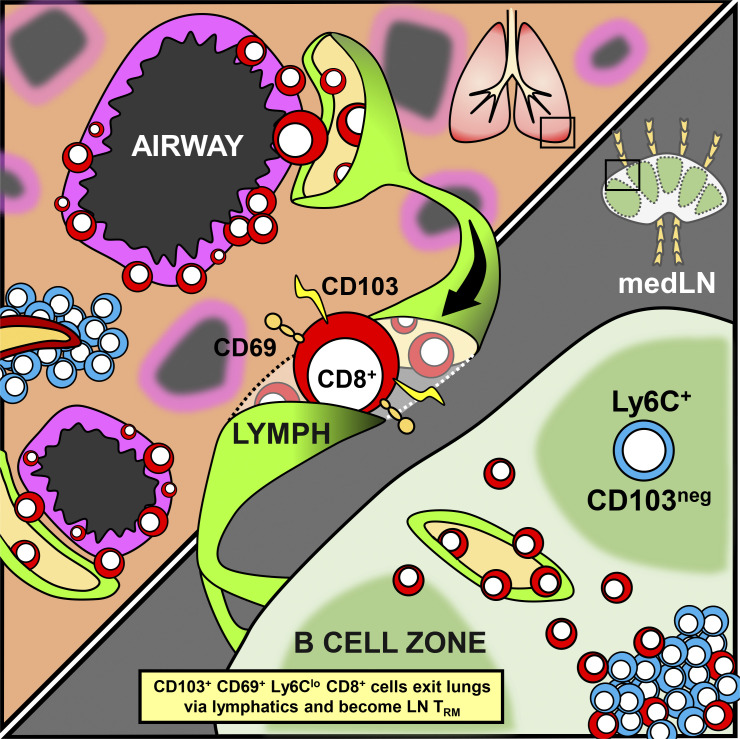
Stolley et al. demonstrate that the decay of lung CD8+ T cell responses after primary influenza virus infection is contemporized by the egress of TRM-phenotype T cells from the lung to the draining mediastinal LN by retrograde migration, where they form more stable residents.
Abstract
Numerous observations indicate that resident memory T cells (TRM) undergo unusually rapid attrition within the lung. Here we demonstrate that contraction of lung CD8+ T cell responses after influenza infection is contemporized with egress of CD69+/CD103+ CD8+ T cells to the draining mediastinal LN via the lymphatic vessels, which we term retrograde migration. Cells within the draining LN retained canonical markers of lung TRM, including CD103 and CD69, lacked Ly6C expression (also a feature of lung TRM), maintained granzyme B expression, and did not equilibrate among immunized parabiotic mice. Investigations of bystander infection or removal of the TCR from established memory cells revealed that the induction of the TRM phenotype was dependent on antigen recognition; however, maintenance was independent. Thus, local lung infection induces CD8+ T cells with a TRM phenotype that nevertheless undergo retrograde migration, yet remain durably committed to the residency program within the draining LN, where they provide longer-lived regional memory while chronicling previous upstream antigen experiences.
Graphical Abstract
Introduction
CD8+ resident memory T cells (TRM) derive from recently activated T cells that migrate to nonlymphoid tissue (NLT), typically during the clonal expansion phase of an immune response (Hofmann and Pircher, 2011; Masopust et al., 2010; Mueller and Mackay, 2016). TRM are defined by migration, remaining parked in situ without recirculating between the tissue of residence and blood. Residency is a mechanism for regionalizing immunosurveillance and provides a means to bias immunity to specific compartments (Farber et al., 2014; Masopust and Soerens, 2019; Mueller et al., 2013; Park and Kupper, 2015; Shin and Iwasaki, 2013). TRM at barrier sites have been shown to accelerate protection against reinfection (Gebhardt et al., 2009; Glennie et al., 2015; Jiang et al., 2012; Teijaro et al., 2011), may be associated with tumor control (Amsen et al., 2018; Ganesan et al., 2017; Malik et al., 2017; Nizard et al., 2017; Park et al., 2019), and may also maintain certain allergic and autoimmune diseases (Clark, 2015; Hondowicz et al., 2016; Riding and Harris, 2019). CD8+ TRM often express CD69 and may also express CD103. However, CD69 is not a perfectly reliable marker for residence (Beura et al., 2018; Walsh et al., 2019), and TRM likely express other tissue-associated markers, although this remains incompletely defined.
Residence is the dominant mechanism by which NLT are patrolled (Sathaliyawala et al., 2013; Steinert et al., 2015; Thome et al., 2014). In contrast, LNs are canonically patrolled by recirculating cells (Gowans and Knight, 1964). This includes naive T cells but also central memory T cells (von Andrian and Mackay, 2000). However, populations of CD4+ and CD8+ TRM have been reported in LNs (Beura et al., 2019; Marriott et al., 2017; Schenkel et al., 2014; Ugur et al., 2014). These appear to be rare after primary infections in mice but are potentially abundant in humans and “dirty” mice with normalized microbial experience (Beura et al., 2018; Buggert et al., 2018; Kumar et al., 2017; Sathaliyawala et al., 2013; Woon et al., 2016). CD69+ memory CD8+ T cells have been demonstrated in mediastinal LN (medLN), which drain the lung, after respiratory infections in mice (Lee et al., 2011; Takamura et al., 2010; Zammit et al., 2006). At the time, this phenomenon was attributed to recent stimulation by antigen depots recognized by recirculating memory T cells and was consistent with a model by which airway memory T cells were maintained by continual recruitment from the circulation (Ely et al., 2006; Takamura et al., 2010). Importantly, these early studies relied on perfusion to ostensibly remove vascular contaminants from the lung, a technique that fails to eliminate abundant memory CD8+ T cells within lung vasculature (Anderson et al., 2012). I.v. labeling is now a widely accepted technique to distinguish cells within blood vessels from those within tissues. Indeed, recent papers using i.v. labeling have challenged the “continual recruitment” hypothesis, demonstrating instead that lung interstitial TRM replenish waning airway epithelial memory (Takamura et al., 2019; Wein et al., 2019). Furthermore, CD8+ T cells isolated from the medLNs of influenza-immune parabiotic mice displayed a strong host bias and were CD69+ (Takamura et al., 2016). These findings raise questions of interpretation about the previously inferred ontogeny of CD69+ memory CD8+ T cells in the medLN.
Reports from our laboratory demonstrated that recall infections in the skin or reproductive mucosa result in expanded populations of TRM in draining LNs. Furthermore, we provided evidence that these cells were at one time present in the upstream NLT (Beura et al., 2018). This study left several open questions: whether cells that had committed to TRM differentiation were capable of tissue egress, whether tonic antigen or TCR stimulation was required to maintain CD69 expression, whether recall responses were required to bias secondary lymphoid organ (SLO) TRM to regional LNs, and whether primary infections could result in the regionalization of immunity within lymphoid tissue.
Several reports indicate that TRM are particularly short lived within the lung compared with other tissues (Hogan et al., 2001; Liang et al.,1994; Slütter et al., 2017; Wu et al., 2014). This impermanence may relate to the unique incompatibility of inflammation with gas exchange, and thus robust T cell responses within the respiratory mucosa may incur a fitness cost. Moreover, the fact that the lung is vascularized by narrow capillaries may afford other means of local immunosurveillance (Sakai et al., 2016). Conventionally, the rapid waning of lung TRM is attributed to death either in situ or via efflux by the mucociliary elevator (Ely et al., 2006; Slütter et al., 2017). Here we demonstrate that contraction of lung CD8+ T cell responses after influenza infection is also a result of the repositioning of lung TRM to the draining medLN via retrograde migration.
Results
A substantive subset of influenza-specific memory CD8+ T cells isolated from medLN express residency markers
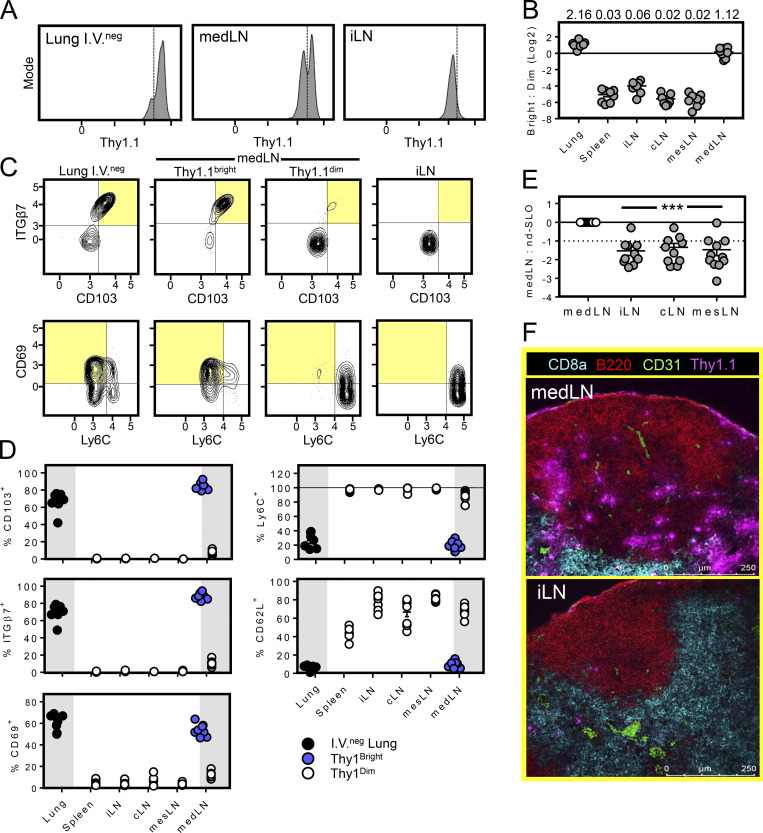
We used an adoptive transfer model to study cellular immunity to influenza virus infection. Here, naive Thy1.1+ P14 cells (transgenic CD8+ T cells recognizing H-2Db/gp33-41) were transferred to SPF C57Bl/6 recipients 1 d before i.n. infection with 500 PFU of recombinant IAV/PR8 expressing the immunodominant gp33 epitope from lymphocytic choriomeningitis virus (LCMV; PR8-gp33). We observed that the identifying P14 marker, Thy1.1, was expressed at higher levels among P14 cells in lung than all nondraining SLOs analyzed (inguinal LN shown, Fig. 1 A). However, the lung-draining medLN contained both Thy1.1bright and Thy1.1dim P14 cells (∼50% of each population 85 d after infection; Fig. 1, A and B). Further analysis revealed that Thy1.1bright P14 cells recovered from medLN expressed flow cytometry staining patterns in common with their counterparts isolated from lung tissue (i.v. staining negative), including CD103hi, ITGb7hi, CD69+, CD62Llo, and Ly6Clo. In contrast, medLN Thy1.1dim P14 cells phenocopied putative recirculating counterparts in distant SLOs (Fig. 1, C and D). Because this phenotype has been associated with residence, this raised the possibility that the medLN harbored a substantive fraction of TRM after respiratory infection and that residence correlated with elevated Thy1.1 expression. In support of this hypothesis, after PR8-gp33 infection, the relative abundance of P14 cells was higher in medLN compared with LNs that did not drain the lung, as visualized by immunofluorescence (IF) microscopy or isolation and subsequent flow cytometric analysis of mechanically dissociated LNs (Fig. 1, E and F). Anecdotally, P14 cells appeared to be overrepresented in the capsule and follicle of the medLN compared with other LNs, and Thy1.1 appeared to stain more brightly in these regions (Fig. 1 F).
Figure 1.
A substantive subset of influenza-specific memory CD8+ T cells isolated from the medLN express residency markers. (A) Mice received Thy1.1 P14 cells 1 d before i.n. infection with PR8-gp33. 85 d later, Thy1.1 expression was measured on P14 cells isolated from medLN by flow cytometry. (B) Ratio of Thy1.1bright to Thy1.1dim P14 cells isolated from the indicated tissues (plots were gated on the i.v.neg fraction of Thy1.1+ CD8+ T cells). Data displayed on a log2 scale, and numbers above bars represent ratio of Thy1.1bright to Thy1.1dim. (C and D) Phenotype of P14 cells described in A. Shaded quadrants highlight TRM phenotypes. (E) Ratio of Thy1.1+ P14 CD8+ T cells isolated from medLN versus lung nondraining SLO (40–60 d after i.n. influenza infection). Frequency of medLN P14 cells among total medLN CD8+ T cells set at 1. iLN, inguinal LN; cLN, cervical LN; mesLN, mesenteric LN. Scale is log2. (F) Histological comparison of P14 cell abundance and Thy1.1 staining intensity within the medLN and iLN 60 d after influenza infection. Data are represented as mean ± SEM. ***, P < 0.001 as determined by unpaired Student’s t test, comparing medLN to any other LN. Data are pooled from two (A–D; n = 8) or three (E; n = 11) independent experiments.
