Abstract
Cholesteryl ester transfer protein (CETP) facilitates the net transfer of cholesteryl esters (CEs) and TGs between lipoproteins, impacting the metabolic fate of these lipoproteins. Previous studies have shown that a CETP antibody can alter CETP’s preference for CE versus TG as transfer substrate, suggesting that CETP substrate preference can be manipulated in vivo. Hamster and human CETPs have very different preferences for CE and TG. To assess the effect of altering CETP’s substrate preference on lipoproteins in vivo, here, we expressed human CETP in hamsters. Chow-fed hamsters received adenoviruses expressing no CETP, hamster CETP, or human CETP. Plasma CETP mass increased 2-fold in both the hamster and human CETP groups. Although the animals expressing human CETP still had low levels of hamster CETP, the CE versus TG preference of their plasma CETP was similar to that of the human ortholog. Hamster CETP overexpression had little impact on lipoproteins. However, expression of human CETP reduced HDL up to 50% and increased VLDL cholesterol 2.5-fold. LDL contained 20% more CE, whereas HDL CE was reduced 40%, and TG increased 6-fold. The HDL3:HDL2 ratio increased from 0.32 to 0.60. Hepatic expression of three cholesterol-related genes (LDLR, SCARB1, and CYP7A1) was reduced up to 40%. However, HDL-associated CE excretion into feces was unchanged. We conclude that expression of human CETP in hamsters humanizes their lipoprotein profile with respect to the relative concentrations of VLDL, LDL, HDL, and the HDL3:HDL2 ratio. Altering the lipid substrate preference of CETP provides a novel approach for modifying plasma lipoproteins.
Keywords: cholesteryl ester transfer protein, lipid preference, cholesterol efflux, proteome, gene expression, reverse cholesterol transport, triglyceride, lipid metabolism, high density lipoprotein
Cholesteryl ester transfer protein (CETP) mediates the transfer of lipids between VLDL, LDL, and HDL lipoproteins. From a metabolic standpoint, perhaps the most important feature of CETP is its ability to facilitate the net movement of cholesteryl ester (CE) from one lipoprotein to another in exchange for TG (1). Through this mechanism, CETP alters the composition of lipoproteins and, consequently, influences their metabolism (2–6).
Reverse cholesterol transport (RCT) is the mechanism whereby cholesterol in nonhepatic cells is transported to the liver for excretion (7). HDL plays a central role in this pathway, serving as a platform for both removing cholesterol from cells and the conversion of this cholesterol to CE by LCAT (7). In humans, much of this CE does not remain in the HDL particle, but is transferred to VLDL and LDL by CETP (8). As a result of this transfer, up to two-thirds of cellular cholesterol that is delivered to the liver for excretion occurs through VLDL and LDL instead of HDL (9, 10).
Given its central role in human lipoprotein metabolism, CETP is a likely target for manipulating plasma lipid metabolism. Inhibiting CETP dramatically increases HDL cholesterol levels, but this has rarely shown clinical benefit (11). As a result, there is a growing consensus that the flux of cholesterol through the HDL compartment, not the absolute concentration of HDL, is most important for its protective role in cardiovascular disease (12). Blocking CETP impairs this flux causing CE, which mostly originates on HDL, to remain there. We previously suggested a novel therapeutic approach that focuses on harnessing the power of CETP to move lipids and influence lipoprotein metabolism (13). This approach capitalizes on the findings that the specificity of CETP for CE or TG as transfer substrate can be altered by small changes in its structure and by compounds that bind to CETP (13, 14). In vitro, modifying this preference altered the ability of CETP to facilitate net CE and TG transfer between VLDL and HDL. The overall objective of the studies described here was to explore the potential therapeutic value of modifying CETP’s substrate preference in vivo. These studies test the consequence of changing the CE versus TG preference of circulating CETP in an animal model where CETP is naturally expressed. This was achieved by expressing human CETP in hamsters. This approach takes advantage of the naturally higher preference of human CETP for CE, whereas hamster CETP prefers TG as a transfer substrate (15). The impact of altering this substrate preference on the levels and physicochemical properties of plasma lipoproteins and on the excretion of cholesterol into feces were measured. The findings illustrate the potential for CETP substrate preference modification as a viable approach for altering plasma lipoproteins.
MATERIALS AND METHODS
Isolation and radiolabeling of lipoproteins
Human and hamster plasma lipoproteins were isolated by sequential ultracentrifugation (16). In some instances, human lipoproteins were doubly labeled with 3H-TG and 14C-CE by a dispersion method (17) before their isolation from plasma. For RCT assays, HDL was labeled with 3H-CE as previously described (18). Alternatively, to radiolabel VLDL, LDL, and HDL with 3H-CE, HDL was initially radiolabeled to high specific activity by CETP-mediated transfer of 3H-CE from 3H-liposomes, followed by reisolation of HDL by ultracentrifugation (19). Subsequently, VLDL, LDL, or HDL were incubated with CETP and a small quantity of 3H-CE HDL (<1.5% of unlabeled lipoprotein) (13). 3H-CE-labeled VLDL, LDL, and HDL were then re-isolated within their original density limits. 3H-CE {[1,2 -3H(N)]cholesteryl oleate}, 14C-CE (cholesteryl-[1-14C]oleate), and 3H-TG {[9,10-3H(N)]triolein} were purchased from PerkinElmer, Inc. (Waltham, MA).
Adenovirus constructs
A vector containing human CETP cDNA (M30185.1) was purchased from Open Biosystems (Pittsburgh, PA). Human CETP cDNA with a single amino acid mutation (H232A) and the cDNA for golden Syrian hamster CETP (Mesocricetus auratus (XM_005078609.2) were synthesized by Gen-Script (Piscataway, NJ). All constructs contain native start and stop codons plus the coding sequence for the 17 amino acid signal peptide. Recombinant E1/E3-deleted adenovirus (serotype 5) constructs containing the CMV promoter alone (Ad-null), hamster CETP (Ad-haCETP), human CETP (Ad-huCETP), or H232A human CETP (Ad-H232A) were custom synthesized by Vector Biolabs (Malvern, PA) using these CETP open reading frames.
In vitro CE mass transfer assays
For CE transfer assays, CETPs were obtained by transforming HepG2 cells with adenoviral constructs expressing hamster CETP, human CETP, or human H232A CETP. Conditioned media from cells grown in Opti-MEM (Life Technologies Corp., Grand Island, NY) for 48 h were used as sources of these CETPs. VLDL, LDL, and HDL were combined in a cholesterol ratio of 1 to 4.1 to 2.3, respectively, approximating the ratio of these lipoproteins in normal human plasma. For each transfer assay, one of these lipoproteins was radiolabeled with 3H-CE. This permitted measurement of CE transfer between all six donor lipoprotein-acceptor lipoprotein pairs in this mixture. Lipoproteins and CETP were combined in Tris-buffered saline containing 1% BSA and incubated at 37°C (17). At 1.5 and 3 h time points, aliquots were placed on ice and combined with 4-fold excess unlabeled lipoproteins. Assay blanks (t = 0) lacked CETP. Triplicate samples were adjusted with NaBr to a density of 1.019 or 1.063 g/ml, and then lipoproteins were fractionated by ultracentrifugation (16). This yielded, VLDL and LDL+HDL fractions (d = 1.019) and VLDL+LDL and HDL fractions (d = 1.063). 3H in each fraction was quantified by liquid scintillation counting. This determined VLDL and HDL radioactivity directly, whereas LDL radioactivity was calculated [LDL = (VLDL + LDL) − VLDL]. The fraction of 3H-CE transferred to a given lipoprotein was multiplied times the CE mass in the donor lipoprotein to determine the mass of CE transferred. Lipid transfers were linear over the 3 h time course for each donor/acceptor pair. In order to compare CETPs with different CE transfer potential, reported lipid transfer values were normalized so that the sum of CE transfers between the six donor/acceptor pairs was the same.
Animals
Male golden Syrian hamsters (101–110 g, ∼7 weeks old) were purchased from Charles River Laboratories (Wilmington, MA). A jugular vein catheter was surgically placed and exteriorized dorsally. Following catheter placement, animals were individually housed. Hamsters were allowed to recover for approximately 1 week before the start of a study. For adenovirus injection, jugular vein catheters were flushed with sterile saline followed by injection of in vivo quality CsCl-gradient purified adenovirus (2–5 × 109 pfu), a second saline flush, and then heparin/glycerol catheter lock solution (Braintree Scientific, Inc., Braintree, MA). This route of administration targets the adenovirus to the liver (20). Chow diet (Envigo, Madison, WI) was provided ad libitum for the duration of the study. On day 4, the jugular vein catheters were flushed with saline, 3H-CE HDL (300 μl containing ∼125 μg protein, 2 μCi 3H) injected, followed by a second saline flush and catheter lock solution. Animals were transferred to cages with wire-bottom platforms to facilitate complete feces collection. After 48 h, blood was collected, and animals were euthanized. A segment of fresh liver was minced and homogenized (100 mg liver per milliliter of water) with a Tissue-Tearor (BioSpec Products, Bartlesville, OK). The remaining portion of liver was immediately snap-frozen in liquid N2 and stored at −80°C for future analysis. Feces were dried overnight at 55°C, pulverized, suspended in 50% ethanol (100 mg feces per milliliter), and homogenized as above. The 3H content of liver and fecal homogenates was determined by liquid scintillation counting of triplicate aliquots. Samples were counted a second time after the addition of a 3H internal standard to quantify quenching caused by sample color. Original sample 3H values were then corrected for differences in counting efficiency. Plasma 3H was determined by direct scintillation counting of whole plasma. Total plasma 3H calculations assumed a plasma volume of 3.5% of body weight. The Cleveland Clinic’s Institutional Animal Care and Use Committee approved all animal studies.
