Abstract
SARS-CoV-2 has devastated the world with its rapid spread and fatality. The researchers across the globe are struggling hard to search a drug to treat this infection. Understanding the time constraint, the best approach is to study clinically approved drugs for control of this deadly pandemic of COVID 19. The repurposing of such drugs can be supported with the study of molecular interactions to enhance the possibility of application. The present work is a molecular docking study of proteins responsible for viral propagation namely 3Clpro, Nsp10/16, Spike protein, SARS protein receptor binding domain, Nsp 9 viral single strand binding protein and viral helicase. The protein through virus enters the host cell-human angiotensin-converting enzyme 2 (ACE2) receptor, is also used as a target for molecular docking. The docking was done with most discussed drugs for SARS-CoV-2 like Ritonavir, Lopinavir, Remdesivir, Chloroquine, Hydroxychloroquine (HCQ), routine antiviral drugs like Oseltamivir and Ribavirin. In addition, small molecules with anti-inflammatory actions like Mycophenolic acid (MPA), Pemirolast, Isoniazid and Eriodictyol were also tested. The generated data confirms the potential of Ritonavir, Lopinavir and Remdesivir as a therapeutic candidate against SARS-CoV-2. It is observed that Eriodictyol binds to almost all selected target proteins with good binding energy, suggesting its importance in treatment of COVID 19. Molecular interactions of Ritonavir, Lopinavir and Remdesivir against SARS-CoV-2 proteins enhanced their potential as a candidate drug for treatment of COVID-19. Eriodictyol had emerged as a new repurposing drug that can be used in COVID-19.
Keywords: Molecular docking, Drug repurposing, SARS-CoV-2, COVID 19
1. Introduction
The current outbreak of the novel coronavirus disease, COVID-19, is in headlines globally because of its threat to public health. This disease is caused by a novel severe acute respiratory syndrome-related coronavirus SARS-CoV-2 that emerged from Wuhan, China, in December 2019 (Lai et al., 2020; Wu et al., 2020). The disease has spread in 215 countries and territories with 31,81,642 confirmed cases and about 2,24,301 deaths to date (www.who.int). The symptoms of COVID-19 range from mild, self-limiting respiratory tract illness to severe progressive pneumonia, multiorgan failure, and death. The virus is highly transmissible among humans, with clusters occurring among close contacts forcing many countries to impose a lockdown on all activities which had already affected the global economy, and the situation will be worse if this disease is not controlled in time. Unfortunately, there is no effective medication for this novel SARS-CoV-2 at present (Liu et al., 2020).
In an attempt to fight against coronavirus, scientists are coming up with different strategies. Drug repurposing, an effective drug discovery strategy, could significantly shorten the time and reduce the cost compared to de novo drug discovery and randomized clinical trials (Cheng et al., 2016, 2017; Cheng, 2019; Zhou et al., 2020). Computational methods to understand the ligand protein interaction is one of the fastest ways to identify the candidate drug and target. Scientists are screening existing molecules from the database, which might be effective against coronavirus as a strategy (Zhou et al., 2020; Chauhan, 2020; Wu et al., 2020). In case of SARS-CoV-2, inputs from this traditional way were lacking because of unavailability of protein structure. But 3D structures of different proteins of this novel SARS-CoV-2 are constantly being deposited in the database and hence in the present study we decided to do molecular docking analysis of these newly deposited proteins against some repeatedly discussed drugs as a repurposing therapeutic approach. We had selected those proteins for docking which play an important role in propagation of virus. We selected 3CLpro which is the main protease of the virus, envelope anchored spike protein from virus and receptor binding domain of this protein, SARS-CoV-2 helicase, Nsp10/16 complex a nonstructural protein and a single strand or RNA binding protein as target because of their importance in viral life cycle (Wu et al., 2020). Virus enters the cell via angiotensin receptor converting enzyme 2 therefore blocking this enzyme could be of immense importance (Li et al., 2003; Han et al., 2006; Ge et al., 2013). This enzyme is also selected as one of the target proteins.
In the present study, some known antiviral drugs like Ritonavir, Lopinavir, Remdesivir, Oseltamivir and Ribavirin, along with some small molecules like Mycophenolic acid (MPA), Chloroquine, Hydroxychloroquine (HCQ), Pemirolast, Isoniazid and Eriodictyol which are being discussed as therapeutic agents against COVID-19, are used as candidate ligands for docking. Molecular docking was done using PyRx version 0.8 (Trott and Olson, 2010).
2. Materials and methods
2.1. Proteins/macromolecules
Different proteins from SARS-CoV-2 were selected which include main protease of SARS-CoV-2 Mpro/3CLpro [PDB ID: 6Y84] (Owen et al., 2020), the crystal structure of COVID-19 main protease in apo form [PDB ID: 6M03] (Zhang et al., 2020) and crystal structure of COVID-19 main protease [PDB ID: 6LU7] (Xu et al., 2020), Nsp10 [PDB ID: 6W75 B and D] and Nsp 16 [PDB ID: 6W75 A and C] (Minasov et al., 2020) the complex which plays a pivotal role in viral transcription; separate chains A, B and C of spike glycoprotein [PDB ID: 6VSB A,B and C chains] (Wrapp et al., 2020)], SARS protein receptor binding domain [PDB ID: 2GHV] ( Cao et al., 2020), Nsp9 RNA binding protein of SARS-CoV-2 [PDB ID: 6W4B] (Tan et al., 2020) and ADP ribose phosphatase of NSP3 from SARS CoV-2 [PDB ID: 6VXS] (Kim et al., 2020). We have also selected the protein from host cell, i.e. human cell, which is responsible for host virus interaction. Virus enters the cell through angiotensin-converting enzyme 2 (ACE2) receptor [PDB ID: 6M18] chains B and D (Yan et al., 2020) by binding with its spike protein. 3D structures were obtained from Protein Data Bank (https://www.rcsb.org/), in .pdb format (Berman et al., 2000).
2.2. Ligand/drug structures
Known antiviral drugs like Ritonavir, Lopinavir, Remdesivir, Oseltamivir and Ribavirin as well as Mycophenolic acid (MPA), Chloroquine, Hydroxychloroquine (HCQ), Pemirolast, Isoniazid and Eriodictyol, which are used to treat other ailments, were selected as a candidate ligands for docking against viral proteins.
The drug/ligand structures were obtained from the DrugBank database which is a comprehensive, freely accessible, online database containing information on drugs and drug targets (Wishart et al., 2018).
2.3. Determination of active sites
The Computed Atlas for Surface Topography of Proteins (CASTp) (Tian et al., 2018) and Biovia Discovery Studio 4.5 (Biovia, D.S 2019) were used to determine the amino acids in the active site of a protein (http://sts.bioe.uic.edu/castp/index.html?201l).
2.4. Molecular docking
Ligand and protein optimization were done using PyMOL version 2.3.3 [The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC]. For ligand optimization, the geometry of ligands was cleaned, whereas for protein, the water was removed. The docking was performed by using PyRx 0.8 (Trott and Olson, 2010). The docking analyses were performed using both Pymol as well as Biovia Discovery Studio 4.5 (Biovia, D.S 2019).
