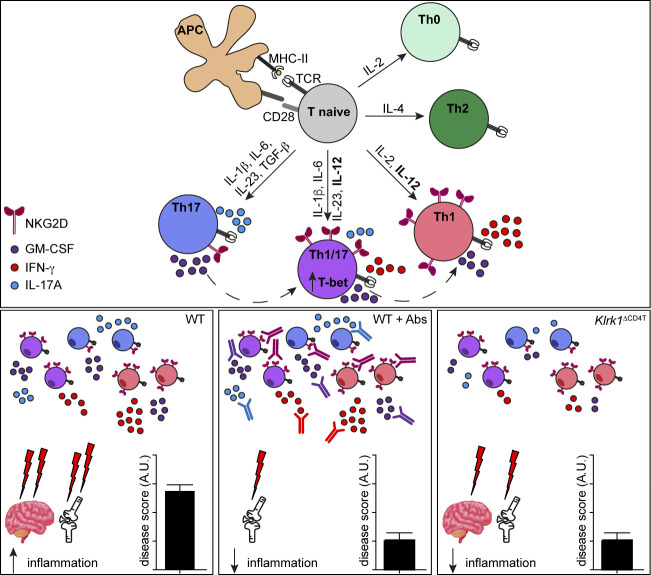
NKG2D is a danger sensor expressed on different subsets of innate and adaptive lymphocytes. This study demonstrates how NKG2D enforces proinflammatory features of Th1 and Th17 cells and promotes inflammation in vivo, identifying NKG2D as an important target for the amelioration of Th1/ Th17-mediated inflammatory diseases.
Abstract
NKG2D is a danger sensor expressed on different subsets of innate and adaptive lymphocytes. Despite its established role as a potent activator of the immune system, NKG2D-driven regulation of CD4+ T helper (Th) cell–mediated immunity remains unclear. In this study, we demonstrate that NKG2D modulates Th1 and proinflammatory T-bet+ Th17 cell effector functions in vitro and in vivo. In particular, NKG2D promotes higher production of proinflammatory cytokines by Th1 and T-bet+ Th17 cells and reinforces their transcription of type 1 signature genes, including Tbx21. Conditional deletion of NKG2D in T cells impairs the ability of antigen-specific CD4+ T cells to promote inflammation in vivo during antigen-induced arthritis and experimental autoimmune encephalomyelitis, indicating that NKG2D is an important target for the amelioration of Th1- and Th17-mediated chronic inflammatory diseases.
Graphical Abstract
Introduction
Differentiation toward distinct CD4+ T helper (Th) effector subsets is driven by antigen recognition via the TCR and modulated by inflammatory signals, mainly sensed through cytokine receptors expressed by these cells. Accumulating evidence shows that during their differentiation, Th lineages can additionally integrate signals from receptors sensing danger or pathogens, some of which are typically expressed on innate cells. For instance, engagement of TLRs, such as TLR2, TLR4, or TLR7, on Th1 and Th17 cells can regulate their development and effector functions, thereby differentially contributing to their pathogenicity, especially in experimental autoimmune encephalomyelitis (EAE; Reynolds et al., 2010, 2012; Ye et al., 2017). In addition to TLRs, natural killer (NK) cell receptors can be found on T cells. CD94/NKG2 is expressed on a subset of CD8+ T cells (Moser et al., 2002) and a subset of Th1 cells, where it contributes to their proliferation and enhanced IFN-γ production (Meyers et al., 2002). DNAX accessory molecule 1 (DNAM-1) is up-regulated upon polarization toward Th1 and Th17 lineages, and its blockade results in both decreased cytokine production and reduced severity of EAE (Dardalhon et al., 2005; Shibuya et al., 2003). Both Th1 and Th17 cells, in particular plastic Th17 cells that acquired type 1 features (Ghoreschi et al., 2010; Hirota et al., 2011), display marked proinflammatory properties and are important players in the development of inflammatory diseases, as shown in samples obtained from patients with rheumatoid arthritis, inflammatory bowel disease, multiple sclerosis, and type 1 diabetes and their corresponding disease models in mice (Bending et al., 2009; Korn et al., 2009; Patel and Kuchroo, 2015; Weaver and Murphy, 2007). Hence, understanding the biology of innate receptors selectively expressed by pathogenic CD4+ T cell effector lineages could represent a promising targeting strategy to modulate the pathogenesis of inflammatory diseases.
NKG2D (NK group 2, member D; encoded by the gene Klrk1) is a molecular sensor of stressed cells and a potent activator of the immune system (Lanier, 2015). It binds to a variety of well-defined danger molecules, such as Rae-1α-ε, H60a-c and mouse UL-16-binding protein-like transcript 1 (MULT-1) in mice, or the MHC class-I-related chain A (MICA)/MICB and the UL-16 binding proteins 1–6 in humans (Raulet et al., 2013). The transcripts of NKG2D ligands are found in many tissues under healthy conditions, but the cell surface expression of the corresponding proteins is kept under tight control by posttranscriptional regulation (Nice et al., 2009). Cellular stress (Hosomi et al., 2017), notably DNA damage (Gasser et al., 2005), TLR signaling (Hamerman et al., 2004), and specific cytokine exposure, can induce NKG2D ligand surface expression, as shown on tumor- or virus-infected cells (Raulet et al., 2013) and during autoimmunity (Groh et al., 2003; Ito et al., 2008; Ogasawara et al., 2004). Ligand recognition by NKG2D can be integrated into a DNAX adaptor protein 12 (DAP12)–dependent activation signal, as seen in NK cells, or into a DAP10-phosphinositide-3 kinase–mediated costimulation signal in CD8+ T cells (Diefenbach et al., 2002; Gilfillan et al., 2002). Based on this, NKG2D expressed by NK, CD8+, and γδ+ T cells plays an important role in the recognition and clearance of infected and transformed cells expressing cognate ligands (Jonjić et al., 2008; Lanier, 2015; Strid et al., 2011).
Expression of NKG2D on CD4+ T cells has been previously reported in humans and mice; however, the role of NKG2D in modulating CD4+ T cell functions and T cell–mediated pathology is to date only poorly understood (Babic and Romagnani, 2018). Whereas low numbers of NKG2D+ CD4+ T cells have been described in peripheral blood of healthy individuals, an increase in frequency has been observed in many different human pathologies (Allez et al., 2007; Dai et al., 2009; Groh et al., 2003; Sáez-Borderías et al., 2006) as well as in mice during inflammatory models in vivo (Andersson et al., 2011; Ito et al., 2008). These data suggest that NKG2D might represent a checkpoint to modulate CD4+ Th cell activation during inflammatory responses. However, the signals driving the expression of NKG2D in CD4+ T cells and the role of this receptor in driving their differentiation and effector functions in vitro and in vivo remain unclear.
Here we show that NKG2D is selectively up-regulated on Th1 and T-box transcription factor 21–positive (T-bet+) proinflammatory Th17 lineage and differentially modulates their cytokine expression. Importantly, we demonstrate that NKG2D marks a population of IFN-γ/IL-17A/GM-CSF–coproducing cells in vivo in a model of antigen-induced arthritis, and that T cell–specific deletion of NKG2D is sufficient to ameliorate the disease.
Results
NKG2D expression is selectively associated with Th1 and Th17 cell differentiation
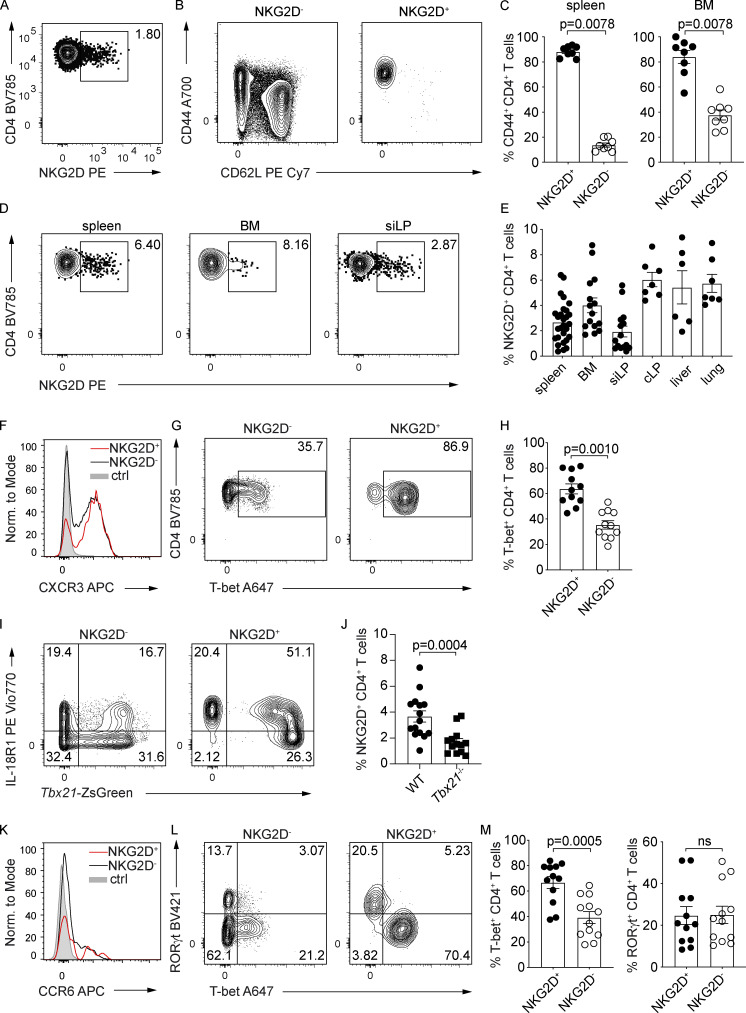
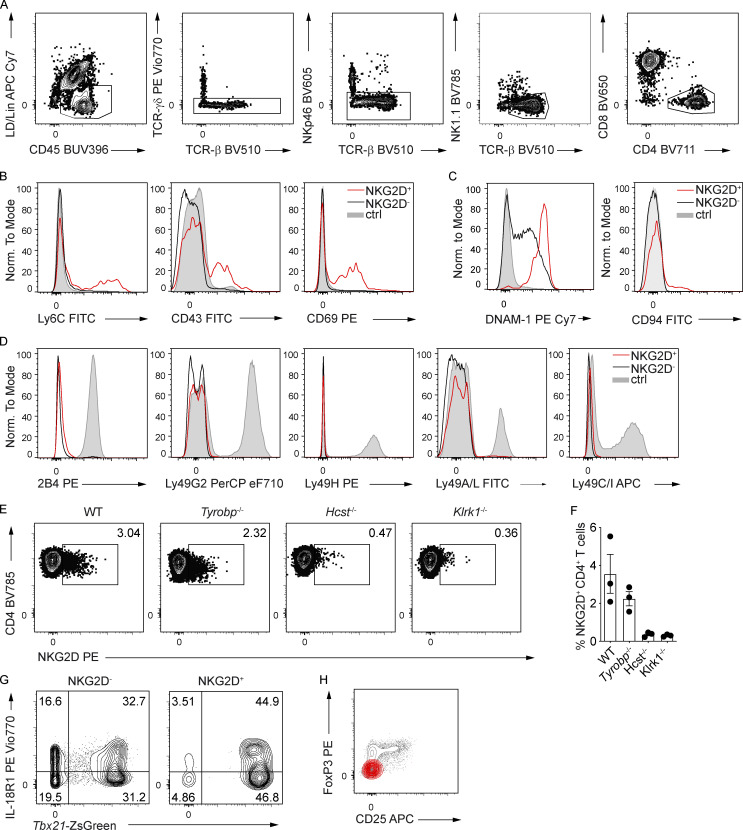
To better understand the role of NKG2D in the regulation of CD4+ T cell–mediated responses, we analyzed the phenotype of NKG2D+ CD4+ T cells present in C57BL/6 (WT) mice under steady-state. NKG2D expression was detectable ex vivo on a small subset of CD4+ TCRβ+ T cells (Fig. 1 A and Fig. S1 A) and selectively enriched within the CD44+ CD62L− cell pool (Fig. 1, B and C). NKG2D+ CD44+ CD4+ T cells were found at comparable frequencies in various organs tested, such as spleen, bone marrow (BM), small intestine lamina propria (siLP), colon lamina propria, liver, and lung (Fig. 1, D and E). In line with their phenotype of antigen experienced cells, NKG2D+ CD44+ CD4+ T cells had enriched expression of activation markers Ly6C, CD69, and CD43 (Fig. S1 B). Intriguingly, although they were enriched in the expression of the NK cell receptor DNAM-1, only a minor subset of NKG2D+ CD44+ CD4+ T cells coexpressed CD94 (Fig. S1 C), whereas no coexpression of other typical NK cell markers such as 2B4, Ly49G2, Ly49H, Ly49A/L, or Ly49C/I was observed (Fig. S1 D). The expression of NKG2D on CD4+ T cells was strongly reduced in Hcst−/− (coding for DAP10), but not in Tyrobp−/− (coding for DAP12) mice (Fig. S1, E and F), indicating its dependence on the adaptor DAP10 and paralleling previous observations in CD8+ T cells (Diefenbach et al., 2002; Gilfillan et al., 2002). Ex vivo analysis of Th lineage-defining transcription factors and chemokine receptors revealed that a large fraction of splenic NKG2D+ CD4+ T cells expressed the Th1-associated markers CXCR3 (Fig. 1 F), IL-18R1, and T-bet, as shown by protein intranuclear staining as well as in Tbx21-ZsGreen reporter mice (Fig. 1, G–I). In line with those observations, the frequency of NKG2D+ CD4+ T cells was significantly reduced in Tbx21-deficient mice (Fig. 1 J). Besides T-bet–expressing cells (Figs. 1 L and S1 G), siLP NKG2D+ CD4+ T cells showcased a population of RAR-related orphan receptor (ROR)γt+ and CCR6+ cells (Fig. 1, K and L). However, only T-bet–expressing, but not RORγt-expressing, cells were enriched within the siLP NKG2D+ T cell pool (Fig. 1, L and M). Of note, NKG2D expression was excluded from Foxp3+ regulatory T cell compartment in the spleen (Fig. S1 H).
Figure 1.
