Abstract
Metastasis, the dispersal of cancer cells from a primary tumor to secondary sites within the body, is the leading cause of cancer-related death. Animal models have been an indispensable tool to investigate the complex interactions between the cancer cells and the tumor microenvironment during the metastatic cascade. The zebrafish (Danio rerio) has emerged as a powerful vertebrate model for studying metastatic events in vivo. The zebrafish has many attributes including ex-utero development, which facilitates embryonic manipulation, as well as optically transparent tissues, which enables in vivo imaging of fluorescently labeled cells in real time. Here, we summarize the techniques which have been used to study cancer biology and metastasis in the zebrafish model organism, including genetic manipulation and transgenesis, cell transplantation, live imaging, and high-throughput compound screening. Finally, we discuss studies using the zebrafish, which have complemented and benefited metastasis research.
The zebrafish (Danio rerio) has become one of the most important vertebrate model organisms in biomedical research. Since the initial studies into genetics and vertebrate development, the zebrafish model has developed into a powerful system for studying many human diseases, including cancer biology, and has been used for anticancer drug discovery. Herein, we will review how the zebrafish model has benefited cancer research with an emphasis on metastasis. We believe that a good representation of the field is provided, however, due to the focus on metastasis we were not able to include every publication on the zebrafish cancer model.
EMERGENCE OF THE ZEBRAFISH AS A VERTEBRATE MODEL ORGANISM
First described by Francis Hamilton in “an account of the fishes found in the river Ganges and its branches” (Hamilton 1822), the zebrafish was originally a popular species for aquarium hobbyists. The potential of the zebrafish as a vertebrate model organism was first recognized by George Streisinger in the late 1960s (Grunwald and Eisen 2002), who published the first paper describing the use of the zebrafish and gynogenetic methods to identify recessive phenotypes from maternal genomes (Streisinger et al. 1981). At the same time, the zebrafish was becoming popular for forward genetic studies of development (Grunwald et al. 1988; Felsenfeld et al. 1990; Hatta et al. 1991). This led to the “big screen,” in the 1990s, in which the zebrafish was the first vertebrate to be used for a large-scale genetic screen to identify developmentally important genes. Christiane Nüsslein-Volhard, in Tübingen and Marc Fishman in Boston performed these screens, in parallel, using N-ethyl-N-nitrosourea (ENU) as a chemical mutagen (Driever et al. 1996; Haffter et al. 1996). Thousands of mutations that affected multiple developmental processes were identified. Further characterization found that many of the zebrafish phenotypes resembled human diseases due to alterations in orthologous genes in the two species. The potential of the zebrafish as a model organism for human disease was now recognized and as a result, the National Institutes of Health established the Trans-NIH Zebrafish Coordinating Committee in 1997 (Grunwald and Eisen 2002). The original zebrafish genetic map (Postlethwait et al. 1994) was expanded (Shimoda et al. 1999) and several zebrafish models of the human disease quickly followed (Brownlie et al. 1998; Wang et al. 1998; Childs et al. 2000). Further development of the model facilitated forward and reverse genetic approaches, genetic manipulation, and transgenesis (Nasevicius and Ekker 2000; Talbot and Hopkins 2000). In the early 2000s, The Sanger Institute embarked upon the Zebrafish Genome Sequencing Project (Howe et al. 2013). In addition, the Zebrafish Information Network (https://zfin.org/) was established (Sprague et al. 2001) as well as the Zebrafish International Resource Center, a central repository for wild-type, mutant, and transgenic zebrafish lines as well as information about zebrafish research. Today, a PubMed search for “zebrafish” will return over 35,000 items. New tools continue to be developed for the zebrafish, including advancements in genome editing using the CRISPR/Cas9 system (Hruscha et al. 2013; Hwang et al. 2013; Stella and Montoya 2016) and automated screening platforms, such as the vertebrate automated screening technology (VAST) (Pardo-Martin et al. 2010; Chang et al. 2012; Early et al. 2018). This will ensure that the zebrafish remains an important vertebrate model organism today and in the future.
ZEBRAFISH DEVELOPMENT AND THE FOUR LIFE STAGES
Zebrafish are small in size and are easy to breed and maintain in large numbers at low costs. Laboratory zebrafish are housed in purpose-built aquaculture systems, maintained at 28.5°C with a controllable light–dark cycle. Zebrafish are highly fecund and a gravid female can produce hundreds of eggs each week through natural spawning. Fertilization and embryo development both occur externally to the female. Embryonic development is rapid, within 24 h the single-celled zygote develops into a motile, transparent embryo with a classical vertebrate body plan (Kimmel et al. 1995). Maintained at 28.5°C, the majority of morphogenesis is complete by 3 d postfertilization (dpf) and the embryo will hatch from the protective chorion. This marks the transition from the embryonic to the larval stage. The larval zebrafish will continue to grow and between 5 and 6 dpf the digestive system and mouth become functional. The yolk sac which has sustained the animal throughout the embryonic and early larval stages is now rapidly depleted and is completely absorbed by 7 dpf (Kimmel et al. 1995). The zebrafish is considered juvenile once it has developed the majority of adult characteristics and an adult when it can produce viable gametes and reproduce (Kimmel et al. 1995; Parichy et al. 2009). Under optimal conditions, laboratory zebrafish will reach sexual maturity during the third month of their development.
THE ADVANTAGES OF USING THE ZEBRAFISH TO MODEL CANCER BIOLOGY
The zebrafish has several attributes that make it a suitable model for investigating human cancer biology and metastasis. Many of the factors involved in tumor progression are highly conserved between zebrafish and humans. The zebrafish genome contains 26,206 protein-coding genes. When compared to the human genome, 71.4% of human genes were found to have at least one zebrafish orthologue, while 82% of human disease-related genes had at least one zebrafish orthologue (Howe et al. 2013). In addition, multiple epigenetic markers regulating gene expression are also conserved across vertebrates including the zebrafish and humans (Long et al. 2013). Although many of the cell cycle genes, tumor suppressors, and oncogenes are conserved, allowing these tumorigenic pathways to be studied and targeted in the fish, it should be noted that zebrafish do not express clear orthologues of several human genes known to be involved in cancer progression. These include leukemia inhibitory factor (LIF), oncostatin M (OSM), and breast cancer 1 early onset (BRCA1) (Howe et al. 2013). However, as the receptors for these proteins are present in the zebrafish genome, it is possible that zebrafish proteins with similar functions to LIF and OSM exist, but sequence diversity is too great to recognize them as orthologues. In addition, a genome duplication has occurred in the Teleost fish genome following the phylogenetic divergence of fish and mammals (Meyer and Schartl 1999). This has resulted in the zebrafish having two copies of many genes (known as ohnologues) of which mammals only have one copy.
Zebrafish are easy to genetically manipulate through gene knockdowns, knockouts, overexpression, and transgenesis allowing the creation and characterization of cancer models. Importantly, there are similarities between the histopathology of human and zebrafish tumors (Amatruda et al. 2002). The rapid ex-utero development of the fertilized egg allows direct visualization and manipulation throughout embryogenesis and larval development. The transparency of the animal at these stages in combination with the generation of transgenic fluorescent reporter lines allows cellular and subcellular in vivo imaging to be performed allowing the tumor and its microenvironment to be monitored in real time. This has been one of the greatest benefits of using zebrafish to study cancer biology.
There is temporal separation in the maturation of the innate and adaptive immune systems in the zebrafish. The innate immune system is functional at 2 dpf, however, the adaptive immune system matures at 28 dpf (Lam et al. 2004). The lack of adaptive immunity in the larval zebrafish makes it an ideal in vivo model for the injection of cancerous cells without risk of rejection. Cells can either be allografted from a donor zebrafish (Langenau et al. 2003) or xenografted from a mammalian source (Nicoli et al. 2007). Proliferation, invasion, and metastasis of the injected cells can be investigated, as well as the interactions between the donor cells and both the host cells and the extracellular environment. However, this may not fully reproduce the behavior of cancer in an immunocompetent host.
