Here Bhattacharyya et al. show that MYC is a molecular point of convergence for microenvironmental and cell-autonomous signals in RAS-transformed pancreatic cancer cells. They identify cancer-associated fibroblast–derived FGF1 as a critical paracrine regulator of MYC, and a novel link between the pancreatic tumor microenvironment and oncogenic transcription.
Abstract
Despite a critical role for MYC as an effector of oncogenic RAS, strategies to target MYC activity in RAS-driven cancers are lacking. In genetically engineered mouse models of lung and pancreatic cancer, oncogenic KRAS is insufficient to drive tumorigenesis, while addition of modest MYC overexpression drives robust tumor formation, suggesting that mechanisms beyond the RAS pathway play key roles in MYC regulation and RAS-driven tumorigenesis. Here we show that acidic fibroblast growth factor (FGF1) derived from cancer-associated fibroblasts (CAFs) cooperates with cancer cell–autonomous signals to increase MYC level, promoter occupancy, and activity. FGF1 is necessary and sufficient for paracrine regulation of MYC protein stability, signaling through AKT and GSK-3β to increase MYC half-life. Patient specimens reveal a strong correlation between stromal CAF content and MYC protein level in the neoplastic compartment, and identify CAFs as the specific source of FGF1 in the tumor microenvironment. Together, our findings demonstrate that MYC is coordinately regulated by cell-autonomous and microenvironmental signals, and establish CAF-derived FGF1 as a novel paracrine regulator of oncogenic transcription.
Introduction
The KRAS oncogene is mutated in >90% of pancreatic ductal adenocarcinoma (PDAC; Waters and Der, 2018), and oncogenic KRAS is critical for PDAC initiation and maintenance (Collins et al., 2012; Ying et al., 2012), making KRAS and its key effectors appealing targets for therapy. The oncogenic transcription factor MYC is well established as a critical effector of oncogenic RAS in multiple tumor types (Saborowski et al., 2014; Soucek et al., 2008, 2013; Walz et al., 2014). In genetically engineered mouse models of lung and pancreatic cancer (Hingorani et al., 2003; Tuveson et al., 2004), oncogenic KRAS is insufficient to drive tumorigenesis, while addition of modest MYC overexpression from the Rosa26 locus drives robust tumor formation (Farrell et al., 2017; Kortlever et al., 2017; Sodir et al., 2020), suggesting that mechanisms beyond the RAS pathway play key roles in MYC regulation and RAS-driven tumorigenesis. We have previously found that stromal cues from PDAC cancer-associated fibroblasts (CAFs) induce a transcriptional program in PDAC cells that significantly overlaps with the transcriptional network regulated by oncogenic KRAS (Sherman et al., 2017; Ying et al., 2012). This overlap suggests a gene-regulatory point of convergence for cell-autonomous and microenvironmental signals. The KRAS-regulated network was previously attributed to MYC-dependent transcription (Ying et al., 2012), but a role for a fibroinflammatory tumor microenvironment in paracrine regulation of MYC has not been established. MYC protein is very short-lived, and its expression and activity are exclusively dependent on mitogenic signals (Farrell and Sears, 2014; Soucek and Evan, 2010). While KRAS mutant PDAC cells exhibit MYC protein stabilization downstream of ERK1/2 (Hayes et al., 2016) or ERK5 (Vaseva et al., 2018), we reasoned that oncogenic levels of MYC in vivo may result from additional signals from the tumor microenvironment, and specifically from stromal CAFs.
Results and discussion
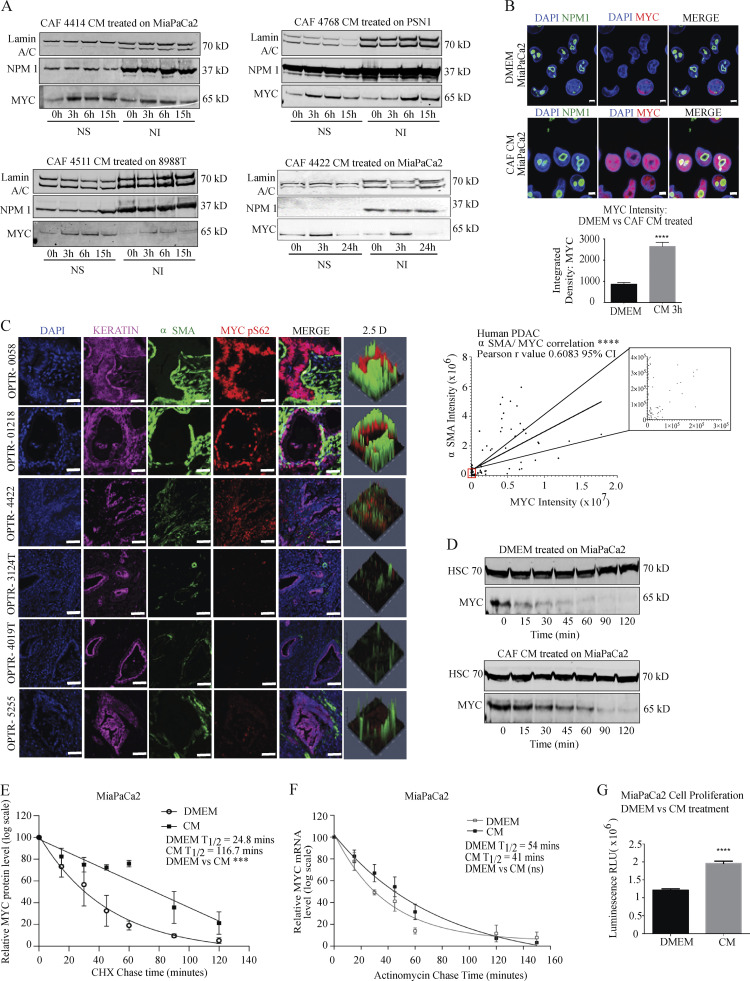
To address a role for CAFs in paracrine regulation of MYC, we applied conditioned media (CM) from primary human PDAC CAFs (validation in Fig. S1, A and B) to PDAC cells, and assessed MYC level across all CAF/PDAC cell combinations tested. Both Western blot and immunofluorescence (IF) microscopy demonstrated that the CAF secretome acted in a paracrine manner to increase MYC protein level (Fig. 1, A and B; and Fig. S1, C–F), peaking by 3 h. Importantly, a noncancer-associated human pancreatic stellate cell (hPSC) line did not induce MYC under the same experimental conditions (Fig. S1 D), suggesting specificity for CAFs and arguing against a nonspecific effect of CM. These increases were more pronounced in the soluble than the insoluble nuclear fraction (at 400 mM NaCl); as MYC is found in both fractions (Myant et al., 2015), we examined total nuclear extracts moving forward. Before performing mechanistic studies, we assessed the relationship between stromal CAF content and MYC level in human PDAC. Immunohistochemical analysis revealed a strong correlation between MYC protein level in keratin (KRT)-positive PDAC cells and α-smooth muscle actin (αSMA)–positive CAF density among human PDAC samples (Fig. 1 C), supporting the notion that CAFs may signal in a paracrine manner to augment MYC expression in the neoplastic compartment. As αSMA was used to mark CAFs, we report this relationship for the previously described myofibroblastic CAF (myCAF) subtype (Öhlund et al., 2017). Importantly, this was not a reflection of increased density of cancer cells among stroma-rich PDAC regions, as we saw no correlation between KRT and αSMA in these tissues (Fig. S1 G). We stained for MYC pS62 as a readout for stable MYC protein in these analyses as total MYC antibodies did not yield consistent, specific staining across our human PDAC tissues (see Materials and methods). To begin to understand the mechanism by which CAFs increase MYC protein levels in PDAC cells, we tested MYC RNA and protein stability under control and CAF CM-treated conditions. CAF CM significantly increased MYC protein stability (Fig. 1, D and E), while RNA stability was not significantly changed (Fig. 1 F), though we noted that MYC RNA at steady-state was increased with CM (Fig. S1 M). CAF CM also increased PDAC cell proliferation (Fig. 1 G), as expected for conditions that augment MYC level and activity and consistent with previous results (Sousa et al., 2016). These results suggest that PDAC CAFs signal to stabilize MYC in the neoplastic compartment in a paracrine manner, an interaction that is reflected in human PDAC samples.
Figure S1.
PDAC CAF-derived FGF1 increases MYC level in PDAC cells. (A) Immunofluorescent staining of the indicated human CAF lines, hPSC, and MiaPaCa2 for common CAF markers and KRT. Scale bar, 20 µm. (B) KRAS genotyping on the indicated CAF lines. (C) Quantification of MYC expression from Western blots in Fig. 1 A, normalized to NPM1 in MiaPaCa2 cells treated with CAF 4414 CM for the indicated duration. *, P < 0.05, **, P < 0.01 by two-way ANOVA (n = 3 independent experiments). (D) Western blots showing MYC levels in PDAC cells (Panc1, top; PSN1, bottom) after treatment with CM from hPSCs for the indicated duration. Lamin A/C and nucleophosmin (NPM1) are loading controls. NS, nuclear soluble fraction; NI, nuclear insoluble fraction (at 400 mM NaCl). Quantification appears below. (E) Western blots showing MYC levels in PDAC cells after treatment with CAF CM for the indicated duration. (F) IF microscopy showing MYC levels in 8988T PDAC cells treated with DMEM or CAF CM for 3 h. NPM1 shows localization of nucleoli; DAPI stains nuclei. Scale bar, 5 µm. (G) Quantification of integrated density of KRT versus αSMA in 50 fields across five different human PDAC patient samples. (H) CAF 4414 CM was incubated alone or with the indicated neutralizing antibodies for 1 h at room temperature, then added to MiaPaCa2 cells for 3 h. MYC levels in nuclear extracts were analyzed by Western blot (representative of three independent experiments). (I) Western blot for MYC in 8988T PDAC cells after treatment with DMEM or CM from the indicated CAF 4414 line for 3 h (representative of n = 3 independent experiments). (J) Top: qPCR for FGF1 in CAF 4422 control and shFGF1 using two independent hairpins. Expression normalized to 36B4. Bottom: MiaPaCa2 cells were treated with CM from control or shFGF1 CAF 4422 for 3 h, and MYC and pAKT levels were measured by Western blot. (K) Western blot for MYC in MIAPaCa2 cells after treatment with DMEM or CM ± FGFR inhibitor Debio 1347 at the indicated concentrations for 3 h. (L) qRT-PCR analysis of MYC mRNA levels (relative to β-actin) in DMEM-treated control MiaPaCa2 cells, MiaPaCa2 cells treated with human recombinant FGF1, and MiaPaCa2 cells treated with a combination of recombinant FGF1 and PD17307. (M) qRT-PCR analysis of MYC mRNA levels (relative to β-actin) in DMEM-treated control MiaPaCa2 cells, MiaPaCa2 cells treated with CAF4414-derived CM, and MiaPaCa2 cells treated with a combination of CAF4414-derived CM and PD173074. **, P < 0.01; ****, P < 0.0001 by one-way ANOVA (n = 3 independent experiments). Ctrl, control.
