Abstract
Too many choices can be problematic. This is certainly the case for human pluripotent stem cells (hPSCs): they harbor the potential to differentiate into hundreds of cell-types, yet it is highly challenging to exclusively differentiate hPSCs into a single desired cell-type. This review focuses on unresolved and fundamental questions regarding hPSC differentiation and critiquing the identity and purity of the resultant cell populations. These are timely issues in view of the fact that hPSC-derived cell populations have or are being transplanted into patients in over 30 ongoing clinical trials. While many in vitro differentiation protocols purport to “mimic development”, the exact number and identity of intermediate steps that a pluripotent cell takes to differentiate into a given cell-type in vivo remains largely unknown. Consequently most differentiation efforts inevitably generate a heterogeneous cellular population, as revealed by single-cell RNA-sequencing and other analyses. The presence of unwanted cell-types in differentiated hPSC populations does not portend well for transplantation therapies. This provides an impetus to precisely control differentiation to desired ends—for instance, by logically blocking the formation of unwanted cell-types or by overexpressing lineage-specifying transcription factors—or by harnessing technologies to selectively purify desired cell-types. Conversely, approaches to differentiate 3-dimensional “organoids” from hPSCs intentionally generate heterogeneous cell populations. While this is intended to mimic the rich cellular diversity of developing tissues, whether all such organoids are spatially organized in a manner akin to native organs (and thus, whether they fully qualify as “organoids”) remains to be fully resolved.
Introduction
The ability of human pluripotent stem cells (hPSCs)—which include embryonic (Thomson et al., 1998) and induced pluripotent stem cells (Park et al., 2008; Takahashi et al., 2007; Yu et al., 2007)—to differentiate into all the hundreds of diverse cell-types within the human body is both a blessing for, and the bane of, regenerative medicine. The blessing is evident: the pluripotency of hPSCs, combined with their ability to prodigiously divide in culture (expanding >10100 fold within several months (Levenstein et al., 2006)), has led to the oft-quoted aspiration that it should be possible to manufacture limitless numbers of a given human cell-type in vitro for transplantation therapies or other applications (Cohen and Melton, 2011; Murry and Keller, 2008; Tabar and Studer, 2014).
However, the very same pluripotency is also a bane: given that hPSCs have a panoply of hundreds of lineage options ahead of them, it has been challenging to exclusively differentiate hPSCs down any one developmental route to yield a pure population of a single lineage. As they differentiate, hPSCs navigate multiple, poorly-understood developmental lineage decisions in stepwise fashion as they progressively segue into more differentiated fates. Directing differentiation is reminiscent of the labors of Odysseus—navigating a narrow course for his ship between the aquatic monstrosities Scylla and Charybdis in Homer’s The Odyssey. Much like Odysseus, hPSCs can easily stray from an intended lineage trajectory, differentiating into unwanted cell-types that could in turn cause deleterious effects in therapeutic settings.
Consequently the field of stem-cell differentiation has seen alternating progress over the past few decades. Most differentiation methods yield a range of lineage outcomes in differing proportions, with the desired lineage often comprising a subset of the whole population (Cohen and Melton, 2011; McKnight et al., 2010). In heterogeneous differentiating cultures, commingled lineages likely reciprocally signal amongst one another, rendering differentiation difficult to control. This has been further complicated by use of undefined animal serum or feeder coculture in some embodiments (Murry and Keller, 2008). Moreover, with suboptimal differentiation protocols, differentiation efficiencies have been reported to vary dramatically between individual hPSC lines (Osafune et al., 2008). Here, we critically assess various challenges in hPSC differentiation, with the view that it should be possible to more precisely guide differentiation towards desired ends by understanding the means through which differentiation occurs.
Finding the right path: accessing desired lineages through the correct intermediate progenitors
Many differentiation approaches purport to “mimic development” to some degree and entail treatment with a sequence of various signaling modulators. However for many mature cell-types, we do not know the exact number or identity of steps through which they develop from pluripotent cells in vivo. Identifying the complete sequence of lineage steps needed to differentiate hPSCs into a desired cell-type remains a major challenge for stem cell and developmental biology. Consequently, certain differentiation protocols claim to yield a terminal cell-type in only a few steps that likely fail to recapitulate the full number and sequence of steps leading to cell-type specification in vivo, with a number of ensuing consequences (see below).
There is thus an urgent need for comprehensive lineage maps of mammalian development to guide hPSC differentiation. These maps may materialize soon, for instance by randomly labeling single progenitors in mouse embryos using Cas9-inflicted genetic barcodes and then creating detailed lineage maps of their progeny (Chan et al., 2019). In any case, precisely mapping intervening developmental intermediates is crucial to effectively differentiate hPSCs into any lineage, as the below vignettes demonstrate.
Primitive streak
Take for instance the initial differentiation of hPSCs into the endoderm, mesoderm and ectoderm germ layers. This was initially conceptualized as a “three way” lineage decision (Fig. 1Ai). Yet it is now widely recognized that these three lineages are instead generated through two nested lineage bifurcations: first, hPSCs bifurcate into primitive streak intermediates or ectoderm, and such primitive streak intermediates subsequently bifurcate into endoderm versus mesoderm intermediates (Fig. 1Ai). Therefore hPSCs cannot be directly differentiated into endoderm or mesoderm in one step. Many differentiation protocols (of which only several are mentioned here for the sake of brevity) now employ some combination of BMP, FGF, TGFβ and/or WNT to differentiate hPSCs towards primitive streak-containing populations as a prelude to downstream endoderm or mesoderm formation (e.g., Ang et al., 2018; Bernardo et al., 2011; Chu et al., 2019; D’Amour et al., 2006; Gertow et al., 2013; Li et al., 2019; Loh et al., 2014; Loh et al., 2016; Mendjan et al., 2014; Rao et al., 2015; Yu et al., 2011). Differentiation of hPSCs through the intermediacy of the primitive streak is paramount to efficiently generate endoderm or mesoderm at later stages of differentiation, as omitting primitive streak induction (e.g., by withholding WNT) leads to a total failure to generate endoderm (Li et al., 2019) or mesoderm from hPSCs (Rao et al., 2015).
Figure 1: Primitive streak differentiation and the importance of the very first steps of hPSC differentiation.
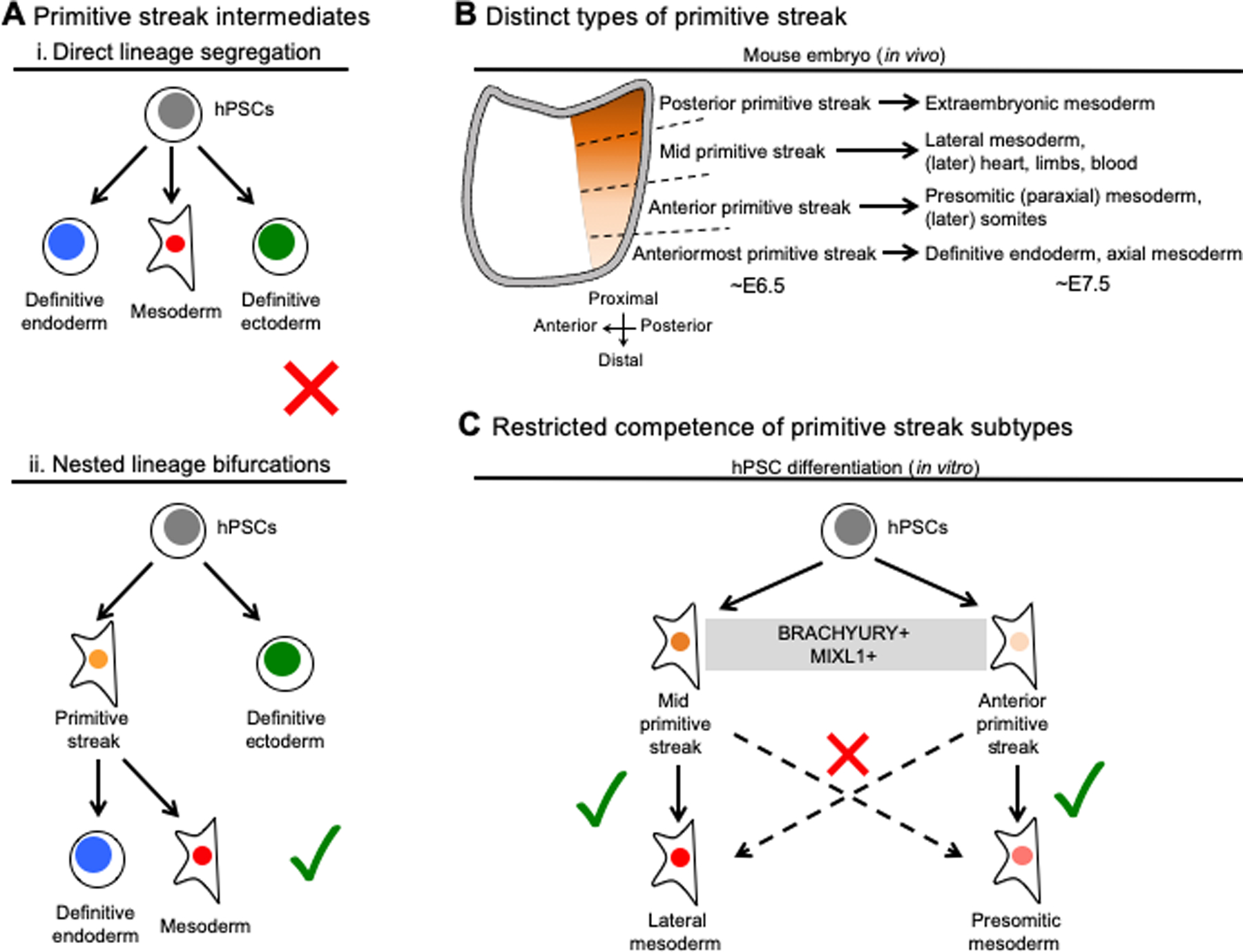
A. Human pluripotent stem cells (hPSCs) do not directly differentiate into definitive endoderm or mesoderm (i), but first must differentiate through a transitory primitive streak intermediate (ii)
B. In the ~6.5-day-old (~E6.5) mouse embryo, there is no “pan-mesoderm” precursor; rather distinct primitive streak lineages give rise to different types of mesoderm (Lawson et al., 1991; Rosenquist, 1970; Tam and Beddington, 1987)
C. hPSC-derived anterior and mid primitive streak populations are broadly marked by both BRACHYURY and MIXL1; however, each primitive streak subtypes has a distinct lineage potential in terms of its ability to further differentiate into downstream cell-types
Yet, even the primitive streak concept is an oversimplification. Historically, the primitive streak has been morphologically defined as a structure within the gastrulating embryo. Nevertheless is now evident that it harbors a spectrum of related yet molecularly and functionally distinct cell-types known as various primitive streak subtypes. Such complexities first surfaced when in vivo analyses failed to discover a singular “primitive streak” intermediate that has the full potential to generate all types of endoderm and mesoderm in vivo. Rather, within the early primitive streak, different subdomains of the primitive streak are each fated to give rise to distinct derivatives in vivo (Fig. 1B). Specifically, anterior-most (distal-most) primitive streak gives rise to definitive endoderm and axial mesoderm; the anterior primitive streak generates presomitic mesoderm; the mid primitive streak conceives lateral mesoderm; and finally, posterior (proximal) primitive streak yields extraembryonic mesoderm (Lawson et al., 1991; Rosenquist, 1970; Tam and Beddington, 1987) (Fig. 1B).
Thus, in a strict sense, hPSCs are not differentiated into “primitive streak”—there is a molecular and functional diversity in primitive streak subtypes, each the starting point for a different downstream cell-type. Within the first 24 hours of differentiation or so, hPSCs initially differentiate into different subtypes of primitive streak (e.g., anterior-most, anterior, mid and posterior primitive streak) (Loh et al., 2016; Mendjan et al., 2014). These primitive streak subtypes are molecularly distinct, with FOXA2, GSC and HHEX enriched in anterior, and CDX2 and FOXF1 enriched in posterior primitive streak populations generated from hPSCs in the first 24 hours of differentiation (Bernardo et al., 2011; Loh et al., 2014; Loh et al., 2016; Mendjan et al., 2014; Sumi et al., 2008). These diverse primitive streak populations are functionally distinct: they have separate lineage potentials. 24 hours later, hPSC-derived anterior-most primitive streak differentiates into definitive endoderm; hPSC-derived anterior primitive streak progresses into presomitic mesoderm; whereas hPSC-derived mid primitive streak forms lateral mesoderm (Loh et al., 2016; Mendjan et al., 2014) (Fig. 1C).
Hence even at the incept of hPSC differentiation, induction of a particular type of primitive streak is imperative for the subsequent generation of endoderm or different types of downstream mesoderm. For instance, if mid primitive streak is inadvertently generated, it cannot be efficiently differentiated into anterior primitive streak derivatives (e.g., presomitic mesoderm) and vice versa (although some degree of differentiation is still possible, suggesting a limited inherent plasticity (Loh et al., 2016)) (Fig. 1C).
The diversity of primitive streak subtypes and their restricted lineage potentials is an important point for hPSC differentiation protocols, because pan-primitive streak markers BRACHYURY and MIXL1 are broadly expressed across all primitive streak subtypes in vivo (Rivera-Pérez and Magnuson, 2005; Robb et al., 2000) and in vitro (Loh et al., 2016; Mendjan et al., 2014). Therefore despite encouraging progress in generating nearly pure cultures of hPSC-derived MIXL1+ “primitive streak” (Chu et al., 2016; Takasato et al., 2014), it is critical to assess whether these protocols actually produce the specific primitive streak subtype (e.g., anterior, mid or posterior) poised to produce a desired downstream differentiation outcome. Broadly speaking, beyond the examples mentioned here, whether other subtypes of primitive streak exist; their respective developmental potentials; and whether they can be efficiently derived from hPSCs remain outstanding questions.
Endoderm
After the endoderm germ layer arises (at embryonic day 7–7.5 [~E7–7.5] of mouse development), it is compartmentalized along the anterior-posterior axis into the anterior endoderm (foregut) and posterior endoderm (midgut/hindgut, ~E8.5), which presages the subsequent emergence of specific endodermal organ progenitors (~E9.5) (Zorn and Wells, 2009) (Fig. 2Ai). Efforts to optimize hPSC differentiation into these endodermal organ progenitors have been availed by markers that identify distinct progenitor domains along the anterior-posterior length of the endoderm in vivo (Grapin-Botton, 2005; Sherwood et al., 2009).
Figure 2: Unexpected complexities in hPSC differentiation towards the endoderm, mesoderm and ectoderm germ layers.