Parabiosis surgery reveals that a substantive fraction of influenza-specific memory CD8+ T cells in the medLN are TRM
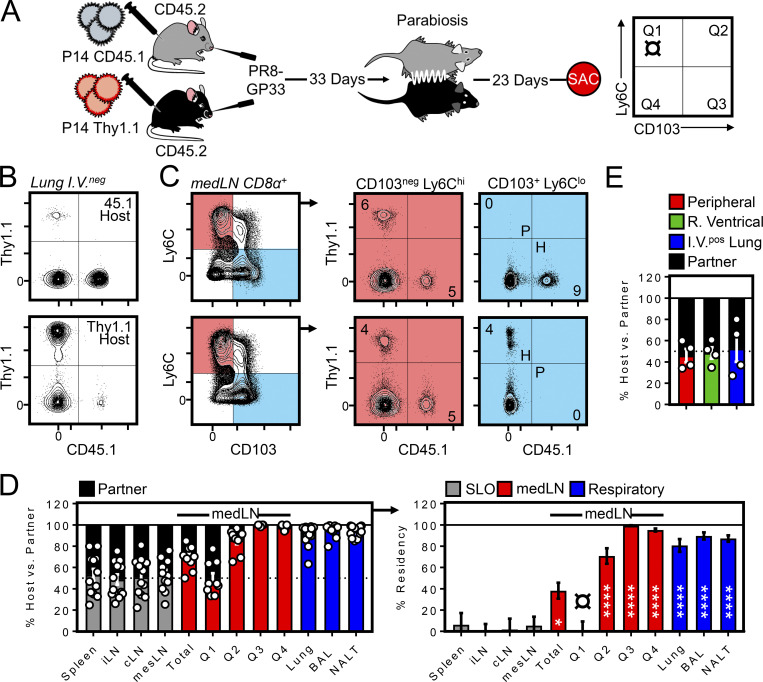
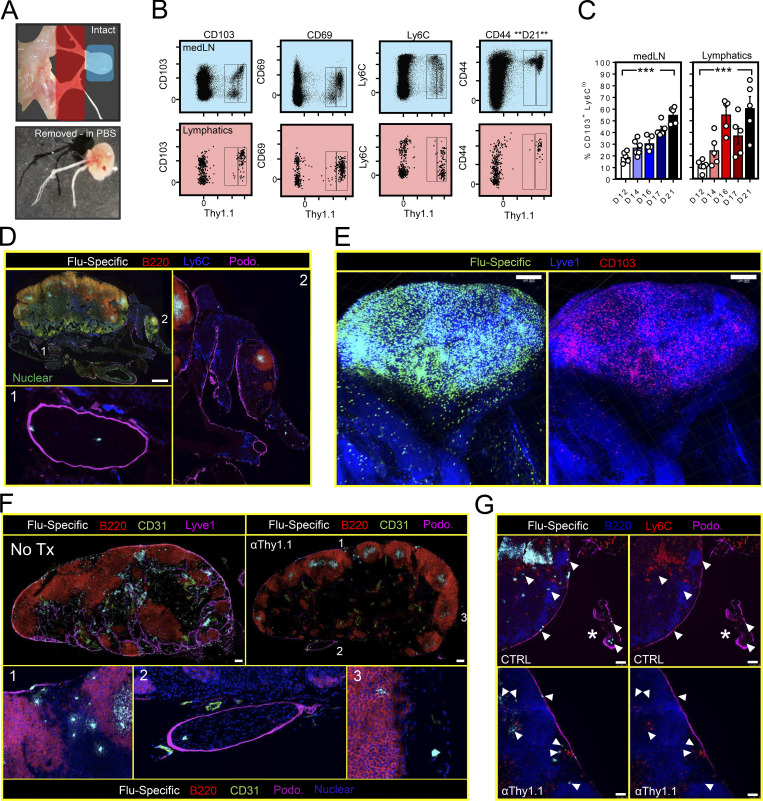
We next used a migration assay to test whether the medLN contained bona fide TRM, and if so, how that property might relate to CD103 and Ly6C expression. For this, CD45.1+ or Thy1.1+ P14 cells were transferred into separate mice followed by i.n. PR8-gp33 infection (Fig. 2 A). 33 d later, mice underwent parabiosis surgery and were conjoined for an additional 23 d, after which various tissues were assessed for the presence of P14 cells of host and partner origin. Corroborating previous reports (Takamura et al., 2019, 2016; Teijaro et al., 2011; Van Braeckel-Budimir et al., 2018; Zammit et al., 2006), the lung parenchyma and airways largely contained P14 cells of host origin (Fig. 2, B and D), indicating that lung immune surveillance was dominated by TRM in this setting. Interestingly, analysis of memory P14 cells in the medLN also revealed a significant host bias specifically attributed to CD103+ Ly6Clo cells (a population accounting for roughly half of the total host-derived medLN P14 cells in these mice; Fig. 2, C and D). In contrast, CD103neg Ly6C+ P14 cells in the medLN, in addition to P14 cells isolated from other nondraining SLOs, were fully equilibrated. The absence of Ly6C was the most accurate single predictor of residency in the medLN, although coexpression of CD103 further refined identification of this population (Fig. 2 D). Notably, P14 cells collected from the right ventricle of the heart and i.v.pos fraction of the lung were fully equilibrated, arguing against a closed migratory circuit between the lungs, medLN, and blood (Fig. 2 E). In conclusion, in addition to conventional LN patrolling by recirculating memory CD8+ T cells, surveillance of the medLNs of influenza immune mice is augmented by the presence of a substantive population of local CD103+ Ly6Clo residents. These data suggest that after local primary lung infections, immunosurveillance can be biased to regional LNs through the mechanism of residence.
Figure 2.
Parabiosis surgery reveals that a substantive fraction of influenza-specific memory CD8+ T cells in the medLN are TRM. (A) Left: C57/Bl6 (CD45.2) recipient mice received CD45.1+ P14 or Thy1.1+ P14 cells 1 d before i.n. infection with PR8-gp33. 33 d later, mice were conjoined by parabiosis for an additional 23 d before sacrifice (SAC) to allow equilibration of the circulating memory compartment between parabiotic partners. Right: Phenotype of cells to be analyzed in subsequent panels: Q1 (CD103− Ly6Chi), Q2 (CD103+ Ly6Chi), Q3 (CD103+ Ly6Clo), and Q4 (CD103− Ly6Clo). (B) I.v.neg P14 cells isolated from lungs. (C) Representative flow cytometry plots demonstrating the degree of equilibration among cells in Q1 (CD103− Ly6Chi) and Q3 (CD103+ Ly6Clo) within host medLNs. H, host-derived; P, partner-derived P14 cells. (D) Enumeration of host and partner P14 cells (left) and percentage resident (right) within the indicated tissues or phenotypic subsets present in the medLN of parabionts. Data are representative of 12 parabiotic partners (6 pairs) analyzed over two independent experiments. *, P < 0.05; ****, P < 0.0001 as determined by one-way ANOVA comparing CD103− Ly6Chi P14 cells in Q1 (denoted by symbol ¤) to any other tissue. Bars represent mean ± SEM. (E) Ratio of host- and partner-derived P14 cells isolated from peripheral blood, right ventricular blood, or the i.v.pos fraction of the lung (n = 4).
TCR-independent maintenance of CD69+ medLN TRM
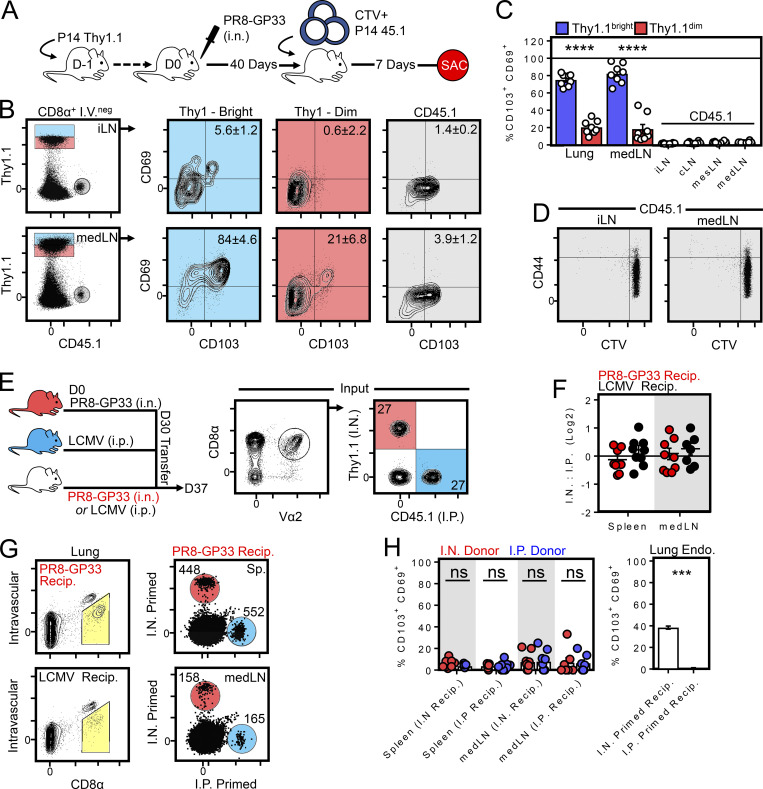
CD69 is transiently up-regulated upon TCR stimulation, and previous reports attributed the presence of CD69+ memory T cells in the medLN to transient TCR activation by a depot of residual antigen (Lee et al., 2011; Takamura et al., 2010; Zammit et al., 2006). It had also been suggested that the route of priming (i.n. vs. i.p.) influences the ability of circulating memory T cells to access residual antigen in the medLN (Takamura et al., 2010). Our observations following parabiosis indicated that CD69 expression (in addition to other markers) on memory P14 cells after influenza infection was consistent with residency but did not address the role of tonic antigen stimulation in maintaining this phenotype. Naive P14 cells were labeled with the cell-division tracking dye CTV and subsequently transferred to mice that had been infected with PR8-gp33 40 d earlier (Fig. 3 A). Fig. 3 (B–D) demonstrates that donor naive T cells (shown in gray) failed to adopt a TRM phenotype or proliferate (as assessed by dilution of CTV) in this setting, which at least suggested that CD103+ Thy1.1bright antigen–experienced P14 cells (shown in blue) could be maintained in an environment lacking sufficient antigen to sensitize naive T cells.
Figure 3.
CD69+/CD103+ memory T cells are maintained in an environment lacking sufficient antigen to sensitize naive or memory T cells. (A) Experimental strategy. 106 naive CD45.1+ P14 cells were labeled with CTV and transferred into mice infected 40 d earlier with PR8-gp33 and containing Thy1.1+ memory P14 cells. SAC, sacrifice. (B) Phenotype of Thy1.1+ (memory) and CD45.1+ (recently transferred naive cells). (C) Enumeration of CD103+ CD69+ expression among Thy1.1+ (memory) and CD45.1+ (transferred naive) cells as in B. Bars represent mean ± SEM. ****, P < 0.0001 as determined by unpaired Student’s t test. (D) CTV retention on CD45.1+ cells isolated from iLN or medLN 7 d after transfer. Data in A–D represent eight mice analyzed over two independent experiments. (E) Left: Experimental strategy to test whether i.n. priming predisposed circulating memory CD8+ T cells to adopt TRM markers by migrating into the medLN. Right: Congenically distinct memory P14s generated from i.n. PR8-gp33 (Thy1.1+) or i.p. LCMV (CD45.1+) infection were cotransferred into mice challenged 30 d earlier with i.n. PR8-gp33 or i.p. LCMV. (F) Ratio of i.n. to i.p. challenged donor cells recovered from the i.v.neg fraction of the lung and medLN of i.n. (red circles) or i.p. (black circles) challenged recipients (Recip.). (G) As in F, representative flow cytometry plots displaying the abundance of donor cells recovered from the spleen and medLN of recipient mice. Histograms are concatenated from five mice from one of two independent experiments with similar results (n = 9). (H) Left: CD103 and CD69 expression among i.n. (red circles) or i.p. (blue circles) primed cells recovered from the spleen or medLN of i.n. or i.p. challenged recipient mice. Right: Fraction of endogenous i.v.neg CD8+ T cells isolated from the lungs of i.n. or i.p. challenged recipients that express CD69 and CD103. Bars represent mean ± SEM. ***, P < 0.001 as determined by unpaired Student’s t test; ns, not significant.
To further test whether i.n. priming predisposed circulating memory CD8+ T cells to adopt TRM markers by migrating to the medLN, memory P14 cells expressing dissimilar congenic markers were isolated from the spleens of mice that were infected either i.n. with PR8-gp33 or i.p. with LCMV (Armstrong strain) and cotransferred into recipient mice challenged 30 d earlier with PR8-gp33 i.n. or LCMV i.p. (Fig. 3 E). Despite a productive lung infection in PR8-gp33 immune recipients (as evidenced by abundant i.v.neg CD8+ T cells in the lung; Fig. 3 G), transferred memory P14 cells failed to accumulate in the medLN (Fig. 3 F) or express markers associated with tissue residency (Fig. 3 H and not depicted), regardless of whether they had been primed i.n. or i.p. before transfer. These results differed from previously reported conclusions (Takamura et al., 2010), which may relate to different influenza infection models (X-31 vs. PR8 strains) or interpretation of previous data in the absence of i.v. labeling. Nevertheless, these data demonstrate that i.n. primed memory T cells do not preferentially return to their LN of origin and further support a model by which medLN TRM are maintained independently of residual influenza antigens.
