Abstract
Cells continuously communicate changes in their microenvironment, both locally and globally, with other cells in the organism. Integration of information arising from signaling networks impart continuous, time-dependent changes of cell function and phenotype. Use of genetically encoded reporters enable researchers to noninvasively monitor time-dependent changes in intercellular and intracellular signaling, which can be interrogated by macroscopic and microscopic optical imaging, nuclear medicine imaging, MRI, and even photoacoustic imaging techniques. Reporters enable noninvasive monitoring of changes in cell-to-cell proximity, transcription, translation, protein folding, protein association, protein degradation, drug action, and second messengers in real time. Because of their positive impact on preclinical research, attempts to improve the sensitivity and specificity of these reporters, and to develop new types and classes of reporters, remain an active area of investigation. A few reporters have migrated to proof-of-principle clinical demonstrations, and recent advances in genome editing technologies may enable the use of reporters in the context of genome-wide analysis and the imaging of complex genomic regulation in vivo that cannot be readily investigated through standard methodologies. The combination of genetically encoded imaging reporters with continuous improvements in other molecular biology techniques may enhance and expedite target discovery and drug development for cancer interventions and treatment.
Keywords: Animal Studies, Fluorescence, Gene Therapy, Molecular Imaging, Molecular Imaging-Reporter Gene Imaging, Molecular Imaging-Stem Cells, Oncology, PET, Photoacoustic Imaging, SPECT
© RSNA, 2020
Essentials
■ Reporter genes generate a measurable signal that can be detected and quantified noninvasively by molecular imaging instrumentation (PET, SPECT, MRI, optical, photoacoustic, etc), allowing real-time imaging of biologic processes.
■ Cell communication involves a network of signaling cascades that are regulated at multiple levels, and genetically encoded imaging reporters can be used to monitor both the activation and regulation of these signaling cascades in vivo.
■ The future for genetically encoded imaging reporters for monitoring cell-to-cell communication includes emerging regulators of signaling cascades, such as superenhancers and long-noncoding RNA, that require more facile integration of reporters directly into the genome.
Introduction
Cell signaling networks enable cells to communicate and respond to changes in the microenvironment through signaling proteins that initiate a series of biochemical reactions within the cell. These networks of reactions are also known as signal transduction cascades (1). At any given time, cells receive numerous internal and external signals, initiating and integrating across multiple signal transduction pathways. These signaling networks change spatially and temporally over the lifespan of a cell, resulting in heterogeneous cell populations with differing and often transient phenotypes. Understanding how cells communicate locally and globally in the context of a living organism continues to yield breakthroughs in our understanding of systems biology and hopefully will enhance and expedite the drug discovery and development process.
Biotechnology and pharmaceutical companies are developing therapeutics that directly target cell-cell communication in vivo. For example, the sonic hedgehog signaling pathway, which includes downstream signaling from Smoothened (SMO) and glioma-associated protein (GLI), is a normal paracrine intercellular signaling cascade that is implicated in cancer to promote unregulated growth of tumors. As such, several therapeutics have been developed to inhibit both SMO and GLI (2). Another example of targeting cell-to-cell communication comes into play with checkpoint blockade inhibitors in immunotherapy. The impact of recent advances in immunotherapy on patient management, clinical trial design, and preclinical interrogation can be hard to overestimate (3). The programmed cell death protein 1 and cytotoxic T-lymphocyte–associated protein 4 are both targets of immunotherapy that are not expressed on the tumor cell itself, but are rather expressed on the immune cell compartment and contribute to cell-to-cell communication in the microenvironment (4–6). Similarly, other biologics act as stimulators of cell-cell communication pathways. Development of methodologies to visualize these processes in vivo can enhance our understanding of these biologic mechanisms for improved target identification.
Classic molecular biology techniques (eg, Western blots, whole-genome sequencing, polymerase chain reaction, etc) only provide a snapshot in time of the cellular genotype and/or phenotype and thus may fail to observe dynamic changes occurring within these signaling networks. To study temporal changes using classic molecular biology techniques, different time points must be evaluated separately and often invasively. Moreover, these molecular changes cannot be easily tracked within the same patient, and thus these studies require many patients to gain statistically significant results due to variations among individuals. Molecular imaging techniques (nuclear, MRI, fluorescence, and bioluminescence) provide a longitudinal, real-time strategy to noninvasively and repetitively monitor, both macroscopically and microscopically, biologic processes at molecular and cellular levels (7).
Molecular imaging is a powerful strategy that enables the visualization of gene expression, biochemical reactions, signal transduction, protein-protein interactions, regulatory pathways, cell trafficking, and drug action within a living system (8–13). Nonetheless, despite modest clinical advancements, genetically encoded reporters represent promising tools that are widely and productively utilized in preclinical research, and the knowledge gained from these reporters has significant translational potential. The genetically encoded strategies outlined herein provide a framework for advanced analysis of cellular communication in the tumor microenvironment (4,6).
Preclinical Assessment of Imaging Genetically Encoded Reporters in Vivo
Overview of Imaging Modalities
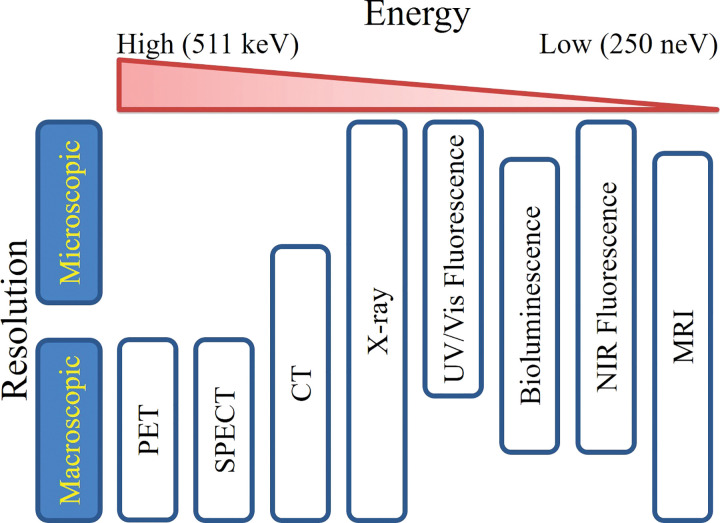
Imaging is an integral part of research, clinical trials, and medical practices related to cancer. Imaging modalities can be classified by spatial resolution (macroscopic or microscopic), information content (anatomic, physiologic, cellular, or molecular), and/or the type of energy used to obtain the images (x-rays, positrons, gamma rays, visible photons, near-infrared photons, short-wave infrared photons, or sound waves). An overview of imaging modalities is shown in Figure 1. In current widespread clinical and preclinical practice, macroscopic imaging systems that provide anatomic and physiologic information, such as the localization of disease and/or injury noninvasively within the body, include radiography, CT, MRI, and US. Molecular imaging modalities, such as PET, SPECT, and MR spectroscopy, are commonly used in clinical practice.
Figure 1:
Clustering of imaging modalities by resolution and energy. Imaging modalities span a range of resolutions and energies, which both contribute to differences in tissue penetration. Very high and very low energy imaging modalities yield high tissue penetration and, in most cases, macroscopic imaging resolution. At intermediate energies, such as fluorescence and bioluminescence, both macroscopic and microscopic imaging resolutions are achievable, but tissue penetration is limited. NIR = near-infrared, UV = ultraviolet, Vis = visible.
Macroscopic and microscopic imaging systems, which are predominantly used in basic and translational research, are currently making a transition into clinical research. These imaging systems allow visualization at the molecular level of the expression and activity of target genes, proteins, cells, and biologic processes that influence disease development and/or responsiveness to intervention. These techniques and systems include MR spectroscopy with and without hyperpolarization, bioluminescence, photoacoustic, and fluorescence imaging. Such molecular imaging techniques often require use of injectable agents or genetically encoded reporters to distinguish different molecular, cellular, and/or tissue types. Specifically, imaging agents generate an output due to a change in state of a target cell, and ideally, the change in signal varies proportionally to the amplitude of change within the cell. Although injectable agents have great potential for clinical translation, the development of these agents is lengthy, requiring significant optimization and characterization for each reporter, and if translated to the clinic may require a regulatory approval process similar to that of therapeutic agents. When high-molar-activity PET tracers are translated into the clinics, the time and cost from preclinical to clinical trials is shortened through microdosing phase 0 registration mechanisms (14,15).
