Abstract
The error-prone replication and life cycle of influenza virus generate a diverse set of genetic variants. Transmission between hosts strictly limits both the number of virus particles and the genetic diversity of virus variants that reach a new host and establish an infection. This sharp reduction in the virus population at transmission––the transmission bottleneck––is significant to the evolution of influenza virus and to its epidemic and pandemic potential. This review describes transmission bottlenecks and their effect on the diversity and evolution of influenza virus. It also reviews the methods for calculating and predicting bottleneck sizes and highlights the host and viral determinants of influenza transmissibility.
The transmission and establishment of an infection in a new host are critical steps in the life cycle of a virus. The host infected by a virus is typically as significant as the virus itself. Human respiratory viruses—notably, influenza virus—are passed on by respiratory droplets, aerosols, or direct contact, unlike arboviruses and most plant viruses for which insects serve as transmission vectors (Weber and Stilianakis 2008; Whitfield et al. 2015; Gallet et al. 2018; Huang et al. 2019; Lefeuvre et al. 2019). The transmission event precipitates a sharp reduction in the size of the virus population from the donor to the recipient host, decreasing overall genetic diversity of the founding population in the recipient. The number and types of virus variants that gain access to the new host determine not only whether an active infection will occur in the recipient but also the rate of virus adaptation. Quantifying the number of virus particles transmitted between donors and recipients—the transmission bottleneck size—provides a measure of the adaptation potential of a virus to new hosts and is therefore a focal point in this field of influenza virus research.
Quantifying bottleneck sizes for influenza virus is, however, a difficult task, as it can rely on noisy short-read, next-generation sequencing data and statistical models that can only track viruses by their genes as single, unlinked alleles, rather than virions carrying eight related genomic segments. Human studies also often lack essential information found to impact transmission in animal studies (e.g., date of infection, number and length of interactions among the donors and recipients, vaccination history, and immune status) (Weinstock et al. 2003; Van Kerkhove et al. 2011; Varble et al. 2014; Yan et al. 2018). Influenza A and B viruses are rapidly evolving with seasonal epidemics associated with a high incidence of morbidity and mortality in the human population each year (Thompson et al. 2003; Reed et al. 2015; Vijaykrishna et al. 2015). High mutation rates and segmented genomes contribute to the genetic diversity of the virus population, ranging from deleterious mutations to novel segment combinations (Scholtissek 1995; Nelson and Holmes 2007; Pauly et al. 2017). Bottleneck sizes inferred from this diversity and transmission events were shown to vary across host species, including horses, humans, and dogs (Murcia et al. 2010; Hughes et al. 2012; McCrone et al. 2018). In humans, acute influenza infections can lead to tight, or stringent, transmission events, with only one to two virus particles predicted to initiate infection in a new host (McCrone et al. 2018). Variations in bottleneck sizes across studies may be due to viral, host, or environmental determinants such as those identified in animal transmission models (Mubareka et al. 2009; Maines et al. 2012; Wu and Webby 2014; Gustin et al. 2015; Lakdawala et al. 2015; Belser et al. 2016; Frise et al. 2016; McMahon et al. 2019). It is also necessary to specify how transmission bottlenecks affect virus adaptation at different scales (i.e., within infected hosts and across the host population). To shed light on these factors, this review examines how bottleneck size estimates are calculated, how transmission influences the diversity and emergence of influenza viruses, and how the host and the virus interact to alter the number of particles transmitted.
INFLUENZA DIVERSITY AND THE IMPACT OF TRANSMISSION BOTTLENECKS
The error-prone replication and life cycle of influenza virus generate a population of distinct but related genetic variants, often referred to as quasispecies (Domingo et al. 1978, 2012; Vignuzzi et al. 2006; Andino and Domingo 2015). Because of the segmented genome, coinfection of a cell with multiple strains or subtypes of influenza virus can lead to reassortment, or swapping, of the segments, producing virus progeny with a mixture of segments (Scholtissek 1995). The large combination of influenza A subtypes found in nature infect only aquatic birds (Webster et al. 1992; Krauss et al. 2004; Munster et al. 2007); and only two of these, H3N2 and H1N1/2009, currently are found cocirculating in the human population during seasonal epidemics. That said, a novel combination of segments can occur through the coinfection with human, pig, or bird strains and induce the emergence of pandemic strains in the human population, a scenario observed in three of the last four recorded influenza pandemics, including the most recent, in 2009 (Garten et al. 2009; Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team et al. 2009; Smith et al. 2009; Taubenberger and Kash 2010; Vijaykrishna et al. 2010).
Influenza viruses also evolve through mutations generated by the RNA-dependent RNA polymerase during replication, in which approximately one mutation per genome occurs in each round of replication (Nelson and Holmes 2007; Pauly et al. 2017). Most mutations generated are deleterious, however, and are eventually removed from the population through purifying selection (Sanjuán et al. 2004). Influenza infections are acute, usually lasting 5–7 d, so selection has a limited time to act on de novo variants that would allow them to achieve detectable frequencies within the host. However, low-frequency mutations within the host are subject to genetic drift after a bottleneck event, in which the size of the overall virus population has been severely reduced (Moya et al. 2004). A dramatic decrease in the population size also leads to a decrease in the overall genetic diversity of the virus population, creating random fluctuations in the frequencies of virus variants observed within hosts and across the host population (Moya et al. 2004; Nelson and Holmes 2007).
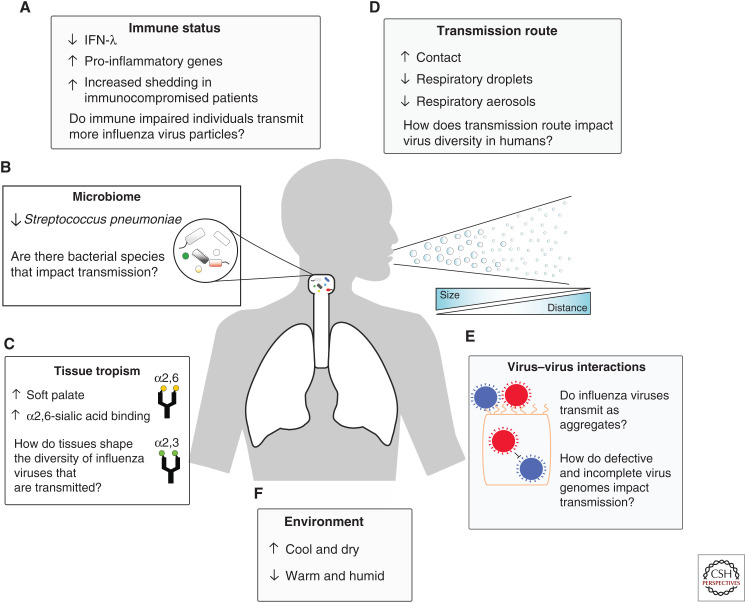
One of the most notable bottleneck events encountered by influenza virus throughout its life cycle is during transmission between hosts, especially when it is cross-species (zoonotic transmission). Even among humans, the transmission mode is a salient determinant of transmission efficiency (Kutter et al. 2018). For example, influenza virus particles are transmitted between humans through large respiratory droplets from sneezing or coughing (5- to 20-µm size); aerosols (<5 µm), produced from breathing or speaking; or through direct contact with infectious secretions (Weber and Stilianakis 2008; Mubareka et al. 2009; Kutter et al. 2018). The distance traveled by the respiratory droplets and aerosols is consequent to the overall size of the secretions, with aerosolized particles traveling further and remaining suspended in the air longer than the larger droplets (Fig. 1; Cowling et al. 2013; Milton et al. 2013). Experiments done in guinea pig and ferret models showed that influenza virus transmits more readily at low temperatures (5°C) compared to high temperatures (30°C). High and intermediate humidity also markedly decreased the efficiency of influenza virus to transmit via respiratory droplets. These findings suggest that the seasonality of influenza is influenced by the environment and efficiency of transmission from host to host (Lowen et al. 2008; Gustin et al. 2015).
