Type 1 conventional dendritic cells are progressively and systemically dysregulated during pancreatic carcinogenesis, subject to semimaturation and IL-6–driven apoptosis. Yet, CD40 agonist synergizes with Flt3L to rescue cDC1 abundance and maturation, enabling a return to full CD8+ T cell priming.
Abstract
Type 1 conventional dendritic cells (cDC1s) are typically thought to be dysregulated secondarily to invasive cancer. Here, we report that cDC1 dysfunction instead develops in the earliest stages of preinvasive pancreatic intraepithelial neoplasia (PanIN) in the KrasLSL-G12D/+ Trp53LSL-R172H/+ Pdx1-Cre–driven (KPC) mouse model of pancreatic cancer. cDC1 dysfunction is systemic and progressive, driven by increased apoptosis, and results in suboptimal up-regulation of T cell–polarizing cytokines during cDC1 maturation. The underlying mechanism is linked to elevated IL-6 concomitant with neoplasia. Neutralization of IL-6 in vivo ameliorates cDC1 apoptosis, rescuing cDC1 abundance in tumor-bearing mice. CD8+ T cell response to vaccination is impaired as a result of cDC1 dysregulation. Yet, combination therapy with CD40 agonist and Flt3 ligand restores cDC1 abundance to normal levels, decreases cDC1 apoptosis, and repairs cDC1 maturation to drive superior control of tumor outgrowth. Our study therefore reveals the unexpectedly early and systemic onset of cDC1 dysregulation during pancreatic carcinogenesis and suggests therapeutically tractable strategies toward cDC1 repair.
Graphical Abstract
Introduction
Solid tumors are typically thought to subvert immune surveillance through evasion of T cell recognition (Schreiber et al., 2011). Yet, immunologically “cold” cancers that do not respond to immune checkpoint blockade (ICB) often exclude antineoplastic T cells from the earliest stages of disease and exhibit no evidence of immunoediting by T cell selective pressure (Clark et al., 2007; Evans et al., 2016). This phenotype is consistent with impaired T cell priming rather than evasion of preexisting T cell immunity as the means of subverting adaptive immune surveillance (Vonderheide, 2018). Suppression of T cell priming may therefore be an early rather than secondary event to tumor formation in such cancers.
Type 1 conventional dendritic cells (cDC1s) are the critical professional APC for T cell priming in spontaneous antitumor adaptive immunity (Böttcher and Reis e Sousa, 2018). cDC1s are necessary for tumor antigen trafficking to draining LNs, antigen cross-presentation, and CD8+ T cell activation (Engelhardt et al., 2012; Broz et al., 2014; Roberts et al., 2016). cDC1s have also been shown to recruit CD8+ T cells into the tumor microenvironment (Spranger et al., 2017). They are required for spontaneous T cell–mediated tumor rejection and response to ICB in a variety of cancer mouse models (Hildner et al., 2008; Broz et al., 2014; Spranger et al., 2015, 2017; Salmon et al., 2016; Sánchez-Paulete et al., 2016; Böttcher and Reis e Sousa, 2018). A recent study in murine pancreatic cancer demonstrates that dendritic cell (DC) paucity can lead to dysfunctional immune surveillance against an engineered model neoantigen, accelerating neoplastic progression (Hegde et al., 2020). Studies of cDC1s in the B-Raf/PTEN−/−–driven genetically engineered mouse model (GEMM) of melanoma have also elucidated cancer cell–intrinsic mechanisms of cDC1 suppression and exclusion such as β-catenin signaling (Spranger et al., 2015, 2017; Salmon et al., 2016). Here, we examine the onset of cDC1 dysregulation during carcinogenesis as it relates to T cell priming.
The KrasLSL-G12D/+ Trp53LSL-R172H/+ Pdx1-Cre–driven (KPC) GEMM of pancreatic ductal adenocarcinoma (PDA) enables the study of immune dynamics in response to developing carcinomas from inception to invasion (Hingorani et al., 2005; Clark et al., 2007; Feig et al., 2013). These mice develop preinvasive pancreatic intraepithelial neoplasias (PanINs) at an early age that progress to metastatic carcinomas with complete penetrance. PanIN formation is accompanied by a variety of changes to the immune milieu of the pancreas, including an influx of tumor-associated macrophages, myeloid-derived suppressor cells (MDSCs), and CD4+ regulatory T cells. These changes persist and intensify upon progression to malignancy with prominent expansion and recruitment of myeloid cells driven by tumor-derived cytokines and chemokines such as GM-CSF and CXCR2 (Bayne et al., 2012; Chao et al., 2016). Antineoplastic T cells are also strongly excluded from KPC tumors, consistent with deficiencies in T cell priming.
In the present study, we use the KPC GEMM to quantify cDC1 abundance and maturation from preinvasive neoplasia to invasive carcinoma. We reveal significant systemic changes in cDC1 biology that impair CD8+ T cell priming from the earliest stages of disease. Elevated serum IL-6 is especially prominent and found to be a key driver of cDC1 apoptosis. Yet, combination therapy with CD40 agonist and Fms-like tyrosine kinase 3 ligand (Flt3L) successfully repairs both quantitative and functional deficits in cDC1s, enabling a return to full CD8+ T cell activation. This combined rescue of cDC1 dysfunction subsequently enables superior control of tumor outgrowth and response to vaccination in tumor-bearing mice.
Results
cDC1 abundance declines progressively and systemically during pancreatic carcinogenesis
To examine cDC1 biology in the KPC GEMM, we defined groups of mice that represent distinct stages of carcinogenesis. Mice homozygous for Pdx1-Cre but lacking mutant Kras and Trp53 were chosen as healthy controls. Littermates from the same colony were chosen to control for potential differences in genetics and microbiota. 8-wk-old KPC mice, confirmed not to have tumors by ultrasound, were used as PanIN-bearing mice. The pancreata of 8-wk-old KPC mice were confirmed to harbor lesions characteristic of PanIN 1A (Fig. 1 A). Finally, KPC mice that were confirmed to have tumors by palpation and ultrasound served as tumor-bearing mice.
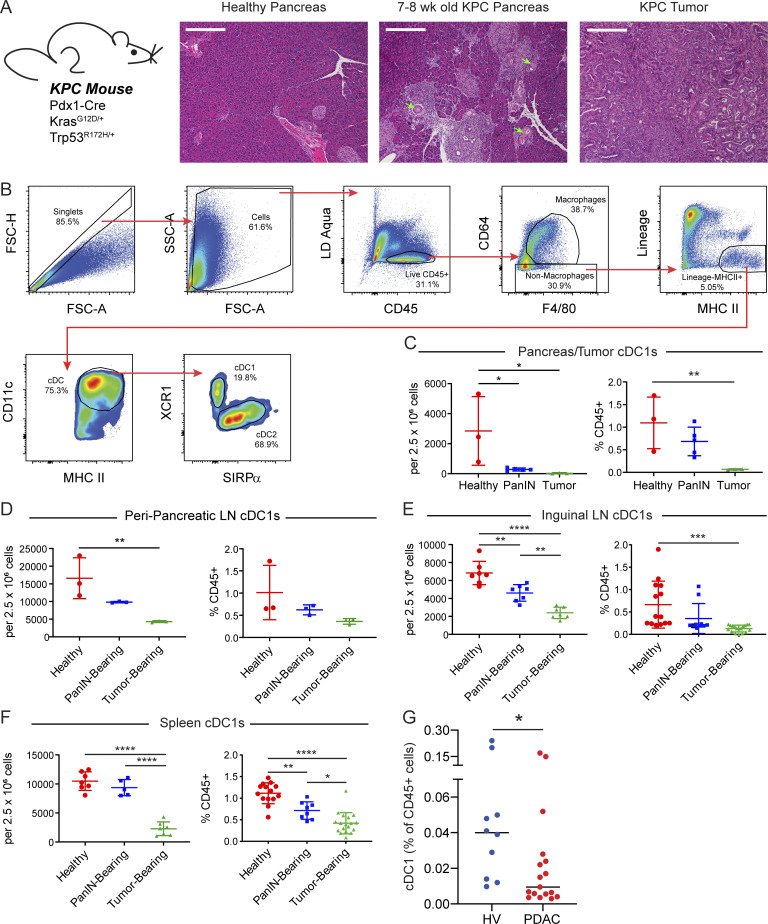
Figure 1.
cDC1 abundance declines systemically during pancreatic carcinogenesis. (A) H&E staining of healthy pancreas, PanIN-bearing pancreas, and PDA. Arrows highlight ducts featuring mucinous metaplasia without dysplasia characteristic of PanIN 1A. All images are taken at 10× magnification. Scale bars denote 300 µm. (B) Flow gating strategy for CD45+CD64−F4/80−Lin−MHC II+CD11c+ cDCs in a representative subcutaneously implanted KPC tumor. Lineage gate is composed of CD3, CD19, B220, NK1.1, and Gr-1. FSC-A, forward scatter-area; FSC-H, forward scatter-height; LD, live/dead. (C–F) Quantification of cDC1s in the (C) pancreas/tumor, (D) ppLNs, (E) iLN, and (F) spleen as a proportion of live cells and CD45+ cells. (G) Frequency of CD141+ cDC1s in peripheral blood of patients with untreated advanced PDA versus healthy volunteers (HV). Error bars indicate mean ± SD. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05 (one-way ANOVA with Tukey’s honest significant difference [HSD] post-test in C–F; Mann–Whitney test in G). Data shown in B–F are representative of at least three independent experiments with at least three mice per group.
To quantify cDCs across tissues, we used a consistent set of phenotypic markers and defined cDCs as live CD45+CD64−Lin−MHC II+CD11c+ cells. We then delineated cDC1s and cDC2s based on XCR1 and SIRPα expression, respectively (Fig. 1 B). This strategy minimizes contamination by B cells, macrophages, monocytes, and MDSCs (Guilliams et al., 2016). cDC1 abundance was found to decline as a proportion of live cells in PanIN-bearing pancreas and KPC tumors (Fig. 1 C). To explore whether cDC1 exclusion was being driven by an influx of myeloid cells, cDC1s were also quantified as a percentage of CD45+ cells. When quantified in this manner, cDC1 abundance was confirmed to decline in KPC tumors with a trend toward decline in PanIN-bearing pancreas (Fig. 1 C), consistent with prior reports (Li et al., 2018). Quantification of cDC1s in the draining peripancreatic LNs (ppLNs) revealed a similar decline in cDC1 abundance in tumor-draining ppLNs with a trend toward decline in PanIN-draining ppLNs (Fig. 1 D).
To determine whether declining cDC1 abundance was occurring systemically or was isolated to the local pancreatic anatomical site, cDC1s were also quantified in the breast pad–draining inguinal LNs (iLNs) and spleen (Fig. 1, E and F). cDC1s in these distant tissues were also observed to decline as a proportion of either total live or CD45+ cells in PanIN- and tumor-bearing mice. However, we noted that when calculated based on tissue weight, cDC1 numbers in the KPC GEMM were not altered across the stages of pancreatic carcinogenesis in pancreas/tumor, ppLNs, iLNs, or spleen (Fig. S1). Thus, our findings show a progressive and systemic decline in cDC1s that is based on cellular proportions and begins in the earliest stages of KPC pancreatic carcinogenesis.
Figure S1.
cDC1 abundance only declines based on cell fractions during pancreatic carcinogenesis. (A–D) Tissue weight, cDC1 number per organ, and cDC1 number per milligram tissue in the (A) pancreas/tumor, (B) ppLNs, (C) iLNs, and (D) spleen from healthy, PanIN-bearing, and tumor-bearing mice. Error bars indicate mean ± SEM. ****, P < 0.0001; *, P < 0.05 (one-way ANOVA with Tukey’s HSD post-test). Data shown are representative of one independent experiment.
cDC1 abundance in patients with pancreatic cancer
To determine if alterations in cDC1 abundance are also present in patients, we isolated peripheral blood leukocytes from a cohort of newly diagnosed, untreated patients with advanced PDA (n = 17) and conducted high-dimensional single-cell mass cytometry to analyze the frequency of cDC1s in circulation. We found a reduced frequency of CD141+ cDC1s in the peripheral blood of patients with PDA compared with healthy volunteers (n = 10; 0.031% vs. 0.068%; P = 0.02; Fig. 1 G). Notably, approximately half of the patients exhibited nearly undetectable levels of circulating cDC1s. Thus, decreased cDC1 abundance is also observed in patients with PDA.
cDC1 maturation is progressively and systemically impaired during pancreatic carcinogenesis
Having observed a progressive decline in cDC1 abundance, we next determined whether cDC1 maturation and function were similarly impacted during carcinogenesis. DCs are considered immature until encountering an activating signal during tissue surveillance and antigen uptake (Merad et al., 2013). DCs then mature, up-regulate CCR7, and home to the draining LN, where antigen presentation and T cell priming occur (Roberts et al., 2016). During this process, cDC1s up-regulate antigen processing and cross-presentation machinery; up-regulate cell surface molecules such as CD40, CD80, CD86, MHC II, and PD-L1; and produce essential T cell–polarizing cytokines such as IL-12 to induce Th1 CD4+ T cell differentiation and CD8+ T cell activation (Dalod et al., 2014).
