Low et al. compare the reactivation of memory CD8+ T cells resident in the lung or draining lymph node after a secondary influenza infection and find that memory T cells in the different anatomical locations have unique reactivation mechanics in terms of reactivation kinetics and efficiency, as well as antigen-presenting cell requirements that drive qualitatively distinct recall responses.
Abstract
CD8+ tissue-resident memory T cells (TRM cells) are poised at the portals of infection and provide long-term protective immunity. Despite their critical roles, the precise mechanics governing TRM cell reactivation in situ are unknown. Using a TCR-transgenic Nur77-GFP reporter to distinguish “antigen-specific” from “bystander” reactivation, we demonstrate that lung CD8+ TRM cells are reactivated more quickly, yet less efficiently, than their counterparts in the draining LNs (TLN cells). Global profiling of reactivated memory T cells revealed tissue-defined and temporally regulated recall response programs. Unlike the reactivation of CD8+ TLN cells, which is strictly dependent on CD11c+XCR1+ APCs, numerous antigen-presenting partners, both hematopoietic and non-hematopoietic, were sufficient to reactivate lung CD8+ TRM cells, but the quality of TRM cell functional responses depended on the identity of the APCs. Together, this work uncovers fundamental differences in the activation kinetics, mechanics, and effector responses between CD8+ memory T cells in peripheral vs. lymphoid organs, revealing a novel tissue-specific paradigm for the reactivation of memory CD8+ T cells.
Introduction
Spatial compartmentalization is a universal strategy to allocate specialization of functional properties to diverse subsets of cells. Memory CD8+ T cells can be compartmentalized into at least two major categories. One category consists of cells that reside within the tissues long-term (tissue-resident memory T cells [TRM cells]; Jiang et al., 2012; Gebhardt et al., 2009; Wakim et al., 2010; Teijaro et al., 2011), and another consists of cells that recirculate within the blood, tissues, and lymphatics (circulating memory T cells [TCIRC cells]). The latter category includes cell subsets conventionally referred to as central memory T cells (TCM cells), effector memory T cells (TEM cells), and peripheral memory T cells (TPM cells; Sallusto et al., 1999; Gerlach et al., 2016). In addition to their varying anatomical distributions, these memory cell subsets also display several unique phenotypical and functional properties (Low and Kaech, 2018; Schenkel and Masopust, 2014). TCM cells preferentially reside within lymphoid organs and are characterized by their superior regenerative potential and IL-2 production; TEM cells (CX3CR1hi) tend to dwell in the vasculature and, upon activation, are able to exert immediate cytotoxic functions; TPM cells (CX3CR1int) are unique patrollers that survey nonlymphoid tissues with a unique migration pattern from blood to tissue to lymph; and TRM cells are tissue sentinels that are able to set up immediate antiviral states locally within infected tissues following reactivation. TRM cells largely reside at barrier tissues and can be found in other peripheral tissues, including secondary (2°) lymphoid organs (Beura et al., 2018b). Collectively, these different subsets of memory T cells cooperate with one another to provide tiered layers of defense during reinfection (Schenkel et al., 2014; Ariotti et al., 2014; Jiang et al., 2012; Wu et al., 2014; Iijima and Iwasaki, 2014).
A hallmark of immunological memory is more rapid induction of effector responses, and thus the presumption was that unlike naive T cells, memory T cells do not require licensing from professional APCs to enable faster reactivation. However, in 2005 this model was brought into question by a study from Dr. Lefrancois’ group which showed that memory CD8+ T cells are paradoxically dependent on CD11c+ dendritic cells (DCs) for their reactivation (Zammit et al., 2005). Subsequent reports also demonstrated that memory CD8+ T cells require CD28 costimulation for their optimal 2° expansion (Borowski et al., 2007; Fuse et al., 2008). These studies challenged the earlier presumption that memory T cells do not require professional APCs for their reactivation; however, these studies were done before the discovery of tissue-dwelling TRM cells and therefore primarily focused on the recall responses of TCIRC cells. With our current appreciation of different subsets of memory T cells that occupy distinct anatomical niches, we revisited this question to assess if the same principles and mechanisms govern the reactivation of all memory CD8+ T cell subsets. That is, do all memory T cells require professional APCs for their reactivation? This is a particularly important question for CD8+ TRM cells that lie at the portals of infection in barrier tissues.
We addressed this question using models of influenza infection, because we need to better understand how protective memory T cell recall responses are regulated in the lung. Many infections are spread through the respiratory tract, such as influenza and coronaviruses, and vaccines that generate lung TRM cells may hold the key to developing potent, durable, and broad immunity to these pathogens. Moreover, the conventional DCs (cDCs) in the lung are well defined, consisting of two main populations of cells, CD103+ XCR1+ and CD11b+ DCs (Heath and Carbone, 2009), whose migration from the lung into the mediastinal LN (medLN) following influenza infection peak around day 2–3 post-infection (p.i.; Helft et al., 2012; Kim and Braciale, 2009). In particular, Batf3-derived CD103+ XCR1+ DCs are required to generate effector CD8+ T cells during priming (Kim et al., 2014). Some of these effector T cells then differentiate into different subsets of memory TCIRC cells and lung-resident TRM cells that are critical for heterologous immunity to influenza infection (Wu et al., 2014; Laidlaw et al., 2014). However, despite their importance, the mechanisms that govern memory CD8+ T cell reactivation within various tissues to kickstart their protective responses remain unclear.
The present study resolves the conundrum between the requirement for APC licensing and rapid memory T cell activation by discovering that the dependence on CD11c+ DCs for memory CD8+ T cell reactivation is location dependent. We found that during a 2° influenza infection, lung CD8+ TRM cells have less-discriminate modes of reactivation and receive the initial antigenic signal from multiple types of APCs, including nonhematopoietic cells. On the other hand, reactivation of memory CD8+ T cells in the medLN was strictly dependent on CD11c+ XCR1+ DCs, even though the source of antigens can be found in other non-CD11c+ cells. Finally, we also revealed that many key genes induced within reactivated lung TRM cells depend on antigen presentation from nonhematopoietic cells, indicating that these non-DC interactions are critical for shaping the quality of the local TRM cell protective response.
Results
