In this study, Szulzewsky et al. show that the YAP1 gene fusions YAP1-MAMLD1, YAP1-FAM118B, YAP1-TFE3, and YAP1-SS18 are oncogenic in mice. Their findings highlight TEAD-dependent YAP activity found in YAP1 gene fusions as critical for oncogenesis and implicate the identified YAP functions as potential therapeutic targets in YAP1 fusion-positive tumors.
Keywords: gene fusion, cancer, YAP1, MAMLD1, TFE3, SS18, TEAD, ependymoma, angiosarcoma, verteporfin
Abstract
YAP1 is a transcriptional coactivator and the principal effector of the Hippo signaling pathway, which is causally implicated in human cancer. Several YAP1 gene fusions have been identified in various human cancers and identifying the essential components of this family of gene fusions has significant therapeutic value. Here, we show that the YAP1 gene fusions YAP1-MAMLD1, YAP1-FAM118B, YAP1-TFE3, and YAP1-SS18 are oncogenic in mice. Using reporter assays, RNA-seq, ChIP-seq, and loss-of-function mutations, we can show that all of these YAP1 fusion proteins exert TEAD-dependent YAP activity, while some also exert activity of the C′-terminal fusion partner. The YAP activity of the different YAP1 fusions is resistant to negative Hippo pathway regulation due to constitutive nuclear localization and resistance to degradation of the YAP1 fusion proteins. Genetic disruption of the TEAD-binding domain of these oncogenic YAP1 fusions is sufficient to inhibit tumor formation in vivo, while pharmacological inhibition of the YAP1–TEAD interaction inhibits the growth of YAP1 fusion-expressing cell lines in vitro. These results highlight TEAD-dependent YAP activity found in these gene fusions as critical for oncogenesis and implicate these YAP functions as potential therapeutic targets in YAP1 fusion-positive tumors.
Most cancer types harbor a highly aberrant genome, including gains and losses of whole chromosomal arms as well as additional activating or inactivating point mutations, leading to multiple concurrent and redundant drivers that make targeting of specific pathways difficult. In contrast, some cancers have a relatively stable genome with a low overall mutational burden but harbor recurrent gene fusions, which are the likely initiating tumorigenic events in oncogenesis (Rowley 1973; Parker and Zhang 2013; Kim et al. 2014), several of which have been shown to be sufficient to cause cancer when expressed in mice (Dash et al. 2002; Oldrini et al. 2018; Ozawa et al. 2018).
Recent cancer genome sequencing studies have identified several gene fusions involving the N′-terminal regions of YAP1 in different tumor types, including supratentorial (ST) ependymoma (YAP1-MAMLD1 and YAP1-FAM118B) (Pajtler et al. 2015, 2019), epithelioid hemangioendothelioma (EHE; YAP1-TFE3) (Antonescu et al. 2013), and cervical squamous cell carcinoma and endocervical adenocarcinoma (YAP1-SS18) (Hu et al. 2018). Additional YAP1 gene fusions have recently been discovered in several human cancers (Kao et al. 2019; Picco et al. 2019; Sekine et al. 2019; Sievers et al. 2019). Since YAP1 gene fusions occur in tumors with relatively low overall mutational burden (Pajtler et al. 2015; Rosenbaum et al. 2019) they might be the oncogenic drivers in these tumors and may pose potential therapeutic targets.
YAP1 is a key transcriptional coactivator that regulates tissue homeostasis, cell fate, and proliferation, that also exerts pro-oncogenic functions, largely attributed to the proproliferative and prosurvival transcriptional program elicited by its interaction with the family of TEAD transcription factors (Zhao et al. 2007; Liu-Chittenden et al. 2012). The activity of YAP1 is negatively regulated by the Hippo pathway, which consists of a series of protein kinases that ultimately phosphorylate several serine residues of the YAP1 protein (most importantly S127 and S397), which in turn leads to both nuclear exclusion and proteasomal degradation of YAP1 (Zhao et al. 2007, 2010).
A multitude of studies have detected high YAP1 abundance being linked to poor survival in several tumor types (Zhang et al. 2018) and inactivating mutations in upstream Hippo pathway members are frequently found in different cancers (Visser and Yang 2010; Deel et al. 2015; Petrilli and Fernández-Valle 2016). In contrast, activating single point mutations within the YAP1 gene are rare events (Wang et al. 2018), suggesting that a single point mutation might not be sufficient to deregulate YAP activity. In turn, gene fusion events that truncate large parts of the YAP1 sequence, but retain the TEAD interaction domain, might generate fusion proteins that are resistant to negative Hippo pathway inhibition and exert deregulated YAP activity.
It remains to be determined which of the recurrent YAP1 fusion genes are sufficient to induce tumor formation and therefore represent potential therapeutic targets. Likewise, little is known about the underlying signaling pathways activated by the different YAP1 fusion proteins, whether there is overlap between the transcriptional programs induced by the different fusions, and whether it is possible to identify a unifying feature that is (1) present in all YAP1 fusion family members required for their ability to drive tumor growth and that can (2) be pharmacologically targeted to inhibit the growth of these tumors.
In this study, we show that the expression of each of the four analyzed YAP1 gene fusions is sufficient to cause tumor formation in mice. We can show that all analyzed YAP1 fusion proteins exert YAP activity. This YAP activity (1) is resistant to negative Hippo pathway signaling, due to constitutive nuclear localization—mediated by a nuclear localization sequence in the C′-terminal fusion half—and resistance to proteasomal degradation, and (2) relies on the interaction with TEAD transcription factors. This TEAD-dependent YAP activity of all four analyzed YAP1 fusions can be pharmacologically inhibited in vitro using small molecule inhibitors (such as verteporfin), which in turn leads to a reduction in the growth of YAP1 fusion-driven cells.
Results
Expression of YAP1-MAMLD1, YAP1-FAM118B, YAP1-TFE3, and YAP1-SS18 is sufficient to cause tumor formation in mice
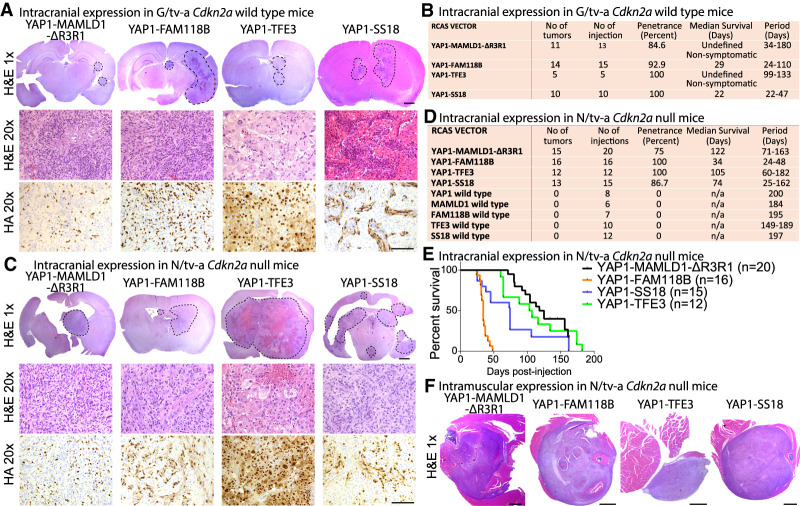
To determine whether the expression of different YAP1 fusions is sufficient to induce tumor formation in vivo, we cloned the coding sequences of YAP1-MAMLD1 (YM), YAP1-FAM118B (YF), YAP1-TFE3 (YT), and YAP1-SS18 (YS), as well as the wild-type versions of all genes into the RCAS retroviral vector (Supplemental Fig. S1A,B). We generated truncated versions of the YM fusion since the RCAS vector has a maximum capacity of ∼2.5 kb and used a version that retained the C′-terminal sequence of MAMLD1 (YMΔR3R1) and displayed an oncogenic potential similar to full-length YM in a second tumor model (Supplemental Fig. S1C–J). We used the RCAS-tv-a model for somatic cell gene transfer, which allowed us to overexpress genes of interest in transgenic mice only in cells that express the viral tv-a receptor under a tissue-specific promoter (Ozawa et al. 2014, 2018). We used mice that expressed tv-a under the control of either the GFAP promoter (G/tv-a) or the Nestin promoter (N/tv-a) (Holland et al. 1998).
We tested whether the expression of the YMΔR3R1, YF (both associated with ST-ependymoma), YT, and YS fusions is sufficient to induce intracranial tumors in G/tv-a wild-type mice, enabling expression in neural stem and progenitor cells (Fig. 1A,B; Supplemental Fig. S1K,L). Expression of YMΔR3R1 (11 out of 13 mice) or YT (five out of five mice) caused the formation of small nonsymptomatic tumors, whereas expression of YF (14 out of 15 mice, median survival 29 d) or YS (10 out of 10 mice, median survival 22 d) resulted in the formation of large symptomatic tumors. All tumors stained positive for the HA tag, indicating the presence of the respective YAP1 fusion proteins.
Figure 1.
YAP1 fusion genes are oncogenic in vivo. (A–D) H&E and HA IHC stainings and summary of tumors caused by intracranial expression of YAP1 fusions in G/tv-a Cdkn2a wild-type (A,B) or N/tv-a Cdkn2a-null (C,D) mice. (E) Symptom-free survival following intracranial expression of YAP1 fusions in N/tv-a Cdkn2a-null mice. (F) H&E images of tumors caused by intramuscular expression of YAP1 fusions in N/tv-a Cdkn2a-null mice. Scale bars: H&E 1×, 1000 µm; H&E 20× and HA 20×, 100 µm.
