Abstract
Background
Transgelin, an actin-binding protein, is associated with cytoskeleton remodeling. Findings from our previous studies demonstrated that transgelin was up-regulated in node-positive colorectal cancer (CRC) versus node-negative disease. Over-expression of TAGLN affected the expression of 256 downstream transcripts and increased the metastatic potential of colon cancer cells in vitro and in vivo. This study aims to explore the mechanisms through which transgelin participates in the metastasis of colon cancer cells.
Methods
Immunofluorescence and immunoblotting analysis were used to determine the cellular localization of endogenous and exogenous transgelin in colon cancer cells. Co-immunoprecipitation and subsequently high-performance liquid chromatography/tandem mass spectrometry were performed to identify the proteins that were potentially interacting with transgelin. The 256 downstream transcripts regulated by transgelin were analyzed with bioinformatics methods to discriminate the specific key genes and signaling pathways. The Gene-Cloud of Biotechnology Information (GCBI) tools were used to predict the potential transcription factors (TFs) for the key genes. The predicted TFs corresponded to the proteins identified to interact with transgelin. The interaction between transgelin and the TFs was verified by co-immunoprecipitation and immunofluorescence.
Results
Transgelin was found to localize in both the cytoplasm and nucleus of the colon cancer cells. Approximately 297 proteins were identified to interact with transgelin. The overexpression of TAGLN led to the differential expression of 184 downstream genes. Network topology analysis discriminated seven key genes, including CALM1, MYO1F, NCKIPSD, PLK4, RAC1, WAS and WIPF1, which are mostly involved in the Rho signaling pathway. Poly (ADP-ribose) polymerase-1 (PARP1) was predicted as the unique TF for the key genes and concurrently corresponded to the DNA-binding proteins potentially interacting with transgelin. The interaction between PARP1 and transgelin in human RKO colon cancer cells was further validated by immunoprecipitation and immunofluorescence assays.
Conclusions
Our results suggest that transgelin binds to PARP1 and regulates the expression of downstream key genes, which are mainly involved in the Rho signaling pathway, and thus participates in the metastasis of colon cancer.
Keywords: Transgelin, PARP1, Colon cancer, Rho signaling, Bioinformatics
Background
Colorectal cancer (CRC) is a frequent malignant tumor in the gastrointestinal tract worldwide. In 2018, it was ranked the third in terms of incidence (1.8 million new cases) and second in terms of mortality rate (881,000 deaths) [1]. Although mechanisms of CRC tumorigenesis and metastasis have been extensively studied, high mortality rates are still reported, especially in patients with advanced disease [1].
Tumor metastasis, the spread of cancer cells from the primary tumor to other areas of the body, is a complex process associated with remodeling of the cytoskeleton. The intracellular cytoskeleton requires a high degree of functional integration and coordination of actin (microfilament), microtubules and intermediate filaments [2–5]. However, abnormal expression of related genes or effector proteins may lead to the activation of various signaling pathways, thus promoting tumor metastasis [2–5].
Transgelin (also known as 22 kDa actin-binding protein, protein WS3-10 or smooth muscle protein 22 alpha) is an actin-binding protein with a molecular weight of 23 kDa and consists of 201 amino acids [6]. It is encoded by the TAGLN gene and composes of an N-terminal calmodulin homologous (CH) domain and a C-terminal calmodulin-like (CLIK) domain, which is closely related to actin binding activity [6]. Transgelin is broadly expressed in the vascular and visceral smooth muscle and is an early marker of smooth muscle differentiation [7]. Furthermore, transgelin is associated with the remodeling of the actin cytoskeleton and promotes the migration and invasion of cancer stem cells [8–10].
Recent studies have shown that besides the involvement in the regulation of actin nucleation, cellulose capping, fragmentation, actin monomer binding and other functions in the cytoplasm, actin-binding proteins are also involved in the formation of transcription complexes [11]. Transgelin has been shown to be a poor prognostic factor associated with advanced CRC [12] and it also promotes transforming growth factor β (TGF β)-dependent tumor growth and migration [13]. Moreover, results from in vitro experiments and a xenograft metastatic mouse model suggest that transgelin may be a promising therapeutic target for treating bladder cancer metastasis [14]. Therefore, we believe that transgelin may serve as a biomarker for tumor metastasis.
In our previous study, transgelin was up-regulated in the node-positive CRC versus node-negative disease [15]. While the up-regulation of transgelin promoted the metastasis of colon cancer cells, down-regulation substantially decreased the ability of cell invasion and metastasis [9, 10, 15]. In addition, gene expression profiling showed that over-expression of TAGLN affected the expression of the 256 downstream transcripts, which were closely related to cell morphology, migration and invasion [9]. We also found that transgelin localized in both the cytoplasm and nucleus of the cultured CRC cells and affected the expression levels of several epithelial to mesenchymal transition (EMT) associated genes [15]. Therefore, we hypothesized that transgelin may be a transcriptional regulator. However, the role of transgelin in colon cancer metastasis remains unknown.
Herein, we verified the nuclear localization of transgelin in different colon cancer cell lines. Approximately 297 proteins that are potentially interacting with transgelin were identified, of which 23 were DNA-binding proteins. Over-expression of TAGLN affected the expression levels of 184 genes. Seven key genes that mainly involved in the Rho signaling pathway were also identified. By analyzing the promoter regions of these key genes, poly (ADP-ribose) polymerase-1 (PARP1), a DNA-binding protein, was predicted to be the transcription factor (TF) of these genes. PARP1 was also among the 23 DNA-binding proteins that were perceived to interact with transgelin. The interaction between transgelin and PARP1 was further verified by immunoprecipitation and immunofluorescence.
Materials and methods
Cell lines
The human CRC cell lines, including RKO, SW480, HCT116, and LOVO were obtained from the Stem Cell Bank, Chinese Academy of Sciences (Shanghai, China). Cells were cultured in a minimum Eagle’s medium (MEM, Gibco, USA), Mccoy’s 5A medium (Gibco, USA) and Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco, USA) with 10% fetal bovine serum (Gibco, USA). Cells were then cultured and incubated at 37 °C with 5% CO2.
Immunofluorescence
Localization of endogenous transgelin in RKO, SW480, HCT116 and LOVO cell lines and the expression of PARP1 in RKO cells were determined by immunofluorescence. The primary antibody (anti-transgelin, 1:500, Abcam, USA; anti-PARP1, 1:500, Cell signaling technology, USA), secondary antibody (Alexa Flour 594 goat anti-rabbit IgG, Alexa Flour 488 goat anti-rabbit IgG, 1:500, Invitrogen, USA), and the VECTASHIELD mounting medium (Vector Laboratories, USA)) with 4′, 6-diamidino-2-phenylindole (DAPI) were used. The immunofluorescence images were taken and preserved under the laser scanning confocal microscope using a 63 × oil-immersion objective lens (Carl Zeiss, USA).
Transfection
The SW480 and RKO cells were cultured in 12-well plates and transfected with pcDNA6/myc-His B-TAGLN-flag and pcDNA6/myc-His B-flag plasmids (Takara, Japan). The RKO cells were transfected with pENTER-TAGLN-Flag and pENTER-Flag control plasmids (Vigene Biosciences, USA) in the co-immunoprecipitation experiment. Transfection was conducted using Lipofectamine 2000/Lipofectamine 3000 (Thermo Fisher Scientific, USA). Cells were then harvested at 48 h after transfection for further analysis.
