Summary:
NADH provides electrons for aerobic ATP production. In cells deprived of oxygen or with impaired electron transport chain activity, NADH accumulation can be toxic. To minimize such toxicity, elevated NADH inhibits the classical NADH producing pathways: glucose, glutamine, and fat oxidation. Here, through deuterium tracing studies in cultured cells and mice, we show that folate-dependent serine catabolism also produces substantial NADH. Strikingly, when respiration is impaired, serine catabolism through methylene tetrahydrofolate dehydrogenase (MTHFD2) becomes a major NADH source. In cells whose respiration is slowed by hypoxia, metformin, or genetic lesions, mitochondrial serine catabolism inhibition partially normalizes NADH levels and facilitates cell growth. In mice with engineered mitochondrial complex I deficiency (NDUSF4−/−), serine’s contribution to NADH is elevated and progression of spasticity is modestly slowed by pharmacological blockade of serine degradation. Thus, when respiration is impaired, serine catabolism contributes to toxic NADH accumulation.
Keywords: NAD, NADH, redox, serine catabolism, serine hydroxymethyltransferase (SHMT2), methylene tetrahydrofolate dehydrogenase (MTHFD2), hypoxia, respiration inhibition, mitochondrial disease, complex I inhibitor
eTOC
Yang et al. identify an unexpected NADH source, mitochondrial serine catabolism. Unlike more classical NADH production routes, serine catabolism persists when respiration is impaired. Both NADH buildup and pathology caused by respiration impairment are ameliorated by blocking serine catabolism.
Graphical Abstract

Mammals break down carbohydrates, amino acids, and fatty acids for energy generation (Chen et al., 2018; Hui et al., 2017; Kalucka et al., 2018; Lundsgaard et al., 2018; Neinast et al., 2019). Through TCA turning in mitochondria, these nutrients are burned into CO2 while producing reducing power in the form of NADH. Electrons from NADH are then transferred to mitochondrial complex I and downstream components of the electron transport chain (ETC), eventually reducing oxygen to water (Titov et al., 2016). By inducing a proton gradient, these reactions power mitochondrial ATP synthase, which provides the bulk of ATP in mammals.
The most common physiological impediment to respiration is hypoxia. Respiration impairment can also be induced by genetic lesions of the ETC (e.g., in mitochondrial disorders) and by pharmaceuticals. Metformin, the first-line treatment for type 2 diabetes, acts at least in part as a complex I inhibitor (El-Mir et al., 2000; Fullerton et al., 2013; Hunter et al., 2018; Liu et al., 2016b; Madiraju et al., 2014; Madiraju et al., 2018; Owen et al., 2000; Wheaton et al., 2014). The consequences of ETC inhibition may vary depending on the respiratory chain component involved. For example, the reduction of complexes III and IV is increased by hypoxia but not by complex I inhibition (Chandel et al., 1998; Wheaton et al., 2014).
Increased NADH and decreased ATP are direct biochemical consequences of ETC impairment. An important question is their relative importance. In cell culture, increasing glycolysis can compensate for impaired mitochondrial ATP generation (DeBerardinis et al., 2008; Lunt and Vander Heiden, 2011). To avoid NADH accumulation, however, requires an alternative electron acceptor, such as pyruvate. Major consequences of NADH accumulation include blockade of TCA turning, impairment of de novo aspartate synthesis, and resulting decreases in protein and nucleotide synthesis (Birsoy et al., 2015; Gameiro et al., 2013; Mullen et al., 2012; Sullivan et al., 2015; Wise et al., 2011; Yoo et al., 2008). Cell growth can be rescued by engineered aspartate assimilation or by expression of LbNOX, an enzyme that oxidizes NADH without generating ATP (Sullivan et al., 2018; Titov et al., 2016). Thus, a major driver of the adverse consequences of ETC impairment is NADH accumulation.
Even though ETC impairment is growth inhibitory, for unclear reasons, genetic ETC lesions have been shown to activate the kinase mTORC1. This induces expression of a wide range of anabolic genes, with the one-carbon (1C) metabolic enzyme methylenetetrahydrofolate dehydrogenase 2 (MTHFD2) strongly up-regulated (Ben-Sahra et al., 2016; Khan et al., 2017; Nikkanen et al., 2016; Tyynismaa et al., 2010). In mitochondria, MTHFD2 oxidizes 5,10-methylene-tetrahydrofolate (THF) to generate the purine precursor 10-formyl-THF, while reducing NAD to NADH. The mTORC1 inhibitor rapamycin decreases MTHFD2 expression, normalizes 1C metabolism, and prolongs the survival of mice with engineered complex I deficiency (NDUSF4−/− mice) (Johnson et al., 2013; Khan et al., 2017; Zheng et al., 2016). The functional importance of MTHFD2 in this context remains, however, unexplored. More generally, the reactions producing the accumulated NADH when ETC activity is impaired remain understudied.
Here, using deuterium labeled tracers, we discover that mitochondrial serine catabolism can produce substantial NADH, especially in pancreas, spleen and tumors. This pathway turns out to be the major source of NADH for cells with impaired ETC activity due to hypoxia, pharmacological ETC inhibition, or genetic ETC lesions. In these contexts, inhibition of mitochondrial serine catabolism improved metabolic homeostasis and enhanced growth of cultured cells. In addition, it slowed onset of spasticity in complex I deficient mice.
Results
Serine catabolism feeds NADH in vivo
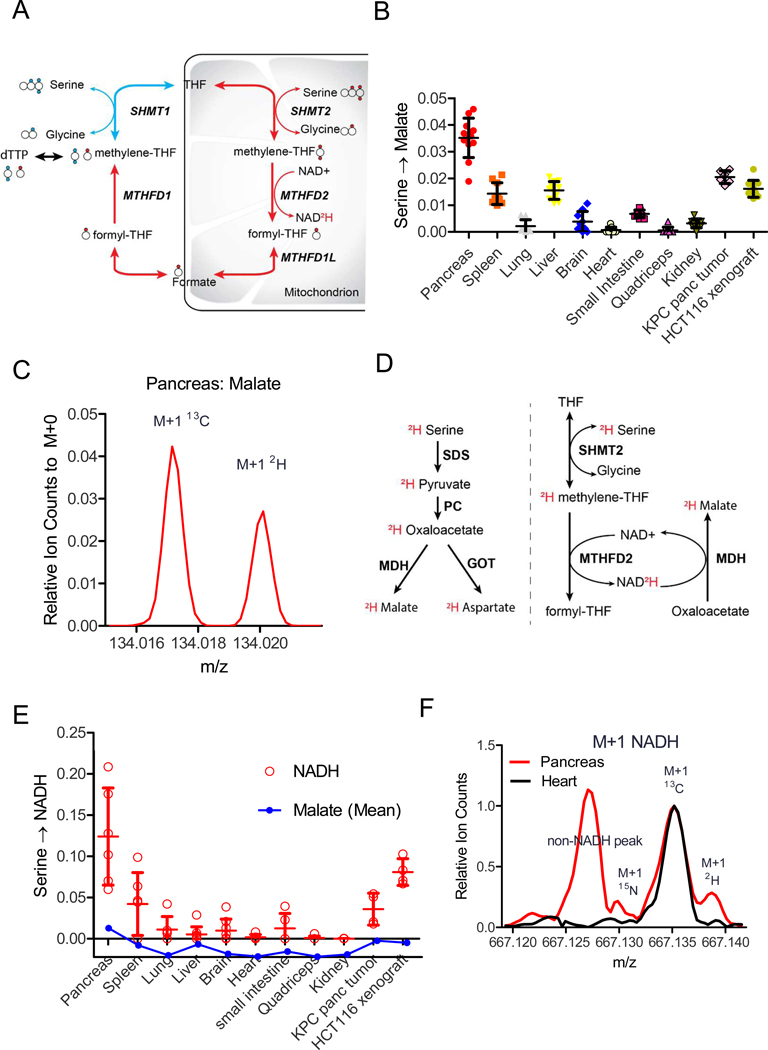
[2,3,3-2H]serine is an isotope tracer that has been used to determine whether 1C units for thymidine synthesis are generated via cytosolic or mitochondrial serine catabolism (Figure 1A) (Ducker et al., 2016; Gregory et al., 2000; Herbig et al., 2002). In studies infusing this tracer into mice, we unexpectedly observed labeling of malate, which is not a canonical product of serine or 1C metabolism (Figure 1B). High-resolution mass spectrometry (orbitrap) confirmed that the labeled peak was 2H-malate (as opposed to natural abundance 13C-malate) (Figure 1C, Supplementary Fig. 1A). While the raw extent of malate labeling was small, due to tracer loss via H-D exchange, even modest 2H labeling is often biologically meaningful (Zhang et al., 2017).
Figure 1:
Serine catabolism feeds NADH in vivo
A: Schematic showing folate-mediated serine catabolism and its tracing with [2,3,3-2H]serine. This tracer has traditionally been used to distinguish cytosolic versus mitochondrial production of 1C units from serine. The larger circles indicate carbons, with serine’s carboxylic acid group on the left. Smaller filled circles indicate labeled hydrogens (deuteriums). The cytosolic pathway (blue) yields M+2 thymidine and the mitochondrial pathway (red) yields M+1 thymidine. B: [2,3,3-2H]serine labels malate in vivo. Data are fractional 2H1-malate relative to tissue serine labeling after 2.5 h [2,3,3-2H]serine infusion (40 nmoles/g/min) (mean ±SD, N≥6). Throughout, all reported labeling fractions are corrected for natural isotope abundance. C: Raw orbitrap mass spectrum of M+1 malate in pancreas after [2,3,3-2H]serine infusion, showing the 2H-peak well resolved from the natural abundance 13C-peak. D: Two possible metabolic pathways by which [2,3,3-2H]serine can produce 2H-malate. E: [2,3,3-2H]serine labels NADH more than malate. Data are fractional 2H-labeling of malate relative to tissue serine labeling (mean ±SD, N≥4). F: Raw FTICR mass spectrum of M+1 NADH in pancreas and heart after [2,3,3-2H]serine infusion. Heart shows only one major peak: the natural abundance 13C-peak. Pancreas shows three peaks: an interfering peak of unknown identity, the natural abundance 13C-peak, and a partially resolved 2H-peak. See also Figure S1
There are two potential pathways for 2H transfer from serine to malate: (i) the 2H remains covalently bound to the serine carbon skeleton, which becomes 2H-pyruvate, 2H-oxaloacetate, and eventually 2H-malate and (ii) the 2H from serine is transferred via 1C metabolism to NAD2H, which reduces unlabeled oxaloacetate to 2H-malate (Figure 1D). The most straightforward way to distinguish these pathways would be measurement of relative labeling of oxaloacetate versus malate, but this is not possible due to the low abundance and instability of oxaloacetate. Aspartate, however, shares the same hydrogens as oxaloacetate. Much more 2H-labeling was observed in malate than aspartate, suggesting that the major route of malate labeling is via NADH (Supplementary Fig. 1B). Consistent with this, 2H-labeling was observed directly in NADH (Figure 1E). Due to the relatively high mass of NADH, standard orbitrap mass spectrometry cannot resolve 2H- from 13C-NADH. Accordingly, we used Fourier-transform ion cyclotron resonance mass spectrometry (FTICR-MS, 260K resolution at m/z 660) to directly confirm 2H-labeled NADH (Figure 1F, Supplementary Fig. 1C). Thus, folate-mediated serine catabolism generates NADH.
