Abstract
Purpose of review
Osteoarthritis is a major source of disability, pain and socioeconomic cost worldwide. The epidemiology of the disorder is multifactorial including genetic, biological and biomechanical components, some of them detectable by MRI. This review provides the most recent update on MRI biomarkers which can provide functional information of the joint structures for diagnosis, prognosis and treatment response monitoring in osteoarthritis trials.
Recent findings
Compositional or functional MRI can provide clinicians with valuable information on glycosaminoglycan content (chemical exchange saturation transfer, sodium MRI, T1ρ) and collagen organization (T2, T2*, apparent diffusion coefficient, magnetization transfer) in joint structures. Other parameters may also provide useful information, such as volumetric measurements of joint structures or advanced image data postprocessing and analysis. Automated tools seem to have a great potential to be included in these efforts providing standardization and acceleration of the image data analysis process.
Summary
Functional or compositional MRI has great potential to provide noninvasive imaging biomarkers for osteoarthritis. Osteoarthritis as a whole joint condition needs to be diagnosed in early stages to facilitate selection of patients into clinical trials and/or to measure treatment effectiveness. Advanced evaluation including machine learning, neural networks and multidimensional data analysis allow for wall-to-wall understanding of parameter interactions and their role in clinical evaluation of osteoarthritis.
Keywords: diagnosis, functional, MRI, osteoarthritis, prognosis
INTRODUCTION
Osteoarthritis is a debilitating disease that causes pain and affects mobility; degeneration of joint structures is a major manifestation of the disease process. Over more than a decade a number of MRI-based biomarkers for collagen and proteoglycan content in articular cartilage affected by osteoarthritis were developed. These compositional (often referred to as functional or biochemical) markers are related to collagen content and organization [1,2], proteoglycan content [3–5] and mechanical properties [6,7] and they can also predict focal cartilage and meniscus disruption [8]. T2 relaxation constant mapping is a well investigated technique reflecting the combination of water content and collagen matrix organization [1,9,10]. Proteoglycans with covalently attached glycosaminoglycan (GAGs) can be potentially assessed with T1ρ (relaxation in rotating frame) [4], glycosaminoglycan chemical exchange saturation transfer (gagCEST) [3] and sodium MRI [11]. All these GAG-specific contrast-free techniques are preferred over conventional delayed Gadolinium Enhanced MRI of Cartilage (dGEMRIC) [12] as the Federal Food and Drug Administration recently warned about gadolinium-based contrast agents which can be potentially harmful for patients due to gadolinium deposition in central nervous system structures [13]. Some joint structures such as ligament, tendons and subchondral bone tendons are characterized by ultrashort relaxation rates resulting in loss of magnetic resonance (MR) signal in conventional MRI images. Ultra-short echo time techniques are able to either acquire signal directly from these structures [14] of even calculate T2* and T1ρ relaxation to describe the collagen fibres organization [15,16]. Diffusion weighted imaging and diffusion tensor imaging also provide an important information about cartilage status by calculating apparent diffusion coefficient [17] and diffusion tensor map [18], respectively. Modern computer science approaches are pushing the field further by implementing machine learning algorithms for image postprocessing and multidimensional data analysis for searching the relations between various biomarkers. Osteoarthritis as a complex multifactorial joint disease which is characterized by structural alteration including GAG loss and collagen remodelling, but is also accompanied by volumetric changes. Here, the deep learning approach helps to automatically segment joint structures such as cartilage and meniscus which is a typically extremely demanding task in terms of man-power [19■]. All these modern methods have a common goal: to establish quantitative MRI biomarkers as reproducible and reliable indicators of joint structures suffering from osteoarthritis. This review summarizes the most recent findings and improvements in the field of MRI-based biomarkers for osteoarthritis assessment and their usage for diagnosis, prognosis and treatment monitoring.
T2 MAPPING
The transversal relaxation constant, T2, in cartilage reflects the interaction of water molecules and the extracellular matrix on a molecular level. Anisotropy of tissue results in different T2 values from deep to superficial cartilage layers depending on the collagen fibre orientation. Previously, T2 was validated as a marker for osteoarthritic alterations in cartilage [20], as a physiological indicator of loading induces changes in cartilage [21] and also as a biomarker for monitoring the patients after surgical treatment of cartilage lesions [22]. Advances in MR hardware, mostly in increasing main magnetic field strength, accompanied with the challenges such specific-absorption limitations and special radiofrequency coils need, initiated using novel MR sequences for T2 mapping, such as triple-echo and double-echo steady-state-based approaches [22,23]. T2 can be also exploited as a predictive marker for focal lesion development which may result in osteoarthritis. Kretzschmar et al. [24] investigated T2 mapping for early osteoarthritis changes in cartilage of 57 patients and they showed that both diffuse and focal changes of cartilage composition precede the development of cartilage lesions. Evaluation of T2 in cartilage can be further enhanced and standardized using human-independent machine learning techniques. Pedoia et al. demonstrated the machine learning capabilities using whole Osteoarthritis Initiative (OAI) Dataset (>4000 individuals). Their results showed that feature learning from T2 maps has a great potential in uncovering information that can prospectively help to better diagnose osteoarthritis [25■■]. This concept can be extended and further improved by combining other features with quantitative MR, such as biomechanical parameters. In another study of Pedoia et al. [26■■], it was shown that the integrated parameter set of demographic, clinical information, gait kinematics and kinetics, cartilage compositional T1ρ and T2 provides an information about their cross-interactions and may serve as a robust early predictor of cartilage lesion progression. A combined network of morphological MRI, biomechanics and quantitative MRI is depicted in Fig. 1.
FIGURE 1.