TECHNIQUES USED TO STUDY CANCER BIOLOGY AND METASTASIS IN THE ZEBRAFISH
Genetic Manipulation
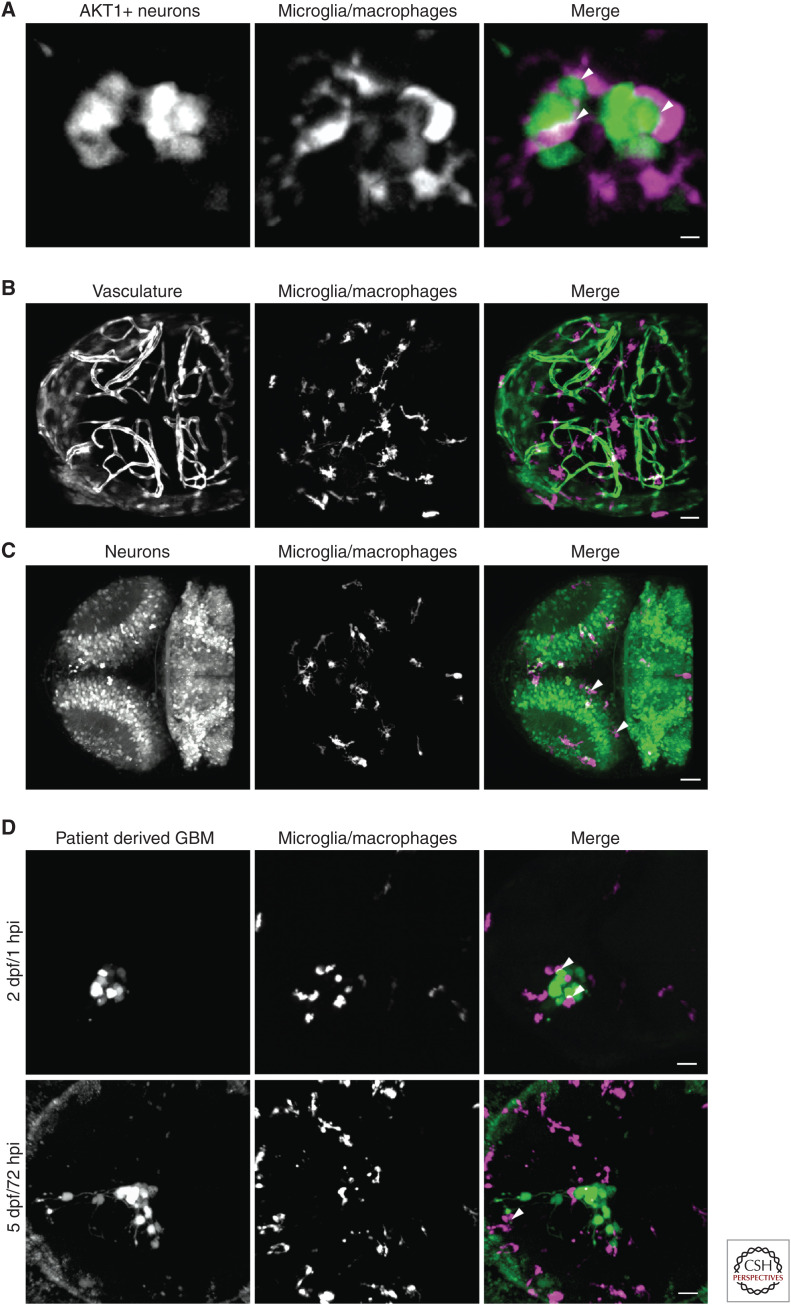
An important method in zebrafish research is the ability to induce cancers by expressing genes, including human oncogenes, in specific cells at specific time points. The transient cell-specific expression of oncogenes can be achieved using Tol2 mediated integration (Kawakami et al. 2004) which allows the early stages of cancer progression to be investigated. We have used this technique to express the human AKT1 oncogene in the neural cells of larval zebrafish. Based on this method, we showed that AKT1 activation in preneoplastic cells attracted macrophages via Sdf1b–Cxcr4b signaling, which in turn promoted oncogenic cell proliferation (Fig. 1A; Chia et al. 2018).
Figure 1.
Confocal fluorescent microscopy enables an analysis of cell interactions, in vivo and in real time. (A) Oncogenesis was achieved in zebrafish neurons (green) by misexpression of a constitutively active form of human AKT1. Direct cellular interactions (white arrowheads) between the oncogenic neurons and microglia/macrophages (magenta) were visualized in vivo in real time. The image is a maximum intensity projection, scale bar is 5 µm. (B,C) By combining fluorescent reporter transgenic lines and transparent zebrafish strains, the zebrafish becomes a powerful system for observing cellular interactions and tissue structures. (B) The Tg(fli1:GFP:mpeg1:mCherry) double transgenic zebrafish allows the vasculature (green) and microglia/macrophages (magenta) to be visualized in the zebrafish brain. (C) Tg(nbt:dsRed:mpeg1:EGFP) double transgenic zebrafish allow visualization of neurons (green) and microglia/macrophages (magenta) in the zebrafish brain. White arrowheads indicate apoptotic neurons that have been phagocytozed by the microglia/macrophages and are being digested within phagosomes. Images are maximum intensity projections showing the dorsal view of representative 3 d postfertilization (dpf) zebrafish embryo brains. Scale bars are 30 µm. (D) The invasiveness and cellular interactions of xenografted patient-derived tumor cells within the zebrafish brain can be investigated over time. Human, patient derived, glioblastoma multiforme cells (GBM; green) were transplanted into the left optic tectum of 2 dpf Tg(mpeg1:mCherry) zebrafish larvae. The larvae were imaged at 1 h postinjection (hpi) and at 72 hpi. Direct cellular interactions between the human tumor cells and the zebrafish microglia/macrophages (magenta) were observed at both time points (white arrowheads). The human cancer cells not only survived and proliferated within the zebrafish brain, but also exhibited an invasive phenotype, becoming more dispersed within the brain and extending long cellular processes into the surrounding brain tissue. Images are maximum intensity projections showing the dorsal view of a representative 2 dpf and a 5 dpf zebrafish embryo brain. Scale bars are 30 µm.
Spatial and temporal control of oncogene expression is often required to prevent oncogene toxicity or untimely death due to tumor burden. This has been achieved in the zebrafish by using the Gal4-UAS expression system (Scheer and Campos-Ortega 1999; Santoriello et al. 2010) the chemically inducible Tet-On system (Gossen and Bujard 1992; Gossen et al. 1995; Li et al. 2013) and LexPR:LexOP system (Emelyanov and Parinov 2008; Nguyen et al. 2012), as well as heat shock induction (Bajoghli et al. 2004; Leacock et al. 2012). Oncogenesis has also been achieved by transgene electroporation in adult zebrafish (TEAZ). Using this technique, oncogenes can be spatiotemporally expressed directly in adult somatic tissue, modeling tumor initiation and progression in a fully immunocompetent adult zebrafish (Callahan et al. 2018). Importantly, many of the zebrafish cancers generated by oncogene misexpression phenotypically copy the human equivalents (Santoriello et al. 2010). Furthermore, techniques are in place to isolate cells from zebrafish larvae followed by purification via flow cytometry to perform downstream applications including transcriptomic profiling (Mazzolini et al. 2018).
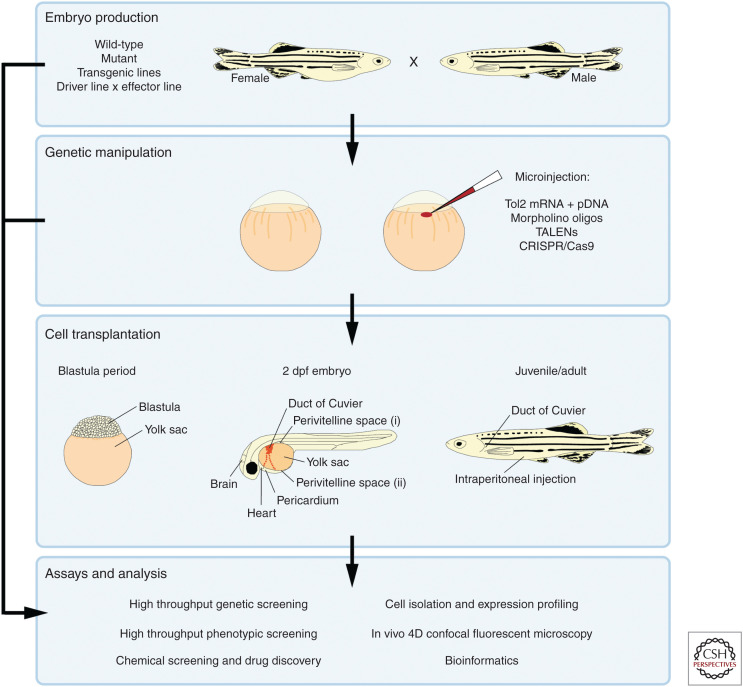
Cancers can also be induced by targeted gene knockdown. Effective gene knockdown can be achieved by microinjection of the one-cell stage embryo (Fig. 2). Morpholino antisense oligonucleotides (Nasevicius and Ekker 2000) can rapidly reveal the function of a gene, however, the phenotypes of morphants do not always correlate with those of stable mutants, implicating morpholino off-target effects (Kok et al. 2015). Synthetic transcription activator-like effector nucleases (TALENs) (Huang et al. 2011b; Sander et al. 2011) and the clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 system (Hruscha et al. 2013; Hwang et al. 2013; Stella and Montoya 2016) can be used to generate inheritable frameshift knockout mutations. However, TALENs have proved difficult to design, synthesize, and validate, preventing widespread adoption of the technique (Gupta and Musunuru 2014). The CRISPR/Cas9 system is robust and can be used to knockout tumor repressor genes to modify the tumor microenvironment. It can also be used to knock-in genes, such as human cytokines or receptors which may be useful for future transplantation studies. However, knockout mutants must be validated to ensure that alternative start sites or splice sites cannot still generate a functional protein or that hypomorphic alleles are generated with reduced gene function. There is also evidence that complete gene knockout can be rescued by genetic compensation from other related genes, which can prevent the appearance of mutant phenotypes (Rossi et al. 2015).
Figure 2.
An overview of the common zebrafish techniques used to study cancer biology and metastasis. Zebrafish embryos are obtained through natural spawning of wild-type, mutant or transgenic fish. Oncogene expression can be achieved by crossing a cell-specific driver line with an effector line carrying the oncogene of interest. Embryos can be genetically manipulated by microinjection of reagents into the single-cell embryo. Tol2 mediated integration can be used to express oncogenes in specific cell types, morpholino antisense oligonucleotides can be used to knockdown gene expression and TALENs or the CRISPR/Cas9 system can be used to knockout target genes. Transplantation of cancer cells into zebrafish has been performed at several developmental stages. Fluorescent labeling allows continuous visualization of the transplanted cells, tumor progression and metastasis. The most common zebrafish developmental stages used for transplantation as well as the most frequently used injection sites are shown. These techniques can be combined to produce zebrafish embryos and larvae for further analysis including high-throughput genetic and phenotypic screening, chemical screening and drug discovery, gene expression profiling and in vivo 4D confocal fluorescent microscopy.
