Highlights
-
•
Novaferon considered as a potential antiviral drug for COVID-19.
-
•
Novaferon inhibited viral replication and protected cells from SARS-CoV-2 attack.
-
•
Antiviral effects of Novaferon for COVID-19 patients observed in randomized trial.
-
•
Inhalation of Novaferon for COVID-19 treatment was safe.
Keywords: COVID-19, SARS-CoV-2, Novaferon, Antiviral drug, Lopinavir/Ritonavir, Viral clearance, Aerosolized inhalation
Abstract
Background
The antiviral effects of Novaferon, a potent antiviral protein drug, on COVID-19 was evaluated in the laboratory, and in a randomized, open-label, parallel-group trial.
Methods
In the laboratory, Novaferon's inhibition of viral replication in cells infected with SARS-CoV-2, and prevention of SARS-CoV-2 entry into healthy cells was determined. Antiviral effects of Novaferon in COVID-19 patients with treatment of Novaferon, Novaferon plus Lopinavir/Ritonavir, or Lopinavir/Ritonavir were evaluated. The primary endpoint was the SARS-CoV-2 clearance rates on day six of treatment, and the secondary endpoint was the time to SARS-CoV-2 clearance.
Results
Novaferon inhibited viral replication (EC50 = 1.02 ng/ml), and prevented viral infection (EC50 = 0.10 ng/ml). Results from the 89 enrolled COVID-19 patients showed that both Novaferon and Novaferon plus Lopinavir/Ritonavir groups had significantly higher viral clearance rates on day six than Lopinavir/Ritonavir group (50.0% vs. 24.1%, p = 0.0400, and 60.0% vs. 24.1%, p = 0.0053). The median time to viral clearance was six days, six days, and nine days for three groups, respectively, a 3-day reduction in both the Novaferon and Novaferon plus Lopinavir/Ritonavir groups compared with the Lopinavir/Ritonavir group.
Conclusions
Novaferon exhibited anti-SARS-CoV-2 effects in vitro and in COVID-19 patients. These data justify further evaluation of Novaferon.
Trial registration number
Number ChiCTR2000029496 at the Chinese Clinical Trial Registry (http://www.chictr.org.cn/).
Introduction
The deadly pandemic of COVID-19 caused by the infection of a novel coronavirus, SARS-CoV-2, represents a major health challenge worldwide (WHO COVID-19 report 76; Zhu et al., 2020, Lu et al., 2020, Wu et al., 2020). The current failure to contain COVID-19 was partially due to the lack of effective antiviral drugs for COVID-19. If administered to early-stage patients or to patients with mild to moderate illness, such antiviral drugs are reasonably expected to speed up viral clearance. Consequently, complete clearance of SARS-CoV-2 will lead to either earlier recovery or a reduction of severe illness. The elimination of viral shedding following viral clearance in patients would also help to reduce viral transmission. Given the immediate availability and established safety profiles, approved antiviral drugs for other indications were repurposed to find effective anti-SARS-CoV-2 drugs in the shortest time possible (Zhou et al., 2020). However, none of the tested or recommended antiviral drugs has been proved effective yet. Most published findings for the antiviral treatment of COVID-19 were based on the individual case reports or cellular antiviral results (Holshue et al., 2020, Li et al., 2014, Li and De Clercq, 2020). Despite the lack of convincing evidence, Lopinavir/Ritonavir was quickly selected and recommended as an antiviral drug for COVID-19 in China since January 2020. So far, only limited observations of Lopinavir/Ritonavir for coronavirus in SARS patients have been reported (Chu et al., 2020). A recently completed trial of Lopinavir/Ritonavir in patients with severe COVID-19 generated disappointing outcomes and revealed no significant antiviral effects (Cao et al., 2020). Health care workers have to rely on supportive and symptomatic treatments to manage COVID-19 patients. Given the daily increase of confirmed COVID-19 cases and mortality, it is even more critical than two months ago to find antiviral drugs with efficacy, supported by data from randomized clinical trials in COVID-19 patients (Xu et al., 2020, Zhang and Liu, 2020).
Novaferon is a novel antiviral protein drug that has been approved for the treatment of chronic hepatitis B in China. It exhibited broad-spectrum antiviral properties (unpublished data, available on request), becoming an obvious candidate to be considered as a potential antiviral drug for COVID-19. The Novaferon molecule is a non-natural protein consisting of 167 amino acids. According to the published information in a US patent (US 7,625,555 B2), this novel protein molecule was created by a modified DNA shuffling technology using cDNA sequences of 12 human interferon subtypes as models and named Novaferon by its inventors (Wang et al., 2011). In addition to its human interferon-like physiological functions, Novaferon exhibits better antiviral activities that are at least 10 times more potent than human interferon-alpha-2b (Li et al., 2014). Novaferon has been shown to enhance and improve the negative conversion of serum HBeAg in clinical studies (Daxian and Deming, 2015). In April 2018, it was approved in China to treat chronic hepatitis B by the former CFDA (Chinese Food and Drug Administration). Novaferon protein's non-proprietary name was temporarily defined as “recombinant cytokine gene-derived protein injection” by the Chinese Pharmacopeia Committee, and the recommended international non-proprietary name (rINN) by WHO is not available yet. For convenience purposes, Novaferon was used as the drug name in our study.