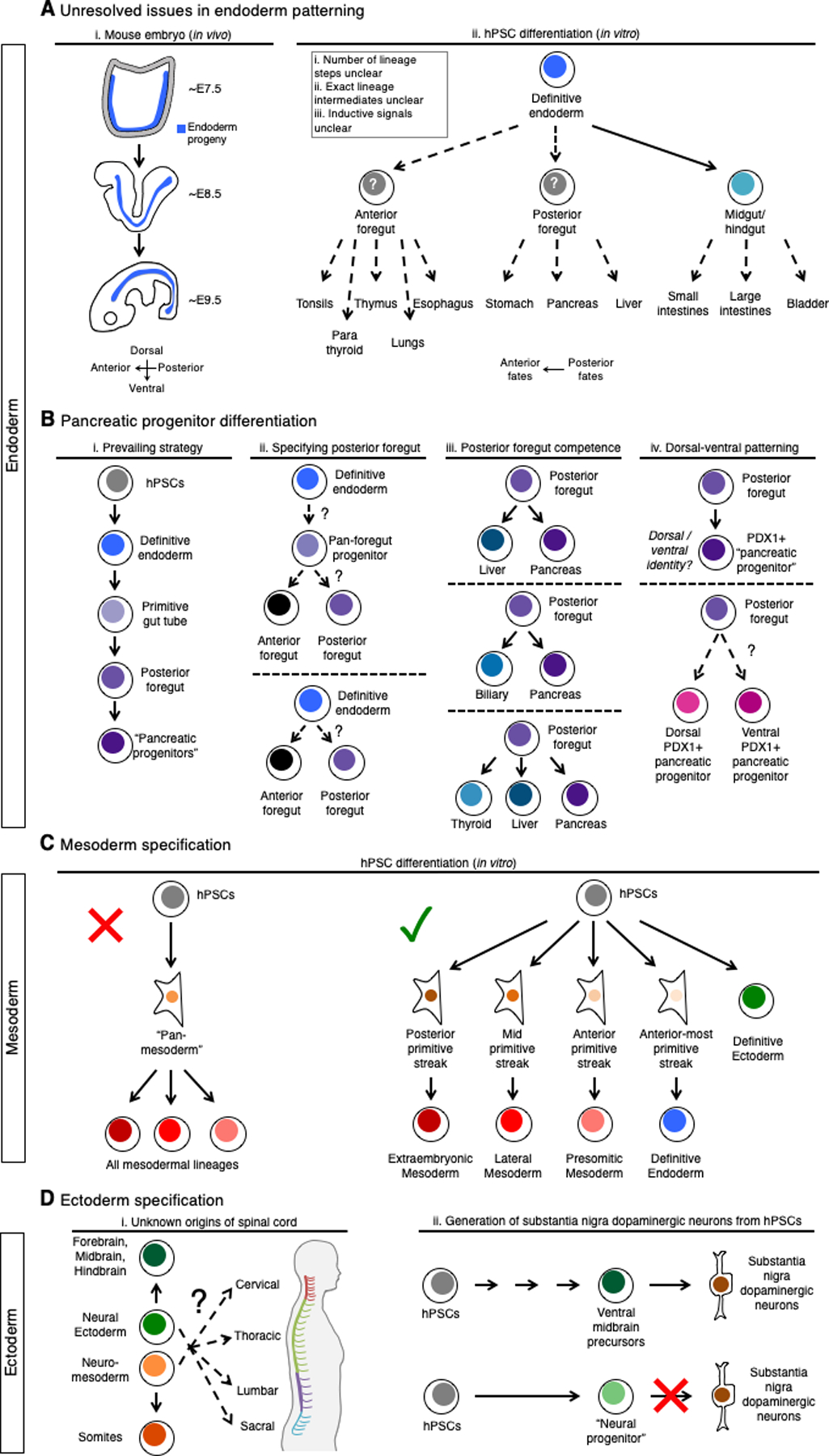
A. The process by which the definitive endoderm germ layer develops into >12 different organs in vivo is poorly understood (left); areas yet to be fully understood stymie the in vitro differentiation of hPSCs into endodermal derivatives (right)
B. Prevailing strategies to differentiate hPSCs towards pancreatic progenitors are ostensibly divided into several intermediate steps (i), yet outstanding questions remain, including: how the posterior foregut is specified (ii); what the exact identity of the posterior foregut is (i.e., what lineages can it differentiate into) (iii); and whether hPSC derived pancreatic progenitors have a dorsal and/or ventral identity (iv)
C. There is no ubiquitous “pan-mesoderm” progenitor that gives rise to all mesoderm lineages (left), but rather hPSCs differentiate into distinct primitive streak subtypes, each of which gives rise to a distinct mesoderm subtype or alternatively the definitive endoderm (right)
D. The developmental origins of the spinal cord remain unknown, with neural ectoderm and neuromesoderm both serving as potential intermediates for generating spinal cord in vitro (i); recent success in generating substantia nigra dopaminergic neurons was made possible by differentiating hPSCs through a ventral midbrain precursor intermediate instead of a “pan-neural progenitor” (ii)
A general concept is that endoderm anterior-posterior patterning must precede organ progenitor specification, both in vivo and in vitro. For instance, endoderm must first be posteriorly patterned into CDX2+ mid/hindgut through the influence of BMP, FGF and/or WNT signals (Loh et al., 2014; Sherwood et al., 2011; Spence et al., 2011) before it can be further differentiated into intestinal cell-types (Fig. 2Aii). Similarly, endoderm must first be anteriorized into anterior foregut endoderm intermediates before one can subsequently access lung or thyroid fates (Green et al., 2011) (Fig. 2Aii). Hence, while the in vitro chronology broadly reflects the in vivo developmental sequence, a number of issues remain unresolved.
There is considerable ambiguity regarding the precise sequence of nested lineage choices and the exact identity of the intermediate progenitors through which the endoderm germ layer eventually becomes diversified into a dozen different endodermal organs (Zorn and Wells, 2009) (Fig. 2Aii). This point is illustrated by the directed differentiation of hPSCs into pancreatic cell-types, which is of immense importance to develop cell therapies for diabetes. Prevailing strategies typically differentiate hPSC-derived endoderm into “primitive gut tube”, “posterior foregut” and then “pancreatic progenitors” (Kroon et al., 2008; Pagliuca et al., 2014; Rezania et al., 2014) (Fig. 2Bi).
Yet, many questions surround the precise developmental origin and identity of posterior foregut endoderm and pancreatic progenitors. First, how is the posterior foregut specified from hPSCs? Does endoderm have to be initially differentiated into a “pan-foregut progenitor” or “primitive gut tube” that bifurcates to form the anterior foregut (Green et al., 2011) vs. the posterior foregut (Fig. 2Bii)? Alternatively, the anterior foregut and posterior foregut might be totally distinct lineages that arise separately from hPSC-derived endoderm without passing through a common bipotent intermediate (Fig. 2Bii). This is not an idle intellectual exercise: in fact, understanding how to appropriately specify hPSC-derived posterior foregut is of paramount importance, as manipulating signals at the posterior foregut stage of differentiation has far-reaching effects on later stages of pancreatic differentiation (Russ et al., 2015; Veres et al., 2019).
Second, while consensus holds that hPSC-derived endoderm has to be differentiated into posterior foregut before generating pancreas, “posterior foregut” progenitors remain to clearly defined in vivo (Fig. 2Biii). Posterior foregut has been variously described as comprising a bipotent liver-pancreas progenitor (Chung et al., 2008), a bipotent pancreas-biliary progenitor (Spence et al., 2009) and/or a multipotent thyroid-liver-pancreatic progenitor (Angelo et al., 2012) in vivo within zebrafish and mouse embryos (Fig. 2Biii). Is there only one “posterior foregut” route to generate pancreas, or are there multiple such routes, and if so, does the choice of intermediate route impact downstream pancreatic differentiation? Indeed hPSC differentiation studies have suggested the existence of at least two types of posterior foregut, one poised for pancreatic, and the other primed for liver, differentiation in vitro (Ang et al., 2018).
Third, while hPSC differentiation protocols endeavor to generate early Pdx1+ pancreatic progenitors, there is no singular “pancreatic progenitor” at this stage in vivo. Instead, at E9.5-E10 of mouse development, two anatomically-distinct dorsal and ventral buds of Pdx1+ pancreatic progenitors emerge (reviewed by Pan and Wright, 2011) (Fig. 2Biv). Do current hPSC-derived pancreatic progenitors in vitro correspond to the dorsal pancreatic progenitor program, ventral pancreatic progenitor program, both, or neither (Fig. 2Biv)? Do dorsal versus ventral pancreatic progenitors differ in their potential to generate downstream pancreatic cell-types (Pan and Wright, 2011), and if so, should hPSC differentiation efforts attempt to specifically generate one or the other? While many efforts have optimized initial anterior-posterior endoderm patterning to enhance downstream production of hPSC-derived organ progenitors, the process of dorsal-ventral endodermal patterning (Sherwood et al., 2009) remains relatively enigmatic—specifically, how is it temporally coordinated with anterior-posterior endodermal patterning and what signals drive it? Incorporating a step emulating dorsal-ventral patterning in future endodermal differentiation strategies will be of interest.
While this vignette has focused on unknowns surrounding pancreatic progenitor specification, the same issues are germane to the generation of most endodermal organ progenitors. By analogy to pancreas, other endodermal organs may also have unexpected developmental progenitors, which may in turn necessitate alterations to hPSC differentiation protocols. For instance, the hindgut (prospective large intestine) emanates from at least two independent progenitors: definitive endoderm as well as extraembryonic endoderm, although the extent of contribution from both types of progenitor and whether their respective progeny last into adulthood remains to be fully resolved (Chan et al., 2019; Kwon et al., 2008; Nowotschin et al., 2019). Taken together, the hierarchy of “pre-organ” lineage intermediates that arise during anterior-posterior and dorsal-ventral patterning of endoderm (or for that matter, all germ layers) remains to be resolved. This knowledge is of paramount importance if we are to efficiently manufacture particular endodermal derivatives from PSCs (Fig. 2Aii).
Mesoderm
Even the notion of “germ layers” has needed revisions and embellishments in recent years. The mesoderm germ layer comprises a range of subtypes—including axial mesoderm, presomitic (paraxial) mesoderm, lateral mesoderm, intermediate mesoderm and extraembryonic mesoderm—and each gives rise to various mature cell-types.
One longstanding supposition of many mesodermal differentiation protocols has been that hPSCs must first be differentiated into a common “pan-mesoderm” progenitor (that can form any type of mesodermal lineage) which can be subsequently directed into a desired mesodermal subtype such as lateral/cardiac mesoderm (Burridge et al., 2012) (Fig. 2C). By contrast, lineage tracing and grafting experiments in mouse and chick embryos (Lawson et al., 1991; Rosenquist, 1970; Tam and Beddington, 1987) have argued against the existence of such a “pan-mesoderm” progenitor in vivo. As aforementioned, the primitive streak does not seem to constitute a “pan-mesoderm” precursor, and rather, distinct anterior, mid and posterior primitive streak subsets are established early, and each primitive streak subset has the restricted competence to only produce a specific mesodermal subtype (Fig. 1C; Fig. 2C).
There is no evidence for a ubiquitous “pan-mesoderm” progenitor that can be diversified into all downstream mesodermal derivatives (Mendjan et al., 2014), and this insight has significant ramifications for hPSC differentiation. Instead of differentiating hPSCs into a common “pan-mesoderm” intermediate before generating cardiac or presomitic mesoderm, hPSCs must be differentiated into the appropriate primitive streak intermediate (e.g., mid primitive streak) before being further differentiated into cardiac mesoderm (Loh et al., 2016; Mendjan et al., 2014) (Fig. 1C). Passage through the wrong primitive streak intermediate precludes the ability to efficiently further differentiate into a desired mesoderm subtype (Loh et al., 2016; Mendjan et al., 2014) (Fig. 1C).
Hence describing mesoderm as a “germ layer” may be a historical anachronism (Baxter, 1977). Axial, presomitic, intermediate, lateral and extraembryonic mesoderm subtypes are anatomically contiguous in the form of a “germ layer”, yet they do not seem to directly emerge from a common “pan-mesoderm” precursor—each mesoderm subtype has its own distinct developmental origin in the primitive streak (Fig. 2C). This raises an interesting concept: mesoderm may constitute a collection of lineages that do not share a common immediate precursor (i.e., they are lineally distinct) yet they are anatomically juxtaposed with one another, which gave rise to the initial assumption that they were lineally-related (Fig. 2C). Mesoderm specification therefore differs from endoderm; in the latter, hPSCs must first be differentiated into a common intermediate precursor (definitive endoderm) that subsequently can be diversified into seemingly all endodermal organ derivatives (see above) (Fig. 2Aii).
That notwithstanding, our understanding of mesoderm patterning remains incomplete. While a number of protocols have been reported to generate presomitic, intermediate and lateral mesoderm derivatives from hPSCs, production of axial or extraembryonic mesoderm in vitro is generally underexplored.
Ectoderm
Unexpected complexities have also surfaced for ectoderm germ layer development. Here we focus on the spinal cord and substantia nigra dopaminergic neurons as two examples to illustrate how identifying intervening developmental intermediates is of paramount importance to guide hPSC differentiation.
The neural ectoderm was previously construed to constitute a uniform precursor population that underwent anterior-posterior patterning to form the forebrain, midbrain, hindbrain and spinal cord (reviewed by Lumsden and Krumlauf, 1996). However, a new emerging hypothesis is that the brain and spinal cord have distinct developmental origins and that they do not originate from a single, monolithic “neural ectoderm” precursor (Henrique et al., 2015; Metzis et al., 2018). It has been hypothesized that neural ectoderm forms the brain, whereas a distinct “neuromesoderm” progenitor (variously referred to as “caudal lateral epiblast” or “axial progenitors”) conceives the spinal cord in addition to the presomitic mesoderm (Takemoto et al., 2011; Tzouanacou et al., 2009) (Fig. 2Di). The existence of a joint spinal cord and presomitic mesoderm progenitor argues against a decisive division in mesoderm and ectoderm “germ layers”; consequently, the germ layer model is potentially oversimplified.