The key redox enzyme of the 1C-NADH pathway is MTHFD2. Labeling of NADH and malate from 2H-serine was highest in pancreas (Figure 1B,E), which has the highest MTHFD2 gene expression (Supplementary Fig. 1D) and greatest serine consumption based on depletion of serine in the draining venous blood (Supplementary Fig. 1E) (Geiger et al., 2013). Substantial malate 2H-labeling was also observed in allografted genetically engineered pancreatic cancer tumors (KPC tumors) and xenografted colon cancer tumors (HCT116) (Figure 1B, E). Collectively, these observations suggest that serine catabolism could be a meaningful mitochondrial NADH source in certain organs (especially pancreas, spleen) and tumors.
Serine catabolism is a major NADH source when respiration is impaired
To better understand the contribution of serine to NADH production, we turned to cell culture. We compared NADH labeling from classical major NADH sources (glutamine, lactate and fat) with that from serine (Figure 2A, Supplementary Fig. 2A-C) (Hui et al., 2017). Under standard tissue culture conditions, NAD2H from serine was less than from glutamine but comparable to lactate and larger than palmitate, verifying that serine can be a substantial NADH source (Figure 2B).
Figure 2:
Serine catabolism is a major NADH source when respiration is impaired
A: Schematic of mitochondrial NADH sources. B: Fractional 2H-labeling of NADH from 2H-labeled glutamine (gln), lactate (lac), palmitate (pal), or serine (ser). HCT116 cells were grown in standard DMEM ± 5mM metformin with the indicated nutrient replaced by its 2H-labeled form (except for palmitate, for which 100 μM was added to standard DMEM) (mean ± SD, N≥3). C: Oxygen consumption rate concentration (mean ±SD, N=4) and fraction NAD2H from [2,3,3-2H]serine as a function of metformin concentration (mean ±SD, N≥2). D: Fraction NAD2H from [2,3,3-2H]serine across different cell lines (mean ±SD, N≥2). E-F: NAD2H from [2,3,3-2H]serine is via the mitochondrial folate pathway both in HCT116 cells (E) and 293T cells (F) (mean ± SD, N=4). ND, none detected; **p<0.01; ***p < 0.001 by two-tailed student’s t test. See also Figure S2
To assess how the contribution of these sources shifts when respiration is impaired, we repeated the measurements in the presence of metformin. Strikingly, metformin (and also the related complex I inhibitor phenformin) dramatically elevated the fractional contribution of serine to NADH (Figure 2B, Supplementary Fig. 2D). This enhancement inversely related to residual oxygen consumption (Figure 2C, Supplementary Fig. 2D) and was seen across a diversity of tumor and normal cell lines (Figure 2D). Both the basal and metformin-induced serine contribution to NADH occurred via folate-mediated mitochondrial serine catabolism, as it was fully blocked by knockout of the mitochondrial folate enzyme SHMT2 or MTHFD2 (Figure 2E-F). SHMT2 re-expression rescued the serine contribution to NADH (Figure2F). Thus, mitochondrial serine catabolism via the SHMT2-MTHFD2 pathway is a major NADH source when respiration is impaired.
NADH/NAD ratio dictates serine’s NADH contribution
We hypothesized that an elevated NADH/NAD ratio disfavors other routes of NADH production, and therefore respiration impairment increases the fractional serine contribution to NADH. As a first step towards exploring this hypothesis, we examined the impact of different respiratory chain inhibitors on serine’s NADH contribution. Loss of mitochondrial DNA (ρ0 cells) or pharmacological inhibition of complexes I, III, or V increased the NADH/NAD ratio and elevated the fractional serine contribution to NADH (Figure 3A-B, Supplementary Fig. 3A). In contrast, inhibition of complex II, which is not required for NADH oxidation and whose inhibition increased succinate but not NADH/NAD, did not (Figure 3A-B, Supplementary Fig. 3B).
Figure 3:
NADH/NAD ratio dictates serine’s NADH contribution
A: Electron transport chain schematic. B: Fraction NAD2H from [2,3,3-2H]serine in the presence of the 1μM rotenone, 500nM atpenin, 1μM antimycin, or 1μM oligomycin or electron transport chain deficient cells (ρ0) (mean ± SD, N≥3). C: Correlation of fraction NAD2H from [2,3,3-2H]serine with intracellular NADH/NAD ratio (relative to DMEM without metformin). D: Duroquinone (DQ) and pyruvate can serve as electron acceptors to convert NADH into NAD. E: Impact of DQ and pyruvate on NADH/NAD and the fraction NAD2H from [2,3,3-2H]serine in HCT116 cells cultured with 5 mM metformin (mean ± SD, N≥4). ** p<0.01, *** p<0.001 by two-tailed student’s t test. See also Figure S3
To further explore the connection between the intracellular NADH/NAD ratio and serine’s NADH contribution, we measured both parameters in response to different dosages of metformin treatment and found that they correlated linearly (Figure 3C). A similar correlation was observed as a function of metformin exposure duration (Supplementary Fig. 3C). As an orthogonal approach to manipulating NADH/NAD, we added duroquinone or pyruvate to metformin-treated cells. These compounds have been previously shown to act as electron acceptors and thereby decrease the NADH/NAD ratio (Figure 3D) (Sullivan et al., 2015). As hypothesized, their addition decreased the fractional contribution of serine to NADH (Figure 3E). This did not appear to require pyruvate entry into mitochondria, as the effect of pyruvate was not blocked by UK5099, an inhibitor of mitochondrial pyruvate carrier (Supplementary Fig. 3D-F). Instead, pyruvate seems to be converted into lactate in the cytosol, with normalization of the cytosolic NAD/NADH ratio indirectly impacting mitochondrial redox status (e.g. via the malate-aspartate shuttle). In sum, across a diversity of manipulations, as the NADH/NAD ratio increases, serine’s fractional contribution to NADH also increases.
High NADH/NAD shuts off other NADH sources but not serine catabolism
The correlation between NADH/NAD and serine’s fractional NADH contribution could be explained by either (i) enhanced serine catabolism when NADH/NAD rises or (ii) shutting off of other routes of NADH production. We considered the latter more likely. To test for it, we measured the contribution of 13C-glucose, glutamine, and palmitate to TCA intermediates as a function of the NADH/NAD ratio induced by respiration inhibition with metformin (for pathway schematics, see Supplementary Fig. 4A-C). As NADH/NAD rose, the TCA contribution from glucose was nearly eliminated. This reflected both pyruvate dehydrogenase inhibition (which was evident as loss of M+2 citrate from glucose) and blocked TCA turning, which was evident as decreased malate and citrate M+4 from glutamine (with glutamine instead contributing to the TCA via reductive carboxylation, Figure 4A-B, Supplementary Fig. 4D-F). As oxidation of glucose and glutamine dropped, initially the fractional fatty acid contribution to TCA rose, but then decreased (Figure 4C, Supplementary Fig. 4G-H). Similarly, de novo serine synthesis, which produces cytoplasmic NADH, nearly ceased (Supplementary Fig. 4I) (Diehl et al., 2019).
Figure 4:
High NADH/NAD shuts off other NADH sources but not serine catabolism
A: Decrease in [U-13C]glucose entry into TCA cycle with increasing cellular NADH/NAD. B: Decrease in [U-13C]glutamine oxidation (glutamine is instead metabolized reductively, see Supplementary Fig. 4F). C: Initial increase, followed by decrease, in [U-13C]palmitate contribution to TCA cycle. D: Maintenance of mitochondrial serine catabolism indicated by M+1 dTTP from [2,3,3-2H] serine (see Fig. 1A). E: Quantification of intracellular NAD(H) concentration with metformin treatment (The increase in NADH/NAD ratio with metformin treatment is largely driven by NADH accumulation). F: Michaelis-Menten plot of pyruvate dehydrogenase (PDH), α-ketoglutarate dehydrogenase (KGDH), malate dehydrogenase (MDH) and MTHFD2 enzyme activity with increasing NADH as the competitive inhibitor (mean ± SD, N=3). See also Figure S4
To examine whether mitochondrial serine catabolism is also shut off, we measured thymidine labeling from [2,3,3-2H]serine. Mitochondrial serine catabolism generates M+1 thymidine, whereas the alternative cytosolic pathway generates M+2 thymidine. There was no decrease in the thymidine M+1 labeling fraction up to a 5-fold increase in NADH/NAD, a level of respiration impairment sufficient to block TCA turning driven by glucose, glutamine and fat and de novo serine synthesis (Figure 4D). Yet higher NADH/NAD ratios did result in a partial shift towards cytosolic serine metabolism. Nevertheless, catabolism of serine is uniquely robust to rising NADH/NAD.
To examine the underlying biochemical mechanism, we examined the sensitivity of pyruvate dehydrogenase (PDH), α-ketoglutarate dehydrogenase (KGDH) and malate dehydrogenase (MDH) versus MTHFD2 to increasing NADH. These assays were conducted in the presence of adequate NAD as substrate, which is appropriate given that physiologically we see large changes in the NADH/NAD ratio driven by increasing NADH, with NAD largely maintained (Figure 4E). While all of the enzymes were subject to product inhibition by NADH, MTHFD2 was at least 10-fold less sensitive, maintaining full activity up to 200 μM NADH, a typical level observed in respiration-impaired cells (Figure 4F). Thus, the key redox enzyme of serine catabolism is NADH-resistant, and therefore serine catabolism persists even after other NADH production pathways shut off.
Loss of mitochondrial serine catabolism paradoxically facilitates cell growth in respiration-impaired cells
Serine catabolism is the primary cellular source of 1C units, which are required for nucleotide synthesis and therefore growth. Inhibition of 1C metabolism with anti-folates is commonly clinically employed to treat proliferative conditions, including cancer and autoimmunity (Chabner and Roberts, 2005; Kremer, 1994; Weinblatt et al., 1985). Thus, serine catabolism is classically pro-growth. Maintenance of redox homeostasis is, however, also critical to cell growth. We wondered if persistent serine catabolism during respiration impairment might induce a pathologically high NADH/NAD ratio and thereby paradoxically impair cell growth.