Figure 1.
Host and virus determinants of influenza transmission. (A) Interferon (IFN)-λ limits the transmission of the influenza virus to recipient mice, whereas subsets of proinflammatory genes are correlated with an increase in virus transmission. Increased shedding of influenza was observed in immunocompromised and obese individuals. (B) Streptococcus pneumoniae limits the transmission of influenza A virus in mice. (C) In ferrets, transmissible influenza variants were enriched in the soft palate. Sialic acid receptors differ by host and tissue location and help limit the cross-species transmission of influenza A virus. (D) Contact transmission in ferret and guinea pig models yields a higher diversity of virus genotypes to recipient hosts; respiratory transmission (aerosol and droplet) reduces the diversity. Respiratory transmission of influenza is categorized by aerosol and droplet. Size of respiratory secretions impacts the distance traveled and the amount of time it remains suspended in the air. (E) Influenza infections contain a diverse collection of genetic variants. Coinfection with multiple variants such as a defective (red viral particle) and functional (blue particle) can lead to interference (shown), cooperation, and complementation. How these interactions act on the transmission of influenza virus is unknown. (F) Influenza viruses transmit better in cold and dry environments than warm and humid ones.
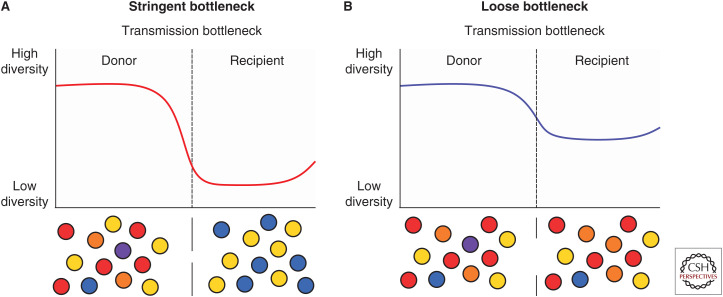
Transmission bottlenecks have a significant impact on the diversity of influenza virus particles that establish an infection in the recipient host. Loose, or large, bottlenecks transmit a higher number of virions, which raises the probability of establishing an infection in the recipient host, as well as increasing the diversity of genetic variants transmitted and on which selection can act (Fig. 2; Geoghegan et al. 2016). In comparison, tight bottlenecks transmit only a few virus particles, which severely limits the genetic diversity of influenza viruses that can generate a productive infection in the recipient host. These small population sizes can be strongly impacted by genetic drift through stochastic fluctuations in the virus population (Geoghegan et al. 2016; McCrone et al. 2018). But note that the decrease in the observed diversity after a transmission event may also be due to other factors, including the removal of nonbeneficial mutants by the host or through clonal competition among multiple virus variants that carry beneficial mutations (Strelkowa and Lässig 2012; Geoghegan et al. 2016).
Figure 2.
Loose and stringent transmission bottlenecks. Transmission of influenza (dashed vertical line) reduces the observed diversity of the virus population in the recipient host. (A) Stringent transmission bottlenecks significantly reduce the diversity observed in the recipient host by transmitting only a few virus particles, making them prone to stochastic fluctuations in variant frequency. (B) Loose transmission bottlenecks transmit a higher diversity of virus particles, allowing selection to act on the virus population in the recipient. In both cases, genetic diversity will increase as the de novo variants are generated through the replication of influenza.
TRANSMISSION OF DEFECTIVE AND INCOMPLETE VIRUS PARTICLES
Most influenza virus particles released from a cell cannot cause a productive infection on their own because replication of the influenza virus can generate defective variants unable to produce essential proteins. Two such common genetic variants are defective viral genomes (DVGs) and incomplete viral genomes (IVGs). DVGs are viruses that contain a truncated form of the viral genome and have been identified in nearly every studied RNA virus, including dengue virus, poliovirus, and human immunodeficiency virus (Cole et al. 1971; Nomoto et al. 1979; Aaskov et al. 2006; Li et al. 2011; Ho et al. 2013). Influenza DVGs, commonly found in both influenza A and B types, are identified by their large internal deletions in at least one of their eight gene segments. They maintain their conserved 5′ and 3′ ends, allowing for their continued replication and packaging. Since their discovery by von Magnus in the 1940s (von Magnus 1947), influenza DVGs have been found to interfere with the replication and packaging of full-length gene segments; they also generate a robust innate immune response and limit disease symptoms of influenza infections in mice and, potentially, humans (Rabinowitz and Huprikar 1979; Davis et al. 1980; Morgan and Dimmock 1992; Baum et al. 2010; Scott et al. 2011; Vasilijevic et al. 2017).
In contrast, IVGs, also called semi-infectious particles, refer to influenza genetic variants that fail to replicate or to express at least one gene segment (Martin and Helenius 1991; Brooke et al. 2013; Jacobs et al. 2019). IVGs are highly abundant within influenza virus populations but differ between influenza type and subtype (Brooke et al. 2013). Theoretical models investigating the spread and replication of IVGs determined that local spatial dispersion of influenza particles reduces the fitness cost of lacking an essential protein, and predicted that approximately 3.6 virions were required to replicate a full set of influenza gene segments (Jacobs et al. 2019). In animal experiments, complementation of laboratory-generated H3N2 IVGs was not observed during transmission of virus particles between guinea pigs, suggesting a requirement for a high multiplicity of infection (MOI) for cooperation and complementation to occur in vivo.
The probability for coinfection at the onset of infection is thought to be low because of the bottleneck at transmission and a high number of cells within the respiratory tract; it rises as the virus begins to replicate to high titers within the host (Baccam et al. 2006). Therefore, we would not expect DVGs and IVGs to add significantly to the effective population size, as they would not contribute progeny to the next generation of viruses without coinfection or complementation from other virus particles. Instances in which a large number of virus particles are shed from a donor host but do not generate a productive infection in the recipient could be due to the transmission of a virus population composed primarily of DVGs and IVGs. Studies have identified both beneficial and detrimental interactions among virus variants that include DVGs, IVGs, and functional viruses (Rabinowitz and Huprikar 1979; Davis et al. 1980; Morgan and Dimmock 1992; Baum et al. 2010; Scott et al. 2011; Brooke 2014; Xue et al. 2016; Vasilijevic et al. 2017; Jacobs et al. 2019). Unexplored to date is how these interactions affect those genetic variants of influenza viruses that are circulating between hosts, and whether the strain-specific generation of defective virus particles determines differences in transmission of influenza subtypes and lineages.
TRANSMISSION AND EMERGING INFLUENZA VIRUSES
Influenza A viruses are often in the spotlight because of their high pandemic potential, incited by their extensive host range and their ability to reassort across subtypes (Webster et al. 1992; Joseph et al. 2017). Currently, the H3N2 and the H1N1/2009 strains are the only two influenza A subtypes that cocirculate yearly within the human population. Since the last influenza A pandemic, in 2009, an important goal of influenza transmission research has been to predict more accurately the next pandemic. We know that most influenza A subtypes infect aquatic birds (Webster et al. 1992) and that, although rare, devastating “spillover” events of avian H7N9 and H5N1 influenza A subtypes in the infection of humans have occurred, with consequent high case mortality rates (de Jong et al. 2006; Gao et al. 2013; Qin et al. 2015; Lipsitch et al. 2016). These “crossover” infections are attributed to close contact between humans and infected birds, and are considered dead-end transmission events (i.e., no onward transmission).