3. Results
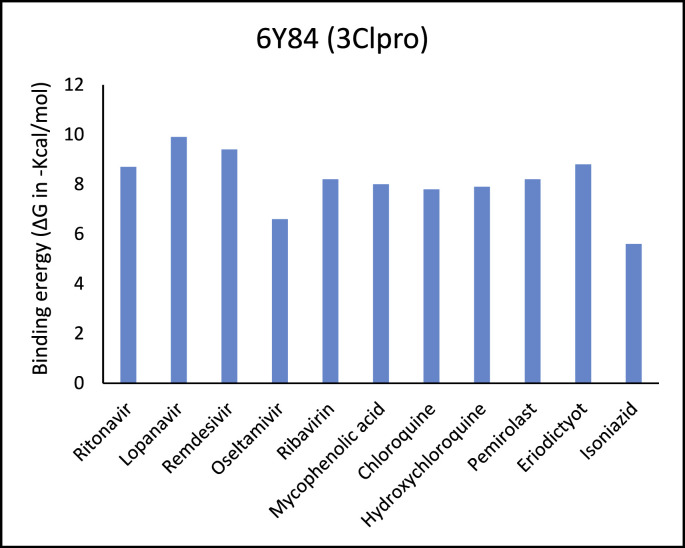
Table 1 represents proteins along with their PDB ID and structure which were used as targets for molecular docking. These proteins play an important role in viral replication and virus entry. Table 2 gives the details of ligands which were selected for molecular docking. Traditional structure-based method of molecular docking was done because of availability of 3D structures of SARS-CoV-2 virus. Table 3A, Table 3B B shows molecular docking data represented in terms of binding energy (ΔG) in Kcal/mole for selected viral target proteins (Table 3 A) and human receptor protein ACE-2 (Table 3 B) with all selected drugs. Result presented in this table clearly show that all these compounds are binding to all viral proteins as well as human ACE 2 to some extent. Different ligands are showing highest binding against different proteins and this data are presented in Table 4 . It was observed that Ritonavir is giving efficient binding energy for Nsp16, 6W75D, 6VSBC and 2GHV whereas Remdesivir is more active against 6M03, 6LU7, 6VSBB, 6W75B. It was observed that Lopinavir shows a highly efficient binding with key viral proteins like 6Y84, 6W4B, 6VSBA, 6VXS and ACE 2 receptor protein 6M18B and 6M18D. Out of the small molecules tested here, Eriodictyol has high binding efficiencies against all selected proteins. Nevertheless, Pemirolast also shows high binding efficiencies against 6M03 and 6W4B. Fig. 1 represents the graph of binding energy in –Kcal/mol of different drug ligands against 6Y84. It was observed that all ligands are binding to this target with good binding energies but binding of Lopinavir, Remdesivir, Eriodictyol and Ritonavir is more favorable that others (Fig. 1). Same drug ligands also show good binding energy against human ACE-2 suggesting their high potential for repurposing. We further analyzed only those docking poses which have binding energy less than −7.5 kcal/mol to understand the molecular interactions. Docking poses with less than −7.5 kcal/mol binding energy were shown in Fig. 2 along with the active site of respective protein. The docking analysis of these poses was done to check the interaction as amino acid level. The comparison of amino acids from specific protein which are binding to ligand and amino acids present in active site of that protein is shown in Table 5 . It was observed that in case of 6Y84 almost all drugs which are binding to this protein are binding to 3 amino acids from active site GLN127, GLU290 and PHE 191. In case of 6VXS, helicase of corona virus, Lopinavir and Pemirolast are binding with −7.9 kcal/mol and −7.6 kcal/mol respectively but both contain only one common amino acid LYS44 with protein active site. Eriodictyol has binding energy of −7.9 kcal/mol against 6VXS and the interactions involves ALA38, ASN40, GLY46, GLY47, GLY48, VAL49, ALA50, PRO125, LEU126, LEU127, SER128, ALA129, ILE131, PHE132 amino acids. All these amino acids are present in the active site of 6VXS protein. Remdesivir and Pemirolast are binding to 6M03, the apo form of main protease, but not involving any amino acid from active site. In case of Ritonavir binding to 2GHV, the receptor binding domain of spike protein of the virus PHE361, SER362 GLY368 amino acids from the active site are involved in the interaction. The 6VSB is a spike protein of the virus to which Eriodictyol is binding with the energy of −7.6 kcal/mol. This interaction involves ASN544, VAL576, PHE543, LEU546, PHE565, ALA522, ARG577, PRO521, PRO579, GLN564 amino acids from the active site.
Table 1.
Structure of viral proteins which were used as a targets for molecular docking with PDB ID (4-character unique identifier of every entry in the Protein Data Bank) and source organism.
| Name of the protein and description | Source | PDB ID | Structure of protein |
|---|---|---|---|
| Mpro/3CLpro (Main protease of SARS CoV 2) | SARS CoV 2 | 6Y84 |  |
| The crystal structure of COVID-19 main protease | SARS CoV 2 | 6LU7 |  |
| The crystal structure of COVID-19 main protease in apo form | SARS CoV 2 | 6M03 |  |
| Nsp10 (Plays a crucial role in viral transcription by stimulating both nsp14 3′-5′ exoribonuclease and nsp16 2′-O-methyltransferase activities.) Nsp 16 (S-adenosylmethionine-dependent (nucleoside-2′-O)-methyltransferase only active in the presence of its activating partner nsp10) |
SARS CoV 2 | 6W75 B 6W75D 6W75 A 6W75D |
 |
| Spike glycoprotein | SARS CoV 2 | 6VSBA 6VSBB 6VSBC |
 |
| SARS protein receptor binding domain | SARS CoV | 2GHV |  |
| Nsp9 (RNA binding protein of SARS CoV 2) and SARS CoV 2 helicase | SARS CoV 2 | 6W4B |  |
| ADP ribose phosphatase of NSP3 from SARS CoV-2 | SARS CoV 2 | 6VXS |  |
| Angiotensin-converting enzyme 2 (ACE2) receptor (Receptor through which virus enters the cell) | Human | 6M18B 6M18D |
 |
Table 2.
Ligands which were selected for molecular docking along with its structure obtained from DrugBank and the important activities.
| Sr. No. | Name | Structure | Function |
|---|---|---|---|
| 1 | Ritonavir |  |
Antiretroviral protease inhibitor, used as common anti HIV in combination with other protease inhibitors |
| 2 | Lopinavir | 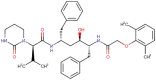 |
Antiretroviral protease inhibitor, used as common anti HIV in combination with other protease inhibitors |
| 3 | Remdesivir | 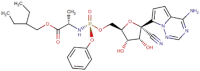 |
Nucleotide analog, novel antiviral treatment for Ebola virus disease and Marburg virus infections, antiviral activity against other single stranded RNA viruses including MERS and SARS viruses |
| 4 | Oseltamivir | 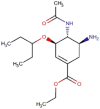 |
An antiviral medication used to treat and prevent influenza A and influenza B (flu) |
| 5 | Ribavirin | 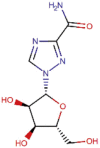 |
An antiviral medication used to treat RSV infection, hepatitis C and some viral hemorrhagic fevers |
| 6 | Mycophenolic acid (MPA) | 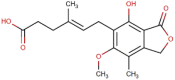 |
An immunosuppressant medication |
| 7 | Chloroquine | 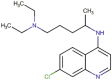 |
Medication primarily used to prevent and treat malaria, used in compassionate treatment of patients with coronavirus infections during Covid19 pandemic |
| 8 | Hydroxychloroquine (HCQ) | 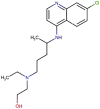 |
Prevent and treat malaria, an experimental treatment for coronavirus disease 2019 (COVID-19) |
| 9 | Pemirolast | 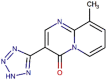 |
Pyrimidinone derivative with anti-allergic activity, anti-inflammatory |
| 10 | Eriodictyol | 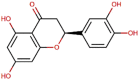 |
Anti-inflammatory |
| 11 | Isoniazid | 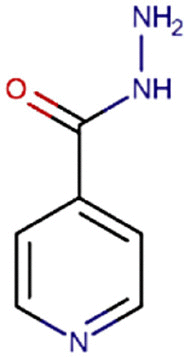 |
Antituberculosis, anti HIV |
Table 3A.
Molecular docking data represented in terms of binding energy (ΔG) in Kcal/mole for viral target proteins with drug ligands.
| Sr. No | Drug | Binding energy ΔG(Kcal/Mol) |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6M03 | 6LU7 | 6Y84 | 6W4b | 6VSBA | 6VSBB | 6VSBC | 2GHV | 6VXS | 6W75A | 6W75C | 6W75B | 6W75D | ||
| 1 | Ritonavir | −7.2 | −6.6 | −8.7 | −5.8 | −6.2 | −5.6 | −7 | −7.6 | −6.3 | −7.3 | −7.2 | −6.1 | −7.3 |
| 2 | Lopinavir | −7.3 | −5.9 | −9.9 | −6.4 | −6.9 | −6.2 | −6.6 | −7.1 | −7.9 | −6.9 | −7.3 | −6.5 | −6.9 |
| 3 | Remdesivir | −7.8 | −7.2 | −9.4 | −6.2 | −6.5 | −6.8 | −6.2 | −6.9 | −6.9 | −6.9 | −7.6 | −7 | −6.9 |
| 4 | Oseltamivir | −6.1 | −5.2 | −6.6 | −4.3 | −5.3 | −6 | −5.4 | −5.8 | −5.7 | −5.7 | −6.2 | −5.6 | −5.7 |
| 5 | Ribavirin | −6 | −6.3 | −8.2 | −5.6 | −5.5 | −5.5 | −5.4 | −6.5 | −6.2 | −6.6 | −6.6 | −5.9 | −6.6 |
| 6 | Mycophenolic acid | −6.4 | −6.4 | −8 | −5.3 | −4.7 | −4.8 | −5.4 | −6.6 | −6.3 | −6.5 | −6.9 | −6.3 | −6.5 |
| 7 | Chloroquine | −6.5 | −5.4 | −7.8 | −5.2 | −4.8 | −5.4 | −4.8 | −5.4 | −6.4 | −6.7 | −7.1 | −5.9 | −6.7 |
| 8 | Hydroxychloroquine | −6.1 | −5.6 | −7.9 | −5.5 | −5.8 | −5.6 | −5.5 | −5.8 | −5.8 | −6.5 | −7 | −6.4 | −6.5 |
| 9 | Pemirolast | −7.7 | −6.4 | −8.2 | −6.5 | −6.6 | −6.2 | −6.3 | −6.7 | −7.6 | −6.7 | −6.9 | −6.4 | −6.7 |
| 10 | Eriodictyol | −7.4 | −6.7 | −8.8 | −6.5 | −7 | −6.7 | −7.6 | −7.3 | −7.8 | −7.5 | −7.5 | −7 | −7.5 |
| 11 | Isoniazid | −4.8 | −4.6 | −5.6 | −4.4 | −5 | −4.4 | −5 | −5.1 | −5 | −5.2 | −5.1 | −4.8 | −5.2 |
Table 3B.
