Abstract
Background
We sought to identify patient subgroups with distinct postprogression overall survival (ppOS) outcomes and investigate the impact of original treatment assignment and initial postprogression treatment (ppRx) on ppOS.
Methods
Recursive partitioning analysis (RPA) was performed to model relationships between prespecified covariates and ppOS in patients with BRAFV600-mutated metastatic melanoma who had experienced progressive disease (PD) following treatment with cobimetinib plus vemurafenib, vemurafenib monotherapy, or dacarbazine in the BRIM-2, BRIM-3, BRIM-7, and coBRIM studies. Prognostic subgroups identified by RPA were then applied to pooled treatment cohorts. The primary endpoint was ppOS, defined as time from first PD to death from any cause.
Results
RPA identified baseline lactate dehydrogenase (LDH), baseline disease stage, Eastern Cooperative Oncology Group performance status at PD, and ppRx as significant prognostic factors for ppOS. Median ppOS was longest in patients with normal baseline LDH, stage M1c disease at baseline, and ppRx with immunotherapy or targeted therapy (12.2 months; 95% CI 10.3–16.1) and shortest in those with elevated baseline LDH > 2 × upper limit of normal (2.3 months; 95% CI 1.8–2.7). Original treatment assignment did not impact ppOS. Across treatment cohorts, patients treated with immunotherapy or targeted therapy after PD had better ppOS than those given other treatments.
Conclusion
A combination of factors at baseline (LDH, disease stage) and PD (performance status, ppRx) impact ppOS outcomes. ppRx with immunotherapy or targeted therapy is an independent prognostic factor for improved overall survival following progression regardless of original treatment.
Trial registration The trials included in this analysis are registered with ClinicalTrials.gov: NCT00949702 (BRIM-2), NCT01006980 (BRIM-3), NCT01271803 (BRIM-7), and NCT01689519 (coBRIM).
Keywords: Vemurafenib, Cobimetinib, Dacarbazine, Melanoma, Survival analysis
Background
Vemurafenib monotherapy and cobimetinib plus vemurafenib have improved survival in patients with BRAFV600-mutated metastatic melanoma [1–7]. Extended follow-up of clinical studies evaluating BRAF inhibitor (BRAFi) monotherapy or combined BRAFi and MEK inhibitor (MEKi) shows a plateau in overall survival (OS) curves after approximately 3 years [8–11]. A similar plateau in OS is observed with ipilimumab [12]. These observations suggest that a subgroup of patients with metastatic melanoma have good long-term survival prognosis. Prognostic factors for survival in patients with metastatic melanoma include disease stage (number/location of metastases), baseline lactate dehydrogenase (LDH), and baseline Eastern Cooperative Oncology Group performance status (ECOG PS) [13, 14]. Recent analyses suggest that LDH remains the most important prognostic factor for survival in patients treated with BRAFi and/or MEKi or immunotherapy, with particularly poor outcomes being observed in patients with LDH elevated > 2 × upper limit of normal (ULN) [15–18]. However, the impact on survival outcomes of clinical and disease-related variables and treatment following progression are poorly understood. Some reports suggest that prior BRAFi treatment is associated with inferior response to subsequent immunotherapy [19–23], whereas others suggest that the efficacy of immunotherapy is unaffected by previous BRAFi therapy [24–27]. Insight into the impact of clinical characteristics and subsequent treatment on survival following progressive disease (PD) may inform treatment decisions in the management of patients with metastatic melanoma.
The objectives of this analysis were to 1) identify patient subgroups with distinct postprogression overall survival (ppOS) outcomes and 2) investigate the impact of original assigned treatment (cobimetinib plus vemurafenib, vemurafenib monotherapy, or dacarbazine) and initial postprogression treatment (ppRx) on ppOS in patients with BRAFV600-mutated metastatic melanoma.
Methods
Study design and participants
Data were pooled from the BRIM-2 (NCT00949702) [1], BRIM-3 (NCT01006980) [3, 4], BRIM-7 (NCT01271803) [5], and coBRIM (NCT01689519) [6, 7] studies. Detailed methods have been previously reported. Briefly, BRIM-2 was an open-label, multicenter phase 2 trial of oral vemurafenib 960 mg twice daily [2]. BRIM-3 was an open-label, multicenter, randomized phase 3 trial of oral vemurafenib 960 mg twice daily compared with intravenous dacarbazine 1000 mg/m2 every 3 weeks [3, 4]. BRIM-7 was an open-label, multicenter phase 1b dose-escalation study of oral cobimetinib 60, 80, or 100 mg once daily on a 14 days on/14 days off schedule (14/14), a 21 days on/7 days off schedule (21/7), or continuously (28/0) combined with oral vemurafenib 720 or 960 mg twice daily [5]. coBRIM was a randomized, double-blind phase 3 study of the combination of oral cobimetinib 60 mg once daily on a 21/7 schedule combined with oral vemurafenib 960 mg twice daily compared with placebo plus vemurafenib [6, 7]. Key eligibility criteria were similar across trials; eligible patients were aged ≥ 18 years with unresectable stage IIIC or IV melanoma harboring a BRAFV600 mutation, ECOG PS of 0–1, and adequate organ function. BRIM-3 and coBRIM enrolled previously untreated patients only, whereas BRIM-2 enrolled patients who had received prior systemic treatment for advanced disease, and BRIM-7 enrolled both previously treated and untreated patients. All four trials allowed enrollment of patients with previously treated, stable brain metastases.
All eligible patients (defined by each study protocol) who had PD at the time of data cutoff were included in analyses regardless of treatment assignment. Patients with missing data for covariates of interest were excluded. For BRIM-7, only BRAFi-naive patients were included in the analysis.
Statistical analysis
Recursive partitioning analysis
Recursive partitioning analysis (RPA) is a statistical method that creates a decision tree by classifying patients into subgroups, defined by independent prognostic factors, with statistically distinct survival probabilities. RPA was undertaken to model relationships between prespecified covariates and ppOS in patients who had experienced PD at the time of data cutoff (BRIM-2: February 1, 2012; BRIM-3: July 8, 2015; BRIM-7: December 2, 2015; coBRIM: August 28, 2015).
Covariates were prespecified in study protocols and/or identified as potential prognostic factors from peer-reviewed medical literature. Prespecified baseline covariates were age (< 65 vs ≥ 65 years), sex (male vs female), race (white vs nonwhite), geographic region (North America, Europe, or Australia/New Zealand/other), ECOG PS (0, 1, or 2), disease stage (unresectable IIIC, M1a, M1b, M1c), LDH (normal, elevated ≤ 2 × ULN or elevated > 2 × ULN), sum of longest diameters (SLD) of target lesions, histologic subtype (acral lentiginous, lentigo maligna, nodular, superficial spreading, or other), prior adjuvant therapy (yes vs no), and original treatment assignment (cobimetinib plus vemurafenib, vemurafenib monotherapy, or dacarbazine). Prespecified covariates at the time of PD were best response before PD, LDH (normal, elevated ≤ 2 × ULN, or elevated > 2 × ULN), ECOG PS (0, 1, or 2), metastasis (new lesion vs no new lesion; central nervous system [CNS] vs non-CNS), SLD, enlargement of baseline lesions, maximum change in SLD, time from maximum change in SLD to PD, absolute lymphocyte count (normal, low, or elevated), ratio of absolute neutrophil count to absolute lymphocyte count, and initial ppRx (immunotherapy/targeted therapy vs other).