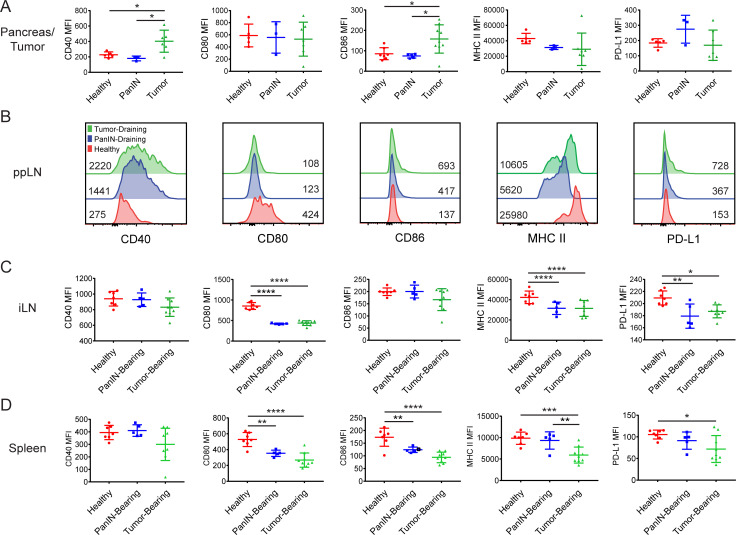
We therefore extended our flow cytometric analysis of cDC1s to include expression of CD40, CD80, CD86, MHC II, and PD-L1. While CD40 and CD86 were found to be increased on cDC1s in KPC tumors relative to healthy and PanIN-bearing pancreas, the expression of CD80, MHC II, and PD-L1 remained unchanged (Fig. 2 A). This partial up-regulation of maturation markers has been previously described as DC semimaturation and is associated with poor T cell priming in cancer patients (Tjomsland et al., 2010; Dudek et al., 2013). Notably, we found in the draining ppLN that cDC1 semimaturation occurred early in pancreatic carcinogenesis and was detected in PanIN-bearing mice (Fig. 2 B). Increases in CD40, CD86, and PD-L1 expression were accompanied by declines in CD80 and MHC II expression that were amplified upon progression to malignancy. cDC1 maturation marker expression also declined systemically as seen by a decrease in the expression of CD80, CD86, MHC II, and PD-L1 in the iLNs and spleen of PanIN- and tumor-bearing mice (Fig. 2, C and D). Thus, cDC1 maturation, like cDC1 abundance, is impacted systemically and progressively beginning in preinvasive carcinogenesis.
Figure 2.
cDC1 maturation marker expression declines systemically during preinvasive neoplasia. Expression of maturation markers CD40, CD80, CD86, MHC II (I-A/I-E), and PD-L1 on cDC1s in the (A) pancreas/tumor, (B) ppLNs, (C) iLNs, and (D) spleen of healthy, PanIN-bearing, and tumor-bearing mice. Geometric mean fluorescence intensities (MFIs) shown. Samples were pooled across three to six mice per treatment group in B. Error bars indicate mean ± SD. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05 (one-way ANOVA with Tukey’s HSD post-test). Data shown are representative of four independent experiments with at least three mice per group.
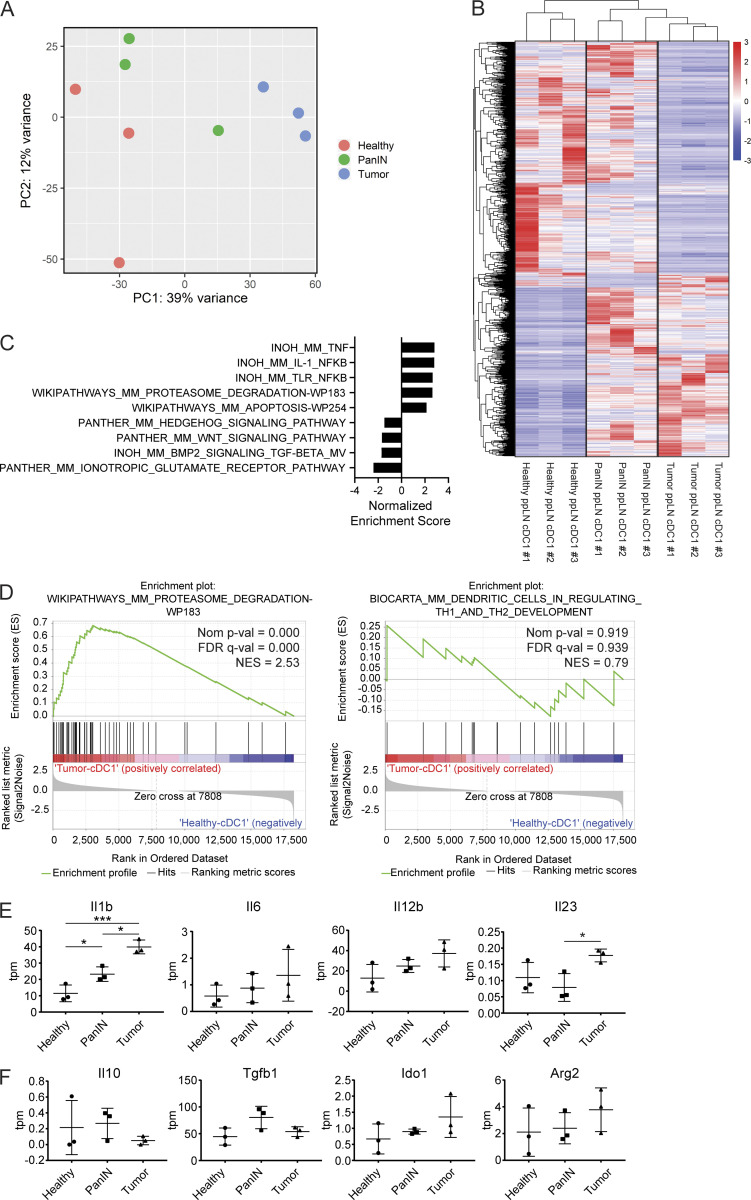
To determine which cDC1 molecular pathways are affected by cDC1 semimaturation, we performed bulk RNA sequencing (RNA-seq) on ppLN cDC1s from healthy, PanIN-bearing, and tumor-bearing mice. Both principal-component and differential gene expression analyses revealed a progressive change in cDC1 gene expression from healthy to tumor-bearing mice, with PanIN-draining ppLN cDC1s representing an intermediate state (Fig. 3, A and B). Gene set enrichment analyses (GSEAs) were performed comparing tumor-draining and PanIN-draining ppLN cDC1s to those of healthy mice (Fig. 3 C). In both comparisons, the proteasome degradation gene set (an aspect of antigen processing machinery that is up-regulated during DC maturation) was up-regulated, while genes encoding T cell–polarizing cytokines such as Il-12b failed to be optimally up-regulated (Fig. 3, D and E). Because cancer cells have been known to exploit DCs to produce immune suppressive factors like indoleamine 2,3-dioxygenase, we determined whether PanIN- and tumor-draining ppLN cDC1s might be directly enforcing adaptive immune tolerance (Munn and Mellor, 2016). Genes encoding known DC-secreted immune suppressive factors were therefore examined (Fig. 3 F). While Ido1 and Arg2 trended toward increased expression in tumor-bearing mice, their transcript abundance remained below five transcripts-per-million (tpm) reads. Thus, it is unlikely that cDC1s in the ppLNs are acquiring immune-suppressive function over the course of KPC carcinogenesis. Rather, suboptimal maturation marker up-regulation coincides with insufficient up-regulation of T cell–polarizing cytokines during cDC1 semimaturation.
Figure 3.
cDC1 maturation is progressively impaired during pancreatic oncogenesis. (A) Principal-component (PC) analysis of cDC1s collected from healthy, PanIN-draining, and tumor-draining ppLNs. (B) Heatmap of differentially expressed genes by z-score across samples. (C) Top hits from GSEA comparing cDC1s from tumor-draining versus healthy ppLNs. (D) Enrichment plots of proteasome degradation and T cell–polarizing cytokine gene sets in cDC1s from GSEA shown in C. FDR, false discovery rate; NES, normalized enrichment score. (E and F) Expression in tpm reads of genes encoding (E) inflammatory cytokines and (F) immune suppressive factors in cDC1s from healthy, PanIN-draining, and tumor-draining ppLNs. n = 3 samples per group. Each sample consists of total RNA collected from 10,000 sorted ppLN cDC1s pooled from three to six mice. Error bars indicate mean ± SD. ***, P < 0.001; *, P < 0.05 (one-way ANOVA with Tukey’s HSD post-test).
cDC1-mediated CD8+ T cell priming is impaired in PanIN- and tumor-bearing mice
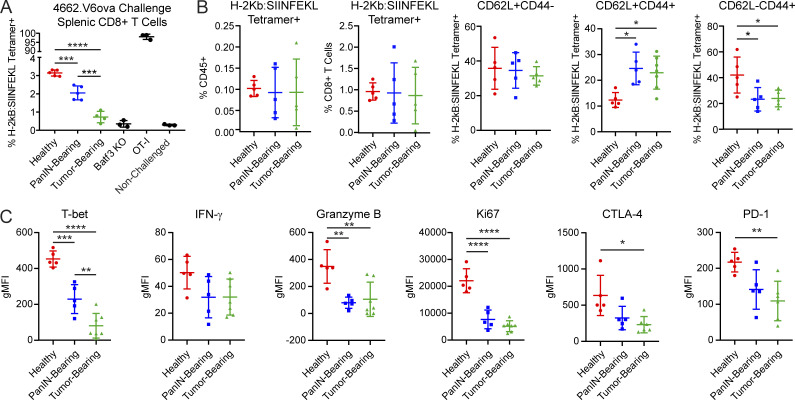
To determine whether cDC1 semimaturation impairs function, we sought to quantify CD8+ T cell priming in response to an antigen-specific challenge. Our group previously demonstrated that a clonal chicken OVA-expressing KPC cell line 4662.V6ova gives rise to spontaneous protective CD8+ T cell immunity following subcutaneous implantation (Evans et al., 2016). Therefore, we subcutaneously implanted 4662.V6ova cells into healthy, PanIN-bearing, and tumor-bearing mice. 7 d after implantation, splenocytes from these mice were stained for OVA-specific H-2Kb:SIINFEKL tetramer–positive CD8+ T cells. Consistent with the early onset of cDC1 semimaturation, the generation of OVA-specific CD8+ T cells progressively declined in PanIN- and tumor-bearing KPC mice (Fig. 4 A). CD8+ T cell priming in tumor-bearing KPC mice was so profoundly impaired that findings were statistically indistinguishable from Batf3−/− mice that lack cDC1s (Hildner et al., 2008).
Figure 4.
cDC1-mediated CD8+ T cell priming is impaired in PanIN- and tumor-bearing mice. (A) Generation of H-2Kb:SIINFEKL tetramer–positive splenic CD8+ T cells in healthy, PanIN-bearing, and tumor-bearing mice 7 d following subcutaneous implantation of 5 × 105 cells from clonal OVA-expressing KPC cell line 4662.V6ova. (B) Quantification of H-2Kb:SIINFEKL tetramer–positive splenic CD8+ T cells from healthy, PanIN-bearing, and tumor-bearing mice 7 d following subcutaneous vaccination with 200 µg OVA + 10 µg CpG (OVA/CpG). (C) Activation/exhaustion marker expression in CD62L−CD44+ H-2Kb:SIINFEKL tetramer–positive CD8+ T cells following vaccination with OVA/CpG. gMFI, geometric mean fluorescence intensity. Error bars indicate mean ± SD. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05 (one-way ANOVA with Tukey’s HSD post-test). Data shown are representative of three independent experiments with at least three mice per group.
Due to the potential for shared suppression between autochthonous KPC neoplasia and 4662.V6ova, we sought to confirm our findings using a nontumor vaccination strategy. Healthy, PanIN-bearing, and tumor-bearing mice were vaccinated with OVA protein and the TLR9 agonist CpG (OVA/CpG). While the total number of tetramer-positive T cells were equivalent across all groups, the proportions of CD62L−CD44+ effector memory T cells were depressed in PanIN- and tumor-bearing mice (Fig. 4 B). Their expression of T-bet, Granzyme B, Ki-67, CTLA-4, and PD-1 declined as well (Fig. 4 C). Thus, like our findings with 4662.V6ova challenge, the CD8+ T cell response to OVA/CpG is defective in PanIN- and tumor-bearing KPC mice.
cDC1 abundance and maturation correlate with increased cytolytic activity in patients with pancreatic adenocarcinoma
To determine whether cDC1 abundance correlates with cytolytic activity in human pancreatic adenocarcinoma, transcript abundance of XCR1, CLEC9A, CD86, HLA-DRA, GZMA, PRF1, and IFNG were quantified from pancreatic carcinoma samples in The Cancer Genome Atlas (TCGA-PAAD; Raphael et al., 2017). Because XCR1 and CLEC9A are known markers of cDC1s in humans, the expression of these genes was used as an indication of cDC1 abundance (Wculek et al., 2020). As a metric of cytolytic activity, cytolytic index was calculated using the geometric mean of GZMA and PRF1, as previously experimentally validated (Rooney et al., 2015). Both XCR1 and CLEC9A gene expression were found to exhibit a strong correlation with cytolytic index (Fig. S2, A and B). Similarly, transcripts of the DC maturation markers HLA-DRA and CD86 also exhibited a strong correlation with cytolytic index (Fig. S2, C and D). Finally, the expression of HLA-DRA and CD86 were compared with intratumoral transcript abundance of IFNG and found to have a moderate correlation (Fig. S2, E and F). Intratumoral cDC1 abundance and maturation, therefore, correlate with cytolytic activity in human pancreatic adenocarcinoma.
Figure S2.
cDC1 abundance and maturation are associated with increased cytolytic activity in human PDA. Correlation analyses of (A) XCR1 gene expression and cytolytic index (CTL), (B) CLEC9A gene expression and cytolytic index, (C) HLA-DRA gene expression and cytolytic index, (D) CD86 gene expression and cytolytic index, (E) HLA-DRA gene expression and IFNG gene expression, and (F) CD86 gene expression and IFNG gene expression in tumors of patients from the TCGA-PAAD. n = 182 total patients in TCGA-PAAD. Regression line, 95% confidence interval, Kendall’s τ rank correlation coefficient, and associated P value shown for all correlation analyses. Cytolytic index is calculated using the geometric mean of PRF1 and GZMA.
