Abstract
Hemagglutinin (HA) is most abundant glycoprotein on the influenza virus surface. Influenza HA promotes viral entry by engaging the receptor and mediating virus–host membrane fusion. At the same time, HA is the major antigen of the influenza virus. HA antigenic shift can result in pandemics, whereas antigenic drift allows human circulating strains to escape herd immunity. Most antibody responses against HA are strain-specific. However, antibodies that have neutralizing activities against multiple strains or even subtypes have now been discovered and characterized. These broadly neutralizing antibodies (bnAbs) target conserved regions on HA, such as the receptor-binding site and the stem domain. Structural studies of such bnAbs have provided important insight into universal influenza vaccine and therapeutic design. This review discusses the HA functions as well as HA–antibody interactions from a structural perspective.
Influenza viruses are classified based on their antigenicity, which is determined by their surface glycoproteins. Four types of influenza viruses, A, B, C, and D, have been isolated and characterized. Influenza A and B viruses have two surface glycoproteins, hemagglutinin (HA) and neuraminidase (NA), whereas influenza C and D viruses have only one surface glycoprotein, hemagglutinin–esterase fusion (HEF). Based on the antigenicity of HA and NA, influenza A viruses are further classified into subtypes. There are 18 known HA subtypes (H1–H18) and 11 known NA subtypes (N1–N11). HA subtypes are further divided into two groups. Group 1 HA includes H1, H2, H5, H6, H8, H9, H11, H12, H13, H16, H17, and H18, whereas group 2 HA includes H3, H4, H7, H10, H14, and H15. Both influenza A and B viruses infect humans and can cause severe illness or death. In contrast, influenza C virus only causes mild symptoms in most cases. Human infection with influenza D virus has not been observed. Therefore, most influenza research has been focused on influenza A and B viruses. A main difference between influenza A and B viruses is that influenza B virus is only found in humans, whereas the primary natural reservoir for influenza A virus is aquatic birds; these avian viruses can give rise to new pandemic viruses in humans through reassortment with human and swine viruses. As a result, influenza A virus usually receives more attention and has been studied more extensively.
The surface of influenza virions is dominated by HA, which outnumbers NA by five- to 10-fold (Harris et al. 2006; Hutchinson et al. 2014). HA confers upon influenza virus the ability to agglutinate red blood cells, which enables rapid quantification of influenza virus (hemagglutination assay) as well as the virus-neutralizing capacity of antibodies and sera (hemagglutination inhibition assay) (Hirst 1942). The ability of HA to agglutinate red blood cells can be attributed to its receptor binding function. HA engages sialylated glycan receptors on host cells to initiate viral entry (Burnet and Stone 1947; Stone 1948). HA also carries the machinery for membrane fusion (Maeda and Ohnishi 1980). HA is a homotrimer consisting of a globular head domain that resides atop a membrane-proximal stem domain (Fig. 1A). Its structure was first reported in 1981 (Wilson et al. 1981), which made it possible to study the structure–function relationships of HA. In a back-to-back article, the major antigenic sites on the H3 HA were also described for the first time (Wiley et al. 1981). Characterization of an HA-peptide antibody in 1984 led to the identification of the HA-tag (Wilson et al. 1984), which is a linear epitope consisting of nine amino acids that is used extensively in protein purification and labeling. Despite being extremely useful as a research tool, the HA-tag epitope is not clinically relevant because it is located in the subunit interface of HA, which is not accessible in the native form of HA. The structure of a neutralizing antibody in complex with HA was first reported in 1995 (Bizebard et al. 1995), which provided important insights into the molecular mechanism of how HA is recognized by the adaptive humoral immune system.
Figure 1.
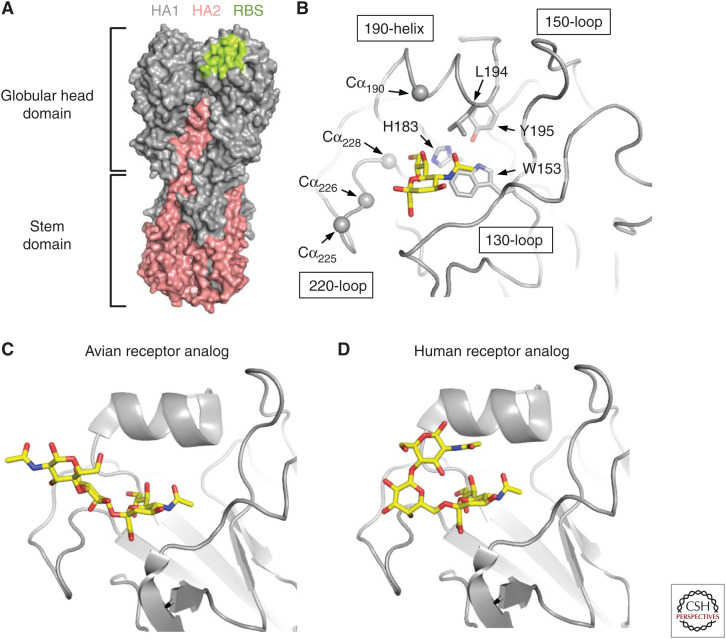
Hemagglutinin (HA) structures and receptor-binding site (RBS). (A) Structure of trimeric HA. The location of the RBS is shown in lime on the HA structure. HA1 is shown in gray and HA2 in salmon. The globular head domain sits on top of the stem domain. (B) The major structural elements, namely 130-loop, 150-loop, 190-helix, and 220-loop, of the HA RBS are shown. Highly conserved residues W153, H183, L194, and Y195 are shown in stick representation. The alpha carbons (Cαs) of HA1 residues 190, 225, 226, and 228, which are involved in the H1, H2, and H3 receptor-specificity switch, are shown in sphere representations. (C) Avian-type receptor (α2,3-linked sialic acid) binds to HA with an extended configuration (Xu et al. 2012). (D) Human-type receptor (α2,6-linked sialic acid) binds to HA with a folded-back configuration (Xu et al. 2012).
As a major antigen of influenza virus, HA constantly evolves to escape herd immunity arising from natural infection and vaccination, while still preserving its functionality (Wu and Wilson 2017). That is why the various components of the seasonal influenza vaccine need to be annually reviewed and updated, when necessary. Most antibodies elicited by seasonal influenza vaccines are strain-specific and the virus can readily escape such antibodies. Nonetheless, many broadly neutralizing antibodies (bnAbs) against HA have been identified and characterized in the past decade (Wu and Wilson 2018), which has enabled structural-based design of universal influenza vaccine candidates (Impagliazzo et al. 2015; Yassine et al. 2015). Understanding the structural biology of HA has been accelerating in recent years, in large part owing to the technical advances in X-ray crystallography (Garman 2014) as well as cryogenic electron microscopy (cryo-EM) (Nogales 2016). This review will describe the biology of HA and antibodies against HA from a structural perspective. H3 numbering is used for HAs throughout, unless indicated otherwise.
RECEPTOR BINDING AND SPECIFICITY OF HEMAGGLUTININ
Each protomer of HA contains a membrane-distal receptor-binding site (RBS) in the globular head domain that binds to sialylated glycan receptors on the host cell surface (Fig. 1A) (Weis et al. 1988). Nevertheless, it should be noted that the H17 and H18 subtypes, which are found only in bats, do not bind to sialic acids and are the exceptions (Sun et al. 2013; Tong et al. 2013; Zhu et al. 2013b). Instead, H17 and H18 HAs utilize the major histocompatibility complex class II (MHC-II) molecule as host receptor (Karakus et al. 2019). The HA RBS is composed of the 130-loop, 150-loop, 190-helix, and 220-loop, named after their relative positions on the HA amino-acid sequence. Several key residues that interact with sialic acid are conserved across influenza A and B HAs (Wang et al. 2007). These include W153, H183, L194, and Y195 (Fig. 1B). However, major structural variations of the HA RBS can also be observed in naturally circulating strains. Examples include a single residue insertion in the 130-loop of some strains from H1 and H5 subtypes and all strains from H6 and H10 subtypes (Lee et al. 2012), a two-residue insertion in the 150-loop of H7, H10, and H15 subtypes (Tzarum et al. 2017), and an eight-residue deletion in the 220-loop of multiple H7N2 strains (Suarez et al. 1999; Yang et al. 2010).
The receptor specificity for avian influenza viruses is α2,3-linked sialic acid (avian-type receptor), whereas for human influenza viruses it is α2,6-linked sialic acid (human-type receptor). When binding to HA, α2,3-linked sialic acid typically displayed an extended configuration (Fig. 1C), whereas α2,6-linked sialic acid typically displayed a folded-back configuration (Fig. 1D) (Shi et al. 2014). This folded-back conformation is in fact the most energetically stable conformation of α2,6-linked sialic acid in sialosides in their unbound form (Sabesan et al. 1991). Switching the receptor specificity from α2,3-linked sialic acid to α2,6-linked sialic acid is required for avian influenza viruses to cause human pandemics and for transmission, as assessed in an animal model between ferrets (Tumpey et al. 2007; Pappas et al. 2010; Roberts et al. 2011). The structural mechanisms for the receptor specificity switch are well-characterized for H1, H2, and H3, all of which are associated with known influenza pandemics—the Spanish flu (H1N1) pandemic in 1918, the Asian flu (H2N2) pandemic in 1957, the Hong Kong flu (H3N2) pandemic in 1968, and the most recent swine flu (H1N1) pandemic in 2009. All avian H1, H2, and H3 HAs predominately encode Glu, Gly, Gln, and Gly at residues 190, 225, 226, and 228, respectively. H1 requires a pair of mutations E190D/G225D to switch receptor specificity from α2,3-linked sialic acid to α2,6-linked sialic acid (Matrosovich et al. 1997; Glaser et al. 2005; Stevens et al. 2006; Tumpey et al. 2007), whereas H2 and H3 require a different pair Q226L/G228S (Rogers et al. 1983; Connor et al. 1994; Pappas et al. 2010; Xu et al. 2010). Recent human H3N2 viruses have further evolved a receptor specificity favoring long α2,6 sialylated glycans with several N-acetyllactosamine (LacNAc) repeats (Peng et al. 2017). Consistently, the receptor-binding mode has subtly evolved in recent human H3N2 viruses (Lin et al. 2012; Wu et al. 2018a).
Other subtypes including H5, H6, H7, H9, and H10 are occasionally transmitted to humans. To date, naturally circulating strains from these subtypes have not acquired the receptor specificity to α2,6-linked sialic acids and, hence, are not able to transmit among humans. Nonetheless, studies have explored the possibility of switching receptor specificity from α2,3-linked sialic acid to α2,6-linked sialic acid in different subtypes that may have pandemic potential. In 2013, two studies independently adapted H5N1 strains to acquire transmissibility among ferrets, which is a commonly used mammalian model for influenza transmission (Herfst et al. 2012; Imai et al. 2012). In both studies, mutations in the HA RBS were observed in the ferret-transmissible H5N1 strains. Common mutations in the HA RBS that were identified by both studies include Q226L and a loss of glycosylation site at residue 158 (Herfst et al. 2012; Imai et al. 2012). Follow-up studies showed that those mutations are important for H5 HA to switch receptor specificity from α2,3-linked sialic acid to α2,6-linked sialic acid (Lu et al. 2013; Xiong et al. 2013; de Vries et al. 2014). Those studies provoked the National Institutes of Health (NIH) to declare a pause on certain influenza gain-of-function studies in 2014 (Casadevall and Imperiale 2014), which was then lifted in late 2017. How other HA subtypes switch receptor specificity from α2,3-linked sialic acid to α2,6-linked sialic acid has also been studied. Different subtypes appear to require different sets of mutations to switch specificity toward α2,6-linked sialic acid—Q226L/G228S for H4 HA (Song et al. 2017), G225D for H6 HA (de Vries et al. 2017b), V186G/K193T/G228S, V186K/K193T/G228S or V186N/N224K/G228S for H7 HA (de Vries et al. 2017a), and K158aA/D193T/Q226L/G228S for H10 HA (Tzarum et al. 2017) (“158a” indicates the insertion of an amino acid after position 158). Because of the structural variability of the RBS in different HA subtypes, it is not surprising that the mutational requirements for switching receptor specificity are different for different subtypes.
