In this study, Hussain et al. demonstrate that cancer-associated fibroblasts (CAFs) in high-grade serous ovarian cancer are heterogeneous, that CAF state drives cancer aggressiveness and patient outcomes, and that TCF21 is a master regulator of CAF state.
Abstract
Recent studies indicate that cancer-associated fibroblasts (CAFs) are phenotypically and functionally heterogeneous. However, little is known about CAF subtypes, the roles they play in cancer progression, and molecular mediators of the CAF “state.” Here, we identify a novel cell surface pan-CAF marker, CD49e, and demonstrate that two distinct CAF states, distinguished by expression of fibroblast activation protein (FAP), coexist within the CD49e+ CAF compartment in high-grade serous ovarian cancers. We show for the first time that CAF state influences patient outcomes and that this is mediated by the ability of FAP-high, but not FAP-low, CAFs to aggressively promote proliferation, invasion and therapy resistance of cancer cells. Overexpression of the FAP-low–specific transcription factor TCF21 in FAP-high CAFs decreases their ability to promote invasion, chemoresistance, and in vivo tumor growth, indicating that it acts as a master regulator of the CAF state. Understanding CAF states in more detail could lead to better patient stratification and novel therapeutic strategies.
Introduction
High-grade serous ovarian cancer (HGSOC) is the most common histological subtype of ovarian cancer and is typically diagnosed at an advanced stage (Ledermann et al., 2013). Optimal surgical debulking and platinum/taxane-based chemotherapy significantly increase the survival of HGSOC patients, but the vast majority relapse and die within 5 yr of diagnosis (Ledermann et al., 2013). Due to early dissemination and implantation of cancer cells within the peritoneal cavity, HGSOC patients typically present at late stage with widespread abdominal disease and nearly invariably develop chemotherapy resistance. In spite of recent advances with targeted therapies such as poly (ADP-ribose) polymerase inhibitors (Moore et al., 2018), bevacizumab (Monk et al., 2016), and immune checkpoint blockade (Hamanishi et al., 2015), these approaches do not currently benefit all patients, and mortality rates remain high. The development of more effective treatments for HGSOC patients thus remains a necessary and important goal.
Cancer-associated fibroblasts (CAFs) are a key component of the tumor microenvironment and have several differences relative to their normal counterparts, including increased proliferation, extracellular matrix (ECM) production, and expression of cytokines and growth factors (Junttila and de Sauvage, 2013). In many cancers, including HGSOC, CAFs have important effects on tumor behavior, including defining the rate and extent of cancer progression through inhibition of cancer cell apoptosis, induction of cancer cell proliferation, promotion of cancer cell migration, and invasion and mediation of chemotherapy resistance (Kalluri, 2016; Mhawech-Fauceglia et al., 2015; Ryner et al., 2015; Thibault et al., 2014; Yeung et al., 2016). More recently, CAFs have also been shown to mediate immune suppression (Fearon, 2014; Kraman et al., 2010; Yang et al., 2016), adding another layer of complexity to their protumorigenic role. A variety of markers have been used to identify CAFs, including α-smooth muscle actin (α-SMA), platelet-derived growth factor receptors, and fibroblast activation protein (FAP), and most studies have focused on CAFs that express these markers. More recent studies have shown that CAFs are heterogeneous, and CAF subtypes with distinct phenotypes have begun to be identified in various malignancies (Costa et al., 2018; Givel et al., 2018; Öhlund et al., 2017; Su et al., 2018; Sugimoto et al., 2006). However, the functional characterization of these cells and their roles in tumor progression and patient outcomes have not yet been revealed, and molecular mechanisms driving epigenetic differences between CAF subtypes remain uncharacterized.
Here, we describe the identification of CD49e as a novel cell surface marker for fibroblasts within HGSOC primary tumor tissues, and we discover two distinct CAF states that exist within the CD49e+ fibroblast compartment and can be distinguished based on FAP expression. We demonstrate that FAP-high and FAP-low CAFs coexist at varying ratios in individual tumors and, importantly, CAF status drives patient outcomes. Purified FAP-high and FAP-low CAFs have distinct transcriptional signatures that are prognostic in The Cancer Genome Atlas (TCGA) cohort, and in vitro and in vivo functional assays reveal differences in their ability to promote cancer cell proliferation, invasion, and chemoresistance. Finally, we show that transcription factor TCF21 is a master regulator of the CAF state. Our extensive molecular and functional characterization of CAFs and analysis of CAF-derived gene signatures in relation to patient outcomes provides novel insights into the significant role of this cell population in HGSOC disease progression and the potential of manipulating the CAF state as a therapeutic strategy.
Results
Isolation and transcriptional profiling of CAFs from primary HGSOC tumor samples
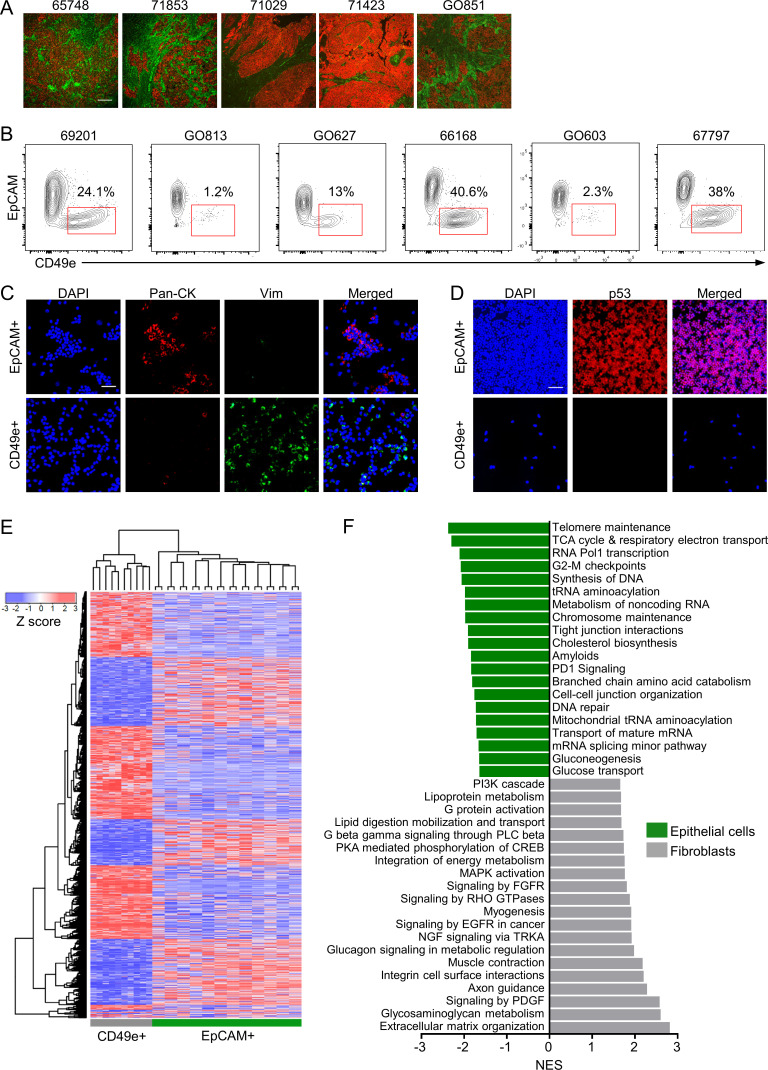
All tumor samples used in this study are listed in Table S1 A. Purification of viable CAFs directly from primary tumors using FACS requires a robust cell surface marker. PDGFR-β and FAP are commonly used CAF markers, but we found these to be either dimly or inconsistently expressed in single-cell suspensions from primary HGSOC samples, in line with other studies showing that expression of established CAF markers is heterogeneous and nonoverlapping (Orimo and Weinberg, 2007; Sugimoto et al., 2006). We therefore used high-throughput flow cytometry (HT-FC) with a panel of 363 antibodies targeting cell surface proteins (Gedye et al., 2014) to analyze cultured CAF lines derived from four HGSOC patients and single-cell suspensions from five primary HGSOC samples. The latter were costained for CD45 and CD31 to allow exclusion of contaminating immune and endothelial cells, respectively. From this screen, we identified several proteins that were uniformly highly expressed on the cultured CAFs but only stained a minority of the CD45−/CD31− cells from primary HGSOC samples, which would be expected to contain a mixture of cancer cells and CAFs (Fig. S1). The greatest difference was seen for CD49e (ITGA5), which was selected for follow up. Immunofluorescence studies of HGSOC sections confirmed that CD49e antibody selectively stained the tumor stromal compartment, while pan-cytokeratin (pan-CK) antibody, as expected, stained tumor cells (Fig. 1 A). We confirmed a similar staining pattern on several other cancers (lung adenocarcinoma, pancreatic ductal adenocarcinoma, colon adenocarcinoma, and prostate adenocarcinoma), but not others (breast carcinoma, lung squamous cell carcinoma, head and neck squamous cell carcinoma, and mesothelioma; Fig. S1 C). Costaining of HGSOC cells with an anti-EpCAM antibody (an epithelial cell marker) enabled clear distinction and isolation of CAFs from HGSOC samples as the CD45−CD31−EpCAM−CD49e+ fraction, which varied in frequency between patients (Fig. 1 B). The identity of isolated CD45−CD31−EpCAM+CD49e− (referred to as EpCAM+) and CD45−CD31−EpCAM−CD49e+ (referred to as CD49e+) fractions as cancer cells and CAFs, respectively, was further validated by generating cytospins of the purified populations and staining them for pan-CK, vimentin, and p53. The CD49e+ fraction was positive for vimentin and negative for pan-CK, as would be expected for a fibroblast population (Fig. 1 C). In patients with strong nuclear p53 staining in the EpCAM+ fraction, indicative of mutant p53 (Köbel et al., 2016), the CD49e+ fraction was negative for p53 staining, demonstrating that CD49e+ cells do not bear the cancer-associated mutation and ruling out the possibility that they are cancer cells that underwent an epithelial-to-mesenchymal transition (Fig. 1 D).
Figure S1.
Identification of CD49e as a CAF-specific cell surface marker. (A) We performed HT-FC with panel of 363 antibodies on four CAF lines and five HGSOC patient samples. Patient samples were costained with CD45 and CD31 to exclude inflammatory and endothelial cells from the analysis. A heatmap of the percentage of positive cells for each antibody on CAFs and the CD45−CD31− fraction of tumor samples was generated using unsupervised hierarchical clustering with a Pearson correlation distance metric and complete linkage. A cluster of differentially expressed markers was found, of which CD49e was the top hit (red arrow). (B) Representative FACS plots of CAFs (left) and a primary HGSOC sample (right) stained for CD49e. The gate for CD49e expression was set based on a fluorescence-minus-one (FMO) control. (C) A tissue microarray containing tissues from nine different types of cancer was stained for CD49e (green) and pan-cytokeratin (CK; red). Cores were scored based on stromal specificity (i.e., staining was exclusive to stroma, or it also stained tumor cells) and on stromal staining pattern (i.e., the stroma was broadly stained, or only a subset of stroma was stained). The data were compiled into a table (left), and representative images are shown (right). Scale bar = 100 µm. FSC-A, forward scatter area; FSC-H, forward scatter height; FSC-W, forward scatter width; SSC-H, side scatter height; SSC-W, side scatter width; ccRCC, clear cell renal cell carcinoma; HN, head and neck.
Figure 1.
Prospective isolation and transcriptional profiling of CAFs from primary HGSOC samples. (A) Representative images of IF on five different HGSOC samples using pan-CK (CK; red) and CD49e (green) antibodies. Scale bar = 100 µm. (B) Representative FACS plots from six different patients showing the viable, CD45−CD31− population stained for EpCAM and CD49e. (C) Representative images of cytospins made from isolated EpCAM+ or CD49e+ cells stained for pan-CK (red), vimentin (green), and Hoechst (blue). Scale bar = 50 µm. (D) Representative images of cytospins made from isolated EpCAM+ or CD49e+ cells stained for p53 (red) and Hoechst (blue). Scale bar = 50 µm. (E) Heatmap showing unsupervised hierarchical clustering of CD49e+ and EpCAM+ cells isolated directly from HGSOC tumor specimens and profiled by Illumina HT-12v4 microarrays. The EpCAM+ cells were separated into CD133+ and CD133− fractions. The heatmap shows EpCAM+ populations from 12 patients and CD49e+ samples from 10 patients due to data being of poor quality from two of the CD49e+ samples. (F) GSEA of the genes differentially expressed in the CD49e+ versus EpCAM+ populations. The top 20 nonredundant gene sets for each population are shown. CREB, cyclic adenosine 3′,5′-monophosphate response element-binding protein; EGFR, epidermal growth factor receptor; FGFR, fibroblast growth factor receptor; NES, normalized enrichment score; NGF, nerve growth factor; PKA, protein kinase A; PLC phospholipase C; RHO, Ras homolog family member; TRKA, tropomyosin receptor kinase A.