Initial ppRx was defined as ≥ 1 dose of ipilimumab, pembrolizumab, nivolumab, or similar therapy within 90 days after PD (immunotherapy); ≥ 1 dose of vemurafenib, cobimetinib, dabrafenib, trametinib, or similar therapy within 90 days after PD (targeted therapy); or receipt of any therapy other than immunotherapy or targeted therapy, both immunotherapy and targeted therapy, or no treatment within 90 days after PD (other).
RPAs used a unified framework for conditional inference (permutation tests) using censored response variables to avoid bias in selection of covariates and minimize overfitting of the data [28]. This approach ensures a right-sized tree with no need for pruning or cross-validation. The global null hypothesis of independence between any of the prespecified input variables and survival was tested. If the null hypothesis could not be rejected, the analysis was stopped; otherwise, the covariate most strongly associated with survival was identified based on univariate P values and a binary split was implemented. These steps were repeated until no further covariates with a significant association with survival could be distinguished. The stop criterion was based on either multiplicity-adjusted univariate P values or a prespecified threshold value of the test statistic (both criteria were maximized using 1—P value).
ppOS
The primary endpoint for this analysis was ppOS, defined as time from first PD to death from any cause, estimated using the Kaplan–Meier method. ppOS was evaluated using updated data in patients who had experienced PD at the time of updated data cutoff (BRIM-2: February 1, 2012; BRIM-3: July 8, 2015; BRIM-7: July 10, 2017; coBRIM: October 13, 2017). Prognostic subgroups identified by RPA were applied to the total pooled population and to pooled treatment cohorts (cobimetinib plus vemurafenib, vemurafenib monotherapy, and dacarbazine). Patients who did not die were censored at data cutoff.
Results
RPA
At the time of data cutoff for the RPA, 1004 patients had experienced PD; after excluding patients with missing data, 809 patients were included in the RPA. Demographic and clinical characteristics were comparable across the pooled population and pooled treatment cohorts (Table 1).
Table 1.
Demographic and clinical characteristics for patients included in the RPA
| Characteristic | All pooled patients N = 1004 | Cobimetinib + vemurafenib n = 184 | Vemurafenib monotherapy n = 555 | Dacarbazine n = 265 |
|---|---|---|---|---|
| Age, n (%) | (n = 1004) | (n = 184) | (n = 555) | (n = 265) |
| < 65 y | 758 (75.5) | 142 (77.2) | 405 (73.0) | 211 (79.6) |
| ≥ 65 y | 246 (24.5) | 42 (22.8) | 150 (27.0) | 54 (20.4) |
| Sex, n (%) | (n = 1004) | (n = 184) | (n = 555) | (n = 265) |
| Male | 600 (59.8) | 117 (63.6) | 335 (60.4) | 148 (55.8) |
| Female | 404 (40.2) | 67 (36.4) | 220 (39.6) | 117 (44.2) |
| Race, n (%) | (n = 1004) | (n = 184) | (n = 555) | (n = 265) |
| White | 971 (96.7) | 167 (90.8) | 539 (97.1) | 265 (100.0) |
| Nonwhite | 33 (3.3) | 17 (9.2) | 16 (2.9) | 0 |
| Region, n (%) | (n = 1004) | (n = 184) | (n = 555) | (n = 265) |
| North America | 270 (26.9) | 47 (25.5) | 163 (29.4) | 60 (22.6) |
| Europe | 590 (58.8) | 108 (58.7) | 314 (56.6) | 168 (63.4) |
| Australia/New Zealand/Others | 144 (14.3) | 29 (15.8) | 78 (14.1) | 37 (14.0) |
| Baseline disease stage, n (%) | (n = 1004) | (n = 184) | (n = 555) | (n = 265) |
| Unresectable IIIC, M1a, or M1b | 332 (33.1) | 57 (31.0) | 184 (33.2) | 91 (34.3) |
| M1c | 672 (66.9) | 127 (69.0) | 371 (66.8) | 174 (65.7) |
| Baseline ECOG PS, n (%) | (n = 1001) | (n = 182) | (n = 554) | (n = 265) |
| 0 | 651 (65.0) | 129 (70.9) | 343 (61.9) | 179 (67.5) |
| 1 | 350 (35.0) | 53 (29.1) | 211 (38.1) | 86 (32.5) |
| Baseline LDH, n (%) | (n = 942) | (n = 173) | (n = 523) | (n = 246) |
| Normal | 495 (52.5) | 82 (47.4) | 276 (52.8) | 137 (55.7) |
| Elevated ≤ 2 × ULN | 290 (30.8) | 61 (35.3) | 154 (29.4) | 75 (30.5) |
| Elevated > 2 × ULN | 157 (16.7) | 30 (17.3) | 93 (17.8) | 34 (13.8) |
| Baseline liver metastasis, n (%) | (n = 1001) | (n = 184) | (n = 552) | (n = 265) |
| Yes | 375 (35.7) | 69 (37.5) | 199 (36.1) | 89 (33.6) |
| No | 644 (64.3) | 115 (62.5) | 353 (63.9) | 176 (66.4) |
| ECOG PS at PD, n (%) | (n = 926) | (n = 166) | (n = 519) | (n = 241) |
| 0 | 489 (52.8) | 108 (65.1) | 266 (51.3) | 115 (47.7) |
| 1 | 356 (38.4) | 50 (30.1) | 209 (40.3) | 97 (40.2) |
| 2 | 55 (5.9) | 4 (2.4) | 29 (5.6) | 22 (9.1) |
| 3 | 22 (2.4) | 4 (2.4) | 13 (2.5) | 5 (2.1) |
| 4 | 4 (0.4) | 0 | 2 (0.4) | 2 (0.8) |
| Postprogression treatment, n (%) | (n = 1004) | (n = 184) | (n = 555) | (n = 265) |
| Immunotherapy | 202 (20.1) | 39 (21.2) | 114 (20.5) | 49 (18.5) |
| Targeted therapy | 58 (5.8) | 14 (7.6) | 27 (4.9) | 17 (6.4) |
| Other | 744 (74.1) | 131 (71.2) | 414 (74.6) | 199 (75.1) |
| Baseline SLD, mm | (n = 995) | (n = 184) | (n = 546) | (n = 265) |
| Mean (SD) | 87.1 (75.2) | 86.2 (63.7) | 91.2 (85.6) | 79.3 (57.2) |
| Median (range) | 70.0 (9–1310) | 72.0 (10–398) | 71.5 (9–1310) | 66.0 (9–295) |
ECOG PS Eastern Cooperative Oncology Group performance status, LDH lactate dehydrogenase, PD progressive disease, ppOS postprogression overall survival, RPA recursive partitioning analysis, SLD sum of longest diameters, ULN upper limit of normal
Median postprogression follow-up durations were 5.4 months (interquartile range [IQR], 2.2–12.5 months) for all patients, 4.8 months (IQR, 1.9–10.7 months) for the cobimetinib plus vemurafenib cohort, 5.2 months (IQR, 2.2–12.0 months) for the vemurafenib monotherapy cohort, and 6.5 months (IQR, 2.6–15.6 months) for the dacarbazine cohort.