Molecular docking data represented in terms of binding energy (ΔG) in Kcal/mole for ACE2 protein with drug ligands.
| Sr. No. | Drug | Binding energy AG(Kcal/Mol) |
|
|---|---|---|---|
| 6M18B | 6M18D | ||
| 1 | Ritonavir | −8.2 | −8.1 |
| 2 | Lopinavir | −8.6 | −8.9 |
| 3 | Remdesivir | −8.2 | −7.7 |
| 4 | Oseltamivir | −6 | −5.8 |
| 5 | Ribavirin | −6.2 | −6.3 |
| 6 | Mycophenolic acid | −6.5 | −6.4 |
| 7 | Chloroquine | −5.8 | −5.2 |
| 8 | Hydroxychloroquine | −6.2 | −6.5 |
| 9 | Pemirolast | −7.3 | −7 |
| 10 | Eriodictyol | −7.5 | −7.4 |
| 11 | Isoniazid | −5.2 | −5.2 |
Table 4.
Drug giving lowest binding energy in Kcal/mole for a particular protein.
| Sr.No. | Protein | Drug | Binding energy in Kcal/mol |
|---|---|---|---|
| 1 | 6M03 | Remdesivir | −7.8 |
| 2 | 6LU7 | Remdesivir | −7.2 |
| 3 | 6Y84 | Lopinavir | −9.9 |
| 4 | 6W4b | Eriodictyol and Pemirolast | −6.5 |
| 5 | 6VSBA | Eriodictyol | −7 |
| 6 | 6VSBB | Remdesivir | −6.8 |
| 7 | 6VSBC | Eriodictyol | −7.6 |
| 8 | 2GHV | Ritonavir | −7.6 |
| 9 | 6VXS | Lopinavir | −7.9 |
| 10 | 6W75A | Eriodictyol | −7.5 |
| 11 | 6W75C | Remdesivir | −7.6 |
| 12 | 6W75B | Remdesivir | −7 |
| 13 | 6W75D | Eriodictyol | −7.5 |
| 14 | 6M18B | Lopinavir | −8.6 |
