(A) Identification of macrophage LD proteins in THP-1 macrophages. Proteins localized to LDs in THP-1 macrophages incubated with oleic acid (0.5 mM) for 6 hours were identified by mass spectrometry. For each protein, intensities in whole-cell lysate and LD fractions were compared. Known LD proteins were used to calculate a fold-change cut-off (for LD enrichment) based on the 99% confidence interval of their distribution (lower boundary is indicated by the dashed line). Results from one representative experiment are shown.
(B) MLX localizes to LDs in multiple cell types. Representative confocal images of mouse primary hepatocytes, HEPG2, U2OS, COS-7, and SUM159 cells transfected with GFP-tagged MLX. Cells were incubated with oleic acid (0.5 mM) for 1 day, stained with LipidTOX Deep Red and thereafter imaged. Scale bars, 10 µm and 2.5 µm (inlay).
(C) Endogenously tagged MLX targets to LDs. MLX was endogenously tagged C-terminally with EGFP using CRISPR/Cas9-mediated engineering in SUM159 cells. Representative images of the localization of MLX in live cells incubated in the presence or absence of oleic acid (0.5 mM) for 1 day are shown. LDs were stained prior to imaging using LipidTOX Deep Red. Scale bars, 10 µm and 2.5 µm (inlay).
(D) Stable binding of MLX to LDs. After transfection with EGFP-tagged MLX, oleation (0.5 mM) for 1 day and staining of LDs with LipidTOX Deep Red, LD-binding properties of MLX were tested using fluorescence recovery after photobleaching. Representative examples of a cell and inlay images are shown in the upper left and right panels, respectively. Recovery kinetics of MLX was quantified from three independent experiments as highlighted in the lower left panel. Scale bars, 5 µm and 2.5 µm (inlay). Abbreviation: OA, oleic acid.
Of the identified LD proteins, four were associated with an LD phenotype in our RNAi screen (Figure 3A). Among these, LPCAT2, an enzyme generating phosphatidylcholine from lysophosphatidylcholine and acyl-CoA, and DHRS3, a retinaldehyde reductase, have been studied in the context of LDs (Deisenroth et al., 2011; Moessinger et al., 2011; Pataki et al., 2018). In contrast, although many Rab GTPases family members have been detected on LDs (Bersuker et al., 2018; Liu et al., 2004), RAB4A, a GTPase required for rapid recycling of proteins from early endosomes to the cell surface (van der Sluijs et al., 1992), has not been associated previously with LD biology.
The fourth protein, MLX, was unexpected as an LD protein. MLX is a transcription factor of the basic helix-loop-helix leucine-zipper family and is thought to be a cytoplasmic protein that binds mitochondria (Billin et al., 1999, 2000; Meroni et al., 2000; Sans et al., 2006). MLX forms a complex and is the obligate partner with either MLXIP or MLXIPL, which contain glucose-sensing domains (Billin et al., 2000; Cairo et al., 2001; Yamashita et al., 2001). Upon glucose stimulation, the complex translocates into the nucleus, where it controls the expression of multiple target genes (Kawaguchi et al., 2001; Ma et al., 2005; Stoltzman et al., 2008). The net result is to coordinate the cellular response to glucose, which depends in part on the cell type (Abdul-Wahed et al., 2017; Richards et al., 2017; Uyeda and Repa, 2006). In our screen, we identified MLX to be a class 5 hit with few, large and eccentric LDs in response to ac-Lipo that contained both TG and cholesterol esters (Figure 2B). This phenotype was readily confirmed and was independent of the uptake route of lipids, as incubation in media supplemented with oleate or cholesterol-rich acetylated low-density lipoprotein (ac-LDL) also resulted in a LD phenotype in MLX-depleted cells (Figure S3B–C).
Transcription Factor MLX Binds to LDs
Our proteomic results indicated that MLX localizes to LDs in THP-1 cells. To determine whether this was a general property, we transfected a variety of cell types with GFP-tagged MLX, incubated them with oleate for 1 day to induce LDs, and analyzed MLX localization by microscopy. As displayed in Figure 3B, MLX efficiently localized to LDs in all cell types tested, including mouse primary hepatocytes. To extend these studies, we also examined endogenous MLX localization in SUM159 mammary carcinoma cells, which are stably diploid and, therefore, well suited for genome engineering, and which efficiently form LDs when cultured with oleate (He et al., 2017; Wang et al., 2016). Similar to the results in THP-1 macrophages, endogenous MLX protein was enriched in the LD fraction isolated from SUM159 cells (Figure S3D, Table S3). To directly visualize endogenous MLX in SUM159 cells, we next GFP-tagged the protein by CRISPR/Cas9-mediated genome engineering (Figure S3E–F). When LD formation was induced by one day of oleate incubation, endogenously GFP-tagged MLX protein was enriched at LD surfaces, with some heterogeneity, possibly due to tangential optical sectioning of some LDs (Figure 3C). Levels of MLX mRNA were not changed by oleate treatment but cellular MLX protein levels increased, likely due to stabilization of the protein by binding to LDs (Figure S3G). To analyze the kinetics of MLX binding to LDs, we expressed GFP-tagged MLX in SUM159 cells and performed a series of fluorescence recovery after photobleaching experiments. Figure 3D and Video S1 show images from a representative time-lapse experiment and a quantification of fluorescence recovery over 1 hour. During this time, we found little recovery of the MLX signal on the subset of cellular LDs bleached at the beginning of the experiment, indicating slow apparent on/off rates and stable binding of MLX to LDs.
MLX Binds LDs Through a C-Terminal Amphipathic Helix
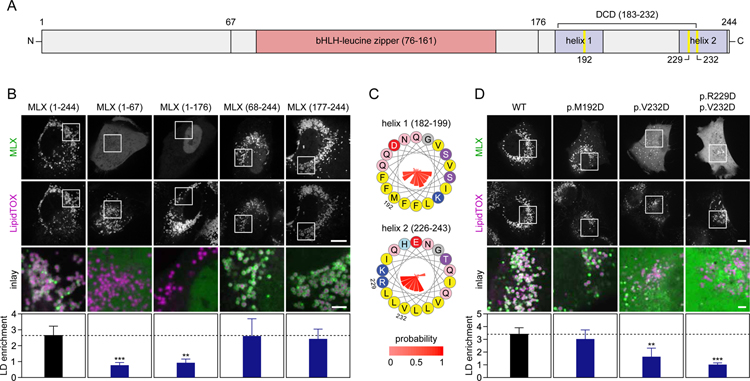
To map the region of MLX required for LD binding, we compared the localization of full-length MLX with truncated versions expressed in SUM159 cells with induced LD formation (Figure 4A). None of the truncations containing the N-terminal portion of MLX targeted to LDs. In contrast, a C-terminal fragment of MLX containing amino acids 177–244 was sufficient and required to target LDs with an efficiency comparable to full-length MLX (Figure 4B).
Figure 4. MLX Binds LDs Through a C-Terminal Amphipathic Helix.
(A) Overview of MLX domains and expression constructs.
(B) The C-terminal domain of MLX is sufficient and required for LD binding. The ability of truncated forms of MLX tagged with EGFP to target to LDs was evaluated in SUM159 cells. Cells were transfected with the indicated constructs and thereafter incubated with oleic acid (0.5 mM) for 1 day. Prior to imaging, LDs were stained with LipidTOX Deep Red. Representative images from one experiment are shown, and results were quantified (n=10–12 cells per construct) using CellProfiler. Results were evaluated using one-way non-parametric ANOVA (Kruskal-Wallis followed by Dunn’s multiple comparisons test). Scale bars, 10 µm and 2.5 µm (inlay).
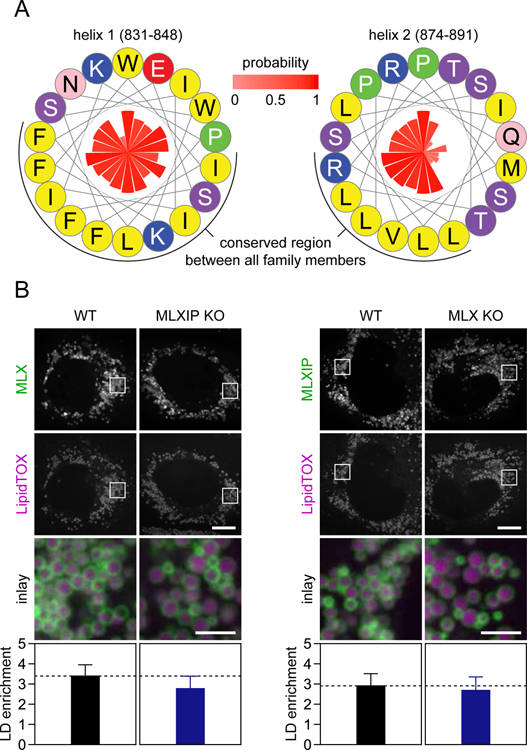
(C) Two C-terminal amphipathic helices are present in MLX. Helical wheels displaying amphipathic helices present in MLX. Amino acid properties and positions are indicated by colors and numbers, respectively, and conservation (expressed as probability %) is indicated by the inner bars.
(D) Point-mutations in the most C-terminal amphipathic helix of MLX abolish LD binding. The experiment was performed exactly as described in Panel (B) with the exception that cells were transfected with full-length constructs containing point-mutations in the amphipathic helices of MLX. Results were evaluated using one-way non-parametric ANOVA (Kruskal-Wallis followed by Dunn’s multiple comparisons test). Scale bars, 10 µm and 2.5 µm (inlay). Abbreviations: bHLH, basic helix-loop-helix; DCD, dimerization and cytoplasmic localization domain.
Proteins target LDs predominantly using either hydrophobic, membrane-embedded sequences or via amphipathic helices (Kory et al., 2016). Cytosolic proteins bind LDs by recognizing phospholipid packing defects that are abundant and persistent on the LD phospholipid monolayer (Čopič et al., 2018; Prévost et al., 2018; Thiam et al., 2013). The LD-targeting sequence of MLX, which spans the dimerization and cytoplasmic localization domain of MLX (Eilers et al., 2002) and the leucine zipper-like domains of MLXIPL (de Luis et al., 2000), contains potential amphipathic helices, shown for MLX in Figure 4C. To test if these motifs in MLX are required for LD binding, we mutated the helices by introducing aspartic acids into the hydrophobic regions. Mutation of the first helix (p.M192D) did not affect LD binding, but mutating the second helix impaired (p.V232D) or abolished (p.R229D combined with p.V232D) LD targeting of the MLX protein (Figure 4D). Thus, our data indicate that the most C-terminal amphipathic helix of MLX is required for its binding to LDs.
Hormone-sensitive lipase (HSL/LIPE), an LD-localized protein, was recently reported to interact with MLXIPL in adipocytes (Morigny et al., 2019). We therefore tested if a protein-protein interaction between HSL and members of the MLX transcription factor family could mediate their binding to LDs in SUM159 cells. However, we did not detect HSL in cell lysates or LD fractions of SUM159 cells (Table S3, Figure S4A), suggesting very low expression of HSL in this cell type. Moreover, knocking out the HSL gene did not alter MLX localization (Figure S4A–C), demonstrating that MLX targeting to LDs is not mediated by HSL. Taken together, our data suggest the MLX transcription factors bind directly to LD surfaces rather than via protein-protein interactions.
MLX and MLXIP Bind LDs Independently of Each Other
In addition to identifying MLX in LD fractions, we also detected MLXIPL and MLXIP in THP-1 macrophages and SUM159 cells, respectively (Table S2–3). This suggests that features required for LD binding are either evolutionarily conserved among the three proteins or that protein interactions between MLX and MLXIP/MLXIPL bring family members to LDs. Sequence alignments across several species combined with secondary structure predictions showed that the C-terminal regions of MLXIP and MLXIPL, including the alpha and beta isoforms of MLXIPL (Herman et al., 2012), contain putative amphipathic helices similar to MLX (Figure 5A, Figure S5A; results not shown for MLXIPL). More specifically, the identified amphipathic helices included evolutionarily conserved stretches of amino acids dominated by hydrophobic residues and more variable hydrophilic regions (Figure 5A, Figure S5A). Based on the sequence similarities between MLX, MLXIP, and MLXIPL, we hypothesized that they each have the capacity to target LDs independently of each other. To test this, we addressed LD binding in SUM159 cells, which according to our proteomics data expressed predominantly the family members MLX and MLXIP. We first confirmed our finding that MLXIP targets to LDs by transfecting cells with GFP-tagged MLXIP (Figure 5B). We next tested whether LD targeting of MLX and MLXIP depends on its interaction partner. For this, we knocked-out MLX or MLXIP by using CRISPR/Cas9-mediated genome engineering (Figure S5B–E). We found no differences in LD targeting for GFP-tagged MLX or MLXIP in cells lacking the interaction partner, indicating that heterocomplex formation is not required for LD binding (Figure 5B).
Figure 5. MLX and MLXIP Bind LDs Independently of Each Other.
(A) Two amphipathic helices, similar to those found in MLX, are present in the C-terminus of MLXIP. Helical wheels displaying amphipathic helices present in MLXIP. Amino acid properties are indicated by colors, and conservation (expressed as probability %) is indicated by the inner bars.
(B) MLX:MLXIP heterocomplex formation is not required for LD targeting of either protein. SUM159 cells were transfected with EGFP-tagged MLX or MLXIP, incubated with oleic acid (0.5 mM) for 1 day, stained with LipidTOX Deep Red and imaged. Left panel; localization of MLX in MLXIP KO cells. Right panel; localization of MLXIP in MLX KO cells. Representative images from one experiment are shown and results were quantified (n=7–12 cells per construct and cell type) using CellProfiler. Results were evaluated using one-way non-parametric ANOVA (Kruskal-Wallis followed by Dunn’s multiple comparisons test). Scale bars, 10 µm and 2.5 µm (inlay).