Zebrafish Transgenics and Pigment Mutants
By combining transgenic lines and transparent zebrafish strains, the zebrafish becomes a powerful system for observing interactions between cancer cells and the surrounding environment. Live confocal fluorescent imaging enables an analysis of cell interactions in vivo and in real time. Hundreds of fluorescent reporter transgenic zebrafish lines have been established, labeling multiple cell types including blood vessels (Fig. 1B; Lawson and Weinstein 2002), immune cells (Fig. 1B,C; Renshaw et al. 2006; Ellett et al. 2011), and neurons (Fig. 1C; Peri and Nüsslein-Volhard 2008). There are also transgenic zebrafish that can be used to drive oncogene expression. For example, the Tg(5XUAS:eGFP-HRASV12)io006 effector line (Santoriello et al. 2010) can be used in combination with a Gal4 driver line to express a constitutively active form of HRas in a specific cell type.
The zebrafish embryo is transparent until 2 dpf, however, this transparency can be prolonged for a further 12 d through the use of melanin synthesis inhibitors (Karlsson et al. 2001). In addition, there are several pigment mutants such as nacre (Lister et al. 1999) and crystal (Antinucci and Hindges 2016). The casper pigment mutant (White et al. 2008) has proved popular for cell transplantation studies as it lacks both iridophores and melanocytes and maintains transparency as an adult fish. Using immunosuppressed adult casper zebrafish and an allografting technique White et al. (2008) were able to assess engraftment and proliferation of either pigmented or fluorescent-labeled melanoma cells. Metastatic melanoma cell invasion from the primary injection site into the host tissues could be followed by repeatedly imaging the same live animal without the need to sacrifice the animal.
Allograft Cancer Cell Transplantation
An allograft is the transfer of cells, tissues or organs from one animal to another of the same species. Transplantation into the embryonic or larval zebrafish prevents transplant rejection and the transparency of the animal at this age allows cancer cells to be visualized during engraftment, proliferation, migration, neovascularization, and therapeutic treatments. Allogeneic cancer cell transplantation into adult zebrafish requires immunosuppression either by chemical ablation using dexamethasone (Langenau et al. 2004; Stoletov et al. 2007) or a sublethal dose of γ-irradiation (Traver et al. 2004). However, both techniques may be hindered by toxicity and incompetent ablation of the immune cells (Smith et al. 2010), while the lack of a complete immune system prevents cancer cell–immune cell interactions from being studied. In addition, recovery of the immune system postirradiation will cause rejection of the transplanted cells and prevents long-term investigations.
To avoid the issues related to immunosuppression, the clonal (syngeneic) zebrafish model (Streisinger et al. 1981) can be used to perform tumor transplantations (Mizgirev and Revskoy 2010). Syngeneic zebrafish have fully competent, matched immune systems. Therefore, cancerous cells from a syngeneic donor can be transferred into a sibling recipient without rejection (Mizgireuv and Revskoy 2006). This removes the need for immunosuppression, allowing long-term tumor progression to be studied in the presence of a fully functional immune system and tumor microenvironment with little variance in the genetic background. Immune compromised mutant zebrafish have also been used for transplantation studies. The rag2E450fs mutant zebrafish has no mature T cells and a variable reduction in functional B cells (Tang et al. 2014). Cells and tissues from unrelated individuals can be transplanted into these fish and malignant cells engraft without the need for immunosuppression or major histocompatibility complex matching (Tenente et al. 2014).
Xenograft Cancer Cell Transplantation
A xenograft is the transfer of cells, tissues or organs from an animal of one species to an animal of a different species. Orthotopic xenografts involve transplanting tumor cells into the same organ type from which the cancer originated. Heterotopic xenografts involve transplanting tumor cells into a different tissue from which the cells originated. The zebrafish larva has proved to be an excellent system for human cancer xenograft studies. Fluorescent labeling allows continuous visualization of the transplanted cells and tumor progression (Ignatius and Langenau 2011). Due to the immature adaptive immune system, the human cancer cells survive within the zebrafish larvae, invade surrounding tissue and metastasize. Importantly, transplanted human cells can communicate with zebrafish cells due to the conservation between human and zebrafish signaling pathways (Tulotta et al. 2016). Furthermore, these pathways can be genetically or chemically targeted. The first zebrafish xenograft involved transplanting human melanoma cells into the zebrafish blastula (Lee et al. 2005), however, cancer cells are more often injected into the zebrafish larva at 2–4 dpf when the body plan has become established. Several injection sites are commonly utilized in the larval xenograft model including the yolk sac, the perivitelline space (the area between the outer membrane of the yolk sac and the skin), the duct of Cuvier (connecting the heart to the trunk vasculature), the pericardium, the heart and the brain (Fig. 2).
Many xenograft studies have used commercially available human (Ren et al. 2017) and murine (Zhao et al. 2011) cancer cell lines. This can be advantageous as it reduces variability and the cells can be manipulated in vitro before transplantation. However, due to the selection pressures of long-term maintenance in tissue culture, established cell lines may no longer resemble primary cancers. Therefore, xenografting studies have also been performed with primary patient-derived cells (Marques et al. 2009; Mercatali et al. 2016; Gaudenzi et al. 2017). As primary patient cells are difficult to obtain and expensive to maintain, the larval xenograft model is again beneficial as only small cell numbers are required for each injection (Fig. 1D). Patient-derived cells from a range of different cancers have also been xenografted into adult immunocompromised zebrafish. The prkdcD3612fs/D3612fs, Il2rgaY91fs/+, casper mutant lacks T, B, and NK cells and the engrafted cancer cells were shown to have similar growth kinetics and histopathological features to those grown in immunodeficient NSG mice (Yan et al. 2019). Various cell-labeling methods were used to follow cell growth, migration and responses to chemotherapeutics at single-cell resolution in vivo and provided proof of principle that this model could be used to develop patient-specific therapeutic approaches in the future.
Human cancer cells have also been transplanted into juvenile zebrafish to study microtumor formation, cell invasion, and angiogenesis (Stoletov et al. 2007). All tissues are fully developed at this stage, therefore, xenografting cells at this time may be more representative of tumor progression in an adult system without interference from developmental signaling in the embryo or larva. However, immunosuppression is still required and microscopy is more challenging at this age as the tissues are opaque. As well as confocal microscopy, several additional noninvasive imaging techniques can be used to identify cancer cells in juvenile and adult zebrafish in vivo. These include ultrasound biomicroscopy (Goessling et al. 2007), 2-photon microscopy and the luciferase based IVIS spectrum in vivo imaging system.
High-Throughput Chemical Genetic Screening
Chemical genetics involves using small molecules to alter biological pathways. The zebrafish is an excellent in vivo animal model for large-scale chemical screening. High fecundity, small size, and ease of handling all facilitate high-throughput screening. The most common format has been the 96-well plate, which allows small volumes of potentially expensive compounds to be tested. The aqueous environment allows the solubilization of compounds allowing easy drug delivery and pathway targeting in a whole organism. The rapid ex-utero development, transparency of the embryo and availability of transgenic lines allows for visualization throughout development and the detection of desired phenotypes within a short time frame. Physiologically relevant events can be evaluated simultaneously with drug efficacy and toxicity within an individual animal.
The first high-throughput forward genetic chemical screen, using wild-type zebrafish, was performed to identify novel compounds that regulate developmental pathways and embryogenesis (Peterson et al. 2000). Since this initial study, in vivo chemical screens utilizing zebrafish have been used to identify novel cancer therapeutic compounds (Wang et al. 2010; Ridges et al. 2012; Astin et al. 2014). Transgenic zebrafish allow phenotypes to be detected by fluorescent microscopy. The Tg(flk1:EGFP) transgenic line, with EGFP labeled vasculature has been used to identify compounds that inhibit angiogenesis (Wang et al. 2010). Tg(lyve1:egfp)nz150, which has EGFP labeled lymphatic vessels, has been used to identify compounds that inhibit lymphatic vessel growth (Astin et al. 2014) and the T-cell reporter line Tg(lck:EGFP) has been used to identify compounds with selective activity against leukemia (Ridges et al. 2012). Automation has increased the efficiency of high-throughput compound screening. Embryos can be automatically sorted and dispensed into multiwell plates (Pulak 2006; Veneman et al. 2013), automated robotic drug dosing and microinjection reduces variability (Wang et al. 2007; Huang et al. 2011a) and automated high-resolution in vivo imaging is achievable using the VAST BioImager (Pardo-Martin et al. 2010; Chang et al. 2012; Early et al. 2018).
ZEBRAFISH STUDIES OF THE METASTATIC PROCESS
The first use of zebrafish in cancer research was reported in 1965. In this study, zebrafish were exposed to carcinogens and were found to develop neoplasms (Stanton 1965). Since this initial publication, the zebrafish has become a powerful vertebrate system for modeling human cancers (Stoletov and Klemke 2008) and is an excellent platform to study all stages of metastasis.
Metastasis is a multistep process that involves the migration of tumor cells away from the primary tumor site, intravasation into the circulatory or lymphatic system, survival in the vasculature, extravasation, and colonization at a secondary site in a foreign environment.
Angiogenesis and Local Invasion from the Primary Tumor
Angiogenesis is important for tumor growth and new vessels may provide a route for cell dissemination and metastasis from the primary tumor. The predictable patterning of the vasculature, optical clarity, and availability of transgenic lines with fluorescently labeled vasculature made the zebrafish an attractive model for studying tumor–vasculature interactions in real time and led to the development of the zebrafish/tumor xenograft angiogenesis assay (Nicoli and Presta 2007). Multiple cancer cell types, including metastatic melanoma, breast adenocarcinoma, and pancreatic cancer have been shown to stimulate angiogenesis when xenografted into the zebrafish (Haldi et al. 2006; Stoletov et al. 2007; Nicoli et al. 2007). Mammalian tumor cells expressing angiogenic vascular endothelial growth factor (VEGF) can rapidly induce microvascularization when xenografted close to the subintestinal vasculature of developing zebrafish embryos (Nicoli et al. 2007), indicating that there is functional signaling between the mammalian and zebrafish cells. In addition to growth factors, modeling tumor-induced angiogenesis in the zebrafish has successfully identified other molecular factors that regulate tumor cell-induced neovascularization in vivo. For example, angiogenesis was significantly reduced in a pancreatic cancer cell xenograft assay following siRNA silencing of either LIMK1 or LIMK2 in the cancer cells (Vlecken and Bagowski 2009).
