Nicola Aceto and colleagues discuss the opportunities and challenges of circulating tumor cells in translational research and precision medicine.
Abstract
Circulating tumor cells are tumor-derived pioneers responsible for the metastatic spread of cancer. Here, we outline recent discoveries, challenges, and future trends for circulating tumor cell investigations, arguing that the time is coming for translation of this work into clinical practice.
We still cannot cure metastatic cancer. Beyond surgery, adjuvant therapy with prevention purposes, and highly successful therapies that hinder cancer cell fitness even in advanced disease stages, cancer evolution and drug resistance mechanisms are major drivers of cancer recurrence and progression, leading to enormous challenges in the identification of curative treatments for metastatic cancers (Welch and Hurst, 2019). Many times, we aim at “precision oncology”; i.e., targeting the treatment to a defined cancer genotype. Yet, this simple equation has a very tricky denominator: how confident are we that the genomic features inferred from a patient’s cancer are correct, updated, and relevant for the treatment? A major obstacle is represented by the discrepancy between the available tumor molecular data and treatment decision time, since in most cases, existing data refer to an outdated tumor biopsy (Simmons et al., 2009).
Tissue biopsies are generally laborious, in some cases stressful from a patient perspective and generally unlikely to be repeated at each cycle of disease progression. For these reasons, most clinical decisions in the metastatic setting tend to rely on past biopsies (typically taken from the primary cancer), which may or may not picture the treatment-relevant genetic makeup of the disease in its metastatic form. How faithfully tissue biopsies comprise the most relevant cancer genotype depends on various well-known factors, such as both intrinsic tumor evolution dynamics and extrinsic selection pressure that results from any given anti-cancer treatment (McGranahan and Swanton, 2017), as well as timing and location of the biopsy itself. This uncertainty has favored the emergence of liquid biopsy–based approaches.
A liquid biopsy is a general term that refers to the withdrawal of biological fluids that may contain cancer-derived material, most frequently blood but also saliva, urine, and pleural and cerebrospinal fluid (Pantel and Alix-Panabières, 2019). Cancer-derived material contained in these fluids may comprise various elements, including extracellular vesicles, circulating tumor DNA (ctDNA), and circulating tumor cells (CTCs). The peculiarities of CTCs as compared with the others are their derivation from living and proliferative tumor regions—for instance, a fraction of CTCs is Ki67 positive (Szczerba et al., 2019)—and the fact that they contain a full repertoire all at once and available for interrogation; i.e., RNA, DNA, proteins, sugars, and lipids. CTCs derive from primary as well as metastatic cancerous lesions and are responsible for the formation of new distant metastases; namely, they play a pivotal and active role in disease progression (Keller and Pantel, 2019). The rarity of CTCs in the peripheral circulation of patients makes their isolation a challenging task. To this end, several platforms have been developed that exploit various features that distinguish CTCs from blood cells, such as marker expression, size, and/or deformability, reviewed in detail elsewhere (Ferreira et al., 2016). Importantly, the presence of CTCs above baseline has been very convincingly shown to be predictive of a poor prognosis in patients with various cancer types and in large clinical trials (Pantel and Alix-Panabières, 2019). Phenotypically, they present as single CTCs or as multicellular aggregates composed of cancer cells only (homotypic CTC clusters) or cancer cells in aggregation with nonneoplastic cells such as platelets, neutrophils, or fibroblasts (heterotypic CTC clusters; Duda et al., 2010; Gkountela et al., 2019; Szczerba et al., 2019). Cell–cell adhesion molecules such as plakoglobin, E-Cadherin, VCAM-1, and Claudins along with paracrine signals such as IL-1β and IL-6 are key to boost the metastatic ability of clustered CTCs (Aceto et al., 2014; Gkountela et al., 2019; Padmanaban et al., 2019; Szczerba et al., 2019), which represent the most efficient metastatic precursors in breast cancer and possibly in other cancer types (Aceto et al., 2014; Brandt et al., 1996; Murlidhar et al., 2017). While CTC phenotypes and enumeration are providing important insights, their functional profiling is key to achieve a better understanding of the fundamental characteristics of the metastatic cascade and provide further input, e.g., for treatment decision making.
