Abstract
Background
The transcription factor Homeobox C8 (HOXC8) is overexpressed and regulates many important genes involved in the proliferation and invasion of many malignant tumors. However, the function of HOXC8 in gliomas remains unclear.
Method
Based on the Chinese Glioma Genome Atlas (CGGA) set, HOXC8 expression is negatively correlated with overall survival (OS). Small interfering RNA (si-HOXC8) was used to downregulate the mRNA and protein expression levels of HOXC8 to assess glioma cell proliferation, migration and invasion.
Results
Patients with higher HOXC8 levels showed poorer prognosis. DAVID analysis results indicated that HOXC8 was related to cell cycle, cell adhesion and immune response. In U251 and LN229 glioma cells treated with small interfering RNA for HOXC8 (si-HOXC8) for gene knockdown, significantly lower cell capacity of growth, migration and invasion was observed. Moreover, HOXC8 knockdown could reduce the protein expression of classical epithelial mesenchymal transition (EMT) related markers.
Conclusion
HOXC8 may play an important role in glioma proliferation, migration and invasion. These findings indicated that HOXC8 may constitute a novel target for glioma treatment.
Keywords: HOXC8, Cell cycle, Migration, Invasion, EMT
Background
Gliomas account for most of malignant tumors of the central nervous system (CNS). Despite the recent progress in surgical resection, radiotherapy and chemotherapy, prognosis for glioma remains poor [1, 2]. Gliomas possessed strong proliferation, migration and invasion abilities, causing high mortality. Therefore, multiple studies have identified key genes that could help achieve a breakthrough in gliomas treatment [3, 4]. In this study, Homeobox C8 (HOXC8), a transcript factor, was selected for in-depth assessment.
HOXC8, located in a cluster on chromosome 12, belongs to the 39-member HOX family of proteins [5]. Under normal circumstances, HOXC8 plays an indispensable role in embryonic morphogenesis and system morphogenesis [6, 7]. However, in various tumors, including ovarian cancer, hepatocellular carcinoma, and breast cancer, HOXC8 induces cell proliferation, migration and invasion, and is inversely correlated with overall survival (OS) through different mechanisms [8-10]. Although HOXC8 is important in tumorigenesis, its mechanism in glioma remains unclear.
Epithelial mesenchymal transition (EMT), a cellular alteration that confers a more invasive and drug-resistant phenotype, was initially observed in embryonic development [11, 12]. EMT was a complex process reflected by epithelial cells acquiring the mesenchymal phenotype and motility through a cascade of biological events [13]. In gliomas, the mechanism by which HOXC8 affected EMT is unclear.
In summary, the aim of our study was to assess the oncogenic function of HOXC8. HOXC8 knockdown could inhibit glioma cell proliferation, migration and invasion. Therefore, HOXC8 could provide novel insights for glioma treatment.
Methods
Dataset and glioma samples
We obtained mRNA samples and follow-up information from the Chinese Glioma Genome Atlas (CGGA). OS was defined as the time from surgery to death. Glioma samples were collected from surgical resection, snap-frozen in liquid nitrogen and stored at − 80 °C until RNA extraction [14]. This study was approved by the Ethics Committee of Beijing Tiantan Hospital, and written informed consent was obtained from all the patients.
Cell culture and siRNA transfection
Two glioma cell lines were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and antibiotics (100 U/ml penicillin and 100 μg/mL streptomycin), in a humidified atmosphere with 5% CO2 at 37 °C [15]. The siRNA segmentand a non-specific control siRNA sequence (NC) were purchased from Genepharma (Shanghai, China), and transfected into LN229 and U251 cells using Lipofectamine™ 2000 (Invitrogen, Shanghai, China), according to the manufacturer’s instructions. After 48 h of incubation, samples were collected for RT-PCR and Western blot assay [16].
Real-time polymerase chain reaction (RT-PCR) and western blot (WB)
RT-PCR and Western blot were performed as previously described [4, 17].The following primers were used for RT-PCR: HOXC8, Forward 5’-ACCGGCCTATTACGACTGC-3′ and Reverse 5′-TGCTGGTAGCCTGAGTTGGA-3′; GAPDH (Forward 5′- GGAGCGAGATCCCTCCAAAAT -3′ and Reverse 5′- GGCTGTTGTCATACTTCTCATGG-3′ was used as an internal control, and fold changes were calculated by the 2 − ΔΔCt method. For Western blotting, densities of specific protein bands were quantified after normalization to Tubulin or Actin level in the same sample.