Application and Implementation of Genetically Encoded Reporters
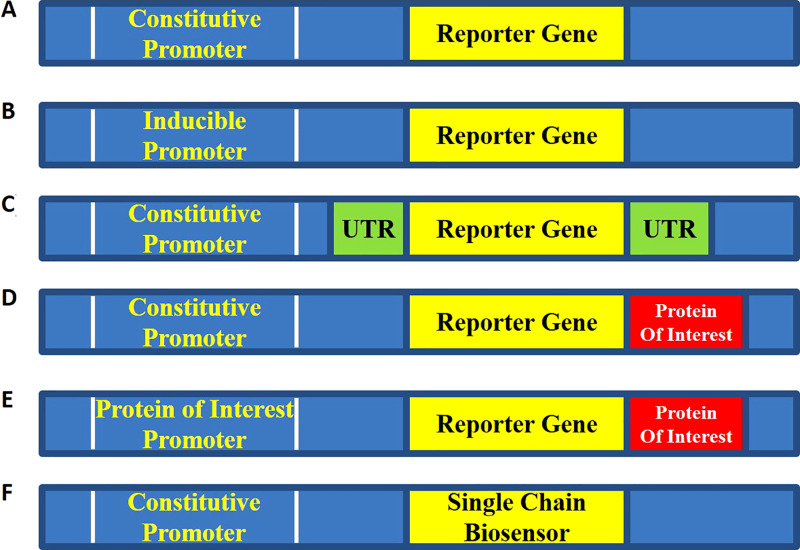
Genetically encoded imaging reporters enable robust and noninvasive methodologies to longitudinally monitor the dynamics of signaling networks with both temporal and spatial resolution (8–13,16–20). Here, the reporter gene generates a measurable signal that is detected and quantified by molecular imaging instrumentation (PET, SPECT, MRI, optical, etc) allowing real-time imaging of biologic processes. The reporter gene is cloned into a constitutive or inducible promoter or enhancer region adjacent to a gene of interest or engineered as a fusion with the protein of interest, wherein signal output indicates reporter expression (Fig 2). The most common genetically encoded reporters produce optical signals; however, reporters for PET/SPECT and MRI, as well as others have been explored as well. Optical genetically encoded reporters produce signal and/or changes in signal: (a) intrinsically by the reporter (eg, fluorescent protein) (12,13,16,21) or (b) reporter-mediated enzymatic activation of an optically silent substrate (eg, the oxidation of d-luciferin by luciferase produces light in the presence of O2, adenosine triphosphate [ATP], and Mg2+) (10,11,16,22). These reporters can then be further modified to study protein-protein interactions, protein conformational changes, proteolysis or environmental changes through fluorescence resonance energy transfer, bioluminescence resonance energy transfer, or complementation strategies (9,12,23–28).
Figure 2:
Typical configurations of genetically encoded imaging reporters. A, Constitutive reporter. The reporter is under the control of a constitutive promoter such as simian virus 40, chicken β-actin, or cytomegalovirus promoter. This design is most often utilized for cell tracking or trafficking experiments. B, Regulated transcriptional reporter. This design is utilized to monitor promoter activity in various cell types under various stimuli. Control constructs such as (A) or constructs with empty promoter regions are required to demonstrate specificity of signal induction. C, Translational or posttranscriptional reporters. The untranslated region (UTR) of interest is included either upstream or downstream of the reporter under the control of a constitutive promoter. Constructs such as (A) or mutated UTR are required to demonstrate specificity of signal change. D, Posttranscriptional reporters. The reporter gene is fused to a protein of interest typically through a glycine-serine linker region. As the protein of interest is degraded or trafficked, the reporter is concomitantly degraded or trafficked through the cell. Key controls involve fusing to a mutated form of the protein of interest that is no longer degraded or appropriately trafficked through the cell. E, Feedback-regulated reporter. In this case, the fusion reporter from (D) is also controlled by the promoter of the gene of interest. The feedback-regulated dynamics of degradation and synthesis can then be studied. F, Biosensor. A modified luciferase or fluorophore that changes either brightness or spectral output in response to changes in the local environment is developed. This construct is placed under the control of a constitutive promoter. Constructs such as (A) or modified biosensors with point mutations no longer capable of sensing environmental changes are required to demonstrate specificity.
Genetically Encoded Reporters at PET Imaging
Genetically encoded reporters for PET imaging have been translated to the clinic. Reporters that use imaging modalities capable of reaching deep tissue sites are the most likely to reach the clinic. Indeed, clinical translation of genetically encoded imaging reporters, such as the mutant herpes simplex 1 thymidine kinase (HSV1-tk) and 9-[4-[(18)F]fluoro-3-(hydroxymethyl)butyl]guanine ([(18)F]FHBG) pair, has been accomplished for PET imaging in human patients (29,30). Broadly, nuclear medicine imaging of genetically encoded reporters involves expression of an enzyme or receptor that selectively modifies a radiolabeled substrate by trapping, binding, or importing the substrate into target cells or tissues (eg, HSV1-tk [31,32] or targeting peptides to cell surface markers [33]).
Genetically Encoded Reporters at MRI
Genetically encoded reporters for MRI use principles similar to that of the nuclear medicine imaging reporters, but signals are instead generated by retention or binding of an MRI contrast agent by the reporter gene protein product (eg, β-galactosidase [34] or transferrin receptor [35]). With the growing exploration of hyperpolarized MRI, a genetically encoded reporter using hyperpolarized xenon was developed using gas vesicles that may be useful for tracking bacteria in vivo (36). However, between eight and 11 bacterial genes must be expressed to achieve robust formation of gas vesicles, which presents a challenge of translating these nanostructures to mammalian cells. In one application, the challenge of utilizing gas vesicles as reporters in mammalian cells appears to have been met (37). More broadly, the unknown immunogenicity of exogenously expressed reporters, bacterial or otherwise, remains one of the primary barriers to translation of genetically encoded reporters into the clinics.
Fluorescent Reporters
Generally, fluorescent reporters are used for single-cell and intravital microscopy applications (38). Applications of fluorescent protein imaging are diverse, including real-time (subsecond) imaging. However, due to the high background (autofluorescence) in standard fluorescence imaging, imaging populations of cells has been limited to in vitro or superficial in vivo studies, as image acquisition is limited by low signal-to-noise ratios, photograph-bleaching, toxicity (reactive oxygen species production), and limited depth penetration of photons (39).
Several far red-shifted fluorescent proteins have been developed to improve limits of detection at depth in vivo (13,40), enabling advanced applications. Decreased scattering and absorbance of photons at the red-shifted wavelengths of excitation and emission of these new fluorescent proteins relative to green fluorescent proteins enhance photon penetration and reduce autofluorescence. The initiation of protein-protein interactions as part of signaling cascades can be detected using bimolecular fluorescence complementation (BiFC) fluorescent assays (41). However, the dynamics (off-rates) of these associations cannot be studied as the fluorescent protein complementation associations are irreversible under normal conditions. The majority of BiFC reporters excite and emit in the visible wavelengths, further limiting their utility in deeper tissues. Near-infrared fluorescent proteins have now also been optimized for BiFC reactions, facilitating their translation into small animal models, but monitoring kinetics of protein dissociation, and by implication, determination of quantitative Kd values, remains a challenge for all BiFC systems (42). Photoacoustic imaging, a hybrid of optical and US imaging, has recently begun to use genetically encoded reporters for cell tracking, cell trafficking, and environmental sensing (43).
Bioluminescent Reporters
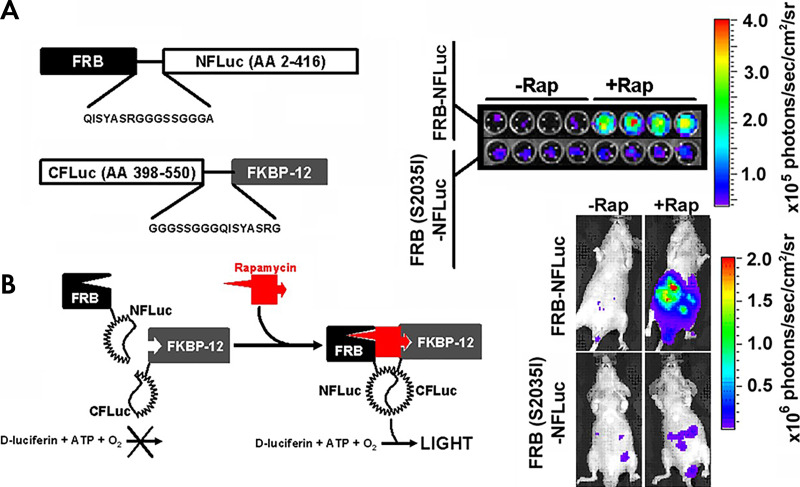
Bioluminescence imaging has proven to be a powerful noninvasive macroimaging technique for both in vitro and in vivo imaging. The major advantages of luciferase reporter systems include ultra-low background signal, high signal-to-noise imaging, modest cost, user-friendly instrumentation, and direct measure of live cell metabolism (ATP-dependent activity) (11). In addition, luciferases have a shorter half-life (approximately 3–5 hours for North American Photinus pyralis firefly and Renilla luciferase) than standard green fluorescent proteins (approximately 12–26 hours), are rapidly folded and activated, and thus, more accurately reflect the endogenous activation on-rates of various processes (10). Thus, to monitor the full dynamics of protein-protein interactions, reversible reporter systems are required. To this end, split luciferases, or luciferase complementation assays, in various colors and utilizing various substrates have been produced and validated (Fig 3) (9,44–46).
Figure 3:
Luciferase complementation imaging. Genetically encoded luciferase complementation strategies enable noninvasive imaging of reversible protein-protein interactions and protein folding events in cellulo and in vivo (44,106). Multiple protein-protein interactions can be monitored through the use of multicolor click beetle luciferases by combining complementation strategies and spectral unmixing (45). A, The two interacting proteins are fused to the N-terminal fragment of the luciferase and the C-terminal fragment of the luciferase, respectively, with an interposed flexible glycine-serine linker. In this example, the two interacting proteins are the rapamycin-binding protein (FKBP) and FKBP rapamycin binding domain (FRB) that associate in the presence of rapamycin. When they associate, the luciferase active site is reconstituted, and light is produced. B, Rapamycin-induced light production is specific both in cellulo (top) and in vivo (bottom). A mutation known to abrogate the binding of rapamycin (S2035I) inhibits light production both in cellulo and in vivo. (Reprinted, with permission, from reference 7.) ATP = adenosine triphosphate, CFLuc = C domain of the luciferase, NFLuc = N domain of the luciferase, Rap = rapamycin.