As a tumor mass grows it must recruit a blood supply to prevent tissue ischemia. However, hypoxia can still occur as tumor-induced vasculature is often disorganized and leaky. It has been suggested that leaky vessels and hypoxia may promote metastasis of tumor cells. To further investigate how hypoxia drives metastasis, the Cao research group used a zebrafish hypoxia model to show that VEGF secreted from tumor cells promoted induction of pathological angiogenesis which increased tumor cell invasion, dissemination, and metastasis under hypoxic conditions (Lee et al. 2009; Rouhi et al. 2010). This was the first animal model that combined the study of tumor hypoxia, angiogenesis, and cancer cell metastasis at the single-cell level.
Macrophages in the tumor microenvironment are also involved in tumor-induced angiogenesis. He et al. reported a macrophage-dependent neovascularization when tumor cells were injected into the duct of Cuvier of 2 dpf Tg(Fli1:GFP) zebrafish, which have fluorescent vasculature (Lawson and Weinstein 2002). This was the first example of de novo vascularization in a zebrafish xenograft system which was essential for the proliferation of both the tumor cells and the stromal cells during tumor progression (He et al. 2012). In addition, it has been shown that macrophage infiltration into glioblastoma xenografts is enhanced by TGF-β1 expression and the JNK pathway (Yang et al. 2013). Britto et al. (2018) found that the angiogenic response to VEGFA secreting xenografted tumor cells can be enhanced by direct interactions between noninflammatory macrophages and the growth tips of the developing blood vessels. Microglia, the resident macrophages of the central nervous system have also been shown to directly interact with human glioblastoma cells promoting the survival and invasiveness of the cancer cells when xenotransplanted into the zebrafish midbrain (Hamilton et al. 2016).
Cancer cells can associate and actively migrate along the outer surface of blood vessels, as has been observed for human glioblastoma when xenotransplanted into the hindbrain ventricle of 2 dpf zebrafish embryos (Gamble et al. 2018). Therefore, angiogenesis may promote cancer cell dispersal from the original cell mass by providing a physical structure along which the cells can travel. In addition, an increased migratory invasive phenotype may aid the initial stages of metastasis. While exploring tumor cell–endothelial cell interactions, Stoletov et al. (2007) found that cancer cells expressing the metastatic gene RhoC were found to disseminate further from the site of xenotransplantation. This may give cancer cells a metastatic advantage by increasing the probability that they will come into contact with the vasculature and undergo intravasation.
Cooperative invasion is a mechanism by which heterogeneous tumor cell populations communicate reciprocally and cooperate to migrate collectively from the initial cell mass. Chapman et al. (2014) found that coinjecting highly invasive melanoma cells, that deposit extracellular matrix and exhibit protease activity, along with less invasive melanoma cells increased the invasive potential of the less invasive cells and promoted melanoma progression.
Immune cells have been shown to be involved in cancer cell dissemination. A study using the zebrafish xenograft model to investigate the interactions between tumor cells and macrophages in association with the vasculature revealed a novel mechanism of intravasation involving tumor-associated macrophages (TAMs) (Wang et al. 2015). A mixture of tumor cells and macrophages were injected into the perivitelline space of 2 dpf Tg(Fli1:EGFP) larvae to partially recapitulate the tumor microenvironment. The findings revealed that IL6- or TNFα-stimulated noninflammatory TAMs enhanced metastasis of the tumor cells at the stage of intravasation and dissemination. Metastasis was dependent upon direct cancer cell–TAM interactions, with the majority of the disseminated tumor cells associated with TAMs in vivo (Wang et al. 2015). This study revealed a novel macrophage-dependent mechanism of metastasis and supported data from mouse tumor models that had previously suggested that TAMs facilitate tumor cell intravasation (Wyckoff et al. 2007; Pollard 2008). In addition, the model provides a functional platform that could be used to test metastatic potential of patient tumor cells and screen for new therapeutic agents.
Intravasation and Survival in the Circulation
The zebrafish xenograft model has been highly valuable when investigating intravasation. To explore tumor cell–endothelial cell interactions, Stoletov et al. (2007) transplanted fluorescent human cancer cells into the peritoneal cavity of 1-mo old immunosuppressed zebrafish and observed the cellular behaviors by high-resolution imaging. RhoC was found to work cooperatively with VEGF to promote intravasation. The injected cells formed microtumors in the zebrafish and VEGF secretion from these cells was found to promote localized angiogenesis, vascular remodeling, and permeablization due to disruptions in the endothelial cell layer. The metastatic gene RhoC regulates the actin–myosin cytoskeleton. Cancer cells expressing RhoC exhibited amoeboid cell migration, promoting dispersal, as well as the formation of long membrane protrusions and blebs. These long cell protrusions were seen to extend through the VEGF induced openings in the vasculature and allowed the tumor cells to enter the vessels.
In a study to determine the mechanisms by which T-lymphoblastic lymphoma (T-LBL) progresses to T-cell acute lymphoblastic leukemia (T-ALL), Feng et al. (2010) studied allografts in Tg(fli:egfp); casper zebrafish and found that overexpression of b-cell lymphoma 2 (bcl2) in combination with c-myc expression prevented MYC-induced apoptosis and increased malignant thymocyte transformation. However, these leukemia cells were unable to transit into the vasculature due to increased adhesion caused by elevated levels of Sphingosine-1-phosphate receptor 1 (S1P1) and Intercellular Adhesion Molecule 1 (ICAM1). This study revealed that reduced intercellular adhesion is required for intravasation and dissemination during the transition from T-LBL to T-ALL.
To investigate the behavior of tumor cells within the circulation, the cells can be injected directly into the duct of Cuvier in the developing zebrafish. Frequent time-lapse confocal microscopy revealed human melanoma cells interacting with zebrafish endothelial cells within the vasculature. Melanoma cells were seen to alternate between rolling along the inside surface of the vessels and being stationary (Hill et al. 2018). This assay can be used to model how tumor cells circulate and disseminate in vivo and how the cells survive the hostile environment of the circulatory system.
Tumor Cell Homing and Extravasation
Metastasis of several hematological cancers, as well as solid tumors, indicates a preference for homing to specific predetermined distant tissues. The bone marrow is a common site for secondary tumor growth during metastasis (Shiozawa et al. 2015). A zebrafish model was established in which the homing of multiple myeloma cells to the caudal hematopoietic tissue (CHT) was investigated (Sacco et al. 2012). The zebrafish CHT is known to be an intermediate site of hematopoiesis and leukocyte differentiation (Chen and Zon 2009) and is equivalent to the mammalian fetal liver (Murayama et al. 2006). The CHT is thought to represent a bone marrow-like hematopoietic stem cell niche which may be involved in the homing of tumor cells to the bone marrow (Shiozawa et al. 2015). Multiple myeloma cells rapidly entered the peripheral circulation and migrated to the CHT after intracardiac injection into 2 dpf casper embryos. Transcriptomic analyses of the multiple myeloma cells that had localized to the CHT revealed enrichment for genes known to regulate IL6 signaling, angiogenesis, and cell adhesion. Furthermore, silencing the known bone marrow homing factors CXCR4 (Alsayed et al. 2007), VLA4 (Sanz-Rodríguez et al. 1999; Ngo et al. 2008), and FAK (Park et al. 2013) in the multiple myeloid cells reduced localization to the CHT. Thus, this zebrafish system complements the murine models, allowing small numbers of valuable patient-derived cancer cells that metastazise to the bone marrow to be studied and used for chemotherapeutic screening. It also provides further evidence that human cells can communicate with the host zebrafish cells. Cancer cell homing and colonizing of the CHT has also been reported in additional xenograft models (Chen et al. 2017).
To investigate the behavior of cancer cells extravasating from the circulation Stoletov et al. (2010) combined xenografting with real-time high-resolution imaging. Human cancer cells were injected directly into the circulation through the pericardium. The cancer cells were found to become lodged in the small vessels (5–8 µm) of the zebrafish head and tail. Extravasation from the vessels was an active process, dependent on the metastatic potential of the cells, and did not cause vasculature damage or leakage. Circulating tumor cells arrested at sites where the nuclei of the endothelial cells protruded into the vessel, suggesting that the arrest of circulating cells was due to size restriction and not due to adhesive mechanisms. Arrested cells were able to migrate along the inside surface of the vessel and could generate enough force to migrate against the blood flow. They also had the ability to alter their shape and move through vessel branching points by adjusting the actin–myosin contractile system and creating rounded membrane protrusions. A thickening of the vasculature cell wall surrounding the arrested tumor cells was observed. This was caused by increased movement and clustering of the endothelial cells and was accompanied by remodeling of the endothelial cell junctions. The prometastatic genes Twist, VEGFA and integrin β1 promoted migration, remodeling of the vasculature and extravasation of the tumor cells (Stoletov et al. 2010).