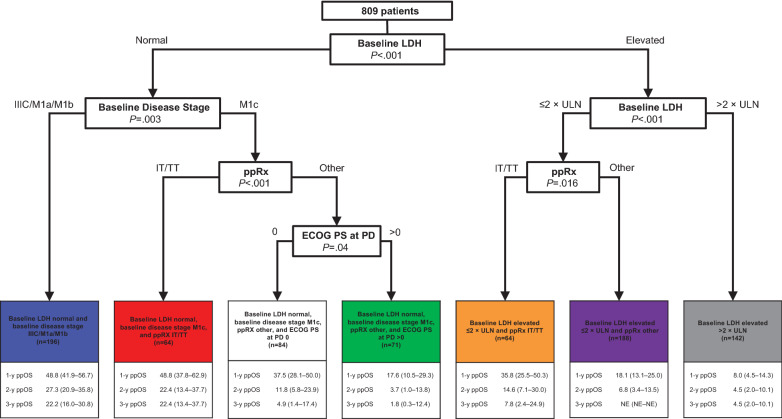
In all pooled patients, the RPA identified baseline LDH, baseline disease stage, ECOG PS at PD, and initial ppRx as significant prognostic factors for ppOS, producing seven subgroups with distinct outcomes (Fig. 1). Notably, CNS metastasis at PD was not identified as a significant prognostic factor for ppOS, although this subgroup was relatively small (n = 174). Baseline LDH level was the strongest determinant of ppOS. Baseline disease stage (IIIC/M1a/M1b vs M1c) was identified as a significant prognostic factor among patients with normal baseline LDH but not among those with elevated LDH. Initial ppRx (immune/targeted therapy vs other) was a significant prognostic factor among patients with normal baseline LDH and baseline disease stage M1c, and among those with baseline LDH elevations ≤ 2 × ULN. ECOG PS at the time of PD was prognostic for ppOS among patients with normal baseline LDH, baseline disease stage M1c, and initial ppRx other than immune/targeted therapy. After adjusting for other covariates, including original treatment assignment, initial ppRx with immunotherapy or targeted therapy was associated with longer ppOS than other ppRx. Original treatment assignment (cobimetinib plus vemurafenib, vemurafenib monotherapy, or dacarbazine) was not identified as a prognostic factor for ppOS.
Fig. 1.
Recursive partitioning analysis for prognostic subgroups in all pooled patients. Recursive partitioning decision tree and ppOS outcomes by identified prognostic subgroups for all pooled patients. Data are presented as percentage (95% CI). ECOG PS Eastern Cooperative Oncology Group performance status, IT immunotherapy, LDH lactate dehydrogenase, PD progressive disease, ppOS postprogression overall survival, ppRx postprogression treatment, TT targeted therapy, ULN upper limit of normal
ppOS
At the time of updated data cutoff for ppOS, 1027 patients had experienced PD; after excluding patients with missing data, 955 patients were included in analyses of ppOS based on RPA-defined prognostic subgroups. Demographic and clinical characteristics remained comparable across cohorts (Additional file 1: Table S1).
Median postprogression follow-up durations were 5.5 months (IQR, 2.2–14.0 months) for all patients, 5.2 months (IQR, 2.1–14.0 months) for the cobimetinib plus vemurafenib cohort, 5.3 months (IQR, 2.2–13.2 months) for the vemurafenib monotherapy cohort, and 6.5 months (IQR, 2.6–15.6 months) for the dacarbazine cohort.
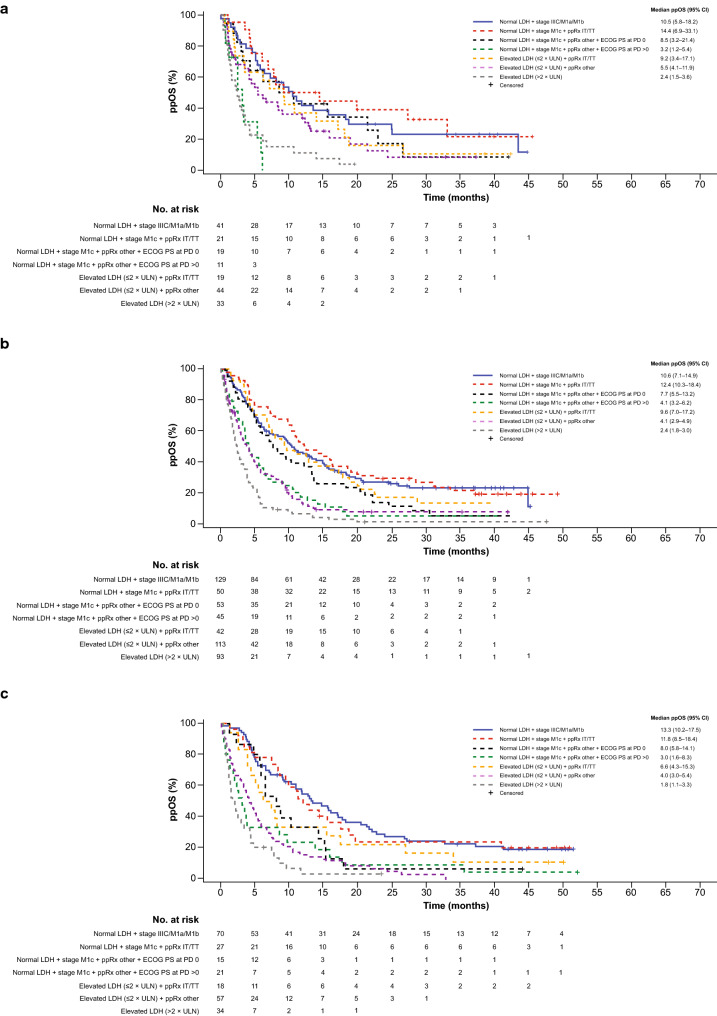
Among all patients, median ppOS was longest in the subgroup with normal baseline LDH, baseline disease stage M1c, and initial ppRx with immunotherapy or targeted therapy (12.2 months; 95% CI 10.3–16.1) and shortest in those with elevated baseline LDH > 2 × ULN (2.3 months; 95% CI 1.8–2.7). Application of prognostic factors identified in the RPA across treatment cohorts (cobimetinib plus vemurafenib, vemurafenib monotherapy, and dacarbazine) revealed similar trends in ppOS (Fig. 2).
Fig. 2.