In the present study, we primarily attempted to observe the antiviral effects of Novaferon on COVID-19. We first determined whether Novaferon was able to inhibit SARS-CoV-2 at the cellular level, and subsequently conducted a randomized, open-label, parallel-group trial to explore the antiviral effects of Novaferon in COVID-19 patients by observing the SARS-CoV-2 clearance rates. The primary endpoint was the SARS-CoV-2 clearance rates on day six, and the secondary endpoint was the median time to SARS-CoV-2 clearance after starting antiviral treatment. As a popular and recommended antiviral drug for COVID-19 in China, Lopinavir/Ritonavir was included in this study to serve as a control for comparison.
Methods
Production of recombinant Novaferon protein
Novaferon is a non-naturally existing protein molecule produced by recombinant technology via inserting a 498-nucleotides cDNA into Escherichia coli. The gene sequence (cDNA) encoding the Novaferon protein molecule was created based on a modified DNA Shuffling technology to mimic the genome's natural evolution, with intentions to invent novel protein molecules that have enhanced natural functions of the model proteins. In brief, cDNA sequences of over 12 human interferon subtype genes were selected as the model genes for DNA shuffling. These model cDNA sequences were cut into fragments by enzymes and then repeatedly amplified to induce randomized nucleotide-mutation of the cDNA fragments. The mutant cDNA fragments in the reaction system randomly and spontaneously connect with each other to form a very large mutant cDNA library. The clones of the newly formed cDNA sequences that encoded protein molecules with broad-spectrum, enhanced antiviral and anti-proliferation activities, were then screened and selected via a proprietary protein-screening method (High-efficient Protein Functional Screen System). A novel protein molecule has been identified after screening more than 100,000 cDNA clones; it exhibits enhanced potency against virus and tumor cells and possesses broad-spectrum antiviral and anti-proliferation properties as well. This novel protein molecule was subsequently named Novaferon. Novaferon is encoded by 498 nucleotides and composed of 166 amino acids with the following amino acid sequence: CNLSQTHSLGSKRTLMLLAQMGKISLFSCLKDRHDFEFPQEEFDGNQFQKAQAISVLHELIQQTFNLFSTKESSAAWDEGLLDKFRTELYRQLNDLEACMMQEVGVEETPLMNADSILAVKKYFQRITLYLMEKKYSPCAWEVVRVEIMRSLSFSTNLQKRLRGKD. As a non-naturally occurring protein, Novaferon is not suitable to be classified according to the terms of human interferon subtypes. The nucleotide and amino-acid sequences of Novaferon are 89% (445/498) and 81% (135/166) homologous to human interferon α-2b, which exhibits the best antiviral activities among all human interferon subtypes (USPTO patent: US 7,625,555 B2) (Wang et al., 2011). The Novaferon drug used in this trial was manufactured in Qingdao city of Shandong Province by Genova Biotech (Qingdao) Company Limited.
In vitro antiviral tests
All the in vitro experiments were conducted in a biosafety level-3 (BSL3/P3) laboratory at the Chinese Center for Disease Control and Prevention (Chinese CDC). We first determined whether Novaferon inhibited the SARS-CoV-2 replication within the Vero E6 cells, which were already infected with SARS-CoV-2. Vero E6 cells (African green monkey kidney cells) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and maintained in a Modified Eagle Medium (MEM; Gibco Invitrogen, Carlsbad, CA, USA) with 10% fetal bovine serum (FBS; Thermo Scientific HyClone, South Logan, UT) at 37 °C in the incubator with 5% CO2. A clinically-isolated strain of SARS-CoV-2 (C-Tan-nCov Wuhan strain 01) was propagated in Vero E6 cells before the experiments, and the plaque assay was used to quantify the titer (plaque-forming unit, PFU) of the SARS-CoV-2. Blank Vero E6 cells in 96-well plates with a density of 1 × 104 per well were incubated with C-Tan-nCov Wuhan strain 01 (100PFU) for 2 h to induce the infection of the cultured cells by SARS-CoV-2. After virus-containing supernatants were removed and cells rinsed, a medium with various concentrations of Novaferon (0, 0.001, 0.01, 0.1, 1, 10, or 100 ng/ml) was added, and the cells were then incubated for 24 h to allow for viral replication. After 24 h, 100 μL of supernatants from each well were collected, and the total viral RNA was extracted (Full-automatic nucleic acid Extraction System from TIANLONG) and measured. Takara Bio's One Step Prime Script Real-time PCR (RT-PCR) Kit (Perfect Real Time) was used to detect the copies of the virus’ RNA.
The Quantitative real-time PCR cycle threshold (Ct) was obtained, and the inhibition rates of virus replication by each Novaferon concentration were calculated. The Ct number of controls obtained in the absence of Novaferon was 22.6 and considered as 100%. The Ct numbers from SARS-CoV-2 infected Vero E6 cells with the addition of various concentrations of Novaferon were measured to calculate the inhibition percentages. The half-maximal effective concentration (EC50) was calculated. The cytotoxicity of Novaferon on Vero E6 cells was assessed, and the half-maximal cytotoxic concentration (CC50) of Novaferon on Vero Cells was determined by observing the cytopathic effects (CPE) of Novaferon. The selectivity index (CC50/EC50) was then calculated.