We observed prominent differences in the histomorphology between YAP1 fusion tumor types (Fig. 1A). Tumor cells generated by expression of either YMΔR3R1, YF, or YS were spindle shaped, grew invasively into the parenchyma, and stained positive for Nestin, but negative for GFAP and Olig2, whereas tumor cells in YT-driven tumors were epithelioid and quite pleomorphic with variation in size and shape and stained strongly positive for GFAP (Supplemental Fig. S1M).
In an attempt to understand general rules of YAP1 fusion-driven oncogenesis, we investigated the impact of additional tumor suppressor losses on the growth of tumors driven by these gene fusions. Dysregulation of cell cycle control is seen in multiple cancers, by functional loss of CDKN2A, p53, or RB1. To mimic this effect, we intracranially expressed each of the different YAP1 fusion genes in N/tv-a and G/tv-a Cdkn2a-null pups. While we did not observe a difference in tumor size or latency of YF- or YS-driven tumors, YMΔR3R1- and YT-driven tumors showed a dramatic increase in size in Cdkn2a-null mice (Fig. 1C–E; Supplemental Fig. S1N,O). In contrast, mice injected with any of the wild-type versions did not develop tumors 200 d after injection (Fig. 1D).
Both YT and YS are found in cancers located outside the central nervous system (CNS). Nestin is widely expressed in peripheral tissues in stem and progenitor cells. We intramuscularly injected N/tv-a Cdkn2a-null mice with YAP1 fusion-expressing RCAS vectors, which enabled us to express the different YAP1 gene fusions in tissues outside the CNS. Similar to the results from intracranial injections, all four YAP1 fusions induced tumor formation. Mice injected with either wild type YAP1, MAMLD1, FAM118B, SS18, or TFE3 did not develop tumors (Fig. 1F; Supplemental Fig. S1P–R). These results suggest that YAP1 gene fusions are the likely oncogenic drivers in the human tumors in which they are.
YAP1 fusion proteins exert YAP1–TEAD activity
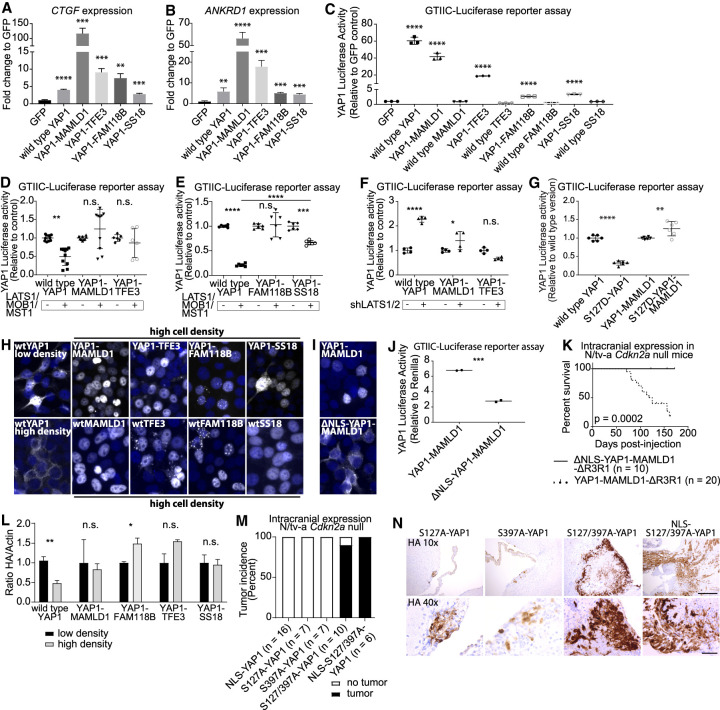
Each of the four YAP1 gene fusions retained the TEAD binding domain near the N′ terminus of wild-type YAP1. To analyze the baseline YAP activity of the different proteins, we transiently transfected HEK293 cells at subconfluency cell densities with either GFP (control), wtYAP1, YM, YF, YT, or YS. We analyzed the expression of known YAP1 target genes (CTGF, CYR61, ANKRD1, AMOT, and AMOTL2) by qRT-PCR and found that each YAP1 fusion significantly enhanced the expression of all five YAP1-responsive genes (Fig. 2A,B; Supplemental Fig. S2A,B).
Figure 2.
YAP1 fusion proteins exert YAP activity that is resistant to negative Hippo pathway signaling. (A–C) YAP1 fusion proteins induce the expression of CTGF (A) and ANKRD1 (B) and activate YAP1-responsive GTIIC-Luc reporter assays (C) (n = 3). (D,E) The effect of additional LATS1/MOB1/MST1 coexpression on the YAP activity of wtYAP1, YM, and YT (n = 8 for wtYAP1 and YM, n = 6 for YT) (D) and wtYAP1, YF, and YS (n = 6) (E). (F) YAP activity of wtYAP1, YM, or YT in control conditions (+GFP) or upon knockdown of LATS1/2 (n = 4). (G) YAP activity of wtYAP1, S127D-YAP1, YM, and S127D-YM (n = 6). (H) HA IF stainings (Hoechst counter stain) of confluent (except wtYAP1 low density) HEK293 cells expressing different YAP1 fusions, or respective wild-type versions. (I) HA IF stainings of HEK293 cells transiently expressing either YM or ΔNLS-YM. (J) YAP activity of YM and ΔNLS-YM (n = 2). (K) Symptom-free survival following intracranial expression of YMΔR3R1 or ΔNLS-YMΔR3R1 in N/tv-a Cdkn2a-null mice. (L) YAP1 fusion proteins are resistant to proteasomal degradation at high cell density conditions. wtYAP1: low density n = 5, high density n = 4. All other conditions: n = 3. (M,N) Tumor incidence of YAP1 point mutants upon intracranial expression in N/tv-a Cdkn2a-null mice (M) and HA IHC stainings of resulting lesions (N). Scale bars: 10×, 250 µm; 40×, 50 µm. Error bars show standard deviation (SD) (A–G) or standard error of mean (SEM) (L). Analysis was done using two-tailed t-test (A–C,J,L) or ordinary one-way ANOVA (D–G). (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001.
We then transfected HEK293 cells grown at subconfluency cell densities with YAP1 reporter plasmids (8xGTIIC-Luc, CTGF-Luc), and either GFP (control), the YAP1 fusions, wild-type YAP1, MAMLD1, FAM118B, TFE3, or SS18. WtYAP1 and all four YAP1 fusions showed activity in both reporter assays, whereas none of the wild-type C′-terminal fusion partners showed activity (Fig. 2C; Supplemental Fig. S2C). Truncation experiments with both YM and wtMAMLD1 showed that the MAMLD1 Mastermind TAD is necessary and sufficient for the YAP activity of YM (Supplemental Fig. S2D–J). These results show that all four YAP1 fusion proteins exert YAP transcriptional activity.
YAP1 fusions are resistant to Hippo pathway-mediated nuclear exclusion and proteasomal degradation
The YAP transcriptional activity of YAP1 gene fusions is resistant to Hippo pathway-mediated inhibition
The activity of wtYAP1 is inhibited by the Hippo pathway at high cell densities in vitro via phosphorylation of the YAP1 protein by LATS1/2 at several serine residues and subsequent nuclear exclusion and proteasomal degradation of the wtYAP1 protein. Since TEAD transcription factors, which mediate YAP activity, become excluded from the nucleus at high cell densities in HEK cells (Supplemental Fig. S2K; Lin et al. 2017), we instead measured the YAP activity of the different YAP1 fusions upon direct modulation of upstream components of the Hippo pathway.
We determined the effect of additional coexpression of the Hippo pathway proteins LATS1, MST1, and MOB1 (compared with GFP control) using GTIIC-Luc reporter assays in transiently transfected HEK293 cells. While the activity of wtYAP1 was significantly reduced, the YAP activity of YM, YT, and YF was not significantly affected (Fig. 2D,E; Supplemental Fig. S2L). The activity of YS was significantly decreased by coexpression of LATS1, MST1, and MOB1; however, it was significantly less affected compared with wtYAP1. In turn, shRNA knockdown of LATS1/2 led to a significant increase of wtYAP1 activity (Padj < 0.0001), whereas there was only a minor increase of the YAP activity of YM and no significant increase in YAP activity of YT (Fig. 2F; Supplemental Fig. S2M). We generated S127D phosphomimetic mutants of both wtYAP1 and YM (the YAP1 sequence of YT is truncated upstream of S127). The YAP activity of S127D-YAP1 was significantly reduced compared with wtYAP1 (Padj < 0.0001), whereas the activity of S127D-YM was significantly increased compared with YM (Padj = 0.0032; Fig. 2G). These results suggest that YAP1 fusion proteins have become constitutively active and resistant to inhibitory Hippo pathway signaling.
YAP1 fusion proteins contain constitutive nuclear localization sequences that are necessary for their oncogenic function
To explore why the YAP activity of YAP1 fusion proteins is resistant to Hippo pathway signaling, we assessed the nuclear localization of the different YAP1 fusion proteins at high cell density conditions. We performed immunofluorescence (IF) stainings of HEK293 expressing HA-tagged versions of wtYAP1, YM, YF, YT, YS, and their respective C′-terminal fusion partners (Fig. 2H). WtYAP1 staining localized mostly to the nucleus at low cell densities but was excluded from the nucleus at high cell densities, whereas YAP1 fusion proteins and their respective wild type C′-terminal fusion partners showed constitutive nuclear staining at high cell densities. Moreover, IHC stainings of our mouse tumors revealed strong nuclear localization of all four YAP1 fusions (Fig. 1A). This suggests the presence of nuclear localization sequences (NLS) in the sequences of the C′-terminal fusion partners, which may contribute to the resistance to nuclear exclusion of the different YAP1 fusions.