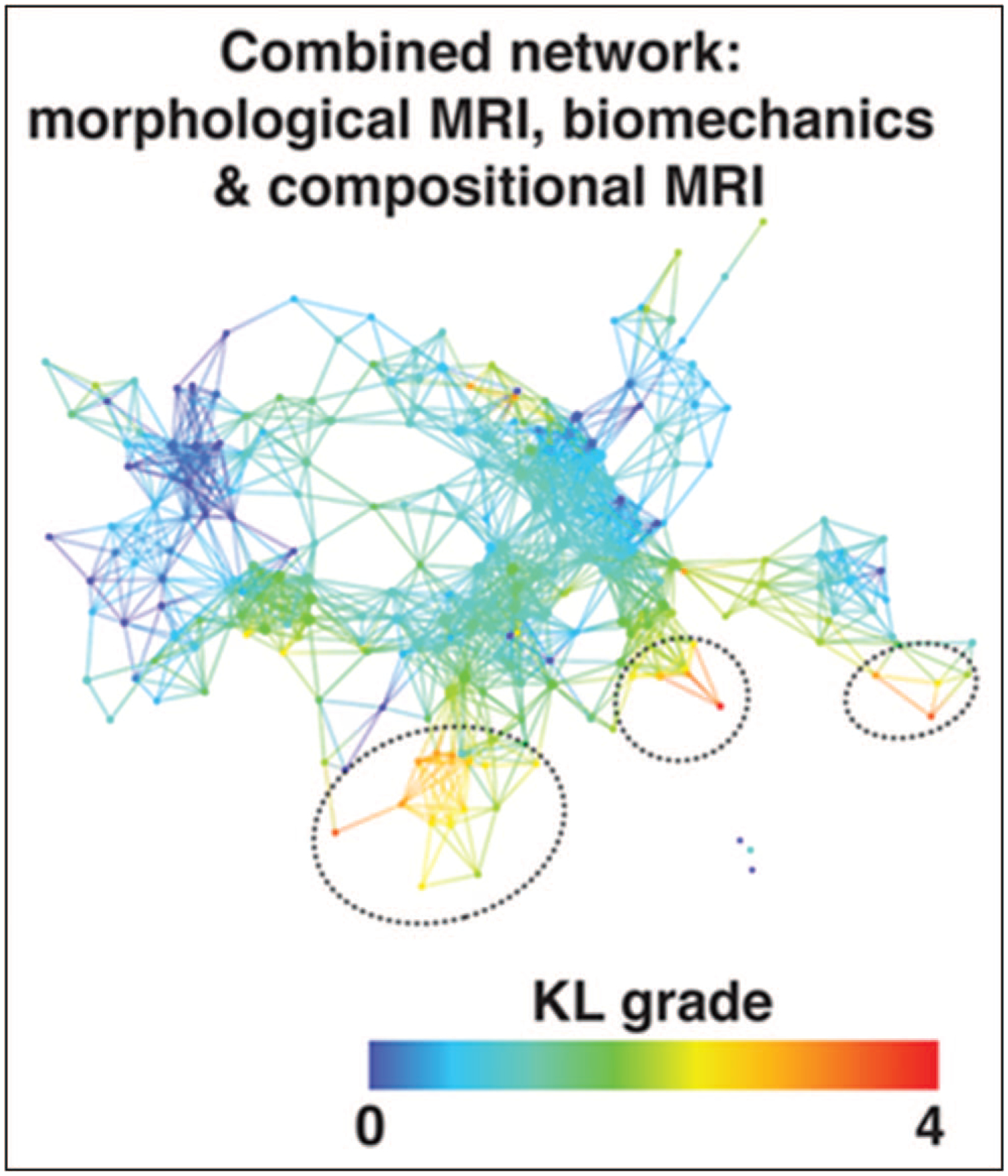
Combined network of morphological MRI, biomechanics and compositional MRI. The network shows a gradient with severe patients appearing in the lower right (marked with dashed black circles) and less severe in the upper left based on Kellgren–Lawrence grading. Reproduced with permission [26■■].
T1ρ MAPPING
T1ρ is sensitive to low-frequency interactions of macromolecular protons and bulk water as well as early biochemical deterioration of cellular matrix [16]. T1ρ imaging of connective tissues affected by osteoarthritis may provide a more systematic assessment of this disease. In the previous studies the correlation was found between both proteoglycan-specific methods like gagCEST [27] and collagen-specific methods like T2 [28]. Atkinson et al. [29] demonstrated using a complex meta-analysis that increased T1ρ and T2 values in cartilage may serve as a risk factors for osteoarthritis. Higher signal-to-noise ratio increases the accuracy of T1ρ calculation, using higher field strengths, for instance; Wyatt et al. [30] found T1ρ at 3 and 7 T significantly higher in the lateral femoral condyle and patella in patients with osteoarthritis; however, more regions were significant at 7 T compared with 3 T. The recent meta-analysis demonstrates that T1ρ provides more discrimination than T2 for osteoarthritis [31]. Advances in MRI techniques and image postprocessing allow for detecting more than a single component of T1ρ in connective tissues which may help to better understand the osteoarthritis pathogenesis in the future [32,33■].
GLYCOSAMINOGLYCAN CHEMICAL EXCHANGE SATURATION TRANSFER
Glycosaminoglycan-specific chemical exchange saturation transfer (gagCEST) is a recently developed technique, which provides information about the amount of GAG in articular cartilage. It is based on selective saturation of the exchangeable hydroxyl protons of GAG, which exchange with water protons [3]. The feasibility of using gagCEST for GAG content determination has been previously validated by number of studies [27,34,35]. Further development aims towards faster clinical MRI acquisition methods and more quantitative acquisition and analysis routines [36]. Stability and feasibility of gagCEST imaging to detection of early cartilage damage was investigated by Brinkhof et al. [37■■]; they developed a 7-min gagCEST protocol and used it to calculate intraclass correlation coefficient (ICC) to show high reproducibility in the medial femoral condyle (ICC = 0.87) and in the lateral femoral condyle (ICC = 0.97) and they also found statistically significant change in GAG between damaged and healthy cartilage. In terms of technical development, Windschuh et al. [38] investigated the effects of a frequency drift on gagCEST contrast in the human knee at 7 T and suggested a retrospective correction method that can eliminate errors. This technique is illustrated in Fig. 2. The water T2, and thus the frequency width of the water peak, may impact the extent of the so-called spillover effect in CEST. Peterson et al. [39■■] demonstrated that T2 substantially influences gagCEST which is obvious especially in femoral cartilage with a wide range of T2 and showed this effect to be more pronounced at 3 T compared with 7 T as the gagCEST effect is weaker at lower field strength.