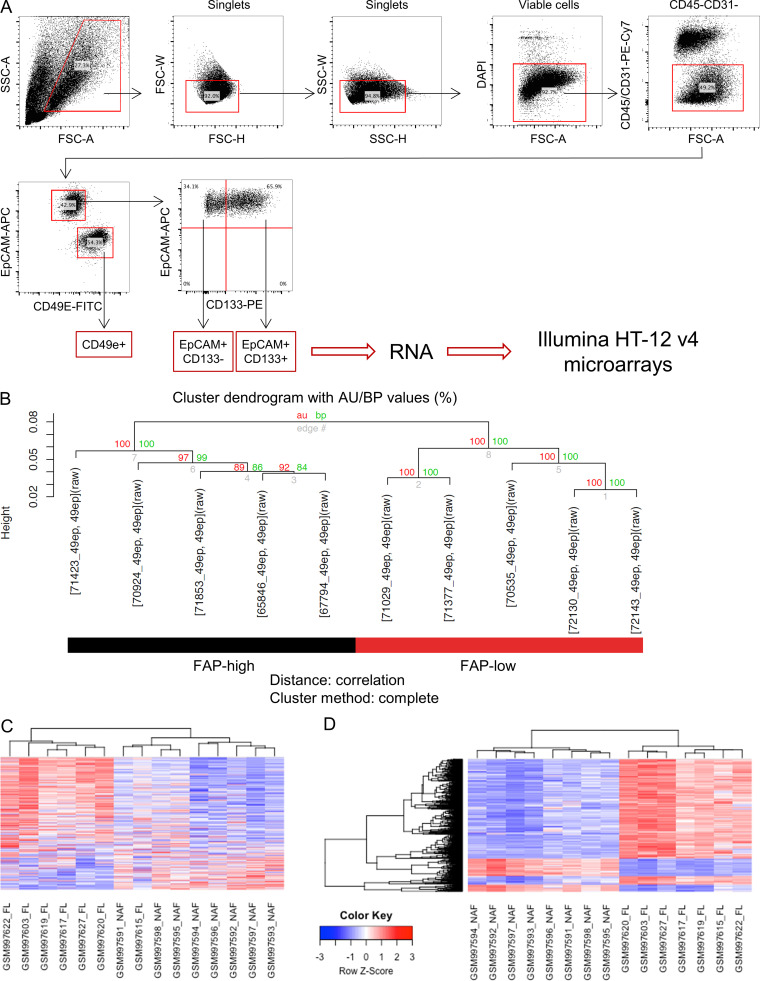
To interrogate the transcriptional profiles of cells isolated directly from primary tumor specimens, FACS was used to isolate the CD49e+ fraction and the EpCAM+ fraction from 12 primary HGSOC samples (Fig. S2 A). The latter was further fractionated into EpCAM+CD133+ and EpCAM+CD133− fractions, as we had previously shown that CD133 is a marker of tumor-initiating cells in the majority of primary HGSOC solid tumors (Stewart et al., 2011). RNA was extracted and analyzed using Illumina HT-12v4 microarrays. Upon unsupervised analysis, CD133+ and CD133− subsets had very similar transcriptional profiles and clustered together, whereas the CD49e+ population formed a distinct cluster (Fig. 1 E). The CD49e+ fraction expressed known CAF-associated genes, including ACTA2, FAP, VIM, POSTN, and SPARC, and multiple collagen genes, among others (Table S2 A). Gene set enrichment analysis (GSEA) was performed on genes differentially expressed between CD49e+ cells and EpCAM+ cells using the MSigDB Reactome database. EpCAM+ cells were enriched for genes related to cell proliferation, metabolism, and epithelial identity (e.g., tight junction interactions and cell–cell junction organization), whereas the CD49e+ subset was enriched for gene sets involved in ECM organization, myogenesis, and known mesenchymal signaling pathways, such as platelet-derived growth factor, fibroblast growth factor, and Ras homolog family member GTPase signaling (Kuzet and Gaggioli, 2016; Zhou et al., 2013). These results further confirm the identity of our purified CD49e+ population as fibroblasts (Fig. 1 F and Table S2 B).
Figure S2.
Isolation and transcriptional profiling of CD49e+ CAFs. (A) Full gating strategy for isolation of cell populations used in microarray analysis. (B) Cluster dendrogram of the CD49e+ CAFs isolated from 10 HGSOC samples (Human Illumina V4 array) after bootstrapping. Red and green numbers are the result of the pvclust function to assess the uncertainty in hierarchical cluster analysis. Red font corresponds to the AU (approximately unbiased) P value and green font to the BP (bootstrap probability) value for each cluster in the dendrogram. 100 (100%) means that the same cluster was obtained for all bootstrapping permutations. (C) Heatmap of the top 500 FL genes (same as in Fig. 2 A) showing only the FL and normal stroma samples from the Leung et al. (2014) study, demonstrating that the majority of the FL genes are higher in FL stroma versus normal adjacent stroma. (D) Heatmap of the top 500 probes from the comparison of the Leung et al. (2014) FL stroma versus normal adjacent stroma samples. FL, FAP low; NAF, normal adjacent fibroblasts; FSC-A, forward scatter area; FSC-H, forward scatter height; FSC-W, forward scatter width; SSC-A, side scatter area; SSC-W, side scatter width.
The CD49e+ fraction separates into two clusters
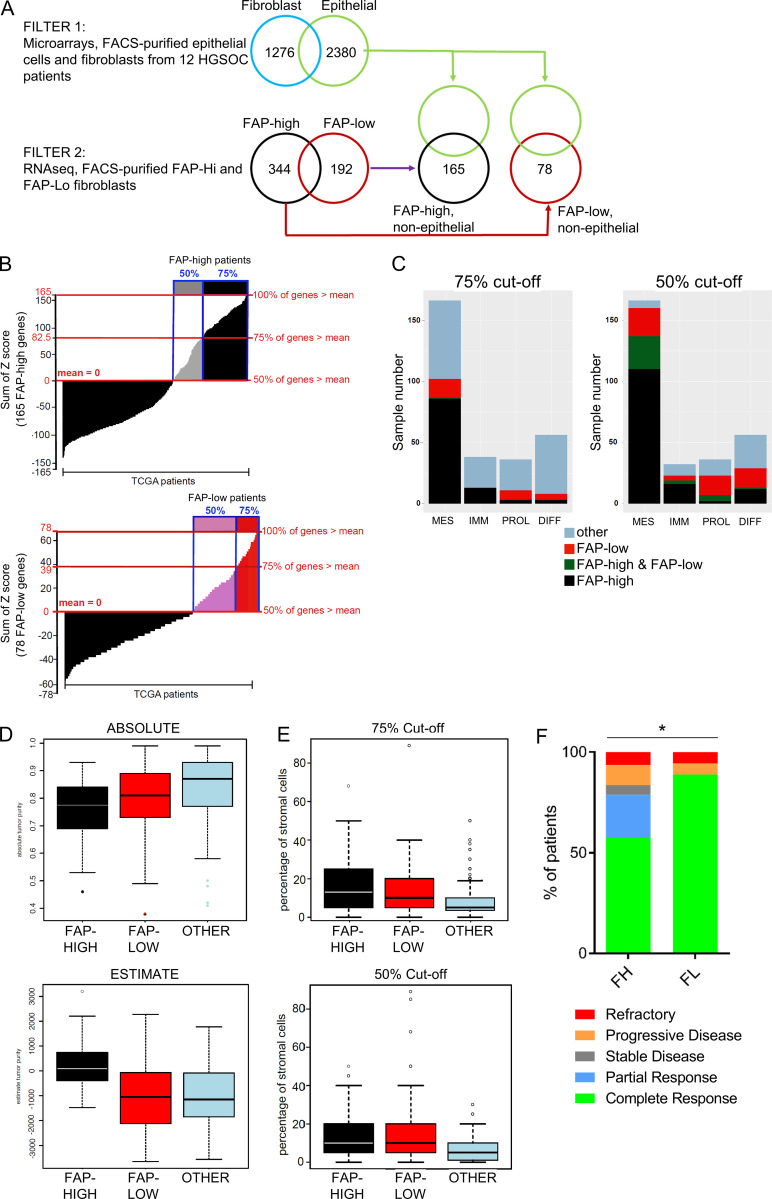
Upon unsupervised hierarchical clustering of the gene expression data, the CD49e+ CAF population segregated into two subclusters (Fig. 1 E and Fig. S2 B), suggesting that CAFs derived from HGSOC patients are heterogeneous. One cluster expressed classical CAF-related genes, such as FAP, TGFβ, COL11A1, SULF1, and inflammatory cytokines (e.g., IL-6 and CXCL12), among others, whereas the other exhibited a distinct gene expression profile that included low expression of the classical CAF marker FAP (Fig. 2 A and Table S3 A). We therefore refer to these two groups of patients as “FAP-high” and “FAP-low”. Several of the FAP-high (FAP, COL11A1, and SULF1)– and FAP-low (DLK1, TCF21, and COLEC11)–specific genes were validated by quantitative RT-PCR (qRT-PCR) on RNA isolated from the CD49e+ fraction of six patients included in the microarray analysis, three from the FAP-high group and three from the FAP-low group (Fig. 2 B). To test whether the two CAF subtypes were a general feature of ovarian cancer stroma, we analyzed the gene expression dataset generated by Leung et al. (2014), who used laser capture microdissection to isolate stromal and epithelial components from 31 HGSOC specimens and 8 normal ovary specimens. We interrogated the expression of the top 500 differentially expressed genes between our FAP-high and FAP-low patients in the stromal samples from this cohort and again found two major clusters (Fig. 2 A). This result verified the ability of our FAP-high versus FAP-low gene list to segregate HGSOC patients into two subtypes based on expression of these genes in their stroma. Importantly, the transcriptional profile of normal ovarian stroma was distinct from the FAP-low cancer stroma, indicating that FAP-low CAFs represent a distinct phenotype of stromal cells within HGSOC and not normal fibroblasts (Fig. S2, C and D; and Table S3 B).
Figure 2.
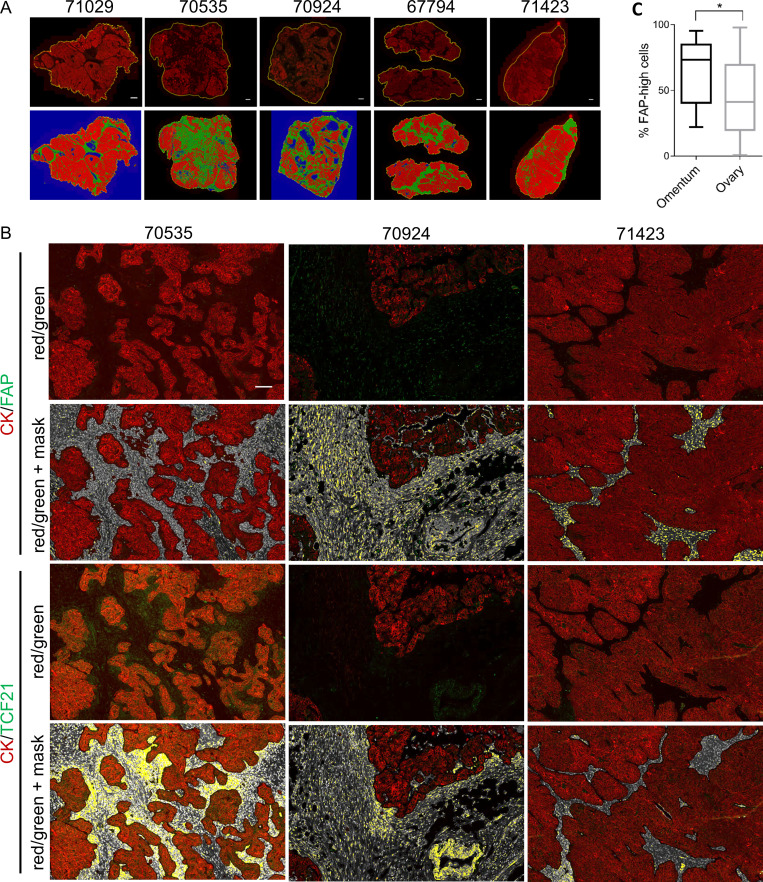
Identification of FAP-high and FAP-low CAFs in HGSOC. (A) Heatmap of the top 500 FAP-high (FH) versus FAP-low (FL) differentially expressed genes (left), and analysis of the same gene list in the Leung et al. (2014) HGSOC and normal ovary stromal samples (right). Black bar, tumor stroma; blue bar, normal stroma; black dendrogram, FAP-high patients; red dendrogram, FAP-low patients. FAP gene expression is enlarged at the bottom of the heatmap. (B) RNA from the CD49e+ fraction of three FAP-high and three FAP-low tumors (as classified by microarray) was analyzed by qRT-PCR for the expression of three FAP-high-specific and three FAP-low-specific genes. Data are presented as mean ± SEM, n = 3. **, P < 0.01; ****, P < 0.0001; Student’s t test. (C) Representative images of HGSOC serial sections stained for FAP or TCF21. Sections were costained for pan-CK and Hoechst. HALO image analysis software was used to quantify antibody staining within the pan-CK− stromal regions. Images show stained sections with HALO analysis mask. Red, epithelial cells; white, nuclei within the stromal region; yellow, FAP+ or TCF21+ cells. Scale bar = 50 µm. See Fig. S3 for additional images. (D) Serial sections from patient 70535 stained for FAP or TCF21. White, FAP-high/TCF21-low region; red, FAP-low/TCF21-high region. Scale bar = 200 µm. (E) Graph comparing the percentage of stromal area positive for FAP and TCF21 within the entire tissue section, for five tumors analyzed; r = 0.844, P = 0.036. (F) Quantification of the proportion of FAP-high cells within the viable CD45−CD31−EpCAM−CD49e+ fraction by FC in 64 primary samples and two matched recurrences. Three representative FACS plots, gated on the CD45−CD31−EpCAM−CD49e+ cells and showing FAP staining, are shown above the graph. The two matched primary (P) and recurrent (R) samples are indicated below the graph. (G) Graph comparing FAP quantification data obtained using FC and IF for samples that were analyzed using both methods; r = 0.969, P = 0.006. SSC-W, side scatter width.
FAP-high and FAP-low fibroblasts coexist in the majority of patients
To further interrogate the FAP-high and FAP-low status of patients classified into these groups by gene expression analysis, we performed immunofluorescence (IF) staining for FAP, TCF21 (a highly expressed FAP-low gene), and pan-CK on serial sections from five tumors that were categorized as FAP-high (n = 3) or FAP-low (n = 2) by transcriptional profiling. The slides were scanned to generate high-resolution digital images of the entire section, and the FAP+ or TCF21+ stained areas within the pan-CK− stromal regions were quantified using HALO image analysis software (Fig. 2 C and Fig. S3). FAP-high patients indeed had more FAP-expressing cells in their tumor stroma than FAP-low patients, whereas TCF21-expressing cells were more abundant in FAP-low patients. However, we also noticed that within tumor samples, FAP+ and TCF21+ cells could both be observed, and regions with predominantly one type of CAF or the other could be identified (Fig. 2 D). Nevertheless, quantification by HALO indicated an anticorrelation between FAP and TCF21 expression (Fig. 2 E). Thus FAP-high and FAP-low CAFs are two distinct subtypes that coexist within HGSOC tumors at varying ratios.
Figure S3.
Quantification of FAP and TCF21 in HGSOC specimens. (A) Low-power images of entire tissue sections showing the pan-CK staining in the red channel (top row) and the HALO software–generated classification of epithelial (red) and stromal (green) compartments. Blue regions are empty space. Scale bars = 1 mm. (B) Representative images of HGSOC specimens at higher magnification showing the epithelial pan-CK+ (red) and FAP+ or TCF21+ (green) staining and the corresponding mask generated by HALO software for quantification of the FAP or TCF21 signal within the stromal regions. Serial sections are shown for FAP versus TCF21 comparison. DAPI+ nuclei within the stroma are white, and FAP or TCF21+ signal in the green channel are overlayed with yellow. Scale bar = 500 µm. (C) Comparison of the %FAP-high cells in tumor tissue obtained from the primary site (ovary, n = 52) versus omental metastases (n = 10). *, P = 0.047, Student’s t test.
To more accurately quantify FAP-high and FAP-low CAFs within patient specimens, we performed FC on 66 HGSOC samples, with the addition of a FAP antibody to the previous combination of CD45, CD31, EpCAM, and CD49e, allowing us to quantify FAP expression within the CD49e+ fraction. The FAP-high subset ranged from 0.6% to 98% within the CD49e+ population (Fig. 2 F). Interestingly, the proportion of FAP-high cells was higher in tissues obtained from the omentum versus the ovary, suggesting a possible link to peritoneal dissemination (Fig. S3 C). Of the five patient samples that were also stained by IF, the FC and IF assays showed a strong positive correlation in the proportion of cells expressing FAP (r = 0.969).