Figure 1.
PDAC CAF-derived factors increase MYC level in PDAC cells. (A) Western blots showing MYC levels in PDAC cells after treatment with CM from primary CAFs for the indicated duration. Lamin A/C and NPM1 (nucleophosmin) are loading controls. NS, nuclear soluble fraction; NI, nuclear insoluble fraction (at 400 mM NaCl). (B) IF microscopy showing MYC levels in MiaPaCa2 PDAC cells treated with DMEM or CAF 4414 CM for 3 h. NPM1 shows localization of nucleoli; DAPI stains nuclei. Scale bar, 5 µm. Quantification of MYC intensity appears below. ****, P < 0.0001 by Student’s t test (n = 3 biological replicates). 78 fields of view were quantified per treatment condition. (C) Left: Fluorescent immunostaining of human PDAC patient tissue samples. DAPI stains nuclei, pan- KRT stains PDAC cells, αSMA stains CAFs. Scale bar, 20 µm. Right: Quantification of integrated density of MYC in KRT+ cells versus αSMA in 100 fields of view from n = 6 human PDAC specimens; ****, P < 0.0001 for Pearson’s correlation. (D) MiaPaCa2 cells were treated with DMEM or CAF 4414 CM for 3 h, then treated with cycloheximide and harvested at the indicated time points to measure MYC protein level by Western blot. HSC70 is a loading control. (E) Quantification of MYC levels in D (n = 3 independent experiments). P value reflects comparison of t1/2 from three independent experiments; ***, P < 0.001 by Student’s t test. (F) MiaPaCa2 cells were treated as in D, then treated with actinomycin D and harvested at the indicated time points for quantitative RT-PCR (qRT-PCR) analysis of MYC mRNA levels (n = 3 independent experiments). Data were normalized to β-actin as a housekeeping gene. For E and F, data were plotted on a semi-log scale, and best-fit lines were calculated using linear regression. (G) Proliferation assay on MiaPaCa2 cells treated with DMEM or CAF 4414 CM for 72 h. ****, P < 0.0001 by Student’s t test (n = 3 independent experiments). Error bars represent SEM from three independent experiments. ns, not significant; RLU, relative luciferase units.
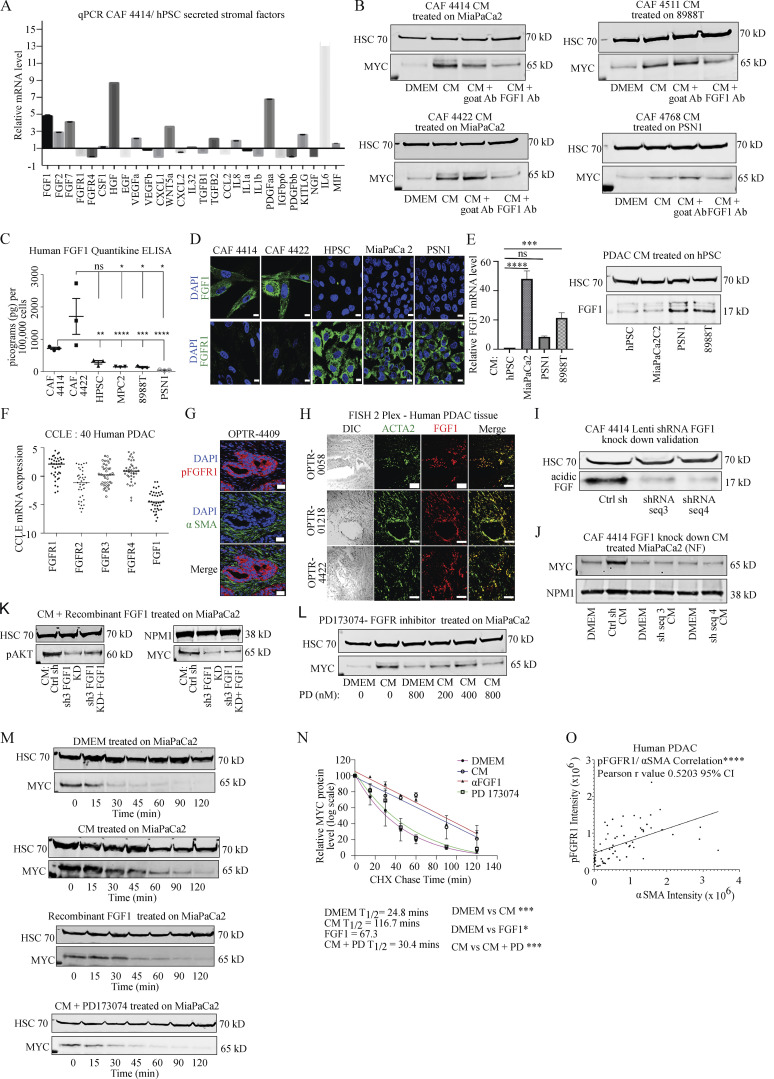
We next aimed to identify the specific CAF-derived factor that stabilizes MYC in PDAC cells. As our hPSC line did not induce MYC via secreted factors, we used this line as a basis for comparison. We extracted a list of secreted growth factors, cytokines, and chemokines expressed in PDAC CAFs from a published RNA sequencing (RNA-seq) dataset (Sherman et al., 2014), and compared expression of each factor in hPSC versus CAFs (Fig. 2 A). This comparison identified six factors with at least threefold higher expression in CAFs than hPSC (FGF2 was included at 2.9-fold). We then tested neutralizing antibodies against each of these six candidate factors in CAF CM to determine whether inhibition of an individual factor could suppress paracrine induction of MYC. Though Sonic Hedgehog (SHH) was not detected in the RNA-seq dataset, it is variably expressed in primary CAFs including the CAF sample used for screening, so we included an antibody against SHH as well as NOTCH1 and NOTCH3 based on prior work linking them to MYC (Albihn et al., 2010). Our top candidate from this neutralizing antibody screen was acidic fibroblast growth factor (FGF1; Fig. 2 B and Fig. S1 H) across all CAF/PDAC cell combinations tested, though we do not exclude potential roles for the other secreted factors in paracrine regulation of MYC, and FGF2 was previously linked to MYC regulation (Lepique et al., 2004). While FGF1 has been linked to cell cycle progression and MYC induction previously (LaVallee et al., 1998; Müller et al., 1984), FGF1 has not previously been linked to MYC regulation in cancer, and has not been studied functionally in the pancreatic tumor microenvironment. We thus confirmed that primary PDAC CAFs secrete FGF1. ELISA assays revealed that PDAC CAFs secrete abundant FGF1, while expression was restricted to basal levels in CM from hPSC and from human PDAC cells (Fig. 2 C), consistent with a role in paracrine signaling. We also examined FGF1 expression by IF microscopy, which similarly showed detectable FGF1 in PDAC CAFs but not in hPSC or PDAC cells (Fig. 2 D). Consistent with a link between exposure to the tumor microenvironment and stromal production of FGF1, PDAC cell CM induced FGF1 expression in hPSC at the RNA and protein levels (Fig. 2 E). FGF1 signals through the four-member family of fibroblast growth factor receptors (FGFRs); to assess FGF1/FGFR signaling more broadly, we analyzed expression of FGF1 and the four FGFRs across 40 PDAC cell lines in the Cancer Cell Line Encyclopedia database and found that PDAC cells consistently express the receptors, with FGFR1 expressing the most highly and consistently, while expression of FGF1 in these lines was low to absent (Fig. 2 F). To confirm FGFR expression in PDAC cells, we used FGFR1 as a readout in light of the results in Fig. 2 F, and because this family member was recently implicated as a target for combination therapy in PDAC (Manchado et al., 2016). We found that this receptor was expressed in PDAC cells; interestingly, this receptor was also highly expressed in hPSC but was found at a very low level in CAFs, showing a reciprocal staining pattern to that of FGF1 (Fig. 2 D). Consistent with results in cultured cells, human PDAC tissues showed robust staining for pFGFR1, confirming its expression and activation in human tumor samples, while CAFs were mostly negative (Fig. 2 G). We next assessed both the expression and the cellular source of FGF1 in human PDAC. As FGF1 is a secreted factor, instead of immunohistochemistry we probed FGF1 expression using RNA fluorescence in situ hybridization, and costained for ACTA2 (encoding αSMA). FGF1 signal was restricted to ACTA2-positive cells, though not all ACTA2-positive cells express FGF1 (Fig. 2 H), suggesting that the αSMA-positive myCAF subset produces FGF1 in PDAC (Öhlund et al., 2017). These results suggest that CAFs are the principal cellular source of FGF1 in the PDAC microenvironment, and that the FGF1–FGFR axis is expressed in human PDAC.
Figure 2.
Stroma-derived FGF1 is necessary and sufficient for paracrine regulation of MYC. (A) qRT-PCR for the indicated secreted factors in CAF 4414 and hPSC (n = 3 independent experiments). Data are plotted as fold change in CAF 4414 compared with hPSC (set to 1); all values were normalized to 36B4 as a housekeeping gene. (B) CAF CM was incubated alone or with FGF1 neutralizing antibody or goat IgG control antibody for 1 h at room temperature, then added to PDAC cells for 3 h. MYC levels in nuclear extracts were analyzed by Western blot (representative of three independent experiments). (C) ELISA for FGF1 in CM from the indicated cell lines. Data are presented as mean ± SEM; *, P < 0.05; **, P < 0.0025; ***, P < 0.001; ****, P < 0.0001 by Student’s t test (n = 3 independent experiments) compared with CAF 4422 CM (upper line and asterisks) or CAF 4414 CM (lower line and asterisks). (D) IF staining for FGF1 or FGFR1 in the indicated cell lines. Scale bar, 10 µm. (E) hPSCs were treated with CM from the indicated PDAC cell lines for 24 h, and FGF1 mRNA was measured by qPCR (left; data were normalized to 36B4 and are presented as mean ± SEM; ***, P < 0.001; ****, P < 0.0001 by Student’s t test) and protein measured by Western blot (right). Results represent three independent experiments. (F) mRNA expression levels of the four FGFRs and FGF1 in 40 human PDAC cells from the Cancer Cell Line Encyclopedia database. (G) Fluorescent immunostaining for pFGFR1 in human PDAC samples (representative of n = 3 patient samples). Scale bar, 20 µm. (H) RNA-FISH for ACTA2 (encoding αSMA) and FGF1 in human PDAC (representative of n = 6 patient samples). Scale bar, 50 µm. Differential interference contrast (left column) shows tissue context. (I) Western blots showing FGF1 knockdown in CAF 4414 knockdown lines (scramble control versus two distinct shFGF1 sequences). (J) Western blot for MYC in MiaPaCa2 cells after treatment with DMEM or CM from the indicated CAF 4414 line for 3 h (representative of n = 3 independent experiments). (K) Western blot for pAKT in MiaPaCa2 cells treated with CAF 4414 control CM, shFGF1 CM, or shFGF1 CM + 50 pg/ml human FGF1 for 3 h. Results represent three independent experiments. (L) Western blot for MYC in MiaPaCa2 cells after treatment with DMEM or CM ± FGFR inhibitor PD173074 at the indicated concentrations for 3 h (representative of n = 3 independent experiments). (M) MiaPaCa2 cells were treated with DMEM or 50 pg/ml FGF1 or CAF 4414 CM for 3 h, then treated with cycloheximide ± 800 nM PD173074 for the indicated duration, and MYC levels were analyzed by Western blot. (N) Quantification of MYC levels in M (n = 3 independent experiments). P value reflects comparison of t1/2 from three independent experiments; *, P < 0.05; ***, P < 0.001 by Student’s t test. (O) Quantification of fluorescent immunostaining of human PDAC patient tissue samples indicating integrated density of pFGFR1 in KRT+ cells and αSMA in 63 fields of view from n = 5 human PDAC specimens; ****, P < 0.0001 for Pearson’s correlation. Ab, antibody; ns, not significant; PD, PD173073.