FIGURE 2.
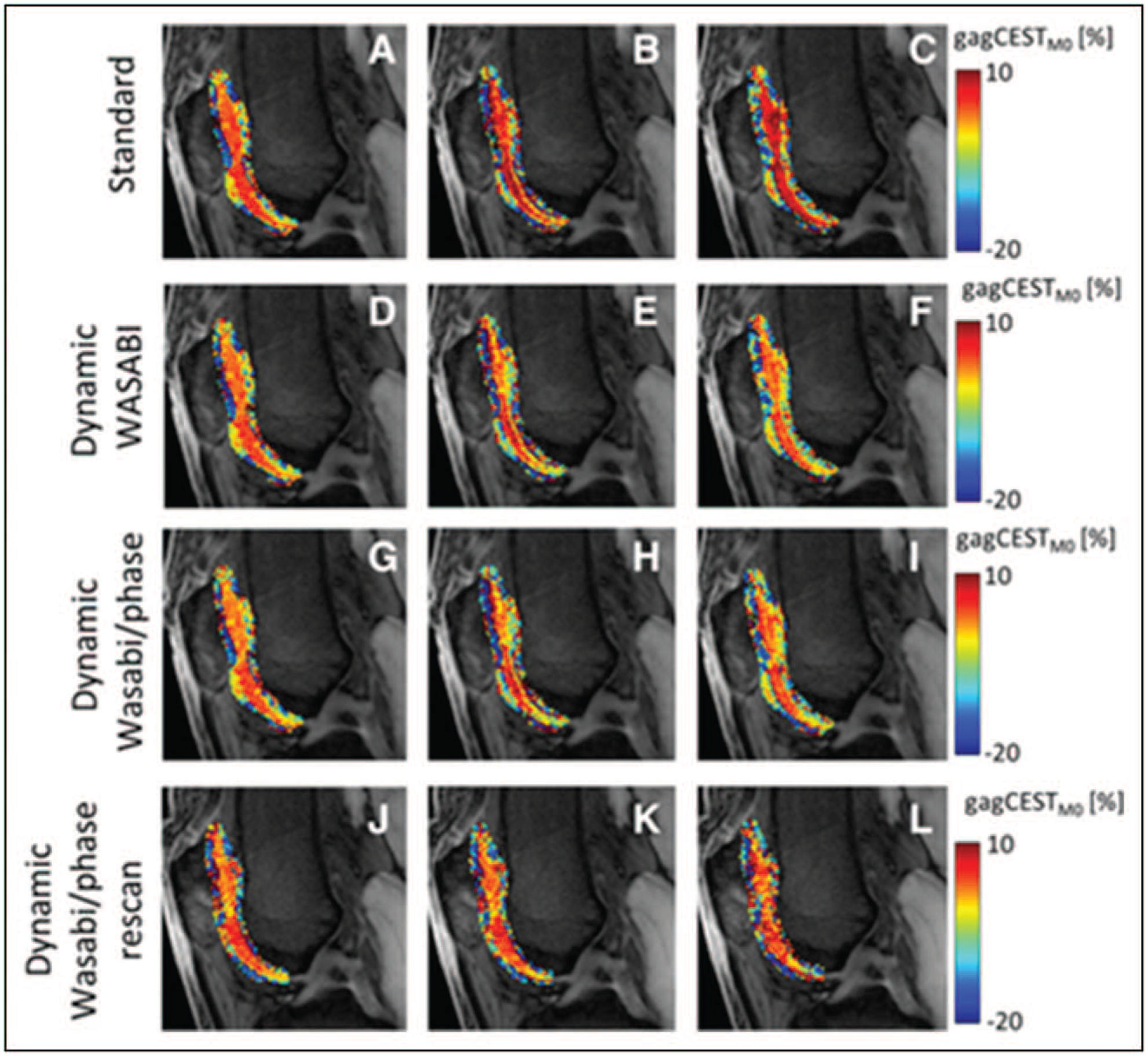
Cropped M0 images superimposed with glycosaminoglycan chemical exchange saturation transferM0 contrast in patellar and femoral cartilage from identical measurements from a 32-year-old healthy volunteer, acquired at t = 5 min (a, d, g, j), t = 17 min (b, e, h, k), and t = 29 min (c, f, i, l). The glycosaminoglycan chemical exchange saturation transferM0 contrast of data using the standard B0 correction (a, b, c) shows an increase over time, whereas the dynamic WASABI (simultaneous mapping of water shift and B1) correction (d, e, f) and the dynamic WASABI/phase correction method (g, h, i) provide a stable contrast. The last row represents data from the same individual acquired 1 month later (j, k, l). Reproduced with permission [38].