HEMAGGLUTININ-MEDIATED MEMBRANE FUSION
Besides engaging the receptor, HA also mediates virus–host membrane fusion, which is essential for viral entry. Comparing the HA structures between prefusion and postfusion conformations shows that HA has to undergo large conformational rearrangements to promote virus–host membrane fusion (Bullough et al. 1994; Chen et al. 1999). Such a large conformational change is only possible after proteolytic processing of the HA trimer. During protein translation, HA trimer is synthesized as a single polypeptide chain (HA0). HA0 is then cleaved by host cell proteases to become mature HA, which consists of HA1 and HA2 subunits that remain cross-linked via a single disulfide bond (Fig. 2A). The mature HA, but not HA0, is fusion-competent. The cleavage site is a surface loop in the stem domain. The amino-acid sequence of the cleavage site varies (Böttcher-Friebertshauser et al. 2013). Although most strains carry a monobasic cleavage site, some strains carry a polybasic cleavage site. Monobasic cleavage site can be processed by proteases such as plasmin (Lazarowitz and Choppin 1975), factor Xa (Gotoh et al. 1990), tryptase Clara (Kido et al. 1992), HAT (Böttcher et al. 2006), and TMRPSS2 (Böttcher et al. 2006). The tissue tropism of influenza virus is strongly influenced by these proteases, which are only present in a limited number of tissues, such as the respiratory or intestinal tract. In contrast, polybasic cleavage sites, mainly found in avian viruses, can be processed by furin, which is ubiquitously expressed. Therefore, influenza strains with polybasic cleavage site are typically highly pathogenic. This proteolytic processing of HA does not induce substantial conformational changes, as shown by the structural comparison of the precursor HA0 (uncleaved form) and mature HA (cleaved form), which are almost identical (Chen et al. 1998), except that the fusion peptide now inserts into the center of the cleaved HA trimer (Wilson et al. 1981).
Figure 2.
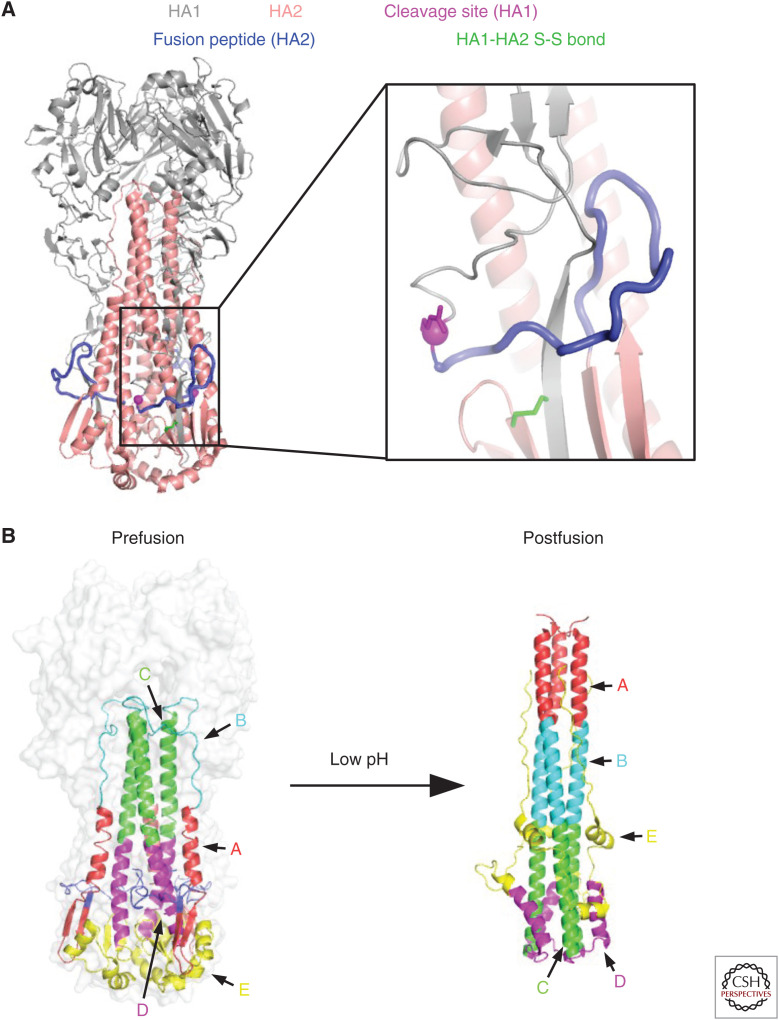
Fusion machinery of hemagglutinin (HA). (A) Several important structural features in the stem domain, including the cleavage site (carboxy-terminal Arg of HA1), fusion peptide (amino-terminal region of HA2), and the disulfide bond that links HA1 and HA2, are highlighted. (B) The structural rearrangements of the HA2 between prefusion and postfusion conformations are shown (Chen et al. 1999).
HA-mediated membrane fusion is a pH-dependent process that occurs in the endosome. The acidic environment inside the endosome triggers the conformational change of HA (Fig. 2B), which in turn induces virus–host membrane fusion. The threshold pH of membrane fusion varies between strains and subtypes (ranging from ∼5.2 to 6.0) (Scholtissek 1985; Puri et al. 1990; Korte et al. 2007; DuBois et al. 2011; Galloway et al. 2013), but is generally lower for HAs from human isolates than for HAs from avian isolates (Galloway et al. 2013). The conformational change of HA during fusion involves substantial structural rearrangements in the stem domain. Consistently, many residues in the stem domain are highly conserved, some of which are determinants of the threshold pH of membrane fusion (Daniels et al. 1985; Rachakonda et al. 2007; Thoennes et al. 2008; Xu and Wilson 2011; Mair et al. 2014; Byrd-Leotis et al. 2015).
ANTIGENIC EVOLUTION OF HEMAGGLUTININ
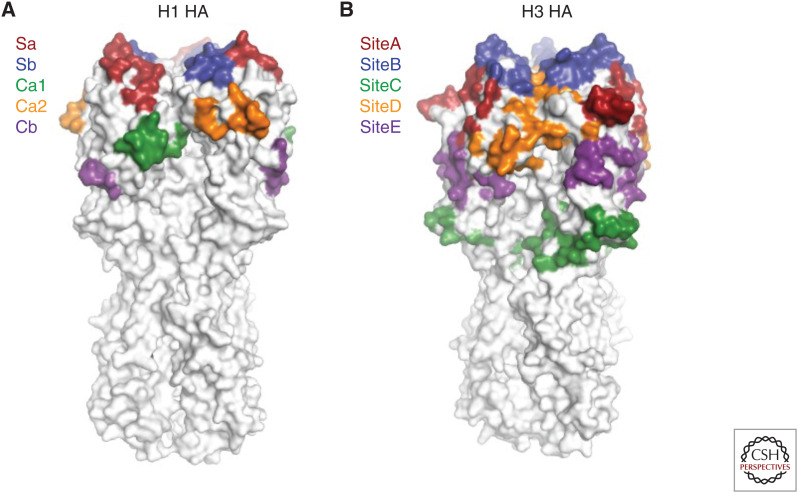
Identifying regions on HA that are targeted by humoral immune response has been a primary focus for influenza research. The antigenicity of H1 and H3 viruses has been studied most extensively among HA subtypes, because of their circulation in the human population. In early studies, five major antigenic sites were proposed for H1 HAs (Sa, Sb, Ca1, Ca2, and Cb) (Gerhard et al. 1981; Caton et al. 1982), as well as for H3 HA (sites A–E) (Wiley et al. 1981; Skehel et al. 1984; Wiley and Skehel 1987). All antigenic sites are located in the globular head domain for both H1 and H3 HAs (Fig. 3). In contrast to the HA globular head domain, the HA stem has a much lower efficiency in eliciting antibody responses and, hence, is less immunogenic (Jegaskanda et al. 2013; Altman et al. 2015; Angeletti et al. 2017; Tan et al. 2019).
Figure 3.
Major antigenic sites of hemagglutinin (HA). (A) The five major antigenic sites on H1 HA are illustrated. (B) The five major antigenic sites on H3 HA are shown.
To continue circulating in human populations, influenza viruses have to constantly evolve to escape herd immunity arising from natural infection and vaccination, via antigenic shift and antigenic drift. Antigenic shift involves the emergence of antigenically novel viral strains, whereas antigenic drift involves incremental changes in circulating viruses via mutation. Antigenic shift is rare as compared with antigenic drift, but can lead to devastating pandemic outbreaks. Antigenic shift often results from reassortment that involves acquisition of antigenically novel HA or NA or both from avian viruses into human or other mammalian viruses (Lowen 2017). Antigenic shift is responsible for known influenza pandemics—the Spanish flu (H1N1) pandemic in 1918, the Asian flu (H2N2) pandemic in 1957, the Hong Kong flu (H3N2) pandemic in 1968, and the most recent swine flu (H1N1) pandemic in 2009. After each pandemic, the progeny influenza strains continue to evolve in the human population and escape herd immunity via antigenic drift. The evolutionary rates of H1N1 and H3N2 in humans are similar and are approximately twofold higher than that of influenza B virus (Furuse et al. 2016). The HA globular head domain evolves faster than the HA stem domain (Kirkpatrick et al. 2018), which is consistent with the notion that the HA globular head domain is immunodominant over the HA stem domain (Jegaskanda et al. 2013; Altman et al. 2015; Angeletti et al. 2017; Tan et al. 2019). Studies have also shown that major antigenic drift in human influenza evolution almost exclusively involves amino-acid substitutions in the major antigenic sites (Smith et al. 2004; Koel et al. 2013). Because the HA RBS partially overlaps with major antigenic sites (Sa, Sb, and Ca2 of H1 HA, and antigenic sites A, B, and D of H3 HA) (Wu and Wilson 2017), antigenic drift may be associated with a change in receptor-binding avidity (Hensley et al. 2009; Li et al. 2013).
Accumulation of N-glycosylation sites is a common immune evasion strategy that is utilized by diverse enveloped viruses (Davis et al. 1990; Simmonds et al. 1991; Cheng-Mayer et al. 1999; Fournillier et al. 2001; Zhang et al. 2004), including human influenza A viruses (Zhang et al. 2004; Blackburne et al. 2008; Igarashi et al. 2008; Cherry et al. 2009; Das et al. 2010; Sun et al. 2011; Lee et al. 2014; Tate et al. 2014; Wu and Wilson 2017; Altman et al. 2019). The accumulation of N-glycosylation sites in the HA globular head domains of human influenza A viruses occurs at discrete 5- to-7-year intervals, until they reach a functional limit (Altman et al. 2019). After reaching this functional limit, which is ∼5 for H1N1 and ∼7 for H3N2, N-glycosylation sites are swapped (i.e., loss of one N-glycosylation site and gain of another at the same time through point mutations) at a longer interval (Altman et al. 2019). If an N-glycosylation site is added next to the RBS, it can negatively influence receptor binding (Tsuchiya et al. 2001, 2002; Das et al. 2011; Kim et al. 2013; Tate et al. 2014; Alymova et al. 2016). Subsequently, the RBS and immediate proximal region become relatively more exposed as N-glycosylation sites accumulate and shield the rest of the HA globular head domain (Wu and Wilson 2017). Consistently, RBS-proximal antigenic site B has become immunodominant in recent human H3N2 HAs (Popova et al. 2012; Chambers et al. 2015), which are highly decorated with N-glycans (Lee et al. 2014; Wu and Wilson 2017).
ANTIBODIES AGAINST THE HEMAGGLUTININ STEM DOMAIN
Most antibody responses focus on epitopes that are highly diverse among strains (Gerhard et al. 1981; Wiley et al. 1981; Caton et al. 1982) and are thus strain-specific (Wang et al. 1986; Wrammert et al. 2008). However, some HA antibodies can neutralize multiple strains within a subtype or even across subtypes by targeting conserved regions. These broadly neutralizing antibodies (bnAbs) target regions that are relatively conserved. The stem domain is one such region on HA (Wu and Wilson 2017). In 1993, a mouse antibody C179 was reported to have cross-subtype neutralization activity and was believed to bind to the stem domain (Okuno et al. 1993). In 2013, the structure of C179 in complex with HA confirmed that C179 indeed binds to the HA stem (Dreyfus et al. 2013). The 20-year delay in the structural characterization of C179 was due to the fact that the research on stem-binding bnAbs did not become active until 2008–2009. In 2008–2009, three sets of pan-group 1 HA antibodies were isolated independently from human subjects in different continents (Kashyap et al. 2008; Throsby et al. 2008; Sui et al. 2009). In 2009, X-ray structural characterization revealed that antibodies CR6261 and F10 bound to a highly conserved epitope in the HA stem domain (Ekiert et al. 2009; Sui et al. 2009). These findings show that influenza bnAbs can be elicited in humans and sparked considerable interest in studying human influenza bnAbs. In 2011, a pan-influenza A stem-binding bnAb FI6v3, which can target both group 1 and group 2 HAs, was reported (Corti et al. 2011). Nevertheless, despite different angles of approach to the HA surface and the use of both heavy and light chains for binding by FI6v3 versus only the heavy chain for CR6261, the epitopes of FI6v3 and CR6261 are highly overlapping. However, unlike CR6261, which utilized the VH1-69 germline gene, FI6v3 arose from the VH3-30 germline gene. In 2012, an antibody called CR9114, which can target both influenza A and B HAs, was reported (Dreyfus et al. 2012). Both CR9114 and CR6261 are encoded by VH1-69 and bind to a very similar epitope. During the late 2000s and subsequent years, many other stem-binding bnAbs from humans were discovered and characterized (Kashyap et al. 2008, 2010; Burioni et al. 2010; Clementi et al. 2011; Ekiert et al. 2011; Nakamura et al. 2013; Yasugi et al. 2013; Friesen et al. 2014; Henry Dunand et al. 2015, 2016; Wu et al. 2015, 2018b; Fu et al. 2016; Joyce et al. 2016; Kallewaard et al. 2016; Andrews et al. 2017; Lang et al. 2017; Yamayoshi et al. 2017), including some that cross-react with both group 1 and 2 HAs (Nakamura et al. 2013; Henry Dunand et al. 2015, 2016; Wu et al. 2015; Fu et al. 2016; Joyce et al. 2016; Kallewaard et al. 2016; Andrews et al. 2017; Lang et al. 2017).