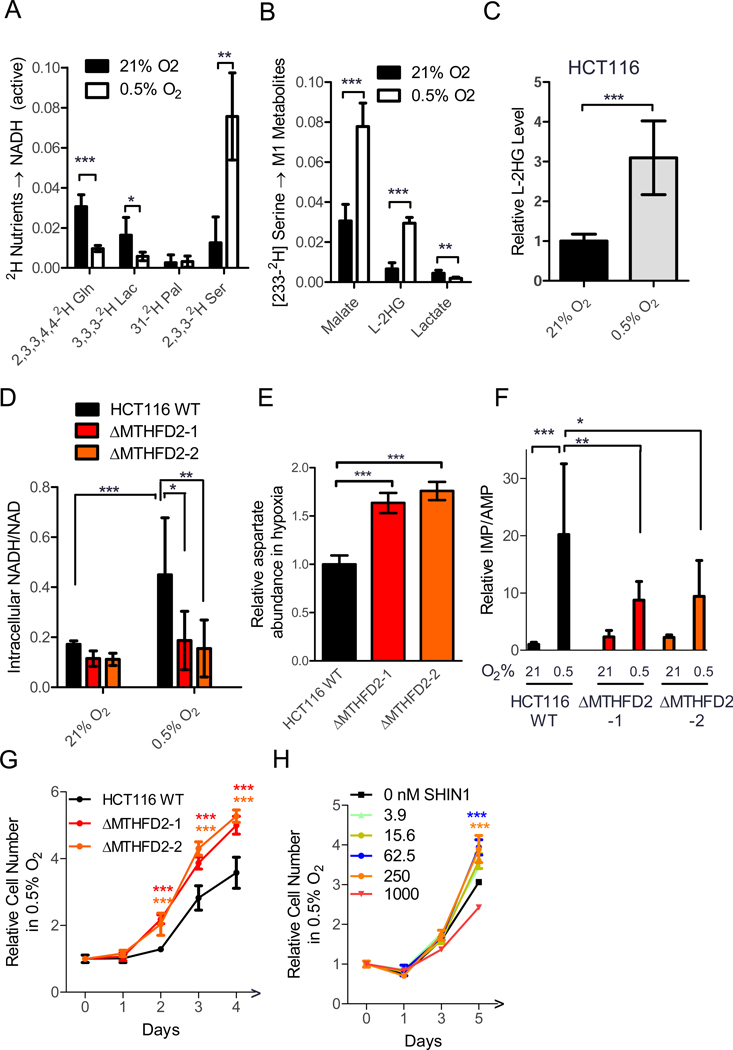
To examine this possibility, we studied metabolite levels and growth of MTHFD2 knockout cells, in the presence and absence of complex I inhibitors. Strikingly, such cells tended to have a lower NADH/NAD ratio under basal conditions, which was significantly lower than wild-type cells after complex I inhibition with metformin, phenformin, or rotenone (Figure 5A, Supplementary Fig. 5A).
Figure 5:
Loss of mitochondrial serine catabolism paradoxically facilitates cell growth in respiration-impaired cells.
A: NADH/NAD for wild type and MTHFD2 knockout HCT116 cells ± metformin (mean ± SD, N≥6). B: Schematic of links between NADH/NAD, aspartate, and nucleotide synthesis. C-E: Intracellular aspartate (C), IMP/AMP ratio (D) and cell number (E) for wild type and MTHFD2 deficient HCT116 in the presence and absence of 1 mM metformin (mean ± SD, panel C&D: N≥6, panel E: N≥6). F: Schematic of SHIN1 as inhibitor of SHMT1/2. G: HCT116 cell growth in normal medium or the presence of 1 mM metformin ± the indicated concentration of the serine catabolism inhibitor SHIN1 (mean ± SD, N=5). **p<0.01, ***p<0.001 by two-tailed student’s t test. See also Figure S5
In response to respiration impairment, elevated NADH/NAD tends to deplete the most oxidized intermediate of the TCA cycle, oxaloacetate, and thereby its transamination partner aspartate. Aspartate is required for protein, purine, and pyrimidine biosynthesis (Figure 5B). Manipulations that restore aspartate can accelerate growth in respiration-impaired conditions (Birsoy et al., 2015; Sullivan et al., 2018). We observed increased aspartate in cells lacking MTHFD2 under both basal and metformin-treated conditions (Figure 5C). This aspartate was mainly M+4 labeled from U-13C-glutamine, indicating its production by oxidative TCA metabolism (Supplementary Fig. 5B).
Aspartate contributes to purine synthesis by donating its amino group to IMP to make AMP. An increased IMP/AMP ratio accordingly indicates aspartate depletion. In the presence of metformin, unlike wild-type cells, MTHFD2 knockout cells maintained a normal IMP/AMP ratio (Figure 5D). Thus, in response to respiration impairment, cells lacking mitochondrial serine catabolism are better at maintaining metabolic homeostasis.
Upon pharmacological complex I inhibition, this improved capacity for metabolic homeostasis resulted in faster growth of MTHFD2 knockout cells, especially at high drug concentrations (Figure 5E, Supplementary Fig. 5C-E). Similarly, removal of serine from the tissue culture medium rendered cell growth partially resistant to metformin (Supplementary Fig. 5F). Given the surprising nature of these findings, to confirm that mitochondrial serine catabolism can be anti-proliferative when respiration is impaired, we treated cells with the small molecule inhibitor of serine catabolism SHIN1 (Fig. 5F) (Ducker et al., 2017). While SHIN1 decreased cell proliferation in standard tissue culture conditions (Fig. 5G), it augmented proliferation in the presence of metformin, phenformin, or rotenone (Figure 5G, Supplementary Fig. 5G-H). Thus, when respiration is impaired, excessive NADH generation via serine catabolism can decrease cell growth.
Hypoxia also induces serine-dependent NADH production
Up to this point, for convenience we relied on complex I inhibitors to induce respiration impairment. While metformin is a very important pharmaceutical, physiologically, hypoxia is the most important cause of respiration impairment. Similar to complex I inhibitors, hypoxia resulted in an elevated fractional contribution of serine to NADH (Figure 6A). Examination of compartmentalized downstream NADH products revealed that the NADH was produced from serine in mitochondria, as label was transferred to malate and L-2 hydroxyglutarate (which can be made in the mitochondria and rose with hypoxia), but not lactate (which is produced from NADH in the cytosol) (Figure 6B-C).
Figure 6:
Hypoxia induces serine-dependent NADH production.
A: Fraction NAD2H from 2H-labeled nutrients in wild-type HCT116 cells in normoxia and hypoxia (tracers as in Fig. 2A, mean ± SD, N≥3). B: 2H-labeled metabolites (M+1) from [2,3,3-2H] serine in normoxia and hypoxia (mean ± SD, N=6). C: Relative L-2 hydroxyglutarate level for HCT116 cell lines in normoxia and hypoxia (mean ± SD, N=9). D: NADH/NAD in normoxia and hypoxia (mean ± SD, N≥8). E: Intracellular aspartate in hypoxia (0.5% O2) (mean ± SD, N=6). F: Relative IMP/AMP ratio (mean ± SD, N=6). G: Cell growth in hypoxia (mean ± SD, N=4). H: HCT116 WT cell growth in hypoxia (0.5% O2) ± the indicated concentrations of the serine catabolism inhibitor SHIN1 (mean ± SD, N=5). P value indicates statistical analysis between WT and KO (G) or no drug vs 62.5 nM or 250 nM SHIN1 (H). * p<0.05, **p<0.01. ***p<0.001 by two-tailed student’s t test.
Similar to metformin treatment, hypoxia induced an increased NADH/NAD ratio, which was blunted by MTHFD2 knockout (Figure 6D). Correspondingly, MTHFD2 knockout cells maintained a higher aspartate concentration, lower IMP/AMP ratio and grew faster than wild-type cells in hypoxia (Figure 6E-G). Low doses of SHIN1 also modestly increased cell growth in hypoxia (Figure 6H). Thus, our observations using metformin generalize to respiration impairment induced by hypoxia.
Serine’s contribution to NADH is enhanced by mitochondrial diseases mutations
Another cause of respiration impairment is genetic deficiencies of the electron transport chain. In humans, such deficiencies manifest in diseases including Leigh Syndrome, a severe neurological disorder that results in cognitive and motor impairment, typically starting during the first year of life (Darin et al., 2001). Leigh Syndrome is caused by a diversity of nuclear and mitochondrial DNA mutations that result in impaired respiratory chain function. We compared fibroblasts from patients with mitochondrial diseases and controls and observed an increased NADH/NAD ratio and 2H-serine contribution to NADH and malate in the primary patient cells (Fig. 7A-C, Supplementary Table 1).
Figure 7:
Serine catabolism provides NADH in fibroblasts derived from Leigh Syndrome patients, and NDUFS4 −/− mice.
A-C: NADH/NAD, fraction NAD2H from [2,3,3-2H]serine and fraction 2H-malate from [2,3,3-2H]serine in cultured fibroblasts from healthy donors (N) and patients with mitochondrial diseases (P) (mean ± SD, N=3 for heathy donors, N=7 from patients with mitochondrial diseases,  : ∆MTL-L1,
: ∆MTL-L1,  : ∆MTO1,
: ∆MTO1,  :∆NDUFS4-male,
:∆NDUFS4-male, : ∆NDUFS4-female,
: ∆NDUFS4-female, : ∆NDUFS3,
: ∆NDUFS3, : ∆NDUFAF2,
: ∆NDUFAF2, : ∆UQCC2; for details, see Table S1) D: Increased malate labeling from [2,3,3-2H]serine in NDUFS4−/− mice in vivo. Data are fractional 2H-malate labeling relative to tissue serine labeling after 2.5 h [2,3,3-2H] serine infusion (40 nmoles/g/min ) (mean ± SD, N=6 for WT, N=9 for NDUFS4 −/−). E: SHIN2 blocks NADH production by serine catabolism in vivo. WT C57BL/6mice were treated with SHIN2 (200mg/kg, ip) immediately before a 2.5 h [2,3,3-2H] serine infusion (mean ± SD, N=3). F: Delayed occurrence of limb clasping in NDUFS4−/− treated with SHIN2 (N=7 for vehicle treatment, N=9 for SHIN2 treatment). *p<0.05, **p<0.01 with two-tailed student’s t test. See also Figure S6–7, Table S1.
: ∆UQCC2; for details, see Table S1) D: Increased malate labeling from [2,3,3-2H]serine in NDUFS4−/− mice in vivo. Data are fractional 2H-malate labeling relative to tissue serine labeling after 2.5 h [2,3,3-2H] serine infusion (40 nmoles/g/min ) (mean ± SD, N=6 for WT, N=9 for NDUFS4 −/−). E: SHIN2 blocks NADH production by serine catabolism in vivo. WT C57BL/6mice were treated with SHIN2 (200mg/kg, ip) immediately before a 2.5 h [2,3,3-2H] serine infusion (mean ± SD, N=3). F: Delayed occurrence of limb clasping in NDUFS4−/− treated with SHIN2 (N=7 for vehicle treatment, N=9 for SHIN2 treatment). *p<0.05, **p<0.01 with two-tailed student’s t test. See also Figure S6–7, Table S1.
One of frequently mutated genes in mitochondrial diseases is MTO1, an enzyme involved in mitochondrial tRNA modification and thereby respiratory chain translation (Fakruddin et al., 2018). We generated colon cancer cells lacking this gene and observed an increased serine contribution to NADH both in cell culture and as xenograft tumors (Supplementary Fig. 6A-B). Moreover, pharmacological blockade of mitochondrial serine metabolism increased these cells’ aspartate level and growth rate (Supplementary Fig. 6C-D).