Transcriptional profiling of purified FAP-high and FAP-low cells
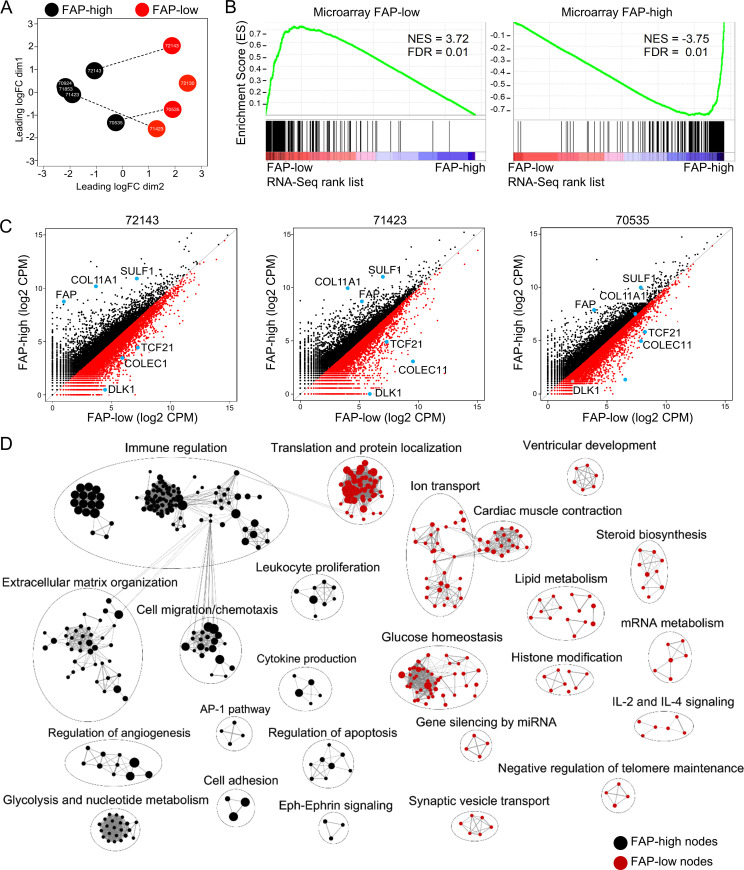
Our initial gene expression analysis was performed on the bulk CD49e+ fraction, which contained mixtures of FAP-high and FAP-low CAFs at varying ratios, suggesting that the observed clustering of patients into FAP-high and FAP-low groups reflected the predominant population, but these populations were not pure. We therefore performed RNA sequencing (RNA-seq) analysis on FAP-high and FAP-low CAFs purified by FACS. For some of these samples, the CD49e+ fraction was predominantly FAP-high or FAP-low, making isolation of both fractions in sufficient numbers difficult; however, for three patients (72143, 70535, and 71423), we successfully generated high-quality RNA-seq data on both fractions. Principal-component analysis of the five FAP-high populations and four FAP-low populations showed that regardless of whether the samples were derived from a FAP-high patient or a FAP-low patient, the samples clustered based on FAP status (Fig. 3 A). GSEA comparing the genes differentially expressed in isolated FAP-high and FAP-low cells showed a very high concordance with the previously obtained microarray-derived gene lists (Fig. 3 B). In addition, FAP, COL11A1, and SULF1 were highly expressed in the FAP-high subset, and TCF21, COLEC11, and DLK1 were highly expressed in the FAP-low subset (Fig. 3 C). These results confirm that HGSOC samples contain mixtures of FAP-high and FAP-low CAFs at varying ratios and that the clustering of patients based on bulk fibroblast profiling was driven by whichever population was dominant in those tumors.
Figure 3.
Transcriptional profiling of purified FAP-high and FAP-low CAFs. (A) Principal-component analysis of RNA-seq data from FACS-purified FAP-high and FAP-low cells. Dotted lines indicate cases for which FAP-high and FAP-low cells were purified from the same tumor specimen. (B) GSEA of the FAP-low (left) and FAP-high (right) gene lists generated from RNA-seq data in comparison to the gene lists previously generated by microarrays. (C) Scatterplots of FAP-high versus FAP-low gene expression in the three patients where both populations were isolated from the same patient. The three FAP-high and three FAP-low genes previously used for validation are indicated as blue dots. CPM, counts per million reads. (D) Cytoscape map of the most significantly differentially expressed Gene Ontology terms and pathways between FAP-high and FAP-low CAFs. GSEA was performed, and the top 300 most differentially expressed Gene Ontology terms and Reactome pathways are shown. The size of individual nodes correlates with the number of genes in each.
There were 800 differentially expressed genes between FAP-high and FAP-low CAFs at a false discovery rate (FDR) value of ≤0.05 (Table S4 A). GSEA of the differentially expressed genes between FAP-high and FAP-low cells (Fig. 3 D) showed that FAP-high cells express genes involved in ECM organization, cell migration and chemotaxis, immune regulation (including neutrophil activation, regulation of defense response, and antigen processing), and regulation of angiogenesis and thus resemble the classical phenotype that is commonly associated with CAFs in the literature (Fearon, 2014; Kalluri and Zeisberg, 2006; Kuzet and Gaggioli, 2016). By contrast, the most dominant gene sets expressed in FAP-low CAFs include glucose/insulin homeostasis, cardiac muscle contraction and ion transport, translation and protein localization, and lipid metabolism. FAP-low CAFs thus represent a previously unrecognized CAF subtype with a distinct gene expression profile from FAP-high CAFs.
FAP-high and FAP-low patients can be identified in TCGA and have distinct clinical outcomes
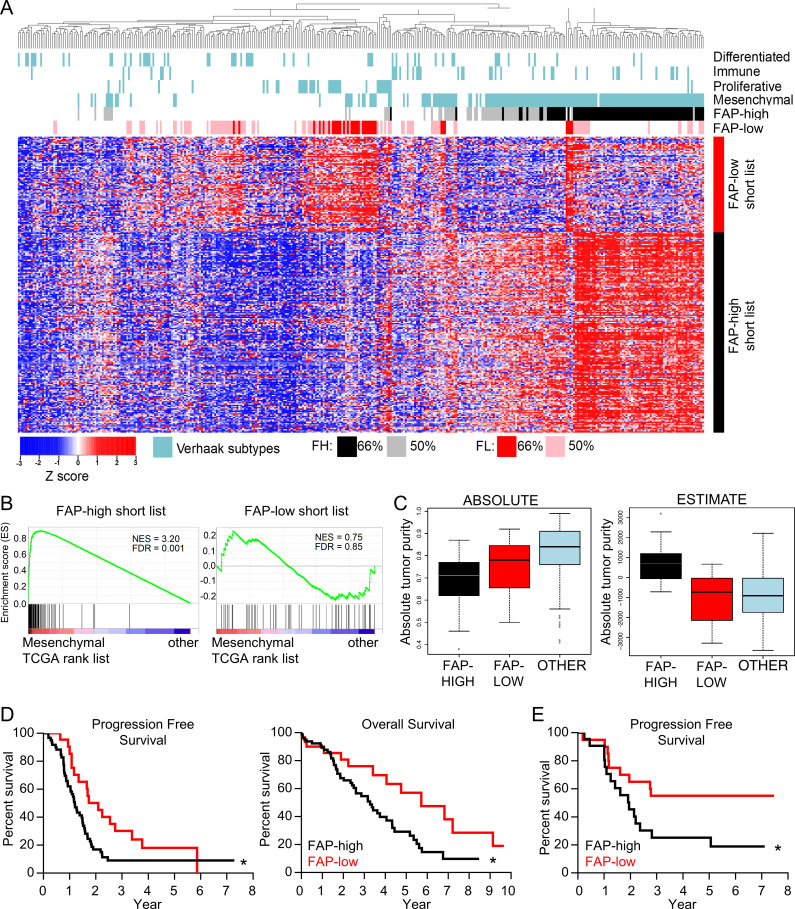
Gene expression profiling of 489 HGSOC samples by TCGA previously led to identification of four molecular subtypes: mesenchymal, proliferative, differentiated, and immunoreactive (Cancer Genome Atlas Research Network, 2011). Refined signatures for these subtypes were subsequently generated, and the mesenchymal subtype was found to be associated with worse outcome (Verhaak et al., 2013). We generated a gene signature based on FAP-high and FAP-low CAF genes by filtering for genes that were both differentially expressed in purified FAP-high versus FAP-low cells, and differentially expressed in CAFs compared with EpCAM+ cells in our original microarray dataset. This analysis resulted in a list of 165 FAP-high–specific genes and 78 FAP-low–specific genes (Fig. S4 A and Table S5). We then interrogated this signature against TCGA RNA-seq data (n = 374 patients) and found that FAP-high and FAP-low transcripts were detectable in a large number of patients, with distinct FAP-high and FAP-low clusters present (Fig. 4 A). A scoring system was established to classify patients (Fig. S4 B). When a patient expressed 75% of the FAP-high genes at a level higher than the population mean, the patient was classified FAP high (shown in black in Fig. 4 A). A second, less stringent threshold classified patients as FAP high if they expressed 50% of the FAP-high genes above the mean (shown in gray in Fig. 4 A). The same rule was applied to classify the FAP-low patients (shown in red and pink, respectively in Fig. 4 A). The majority of the FAP-high patients overlapped with the TCGA mesenchymal subtype, whereas the FAP-low patients were distributed among the mesenchymal, proliferative, and differentiated subtypes (Fig. 4 A and Fig. S4 C). GSEA shows that the FAP-high genes were highly enriched in the mesenchymal signature (Verhaak et al., 2013), whereas the FAP-low genes were not (Fig. 4 B), suggesting that that the mesenchymal subtype is largely driven by the presence of FAP-high fibroblasts.
Figure S4.
FAP-high and FAP-low gene lists and TCGA patient classification. (A) Schematic of filtering strategy used to generate a gene signature for interrogation of TCGA data. Filter 1 is selecting genes that are differentially expressed in CD49e+ cells compared with cancer epithelial cells (FDR ≤ 0.05), and filter 2 is selecting genes based on the comparison of purified FAP-high and FAP-low CAFs (FDR ≤ 0.05 and logFC ≥ 2). (B) TCGA patients were ranked using a score that counts how many genes from the FAP-high gene list (top) or FAP-low gene list (bottom) have a normalized value greater than the patient mean (i.e., a z-score). Patients with positive scores in at least 75% of the gene list (corresponding to a sum of z-scores ≥ gene list length/2) are shown in black and red for FAP high and FAP low, respectively. Patients with positive scores in at least 50% of the gene list (corresponding to a sum of z-scores ≥ 0) are shown in gray and pink for FAP high and FAP low, respectively. (C) Distribution of FAP-high and FAP-low patients in TCGA across the Verhaak et al. (2013) subtypes. Bar plots showing the number of FAP-high and FAP-low samples that fell in each Verhaak et al. (2013) category. “Other” indicates patients that fell into neither category; FAP-high and FAP-low indicate patients who expressed both FAP-high and FAP-low genes. Diff, differentiated subtype; Imm, immune subtype; Mes, mesenchymal subtype; Pro, proliferative subtype. (D) ABSOLUTE (top) and ESTIMATE (bottom) algorithms applied to patients falling into the FAP-high (black and gray), FAP-low (red and pink) and other (blue) categories of patients using the 50% cutoff. At this less stringent cutoff, the same results are obtained as shown in Fig. 4 C using the 75% cutoff. (E) Stromal content of patients falling into the FAP-high, FAP-low, or other categories based on histopathology provided in the TCGA dataset. As with the ABSOLUTE algorithm, patients falling into the “other” category have lower stromal content than patients in the FAP-high and FAP-low categories. The same is true regardless of whether the 75% (top) or 50% cutoff (bottom) is applied. (F) A subset of TCGA patients could be classified into groups based on their response to chemotherapy, as described in Villalobos et al. (2018). Comparison of these classifications with the FAP-high (n = 61) and FAP-low (n = 18) patients revealed that complete responses were more frequent in FAP-low patients (*, P = 0.023, Fisher’s exact test).
Figure 4.
FAP-high patients correspond to the mesenchymal molecular subtype and have worse clinical outcomes than FAP-low patients. (A) Heatmap of TCGA HGSOC data showing unsupervised clustering of patients using the FAP-high (FH) and FAP-low (FL) specific CAF gene lists. The TCGA molecular subtypes (Verhaak et al., 2013) are shown above the heatmap in blue. Patients expressing either 75% or 50% of the FAP-high signature genes are indicated in black and gray, respectively. Patients expressing either 75% or 50% of the FAP-low signature genes are indicated in red and pink, respectively. (B) GSEA comparing the FAP-high (left) and FAP-low (right) gene lists to the genes expressed in TCGA mesenchymal subtype. (C) The ABSOLUTE (left) and ESTIMATE (right) algorithms were applied to patients falling into the FAP-high (black), FAP-low (red), and “other” (blue) categories of patients selected using the more stringent 75% cutoff. (D) Kaplan–Meier survival curves of patients falling into the FAP-high (n = 80) and FAP-low (n = 30) groups (based on the 75% cutoff). Median progression-free survival was 1.2 yr for FAP-high and 2.1 yr for FAP-low patients; log-rank P value = 0.0204; hazard ratio (HR) = 1.89; 95% confidence interval (CI), 1.11–2.97. Median overall survival was 3.3 yr for FAP-high and 5.7 yr for FAP-low patients; log-rank P value = 0.0236, HR (95% CI) = 1.95 (1.11–3.24). (E) Kaplan–Meier progression-free survival curve of patients from our center that were profiled by FC for the proportion of CD49e+ CAFs that were positive for FAP staining. Patients were separated into FAP-high and FAP-low based on the median FAP expression. Log-rank P value = 0.041, HR (95% CI) = 2.28 (1.04–5.05). See Table S1 B for recurrence data.
Using the ABSOLUTE algorithm, which uses somatic copy number data to estimate the cellularity of tumor samples (Carter et al., 2012), it was shown previously that the mesenchymal subtype in the TCGA HGSOC study had the lowest tumor purity (Zhang et al., 2015). Using the same algorithm, FAP-high and FAP-low TCGA patients both had lower tumor purities than the remaining “other” patients that did not fall into either category (Fig. 4 C and Fig. S4 D). Histopathology data for the TCGA samples also indicates that tumor samples in the “other” category had a lower stromal content than both the FAP-high and FAP-low categories (Fig. S4 E). Taken together, these findings suggest that the unclassified samples could not be classified as FAP-high or FAP-low due to low stromal content within the tumor specimen analyzed.