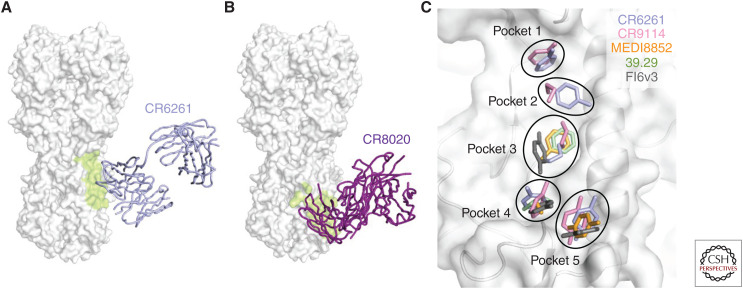
Structural characterization of stem-binding bnAbs antibodies illustrated that most stem-binding bnAbs have an epitope that highly overlaps with that of CR6261 (Fig. 4A) (Wu and Wilson 2018), although group 2–specific stem-binding bnAbs bind slightly lower down the stem domain (Fig. 4B) (Ekiert et al. 2011; Friesen et al. 2014). Five pockets in the stem domain are often targeted by stem-binding bnAbs, mainly using hydrophobic and aromatic residues (Fig. 4C) (Kadam et al. 2017). Identification of these common binding features facilitated structure-based design of stem-binding proteins (Fleishman et al. 2011; Whitehead et al. 2012; Chevalier et al. 2017), peptides (Kadam et al. 2017), and small molecules (van Dongen et al. 2019) with neutralizing activity akin to the bnAbs.
Figure 4.
Hemagglutinin (HA) stem-binding antibodies. (A) Binding of group 1–specific broadly neutralizing antibody (bnAb) CR6261 to HA (Ekiert et al. 2009). (B) Binding of group 2–specific bnAb CR8020 to HA (Ekiert et al. 2011). (A,B) The HA surface is colored in white and bnAb epitope colored in lime. (C) Pockets that are commonly targeted by HA stem-binding antibodies are shown. Side chains of key binding residues in the paratopes (i.e., the parts of an antibody that bind to the epitope, usually the complementarity determining regions [CDRs], but can also include some framework residues) of CR6261 (Ekiert et al. 2009), CR9114 (Dreyfus et al. 2012), MEDI8852 (Kallewaard et al. 2016), 39.29 (Nakamura et al. 2013), and FI6v3 (Corti et al. 2011) are shown in stick representation. The HA surface and underlying backbone structure are colored in white.
Although the stem-binding bnAb epitopes are highly conserved, variations can be observed in some of the epitope or epitope-proximal residues and can limit the breadth of stem-binding bnAbs (Zost et al. 2019). A well-known example is the N-glycosylation site at HA1 Asn38, which is adjacent to the conserved stem epitope (Ekiert et al. 2009; Sui et al. 2009). The N-glycosylation site at HA1 Asn38 is not present among group 1 HAs, but is highly conserved among group 2 HAs. The N-glycan at HA1 Asn38 can block binding of stem-binding bnAbs to group 2 HAs. Binding to group 2 HAs often involves a reorientation of the group 2–specific glycan at HA1 Asn38 and other residues, including HA2 Trp 21 (Corti et al. 2011; Dreyfus et al. 2012). Structural comparison of VH1-69-encoded stem-binding bnAbs suggests that the precise approach angle of stem-binding bnAbs is a determinant of whether the N-glycan at HA1 Asn38 can be accommodated (Lang et al. 2017).
ANTIBODIES AGAINST THE HEMAGGLUTININ RECEPTOR-BINDING SITE
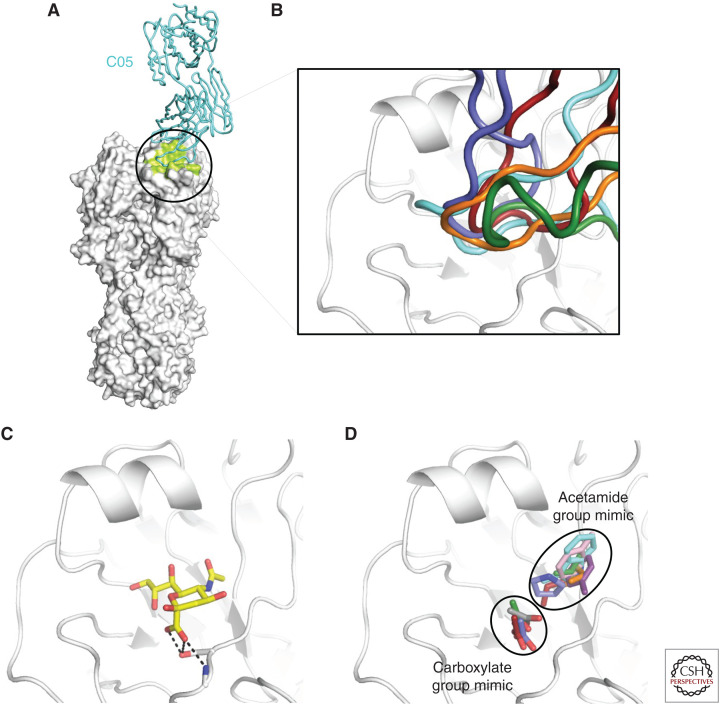
Another conserved region on HA is the RBS, despite being more variable than the stem domain (Wu and Wilson 2017). A number of RBS-targeting bnAbs have been identified and characterized (Fleury et al. 1998; Barbey-Martin et al. 2002; Yoshida et al. 2009; Krause et al. 2011, 2012; Ohshima et al. 2011; Whittle et al. 2011; Ekiert et al. 2012; Lee et al. 2012, 2014; Tsibane et al. 2012; Hong et al. 2013; Schmidt et al. 2013; Xu et al. 2013; Lee and Wilson 2015; McCarthy et al. 2018). By targeting the receptor-binding sites, these bnAbs prevent HA from binding to the receptor. Unlike the stem domain, which is close to the viral membrane, the RBS is highly exposed. Therefore, RBS-targeted bnAbs can adopt a much wider range of approach angles when engaging the HA as compared with stem-binding bnAbs (Wu and Wilson 2017). By comparing the structures of RBS-targeting bnAbs, several common features were revealed. RBS-targeting bnAbs often insert a single complementarity determining region (CDR) loop, mostly CDR H3 but occasionally H2, into the RBS (Fig. 5A,B) (Lee and Wilson 2015). The tips of these CDR loops mimic the binding mode of sialic acid (Lee and Wilson 2015). The most common strategy is to mimic the acetamide group of sialic acid using a hydrophobic, often aromatic, amino acid, such as Trp (Ekiert et al. 2012), Tyr (Fleury et al. 1998; Xu et al. 2013), Phe (Xu et al. 2013), Met (Lee et al. 2012), Leu (Lee et al. 2014), Val (Whittle et al. 2011; Schmidt et al. 2015), and Pro (Fig. 5C) (Hong et al. 2013; Schmidt et al. 2015). Mimicking the sialic acid carboxylate group is another strategy that is utilized by some RBS-targeting bnAbs to enable formation of an extensive hydrogen bond network. A number of RBS-targeting bnAbs are able to completely mimic the carboxylate group using an Asp at the tip of CDR H3 (Fig. 5D) (Whittle et al. 2011; Hong et al. 2013; Schmidt et al. 2013, 2015; Lee et al. 2014), whereas some other RBS-targeting bnAbs partially mimic the carboxylate group using a backbone carbonyl group (Ekiert et al. 2012; Lee et al. 2012; McCarthy et al. 2018). Structural characterization of RBS-targeting bnAbs has also inspired the development of anti-influenza protein binders against the RBS (Strauch et al. 2017). In addition, by harnessing the knowledge of RBS-targeting and stem-binding bnAbs, a multidomain llama antibody has been developed, which can broadly cross-neutralize influenza A and B viruses (Laursen et al. 2018).
Figure 5.
Hemagglutinin (HA) receptor-binding site (RBS)-targeting antibodies. (A) Structure of a prototypic RBS-targeting antibody C05 in complex with HA (Ekiert et al. 2012). HA is colored in white and the C05 epitope is colored in lime. (B) bnAbs C05 (cyan), F045-092 (green), 5J8 (slate blue), and CH65 (red) target the HA RBS using their complementarity determining region (CDR) H3 (Whittle et al. 2011; Ekiert et al. 2012; Hong et al. 2013; Lee et al. 2014). S139/1 (orange) targets the HA RBS using its CDR H2 (Lee et al. 2012). (C) Sialic acid in complex with HA. Hydrogen bonds between the carboxylate group of sialic acid and HA are shown in black dashed lines. (D) The carboxylate and acetamide groups of sialic acid are mimicked by RBS-targeting antibodies, including C05 (cyan) (Ekiert et al. 2012), S139/1 (orange) (Lee et al. 2012), F045-092 (green) (Lee et al. 2014), 2G1 (purple) (Xu et al. 2013), 8M2 (pink) (Xu et al. 2013), 5J8 (slate blue) (Hong et al. 2013), HC63 (gray) (Barbey-Martin et al. 2002), and CH65 (red) (Whittle et al. 2011). The side chains that act as mimics of sialic acid receptor moieties are shown in stick representation.
Because small variations can be found within the HA RBS as well as high variability in the RBS-proximal regions (Wu and Wilson 2017), it is difficult for RBS-targeted bnAbs to completely avoid contacting variable regions, despite their small binding footprints using a single CDR. As a result, the reactivity of RBS-targeted bnAbs is generally not as broad as stem-binding bnAbs. Many RBS-targeted bnAbs are subtype-specific (Whittle et al. 2011; Hong et al. 2013; Schmidt et al. 2013, 2015), and some can cross-react with a limited number of subtypes (Yoshida et al. 2009; Ohshima et al. 2011; Ekiert et al. 2012; Krause et al. 2012; Lee et al. 2012, 2014; Xu et al. 2013; McCarthy et al. 2018). It has been proposed that the bivalence of immunoglobulin G (IgG) confers RBS-targeted bnAbs with an avidity effect, by binding to two adjacent trimers simultaneously (Ekiert et al. 2012; Lee et al. 2012). As a result, the potency of RBS-targeting bnAbs is usually higher than stem-binding bnAbs. Cross-linking HA trimers on the viral surface by RBS-targeting bnAbs may also prevent HA conformational changes that normally occur during membrane fusion (Barbey-Martin et al. 2002).
OTHER BROADLY REACTIVE ANTIBODIES AGAINST HEMAGGLUTININ
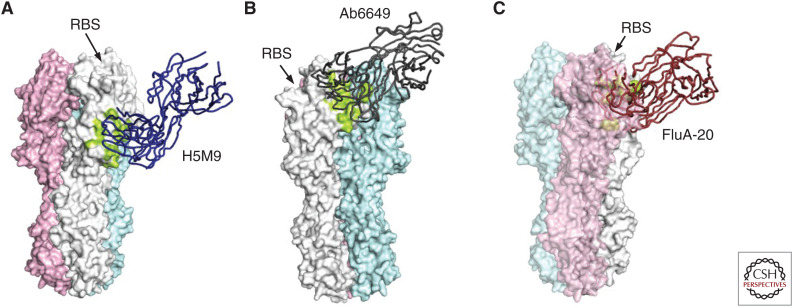
Other regions on HA besides the RBS and stem domain are also targeted by bnAbs. Because most regions on HA besides the RBS and stem domain are highly diverse across HA subtypes, bnAbs that do not target the RBS or the stem domain are usually subtype-specific. One example is the vestigial esterase subdomain on HA1, which is conserved among strains within a certain subtype but not across subtypes (Fig. 6A). Several protective antibodies that target the vestigial esterase subdomain have been characterized (Li et al. 2009; Dreyfus et al. 2012; Zhu et al. 2013a; Zuo et al. 2015; Chai et al. 2017; Bangaru et al. 2018), including H5M9 (Li et al. 2009; Zhu et al. 2013a), CR8071 (Dreyfus et al. 2012), 46B8 (Chai et al. 2017), 100F4 (Zuo et al. 2015), F005-126 (Iba et al. 2014), and H3v-47 (Bangaru et al. 2018). Both H5M9, which is a mouse antibody, and 100F4, which is a human antibody, target multiple clades of H5N1 viruses. CR8071 and 46B8 are human antibodies that target influenza B viruses, whereas F005-126 and H3v-47 are human antibodies that target diverse strains of H3N2 viruses. Although these bnAbs all target the vestigial esterase subdomain, their mechanisms of action are different. H5M9, 46B8, and F005-126 prevent the pH-dependent conformational changes of the HA, which is required for virus–host membrane fusion (Li et al. 2009; Zhu et al. 2013a), whereas CR8071 and H3v-47 primarily function via blocking the viral budding and progeny release process (Brandenburg et al. 2013; Bangaru et al. 2018).