Clone formation assay
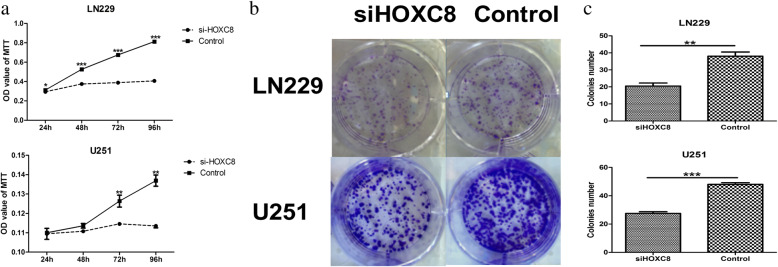
A total of 1000 cells were seeded per well in a 6-well plate in DMEM and 10% FBS, and cultured for 2 weeks before Crystal violet stain. Then, clone numbers were recorded in siRNA and control groups, respectively.
Transwell migration assay
Transwell plates were used to study cell migration and invasion. A total of 1 × 105 LN229 or U251 cells were seeded in each well in DMEM only. Control medium was added to the lower chamber of the transwell plates. After 36 h (invasion assay) or 12 h (migration assay) of incubation, the cells in upper chambers were removed carefully before Crystal violet staining. Analysis was carried out by microscopy, counting cells in siRNA and control groups, respectively [18].
Statistical analysis
We used Kaplan-Meier analysis (log-rank test) to evaluate predictive value of HOXC8 expression for OS in different groups. Univariate and multivariate Cox regression analyses were used to determine that HOXC8 was an independent factor to predict OS. Pearson relation analysis was performed in CGGA array set using R language package. All values were considered statistically significant at p < 0.05.
Results
HOXC8 was associated with overall survival in gliomas
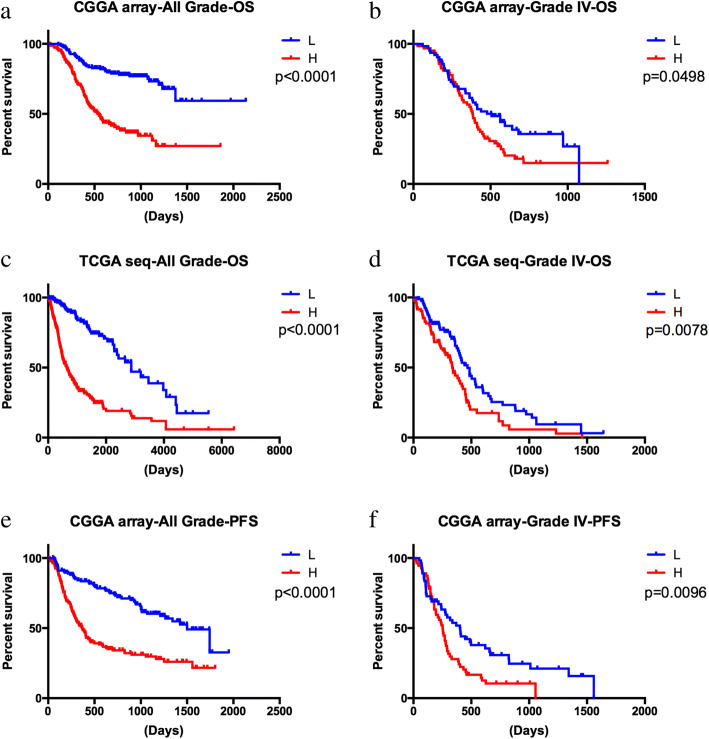
We obtained the HOXC8 mRNA expression and prognosis information from the CGGA mRNA array and TCGA RNA-seq datasets, and assessed the association of HOXC8 expression with overall survival (OS). As shown in Fig. 1a and b, the level of HOXC8 mRNA expression was negatively related with OS in all grade gliomas and grade IV gliomas (p < 0.05). Moreover, we further validated the results in TCGA RNA-seq set (Fig. 1c and d, p < 0.05). Moreover, in Fig. 1e and f, in CGGA array set, progression-free survival time of was shorter in higher HOXC8 expression group (p < 0.05). To further validation our results, we performed a univariate Cox regression analysis, the results showed that HOXC8 was showed a risk factor for glioma patients (p < 0.0001, HR = 1.545, Table 1). What is more, age at diagnosis, IDH1 status, gliomas grade, TCGA subtype was significantly related with survival time of glioma patients (p < 0.0001, Table 1). Then, the results from multivariate Cox regression analysis including HOXC8 expression, age at diagnosis, IDH1 status, gliomas grade and TCGA subtype showed that HOXC8 was an independent prognostic factor for glioma patients (p < 0.0001, HR = 1.247, Table 1).