ESTIMATE is another algorithm designed to estimate the quantity of infiltrating fibroblasts and immune cells using gene expression data on tumor tissues (Yoshihara et al., 2013). While the ESTIMATE algorithm generated a high “stromal” score for the FAP-high samples, it failed to identify higher stromal content in tumor samples falling into the FAP-low category (Fig. 4 C and Fig. S4 D). Deeper analysis of this discrepancy showed that the list of 141 genes used to define “stroma” in the ESTIMATE algorithm is enriched for FAP-high genes (21-gene overlap), but not for FAP-low genes (two-gene overlap). As a result, patients with a high fraction of FAP-low CAFs were not identified as having higher stromal content using this algorithm.
To determine if the CAF subtype has an impact on survival, we performed a Kaplan–Meier analysis of the TCGA patients that were classified as FAP-high or FAP-low (using the more stringent cutoff of 75%). FAP-high patients had significantly shorter progression-free and overall survival than FAP-low patients; the median overall survival of FAP-low patients was 2 yr longer than that of FAP-high patients (Fig. 4 D). To validate this finding, we compared the percentage of FAP+ CAFs from patients that were analyzed by FC in Fig. 2 F to progression-free survival for 46 of the patients for which these data were available. We defined FAP-high patients as those with a percentage of FAP+ CAFs above the median, and FAP-low patients as those with a percentage of FAP+ CAFs below the median. We found that FAP-high patients had a significantly worse progression-free survival compared with FAP-low patients (Fig. 4 E). To specifically investigate the relationship between FAP-high and FAP-low CAFs and chemotherapy response, we used a study that annotated TCGA patients into the following categories based on their response to treatment: complete response, partial response, stable disease, progressive disease, or refractory (Villalobos et al., 2018). A comparison of FAP-high and FAP-low patients showed that a significantly higher proportion of FAP-low patients demonstrated a complete response to chemotherapy (Fig. S4 F).
FAP-low cells require different culture conditions than FAP-high cells
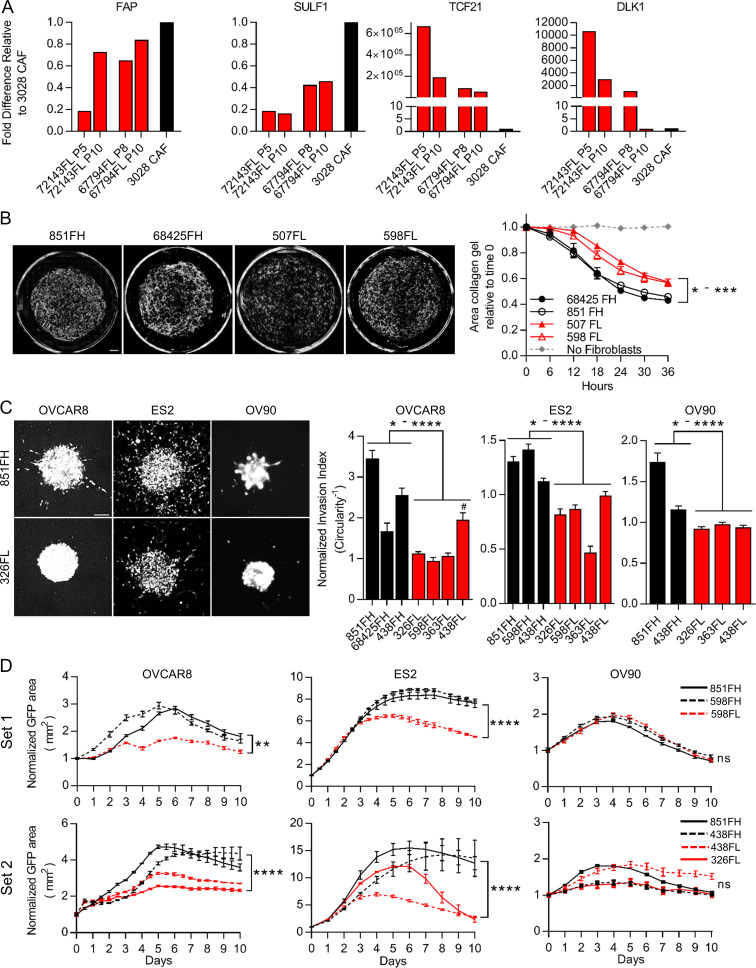
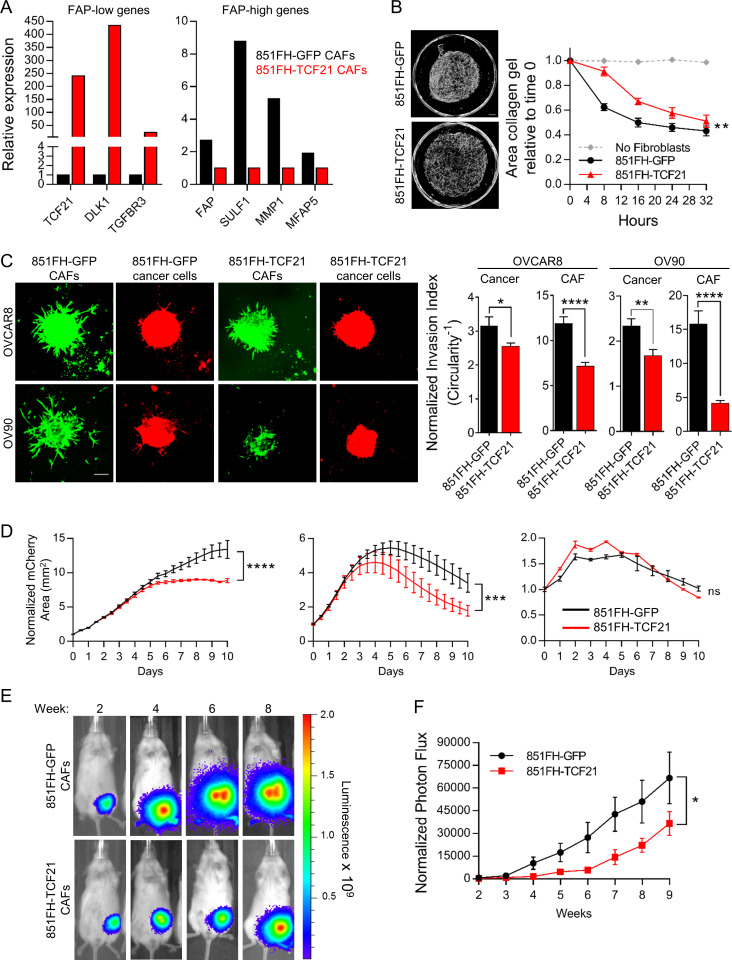
Most studies of CAFs use cultures established by plating tumor cell suspensions onto tissue culture plastic in the presence of 10% FBS. In these conditions, CAFs rapidly adhere and proliferate, allowing for their selection and outgrowth. Our identification of CD49e as a CAF marker was facilitated by analysis of such patient-derived cultured CAF lines. Analysis of several of our cultured CAF lines indicated that they are FAP high by FC and express FAP-high, but not FAP-low, genes (data not shown). When we placed FAP-high and FAP-low cells purified by FACS directly from patient samples into standard 10% FBS conditions, we found that FAP-high cells had a significantly higher growth rate than FAP-low cells and that FAP-low cells in these conditions did not reach confluence and could not be successfully passaged (Fig. S5 A); thus, when bulk cells are placed in culture to derive CAF lines, FAP-high cells will outcompete FAP-low cells over time due to their growth advantage. We therefore sought alternative culture conditions for FAP-low cells; based on the expression of some genes related to adipogenesis in our FAP-low population (e.g., DLK1 and PPARG), we tested commercially available preadipocyte media and found that this media supported the expansion of FAP-low cells over multiple passages (Fig. S5 B). qRT-PCR analysis of FAP-high and FAP-low genes indicated that FAP-low cells continued to express FAP-low genes (TCF21 and DLK1) at very high levels for up to eight passages. However, we did see a decrease in these genes with increasing passage number, suggesting that even in preadipocyte media FAP-low cells drift toward a FAP-high phenotype with increasing time in culture (Fig. 5 A). We also saw an increase in FAP and SULF1 gene expression with passage in some cases (Fig. 5 A). Thus, to carry out the functional assays described below, it was necessary to repeatedly isolate FAP-low cells from patient samples and use them at passage 5 or less.
Figure S5.
Supplemental data relating to functional assays. (A) Growth curves of FAP-high and FAP-low CAFs isolated from three patients cultured in 10% FBS. Data are presented as mean ± SEM, n = 3. **, P < 0.01; ***, P < 0.001, two-way ANOVA. (B) FAP-low cells can be expanded in preadipocyte media. Scale bar = 50 µm. (C) Dose–response curves of OVCAR8, ES2, and OV90 cells in response to carboplatin, in adherent growth conditions. IC50s were calculated using GraphPad software. (D) Representative images of OVCAR8, ES2, or OV90 cells co-cultured with 598FH (top) or 598FL (bottom) CAFs treated with 10 µM carboplatin at the 7-d time point. Pink indicates mask used to quantify GFP+ cell area. Scale bar = 100 µM. (E) Quantification of cells remaining after 7 d of treatment with 10 µM carboplatin, normalized to day 0. n = 3; *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, Student’s t test. (F) Western blot for TCF21 expression in 851FH-GFP and 851FH-TCF21 CAFs. (G) Growth curves of mCherry-labeled OVCAR8 or ES2 cells co-cultured with 438FH-GFP or 438FH-TCF21 CAFs after treatment with 10 µM carboplatin. Data were normalized to the mCherry+ area at day 0 and are presented as mean ± SEM, n = 3. Asterisks reflect differences at the end of 10 d. ***, P < 0.001; ****, P < 0.0001, Student’s t test.
Figure 5.
FAP-high and FAP-low CAFs have distinct functional properties. (A) qRT-PCR for FAP-high genes (FAP and SULF1) and FAP-low genes (TCF21 and DLK1) in passaged FAP-low cells (red; passage number indicated below each bar). A cultured patient-derived CAF line was used as a control (black bar). Fold-differences in gene expression were determined using the ΔΔ−Ct method, using UBC as a housekeeping gene, with 3 technical replicates per sample assayed. (B) Representative images of collagen gels cultured for 36 h with two FAP-high derived CAF lines and two FAP-low-derived CAF lines (left); quantification of gel contraction over time (right). Data are presented as mean ± SEM, n = 4. P values for comparisons between FAP-high and FAP-low lines ranged from 0.02 to 0.002, two-way ANOVA. Scale bar = 500 µm. (C) Representative images of spheroids generated using GFP-expressing HGSOC cell lines OVCAR8, ES2 or OV90 combined with FAP-high or FAP-low fibroblasts (left). Circularity was quantified for OVCAR8 and OV90 cells, and the number of cells outside the spheroid was quantified for ES2 cells (right). Data are normalized to spheroids containing no CAFs and are presented as mean ± SEM, n = 5–10 spheroids per condition. P values, calculated using Student’s t test, ranged between 0.039 to <0.0001. Scale bar = 100 µm. The difference between 438FL and 68425FH in OVCAR8 cells was not significant. (D) Growth curves of GFP-labeled cancer cells co-cultured with FAP-high or FAP-low CAFs during treatment with 10 µM carboplatin. Experiments were done in two separate batches (“set 1” and “set 2”). CAF line 851FH was used in both experiments. Data were normalized to the GFP+ area at day 0 and are presented as mean ± SEM, n = 3. Asterisks reflect differences at the end of 10 d. *, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001, Student’s t test. FH, FAP high; FL, FAP low; ns, not significant.
FAP-high cells promote more gel contraction, cancer cell invasion, and chemotherapy resistance than FAP-low cells
A hallmark of CAFs is their ability to contract collagen gels (Ngo et al., 2006). Notably, two early-passage FAP-high CAF lines (851FH and 68425FH) showed a greater ability to contract collagen gels than two early-passage FAP-low CAF lines (507FL and 598FL; Fig. 5 B). Another property commonly attributed to CAFs is the ability to promote cancer cell invasion (Goetz et al., 2011; Kalluri, 2016; Karnoub et al., 2007). To compare the ability of FAP-high and FAP-low CAFs to promote invasion, spheroids were established with the GFP-expressing HGSOC cell lines OVCAR8, ES2, and OV90, either alone or together with FAP-high CAFs or FAP-low CAFs at a ratio of 5:1 (CAFs/cancer cells). The spheroids were embedded in Matrigel and imaged by fluorescence microscopy after a period of incubation at 37°C that was optimized for individual cell lines and different batches of Matrigel. For OVCAR8 and OV90, invasion was quantified by measuring the circularity of at least five spheroids per condition; a decrease in circularity is indicative of cells invading into the Matrigel, generating branches radiating away from the spheroids and thus causing spheroids to deviate from a circular shape. The invasion pattern of ES2 cells was distinct and consisted of individual cells migrating outwards into the Matrigel rather than formation of branches. Invasion was therefore quantified by counting the number of cells outside of the sphere rather than measuring circularity. Upon testing of FAP-high and FAP-low CAFs from multiple patients, FAP-high CAFs had an overall significantly greater ability to promote the invasion of OVCAR8, ES2, and OV90 cells compared with FAP-low CAFs. Notably, in cases where it was possible to assess patient-matched FAP-high and FAP-low CAFs (patient 438 on all three cell lines and patient 598 on ES2 cells), there was a statistically significant difference, with FAP-high CAFs inducing more invasion than FAP-low CAFs (Fig. 5 C). In assays where OVCAR8 and OV90 monoculture spheroids were treated with conditioned media from FAP-high or FAP-low CAFs (n = 3 of each), no invasion was seen (data not shown).
The standard of care for patients with HGSOC includes treatment with platinum-based chemotherapy, and chemoresistance is a major factor leading to poor outcome in this disease. We determined the half maximal inhibitory concentration (IC50) for carboplatin in OVCAR8, ES2, and OV90 cells growing under adherent conditions on plastic and showed that they ranged from 6 to 24 µM at 5 d after treatment (Fig. S5 C). Based on this information, subsequent experiments were performed using 10 µM carboplatin. To determine if FAP-high and/or FAP-low CAFs could influence cancer cell responses to chemotherapy, GFP-labeled OVCAR8, ES2, or OV90 cells were seeded onto confluent feeder layers of FAP-high CAFs or FAP-low CAFs in flat-bottom 96-well plates. The next day, 10 µM carboplatin was added, and cells were cultured for an additional 10 d. GFP+ cancer cells were quantified daily using an Incucyte Zoom live-cell imaging system (Fig. S5 D). Both OVCAR8 and ES2 cells were rendered more resistant to carboplatin treatment in the presence of FAP-high CAFs compared with FAP-low CAFs, as indicated by more robust growth and a larger number of cells remaining at the end of the 10-d treatment period in FAP-high co-cultures (Fig. 5 D and Fig. S5 E). This included two pairs of patient-matched FAP-high and FAP-low CAFs (598FH and FL; 438FH and FL) that showed distinct outcomes. OV90 cells did not display any differences. Notably, for the two separate batches of experiments performed (Fig. 5 D) 851FH CAFs were used in both and displayed very reproducible growth curves in all three cell lines. The difference in behavior in co-cultures with FAP-high or FAP-low CAFs could not be accounted for by differences in viability of the CAFs themselves, as both FAP-high and FAP-low CAFs remained >98% viable in the presence of 10 µM carboplatin for 10 d (data not shown). In addition, no difference in carboplatin response was seen when OVCAR8 cells were cultured in conditioned medium from FAP-high or FAP-low CAFs (n = 3 of each; data not shown).