We further observed whether the previous treatment of Vero E6 cells with Novaferon protected the cells from viral entry through the later exposure of the pre-treated cells to SARS-CoV-2. Detailed operation procedures were identical to the above description, except that the step orders were changed to observe the preventive effects of Novaferon. Briefly, blank Vero E6 cells were incubated with series concentrations of Novaferon for 2 h, and the supernatants containing Novaferon were then removed. The pre-treated Vero E6 cells were exposed to SARS-CoV-2 by incubation with C-Tan-nCov Wuhan strain 01 (100PFU) for 2 h; supernatants containing SARS-CoV-2 were then removed. Fresh medium was added, and the cells were incubated for 48 h. 100 μL of supernatants were taken from each well, and the total viral RNA in the supernatants was measured using the same methods described above. The Ct number obtained from Vero E6 cells without pre-treatment of Novaferon was considered 100%, and the decreased Ct numbers obtained from the pre-treated Vero E6 cells with various concentrations of Novaferon were used to calculate the inhibition percentages. The preventive effects of Novaferon were then determined by observing the viral RNA reduction in the cells pre-treated with Novaferon. The EC50 of Novaferon for the observed preventive effects was decided accordingly.
Laboratory detection of SARS-CoV-2 nucleic acids by RT-PCR in nasopharyngeal swab samples
SARS-CoV-2 virus nucleic acids were detected by RT-PCR using the SLAN-96P automatic medical PCR analysis system. The SARS-CoV-2 nucleic acid detection kit was obtained from Sensure Biotechnology Co. Ltd (Hunan province, China, which had been approved for a clinical test of SARS-CoV-2 by the NMPA (National Medical Products Administration). The lowest detection limit (sensitivity) of this RT-PCR assay kit was 200 copies of SARS-CoV-2RNA in specimens. The specificity of this RT-PCR assay kit was determined by the failure of detecting viral RNA (cross-reaction) in specimens containing other coronaviruses, rotavirus, astrovirus, and adenovirus, etc.
Procedures of the tests strictly followed the protocol of the kit. Samples from nasopharyngeal swabs were collected following the New Coronavirus Infection Pneumonia Laboratory Test Guide's standard procedures. FAM (ORF-1ab region) was used as a fluorescent detection channel, and ROX (N gene) channels were used to detect the SARS-CoV-2 nucleic acids, while the HEX channel was used as the internal standard. Cycle parameter steps were in the following order: (1) reverse transcription at 50 °C for 30 min for 1 cycle; (2) pre-denaturation of cDNA at 95 °C for 1 min for 1 cycle; (3) denaturation at 95 °C for 15 s, and annealing, extension and fluorescence acquisition at 60 °C for 30 s for 45 cycles; (4) cooling at 25 °C for 10 s for 1 cycle. Positive results were determined by comparing the Ct numbers of the testing samples and the standard Ct number (40 in this assay).
Clinical study
Patients
The study was initially designed as a multi-center study across hospitals in Changsha city and in other cities of Hunan Province, China. However, per a government order, all patients from hospitals in Changsha city had to be relocated to the First Hospital of Changsha, a designated treatment center for all COVID-19 patients in Changsha city, and hospitals in other cities of Hunan Province were not able to participate due to various reasons. The study was changed to a single-center study. This study was approved by the ethics committee of the First Hospital of Changsha (file number KX-2020002) and was conducted at the hospital. The study was also registered in the Chinese Clinical Trial Registry (http://www.chictr.org.cn/), number ChiCTR2000029496.
Hospitalized COVID-19 patients with confirmed SARS-CoV-2 detection, clinically classified as moderate or severe, at an age over 18 years, and without comorbidity of severe heart, lung, or brain diseases, were eligible for enrolling into this study. Moderate patients were defined as “patients with fever, symptoms of the respiratory system and pneumonia changes in CT images, and severe patients were defined as “patients with any of the following: (1) Respiratory distress, respiratory frequency ≥ 30/min; (2) Under rest status, arterial oxygen saturation (SaO2) ≤ 93%; (3) Arterial partial pressure of oxygen (PaO2)/fraction of inspired oxygen (FiO2) ≤ 300 mmHg. In this study, we aimed to observe moderate and severe COVID-19 patients as these patients would likely benefit more from antiviral treatments.
Trial design and treatments
This was a randomized, open-label, parallel-group study. Patients eligible for the study were assigned, in a 1:1:1 ratio, to Novaferon, Novaferon plus Lopinavir/Ritonavir, or Lopinavir/Ritonavir group. An SAS generated simple randomization schedule was prepared by a statistician not involved in the trial. Using the order in which they enrolled in the study, patients were assigned to a treatment group that was implemented by a research assistant. Informed consent was obtained from all enrolled patients. Antiviral effects were assessed on day three, day six, and day nine after starting drug administration.
The approved dosage of Novaferon for hepatitis B application is the daily injection of 10 μg of protein in 1.0 ml volume per vial. Lopinavir/Ritonavir (Kaletra) was manufactured by AbbVie Inc.; each tablet contained 200 mg of Lopinavir and 50 mg of Ritonavir. The total daily doses (40 μg) of Novaferon were administered to patients twice per day by oxygen-driven aerosolized inhalation for 15 min of 20 μg of Novaferon (2 × 1 ml vials) diluted with saline. For patients receiving Lopinavir/Ritonavir (Kaletra), two tablets were orally taken twice per day. The aerosolized inhalation was administrated to hospitalized patients in the negative-pressure wards at the designated COVID-19 center to minimize the risk of disease transmission.