Figure 6.
Other neutralizing epitopes on hemagglutinin (HA). (A) Binding of 5HM9 to HA in the vestigial esterase region (Zhu et al. 2013a). (B) Binding of Ab6649 to HA in the “lateral patch” on the globular head domain, which is next to the receptor-binding site (RBS) and above the vestigial esterase subdomain (Raymond et al. 2018). (C) Binding of FluA-20 to the HA trimer interface (Bangaru et al. 2019). The epitope of FluA-20 is located in a conserved region in the HA protomer–protomer interface. Therefore, binding of FluA-20 to HA requires dissociation or breathing of the HA trimer. (A–C) Three protomers of the HA trimer are colored in white, pink, and cyan, respectively. The epitope is colored in lime. The location of the RBS is indicated.
Besides the vestigial esterase subdomain, some other regions on HA are also bnAb epitopes. For example, Ab6649 is a human H1-specific bnAb that targets an epitope above the vestigial esterase subdomain on HA1 and is highly conserved among H1N1 strains (Fig. 6B) (Raymond et al. 2018). Antibodies H7.137, H7.167, and H7.169 are H7-specific bnAbs that bind to a similar epitope that is highly conserved within the H7 subtype (Thornburg et al. 2016). An H1-specific bnAb FISW84 binds to an epitope that is near the ectodomain membrane anchor junction and is conserved among H1 HAs (Benton et al. 2018). Antibodies D1 H1-3/H3-3, D1 H1-17/H3-14, and D2 H1-1/H3-1, which are nonneutralizing but offer in vivo protection, target a conserved epitope that is slightly below the RBS and can cross-react with both H1 and H3 subtypes (Lee et al. 2016).
Recent studies have revealed that antibodies can target conserved epitopes in HA1 that involve the trimer interface, which is not solvent-accessible in the conventional HA trimer configuration (Bajic et al. 2019; Bangaru et al. 2019; Turner et al. 2019; Watanabe et al. 2019). H7.5 is a human H7-specific bnAb that targets an epitope that involves adjacent protomers in the HA trimer (Thornburg et al. 2016; Turner et al. 2019). Part of the H7.5 epitope involves the trimer interface, suggesting that HA1 globular head is structurally dynamic in its prefusion state (Turner et al. 2019). Single-molecule Förster resonance energy transfer (FRET) also reveals that HA undergoes reversible conformational changes (Garcia et al. 2015). The epitopes of other HA interface-targeting antibodies, such as FluA-20 (Bangaru et al. 2019), S5V2-29 (Watanabe et al. 2019), H2214 (Watanabe et al. 2019), and 8H10 (Bajic et al. 2019), are mainly composed of residues in the trimer interface (Fig. 6C). FluA-20 has the largest breadth among these antibodies, with the ability to bind to 14 out of 15 tested HA subtypes. Electron microscopy (EM) analysis indicated that these HA interface-targeting antibodies can induce dissociation of the HA trimer (Bangaru et al. 2019; Turner et al. 2019). Nonetheless, the mechanisms of action of these interface-targeting antibodies are different. Although H7.5 has neutralizing activity (Thornburg et al. 2016), FluA-20, S5V2-29, H2214, and 8H10 have no direct neutralizing activity (Bajic et al. 2019; Bangaru et al. 2019; Watanabe et al. 2019). These interface-targeting antibodies appear to offer in vivo protection through antibody-dependent cellular cytotoxicity (ADCC) (Bangaru et al. 2019; Watanabe et al. 2019).
VACCINES AGAINST HEMAGGLUTININ
Structural characterization of bnAbs has been a major driver of universal influenza vaccine development. Antibodies elicited by seasonal influenza vaccine are typically strain-specific. As a result, the components of seasonal influenza vaccine have to be annually reviewed and updated, if necessary, to account for the antigenic drift of the circulating strains. However, antigenic mismatch between the vaccine strains and the circulating strains occur occasionally (Tricco et al. 2013). In addition, most seasonal influenza vaccines are produced in embryonated chicken eggs, which may result in HA egg-adaptive mutations that change the antigenicity of the vaccine strains (Robertson et al. 1987; Kodihalli et al. 1995; Chen et al. 2010; Popova et al. 2012; Parker et al. 2016; Raymond et al. 2016; Wu et al. 2017; Zost et al. 2017). Egg-adaptation mutations allow HAs from human influenza viruses, which has a receptor specificity toward α2,6-linked sialic acid, to increase affinity to α2,3-linked sialic acid (Wu et al. 2017, 2019), which is abundant on the chorioallantoic membrane (Ito et al. 1997; Sriwilaijaroen et al. 2009). As a result, the effectiveness of seasonal influenza vaccine is far from satisfying (Belongia et al. 2016).
A major direction of influenza vaccine research is to develop immunogens that can elicit protective antibodies against multiple strains or even subtypes, such that annual updates of the influenza vaccine are not needed. HA stem-binding bnAbs have provided significant insight into structural-based design of such immunogens (Wu and Wilson 2018). Several stem immunogens that can elicit heterosubtypic antibodies have been described (Steel et al. 2010; Bommakanti et al. 2012; Mallajosyula et al. 2014, 2015; Impagliazzo et al. 2015; Wohlbold et al. 2015; Yassine et al. 2015; Valkenburg et al. 2016; Corbett et al. 2019). Some of these stem immunogens are referred to as “headless HA” (Steel et al. 2010; Valkenburg et al. 2016; Corbett et al. 2019), because they are basically HA without the globular head domain. The general framework for stem immunogen design is to link the amino- and carboxy-terminal regions of the HA1 chain with the entire HA2 chain. Displaying these stem immunogens on nanoparticles can enhance their immunogenicity (Yassine et al. 2015; Corbett et al. 2019). Thus, these stem immunogens are promising candidates for a universal influenza vaccine.
CONCLUDING REMARKS
Although the first structure of HA was reported almost 40 years ago, knowledge of HA recognition by antibodies had remained limited for many years. Because the major antigenic sites on HA are highly variable, it has been long believed that human antibodies with heterosubtypic activity against HA did not exist. Nonetheless, over the past decade, many human bnAbs to HA have been discovered and characterized. These bnAbs can neutralize multiple strains within a given subtype or across multiple subtypes. Structural characterization has shown that the epitopes of these bnAbs are distinct from the classic antigenic sites. Most HA bnAbs target the RBS or the stem domain, which are both evolutionarily conserved owing to functional constraints. These bnAbs have provided important insight into universal influenza vaccine design and also for the design of multidomain antibody, small protein, peptide, and small molecule therapeutic candidates.
ACKNOWLEDGMENTS
This work was supported by the Bill and Melinda Gates Foundation Post-Doctoral Fellowship in Global Health (N.C.W.) and the Bill and Melinda Gates Foundation OPP1170236 (I.A.W.).
This article has been made freely available online courtesy of TAUNS Laboratories.
Footnotes
Editors: Gabriele Neumann and Yoshihiro Kawaoka
Additional Perspectives on Influenza: The Cutting Edge available at www.perspectivesinmedicine.org
REFERENCES
- Altman MO, Bennink JR, Yewdell JW, Herrin BR. 2015. Lamprey VLRB response to influenza virus supports universal rules of immunogenicity and antigenicity. eLlife 4: e07467 10.7554/eLife.07467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altman MO, Angel M, Košik I, Trovão NS, Zost SJ, Gibbs JS, Casalino L, Amaro RE, Hensley SE, Nelson MI, et al. 2019. Human influenza A virus hemagglutinin glycan evolution follows a temporal pattern to a glycan limit. mBio 10: e00204–e00219. 10.1128/mBio.00204-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alymova IV, York IA, Air GM, Cipollo JF, Gulati S, Baranovich T, Kumar A, Zeng H, Gansebom S, McCullers JA. 2016. Glycosylation changes in the globular head of H3N2 influenza hemagglutinin modulate receptor binding without affecting virus virulence. Sci Rep 6: 36216 10.1038/srep36216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews SF, Joyce MG, Chambers MJ, Gillespie RA, Kanekiyo M, Leung K, Yang ES, Tsybovsky Y, Wheatley AK, Crank MC, et al. 2017. Preferential induction of cross-group influenza A hemagglutinin stem-specific memory B cells after H7N9 immunization in humans. Sci Immunol 2: eaan2676 10.1126/sciimmunol.aan2676 [DOI] [PubMed] [Google Scholar]
- Angeletti D, Gibbs JS, Angel M, Kosik I, Hickman HD, Frank GM, Das SR, Wheatley AK, Prabhakaran M, Leggat DJ, et al. 2017. Defining B cell immunodominance to viruses. Nat Immunol 18: 456–463. 10.1038/ni.3680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic G, Maron MJ, Adachi Y, Onodera T, McCarthy KR, McGee CE, Sempowski GD, Takahashi Y, Kelsoe G, Kuraoka M, et al. 2019. Influenza antigen engineering focuses immune responses to a subdominant but broadly protective viral epitope. Cell Host Microbe 25: 827–835.e6. 10.1016/j.chom.2019.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangaru S, Zhang H, Gilchuk IM, Voss TG, Irving RP, Gilchuk P, Matta P, Zhu X, Lang S, Nieusma T, et al. 2018. A multifunctional human monoclonal neutralizing antibody that targets a unique conserved epitope on influenza HA. Nat Commun 9: 2669 10.1038/s41467-018-04704-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bangaru S, Lang S, Schotsaert M, Vanderven HA, Zhu X, Kose N, Bombardi R, Finn JA, Kent SJ, Gilchuk P, et al. 2019. A site of vulnerability on the influenza virus hemagglutinin head domain trimer interface. Cell 177: 1136–1152.e18. 10.1016/j.cell.2019.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbey-Martin C, Gigant B, Bizebard T, Calder LJ, Wharton SA, Skehel JJ, Knossow M. 2002. An antibody that prevents the hemagglutinin low pH fusogenic transition. Virology 294: 70–74. 10.1006/viro.2001.1320 [DOI] [PubMed] [Google Scholar]
- Belongia EA, Simpson MD, King JP, Sundaram ME, Kelley NS, Osterholm MT, McLean HQ. 2016. Variable influenza vaccine effectiveness by subtype: a systematic review and meta-analysis of test-negative design studies. Lancet Infect Dis 16: 942–951. 10.1016/S1473-3099(16)00129-8 [DOI] [PubMed] [Google Scholar]
- Benton DJ, Nans A, Calder LJ, Turner J, Neu U, Lin YP, Ketelaars E, Kallewaard NL, Corti D, Lanzavecchia A, et al. 2018. Influenza hemagglutinin membrane anchor. Proc Natl Acad Sci 115: 10112–10117. 10.1073/pnas.1810927115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bizebard T, Gigant B, Rigolet P, Rasmussen B, Diat O, Bösecke P, Wharton SA, Skehel JJ, Knossow M. 1995. Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nature 376: 92–94. 10.1038/376092a0 [DOI] [PubMed] [Google Scholar]
- Blackburne BP, Hay AJ, Goldstein RA. 2008. Changing selective pressure during antigenic changes in human influenza H3. PLoS Pathog 4: e1000058 10.1371/journal.ppat.1000058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bommakanti G, Lu X, Citron MP, Najar TA, Heidecker GJ, ter Meulen J, Varadarajan R, Liang X. 2012. Design of Escherichia coli–expressed stalk domain immunogens of H1N1 hemagglutinin that protect mice from lethal challenge. J Virol 86: 13434–13444. 10.1128/JVI.01429-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher E, Matrosovich T, Beyerle M, Klenk HD, Garten W, Matrosovich M. 2006. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J Virol 80: 9896–9898. 10.1128/JVI.01118-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher-Friebertshauser E, Klenk HD, Garten W. 2013. Activation of influenza viruses by proteases from host cells and bacteria in the human airway epithelium. Pathog Dis 69: 87–100. 10.1111/2049-632X.12053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandenburg B, Koudstaal W, Goudsmit J, Klaren V, Tang C, Bujny MV, Korse HJ, Kwaks T, Otterstrom JJ, Juraszek J, et al. 2013. Mechanisms of hemagglutinin targeted influenza virus neutralization. PLoS ONE 8: e80034 10.1371/journal.pone.0080034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullough PA, Hughson FM, Treharne AC, Ruigrok RW, Skehel JJ, Wiley DC. 1994. Crystals of a fragment of influenza haemagglutinin in the low pH induced conformation. J Mol Biol 236: 1262–1265. 10.1016/0022-2836(94)90027-2 [DOI] [PubMed] [Google Scholar]