Quantification of CETP in hamster plasma
The assay conditions below were established following optimization of the quantity the TP2 antibody and the length of its incubation with plasma, and the quantity of immobilized anti mouse IgG and the time it was incubated with TP2-CETP complexes. Hamster plasma was diluted 10-fold with PBS. Diluted plasma (30 μl) was combined with 300 ng TP2 anti-CETP antibody (Ottawa Heart Institute, Ottawa, Ontario, Canada) and incubated at room temperature for 1 h. Fifty microliters of M-280 sheep anti mouse IgG magnetic Dynabeads (Invitrogen, Carlsbad, CA) were washed twice with PBS containing 0.1% BSA, and then resuspended in 1 ml of the same buffer. Plasma incubated with TP2 was then combined with the washed Dynabead suspension and mixed end-over-end for 1 h at room temperature. Incubated beads were washed twice with 1 ml PBS. After the second wash, residual buffer was carefully removed, and then 50 μl of 1× SDS-PAGE buffer [62.5 mM Tris-HCl (pH 8.3), 2% sodium dodecyl sulfate, 10% glycerol, 5% β-mercaptoethanol, and 0.5 mg/ml bromophenol blue] were added and samples heated at 100°C for 5 min. Beads were removed by a magnet and the supernatant containing CETP was collected. Samples were fractionated by SDS-PAGE on 8% gels (Invitrogen). To adequately resolve hamster and human CETP, gels were electrophoresed until the 53k Da molecular weight standard (Lonza, Rockland, ME) was near the bottom of the gel. Proteins were transferred to PVDF, and CETP was detected with TP2 antibody followed by a goat anti-mouse IgG HRP secondary antibody (21). Bands were visualized by Western Lightning Plus ECL reagent (Perkin-Elmer Life Sciences). Chemiluminescence was captured on a digital imager (GE Healthcare, Marlborough MA) and quantified by ImageJ (https://imagej.nih.gov/ij/).
For CETP quantitation, plasma from three hamsters that developed high levels of plasma CETP in response to Ad-haCETP were pooled and stored at −20°C. This standard was serially diluted and immunoprecipitated as described above to create a four point standard curve that was run on every gel along with unknowns. Relative CETP concentrations determined from this standard curve were then normalized to the average CETP value for Ad-null animals.
Plasma CETP activity
Hamster plasma (10 μl) was combined with 100 μg 3H-TG, 14C-CE- labeled human LDL, 100 μg unlabeled human HDL, and Tris-buffered 1% BSA in 0.7 ml final volume as previously described (22). After 5 h at 37°C, lipid transfer was stopped by the addition of 200 μl of 0.45 M sodium phosphate (pH 7.4) and 300 μl of 0.1 M MnCl2. After centrifugation to pellet the VLDL/LDL precipitate, radiolabel in the HDL-containing supernatant was determined by scintillation counting. Transfer was calculated as previously described (23).
mRNA qPCR
Liver tissues were homogenized by a Tissuelyser II (Qiagen, Germantown, MD). Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA). First-strand cDNAs were synthesized using random primers and reverse transcriptase (Promega, Madison, WI). qPCR was performed using Power SYBR™ Green PCR Master Mix (Thermo Fisher Scientific, Waltham, MA) and a StepOnePlus RT-PCR system (Life Technologies Corp.). qPCR primers are listed in supplemental Table S1. mRNA values were normalized to ACTB. Gene expression was calculated using the 2−ΔΔCT method (24) and reported relative to control cells.
Cholesterol efflux assay
RAW264.7 mouse macrophages (TIB-71, American Type Culture Collection, Manassas, VA) were labeled overnight with 3H-cholesterol in DMEM + 0.2% BSA. Labeled cells were then incubated ±0.3 mM 8-bromo cAMP (Sigma-Aldrich, St. Louis, MO) in DMEM + 0.2% BSA to upregulate ABCA1 expression. The following day, cells were washed and incubated in the same medium plus 2.8% ApoB-depleted serum (= 2% of original serum) (25). After 4 h, medium was removed and centrifuged to remove cell debris. Cells were solubilized with RIPA buffer. 3H in both fractions was quantified by scintillation counting. 3H-cholesterol efflux was calculated as the percent of total counts per minute recovered in the media. ABCA1-dependent efflux was determined from the difference between total efflux (plus 8-bromo cAMP) and ABCA1-independent efflux (minus 8-bromo cAMP) cells.
MS
HDL proteins were reduced with DTT and alkylated with iodoacetamide prior to trypsin digestion. Samples were desalted (PepClean C18 spin column; Thermo Fisher Scientific), evaporated, and reconstituted in 1% acetic acid for LC/MS analysis on a Thermo Fisher LTQ-Obitrap Elite hybrid mass spectrometer system. The HPLC column was a Dionex 15 cm × 75 μm id Acclaim Pepmap C18, 2 μm, 100 Å reversed phase capillary chromatography column. Peptides eluted from the column by an acetonitrile/0.1% formic acid gradient at a flow rate of 0.3 μl/min were introduced into the mass spectrometer on-line.
Data were analyzed using the Proteome Discoverer V2.3 software package and MS/MS data were searched using the Sequest component of this software and the UniProtKB Unreviewed (TrEMBL) protein sequence database of golden hamster [10036] containing 31,951 entries. The MS/MS spectra were used for peptide sequencing and protein identification, and the full scans were used for peptide precursor intensity calculations. Two missed and/or nonspecific cleavages were permitted. Carbamidomethylation of C and oxidation of M were considered. Mass tolerance for precursor ions was 10 ppm and tolerance for fragment ions was 0.8 Da. The estimated false discovery rate was <1% based on a decoy database search. The threshold score value for accepting individual spectra was 1% false discovery rate. Single-peptide identification of proteins was not used. Relative protein abundance was determined using the Minora Feature Detector node of Proteome Discoverer and normalized by the total peptide amount in each sample. The peptides used in this quantitation corresponded to unique + razor.
Other analytical methods
Plasma lipoproteins were fractionated by tandem Superose 6 columns as previously described (22, 26). The column eluate was continuously combined with Infinity cholesterol detection reagent (Thermo Fisher Scientific) and the reaction product was monitored at 505 nm. VLDL, LDL, and HDL peaks were identified based on the elution profile of hamster lipoproteins isolated by ultracentrifugation.
Protein was measured by a modification of the Lowry et al. method (27) with BSA as standard. Total cholesterol (TC), free cholesterol (FC), and TG were quantified by enzyme-based kits from Thermo Fisher Scientific (TC, TG) or from Wako Diagnostics Inc., Mountain View, CA (FC). CE was calculated as TC minus FC times 1.69 to adjust for the fatty acid contained in this molecule. Phospholipid phosphorus was determined chemically by the method of Bartlett (28). HDL particle size distribution was determined by native gradient gel electrophoresis (29, 30).
To quantify tissue cholesterol, liver homogenates were saponified with 2% ethanolic KOH at 60°C for 1 h, and then lipids were extracted (31). Following removal of solvent under N2, cholesterol was measured by a ferric chloride method (32). To measure liver TG, homogenate lipids were extracted (31), solvent removed, and lipids solubilized with 2.5% Triton X-100. TG was quantified by an enzymatic kit (Thermo Fisher Scientific).
Statistical analysis was performed by unpaired t-test (Instat 3, GraphPad Software, San Diego, CA). P values <0.05 were considered statistically significant. Group sizes are indicated in the tables and figures.
RESULTS
Impact of CETP substrate preference on CE net transfer in vitro
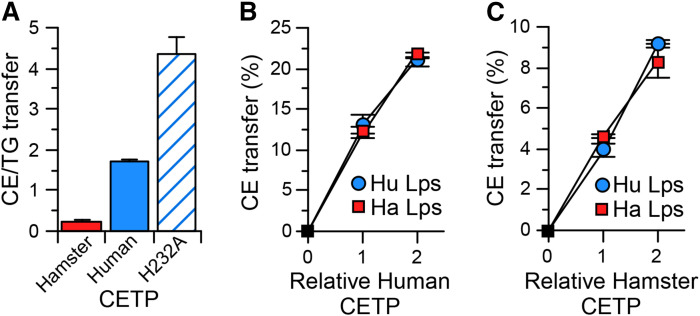
We previously observed that hamster CETP, unlike human CETP, prefers TG over CE as a substrate (15). This difference in substrate preference altered the net transfer of CE and TG between isolated HDL and VLDL (13, 15). To assess how differences in CETP substrate preference might impact the overall distribution of CE between plasma lipoproteins, we measured the CETP-mediated net transfer of CE between a plasma-like mixture of VLDL, LDL, and HDL. For these studies, recombinant hamster CETP, human CETP, and the H232A mutant of human CETP, which has a very high preference for CE as a substrate (13, 14), were expressed in HepG2 cells. This provided CETPs of widely differing CE versus TG substrate preference (Fig. 1A). Rates of CE mass transfer between each pair of lipoproteins within this mixture were measured. To facilitate comparisons, transfers were normalized so that the sum of CE transfers between all lipoproteins was the same for each CETP. CETP substrate preference had no detectable impact on CE transfer between LDL and HDL (Table 1). Compared with native human CETP’s activity with these human lipoproteins, hamster CETP promoted greater net transfer of CE from LDL and HDL into VLDL, whereas H232A human CETP promoted lower CE transfer to VLDL (Table 2). CETP preference had a relatively small impact on the relative contribution of HDL versus LDL as CE donors to VLDL. Overall, these data show that CETP substrate preference directly impacts the net transfer of CE from CE-rich lipoproteins to TG-rich lipoproteins.