Assessments
Samples of nasopharyngeal swabs on day three, day six, and day nine during the seven to ten-day course of antiviral treatment were collected from the patients and tested for SARS-CoV-2 nucleic acids by RT-PCR. SARS-CoV-2 clearance in COVID-19 patients was defined as two consecutive negative-detection of SARS-CoV-2 RNA in nasopharyngeal swab samples with an interval of over 24 h. Adverse events were monitored throughout the trial, reported and graded based on the WHO Toxicity Grading Scale for Determining the Severity.
The peak levels of the SARS virus were around ten days after onset, and then the viral level began to decrease without effective antiviral treatment in SARS patients (Peiris et al., 2003). Considering the homology of gene sequences of SARS-CoV-2 and SARS was over 90% (Zhu et al., 2020), we assumed that the intervention of antiviral drugs in COVID-19 patients would likely enhance or shorten the time to viral clearance. In this regard, this study's primary endpoint was chosen based on the SARS-CoV-2 clearance rates in COVID-19 patients assessed on day six of antiviral treatment. The secondary endpoint was the median time to SARS-CoV-2 clearance.
Statistical analysis
Statistical analysis was performed on an intent-to-treat basis, and all patients randomized and treated at least once with the study medications were included in the primary analysis. For patient demographics information and baseline disease characteristics, qualitative variables were compared among treatment groups using the Chi-square test, and quantitative variables were compared with the use of an ANOVA model. Only the overall differences among the three treatment groups were tested (based on the null hypothesis, “all three groups were the same,” against an alternative hypothesis, “at least one group was different”). Therefore, no pairwise comparison was performed for baseline characteristics.
For the primary endpoint, SARS-CoV-2 clearance rate, estimates of the rates were calculated based on a binomial distribution. The difference between treatment groups was tested using the Chi-square test. To control the study's overall significance level, the three pairwise comparisons for the primary endpoint were performed at the two-sided alpha = 0.05, using a closed testing procedure according to the following order: (1) Novaferon plus Lopinavir/Ritonavir vs. Lopinavir/Ritonavir alone; (2) Novaferon alone vs. Lopinavir/Ritonavir alone; (3) Novaferon plus Lopinavir/Ritonavir vs. Novaferon alone. For the secondary endpoint, time to SARS-CoV-2 clearance, the median time for each group was estimated using the Kaplan–Meier method, and treatment differences were tested using the log-rank test. All tests were two-sided, with a P-value of less than 0.05 considered to indicate statistical significance. The analysis was conducted using SAS V9.2.
For missing SARS-CoV-2 clearance status, the Last Observation Carried Forward (LOCF) analysis was presented as the primary analysis. For the purpose of sensitivity analyses, complete case analysis, and worst-case imputation methods were also performed. For the worst-case imputation, missing SARS-CoV-2 status was replaced with ‘positive.’
The planned sample size of 90 patients (30 patients per group) was not determined based on statistical consideration.
Adverse events were reported and graded using the WHO Toxicity Grading Scale for determining the severity. The incidence of adverse events was summarized descriptively without a formal statistical test.
Results
Inhibitory effects of Novaferon on SARS-CoV-2 at the cellular level
Incubation of Novaferon (0.1–100 ng/ml) with SARS-CoV-2-infected Vero E6 cells resulted in the dose-dependent reductions of the SARS-CoV-2 RNA that was released from the infected Vero E6 cells. The half-maximal effective concentration (EC50) of Novaferon was 1.02 ng/ml. The tested Novaferon concentrations showed minimal cytotoxicity to Vero E6 cells, and the half-maximal cytotoxic concentration (CC50) was over 100 ng/ml. The selectivity index (CC50/EC50) was over 98. These data indicated that Novaferon effectively inhibited viral replication within SARS-CoV-2-infected cells. Healthy Vero E6 cells that were previously incubated with Novaferon resisted the entry of SARS-CoV-2 into cells, as indicated by the reduction of cellular viral RNA after Novaferon was removed; treated cells were exposed to SARS-CoV-2 later. Novaferon exhibited this preventive effect efficiently with the EC50 (0.1 ng/ml) lower than the EC50 for inhibiting SARS-CoV-2 replication in the infected cells (supplementary figure). These data suggested that Novaferon inhibited viral replication in SARS-CoV-2-infected cells and enabled healthy cells to resist the viral attack.