In light of this, understanding the potentially unique developmental origins of spinal cord is critical to efficiently generate sought-after spinal cord motor neurons from hPSCs. Prevailing methods differentiate hPSCs into spinal cord motor neurons proceed through neural ectoderm intermediates (reviewed by Sances et al., 2016), or more recently, presumptive neuromesoderm intermediates (Denham et al., 2015; Gouti et al., 2014; Lippmann et al., 2015; Verrier et al., 2018). Can spinal cord progenitors truly be derived via two independent routes (Tao and Zhang, 2016)? In vivo analyses are warranted to test whether both neural ectoderm and/or neuromesoderm are legitimate progenitors to the spinal cord; to quantify how extensively each might contribute to the spinal cord; and to assess whether each progenitor pool might preferentially contribute only to specific spinal cord regions (e.g., cervical, thoracic, lumbar and sacral) (Fig. 2Di). Indeed, though spinal cord differentiation is rightly regarded as the exemplar for directed differentiation efforts (Wichterle et al., 2002), the current controversies over spinal cord origins underscore the dire need for comprehensive lineage maps as the field revisits the cellular origins of seemingly “well-understood” cell-types.
A recent success in the field has been the successful generation of substantia nigra dopaminergic neurons from hPSCs. There are multiple dopaminergic neuron subtypes, including those in the substantia nigra that developmentally originate from the ventral midbrain and principally control motor functions; these neurons are lost in Parkinson’s Disease and are priority targets for cell replacement therapies (reviewed by Arenas et al., 2015). While many efforts endeavored to differentiate hPSCs into “pan-neural progenitors” and then into dopaminergic neurons, the resultant neurons often lacked archetypic substantia nigra transcription factors and failed to properly engraft in vivo (reviewed by Arenas et al., 2015) (Fig. 2Dii). Access to hPSC-derived substantia nigra dopaminergic neurons was only made possible by identifying crucial intervening lineage intermediates—namely, recognizing their specific origin in the ventral midbrain. Recent protocols have now differentiated hPSCs into ventral midbrain precursors through the manipulation of HEDGEHOG and WNT signals, and such precursors were subsequently competent to differentiate into substantia nigra dopaminergic neurons (Fasano et al., 2010; Kirkeby et al., 2012; Kriks et al., 2011) (Fig. 2Dii). In sum, production of substantia nigra dopaminergic neurons proved intractable until these cells were accessed through the correct developmental intermediate.
Making a firm decision: accurately directing cell fate at lineage segregation points
How can hPSCs be coerced down a desired lineage route in preference to other possible developmental endpoints? The realization that development is organized as a cascade of “branching track” lineage choices (Waddington, 1940) has important corollaries. One recurrent principle is that at each binary lineage choice (also known as a lineage bifurcation), it is possible to exclusively differentiate progenitors into a given cell-type through a two-pronged approach to promote formation of a desired lineage while actively inhibiting formation of the alternative fate. This can be achieved by providing the relevant inductive signal(s) to specify the desired outcome, while of equal importance inhibiting signal(s) that would otherwise promote the alternate fate.
Indeed this strategy has enabled efficient negotiation of the lineage choices leading from pluripotency to early germ layer fates. The first lineage bifurcation encountered by hPSCs leads them to either differentiate into primitive streak (which is the developmental precursor of endoderm and mesoderm) or ectoderm. BMP, FGF, TGFβ and WNT promote primitive streak while inhibiting ectoderm formation (Bernardo et al., 2011; Blauwkamp et al., 2012; Chambers et al., 2009; Gadue et al., 2006; Loh et al., 2014; Loh et al., 2016; Yu et al., 2011). Hence, simultaneous application of these four signals (or combinations thereof) can generate a >98% pure MIXL1+ primitive streak population within 24 hours of hESC differentiation by blocking ectoderm formation (Loh et al., 2014; Loh et al., 2016). (Different levels of these four signals generates the graded primitive subtypes described above (Loh et al., 2016; Mendjan et al., 2014)). By contrast, complete blockade of primitive streak-inducing BMP and TGFβ signals suppresses primitive streak formation and instead diverts cells into ectoderm (Chambers et al., 2009).
Subsequently, primitive streak cells face another fork in the road to become either definitive endoderm or different subtypes of mesoderm. High levels of TGFβ specify endoderm (D’Amour et al., 2005; Loh et al., 2014) whereas BMP and WNT respectively promote lateral and presomitic mesoderm formation (Cheung et al., 2012; Loh et al., 2016; Umeda et al., 2012) (Fig. 3ai). Hence treating primitive streak intermediates with high TGFβ while concurrently inhibiting pro-mesodermal BMP signaling can “force” primitive streak cells to exclusively differentiate into SOX17+ endoderm and not mesoderm (Loh et al., 2014; Sumi et al., 2008) with up to 99% purity by day 2 of hESC differentiation (Loh et al., 2014; Loh et al., 2016) (Fig. 3Ai). Importantly, since differentiating hPSCs seemingly produce BMP, active inhibition of endogenous BMP signaling is crucial to fully suppress mesoderm formation and consolidate endodermal fate (Loh et al., 2014) (Fig. 3Ai). Hence while the factors exogenously added to coax differentiation are important, differentiating cells also endogenously signal to one another and manipulating such endogenous signals is critical. Differentiation protocols that principally specify endoderm through TGFβ addition (but do not include a BMP inhibitor to repress mesoderm) tend to yield endoderm with lower efficiency or consistency (Rostovskaya et al., 2015).
Figure 3: Reconstituting cell differentiation and assaying the products thereof.
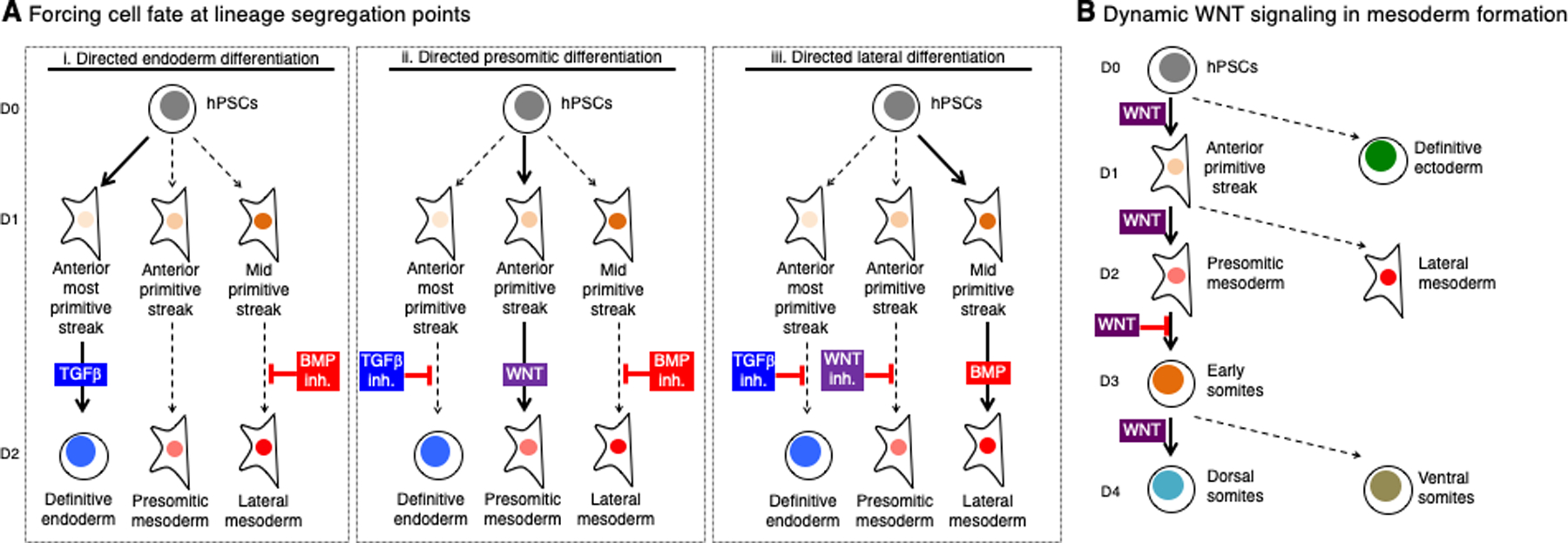
A. In the first day of hPSC differentiation to endoderm and mesoderm lineages, hPSCs differentiate into anteriormost primitive streak, anterior primitive streak and mid primitive streak, which respectively have the competence to further differentiate into definitive endoderm, presomitic mesoderm or lateral mesoderm, respectively. Subsequent to the primitive streak, manipulation of BMP, TGFβ and WNT allows guided differentiation into one of these three lineages while suppressing differentiation into unwanted fates (i-iii) (Loh et al., 2016).
B. Over the course of four days of hPSC differentiation into dorsal somites (dermomyotome/future skeletal muscle precursors), WNT specifies four distinct cell-types (Chal et al., 2015; Loh et al., 2016) and hence dynamic control of WNT signals every 24 hours of differentiation is crucial to progressively differentiate cells along this developmental trajectory
Conversely, blockade of endoderm-inducing signals can instead steer differentiating primitive streak cells into mesoderm. Inhibiting TGFβ suppresses endoderm differentiation from primitive streak and instead broadly promotes mesodermal fate. WNT activation together with simultaneous blockade of TGFβ and BMP pathways efficiently differentiates primitive streak into presomitic mesoderm while respectively inhibiting differentiation towards endoderm and lateral mesoderm (Chu et al., 2019; Loh et al., 2016) (Fig. 3Aii). Along the alternate developmental route, BMP activation together with dual inhibition of TGFβ and WNT pathways specifies lateral mesoderm from primitive streak (Loh et al., 2016) (Fig. 3Aiii). The need to concomitantly manipulate these three pathways (BMP, TGFβ and WNT) at this developmental stage emphasizes that, at any given point of development, there is no “single” dominant signaling pathway but rather the combinatorial integration of multiple cues is needed for the specification of most lineages.
In sum, by reducing the complex process of development into a sequence of simple lineage choices, it is possible at each stage to “force” progenitors at each juncture to exclusively differentiate towards a single desired outcome while suppressing extraneous differentiation to mutually-exclusive, unwanted fates. By understanding the signals that specify one fate or the other at each lineage choice it is possible to apply “paired inhibitor/agonist combinations” (Kyba, 2016) to more precisely guide differentiation towards a given lineage while deferring the competing dangers of other potential lineage outcomes—analogous to Odysseus precisely charting a path between Scylla and Charybdis. However, in vertebrate development it is evident that developmental lineage choices are usually, but not always, binary (Davidson, 2010; Graf and Enver, 2009). Single-cell transcriptional analyses have suggested that certain lineage decisions may not be sharp segregations, but rather may be construed as gradually diverging continua (Laurenti and Göttgens, 2018; Wagner et al., 2018). Validation of whether complex “multi-way” lineage decision points truly exist—and whether cellular fate can be precisely directed at such junctures—warrants further attention.
Temporally dynamic signaling and rapid cell-fate transitions
Another prevailing principle is how single developmental signals are dynamically re-interpreted to specify distinct lineages within a short span of time during in vitro differentiation (Rao and Greber, 2016). By way of example, while in the first 24 hours of differentiation WNT initially promotes hPSC differentiation into primitive streak, 24 hours later it promotes the progression of primitive streak into presomitic mesoderm (Chal et al., 2015; Loh et al., 2016). Then 24 hours later it inhibits presomitic mesoderm differentiation into early somites, and finally 24 hours later it blocks somite differentiation into dorsal somites while promoting a ventral somite fate (Chal et al., 2015; Loh et al., 2016). Hence, during the short span of 4 days of in vitro differentiation, WNT specifies four distinct lineages (Fig. 3B). This closely parallels the respective emergence of these lineages in the mouse embryo, wherein ~E5.5 post-implantation epiblast differentiates into ~E6.5 primitive streak, forming presomitic mesoderm by ~E7.0-E7.5 which then segments into early somites (~E8.0) and then forms dorsal somites (~E8.0-E8.5), with lineages segueing into one another every ~12–24 hours.
Yet in certain differentiation schema, the same developmental signals are continuously applied for several consecutive days, weeks or even months (especially in the case of organoid differentiation [see below]). This might explain why such protocols yield a heterogeneous assembly of lineages: the same signal is dynamically re-interpreted to yield a multitude of outcomes as time progresses.
How is a single signal re-interpreted over such a short span of time by a developing cell to signify different outcomes (Rankin et al., 2017; Wandzioch and Zaret, 2009)? The answer may lie in cell-intrinsic changes in competence to respond to the same signal. Additionally, interpretation of any given signal also likely depends on the context of combinatorial signals delivered in parallel. Whatever the underlying mechanisms, the rapid “re-utilization” of a single signal across consecutive lineage decisions to specify extremely different fates is likely how evolution succeeded in efficiently utilizing a common developmental toolkit of roughly a dozen major pathways (De Robertis, 2008) to encode hundreds of distinct possible fates. In any case, the temporal dynamism with which a single signal (e.g., WNT) is re-interpreted in turn demands equally dynamic modulation of developmental signals to specify desired outcomes as differentiating cells dynamically pass through successive fate transitions (Fig. 3B).
Fast forwarding differentiation: transcription factor overexpression
Given that it has been so vexing to efficiently differentiate hPSCs using extracellular signals, others have eschewed the use of such signals and have instead directly overexpressed transcription factors (TFs) of the desired cell-type within hPSCs. The goal is to forcibly instantiate the transcriptional program of the desired cell-type, thus “short cutting” normal development. This is reminiscent of how in Greek mythology, Athena was reputed to be born “fully-fledged” from the head of Zeus, dispensing with the usual protracted process of postnatal development.
While it typically takes weeks or months for extracellular signals to differentiate hPSCs into electrophysiologically-active neurons, remarkably the overexpression of the neuronal TF NGN2 in hPSCs generated neurons within ~1 week and with nearly 100% efficiency (Zhang et al., 2013) (Fig. 4A). The unprecedented speed and efficiency of neuronal specification in this system speaks to the power of TF overexpression as a potential strategy to generate desired cell-types; other early successes included the rapid derivation of megakaryocytes (Elcheva et al., 2014; Moreau et al., 2016). The surprising ability to “fast forward” differentiation from hPSCs into desired cell-types—while skipping intervening steps once thought to be crucial (see above)—raises a host of questions.
Figure 4: Expedited differentiation via the overexpression of lineage-specifying transcription factors.