Further work by Kanada et al. (2014) detected two distinct mechanisms of extravasation in the zebrafish. RFP-labeled Hela cells were injected into the circulation of 2 dpf Tg(flk1:EGFP) embryos and the process of extravasation was observed by long-time fluorescent time-lapse imaging. As previously observed (Stoletov et al. 2010), some of the Hela cells arrested in the vasculature and then underwent VEGF-dependent extravasation by actively penetrating the vessel walls. The second mechanism of extravasation occurred independently of VEGF signaling and involved stationary clusters of noninvasive Hela cells being progressively covered by a layer of endothelial cells. This continued until the Hela cells were excluded from the vessel, at which point the original endothelial cell layer disappeared and the Hela cells dispersed into the surrounding tissues (Kanada et al. 2014).
As well as the signaling between tumor cells and epithelial cells, there is evidence that immune cells can also have a role in promoting extravasation in vivo. Another study into tumor cell dispersal found that neutrophils can promote extravasation and invasion into secondary sites. He et al. (2012) injected fluorescent tumor cells into the duct of Cuvier of 2 dpf Tg(Fli1:GFP) transgenic zebrafish. Some of the injected cells were disseminated through the vasculature by the blood circulation. Extravasation of the dispersed tumor cells was observed at different locations throughout the larvae, however, only cells that had localized to the CHT survived and successfully invaded the surrounding tissue. This was found to be due to the high numbers of neutrophils in the CHT. Migration of neutrophils between the CHT and tail fin distorted the extracellular fibrillary collagen matrix forming a metastatic niche that promoted tumor cell invasion. As had been reported in mouse models and human patients treated with VEGFR inhibitors (Ebos et al. 2009; Loges et al. 2009; Pàez-Ribes et al. 2009), VEGFR inhibition was found to reduce tumor growth at the primary site, but promoted metastasis and cell invasion. This may be due to increased neutrophil migration generating pathways in the collagen, through which the tumor cells can migrate (He et al. 2012). Further study of the involvement of neutrophils during metastasis found that neutrophils in zebrafish larvae express high levels of cxcr4 (cxcr4b), which controlled the neutrophil number, adhesion, and motility (Tulotta et al. 2019). Tumor cells xenografted into cxcr4b homozygote mutant zebrafish failed to form micrometastazes in the CHT. This was due to either neutrophil migratory impairment, resulting in reduced metastatic niche formation, or the absence of cxcr4b-dependent tumor cell homing. Once the human tumor cells had extravasated through the CHT and invaded the surrounding tissue, a cxcr4b-dependent migration of neutrophils out of the CHT was observed. These neutrophils were seen to slow down and interact with the metastasizing cancer cells. These studies suggest that targeting cxcr4 on both tumor cells and neutrophils could have a therapeutic benefit to constrain cancer progression (Tulotta et al. 2019).
Colonization at a Secondary Site
The final stage of metastasis involves colonization of a secondary site in a foreign environment. Therefore, a metastatic niche capable of sustaining tumor growth must be established. To investigate this process further Ignatius et al. used the KRASG12D-induced embryonal rhabdomyosarcoma (ERMS) zebrafish model of an aggressive pediatric muscle sarcoma (Langenau et al. 2007) to identify tumor-propagating cells (Ignatius et al. 2012). ERMS-propagating cells expressing myf5 exhibited enhanced proliferative capacity but minimal migratory potential. These cells were unable to undergo intravasation and were often restricted to the primary tumor mass. As differentiation progressed, a subset of ERMS cells expressing myogenin became highly migratory. These cells lack tumor-propagating potential, but, can transverse the vasculature and seed secondary sites for tumor growth. These secondary sites were later colonized by the slower moving ERMS-propagating cells (Ignatius et al. 2012). This work suggests that nontumor propagating cells may have important roles in facilitating tumor spread as well as maintaining the primary tumor microenvironment, which could have therapeutic implications.
After metastatic dissemination, the microenvironment of the secondary site can produce factors to promote phenotypic switching and survival of the tumor cells at the new location. The White laboratory used a zebrafish melanoma model to observe changes in cell differentiation during metastatic engraftment (Kim et al. 2017). This study identified that after extravasation melanoma cells undergo melanocytic differentiation and proliferation, regulated by the microenvironmental factor Edn3. CRISPR/Cas9 mediated knockdown of this developmental morphogen prolonged survival of zebrafish transplanted with melanoma cells. This study emphasizes the significant role that the microenvironment plays in metastatic success and disease progression (Kim et al. 2017).
CONCLUSION
The zebrafish has proved itself as a suitable and useful model vertebrate to study tumor biology and complements the in vitro and in vivo mammalian cancer models. Techniques such as tumor cell transplantation assays, genetic manipulation, high-throughput chemical screening and in vivo live imaging have all enhanced our knowledge of cancer progression and metastasis. The zebrafish has been invaluable for single-cell high-resolution imaging and observing direct tumor cell interactions with the microenvironment during the more elusive stages of metastasis. The model is continuously being improved and advances in automation as well as the use of patient-derived xenografting assays can be used to screen in vivo drug responses and the kinetics of combinational therapies. In turn, this could lead to rapid patient-specific preclinical screening of cancer cell responses to specific drug combinations and guide treatment plans to target and prevent metastasis.
Footnotes
Editors: Jeffrey W. Pollard and Yibin Kang
Additional Perspectives on Metastasis: Mechanism to Therapy available at www.perspectivesinmedicine.org
REFERENCES
- Alsayed Y, Ngo H, Runnels J, Leleu X, Singha UK, Pitsillides CM, Spencer JA, Kimlinger T, Ghobrial JM, Jia X, et al. 2007. Mechanisms of regulation of CXCR4/SDF-1 (CXCL12)-dependent migration and homing in multiple myeloma. Blood 109: 2708–2717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amatruda JF, Shepard JL, Stern HM, Zon LI. 2002. Zebrafish as a cancer model system. Cancer Cell 1: 229–231. 10.1016/S1535-6108(02)00052-1 [DOI] [PubMed] [Google Scholar]
- Antinucci P, Hindges R. 2016. A crystal-clear zebrafish for in vivo imaging. Sci Rep 6: 29490 10.1038/srep29490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Astin JW, Jamieson SMF, Eng TCY, Flores MV, Misa JP, Chien A, Crosier KE, Crosier PS. 2014. An in vivo antilymphatic screen in zebrafish identifies novel inhibitors of mammalian lymphangiogenesis and lymphatic-mediated metastasis. Mol Cancer Ther 13: 2450–2462. 10.1158/1535-7163.MCT-14-0469-T [DOI] [PubMed] [Google Scholar]
- Bajoghli B, Aghaallaei N, Heimbucher T, Czerny T. 2004. An artificial promoter construct for heat-inducible misexpression during fish embryogenesis. Dev Biol 271: 416–430. 10.1016/j.ydbio.2004.04.006 [DOI] [PubMed] [Google Scholar]
- Britto DD, Wyroba B, Chen W, Lockwood RA, Tran KB, Shepherd PR, Hall CJ, Crosier KE, Crosier PS, Astin JW. 2018. Macrophages enhance Vegfa-driven angiogenesis in an embryonic zebrafish tumour xenograft model. Dis Model Mech 11: dmm035998 10.1242/dmm.035998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlie A, Donovan A, Pratt SJ, Paw BH, Oates AC, Brugnara C, Witkowska HE, Sassa S, Zon LI. 1998. Positional cloning of the zebrafish sauternes gene: a model for congenital sideroblastic anaemia. Nat Genet 20: 244–250. 10.1038/3049 [DOI] [PubMed] [Google Scholar]
- Callahan SJ, Tepan S, Zhang YM, Lindsay H, Burger A, Campbell NR, Kim IS, Hollmann TJ, Studer L, Mosimann C, et al. 2018. Cancer modeling by transgene electroporation in adult zebrafish (TEAZ). Dis Model Mech 11: dmm034561 10.1242/dmm.034561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang TY, Pardo-Martin C, Allalou A, Wählby C, Yanik MF. 2012. Fully automated cellular-resolution vertebrate screening platform with parallel animal processing. Lab Chip 12: 711–716. 10.1039/C1LC20849G [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman A, Fernandez del Ama L, Ferguson J, Kamarashev J, Wellbrock C, Hurlstone A. 2014. Heterogeneous tumor subpopulations cooperate to drive invasion. Cell Rep 8: 688–695. 10.1016/j.celrep.2014.06.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AT, Zon LI. 2009. Zebrafish blood stem cells. J Cell Biochem 108: 35–42. 10.1002/jcb.22251 [DOI] [PubMed] [Google Scholar]