NKG2D is expressed on antigen-experienced CD4+ T cells in steady-state C57BL/6 mice. (A) Representative FC analysis of NKG2D expression on single-cell suspensions from the spleen of C57BL/6 mice. (B) Representative FC analysis of CD44 and CD62L expression on NKG2D+ and NKG2D− CD4+ T cells in the spleen. (C) Frequency of CD44+ CD62L− T cells; quantification from B; n = 8; Wilcoxon test. (D) Representative FC analysis of NKG2D expression on CD44+ CD4+ T cell from single-cell suspensions of C57BL/6 mice. (E) Frequency of NKG2D+ CD44+ CD4+ T cells in indicated organs, spleen (n = 29), BM (n = 15), siLP (n = 15), colon lamina propria (cLP; n = 7), liver (n = 6), and lung (n = 7). (F and K) Representative FC analysis of CXCR3 (F) and CCR6 (K) expression on CD4+ CD44+ NKG2D− (black line) or NKG2D+ (red line) in spleen and siLP, respectively. Shaded histogram represents live CD19+ (F) or live CD8+ (K) cells; n = 6. (G and L) Representative FC analysis of T-bet expression in spleen (G) and RORγt and T-bet expression in siLP (L) in CD4+ CD44+ NKG2D− or NKG2D+ T cells. (H and M) Frequency of T-bet+ CD44+ CD4+ T cells in spleen (H), quantification from G (n = 11) and T-bet+ or RORγt+ CD44+ CD4+ T cells in siLP (M), quantification from L (n = 12); Wilcoxon test. ns, not significant. (I) Representative FC analysis of IL18R1 and ZsGreen (Tbx21) expression on CD4+ CD44+ NKG2D− and CD4+ CD44+ NKG2D+ in spleen of Tbx21-ZsGreen mice (n = 3). (J) Frequency of splenic NKG2D+ CD44+ CD4+ T cells from WT (n = 15) and Tbx21−/− mice (n = 13); Mann–Whitney U test. In C, E, H, J, and M, each symbol represents a mouse, lines represent mean ± SEM, and data are pooled from at least three independent experiments.
Figure S1.
NKG2D is expressed on antigen-experienced CD4+ T cells in steady-state C57BL/6 mice. (A) FC gating strategy for the characterization of NKG2D+ CD4+ T cells in C57BL/6 mice (spleen shown). Lineage (Lin) includes CD19, FcεRI, Gr-1, and F4/80. (B) Representative FC analysis of indicated surface marker expression on NKG2D+ (red line) and NKG2D− (black line) CD44+ CD4+ T cells in the spleen. Shaded histogram represents live CD19+ cells (n = 3). (C) Representative FC analysis of DNAM-1 and CD94 expression on splenic CD4+ CD44+ NKG2D− (black line) and CD4+ CD44+ NKG2D+ (red line). Shaded histogram represents live CD19+ cells (n = 6). (D) FC analysis of selected surface marker expression on NKG2D+ (red line) and NKG2D− (black line) CD44+ CD4+ T cells in spleen. Shaded histogram represents NK cells (n = 3). (E) Representative FC analysis of NKG2D expression on CD44+ CD4+ T cells from spleen of WT, Tyrobp−/−, Hcst−/−, and Klrk1−/− mice. (F) Frequency of NKG2D+ CD44+ CD4+ T cells, quantification of E. Each symbol represents a mouse; line represents the mean ± SEM (n = 3). Data are pooled from two independent experiments. (G) Representative FC analysis of indicated marker expression on NKG2D+ and NKG2D− CD44+ CD4+ T cells in the siLP of Tbx21-ZsGreen mice (n = 3). (H) Representative FC analysis of FoxP3 and CD25 expression on NKG2D+ (red) and NKG2D− (gray) CD4+ T cells from the spleen of WT mice (n = 3). In A–D, G, and H, data are representative of at least two independent experiments.
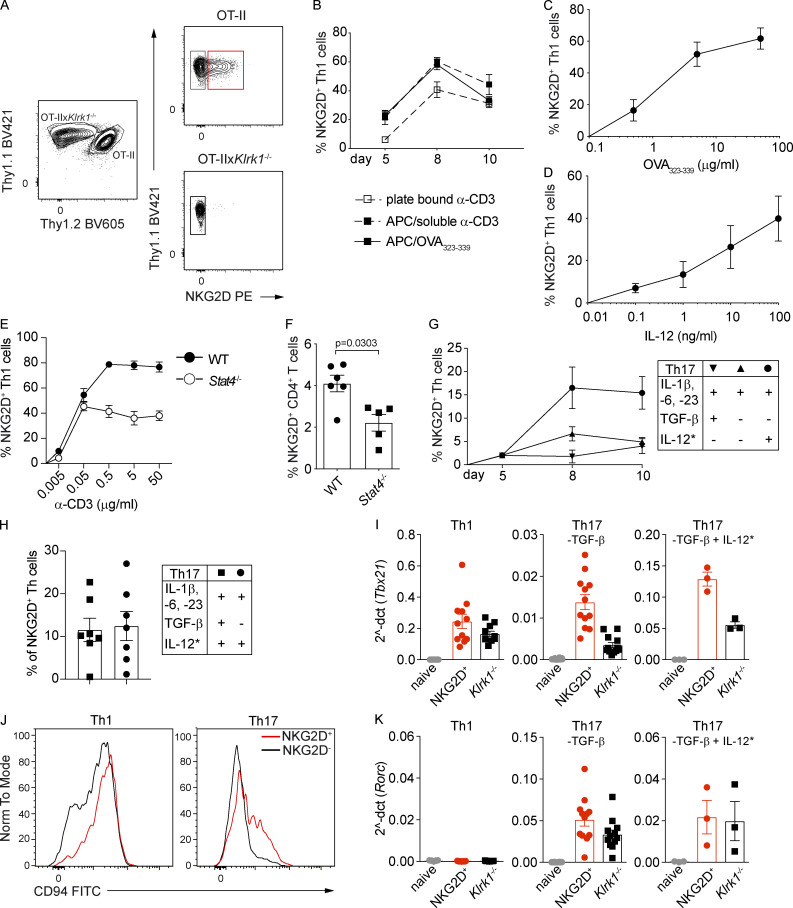
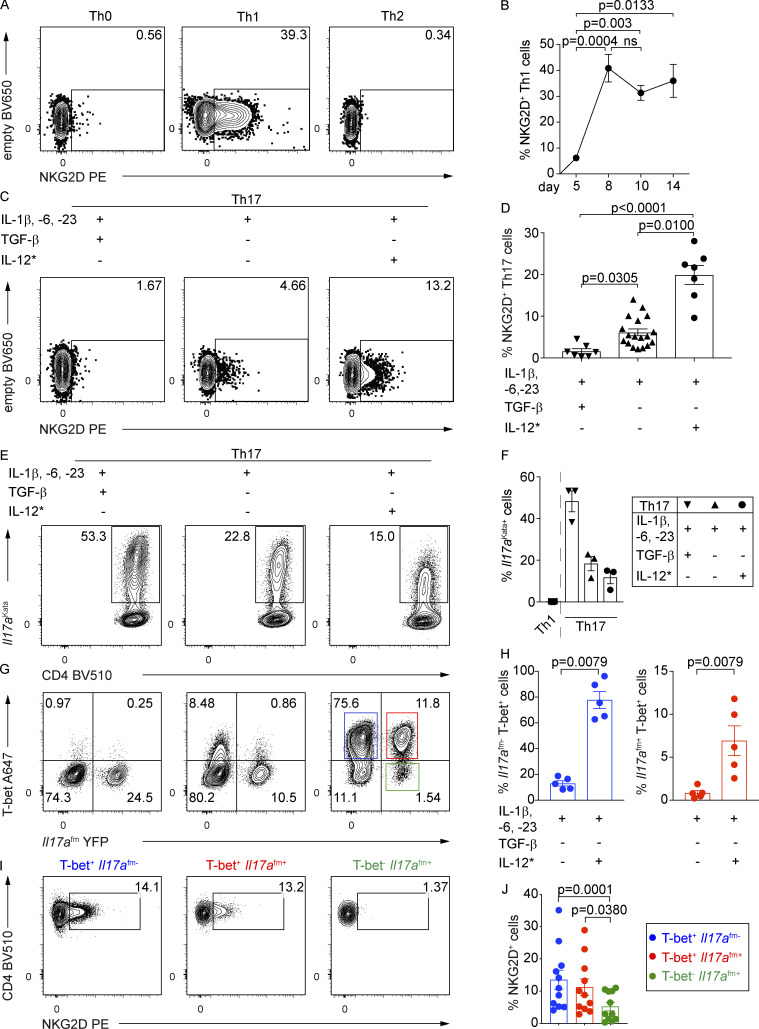
We next evaluated whether NKG2D can be induced during in vitro Th polarization of naive (NKG2D−) CD4+ T cells, by using a coculture system of OT-II and OT-IIxKlrk1−/− cells (Fig. S2 A). NKG2D was strongly up-regulated on Th1 and absent on Th0 and Th2 cells (Fig. 2 A). The percentage of NKG2D+ Th1 cells peaked at day 8 and remained stable until day 14 (Fig. 2 B), showing similar kinetics of expression in cultures generated in both the presence and absence of antigen-presenting cells (APCs; Fig. S2 B). By dissecting the signals promoting NKG2D up-regulation, we observed a dose-dependent role for IL-12 and antigen stimulation (Fig. S2, C and D). Although NKG2D could be induced in Stat4−/− Th1 cells, these cells failed to induce further enhancement of NKG2D expression upon increasing strength of TCR signaling, suggesting a synergistic role for these two pathways (Fig. S2 E). Further highlighting the contribution of IL-12 in the up-regulation of NKG2D, the frequency of NKG2D+ CD4+ T cells observed ex vivo was reduced in Stat4−/− mice (Fig. S2 F).
Figure S2.
NKG2D is induced de novo on naive CD4+ T cells under Th1- and Th17-polarizing conditions. (A) Thy1.2+ OT-II and Thy1.2− OT-IIxKlrk1−/− naive CD4+ T cells were cocultured and polarized toward different Th lineages as described in Materials and methods. (B) Frequency of NKG2D+ Th1 cells generated from WT (α-CD3) or OT-II (OVA323–339) mice in the presence or absence of APCs. Data shown are mean ± SEM, n = 15 (pCD3), n = 6 (sCD3/APC), and n = 16 (OVA323–339/APC). (C) Frequency of NKG2D+ Th1 cells in response to increasing doses of OVA323–339. Data shown are mean ± SEM; line connects the mean (n = 5). (D) Frequency of NKG2D+ Th1 cells in response to increasing doses of IL-12. Data shown are mean ± SEM (n = 6). (E) Frequency of NKG2D+ Th1 cells derived from WT or Stat4−/− mice in response to increasing doses of α-CD3. Mean ± SEM is shown; line connects the means (n = 3). (F) Frequency of NKG2D+ CD4+ T cells after gating on CXCR3+ CD44+ population. Each symbol represents a mouse. Data shown are mean ± SEM; n = 6 (WT) and n = 5 (Stat4−/−). Mann–Whitney U test. ns, not significant. (G) Frequency of NKG2D+ Th17 polarized as indicated, at different time points of the culture. Data show mean ± SEM (n = 3–7). Asterisk (*) indicates that IL-12 was added in the second week of the culture. (H) Frequency of NKG2D+ Th17 polarized as indicated. Data show mean ± SEM (n = 7). (I) qPCR analysis of Tbx21 expression in Th1 and Th17 cells sorted as NKG2D+ or Klrk1−/− cells and in naive CD4+ T cells. Values are normalized to housekeeping genes Eef1a1 and Ube2g1. Each symbol represents an individual culture. Data show mean ± SEM; n = 10 (Th1), n = 12 (Th17; -TGF-β), and n = 3 (Th17; -TGF-β + IL-12*). (J) Representative FC analysis of CD94 expression on NKG2D+ and Klrk1−/− Th1 or Th17 (-TGF-β + IL-12*) cells on day 10 of culture (n = 6). (K) qPCR analysis of Rorc expression in Th1 and Th17 cells sorted as NKG2D+ or Klrk1−/− cells and in naive CD4+ T cells. Values are normalized to housekeeping genes Eef1a1 and Ube2g1. Each symbol represents an individual culture. Data show mean ± SEM; n = 10 (Th1), n = 11 (Th17; -TGF-β), and n = 3 (Th17; -TGF-β + IL-12*). In A–K, data are representative of at least three independent experiments.
Figure 2.
NKG2D is induced de novo on naive CD4+ T cells under Th1- and Th17-polarizing conditions. (A) Representative FC analysis of NKG2D expression on Th0, Th1, or Th2 cells on day 10 of culture (n = 6). (B) Frequency of NKG2D+ Th1 cells, differentiated using plate-bound α-CD3 stimulation, at indicated time points of the polarizing culture. Data shown are mean ± SEM (n = 5–15), Kruskal–Wallis test with multiple comparison correction. ns, not significant. (C) Representative FC analysis of NKG2D expression on Th17 cells, polarized as indicated, on day 10. Asterisk (*) indicates that IL-12 was added in the second week of culture. (D) Frequency of NKG2D+ Th17 cells, polarized as indicated, as measured on day 8 or 10 of culture; black inverted triangle (n = 7), black triangle (n = 18), and black circle (n = 7). Each symbol represents an individual culture replicate; lines represent mean ± SEM. Kruskal–Wallis test with multiple comparison correction. (E and F) Representative FC analysis (E) and frequency of Il17aKata expression in Th17 cells (F), from Il17afm(YFP) × Il17aKata double reporter mice–derived naive CD4+ T cells, polarized as indicated, at day 10 of culture (n = 3). Each symbol represents an individual culture replicate; lines represent mean ± SEM. (G) Representative FC analysis of T-bet and Il17a(fm)YFP expression in Th17 cells, polarized as in E. (H) Frequency of Il17afm-T-bet+ and Il17afm+T-bet+ cells among Th17 cells polarized as indicated (n = 5). Data shown as mean ± SEM; Mann–Whitney U test. (I) Representative FC analysis of NKG2D expression in Th17 cell subsets, polarized with IL-1β, IL-6, IL-23, and IL-12*, indicated in G; blue square, T-bet+fm−; red square, T-bet+fm+; green square, T-bet−fm+ cells. (J) Frequency of NKG2D+ Th17 cells, quantification of I. Each symbol represents an individual culture replicate; data shown as mean ± SEM (n = 11). Kruskal–Wallis test with multiple comparison correction. In A–J, data are pooled from at least three independent experiments.
To test whether NKG2D was also expressed during Th17 differentiation, naive T cells were cultured with IL-1β, IL-6, and IL-23: (a) in the presence of TGF-β (Mangan et al., 2006; Veldhoen et al., 2006), (b) in the absence of TGF-β (Ghoreschi et al., 2010), and (c) in the absence of TGF-β but with the addition of IL-12 during the second week of culture (Lexberg et al., 2010; Wang et al., 2014; Fig. 2 C). Extending the data of Th1 cells, NKG2D expression was up-regulated at approximately day 8 (and remained stable up to day 10) on a small fraction of Th17 cells cultured preferentially in the absence of TGF-β, and was strongly enhanced in the presence of IL-12 (Fig. 2, C and D; and Fig. S2 G). Addition of IL-12 resulted in comparable up-regulation of NKG2D on Th17 cells generated in both the presence and absence of TGF-β (Fig. S2 H). Given the already reported plasticity of Th17 cells and their ability to acquire T-bet and Th1 features, especially in the absence of TGF-β and in the presence of IL-12 (Ghoreschi et al., 2010; Hirota et al., 2011; Lexberg et al., 2010), we also assessed the expression of NKG2D in Th17 cultures using cells from the double reporter mouse model, obtained by crossing Il17a fate reporter (Il17a(fm)YFP) with Il17aKatushka (Il17aKata) reporter mice (Gagliani et al., 2015; Hirota et al., 2011). In this system, Katushka (Kata) marked cells with actual transcription of Il17a, whereas YFP permanently marked cells that previously expressed Il17a, thus enabling the identification of T-bet+ Th17 cells that had lost the expression of IL-17A. All three Th17 polarizing conditions enabled the generation of Il17aKata-expressing cells, although the absence of TGF-β decreased their frequency (Fig. 2, E and F), as expected (Ghoreschi et al., 2010). Addition of IL-12 strongly promoted induction of T-bet+ Il17a(fm)+ as well as T-bet+ Il17a(fm)− cells (Fig. 2, G and H). NKG2D expression was comparable between T-bet+ Il17a(fm)− and T-bet+ Il17a(fm)+ but almost absent in T-bet− Il17a(fm)+, suggesting that NKG2D is preferentially up-regulated on Th17 cells that had acquired T-bet (Fig. 2, I and J). Altogether, our results indicate that NKG2D expression is a selective feature associated ex vivo and in vitro with Th1 and Th17 cell differentiation.