Systemic deficits in cDC1 abundance and maturation are specific to pancreatic neoplasia
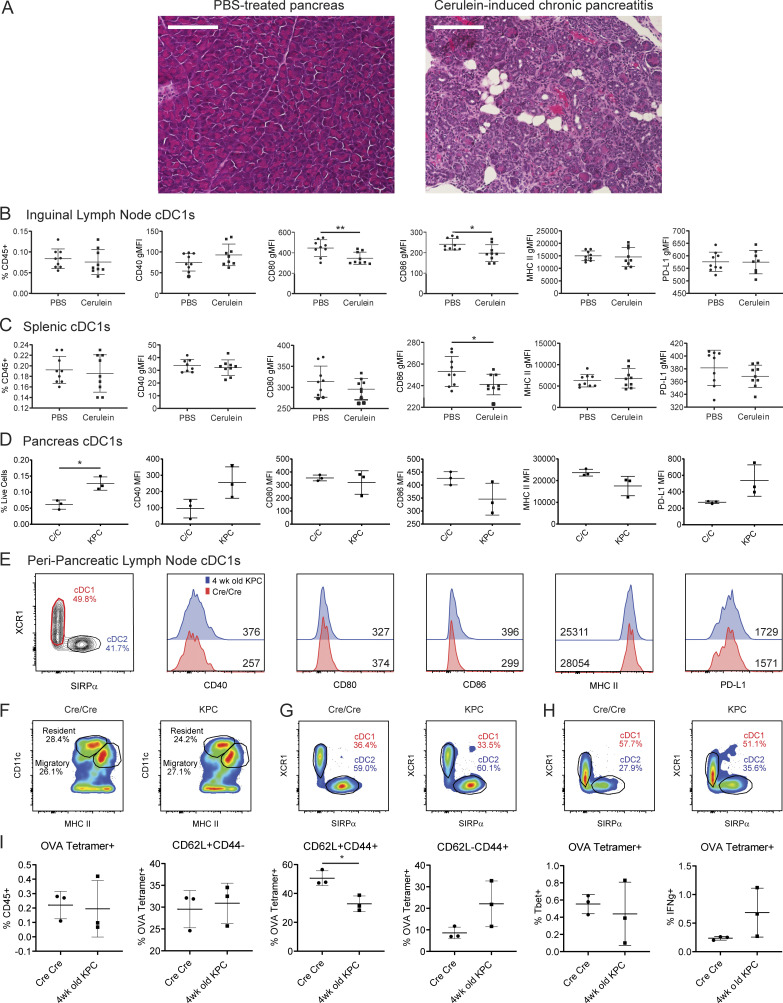
PanIN development in KPC mice occurs in the setting of chronic mutant Kras–driven inflammation (Clark et al., 2007; McAllister et al., 2014). To determine if systemic declines in cDC1 abundance and maturation could be reproduced in the setting of chronic pancreatitis, C57BL/6J mice were treated for 11 wk with supraphysiologic levels of the cholecystokinin analogue cerulein (Elsässer et al., 1992). Repeated administration resulted in significant intrapancreatic edema, inflammatory infiltrate, acinar atrophy, ductal dilation, and parenchymal fibrosis characteristic of cerulein-induced chronic pancreatitis (Fig. S3 A). In both the iLNs and spleen, cDC1 abundance remained unchanged while maturation marker expression either remained unchanged or changed minimally compared with the declines seen in PanIN- and tumor-bearing mice (Fig. S3, B and C). Cerulein-induced chronic pancreatitis, therefore, fails to recapitulate the systemic cDC1 deficits seen in preinvasive pancreatic carcinogenesis.
Figure S3.
Systemic cDC1 dysregulation requires neoplastic development. (A) H&E staining of pancreas from mice treated for 11 wk with PBS or cerulein. All images taken at 20× magnification. Scale bars denote 150 µm. (B and C) Enumeration of and expression of maturation markers CD40, CD80, CD86, MHC II, and PD-L1 on (B) iLN and (C) splenic cDC1s from PBS-treated and cerulein-treated mice. (D and E) Enumeration of and maturation marker expression on cDC1s from (D) pancreas and (E) ppLN cDC1s from 4-wk-old Cre/Cre (C/C) and KPC mice. Geometric MFIs are shown in E. (F) Proportions of CD11chiMHCIIint resident/resting versus CD11cintMHCIIhi migratory/activated ppLN cDCs. (G and H) Proportion of cDC1s and cDC2s among (G) resident/resting and (H) migratory/activated ppLN cDCs shown in F. (I) Quantification of and Tbet and IFN-γ expression in H-2Kb:SIINFEKL tetramer–positive splenic CD8+ T cells 7 d following vaccination with 200 µg OVA + 10 µg CpG. Samples pooled across three mice per group in E. Error bars indicate mean ± SD. **, P < 0.01; *, P < 0.05 (two-tailed Student’s t test). Data shown in A-C are representative of one independent experiment. Data shown in D-E are representative of three independent experiments with at least three mice per group.
We next sought to confirm that neoplastic development was required for systemic cDC1 dysregulation to occur. To address this, cDC1s were compared between 4-wk-old Cre/Cre mice and 4-wk-old KPC mice that have not yet developed PanINs. While cDC1 abundance was slightly increased in KPC pancreas, cDC1 maturation marker expression did not differ between KPC and Cre/Cre pancreas (Fig. S3 D). Maturation marker expression likewise remained unchanged on cDC1s from KPC versus Cre/Cre ppLNs (Fig. S3 E). KPC and Cre/Cre ppLNs did not differ in their proportions of CD11chiMHC IIint resident versus CD11cintMHC IIhi migratory cDCs or their proportions of cDC1s vs. cDC2s (Fig. S3, F–H).
To rule out the possibility that defective response to vaccination is an inherent feature of the KPC genotype, independent of neoplasia, 4-wk-old KPC mice (that lack PanINs and have normal cDC1s at this age) were also vaccinated with OVA/CpG. Both groups generated similar numbers of H-2Kb:SIINFEKL tetramer–positive CD8+ T cells in response to vaccination (Fig. S3 I). The proportion of CD62L−CD44+ T cells was the same in 4-wk-old KPC mice and Cre/Cre mice, though the proportion of CD62L+CD44+ T cells was decreased in 4-wk-old KPC mice. Expression of Tbet and IFN-γ were also similar between both groups. Thus, systemic cDC1 dysfunction is seen in KPC mice only after the initiation of neoplasia.
Evaluation of cDC1s in the KP GEMM of lung adenocarcinoma
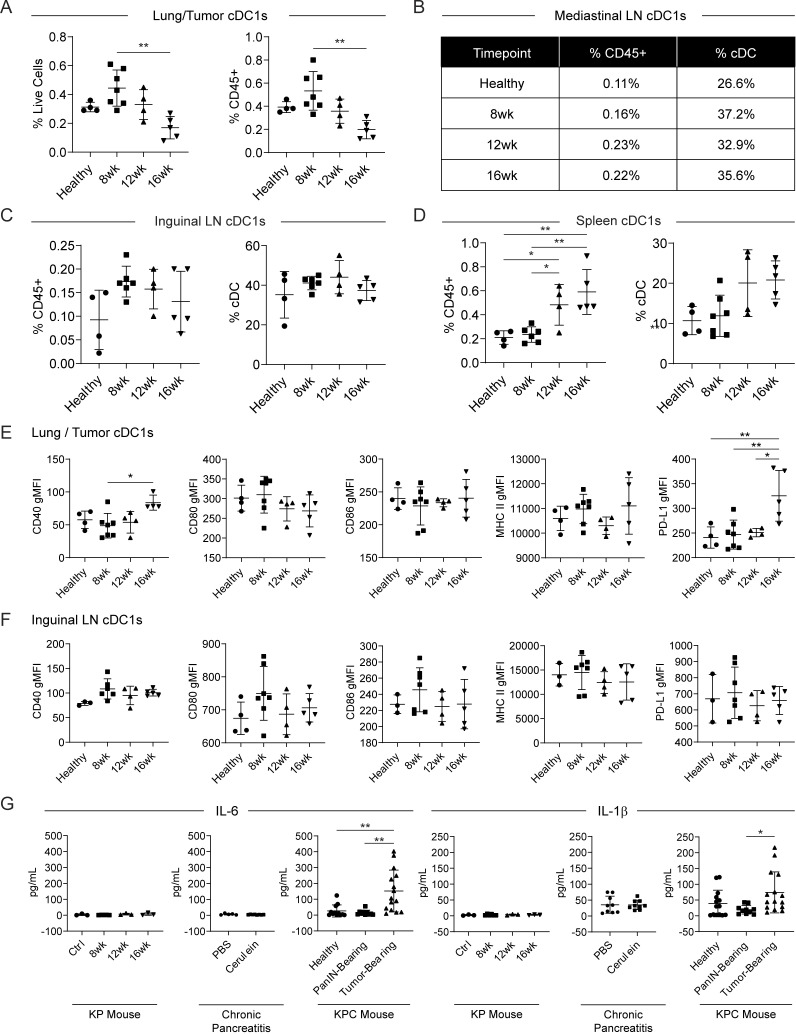
To study cDC1 biology in another mouse model of carcinoma, cDC1 abundance and maturation marker expression were quantified in the KrasLSL-G12D/+Trp53fl/fl (KP) mouse model of lung adenocarcinoma (DuPage et al., 2009). Expression of Cre recombinase was induced through endotracheal instillation of Ad:SPC-Cre adenovirus. Tissues were harvested from KP mice at 8, 12, and 16 wk after adenoviral induction of Cre recombinase. Control mice were sacrificed 16 wk after infection with Ad:CMV-FlpO. While cDC1 abundance declined progressively in the lung/tumor, cDC1 abundance was observed to increase in the mediastinal LN, iLNs, and spleen of Cre-infected KP mice (Fig. S4, A–D). cDC1 maturation marker expression in the lung/tumor also remained largely unchanged apart from increases in CD40 and PD-L1 at 16 wk after induction (Fig. S4 E). cDC1 maturation marker expression did not change in the iLNs (Fig. S4 F). Thus, although declines in cDC1 abundance and cDC1 semimaturation are present in the tumor microenvironment of both Kras/p53-driven mouse models, systemic declines in cDC1 abundance, maturation, and function were unique to the KPC GEMM of pancreatic adenocarcinoma.
Figure S4.
Systemic cDC1 dysfunction does not occur in the KP mouse model of lung adenocarcinoma. (A) Enumeration of cDC1s as a proportion of live cells and CD45+ cells in the lung/tumor of AdFlp-treated controls and KP mice 8, 12, or 16 wk after inhalation of adenoviral Cre recombinase. (B–D) Enumeration of cDC1s as a proportion of CD45+ cells and total cDCs in the (B) mediastinal LN, (C) iLNs, and (D) spleen. (E and F) Expression of maturation markers CD40, CD80, CD86, MHC II, and PD-L1 on cDC1s from the (E) lung/tumor and (F) iLNs. (G) Serum levels of IL-6 and IL-1β as determined by cytokine bead array in the KP and KPC cancer mouse models, as well as cerulein-induced chronic pancreatitis. Samples pooled across four to seven mice per group in B. Error bars indicate mean ± SD. **, P < 0.01; *, P < 0.05 (one-way ANOVA with Tukey’s HSD post-test). Data shown are representative of one independent experiment.
cDC1 abundance declines as a result of apoptosis
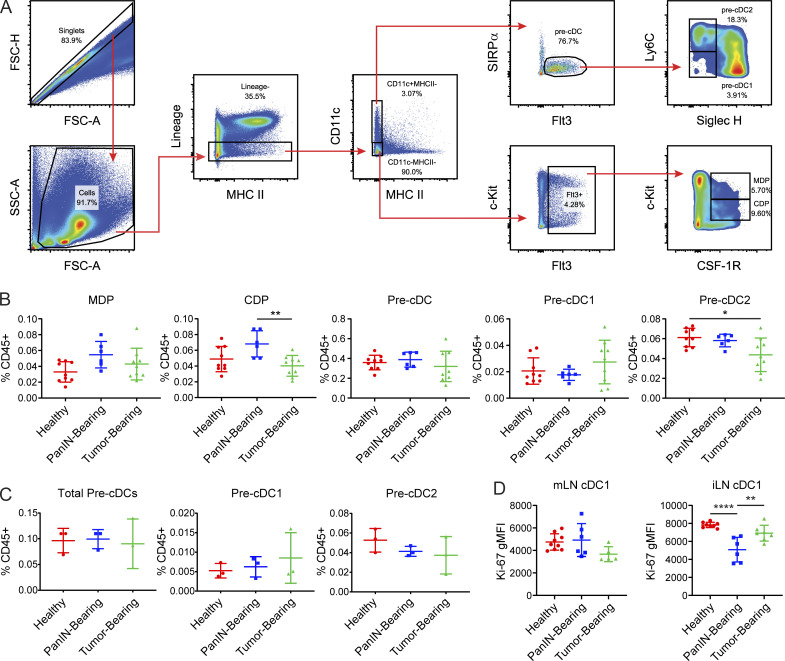
We next determined whether declining cDC1 abundance was due to impaired bone marrow generation. cDC1 progenitors consist of pre-cDCs, common DC progenitors (CDPs), and monocyte DC precursors (MDPs; Merad et al., 2013). MDPs have the potential to generate CDPs, monocytes, and monocyte-derived DCs, while CDPs give rise to pre-cDCs, which include pre-cDC1s and pre-cDC2s. Pre-cDC1s then circulate to peripheral tissues, where they differentiate into cDC1s. Using flow cytometry, MDPs, CDPs, and pre-cDCs were quantified in the bone marrow of healthy, PanIN-bearing, and tumor-bearing mice (Fig. 5 A). Pre-cDC1s and pre-cDC2s were distinguished based on their expression of Ly6C and Siglec H (Schlitzer et al., 2015). Numbers of bone marrow precDC1s did not decline over the course of pancreatic oncogenesis (Fig. 5 B). Pre-cDC1s in the peripheral blood were similarly unchanged (Fig. 5 C). Ki-67 levels in mesenteric LN (mLN) and iLN cDC1s were not significantly decreased in tumor-bearing mice compared with healthy controls, though Ki-67 was transiently decreased in the iLN cDC1s of PanIN-bearing mice (Fig. 5 D). Thus, we conclude that cDC1 generation in this model is not affected at the level of the bone marrow, peripheral blood, or proliferation during pancreatic carcinogenesis.