LD Binding Modulates MLX:MLXIP Transcriptional Activity
The ability of MLX transcription factors to target LDs raises the question of whether LD binding affects their transcriptional activity. To address this question, we first analyzed the localization of endogenously tagged MLX after shifting cells from glucose-free to glucose-containing medium, in either the absence or presence of LDs induced by oleic acid. Quantitation of the fluorescence signal revealed that, in the presence of LDs, the concentration of MLX in the nucleus remained lower, with more of the protein accumulating on LDs (Figure 6A, Figure S6A). To deplete LDs from cells, we incubated them with highly specific inhibitors of acyl CoA:diacylglycerol O-acyltransferase (DGAT) 1 (Liu et al., 2013) and DGAT2 (Imbriglio et al., 2015), the two enzymes synthesizing the TG cores of LDs (Cases et al., 1998, 2001; Lardizabal et al., 2001). Under these conditions, the addition of oleic acid did not result in LD formation, and when these cells were shifted from glucose starvation to high glucose-containing medium, MLX translocation was further increased (Figure 6B, Figure S6B). These findings indicate that MLX binding to LDs sequesters it away from the nuclei. Consistent with this, an N-terminally truncated version of MLX was highly enriched in the nuclear fraction in the absence of oleate, but this nuclear localization was dramatically reduced when LDs were induced (Figure S6C). Additionally, the nuclear enrichment of mutants of MLX was inversely correlated with defective LD binding (Figure 4D, Figure S6D).
Figure 6. LD Binding Modulates MLX:MLXIP Transcriptional Activity.
(A–B) LD binding sequesters MLX away from the nuclei. Effects of alterations in lipid storage and glucose on endogenous MLX localization were determined in SUM159 cells by confocal microscopy. (A) In the presence or absence of oleic acid (0.5 mM), cells were starved from glucose for 2 days and thereafter incubated for 1 day with 0 or 25 mM glucose-containing media. (B) Cells were starved from glucose for 2 days and thereafter incubated overnight with DMSO or DGAT1/DGAT2 inhibitors as well as glucose (25 mM) and oleic acid (0.5 mM). Prior to live cell imaging, LDs and nuclei were stained with LipidTOX Deep Red and Hoechst, respectively. For panels A-B, representative images from one experiment are shown, and results were quantified (n=19–25 cells per condition) by CellProfiler. Results were evaluated using two-way ANOVA followed by Tukey-Kramer test. Scale bars, 10 µm and 2.5 µm (inlay).
(C) TXNIP mRNA levels are upregulated by glucose. Genes regulated by glucose were identified in SUM159 cells using RNA sequencing. Prior to lysis, cells were starved from glucose for 2 days and thereafter incubated with or without glucose (25 mM) for 6 hours. Results are based on three replicates per condition and were evaluated using DESeq2.
(D–E) Glucose-mediated induction of TXNIP mRNA requires MLX and MLXIP. Messenger RNA levels of TXNIP were measured by qPCR in WT, MLXIP (D), and MLX (E) knockout cells. The cells were starved from glucose for 2 days and thereafter incubated in the presence/absence of glucose (25 mM) for 1 day. (E) As indicated in the panel, MLX-EGFP was stably expressed in MLX knockout cells from the AAVS1 locus (add-back). Results from one representative experiment containing three replicates are shown.
(F–G) LDs regulate MLX:MLXIP activity. TXNIP mRNA levels were measured in SUM159 cells with or without LDs. (F) Cells were starved from glucose in the presence or absence of oleic acid (0.5 mM) for one day and thereafter washed and starved from glucose for another 12 hours. After this, cells were incubated in with or without glucose (25 mM) and oleic acid (0.5 mM) for 1 day. (G) Cells were starved from glucose for 2.5 days and thereafter incubated with or without glucose (25 mM) in the presence of DMSO or DGAT1/DGAT2 inhibitors overnight. Results are based on two independent experiments, each containing three replicates and were evaluated using two-way ANOVA followed by Tukey-Kramer test.
(H) LDs regulate MLX promoter binding. TXNIP promoter binding by endogenously EGFP-tagged MLX was measured in SUM159 cells. Cells were starved from glucose for 1.5 days and thereafter incubated OA-containing media with or without glucose (25 mM) in the presence of DMSO or DGAT1/DGAT2 inhibitors for 12 hours. Results from three replicates are shown using an anti-MLX antibody (left panel) or an anti-GFP antibody (right panel). Abbreviations: DGATi, DGAT1/2 inhibition; KO, knockout; OA, oleic acid.
The regulation of MLX, MLXIP and MLXIPL transcriptional activity is complex and does not depend only on cellular localization. For instance, trapping MLXIPL in the nucleus by inhibiting its export does not activate, but actually inhibits its transcriptional activity (Davies et al., 2008). Additionally, target genes for these transcription factors vary among different cell types (Richards et al., 2018; Sae-Lee et al., 2016). To monitor the transcriptional output of MLX in SUM159 cells, we therefore first sought to identify a robust transcriptional target of its activity. RNA sequencing analyses showed that mRNA encoding the thioredoxin-interacting protein (TXNIP) responded most robustly (six-fold upregulation) after the shift from glucose-free to glucose-containing medium (Figure 6C). TXNIP is known as an MLX:MLXIP target gene. It is a member of the α-arrestin family that inhibits the antioxidative function of thioredoxin and effects the endocytosis-mediated downregulation of plasma membrane glucose transporters (Minn et al., 2005; Wu et al., 2013). In SUM159 cells, TXNIP mRNA induction by glucose depended on MLX and MLXIP, as it was absent in cells deleted for either of the two proteins. In addition, re-expressing MLX in MLX knockout cells restored the glucose-dependent induction of TXNIP mRNA expression (Figure 6D–E, Figure S6E–F).
An MLX mutant that does not bind LDs but is transcriptionally active would be an informative tool. However, by using TXNIP as a reporter, we tested several LD binding-mutants of MLX and found that they were unable to induce TXNIP expression under any condition when re-introduced into MLX-deficient cells (Figure S6G), indicating that these mutations also disrupt transcriptional activity. As an alternative approach to address the question how LD binding impacts MLX:MLXIP transcriptional activity, we assayed for the transcriptional response to glucose in the presence or absence of LDs. In line with endogenous MLX localization, we found that cells with abundant LDs induced the MLX:MLXIP target less than those with only a few LDs (Figure 6F). In contrast, cells lacking LDs due to DGAT1 and DGAT2 inhibition displayed increased MLX:MLXIP activity (Figure 6G). Consistent with these results, we found by chromatin immunoprecipitation and qPCR increased glucose-stimulated TXNIP promoter occupancy by MLX in cells depleted of LDs (Figure 6H).
LDs Modulate MLX:MLXIPL Gene Regulation and Glucose Uptake in Human Macrophages
Our findings in SUM159 cells indicated that binding of MLX transcription factors to LDs alters target gene expression. We next asked whether similar paradigms operate in human macrophages to modulate target gene expression and cellular physiology. First, we identified both direct and indirect target genes of MLX in THP-1 macrophages by sequencing cellular RNAs after glucose stimulation of cells treated with MLX siRNA and comparing them with controls. As in SUM159 cells, this showed TXNIP as a strongly glucose regulated gene, whose induction was dependent on MLX (Figure 7A). However, unlike in SUM159 cancer cells, additional genes were MLX-regulated, including ARRDC4, another validated MLX target gene, which also functions as an arrestin (Stoltzman et al., 2008) (Table S4).
Figure 7. LDs modulate MLX:MLXIPL Gene Regulation in Human Macrophages.
(A–B) MLX controls a specific set of putative lipid storage response genes. (A) Genes depending on MLX for induction after glucose stimulus were identified by RNA sequencing. Control and MLX-depleted THP-1 macrophages were starved from glucose for 2 days and thereafter incubated in the presence or absence of glucose (25 mM) for 1 day. Known targets of MLX (TXNIP and ARRDC4) and genes displaying a similar LD phenotype as MLX in the RNAi screen (r>0.5) are highlighted. Results are based on three replicates per condition and were evaluated using DESeq2. (B) Heatmap and distribution chart of the effects of lipid storage induction by ac-Lipo on putative target gene expression. Counts from the RNA sequencing were scaled prior to clustering (left panel) and the relative position among all measured genes highlighted (right panel). Results are based on two replicates per condition.
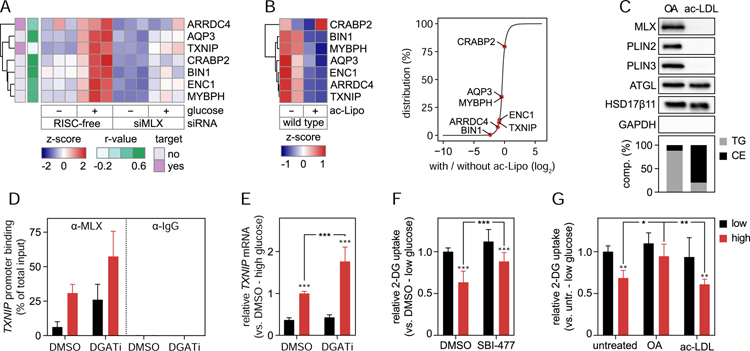
(C) MLX binds specifically to TG-containing LDs. Based on fractionated THP-1 cells, protein and lipid composition of isolated LDs induced by incubations with OA (0.5 mM) or ac-LDL (25 µg/mL) for 2 days were determined using western blotting and TLC, respectively.
(D–E) LDs regulate MLX activity. TXNIP promoter binding of MLX and TXNIP mRNA levels were measured in THP-1 macrophages. Cells were starved from glucose for 1.5 days and thereafter incubated OA-containing media with or without glucose (10 mM in panel D and 25 mM in panel E) in the presence of DMSO or DGAT1/DGAT2 inhibitors for 12–16 hours. Results are based on one (D) or two (E) independent experiments, each containing three replicates and (E) were evaluated using two-way ANOVA followed by Tukey-Kramer test.
(F–G) Alterations in MLX:MLXIPL activity control 2-DG uptake. Uptake of [3H] 2-Deoxy-D-glucose in THP-1 macrophages was determined in cells (F) starved from glucose for 1 day and thereafter incubated with or without glucose in the presence of DMSO or 10 µM SBI-477 for 1 day and (G) incubated in varying concentrations of glucose in the presence or absence of OA (0.5 mM) or ac-LDL (25 µg/mL) for 2 days. Results are based on three (F) or two (G) independent experiments, each containing three replicates and were evaluated using two-way ANOVA followed by Tukey-Kramer test. Abbreviations: 2-DG, 2-deoxyglucose; ac-LDL, acetylated low-density lipoprotein; ac-Lipo, acetylated apolipoprotein B-containing lipoprotein; CE, cholesterol ester; comp., composition; OA, oleic acid; TG, triacylglycerol; untr., untreated.
Among the putative target genes, several had phenotypes in our genome-wide RNAi screen. Depletion of either the glycerol transporter AQP3, the endocytic protein BIN1, the fatty acid–binding protein CRABP2, or the transcriptional regulators ENC1 and MYBPH in THP-1 cells resulted in phenotypes similar to that of MLX depletion (Figure S7A, Table S4). These findings suggest that MLX executes a transcriptional response induced by glucose that regulates lipid storage in LDs via several of the identified genes.
We utilized the genes regulated by MLX to determine if the presence of LDs modulates the MLX transcriptional response also in THP-1 macrophages. Consistent with our results in SUM159 cells, we found that all but one of the MLX-dependent genes of this network exhibited reduced mRNA expression when LDs were induced by incubation with ac-Lipo (Figure 7B, Table S5).
Because THP-1 cells can form either CE- or TG-rich LDs, this system afforded us the possibility to dissect the MLX response to the storage of different lipid types. Strikingly, subcellular fractionation of THP-1 cells induced to harbor CE- or TG-rich LDs, revealed that binding of MLX (and PLIN2 and PLIN3/TIP47) only occurred for TG-rich LDs, whereas other LD proteins, such as ATGL/PNPLA2 and HSD17β11, were bound to LDs containing either neutral lipid (Figure 7C). Consistent with this finding, formation of TG-rich LDs blunted the MLX occupancy on the TXNIP promoter and reduced TXNIP mRNA levels after glucose stimulation (Figure 7D–E). We also measured by ChIP-qPCR binding of MLX to other putative target genes (Figure S7B). In these experiments, ARRDC4 displayed the same pattern of regulation as TXNIP, and other promoters displayed much lower occupancy compared to TXNIP, consistent with a recent report (Wilde et al., 2019).
To test whether MLX binding to LDs modulates cellular physiology, we next analyzed cellular glucose uptake regulation, a salient feature of the MLX response across cell types that is regulated by the TXNIP and ARRDC4 arrestins (Ahn et al., 2016; Richards et al., 2018; Stoltzman et al., 2008). Consistent with this, stimulating cells with high levels of glucose for one day down-regulated uptake of 2-deoxyglucose. This was MLX-dependent as an inhibitor of the transcription factor, SBI-477 (Ahn et al., 2016), blunted this inhibition (Figure 7F). Similarly, the presence of TG-rich, but not CE-rich, LDs abolished the inhibition of glucose uptake as incubating cells with oleate abrogated the effect of glucose stimulation on 2-deoxyglucose uptake, whereas incubation with ac-LDL left the response intact (Figure 7G). Taken together, these data indicate that TG-containing LDs, which bind MLX, attenuate a MLX-regulated physiological response.