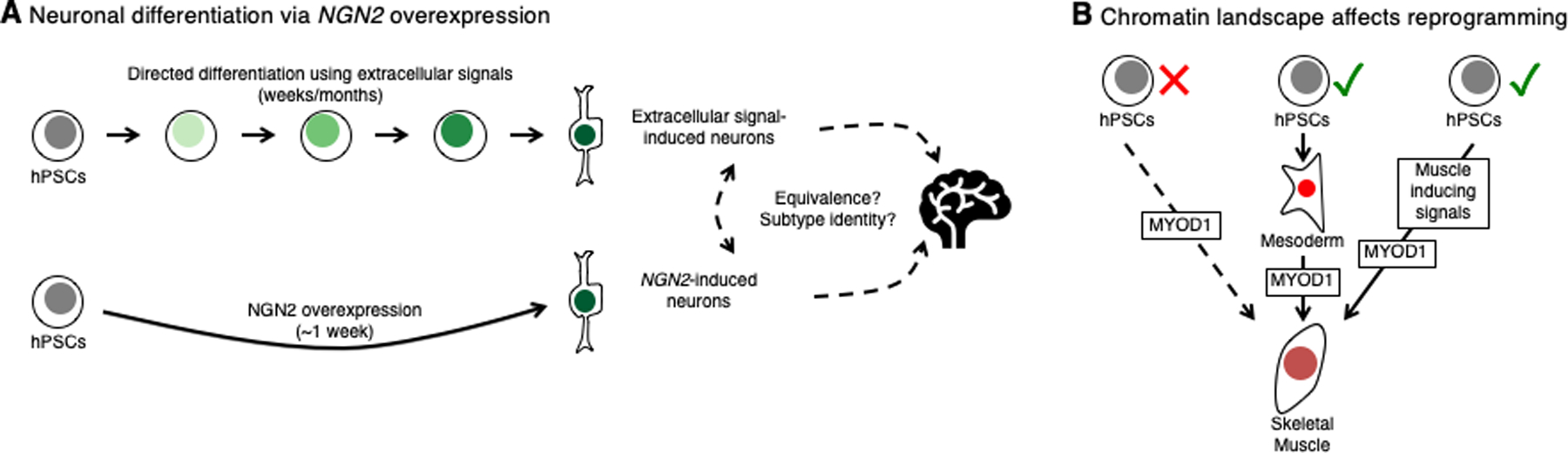
A. Electrophysiologically-active neurons can be generated from hPSCs via directed differentiation using extracellular signals or via overexpression of a neuron-specifying TF (NGN2) (Zhang et al., 2013). Do these two differentiation strategies generate equivalent neurons, do the resultant neurons have a clear subtype identity, and how comparable are they to their in vivo counterparts?
B. Overexpression of MYOD1 in hPSCs fails to induce skeletal muscle in the absence of either first differentiating the cells through a mesoderm intermediate (Albini et al., 2013) or concurrently adding muscle-inducing extracellular signals (Pawlowski et al., 2017), suggesting the chromatin landscape decisively dictates the success of TF-based hPSC differentiation.
First, does developmental history matter: are the TF-induced lineages “normal”, though they have forsaken their normal developmental progenitors? That is, do the ends justify the (unusual) means? For instance, while NGN2-induced neurons were electrophysiologically functional, it remains to be determined whether they transcriptionally correspond to any neuronal subtype found in the native brain (Zhang et al., 2013) (Fig. 4A). Perhaps NGN2 overexpression may directly shortcut to the end of neuronal commitment by directly transactivating core “pan-neuronal” genes without affecting developmental genes linked with regional identity, which would be normally induced during the natural course of brain anterior-posterior and dorsal-ventral patterning (Fig. 4A). This concept that TF-induced neurons are a tabula rasa partially lacking regional identity was supported by a recent analysis of motor neurons derived by overexpressing the TFs Ngn2, Isl1 and Lhx3 in mouse PSCs (Briggs et al., 2017). These TF-induced motor neurons seemingly lacked an anterior-posterior identity (as indicated by Hox genes) that was typically found in motor neurons that were derived from embryos or that were differentiated from PSCs by extracellular signals (Briggs et al., 2017).
Second, what happens when a TF that is typically expressed in a mature cell-type is mis-expressed in a hPSC—will it appropriately engage its appropriate target genes within the foreign chromatin landscape of a pluripotent cell? While some TFs constitute versatile “pioneer factors” that can activate their target genes irrespective of their local chromatin state (Iwafuchi-Doi and Zaret, 2016), the binding of most TFs is thought to be constrained by the cell’s chromatin landscape (Lambert et al., 2018), which is in turn determined by its lineage. That is, forced expression of a lineage-specifying master TF in hPSCs may fail to elicit the desired lineage, if same TF exerts different effects in different cell states. Indeed, ectopic overexpression of the muscle-specifying TF MYOD1 in undifferentiated hPSCs fails to drive skeletal muscle differentiation, unless hPSCs are first differentiated into mesoderm prior to MYOD1 induction (Albini et al., 2013) or are concurrently treated with skeletal muscle-inducing extracellular signals (Pawlowski et al., 2017) (Fig. 4B). Nonetheless, the majority of cell-types have not yet been successfully generated from hPSCs via TF overexpression, and it remains an open question as to why this is the case.
Perhaps one could obtain the best of both worlds through a collaboration of the two approaches, using TF overexpression to efficiently and rapidly engender the core transcriptional program of a desired cell-type while using extracellular signals to concomitantly entrain or refine their regional (anterior-posterior or dorsal-ventral) identity. For instance, neurons have recently been induced from hPSCs by combining NGN2 overexpression with neural ectoderm-specifying signals (dual BMP and TGFβ blockade) (Nehme et al., 2018). While this broadly increased the expression of pan-forebrain markers (Nehme et al., 2018), it remains unclear whether this strategy could be targeted to precisely induce a specific neuronal subtype in preference to others. One general limitation of TF-based hPSC differentiation is that TF overexpression is typically accomplished using genomically-integrated transgenes in hPSCs. This might be less suitable for clinical applications unless mRNA transfection (Warren et al., 2010) or similarly transient transgene delivery techniques were used.
The virtues of purity
We have thus far emphasized the production of a single cell-type in isolation from hPSCs, but is this even worthwhile or practicable to pursue? We opine that generating a pure population of a given lineage from hPSCs is a meaningful pursuit, at least for certain applications. Pioneering early studies differentiated hPSCs into an impure population containing a subpopulation of pancreatic cells. Transplantation of these impure cell populations into mouse models yielded not only human pancreatic tissue, but occasionally also mesodermal derivatives such as bone and cartilage in vivo (Kroon et al., 2008; Rezania et al., 2012; Rostovskaya et al., 2015) (Fig. 5Ai). Tumors were also observed after transplantation of PSC-derived, heterogeneous cell populations containing either a subset of liver cells (Haridass et al., 2010) or neural cells (Ganat et al., 2012). While subsequent advances have increased the uniformity of these cell populations, it is evident that the presence of unwanted cell-type(s) in heterogeneous cell populations can lead to untoward consequences after transplantation.
Figure 5: The virtues of purity and identity during hPSC differentiation.
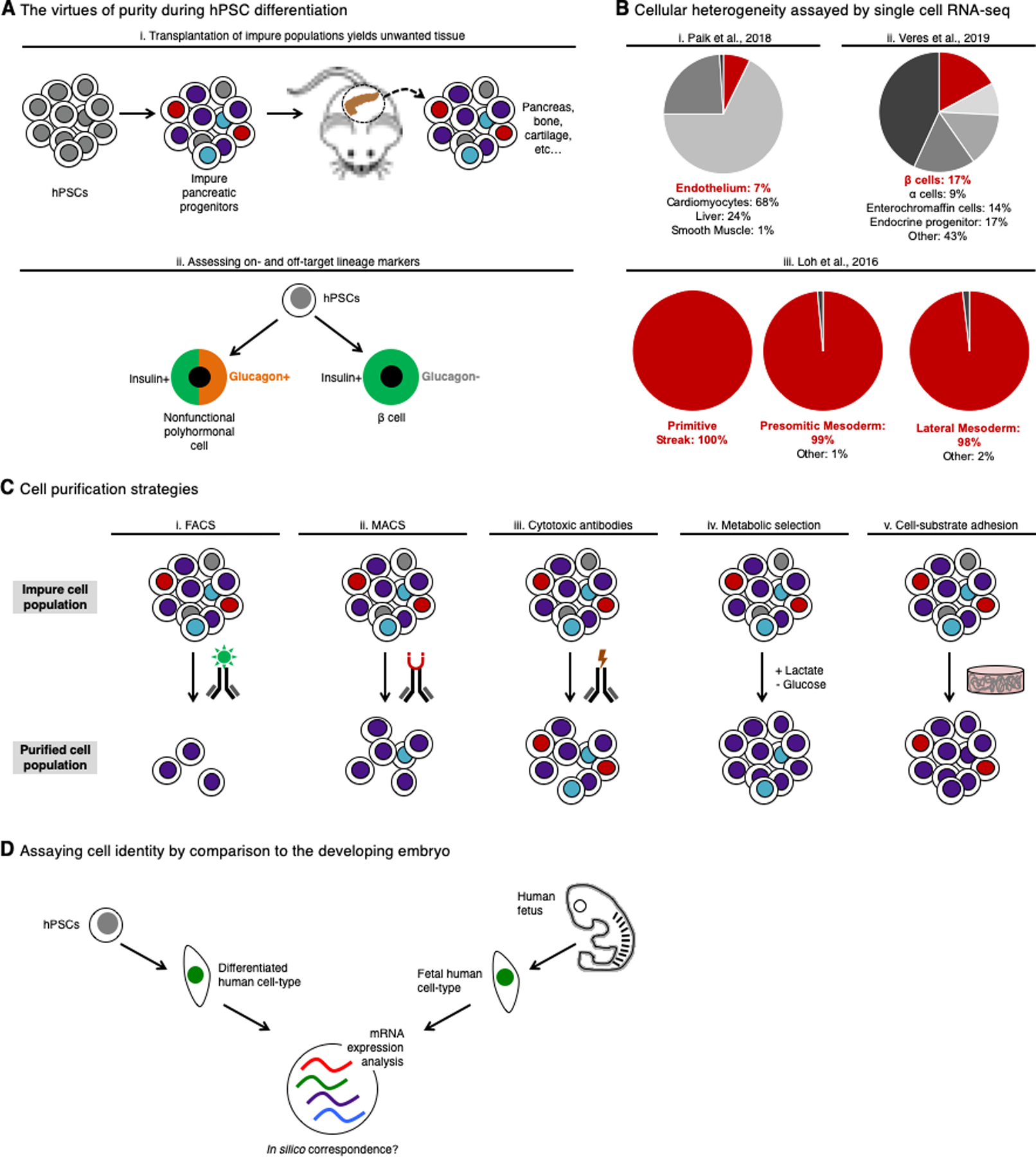
A. Transplantation of hPSC-derived heterogeneous populations containing a subset of pancreatic progenitors into rodent models yielded a variety of unwanted cell types including bone and cartilage (Kroon et al., 2008; Rezania et al., 2012; Rostovskaya et al., 2015) (i); at later differentiation stages, assessing the expression of both insulin (an “on-target” marker) and glucagon (an “off-target marker) allows for the distinction between nonfunctional polyhormonal cells and β-cells (ii) (Pagliuca et al., 2014; Rezania et al., 2014) (Russ et al., 2015)
B. Single-cell RNA-sequencing of hPSC-derived endothelial (Paik et al., 2018) (i), pancreatic (Veres et al., 2019) (ii), primitive streak (Loh et al., 2016) (iii), presomitic mesoderm (Loh et al., 2016) (iii) or lateral mesoderm (Loh et al., 2016) (iii) populations estimates the purity and the composition of the respective cultures; percentages and cell-type identities are reported here as indicated in each of the published papers
C. Enrichment of a particular cell-type from a heterogeneous cell population can be accomplished using: (i) FACS, which strictly purifies cell-types while adversely affecting cell yield and survival; (ii) MACS (magnetic activated cell sorting), which enriches for cell-types with lower purity, but maintains higher cell yield and survival; (iii) cytotoxic antibodies, which generally can deplete one unwanted cell type (e.g., hPSCs), but spare other contaminating cell-types; (iv) metabolic selection, which facilitates the selective growth of a desired cell-type while also potentially maintaining other contaminating cell types that share the same metabolic growth advantage; (v) and cell-substrate adhesion which provides favorable conditions for target cells to survive while some, but not all, contaminating cells die.
D. The developmental potential of early hPSC-derived cell-types might be tested by gene expression comparisons with the analogous cell-type derived from the human fetus
The identity and purity of hPSC-derived cell populations is often assessed by quantifying the percentage of cells expressing a “cell-type-specific” marker gene (or several such markers), but classifying cells in such a way inevitably piques questions about the choice and specificity of the marker gene(s). By way of example, while efforts to generate hPSC-derived β-cells routinely relied on INSULIN as a diagnostic marker, an incisive realization was that INSULIN+ cells could spuriously coexpress the α-cell marker GLUCAGON and that these “polyhormonal” cells did not constitute functional β-cells (reviewed by Shahjalal et al., 2018) (Fig. 5Aii). It is thus imperative to examine expression of unwanted lineage markers to ensure that they are not coexpressed with markers of the desired lineage. Taken together, the full battery of markers necessary to confidently assign cell-type identity is debatable.
Single-cell RNA-seq (scRNAseq)—which allows one to determine the coexpression of many genes in individual cells and to estimate the proportions of marker-positive cells—is becoming an objective measure of the composition and heterogeneity of differentiated populations (reviewed by Camp et al., 2018). For instance, scRNAseq analysis of hPSC-derived endothelial cell populations revealed that less than 7% of cells were endothelial cells, with the bulk of the culture resembling cardiomyocytes, liver cells or smooth muscle cells (Paik et al., 2018) (Fig. 5Bi). scRNAseq analysis of hPSC-derived pancreatic populations revealed that less than 20% of cells were β-cells, with other pancreatic cell-types predominating in the cultures (Veres et al., 2019) (Fig. 5Bii). scRNAseq analysis of hPSC-derived neural populations revealed that both forebrain and midbrain/hindbrain cell-types were simultaneously generated, and intriguingly, that differentiation was asynchronous, with both neural progenitors and neurons co-existing even at later stages of differentiation (Yao et al., 2016). In another instance, scRNAseq was used to quantify the purity of hPSC-derived primitive streak, presomitic mesoderm and lateral mesoderm, revealing them to be 96–100% pure with regard to expression of selected lineage markers (Loh et al., 2016) (Fig. 5Biii).
However, an oft-overlooked limitation of scRNAseq is that it is an imperfect measurement of a single cell’s transcriptome due to dropout: a technical limitation whereby lowly-expressed marker genes in a single cell spuriously evade detection (Kharchenko et al., 2014). Despite dropout, transcriptome-wide measurements of highly-expressed genes can be used to assign putative cell-types from scRNAseq data (Pollen et al., 2014), although the details of the clustering algorithm and other computational parameters can affect the number of “cell-types” identified in the population. Another drawback is that scRNAseq measures mRNA, not protein levels. Consequently scRNAseq is an estimation, but not a precise measurement, of cellular identity or purity.