Fig. 1.
a-d HOXC8 expression was negatively related with OS in all grade gliomas and grade IV gliomas repectively in CGGA and TCGA sets (p < 0.05); e-f HOXC8 expression was negatively related with PFS in all grade gliomas and grade IV gliomas repectively in CGGA set (p < 0.05); OS: overall survival; PFS: progression-free survival
Table 1.
Univariate and multivariate Cox analysis in CGGA glioma samples
| Clinical factors | Univariate | P | Multivariate | P | ||||
|---|---|---|---|---|---|---|---|---|
| HR | 95%CI | HR | 95%CI | |||||
| Lower | Upper | Lower | Upper | |||||
| Age at diagnosis | 1.044 | 1.026 | 1.062 | < 0.0001 | 1.011 | 0.993 | 1.029 | 0.240 |
| Gender (male) | 1.162 | 0.792 | 1.704 | 0.443 | ||||
| IDH1 status (mutation) | 0.335 | 0.219 | 0.511 | < 0.0001 | 0.932 | 0.544 | 1.597 | 0.799 |
| High HOXC8 expression | 1.545 | 1.397 | 1.709 | < 0.0001 | 1.247 | 1.110 | 1.402 | < 0.0001 |
| Grade (high grade) | 3.960 | 2.960 | 5.299 | < 0.0001 | 3.176 | 2.307 | 4.371 | < 0.0001 |
| TCGA subtype (mesenchymal type) | 1.605 | 1.362 | 1.890 | < 0.0001 | 1.048 | 0.866 | 1.267 | 0.631 |
IDH1: isocitrate dehydrogenase 1; CGGA: Chinese Glioma Genome Atlas; TCGA: The Cancer Genome Atlas; CI: Confidence Interval
In conclusion, we hypothesized that HOXC8 could serve as a novel biomarker for predicting glioma OS.
Gene functional analysis
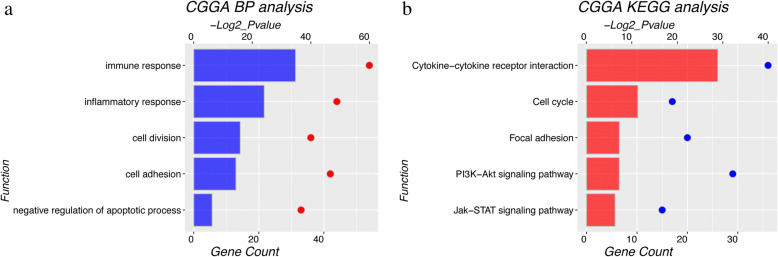
The function of HOXC8 in glioma remains unclear. We performed Pearson relation analysis in CGGA array set using R language package. To explore differential biological function of HOXC8, we selected top 1000 genes positively or negatively related with HOXC8 for DAVID analysis respectively. In Fig. 2a and b, Gene Ontology (GO) and KEGG pathways analysis showed that cell cycle, cell adhesion, immune response and inflammatory response were high expression in high-HOXC8 expression group. Meanwhile, taken together, gliomas in high-HOXC8 group had a greater capacity for proliferation, migration and invasion than these in low-HOXC8 group.
Fig. 2.