For influenza virus to attach and enter a cell, it must be able to recognize the sialic acids linked to glycoproteins and glycolipids on the surface of the host epithelial cells (for review, see Long et al. 2019). Human and avian influenza viruses bind preferentially to different sialic acid receptors, with human influenza strains preferentially binding to α2,6-linked sialic acid residues; avian subtypes bind to α2,3 (Rogers et al. 1983; Long et al. 2019). The differences in abundance and distribution of sialic acid receptors are potentially limiting factors in human-to-human transmission of avian influenza subtypes, so influenza researchers seek to understand how reassortment between avian and human influenza subtypes or amino acid substitutions can broaden receptor specificity. Jackson et al. (2009) considered whether reassortment between avian H5N1 and human H3N2 strains would improve the transmissibility of the virus between ferret hosts. Ferrets were coinfected with both subtypes of influenza, and virus plaques were assessed from the nasal wash secretions collected from the animals. Although reassortment did happen within the hosts, no transmission was observed, highlighting the genetic complexity of influenza transmissibility. Similarly, reassortants of avian H9N2 and human H3N2 strains failed to transmit between donor and recipient ferrets even with high levels of sneezing and viral shedding by the donor ferrets (Wan et al. 2008).
Efficient transmission of H7N2 and H9N2 avian subtypes between ferrets did take place and was associated with a change in the virus receptor specificity to α2,6 through substitutions in hemagglutinin (HA), the surface protein of influenza that is essential for binding and entering the host cell. But this was insufficient for avian influenza viruses to transmit between ferrets, suggesting that other substitutions are necessary for efficient transmission (Belser et al. 2008). Using previously identified substitutions found in ferret transmissible H5N1 (Herfst et al. 2012; Imai et al. 2012), Linster et al. (2014) identified the minimal set of substitutions needed to generate an avian H5N1 strain capable of efficiently replicating and transmitting between ferrets. These substitutions were found in the PB2, PB1, and HA segments, which allowed for efficient replication and transcription, preferences in sialic acid receptors, and greater stability of the HA (Herfst et al. 2012; Imai et al. 2012; Linster et al. 2014). Together, these studies highlight how the genetic composition of the viruses can limit intraspecies transmission of influenza A subtypes.
Also potentially important for the generation of highly transmissible influenza virus strains is tissue tropism in ferrets. In a study in which an H1N1/2009 influenza virus strain was engineered to preferentially bind to α2,3-linked sialic acid receptors (rather than its usual α2,6), a single amino acid change in the HA protein leading to the preferential binding of the virus from α2,3 to α2,6 (Lakdawala et al. 2015) was primarily enriched in the soft palate of ferrets—a site important both for the establishment and transmission of the infection. The mutational change conferred binding to α2,6 without impairing the virus from binding to α2,3-linked sialic receptors, suggesting that specific tissues or sites within the host may be essential for generating virus variants that can be transmitted to recipient hosts.
Influenza B virus, unlike A, is believed to be chiefly limited to humans, with a few reports of infections in seals (Joseph et al. 2017); but studies on influenza B transmission, bottleneck size, and the impact of the host, vaccines, and antivirals on transmission are limited. To date, its narrow host range and small number of reassortants are thought to preclude it as a pandemic threat.
QUANTIFYING TRANSMISSION BOTTLENECKS
The number of virus particles transmitted to a host is an essential factor of virus adaptability; thus, accurately quantifying bottleneck size is a crucial measurement, but one that has proved difficult to determine. Multiple methods, both experimental and theoretical, have been enlisted to calculate bottleneck size and often result in different results. One experimental method, for example, used barcoded influenza A viruses (H1N1/2009 strains) and small mammalian transmission models to quantify the number of barcoded genotypes that transfer from the donor to the recipient host (Varble et al. 2014). According to the study, the transmission route acts on the number of genotypes transmitted, with contact transmission producing more genotypes (seven to 24) than respiratory droplet transmission (three to five early in the infection, up to 25 with increased recipient exposure to the donor) (Varble et al. 2014). A separate study calculated the number of influenza virus shed in the breath of naturally infected college students (Yan et al. 2018). Exhalations were collected in fine and coarse aerosol fractions over 30 min; the number of infectious particles released and the number of RNA molecules (from both infectious and noninfectious particles) were quantified by plaque assay and qPCR, respectively. Results indicated that breathing for 30 min effectively transmitted influenza virus particles from aerosol samples containing an average of 3.8 × 104 viral RNA molecules or from coarse aerosol fractions with an average of 1.2 × 104 viral RNA molecules (Yan et al. 2018). Both studies propose route of transmission as a determinant of the number of virus particles transmitted to the recipient host. However, measuring the number of barcoded genotypes transmitted can only infer the stringency of the bottleneck size, not how many particles of each genotype are actually transmitted.
Fortunately, measuring bottleneck sizes in naturally infected individuals has improved thanks to next-generation sequencing. By sequencing samples from the donor and recipient of transmission pairs, virus variants that occur at low frequency within the host can be quantified and characterized (Wang et al. 2010; Debbink et al. 2017; Sobel Leonard et al. 2017; McCrone et al. 2018). Similar to the barcoded viruses used by Varble et al. (2014), early methods of measuring viral transmission in human immunodeficiency and hepatitis C virus studies, among others, factored in changes in genetic diversity between the donor and recipient hosts to estimate the number of transmitted genotypes. Both viruses were estimated to need only one to two genotypes to generate an infection in a recipient host (Edwards et al. 2006; Keele et al. 2008; Bull et al. 2011; Russell et al. 2011). Determining the number of genotypes necessary to establish an infection is, however, considered an underestimate of the bottleneck size because it does not factor in either the number of virus particles transmitted or quickly lost in the recipient host (Sobel Leonard et al. 2017).
To improve the accuracy of determining bottleneck sizes, two recent methods have targeted the total number of virions in the founding virus population at the onset of the infection in the recipient after transmission from the donor. These methods combine statistical models and next-generation sequencing data and feature the presence–absence and β-binomial models (Sobel Leonard et al. 2017; McCrone et al. 2018). The models assume that observed minor genetic variants are independent and view transmission as a random sampling event. The simplified presence–absence model considers only those variants present within both the donor and recipient samples. The probability of the donor transmitting the variant is related to the frequency of the observed variant in the sequencing data of the donor's virus population in which a variant at high frequency in the donor is more likely to transmit to the recipient. To account for the inherent noise associated with sequencing data, thresholds for identifying minor variants are set to ensure that sequencing artifacts are not factored when estimating bottleneck sizes. Often, though, these cutoffs are conservative, and consequently miss variants present at low frequency within the recipient hosts. The simplicity of the presence–absence model also means it misses instances in which a variant is present at below-detectable levels in the recipient, resulting in low bottleneck size estimates. The β-binomial approach circumvents these limitations by considering the frequencies of shared variants within the donor and recipient hosts, as well as the frequency of variants found only within the donor. And to account for viral replication dynamics that occur between transmission of the virus and sampling of the individual, frequencies of shared variants are allowed to change according to the β distribution.