We next performed functional studies to assess the significance of FGF1 in paracrine regulation of MYC. To this end, we stably knocked down FGF1 in primary PDAC CAFs using two independent hairpin sequences (Fig. 2 I). CM from these knockdown lines resulted in markedly reduced MYC induction in PDAC cells compared with controls (Fig. 2 J and Fig. S1 I). Similar results were seen with FGF1 knockdown in an independent CAF line (Fig. S1 J). Recombinant FGF1 partially restored MYC stabilization in PDAC cells treated with shFGF1 CM (Fig. 2 K), suggesting specificity for FGF1 signaling. To further examine this functional connection with pharmacologic inhibitors, and to probe a role for FGFRs in MYC regulation, we incubated PDAC cells in CAF CM in the presence or absence of FGFR inhibitor PD173074 or Debio-1347. Both inhibitors blocked induction of MYC by CAF-derived signals (Fig. 2 L and Fig. S1 K). FGFR inhibitor prevented the stroma-inducible increase in MYC half-life, such that CM containing FGFR inhibitor yielded MYC half-life measurements similar to untreated controls (Fig. 2, M and N). While these results together suggested that FGF1 is necessary to stabilize MYC, we next tested whether this factor is sufficient for MYC stabilization. Using the lowest concentration of FGF1 found in CAF CM in our ELISA assays (Fig. 2 C), we found that recombinant human FGF1 was sufficient to increase MYC protein in PDAC cells and significantly increased MYC stability (Fig. 2, M and N), though we cannot exclude that additional secreted factors contribute to paracrine MYC regulation. FGF1 alone did not induce MYC RNA, and FGFR inhibition did not suppress the induction of MYC mRNA by CAF CM (Fig. S1, L and M), suggesting that MYC protein stabilization is the relevant mechanism of MYC induction by stroma-derived FGF1. Importantly, like MYC, we found that pFGFR1 correlated with αSMA staining in human PDAC (Fig. 2 O), supporting a relationship between stromal CAF (myCAF) density and activation of FGF1–FGFR signaling. Together, these results suggest that CAF-derived FGF1 is necessary and sufficient for paracrine stabilization of MYC protein in PDAC cells.
We next queried the pathway downstream of FGF1–FGFR signaling that results in stabilization of MYC. In RAS mutant cancer cells, MYC is classically stabilized by phosphorylation on S62 downstream of the MAPK pathway; further phosphorylation on T58 targets MYC for S62 dephosphorylation and subsequent proteasomal degradation (Sears et al., 1999). AKT activation can result in an inhibitory phosphorylation event on GSK-3β, preventing phosphorylation of MYC T58 by GSK-3β and further promoting MYC protein stability (Sears et al., 2000). We found that CAF CM induced phosphorylation of FGFR1, as well as downstream AKT and GSK-3β, across all CAF/PDAC cell combinations tested (Fig. 3 A and Fig. S2, A–D). ERK phosphorylation was high but unchanged by CM at this time point, consistent with previous studies (Hwang et al., 2008), suggesting that MYC stabilization by the stroma is distinct from cell-autonomous RAS-MAPK signaling. Total MYC and pS62 MYC were increased to a similar extent by CAF CM, while pT58 MYC was induced to a lesser extent (Fig. 3 A and Fig. S2, B–D), raising the possibility that CAF CM suppresses MYC T58 phosphorylation. The relevant concentration of recombinant FGF1 was sufficient to induce phosphorylation of AKT and GSK-3β at time points consistent with MYC stabilization (Fig. 3 B and Fig. S2 E), though we note that the effect of bulk CM on these signaling events was generally more pronounced. Addition of FGFR inhibitor to CM blunted paracrine induction of AKT and GSK-3β phosphorylation and increased pT58 MYC levels relative to total MYC, while ERK phosphorylation was increased (Fig. 3 C). To determine whether FGF1 is necessary for CAFs to activate this axis in PDAC cells, we looked at signaling after incubating PDAC cells with CM from our FGF1 knockdown CAFs. Surprisingly, paracrine induction of AKT and GSK-3β phosphorylation were suppressed in the absence of FGF1 (Fig. 3 D, Fig. S1 J, and Fig. S2 F). Though CAFs secrete numerous growth factors that can presumably activate AKT, these results suggest that stromal FGF1 creates a permissive context needed for robust paracrine regulation of AKT. To further test the role of AKT and GSK-3β in regulation of MYC by stromal signaling, we added an AKT inhibitor to CAF CM, which reduced GSK-3β phosphorylation and MYC induction (Fig. 3 E). To provide genetic evidence for the roles of these kinases, we transduced PDAC cells with dominant-negative AKT, or with constitutively active GSK-3β. Both interventions reduced paracrine induction of MYC (Fig. S2, G and H). To further assess the significance of MYC T58 phosphorylation in paracrine regulation of MYC stability, we transduced PDAC cells with Flag-tagged WT or T58A MYC, then treated these cells with CAF CM. While WT MYC levels were increased by CM, T58A MYC was not affected (Fig. 3 F), consistent with stromal suppression of T58 phosphorylation as a mechanism underlying the increase in MYC protein stability. Together, these results provide support for a central role for AKT and GSK-3β in FGF1-mediated stabilization of MYC.
Figure 3.
Paracrine FGF1 signaling augments MYC expression and stability via AKT/GSK3β signaling. (A) Western blots for the indicated signaling events in PDAC cells after 3 h treatment with DMEM or CAF CM. WCL, whole cell lysate; NF, nuclear fraction. (B) Western blots for the indicated signaling events in PDAC cells (MiaPaCa2, top; PSN1, bottom) after a time course of treatment with 50 pg/ml recombinant human FGF1. (C) Western blots for the indicated signaling events in MiaPaCa2 cells after 3 h treatment with CAF 4414 CM ± 800 nM PD173074. (D) Western blots for the indicated signaling events in MiaPaCa2 cells treated with CM from control or FGF1 knockdown CAFs for 3 h. (E) Western blots for the indicated signaling events in MiaPaCa2 cells treated with CAF 4414 CM ± AKT inhibitor MK2206 (10 µM) for 3 h. (F) Western blots for Flag and HSC70 (loading control) in MIAPaCa2 cells transduced with WT or T58A Flag-tagged MYC and treated with CAF 4414 CM for 3 h. All results represent at least three independent experiments. SF, serum-free.
Figure S2.
Paracrine FGF1 signaling augments MYC expression and stability via AKT/GSK-3β signaling. (A) Quantification of Western blots in Fig. 3 A (top), CAF 4414 CM on MiaPaCa2 (n = 3 independent experiments); ****, P < 0.0001 by Student’s t test. (B) Quantification of the Western blots in Fig. 3 A (bottom), CAF 4414 CM on MiaPaCa2 (n = 3 independent experiments); **, P < 0.01 by Student’s t test. (C) Quantification of the Western blots in Fig. 3 C (top) and Fig. 3 E (bottom), CAF 4414 CM on MiaPaCa2 (n = 3 independent experiments); *, P < 0.05; **, P < 0.01 by Student’s t test. (D) Western blots for the indicated signaling events in PDAC cells after 3 h treatment with DMEM or CAF CM. (E) Western blots for the indicated signaling events in 8988T PDAC cells after a time course of treatment with 50 pg/ml recombinant human FGF1. (F) Western blots for the indicated signaling events in 8988T PDAC cells treated with CM from control or FGF1 knockdown CAFs for 3 h. (G) Western blots for the indicated signaling events in MiaPaCa2 cells transfected with control plasmid or dominant-negative AKT and treated with DMEM or CAF 4414 CM for 3 h. (H) Western blots for the indicated signaling events in MiaPaCa2 cells transfected with control plasmid or constitutively active GSK-3β S9A and treated with DMEM or CAF 4414 CM for 3 h. Western blots for the indicated MYC species were on nuclear lysates from cells under the same treatment conditions as for whole cell lysate analysis. MK, MK2206; DN, dominant negative.