NKG2D+ Th1 and Th17 cells express a reinforced type 1 response gene signature
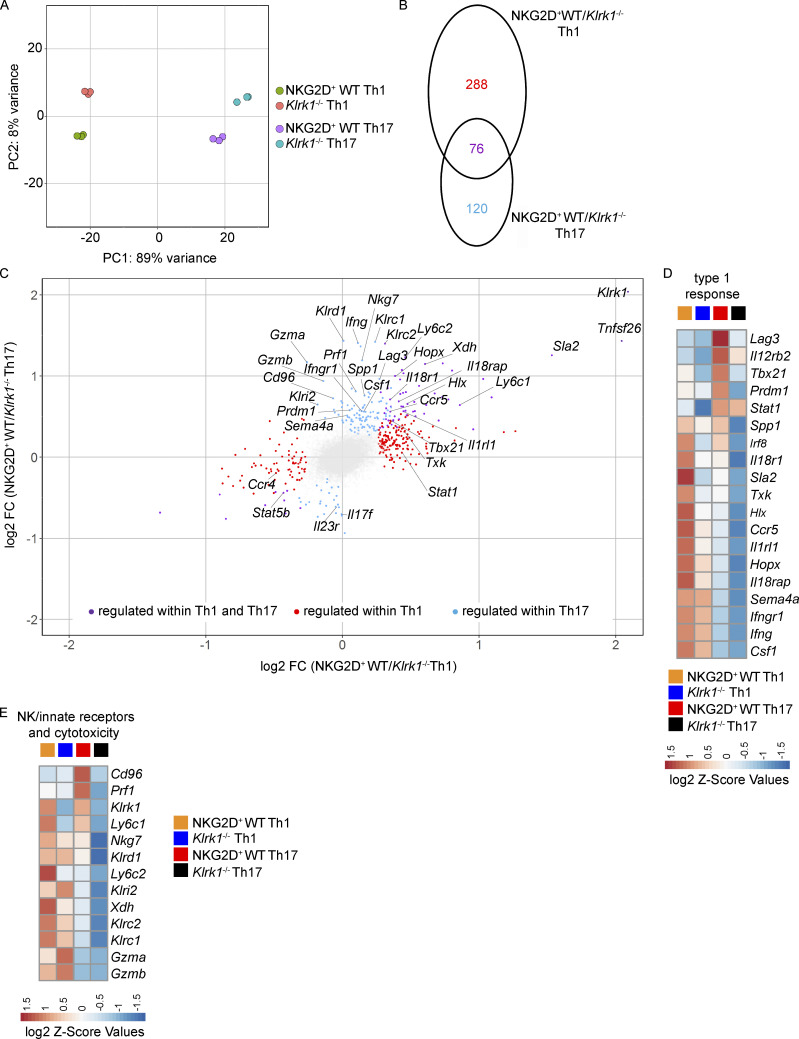
We further performed comparative analysis of global gene expression between in vitro–generated NKG2D+ and Klrk1−/− Th1 and Th17 cells. Principal component analysis of the data revealed clear separate clustering of Th1 and Th17 cells (showing 89% of the variance in principal component 1 [PC1]) irrespective of NKG2D expression (Fig. 3 A). Genes associated with the expression of NKG2D appeared to drive PC2, showing 8% of variance in each subset. We obtained 484 genes significantly differentially regulated between NKG2D+ and Klrk1−/− subsets, from which 288 were regulated by NKG2D exclusively within Th1 cells and 120 among Th17; 76 were coregulated (Fig. 3 B and Table S1). Transcripts for Ifng, as well as for a large set of genes associated with the regulation of IFN-γ production, IL-12–mediated signaling pathways, and type 1 immune responses, including Tbx21, Il18r1, Il18rap, Ifngr1, Hlx, and Hopx, were differentially regulated between NKG2D+ and Klrk1−/− Th cells (Fig. 3, C and D; and Fig. S2 I). Moreover, NKG2D+ cells were enriched in the expression of a variety of NK/innate receptors and molecules associated with cytotoxicity, such as Klrc1 (NKG2A/B), Klrc2 (NKG2C), Klrd1 (CD94), Gzma, Gzmb, and Prf1 compared with Klrk1−/− cells, particularly within Th17 cells (Fig. 3, C and E; and Fig. S2 J). While significantly different between Th1 and Th17 cultures, Rorc and Il17a transcript expression was not differentially regulated between NKG2D+ and Klrk1−/− Th17 cells (Figs. 3 C) and S2 K). Collectively, these data show that NKG2D reinforces type 1 response gene signature, especially in Th17 cells.
Figure 3.
NKG2D is associated with reinforced type 1 response gene signature in in vitro–generated Th1 and Th17 cells. (A–E) Transcriptome analysis of Th1 and Th17 (IL-1β, -6, -23, and IL-12*) cells sorted as NKG2D+ or Klrk1−/−. (A) Principal component analysis of Th cell subsets based on differentially expressed genes for each of the comparisons (filtered for padj < 0.05, Mann–Whitney U test). Each dot represents an independent culture (n = 3). (B and C) Venn diagram showing number of transcripts (filtered for padj < 0.05; B) and scatter plot showing transcripts of genes differentially expressed in NKG2D+ and Klrk1−/− cells (filtered for padj < 0.05 and FC >1.2; C) preferentially in Th1 (red), Th17 (blue), or both Th1 and Th17 lineages (purple). Gray dots represent the genes below the selected threshold. (D and E) Z-score heat maps showing relative expression of selected genes in the four T cell subsets. Selected genes are filtered for padj < 0.05 and FC >1.2 in at least one comparison.
NKG2D promotes the expression of proinflammatory cytokines
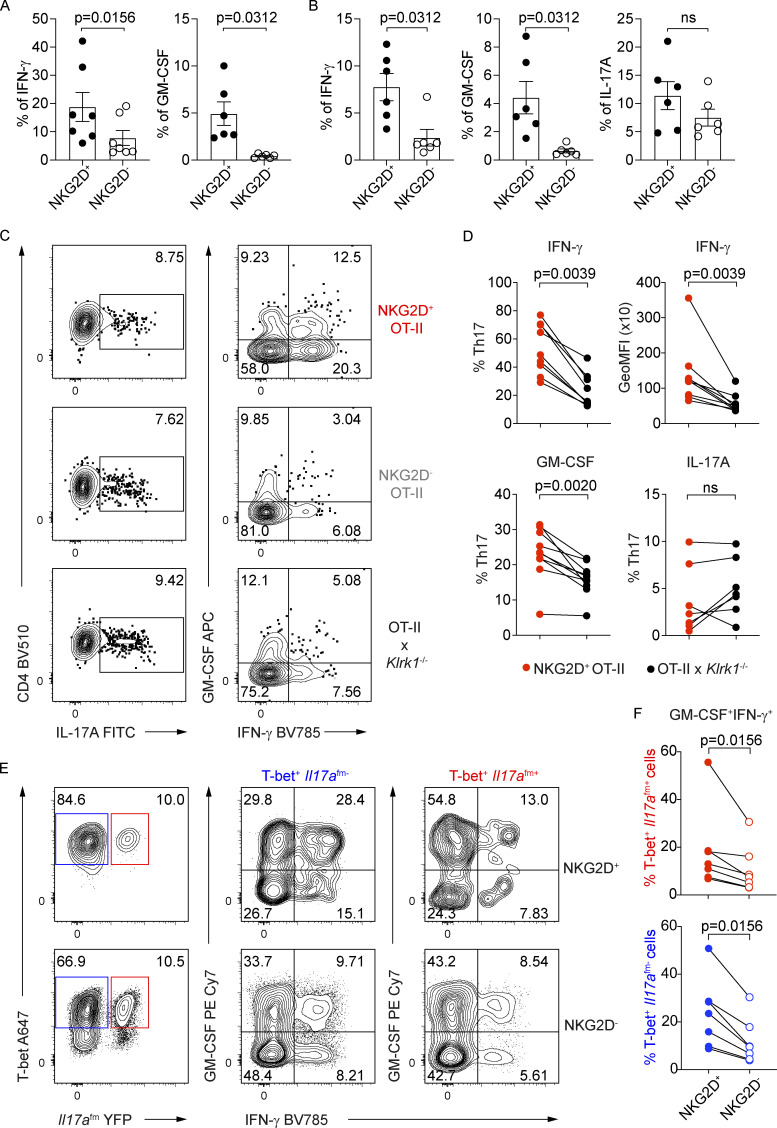
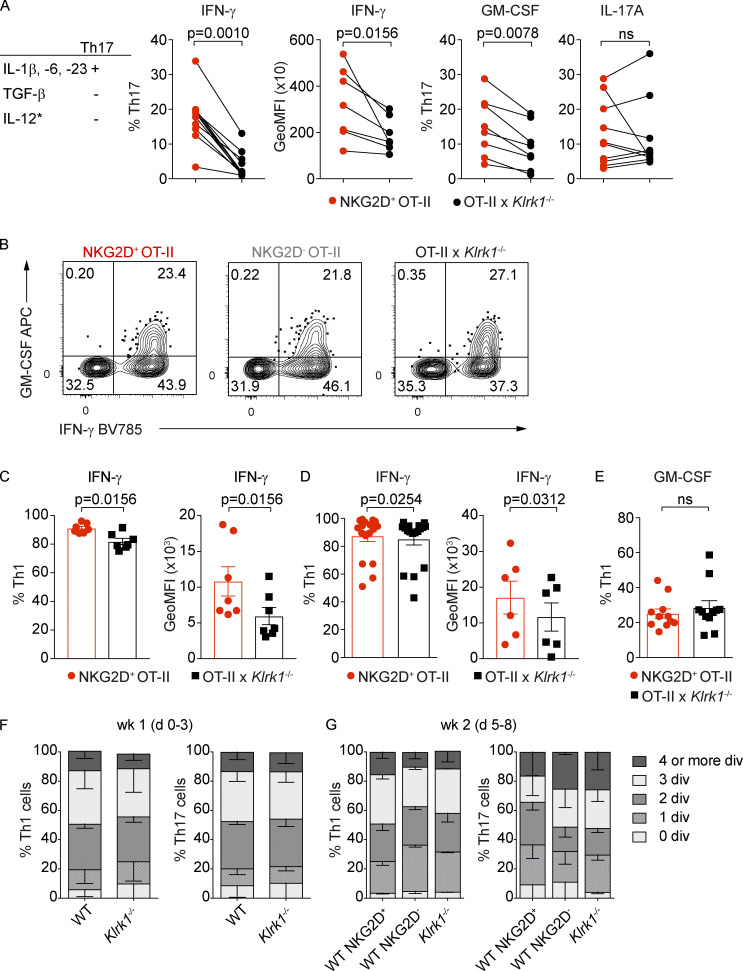
Given their phenotype of antigen-experienced cells ex vivo and their association with Th1 and Th17 lineages in vitro, we wanted to assess the functional potential of NKG2D+ Th cells, both at steady-state and after in vitro differentiation. Intracellular cytokine expression analysis after restimulation showed that the expression of IFN-γ and GM-CSF was increased in NKG2D+ compared with NKG2D− Th cells isolated ex vivo from spleen or siLP (Fig. 4, A and B). Restimulated NKG2D+ Th17 cells, generated in vitro, showcased higher IFN-γ and GM-CSF expression compared with their NKG2D− or Klrk1−/− counterparts (Fig. 4, C and D; and Fig. S3 A), whereas the effect was less visible or even absent under Th1 polarizing conditions (Fig. S3, B–E). However, supporting the role for NKG2D in modulating IFN-γ and GM-CSF expression in T-bet+ Th cells, NKG2D expression was associated with a higher frequency of IFN-γ+ GM-CSF+ cells in both T-bet+ Il17a(fm)- and T-bet+ Il17a(fm)+ Th cells (Fig. 4, E and F). A significant difference in IL-17A protein expression was not observed between NKG2D+ and NKG2D− Th cells isolated ex vivo from siLP (Fig. 4 B) or between NKG2D+, NKG2D−, and Klrk1−/− Th17 cells generated in vitro (Fig. 4, C and D). Differential expression of cytokines did not reflect distinct proliferative ability, as NKG2D+ Th17 and Th1 cells underwent a comparable number of divisions as their NKG2D− and Klrk1−/− counterparts (Fig. S3, F and G).
Figure 4.
NKG2D+ Th17 and Th1 cells are enriched in IFN-γ and GM-CSF producers. (A and B) Frequency of IFN-γ+ and GM-CSF+ CD44+ CD4+ T cells in spleen (A) and of IFN-γ+, GM-CSF+, and IL-17A+ CD44+ CD4+ T cells in siLP (B) of C57BL/6 mice (n = 6–7), Wilcoxon test. Each symbol represents a mouse; lines represent mean ± SEM; data are pooled from at least three independent experiments. (C) Representative FC analysis of intracellular cytokine expression in Th17 cells, polarized with IL-1β, IL-6, IL-23, or IL-12*, on day 10 after sorting for NKG2D+ OT-II, NKG2D− OT-II, and OT-IIxKlrk1−/− and PMA/Iono restimulation. (D) Frequency of indicated cytokine-producing cells and per-cell content of IFN-γ among Th17 cells, quantification of C. GeoMFI, geometric mean fluorescence intensity; ns, not significant. (E) Representative FC analysis of Il17afm(YFP), T-bet, and intracellular cytokine expression in NKG2D+ and NKG2D− Th17 cells, polarized as in C. (F) Frequency of Th17 cell subsets, expressing indicated cytokines, quantification of E. In D and F, each symbol represents the individual culture replicate; lines connect matching samples from individual cocultures (n = 7–10); Wilcoxon test. Data are pooled from at least three independent experiments.
Figure S3.