FAP-high CAFs promote in vivo tumor growth and metastasis to lymph nodes
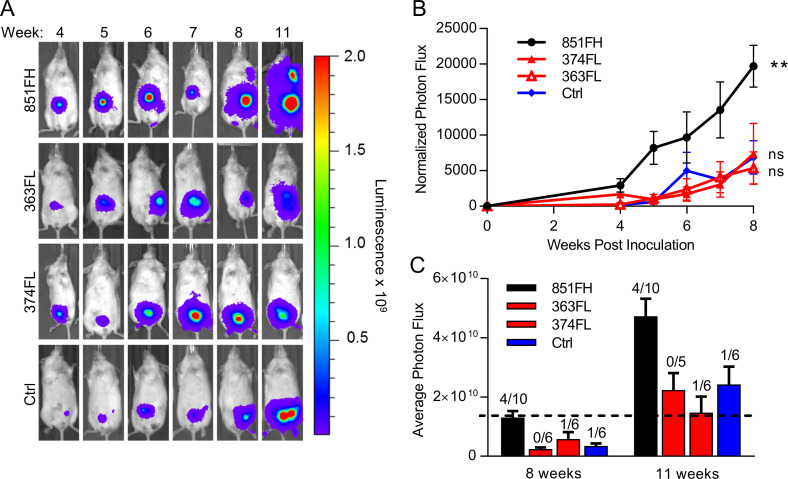
To ask if FAP-high and FAP-low CAFs have distinct influences over cancer cell behavior in vivo, we generated spheroids as above using luciferase-tagged OVCAR8 cells and implanted single spheroids into the mammary fat pads of nonobese diabetic/severe combined immunodeficient/IL2R-γ double KO (NSG) mice. Tumor growth was monitored by serial imaging of luciferase activity (Fig. 6 A). Of 10 mice implanted in each group, take rates were 6/10, 5/10, 6/10, and 9/10 for the no CAF control, 363FL CAFs, 374FL CAFs, and 851FH CAFs, respectively, suggesting a possible enhancement of take rate in spheroids containing FAP-high CAFs, although differences did not reach statistical significance. Tumors that contained FAP-high CAFs (851FH) grew more rapidly than tumors containing FAP-low CAFs (363FL or 374FL; Fig. 6 B). Furthermore, 4 of 10 mice in the FAP-high group had axillary lymph node or abdominal metastases compared with 0 of 5 mice and 1 of 6 mice in the two FAP-low groups and 1 of 6 mice in the no CAF control group (Fig. 6 C). Importantly, this held true when the mice injected with FAP-low CAF-containing spheroids were maintained for an additional 3 wk, allowing the size of their tumors to reach a size exceeding the FAP-high tumors at week 8, indicating that the difference in metastasis was not simply a function of larger tumor size in the FAP-high group.
Figure 6.
FAP-high, but not FAP-low, CAFs promote in vivo tumor growth and metastasis. (A) Spheroids of luciferase-expressing OVCAR8 cells alone or combined with FAP-high or FAP-low CAFs were generated as for invasion assays. After 72 h of spheroid formation, individual spheroids were implanted into the mammary fat pads of NSG mice. Tumor growth was monitored over time using the Xenogen IVIS Imaging System 100. Representative images of a single mouse from each condition over time are shown. All mice were exposed for the same amount of time to allow visualization of differences in tumor size over time. (B) Quantification of luciferase signal, normalized to an unimplanted mouse over a period of 8 wk. Data are presented as mean ± SEM, n = 5–10 (animals in which tumors failed to grow were not included). **, P < 0.01 versus control group, linear regression. (C) Average tumor sizes in each group at 8 and 11 wk after implantation. Numbers above the bars indicate the number of mice with metastases over the total number of tumor-bearing mice in each group. Data are presented as mean ± SEM, n = 5–10. FH, FAP high; FL, FAP low; ns, not significant.
Overexpression of TCF21 in FAP-high CAFs inhibits their protumorigenic functions
TCF21 is the most highly expressed transcription factor in FAP-low CAFs and is essential for the formation of cardiac fibroblasts during embryonic development (Acharya et al., 2012). TCF21 has differentiation inhibiting function in skeletal muscle and smooth muscle cells (Braitsch et al., 2012; Funato et al., 2003; Nurnberg et al., 2015) and is also highly expressed in white adipose tissues (de Jong et al., 2015). Because lipid metabolism, ventricular development, and cardiac related pathways were enriched in FAP-low CAFs (Fig. 3 D), we hypothesized that TCF21 might be a master regulator of FAP-low CAF identity. To test this possibility, 851FH CAFs were transduced with lentiviral vectors expressing either TCF21 and GFP (851FH-TCF21) or GFP alone (851FH-GFP) and GFP+ cells were FACS purified and briefly expanded. TCF21 expression was validated by Western blot (Fig. S5 F), and qRT-PCR demonstrated up-regulation of TCF21 and two additional FAP-low–specific transcripts (DLK and TGFBR3), as well as down-regulation of four FAP-high–specific genes (FAP, SULF1, MMP1, and MFAP5; Fig. 7 A). We then compared the functional properties of 851FH-TCF21 and 851FH-GFP cells. The ability of 851FH-TCF21 CAFs to contract collagen gels was decreased in comparison to 851FH-GFP cells (Fig. 7 B). The spheroid invasion assay was performed using mCherry-labeled OVCAR8 and OV90 cells (because the 851FH-TCF21 and 851FH-GFP CAFs were GFP+), allowing imaging of both cancer and CAF cells in this assay. The invasion of both cell types was significantly reduced upon overexpression of TCF21 in 851FH CAFs (Fig. 7 C). We next performed co-cultures of OVCAR8, OV90, and ES2 cells with 851FH-TCF21 or 851FH-GFP CAFs in the presence of 10 µM carboplatin. OVCAR8 and ES2 cells grew more robustly in co-cultures with 851FH-GFP CAFs than in co-cultures with 851FH-TCF21 CAFs, suggesting that TCF21 expression reduced the ability of 851FH CAFs to promote the survival of cancer cells in the presence of carboplatin (Fig. 7 D). Once again, no effect was seen with OV90 cells. A similar result was obtained upon overexpression of TCF21 in 438 FH CAFs (Fig. S5 G). Finally, heterospheroids composed of OVCAR8 cancer cells and 851FH-TCF21 or 851FH-GFP fibroblasts were implanted in the mammary fat pad of NSG mice, and growth was monitored by serial imaging of luciferase activity (Fig. 7 E). 15 of 15 mice implanted with 851FH-GFP–containing spheres grew tumors, whereas only 10 of 15 mice implanted with 851FH-TCF21–containing spheres grew tumors (P = 0.042, Fisher’s exact test). In addition, overexpression of TCF21 in 851FH CAFs led to a significant growth delay compared with the 851FH-GFP control (Fig. 7 F).
Figure 7.
Overexpression of TCF21 in FAP-high CAFs inhibits their protumorigenic functions. (A) qRT-PCR for FAP-low genes (TCF21, DLK1, and TGFBR3) and FAP-high genes (FAP, SULF1, MMP1, and MFAP5) in control versus TCF21 overexpressing 851FH CAFs. Fold-differences in gene expression were determined using the ΔΔ−Ct method, using UBC as a housekeeping gene, with three technical replicates per sample assayed. (B) Representative images of collagen gels cultured for 32 h with 851FH-GFP CAFs or 851-TCF21 CAFs (left); quantification of gel contraction over time (right). Data are presented as mean ± SEM, n = 3. **, P < 0.01, two-way ANOVA. Scale bar = 500 µm. (C) Representative images of spheroids generated using mCherry-expressing HGSOC cell lines OVCAR8 or OV90 with 851FH-GFP or 851FH-TCF21 CAFs mixed in. Spheroids were embedded in Matrigel and imaged after 4 d (left). Circularity was quantified in both channels (green, CAFs; red, cancer cells) using ImageJ (right). Data are presented as mean ± SEM, n = 10 spheroids per condition. *, P < 0.05; **, P < 0.01; ****, P < 0.0001, Student’s t test. Scale bar = 100 µm. (D) Growth curves of mCherry-labeled cancer cells co-cultured with 851FH-GFP or 851FH-TCF21 CAFs after treatment with 10 µM carboplatin. Data were normalized to the mCherry+ area at day 0 and are presented as mean ± SEM, n = 3. Asterisks reflect differences at the end of 10 d. ***, P < 0.001; ****, P < 0.0001, Student’s t test. (E) Spheroids of luciferase-expressing OVCAR8 cells alone or with 851FH-GFP or 851FH-TCF21 CAFs mixed in were generated as for invasion assays. After 72 h of spheroid formation, individual spheroids were implanted into the mammary fat pads of NSG mice. Representative images of individual mice are shown. (F) Quantification of luciferase signal, normalized to an unimplanted mouse over a period of 9 wk. Data are presented as mean ± SEM, n = 10–15 (animals in which tumors failed to grow were not included). *, P < 0.05, two-way ANOVA. FH, FAP high; FL, FAP low; ns, not significant.
Discussion
In this study, we demonstrate that CAFs in HGSOC are heterogeneous, that different subtypes have distinct influences on cancer aggressiveness and patient outcomes, and that TCF21 is a master regulator of CAF state. The identification of distinct CAF subtypes in HGSOC was facilitated by the identification of CD49e as a novel pan-fibroblast marker and the resulting ability to profile CAFs isolated directly from primary tumor specimens. We showed that patients with predominantly FAP-high CAFs in their stroma have shorter disease-free and/or overall survival. Functional assays demonstrated that FAP-high CAFs promote cancer cell invasion, resistance to carboplatin, and proliferation and metastasis in vivo, thus explaining the negative role that this CAF subtype plays in patient outcomes. By contrast, FAP-low CAFs do not promote these behaviors in cancer cells. Finally, we show that TCF21 expression in FAP-high CAFs suppresses their protumorigenic phenotype.
While it has been known for some time that CAFs are heterogeneous (Kalluri, 2016; Öhlund et al., 2014; Sugimoto et al., 2006), only recently have distinct CAF subtypes begun to be identified and characterized. For example, a recent study showed that CAFs in close proximity to cancer cells in pancreatic cancer are α-SMA+ myofibroblasts, whereas fibroblasts more distant from cancer cells lack elevated α-SMA and instead secrete inflammatory cytokines (Öhlund et al., 2017). A CAF subset defined by expression of CD10 and GPR77 varied in frequency in breast and lung cancers and was associated with resistance to chemotherapy and shorter patient survival (Su et al., 2018). Importantly, in both of these studies, the identified subtypes were all FAP+, suggesting that they represent heterogeneity within the FAP-high fraction. An additional study of breast cancer used multiple fibroblast markers to identify four CAF subsets, of which only one was FAP+ (S1) and three were FAP− (S2–S4; Costa et al., 2018). Interestingly, the S1 and S4 subtypes were associated with aggressive HER2 and triple-negative breast cancers, and triple-negative breast cancer could be divided into two subgroups based on the presence of either S1 or S4 CAF subtypes. In a separate study, the same group identified S1 and S4 CAFs in HGSOC (Givel et al., 2018). They showed that “mesenchymal” samples are enriched for S1 CAFs, suggesting that S1 CAFs resemble our FAP-high CAFs. However, the S4 CAFs quickly died and could not be maintained in culture; thus, the authors were unable to carry out significant characterization of these cells. In addition, no analyses were done to evaluate the clinical significance of the CAF subtypes they identified. Thus, our work represents a significant advance beyond these important early studies, through demonstration of the clinical significance of CAF heterogeneity in HGSOC, as well the extensive functional and molecular characterization of isolated CAF subtypes.
The seminal studies by Givel et al. (2018) and Costa et al. (2018) showed that FAP+ CAFs have immunosuppressive functions in HGSOC and breast cancer. Other studies have also demonstrated an immune-suppressive function of FAP-expressing stromal cells (Fearon, 2014; Kraman et al., 2010; Yang et al., 2016). We find a large cluster of pathways involved in immune regulation in FAP-high CAFs (Fig. 3 D andTable S4 A), which include many chemokines and cytokines involved in immune processes, such as generation of myeloid-derived suppressor cells (e.g., CXCL12, IL11, and VEGF), macrophage polarization (e.g., CXCL12, IL10, and Chi3L1), differentiation of immune suppressive T regulatory cells (e.g., IL1B, IL10, and IL11) and inhibition of CD8+ cytotoxic T cells (e.g., PDCD1LG2 and LGALS1). Thus, future work should include analysis of immune subtypes present in FAP-high versus FAP-low patients and understanding the cross-talk between subtypes of CAFs and immune cells in this cancer. By contrast, the FAP-low CAFs have a distinct gene expression profile that lacks the secretory phenotype seen in FAP-high cells. Prominent networks include translation/protein localization, ion transport/cardiac muscle contraction, and lipid metabolism and steroid biosynthesis. The latter is in agreement with our finding that FAP-low cells grew preferentially in preadipocyte media, which contains low serum plus supplements that include epidermal growth factor and compounds that are proadipogenic such as dexamethasone, 3-isobutyl-1-methylxanthine and ciglitazone, a PPARγ agonist. However, the prominent muscle contraction network suggests that these cells may have a more primitive mesenchymal progenitor phenotype that has both myogenic and adipogenic potential. Comparison of the FAP-low gene signature to published mesenchymal stem cell (MSC) signatures, however, suggests that these cells are not MSCs. Indeed, MSCs more closely resemble FAP-high CAFs, as they have elevated expression of FAP as well as multiple ECM proteins and ECM remodeling enzymes (Jeong et al., 2005; Wagner et al., 2005); thus, further investigations, including functional assays, will be required to better elucidate the origin and/or identity of FAP-low CAFs. For example, it will be of interest to compare the abilities of FAP-high and FAP-low CAFs to differentiate into various mesenchymal lineages and determine if additional manipulations of the culture conditions can identify a condition that can maintain FAP-low CAFs indefinitely in vitro.