Unlike fluorescent reporters, bioluminescence reporters have a more limited number of luciferase proteins, although the development of red-emitting firefly and click beetle luciferases allow up to three different luciferases to be spectrally unmixed in live cells in culture (Fig 4) (47). A limitation of bioluminescent imaging in vivo is the substrate dependence of the luciferase enzymes as the pharmacokinetics of substrate delivery can confound analysis, and the potential exists for small molecule inhibitors to directly inhibit the luciferase reporter. Bioluminescence has been most useful for macroimaging, but recent advances in low-light microscopy technologies have extended bioluminescence applications to single cell imaging (48–52).
Figure 4:
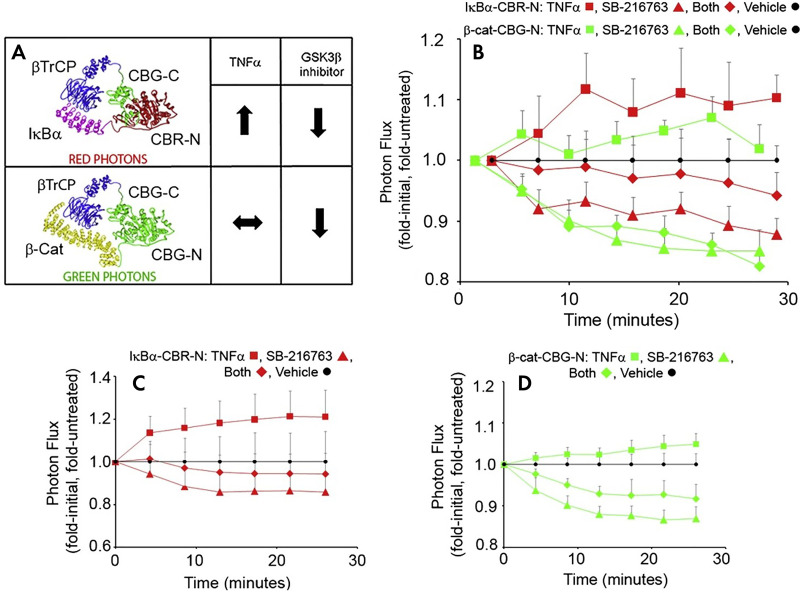
Multispectral luciferase complementation. A, The C-terminal fragment of click beetle green (CBG-C) was fused to β transducin repeats-containing proteins (βTrCP). The N-terminal fragment of click beetle red (CBR-N) was fused to IκBα. The N-terminal fragment of click beetle green was fused to β-catenin. The spectral emission of the reconstituted click-beetle luciferases maps to the N-terminal portion. Thus, light produced from the β-catenin/βTrCP interaction (green) can be resolved from the IκBa/βTrCP interaction (red) through spectral unmixing. B–D, The simultaneous quantification of the real-time switching of protein-protein interactions with βTrCP can be measured, depending on the exogenous stimulus or small molecule inhibitor. Data in red indicate IκBα/βTrCP interaction, and data in green indicate β-catenin/βTrCP interaction. (Reprinted, with permission, from reference 45.) GSK3β = glycogen synthase kinese 3 beta, SB-216763 = 3-(2,4-Dichlorophenyl)-4-(1-methyl-1H-indol-3-yl)-1H-pyrrole-2,5-dione.
Monitoring Cell-Cell Communication in the Microenvironment
Cell Tracking and Trafficking
Early genetically encoded imaging reporters were intended for labeling and tracking the distribution of cells, bacteria, viral infection, tumor cells, and metastatic burden in vivo by expressing reporter genes under the control of a constitutive promoter, such as cytomegalovirus or simian vacuolating virus 40 immediate early promoters. The choice of cell type is primarily driven by the desired application. For example, when studying the interaction of autochthonous cells, murine mammalian cells might be the best choice. If studying allochthonous cells or particles, such as tumor invasive bacteria or virus, one might choose to label either the virus or bacteria or the surrounding mammalian cells.
The most straightforward and powerful application of these genetically encoded reporters enabled monitoring of spatiotemporal changes in signal after implantation of cells where stable expression of the reporter was engineered ex vivo. Because firefly luciferase requires both ATP and luciferin to produce light, constitutively active imaging reporters provide a more refined readout of tumor growth and response. However, bioluminescence imaging of luciferase expression may be considered superior to caliper measurements because bioluminescence reports the quantity of viable tumor cells rather than total mass and volume. When cells are dead, the intracellular ATP concentration is reduced to below the levels needed for luciferase activity, and therefore dead cells no longer emit light. The total tumor volume can include stroma and immune cell infiltrates that may lead to misleading conclusions about tumor growth and drug response, particularly in the context of immunotherapy. Thus, for preclinical immunotherapy studies, bioluminescent reporters may help disambiguate pseudoprogression (increase in tumor volume due to immune cell infiltrate) from progression.
Indeed, the proximity of immune cells to tumor cells has been studied with a clever twist on the use of genetically encoded reporters for cell tracking (53). Bone marrow harvested from a transgenic mouse that expresses β-galactosidase, a genetically encoded reporter completely orthogonal to firefly luciferase, is implanted into a nu/nu mouse. Next, luciferase positive breast tumor cells are implanted orthotopically into the mammary fat pad of the same mouse. A galactose-caged luciferin, lugal, is then systemically injected into the mouse. Firefly luciferase cannot utilize the galactose-caged luciferin as a substrate for light production. Only when the β-galactosidase expressing immune cells are near the tumor cells will the galactose be cleaved from lugal and luciferin be released (Fig 5). The released luciferin can then diffuse in sufficiently high local concentration to serve as a substrate for the firefly luciferase expressed in tumor cells and then produce light. Thus, in principle, cell proximity studies in the tumor microenvironment can be explored noninvasively with bioluminescence imaging.
Figure 5:
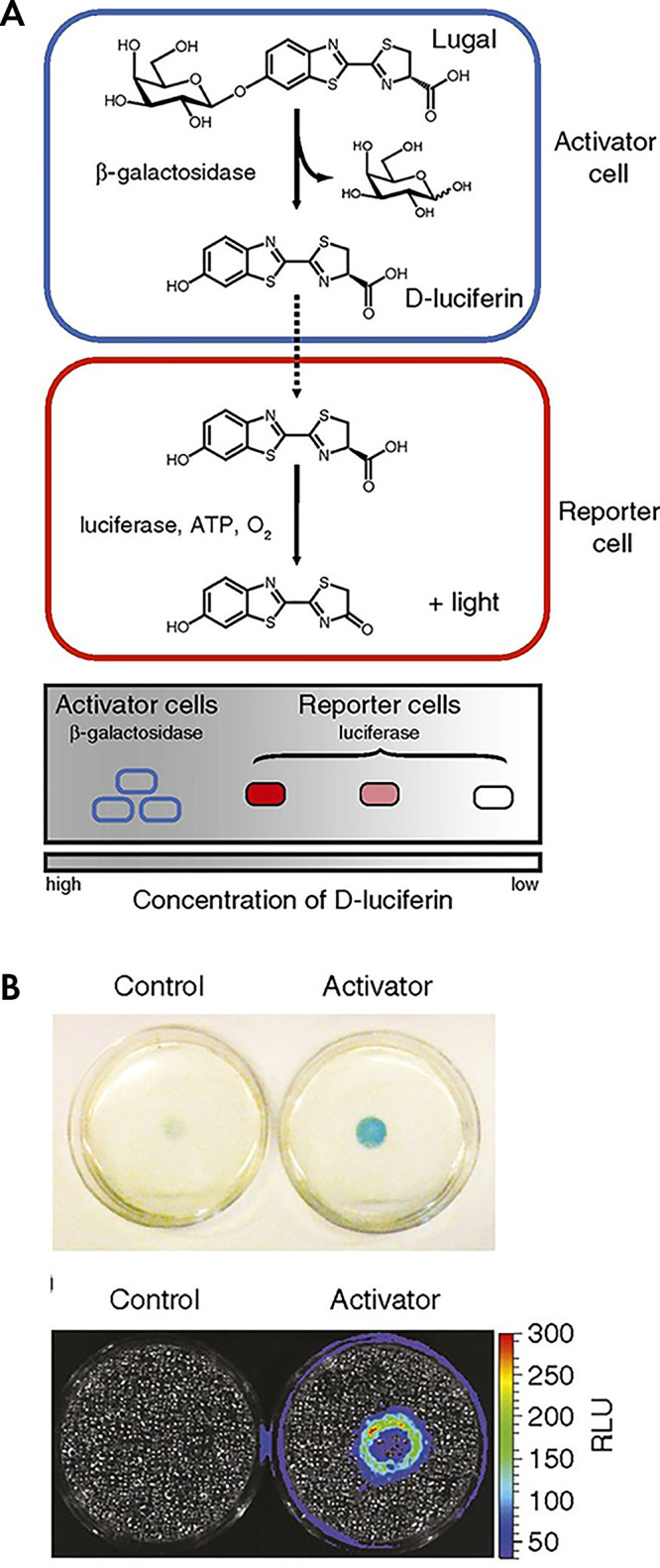
Dual enzyme–activated proximity sensor. A, Activator cells (expressing β-galactosidase) catalyze the cleavage of lugal, ultimately releasing d-luciferin. The liberated substrate enters nearby reporter cells, where it is used by luciferase to produce light. B, Reporter cells surrounding either control (left) or activator (right) cells were incubated with X-gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) for 4 hours and imaged. The blue color correlates with β-gal activity. Representative bioluminescence image of cocultures after 48 hours of incubation and subsequent incubation with lugal. Each dish was incubated with lugal (100 μg/mL) for 1 hour before image acquisition. (Reprinted, with permission, from reference 53.) ATP = adenosine triphosphate, RLU = relative light units.