CD8+ lung TRM cells are reactivated more rapidly but less efficiently than LN memory T cells (TLN cells)
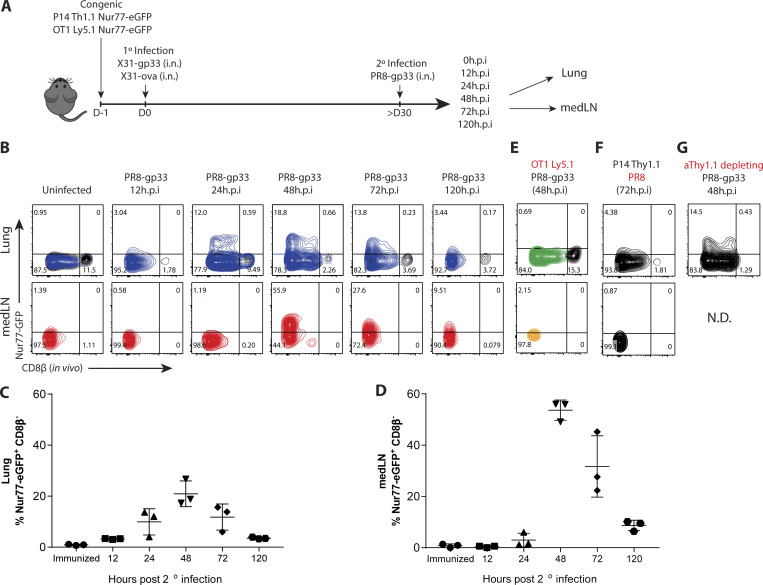
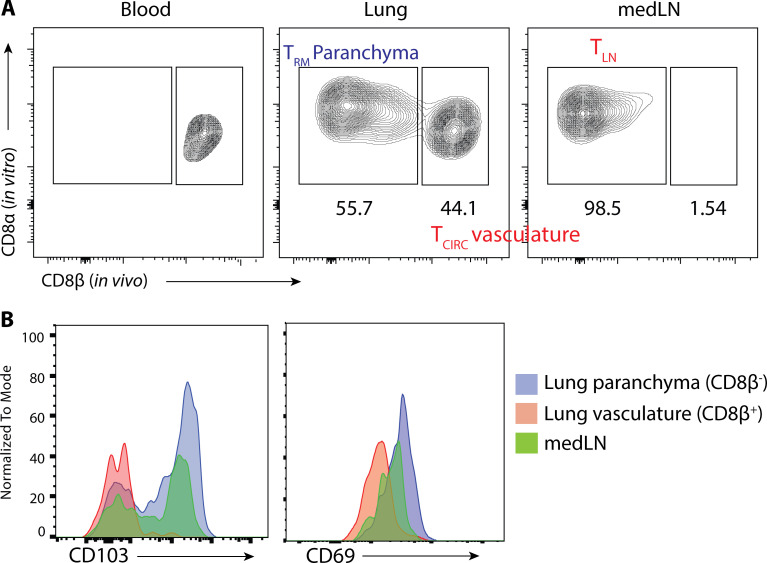
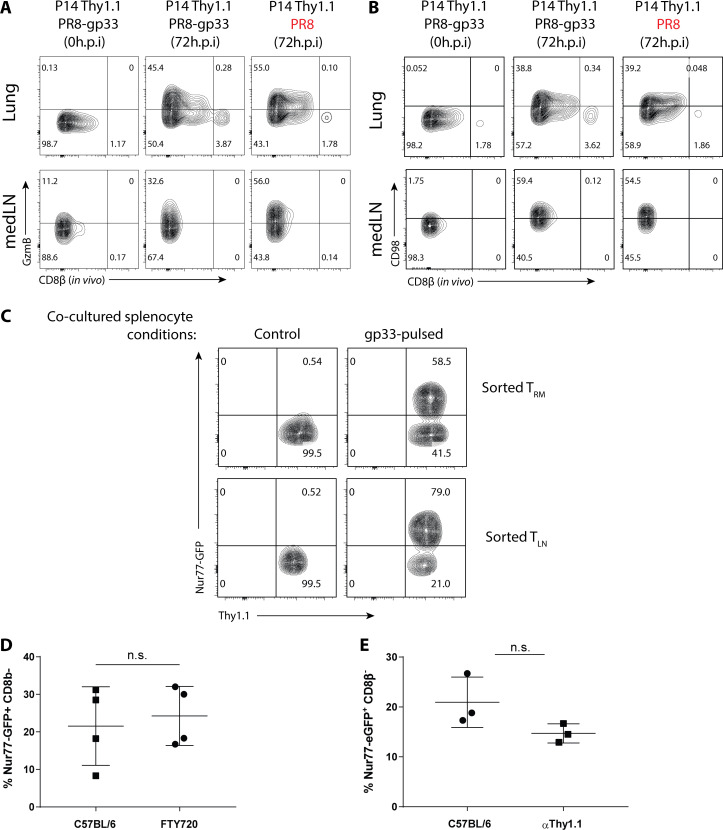
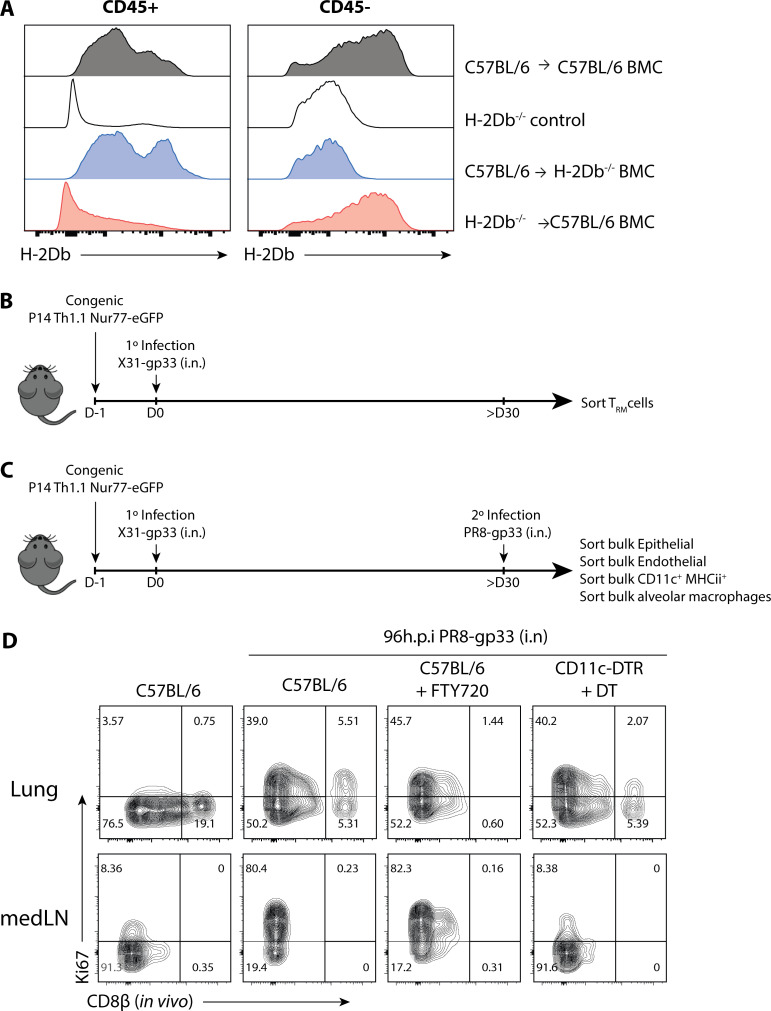
We began to investigate the mechanics that govern memory T cell reactivation using influenza infection, because local CD8+ TRM cells, recirculating memory CD8+ cells, and inflammation-driven bystander responses collectively contribute to protection against influenza reinfection (Wu et al., 2014; Slütter et al., 2013; Ely et al., 2003). We developed a system wherein we could distinguish between antigen-specific and “bystander” TRM cell reactivation using the Nur77 (Nr4a1)-eGFP transgene (referred to as Nur77-GFP), a downstream reporter of TCR signaling (Moran et al., 2011). We generated Nur77-GFP P14+ (lymphocytic choriomeningitis virus [LCMV] H-2Db GP33–41–specific TCR) and Nur77-GFP OT-I+ (OVA257–264–specific TCR) transgenic mice and then transferred 50,000 naive P14+ (Thy1.1+) and OT-I+ (Ly5.1+) cells in combination into C57BL/6 mice that were later infected simultaneously with influenza viruses X31-gp33 and X31-ova i.n. At 30 d p.i., we rechallenged the immunized mice with a heterosubtypic strain of influenza PR8-gp33 to selectively reactivate P14+ memory CD8+ T cells in a TCR-dependent manner, while the neighboring OT-I+ memory T cells allowed for the assessment of bystander activation (Fig. 1 A). 5 min before euthanasia, we administered fluorescently labeled anti-CD8β intravascularly to distinguish CD8+ cells in the lung tissue parenchyma from those in the vasculature (Fig. S1 A). While resident memory CD8+ T cells have been described in draining LNs (Beura et al., 2018b), in our studies, medLNs contained a mixture of memory cells with resident and circulating phenotypes, defined by their surface expression of CD69 and CD103 (Fig. S1 B). Therefore, we coined these CD8+ LN memory T cells collectively TLN cells. Kinetic profiling of Nur77-GFP expression showed that P14+ lung TRM cells were reactivated first, as early as 12–24 h p.i. (h.p.i.), and peaked at 48 h.p.i., whereas reactivation of P14+ TLN cells in the medLN was delayed by an entire day and was only evident at 48 h.p.i. (Fig. 1, B–D). Notably, the efficiency of medLN memory T cell reactivation was superior to that in the lung based on the frequency of Nur77-GFP+ cells at the peak of the response (Fig. 1, B–D). This result demonstrated that TRM cells are indeed the “first responders” to a 2° infection, which has always been inferred but never formally shown. Internal controls validated that Nur77-GFP was a selective marker for antigen-specific memory T cell reactivation, as only the P14+ and not the OT-I+ memory CD8+ T cells increased Nur77-GFP expression following PR8-gp33 challenge (Fig. 1 E). Further, challenge with the parental PR8 strain that lacks the gp33 epitope also did not augment GFP expression in P14+ Nur77-GFP memory T cells (Fig. 1 F), but it notably induced the expression of GranzymeB (GzmB) and CD98 on the memory cells, indicating these are markers of bystander and not antigen-dependent activation (Fig. S2, A and B). Additionally, when ex vivo–sorted P14+ TRM or TLN were cultured in vitro with gp33 peptide–pulsed splenocytes, no discernible difference was observed in the kinetics of Nur77-GFP upregulation (Fig. S2 C). Therefore, we specifically chose Nur77-GFP for our subsequent analysis to investigate which cells present antigen to TRM cells in the lung, because it was the only readout we identified that selectively differentiated between TCR and bystander activation early after 2° infection. To ensure we were assessing the reactivation of preexisting TRM cells and not infiltrating TLN cells that were recruited from the blood into the lung tissue early after 2° infection, we administered low-dose αThy1.1 mAb to deplete P14+ TLN cells or gave fingolimod (FTY720) to prevent lymphocyte recirculation. While the P14+ TLN cells in the blood were eliminated by both treatments, neither had any significant effect on the kinetics or frequency of Nur77-GFP expression in the P14+ TRM cells in the lungs at the early time points analyzed (Fig. 1 G and Fig. S2, D and E).
Figure 1.
Lung TRM cells are reactivated more rapidly but less efficiently than TLN cells in medLNs. (A) P14/OT-I Nur77-GFP immune chimeras were generated by cotransferring naive P14+ (Thy1.1+) and OT-I+ (Ly5.1+) Nur77-GFP CD8+ T cells (5 × 104 cells each) into C57BL/6 (Th1.2+/Ly5.2+) mice 1 d before i.n. coinfection with recombinant influenza X31-gp33 and X31-ova. 30 d later, the P14/OT-I Nur77-GFP immune chimeras were reinfected with PR8-gp33 i.n., and reactivation of P14+ lung TRM and medLN TLN cells was assessed at 0, 12, 24, 48, and 120 h.p.i. (B) Representative flow plots of Nur77-GFP expression in P14+ lung TRM cells and medLN TLN cells. In vivo labeling with αCD8β antibody was used to distinguish CD8+ TRM cells in the lung parenchyma from CD8+ T cells in the vasculature. (C and D) Frequency of Nur77-GFP+ CD8β− P14+ cells (i.e., top left quadrant of flow plots) in B was quantified. Data are expressed as mean ± SD. (E and F) As negative controls, Nur77-GFP expression in OT-I+ and P14+ cells was examined after PR8-gp33 and PR8 2° infection, respectively. (G) To limit the contribution of TCIRC cells, a low dose of αThy1.1 (clone 19E12)–depleting antibody was administered i.p., and the frequency of Nur77-GFP+ P14+ TRM cell reactivation was examined. N.D., not detected. Antigen-activated P14+ lung TRM (blue) and medLN TLN (red) cells are distinguished from the bystander-activated OT-I+ lung TRM (green) and medLN TLN (gold) cell samples by color. Data shown are representative of two independent experiments (n = 3–5 mice/group).
Figure S1.
Intravascular staining to distinguish parenchymal TRM cells from vasculature TLN cells. (A) Flu-immunized mice were injected i.v. with fluorescently labeled αCD8β antibody 5 min before sacrifice. Blood, lung, and medLN were collected and stained with αCD8α antibody ex vivo. Shown are gated on total CD8α+ T cells. (B) Expression of CD69 and CD103 in medLN, lung parenchyma (CD8a+ CD8β−), and lung vasculature (CD8a+ CD8β+) are shown.
Figure S2.
GzmB and CD98 are markers of bystander inflammation, and recruitment of circulating T cells is minimal at 48 h.p.i. (A and B) P14+ immunized mice were infected with PR8-gp33 (TCR-activation) or PR8 (bystander-activation) i.n., and the expression of GzmB (A) and CD98 (B) at 72 h.p.i. in the lung (top panel) and medLN (bottom panel) is shown. (C) P14+ TRM and TLN cells were sorted from resting P14+ chimera and co-cultured with control- or gp33-pulsed splenocytes, and Nur77-GFP expression was accessed 24 h later. (D and E) To limit the contribution of TCIRC cells to the readout of Nur77-GFP in cells in the lung, FTY720 (D) and a low dose of αThy1.1-depleting antibody (E) were administered i.p., and the effects on P14+ TRM cell reactivation was examined 48 h.p.i. n.s., not significant. Data are expressed as mean ± SD.