NKG2D modulates the cytokine production of CD4+ Th cells in vitro. (A) Frequency of indicated cytokine-producing cells and per-cell content of IFN-γ among Th17 polarized with IL-1β, IL-6, or IL-23. Each symbol represents the individual coculture replicate; line connects matching samples (n = 7–10); Wilcoxon test. Data are pooled from at least three individual experiments. GeoMFI, geometric mean fluorescence intensity. (B) Representative FC analysis of intracellular cytokine expression in Th1 cells on day 10 after sorting for NKG2D+ OT-II, NKG2D− OT-II, and OT-IIxKlrk1−/− and PMA/Iono restimulation. (C and D) Frequency of IFN-γ+ and per-cell content of IFN-γ in NKG2D+ (red) and Klrk1−/− (black) 1-wk (C) or 2-wk (D) differentiated Th1 cells. Each symbol represents an individual culture; line represents mean ± SEM, n = 7 (C), n = 17 (%), or 6 (GeoMFI; D). Wilcoxon test. Data are pooled from at least three individual experiments. (E) Frequency of GM-CSF+ among NKG2D+ (red) and Klrk1−/− (black) Th1 cells. Each symbol represents an individual culture from at least three independent experiments; line represents mean ± SEM; n = 10. (F and G) Frequency of cells undergoing 0, 1, 2, 3, 4, and more divisions (div.) among WT and Klrk1−/− Th1 or Th17 cells (day 3) or among NKG2D+ WT, NKG2D− WT and Klrk1−/− Th1 or Th17 cells (day 8). Th17 cells were polarized as in A. Data shown are mean ± SEM; n = 3; pooled from at least two independent experiments.
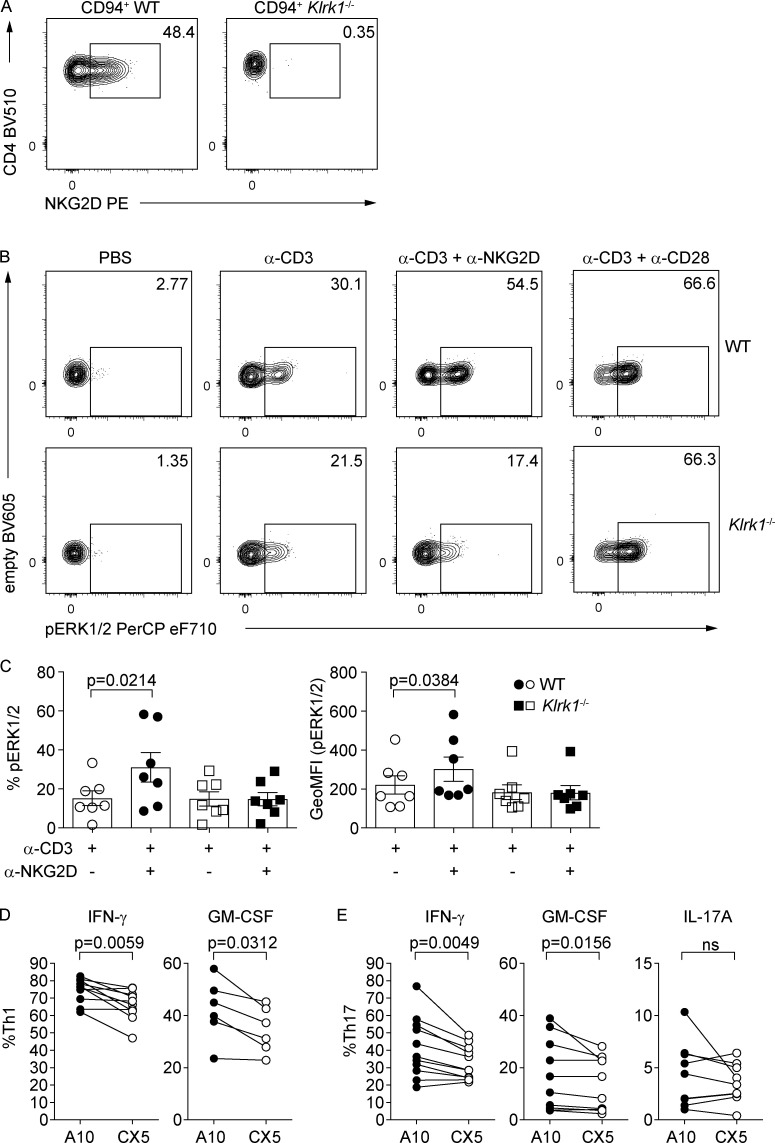
Because NKG2D acts as a costimulatory molecule in CD8+ T cells (Diefenbach et al., 2002), we investigated whether the differences in cytokine expression observed in our experiments could be linked to direct NKG2D engagement and downstream signaling also in CD4+ Th cells. To this aim, levels of ERK1/2 phosphorylation (pERK1/2) were analyzed in WT and Klrk1−/− Th1 cells, after sorting for CD94 expression, which allows for the enrichment of NKG2D+ Th1 cells (Fig. 5 A). Combined cross-linking of α-CD3 and α-NKG2D resulted in significant pERK1/2 increase in WT but not Klrk1−/− Th1 cells, indicating an active contribution of NKG2D signaling in CD4+ Th cells (Fig. 5, B and C). Of note, no differences in pERK1/2 levels could be observed between WT and Klrk1−/− Th1 cells upon α-CD3 or α-CD3 + α-CD28 cross-linking (Fig. 5, B and C), demonstrating that NKG2D deficiency is not associated with impaired TCR signaling. Next, we tested whether NKG2D engagement could directly contribute to cytokine modulation in Th1 and Th17 cells. NKG2D cross-linking by the activating α-NKG2D antibody (A10) in the presence of suboptimal levels of α-CD3 selectively resulted in elevated expression of IFN-γ and GM-CSF in Th1 and Th17 cells, compared with the engagement by α-NKG2D blocking antibody (CX5; Fig. 5, D and E), whereas IL-17A levels in Th17 cells were not significantly impacted. Altogether, our data suggest that NKG2D expression is associated with higher IFN-γ and GM-CSF expression, and that NKG2D costimulation contributes to modulate cytokine production in Th1 and Th17 cells.
Figure 5.
NKG2D signaling promotes IFN-γ and GM-CSF expression. (A) Representative FC analysis of NKG2D on WT or Klrk1−/− Th1 cells after gating on CD94+ cells on day 10 of culture. (B) Representative FC analysis of ERK1/2 phosphorylation after antibody cross-linking of CD3, CD3 and NKG2D (clone A10), or CD3 and CD28 in WT and Klrk1−/− Th1 cells. (C) Frequency of pERK1/2+ Th1 cells (left) and per-cell content (right) of pERK1/2, quantification of B. Data shown are pooled from individual cocultures; line shows mean ± SEM (n = 7); Friedman test with Dunn´s multiple comparison test. GeoMFI, geometric mean fluorescence intensity. (D and E) Th1 and Th17 (IL-1β, IL-6, IL-23, or IL-12*) cells were sorted as NKG2D+ using A10 (activating; black circles) or CX5 (blocking; open circles) α-NKG2D antibody, cross-linked by plate-bound secondary antibody in the presence of α-CD3 for 6 h, and analyzed by FC for intracellular cytokine expression. Frequency of IFN-γ+ (n = 10) and GM-CSF+ Th1 (n = 6; D) and IFN-γ+ (n = 11), GM-CSF+ (n = 10), and IL-17A+ (n = 9) Th17 (E). Each symbol represents the value of the individual coculture; lines connect matching samples; Wilcoxon test. ns, not significant. Data are pooled from at least three independent experiments.
NKG2D marks proinflammatory Th1/Th17 cells in vivo
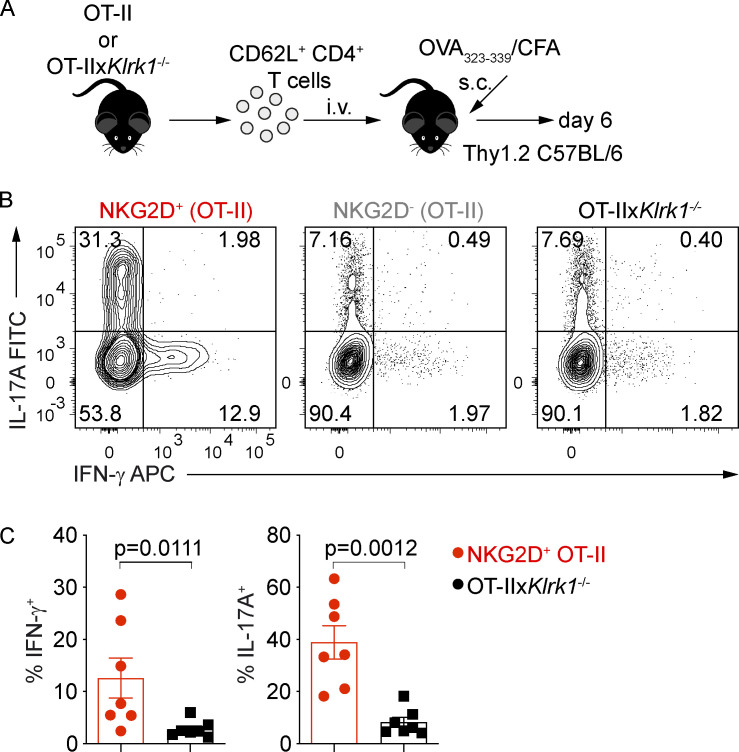
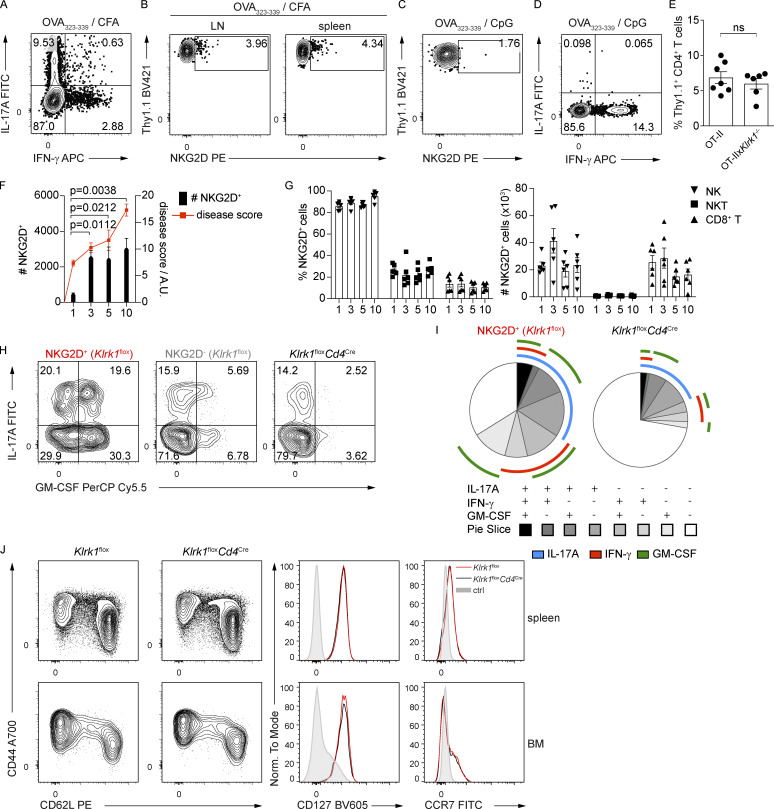
We further investigated whether NKG2D could be induced in CD4+ Th cells under inflammatory conditions in vivo and intrinsically modulate their cytokine production and pathogenicity. De novo up-regulation of NKG2D expression was tested on naive NKG2D− OT-II+ CD4+ T cells adoptively transferred into C57BL/6 recipients immunized with OVA323–339 peptide in CFA (Fig. 6 A), which is able to efficiently induce a Th1/Th17 response in vivo (Coffman et al., 2010; Fig. S4 A). Whereas under these conditions, cells recovered from LN and spleen of recipient mice expressed NKG2D (Fig. S4 B), the same was not observed if mice were immunized using the same antigen mixed with CpG (Fig. S4 C), a condition that preferentially induced a Th1 response (Saraiva et al., 2009; Fig. S4 D). Of note, Klrk1 deficiency did not impact the recovery of cells upon adoptive transfer and OVA323–339/CFA immunization (Fig. S4 E). In line with our in vitro data, NKG2D+ OT-II+ CD4+ T cells generated in vivo upon OVA323–339/CFA immunization were highly enriched in IFN-γ producers compared with their NKG2D− and Klrk1−/− counterparts (Fig. 6, B and C). Interestingly, and in contrast to our in vitro data, NKG2D+ OT-II+ CD4+ T cells were also enriched in IL-17A (Fig. 6, B and C).
Figure 6.
NKG2D marks proinflammatory Th17 cells in vivo. (A) Schematic representation of the experimental workflow during antigen/adjuvant immunizations. (B and C) Representative FC analysis (B) and quantification of intracellular cytokine expression (C) in transferred OT-II cells gated as NKG2D+ and NKG2D−, or Klrk1−/− OT-II cells, from draining lymph nodes of recipient mice at day 6 after immunization with OVA323–339/CFA and after restimulation with PMA/Iono. Each symbol represents a mouse. Line represents mean ± SEM (n = 7), Mann–Whitney U test. Data are pooled from two independent experiments.
Figure S4.
NKG2D modulates the cytokine production in CD4+ T cells in vivo and promotes immunopathology in antigen-induced arthritis. (A) Representative FC analysis of intracellular cytokine expression in transferred OT-II cells, from draining lymph nodes of recipient mice at day 6 after immunization with OVA323–339/CFA and after restimulation with PMA/Iono (n = 7). Data are representative of two independent experiments. (B) Representative FC analysis showing NKG2D expression on transferred WT OT-II+ CD4+ T cells 6 d after OVA323–339/CFA immunization, n = 7. Data are representative of two independent experiments. (C and D) Representative FC analysis of NKG2D expression (C) and intracellular cytokine expression (D) in OT-II+ CD4+ T cells in dLN after OVA323–339/CpG immunization (n = 5). Data are representative of two independent experiments. (E) Frequency of recovered OT-II and OT-IIxKlrk1−/− transferred cells, from dLN of recipient mice at day 6 after immunization with OVA323–339/CFA. Data show mean ± SEM; n = 7 (OT-II) and 6 (OT-IIxKlrk1−/−). ns, not significant. Data are pooled from two independent experiments. (F) Graph representing absolute numbers of NKG2D+ CD40L+ CD4+ T cells (black bars) in the dLN of mice and histological disease score (red dots) in the knee joints of mice at indicated time points after arthritis induction. Bar graph data are mean ± SEM (n = 6), Kruskal–Wallis test; red line connects means of score values (n = 6). Data are pooled from two independent experiments. A.U., arbitrary units. (G) Frequency and absolute numbers of NKG2D-expressing cells analyzed on indicated cell subsets from dLN during the course of OIA; data represent mean ± SEM; n = 6 for each time point. Data are pooled from two independent experiments. (H) Representative FC analysis of intracellular cytokine expression in OVA-restimulated NKG2D+ and NKG2D− CD40L+ CD4+ T cells in Klrk1flox mice and CD40L+ CD4+ T cells in Klrk1floxCd4Cre mice from dLN at day 10 after the OIA induction. Data are representative of three independent experiments. (I) SPICE charts depict combinatorial expression of IL-17A, IFN-γ, and GM-CSF in NKG2D+ CD40L+ CD4+ T cells in dLN of Klrk1flox mice and CD40L+ CD4+ T cells in Klrk1floxCd4Cre mice, at day 10 after OIA induction (n = 9, Klrk1flox; n = 7, Klrk1floxCd4Cre). Data are pooled from two independent experiments. (J) Representative FC analysis of CD44, CD62L, CD127, and CCR7 expression on CD4+ T cells from the spleen and BM of Klrk1flox (red line) and Klrk1floxCd4Cre (black line) mice. Shaded histogram represents live CD19+ cells (n = 3). Data are representative of two independent experiments.