Mutations in the complex I accessory subunit NDUFS4 also cause mitochondrial deficiency. NDUFS4−/− mice have emerged as a useful model for mitochondrial diseases including the neuromuscular disorder Leigh Syndrome (Jain et al., 2016) Consistent with defective oxidative metabolic capacity and decreased whole body oxygen uptake, NDUFS4−/− mice manifested lower circulatory turnover flux of glutamine (Supplementary Fig. 6E) (Jain et al., 2019). The fractional contribution of carbohydrate (as measured by TCA labeling from U-13C-glucose) and U-13C-glutamine to TCA metabolism was not dramatically altered in the complex I deficient mice, with the notable exception of a decreased glutamine TCA contribution in pancreas and liver (Supplementary Fig. 6F). In addition, TCA isotope labeling patterns did not shift much, with no evidence for decreased PDH flux (as measured by M+2/M+3 malate labeling from U-13C-glucose) or increased reductive carboxylation (as measured by increased M+3/M+4 malate labeling from U-13C-glutamine) (Supplementary Fig. 6G). [2,3,3-2H]serine tracing revealed increased 2H-malate labeling in the mutant mice, which was most evident in pancreas but also occurred with incomplete penetrance in liver and spleen, the other two major organs to use circulating serine as an NADH supplier (Fig. 1B, 7D). Upon in vitro stimulation, T cells from NDUFS4−/− mice also showed increased NADH/NAD and serine contribution to NADH (Supplementary Fig. 7A). Thus, mitochondrial genetic mutations enhance serine’s contribution to NADH both in vitro and in vivo.
One of frequently observed symptoms for the neurological disorder in NDUFS4 −/−mice is limb clasping, which reflects mitochondrial disease progression (Johnson et al., 2013). As cancer cells are partially protected from complex I inhibition by serine catabolism inhibition, we were curious if such protection extends to muscle, where it could potentially be therapeutically advantageous. Indeed, C2C12 myoblasts better tolerated metformin in the presence of pharmacological serine catabolism inhibitor (SHIN1) (Supplementary Fig. 7B-D). We then tested whether a dual SHMT1/2 inhibitor with improved pharmacokinetics, SHIN2, could decrease NADH production from serine in vivo. Single intraperitoneal injection of SHIN2 (200 mg/kg) before tracer infusion blocked most malate labeling from [2,3,3-2H]serine (Fig. 7E). We then proceeded to test whether SHIN2 could partially ameliorate the long-term effects of complex I deficiency. Although not significantly altering survival, treatment NDUFS4−/− mice with SHIN2 (50mg/kg b.i.d. continuously starting from postnatal day 25±4) delayed pathological clasping behavior (Figure 7E, Supplementary Fig. 7E). Thus, serine catabolism can contribute to mitochondrial disease-induced spasticity in vivo.
Discussion:
NAD is essential for metabolism. Oxidative energy production correlates with flux through mitochondrial NADH. Accordingly, there has been extensive research on NADH sources in different physiological and pathological contexts (Chen et al., 2018; Gaude et al., 2018; Guarente, 2008; McKenna et al., 2006; Owen et al., 2002; Solaini et al., 2010; Yang et al., 2014; Yaniv et al., 2013). A limitation of these studies has been inferring NADH sources based on tracking carbon flows, even though the key energy-carrying functional group in NADH is actually the redox-active hydrogen. Here we directly measure NADH sources using heavy hydrogen (deuterium), revealing that serine is a substantial NADH source.
Serine contributes to NADH independent from the TCA cycle, via folate-mediated mitochondrial serine catabolism. The enzyme SHMT2 converts serine to glycine with concomitant production of methylene-THF. The enzyme MTHFD2 then transfers electrons from methylene-THF to NAD, producing 10-formyl-THF and NADH. We prove genetically that both of these enzymes are essential to serine-dependent NADH production. Because MTHFD2 is resistant to product inhibition by NADH, the activity of these enzymes persists when respiration is impaired. Accordingly, when respiration is impaired, even though the absolute flux through serine catabolism likely does not increase, the fraction of NADH made by serine catabolism rises.
Is this flux physiologically significant? In normal physiological conditions, 4% of NADH in pancreas is labeled by 2H-serine infusion. The real contribution of serine catabolism is, however, larger, as labeling is limited by the deuterium kinetic isotope effect (slower reactivity of deuterated, compared to unlabeled, methylene-THF) and by 1H-2H exchange between NADH and water (Ducker et al., 2016; Schmidt et al., 2000; Wiberg, 1955; Zhang et al., 2017). The magnitude of both of these effects is on the order of 2-fold (perhaps substantially larger for 1H-2H exchange), suggesting that ~20% or more of pancreas NADH is made by serine catabolism, with serine’s contribution less in other tissues.
When respiration is impaired, the importance of serine catabolism as NADH source further increases. This reflects shutting off of other routes of NADH production, which in cell culture occurs in a hierarchical manner. Relatively modest elevations in NADH inhibit serine synthesis from glucose, pyruvate dehydrogenase, and/or NADH-dependent isocitrate dehydrogenase, preventing oxidation of pyruvate, lactate, and glucose. Further elevations re-direct glutamine metabolism, from oxidative TCA turning to reductive carboxylation. Yet stronger NADH elevations decrease fatty acid oxidation. These mechanisms collectively help to prevent pathological NADH elevation. Interestingly, in complex I deficient mice, overall glutamine flux was decreased, but the relative contribution of glucose and glutamine to TCA did not shift markedly in most tissues, nor was there evidence for dramatic shifts in their handling within the TCA cycle. This is consistent with a requirement to maintain mass balance at the whole-body level: ultimately, the contents of ingested food must be oxidized. This contrasts with cultured cells, which can pick their preferred substrates from diverse media constituents.
Not only does serine catabolism persist when respiration is impaired, certain regulatory mechanisms actively promote it. Hypoxia induces SHMT2 via MYC and HIF, which can also upregulate the serine synthesis pathway and other mitochondrial 1C enzymes (MTHFD2, MTHFD1L) (Heng et al., 2019; Martinez-Reyes and Chandel, 2014; Papandreou et al., 2006; Samanta et al., 2016; Ye et al., 2014). We find that this induction of serine catabolism is, at least at the cellular level, maladaptive: when respiration is impaired, genetic or pharmacological inhibition of serine catabolism enhances cell growth. Further research is merited to determine whether this is a quirk of evolution or reflects a higher organismal objective, such as to prevent the proliferation of respiration deficient cells or to attain other products of 1C metabolism at a poorly vascularized wound site.
In mitochondrial diseases, additional physiological signaling mechanisms result in counter-productive responses. Mitochondrial deficiency turns on amino acid starvation response signaling by ATF4, likely via aspartate depletion. For unclear reasons, it also activates mTOR. Both ATF4 and mTOR promote MTHFD2 expression and thus mitochondrial serine catabolism (Ben-Sahra et al., 2016; Khan et al., 2017; Nikkanen et al., 2016; Tyynismaa et al., 2010; Zheng et al., 2016). The activation of mTOR in mitochondrial disorders is maladaptive, as rapamycin improves mitochondrial myopathy and prolongs life span (Johnson et al., 2013; Khan et al., 2017; Zheng et al., 2016). Moreover, we find that serine catabolism inhibitor modestly ameliorates disease progression in the NDUFS4 Leigh Syndrome mouse model.
Clinically, there is no curative therapy for mitochondrial diseases (Murayama et al., 2019; Suomalainen and Battersby, 2018). “Mitochondrial cocktails” of vitamins and cofactors including coenzyme Q are widely used but have no proven clinical efficacy (Kerr, 2013; Pfeffer et al., 2012). Massive pyruvate intake offers a non-mitochondrial pathway to NADH oxidation, but is far from a robust solution (Fujii et al., 2014; Saito et al., 2012; Sullivan et al., 2015). Chronic hypoxia slows disease progression in mice, by suppressing reactive oxygen species, but is hard to implement clinically (Ferrari et al., 2017; Jain et al., 2019; Jain et al., 2016). Each of these strategies might work better when combined with inhibition of serine catabolism, to decrease serine-driven NADH accumulation.
More generally, it may be possible to use the serine pathway to modulate NADH status. For example, one can envision feeding excess serine and thereby overloading hypoxic tumors with NADH. Conversely, serine catabolism inhibitors might mitigate NADH buildup during ischemic events, reducing suffering in diseases of chronic vascular insufficiency and/or enabling safer reperfusion in acute events. These ideas highlight the many new directions opened by the discovery of a previously unappreciated route to NADH.
Limitation of Study:
The present work examines NADH sources in cultured cells and mice. The measurements rely on deuterium (2H) tracing, which makes quantitative determination of pathway contributions difficult due to the deuterium kinetic isotope effect and 1H-2H exchange with water. In cultured cells, we nevertheless obtain strong NADH labeling from 2H-serine, demonstrating that folate-mediated serine catabolism is a major NADH contributor, especially when respiration is impaired. In mice, however, the extent of measured labeling is substantially less, with a material contribution evident mainly in pancreas and tumors. The lesser NADH labeling in vivo may reflect technical issues and/or the serine pathway being a comparatively unimportant NADH source in many tissues.
Consistent with the large serine catabolism contribution to NADH in cell culture, we find that serine catabolism inhibition can substantially ameliorate NADH buildup and promote cell growth during respiration impairment. To examine respiration impairment in vivo, we took advantage of the NDUSF4 knockout mitochondrial disease model. We found that pharmacological serine catabolism inhibition modestly ameliorated disease progression. The extent of NADH buildup in this model, however, was modest (no more than 2-fold at the whole tissue level) and we did not prove that serine catabolism inhibition decreases NADH levels. It is possible that the observed health benefits may occur via routes other than decreasing NADH. For example, serine catabolism inhibition can alter nucleotide precursor levels (and thereby activate AMPK) or the serine/alanine ratio (and thereby prevent pathological shifts towards 1-deoxy-sphingolipids)(Ducker et al., 2016; Esaki et al., 2015; Gantner et al., 2019; Garcia and Shaw, 2017).
STAR Methods:
LEAD CONTACT AND MATERIALS AVAILABILITY
Requests for further information and resources should be directed to and will be fulfilled by the Lead Contact, Joshua Rabinowitz (joshr@princeton.edu). This study did not generate new unique reagents
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell lines and primary cell culture:
Cancer cell lines were incubated with DMEM + 10% FBS, 37 °C, 5% CO2 Cells in hypoxia chamber were incubated with DMEM + 10% FBS, 37 °C, 5% CO2, 0.5%O2. For primary patients’ derived fibroblasts were incubated with DMEM+10% FBS+1mM sodium pyruvate+100 μM hypoxanthine and 16 μM thymidine, 37 °C, 5% CO2. Jurkat was incubated with RPMI+10% FBS. HCT116, HepG2, Panc1, A549, Jurkat, 293T, C2C12 were purchased from ATCC, HAP1 were purchased from Horizon Discovery, U2OS is kindly provided by Reuben Shaw (Salk Institute). Primary patients’ derived fibroblasts were provided by Johannes A. Mayr (Department of Pediatrics, Salzburger Landeskliniken and Paracelsus Medical University, Salzburg 5020, Austria).