| 15 | 6m18D | Lopinavir | −8.9 |
Fig. 1.
Represents the binding energy in – Kcal/mole of all ligand molecules against 6Y84, the main protease of SARS-CoV-2.
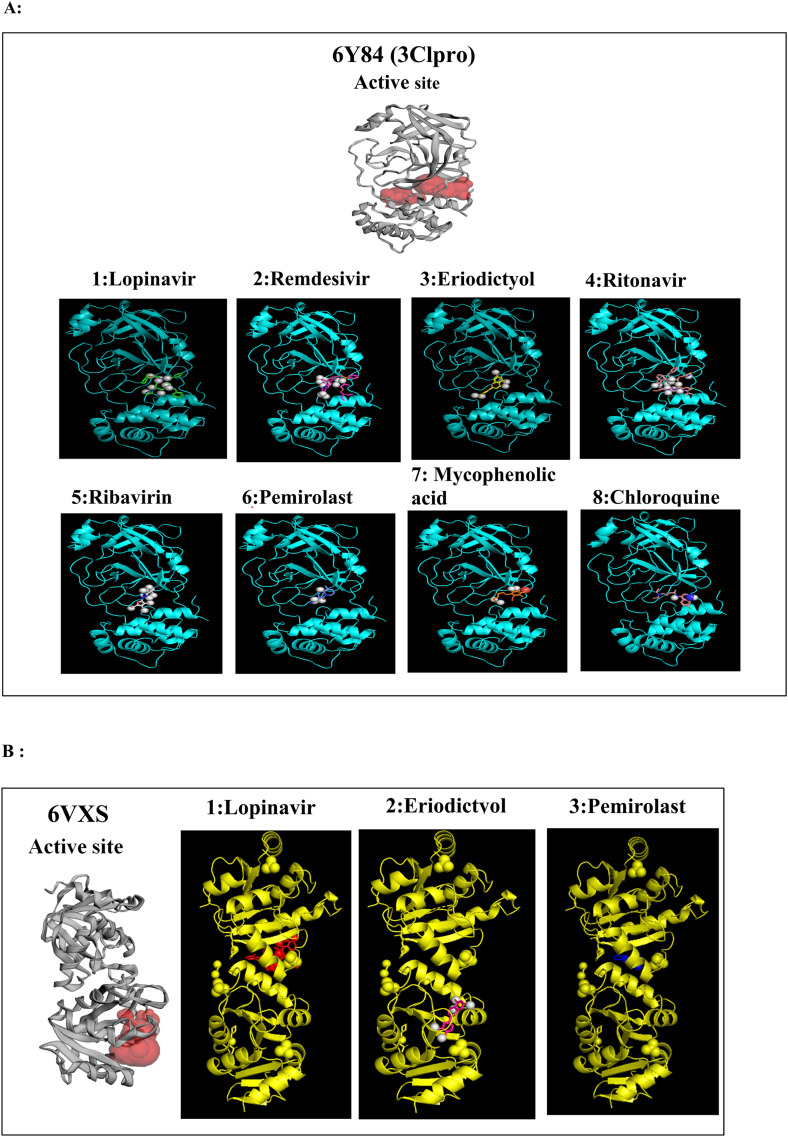
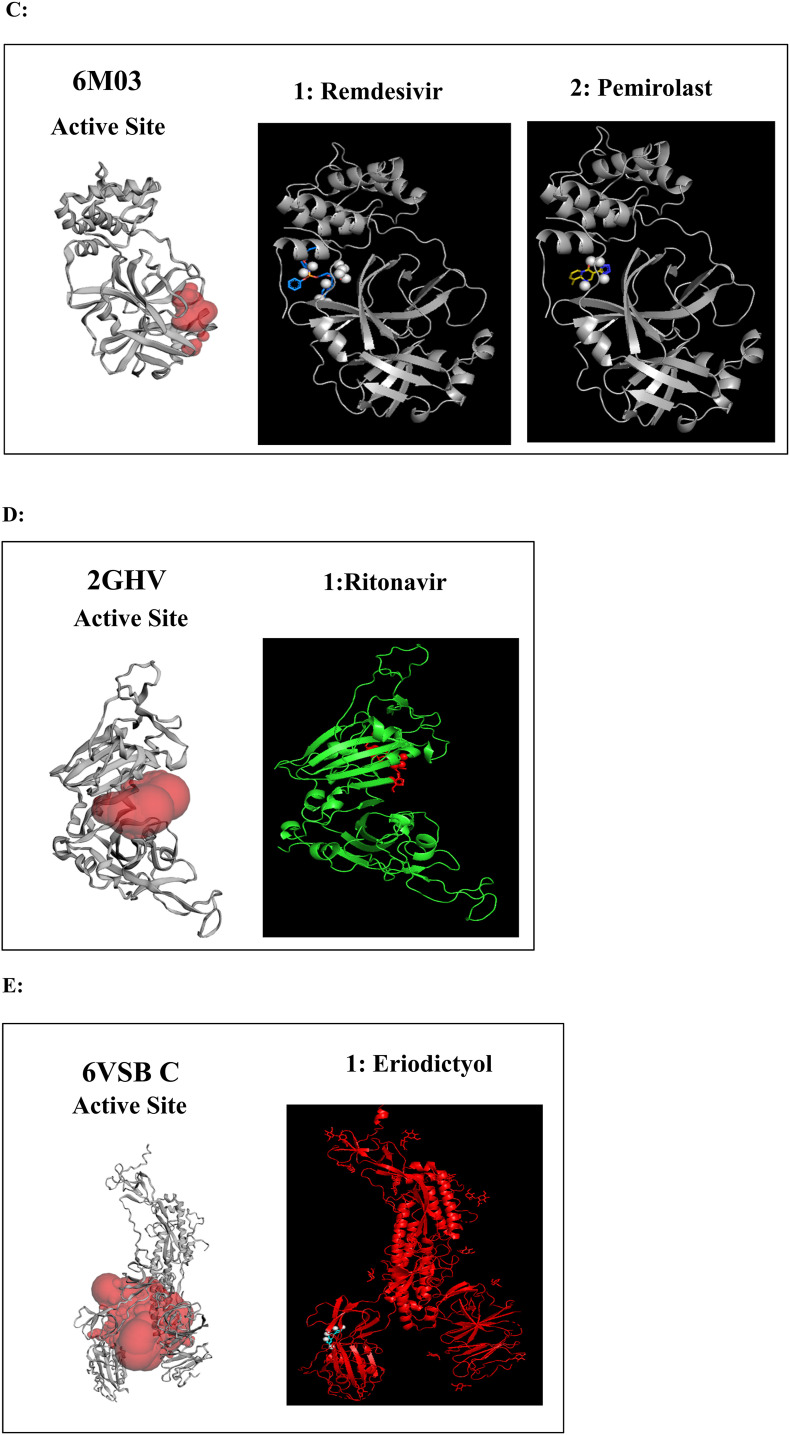
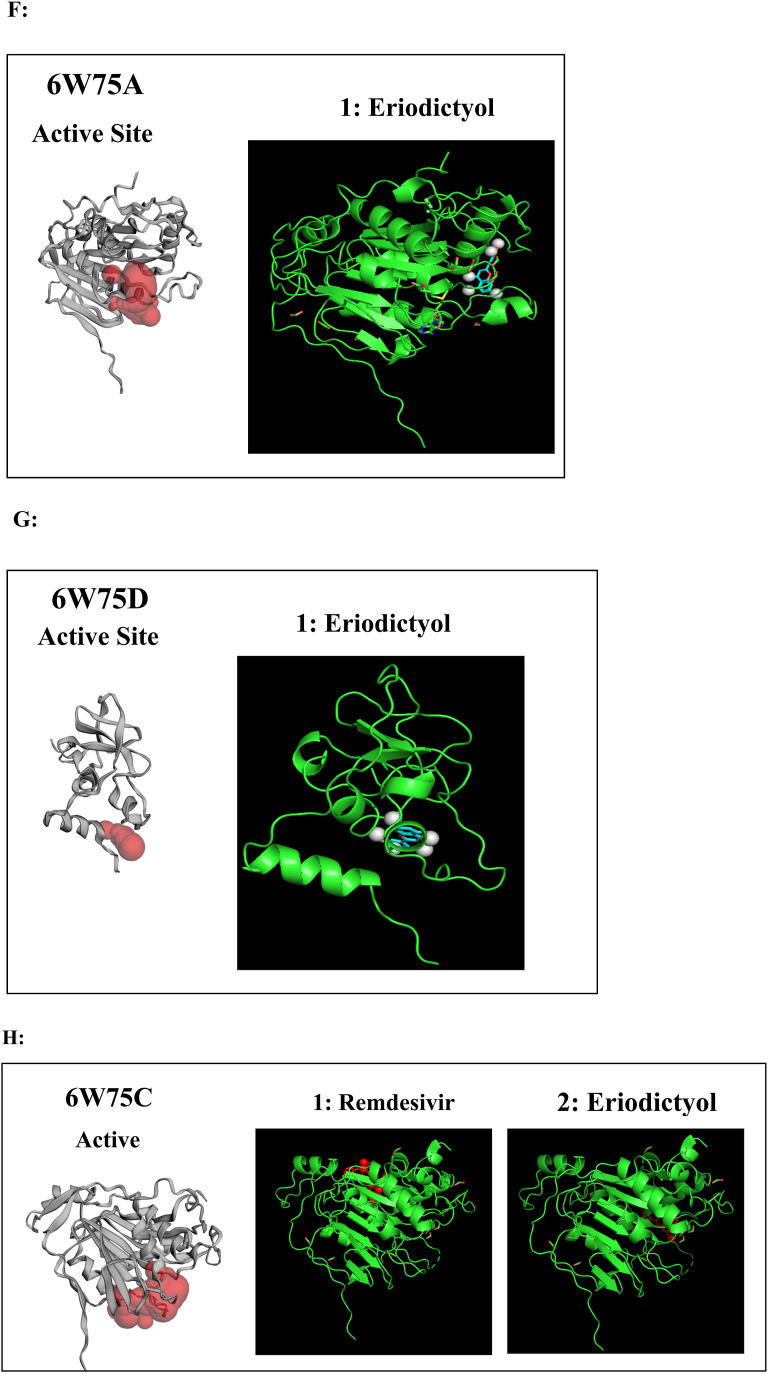
Fig. 2.
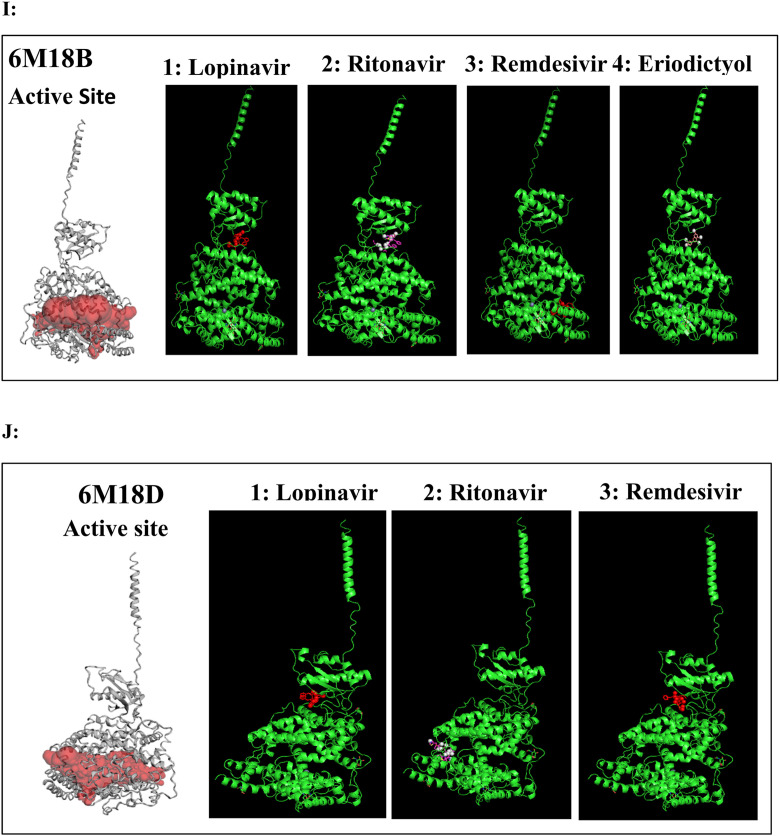
A to J: Represents the active site of a particular protein and binding poses of drug ligands showing binding energy less than -7.5 Kcal/mol. A: 3Clpro (6Y84) with 1: Lopinavir, 2: Remdesivir, 3: Eriodictyol, 4: Ritonavir, 5: Ribavirin, 6: Pemirolast, 7: Mycophenolic acid (MPA), 8: Chloroquine, B: Helicase (6VXS) with 1: Lopinavir, 2: Eriodictyol 3: Pemirolast, C: 3Clpro (6M03) with 1: Remdesivir, 2: Pemirolast, D: Receptro binding domain of S protein (2GHV) with 1: Ritonavir, E : S protein (VSB C chain) with 1: Eriodictyol, F: NsP10/16 complex (6W75A) with 1: Eriodictyol, G: NsP10/16 complex (6W75D) with 1: Eriodictyol, H: NsP10/16 complex (6W75C) with 1: Remdesivir, 2: Eriodictyol, I: ACE 2 receptor (6M18B) with 1: Lopinavir, 2: Ritonavir, 3: Remdesivir, 4: Eriodictyol, J: ACE 2 receptor (6M18D) with 1: Lopinavir, 2: Ritonavir, 3: Remdesivir.
Table 5.