SODIUM MRI AND PET/MAGNETIC RESONANCE
Sodium MRI is another technique which has potential to noninvasively determine GAG content in human articular cartilage; however, it requires special radiofrequency hardware (dedicated coils, ultra-high field scanners) and complicated postprocessing [40]. Despite that, the number of studies was performed demonstrating a high sensitivity and reproducibility of sodium MRI in detecting GAG content levels in cartilage either in osteoarthritis [5,41] or in cartilage repair [42,43]. In a recent study, Madelin et al. [44■■] used two sodium MR sequences (radial three-dimensional and fluid suppression by adiabatic inversion recovery) to investigate whether they can detect the sodium volume changes in patients with osteoarthritis after 16 months. A significant decrease of measured apparent sodium concentration in cartilage using IR in both femoral condyles and patella proved the sensitivity of sodium MRI to detect subtle changes of GAG concentration. Fluid suppression in sodium MRI was also investigated by Xia et al. [45] they developed quadrupolar jump-and-return pulse sequence to suppress the fluid signal from the artery and enhance the contrast of knee cartilage in vivo. Another modern approach towards the advanced evaluation of osteoarthritic alterations in joints is the hybrid PET/MRI scanning. Wandler et al. [46] investigated the potential of 18F-fluoro-deoxyglucose uptake pattern within the shoulder cartilage and they found out it can be is associated with signs and symptoms of osteoarthritis or bursitis. Kogan et al. [47] used PET/MR to assess the metabolic activity in osteoarthritis and concluded that PET/MR may detect metabolic abnormalities in subchondral bone, which appear normal on MRI. In another study by Savic et al. [48], significant cartilage and bone interactions were demonstrated in osteoarthritis of the knee joint using simultaneous [18F]-sodium fluoride PET/MR.
CARTILAGE VOLUMETRIC MEASUREMENTS
Cartilage tissue loss is a hallmark of osteoarthritis. Thinning and cartilage/bone deformation can be manually assessed from high-resolved morphological MR images, however, this approach requires enormous workload. There are many techniques for automated cartilage segmentation which were developed in recent years, including intensity-based [49] and edge-based [50] approaches, deformable models [51], clustering [52] and atlas/graph-based methods [53]. Machine learning has vast ranging applications including automated segmentation of cartilage allowing for reliable segmentation using convolutional neural networks (CNN) [19■,54–56]. In addition to knee cartilage, other joints have been segmented using automated techniques such as hip cartilage [57], shoulder [58], bones [59] or even wrist cartilage [60]. The logical translation of automated cartilage segmentation is the use for quantitative MR assessment which is also often related to tedious manual segmentation. Hesper et al. [61■■] found automated hip cartilage segmentation for dGEMRIC assessment to be reliable and reader-independent approach for assessment of biochemical cartilage status. Norman et al. used a CNN-based approach for automated T1ρ assessment demonstrating the ability to quantify, in a longitudinally repeatable way, relaxometry and morphology in a single session [55].
ADVANCES IN IMAGE POSTPROCESSING
Machine learning is more and more employed in extracting the predictive or diagnostic value from the existing multidimensional patient data. Up-to-now, there were many attempts to developed patient-specific prediction models for knee osteoarthritis by analyzing multivariable ‘big data’ using either conventional statistical approach [62] or machine learning [63]. To classify early stages of osteoarthritis is also of interest for machine learning; Ashinsky et al. [64] used such an approach to predict early symptomatic osteoarthritis by combining T2 mapping and clinical outcome variables and they found the classification accuracy to be 0.75 ± 0.9%. Lazzarini et al. [65] proposed five highly predictive small models that can be possibly adopted for an early prediction of knee osteoarthritis where all the models showed high performance (area under curve >0.7). Sodium imaging was also tested with various machine learning to classify osteoarthritis patients with accuracy ranging from 63 to 78% [66]. Du et al. [67■■] suggested a novel approach to predict knee osteoarthritis combining four machine learning methods and the clinical data. In general, selecting the best variables to incorporate into prediction models can be a complex task owing to the great number of variables to choose from [68].
Quantitative MR analysis can be extended using texture analysis techniques, typically feature extraction from grey-scale co-occurrence matrix (GLCM). Texture analysis provides a reliable tool for assessing knee osteoarthritis with more sensitive detection of cartilage degeneration compared with the simple mean T2 or T1ρ value in an identical region of interest. As cartilage is a complex-shaped complex organ, original texture analysis suggested by Haralick [69] needs to be modified. Peuna et al. [70■■] developed a cartilage-specific GLCM analysis algorithm taking into account-specific cartilage features such as curvature and angulation. Joseph et al. [71] studied osteoarthritis patients using conventional T2 maps and texture analysis; they found out individuals at risk for osteoarthritis have both higher and more heterogeneous T2 values than controls and that individuals with cartilage abnormalities have elevated cartilage T2 parameters compared with individuals without abnormalities (Fig. 3) [71]. Heilmeier et al. [72■■] analyzed more than 300 individuals from the OAI and according to their findings combined clinical score, quantitative MR and texture can help predict the patient’s individual risk for an incident total knee replacement 4–7 years later.
FIGURE 3.