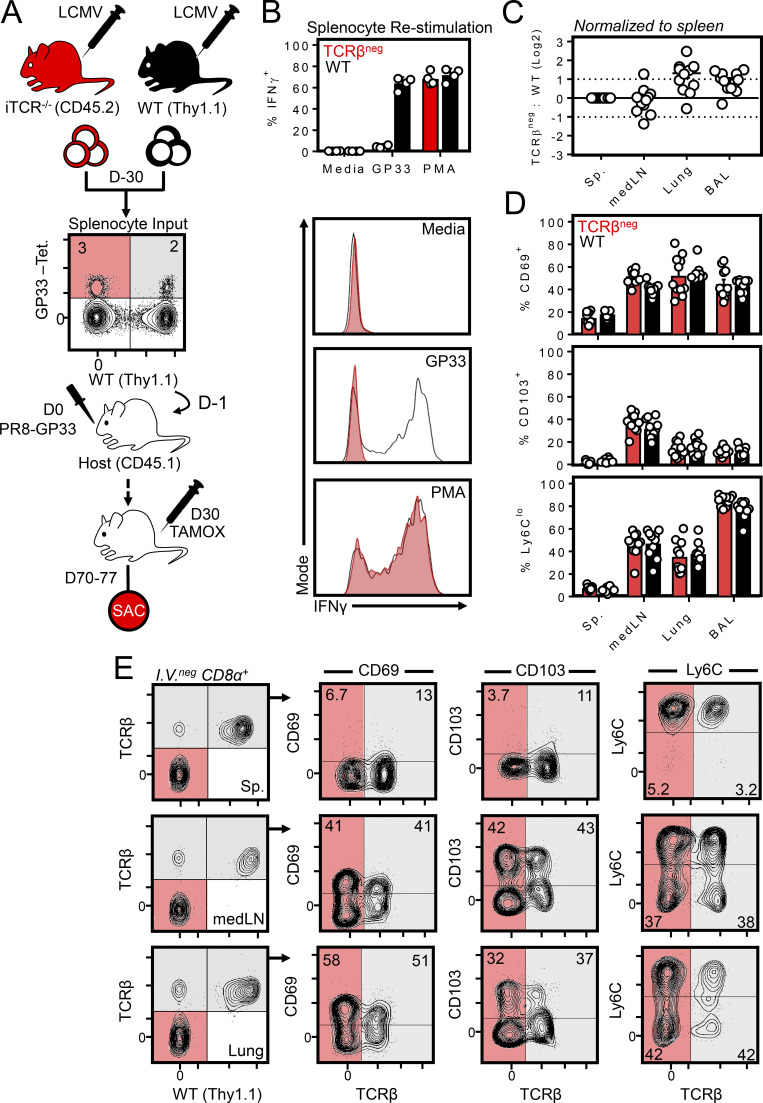
To categorically eliminate a role for residual antigen in the maintenance of CD69 expression, we used inducible ub-CreERT2 × TracFL mice (hereafter iTCR−/− mice), enabling the temporal elimination of the TCR in vivo (Levine et al., 2014). Here, it was essential to design an experiment that allowed identification of virus-specific CD8+ T cells, albeit without the use of an MHC class I tetramer (Fig. 4 A). Thy1.1+/CD45.2+ WT and Thy1.2+/CD45.2+ iTCR−/− mice were each infected with LCMV, resulting in the expansion of polyclonal gp33-specific CD8+ T cells. 30 d later, splenocytes from both mice were combined such that the ratio of gp33-specific cells of WT or iTCR−/− origin was ∼1:1 (as determined by tetramer staining). Cells were then cotransferred into naive CD45.1+ recipients and infected 1 d later with PR8-gp33 (i.n.) to induce a secondary response among only gp33-specific donor CD8+ T cells. 30 d later, tamoxifen was administered over the course of a week to selectively ablate the TCR among iTCR−/−-derived cells, although these CD8+ cells could still be identified by Thy1.1+/CD45.2+ phenotype and lack of TCRβ (Fig. 4 E). In vitro peptide stimulation confirmed that iTCR−/− cells were refractory to peptide-induced IFNγ production and thus lacked a functional TCR, although they did produce IFNγ in response to PMA (Fig. 4 B). Despite this, 40 d after tamoxifen treatment, iTCR−/− and WT cells were present at equivalent ratios (Fig. 4 C) and expressed identical levels of TRM markers including CD69, CD103, and Ly6Clo (Fig. 4, D and E). These data indicated that medLN TRM could be maintained independently of tonic antigen or TCR stimulation.
Figure 4.
MedLN TRM are maintained independently of TCR. (A) Experimental strategy for generating gp33-specific WT and TCRβneg cells from a polyclonal repertoire. WT and ub-CreERT2 × TracFL mice (referred to here as iTCR−/− mice) were infected with LCMV i.p., and splenocytes were isolated 30 d later and combined such that the frequency of tetramer+ gp33-specific CD8+ T cells of WT or iTCR−/− origin were ∼1:1. Red shaded quadrants indicate iTCR−/−-derived tetramer+ cells; gray shaded quadrants represent WT-derived tetramer+ cells. Cells were cotransferred into congenically distinct recipient mice subsequently infected with PR8-gp33 i.n. to establish secondary memory medLN TRM from both donor sources. 30 d later, mice were treated with tamoxifen daily for 7 d to ablate the TCR specifically from iTCR−/−-derived cells. Mice were sacrificed (SAC) 40 d after the induction of tamoxifen treatment. (B) In vitro peptide stimulation of WT and iTCR−/− cocultures confirmed TCRβneg cells were refractory to peptide reactivation but could produce IFNγ through a TCR-independent mechanism (PMA). Data are from one of two independent experiments with similar results. (C) Ratio of TCRβneg: WT cells in the spleen, medLN, lung, and brochoalveolar lavage. (D and E) Frequency of CD103+, CD69+, and Ly6Clo cells among transferred WT or TCRβneg memory CD8+ T cells within the indicated tissues. Gray shade indicates WT cells; red indicates iTCR−/− cells in which the TCR was successfully ablated (as indicated by an absence of TCRβ). iTCR−/−-derived cells that did not undergo loss of TCR expression after tamoxifen treatment were included within analysis of WT cells. C and D pooled from 10–12 mice from three independent experiments; E shows representative flow cytometry plots. Bars represent mean ± SEM.
Evidence that antigen in the lung is required for medLN TRM formation
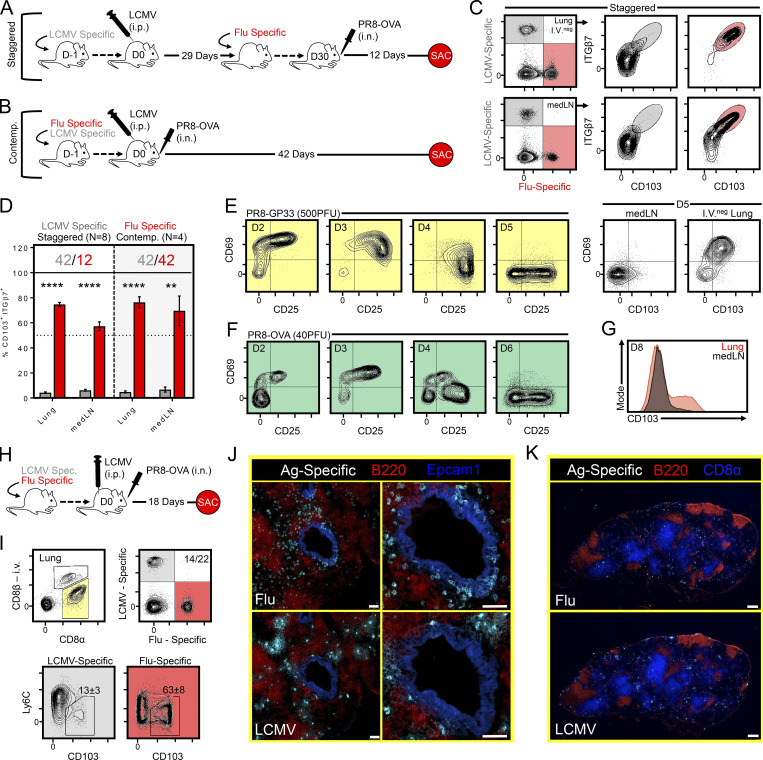
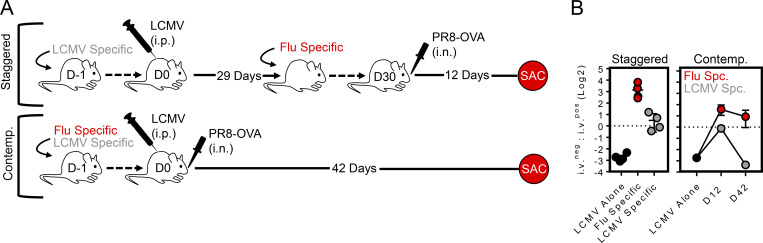
We next asked if strong local inflammation and recruitment into the lung parenchyma was sufficient to induce the formation of medLN TRM. Two infection modalities, termed “staggered” and “contemporaneous,” were applied to elicit cells of an irrelevant specificity (in this case, LCMV-specific P14 cells) into the inflamed lung. LCMV Armstrong delivered i.p. was chosen because it induces little infection of the lung, and it does not generate an appreciable population of lung-resident memory CD8+ T cells (Anderson et al., 2012; Beura et al., 2015; Steinert et al., 2015). With the staggered approach, LCMV immune mice (29 d after infection) harboring circulating memory P14 cells were seeded with OT-I cells (transgenic CD8+ T cells recognizing H-2Kb/SIINFEKL) and subsequently infected with 40 PFU of recombinant PR8 expressing the SIINFEKL epitope from ovalbumin (PR8-ova; Fig. 5 A). 12 d later, while numerous P14 cells gained access to the lung parenchyma (approximately nine times more than i.p. LCMV infection alone; see Fig. S1), only OT-I cells acquired a TRM phenotype (Fig. 5, C and D), consistent with a previously proposed model for lung TRM differentiation (McMaster et al., 2018). Alternatively, mice were coseeded with P14 and OT-I cells and contemporaneously infected with LCMV i.p. and PR8-ova i.n. (Fig. 5 B). This experimental design similarly recruited abundant P14 cells into the lung, presumably because of inflammation-induced bystander recruitment, and thus tested the hypothesis that recently primed (LCMV-specific CD8+ T cells in this case) cells would be permissive to TRM differentiation simply by migrating to an infected lung. However, only OT-I cells became TRM in both the lung and medLN when assessed 42 d later (Fig. 5 D). The provision of i.n. gp33 antigen during contemporaneous infection did not enhance the ability of P14 cells to accumulate or convert to a TRM phenotype in the medLN (Fig. S2). Moreover, activation-induced expression of CD69 (and CD25) was transient on recently activated influenza-specific CD8+ T cells within the medLN, and CD103 expression first appeared on CD8+ T cells within the i.v.neg fraction of the lung, rather than the medLN (Fig. 5, E and G). These observations indicate that the TRM differentiation program is induced in the lung rather than the draining LN. Collectively, these experiments indicated several criteria regarding the formation of medLN TRM: (a) local inflammation is insufficient to generate medLN TRM, (b) antigen specificity played an essential role in TRM differentiation but not maintenance, (c) recruitment into the lung does not forecast medLN TRM formation, and (d) medLN TRM develop only after corresponding TRM-phenotype CD8+ T cells are present in the upstream tissue, suggesting that this differentiation program is not acquired in the LN.
Figure 5.