DISCUSSION
In this study, we systematically investigated the determinants of lipid storage in human macrophages. Our results provide a comprehensive analysis of the genetic landscape impacting LDs and report the LD protein composition for human macrophages. With this integrative and unbiased approach, we defined ~550 genes governing LD biology and identified some 80 bona fide LD proteins in human macrophages, providing a wealth of new information for research on LD biology.
Most importantly, we utilized this dataset to uncover an unexpected paradigm in which cells coordinate lipid storage with metabolic gene regulation by LD binding of the MLX family of glucose-sensing transcription factors: MLX, MLXIP and MLXIPL. Few, if any, transcription factors have been shown previously to localize to LDs. We find that binding of MLX family of transcription factors to LDs does not require heterocomplex formation but instead appears to be mediated by highly conserved sequences in each protein that have the propensity to form amphipathic helices. Our previous results show that such sequences detect LD surfaces through large hydrophobic residues interacting with phospholipid packing defects in the phospholipid monolayer, induced by the underlying oil phase, which are larger, more prevalent and persistent than for instance in flat membranes (Prévost et al., 2018). Because this leads to folding of the sequence into an amphipathic helix, targeting becomes effectively irreversible, at least until LDs are consumed sufficiently for proteins to be displaced from the LD surface via macromolecular crowding (Kory et al., 2015). Consistent with this, we find that MLX binds stably to LDs and, once bound, has a low apparent off-rate, preventing its translocation into the nucleus and thereby potentially inactivating the protein. Collectively, our data suggest that MLX family proteins utilize these amphipathic helical sequences to bind directly to the phospholipid monolayer of LDs. Supporting our findings of MLX transcription factors binding to LDs, MLX was found with a recent proximity labeling strategy for identifying bona fide LD proteins (Bersuker et al., 2018) and MLX and MLXIPL were found in the LD proteome of murine liver (Krahmer et al., 2018).
A recent study suggested that MLXIPL interacts with HSL (Morigny et al., 2019), an LD protein suggesting that MLX family members could localize to LDs via HSL binding. However, we found that MLX binds LDs in SUM159 cells, which do not express HSL. Thus an alternative explanation for the reported HSL and MLXIPL interaction could be that these proteins are bridged by LDs, as both proteins bind tightly to LD surfaces.
A key finding from our studies is that LD binding of MLX transcription factors attenuates their transcriptional responses. In support of this, we show that the presence of LDs in two different cell types blunts MLX:MLXIP or MLX:MLXIPL activities, as measured by target genes, in response to glucose. Conversely, abolishing LD formation by inhibiting TG synthesis results in an increased activation of MLX targets to glucose. Further, we show that these transcriptional effects are manifested by changes in glucose uptake in human macrophages. We posit that this mechanism will ensure the correct response of cells to fluctuating concentrations of glucose, depending on whether abundant cellular energy stores are present in the form of LDs. The response for these transcription factors may be contextual, given that they regulate different genes in different cell types. For example, MLX and MLXIP are highly expressed in skeletal muscle, where they regulate predominantly the processes of glucose uptake and neutral lipid storage (Ahn et al., 2016). In contrast, MLX and MLXIPL are predominantly expressed in white adipose tissue and liver where they control the transcription of enzymes regulating de novo lipogenesis (Benhamed et al., 2012; Dentin et al., 2006; Herman et al., 2012; Iizuka et al., 2004; Vijayakumar et al., 2017). In support of these possible physiological outcomes predicted by our data, the expression of a dominant-negative MLX in murine liver results in a reduction in glucose-regulated genes and liver fat, and also improves glucose tolerance (Iizuka et al., 2009). Our findings provide an opportunity for further in vivo studies to examine the functional role of LD binding of MLX factors in modulating metabolism.
Based on our results, we propose a model in which the binding of MLX, MLXIP and MLXIPL to accumulating LDs limits glucose-stimulated gene transcription. The net result of this sequestration may be to signal to the cell that energy stores are becoming replete, thereby initiating a feedback circuit of pathways that alter energy uptake or storage. Possibly a similar system exists in yeast, where at least under some conditions the transcriptional repressor Opi1 can be found on LDs (Romanauska and Köhler, 2018). We suggest that the mechanism regulating MLX-family transcription factors may be one component of a network of responses for coordination of a more global lipid storage response (LSR), similar to homeostatic paradigms that have been described for transcription responses linked to other organelles, such as the Ire1 and the UPR for ER proteostasis (Cox et al., 1993; Mori et al., 1993), SREBP1/2 for ER lipid homeostasis (Briggs et al., 1993; Hua et al., 1993; Wang et al., 1993, 1994), or TFEB for homeostasis in lysosomes (Sardiello et al., 2009; Settembre et al., 2011).
Our screen identified MLX to be a class 5 hit, with few, large and eccentric LDs in response to ac-Lipo lipid loading. It is currently unclear how MLX deficiency results in the class 5 phenotype, but our data suggest that this is not due to changing the expression of a single gene, as we found that deficiency of several disparate MLX target genes (e.g. AQP3, BIN1, and CRABP2) in macrophages result in an LD phenotype that is similar to MLX deficiency. Of these, AQP3 is perhaps easiest to connect to glycerolipid metabolism, since its product is a mediator of glycerol transport (Ishibashi et al., 1994). However, each of these MLX targets yielding similar phenotype may also be components of the LSR.
Although we focus here on the biology of one particular hit from our studies, the data from this genome-perturbation screen and LD proteome should be a boon to studies of LD biology. Our screen is rich in image content, with a pipeline of 133 image parameters utilized in classifying LD changes in response to lipid loading with both cholesterol and fatty acids. Importantly, our screen confirmed key functions for seipin, retromer proteins, endocytic recycling pathways, and proteasomal activity in lipid storage (Fei et al., 2008; Fröhlich et al., 2015; Guo et al., 2008; Szymanski et al., 2007; Wang et al., 2016). The hits from our screen binned into six distinct phenotypic classes, which will enable further identification of pathways governing LD biology that are common to the different classes, as we showed previously for an LD screen in D. melanogaster cells (Guo et al., 2008). Additionally, we identify many novel genes affecting human LD biology in a cell type relevant to the inflammation and to the pathogenesis of many common metabolic diseases. Thus, we expect further data analyses of our results will lead to many more insights into LD biology and diseases of lipid storage.
STAR METHODS
Lead contact and materials availability
The Lead Contact for this study is Tobias C. Walther (twalther@hsph.harvard.edu). All unique/stable reagents generated in this study are available from the Lead Contact with a completed Materials Transfer Agreement.
Experimental model and subject details
Cell culture
THP-1 (monocytes derived from a male infant), HEPG2 (hepatocytes derived from a male subject), and COS-7 (fibroblast-like cells derived from African green monkey kidney tissue) cells were bought from American Type Culture Collection. SUM159 (triple negative breast cancer cells derived from a female subject) and U2OS (osteosarcoma cells derived from a female subject) cells were kindly provided from the laboratories of Tomas Kirchhausen (Harvard Medical School) and Peter Walter (University of California, San Francisco), respectively. Primary hepatocyte isolation and cultures were carried out as previously described with a two-step digestion process (Matsumoto et al., 2007). Briefly, livers from 8–12 week old male mice were drained of blood by perfusion via vena cava with 42°C perfusion buffer (0.4 g/L KCl, 1.0 g/L dextrose, 1.8 g/L NaHCO3, 0.2 g/L EDTA ) for 3 minutes. Connective tissue within the liver was digested by perfusion with 42°C liver digest media (0.4 g/L KCl, 1.0 g/L dextrose, 1.8 g/L NaHCO3, 0.5 g/L CaCl2, 10 g/L BSA (NC9227912, Fischer Scientific), 30 mg/L Collagenase Type IV (C5138, Sigma-Aldrich) for 10 minutes. The liver was mechanically dissociated in plating media (DMEM no glucose (11966025, Gibco) containing 10% FBS (100–106, lot A16E00E, Gemini), 2 mM sodium pyruvate (11360070, Gibco), 2% streptomycin and penicillin (15070063, Gibco), 1 µm dexamethasone (D-2915, Sigma-Aldrich), 0.1 µM insulin (I6634, Sigma-Aldrich) at 4°C, strained through 70 micron cell strainer, and hepatocytes were collected by centrifugation at 50 × g for 3 minutes. Hepatocytes were further isolated from other cells on a percoll gradient (P4937, Sigma-Aldrich) with centrifugation at 650 × rpm for 5 minutes. The cell pellet was washed and resuspended in plating media, and cells were plated in 6-well plates at a density of 4×105 cells/well. Unless otherwise stated, THP-1 cells were cultured in RPMI 1640 medium containing HEPES (22400105, Gibco) supplemented with 10% FBS (10438026, Gibco), 100 µg/mL streptomycin, and 100 U/mL penicillin (15140122, Gibco), and 0.05 mM 2-mercaptoethanol (AB01340, AmericanBio) and SUM159 were cultured as described in Wang et al. (Wang et al., 2016). HEPG2, U2OS, and COS-7 cells were grown in DMEM medium (11995073, Gibco) supplemented with 10% FBS (10438026, Gibco), and 100 µg/mL streptomycin, and 100 U/mL penicillin (15140122, Gibco). Primary hepatocytes were cultured in DMEM no glucose (11966025, Gibco) containing 10% FBS (100–106 lot A16E00E, Gemini), 2 mM sodium pyruvate (Gibco, 11360070), 2% streptomycin and penicillin (15070063, Gibco), 0.1 µM dexamethasone (D-2915, Sigma-Aldrich), 1 nM insulin (I6634, Sigma-Aldrich). DMEM and RPMI with/without glucose were mixed to obtain indicated concentrations of glucose. To induce LDs, cells were incubated with acetylated apolipoprotein B-containing lipoproteins (ac-Lipo, inducing TG and CE storage), acetylated low-density lipoprotein (ac-LDL, inducing mostly CE storage) or oleic acid (inducing TG storage) as stated in the following method sections. Human ac-Lipo (A6961, PanReac Applichem) was acetylated as previously described (Basu et al., 1976) and oleic acid (O1383, Sigma-Aldrich) was complexed with essentially fatty acid free BSA (A6003, Sigma-Aldrich) at a fatty acid/albumin molar ratio of 3:1. Human ac-LDL was purchased from Alfa Aesar (BT-906). To block TG synthesis, DGAT1 (Liu et al., 2013) and DGAT2 inhibitors (Imbriglio et al., 2015) from Merck & Co. were dissolved in DMSO (D2650, Sigma-Aldrich) and used at a final concentration of 10 µM. The small-molecule probe, SBI-477 (PC-61673, ProbeChem), was used to inhibit MLX-family member activity as described in Ahn et al. (Ahn et al., 2016). All cells were cultured at 37 °C in the presence of 5% CO2.
Method details
Antibodies
Primary antibodies targeting the following proteins were used in the present study: ATGL (2138S, Cell Signaling Technology), CANX (sc-46669, Santa Cruz Biotechnology), GAPDH (5174S, Cell Signaling Technology), GFP (ab290, Abcam), GM130 (12480S, Cell Signaling Technology), HSD17β11 (ab136109, Abcam), HSP60 (4870S, Cell Signaling Technology), IgG (2729S, Cell Signaling Technology), MLX (85570S, Cell Signaling Technology), MLXIP (A303–195A, Bethyl Laboratories), NFkB p65 (33-9900, Invitrogen), SR-AI/MSR (MAB2708, R&D systems), PLIN2 (ab108323, Abcam), PLIN3 (20R-2598, Fitzgerald), and TUBA4A (T5168, Sigma-Aldrich). HRP-conjugated secondary antibodies against mouse (sc-516102), rabbit (sc-2357) and guinea pig (sc-2438) were from Santa Cruz Biotechnology. For immunofluorescence, Alexa Flour 647 goat anti-mouse IgG (A21236, Invitrogen) was used.
Plasmid construction
Genomic DNA was isolated using GenElute™ Mammalian Genomic DNA Miniprep Kit (G1N350, Sigma-Aldrich), PCRs were performed using Q5® High-Fidelity PCR Kit or Phusion® High-Fidelity PCR Kit (M0493L and M0530L, New England Biolabs), and restriction enzymes were from New England Biolabs. To generate expression plasmids regulated by the relatively weak HSV-thymidine kinase gene promoter (pTK), the CMV-promoter of pEGFP-C1 and pEGFP-N1 (Takara) were removed by restriction enzyme digestion with AseI/NheI. The resulting backbones were then isolated by electrophoresis, followed by gel extraction of the DNA using NucleoSpin® Gel and PCR Clean-up (740609.250, Macherey-Nagel). DNA fragments encoding the pTK promoter flanked by AseI/NheI were then ligated with T4 DNA ligase (M0202L, New England Biolabs). Repeating the same procedure combined with PCR, MLX (SC322389, OriGene Technologies) and MLXIP (EX-A1755-M02, GeneCopoeia) were cloned into these backbones using SacI/AgeI (pTK-MLX-EGFP), XhoI/BamHI (pTK-EGFP-MLX), or HindIII/KpnI (pTK-EGFP-MLXIP). Similarly, truncated forms of MLX, generated using PCR, were ligated in using XhoI/BamHI (pTK-EGFP-MLX-1-67 and pTK-EGFP-MLX-1-176) or SacI/AgeI (pTK-MLX-68-244-EGFP and pTK-MLX-177-244-EGFP). Constructs containing point-mutations in MLX were generated by ligating in gBlocks gene fragments (synthesized by Integrated DNA Technologies) flanked by SacI/AgeI.