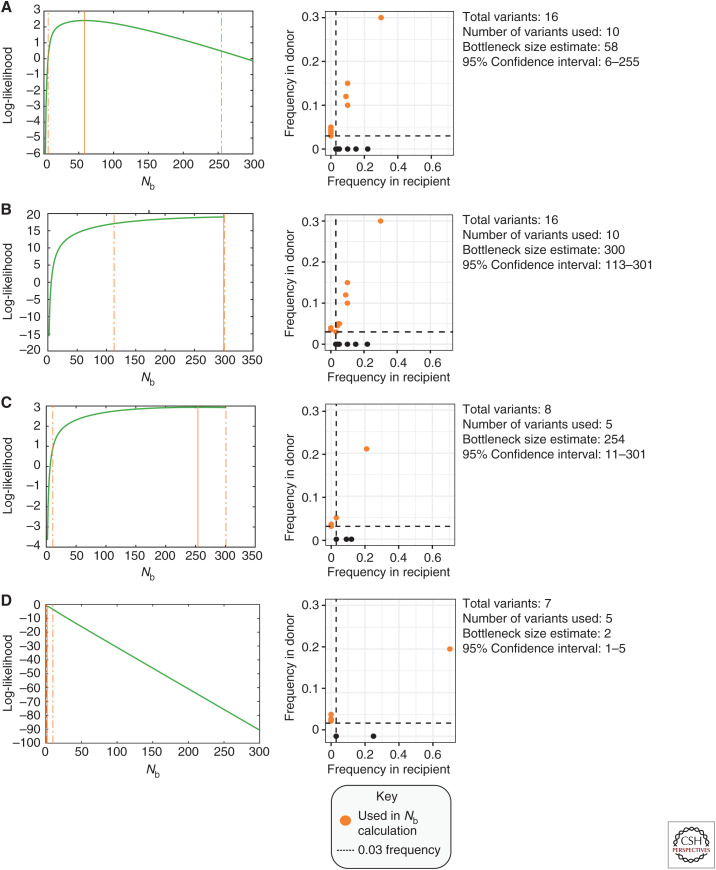
The presence–absence and β-binomial models were each used to estimate the bottleneck size of influenza transmission during the 2010–2015 influenza seasons and produced similar results: one to two influenza particles transmitted between human hosts (McCrone et al. 2018). But these estimates were variable among transmission pairs and often yielded wide confidence intervals ranging from the minimum number of virus particles necessary to establish an infection (one particle) to the highest estimate used in the model (200) (Fig. 3). Furthermore, the high prevalence of reassortment observed in the global virus population suggests the one to two range to be an underestimate. One theory to explain how influenza virus adapts at a global scale if bottleneck sizes are highly stringent is that rapid replication within the host, and a large number of potential hosts, may offset the cost of a sharp decrease in diversity due to a severe bottleneck (Manrubia et al. 2005; McCrone et al. 2018).
Figure 3.
β-binomial bottleneck size estimates (Nb). The frequency and number of minor variants shared between donor and recipient are factors in the transmission bottleneck size estimate. Orange points highlight variants used for the β-binomial calculation. Black points indicate de novo mutations identified in the recipient but not the donor. Dashed lines indicate the 3% frequency cutoff used to identify minor variants in sequencing data. (A,B) Examples of donors and recipients with high shared diversity. Increasing the number of shared variants (B) increases the overall bottleneck size estimate. (C,D) Examples of donors and recipients with low numbers of virus variants, commonly observed in acute influenza infections. (C) Variants shared at a similar frequency within the donor and recipient generate a large bottleneck estimate with wide confidence intervals. (D) Stringent bottlenecks are observed when few minor variants are shared between donor and recipient, and the frequency of a shared variant becomes dominant (>50% frequency) in the recipient host.
INFLUENZA TRANSMISSION IN ANIMAL MODELS
Animal models—mice, ferrets, and guinea pigs—have been instrumental in the study of host and virus factors that impact the transmission of influenza virus. None of these models can accurately replicate human infections and transmission, yet each has helped expand our understanding of those factors that influence influenza transmissibility.
Mice were the first animals used to study how the host influences influenza infections (Schulman and Kilbourne 1963), revealing that infection is highly dependent on both the strain of the virus and the strain of the mouse model (Schulman and Kilbourne 1963; Lowen et al. 2006; Haller et al. 2015; Ivinson et al. 2017). Most studies have found that H3N2 strains can transmit when mice are in direct contact with each other, whereas H1N1 strains do not transmit efficiently unless adapted to do so (Lowen et al. 2006; Edenborough et al. 2012). Currently, establishing standard laboratory procedures is underway to produce mice that are more effective transmission models of influenza (Edenborough et al. 2012; Ivinson et al. 2017).
The ferret is perhaps the best-suited animal model for studying influenza pathogenesis, tropism, and transmission (Maher and DeStefano 2004; Belser et al. 2011). Ferrets do not require host-adapted strains and are susceptible to a broad range of influenza virus strains (Herlocher et al. 2001; Maines et al. 2005; Wan et al. 2008; Itoh et al. 2009; Jackson et al. 2009; Kim et al. 2009; Munster et al. 2009). Furthermore, ferrets possess a similar distribution of sialic acid receptors to that in humans (van Riel et al. 2007; de Graaf and Fouchier 2014); thus, like humans, ferrets sneeze and cough during influenza infection, making them prime candidates for the study of respiratory droplet and aerosol transmission (Belser et al. 2016). On the downside, ferrets incur intensive care requirements at associated high costs, restricting the design of experiments for a statistically significant number of animals. Constraints aside, ferrets have been proven influential in determining how tissue tropism, environment, and immune response affect the transmission of influenza virus (Maines et al. 2012; Gustin et al. 2015; Lakdawala et al. 2015; Belser et al. 2016; Frise et al. 2016).
For a number of reasons, guinea pigs are gaining in popularity for the study of influenza virus transmission (Lowen et al. 2006). They are small, easy to maintain, cheaper than ferrets, and susceptible to human influenza virus strains. Like ferrets, guinea pigs can be used to study aerosol, droplet, and contact transmission, depending on how they are caged. First described in 2006 by Lowen et al., guinea pig studies have since produced a number of essential findings regarding how the host and environmental factors affect transmission (Lowen et al. 2008; Varble et al. 2014; McMahon et al. 2019).
Route of Transmission
The route influenza virus takes to its destination—whether aerosol, droplet, or direct contact—will determine in part the number and diversity of virus particles (Mubareka et al. 2009; Varble et al. 2014). Loose bottlenecks, associated with contact transmission, allowed for the transmission of drug-resistant variants between ferrets that were cocaged, and mixed infections in guinea pigs, in which a high incidence of reassortment was also observed (Frise et al. 2016). In spillover events, avian influenza virus strains have infected humans through direct contact (Koopmans et al. 2004; Plowright et al. 2017). Together, these findings lend credence to the theory that the circulation of drug-resistant or other rare genetic variants is likely due to close contact with infected individuals or infectious secretions (Edenborough et al. 2012; Varble et al. 2014). Needed now is more research into characterizing and quantifying interactions between individuals within human cohort transmission studies, along with sequencing of virus genomes collected from transmission pairs.
Immune Status and Transmission
Innate and adaptive immunity in hosts figures heavily in limiting viral replication and transmission (Iwasaki and Pillai 2014; Chen et al. 2018). Notably, Klinkhammer et al. (2018) found that interferon (IFN)-λ limited the transmission of influenza virus between mice lacking functional innate immune receptors. This study examined a mouse-adapted H7N7 strain (A/seal/Massachusetts/1/1980), and H3N2 strains (A/Hong Kong/8/68 and A/Udorn/72) in donor mice that lacked either a functional type I IFN receptor or a functional type III IFN receptor (the IFN-λ receptor). To mimic upper respiratory tract infections observed commonly in humans, mice were inoculated with a smaller volume of viral inoculum and were cocaged with double-knockout recipients. Results showed that mice lacking a functional IFN-λ receptor not only shed more virus but also transmitted more readily to the recipient mice. When the donor mice were treated prophylactically with IFN-λ, transmission to naive mice was reduced (Klinkhammer et al. 2018). In another study, IFN-λ was identified as inducing an adaptive immune response to viruses when vaccines were administered intranasally (Ye et al. 2019). Intranasal vaccines using a recombinant influenza B neuraminidase protein were also found to limit the spread of influenza B viruses in guinea pigs (McMahon et al. 2019).
Research in ferrets profiled the expression of innate immune genes during peak viral shedding periods and transmission and found proinflammatory genes that were up-regulated and correlated with increased transmissibility of the influenza virus (Maines et al. 2012). Collectively, these studies raise the question: Do individuals with impaired immune function differ in their ability to transmit influenza virus to others? When influenza virus evolution has been tracked within immunocompromised patients with prolonged influenza infections, those with impaired immune function were shown to act as reservoirs for adapted influenza virus variants and, possibly, to transmit higher numbers of virus particles (Weinstock et al. 2003; Rogers et al. 2015; Xue et al. 2017).