- Burioni R, Canducci F, Mancini N, Clementi N, Sassi M, De Marco D, Diotti RA, Saita D, Sampaolo M, Sautto G, et al. 2010. Monoclonal antibodies isolated from human B cells neutralize a broad range of H1 subtype influenza A viruses including swine-origin Influenza virus (S-OIV). Virology 399: 144–152. 10.1016/j.virol.2009.12.014 [DOI] [PubMed] [Google Scholar]
- Burnet FM, Stone JD. 1947. The receptor-destroying enzyme of V. cholerae. Aust J Exp Biol Med Sci 25: 227–233. 10.1038/icb.1947.33 [DOI] [PubMed] [Google Scholar]
- Byrd-Leotis L, Galloway SE, Agbogu E, Steinhauer DA. 2015. Influenza hemagglutinin (HA) stem region mutations that stabilize or destabilize the structure of multiple HA subtypes. J Virol 89: 4504–4516. 10.1128/JVI.00057-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadevall A, Imperiale MJ. 2014. Risks and benefits of gain-of-function experiments with pathogens of pandemic potential, such as influenza virus: a call for a science-based discussion. mBio 5: e01730–e01714. 10.1128/mBio.01730-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caton AJ, Brownlee GG, Yewdell JW, Gerhard W. 1982. The antigenic structure of the influenza virus A/PR/8/34 hemagglutinin (H1 subtype). Cell 31: 417–427. 10.1016/0092-8674(82)90135-0 [DOI] [PubMed] [Google Scholar]
- Chai N, Swem LR, Park S, Nakamura G, Chiang N, Estevez A, Fong R, Kamen L, Kho E, Reichelt M, et al. 2017. A broadly protective therapeutic antibody against influenza B virus with two mechanisms of action. Nat Commun 8: 14234 10.1038/ncomms14234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers BS, Parkhouse K, Ross TM, Alby K, Hensley SE. 2015. Identification of hemagglutinin residues responsible for H3N2 antigenic drift during the 2014–2015 influenza season. Cell Rep 12: 1–6. 10.1016/j.celrep.2015.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Lee KH, Steinhauer DA, Stevens DJ, Skehel JJ, Wiley DC. 1998. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 95: 409–417. 10.1016/S0092-8674(00)81771-7 [DOI] [PubMed] [Google Scholar]
- Chen J, Skehel JJ, Wiley DC. 1999. N- and C-terminal residues combine in the fusion-pH influenza hemagglutinin HA2 subunit to form an N cap that terminates the triple-stranded coiled coil. Proc Natl Acad Sci 96: 8967–8972. 10.1073/pnas.96.16.8967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Zhou H, Jin H. 2010. The impact of key amino acid substitutions in the hemagglutinin of influenza A (H3N2) viruses on vaccine production and antibody response. Vaccine 28: 4079–4085. 10.1016/j.vaccine.2010.03.078 [DOI] [PubMed] [Google Scholar]
- Cheng-Mayer C, Brown A, Harouse J, Luciw PA, Mayer AJ. 1999. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J Virol 73: 5294–5300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry JL, Lipman DJ, Nikolskaya A, Wolf YI. 2009. Evolutionary dynamics of N-glycosylation sites of influenza virus hemagglutinin. PLoS Curr 1: RRN1001 10.1371/currents.RRN1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevalier A, Silva DA, Rocklin GJ, Hicks DR, Vergara R, Murapa P, Bernard SM, Zhang L, Lam KH, Yao G, et al. 2017. Massively parallel de novo protein design for targeted therapeutics. Nature 550: 74–79. 10.1038/nature23912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clementi N, De Marco D, Mancini N, Solforosi L, Moreno GJ, Gubareva LV, Mishin V, Di Pietro A, Vicenzi E, Siccardi AG, et al. 2011. A human monoclonal antibody with neutralizing activity against highly divergent influenza subtypes. PLoS ONE 6: e28001 10.1371/journal.pone.0028001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connor RJ, Kawaoka Y, Webster RG, Paulson JC. 1994. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology 205: 17–23. 10.1006/viro.1994.1615 [DOI] [PubMed] [Google Scholar]
- Corbett KS, Moin SM, Yassine HM, Cagigi A, Kanekiyo M, Boyoglu-Barnum S, Myers SI, Tsybovsky Y, Wheatley AK, Schramm CA, et al. 2019. Design of nanoparticulate group 2 influenza virus hemagglutinin stem antigens that activate unmutated ancestor B cell receptors of broadly neutralizing antibody lineages. mBio 10: e02810–e02818. 10.1128/mBio.02810-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corti D, Voss J, Gamblin SJ, Codoni G, Macagno A, Jarrossay D, Vachieri SG, Pinna D, Minola A, Vanzetta F, et al. 2011. A neutralizing antibody selected from plasma cells that binds to group 1 and group 2 influenza A hemagglutinins. Science 333: 850–856. 10.1126/science.1205669 [DOI] [PubMed] [Google Scholar]
- Daniels RS, Downie JC, Hay AJ, Knossow M, Skehel JJ, Wang ML, Wiley DC. 1985. Fusion mutants of the influenza virus hemagglutinin glycoprotein. Cell 40: 431–439. 10.1016/0092-8674(85)90157-6 [DOI] [PubMed] [Google Scholar]
- Das SR, Puigbò P, Hensley SE, Hurt DE, Bennink JR, Yewdell JW. 2010. Glycosylation focuses sequence variation in the influenza A virus H1 hemagglutinin globular domain. PLoS Pathog 6: e1001211 10.1371/journal.ppat.1001211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das SR, Hensley SE, David A, Schmidt L, Gibbs JS, Puigbo P, Ince WL, Bennink JR, Yewdell JW. 2011. Fitness costs limit influenza A virus hemagglutinin glycosylation as an immune evasion strategy. Proc Natl Acad Sci 108: E1417–E1422. 10.1073/pnas.1108754108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis D, Stephens DM, Willers C, Lachmann PJ. 1990. Glycosylation governs the binding of antipeptide antibodies to regions of hypervariable amino acid sequence within recombinant gp120 of human immunodeficiency virus type 1. J Gen Virol 71: 2889–2898. 10.1099/0022-1317-71-12-2889 [DOI] [PubMed] [Google Scholar]
- de Vries RP, Zhu X, McBride R, Rigter A, Hanson A, Zhong G, Hatta M, Xu R, Yu W, Kawaoka Y, et al. 2014. Hemagglutinin receptor specificity and structural analyses of respiratory droplet-transmissible H5N1 viruses. J Virol 88: 768–773. 10.1128/JVI.02690-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries RP, Peng W, Grant OC, Thompson AJ, Zhu X, Bouwman KM, de la Pena ATT, van Breemen MJ, Ambepitiya Wickramasinghe IN, de Haan CAM, et al. 2017a. Three mutations switch H7N9 influenza to human-type receptor specificity. PLoS Pathog 13: e1006390 10.1371/journal.ppat.1006390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries RP, Tzarum N, Peng W, Thompson AJ, Ambepitiya Wickramasinghe IN, de la Pena ATT, van Breemen MJ, Bouwman KM, Zhu X, McBride R, et al. 2017b. A single mutation in Taiwanese H6N1 influenza hemagglutinin switches binding to human-type receptors. EMBO Mol Med 9: 1314–1325. 10.15252/emmm.201707726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus C, Laursen NS, Kwaks T, Zuijdgeest D, Khayat R, Ekiert DC, Lee JH, Metlagel Z, Bujny MV, Jongeneelen M, et al. 2012. Highly conserved protective epitopes on influenza B viruses. Science 337: 1343–1348. 10.1126/science.1222908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus C, Ekiert DC, Wilson IA. 2013. Structure of a classical broadly neutralizing stem antibody in complex with a pandemic H2 influenza virus hemagglutinin. J Virol 87: 7149–7154. 10.1128/JVI.02975-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBois RM, Zaraket H, Reddivari M, Heath RJ, White SW, Russell CJ. 2011. Acid stability of the hemagglutinin protein regulates H5N1 influenza virus pathogenicity. PLoS Pathog 7: e1002398 10.1371/journal.ppat.1002398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Bhabha G, Elsliger MA, Friesen RH, Jongeneelen M, Throsby M, Goudsmit J, Wilson IA. 2009. Antibody recognition of a highly conserved influenza virus epitope. Science 324: 246–251. 10.1126/science.1171491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Friesen RH, Bhabha G, Kwaks T, Jongeneelen M, Yu W, Ophorst C, Cox F, Korse HJ, Brandenburg B, et al. 2011. A highly conserved neutralizing epitope on group 2 influenza A viruses. Science 333: 843–850. 10.1126/science.1204839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekiert DC, Kashyap AK, Steel J, Rubrum A, Bhabha G, Khayat R, Lee JH, Dillon MA, O'Neil RE, Faynboym AM, et al. 2012. Cross-neutralization of influenza A viruses mediated by a single antibody loop. Nature 489: 526–532. 10.1038/nature11414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleishman SJ, Whitehead TA, Ekiert DC, Dreyfus C, Corn JE, Strauch EM, Wilson IA, Baker D. 2011. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science 332: 816–821. 10.1126/science.1202617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleury D, Wharton SA, Skehel JJ, Knossow M, Bizebard T. 1998. Antigen distortion allows influenza virus to escape neutralization. Nat Struct Biol 5: 119–123. 10.1038/nsb0298-119 [DOI] [PubMed] [Google Scholar]
- Fournillier A, Wychowski C, Boucreux D, Baumert TF, Meunier JC, Jacobs D, Muguet S, Depla E, Inchauspe G. 2001. Induction of hepatitis C virus E1 envelope protein-specific immune response can be enhanced by mutation of N-glycosylation sites. J Virol 75: 12088–12097. 10.1128/JVI.75.24.12088-12097.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesen RH, Lee PS, Stoop EJ, Hoffman RM, Ekiert DC, Bhabha G, Yu W, Juraszek J, Koudstaal W, Jongeneelen M, et al. 2014. A common solution to group 2 influenza virus neutralization. Proc Natl Acad Sci 111: 445–450. 10.1073/pnas.1319058110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Zhang Z, Sheehan J, Avnir Y, Ridenour C, Sachnik T, Sun J, Hossain MJ, Chen LM, Zhu Q, et al. 2016. A broadly neutralizing anti-influenza antibody reveals ongoing capacity of haemagglutinin-specific memory B cells to evolve. Nat Commun 7: 12780 10.1038/ncomms12780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuse Y, Matsuzaki Y, Nishimura H, Oshitani H. 2016. Analyses of evolutionary characteristics of the hemagglutinin-esterase gene of influenza C virus during a period of 68 years reveals evolutionary patterns different from influenza A and B viruses. Viruses 8: 321 10.3390/v8120321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galloway SE, Reed ML, Russell CJ, Steinhauer DA. 2013. Influenza HA subtypes demonstrate divergent phenotypes for cleavage activation and pH of fusion: implications for host range and adaptation. PLoS Pathog 9: e1003151 10.1371/journal.ppat.1003151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia NK, Guttman M, Ebner JL, Lee KK. 2015. Dynamic changes during acid-induced activation of influenza hemagglutinin. Structure 23: 665–676. 10.1016/j.str.2015.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garman EF. 2014. Developments in X-ray crystallographic structure determination of biological macromolecules. Science 343: 1102–1108. 10.1126/science.1247829 [DOI] [PubMed] [Google Scholar]
- Gerhard W, Yewdell J, Frankel ME, Webster R. 1981. Antigenic structure of influenza virus haemagglutinin defined by hybridoma antibodies. Nature 290: 713–717. 10.1038/290713a0 [DOI] [PubMed] [Google Scholar]