Fig. 1.
CETP transfer properties. A: The ratio of CE transfer activity to TG transfer activity mediated by the indicated CETP. H232A is the H232A mutation of human CETP. Values are mean ± SD, n = 4. The capacity of human (B) or hamster (C) CETP to facilitate CE transfer from LDL to HDL was measured using equal amounts of lipoproteins (Lps) isolated from human (Hu) and from hamster (Ha) plasma. Values are mean ± SD, n = 3.
TABLE 1.
CE transfer between lipoproteins by different CETPs: unidirectional transfer
| CETP | CE Transfer from/to the Indicated Lipoprotein Pair | |||||
| VLDL to LDL | LDL to VLDL | VLDL to HDL | HDL to VLDL | LDL to HDL | HDL to LDL | |
| Hamster | 23 ± 4 | 461 ± 16 | 59 ± 5 | 1,153 ± 16 | 659 ± 22 | 659 ± 22 |
| Human | 37 ± 7a | 392 ± 22a | 128 ± 4a | 1,147 ± 17 | 676 ± 93 | 646 ± 37 |
| H232A | 53 ± 10a,b | 379 ± 16a | 173 ± 5a,b | 1,116 ± 27 | 615 ± 26 | 677 ± 50 |
VLDL, LDL, and HDL were combined in a physiologic ratio and incubated with the indicated CETP. To measure a given unidirectional transfer, the indicated donor lipoprotein was labeled with 3H-CE. Shown are the rates of CE transfer from the indicated donor to the indicated acceptor. H232A is a mutated form of human CETP. The values shown are in nanograms of CE per hour. Mean ± SD, n = 6.
P < 0.01 versus hamster.
P < 0.01 versus. human.
TABLE 2.
CE transfer between lipoproteins by different CETPs: net mass transfer
| CETP | Net CE Transfer | |||
| LDL to VLDL | HDL to VLDL | Total Net CE to VLDL | HDL to VLDL/LDL to VLDL | |
| Hamster | 439 ± 16 | 1,094 ± 17 | 1,533 ± 40 | 2.49 ± 0.10 |
| Human | 354 ± 23a | 1,019 ± 17a | 1,373 ± 55a | 2.88 ± 0.19a |
| H232A | 327 ± 19a | 943 ± 28a,b | 1,270 ± 56a,b | 2.89 ± 0.19a |
VLDL, LDL, and HDL were combined in a physiologic ratio and incubated with the indicated CETP. Shown are the rates of net CE transfer from the indicated donor to the indicated acceptor mediated by CETP. CE net transfer was calculated from the difference between unidirectional transfers reported in Table 1 (for example, LDL to VLDL minus VLDL to LDL). H232A is a mutated form of human CETP. The values shown are in nanograms of CE per hour. Mean ± SD, n = 6.
P < 0.01 versus hamster.
P < 0.01 versus. human.
Expression of human CETP in hamsters
To extend these findings, we examined the consequence of expressing human CETP in hamsters. The in vitro studies above compared these CETPs as they interacted with human lipoproteins. Therefore, we initially sought to verify in vitro that these transfer proteins interact similarly with human and hamster lipoproteins. CE transfer facilitated by human CETP was the same whether human or hamster lipoproteins were used in the assay (Fig. 1B). Likewise, hamster CETP activity was the same with human or hamster lipoproteins (Fig. 1C).
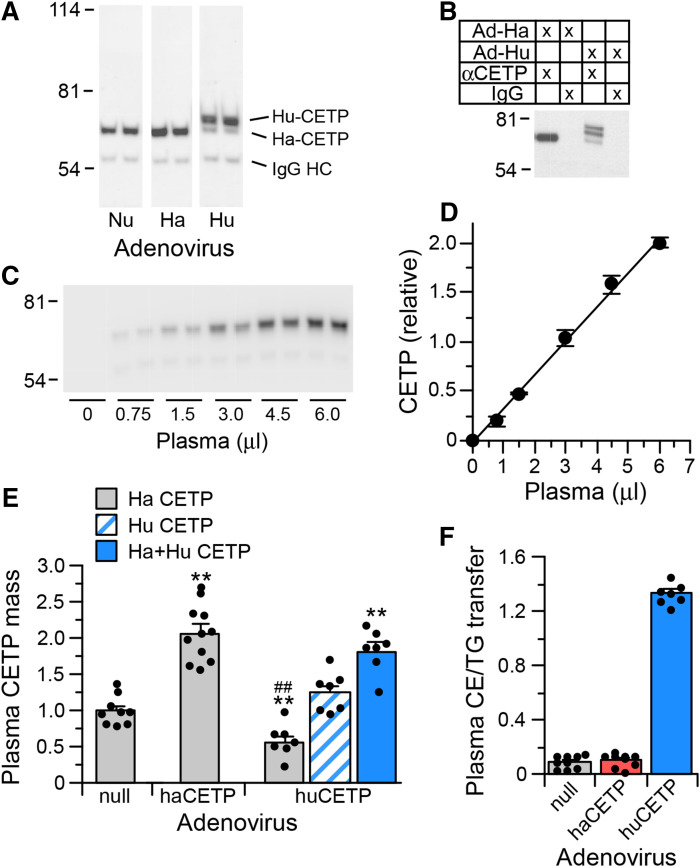
CETP expression in hamsters was achieved by recombinant adenovirus infection. Hamsters received either control adenovirus (Ad-null) or an adenovirus expressing either hamster CETP (Ad-haCETP) or human CETP (Ad-huCETP). To quantify hamster and human CETP in plasma, an assay was developed using the anti-human CETP TP2 antibody. The TP2 epitope is fully conserved in hamster CETP (13, 33). Hamster CETP has a smaller apparent molecular weight due to reduced carbohydrate content (15, 34). This difference facilitated separation of hamster and human CETP by SDS-PAGE. Immunoprecipitation of CETP in the plasma from Ad-haCETP and Ad-huCETP animals with the TP2 antibody isolated both CETPs (Fig. 2A). An IgG1 kappa isotype control antibody did not precipitate these proteins (Fig. 2B). As illustrated here, in some instances the electrophoretic conditions resolved human CETP into two bands, reflecting the two previously recognized glycosylation states of this protein (34). Immunoprecipitation conditions were established such that the amount of CETP precipitated was linearly related to plasma dosage (Fig. 2C, D). CETP mass values from this assay were highly correlated with TG transfer activity measured in vitro in Ad-null and Ad-haCETP plasmas (r = 0.93) and with CE transfer activity measured in Ad-huCETP plasmas (r = 0.95).
Fig. 2.
CETP mass assay. A: Western blot of CETP immunoprecipitated from the plasma of hamsters receiving Ad-null (Nu), Ad-haCETP (Ha), or Ad-huCETP (Hu) recombinant adenovirus. IgG HC, IgG heavy chain. B: CETP Western blot of hamster plasma immunoprecipitated with anti-CETP or an IgG isotype control immunoglobulin. C: Representative Western blot of CETP in the indicated amount of hamster plasma. D: CETP mass dose response curve of blot shown in C. Mean ± SD, n = 2. E: Relative CETP mass in plasma from hamsters injected with Ad-null (9), Ad-haCETP (11), or Ad-huCETP (7). Mean ± SEM of the indicated group sizes. F: Ratio of CE transfer activity to TG transfer activity mediated by CETP contained in the plasma of the indicated adenovirus groups. Mean ± SEM (n = same as in E). **P < 0.01 versus Ad-null; ##P < 0.01 versus Ad-haCETP.
With this Western-blot based assay, plasma CETP levels were determined to be increased 2-fold in Ad-haCETP animals (Fig. 2E). In Ad-huCETP animals, endogenous hamster CETP levels were reduced to half of Ad-null levels, and human CETP was the predominant plasma CETP form present (Fig. 2E). Importantly, total plasma CETP mass in Ad-haCETP and Ad-huCETP animals was the same. As a result of the decreased hamster CETP and expressed human CETP in the plasma of Ad-huCETP animals, the overall preference of plasma CETP for CE versus TG as a transfer substrate in these animals was markedly elevated compared with animals expressing only hamster CETP (Fig. 2F). This substrate preference is almost the same as that measured for human plasma CETP (CE/TG = 1.5). Therefore, Ad-haCETP and Ad-huCETP animals represent conditions where plasma CETP levels are equivalent but the functional properties of CETP in these animals reflect that of hamster and human plasma, respectively.