HOXC8 associated genes and their potential function a GO analysis; b KEGG pathway analysis. GO: Gene Ontology
LN229 and U251 glioma cell lines were used to investigate biological processes
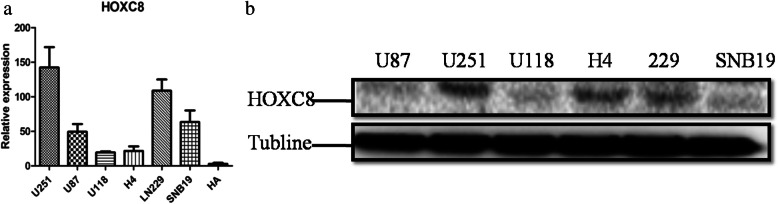
Then in Fig. 3a and b, seven (six glioma cell lines and one normal human astrocyte) frequently-studied cell lines (LN229, U87, U118, U251, SNB19, H4, and HA) were selected for HOXC8 mRNA and protein expression assessment. Interestingly, the six glioma cell lines showed higher HOXC8 expression levels compared with HA cells (human astrocytes). More importantly, LN229 and U251 cells had highest endogenous mRNA and protein levels of HOXC8. Based on the above results, LN229 and U251 were selected for subsequent experiments.
Fig. 3.
HOXC8 expression in different glioma cell lines. a mRNA expression level of HOXC8 in six glioma cell lines (LN229, U87, U118, U251, SNB19, H4) and one normal human astrocyte (HA); b Protein expression level of HOXC8 in six glioma cell lines
HOXC8 could significantly promote glioma proliferation
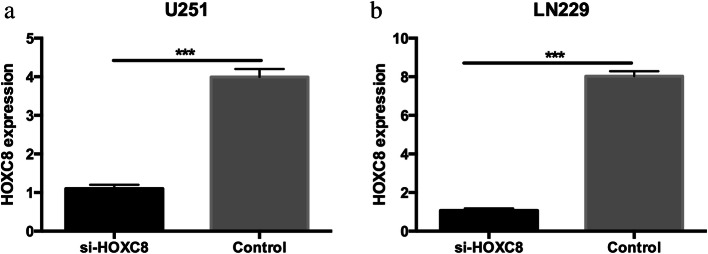
To assess the effect of HOXC8 on cell proliferation, HOXC8 was silenced in LN229 and U251 cells by siRNA; 48 h after transfection, siRNA successfully knocked down HOXC8 at the mRNA levels (Fig. 4a and b). Cell viability was measured by MTT and clonogenic assays, as shown in Fig. 5a-c, antagonism of HOXC8 expression suppressed the cell growth and the formation of cell clones (p < 0.05).
Fig. 4.
Small interfering RNA of HOXC8 (si-HOXC8) could significantly knocked down HOXC8 mRNA expression in U251 (a) and LN229 (b) glioma cell lines (p < 0.05)
Fig. 5.
a MTT assay indicates that si-HOXC8 can suppress LN229 and U251 cell proliferation (p < 0.05); b & c Clonogenic assay indicates that si-HOXC8 can suppress LN229 and U251cell clone formation (p < 0.05)
Downregulation of HOXC8 inhibits cell migration and invasion by blocking the EMT pathway
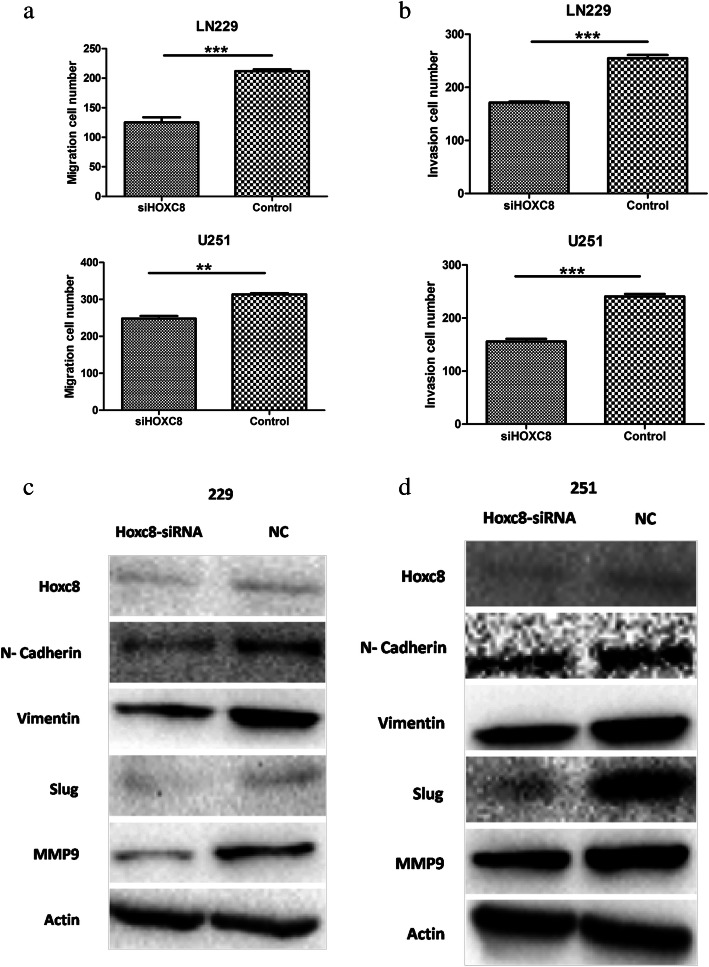
To assess whether the migration and invasion capacity of glioma cells could be inhibited by si-HOXC8, we performed migration and Matrigel invasion assays using LN229 and U251 cells. Crystal violet staining showed markedly less migration and invasion glioma cells after HOXC8 silencing compared with negative-control values (p < 0.01; Fig. 6a and b). In addition, EMT was a novel mechanism involved in metastasis [19, 20]. The protein expression levels of mesenchymal markers (N-Cadherin, Slug, Vimentin and MMP9) were reduced in si-HOXC8 LN229 and U251 cells compared with the control group, as assessed by Western blot (Fig. 6c and d). Taken together, si-HOXC8 suppressed cell migration and invasion by reversing EMT.
Fig. 6.