Effects of Age and the Microbiome on Transmission
To address the inconsistency of transmission in adult mouse models, Ortigoza et al. (2018) began with infant mice to track the transmission of influenza virus. Infant mice ∼7 days old efficiently transmitted the laboratory strain A/X-31(H3N2) to their cocaged infant recipients, whereas newly weaned and adult mice (>28 d) failed to transmit the virus altogether. Mothers pre-exposed to the influenza A virus were able to transfer immunoglobulin G to the infant mice pre- and postnatally, which protected the infant mice and reduced transmission. From this, the researchers concluded that age of the host and pre-exposure status of the mother mouse had a role in the transmissibility of influenza virus among the infant mice.
The infant mouse transmission model was similarly used to study effects on transmission of the precolonization of Streptococcus pneumoniae, because in human children, S. pneumoniae often precedes influenza A virus infections (McCullers 2006; Morris et al. 2017). Here, recipient mice showed a lower incidence of influenza infection when prior colonization of S. pneumoniae occurred, further highlighting the potential importance of the host's microbiome in limiting the transmission of influenza (Ortigoza et al. 2018). It is reasonable to speculate, however, that other bacterial species may help influenza transmit to other hosts. As a case in point, polio viruses adhere to bacterial cells and travel as “collective infectious units” when transmitting to new cells within the host (Erickson et al. 2018). These aggregates of virus particles effectively increase the multiplicity of infection and could potentially allow for the transmission of defective or incomplete influenza particles with functional particles (Hirst and Pons 1973; Sanjuán 2017). Although influenza aggregates have been identified in cell culture studies, it is unclear whether influenza transmits as aggregates (Campbell et al. 2004).
CONCLUSION
To identify new and more accurate control methods to improve our ability to predict which virus strains will circulate in the next influenza season, we must first identify what may limit the transmission of the influenza virus. Virus–virus interactions, host immune status, the host microbiome, virus shedding, and the environment all contribute to the size of the transmission bottleneck (Fig. 1) and to variations across data sets from human and animal studies, as do the techniques used to derive the data. Even as animal models help us to better understand the dynamic interplay between the host and the virus that contributes to greater transmissibility, we remain in the dark about how exactly this information relates to the human experience of influenza infection. We lack, too, critical background information such as immune status, weight, age, and degree of interaction between donor and recipient. Next-generation sequencing data from natural influenza infections in humans and statistical models estimate a narrow bottleneck size, but estimates vary between different transmission pairs, underscoring the need for more, and more effective, methods of tracking and quantifying interactions among human cohorts in the study of influenza virus transmission. Future models for estimating bottleneck sizes will need to incorporate pathogen interactions, including those between functional and defective viruses and between influenza and other microbial pathogens.
ACKNOWLEDGMENT
This review has been made freely available online courtesy of TAUNS Laboratories.
Footnotes
Editors: Gabriele Neumann and Yoshihiro Kawaoka
Additional Perspectives on Influenza: The Cutting Edge available at www.perspectivesinmedicine.org
REFERENCES
- Aaskov J, Buzacott K, Thu HM, Lowry K, Holmes EC. 2006. Long-term transmission of defective RNA viruses in humans and Aedes mosquitoes. Science 311: 236–238. 10.1126/science.1115030 [DOI] [PubMed] [Google Scholar]
- Andino R, Domingo E. 2015. Viral quasispecies. Virology 479-480: 46–51. 10.1016/j.virol.2015.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baccam P, Beauchemin C, Macken CA, Hayden FG, Perelson AS. 2006. Kinetics of influenza A virus infection in humans. J Virol 80: 7590–7599. 10.1128/JVI.01623-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baum A, Sachidanandam R, García-Sastre A. 2010. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc Natl Acad Sci 107: 16303–16308. 10.1073/pnas.1005077107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, Blixt O, Chen LM, Pappas C, Maines TR, Van Hoeven N, Donis R, Busch J, McBride R, Paulson JC, et al. 2008. Contemporary North American influenza H7 viruses possess human receptor specificity: implications for virus transmissibility. Proc Natl Acad Sci 105: 7558–7563. 10.1073/pnas.0801259105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, Katz JM, Tumpey TM. 2011. The ferret as a model organism to study influenza A virus infection. Dis Model Mech 4: 575–579. 10.1242/dmm.007823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belser JA, Eckert AM, Tumpey TM, Maines TR. 2016. Complexities in ferret influenza virus pathogenesis and transmission models. Microbiol Mol Biol Rev 80: 733–744. 10.1128/MMBR.00022-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke CB. 2014. Biological activities of “noninfectious” influenza A virus particles. Future Virol 9: 41–51. 10.2217/fvl.13.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke CB, Ince WL, Wrammert J, Ahmed R, Wilson PC, Bennink JR, Yewdell JW. 2013. Most influenza a virions fail to express at least one essential viral protein. J Virol 87: 3155–3162. 10.1128/JVI.02284-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull RA, Luciani F, McElroy K, Gaudieri S, Pham ST, Chopra A, Cameron B, Maher L, Dore GJ, White PA, et al. 2011. Sequential bottlenecks drive viral evolution in early acute hepatitis C virus infection. PLoS Pathog 7: e1002243 10.1371/journal.ppat.1002243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell JN, Epand RM, Russo PS. 2004. Structural changes and aggregation of human influenza virus. Biomacromolecules 5: 1728–1735. 10.1021/bm049878z [DOI] [PubMed] [Google Scholar]
- Chen X, Liu S, Goraya MU, Maarouf M, Huang S, Chen JL. 2018. Host immune response to influenza A virus infection. Front Immunol 9: 320 10.3389/fimmu.2018.00320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole CN, Smoler D, Wimmer E, Baltimore D. 1971. Defective interfering particles of poliovirus. I. Isolation and physical properties. J Virol 7: 478–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowling BJ, Ip DKM, Fang VJ, Suntarattiwong P, Olsen SJ, Levy J, Uyeki TM, Leung GM, Malik Peiris JS, Chotpitayasunondh T, et al. 2013. Aerosol transmission is an important mode of influenza A virus spread. Nat Commun 4: 1935 10.1038/ncomms2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis AR, Hiti AL, Nayak DP. 1980. Influenza defective interfering viral RNA is formed by internal deletion of genomic RNA. Proc Natl Acad Sci 77: 215–219. 10.1073/pnas.77.1.215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debbink K, McCrone JT, Petrie JG, Truscon R, Johnson E, Mantlo EK, Monto AS, Lauring AS. 2017. Vaccination has minimal impact on the intrahost diversity of H3N2 influenza viruses. PLoS Pathog 13: e1006194 10.1371/journal.ppat.1006194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf M, Fouchier RAM. 2014. Role of receptor binding specificity in influenza A virus transmission and pathogenesis. EMBO J 33: 823–841. 10.1002/embj.201387442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJD, Chau TNB, Hoang DM, Chau NVV, Khanh TH, Dong VC, et al. 2006. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med 12: 1203–1207. 10.1038/nm1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo E, Sabo D, Taniguchi T, Weissmann C. 1978. Nucleotide sequence heterogeneity of an RNA phage population. Cell 13: 735–744. 10.1016/0092-8674(78)90223-4 [DOI] [PubMed] [Google Scholar]