Separating wheat from the chaff: cell purification strategies
A sobering realization is that even if directed differentiation protocols produce increasingly homogeneous cell populations from hPSCs, even a “99% pure” cell population may not be apropos for therapeutic transplantation. Even a frighteningly small number of undifferentiated hPSCs (10,000 cells) can form a tumor (a teratoma) upon transplantation (Lee et al., 2009). Certain hPSC-derived cell therapies may entail the transplantation of billions of cells into a given patient. To attain a desirable safety profile, by inference differentiation may have to exceed 99.99999% efficiency, which is difficult to fathom with extant directed differentiation schema (despite the developmental biology-based improvements detailed above).
Strategies to selectively purify a desired cell-type and to eliminate all traces of unwanted cell-types—especially undifferentiated hPSCs—from a heterogeneous population urgently warrant further exploration. Fluorescence-activated cell sorting (FACS) can yield highly pure populations but can be lengthy and strenuous on “sensitive” cell-types (e.g., hPSC-derived pancreatic progenitors) (Kelly et al., 2011) (Fig. 5Ci). By contrast, magnetic enrichment is faster and gentler, but can yield lower cell purities (Kelly et al., 2011) (Fig. 5Cii). Both of these strategies sort cells by virtue of their expression of surface markers and their efficacy is thus limited by the cell-type-specificity of the chosen markers; moreover, they entail cell dissociation and fundamentally rely on technologies that sort cells.
Recently, three alternate purification schema have emerged that do not entail the physical sorting of cells. First, treatment of heterogeneous differentiated populations with a cytotoxic anti-PODXL antibody efficiently lyses residual PODXL+ hPSCs, thus depleting hPSCs without recourse to cell sorting (Choo et al., 2008) (Fig. 5Ciii). Second, specialized media can be used to selectively ablate certain cell-types in culture by exploiting cell type-specific metabolic vulnerabilities. For instance, differentiation in lactate-supplemented media in the absence of glucose significantly enriches for hPSC-derived cardiomyocytes, as cardiomyocytes can efficiently utilize lactate whereas undifferentiated hPSCs and some other cell-types cannot (Tohyama et al., 2016; Tohyama et al., 2013) (Fig. 5Civ). Third, the culture substrate itself can be used to select for certain lineages—for instance, while neural cells can adhere to laminin-111, while hPSCs cannot (Kirkeby et al., 2017) (Fig. 5Cv). Cytotoxic antibodies, metabolic selection and cell-substrate adhesion represent means to enrich for desired cell-types without physical cell sorting, but have yet to be broadly applied to the enrichment of diverse cell-types.
Identifying the target: benchmarking hPSC-derived cell-types
Even if the field can generate a “pure” population of a given cell-type, how closely will hPSC-derived cell-types approximate their in vivo counterparts, as pertains to a combination of molecular and/or functional criteria?
To this end, we must benchmark hPSC-derived cell-types against freshly-derived primary human tissue. It is possible to obtain cells from mid- to late-gestation human embryos or from adult humans as comparators for terminally-differentiated cell-types from hPSCs. For instance, scRNAseq and other transcriptional assays have been used to directly compare neural cells isolated from a human fetus against their hPSC-derived counterparts (Kirkeby et al., 2012; La Manno et al., 2016; Pollen et al., 2019) (Fig. 5D).
While it is possible to directly compare more developmentally advanced cell-types in this way, molecularly benchmarking hPSC-derived germ layer or early tissue progenitors is currently impossible. It is technically and ethically infeasible to obtain their in vivo counterparts—which arise in weeks 2–4 of human embryonic development (O’Rahilly and Müller, 1987)—for molecular comparisons. Even if a given cell-type can be isolated from the human fetus or adult, any comparison against in vitro-derived cell-types will likely be imperfect, as the genetic background of the hPSC line under investigation will almost certainly differ from that of the tissue donor unless an isogenic hiPSC line was derived and used for differentiation. Comparisons matched in both genotype and developmental stage are logistically easier for mouse PSC-derived cell-types, as recently performed for in vivo- vs. in vitro-derived mouse motor neurons (Ichida et al., 2018).
But what degree of concordance would one expect to see between in vivo cell-types that have experienced a physiological environment by comparison to in vitro lineages mostly grown on plastic (Fig. 5D)? Even naïve mouse PSCs (an extremely well-characterized in vitro cell-type) have transcriptional differences by comparison to their in vivo counterparts within the pre-implantation blastocyst, mostly related to metabolism (Boroviak et al., 2014). This might reflect metabolic adaptation to cell culture and as such may be less significant. Nonetheless it raises an interesting precedent as to how precisely in vitro-differentiated counterparts (or, for that matter, any cultured cell) will resemble their presumed in vivo counterparts. Indeed scRNAseq recently revealed that while hPSC-derived substantia nigra dopaminergic neurons approximated their in vivo fetal counterparts, they also shared certain attributes with other mutually-exclusive lineages (La Manno et al., 2016). Expanding this scRNAseq analysis to additional hPSC-derived neural lineages revealed that in vitro-derived cell-types consistently displayed a perturbed metabolic signature entailing elevated glycolysis and an unfolded protein response by comparison to primary human fetal cell-types (Pollen et al., 2019).
Of course, it is likely that hPSC-derived lineages will never be a perfect replica of authentic cell-types in vivo; at best, they will constitute a facsimile. From a pragmatic point of view, minor transcriptional differences between in vivo- and in vitro-derived lineages may be inconsequential. The unresolved question is whether any discrepancies from the in vivo transcriptional program might compromise the functionality of differentiated cells. By way of example, current-generation hPSC-derived β-cells express cornerstone markers of β-cell identity, but minimally express transcription factors that delineate mature, adult β-cells (e.g., MAFA and SIX3). (Veres et al., 2019). It remains to be determined whether the lack of these “mature” β-cell transcription factors might underscore functional defects in hPSC-derived β-cells.
Dividends from diversity: deliberately generating a heterogenous cell population and what it means to be an “organoid”
While many in vitro differentiation strategies endeavor to create a homogeneous cell population, actual tissues are not homogeneous: rather, they comprise a rich diversity of cell-types. To imitate this diversity, some hPSC differentiation efforts intentionally generate a heterogeneous cell population, often in the form of 3-dimensional “organoids” (defined below) (Fig. 6A).
Figure 6: The merits of heterogeneous cell populations and their relationship to organoids.
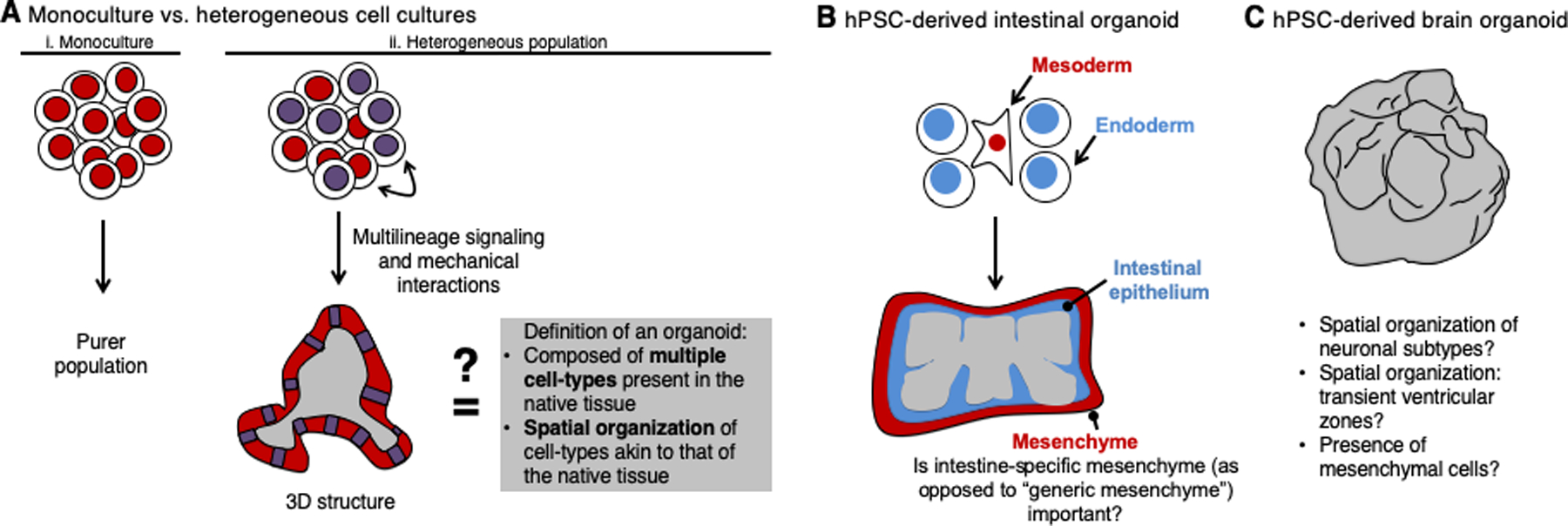
A. Differentiating hPSCs into a homogenous monoculture creates purer populations of a given cell-type (i), while differentiating cells in a heterogenous 3D culture provides different cell-types the opportunity to reciprocally signal, and mechanically interact, with one another (ii); however whether all 3D cultures meet the strict definition of an “organoid” (Lancaster and Knoblich, 2014) remains to be determined
B. Current-generation hPSC-derived intestinal organoids rely on the codifferentiation of endoderm and mesoderm derivatives to generate appropriate cellular diversity and spatial organization akin to the native intestine
C. Current-generation hPSC-derived brain organoids possess some key features of early brain development, but various questions remain
Cellular heterogeneity has its advantages: development entails reciprocal signaling between multiple cell-types that decide cellular fate in addition to mechanical interactions between multiple cell-types that fashion a tissue’s final shape (Fig. 6A). Hence, a single cell-type in isolation might be unlikely to fully develop in monoculture. This speaks to the importance of cellular heterogeneity in some contexts, as embodied by hPSC-derived intestinal and kidney organoids.
The intestine contains commingled epithelial and mesenchymal cell-types of endodermal and mesodermal origin, respectively. In vivo, intestinal mesenchyme is strictly required to instruct proper development of the adjacent intestinal epithelium, both by sculpting its morphogenetic shape and by serving as a source of critical extracellular signals (Roberts et al., 1998; Shyer et al., 2015). Thus efforts to differentiate hPSC-derived, 3-dimensional intestinal organoids deliberately produce a heterogeneous population of endoderm together with a subset of mesoderm at an early stage of differentiation, with the objective of generating intestinal epithelial cells as well as their ensconcing mesenchyme (Spence et al., 2011) (Fig. 6B). While it remains to be formally proven, it seems likely that coexistence of endodermal and mesodermal components is indispensable for the formation of spatially-complex intestinal organoids.
The kidney is also constructed from the spatial juxtaposition of two lineages—the uretic epithelium and metanephric mesenchyme—whose coalescence forms a ramified tree of collecting ducts decorated with nephrons. hPSC differentiation into a heterogeneous population comprising both uretic epithelium and metanephric mesenchyme eventually generates 3-dimensional kidney organoids with convoluted tubules resembling part of the kidney (Takasato et al., 2015). Again, the simultaneous generation of both uretic epithelium and metanephric mesenchyme in vitro harkens to the developmental co-existence of these cell-types and how reciprocal signaling between them is required for proper kidney formation in vivo.
With such remarkable progress in generating organoids, it is worth querying how closely they approximate actual tissues. This speaks to the deeper question of what is the precise definition of the term “organoid”? If, as their namesake implies, “organoids” are to be reminiscent of organs, we suggest that “organoids” meet two critical criteria. First, they should comprise multiple cell-types that are found in the native organ (Lancaster and Knoblich, 2014). Second, these cell-types should be spatially organized in a manner analogous to that of the native organ (Lancaster and Knoblich, 2014) (Fig. 6A).
We therefore urge caution that not all 3-dimensional cell cultures should be referred to as organoids: while many 3-dimensional cultures contain multiple cell-types, if they are not spatially organized analogous to the native tissue, they should not be given the moniker “organoid”. For instance, certain 3-dimensional differentiation strategies generate heterogeneous cell populations that do not show appropriate spatial organization and therefore are better classified as “spheroids” or “aggregates”. Such deviations from normal tissue architecture are fascinating and informative, as they suggest gaps in our understanding of tissue assembly and potential strategies for improvement. For instance, hPSC-derived brain organoids often harbor neurons radially organized around ventricular zone-like progenitor regions (Kadoshima et al., 2013; Lancaster et al., 2013; Pasca et al., 2015). However these ventricular zone regions have been reported to disperse after several weeks (Velasco et al., 2019) (Fig. 6C). Moreover the positions of TBR1+/SATB2+ deep and BRN2+ middle cortical layer neurons appears to be reversed in hPSC-derived brain organoids (Qian et al., 2016) (Fig. 6C). Therefore an improved understanding of cortical neuron subtype migration and/or cell sorting could be harnessed to rationally engineer better-organized organoids.
As a key step towards generating organoids, how can one simultaneously generate multiple cell-types in the same culture and control the relative frequencies of these cell-types? To meet this challenge, a recent method generated hPSC-derived liver precursors and then, at an intermediate step of differentiation, introduced defined numbers of generic endothelial cells (e.g., human umbilical vein endothelial cells) and generic mesenchymal cells to produce 3-dimensional cultures (Takebe et al., 2013; Takebe et al., 2017). Incorporation of endothelial and mesenchymal cells is an important step to engineer increasingly sophisticated organ simulacra.
However, such multilineage coculture strategies also bring their own attendant difficulties, because it has been recently appreciated that there are no “generic” endothelial cells and no “generic” mesenchymal cells in vivo. Instead, endothelium and mesenchyme adopt organ-specific subspecializations and considerably vary in their transcriptomes and functions across different organs (Han et al., 2018; Potente and Mäkinen, 2017). It is unclear whether “generic” endothelial and mesenchymal cells can productively interact with liver progenitors, or whether liver-specific endothelial cells and liver-specific mesenchymal cells might fare better for this application. The same applies for the aforementioned hPSC-derived intestinal organoids (Spence et al., 2011): while they clearly harbor mesenchyme, do these constitute intestinal-specific mesenchyme that might be uniquely suited to promote intestinal epithelium development (Fig. 6B)?
Finally, while cellular heterogeneity is integral to the very definition of organoids, excessive or unwanted heterogeneity may be deleterious. By way of example, scRNAseq has revealed the coexistence of dorsal forebrain, ventral forebrain, mesenchymal and other cell-types in hPSC-derived brain organoids (Camp et al., 2015; Pollen et al., 2019; Quadrato et al., 2017). Such scRNAseq analyses have revealed substantial variation between individual experiments and amongst organoids from the same experiment (Quadrato et al., 2017) or among organoids derived from different hPSC lines (Pollen et al., 2019), although other analyses suggested less variability (Velasco et al., 2019; Yoon et al., 2018). In any case, the presence of mesenchymal-like cells that express MYOSIN, COLLAGEN and DECORIN genes (Camp et al., 2015; Pollen et al., 2019; Quadrato et al., 2017) in current-generation brain organoids is unexpected (Fig. 6C). It remains to be determined whether these mesenchymal cells foster, or might occlude, proper brain development and the spatial organization of such organoids.