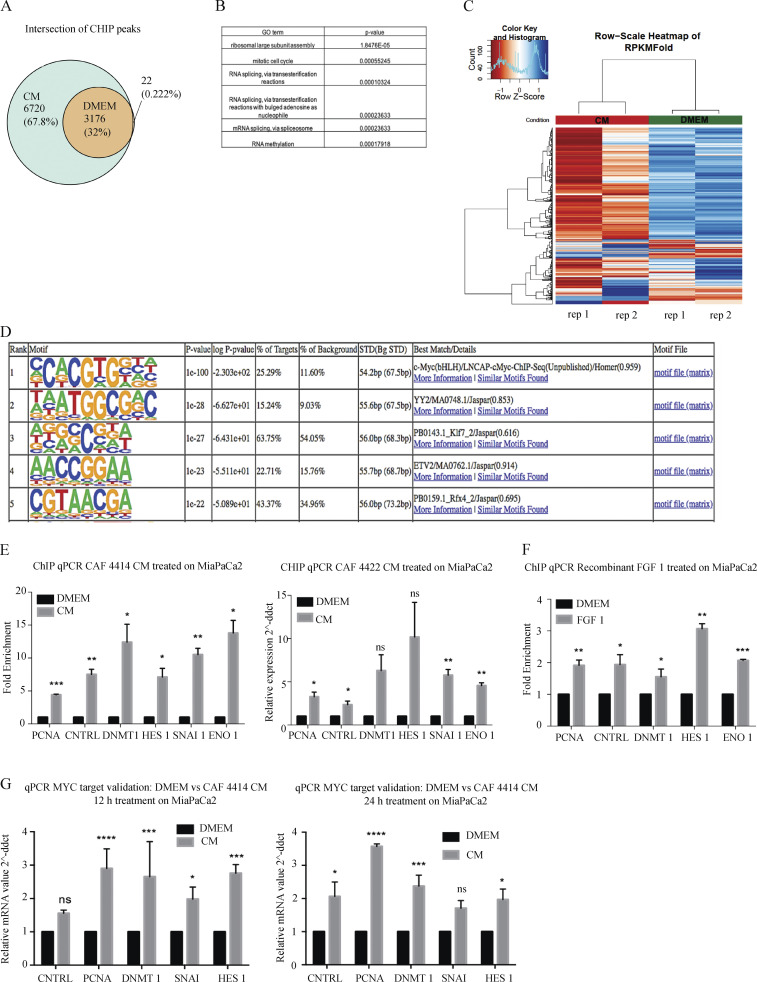
We next analyzed MYC genomic localization to better understand the consequences of increased MYC protein downstream of stromal FGF1. To this end, we performed chromatin immunoprecipitation sequencing (ChIP-seq) for MYC in PDAC cells incubated in control medium or CAF CM. As in previous studies featuring varying levels of MYC (Fernandez et al., 2003; Sabò et al., 2014), we find that nearly all MYC-bound sites under control conditions were also occupied under CM-stimulated conditions; CM resulted in 6,720 additional MYC binding sites (Fig. 4 A). Unique MYC-bound promoters upon CM treatment were associated with genes involved in post-transcriptional regulatory mechanisms including RNA processing and ribosome biogenesis (Fig. 4 B). Further, commonly bound peaks generally had a higher ChIP-seq signal under CM-treated conditions (Fig. 4 C). Motif analysis showed that a consensus MYC binding site was the top enriched motif (P = 10−100; Fig. 4 D), supporting the specificity of the immunoprecipitation for MYC. ChIP–quantitative PCR (qPCR) for differentially bound loci validated our ChIP-seq results with CM from two independent CAF lines (Fig. 4 E), and importantly, these increases in MYC promoter occupancy were recapitulated with FGF1 (Fig. 4 F). These differentially bound genes also revealed significant changes in gene expression upon CM treatment (Fig. 4 G). These results demonstrate that stromal cues increase MYC promoter occupancy in PDAC cells, consistent with increased protein levels.
Figure 4.
Stromal stimulation alters MYC chromatin occupancy and target gene expression. (A) Venn diagram showing overlap of significant MYC-bound peaks from ChIP-seq in MiaPaCa2 cells treated with DMEM or CAF 4414 CM for 3 h (n = 2 biological replicates). (B) Gene ontology (GO) terms enriched among differentially bound loci by ChIP-seq (significantly enriched among CM-restricted binding sites). (C) Heatmap showing hierarchical clustering of MYC binding sites identified under both DMEM and CM conditions by ChIP-seq as described in A. (D) Results of HOMER analysis of top motifs among bound regions in MYC ChIP-seq (assessing all bound peaks, including DMEM and CM treatments). (E) ChIP-qPCR for MYC binding sites in the indicated differentially bound promoters, as determined by ChIP-seq, in MiaPaCa2 cells treated with DMEM or CAF 4414 CM or CAF 4422 CM for 3 h (n = 2 independent experiments). (F) ChIP-qPCR for MYC binding sites in the indicated differentially bound promoters, as determined by ChIP-seq, in MiaPaCa2 cells treated with DMEM or 50 pg/ml FGF1 for 3 h (n = 2 independent experiments). (G) qRT-PCR for the indicated genes in MiaPaCa2 cells treated for 12 h or 24 h with CAF 4414 CM. Results were normalized to 36B4 as a housekeeping gene (n = 3 independent experiments). For E–G, *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by Student’s t test. ns, not significant; ddct, ΔΔcycle threshold; STD, standard.
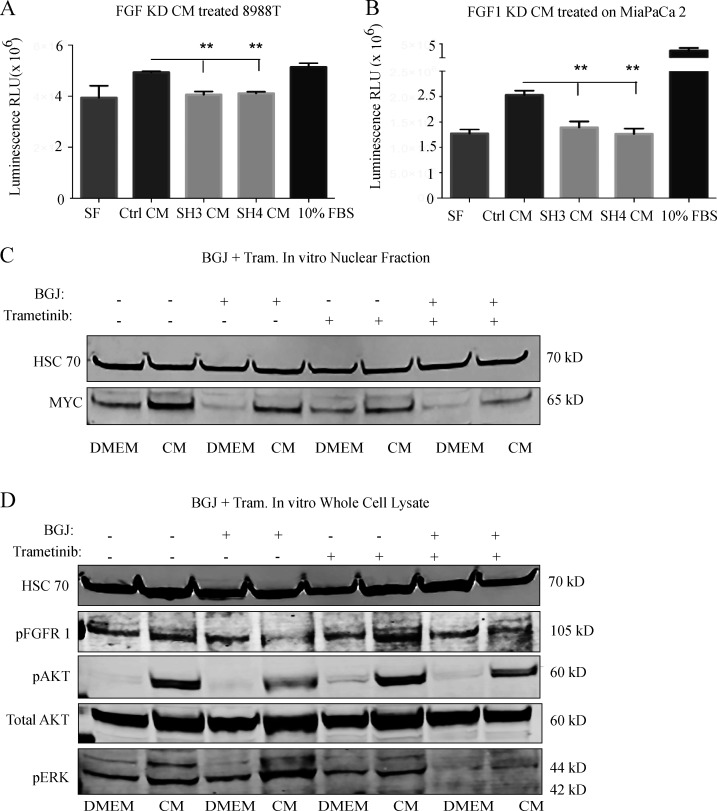
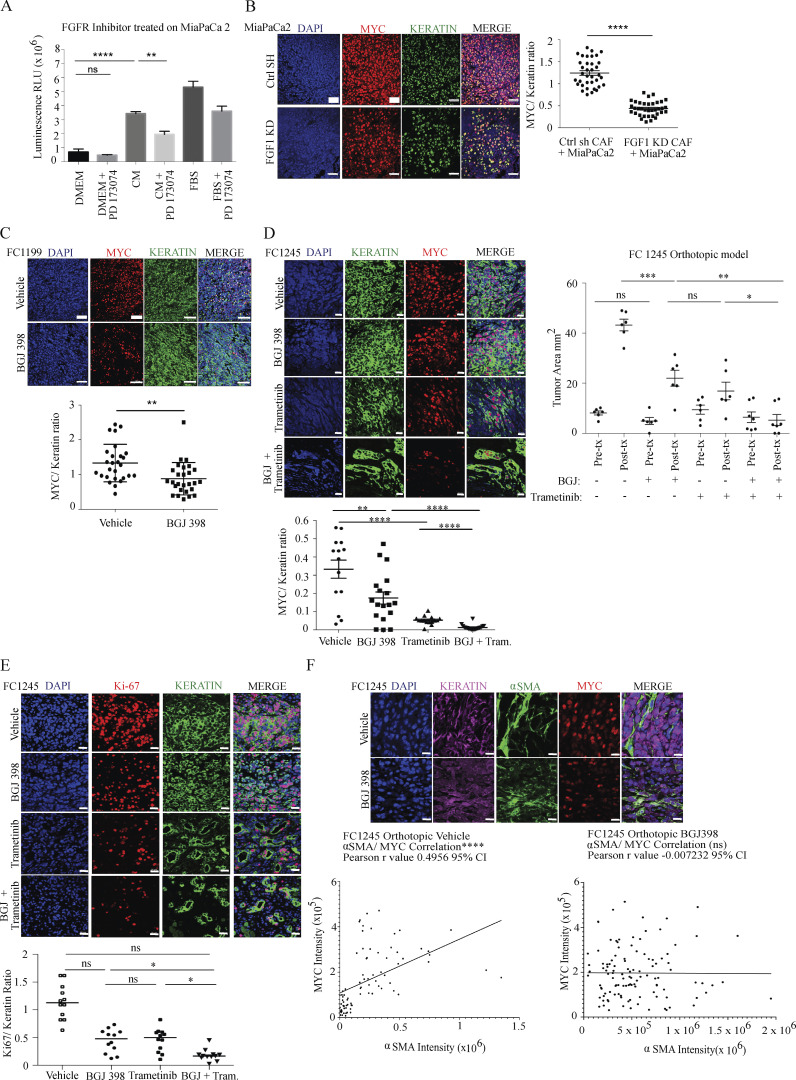
As MYC activity is principally associated with cellular growth control, we next assessed a role for the FGF1–FGFR axis in PDAC cell proliferation. While control CAF CM increased PDAC cell proliferation, this effect was significantly reduced with CM from FGF1 knockdown CAFs (Fig. S3, A and B). Similarly, FGFR inhibition significantly reduced stroma-inducible proliferation (Fig. 5 A). To address the link between FGF1–FGFR signaling and MYC levels in vivo, we suppressed this signaling axis in three distinct models. First, we performed subcutaneous transplantation of human PDAC cells together with control or FGF1 knockdown CAFs into the flanks of nude mice. Quantification of MYC intensity within KRT-positive cells showed that MYC levels in PDAC cells were significantly reduced when cotransplanted with FGF1-knockdown CAFs compared with controls (Fig. 5 B). Next, we performed orthotopic transplantation of the murine KrasLSL-G12D/+;Trp53LSL-R172H/+;Pdx1-Cre cell lines FC1199 and FC1245 into pancreata of immune-competent C57BL/6J hosts. Once tumors were detectable by ultrasound, mice were treated with vehicle or bioavailable FGFR inhibitor BGJ 398 for 9 d. FGFR inhibition resulted in a significant reduction in MYC protein level in PDAC cells in both models (Fig. 5 C). While FGFR inhibition significantly suppressed tumor growth compared with controls (Fig. 5 D), tumors did continue to grow. We hypothesized that inhibition of parallel MAPK signaling and FGF1–FGFR signaling would further reduce MYC levels by blocking two MYC-stabilizing pathways and further reduce tumor growth. We first tested this hypothesis in vitro and found that the combination of MEK inhibitor trametinib and BGJ 398 reduced paracrine induction of AKT and MYC compared with either single agent (Fig. S3 C). Combined treatment with trametinib and BGJ 398 significantly reduced tumor growth in vivo compared with either drug alone (Fig. 5 D), consistent with a reduction in MYC levels in PDAC cells and with reduced proliferation (Fig. 5, C and E) and consistent with a previous study of trametinib and FGFR1 inhibition (Manchado et al., 2016). Importantly, while a significant correlation between PDAC cell MYC levels and αSMA-positive CAF density was seen in control tumors, this association was abolished with FGFR inhibition (Fig. 5 F), implicating FGFR signaling as a mediator of stroma-inducible MYC stabilization. Taken together, our results highlight a specific role for CAF-derived FGF1 as a paracrine regulator of MYC in pancreatic cancer.