- Domingo E, Sheldon J, Perales C. 2012. Viral quasispecies evolution. Microbiol Mol Biol Rev 76: 159–216. 10.1128/MMBR.05023-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenborough KM, Gilbertson BP, Brown LE. 2012. A mouse model for the study of contact-dependent transmission of influenza A virus and the factors that govern transmissibility. J Virol 86: 12544–12551. 10.1128/JVI.00859-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards CTT, Holmes EC, Wilson DJ, Viscidi RP, Abrams EJ, Phillips RE, Drummond AJ. 2006. Population genetic estimation of the loss of genetic diversity during horizontal transmission of HIV-1. BMC Evol Biol 6: 28 10.1186/1471-2148-6-28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson AK, Jesudhasan PR, Mayer MJ, Narbad A, Winter SE, Pfeiffer JK. 2018. Bacteria facilitate enteric virus coinfection of mammalian cells and promote genetic recombination. Cell Host Microbe 23: 77–88.e5. 10.1016/j.chom.2017.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frise R, Bradley K, van Doremalen N, Galiano M, Elderfield RA, Stilwell P, Ashcroft JW, Fernandez-Alonso M, Miah S, Lackenby A, et al. 2016. Contact transmission of influenza virus between ferrets imposes a looser bottleneck than respiratory droplet transmission allowing propagation of antiviral resistance. Sci Rep 6: 29793 10.1038/srep29793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallet R, Michalakis Y, Blanc S. 2018. Vector-transmission of plant viruses and constraints imposed by virus–vector interactions. Curr Opin Virol 33: 144–150. 10.1016/j.coviro.2018.08.005 [DOI] [PubMed] [Google Scholar]
- Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, et al. 2013. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 368: 1888–1897. 10.1056/NEJMoa1304459 [DOI] [PubMed] [Google Scholar]
- Garten RJ, Davis CT, Russell CA, Shu B, Lindstrom S, Balish A, Sessions WM, Xu X, Skepner E, Deyde V, et al. 2009. Antigenic and genetic characteristics of swine-origin 2009 A(H1N1) influenza viruses circulating in humans. Science 325: 197–201. 10.1126/science.1176225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geoghegan JL, Senior AM, Holmes EC. 2016. Pathogen population bottlenecks and adaptive landscapes: overcoming the barriers to disease emergence. Proc Biol Sci 283: 20160727 10.1098/rspb.2016.0727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustin KM, Belser JA, Veguilla V, Zeng H, Katz JM, Tumpey TM, Maines TR. 2015. Environmental conditions affect exhalation of H3N2 seasonal and variant influenza viruses and respiratory droplet transmission in ferrets. PLoS ONE 10: e0125874 10.1371/journal.pone.0125874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller O, Staeheli P, Schwemmle M, Kochs G. 2015. Mx GTPases: dynamin-like antiviral machines of innate immunity. Trends Microbiol 23: 154–163. 10.1016/j.tim.2014.12.003 [DOI] [PubMed] [Google Scholar]
- Herfst S, Schrauwen EJA, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, et al. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336: 1534–1541. 10.1126/science.1213362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlocher ML, Elias S, Truscon R, Harrison S, Mindell D, Simon C, Monto AS. 2001. Ferrets as a transmission model for influenza: sequence changes in HA1 of type A (H3N2) virus. J Infect Dis 184: 542–546. 10.1086/322801 [DOI] [PubMed] [Google Scholar]
- Hirst GK, Pons MW. 1973. Mechanism of influenza recombination: II. Virus aggregation and its effect on plaque formation by so-called noninfective virus. Virology 56: 620–631. 10.1016/0042-6822(73)90063-9 [DOI] [PubMed] [Google Scholar]
- Ho YC, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DIS, Lai J, Blankson JN, Siliciano JD, Siliciano RF. 2013. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155: 540–551. 10.1016/j.cell.2013.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang YJS, Higgs S, Vanlandingham DL. 2019. Arbovirus-mosquito vector–host interactions and the impact on transmission and disease pathogenesis of arboviruses. Front Microbiol 10: 22 10.3389/fmicb.2019.00022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes J, Allen RC, Baguelin M, Hampson K, Baillie GJ, Elton D, Newton JR, Kellam P, Wood JLN, Holmes EC, et al. 2012. Transmission of equine influenza virus during an outbreak is characterized by frequent mixed infections and loose transmission bottlenecks. PLoS Pathog 8: e1003081 10.1371/journal.ppat.1003081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, et al. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486: 420–428. 10.1038/nature10831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y, Shinya K, Kiso M, Watanabe T, Sakoda Y, Hatta M, Muramoto Y, Tamura D, Sakai-Tagawa Y, Noda T, et al. 2009. In vitro and in vivo characterization of new swine-origin H1N1 influenza viruses. Nature 460: 1021–1025. 10.1038/nature08260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivinson K, Deliyannis G, McNabb L, Grollo L, Gilbertson B, Jackson D, Brown LE. 2017. Salivary blockade protects the lower respiratory tract of mice from lethal influenza virus infection. J Virol 91: e00624-17 10.1128/JVI.00624-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki A, Pillai PS. 2014. Innate immunity to influenza virus infection. Nat Rev Immunol 14: 315–328. 10.1038/nri3665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S, Van Hoeven N, Chen LM, Maines TR, Cox NJ, Katz JM, Donis RO. 2009. Reassortment between avian H5N1 and human H3N2 influenza viruses in ferrets: a public health risk assessment. J Virol 83: 8131–8140. 10.1128/JVI.00534-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs NT, Onuoha NO, Antia A, Steel J, Antia R, Lowen AC. 2019. Incomplete influenza A virus genomes occur frequently but are readily complemented during localized viral spread. Nat Commun 10: 3526 10.1038/s41467-019-11428-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph U, Su YCF, Vijaykrishna D, Smith GJD. 2017. The ecology and adaptive evolution of influenza A interspecies transmission. Influenza Other Respi Viruses 11: 74–84. 10.1111/irv.12412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, Salazar MG, Sun C, Grayson T, Wang S, Li H, et al. 2008. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci 105: 7552–7557. 10.1073/pnas.0802203105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YH, Kim HS, Cho SH, Seo SH. 2009. Influenza B virus causes milder pathogenesis and weaker inflammatory responses in ferrets than influenza A virus. Viral Immunol 22: 423–430. 10.1089/vim.2009.0045 [DOI] [PubMed] [Google Scholar]
- Klinkhammer J, Schnepf D, Ye L, Schwaderlapp M, Gad HH, Hartmann R, Garcin D, Mahlakõiv T, Staeheli P. 2018. IFN-λ prevents influenza virus spread from the upper airways to the lungs and limits virus transmission. eLife 7: e33354 10.7554/eLife.33354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koopmans M, Wilbrink B, Conyn M, Natrop G, van der Nat H, Vennema H, Meijer A, van Steenbergen J, Fouchier R, Osterhaus A, et al. 2004. Transmission of H7N7 avian influenza A virus to human beings during a large outbreak in commercial poultry farms in the Netherlands. Lancet 363: 587–593. 10.1016/S0140-6736(04)15589-X [DOI] [PubMed] [Google Scholar]
- Krauss S, Walker D, Pryor SP, Niles L, Chenghong L, Hinshaw VS, Webster RG. 2004. Influenza A viruses of migrating wild aquatic birds in North America. Vector Borne Zoonotic Dis 4: 177–189. 10.1089/vbz.2004.4.177 [DOI] [PubMed] [Google Scholar]
- Kutter JS, Spronken MI, Fraaij PL, Fouchier RA, Herfst S. 2018. Transmission routes of respiratory viruses among humans. Curr Opin Virol 28: 142–151. 10.1016/j.coviro.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakdawala SS, Jayaraman A, Halpin RA, Lamirande EW, Shih AR, Stockwell TB, Lin X, Simenauer A, Hanson CT, Vogel L, et al. 2015. The soft palate is an important site of adaptation for transmissible influenza viruses. Nature 526: 122–125. 10.1038/nature15379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefeuvre P, Martin DP, Elena SF, Shepherd DN, Roumagnac P, Varsani A. 2019. Evolution and ecology of plant viruses. Nat Rev Microbiol 17: 632–644. 10.1038/s41579-019-0232-3 [DOI] [PubMed] [Google Scholar]