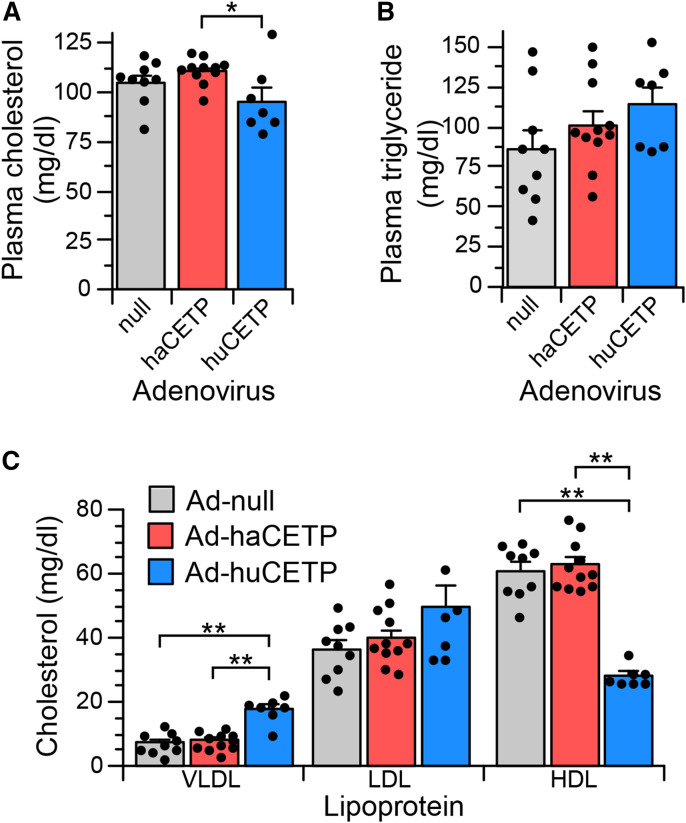
Effect of human CETP on hamster lipoproteins
Plasma cholesterol concentrations were modestly reduced in Ad-huCETP animals compared with Ad-haCETP animals but unchanged compared with Ad-null animals (Fig. 3A). Plasma TG levels were not different among study groups (Fig. 3B). The 2-fold overexpression of CETP by Ad-haCETP did not alter the distribution of cholesterol among lipoproteins compared with Ad-null controls (Fig. 3C). However, expression of human CETP had significant effects. In Ad-huCETP animals, plasma VLDL cholesterol increased 2.5-fold and HDL cholesterol decreased more than 2-fold, whereas LDL cholesterol levels were unchanged. Plasma VLDL, LDL, and HDL concentrations, quantified by their protein content, were also not different between Ad-haCETP and Ad-null groups. Similarly, plasma VLDL (13.9 ± 1.1 mg/dl) and LDL (46.3 ± 5.3 mg/dl) protein levels in Ad-huCETP animals were not significantly different from those in Ad-haCETP animals (9.5 ± 0.8 and 37.7 ± 2.2 mg/dl, respectively). However, plasma HDL protein levels in Ad-huCETP animals, like cholesterol, were reduced (145.9 ± 2.8 mg/dl, P < 0.01) compared with that in Ad-null and Ad-haCETP (186.2 ± 9.7 and 192.1 ± 6.0 mg/dl, respectively). Thus, a 2-fold increase in hamster CETP by Ad-haCETP adenovirus did not alter plasma lipoprotein cholesterol or protein concentrations, whereas expression of human CETP decreased HDL protein and cholesterol, but increased the cholesterol content of VLDL such that its cholesterol/protein ratio increased from 0.66 to 1.40.
Fig. 3.
Total plasma lipids and cholesterol distribution among lipoproteins in adenovirus-treated hamsters. A: Plasma cholesterol concentration. B: Plasma TG concentration. C: VLDL, LDL, and HDL cholesterol levels in the indicated groups. Values are mean ± SEM. See Fig. 2 for group sizes. *P < 0.05, **< P < 0.01.
Hamster CETP overexpression induced small changes in the CE and TG content of LDL with no apparent change in LDL size as estimated by surface/core calculations (Table 3). Expression of human CETP affected the relative levels of most lipids in LDL, resulting in a decrease in the surface to core ratio, indicative of larger LDL particles. For HDL, overexpression of hamster CETP only reduced FC (Table 3). Human CETP expression also reduced HDL FC, but additionally decreased CE levels by 40% and markedly increased its TG content, resulting in a 9-fold change in the CE/TG ratio of these particles. The calculated ratio of surface components to core components for Ad-huCETP HDL indicates they are significantly smaller.
TABLE 3.
Lipid composition of LDL and HDL
| Adenovirus | TC | FC | CE | TG | PL | S/C |
| μg / mg protein | ||||||
| LDL | ||||||
| Null | 994.5 ± 41.8 | 298.8 ± 23.6 | 1,175.8 ± 44.5 | 539.3 ± 51.5 | 920.7 ± 14.7 | 1.30 ± 0.04 |
| haCETP | 1,058.9 ± 28.4 | 277.0 ± 13.3 | 1,321.5 ± 43.5a | 382.5 ± 21.8b | 973.5 ± 31.2 | 1.33 ± 0.04 |
| huCETP | 1,054.6 ± 49.5 | 227.5 ± 16.0a,c | 1,397.9 ± 75.5a | 633.4 ± 81.9d | 1,108.2 ± 36.1b,c | 1.15 ± 0.02b,d |
| HDL | ||||||
| Null | 328.6 ± 5.3 | 38.8 ± 2.7 | 489.7 ± 10.7 | 3.1 ± 0.3 | 583.1 ± 21.2 | 3.17 ± 0.07 |
| haCETP | 326.5 ± 5.7 | 28.7 ± 2.2b | 503.2 ± 11.4 | 2.9 ± 0.4 | 578.0 ± 10.6 | 3.09 ± 0.05 |
| huCETP | 193.5 ± 7.3b,d | 15.4 ± 2.0 b,d | 301.1 ± 13.5b,d | 20.9 ± 3.4b,d | 520.7 ± 23.1 | 4.69 ± 0.15b,d |
The cholesterol (TC), FC, CE, TG, and phospholipid (PL) content of LDL and HDL were quantified. The ratio of components residing in the LDL surface (S) and core (C) was calculated as: (Protein + PL + FC)/(CE + TG). For HDL, this calculation assumes 40% of FC resides in the core (59). Mean ± SEM, n = 8 (null), 11 (haCETP), 7 (huCETP).
P < 0.05 versus Ad-null.
P < 0.01 versus Ad-null.
P < 0.05 versus Ad-haCETP.
P < 0.01 versus Ad-haCETP.
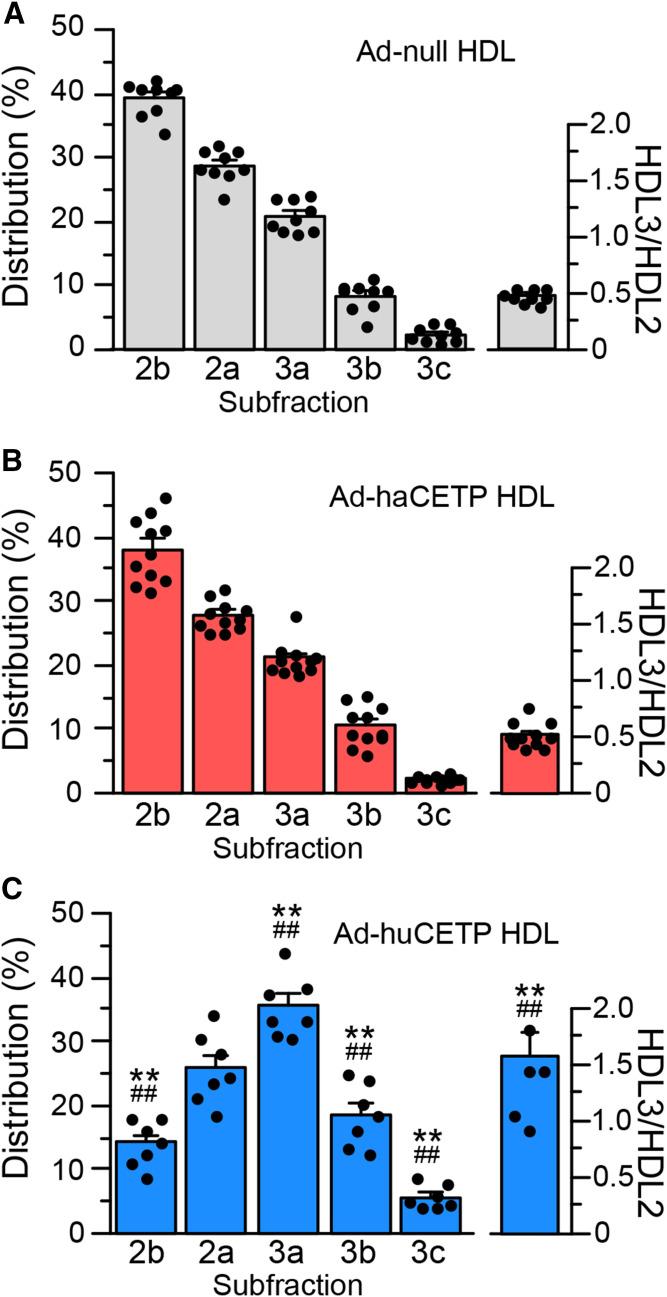
To investigate changes in HDL further, the size distribution of HDL was measured by native gradient gel electrophoresis. Consistent with the compositional analysis, overexpression of CETP in Ad-haCETP animals did not alter HDL size (Fig. 4A, B). In Ad-null and Ad-haCETP animals, the most abundant HDL subfraction was HDL2b, and total HDL2 particles exceeded HDL3 particles by 2-fold. In contrast, in Ad-huCETP animals, the HDL2b subclass was reduced to 35% of control levels and there was an overall shift in HDL particles toward smaller sizes (Fig. 4C). In these animals, the three HDL3 subfractions increased up to 2.3-fold, causing particles in the HDL3 subclass to become the predominant HDL species.
