This Outlook discusses the findings by Reizel et al. describing FOXA as a key, opening regions of chromatin during development, and as a doorstep, maintaining the established euchromatic structure in adult tissues.
Keywords: pioneer factor, transcriptional network, winged helix protein
Abstract
Pioneer factors are transcriptional regulators with the capacity to bind inactive regions of chromatin and induce changes in accessibility that underpin cell fate decisions. The FOXA family of transcription factors is well understood to have pioneer capacity. Indeed, researchers have uncovered numerous examples of FOXA-dependent epigenomic modulation in developmental and disease processes. Despite the presence of FOXA being essential for correct epigenetic patterning, the need for continued FOXA presence postchromatin modulation has been debated. In a recent study in this issue of Genes & Development, Reizel and colleagues (pp. 1039–1050) show that the tissue-specific ablation of FOXA1/2/3 in the adult mouse liver results in the collapse of the epigenetic profile that maintains the hepatic gene expression profile. Thus, FOXA functions as a key, opening regions of chromatin during development, and as a doorstep, maintaining the established euchromatic structure in adult tissue.
The changes in cell identify that occur during development, reprograming, and the progression of disease states are underpinned by modifications to the epigenomic profile of the cell. Pioneer factors are a subset of transcription factors with the capacity to bind nucleosomal DNA and induce changes in the accessibility of chromatin at specific loci. Pioneer factors may therefore be thought of as a key, unlocking regions of chromatin for subsequent binding by transcriptional activators and repressors. Once unlocked, the continued presence of pioneer factors such as Zelda, a Drosophila transcription factor from which much of the initial understanding of pioneer factor function was derived, is no longer required (Liang et al. 2008). Instead, the euchromatic structure is maintained by the binding of nonpioneer “settler” transcription factors and the recruitment of the basic transcriptional machinery.
The FoxA transcription factor family contains paradigm mammalian pioneer factors. Since the capacity of FoxA to bind and modify nucleosomal DNA was first described (Gualdi et al. 1996), much work has focused on how FoxA induces changes in chromatin accessibility during development and disease. Indeed, FoxA factors have recently been shown to bind and induce changes in chromatin accessibility during early liver and pancreatic development (Cernilogar et al. 2019; Genga et al. 2019; Lee et al. 2019; Meers et al. 2019). However, it is not clear whether continued FoxA expression is required to maintain these changes in chromatin accessibility, with recent reports suggesting that HNF4α, but not FoxA2, is required for maintaining enhancer activity in the adult liver (Thakur et al. 2019).
Reizel et al. (2020) investigated the importance of continued FoxA expression in the liver. As significant redundancy exists between the FoxA factors, investigation of the individual proteins precludes identification of the role played by the family as a whole. Moreover, FoxA2 deletion is embryonic-lethal. To circumvent these difficulties, the investigators developed a conditional triple-knockout mouse model using a hepatocyte-specific promoter, inducing the deletion of FoxA1/2/3 expression specifically in the parenchymal cells of the adult liver. Upon induction of FoxA deletion, the FoxA-null mice showed declining liver function over a period of 15–20 d, culminating in liver failure.
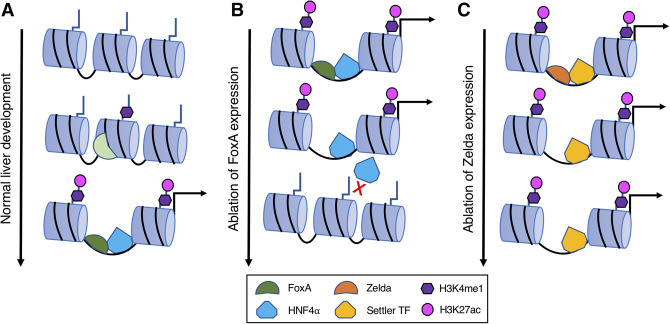
Investigation of the pathophysiology revealed significant reduction of mature liver markers. The investigators reported that the loss of gene expression most commonly occurred at genes that neighbored HNF4α–FoxA sites, rather than FoxA alone. Further analyses revealed that the loss of hepatic gene expression was associated with the reduced capacity of HNF4α to bind to sites that were cobound by FoxA. Importantly, at the sites of HNF4α–FoxA cobinding, FoxA deletion also resulted in the loss of H3K4me1 and H3K27ac deposition—posttranslational histone tail modifications associated with active enhancers. In addition, ATAC-seq analysis revealed that the HNF4α–FoxA-cobound sites demonstrated reduced chromatin accessibility in FoxA-null livers. Taken together with known FoxA roles in development, FoxA expression is required for both the acquisition and maintenance of accessible chromatin. Therefore, FoxA functions as the key to open chromatin, which facilitates the binding of transcriptional regulators, as well as a doorstop that maintains the open euchromatic state. Once the FoxA doorstop is removed, chromatin closes, preventing the binding of transcriptional regulators, such as HNF4α, that drive expression of genes that define liver function (Fig. 1). Thus, the expression of a significant cluster of hepatic genes is likely dependent on FoxA to induce and maintain chromatin accessibility and for HNF4α to recruit transcriptional complexes (Delaforest et al. 2019; Thakur et al. 2019).