Figure S3.
The FGF1/FGFR axis augments MYC levels and PDAC growth in vitro. (A) Proliferation assay on 8988T cells treated with CM from control or FGF1 knockdown CAF 4414 for 72 h. **, P < 0.01 by one-way ANOVA (n = 3 independent experiments). (B) Proliferation assay for MiaPaCa2 cells treated with CM from control or FGF1 knockdown CAF 4414 for 72 h. **, P < 0.01 by one-way ANOVA (n = 3 independent experiments). (C and D) Western blots for (C) MYC and (D) the indicated signaling events in PSN1 cells treated with CAF 4414 CM, BGJ 398 (2 µM), and trametinib (20 nM) for 3 h.
Figure 5.
The FGF1/FGFR axis regulates MYC levels and PDAC growth in vivo. (A) Proliferation assay for MiaPaCa2 cells treated with CAF 4414 CM ± PD173074 (800 nM) for 72 h. **, P < 0.01; ****, P < 0.0001 by one-way ANOVA (n = 3 biological replicates). (B) Fluorescent immunostaining of cotransplanted subcutaneous xenografts. Scale bar, 50 µm. Quantification appears to the right to determine the ratio of MYC intensity within KRT+ cells over KRT signal. 36 fields of view were analyzed from n = 5 mice per condition. (C) Left: Fluorescent immunostaining of orthotopic transplants of FC1199 cells treated with vehicle or BGJ 398. Quantification appears below to determine the ratio of MYC intensity within KRT+ cells over KRT signal. 28 fields of view were analyzed from n = 3 mice per condition. For B and C (left), **, P < 0.01; ****, P < 0.0001 by Student’s t test. Scale bar, 50 µm. Right: Fluorescent immunostaining of orthotopic transplants of FC1245 cells from the indicated treatment groups. Quantification appears below to determine the ratio of MYC intensity within KRT+ cells over KRT signal (n = 3 mice per group, six fields imaged per mouse). **, P < 0.01; ****, P < 0.0001 by one-way ANOVA. Scale bar, 20 µm. (D) Tumor measurements by high-resolution ultrasound at the indicated time points in FC1245 orthotopic transplants (vehicle, BGJ 398, trametinib: n = 6; BGJ 398 + trametinib, n = 7). *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001 by one-way ANOVA. (E) Fluorescent immunostaining of Ki67 in orthotopic transplants of FC1245 cells from the indicated treatment groups. Quantification appears below to determine the ratio of Ki67 intensity within KRT+ cells over KRT signal (n = 3 mice per group, four fields imaged per mouse). *, P < 0.05 by one-way ANOVA. Scale bar, 20 µm. (F) Fluorescent immunostaining of KRT, MYC, and αSMA in orthotopic FC1245 tumors treated with vehicle or BGJ 398. Quantification of MYC integrated density in KRT+ cells and of αSMA integrated density appear below in 80 fields of view from n = 5 vehicle samples, and 114 fields of view from n = 8 BGJ 398 samples; ****, P < 0.0001 for Pearson’s correlation. Scale bar, 10 µm. ns, not significant.
Though compelling prior studies have highlighted cell-autonomous mechanisms of MYC regulation in RAS-driven cancers (Hayes et al., 2016; Sears et al., 1999, 2000; Vaseva et al., 2018), here we show that MYC is further subject to regulation by soluble cues from stromal CAFs in pancreatic cancer. As fibroblasts evolved in part to facilitate wound repair, this may reflect a conserved mechanism by which mesenchymal cells signal to the epithelial compartment to promote regeneration and healing, as MYC has been previously implicated in the wound-healing process (Schäfer and Werner, 2007; Su et al., 2018; Zanet et al., 2005), as have MYC-regulated bioenergetics (Shyh-Chang et al., 2013). While previous studies have demonstrated a critical role for MYC in regulation of the tumor microenvironment (Kortlever et al., 2017; Sodir et al., 2011, 2020), our findings suggest the existence of a feedback loop, in which cues from the fibroinflammatory microenvironment in turn augment MYC protein level and oncogenic activity. As PDAC rarely harbors PI-3K mutations (Cancer Genome Atlas Research Network, 2017), it was not entirely surprising that external signals can increase AKT phosphorylation in PDAC cells. However, we were surprised to find that FGF1 is required for paracrine AKT activation by CAFs, given the abundance of additional secreted factors from CAFs that may signal through growth factor receptors to activate AKT. These findings suggest that FGF1 creates a permissive context for activation of the AKT/GSK-3β axis, and future studies will aim to understand the broader proteomic network regulated by FGF1 in PDAC cells. Future efforts will also aim to discover additional therapeutic strategies that, together with FGF1/FGFR inhibition, improve outcome in RAS/MYC-driven cancers.
Materials and methods
Animals
All experiments were reviewed and overseen by the institutional animal use and care committee at Oregon Health and Science University (OHSU) in accordance with National Institutes of Health guidelines for the humane treatment of animals. C57BL/6J (000664) or NU/J (002019) mice from Jackson Laboratory were used for orthotopic transplant and xenograft experiments, respectively, at the ages denoted in the paper.
Human tissue samples
Human patient PDAC tissue samples donated to the Oregon Pancreas Tissue Registry program with informed written patient consent (Institutional Review Board approved, IRB00003609) in accordance with full ethical approval by the OHSU Institutional Review Board were kindly shared by J. Link and R.C. Sears.
Generation of human PDAC primary CAFs
All of the primary fibroblast strains were derived in the same way from pancreatic tumor explant cultures. Tissue samples from primary pancreatic tumor were cut into small pieces ∼2–3 mm3 in size and seeded in FCS with 200 U/ml penicillin, 200 μg/ml streptomycin, 100 μg/ml gentamicin, and 2.5 g/ml amphotericin B. When outgrowths of fibroblasts appeared, the culture medium was replaced with complete F medium to facilitate fibroblast growth. The remnants of the tissue were carefully washed away, and CAFs were routinely maintained in complete F medium at 37°C in a humid atmosphere containing 5% CO2. Immunohistochemistry and RNA-seq were used to confirm minimal expression of cytokeratin and abundant expression of αSMA in primary CAF lines. Wild-type KRAS codon 12 sequence was confirmed for all CAF lines, and mutant KRAS-G12 was confirmed in all corresponding parent tumors. All experiments in this study were performed with CAFs at seven passages at most, except for CAF 4511 and 4768, which were cultured to a maximum of 20 passages.
Cell lines
Human pancreatic cancer cell lines MIAPaCa-1, PA-TU-8988T, Panc1, and PSN-1 were obtained from the American Type Culture Collection and grown in DMEM containing 10% fetal bovine serum, and the hPSC line was obtained from ScienCell as previously described (Langer et al., 2019).
CM treatments
CM were generated by adding fresh serum-free DMEM and incubating cells for 48 h as previously described (Sherman et al., 2014). CM was harvested, spun at 300 g for 5 min to pellet debris, and added to PDAC cells for the indicated duration. Serum-free DMEM served as a control for these experiments.
Western blotting
PDAC cells were serum-starved for 48 h and treated as mentioned in the text. Sub-cellular fractions were prepared using the NE-PER Nuclear and Cytoplasmic Extraction reagents (Thermo Fisher Scientific) as per the manufacturer’s instructions. Where indicated, the nuclear insoluble fraction was prepared by lysing the nuclear pellet in radioimmunoprecipitation assay buffer buffer and sonicating for five rounds of 30 s. Protein concentration was quantitated using the bicinchoninic protein assay kit (Pierce). Equal amounts of protein were loaded in each lane and separated on a 4–12% Bis-Tris NuPAGE gel (Invitrogen), then transferred onto a PVDF membrane. Membranes were probed with primary antibodies and infrared secondary antibodies: IRDye 700 goat anti-rabbit IgG or IRDye 800 goat anti-mouse IgG (LI-COR Biosciences). For protein band quantitation, infrared signals were detected using the Odyssey CLx infrared imaging system and bands quantified using Image Studio software (LICOR Biosciences).
Neutralizing antibody screen
Specific neutralizing antibodies were added to CAF-derived CM and rocked at room temperature for 1 h for effective neutralization of CAF secreted factors. PDAC target cells were serum-starved for 48 h and then treated with CAF-derived CM only or CAF-derived CM with specific neutralizing antibodies. The complete list of neutralizing antibodies used is described under the Antibodies section of Materials and methods.
Immunostaining
Cells
Cells plated on coverslips were fixed in 4% paraformaldehyde for 15 min at room temperature, washed three times with PBS, and permeabilized with 0.1% Triton X-100 for 10 min at room temperature. Following permeabilization, coverslips were blocked for 1 h at room temperature in blocking solution (8% BSA solution) and then transferred to a carrier solution (8% BSA solution) containing diluted antibodies (described in the Antibodies section of Materials and methods). Coverslips were incubated with the primary antibody for 3 h at room temperature and then washed five times for 5 min each in PBS, following which secondary Alexa Fluor–conjugated antibodies diluted in the same carrier solution (1:400) were added to the coverslips for 1 h at room temperature. After the secondary antibody incubation, coverslips were washed five times for 5 min each in PBS and mounted with Vectashield mounting media containing DAPI.
Mouse and human tissue sections
Mice were anesthetized and euthanized according to institutional guidelines. Pancreatic tumors/subcutaneous tumors were excised carefully and drop-fixed overnight in 4% paraformaldehyde. Tissue samples were paraffin-embedded, sectioned, and H&E-stained at the OHSU Histopathology Core. In brief, tissue sections were de-paraffinized and rehydrated through an ethanol series and ultimately in PBS. Following antigen retrieval, tissue samples were blocked for 2 h at room temperature in blocking solution (Aqua Block buffer from Abcam) and then transferred to a carrier solution (Aqua Block buffer from Abcam) containing diluted antibodies (described in Antibodies section of Materials and methods). Sections were incubated overnight at room temperature and then washed five times for 5 min each in PBS, following which secondary Alexa Fluor–conjugated antibodies diluted in the same carrier solution (1:400) were added to the sections for 1 h at room temperature. Sections were then washed five times for 5 min each in PBS and were mounted with Vectashield mounting media containing DAPI. To stain for MYC, human PDAC sections were stained with a MYC pS62 antibody (Abcam, ab185656), while subcutaneous and orthotopic tumors in mice were stained with a total MYC antibody (Cell Signaling Technology, 5605). Additional antibodies are listed in the Antibodies section.