Postprogression overall survival by treatment cohort. Kaplan–Meier curves of ppOS in a cobimetinib plus vemurafenib cohort, b vemurafenib monotherapy cohort, and c dacarbazine cohort. CI confidence interval, ECOG PS Eastern Cooperative Oncology Group performance status, IT immunotherapy, LDH lactate dehydrogenase, NE not estimable, PD progressive disease, ppOS postprogression overall survival, ppRx initial postprogression treatment, TT targeted therapy, ULN upper limit of normal
ppOS outcomes, stratified by prognostic subgroups identified in the RPA, appeared to be similar across treatment cohorts (Table 2), consistent with the initial RPA demonstrating no impact of original treatment assignment on ppOS. Among patients with normal baseline LDH, baseline disease stage M1c, and initial ppRx with immunotherapy or targeted therapy (the most favorable subgroup), 3-year ppOS rates were 21.7% (95% CI 0.0–44.2) in the cobimetinib plus vemurafenib cohort, 22.3% (95% CI 10.3–34.3) in the vemurafenib monotherapy cohort, and 24.4% (95% CI 7.8–41.1) in the dacarbazine cohort. Among patients with baseline LDH elevations > 2 × ULN (the least favorable subgroup), 3-year ppOS rates were not estimable (NE; 95% CI NE–NE) in the cobimetinib plus vemurafenib cohort, 2.5% (95% CI 0.0–5.8) in the vemurafenib monotherapy cohort, and NE (95% CI NE–NE) in the dacarbazine cohort.
Table 2.
Postprogression overall survival among all pooled patients and by treatment cohort according to prognostic subgroup
| Prognostic subgroup | All patients | Cobimetinib + vemurafenib | Vemurafenib monotherapy | Dacarbazine | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| N (events) | ppOS, median, mo (95% CI) | 3-year ppOS, % (95% CI) | N (events) | ppOS, median, mo (95% CI), | 3-year ppOS, % (95% CI) | N (events) | ppOS, median, mo (95% CI) | 3-year ppOS, % (95% CI) | N (events) | ppOS, median, mo (95% CI) | 3-year ppOS, % (95% CI) | |
| Normal LDH + stage IIIC/M1a/M1b | 240 (170) |
11.2 (9.7–14.0) |
23.1 (17.3–28.9) |
41 (28) |
10.5 (5.8–18.2) |
23.1 (8.8–37.5) |
129 (88) |
10.6 (7.1–14.9) |
23.8 (15.5–32.1) |
70 (54) |
13.3 (10.2–17.5) |
22.8 (12.6–32.9) |
| Normal LDH + stage M1c + ppRx IT/TT | 98 (74) |
12.2 (10.3–16.1) |
23.4 (14.5–32.3) |
21 (14) |
14.4 (6.9–33.1) |
21.7 (0.0–44.2) |
50 (39) |
12.4 (10.3–18.4) |
22.3 (10.3–34.3) |
27 (21) |
11.8 (8.5–18.4) |
24.4 (7.8–41.1) |
| Normal LDH + stage M1c + ppRx other + ECOG PS at PD 0 | 87 (72) |
8.2 (6.0–10.6) |
6.8 (0.7–13.0) |
19 (13) |
8.5 (3.2–21.4) |
8.6 (0.0–24.4) | 53 (45) |
7.7 (5.5–13.2) |
6.0 (0.0–13.6) |
15 (14) |
8.1 (5.8–14.1) |
6.7 (0.0–19.3) |
| Normal LDH + stage M1c + ppRx other + ECOG PS at PD > 0 | 77 (71) |
3.6 (2.6–4.6) |
4.5 (0.0–9.4) |
11 (10) |
3.2 (1.3–5.4) |
0.0 (0.0–0.0) |
45 (41) |
4.1 (3.2–6.2) |
5.7 (0.0–13.0) |
21 (20) |
3.0 (1.6–8.3) |
4.8 (0.0–13.9) |
| Elevated LDH (≤ 2 × ULN) + ppRx IT/TT | 79 (67) |
8.1 (6.7–12.9) |
11.4 (3.7–19.2) |
19 (17) |
9.2 (3.4–17.1) |
10.5 (0.0–24.3) |
42 (34) |
9.6 (7.0–17.2) |
14.1 (2.8–25.5) |
18 (16) |
6.6 (4.3–15.3) |
11.1 (0.0–25.6) |
| Elevated LDH (≤ 2 × ULN) + ppRx other | 214 (186) |
4.2 (3.5–5.0) |
5.0 (1.1–8.8) |
44 (35) |
5.5 (4.1–11.9) |
8.4 (0.0–18.9) |
113 (95) |
4.1 (2.9–4.9) |
8.6 (2.8–14.3) |
57 (56) |
4.0 (3.0–5.4) |
0.0 (0.0–0.0) |
| Elevated LDH (> 2 × ULN) | 160 (150) |
2.3 (1.8–2.7) |
2.7 (0.0–5.5) |
33 (29) |
2.4 (1.5–3.6) |
NE (NE–NE) |
93 (89) |
2.4 (1.8–3.0) |
2.5 (0.0–5.8) |
34 (32) |
1.8 (1.2–3.3) |
NE (NE–NE) |
CI confidence interval, ECOG PS Eastern Cooperative Oncology Group performance status, IT immunotherapy, LDH lactate dehydrogenase, NE not estimable, PD progressive disease, ppOS postprogression overall survival, ppRx postprogression treatment, TT targeted therapy, ULN upper limit of normal
Across treatment cohorts, patients who received immunotherapy or targeted therapy as initial treatment after progression had better survival outcomes than those given other treatments, even among those with LDH elevations.
Impact of Initial ppRx on ppOS
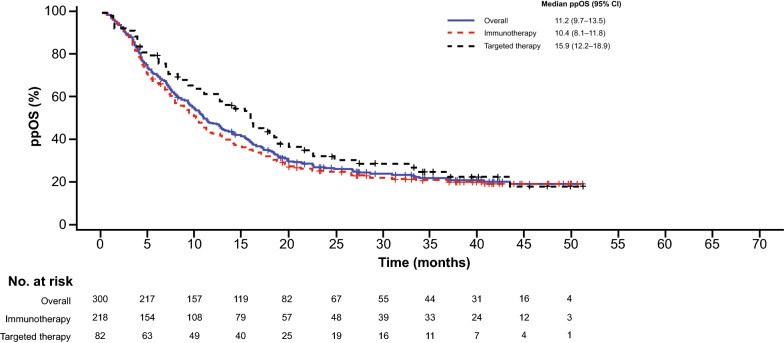
Among 1027 patients with PD at the time of data cutoff, 218 received initial ppRx with immunotherapy and 82 received targeted therapy (Additional file 1: Table S2). Median ppOS was 10.4 months (95% CI 8.1–11.8) in patients who received initial ppRx with immunotherapy and 15.9 months (95% CI 12.2–18.9) in those who received targeted therapy (Fig. 3). ppOS rates at 3 years were 21.1% (95% CI 15.5–26.7) and 25.4% (95% CI 15.0–35.9), respectively.
Fig. 3.