Mice Models:
Animal studies followed protocols approved by the Princeton University Institutional Animal Care and Use Committee and the Rutgers University Institutional Animal Care and Use Committee. NDUFS4 +/−mice were purchased from Jackson Laboratory, and homozygous litters were breed in house with PicoLab 5058. Male C57BL/6 mice and CD1 nude mice were purchased from Charles River at the age of 6–8 week. The mice were on normal light cycle (7 AM – 7 PM) at controlled temperature (23 ° C±2 ° C). PicoLab 5053 was provided as normal chow for C57BL/6 mice/CD1 and NDUFS4 mice after postnatal day 21.
METHODS DETAILS
Isolation, culture and stimulation of naïve CD8+ or Total T cells
To isolate naïve CD8+ T or total T cells, spleens were harvested and single cell suspensions prepared by manual disruption and passage through a 70 μm cell strainer in PBS supplemented with 0.5% BSA and 2 mM EDTA. After red blood cell lysis, naïve CD8+ T or total T cells were purified by magnetic bead separation using commercially available kits following vendor instructions (Naive CD8a+ T Cell Isolation Kit, mouse or Pan T Cell Isolation Kit II, mouse, Miltenyi Biotec Inc).
Cells were cultured in complete RPMI media (11875–093, supplemented with 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 50 μM 2-mercaptoethanol). For activation, T cells were stimulated for 48 h with plate-bound anti-CD3 (10 μg/ml) and anti-CD28 (5 μg/ml) in complete media supplemented with 100 μM alanine and recombinant IL-2 (100 U/mL). Cells were maintained in complete RPMI media supplemented with 100 U/mL recombinant IL-2. Metabolomics experiments were performed at day 4–5 post-activation.
Mice Experiments:
In vivo infusion was performed on 9–10 week old C57BL/6 mice pre-catheterized on the right jugular vein for general [2,3,3-2H]serine tracing experiment, and 4-week old wild type NDUFS4 −/− or homozygous NDUFS4+/+ mice pre-catheterized on the jugular vein (jugular vein catheterization surgery was performed one week before infusion in mice procedure room) for [2,3,3-2H]serine/[U-13C]glucose/[U-13C]glutamine tracing. Infusion was performed for 2.5 h to achieve isotopic pseudo-steady state. The mouse infusion setup included a tether and swivel system, connecting to the button pre-implanted under the back skin of mice. Mice were fasted from 9:00 am to 2 pm, and infused from 2pm till 4:30 pm. 400mM [2,3,3-2H]serine/200mM [U-13C]glucose/100mM [U-13C]glutamine was dissolved in saline, and infused via the catheter at a constant rate (0.1 μl/min/g mouse weight) using a Just infusion Syringe Pump. At the end of infusion, mice were dissected and tissues were clamped in aluminum foil and stored in liquid nitrogen.
NDUFS4 −/− mice were housed with its wild type/heterozygous littermates. Food was placed on the bedding to guarantee the accessibility of food. Mice were randomly grouped as vehicle control group and SHIN2 treatment groups. SHIN2 was dissolved in 20% 2-hydroxypropyl-cyclodextrin solution and delivered by intraperitoneal (50 μg/g mice) b.i.d. Mice were treated from postnatal day 25±4 till humane endpoint. Mice weights were recorded every day. Limb clasps were recorded daily, with mice annotated as having the pathological limb clasping phenotype on the third consecutive day of clasping. 20% weight loss of the peak weight for each mouse is considered as the humane endpoint.
For tumor xenograft tracing experiments, 7–8 week-old male CD1/nude mice were injected in the rear flanks with HCT-116 (3 × 106 cells in 100 μL 1:1 DMEM:Matrigel). For tumor allograft tracing experiment, 6- to 8-week-old male C57BL6 mice were injected in the rear flank with KrasLSL.G12D/+ x p53R172H/+ x PdxCretg/+ pancreatic tumor tissue suspension (200 μL 1:1 DMEM:Matrigel). Jugular vein catheterization surgery was performed when tumor size reached 250 mm3 [volume = 1∕2 (length × width × height)]. Animals were euthanized when tumors reached 1,000 mm3 or if they displayed any signs of distress or morbidity. Tracing experiments were performed after 1 week of post-surgery monitoring. Tumors were removed and immediately clamped to aluminum foil using Wollenberger clamp and stored in liquid nitrogen. For Long term storage, tissues were kept in −80°C until future sample preparation.
Blood sampling from pigs:
Pig studies followed protocols approved by the University of Pennsylvania Institutional Animal Care and Use Committee. Five-month old male Yorkshire pigs were fed at 4 PM and then overnight fasted. At 7 AM in the next morning, anesthesia was initiated with a ketamine IM injection (15~35 mg/kg), followed by intubation and maintenance of anesthesia with inhaled isoflurane at 0.5~5%. Arterial and venous access was obtained to enable fluid infusion (NaCl 4 – 10 mcg/kg/min), blood pressure monitoring, pharmacological control of blood pressure (0.2 – 2.0 mcg/kg/min dobutamine and 0.1 – 5.0 mcg/kg/min phenylephrine, as required), and blood sampling. The primary initiative venous access was via ultrasound-guided percutaneous puncture of the external jugular vein. Arterial access was through the neck to the carotid artery. The surgical site was blocked with Bupivacaine SQ just after the first incision. The subcutaneous tissues and muscle layers were carefully dissected, with hemostasis achieved with electrocautery. For blood sampling from the hepatic and the portal vein, a catheter was placed on each vein. Blood from inferior vena cava, internal jugular vein, femoral vein, renal vein, splenic vein, inferior pancreatic vein, inferior mesenteric vein, lateral auricular vein, and coronary sinus were obtained by drawing with a 27G needle connected to 1 mL syringe. Blood was drawn from each vessel twice at 1 – 2 min intervals, except for the coronary sinus (only a single draw). After completion of blood sampling, the animal was euthanized. Blood samples were placed at room temperature for 10 – 20 min, and centrifuged at 1,000 x g for 10 min to obtain serum. Data represents the median value for triplicate technical replicates.
Metabolite extraction from cell culture:
Adherent cells:
Cells were seeded in 6 well plates overnight and then were treated with ETC inhibitors. Serine-free DMEM with 10% dialyzed FBS was supplemented with 400 μM [2,3,3, 2H]serine. Medium was replaced by the isotope tracing medium with corresponding drugs, and cells were incubated for 3 h. Medium was aspirated, and metabolism was quenched with extraction buffer (40:40:20 acetonitrile: methanol: water with 0.5% formic acid; 40 μl of extraction solvent per 1 μl packed cell volume). Plates were placed on dry ice for 10 mins, and thereafter the extraction solution was neutralized with 15% NH4HCO3 (70 μl for 800 μl extraction buffer). Cells were then scraped off from plates by using a cell lifter (Corning 3008), transferred into Eppendorf tubes and centrifuged in a benchtop microfuge at maximum speed for 30 min at 4°C. Supernatant was transferred to LC-MS vials for analysis.
T cells:
RPMI-1640 media without glucose, glycine or serine was supplemented with 11 mM glucose, 0.133 mM glycine, 0.286 mM [2,3,3–2H]serine, 10% dFBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 50 μM 2-mercaptoethanol and 100 U/mL recombinant IL-2. Cells were seeded at 106 cells/mL and incubated for 3 hours. They were then transferred to 1.5mL Eppendorf tubes and pelleted (30 s, 6000 g, RT). Media was removed by aspiration and metabolome extraction was performed by the addition of 75 μL of extraction buffer. After a 5-min incubation on ice, the extraction solution was neutralized by the addition of NH4HCO3 as above. After centrifugation (15 min, benchtop microfuge maximum speed, 4°C), the clean supernatant was transferred to LC-MS vial for analysis.
Metabolite extraction from tissue, tumors and serum:
Tissues and tumors were collected from mice and immediately clamped into liquid nitrogen using Wollenberger clamp, tissue were stored in −80°C. Frozen tissues were transferred into 2 ml Eppendorf tubes, which are precooled on dry ice, and pulverized by using cyromill. The resulting tissue powder was weighed (around 10mg) and mixed well by vortexing in extraction buffer (40 μL extraction buffer per mg tissue). The extraction solution was neutralized with NH4HCO3 as above and centrifuged in a microfuge at maximum speed for 30 min at 4°C. Supernatant was transferred to LC-MS vials for analysis.
Blood samples were drawn from mice tail, using microvette CB300 Z, and kept on ice. After centrifugation (10 min, benchtop microfuge maximum speed, 4°C), serum were collected to 1.5ml tube and store in −80 °C for future analysis. 5 μl of serum were mixed with 200 μl of extraction buffer (40:40:20 acetonitrile: methanol: water with 0.5% formic acid) and neutralized with 15% NH4HCO3. After centrifugation (30 min, benchtop microfuge maximum speed, 4°C), supernatant were transferred into LC-MS vial for analysis.
LC-MS:
LC separation was achieved using a Vanquish UHPLC system (Thermo Fisher Scientific) and an Xbridge BEH Amide column (150×2mm, 2.5 μm particle size; Waters, Milford, MA), column temperature 25°C. Solvent A is 95:5 water: acetonitrile with 20 mM ammonium acetate and 20 mM ammonium hydroxide at pH 9.4, and solvent B is acetonitrile. The gradient was 0 min, 85% B; 2 min, 85% B; 3 min, 60% B; 9 min, 60% B; 9.5 min,35% B; 13 min, 5% B; 15.5 min, 5% B; 16 min, 85% B; 20 min, stop run. Injection volume was 15 μL. The Q-Exactive Plus mass spectrometer was operated in negative ion mode scanning from m/z 70–1000 with a resolution (at m/z 200) of 140,000 (AGC target 3e6, Maximum IT 200ms) (Du et al., 2019). A second scan from m/z 640–765 with a resolution (at m/z 200) of 35,000 (AGC target 5e5, maximum IT 200ms) was used to measure NAD(P), NAD(P)H, and their labeling. The purpose of the second scan is to enhance sensitivity for NAD(P)(H) by restricting the range of ions in the orbitrap and to maximize the accuracy of isotope ratios by decreasing the scan range and mass resolution (Su et al., 2017).
Natural isotope abundance correction:
Accucor was used to correct all of data except NADH from mice (Figure 1E) for natural isotope abundance (Su et al., 2017) . For NADH data from mice, it was necessary to correct also for the interfering peak (non-NADH peak in Figure 1F). For this purpose, data were corrected using Elemcor, using unlabeled tissue data as the background (Du et al., 2019; Millard et al., 2012). The background correction approach is necessary because orbitrap (Q-Exactive, resolution 78K at 660 m/z) cannot resolve the 2H-NADH peak neatly from either the 13C-NADH peak or the non-NADH interfering peak.
Calculation of 2H-serine contribution to malate, aspartate, and NADH:
For malate and aspartate, the 2H-labeling fraction was measured directly (fractional size of 2H-peak relative to sum of all peaks, after isotope correction as above). For NADH, the 2H-labeling fraction was measured based on the active hydride labeling (x), which was determined from the labeling pattern of NAD+ and NADH by least square fitting in MATLAB of the observed experimental data (after natural isotope correction) to the below vectors (Liu et al., 2016a).