For knockout cell generation, guide RNAs were cloned into pSpCas9(BB)-2A-Puro (PX459) V2.0 (plasmid #62988, Addgene) as described in section 5B of Ran et al. (Ran et al., 2013). For HSL, MLX and MLXIP, the following guide RNAs were used: 5′-TGC TGG TTA CCA ATC GGC CG-3′, 5′-ACA AAG ACC GGCGGC GGC GC-3′and 5′-TGG ACG TAG ACG AGC ACC GC-3′, respectively. For gene editing of the AAVS1 locus, the T2 target guide RNA (5′-GGG GCC ACT AGG GAC AGG AT-3′) was cloned into PX330-U6-Chimeric_BB-CBh-hSpCas9 (plasmid #42230, Addgene) (Cong et al., 2013) as described above for HSL, MLX and MLXIP guide RNAs. pTK-MLX-EGFP was PCR amplified and ligated into AAVS1_Puro_PGK1_3xFLAG_Twin_Strep (plasmid #68375, Addgene) (Dalvai et al., 2015) with NsiI/XbaI. For endogenous C-terminal tagging of MLX with EGFP, a guide RNA (5′-CAG CTT TAC TGA CCG GTT CT-3′) targeting closely to the stop codon of MLX was cloned into pSpCas9(BB)-2A-Puro (PX459) V2.0 as described above. A homologous repair template was generated in two steps: Step 1) 5′-phosphorylated oligonucleotides (synthesized by Integrated DNA Technologies) containing SpeI, NgoMIV, KpnI and NotI restriction enzyme sites were ligated into pSMART vector (Lucigen) using HindIII/XbaI to extend the multiple cloning site. In brief, two µg of each oligo was resuspended in 50 µL annealing buffer (10 mM Tris-HCl, pH7.4, 50 mM NaCl, 1 mM EDTA, pH 8.0). The mix was heated to 95°C for 2 minutes and then gradually cooled down to 25°C over 45 minutes. To ligate the annealed oligos into the pSMART vector, the oligos were first diluted 1:10 in nuclease-free water and then ligated as described above using T4 DNA ligase. Step 2) Left and right homology arms, as well as EGFP, were amplified by PCR from genomic/plasmid DNA, respectively. The following primers were used: left homology arm fw, 5′-CTG TCT GGT AAC TAG TGG TTC ACG CCA TTC TCC TGC-3′; left homology arm rv, 5′-CTG TCT GGT AGC CGG CGT AAA GCT GGT TTT TCA ATT GG-3′; EGFP fw, 5′-CTG TCT GGT AGC CGG CCG ACC GGT CGC CAC CAT GGT G-3′; EGFP rv, 5′-CTG TCT GGT AGG TAC CTT ACT TGT ACA GCT CGT CCA TGC-3′; right homology arm fw, 5′-CTG TCT GGT AGG TAC CCC GGT TCT TGT AAA CCT GGA G-3′; right homology arm rv, 5′-CTG TCT GGT AGC GGC CGC CCA AGT CCT GGG AGA AAT GC-3′. The PCR products were purified, digested using SpeI/NgoMIV (left homology arm), NgoMIV/KpnI (EGFP) or KpnI/NotI (right homology arm) and then ligated into the modified pSMART vector as described above. To avoid re-cutting of the target sequence after homology-directed repair, a silent point-mutation in the protospacer adjacent motif was introduced in the forward primer of the right homology arm.
siRNA transfection
In addition to the genome-wide RNAi screen, siRNA transfection in THP-1 cells was performed using duplexes targeting RELA (M-003533-02-0005, Dharmacon), a gene which encodes the NFκB p65 subunit, and MLX (M-009724-00-0005, Dharmacon). Cells were plated and transfected as described in “Genome-wide RNAi screen”. For optimization of transfection experiments using RELA, cells were fixed in 4% paraformaldehyde (15710, Electron Microscopy Sciences) three days after transfection and p65 protein levels determined using immunofluorescence. For knockdown of MLXIP, SUM159 cells were grown on 100 mm cell culture dishes and transfected with 20 nM of duplexes targeting MLXIP (M-008976-01-0005, Dharmacon). Transfection complexes were generated by mixing siRNAs diluted in 500 µL Opti-MEM™ I Reduced Serum Medium (31985070, Gibco) with 25 µL Lipofectamine RNAiMAX (13778150, Invitrogen) diluted in 500 µL Opti-MEM™ I Reduced Serum Medium. siGENOME RISC-Free Control (D-001220-01-05, Dharmacon) was used as a control for all experiments.
Pre-screen
To identify controls for the genome-wide RNAi screen, a small pre-screen was performed using a panel of siRNAs (Dharmacon) targeting genes regulating LD function (Guo et al., 2008). Each siRNA was evaluated with six replicates. Cells were treated, and data were extracted exactly as described in “Genome-wide RNAi screen” and results were evaluated using hierarchical clustering.
Genome-wide RNAi screen
The genome-wide screen was run in triplicate using the Dharmacon siGENOME SMARTpool siRNA library comprising 18,119 target genes (G-005005-025, Dharmacon). The library consists of pools of four different oligos per target gene. Based on our pre-screen results, all library plates were designed to contain wells with control siRNAs (RISC-free non-targeting control, BSCL2 and FITM1), which served to control the quality of every plate tested in the screen (D-001220-01-05, M-016749-00-0005, M-031931-00-0005, Dharmacon). The screen was performed as follows: THP-1 cells were plated in 384-well plates at 12 × 103 cells/well and differentiated in RPMI 1640 medium supplemented with 50 nM phorbol 12-myristate 13-acetate (P1585, Sigma-Aldrich) for 1 day. Adherent cells were washed twice with serum-free RPMI 1640 medium and maintained in serum-free media for the remaining assay period. To generate transfection complexes, 10 µL of 100 nM siRNA in siRNA buffer (B-002000-UB-100, ThermoFisher Scientific) was preincubated with 10 µL of Lipofectamine RNAiMAX (13778150, Invitrogen) diluted 1:100 in Opti-MEM™ I Reduced Serum Medium (31985070, Gibco) for 20 minutes. The mixture was subsequently added to cells using a Freedom Evo 200 platform (Tecan). After 3 days of transfection, all cells were incubated with 25 µg/mL of acetylated LDL for 2 days except for one column per plate that was incubated without lipids. To stain LDs and nuclei, cells were fixed with 4% paraformaldehyde (15710, Electron Microscopy Sciences) for 15 minutes, washed in PBS, incubated with 1 µg/mL BODIPY 493/503 (D3922, Molecular Probes) and 1 µg/mL Hoechst 33342 (D3922, Molecular Probes) for 20 minutes and washed again in PBS. Addition of reagents to plates and PBS washes were performed using a Multidrop Combi (ThermoFisher Scientific) and a Power Washer 384 (Tecan), respectively. Cells were imaged at 60X magnification using an Opera High Content microscope (Perkin Elmer). Seven images per well were acquired for each channel.
Validation screen
To validate the genome-wide RNAi screen, 51 out of the 556 (~9%) screen hits were randomly selected and re-screened using four individual siRNAs per gene (Dharmacon). Data were generated as described in “Genome-wide RNA screen,” and results were compared between the genome-wide RNAi screen and the validation study, using Spearman’s rank correlations using the set of high-confidence image features.
Immunofluorescence
THP-1 cells were washed with PBS and fixed with 4% paraformaldehyde (15710, Electron Microscopy Sciences) for 15 minutes at room temperature. Blocking and permeabilization were performed in blocking buffer (PBS supplemented with 0.1% Triton X-100 (T8787, Sigma-Aldrich)) and 3% bovine serum albumin (A7906, Sigma-Aldrich) for 30 minutes at room temperature. Cells were subsequently incubated with primary antibodies diluted 1:1000 in blocking buffer for 1 hour at room temperature. Cells were then washed three times in PBS and incubated with Alexa Flour secondary 647 antibody and 1 µg/mL of Hoechst 33342 for 1 hour in the dark. After three washes with PBS, the cells were imaged at 40X magnification on the Opera high-content microscope (Perkin Elmer).
CRISPR/Cas9-mediated genome editing
To generate HSL, MLX and MLXIP knockout clones, SUM159 cells were transfected with 1 µg of PX459 plasmids (containing the appropriate guide RNA) using 4 µL of FuGENE® HD Transfection Reagent (E2311, Promega) and 100 µL of Opti-MEM™ I Reduced Serum Medium (31985070, Gibco). Approximately 2–3 days post-transfection, cells were washed with PBS and switched to medium supplemented with 1.5 µg/mL of puromycin (A1113803, Gibco). Selection was stopped when untransfected control cells were dead (~ 2–3 days). To add back MLX in MLX knockout cells, 0.5 µg of PX330 and 0.5 µg of repair template (containing pTK-MLX-EGFP) were transfected as described above. As a successful repair integrates the pac gene, cells were selected using puromycin a few days after transfection as described above. Endogenous tagging of MLX was performed by transfecting SUM159 cells with 0.5 µg of PX459 and 0.5 µg of repair template as described above, and cells were switched to puromycin-containing medium as described for knockout generation. For all of the above, single-cell cultures were obtained after puromycin selection by fluorescence-activated cell sorting (FACS-Aria II; BD Biosciences). Clones were screened by Sanger sequencing (GENEWIZ) of purified genomic PCR amplicons around the target region of the guide RNA, qPCR and/or western blot. For knockout cells, allelic information was obtained by subcloning the PCR products into pCR™Blunt II-TOPO® vector (450245, Invitrogen), followed by Sanger sequencing of individual colonies (GENEWIZ). Primers used for screening were as follows: HSL knockout fw, 5′-CAC AAA TCC CGC TAT GTG GC-3′; HSL knockout rv, 5′-TAC CTG CTG TTT GCT GAG TCC-3′; MLX knockout fw, 5′-TGA GGG TCT GGC AAT GTT CC-3′; MLX knockout rv, 5′-GCA CAA AGT TCC TCC ATG ACA CC-3′; MLXIP knockout fw, 5′-CCT GTG TGG CTC TGA AGA AAT CC-3′; MLXIP knockout rv, 5′-GTA TGT TTC CAC TCT CAG CCA CC-3′; MLX endogenous tagging fw, 5′-GGC AGG CAT CTT GGA AAC TAC TC-3′; MLX endogenous tagging rv, 5′-GGA GAA CTA GGG TAG AGA GAG GTT G-3′.
Fractionation of cells
Cells were first washed with ice-cold PBS and thereafter with homogenization buffer (20 mM Tris-HCl pH 7.4, 250 mM sucrose, 1 mM EDTA pH 8) supplemented with cOmplete™, EDTA-free Protease Inhibitor Cocktail (11873580001, Roche). After scraping into homogenization buffer and transfer from cell culture dishes to Eppendorf tubes, 500 units of Benzonase® Nuclease (E1014, Millipore) were added, and the volumes of the cell suspensions were set to 1.0 mL. The suspensions were subsequently passed through 23G needles 30 times, and NaCl was added to a final concentration of 100 mM. To remove intact cells and nuclei, the lysates were subjected to 1,000 × g centrifugation for 10 minutes at 4°C. The supernatants of this spin were subjected to 20,000 × g ultracentrifugation for 40 minutes at 4°C to obtain membrane pellets and soluble supernatant fractions. To purify different fractions, including LDs, the supernatants were mixed 1:1 with 50% OptiPrep™ Density Gradient Medium (D1556, Sigma-Aldrich) diluted in homogenization buffer and layered with 400 µL of the following OptiPrep gradient solutions: 22, 16, 12, 8, 5, 2, and 0%. Gradients were subjected to 150,000 × g ultracentrifugation in a swinging bucket rotor (TLS55, Beckman-Coulter) overnight at 4°C. Fractions were collected using a tube slicer, and proteins were precipitated as described (Wessel and Flügge, 1984). For western blots, precipitates were re-solublized with RIPA Lysis and Extraction Buffer (89900, Thermo Scientific), supplemented with 2% SDS (S0294, Teknova) and cOmplete™, EDTA-free Protease Inhibitor Cocktail (11873580001, Roche). Protein concentrations were determined using Pierce™ BCA Protein Assay Kit (23225, Thermo Scientific). For mass spectrometry analyses of total cell lysates and purified LDs, see immediately below.
Mass spectrometry
Mass spectrometry analyses were performed as described (Jayson et al., 2018). Briefly, precipitated proteins from total cell lysates and purified LDs were resolubilized in 100 mM NaOH, aided by sonication at 4°C, and the solution was brought to pH 7.5 with 200 mM HEPES (4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid). Proteins were reduced using 5 mM dithiothreitol (10197777001, MilliporeSigma) for 1 hour at 37°C, followed by alkylation of cysteine residues using 15 mM iodoacetamide (I1149, MilliporeSigma) in the dark for 1 hour at room temperature. Excessive iodoacetamide was quenched using 10 mM dithiothreitol. Protein mixtures were diluted in 1:6 ratio (v/v) using ultrapure water prior to digestion using sequencing grade trypsin (LS02120, Worthington Biochemical) for 16 hours at 37°C. Digested peptides were subsequently desalted using self-packed C18 STAGE tips (2215, 3M Empore™) for LC-MS/MS analysis (Rappsilber et al., 2003). After desalting, peptides were resuspended in 0.1% (v/v) formic acid and loaded onto HPLC-MS/MS system for analysis on an Orbitrap Q-Exactive HF mass spectrometer (ThermoFisher Scientific) coupled to an Easy-n LC 1000 (ThermoFisher Scientific) with a flow rate of 300 nL per minute. The stationary phase was 0.1% formic acid, and the mobile phase was 0.1% (v/v) formic acid in 80% (v/v) acetonitrile. Chromatography for peptide separation was performed using increasing organic proportion of acetonitrile (5–40% (v/v)) over a 265-minute gradient) on a self-packed analytical column with a PicoTip™ emitter (New Objective) and a Reprosil Gold 120 C-18, 1.9-µm particle-size resin (Dr. Maisch). The mass spectrometry analyzer operated in data-dependent acquisition mode with a top 10 method at a mass range of 300–2000 Da.