Fig. 4.
HDL particle size. HDL isolated from the indicated adenovirus group was fractionated by nondenaturing gradient gel electrophoresis. HDL protein was detected with Coomassie blue stain. A–C: HDL subfraction distribution. Values are mean ± SEM. See Fig. 2 for group sizes. **P < 0.01 versus Ad-null; ##P < 0.01 versus Ad-haCETP.
Altered CETP expression also modified the HDL proteome. Although hamster CETP overexpression did not measurably alter HDL size and minimally altered its lipid composition, levels of ApoD and LCAT were increased more than 2-fold (Table 4, supplemental Table S2). As expected, the CETP content of Ad-huCETP HDL was also elevated. By contrast, the reduced size of Ad-huCETP HDL was associated with large changes in apolipoprotein composition. With a 2-fold change as the cut-off for significance, Ad-huCETP HDL had reduced ApoA-IV, ApoC-II, ApoC-III, ApoC-IV, and ApoE content, but elevated ApoD (Table 4). Notably, the levels of ApoC-I and ApoF in HDL, known regulators of CETP (18, 35), were not changed by expression of either CETP species.
TABLE 4.
HDL proteome
| Relative Abundance | Protein | Fold Change (LFQ Ratio) | ||
| Ha/Nu | Hu/Nu | Hu/Ha | ||
| Apolipoproteins | ||||
| 1 | ApoA-I | — | — | — |
| 4 | ApoA-II | — | — | 0.67 |
| 5 | ApoA-IV | 1.90 | 0.18 | 0.10 |
| 9 | ApoC-I | — | — | — |
| 6 | ApoC-II | 1.36 | 0.41 | 0.30 |
| 3 | ApoC-III | — | 0.29 | 0.28 |
| 14 | ApoC-IV | — | 0.24 | 0.15 |
| 20 | ApoD | 2.41 | 2.26 | — |
| 2 | ApoE | — | 0.14 | 0.12 |
| 13 | ApoF | — | — | — |
| 22 | ApoH | — | 1.90 | — |
| 7 | ApoM | — | — | — |
| Enzymes/transfer proteins | ||||
| 17 | CETP | 2.37 | 0.43 | 0.18 |
| 24 | Glutathione peroxidase 3 | — | — | — |
| 18 | LCAT | 2.10 | 1.97 | — |
| 23 | Phospholipid transfer protein | — | — | — |
| 8 | Paraoxonase 1 | 1.78 | 1.87 | — |
| Other proteins | ||||
| 11 | α2-HS-glycoprotein | — | 1.85 | — |
| 19 | Fibrin α-chain | — | — | — |
| 21 | Complement C3 | — | — | — |
| 25 | Hemopexin | — | — | — |
| 10 | Serum amyloid A protein | — | — | — |
| 16 | Serotransferrin | — | 1.83 | 1.66 |
| 12 | Transthyretin | — | — | — |
| 15 | Vitamin D-binding protein | — | 1.96 | — |
HDL proteins were quantified by MS. Four biological replicates, with a single LC-MS/MS performed for each sample, were analyzed in each group. Hamster proteins that have also been identified as predominate components of human HDL are shown (60). ApoJ, ApoL-1, PAF-AH, and α-1B-glycoprotein, present on human HDL, were not detected in hamster HDL. A two-sided Student’s t-test was performed between groups. The values shown are the fold changes in abundance between groups with P < 0.05. Fold changes ≥2-fold were considered significant. The order of protein relative abundance in HDL is based on Ad-null. The hamster sequence database reports two proteins with a gene ID of ApoC-IV but are designated as ApoC-II isoforms X2 (accession A0A1U8CN60) and X5 (accession A0A1U7R5I0). Based on sequence analysis, the X2 isoform is highly homologous to ApoC-II in other species, whereas the X5 isoform is highly homologous to ApoC-IV. Based on this, the X2 isoform is reported here as ApoC-II and the X5 isoform is reported as ApoC-IV. Nu, Ad-null; Ha, Ad-haCETP; Hu, Ad-huCETP.
Impact of human CETP on hamster hepatic gene expression
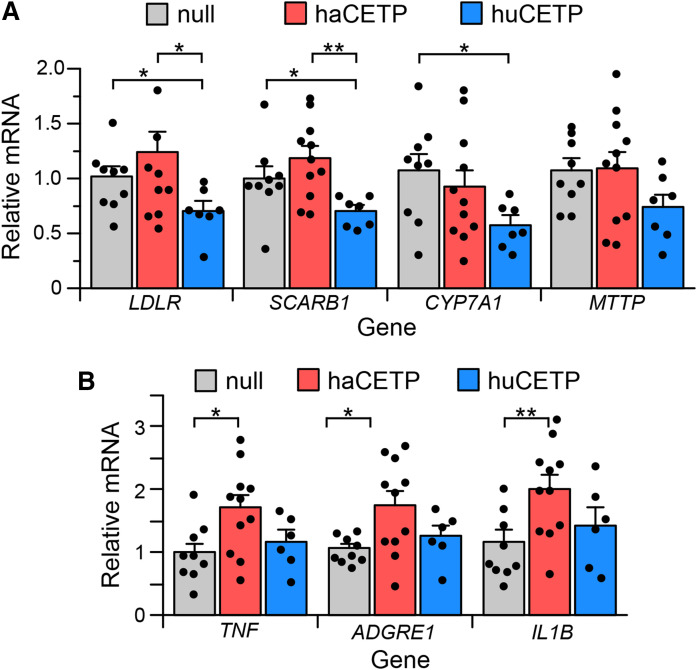
The redistribution of cholesterol among lipoproteins in Ad-huCETP animals changes how cholesterol is presented to the liver and may alter hepatic gene expression. Indeed, in hamsters expressing human CETP, hepatic mRNA levels for three genes involved in cholesterol uptake and metabolism (LDLR, SCARB1, and CYP7A1) were decreased whereas expression of the MTTP gene involved in TG-rich lipoprotein assembly was unchanged (Fig. 5A). In Ad-haCETP animals, which have the same plasma level of CETP as Ad-huCETP animals, there were no changes in the mRNA levels for any of these genes.
Fig. 5.
Hepatic mRNA. A, B: Liver mRNA levels of the indicated gene in Ad-null, Ad-haCETP, and Ad-huCETP animals. mRNA levels are normalized by ACTB mRNA content. Mean ± SEM. See Fig. 2 for group sizes. *P < 0.05; **P < 0.01.
Links between CETP expression and inflammation have been previously reported (36). Hepatic expression of human CETP may also elicit an immune response. We considered that differences in hepatic response to the expression of hamster versus human CETP expression may reflect altered inflammatory status. Hepatic inflammatory status was assessed by expression levels of three inflammatory gene markers, TNF, ADGRE1 (F4/80 protein), and IL1B. Expression of the foreign protein, human CETP, in hamsters did not increase expression of any of these genes (Fig. 5B). Overexpression of hamster CETP did modestly elevate the expression of all three markers. However, other indicators of hepatic inflammation (37), including liver weight (4.81 ± 0.12% body weight), and hepatic cholesterol (2.75 ± 0.03 mg/g liver wet weight) and TG (1.18 ± 0.10 mg/g liver wet weight) content, in Ad-haCETP and Ad-huCETP animals were not different from these Ad-null values.
Effect of CETP overexpression on HDL function and RCT
A major beneficial function of HDL is to promote cholesterol clearance by the RCT pathway. The changes in HDL concentration, lipid and protein composition, and particle size that occur in hamsters expressing human CETP may alter this pathway. HDL-dependent removal of cholesterol from cells is an initial step in RCT. Despite its significantly lower HDL content, TC efflux mediated by ApoB-depleted serum from Ad-huCETP animals was not different from that of controls (supplemental Table S3). The ABCA1-receptor-independent component of this efflux was modestly reduced. These data suggest that changes in Ad-huCETP HDL structure compensate for its lower plasma concentration to maintain cholesterol efflux potential.
Tissue-derived cholesterol in HDL is subsequently converted to CE by HDL-associated LCAT. Although endogenous LCAT activity was not measured directly in these plasmas, other studies show that altered LCAT activity is evidenced by changes in the plasma ratio of FC to TC (38, 39). This ratio in Ad-huCETP plasma (0.210 ± 0.021) was not different from that in Ad-null (0.244 ± 0.008) or Ad-haCETP (0.236 ± 0.008) animals. Also, the FC/TC ratios of HDL isolated from Ad-haCETP and Ad-huCETP plasma were the same (0.089 ± 0.007 vs. 0.081 ± 0.012, respectively). Consistent with their higher LCAT content (Table 4), these HDL FC/TC values were lower than in Ad-null HDL (0.119 ± 0.009, P < 0.05).