NLS mapping predicted two NLS located between amino acid 50 and amino acid 86 of wtMAMLD1. We confirmed these results by generating a series of C′-terminal YM truncations (Supplemental Fig. S2N). Mutated versions of YM lacking these two NLS (ΔNLS-YM and ΔNLS-YMΔR3R1) displayed purely cytoplasmic localization and reduced activity in the GTIIC-Luc reporter assay (Fig. 2I,J). ΔNLS-YMΔR3R1 failed to induce tumor formation in N/tv-a Cdkn2a-null mice (Fig. 2K), suggesting that the nuclear localization of YM is necessary for its oncogenic potential.
YAP1 fusion proteins are resistant to Hippo pathway-mediated proteasomal degradation
Since the YAP1 sequence of all four YAP1 fusion proteins is truncated upstream of the S397 residue, we verified that the different YAP1 fusion proteins are functionally resistant to Hippo pathway-mediated proteasomal degradation. We transduced NIH3T3 cells to stably express wtYAP1 or either of the four YAP1 fusions (all constructs contained an N′-terminal HA tag). We isolated total protein from cells at low and high confluency conditions and quantified the abundance of HA-tagged protein normalized to Actin via Western blot. While we observed a significant reduction in wtYAP1 protein at high cell density states compared with low cell density states (P = 0.0029), we did not observe a significant reduction of YM, YT, or YS protein and observed a significant increase of YF protein (P = 0.027) at high density conditions (Fig. 2L; Supplemental Fig. S2O). These results suggest that all four YAP1 fusion proteins have become resistant to Hippo pathway-mediated proteasomal degradation.
Combined inhibition of nuclear exclusion and proteasomal degradation is sufficient to render wild-type YAP1 oncogenic
Our results suggest that the different YAP1 fusion proteins deregulate YAP activity and have become insensitive to negative Hippo pathway regulation. To test whether resistance to negative Hippo pathway signaling is sufficient to render nonfusion YAP1 oncogenic we generated mutant versions of wtYAP1 resistant to nuclear exclusion (S127A) and/or proteasomal degradation (S397A in YAP1 transcript variant 1), as previously described (Zhao et al. 2007, 2010). In addition, we added a myc NLS to the N′ terminus of wtYAP1 (NLS-YAP1) to mimic the presence of a YAP1 fusion NLS (Supplemental Fig. S2P–R).
We intracranially expressed our collection of YAP1 point mutants in N/tv-a Cdkn2a-null mice (Fig. 2M,N). Expression of NLS-YAP1 was unable to induce detectable tumors 200 d after injection (zero out of 16 mice) suggesting that nuclear localization is insufficient. Similarly, expression of single mutants S127A-YAP1 and S397A-YAP1 did not cause detectable tumors 50 d after injection (both zero out of seven mice), although we detected small HA-positive clusters along the ventricles for both groups. Intracranial expression of S127/397A-YAP1 (2SA-YAP1) and NLS-2SA-YAP1, that are resistant to both nuclear exclusion and proteasomal degradation, lead to the formation of detectable tumors in nine out of 10 mice (0.276 mm2 ± 0.097 SEM) and six out of six mice (1.20 mm2 ± 0.53 SEM), respectively, 50 d after injection. This suggests that the combined resistance to nuclear exclusion and proteasomal degradation is necessary and sufficient to render YAP1 oncogenic (Supplemental Fig. S2S).
Thus, constitutive activation of YAP1 in vivo is sufficient to cause tumor formation. Interestingly, we noticed that the resulting tumors did not histomorphologically resemble any of the YAP1 fusion gene-driven mouse tumors, suggesting that the additional function of the different C′-terminal fusion partners contributes to the tumor histology (Supplemental Fig. S2T).
YAP1 fusion proteins induce a shared core gene expression signature that overlaps with YAP1 signaling
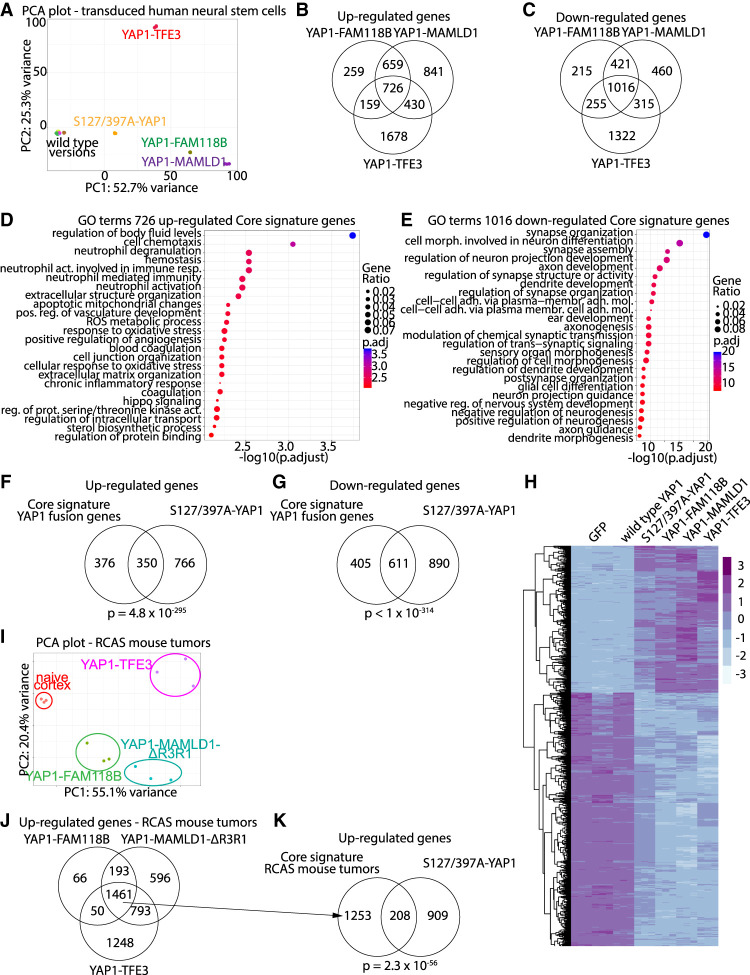
We transduced U5 human neural stem cells to express either GFP (control), wtYAP1, 2SA-YAP1, YM, YF, YT, wtMAMLD1, wtFAM118B, or wtTFE3 and performed RNA sequencing (RNA-seq) analysis (Supplemental Fig. S3A). Principle component analysis (PCA) clearly separated samples expressing oncogenic transcripts (2SA-YAP1, YM, YF, or YT) from nononcogenic wild-type transcripts (Fig. 3A; Supplemental Fig. S3B,C). We calculated differentially expressed genes (DEGs; FC ≥ 1.5 or ≤−1.5, FDR ≤ 0.05) compared with GFP controls and observed dramatic changes in gene expression in samples expressing oncogenic transcripts (Supplemental Fig. S3D; Supplemental Table S1A). We identified a core signature of 726 up-regulated DEGs and 1016 down-regulated DEGs shared by all three YAP1 gene fusions (Fig. 3B–E; Supplemental Table S1B–L) that shared a significant overlap with the up-regulated (350 overlapping genes, P = 4.8 × 10−295) and down-regulated DEGs (611 overlapping DEGs, P < 1 × 10−314) of 2SA-YAP1-expressing cells (Fig. 3F–H; Supplemental Fig. S3E,F; Supplemental Table S1M).
Figure 3.
RNA-seq analysis of human neural stem cells expressing YAP1-MAMLD1, YAP1-FAM118B, or YAP1-TFE3. (A) PCA plot of RNA-seq samples. (B,C) Venn diagrams showing the overlaps of up-regulated (B) and down-regulated (C) genes of YM-, YF-, or YT-expressing cells. (D,E) Biological process GO terms of the up-regulated (D) and down-regulated (E) YAP1 fusion core genes shared by all three YAP1 fusion genes. (F,G) Overlap between up-regulated (F) and down-regulated (G) YAP1 fusion core gene signatures and respective DEGs in 2SA-YAP1-expressing cells. (H) Heat map showing expression levels of core genes significantly regulated in 2SA-YAP1-, YM-, YF-, or YT-expressing samples. (I) PCA plot of RNA-seq samples from naïve mouse cortex tissue and RCAS tumors. (J) Overlap of up-regulated genes between the different RCAS mouse tumor types. (K) Overlap between the up-regulated genes shared by all three RCAS mouse tumor types and genes up-regulated in 2SA-YAP1-expressing cells. (B,C,F,G,J,K) Analysis was done using a hypergeometric test.