Figure 5.
cDC1 generation is unaffected by pancreatic neoplastic development. (A) Flow gating strategy for MDPs, CDPs, pre-cDCs, pre-cDC1s, and pre-cDC2s in RBC-lysed bone marrow suspension from a wild-type C57BL/6J mouse. Lineage gate consists of CD3, CD19, B220, NK1.1, and Gr-1. FSC-A, forward scatter-area; FSC-H, forward scatter-height; SSC-A, side scatter-area. (B) Enumeration of MDPs, CDPs, pre-cDCs, pre-cDC1s, and pre-cDC2s in the bone marrow of healthy, PanIN-bearing, and tumor-bearing mice. (C) Enumeration of pre-cDCs, pre-cDC1s, and pre-cDC2s in peripheral blood. (D) Expression of Ki67 in cDC1s from the mLN and iLN of healthy, PanIN-bearing, and tumor-bearing mice. Error bars indicate mean ± SD. ****, P < 0.0001; **, P < 0.01; *, P < 0.05 (one-way ANOVA with Tukey’s HSD post-test). Data shown are representative of at least two independent experiments with at least three mice per group. gMFI, geometric mean fluorescence intensity.
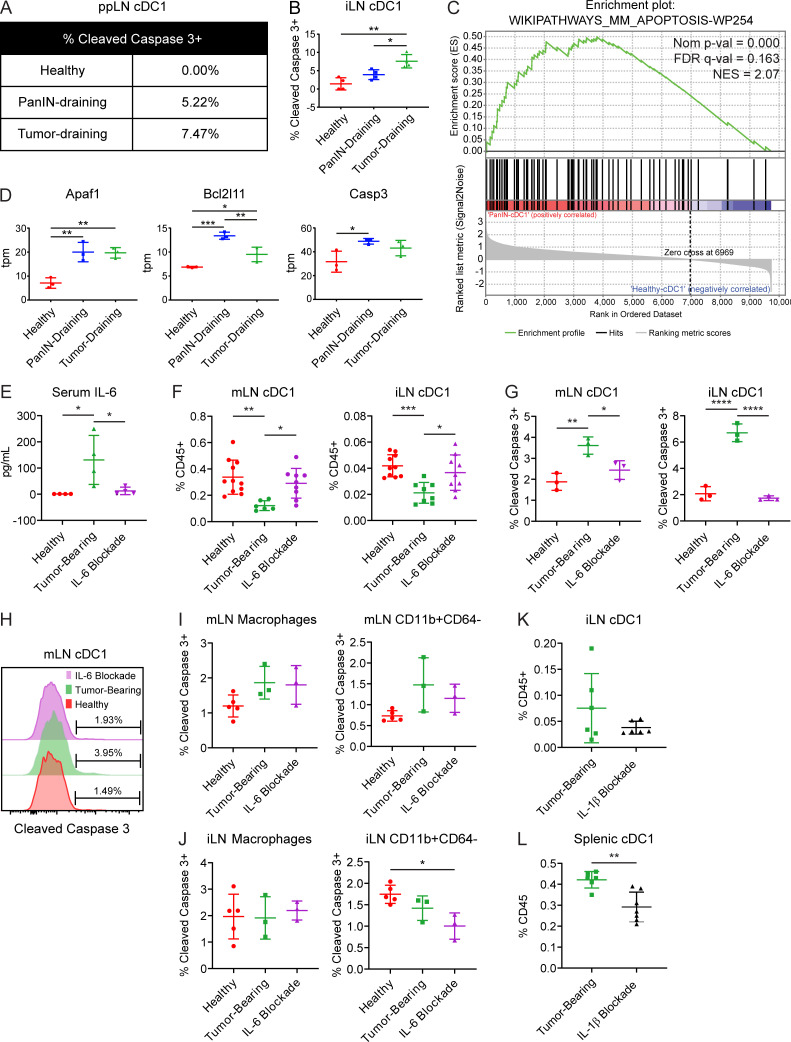
We next considered that systemic declines in cDC1 number might instead be driven by increased apoptosis. Therefore, we examined cDC1 apoptosis in the ppLNs and iLNs by staining for active cleaved caspase-3 (Fig. 6, A and B). We found that active cleaved caspase-3 increased progressively during pancreatic carcinogenesis in both ppLNs and iLNs. Furthermore, transcriptomic analysis of ppLN cDC1s from PanIN- and tumor-bearing mice revealed a positive enrichment for genes involved in apoptosis, including Apaf1, Bcl2l11, and Casp3 (Fig. 6, C and D; and Fig. 3 C).
Figure 6.
Increased serum IL-6 drives cDC1 apoptosis systemically in tumor-bearing KPC mice. (A and B) Percentage of (A) ppLN and (B) iLN cDC1s positive for expression of active cleaved caspase-3 in healthy, PanIN-bearing, and tumor-bearing mice. (C) Enrichment plot of apoptosis gene set in cDC1s from PanIN-draining versus healthy ppLNs. FDR, false discovery rate; NES, normalized enrichment score. (D) Expression of select genes in tpm reads from the gene set shown in C. (E) Serum IL-6 levels as determined by cytokine bead array in healthy mice, tumor-bearing mice, and tumor-bearing mice treated with IL-6–neutralizing antibody (MP5-20F3). (F) Enumeration of cDC1s in the mLN and iLN. (G) Percentage of mLN and iLN cDC1s positive for expression of cleaved caspase-3. (H) Representative histogram of cleaved caspase-3 expression in mLN cDC1s from G. (I and J) Percentage of CD64+F4/80+ macrophages and CD64−CD11b+ myeloid cells positive for expression of cleaved caspase-3 in the (I) mLN and (J) iLN. (K and L) Quantification of cDC1s as a percentage of live CD45+ cells in (K) iLN cDC1s and (L) splenic cDC1s from tumor-bearing KPC mice treated with IL-1β blocking monoclonal antibody (AF-401-NA). Samples pooled across at least four mice per treatment group in A. (C and D) Each sample consists of total RNA collected from 10,000 sorted ppLN cDC1s pooled across three to six mice. Error bars indicate mean ± SD. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05 (one-way ANOVA with Tukey’s HSD post-test in B, D, E–G, I, and J; two-tailed Student’s t test in K and L). Data shown are representative of at least two independent experiments with at least three mice per group.
IL-6 drives increased cDC1 apoptosis
To define mechanisms of cDC1 dysfunction that might be unique to the KPC model, serum levels of 13 chemokines and cytokines were quantified and compared in KPC pancreatic carcinogenesis, KP pulmonary carcinogenesis, and cerulein-induced chronic pancreatitis. Serum IL-6 and IL-1β levels were found to be significantly higher in KPC pancreatic adenocarcinoma than in KP lung adenocarcinoma and cerulein-induced chronic pancreatitis (Fig. S4 G). To assess whether these cytokines drive systemic declines in cDC1 survival, we first confirmed that serum IL-6 could be experimentally neutralized following administration of an IL-6–depleting antibody, MP5-20F3 (Fig. 6 E). Following 6 d of treatment with MP5-20F3 in tumor-bearing mice, quantification of cDC1s in the mLNs and iLNs revealed a rebound in cDC1 abundance (Fig. 6 F). To determine whether this rebound was being driven by decreased cDC1 apoptosis, active cleaved caspase-3 was quantified in mLN and iLN cDC1s (Fig. 6, G and H). Levels of active cleaved caspase-3 in cDC1s from tumor-bearing mice declined to levels close to those of healthy mice following IL-6 neutralization. Quantification of cleaved caspase-3 in macrophages and nonmacrophage CD11b+ cells showed no increased apoptosis in tumor-bearing mice and no effect with IL-6 neutralization; thus, the observed phenotype is specific to cDC1s (Fig. 6, I and J). Administration of an IL-1β blocking antibody, AF-401-NA, in contrast, failed to alleviate declines in cDC1 abundance and in fact seemed to worsen these deficits (Fig. 6, K and L). Together, these data suggest that declines in systemic cDC1 abundance in the KPC GEMM are attributable to increased apoptosis driven by IL-6.
CD40 activation rescues cDC1 maturation
CD40 is a receptor expressed on APCs that licenses them to mature upon contact with CD40 ligand on activated CD4+ T cells (Grewal and Flavell, 1997; van Kooten and Banchereau, 2000). To determine whether CD40 activation can reverse cDC1 dysfunction, a KPC cell line 6419.c5 was subcutaneously implanted into C57BL/6J mice, which were then treated with FGK45, an agonistic monoclonal rat antibody directed against murine CD40 (Fig. 7 A). cDC1 abundance declined in the tumor microenvironment following treatment (Fig. 7 B). This was found to be driven by cDC1 migration to the tumor-draining iLNs (TdLNs), as numbers of CD11cintMHC IIhi activated/migratory cDC1s increased in the TdLNs following treatment (Fig. 7 C). The expression of Ccr7 in TdLN CD11c+ cells also increased, further supporting the migration and maturation of activated cDC1s from the tumor microenvironment (Fig. 7 D). Maturation marker expression increased universally on cDC1s in the tumor microenvironment following FGK45 administration, consistent with fully repaired cDC1 maturation (Fig. 7 E). In the TdLN, the expression of CD40, CD86, and PD-L1 also increased in response to FGK45 (Fig. 7 F). In the spleen, maturation marker expression increased, except for MHC II, which declined (Fig. 7 G).
Figure 7.
CD40 activation repairs cDC1 maturation in KPC tumors. (A) Timeline of subcutaneous implantation of KPC cell line 6419.c5, administration of CD40 agonist (FGK45), and harvest of tissues for flow cytometric analysis. (B) Enumeration of cDC1s per live cells in subcutaneous KPC tumors from untreated and FGK45-treated mice. (C) Enumeration of CD11cintMHCIIhi migratory/activated cDC1s in the TdLN. (D) Expression of Ccr7 in CD11c+ cells purified from the iLNs of healthy mice and TdLNs of untreated and FGK45-treated mice bearing subcutaneously implanted KPC tumors. (E) Expression of maturation markers CD40, CD80, CD86, MHC II (I-A/I-E), and PD-L1 on cDC1s from the tumors of untreated and FGK45-treated mice. MFI, mean fluorescence intensity. (F and G) Maturation marker expression on cDC1s from the (F) TdLN and (G) spleen of healthy mice, untreated tumor-bearing mice, and FGK45-treated tumor-bearing mice. (H) Enumeration of H-2Kb:SIINFEKL tetramer–positive splenic CD8+ T cells from healthy mice, untreated tumor-bearing KPC mice, and FGK45-treated tumor-bearing KPC mice 12 d following subcutaneous implantation of OVA-expressing clonal KPC cell line 4662.V6ova. 100 µg FGK45 was administered on day 9 after implantation. Error bars indicate mean ± SD. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05 (two-tailed Student’s t test in B, C, E; one-way ANOVA with Tukey’s HSD post-test in D, F, G, H). Data shown are representative of four independent experiments with at least three mice per group.
Tumor-bearing KPC mice were then subcutaneously implanted with 4662.V6ova and treated with FGK45 to determine whether cDC1-mediated CD8+ T cell priming can be restored. Indeed, administration of FGK45 restored the generation of H-2Kb:SIINFEKL tetramer–positive CD8+ T cells to levels seen in healthy mice (Fig. 7 H).
To determine which cDC1 molecular pathways are up-regulated by treatment with FGK45, RNA-seq was performed on TdLN cDC1s from mice bearing subcutaneous tumors. Principal-component analysis revealed a broad transcriptomic change in cDC1s along PC1 following FGK45 administration (Fig. 8 A). However, differential gene expression analysis revealed that treatment with FGK45 induced a significantly different transcriptomic signature depending on whether tumor was present (Fig. 8 B). GSEA comparing cDC1s in FGK45-treated versus untreated tumor-bearing mice showed an induction of genes associated with type II interferon signaling, including Stat1 and Stat2 (Fig. 8, C–E). Thus, our data demonstrate that cDC1 maturation and function are rescued by CD40 activation and associated with induction of a type II interferon transcriptomic signature.
Figure 8.
CD40-driven cDC1 maturation is associated with an IFN-γ response signature. (A) Principal-component (PC) analysis of iLN cDC1 transcriptomes in the presence or absence of subcutaneously implanted KPC tumor, either treated or untreated with CD40 agonist (FGK45). (B) Heatmap comparing expression of differentially expressed genes across samples, scaled by z-score. (C) Top hits from GSEA of TdLN cDC1s from FGK45-treated versus untreated mice. (D) Enrichment plot of type II interferon response gene set from GSEA shown in C. FDR, false discovery rate; NES, normalized enrichment score. (E) Expression of Stat1 and Stat2 in tpm reads from gene set shown in D. n = 3 samples per group. Each sample consisted of total RNA collected from 10,000 sorted iLN cDC1s pooled from five mice per group. Error bars indicate mean ± SD. **, P < 0.01; *, P < 0.05 (one-way ANOVA with Tukey’s HSD post-test).