Si-HOXC8 inhibited glioma cell migration, invasion and EMT process a & b The number of migration and invasion LN229 and U251 cells were decreased in si-HOXC8 group (p < 0.05); c & d Si-HOXC8 could significantly decrease EMT related marker including N-cadherin, SLUG, VIMENTIN, MMP9. EMT: epithelial mesenchymal transition
Discussion
Massive evidence supports that HOXC8 plays an important role in malignant tumor formation and progression [21, 22]. As a transcription factor, HOXC8 is able to regulate and coordinate multiplevital genes (e.g. Mgl1, Embigin, Meis1, and Fyn) involved in cancer development and progression [8, 22, 23]. Previous findings demonstrated that HOXC8 depletion by small interfering RNA suppresses epithelial ovarian cancer proliferation and migration, and induces apoptosis by increasing ZAC1 expression [24]. HOXC8, overexpressed in hepatocellular carcinoma (HCC) compared with adjacent non-tumor tissues, is associated with poor prognosis. Si-HOXC8 blocks G1-S phase transition, inhibits cell proliferation and renders cells more sensitive to oxaliplatin [9]. Therefore, HOXC8 as a potential oncogenic driver, plays an important role in cancer cell proliferation, migration and invasion, by overregulating many oncogenes and downregulating tumor suppressor genes. In this study, we demonstrated that si-HOXC8 blunted glioma cell proliferation, migration and invasion by reversing EMT.
EMT is known to be implicated in cancer progression, metastasis and drug resistance [25]. EMT reversibly enables polarized epithelial cells to lose their epithelial characteristics and to acquire mesenchymal properties [13]. The availability of antibodies targeting epithelial (E-cadherin, β-catenin, and Claudin-1) and mesenchymal (N-cadherin, Slug, Vimentin and MMP9) markers makes it convenient to assess EMT [26]. Multiple genes and pathways were reported to be involved in EMT [26-28]. However, the mechanisms by which HOXC8 affects EMT remains unclear. This study found that the HOXC8 was involved in EMT. Moreover, si-HOXC8 decreased the expression of classical mesenchymal markers (N-cadherin, Slug, Vimentin and MMP9). Hence, the present findings provide additional insights regarding the implication of HOXC8 and target genes in EMT as well as malignant tumor progression.
In summary, this study demonstrated that higher HOXC8 expression resulted in shorter OS. In agreement, HOXC8 knockdown inhibited glioma cell proliferation. Meanwhile, reduction of HOXC8 could also affected migration and invasion through blocking EMT pathway. The present findings suggested that HOXC8 should be considered a novel biomarker and target for glioma treatment.
Acknowledgements
Thanks for Chinese Glioma Genome Atlas dataset and The Cancer Genome Atlas dataset for data share.