We performed RNA-seq on whole tumor tissues derived from intracranial RCAS-YAP1 fusion-driven mouse tumors (YMΔR3R1, YF, or YT) and naïve mouse cortex derived from N/tv-a Cdkn2a-null mice (n = 3 each) (Supplemental Table S2A–I). Similar to the in vitro samples, the RCAS-YAP1 fusion-driven tumors clearly separated from naïve cortex samples by PCA (Fig. 3I; Supplemental Fig. S3G). We detected highly significant overlaps between the DEGs of respective in vivo and in vitro samples (Supplemental Table S2J). Lastly, we found that 1461 genes were jointly up-regulated in all three RCAS mouse tumor types (Fig. 3J; Supplemental Table S2B). These 1461 genes were mostly related to immune system processes (Supplemental Table S2C); however, we detected a significant overlap with the up-regulated DEGs of 2SA-YAP1-expressing cells (208 genes, P = 2.27 × 10−56) (Fig. 3K). This suggests that all three tumor types share a gene signature related to oncogenic YAP1 signaling. We compared our RNA-seq data from the YAP1 core signature, YM-, and S2A-YAP1-expressing cells, as well as RCAS-YMΔR3R1 mouse tumors to published microarray data from human ST-EPN-YAP1 tumors (Pajtler et al. 2015) and detected significant overlaps, indicating that the YAP1 expression signature is conserved in these human tumors (Supplemental Table S2K).
CUT&RUN identifies overlap in occupied genomic regions between oncogenic YAP1 fusions and wild-type YAP1
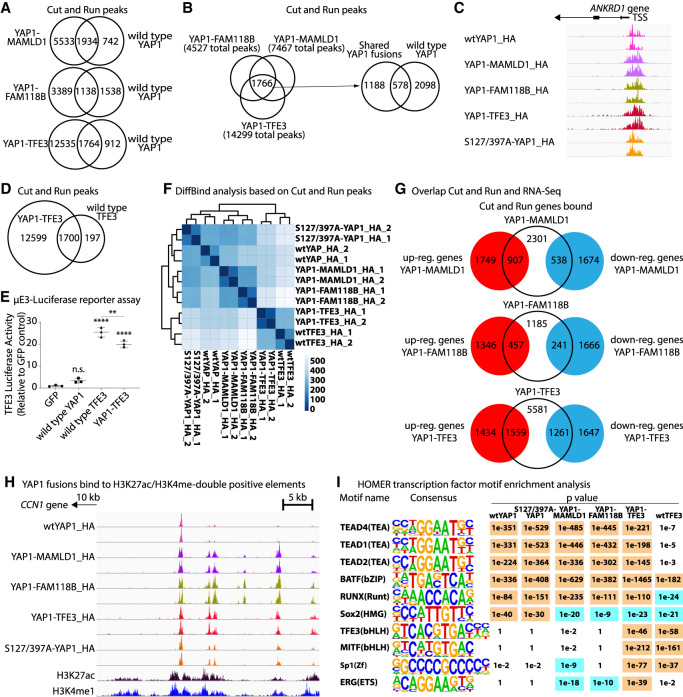
To gain a deeper insight on which genomic regions are occupied by the different YAP1 fusion proteins, we expressed wtYAP1, 2SA-YAP1, YM, YF, and YT (all containing an N′-terminal HA tag) in human neural stem cells and performed CUT&RUN (C&R) experiments using an anti-HA antibody. We detected robust signals for all samples (Supplemental Table S3A–H) and assessed the extent to which the different YAP1 fusion proteins occupied YAP1-specific target regions. Each individual YAP1 fusion protein shared significant overlaps in the occupied genomic regions with both wtYAP1 and 2SA-YAP1 (Fig. 4A; Supplemental Table S3B). We identified a core signature of 1766 peaks (corresponding to 1153 genes) that overlapped between YM, YF, and YT samples and overlapped with wtYAP1 (578 peaks, 21.6% of all wtYAP1 peaks) and 2SA-YAP1 peaks (749 peaks, 21.5% of all 2SA-YAP1 peaks) (Fig. 4B,C). These results suggest that the different YAP1 fusion proteins occupy a common set of YAP1 target regions.
Figure 4.
CUT&RUN identifies overlap in occupied genomic regions between oncogenic YAP1 fusions and wild-type YAP1. (A) Overlap of C&R peaks between wtYAP1_HA and YM_HA, YF_HA, or YT_HA. (B) Overlap of C&R peaks between YM_HA, YF_HA, and YT_HA. (C) WtYAP1, 2SA-YAP1, and YAP1 fusion proteins occupy the ANKRD1 promoter. (D) Overlap of C&R peaks between YT_HA and wtTFE3_HA. (E) µE3-Luc reporter assay showing the TFE3 activity of YT, wtYAP1, and wtTFE3 (n = 3). (F) DiffBind clustering of C&R samples. (G) Overlaps between C&R and RNA-seq. (H) YAP1 fusions occupy H3K27ac/H3K4me1-double positive regulatory elements located in distal intergenic regions. (I) Enriched motifs from HOMER analysis. (E) Error bars show SD. (E) Analysis was done using ordinary one-way ANOVA. (**) P < 0.01; (****) P < 0.0001.
We determined whether the different YAP1 fusion proteins share overlapping functions with their respective C′-terminal fusion partners and performed C&R on human neural stem cells expressing wtTFE3, MAMLD1, or FAM118B (containing N′-terminal HA tags). Direct pull-down of wtTFE3 yielded 1897 peaks, 1700 (89.6%) of which were also present in YT peaks (Fig. 4D). HOMER motif analysis further confirmed a highly significant enrichment of a TFE3-binding motif in YT peaks (P = 1 × 10−46). We confirmed the C′-terminal TFE3 activity of YT using a TFE3-responsive Luciferase reporter assay (Fig. 4E). DiffBind analysis of all samples based on similarities of peak sequences showed that wtTFE3 and YT samples clustered closely together (Fig. 4F). We did not observe consistent peaks upon direct pull-down of either wtMAMLD1 or wtFAM118B (n = 4 for each).
We then analyzed the overlap between C&R peaks and RNA-seq DEGs and found that YM-associated peaks mapped to 907 (34.1%) up-regulated and 538 (24.3%) down-regulated genes in YM-expressing cells, suggesting a direct regulation of these genes. Similarly, YF-associated peaks mapped to 457 (25.3%) up-regulated and 241 (12.6%) down-regulated genes in YF-expressing cells, whereas YT-associated peaks mapped to 1559 (52.1%) up-regulated and 1261 (43.4%) down-regulated genes (Fig. 4G; Supplemental Fig. S4A; Supplemental Table S3I). WtYAP1, 2SA-YAP1, YM, and YF peaks were mostly located in distal intergenic regions (42%–46% of peaks) or introns (38%–43% of peaks), followed by promoter regions (7%–13% of peaks). In turn, both wtTFE3 and YT peaks were predominantly located in promoter regions (47% and 33% of peaks, respectively), suggesting that the TFE3 part of the fusion directs YT to TFE3-specific promoter regions (Supplemental Fig. S4B,C). Since a large fraction of these peaks were located in nonpromoter regions, we compared our C&R data with published H3K27ac/H3K4me1 ChIP-seq data (Galli et al. 2015) and found large overlaps between both data sets, indicating that YAP1 fusions occupy H3K27ac/H3K4me1-double-positive regulatory elements located in distal intergenic and intronic regions (Fig. 4H; Supplemental Fig. S4D). Furthermore, we compared our C&R data for YM with published YAP1 ChIP-seq data from three human ST-EPN-YAP1 tumors (Pajtler et al. 2019) and observed robust overlaps between both data sets (Supplemental Fig. S4E).
YAP1 fusion target DNA sequences are enriched for TEAD transcription factors motifs
The interaction between YAP1 and different TEADs is facilitated through the YAP1 TEAD-interacting domain (TID) (Zhao et al. 2008; Galli et al. 2015; Stein et al. 2015) that is retained in all four YAP1 fusions. We performed HOMER motif analysis to identify transcription factor-binding motifs in the peak sequences of wtYAP1, 2SA-YAP1, and YAP1 fusion samples and detected a highly significant enrichment for TEAD1, TEAD2, and TEAD4 motifs in all five samples. Motifs for BATF, ATF3, FRA1/2, BACH2, AP-1, FOSL2, and JUNB (all sharing a similar core motif), as well as RUNX1/2 and different members of the SOX family, were also highly enriched in all five samples. In addition, we detected significant enrichments of MITF, BMAL1, USF1/2, CLOCK, and SP1 motifs in YT and wtTFE3 peaks, as well as ETS family proteins ERG-, SPDEF-, and ETV2-binding motifs in all three YAP1 fusion samples, but not in wtYAP1 or 2SA-YAP1 samples (Fig. 4I; Supplemental Table S3J).
We performed additional C&R experiments using an antibody against TEAD4 (Supplemental Table S3K–O). We observed robust overlaps between HA and TEAD4 peaks, suggesting that, similar to wtYAP1, YAP1 fusion proteins interact with TEAD transcription factors to bind DNA (Supplemental Fig. S4F–I; Supplemental Table S3B). DiffBind analysis of all samples based on peak sequences showed that the respective HA and TEAD4 samples of each sample group clustered closely together (Supplemental Fig. S4H). These results suggest that the different YAP1 fusion proteins interact with TEAD transcription factors and occupy TEAD target regions.