A human is not a mouse (or a frog, fish or chicken)
A common critique of current “developmentally-guided” hPSC differentiation efforts is that such strategies are not actually guided by human development; rather, they are informed by our knowledge of how model organisms such as mouse, frog, fish or chicken develop (as emphasized by this very review). Yet it is evident that a human is not a mouse. Human and mouse evolutionarily diverged tens of millions of years ago (Perlman, 2016), since which their exact mode of development—including the molecular identity of various embryonic progenitors and the signals that specify them—have diverged.
This issue of evolutionary differences is not simply theoretical; take for instance the very practical issue of how long differentiation should take. hPSC differentiation protocols often generate target cell-types in weeks or months, and this protracted time course is assumed to arise from, and parallel, the prolonged duration of human embryogenesis, which takes months in vivo (Tao and Zhang, 2016). Speaking to this point, hPSCs differentiating into a specific cell-type sometimes demonstrate exquisitely species-specific timings (Chu et al., 2019), which implies that relative slowness might be deeply and genetically ingrained into the process of hPSC differentiation. Yet in other cases, differentiation can be externally accelerated. As aforementioned, alternating activation and inhibition of WNT (and other signals) sequentially differentiates hPSCs into primitive streak, presomitic mesoderm, somites and finally dorsal somites within 4 days in vitro (Loh et al., 2016) (Fig. 3b). While this respects the developmental timing of the mouse embryo—wherein the same cell-types are produced over the course of 4 days in vivo—it is unclear why human cells have the capacity to differentiate along the “mouse” schedule when human development is ostensibly many times longer (O’Rahilly and Müller, 1987). Perhaps the changing extrinsic signals that induce these respective cell-types are provided by the human embryo over a protracted duration, but if they are delivered rapidly in quick succession, the pace of differentiation can be artificially accelerated. This point notwithstanding, timing is only one out of many possible divergences between human vs. model organism development: hence, analyses of human embryogenesis are direly needed to guide hPSC differentiation.
Frankly, the fundamental issue is that there is a paucity of knowledge surrounding early human embryonic development. As aforementioned, human germ layer specification, germ layer patterning, and organ progenitor specification unfolds in weeks 2–4 of human embryogenesis, when it is technically and ethically impossible to obtain human embryos for analysis (O’Rahilly and Müller, 1987). Heroic efforts have analyzed the gastrulating embryos of pig (Kobayashi et al., 2017) or monkeys (Sasaki et al., 2016), and in rare cases even somite-stage human embryos were obtained (Xi et al., 2017; Zhang et al., 2010). However, we still lack a precise understanding of how germ layer specification and patterning occurs in human embryos, although the core principles may be reminiscent of model organisms.
Recent work has revealed partial differences in the expression and function of genes between the human and mouse blastocyst (reviewed by Wamaitha and Niakan, 2018), raising the question of whether later developmental events might also diverge in significant ways. Indeed, while primordial germ cells in mouse express Sox2, their human counterparts do not express SOX2 but rather express the related family member SOX17 (Irie et al., 2014; Sasaki et al., 2015). This evolutionary divergence significantly impacts in vitro differentiation, as efforts to generate primordial germ cells from hPSCs leverage SOX17, but not SOX2, as a diagnostic marker for such cells (Irie et al., 2014; Sasaki et al., 2015). PAX6, a marker of early neural commitment, has been reported to have a different onset of expression in human versus mouse (Zhang et al., 2010). However whether most marker genes (including the ones listed above) show different expression patterns or functions in human versus mouse still awaits unambiguous confirmation owing to the dearth of appropriately stage-matched human embryos for analysis.
Future perspectives
On the face of it, the challenge of exclusively differentiating hPSCs into any one out of hundreds of potential fates seems bewildering. This task is evocative of how Odysseus too had to chart a convoluted path back home with incomplete knowledge of the landscape before him, sometimes accessing the correct waypoints and at other times narrowly avoiding side trips leading to unsavory destinations. Despite alternating progress over the past few decades, it is now possible to more effectively generate certain lineages from hPSCs, in part by drawing on the principles outlined above.
One emergent principle to enhance the efficiency of hPSC differentiation is to suppress the production of unwanted cell-types. This can be accomplished by logically blocking the extracellular signals that specify unwanted cell-types at each developmental lineage decision, thus coercing hPSCs towards a desired developmental end (Fig. 3A). A second issue is that as differentiating cells navigate successive lineage choices, the extracellular signals that induce a given cell-type change rapidly, even within 24 hours. The same signal can initially promote, and then repress, the formation of a given lineage. These signals must thus be manipulated with equal temporal dynamism to enable cells to segue between successive differentiation states (Fig. 3B). However the coming years will continue to bring ample challenges, four of which we detail below and others that we cannot foresee.
First, there is an ever-increasing proliferation of differentiation protocols to generate various cell-types. There will be a need for systematic side-by-side comparisons of these protocols across diverse hPSC lines to assess reproducibility and achieve standardization, as recently done for definitive endoderm specification (Rostovskaya et al., 2015).
Second, developmental roadmaps must be expanded to encompass new terra incognita. As this review has emphasized, we are still ignorant of the complete number and identity of steps through which pluripotent cells differentiate into most cell-types in vivo. Extant differentiation protocols may entail transition through some, but perhaps not all, of the requisite steps. While it may seem obvious that we must map the steps leading to desired lineages, to logically block the formation of unwanted cell-types from hPSCs (Fig. 3A), ironically we also have to map the routes and signals leading to these unwanted lineages as well. We also must understand, at the last steps of development, how fetal-like differentiated cell-types mature into fully-fledged, functional cell-types (Cohen and Melton, 2011). New high-throughput strategies to determine progenitor-progeny differentiation paths in the mouse embryo (Chan et al., 2019) could revolutionize this field.
Third, beyond measurement of several molecular markers, it will be important to develop new ways to benchmark hPSC-derived cell-types against their in vivo counterparts, especially entailing assays of physiologic functions. This review has emphasized transcriptional comparisons of hPSC-derived cell-types to their in vivo counterparts in the human fetus (Fig. 5D). However, orthotopic transplantation of hPSC-derived cell-types into non-human embryos (within technical and ethical limits) could constitute a new functional assay (Mascetti and Pedersen, 2016). Indeed, in utero transplantation of hPSC-derived neural crest cells into E8.5 mouse embryos led to engraftment in roughly one-quarter of mice (Cohen et al., 2016). However, it will not be trivial to precisely transplant hPSC-derived tissue progenitors into their counterpart tissues within the mouse embryo at most developmental times and places. Interspecific incompatibility could also obscure the ability of hPSC-derived tissue progenitors to develop in vivo: transplanted human cells may intrinsically differentiate at a different pace than the recipient non-human embryo or may not be able to respond to certain developmental signals emanating from it as well (Suchy et al., 2018).
Fourth, is it possible to generate hPSC-derived cultures that contain multiple cell-types and are also spatially organized in ways that approximate native organs? There is tremendous enthusiasm in creating hPSC-derived 3D multi-lineage cultures, but in certain cases, the spatial organization of their constituent cells and the reproducibility of such spatial organization warrants further attention. This is paramount to assess whether specific 3D cultures satisfy the complete definition of being “organoids” (Lancaster and Knoblich, 2014) (Fig. 6B). Or, if not, how can we reproducibly program their spatial organization? This is especially important because some definitions of “organoids” suggest that they must imitate some functions of their native organ (Lancaster and Knoblich, 2014). Adhering to the general biological principle that form prescribes function, it is likely that the correct spatial organization of multiple cell-types relative to one another is necessary for organs (or organoids) to execute many physiological functions. We therefore urgently require high-throughput methods to systematically map the spatial architecture of 3D cultures. This is currently accomplished using immunostaining; yet, analyses that entail cell dissociation (e.g., scRNAseq (Camp et al., 2018)) do not capture spatial information. However, for a given tissue section it is now possible to simultaneously image the expression of hundreds of different mRNAs with great breadth (across large fields of cells) and precision (at single-cell resolution) (Wang et al., 2018). Such technologies may enable the high-throughput interrogation of organoid architecture, thus enabling the optimization of future organoid differentiation protocols that lead to improved spatial organization.
Answers to these questions are of practical import. There are over 30 completed or ongoing clinical trials that have, or are, transplanting patients with hPSC-derived cell populations (Guhr et al., 2018). It stands to reason that the efficacy and safety of future cell replacement therapies will be considerably advanced by understanding, and further improving, the process of hPSC differentiation.
Acknowledgments
We apologize to all our developmental and stem-cell biology colleagues whose work we could not cite in this review. We are grateful for discussions with Renata Martin (on safety), Marius Wernig (on differentiation), James Wells (on heterogeneity) and Will Allen (on spatial organization). J.L.F. is supported by the National Defense Science and Engineering Graduate (NDSEG) and Stanford Honorary Bio-X Fellowships. L.T.A. and K.M.L. are supported by the California Institute for Regenerative Medicine (DISC2-10679 and DISC2-11105) and the Stanford-UC Berkeley Siebel Stem Cell Institute. K.M.L. is supported by the NIH Director’s Early Independence Award (DP5OD024558), Stanford Beckman Center, the Anonymous Family and as a Packard Foundation Fellow, Pew Scholar, Human Frontier Science Program Young Investigator (RGY0069/2019), Baxter Foundation Faculty Scholar and The Anthony DiGenova Endowed Faculty Scholar.
References
- Albini S, Coutinho P, Malecova B, Giordani L, Savchenko A, Forcales SV, and Puri PL (2013). Epigenetic reprogramming of human embryonic stem cells into skeletal muscle cells and generation of contractile myospheres. Cell Reports 3, 661–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang L, Tan A, Autio M, Goh S, Choo S, Lee K, Tan J, Pan B, Lee J, Lum J, et al. (2018). A roadmap for human liver differentiation from pluripotent stem cells. Cell Reports 22, 2190–2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelo JR, Guerrero-Zayas M-I, and Tremblay KD (2012). A fate map of the murine pancreas buds reveals a multipotent ventral foregut organ progenitor. PLoS ONE 7, e40707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arenas E, Denham M, and Villaescusa JC (2015). How to make a midbrain dopaminergic neuron. Development 142, 1918–1936. [DOI] [PubMed] [Google Scholar]
- Baxter AL (1977). B. B. Wilson’s “Destruction” of the Germ- Layer Theory. Isis 68, 363–374. [DOI] [PubMed] [Google Scholar]
- Bernardo AS, Faial T, Gardner L, Niakan KK, Ortmann D, Senner CE, Callery EM, Trotter MW, Hemberger M, Smith JC, et al. (2011). BRACHYURY and CDX2 mediate BMP-induced differentiation of human and mouse pluripotent stem cells into embryonic and extraembryonic lineages. Cell Stem Cell 9, 144–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blauwkamp TA, Nigam S, Ardehali R, Weissman IL, and Nusse R (2012). Endogenous Wnt signalling in human embryonic stem cells generates an equilibrium of distinct lineage-specified progenitors. Nat Commun 3, 1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boroviak T, Loos R, Bertone P, Smith A, and Nichols J (2014). The ability of inner-cell-mass cells to self-renew as embryonic stem cells is acquired following epiblast specification. Nature Cell Biology 16, 516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs JA, Li VC, Lee S, Woolf CJ, Klein A, and Kirschner MW (2017). Mouse embryonic stem cells can differentiate via multiple paths to the same state. eLife 6, e26945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge PW, Keller G, Gold JD, and Wu JC (2012). Production of De Novo Cardiomyocytes: Human Pluripotent Stem Cell Differentiation and Direct Reprogramming. Cell Stem Cell 10, 16–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp JG, Badsha F, Florio M, Kanton S, Gerber T, Wilsch-Bräuninger M, Lewitus E, Sykes A, Hevers W, Lancaster M, et al. (2015). Human cerebral organoids recapitulate gene expression programs of fetal neocortex development. Proceedings of the National Academy of Sciences of the United States of America 112, 15672–15677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camp JG, Wollny D, and Treutlein B (2018). Single-cell genomics to guide human stem cell and tissue engineering. Nature Methods, 1–7. [DOI] [PubMed] [Google Scholar]
- Chal J, Oginuma M, Al Tanoury Z, Gobert B, Sumara O, Hick A, Bousson F, Zidouni Y, Mursch C, Moncuquet P, et al. (2015). Differentiation of pluripotent stem cells to muscle fiber to model Duchenne muscular dystrophy. Nature Biotechnology 33, 962–969. [DOI] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, and Studer L (2009). Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 27, 275–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MM, Smith ZD, Grosswendt S, Kretzmer H, Norman TM, Adamson B, Jost M, Quinn JJ, Yang D, Jones MG, et al. (2019). Molecular recording of mammalian embryogenesis. Nature, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung C, Bernardo AS, Trotter MWB, Pedersen RA, and Sinha S (2012). Generation of human vascular smooth muscle subtypes provides insight into embryological origin-dependent disease susceptibility. Nat Biotechnol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choo AB, Tan HL, Ang SN, Fong WJ, Chin A, Lo J, Zheng L, Hentze H, Philp RJ, Oh SKW, et al. (2008). Selection against undifferentiated human embryonic stem cells by a cytotoxic antibody recognizing podocalyxin-like protein-1. Stem Cells 26, 1454–1463. [DOI] [PubMed] [Google Scholar]