- Chen L, Groenewoud A, Tulotta C, Zoni E, Kruithof-de Julio M, van der Horst G, van der Pluijm G, Ewa Snaar-Jagalska B. 2017. A zebrafish xenograft model for studying human cancer stem cells in distant metastasis and therapy response. Methods Cell Biol 138: 471–496. 10.1016/bs.mcb.2016.10.009 [DOI] [PubMed] [Google Scholar]
- Chia K, Mazzolini J, Mione M, Sieger D. 2018. Tumor initiating cells induce Cxcr4-mediated infiltration of pro-tumoral macrophages into the brain. eLife 7: e31918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs S, Weinstein BM, Mohideen MA, Donohue S, Bonkovsky H, Fishman MC. 2000. Zebrafish dracula encodes ferrochelatase and its mutation provides a model for erythropoietic protoporphyria. Curr Biol 10: 1001–1004. 10.1016/S0960-9822(00)00653-9 [DOI] [PubMed] [Google Scholar]
- Driever W, Solnica-Krezel L, Schier AF, Neuhauss SC, Malicki J, Stemple DL, Stainier DY, Zwartkruis F, Abdelilah S, Rangini Z, et al. 1996. A genetic screen for mutations affecting embryogenesis in zebrafish. Development 123: 37–46. [DOI] [PubMed] [Google Scholar]
- Early JJ, Cole KL, Williamson JM, Swire M, Kamadurai H, Muskavitch M, Lyons DA. 2018. An automated high-resolution in vivo screen in zebrafish to identify chemical regulators of myelination. eLife 7: e35136 10.7554/eLife.35136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebos JML, Lee CR, Cruz-Munoz W, Bjarnason GA, Christensen JG, Kerbel RS. 2009. Accelerated metastasis after short-term treatment with a potent inhibitor of tumor angiogenesis. Cancer Cell 15: 232–239. 10.1016/j.ccr.2009.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellett F, Pase L, Hayman JW, Andrianopoulos A, Lieschke GJ. 2011. mpeg1 promoter transgenes direct macrophage-lineage expression in zebrafish. Blood 117: e49–e56. 10.1182/blood-2010-10-314120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelyanov A, Parinov S. 2008. Mifepristone-inducible LexPR system to drive and control gene expression in transgenic zebrafish. Dev Biol 320: 113–121. 10.1016/j.ydbio.2008.04.042 [DOI] [PubMed] [Google Scholar]
- Felsenfeld AL, Walker C, Westerfield M, Kimmel C, Streisinger G. 1990. Mutations affecting skeletal muscle myofibril structure in the zebrafish. Development 108: 443–459. [DOI] [PubMed] [Google Scholar]
- Feng H, Stachura DL, White RM, Gutierrez A, Zhang L, Sanda T, Jette CA, Testa JR, Neuberg DS, Langenau DM, et al. 2010. T-lymphoblastic lymphoma cells express high levels of BCL2, S1P1, and ICAM1, leading to a blockade of tumor cell intravasation. Cancer Cell 18: 353–366. 10.1016/j.ccr.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamble JT, Reed-Harris Y, Barton CL, La Du J, Tanguay R, Greenwood JA. 2018. Quantification of glioblastoma progression in zebrafish xenografts: adhesion to laminin alpha 5 promotes glioblastoma microtumor formation and inhibits cell invasion. Biochem Biophys Res Commun 506: 833–839. 10.1016/j.bbrc.2018.10.076 [DOI] [PubMed] [Google Scholar]
- Gaudenzi G, Albertelli M, Dicitore A, Würth R, Gatto F, Barbieri F, Cotelli F, Florio T, Ferone D, Persani L, et al. 2017. Patient-derived xenograft in zebrafish embryos: a new platform for translational research in neuroendocrine tumors. Endocrine 57: 214–219. 10.1007/s12020-016-1048-9 [DOI] [PubMed] [Google Scholar]
- Goessling W, North TE, Zon LI. 2007. Ultrasound biomicroscopy permits in vivo characterization of zebrafish liver tumors. Nat Methods 4: 551–553. 10.1038/nmeth1059 [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H. 1992. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci 89: 5547–5551. 10.1073/pnas.89.12.5547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossen M, Freundlieb S, Bender G, Müller G, Hillen W, Bujard H. 1995. Transcriptional activation by tetracyclines in mammalian cells. Science 268: 1766–1769. 10.1126/science.7792603 [DOI] [PubMed] [Google Scholar]
- Grunwald DJ, Eisen JS. 2002. Headwaters of the zebrafish—emergence of a new model vertebrate. Nat Rev Genet 3: 717–724. 10.1038/nrg892 [DOI] [PubMed] [Google Scholar]
- Grunwald DJ, Kimmel CB, Westerfield M, Walker C, Streisinger G. 1988. A neural degeneration mutation that spares primary neurons in the zebrafish. Dev Biol 126: 115–128. 10.1016/0012-1606(88)90245-X [DOI] [PubMed] [Google Scholar]
- Gupta RM, Musunuru K. 2014. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J Clin Invest 124: 4154–4161. 10.1172/JCI72992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haffter P, Granato M, Brand M, Mullins MC, Hammerschmidt M, Kane DA, Odenthal J, van Eeden FJ, Jiang YJ, Heisenberg CP, et al. 1996. The identification of genes with unique and essential functions in the development of the zebrafish, Danio rerio. Development 123: 1–36. [DOI] [PubMed] [Google Scholar]
- Haldi M, Ton C, Seng WL, McGrath P. 2006. Human melanoma cells transplanted into zebrafish proliferate, migrate, produce melanin, form masses and stimulate angiogenesis in zebrafish. Angiogenesis 9: 139–151. 10.1007/s10456-006-9040-2 [DOI] [PubMed] [Google Scholar]
- Hamilton F. 1822. An account of the fishes found in the river ganges and its branches. Archibald Constable and Company, Edinburgh. [Google Scholar]
- Hamilton L, Astell KR, Velikova G, Sieger D. 2016. A zebrafish live imaging model reveals differential responses of microglia toward glioblastoma cells in vivo. Zebrafish 13: 523–534. 10.1089/zeb.2016.1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatta K, Kimmel CB, Ho RK, Walker C. 1991. The cyclops mutation blocks specification of the floor plate of the zebrafish central nervous system. Nature 350: 339–341. 10.1038/350339a0 [DOI] [PubMed] [Google Scholar]
- He S, Lamers GE, Beenakker JWM, Cui C, Ghotra VP, Danen EH, Meijer AH, Spaink HP, Snaar-Jagalska BE. 2012. Neutrophil-mediated experimental metastasis is enhanced by VEGFR inhibition in a zebrafish xenograft model. J Pathol 227: 431–445. 10.1002/path.4013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill D, Chen L, Snaar-Jagalska E, Chaudhry B. 2018. Embryonic zebrafish xenograft assay of human cancer metastasis. F1000Res 7: 1682 10.12688/f1000research.16659.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, et al. 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496: 498–503. 10.1038/nature12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hruscha A, Krawitz P, Rechenberg A, Heinrich V, Hecht J, Haass C, Schmid B. 2013. Efficient CRISPR/Cas9 genome editing with low off-target effects in zebrafish. Development 140: 4982–4987. 10.1242/dev.099085 [DOI] [PubMed] [Google Scholar]
- Huang H, Mills JK, Lu C, Sun D. 2011a. A universal piezo-driven ultrasonic cell microinjection system. Biomed Microdevices 13: 743–752. 10.1007/s10544-011-9544-4 [DOI] [PubMed] [Google Scholar]
- Huang P, Xiao A, Zhou M, Zhu Z, Lin S, Zhang B. 2011b. Heritable gene targeting in zebrafish using customized TALENs. Nat Biotechnol 29: 699–700. 10.1038/nbt.1939 [DOI] [PubMed] [Google Scholar]
- Hwang WY, Fu Y, Reyon D, Maeder ML, Kaini P, Sander JD, Joung JK, Peterson RT, Yeh JRJ. 2013. Heritable and precise zebrafish genome editing using a CRISPR-Cas system. PLoS One 8: e68708 10.1371/journal.pone.0068708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatius MS, Langenau DM. 2011. Fluorescent imaging of cancer in zebrafish. Methods Cell Biol 105: 437–459. 10.1016/B978-0-12-381320-6.00019-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatius MS, Chen E, Elpek NM, Fuller AZ, Tenente IM, Clagg R, Liu S, Blackburn JS, Linardic CM, Rosenberg AE, et al. 2012. In vivo imaging of tumor-propagating cells, regional tumor heterogeneity, and dynamic cell movements in embryonal rhabdomyosarcoma. Cancer Cell 21: 680–693. 10.1016/j.ccr.2012.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanada M, Zhang J, Yan L, Sakurai T, Terakawa S. 2014. Endothelial cell-initiated extravasation of cancer cells visualized in zebrafish. Peer J 2: e688 10.7717/peerj.688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsson J, von Hofsten J, Olsson PE. 2001. Generating transparent zebrafish: a refined method to improve detection of gene expression during embryonic development. Mar Biotechnol (NY) 3: 522–527. 10.1007/s1012601-0053-4 [DOI] [PubMed] [Google Scholar]
- Kawakami K, Takeda H, Kawakami N, Kobayashi M, Matsuda N, Mishina M. 2004. A transposon-mediated gene trap approach identifies developmentally regulated genes in zebrafish. Dev Cell 7: 133–144. 10.1016/j.devcel.2004.06.005 [DOI] [PubMed] [Google Scholar]
- Kim IS, Heilmann S, Kansler ER, Zhang Y, Zimmer M, Ratnakumar K, Bowman RL, Simon-Vermot T, Fennell M, Garippa R, et al. 2017. Microenvironment-derived factors driving metastatic plasticity in melanoma. Nat Commun 8: 14343 10.1038/ncomms14343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF. 1995. Stages of embryonic development of the zebrafish. Dev Dyn 203: 253–310. 10.1002/aja.1002030302 [DOI] [PubMed] [Google Scholar]