- Li D, Lott WB, Lowry K, Jones A, Thu HM, Aaskov J. 2011. Defective interfering viral particles in acute dengue infections. PLoS ONE 6: e19447 10.1371/journal.pone.0019447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linster M, van Boheemen S, de Graaf M, Schrauwen EJA, Lexmond P, Mänz B, Bestebroer TM, Baumann J, van Riel D, Rimmelzwaan GF, et al. 2014. Identification, characterization, and natural selection of mutations driving airborne transmission of A/H5N1 virus. Cell 157: 329–339. 10.1016/j.cell.2014.02.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipsitch M, Barclay W, Raman R, Russell CJ, Belser JA, Cobey S, Kasson PM, Lloyd-Smith JO, Maurer-Stroh S, Riley S, et al. 2016. Viral factors in influenza pandemic risk assessment. eLife 5: e18491 10.7554/eLife.18491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JS, Mistry B, Haslam SM, Barclay WS. 2019. Host and viral determinants of influenza A virus species specificity. Nat Rev Microbiol 17: 67–81. 10.1038/s41579-018-0115-z [DOI] [PubMed] [Google Scholar]
- Lowen AC, Mubareka S, Tumpey TM, García-Sastre A, Palese P. 2006. The guinea pig as a transmission model for human influenza viruses. Proc Natl Acad Sci 103: 9988–9992. 10.1073/pnas.0604157103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowen AC, Steel J, Mubareka S, Palese P. 2008. High temperature (30°C) blocks aerosol but not contact transmission of influenza virus. J Virol 82: 5650–5652. 10.1128/JVI.00325-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher JA, DeStefano J. 2004. The ferret: an animal model to study influenza virus. Lab Anim 33: 50–53. 10.1038/laban1004-50 [DOI] [PubMed] [Google Scholar]
- Maines TR, Lu XH, Erb SM, Edwards L, Guarner J, Greer PW, Nguyen DC, Szretter KJ, Chen LM, Thawatsupha P, et al. 2005. Avian influenza (H5N1) viruses isolated from humans in Asia in 2004 exhibit increased virulence in mammals. J Virol 79: 11788–11800. 10.1128/JVI.79.18.11788-11800.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maines TR, Belser JA, Gustin KM, van Hoeven N, Zeng H, Svitek N, von Messling V, Katz JM, Tumpey TM. 2012. Local innate immune responses and influenza virus transmission and virulence in ferrets. J Infect Dis 205: 474–485. 10.1093/infdis/jir768 [DOI] [PubMed] [Google Scholar]
- Manrubia SC, Escarmís C, Domingo E, Lázaro E. 2005. High mutation rates, bottlenecks, and robustness of RNA viral quasispecies. Gene 347: 273–282. 10.1016/j.gene.2004.12.033 [DOI] [PubMed] [Google Scholar]
- Martin K, Helenius A. 1991. Nuclear transport of influenza virus ribonucleoproteins: the viral matrix protein (M1) promotes export and inhibits import. Cell 67: 117–130. 10.1016/0092-8674(91)90576-K [DOI] [PubMed] [Google Scholar]
- McCrone JT, Woods RJ, Martin ET, Malosh RE, Monto AS, Lauring AS. 2018. Stochastic processes constrain the within and between host evolution of influenza virus. eLife 7: e35962 10.7554/eLife.35962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullers JA. 2006. Insights into the interaction between influenza virus and pneumococcus. Clin Microbiol Rev 19: 571–582. 10.1128/CMR.00058-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon M, Kirkpatrick E, Stadlbauer D, Strohmeier S, Bouvier NM, Krammer F. 2019. Mucosal immunity against neuraminidase prevents influenza B virus transmission in guinea pigs. MBio 10: e00560-19 10.1128/mBio.00560-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milton DK, Fabian MP, Cowling BJ, Grantham ML, McDevitt JJ. 2013. Influenza virus aerosols in human exhaled breath: particle size, culturability, and effect of surgical masks. PLoS Pathog 9: e1003205 10.1371/journal.ppat.1003205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan DJ, Dimmock NJ. 1992. Defective interfering influenza virus inhibits immunopathological effects of infectious virus in the mouse. J Virol 66: 1188–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris DE, Cleary DW, Clarke SC. 2017. Secondary bacterial infections associated with influenza pandemics. Front Microbiol 8: 1041 10.3389/fmicb.2017.01041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moya A, Holmes EC, González-Candelas F. 2004. The population genetics and evolutionary epidemiology of RNA viruses. Nat Rev Microbiol 2: 279–288. 10.1038/nrmicro863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mubareka S, Lowen AC, Steel J, Coates AL, García-Sastre A, Palese P. 2009. Transmission of influenza virus via aerosols and fomites in the guinea pig model. J Infect Dis 199: 858–865. 10.1086/597073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster VJ, Baas C, Lexmond P, Waldenström J, Wallensten A, Fransson T, Rimmelzwaan GF, Beyer WEP, Schutten M, Olsen B, et al. 2007. Spatial, temporal, and species variation in prevalence of influenza A viruses in wild migratory birds. PLoS Pathog 3: e61 10.1371/journal.ppat.0030061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munster VJ, de Wit E, van den Brand JMA, Herfst S, Schrauwen EJA, Bestebroer TM, van de Vijver D, Boucher CA, Koopmans M, Rimmelzwaan GF, et al. 2009. Pathogenesis and transmission of swine-origin 2009 A(H1N1) influenza virus in ferrets. Science 325: 481–483. 10.1126/science.1177127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murcia PR, Baillie GJ, Daly J, Elton D, Jervis C, Mumford JA, Newton R, Parrish CR, Hoelzer K, Dougan G, et al. 2010. Intra- and interhost evolutionary dynamics of equine influenza virus. J Virol 84: 6943–6954. 10.1128/JVI.00112-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MI, Holmes EC. 2007. The evolution of epidemic influenza. Nat Rev Genet 8: 196–205. 10.1038/nrg2053 [DOI] [PubMed] [Google Scholar]
- Nomoto A, Jacobson A, Lee YF, Dunn J, Wimmer E. 1979. Defective interfering particles of poliovirus: mapping of the deletion and evidence that the deletions in the genomes of DI(1), (2) and (3) are located in the same region. J Mol Biol 128: 179–196. 10.1016/0022-2836(79)90125-6 [DOI] [PubMed] [Google Scholar]
- Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team, Dawood FS, Jain S, Finelli L, Shaw MW, Lindstrom S, Garten RJ, Gubareva LV, Xu X, Bridges CB, et al. 2009. Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N Engl J Med 360: 2605–2615. 10.1056/NEJMoa0903810 [DOI] [PubMed] [Google Scholar]
- Ortigoza MB, Blaser SB, Zafar MA, Hammond AJ, Weiser JN. 2018. An infant mouse model of influenza virus transmission demonstrates the role of virus-specific shedding, humoral immunity, and sialidase expression by colonizing Streptococcus pneumoniae. MBio 9: e02359-18 10.1128/mBio.02359-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauly MD, Procario MC, Lauring AS. 2017. A novel twelve class fluctuation test reveals higher than expected mutation rates for influenza A viruses. eLife 6: e26437 10.7554/eLife.26437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plowright RK, Parrish CR, McCallum H, Hudson PJ, Ko AI, Graham AL, Lloyd-Smith JO. 2017. Pathways to zoonotic spillover. Nat Rev Microbiol 15: 502–510. 10.1038/nrmicro.2017.45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Horby PW, Tsang TK, Chen E, Gao L, Ou J, Nguyen TH, Duong TN, Gasimov V, Feng L, et al. 2015. Differences in the epidemiology of human cases of avian influenza A(H7N9) and A(H5N1) viruses infection. Clin Infect Dis 61: 563–571. 10.1093/cid/civ345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabinowitz SG, Huprikar J. 1979. The influence of defective-interfering particles of the PR-8 strain of influenza A virus on the pathogenesis of pulmonary infection in mice. J Infect Dis 140: 305–315. 10.1093/infdis/140.3.305 [DOI] [PubMed] [Google Scholar]