Evidence that antigen in the lung is required for medLN TRM formation. (A and B) Staggered (A) and contemporaneous (Contemp.; B) approach for recruiting LCMV-specific cells into the inflamed lung. (C) Representative flow cytometry plots of LCMV-specific (gray) or influenza-specific (red) CD8+ T cells gated on the i.v.neg fraction isolated from lung or medLN after staggered infection. (D) Frequency of LCMV-specific (gray) and influenza-specific (red) cells within the i.v.neg fraction of the lung or medLN that coexpressed integrin-β7 and CD103 following staggered or contemporaneous infection modalities. Numbers above the bars represent days after infection for either LCMV-specific (gray) or PR8-ova–specific (red) CD8+ T cells. Data collected over two independent experiments (n = 8). Bars represent mean ± SEM. **, P < 0.01; ****, P < 0.0001 as determined by unpaired Student’s t test. (E–G) Dynamics of CD69 and CD25 expression on P14 and OT-1 cells within the medLN following infection with PR8-gp33 (E) or PR8-ova (F) at the indicated time points. Expression of CD69 and/or CD103 in the indicated tissue among transgenic i.v.neg CD8+ T cells after PR8-gp33 (E, right plots, day 5) or PR8-ova (G, day 8). Mice received 5 × 105 transgenic CD8+ T cells before infection to detect cells in the medLN at early time points. (H–K) Mice were contemporaneously challenged with LCMV i.p. and PR8-ova i.n. (as in B), and OT-I (red) and P14 (gray) cells were examined 18 d later. Spec., specific. (I) Phenotype by flow cytometry. Numbers in quadrants represent mean ± SEM from a three-mouse experiment performed twice with similar results (Fig. S2). (J and K) Distribution of influenza- and LCMV-specific cells by IF microscopy in the lung (J; enlarged images on right) and medLN (K). Scale bars represent 50 µm in J and 300 µm in K.
Figure S1.
PR8 infection recruits LCMV-specific cells to the lung. (A) Staggered and contemporaneous (Contemp.) infection strategy. SAC, sacrifice. (B) LCMV alone (black circles) indicate i.v.neg P14 cells in the lung 40–60 d after i.p. LCMV. After the staggered infection modality, 30 d after LCMV infection, mice received OT-I cells and PR8-ova i.n. 12 d later. OT-I (red) and P14 (gray) cells were enumerated in lung. Contemporaneous infection similarly recruited abundant LCMV-specific P14 cells into the lung early after infection (day 12); however, their maintenance was not sustained by 42 d after infection. Data representative of four mice per group from one of two independent experiments. Spc, specific.
Figure S2.
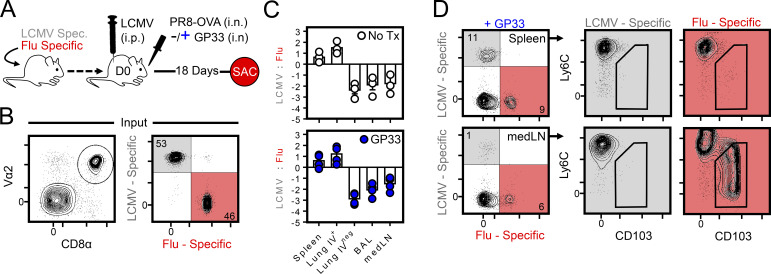
Intranasal LCMV antigen was not sufficient to induce medLN TRM after i.p. LCMV infection. (A) Strategy for determining if local antigen presentation in the medLN could drive the formation of medLN TRM de novo. Mice were seeded with P14 and OT-I cells and contemporaneously infected with LCMV i.p. and PR8-ova i.n. ± 3 µg gp33 peptide within the i.n. PR8-ova inoculum. SAC, sacrifice; Spec., specific. (B) Cotransferred transgenic T cells (both P14 and OT-I are Vα2+). (C) Ratio of P14 to OT-I cells ± i.n. gp33 peptide (log2 scale). Solid line indicates 1:1 ratio. While P14 and OT-I cells were similarly expanded in circulation, LCMV-specific cells were underrepresented in the medLN and i.v.neg respiratory tissues. (D) Phenotyping of P14 and OT-I cells recovered from the spleen or medLN of contemporaneously challenged mice given i.n. gp33 peptide at time of infection as in C. n = 3 without gp33; n = 4 with gp33. Bars represent mean ± SEM.
Using IF microscopy, we next asked whether P14 cells elicited into the lungs 18 d after contemporaneous infection were differentially localized with respect to their OT-I cell counterparts (Fig. 5 H). A portion of the lung was reserved for flow cytometry to confirm that P14 and OT-1 cells were phenotypically dissimilar with respect to canonical TRM markers, as previously observed (Fig. 5 I). When analyzing serial lung sections, we identified abundant influenza-specific OT-I cells interspersed within Epcam1+ airway epithelium. Comparatively, LCMV-specific P14 cells were underrepresented in airway epithelium, despite distribution throughout other regions of the lung (Fig. 5 J and not depicted). No apparent differences in localization were observed in the medLN after contemporaneous infection (Fig. 5 K). These observations inspired the hypothesis that the decision to become medLN TRM may have occurred in the lung; if so, that would suggest migration of TRM or their precursors occurs from lung to draining LN following a primary immune response.
CD8+ T cells bearing residency markers are abundant within airway epithelium and appear to migrate to medLN via lymphatics
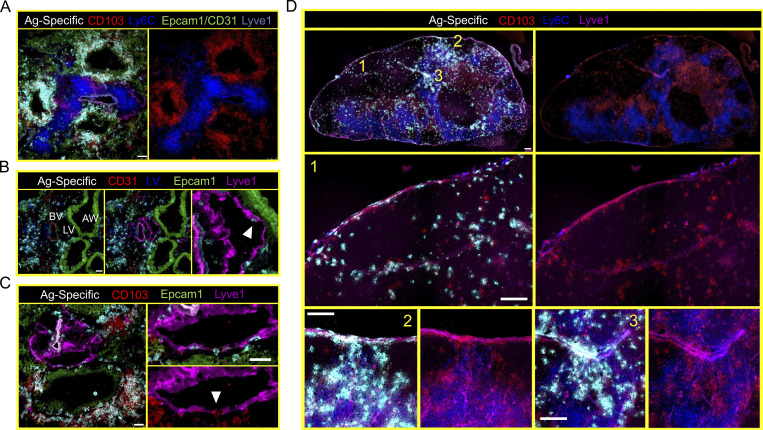
The observation that LCMV-specific cells that were recruited into the lung failed to access airway epithelium or express CD103 suggested that the airway epithelium might be a crucial TRM induction site. Supporting this hypothesis, dense aggregates of CD103+ Ly6Clo influenza-specific cells could be found within and surrounding Epcam1+ airway epithelium by IF microscopy 18 d after infection (Fig. 6 A). In contrast, CD103− Ly6Chi cells were localized away from the epithelium, in stromal regions also occupied by both blood (CD31+) and lymphatic (Lyve1+) vessels. Lyve1+ lymphatic vessels abutted Epcam1+ airway epithelium, and we observed occasional influenza-specific CD8+ T cells within these vessels (Fig. 6 B). Intralymphatic cells were often CD103+ and Ly6Clo; however, we did not analyze enough cells to confidently enumerate this phenotype by IF microscopy (Fig. 6 C). Nevertheless, these data suggested to us that TRM-phenotype cells might egress from the lung through lymphatic conduits to later occupy the draining medLN. Indeed, abundant CD103+ Ly6Clo influenza-specific CD8+ T cells could be found in or near Lyve1+ vessels within the subcapsular sinus and interfollicular areas of the medLN (Fig. 6 D). The location of these Lyve1+ vessels corresponded with regions of the LN associated with afferent (tissue-draining) lymphatics (Reynoso et al., 2019).
Figure 6.
CD8+ T cells bearing residency markers are abundant within airway epithelium and appear to migrate to medLN via lymphatics. (A) Localization of antigen-specific CD8+ T cells in the lungs of mice infected 18 d earlier with influenza virus revealed abundant CD103+ Ly6Clo cells within and surrounding Epcam1+ airway epithelium. (B) Characterization of blood vessels (BV), lymphatic vessels (LV), and airway epithelium (AW) in the lungs of influenza-immune mice. Blood vessels typically costained with CD31 and Lyve1, while lymphatic vessels stained solely with Lyve1. Arrow highlights an antigen-specific CD8+ T cell within the lumen compartment of the lymphatic vessel. (C) Lymphatic vessels were commonly found abutting airway epithelium (also observed in B). Arrow highlights an antigen-specific CD103+ CD8+ T cell traversing the lymphatic endothelium. Regions of airway epithelium contacting Lyve1+ lymphatic vessels were often sparse for antigen-specific CD8+ T cells. (D) Representative IF microscopy of the medLN of a mouse infected 18 d earlier with PR8-ova. Note antigen-specific CD103+ Ly6Clo CD8+ T cells potentially infiltrating medLN through regions of the LN associated with afferent lymphatics and colocalizing with Lyve1 staining. Numbers orient regions of the medLN enlarged below. Antigen-specific cell staining was removed from the right duplicate images to better display the CD103+ Ly6Clo phenotype. Scale bars represent 50 µm (A–C) or 200 µm (D). Representative images are from at least three sections per tissue, per mouse, of three or more individual mice.
Retrograde migration from the lung supplies medLN TRM
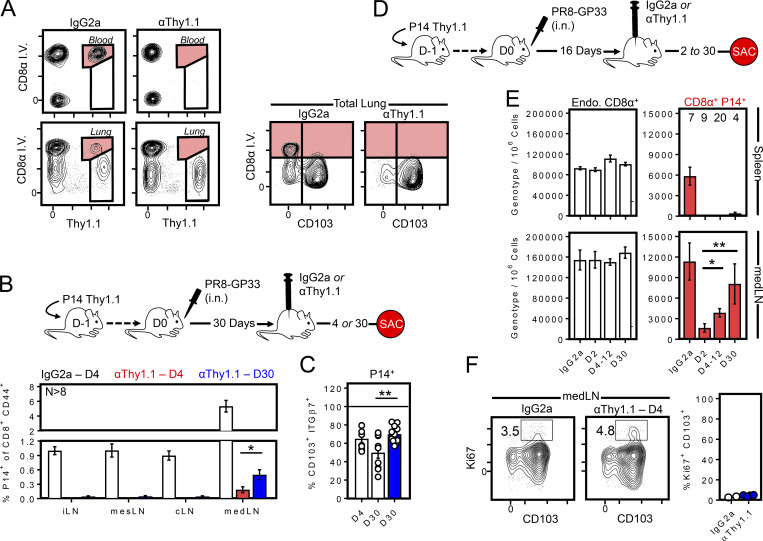
Histological analysis suggested that medLN were emigrating from the upstream tissue via lymphatic conduits. However, we aimed to formally test whether T cells within the lung engendered medLN TRM through a more stringent flow cytometry–based approach. We found that administration of 1 µg of α-Thy1.1 antibody i.p. (HIS51, BD Pharmagen) efficiently eliminated memory P14 cells from blood, lung vasculature, spleen, and all LNs (regardless of TRM phenotype) while leaving those in the lung parenchyma protected (Fig. 7, A and B; and not depicted). As in previous studies using this approach (Schenkel et al., 2013), we used MHC class I tetramer staining to confirm that the inability to detect blood-borne P14 cells 4 d after treatment was a consequence of P14 cell depletion rather than blocking Thy1.1 staining (data not shown). This approach allowed us to test whether depleted medLN TRM would be reconstituted over time, which would be congruent with migration from the undepleted lung. Thy1.1+ cells were depleted 30 d after PR8-gp33 infection. We found that P14 cells remained efficiently depleted from all LNs examined, including the medLN, 4 d after α-Thy1.1 antibody treatment. However, over the following month, P14 cells reappeared almost exclusively in the medLN. It was notable that a greater fraction of these newcomers were CD103+ compared with the total P14 cell population isolated from the medLN of undepleted infection-matched mice (Fig. 7 C, day 30 white vs. blue bar). These observations lent further support to the hypothesis that medLN TRM arise from T cells that emigrate from the lung and indicate that this process occurs for weeks to possibly months after clearance of the infection. We term this retrograde migration. Moreover, many emigrants appear to be retained within the draining medLN rather than immediately rejoining the circulation. In other words, a residence program was preserved. We next treated mice with α-Thy1.1 antibody (as in Fig. 7, B and C) earlier (16 d) after infection to test whether retrograde migration was more pronounced during the contraction phase of the primary immune response (Fig. 7 D). Here we noted significant repopulation of medLN (but not other LNs or spleen) between day 18 and 20–30 of PR8 infection (days 2 to 4–12 after P14 cell depletion; Fig. 7 E and not depicted). P14 cells continued to accumulate exclusively in the medLN between days 12 and 30 after infection. Ki67 staining indicated that repopulating cells had not undergone recent cell division, further indicating a process of retrograde migration, and highlighting that this process was even more pronounced early after infection than later (Fig. 7, B–F).
Figure 7.
Evidence that retrograde migration from lung supplies medLN TRM. (A) 30 d after influenza infection, Thy1.1+ P14-bearing mice were treated with 1 µg α-Thy1.1 depleting antibody or IgG2a (control), then analyzed 4 d later for the compartmentalization and phenotype of remaining P14 cells. (B) As in A, except the frequency of P14 cells among total activated CD8+ T cells was assessed 4 or 30 d later. Data are graphed as percentage Thy1.1+ (P14) of CD44+ CD8+ T cells. *, P < 0.05 as determined by Student’s t test. (C) CD103 expression among P14 cells isolated from medLN in B, comparing Thy1.1-treated mice (blue) to control IgG2a-treated mice at the indicated time points after antibody treatment. Data are combined from three independent experiments with 8–12 mice per group. **, P < 0.01 as determined by one-way ANOVA. (D and E) Mice were treated with α-Thy1.1 antibody 16 d after influenza infection, and P14 cells were enumerated 2, 4–12, and 30 d later. Data are pooled from two independent experiments. Numbers above bars indicate replicates per group. Endo. (total non-P14 endogenous CD8+ T cells) serves as a control Thy1.1neg population. *, P < 0.05; **, P < 0.01 as determined by an unpaired Student’s t test comparing D2 after depletion to days 4–12 and day 30 after depletion. Similar results were observed in a third independent experiment comparing medLN P14 cell abundance 4 and 30 d after D16 depletion (n ≥ 4, P = 0.038; not depicted). (F) Ki67 staining of P14 cells recovered from the medLN in α-Thy1.1–treated and control mice. Bars represent mean ± SEM.