Altogether, our functional assays suggest that the worse survival outcomes of patients with a FAP-high gene signature and/or a predominance of FAP-high CAFs within their stroma are mediated by CAF–cancer cell interactions that promote cancer cell proliferation, invasion, and therapy resistance, as well as immune suppression, as shown by others (Givel et al., 2018). Interestingly, conditioned media from FAP-high CAFs did not promote cancer cell invasion or chemotherapy resistance, suggesting that secreted factors alone are insufficient to induce these behaviors. Furthermore, there was some variability between individual FAP-high CAF lines in their ability to promote invasion and chemotherapy resistance and also variability in cell lines in their responses (e.g., the carboplatin response of OV90 cells was not affected by FAP-high CAFs). This suggests different mechanisms for each of the induced behaviors, as well as different responses between individual tumors. In spite of this variation, our results indicate that future studies focused on targeting FAP-high CAFs in order to improve outcomes and/or responses to standard chemotherapy or immunotherapy are warranted.
The unique transcriptional programs of FAP-high and FAP-low CAFs prompted us to more closely examine differentially expressed transcription factors between the two CAF subtypes, and TCF21 was the most highly differentially expressed transcription factor in FAP-low cells. TCF21 is expressed in epicardial progenitor cells that give rise to coronary artery smooth muscle cells and cardiac fibroblasts (Nurnberg et al., 2015), the latter of which are a source of activated myofibroblasts in the infarcted heart (Kanisicak et al., 2016). TCF21 is also a marker for white adipose tissue and is abundantly expressed in visceral fat-derived stem cells (Akama and Chun, 2018). When we overexpressed TCF21 in FAP-high CAFs, our results indicated that TCF21 on its own can significantly dampen the ability FAP-high CAFs to promote gel contraction, invasion, chemoresistance, and in vivo tumor growth. However, additional transcription factors or coregulators of TCF21 and/or epigenetic regulators of chromatin accessibility are likely required to completely reprogram FAP-high CAFs to a state that lacks protumorigenic properties. Future epigenomic profiling studies will be required to compare the epigenetic states of FAP-high and FAP-low CAFs and identify potential avenues to “reprogram” FAP-high CAFs to a state that is not supportive of cancer cell invasion, chemoresistance, or immune suppression.
Materials and methods
Primary CAF cultures
CAFs were derived from either bulk tumor cell suspensions or FACS-purified cells (see detailed methods below). Bulk tumor cell suspensions were seeded into tissue culture plates in IMDM with 10% FBS. Fibroblasts adhered and grew out preferentially under these conditions over multiple passages. Fibroblast identity was verified based on cell morphology, as well as expression of vimentin and lack of expression of CK. CAFs were also verified to be negative for p53 staining (i.e., wild-type) in patients with positive p53 staining (i.e., mutant) in their tumor tissues. FAP-high CAFs isolated by FC (viable, CD45−, CD31−, EpCAM−, CD49e+, FAP high) were seeded into wells of 96-well culture plates in IMDM with 10% FBS. FAP-low CAFs isolated by FC (viable, CD45−, CD31−, EpCAM−, CD49e+, FAP low) were seeded into wells of 96-well culture plates in preadipocyte media (PromoCell). Cells were passaged 1:2 when they reached confluence using 0.25% Trypsin/1 mM EDTA. All CAF experiments were done at passage ≤10 for cultured CAFs and FAP-high CAFs and passage ≤5 for FAP-low CAFs.
Cell lines
Three HGSOC cell lines, OVCAR8, OV90, and ES2, were obtained from the ATCC, and their identity was verified by short tandem repeat profiling (Life Technologies; AmpFLSTR Identifiler), performed by The Centre for Applied Genomics, The Hospital for Sick Children, Toronto, Canada, and tested negative for Mycoplasma infection by MycoAlert (Lonza) according to the manufacturer’s instructions. Lines were maintained as recommended by ATCC, as follows: OVCAR8 in IMDM with 10% FBS, ES2 in McCoy’s 5A media with 10% FBS, and OV90 in 1:1 M199:MCDB105 media with 15% FBS.
Primary tumor dissociation
Bulk tumors were mechanically minced into a slurry with sterile scalpels and then enzymatically digested in Media 199 containing 300 U/ml collagenase and 100 U/ml hyaluronidase mixture (Stem Cell Technologies) and 125 U/ml DNase-I (Cedarlane) for 1–2 h at 37°C. Following digestion, samples were centrifuged at 350 ×g for 5 min before treatment with 1-2 ml of ACK lysing buffer (ThermoFisher Scientific) for 5 min on ice. Following red cell lysis, cells were pelleted, resuspended, and filtered through a 70-µm sterile nylon mesh and viable cells defined by trypan blue exclusion. Cells were cryopreserved in 90% FBS/10% dimethyl sulfoxide.
HT-FC
HT-FC was performed on cultured CAFs and primary HGSOC tumor cell suspensions as previously described (Gedye et al., 2014). Briefly, 363 commercially available antibodies to cell surface antigens conjugated to PE, FITC, or APC were aliquoted into round bottom 96-well plates, 2 µl per well into 48 µl of FC buffer (Hanks balanced salt solution + 2% FBS). Cell suspensions of 0.5 to 1 million cells/ml in FC buffer were aliquoted by multichannel pipette into preprepared HT-FC plates (50 µl per well), for a final volume of 100 µl per well and a final antibody dilution of 1:50. Plates were incubated for 20 min on ice in the dark, centrifuged for 5 min at 350 ×g, washed twice with 200 µl FC buffer, and resuspended in 50 to 80 µl FC buffer containing 0.1 µg/ml DAPI (Sigma-Aldrich). Primary tumor samples were stained with CD45-APC-Cy7 (BioLegend; 1:200) and CD31-PE-Cy7 (BioLegend; 1:200) before aliquoting into plates. Fluorescence-minus-one controls were generated for each fluorochrome used and compensations were set using BD Plus CompBeads and FACSDiva software. Data collection was performed on a Becton Dickinson LSR II flow cytometer with ultraviolet (20 mW), violet (25 mW), blue (20 mW), and red (17 mW) lasers, with default filter configuration, using the high-throughput sampler attachment. At least 10,000 events were collected per well. The gating strategy based on fluorescence-minus-one controls is illustrated in Fig. S1 B.
FC and FACS
Cryopreserved or fresh HGSOC single-cell suspensions were washed and resuspended at ≤107 cells/ml in FC buffer and incubated with 10 µg/ml mouse IgG in FC buffer on ice for 10 min, followed by incubation with the following primary antibodies: CD45-PECy7 (BioLegend; 1:200), CD31-PECy7 (BioLegend; 1:200), CD49e-PE (BD Biosciences; 1:100), EpCAM-APC (BioLegend; 1:100). For experiments including CD133, an unconjugated mouse anti-human CD133 antibody was used (Miltenyi Biotec; 1:20), and for experiments including FAP, an unconjugated mouse anti-FAP antibody was used (R&D Systems; 1:50). Cells were incubated on ice for 15 min, washed and resuspended in FC buffer with goat-anti-mouse Alexa488 (Invitrogen; 1:400) for an additional 15 min, and then washed and stained with the remaining directly conjugated antibodies as described above. Fluorescence-minus-one controls were generated for each antibody and used as gating controls. Single-color stained compensation beads (BD Biosciences) were used as compensation controls. Cells were analyzed on a BD LSR II flow cytometer or sorted using a BD FACS Aria.
Immunofluorescence: Cytospins
FACs-sorted cells were suspended in PBS+ 2% FBS at a concentration of 5 × 104 cells/ml, and 200 µl of cell suspension was spun onto each glass slide using a cytocentrifuge at 800 rpm for 5 min. Slides were air dried, fixed in 100% ice-cold acetone, and air dried again. Cells were permeabilized in Tris-buffered saline (TBS) containing 0.1% Tween for CK and vimentin or 0.3% Triton X-100 for p53 (TBS-T) for 10 min and then incubated for 30 min in TBS-T/5% BSA/5% goat serum, followed by incubation with the following antibodies: rabbit anti-wide spectrum CK (Abcam; 1:100), mouse-anti-human vimentin (Abcam; 1:100), mouse-anti-human p53 (Santa Cruz; 1:100) overnight at 4°C. The following day, slides were washed three times in TBS and incubated with the appropriate secondary antibodies: goat-anti-rabbit-Alexa594 (Invitrogen; 1:1,000) plus goat-anti-mouse-Alexa488 (Invitrogen; 1:400) for pan-CK and vimentin; or goat-anti-mouse-Alexa594 (Invitrogen; 1:200) for p53. Secondaries were incubated for 1 h at room temperature. Slides were then washed and incubated for 1 min in TBS with 1 µg/ml Hoechst 333258, then coverslipped with Mowiol 4–88 (Sigma-Aldrich).
Immunofluorescence: FFPE tissue sections
Paraffin blocks of HGSOC tissues from five patients were obtained from the University Health Network (UHN) Biospecimen Sciences Program in accordance with regulations for excess tissue use stipulated by the UHN research ethics board. 4-µm sections were transferred onto positively charged slides. Sections were deparaffinized using xylene and ethanol. For antigen retrieval, slides were incubated in 0.01 M citrate buffer (pH 6.0) with 0.05% Tween in a glass vessel submerged in boiling water for 20 min. Sections were then permeabilized in TBS-T (containing 0.1% Tween for CK, CD49e, and FAP, and 0.3% Triton X-100 for TCF21) for 10 min. Sections were incubated for 2 h in TBS-T/0.5% BSA/5% goat serum, followed by incubation with rabbit anti-human TCF21 (Sigma-Aldrich; 1:100), mouse-anti-human-CD49e (BD Biosciences; 1:100), mouse-anti-human-FAP (R&D Systems; 1:50), and either mouse-anti-pan-CK (Abcam; 1:100) or rabbit anti-wide spectrum CK (1:100) overnight at 4°C. The following day, slides were washed three times and incubated with goat-anti-rabbit-Alexa Fluor 594 (Invitrogen; 1:1,000) plus goat-anti-mouse-Alexa Fluor 488 (Invitrogen; 1:400) for 1 h at room temperature. Slides were then washed and incubated for 1 min in TBS with 1 µg/ml Hoechst 333258 and then coverslipped with Mowiol 4–88 (Sigma-Aldrich). Slides were scanned using a Zeiss Axio Slide Scanner, and images were analyzed using HALO software (Indica Labs).
Microarrays
HGSOC single-cell suspensions were stained for FACS as described above. Doublets and dead cells were excluded, and CD31−CD45−EpCAM−CD49e+ cells, CD31−CD45−EpCAM+CD133− cells, and CD31−CD45−EpCAM+CD133+ cells were gated for sorting based on fluorescence-minus-one controls. Postsort purity checks for each sample confirmed >98% purity for each population. 12 patient samples were sorted into tubes containing IMDM with 10% FBS. Sorted populations were washed with PBS, and then RNA was extracted immediately using the RNeasy Plus Micro kit (Qiagen). RNA quality was verified using a Bioanalyzer 2100 (Agilent Technologies). All samples had an RNA integrity number (RIN) >8. 5 ng of RNA per sample were amplified using the Ovation pica WTA V2 kit (Nugen) as per the manufacturer’s instructions. Amplified cDNA from each sample was labeled following Nugen Illumina solution application note #2. 750 ng amplified biotin-labeled cDNA generated from these samples was randomized and hybridized onto three Illumina Human HT-12 v4 BeadChips. BeadChips were incubated at 48°C at rotation speed 5 for 15 h for hybridization. The BeadChips were washed and stained as per Illumina protocol and scanned on the iScan (Illumina). Data files were quantified in GenomeStudio Version 2011.1 (Illumina) and passed sampled-dependent and independent quality control metrics.
Probe intensities were normalized between arrays using quantile normalization and transformed using the logarithm of base 2. Differential expression between the different groups of samples (CD49e+ and EpCAM+, FAP-high and FAP-low) was estimated using limma 3.28.21. HUGO Gene Nomenclature Committee gene names were associated with probe identities using biomaRt 2.28.0. Two-color heatmaps were generated using the heatmap.2 function of the gplots R package 3.0.1 and pvclust 2.0.0 was used to cluster the CAF FAP-high and FAP-low samples with bootstrapping.
To carry out pathway analysis of CD49e+ and EpCAM+ cells, an expression score was created to rank all genes from top up-regulated to top down-regulated using the formula sign(logFC) × −log10(P value). GSEA (Broad Institute) was applied on this rank list using 1,000 permutations. The gene sets tested by GSEA were the Reactome database included in the MSig c2.cp.reactome.v6.1.symbols.gmt file. Results were visualized in Cytoscape 3.6.1 using EnrichmentMap 3.1, clustered and annotated using AutoAnnotate 1.2. Cluster labels were manually edited for clarity.
qRT-PCR
qRT-PCR was performed in triplicate using Power SYBR Green PCR Master Mix (Life Technologies). Samples were loaded into a Bio-Rad CFX96 real-time PCR detection system following the manufacturer’s protocols. Relative amounts of mRNA were calculated by the ΔΔCt method and normalized to expression levels of UBC. The following primer sequences were used: FAP, forward 5′-TGGCGATGAACAATATCCTAGA-3′, reverse 5′-ATCCGAACAACGGGATTCTT-3′; COL11A1, forward 5′-TTTTCCAGGATTCAAAGGTGA-3′, reverse 5′-TGGGCCAATTTGACCAAC-3′; SULF1, forward 5′-ACCAGACAGCCTGTGAACAA-3′, reverse 5′-ATTCGAAGCTTGCCAGATGT-3′; TCF21, forward 5′-CGACAAATACGAGAACGGGTA-3′, reverse 5′-TCAGGTCACTCTCGGGTTTC-3′; COLEC11, forward 5′-CCCCTGGTCCTAATGGAGA-3′, reverse 5′-TCAGCTGAGAGACCTGGTTGT-3′; DLK1, forward 5′-GACGGGGAGCTCTGTGATAG-3′, reverse 5′-GGGCACAGGAGCATTCATA-3′; and UBC, forward 5′-AGGCAAAGATCCAAGATAAGGA-3′, reverse 5′-GGACCAAGTGCAGAGTGGAC-3′.