RNA isolation, reverse transcription and polymerase chain reaction (RT-PCR)
Extraction of total RNA was performed using Trizol (Invitrogen) followed by reverse transcription (RT). Real-time polymerase chain reaction (PCR) was carried out using a Light Cycler 480 SYBR Green I Master mix (Roche, USA) on a Light Cycler 480 System (Roche, USA) according to the manufacturer’s instructions. The PCR conditions were as follows: 95 °C for 30 s, 35 cycles at 95 °C for 5 s, then 60 °C for 30 s. PCR primers are listed in Additional file 1: Table S2.
Immunoblotting
The nuclear and plasma proteins from HCT116, SW480, LOVO and RKO cell lines were extracted using the NE-PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific, USA). The protein concentration of the extracted cytoplasmic and nuclear proteins was determined. Immunoblotting was performed with the primary antibody anti-transgelin (1:500, Abcam, USA, or 1:500, R&D, USA), anti-GADPH (1:400, Abcam, USA or 1:500, Cell signaling technology, USA), anti-PARP1 (1:500, Cell signaling technology, USA), anti-Lamin B1(1:1000, Cell signaling technology, USA), anti-flag (1:500, Cell signaling technology, USA) and the secondary antibody (horseradish peroxidase (HRP)-conjugated goat anti-rabbit or anti-mouse IgG, 1:30,000, Sigma-Aldrich, USA) or IgG Detector (IgG Detector Solution v2, HRP labeled, 1:1000, Takara, Japan). Antibody detection was performed using a chemiluminescence substrate and the protein bands were visualized with Syngene G: BOX Chemi XT4 fluorescence and chemiluminescence gel imaging system (Cambridge, UK).
Immunoprecipitation
The RKO cells were cultured conventionally and transfected with pcDNA6/myc-His B-TAGLN-flag and pcDNA6/myc-His B-flag plasmids. The RKO cells were transfected with pENTER-TAGLN-Flag and pENTER-Flag control plasmids in the validation experiment. Cells were then harvested at 48 h after transfection for further analysis. Antibody immobilization, cell lysis, pretreatment of cell lysate with control agarose resin, immunoprecipitation, immunoprecipitation elution, and immunoblotting analysis were performed in sequence according to the protocol of the Pierce Crosslink Immunoprecipitation Kit (Thermo Fisher Scientific, USA). Anti-flag antibody (10ug, Sigma-Aldrich, USA, for the subsequent mass spectrometry; 1:50, Cell signaling technology, USA, for the validation experiment) and the control rabbit IgG (1:50, Cell signaling technology, USA) were used.
Mass spectrometry
A fraction of the protein samples after immunoprecipitation was analyzed using SDS-PAGE and silver staining (Invitrogen, USA). Another fraction of the samples was used for high-performance liquid chromatography assay (EASY-nLC™, Thermo Fisher Scientific, USA) after filter-aided sample preparation (FASP) and enzymatic hydrolysis. The samples were then analyzed with a Q-Exactive Mass Spectrometer (Thermo Finnigan, USA). The mass/charge ratios of peptides and fragments of peptides were collected. Maxquant 1.3.0.5 software was used to retrieve the Uniprot database by using the raw file as source. The search in the database was set up with specific parameters (Enzyme, trypsin; De-Isotopic, True; Max Missed Cleavages, 2; Fixed modifications, Carbamidomethyl (C); Variable modifications, Oxidation (M); First search ppm, 20 ppm; Main search ppm, 6 ppm; Decoy database pattern, reverse; Min. Reporter PIF, 0.75; Peptides false discovery rate (FDR) ≤ 0.01; Protein FDR ≤ 0.01).
Bioinformatics
Identification of differential expression genes (DEGs), functional enrichment and signaling pathway enrichment analysis
The relevant cDNA microarray data were obtained using the Affymetrix microarray technique based on our previous work [9]. Over-expression of TAGLN in RKO human colon cancer cells resulted in 256 downstream transcripts that were differentially expressed with at least a twofold change (P < 0.05). Among these, transcripts without gene symbols, gene database codes and duplicates were excluded. The remaining DEGs were screened for further bioinformatics analysis.
Using the Metascape tool (http://www.metascape.org/), the screening parameters were set as follows: P < 0.01 or 0.001 (Biological Process), participating genes ≥ 3 and enrichment factor > 1.5. We conducted functional and signaling pathway enrichment analysis of the DEGs referring to the gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway and Reactome databases.
Construction of the protein–protein interaction (PPI) network, topological analysis and key gene screening
The DEGs were simultaneously translated into proteins and the search tool for retrieval of interacting genes (STRING 10.0, https://string-db.org/) [16] was used for PPI analysis. Subsequently, relevant data was imported into the Cytoscape online software (http://www.cytoscape.org/) [17] and a PPI network was constructed. In this study, the degree centrality and intermediate centrality of the DEGs were calculated using the CytoHubba plug-ins. Those with values twofold higher than the overall average value were selected as the core genes in the network. In addition, the core modules were obtained with an MCODE plug-in (k-core = 2). The core genes and the genes included in the core modules were defined as the key genes. Key genes were further analyzed with Metascape for signaling pathway enrichment in KEGG and Reactome database using the same parameters previously mentioned.
Prediction of the TFs for the key genes
The TF evaluation model within the Gene-cloud of biotechnology information (GCBI) tools (https://www.gcbi.com.cn/) was used to predict the TFs for the key genes. Those with medium or high recommendations were selected and potential TFs were selected for further analysis. We then compared these potential TFs to the DNA-binding proteins identified in the mass spectrometry analysis.
Nuclear localization signal analysis
The sequences of the selected potential TF(s) were obtained from the Uniprot database (https://www.uniprot.org/) [18]. The classical nuclear localization signals (cNLS) Mapper (http://www.nls-mapper.iab.keio.ac.jp/) [19] was used to detect the nuclear localization signal of the potential TF(s).
Statistical analysis
Statistical analysis was performed using the SPSS 20.0 software (IBM Corp., Armonk, USA). The relevant values were expressed as mean ± standard deviation (SD), and the significance of the difference between two groups was determined with the Student’s t test. Pearson correlation analysis was used to analyze the expression level of the key genes obtained from cDNA microarray and RT-PCR. P < 0.05 (bilateral) was considered statistically significant.