Funding
This study was funded by National Natural Science Fund (Grant No. 81301112) and Yu Miao Plan of Beijing Ditan Hospital (Grant No. DTYM201603) and Beijing Medical Management Bureau Cultivation Plan (Grant No. PX2018079).
Availability of data and materials
All the data in this study were obtained from The Cancer Genome Atlas dataset (TCGA; http://cancergenome.nih.gov/) and Chinese Glioma Genome Atlas dataset (CGGA; http://www.cgga.org.cn/).
Authors’ contributions
Conception and design: ESF and GY; Performed the experiments: TYL and XXW; Data collection and analysis: TYL; Order reagents/materials: TYL and PLL; Wrote the paper: YC. All authors have read and approved the final manuscript.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of Beijing Tiantan Hospital, and written informed consent was obtained from all the patients.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interest.
Footnotes
Tingyu Liang and Xiaoxuan Wang contributed equally to this work.
Contributor Information
Enshan Feng, Email: enshanfeng@126.com.
Gan You, Phone: +86-10-67098431, Email: Ganyou2016@126.com.
References
- 1.Norden AD, Drappatz J, Wen PY. Antiangiogenic therapies for high-grade glioma. Nat Rev Neurol. 2009;5:610–620. doi: 10.1038/nrneurol.2009.159. [DOI] [PubMed] [Google Scholar]
- 2.Zhou X, Ren Y, Zhang J, et al. HOTAIR is a therapeutic target in glioblastoma. Oncotarget. 2015;6:8353–8365. doi: 10.18632/oncotarget.3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen PH, Chang CK, Shih CM, et al. The miR-204-3p-targeted IGFBP2 pathway is involved in xanthohumol-induced glioma cell apoptotic death. Neuropharmacology. 2016;110:362–375. doi: 10.1016/j.neuropharm.2016.07.038. [DOI] [PubMed] [Google Scholar]
- 4.You G, Feng L, Yan W, et al. BCL2A1 is a potential biomarker for postoperative seizure control in patients with low-grade gliomas. CNS Neurosci Ther. 2013;19:882–888. doi: 10.1111/cns.12148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gehring WJ, Hiromi Y. Homeotic genes and the homeobox. Annu Rev Genet. 1986;20:147–173. doi: 10.1146/annurev.ge.20.120186.001051. [DOI] [PubMed] [Google Scholar]
- 6.Gao Y, Hyttel P, Hall VJ. Regulation of H3K27me3 and H3K4me3 during early porcine embryonic development. Mol Reprod Dev. 2010;77:540–549. doi: 10.1002/mrd.21180. [DOI] [PubMed] [Google Scholar]
- 7.Kalyani R, Lee JY, Min H, Yoon H, Kim MH. Genes frequently Coexpressed with Hoxc8 provide insight into the discovery of target genes. Mol Cells. 2016;39:395–402. doi: 10.14348/molcells.2016.2311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao F, Zhang J, Zhang Y, et al. Embigin, regulated by HOXC8, plays a suppressive role in breast tumorigenesis. Oncotarget. 2015;6:23496–23509. doi: 10.18632/oncotarget.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu P, Zhang X, Ni W, et al. Upregulated HOXC8 expression is associated with poor prognosis and Oxaliplatin resistance in hepatocellular carcinoma. Dig Dis Sci. 2015;60:3351–3363. doi: 10.1007/s10620-015-3774-x. [DOI] [PubMed] [Google Scholar]
- 10.Zhou W, Ke SQ, Huang Z, et al. Periostin secreted by glioblastoma stem cells recruits M2 tumour-associated macrophages and promotes malignant growth. Nat Cell Biol. 2015;17:170–182. doi: 10.1038/ncb3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zeisberg M, Neilson EG. Biomarkers for epithelial-mesenchymal transitions. J Clin Invest. 2009;119:1429–1437. doi: 10.1172/JCI36183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang L, Zhang W, Li Y, et al. SHP-2-upregulated ZEB1 is important for PDGFRalpha-driven glioma epithelial-mesenchymal transition and invasion in mice and humans. Oncogene. 2016;35:5641–5652. doi: 10.1038/onc.2016.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felipe Lima J, Nofech-Mozes S, Bayani J, Bartlett JM. EMT in breast carcinoma-a review. J Clin Med. 2016;5 [DOI] [PMC free article] [PubMed]