For cell culture experiments, serine was nearly completely labeled and fractional labeling as above is reported in the manuscript without further correction. For the in vivo experiments, measured labeling fractions were normalized to the fractional labeling of serine in the tissue of interest. Serine labeling is measured as the fraction M+2 or M+3 serine, as both M+2 and M+3 serine (from [2,3,3-2H]serine) are labeled at the hydrogen that is transfer to NADH by MTHFD2.
FT-ICR MS
We used the FT-ICR MS to resolve M+1 2H NADH from M+1 13C NADH (only for Figure 1F). Analysis was carried out using a 9.4 T Bruker SolariX FT-ICR MS (Bruker Daltonics Inc., MA, USA) with an ESI source in positive mode. Continuous accumulation of selected ions (CASI) was utilized to increase sensitivity. The CASI window was 50 mass units with a center mass of 660. The ion accumulation time was set to 60 ms. The mass spectra were calibrated externally with 1mM arginine solution. Resolving power at the FT-ICR was measured to be 260 K at m/z 660, which is sufficient to isolate M+1 2H NADH from M+1 13C NADH, as well as the similar mass non-NADH peak (which we discovered via this analysis).
Seahorse XF96e:
Cells were seeded to Seahorse XF96 cell seeding plates (101085–004) for overnight attachment. Metformin was added to DMEM+ 10% dialyzed FBS, and cells were treated for 24 h and oxygen consumption measured. Oxygen consumption rates were normalized to DNA content by using DNA qualification kits (Thermo Fisher C7206).
Enzymatic assay for MTHFD2:
NAD (Sigma N0632), NADH (Sigma N8129) and 5, 10-meTHF (Schircks Laboratories, 16.226) solution were freshly prepared each time in water. Reaction mixtures consisted of 50 mM KH2PO4 pH 7.4, 5 mM dithiothreitol (DTT), 5% DMSO, and 0.005 mg/mL MTHFD2 (obtained from Raze Therapeutics), and NAD and NADH as indicated in the Figures. Reaction mixtures were incubated for 5 min at 37℃ before initiation of the reaction by addition of 5,10-meTHF to a final concentration of 0.4 mM using multichannel. The plate was mixed for 5 s in the plate reader (Biotek Synergy HT), and the rate of change in absorbance at 340 nm due to NADH accumulation was recorded over the duration of linear NADH accumulation.
Enzymatic assay for MDH:
NAD and NADH solutions were freshly prepared in water; malate solution was prepared in 50 mM Tris-HCl and adjusted pH to 7.6 with 10 M KOH. Reaction mixtures consisted of 30 mM Tris-HCl pH 7.6, 0.00047 mg/ml malate dehydrogenase (Sigma M1567), and NAD and NADH as indicated in the Figures. Reaction mixtures were incubated for 5 min at 25℃ before initiation of the reaction by addition of malate to a final concentration of 20 mM using multichannel. The plate was mixed for 5 s in the plate reader, and absorbance at 340 nm recorded as above.
Enzymatic assay for PDH and KGDH:
Pyruvate dehydrogenase and alpha-ketoglutarate dehydrogenase solutions were freshly prepared to a concentration 0.075unit/ml with buffer containing 20 mM sodium phosphate and 1 mM MgCl2, pH7.2. A mixture of thiamine pyrophosphate and Coenzyme A was freshly prepared in the same buffer and diluted by 5 times before use. NAD and NADH solutions were freshly prepared in the buffer through a serial dilution; sodium pyruvate and alpha-ketoglutarate solutions were prepared in buffer in a stock concentration of 100mM. All solutions were prepared on ice. Reaction mixtures consisted of a final concentration of 0.015 unit/ml of each enzyme, 0.2mM TPP and 0.5mM CoA, and NAD and NADH as indicated in the Figures. Reaction mixtures were incubated for 5 min at room temperature before initiation of the reaction by addition of substrate to a final concentration of 5 mM using multichannel. The reaction took place at 37°C. The plate was mixed for 5 s in the plate reader, and absorbance at 340 nm recorded as above.
Cell proliferation assays:
Cells were seeded in seeded in 96 well plates (Corning 3603) for overnight attachment. Medium (with or without drugs) was changed next day, and cell numbers were recorded using Thermofisher CyQUANT kit.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data are displayed as mean ± SE with N the number of biological replicates. Unless otherwise specified, p-value were determined by two-tailed unpaired Student’s t-test. For Supplementary Figure 6B, p-values were determined by two-tailed paired Students’s t-test, and for Supplementary Figure 7E by log-rank test.
DATA AND CODE AVAILABILITY
LC-MS files have been deposited at Mendeley data: http://dx.doi.org/10.17632/k27cy7fwh8.3
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-mouse CD3 | BioXCell | Cat# Clone 145–2C11, BE0001–1; RRID:AB_1107634 |
| Anti-mouse CD28 | BioXCell | Cat# Clone 37.51, BE0015–1; RRID:AB_1107624 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| MDH | Sigma Aldrich | Cat# M1567 |
| PDH | Sigma Aldrich | Cat# P7032 |
| KGDH | Sigma Aldrich | Cat# K1502 |
| MTHFD2 | Gift from Raze Therapeutics | N/A |
| [2,3,3-2H]Serine | Cambridge Isotope | Cat# DLM-582 |
| [U-13C]Glucose | Cambridge Isotope | Cat# CLM-1396 |
| [U-13C]Glutamine | Cambridge Isotope | Cat# CLM-1822 |
| [U-13C]Palmitate | Cambridge Isotope | Cat# CLM-6059 |
| [2,3,3,4,4-2H]Glutamine | Cambridge Isotope | Cat# DLM-1826 |
| [3,3,3-2H]Lactate | Cambridge Isotope | Cat# DLM-9071 |
| [U-31-2H]Palmitate | Sigma Aldrich | Cat# 366897 |
| Metformin | Sigma Aldrich | Cat# PHR 1084 |
| Phenformin | Sigma Aldrich | Cat# PHR1573 |
| Atepnin | Cayman chemicals | Cat# 119509–24-9 |
| Rotenone | Sigma Aldrich | Cat# 557368 |
| Oligomycin | Sigma Aldrich | Cat# 75351 |
| Antimycin | Sigma Aldrich | Cat# A8674 |
| Duroquinone | Sigma Aldrich | Cat# D223204 |
| UK5099 | Cayman Chemicals | Cat# 16980 |
| NAD | Sigma Aldrich | Cat# N0632 |
| NADH | Sigma Aldrich | Cat# N8129 |
| 5, 10 – meTHF | Schircks Laboratories | Cat# 16.226 |
| CoA | Sigma Aldrich | Cat# A2056 |
| Thiamine pyrophosphate | Fisher | Cat# T10835G |
| 2-hydroxypropyl-cyclodextrin | Cayman Chemicals | Cat# 16169 |
| Matrigel | Corning | Cat# 354234 |
| HT Supplement | Thermo Fisher Scientific | Cat# 11067–030 |
| Sodium Pyruvate | Gibco | Cat# 11360070 |
| Murine IL-2 | Peprotech | Cat# 212–12 |
| SHIN1 | Synthesized in house | N/A |
| SHIN2 | Synthesized in house | N/A |
| Critical Commercial Assays | ||
| Naïve CD8a+ T cell isolation kit | Miltenyi Biotec | Cat# 130–096-543 |
| Pan T cell isolation kit II | Miltenyi Biotec | Cat# 130–095-130 |
| Seahorse Mitostress Kit | Agilent | Cat# 103015–100 |
| CyQUANT NF cell proliferation assay | Thermofisher | Cat# C35006 |
| CyQUANT cell proliferation for seahorse assay | Thermofisher | Cat# C7026 |
| Experimental Models: Cell Lines | ||
| Human: HCT116 | ATCC | Cat# CCL-247; RRID:CVCL_0291 |
| Human: U2OS | Gift from Reuben Shaw (Salk Institute) | N/A |
| Human: HepG2 | ATCC | Cat# HB-8065; RRID:CVCL_0027 |
| Human: HAP1 | Horizon Discovery | N/A |
| Human: Panc1 | ATCC | Cat# CRL-1469; RRID:CVCL_0480 |
| Human: A549 | ATCC | Cat# CCL-185; RRID:CVCL_0023 |
| Human: C2C12 | ATCC | Cat# CRL-1772; RRID:CVCL_0188 |
| Human: HCT116 ΔMTHFD2 | Morscher et al., 2018 | N/A |
| Human: HCT116 ΔSHMT1 | Morscher et al., 2018 | N/A |
| Human: HCT116 ΔSHMT2 | Morscher et al., 2018 | N/A |
| Human: HEK 293T/17 | ATCC | Cat# CRL-11268; RRID:CVCL_1926 |
| Human: HEK 293T/17 ΔSHMT2 |
Morscher et al., 2018 | N/A |
| Human: HEK 293T/17 ΔSHMT2 Reexpressed | Morscher et al., 2018 | N/A |
| KrasLSL.G12D/+ x p53R172H/+ x PdxCretg/+ pancreatic tumor | Gift from Haiyong Han (TGEN) | N/A |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6 | Charles River Laboratories | Cat# 027; RRID:IMSR_CRL:027 |
| Mouse: NDUFS4+/− | Jackson Laboratory | Cat# 027058; RRID:IMSR_JAX:027058 |
| Mouse: CD1 nude | Charles River Laboratories | Cat# 086; RRID:IMSR_CRL:086 |
| Software and Algorithms | ||
| El-Maven | Elucidata | https://resources.elucidata.io/elmaven |
| GraphPad Prism 7 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Deposited Data | Mendeley data | http://dx.doi.org/10.17632/k27cy7fwh8.3 |
| Other | ||
| LS columns | Miltenyi Biotec | Cat# 130–042-401 |
| Pre-seperation filters (70µm) | Miltenyi Biotec | Cat# 130–095-823 |
| 6 well plates | Corning | Cat# 353046 |
| 96 well plates | Corning | Cat# 278743 |
| Seahorse XF96e | Agilent | https://www.agilent.com/en/products/cell-analysis/seahorse-analyzers/seahorse-xfe96-analyzer |
| LC-MS Q-Exactive plus | Thermofisher | Cat# IQLAAEGAAPFALGMBDK |
| Biotek Synergy HT | Biotek | N/A |
| Mouse Jugular Vein Catheter | SAI | Cat# C20PU-MJV1301 |
| Mouse Button | Instech | Cat# VABM1B/25 |
| XBridge BEH Amide XP column | Waters | Cat# 186006725 |
| Surgery tools | Fine Science Tools | N/A |
| Cyromill | Retsch | Cat# 20.749.0001 |
| Beads | Inframat | Cat# 4039GM-S005 |
| microvette CB300 Z | Sarstedt | Cat# 16.440 |
| SolariX XR MS | Bruker | https://www.bruker.com/products/mass-spectrometry-and-separations/mrms/solarix.html |
| Just infusion Syringe Pump | Just infusion | Cat# NE-300 |
| Hypoxia Chamber | COY Laboratory | https://coylab.com/ |
| DMEM | Corning | Cat# 10–017-CV |
| RPMI | Gibco | Cat# 11875–093 |
| DMEM without glucose, glutamine, serine,glycine | US Biological | Cat# D9802–01 |
| RPMI without glucose serine/glycine | Teknova | Cat# R9660 |
| FBS | Sigma Aldrich | Cat# F2442 |
| Dialyzed FBS | Sigma Aldrich | Cat# FD392 |
Context and Significance:
Energy generation involves breaking down food to release electrons. These electrons are stored in the cofactor NADH, before being used to make the energy currency molecule ATP. While NADH is critical to energy metabolism, too much NADH is toxic. NADH accumulation occurs when cells have too little oxygen or can’t use it efficiently, for example, in mitochondrial disorders. The B vitamin folate plays a central role in growth, development, autoimmunity and cancer. Here Princeton University researchers report that folate metabolism unexpectedly makes substantial NADH. When cells have too little oxygen, most other NADH producing pathways shut down, but folate-mediated NADH production continues, driving toxic NADH accumulation. Modulating folate metabolism may have benefits in mitochondrial diseases and other redox disorders.