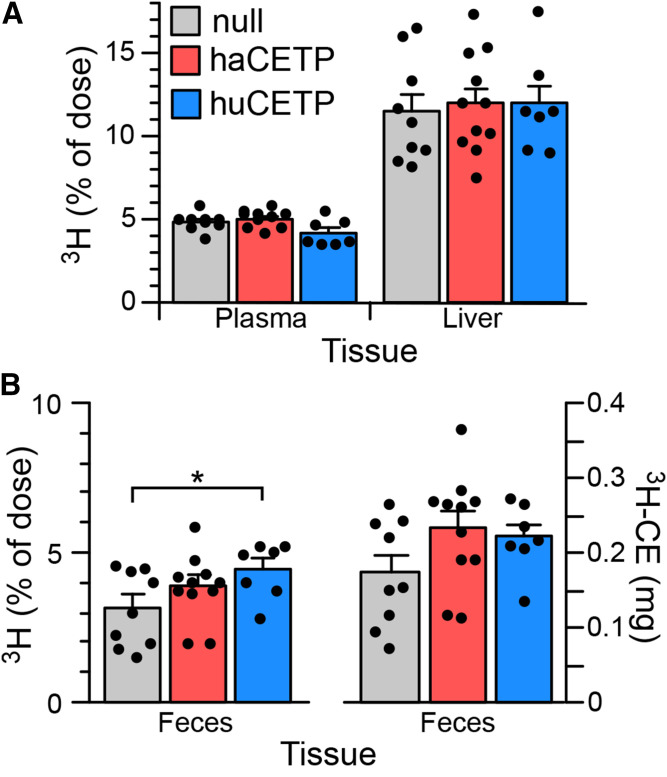
3H-CE-labeled HDL was injected into hamsters to measure hepatic uptake and excretion of HDL-associated CE. After 48 h, 3H remaining in plasma and that accumulated in the liver were the same for all three adenovirus groups (Fig. 6A). In Ad-huCETP animals, the percentage of injected 3H present in feces was 40% higher than in Ad-null animals but not different from that in Ad-haCETP animals (Fig. 6B). However, because HDL levels are lower in Ad-huCETP animals, and CETP transfers the 3H-CE initially contained in HDL to other lipoproteins, a more appropriate calculation of fecal RCT assumes that the injected 3H-CE equilibrates with the total plasma CE pool during the RCT experiment. When calculated in this way, the mass of CE delivered to feces in Ad-huCETP animals was not different (Fig. 6B). Thus, despite large changes in the plasma concentration and physicochemical properties of HDL in Ad-huCETP animals, the excretion of HDL-derived CE to feces remained the same.
Fig. 6.
RCT assay. Animals were injected with 3H-CE-labeled HDL. After 48 h, the 3H content of plasma, liver, and feces was determined. A: Percentage of injected dose recovered in plasma and liver. B: Fecal 3H calculated as the percent of injected dose and as the equivalent amount of plasma CE mass. See the legend in A for group identification. Mean ± SEM. See Fig. 2 for group sizes. *P < 0.05.
DISCUSSION
The role of CETP in lipoprotein metabolism has been investigated in multiple species. For example, many studies have examined the effect of CETP expression in species that do not naturally express this protein, including mice [for example, (40–43)], rats (44–46), and pigs (47). One study reported the expression of human CETP in rabbits, a species where CETP is native (48). In these cholesterol-fed rabbits, a 2-fold increase in plasma CETP mass reduced HDL cholesterol by 30%, but it did not change plasma TC or LDL cholesterol or alter atherosclerotic lesion size. However, because rabbit and human CETPs prefer CE as substrate and have similar relative preferences for CE versus TG (15), these findings largely reflect the effect of CETP overexpression in a cholesterol-fed animal model. CETP expression studies where the relative preference of CETP for CE versus TG has been altered have not been reported.
To test the capacity of altered CETP substrate specificity to regulate lipoprotein metabolism in vivo, we compared the effects of expressing hamster CETP and human CETP in chow-fed hamsters. Hamster CETP prefers TG as a substrate, whereas human CETP prefers CE (15). In a control group where hamster CETP was overexpressed, increasing plasma CETP mass and activity 2-fold had no effect on HDL lipid composition, the distribution of cholesterol among plasma lipoproteins, lipoprotein size, the expression of lipoprotein-related genes in the liver, or on cholesterol RCT. The primary mechanism for lipoprotein remodeling by CETP is the heteroexchange of CE in CE-rich lipoproteins for TG in TG-rich lipoproteins (1). In normolipidemic humans, the low plasma TG levels in these subjects limit the extent to which CETP can remodel lipoproteins (49). Similarly, the low level of TG-rich VLDL in chow-fed hamster plasma likely limits the extent to which lipoprotein composition can be modified by CETP. This would also explain the failure of hamster CETP overexpression to significantly alter lipoproteins.
It is notable that the lack of effect of hamster CETP overexpression was different from that observed in studies where ApoF, a CETP regulator, was knocked down in chow-fed hamsters (18). The increase in plasma CETP in Ad-haCETP animals impacts lipid transfer between all lipoproteins equally. By contrast, although depletion of ApoF increases CETP activity to a similar extent, this increase only occurs for lipid transfers involving LDL (18). Although these two methods of altering plasma CETP activity did not modify lipoprotein levels or their composition in chow-fed animals due to limited VLDL, they had different effects on RCT. In Ad-haCETP animals, overexpression of CETP did not alter RCT, whereas in ApoF-deficient hamsters, RCT was increased (18). These results show that selective augmentation of CETP activity on LDL by ApoF knockdown is not replicated by a similar rise in lipid transfer activity among all plasma lipoproteins. This emphasizes the importance of ApoF as a selective regulator of CETP activity.
The plasma of hamsters infected with adenovirus expressing human CETP contained human CETP plus low levels of endogenous hamster CETP. Total CETP in these plasmas was the same as in the hamster CETP overexpression group (Ad-haCETP), but the overall preference of CETP for CE versus TG in the plasma of Ad-huCETP animals was similar to that of human plasma. This resulted in pronounced changes in levels and composition of lipoproteins. Although LDL composition was modestly altered and VLDL cholesterol increased, the predominant phenotype was marked lower HDL cholesterol (50%), HDL protein (25%), and reduced HDL size. Remarkably, the expression of human CETP in hamsters humanized their lipoprotein profile. In animals receiving Ad-null and Ad-haCETP adenoviruses, HDL was the major cholesterol-carrying lipoprotein. However, in Ad-huCETP animals the percent distribution of cholesterol between VLDL, LDL, and HDL was 19:51:30, respectively. This is similar to a typical ratio of these lipoproteins in adult male humans (50). Also, the ratio of HDL3 to HDL2 in hamsters was ∼0.5, but in animals expressing human CETP this ratio increased to 1.6, similar to the 2.1 ratio typical of humans (51). These studies show that even though plasma lipoprotein concentrations and properties are controlled by the interaction of multiple enzymes, transfer proteins, and cellular receptors, the lipoprotein profile of hamsters could be converted to a human-like profile by simply altering the substrate preference of circulating CETP. These findings underscore the importance of CETP in the steady-state properties of plasma lipoproteins. These data also suggest a novel animal model for future intervention studies that more closely reflects the lipoproteins in humans.
Cholesterol efflux mediated by ApoB-depleted serum from Ad-huCETP animals was not different from that of control animals even though its HDL concentration was much lower. Efflux by ApoB-depleted serum depends on its HDL and preβ-HDL content (52, 53). Because ABCA1-dependent efflux, a preβ-HDL-driven process, was not different, this suggests that HDLs from Ad-huCETP animals are more effective in promoting cholesterol efflux from cells. This may relate to their smaller size and altered lipid composition, including a much lower ratio of FC/phospholipid (0.067 ± 0.005 vs. 0.030 ± 0.005) or altered proteome. Ad-huCETP HDLs were depleted of multiple apolipoproteins, including ApoA-IV, ApoC-II, ApoC-III, and ApoE. These proteins influence multiple metabolic pathways that impact the properties of HDL (54–57), including direct modulation of cholesterol efflux (54).
An interesting finding of these studies is that, despite markedly reduced plasma HDL levels, the excretion of HDL-associated CE into feces was not altered in huCETP animals. It is possible that the increased transfer of HDL CE to other lipoproteins by huCETP provides alternative routes for hepatic cholesterol delivery that compensate for reduced HDL delivery. The altered hepatic gene expression in Ad-huCETP animals may reflect this change in cholesterol delivery pathways. Alternatively, it may be that, in chow-fed hamsters, most HDL CE is normally delivered to the liver via VLDL- and LDL-dependent pathways instead of direct HDL uptake. As a result, a reduction in plasma HDL concentration may not appreciably affect TC clearance. This is consistent with kinetic studies in other CETP-expressing species (humans and rabbits), showing that the vast majority of plasma CE recovered in bile is derived from VLDL and LDL, not HDL (9). This contrasts with CETP-deficient species where hepatic cholesterol delivery via HDL is most important. It seems likely that the enhanced transfer of HDL CE to ApoB-containing lipoproteins by human CETP expression may have different consequences on cholesterol excretion in fat-fed animals where hepatic LDLR gene expression is extensively downregulated and LDL accumulates in plasma (18).