Lung CD8+ TRM cells and TLN cells exert tissue-defined functional programs that are temporally regulated
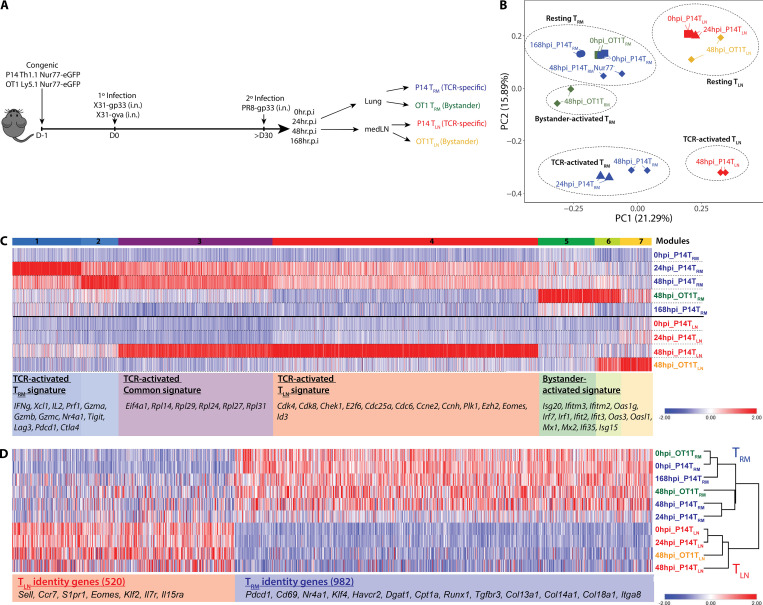
Using the established Nur77-GFP system, we investigated whether the quality of the 2° recall responses between TRM and TLN cells was influenced by their anatomical locations and/or their differentiation states by profiling the global gene expression changes via RNA sequencing (RNA-seq) in P14+ and OT-I+ TRM and TLN cells before and 24 and 48 h after in vivo PR8-gp33 reinfection. Activated P14+ T cells were sorted based on Nur77-GFP+ expression, and bystander-activated OT-I T cells were sorted based on CD98hi expression (Fig. 2 A). Unbiased clustering using k-means analysis (k = 5) and principal-component analysis (PCA) identified five clusters (drawn ellipses) of samples broadly categorized into resting TRM cells, resting TLN cells, TCR-activated TRM cells, TCR-activated TLN cells, and bystander TRM cells (Fig. 2 B). Several points emerged from this analysis. First, the resting TRM and TLN cell samples segregated from each other before and after 2° infection, indicating distinct gene expression profiles between the two memory T cell populations at baseline as well as after TCR activation (Nur77-GFP+). Second, as one would predict if Nur77-GFP was a faithful readout of TCR activation, GFP-negative (Nur77-GFP−) P14+ TRM cells at 48 h.p.i. had gene expression profiles similar to resting lung TRM P14+ cells but also shared some properties with bystander-activated OT-I+ TRM cells. Further, the lung TRM cells showed a TCR-activated gene signature by 24 h.p.i., which was not evident in the medLN TLN cells until 48 h.p.i., supporting TRM cells are first responders transcriptionally (Fig. 2, B and C). Third and most revealing, tissue-specific responses indicative of a division of labor were observed between the memory T cells reactivated in the lung (Fig. 2 C, modules 1 and 2) and the medLN (module 4). Specifically, reactivated TRM cells in the lung up-regulated antiviral and cytotoxic molecules such as Ifng, Gzma, and Gzmb. In contrast, reactivated memory T cells in the medLN more robustly up-regulated genes involved in proliferation such as Cdk4, Cdk8, Chek1, and E2f7, in line with the well-known superior proliferative capacity of TCM cells. There was a common gene set “reactivation genes” including ribosomal genes (Rpl family genes) induced in both TRM and TLN populations, demonstrating core protein synthesis pathways underling memory T cell recall responses (Fig. 2 C, module 3). Fourth, there was a distinct bystander-activated signature enriched in IFN-stimulated genes (ISGs; Isg20, Ifitm3, and Mx1) that were predominantly up-regulated in OT-I+ TRM cells, likely due to local production of type I IFNs (Stetson and Medzhitov, 2006; Fig. 2 C, clusters 5–7). Interestingly, activated (Nur77-GFP+) P14+ TRM cells at 48 h.p.i. exhibited a dampened bystander-driven interferon signature relative to their Nur77-GFP− counterparts in the lung (Fig. 2 B), demonstrating that TCR-driven signals override instructions set up by bystander inflammation. Lastly, hierarchical clustering using Spearman’s rank correlation revealed that the unique genetic signatures that define resting TRM and TLN cells (at 0 h.p.i.; Mackay et al., 2013; Fig. S3) were not lost upon reactivation, indicating maintenance of their distinct memory T cell identities shortly after reactivation (Fig. 2 D).
Figure 2.
CD8+ memory T cells in the lung and medLNs display tissue-defined functional programs that are temporally regulated following reactivation in situ. (A) P14+ and OT-I+ memory CD8+ T cells were purified at 0, 24, 48, and 168 h after 2° infection from the lung and medLNs, and RNA-seq was performed. Activated P14+ T cells were sorted based on Nur77-GFP+ expression, and bystander-activated OT-I T cells were sorted based on CD98hi expression. (B) PCA of global gene expression showed five distinct clusters reflected by dotted ellipses. (C) Heatmap of differentially expressed TCR-activated genes and bystander-activated genes (log2FC ≥2 and P < 0.01). Seven modules were distinguished and several representative genes are shown. TCR-activated P14+ Nur77-GFP+ lung TRM (blue) and medLN TLN (red) are distinguished from the bystander-activated (Nur77-GFP− CD98hi) OT-I+ lung TRM (green) and medLN TLN (gold) samples by color. Squares, 0 h.p.i; triangles, 24 h.p.i.; diamonds, 48 h.p.i; circles, 168 h.p.i. (D) TRM and TLN identity genes were identified by comparing 0 h resting P14+ memory CD8+ T cells from the lung and medLNs, based on 1.5-log2FC and P < 0.05, and the temporal changes of these identity genes following reactivation in TRM and TLN cell populations are shown in this heatmap.
Figure S3.
TRM and TLN identity genes are maintained following reactivation. TRM and TLN identity genes were identified by comparing 0-h resting P14+ TRM cells with 0-h resting P14+ TLN cells based on 1.5-log2FC and P < 0.05, as represented in this volcano plot. 982 TRM identity genes and 520 TLN identity genes were identified.
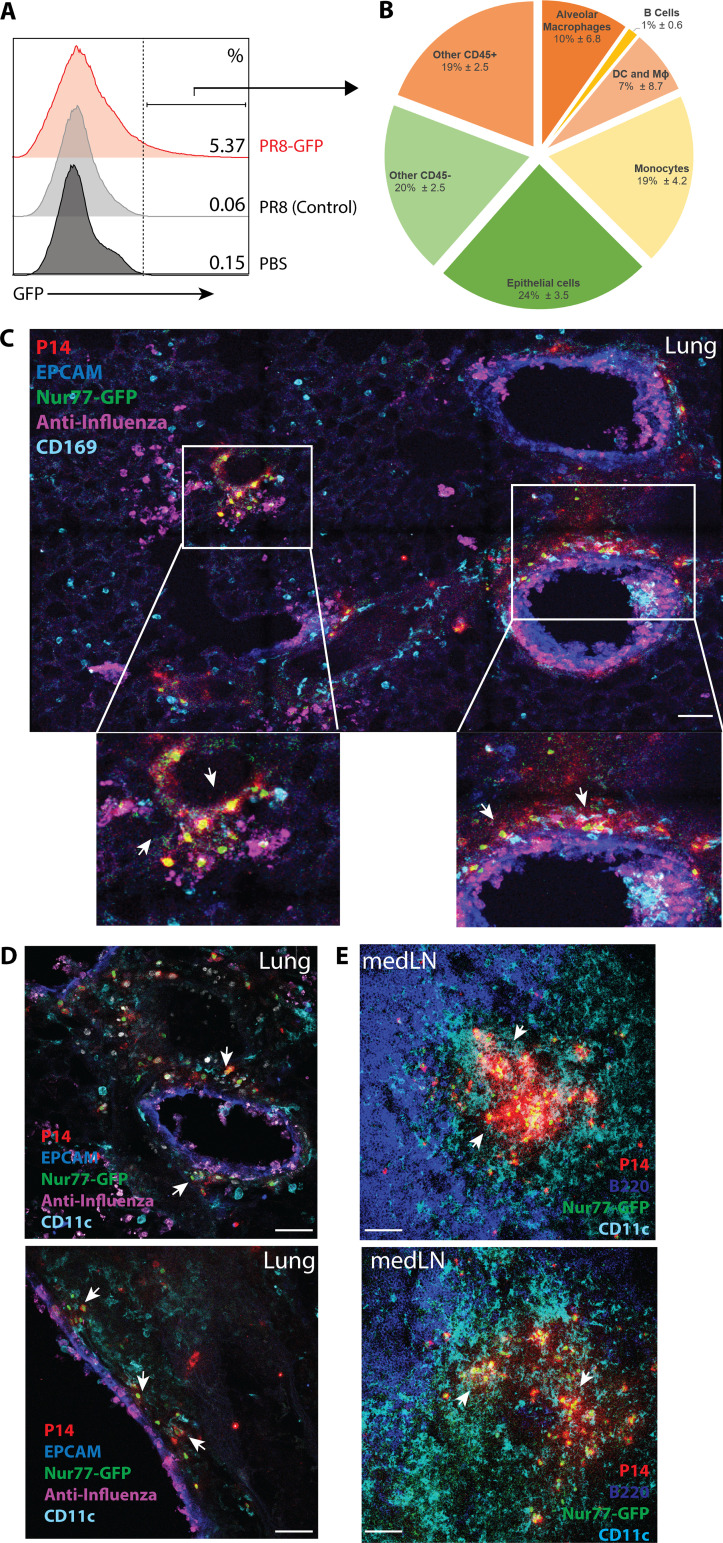
Reactivated TRM cells are in close proximity to different infected cell types
How are these immediate protective CD8+ TRM cell responses triggered in the local environment? The current paradigm is that memory CD8 T cell 2° responses require licensing by CD11c+ DCs (Zammit et al., 2005); however, it is not known if this also applies to TRM cells reactivated locally in peripheral tissues. To begin uncovering the mechanics that govern TRM cell reactivation in situ, we first sought to identify the potential APCs by infecting X31-immune mice with a PR8 strain that expresses GFP (PR8-GFP) or PR8 as negative staining control. Infected GFP+ cells were assessed using flow cytometry (Fig. 3 A), which showed that the primary cell types infected by influenza during 2° infection were CD11c+ cells, CD169+ macrophages, Ly6C+ monocytes, and EPCAM+ epithelial cells (Fig. 3 B), similar to previous studies on PR8-GFP infection on naive mice (Manicassamy et al., 2010; De Baets et al., 2015). Second, immunofluorescent microscopy of lungs containing P14+ Nur77-GFP TRM cells shortly after 2° infection with PR8-gp33 showed that the activated Nur77-GFP+ TRM cells were typically in close proximity to infected cells, such as epithelial cells, CD169+ cells, and CD11c+ cells (Fig. 3, C and D). In contrast, Nur77-GFP+ in the medLN were much more densely aggregated in close proximity to CD11c+ cells in the T cell zone (Fig. 3 E), supporting the higher efficiency of memory T cell reactivation in the draining LNs observed in Fig. 1 D.