SDS-PAGE and western blot
Protein lysates were denatured in Laemmli buffer at 37°C for 20 minutes, and proteins were thereafter separated on polyacrylamide gels in the presence of SDS and transferred to Immun-Blot PVDF membranes (1620177, Bio-Rad) using the Mini-PROTEAN Tetra electrophoresis system (Bio-Rad). After blocking in TBS-T supplemented with 5% Blotto, non-fat dry milk (sc-2325, Santa Cruz Biotechnology) at room temperature for 60 minutes, primary antibodies were added, and the membranes were incubated at 4°C with gentle shaking overnight. Primary antibody diluents and concentrations were as suggested by the manufacturer. Membranes were washed three times in TBS-T for 10 minutes each and incubated at room temperature for 60 minutes with appropriate horseradish peroxidase–conjugated secondary antibodies (Santa Cruz Biotechnology) diluted 1:5000 in blocking buffer. Subsequently, membranes were washed again as described above and developed using SuperSignal West Pico or Femto chemiluminescent substrates (34580 and 34095, ThermoFisher Scientific). Signals were detected using ChemiDoc XRS+ (BioRad).
RNA isolation, cDNA synthesis and qPCR
Total RNA was isolated from cells using RNeasy Mini Kit (74106, QIAGEN). Complete lysis was ensured using QIAshredder and DNA digested with RNase-Free DNase Set (79656 and 79254, QIAGEN). cDNA was synthesized using iScript™ cDNA Synthesis Kit (1708891, Bio-Rad) and real-time qPCR was performed with Power SYBR™ Green PCR Master Mix (4367659, Applied Biosystems). Forward and reverse primers (500 nM of each) were as follows: MLX (Fw 5′-CTA CAA GGA GTC CTA CAA AGA C-3′, Rv 5′-CAT CAT AGC CTC TCT TGA TGG-3′), MLXIP (Fw 5′-CCT CTT CTC CAC ACT TTC TTC-3′, Rv 5′-CAT TTC CCA GAT GTG CTA TTT C-3′), PPIA (Fw 5′-ATG CTG GAC CCA ACA CAA AT-3′, Rv 5′-TTT CAC TTT GCC AAA CAC CA-3′) and TXNIP (Fw 5′-GTC ATC AGT CAG AGG CAA TC-3′, Rv 5′-GGA ACG CTA ACA TAG ATC AGT AA-3′). Results were normalized to the reference gene PPIA and evaluated using the delta-delta Ct method.
RNA sequencing
Total RNA was isolated from SUM159 and THP-1 cells as described in “total RNA isolation, cDNA synthesis and real-time qPCR” and samples were submitted to the Genomics Core at Tufts University or to the Yale Stem Cell Center for RNA sequencing where RNA quality was tested with a Fragment Analyzer (Advanced Analytical Technologies). After quality controls, 100–1000 ng total RNA was used as input for library preparation using TruSeq® Stranded mRNA Library Prep kit (20020594, Illumina). In brief, mRNAs were enriched with polyA selection. The enriched mRNAs were then chemically fragmented, followed by double-strand cDNA synthesis, A-tailing, adaptor ligation size-selection, and finally PCR amplification. The resulting libraries were individually quantified using a Fragment Analyzer, mixed (equal molar) into a library pool, loaded on a cBot (Illumina) for clustering onto a flow cell, and sequenced on a HiSeq 2500 using V4 chemistry (Illumina).
Chromatin immunoprecipitation
For THP-1 cells, chromatin immunoprecipitation was performed using SimpleChIP® Enzymatic Chromatin IP Kit (9003, Cell Signaling Technology) as described by the manufacturer. SUM159 cells were trypsinized, collected and cross-linked by incubating cells with PBS supplemented with 1% formaldehyde (15710, Electron Microscopy Sciences) for 10 minutes at room temperature. Subsequently, cells were incubated for 5 minutes with glycine (final concentration: 0.125 M), washed three times with ice-cold PBS and incubated in cell lysis buffer (20 mM Tris-HCl pH 8.0, 85 mM KCl and 0.5% NP-40) for 10 minutes on ice. The lysate was subjected to 5,000 × g centrifugation for 3.5 minutes at 4°C and the resulting pellet was incubated one more time in cell lysis buffer. Following centrifugation and removal of the supernatant, nuclear lysis buffer (10 mM Tris-HCl pH 7.5, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS) was added to the nuclear pellet and the lysate was incubated on ice for 10 minutes. All lysis buffers were supplemented with cOmplete™, EDTA-free Protease Inhibitor Cocktail (11873580001, Roche). Solubilized chromatin was fragmented to a size range of ~200 to 600 bp using a Branson 250 digital sonifier for 6 minutes and the sheared chromatin was subjected to immunoprecipitation with antibody overnight at 4°C. A 50/50 slurry of protein A and protein G Dynabeads (10001D/10009D, Invitrogen) was used to capture enriched chromatin. Reverse cross-linking and proteinase K digestion were performed at 65°C for 6 hours. DNA was subsequently extracted using AMPure XP beads (A63882, Beckman Coulter) in accordance with the manufacturer’s manual. Occupancy was quantified as a percent of total input by qPCR and as a negative control immunoprecipitations were carried out using an anti-IgG antibody. For these control experiments, we did not observe any regulation between samples groups and the occupancy of each promoter was minimal (less than 0.3% of total input for all primers tested). For TXNIP, ARRDC4 and BIN1, previously validated sets of primers were used (McKenna et al., 2012; Wilde et al., 2019). For AQP3 (Fw 5′-CAG GTA CAC GTG TGA CAA GCT A-3′, Rv 5′-GGA TGT CCT GTG TGC CTA ATT G-3′), CRABP2 (Fw 5′-GAA TCA CGT AGA AAC CAG AAG CG-3′, Rv 5′-AAG GAG ATT GGA ATG TCT CCG A-3′), ENC1 (Fw 5′-CAT GCT GGA GTT TCA AGA CAT C-3′, Rv 5′-CAT TCT CCA AGA TAG TTC GTA CAG-3′) and MYBPH (Fw 5′-CAG TCC TCC TGC TTG ACC TG-3′, Rv 5′-ATC ATT GCT GGA CTG GCT GG-3′) primers were designed to span putative e-boxes located closely to the transcription start site of each gene.
Lipid extraction and thin layer chromatography
After incubations with ac-Lipo, ac-LDL or OA for the indicated time, lipid composition of intact cells and isolated LDs were determined. Cells were washed with PBS, harvested in 1.0 mL of ice-cold PBS and immediately transferred to 4.0 mL of chloroform:methanol (2:1) to extract lipids (Folch et al., 1957). For determination of LD lipid composition, lipids were collected at the last step of the protein precipitation procedure (Wessel and Flügge, 1984). The lipid phase was evaporated under a stream of N2, dissolved in chloroform and applied onto Silica gel 60 TLC plates (105721, Supelco). Separation of neutral lipids were performed by running the plates in a neutral lipid solvent (heptane/isopropyl ether/acetic acid, 60:40:4, v/v/v) as described (Lehner and Vance, 1999). Bands were detected by cerium molybdate staining, and lipids identified by comparing with lipid standards. Quantifications were performed in Fiji (Schindelin et al., 2012).
Sequence analyses and structure predictions
MLX and MLXIP homologous proteins were identified across species using a basic local alignment search tool (Altschul et al., 1990) and alignment generation from EVcouplings (Marks et al., 2011). Sequences displayed in Figure S5A were imported into Jalview (Waterhouse et al., 2009) and aligned using Clustal with default settings. Protein secondary structure predictions were generated using PSIPRED version 3.3 (McGuffin et al., 2000) and Phyre2 (Kelley et al., 2015). Based on these results, amino acid compositions and physicochemical properties of the predicted alpha helical regions were calculated using HELIQUEST (Gautier et al., 2008). Conservation scores were generated by uploading alignment results from EVcouplings to WebLogo 3 (Schneider and Stephens, 1990).
Live cell imaging
To test the localization of MLX and MLXIP, including full-length, truncated and point-mutated forms, SUM159 cells were transfected with 1.0 µg of plasmid DNA as described in “CRISPR/Cas9-mediated genome editing”. To monitor full-length MLX localization, COS-7 and HEPG2 were transfected similarly to SUM159 cells, and U2OS cells were transfected with 1.0 µg of plasmid DNA using Lipofectamine™ 3000 Transfection Reagent (L3000008, Invitrogen), according to the manufacturer’s instructions. Mouse primary hepatocytes were transfected with 3.0 µg of plasmid DNA using Targefect Hepatocyte (HEP-01, Targeting Systems) transfection reagent using 6 µL of targefect and 12 µL of virofect to form transfection complexes in 1 mL of Opti-MEM™ I Reduced Serum Medium (31985070, Gibco). Five hours after transfection, cells were incubated in the presence or absence of 0.5 mM oleic acid for 24 hours. Approximately 20 minutes before imaging, cells were stained with HCS LipidTOX™ Deep Red Neutral Lipid Stain (H34477, Invitrogen) at a 1:2000 dilution, and 1.0 µg/mL of Hoechst 33342 (H3570, Invitrogen). Endogenously tagged MLX was incubated as described above with the exception that cells were not transfected with plasmid DNA prior to imaging. Live cell imaging was performed using a Nikon Eclipse Ti inverted microscope equipped with a CSU-X1 spinning disk confocal head (Yokogawa); 405-, 488-, or 639-nm laser lines; Plan Apochromat VC 100x Oil objective (Nikon); and iXon Ultra 897 electron-multiplying charge-coupled device (EMCCD) or Zyla 4.2 Plus scientific complementary metal-oxide semiconductor (sCMOS) cameras (Andor). To control 85% humidity, 37 °C and 5% C02 levels, a stage top chamber was used (Okolab). Blue, green, red, and far-red fluorescence was excited by 405 nm, 488 nm, 560 nm, 637 nm (solid state; Andor, Andor, Cobolt, Coherent, respectively) lasers. All laser lines shared a quad-pass dichroic beamsplitter (Di01-T405/488/568/647, Semrock). Blue, green, red, and far-red emission was selected with FF01–452/45, FF03–525/50, FF01–607/36, or FF02–685/40 filters (Semrock) respectively, mounted in an external filter wheel. Multicolor images were acquired sequentially using NIS-Elements (Nikon) and processed using Fiji (Schindelin et al., 2012).
Fluorescence recovery after photobleaching
Kinetics of MLX diffusion was determined using fluorescence recovery after photobleaching. Cells were transfected with pTK-MLX-EGFP and incubated with oleic acid and HCS LipidTOX™ Deep Red Neutral Lipid Stain as described in “Live cell imaging”. Experiments (n=3) were performed using the Nikon Galvo miniscanner (Bruker) in which a region of interest (square of 7.60–8.48 µm in length) was photobleached using the 405 nm laser line at 15% laser power, 200 µs dwell time. Recovery of fluorescence was monitored immediately after photobleaching and thereafter at 60-second intervals for 60 minutes.
2-Deoxyglucose uptake
Uptake of [3H] 2-Deoxy-D-glucose was determined in THP-1 macrophages. Following indicated treatments, cells were washed twice in PBS and thereafter incubated for 7 minutes at 37°C with glucose-free media supplemented with 10 mM 2-Deoxy-D-glucose (D3179, Sigma-Aldrich) and 2.5 µCi/mL [3H] 2-Deoxy-D-glucose (NET549001MC, PerkinElmer). Assays were terminated by washing cells two times with ice-cold PBS. Subsequently, cells were solubilized on ice with cold lysis buffer containing 0.1% SDS, cOmplete™, EDTA-free Protease Inhibitor Cocktail (11873580001, Roche) and 70 units/mL Benzonase® Nuclease (E1014, Millipore). 3H was detected in 4 mL of Ultima Gold™ liquid scintillation cocktail (6013326, PerkinElmer) using a Hidex 300 SL scintillation counter and protein concentrations were determined using Pierce™ BCA Protein Assay Kit (23225, Thermo Scientific). Counts from each well were normalized by protein content.
Quantification and statistical analysis
Statistics
Unless otherwise stated, results are presented as mean ± standard deviation. Visualizations and statistical analyses of results were performed using appropriate packages in RStudio (version 1.0.143) and GraphPad Prism 8 (for statistical details of each experiment, see figure legends and below). Outliers were identified with the Grubbs’ method test using GraphPad Prism, setting alpha to 0.2. Statistically significant differences are denoted as follows: *p < 0.05, **p < 0.01, ***p < 0.001.
Analyses of genome-wide RNAi screen data
CellProfiler (Carpenter et al., 2006) was used to extract features from the RNAi screen images. For each extracted image feature, the median rz-score was calculated per gene, and the resulting matrix was filtered to exclude rows with a median |cell count rz-score| > 3 (n=547). Based on this dataset, image feature replicates were compared pair-wise across the genome-wide screen and non-reproducible parameters (median r-value < 0.3) were excluded. After this, a correlation matrix was generated by correlating all included image features with each other and the dimensionality of the matrix was tested using hierarchical clustering. Starting from the most reproducible image parameter in each major dimension (from the clustering), features were excluded if they covaried (|rho|>0.7) with the selected parameters and displayed low median reproducibility (r-value<0.4). This generated a dataset containing 21 high-confidence image parameters from which hits were identified. As the rz-scores for the different features varied in symmetry and dispersion (results not shown), a ranked-based approach was applied in which each image parameter was ranked from top to bottom by their respective rz-score, and hits were selected if they were distributed top or bottom 15 for one image parameter and/or top or bottom 50 for more than one image parameter. This resulted in 558 hits. Cluster 3 (de Hoon et al., 2004) and Java TreeView (Saldanha, 2004) were used to cluster and visualize hits without any cutoffs. To create a classification system, all hits were pair-wise correlated with each other across the 21 high-confidence image parameters. The resulting matrix was used as input for Cytoscape (Shannon et al., 2003) in which a hit network was constructed by filtering out low/modest pair-wise similarities (rho<0.9). After filtration, 292 hits interconnected by 630 edges remained. The topology of the network was generated using a prefuse force-directed layout and classification was performed manually based on visual inspection.