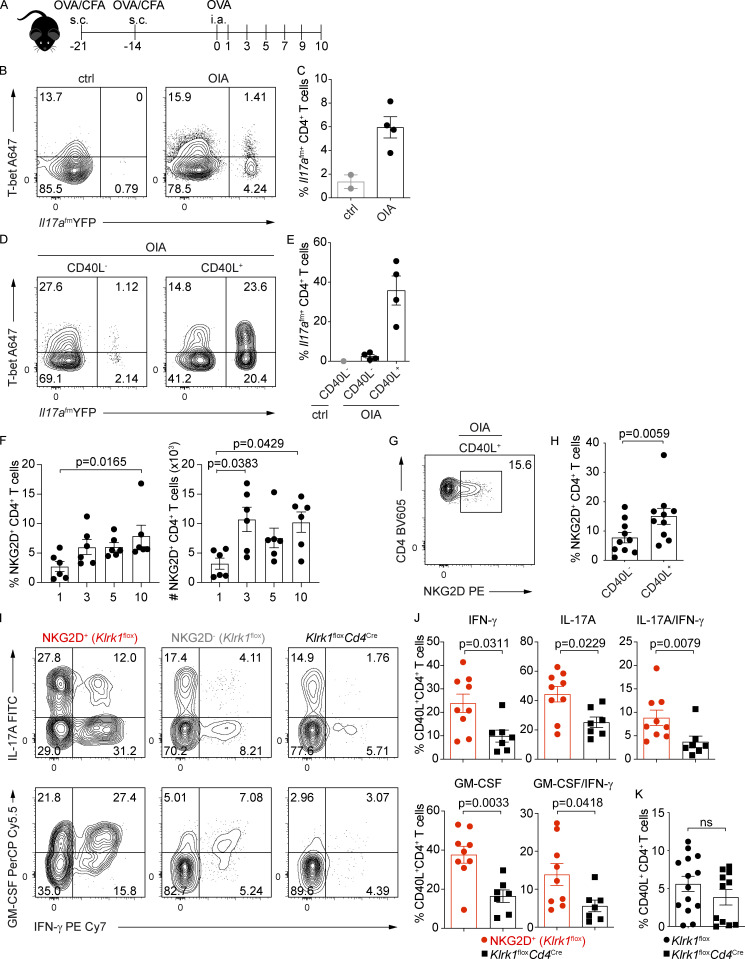
To test NKG2D expression and the role of NKG2D+ Th1/Th17 cells in a relevant model of inflammation, we analyzed Th1/Th17 cell–mediated responses in a model of OVA-induced arthritis (OIA; Fig. 7 A). By using the Il17a fate map reporter mice, we could demonstrate significant accumulation of Il17a(fm)+ CD4+ T cells, both T-bet+ and T-bet− ones, in the draining LN of diseased mice (Fig. 7, B and C). Enrichment of Il17a(fm)+ CD4+ T cells was most prominent within the pool of antigen-specific cells, as measured by their expression of CD40L after antigen restimulation (Frentsch et al., 2005; Fig. 7, D and E). CD4+ T cells expressing NKG2D accumulated in the draining LN of immunized WT mice (Fig. 7 F) and particularly within antigen-specific cells (Fig. 7, G and H), paralleling the increasing disease score (Fig. S4 F). Conversely, we did not observe major perturbations in the frequency of NKG2D-expressing cells among NK, NKT, or CD8+ T cells (Fig. S4 G).
Figure 7.
NKG2D-expressing antigen-specific CD4+ T cells accumulate in vivo during OIA. (A) Schematic representation of the experimental workflow during OIA. Mice were s.c. immunized with catOVA/CFA. Disease was induced with i.a. injection of catOVA (OIA). Control animals received only i.a. injection of catOVA (ctrl). Cells from draining LN were analyzed on day 10 after disease induction and after in vitro restimulation with catOVA. (B and C) Representative FC analysis (B) and quantification of the frequency of Il17a(fm)+ cells among CD44+ CD4+ T cells (C) in control and OIA animals 10 d after disease induction. (D and E) Representative FC analysis (D) and quantification of the frequency of Il17a(fm)+ cells (E) among CD44+ CD40L− CD4+ T cells in control and CD44+ CD40L− and CD44+ CD40L+ CD4+ T cells in OIA animals 10 d after disease induction. In C and E, each symbol represents an individual mouse; data shown as mean ± SEM; n = 2 for control and n = 4 for OIA animals. Data are pooled from two independent experiments. (F) Frequency and absolute numbers of NKG2D+CD44+CD4+ T cells in the dLN of mice with OIA at indicated time points after disease induction. Data show mean ± SEM from two independent experiments (n = 6); Kruskal–Wallis test with multiple comparison correction. (G) Representative FC analysis of NKG2D expression on CD44+ CD40L+ CD4+ T cells from Klrk1flox mice 10 d after disease induction. (H) Frequency of NKG2D-expressing CD44+ CD4+ T cells among indicated subsets in mice with OIA (n = 10); Wilcoxon test. Line shows mean ± SEM pooled from three independent experiments. (I and J) Representative FC analysis (I) and quantification of intracellular cytokine expression in NKG2D+ and NKG2D− CD40L+ CD4+ T cells of Klrk1flox mice (n = 9) and CD40L+ CD4+ T cells from Klrk1floxCd4Cre mice (n = 7; J); Mann–Whitney U test. Each symbol represents an individual mouse; data show mean ± SEM from three independent experiments. (K) Frequency of CD40L+ CD4+ T cells in Klrk1flox (n = 14) and Klrk1floxCd4Cre (n = 11) mice; Mann–Whitney U test. ns, not significant. Each symbol represents an individual mouse; data show mean ± SEM from three independent experiments.
NKG2D expression marked IL-17A, IFN-γ, and GM-CSF producers (Fig. 7, I and J; and Fig. S4, H and I). IL-17A, IFN-γ, and GM-CSF expression in NKG2D-deficient CD40L+ CD4+ T cells, i.e., from Klrk1floxCd4Cre mice, was strongly reduced compared with NKG2D+ CD40L+ CD4+ T cells from Klrk1flox mice (Fig. 7, I and J; and Fig. S4, H and I). This did not reflect an impact of NKG2D on the size of the CD4+ T cell pool, as the frequency of antigen-specific T cells generated was not affected by NKG2D deficiency (Fig. 7 K). Accordingly, the phenotype of CD4+ T cells, as measured by CD44, CD62L, CCR7, and CD127 expression, was comparable in Klrk1floxCd4Cre and Klrk1flox mice (Fig. S4 J), indicating that NKG2D deficiency does not impact the formation of primary CD4+ Th cell responses.
NKG2D intrinsically promotes T cell pathogenicity during antigen-induced arthritis and EAE
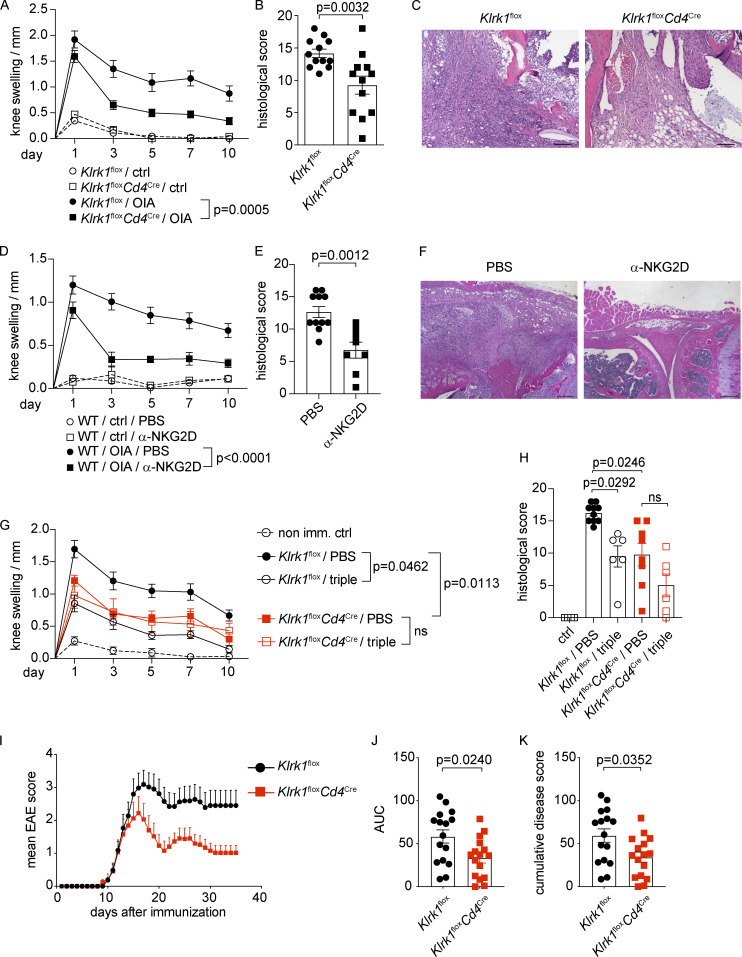
Finally, we evaluated whether conditional NKG2D deficiency in CD4+ T cells could affect disease severity and pathology. Klrk1floxCd4Cre mice displayed significantly decreased OIA disease severity and pathology compared with littermate control (Klrk1flox) mice, as measured by reduced knee swelling as well as improved histological disease score (Fig. 8, A–C). Interestingly, disease amelioration in Klrk1floxCd4Cre mice was comparable to that observed after treatment of C57BL/6 mice with a blocking α-NKG2D antibody (Fig. 8, D–F), indicating that selective deficiency of NKG2D in T cells is sufficient to ameliorate antigen-induced arthritis.
Figure 8.
NKG2D promotes immunopathology in antigen-induced arthritis and EAE. (A–H) C57BL/6, Klrk1flox or Klrk1floxCd4Cre were s.c. immunized with catOVA/CFA. Disease was induced with i.a. injection of catOVA (OIA). Control animals received only i.a. injection of catOVA (ctrl). (A) Knee swelling measurement analysis at indicated time points after induction of the disease (n = 12). (B) Disease score analysis in arthritis-induced mice 10 d after induction of the disease (n = 12); Mann–Whitney U test. (C) Representative H&E staining of the knee joint area. Scale bar 100 µm. (D and E) Knee swelling measurement analysis (D) and disease score analysis (E) in PBS-treated (n = 11) or α-NKG2D blocking antibody–treated (n = 8) arthritis-induced C57BL/6 mice 10 d after induction of the disease; Mann–Whitney U test. (F) Representative H&E staining of the knee joint area. Scale bar 200 µm. (G and H) Knee swelling measurement analysis (G) and disease score analysis (H) in PBS-treated or α-IL-17A/α-GM-CSF/α-IFN-γ blocking antibody–treated arthritis-induced Klrk1flox (n = 10/6) or Klrk1floxCd4Cre (n = 8/6) mice; Kruskal–Wallis test with multiple comparison test. In A, D, and G: ctrl, nonimmunized mice; OIA, OVA/CFA immunized (imm.) mice; value for each individual mouse is a difference in the knee thickness between the injected and noninjected knee. Data show mean ± SEM, pooled from at least two independent experiments. Two-way ANOVA with multiple comparison test. ns, not significant. In B, E, and H, each symbol represents an individual mouse; lines represent mean ± SEM. Data are pooled from at least two individual experiments. (I–K) Clinical scores of EAE in Klrk1flox or Klrk1floxCd4Cre mice (n = 16), Mann–Whitney U test. AUC, area under the curve. Data are pooled from three independent experiments.
Directly mirroring NKG2D-associated modulation of CD4+ T cell cytokines, we observed that the triple antibody (α-IL-17A/α-IFN-γ/α-GM-CSF) blockade treatment of Klrk1flox mice resulted in disease severity reduction to the levels obtained in Klrk1floxCd4Cre mice (Fig. 8, G and H) but did not induce any further significant amelioration of disease course and severity when performed in Klrk1floxCd4Cre mice. Altogether, these data suggest that NKG2D may enhance pathology mainly by modulating the cytokine expression in CD4+ T cells.
Because GM-CSF and IFN-γ expression has been linked to the encephalitogenic potential of Th17 cells, we assessed whether NKG2D deficiency affected the severity of EAE. Klrk1floxCd4Cre showed significant resistance to the development of EAE (Fig. 8, I–K) compared with Klrk1flox littermate mice, further highlighting an important role for NKG2D in promoting Th1/Th17 pathogenicity. Altogether, our data indicate that the deficiency of NKG2D in T cells affects the expression of proinflammatory cytokines in antigen-specific Th1/Th17 cells and is sufficient to ameliorate antigen-induced arthritis and EAE.
Discussion
The NKG2D+ CD4+ T cell population was previously described in both humans and mice, mostly in association with inflammatory diseases, while harboring different functional properties. Our study clearly shows that the expression of NKG2D on CD4+ T cells is a selective feature of Th1/Th17 lineages. Although transcripts of NKG2D were reported to be present also in Th2 cells (Meyers et al., 2002), we could not observe any protein expression on the surface of these cells by flow cytometry (FC), suggesting that these cells did not receive appropriate signals leading to its up-regulation. Our data show that the IL-12–STAT4–T-bet axis is important for the expression of NKG2D, mainly supported by elevated expression of NKG2D upon addition of IL-12, as well as by reduced frequency of NKG2D+ CD4+ T cells in Tbx21−/− and Stat4−/− mice. Moreover, chromatin immunoprecipitation sequencing analysis performed on Th1 cells deriving from WT, Stat4−/− (Wei et al., 2010), and Tbx21−/− (Zhu et al., 2012) mice shows that STAT4 and T-bet bind in close proximity to the Klrk1 gene, suggesting a direct role for these transcription factors in regulating NKG2D expression. Interestingly, a recent report showed that NKG2D marks IFN-γ–expressing T follicular helper cells that previously expressed T-bet; however, it remains unclear whether NKG2D plays any functional role in this T cell subset (Fang et al., 2018). Moreover, we observed that the expression of NKG2D is dependent not only on IL-12, but also on the strength of TCR engagement, as the expression is significantly enhanced in the second week of culture after TCR restimulation, as well as in the presence of increasing doses of antigen. Indeed, our data suggest that although the TCR triggering is sufficient for baseline expression of NKG2D, the two pathways most likely synergize to achieve high expression. In vivo, the induction of NKG2D on transferred naive CD4+ T cells is critically modulated by the adjuvant: NKG2D expression is strongly up-regulated on CD4+ T cells when mice are immunized with antigen in CFA emulsion, but not with antigen aqueous solutions in the presence of CpG. As CFA can both promote antigen persistence and shape the inflammatory microenvironment favoring Th1/Th17 cell differentiation (Coffman et al., 2010), it is likely that both effects contribute to the generation and activation of NKG2D+ Th cells in vivo.