IF image analysis was done using the global thresholding method using ImageJ software. Global thresholding allowed us to segment fluorescently labeled pixels from the background in an image. In global thresholding, a value cutoff is chosen, such that every pixel less than that value is considered one class, while every pixel greater than that value is considered the other class. This technique of image segmentation allowed us to divide an image into two classes of pixels, “foreground” and “background.” In our application, the pixels classified as foreground, or segmented pixels, are also referred to as fluorescent pixels as they are assumed to carry the fluorescent signal of the image. For our analyses, we specifically used the original method of auto thresholding available in ImageJ, the Default image segmentation algorithm. This plugin binaries 8 and 16-bit images using the Default (histogram-derived algorithm) method.
Two-plex FISH
Human PDAC tumor tissue samples were subjected to FISH using the ViewRNA ISH Tissue Assay Kit (two-plex) from Thermo Fisher Scientific according to manufacturer’s guidelines. In brief, samples were subjected to controlled protease digestion for initial permeabilization, following which the samples were incubated with the proprietary probe-containing solution according to the manufacturer’s guidelines. It was crucial that samples remained fully submerged during the entire incubation. Following probe hybridization, samples were washed and then subjected to sequential hybridization with preamplifier and amplifier DNA and fluorophore. Hybridizations with preamplifier, amplifier, and fluorophore were performed as indicated by the manufacturer. Samples were mounted using DAKO ultramount mounting medium as indicated by the manufacturer.
Microscopy
Imaging of fluorescence staining was done by confocal imaging of fixed cells and tissues with a laser-scanning confocal inverted microscope (Carl Zeiss, Inc., LSM 880), and a 40×/ 1.1 numerical aperture water objective or 63×/1.4 numerical aperture oil objective was used to image the samples. A Zeiss Axio Scan automated slide scanning microscope was used to take 20× tiled images of entire H&E-stained mouse tumor sections.
ChIP-seq
For immunoprecipitation and sequencing, chromatin immunoprecipitation was performed as described previously (Sherman et al., 2014). Briefly, PDAC cells were fixed in 1% formaldehyde, and nuclei were isolated and lysed in buffer containing 1% SDS, 10 mM EDTA, 50 mM Tris-HCl, pH 8.0, and protease inhibitors, and sheared with a Diagenode Bioruptor to chromatin fragment sizes of 200–1,000 base pairs. Chromatin was immunoprecipitated with antibodies to MYC (Cell Signaling Technology, 9402), or rabbit IgG (Santa Cruz Biotechnology). ChIP-seq libraries were constructed using Illumina’s TruSeq ChIP library preparation protocol, using 5 ng input DNA. Libraries were sequenced using 100-cycle single-read sequencing on a HiSeq 2500.
For alignment and peak calling, reads were first trimmed of adapter sequences using default parameters of Trim Galore! (version 0.4.3; https://www.bioinformatics.babraham.ac.uk/projects/trim_galore/). Adapter-trimmed reads were aligned to the human reference genome (GRCh38, release 87) using bowtie2 (version 2.3.4; Langmead and Salzberg, 2012). The resulting alignment files were further filtered to contain reads with a MAPQ score of 11 or greater using samtools (version 1.3.1; Li et al., 2009), and all duplicates were removed using picard tools (version 2.9.0; http://broadinstitute.github.io/picard). Peaks were called using the “callpeak” method of MACS2 (version 2.1.1.20160309; Zhang et al., 2008) with a q-value cut-off of 0.05.
For overlapping peaks, peaks output by MACS2 was filtered for known chromosomes only. The findOverlapsOfPeaks() function from the ChIPpeakAnno R package (version 3.14.2; Zhu et al., 2010) was used to determine peaks shared between replicates within a treatment. The within-treatment consensus peaks were similarly evaluated in order to create a final set of peaks shared between both treatments.
The resulting between-treatment consensus peaks were evaluated for motif enrichment using the findMotifsGenome.pl script from the homer package (version 4.10.1; Heinz et al., 2010). The read counts of the ChIP reads relative to the control reads were used to visualize the binding density of the between-treatment consensus peaks using the dba.plotHeatmap() function from the diffBind R package (version 2.8.0; https://bioconductor.org/packages/release/bioc/html/DiffBind.html).
Gain of MYC binding at known MYC target gene promoter sequences was confirmed by ChIP-qPCR. Enrichment was calculated against DMEM-treated samples, and enrichment values were normalized to a control intergenic region of the genome. The primer sequences used are described in the Primer sequences section of Materials and methods.
Gene expression analysis by qPCR
The isolated total RNA (1 µg) was reverse-transcribed to produce cDNA using iScript Reverse Transcription Supermix kit (Bio-Rad). Real-time PCR was performed using SYBR Green Supermix (Bio-Rad). The cDNA sequences for specific gene targets were obtained from the human genome assembly (http://genome.ucsc.edu), and gene-specific primer pairs were designed using the Primer3 program (http://bioinfo.ut.ee/primer3/). Relative gene expression was expressed as fold change in gene expression between nontumor pancreatic stellate cells and primary pancreatic tumor–associated CAFs or DMEM-treated cells and CM-treated cells, with threshold cycle values normalized using the 36B4 housekeeping gene. Gene-specific primer pair sequences are provided in the Primer sequences section of Materials and methods.
ELISA
Secreted human acidic FGF in CM was measured using a sandwich immune-luminometric assay based on the manufacturer’s instructions (Human FGF acidic Quantikine Elisa Kit; R&D Systems, DFA00B). The concentration of secreted acidic FGF was compared across a subset of primary pancreatic tumor–derived CAF strains and a panel of PDAC cell lines.
Stable and transient plasmid transfections
The vectors containing shRNA against human acidic FGF were purchased from Dharmacon. In brief, 15 μg of vector, together with 7.5 µg of each packaging vector (pMD2.G and psPAX2), were cotransfected into 293T cells. Supernatant-containing lentivirus particles were harvested 48 h and 72 h after transfection, passed through a 0.45-µm membrane filter, and directly used to infect primary pancreatic tumor–associated CAFs in the presence of 6 µg/ml polybrene. After 48 h, the infected cells were maintained in the medium with 2 µg/ml puromycin for 2 wk before knockdown assessment.
For transient transfections with purified plasmid DNA, transfections were performed using Lipofectamine 2000 following the manufacturer’s instructions.
Proliferation assay
For the growth assays, PDAC cells were seeded into 96-well plates at 2 × 103 cells per well in DMEM containing 10% FBS. The next day, cells were washed with PBS and changed to serum-free DMEM containing 25 mM glucose and 4 mM glutamine for 72 h. After serum starvation, cells were treated with CM ± pharmacological inhibitors (as mentioned) at the indicated concentrations for another 72 h. After 72 h, cells were lysed with CellTiter-Glo Luminescent Cell Viability Assay reagent (Promega), and luminescence was read using the GloMax plate reader.
Antibodies
IF studies
IF studies were conducted with the following: smooth muscle actin mAb (1A4 [asm-1]; Invitrogen, MA5-11547); anti–c-Myc antibody (Y69; Abcam, ab32072); anti–c-Myc (pS62) antibody (EPR17924; Abcam, ab185656); anti–c-Myc (D84C12) rabbit mAb (Cell Signaling Technology, 5605); anti-FGFR1 (phospho Y653) antibody (EPR843 [N]; Abcam, ab173305); anti-FGF1 antibody (R&D Systems, AF232); FGFR1 mAb (VBS-7; Invitrogen, 13–3100); NPM1 mAb (FC-61991; Invitrogen, 32–5200); pan-KRT (C11) mouse mAb (Alexa Fluor 647 Conjugate; Cell Signaling Technology, 4528); and Cytokeratin Pan Type I/II Antibody Cocktail (Thermo Fisher Scientific, MA5-13156).
Western blots
Western blots were conducted with the following: c-Myc (D84C12) rabbit mAb (Cell Signaling Technology, 5605); anti–c-Myc (pS62) antibody (EPR17924; Abcam, ab185656); anti–c-Myc (pT58) antibody (EPR17924; Abcam, ab28842); phospho-Akt (Ser473; D9E) XP rabbit mAb (Cell Signaling Technology, 4060); Akt (pan; C67E7) rabbit mAb (Cell Signaling Technology, 4691); phospho-GSK-3β (Ser9) antibody (Cell Signaling Technology, 9336); GSK-3β (27C10) rabbit mAb (Cell Signaling Technology, 9315); Lamin A/C (4C11) mouse mAb (Cell Signaling Technology, 4777); HSC 70 mouse antibody (B-6; Selleck Chemicals, 7298); NPM1 mAb (FC-61991; Invitrogen, 32–5200); and FGF1 antibody (R&D Systems, AF232).
Neutralizing antibodies
The following were used as neutralizing antibodies: human FGF acidic neutralizing antibody (R&D Systems, AF232); human FGF basic neutralizing antibody (R&D Systems, AF232); human FGF-7 neutralizing antibody (R&D Systems, MAB251); human Il-6 neutralizing antibody (R&D Systems, MAB2061); human HGF neutralizing antibody (R&D Systems, AB-294-NA); human PDGF-AA neutralizing antibody (R&D Systems, MAB221); human PDGF-BB neutralizing antibody (R&D Systems, AB-220-NA); and human/mouse Wnt-3a neutralizing antibody (R&D Systems, MAB9025).
Plasmids
HA-AKT DN (K17M) was a gift from Mien-Chie Hung (University of Texas MD Anderson Cancer Center, Houston, TX; Addgene plasmid 16243). HA-GSK3 β S9A pcDNA3 was a gift from Jim Woodgett (Lunenfeld-Tanenbaum Research Institute, Toronto, Ontario, Canada; Addgene plasmid 14754). SMARTvector lentiviral human shRNA constructs were purchased from Dharmacon (224940601, 225482233, 226573540, and 228128665). Flag-tagged MYC WT and T58A plasmids were kindly provided by Mushui Dai (OHSU, Portland, OR).