In summary, this is the first in vivo study demonstrating the consequences of altering the CE versus TG substrate preference of CETP on plasma lipoproteins. In hamsters, changing the substrate preference of circulating CETP to one similar to that of human CETP created a novel phenotype primarily characterized by low plasma HDL levels and HDL particles with markedly altered physicochemical properties. Even though lipoprotein metabolism is complex, simply making this change in the substrate preference of circulating CETP was sufficient to create a human-like lipoprotein profile in chow-fed hamsters. We suggest that manipulating the substrate properties of CETP, perhaps by pharmacologic means, may be a valuable approach for altering lipid metabolism and modifying cardiovascular disease risk. In humans, shifting the CE versus TG substrate preference of endogenous CETP toward TG may provide a novel way to lower the cholesterol content of ApoB-containing lipoproteins.
Data availability
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (58) partner repository with the dataset identifier PXD017998 and 10.6019/PXD017998.
Supplementary Material
Acknowledgments
The authors thank Dani Mihna and Lahoucine Izem for their technical assistance in these studies. The Orbitrap Elite mass spectrometer was purchased via National Institutes of Health shared instrument Grant 1S10RR031537-01.
Footnotes
Abbreviations:
- Ad
- adenovirus
- CE
- cholesteryl ester
- CETP
- cholesteryl ester transfer protein
- FC
- free cholesterol
- haCETP
- hamster cholesteryl ester transfer protein
- huCETP
- human cholesteryl ester transfer protein
- RCT
- reverse cholesterol transport
- TC
- total cholesterol
This research was supported in part by American Heart Association Grant 16GRNT31000002 and National Heart, Lung, and Blood Institute, National Institutes of Health Grant HL130041. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health or the American Heart Association. The authors declare that they have no conflicts of interest with the contents of this article.
The online version of this article (available at https://www.jlr.org) contains a supplement.
REFERENCES
- 1.Morton R. E., and Zilversmit D. B.. 1983. Inter-relationship of lipids transferred by the lipid-transfer protein isolated from human lipoprotein-deficient plasma. J. Biol. Chem. 258: 11751–11757. [PubMed] [Google Scholar]
- 2.Morton R. E. 1999. Cholesteryl ester transfer protein and its plasma regulator: lipid transfer inhibitor protein. Curr. Opin. Lipidol. 10: 321–327. [DOI] [PubMed] [Google Scholar]
- 3.Tall A. R. 1993. Plasma cholesteryl ester transfer protein. J. Lipid Res. 34: 1255–1274. [PubMed] [Google Scholar]
- 4.Oliveira H. C. F., and de Faria E. C.. 2011. Cholesteryl ester transfer protein: the controversial relation to atherosclerosis and emerging new biological roles. IUBMB Life. 63: 248–257. [DOI] [PubMed] [Google Scholar]
- 5.Barter P. J., Nicholls S. J., Kastelein J. J. P., and Rye K-A.. 2015. CETP inhibition as a strategy to reduce cardiovascular risk: the pro case. Circulation. 132: 423–432. [DOI] [PubMed] [Google Scholar]
- 6.Hovingh G. K., Ray K. K., and Boekholdt S. M.. 2015. CETP as a target to lower CVD risk: suspension of disbelief? Circulation. 132: 433–440. [DOI] [PubMed] [Google Scholar]
- 7.Tall A. R. 1998. An overview of reverse cholesterol transport. Eur. Heart J. 19 (Suppl. A): A31–A35. [PubMed] [Google Scholar]
- 8.Yen F. T., Deckelbaum R. J., Mann C. J., Marcel Y. L., Milne R. W., and Tall A. R.. 1989. Inhibition of cholesteryl ester transfer protein activity by monoclonal antibody. Effects on cholesteryl ester formation and neutral lipid mass transfer in human plasma. J. Clin. Invest. 83: 2018–2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz C. C., VandenBroek J. M., and Cooper P. S.. 2004. Lipoprotein cholesteryl ester production, transfer, and output in vivo in humans. J. Lipid Res. 45: 1594–1607. [DOI] [PubMed] [Google Scholar]
- 10.Rader D. J., Alexander E. T., and Rothblat G. H.. 2009. The role of reverse cholesterol transport in animals and humans and relationship to atherosclerosis. J. Lipid Res. 50 (Suppl.): S189–S194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Z., Niimi M., Ding Q., Liu Z., Wang L., Zhang J., Xu J., and Fan J.. 2017. Comparative studies of three cholesteryl ester transfer proteins and their interactions with known inhibitors. PLoS One. 12: e0180772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rader D. J., and Tall A. R.. 2012. Is it time to revise the HDL cholesterol hypothesis? Nat. Med. 18: 1344–1346. [DOI] [PubMed] [Google Scholar]
- 13.Morton R. E., and Izem L.. 2015. Modification of CETP function by changing its substrate preference: a new paradigm for CETP drug design. J. Lipid Res. 56: 612–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qiu X., Mistry A., Ammirati M., Chrunyk B., Clark R., Cong Y., Culp J., Danley D., Freeman T., Geoghegan K., et al. 2007. Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules. Nat. Struct. Mol. Biol. 14: 106–113. [DOI] [PubMed] [Google Scholar]
- 15.Morton R. E., and Izem L.. 2014. Cholesteryl ester transfer proteins from different species do not have equivalent activities. J. Lipid Res. 55: 258–265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Havel R. J., Eder H. A., and Bragdon J. H.. 1955. The distribution and chemical composition of ultracentrifugally separated lipoproteins in human serum. J. Clin. Invest. 34: 1345–1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morton R. E., and Zilversmit D. B.. 1981. A plasma inhibitor of triglyceride and cholesteryl ester transfer activities. J. Biol. Chem. 256: 11992–11995. [PubMed] [Google Scholar]
- 18.Morton R. E., Liu Y., and Izem L.. 2019. ApoF knockdown increases cholesteryl ester transfer to LDL and impairs cholesterol clearance in fat-fed hamster. J. Lipid Res. 60: 1868–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Serdyuk A. P., and Morton R. E.. 1999. Lipid transfer inhibitor protein defines the participation of lipoproteins in lipid transfer reactions: CETP has no preference for cholesteryl esters in HDL versus LDL. Arterioscler. Thromb. Vasc. Biol. 19: 718–726. [DOI] [PubMed] [Google Scholar]
- 20.Zinn K. R., Douglas J. T., Smyth C. A., Liu H-G., Wu Q., Krasnykh V. N., Mountz J. D., Curiel D. T., and Mountz J. M.. 1998. Imaging and tissue biodistribution of 99mTc-labeled adenovirusknob (serotype 5). Gene Ther. 5: 798–808. [DOI] [PubMed] [Google Scholar]
- 21.Izem L., Greene D. G., Bialkowska K., and Morton R. E.. 2015. Overexpression of full-length cholesteryl ester transfer protein in SW872 cells reduces lipid accumulation. J. Lipid Res. 56: 515–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Izem L., and Morton R. E.. 2009. Molecular cloning of hamster lipid transfer inhibitor protein (apolipoprotein F) and regulation of its expression by hyperlipidemia. J. Lipid Res. 50: 676–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pattnaik N. M., Montes A., Hughes L. B., and Zilversmit D. B.. 1978. Cholesteryl ester exchange protein in human plasma: Isolation and characterization. Biochim. Biophys. Acta. 530: 428–438. [DOI] [PubMed] [Google Scholar]
- 24.Livak K. J., and Schmittgen T. D.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 25.Asztalos B. F., de la Llera-Moya M., Dallal G. E., Horvath K. V., Schaefer E. J., and Rothblat G. H.. 2005. Differential effects of HDL subpopulations on cellular ABCA1- and SR-BI-mediated cholesterol efflux. J. Lipid Res. 46: 2246–2253. [DOI] [PubMed] [Google Scholar]
- 26.Garber D. W., Kulkarni K. R., and Anantharamaiah G. M.. 2000. A sensitive and convenient method for lipoprotein profile analysis of individual mouse plasma samples. J. Lipid Res. 41: 1020–1026. [PubMed] [Google Scholar]
- 27.Peterson G. L. 1977. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal. Biochem. 83: 346–356. [DOI] [PubMed] [Google Scholar]
- 28.Bartlett G. R. 1959. Phosphorus assay in column chromatography. J. Biol. Chem. 234: 466–468. [PubMed] [Google Scholar]
- 29.Skeggs J. W., and Morton R. E.. 2002. LDL and HDL enriched in triglyceride promote abnormal cholesterol transport. J. Lipid Res. 43: 1264–1274. [PubMed] [Google Scholar]
- 30.Nichols A. V., Krauss R. M., and Musliner T. A.. 1986. Nondenaturing polyacrylamide gradient gel electrophoresis. Methods Enzymol. 128: 417–431. [DOI] [PubMed] [Google Scholar]