Flt3L synergizes with CD40 agonist to rescue cDC1 abundance and maturation
While CD40 agonism successfully rescued cDC1 maturation in tumor-bearing mice, it failed to increase cDC1 abundance in the tumor microenvironment. Therefore, to increase cDC1 abundance, we considered Flt3, which is a receptor tyrosine kinase that is highly expressed on hematopoietic progenitors, including MDPs, CDPs, and pre-cDCs (Lyman and Jacobsen, 1998). Administration of Flt3L has been shown to expand these progenitor populations and specifically promote cDC1 differentiation and survival (Karsunky et al., 2003). As a result, we hypothesized that Flt3L would synergize effectively with CD40 activation to increase cDC1 abundance in tumor-bearing mice.
To explore a role for Flt3L in enhancing the efficacy of CD40 agonist, we subcutaneously implanted C57BL/6J mice with cells derived from the clonal KPC cell line 4662.MD10. 14 d after implantation, mice were treated with FGK45 in combination with Flt3L (Fig. 9 A). Tissues were then harvested after 9 d of daily treatment with Flt3L. Quantification of cDC1s demonstrated that while Flt3L alone increased cDC1 abundance in the spleen, it decreased the abundance in the tumor and had no effect on cDC1 abundance in TdLN (Fig. 9 B). We again found that FGK45 alone decreased cDC1 abundance in the tumor microenvironment with no effect on spleen or TdLN cDC1s. In contrast, we observed a strong increase in cDC1 abundance in the TdLN with combination FGK45 and Flt3L. Combination therapy also increased cDC1 abundance in the spleen and increased cDC1 abundance in the tumor compared with Flt3L or FGK45 alone. Combination treatment also potentiated cDC1 expression of MHC II in the TdLN beyond levels seen with FGK45 alone (Fig. 9 C). Expression of CD80 and CD86 also trended higher in combination-treated mice compared with mice treated with FGK45 alone. Because Flt3L also serves as a survival factor for cDC1s, active cleaved caspase-3 was quantified in ppLN and iLN cDC1s from tumor-bearing KPC mice treated with combination FGK45 and Flt3L. Levels of active cleaved caspase-3 in ppLN and iLN cDC1s were significantly reduced after combination treatment (Fig. 9, D and E). Combination FGK45 and Flt3L, therefore, reverses the increased cDC1 apoptosis seen in PanIN- and tumor-bearing KPC mice while further potentiating cDC1 maturation beyond levels seen with CD40 agonist alone.
Figure 9.
Flt3L synergizes with CD40 activation to promote cDC1 survival and function. (A) Timeline of treatment of mice subcutaneously implanted with 3 × 105 KPC cell line 4662.MD10 with CD40 agonist (FGK45) and Flt3L. Treatment was initiated 14 d after transplant. (B) Enumeration of cDC1s in the tumor microenvironment, TdLN, and spleen of untreated, Flt3L-treated, FGK45-treated, and combination-treated mice. (C) Expression of MHC II, CD80, and CD86 on TdLN cDC1s. gMFI, geometric mean fluorescence intensity. (D and E) Percentage of cDC1s positive for expression of active cleaved caspase-3 in the (D) ppLN (percentages in healthy and tumor-bearing mice are also reported in Fig. 6 A) and (E) iLNs of healthy mice, tumor-bearing KPC mice, and tumor-bearing KPC mice treated with FGK45 and Flt3L. (F) Enumeration of and IFN-γ expression in H-2Kb:SIINFEKL tetramer–positive splenic CD8+ T cells 7 d following subcutaneous vaccination with 200 µg OVA + 10 µg CpG in tumor-bearing KPC mice treated with FGK45 and Flt3L. Samples were pooled across at least four mice per treatment group in D. Error bars indicate mean ± SD. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05 (one-way ANOVA with Tukey’s HSD post-test). Data shown are representative of at least two independent experiments with at least three mice per group.
To determine whether this combined rescue of cDC1 abundance and maturation results in improved cDC1-mediated CD8+ T cell priming, tumor-bearing mice were vaccinated with OVA/CpG and treated with FGK45 and Flt3L. The generation of H-2Kb:SIINFEKL tetramer–positive CD8+ T cells was significantly increased in combination-treated tumor-bearing mice beyond levels seen in healthy mice (Fig. 9 F). Likewise, the proportion of effector memory CD62L−CD44+ vaccine-responsive T cells and their expression of IFN-γ also reflected greater activation than in healthy mice. Thus, combination treatment with FGK45 and Flt3L reverses the quantitative and functional cDC1 deficits seen in PanIN- and tumor-bearing KPC mice, enabling productive CD8+ T cell priming and activation.
Combination CD40 agonist and Flt3L results in superior T cell activation
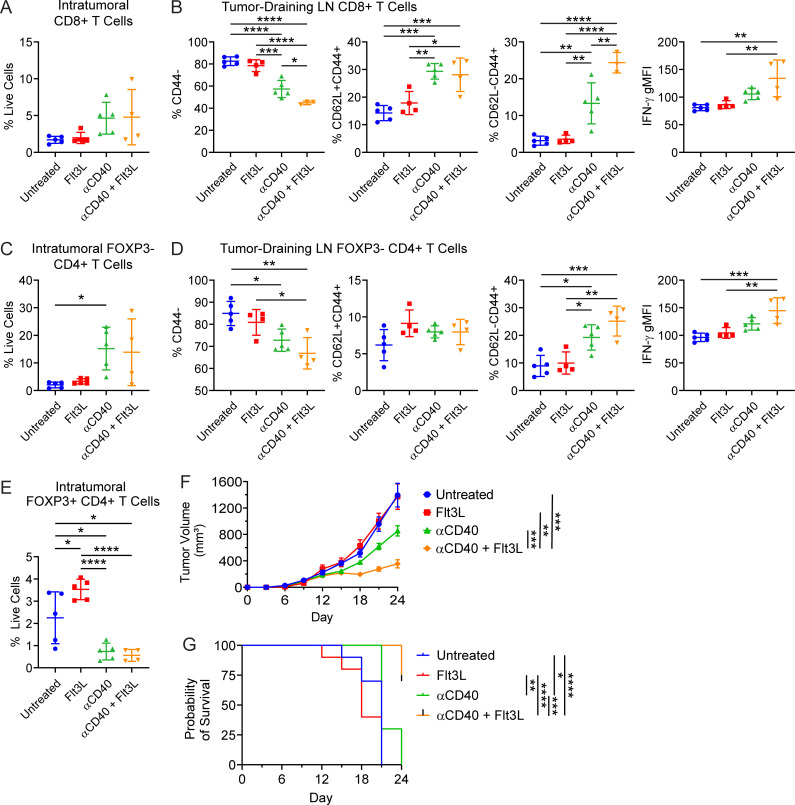
To determine whether combination FGK45 and Flt3L can enhance anti-tumor adaptive immunity, CD8+ and CD4+ T cells in the tumor microenvironment and TdLN were examined using flow cytometry. Consistent with our prior studies, CD8+ T cells trended toward an increase in the tumor microenvironment following CD40 agonism (Winograd et al., 2015; Byrne and Vonderheide, 2016; Fig. 10 A). The addition of Flt3L did not further enhance CD8+ T cell enrichment in the tumor microenvironment. However, based on proportions of CD62L−CD44+ T cells and expression of IFN-γ, combination CD40 agonist and Flt3L improved CD8+ T cell activation in the TdLN (Fig. 10 B). CD40 agonism also resulted in an influx of CD4+FOXP3− T cells into the tumor microenvironment (Fig. 10 C). Combination FGK45 and Flt3L enhanced the activation of FOXP3−CD4+ T cells in the TdLN (Fig. 10 D), and FOXP3+ CD4+ T regulatory cells in the tumor microenvironment decreased after CD40 agonism and combination treatment (Fig. 10 E).
Figure 10.
Combination therapy with CD40 agonist and Flt3L results in superior T cell activation in the tumor-draining LN. (A) Enumeration of CD8+ T cells in the tumor microenvironment of untreated, Flt3L-treated, CD40 agonist (FGK45)-treated, or combination-treated subcutaneously implanted KPC tumors as shown in Fig. 9 A. (B) Enumeration of and IFN-γ production in CD8+ T cells from the TdLN. (C) Enumeration of FOXP3− CD4+ T cells in the tumor microenvironment. (D) Enumeration of and IFN-γ production in FOXP3− CD4+ T cells from the TdLN. (E) Enumeration of FOXP3+ CD4+ T cells in the tumor microenvironment. (F and G) Tumor growth (F) and survival curves (G) from mice subcutaneously implanted with 5 × 105 KPC cell line 6419c5. Mice were treated with CD40 agonist and Flt3L beginning on day 12 after implantation using the treatment schedule shown in Fig. 9 A (corresponds to Fig. S5). n = 10 mice per group (F and G). Error bars indicate mean ± SEM. ****, P < 0.0001; ***, P < 0.001; **, P < 0.01; *, P < 0.05 (one-way ANOVA with Tukey’s HSD post-test in A–E; two-way ANOVA with Tukey’s HSD post-test in F; pairwise Kaplan–Meier survival log-rank test in G). Data shown are representative of three independent experiments with at least five mice per group.
To determine whether enhanced T cell priming produced with FGK45 and Flt3L in the draining LN might drive improved immune control of tumor outgrowth, a T cell–low KPC cell line, 6419c5, was implanted subcutaneously into C57BL/6J mice (Li et al., 2018). Beginning on day 12 after implantation, combination treatment with CD40 agonist and Flt3L was initiated according to the schedule described in Fig. 9 A. CD40 agonist and Flt3L induced superior control of tumor outgrowth compared with CD40 agonist monotherapy (Fig. 10 F and Fig. S5). Notably, Flt3L monotherapy had no discernable effect. Furthermore, the combination treatment extended overall survival of tumor-bearing mice (Fig. 10 G). Thus, we conclude that combined rescue of cDC1 abundance and maturation through CD40 agonist and Flt3L results in superior T cell activation in the draining LN and improved immune control of KPC tumor outgrowth.
Figure S5.
Tumor growth curves from subcutaneous implantation of 6419c5 and combination treatment with CD40 agonist and Flt3L. Individual tumor growth curves following subcutaneous implantation of 5 × 105 T cell low KPC cell line 6419c5 in (A) untreated, (B) Flt3L-treated, (C) CD40 agonist-treated, and (D) combination-treated mice. CD40 agonist and Flt3L were administered beginning day 12 after implantation using the treatment schedule shown in Fig. 9 A. Data shown are representative of three independent experiments with at least five mice per group (corresponds to Fig. 10, F and G).
Discussion
In this study, we aimed to decipher the nature and mechanism of cDC1 dysfunction during cancer progression. Using the KPC GEMM of pancreatic adenocarcinoma, we report that cDC1 dysregulation develops systemically and with early onset, before invasive tumor formation in mice bearing PanIN 1A. Elevated serum IL-6 in the setting of cancer development resulted in increased cDC1 apoptosis and systemically decreased cDC1 abundance. cDC1 maturation was also uniquely impacted in KPC mice, resulting in impaired T cell response to vaccination from the earliest stage of preinvasive neoplasia. Yet, these deficits were reversible in vivo. Combination treatment of tumor-bearing mice with CD40 agonist and Flt3L reversed deficits in cDC1 abundance and maturation, ameliorated apoptosis, and superior CD8+ T cell activation driving improved response to vaccination and immune control of tumor outgrowth.
A key conclusion of our study is that systemically decreased cDC1 abundance in the KPC model results from increased cDC1 apoptosis driven by IL-6. Antibody-based neutralization of elevated serum IL-6 abrogated increased expression of active cleaved caspase-3 in cDC1s from tumor-bearing mice and restored cDC1 abundance to levels seen in healthy controls. IL-6 in both murine and human pancreatic adenocarcinoma has been shown to be primarily produced by tumor-associated macrophages and inflammatory cancer-associated fibroblasts (Martignoni et al., 2005; Öhlund et al., 2017; Lee et al., 2019). In KrasG12D mice, IL-6 signaling promotes PanIN progression and development of pancreatic cancer (Lesina et al., 2011). Patients with pancreatic cancer also have elevated levels of serum IL-6 compared with age-matched healthy controls (Long et al., 2017). Overproduction of IL-6 has been strongly associated with chemoresistance, decreased survival, poor performance status, and cachexia in patients (Ebrahimi et al., 2004). Here, we argue that IL-6 is linked to cDC1 dysfunction in cancer. Serum IL-6 is found to be elevated in KPC pancreatic adenocarcinoma but not KP lung adenocarcinoma or cerulein-induced chronic pancreatitis. Likewise, out of these three models, systemic cDC1 dysfunction is only observed in the KPC GEMM. In non–tumor-bearing mice, IL-6 has been shown to play a major role in maintaining immature DCs. IL-6 knockout mice have increased numbers of mature DCs, indicating that IL-6 blocks DC maturation in vivo (Park et al., 2004). In addition, autocrine IL-6 and IL-10 promote differentiation of IL-10–producing immunosuppressive DCs (Tang et al., 2015). Interestingly, targeted inhibition of IL-6 with antibodies enables sensitivity to PD-L1 blockade and cooperates with chemotherapy to drive tumor regression in mouse models of pancreatic cancer (Long et al., 2017; Mace et al., 2018). It will be essential to perform cDC1 immunohistochemistry in future studies to determine whether cDC1 spatial distribution is also altered during pancreatic carcinogenesis or in tumor-bearing mice following IL-6 blockade. Overall, data in mice indicate that IL-6 plays a major role in DC biology. Our findings here point to a previously unappreciated role for IL-6 in cDC1 apoptosis in cancer.