In line with a previous study reporting neither a difference in the thymic development nor perturbations of CD4+ and CD8+ T cells in spleen, liver, and LN (Kavazović et al., 2017), our data show that NKG2D deficiency does not have a major impact on the formation and the size of primary CD4+ T cell responses and likewise does not affect proliferation or survival of CD4+ T cells. In contrast, NKG2D regulates effector functions for selected proinflammatory programs. One of the most striking features of NKG2D-mediated regulation is the enhancement of IFN-γ expression in Th17. Heightened expression of IFN-γ is likely a combined effect of direct and indirect mechanisms, as engagement of NKG2D results in increased expression of IFN-γ, on both transcript and protein levels. Moreover, NKG2D expression in Th cells is strongly associated with reinforcement of type 1 signature genes, among which are Tbx21, Hlx, Hopx, Ifngr1, and Il18r1, with Tbx21 being one of the most prominently regulated transcripts. Interestingly, we could show that NKG2D clearly regulates cytokine expression in Th1 and Th17 cells, although the effect is much more pronounced in vivo, especially during inflammation, than in vitro. In particular, enhancement of IFN-γ and GM-CSF production under Th1-polarizing conditions is very subtle in vitro. Similarly, the effect on IL-17A expression is not visible in Th17 cultures, while it is clearly observed in vivo both in OIA and after OVA/CFA immunization. This discrepancy between in vitro polarizing conditions and in vivo data might reflect the intrinsic limitations of in vitro culture systems which, despite their indisputable validity, cannot faithfully reproduce the complex inflammatory milieu formed in vivo. However, our in vivo data suggest that cytokine modulation by NKG2D represents a central node of its regulation of inflammatory responses, as shown by cytokine blocking in Klrk1flox versus Klrk1floxCd4Cre mice. Although we showed that NKG2D cross-linking induces signaling in CD4+ T cells and can directly contribute to enhance IFN-γ and GM-CSF production in these cells, further experiments are needed to clarify the molecular mechanisms through which NKG2D reinforces cytokine production and type 1 signature in Th1/Th17 cells.
NKG2D+ Th cells coexpress a variety of other innate receptors, such as Klrc1, Klrc2, and Klrd1, along with Cd96 and Lag3, possibly reflecting a phenotype of chronically stimulated T cells (Blackburn et al., 2009). Although expression of Lag-3 in particular has also been associated with an exhausted T cell phenotype (Crawford et al., 2014), we show that NKG2D+ Th cells are highly functional in vitro and in vivo. Whether these receptors might play a functional role in regulating NKG2D+ Th cell–mediated responses during chronic inflammation remains to be elucidated. In a recent report by Marshall et al. (2017), the authors showed that after influenza A virus infection, a subset of NKG2C/E+ CD4+ T cells with cytotoxic properties and high IFN-γ potential emerge in the lung. However, the activating signals deriving from NKG2C/E do not contribute to the generation of this specialized subset of cytotoxic Th cells or to their ability to kill, suggesting involvement of another receptor. In our gene expression analysis, differential regulation of cytotoxicity-associated genes was observed in NKG2D+ compared with Klrk1−/− Th cells, with more pronounced impact observed in Th17 cells. Nonetheless, the cytotoxic ability of in vitro–generated WT and Klrk1−/− Th1 cells against NKG2D ligand–expressing target cells was comparable (not depicted), suggesting that NKG2D does not directly promote Th1 cell killing, at least under tested conditions.
Consequently, CD4+ T cell expression of innate receptors, including NKG2D, represents an additional module regulating proinflammatory properties of these cells, making these receptors promising therapeutic targets in CD4+ T cell–mediated diseases. In mice, most data on the function of NKG2D comes from experiments using blocking antibody treatments in models of autoimmune diseases (Andersson et al., 2011; Ito et al., 2008; Ruck et al., 2013). However, using this approach, targeting of other NKG2D-expressing cell types possibly contributing to the outcome of the disease cannot be avoided. By using mice with T cell–specific deficiency of NKG2D (Klrk1floxCd4Cre), we showed that the selective deletion of NKG2D in T cells was sufficient to ameliorate EAE and arthritis, where it also mirrored the effects obtained using α-NKG2D blocking antibody. Although we cannot exclude that in EAE, within a pool of NKG2D+ cells, CD8+ T cells also contribute to the pathology (Legroux et al., 2019), OIA is predominantly a CD4+ T cell–mediated disease, in which CD8+ T cells play no role for the establishment of pathology (Wong et al., 2006). In line with these data, the frequency of NKG2D+ CD8+ T cells remained unaltered during the course of disease. Conversely, along with a prominent differentiation toward Th1/Th17, we detected significant accumulation of proinflammatory NKG2D+ antigen-specific Th1/Th17 cells after OIA induction.
Taken together, our data identify NKG2D as a marker of Th1 cells, as well as T-bet+ Th17 cells with high inflammatory properties, and show that the absence of NKG2D in T cells is sufficient to dampen inflammation during antigen-induced arthritis and EAE. These findings suggest that NKG2D might represent an important checkpoint target for Th1/Th17-mediated diseases.
Materials and methods
Mice
Klrk1−/− and Klrk1flox mice were generated as described (Zafirova et al., 2009, 2011). Klrk1flox were crossed to Cd4Cre Tg mice for the generation of mice with T cell–specific deficiency of NKG2D (hereafter Klrk1floxCd4Cre; Kavazović et al., 2017). Hcst−/− (Gilfillan et al., 2002), Tyrobp−/− (Bakker et al., 2000), Tbx21−/− (Finotto et al., 2002), Stat4−/− (Kaplan et al., 1996), and Tbx21-ZsGreen mice (Zhu et al., 2012) were described previously. B6PLxOT-II mice were described previously (Weber et al., 2015) and were crossed to C57BL/6J or Klrk1−/− for establishing cocultures and to track cells in adoptive transfer experiments. Il17aCre mice were described before (Hirota et al., 2011). Il17aCre crossed to R26RRFP (Il17aCre/wt × R26RRFP) and Il17aCre crossed to R26ReYFP and to Il17aKatushka (Il17aCre/wt × R26ReYFP × Il17aKata; Gagliani et al., 2015) were used for Il17a-fate mapping studies. All mice were bred under specific pathogen–free conditions in the animal facility of the Federal Institute for Risk Assessment (Berlin, Germany). For the experiments, both male and female mice at 8–16 wk of age were used in a sex- and age-matched fashion. Where appropriate, littermate controls were used. For sample size, see corresponding figure legends. Animal handling and experiments were conducted according to the German animal protection laws and approved by the responsible governmental authority (das Landesamt für Gesundheit und Soziales).
Cell isolation and FC
Single-cell suspensions from lung and siLP were obtained using the gentle magnetic activated cell sorting dissociator (Miltenyi Biotec) as described before (Paclik et al., 2015). Single-cell suspensions from BM were obtained by flushing of femur and tibia with PBS/BSA/EDTA. Liver lymphocytes were enriched on a Percoll gradient. For the purpose of ex vivo analysis, cells were stained using the following antibodies: APC Vio770 anti-mouse CD19 (1D3), APC Fire780 anti-mouse F4/80 (BM8), APC Fire780 anti-mouse Gr-1 (RB6-8C5), APC Cy7 anti-mouse FcεRIα (MAR-1), V500 anti-mouse CD45 (30-F11), BUV396 anti mouse CD45 (30-F11), APC anti-mouse γδTCR (GL-3), FITC anti-mouse γδTCR (GL-3), PerCP Cy5.5 anti-mouse γδTCR (GL-3), PE Cy7 anti-mouse TCRβ (H57-597), PerCP Cy5.5 anti-mouse TCRβ (H57-597), BV510 anti-mouse TCRβ (H57-597), BV605 anti-mouse NKp46 (29A1.4), BV650 anti-mouse NK1.1 (PK136), BV785 anti-mouse NK1.1 (PK136), BV650 anti-mouse CD8 (53–6.7), Alexa Fluor 700 anti-mouse CD8 (53–6.7), BV785 anti-mouse CD4 (RM4-5), BV711 anti-mouse CD4 (RM4-5), PE anti-mouse NKG2D (CX5), PE eF615 anti-mouse NKG2D (CX5), PE Cy7 anti-mouse NKG2D (CX5), biotin-labeled anti-mouse NKG2D (CX5), Alexa Fluor 700 anti-mouse/human CD44 (IM7), PE Cy7 anti-mouse 4-1BB (17B5), PE Cy7 anti-mouse CD62L (MEL14), PE anti-mouse CD62L (MEL14), FITC anti-mouse CD94 (18D3), PE Cy7 anti-mouse DNAM-1 (10E5), FITC anti-mouse Ly6C (HK1.4), FITC anti-mouse CD43 (1B11), PE anti-mouse CD69 (H1.2F3), PE anti-mouse 2B4 (m2B4458.1), PerCP eFluor710 anti-mouse Ly49G2 (4D11), PE anti-mouse Ly49H (3D10), FITC anti-mouse Ly49A/L (YE1/48), AF647 anti-mouse Ly49C/I (5E6), Alexa Fluor 488 anti-mouse CCR7 (4B12), BV605 anti-mouse CD127 (A7R34), APC anti-mouse CXCR3 (CXCR3-173), APC anti-mouse CCR6 (29-2L17), APC anti-mouse CD25 (PC61), and BV421-labeled streptavidin. BV421-labeled PBS-57–loaded CD1d tetramer was provided by National Institutes of Health Tetramer facility. To exclude dead cells, cells were stained with Fixable Viability Dye (LD; eBioscience) before fixation.
For intranuclear staining, cells were fixed with transcription factor staining buffer set (BD Bioscience) and stained with PE-Vio615 anti-mouse T-bet (REA102), Alexa Fluor 647 anti-mouse T-bet (4B10), PE anti-mouse RORγt (Q31-378), BV421 anti-mouse RORγt (Q31-378), and PE anti-mouse FoxP3 (FJK-16s). Unless otherwise stated, all antibodies were purchased at Thermo Fisher Scientific, BioLegend UK, Miltenyi Biotec, or BD Biosciences.
Cells were acquired on an LSR Fortessa and BD FACSymphony flow cytometer (BD Biosciences). Data were further analyzed with FlowJo Software v10 and SPICE (Roederer et al., 2011). All FC analysis was performed according to the Guidelines for the Use of Flow Cytometry and Cell Sorting in Immunological Studies (Cossarizza et al., 2017).
Cell culture, stimulation, and intracellular staining
Single-cell suspensions from spleen and inguinal lymph nodes were enriched for CD4+ T cell fraction using magnetic activated cell sorting, followed by staining with fluorochrome-conjugated antibody mixture, and sorted on BD FACSAria II (BD Bioscience) as live CD4+ CD44− CD62L+ cells.
Sorted naive murine CD4+ CD62L+ lymphocytes were resuspended in IMDM (Gibco BRL Life Technologies) supplemented with FCS (10%), 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 50 µM β-mercaptoethanol (Gibco BRL Life Technologies) and plated with irradiated splenocytes in a 1:3 T cell:APC ratio. Where indicated, WT (OT-II) and Klrk1−/− (OT-IIxKlrk1−/−) cells or WT and Stat4−/− cells were cocultured in a 1:1 ratio and distinguished by the expression of Thy1.1 and Thy1.2 markers during surface staining and sorting.
Cells were activated in the presence of soluble low endotoxin, azide-free (LEAF)–purified α-CD3 (5 µg/ml; 145-2C11) and LEAF-purified α-CD28 (6 µg/ml; 37.51) antibodies (or for cells expressing rearranged TCR specific for OVA peptide [OT-II] in the presence of OVA323–339 [5 µg/ml; custom synthesized at the peptide synthesis facility of the Charité Crossover] and α-CD28). For APC-free system, plate bound α-CD3 (5 µg/ml) and α-CD28 (6 µg/ml) were used. Cells were polarized in the presence of IL-2 (10 ng/ml), LEAF-purified α-IL-4 (10 µg/ml; 11B11), LEAF-purified α-IFN-γ (10 µg/ml; XMG1.2), LEAF-purified α-IL-12/IL-23 (10 µg/ml; C17.8; Th0), IL-2 (10 ng/ml), IL-12 (20 ng/ml), α-IL-4 (10 µg/ml; Th1), IL-2 (10 ng/ml), IL-4 (4 ng/ml), α-IFN-γ (10 µg/ml; Th2), IL-1β (20 ng/ml), IL-6 (50 ng/ml), IL-23 (5 ng/ml), TGF-β (1 ng/ml), α-IL-4 (10 µg/ml), and α-IFN-γ (10 µg/ml; Th17). On day 5, cells were purified using Histopaque-1083 (Sigma-Aldrich) and restimulated as described for day 0. Where indicated, 1-wk-old Th17 cells were cultured in the presence of IL-12 (20 ng/ml) for another week. All cytokines were purchased from Miltenyi Biotec, and all LEAF-purified antibodies were purchased from BioLegend UK.
Intracellular cytokine production (ICC) of CD4+ T lymphocytes upon polyclonal stimulation was assessed either ex vivo from single-cell suspensions or on day 10 of in vitro Th coculture, in bulk or, where indicated, after sorting. Up to 5 × 106 cells were washed and stimulated with 10 ng/ml of PMA (Sigma-Aldrich) and 500 ng/ml ionomycin (Iono; Sigma-Aldrich) in 10% IMDM in the presence of Brefeldin A containing GolgiPlug (1 µg/ml; BD Biosciences) for 4 h at 37°C. To exclude dead cells, cells were stained with Live/Dead Fixable Near IR Dead Cell Stain Kit (Life Technologies) before fixation. Cells were fixed in 1× InsideFix (Miltenyi Biotec) and stained, using the following antibodies in addition to those listed above: BV510 anti-mouse CD4 (RM4-5), BV421 anti-mouse Thy1.1 (OX-7), BV605 anti-mouse Thy1.2 (30-H12), APC anti-mouse Thy1.2 (30-H12), Pacific Blue anti-mouse Thy1.2 (30-H12), PE anti-mouse NKG2D (CX5), PE Cy7 anti-mouse NKG2D (CX5), FITC anti-mouse IL-17A (eBio17B7), APC anti-mouse IL-17A (eBio17B7), APC anti-mouse IFN-γ (XMG1.2), PerCP Cy5.5 anti-mouse IFN-γ (XMG1.2), PE Cy7 anti-mouse IFN-γ (XMG1.2), BV785 anti-mouse IFN-γ (XMG1.2), PerCP Cy5.5 anti-mouse GM-CSF (MP1-22E9), APC anti-mouse GM-CSF (MP1-22E9), PE Cy7 anti-mouse GM-CSF (MP1-22E9), and Alexa Fluor 647 anti-mouse T-bet (4B10). In the case of cells deriving from Il17a fate map reporter mice, cells were fixed in 2% paraformaldehyde (Electron Microscopy Science) and stained in 1× Perm Buffer (eBioscience).