Figure 3.
Reactivated TRM cells are in close proximity to different infected cell types. (A) Influenza-immunized mice were infected with PR8-GFP and PR8 (as control), and the lungs were analyzed by flow cytometry 48 h.p.i. (B) The pie chart shows the types of infected GFP+ cells identified as a fraction of total GFP+ cells (pie chart: alveolar macrophages [CD45+ CD169+ SiglecF+], B cells [CD45+ B220+], DCs and macrophages [CD45+ SiglecF− B220− CD11c+ Ly6C−], monocytes [CD45+ SiglecF− B220− CD11c+ Ly6C+], and epithelial cells [CD45− EPCAM+]). (C–E) P14+ immune chimeras were rechallenged with PR8-gp33, and the localization of activated P14+ Nur77-GFP+ cells in the lung (C and D) and medLN (E) at 48 h.p.i. was analyzed by immunofluorescence confocal imaging. Arrows indicate TCR-activated regions of P14+ Nur77-GFP+ cells and their interactions with infected cells and different immune subsets. Data shown are representative of two to three independent experiments (n = 3–5 mice/group). All scale bars indicate 50 µm.
cDCs are dispensable for lung CD8+ TRM reactivation, unlike in the medLNs
Next, to identify relevant APCs in TRM and TLN reactivation, we made P14+ Nur77-GFP immune chimeras in CD11c-DTR, Xcr1-DTR, and Zbtb46-DTR hosts that were subsequently depleted of DTR-expressing cells by diphtheria toxin (DT) treatment before 2° infection (Fig. S4 A). First, we examined the medLN CD8+ memory T cells and observed, as predicted, there was a major defect in their reactivation in the absence of CD11c+ and Zbtb46+ cells which depletes cDCs (Fig. 4 G and Fig. S4 B). Further, using XCR1-DTR hosts to distinguish between the two major populations of cDCs in the lung (CD103+ XCR1+ and CD11b+; Ballesteros-Tato et al., 2010; Kim and Braciale, 2009; Ohta et al., 2016), we found that the migratory XCR1+ DCs were critical to reactivate medLN memory T cells (Fig. 4 B). In stark contrast, lung TRM cell reactivation was not affected by depletion of CD11c+, XCR1+, or Zbtb46+ cDCs (Fig. 4 C), and the viral titer in the CD11c-depleted group was also not affected (Fig. S4 C). Second, we examined other candidate APC populations including CD169+ macrophages, B cells or GR-1+ monocytes by using CD169-DTR or uMT−/− hosts or depleting αGR-1 mAbs, respectively, to remove these cell populations from lungs during 2° infection (Fig. S4 A). However, neither lung TRM nor medLN TLN reactivation was affected by deletion of these cell types indicating they were not necessary APCs during influenza reinfection (Fig. 4, B and C). These latter experiments also reaffirmed that DT treatment alone does not induce spurious, nonspecific Nur77-GFP expression. Since influenza induces a productive local infection, we also investigated the APC requirements for memory T cell reactivation following intranasal infection with LCMV, a systemic pathogen that not only has different viral tropism but also has previously been shown to infect multiple cell types, including subcapsular macrophages in the draining LN (Sung et al., 2012). There were no major differences in the results between 2° infection with LCMV or influenza. That is, lung TRM cells could still be reactivated by LCMV-infected cells in the absence of CD11c+ cells, whereas the memory T cells in the medLN could not (Fig. 4 D; note that there was a modest reduction in percentage of Nur77-GFP+ cells in the lungs of LCMV-infected CD11c-DTR mice, suggesting that cDCs may be a more prominent APC in this tissue during LCMV infection relative to influenza). To verify that LCMV infected multiple cell types in the medLN after intranasal infection, we infected influenza-immune mice with LCMV-ZsGreen to trace LCMV-infected cells and quantified the overlap with CD11c+ cells, F4/80+ cells, or double-negative (“other”) cells in medLNs using Imaris software (Fig. 4, E and F). Consistent with a previous study (Sung et al., 2012), of the LCMV infected cells, ∼40% were CD11c+ F4/80−, ∼10% were F4/80+ CD11c−, and the remaining were other cell types (Fig. 4, E and F). Thus, even though there were multiple putative APCs in the medLN, memory T cells in the medLN were still reliant on CD11c+ cells for their reactivation. Taken together, in both influenza and LCMV reinfection, lung TRM CD8 T cell reactivation occurred independent of cDCs, demonstrating that the mechanisms governing memory T cell reactivation operate in a tissue-specific manner.
Figure S4.
Depletion validation and Zbtb46+ cells are required for medLN TLN cell, but not lung TRM cell, reactivation. P14+ immune chimeras were generated in various genetic hosts, and DT was administered i.t. 1 d before 2° infection to deplete DTR-expressing cells; for αGR-1 depletion, 200 μg αGR-1 (RB6-9C5) was administered i.p. daily starting the day before 2° rechallenge. (A) Validation of depletion strategies across different immunized-genetic hosts 48 h after the initial treatments in the lung or bronchoalveolar lavage. (B) Frequency of Nur77-GFP+ P14+ TRM cells in the lung and TLN cells in the medLN were quantified at 48 h.p.i. in Zbtb46-DTR hosts. Statistical analysis was performed using Student’s t test (two tailed); ****, P < 0.0001. (C) Flu viral titer was obtained by quantitative PCR of whole lung tissue for PR8 polymerase acidic protein (PA) 48 h.p.i. Data are expressed as mean ± SD.
Figure 4.
CD11c+ XCR1+ cells are strictly required for medLN CD8+ TLN reactivation, but conventional APCs are dispensable for lung CD8+ TRM reactivation. (A) P14+ immune chimeras were generated in various genetic hosts as shown. For DTR mice, DT was administered i.t. 1 d before 2° infection to deplete DTR-expressing cells; for αGR-1 depletion, 200 μg αGR-1 (RB6-9C5) was administered i.p. daily starting the day before 2° rechallenge. For LCMV rechallenge experiments, flu-immunized mice were 2° infected with Armstrong strain of LCMV i.n. or LCMV-GFP. (B–D) Quantification of the frequency of Nur77-GFP+ P14+ TLN cells in the medLN (B) and P14+ TRM cells in the lung (C) at 48 h after 2° PR8-gp33 infection (B and C) or 48 h after 2° LCMV infection (D). Data shown are a collection of two or more independent experiments (n = 3–5 mice/group). (E and F) P14+ immune chimeras were rechallenged i.n. with LCMV-ZsGreen (E), and the colocalizations of ZsGreen signal with various immune cells in the medLN was quantified by Imaris (F). Top scale bars indicate 150 µm, and bottom scale bars indicate 20 µm. Data are expressed as mean ± SEM. Statistical analysis was performed using Student’s t test (two tailed) comparing immunized C57BL/6 to various treatment groups. ****, P < 0.0001.