We next determined whether we could visualize cells in the act of retrograde migration. We first isolated medLN-associated lymphatics from mice infected 16 d earlier with PR8-gp33 (Fig. 8 A). By flow cytometry, these vessels were highly enriched for the subset of P14 cells that were Thy1.1bright, expressing canonical markers of TRM including CD103+, CD69+, and Ly6Clo (Fig. 8 B). Additionally, the abundance of TRM-phenotype cells significantly increased within the medLN and associated lymphatics over a 9-d period spanning days 12–21 after infection (Fig. 8 C). In thin histological sections, Ly6Clo P14 cells were also identified within podoplanin+ lymphatic vessels surrounding the medLN (Fig. 8 D). We next used clearing-enhanced 3D imaging (Ce3D) of the medLN and intact associated lymphatic vessels (Li et al., 2017). As shown in Fig. 8 E, CD103+ P14 cells were present within medLN-associated lymphatics (Video 1). These further supported the hypothesis that medLN TRM result from retrograde migration from the lung. Confirming this interpretation, when P14 cells were depleted from mice infected 16 d earlier with PR8, newcomer P14 cells (present 4 d later) could be observed in or near podoplanin+ vessels and within subcapsular sinus or interfollicular regions that, based on LN orientation, are associated with afferent lymphatics (Fig. 8 F; Reynoso et al., 2019). Fig. 8 G shows that P14 cells putatively repopulating medLN are Ly6Clo (top row undepleted, bottom row 4 d after depletion). We conclude that lung T cells that have adopted (or have become committed to adopt) signatures of TRM differentiation undergo retrograde migration from the lung and establish residence within the draining medLN.
Figure 8.
Visualizing retrograde migration. (A and B) Representative example of the medLN (blue shade) and medLN-associated lymphatics (red shade) dissected out for flow cytometric analysis 16 d after P14 transfer and i.n. PR8 infection. (C) Frequency of P14 cells expressing a CD103+ Ly6Clo phenotype isolated from medLN (left) or associated lymphatic vessels (right) 12–21 d after infection with PR8-gp33 (n = 5 or 6 mice per time point). Bars represent mean ± SEM. ***, P < 0.001 as determined by Student’s t test comparing day 12 to day 21. (D) 18 d after P14 cell transfer and i.n. PR8-gp33 infection, IF microscopy of the medLN and associated lymphatic vessels. Arrows indicates Ly6Clo P14 cell in lymphatic vessels. Scale bar represents 500 µm. (E) Ce3D and two-photon microscopy of the medLN and associated lymphatics 16 d after PR8 infection (Video 1). Scale bar represents 200 µm. (F) Mice infected with influenza virus 16 d earlier were treated with α-Thy1.1–depleting antibody and compared with untreated (No Tx) controls 4 d later. Numbers in top right image indicate regions enlarged in the panels below. Scale bars represent 200 µm. (G) As in F, Ly6Clo P14 cells could be identified at the medLN periphery and directly within medLN-associated lymphatics (marked by asterisk) in both untreated mice (top) and mice treated 4 d earlier with α-Thy1.1–depleting antibody (bottom). Arrows highlight influenza-specific cells that are Ly6Clo/neg. Scale bars represent 50 µm. Representative images are from at least three sections per tissue, per mouse, of three or more individual mice.
Video 1.
Ce3D of the medLN and associated lymphatics of a mouse infected 16 d earlier with PR8-gp33. Isolated medLN and associated lymphatics were made optically transparent as previously described (Li et al., 2017). Cleared tissue was imaged by tiling Z-stacks with confocal laser scanning microscopy (Leica SP5). 3D reconstructions were done using Imaris (Bitplane) software v9.2.1, and GFP+ P14 cells were displayed through a surface mask. Green, GFP+ P14 CD8+ T cells; magenta, CD103; blue, Lyve1. Corresponding still images from this video are displayed in Fig. 8 E. Images are representative of three Ce3D-processed tissue specimens comprising the lung, medLN, and medLN-associated lymphatics.
medLN TRM durably increase regional memory
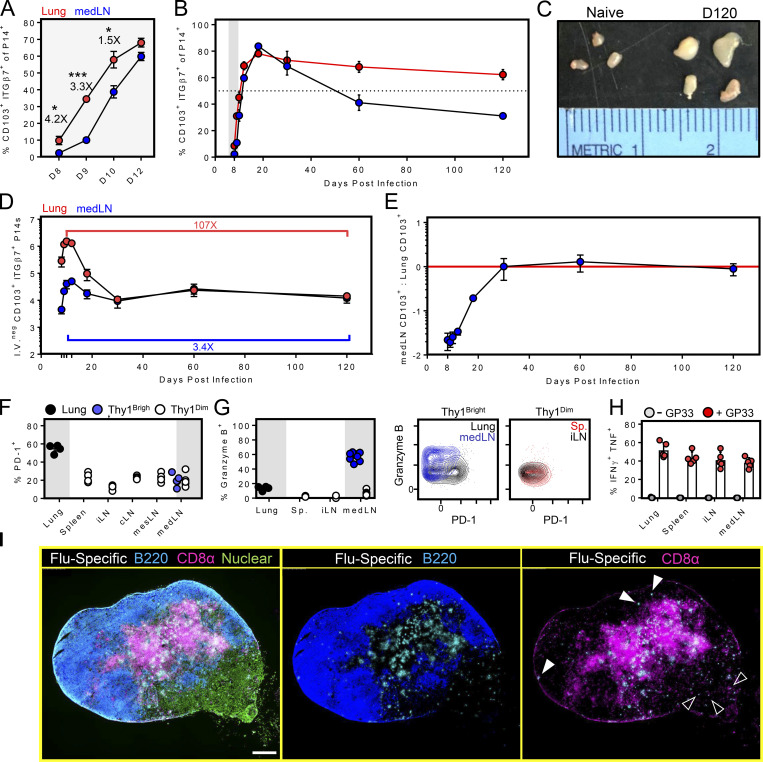
Durable maintenance is a defining function of memory T cells. We found that there was a lag between the appearance of CD103+ cells within the medLN compared with the lung (Fig. 9 A) consistent with the origin of medLN TRM in the upstream tissue. However, once present in the medLN, CD103 expression was durably maintained (Fig. 9 B). medLN hypertrophy was also noted at all time points analyzed (Fig. 9 C). We observed a precipitous contraction of virus-specific memory CD8+ T cells within the lung, in agreement with other reports, but little contraction in the medLN (Fig. 9, D and E). These observations support a model by which retrograde migration results in the displacement of T cell immunity from the lung to the afferent LN. This mechanism may preserve a component of regional immunity, albeit not directly at the site of influenza infection of respiratory epithelium. It also indicates pliancy in the homeostasis of lung-draining lymphoid tissue. medLN TRM were not senescent or terminally differentiated, as they proliferated and lost CD103 expression following either in vitro peptide restimulation or adoptive transfer and subsequent i.p. LCMV infection (Fig. S3). medLN P14 TRM also underwent robust proliferation after secondary i.n. infection with vaccinia virus expressing gp33 (Fig. S4).
Figure 9.
medLN TRM durably increase regional memory. P14 cells were transferred to naive mice, followed by PR8-gp33 influenza virus infection i.n. (A and B) Dynamics of CD103 and ITGβ7 expression among P14 cells isolated from medLN and lung. Numbers in A represent fold difference between lung and medLN. *, P < 0.05; ***, P < 0.001 as determined by an unpaired Student’s t test. (C) Comparison of medLN size between naive mice and mice infected 120 d earlier with PR8-gp33. (D and E) Enumeration of ITGβ7+ CD103+ P14 cells isolated from the lung and medLN. Data representative of at least four mice per time point. Error bars represent mean ± SEM. (F and G) PD-1 and granzyme B expression on memory P14 cells isolated from the indicated tissues 85 d after infection, as determined by flow cytometry. Similar results were obtained from mice analyzed 30 d earlier. (H) As in F, percentage of P14 cells that produce both IFNγ and TNFα after 4-h ex vivo peptide restimulation. Bars represent mean ± SEM. (I) IF imaging of the medLN 120 d after PR8-gp33 infection. Closed arrowheads indicate bright Thy1.1 staining; open arrowheads indicate less intense Thy1.1 staining.
Figure S3.
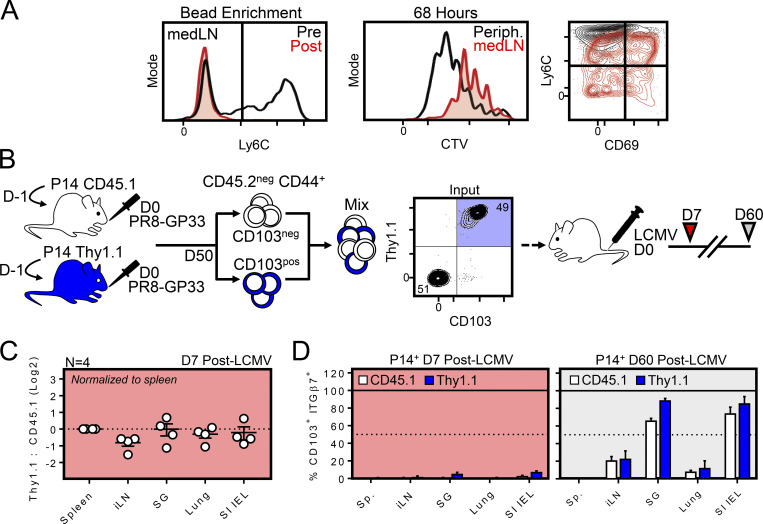
Evidence that medLN TRM can participate in a secondary response. (A) In vitro proliferation of bead-enriched, Ly6Cneg medLN P14 cells compared with Ly6C+ P14 cells pooled from peripheral (Periph.) LNs. Left: Ly6C phenotype of medLN P14 cells before (black) and after (red) bead enrichment. Right: CTV dilution and phenotype of medLN TRM 68 h after in vitro peptide stimulation. Data are representative of two independent experiments. (B) Congenically distinct circulating (CD103neg CD45.1) and resident (CD103+ Thy1.1) memory P14 cells were sorted from the medLN using an approach obviating Thy1.1 (depleting) antibody. Host medLN CD8+ T cells were excluded based on CD45.2 expression. (C) Abundance of Thy1.1 and CD45.1 P14 cells in the indicated tissues 7 d after systemic LCMV infection. (D) CD103 and integrin-β7 phenotype on CD45.1 and Thy1.1 P14 cells isolated from the indicated tissues 7 and 60 d after systemic LCMV infection (n = 4). Error bars represent mean ± SEM.
Figure S4.
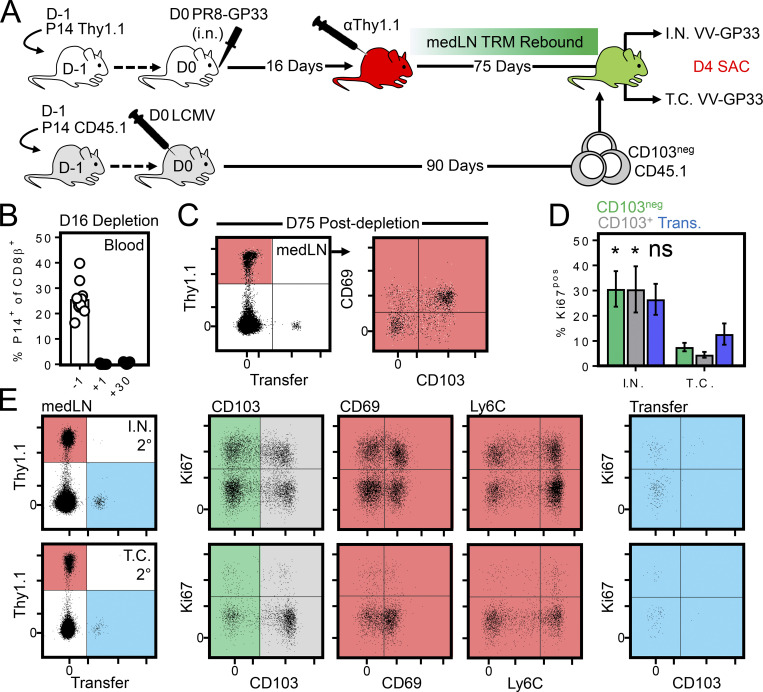
Unmanipulated medLN TRM proliferate after secondary lung challenge. (A) Influenza-immune mice were depleted of circulating P14 cells 16 d after infection by injection of αThy1.1-depleting antibody. 75 d later, mice received infection-matched congenically distinct circulating memory P14 cells. Mice were then rechallenged i.n. or transcervically with VV-gp33 1 d later. (B) P14 Thy1.1 cell abundance within blood 1 d before, 1 d after, and 30 d after αThy1.1 antibody depletion. (C) Phenotype of medLN P14 cells 75 d after αThy1.1 antibody depletion. Mice had received circulating CD45.1 P14 cells 1 d earlier. (D) Percentage of Ki67+ cells among CD103neg, CD103+, and CD45.1 (circulating) medLN P14 cells 4 d after i.n. or t.c. VV-gp33 challenge. *, P < 0.05, as determined by an unpaired Student’s t test comparing cells of the indicated phenotype after i.n. or transcervical rechallenge; ns, not significant. (E) As in D, representative flow cytometry plots displaying the degree of proliferation among medLN P14 cells bearing a CD103+, CD69+, or Ly6Clo phenotype. Data are pooled from two independent experiments (n = 9 for i.n. rechallenge; n = 6 for transcervical rechallenge). Error bars represent mean ± SEM.