For the assessment of Ifng as well as intracellular cytokine expression levels after NKG2D stimulation, Th1 and Th17 cells were sorted using PE-labeled α-NKG2D (clone A10 and clone CX5, 2 µg/ml, Thermo Fisher Scientific) and plated at a concentration of 105 cells/condition on tubes precoated with goat-anti–Armenian hamster secondary antibodies (10 µg/ml; JIR) in the presence of α-CD3 (0.1–0.2 µg/ml) or isotype control. After 6 h of stimulation in 10% IMDM in the presence (ICC) or absence (quantitative PCR [qPCR]) of Brefeldin A, cells were collected, washed, and further used for ICC or RNA isolation, as described.
Cell proliferation assay
To study the proliferative capacity of T cells in polarizing conditions, sorted naive T cells were resuspended in FCS-free RPMI medium at concentration of 106 cells/ml and stained with 1 µl/ml of CellTrace Violet Cell Proliferation Kit (Life Technologies) dye to a final concentration of 5 µM. Labeled cells were plated under polarizing conditions as described above. Dilution of the dye was measured daily by multicolor FC. To monitor the proliferation of the cells after the second TCR stimulation, a portion of cells were labeled after the Histopaque-1083 purification on day 5.
Phosphorylation assays
On day 10 of Th1 culture, NKG2D downstream signaling was assessed in WT and Klrk1−/− cells sorted as viable CD4+ CD94+ lymphocytes to reach ≥60% expression of NKG2D in WT cells. After sorting, cells were incubated with α-MULT-1 antibody (10 µg/ml; Thermo Fisher Scientific) and rested for 3 h at 37°C in FCS-free RPMI 1640 supplemented with 100 U/ml penicillin, 0.1 mg/ml streptomycin, and 50 µM β-mercaptoethanol. 105 cells/condition were stained with different combinations of the biotinylated α-CD3 (2–10 µg/ml, BioLegend), α-CD28 (10 µg/ml, Miltenyi Biotec), and α-CD314 (α-NKG2D, clone A10, 10 µg/ml, Thermo Fisher Scientific) antibodies, rested for 30 min at 37°C, and cross-linked by the addition of 20 µg/ml Streptavidin-UNLB (Southern Biotec). Cells were fixed with BD Cytofix buffer and permeabilized with BD Phosflow Perm III buffer (both BD Bioscience) and subsequently stained for pERK1/2 (PerCP-eFluor710 anti-mouse pERK1/2, clone MILAN8R, Thermo Fisher Scientific).
RT-PCR
mRNA was isolated from cells by using NucleoSpin RNA (Macherey-Nagel) and following the manufacturer’s protocol. First-strand cDNA was synthesized by using Reverse Transcription Reagents and assayed by qPCR in triplicates by using a StepOne Plus real-time PCR system and TaqMan Gene expression assays (all Thermo Fisher Scientific): Ifng (Mm01168134_m1), Rorc (Mm01261022_m1), Tbx21 (Mm00450960_m1), Eef1a1 (Mm01973893_g1), Ube2g1 (Mm00476128_m1), and Gapdh (Mm99999915_g1). mRNA content was normalized to the mean expression of Eef1a1 and Ube2g1 or, where indicated, to Gapdh expression, and mean relative gene expression was determined by using the ΔCT method.
Gene expression analysis
cDNA libraries were prepared from 100 ng total RNA using the TruSeq Stranded Total RNA Library Prep Kit with Ribo-Zero (Illumina) according to the low sample protocol. A paired-end (2 × 75 nt) sequencing of the resulting cDNA libraries was performed on a NextSeq500 Illumina device using the NextSeq 500/550 Mid output Kit v2 (150 cycles). Sequence reads were mapped to mouse GRCm38/mm10 genome with TopHat2 (Kim et al., 2013) in very-sensitive settings for Bowtie2 (Langmead and Salzberg, 2012) and EnsEMBL annotation release 67. Gene expression was quantified by featureCounts (Liao et al., 2014) and analyzed using the DESeq2 (Love et al., 2014) within R based on adjusted P (padj) < 0.05 and, where applied, fold change > 1.2.
Adoptive transfer and T cell tracking studies
Recipient mice received up to 107 OT-II or OT-IIxKlrk1−/− naive CD4+ T cells intravenously 1 d before being immunized subcutaneously in the flank with 100 µg OVA323–339 in CFA (Sigma-Aldrich) or 100 µg OVA323–339 in 30 µg CpG (ODN 1826, InvivoGen). The phenotype of OT-II+ cells was assessed by FC with gating on live CD4+ Thy1.1+ cells. Intracellular cytokine staining was performed after in vitro restimulation with PMA (10 ng/ml) and Iono (500 ng/ml) in the presence of Brefeldin A (1 µg/ml).
OIA
C57BL/6J (WT), Il17a fate map reporter mice, or littermate Klrk1flox and Klrk1floxCd4Cre mice were immunized subcutaneously twice, at day −21 and day −14, with 100 µg of cationized OVA (catOVA) in CFA. At day 0, arthritis was induced by intra-articular (i.a.) injection of 60–100 µg catOVA in PBS in one knee joint. Animals that were not immunized but did receive i.a. injection of catOVA on day 0 served as controls. Where indicated, treatment of animals was performed on days −1, 1, and 5 with blocking α-NKG2D antibody (clone C7; 300 µg/animal), α-IL-17A (clone: MM17F3, 200 µg/animal), α-GM-CSF (clones: A7.39 PAM751 and B2.6 PAM568, 200 µg/animal), or α-IFN-γ (clone: XMG1.2, 200 µg/animal). Joint swelling was monitored at indicated time points with an Oditest micrometer gauge (Kroeplin) and calculated as the difference between injected and noninjected joints. Mice were sacrificed 10 d after arthritis induction, and the knee joints were fixed in 10% formalin, decalcified in EDTA solution, and embedded in paraffin. Sections were stained with H&E and analyzed in a blind manner for the following histological parameters: exudates, granulocyte infiltration, hyperplasia of synovial lining, mononuclear cell infiltration/fibroblast activation, and peri-articular infiltration (each scoring 0–3); bone and cartilage destruction (0–4 points); and one additional point for fibrin exudates, adding up to a maximum score of 20. Single-cell suspensions from draining lymph nodes were restimulated in vitro with 100 µg/ml catOVA in the presence of Brefeldin A (1 µg/ml) before assessment of intracellular cytokine expression. Antigen-specific cells were defined according to CD154 (CD40L; PE anti-mouse CD154, APC anti-mouse CD154, clone MR1) expression induced after stimulation in vitro (Frentsch et al., 2005).
EAE
EAE was induced in littermate Klrk1flox and Klrk1floxCd4Cre female mice by subcutaneous immunization with 50 µg of MOG35–55 in CFA (Sigma-Aldrich). Mice additionally received intravenous injection of 240 ng of pertussis toxin (Sigma-Aldrich) on days 0 and 2. Mice were monitored daily for 35 d and scored for symptoms of the disease as following: 0, no signs of the disease; 1, limp tail; 2, limp tail and weakness of hind legs; 3, limp tail and paralysis of hind legs; 4, limp tail, complete hind leg and partial front leg paralysis; 5, dead or euthanized. Moreover, weight of the animals was monitored daily, and mice with >20% of weight reduction were excluded from the study.
Statistical analysis
Two-tailed Wilcoxon signed rank, two-tailed Mann–Whitney U test, Kruskal–Wallis, and Friedman’s test with multiple comparison corrections were employed for statistical analysis of datasets. Statistical analyses were performed with Prism 8 (GraphPad Software) using a confidence level of 0.95, and P values >0.05 were considered not significant.
Data availability
Raw transcriptome data reported in this paper are deposited and available from the Gene Expression Omnibus under accession code GSE143522. Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding authors.
Online supplemental material
Fig. S1 shows the gating strategy for the identification of NKG2D+ CD4+ T cell population in different tissues of C57BL/6 mice at steady state and its phenotype analysis. Fig. S2 shows kinetics of NKG2D expression in Th cultures generated in different polarizing conditions, dependence of expression on IL-12–STAT4–T-bet axis and antigen dose, and expression of Tbx21 and Rorc in in vitro–generated NKG2D+ and Klrk1−/− Th subsets. Fig. S3 displays effector cytokine expression and proliferation capacity of NKG2D+ and Klrk1−/− Th subsets. Fig. S4 shows cytokine and NKG2D expression in adoptively transferred CD4+ T cells in mice immunized with OVA323–339/CFA, frequency and absolute numbers of NKG2D-expressing cell subsets, and effector signature of NKG2D-expressing antigen-specific CD4+ T cells during OIA and phenotype analysis of CD4+ T cells from steady-state Klrk1flox and Klrk1floxCd4Cre mice. Table S1 lists all differentially expressed genes in in vitro–generated NKG2D+ and Klrk1−/− Th cells.
Supplementary Material
lists differentially expressed genes (DEG; n = 364) in Th1 (NKG2D+ vs. Klrk1−/−).
Acknowledgments
We thank J. Kirsch and T. Kaiser (Flow Cytometry Core Facility, Deutsches Rheuma-Forschungszentrum, Berlin) for the expert cell sorting services; A. Kühl (iPATH, Charité) for histological scoring of arthritic mice; K. Lehmann (Deutsches Rheuma-Forschungszentrum) for technical help; M. Colonna (University of Washington, St. Louis, MO) for providing Hcst−/− mice; Lewis L. Lanier (University of California, San Francisco, San Francisco, CA) for providing Tyrobp−/− mice; and S. Jonjic (University of Rijeka, Croatia) and Daniela Hernández-Torres (Deutsches Rheuma-Forschungszentrum) for critically reading the manuscript.
This research was supported by the European Union Seventh Framework Programme, Marie Curie Intra-European Fellowship PIEF-GA-2012-327643, and Deutsche Forschungsgemeinschaft grant BA5642/1-1 to M. Babic; the State of Berlin and European Regional Development Fund (ERDF 2014-2020, EFRE 1.8/11) to F. Heinrich and M-F. Mashreghi; Croatian Science Foundation grant HrZZ-IP-2016-06-9306 to B. Polic; and Deutsche Forschungsgemeinschaft grants RO3565/2-1, RO3565/4-1, and SFB TRR241 to C. Romagnani. C. Romagnani is supported by the Deutsche Forschungsgemeinschaft Heisenberg Program (RO 3565/1-1) and Leibniz ScienceCampus Chronic Inflammation.
Author contributions: M. Babic initiated the study, designed and performed most of the experiments, and analyzed data. C. Dimitropoulos, C. Stehle, Q. Hammer, A. Eisele, A. Sarsenbayeva, F. Heinrich, P. Durek, and M-F. Mashreghi performed and analyzed experiments. B. Lisnic and J. Van Snick provided reagents. S. Fillatreau provided help with EAE experiments. B. Polic, R.A. Flavell, S. Huber, N. Gagliani, D.R. Withers, and M. Löhning provided mouse mutants. C. Romagnani initiated and supervised the study and wrote the paper together with M. Babic, with input from all co-authors.