- Kok FO, Shin M, Ni CW, Gupta A, Grosse AS, van Impel A, Kirchmaier BC, Peterson-Maduro J, Kourkoulis G, Male I, et al. 2015. Reverse genetic screening reveals poor correlation between morpholino-induced and mutant phenotypes in zebrafish. Dev Cell 32: 97–108. 10.1016/j.devcel.2014.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SH, Chua HL, Gong Z, Lam TJ, Sin YM. 2004. Development and maturation of the immune system in zebrafish, Danio rerio: a gene expression profiling, in situ hybridization and immunological study. Dev Comp Immunol 28: 9–28. 10.1016/S0145-305X(03)00103-4 [DOI] [PubMed] [Google Scholar]
- Langenau DM, Traver D, Ferrando AA, Kutok JL, Aster JC, Kanki JP, Lin S, Prochownik E, Trede NS, Zon LI, et al. 2003. Myc-induced T cell leukemia in transgenic zebrafish. Science 299: 887–890. 10.1126/science.1080280 [DOI] [PubMed] [Google Scholar]
- Langenau DM, Ferrando AA, Traver D, Kutok JL, Hezel JPD, Kanki JP, Zon LI, Look AT, Trede NS. 2004. In vivo tracking of T cell development, ablation, and engraftment in transgenic zebrafish. Proc Natl Acad Sci 101: 7369–7374. 10.1073/pnas.0402248101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langenau DM, Keefe MD, Storer NY, Guyon JR, Kutok JL, Le X, Goessling W, Neuberg DS, Kunkel LM, Zon LI. 2007. Effects of RAS on the genesis of embryonal rhabdomyosarcoma. Genes Dev 21: 1382–1395. 10.1101/gad.1545007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson ND, Weinstein BM. 2002. In vivo imaging of embryonic vascular development using transgenic zebrafish. Dev Biol 248: 307–318. 10.1006/dbio.2002.0711 [DOI] [PubMed] [Google Scholar]
- Leacock SW, Basse AN, Chandler GL, Kirk AM, Rakheja D, Amatruda JF. 2012. A zebrafish transgenic model of Ewing's sarcoma reveals conserved mediators of EWS-FLI1 tumorigenesis. Dis Model Mech 5: 95–106. 10.1242/dmm.007401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee LMJ, Seftor EA, Bonde G, Cornell RA, Hendrix MJC. 2005. The fate of human malignant melanoma cells transplanted into zebrafish embryos: assessment of migration and cell division in the absence of tumor formation. Dev Dyn 233: 1560–1570. 10.1002/dvdy.20471 [DOI] [PubMed] [Google Scholar]
- Lee SLC, Rouhi P, Dahl Jensen L, Zhang D, Ji H, Hauptmann G, Ingham P, Cao Y. 2009. Hypoxia-induced pathological angiogenesis mediates tumor cell dissemination, invasion, and metastasis in a zebrafish tumor model. Proc Natl Acad Sci 106: 19485–19490. 10.1073/pnas.0909228106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zheng W, Wang Z, Zeng Z, Zhan H, Li C, Zhou L, Yan C, Spitsbergen JM, Gong Z. 2013. A transgenic zebrafish liver tumor model with inducible Myc expression reveals conserved Myc signatures with mammalian liver tumors. Dis Model Mech 6: 414–423. 10.1242/dmm.010462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister JA, Robertson CP, Lepage T, Johnson SL, Raible DW. 1999. nacre encodes a zebrafish microphthalmia-related protein that regulates neural-crest-derived pigment cell fate. Development 126: 3757–3767. [DOI] [PubMed] [Google Scholar]
- Loges S, Mazzone M, Hohensinner P, Carmeliet P. 2009. Silencing or fueling metastasis with VEGF inhibitors: antiangiogenesis revisited. Cancer Cell 15: 167–170. 10.1016/j.ccr.2009.02.007 [DOI] [PubMed] [Google Scholar]
- Long HK, Sims D, Heger A, Blackledge NP, Kutter C, Wright ML, Grützner F, Odom DT, Patient R, Ponting CP, et al. 2013. Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. eLife 2: e00348 10.7554/eLife.00348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques IJ, Weiss FU, Vlecken DH, Nitsche C, Bakkers J, Lagendijk AK, Partecke LI, Heidecke CD, Lerch MM, Bagowski CP. 2009. Metastatic behaviour of primary human tumours in a zebrafish xenotransplantation model. BMC Cancer 9: 128 10.1186/1471-2407-9-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzolini J, Chia K, Sieger D. 2018. Isolation and RNA extraction of neurons, macrophages and microglia from larval zebrafish brains. J Vis Exp. 134: e57431. doi:10.3791/57431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercatali L, La Manna F, Groenewoud A, Casadei R, Recine F, Miserocchi G, Pieri F, Liverani C, Bongiovanni A, Spadazzi C, et al. 2016. Development of a patient-derived xenograft (PDX) of breast cancer bone metastasis in a zebrafish model. Int J Mol Sci 17: E1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Schartl M. 1999. Gene and genome duplications in vertebrates: the one-to-four (-to-eight in fish) rule and the evolution of novel gene functions. Curr Opin Cell Biol 11: 699–704. 10.1016/S0955-0674(99)00039-3 [DOI] [PubMed] [Google Scholar]
- Mizgireuv IV, Revskoy SY. 2006. Transplantable tumor lines generated in clonal zebrafish. Cancer Res 66: 3120–3125. 10.1158/0008-5472.CAN-05-3800 [DOI] [PubMed] [Google Scholar]
- Mizgirev I, Revskoy S. 2010. Generation of clonal zebrafish lines and transplantable hepatic tumors. Nat Protoc 5: 383–394. 10.1038/nprot.2010.8 [DOI] [PubMed] [Google Scholar]
- Murayama E, Kissa K, Zapata A, Mordelet E, Briolat V, Lin HF, Handin RI, Herbomel P. 2006. Tracing hematopoietic precursor migration to successive hematopoietic organs during zebrafish development. Immunity 25: 963–975. 10.1016/j.immuni.2006.10.015 [DOI] [PubMed] [Google Scholar]
- Nasevicius A, Ekker SC. 2000. Effective targeted gene “knockdown” in zebrafish. Nat Genet 26: 216–220. 10.1038/79951 [DOI] [PubMed] [Google Scholar]
- Ngo HT, Leleu X, Lee J, Jia X, Melhem M, Runnels J, Moreau AS, Burwick N, Azab AK, Roccaro A, et al. 2008. SDF-1/CXCR4 and VLA-4 interaction regulates homing in Waldenstrom macroglobulinemia. Blood 112: 150–158. 10.1182/blood-2007-12-129395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen AT, Emelyanov A, Koh CHV, Spitsbergen JM, Parinov S, Gong Z. 2012. An inducible krasV12 transgenic zebrafish model for liver tumorigenesis and chemical drug screening. Dis Model Mech 5: 63–72. 10.1242/dmm.008367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoli S, Presta M. 2007. The zebrafish/tumor xenograft angiogenesis assay. Nat Protoc 2: 2918–2923. 10.1038/nprot.2007.412 [DOI] [PubMed] [Google Scholar]
- Nicoli S, Ribatti D, Cotelli F, Presta M. 2007. Mammalian tumor xenografts induce neovascularization in zebrafish embryos. Cancer Res 67: 2927–2931. 10.1158/0008-5472.CAN-06-4268 [DOI] [PubMed] [Google Scholar]
- Pàez-Ribes M, Allen E, Hudock J, Takeda T, Okuyama H, Viñals F, Inoue M, Bergers G, Hanahan D, Casanovas O. 2009. Antiangiogenic therapy elicits malignant progression of tumors to increased local invasion and distant metastasis. Cancer Cell 15: 220–231. 10.1016/j.ccr.2009.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Martin C, Chang TY, Koo BK, Gilleland CL, Wasserman SC, Yanik MF. 2010. High-throughput in vivo vertebrate screening. Nat Methods 7: 634–636. 10.1038/nmeth.1481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parichy DM, Elizondo MR, Mills MG, Gordon TN, Engeszer RE. 2009. Normal table of postembryonic zebrafish development: staging by externally visible anatomy of the living fish. Dev Dyn 238: 2975–3015. 10.1002/dvdy.22113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Wolfram P, Canty K, Harley B, Nombela-Arrieta C, Pivarnik G, Manis J, Beggs HE, Silberstein LE. 2013. Focal adhesion kinase regulates the localization and retention of pro-B cells in bone marrow microenvironments. J Immunol 190: 1094–1102. 10.4049/jimmunol.1202639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peri F, Nüsslein-Volhard C. 2008. Live imaging of neuronal degradation by microglia reveals a role for v0-ATPase a1 in phagosomal fusion in vivo. Cell 133: 916–927. 10.1016/j.cell.2008.04.037 [DOI] [PubMed] [Google Scholar]
- Peterson RT, Link BA, Dowling JE, Schreiber SL. 2000. Small molecule developmental screens reveal the logic and timing of vertebrate development. Proc Natl Acad Sci 97: 12965–12969. 10.1073/pnas.97.24.12965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard JW. 2008. Macrophages define the invasive microenvironment in breast cancer. J Leukoc Biol 84: 623–630. 10.1189/jlb.1107762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postlethwait JH, Johnson SL, Midson CN, Talbot WS, Gates M, Ballinger EW, Africa D, Andrews R, Carl T, Eisen JS, et al. 1994. A genetic linkage map for the zebrafish. Science 264: 699–703. 10.1126/science.8171321 [DOI] [PubMed] [Google Scholar]
- Pulak R. 2006. Techniques for analysis, sorting, and dispensing of C. elegans on the COPAS flow-sorting system. Methods Mol Biol 351: 275–286. [DOI] [PubMed] [Google Scholar]
- Ren J, Liu S, Cui C, Ten Dijke P. 2017. Invasive behavior of human breast cancer cells in embryonic zebrafish. J Vis Exp 122: e55459 10.3791/55459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renshaw SA, Loynes CA, Trushell DMI, Elworthy S, Ingham PW, Whyte MKB. 2006. A transgenic zebrafish model of neutrophilic inflammation. Blood 108: 3976–3978. 10.1182/blood-2006-05-024075 [DOI] [PubMed] [Google Scholar]