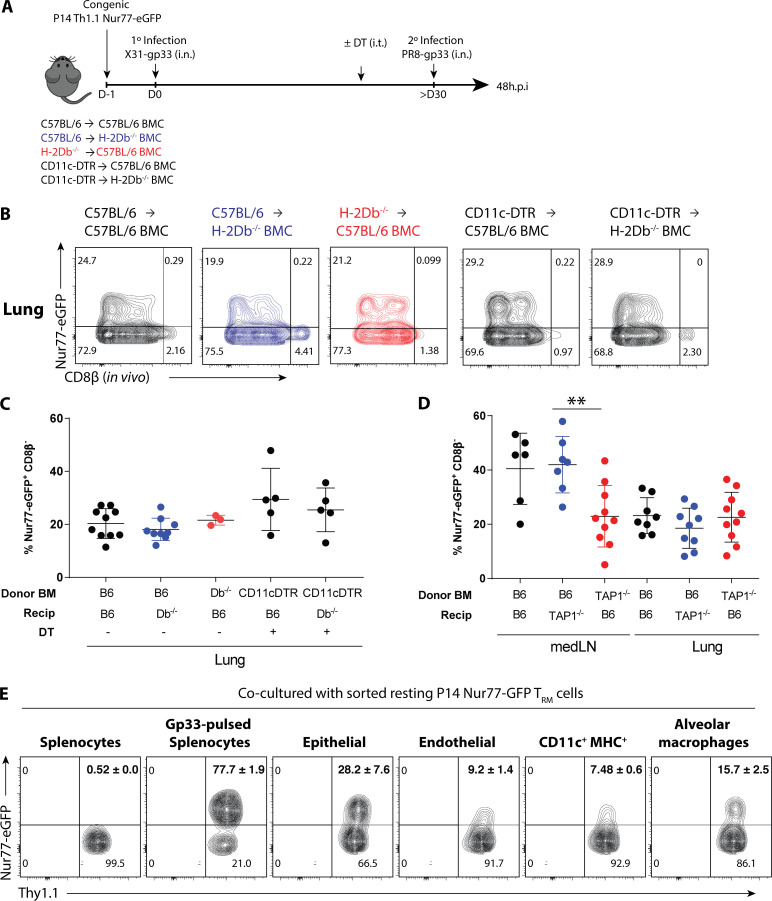
CD8+ lung TRM cells can be reactivated by both hematopoietic and nonhematopoietic APCs
Since the airway epithelium is a major reservoir for influenza infection and many lung CD8+ TRM cells are intraepithelial due to their expression of CD103 (Wu et al., 2014; Laidlaw et al., 2014; Fig. 3 C and Fig. S1 B), we considered that epithelial cells may play a key role in reactivating CD8+ TRM cells. To address this question, three groups of bone marrow chimeras (BMCs) were generated by transferring (1) H-2Db+/+ BM → C57BL/6 recipients, (2) H-2Db+/+ BM→ H2-Db−/− recipients, or (3) H2-Db−/− BM → C57BL/6 recipients (Fig. 5 A and Fig. S5 A). After BM reconstitution, P14+ Nur77-GFP immune chimeras were generated as previously described, and subsequently reinfected with PR8-gp33. Unexpectedly, lung CD8+ TRM reactivation (based on Nur77-GFP expression at 48 h.p.i.) occurred regardless if antigen was presented on hematopoietic or nonhematopoietic cells (Fig. 5, B and C). Furthermore, we tested the possibility of redundancy between nonhematopoietic cells and classical antigen presenting CD11c+ cells by generating BMCs with CD11c-DTR BM → C57BL/6 or H2-Db−/− recipients. After BM reconstitution, P14+ Nur77-GFP immune chimeras were generated as previously described, and before PR8-gp33 reinfection, the hosts were treated with DT to delete CD11c+ DCs. Surprisingly, the absence of antigen presentation from both nonhematopoietic cells and CD11c+ cells did not reduce the frequency of Nur77-GFP+ P14+ lung TRM cells, indicating alternative APCs such as monocytes and/or macrophages were sufficient for TRM cell reactivation (Fig. 5, B and C). We further validated our finding in TAP1−/− BMCs, which lack the machinery to load peptide onto MHC class I in either the hematopoietic (TAP1−/− BM → C57BL/6 recipients; TAP1BM−/−) or nonhematopoietic (C57BL/6 BM→ TAP1−/− recipients; TAP1nonhemo−/−) compartments. Consistent to the results of H-2Db−/− BMCs, CD8+ TRM cell reactivation was unaffected in all three BMC groups, while the activation of medLN TLN cells were significantly impaired if the hematopoietic cells lack TAP1 expression (Fig. 5 D). Altogether, these results indicate considerable redundancy between the major APC populations in reactivating lung CD8+ TRM cells in vivo during 2° infection. To more closely examine this apparent redundancy in the cell types that can reactivate lung TRM cells, Nur77-GFP P14+ CD8+ TRM cells were isolated from the lungs of X31-gp33 immune mice and co-cultured with bulk epithelial cells, endothelial cells, CD11c+ MHCII+ cells, and alveolar macrophages purified from mice infected with PR8-gp33 48 h prior or splenocytes loaded with and without gp33 peptide as controls (Fig. 5 E and Fig. S5, B and C). This experiment supported the above genetic data, because all of these lung-derived cell populations could reactivate CD8+ TRM cells ex vivo (Fig. 5 E).
Figure 5.
CD8+ lung TRM cells can be reactivated by both hematopoietic and nonhematopoietic APCs. (A) P14+ immune chimeras were generated in different H-2Db−/− BMCs as outlined. For CD11c-DTR donor bone marrow groups, DT was administered 500 ng/mouse (i.p.) 1 d before 2° infection. (B) Representative flow plots of Nur77-GFP expression in the P14+ lung TRM cells at 48 h.p.i. and quantified in C. (D) P14+ immune chimeras were generated in TAP1−/− BMCs similar to A and the frequency of reactivated medLN TLN and lung TRM cells are quantified. (E) P14+ TRM cells were sorted from steady-state memory lung at >30 d p.i. and co-cultured with epithelial cells (EPCAM+), endothelial cells (CD31+), CD11c+ MHCII+ cells, and alveolar macrophages (CD169+) sorted from influenza-immunized mice that were 2°-reinfected with PR8-gp33 24 h prior. Flow plots of Nur77-GFP expression in P14+ TRM cells following overnight co-culture. Data shown are expressed as mean ± SD, representative of two independent experiments (n = 3–5 mice/group). Statistical analysis was performed using Student’s t test (two tailed). **, P < 0.01.
Figure S5.
CD8+ lung TRM cells are reactivated by both hematopoietic and nonhematopoietic APCs, and consequentially, their 2° expansion in situ is independent of cDCs. (A) Validation of H-2Db expression in CD45+ and CD45− compartments of BMCs. (B) Schematic of ex vivo co-culture. P14+ TRM cells were sorted from steady-state memory lung at >30 d p.i. and co-cultured with epithelial cells (EPCAM+), endothelial cells (CD31+), CD11c+ MHCII+ cells, and alveolar macrophages (CD169+) sorted from influenza-immunized mice that were 2°-reinfected with PR8-gp33 24 h prior. (C) P14+ immune chimeras were generated in C57BL/6 or CD11c-DTR hosts and treated with either FTY720 (i.p., daily 2 days prior) or 100 ng DT (i.t., every 2–3 d) the day before 2° reinfection with PR8-GP33 i.n. Shown are representative flow plots of the expression of Ki67 on P14+ cells 96 h.p.i. (D) P14+ immune chimeras were generated in C57BL/6 or CD11c-DTR hosts and treated with either FTY720 (i.p.; daily 2 d prior) or 100 ng DT (i.t.; every 2–3 d) the day prior to 2o reinfection with PR8-GP33 i.n. 96 h.p.i. the expression of Ki67 on P14+ cells were examined and quantified. After infection., the expression of Ki67 on P14+ cells were examined and quantified.
Activation by hematopoietic and nonhematopoietic cells induces differential functional output in TRM cells
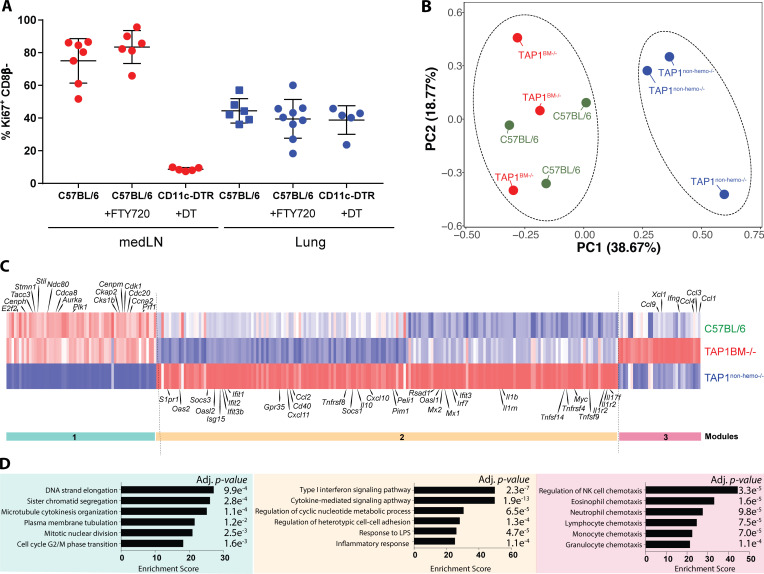
Since our data showed that TRM cells can be activated by both hematopoietic and nonhematopoietic cells, we sought to investigate if the quality of the 2° TRM response was affected by which APC type the TRM cell first encountered. First, the promiscuity in CD8+ TRM cell reactivation had apparent beneficial consequences as the proliferative burst of TRM cells in situ (Park et al., 2018; Beura et al., 2018a) was not constrained by CD11c+ cells. This contrasts with the medLN TLN cells, whose 2° expansion was strictly dependent on CD11c+ XCR1+ DCs (Fig. 6 A and Fig. S5 D). Second, we performed RNA-seq on 48-h reactivated Nur77-GFP+ TRM cells sorted from the lungs of C57BL/6 TAP1+/+, TAP1BM−/−, and TAP1nonhemo−/− BMCs. PCA and k-means analysis (k = 2) identified TRM cells reactivated in the absence of TAP1 on nonhematopoietic cells (TAP1nonhemo−/− BMCs) clustered distinctly from the control or TAP1BM−/− samples (Fig. 6 B). This indicated that overall, the antigen-driven interactions with nonhematopoietic cells had a large effect size on the quality of the TRM 2° responses. Interestingly, disruption of antigen presentation by either hematopoietic or nonhematopoietic cells differentially impacted the TRM activation signatures described in Fig. 2 (Fig. 6 C), indicating that distinct types of signals or instructions were sent by each type of APC to influence CD8+ TRM responses. In particular, three gene signature modules were most affected: (1) antigen presentation by nonhematopoietic cells was needed to induce genes involved in cell cycle and proliferation (e.g., Cdca8, Cdc20, and Cdk1 in module 1) and (2) restrain type I ISGs (e.g., Ifit1, Ifit2, and Mx1 in module 2) in the reactivated TRM cells. The latter result indicated this antigen-driven interaction with nonhematopoietic cells prevented the induction of a bystander-activated TRM cell signature (Fig. 2 C). (3) In contrast, antigen presentation by hematopoietic cells tempered the expression of several chemokines and cytokines (e.g., Ccl1, Ccl3, Ccl9, Ifng, and Xcl1 in module 3) in TRM cells (Fig. 6, C and D). This result suggested that antigen-dependent interactions with hematopoietic cells help tame inflammation and leukocyte recruitment, perhaps to keep the TRM response in check by provision of inhibitory ligands or cytokines to prevent excessive inflammation in the lung. In summary, these findings demonstrate that TRM cells not only recognize their cognate antigen from multiple cell types during reinfection, but their functional outputs are qualitatively different depending on the nature of the APC encountered.
Figure 6.