References
- Allez M., Tieng V., Nakazawa A., Treton X., Pacault V., Dulphy N., Caillat-Zucman S., Paul P., Gornet J.M., Douay C., et al. 2007. CD4+NKG2D+ T cells in Crohn’s disease mediate inflammatory and cytotoxic responses through MICA interactions. Gastroenterology. 132:2346–2358. 10.1053/j.gastro.2007.03.025 [DOI] [PubMed] [Google Scholar]
- Andersson A.K., Sumariwalla P.F., McCann F.E., Amjadi P., Chang C., McNamee K., Tornehave D., Haase C., Agersø H., Stennicke V.W., et al. 2011. Blockade of NKG2D ameliorates disease in mice with collagen-induced arthritis: a potential pathogenic role in chronic inflammatory arthritis. Arthritis Rheum. 63:2617–2629. 10.1002/art.30460 [DOI] [PubMed] [Google Scholar]
- Babic M., and Romagnani C.. 2018. The Role of Natural Killer Group 2, Member D in Chronic Inflammation and Autoimmunity. Front. Immunol. 9:1219 10.3389/fimmu.2018.01219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker A.B., Hoek R.M., Cerwenka A., Blom B., Lucian L., McNeil T., Murray R., Phillips L.H., Sedgwick J.D., and Lanier L.L.. 2000. DAP12-deficient mice fail to develop autoimmunity due to impaired antigen priming. Immunity. 13:345–353. 10.1016/S1074-7613(00)00034-0 [DOI] [PubMed] [Google Scholar]
- Bending D., De la Peña H., Veldhoen M., Phillips J.M., Uyttenhove C., Stockinger B., and Cooke A.. 2009. Highly purified Th17 cells from BDC2.5NOD mice convert into Th1-like cells in NOD/SCID recipient mice. J. Clin. Invest. 119:565–572. 10.1172/JCI37865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn S.D., Shin H., Haining W.N., Zou T., Workman C.J., Polley A., Betts M.R., Freeman G.J., Vignali D.A., and Wherry E.J.. 2009. Coregulation of CD8+ T cell exhaustion by multiple inhibitory receptors during chronic viral infection. Nat. Immunol. 10:29–37. 10.1038/ni.1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coffman R.L., Sher A., and Seder R.A.. 2010. Vaccine adjuvants: putting innate immunity to work. Immunity. 33:492–503. 10.1016/j.immuni.2010.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cossarizza A., Chang H.D., Radbruch A., Akdis M., Andrä I., Annunziato F., Bacher P., Barnaba V., Battistini L., Bauer W.M., et al. 2017. Guidelines for the use of flow cytometry and cell sorting in immunological studies. Eur. J. Immunol. 47:1584–1797. 10.1002/eji.201646632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford A., Angelosanto J.M., Kao C., Doering T.A., Odorizzi P.M., Barnett B.E., and Wherry E.J.. 2014. Molecular and transcriptional basis of CD4+ T cell dysfunction during chronic infection. Immunity. 40:289–302. 10.1016/j.immuni.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z., Turtle C.J., Booth G.C., Riddell S.R., Gooley T.A., Stevens A.M., Spies T., and Groh V.. 2009. Normally occurring NKG2D+CD4+ T cells are immunosuppressive and inversely correlated with disease activity in juvenile-onset lupus. J. Exp. Med. 206:793–805. 10.1084/jem.20081648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardalhon V., Schubart A.S., Reddy J., Meyers J.H., Monney L., Sabatos C.A., Ahuja R., Nguyen K., Freeman G.J., Greenfield E.A., et al. 2005. CD226 is specifically expressed on the surface of Th1 cells and regulates their expansion and effector functions. J. Immunol. 175:1558–1565. 10.4049/jimmunol.175.3.1558 [DOI] [PubMed] [Google Scholar]
- Diefenbach A., Tomasello E., Lucas M., Jamieson A.M., Hsia J.K., Vivier E., and Raulet D.H.. 2002. Selective associations with signaling proteins determine stimulatory versus costimulatory activity of NKG2D. Nat. Immunol. 3:1142–1149. 10.1038/ni858 [DOI] [PubMed] [Google Scholar]
- Fang D., Cui K., Mao K., Hu G., Li R., Zheng M., Riteau N., Reiner S.L., Sher A., Zhao K., et al. 2018. Transient T-bet expression functionally specifies a distinct T follicular helper subset. J. Exp. Med. 215:2705–2714. 10.1084/jem.20180927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finotto S., Neurath M.F., Glickman J.N., Qin S., Lehr H.A., Green F.H., Ackerman K., Haley K., Galle P.R., Szabo S.J., et al. 2002. Development of spontaneous airway changes consistent with human asthma in mice lacking T-bet. Science. 295:336–338. 10.1126/science.1065544 [DOI] [PubMed] [Google Scholar]
- Frentsch M., Arbach O., Kirchhoff D., Moewes B., Worm M., Rothe M., Scheffold A., and Thiel A.. 2005. Direct access to CD4+ T cells specific for defined antigens according to CD154 expression. Nat. Med. 11:1118–1124. 10.1038/nm1292 [DOI] [PubMed] [Google Scholar]
- Gagliani N., Amezcua Vesely M.C., Iseppon A., Brockmann L., Xu H., Palm N.W., de Zoete M.R., Licona-Limón P., Paiva R.S., Ching T., et al. 2015. Th17 cells transdifferentiate into regulatory T cells during resolution of inflammation. Nature. 523:221–225. 10.1038/nature14452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasser S., Orsulic S., Brown E.J., and Raulet D.H.. 2005. The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature. 436:1186–1190. 10.1038/nature03884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoreschi K., Laurence A., Yang X.P., Tato C.M., McGeachy M.J., Konkel J.E., Ramos H.L., Wei L., Davidson T.S., Bouladoux N., et al. 2010. Generation of pathogenic T(H)17 cells in the absence of TGF-β signalling. Nature. 467:967–971. 10.1038/nature09447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilfillan S., Ho E.L., Cella M., Yokoyama W.M., and Colonna M.. 2002. NKG2D recruits two distinct adapters to trigger NK cell activation and costimulation. Nat. Immunol. 3:1150–1155. 10.1038/ni857 [DOI] [PubMed] [Google Scholar]
- Groh V., Bruhl A., El-Gabalawy H., Nelson J.L., and Spies T.. 2003. Stimulation of T cell autoreactivity by anomalous expression of NKG2D and its MIC ligands in rheumatoid arthritis. Proc. Natl. Acad. Sci. USA. 100:9452–9457. 10.1073/pnas.1632807100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamerman J.A., Ogasawara K., and Lanier L.L.. 2004. Cutting edge: Toll-like receptor signaling in macrophages induces ligands for the NKG2D receptor. J. Immunol. 172:2001–2005. 10.4049/jimmunol.172.4.2001 [DOI] [PubMed] [Google Scholar]
- Hirota K., Duarte J.H., Veldhoen M., Hornsby E., Li Y., Cua D.J., Ahlfors H., Wilhelm C., Tolaini M., Menzel U., et al. 2011. Fate mapping of IL-17-producing T cells in inflammatory responses. Nat. Immunol. 12:255–263. 10.1038/ni.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosomi S., Grootjans J., Tschurtschenthaler M., Krupka N., Matute J.D., Flak M.B., Martinez-Naves E., Gomez Del Moral M., Glickman J.N., Ohira M., et al. 2017. Intestinal epithelial cell endoplasmic reticulum stress promotes MULT1 up-regulation and NKG2D-mediated inflammation. J. Exp. Med. 214:2985–2997. 10.1084/jem.20162041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Kanai T., Totsuka T., Okamoto R., Tsuchiya K., Nemoto Y., Yoshioka A., Tomita T., Nagaishi T., Sakamoto N., et al. 2008. Blockade of NKG2D signaling prevents the development of murine CD4+ T cell-mediated colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 294:G199–G207. 10.1152/ajpgi.00286.2007 [DOI] [PubMed] [Google Scholar]
- Jonjić S., Babić M., Polić B., and Krmpotić A.. 2008. Immune evasion of natural killer cells by viruses. Curr. Opin. Immunol. 20:30–38. 10.1016/j.coi.2007.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan M.H., Sun Y.L., Hoey T., and Grusby M.J.. 1996. Impaired IL-12 responses and enhanced development of Th2 cells in Stat4-deficient mice. Nature. 382:174–177. 10.1038/382174a0 [DOI] [PubMed] [Google Scholar]
- Kavazović I., Lenartić M., Jelenčić V., Jurković S., Lemmermann N.A.W., Jonjić S., Polić B., and Wensveen F.M.. 2017. NKG2D stimulation of CD8+ T cells during priming promotes their capacity to produce cytokines in response to viral infection in mice. Eur. J. Immunol. 47:1123–1135. 10.1002/eji.201646805 [DOI] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., and Salzberg S.L.. 2013. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36 10.1186/gb-2013-14-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korn T., Bettelli E., Oukka M., and Kuchroo V.K.. 2009. IL-17 and Th17 Cells. Annu. Rev. Immunol. 27:485–517. 10.1146/annurev.immunol.021908.132710 [DOI] [PubMed] [Google Scholar]
- Langmead B., and Salzberg S.L.. 2012. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 9:357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanier L.L. 2015. NKG2D Receptor and Its Ligands in Host Defense. Cancer Immunol. Res. 3:575–582. 10.1158/2326-6066.CIR-15-0098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legroux L., Moratalla A.C., Laurent C., Deblois G., Verstraeten S.L., and Arbour N.. 2019. NKG2D and Its Ligand MULT1 Contribute to Disease Progression in a Mouse Model of Multiple Sclerosis. Front. Immunol. 10:154 10.3389/fimmu.2019.00154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lexberg M.H., Taubner A., Albrecht I., Lepenies I., Richter A., Kamradt T., Radbruch A., and Chang H.D.. 2010. IFN-γ and IL-12 synergize to convert in vivo generated Th17 into Th1/Th17 cells. Eur. J. Immunol. 40:3017–3027. 10.1002/eji.201040539 [DOI] [PubMed] [Google Scholar]
- Liao Y., Smyth G.K., and Shi W.. 2014. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30:923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangan P.R., Harrington L.E., O’Quinn D.B., Helms W.S., Bullard D.C., Elson C.O., Hatton R.D., Wahl S.M., Schoeb T.R., and Weaver C.T.. 2006. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 441:231–234. 10.1038/nature04754 [DOI] [PubMed] [Google Scholar]
- Marshall N.B., Vong A.M., Devarajan P., Brauner M.D., Kuang Y., Nayar R., Schutten E.A., Castonguay C.H., Berg L.J., Nutt S.L., et al. 2017. NKG2C/E Marks the Unique Cytotoxic CD4 T Cell Subset, ThCTL, Generated by Influenza Infection. J. Immunol. 198:1142–1155. 10.4049/jimmunol.1601297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers J.H., Ryu A., Monney L., Nguyen K., Greenfield E.A., Freeman G.J., and Kuchroo V.K.. 2002. Cutting edge: CD94/NKG2 is expressed on Th1 but not Th2 cells and costimulates Th1 effector functions. J. Immunol. 169:5382–5386. 10.4049/jimmunol.169.10.5382 [DOI] [PubMed] [Google Scholar]
- Moser J.M., Gibbs J., Jensen P.E., and Lukacher A.E.. 2002. CD94-NKG2A receptors regulate antiviral CD8(+) T cell responses. Nat. Immunol. 3:189–195. 10.1038/ni757 [DOI] [PubMed] [Google Scholar]
- Nice T.J., Coscoy L., and Raulet D.H.. 2009. Posttranslational regulation of the NKG2D ligand Mult1 in response to cell stress. J. Exp. Med. 206:287–298. 10.1084/jem.20081335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogasawara K., Hamerman J.A., Ehrlich L.R., Bour-Jordan H., Santamaria P., Bluestone J.A., and Lanier L.L.. 2004. NKG2D blockade prevents autoimmune diabetes in NOD mice. Immunity. 20:757–767. 10.1016/j.immuni.2004.05.008 [DOI] [PubMed] [Google Scholar]
- Paclik D., Stehle C., Lahmann A., Hutloff A., and Romagnani C.. 2015. ICOS regulates the pool of group 2 innate lymphoid cells under homeostatic and inflammatory conditions in mice. Eur. J. Immunol. 45:2766–2772. 10.1002/eji.201545635 [DOI] [PubMed] [Google Scholar]
- Patel D.D., and Kuchroo V.K.. 2015. Th17 Cell Pathway in Human Immunity: Lessons from Genetics and Therapeutic Interventions. Immunity. 43:1040–1051. 10.1016/j.immuni.2015.12.003 [DOI] [PubMed] [Google Scholar]
- Raulet D.H., Gasser S., Gowen B.G., Deng W., and Jung H.. 2013. Regulation of ligands for the NKG2D activating receptor. Annu. Rev. Immunol. 31:413–441. 10.1146/annurev-immunol-032712-095951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J.M., Martinez G.J., Chung Y., and Dong C.. 2012. Toll-like receptor 4 signaling in T cells promotes autoimmune inflammation. Proc. Natl. Acad. Sci. USA. 109:13064–13069. 10.1073/pnas.1120585109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds J.M., Pappu B.P., Peng J., Martinez G.J., Zhang Y., Chung Y., Ma L., Yang X.O., Nurieva R.I., Tian Q., et al. 2010. Toll-like receptor 2 signaling in CD4(+) T lymphocytes promotes T helper 17 responses and regulates the pathogenesis of autoimmune disease. Immunity. 32:692–702. 10.1016/j.immuni.2010.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roederer M., Nozzi J.L., and Nason M.C.. 2011. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A. 79:167–174. 10.1002/cyto.a.21015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruck T., Bittner S., Gross C.C., Breuer J., Albrecht S., Korr S., Göbel K., Pankratz S., Henschel C.M., Schwab N., et al. 2013. CD4+NKG2D+ T cells exhibit enhanced migratory and encephalitogenic properties in neuroinflammation. PLoS One. 8 e81455 10.1371/journal.pone.0081455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sáez-Borderías A., Gumá M., Angulo A., Bellosillo B., Pende D., and López-Botet M.. 2006. Expression and function of NKG2D in CD4+ T cells specific for human cytomegalovirus. Eur. J. Immunol. 36:3198–3206. 10.1002/eji.200636682 [DOI] [PubMed] [Google Scholar]
- Saraiva M., Christensen J.R., Veldhoen M., Murphy T.L., Murphy K.M., and O’Garra A.. 2009. Interleukin-10 production by Th1 cells requires interleukin-12-induced STAT4 transcription factor and ERK MAP kinase activation by high antigen dose. Immunity. 31:209–219. 10.1016/j.immuni.2009.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya K., Shirakawa J., Kameyama T., Honda S., Tahara-Hanaoka S., Miyamoto A., Onodera M., Sumida T., Nakauchi H., Miyoshi H., et al. 2003. CD226 (DNAM-1) is involved in lymphocyte function-associated antigen 1 costimulatory signal for naive T cell differentiation and proliferation. J. Exp. Med. 198:1829–1839. 10.1084/jem.20030958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strid J., Sobolev O., Zafirova B., Polic B., and Hayday A.. 2011. The intraepithelial T cell response to NKG2D-ligands links lymphoid stress surveillance to atopy. Science. 334:1293–1297. 10.1126/science.1211250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhoen M., Hocking R.J., Atkins C.J., Locksley R.M., and Stockinger B.. 2006. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 24:179–189. 10.1016/j.immuni.2006.01.001 [DOI] [PubMed] [Google Scholar]
- Wang Y., Godec J., Ben-Aissa K., Cui K., Zhao K., Pucsek A.B., Lee Y.K., Weaver C.T., Yagi R., and Lazarevic V.. 2014. The transcription factors T-bet and Runx are required for the ontogeny of pathogenic interferon-γ-producing T helper 17 cells. Immunity. 40:355–366. 10.1016/j.immuni.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver C.T., and Murphy K.M.. 2007. The central role of the Th17 lineage in regulating the inflammatory/autoimmune axis. Semin. Immunol. 19:351–352. 10.1016/j.smim.2008.01.001 [DOI] [PubMed] [Google Scholar]
- Weber J.P., Fuhrmann F., Feist R.K., Lahmann A., Al Baz M.S., Gentz L.J., Vu Van D., Mages H.W., Haftmann C., Riedel R., et al. 2015. ICOS maintains the T follicular helper cell phenotype by down-regulating Krüppel-like factor 2. J. Exp. Med. 212:217–233. 10.1084/jem.20141432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei L., Vahedi G., Sun H.W., Watford W.T., Takatori H., Ramos H.L., Takahashi H., Liang J., Gutierrez-Cruz G., Zang C., et al. 2010. Discrete roles of STAT4 and STAT6 transcription factors in tuning epigenetic modifications and transcription during T helper cell differentiation. Immunity. 32:840–851. 10.1016/j.immuni.2010.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong P.K., Quinn J.M., Sims N.A., van Nieuwenhuijze A., Campbell I.K., and Wicks I.P.. 2006. Interleukin-6 modulates production of T lymphocyte-derived cytokines in antigen-induced arthritis and drives inflammation-induced osteoclastogenesis. Arthritis Rheum. 54:158–168. 10.1002/art.21537 [DOI] [PubMed] [Google Scholar]
- Ye J., Wang Y., Liu X., Li L., Opejin A., Hsueh E.C., Luo H., Wang T., Hawiger D., and Peng G.. 2017. TLR7 Signaling Regulates Th17 Cells and Autoimmunity: Novel Potential for Autoimmune Therapy. J. Immunol. 199:941–954. 10.4049/jimmunol.1601890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafirova B., Mandarić S., Antulov R., Krmpotić A., Jonsson H., Yokoyama W.M., Jonjić S., and Polić B.. 2009. Altered NK cell development and enhanced NK cell-mediated resistance to mouse cytomegalovirus in NKG2D-deficient mice. Immunity. 31:270–282. 10.1016/j.immuni.2009.06.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafirova B., Wensveen F.M., Gulin M., and Polić B.. 2011. Regulation of immune cell function and differentiation by the NKG2D receptor. Cell. Mol. Life Sci. 68:3519–3529. 10.1007/s00018-011-0797-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Jankovic D., Oler A.J., Wei G., Sharma S., Hu G., Guo L., Yagi R., Yamane H., Punkosdy G., et al. 2012. The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses. Immunity. 37:660–673. 10.1016/j.immuni.2012.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists differentially expressed genes (DEG; n = 364) in Th1 (NKG2D+ vs. Klrk1−/−).
Data Availability Statement
Raw transcriptome data reported in this paper are deposited and available from the Gene Expression Omnibus under accession code GSE143522. Further information and requests for resources and reagents should be directed to and will be fulfilled by the corresponding authors.