Specific binding site for a particular drug ligand for a specific protein with binding energy less than −7.5 kcal/mol. Binding site was compared with active site and common amino acids are listed.
| Sr. No. | Active site | Name of the drug with binding energy in –Kcal/mol | Binding with drug with binding energy |
|---|---|---|---|
| 1. |
6Y84 (Mpro/3CLpro (Main protease of SARS-CoV-2)) MET6, ALA7, PHE8, PRO9, VAL104, ILE106, GLN107, PRO108, GLY109, GLN110, THR111, GLN 127, PRO132, ASN151, ILE152, ASP153, TYR154, CYS156, SER158, ILE200, THR201, VAL202, ASN203, GLU240, PRO241, ASP245, HIS246, ILE 249, GLU290, PHE291, THR292, PRO293, PHE294, ASP295, ARG298, GLN299, GLY302, VAL303, THR304 |
Lopinavir (−9.9) GLN 127, GLU290, PHE291 |
 |
| Remdesivir (−9.4) GLN 127, GLU290, PHE291 |
 |
||
| Eriodictyol (−8.8) | 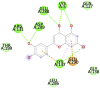 |
||
| GLN 127, GLU290 | |||
| Ritonavir (−8.7) GLN 127, GLU290, PHE291 |
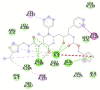 |
||
| Ribavirin (−8.2) GLN127, GLU290 |
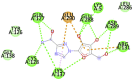 |
||
| Pemirolast (−8.2) GLN127, GLU290 |
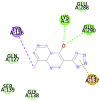 |
||
| Mycophenolic acid (−8) GLN127, GLU290 |
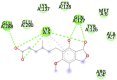 |
||
| Hydroxychloroquine (−7.9) GLN 127, GLU290, PHE291 |
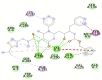 |
||
| Chloroquine (- 7.8) GLU290, PHE291 |
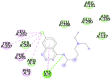 |
||
| 2 |
6VXS (ADP ribose phosphatase of NSP3 from SARS CoV-2) ALA21, ASP22, ILE23, VAL24, ALA38, ALA39, ASN40, LYS44, GLY46, GLY47, GLY48, VAL49, ALA50, ALA52, VAL95, GLY97, PRO125, LEU126, LEU127, SER128, ALA129, GLY130, ILE131, PHE132, PRO136, ALA154, VAL155, PHE156, ASP157, LEU160 |
Lopinavir(-7.9) LYS44 |
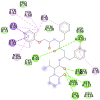 |
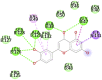
| Eriodictyol (−7.8) ALA38, ASN40, GLY46, GLY47, GLY48,VAL49, ALA50, PRO125, LEU126, LEU127, SER128, ALA129, ILE131, PHE132 |
 |
||
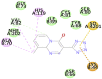
| Pemirolast (−7.6) LYS44 |
 |
||
| 3 |
6M03 (COVID-19 main protease in apo form) CYS 22, GLY23, THR24, THR25, THR26, LEU27, HIS41, VAL42, ILE43, CYS44, THR45, SER46, MET49, LEU50, LYS61, PHE140, LEU141, ASN142, GLY143, SER144, CYS145, HIS163, HIS164, MET165, GLU166, LEU167, PRO168, ARG188, GLN189, THR190, GLN192 |
Remdesivir (−7.8) | 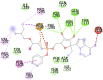 |
| Pemirolast (−7.7) | 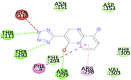 |
||
| 4 |
2GHV (SARS spike protein receptor binding domain) Chain E PHE361, SER362, THR363, PHE364, LYS365, CYS366,TYR367, GLY368, GLY391, ASP392, VAL394, ARG395, ILE397, ALA398, PRO399, GLN401, ASP414, PHE416, VAL420, ALA422, ILE489, GLY490, TYR494 Chain C PHE361, SER362, THR363, PHE364, LYS365, CYS366, TYR367, GLY368, GLY391, ASP392, VAL394, ARG395, ILE397, ALA398, PRO399,GLY400, GLN401, ASP414, PHE416, VAL420, ALA422, ILE489, GLY490, TYR494 |
Ritonavir (−7.6) Chain E PHE361, SER362 Chain C GLY368 |
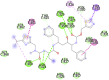 |
| 5 |
6VSB C (Spike glycoprotein) Active site contains many amino acids |
Eriodictyol (−7.6) ASN544, VAL576, PHE543, LEU546, PHE565, ALA522, ARG577, PRO521, PRO579, GLN564 |
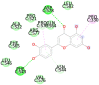 |
| 6 |
6W75A (NSP10 – NSP16 Complex from SARS-CoV-2) ASN6841, LYS6844, TYR6845, HIS6867, PHE6868, GLY6869, ALA6870, GLY6871, SER6872, ASP6873, PRO6878, GLY6879, SER6896, ASP6897, LEU6898, ASN6899, ASP6900, PHE6901, GLY6911, ASP6912, CYS6913, ASP6928, MET6929, TYR6930, ASP6931, PRO6932, LYS6933, GLY6946, PHE6947, LYS6968 |
Eriodictyol (−7.8) | 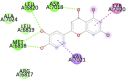 |
| 7 |
6W75C (NSP10 – NSP16 Complex from SARS-CoV-2) ASN6841, LYS6844, TYR6845, HIS6867, PHE6868, GLY6869, ALA6870, GLY6871, SER6872, ASP6873, PRO6878, GLY6879, SER6896, ASP6897, LEU6898, ASN6899, ASP6900, PHE6901, GLY6911, ASP6912, CYS6913, ASP6928, MET6929, TYR6930, ASP6931, PRO6932, LYS6933, GLY6946, PHE6947, LYS6968 |
Remdesivir (−7.6) | 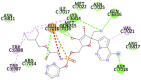 |
| Eriodictyol (−7.5) | 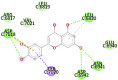 |
||
| 8. | 6W75D (NSP10 – NSP16 Complex from SARS-CoV-2) ALA4273, VAL4274, ASP4275, ALA4276, ALA4277, TYR4329, ILE4334, ASP4335, HIS 4336, PRO4337 | Eriodictyol (−7.5) TYR4329, HIS 4336, PRO4337 | 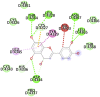 |
| 9 |
6M18B (Angiotensin-converting enzyme 2 (ACE2) receptor) SER623, LYS625, SER626, ALA627, LEU628, GLY629, ASP630, LYS631, ARG678, ARG705, ARG708, SER709, ASN712, ASP713, ARG716, LEU717, ASN718, ASP719, ASN720, SER721, GLU723, ILE727, GLN728, PRO729, THR730, LEU731, GLY732, PRO733, PRO734, ASN735, GLN736, PRO737, PRO738, VAL739, ILE741, ILE744, VAL745, VAL748, VAL749, VAL752, ILE753, VAL755, GLY756, VAL758, ILE759, LEU760, PHE762, THR763, ARG766, ASP767, ARG768, SER623 |
Lopinavir (−8.6) | 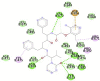 |
| Ritonavir(-8.2) | 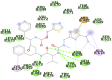 |
||
| Remdesivir (−8.2) | 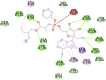 |
||
| Eriodictyol (−7.5) | 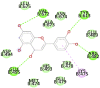 |
||
| 10 |
6M18D (Angiotensin-converting enzyme 2 (ACE2) receptor) SER623, LYS625, SER626, ALA627, LEU628, GLY629, ASP630, LSY631, ARG678, ARG705, ARG708, SER709, ASN712, ASP713, ARG716, LEU717, ASN718, ASP719, ASN720, SER721, GLU723, ILE727, GLN728, PRO729, THR730, LEU731, GLY732, PRO733, PRO734, ASN735, GLN736, PRO737, PRO738, VAL739, ILE741, ILE744, VAL745, VAL748, VAL749, VAL752, ILE753, VAL755, GLY756, VAL758, ILE759, LEU760, PHE762, THR763, ARG766, ASP767, ARG768 |
Lopinavir (−8.9) | 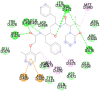 |
| Ritonavir (−8.1) | 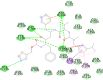 |
||
| Remdesivir (−7.7) | 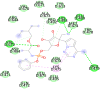 |
In case of 6W75 Nsp10/16 complex, Remdesivir and Eriodictyol are binding to the protein but only Eriodictyol is directly interacting with amino acids TYR4329, HIS 4336, PRO4337 which are present in active site of 6W75 chain C. Lopinavir, Ritonavir, Eriodictyol are binding at 6M18B and Lopinavir, Ritonavir, Remdesivir are binding at 6M18D but their binding but is not in the active site.
The comparison of anti SARS-CoV-2 potential of different drugs is summarized in Table 6 . It was observed that all drug ligands except Oseltamivir and Isoniazid are binding to 6Y84, the main protease in virus, in active site. Based on this study all drug ligands except Oseltamivir and Isoniazid have anti-SARS-CoV-2 potential. Lopinavir and Pemirolast are binding in the active site of 6VXS other than 6Y84 and Ritonavir is binding in active site of 2GHV along with 6Y84. Among all these ligands, Eriodictyol has the highest potential to be repurposed as an antiviral against SARS-CoV-2 since it is binding to 6VXS, 6VSB C, 6W75D along with 6Y84.
Table 6.