A core component of cDC1 dysfunction in KPC mice is DC semimaturation, again evident from the earliest stage of preinvasive neoplasia. DC semimaturation is currently understood as the inconsistent up-regulation of maturation markers on peripheral blood DCs associated with suboptimal T cell priming (Tjomsland et al., 2010; Dudek et al., 2013). As noted above, IL-6 signaling can enforce such a phenotype physiologically (Park et al., 2004; Tang et al., 2015). In the present study, high-throughput RNA-seq demonstrates that cDC1 semimaturation coincides with induction of genes involved in proteasomal degradation and antigen processing whereas genes encoding T cell–polarizing cytokines fail to be appropriately up-regulated. This results in a suspended state of cDC1 semimaturation during pancreatic carcinogenesis. In KPC mice, OVA-specific CD8+ T cell priming following challenge with OVA as a model tumor antigen or vaccination with OVA/CpG are significantly reduced. cDC1 semimaturation is therefore associated with impaired induction of T cell–polarizing cytokines and defective T cell priming in PanIN- and tumor-bearing mice.
Despite the early onset of cDC1 dysregulation during carcinogenesis, we find that deficits in cDC1 abundance, maturation, and function remain reversible through malignancy. Prior studies from our group have shown that CD40 agonist effectively promotes T cell infiltration into KPC tumors, enabling response to ICB in a cDC1-dependent manner (Winograd et al., 2015; Twyman-Saint Victor et al., 2015; Byrne and Vonderheide, 2016). Here, we reveal CD40 agonism specifically induces an IFN-γ response signature in cDC1s that rescues their maturation. It remains unclear whether this is driven by direct ligation of CD40 on cDC1s or IFN-γ secretion by another CD40-expressing cell type (Grewal and Flavell, 1997; Schoenberger et al., 1998). We also find that to rescue cDC1 abundance as well as maturation, addition of Flt3L is needed and in fact potentiates cDC1 maturation beyond levels seen with CD40 agonist alone. This boosts CD8+ T cell activation and improves response to vaccination and immune control of tumor outgrowth. However, while combination treatment with CD40 agonist and Flt3L drives superior T cell priming in tumor-bearing mice, we cannot rule out that these differences may be due at least in part to changes in the MDSC compartment. Our group has previously described a significant increase in MDSCs that occurs in the KPC GEMM and underlies tumor-intrinsic mechanisms of immune suppression, particularly evident at the invasive stage (Bayne et al., 2012). It remains possible that the rescued T cell priming following combination CD40 agonist and Flt3L is at least partially driven by changes in MDSC abundance and function in addition to the increased cDC1 abundance and maturation described in our study.
A decline in cDC1 abundance has previously been reported in tumor-bearing KPC mice (Meyer et al., 2018). However, the conclusion provided in that study contrasts with our findings. While Meyer et al. attribute decreased cDC1 abundance to impaired bone marrow cDC1 generation caused by G-CSF–mediated suppression of IRF8, we observe that cDC1 generation is unaffected during pancreatic carcinogenesis. Rather, we focus on very early events in cDC1 dysfunction and show prominent apoptosis and semimaturation of cDC1s in PanIN-bearing mice that have not previously been reported. In addition, the same group recently used endogenously expressed OVA as a model neoantigen in the KPC model to demonstrate that Flt3L reverses cDC paucity and restores T cell priming upon combination with CD40 agonist (Hegde et al., 2020). While their results with combination CD40 agonist and Flt3L in neoantigen-positive KPC closely mirror ours, we find that Flt3L monotherapy is ineffective in neoantigen-negative KPC mice for increasing the cDC1 content of tumors and tumor-draining LNs. Our group has demonstrated in prior studies that cDC1s are necessary for response to CD40 agonism. Therefore, it will be critical in future studies to experimentally establish whether cDC1s are required for therapeutic response to combination CD40 agonist and Flt3L (Byrne and Vonderheide, 2016; Morrison et al., 2020).
Our findings in KPC mice have relevance to patients with pancreatic cancer. As the critical APC for antigen cross-presentation, cDC1s in humans are critical for CD8+ T cell responses against necrotic cell antigens (Jongbloed et al., 2010; Broz et al., 2014; Roberts et al., 2016). Here, we show that peripheral blood cDC1s are significantly reduced in patients with newly diagnosed and untreated metastatic pancreatic cancer compared with healthy volunteers. Furthermore, we analyzed transcriptomic data from 182 patients in TCGA-PAAD. Using expression of known human cDC1 markers XCR1 and CLEC9A as an indication of cDC1 abundance in the tumor microenvironment (Böttcher and Reis e Sousa, 2018; Wculek et al., 2020), we found a statistically significant correlation between cDC1 markers and cytolytic activity. Furthermore, expression of the maturation markers HLA-DR and CD86 also correlated strongly with cytolytic index in this human dataset. Thus, like the KPC GEMM, cDC1 abundance and maturation correlate with cytolytic activity in human pancreatic tumors (Li et al., 2018). It was recently shown that cDC1 abundance is significantly reduced as a proportion of CD45+ cells in the tumor microenvironment of human PDA relative to non–small cell lung adenocarcinoma (Hegde et al., 2020). This recent finding is consistent with a previous observation that total DC abundance as measured by immunohistochemistry declines progressively in human premalignant PanINs, consistent with our findings in the KPC GEMM (Hiraoka et al., 2011).
Systemic DC dysfunction has been reported in the past in advanced-stage cancer patients (Gabrilovich et al., 1997; Almand et al., 2000; Hoffmann et al., 2002). Although cancer patients generally do not suffer opportunistic infections like patients with AIDS, there is evidence for cancer patients having immunodeficiencies. One example is the higher risk of Varicella zoster, a classically T cell–controlled pathogen, across multiple liquid and solid malignancies compared with age-matched controls (Hansson et al., 2017). Pancreatic cancer patients also exhibit abnormalities in T cell subsets and activation at the time of diagnosis before therapy (Alanio et al., 2019). It seems likely that progressive cancer itself reflects, to a greater or lesser extent, failed immune surveillance, even in pancreatic cancer (Balachandran et al., 2017). With these new insights into cDC1 dysfunction in KPC mice, it will be important to examine T cell immunity in cancer patients more deeply with a mindful eye toward clinical and immune phenotypes in the future.
Finally, the reversal of cDC1 dysfunction through CD40 agonism is interesting in light of recent efforts to use agonist CD40 antibodies as cancer immunotherapy in patients (Vonderheide, 2018). In metastatic pancreatic adenocarcinoma, preliminary results are promising (O’Hara et al., 2019). These and other trials of agonistic CD40 antibody provide the opportunity to study treatment effects on patient DCs in tissue and blood, using strategies informed by our mouse studies here. Our findings suggest that CD40 agonist may synergize with Flt3L clinically to enable T cell responses in cancer patients for whom T cell priming is deficient.
Materials and methods
Human subjects research
Blood samples from patients with advanced pancreatic cancer and healthy volunteers were collected and enriched using Ficoll centrifugation and cryopreserved in liquid nitrogen until analysis. Samples were obtained after informed consent and institutional review board approval from the University of Pennsylvania. A total of 17 patients (40–81 yr old, males and females) with untreated advanced PDA (2 locally advanced, 15 metastatic) and 10 healthy volunteers (54–75 yr old) were included in the study. Patients with PDA and healthy volunteers were comparable (median age of patients, 59 yr; median age of healthy volunteers, 65 yr; P = 0.059 by two-tailed Student’s t test).
Animal studies
All mouse experiments were done at the University of Pennsylvania Perelman School of Medicine, approved by the UPenn Institutional Animal Care and Use Committee, and performed in strict compliance with protocols 804666 and 804774. Mice were housed under pathogen-free conditions in a barrier facility. C57BL/6 mice were purchased from Jackson Laboratories or bred in-house. The size of each animal cohort was determined by estimating biologically relevant effect sizes between control and experimental groups and then using the minimum number that could reveal statistical significance.
Subcutaneous tumor implantation
Subcutaneously implanted KPC tumors were generated by injecting 3 × 105 cells in sterile DMEM into the right flank of female C57BL/6 mice unless otherwise specified. Cre/Cre and KPC mice were bred in-house. Tumor volume was calculated as greater diameter × smaller diameter2. Mice were considered to have reached endpoint in survival analyses upon reaching a tumor volume of 500 mm3.
Antibody-based experiments
IL-6 blockade in tumor-bearing KPC mice was performed by injecting 200 µg IL-6 depleting antibody (InVivomAb; MP5-20F3) in 100 µl PBS intraperitoneally on days 0 and 3 before flow cytometric analysis on day 6. IL-1β blockade in tumor-bearing KPC mice was performed by injecting 10 µg IL-1β blocking antibody (InVivomAb; AF-401-NA) in 100 µl PBS intraperitoneally on days 0, 2, and 4 before flow cytometric analysis on day 6. CD40 agonist studies were performed via a single intraperitoneal injection of 100 µg of monoclonal CD40 agonistic antibody (InVivomAb; FGK45) in 100 µl PBS. Flt3L studies were performed with once-daily injections of 10 µg Flt3L in 100 µl PBS subcutaneously at the nape of the neck.
Vaccination studies
Vaccination with OVA/CpG was performed by subcutaneous injection of 200 µg endotoxin-free OVA (Invivogen; vac-pova-100) + 10 µg endotoxin-free ODN1826 CpG (Invivogen; tlrl-1826-1) in 200 µl PBS subcutaneously into the right flank.
Cerulein chronic pancreatitis
Cerulein-induced chronic pancreatitis was performed via intraperitoneal injection of cerulein (Sigma-Aldrich; C9026) at 50 µg/kg/h × 6 h twice a week for 11 wk.
KPC mouse model
The KPC GEMM of PDA is driven by Pdx1-Cre KrasLSL-G12D/+Trp53LSL-R172H/+. As previously published, KPC mice in our colony are fully backcrossed to C57BL/6 based on the DartMouse Illumina GoldenGate Genotyping Assay, which interrogated 1,449 single-nucleotide proteins spread throughout the genome (Evans et al., 2016).
KP mouse model
KrasLSL-G12D mice (JAX; stock number 008179) and Trp53fl/fl mice (JAX; stock number 008462) have previously been described (Jackson et al., 2001, 2005). Mice are mixed B6J/129S4vJae. Non–tumor-bearing control mice were transduced with 2.5 × 107 PFUs of Ad:CMV-FlpO 16 wk before sacrifice, while tumor-bearing mice were given 2 × 108 PFUs of Ad:SPC-Cre at 16, 12, or 8 wk before sacrifice. Viral particles were obtained from University of Iowa Viral Vector Core, and mice were transduced by endotracheal instillation as previously described (DuPage et al., 2009).
KPC cell lines and cell culture
Tumor cell lines were derived from spontaneous tumors in the KPC GEMM. 4662.V6ova is an OVA-transduced clonal KPC cell line (Evans et al., 2016). 4662.MD10 and 6419.c5 are clonal KPC cell lines (Li et al., 2018). Cell culture was performed using DMEM supplemented with 10% FBS, L-glutamine, and penicillin/streptomycin. Cell lines were tested for mycoplasma contamination once every 6 mo.
Pancreas and tumor histology
Pancreas and KPC tumor were harvested and fixed in 4% PFA overnight and then paraffin processed and stained with H&E following standard protocols. Images were obtained using a Nikon Eclipse 50i microscope and Nikon Elements BR v5.01.01 software.
Tissue processing and cell isolation
Tumors and pancreas were dissected and minced in DMEM-F12 + 10% FBS at 4°C and then digested in DMEM-F12 with 1 mg/ml collagenase with protease inhibitor (Sigma-Aldrich; C6079) for 30 min at 37°C. Tissues were filtered through a 70-µm cell strainer and then a 40-µm cell strainer with 9 ml FACS buffer (PBS with 0.2% BSA + 2 mM EDTA). LNs, spleens, and bone marrow were dissected and minced in RPMI + 5% FBS at 4°C and then digested in RPMI with 1 mg/ml collagenase (Sigma-Aldrich; C5138) for 20 min at 37°C. Spleens and bone marrow were subject to two rounds of RBC lysis using 1 mL ACK Lysis Buffer (Gibco; A1049201). Samples were then filtered through a 40-µm cell straining and rinsed with 9 ml FACS buffer. Due to the small size of ppLNs (especially in healthy mice), ppLN samples were always pooled across all mice per experimental group to achieve sufficient cDC1 quantities for downstream analysis.
Flow cytometric analysis
All stainings were performed in the dark. Tissue-derived cells were washed with PBS before viability stain with LIVE/DEAD Fixable Aqua (Invitrogen; L34957) for 20 min at room temperature. Samples for DC analysis were then washed with FACS Buffer before being stained for immune markers CD45, CD64, F4/80, CD3, CD19, B220, NK1.1, Gr-1, I-A/I-E, CD11c, XCR1, SIRPα, CD103, CD11b, CD40, CD80, CD86, and PD-L1 for 30 min at 4°C. Where appropriate, cDC1s were intracellularly stained for Ki-67 and cleaved caspase-3 overnight at 4°C. Samples for T cell analysis were stained for immune markers CD45, CD3, CD8, CD4, H-2Kb:SIINFEKL tetramer, TIM-3, LAG3, CTLA-4, PD-1, CD62L, and CD44 extracellularly for 30 min at 4°C and FOXP3, CTLA-4, Eomes, Granzyme B, Tbet, Ki-67, and IFN-γ intracellularly overnight at 4°C. Bone marrow samples were stained extracellularly for Siglec H, c-Kit, CSF-1R, Flt3, SIRPα, I-A/I-E, CD45, CD11b, Ly6C, CD11c, CD3, CD19, B220, NK1.1, and Gr-1 at 4°C for 30 min. To aid in obtaining an accurate quantification of cells in tumor samples, target events were normalized using CountBright Absolute Counting Beads (Life Technologies; C36950) per manufacturer’s instructions. Samples were analyzed on a BD Biosciences LSR Fortessa. All flow panels are provided in Table S1.