- Glaser L, Stevens J, Zamarin D, Wilson IA, García-Sastre A, Tumpey TM, Basler CF, Taubenberger JK, Palese P. 2005. A single amino acid substitution in 1918 influenza virus hemagglutinin changes receptor binding specificity. J Virol 79: 11533–11536. 10.1128/JVI.79.17.11533-11536.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotoh B, Ogasawara T, Toyoda T, Inocencio NM, Hamaguchi M, Nagai Y. 1990. An endoprotease homologous to the blood clotting factor X as a determinant of viral tropism in chick embryo. EMBO J 9: 4189–4195. 10.1002/j.1460-2075.1990.tb07643.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris A, Cardone G, Winkler DC, Heymann JB, Brecher M, White JM, Steven AC. 2006. Influenza virus pleiomorphy characterized by cryoelectron tomography. Proc Natl Acad Sci 103: 19123–19127. 10.1073/pnas.0607614103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry Dunand CJ, Leon PE, Kaur K, Tan GS, Zheng NY, Andrews S, Huang M, Qu X, Huang Y, Salgado-Ferrer M, et al. 2015. Preexisting human antibodies neutralize recently emerged H7N9 influenza strains. J Clin Invest 125: 1255–1268. 10.1172/JCI74374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henry Dunand CJ, Leon PE, Huang M, Choi A, Chromikova V, Ho IY, Tan GS, Cruz J, Hirsh A, Zheng NY, et al. 2016. Both neutralizing and non-neutralizing human H7N9 influenza vaccine-induced monoclonal antibodies confer protection. Cell Host Microbe 19: 800–813. 10.1016/j.chom.2016.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley SE, Das SR, Bailey AL, Schmidt LM, Hickman HD, Jayaraman A, Viswanathan K, Raman R, Sasisekharan R, Bennink JR, et al. 2009. Hemagglutinin receptor binding avidity drives influenza A virus antigenic drift. Science 326: 734–736. 10.1126/science.1178258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herfst S, Schrauwen EJ, Linster M, Chutinimitkul S, de Wit E, Munster VJ, Sorrell EM, Bestebroer TM, Burke DF, Smith DJ, et al. 2012. Airborne transmission of influenza A/H5N1 virus between ferrets. Science 336: 1534–1541. 10.1126/science.1213362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirst GK. 1942. The quantitative determination of influenza virus and antibodies by means of red cell agglutination. J Exp Med 75: 49–64. 10.1084/jem.75.1.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong M, Lee PS, Hoffman RM, Zhu X, Krause JC, Laursen NS, Yoon SI, Song L, Tussey L, Crowe JE Jr, et al. 2013. Antibody recognition of the pandemic H1N1 Influenza virus hemagglutinin receptor binding site. J Virol 87: 12471–12480. 10.1128/JVI.01388-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson EC, Charles PD, Hester SS, Thomas B, Trudgian D, Martínez-Alonso M, Fodor E. 2014. Conserved and host-specific features of influenza virion architecture. Nat Commun 5: 4816 10.1038/ncomms5816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iba Y, Fujii Y, Ohshima N, Sumida T, Kubota-Koketsu R, Ikeda M, Wakiyama M, Shirouzu M, Okada J, Okuno Y, et al. 2014. Conserved neutralizing epitope at globular head of hemagglutinin in H3N2 influenza viruses. J Virol 88: 7130–7144. 10.1128/JVI.00420-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi M, Ito K, Kida H, Takada A. 2008. Genetically destined potentials for N-linked glycosylation of influenza virus hemagglutinin. Virology 376: 323–329. 10.1016/j.virol.2008.03.036 [DOI] [PubMed] [Google Scholar]
- Imai M, Watanabe T, Hatta M, Das SC, Ozawa M, Shinya K, Zhong G, Hanson A, Katsura H, Watanabe S, et al. 2012. Experimental adaptation of an influenza H5 HA confers respiratory droplet transmission to a reassortant H5 HA/H1N1 virus in ferrets. Nature 486: 420–428. 10.1038/nature10831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Impagliazzo A, Milder F, Kuipers H, Wagner MV, Zhu X, Hoffman RM, van Meersbergen R, Huizingh J, Wanningen P, Verspuij J, et al. 2015. A stable trimeric influenza hemagglutinin stem as a broadly protective immunogen. Science 349: 1301–1306. 10.1126/science.aac7263 [DOI] [PubMed] [Google Scholar]
- Ito T, Suzuki Y, Takada A, Kawamoto A, Otsuki K, Masuda H, Yamada M, Suzuki T, Kida H, Kawaoka Y. 1997. Differences in sialic acid-galactose linkages in the chicken egg amnion and allantois influence human influenza virus receptor specificity and variant selection. J Virol 71: 3357–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jegaskanda S, Weinfurter JT, Friedrich TC, Kent SJ. 2013. Antibody-dependent cellular cytotoxicity is associated with control of pandemic H1N1 influenza virus infection of macaques. J Virol 87: 5512–5522. 10.1128/JVI.03030-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce MG, Wheatley AK, Thomas PV, Chuang GY, Soto C, Bailer RT, Druz A, Georgiev IS, Gillespie RA, Kanekiyo M, et al. 2016. Vaccine-induced antibodies that neutralize group 1 and group 2 influenza A viruses. Cell 166: 609–623. 10.1016/j.cell.2016.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadam RU, Juraszek J, Brandenburg B, Buyck C, Schepens WBG, Kesteleyn B, Stoops B, Vreeken RJ, Vermond J, Goutier W, et al. 2017. Potent peptidic fusion inhibitors of influenza virus. Science 358: 496–502. 10.1126/science.aan0516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kallewaard NL, Corti D, Collins PJ, Neu U, McAuliffe JM, Benjamin E, Wachter-Rosati L, Palmer-Hill FJ, Yuan AQ, Walker PA, et al. 2016. Structure and function analysis of an antibody recognizing all influenza A subtypes. Cell 166: 596–608. 10.1016/j.cell.2016.05.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karakus U, Thamamongood T, Ciminski K, Ran W, Günther SC, Pohl MO, Eletto D, Jeney C, Hoffmann D, Reiche S, et al. 2019. MHC class II proteins mediate cross-species entry of bat influenza viruses. Nature 567: 109–112. 10.1038/s41586-019-0955-3 [DOI] [PubMed] [Google Scholar]
- Kashyap AK, Steel J, Oner AF, Dillon MA, Swale RE, Wall KM, Perry KJ, Faynboym A, Ilhan M, Horowitz M, et al. 2008. Combinatorial antibody libraries from survivors of the Turkish H5N1 avian influenza outbreak reveal virus neutralization strategies. Proc Natl Acad Sci 105: 5986–5991. 10.1073/pnas.0801367105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashyap AK, Steel J, Rubrum A, Estelles A, Briante R, Ilyushina NA, Xu L, Swale RE, Faynboym AM, Foreman PK, et al. 2010. Protection from the 2009 H1N1 pandemic influenza by an antibody from combinatorial survivor-based libraries. PLoS Pathog 6: e1000990 10.1371/journal.ppat.1000990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kido H, Yokogoshi Y, Sakai K, Tashiro M, Kishino Y, Fukutomi A, Katunuma N. 1992. Isolation and characterization of a novel trypsin-like protease found in rat bronchiolar epithelial Clara cells. A possible activator of the viral fusion glycoprotein. J Biol Chem 267: 13573–13579. [PubMed] [Google Scholar]
- Kim JI, Lee I, Park S, Hwang MW, Bae JY, Lee S, Heo J, Park MS, García-Sastre A, Park MS. 2013. Genetic requirement for hemagglutinin glycosylation and its implications for influenza A H1N1 virus evolution. J Virol 87: 7539–7549. 10.1128/JVI.00373-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkpatrick E, Qiu X, Wilson PC, Bahl J, Krammer F. 2018. The influenza virus hemagglutinin head evolves faster than the stalk domain. Sci Rep 8: 10432 10.1038/s41598-018-28706-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodihalli S, Justewicz DM, Gubareva LV, Webster RG. 1995. Selection of a single amino acid substitution in the hemagglutinin molecule by chicken eggs can render influenza A virus (H3) candidate vaccine ineffective. J Virol 69: 4888–4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koel BF, Burke DF, Bestebroer TM, van der Vliet S, Zondag GC, Vervaet G, Skepner E, Lewis NS, Spronken MI, Russell CA, et al. 2013. Substitutions near the receptor binding site determine major antigenic change during influenza virus evolution. Science 342: 976–979. 10.1126/science.1244730 [DOI] [PubMed] [Google Scholar]
- Korte T, Ludwig K, Huang Q, Rachakonda PS, Herrmann A. 2007. Conformational change of influenza virus hemagglutinin is sensitive to ionic concentration. Eur Biophys J 36: 327–335. 10.1007/s00249-006-0116-0 [DOI] [PubMed] [Google Scholar]
- Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Basler CF, Crowe JE Jr. 2011. A broadly neutralizing human monoclonal antibody that recognizes a conserved, novel epitope on the globular head of the influenza H1N1 virus hemagglutinin. J Virol 85: 10905–10908. 10.1128/JVI.00700-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause JC, Tsibane T, Tumpey TM, Huffman CJ, Albrecht R, Blum DL, Ramos I, Fernandez-Sesma A, Edwards KM, García-Sastre A, et al. 2012. Human monoclonal antibodies to pandemic 1957 H2N2 and pandemic 1968 H3N2 influenza viruses. J Virol 86: 6334–6340. 10.1128/JVI.07158-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang S, Xie J, Zhu X, Wu NC, Lerner RA, Wilson IA. 2017. Antibody 27F3 broadly targets influenza A group 1 and 2 hemagglutinins through a further variation in VH1-69 antibody orientation on the HA stem. Cell Rep 20: 2935–2943. 10.1016/j.celrep.2017.08.084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laursen NS, Friesen RHE, Zhu X, Jongeneelen M, Blokland S, Vermond J, van Eijgen A, Tang C, van Diepen H, Obmolova G, et al. 2018. Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin. Science 362: 598–602. 10.1126/science.aaq0620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarowitz SG, Choppin PW. 1975. Enhancement of the infectivity of influenza A and B viruses by proteolytic cleavage of the hemagglutinin polypeptide. Virology 68: 440–454. 10.1016/0042-6822(75)90285-8 [DOI] [PubMed] [Google Scholar]
- Lee PS, Wilson IA. 2015. Structural characterization of viral epitopes recognized by broadly cross-reactive antibodies. Curr Top Microbiol Immunol 386: 323–341. doi: 10.1007/82_2014_413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PS, Yoshida R, Ekiert DC, Sakai N, Suzuki Y, Takada A, Wilson IA. 2012. Heterosubtypic antibody recognition of the influenza virus hemagglutinin receptor binding site enhanced by avidity. Proc Natl Acad Sci 109: 17040–17045. 10.1073/pnas.1212371109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PS, Ohshima N, Stanfield RL, Yu W, Iba Y, Okuno Y, Kurosawa Y, Wilson IA. 2014. Receptor mimicry by antibody F045-092 facilitates universal binding to the H3 subtype of influenza virus. Nat Commun 5: 3614 10.1038/ncomms4614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Boutz DR, Chromikova V, Joyce MG, Vollmers C, Leung K, Horton AP, DeKosky BJ, Lee CH, Lavinder JJ, et al. 2016. Molecular-level analysis of the serum antibody repertoire in young adults before and after seasonal influenza vaccination. Nat Med 22: 1456–1464. 10.1038/nm.4224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wang Y, Liang Y, Ni B, Wan Y, Liao Z, Chan KH, Yuen KY, Fu X, Shang X, et al. 2009. Fine antigenic variation within H5N1 influenza virus hemagglutinin's antigenic sites defined by yeast cell surface display. Eur J Immunol 39: 3498–3510. 10.1002/eji.200939532 [DOI] [PubMed] [Google Scholar]
- Li Y, Bostick DL, Sullivan CB, Myers JL, Griesemer SB, Stgeorge K, Plotkin JB, Hensley SE. 2013. Single hemagglutinin mutations that alter both antigenicity and receptor binding avidity influence influenza virus antigenic clustering. J Virol 87: 9904–9910. 10.1128/JVI.01023-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YP, Xiong X, Wharton SA, Martin SR, Coombs PJ, Vachieri SG, Christodoulou E, Walker PA, Liu J, Skehel JJ, et al. 2012. Evolution of the receptor binding properties of the influenza A(H3N2) hemagglutinin. Proc Natl Acad Sci 109: 21474–21479. 10.1073/pnas.1218841110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowen AC. 2017. Constraints, drivers, and implications of influenza A virus reassortment. Annu Rev Virol 4: 105–121. 10.1146/annurev-virology-101416-041726 [DOI] [PubMed] [Google Scholar]
- Lu X, Shi Y, Zhang W, Zhang Y, Qi J, Gao GF. 2013. Structure and receptor-binding properties of an airborne transmissible avian influenza A virus hemagglutinin H5 (VN1203mut). Protein Cell 4: 502–511. 10.1007/s13238-013-3906-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Ohnishi S. 1980. Activation of influenza virus by acidic media causes hemolysis and fusion of erythrocytes. FEBS Lett 122: 283–287. 10.1016/0014-5793(80)80457-1 [DOI] [PubMed] [Google Scholar]