Postprogression overall survival by initial postprogression treatment. Kaplan–Meier curves of ppOS in all pooled patients receiving initial postprogression treatment with immunotherapy or targeted therapy (n = 300). ppOS postprogression overall survival
ppRx patterns
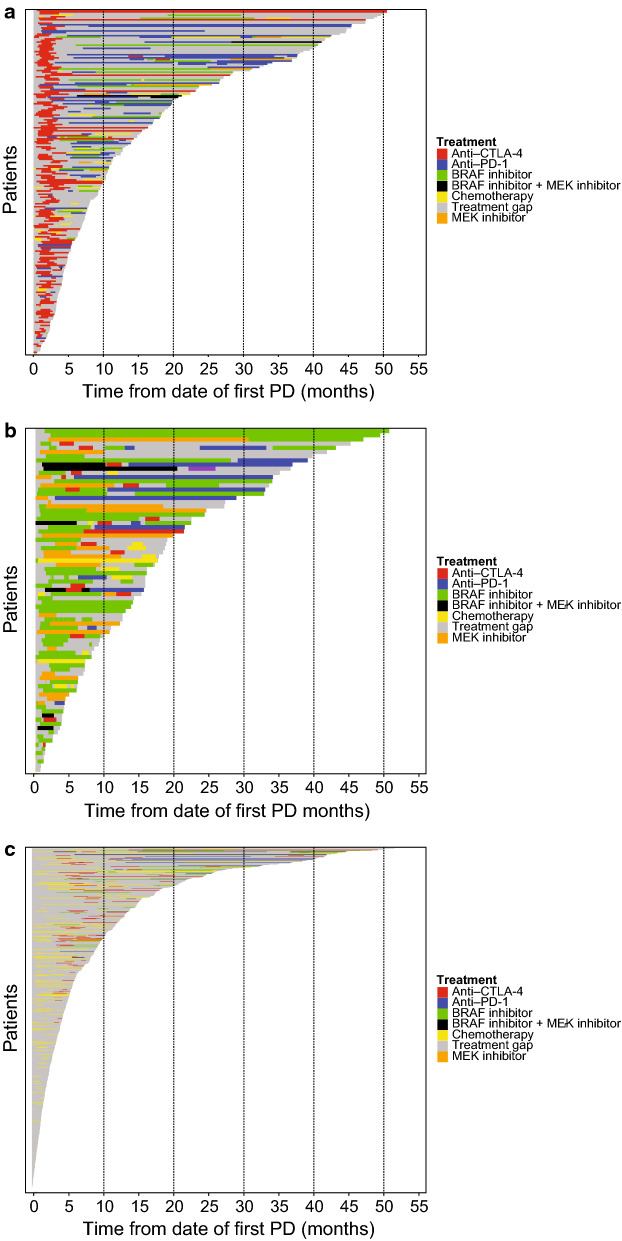
Among patients who received initial ppRx with immunotherapy, initial treatment was ipilimumab in most patients (200 of 218 patients; 91.7%) (Additional file 1: Table S2); subsequent treatment frequently comprised anti–PD-1 therapy or BRAF and/or MEK inhibition (Fig. 4; Additional file 1: Table S3). Among patients who received initial ppRx with targeted therapy, initial treatment was most commonly single-agent BRAFi (61 of 82 patients [74.4%]; Additional file 1: Table S2); targeted therapy was frequently followed by anti–PD-1 or anti–cytotoxic T-lymphocyte–associated protein 4 (CTLA-4) therapy (Fig. 4; Additional file 1: Table S3). Among patients who received other initial ppRx, 220 received chemotherapy and 507 received no treatment (Additional file 1: Table S2). Among the 220 patients who received initial ppRx with chemotherapy, subsequent treatment was most commonly chemotherapy or anti–CTLA-4 therapy (Fig. 4; Additional file 1: Table S3).
Fig. 4.
Postprogression treatment patterns by initial postprogression treatment. Postprogression treatment patterns in patients who received initial postprogression treatment with a immunotherapy (n = 218), b targeted therapy (n = 82), and c other (n = 727). CTLA-4 cytotoxic T lymphocyte–associated antigen 4, PD-1 programmed death receptor 1
Among patients with evidence of additional treatment, median time to next treatment (time from start of initial ppRx to start of next line of ppRx or death) was 4.4 months (95% CI 4.0–4.9) in patients who received initial ppRx with immunotherapy, 6.0 months (95% CI 4.3–7.6) in those who received targeted therapy, and 3.8 months (95% CI 3.3–4.3) in those who received chemotherapy.
Discussion
In this exploratory analysis of data pooled from four key trials of vemurafenib or cobimetinib plus vemurafenib, a combination of baseline LDH and disease stage, initial ppRx, and ECOG PS at PD were identified as key prognostic factors for ppOS in patients with metastatic melanoma. Original treatment regimen (cobimetinib plus vemurafenib, vemurafenib monotherapy, or dacarbazine) did not appear to impact survival outcomes associated with ppRx.
Previous analyses in patients with metastatic melanoma treated with BRAFi and/or MEKi have consistently identified baseline LDH, ECOG performance status, and extent of disease as prognostic factors for OS [15–18]. In the current analysis, baseline LDH remained the most important prognostic factor for OS following progression. Among patients with normal baseline LDH, baseline disease stage and ECOG PS at the time of PD were also identified as key prognostic factors for ppOS. Additionally, initial ppRx was identified as a statistically significant independent prognostic factor for ppOS on both sides of the regression tree (among patients with normal baseline LDH and those with LDH elevations ≤ 2 × ULN). Interestingly, among patients with normal baseline LDH, patients with stage M1c disease who received initial ppRx with immunotherapy or targeted therapy were able to achieve ppOS that was at least as good as that obtained in patients with stage IIIC/M1a/M1b disease.
Considerable debate exists regarding the optimal sequence of treatment with targeted therapies and immunotherapies for patients with BRAF-mutated metastatic melanoma. To date, only low-quality evidence is available to inform treatment sequencing decisions, but randomized studies are currently underway. Retrospective analyses, mostly involving ipilimumab, have suggested that prior treatment with a BRAFi is associated with an inferior response to subsequent immunotherapy [19–21], whereas recent analyses involving nivolumab and pembrolizumab are more controversial [22, 23, 25–27]. In the current analysis, patients who received initial ppRx with immunotherapy or targeted therapy had similar OS outcomes across the cobimetinib plus vemurafenib, vemurafenib monotherapy, and dacarbazine cohorts. After adjusting for other covariates, ppRx with immunotherapy or targeted therapy was associated with longer ppOS than ppRx with other treatments regardless of original treatment assignment. Furthermore, observed ppOS outcomes in patients receiving initial ppRx with immunotherapy in this study (92% ipilimumab) appeared to be comparable to reported OS outcomes with ipilimumab in previously treated patients (median OS, 10.4 vs 10.7 months; 3-year OS, 21% vs 20%) [12]. Taken together, these results suggest that clinically relevant responses can be achieved with immunotherapy following progression on targeted therapy. In addition, observation of longer ppOS among patients who received initial ppRx with targeted therapy (74% single-agent BRAFi) compared with initial ppRx with treatments other than immunotherapy or targeted therapy is consistent with the findings of recent studies suggesting that reinitiation of BRAFi and/or MEKi following PD can provide clinical benefit [29].