- Ridges S, Heaton WL, Joshi D, Choi H, Eiring A, Batchelor L, Choudhry P, Manos EJ, Sofla H, Sanati A, et al. 2012. Zebrafish screen identifies novel compound with selective toxicity against leukemia. Blood 119: 5621–5631. 10.1182/blood-2011-12-398818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A, Kontarakis Z, Gerri C, Nolte H, Hölper S, Krüger M, Stainier DYR. 2015. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature 524: 230–233. 10.1038/nature14580 [DOI] [PubMed] [Google Scholar]
- Rouhi P, Jensen LD, Cao Z, Hosaka K, Länne T, Wahlberg E, Steffensen JF, Cao Y. 2010. Hypoxia-induced metastasis model in embryonic zebrafish. Nat Protoc 5: 1911–1918. 10.1038/nprot.2010.150 [DOI] [PubMed] [Google Scholar]
- Sacco E, Hasan MM, Alberghina L, Vanoni M. 2012. Comparative analysis of the molecular mechanisms controlling the initiation of chromosomal DNA replication in yeast and in mammalian cells. Biotechnol Adv 30: 73–98. 10.1016/j.biotechadv.2011.09.009 [DOI] [PubMed] [Google Scholar]
- Sander JD, Cade L, Khayter C, Reyon D, Peterson RT, Joung JK, Yeh JRJ. 2011. Targeted gene disruption in somatic zebrafish cells using engineered TALENs. Nat Biotechnol 29: 697–698. 10.1038/nbt.1934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoriello C, Gennaro E, Anelli V, Distel M, Kelly A, Köster RW, Hurlstone A, Mione M. 2010. Kita driven expression of oncogenic HRAS leads to early onset and highly penetrant melanoma in zebrafish. PLoS One 5: e15170 10.1371/journal.pone.0015170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz-Rodríguez F, Ruiz-Velasco N, Pascual-Salcedo D, Teixidó J. 1999. Characterization of VLA-4-dependent myeloma cell adhesion to fibronectin and VCAM-1. Br J Haematol 107: 825–834. 10.1046/j.1365-2141.1999.01762.x [DOI] [PubMed] [Google Scholar]
- Scheer N, Campos-Ortega JA. 1999. Use of the Gal4-UAS technique for targeted gene expression in the zebrafish. Mech Dev 80: 153–158. 10.1016/S0925-4773(98)00209-3 [DOI] [PubMed] [Google Scholar]
- Shimoda N, Knapik EW, Ziniti J, Sim C, Yamada E, Kaplan S, Jackson D, de Sauvage F, Jacob H, Fishman MC. 1999. Zebrafish genetic map with 2000 microsatellite markers. Genomics 58: 219–232. 10.1006/geno.1999.5824 [DOI] [PubMed] [Google Scholar]
- Shiozawa Y, Eber MR, Berry JE, Taichman RS. 2015. Bone marrow as a metastatic niche for disseminated tumor cells from solid tumors. Bonekey Rep 4: 689 10.1038/bonekey.2015.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith ACH, Raimondi AR, Salthouse CD, Ignatius MS, Blackburn JS, Mizgirev IV, Storer NY, de Jong JLO, Chen AT, Zhou Y, et al. 2010. High-throughput cell transplantation establishes that tumor-initiating cells are abundant in zebrafish T-cell acute lymphoblastic leukemia. Blood 115: 3296–3303. 10.1182/blood-2009-10-246488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague J, Doerry E, Douglas S, Westerfield M. 2001. The Zebrafish Information Network (ZFIN): a resource for genetic, genomic and developmental research. Nucleic Acids Res 29: 87–90. 10.1093/nar/29.1.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanton MF. 1965. Diethylnitrosamine-induced hepatic degeneration and neoplasia in the aquarium fish, brachydanio rerio. J Natl Cancer Inst 34: 117–130. 10.1093/jnci/34.1.117 [DOI] [PubMed] [Google Scholar]
- Stella S, Montoya G. 2016. The genome editing revolution: a CRISPR-Cas TALE off-target story. Bioessays 38: S4–S13. 10.1002/bies.201670903 [DOI] [PubMed] [Google Scholar]
- Stoletov K, Klemke R. 2008. Catch of the day: zebrafish as a human cancer model. Oncogene 27: 4509–4520. 10.1038/onc.2008.95 [DOI] [PubMed] [Google Scholar]
- Stoletov K, Montel V, Lester RD, Gonias SL, Klemke R. 2007. High-resolution imaging of the dynamic tumor cell vascular interface in transparent zebrafish. Proc Natl Acad Sci 104: 17406–17411. 10.1073/pnas.0703446104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoletov K, Kato H, Zardouzian E, Kelber J, Yang J, Shattil S, Klemke R. 2010. Visualizing extravasation dynamics of metastatic tumor cells. J Cell Sci 123: 2332–2341. 10.1242/jcs.069443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streisinger G, Walker C, Dower N, Knauber D, Singer F. 1981. Production of clones of homozygous diploid zebra fish (Brachydanio rerio). Nature 291: 293–296. 10.1038/291293a0 [DOI] [PubMed] [Google Scholar]
- Talbot WS, Hopkins N. 2000. Zebrafish mutations and functional analysis of the vertebrate genome. Genes Dev 14: 755–762. [PubMed] [Google Scholar]
- Tang Q, Abdelfattah NS, Blackburn JS, Moore JC, Martinez SA, Moore FE, Lobbardi R, Tenente IM, Ignatius MS, Berman JN, et al. 2014. Optimized cell transplantation using adult rag2 mutant zebrafish. Nat Methods 11: 821–824. 10.1038/nmeth.3031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenente IM, Tang Q, Moore JC, Langenau DM. 2014. Normal and malignant muscle cell transplantation into immune compromised adult zebrafish. J Vis Exp 94: e52597. doi:10.3791/52597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traver D, Winzeler A, Stern HM, Mayhall EA, Langenau DM, Kutok JL, Look AT, Zon LI. 2004. Effects of lethal irradiation in zebrafish and rescue by hematopoietic cell transplantation. Blood 104: 1298–1305. 10.1182/blood-2004-01-0100 [DOI] [PubMed] [Google Scholar]
- Tulotta C, Stefanescu C, Beletkaia E, Bussmann J, Tarbashevich K, Schmidt T, Snaar-Jagalska BE. 2016. Inhibition of signaling between human CXCR4 and zebrafish ligands by the small molecule IT1t impairs the formation of triple-negative breast cancer early metastases in a zebrafish xenograft model. Dis Model Mech 9: 141–153. 10.1242/dmm.023275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulotta C, Stefanescu C, Chen Q, Torraca V, Meijer AH, Snaar-Jagalska BE. 2019. CXCR4 signaling regulates metastatic onset by controlling neutrophil motility and response to malignant cells. Sci Rep 9: 2399 10.1038/s41598-019-38643-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veneman WJ, Stockhammer OW, de Boer L, Zaat SAJ, Meijer AH, Spaink HP. 2013. A zebrafish high throughput screening system used for Staphylococcus epidermidis infection marker discovery. BMC Genomics 14: 255 10.1186/1471-2164-14-255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlecken DH, Bagowski CP. 2009. LIMK1 and LIMK2 are important for metastatic behavior and tumor cell-induced angiogenesis of pancreatic cancer cells. Zebrafish 6: 433–439. 10.1089/zeb.2009.0602 [DOI] [PubMed] [Google Scholar]
- Wang H, Long Q, Marty SD, Sassa S, Lin S. 1998. A zebrafish model for hepatoerythropoietic porphyria. Nat Genet 20: 239–243. 10.1038/3041 [DOI] [PubMed] [Google Scholar]
- Wang W, Liu X, Gelinas D, Ciruna B, Sun Y. 2007. A fully automated robotic system for microinjection of zebrafish embryos. PLoS One 2: e862 10.1371/journal.pone.0000862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Tao W, Wang Y, Bikow J, Lu B, Keating A, Verma S, Parker TG, Han R, Wen XY. 2010. Rosuvastatin, identified from a zebrafish chemical genetic screen for antiangiogenic compounds, suppresses the growth of prostate cancer. Eur Urol 58: 418–426. 10.1016/j.eururo.2010.05.024 [DOI] [PubMed] [Google Scholar]
- Wang J, Cao Z, Zhang XM, Nakamura M, Sun M, Hartman J, Harris RA, Sun Y, Cao Y. 2015. Novel mechanism of macrophage-mediated metastasis revealed in a zebrafish model of tumor development. Cancer Res 75: 306–315. 10.1158/0008-5472.CAN-14-2819 [DOI] [PubMed] [Google Scholar]
- White RM, Sessa A, Burke C, Bowman T, LeBlanc J, Ceol C, Bourque C, Dovey M, Goessling W, Burns CE, et al. 2008. Transparent adult zebrafish as a tool for in vivo transplantation analysis. Cell Stem Cell 2: 183–189. 10.1016/j.stem.2007.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyckoff JB, Wang Y, Lin EY, Li J, Goswami S, Stanley ER, Segall JE, Pollard JW, Condeelis J. 2007. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res 67: 2649–2656. 10.1158/0008-5472.CAN-06-1823 [DOI] [PubMed] [Google Scholar]
- Yan C, Brunson DC, Tang Q, Do D, Iftimia NA, Moore JC, Hayes MN, Welker AM, Garcia EG, Dubash TD, et al. 2019. Visualizing engrafted human cancer and therapy responses in immunodeficient zebrafish. Cell 177: 1903–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Chen G, Yu S, Xu C, Xin Y, Li T, Shi Y, Gu A, Duan J, Qian C, et al. 2013. TGF-β1 enhances tumor-induced angiogenesis via JNK pathway and macrophage infiltration in an improved zebrafish embryo/xenograft glioma model. Int Immunopharmacol 15: 191–198. 10.1016/j.intimp.2012.12.002 [DOI] [PubMed] [Google Scholar]
- Zhao C, Yang H, Shi H, Wang X, Chen X, Yuan Y, Lin S, Wei Y. 2011. Distinct contributions of angiogenesis and vascular co-option during the initiation of primary microtumors and micrometastases. Carcinogenesis 32: 1143–1150. 10.1093/carcin/bgr076 [DOI] [PubMed] [Google Scholar]