Primer sequences
Gene expression analyses
Gene expression analyses were conducted with the following primers: human MYC F: 5′-CAGCTGCTTAGACGCTGGATT-3′; R: 5′-GTAGAAATACGGCTGCACCGA-3′; human FGF1 F: 5′-CAATGTTTGGGCTAAGACCTG-3′; R: 5′-GGCTGTGAAGGTGGTGATTT-3′; human FGF2 F: 5′-GTGTGTGCTAACCGTTACCT-3′; R: 5′-GCTCTTAGCAGACATTGGAAG-3′; human FGF7F: 5′-ATCAGGACAGTGGCAGTTGGA-3′; R: 5′-AACATTTCCCCTCCGTTGTGT-3′; human CSF1 F: 5′-GCTCTCCCAGGATCTCATCAC-3′; R: 5′-TCAAAGGAACGGAGTTAAAACGG-3′; human EGF F: 5′-CTGTGGTGCTGTCATCTGTC-3′; R: 5′-GTCCCCAGCCGATTCCTTG-3′; human HGF F: 5′-AAGGTGACTCTGAATGAGTC-3′; R: 5′-GGCACATCCACGACCAGGAACAATG-3′; human NGF F: 5′-CACACTGAGGTGCATAGCGT-3′; R: 5′-TGATGACCGCTTGCTCCTGT-3′; human PDGFAA F: 5′-CACACCTCCTCGCTG TAGTATTTA-3′; R: 5′-GTTATCGGTGTAAATGTCATCCAA-3′; human PDGFBB F: 5′-TCCCGAGGAGCTTTATGAGA-3′; R: 5′-ACTGCACGTTGCGGTTGT-3′; human KITLG F: 5′-CAGAGTCAGTGTCACAAAACCATT-3′; R: 5′-TTGGCCTTCCTATTACTGCTACTG-3′; human TGFB 1 F: 5′-GCAACAATTCCTGGCGATACCTC-3′; R: 5′-AGTTCTTCTCCGTGGAGCTGAAG-3′; human TGFB 2 F: 5′-AGAGTGCCTGAACAACGGATT-3′; R: 5′-CCATTCGCCTTCTGCTCTT-3′; human VEGFA F: 5′-GCCTTGCTGCTCTACCTCCA-3′; R: 5′-CAAGGCCCACAGGGATTTT-3′; human VEGFB F: 5′-AGCACCAAGTCCGGATG-3′; R: 5′-GTCTGGCTTCACAGCACTG-3′; human IL-6 F: 5′-AAAGAGGCACTGGCAGAAAA-3′; R: 5′-AGCTCTGGCTTGTTCCTCAC-3′; human IL-8 F: 5′-CTGGCCGTGGCTCTCTTG-3′; R: 5′-CCTTGGCAAAACTGCACCTT-3′; human IL-1a F: 5′-AGGGAATTCACCCCAAGAAC-3′; R: 5′-ACTATGGGGGATGCAGGATT-3′; human IL-1b F: 5′-AAGCTGAGGAAGATGCTG-3′; R: 5′-ATCTACACTCTCCAGCTG-3′; human IL-32 F: 5′-ATGTGCTTCCCGAAGGTCCTCTCTGA-3′; R: 5′-TCATTTTGAGGATTGGGGTTCAGAGC-3′; human CXCL1 F: 5′-AGGGAATTCACCCCAAGAAC-3′; R: 5′-ACTATGGGGGATGCAGGATT-3′; human WNT5a F: 5′-AGAAGAAACTGTGCCACTTGTATCAG-3′; R: 5′-CCTTCGATGTCGGAATTGATACT-3′; human MIF F: 5′-CGCAGAACCGCTCCTACAG-3′; R: 5′-GGAGTTGTTCCAGCCCACAT-3′; human β actin F: 5′-ATTGGCAATGAGCGGTTCCGC-3′; R: 5′-CTCCTGCTTGCTGATCCACATC-3′; human 36B4 F: 5′-GTGCTGATGGGCAAGAAC-3′; and R: 5′-AGGTCCTCCTTGGTGAAC-3′.
MYC target validation
MYC target validation was conducted with the following primers: human CNTRL F: 5′-CAGATGAAAGCCCTTACATTGGC-3′; R: 5′-CTGCCTGAGCACTGTCAATAAT-3′; human PCNA F: 5′-AGGGCTCCATCCTCAAGAAG-3′; R: 5′-GTAGGTGTCGAAGCCCTCAG-3′; human DNMT1 F: 5′-TCAGGGACCACATCTGTAAGG-3′; R: 5′-GCCGTTCTTCCTGTCATGG-3′; human SNAI1 F: 5′-CTAGAGTCTGAGATGCCCCG-3′; R: 5′-AGTTCTGGGAGACACATCGG-3′; human EIF2B5 F: 5′-AGAGGCGAACTTCACTGACA-3′; R: 5′-GTCCAGCACCACGTTATCAC-3′; human HES1 F: 5′-GCTTTCCTCATTCCCAACGG-3′; and R: 5′-GTGGGTTGGGGAGTTTAGGA-3′.
ChIP-qPCR validation
ChIP-qPCR validation was conducted with the following primers: human DNMT1 F: 5′-TGCAATGAATTCCAGATGTG-3′; R: 5′-GGAGGGGCAGAGTGAGAGAT-3′; human PCNA F: 5′-TTGGCTAATCGCACACTGA-3′; R: 5′-GTCCGGAATATCCACCAATG-3′; human SNAI1 F: 5′-GGGCCTTTTCCCTTGATAAT-3′; R: 5′-AAGGGAAGTGTGCTTTGGTG-3′; human HES1 F: 5′-AAGTTTCACACGAGCCGTTC-3′; R: 5′-GAGAGGTAGACGGGGGATC-3′; human CNTRL F: 5′-CCTTCCAACAGTACCGGAGA-3′; and R: 5′-GCAGCCATTTTGTTGTGTTG-3′.
Pharmacological compounds
Cycloheximide was purchased from Cell Signaling Technology (CST 2112). Actinomycin D was purchased from Sigma-Aldrich (A1410-5MG). PD173074 was purchased from Selleck Chemicals (S1264). NVP-BGJ398 was purchased from Selleck Chemicals (S2183). MK-2206 2HCl was purchased from Selleck Chemicals (S1078). Trametinib was purchased from MedChem Express (HY-10999A). Purified recombinant human FGF acidic was purchased from Stem Cell Technologies (78187.1). All pharmacological compounds were resuspended and stored according to the manufacturer’s guidelines. Complete F media for growing and maintenance of primary human PDAC-associated CAFs were as follows: 3:1 DMEM/F12, 5% FBS, 0.4 μg/ml hydrocortisone, 5 μg/ml insulin, 8.4 ng/ml cholera toxin, 10 ng/ml epidermal growth factor (EGF), 24 μg/ml Adenine, and 10 μM Rho-associated protein kinase inhibitor.
Cycloheximide chase
Cells were serum-starved for 48 h in serum-free DMEM, and following serum starvation, cells were treated with either DMEM or CAF-derived CM for 3 h, following which cells were treated with 50 µg/ml cycloheximide for the indicated times. Then, the cells were lysed, and whole cell lysates or nuclear lysates were prepared for Western blotting analysis. MYC levels relative to time point 0 are shown in all graphs of cycloheximide experiments.
Actinomycin D chase
Cells were serum-starved for 48 h in serum-free DMEM, and following serum starvation, cells were treated with either DMEM or CAF-derived CM for 3 h, following which actinomycin D (5 µg/ml final concentration) was added and RNA was isolated at the times indicated. Analysis of MYC mRNA from PDAC cells was done by quantitative PCR using MYC-specific primer pairs and threshold cycle values normalized using ACTB as a housekeeping gene. MYC mRNA levels relative to time point 0 are shown in all graphs of actinomycin D chase experiments.
Data availability
All sequence data from this study have been deposited in the publicly available Gene Expression Omnibus under accession no. GSE143804.
Orthotopic transplant/allograft model
The orthotopic transplant model used here was described previously (Collisson et al., 2012). In brief, 8-wk-old wild-type male C57BL/6J mice were orthotopically transplanted as described previously with 10,000 FC1199 cells or FC1245 cells, isolated from PDAC in LSL-KrasG12D/+; Trp53R172H/+; Pdx1-Cre C57B6/J mice, obtained from the David Tuveson laboratory (Cold Spring Harbor Laboratory, Cold Spring Harbor, NY), in 50% Matrigel. After ultrasound imaging to confirm the presence of PDAC, mice were randomized into treatment groups: mice that were treated by oral gavage daily with vehicle alone, with BGJ 398 (50 mg/kg body weight), with trametinib (1 mg/kg body weight), or BGJ 398 + trametinib. Tumors were measured and mice euthanized after 9 d of treatment, and pancreata were harvested, sliced, and flash-frozen in liquid nitrogen or immediately fixed in formalin.
Subcutaneous PDAC transplants
All surgical procedures and care of the animals were in accordance with institutional guidelines. A 100 µl volume of 1:1 mixture of Matrigel (BD Biosciences, 354248) and 106 MiaPaCa2 cells alone, or 106 MiaPaCa2 cells plus 5 × 106 control or shFGF1 CAFs were subcutaneously injected into NU/J (002019) mice flanks. Tumor size was measured every other day using digital calipers, and tumor volume was estimated using the following formula: V = (LW2)/2, where V, volume (mm3); L, largest diameter (mm); and W, smallest diameter (mm). Mice were euthanized and tumors harvested on day 40 after transplantation for analysis.
Statistical analysis
All statistical analyses were performed using GraphPad Prism 5.0 Software.
Online supplemental material
Fig. S1 shows that PDAC CAF-secreted factors, and stroma-derived FGF1 in particular, increase MYC protein level in PDAC cells. Fig. S2 shows that paracrine FGF1 signaling from stromal CAFs augments MYC expression and protein stability via AKT/GSK-3β signaling in PDAC cells. Fig. S3 shows that the FGF1/FGFR axis augments MYC protein level and PDAC cell proliferation in vitro.
Acknowledgments
We thank the OHSU Massively Parallel Sequencing Shared Resource, Advanced Light Microscopy Shared Resource, and Histopathology Shared Resource for technical help; the OHSU Brenden-Colson Center for Pancreatic Care for providing primary PDAC reagents; Mark Berry and Cameron Roberts for technical support; and Sara Courtneidge and members of the Sherman laboratory for helpful discussion and feedback.
This work was supported by funds from the National Cancer Institute (R00CA188259 and R01CA229580) and a Medical Research Foundation New Investigator Grant (to M.H. Sherman).
Author contributions: S. Bhattacharyya designed, performed, and analyzed all experiments with technical support from C. Oon and A. Kothari, and under the guidance of M.H. Sherman. W. Horton analyzed the ChIP-seq data. J. Link and R.C. Sears assisted with generation of primary PDAC CAFs and tissue samples. S. Bhattacharyya and M.H. Sherman wrote the manuscript.