These findings introduce an additional layer of complexity in the interpretation of treatment effects related to OS outcomes reported in randomized controlled trials. It should be noted that a significant proportion of patients across treatment cohorts received multiple lines of ppRx. Among patients who received ppRx with immunotherapy, more than 90% received ipilimumab as their initial ppRx, whereas anti–PD-1 therapy was predominantly used after failure of initial ppRx. Results from a recent study, albeit in the treatment-naive setting, suggest more favorable outcomes with anti–PD-1 followed by ipilimumab compared with the reverse sequence [30]. Proper interpretation of OS outcomes will require consideration of patient and disease characteristics as well as the distribution and sequence of treatments received following progression.
Strengths of this pooled analysis include the large number of patients from studies conducted in similar populations, sufficient follow-up for robust survival analyses, and prospective data collection using standardized schedules and methods. Further, the current analysis is largely based on data from randomized studies, wherein patients with similar baseline prognostic factors received a reference first-line and experimental treatment; this analysis is, therefore, less subject to the bias inherent in retrospective analyses of postprogression survival using data from patients treated in routine clinical practice where treatment decisions were based on physician and individual patient preferences [19–22]. However, as a retrospective exploratory analysis our analysis is also subject to limitations. The pooled dataset included patients enrolled in nonrandomized studies and original treatment assignment may be subject to selection bias. Also, choice of ppRx was not randomized or dictated by study protocols and may have been influenced in a nonuniform manner by changes in access to new treatments across the approximate 8-year time period over which the included studies were conducted.
Conclusions
LDH, extent of disease, and ECOG PS remain significant prognostic factors following progression after treatment with cobimetinib plus vemurafenib, vemurafenib monotherapy, or dacarbazine in patients with BRAFV600-mutated metastatic melanoma. ppRx was also identified as an additional independent prognostic factor for OS following progression. Patients who progress following treatment with BRAFi and/or MEKi derive an OS benefit from subsequent treatment with immunotherapy or additional targeted therapy.
Supplementary information
Additional file 1: Table S1. Demographic and clinical characteristics for patients included in the updated analysis of ppOS (N = 1027). Table S2. Initial postprogression treatment in all pooled patients. Table S3. Distribution of subsequent postprogression treatments according to initial postprogression treatment.
Acknowledgements
Medical writing and editorial support for this manuscript was provided by Melanie Sweetlove, MSc (ApotheCom, San Francisco, CA), and was funded by F. Hoffmann-La Roche Ltd.
Abbreviations
- BRAFi
BRAF inhibitor
- CI
Confidence interval
- CNS
Central nervous system
- CTLA-4
Cytotoxic T-lymphocyte-associated protein 4
- ECOG PS
Eastern Cooperative Oncology Group performance status
- IQR
Interquartile range
- LDH
Lactate dehydrogenase
- MEKi
MEK inhibitor
- NE
Not estimable
- OS
Overall survival
- PD
Progressive disease
- PD-1
Programmed death 1
- ppOS
Postprogression overall survival
- ppRx
Postprogression treatment
- RPA
Recursive partitioning analysis
- SLD
Sum of longest diameters
- ULN
Upper limit of normal
Authors’ contributions
PAA, GAM, AH, KTF, EM, and BD contributed to study design and concepts. PAA, AR, JL, GAM, KDL, AH, KTF, and BD collected the data. All authors had full access to the data output and interpreted the data. QZ and YM performed statistical analyses. PAA and EM collaboratively wrote the first draft of the manuscript. All authors read and approved the final manuscript.
Funding
The BRIM-2, BRIM-3, BRIM-7, and coBRIM studies were designed and funded by the sponsor, F. Hoffmann-La Roche Ltd.
Availability of data and materials
Qualified researchers requesting access to de-identified patient-level data through the Clinical Study Data Request platform will be provided with accompanying clinical study documentation (protocol and any associated amendments, annotated case report form, reporting and analysis plan, dataset specifications, clinical study report). Researchers requesting access to clinical study documentation only, can do so via the following link: http://www.roche.com/research_and_development/who_we_are_how_we_work/clinical_trials/our_commitment_to_data_sharing/clinical_study_documents_request_form.htm
Ethics approval and consent to participate
Each trial was conducted in accordance with the Declaration of Helsinki and the principles of Good Clinical and Laboratory Practice and with the approval of appropriate ethics committees. All participants provided written informed consent.
Consent for publication
Not applicable.
Competing interests
PAA reports consulting/advisory role for Bristol-Myers Squibb, Roche/Genentech, Merck Sharp & Dohme, Array, Novartis, Merck Serono, Pierre Fabre, Incyte, Genmab, NewLink Genetics, MedImmune, AstraZeneca, Syndax, Sun Pharma, Sanofi, Idera, Ultimovacs, Sandoz, and Immunocore; research funding from Bristol-Myers Squibb, Roche/Genentech, and Array; and travel, accommodations, or expenses from Merck Sharp & Dohme. AR reports honoraria from Amgen, Chugai, Genentech, Merck, and Novartis; consulting/advisory role for Amgen, Chugai, Genentech, Merck, and Novartis; and stock or other ownership in Advaxis, Arcus, Bioncotech, Compugen, CytomX, Five Prime, FLX-Bio, ImaginAb, IsoPlexis, Kite-Gilead, Merus, Rgenix, Lutris, PACT Pharma, and Tango Therapeutics. JL reports honoraria from Achilles Therapeutics, AstraZeneca, Boston Biomedical, Bristol-Myers Squibb, Eisai, EUSA Pharma, GlaxoSmithKline, Ipsen, Imugene, Incyte, Onctura, Kymab, Merck Serono, Merck Sharp & Dohme, Nektar, Novartis, Pierre Fabre, Pfizer, Roche/Genentech, Secarna, and Vitaccess; consulting/advisory role for Achilles Therapeutics, AstraZeneca, Boston Biomedical, Bristol-Myers Squibb, Eisai, EUSA Pharma, GlaxoSmithKline, Ipsen, Imugene, Incyte, Onctura, Kymab, Merck Serono, Merck