- Reed C, Chaves SS, Daily Kirley P, Emerson R, Aragon D, Hancock EB, Butler L, Baumbach J, Hollick G, Bennett NM, et al. 2015. Estimating influenza disease burden from population-based surveillance data in the United States. PLoS ONE 10: e0118369 10.1371/journal.pone.0118369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, Wiley DC. 1983. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature 304: 76–78. 10.1038/304076a0 [DOI] [PubMed] [Google Scholar]
- Rogers MB, Song T, Sebra R, Greenbaum BD, Hamelin M-E, Fitch A, Twaddle A, Cui L, Holmes EC, Boivin G, et al. 2015. Intrahost dynamics of antiviral resistance in influenza A virus reflect complex patterns of segment linkage, reassortment, and natural selection. MBio 6: e02464-14 10.1128/mBio.02464-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell ES, Kwiek JJ, Keys J, Barton K, Mwapasa V, Montefiori DC, Meshnick SR, Swanstrom R. 2011. The genetic bottleneck in vertical transmission of subtype C HIV-1 is not driven by selection of especially neutralization-resistant virus from the maternal viral population. J Virol 85: 8253–8262. 10.1128/JVI.00197-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán R. 2017. Collective infectious units in viruses. Trends Microbiol 25: 402–412. 10.1016/j.tim.2017.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanjuán R, Moya A, Elena SF. 2004. The distribution of fitness effects caused by single-nucleotide substitutions in an RNA virus. Proc Natl Acad Sci 101: 8396–8401. 10.1073/pnas.0400146101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtissek C. 1995. Molecular evolution of influenza viruses. Virus Genes 11: 209–215. 10.1007/BF01728660 [DOI] [PubMed] [Google Scholar]
- Schulman JL, Kilbourne ED. 1963. Experimental transmission of influenza virus infection in mice. I. The period of transmissibility. J Exp Med 118: 257–266. 10.1084/jem.118.2.257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott PD, Meng B, Marriott AC, Easton AJ, Dimmock NJ. 2011. Defective interfering influenza virus confers only short-lived protection against influenza virus disease: evidence for a role for adaptive immunity in DI virus-mediated protection in vivo. Vaccine 29: 6584–6591. 10.1016/j.vaccine.2011.06.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GJD, Vijaykrishna D, Bahl J, Lycett SJ, Worobey M, Pybus OG, Ma SK, Cheung CL, Raghwani J, Bhatt S, et al. 2009. Origins and evolutionary genomics of the 2009 swine-origin H1N1 influenza A epidemic. Nature 459: 1122–1125. 10.1038/nature08182 [DOI] [PubMed] [Google Scholar]
- Sobel Leonard A, Weissman D, Greenbaum B, Ghedin E, Koelle K. 2017. Transmission bottleneck size estimation from pathogen deep-sequencing data, with an application to human influenza A virus. J Virol 91: e00171-17 10.1128/JVI.00171-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strelkowa N, Lässig M. 2012. Clonal interference in the evolution of influenza. Genetics 192: 671–682. 10.1534/genetics.112.143396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taubenberger JK, Kash JC. 2010. Influenza virus evolution, host adaptation, and pandemic formation. Cell Host Microbe 7: 440–451. 10.1016/j.chom.2010.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K. 2003. Mortality associated with influenza and respiratory syncytial virus in the United States. J Am Med Assoc 289: 179–186. 10.1001/jama.289.2.179 [DOI] [PubMed] [Google Scholar]
- Van Kerkhove MD, Vandemaele KAH, Shinde V, Jaramillo-Gutierrez G, Koukounari A, Donnelly CA, Carlino LO, Owen R, Paterson B, Pelletier L, et al. 2011. Risk factors for severe outcomes following 2009 influenza A (H1N1) infection: a global pooled analysis. PLoS Med 8: e1001053 10.1371/journal.pmed.1001053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Riel D, Munster VJ, de Wit E, Rimmelzwaan GF, Fouchier RAM, Albert D M, Kuiken T. 2007. Human and avian influenza viruses target different cells in the lower respiratory tract of humans and other mammals. Am J Pathol 171: 1215–1223. 10.2353/ajpath.2007.070248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varble A, Albrecht RA, Backes S, Crumiller M, Bouvier NM, Sachs D, García-Sastre A, tenOever BR. 2014. Influenza A virus transmission bottlenecks are defined by infection route and recipient host. Cell Host Microbe 16: 691–700. 10.1016/j.chom.2014.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilijevic J, Zamarreño N, Oliveros JC, Rodriguez-Frandsen A, Gómez G, Rodriguez G, Pérez-Ruiz M, Rey S, Barba I, Pozo F, et al. 2017. Reduced accumulation of defective viral genomes contributes to severe outcome in influenza virus infected patients. PLoS Pathog 13: e1006650 10.1371/journal.ppat.1006650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. 2006. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature 439: 344–348. 10.1038/nature04388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaykrishna D, Poon LLM, Zhu HC, Ma SK, Li OTW, Cheung CL, Smith GJD, Peiris JSM, Guan Y. 2010. Reassortment of pandemic H1N1/2009 influenza A virus in swine. Science 328: 1529 10.1126/science.1189132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijaykrishna D, Holmes EC, Joseph U, Fourment M, Su YCF, Halpin R, Lee RTC, Deng Y-M, Gunalan V, Lin X, et al. 2015. The contrasting phylodynamics of human influenza B viruses. eLife 4: e05055 10.7554/eLife.05055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Magnus P. 1947. Studies on interference in experimental influenza. Ark Kemi Mineral Geol 24: 1–6. [Google Scholar]
- Wang GP, Sherrill-Mix SA, Chang K-M, Quince C, Bushman FD. 2010. Hepatitis C virus transmission bottlenecks analyzed by deep sequencing. J Virol 84: 6218–6228. 10.1128/JVI.02271-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan H, Sorrell EM, Song H, Hossain MJ, Ramirez-Nieto G, Monne I, Stevens J, Cattoli G, Capua I, Chen LM, et al. 2008. Replication and transmission of H9N2 influenza viruses in ferrets: evaluation of pandemic potential. PLoS ONE 3: e2923 10.1371/journal.pone.0002923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber TP, Stilianakis NI. 2008. Inactivation of influenza A viruses in the environment and modes of transmission: a critical review. J Infect 57: 361–373. 10.1016/j.jinf.2008.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. 1992. Evolution and ecology of influenza A viruses. Microbiol Rev 56: 152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock DM, Gubareva LV, Zuccotti G. 2003. Prolonged shedding of multidrug-resistant influenza A virus in an immunocompromised patient. N Engl J Med 348: 867–868. 10.1056/NEJM200302273480923 [DOI] [PubMed] [Google Scholar]
- Whitfield AE, Falk BW, Rotenberg D. 2015. Insect vector-mediated transmission of plant viruses. Virology 479-480: 278–289. 10.1016/j.virol.2015.03.026 [DOI] [PubMed] [Google Scholar]
- Wu G, Webby RJ. 2014. Barcoding influenza virus to decode transmission. Cell Host Microbe 16: 559–561. 10.1016/j.chom.2014.10.016 [DOI] [PubMed] [Google Scholar]
- Xue KS, Hooper KA, Ollodart AR, Dingens AS, Bloom JD. 2016. Cooperation between distinct viral variants promotes growth of H3N2 influenza in cell culture. eLife 5: e13974 10.7554/eLife.13974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue KS, Stevens-Ayers T, Campbell AP, Englund JA, Pergam SA, Boeckh M, Bloom JD. 2017. Parallel evolution of influenza across multiple spatiotemporal scales. eLife 6: e26875 10.7554/eLife.26875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Grantham M, Pantelic J, Bueno de Mesquita PJ, Albert B, Liu F, Ehrman S, Milton DK, EMIT Consortium. 2018. Infectious virus in exhaled breath of symptomatic seasonal influenza cases from a college community. Proc Natl Acad Sci 115: 1081–1086. 10.1073/pnas.1716561115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye L, Schnepf D, Becker J, Ebert K, Tanriver Y, Bernasconi V, Gad HH, Hartmann R, Lycke N, Staeheli P. 2019. Interferon-λ enhances adaptive mucosal immunity by boosting release of thymic stromal lymphopoietin. Nat Immunol 20: 593–601. 10.1038/s41590-019-0345-x [DOI] [PubMed] [Google Scholar]