Paracrine and Autocrine Signaling
Location of cells and their relative abundance only relays a part of the complexity of cellular communication in vivo. Paracrine signaling and cell-to-cell communication involves a network of signaling cascades that are regulated at multiple levels, and genetically encoded imaging reporters can be used to monitor both the activation and regulation of these signaling cascades in vivo. Transcriptional reporters provide a window into the change in activity of transcription factors, epigenetic regulation, and promoter activation in cells. Translational reporters can monitor processes and pathways that affect the ability of the cell to convert messenger RNA into protein, including mRNA stability, folding, and processing. Posttranslational reporters monitor changes in the primary, secondary, tertiary, and quaternary structure of proteins as a result of upstream signaling events. Finally, metabolic reporters, or single-chain biosensors, can monitor changes in the local environment, such as pH, membrane potential, cyclic adenosine monophosphate (cAMP), or Ca2+ concentrations.
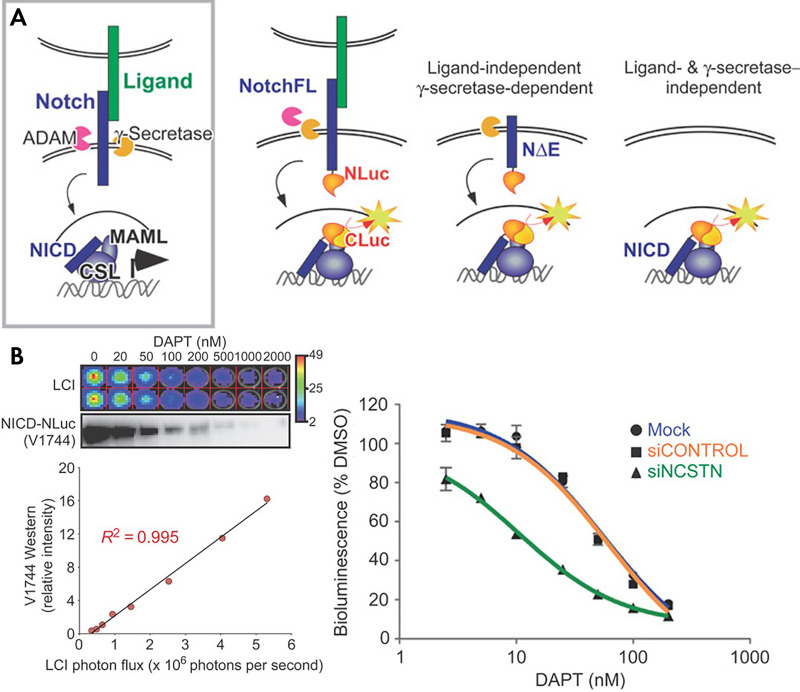
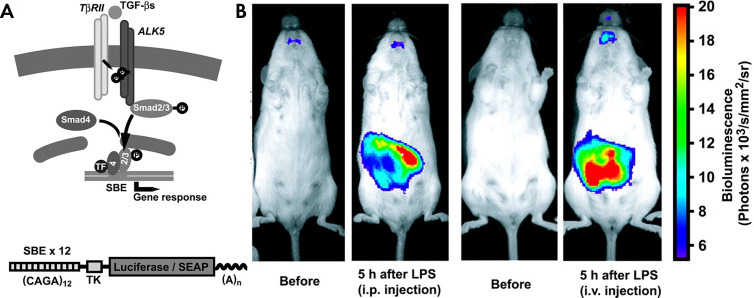
Geneticists have analyzed transcription factor binding and activation with luciferase reporters in vitro for many years (54). As an example, platelet-derived growth factor (PDGF) is a locally secreted growth factor that affects tissue development and tumor progression. More recently, cell lines and transgenic mice have been developed that can report on the activation of the PDGF pathway by coupling the transcription of firefly luciferase to the GLI1 promoter (55). Similarly, paracrine signaling by transforming growth factor β (TGFβ) can be monitored noninvasively with genetically engineered mice that express luciferase under the control of repeating SMAD binding consensus sequences (Fig 6) (56). Direct cell-to-cell signaling through activation of the Notch pathway can also be imaged in both whole animals and living cells (49). In the case of living animals, a transgenic mouse has been engineered to express CRE when the Notch signaling pathway is activated (57). To study the dynamics of Notch signaling, a luciferase complementation assay (split luciferase) was used to report activation and subsequent nuclear translocations of the cleaved intracellular domain of Notch (Fig 7) (49).
Figure 6:
Reversible notch signaling reporter. A, In the endogenous Notch pathway, ligand binding leads to ectodomain shedding due to cleavage at the S2 site by ADAM, followed by intramembrane proteolysis at the S3 site by γ-secretase. The released Notch intracellular domain (NICD) translocates to the nucleus and interacts with the DNA-binding protein CSL to recruit MAML proteins and other coactivators to activate gene expression. In the split luciferase construct, each half of the luciferase is kept separated and dark until the pathway is fully engaged in the nucleus. B, The reporter recapitulates the dose response curve of the Notch pathway to DAPT both linearly and over a large dynamic range. This inhibition could be specifically sensitized with siRNA targeting NCSTN. (Reprinted, with permission, from reference 49.) CSL = CBF1/RBPjκ/Su(H)/Lag1, DMSO = dimethyl sulfoxide, LCI = luciferase complementation imaging, MAML = mastermind like.
Figure 7:
Transcriptional TGF reporter. A, TβRII = TGF-β-type II receptor, TF = transcription factor. B, SBE-luc reporter gene construct consisting of 12 SBE repeats, a herpes simplex virus thymidine kinase minimal promoter (TK), firefly luciferase or SEAP, and an SV40 late 5-bromo-4-chloro-3-inodolyl-β-d-galactopyranoside signal (A). B, LPS administration results in tissue-specific activation of the SBE-luc reporter in vivo, reflecting in vivo induction of SMAD pathways by the innate immune system. (Reprinted, with permission, from reference 56.) SBE = Smad-binding element, SEAP = secreted alkaline phosphatase, TGF = transforming growth factor.
The Wnt and β-catenin, as well as the nuclear factor κB(NFκB) and IκB kinase (IKK), pathways are activated and repressed through paracrine signaling pathways. Both pathways signal through phosphorylation and degradation of key signaling proteins. Phosphorylated proteins are recognized by E3 ligases, and are subsequently ubiquitinated, which are then targeted by the proteasome for degradation. This process can be studied using reporters of posttranslational modification. By fusing luciferase to key proteins in the pathway and monitoring their degradation, the activation of the protein in the respective pathways can be monitored (8,58,59). Indeed, the IKK/IκBα reporters can be coupled to the NFκB response element to mimic and study the endogenous feedback loop inherent to the NFκB pathway (52).
Chemokines and cytokines are utilized by cells to remotely and locally signal to other cells, often immune cells or cancer cells, to relocate to a new area and/or to divide more rapidly. Luciferase complementation strategies also have been utilized to study the engagement of growth factor and chemokine receptors upstream of second messengers. In one strategy, the activation of chemokine receptors, such as C-X-C chemokine receptor 4 (CXCR4) and CXCR7, have been imaged in vivo via the heterodimerization of their intracellular domain with β-arrestin 2 upon ligand binding. This dimerization subsequently reconstitutes luciferase activity (60,61). On other fronts, monitoring extracellular protein interactions requires the use of a luciferase that is ATP independent because ATP concentrations in healthy extracellular spaces are near zero. Luker et al designed a split Gaussia luciferase to monitor these types of interactions (46,62). Gaussia luciferase is an ideal candidate as it is small and only requires O2. While it does use coelenterazine (a luciferin), these signals should be independent of the multidrug resistance status (63) of cells since the reconstituted luciferase remains in the extracellular space. In addition to dimerization (64), luciferase complementation can also be utilized to detect subtle changes in protein conformation, for example, of growth factor receptors (65). For these applications, transient binding of epidermal growth factor (EGF) to EGF receptor (EGFR) combined with luciferase complementation has been used to identify rapid changes (< 1 min) in conformation of the EGFR protein through real-time monitoring of bioluminescence (66).
Imaging MicroRNA Trafficking
Intercellular communication through the passage of microRNA between cells both directly and through extracellular vesicle trafficking is an active area of investigation (67). Translational reporters can be utilized to study the effects of microRNA on repression or activation (68). For example, to validate the effect of mir-134 on the 3ʹUTR (untranslated region) of the cAMP response-element binding protein, the 3ʹUTR of CREB (cAMP-response element binding protein) was cloned immediately upstream of luciferase, and differences in luciferase activity were used to validate the binding and suppression of translation (69). In a more direct study of cell-cell communication, a luciferase assay was used to validate and study the transfer of microRNA between glioma cells in culture (70). In this study, it was determined that microRNA was transferred between cells using gap junctions rather than by exosomes, providing mechanistic information on cellular processes. Indeed, a similar luciferase reporter was utilized to test and validate that packaged microRNA could affect translation of proteins in a target cell. While this assay was conducted in living cells in culture, there is no reason this strategy could not be extended in vivo (71).