RNA-seq
HGSOC single-cell suspensions were stained for FACS as described above. Doublets and dead cells were excluded, and CD45−CD31−EpCAM−CD49e+FAP-high and CD45−CD31−EpCAM−CD49e+FAP-low populations were gated for sorting. At least 10,000 cells were sorted from each population. 12 patient samples were sorted into tubes containing IMDM with 10% FBS. Sorted populations were washed with PBS, and then RNA was extracted immediately using the RNeasy Plus Micro kit (Qiagen). RNA samples were assessed on an RNA 6000 Pico chip (Agilent Technologies) using the Agilent Bioanalyzer to determine sample RIN and quantified by the Qubit RNA HS assay kit (Life Technologies). All samples used had RIN values >8.5. 4 ng of RNA were used to prepare RNA libraries using the SMARTer Stranded Total RNA-seq Kit Pico Input Mammalian (Takara Bio). Briefly, the samples underwent first-strand synthesis via random priming oligos on the 3′ end. A template-switching oligo mix was then incorporated to allow the RT reaction to continue replicating the 5′ of the RNA strand. Following this, the samples were PCR amplified to incorporate full-length Illumina adapters and sample barcodes by binding to either the template-switching oligo mix stretch on the 5′ end, or the random priming oligo sequence on the 3′ end. The amplified cDNA is treated with ZapR, which specifically targets ribosomal RNA in the presence of mammalian-specific R-Probes. This process leaves nonribosomal RNA untouched, while ribosomal RNAs are cleaved, leaving them nonamplifiable. A final PCR reaction was done to enrich the uncut strands of cDNA to generate the final RNA library. Final RNA library sizing was verified on the Agilent high-sensitivity DNA kit (Agilent Technologies) using the Agilent Bioanalyzer while library concentration was quantified by qPCR using the KAPA SYBR FAST qPCR kit (Kapa Biosystems). Libraries were normalized to 10 nM, diluted to 2 nM, denatured using 0.2 N NaOH, and diluted again to 1.7 pM before loading onto the NextSeq 500 system. The samples were sequenced using a paired-end 75 cycle sequencing run to achieve a minimum of ∼40 million reads per sample. The reads were mapped using STAR/2.5.2 to the hg38 reference genome. Read counts per gene were obtained through htseq-count v.0.6.1. After removing low count genes whose counts per million reads were <0.75 to in one third of the total number of samples, the edgeR R package v.3.8.6 was used to normalize the data using the trimmed mean of M values method and to estimate differential expression by applying a generalized linear model between the FAP-low fraction samples and the FAP-high fraction samples. The multidimensional scaling plot was created using the edgeR plotMDS.DGEList() function.
For pathway analysis of FAP-high and FAP-low CAFs, an expression score was created to rank all genes from top up-regulated to top down-regulated using the formula sign(logFC) × −log10(P value). GSEA (Broad Institute) was applied on this rank list using 1,000 permutations. The gene sets tested by GSEA were first the microarray FAP-high and FAP-low gene lists and second gene sets from the Reactome database included in the MSig c2.cp.reactome.v6.1.symbols.gmt file. Results were visualized in Cytoscape 3.6.1 using EnrichmentMap 3.1, clustered and annotated using AutoAnnotate 1.2. Cluster labels were manually edited for clarity.
TCGA data analysis
To identify genes specific to FAP-high and FAP-low CAFs, genes differentially expressed between FAP-high and FAP-low fractions at an FDR cutoff of 0.05 and with a logFC greater than twofold (logFC >2 for FAP high and <−2 for FAP low) were selected. These genes were then filtered to include only those that have a higher expression in CD49e+ cells compared with EpCAM+ CD133+ at an FDR cutoff of 0.05 in the Human Illumina HT-12 V4 array. This generated a list of 165 FAP-high CAF-specific genes and 78 FAP-low–specific genes.
HGSOC data were downloaded from the Genomic Data Commons Data Portal (https://portal.gdc.cancer.gov/). HiSeq gene level counts (level 2 RNA-seq data) and corresponding clinical data were downloaded for 374 samples included in this analysis.
Trimmed mean of M values followed by count per million and logarithm of base 2 transformation was used to normalize the data within the edgeR package. Data were selected to only contain the FAP-high and FAP-low gene list, and a heatmap was created using R heatmap.2. Color bars were added to the heatmap to identify patients based on molecular phenotype described by Verhaak et al. (2013). These phenotypes include the categories “mesenchymal,” “immune,” “proliferative,” and “differentiated,” and they were extracted from supplemental table 1 of Verhaak et al. (2013). Differential expression was calculated within edgeR for samples defined as mesenchymal versus all the other samples and a list ranking genes from top up-regulated to down-regulated was generated using the formula sign(logFC) × −log10(P value).
For the FAP-high and FAP-low color bar, TCGA patients were ranked using a score that counts how many genes from the FAP-high gene list or FAP-low gene list have a normalized value greater than the patient mean (i.e., a z-score). Patients were considered FAP-high if they had positive scores for at least 75% of the gene list (corresponding to a sum of z-scores > gene list length/2). A less stringent threshold classified patients as FAP high or FAP low if they had positive scores in at least 50% of the gene list (corresponding to a sum of z-scores > 0). The FAP-high and FAP-low patient categories were added to the heatmap color bar and used for further analysis.
TCGA patients were then grouped into FAP-high, FAP-low, and “other” categories using either the 75% or the 50% thresholds, and ESTIMATE and ABSOLUTE values, which are known to measure percentage of stromal cell and tumor purity content, respectively (Carter et al., 2012; Yoshihara et al., 2013), were determined using the respective R packages. Data were plotted on a whisker boxplot for each category. Clinical data, including overall survival and progression-free events, were retrieved for TCGA patients falling into the FAP-high and FAP-low categories based on the more stringent 75% cutoff, and Kaplan–Meier curves were generated and associated with a log-rank test using GraphPad Prism software.
For GSEA testing of the short FAP-high and FAP-low lists in the mesenchymal subtype, the mesenchymal rank list was tested against the short FAP-high and FAP-low gene lists using default parameters.
Gel contraction
FAP-high and FAP-low CAFs were suspended in IMDM with 10% FBS at a concentration of 100,000 cells/ml and kept on ice. 3 mg/ml Collagen-I solution (ThermoFisher Scientific, Corning; 354236) and 1 M NaOH were then added to the cell suspension to give a final concentration of 1 mg/ml Collagen-I and a neutral pH, and 100 µl was aliquoted into non–tissue culture–treated flat-bottom 96-well plates. Plates were left for 20 min at room temperature to solidify, then 120 µl IMDM + 10% FBS was added to the wells. A small pipet tip was gently run around the perimeter of each well before imaging on the Incucyte ZOOM every 6 h for 3 d. The areas of the gels at different time points were quantified using ImageJ (Fiji) software.
3D spheroid invasion assays
GFP-expressing HGSOC cell lines (OVCAR8, OV90, and ES2) were mixed with FAP-high, FAP-low, 851FH-TCF21, or 851FH-GFP CAFs at a ratio of 5:1 (CAF/cancer cells). 6,000 total cells per well were then plated in 90 µl basal media (IMDM for OVCAR8, McCoy’s 5A for ES2, or MCDB105:M199 for OV90) supplemented with 2% FBS in ultra-low-attachment round-bottom 96-well plates (Corning; 7007). Plates were centrifuged at low speed to center the cell suspension before incubation at 37°C for 3 d. At that point, spheroids were formed and 30 µl media was removed from each well and replaced with 30 µl growth factor reduced Matrigel (Corning; 354230) to give a final Matrigel concentration of 33% vol/vol. Plates were incubated for 60 min at 37°C, and then 100 µl per well of the appropriate basal media supplemented with 2% FBS was added and plates were returned to the incubator. The time point used to quantify invasion was optimized for individual cell lines and ranged from 1 d (for ES2 cells) to 4 d (for OVCAR8 and OV90 cells). Spheroids were imaged with a Zeiss LSM700 confocal microscope. Circularity of spheroids (OVCAR 8, OV90) and cell counts outside the spheroid (ES2) were analyzed using ImageJ (Fiji) software. In some cases, multiple spheroids formed per well, or the spheroids were not centered in the well and thus could not be imaged. However, a minimum of 5 and up to 10 spheroids per experimental group were analyzed in all cases.
In vitro carboplatin treatment
1,000 cells per well of GFP or mCherry-expressing OVCAR8, OV90, or ES2 cells were seeded in their respective media onto a monolayer of CAFs in 96-well flat-bottom tissue culture plates. The plates were incubated at 37°C for 24 h, and then carboplatin (Hospira) was added to reach a final concentration of 10 µM. Cancer cell proliferation was monitored using Incucyte ZOOM live cell imaging system for up to 10 d.
In vivo assays
NSG mice were bred in-house at the UHN Animal Resources Centre. HGSOC OVCAR8 cells constitutively expressing firefly luciferase were mixed with FAP-high, FAP-low, 851FH-TCF21, or 851FH-GFP CAFs at a ratio of 5:1 (CAF/cancer cells). OVCAR8 cells alone were used as a control. Spheroids were formed as described above and suspended in a final volume of 100 µl IMDM media supplemented with 2% FBS. 100 µl of growth factor reduced Matrigel was added to the wells, mixed gently, and then immediately loaded into blunt-end 16-gauge syringes (StemCell Technologies; 28110). 6- to 8-wk-old female NSG mice were anaesthetized with isoflurane, and an incision was made near the fourth mammary fat pad. Spheroids were then implanted into the fat pad directly, and the incision was stapled. Mice were injected weekly with 30 mg/ml luciferin (Cedarlane) and imaged using a Xenogen IVIS Imaging System 100. Signal was quantified using IVIS Living Image software.
Lentiviral constructs
A PGK promoter-driven TCF21-P2A-GFP lentiviral vector and control PGK-GFP and PGK-mCherry vectors were designed and purchased from Vectorbuilder (Cyagen Biosciences). For lentiviral infections, cells were plated in 6-well plates at 1.0 × 105 cells per well and incubated with viral supernatants for 48 h at 37°C. Infected cells were purified by FACS on the basis of GFP or mCherry expression and expanded for further use.
TCF21 Western blot
851FH-GFP and 851FH-TCF21 CAFs were lysed in RIPA lysis buffer supplemented with EDTA-free protease inhibitor cocktail tablets (Roche) and normalized for total protein amount. 35 µg protein from each sample was resolved in a 12% SDS-PAGE gel and transferred onto Immobilon-P membranes (Millipore) using a semidry transfer method (Bio-Rad). Blots were probed overnight at 4°C using a mouse-anti-human ERK2 antibody (Santa Cruz; 1:1,000) and a rabbit-anti-human TCF21 antibody (Sigma-Aldrich; 1:250), followed by a 45-min incubation at room temperature with a goat anti-mouse IgG HRP-linked secondary antibody (Invitrogen; 1:1,000), and goat-anti-rabbit IgG HRP-linked secondary antibody (Cell Signaling; 1:2,500). Proteins were detected using enhanced chemiluminescence reagent (ThermoFisher Scientific) and autoradiograph exposure (Sigma-Aldrich).
Quantification and statistical analysis
Information about statistical details and analysis of microarray and RNA-seq data are indicated in text, figure legends, or method details. Graphs and statistical values (P values, correlation coefficients, and hazard ratios) were generated using GraphPad Prism 6.03. Error bars indicate SEM or SD for a minimum of three independent experiments.
Data availability
Microarray and RNA-seq data that support the findings of this study have been deposited at the National Center for Biotechnology Information’s Gene Expression Omnibus and are accessible through accession no. GSE126133.
Study approval
Tumor samples were obtained from 69 patients with HGSOC who underwent surgery at the UHN (Table S1 A). All patient tumor samples were collected after obtaining written informed consent according to the research protocol #06-0903, approved by the UHN Research Ethics Board, Toronto, Canada. Animal experiments were performed in accordance with national and institutional guidelines approved by the Canadian Counsel on Animal Care and approved by the UHN Animal Care Committee protocol #1542.
Online supplemental material
Fig. S1 shows HT-FC data on fibroblasts and HGSOC samples, representative FACS plots stained for CD49e, and immunofluorescent staining of other tumor types for CD49e. Fig. S2 shows the gating strategy for isolation of cell populations from primary HGSOC samples used for RNA profiling, the cluster dendrogram of the CD49e+ CAFs, and heatmaps comparing FAP-low versus normal adjacent fibroblasts profiled in the study by Leung et al. (2014). Fig. S3 shows additional images of IF staining of HGSOC tissues for pan-CK, FAP, and TCF21 to illustrate the classification of stroma and epithelia, FAP, and TCF21 quantification, as well as the quantification of FAP-high CAFs in omentum versus ovary tissue sites. Fig. S4 illustrates how FAP-high and FAP-low gene lists were generated and provides additional information on TCGA patient classification. Fig. S5 shows additional data relating to functional assays, including growth of FAP-high and FAP-low cells in vitro, determining IC50s of HGSOC cell lines in response to carboplatin, and the effects of TCF21 overexpression in FAP-high CAFs on carboplatin response of HGSOCs in co-culture. Table S1 shows clinical data and the percentage of FAP+ CAFs in primary tumor specimens used in the study, as well as recurrence data for the subset of patients analyzed in Fig. 4 E. Table S2 shows a list differentially expressed genes between FACS-isolated CD49e+ CAFs and EpCAM+CD133+ epithelial cells and the gene sets associated with these two cell populations. Table S3 shows a list of differentially expressed genes between FAP-high and FAP-low clusters of isolated CD49e+ CAFs and differentially expressed genes between FAP-low CAFs and normal stroma in the Leung et al. (2014) dataset. Table S4 shows a list of differentially expressed genes between FACS-purified FAP-high and FAP-low CAFs and the gene sets associated with these two cell populations. Table S5 shows the genes contained within the gene signature used to interrogate TCGA data.
Supplementary Material
lists clinical data and the percentage of FAP+ CAFs in primary tumor specimens.
lists differentially expressed genes between FACS-isolated CD49e+ CAFs and EpCAM+CD133+ epithelial cells.
lists differentially expressed genes between FH and FL clusters of isolated CD49e+ CAFs.
lists differentially expressed genes between FACS-purified FH and FL CAFs.
contains the gene signature used to interrogate TCGA data.
Acknowledgments
We thank the patients who donated their cancer tissues for research and the UHN Program for Biospecimen Sciences for facilitating fresh tissue acquisition. We are grateful to the UHN Animal Resources Centre staff for animal care. We also thank Catherine O’Brien and Evelyne Lima Fernandes for critical reading of the manuscript.
This work was supported by grants from the Canadian Institutes of Health Research (PJT153147), the Ontario Institute for Cancer Research (IA-016), and the Princess Margaret Cancer Foundation, thanks to the generosity of the Westaway Charitable Foundation. A. Hussain was supported by Ontario Student Opportunity Trust Funds and the University of Toronto Medical Biophysics Excellence awards.
Author contributions: A. Hussain, B.G. Neel, and L.E. Ailles designed research studies; A. Hussain, S. Poon, C. Karamboulas, N.H.B. Bui, J. Meens, J. Dmytryshyn, and J. Paterson conducted experiments and acquired data; A. Hussain, V. Voisin, G.D. Bader, and M.Q. Bernardini analyzed data; V.W. Ho, K.H. Tang, B.A. Clarke, and M.Q. Bernardini provided reagents, including clinical specimens; and A. Hussain, B.G. Neel, and L.E. Ailles wrote the manuscript.