References
- Albihn A., Johnsen J.I., and Henriksson M.A.. 2010. MYC in oncogenesis and as a target for cancer therapies. Adv. Cancer Res. 107:163–224. 10.1016/S0065-230X(10)07006-5 [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network 2017. Integrated Genomic Characterization of Pancreatic Ductal Adenocarcinoma. Cancer Cell. 32:185–203.e13. 10.1016/j.ccell.2017.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins M.A., Bednar F., Zhang Y., Brisset J.C., Galbán S., Galbán C.J., Rakshit S., Flannagan K.S., Adsay N.V., and Pasca di Magliano M.. 2012. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J. Clin. Invest. 122:639–653. 10.1172/JCI59227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collisson E.A., Trejo C.L., Silva J.M., Gu S., Korkola J.E., Heiser L.M., Charles R.P., Rabinovich B.A., Hann B., Dankort D., et al. 2012. A central role for RAF→MEK→ERK signaling in the genesis of pancreatic ductal adenocarcinoma. Cancer Discov. 2:685–693. 10.1158/2159-8290.CD-11-0347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell A.S., and Sears R.C.. 2014. MYC degradation. Cold Spring Harb. Perspect. Med. 4 a014365 10.1101/cshperspect.a014365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrell A.S., Joly M.M., Allen-Petersen B.L., Worth P.J., Lanciault C., Sauer D., Link J., Pelz C., Heiser L.M., Morton J.P., et al. 2017. MYC regulates ductal-neuroendocrine lineage plasticity in pancreatic ductal adenocarcinoma associated with poor outcome and chemoresistance. Nat. Commun. 8:1728 10.1038/s41467-017-01967-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez P.C., Frank S.R., Wang L., Schroeder M., Liu S., Greene J., Cocito A., and Amati B.. 2003. Genomic targets of the human c-Myc protein. Genes Dev. 17:1115–1129. 10.1101/gad.1067003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes T.K., Neel N.F., Hu C., Gautam P., Chenard M., Long B., Aziz M., Kassner M., Bryant K.L., Pierobon M., et al. 2016. Long-Term ERK Inhibition in KRAS-Mutant Pancreatic Cancer Is Associated with MYC Degradation and Senescence-like Growth Suppression. Cancer Cell. 29:75–89. 10.1016/j.ccell.2015.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingorani S.R., Petricoin E.F., Maitra A., Rajapakse V., King C., Jacobetz M.A., Ross S., Conrads T.P., Veenstra T.D., Hitt B.A., et al. 2003. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 4:437–450. 10.1016/S1535-6108(03)00309-X [DOI] [PubMed] [Google Scholar]
- Hwang R.F., Moore T., Arumugam T., Ramachandran V., Amos K.D., Rivera A., Ji B., Evans D.B., and Logsdon C.D.. 2008. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Res. 68:918–926. 10.1158/0008-5472.CAN-07-5714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kortlever R.M., Sodir N.M., Wilson C.H., Burkhart D.L., Pellegrinet L., Brown Swigart L., Littlewood T.D., and Evan G.I.. 2017. Myc Cooperates with Ras by Programming Inflammation and Immune Suppression. Cell. 171:1301–1315.e14. 10.1016/j.cell.2017.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langer E.M., Allen-Petersen B.L., King S.M., Kendsersky N.D., Turnidge M.A., Kuziel G.M., Riggers R., Samatham R., Amery T.S., Jacques S.L., et al. 2019. Modeling Tumor Phenotypes In Vitro with Three-Dimensional Bioprinting. Cell Rep. 26:608–623.e6. 10.1016/j.celrep.2018.12.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaVallee T.M., Prudovsky I.A., McMahon G.A., Hu X., and Maciag T.. 1998. Activation of the MAP kinase pathway by FGF-1 correlates with cell proliferation induction while activation of the Src pathway correlates with migration. J. Cell Biol. 141:1647–1658. 10.1083/jcb.141.7.1647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepique A.P., Moraes M.S., Rocha K.M., Eichler C.B., Hajj G.N., Schwindt T.T., and Armelin H.A.. 2004. c-Myc protein is stabilized by fibroblast growth factor 2 and destabilized by ACTH to control cell cycle in mouse Y1 adrenocortical cells. J. Mol. Endocrinol. 33:623–638. 10.1677/jme.1.01485 [DOI] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R.; 1000 Genome Project Data Processing Subgroup . 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 25:2078–2079. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manchado E., Weissmueller S., Morris J.P. IV, Chen C.C., Wullenkord R., Lujambio A., de Stanchina E., Poirier J.T., Gainor J.F., Corcoran R.B., et al. 2016. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature. 534:647–651. 10.1038/nature18600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R., Bravo R., Burckhardt J., and Curran T.. 1984. Induction of c-fos gene and protein by growth factors precedes activation of c-myc. Nature. 312:716–720. 10.1038/312716a0 [DOI] [PubMed] [Google Scholar]
- Myant K., Qiao X., Halonen T., Come C., Laine A., Janghorban M., Partanen J.I., Cassidy J., Ogg E.L., Cammareri P., et al. 2015. Serine 62-Phosphorylated MYC Associates with Nuclear Lamins and Its Regulation by CIP2A Is Essential for Regenerative Proliferation. Cell Rep. 12:1019–1031. 10.1016/j.celrep.2015.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öhlund D., Handly-Santana A., Biffi G., Elyada E., Almeida A.S., Ponz-Sarvise M., Corbo V., Oni T.E., Hearn S.A., Lee E.J., et al. 2017. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 214:579–596. 10.1084/jem.20162024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabò A., Kress T.R., Pelizzola M., de Pretis S., Gorski M.M., Tesi A., Morelli M.J., Bora P., Doni M., Verrecchia A., et al. 2014. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 511:488–492. 10.1038/nature13537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saborowski M., Saborowski A., Morris J.P. IV, Bosbach B., Dow L.E., Pelletier J., Klimstra D.S., and Lowe S.W.. 2014. A modular and flexible ESC-based mouse model of pancreatic cancer. Genes Dev. 28:85–97. 10.1101/gad.232082.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer M., and Werner S.. 2007. Transcriptional control of wound repair. Annu. Rev. Cell Dev. Biol. 23:69–92. 10.1146/annurev.cellbio.23.090506.123609 [DOI] [PubMed] [Google Scholar]
- Sears R., Leone G., DeGregori J., and Nevins J.R.. 1999. Ras enhances Myc protein stability. Mol. Cell. 3:169–179. 10.1016/S1097-2765(00)80308-1 [DOI] [PubMed] [Google Scholar]
- Sears R., Nuckolls F., Haura E., Taya Y., Tamai K., and Nevins J.R.. 2000. Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 14:2501–2514. 10.1101/gad.836800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman M.H., Yu R.T., Engle D.D., Ding N., Atkins A.R., Tiriac H., Collisson E.A., Connor F., Van Dyke T., Kozlov S., et al. 2014. Vitamin D receptor-mediated stromal reprogramming suppresses pancreatitis and enhances pancreatic cancer therapy. Cell. 159:80–93. 10.1016/j.cell.2014.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman M.H., Yu R.T., Tseng T.W., Sousa C.M., Liu S., Truitt M.L., He N., Ding N., Liddle C., Atkins A.R., et al. 2017. Stromal cues regulate the pancreatic cancer epigenome and metabolome. Proc. Natl. Acad. Sci. USA. 114:1129–1134. 10.1073/pnas.1620164114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyh-Chang N., Zhu H., Yvanka de Soysa T., Shinoda G., Seligson M.T., Tsanov K.M., Nguyen L., Asara J.M., Cantley L.C., and Daley G.Q.. 2013. Lin28 enhances tissue repair by reprogramming cellular metabolism. Cell. 155:778–792. 10.1016/j.cell.2013.09.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodir N.M., Swigart L.B., Karnezis A.N., Hanahan D., Evan G.I., and Soucek L.. 2011. Endogenous Myc maintains the tumor microenvironment. Genes Dev. 25:907–916. 10.1101/gad.2038411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sodir N.M., Kortlever R.M., Barthet V.J.A., Campos T., Pellegrinet L., Kupczak S., Anastasiou P., Swigart L.B., Soucek L., Arends M.J., et al. 2020. Myc instructs and maintains pancreatic adenocarcinoma phenotype. Cancer Discov. 10:588–607. 10.1158/2159-8290.CD-19-0435 [DOI] [PubMed] [Google Scholar]
- Soucek L., and Evan G.I.. 2010. The ups and downs of Myc biology. Curr. Opin. Genet. Dev. 20:91–95. 10.1016/j.gde.2009.11.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L., Whitfield J., Martins C.P., Finch A.J., Murphy D.J., Sodir N.M., Karnezis A.N., Swigart L.B., Nasi S., and Evan G.I.. 2008. Modelling Myc inhibition as a cancer therapy. Nature. 455:679–683. 10.1038/nature07260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soucek L., Whitfield J.R., Sodir N.M., Massó-Vallés D., Serrano E., Karnezis A.N., Swigart L.B., and Evan G.I.. 2013. Inhibition of Myc family proteins eradicates KRas-driven lung cancer in mice. Genes Dev. 27:504–513. 10.1101/gad.205542.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sousa C.M., Biancur D.E., Wang X., Halbrook C.J., Sherman M.H., Zhang L., Kremer D., Hwang R.F., Witkiewicz A.K., Ying H., et al. 2016. Pancreatic stellate cells support tumour metabolism through autophagic alanine secretion. Nature. 536:479–483. 10.1038/nature19084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y., Pelz C., Huang T., Torkenczy K., Wang X., Cherry A., Daniel C.J., Liang J., Nan X., Dai M.S., et al. 2018. Post-translational modification localizes MYC to the nuclear pore basket to regulate a subset of target genes involved in cellular responses to environmental signals. Genes Dev. 32:1398–1419. 10.1101/gad.314377.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuveson D.A., Shaw A.T., Willis N.A., Silver D.P., Jackson E.L., Chang S., Mercer K.L., Grochow R., Hock H., Crowley D., et al. 2004. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 5:375–387. 10.1016/S1535-6108(04)00085-6 [DOI] [PubMed] [Google Scholar]
- Vaseva A.V., Blake D.R., Gilbert T.S.K., Ng S., Hostetter G., Azam S.H., Ozkan-Dagliyan I., Gautam P., Bryant K.L., Pearce K.H., et al. 2018. KRAS Suppression-Induced Degradation of MYC Is Antagonized by a MEK5-ERK5 Compensatory Mechanism. Cancer Cell. 34:807–822.e7. 10.1016/j.ccell.2018.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walz S., Lorenzin F., Morton J., Wiese K.E., von Eyss B., Herold S., Rycak L., Dumay-Odelot H., Karim S., Bartkuhn M., et al. 2014. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 511:483–487. 10.1038/nature13473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters A.M., and Der C.J.. 2018. KRAS: The Critical Driver and Therapeutic Target for Pancreatic Cancer. Cold Spring Harb. Perspect. Med. 8 a031435 10.1101/cshperspect.a031435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying H., Kimmelman A.C., Lyssiotis C.A., Hua S., Chu G.C., Fletcher-Sananikone E., Locasale J.W., Son J., Zhang H., Coloff J.L., et al. 2012. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 149:656–670. 10.1016/j.cell.2012.01.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanet J., Pibre S., Jacquet C., Ramirez A., de Alborán I.M., and Gandarillas A.. 2005. Endogenous Myc controls mammalian epidermal cell size, hyperproliferation, endoreplication and stem cell amplification. J. Cell Sci. 118:1693–1704. 10.1242/jcs.02298 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9:R137 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L.J., Gazin C., Lawson N.D., Pagès H., Lin S.M., Lapointe D.S., and Green M.R.. 2010. ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics. 11:237 10.1186/1471-2105-11-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All sequence data from this study have been deposited in the publicly available Gene Expression Omnibus under accession no. GSE143804.