Functional profiling of CTCs through molecular analysis has one major challenge: low cell number. This has two main consequences. First, CTC profiling requires single-cell resolution technologies, which are largely available within the community and relatively well established, yet still prone to biases such as strong stochastic variation, low (and/or uneven) coverage, and high dropout and error rates (Gawad et al., 2016). Recent technological breakthroughs further allow parallel multi-omics assays from individual cells, thus providing comprehensive profiles (Macaulay et al., 2017). In all cases, the development of tailored, standardized computational methods that incorporate bias correction and (noisy) data integration from multiple layers will be crucial for the implementation of CTC molecular analysis in the clinic. Second, is a low CTC number representative enough of the disease complexity in a patient? The easy answer would seem to be no. However, we should put this to the test in comparison with current standard methods aimed at the interrogation of cancer genotypes. Standard tissue biopsies typically contain only a minute amount of cancer cells, and as discussed above, they present with some limitations. Additionally, regarding the fact that they may not be timed with treatment decisions (i.e., be outdated), there is no guarantee that the most aggressive tumor clones are sampled during the procedure. Concerning ctDNA, per definition it is derived from cells that underwent necrosis or apoptosis. While this is certainly informative in terms of (early) genetic alterations that are common to all tumor cells, it is conceivable that the most aggressive tumor clones—highly proliferative, resistant to therapy-induced cell death—may be underrepresented. Experimental approaches to compare ctDNA composition across various tumor clones along with therapy sensitivity would be very interesting to gain quantitative insights into this aspect. In contrast, while only a handful of CTCs is available in a given patient, these are derivatives of living—and in some cases, proliferating—tumor regions, can be timed to treatment decisions, and have been shown to be one step ahead, i.e., carry those genetic features that will be prominent in the subsequent disease relapse (Chemi et al., 2019). Together, evidence is accumulating that CTC interrogation through molecular analysis may provide important cues to support clinical decision making (Fig. 1), not only concerning the discernment between high-risk and low-risk patients, but also regarding a cancer’s molecular profile.
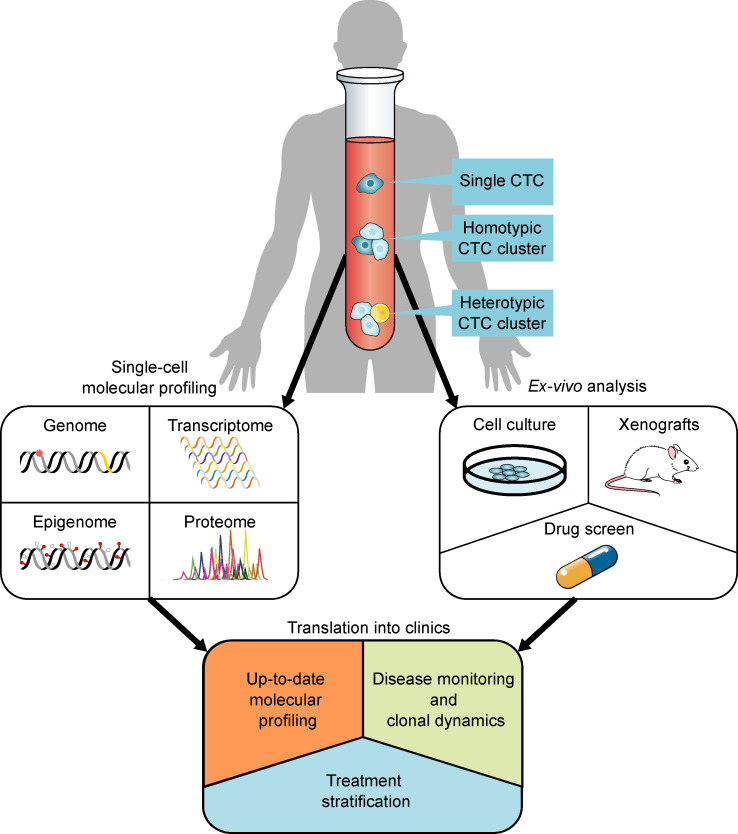
Figure 1.
Schematic representation of CTC-based analyses with translational potential. CTCs are present in blood samples from patients with cancer in various forms: as single CTCs and as homotypic or heterotypic CTC clusters. Direct single-cell resolution molecular profiling can provide information on their genome, transcriptome, epigenome, or proteome. Additionally, ex vivo functional approaches such as expansion in culture, transplantation in xenograft models, and phenotypic drug screens provide new means to address properties such as tumorigenicity and drug sensitivity. Together, both direct molecular profiling and ex vivo analyses allow the generation of data with translational perspective, such as the possibility to retrieve an up-to-date molecular profiling of a patient’s cancer, to monitor disease evolution and clonal dynamics, and to explore biomarkers for treatment stratification.
Another work-in-progress CTC application with potential for treatment decision making is linked to recent advancements in ex vivo CTC culture and drug screening (Fig. 1). A major motivation to push this forward is the fact that the concept of “precision medicine” relies on associations between genotype and drug sensitivity; however, this may not always be immediately evident. Particularly during advanced disease stages and after failure of multiple treatment lines, a cancer’s genetic makeup displaying high complexity (e.g., multiple genetic aberrations and driver mutations across various pathways) may not be unequivocally associated with one treatment. In this context, having the possibility to conduct a small-scale phenotypic drug screen on patient cells could favor the identification and prioritization of the next treatment line that is more likely to be successful. To this end, an example is provided for breast cancer. As a proof of concept, upon the identification of a suitable growth medium that enables ex vivo breast CTC expansion, multiple CTC lines were obtained from breast cancer patients with advanced progressive disease and were used to execute a drug screen with approved agents (Yu et al., 2014). In vitro results followed by in vivo validation with CTC-derived xenografts clearly allowed the identification of compounds (and combinations) with increased tumor-killing activity, highlighting the suitability of this approach in principle. Two main challenges need to be overcome here to enable translation into the clinic. First, ex vivo CTC expansion is currently a lengthy process, typically requiring months to obtain a stable cell line derived from freshly isolated cells. This is incompatible with treatment decision requirements, such as the need to decide rapidly for the benefit of the patient. Several methods are being currently tested in various laboratories in this regard, such as innovative medium compositions that favor rapid cell growth in combination with microfluidic tools that are tuned to enhance cell survival. Second, we need to understand whether a drug screen on a population of cells that is derived from few CTCs can be truly meaningful. This relates to the degree of representativeness of CTCs in regard to the entire tumor complexity as discussed above, and clinical trials will be needed to conclusively address this point.