Monitoring Intracellular Inositol Triphosphate and Calcium Concentrations during Signal Transduction
Cytokine and chemokine receptors as well as various cell surface receptors utilize inositol triphosphate (IP3) as a secondary messenger. The inositol diphosphate (IP2) is phosphorylated to IP3 by phosphoinositide 3-kinase (PI3 K), resulting in a host of downstream signaling cascades, including release of intracellular calcium on short and long time scales. Intracellular calcium concentrations can slowly increase or begin a transient wave of oscillating concentrations (72). Depending on the context of the signaling event, intracellular calcium fluxes can initiate NFκB pathway signaling, expression of survival genes, cell cycle progression, or apoptosis.
To study changes in intracellular IP3 concentration, researchers have leveraged the internal core of the IP3 receptor type 1, IP3R1, gene combined with a tethered split firefly luciferase construct to generate a single chain biosensor (72). When IP3 is not bound to the IP3R1 core, the luciferase domains are held sufficiently far apart to prevent interaction resulting in low levels of light production. When IP3 is elevated inside of the cell and binds to the core domain, the conformation changes (similar to a clamshell closing) drawing the two luciferase domains together. This reconstitutes the active site enabling the production of light. This reporter was validated both in vitro and in live cells through activation of the IP3 pathway with bradykinin and ATP.
As might be expected, there has been significant and early effort directed toward studying and quantifying changes in intracellular calcium concentrations with genetically encoded reporters. As indicated above, these changes can occur downstream of IP3 signaling, but they can also be induced by mechanical changes and physical stresses on the cell induced by interactions with the microenvironment (73). Additionally, calcium transients can propagate from cell to cell, particularly when joined by tight junctions. Miyawaki and colleagues developed a genetically encoded fluorescence resonance energy transfer reporter for studying changes in intracellular calcium in live cells (23). Due to the wavelengths involved, use of this reporter is limited to cells in culture, transparent thin model organisms such as zebrafish (74), or window chambers in mice (75).
Extracellular pH and Action Potentials
In tumor microenvironments, cancer cells can communicate with and suppress the immune system by lowering of the extracellular pH through the production and excretion of lactic acid (76). This lactate can be accumulated through production both by the tumor and by the stroma, depending on the tumor type and location. Lactate accumulation lowers the local pH from 7.4 to approximately 6–6.5. This cellular physiology characteristic spurred the development of therapies that seek to exploit this pH gradient to generate a therapeutic window (77) or decrease off-target toxicities (78). However, a pH gradient in tumors is not universal and therefore not a panacea.
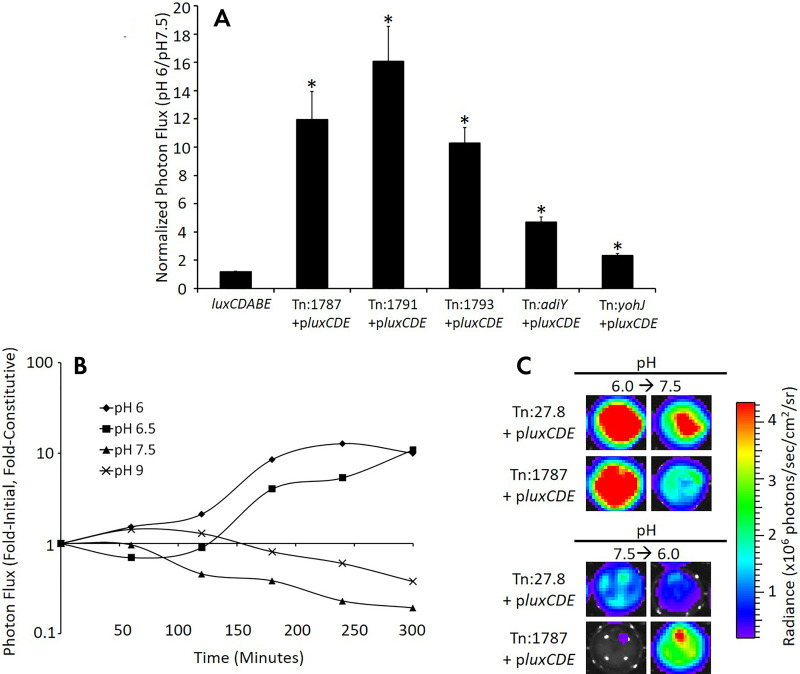
To study this heterogeneity, investigators have developed genetically encoded reporters to study the causes of and effects on therapy of local pH. Through an unbiased bioluminescent reporter transposon trap screen, a promoter was identified that when coupled with bacterial luciferase would act as a transcriptional reporter for the low pH found near tumors. As mentioned above, this is a case where a reporter blurs the line between a metabolic sensor, in this case for [H+], and a transcriptional reporter where the luciferase reports on the activation of the bacterial promoter STM1787, that is in turn reporting on the extracellular pH sensed by the Salmonella (Fig 8). Furthermore, the acidic promoter-reporter cassette could be converted into a potential therapeutic by delivering toxin, for example, Shiga toxin 2 gene (Stx2), when the promoter is activated in a low pH environment, killing the tumor. While the pH responsive bioluminescent strategy works well for whole organisms, pH-sensitive fluorescent proteins lead the way for studying changes on a microscopic scale both in vitro and in live animals. A library screen for ratiometric pH-responsive green fluorescent proteins yielded the variant pHluorin, wherein the ratio of the fluorescence emitted when excited by 410-nm light to 470-nm light reports on the local pH experienced by the fluorescent (79). An independent genetic screen of red fluorescent protein mutants yielded a red fluorescent, pH-sensitive mutant, pHTomato (80), and recently, monomeric RFP mutants with good dynamic range and physiologically relevant pKa have been developed. Finally, a new reporter that combines pH sensitivity with chloride sensitivity, when carefully calibrated, can be utilized to simultaneously monitor pH and chloride concentrations (81). The subcellular localization of these pH sensors can then be changed by selecting the appropriate leader sequence in the fusion protein to better understand local transient changes in pH in response to local stimuli (82). The propagation of action potentials between cells via cell-cell junctions and their subsequent integration and further dissemination across neural networks might be considered the ultimate in rapid cell-cell communication.
Figure 8:
Acidic pH specifically and reversibly stimulates the STM Tn:1787 promoter. A, Bacteria were cultured in media of different pH values, and reporter activation by Salmonella library clones in low pH media (pH 6) were compared with reporter activation in normal pH (pH 7.5). Genes identified in the tumor cell coculture screen were activated in the context of acidic pH compared with pH 7.5. pMAAC001 and luxCDABE constitutively express plasmid-encoded and chromosomally encoded luxCDABE imaging reporters, respectively. Data were normalized as the ratio of the signal in media pH 6.0 to signal in media pH 7.5. Error bars correspond to standard error of the mean. B, Mice bearing B16F10 flank tumor xenografts were injected intratumorally with tumor-activated bioluminescent (Tn:1787+pluxCDE) or constitutively bioluminescent (Tn:27.8+pluxCDE) Salmonella. The excised tumors were then imaged hourly, and data are presented as the normalized signal at each time point. The normalized signal represents the ratio of the mean of the fold-initial signal of two Tn:1787+pluxCDE-colonized tumors to the mean of the fold-initial signal of two constitutive Tn:27.8+pluxCDE-colonized tumors. C, Representative tumor imaging ex vivo shows reversibility of the bioluminescent signal in the tumor-activated Salmonella. Images on the left show Salmonella-infected tumor explants after 6 hours of incubation at the indicated pH (pH 6.0, top; pH 7.5, bottom). Two hours later (8 hours total), media were removed and replaced with media of the indicated pH (pH 7.5, top; pH 6.0, bottom). Images on the right show Salmonella-infected tumor explants 4 hours after the pH of the media was changed. Note the reversibility of the bioluminescent signal (107). (Reprinted, with permission, from reference 107.)
Direct measure of changes in membrane potential has long been the domain of the electrophysiologist. However, there have been recent inroads into the development of genetically encoded reporters for measuring membrane potentials with fluorescent techniques (83,84). Local membrane potentials were measured by quantifying and fitting the fluctuations in the data to a kinetic model using principle component analysis. These images were not based upon simple ratios but rather regression of time series data. While promising, significant simplification of the process will be required for broad-scale adoption. Second, as the authors identify, improperly folded or trafficked reporters in the cytosol confound the imaging data, so confocal microscopy is required. Because these require fitting of data kinetic at high frequency, it will be challenging to migrate into any in vivo setting where motion artifacts and autofluorescence are significant (85).