Sharp & Dohme, Nektar, Novartis, Pierre Fabre, Pfizer, Roche/Genentech, Secarna, and Vitaccess; research funding from Achilles Therapeutics, Aveo, Bristol-Myers Squibb, Merck Sharp & Dohme, Nektar, Novartis, Pfizer, Roche/Genentech, Covance, and Immunocore. GAM reports steering committee membership and principal investigator roles for Roche/Genentech. KDL reports a consulting/advisory role for Roche, Merck, Regeneron, and Array; and research funding from Roche, Merck, Array, Regeneron, Iovance, Incyte, Neon, Nektar, and Bristol-Myers Squibb. AH reports honoraria from Amgen, Bristol-Myers Squibb, Merck Serono, Merck Sharp & Dohme, Philogen, Pierre Fabre, Provectus, Regeneron, Roche, OncoSec, Sanofi-Genzyme, and Novartis Pharma; consulting/advisory role for Amgen, Bristol-Myers Squibb, Merck Serono, Merck Sharp & Dohme, Philogen, Pierre Fabre, Provectus, Regeneron, Roche, OncoSec, Sanofi-Genzyme, and Novartis Pharma; research funding from Amgen, Bristol-Myers Squibb, Merck Serono, Merck Sharp & Dohme, Philogen, Pierre Fabre, Provectus, Regeneron, Roche, Sanofi-Genzyme, and Novartis Pharma. KTF reports consulting/advisory role for Roche, X4 Pharmaceutical, Apricity, PIC Therapeutics, Sanofi, Amgen, Asana, Adaptimmune, Fount Therapeutics, Aeglea, Array BioPharma, Shattuck Labs, Arch Oncology, Tolero Pharmaceuticals, Oncoceutics, Fog Pharma, Neon Therapeutics, Tvardi, Novartis, Genentech, Bristol-Myers Squibb, Merck, Takeda, Verastem, Checkmate, Boston Biomedical, Pierre Fabre, Cell Medica, and Debiopharm; research funding from Roche; stock ownership for Loxo Oncology, Clovis Oncology, Strata Oncology, Vivid Biosciences, X4 Pharmaceuticals, Apricity, PIC Therapeutics, Fount Therapeutics, Shattuck Labs, Oncoceutics, Fog Pharma, and Tvardi; and served on the board of directors for Loxo Oncology, Clovis Oncology, Strata Oncology, and Vivid Biosciences. EM and YM report employment with Genentech and stock or other ownership in Genentech. QZ reports employment with Edwards Lifesciences; stock or other ownership in Edwards Lifesciences; and a consulting/advisory role with Genentech. BD reports honoraria from Bristol-Myers Squibb, Roche, Novartis, and Pierre Fabre; consulting/advisory roles for Bristol-Myers Squibb, Roche, Novartis, and Pierre Fabre; research funding from Bristol-Myers Squibb and Roche; and travel, accommodations, or expenses from Roche and Bristol-Myers Squibb.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Paolo A. Ascierto, Email: paolo.ascierto@gmail.com, Email: p.ascierto@istitutotumori.na.it
Antoni Ribas, Email: ARibas@mednet.ucla.edu.
James Larkin, Email: James.Larkin@rmh.nhs.uk.
Grant A. McArthur, Email: grant.mcarthur@petermac.org
Karl D. Lewis, Email: karl.lewis@ucdenver.edu
Axel Hauschild, Email: ahauschild@dermatology.uni-kiel.de.
Keith T. Flaherty, Email: kflaherty@partners.org
Edward McKenna, Email: mckenna.edward@gene.com.
Qian Zhu, Email: zhu.qian@gene.com.
Yong Mun, Email: mun.yong@gene.com.
Brigitte Dréno, Email: brigitte.dreno@wanadoo.fr.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12967-020-02458-x.
References
- 1.Rozeman EA, Dekker TJA, Haanen J, Blank CU. Advanced melanoma: current treatment options, biomarkers, and future perspectives. Am J Clin Dermatol. 2018;19:303–317. doi: 10.1007/s40257-017-0325-6. [DOI] [PubMed] [Google Scholar]
- 2.Sosman JA, Kim KB, Schuchter L, Gonzalez R, Pavlick AC, Weber JS, et al. Survival in BRAFV600-mutant advanced melanoma treated with vemurafenib. N Engl J Med. 2012;366:707–714. doi: 10.1056/NEJMoa1112302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chapman PB, Hauschild A, Robert C, Haanen JB, Ascierto P, Larkin J, et al. Improved survival with vemurafenib in melanoma with BRAF V600E mutation. N Engl J Med. 2011;364:2507–2516. doi: 10.1056/NEJMoa1103782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McArthur GA, Chapman PB, Robert C, Larkin J, Haanen JB, Dummer R, et al. Safety and efficacy of vemurafenib in BRAFv600E and BRAFv600K mutation-positive melanoma (BRIM-3): extended follow-up of a phase 3, randomised, open-label study. Lancet Oncol. 2014;15:323–332. doi: 10.1016/S1470-2045(14)70012-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ribas A, Gonzalez R, Pavlick A, Hamid O, Gajewski TF, Daud A, et al. Combination of vemurafenib and cobimetinib in patients with advanced BRAF(V600)-mutated melanoma: a phase 1b study. Lancet Oncol. 2014;15:954–965. doi: 10.1016/S1470-2045(14)70301-8. [DOI] [PubMed] [Google Scholar]
- 6.Larkin J, Ascierto PA, Dreno B, Atkinson V, Liszkay G, Maio M, et al. Combined vemurafenib and cobimetinib in BRAF-mutated melanoma. N Engl J Med. 2014;371:1867–1876. doi: 10.1056/NEJMoa1408868. [DOI] [PubMed] [Google Scholar]
- 7.Ascierto PA, McArthur GA, Dreno B, Atkinson V, Liszkay G, Di Giacomo AM, et al. Cobimetinib combined with vemurafenib in advanced BRAF V600-mutant melanoma (coBRIM): updated efficacy results from a randomised, double-blind, phase 3 trial. Lancet Oncol. 2016;17:1248–1260. doi: 10.1016/S1470-2045(16)30122-X. [DOI] [PubMed] [Google Scholar]
- 8.Chapman PB, Robert C, Larkin J, Haanen JB, Ribas A, Hogg D, et al. Vemurafenib in patients with BRAFV600 mutation-positive metastatic melanoma: final overall survival results of the randomized BRIM-3 study. Ann Oncol. 2017;28:2581–2587. doi: 10.1093/annonc/mdx339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dréno B, Ascierto PA, McArthur GA, Atkinson V, Liszkay G, Di Giacomo AM, et al. Efficacy and safety of cobimetinib (C) combined with vemurafenib (V) in patients (pts) with BRAF V600-mutation–positive metastatic melanoma: analysis from the 4-year extended follow-up follow-up of the phase 3 coBRIM study. J Clin Oncol. 2018;36:9522. doi: 10.1200/JCO.2018.36.15_suppl.9522. [DOI] [Google Scholar]
- 10.Menzies AM, Wilmott JS, Drummond M, Lo S, Lyle M, Chan MM, et al. Clinicopathologic features associated with efficacy and long-term survival in metastatic melanoma patients treated with BRAF or combined BRAF and MEK inhibitors. Cancer. 2016;121:3826–3835. doi: 10.1002/cncr.29586. [DOI] [PubMed] [Google Scholar]
- 11.Long GV, Weber JS, Infante JR, Kim KB, Daud A, Gonzalez R, et al. Overall survival and durable responses in patients with BRAF V600-mutant metastatic melanoma receiving dabrafenib combined with trametinib. J Clin Oncol. 2016;34:871–878. doi: 10.1200/JCO.2015.62.9345. [DOI] [PubMed] [Google Scholar]