Patients and treatments
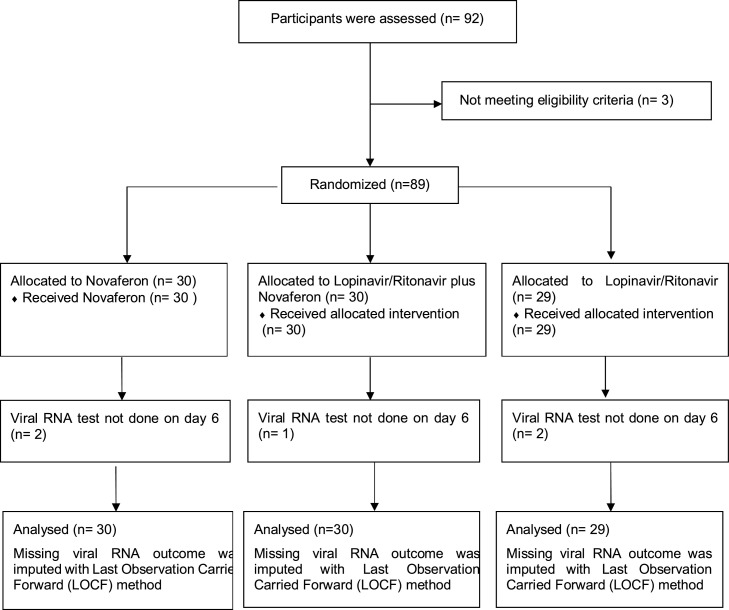
As presented in Figure 1 , a total of 92 patients with moderate or severe illness were assessed for the eligibility criteria, and three patients were excluded. 89 patients were randomized into the study from February 1–20, 2020. Of the 89 patients, 30, 30, and 29 patients were assigned into the Novaferon group, the Novaferon plus Lopinavir/Ritonavir group or the Lopinavir/Ritonavir group respectively, among whom 84 had a moderate illness and five had severe illness. Supported by the government policy of reimbursing COVID-19-related expenses, patients were diagnosed, screened, and enrolled shortly after symptom onset. The median time (IQR) from symptom onset to antiviral drug administration was 4.0 days (3.0–6.5), 7.0 days (3.3–11.3), and 4.0 days (3.0–6.0) in the Novaferon group, the Novaferon plus Lopinavir/Ritonavir group or the Lopinavir/Ritonavir group, respectively. Enrollment screening excluded patients with co-existing severe cardiac, kidney, or liver diseases as described in the exclusion criteria; none of the enrolled patients had been given steroid treatment. The baseline demographic and clinical characteristics of the 89 patients are summarized in Table 1 . Except for some imbalances between the groups, there were no major differences between groups in demographic characteristics, baseline laboratory test results, and disease severity at enrollment (Table 1).
Figure 1.
Randomization and treatment assignment.
Table 1.
Demographics and baseline clinical characteristics.
| Variables | Novaferon N = 30, n (%) |
LPV/ra + Novaferon N = 30, n (%) |
LPV/rN = 29, n (%) |
|---|---|---|---|
| Age, median (IQR) | 46.5 (40.0–63.8) | 50.0 (37.8–62.8) | 37.0 (26.0–54.0) |
| Male, n (%) | 17 (56.7%) | 13 (43.3%) | 12 (41.4%) |
| Median time from symptoms to therapy, days (IQR) | 4.0 (3.0–6.5) | 7.0 (3.3–11.3) | 4.0 (3.0–6.0) |
| Moderate cases, n (%) | 28 (93.3%) | 28 (93.3%) | 28 (96.6%) |
| Severe cases, n (%) | 2 (6.7%) | 2 (6.7%) | 1 (3.4%) |
| Comorbidity, n (%) | 7 (23.33) | 6 (20.00) | 5 (17.24) |
| Diabetes | 3 (10.00) | 3 (10.00) | 2 (6.90) |
| Hypertension | 2 (6.67) | 3 (10.00) | 1 (3.34) |
| Coronary heart disease | 1 (3.33) | 1 (3.33) | 1 (3.34) |
| Chronic hepatitis B | 1 (3.33) | 0 (0) | 1 (3.34) |
| Chronic bronchitis | 1 (3.33) | 0 (0) | 1 (3.34) |
| Fever, n (%) | 17 (56.67) | 20 (66.67) | 20 (68.97) |
| Cough, n (%) | 16 (53.33) | 16 (53.33) | 13 (44.83) |
| Fatigue, n (%) | 6 (20.00) | 8 (26.67) | 12 (41.38) |
| Sore throat, n (%) | 3 (10.00) | 2 (6.67) | 4 (13.79) |
| Headache, n (%) | 3 (10.00) | 2 (6.67) | 2 (6.90) |
| Myalgia, n (%) | 3 (10.00) | 1 (3.33) | 4 (13.79) |
| Dizziness, n (%) | 1 (3.33) | 3 (10.00) | 1 (3.45) |
| Diarrhea, n (%) | 1 (3.33) | 3 (10.00) | 1 (3.45) |
| Rhinorrhoea, n (%) | 1 (3.33) | 1 (3.33) | 1 (3.45) |
| Nausea, n (%) | 0 (0) | 2 (6.67) | 1 (3.45) |
| Vomiting, n (%) | 0 (0) | 2 (6.67) | 1 (3.45) |
| Dyspnea, n (%) | 2 (6.67) | 2 (6.67) | 1 (3.33) |
| Loss of appetite, n (%) | 0 (0) | 0 (0) | 4 (13.79) |
| Chill, n (%) | 0 (0) | 0 (0) | 2 (6.90) |
| Leukopenia, n (%) | 13 (43.33) | 10 (33.33) | 13 (44.83) |
| Neutropenia, n (%) | 11 (36.67) | 4 (13.33) | 6 (20.69) |
| Lymphopenia, n (%) | 4 (13.33) | 7 (23.33) | 6 (20.69) |
| Thrombocytopenia, n (%) | 4 (13.33) | 1 (3.33) | 2 (6.90) |
| Hemoglobin decreased, n (%) | 1 (3.33) | 2 (6.67) | 2 (6.90) |
| ALT increased, n (%) | 0 (0) | 2 (6.67) | 1 (3.45) |
| AST increased, n (%) | 1 (3.33) | 1 (3.33) | 3 (10.35) |
LPV/r: Lopinavir/Ritonavir.