Results
Localization of transgelin in human colon cancer cell lines
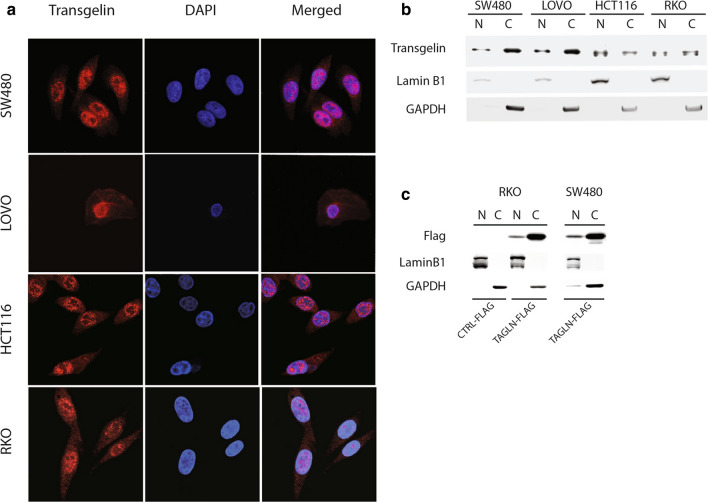
The expression of transgelin in colon cancer cell lines (HCT116, SW480, RKO and LOVO) was detected by immunofluorescence and immunoblotting assays. Both cytoplasmic and nuclear localization of endogenous transgelin were observed (Fig. 1a, b). Further, pcDNA6/myc-His B-TAGLN-flag and pcDNA6/myc-His B-flag control plasmids were transiently transfected into RKO and SW480 cells. Immunoblotting analysis showed detectable levels of exogenous transgelin-flag protein in both the cytoplasm and nucleus (mainly in the cytoplasm) of the associated RKO and SW480 cells (Fig. 1c). The expression level of transgelin-flag protein (1.00 ± 0.05) was significantly increased in the RKO-TAGLN-FLAG cells compared with the RKO-CTRL-FLAG cells (0.13 ± 0.03, P < 0.0001, Fig. 2a) and the wild type (WT) RKO cells (0.08 ± 0.02, P < 0.0001).
Fig. 1.
The localization of transgelin in different human colon cancer cell lines. a Transgelin in colon cancer cell lines RKO, SW480, HCT116 and LOVO was observed via immunofluorescence. The panels show transgelin immunostaining (red), 4′,6-diamidino-2-phenylindole DNA staining (DAPI), and a merged image is shown. b The distribution of transgelin in SW480, LOVO, HCT116 and RKO cells were identified via immunoblotting. c The distribution of transgelin-flag fusion protein in RKO and SW480 cells that were transiently transfected with pcDNA6/myc-His B-TAGLN-flag plasmid and control plasmid as detected via immunoblotting. N refers to the nuclear protein fraction, and C is the cytoplasmic protein fraction. Lamin B1 is a nuclear protein marker, GAPDH is a cytoplasmic protein marker
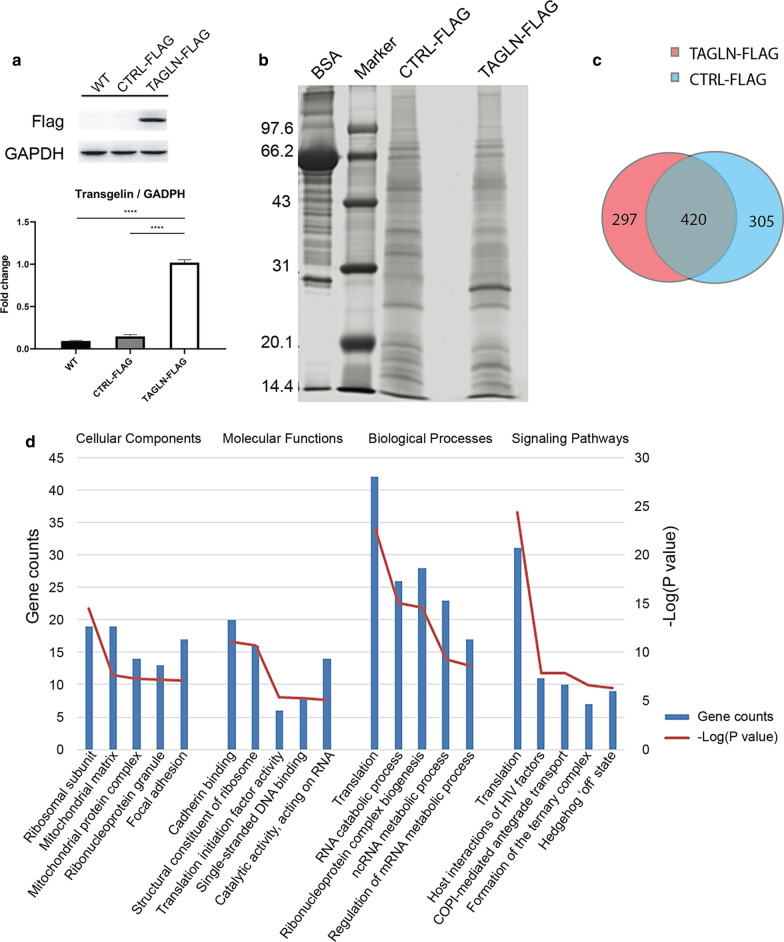
Fig. 2.
Identification of proteins potentially interacting with transgelin in the RKO cells. a Transgelin-flag protein was expressed in RKO cells transiently transfected with plasmids. Transgelin expression in RKO wild-type (WT), RKO-CTRL-FLAG and RKO-TAGLN-FLAG cells was detected via immunoblotting. **** P < 0.0001. b Proteins extracted from RKO-CTRL-FLAG and RKO-TAGLN-FLAG cells were immunoprecipitated with corresponding anti-flag antibody and visualized via silver staining. c The relationship between immunoprecipitated proteins by anti-flag antibody in RKO-CTRL-FLAG and RKO-TAGLN-FLAG cells. d Functional enrichment analysis of the 297 proteins potentially interacting with transgelin-flag fusion protein
Identification of proteins potentially interacting with transgelin in the RKO cells
To explore the proteins that were potentially interacting with transgelin, we performed immunoprecipitation in the RKO-TAGLN-FLAG cells and the RKO-CTRL-FLAG cells using an anti-flag monoclonal antibody. We observed a clear band in the RKO-TAGLN-FLAG group, ranging from 20.1 to 31 kb in the silver staining gel (Fig. 2b). To identify the proteins in the samples, we performed the high-performance liquid chromatography coupled with the tandem mass spectrometry. Approximately 725 proteins were identified in the RKO-CTRL-FLAG group, while 717 were in the RKO-TAGLN-FLAG group (Additional files 2 and 3). However, about 297 proteins were uniquely present in the RKO-TAGLN-FLAG group (Fig. 2c, Additional file 4: Table S1). Gene ontology (GO) functional enrichment analysis suggested that the 297 proteins in the RKO-TAGLN-FLAG group were mainly involved in translation, RNA processing, enzyme activity and cell junction adherence (Fig. 2d). In addition, among these, 23 proteins were DNA-binding proteins (Table 1).
Table 1.