Activation by hematopoietic and nonhematopoietic cells induces differential functional output in TRM cells. (A) P14+ immune chimeras were generated in C57BL/6 or CD11c-DTR hosts and treated with either FTY720 (i.p., daily 2 d prior) or 100 ng DT (i.t., every 2–3 d) the day before 2° reinfection with PR8-GP33 i.n. 96 h.p.i. the expression of Ki67 on P14+ cells was examined and quantified. Data are expressed as mean ± SEM. (B) Activated P14+ TRM cells were sorted based on Nur77-GFP+ expression in C57BL/6 TAP1+/+, TAP1BM−/−, TAP1nonhemo−/− BMCs, and RNA-seq was performed. PCA of global gene expression showed two distinct clusters reflected by dotted ellipses. (C) Heatmap of differentially expressed genes between the control and TAP1−/− BMCs (≥ or ≤1.5 log2FC and P < 0.05), filtered on all the differentially expressed genes of TCR and bystander TRM cell signature from Fig. 2 C. Three main modules were observed. (D) Genes from each module were parsed for enrichment of biological pathways using Enrichr. Top pathways from Gene Ontology Biological Processes with corresponding adjusted P values are shown. Relevant gene names are highlighted in heatmap in C.
Discussion
Since the conceptualization of memory T cell subsets 21 yr ago (Sallusto et al., 1999), it is now well appreciated that there are different subsets of memory T cells that occupy distinct anatomical locations, have different functional properties, and collectively are critical in mounting a tiered layer of response during 2° infection (Schenkel and Masopust, 2014; Low and Kaech, 2018). Our study furthers the understanding of memory T cell biology by reshaping the current paradigm of memory CD8+ T cell reactivation, demonstrating that the mechanics that govern their reactivation are contextual and dependent on their anatomical locations. CD8+ TRM cells are tissue sentinels that provide protective antigen-specific and bystander-inflammatory responses, and we propose that their reactivation promiscuity by multiple types of APCs allows for more rapid and sensitive pathogen sensing. Conversely, CD8+ memory T cells in the draining LNs were strictly dependent on migratory CD11c+ DCs for their reactivation, even in cases such as LCMV infection, where more than half of the putative APCs in the medLN were not CD11c+ cells. Although temporally slower, interestingly, the organized architecture of the draining LN allowed for more concentrated and efficient memory T reactivation. Thus, our findings demonstrate major differences in tissue-dependent control of memory T cell reactivation between peripheral tissues and 2° lymphoid organs. Extension of this work will be needed to assess how common this principle is to other peripheral tissues and pathogens because viral tropism, antigen processing and tissue injury are additional factors to consider for assessing memory T cell reactivation, which is why we decidedly used live replicating virus for all our infections instead of peptide stimulation. We employed two different viruses, influenza A virus PR8 and LCMV, which are naturally transmitted via the respiratory route and can replicate to similar viral titers, yet induce different immunopathologies in the lung (Knudson et al., 2014; Chen et al., 2003). For example, influenza is cytopathic to lung epithelium whereas LCMV is not (Dylla et al., 2008; Pythoud et al., 2015). Nonetheless, in both infections, lung TRM cell reactivation could occur independent of CD11c+ cDC antigen presentation (albeit, there was greater reliance on CD11c+ APCs following LCMV compared with influenza, suggesting that the type of pathogen will likely also play a role in governing the mechanics of TRM cell reactivation in peripheral tissues). Relatedly, two studies on HSV in the skin (Wakim et al., 2008) and the female reproductive tract (Shin et al., 2016) found that DCs appear important for reactivation of CD8+ TRM cells, although these conclusions were based on the measurements of T cell expansion, disease scores, weight loss, survival, and viral titers many days following infection, as opposed to our study, which concentrated on the first phase of TRM cell reactivation. Nonetheless, it is possible that tissue-specific principles will govern TRM cell reactivation in different tissues, and some types of tissues (or pathogens) will display a dependence on cDCs for TRM cell reactivation; our study outlines an approach by which to assess this.
Elegant studies have shown that activation of TRM cells can lead to multiple functional outputs, including innate cell activation, immune cell recruitment, and in situ proliferation (Schenkel et al., 2014; Beura et al., 2018a; Park et al., 2018), to rapidly set up antiviral states across a broad range of tissue territory, but little is known about how these different functions are initiated and regulated. Notably, we observed distinct tissue-specific compartmentalization of the functional recall responses between memory T cells in the lung vs. medLN. This is important given that recent data show that CD69+ resident memory T cells can also persist in the draining LN (Beura et al., 2018b). Nonetheless, the transcriptional program of the memory T cells in the medLN were quite distinct from those in the lung both before and after 2° infection, suggesting a division of labor between the memory T cells in peripheral vs. 2° lymphoid tissues. Within 24–48 h, lung TRM cells expressed various cytotoxic, costimulatory and coinhibitory molecules including Ifng, Gzmb, Xcl1, Il2, Tnfrsf9 (4-1BB), Icos, Tigit, Pdcd1, Tnfrsf18 (GITR), Ctla4, and Lag3 to tightly balance the cytotoxic activities of the CD8+ T cells. Alternatively, the memory T cells in the medLN dominantly up-regulated cell cycle genes. It is important to point out that the lung TRM cells up-regulated these genes too, albeit to a substantially lower extent, in line with their ability to proliferate in situ (Beura et al., 2018a; Park et al., 2018). These results suggest that lung TRM cells exert a robust antiviral effector program immediately after reactivation, whereas those in the medLN dedicate their responses to a proliferative program. Both populations appear to engage anabolic programs by increasing protein synthesis.
Our findings also revealed that the different CD8+ TRM cell functional responses can be modulated by the nature of antigen-presenting partners, i.e., whether CD8+ TRM cells received antigen from hematopoietic cells or nonhematopoietic cells. Presentation by nonhematopoietic cells induced programs of cell cycle proliferation while simultaneously repressing the bystander-like IFN signature. Given that IFN signaling can be antiproliferative (Petricoin et al., 1997; Bromberg et al., 1996), this indicates that the interaction between TRM cells and nonhematopoietic APCs is important in finetuning the proliferative burst of TRM cells locally within the tissue. In agreement with our study, a prior study showed that the TRM cells in the female reproductive tract can also undergo in situ proliferation independent of CD11c+ cells (Beura et al., 2018a). Perhaps one way in which TCR signaling may suppress type I IFN signature is through T-bet induction, which has been previously shown to repress type I IFN transcription factor and ISGs in CD4 Th1 cells (Iwata et al., 2017). On the other hand, antigen presentation by hematopoietic cells was also important in regulating chemokine and cytokine production in TRM cells. Relatedly, another study showed that HSV-infected APCs could stimulate IFN-γ production by HSV-specific effector CD8+ T cells, whereas uninfected cross-presenting DCs could not (despite the fact that such DCs could activate naive T cells; Macleod et al., 2014); this is another demonstration of APC-mediated tuning of functional responses in effector and memory CD8+ T cells. A prominent feature of DC–T cell interactions is costimulation; however, our study highlights that perhaps the more dominant feature during a recall response is the restraint of memory T cell effector functions. Lung CD8+ TRM cells express PD-1 (Wu et al., 2014), and thus, possibly, this suppression operates through coinhibitory molecules such as PD-L1. In fact, PD-L1 expression on hematopoietic cells has previously been shown to limit CD8+ T cell functional responses during a chronic infection (Mueller et al., 2010).
Mounting human studies have correlated CD8+ TRM-like cells with better protection against infection and cancer, but these cells may also cause immunopathology in certain inflammatory and autoimmune diseases (Boyman et al., 2004; Ganesan et al., 2017; Wang et al., 2016; Edwards et al., 2018; Webb et al., 2014; Cheuk et al., 2017). These paradoxical roles in human health highlight the need to better understand the principles and cell types that govern memory T cell reactivation in situ, how their responses are regulated by antigen or bystander inflammation in different tissues, and how their functional outputs are modulated by different APCs. The results of this study bring forth new knowledge for how memory CD8+ T cell recall responses are differentially regulated in tissue-dependent contexts, which fundamentally enlightens our understanding of the different modes of memory T cell reactivation in situ.
Materials and methods
Mice, infections, and treatments
C57BL/6 (B6) mice were purchased from Charles River Laboratories. P14+ (LCMV H-2Db gp33–41–specific) TCR transgenic mice, OT-I+ (H-2Kb OVA257–264–specific) TCR transgenic mice, CD11c-DTR mice, and uMT−/− mice were purchased from The Jackson Laboratory. CD169-DTR were kindly provided by Dr. Masato Tanaka at the Tokyo University of Pharmacy and Life Sciences (Tokyo, Japan). H-2Db−/− mice were kindly provided by Linda Cauley (University of Connecticut, Farmington, CT). Xcr1-DTR mice were kindly provided by Dr. Tsuneyasu Kaisho (Wakayama Medical University, Wakayama, Japan). “P14+/OT-I+ immune chimera” were generated by transferring 5 × 104 Thy1.1+ P14+ CD8+ T cells and/or 5 × 104 Ly5.1+ CD8+ T cells i.v. into naive Thy1.2+/Ly5.2+ B6 recipients and subsequently inoculated i.n. with 0.8 × 105 TCID50 recombinant X31 influenza expressing the LCMV gp33–41 epitope (X31-gp33) and recombinant X31 influenza expressing OVA (X31-ova). For 2° LCMV infections, P14+ immune chimeras were infected with 5 × 104 PFU LCMV Armstrong or 105 LCMV-ZsGreen i.n. kindly provided by Prof. Juan Carlos de la Torre (Scripps Research, La Jolla, CA). In vivo depletion of circulating P14+ cells was achieved with i.p. injections of low dose (0.7 μg) αThy1.1 (clone 19E12) depleting antibody. 10 μg FTY720 (Cayman Chemicals) was administered i.p. for 2 d consecutively before reinfection and maintained daily treatment until end point. DT treatments were done by i.t. administration of 100 ng DT (Sigma) or 500 ng DT (Sigma) for BMCs. αGr-1 antibody (BioXcell; RB6-8C5) was administered i.p. for 3 d consecutively, starting 1 d before reinfection. For rechallenge experiments, mice were infected with 1,500 TCID50 recombinant PR8 influenza virus expressing the LCMV gp33 epitope (PR8-gp33) i.n. Prior to all intranasal infections, mice were anesthetized by i.p. injection of ketamine hydrochloride and xylazine in 0.2 ml PBS. For in vivo labeling, 0.8 µg αCD8β-PE antibody was administered i.v. All animal studies and procedures were performed in accordance with the Yale University Institution of Animal Care and Use Committee.