- Chu L-F, Leng N, Zhang J, Hou Z, Mamott D, Vereide DT, Choi J, Kendziorski C, Stewart R, and Thomson JA (2016). Single-cell RNA-seq reveals novel regulators of human embryonic stem cell differentiation to definitive endoderm. Genome Biology 17, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu L-F, Mamott D, Ni Z, Bacher R, Liu C, Swanson S, Kendziorski C, Stewart R, and Thomson JA (2019). An In Vitro Human Segmentation Clock Model Derived from Embryonic Stem Cells. Cell Reports 28, 2247–2255. e2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung W-S, Shin CH, and Stainier DYR (2008). Bmp2 signaling regulates the hepatic versus pancreatic fate decision. Developmental cell 15, 738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen DE, and Melton D (2011). Turning straw into gold: directing cell fate for regenerative medicine. Nature Reviews Genetics. [DOI] [PubMed] [Google Scholar]
- Cohen MA, Wert KJ, Goldmann J, Markoulaki S, Buganim Y, Fu D, and Jaenisch R (2016). Human neural crest cells contribute to coat pigmentation in interspecies chimeras after in utero injection into mouse embryos. Proceedings of the National Academy of Sciences of the United States of America, 201525518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, and Baetge EE (2005). Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol 23, 1534–1541. [DOI] [PubMed] [Google Scholar]
- D’Amour KA, Bang AG, Eliazer S, Kelly OG, Agulnick AD, Smart NG, Moorman MA, Kroon E, Carpenter MK, and Baetge EE (2006). Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat Biotechnol 24, 1392–1401. [DOI] [PubMed] [Google Scholar]
- Davidson EH (2010). Emerging properties of animal gene regulatory networks. Nature 468, 911–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Robertis EM (2008). Evo-Devo: Variations on Ancestral Themes. Cell 132, 185–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denham M, Hasegawa K, Menheniott T, Rollo B, Zhang D, Hough S, Alshawaf A, Febbraro F, Ighaniyan S, Leung J, et al. (2015). Multipotent caudal neural progenitors derived from human pluripotent stem cells that give rise to lineages of the central and peripheral nervous system. STEM CELLS 33, 1759–1770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elcheva I, Brok-Volchanskaya V, Kumar A, Liu P, Lee J-H, Tong L, Vodyanik M, Swanson S, Stewart R, Kyba M, et al. (2014). Direct induction of haematoendothelial programs in human pluripotent stem cells by transcriptional regulators. Nat Commun 5, 4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fasano CA, Chambers SM, Lee G, Tomishima MJ, and Studer L (2010). Efficient derivation of functional floor plate tissue from human embryonic stem cells. Cell Stem Cell 6, 336–347. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gadue P, Huber TL, Paddison PJ, and Keller GM (2006). Wnt and TGF-β signaling are required for the induction of an in vitro model of primitive streak formation using embryonic stem cells. Proc Natl Acad Sci USA 103, 16806–16811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganat YM, Calder EL, Kriks S, Nelander J, Tu EY, Jia F, Battista D, Harrison N, Parmar M, Tomishima MJ, et al. (2012). Identification of embryonic stem cell-derived midbrain dopaminergic neurons for engraftment. Journal of Clinical Investigation 122, 2928–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gertow K, Hirst CE, Yu QC, Ng ES, Pereira LA, Davis RP, Stanley EG, and Elefanty AG (2013). WNT3A Promotes Hematopoietic or Mesenchymal Differentiation from hESCs Depending on the Time of Exposure. Stem Cell Reports 1, 53–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouti M, Tsakiridis A, Wymeersch FJ, Huang Y, Kleinjung J, Wilson V, and Briscoe J (2014). In Vitro Generation of Neuromesodermal Progenitors Reveals Distinct Roles for Wnt Signalling in the Specification of Spinal Cord and Paraxial Mesoderm Identity. PLoS Biology 12, e1001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf T, and Enver T (2009). Forcing cells to change lineages. Nature 462, 587–594. [DOI] [PubMed] [Google Scholar]
- Grapin-Botton A (2005). Antero-posterior patterning of the vertebrate digestive tract: 40 years after Nicole Le Douarin’s PhD thesis. Int J Dev Biol 49, 335–347. [DOI] [PubMed] [Google Scholar]
- Green MD, Chen A, Nostro M-C, D’Souza SL, Schaniel C, Lemischka IR, Gouon-Evans V, Keller G, and Snoeck H-W (2011). Generation of anterior foregut endoderm from human embryonic and induced pluripotent stem cells. Nat Biotechnol. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guhr A, Kobold S, Seltmann S, Wulczyn AEMS, Kurtz A, and LOser P (2018). Recent Trends in Research with Human Pluripotent Stem Cells: Impact of Research and Use of Cell Lines in Experimental Research and Clinical Trials. Stem Cell Reports 11, 485–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Wang R, Zhou Y, Fei L, Sun H, Lai S, Saadatpour A, Zhou Z, Chen H, Ye F, et al. (2018). Mapping the Mouse Cell Atlas by Microwell-Seq. Cell 172, 1091–1107. e1017. [DOI] [PubMed] [Google Scholar]
- Haridass D, Yuan Q, Becker PD, Cantz T, Iken M, Rothe M, Narain N, Bock M, Nörder M, Legrand N, et al. (2010). Repopulation Efficiencies of Adult Hepatocytes, Fetal Liver Progenitor Cells, and Embryonic Stem Cell- Derived Hepatic Cells in Albumin-Promoter-Enhancer Urokinase-Type Plasminogen Activator Mice. The American Journal of Pathology 175, 1483–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrique D, Abranches E, Verrier L, and Storey KG (2015). Neuromesodermal progenitors and the making of the spinal cord. Development 142, 2864–2875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichida JK, Staats KA, Davis-Dusenbery BN, Clement K, Galloway KE, Babos KN, Shi Y, Son EY, Kiskinis E, Atwater N, et al. (2018). Comparative genomic analysis of embryonic, lineage-converted and stem cell-derived motor neurons. Development 145, dev168617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irie N, Weinberger L, Tang WWC, Kobayashi T, Viukov S, Manor YS, Dietmann S, Hanna JH, and Surani MA (2014). SOX17 Is a Critical Specifier of Human Primordial Germ Cell Fate. Cell, 1–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwafuchi-Doi M, and Zaret KS (2016). Cell fate control by pioneer transcription factors. Development 143, 1833–1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadoshima T, Sakaguchi H, Nakano T, Soen M, Ando S, Eiraku M, and Sasai Y (2013). Self-organization of axial polarity, inside-out layer pattern, and species-specific progenitor dynamics in human ES cell-derived neocortex. Proceedings of the National Academy of Sciences of the United States of America 110, 20284–20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly OG, Chan MY, Martinson LA, Kadoya K, Ostertag TM, Ross KG, Richardson M, Carpenter MK, D’amour KA, Kroon E, et al. (2011). Cell-surface markers for the isolation of pancreatic cell types derived from human embryonic stem cells. Nat Biotechnol 29, 750–756. [DOI] [PubMed] [Google Scholar]
- Kharchenko PV, Silberstein L, and Scadden DT (2014). Bayesian approach to single-cell differential expression analysis. Nature Methods 11, 740–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkeby A, Grealish S, Wolf DA, Nelander J, Wood J, Lundblad M, Lindvall O, and Parmar M (2012). Generation of regionally specified neural progenitors and functional neurons from human embryonic stem cells under defined conditions. Cell Reports 1, 703–714. [DOI] [PubMed] [Google Scholar]
- Kirkeby A, Nolbrant S, Tiklova K, Heuer A, Kee N, Cardoso T, Ottosson DR, Lelos MJ, Rifes P, Dunnett SB, et al. (2017). Predictive Markers Guide Differentiation to Improve Graft Outcome in Clinical Translation of hESC-Based Therapy for Parkinson’s Disease. Cell Stem Cell 20, 135–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Zhang H, Tang WWC, Irie N, Withey S, Klisch D, Sybirna A, Dietmann S, Contreras DA, Webb R, et al. (2017). Principles of early human development and germ cell program from conserved model systems. Nature, 1–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriks S, Shim J-W, Piao J, Ganat YM, Wakeman DR, Xie Z, Carrillo-Reid L, Auyeung G, Antonacci C, Buch A, et al. (2011). Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon E, Martinson LA, Kadoya K, Bang AG, Kelly OG, Eliazer S, Young H, Richardson M, Smart NG, Cunningham J, et al. (2008). Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat Biotechnol 26, 443–452. [DOI] [PubMed] [Google Scholar]
- Kwon GS, Viotti M, and Hadjantonakis A-K (2008). The endoderm of the mouse embryo arises by dynamic widespread intercalation of embryonic and extraembryonic lineages. Developmental Cell 15, 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyba M (2016). Mesoderm, Cooked Up Fast and Served to Order. Cell Stem Cell 19, 146–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Gyllborg D, Codeluppi S, Nishimura K, Salto C, Zeisel A, Borm LE, Stott SRW, Toledo EM, Villaescusa JC, et al. (2016). Molecular Diversity of Midbrain Development in Mouse, Human, and Stem Cells. Cell 167, 566–580. e519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, Chen X, Taipale J, Hughes TR, and Weirauch MT (2018). The Human Transcription Factors. Cell 172, 650–665. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, and Knoblich JA (2014). Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 345, 1247125–1247125. [DOI] [PubMed] [Google Scholar]
- Lancaster MA, Renner M, Martin C-A, Wenzel D, Bicknell LS, Hurles ME, Homfray T, Penninger JM, Jackson AP, and Knoblich JA (2013). Cerebral organoids model human brain development and microcephaly. Nature 501, 373–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti E, and Göttgens B (2018). From haematopoietic stem cells to complex differentiation landscapes. Nature 553, 418–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson KA, Meneses JJ, and Pedersen RA (1991). Clonal analysis of epiblast fate during germ layer formation in the mouse embryo. Development 113, 891–911. [DOI] [PubMed] [Google Scholar]
- Lee AS, Tang C, Cao F, Xie X, van der Bogt K, Hwang A, Connolly AJ, Robbins RC, and Wu JC (2009). Effects of cell number on teratoma formation by human embryonic stem cells. Cell Cycle 8, 2608–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levenstein ME, Ludwig TE, Xu R-H, Llanas RA, VanDenHeuvel-Kramer K, Manning D, and Thomson JA (2006). Basic fibroblast growth factor support of human embryonic stem cell self-renewal. Stem Cells 24, 568–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QV, Dixon G, Verma N, Rosen BP, Gordillo M, Luo R, Xu C, Wang Q, Soh C-L, Yang D, et al. (2019). Genome-scale screens identify JNK-JUN signaling as a barrier for pluripotency exit and endoderm differentiation. Nature Genetics 33, 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippmann ES, Williams CE, Ruhl DA, Estevez-Silva MC, Chapman ER, Coon JJ, and Ashton RS (2015). Deterministic HOX patterning in human pluripotent stem cell-derived neuroectoderm. Stem Cell Reports 4, 632–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh KM, Ang LT, Zhang J, Kumar V, Ang J, Auyeong JQ, Lee KL, Choo SH, Lim CYY, Nichane M, et al. (2014). Efficient Endoderm Induction from Human Pluripotent Stem Cells by Logically Directing Signals Controlling Lineage Bifurcations. Cell Stem Cell 14, 237–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh KM, Chen A, Koh PW, Deng TZ, Sinha R, Tsai JM, Barkal AA, Shen KY, Jain R, Morganti RM, et al. (2016). Mapping the Pairwise Choices Leading from Pluripotency to Human Bone, Heart, and Other Mesoderm Cell Types. Cell 166, 451–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lumsden A, and Krumlauf R (1996). Patterning the vertebrate neuraxis. Science 274, 1109–1115. [DOI] [PubMed] [Google Scholar]
- Mascetti VL, and Pedersen RA (2016). Contributions of Mammalian Chimeras to Pluripotent Stem Cell Research. Cell Stem Cell 19, 163–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKnight K, Wang P, and Kim SK (2010). Deconstructing pancreas development to reconstruct human islets from pluripotent stem cells. Cell Stem Cell 6, 300–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendjan S, Mascetti VL, Ortmann D, Ortiz M, Karjosukarso DW, Ng Y, Moreau T, and Pedersen RA (2014). NANOG and CDX2 Pattern Distinct Subtypes of Human Mesoderm during Exit from Pluripotency. Cell Stem Cell. [DOI] [PubMed] [Google Scholar]
- Metzis V, Steinhauser S, Pakanavicius E, Gouti M, Stamataki D, Ivanovitch K, Watson T, Rayon T, Mousavy Gharavy SN, Lovell-Badge R, et al. (2018). Nervous System Regionalization Entails Axial Allocation before Neural Differentiation. Cell 175, 1105–1118. e1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau T, Evans AL, Vasquez L, Tijssen MR, Yan Y, Trotter MW, Howard D, Colzani M, Arumugam M, Wu WH, et al. (2016). Large-scale production of megakaryocytes from human pluripotent stem cells by chemically defined forward programming. Nature Communications 7, 11208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murry CE, and Keller G (2008). Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 132, 661–680. [DOI] [PubMed] [Google Scholar]
- Nehme R, Zuccaro E, Ghosh SD, Li C, Sherwood JL, Pietilainen O, Barrett LE, Limone F, Worringer KA, Kommineni S, et al. (2018). Combining NGN2 Programming with Developmental Patterning Generates Human Excitatory Neurons with NMDAR-Mediated Synaptic Transmission. Cell Reports 23, 2509–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowotschin S, Setty M, Kuo Y-Y, Liu V, Garg V, Sharma R, Simon CS, Saiz N, Gardner R, Boutet SC, et al. (2019). The emergent landscape of the mouse gut endoderm at single-cell resolution. Nature 569, 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rahilly R, and Müller F (1987). Developmental stages in human embryos (Baltimore, Maryland: Carnegie Institute of Washington; ). [Google Scholar]
- Osafune K, Caron L, Borowiak M, Martinez RJ, Fitz-Gerald CS, Sato Y, Cowan CA, Chien KR, and Melton DA (2008). Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol 26, 313–315. [DOI] [PubMed] [Google Scholar]