Primary endpoint
Table 2 summarizes the complete RT-PCR test results of all 89 patients on day three, day six, and day nine after starting drug administration. The negative results of SARS-CoV-2 nucleic acid detection in the tested samples served as the indicator of in vivo SARS-CoV-2 clearance in patients. The SARS-CoV-2 clearance rates on day three, day six, and day nine in three treatment groups were presented and compared (Table 2). On day three, SARS-CoV-2 clearance rates were 16.7% (5/30) in the Novaferon group, 36.7% (11/30) in Novaferon plus Lopinavir/Ritonavir group, and 10.3% (3/29) in Lopinavir/Ritonavir group respectively. The SARS-CoV-2 clearance rate in Novaferon plus Lopinavir/Ritonavir group was significantly higher than in Lopinavir/Ritonavir group on day three (36.7% vs. 10.3%, p = 0.0175). No significant difference between the Novaferon group and Novaferon plus Lopinavir/Ritonavir group was observed. On day six, SARS-CoV-2 clearance rates in the Novaferon group and the Novaferon plus Lopinavir/Ritonavir group reached 50.0% (15/30) and 60.0% (18/30), respectively, and were significantly higher than in the Lopinavir/Ritonavir group (50.0% vs. 24.1%, p = 0.0400, and 60.0% vs. 24.1%, p = 0.0053, respectively). There was no statistically significant difference between the Novaferon group and the Novaferon plus Lopinavir/Ritonavir group, suggesting the similar extent of enhanced SARS-CoV-2 clearance on day six by Novaferon alone or together with Lopinavir/Ritonavir. On day nine, SARS-CoV-2 clearance rates were 56.7% (17/30) in the Novaferon group, 70.0% (21/30) in the Novaferon plus Lopinavir/Ritonavir group, and 51.7% (15/29) in the Lopinavir/Ritonavir group. There were no statistically significant differences between the groups.
Table 2.
Summary of SARS-CoV-2 clearance rates.
|
p valueb |
||||||
|---|---|---|---|---|---|---|
| Novaferon (N = 30) | LPV/ra + Novaferon (N = 30) | LPV/r (N = 29) | LPV/r vs. Novaferon | LPV/r vs. LPV/r + Novaferon | Novaferon vs. LPV/r + Novaferon | |
| Day 3 | 16.7% (5/30) | 36.7% (11/30) | 10.3% (3/29) | 0.4783 | 0.0175 | 0.0798 |
| Day 6 | 50.0% (15/30) | 60.0% (18/30) | 24.1% (7/29) | 0.0400 | 0.0053 | 0.4363 |
| Day 9 | 56.7% (17/30) | 70.0% (21/30) | 51.7% (15/29) | 0.7032 | 0.1502 | 0.2839 |
LPV/r: Lopinavir/Ritonavir.
p values for comparisons between treatment groups using Chi-square test; At any visit, missing viral RNA outcome was imputed with Last Observation Carried Forward (LOCF) method.
Secondary endpoint
The median time to SARS-CoV-2 clearance was six days, six days, and nine days for the Novaferon group, the Novaferon plus Lopinavir/Ritonavir group, and the Lopinavir/Ritonavir group respectively, indicating a 3-day reduction of time to SARS-CoV-2 clearance in both Novaferon and Novaferon plus Lopinavir/Ritonavir groups compared with Lopinavir/Ritonavir group (Table 3 ). During the observation period, none of the moderately ill patients in the Novaferon group and Novaferon plus Lopinavir/Ritonavir group progressed to severe illness. In contrast, four moderately ill patients in the Lopinavir/Ritonavir group progressed to severe illness.
Table 3.
Analysis for time to SARS-CoV-2 clearance.
|
p-value |
||||||
|---|---|---|---|---|---|---|
| Novaferon | LPV/ra + Novaferon | LPV/r | LPV/r vs. Novaferon | LPV/r vs. LPV/r + Novaferon | Novaferon vs. LPV/r + Novaferon | |
| N (Censored) | 30 (13) | 30 (9) | 29 (14) | |||
| Mean (days) | 7.0 | 6.1 | 8.0 | |||
| Median (days)b | 6 | 6 | 9 | 0.417 | 0.036 | 0.183 |
LPV/r: Lopinavir/Ritonavir.
Median time for each group was estimated with the use of the Kaplan–Meier method and treatment differences were tested using the log-rank test.
Sensitivity analysis for missing data
Analyses based on both complete case analysis and worst-case imputation for SARS-CoV-2 clearance rates showed little differences with the LOCF analysis, and the statistical conclusions for all the treatment comparisons remained the same.
Adverse events
No severe adverse events (SAE) associated with the tested antiviral drugs were reported. The observed adverse events (AE) were grade 1, or grade 2, and are summarized in Table 4 . No specific AEs were related to Novaferon treatment, and certain reported adverse events overlapped with the disease symptoms and laboratory findings.
Table 4.