The interaction with TEAD transcription factors is necessary for the oncogenic function of YAP1 fusion proteins
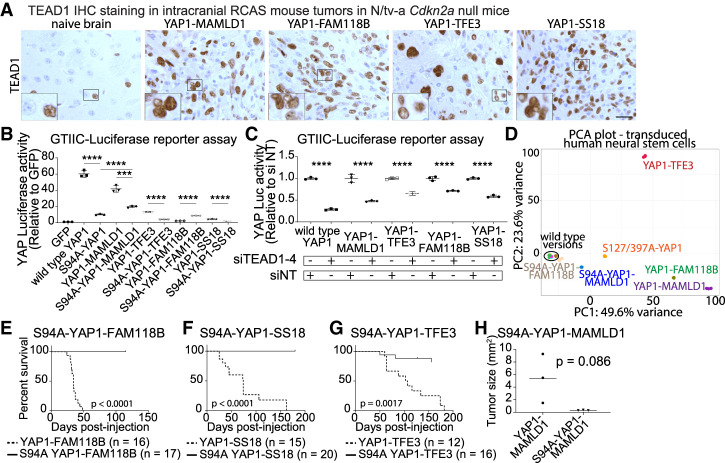
RNA-seq data confirmed the expression of all four TEADs in YAP1 fusion-driven mouse tumors (Supplemental Fig. S5A,B) and IHC stainings confirmed nuclear TEAD1 staining in each tumor type (Fig. 5A). Serine 94 of YAP1 is a key residue for the YAP1–TEAD interaction and S94A mutation of YAP1 strongly reduces YAP activity (Zhao et al. 2008; Mo et al. 2015). We generated S94A mutant versions of all YAP1 fusions and performed GTIIC-Luc reporter assays (Fig. 5B). We found that all S94A mutants (except S94A-YF) showed a significant reduction of YAP activity. We detected a significant increase in the YAP activity of S94A-YF compared with YF. Similarly, combined siRNA-mediated knockdown of TEAD1-4 resulted in a significant reduction of YAP activity, while separate knockdown of individual TEADs did not (Fig. 5C; Supplemental Fig. S5C–F).
Figure 5.
Genetic disruption of YAP1–TEAD binding ablates tumor forming ability of YAP1 fusions. (A) TEAD1 IHC stainings of intracranial RCAS tumors in N/tv-a Cdkn2a-null mice. Scale bar, 25 µm. (B) YAP activity of S94A mutant YAP1 fusions (n = 3). (C) Combined knockdown of TEAD1–4 leads to reduced YAP activity of YAP1 fusions (n = 3). (D) PCA plot of S94A-YM and S94A-YF RNA-seq samples. (E–G) Symptom-free survival of N/tv-a Cdkn2a-null mice after intracranial expression of S94A mutant versions of YF (E), YS (F), or YT (G). (H) Tumor sizes after intracranial injection of murine neural stem cells expressing either unmutated or S94A mutant YM (n = 3 each). Error bars show SD. Analysis was done using two-tailed t-test (B,H), ordinary one-way ANOVA (C), and log rank (Mantel-Cox) test (E–G). (***) P < 0.001; (****) P < 0.0001.
We performed RNA-seq on human neural stem cells expressing either S94A-YM or S94A-YF (n = 3 each) to analyze the transcriptional effects of ablation of YAP1–TEAD interaction. Both S94A-YM (1256 DEGs) and S94A-YF (343 DEGs) were unable to cause the dramatic gene expression changes induced by their respective nonmutated counterparts (Fig. 5D). We detected significant overlaps between the gene expression profiles of S94A-YM and YM (871 DEGs, P = 3.74 × 10−312) (Supplemental Fig. S5G). These results suggest that the function of YM and YF greatly depends on the interaction with TEAD transcription factors.
We intracranially expressed S94A-YT, S94A-YF, or S94A-YS mutant versions (Supplemental Fig. S5H) in N/tv-a Cdkn2a-null mice and observed a complete lack of oncogenic functions for S94A-YF or S94A-YS (Fig. 5E,F) and a significantly reduced oncogenic potential for S94A-YT (three out of 17 mice, P = 0.0017) (Fig. 5G; Supplemental Fig. S5I). To assess the oncogenic potential of full-length S94A-YM, we facilitated a second model system. We intracranially implanted in vitro-transduced murine Cdkn2a-null neural stem cells expressing either full-length YM or S94A-YM (n = 3) (Supplemental Fig. S1E). We observed the formation of small nonsymptomatic tumors in mice injected with S94A-YM-expressing cells 14 d after injection. The tumor size was reduced compared with mice injected with cells expressing YM (5.42 mm2 ± 2.25 SEM vs. 0.32 mm2 ± 0.02 SEM, P = 0.086) (Fig. 5H; Supplemental Fig. S5J–L). These findings suggest that the interaction with TEAD transcription factors is essential for the oncogenic functions of all four YAP1 fusion proteins.
The oncogenic functions of YAP1 fusion proteins can be pharmacologically inhibited in vitro
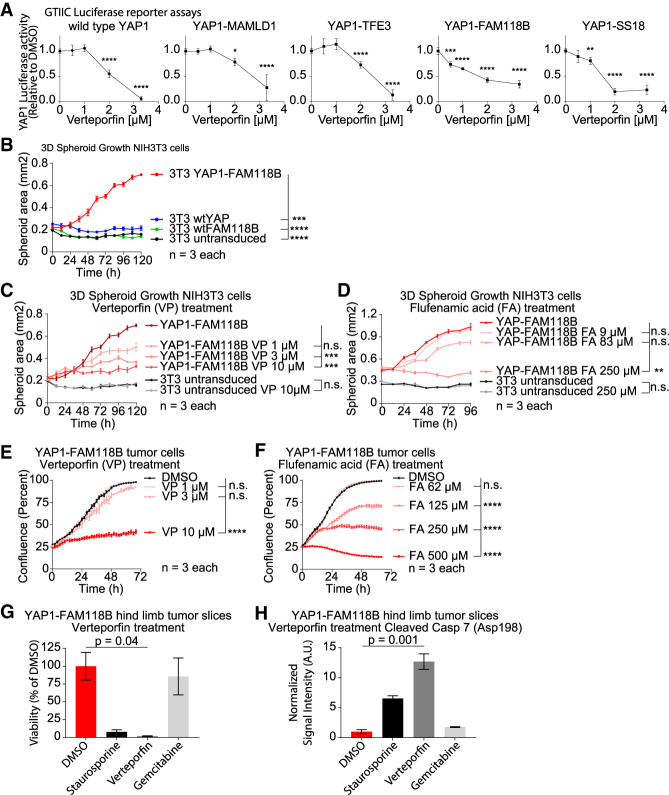
Verteporfin (VP) is a small molecule inhibitor that has been shown to inhibit YAP1 functions by blocking the interaction between YAP1 and TEAD (Liu-Chittenden et al. 2012; Brodowska et al. 2014). To determine whether VP can inhibit the YAP activity of the different YAP1 fusion proteins, we performed GTIIC-Luc reporter assays with transiently transfected HEK293 cells at different VP concentrations and observed a dose-dependent reduction of the YAP activity of wtYAP1 and all of the YAP1 fusions (Fig. 6A).
Figure 6.
The oncogenic functions of YAP1 fusion proteins can be pharmacologically inhibited in vitro. (A) GTIIC-Luc reporter assays showing the YAP activity of wtYAP1, YM, YT, YF, and YS at different concentrations of verteporfin in comparison with the DMSO control (0 µM verteporfin) (n = 4). (B) Spheroid growth of stably transduced NIH3T3 cells (untransduced control, wtYAP1, wtFAM118B, or YF) (n = 3). (C,D) Spheroid growth of NIH3T3 cells (untransduced control or expressing YF) when treated with VP (C) or FA (D) at the indicated concentrations or DMSO only (n = 3). (E,F) Growth of YF tumor cells when treated with either VP (E) or FA (F) at indicated concentrations or DMSO only (n = 3). (G,H) Viability (G) and cleaved caspase 7 (Asp198) levels (H) of YF hindlimb organotypic tumor slices after treatment with DMSO (control), staurosporine, verteporfin, or gemcitabine (n = 2). Error bars show SD (A) or SEM (B–H). Analysis was done using ordinary two-way ANOVA (A–F) or ordinary one-way ANOVA (G,H). P-values for B–F are for the final time point. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001.
We generated stably transduced NIH3T3 cells (untransduced control, wtYAP1, wtFAM188B, or YAP1-FAM118B) (Supplemental Fig. S6A–D). In 3D culturing conditions neither untransduced control cells, wtYAP1-expressin, nor wtFAM188B-expressing cells were able to grow into spheroids, likely due to high contact inhibition (Holley and Kiernan 1968), whereas YAP1-FAM118B-expressing cells were able to grow as spheroids (Fig. 6B). Using two YAP1–TEAD inhibitors (VP and flufenamic acid [FA]) we found that the spheroid growth of YAP1-FAM118-expressing NIH3T3 cells could be inhibited by VP and FA treatment in a dose-dependent manner, whereas control cells were not affected (Fig. 6C,D; Supplemental Fig. S6E,F).
We established a cell line from an RCAS-YAP1-FAM118B mouse hindlimb tumor. The presence of YAP1-FAM118B in the cell line was confirmed by IF stainings for the N′-terminal HA tag (Supplemental Fig. S6G). Treatment with VP and FA was able to inhibit the growth of this YAP1-FAM118B tumor cell line and was accompanied by a dose-dependent decrease in the expression of the YAP1 target genes Ctgf and Cyr61 (Fig. 6E,F; Supplemental Fig. S6H,I). We then generated organotypic slice cultures from an RCAS-YAP1-FAM118B hindlimb mouse tumor and treated the slice cultures with either DMSO (control), staurosporine, VP, or gemcitabine for 6 d. We found that VP treatment leads to a significant decrease in viability (P = 0.04) and increases in apoptosis (P = 0.001) in the tumor slices compared with DMSO control (Fig. 6G,H; Supplemental Fig. S6J).