Highlights:
Serine catabolism by the mitochondrial folate pathway produces substantial NADH
Respiration inhibition blocks NADH production by TCA cycle, but not serine catabolism
In respiration-impaired cells, serine catabolism drives NADH buildup
Serine pathway modulation can impact cellular redox state
Acknowledgements:
We thank Reuben Shaw (Salk Institute), Greg Ducker (The University of Utah), Serina NG(TGen), Haiyong Han (TGen) and Michel Nofal and all other members of the Rabinowitz laboratory. This work was supported by funding to J.D.R. from the US National Institutes of Health (NIH) (R01CA163591 and DP1DK113643) and Stand Up to Cancer (SU2CAACR-DT-20–16). S. J, Z. S. H., and L. Z. were supported by NIH grants to E.W. (R01CA163591 and R01 CA130893). L.Y. was supported by a postdoctoral fellowship from the New Jersey Commission on Cancer Research. J.C.G.C. is supported by funding from the European Union’s Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement No 751423. C.J. is supported by fellowship from American Diabetes Association. M.R.M is supported by both the Hanna H. Gray Fellows Program of the Howard Hughes Medical Institute and the Burroughs Wellcome Fund Postdoctoral Enrichment Program.
Footnotes
Declaration of Interests:
Joshua Rabinowitz is a member of the Rutgers Cancer Institute of New Jersey and the University of Pennsylvania Diabetes Research Center and a consultant or advisor to Pfizer, Agios Pharmaceuticals, Kadmon Pharmaceuticals, L.E.A.F. Pharmaceuticals, and Rafael Pharmaceuticals, and co-founder of Raze Therapeutics. Eileen White is an SAB member and founder of Vescor Therapeutics and SAB member of Forma Therapeutics. Princeton University has filed patents related to SHMT inhibitors and their uses.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- Ben-Sahra I, Hoxhaj G, Ricoult SJH, Asara JM, and Manning BD (2016). mTORC1 induces purine synthesis through control of the mitochondrial tetrahydrofolate cycle. Science 351, 728–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, and Sabatini DM (2015). An Essential Role of the Mitochondrial Electron Transport Chain in Cell Proliferation Is to Enable Aspartate Synthesis. Cell 162, 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabner BA, and Roberts TG Jr. (2005). Timeline: Chemotherapy and the war on cancer. Nature reviews Cancer 5, 65–72. [DOI] [PubMed] [Google Scholar]
- Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, and Schumacker PT (1998). Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proceedings of the National Academy of Sciences of the United States of America 95, 11715–11720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen QY, Kirk K, Shurubor YI, Zhao DZ, Arreguin AJ, Shahi I, Valsecchi F, Primiano G, Calder EL, Carelli V, et al. (2018). Rewiring of Glutamine Metabolism Is a Bioenergetic Adaptation of Human Cells with Mitochondrial DNA Mutations. Cell metabolism 27, 1007-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darin N, Oldfors A, Moslemi AR, Holme E, and Tulinius M. (2001). The incidence of mitochondrial encephalomyopathies in childhood: clinical features and morphological, biochemical, and DNA abnormalities. Annals of neurology 49, 377–383. [PubMed] [Google Scholar]
- DeBerardinis RJ, Lum JJ, Hatzivassiliou G, and Thompson CB (2008). The biology of cancer:Metabolic reprogramming fuels cell growth and proliferation. Cell metabolism 7, 11–20. [DOI] [PubMed] [Google Scholar]
- Diehl FF, Lewis CA, Fiske BP, and Vander Heiden MG (2019). Cellular redox state constrains serine synthesis and nucleotide production to impact cell proliferation. Nature metabolism 1, 861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du D, Tan L, Wang Y, Peng B, Weinstein JN, Wondisford FE, Su X, and Lorenzi PL (2019). ElemCor: accurate data analysis and enrichment calculation for high-resolution LC-MS stable isotope labeling experiments. BMC bioinformatics 20, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducker GS, Chen L, Morscher RJ, Ghergurovich JM, Esposito M, Teng X, Kang Y, and Rabinowitz JD (2016). Reversal of Cytosolic One-Carbon Flux Compensates for Loss of the Mitochondrial Folate Pathway. Cell Metab 24, 640–641. [DOI] [PubMed] [Google Scholar]
- Ducker GS, Ghergurovich JM, Mainolfi N, Suri V, Jeong SK, Hsin-Jung Li S, Friedman A, Manfredi MG, Gitai Z, Kim H, et al. (2017). Human SHMT inhibitors reveal defective glycine import as a targetable metabolic vulnerability of diffuse large B-cell lymphoma. Proceedings of the National Academy of Sciences of the United States of America 114, 11404–11409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Mir MY, Nogueira V, Fontaine E, Averet N, Rigoulet M, and Leverve X. (2000). Dimethylbiguanide inhibits cell respiration via an indirect effect targeted on the respiratory chain complex I. J Biol Chem 275, 223–228. [DOI] [PubMed] [Google Scholar]
- Esaki K, Sayano T, Sonoda C, Akagi T, Suzuki T, Ogawa T, Okamoto M, Yoshikawa T, Hirabayashi Y, and Furuya S. (2015). L-Serine Deficiency Elicits Intracellular Accumulation of Cytotoxic Deoxysphingolipids and Lipid Body Formation. The Journal of biological chemistry 290, 14595–14609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakruddin M, Wei FY, Suzuki T, Asano K, Kaieda T, Omori A, Izumi R, Fujimura A, Kaitsuka T, Miyata K, et al. (2018). Defective Mitochondrial tRNA Taurine Modification Activates Global Proteostress and Leads to Mitochondrial Disease. Cell reports 22, 482–496. [DOI] [PubMed] [Google Scholar]
- Ferrari M, Jain IH, Goldberger O, Rezoagli E, Thoonen R, Chen KH, Sosnovik DE, Scherrer-Crosbie M, Mootha VK, and Zapol WM (2017). Hypoxia treatment reverses neurodegenerative disease in a mouse model of Leigh syndrome. Proceedings of the National Academy of Sciences of the United States of America 114, E4241–E4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii T, Nozaki F, Saito K, Hayashi A, Nishigaki Y, Murayama K, Tanaka M, Koga Y, Hiejima I, and Kumada T. (2014). Efficacy of pyruvate therapy in patients with mitochondrial disease: A semi-quantitative clinical evaluation study. Mol Genet Metab 112, 133–138. [DOI] [PubMed] [Google Scholar]
- Fullerton MD, Galic S, Marcinko K, Sikkema S, Pulinilkunnil T, Chen ZP, O’Neill HM, Ford RJ, Palanivel R, O’Brien M, et al. (2013). Single phosphorylation sites in Acc1 and Acc2 regulate lipid homeostasis and the insulin-sensitizing effects of metformin. Nat Med 19, 1649–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gameiro PA, Yang JJ, Metelo AM, Perez-Carro R, Baker R, Wang ZW, Arreola A, Rathmell WK, Olumi A, Lopez-Larrubia P, et al. (2013). In Vivo HIF-Mediated Reductive Carboxylation Is Regulated by Citrate Levels and Sensitizes VHL-Deficient Cells to Glutamine Deprivation. Cell Metab 17, 372–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gantner ML, Eade K, Wallace M, Handzlik MK, Fallon R, Trombley J, Bonelli R, Giles S, Harkins-Perry S, Heeren TFC, et al. (2019). Serine and Lipid Metabolism in Macular Disease and Peripheral Neuropathy. The New England journal of medicine 381, 1422–1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia D, and Shaw RJ (2017). AMPK: Mechanisms of Cellular Energy Sensing and Restoration of Metabolic Balance. Molecular cell 66, 789–800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaude E, Schmidt C, Gammage PA, Dugourd A, Blacker T, Chew SP, Saez-Rodriguez J, O’Neill JS, Szabadkai G, Minczuk M, et al. (2018). NADH Shuttling Couples Cytosolic Reductive Carboxylation of Glutamine with Glycolysis in Cells with Mitochondrial Dysfunction. Mol Cell 69, 581–593 e587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T, Velic A, Macek B, Lundberg E, Kampf C, Nagaraj N, Uhlen M, Cox J, and Mann M. (2013). Initial quantitative proteomic map of 28 mouse tissues using the SILAC mouse. Molecular & cellular proteomics : MCP 12, 1709–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregory JF 3rd, Cuskelly GJ, Shane B, Toth JP, Baumgartner TG, and Stacpoole PW (2000). Primed, constant infusion with [2H3]serine allows in vivo kinetic measurement of serine turnover, homocysteine remethylation, and transsulfuration processes in human one-carbon metabolism. Am J Clin Nutr 72, 1535–1541. [DOI] [PubMed] [Google Scholar]
- Guarente L. (2008). Mitochondria--a nexus for aging, calorie restriction, and sirtuins? Cell 132, 171–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heng JS, Rattner A, Stein-O’Brien GL, Winer BL, Jones BW, Vernon HJ, Goff LA, and Nathans J. (2019). Hypoxia tolerance in the Norrin-deficient retina and the chronically hypoxic brain studied at single-cell resolution. P Natl Acad Sci USA 116, 9103–9114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbig K, Chiang EP, Lee LR, Hills J, Shane B, and Stover PJ (2002). Cytoplasmic serine hydroxymethyltransferase mediates competition between folate-dependent deoxyribonucleotide and S-adenosylmethionine biosyntheses. J Biol Chem 277, 38381–38389. [DOI] [PubMed] [Google Scholar]
- Hui S, Ghergurovich JM, Morscher RJ, Jang C, Teng X, Lu W, Esparza LA, Reya T, Le Z, Yanxiang Guo J, et al. (2017). Glucose feeds the TCA cycle via circulating lactate. Nature 551, 115–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter RW, Hughey CC, Lantier L, Sundelin EI, Peggie M, Zeqiraj E, Sicheri F, Jessen N, Wasserman DH, and Sakamoto K. (2018). Metformin reduces liver glucose production by inhibition of fructose-1–6-bisphosphatase. Nat Med 24, 1395-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain IH, Zazzeron L, Goldberger O, Marutani E, Wojtkiewicz GR, Ast T, Wang H, Schleifer G, Stepanova A, Brepoels K, et al. (2019). Leigh Syndrome Mouse Model Can Be Rescued by Interventions that Normalize Brain Hyperoxia, but Not HIF Activation. Cell Metab 30, 824-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain IH, Zazzeron L, Goli R, Alexa K, Schatzman-Bone S, Dhillon H, Goldberger O, Peng J, Shalem O, Sanjana NE, et al. (2016). Hypoxia as a therapy for mitochondrial disease. Science 352, 54–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, Yanos ME, Kayser EB, Quintana A, Sangesland M, Castanza A, Uhde L, Hui J, Wall VZ, Gagnidze A, et al. (2013). mTOR inhibition alleviates mitochondrial disease in a mouse model of Leigh syndrome. Science 342, 1524–1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalucka J, Bierhansl L, Conchinha NV, Missiaen R, Elia I, Bruning U, Scheinok S, Treps L, Cantelmo AR, Dubois C, et al. (2018). Quiescent Endothelial Cells Upregulate Fatty Acid (beta-Oxidation for Vasculoprotection via Redox Homeostasis. Cell metabolism 28, 881-+. [DOI] [PubMed] [Google Scholar]