References
- Acharya A., Baek S.T., Huang G., Eskiocak B., Goetsch S., Sung C.Y., Banfi S., Sauer M.F., Olsen G.S., Duffield J.S., et al. 2012. The bHLH transcription factor Tcf21 is required for lineage-specific EMT of cardiac fibroblast progenitors. Development. 139:2139–2149. 10.1242/dev.079970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akama T., and Chun T.H.. 2018. Transcription factor 21 (TCF21) promotes proinflammatory interleukin 6 expression and extracellular matrix remodeling in visceral adipose stem cells. J. Biol. Chem. 293:6603–6610. 10.1074/jbc.RA117.000456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braitsch C.M., Combs M.D., Quaggin S.E., and Yutzey K.E.. 2012. Pod1/Tcf21 is regulated by retinoic acid signaling and inhibits differentiation of epicardium-derived cells into smooth muscle in the developing heart. Dev. Biol. 368:345–357. 10.1016/j.ydbio.2012.06.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cancer Genome Atlas Research Network 2011. Integrated genomic analyses of ovarian carcinoma. Nature. 474:609–615. 10.1038/nature10166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter S.L., Cibulskis K., Helman E., McKenna A., Shen H., Zack T., Laird P.W., Onofrio R.C., Winckler W., Weir B.A., et al. 2012. Absolute quantification of somatic DNA alterations in human cancer. Nat. Biotechnol. 30:413–421. 10.1038/nbt.2203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa A., Kieffer Y., Scholer-Dahirel A., Pelon F., Bourachot B., Cardon M., Sirven P., Magagna I., Fuhrmann L., Bernard C., et al. 2018. Fibroblast Heterogeneity and Immunosuppressive Environment in Human Breast Cancer. Cancer Cell. 33:463–479.e10. 10.1016/j.ccell.2018.01.011 [DOI] [PubMed] [Google Scholar]
- de Jong J.M., Larsson O., Cannon B., and Nedergaard J.. 2015. A stringent validation of mouse adipose tissue identity markers. Am. J. Physiol. Endocrinol. Metab. 308:E1085–E1105. 10.1152/ajpendo.00023.2015 [DOI] [PubMed] [Google Scholar]
- Fearon D.T. 2014. The carcinoma-associated fibroblast expressing fibroblast activation protein and escape from immune surveillance. Cancer Immunol. Res. 2:187–193. 10.1158/2326-6066.CIR-14-0002 [DOI] [PubMed] [Google Scholar]
- Funato N., Ohyama K., Kuroda T., and Nakamura M.. 2003. Basic helix-loop-helix transcription factor epicardin/capsulin/Pod-1 suppresses differentiation by negative regulation of transcription. J. Biol. Chem. 278:7486–7493. 10.1074/jbc.M212248200 [DOI] [PubMed] [Google Scholar]
- Gedye C.A., Hussain A., Paterson J., Smrke A., Saini H., Sirskyj D., Pereira K., Lobo N., Stewart J., Go C., et al. 2014. Cell surface profiling using high-throughput flow cytometry: a platform for biomarker discovery and analysis of cellular heterogeneity. PLoS One. 9 e105602 10.1371/journal.pone.0105602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Givel A.M., Kieffer Y., Scholer-Dahirel A., Sirven P., Cardon M., Pelon F., Magagna I., Gentric G., Costa A., Bonneau C., et al. 2018. miR200-regulated CXCL12β promotes fibroblast heterogeneity and immunosuppression in ovarian cancers. Nat. Commun. 9:1056 10.1038/s41467-018-03348-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz J.G., Minguet S., Navarro-Lérida I., Lazcano J.J., Samaniego R., Calvo E., Tello M., Osteso-Ibáñez T., Pellinen T., Echarri A., et al. 2011. Biomechanical remodeling of the microenvironment by stromal caveolin-1 favors tumor invasion and metastasis. Cell. 146:148–163. 10.1016/j.cell.2011.05.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamanishi J., Mandai M., Ikeda T., Minami M., Kawaguchi A., Murayama T., Kanai M., Mori Y., Matsumoto S., Chikuma S., et al. 2015. Safety and Antitumor Activity of Anti-PD-1 Antibody, Nivolumab, in Patients With Platinum-Resistant Ovarian Cancer. J. Clin. Oncol. 33:4015–4022. 10.1200/JCO.2015.62.3397 [DOI] [PubMed] [Google Scholar]
- Jeong J.A., Hong S.H., Gang E.J., Ahn C., Hwang S.H., Yang I.H., Han H., and Kim H.. 2005. Differential gene expression profiling of human umbilical cord blood-derived mesenchymal stem cells by DNA microarray. Stem Cells. 23:584–593. 10.1634/stemcells.2004-0304 [DOI] [PubMed] [Google Scholar]
- Junttila M.R., and de Sauvage F.J.. 2013. Influence of tumour micro-environment heterogeneity on therapeutic response. Nature. 501:346–354. 10.1038/nature12626 [DOI] [PubMed] [Google Scholar]
- Kalluri R. 2016. The biology and function of fibroblasts in cancer. Nat. Rev. Cancer. 16:582–598. 10.1038/nrc.2016.73 [DOI] [PubMed] [Google Scholar]
- Kalluri R., and Zeisberg M.. 2006. Fibroblasts in cancer. Nat. Rev. Cancer. 6:392–401. 10.1038/nrc1877 [DOI] [PubMed] [Google Scholar]
- Kanisicak O., Khalil H., Ivey M.J., Karch J., Maliken B.D., Correll R.N., Brody M.J., J Lin S.C., Aronow B.J., Tallquist M.D., et al. 2016. Genetic lineage tracing defines myofibroblast origin and function in the injured heart. Nat. Commun. 7:12260 10.1038/ncomms12260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karnoub A.E., Dash A.B., Vo A.P., Sullivan A., Brooks M.W., Bell G.W., Richardson A.L., Polyak K., Tubo R., and Weinberg R.A.. 2007. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 449:557–563. 10.1038/nature06188 [DOI] [PubMed] [Google Scholar]
- Köbel M., Piskorz A.M., Lee S., Lui S., LePage C., Marass F., Rosenfeld N., Mes Masson A.M., and Brenton J.D.. 2016. Optimized p53 immunohistochemistry is an accurate predictor of TP53 mutation in ovarian carcinoma. J. Pathol. Clin. Res. 2:247–258. 10.1002/cjp2.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraman M., Bambrough P.J., Arnold J.N., Roberts E.W., Magiera L., Jones J.O., Gopinathan A., Tuveson D.A., and Fearon D.T.. 2010. Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science. 330:827–830. 10.1126/science.1195300 [DOI] [PubMed] [Google Scholar]
- Kuzet S.E., and Gaggioli C.. 2016. Fibroblast activation in cancer: when seed fertilizes soil. Cell Tissue Res. 365:607–619. 10.1007/s00441-016-2467-x [DOI] [PubMed] [Google Scholar]
- Ledermann J.A., Raja F.A., Fotopoulou C., Gonzalez-Martin A., Colombo N., and Sessa C.; ESMO Guidelines Working Group . 2013. Newly diagnosed and relapsed epithelial ovarian carcinoma: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 24(Suppl 6):vi24-vi32. 10.1093/annonc/mdt333 [DOI] [PubMed] [Google Scholar]
- Leung C.S., Yeung T.L., Yip K.P., Pradeep S., Balasubramanian L., Liu J., Wong K.K., Mangala L.S., Armaiz-Pena G.N., Lopez-Berestein G., et al. 2014. Calcium-dependent FAK/CREB/TNNC1 signalling mediates the effect of stromal MFAP5 on ovarian cancer metastatic potential. Nat. Commun. 5:5092 10.1038/ncomms6092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mhawech-Fauceglia P., Yan L., Sharifian M., Ren X., Liu S., Kim G., Gayther S.A., Pejovic T., and Lawrenson K.. 2015. Stromal Expression of Fibroblast Activation Protein Alpha (FAP) Predicts Platinum Resistance and Shorter Recurrence in patients with Epithelial Ovarian Cancer. Cancer Microenviron. 8:23–31. 10.1007/s12307-014-0153-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monk B.J., Minion L.E., and Coleman R.L.. 2016. Anti-angiogenic agents in ovarian cancer: past, present, and future. Ann. Oncol. 27(Suppl 1):i33–i39. 10.1093/annonc/mdw093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore K., Colombo N., Scambia G., Kim B.G., Oaknin A., Friedlander M., Lisyanskaya A., Floquet A., Leary A., Sonke G.S., et al. 2018. Maintenance Olaparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. N. Engl. J. Med. 379:2495–2505. 10.1056/NEJMoa1810858 [DOI] [PubMed] [Google Scholar]
- Ngo P., Ramalingam P., Phillips J.A., and Furuta G.T.. 2006. Collagen gel contraction assay. Methods Mol. Biol. 341:103–109. [DOI] [PubMed] [Google Scholar]
- Nurnberg S.T., Cheng K., Raiesdana A., Kundu R., Miller C.L., Kim J.B., Arora K., Carcamo-Oribe I., Xiong Y., Tellakula N., et al. 2015. Coronary Artery Disease Associated Transcription Factor TCF21 Regulates Smooth Muscle Precursor Cells That Contribute to the Fibrous Cap. PLoS Genet. 11 e1005155 10.1371/journal.pgen.1005155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öhlund D., Elyada E., and Tuveson D.. 2014. Fibroblast heterogeneity in the cancer wound. J. Exp. Med. 211:1503–1523. 10.1084/jem.20140692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Öhlund D., Handly-Santana A., Biffi G., Elyada E., Almeida A.S., Ponz-Sarvise M., Corbo V., Oni T.E., Hearn S.A., Lee E.J., et al. 2017. Distinct populations of inflammatory fibroblasts and myofibroblasts in pancreatic cancer. J. Exp. Med. 214:579–596. 10.1084/jem.20162024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orimo A., and Weinberg R.A.. 2007. Heterogeneity of stromal fibroblasts in tumors. Cancer Biol. Ther. 6:618–619. 10.4161/cbt.6.4.4255 [DOI] [PubMed] [Google Scholar]
- Ryner L., Guan Y., Firestein R., Xiao Y., Choi Y., Rabe C., Lu S., Fuentes E., Huw L.Y., Lackner M.R., et al. 2015. Upregulation of Periostin and Reactive Stroma Is Associated with Primary Chemoresistance and Predicts Clinical Outcomes in Epithelial Ovarian Cancer. Clin. Cancer Res. 21:2941–2951. 10.1158/1078-0432.CCR-14-3111 [DOI] [PubMed] [Google Scholar]
- Stewart J.M., Shaw P.A., Gedye C., Bernardini M.Q., Neel B.G., and Ailles L.E.. 2011. Phenotypic heterogeneity and instability of human ovarian tumor-initiating cells. Proc. Natl. Acad. Sci. USA. 108:6468–6473. 10.1073/pnas.1005529108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su S., Chen J., Yao H., Liu J., Yu S., Lao L., Wang M., Luo M., Xing Y., Chen F., et al. 2018. CD10+GPR77+ Cancer-Associated Fibroblasts Promote Cancer Formation and Chemoresistance by Sustaining Cancer Stemness. Cell. 172:841–856.e16. 10.1016/j.cell.2018.01.009 [DOI] [PubMed] [Google Scholar]
- Sugimoto H., Mundel T.M., Kieran M.W., and Kalluri R.. 2006. Identification of fibroblast heterogeneity in the tumor microenvironment. Cancer Biol. Ther. 5:1640–1646. 10.4161/cbt.5.12.3354 [DOI] [PubMed] [Google Scholar]
- Thibault B., Castells M., Delord J.P., and Couderc B.. 2014. Ovarian cancer microenvironment: implications for cancer dissemination and chemoresistance acquisition. Cancer Metastasis Rev. 33:17–39. 10.1007/s10555-013-9456-2 [DOI] [PubMed] [Google Scholar]
- Verhaak R.G., Tamayo P., Yang J.Y., Hubbard D., Zhang H., Creighton C.J., Fereday S., Lawrence M., Carter S.L., Mermel C.H., et al. ; Cancer Genome Atlas Research Network . 2013. Prognostically relevant gene signatures of high-grade serous ovarian carcinoma. J. Clin. Invest. 123:517–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalobos V.M., Wang Y.C., and Sikic B.I.. 2018. Reannotation and Analysis of Clinical and Chemotherapy Outcomes in the Ovarian Data Set From The Cancer Genome Atlas. JCO Clin. Cancer Inform. 2:1–16. 10.1200/CCI.17.00096 [DOI] [PubMed] [Google Scholar]
- Wagner W., Wein F., Seckinger A., Frankhauser M., Wirkner U., Krause U., Blake J., Schwager C., Eckstein V., Ansorge W., et al. 2005. Comparative characteristics of mesenchymal stem cells from human bone marrow, adipose tissue, and umbilical cord blood. Exp. Hematol. 33:1402–1416. 10.1016/j.exphem.2005.07.003 [DOI] [PubMed] [Google Scholar]
- Yang X., Lin Y., Shi Y., Li B., Liu W., Yin W., Dang Y., Chu Y., Fan J., and He R.. 2016. FAP Promotes Immunosuppression by Cancer-Associated Fibroblasts in the Tumor Microenvironment via STAT3-CCL2 Signaling. Cancer Res. 76:4124–4135. 10.1158/0008-5472.CAN-15-2973 [DOI] [PubMed] [Google Scholar]
- Yeung T.L., Leung C.S., Li F., Wong S.S., and Mok S.C.. 2016. Targeting Stromal-Cancer Cell Crosstalk Networks in Ovarian Cancer Treatment. Biomolecules. 6:3 10.3390/biom6010003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshihara K., Shahmoradgoli M., Martínez E., Vegesna R., Kim H., Torres-Garcia W., Treviño V., Shen H., Laird P.W., Levine D.A., et al. 2013. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 4:2612 10.1038/ncomms3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Jing Y., Zhang M., Zhang Z., Ma P., Peng H., Shi K., Gao W.Q., and Zhuang G.. 2015. Stroma-associated master regulators of molecular subtypes predict patient prognosis in ovarian cancer. Sci. Rep. 5:16066 10.1038/srep16066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Huang X., Hecker L., Kurundkar D., Kurundkar A., Liu H., Jin T.H., Desai L., Bernard K., and Thannickal V.J.. 2013. Inhibition of mechanosensitive signaling in myofibroblasts ameliorates experimental pulmonary fibrosis. J. Clin. Invest. 123:1096–1108. 10.1172/JCI66700 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
lists clinical data and the percentage of FAP+ CAFs in primary tumor specimens.
lists differentially expressed genes between FACS-isolated CD49e+ CAFs and EpCAM+CD133+ epithelial cells.
lists differentially expressed genes between FH and FL clusters of isolated CD49e+ CAFs.
lists differentially expressed genes between FACS-purified FH and FL CAFs.
contains the gene signature used to interrogate TCGA data.
Data Availability Statement
Microarray and RNA-seq data that support the findings of this study have been deposited at the National Center for Biotechnology Information’s Gene Expression Omnibus and are accessible through accession no. GSE126133.