DNA-binding proteins that were potentially interacted with Transgelin (FDR ≤ 0.01)
| gi number | Name of the protein | Molecular weight (Dalton) | |
|---|---|---|---|
| 1 | gi|124494254 | Proliferation-associated protein2G4 | 43,786 |
| 2 | gi|114205460 | HIST1H2BC protein | 13,833 |
| 3 | gi|21361745 | Spermatid perinuclear RNA-binding protein | 73,651 |
| 4 | gi|4827071 | Cellular nucleic acid-binding protein | 19,462 |
| 5 | gi|156523968 |
poly (ADP-ribose) polymerase family, member 1 (PARP1) |
113,084 |
| 6 | gi|29612542 | Histone H2A | 13,162 |
| 7 | gi|6912616 | Histone H2A | 13,508 |
| 8 | gi|323650782 | HMGA2 fusion protein | 13,811 |
| 9 | gi|297262894 |
High mobility group protein HMGI-C |
12,714 |
| 10 | gi|4506491 | Replication factor C subunit 4 | 36,877 |
| 11 | gi|4502747 | Cyclin-dependent kinase 9 | 42,777 |
| 12 | gi|345783096 | Barrier-to-autointegration factor | 10,058 |
| 13 | gi|7661672 | Polymerase delta-interacting | 42,032 |
| 14 | gi|98986457 | Host cell factor 1 | 208,730 |
| 15 | gi|32129199 | SAP domain-containing Ribonucleo protein | 23,670 |
| 16 | gi|57530065 | CCR4-NOT transcription complex subunit 7 | 32,744 |
| 17 | gi|302699237 | Eukaryotic translation initiation factor 4 gamma 1 | 158,643 |
| 18 | gi|5730027 |
KH domain-containing, RNA-binding, signal transduction-associated protein 1 |
48,226 |
| 19 | gi|238066755 | Disrupted in schizophrenia 1isoform 49 | 21,427 |
| 20 | gi|351694577 |
Activated RNA polymerase II transcriptional coactivator p15 |
13,993 |
| 21 | gi|119607091 | DNA replication licensing factor MCM4 | 11,656 |
| 22 | gi|7673373 | SCAN-related protein RAZ1 | 23,430 |
| 23 | gi|4758356 | Flap endonuclease 1 | 42,592 |
Effects of TAGLN over-expression on downstream genes and signaling pathways
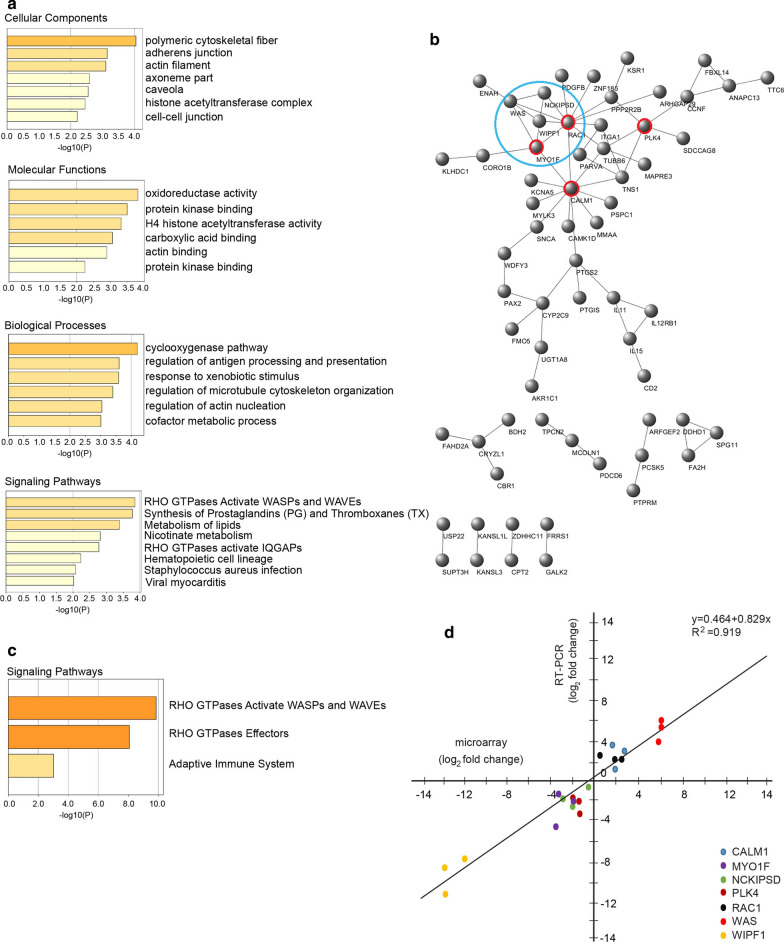
In our previous study, over-expression of TAGLN in RKO cells led to differential expression of the 256 transcripts in the Affymetrix cDNA microarray [9]. Of these, 68 transcripts with undefined gene symbols and gene database codes and 4 duplicates were eliminated. Finally, 184 DEGs (92 up-regulated and 92 down-regulated DEGs) were obtained for further analysis. Functional enrichment and signaling pathway enrichment analyses were performed using the Metascape tool (Fig. 3a). The results showed that the 184 DEGs were mainly associated with the cytoskeleton, protein kinase binding, regulation of cytoskeleton remodeling and Rho GTPase activation.
Fig. 3.
Effects of TAGLN overexpression on other genes and signaling pathways in RKO cells. Over-expression of TAGLN in RKO human colon cancer cells resulted in 184 genes differentially expressed with at least a twofold change (P < 0.05). a Functional enrichment (including cellular components, molecular functions, biological processes) and signaling pathway analysis of the DEGs were performed. b Topological analysis of the network illustrating the relationship between the proteins encoded by the DEGs. Genes in the red circle are core genes. The blue circle is the core module. The combinations of genes in the red and the blue circle are the key genes. c Signaling pathway enrichment analysis of the key genes identified the Rho signaling pathway. d Levels of the identified key genes as measured by cDNA microarray and real time RT-PCR. Relative gene expression was illustrated on a log-transformed scale. Data was obtained from three biological replicates
A PPI network was constructed with proteins encoded by the 184 DEGs using the STRING tool. The topological properties of the network were analyzed, which were composed of 167 nodes and 70 edges. The PPI network data were introduced into the Cytoscape (Fig. 3b). CytoHubba plug-ins were used to calculate the degree centrality and intermediate centrality of the DEGs. The mean values of global centrality and intermediate centrality were 4.375 and 153.375, respectively. Four genes, including CALM1, RAC1, PLK4 and MYO1F, were selected as the core genes in this network (Fig. 3b in red circles). An MCODE plug-in was then utilized to analyze interactions within the network. A core module with 4.5 points was selected (Fig. 3b in blue circle) by the k kernel analysis (k = 2), consisting of five nodes (RAC1, WAS, WIPF1, NCKIPSD, MYO1F) and nine edges. Based on the STRING tool, there were also complex interactions among gene-encoded proteins.
The core genes and the genes included in the core module were combined and seven discrete genes, including CALM1, PLK4, RAC1, WAS, WIPF1, NCKIPSD and MYO1F, were selected as key genes. Signaling pathway enrichment analysis of the key genes was performed. Three entries, mainly involving the Rho GTPase signaling pathway with significant differences, were obtained (Fig. 3c). We validated the expression levels of the key genes using RNA from the RKO cells, which were used for microarray, by real time RT-PCR (Fig. 3d). A scatter plot illustrated the agreement between the cDNA microarray and RT-PCR, with a coefficient of determination, R2, of 0.919. The correlation was significant based on Pearson correlation analysis (P < 0.01).