Additional YAP1 fusions share characteristics with established YAP1 fusions
Several additional YAP1 fusion genes have been discovered recently in different human cancers, such as NF2 wild-type meningioma (YAP1-MAML2, YAP1-LMO1, and YAP1-PYGO1), poroma/porocarcinoma (YAP1-MAML2 and YAP1-NUTM1), and sclerosing epithelioid fibrosarcoma (YAP1-KMT2A) (Kao et al. 2019; Picco et al. 2019; Sekine et al. 2019; Sievers et al. 2019). We found that all of these additional YAP1 fusions shared key features with the YAP1 fusions investigated in this study, (1) retention of the YAP1 TID domain, including S94; (2) loss of the S397 residue important for proteasomal degradation; (3) the presence of an NLS; and (4) the presence of putative TADs in the C′-terminal fusion partner sequences that are retained in the resulting YAP1 fusion proteins (Supplemental Fig. S6K; Boehm et al. 1990; Lin et al. 2002; Yokoyama et al. 2002; Städeli and Basler 2005; Reynoird et al. 2010). These results indicate that the newly identified YAP1 fusion proteins share similar features to the YAP1 fusion proteins analyzed in this study.
Discussion
A series of YAP1 gene fusions have been identified in several human tumor types. These YAP1 fusion gene-positive tumors have an overall low mutational burden, suggesting that the YAP1 fusion proteins might be the oncogenic drivers in these cancers (Pajtler et al. 2015; Rosenbaum et al. 2019). Here, we show that all four investigated YAP1 fusion genes are oncogenic when expressed in mice, indicating their causal role in tumor initiation. Our results indicate that additional loss of Cdkn2a enhanced the growth of some YAP1 fusion-driven RCAS mouse tumors (YM and YT). Genomic Cdkn2a loss has not been reported for ST-EPN-YAP1 ependymoma (harboring YM) or EHE (harboring YT) (Pajtler et al. 2015; Rosenbaum et al. 2019); however, it is unclear whether functional inactivation of Cdkn2a or associated pathways (such as p53) occurs in other ways in these cancers, for example, by epigenetic silencing or inactivation of downstream mediators. However, we are not proposing that these mutations actually occur in the human cancers in which these YAP1 fusions are found.
Our results demonstrate that all four analyzed YAP1 fusion proteins exert canonical YAP transcriptional activity and occupy YAP1 target regions. Although the baseline YAP activity (as measured by GTIIC-Luc reporter assays) of some YAP1 fusions was below that of wtYAP1, we found that, in contrast to wtYAP1, the YAP activity of the YAP1 fusion proteins is resistant to Hippo pathway-mediated inhibition. The different YAP1 fusion proteins achieve this in part by constant nuclear localization and resistance to proteasomal degradation, mediated by a nuclear localization sequence located in the sequences of the C′-terminal fusion partner and by loss of the S397 residue of wtYAP1, respectively. These data suggest that the oncogenic activity of the different YAP1 fusions is in part achieved by generating an unregulatable constitutively active YAP1. This is also supported by our finding that a double point mutant constitutively active full-length YAP1 (2SA-YAP1), which is resistant to both nuclear exclusion and proteasomal degradation, was oncogenic, indicating that constitutive activation of YAP1 signaling is sufficient to induce oncogenic transformation in our in vivo model system. These results are supported by a recent study that showed that germline expression of 5SA-YAP1 (S61A, S109A, S127A, S164A, and S397A) as well as combined deletion of Lats1/2 in Neuro6-positive cells leads to the formation of Ependymoma-like tumors in mice shortly after birth (Eder et al. 2020).
Our data suggest that, in addition to the YAP activity present in all four analyzed YAP1 fusion proteins, at least some of the YAP1 fusions also retain activity of the C′-terminal fusion partners. Both MAMLD1 and TFE3 are known transcriptional coactivators and transcription factors, respectively (Huan et al. 2005; Fukami et al. 2008) and we could show that both YT and YM activate TFE3- and MAMLD1-responsive reporter assays, respectively. In addition, using CUT&RUN we can show that YT also occupies TFE3 target regions. Furthermore, a truncated YM variant that showed reduced MAMLD1 activity showed diminished oncogenic activity. The oncogenic activity is therefore likely a combination of YAP1 and non-YAP1 effects. This is further exemplified by the noticeable differences in the histomorphology between the tumors generated by different YAP1 fusions and 2SA-YAP1, suggesting that the C′ terminal partners contribute YAP1-independent activity that may affect the tumor histomorphology and behavior.
Because we are particularly interested in identifying a potential common therapeutic strategy for tumors harboring these fusions, we are highlighting the YAP activity that is common between all analyzed YAP1 gene fusions and is essential for their oncogenic functions. To identify a possible approach for inhibiting this YAP activity, we show that, similar to wtYAP1 (Zhao et al. 2008; Galli et al. 2015; Stein et al. 2015), the YAP activity of all analyzed YAP1 fusion products is dependent on the interaction with TEAD transcription factors and that genetic ablation of this YAP1–TEAD interaction inhibits the ability of all four analyzed different YAP1 fusions to cause tumor formation in vivo. Our results are supported by Pajtler et al. (2019), who showed that genetic ablation of TEAD binding inhibits the oncogenic functions of YAP1-MAMLD1 in a different mouse model. Toward this goal, we demonstrated that pharmacological inhibition of the YAP1–TEAD interaction using small molecule inhibitors is sufficient to reduce the YAP activity of the four YAP1 fusion proteins and reduce the growth of YAP1-FAM118B-driven cells in vitro without affecting the growth of control cells. This data with verteporfin and flufenamic acid as examples of that class of molecules suggests that pharmacological inhibitors targeting YAP1–TEAD binding might be a feasible approach to inhibit the growth of these cancers. Future studies will be necessary to determine whether such an approach is sufficient to induce the regression of established YAP1 fusion-driven tumors in vivo.
A growing number of YAP1 fusion genes has been discovered in different human cancers. We hypothesize that most YAP1 fusion proteins are oncogenic (at least in part) through stabilizing TEAD-dependent oncogenic YAP activity and might be susceptible to YAP1–TEAD disruption in a similar way as the four YAP1 fusions we investigated. In addition, gene fusions involving the YAP1 homolog TAZ have been reported in several cancers and might be susceptible to disruption of TEAD binding in a way similar to YAP1 fusion proteins (Tanas et al. 2016; Sekine et al. 2019).
In summary, our results show that YAP1 gene fusions are oncogenic and the likely tumor-driving events in YAP1 fusion-positive cancers. The general rules for oncogenesis of these YAP1 fusion proteins are (1) the presence of the YAP1–TEAD-binding domain; (2) loss of the Hippo target sequence for degradation; (3) the presence of a nuclear localization sequence that is not regulated by Hippo signaling, often brought in by the C′ partner gene; and (4) the presence of a TAD that is either from YAP1 or brought in by the C′ partner gene. Finally, disruption of YAP1–TEAD interaction could be a general therapeutic approach to block the oncogenic activities of these YAP1 fusion proteins.
Materials and methods
Generation of RCAS mouse tumors
All animal experiments were done in accordance with protocols approved by the Institutional Animal Care and Use Committees of Fred Hutchinson Cancer Research Center (protocol no. 50842) and followed National Institutes of Health guidelines for animal welfare. The RCAS/tv-a system used in this work has been described previously (Holland et al. 2000; Ozawa et al. 2018). GFAP (G)/tv-a Cdkn2a wild type, G/tv-a;Cdkn2a-null, or Nestin (N)/tv-a;Cdkn2a-null mice were used for RCAS-mediated brain tumor formation in this study and have been described previously (Ozawa et al. 2018). N/tv-a;Cdkn2a-null mice were used for RCAS-mediated intramuscular tumor formation in hindlimbs. DF1 cells (1 × 105) in a volume of 1 μL were injected into newborn pup brains or hindlimbs (within 3 d after birth). The mice were monitored until they developed symptoms of disease, such as visible tumors, lethargy, poor grooming, weight loss, dehydration, macrocephaly, seizures, jumping, or paralysis, or until 200 d after injection.
Mouse survival data
Kaplan-Meier analysis demonstrating symptom-free survival of murine brain tumors was performed using log rank test in the Prism 8 software (GraphPad) for all mice injected with the relevant RCAS virus. Log rank P-values were measured with the Mantel-Cox test. A value of P < 0.05 was considered significant in this study.
Plasmid generation
Primers used for plasmid generation are listed in Supplemental Table S4A. Additional plasmids used in this study are listed in Supplemental Table S4D.
Tissue slice preparation and drug treatments
Tumor slices were prepared as described previously (Sivakumar et al. 2019; Nishida-Aoki et al. 2020). Briefly, dissected tumor tissues were molded into a 6-mm core using a biopsy punch. The cores were cut into 250-µm slices using Leica Vibratome VT1200. Slices were immediately placed on inserts in 24-well plates and incubated with Williams’ medium containing 12 mM nicotinamide, 150 nM ascorbic acid, 2.25 mg/mL sodium bicarbonate, 20 mM HEPES, 50 mg/mL additional glucose, 1 mM sodium pyruvate, 2 mM L-glutamine, 1% (v/v) ITS, 20 ng/mL EGF, 40 IU/mL penicillin, and 40 µg/mL streptomycin. After 24 h, slices were exposed to either DMSO (control), 200 nM staurosporine, 5 µM verteporfin, or 5 µM gemcitabine for 6 d. Overall tumor tissue viability was measured using real-time Glo (Promega) according to the manufacturer's instructions. Viability measurement was taken before (day 0) and after (day 6) drug treatment using a Synergy H4 instrument (Biotek).