- Mair CM, Meyer T, Schneider K, Huang Q, Veit M, Herrmann A. 2014. A histidine residue of the influenza virus hemagglutinin controls the pH dependence of the conformational change mediating membrane fusion. J Virol 88: 13189–13200. 10.1128/JVI.01704-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallajosyula VV, Citron M, Ferrara F, Lu X, Callahan C, Heidecker GJ, Sarma SP, Flynn JA, Temperton NJ, Liang X, et al. 2014. Influenza hemagglutinin stem-fragment immunogen elicits broadly neutralizing antibodies and confers heterologous protection. Proc Natl Acad Sci 111: E2514–E2523. 10.1073/pnas.1402766111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallajosyula VV, Citron M, Ferrara F, Temperton NJ, Liang X, Flynn JA, Varadarajan R. 2015. Hemagglutinin sequence conservation guided stem immunogen design from influenza A H3 subtype. Front Immunol 6: 329 10.3389/fimmu.2015.00329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich MN, Gambaryan AS, Teneberg S, Piskarev VE, Yamnikova SS, Lvov DK, Robertson JS, Karlsson KA. 1997. Avian influenza A viruses differ from human viruses by recognition of sialyloligosaccharides and gangliosides and by a higher conservation of the HA receptor-binding site. Virology 233: 224–234. 10.1006/viro.1997.8580 [DOI] [PubMed] [Google Scholar]
- McCarthy KR, Watanabe A, Kuraoka M, Do KT, McGee CE, Sempowski GD, Kepler TB, Schmidt AG, Kelsoe G, Harrison SC. 2018. Memory B cells that cross-react with group 1 and group 2 influenza A viruses are abundant in adult human repertoires. Immunity 48: 174–184.e9. 10.1016/j.immuni.2017.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura G, Chai N, Park S, Chiang N, Lin Z, Chiu H, Fong R, Yan D, Kim J, Zhang J, et al. 2013. An in vivo human-plasmablast enrichment technique allows rapid identification of therapeutic influenza A antibodies. Cell Host Microbe 14: 93–103. 10.1016/j.chom.2013.06.004 [DOI] [PubMed] [Google Scholar]
- Nogales E. 2016. The development of cryo-EM into a mainstream structural biology technique. Nat Methods 13: 24–27. 10.1038/nmeth.3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima N, Iba Y, Kubota-Koketsu R, Asano Y, Okuno Y, Kurosawa Y. 2011. Naturally occurring antibodies in humans can neutralize a variety of influenza virus strains, including H3, H1, H2, and H5. J Virol 85: 11048–11057. 10.1128/JVI.05397-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okuno Y, Isegawa Y, Sasao F, Ueda S. 1993. A common neutralizing epitope conserved between the hemagglutinins of influenza A virus H1 and H2 strains. J Virol 67: 2552–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pappas C, Viswanathan K, Chandrasekaran A, Raman R, Katz JM, Sasisekharan R, Tumpey TM. 2010. Receptor specificity and transmission of H2N2 subtype viruses isolated from the pandemic of 1957. PLoS ONE 5: e11158 10.1371/journal.pone.0011158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker L, Wharton SA, Martin SR, Cross K, Lin Y, Liu Y, Feizi T, Daniels RS, McCauley JW. 2016. Effects of egg-adaptation on receptor-binding and antigenic properties of recent influenza A (H3N2) vaccine viruses. J Gen Virol 97: 1333–1344. 10.1099/jgv.0.000457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng W, de Vries RP, Grant OC, Thompson AJ, McBride R, Tsogtbaatar B, Lee PS, Razi N, Wilson IA, Woods RJ, et al. 2017. Recent H3N2 viruses have evolved specificity for extended, branched human-type receptors, conferring potential for increased avidity. Cell Host Microbe 21: 23–34. 10.1016/j.chom.2016.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popova L, Smith K, West AH, Wilson PC, James JA, Thompson LF, Air GM. 2012. Immunodominance of antigenic site B over site A of hemagglutinin of recent H3N2 influenza viruses. PLoS ONE 7: e41895 10.1371/journal.pone.0041895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri A, Booy FP, Doms RW, White JM, Blumenthal R. 1990. Conformational changes and fusion activity of influenza virus hemagglutinin of the H2 and H3 subtypes: effects of acid pretreatment. J Virol 64: 3824–3832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachakonda PS, Veit M, Korte T, Ludwig K, Böttcher C, Huang Q, Schmidt MF, Herrmann A. 2007. The relevance of salt bridges for the stability of the influenza virus hemagglutinin. FASEB J 21: 995–1002. 10.1096/fj.06-7052hyp [DOI] [PubMed] [Google Scholar]
- Raymond DD, Stewart SM, Lee J, Ferdman J, Bajic G, Do KT, Ernandes MJ, Suphaphiphat P, Settembre EC, Dormitzer PR, et al. 2016. Influenza immunization elicits antibodies specific for an egg-adapted vaccine strain. Nat Med 22: 1465–1469. 10.1038/nm.4223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raymond DD, Bajic G, Ferdman J, Suphaphiphat P, Settembre EC, Moody MA, Schmidt AG, Harrison SC. 2018. Conserved epitope on influenza-virus hemagglutinin head defined by a vaccine-induced antibody. Proc Natl Acad Sci 115: 168–173. 10.1073/pnas.1715471115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts KL, Shelton H, Scull M, Pickles R, Barclay WS. 2011. Lack of transmission of a human influenza virus with avian receptor specificity between ferrets is not due to decreased virus shedding but rather a lower infectivity in vivo. J Gen Virol 92: 1822–1831. 10.1099/vir.0.031203-0 [DOI] [PubMed] [Google Scholar]
- Robertson JS, Bootman JS, Newman R, Oxford JS, Daniels RS, Webster RG, Schild GC. 1987. Structural changes in the haemagglutinin which accompany egg adaptation of an influenza A(H1N1) virus. Virology 160: 31–37. 10.1016/0042-6822(87)90040-7 [DOI] [PubMed] [Google Scholar]
- Rogers GN, Paulson JC, Daniels RS, Skehel JJ, Wilson IA, Wiley DC. 1983. Single amino acid substitutions in influenza haemagglutinin change receptor binding specificity. Nature 304: 76–78. 10.1038/304076a0 [DOI] [PubMed] [Google Scholar]
- Sabesan S, Bock K, Paulson JC. 1991. Conformational analysis of sialyloligosaccharides. Carbohydr Res 218: 27–54. 10.1016/0008-6215(91)84084-R [DOI] [PubMed] [Google Scholar]
- Schmidt AG, Xu H, Khan AR, O'Donnell T, Khurana S, King LR, Manischewitz J, Golding H, Suphaphiphat P, Carfi A, et al. 2013. Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody. Proc Natl Acad Sci 110: 264–269. 10.1073/pnas.1218256109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt AG, Therkelsen MD, Stewart S, Kepler TB, Liao HX, Moody MA, Haynes BF, Harrison SC. 2015. Viral receptor-binding site antibodies with diverse germline origins. Cell 161: 1026–1034. 10.1016/j.cell.2015.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholtissek C. 1985. Stability of infectious influenza A viruses at low pH and at elevated temperature. Vaccine 3: 215–218. 10.1016/0264-410X(85)90109-4 [DOI] [PubMed] [Google Scholar]
- Shi Y, Wu Y, Zhang W, Qi J, Gao GF. 2014. Enabling the “host jump”: structural determinants of receptor-binding specificity in influenza A viruses. Nat Rev Microbiol 12: 822–831. 10.1038/nrmicro3362 [DOI] [PubMed] [Google Scholar]
- Simmonds P, Zhang LQ, McOmish F, Balfe P, Ludlam CA, Brown AJ. 1991. Discontinuous sequence change of human immunodeficiency virus (HIV) type 1 env sequences in plasma viral and lymphocyte-associated proviral populations in vivo: implications for models of HIV pathogenesis. J Virol 65: 6266–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skehel JJ, Stevens DJ, Daniels RS, Douglas AR, Knossow M, Wilson IA, Wiley DC. 1984. A carbohydrate side chain on hemagglutinins of Hong Kong influenza viruses inhibits recognition by a monoclonal antibody. Proc Natl Acad Sci 81: 1779–1783. 10.1073/pnas.81.6.1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DJ, Lapedes AS, de Jong JC, Bestebroer TM, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. 2004. Mapping the antigenic and genetic evolution of influenza virus. Science 305: 371–376. 10.1126/science.1097211 [DOI] [PubMed] [Google Scholar]
- Song H, Qi J, Xiao H, Bi Y, Zhang W, Xu Y, Wang F, Shi Y, Gao GF. 2017. Avian-to-human receptor-binding adaptation by influenza A virus hemagglutinin H4. Cell Rep 20: 1201–1214. 10.1016/j.celrep.2017.07.028 [DOI] [PubMed] [Google Scholar]
- Sriwilaijaroen N, Kondo S, Yagi H, Wilairat P, Hiramatsu H, Ito M, Ito Y, Kato K, Suzuki Y. 2009. Analysis of N-glycans in embryonated chicken egg chorioallantoic and amniotic cells responsible for binding and adaptation of human and avian influenza viruses. Glycoconj J 26: 433–443. 10.1007/s10719-008-9193-x [DOI] [PubMed] [Google Scholar]
- Steel J, Lowen AC, Wang TT, Yondola M, Gao Q, Haye K, García-Sastre A, Palese P. 2010. Influenza virus vaccine based on the conserved hemagglutinin stalk domain. mBio 1: e00018–e00010. 10.1128/mBio.00018-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens J, Blixt O, Glaser L, Taubenberger JK, Palese P, Paulson JC, Wilson IA. 2006. Glycan microarray analysis of the hemagglutinins from modern and pandemic influenza viruses reveals different receptor specificities. J Mol Biol 355: 1143–1155. 10.1016/j.jmb.2005.11.002 [DOI] [PubMed] [Google Scholar]
- Stone JD. 1948. Prevention of virus infection with enzyme of V. cholerae. II: studies with influenza virus in mice. Aust J Exp Biol Med Sci 26: 287–298. 10.1038/icb.1948.30 [DOI] [PubMed] [Google Scholar]
- Strauch EM, Bernard SM, La D, Bohn AJ, Lee PS, Anderson CE, Nieusma T, Holstein CA, Garcia NK, Hooper KA, et al. 2017. Computational design of trimeric influenza-neutralizing proteins targeting the hemagglutinin receptor binding site. Nat Biotechnol 35: 667–671. 10.1038/nbt.3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez DL, Garcia M, Latimer J, Senne D, Perdue M. 1999. Phylogenetic analysis of H7 avian influenza viruses isolated from the live bird markets of the Northeast United States. J Virol 73: 3567–3573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Hwang WC, Perez S, Wei G, Aird D, Chen LM, Santelli E, Stec B, Cadwell G, Ali M, et al. 2009. Structural and functional bases for broad-spectrum neutralization of avian and human influenza A viruses. Nat Struct Mol Biol 16: 265–273. 10.1038/nsmb.1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S, Wang Q, Zhao F, Chen W, Li Z. 2011. Glycosylation site alteration in the evolution of influenza A (H1N1) viruses. PLoS ONE 6: e22844 10.1371/journal.pone.0022844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Shi Y, Lu X, He J, Gao F, Yan J, Qi J, Gao GF. 2013. Bat-derived influenza hemagglutinin H17 does not bind canonical avian or human receptors and most likely uses a unique entry mechanism. Cell Rep 3: 769–778. 10.1016/j.celrep.2013.01.025 [DOI] [PubMed] [Google Scholar]
- Tan HX, Jegaskanda S, Juno JA, Esterbauer R, Wong J, Kelly HG, Liu Y, Tilmanis D, Hurt AC, Yewdell JW, et al. 2019. Subdominance and poor intrinsic immunogenicity limit humoral immunity targeting influenza HA stem. J Clin Invest 129: 850–862. 10.1172/JCI123366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate MD, Job ER, Deng YM, Gunalan V, Maurer-Stroh S, Reading PC. 2014. Playing hide and seek: how glycosylation of the influenza virus hemagglutinin can modulate the immune response to infection. Viruses 6: 1294–1316. 10.3390/v6031294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoennes S, Li ZN, Lee BJ, Langley WA, Skehel JJ, Russell RJ, Steinhauer DA. 2008. Analysis of residues near the fusion peptide in the influenza hemagglutinin structure for roles in triggering membrane fusion. Virology 370: 403–414. 10.1016/j.virol.2007.08.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornburg NJ, Zhang H, Bangaru S, Sapparapu G, Kose N, Lampley RM, Bombardi RG, Yu Y, Graham S, Branchizio A, et al. 2016. H7N9 influenza virus neutralizing antibodies that possess few somatic mutations. J Clin Invest 126: 1482–1494. 10.1172/JCI85317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Throsby M, van den Brink E, Jongeneelen M, Poon LL, Alard P, Cornelissen L, Bakker A, Cox F, van Deventer E, Guan Y, et al. 2008. Heterosubtypic neutralizing monoclonal antibodies cross-protective against H5N1 and H1N1 recovered from human IgM+ memory B cells. PLoS ONE 3: e3942 10.1371/journal.pone.0003942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong S, Zhu X, Li Y, Shi M, Zhang J, Bourgeois M, Yang H, Chen X, Recuenco S, Gomez J, et al. 2013. New world bats harbor diverse influenza A viruses. PLoS Pathog 9: e1003657 10.1371/journal.ppat.1003657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tricco AC, Chit A, Soobiah C, Hallett D, Meier G, Chen MH, Tashkandi M, Bauch CT, Loeb M. 2013. Comparing influenza vaccine efficacy against mismatched and matched strains: a systematic review and meta-analysis. BMC Med 11: 153 10.1186/1741-7015-11-153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsibane T, Ekiert DC, Krause JC, Martinez O, Crowe JE Jr, Wilson IA, Basler CF. 2012. Influenza human monoclonal antibody 1F1 interacts with three major antigenic sites and residues mediating human receptor specificity in H1N1 viruses. PLoS Pathog 8: e1003067 10.1371/journal.ppat.1003067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuchiya E, Sugawara K, Hongo S, Matsuzaki Y, Muraki Y, Li ZN, Nakamura K. 2001. Antigenic structure of the haemagglutinin of human influenza A/H2N2 virus. J Gen Virol 82: 2475–2484. 10.1099/0022-1317-82-10-2475 [DOI] [PubMed] [Google Scholar]