Figure 1.
Overview of the dual role of FoxA as a pioneer and settler transcription factor (A) FoxA functions as a pioneer factor during normal development, inducing nucleosome repositioning, which allows for binding of transcriptional regulators, such as HNF4α. (B) Ablation of FoxA factors in the adult liver parenchyma results in the loss of accessibility and active histone modifications at FoxA-binding sites. The changes in epigenetic structure prevent the continued binding of HNF4α, manifesting in the loss of hepatic identity. (C) In contrast to FoxA ablation, loss of Zelda transcription factor expression does not result in a loss of accessibility at sites previously “opened” by Zelda-dependent pioneer activity. Instead, the euchromatic chromatin state is maintained by settler transcription factors and the recruitment of transcriptional machinery.
The importance of epigenetic structure to define cell identity and the capacity of pioneer factors to induce wholesale changes to the chromatin profile mean that pioneer factor misregulation contributes to numerous disease states. For example, overexpression of FoxA factors is associated with progression of cancer (Dobersch et al. 2019). It will therefore be of interest to determine whether the reduced expression of FoxA factors in adult tissues can cause epigenomic remodeling that has implications for pathogenesis, including in chronic liver diseases and cancer progression.
Understanding the role of FoxA as both a pioneer and settler factor is an important advance that impacts our understanding of pioneer factor function. It will be interesting to see whether similar FoxA–HNF4α relationships exist between other transcription factors within different organs, such as the pancreas, or during specific developmental stages. Moreover, it will be important to understand whether the dual pioneer/settler role of FoxA factors is the exception or the rule for mammalian pioneer factors and how the cell orchestrates reduction of accessibility at sites previously bound by FoxA.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.340570.120.
References
- Cernilogar FM, Hasenöder S, Wang Z, Scheibner K, Burtscher I, Sterr M, Smialowski P, Groh S, Evenroed IM, Gilfillan GD, et al. 2019. Pre-marked chromatin and transcription factor co-binding shape the pioneering activity of Foxa2. Nucleic Acids Res 47: 9069–9086. 10.1093/nar/gkz627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaforest A, Di Furio F, Jing R, Ludwig-Kubinski A, Twaroski K, Urick A, Pulakanti K, Rao S, Duncan SA. 2019. HNF4A regulates the formation of hepatic progenitor cells from human iPSC-derived endoderm by facilitating efficient recruitment of RNA pol II. Genes (Basel) 10: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobersch S, Rubio K, Barreto G. 2019. Pioneer factors and architectural proteins mediating embryonic expression signatures in cancer. Trends Mol Med 25: 287–302. 10.1016/j.molmed.2019.01.008 [DOI] [PubMed] [Google Scholar]
- Genga RMJ, Kernfeld EM, Parsi KM, Parsons TJ, Ziller MJ, Maehr R. 2019. Single-cell RNA-sequencing-based CRISPRi screening resolves molecular drivers of early human endoderm development. Cell Rep 27: 708–718.e10. 10.1016/j.celrep.2019.03.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gualdi R, Bossard P, Zheng M, Hamada Y, Coleman JR, Zaret KS. 1996. Hepatic specification of the gut endoderm in vitro: cell signaling and transcriptional control. Genes Dev 10: 1670–1682. 10.1101/gad.10.13.1670 [DOI] [PubMed] [Google Scholar]
- Lee K, Cho H, Rickert RW, Li QV, Pulecio J, Leslie CS, Huangfu D. 2019. FOXA2 is required for enhancer priming during pancreatic differentiation. Cell Rep 28: 382–393.e7. 10.1016/j.celrep.2019.06.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang HL, Nien CY, Liu HY, Metzstein MM, Kirov N, Rushlow C. 2008. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456: 400–403. 10.1038/nature07388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meers MP, Janssens DH, Henikoff S. 2019. Pioneer factor-nucleosome binding events during differentiation are motif encoded. Mol Cell 75: 562–575.e5. 10.1016/j.molcel.2019.05.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizel Y, Morgan A, Gao L, Lan Y, Manduchi E, Waite EL, Wang AW, Wells A, Kaestner KH. 2020. Collapse of the hepatic gene regulatory network in the absence of FoxA factors. Genes Dev (this issue) 10.1101/gad.337691.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur A, Wong JCH, Wang EY, Lotto J, Kim D, Cheng J, Mingay M, Cullum R, Moudgil V, Ahmed N, et al. 2019. Hepatocyte nuclear factor 4-α is essential for the active epigenetic state at enhancers in mouse liver. Hepatology 70: 1360–1376. 10.1002/hep.30631 [DOI] [PMC free article] [PubMed] [Google Scholar]