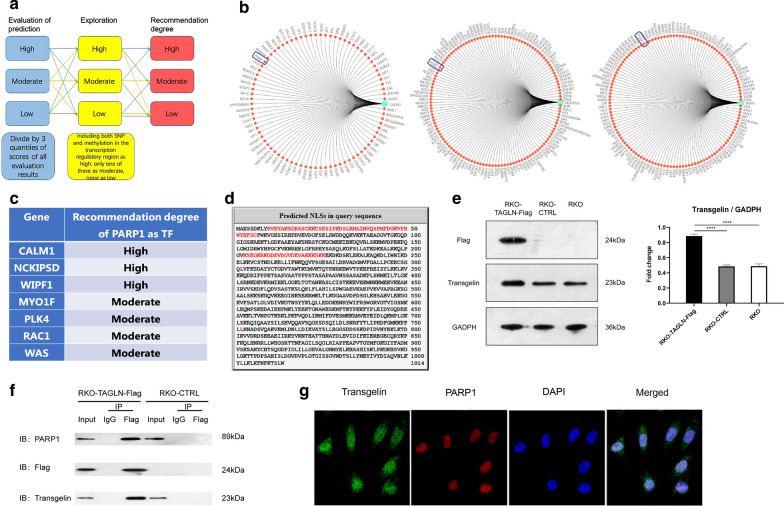
Prediction of the TF for the key genes and validation of its interaction with transgelin
We then analyzed the promoter regions of the seven key genes (CALM1, PLK4, RAC1, WAS, WIPF1, NCKIPSD, MYO1F) to explore if they share the same transcription factor(s) using the GCBI tools in Ensembl, Transfac, COSMIC and dbSNP databases. A computational model was utilized as described from the GCBI website (http://college.gcbi.com.cn/archives/2437) (Fig. 4a). PARP1 was predicted as the transcription factor for the seven key genes (Fig. 4b, c). It was also included in the 23 DNA-binding proteins potentially interacting with the transgelin-flag fusion protein (Table 1). Besides, the cNLS Mapper identified nuclear localization signals in the PARP1 protein (Fig. 4d).
Fig. 4.
Prediction of the transcription factor(s) of the key genes and validation of transgelin-PARP1 interaction. a An illustration of the computational model for predicting the transcription factors of the key genes. b The prediction of the transcription factors of the key genes (partially illustrated). PARP1 protein is circled in a blue frame. c The recommendation degree of PARP1 as the transcription factor of the seven key genes downstream of transgelin. d The sequence of PARP1 protein. The red fonts represent the corresponding sequences of possible nuclear localization signal in PARP1 protein. e Immunoblotting analysis of transgelin and flag protein expression in RKO-TAGLN-FLAG, RKO-CTRL and wild type RKO cells, ****P < 0.0001. f Interaction between the transgelin-flag fusion protein and PARP1 was validated by co-immunoprecipitation. RKO-CTRL was the control group and normal rabbit IgG was used as the control antibody. g Immunofluorescence analysis of transgelin and PARP1 in RKO cells. The panels show transgelin immunostaining (green), PARP1 immunostaining (red), 4′,6-diamidino-2-phenylindole DNA staining (DAPI), and a merged image is indicated
To validate the interaction between PARP1 and transgelin, we transiently transfected pENTER-TAGLN-FLAG and pENTER-Flag control plasmids into the RKO cells. Immunoblotting analysis showed the over-expression of transgelin in RKO-TAGLN-FLAG cells as compared to the RKO-CTRL-FLAG cells (P < 0.0001, Fig. 4e). Immunoprecipitation followed by immunoblotting assays showed that the anti-flag antibody specifically immunoprecipitated PARP1 in the RKO-TAGLN-FLAG cells, validating its binding to the transgelin-flag fusion protein (Fig. 4f). In addition, immunofluorescence analysis indicated that endogenous transgelin was co-localized with PARP1 in the RKO cells (Fig. 4g).
Discussion
Transgelin in colon cancer metastasis
Transgelin is an actin-binding protein presumably existing in the cytoplasm of smooth muscle cells. Findings from our previous study showed that transgelin increased the metastatic potential of colon cancer cells by remodeling the cytoskeleton in the cytoplasm [10]; it also altered the expression of metastasis-related genes, thereby promoting the formation of metastatic phenotypes in the tumor cells [9]. Since many actin-binding proteins have been proven to exert different biological functions in the cytoplasm and nucleus [20–23], we hypothesized that transgelin could play a central role in the invasion and metastasis of colon cancer cells through specific mechanisms in different cellular localization.
In the present study, we found that both endogenous and exogenous transgelin were expressed in the cytoplasm and nucleus of the colon cancer cells (Fig. 1). In addition, transgelin was shown to interact with a variety of metabolic-related enzymes, transport proteins, transcription factors, and cytoskeletal proteins (Additional file 4: Table S1). These results indicate that transgelin is likely to have a nuclear-cytoplasmic shuttling and perform its biological functions in different cellular compartments that collaboratively participate in the invasion and metastasis of the colon cancer cells.
Transgelin and Rho signaling pathway in colon cancer cells
At present, studies on actin and its interacting molecules largely focus on specific signaling pathways, such as the Rho GTPases and its downstream effector proteins, which mediate tumor cell migration, invasion and metastasis through cytoskeleton (reviewed in [24]).
Rho GTPase, a family of 20 small G proteins, interacts with downstream proteins to influence cell cycle, polarity, and migration by regulating the cytoskeleton [25]. In addition, various studies have suggested that an increase in the expression level of the Rho GTPase gene is associated with an increase in cell invasiveness and metastatic phenotype (reviewed in [24]). Rho GTPase interacts with Rho, Rac and Cdc42 in the eukaryotic cells to regulate the assembly and remodeling of the actin cytoskeleton (reviewed in [24]). Rho recruits Rho kinase (ROCK) and phosphorylates various cytoskeletal proteins, thus promoting the formation of actin fiber stress and generating contractile force [26, 27]. ROCK, a major downstream effector of the Rho GTPase family proteins, participates in the regulation of actin remodeling by phosphorylation of the cofilin and myosin light chain (MLC) [26, 27].
Based on the expression profiling data from our previous study [9], we obtained 184 DEGs and identified seven key genes, including CALM1, MYO1F, NCKIPSD, PLK4, RAC1, WAS and WIPF1, downstream of TAGLN using bioinformatics methods. These genes are associated with tumor formation and metastasis [28–36]. Moreover, they have been implicated in the Rho GTPases activation pathway, which could be a major pathway for transgelin to participate in colon cancer metastasis. Although some of the DEGs identified from the same cDNA microarray were validated by quantitative RT-PCR in another cell line (DLD-1) [9], the effects of transgelin on Rho signaling pathway warrant further investigation to fully uncover the underlying mechanisms.
Transgelin interacts with PARP1 in colon cancer cells
Findings from this study confirmed the localization of transgelin in the nucleus of the colon cancer cells. Manipulation of transgelin expression resulted in differential expression of a variety of genes and affected the biological behaviors of the colon cancer cells in vitro and in vivo [9]. Although transgelin potentially interacted with 297 proteins, neither it bound directly to the RNA polymerase II (Additional file 4: Table S1) nor had a nuclear localization signal. Therefore, we speculate that transgelin may interact with other partner(s) to regulate the downstream target genes, thereby affecting colon cancer metastasis. After analyzing the promoter regions of the key genes downstream of TAGLN to predict their potential TF(s) (Fig. 4a–c) and comparing them with the 23 DNA-binding proteins that were interacting with transgelin (Table 1), PARP1 was found to be the only one mapping to both.