- Tsuchiya E, Sugawara K, Hongo S, Matsuzaki Y, Muraki Y, Li ZN, Nakamura K. 2002. Effect of addition of new oligosaccharide chains to the globular head of influenza A/H2N2 virus haemagglutinin on the intracellular transport and biological activities of the molecule. J Gen Virol 83: 1137–1146. 10.1099/0022-1317-83-5-1137 [DOI] [PubMed] [Google Scholar]
- Tumpey TM, Maines TR, Van Hoeven N, Glaser L, Solorzano A, Pappas C, Cox NJ, Swayne DE, Palese P, Katz JM, et al. 2007. A two-amino acid change in the hemagglutinin of the 1918 influenza virus abolishes transmission. Science 315: 655–659. 10.1126/science.1136212 [DOI] [PubMed] [Google Scholar]
- Turner HL, Pallesen J, Lang S, Bangaru S, Urata S, Li S, Cottrell CA, Bowman CA, Crowe JE Jr, Wilson IA, et al. 2019. Potent anti-influenza H7 human monoclonal antibody induces separation of hemagglutinin receptor-binding head domains. PLoS Biol 17: e3000139 10.1371/journal.pbio.3000139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzarum N, de Vries RP, Peng W, Thompson AJ, Bouwman KM, McBride R, Yu W, Zhu X, Verheije MH, Paulson JC, et al. 2017. The 150-loop restricts the host specificity of human H10N8 influenza virus. Cell Rep 19: 235–245. 10.1016/j.celrep.2017.03.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valkenburg SA, Mallajosyula VV, Li OT, Chin AW, Carnell G, Temperton N, Varadarajan R, Poon LL. 2016. Stalking influenza by vaccination with pre-fusion headless HA mini-stem. Sci Rep 6: 22666 10.1038/srep22666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dongen MJP, Kadam RU, Juraszek J, Lawson E, Brandenburg B, Schmitz F, Schepens WBG, Stoops B, van Diepen HA, Jongeneelen M, et al. 2019. A small-molecule fusion inhibitor of influenza virus is orally active in mice. Science 363: eaar6221 10.1126/science.aar6221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ML, Skehel JJ, Wiley DC. 1986. Comparative analyses of the specificities of anti-influenza hemagglutinin antibodies in human sera. J Virol 57: 124–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Tian X, Chen X, Ma J. 2007. Structural basis for receptor specificity of influenza B virus hemagglutinin. Proc Natl Acad Sci 104: 16874–16879. 10.1073/pnas.0708363104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, McCarthy KR, Kuraoka M, Schmidt AG, Adachi Y, Onodera T, Tonouchi K, Caradonna TM, Bajic G, Song S, et al. 2019. Antibodies to a conserved influenza head interface epitope protect by an IgG subtype-dependent mechanism. Cell 177: 1124–1135.e16. 10.1016/j.cell.2019.03.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weis W, Brown JH, Cusack S, Paulson JC, Skehel JJ, Wiley DC. 1988. Structure of the influenza virus haemagglutinin complexed with its receptor, sialic acid. Nature 333: 426–431. 10.1038/333426a0 [DOI] [PubMed] [Google Scholar]
- Whitehead TA, Chevalier A, Song Y, Dreyfus C, Fleishman SJ, De Mattos C, Myers CA, Kamisetty H, Blair P, Wilson IA, et al. 2012. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat Biotechnol 30: 543–548. 10.1038/nbt.2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittle JR, Zhang R, Khurana S, King LR, Manischewitz J, Golding H, Dormitzer PR, Haynes BF, Walter EB, Moody MA, et al. 2011. Broadly neutralizing human antibody that recognizes the receptor-binding pocket of influenza virus hemagglutinin. Proc Natl Acad Sci 108: 14216–14221. 10.1073/pnas.1111497108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley DC, Skehel JJ. 1987. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem 56: 365–394. 10.1146/annurev.bi.56.070187.002053 [DOI] [PubMed] [Google Scholar]
- Wiley DC, Wilson IA, Skehel JJ. 1981. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 289: 373–378. 10.1038/289373a0 [DOI] [PubMed] [Google Scholar]
- Wilson IA, Skehel JJ, Wiley DC. 1981. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature 289: 366–373. 10.1038/289366a0 [DOI] [PubMed] [Google Scholar]
- Wilson IA, Niman HL, Houghten RA, Cherenson AR, Connolly ML, Lerner RA. 1984. The structure of an antigenic determinant in a protein. Cell 37: 767–778. 10.1016/0092-8674(84)90412-4 [DOI] [PubMed] [Google Scholar]
- Wohlbold TJ, Nachbagauer R, Margine I, Tan GS, Hirsh A, Krammer F. 2015. Vaccination with soluble headless hemagglutinin protects mice from challenge with divergent influenza viruses. Vaccine 33: 3314–3321. 10.1016/j.vaccine.2015.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrammert J, Smith K, Miller J, Langley WA, Kokko K, Larsen C, Zheng NY, Mays I, Garman L, Helms C, et al. 2008. Rapid cloning of high-affinity human monoclonal antibodies against influenza virus. Nature 453: 667–671. 10.1038/nature06890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu NC, Wilson IA. 2017. A perspective on the structural and functional constraints for immune evasion: insights from influenza virus. J Mol Biol 429: 2694–2709. 10.1016/j.jmb.2017.06.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu NC, Wilson IA. 2018. Structural insights into the design of novel anti-influenza therapies. Nat Struct Mol Biol 25: 115–121. 10.1038/s41594-018-0025-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y, Cho M, Shore D, Song M, Choi J, Jiang T, Deng YQ, Bourgeois M, Almli L, Yang H, et al. 2015. A potent broad-spectrum protective human monoclonal antibody crosslinking two haemagglutinin monomers of influenza A virus. Nat Commun 6: 7708 10.1038/ncomms8708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu NC, Zost SJ, Thompson AJ, Oyen D, Nycholat CM, McBride R, Paulson JC, Hensley SE, Wilson IA. 2017. A structural explanation for the low effectiveness of the seasonal influenza H3N2 vaccine. PLoS Pathog 13: e1006682 10.1371/journal.ppat.1006682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu NC, Thompson AJ, Xie J, Lin CW, Nycholat CM, Zhu X, Lerner RA, Paulson JC, Wilson IA. 2018a. A complex epistatic network limits the mutational reversibility in the influenza hemagglutinin receptor-binding site. Nat Commun 9: 1264 10.1038/s41467-018-03663-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu NC, Yamayoshi S, Ito M, Uraki R, Kawaoka Y, Wilson IA. 2018b. Recurring and adaptable binding motifs in broadly neutralizing antibodies to influenza virus are encoded on the D3-9 segment of the Ig gene. Cell Host Microbe 24: 569–578.e4. 10.1016/j.chom.2018.09.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu NC, Lv H, Thompson AJ, Wu DC, Ng WWS, Kadam RU, Lin CW, Nycholat CM, McBride R, Liang W, et al. 2019. Preventing an antigenically disruptive mutation in egg-based H3N2 seasonal influenza vaccines by mutational incompatibility. Cell Host Microbe 25: 836–844.e5. 10.1016/j.chom.2019.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong X, Coombs PJ, Martin SR, Liu J, Xiao H, McCauley JW, Locher K, Walker PA, Collins PJ, Kawaoka Y, et al. 2013. Receptor binding by a ferret-transmissible H5 avian influenza virus. Nature 497: 392–396. 10.1038/nature12144 [DOI] [PubMed] [Google Scholar]
- Xu R, Wilson IA. 2011. Structural characterization of an early fusion intermediate of influenza virus hemagglutinin. J Virol 85: 5172–5182. 10.1128/JVI.02430-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, McBride R, Paulson JC, Basler CF, Wilson IA. 2010. Structure, receptor binding, and antigenicity of influenza virus hemagglutinins from the 1957 H2N2 pandemic. J Virol 84: 1715–1721. 10.1128/JVI.02162-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, McBride R, Nycholat CM, Paulson JC, Wilson IA. 2012. Structural characterization of the hemagglutinin receptor specificity from the 2009 H1N1 influenza pandemic. J Virol 86: 982–990. 10.1128/JVI.06322-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Krause JC, McBride R, Paulson JC, Crowe JE Jr, Wilson IA. 2013. A recurring motif for antibody recognition of the receptor-binding site of influenza hemagglutinin. Nat Struct Mol Biol 20: 363–370. 10.1038/nsmb.2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamayoshi S, Uraki R, Ito M, Kiso M, Nakatsu S, Yasuhara A, Oishi K, Sasaki T, Ikuta K, Kawaoka Y. 2017. A broadly reactive human anti-hemagglutinin stem monoclonal antibody that inhibits influenza A virus particle release. EBioMedicine 17: 182–191. 10.1016/j.ebiom.2017.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Chen LM, Carney PJ, Donis RO, Stevens J. 2010. Structures of receptor complexes of a North American H7N2 influenza hemagglutinin with a loop deletion in the receptor binding site. PLoS Pathog 6: e1001081 10.1371/journal.ppat.1001081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yassine HM, Boyington JC, McTamney PM, Wei CJ, Kanekiyo M, Kong WP, Gallagher JR, Wang L, Zhang Y, Joyce MG, et al. 2015. Hemagglutinin-stem nanoparticles generate heterosubtypic influenza protection. Nat Med 21: 1065–1070. 10.1038/nm.3927 [DOI] [PubMed] [Google Scholar]
- Yasugi M, Kubota-Koketsu R, Yamashita A, Kawashita N, Du A, Sasaki T, Nishimura M, Misaki R, Kuhara M, Boonsathorn N, et al. 2013. Human monoclonal antibodies broadly neutralizing against influenza B virus. PLoS Pathog 9: e1003150 10.1371/journal.ppat.1003150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R, Igarashi M, Ozaki H, Kishida N, Tomabechi D, Kida H, Ito K, Takada A. 2009. Cross-protective potential of a novel monoclonal antibody directed against antigenic site B of the hemagglutinin of influenza A viruses. PLoS Pathog 5: e1000350 10.1371/journal.ppat.1000350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Gaschen B, Blay W, Foley B, Haigwood N, Kuiken C, Korber B. 2004. Tracking global patterns of N-linked glycosylation site variation in highly variable viral glycoproteins: HIV, SIV, and HCV envelopes and influenza hemagglutinin. Glycobiology 14: 1229–1246. 10.1093/glycob/cwh106 [DOI] [PubMed] [Google Scholar]
- Zhu X, Guo YH, Jiang T, Wang YD, Chan KH, Li XF, Yu W, McBride R, Paulson JC, Yuen KY, et al. 2013a. A unique and conserved neutralization epitope in H5N1 influenza viruses identified by an antibody against the A/Goose/Guangdong/1/96 hemagglutinin. J Virol 87: 12619–12635. 10.1128/JVI.01577-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Yu W, McBride R, Li Y, Chen LM, Donis RO, Tong S, Paulson JC, Wilson IA. 2013b. Hemagglutinin homologue from H17N10 bat influenza virus exhibits divergent receptor-binding and pH-dependent fusion activities. Proc Natl Acad Sci 110: 1458–1463. 10.1073/pnas.1218509110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost SJ, Parkhouse K, Gumina ME, Kim K, Diaz Perez S, Wilson PC, Treanor JJ, Sant AJ, Cobey S, Hensley SE. 2017. Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc Natl Acad Sci 114: 12578–12583. 10.1073/pnas.1712377114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zost SJ, Wu NC, Hensley SE, Wilson IA. 2019. Immunodominance and antigenic variation of influenza virus hemagglutinin: implications for design of universal vaccine immunogens. J Infect Dis 219: S38–S45. 10.1093/infdis/jiy696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo T, Sun J, Wang G, Jiang L, Zuo Y, Li D, Shi X, Liu X, Fan S, Ren H, et al. 2015. Comprehensive analysis of antibody recognition in convalescent humans from highly pathogenic avian influenza H5N1 infection. Nat Commun 6: 8855 10.1038/ncomms9855 [DOI] [PMC free article] [PubMed] [Google Scholar]