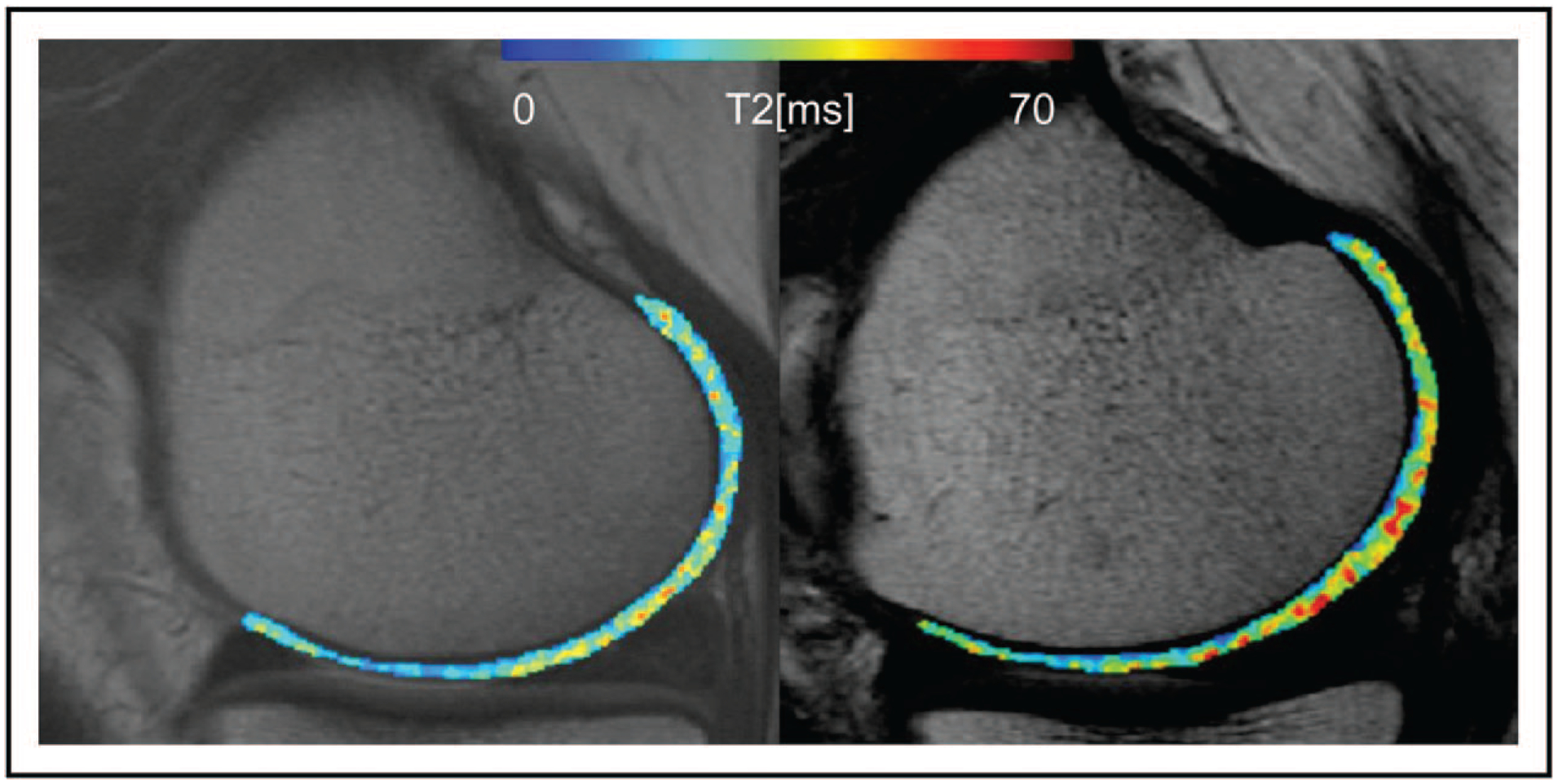
Representative T2 maps from an individual from the control cohort (left) and an individual from the incidence cohort (right). Cartilage T2 maps are median-filtered with a 3 × 3 kernel for visualization. Both individuals have no cartilage abnormalities and no pain; however, the individual from the incidence cohort has elevated mean T2 (39.12 versus 33.39 ms), elevated grey level co-occurrence matrix variance (311.63 versus 190.50), elevated grey-scale co-occurrence matrix contrast (466.16 versus 266.82), and elevated grey-scale co-occurrence matrix entropy (7.17 versus 6.80). Reproduced with permission [71].
CONCLUSION
Functional compositional MR techniques provide an important extension to clinical, morphological and biochemical assessment of osteoarthritis diagnosis and progression staging. There are still remaining issues to be addressed such as lack of standardization and tedious image postprocessing; however, the clinical value of quantitative magnetic resonance imaging is undeniable already in the presence. Future challenges include advancements in imaging techniques, searching relationships between various markers by machine learning and multidimensional data analysis, and parameter validation using large freely available database such as the OAI.
KEY POINTS.
Advances in image-processing, image reconstruction and data analysis yield better understanding of MRI-based markers for joint structure status.
Quantitative MRI provides noninvasive markers for osteoarthritis diagnosis and prognosis.
Recent developments allow for faster magnetic resonance scanning, reliable cartilage and meniscus segmentation and complex multidimensional data analysis.
Financial support and sponsorship
The current work was supported by Austrian Science Fund (FWF) KLI541-B30, Slovak Grant Agency APVV-15-0029 and Fulbright Commission Grant 18-22-04.
Footnotes
Conflicts of interest
There are no conflicts of interest.