Summary of anti-SARS-CoV-2 potential of drug ligands.
| Sr. No | Name | Name of Protein to which binding is in active site | anti-SARC-CoV-2 potential |
|---|---|---|---|
| 1 | Ritonavir | 6Y84 (Mpro/3CLpro (Main protease of SARS-CoV-2) 2GHV (SARS spike protein receptor binding domain) |
Yes |
| 2 | Lopinavir | 6Y84 (Mpro/3CLpro (Main protease of SARS-CoV-2) 6VXS (ADP ribose phosphatase of NSP3 from SARS CoV-2) |
Yes |
| 3 | Remdesivir | 6Y84 (Mpro/3CLpro (Main protease of SARS-CoV-2) | Yes |
| 4 | Oseltamivir | – | No |
| 5 | Ribavirin | 6Y84 (Mpro/3CLpro (Main protease of SARS-CoV-2) | Yes |
| 6 | Mycophenolic acid (MPA) | 6Y84 (Mpro/3CLpro (Main protease of SARS-CoV-2) | Yes |
| 7 | Chloroquine | 6Y84 (Mpro/3CLpro (Main protease of SARS-CoV-2) | Yes |
| 8 | Hydroxychloroquine (HCQ) | 6Y84 (Mpro/3CLpro (Main protease of SARS-CoV-2) | Yes |
| 9 | Pemirolast | 6Y84 (Mpro/3CLpro (Main protease of SARS-CoV-2) 6VXS (ADP ribose phosphatase of NSP3 from SARS CoV-2) |
Yes |
| 10 | Eriodictyol | 6Y84 (Mpro/3CLpro (Main protease of SARS-CoV-2) 6VXS (ADP ribose phosphatase of NSP3 from SARS CoV-2) 6VSB C (Spike glycoprotein) 6W75D (NSP10 – NSP16 Complex from SARS-CoV-2) |
Yes |
| 11 | Isoniazid | – | No |
4. Discussion
Since the current outbreak of viral epidemic, scientists around the world are taking enormous efforts to understand this new virus and pathophysiology of the disease. Overall current knowledge of the SARS-CoV-2 virus as well as previous understanding of SARS-CoV 2003 and MERS-CoV 2012 is helpful to discover new therapeutic agents, targets and vaccines (Wu et al., 2020; Jiang et al., 2020). Considering the nature of SARS-CoV-2 and its high infection rate, finding new therapeutic target and development of a vaccine is actually a race against time. There is no treatment available to kill the virus or to stop the infection till date. Therefore, finding broad-spectrum inhibitors which may reduce the effects of human coronavirus infection is challenging (Liu et al., 2020). The fastest strategy for such a widely spreading infectious diseases is to utilize the existing medication of other diseases i.e. repurposing of drug. Such drugs are already approved for their safety and it's ADME properties are already known (Smith and Smith, 2020). Unavailability of protein structures of SARS-CoV-2 was one of the major hurdles to curate the huge bulk of antivirals for studying molecular interaction. As 3D structures of many proteins from SARS-CoV-2 are reconstructed and deposited in protein databases, we decided to use these structures to study the molecular interactions of known antivirals.
We have selected viral proteins which are important for viral replication and viral entry to host cell. One such key protein is 3CLpro which is the main protease of the virus. It is continuously discussed as a potential target for drug development (Khaerunnisa et al., 2020; Xu et al., 2020; Zhang et al., 2020). The first long polypeptides of virus become functional only when they are cut into smaller pieces by 3CLpro proteases. Thus, the propagation of Coronavirus is highly dependent on proteases. This protease cuts the long polypeptide at the recognition sequence at most sites is Leu-Gln↓(Ser,Ala,Gly) (↓ marks the cleavage site). Since no human proteases with a similar cleavage specificity are known, blocking this protease would be beneficial without disturbing host cell activities (Zhang et al., 2020). Another important target we selected was the envelope anchored spike protein from the virus and receptor binding domain of this protein The spike protein is a glycoprotein which binds with human cell receptor by its receptor binding domain, and enters in the cells (Wan et al., 2020; Li, 2016). SARS-CoV-2 helicase is used as another target. This is responsible for unwinding RNA-DNA duplex during virus replication (Jiang et al., 2020). Nsp10/16 complex is a nonstructural protein that plays an important role in viral translation and could be of importance as a target for drug development (Wang et al., 2015; Decroly et al., 2011). A single strand or RNA binding protein was also selected as target. Virus enters the cell via angiotensin receptor converting enzyme 2 therefore blocking this protein could be of immense importance (Wan et al., 2020).
During docking it was observed that all drug ligands except Oseltamivir and Isoniazid are binding to 6Y84 with binding energy between binding energy between −7.5 kcal/mol and −9.9 kcal/mol but interaction includes only 3 amino acids. As expected, Lopinavir is showing minimum energy for binding the 6Y84 target followed by Remdesivir. This supports the idea of repurposing the two drugs for the treatment of COVID-19. It was reported that Remdesivir is more effective as compared to the combination of Lopinavir, Ritonavir and interferon beta in improvement of pulmonary function in MERS-CoV in mouse model (Sheahan et al., 2020). Lopinavir and Ritonavir combination treatment trials were reported by Cao et al., (2020) in total 99 patients with no benefit (Cao et al., 2020). In our study we observed that Ritonavir, Lopinavir as well as Remdesivir are binding with main protease of SARS-CoV-2 virus. High quality randomized clinical trials are needed to find the efficacy of these drugs in COVID-19. Ritonavir, Lopinavir and Remdesivir are also binding with 6M18 i.e. ACE-2 receptor of human cell but this binding is not at active site. As all three drugs are binding to viral main protease and ACE 2, these drugs might be the important candidates for repurposing if mechanism of action is known. Wan et al. recently reported that Remdesivir and Chloroquine effectively inhibit the recently emerged novel coronavirus (2019-nCoV) in vitro (Wang et al., 2020). Ritonavir, apart from having the individual protease inhibitor activity, is also a protease inhibitor booster which increases the pharmacokinetics of another protease inhibitor if used in combination (Hull and Montaner, 2011). Pulmonary discomfort is one of the important symptoms in COVID-19. Pemirolast, is a mast cell stabilizer used in anti-allergy medication for treating chronic asthma (Kemp et al., 1992). Eriodictyol, is a flavanone found in Yerba Santa a plant native to North America and is a traditional herbal remedy used for asthma and treating common colds (Bown and America, 1995). Asthma drugs may act locally to get control over the respiratory system. This will reduce the hospitalization time and improve the general health of the patient. Therefore, these drugs were tested to check their potential role in suppressing the viral replication via interacting with viral proteins. One interesting observation here is that the drug Eriodictyol is binding with all selected proteins with the binding energy between −6.7 kcal/mol and 8.8 kcal/mol. In case of 6VXS, the viral helicase, the Eriodictyol is binding with amino acids ALA38, ASN40, GLY46, GLY47, GLY48, VAL49, ALA50, PRO125, LEU126, LEU127, SER128, ALA129, ILE131, PHE132. All these amino acids are present in the active site of helicase. This binding has 2 van der Waals interactions and 6 hydrogen bonds. There is locking at left terminal oxygen with amino acids 126, 127, 128 and 129 making it a strong interaction (Table 5). This strong interaction may affect the activity of enzyme. Eriodictyol is binding to 6VSB C viral spike protein, 6W75D Nsp10/16 complex and 6Y84 main protease along with 6VXS viral helicase with efficient binding energy and within active site. The prediction of protein functioning based on mere molecular docking in case of small molecules is difficult. Eriodictyol, bitter taste masker and is known to have anti-inflammatory, antioxidant, cytotoxic, antidiabetic properties (Shukla et al., 2019; Rossato et al., 2011; Habtemariam and Dagne, 2009; Zhang et al., 2012) It is reported to suppress the lipopolysaccharide (LPS)-induced inflammatory responses in macrophages by reducing nitric oxygen, pro-inflammatory cytokines in Raw 264.7 cells (Lee, 2011). Zhu et al. had reported anti-inflammatory action of it against LPS induced acute lung injury (Zhu et al., 2015). This data definitely suggests that Eriodictyol might affect the virus lifecycle by interacting with multiple proteins. With it's anti-inflammatory, antioxidant activity and positive drug score Eriodictyol could be a strong candidate for repurposing in case of SARS-CoV-2. Antimalarial drugs Hydroxychloroquine and Chloroquine are discussed for the treatment against this virus (Al-bari, 2017; Lai et al., 2020; Kumar et al., 2020). A recent preprint in BioRxiv indicates that Chloroquine, and azithromycin, another drug of interest in COVID-19 context, work by altering pH of intracellular compartments in respiratory epithelial cells with potential effects on viral life cycle and additional anti-inflammatory effect. Chloroquine also seems to act as a zinc ionophore, that allows extracellular zinc to enter the cell and inhibit viral RNA-dependent RNA polymerase (Xue et al., 2014; Velthuis et al., 2010). Hydroxychloroquine is also suggested to work in the same manner as that of Chloroquine. In this study Chloroquine and Hydroxychloroquine drugs are interacting with main protease 6Y84 in active site but are not showing efficient binding energy with any selected proteins. This suggests that their role is not to kill the virus directly but to change the cells environment which is consistent with previous reports (Vincent et al., 2005; Salata et al., 2017).