Spheroid assay
NIH3T3 cells expressing wtYAP1, wtFAM118B, or YAP1-FAM118B were seeded at 5 × 103 cells per well in a 96-well ultralow adherence plates (Costar) in DMEM with 10% FBS and briefly spun down at 1000 rpm for 10 min. After 1 d, cells were treated with VP or FA at varying concentrations. Growth of spheroids was monitored using live-cell imaging every 2 h for 4 d in the Incucyte Zoom system (Essen).
Kinetic cell growth assay
YAP-FAM118B-expressing mouse tumor cells were plated on 96-well plates (Essen ImageLock, Essen Instruments) at 5 × 103 density. Cells were treated with VP or FA at different doses 24 h after plating and cell confluence was monitored with Incucyte live-cell imaging system and software (Essen Instruments). Confluence was observed every 2 h for 48–144 h or until the control (DMSO only) samples reached 100% confluence.
RNA-seq
For in vitro gene expression profiles, U5 human neural stem cells were cultured and transduced with pLJM1 VSV-G pseudotyped lentiviral particles as described above. For in vivo gene expression data from RCAS tumors, brain tumor tissues and half normal forebrains were dissected, flash-frozen in liquid nitrogen, and subsequently crushed on dry ice. RNA was extracted using the Qiagen RNeasy minikit according to the manufacturer's instructions. Genomic DNA was removed by on-column DNase digestion. To confirm overexpression in U5 cells, we generated cDNA using the SuperScript III first strand synthesis system (Invitrogen) and performed quantitative RT-PCR on a Quantstudio 7 Flex (Applied Biosystems) using PowerUp SYBR Green master mix (Applied Biosystems). For a list of primers, see Supplemental Table S4A. Total RNA was then labeled and amplified with the Illumina protocol, followed by sequencing on an Illumina HiSeq2500 and used a paired-end, 50-base read length (PE50) sequencing strategy at the Genomics Shared Resource at Fred Hutchinson Cancer Research Center.
CUT&RUN
U5 human neural stem cells were cultured and transduced with pLJM1 VSV-G pseudotyped lentiviral particles as described (Supplemental Material). Cells were processed as described elsewhere (Janssens et al. 2018). For a list of antibodies used, see Supplemental Table S4B. Sequencing was performed using an Illumina HiSeq 2500 in Rapid Run mode and used a paired-end, 25-base read length (PE50) sequencing strategy at the Genomics Shared Resource at Fred Hutchinson Cancer Research Center.
Data availability
RNA-seq and CUT&RUN sequencing data can be accessed at NCBI Gene Expression Omnibus GSE137040. See also the Supplemental Material.
Data and material availability
The data that support the findings of this study are included here and in the Supplemental Material and are also available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank Deby Kumasaka, James Yan, Casey Love, Jenny Zhang, and Denis Adair for continued technical and administrative assistance and support throughout these experiments. We thank Alyssa Dawson, Elizabeth Jensen, and Philip Corrin at the Fred Hutchinson Genomics Core for help with DNA sequencing, RNA/DNA-seq, and Auto CUT&RUN. We thank Christa Flueck, Ryoichiro Kageyama, and Christopher Roman for providing plasmids. Funding for this research was provided by the following: National Institutes of Health R01 CA195718 (to E.C.H.), U54 CA193461 (to E.C.H. and F.S.), R01 CA100688 (to E.C.H.), U54 CA243125 (to E.C.H.), Fred Hutch Pilot Project funding (to E.C.H. and F.S.), R01 CA190957 (to P.J.P.), American Cancer Society ACS-RSG-14-056-01 (to P.J.P.), and T32 CA080416 (to P.H.). This work was funded by the Discovery Award from the American Lung Association (to T.S.G.).
Author contributions: F.S., P.J.C., T.O., T.S.G., V.V., and E.C.H. conceived the study. F.S., P.H., C.K., M.C., E.N., and T.O. performed the experiments. F.S., S.A., M.C., T.S.G., and P.J.C. analyzed the data. F.S., P.J.C., T.S.G., and E.C.H. wrote the manuscript. F.S., P.J.C., T.S.G., K.W.P., D.K., R.J.G., V.V., P.J.P., and E.C.H. reviewed and edited the manuscript. F.S., P.J.P., and E.C.H. acquired the funding. T.S.G., R.J.G., P.J.P., V.V., and E.C.H. supervised the study. All authors read, reviewed, and approved the manuscript.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.338681.120.
References
- Antonescu CR, Le Loarer F, Mosquera JM, Sboner A, Zhang L, Chen CL, Chen HW, Pathan N, Krausz T, Dickson BC, et al. 2013. Novel YAP1-TFE3 fusion defines a distinct subset of epithelioid hemangioendothelioma. Genes Chromosomes Cancer 52: 775–784. 10.1002/gcc.22073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm T, Foroni L, Kennedy M, Rabbitts TH. 1990. The rhombotin gene belongs to a class of transcriptional regulators with a potential novel protein dimerisation motif. Oncogene 5: 1103–1105. [PubMed] [Google Scholar]
- Brodowska K, Al-Moujahed A, Marmalidou A, Meyer Zu Horste M, Cichy J, Miller JW, Gragoudas E, Vavvas DG. 2014. The clinically used photosensitizer verteporfin (VP) inhibits YAP–TEAD and human retinoblastoma cell growth in vitro without light activation. Exp Eye Res 124: 67–73. 10.1016/j.exer.2014.04.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash AB, Williams IR, Kutok JL, Tomasson MH, Anastasiadou E, Lindahl K, Li S, Van Etten RA, Borrow J, Housman D, et al. 2002. A murine model of CML blast crisis induced by cooperation between BCR/ABL and NUP98/HOXA9. Proc Natl Acad Sci 99: 7622–7627. 10.1073/pnas.102583199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deel MD, Li JJ, Crose LE, Linardic CM. 2015. A review: molecular aberrations within Hippo signaling in bone and soft-tissue sarcomas. Front Oncol 5: 190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder N, Roncaroli F, Dolmart MC, Horswell S, Andreiuolo F, Flynn HR, Lopes AT, Claxton S, Kilday JP, Collinson L, et al. 2020. YAP1/TAZ drives ependymoma-like tumour formation in mice. Nat Commun 11: 2380 10.1038/s41467-020-16167-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukami M, Wada Y, Okada M, Kato F, Katsumata N, Baba T, Morohashi K, Laporte J, Kitagawa M, Ogata T. 2008. Mastermind-like domain-containing 1 (MAMLD1 or CXorf6) transactivates the Hes3 promoter, augments testosterone production, and contains the SF1 target sequence. J Biol Chem 283: 5525–5532. 10.1074/jbc.M703289200 [DOI] [PubMed] [Google Scholar]
- Galli GG, Carrara M, Yuan WC, Valdes-Quezada C, Gurung B, Pepe-Mooney B, Zhang T, Geeven G, Gray NS, de Laat W, et al. 2015. YAP drives growth by controlling transcriptional pause release from dynamic enhancers. Mol Cell 60: 328–337. 10.1016/j.molcel.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland EC, Hively WP, DePinho RA, Varmus HE. 1998. A constitutively active epidermal growth factor receptor cooperates with disruption of G1 cell-cycle arrest pathways to induce glioma-like lesions in mice. Genes Dev 12: 3675–3685. 10.1101/gad.12.23.3675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland EC, Celestino J, Dai C, Schaefer L, Sawaya RE, Fuller GN. 2000. Combined activation of Ras and Akt in neural progenitors induces glioblastoma formation in mice. Nat Genet 25: 55–57. 10.1038/75596 [DOI] [PubMed] [Google Scholar]
- Holley RW, Kiernan JA. 1968. ‘Contact inhibition’ of cell division in 3T3 cells. Proc Natl Acad Sci 60: 300–304. 10.1073/pnas.60.1.300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X, Wang Q, Tang M, Barthel F, Amin S, Yoshihara K, Lang FM, Martinez-Ledesma E, Lee SH, Zheng S, et al. 2018. TumorFusions: an integrative resource for cancer-associated transcript fusions. Nucleic Acids Res 46: D1144–D1149. 10.1093/nar/gkx1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huan C, Sashital D, Hailemariam T, Kelly ML, Roman CA. 2005. Renal carcinoma-associated transcription factors TFE3 and TFEB are leukemia inhibitory factor-responsive transcription activators of E-cadherin. J Biol Chem 280: 30225–30235. 10.1074/jbc.M502380200 [DOI] [PubMed] [Google Scholar]