Processing of mass spectrometry data
Mass spectrometry data from proteome analyses were processed by MaxQuant software version 1.5.2.8 (Cox and Mann, 2008) using the following settings: oxidized methionine residues and protein N-terminal acetylation as variable modification, cysteine carbamidomethylation as fixed modification, first search peptide tolerance 20 ppm, main search peptide tolerance 4.5 ppm. Protease specificity was set to trypsin with up to two missed cleavages were allowed. Only peptides longer than six amino acids were analyzed, and the minimal ratio count to quantify a protein was set to two. The false discovery rate was set to 5% for peptide and protein identifications. Database searches were performed using the Andromeda search engine integrated into the MaxQuant environment (Cox et al., 2011) against the UniProt-human database containing 73,593 entries (November, 2018). “Matching between runs” algorithm with a time window of 0.7 minutes was employed to transfer identifications between samples processed using the same nanospray conditions. Protein tables were filtered to eliminate identifications from the reverse database and also common contaminants. Fold changes comparing LD fractions with total cell lysates were based on label-free quantification. To calculate 99% confidence intervals for canonical LD proteins, the top 50 high-confidence proteins targeting to LDs were extracted (Bersuker et al., 2018) and overlapped with the results generated within this study.
Processing of RNA sequencing data
Raw sequence data were converted to FASTQ format using bcl2fastq software (Illumina), and transcript abundance was quantified using Salmon (Patro et al., 2017). Results were imported into RStudio using tximport (Soneson et al., 2015), and differentially expressed genes were identified using DESeq2 (Love et al., 2014). In brief, the dataset was filtered to remove rows with only a single count across all samples and differentially expressed genes were identified using alpha < 0.1. Pathway analyses were performed using ToppFun (Chen et al., 2009). MLX target genes in THP-1 cells were identified using the following criteria: 1) induced by glucose (log2 fold change >1, alpha<0.1) in RISC-free transfected cells and 2) differentially expressed comparing RISC-free vs. siMLX transfected cells stimulated with 25 mM glucose (alpha<0.1) and 3) clustering next to known MLX target genes (ARRDC4 and TXNIP) and 4) displaying a similar LD phenotype (r>0.5) as MLX depletion across the 21 image parameters of the RNAi screen.
Image analyses
MLX LD enrichment was calculated as the average MLX signal identified around the LDs (± 1 pixel) divided by the average MLX signal of the cell minus LDs. Prior to calculations of nuclear MLX enrichment, nuclei were shrunken by 10 pixels. Prior to calculating the enrichment scores, background subtractions were performed. For FRAP experiments, median intensity of MLX around LDs was quantified. As a control, an unbleached region within the same cells was processed identically to the bleached region. All images were quantified using CellProfiler (Carpenter et al., 2006).
Data and code availability
Mass spectrometry source files were deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al., 2016) partner repository with the dataset identifier PXD012640. RNA sequencing data are deposited in the National Center for Biotechnology Information Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/), accession number GSE126002.
Supplementary Material
Table S1 (related to Figure 1). Robust z-scores for the identified RNAi screen hits.
Table S2 (related to Figure 3). THP-1 macrophage lipid droplet proteome.
Table S3 (related to Figure 3). SUM159 cell lipid droplet proteome.
Table S4 (related to Figure 7). Genes dependent on MLX for induction after glucose stimulus.
Table S5 (related to Figure 7). Genes regulated by ac-Lipo in THP-1 macrophages.
Data S1 (related to Figure 2). Clustering of RNAi screen hits.
Video S1 (related to Figure 3). MLX binds stably to LDs
After transfection with EGFP-tagged MLX, oleation (0.5 mM) for 1 day and staining of LDs with LipidTOX Deep Red, LD-binding properties of MLX were tested using fluorescence recovery after photobleaching. A representative time-lapse video of a cell is shown pre- and post-bleach.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-ATGL | Cell Signaling Techn. | Cat# 2138S |
| Mouse monoclonal anti-Calnexin | Santa Cruz Biotechn. | Cat# sc-46669 |
| Rabbit monoclonal anti-GAPDH | Cell Signaling Techn. | Cat# 5174S |
| Rabbit polyclonal anti-GFP | Abcam | Cat# ab290 |
| Rabbit monoclonal anti-GM130 | Cell Signaling Techn. | Cat# 12480S |
| Rabbit polyclonal anti-HSD17β11 | Abcam | Cat# ab136109 |
| Rabbit polyclonal anti-HSP60 | Cell Signaling Techn. | Cat# 4870S |
| Rabbit polyclonal anti-IgG | Cell Signaling Techn. | Cat# 2729S |
| Rabbit monoclonal anti-MLX | Cell Signaling Techn. | Cat# 85570S |
| Rabbit polyclonal anti-MLXIP | Bethyl Laboratories | Cat# A303–195A |
| Mouse monoclonal anti-NFkB p65 | Invitrogen | Cat# 33–9900 |
| Mouse monoclonal anti-SR-AI/MSR | R&D Systems | Cat# MAB2708 |
| Rabbit monoclonal anti-PLIN2 | Abcal | Cat# ab108323 |
| Guinea pig polyclonal anti-PLIN3 | Fitzgerald Industries | Cat# 20R-2598 |
| Mouse monoclonal anti-TUBA4A | Sigma-Aldrich | Cat# T5168 |
| Mouse anti-IgG kappa binding protein-HRP | Santa Cruz Biotechn. | Cat# sc-516102 |
| Mouse monoclonal anti-rabbit IgG-HRP | Santa Cruz Biotechn. | Cat# sc-2357 |
| Goat anti-guinea pig IgG-HRP | Santa Cruz Biotechn. | Cat# sc-2438 |
| Goat polyclonal anti-mouse IgG (H+L)-Alexa Fluor 647 | Invitrogen | Cat# A21236 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Human apolipoprotein B-containing lipoprotein | PanReac Applichem | Cat# A6961 |
| Oleic acid | Sigma-Aldrich | Cat# O1383 |
| Human acetylated low density lipoprotein | Alfa Aesar | Cat# BT-906 |
| DGAT1 inhibitor | Merck & Co | Liu et al., 2013 |
| DGAT2 inhibitor | Merck & Co | Imbriglio et al., 2015 |
| SBI-477 inhibitor | ProbeChem | Cat# PC-61673 |
| Phorbol 12-myristate 13-acetate | Sigma-Aldrich | Cat# P1585 |
| Lipofectamine RNAiMAX | Invitrogen | Cat# 13778150 |
| BODIPY 493/503 | Molecular Probes | Cat# D3922 |
| Hoechst 33342 | Molecular Probes | Cat# D3922 |
| FuGENE® HD Transfection Reagent | Promega | Cat# E2311 |
| Puromycin | Gibco | Cat# A1113803 |
| Benzonase® Nuclease | Millipore | Cat# E1014 |
| OptiPrep™ Density Gradient Medium | Sigma-Aldrich | Cat# D1556 |
| Sequencing grade Trypsin | Worthington Biochemical | Cat# LS02120 |
| Lipofectamine™ 3000 Transfection Reagent | Invitrogen | Cat# L3000008 |
| Targefect Hepatocyte | Targeting Systems | Cat# HEP-01 |
| HCS LipidTOX™ Deep Red Neutral Lipid Stain | Invitrogen | Cat# H34477 |
| [3H] 2-Deoxy-D-glucose | PerkinElmer | Cat# NET549001MC |
| Critical Commercial Assays | ||
| GenElute™ Mammalian Genomic DNA Miniprep Kit | Sigma-Aldrich | Cat# G1N350 |
| SimpleChIP® Enzymatic Chromatin IP Kit | Cell Signaling Techn. | Cat# 9003 |
| Q5® High-Fidelity PCR Kit | New England Biolabs | Cat# M0493L |
| Phusion® High-Fidelity PCR Kit | New England Biolabs | Cat# M0530L |
| NucleoSpin® Gel and PCR Clean-up Kit | Macherey-Nagel | Cat# 740609.250 |
| Zero Blunt™ TOPO™ PCR Cloning Kit | Invitrogen | Cat# 450245 |
| Pierce™ BCA Protein Assay Kit | Thermo Scientific | Cat# 23225 |
| SuperSignal West Pico | ThermoFisher Scientific | Cat# 34580 |
| SuperSignal West Femto | ThermoFisher Scientific | Cat# 34095 |
| RNeasy Mini Kit | QIAGEN | Cat# 74106 |
| QIAshredder | QIAGEN | Cat# 79656 |
| RNase-Free DNase Set | QIAGEN | Cat# 79254 |
| iScript™ cDNA Synthesis Kit | Bio-Rad | Cat# 1708891 |
| Power SYBR™ Green PCR Master Mix | Applied Biosystems | Cat# 4367659 |
| TruSeq® Stranded mRNA Library Prep kit | Illumina | Cat# 20020594 |
| Deposited Data | ||
| Proteomics | This paper | PXD012640 |
| RNA sequencing | This paper | GSE126002 |
| Experimental Models: Cell Lines | ||
| African green monkey COS-7 cells | ATCC | Cat# CRL-1651 |
| Human HEPG2 cells | ATCC | Cat# HB-8065 |
| Mouse primary hepatocytes | Nika Danial lab | N/A |
| Human SUM159 cells | Tomas Kirchhausen lab | N/A |
| Human THP-1 cells | ATCC | Cat# TIB-202 |
| Human U2OS cells | Peter Walter lab | N/A |
| Oligonucleotides | ||
| Guide RNA for HSL KO: TGC TGG TTA CCA ATC GGC CG |
This paper | N/A |
| Guide RNA for MLX KO: ACA AAG ACC GGCGGC GGC GC |
This paper | N/A |
| Guide RNA for MLXIP KO: TGG ACG TAG ACG AGC ACC GC |
This paper | N/A |
| Guide RNA for AAVS1 safe harbor: GGG GCC ACT AGG GAC AGG AT |
This paper | N/A |
| Guide RNA for MLX KI: CAG CTT TAC TGA CCG GTT CT |
This paper | N/A |
| siGENOME duplexes targeting RELA | Dharmacon | Cat# M-003533-02-0005 |
| siGENOME duplexes targeting MLXIP | Dharmacon | Cat# M-008976-01-0005 |
| siGENOME RISC-Free Control | Dharmacon | Cat# D-001220-01-05 |
| siGENOME duplexes targeting MLX | Dharmacon | Cat# M-009724-00-0005 |
| Dharmacon siGENOME SMARTpool siRNA library | Dharmacon | Cat# G-005005-025 |
| siGENOME duplexes targeting BSCL2 | Dharmacon | Cat# M-016749-00-0005 |
| siGENOME duplexes targeting FITM1 | Dharmacon | Cat# M-031931-00-0005 |
| For primer for qPCR and cloning, see STAR methods | This paper | N/A |
| Recombinant DNA | ||
| Plasmid: MLX (NM_198204) Human Untagged Clone | OriGene Technologies | Cat# SC322389 |
| Plasmid: MLXIP ORF cDNA clone | GeneCopoeia | Cat# EX-A1755-M02 |
| Plasmid: pSpCas9(BB)-2A-Puro (PX459) V2.0 | Ran et al., 2013 | Addgene cat# 62988 |
| Plasmid: PX330-U6-Chimeric_BB-CBh-hSpCas9 | Cong et al., 2013 | Addgene cat# 42230 |
| Plasmid: AAVS1_Puro_PGK1_3xFLAG_Twin_Strep | Dalvai et al., 2015 | Addgene cat# 68375 |
| Double-stranded, linear, nucleic acids: gBlocks gene fragments | Integrated DNA Technologies | N/A |
| Software and Algorithms | ||
| EVcouplings | Marks et al., 2011 | http://evfold.org/evfold-web/evfold.do |
| Jalview | Waterhouse et al., 2009 | http://www.jalview.org/ |
| PSIPRED | McGuffin et al., 2000 | http://bioinf.cs.ucl.ac.uk/psipred/ |
| Phyre2 | Kelley et al., 2015 | http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index |
| HELIQUEST | Gautier et al., 2008 | http://heliquest.ipmc.cnrs.fr/ |
| WebLogo 3 | Schneider and Stephens, 1990 | http://weblogo.threeplusone.com/create.cgi |
| NIS-Elements | Nikon | N/A |
| Fiji | Schindelin et al., 20112 | https://fiji.sc/ |
| RStudio (version 1.0.143) | RStudio Team | https://www.rstudio.com/ |
| GraphPad Prism 8 | GraphPad | www.graphpad.com |
| CellProfiler | Carpenter et al., 2006 | https://cellprofiler.org/ |
| Cluster 3 | de Hoon et al., 2004 | http://bonsai.hgc.jp/~mdehoon/software/cluster/software.htm |
| Java TreeView | Saldanha, 2004 | http://jtreeview.sourceforge.net/ |
| Cytoscape | Shannon et al., 2003 | https://cytoscape.org/ |
| MaxQuant | Cox and Mann, 2008 | https://www.maxquant.org/ |
| bcl2fastq | Illumina | https://support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software/downloads.html |
| Salmon | Patro et al., 2017 | https://combine-lab.github.io/salmon/ |
| ToppFun | Chen et al., 2009 | https://toppgene.cchmc.org/ |
Highlights.