- 12.Schadendorf D, Hodi FS, Robert C, Weber JS, Margolin K, Hamid O, et al. Pooled analysis of long-term survival data from phase II and phase III trials of ipilimumab in unresectable or metastatic melanoma. J Clin Oncol. 2015;33:1889–1894. doi: 10.1200/JCO.2014.56.2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dickson PV, Gershenwald JE. Staging and prognosis of cutaneous melanoma. Surg Oncol Clin N Am. 2011;20:1–17. doi: 10.1016/j.soc.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27:6199–6206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Long GV, Grob JJ, Nathan P, Ribas A, Robert C, Schadendorf D, et al. Factors predictive of response, disease progression, and overall survival after dabrafenib and trametinib combination treatment: a pooled analysis of individual patient data from randomised trials. Lancet Oncol. 2016;17:1743–1754. doi: 10.1016/S1470-2045(16)30578-2. [DOI] [PubMed] [Google Scholar]
- 16.Long GV, Blank C, Ribas A, Mortier L, Carlino MS, Lotem M, et al. Impact of baseline serum lactate dehydrogenase concentration on the efficacy of pembrolizumab and ipilimumab in patients with advanced melanoma: data from KEYNOTE-006. Eur J Cancer. 2017;72:S122–S123. doi: 10.1016/S0959-8049(17)30482-3. [DOI] [Google Scholar]
- 17.Schadendorf D, Long GV, Stroiakovski D, Karaszewska B, Hauschild A, Levchenko E, et al. Three-year pooled analysis of factors associated with clinical outcomes across dabrafenib and trametinib combination therapy phase 3 randomised trials. Eur J Cancer. 2017;82:45–55. doi: 10.1016/j.ejca.2017.05.033. [DOI] [PubMed] [Google Scholar]
- 18.Hauschild A, Larkin J, Ribas A, Dreno B, Flaherty KT, Ascierto PA, et al. Modeled prognostic subgroups for survival and treatment outcomes in BRAF V600-mutated metastatic melanoma: pooled analysis of 4 randomized clinical trials. JAMA Oncol. 2018;4:1382–1388. doi: 10.1001/jamaoncol.2018.2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ackerman A, Klein O, McDermott DF, Wang W, Ibrahim N, Lawrence DP, et al. Outcomes of patients with metastatic melanoma treated with immunotherapy prior to or after BRAF inhibitors. Cancer. 2014;120:1695–1701. doi: 10.1002/cncr.28620. [DOI] [PubMed] [Google Scholar]
- 20.Ascierto PA, Simeone E, Sileni VC, Del Vecchio M, Marchetti P, Cappellini GC, et al. Sequential treatment with ipilimumab and BRAF inhibitors in patients with metastatic melanoma: data from the Italian cohort of the ipilimumab expanded access program. Cancer Invest. 2014;32:144–149. doi: 10.3109/07357907.2014.885984. [DOI] [PubMed] [Google Scholar]
- 21.Polkowska M, Ekk-Cierniakowski P, Czepielewska E, Wysoczanski W, Matusewicz W, Kozlowska-Wojciechowska M. Survival of melanoma patients treated with novel drugs: retrospective analysis of real-world data. J Cancer Res Clin Oncol. 2017;143:2087–2094. doi: 10.1007/s00432-017-2453-z. [DOI] [PubMed] [Google Scholar]
- 22.Simeone E, Grimaldi AM, Festino L, Giannarelli D, Vanella V, Palla M, et al. Correlation between previous treatment with BRAF inhibitors and clinical response to pembrolizumab in patients with advanced melanoma. Oncoimmunology. 2017;6:e1283462. doi: 10.1080/2162402X.2017.1283462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Long GV, Atkinson V, Lo S, Sandhu S, Guminski AD, Brown MP, et al. Combination nivolumab and ipilimumab or nivolumab alone in melanoma brain metastases: a multicentre randomised phase 2 study. Lancet Oncol. 2018;19:672–681. doi: 10.1016/S1470-2045(18)30139-6. [DOI] [PubMed] [Google Scholar]
- 24.Aya F, Fernandez-Martinez A, Gaba L, Victoria I, Tosca M, Pineda E, et al. Sequential treatment with immunotherapy and BRAF inhibitors in BRAF-mutant advanced melanoma. Clin Transl Oncol. 2017;19:119–124. doi: 10.1007/s12094-016-1514-0. [DOI] [PubMed] [Google Scholar]
- 25.Larkin J, Lao CD, Urba WJ, McDermott DF, Horak C, Jiang J, et al. Efficacy and safety of nivolumab in patients with BRAF V600 mutant and BRAF wild-type advanced melanoma: a pooled analysis of 4 clinical trials. JAMA Oncol. 2015;1:433–440. doi: 10.1001/jamaoncol.2015.1184. [DOI] [PubMed] [Google Scholar]
- 26.Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, et al. Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med. 2015;372:2521–2532. doi: 10.1056/NEJMoa1503093. [DOI] [PubMed] [Google Scholar]
- 27.Johnson DB, Pectasides E, Feld E, Ye F, Zhao S, Johnpulle R, et al. Sequencing treatment in BRAFV600 mutant melanoma: anti-PD-1 before and after BRAF inhibition. J Immunother. 2017;40:31–35. doi: 10.1097/CJI.0000000000000148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hothorn T, Hornik K, Zeileis A. Unbiased recursive partitioning: a conditional inference framework. J Comput Graph Stat. 2006;15:651–674. doi: 10.1198/106186006X133933. [DOI] [Google Scholar]
- 29.Chan MM, Haydu LE, Menzies AM, Azer MW, Klein O, Lyle M, et al. The nature and management of metastatic melanoma after progression on BRAF inhibitors: effects of extended BRAF inhibition. Cancer. 2014;120:3142–3153. doi: 10.1002/cncr.28851. [DOI] [PubMed] [Google Scholar]
- 30.Weber JS, Gibney G, Sullivan RJ, Sosman JA, Slingluff CL, Jr, Lawrence DP, et al. Sequential administration of nivolumab and ipilimumab with a planned switch in patients with advanced melanoma (CheckMate 064): an open-label, randomised, phase 2 trial. Lancet Oncol. 2016;17:943–955. doi: 10.1016/S1470-2045(16)30126-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S1. Demographic and clinical characteristics for patients included in the updated analysis of ppOS (N = 1027). Table S2. Initial postprogression treatment in all pooled patients. Table S3. Distribution of subsequent postprogression treatments according to initial postprogression treatment.
Data Availability Statement
Qualified researchers requesting access to de-identified patient-level data through the Clinical Study Data Request platform will be provided with accompanying clinical study documentation (protocol and any associated amendments, annotated case report form, reporting and analysis plan, dataset specifications, clinical study report). Researchers requesting access to clinical study documentation only, can do so via the following link: http://www.roche.com/research_and_development/who_we_are_how_we_work/clinical_trials/our_commitment_to_data_sharing/clinical_study_documents_request_form.htm