Poly (ADP-ribose) polymerase-1, a 113 kDa nuclear enzyme, is encoded by the PARP1 gene [37]. Its N-terminal contains a DNA binding domain consisting of two zinc finger motifs and a nuclear localization sequence [37]. It is involved in DNA repair, cell cycle, cell death, tumorigenesis and other cellular processes [38–41]. PARP1 has been reported to play an important role in the early development and progression of CRC [42, 43]. It has also been found to promote tumor metastasis in soft tissue sarcoma [44] and non-small cell lung cancer [45]. Moreover, Dorsam et al. [46] have shown that PARP1 reduced the N-nitroso compounds (NOC)-induced tumorigenesis, regulated intestinal inflammation through innate immune response and promoted colorectal tumor growth. PARP1 has also been suggested to regulate the transcription of genes by directly binding to their promoters [47–49]. Taken together, these findings imply that PARP1 could be a promising target for malignant tumor intervention.
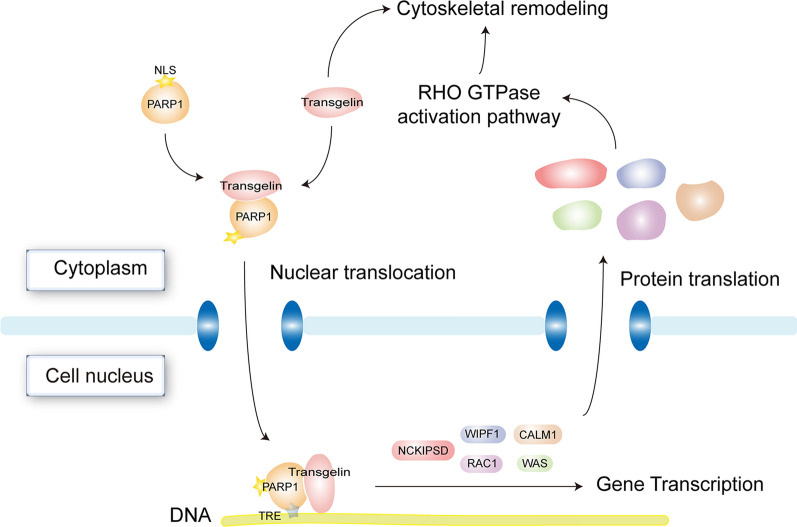
In the current study, we validated the interaction between transgelin and PARP1 with immunoprecipitation and immunofluorescence assays (Fig. 4f, g). Although this study endeavors to delineate the mechanisms of how transgelin and PARP1 interaction influences the Rho signaling pathway and participates in colon cancer metastasis, a proper understanding of these mechanisms warrants more comprehensive analysis. We believe that transgelin functions through a dual mechanism. Firstly, we suppose that transgelin directly takes part in the cytoskeletal remodeling in the cytoplasm following cancer cells signaling from the tumor microenvironment. Secondly, transgelin binds to PARP1 forming a complex that translocates into the nucleus, where the complex regulates the expression of the key genes and subsequently affects the Rho GTPase activation pathway, initiating cytoskeletal remodeling (Fig. 5). The dual mechanism may simultaneously promote colon cancer metastasis.
Fig. 5.
Proposed model of the transgelin mechanisms involved in promoting colon cancer metastasis. Once the cancer cells receive signals from the tumor micro-environment, transgelin directly remodels the cytoskeleton. It also binds to the PARP1 forming a complex which translocates into the nucleus where it regulates the expression of the key genes. Subsequently, the Rho signaling pathway is stimulated and initiates cytoskeletal remodeling which promotes colon cancer metastasis
Conclusions
Our results support a hypothesis that transgelin interacts with PARP1 and regulates the expression of downstream key genes (CALM1, MYO1F, NCKIPSD, PLK4, RAC1, WAS and WIPF1), which are mainly involved in the Rho signaling pathway in the human RKO colon cancer cells.
Supplementary information
Additional file 1: Table S2. PCR primers of the identified key genes.
Additional file 2. The metadata of protein identification with high-performance liquid chromatography/tandem mass spectrometry. The dataset included two sheets; the NC sheet contained proteins identified in the RKO-CTRL-FLAG group, while the TAGLN sheet contained proteins identified in the RKO-TAGLN-FLAG group.
Additional file 3. The metadata of peptide identified with high-performance liquid chromatography/tandem mass spectrometry. The dataset included two sheets; the NC sheet contained peptides identified in the RKO-CTRL-FLAG group, while the TAGLN sheet contained peptides identified in the RKO-TAGLN-FLAG group.
Additional file 4: Table S1. Proteins potentially interacting with a transgelin-flag fusion protein (FDR ≤ 0.01). Proteins that were uniquely present in the RKO-TAGLN-FLAG group were listed after the exclusion of those present both in the RKO-CTRL-FLAG and RKO-TAGLN-FLAG groups.
Acknowledgements
None.
Abbreviations
- CRC
Colorectal cancer
- GCBI
Gene-cloud of biotechnology and information
- TF
Transcription factor
- PARP1
Poly ADP-ribose polymerase-1
- CH
Calmodulin homologous
- CLIK
C-terminal calmodulin like
- TGF β
Transforming growth factor β
- EMT
Epithelial to mesenchymal transition
- MEM
Minimum Eagle’s medium
- RPMI
Roswell Park Memorial Institute
- DAPI
4′,6-diamidino-2-phenylindole
- RT-PCR
Reverse transcription-polymerase chain reaction
- HRP
Horseradish peroxidase
- FASP
Filtered aided proteome preparation
- FDR
False discovery rate
- DEGs
Differential expression genes
- GO
Gene ontology
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- PPI
Protein–protein interaction
- NLS
Nuclear localization signal
- SD
Standard deviation
- WT
Wild type
- ROCK
Rho kinase
- MLC
Myosin light chain
- NOC
Nitroso compound
Authors’ contributions
Substantial contribution to the conception and design of the work: YL, HZ; analysis and interpretation of the data: ZL, YF, ZY; drafting the manuscript: ZL, WZ, and ZY; revising the work critically for important intellectual content: XY, DC, SL, and LC; collecting of grants: YL and HZ. All authors read and approval the final manuscript.
Funding
The National Natural Science Foundation of China (Grant No. 81641179, YL; No. 81602110, HZ) and the Natural Science Foundation of Guangdong Province (Grant No. 2017A030313603, YL) supported the present study. Grant (2013) 163 from the Key Laboratory of Malignant Tumor Molecular Mechanism and Translational Medicine of Guangzhou Bureau of Science and Information Technology, and Grant No. KLB09001 from the Key Laboratory of Malignant Tumor Gene Regulation and Target Therapy of Guangdong Higher Education Institutes also supported the present study.
Availability of data and materials
The microarray datasets analyzed in this manuscript have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE48998 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE48998). Other datasets generated and/or analyzed during the current study are included within the article and its additional files.