Global RNAi screening identifies determinants of lipid storage in human macrophages
RNAi screen hits belong to six major classes with distinct lipid droplet morphology
MLX family of transcription factors bind to lipid droplets via amphipathic helices
Lipid droplet binding reduces MLX/MLXIP/MLXIPL transcriptional activity
ACKNOWLEDGMENTS
We thank Deepti Pant, Linus T. Tsai and Evan D. Rosen at the Functional Genomics and Bioinformatics Core (Boston Nutrition Obesity Research Center) for performing chromatin immuprecipitation experiments in SUM159 cells, Michael Wyler for contributions in assay development prior to screening, Nicholas Carreiro for data processing, David Logan and Jason Ignatius for help with CellProfiler, Jane Lee for technical assistance, Gary Howard for manuscript editing, Tomas Kirchhausen and Peter Walter for providing SUM159 and U2OS cells (respectively), and Merck & Co. for providing DGAT1 and DGAT2 inhibitors. This work was supported by the NIH [Mechanisms of Lipid Droplet Protein Targeting (R01GM097194) to T.C.W. and Mechanisms of Lipid Droplet Formation (5R01GM124348) to R.V.F.], Marie Skłodowska-Curie Actions (to N.M.), and The G. Harold and Leila Y. Mathers Foundation (to T.C.W.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTERESTS
The authors declare that no competing interests exist.
REFERENCES
- Abdul-Wahed A, Guilmeau S, and Postic C (2017). Sweet Sixteenth for ChREBP: Established Roles and Future Goals. Cell Metab 26, 324–341. [DOI] [PubMed] [Google Scholar]
- Accad M, Smith SJ, Newland DL, Sanan DA, King LE, Linton MF, Fazio S, and Farese RV (2000). Massive xanthomatosis and altered composition of atherosclerotic lesions in hyperlipidemic mice lacking acyl CoA:cholesterol acyltransferase 1. J. Clin. Invest 105, 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn B, Soundarapandian MM, Sessions H, Peddibhotla S, Roth GP, Li J-L, Sugarman E, Koo A, Malany S, Wang M, et al. (2016). MondoA coordinately regulates skeletal myocyte lipid homeostasis and insulin signaling. J. Clin. Invest 126, 3567–3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, and Lipman DJ (1990). Basic local alignment search tool. J. Mol. Biol 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Basu SK, Goldstein JL, Anderson GW, and Brown MS (1976). Degradation of cationized low density lipoprotein and regulation of cholesterol metabolism in homozygous familial hypercholesterolemia fibroblasts. Proc. Natl. Acad. Sci. U. S. A 73, 3178–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benhamed F, Denechaud P-D, Lemoine M, Robichon C, Moldes M, Bertrand-Michel J, Ratziu V, Serfaty L, Housset C, Capeau J, et al. (2012). The lipogenic transcription factor ChREBP dissociates hepatic steatosis from insulin resistance in mice and humans. J. Clin. Invest 122, 2176–2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bersuker K, Peterson CWH, To M, Sahl SJ, Savikhin V, Grossman EA, Nomura DK, and Olzmann JA (2018). A Proximity Labeling Strategy Provides Insights into the Composition and Dynamics of Lipid Droplet Proteomes. Dev. Cell 44, 97–112.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billin AN, Eilers AL, Queva C, and Ayer DE (1999). Mlx, a novel Max-like BHLHZip protein that interacts with the Max network of transcription factors. J. Biol. Chem 274, 36344–36350. [DOI] [PubMed] [Google Scholar]
- Billin AN, Eilers AL, Coulter KL, Logan JS, and Ayer DE (2000). MondoA, a novel basic helix-loop-helix-leucine zipper transcriptional activator that constitutes a positive branch of a max-like network. Mol. Cell. Biol 20, 8845–8854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brasaemle DL, Barber T, Wolins NE, Serrero G, Blanchette-Mackie EJ, and Londos C (1997). Adipose differentiation-related protein is an ubiquitously expressed lipid storage droplet-associated protein. J. Lipid Res 38, 2249–2263. [PubMed] [Google Scholar]
- Briggs MR, Yokoyama C, Wang X, Brown MS, and Goldstein JL (1993). Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. I. Identification of the protein and delineation of its target nucleotide sequence. J. Biol. Chem 268, 14490–14496. [PubMed] [Google Scholar]
- Cairo S, Merla G, Urbinati F, Ballabio A, and Reymond A (2001). WBSCR14, a gene mapping to the Williams--Beuren syndrome deleted region, is a new member of the Mlx transcription factor network. Hum. Mol. Genet 10, 617–627. [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. (2006). CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7, R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases S, Smith SJ, Zheng YW, Myers HM, Lear SR, Sande E, Novak S, Collins C, Welch CB, Lusis AJ, et al. (1998). Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. U. S. A 95, 13018–13023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cases S, Stone SJ, Zhou P, Yen E, Tow B, Lardizabal KD, Voelker T, and Farese RV (2001). Cloning of DGAT2, a Second Mammalian Diacylglycerol Acyltransferase, and Related Family Members. J. Biol. Chem 276, 38870–38876. [DOI] [PubMed] [Google Scholar]
- Chen J, Bardes EE, Aronow BJ, and Jegga AG (2009). ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 37, W305–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chitraju C, Mejhert N, Haas JT, Diaz-Ramirez LG, Grueter CA, Imbriglio JE, Pinto S, Koliwad SK, Walther TC, and Farese RV (2017). Triglyceride Synthesis by DGAT1 Protects Adipocytes from Lipid-Induced ER Stress during Lipolysis. Cell Metab 26, 407–418.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. (2013). Multiplex Genome Engineering Using CRISPR/Cas Systems. Science (80-.) 339, 819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Čopič A, Antoine-Bally S, Giménez-Andrés M, La Torre Garay C, Antonny B, Manni MM, Pagnotta S, Guihot J, and Jackson CL (2018). A giant amphipathic helix from a perilipin that is adapted for coating lipid droplets. Nat. Commun 9, 1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J, and Mann M (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol 26, 1367–1372. [DOI] [PubMed] [Google Scholar]
- Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, and Mann M (2011). Andromeda: a peptide search engine integrated into the MaxQuant environment. J. Proteome Res 10, 1794–1805. [DOI] [PubMed] [Google Scholar]
- Cox JS, Shamu CE, and Walter P (1993). Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73, 1197–1206. [DOI] [PubMed] [Google Scholar]
- Dalvai M, Loehr J, Jacquet K, Huard CC, Roques C, Herst P, Côté J, and Doyon Y (2015). A Scalable Genome-Editing-Based Approach for Mapping Multiprotein Complexes in Human Cells. Cell Rep 13, 621–633. [DOI] [PubMed] [Google Scholar]
- Davies MN, O’Callaghan BL, and Towle HC (2008). Glucose activates ChREBP by increasing its rate of nuclear entry and relieving repression of its transcriptional activity. J. Biol. Chem 283, 24029–24038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deisenroth C, Itahana Y, Tollini L, Jin A, and Zhang Y (2011). p53-inducible DHRS3 Is an Endoplasmic Reticulum Protein Associated with Lipid Droplet Accumulation. J. Biol. Chem 286, 28343–28356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dentin R, Benhamed F, Hainault I, Fauveau V, Foufelle F, Dyck JRB, Girard J, and Postic C (2006). Liver-Specific Inhibition of ChREBP Improves Hepatic Steatosis and Insulin Resistance in ob/ob Mice. Diabetes 55, 2159–2170. [DOI] [PubMed] [Google Scholar]
- Eilers AL, Sundwall E, Lin M, Sullivan AA, and Ayer DE (2002). A novel heterodimerization domain, CRM1, and 14–3-3 control subcellular localization of the MondoA-Mlx heterocomplex. Mol. Cell. Biol 22, 8514–8526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazio S, Major AS, Swift LL, Gleaves LA, Accad M, Linton MF, and Farese RV (2001). Increased atherosclerosis in LDL receptor-null mice lacking ACAT1 in macrophages. J. Clin. Invest 107, 163–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fei W, Shui G, Gaeta B, Du X, Kuerschner L, Li P, Brown AJ, Wenk MR, Parton RG, and Yang H (2008). Fld1p, a functional homologue of human seipin, regulates the size of lipid droplets in yeast. J. Cell Biol 180, 473–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folch J, Lees M, and Sloane Stanley GH (1957). A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem 226, 497–509. [PubMed] [Google Scholar]
- Fröhlich F, Petit C, Kory N, Christiano R, Hannibal-Bach H-K, Graham M, Liu X, Ejsing CS, Farese RV, and Walther TC (2015). The GARP complex is required for cellular sphingolipid homeostasis. Elife 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier R, Douguet D, Antonny B, and Drin G (2008). HELIQUEST: a web server to screen sequences with specific alpha-helical properties. Bioinformatics 24, 2101–2102. [DOI] [PubMed] [Google Scholar]
- Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, and Farese RV (2008). Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature 453, 657–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He K, Marsland R, Upadhyayula S, Song E, Dang S, Capraro BR, Wang W, Skillern W, Gaudin R, Ma M, et al. (2017). Dynamics of phosphoinositide conversion in clathrin-mediated endocytic traffic. Nature 552, 410–414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Reese ML, and Goodman JM (2018). The assembly of lipid droplets and their roles in challenged cells. EMBO J 37, e98947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman MA, Peroni OD, Villoria J, Schön MR, Abumrad NA, Blüher M, Klein S, and Kahn BB (2012). A novel ChREBP isoform in adipose tissue regulates systemic glucose metabolism. Nature 484, 333–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hoon MJL, Imoto S, Nolan J, and Miyano S (2004). Open source clustering software. Bioinformatics 20, 1453–1454. [DOI] [PubMed] [Google Scholar]
- Hua X, Yokoyama C, Wu J, Briggs MR, Brown MS, Goldstein JL, and Wang X (1993). SREBP-2, a second basic-helix-loop-helix-leucine zipper protein that stimulates transcription by binding to a sterol regulatory element. Proc. Natl. Acad. Sci. U. S. A 90, 11603–11607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka K, Bruick RK, Liang G, Horton JD, and Uyeda K (2004). Deficiency of carbohydrate response element-binding protein (ChREBP) reduces lipogenesis as well as glycolysis. Proc. Natl. Acad. Sci 101, 7281–7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iizuka K, Takeda J, and Horikawa Y (2009). Hepatic overexpression of dominant negative Mlx improves metabolic profile in diabetes-prone C57BL/6J mice. Biochem. Biophys. Res. Commun 379, 499–504. [DOI] [PubMed] [Google Scholar]
- Imbriglio JE, Shen D-M, Liang R, Marby K, You M, Youm HW, Feng Z, London C, Xiong Y, Tata J, et al. (2015). Discovery and Pharmacology of a Novel Class of Diacylglycerol Acyltransferase 2 Inhibitors. J. Med. Chem 58, 9345–9353. [DOI] [PubMed] [Google Scholar]
- Ishibashi K, Sasaki S, Fushimi K, Uchida S, Kuwahara M, Saito H, Furukawa T, Nakajima K, Yamaguchi Y, and Gojobori T (1994). Molecular cloning and expression of a member of the aquaporin family with permeability to glycerol and urea in addition to water expressed at the basolateral membrane of kidney collecting duct cells. Proc. Natl. Acad. Sci. U. S. A 91, 6269–6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayson CBK, Arlt H, Fischer AW, Lai ZW, Farese RV, and Walther TC (2018). Rab18 is not necessary for lipid droplet biogenesis or turnover in human mammary carcinoma cells. Mol. Biol. Cell 29, 2045–2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi T, Takenoshita M, Kabashima T, and Uyeda K (2001). Glucose and cAMP regulate the L-type pyruvate kinase gene by phosphorylation/dephosphorylation of the carbohydrate response element binding protein. Proc. Natl. Acad. Sci. U. S. A 98, 13710–13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley LA, Mezulis S, Yates CM, Wass MN, and Sternberg MJE (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc 10, 845–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koliwad SK, Streeper RS, Monetti M, Cornelissen I, Chan L, Terayama K, Naylor S, Rao M, Hubbard B, and Farese RV (2010). DGAT1-dependent triacylglycerol storage by macrophages protects mice from diet-induced insulin resistance and inflammation. J. Clin. Invest 120, 756–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kory N, Thiam A-R, Farese RV, and Walther TC (2015). Protein Crowding Is a Determinant of Lipid Droplet Protein Composition. Dev. Cell 34, 351–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kory N, Farese RV, and Walther TC (2016). Targeting Fat: Mechanisms of Protein Localization to Lipid Droplets. Trends Cell Biol 26, 535–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N, Farese RV, and Walther TC (2013). Balancing the fat: lipid droplets and human disease. EMBO Mol. Med 5, 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krahmer N, Najafi B, Schueder F, Quagliarini F, Steger M, Seitz S, Kasper R, Salinas F, Cox J, Uhlenhaut NH, et al. (2018). Organellar Proteomics and Phospho-Proteomics Reveal Subcellular Reorganization in Diet-Induced Hepatic Steatosis. Dev. Cell 47, 205–221.e7. [DOI] [PubMed] [Google Scholar]
- Lardizabal KD, Mai JT, Wagner NW, Wyrick A, Voelker T, and Hawkins DJ (2001). DGAT2 Is a New Diacylglycerol Acyltransferase Gene Family. J. Biol. Chem 276, 38862–38869. [DOI] [PubMed] [Google Scholar]