BMC
C57BL/6, H-2Db−/−, or TAP1−/− recipients were irradiated with 600 rad twice in 3–4 h intervals and reconstituted with 5–10 million bone marrow cells from C57BL/6, H-2Db−/−, TAP1−/−, or CD11c-DTR donors isolated from tibias and femurs. After 6–8 weeks of reconstitution, P14+ immune chimeras were generated as previously described.
RNA-seq
P14+ and OT-I+ TRM cells and TLN cells were sorted from the lungs and medLN of P14+ and OT-I+ immune chimeras at indicated hours after infection. P14+ cells were sorted based on Nur77-GFP− for 0 h.p.i. and 168 h.p.i.; Nur77-GFP+ for 24 h.p.i. and 48 h.p.i. (24 h.p.i. TLN cells were sorted based on Nur77-GFP−). OT-I cells were sorted based on CD98lo for 0 h.p.i. and CD98hi for 48 h.p.i. Total RNA was isolated using Trizol–Qiagen RNAeasy Micro kit and sent to Yale Keck Sequencing Facility for library prep and sequencing run on Illumina Hiseq2500. Raw data were processed using the STAR and HTseq pipeline. Subsequent analyses were performed using R-studio. Pathway analysis was done with Enrichr (Chen et al., 2013; Kuleshov et al., 2016). Data are available online at the Gene Expression Omnibus (accession number GSE147908).
Immunofluorescent confocal imaging
Lung were inflated with 1 ml 1% paraformaldehyde (PFA)/0.5% optimal cutting temperature compound ratio and the entire lung placed in 1% PFA for 1 h. Samples were washed and then subjected to sucrose gradient before freezing in optimal cutting temperature compound. 25-µm sections were fixed in 1% PFA, blocked in anti-CD16/CD32, and stained with staining mix for at least 2 h. Endogenous GFP signaling is amplified by anti-GFP polyclonal antibody (Thermo; A11122). Slides were mounted in ProLong Gold antifade reagent, and the images were taken from Leica SP8 confocal microscope. For imaging of LCMV-ZsGreen-infected mLNs, mLNs were harvested and fixed in 2% PFA at 4°C for 6 h. mLNs were then embedded in 4% low-gelling agarose (Sigma; A9414). 4050-µm sections were cut using microtome (Leica; VT1000S). After blocking, the sections were stained overnight at 4°C with indicated antibodies in stain buffer (2% BSA and 0.1% Tween-20). Immunofluorescence confocal microscopy was performed using a Zeiss LSM 880 microscope. Micrographs were acquired in sequential scans and merged to obtain a multicolor image. Images were analyzed using Imaris software (Bitplane). Uninfected LN, labeled with the same antibody mix, was used to determine the baseline GFP+ signal. ZsGreen has similar excitation and emission properties as eGFP. Surfaces were made for GFP+ signal in infected LNs, which were then used to create masked channels for CD11c or F4/80 signals to determine GFP+CD11c+ or GFP+F4/80+ signals, respectively. GFP+ cells that were neither CD11c+ nor F4/80+ were referred to as “others.”
In vitro co-culturing
Bulk epithelial cells, endothelial cells, CD11cHiMHCHi cells, and alveolar macrophages were sorted from influenza-immunized mice that were infected with PR8-gp33 48 h prior. P14+ TRM cells were sorted from the lungs of P14+ immune chimera >30 d p.i.. These cells were co-cultured overnight before flow cytometry analysis.
Online supplemental material
Fig. S1 shows the identification and surface marker phenotyping of memory T cells in lung tissue and blood. Fig. S2 describes validation of Nur77-GFP reporter as a reliable readout for antigen-specific activation. Fig. S3 displays the criteria to identify TRM cell and TLN cell identity genes, respectively. Fig. S4 showcases validation of various genetic/antibody depletion strategies. Fig. S5 shows validation of BMC models and that one functional output of TRM cells is not impaired in the absence of CD11c+ cells.
Acknowledgments
We thank members of the Kaech and Flavell laboratories for helpful discussions and comments. We thank Juan Carlos de la Torre for the kind provision of LCMV-ZsGreen.
This study was supported in part by the National Institutes of Health R01 AI123864 (L.S. Cauley and S.M. Kaech), R37 AI066232 (S.M. Kaech), S10 OD020142 (Yale Center for Cellular and Molecular Imaging), and P30 CA106359-39 (Yale Cancer Center Grant); an A*STAR National Science Scholarship PhD (J.S. Low); a Swiss National Science Foundation early postdoc mobility fellowship (P2BEP3_178444); and the George E. Hewitt Foundation fellowship (Y. Farsakoglu). R.A. Flavell is an investigator with the Howard Hughes Medical Institute. This work was supported by the Yale Center for Research Computing, the Yale Center for Genome Analysis, and the Waitt Advanced Biophotonics Core at the Salk Institute for Biological Studies.
Author contributions: J.S. Low, Y. Farsakoglu, M.C. Amezcua Vesely, R. Jackson, X. Jiang, J.A. Shyer, L.S. Cauley, R.A. Flavell, and S.M. Kaech designed the research. J.S. Low performed the experiments. Y. Farsakoglu performed the LCMV-ZsGreen imaging experiments and analysis. J.S. Low performed the bioinformatics analyses under the advisement of E. Sefik, J.B. Kelly, and C.C.D. Harman. J.S. Low and S.M. Kaech wrote the manuscript.
References
- Ariotti S., Hogenbirk M.A., Dijkgraaf F.E., Visser L.L., Hoekstra M.E., Song J.-Y., Jacobs H., Haanen J.B., and Schumacher T.N.. 2014. T cell memory. Skin-resident memory CD8+ T cells trigger a state of tissue-wide pathogen alert. Science. 346:101–105. 10.1126/science.1254803 [DOI] [PubMed] [Google Scholar]
- De Baets S., Verhelst J., Van den Hoecke S., Smet A., Schotsaert M., Job E.R., Roose K., Schepens B., Fiers W., and Saelens X.. 2015. A GFP expressing influenza A virus to report in vivo tropism and protection by a matrix protein 2 ectodomain-specific monoclonal antibody. PLoS One. 10 e0121491 10.1371/journal.pone.0121491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballesteros-Tato A., León B., Lund F.E., and Randall T.D.. 2010. Temporal changes in dendritic cell subsets, cross-priming and costimulation via CD70 control CD8(+) T cell responses to influenza. Nat. Immunol. 11:216–224. 10.1038/ni.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura L.K., Mitchell J.S., Thompson E.A., Schenkel J.M., Mohammed J., Wijeyesinghe S., Fonseca R., Burbach B.J., Hickman H.D., Vezys V., et al. 2018a Intravital mucosal imaging of CD8+ resident memory T cells shows tissue-autonomous recall responses that amplify secondary memory. Nat. Immunol. 19:173–182. 10.1038/s41590-017-0029-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura L.K., Wijeyesinghe S., Thompson E.A., Macchietto M.G., Rosato P.C., Pierson M.J., Schenkel J.M., Mitchell J.S., Vezys V., Fife B.T., et al. 2018b T Cells in Nonlymphoid Tissues Give Rise to Lymph-Node-Resident Memory T Cells. Immunity. 48:327–338.e5. 10.1016/j.immuni.2018.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borowski A.B., Boesteanu A.C., Mueller Y.M., Carafides C., Topham D.J., Altman J.D., Jennings S.R., and Katsikis P.D.. 2007. Memory CD8+ T cells require CD28 costimulation. J. Immunol. 179:6494–6503. 10.4049/jimmunol.179.10.6494 [DOI] [PubMed] [Google Scholar]
- Boyman O., Hefti H.P., Conrad C., Nickoloff B.J., Suter M., and Nestle F.O.. 2004. Spontaneous development of psoriasis in a new animal model shows an essential role for resident T cells and tumor necrosis factor-alpha. J. Exp. Med. 199:731–736. 10.1084/jem.20031482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bromberg J.F., Horvath C.M., Wen Z., Schreiber R.D., and Darnell J.E. Jr.. 1996. Transcriptionally active Stat1 is required for the antiproliferative effects of both interferon alpha and interferon gamma. Proc. Natl. Acad. Sci. USA. 93:7673–7678. 10.1073/pnas.93.15.7673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.D., Fraire A.E., Joris I., Welsh R.M., and Selin L.K.. 2003. Specific history of heterologous virus infections determines anti-viral immunity and immunopathology in the lung. Am. J. Pathol. 163:1341–1355. 10.1016/S0002-9440(10)63493-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R., and Ma’ayan A.. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 14:128 10.1186/1471-2105-14-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheuk S., Schlums H., Gallais Sérézal I., Martini E., Chiang S.C., Marquardt N., Gibbs A., Detlofsson E., Introini A., Forkel M., et al. 2017. CD49a Expression Defines Tissue-Resident CD8+ T Cells Poised for Cytotoxic Function in Human Skin. Immunity. 46:287–300. 10.1016/j.immuni.2017.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dylla D.E., Michele D.E., Campbell K.P., and McCray P.B. Jr.. 2008. Basolateral entry and release of New and Old World arenaviruses from human airway epithelia. J. Virol. 82:6034–6038. 10.1128/JVI.00100-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards J., Wilmott J.S., Madore J., Gide T.N., Quek C., Tasker A., Ferguson A., Chen J., Hewavisenti R., Hersey P., et al. 2018. CD103+ Tumor-Resident CD8+ T Cells Are Associated with Improved Survival in Immunotherapy-Naïve Melanoma Patients and Expand Significantly During Anti-PD-1 Treatment. Clin. Cancer Res. 24:3036–3045. 10.1158/1078-0432.CCR-17-2257 [DOI] [PubMed] [Google Scholar]
- Ely K.H., Cauley L.S., Roberts A.D., Brennan J.W., Cookenham T., and Woodland D.L.. 2003. Nonspecific recruitment of memory CD8+ T cells to the lung airways during respiratory virus infections. J. Immunol. 170:1423–1429. 10.4049/jimmunol.170.3.1423 [DOI] [PubMed] [Google Scholar]
- Fuse S., Zhang W., and Usherwood E.J.. 2008. Control of memory CD8+ T cell differentiation by CD80/CD86-CD28 costimulation and restoration by IL-2 during the recall response. J. Immunol. 180:1148–1157. 10.4049/jimmunol.180.2.1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan A.P., Clarke J., Wood O., Garrido-Martin E.M., Chee S.J., Mellows T., Samaniego-Castruita D., Singh D., Seumois G., Alzetani A., et al. 2017. Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat. Immunol. 18:940–950. 10.1038/ni.3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt T., Wakim L.M., Eidsmo L., Reading P.C., Heath W.R., and Carbone F.R.. 2009. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat. Immunol. 10:524–530. 10.1038/ni.1718 [DOI] [PubMed] [Google Scholar]