RNA viruses are subject to genetic mutations and recombination which leads to cross contamination and emergence of novel infectious diseases. The world is still not prepared for effective management of such diseases. This increases the need of research and development of new drugs for coronaviruses. This work is a small attempt in the process of repurposing the existing drugs for treatment of SARS-CoV-2. The data shows efficient binding of Ritonavir, Lopinavir and Remdesivir with 3CLpro, the main protease of the virus and ACE 2 receptor of human cell. This supports the need of more randomized trials of these drugs against COVID-19 in spite of discouraging results in the preliminary clinical trial. We also impart the need of clinical trials of one more flavonoid drug - Eriodictyol which is showing very good binding energies with all viral proteins as well as ACE 2. In case of SARS-CoV-2 helicase Eriodictyol binding is stronger as compared to other interactions. .SARS-CoV-2 helicase is a potential drug target as it recognizes the RNA: DNA duplex which is unique feature of virus (Jang et al., 2020). It also binds within the active site of viral spike glycoprotein C chain with 10 amino acids. It is binding with ACE 2 receptor on human cells. Since all these proteins are important in viral replication, Eriodictyol can become a multi-target molecule which can be a strong candidate for repurposing against SARS- CoV- 2.
5. Conclusion
Drug repurposing is the best approach for finding out solution for treatment of novel coronavirus infection. If the targets and mechanism of action of these drugs are known then formulating treatment would be easier. Therefore, the extensive work carried out in this study to understand the interaction of multiple viral proteins and different drug ligands which are discussed for treatment in COVID-19. Proteins important in viral replication were selected in the study because inhibiting these proteins might be useful to block the initiation of infection and chain of replication. In silico molecular docking study enhanced the potential of Lopinavir, Ritonavir, Remdesivir as a candidate drugs for treatment of COVID-19. Eriodictyol is binding to multiple targets like 6Y84, 6VXS, 6VSB C, 6W75D with efficient binding energy and within the active site. Its strong interaction in active site of SARS-CoV-2 helicase (6VXS) reinforces its application. This small molecule with its anti-inflammatory activity has emerged as a promising candidate for treatment of corona infection.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Rujuta R. Deshpande: Methodology, Software, Validation, Data curation, Writing - original draft, Visualization. Arpita Pandey Tiwari: Conceptualization, Investigation, Resources, Writing - original draft. Narendra Nyayanit: Conceptualization, Methodology, Formal analysis, Data curation. Manisha Modak: Conceptualization, Methodology, Formal analysis, Writing - original draft, Writing - review & editing, Visualization, Supervision.
Declaration of competing interest
The authors declare ‘‘No conflict of interest”.
Acknowledgement
All authors are thankful to Dr. Abhijit Chavan, Department of Chemistry, Sir Parashurambhau College, Pune for discussions regarding molecular interactions. Authors also extend thanks to Mr. Bhupendra Dandekar JRF, TIFR, Hyderabad, for resolving the queries in computational studies.
References
- Al-bari A.A. Targeting endosomal acidification by Chloroquine analogs as a promising strategy for the treatment of emerging viral diseases. Pharmacol. Res. Perspect. 2017;5:1–13. doi: 10.1002/prp2.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bown D., America H.S. Dorling Kindersley; 1995. Encyclopedia of Herbs and Their Uses. [Google Scholar]
- Cao B., Wang Y., Wen D., Liu W., Wang Jingli, Fan G., Ruan L., Song B., Cai Y., Wei M., Li X., Xia J., Chen N., Xiang J., Yu T., Bai T., Xie X., Zhang L., Li C., Yuan Y., Chen H., Li Huadong, Huang H., Tu S., Gong F., Liu Y., Wei Y., Dong C., Zhou F., Gu X., Xu J., Liu Z., Zhang Y., Li Hui, Shang L., Wang K., Li K., Zhou X., Dong X., Qu Z., Lu S., Hu X., Ruan S., Luo S., Wu J., Peng L., Cheng F., Pan L., Zou J., Jia C., Wang Juan, Liu X., Wang S., Wu X., Ge Q., He J., Zhan H., Qiu F., Guo L., Huang C., Jaki T., Hayden F.G., Horby P.W., Zhang D., Wang C. A trial of lopinavir–ritonavir in adults hospitalized with severe Covid-19. N. Engl. J. Med. 2020;382:1787–1799. doi: 10.1056/NEJMoa2001282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan N. 2020. Possible Drug Candidates for COVID-19 Possible Drug Candidates for COVID-19. ChemRxiv. [DOI] [Google Scholar]
- Cheng F. 2019. In Silico Oncology Drug Repositioning and Polypharmacology. [DOI] [PubMed] [Google Scholar]
- Cheng F., Hong H., Yang S., Wei Y. Individualized network-based drug repositioning infrastructure for precision oncology in the panomics era. Brief. Bioinform. 2017;18:682–697. doi: 10.1093/bib/bbw051. [DOI] [PubMed] [Google Scholar]
- Cheng F., Murray J.L., Rubin D.H. Drug Repurposing : new treatments for Zika virus Infection ? Trends Mol. Med. 2016 doi: 10.1016/j.molmed.2016.09.006. [DOI] [PubMed] [Google Scholar]
- Decroly E., Debarnot C., Ferron F., Bouvet M., Coutard B., Imbert I., Gluais L., Papageorgiou N., Sharff A., Bricogne G., Ortiz-Lombardia M., Lescar J., Canard B. Crystal structure and functional analysis of the SARS- Nsp16 complex. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X., Li J., Yang X., Chmura A.A., Zhu G., Epstein J.H., Mazet J.K., Hu B., Zhang W., Peng C., Zhang Y., Luo C., Tan B., Wang N., Zhu Y., Crameri G., Zhang S., Wang L., Daszak P., Shi Z. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. doi: 10.1038/nature12711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habtemariam S., Dagne E. Comparative antioxidant, prooxidant and cytotoxic activity of Sigmoidin A and Eriodictyol. Planta Med. 2009;76:589–594. doi: 10.1055/s-0029-1240604. [DOI] [PubMed] [Google Scholar]
- Han D.P., Penn-nicholson A., Cho M.W. Identification of critical determinants on ACE2 for SARS-CoV entry and development of a potent entry inhibitor. Virology. 2006 doi: 10.1016/j.virol.2006.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hull M.W., Montaner J.S.G. Ritonavir-boosted protease inhibitors in HIV therapy. Ann. Med. 2011;43:375–388. doi: 10.3109/07853890.2011.572905. [DOI] [PubMed] [Google Scholar]
- Jang K., Jeong S., Kang D.Y., Sp N., Yang Y.M., Kim D.-E. A high ATP concentration enhances the cooperative translocation of the SARS coronavirus helicase Nsp13 in the unwinding of duplex RNA. Nat. Sci. Rep. 2020:1–13. doi: 10.1038/s41598-020-61432-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang S., Xia S., Ying T., Lu L. A novel coronavirus ( 2019-nCoV) causing pneumonia-associated respiratory syndrome. Cell. Mol. Immunol. 2020 doi: 10.1038/s41423-020-0372-4. 200032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemp J.P., Bernstein I.L., Bierman C.W., Li J.T., Siegel S.C., Spangenberg R.D., Tinkelman D.G. Pemirolast, a new oral nonbronchodilator drug for chronic asthma. Ann. Allergy. 1992;68:488–491. [PubMed] [Google Scholar]
- Khaerunnisa S., Kurniawan H., Awaluddin R., Suhartati S., Soetjipto S. Potential inhibitor of COVID-19 main protease (M pro) from several medicinal plant compounds by molecular docking study. 2020. Preprints 1–14. [DOI]
- Kim Y., Jedrzejczak R., Maltseva N., Endres M., Mececar A., Michalska K., Joachimiak A. 2020. Crystal Structure of ADP ribose phosphatase of NSP3 from SARS CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Singh A., Shaikh A., Singh R., Misra A. Chloroquine and Hydroxychloroquine in the treatment of COVID-19 with or without diabetes: a systematic search and a narrative review with a special reference to India and other developing countries. Diabetes Metab. Syndr. Clin. Res. Rev. 2020:241–246. doi: 10.1016/j.dsx.2020.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai C., Hung Y., Wang C., Wang Y.-H., Hsueh S.-C., Yen M.-Y., Ko W.-C., Hsueh P.-R. Asymptomatic carrier state, acute respiratory disease, and pneumonia due to severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): facts and myths. J. Microbiol. Immunol. Infect. 2020;53:404–412. doi: 10.1016/j.jmii.2020.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.K. Anti-inflammatory effects of Eriodictyol in lipopolysaccharide- stimulated raw 264.7 murine macrophages. Arch Pharm. Res. (Seoul) 2011;34:671–679. doi: 10.1007/s12272-011-0418-3. [DOI] [PubMed] [Google Scholar]