- Lee YS, Wollam J, and Olefsky JM (2018). An Integrated View of Immunometabolism. Cell 172, 22–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner R, and Vance DE (1999). Cloning and expression of a cDNA encoding a hepatic microsomal lipase that mobilizes stored triacylglycerol. Biochem. J 343 Pt 1, 1–10. [PMC free article] [PubMed] [Google Scholar]
- Listenberger LL, Han X, Lewis SE, Cases S, Farese RV, Ory DS, and Schaffer JE (2003). Triglyceride accumulation protects against fatty acid-induced lipotoxicity. Proc. Natl. Acad. Sci. U. S. A 100, 3077–3082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Gorski JN, Gold SJ, Chen D, Chen S, Forrest G, Itoh Y, Marsh DJ, McLaren DG, Shen Z, et al. (2013). Pharmacological inhibition of diacylglycerol acyltransferase 1 reduces body weight and modulates gut peptide release-Potential insight into mechanism of action. Obesity 21, 1406–1415. [DOI] [PubMed] [Google Scholar]
- Liu P, Ying Y, Zhao Y, Mundy DI, Zhu M, and Anderson RGW (2004). Chinese Hamster Ovary K2 Cell Lipid Droplets Appear to Be Metabolic Organelles Involved in Membrane Traffic. J. Biol. Chem 279, 3787–3792. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, and Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Luis O, Valero MC, and Pérez Jurado LA (2000). WBSCR14, a putative transcription factor gene deleted in Williams-Beuren syndrome: complete characterisation of the human gene and the mouse ortholog. Eur. J. Hum. Genet 8, 215–222. [DOI] [PubMed] [Google Scholar]
- Ma L, Tsatsos NG, and Towle HC (2005). Direct role of ChREBP.Mlx in regulating hepatic glucose-responsive genes. J. Biol. Chem 280, 12019–12027. [DOI] [PubMed] [Google Scholar]
- Marks DS, Colwell LJ, Sheridan R, Hopf TA, Pagnani A, Zecchina R, and Sander C (2011). Protein 3D structure computed from evolutionary sequence variation. PLoS One 6, e28766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S, Driessen K, Nixon SJ, Zerial M, and Parton RG (2005). Regulated Localization of Rab18 to Lipid Droplets. J. Biol. Chem 280, 42325–42335. [DOI] [PubMed] [Google Scholar]
- Matsumoto M, Pocai A, Rossetti L, Depinho RA, and Accili D (2007). Impaired regulation of hepatic glucose production in mice lacking the forkhead transcription factor Foxo1 in liver. Cell Metab 6, 208–216. [DOI] [PubMed] [Google Scholar]
- McGuffin LJ, Bryson K, and Jones DT (2000). The PSIPRED protein structure prediction server. Bioinformatics 16, 404–405. [DOI] [PubMed] [Google Scholar]
- McKenna ES, Tamayo P, Cho Y-J, Tillman EJ, Mora-Blanco EL, Sansam CG, Koellhoffer EC, Pomeroy SL, and Roberts CWM (2012). Epigenetic inactivation of the tumor suppressor BIN1 drives proliferation of SNF5-deficient tumors. Cell Cycle 11, 1956–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meroni G, Cairo S, Merla G, Messali S, Brent R, Ballabio A, and Reymond A (2000). Mlx, a new Max-like bHLHZip family member: the center stage of a novel transcription factors regulatory pathway? Oncogene 19, 3266–3277. [DOI] [PubMed] [Google Scholar]
- Minn AH, Hafele C, and Shalev A (2005). Thioredoxin-Interacting Protein Is Stimulated by Glucose through a Carbohydrate Response Element and Induces β-Cell Apoptosis. Endocrinology 146, 2397–2405. [DOI] [PubMed] [Google Scholar]
- Moessinger C, Kuerschner L, Spandl J, Shevchenko A, and Thiele C (2011). Human lysophosphatidylcholine acyltransferases 1 and 2 are located in lipid droplets where they catalyze the formation of phosphatidylcholine. J. Biol. Chem 286, 21330–21339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore KJ, Sheedy FJ, and Fisher EA (2013). Macrophages in atherosclerosis: a dynamic balance. Nat. Rev. Immunol 13, 709–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori K, Ma W, Gething MJ, and Sambrook J (1993). A transmembrane protein with a cdc2+/CDC28-related kinase activity is required for signaling from the ER to the nucleus. Cell 74, 743–756. [DOI] [PubMed] [Google Scholar]
- Morigny P, Houssier M, Mairal A, Ghilain C, Mouisel E, Benhamed F, Masri B, Recazens E, Denechaud P-D, Tavernier G, et al. (2019). Interaction between hormone-sensitive lipase and ChREBP in fat cells controls insulin sensitivity. Nat. Metab 1, 133–146. [DOI] [PubMed] [Google Scholar]
- Olzmann JA, and Carvalho P (2018). Dynamics and functions of lipid droplets. Nat. Rev. Mol. Cell Biol [DOI] [PMC free article] [PubMed]
- Ozeki S, Cheng J, Tauchi-Sato K, Hatano N, Taniguchi H, and Fujimoto T (2005). Rab18 localizes to lipid droplets and induces their close apposition to the endoplasmic reticulum-derived membrane. J. Cell Sci 118, 2601–2611. [DOI] [PubMed] [Google Scholar]
- Pataki CI, Rodrigues J, Zhang L, Qian J, Efron B, Hastie T, Elias JE, Levitt M, and Kopito RR (2018). Proteomic analysis of monolayer-integrated proteins on lipid droplets identifies amphipathic interfacial α-helical membrane anchors. Proc. Natl. Acad. Sci. U. S. A 115, E8172–E8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patro R, Duggal G, Love MI, Irizarry RA, and Kingsford C (2017). Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévost C, Sharp ME, Kory N, Lin Q, Voth GA, Farese RV, and Walther TC (2018). Mechanism and Determinants of Amphipathic Helix-Containing Protein Targeting to Lipid Droplets. Dev. Cell 44, 73–86.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, and Zhang F (2013). Genome engineering using the CRISPR-Cas9 system. Nat. Protoc 8, 2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Ishihama Y, and Mann M (2003). Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem 75, 663–670. [DOI] [PubMed] [Google Scholar]
- Richards P, Ourabah S, Montagne J, Burnol A-F, Postic C, and Guilmeau S (2017). MondoA/ChREBP: The usual suspects of transcriptional glucose sensing; Implication in pathophysiology. Metabolism 70, 133–151. [DOI] [PubMed] [Google Scholar]
- Richards P, Rachdi L, Oshima M, Marchetti P, Bugliani M, Armanet M, Postic C, Guilmeau S, and Scharfmann R (2018). MondoA Is an Essential Glucose-Responsive Transcription Factor in Human Pancreatic β-Cells. Diabetes 67, 461–472. [DOI] [PubMed] [Google Scholar]
- Romanauska A, and Köhler A (2018). The Inner Nuclear Membrane Is a Metabolically Active Territory that Generates Nuclear Lipid Droplets. Cell 174, 700–715.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, and Spiegelman BM (2014). What We Talk About When We Talk About Fat. Cell 156, 20–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sae-Lee C, Moolsuwan K, Chan L, and Poungvarin N (2016). ChREBP Regulates Itself and Metabolic Genes Implicated in Lipid Accumulation in β-Cell Line. PLoS One 11, e0147411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ (2004). Java Treeview--extensible visualization of microarray data. Bioinformatics 20, 3246–3248. [DOI] [PubMed] [Google Scholar]
- Sans CL, Satterwhite DJ, Stoltzman CA, Breen KT, and Ayer DE (2006). MondoA-Mlx Heterodimers Are Candidate Sensors of Cellular Energy Status: Mitochondrial Localization and Direct Regulation of Glycolysis. Mol. Cell. Biol 26, 4863–4871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. (2009). A Gene Network Regulating Lysosomal Biogenesis and Function. Science (80-. ) 325, 473–477. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider TD, and Stephens RM (1990). Sequence logos: a new way to display consensus sequences. Nucleic Acids Res 18, 6097–6100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Settembre C, Di Malta C, Polito VA, Arencibia MG, Vetrini F, Erdin S, Erdin SU, Huynh T, Medina D, Colella P, et al. (2011). TFEB Links Autophagy to Lysosomal Biogenesis. Science (80-. ) 332, 1429–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003). Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res 13, 2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluijs P, Hull M, Webster P, Mâle P, Goud B, and Mellman I (1992). The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell 70, 729–740. [DOI] [PubMed] [Google Scholar]
- Soneson C, Love MI, and Robinson MD (2015). Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Research 4, 1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoltzman CA, Peterson CW, Breen KT, Muoio DM, Billin AN, and Ayer DE (2008). Glucose sensing by MondoA:Mlx complexes: A role for hexokinases and direct regulation of thioredoxin-interacting protein expression. Proc. Natl. Acad. Sci 105, 6912–6917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymanski KM, Binns D, Bartz R, Grishin NV, Li W-P, Agarwal AK, Garg A, Anderson RGW, and Goodman JM (2007). The lipodystrophy protein seipin is found at endoplasmic reticulum lipid droplet junctions and is important for droplet morphology. Proc. Natl. Acad. Sci. U. S. A 104, 20890–20895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabas I, and Bornfeldt KE (2016). Macrophage Phenotype and Function in Different Stages of Atherosclerosis. Circ. Res 118, 653–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiam AR, Antonny B, Wang J, Delacotte J, Wilfling F, Walther TC, Beck R, Rothman JE, and Pincet F (2013). COPI buds 60-nm lipid droplets from reconstituted water-phospholipid-triacylglyceride interfaces, suggesting a tension clamp function. Proc. Natl. Acad. Sci. U. S. A 110, 13244–13249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uyeda K, and Repa JJ (2006). Carbohydrate response element binding protein, ChREBP, a transcription factor coupling hepatic glucose utilization and lipid synthesis. Cell Metab 4, 107–110. [DOI] [PubMed] [Google Scholar]
- Vijayakumar A, Aryal P, Wen J, Syed I, Vazirani RP, Moraes-Vieira PM, Camporez JP, Gallop MR, Perry RJ, Peroni OD, et al. (2017). Absence of Carbohydrate Response Element Binding Protein in Adipocytes Causes Systemic Insulin Resistance and Impairs Glucose Transport. Cell Rep 21, 1021–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva CJ, Monetti M, Shih M, Zhou P, Watkins SM, Bhanot S, and Farese RV (2009). Specific role for acyl CoA:Diacylglycerol acyltransferase 1 (Dgat1) in hepatic steatosis due to exogenous fatty acids. Hepatology 50, 434–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno JA, Csordas A, Del-Toro N, Dianes JA, Griss J, Lavidas I, Mayer G, Perez-Riverol Y, Reisinger F, Ternent T, et al. (2016). 2016 update of the PRIDE database and its related tools. Nucleic Acids Res 44, 11033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther TC, Chung J, and Farese RV (2017). Lipid Droplet Biogenesis. Annu. Rev. Cell Dev. Biol 33, 491–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Becuwe M, Housden BE, Chitraju C, Porras AJ, Graham MM, Liu XN, Thiam AR, Savage DB, Agarwal AK, et al. (2016). Seipin is required for converting nascent to mature lipid droplets. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Briggs MR, Hua X, Yokoyama C, Goldstein JL, and Brown MS (1993). Nuclear protein that binds sterol regulatory element of low density lipoprotein receptor promoter. II. Purification and characterization. J. Biol. Chem 268, 14497–14504. [PubMed] [Google Scholar]
- Wang X, Sato R, Brown MS, Hua X, and Goldstein JL (1994). SREBP-1, a membrane-bound transcription factor released by sterol-regulated proteolysis. Cell 77, 53–62. [DOI] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, and Barton GJ (2009). Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics 25, 1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D, and Flügge UI (1984). A method for the quantitative recovery of protein in dilute solution in the presence of detergents and lipids. Anal. Biochem 138, 141–143. [DOI] [PubMed] [Google Scholar]
- Wilde BR, Ye Z, Lim T-Y, and Ayer DE (2019). Cellular acidosis triggers human MondoA transcriptional activity by driving mitochondrial ATP production. Elife 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu N, Zheng B, Shaywitz A, Dagon Y, Tower C, Bellinger G, Shen C-H, Wen J, Asara J, McGraw TE, et al. (2013). AMPK-dependent degradation of TXNIP upon energy stress leads to enhanced glucose uptake via GLUT1. Mol. Cell 49, 1167–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita H, Takenoshita M, Sakurai M, Bruick RK, Henzel WJ, Shillinglaw W, Arnot D, and Uyeda K (2001). A glucose-responsive transcription factor that regulates carbohydrate metabolism in the liver. Proc. Natl. Acad. Sci 98, 9116–9121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 (related to Figure 1). Robust z-scores for the identified RNAi screen hits.
Table S2 (related to Figure 3). THP-1 macrophage lipid droplet proteome.
Table S3 (related to Figure 3). SUM159 cell lipid droplet proteome.
Table S4 (related to Figure 7). Genes dependent on MLX for induction after glucose stimulus.
Table S5 (related to Figure 7). Genes regulated by ac-Lipo in THP-1 macrophages.
Data S1 (related to Figure 2). Clustering of RNAi screen hits.
Video S1 (related to Figure 3). MLX binds stably to LDs
After transfection with EGFP-tagged MLX, oleation (0.5 mM) for 1 day and staining of LDs with LipidTOX Deep Red, LD-binding properties of MLX were tested using fluorescence recovery after photobleaching. A representative time-lapse video of a cell is shown pre- and post-bleach.
Data Availability Statement
Mass spectrometry source files were deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al., 2016) partner repository with the dataset identifier PXD012640. RNA sequencing data are deposited in the National Center for Biotechnology Information Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo/), accession number GSE126002.