Emerging Reporter Technologies
While not yet ready for use in live animals or deep tissue, novel reporters and labeling strategies continue to emerge, generally inspired by nature’s own pool of biosensors. In one case, researchers leveraged the co-operative folding of mRNA aptamers to rapidly illuminate the presence of a small molecule target in the cytosol of Escherichia coli. Herein, the aptamer contains two binding domains, one for the target, and one for a fluorophore. When the small molecule is bound by the aptamer, the aptamer adopts a conformation that enables the binding of the fluorophore. If there is a concomitant increase in either the quantum yield or the extinction coefficient of the fluorophore upon binding to the aptamer, the system now acts as a molecular beacon. Upon binding to the aptamer, the fluorescence is enhanced, thus reporting on the presence of the small molecule of interest (86). The challenge of adapting this system to in vivo is twofold; the first challenge will be matching the uptake and retention of the fluorophore to the dynamics of the metabolite under study. The second, currently underway, will be shifting the fluorophore under study into the near-infrared (87). Neither are insurmountable challenges.
Other new strategies face similar challenges. One such strategy involves first modifying cells with a transfer RNA (tRNA) and paired tRNA transferase that will accept a nonnatural amino acid with a fluorophore-modified side chain. This has enabled the labeling of proteins with a nonnatural, fluorescent amino acids at high efficiency in culture (88). Similarly, in zebrafish embryos, one can utilize a pulse-chase strategy combined with copperless click chemistry to selectively label glycans in vivo (89). Like other reporters that start in smaller organisms, significant effort has been spent in extending these reporters into the near-infrared from the visible (90). This strategy might enable the study of other cell surface molecules, but when adapted to larger animals, the pharmacokinetics of both substrates will need to be well matched for easily interpretable images.
Looking to the Future
The future of genetically encoded imaging reporters for monitoring cell-to-cell communication include monitoring emerging regulators of signaling cascades such as superenhancers and long-noncoding RNA. Genetically encoded reporters delivered into the native context of the genome will be required for studying how these new regulators function. Plasmid-based extrachromosomal reporters that are randomly integrated into the genome do not provide physiologically faithful information as their genetic function is dependent upon its context in the whole genome. Additionally, superenhancers are long (> 1 kb) clusters of enhancers that produce strong cooperativity at promoter sites and are under the control of genes that are important in development, stem cell biology, and oncology. Areas of the genome that require tight regulation and stability of expression (or lack thereof) appear to yield more of these superenhancer regions (91). Since superenhancers are far too long to include in a plasmid-based system, noninvasive imaging of these superenhancers will need to occur in the context of genomic engineering of native organisms and cell lines.
Improved Methodologies for Improved Genomic Integration of Reporter Genes
Targeted genomic reporters show great promise to enable the evaluation of genes in their native genomic context and requires precisive planning. Depending on their design, reporters might still inevitably affect the production of noncoding RNAs found in introns, and potentially, regulation by other microRNAs including mirtons and snoRNA (92–94). The regulation of cell-to-cell communication by such microRNAs is no longer hypothetical. For example, exosomes, nanoscale lipid vesicles containing microRNAs, as well as DNA fragments, proteins, and other cytosolic components, are utilized regularly for long-range communication in the body (95,96). In addition, cells in direct contact can also exchange microRNA by which to communicate and influence their neighbors.
Many current strategies for incorporating genetically encoded reporters typically result in random integration of the target into the host genome. For more efficient targeted integration of reporters, researchers have turned to the field of genome editing. Genome-editing technologies and targeted viral vectors yield superior specificity and the opportunity to monitor pathways in their true genomic context. All genome-editing technologies generate double-stranded DNA breaks in the cell nucleus and then rely on endogenous homologous recombination mechanisms to integrate a gene or reporter cite specifically. Microbial research has yielded the most recent and most widely used addition to the genome-editing family, the CRISPR/Cas9 system (97–99). While the specificity of the original version has been questioned (100), the low cost, ease of use, and multiplex design drove researchers to rapidly improve the specificity of the approach. However, given the low probabilities of success, direct in-frame fusions with endogenous genes/proteins in vivo will be challenging. Nonetheless, promising results have been achieved for site-directed insertion of genetically encoded reporters into generally active sites, such as with adeno-associated virus (AAV1) (101). However, the cost and continued risk of off-target integration makes the use these reporters in patients purely for diagnostic purposes an unlikely proposition (102).
Future Applications for Genetically Encoded Reporters
The most likely role for genetically encoded reporters in the clinic will likely remain cell-tracking and cell-trafficking applications. While chimeric antigen receptor (CAR-T) cells have shown promise in liquid tumors, much work remains to be done in solid tumors (103). Indeed, some groups have turned to modifying natural killer (NK) cells to target and kill solid tumors (104). Others are turning to a different subset of T cells, gamma-delta T cells, to attempt to increase tumor lysis while suppressing adverse effects (105). While there are already several reporters available, the key will be identifying the critical balance between the limit of detection of the reporter-labeled cells and preventing the patient’s immune system from attacking these cells due to an immune response against the reporter or selection agent. Unfortunately, preclinical models are poor predictors of human immunogenicity, and these competing requirements will only be managed through carefully designed clinical trials.
Concluding Remarks
There will be a continual need to study the communication of cells at different scales and in different contexts as we continue to understand more about cellular physiology in cancer. Different challenges and opportunities arise as one moves from studying groups of cells, to tissues, to living mice, to humans. As a result, both fluorescent and bioluminescent genetically encoded imaging reporters will continue to be developed depending on the resolution required and the depth of target tissue. These reporters may prove invaluable for understanding intercellular communications, yielding a better fundamental understanding of complex biologic systems that hopefully will in turn yield better cancer diagnostics and therapeutics.
Work was funded by a grant to the Washington University-MD Anderson Cancer Center Inter-Institutional Molecular Imaging Center (National Cancer Institute P50 CA94056), an Odyssey Fellowship to T.W.L., and a Canadian Institute of Health Research Postdoctoral Fellowship to T.W.L.
Disclosures of Conflicts of Interest: S.T.G. Activities related to the present article: institution received grant from National Cancer Institute (Washington University-MD Anderson Cancer Center Inter-Institutional Molecular Imaging Center (NCI P50 CA94056). Activities not related to the present article: disclosed no relevant relationships. Other relationships: disclosed no relevant relationships. T.W.L. disclosed no relevant relationships. D.P.W. disclosed no relevant relationships.
Abbreviations:
- ATP
- adenosine triphosphate
- cAMP
- cyclic adenosine monophosphate
- IP3
- inositol triphosphate
References
- 1.O’Connor CM, Adams JU. Essentials of Cell Biology. Cambridge, Mass: NPG Education, 2010. [Google Scholar]
- 2.Carpenter RL, Ray H. Safety and Tolerability of Sonic Hedgehog Pathway Inhibitors in Cancer. Drug Saf 2019;42(2):263–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Couzin-Frankel J. Breakthrough of the year 2013. Cancer immunotherapy. Science 2013;342(6165):1432–1433. [DOI] [PubMed] [Google Scholar]
- 4.Wei SC, Duffy CR, Allison JP. Fundamental mechanisms of immune checkpoint blockade therapy. Cancer Discov 2018;8(9):1069–1086. [DOI] [PubMed] [Google Scholar]
- 5.Sharma P, Allison JP. Immune checkpoint targeting in cancer therapy: toward combination strategies with curative potential. Cell 2015;161(2):205–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iwai Y, Hamanishi J, Chamoto K, Honjo T. Cancer immunotherapies targeting the PD-1 signaling pathway. J Biomed Sci 2017;24(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gross S, Piwnica-Worms D. Spying on cancer: molecular imaging in vivo with genetically encoded reporters. Cancer Cell 2005;7(1):5–15. [DOI] [PubMed] [Google Scholar]
- 8.Gross S, Piwnica-Worms D. Real-time imaging of ligand-induced IKK activation in intact cells and in living mice. Nat Methods 2005;2(8):607–614. [DOI] [PubMed] [Google Scholar]
- 9.Villalobos V, Naik S, Piwnica-Worms D. Current state of imaging protein-protein interactions in vivo with genetically encoded reporters. Annu Rev Biomed Eng 2007;9(1):321–349. [DOI] [PubMed] [Google Scholar]