- Gerlach C., Moseman E.A., Loughhead S.M., Alvarez D., Zwijnenburg A.J., Waanders L., Garg R., de la Torre J.C., von Andrian U.H., Anderson K.G., et al. 2016. The Chemokine Receptor CX3CR1 Defines Three Antigen-Experienced CD8 T Cell Subsets with Distinct Roles in Immune Surveillance and Homeostasis. Immunity. 45:1270–1284. 10.1016/j.immuni.2016.10.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath W.R., and Carbone F.R.. 2009. Dendritic cell subsets in primary and secondary T cell responses at body surfaces. Nat. Immunol. 10:1237–1244. 10.1038/ni.1822 [DOI] [PubMed] [Google Scholar]
- Helft J., Manicassamy B., Guermonprez P., Hashimoto D., Silvin A., Agudo J., Brown B.D., Schmolke M., Miller J.C., Leboeuf M., et al. 2012. Cross-presenting CD103+ dendritic cells are protected from influenza virus infection. J. Clin. Invest. 122:4037–4047. 10.1172/JCI60659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima N., and Iwasaki A.. 2014. T cell memory. A local macrophage chemokine network sustains protective tissue-resident memory CD4 T cells. Science. 346:93–98. 10.1126/science.1257530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwata S., Mikami Y., Sun H.W., Brooks S.R., Jankovic D., Hirahara K., Onodera A., Shih H.Y., Kawabe T., Jiang K., et al. 2017. The Transcription Factor T-bet Limits Amplification of Type I IFN Transcriptome and Circuitry in T Helper 1 Cells. Immunity. 46:983–991.e4. 10.1016/j.immuni.2017.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Clark R.A., Liu L., Wagers A.J., Fuhlbrigge R.C., and Kupper T.S.. 2012. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature. 483:227–231. 10.1038/nature10851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.S., and Braciale T.J.. 2009. Respiratory dendritic cell subsets differ in their capacity to support the induction of virus-specific cytotoxic CD8+ T cell responses. PLoS One. 4 e4204 10.1371/journal.pone.0004204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T.S., Gorski S.A., Hahn S., Murphy K.M., and Braciale T.J.. 2014. Distinct dendritic cell subsets dictate the fate decision between effector and memory CD8(+) T cell differentiation by a CD24-dependent mechanism. Immunity. 40:400–413. 10.1016/j.immuni.2014.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knudson C.J., Weiss K.A., Hartwig S.M., and Varga S.M.. 2014. The pulmonary localization of virus-specific T lymphocytes is governed by the tissue tropism of infection. J. Virol. 88:9010–9016. 10.1128/JVI.00329-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A., et al. 2016. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44(W1):W90-7. 10.1093/nar/gkw377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laidlaw B.J., Zhang N., Marshall H.D., Staron M.M., Guan T., Hu Y., Cauley L.S., Craft J., and Kaech S.M.. 2014. CD4+ T cell help guides formation of CD103+ lung-resident memory CD8+ T cells during influenza viral infection. Immunity. 41:633–645. 10.1016/j.immuni.2014.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Low J.S., and Kaech S.M.. 2018. Trials and Tribble-ations of tissue TRM cells. Nat. Immunol. 19:102–103. 10.1038/s41590-017-0031-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay L.K., Rahimpour A., Ma J.Z., Collins N., Stock A.T., Hafon M.-L., Vega-Ramos J., Lauzurica P., Mueller S.N., Stefanovic T., et al. 2013. The developmental pathway for CD103(+)CD8+ tissue-resident memory T cells of skin. Nat. Immunol. 14:1294–1301. 10.1038/ni.2744 [DOI] [PubMed] [Google Scholar]
- Macleod B.L., Bedoui S., Hor J.L., Mueller S.N., Russell T.A., Hollett N.A., Heath W.R., Tscharke D.C., Brooks A.G., and Gebhardt T.. 2014. Distinct APC subtypes drive spatially segregated CD4+ and CD8+ T-cell effector activity during skin infection with HSV-1. PLoS Pathog. 10 e1004303 10.1371/journal.ppat.1004303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manicassamy B., Manicassamy S., Belicha-Villanueva A., Pisanelli G., Pulendran B., and García-Sastre A.. 2010. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc. Natl. Acad. Sci. USA. 107:11531–11536. 10.1073/pnas.0914994107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran A.E., Holzapfel K.L., Xing Y., Cunningham N.R., Maltzman J.S., Punt J., and Hogquist K.A.. 2011. T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208:1279–1289. 10.1084/jem.20110308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S.N., Vanguri V.K., Ha S.J., West E.E., Keir M.E., Glickman J.N., Sharpe A.H., and Ahmed R.. 2010. PD-L1 has distinct functions in hematopoietic and nonhematopoietic cells in regulating T cell responses during chronic infection in mice. J. Clin. Invest. 120:2508–2515. 10.1172/JCI40040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta T., Sugiyama M., Hemmi H., Yamazaki C., Okura S., Sasaki I., Fukuda Y., Orimo T., Ishii K.J., Hoshino K., et al. 2016. Crucial roles of XCR1-expressing dendritic cells and the XCR1-XCL1 chemokine axis in intestinal immune homeostasis. Sci. Rep. 6:23505 10.1038/srep23505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.L., Zaid A., Hor J.L., Christo S.N., Prier J.E., Davies B., Alexandre Y.O., Gregory J.L., Russell T.A., Gebhardt T., et al. 2018. Local proliferation maintains a stable pool of tissue-resident memory T cells after antiviral recall responses. Nat. Immunol. 19:183–191. 10.1038/s41590-017-0027-5 [DOI] [PubMed] [Google Scholar]
- Petricoin E.F. III, Ito S., Williams B.L., Audet S., Stancato L.F., Gamero A., Clouse K., Grimley P., Weiss A., Beeler J., et al. 1997. Antiproliferative action of interferon-alpha requires components of T-cell-receptor signalling. Nature. 390:629–632. 10.1038/37648 [DOI] [PubMed] [Google Scholar]
- Pythoud C., Rothenberger S., Martínez-Sobrido L., de la Torre J.C., and Kunz S.. 2015. Lymphocytic Choriomeningitis Virus Differentially Affects the Virus-Induced Type I Interferon Response and Mitochondrial Apoptosis Mediated by RIG-I/MAVS. J. Virol. 89:6240–6250. 10.1128/JVI.00610-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F., Lenig D., Förster R., Lipp M., and Lanzavecchia A.. 1999. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 401:708–712. 10.1038/44385 [DOI] [PubMed] [Google Scholar]
- Schenkel J.M., and Masopust D.. 2014. Tissue-resident memory T cells. Immunity. 41:886–897. 10.1016/j.immuni.2014.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel J.M., Fraser K.A., Beura L.K., Pauken K.E., Vezys V., and Masopust D.. 2014. T cell memory. Resident memory CD8 T cells trigger protective innate and adaptive immune responses. Science. 346:98–101. 10.1126/science.1254536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., Kumamoto Y., Gopinath S., and Iwasaki A.. 2016. CD301b+ dendritic cells stimulate tissue-resident memory CD8+ T cells to protect against genital HSV-2. Nat. Commun. 7:13346 10.1038/ncomms13346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slütter B., Pewe L.L., Kaech S.M., and Harty J.T.. 2013. Lung airway-surveilling CXCR3(hi) memory CD8(+) T cells are critical for protection against influenza A virus. Immunity. 39:939–948. 10.1016/j.immuni.2013.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson D.B., and Medzhitov R.. 2006. Type I interferons in host defense. Immunity. 25:373–381. 10.1016/j.immuni.2006.08.007 [DOI] [PubMed] [Google Scholar]
- Sung J.H., Zhang H., Moseman E.A., Alvarez D., Iannacone M., Henrickson S.E., de la Torre J.C., Groom J.R., Luster A.D., and von Andrian U.H.. 2012. Chemokine guidance of central memory T cells is critical for antiviral recall responses in lymph nodes. Cell. 150:1249–1263. 10.1016/j.cell.2012.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro J.R., Turner D., Pham Q., Wherry E.J., Lefrançois L., and Farber D.L.. 2011. Cutting edge: Tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J. Immunol. 187:5510–5514. 10.4049/jimmunol.1102243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakim L.M., Waithman J., van Rooijen N., Heath W.R., and Carbone F.R.. 2008. Dendritic cell-induced memory T cell activation in nonlymphoid tissues. Science. 319:198–202. 10.1126/science.1151869 [DOI] [PubMed] [Google Scholar]
- Wakim L.M., Woodward-Davis A., and Bevan M.J.. 2010. Memory T cells persisting within the brain after local infection show functional adaptations to their tissue of residence. Proc. Natl. Acad. Sci. USA. 107:17872–17879. 10.1073/pnas.1010201107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Q., Milne K., Derocher H., Webb J.R., Nelson B.H., and Watson P.H.. 2016. CD103 and Intratumoral Immune Response in Breast Cancer. Clin. Cancer Res. 22:6290–6297. 10.1158/1078-0432.CCR-16-0732 [DOI] [PubMed] [Google Scholar]
- Webb J.R., Milne K., Watson P., Deleeuw R.J., and Nelson B.H.. 2014. Tumor-infiltrating lymphocytes expressing the tissue resident memory marker CD103 are associated with increased survival in high-grade serous ovarian cancer. Clin. Cancer Res. 20:434–444. 10.1158/1078-0432.CCR-13-1877 [DOI] [PubMed] [Google Scholar]
- Wu T., Hu Y., Lee Y.-T., Bouchard K.R., Benechet A., Khanna K., and Cauley L.S.. 2014. Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J. Leukoc. Biol. 95:215–224. 10.1189/jlb.0313180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit D.J., Cauley L.S., Pham Q.-M., and Lefrançois L.. 2005. Dendritic cells maximize the memory CD8 T cell response to infection. Immunity. 22:561–570. 10.1016/j.immuni.2005.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]