- 31.Thompson J. N., Erdody P., Brien R., and Murray T. K.. 1971. Fluorometric determination of vitamin A in human blood and liver. Biochem. Med. 5: 67–89. [DOI] [PubMed] [Google Scholar]
- 32.Zak B., Moss N., Boyle A. J., and Zlatkis A.. 1954. Reactions of certain unsaturated steroids with acid iron reagent. Anal. Chem. 26: 776–777. [Google Scholar]
- 33.Wang S., Deng L., Milne R. W., and Tall A. R.. 1992. Identification of a sequence within the C-terminal 26 amino acids of cholesteryl ester transfer protein responsible for binding a neutralizing monoclonal antibody and necessary for neutral lipid transfer activity. J. Biol. Chem. 267: 17487–17490. [PubMed] [Google Scholar]
- 34.Stevenson S. C., Wang S., Deng L., and Tall A. R.. 1993. Human plasma cholesteryl ester transfer protein consists of a mixture of two forms reflecting variable glycosylation at asparagine 341. Biochemistry. 32: 5121–5126. [DOI] [PubMed] [Google Scholar]
- 35.Gautier T., Masson D., Athias A., Gambert P., Aunis D., Metz-Boutigue M. H., and Lagrost L.. 2000. Human apolipoprotein C–I accounts for the ability of plasma high density lipoproteins to inhibit the cholesteryl ester transfer protein activity. J. Biol. Chem. 275: 37504–37509. [DOI] [PubMed] [Google Scholar]
- 36.van der Tuin S. J. L., Li Z., Berbée J. F. P., Verkouter I., Ringnalda L. E., Neele A. E., van Klinken J. B., Rensen S. S., Fu J., de Winther M. P. J., et al. 2018. Lipopolysaccharide lowers cholesteryl ester transfer protein by activating F4/80+Clec4f+Vsig4+Ly6C− Kupffer cell subsets. J. Am. Heart Assoc. 7: e008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lefort C., van Hul M., Delzenne N. M., Everard A., and Cani P. D.. 2019. Hepatic MyD88 regulates liver inflammation by altering synthesis of oxysterols. Am. J. Physiol. Endocrinol. Metab. 317: E99–E108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brousseau M. E., Santamarina-Fojo S., Zech L. A., Bérard A. M., Vaisman B., Meyn S. M., Powell D., Brewer H. B. J., and Hoeg J. M.. 1996. Hyperalphalipoproteinemia in human lecithin cholesterol acyltransferase transgenic rabbits. In vivo apolipoprotein A-I catabolism is delayed in a gene dose-dependent manner. J. Clin. Invest. 97: 1844–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakai N., Vaisman B. L., Koch C. A., Hoyt R. F. J., Meyn S. M., Talley G. D., Paiz J. A., Brewer H. B. J., and Santamarina-Fojo S.. 1997. Targeted disruption of the mouse lecithin-cholesterol acyltransferase (LCAT) gene. Generation of a new animal model for human LCAT deficiency. J. Biol. Chem. 272: 7506–7510. [DOI] [PubMed] [Google Scholar]
- 40.Hayek T., Azrolan N., Verdery R. B., Walsh A., Chajek-Shaul T., Agellon L. B., Tall A. R., and Breslow J. L.. 1993. Hypertriglyceridemia and cholesteryl ester transfer protein interact to dramatically alter high density lipoprotein levels, particle sizes, and metabolism. Studies in transgenic mice. J. Clin. Invest. 92: 1143–1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou H., Li Z., Silver D. L., and Jiang X. C.. 2006. Cholesteryl ester transfer protein (CETP) expression enhances HDL cholesteryl ester liver delivery, which is independent of scavenger receptor BI, LDL receptor related protein and possibly LDL receptor. Biochim. Biophys. Acta. 1761: 1482–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tchoua U., D’Souza W., Mukhamedova N., Blum D., Niesor E., Mizrahi J., Maugeais C., and Sviridov D.. 2008. The effect of cholesteryl ester transfer protein overexpression and inhibitonon reverse cholesterol transport. Cardiovasc. Res. 77: 732–739. [DOI] [PubMed] [Google Scholar]
- 43.Marotti K. R., Castle C. K., Boyle Y. P., Lin A. H., Murray R. W., and Melchior G. W.. 1993. Severe atherosclerosis in transgenic mice expressing simian cholesteryl ester transfer protein. Nature. 364: 73–75. [DOI] [PubMed] [Google Scholar]
- 44.Zak Z., Gautier T., Dumont L., Masson D., Deckert V., Duverneuil L., Pais De Barros J. P., Le Guern N., Schneider M., Moulin P., et al. 2005. Effect of cholesteryl ester transfer protein (CETP) expression on diet-induced hyperlipidemias in transgenic rats. Atherosclerosis. 178: 279–286. [DOI] [PubMed] [Google Scholar]
- 45.Masson D., Pais de Barros J. P., Zak Z., Gautier T., Le Guern N., Assem M., Chisholm J. W., Paterniti J. R. Jr., and Lagrost L.. 2006. Human apoA-I expression in CETP transgenic rats leads to lower levels of apoC-I in HDL and to magnification of CETP-mediated lipoprotein changes. J. Lipid Res. 47: 356–365. [DOI] [PubMed] [Google Scholar]
- 46.Zak Z., Lagrost L., Gautier T., Masson D., Deckert V., Duverneuil L., De Barros J. P., Le Guern N., Dumont L., Schneider M., et al. 2002. Expression of simian CETP in normolipidemic Fisher rats has a profound effect on large sized apoE-containing HDL. J. Lipid Res. 43: 2164–2171. [DOI] [PubMed] [Google Scholar]
- 47.Chen T., Sun M., Wang J-Q., Cui J-J., Liu Z-H., and Yu B.. 2017. A novel swine model for evaluation of dyslipidemia and athersclerosis induced by human CETP overexpression. Lipids Health Dis. 16: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gao S., Wang X., Cheng D., Li J., Li L., Ran L., Zhao S., Fan J., and Liu E.. 2017. Overexpression of cholesteryl ester transfer protein increases macrophage-derived foam cell accumulation in atherosclerotic lesions of transgenic rabbits. Mediators Inflamm. 2017: 3824276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sammett D., and Tall A. R.. 1985. Mechanisms of enhancement of cholesteryl ester transfer protein activity by lipolysis. J. Biol. Chem. 260: 6687–6697. [PubMed] [Google Scholar]
- 50.Jones G. J. L., Hewitt D., Godin G. J., Breckenridge W. C., Bird J., Mishkel M. A., Steiner G., and Little J. A.. 1980. Plasma lipoprotein levels and the prevalence of hyperlipoproteinemia in a Canadian working population. Can. Med. Assoc. J. 122: 37–38. [PMC free article] [PubMed] [Google Scholar]
- 51.Verdery R. B., Benham D. F., Baldwin H. L., Goldberg A. P., and Nichols A. V.. 1989. Measurement of normative HDL subfraction cholesterol levels by Gaussian summation analysis of gradient gels. J. Lipid Res. 30: 1085–1095. [PubMed] [Google Scholar]
- 52.de la Llera-Moya M., Drazul-Schrader D., Asztalos B. F., Cuchel M., Rader D. J., and Rothblat G. H.. 2010. The ability to promote efflux via ABCA1 determines the capacity of serum specimens with similar high-density lipoprotein cholesterol to remove cholesterol from macrophages. Arterioscler. Thromb. Vasc. Biol. 30: 796–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Asztalos B. F., Horvath K. V., Mehan M., Yokota Y., and Schaefer E. J.. 2017. Influence of HDL particles on cell-cholesterol efflux under various pathological conditions. J. Lipid Res. 58: 1238–1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qu J., Ko C-W., Tso P., and Bhargava A.. 2019. Apolipoprotein A-IV: A multifunctional protein involved in protection against atherosclerosis and diabetes. Cells. 8: 319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kei A. A., Filippatos T. D., Tsimihodimos V., and Elisaf M. S.. 2012. A review of the role of apolipoprotein C–II in lipoprotein metabolism and cardiovascular disease. Metabolism. 61: 906–921. [DOI] [PubMed] [Google Scholar]
- 56.Kohan A. B. 2015. ApoC-III: a potent modulator of hypertriglyceridemia and cardiovascular disease. Curr. Opin. Endocrinol. Diabetes Obes. 22: 119–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeon H., and Blacklow S. C.. 2005. Structure and physiologic function of the low-density lipoprotein receptor. Annu. Rev. Biochem. 74: 535–562. [DOI] [PubMed] [Google Scholar]
- 58.Perez-Riverol Y., Csordas A., Bai J., Bernal-Llinares M., Hewapathirana S., Kundu D. J., Inuganti A., Griss J., Mayer G., Eisenacher M., et al. 2019. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 47: D442–D450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lund-Katz S., and Phillips M. C.. 1984. Packing of cholesterol molecules in human high-density lipoproteins. Biochemistry. 23: 1130–1138. [DOI] [PubMed] [Google Scholar]
- 60.Kontush A., Lindahl M., Lhomme M., Calabresi L., Chapman M. J., and Davidson W. S.. 2015. Structure of HDL: Particle subclasses and molecular components. Handb. Exp. Pharmacol. 224: 3–51. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The MS proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (58) partner repository with the dataset identifier PXD017998 and 10.6019/PXD017998.