- Li F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016;3:237–261. doi: 10.1146/annurev-virology-110615-042301.Structure. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Moore M.J., Vasilieva N., Sui J., Wong S.K., Berne M.A., Somasundaran M., Sullivan J.L., Luzuriaga K., Greenough T.C., Hyeryun C., Farzan M. Angiotensin-converting enzyme 2 is a functional receptor for the SARS coronavirus. Lett. Nat. 2003;426 doi: 10.1038/nature02145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Zhou Q., Li Y., Garner L.V., Watkins S.P., Carter L.J., Smoot J., Gregg A.C., Daniels A.D., Jervey S., Albaiu D. Research and development on therapeutic agents and vaccines for COVID-19 and related human coronavirus diseases. ACS Cent. Sci. 2020;6:315–331. doi: 10.1021/acscentsci.0c00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minasov G., Shuvalova L., Rosas-Lemus M., Kiryukhina O., Wiersum G., Godzik A., Jaroszewski L., Stogios P.J., Skarina T., Satchell K.J.F. 2020. 1.95 Angstrom Resolution Crystal Structure of NSP10-NSP16 Complex from SARS-CoV-2. [DOI] [Google Scholar]
- Owen C.D., Lukacik P., Strain-Damerell C.M., Douangamath A., Powell A.J., Fearon D., Brandao-Neto J., Crawshaw A.D., Aragao D., Williams M., Flaig R., Hall D.R., McAuley K.E., Mazzorana M., Stuart D.I., von Delft F., Walsh M.A. 2020. SARS-CoV-2 main protease with unliganded active site (2019-nCoV, coronavirus disease COVID-19) [DOI] [Google Scholar]
- Rossato M.F., Trevisan G., Walker C.I.B., Klafke J.Z., de Oliveira A.P., Villarinho J.G., Zanon R.B., Royes L.F.F., Athayde M.L., Gomez M.V., Ferreira J. Eriodictyol: a flavonoid antagonist of the TRPV1 receptor with antioxidant activity. Biochem. Pharmacol. 2011;81:544–551. doi: 10.1016/j.bcp.2010.11.004. [DOI] [PubMed] [Google Scholar]
- Salata C., Calistri A., Parolin C., Baritussio A., Palu G. Antiviral activity of cationic amphiphilic drugs. Expert Rev. Anti Infect. Ther. 2017;15:483–492. doi: 10.1080/14787210.2017.1305888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheahan T.P., Sims A.C., Leist S.R., Alexandra S., John W., Ariane J.B., Stephanie A.M., Alison H., Darius B., Clarke M.O., Spahn E.J., Bauer L., Sellers S., Porter D., Feng J.Y., Cihlar T., Jordan R., Denison M.R., Baric R.S. Comparative therapeutic efficacy of remdesivir and combination lopinavir, ritonavir, and interferon beta against MERS-CoV. Nat. Commun. 2020;11:222. doi: 10.1038/s41467-019-13940-6|w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla R., Pandey V., Vadnere G.P., Lodhi S. In: Bioactive Food as Dietary Interventions for Arthritis and Related Inflammatory Diseases. second ed.. Watson R.R., Preedy V., editors. Academic Press; 2019. Chapter 18 - Role of Flavonoids in Management of Inflammatory Disorders; pp. 293–322. [DOI] [Google Scholar]
- Smith M.D., Smith J.C. 2020. Repurposing Therapeutics for COVID-19 : Supercomputer-Based Docking to the SARS-CoV-2 Viral Spike Protein and Viral Spike Protein-Human ACE2 Interface. [Google Scholar]
- Tan K., Kim Y., Jedrzejczak R., Maltseva N., Endres M., Michalska K., Joachimiak A. 2020. The crystal structure of Nsp9 RNA binding protein of SARS CoV-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W., Chen C., Lei X., Zhao J., Liang J. CASTp 3.0 : computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46:363–367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trott O., Olson A.J. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading Oleg Public Access. J. Comput. Chem. 2010;31:455–461. doi: 10.1002/jcc.21334.AutoDock. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velthuis A.J.W., Van Den Worm S.H.E., Sims A.C., Baric R.S., Snijder E.J., Van Hemert M.J. Zn 2 + inhibits coronavirus and arterivirus RNA polymerase activity in vitro and zinc ionophores block the replication of these viruses in cell culture. PLoS Pathog. 2010;6:1–10. doi: 10.1371/journal.ppat.1001176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincent M.J., Bergeron E., Benjannet S., Erickson B.R., Rollin P.E., Ksiazek T.G., Seidah N.G., Nichol S.T. Chloroquine is a potent inhibitor of SARS coronavirus infection and spread. Virol. J. 2005;2:1–10. doi: 10.1186/1743-422X-2-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Shang J., Graham R., Baric R.S., Li F. Receptor recognition by novel coronavirus from Wuhan: an analysis based on decade-long structural studies of SARS. J. Virol. 2020 doi: 10.1128/JVI.00127-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cao R., Zhang L., Yang X., Liu J., Xu M., Shi Z., Hu Z., Zhong W., Xiao G. Remdesivir and Chloroquine effectively inhibit the recently emerged novel coronavirus ( 2019-nCoV) in vitro. Cell Res. 2020 doi: 10.1038/s41422-020-0282-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Sun Y., Wu A., Xu S., Pan R., Zeng C., Jin X., Ge X., Shi Z., Aloha T., Chen Y., Guo D. Coronavirus Nsp10/Nsp16 methyltransferase can be targeted by Nsp10-derived peptide in vitro and in vivo to reduce replication and pathogenesis. J. Virol. 2015;232 doi: 10.1128/JVI.00948-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wishart D.S., Feunang Y.D., Guo A.C., Lo E.J., Marcu A., Grant R., Sajed T., Johnson D., Li C., Sayeeda Z., Assempour N., Iynkkaran I., Liu Y., Maciejewski A., Gale N., Wilson A., Chin L., Cummings R., Le D., Pon A., Knox C., Wilson M. DrugBank 5 . 0 : a major update to the DrugBank database for 2018. Nucleic Acids Res. 2018;46:1074–1082. doi: 10.1093/nar/gkx1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrapp D., Wang N., Corbett K.S., Goldsmith J.A., Hsieh C., Abiona O., Graham B.S., McLellan J.S. Prefusion 2019-nCoV spike glycoprotein with a single receptor-binding domain up. Science. 2020;367 doi: 10.2210/pdb6VSB/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C., Liu Y., Yang Y., Zhang P., Li X., Zheng M., Chen L., Li H. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm. Sin. B. 2020;10:766–788. doi: 10.1016/j.apsb.2020.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Peng C., Shi Y., Zhu Z., Mu K., Wang X., Zhu W. bioRxiv; 2020. Nelfinavir was predicted to be a potential inhibitor of 2019-nCov main protease by an integrative approach combining homology modelling, molecular docking and binding free energy calculation. Prepr. 1201, 0–2, Submitted for publication. [Google Scholar]
- Xue J., Moyer A., Peng B., Wu J., Hannafon B.N., Ding W. Chloroquine is a zinc ionophore. PloS One. 2014;9:1–6. doi: 10.1371/journal.pone.0109180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan R.H., Zhang Y.Y., Li Y.N., Xia L., Zhou Q. 2020. ACE2–B0AT1 complex. [DOI] [Google Scholar]
- Zhang L., Lin D., Sun X., Curth U., Drosten C., Sauerhering L., Becker S., Rox K., Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved a -ketoamide inhibitors. Science (80-.) 2020;412:409–412. doi: 10.1126/science.abb3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W.-Y., Lee J.-J., Kim Y., Kim I.-S., Han J.-H., Lee S.-G., Ahn M.-J., Jung S.-H., Myung C.-S. Effect of Eriodictyol on glucose uptake and insulin resistance in vitro. J. Agric. Food Chem. 2012;60:7652–7658. doi: 10.1021/jf300601z. [DOI] [PubMed] [Google Scholar]
- Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov. 2020 doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu G.F.A., Guo H.J., Huang Y.A.N., Wu C.T., Zhang X.F. Eriodictyol , a plant flavonoid , attenuates LPS - induced acute lung injury through its antioxidative and anti - inflammatory activity. Exp. Ther. Med. 2015;10:2259–2266. doi: 10.3892/etm.2015.2827. [DOI] [PMC free article] [PubMed] [Google Scholar]