REFERENCES AND RECOMMENDED READING
Papers of particular interest, published within the annual period of review, have been highlighted as:
■ of special interest
■■ of outstanding interest
- 1.Mosher TJ, Dardzinski BJ. Cartilage MRI T2 relaxation time mapping: overview and applications. Semin Musculoskelet Radiol 2004; 8:355–368. [DOI] [PubMed] [Google Scholar]
- 2.Liess C, Lusse S, Karger N, et al. Detection of changes in cartilage water content using MRI T2-mapping in vivo. Osteoarthritis Cartilage 2002; 10:907–913. [DOI] [PubMed] [Google Scholar]
- 3.Ling W, Regatte RR, Navon G, et al. Assessment of glycosaminoglycan concentration in vivo by chemical exchange-dependent saturation transfer (gagCEST). Proc Natl Acad Sci U S A 2008; 105:2266–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Duvvuri U, Reddy R, Patel SD, et al. T1rho-relaxation in articular cartilage: effects of enzymatic degradation. Magn Reson Med 1997; 38:863–867. [DOI] [PubMed] [Google Scholar]
- 5.Wheaton AJ, Borthakur A, Shapiro EM, et al. Proteoglycan loss in human knee cartilage: quantitation with sodium MR imaging – feasibility study. Radiology 2004; 231:900–905. [DOI] [PubMed] [Google Scholar]
- 6.Juras V, Bittsansky M, Majdisova Z, et al. In vitro determination of biomechanical properties of human articular cartilage in osteoarthritis using multipara-metric MRI. J Magn Reson 2009; 197:40–47. [DOI] [PubMed] [Google Scholar]
- 7.Nissi MJ, Toyras J, Laasanen MS, et al. Proteoglycan and collagen sensitive MRI evaluation of normal and degenerated articular cartilage. J Orthop Res 2004; 22:557–564. [DOI] [PubMed] [Google Scholar]
- 8.Prasad AP, Nardo L, Schooler J, et al. T(1)rho and T(2) relaxation times predict progression of knee osteoarthritis. Osteoarthritis Cartilage 2013; 21:69–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Welsch GH, Mamisch TC, Hughes T, et al. In vivo biochemical 7.0 Tesla magnetic resonance: preliminary results of dGEMRIC, zonal T2, and T2* mapping of articular cartilage. Invest Radiol 2008; 43:619–626. [DOI] [PubMed] [Google Scholar]
- 10.Nieminen MT, Rieppo J, Toyras J, et al. T2 relaxation reveals spatial collagen architecture in articular cartilage: a comparative quantitative MRI and polarized light microscopic study. Magn Reson Med 2001; 46:487–493. [DOI] [PubMed] [Google Scholar]
- 11.Reddy R, Insko EK, Noyszewski EA, et al. Sodium MRI of human articular cartilage in vivo. Magn Reson Med 1998; 39:697–701. [DOI] [PubMed] [Google Scholar]
- 12.Burstein D, Velyvis J, Scott KT, et al. Protocol issues for delayed Gd(DTPA)(2-)-enhanced MRI (dGEMRIC) for clinical evaluation of articular cartilage. Magn Reson Med 2001; 45:36–41. [DOI] [PubMed] [Google Scholar]
- 13.Gulani V, Calamante F, Shellock FG, et al. Gadolinium deposition in the brain: summary of evidence and recommendations. Lancet Neurol 2017; 16:564–570. [DOI] [PubMed] [Google Scholar]
- 14.Bae WC, Du J, Bydder GM, et al. Conventional and ultrashort time-to-echo magnetic resonance imaging of articular cartilage, meniscus, and intervertebral disk. Top Magn Reson Imaging 2010; 21:275–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Juras V, Zbyn S, Pressl C, et al. Regional variations of T(2)* in healthy and pathologic achilles tendon in vivo at 7 Tesla: preliminary results. Magn Reson Med 2012; 68:1607–1613. [DOI] [PubMed] [Google Scholar]
- 16.Du J, Carl M, Diaz E, et al. Ultrashort TE T1rho (UTE T1rho) imaging of the Achilles tendon and meniscus. Magn Reson Med 2010; 64:834–842. [DOI] [PubMed] [Google Scholar]
- 17.Mlynarik V, Sulzbacher I, Bittsansky M, et al. Investigation of apparent diffusion constant as an indicator of early degenerative disease in articular cartilage. J Magn Reson Imaging 2003; 17:440–444. [DOI] [PubMed] [Google Scholar]
- 18.Raya JG, Horng A, Dietrich O, et al. Articular cartilage: in vivo diffusion-tensor imaging. Radiology 2012; 262:550–559. [DOI] [PubMed] [Google Scholar]
- 19.■.Ambellan F, Tack A, Ehlke M, et al. Automated segmentation of knee bone and cartilage combining statistical shape knowledge and convolutional neural networks: data from the osteoarthritis initiative. Med Image Anal 2019; 52:109–118. [DOI] [PubMed] [Google Scholar]; Current status of automated cartilage segmentation.
- 20.Li X, Benjamin Ma C, Link TM, et al. In vivo T(1rho) and T(2) mapping of articular cartilage in osteoarthritis of the knee using 3 T MRI. Osteoarthritis Cartilage 2007; 15:789–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishii T, Kuroda K, Matsuoka Y, et al. Change in knee cartilage T2 in response to mechanical loading. J Magn Reson Imaging 2008; 28:175–180. [DOI] [PubMed] [Google Scholar]
- 22.Juras V, Schreiner M, Laurent D, et al. The comparison of the performance of 3T and 7T T2 mapping for untreated low-grade cartilage lesions. Magn Reson Imaging 2019; 55:86–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaudhari AS, Black MS, Eijgenraam S, et al. Five-minute knee MRI for simultaneous morphometry and T2 relaxometry of cartilage and meniscus and for semiquantitative radiological assessment using double-echo in steady-state at 3T. J Magn Reson Imaging 2018; 47:1328–1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kretzschmar M, Nevitt MC, Schwaiger BJ, et al. Spatial distribution and temporal progression of T2 relaxation time values in knee cartilage prior to the onset of cartilage lesions – data from the osteoarthritis initiative (OAI). Osteoarthritis Cartilage 2019; 27:737–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.■■.Pedoia V, Lee J, Norman B, et al. Diagnosing osteoarthritis from T2 maps using deep learning: an analysis of the entire osteoarthritis initiative baseline cohort. Osteoarthritis Cartilage 2019; 27:1002–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]; An interesting study demonstrating that feature learning from T2 maps has potential in uncovering information that can potentially better diagnose osteoarthritis than simple averages or linear patterns decomposition.
- 26.■■.Pedoia V, Haefeli J, Morioka K, et al. MRI and biomechanics multidimensional data analysis reveals R2-R1rho as an early predictor of cartilage lesion progression in knee osteoarthritis. J Magn Reson Imaging 2018; 47:78–90. [DOI] [PMC free article] [PubMed] [Google Scholar]; An interesting study where the complex multitissue biochemical and biomechanical interactions were investigated showing a potential of R2-R1r to serve as a maging biomarker for early osteoarthritis.