Serum cytokine analysis
1 ml blood was collected from each mouse via eye enucleation into 1.5-ml Eppendorf tubes. Once blood had been allowed to clot at room temperature for at least 30 min, Eppendorf tubes were centrifuged at 2,000 ×g for 10 min at 4°C. Serum was then collected and frozen at −80°C. Cytokine bead array was then performed using the LEGENDplex Mouse Inflammation Panel (13-plex) with V-bottom plate (BioLegend; 740446) per the manufacturer’s instructions.
RNA-seq analysis, differential gene expression, and GSEA
cDC1s were sorted using a BD Biosciences Aria II cell sorter with 100-µm nozzle into an Eppendorf tube with 350 µl Buffer RLT Plus at 4°C using the gating strategy shown in Fig. 1 B. RNA was isolated from sorted cDC1s using the Qiagen RNeasy Plus Micro Kit per the manufacturer’s instructions. RNA purity and integrity were measured with an Agilent TapeStation before poly(A) selection and library construction followed by single-end 100-bp sequencing on an Illumina HiSeq4000 high-throughput sequencer at a depth of 20 million reads per sample by the UPenn Next-Generation Sequencing Core. The curated RNA-seq analysis pipeline from bcbio-nextgen was used for downstream analysis (https://github.com/chapmanb/bcbio-nextgen). FASTQ files were checked for quality using FastQC and qualimap. Alignment was performed with STAR under default settings using the mm10 reference genome. Raw counts of gene transcripts were obtained from BAM files using featureCounts (Liao et al., 2014). The resulting count matrix was then imported into R (version 3.6.1) and used as input to DESeq2 for normalization and differential gene expression analysis (Love et al., 2014). Salmon/Sailfish quasialignment was used to normalize and quantify gene expression and generate a tpm matrix to be used as input for GSEA (Mootha et al., 2003). Pathway and gene ontology analyses were performed using GSEA and Gene Set Knowledgebase, a curated functional genomics database for murine transcriptomes (Bares, 2015). RNA-seq data have been submitted to (and may be accessed at) the Gene Expression Omnibus database repository (accession no. GSE126389).
TCGA
RNA-seq datasets were downloaded with authorization for all TCGA-PAAD patients on the National Cancer Institute’s Genomic Data Commons Portal (Raphael et al., 2017).
Mass cytometry antibodies
Metal-conjugated antibodies were purchased from Fluidigm. Antibody, metal conjugate, and clone information is available in Table S2.
Mass cytometry sample preparation and data acquisition
Samples were thawed for analysis and washed with FACS buffer. Total cell concentration was determined using a TC20 automated cell counter (Bio-Rad). A 1-µM solution of 198Pt monoisotopic cisplatin (Fluidigm) was added to at most 4 × 106 cells for 1 min at room temperature. Cells were immediately washed twice with FACS buffer and incubated with Cytofix fixation buffer (BD Biosciences) for 25 min on ice. Samples were washed twice in FACS buffer and then split. 1.5 × 106 cells were cryopreserved for future use and the remaining cells were labeled using palladium barcoding per the manufacturer’s protocol (Fluidigm). Following barcoding, samples were pooled and incubated with Human TruStain FcX (BioLegend) for 10 min at room temperature. Then, a 2× master mix of metal-tagged antibodies was added directly to the samples for 30 min at room temperature. After washing with permeabilization working solution (eBioscience) samples were fixed again with 2.4% formaldehyde in PBS containing 125 nM iridium nucleic acid intercalator (Fluidigm) and left overnight. Samples were cryopreserved in 10% DMSO in FBS and stored at −80°C until thawing immediately before acquisition. Samples were washed twice with PBS + 0.2% BSA, once with cell acquisition solution and then resuspended at a concentration of 106 cells/ml in cell acquisition solution containing 5% EQ Four Element Calibration beads (Fluidigm). Samples were acquired on a Helios mass cytometer (Fluidigm) using a standardized acquisition template following routine tuning and optimization.
Mass cytometry data analysis
Flow cytometry standard data files were bead-normalized using CyTOF Software v6.7 (Fluidigm) and debarcoded using Astrolabe (Astrolabe Diagnostics). Manual gating in FlowJo (BD Biosciences) was used to exclude debris, dead cells, and doublets. The frequency of CD141+ cDC1s was defined by manual gating as follows: exclusion of CD3+, CD19+, CD14+, and CD56+ cells and selection for HLA-DR+ and CD11c+ cells followed by exclusion of CD1b and positive selection for CD141. The frequency of cDC1s among patients and healthy volunteers was compared by Mann–Whitney test using Prism 8.0 software (GraphPad).
Statistical analysis
Data points that were more than two standard deviations from the mean were removed as outliers. All statistical analyses of flow cytometry were performed using GraphPad Prism 7 or 8. Statistics in GSEA were performed using the gene set permutation setting within the Broad Institute GSEA software. Adjusted P values <0.05 and false discovery rate q-values <0.25 were considered statistically significant. Correlation analyses of TCGA-PAAD gene expression were performed using Kendall’s τ rank correlation coefficient due to a lack of bivariate normality as determined using the Shapiro–Wilks test in R v3.6.1.
Online supplemental material
Fig. S1 depicts tissue weights, absolute cDC1 number per organ, and enumeration of cDC1s based on tissue weight. Fig. S2 provides evidence of cDC1 dependency in CD8+ T cell activity patients with PDA from TCGA. Fig. S3 demonstrates the lack of systemic cDC1 dysfunction in mice with cerulein-induced chronic pancreatitis and KPC mice before PanIN-bearing age, thus establishing that systemic cDC1 dysregulation is specific to neoplastic development. Fig. S4 reveals the lack of systemic cDC1 dysregulation in the KP mouse model of lung adenocarcinoma. Fig. S5 contains individual tumor growth curves for all mice shown in Fig. 10, F and G. Table S1 provides reagents and antibodies used in murine studies. Table S2 provides reagents and antibodies used in human studies.
Supplementary Material
lists reagents and antibodies used in murine studies.
lists reagents and antibodies used in human studies.
Acknowledgments
We would like to acknowledge Liz Quinones and Dr. Cynthia Clendenin of the Abramson Cancer Center Pancreatic Cancer Mouse Hospital, who generated the KPC mice used in this study. We also thank Dr. Ben Stanger for valuable discussions.
This study was supported by the American Medical Association (Pancreatic Cancer Seed Grant to J.H. Lin), the Parker Institute for Cancer Immunotherapy (R.H. Vonderheide), the National Institutes of Health (grants R01 CA229803, P30 CA016520, P01 CA210944 [to R.H. Vonderheide], T32 HL007439-41 [to M.M. Wattenberg], and R01 CA197916 [to G.L. Beatty]), and the Penn Pancreatic Cancer Research Center (E.L. Carpenter).
Author contributions: J.H. Lin and R.H. Vonderheide conceived and designed the study. J.H. Lin, A.P. Huffman, and D.M. Walter performed mouse experiments. E.L. Carpenter, M.M. Wattenberg, and G.L. Beatty performed human studies. J.H. Lin and M.M. Wattenberg conducted bioinformatic analyses. All authors helped design experiments and analyzed and interpreted data. J.H. Lin and R.H. Vonderheide wrote the manuscript with contributions from A.P. Huffman, M.M. Wattenberg, G.L. Beatty, and E.E. Furth. R.H. Vonderheide supervised the overall conduct of the study. All authors read and approved the final manuscript.
References
- Alanio C., Bengsch B., Gies J.R., Henrickson S., Weng N.P., Ritz-Romeo J.A., O’Hara M., Melenhorst J.J., Lacey S., Young R.M., et al. 2019. Abstract A123: Skewed CD4 and CD8 T-cell differentiation in pancreatic cancer patients. Cancer Immunol. Res. 7:A123–A123. [Google Scholar]
- Almand B., Resser J.R., Lindman B., Nadaf S., Clark J.I., Kwon E.D., Carbone D.P., and Gabrilovich D.I.. 2000. Clinical significance of defective dendritic cell differentiation in cancer. Clin. Cancer Res. 6:1755–1766. [PubMed] [Google Scholar]
- Balachandran V.P., Łuksza M., Zhao J.N., Makarov V., Moral J.A., Remark R., Herbst B., Askan G., Bhanot U., Senbabaoglu Y., et al. ; ARC-Net Centre for Applied Research on Cancer . 2017. Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature. 551:512–516. 10.1038/nature24462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bares V, G.X. 2015. gskb: Gene Set data for pathway analysis in mouse. R package version 1.12.0.
- Bayne L.J., Beatty G.L., Jhala N., Clark C.E., Rhim A.D., Stanger B.Z., and Vonderheide R.H.. 2012. Tumor-derived granulocyte-macrophage colony-stimulating factor regulates myeloid inflammation and T cell immunity in pancreatic cancer. Cancer Cell. 21:822–835. 10.1016/j.ccr.2012.04.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böttcher J.P., and Reis e Sousa C.. 2018. The role of type 1 conventional dendritic cells in cancer immunity. Trends Cancer. 4:784–792. 10.1016/j.trecan.2018.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broz M.L., Binnewies M., Boldajipour B., Nelson A.E., Pollack J.L., Erle D.J., Barczak A., Rosenblum M.D., Daud A., Barber D.L., et al. 2014. Dissecting the tumor myeloid compartment reveals rare activating antigen-presenting cells critical for T cell immunity. Cancer Cell. 26:638–652. 10.1016/j.ccell.2014.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne K.T., and Vonderheide R.H.. 2016. CD40 stimulation obviates innate sensors and drives T cell immunity in cancer. Cell Rep. 15:2719–2732. 10.1016/j.celrep.2016.05.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao T., Furth E.E., and Vonderheide R.H.. 2016. CXCR2-dependent accumulation of tumor-associated neutrophils regulates T cell immunity in pancreatic ductal adenocarcinoma. Cancer Immunol. Res. 4:968–982. 10.1158/2326-6066.CIR-16-0188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark C.E., Hingorani S.R., Mick R., Combs C., Tuveson D.A., and Vonderheide R.H.. 2007. Dynamics of the immune reaction to pancreatic cancer from inception to invasion. Cancer Res. 67:9518–9527. 10.1158/0008-5472.CAN-07-0175 [DOI] [PubMed] [Google Scholar]
- Dalod M., Chelbi R., Malissen B., and Lawrence T.. 2014. Dendritic cell maturation: functional specialization through signaling specificity and transcriptional programming. EMBO J. 33:1104–1116. 10.1002/embj.201488027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek A.M., Martin S., Garg A.D., and Agostinis P.. 2013. Immature, semi-mature, and fully mature dendritic cells: Toward a DC-cancer cells interface that augments anticancer immunity. Front. Immunol. 4:438 10.3389/fimmu.2013.00438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuPage M., Dooley A.L., and Jacks T.. 2009. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc. 4:1064–1072. 10.1038/nprot.2009.95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi B., Tucker S.L., Li D., Abbruzzese J.L., and Kurzrock R.. 2004. Cytokines in pancreatic carcinoma: correlation with phenotypic characteristics and prognosis. Cancer. 101:2727–2736. 10.1002/cncr.20672 [DOI] [PubMed] [Google Scholar]
- Elsässer H.P., Haake T., Grimmig M., Adler G., and Kern H.F.. 1992. Repetitive cerulein-induced pancreatitis and pancreatic fibrosis in the rat. Pancreas. 7:385–390. 10.1097/00006676-199205000-00017 [DOI] [PubMed] [Google Scholar]
- Engelhardt J.J., Boldajipour B., Beemiller P., Pandurangi P., Sorensen C., Werb Z., Egeblad M., Krummel M.F., Adler A.J., Marsh D.W., et al. 2012. Marginating dendritic cells of the tumor microenvironment cross-present tumor antigens and stably engage tumor-specific T cells. Cancer Cell. 21:402–417. 10.1016/j.ccr.2012.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans R.A., Diamond M.S., Rech A.J., Chao T., Richardson M.W., Lin J.H., Bajor D.L., Byrne K.T., Stanger B.Z., Riley J.L., et al. 2016. Lack of immunoediting in murine pancreatic cancer reversed with neoantigen. JCI Insight. 1:88328 10.1172/jci.insight.88328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig C., Jones J.O., Kraman M., Wells R.J.B., Deonarine A., Chan D.S., Connell C.M., Roberts E.W., Zhao Q., Caballero O.L., et al. 2013. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti-PD-L1 immunotherapy in pancreatic cancer. Proc. Natl. Acad. Sci. USA. 110:20212–20217. 10.1073/pnas.1320318110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabrilovich D.I., Corak J., Ciernik I.F., Kavanaugh D., and Carbone D.P.. 1997. Decreased antigen presentation by dendritic cells in patients with breast cancer. Clin. Cancer Res. 3:483–490. [PubMed] [Google Scholar]
- Grewal I.S., and Flavell R.A.. 1997. The CD40 ligand. At the center of the immune universe? Immunol. Res. 16:59–70. 10.1007/BF02786323 [DOI] [PubMed] [Google Scholar]