- 10.Kocher B, Piwnica-Worms D. Illuminating cancer systems with genetically engineered mouse models and coupled luciferase reporters in vivo. Cancer Discov 2013;3(6):616–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dothager RS, Flentie K, Moss B, Pan MH, Kesarwala A, Piwnica-Worms D. Advances in bioluminescence imaging of live animal models. Curr Opin Biotechnol 2009;20(1):45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weissleder R, Pittet MJ. Imaging in the era of molecular oncology. Nature 2008;452(7187):580–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Condeelis J, Weissleder R. In vivo imaging in cancer. Cold Spring Harb Perspect Biol 2010;2(12):a003848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schwarz SW, Dick D, VanBrocklin HF, Hoffman JM. Regulatory Requirements for PET Drug Production. J Nucl Med 2014;55(7):1132–1137. [DOI] [PubMed] [Google Scholar]
- 15.Mosessian S, Duarte-Vogel SM, Stout DB, et al. INDs for PET molecular imaging probes-approach by an academic institution. Mol Imaging Biol 2014;16(4):441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singer RH, Lawrence DS, Ovryn B, Condeelis J. Imaging of gene expression in living cells and tissues. J Biomed Opt 2005;10(5):051406. [DOI] [PubMed] [Google Scholar]
- 17.Bhatnagar A, Wang Y, Mease RC, et al. AEG-1 promoter-mediated imaging of prostate cancer. Cancer Res 2014;74(20):5772–5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Röck R, Bachmann V, Bhang HE, et al. In-vivo detection of binary PKA network interactions upon activation of endogenous GPCRs. Sci Rep 2015;5(1):11133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Palangat M, Larson DR. Single-gene dual-color reporter cell line to analyze RNA synthesis in vivo. Methods 2016;103:77–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ahn BC, Parashurama N, Patel M, et al. Noninvasive reporter gene imaging of human Oct4 (pluripotency) dynamics during the differentiation of embryonic stem cells in living subjects. Mol Imaging Biol 2014;16(6):865–876. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez EA, Campbell RE, Lin JY, et al. The growing and glowing toolbox of fluorescent and photoactive proteins. Trends Biochem Sci 2017;42(2):111–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prescher JA, Contag CH. Guided by the light: visualizing biomolecular processes in living animals with bioluminescence. Curr Opin Chem Biol 2010;14(1):80–89. [DOI] [PubMed] [Google Scholar]
- 23.Miyawaki A, Llopis J, Heim R, et al. Fluorescent indicators for Ca2+ based on green fluorescent proteins and calmodulin. Nature 1997;388(6645):882–887. [DOI] [PubMed] [Google Scholar]
- 24.Akemann W, Mutoh H, Perron A, Rossier J, Knöpfel T. Imaging brain electric signals with genetically targeted voltage-sensitive fluorescent proteins. Nat Methods 2010;7(8):643–649. [DOI] [PubMed] [Google Scholar]
- 25.Romoser VA, Hinkle PM, Persechini A. Detection in living cells of Ca2+-dependent changes in the fluorescence emission of an indicator composed of two green fluorescent protein variants linked by a calmodulin-binding sequence. A new class of fluorescent indicators. J Biol Chem 1997;272(20):13270–13274. [DOI] [PubMed] [Google Scholar]
- 26.Kuner T, Augustine GJ. A genetically encoded ratiometric indicator for chloride: capturing chloride transients in cultured hippocampal neurons. Neuron 2000;27(3):447–459. [DOI] [PubMed] [Google Scholar]
- 27.Gammon ST, Villalobos VM, Roshal M, Samrakandi M, Piwnica-Worms D. Rational design of novel red-shifted BRET pairs: Platforms for real-time single-chain protease biosensors. Biotechnol Prog 2009;25(2):559–569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dothager RS, Goiffon RJ, Jackson E, Harpstrite S, Piwnica-Worms D. Cerenkov radiation energy transfer (CRET) imaging: a novel method for optical imaging of PET isotopes in biological systems. PLoS One 2010;5(10):e13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eissenberg LG, Rettig MP, Ritchey JK, et al. [(18)F]FHBG PET/CT Imaging of CD34-TK75 Transduced Donor T Cells in Relapsed Allogeneic Stem Cell Transplant Patients: Safety and Feasibility. Mol Ther 2015;23(6):1110–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Keu KV, Witney TH, Yaghoubi S, et al. Reporter gene imaging of targeted T cell immunotherapy in recurrent glioma. Sci Transl Med 2017;9(373):eaag2196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gambhir SS, Barrio JR, Phelps ME, et al. Imaging adenoviral-directed reporter gene expression in living animals with positron emission tomography. Proc Natl Acad Sci U S A 1999;96(5):2333–2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luker GD, Sharma V, Pica CM, et al. Noninvasive imaging of protein-protein interactions in living animals. Proc Natl Acad Sci U S A 2002;99(10):6961–6966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buchsbaum DJ. Imaging and therapy of tumors induced to express somatostatin receptor by gene transfer using radiolabeled peptides and single chain antibody constructs. Semin Nucl Med 2004;34(1):32–46. [DOI] [PubMed] [Google Scholar]
- 34.Louie AY, Hüber MM, Ahrens ET, et al. In vivo visualization of gene expression using magnetic resonance imaging. Nat Biotechnol 2000;18(3):321–325. [DOI] [PubMed] [Google Scholar]
- 35.Weissleder R, Moore A, Mahmood U, et al. In vivo magnetic resonance imaging of transgene expression. Nat Med 2000;6(3):351–355. [DOI] [PubMed] [Google Scholar]
- 36.Shapiro MG, Ramirez RM, Sperling LJ, et al. Genetically encoded reporters for hyperpolarized xenon magnetic resonance imaging. Nat Chem 2014;6(7):629–634. [DOI] [PubMed] [Google Scholar]
- 37.Farhadi A, Ho GH, Sawyer DP, Bourdeau RW, Shapiro MG. Ultrasound imaging of gene expression in mammalian cells. Science 2019;365(6460):1469–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods 2005;2(12):905–909. [DOI] [PubMed] [Google Scholar]
- 39.Giepmans BN, Adams SR, Ellisman MH, Tsien RY. The fluorescent toolbox for assessing protein location and function. Science 2006;312(5771):217–224. [DOI] [PubMed] [Google Scholar]
- 40.Shen Y, Chen Y, Wu J, Shaner NC, Campbell RE. Engineering of mCherry variants with long Stokes shift, red-shifted fluorescence, and low cytotoxicity. PLoS One 2017;12(2):e0171257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kerppola TK. Bimolecular fluorescence complementation (BiFC) analysis of protein interactions in live cells. Cold Spring Harb Protoc 2013;2013(8):727–731. [DOI] [PubMed] [Google Scholar]
- 42.Filonov GS, Verkhusha VV. A near-infrared BiFC reporter for in vivo imaging of protein-protein interactions. Chem Biol 2013;20(8):1078–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brunker J, Yao J, Laufer J, Bohndiek SE. Photoacoustic imaging using genetically encoded reporters: a review. J Biomed Opt 2017;22(7):070901. [DOI] [PubMed] [Google Scholar]
- 44.Luker KE, Smith MC, Luker GD, Gammon ST, Piwnica-Worms H, Piwnica-Worms D. Kinetics of regulated protein-protein interactions revealed with firefly luciferase complementation imaging in cells and living animals. Proc Natl Acad Sci U S A 2004;101(33):12288–12293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villalobos V, Naik S, Bruinsma M, et al. Dual-color click beetle luciferase heteroprotein fragment complementation assays. Chem Biol 2010;17(9):1018–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Luker KE, Mihalko LA, Schmidt BT, et al. In vivo imaging of ligand receptor binding with Gaussia luciferase complementation. Nat Med 2011;18(1):172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gammon ST, Leevy WM, Gross S, Gokel GW, Piwnica-Worms D. Spectral unmixing of multicolored bioluminescence emitted from heterogeneous biological sources. Anal Chem 2006;78(5):1520–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tinkum KL, Marpegan L, White LS, et al. Bioluminescence imaging captures the expression and dynamics of endogenous p21 promoter activity in living mice and intact cells. Mol Cell Biol 2011;31(18):3759–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ilagan MX, Lim S, Fulbright M, Piwnica-Worms D, Kopan R. Real-time imaging of notch activation with a luciferase complementation-based reporter. Sci Signal 2011;4(181):rs7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spiller DG, Wood CD, Rand DA, White MR. Measurement of single-cell dynamics. Nature 2010;465(7299):736–745. [DOI] [PubMed] [Google Scholar]
- 51.Negrin RS, Contag CH. In vivo imaging using bioluminescence: a tool for probing graft-versus-host disease. Nat Rev Immunol 2006;6(6):484–490. [DOI] [PubMed] [Google Scholar]
- 52.Moss BL, Elhammali A, Fowlkes T, et al. Interrogation of inhibitor of nuclear factor κB α/nuclear factor κB (IκBα/NF-κB) negative feedback loop dynamics: from single cells to live animals in vivo. J Biol Chem 2012;287(37):31359–31370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sellmyer MA, Bronsart L, Imoto H, Contag CH, Wandless TJ, Prescher JA. Visualizing cellular interactions with a generalized proximity reporter. Proc Natl Acad Sci U S A 2013;110(21):8567–8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ow DW, Jacobs JD, Howell SH. Functional regions of the cauliflower mosaic virus 35S RNA promoter determined by use of the firefly luciferase gene as a reporter of promoter activity. Proc Natl Acad Sci U S A 1987;84(14):4870–4874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Becher OJ, Hambardzumyan D, Fomchenko EI, et al. Gli activity correlates with tumor grade in platelet-derived growth factor-induced gliomas. Cancer Res 2008;68(7):2241–2249. [DOI] [PubMed] [Google Scholar]
- 56.Lin AH, Luo J, Mondshein LH, et al. Global analysis of Smad2/3-dependent TGF-beta signaling in living mice reveals prominent tissue-specific responses to injury. J Immunol 2005;175(1):547–554. [DOI] [PubMed] [Google Scholar]
- 57.Liu Z, Turkoz A, Jackson EN, et al. Notch1 loss of heterozygosity causes vascular tumors and lethal hemorrhage in mice. J Clin Invest 2011;121(2):800–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Naik S, Piwnica-Worms D. Real-time imaging of beta-catenin dynamics in cells and living mice. Proc Natl Acad Sci U S A 2007;104(44):17465–17470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kesarwala AH, Samrakandi MM, Piwnica-Worms D. Proteasome inhibition blocks ligand-induced dynamic processing and internalization of epidermal growth factor receptor via altered receptor ubiquitination and phosphorylation. Cancer Res 2009;69(3):976–983. [DOI] [PubMed] [Google Scholar]