- 14.Yang P, Liang T, Zhang C, et al. Clinicopathological factors predictive of postoperative seizures in patients with gliomas. Seizure. 2016;35:93–99. doi: 10.1016/j.seizure.2015.12.013. [DOI] [PubMed] [Google Scholar]
- 15.Kaur H, Ali SZ, Huey L, et al. The transcriptional modulator HMGA2 promotes stemness and tumorigenicity in glioblastoma. Cancer Lett. 2016;377:55–64. doi: 10.1016/j.canlet.2016.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang X, Shi W, Shi H, et al. TRIM11 overexpression promotes proliferation, migration and invasion of lung cancer cells. J Exp Clin Cancer Res: CR. 2016;35:100. doi: 10.1186/s13046-016-0379-y. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Li MY, Yang P, Liu YW, et al. Low c-met expression levels are prognostic for and predict the benefits of temozolomide chemotherapy in malignant gliomas. Sci Rep. 2016;6:21141. doi: 10.1038/srep21141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao T, Yang H, Tian Y, et al. SOX7 is associated with the suppression of human glioma by HMG-box dependent regulation of Wnt/beta-catenin signaling. Cancer Lett. 2016;375:100–107. doi: 10.1016/j.canlet.2016.02.044. [DOI] [PubMed] [Google Scholar]
- 19.Li J, Yuan J, Yuan X, et al. MicroRNA-200b inhibits the growth and metastasis of glioma cells via targeting ZEB2. Int J Oncol. 2016;48:541–550. doi: 10.3892/ijo.2015.3267. [DOI] [PubMed] [Google Scholar]
- 20.Thiyagarajan V, Tsai MJ, Weng CF. Antroquinonol targets FAK-signaling pathway suppressed cell migration, invasion, and tumor growth of C6 glioma. PLoS One. 2015;10:e0141285. doi: 10.1371/journal.pone.0141285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Du YB, Dong B, Shen LY, et al. The survival predictive significance of HOXC6 and HOXC8 in esophageal squamous cell carcinoma. J Surg Res. 2014;188:442–450. doi: 10.1016/j.jss.2014.01.017. [DOI] [PubMed] [Google Scholar]
- 22.Ruthala K, Gadi J, Lee JY, Yoon H, Chung HJ, Kim MH. Hoxc8 downregulates Mgl1 tumor suppressor gene expression and reduces its concomitant function on cell adhesion. Molecules Cells. 2011;32:273–279. doi: 10.1007/s10059-011-0069-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lei H, Wang H, Juan AH, Ruddle FH. The identification of Hoxc8 target genes. Proc Natl Acad Sci U S A. 2005;102:2420–2424. doi: 10.1073/pnas.0409700102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu S, Liu R, Su M, et al. Overexpression of HOXC8 is associated with poor prognosis in epithelial ovarian Cancer. Reprod Sci. 2016;23:944–954. doi: 10.1177/1933719115625845. [DOI] [PubMed] [Google Scholar]
- 25.Du B, Shim JS. Targeting epithelial-mesenchymal transition (EMT) to overcome drug resistance in Cancer. Molecules. 2016;21 [DOI] [PMC free article] [PubMed]
- 26.Wang H, Tao T, Yan W, et al. Upregulation of miR-181s reverses mesenchymal transition by targeting KPNA4 in glioblastoma. Sci Rep. 2015;5:13072. doi: 10.1038/srep13072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ding Y, Li X, Hong D, Jiang L, He Y, Fang H. Silence of MACC1 decreases cell migration and invasion in human malignant melanoma through inhibiting the EMT. Biosci Trends. 2016;10:258–264. doi: 10.5582/bst.2016.01091. [DOI] [PubMed] [Google Scholar]
- 28.Tang H, Fan D, Lei CT, et al. MAD2B promotes tubular epithelial-to-mesenchymal transition and renal tubulointerstitial fibrosis via Skp2. J Mol Med. 2016;94:1297–1307. doi: 10.1007/s00109-016-1448-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data in this study were obtained from The Cancer Genome Atlas dataset (TCGA; http://cancergenome.nih.gov/) and Chinese Glioma Genome Atlas dataset (CGGA; http://www.cgga.org.cn/).