It has been proposed that lung immunity wanes. It has also been reported that lung memory T cells express elevated levels of inhibitory receptors that limit their pathogenic capacity (Hombrink et al., 2016; Wang et al., 2019); indeed, the potential for immunopathology at delicate sites of gas exchange is one proposed explanation for why the lung may not durably retain TRM. We confirmed PD-1 expression on lung memory P14 cells even 85 d after PR8 infection. However, medLN P14 cells did not retain PD-1, suggesting they may be alternatively regulated with respect to their lung counterparts (Fig. 9 F). Indeed, while cells from all locations appeared competent to express IFNγ and TNFα upon restimulation, medLN TRM uniquely maintained granzyme B expression (unlike lung or other SLOs; Fig. 9, F–H). We addressed sublocalization within the medLN histologically. Although P14 cells were evident throughout the medLN in mice infected 120 d earlier with influenza virus (Fig. 9 I), those near the follicles or in the subcapsular region appeared to be Thy1.1bright (closed arrowheads), while those associated with the medullary sinus appeared to be Thy1.1dim (open arrowheads). This may indicate even further complexity of regional immune surveillance by CD8+ T cells within the medLN.
Taken together, these data support a model by which contraction of lung TRM may be partially counterbalanced by their repositioning into the draining LN, resulting in a durable augmentation in regional, but not frontline, mucosal immunity.
Discussion
Tissue residency (TRM) is a mechanism for regionalizing immunosurveillance through the biasing of memory T cell distribution to specific sites most associated with exposure to infection. While CD8+ TRM are hypothesized to be long-lived in most tissues, it has been well documented that lung TRM undergo rapid attrition after resolution of influenza infection in mice. Similar observations have been made after Sendai infection in mice (Ely et al., 2006; Hikono et al., 2007; Hogan et al., 2001) or after respiratory syncytial virus infection in humans (Jozwik et al., 2015), suggesting this may be a general feature of the lung. Proposed explanations include a predilection for local T cell death, removal of T cells via the mucociliary elevator, limited interstitial space that may constrain cellular carrying capacity, an extensive vascular network that may afford additional mechanisms of pulmonary surveillance, and an immunosuppressive environment that discourages local T cell proliferation and perhaps sustained retention of memory cells (Borron et al., 1998; Reagin and Klonowski, 2018; Van Braeckel-Budimir and Harty, 2017). Indeed, efficient gas exchange depends on delicate architecture and is inflammation intolerant. Waning T cell immunity within the respiratory tract may safeguard a particularly vulnerable tissue against damaging immune responses directed against microbes and innocuous environmental antigens that access the airways in the estimated 4 × 108 liters of air inhaled over the average human lifespan.
The results of our current study, based on parabiosis, subset-specific depletion, imaging, and lymphatic cell isolation and phenotyping, demonstrate an additional mechanism by showing that attrition of T cell memory in the lung is contemporized by retrograde migration, and that a substantive fraction of emigrants retain a residency program within the draining medLN. This process may provide a means for durably augmenting regionalized T cell memory, albeit not at the immediate front lines of typical pathogen exposure within the respiratory mucosa. This strategy may be less effective at contributing to rapid protection but may be an evolved compromise that mitigates the potential for reactivated T cell–driven immunopathology after elevated pulmonary innate immunity subsides.
Our data indicate that TRM precursors leave the lung despite having up-regulated markers associated with retention. Moreover, bystander cells recruited into the inflamed lung failed to form TRM in either the lung or medLN. In our interpretation, these data favor a model in which recently activated T cells are programmed to undergo TRM differentiation within regions of the lung itself (perhaps including infected sites within epithelium), rather than adopting a TRM differentiation program in the LN. It should be noted that this model is different from previously proposed models attributing the presence of CD69+ cells in the medLN to circulating memory cells transiently activated by a depot of residual antigen (Lee et al., 2011; Takamura et al., 2010; Zammit et al., 2006). While respiratory infections can result in persistent antigen, in our study, removal of TCR expression did not diminish maintenance of CD69 expression or the retention of memory CD8+ T cells within the medLN. Our findings also suggest that medLN TRM are durably maintained in the medLN without significant displacement or turnover from new retrograde migrants, because P14 cells did not repopulate other LNs over a 30-d period following α-Thy1.1 antibody depletion. With enough time, it may be possible that medLN TRM lose expression of canonical residency markers and rejoin the circulating pool. However, we failed to observe any significant attrition of CD103+ P14 cells in either the lung or medLN 30–120 d after PR8-gp33 infection, suggesting durable retention within the medLN for at least 4 mo after infection.
This study raises several questions related to (a) the mechanisms of tissue egress, (b) the dynamics of commitment to tissue residency, and (c) the function of LN TRM. Addressing the last question would benefit from the development of techniques that deplete all circulating cells and lung TRM while preserving medLN TRM. We did observe that medLN TRM retained granzyme B, suggesting that they may be poised for cytolytic function in LN upon reinfection. This does not rule out the hypothesis that medLN TRM might have an enhanced capacity to repopulate TRM within the lung after restimulation, contribute to protection of the LN itself, or help accelerate recall responses by recirculating T cells (Brewitz et al., 2017).
This study began with the observation that a substantial portion of influenza-specific memory CD8+ T cells in the medLN bearing canonical residency markers expressed Thy1.1bright, similarly to those in upstream respiratory tissues. We found additional examples in the literature of discrepancies in Thy1.1 staining on cells isolated from NLT versus SLO (Khan et al., 2016; Wein et al., 2019). The biological significance of expressing elevated Thy1.1 requires further investigation, but nevertheless, a Thy1.1bright phenotype might provide a useful tool for discriminating TRM from i.v.neg TEM transiently occupying NLT. Indeed, influenza-specific Thy1.1dim memory P14 cells recovered from the i.v.neg fraction of the lung lacked TRM-associated markers and were CD62Lneg.
It remains unclear whether retrograde migration is a lung-specific phenomenon or a more generalizable feature of local infection. The propinquity of airway epithelium harboring abundant CD103+ Ly6Clo CD8+ T cells to lymphatic vessels serving as a means of tissue egress suggests that the proximity of locally infected parenchymal cells with draining lymphatics may be required for retrograde migration to occur. However, CD8+ TRM have been observed in many LNs throughout the mouse after primary viscerotropic LCMV infection (Schenkel et al., 2014), albeit at much lower frequencies than observed in the medLN in this study (the proportion of medLN TRM was quite high). Local recall in the skin or reproductive mucosa enhances TRM in the draining node, and it was shown that these cells did emigrate from NLT, although whether they were formally resident could not be concluded (Beura et al., 2018). This suggests a common upstream NLT origin for LN TRM, perhaps particularly notable in the lung after primary infections, but expansible in any downstream LN after iterative T cell stimulation. This biology may have convenient ramifications that could be exploited as a means to better understand the antigenic landscape of a particular tissue by cataloguing the specificity of LN TRM contained within its draining LN. That is, analyzing TRM-phenotype T cells may provide a TCR fingerprint of the upstream tissue’s infectious past. Unlike circulating cells that exhibit unbiased distribution, LN TRM would reflect regional experience. Thus, LN TRM could accelerate epitope discovery for pathogens, and this may apply to tumors and the development of chimeric antigen receptor T cells.
In summary, this study demonstrates that contraction of lung CD8+ T cell responses after influenza virus infection is contemporized by the repositioning of TRM to regional lymphoid tissue, where they can be maintained independently of tonic antigen. Models of LN immune surveillance, which has long focused on recirculation, will have to incorporate the phenomenon of residence that augments the preponderance of memory T cells that responded to regional infections (Butcher and Picker, 1996; Gowans and Knight, 1964; von Andrian and Mackay, 2000). Future studies should explore the fate and functions of these populations in the event of local reinfection and evaluate their usefulness as a tool for understanding the antigenic history of the upstream tissue they once occupied.
Materials and methods
Mice
C57BL/6J (B6) mice were purchased from the Jackson Laboratory and were maintained in specific pathogen–free conditions at the University of Minnesota. Thy1.1+ P14 (Pircher et al., 1989) and CD45.1+ OT-I (Hogquist et al., 1994) mice were fully backcrossed to C57BL/6J mice and maintained in our animal colony. TracFL mice (Polic et al., 2001) and ub-CreERT2 mice (Ruzankina et al., 2007) were provided by the laboratory of Dr. Kristin Hogquist (University of Minnesota, Minneapolis, MN). P14 CD8+ T cell transgenic mice, Thy1.1 B6 mice, and CD45.1 mice were maintained in-house. Both female and male mice were used in experiments. All mice were used in accordance with the Institutional Animal Care and Use Committees guidelines at the University of Minnesota.
Adoptive transfers, infections, bead enrichment, and in vitro peptide stimulations
P14 and OT-1 immune chimeras were generated by transferring 5 × 104 CD8+ T cells of either genotype into naive C57BL/6J mice, followed by i.n. infection with 500 PFU PR8-gp33 or 40 PFU PR8-ova (i.n.) 1 d later. R.A. Langlois made recombinant influenza viruses. Alternatively, mice were infected with 2 × 105 PFU Armstrong (i.p.) 1 d after cell transfer. For contemporaneous infections, mice were given 2.5 × 104 P14 and OT-1 cells and infected 1 d later. Where noted, CD8+ T cells were enriched using a negative-isolation kit (StemCell Technologies) supplemented with biotinylated Ly6C (1A8) antibody to remove circulating memory P14 cells. For in vitro peptide stimulations, cells were cultured for 4 h at 37°C in restimulation buffer (RPMI 1640 supplemented with 10% FBS, l-glutamine, sodium pyruvate, penicillin/streptomycin, Hepes, nonessential amino acids, and β-mercaptoethanol) containing 1× Golgi-plug (BD) and gp33 peptide (New England Peptide) at a final concentration of 0.2 µg/ml. For in vitro proliferation assays, cells were labeled with the cell tracking dye CellTracker Violet (Thermo Fisher Scientific) and cultured for 68 h at 37°C with 2 × 105 congenically distinct feeder cells in medium containing 0.66 µg/ml gp33 peptide.
Parabiosis, tamoxifen treatment, and Thy1.1 depletion
Parabiosis surgery was done as previously described (Steinert et al., 2015). Percentage residency was calculated using the formula % residency = 100 × [1 – (no. of P14 cells in naive parabiont organ × 2)/(no. of P14 cells in naive + immune parabiont organ)]. Tamoxifen was purchased from Sigma-Aldrich (T5648) and administered to mice i.p. at a dose of 75 mg/kg every 24 h over five consecutive days. αThy1.1 depleting antibody (HIS51, BD Pharmagen; 554892) was delivered i.p. at a dose of 1 µg/mouse in 200 µl of PBS. Depletion was confirmed by flow cytometric analysis of peripheral blood 18 h after treatment.