- 27.Kogan F, Hargreaves BA, Gold GE. Volumetric multislice gagCEST imaging of articular cartilage: optimization and comparison with T1rho. Magn Reson Med 2017; 77:1134–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keenan KE, Besier TF, Pauly JM, et al. Prediction of glycosaminoglycan content in human cartilage by age, T1rho and T2 MRI. Osteoarthritis Cartilage 2011; 19:171–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Atkinson HF, Birmingham TB, Moyer R, et al. T1rho and T2 relaxation of knee articular cartilage in patients with and at risk for knee osteoarthritis: a systematic review and meta-analysis. Osteoarthritis Cartilage Suppl 2017; 25:S236–S237. [Google Scholar]
- 30.Wyatt C, Guha A, Venkatachari A, et al. Improved differentiation between knees with cartilage lesions and controls using 7T relaxation time mapping. J Orthop Translat 2015; 3:197–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.MacKay JW, Low SBL, Smith TO, et al. Systematic review and meta-analysis of the reliability and discriminative validity of cartilage compositional MRI in knee osteoarthritis. Osteoarthritis Cartilage 2018; 26:1140–1152. [DOI] [PubMed] [Google Scholar]
- 32.Sharafi A, Baboli R, Chang G, et al. 3D-T1rho prepared zero echo time-based PETRA sequence for in vivo biexponential relaxation mapping of semisolid short-T2 tissues at 3 T. J Magn Reson Imaging 2019; 50:1207–1218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.■.Baboli R, Sharafi A, Chang G, et al. Biexponential T1rho relaxation mapping of human knee menisci. J Magn Reson Imaging 2019; 50:824–835. [DOI] [PMC free article] [PubMed] [Google Scholar]; The first time when an appearance of bi-component T1rho behavior was described.
- 34.Krishnamoorthy G, Nanga RPR, Bagga P, et al. High quality three-dimensional gagCEST imaging of in vivo human knee cartilage at 7 Tesla. Magn Reson Med 2017; 77:1866–1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krusche-Mandl I, Schmitt B, Zak L, et al. Long-term results 8 years after autologous osteochondral transplantation: 7 T gagCEST and sodium magnetic resonance imaging with morphological and clinical correlation. Osteoarthritis Cartilage 2012; 20:357–363. [DOI] [PubMed] [Google Scholar]
- 36.Jones KM, Pollard AC, Pagel MD. Clinical applications of chemical exchange saturation transfer (CEST) MRI. J Magn Reson Imaging 2018; 47:11–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.■■.Brinkhof S, Nizak R, Khlebnikov V, et al. Detection of early cartilage damage: feasibility and potential of gagCEST imaging at 7T. Eur Radiol 2018; 28:2874–2881. [DOI] [PMC free article] [PubMed] [Google Scholar]; An application of glycosaminoglycan chemical exchange saturation transfer (gagCEST) in deteecting an early cartilage impairment.
- 38.Windschuh J, Zaiss M, Ehses P, et al. Assessment of frequency drift on CEST MRI and dynamic correction: application to gagCEST at 7 T. Magn Reson Med 2019; 81:573–582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.■■.Peterson P, Olsson E, Svensson J. T2 relaxation time bias in gagCEST at 3T and 7T: comparison of saturation schemes. Magn Reson Med 2019; 81:1044–1051. [DOI] [PubMed] [Google Scholar]; An important analysis of T2 impact on gagCEST.
- 40.Guermazi A, Alizai H, Crema MD, et al. Compositional MRI techniques for evaluation of cartilage degeneration in osteoarthritis. Osteoarthritis Cartilage 2015; 23:1639–1653. [DOI] [PubMed] [Google Scholar]
- 41.Borthakur A, Shapiro EM, Beers J, et al. Sensitivity of MRI to proteoglycan depletion in cartilage: comparison of sodium and proton MRI. Osteoarthritis Cartilage 2000; 8:288–293. [DOI] [PubMed] [Google Scholar]
- 42.Trattnig S, Welsch GH, Juras V, et al. 23Na MR imaging at 7 T after knee matrix-associated autologous chondrocyte transplantation preliminary results. Radiology 2010; 257:175–184. [DOI] [PubMed] [Google Scholar]
- 43.Zbyn S, Stelzeneder D, Welsch GH, et al. Evaluation of native hyaline cartilage and repair tissue after two cartilage repair surgery techniques with 23Na MR imaging at 7T: initial experience. Osteoarthritis Cartilage 2012; 20:837–845. [DOI] [PubMed] [Google Scholar]
- 44.■■.Madelin G, Xia D, Brown R, et al. Longitudinal study of sodium MRI of articular cartilage in patients with knee osteoarthritis: initial experience with 16-month follow-up. Eur Radiol 2018; 28:133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]; A clinical application of sodium magnetic resonance (MR) in patients with osteoarthritis.