Lastly, recent work aimed at a better understanding of CTC biology has allowed direct translation of preclinical findings into clinical trials. While attempting to gain insights into the molecular features of CTC clusters, we realized that cardiac glycosides, which are typically used for the treatment of cardiac arrhythmias in patients, could dissociate clusters of patient-derived CTCs ex vivo and prevent spontaneous CTC cluster formation in mouse models (Gkountela et al., 2019). Dissociation of CTC clusters not only resulted in molecular changes that decreased their stem-like traits but also suppressed their direct metastatic ability in preclinical in vivo models (Gkountela et al., 2019). Based on these results and given that cardiac glycosides characterized by well-known safety profiles are routinely given to patients, a proof-of-concept clinical trial has been set up to evaluate their effects on CTC clusters in breast cancer patients with progressive disease (NCT03928210). Along with this example, many other clinical trials are currently ongoing, not only using CTCs as biomarkers for the identification of high-risk subsets of patients, but also to assess specific molecular features of CTCs (e.g., PDL-1 expression in CTCs of non–small cell lung cancer, AR-V7 splice variant detection in CTCs of metastatic prostate cancer, and HER2 level assessment in CTCs of metastatic breast cancer considered to be HER2 negative) and in relation to drug sensitivity. The results of these trials, together with technological improvements in CTC functional profiling and ex vivo expansion for drug susceptibility testing, are now key to measure the potential of CTC analysis for clinical practice.
Acknowledgments
We thank all members and collaborators of the Aceto laboratory for valuable input across many discussions, fuel for the ideas and considerations presented here.
Research in the Aceto laboratory is supported by the European Research Council (grants 678834 and 840636), the European Union (grant 801159-B2B), the Swiss National Science Foundation (grants PP0P3_163938, PP00P3_190077, and IZLIZ3_182962), the Swiss Cancer League (grants KFS-3811-02-2016, KLS-4222-08-2017, and KLS-4834-08-2019), the Basel Cancer League (grants KLbB-4173-03-2017 and KLbB-4763-02-2019), the two Cantons of Basel through the ETH Zürich (grant PMB-01-16), and the University of Basel.
N. Aceto is listed as inventor in patent applications related to CTCs and is a paid consultant for companies with an interest in liquid biopsy. The remaining authors declare no competing financial interests.
References
- Aceto N., et al. 2014. Cell. 10.1016/j.cell.2014.07.013 [DOI] [Google Scholar]
- Brandt B., et al. 1996. Cancer Res. 56:4556–4561. [PubMed] [Google Scholar]
- Chemi F., et al. ; TRACERx Consortium . 2019. Nat. Med. 10.1038/s41591-019-0593-1 [DOI] [Google Scholar]
- Duda D.G., et al. 2010. Proc. Natl. Acad. Sci. USA. 10.1073/pnas.1016234107 [DOI] [Google Scholar]
- Ferreira M.M., et al. 2016. Mol. Oncol. 10.1016/j.molonc.2016.01.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawad C., et al. 2016. Nat. Rev. Genet. 10.1038/nrg.2015.16 [DOI] [PubMed] [Google Scholar]
- Gkountela S., et al. 2019. Cell. 10.1016/j.cell.2018.11.046 [DOI] [Google Scholar]
- Keller L., and Pantel K.. 2019. Nat. Rev. Cancer. 10.1038/s41568-019-0180-2 [DOI] [PubMed] [Google Scholar]
- Macaulay I.C., et al. 2017. Trends Genet. 10.1016/j.tig.2016.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGranahan N., and Swanton C.. 2017. Cell. 10.1016/j.cell.2017.01.018 [DOI] [Google Scholar]
- Murlidhar V., et al. 2017. Cancer Res. 10.1158/0008-5472.CAN-16-2072 [DOI] [Google Scholar]
- Padmanaban V., et al. 2019. Nature. 10.1038/s41586-019-1526-3 [DOI] [Google Scholar]
- Pantel K., and Alix-Panabières C.. 2019. Nat. Rev. Clin. Oncol. 10.1038/s41571-019-0187-3 [DOI] [PubMed] [Google Scholar]
- Simmons C., et al. 2009. Ann. Oncol. 10.1093/annonc/mdp028 [DOI] [Google Scholar]
- Szczerba B.M., et al. 2019. Nature. 10.1038/s41586-019-0915-y [DOI] [Google Scholar]
- Welch D.R., and Hurst D.R.. 2019. Cancer Res. 10.1158/0008-5472.CAN-19-0458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M., et al. 2014. Science. 10.1126/science.1253533 [DOI] [Google Scholar]