- Kerr DS (2013). Review of clinical trials for mitochondrial disorders: 1997–2012. Neurotherapeutics 10, 307–319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan NA, Nikkanen J, Yatsuga S, Jackson C, Wang LY, Pradhan S, Kivela R, Pessia A, Velagapudi V, and Suomalainen A. (2017). mTORC1 Regulates Mitochondrial Integrated Stress Response and Mitochondrial Myopathy Progression. Cell metabolism 26, 419-+. [DOI] [PubMed] [Google Scholar]
- Kremer JM (1994). The mechanism of action of methotrexate in rheumatoid arthritis: the search continues. The Journal of rheumatology 21, 1–5. [PubMed] [Google Scholar]
- Liu L, Shah S, Fan J, Park JO, Wellen KE, and Rabinowitz JD (2016a). Malic enzyme tracers reveal hypoxia-induced switch in adipocyte NADPH pathway usage. Nature chemical biology 12, 345–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XJ, Romero IL, Litchfield LM, Lengyel E, and Locasale JW (2016b). Metformin Targets Central Carbon Metabolism and Reveals Mitochondrial Requirements in Human Cancers. Cell metabolism 24, 728–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundsgaard AM, Fritzen AM, and Kiens B. (2018). Molecular Regulation of Fatty Acid Oxidation in Skeletal Muscle during Aerobic Exercise. Trends Endocrin Met 29, 18–30. [DOI] [PubMed] [Google Scholar]
- Lunt SY, and Vander Heiden MG (2011). Aerobic Glycolysis: Meeting the Metabolic Requirements of Cell Proliferation. Annu Rev Cell Dev Bi 27, 441–464. [DOI] [PubMed] [Google Scholar]
- Madiraju AK, Erion DM, Rahimi Y, Zhang XM, Braddock DT, Albright RA, Prigaro BJ, Wood JL, Bhanot S, MacDonald MJ, et al. (2014). Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature 510, 542-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madiraju AK, Qiu Y, Perry RJ, Rahimi Y, Zhang XM, Zhang DY, Camporez JPG, Cline GW, Butrico GM, Kemp BE, et al. (2018). Metformin inhibits gluconeogenesis via a redox-dependent mechanism in vivo. Nat Med 24, 1384-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Reyes I, and Chandel NS (2014). Mitochondrial One-Carbon Metabolism Maintains Redox Balance during Hypoxia. Cancer discovery 4, 1371–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna MC, Waagepetersen HS, Schousboe A, and Sonnewald U. (2006). Neuronal and astrocytic shuttle mechanisms for cytosolic-mitochondrial transfer of reducing equivalents: Current evidence and pharmacological tools. Biochem Pharmacol 71, 399–407. [DOI] [PubMed] [Google Scholar]
- Millard P, Letisse F, Sokol S, and Portais JC (2012). IsoCor: correcting MS data in isotope labeling experiments. Bioinformatics 28, 1294–1296. [DOI] [PubMed] [Google Scholar]
- Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang YF, Linehan WM, Chandel NS, and DeBerardinis RJ (2012). Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature 481, 385–U171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama K, Shimura M, Liu Z, Okazaki Y, and Ohtake A. (2019). Recent topics: the diagnosis, molecular genesis, and treatment of mitochondrial diseases. J Hum Genet 64, 113–125. [DOI] [PubMed] [Google Scholar]
- Neinast MD, Jang C, Hui S, Murashige DS, Chu Q, Morscher RJ, Li X, Zhan L, White E, Anthony TG, et al. (2019). Quantitative Analysis of the Whole-Body Metabolic Fate of Branched-Chain Amino Acids. Cell metabolism 29, 417–429 e414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikkanen J, Forsstrom S, Euro L, Paetau I, Kohnz RA, Wang LY, Chilov D, Viinamaki J, Roivainen A, Marjamaki P, et al. (2016). Mitochondrial DNA Replication Defects Disturb Cellular dNTP Pools and Remodel One-Carbon Metabolism. Cell Metab 23, 635–648. [DOI] [PubMed] [Google Scholar]
- Owen MR, Doran E, and Halestrap AP (2000). Evidence that metformin exerts its anti-diabetic effects through inhibition of complex 1 of the mitochondrial respiratory chain. Biochem J 348, 607–614. [PMC free article] [PubMed] [Google Scholar]
- Owen OE, Kalhan SC, and Hanson RW (2002). The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem 277, 30409–30412. [DOI] [PubMed] [Google Scholar]
- Papandreou I, Cairns RA, Fontana L, Lim AL, and Denko NC (2006). HIF-1 mediates adaptation to hypoxia by actively downregulating mitochondrial oxygen consumption. Cell Metab 3, 187–197. [DOI] [PubMed] [Google Scholar]
- Pfeffer G, Majamaa K, Turnbull DM, Thorburn D, and Chinnery PF (2012). Treatment for mitochondrial disorders. Cochrane Database Syst Rev, CD004426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Kimura N, Oda N, Shimomura H, Kumada T, Miyajima T, Murayama K, Tanaka M, and Fujii T. (2012). Pyruvate therapy for mitochondrial DNA depletion syndrome. Bba-Gen Subjects 1820, 632–636. [DOI] [PubMed] [Google Scholar]
- Samanta D, Park Y, Andrabi SA, Shelton LM, Gilkes DM, and Semenza GL (2016). PHGDH Expression Is Required for Mitochondrial Redox Homeostasis, Breast Cancer Stem Cell Maintenance, and Lung Metastasis. Cancer Res 76, 4430–4442. [DOI] [PubMed] [Google Scholar]
- Schmidt A, Wu HP, MacKenzie RE, Chen VJ, Bewly JR, Ray JE, Toth JE, and Cygler M. (2000). Structures of three inhibitor complexes provide insight into the reaction mechanism of the human methylenetetrahydrofolate dehydrogenase/cyclohydrolase. Biochemistry-Us 39, 6325–6335. [DOI] [PubMed] [Google Scholar]
- Solaini G, Baracca A, Lenaz G, and Sgarbi G. (2010). Hypoxia and mitochondrial oxidative metabolism. Biochimica et biophysica acta 1797, 1171–1177. [DOI] [PubMed] [Google Scholar]
- Su X, Lu W, and Rabinowitz JD (2017). Metabolite Spectral Accuracy on Orbitraps. Analytical chemistry 89, 5940–5948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Gui DY, Hosios AM, Bush LN, Freinkman E, and Vander Heiden MG (2015). Supporting Aspartate Biosynthesis Is an Essential Function of Respiration in Proliferating Cells. Cell 162, 552–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan LB, Luengo A, Danai LV, Bush LN, Diehl FF, Hosios AM, Lau AN, Elmiligy S, Malstrom S, Lewis CA, et al. (2018). Aspartate is an endogenous metabolic limitation for tumour growth. Nat Cell Biol 20, 782-+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suomalainen A, and Battersby BJ (2018). Mitochondrial diseases: the contribution of organelle stress responses to pathology. Nat Rev Mol Cell Biol 19, 77–92. [DOI] [PubMed] [Google Scholar]
- Titov DV, Cracan V, Goodman RP, Peng J, Grabarek Z, and Mootha VK (2016). Complementation of mitochondrial electron transport chain by manipulation of the NAD(+)/NADH ratio. Science 352, 231–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyynismaa H, Carroll CJ, Raimundo N, Ahola-Erkkila S, Wenz T, Ruhanen H, Guse K, Hemminki A, Peltola-Mjosund KE, Tulkki V, et al. (2010). Mitochondrial myopathy induces a starvation-like response. Hum Mol Genet 19, 3948–3958. [DOI] [PubMed] [Google Scholar]
- Weinblatt ME, Coblyn JS, Fox DA, Fraser PA, Holdsworth DE, Glass DN, and Trentham DE (1985). Efficacy of low-dose methotrexate in rheumatoid arthritis. The New England journal of medicine 312, 818–822. [DOI] [PubMed] [Google Scholar]
- Wheaton WW, Weinberg SE, Hamanaka RB, Soberanes S, Sullivan LB, Anso E, Glasauer A, Dufour E, Mutlu GM, Budigner GRS, et al. (2014). Metformin inhibits mitochondrial complex I of cancer cells to reduce tumorigenesis. Elife 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiberg KB (1955). The Deuterium Isotope Effect. Chem Rev 55, 713–743. [Google Scholar]
- Wise DR, Ward PS, Shay JES, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, and Thompson CB (2011). Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. P Natl Acad Sci USA 108, 19611–19616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang CD, Ko B, Hensley CT, Jiang L, Wasti AT, Kim J, Sudderth J, Calvaruso MA, Lumata L, Mitsche M, et al. (2014). Glutamine Oxidation Maintains the TCA Cycle and Cell Survival during Impaired Mitochondrial Pyruvate Transport. Mol Cell 56, 414–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaniv Y, Juhaszova M, and Sollott SJ (2013). Age-related changes of myocardial ATP supply and demand mechanisms. Trends Endocrinol Metab 24, 495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Fan J, Venneti S, Wan YW, Pawel BR, Zhang J, Finley LW, Lu C, Lindsten T, Cross JR, et al. (2014). Serine catabolism regulates mitochondrial redox control during hypoxia. Cancer discovery 4, 1406–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H, Antoniewicz MR, Stephanopoulos G, and Kelleher JK (2008). Quantifying reductive carboxylation flux of glutamine to lipid in a brown adipocyte cell line. J Biol Chem 283, 20621–20627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Chen L, Liu L, Su X, and Rabinowitz JD (2017). Chemical Basis for Deuterium Labeling of Fat and NADPH. Journal of the American Chemical Society 139, 14368–14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng XD, Boyer L, Jin MJ, Kim Y, Fan WW, Bardy C, Berggren T, Evans RM, Gage FH, and Hunter T. (2016). Alleviation of neuronal energy deficiency by mTOR inhibition as a treatment for mitochondria-related neurodegeneration. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
LC-MS files have been deposited at Mendeley data: http://dx.doi.org/10.17632/k27cy7fwh8.3