Intravascular labeling, cell isolation, flow cytometry, and cell sorting
Intravascular staining was used to discriminate cells present in the vasculature from tissue parenchyma, as previously described (Anderson et al., 2012). Briefly, 3 µg of biotinylated αCD8β antibody (YTS156.7.7) was injected retroorbitally into mice 3 min before sacrifice. Mice were bled via saphenous vein puncture 30 s before sacrifice to confirm productive i.v. labeling. For lung cell isolation, bronchoalveolar lavage fluid was first collected in three 1-ml lavages with cold PBS followed by removal of the medLN and any associated lymphatic vessels. Lungs were excised, minced, and digested in Collagenase I (Sigma-Aldrich) for 1 h before mechanical dissociation via gentleMACS Dissociator (Miltenyi Biotec) twice. Lung homogenates were passed through a 70-µm filter and enriched on a Percoll gradient. Isolated mouse cells were stained with antibodies against CD8α (53-6.7), CD8β (YTS156.7.7), CD90.1 (OX-7), CD90.1 (HIS51), CD45.1 (A20), CD103 (M290), CD69 (H1.2F3), CD25 (PC61), CD44 (IM7), CD62L (MEL-14), Ly6C (1A8), PD1 (RMP1-30), and integrin-β7 (FIB504). Intracellular staining was performed using the Foxp3/Transcription Factor Staining Buffer Kit (Tonbo Biosciences) with antibodies against Ki67 (SolA15), IFNγ (XMG1.2), TNFα (MP6-XT22), and Granzyme B (GB12). Ghost Dye (Tonbo Biosciences) was used to discriminate dead cells by flow cytometry. Enumeration of cells was done using Polybead Polystyrene 10-µm microspheres (Polysciences). Stained samples were acquired with LSRII or LSR Fortessa flow cytometers (BD) and analyzed with FlowJo software (TreeStar). TRM (CD45.2neg CD8+ CD44+ CD103+) and central memory T cells (CD45.2neg CD8+ CD44+ CD103neg) populations were sorted from the medLN of PR8-gp33 immune chimeras using a BD FACSAria II.
IF microscopy
Harvested murine tissues were embedded in the optimum cutting temperature tissue freezing medium and frozen in a 2-methyl butane liquid bath. Frozen blocks were cut on a Leica cryostat to prepare 7-µm-thick sections. Slides were stained with antibodies against CD8β (YTS156.7.7), CD8α (53-6.7), CD90.1 (OX-7), CD45.1 (A20), CD103 (M290) Lyve1 (polyclonal; Novus), Podoplanin (8.1.1), CD31 (MEC13.3), Epcam1 (G8.8), and Ly6C (1A8). IF microscopy was performed using a Leica DM6000 B microscope. Counterstaining with Sytox Green (Thermo Fisher Scientific) or DAPI was done to detect nuclei where indicated.
Ce3D and two-photon microscopy
LN and lymphatics were harvested and fixed with BD Cytofix/Cytoperm diluted 1:4 in PBS overnight at 4°C. Fixed tissues were incubated for ≥8 h in blocking buffer containing 1% BSA and 0.3% Triton X-100 in PBS at 37°C. Tissues were then incubated with directly conjugated antibodies diluted 1:100 in blocking buffer for ≤2 d at 37°C on a shaker. Stained samples were washed with PBS containing 0.2% Triton X-100 and 1-thioglycerol (0.5%) for 12–24 h at 24°C or 37°C. Tissue was cleared using Ce3D as previously described (Li et al., 2017). In brief, N-methylacetamide prepared to 40% (vol/vol) in PBS was used to dissolve Histodenz to 86% (wt/vol) concentration (∼1.455 g Histodenz per 1 ml of 40% N-methylacetamide) inside a chemical fume hood, with the mixture incubated at 37°C, and dissolution was expedited using a stir bar and plate. Triton X-100 (0.1% vol/vol) and 1-thioglycerol (0.5% vol/vol) were added to the clearing solution. Stained tissues were placed in the Ce3D medium and incubated for ∼24 h at room temperature on a rotor. Stained and cleared tissue was mounted in fresh Ce3D medium with an adhesive silicone rubber spacer and tightly covered with a glass coverslip. Cleared tissue was imaged by tiling Z-stacks with confocal laser scanning microscopy on a Leica SP5. A 25× water-immersion coverslip-corrected objective was used to capture images with an XY voxel size of 971 nm, 5 µm step size, and 1-A.U. pinhole. 3D reconstructions were done using Imaris (Bitplane) software v9.2.1, and GFP+ P14 cells were displayed through a surface mask.
Statistical analysis
An unpaired Student’s t test was used when comparing two groups. One-way ANOVA with Tukey’s multiple comparison test was used when comparing more than two groups. All bar graphs represent mean ± SEM. P < 0.05 was considered significant. Statistical analysis was done in Prism (GraphPad Software). Sample size was chosen based on previous experience. No sample exclusion criteria were applied, and investigators were not blinded.
Online supplemental material
Fig. S1 demonstrates that both staggered and contemporaneous infection modalities recruit abundant LCMV-specific P14 cells into the lung parenchyma 12 d after PR8-ova infection. Fig. S2 depicts an experiment to test whether cognate antigen recognition within the medLN was sufficient to drive the formation of medLN TRM during priming. Fig. S3 shows that medLN TRM can proliferate after in vitro or in vivo antigen restimulation. Fig. S4 demonstrates that unmanipulated medLN TRM proliferate after secondary i.n. infection. Video 1 shows the medLN and associated lymphatics from a mouse infected 16 d earlier with PR8-gp33. Corresponding still images are displayed in Fig. 8 E.
Acknowledgments
We thank the University of Minnesota flow cytometry and microscopy cores. Dr. Vaiva Vezys (University of Minnesota, Minneapolis, MN) provided helpful input, mice, and reagents.
This work was funded by the Howard Hughes Medical Institute Faculty Scholars program, the National Institutes of Health (R01AI084913 to D. Masopust and T90 DE 022732 to J.M. Stolley), and Sinai-Emory Multi-institutional Collaborative Influenza Vaccine Innovation Centers (75N93019C00051 to D. Masopust).
Author contributions: J.M. Stolley and D. Masopust conceived of the study, designed experiments, and wrote the manuscript; J.M. Stolley performed experiments and analyzed data; T.S. Johnston, A.G. Soerens, L.K. Beura, P.C. Rosato, V. Joag, and S.P. Wijeyesinghe assisted with experiments; R.A. Langlois made and provided recombinant influenza viruses; and K.C. Osum and J.S. Mitchell performed Ce3D.
References
- Amsen D., van Gisbergen K.P.J.M., Hombrink P., and van Lier R.A.W.. 2018. Tissue-resident memory T cells at the center of immunity to solid tumors. Nat. Immunol. 19:538–546. 10.1038/s41590-018-0114-2 [DOI] [PubMed] [Google Scholar]
- Anderson K.G., Sung H., Skon C.N., Lefrancois L., Deisinger A., Vezys V., and Masopust D.. 2012. Cutting edge: intravascular staining redefines lung CD8 T cell responses. J. Immunol. 189:2702–2706. 10.4049/jimmunol.1201682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura L.K., Anderson K.G., Schenkel J.M., Locquiao J.J., Fraser K.A., Vezys V., Pepper M., and Masopust D.. 2015. Lymphocytic choriomeningitis virus persistence promotes effector-like memory differentiation and enhances mucosal T cell distribution. J. Leukoc. Biol. 97:217–225. 10.1189/jlb.1HI0314-154R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura L.K., Fares-Frederickson N.J., Steinert E.M., Scott M.C., Thompson E.A., Fraser K.A., Schenkel J.M., Vezys V., and Masopust D.. 2019. CD4+ resident memory T cells dominate immunosurveillance and orchestrate local recall responses. J. Exp. Med. 216:1214–1229. 10.1084/jem.20181365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beura L.K., Wijeyesinghe S., Thompson E.A., Macchietto M.G., Rosato P.C., Pierson M.J., Schenkel J.M., Mitchell J.S., Vezys V., Fife B.T., et al. 2018. T Cells in Nonlymphoid Tissues Give Rise to Lymph-Node-Resident Memory T Cells. Immunity. 48:327–338.e5. 10.1016/j.immuni.2018.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borron P.J., Crouch E.C., Lewis J.F., Wright J.R., Possmayer F., and Fraher L.J.. 1998. Recombinant rat surfactant-associated protein D inhibits human T lymphocyte proliferation and IL-2 production. J. Immunol. 161:4599–4603. [PubMed] [Google Scholar]
- Brewitz A., Eickhoff S., Dähling S., Quast T., Bedoui S., Kroczek R.A., Kurts C., Garbi N., Barchet W., Iannacone M., et al. 2017. CD8+ T Cells Orchestrate pDC-XCR1+ Dendritic Cell Spatial and Functional Cooperativity to Optimize Priming. Immunity. 46:205–219. 10.1016/j.immuni.2017.01.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buggert M., Nguyen S., Salgado-Montes de Oca G., Bengsch B., Darko S., Ransier A., Roberts E.R., Del Alcazar D., Brody I.B., Vella L.A., et al. 2018. Identification and characterization of HIV-specific resident memory CD8+ T cells in human lymphoid tissue. Sci. Immunol. 3 eaar4526 10.1126/sciimmunol.aar4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher E.C., and Picker L.J.. 1996. Lymphocyte homing and homeostasis. Science. 272:60–67. 10.1126/science.272.5258.60 [DOI] [PubMed] [Google Scholar]
- Clark R.A. 2015. Resident memory T cells in human health and disease. Sci. Transl. Med. 7 269rv1 10.1126/scitranslmed.3010641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ely K.H., Cookenham T., Roberts A.D., and Woodland D.L.. 2006. Memory T cell populations in the lung airways are maintained by continual recruitment. J. Immunol. 176:537–543. 10.4049/jimmunol.176.1.537 [DOI] [PubMed] [Google Scholar]
- Farber D.L., Yudanin N.A., and Restifo N.P.. 2014. Human memory T cells: generation, compartmentalization and homeostasis. Nat. Rev. Immunol. 14:24–35. 10.1038/nri3567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan A.-P., Clarke J., Wood O., Garrido-Martin E.M., Chee S.J., Mellows T., Samaniego-Castruita D., Singh D., Seumois G., Alzetani A., et al. 2017. Tissue-resident memory features are linked to the magnitude of cytotoxic T cell responses in human lung cancer. Nat. Immunol. 18:940–950. 10.1038/ni.3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt T., Wakim L.M., Eidsmo L., Reading P.C., Heath W.R., and Carbone F.R.. 2009. Memory T cells in nonlymphoid tissue that provide enhanced local immunity during infection with herpes simplex virus. Nat. Immunol. 10:524–530. 10.1038/ni.1718 [DOI] [PubMed] [Google Scholar]
- Glennie N.D., Yeramilli V.A., Beiting D.P., Volk S.W., Weaver C.T., and Scott P.. 2015. Skin-resident memory CD4+ T cells enhance protection against Leishmania major infection. J. Exp. Med. 212:1405–1414. 10.1084/jem.20142101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowans J.L., and Knight E.J.. 1964. The route of re-circulation of lymphocytes in the rat. Proc. R. Soc. Lond. B Biol. Sci. 159:257–282. 10.1098/rspb.1964.0001 [DOI] [PubMed] [Google Scholar]
- Hikono H., Kohlmeier J.E., Takamura S., Wittmer S.T., Roberts A.D., and Woodland D.L.. 2007. Activation phenotype, rather than central- or effector-memory phenotype, predicts the recall efficacy of memory CD8+ T cells. J. Exp. Med. 204:1625–1636. 10.1084/jem.20070322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M., and Pircher H.. 2011. E-cadherin promotes accumulation of a unique memory CD8 T-cell population in murine salivary glands. Proc. Natl. Acad. Sci. USA. 108:16741–16746. 10.1073/pnas.1107200108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan R.J., Usherwood E.J., Zhong W., Roberts A.A., Dutton R.W., Harmsen A.G., and Woodland D.L.. 2001. Activated antigen-specific CD8+ T cells persist in the lungs following recovery from respiratory virus infections. J. Immunol. 166:1813–1822. 10.4049/jimmunol.166.3.1813 [DOI] [PubMed] [Google Scholar]
- Hogquist K.A., Jameson S.C., Heath W.R., Howard J.L., Bevan M.J., and Carbone F.R.. 1994. T cell receptor antagonist peptides induce positive selection. Cell. 76:17–27. 10.1016/0092-8674(94)90169-4 [DOI] [PubMed] [Google Scholar]
- Hombrink P., Helbig C., Backer R.A., Piet B., Oja A.E., Stark R., Brasser G., Jongejan A., Jonkers R.E., Nota B., et al. 2016. Programs for the persistence, vigilance and control of human CD8+ lung-resident memory T cells. Nat. Immunol. 17:1467–1478. 10.1038/ni.3589 [DOI] [PubMed] [Google Scholar]