Ethics approval and consent to participate
All procedures performed in studies were in accordance with the ethical standards of Sun Yat-sen Memorial Hospital. Written informed consents were obtained from all individual participants included in the study.
Consent for publication
Not applicable.
Competing interests
The authors declare that there are no competing interests associated with the manuscript.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Zhen-xian Lew and Hui-min Zhou contributed equally to this work
Contributor Information
Zhen-xian Lew, Email: zhenxian.liu@concordmedical.com.
Hui-min Zhou, Email: 453867737@qq.com.
Yuan-yuan Fang, Email: 517996747@qq.com.
Zhen Ye, Email: yezh6@mail2.sysu.edu.cn.
Wa Zhong, Email: zhongwazs@163.com.
Xin-yi Yang, Email: xinyi_65@hotmail.com.
Zhong Yu, Email: yuzhong.gd@163.com.
Dan-yu Chen, Email: 994402997@qq.com.
Si-min Luo, Email: luo950711@163.com.
Li-fei Chen, Email: lifei.chen111@outlook.com.
Ying Lin, Email: linwy@mail.sysu.edu.cn.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s12935-020-01461-y.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 3.Fife CM, McCarroll JA, Kavallaris M. Movers and shakers: cell cytoskeleton in cancer metastasis. Br J Pharmacol. 2014;171(24):5507–5523. doi: 10.1111/bph.12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139(5):871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15(3):178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fu Y, Liu HW, Forsythe SM, Kogut P, McConville JF, Halayko AJ, et al. Mutagenesis analysis of human SM22: characterization of actin binding. J Appl Physiol. 2000;89(5):1985–1990. doi: 10.1152/jappl.2000.89.5.1985. [DOI] [PubMed] [Google Scholar]
- 7.Assinder SJ, Stanton JA, Prasad PD. Transgelin: an actin-binding protein and tumour suppressor. Int J Biochem Cell Biol. 2009;41(3):482–486. doi: 10.1016/j.biocel.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 8.Lee EK, Han GY, Park HW, Song YJ, Kim CW. Transgelin promotes migration and invasion of cancer stem cells. J Proteome Res. 2010;9(10):5108–5117. doi: 10.1021/pr100378z. [DOI] [PubMed] [Google Scholar]
- 9.Zhou HM, Fang YY, Weinberger PM, Ding LL, Cowell JK, Hudson FZ, et al. Transgelin increases metastatic potential of colorectal cancer cells in vivo and alters expression of genes involved in cell motility. BMC Cancer. 2016;16:55. doi: 10.1186/s12885-016-2105-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou H, Zhang Y, Chen Q, Lin Y. AKT and JNK signaling pathways increase the metastatic potential of colorectal cancer cells by altering transgelin expression. Dig Dis Sci. 2016;61(4):1091–1097. doi: 10.1007/s10620-015-3985-1. [DOI] [PubMed] [Google Scholar]
- 11.Zheng B, Han M, Bernier M, Wen JK. Nuclear actin and actin-binding proteins in the regulation of transcription and gene expression. FEBS J. 2009;276(10):2669–2685. doi: 10.1111/j.1742-4658.2009.06986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu L, Gao Y, Chen Y, Xiao Y, He Q, Qiu H, et al. Quantitative proteomics reveals that distant recurrence-associated protein R-Ras and Transgelin predict post-surgical survival in patients with Stage III colorectal cancer. Oncotarget. 2016;7(28):43868–43893. doi: 10.18632/oncotarget.9701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elsafadi M, Manikandan M, Almalki S, Mahmood A, Shinwari T, Vishnubalaji R, et al. Transgelin is a poor prognostic factor associated with advanced colorectal cancer (CRC) stage promoting tumor growth and migration in a TGFbeta-dependent manner. Cell Death Dis. 2020;11(5):341. doi: 10.1038/s41419-020-2529-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li WM, Wu WJ. Transgelin in bladder cancer: a potential biomarker and therapeutic target. EBioMedicine. 2019;48:16–17. doi: 10.1016/j.ebiom.2019.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin Y, Buckhaults PJ, Lee JR, Xiong H, Farrell C, Podolsky RH, et al. Association of the actin-binding protein transgelin with lymph node metastasis in human colorectal cancer. Neoplasia. 2009;11(9):864–873. doi: 10.1593/neo.09542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43(Database issue):D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.UniProt C. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kosugi S, Hasebe M, Tomita M, Yanagawa H. Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci USA. 2009;106(25):10171–10176. doi: 10.1073/pnas.0900604106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aksenova V, Turoverova L, Khotin M, Magnusson KE, Tulchinsky E, Melino G, et al. Actin-binding protein alpha-actinin 4 (ACTN4) is a transcriptional co-activator of RelA/p65 sub-unit of NF-kB. Oncotarget. 2013;4(2):362–372. doi: 10.18632/oncotarget.901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng B, Wen JK, Han M, Zhou AR. hhLIM protein is involved in cardiac hypertrophy. Biochim Biophys Acta. 2004;1690(1):1–10. doi: 10.1016/j.bbadis.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 22.Zheng B, Han M, Wen JK, Zhang R. Human heart LIM protein activates atrial-natriuretic-factor gene expression by interacting with the cardiac-restricted transcription factor Nkx2.5. Biochem J. 2008;409(3):683–690. doi: 10.1042/BJ20070977. [DOI] [PubMed] [Google Scholar]
- 23.Yang X, Lin Y. Functions of nuclear actin-binding proteins in human cancer. Oncol Lett. 2018;15(3):2743–2748. doi: 10.3892/ol.2017.7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karlsson R, Pedersen ED, Wang Z, Brakebusch C. Rho GTPase function in tumorigenesis. Biochim Biophys Acta. 2009;1796(2):91–98. doi: 10.1016/j.bbcan.2009.03.003. [DOI] [PubMed] [Google Scholar]
- 25.Jaffe AB, Hall A. Rho GTPases: biochemistry and biology. Annu Rev Cell Dev Biol. 2005;21:247–269. doi: 10.1146/annurev.cellbio.21.020604.150721. [DOI] [PubMed] [Google Scholar]
- 26.Alexander SP, Benson HE, Faccenda E, Pawson AJ, Sharman JL, Spedding M, et al. The concise guide to pharmacology 2013/14: enzymes. Br J Pharmacol. 2013;170(8):1797–1867. doi: 10.1111/bph.12451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hall A. Rho family GTPases. Biochem Soc Trans. 2012;40(6):1378–1382. doi: 10.1042/BST20120103. [DOI] [PubMed] [Google Scholar]