- 60.Luker KE, Gupta M, Luker GD. Imaging CXCR4 signaling with firefly luciferase complementation. Anal Chem 2008;80(14):5565–5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Luker KE, Gupta M, Steele JM, Foerster BR, Luker GD. Imaging ligand-dependent activation of CXCR7. Neoplasia 2009;11(10):1022–1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Luker KE, Luker GD. Split Gaussia luciferase for imaging ligand-receptor binding. Methods Mol Biol 2014;1098:59–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pichler A, Prior JL, Piwnica-Worms D. Imaging reversal of multidrug resistance in living mice with bioluminescence: MDR1 P-glycoprotein transports coelenterazine. Proc Natl Acad Sci U S A 2004;101(6):1702–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kobayashi D, Endo M, Ochi H, Hojo H, Miyasaka M, Hayasaka H. Regulation of CCR7-dependent cell migration through CCR7 homodimer formation. Sci Rep 2017;7(1):8536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Macdonald-Obermann JL, Adak S, Landgraf R, Piwnica-Worms D, Pike LJ. Dynamic analysis of the epidermal growth factor (EGF) receptor-ErbB2-ErbB3 protein network by luciferase fragment complementation imaging. J Biol Chem 2013;288(42):30773–30784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ronan T, Macdonald-Obermann JL, Huelsmann L, Bessman NJ, Naegle KM, Pike LJ. Different epidermal growth factor receptor (EGFR) agonists produce unique signatures for the recruitment of downstream signaling proteins. J Biol Chem 2016;291(11):5528–5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ressel S, Rosca A, Gordon K, Buck AH. Extracellular RNA in viral-host interactions: Thinking outside the cell. Wiley Interdiscip Rev RNA; 2019;10(4):e1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Olanich ME, Moss BL, Piwnica-Worms D, Townsend RR, Weber JD. Identification of FUSE-binding protein 1 as a regulatory mRNA-binding protein that represses nucleophosmin translation. Oncogene 2011;30(1):77–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Huang W, Liu X, Cao J, et al. miR-134 regulates ischemia/reperfusion injury-induced neuronal cell death by regulating CREB signaling. J Mol Neurosci 2015;55(4):821–829. [DOI] [PubMed] [Google Scholar]
- 70.Katakowski M, Buller B, Wang X, Rogers T, Chopp M. Functional microRNA is transferred between glioma cells. Cancer Res 2010;70(21):8259–8263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Singh R, Pochampally R, Watabe K, Lu Z, Mo YY. Exosome-mediated transfer of miR-10b promotes cell invasion in breast cancer. Mol Cancer 2014;13(1):256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ataei F, Torkzadeh-Mahani M, Hosseinkhani S. A novel luminescent biosensor for rapid monitoring of IP3 by split-luciferase complementary assay. Biosens Bioelectron 2013;41:642–648. [DOI] [PubMed] [Google Scholar]
- 73.Lam AK, Galione A. The endoplasmic reticulum and junctional membrane communication during calcium signaling. Biochim Biophys Acta 2013;1833(11):2542–2559. [DOI] [PubMed] [Google Scholar]
- 74.Chen TW, Wardill TJ, Sun Y, et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 2013;499(7458):295–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Atkin SD, Patel S, Kocharyan A, et al. Transgenic mice expressing a cameleon fluorescent Ca2+ indicator in astrocytes and Schwann cells allow study of glial cell Ca2+ signals in situ and in vivo. J Neurosci Methods 2009;181(2):212–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Pilon-Thomas S, Kodumudi KN, El-Kenawi AE, et al. Neutralization of tumor acidity improves antitumor responses to immunotherapy. Cancer Res 2016;76(6):1381–1390 [Published correction appears in Cancer Res 2017;77(9):2552.]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chauhan VP, Chen IX, Tong R, et al. Reprogramming the microenvironment with tumor-selective angiotensin blockers enhances cancer immunotherapy. Proc Natl Acad Sci U S A 2019;116(22):10674–10680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chakraborty P, Bouchlaka MN, Capoccia BJ, et al. AO-176, a normal cell sparing humanized anti-CD47 antibody. Cancer Res 2019;79(13 Suppl):540. [Google Scholar]
- 79.Mahon MJ. pHluorin2: an enhanced, ratiometric, pH-sensitive green florescent protein. Adv Biosci Biotechnol 2011;2(3):132–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li Y, Tsien RW. pHTomato, a red, genetically encoded indicator that enables multiplex interrogation of synaptic activity. Nat Neurosci 2012;15(7):1047–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zhong S, Navaratnam D, Santos-Sacchi J. A genetically-encoded YFP sensor with enhanced chloride sensitivity, photostability and reduced ph interference demonstrates augmented transmembrane chloride movement by gerbil prestin (SLC26a5). PLoS One 2014;9(6):e99095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Rathje M, Fang H, Bachman JL, et al. AMPA receptor pHluorin-GluA2 reports NMDA receptor-induced intracellular acidification in hippocampal neurons. Proc Natl Acad Sci U S A 2013;110(35):14426–14431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mutoh H, Akemann W, Knöpfel T. Genetically engineered fluorescent voltage reporters. ACS Chem Neurosci 2012;3(8):585–592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hou JH, Venkatachalam V, Cohen AE. Temporal dynamics of microbial rhodopsin fluorescence reports absolute membrane voltage. Biophys J 2014;106(3):639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Xu Y, Zou P, Cohen AE. Voltage imaging with genetically encoded indicators. Curr Opin Chem Biol 2017;39:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Strack RL, Song W, Jaffrey SR. Using Spinach-based sensors for fluorescence imaging of intracellular metabolites and proteins in living bacteria. Nat Protoc 2014;9(1):146–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tan X, Constantin TP, Sloane KL, Waggoner AS, Bruchez MP, Armitage BA. Fluoromodules consisting of a promiscuous RNA aptamer and red or blue fluorogenic cyanine dyes: selection, characterization, and bioimaging. J Am Chem Soc 2017;139(26):9001–9009. [DOI] [PubMed] [Google Scholar]
- 88.Young DD, Schultz PG. Playing with the molecules of life. ACS Chem Biol 2018;13(4):854–870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Beahm BJ, Dehnert KW, Derr NL, et al. A visualizable chain-terminating inhibitor of glycosaminoglycan biosynthesis in developing zebrafish. Angew Chem Int Ed Engl 2014;53(13):3347–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Huang J, Brenna C, Khan AUM, et al. A cationic near infrared fluorescent agent and ethyl-cinnamate tissue clearing protocol for vascular staining and imaging. Sci Rep 2019;9(1):521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.He Y, Long W, Liu Q. Targeting super-enhancers as a therapeutic strategy for cancer treatment. Front Pharmacol 2019;10:361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Westholm JO, Lai EC. Mirtrons: microRNA biogenesis via splicing. Biochimie 2011;93(11):1897–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature 2007;448(7149):83–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ooi SL, Samarsky DA, Fournier MJ, Boeke JD. Intronic snoRNA biosynthesis in Saccharomyces cerevisiae depends on the lariat-debranching enzyme: intron length effects and activity of a precursor snoRNA. RNA 1998;4(9):1096–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pegtel DM, Peferoen L, Amor S. Extracellular vesicles as modulators of cell-to-cell communication in the healthy and diseased brain. Philos Trans R Soc Lond B Biol Sci 2014;369(1652):20130516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Salido-Guadarrama I, Romero-Cordoba S, Peralta-Zaragoza O, Hidalgo-Miranda A, Rodríguez-Dorantes M. MicroRNAs transported by exosomes in body fluids as mediators of intercellular communication in cancer. OncoTargets Ther 2014;7:1327–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Cong L, Ran FA, Cox D, et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013;339(6121):819–823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mali P, Yang L, Esvelt KM, et al. RNA-guided human genome engineering via Cas9. Science 2013;339(6121):823–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012;337(6096):816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Kuscu C, Arslan S, Singh R, Thorpe J, Adli M. Genome-wide analysis reveals characteristics of off-target sites bound by the Cas9 endonuclease. Nat Biotechnol 2014;32(7):677–683. [DOI] [PubMed] [Google Scholar]
- 101.Jiang Y, Zhou Y, Bao X, et al. An ultrasensitive calcium reporter system via CRISPR-Cas9-mediated genome editing in human pluripotent stem cells. iScience 2018;9:27–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liang P, Xu Y, Zhang X, et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell 2015;6(5):363–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Emami-Shahri N, Papa S. Dynamic imaging for CAR-T-cell therapy. Biochem Soc Trans 2016;44(2):386–390. [DOI] [PubMed] [Google Scholar]
- 104.Daher M, Rezvani K. Next generation natural killer cells for cancer immunotherapy: the promise of genetic engineering. Curr Opin Immunol 2018;51:146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Silva-Santos B, Mensurado S, Coffelt SB. gδ T cells: pleiotropic immune effectors with therapeutic potential in cancer. Nat Rev Cancer 2019;19(7):392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Macdonald-Obermann JL, Piwnica-Worms D, Pike LJ. Mechanics of EGF receptor/ErbB2 kinase activation revealed by luciferase fragment complementation imaging. Proc Natl Acad Sci U S A 2012;109(1):137–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Flentie K, Kocher B, Gammon ST, Novack DV, McKinney JS, Piwnica-Worms D. A bioluminescent transposon reporter-trap identifies tumor-specific microenvironment-induced promoters in Salmonella for conditional bacterial-based tumor therapy. Cancer Discov 2012;2(7):624–637. [DOI] [PMC free article] [PubMed] [Google Scholar]