- Janssens DH, Wu SJ, Sarthy JF, Meers MP, Myers CH, Olson JM, Ahmad K, Henikoff S. 2018. Automated in situ chromatin profiling efficiently resolves cell types and gene regulatory programs. Epigenetics Chromatin 11: 74 10.1186/s13072-018-0243-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao YC, Lee JC, Zhang L, Sung YS, Swanson D, Hsieh TH, Liu YR, Agaram NP, Huang HY, Dickson BC, et al. 2019. Recurrent YAP1 and KMT2A gene rearrangements in a subset of MUC4-negative sclerosing epithelioid fibrosarcoma. Am J Surg Pathol 44: 368–377. 10.1097/PAS.0000000000001382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Lee Y, Cho HJ, Lee YE, An J, Cho GH, Ko YH, Joo KM, Nam DH. 2014. NTRK1 fusion in glioblastoma multiforme. PLoS One 9: e91940 10.1371/journal.pone.0091940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SE, Oyama T, Nagase T, Harigaya K, Kitagawa M. 2002. Identification of new human mastermind proteins defines a family that consists of positive regulators for notch signaling. J Biol Chem 277: 50612–50620. 10.1074/jbc.M209529200 [DOI] [PubMed] [Google Scholar]
- Lin KC, Moroishi T, Meng Z, Jeong HS, Plouffe SW, Sekido Y, Han J, Park HW, Guan KL. 2017. Regulation of Hippo pathway transcription factor TEAD by p38 MAPK-induced cytoplasmic translocation. Nat Cell Biol 19: 996–1002. 10.1038/ncb3581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu-Chittenden Y, Huang B, Shim JS, Chen Q, Lee SJ, Anders RA, Liu JO, Pan D. 2012. Genetic and pharmacological disruption of the TEAD-YAP complex suppresses the oncogenic activity of YAP. Genes Dev 26: 1300–1305. 10.1101/gad.192856.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo JS, Meng Z, Kim YC, Park HW, Hansen CG, Kim S, Lim DS, Guan KL. 2015. Cellular energy stress induces AMPK-mediated regulation of YAP and the Hippo pathway. Nat Cell Biol 17: 500–510. 10.1038/ncb3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida-Aoki N, Bondesson AJ, Gujral TS. 2020. Measuring real-time drug response in organotypic tumor tissue slices. J Vis Exp 159: e61036 10.3791/61036 [DOI] [PubMed] [Google Scholar]
- Oldrini B, Curiel-García A, Marques C, Matia V, Uluçkan O, Graña-Castro O, Torres-Ruiz R, Rodriguez-Perales S, Huse JT, Squatrito M. 2018. Somatic genome editing with the RCAS–TVA–CRISPR–Cas9 system for precision tumor modeling. Nat Commun 9: 1466 10.1038/s41467-018-03731-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T, Riester M, Cheng YK, Huse JT, Squatrito M, Helmy K, Charles N, Michor F, Holland EC. 2014. Most human non-GCIMP glioblastoma subtypes evolve from a common proneural-like precursor glioma. Cancer Cell 26: 288–300. 10.1016/j.ccr.2014.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozawa T, Arora S, Szulzewsky F, Juric-Sekhar G, Miyajima Y, Bolouri H, Yasui Y, Barber J, Kupp R, Dalton J, et al. 2018. A de novo mouse model of C11orf95-RELA fusion-driven ependymoma identifies driver functions in addition to NF-κB. Cell Rep 23: 3787–3797. 10.1016/j.celrep.2018.04.099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajtler KW, Witt H, Sill M, Jones DT, Hovestadt V, Kratochwil F, Wani K, Tatevossian R, Punchihewa C, Johann P, et al. 2015. Molecular classification of ependymal tumors across All CNS compartments, histopathological grades, and age groups. Cancer Cell 27: 728–743. 10.1016/j.ccell.2015.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajtler KW, Wei Y, Okonechnikov K, Silva PBG, Vouri M, Zhang L, Brabetz S, Sieber L, Gulley M, Mauermann M, et al. 2019. YAP1 subgroup supratentorial ependymoma requires TEAD and nuclear factor I-mediated transcriptional programmes for tumorigenesis. Nat Commun 10: 3914 10.1038/s41467-019-11884-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker BC, Zhang W. 2013. Fusion genes in solid tumors: an emerging target for cancer diagnosis and treatment. Chin J Cancer 32: 594–603. 10.5732/cjc.013.10178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrilli AM, Fernández-Valle C. 2016. Role of Merlin/NF2 inactivation in tumor biology. Oncogene 35: 537–548. 10.1038/onc.2015.125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picco G, Chen ED, Alonso LG, Behan FM, Gonçalves E, Bignell G, Matchan A, Fu B, Banerjee R, Anderson E, et al. 2019. Functional linkage of gene fusions to cancer cell fitness assessed by pharmacological and CRISPR-Cas9 screening. Nat Commun 10: 2198 10.1038/s41467-019-09940-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynoird N, Schwartz BE, Delvecchio M, Sadoul K, Meyers D, Mukherjee C, Caron C, Kimura H, Rousseaux S, Cole PA, et al. 2010. Oncogenesis by sequestration of CBP/p300 in transcriptionally inactive hyperacetylated chromatin domains. EMBO J 29: 2943–2952. 10.1038/emboj.2010.176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum E, Jadeja B, Xu B, Zhang L, Agaram NP, Travis W, Singer S, Tap WD, Antonescu CR. 2019. Prognostic stratification of clinical and molecular epithelioid hemangioendothelioma subsets. Mod Pathol 33: 1–12. 10.1038/s41379-019-0368-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley JD. 1973. Letter: a new consistent chromosomal abnormality in chronic myelogenous leukaemia identified by quinacrine fluorescence and Giemsa staining. Nature 243: 290–293. 10.1038/243290a0 [DOI] [PubMed] [Google Scholar]
- Sekine S, Kiyono T, Ryo E, Ogawa R, Wakai S, Ichikawa H, Suzuki K, Arai S, Tsuta K, Ishida M, et al. 2019. Recurrent YAP1-MAML2 and YAP1-NUTM1 fusions in poroma and porocarcinoma. J Clin Invest 129: 3827–3832. 10.1172/JCI126185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers P, Chiang J, Schrimpf D, Stichel D, Paramasivam N, Sill M, Gayden T, Casalini B, Reuss DE, Dalton J, et al. 2019. YAP1-fusions in pediatric NF2-wildtype meningioma. Acta Neuropathol 139: 215–218. 10.1007/s00401-019-02095-9 [DOI] [PubMed] [Google Scholar]
- Sivakumar R, Chan M, Shin JS, Nishida-Aoki N, Kenerson HL, Elemento O, Beltran H, Yeung R, Gujral TS. 2019. Organotypic tumor slice cultures provide a versatile platform for immuno-oncology and drug discovery. Oncoimmunology 8: e1670019 10.1080/2162402X.2019.1670019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Städeli R, Basler K. 2005. Dissecting nuclear Wingless signalling: recruitment of the transcriptional co-activator Pygopus by a chain of adaptor proteins. Mech Dev 122: 1171–1182. 10.1016/j.mod.2005.07.004 [DOI] [PubMed] [Google Scholar]
- Stein C, Bardet AF, Roma G, Bergling S, Clay I, Ruchti A, Agarinis C, Schmelzle T, Bouwmeester T, Schübeler D, et al. 2015. YAP1 exerts its transcriptional control via TEAD-mediated activation of enhancers. PLoS Genet 11: e1005465 10.1371/journal.pgen.1005465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanas MR, Ma S, Jadaan FO, Ng CK, Weigelt B, Reis-Filho JS, Rubin BP. 2016. Mechanism of action of a WWTR1(TAZ)-CAMTA1 fusion oncoprotein. Oncogene 35: 929–938. 10.1038/onc.2015.148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visser S, Yang X. 2010. LATS tumor suppressor: a new governor of cellular homeostasis. Cell Cycle 9: 3892–3903. 10.4161/cc.9.19.13386 [DOI] [PubMed] [Google Scholar]
- Wang Y, Xu X, Maglic D, Dill MT, Mojumdar K, Ng PK, Jeong KJ, Tsang YH, Moreno D, Bhavana VH, et al. 2018. Comprehensive molecular characterization of the hippo signaling pathway in cancer. Cell Rep 25: 1304–1317.e5. 10.1016/j.celrep.2018.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Kitabayashi I, Ayton PM, Cleary ML, Ohki M. 2002. Leukemia proto-oncoprotein MLL is proteolytically processed into 2 fragments with opposite transcriptional properties. Blood 100: 3710–3718. 10.1182/blood-2002-04-1015 [DOI] [PubMed] [Google Scholar]
- Zhang X, Abdelrahman A, Vollmar B, Zechner D. 2018. The ambivalent function of YAP in apoptosis and cancer. Int J Mol Sci 19: 3770 10.3390/ijms19123770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Wei X, Li W, Udan RS, Yang Q, Kim J, Xie J, Ikenoue T, Yu J, Li L, et al. 2007. Inactivation of YAP oncoprotein by the Hippo pathway is involved in cell contact inhibition and tissue growth control. Genes Dev 21: 2747–2761. 10.1101/gad.1602907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Ye X, Yu J, Li L, Li W, Li S, Yu J, Lin JD, Wang CY, Chinnaiyan AM, et al. 2008. TEAD mediates YAP-dependent gene induction and growth control. Genes Dev 22: 1962–1971. 10.1101/gad.1664408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B, Li L, Tumaneng K, Wang CY, Guan KL. 2010. A coordinated phosphorylation by Lats and CK1 regulates YAP stability through SCF(β-TRCP). Genes Dev 24: 72–85. 10.1101/gad.1843810 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and CUT&RUN sequencing data can be accessed at NCBI Gene Expression Omnibus GSE137040. See also the Supplemental Material.
Data and material availability
The data that support the findings of this study are included here and in the Supplemental Material and are also available from the corresponding author upon reasonable request.