Summary of common adverse events.
| Event | Novaferon (N = 30), n (%) |
Novaferon + LPV/r( N = 30), n (%) |
LPV/r(N = 29), n (%) |
|---|---|---|---|
| Any adverse event | 25 (83.3) | 25 (83.3) | 26 (89.6) |
| Lymphopenia | 8 (26.7) | 14 (46.7) | 9 (31.0) |
| Loss of appetite | 8 (26.7) | 10 (33.3) | 9 (31.0) |
| Cough | 7 (23.3) | 14 (46.7) | 4 (13.8) |
| Fatigue | 7 (23.3) | 9 (30.0) | 10 (34.5) |
| Neutropenia | 5 (16.7) | 7 (23.3) | 7 (24.1) |
| Dizziness | 5 (16.7) | 5 (16.7) | 3 (10.3) |
| Diarrhea | 4 (13.3) | 4 (13.3) | 5 (17.2) |
| Abdominal discomfort | 4 (13.3) | 1 (3.3) | 4 (13.8) |
| Anemia | 3 (10.0) | 4 (13.3) | 3 (10.3) |
| Sleep disorders | 3 (10.0) | 3 (10.0) | 4 (13.8) |
| Nausea | 2 (6.7) | 5 (16.7) | 1 (3.4) |
| Dyspnea | 2 (6.7) | 2 (6.67) | 3 (10.35) |
| Vomiting | 1 (3.33) | 4 (13.3) | 1 (3.3) |
| Hepatic injury | 1 (3.3) | 3 (10.0) | 3 (10.3) |
| Chest tightness | 1 (3.3) | 2 (6.7) | 6 (20.7) |
Adverse events occurred in 25 of 30 (83.3%) patients in the Novaferon group, 25 of 30 (83.3%) patients in Novaferon plus Lopinavir/Ritonavir group, and 26 of 29 (89.6%) patients in Lopinavir/Ritonavir group. The most common adverse events were lymphopenia and loss of appetite in the Novaferon group, lymphopenia and cough in Novaferon plus Lopinavir/Ritonavir group, and fatigue and lymphopenia in Lopinavir/Ritonavir group. The observed adverse reactions did not need new medical interventions or cause termination of antiviral treatment.
Discussion
No matter whether exhibiting good, poor, or none anti-COVID-19 effects, Lopinavir/Ritonavir in this study served as the control and allowed us to assess the antiviral effects of Novaferon by analyzing the differences between Novaferon and Lopinavir/Ritonavir. In this regard, the significantly higher SARS-CoV-2 clearance rates on day six (the primary endpoint) in patients with treatment of Novaferon alone or together with Lopinavir/Ritonavir comparing with Lopinavir/Ritonavir alone indicated that Novaferon indeed exhibited antiviral effects in COVID-19patients. The 3-day reduction of time to SARS-CoV-2 clearance in patients with Novaferon treatment further supported the antiviral effects of Novaferon.
As viral shedding in the early stages of COVID-19 represents a significant challenge for controlling the transmission of SARS-CoV-2 (Wölfel et al., 2020), the sufficient viral clearance in patients undergoing Novaferon treatment in the early course of the disease was valuable in the clinical setting. The negative detection of SARS-CoV-2 in samples from the respiratory system indicated the elimination of viral shedding in patients. In turn, this would contribute to the effective reduction of virus transmission by early-stage patients who have been found to have the highest viral loads (Pan et al., 2020).
The antiviral effects of Novaferon on COVID-19 patients were consistent with the laboratory findings. Inhibition of the viral replication by Novaferon at the cellular level was very efficient, as indicated by the low EC50 (1.02 ng/ml). More interestingly, healthy cells that were pre-treated with Novaferon retained the ability, in the absence of Novaferon, to resist viral entry into cells when the treated cells were exposed to SARS-CoV-2 later (EC50 0.1 ng/ml). It might be worth exploring the potential use of Novaferon as a preventive agent for high-risk populations, especially for health care workers who have to routinely contact COVID-19 patients.
The viral loads in COVID-19 patients were reported to reach peak levels around 5–6 days after symptom onset (Pan et al., 2020), and for severe patients, the average time from the onset of symptoms to severe illness took about one week (WHO, Report of the WHO-China joint mission, 2020). The early clearance of SARS-CoV-2 might help shorten the disease course or prevent disease progression in patients with mild to moderate COVID-19. Considering that high viral loads and peak levels of SARS-CoV-2 were found in the first week of symptom onset, the increased SARS-CoV-2 clearance rates on day six in COVID-19 patients with Novaferon treatment were unlikely related to the natural disease course. Instead, the observed enhancement of SARS-CoV-2 clearance apparently reflected the antiviral effects of Novaferon in observed COVID-19 patients.
Limitations of this study
Our study has several limitations. First, all observations were done at one hospital in one city. Second, the sample size was relatively small and was not based on statistical analysis, as limited by the availability of COVID-19 patients in Changsha city. Third, the unexpected difficulties associated with the COVID-19 outbreak compromised the quality of this study. For example, adverse events were likely under-reported due to the emergent and risk situation. However, these limitations should not change the overall conclusion for this randomized trial because the antiviral assessments were strictly performed according to a rigorous standard.
Conclusions
Novaferon exhibited anti-SARS-CoV-2 effects at the cellular level and in patients with COVID-19. Data obtained from this randomized, open-label, parallel-group trial, preliminarily demonstrated the anti-SARS-CoV-2 effects of Novaferon for COVID-19 and justified large-scale clinical studies to verify the efficacy of Novaferon as a potential antiviral drug for COVID-19.