- Guilliams M., Dutertre C.-A., Scott C.L., McGovern N., Sichien D., Chakarov S., Van Gassen S., Chen J., Poidinger M., De Prijck S., et al. 2016. Unsupervised high-dimensional analysis aligns dendritic cells across tissues and species. Immunity. 45:669–684. 10.1016/j.immuni.2016.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansson E., Forbes H.J., Langan S.M., Smeeth L., and Bhaskaran K.. 2017. Herpes zoster risk after 21 specific cancers: population-based case-control study. Br. J. Cancer. 116:1643–1651. 10.1038/bjc.2017.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde S., Krisnawan V.E., Herzog B.H., Zuo C., Breden M.A., Knolhoff B.L., Hogg G.D., Tang J.P., Baer J.M., Mpoy C., et al. 2020. Dendritic cell paucity leads to dysfunctional immune surveillance in pancreatic cancer. Cancer Cell. 37:289–307.e9. 10.1016/j.ccell.2020.02.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildner K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S., et al. 2008. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 322:1097–1100. 10.1126/science.1164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani S.R., Wang L., Multani A.S., Combs C., Deramaudt T.B., Hruban R.H., Rustgi A.K., Chang S., and Tuveson D.A.. 2005. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell. 7:469–483. 10.1016/j.ccr.2005.04.023 [DOI] [PubMed] [Google Scholar]
- Hiraoka N., Yamazaki-Itoh R., Ino Y., Mizuguchi Y., Yamada T., Hirohashi S., and Kanai Y.. 2011. CXCL17 and ICAM2 are associated with a potential anti-tumor immune response in early intraepithelial stages of human pancreatic carcinogenesis. Gastroenterology. 140:310–321. 10.1053/j.gastro.2010.10.009 [DOI] [PubMed] [Google Scholar]
- Hoffmann T.K., Müller-Berghaus J., Ferris R.L., Johnson J.T., Storkus W.J., and Whiteside T.L.. 2002. Alterations in the frequency of dendritic cell subsets in the peripheral circulation of patients with squamous cell carcinomas of the head and neck. Clin. Cancer Res. 8:1787–1793. [PubMed] [Google Scholar]
- Jackson E.L., Willis N., Mercer K., Bronson R.T., Crowley D., Montoya R., Jacks T., and Tuveson D.A.. 2001. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes Dev. 15:3243–3248. 10.1101/gad.943001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson E.L., Olive K.P., Tuveson D.A., Bronson R., Crowley D., Brown M., and Jacks T.. 2005. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer Res. 65:10280–10288. 10.1158/0008-5472.CAN-05-2193 [DOI] [PubMed] [Google Scholar]
- Jongbloed S.L., Kassianos A.J., McDonald K.J., Clark G.J., Ju X., Angel C.E., Chen C.J.J., Dunbar P.R., Wadley R.B., Jeet V., et al. 2010. Human CD141+ (BDCA-3)+ dendritic cells (DCs) represent a unique myeloid DC subset that cross-presents necrotic cell antigens. J. Exp. Med. 207:1247–1260. 10.1084/jem.20092140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karsunky H., Merad M., Cozzio A., Weissman I.L., and Manz M.G.. 2003. Flt3 ligand regulates dendritic cell development from Flt3+ lymphoid and myeloid-committed progenitors to Flt3+ dendritic cells in vivo. J. Exp. Med. 198:305–313. 10.1084/jem.20030323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.W., Stone M.L., Porrett P.M., Thomas S.K., Komar C.A., Li J.H., Delman D., Graham K., Gladney W.L., Hua X., et al. 2019. Hepatocytes direct the formation of a pro-metastatic niche in the liver. Nature. 567:249–252. 10.1038/s41586-019-1004-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesina M., Kurkowski M.U., Ludes K., Rose-John S., Treiber M., Klöppel G., Yoshimura A., Reindl W., Sipos B., Akira S., et al. 2011. Stat3/Socs3 activation by IL-6 transsignaling promotes progression of pancreatic intraepithelial neoplasia and development of pancreatic cancer. Cancer Cell. 19:456–469. 10.1016/j.ccr.2011.03.009 [DOI] [PubMed] [Google Scholar]
- Li J., Byrne K.T., Yan F., Yamazoe T., Chen Z., Baslan T., Richman L.P., Lin J.H., Sun Y.H., Rech A.J., et al. 2018. Tumor cell-intrinsic factors underlie heterogeneity of immune cell infiltration and response to immunotherapy. Immunity. 49:178–193.e7. 10.1016/j.immuni.2018.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., and Shi W.. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Long K.B., Tooker G., Tooker E., Luque S.L., Lee J.W., Pan X., and Beatty G.L.. 2017. IL6 receptor blockade enhances chemotherapy efficacy in pancreatic ductal adenocarcinoma. Mol. Cancer Ther. 16:1898–1908. 10.1158/1535-7163.MCT-16-0899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyman S.D., and Jacobsen S.E.W.. 1998. c-kit ligand and Flt3 ligand: stem/progenitor cell factors with overlapping yet distinct activities. Blood. 91:1101–1134. 10.1182/blood.V91.4.1101 [DOI] [PubMed] [Google Scholar]
- Mace T.A., Shakya R., Pitarresi J.R., Swanson B., McQuinn C.W., Loftus S., Nordquist E., Cruz-Monserrate Z., Yu L., Young G., et al. 2018. IL-6 and PD-L1 antibody blockade combination therapy reduces tumour progression in murine models of pancreatic cancer. Gut. 67:320–332. 10.1136/gutjnl-2016-311585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martignoni M.E., Kunze P., Hildebrandt W., Künzli B., Berberat P., Giese T., Klöters O., Hammer J., Büchler M.W., Giese N.A., et al. 2005. Role of mononuclear cells and inflammatory cytokines in pancreatic cancer-related cachexia. Clin. Cancer Res. 11:5802–5808. 10.1158/1078-0432.CCR-05-0185 [DOI] [PubMed] [Google Scholar]
- McAllister F., Bailey J.M., Alsina J., Nirschl C.J., Sharma R., Fan H., Rattigan Y., Roeser J.C., Lankapalli R.H., Zhang H., et al. 2014. Oncogenic Kras activates a hematopoietic-to-epithelial IL-17 signaling axis in preinvasive pancreatic neoplasia. Cancer Cell. 25:621–637. 10.1016/j.ccr.2014.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merad M., Sathe P., Helft J., Miller J., and Mortha A.. 2013. The dendritic cell lineage: ontogeny and function of dendritic cells and their subsets in the steady state and the inflamed setting. Annu. Rev. Immunol. 31:563–604. 10.1146/annurev-immunol-020711-074950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M.A., Baer J.M., Knolhoff B.L., Nywening T.M., Panni R.Z., Su X., Weilbaecher K.N., Hawkins W.G., Ma C., Fields R.C., et al. 2018. Breast and pancreatic cancer interrupt IRF8-dependent dendritic cell development to overcome immune surveillance. Nat. Commun. 9:1250 10.1038/s41467-018-03600-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. 2003. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34:267–273. 10.1038/ng1180 [DOI] [PubMed] [Google Scholar]
- Morrison A.H., Diamond M.S., Hay C.A., Byrne K.T., and Vonderheide R.H.. 2020. Sufficiency of CD40 activation and immune checkpoint blockade for T cell priming and tumor immunity. Proc. Natl. Acad. Sci. USA. 117:8022–8031. 10.1073/pnas.1918971117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn D.H., and Mellor A.L.. 2016. IDO in the tumor microenvironment: inflammation, counter-regulation, and tolerance. Trends Immunol. 37:193–207. 10.1016/j.it.2016.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hara M.H., O’Reilly E.M., Rosemarie M., Varadhachary G., Wainberg Z.A., Ko A., Fisher G.A. Jr., Rahma O., Lyman J.P., Cabanski C.R., et al. 2019. A Phase Ib study of CD40 agonistic monoclonal antibody APX005M together with gemcitabine and nab-paclitaxel with or without nivolumab in untreated metastatic ductal pancreatic adenocarcinoma patients. In AACR Annual Meeting 2019. Atlanta, GA. [Google Scholar]
- Öhlund D., Handly-Santana A., Biffi G., Elyada E., Almeida A.S., Ponz-Sarvise M., Corbo V., Oni T.E., Hearn S.A., Lee E.J., et al. 2017. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 214:579–596. 10.1084/jem.20162024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.-J., Nakagawa T., Kitamura H., Atsumi T., Kamon H., Sawa S., Kamimura D., Ueda N., Iwakura Y., Ishihara K., et al. 2004. IL-6 regulates in vivo dendritic cell differentiation through STAT3 activation. J. Immunol. 173:3844–3854. 10.4049/jimmunol.173.6.3844 [DOI] [PubMed] [Google Scholar]
- Raphael B.J., Hruban R.H., Aguirre A.J., Moffitt R.A., Yeh J.J., Stewart C., Robertson A.G., Cherniack A.D., Gupta M., Getz G., et al. ; Cancer Genome Atlas Research Network . 2017. Integrated genomic characterization of pancreatic ductal adenocarcinoma. Cancer Cell. 32:185–203.e13. 10.1016/j.ccell.2017.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts E.W., Broz M.L., Binnewies M., Headley M.B., Nelson A.E., Wolf D.M., Kaisho T., Bogunovic D., Bhardwaj N., and Krummel M.F.. 2016. Critical role for CD103+/CD141+ dendritic cells bearing CCR7 for tumor antigen trafficking and priming of T cell immunity in melanoma. Cancer Cell. 30:324–336. 10.1016/j.ccell.2016.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rooney M.S., Shukla S.A., Wu C.J., Getz G., and Hacohen N.. 2015. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 160:48–61. 10.1016/j.cell.2014.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salmon H., Idoyaga J., Rahman A., Leboeuf M., Remark R., Jordan S., Casanova-Acebes M., Khudoynazarova M., Agudo J., Tung N., et al. 2016. Expansion and activation of CD103+ dendritic cell progenitors at the tumor site enhances tumor responses to therapeutic PD-L1 and BRAF inhibition. Immunity. 44:924–938. 10.1016/j.immuni.2016.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Paulete A.R., Cueto F.J., Martínez-López M., Labiano S., Morales-Kastresana A., Rodríguez-Ruiz M.E., Jure-Kunkel M., Azpilikueta A., Aznar M.A., Quetglas J.I., et al. 2016. Cancer immunotherapy with immunomodulatory anti-CD137 and anti-PD-1 monoclonal antibodies requires BATF3-dependent dendritic cells. Cancer Discov. 6:71–79. 10.1158/2159-8290.CD-15-0510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlitzer A., Sivakamasundari V., Chen J., Sumatoh H.R., Schreuder J., Lum J., Malleret B., Zhang S., Larbi A., Zolezzi F., et al. 2015. Identification of cDC1- and cDC2-committed DC progenitors reveals early lineage priming at the common DC progenitor stage in the bone marrow. Nat. Immunol. 16:718–728. 10.1038/ni.3200 [DOI] [PubMed] [Google Scholar]
- Schoenberger S.P., Toes R.E.M., van der Voort E.I.H., Offringa R., and Melief C.J.M.. 1998. T-cell help for cytotoxic T lymphocytes is mediated by CD40-CD40L interactions. Nature. 393:480–483. 10.1038/31002 [DOI] [PubMed] [Google Scholar]
- Schreiber R.D., Old L.J., and Smyth M.J.. 2011. Cancer immunoediting: integrating immunity’s roles in cancer suppression and promotion. Science. 331:1565–1570. 10.1126/science.1203486 [DOI] [PubMed] [Google Scholar]
- Spranger S., Bao R., and Gajewski T.F.. 2015. Melanoma-intrinsic β-catenin signalling prevents anti-tumour immunity. Nature. 523:231–235. 10.1038/nature14404 [DOI] [PubMed] [Google Scholar]
- Spranger S., Dai D., Horton B., and Gajewski T.F.. 2017. Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell. 31:711–723.e4. 10.1016/j.ccell.2017.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang M., Diao J., Gu H., Khatri I., Zhao J., and Cattral M.S.. 2015. Toll-like receptor 2 activation promotes tumor dendritic cell dysfunction by regulating IL-6 and IL- 10 receptor signaling. Cell Rep. 13:2851–2864. 10.1016/j.celrep.2015.11.053 [DOI] [PubMed] [Google Scholar]
- Tjomsland V., Spångeus A., Sandström P., Borch K., Messmer D., and Larsson M.. 2010. Semi mature blood dendritic cells exist in patients with ductal pancreatic adenocarcinoma owing to inflammatory factors released from the tumor. PLoS One. 5 e13441 10.1371/journal.pone.0013441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twyman-Saint Victor C., Rech A.J., Maity A., Rengan R., Pauken K.E., Stelekati E., Benci J.L., Xu B., Dada H., Odorizzi P.M., et al. 2015. Radiation and dual checkpoint blockade activate non-redundant immune mechanisms in cancer. Nature. 520:373–377. 10.1038/nature14292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kooten C., and Banchereau J.. 2000. CD40-CD40 ligand. J. Leukoc. Biol. 67:2–17. 10.1002/jlb.67.1.2 [DOI] [PubMed] [Google Scholar]
- Vonderheide R.H. 2018. The immune revolution: A case for priming, not checkpoint. Cancer Cell. 33:563–569. 10.1016/j.ccell.2018.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wculek S.K., Cueto F.J., Mujal A.M., Melero I., Krummel M.F., and Sancho D.. 2020. Dendritic cells in cancer immunology and immunotherapy. Nat. Rev. Immunol. 20:7–24. 10.1038/s41577-019-0210-z [DOI] [PubMed] [Google Scholar]
- Winograd R., Byrne K.T., Evans R.A., Odorizzi P.M., Meyer A.R.L., Bajor D.L., Clendenin C., Stanger B.Z., Furth E.E., Wherry E.J., et al. 2015. Induction of T cell immunity overcomes complete resistance to PD-1 and CTLA-4 blockade and improves survival in pancreatic carcinoma. Cancer Immunol. Res. 3:399–411. 10.1158/2326-6066.CIR-14-0215 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists reagents and antibodies used in murine studies.
lists reagents and antibodies used in human studies.