- Hondowicz B.D., An D., Schenkel J.M., Kim K.S., Steach H.R., Krishnamurty A.T., Keitany G.J., Garza E.N., Fraser K.A., Moon J.J., et al. 2016. Interleukin-2-Dependent Allergen-Specific Tissue-Resident Memory Cells Drive Asthma. Immunity. 44:155–166. 10.1016/j.immuni.2015.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang X., Clark R.A., Liu L., Wagers A.J., Fuhlbrigge R.C., and Kupper T.S.. 2012. Skin infection generates non-migratory memory CD8+ T(RM) cells providing global skin immunity. Nature. 483:227–231. 10.1038/nature10851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jozwik A., Habibi M.S., Paras A., Zhu J., Guvenel A., Dhariwal J., Almond M., Wong E.H.C., Sykes A., Maybeno M., et al. 2015. RSV-specific airway resident memory CD8+ T cells and differential disease severity after experimental human infection. Nat. Commun. 6:10224 10.1038/ncomms10224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan T.N., Mooster J.L., Kilgore A.M., Osborn J.F., and Nolz J.C.. 2016. Local antigen in nonlymphoid tissue promotes resident memory CD8+ T cell formation during viral infection. J. Exp. Med. 213:951–966. 10.1084/jem.20151855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar B.V., Ma W., Miron M., Granot T., Guyer R.S., Carpenter D.J., Senda T., Sun X., Ho S.-H., Lerner H., et al. 2017. Human Tissue-Resident Memory T Cells Are Defined by Core Transcriptional and Functional Signatures in Lymphoid and Mucosal Sites. Cell Rep. 20:2921–2934. 10.1016/j.celrep.2017.08.078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.-T., Suarez-Ramirez J.E., Wu T., Redman J.M., Bouchard K., Hadley G.A., and Cauley L.S.. 2011. Environmental and antigen receptor-derived signals support sustained surveillance of the lungs by pathogen-specific cytotoxic T lymphocytes. J. Virol. 85:4085–4094. 10.1128/JVI.02493-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine A.G., Arvey A., Jin W., and Rudensky A.Y.. 2014. Continuous requirement for the TCR in regulatory T cell function. Nat. Immunol. 15:1070–1078. 10.1038/ni.3004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Germain R.N., and Gerner M.Y.. 2017. Multiplex, quantitative cellular analysis in large tissue volumes with clearing-enhanced 3D microscopy (Ce3D). Proc. Natl. Acad. Sci. USA. 114:E7321–E7330. 10.1073/pnas.1708981114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S., Mozdzanowska K., Palladino G., and Gerhard W.. 1994. Heterosubtypic immunity to influenza type A virus in mice. Effector mechanisms and their longevity. J. Immunol. 152:1653–1661. [PubMed] [Google Scholar]
- Malik B.T., Byrne K.T., Vella J.L., Zhang P., Shabaneh T.B., Steinberg S.M., Molodtsov A.K., Bowers J.S., Angeles C.V., Paulos C.M., et al. 2017. Resident memory T cells in the skin mediate durable immunity to melanoma. Sci. Immunol. 2 eaam6346 10.1126/sciimmunol.aam6346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marriott C.L., Dutton E.E., Tomura M., and Withers D.R.. 2017. Retention of Ag-specific memory CD4+ T cells in the draining lymph node indicates lymphoid tissue resident memory populations. Eur. J. Immunol. 47:860–871. 10.1002/eji.201646681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D., and Soerens A.G.. 2019. Tissue-Resident T Cells and Other Resident Leukocytes. Annu. Rev. Immunol. 37:521–546. 10.1146/annurev-immunol-042617-053214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masopust D., Choo D., Vezys V., Wherry E.J., Duraiswamy J., Akondy R., Wang J., Casey K.A., Barber D.L., Kawamura K.S., et al. 2010. Dynamic T cell migration program provides resident memory within intestinal epithelium. J. Exp. Med. 207:553–564. 10.1084/jem.20090858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster S.R., Wein A.N., Dunbar P.R., Hayward S.L., Cartwright E.K., Denning T.L., and Kohlmeier J.E.. 2018. Pulmonary antigen encounter regulates the establishment of tissue-resident CD8 memory T cells in the lung airways and parenchyma. Mucosal Immunol. 11:1071–1078. 10.1038/s41385-018-0003-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S.N., and Mackay L.K.. 2016. Tissue-resident memory T cells: local specialists in immune defence. Nat. Rev. Immunol. 16:79–89. 10.1038/nri.2015.3 [DOI] [PubMed] [Google Scholar]
- Mueller S.N., Gebhardt T., Carbone F.R., and Heath W.R.. 2013. Memory T cell subsets, migration patterns, and tissue residence. Annu. Rev. Immunol. 31:137–161. 10.1146/annurev-immunol-032712-095954 [DOI] [PubMed] [Google Scholar]
- Nizard M., Roussel H., Diniz M.O., Karaki S., Tran T., Voron T., Dransart E., Sandoval F., Riquet M., Rance B., et al. 2017. Induction of resident memory T cells enhances the efficacy of cancer vaccine. Nat. Commun. 8:15221 10.1038/ncomms15221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park C.O., and Kupper T.S.. 2015. The emerging role of resident memory T cells in protective immunity and inflammatory disease. Nat. Med. 21:688–697. 10.1038/nm.3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S.L., Buzzai A., Rautela J., Hor J.L., Hochheiser K., Effern M., McBain N., Wagner T., Edwards J., McConville R., et al. 2019. Tissue-resident memory CD8+ T cells promote melanoma-immune equilibrium in skin. Nature. 565:366–371. 10.1038/s41586-018-0812-9 [DOI] [PubMed] [Google Scholar]
- Pircher H., Bürki K., Lang R., Hengartner H., and Zinkernagel R.M.. 1989. Tolerance induction in double specific T-cell receptor transgenic mice varies with antigen. Nature. 342:559–561. 10.1038/342559a0 [DOI] [PubMed] [Google Scholar]
- Polic B., Kunkel D., Scheffold A., and Rajewsky K.. 2001. How alpha beta T cells deal with induced TCR alpha ablation. Proc. Natl. Acad. Sci. USA. 98:8744–8749. 10.1073/pnas.141218898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reagin K.L., and Klonowski K.D.. 2018. Incomplete Memories: The Natural Suppression of Tissue-Resident Memory CD8 T Cells in the Lung. Front. Immunol. 9:17 10.3389/fimmu.2018.00017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynoso G.V., Weisberg A.S., Shannon J.P., McManus D.T., Shores L., Americo J.L., Stan R.V., Yewdell J.W., and Hickman H.D.. 2019. Lymph node conduits transport virions for rapid T cell activation. Nat. Immunol. 20:602–612. 10.1038/s41590-019-0342-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riding R.L., and Harris J.E.. 2019. The Role of Memory CD8+ T Cells in Vitiligo. J. Immunol. 203:11–19. 10.4049/jimmunol.1900027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzankina Y., Pinzon-Guzman C., Asare A., Ong T., Pontano L., Cotsarelis G., Zediak V.P., Velez M., Bhandoola A., and Brown E.J.. 2007. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 1:113–126. 10.1016/j.stem.2007.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai S., Kauffman K.D., Sallin M.A., Sharpe A.H., Young H.A., Ganusov V.V., and Barber D.L.. 2016. CD4 T Cell-Derived IFN-γ Plays a Minimal Role in Control of Pulmonary Mycobacterium tuberculosis Infection and Must Be Actively Repressed by PD-1 to Prevent Lethal Disease. PLoS Pathog. 12 e1005667 10.1371/journal.ppat.1005667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathaliyawala T., Kubota M., Yudanin N., Turner D., Camp P., Thome J.J.C., Bickham K.L., Lerner H., Goldstein M., Sykes M., et al. 2013. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity. 38:187–197. 10.1016/j.immuni.2012.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel J.M., Fraser K.A., Vezys V., and Masopust D.. 2013. Sensing and alarm function of resident memory CD8+ T cells. Nat. Immunol. 14:509–513. 10.1038/ni.2568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkel J.M., Fraser K.A., and Masopust D.. 2014. Cutting edge: resident memory CD8 T cells occupy frontline niches in secondary lymphoid organs. J. Immunol. 192:2961–2964. 10.4049/jimmunol.1400003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H., and Iwasaki A.. 2013. Tissue-resident memory T cells. Immunol. Rev. 255:165–181. 10.1111/imr.12087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slütter B., Van Braeckel-Budimir N., Abboud G., Varga S.M., Salek-Ardakani S., and Harty J.T.. 2017. Dynamics of influenza-induced lung-resident memory T cells underlie waning heterosubtypic immunity. Sci. Immunol. 2 eaag2031 10.1126/sciimmunol.aag2031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinert E.M., Schenkel J.M., Fraser K.A., Beura L.K., Manlove L.S., Igyártó B.Z., Southern P.J., and Masopust D.. 2015. Quantifying Memory CD8 T Cells Reveals Regionalization of Immunosurveillance. Cell. 161:737–749. 10.1016/j.cell.2015.03.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura S., Kato S., Motozono C., Shimaoka T., Ueha S., Matsuo K., Miyauchi K., Masumoto T., Katsushima A., Nakayama T., et al. 2019. Interstitial-resident memory CD8+ T cells sustain frontline epithelial memory in the lung. J. Exp. Med. 216:2736–2747. 10.1084/jem.20190557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura S., Roberts A.D., Jelley-Gibbs D.M., Wittmer S.T., Kohlmeier J.E., and Woodland D.L.. 2010. The route of priming influences the ability of respiratory virus-specific memory CD8+ T cells to be activated by residual antigen. J. Exp. Med. 207:1153–1160. 10.1084/jem.20090283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura S., Yagi H., Hakata Y., Motozono C., McMaster S.R., Masumoto T., Fujisawa M., Chikaishi T., Komeda J., Itoh J., et al. 2016. Specific niches for lung-resident memory CD8+ T cells at the site of tissue regeneration enable CD69-independent maintenance. J. Exp. Med. 213:3057–3073. 10.1084/jem.20160938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teijaro J.R., Turner D., Pham Q., Wherry E.J., Lefrançois L., and Farber D.L.. 2011. Cutting edge: Tissue-retentive lung memory CD4 T cells mediate optimal protection to respiratory virus infection. J. Immunol. 187:5510–5514. 10.4049/jimmunol.1102243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thome J.J.C., Yudanin N., Ohmura Y., Kubota M., Grinshpun B., Sathaliyawala T., Kato T., Lerner H., Shen Y., and Farber D.L.. 2014. Spatial map of human T cell compartmentalization and maintenance over decades of life. Cell. 159:814–828. 10.1016/j.cell.2014.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugur M., Schulz O., Menon M.B., Krueger A., and Pabst O.. 2014. Resident CD4+ T cells accumulate in lymphoid organs after prolonged antigen exposure. Nat. Commun. 5:4821 10.1038/ncomms5821 [DOI] [PubMed] [Google Scholar]
- Van Braeckel-Budimir N., and Harty J.T.. 2017. Influenza-induced lung Trm: not all memories last forever. Immunol. Cell Biol. 95:651–655. 10.1038/icb.2017.32 [DOI] [PubMed] [Google Scholar]
- Van Braeckel-Budimir N., Varga S.M., Badovinac V.P., and Harty J.T.. 2018. Repeated Antigen Exposure Extends the Durability of Influenza-Specific Lung-Resident Memory CD8+ T Cells and Heterosubtypic Immunity. Cell Rep. 24:3374–3382.e3. 10.1016/j.celrep.2018.08.073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Andrian U.H., and Mackay C.R.. 2000. T-cell function and migration. Two sides of the same coin. N. Engl. J. Med. 343:1020–1034. 10.1056/NEJM200010053431407 [DOI] [PubMed] [Google Scholar]
- Walsh D.A., Borges da Silva H., Beura L.K., Peng C., Hamilton S.E., Masopust D., and Jameson S.C.. 2019. The Functional Requirement for CD69 in Establishment of Resident Memory CD8+ T Cells Varies with Tissue Location. J. Immunol. 203:946–955. 10.4049/jimmunol.1900052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Wang S., Goplen N.P., Li C., Cheon I.S., Dai Q., Huang S., Shan J., Ma C., Ye Z., et al. 2019. PD-1hi CD8+ resident memory T cells balance immunity and fibrotic sequelae. Sci. Immunol. 4 eaaw1217 10.1126/sciimmunol.aaw1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wein A.N., McMaster S.R., Takamura S., Dunbar P.R., Cartwright E.K., Hayward S.L., McManus D.T., Shimaoka T., Ueha S., Tsukui T., et al. 2019. CXCR6 regulates localization of tissue-resident memory CD8 T cells to the airways. J. Exp. Med. 216:2748–2762. 10.1084/jem.20181308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woon H.G., Braun A., Li J., Smith C., Edwards J., Sierro F., Feng C.G., Khanna R., Elliot M., Bell A., et al. 2016. Compartmentalization of Total and Virus-Specific Tissue-Resident Memory CD8+ T Cells in Human Lymphoid Organs. PLoS Pathog. 12 e1005799 10.1371/journal.ppat.1005799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Hu Y., Lee Y.-T., Bouchard K.R., Benechet A., Khanna K., and Cauley L.S.. 2014. Lung-resident memory CD8 T cells (TRM) are indispensable for optimal cross-protection against pulmonary virus infection. J. Leukoc. Biol. 95:215–224. 10.1189/jlb.0313180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zammit D.J., Turner D.L., Klonowski K.D., Lefrançois L., and Cauley L.S.. 2006. Residual antigen presentation after influenza virus infection affects CD8 T cell activation and migration. Immunity. 24:439–449. 10.1016/j.immuni.2006.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]