- Pagliuca FW, Millman JR, Gürtler M, Segel M, Van Dervort A, Ryu JH, Peterson QP, Greiner D, and Melton DA (2014). Generation of functional human pancreatic β cells in vitro. Cell 159, 428–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paik DT, Tian L, Lee J, Sayed N, Chen IY, Rhee S, Rhee J-W, Kim Y, Wirka RC, Buikema JW, et al. (2018). Large-Scale Single-Cell RNA-Seq Reveals Molecular Signatures of Heterogeneous Populations of Human Induced Pluripotent Stem Cell-Derived Endothelial Cells. Circulation Research. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan FC, and Wright C (2011). Pancreas organogenesis: from bud to plexus to gland. Dev Dyn 240, 530–565. [DOI] [PubMed] [Google Scholar]
- Park I-H, Zhao R, West JA, Yabuuchi A, Huo H, Ince TA, Lerou PH, Lensch MW, and Daley GQ (2008). Reprogramming of human somatic cells to pluripotency with defined factors. Nature 451, 141–146. [DOI] [PubMed] [Google Scholar]
- Pasca AM, Sloan SA, Clarke LE, Tian Y, Makinson CD, Huber N, Kim CH, Park J-Y, O’Rourke NA, Nguyen KD, et al. (2015). Functional cortical neurons and astrocytes from human pluripotent stem cells in 3D culture. Nature Methods 12, 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski M, Ortmann D, Bertero A, Tavares JM, Pedersen RA, Vallier L, and Kotter MRN (2017). Inducible and Deterministic Forward Programming of Human Pluripotent Stem Cells into Neurons, Skeletal Myocytes, and Oligodendrocytes. Stem Cell Reports, 1–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlman RL (2016). Mouse models of human disease: An evolutionary perspective. Evolution, medicine, and public health 2016, 170–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Bhaduri A, Andrews MG, Nowakowski TJ, Meyerson OS, Mostajo-Radji MA, Di Lullo E, Alvarado B, Bedolli M, Dougherty ML, et al. (2019). Establishing Cerebral Organoids as Models of Human-Specific Brain Evolution. Cell 176, 743–756. e717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Shuga J, Wang X, Leyrat AA, Lui JH, Li N, Szpankowski L, Fowler B, Chen P, et al. (2014). Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nature Biotechnology 32, 1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potente M, and Mäkinen T (2017). Vascular heterogeneity and specialization in development and disease. Nature Reviews Molecular Cell Biology 18, 477–494. [DOI] [PubMed] [Google Scholar]
- Qian X, Nguyen HN, Song MM, Hadiono C, Ogden SC, Hammack C, Yao B, Hamersky GR, Jacob F, Zhong C, et al. (2016). Brain-Region-Specific Organoids Using Mini- bioreactors for Modeling ZIKV Exposure. Cell 165, 1238–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadrato G, Nguyen T, Macosko EZ, Sherwood JL, Min Yang S, Berger DR, Maria N, Scholvin J, Goldman M, Kinney JP, et al. (2017). Cell diversity and network dynamics in photosensitive human brain organoids. Nature 18, 736–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankin SA, McCracken KW, Luedeke DM, Han L, Wells JM, Shannon JM, and Zorn AM (2017). Timing is everything: Reiterative Wnt, BMP and RA signaling regulate developmental competence during endoderm organogenesis. Developmental Biology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao J, and Greber B (2016). Concise Review: Signaling Control of Early Fate Decisions Around the Human Pluripotent Stem Cell State. STEM CELLS. [DOI] [PubMed] [Google Scholar]
- Rao J, Pfeiffer MJ, Frank S, Adachi K, Piccini I, Quaranta R, Araúzo-Bravo M, Schwarz J, Schade D, Leidel S, et al. (2015). Stepwise Clearance of Repressive Roadblocks Drives Cardiac Induction in Human ESCs. Cell Stem Cell. [DOI] [PubMed] [Google Scholar]
- Rezania A, Bruin JE, Arora P, Rubin A, Batushansky I, Asadi A, O’Dwyer S, Quiskamp N, Mojibian M, Albrecht T, et al. (2014). Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nature Biotechnology 32, 1121–1133. [DOI] [PubMed] [Google Scholar]
- Rezania A, Bruin JE, Riedel MJ, Mojibian M, Asadi A, Xu J, Gauvin R, Narayan K, Karanu F, O’Neil JJ, et al. (2012). Maturation of human embryonic stem cell-derived pancreatic progenitors into functional islets capable of treating pre-existing diabetes in mice. Diabetes. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Pérez JA, and Magnuson T (2005). Primitive streak formation in mice is preceded by localized activation of Brachyury and Wnt3. Developmental Biology 288, 363–371. [DOI] [PubMed] [Google Scholar]
- Robb L, Hartley L, Begley CG, Brodnicki TC, Copeland NG, Gilbert DJ, Jenkins NA, and Elefanty AG (2000). Cloning, expression analysis, and chromosomal localization of murine and human homologues of a Xenopus mix gene. Dev Dyn 219, 497–504. [DOI] [PubMed] [Google Scholar]
- Roberts DJ, Smith DM, Goff DJ, and Tabin CJ (1998). Epithelial-mesenchymal signaling during the regionalization of the chick gut. Development 125, 2791–2801. [DOI] [PubMed] [Google Scholar]
- Rosenquist GC (1970). Location and movements of cardiogenic cells in the chick embryo: the heart-forming portion of the primitive streak. Developmental Biology 22, 461–475. [DOI] [PubMed] [Google Scholar]
- Rostovskaya M, Bredenkamp N, and Smith A (2015). Towards consistent generation of pancreatic lineage progenitors from human pluripotent stem cells. Philosophical transactions of the Royal Society of London Series B, Biological sciences 370, 20140365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russ HA, Parent AV, Ringler JJ, Hennings TG, Nair GG, Shveygert M, Guo T, Puri S, Haataja L, Cirulli V, et al. (2015). Controlled induction of human pancreatic progenitors produces functional beta-like cells in vitro. The EMBO Journal 34, 1759–1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sances S, Bruijn LI, Chandran S, Eggan K, Ho R, Klim JR, Livesey MR, Lowry E, Macklis JD, Rushton D, et al. (2016). Modeling ALS with motor neurons derived from human induced pluripotent stem cells. Nature Neuroscience 16, 542–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K, Nakamura T, Okamoto I, Yabuta Y, Iwatani C, Tsuchiya H, Seita Y, Nakamura S, Shiraki N, Takakuwa T, et al. (2016). The Germ Cell Fate of Cynomolgus Monkeys Is Specified in the Nascent Amnion. Developmental Cell 39, 169–185. [DOI] [PubMed] [Google Scholar]
- Sasaki K, Yokobayashi S, Nakamura T, Okamoto I, Yabuta Y, Kurimoto K, Ohta H, Moritoki Y, Iwatani C, Tsuchiya H, et al. (2015). Robust In Vitro Induction of Human Germ Cell Fate from Pluripotent Stem Cells. Cell Stem Cell 17, 178–194. [DOI] [PubMed] [Google Scholar]
- Shahjalal HM, Abdal Dayem A, Lim KM, Jeon T-I, and Cho S-G (2018). Generation of pancreatic β cells for treatment of diabetes: advances and challenges. Stem Cell Research & Therapy 9, 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Chen T-YA, and Melton DA (2009). Transcriptional dynamics of endodermal organ formation. Dev Dyn 238, 29–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherwood RI, Maehr R, Mazzoni EO, and Melton DA (2011). Wnt signaling specifies and patterns intestinal endoderm. Mech Dev 128, 387–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyer AE, Huycke TR, Lee C, Mahadevan L, and Tabin CJ (2015). Bending Gradients: How the Intestinal Stem Cell Gets Its Home. Cell 161, 569–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JR, Lange AW, Lin S-CJ, Kaestner KH, Lowy AM, Kim I, Whitsett JA, and Wells JM (2009). Sox17 regulates organ lineage segregation of ventral foregut progenitor cells. Developmental Cell 17, 62–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JR, Mayhew CN, Rankin SA, Kuhar MF, Vallance JE, Tolle K, Hoskins EE, Kalinichenko VV, Wells SI, Zorn AM, et al. (2011). Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470, 105–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suchy F, Yamaguchi T, and Nakauchi H (2018). iPSC-Derived Organs In Vivo: Challenges and Promise. Cell Stem Cell 22, 21–24. [DOI] [PubMed] [Google Scholar]
- Sumi T, Tsuneyoshi N, Nakatsuji N, and Suemori H (2008). Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical Wnt/β-catenin, Activin/Nodal and BMP signaling. Development 135, 2969–2979. [DOI] [PubMed] [Google Scholar]
- Tabar V, and Studer L (2014). Pluripotent stem cells in regenerative medicine: challenges and recent progress. Nature Reviews Genetics 15, 82–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, and Yamanaka S (2007). Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131, 861–872. [DOI] [PubMed] [Google Scholar]
- Takasato M, Er PX, Becroft M, Vanslambrouck JM, Stanley EG, Elefanty AG, and Little MH (2014). Directing human embryonic stem cell differentiation towards a renal lineage generates a self-organizing kidney. Nature Cell Biology 16, 118–126. [DOI] [PubMed] [Google Scholar]
- Takasato M, Er PX, Chiu HS, Maier B, Baillie GJ, Ferguson C, Parton RG, Wolvetang EJ, Roost MS, Chuva de Sousa Lopes SM, et al. (2015). Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526, 564–568. [DOI] [PubMed] [Google Scholar]
- Takebe T, Sekine K, Enomura M, Koike H, Kimura M, Ogaeri T, Zhang R-R, Ueno Y, Zheng Y-W, Koike N, et al. (2013). Vascularized and functional human liver from an iPSC-derived organ bud transplant. Nature 499, 481–484. [DOI] [PubMed] [Google Scholar]
- Takebe T, Sekine K, Kimura M, Yoshizawa E, Ayano S, Koido M, Funayama S, Nakanishi N, Hisai T, Kobayashi T, et al. (2017). Massive and Reproducible Production of Liver Buds Entirely from Human Pluripotent Stem Cells. Cell Reports 21, 2661–2670. [DOI] [PubMed] [Google Scholar]
- Takemoto T, Uchikawa M, Yoshida M, Bell DM, Lovell-Badge R, Papaioannou VE, and Kondoh H (2011). Tbx6-dependent Sox2 regulation determines neural or mesodermal fate in axial stem cells. Nature 470, 394–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam PP, and Beddington RS (1987). The formation of mesodermal tissues in the mouse embryo during gastrulation and early organogenesis. Development 99, 109–126. [DOI] [PubMed] [Google Scholar]
- Tao Y, and Zhang S-C (2016). Neural Subtype Specification from Human Pluripotent Stem Cells. Cell Stem Cell 19, 573–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, and Jones JM (1998). Embryonic stem cell lines derived from human blastocysts. Science 282, 1145–1147. [DOI] [PubMed] [Google Scholar]
- Tohyama S, Fujita J, Hishiki T, Matsuura T, Hattori F, Ohno R, Kanazawa H, Seki T, Nakajima K, Kishino Y, et al. (2016). Glutamine Oxidation Is Indispensable for Survival of Human Pluripotent Stem Cells. Cell Metabolism 23, 663–674. [DOI] [PubMed] [Google Scholar]
- Tohyama S, Hattori F, Sano M, Hishiki T, Nagahata Y, Matsuura T, Hashimoto H, Suzuki T, Yamashita H, Satoh Y, et al. (2013). Distinct metabolic flow enables large-scale purification of mouse and human pluripotent stem cell-derived cardiomyocytes. Cell Stem Cell 12, 127–137. [DOI] [PubMed] [Google Scholar]
- Tzouanacou E, Wegener A, Wymeersch FJ, Wilson V, and Nicolas J-F (2009). Redefining the progression of lineage segregations during mammalian embryogenesis by clonal analysis. Developmental Cell 17, 365–376. [DOI] [PubMed] [Google Scholar]
- Umeda K, Zhao J, Simmons P, Stanley E, Elefanty A, and Nakayama N (2012). Human chondrogenic paraxial mesoderm, directed specification and prospective isolation from pluripotent stem cells. Sci Rep 2, 455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco S, Kedaigle AJ, Simmons SK, Nash A, Rocha M, Quadrato G, Paulsen B, Nguyen L, Adiconis X, Regev A, et al. (2019). Individual brain organoids reproducibly form cell diversity of the human cerebral cortex. Nature, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veres A, Faust AL, Bushnell HL, Engquist EN, Kenty JH-R, Harb G, Poh Y-C, Sintov E, Gürtler M, Pagliuca FW, et al. (2019). Charting cellular identity during human in vitro β-cell differentiation. Nature 569, 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verrier L, Davidson L, Gierliński M, Dady A, and Storey KG (2018). Neural differentiation, selection and transcriptomic profiling of human neuromesodermal progenitor-like cells in vitro. Development 145, dev166215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waddington CH (1940). Organisers and Genes (Cambridge, UK: Cambridge University Press; ). [Google Scholar]
- Wagner DE, Weinreb C, Collins ZM, Briggs JA, Megason SG, and Klein AM (2018). Single-cell mapping of gene expression landscapes and lineage in the zebrafish embryo. Science 360, 981–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wamaitha SE, and Niakan KK (2018). Human Pre-gastrulation Development. Current topics in developmental biology 128, 295–338. [DOI] [PubMed] [Google Scholar]
- Wandzioch E, and Zaret KS (2009). Dynamic signaling network for the specification of embryonic pancreas and liver progenitors. Science 324, 1707–1710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Allen WE, Wright MA, Sylwestrak EL, Samusik N, Vesuna S, Evans K, Liu C, Ramakrishnan C, Liu J, et al. (2018). Three-dimensional intact-tissue sequencing of single-cell transcriptional states. Science, eaat5691–5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren L, Manos PD, Ahfeldt T, Loh Y-H, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, et al. (2010). Highly Efficient Reprogramming to Pluripotency and Directed Differentiation of Human Cells with Synthetic Modified mRNA. Cell Stem Cell, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterle H, Lieberam I, Porter JA, and Jessell TM (2002). Directed differentiation of embryonic stem cells into motor neurons. Cell 110, 385–397. [DOI] [PubMed] [Google Scholar]
- Xi H, Fujiwara W, Gonzalez K, Jan M, Liebscher S, Van Handel B, Schenke-Layland K, and Pyle AD (2017). In Vivo Human Somitogenesis Guides Somite Development from hPSCs. Cell Reports 18, 1573–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao Z, Mich JK, Ku S, Menon V, Krostag A-R, Martinez RA, Furchtgott L, Mulholland H, Bort S, Fuqua MA, et al. (2016). A Single-Cell Roadmap of Lineage Bifurcation in Human ESC Models of Embryonic Brain Development. Cell Stem Cell, 1–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon S-J, Elahi LS, Pasca AM, Marton RM, Gordon A, Revah O, Miura Y, Walczak EM, Holdgate GM, Fan HC, et al. (2018). Reliability of human cortical organoid generation. Nature Methods, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, Nie J, Jonsdottir GA, Ruotti V, Stewart R, et al. (2007). Induced pluripotent stem cell lines derived from human somatic cells. Science 318, 1917–1920. [DOI] [PubMed] [Google Scholar]
- Yu P, Pan G, Yu J, and Thomson JA (2011). FGF2 sustains Nanog and switches the outcome of BMP4-induced human embryonic stem cell differentiation. Cell Stem Cell 8, 326–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Huang CT, Chen J, Pankratz MT, Xi J, Li J, Yang Y, Lavaute TM, Li X-J, Ayala M, et al. (2010). Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell 7, 90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Pak C, Han Y, Ahlenius H, Zhang Z, Chanda S, Marro S, Patzke C, Acuna C, Covy J, et al. (2013). Rapid Single-Step Induction of Functional Neurons from Human Pluripotent Stem Cells. Neuron 78, 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorn AM, and Wells JM (2009). Vertebrate endoderm development and organ formation. Annu Rev Cell Dev Biol 25, 221–251. [DOI] [PMC free article] [PubMed] [Google Scholar]