- 28.Taylor MD, Sadhukhan S, Kottangada P, Ramgopal A, Sarkar K, D’Silva S, et al. Nuclear role of WASp in the pathogenesis of dysregulated TH1 immunity in human Wiskott-Aldrich syndrome. Sci Transl Med. 2010;2(37):37ra44. doi: 10.1126/scitranslmed.3000813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schrank BR, Aparicio T, Li Y, Chang W, Chait BT, Gundersen GG, et al. Nuclear ARP2/3 drives DNA break clustering for homology-directed repair. Nature. 2018;559(7712):61–66. doi: 10.1038/s41586-018-0237-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cao R, Chen J, Zhang X, Zhai Y, Qing X, Xing W, et al. Elevated expression of myosin X in tumours contributes to breast cancer aggressiveness and metastasis. Br J Cancer. 2014;111(3):539–550. doi: 10.1038/bjc.2014.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arjonen A, Kaukonen R, Mattila E, Rouhi P, Hognas G, Sihto H, et al. Mutant p53-associated myosin-X upregulation promotes breast cancer invasion and metastasis. J Clin Invest. 2014;124(3):1069–1082. doi: 10.1172/JCI67280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rahimi N, Rezazadeh K, Mahoney JE, Hartsough E, Meyer RD. Identification of IGPR-1 as a novel adhesion molecule involved in angiogenesis. Mol Biol Cell. 2012;23(9):1646–1656. doi: 10.1091/mbc.E11-11-0934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berchtold MW, Villalobo A. The many faces of calmodulin in cell proliferation, programmed cell death, autophagy, and cancer. Biochim Biophys Acta. 2014;1843(2):398–435. doi: 10.1016/j.bbamcr.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 34.Chin D, Means AR. Calmodulin: a prototypical calcium sensor. Trends Cell Biol. 2000;10(8):322–328. doi: 10.1016/s0962-8924(00)01800-6. [DOI] [PubMed] [Google Scholar]
- 35.Kazazian K, Go C, Wu H, Brashavitskaya O, Xu R, Dennis JW, et al. Plk4 promotes cancer invasion and metastasis through Arp2/3 complex regulation of the actin cytoskeleton. Cancer Res. 2017;77(2):434–447. doi: 10.1158/0008-5472.CAN-16-2060. [DOI] [PubMed] [Google Scholar]
- 36.Rosario CO, Kazazian K, Zih FS, Brashavitskaya O, Haffani Y, Xu RS, et al. A novel role for Plk4 in regulating cell spreading and motility. Oncogene. 2015;34(26):3441–3451. doi: 10.1038/onc.2014.275. [DOI] [PubMed] [Google Scholar]
- 37.Ha HC, Snyder SH. Poly(ADP-ribose) polymerase-1 in the nervous system. Neurobiol Dis. 2000;7(4):225–239. doi: 10.1006/nbdi.2000.0324. [DOI] [PubMed] [Google Scholar]
- 38.Jubin T, Kadam A, Jariwala M, Bhatt S, Sutariya S, Gani AR, et al. The PARP family: insights into functional aspects of poly (ADP-ribose) polymerase-1 in cell growth and survival. Cell Prolif. 2016;49(4):421–437. doi: 10.1111/cpr.12268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matveeva EA, Al-Tinawi QMH, Rouchka EC, Fondufe-Mittendorf YN. Coupling of PARP1-mediated chromatin structural changes to transcriptional RNA polymerase II elongation and cotranscriptional splicing. Epigenetics Chromatin. 2019;12(1):15. doi: 10.1186/s13072-019-0261-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ke Y, Wang C, Zhang J, Zhong X, Wang R, Zeng X, et al. The role of PARPs in inflammation-and metabolic-related diseases: molecular mechanisms and beyond. Cells. 2019;8(9):1047. doi: 10.3390/cells8091047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Alshammari AH, Shalaby MA, Alanazi MS, Saeed HM. Novel mutations of the PARP-1 gene associated with colorectal cancer in the Saudi population. Asian Pac J Cancer Prev. 2014;15(8):3667–3673. doi: 10.7314/apjcp.2014.15.8.3667. [DOI] [PubMed] [Google Scholar]
- 42.Li M, Threadgill MD, Wang Y, Cai L, Lin X. Poly(ADP-ribose) polymerase inhibition down-regulates expression of metastasis-related genes in CT26 colon carcinoma cells. Pathobiology. 2009;76(3):108–116. doi: 10.1159/000209388. [DOI] [PubMed] [Google Scholar]
- 43.Nosho K, Yamamoto H, Mikami M, Taniguchi H, Takahashi T, Adachi Y, et al. Overexpression of poly(ADP-ribose) polymerase-1 (PARP-1) in the early stage of colorectal carcinogenesis. Eur J Cancer. 2006;42(14):2374–2381. doi: 10.1016/j.ejca.2006.01.061. [DOI] [PubMed] [Google Scholar]
- 44.Bertucci F, Finetti P, Monneur A, Perrot D, Chevreau C, Le Cesne A, et al. PARP1 expression in soft tissue sarcomas is a poor-prognosis factor and a new potential therapeutic target. Mol Oncol. 2019;13(7):1577–1588. doi: 10.1002/1878-0261.12522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen K, Li Y, Xu H, Zhang C, Li Z, Wang W, et al. An analysis of the gene interaction networks identifying the role of PARP1 in metastasis of non-small cell lung cancer. Oncotarget. 2017;8(50):87263–87275. doi: 10.18632/oncotarget.20256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dorsam B, Seiwert N, Foersch S, Stroh S, Nagel G, Begaliew D, et al. PARP-1 protects against colorectal tumor induction, but promotes inflammation-driven colorectal tumor progression. Proc Natl Acad Sci USA. 2018;115(17):E4061–E4070. doi: 10.1073/pnas.1712345115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ding L, Chen X, Xu X, Qian Y, Liang G, Yao F, et al. PARP1 suppresses the transcription of PD-L1 by Poly(ADP-Ribosyl)ating STAT3. Cancer Immunol Res. 2019;7(1):136–149. doi: 10.1158/2326-6066.CIR-18-0071. [DOI] [PubMed] [Google Scholar]
- 48.Tokarz P, Ploszaj T, Regdon Z, Virag L, Robaszkiewicz A. PARP1-LSD1 functional interplay controls transcription of SOD2 that protects human pro-inflammatory macrophages from death under an oxidative condition. Free Radic Biol Med. 2019;131:218–224. doi: 10.1016/j.freeradbiomed.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 49.Wang C, Xu W, Zhang Y, Zhang F, Huang K. PARP1 promote autophagy in cardiomyocytes via modulating FoxO3a transcription. Cell Death Dis. 2018;9(11):1047. doi: 10.1038/s41419-018-1108-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Table S2. PCR primers of the identified key genes.
Additional file 2. The metadata of protein identification with high-performance liquid chromatography/tandem mass spectrometry. The dataset included two sheets; the NC sheet contained proteins identified in the RKO-CTRL-FLAG group, while the TAGLN sheet contained proteins identified in the RKO-TAGLN-FLAG group.
Additional file 3. The metadata of peptide identified with high-performance liquid chromatography/tandem mass spectrometry. The dataset included two sheets; the NC sheet contained peptides identified in the RKO-CTRL-FLAG group, while the TAGLN sheet contained peptides identified in the RKO-TAGLN-FLAG group.
Additional file 4: Table S1. Proteins potentially interacting with a transgelin-flag fusion protein (FDR ≤ 0.01). Proteins that were uniquely present in the RKO-TAGLN-FLAG group were listed after the exclusion of those present both in the RKO-CTRL-FLAG and RKO-TAGLN-FLAG groups.
Data Availability Statement
The microarray datasets analyzed in this manuscript have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE48998 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE48998). Other datasets generated and/or analyzed during the current study are included within the article and its additional files.