Authors’ contribution
Study design: Guozhong Gong, Wenjie Tan, Yuanlin Xie, Yongfang Jiang. Data collection: Fang Zheng, Yanwen Zhou, Zhiguo Zhou, Fei Ye, Baoying Huang, Yaxiong Huang, Jing Ma, Qi Zuo, Xin Tan, Jun Xie, Peihua Niu, Wenlong Wang. Data analysis: Yun Xu, Feng Peng, Ning Zhou, Chunlin Cai, Wei Tang, Xinqiang Xiao, Yi Li, Zhiguang Zhou. Writing: Yongfang Jiang, Yuanlin Xie, Wenjie Tan, Guozhong Gong. All authors read and approved the final manuscript. Fang Zheng, Yanwen Zhou, Zhiguo Zhou, Fei Ye contributed equally to this work.
Competing interests
The authors declare no competing interests.
Ethical approval
The study was anonymous; the protocol was approved by the Ethics Committee of the First Hospital of Changsha, according to the Declaration of Helsinki, 2013. Written informed consent was obtained from all participants.
Acknowledgments
We thank all medical and management staff, who came from hospitals across Changsha City and worked at the First Hospital of Changsha, for their courage and dedication to COVID-19 patient care and their help in the hospital's overall operations during this difficult time. We especially thank Dr. Charlie Chen of SRD ClinMax Corporation for conducting the statistical analysis. This work was supported by the National Science and Technology Major Project (2017ZX10202201, 2017ZX10202203), the National Key Research and Development Program of China (No. 2016YFD0500301), the Natural Science Foundation of Hunan Province (2018JJ2452) and Specialized Science and Technology Project of Hunan Province (2020SK3013).
Footnotes
Supplementary material related to this article can be found, in the online version, at https://doi.org/10.1016/j.ijid.2020.07.053.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Cao B., Wang Y., Wen D., Liu W., Wang J., Fan G. A trial of lopinavir–ritonavir in adults hospitalized with severe COVID-19. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu C.M., Cheng V.C., Hung I.F., Wong M.M.L., Chan K.H., Chan K.S. Role of lopinavir/ritonavir in the treatment of SARS: initial virological and clinical findings. Thorax. 2020;59:252–256. doi: 10.1136/thorax.2003.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daxian W., Deming T. The impacts of baseline clinical characteristics and hepatitis B Virus mutations on curative effects chronic hepatitis B treatment with Novaferon. J Hepatol. 2015;62:S263–S264. [Google Scholar]
- Holshue M.L., DeBolt C., Lindquist C., Lofy K.H., Wiesman J., Bruce H. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020 doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G., De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV) Nat Rev Drug Discov. 2020;19:149–150. doi: 10.1038/d41573-020-00016-0. [DOI] [PubMed] [Google Scholar]
- Li M., Rao C., Pei D., Wang L., Li Y., Gao K. Novaferon, a novel recombinant protein produced by DNA-shuffling of IFN-alpha, shows antitumor effect in vitro and in vivo. Cancer Cell Int. 2014;14:1–9. doi: 10.1186/1475-2867-14-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu R., Zhao X., Li J., Niu P., Yang B., Wu H. Genomic characterization, and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020;395:565–574. doi: 10.1016/S0140-6736(20)30251-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y., Zhang D., Yang P., Poon L.L.M., Wang Q. Viral load of SARS-CoV-2 in clinical samples. Lancet. 2020 doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiris J.S.M., Chu C.M., Cheng V.C.C., Chan K.S., Hung I.F.N., Poon L.L.M. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet. 2003;361:1767–1772. doi: 10.1016/S0140-6736(03)13412-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Mao C, Li J, Xu J, Zhang R, Wang L, et al., Recombinant human interferon-like proteins (Patent No. US 7,625,555 B2). In Vol US 7,868,151; 2011, B2:37.
- Wölfel R., Corman V.M., GuggemosW, Seilmaier M., Zange S., Muller M.A. Virologic assessment of hospitalized patients with COVID-2019. Nature. 2020 doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- World Health Organization. Coronavirus disease 2019 (COVID-19) situation Report-76. Available from: http://www.who.int/docs/default-source/coronaviruse/situation-reports/20200405-sitrep-76-covid-19.pdf?sfvrsn=6ecf0977_2.
- World Health Organization. Report of the WHO-China joint mission on coronavirus disease 2019 (COVID-19). Available from: https://www.who.int/docs/default-source/coronaviruse/who-china-joint-mission-on-covid-19-final-report.pdf.
- Wu F., Zhao S., Yu B., Chen Y.M., Wang W., Song Z.G. A new coronavirus associated with human respiratory disease in China. Nature. 2020 doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K., Cai H., Shen Y., Ni Q., Chen Y., Hu S. Management of coronavirus disease-19 (COVID-19): the Zhejiang experience. Zhejiang Da XueXue Bao Yi Xue Ban. 2020 doi: 10.3785/j.issn.1008-9292.02.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Liu Y. Potential interventions for novel coronavirus in China: a systematic review. J Med Virol. 2020 doi: 10.1002/jmv.2570.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020 doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu N., Zhang D., Wang W., Li X., Yang B., Song J. A novel coronavirus from patients with pneumonia in China 2019. N Engl J Med. 2020;382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.