- 45.Xia D, Lee JS, Regatte RR. Quadrupolar jump-and-return pulse sequence for fluid-suppressed sodium MRI of the knee joint at 7T. Magn Reson Med 2018; 80:641–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wandler E, Kramer EL, Sherman O, et al. Diffuse FDG shoulder uptake on PET is associated with clinical findings of osteoarthritis. AJR Am J Roentgenol 2005; 185:797–803. [DOI] [PubMed] [Google Scholar]
- 47.Kogan F, Fan AP, McWalter EJ, et al. PET/MRI of metabolic activity in osteoarthritis: a feasibility study. J Magn Reson Imaging 2017; 45:1736–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Savic D, Pedoia V, Seo Y, et al. Imaging bone-cartilage interactions in osteoarthritis using [(18)F]-NaF PET-MRI. Mol Imaging 2016; 15:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cashman PM, Kitney RI, Gariba MA, et al. Automated techniques for visualization and mapping of articular cartilage in MR images of the osteoarthritic knee: a base technique for the assessment of microdamage and submicro damage. IEEE Trans Nanobiosci 2002; 1:42–51. [DOI] [PubMed] [Google Scholar]
- 50.Kshirsagar AA, Watson PJ, Tyler JA, et al. Measurement of localized cartilage volume and thickness of human knee joints by computer analysis of three-dimensional magnetic resonance images. Invest Radiol 1998; 33: 289–299. [DOI] [PubMed] [Google Scholar]
- 51.Tang J, Millington S, Acton ST, et al. Surface extraction and thickness measurement of the articular cartilage from MR images using directional gradient vector flow snakes. IEEE Trans Biomed Eng 2006; 53:896–907. [DOI] [PubMed] [Google Scholar]
- 52.Folkesson J, Dam EB, Olsen OF, et al. Segmenting articular cartilage automatically using a voxel classification approach. IEEE Trans Med Imaging 2007; 26:106–115. [DOI] [PubMed] [Google Scholar]
- 53.Shim H, Chang S, Tao C, et al. Knee cartilage: efficient and reproducible segmentation on high-spatial-resolution MR images with the semiautomated graph-cut algorithm method. Radiology 2009; 251:548–556. [DOI] [PubMed] [Google Scholar]
- 54.Liu F, Zhou Z, Samsonov A, et al. Deep learning approach for evaluating knee MR images: achieving high diagnostic performance for cartilage lesion detection. Radiology 2018; 289:160–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Norman B, Pedoia V, Majumdar S. Use of 2D U-net convolutional neural networks for automated cartilage and meniscus segmentation of knee MR imaging data to determine relaxometry and morphometry. Radiology 2018; 288:177–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou X, Ju MJ, Huang L, et al. Slope-based segmentation of articular cartilage using polarization-sensitive optical coherence tomography phase retardation image. J Biomed Opt 2019; 24:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Fernquest S, Park D, Marcan M, et al. Segmentation of hip cartilage in compositional magnetic resonance imaging: a fast, accurate, reproducible, and clinically viable semi-automated methodology. J Orthop Res 2018; 36:2280–2287. [DOI] [PubMed] [Google Scholar]
- 58.Ran LYCWHGWRJ Automatic segmentation of shoulder joint in MRI using patch-based and fully convolutional networks In 25th IEEE International Conference on Image Processing (ICIP). 2018. Athens, Greece: IEEE. [Google Scholar]
- 59.Deniz CM, Xiang S, Hallyburton RS, et al. Segmentation of the proximal femur from mr images using deep convolutional neural networks. Sci Rep 2018; 8:16485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Foster B, Joshi AA, Borgese M, et al. WRIST: a WRist Image Segmentation Toolkit for carpal bone delineation from MRI. Comput Med Imaging Graph 2018; 63:31–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.■■.Hesper T, Bittersohl B, Schleich C, et al. Automatic cartilage segmentation for delayed gadolinium-enhanced magnetic resonance imaging of hip joint cartilage: a feasibility study. Cartilage 2018; doi: 10.1177/1947603518783481. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]; An application of automated cartilage segmentation and quantitative MR.
- 62.Zhang W, McWilliams DF, Ingham SL, et al. Nottingham knee osteoarthritis risk prediction models. Ann Rheum Dis 2011; 70:1599–1604. [DOI] [PubMed] [Google Scholar]
- 63.Yoo TK, Kim DW, Choi SB, et al. Simple scoring system and artificial neural network for knee osteoarthritis risk prediction: a cross-sectional study. PLoS One 2016; 11:e0148724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ashinsky BG, Bouhrara M, Coletta CE, et al. Predicting early symptomatic osteoarthritis in the human knee using machine learning classification of magnetic resonance images from the osteoarthritis initiative. J Orthop Res 2017; 35:2243–2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lazzarini N, Runhaar J, Bay-Jensen AC, et al. A machine learning approach for the identification of new biomarkers for knee osteoarthritis development in overweight and obese women. Osteoarthritis Cartilage 2017; 25:2014–2021. [DOI] [PubMed] [Google Scholar]
- 66.Madelin G, Poidevin F, Makrymallis A, et al. Classification of sodium MRI data of cartilage using machine learning. Magn Reson Med 2015; 74:1435–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.■■.Du YD, Almajalid R, Shan J, et al. A novel method to predict knee osteoarthritis progression on MRI using machine learning methods. IEEE Trans Nanobiosci 2018; 17:228–236. [DOI] [PubMed] [Google Scholar]; An interesting application of machine learnig to predict osteaorthritis progression.
- 68.Jamshidi A, Pelletier JP, Martel-Pelletier J. Machine-learning-based patient-specific prediction models for knee osteoarthritis. Nat Rev Rheumatol 2019; 15:49–60. [DOI] [PubMed] [Google Scholar]
- 69.Haralick RM. Statistical and structural approaches to texture. Proc IEEE 1979; 67:786–804. [Google Scholar]
- 70.■■.Peuna A, Hekkala J, Haapea M, et al. Variable angle gray level co-occurrence matrix analysis of T2 relaxation time maps reveals degenerative changes of cartilage in knee osteoarthritis: Oulu Knee Osteoarthritis Study. J Magn Reson Imaging 2018; 47:1316–1327. [DOI] [PubMed] [Google Scholar]; Advanced texture analysis of cartilage in osteaorthritis.
- 71.Joseph GB, Baum T, Carballido-Gamio J, et al. Texture analysis of cartilage T2 maps: individuals with risk factors for OA have higher and more heterogeneous knee cartilage MR T2 compared to normal controls – data from the osteoarthritis initiative. Arthritis Res Ther 2011; 13:R153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.■■.Heilmeier U, Wamba JM, Joseph GB, et al. Baseline knee joint effusion and medial femoral bone marrow edema, in addition to MRI-based T2 relaxation time and texture measurements of knee cartilage, can help predict incident total knee arthroplasty 4–7 years later: data from the Osteoarthritis Initiative. Skeletal Radiol 2019; 48:89–101. [DOI] [PubMed] [Google Scholar]; Current status of advanced evaluation quantitative MR in osteoarthritis and employment of artificial intelligence.


