Abstract
Objective:
In AIDS Clinical Trials Group study A5316, efavirenz significantly lowered plasma concentrations of etonogestrel and ethinyl estradiol, given as a vaginal ring, while atazanavir/ritonavir increased etonogestrel and lowered ethinyl estradiol concentrations. We characterized the pharmacogenetics of these interactions.
Methods:
In A5316, women with HIV enrolled into control (no antiretrovirals), efavirenz (600 mg daily with nucleoside/nucleotide reverse transcriptase inhibitors (NRTIs)), and atazanavir/ritonavir (300/100mg daily with NRTIs) groups. On day 0, a vaginal ring was inserted, releasing etonogestrel/ethinyl estradiol 120/15 mcg/day. Intensive plasma sampling for antiretrovirals was obtained on days 0 and 21, and single samples for etonogestrel and ethinyl estradiol were obtained on days 7, 14 and 21. Seventeen genetic polymorphisms were analyzed.
Results:
The 72 participants in this analysis included 25, 24 and 23 in the control, efavirenz and atazanavir/ritonavir group, respectively. At day 21 in the efavirenz group, CYP2B6 genotype was associated with increased plasma efavirenz exposure (p=3.2×10−3), decreased plasma concentrations of etonogestrel (p=1.7×10−3), and decreased ethinyl estradiol (p=6.7×10−4). Compared to controls, efavirenz reduced median etonogestrel concentrations by at least 93% in CYP2B6 slow metabolizers versus ~75% in normal and intermediate metabolizers. Efavirenz reduced median ethinyl estradiol concentrations by 75% in CYP2B6 slow metabolizers versus ~41% in normal and intermediate metabolizers. No other polymorphisms were significantly associated with hormone or antiretroviral pharmacokinetics.
Conclusions:
CYP2B6 slow metabolizer genotype worsens the pharmacokinetic interaction of efavirenz with hormonal contraceptives administered by vaginal ring. Efavirenz dose reduction in CYP2B6 slow metabolizers may reduce, but will likely not eliminate, this interaction.
Introduction
It is important that women of reproductive potential who are living with HIV be provided with effective contraceptive options. Unfortunately, drug-drug interactions between some antiretroviral therapies (ART) and hormonal contraceptives hinder available options for effective family planning. Efavirenz-containing ART significantly lowers plasma exposure to progestin contraceptives given orally or via subdermal implant, [1–5] which may reduce hormonal contraception effectiveness.[6] HIV protease inhibitors that are combined with ritonavir increase progestin exposure, but may decrease estrogen exposure.[7, 8] These drug-induced changes in plasma hormone exposure are likely due to antiretroviral effects on hormone metabolism by cytochrome P450 (CYP) 3A4, the isoenzyme believed to mediate progestin metabolism, and multiple CYP and glucuronidation pathways associated with estrogen metabolism.[8] In addition, some studies have identified somewhat lower antiretroviral exposure when combined with hormonal contraceptives,[7, 8] possibly due to progestin and estrogen influences on drug metabolizing enzymes.[9, 10]
Vaginal rings are currently available for hormone replacement therapy and for contraception, and in development as vehicles to simultaneously deliver antiretrovirals for HIV prevention and progestins for contraception. Progestins delivered by combined hormonal contraceptive vaginal rings reach systemic concentrations sufficient to prevent ovulation, similar to oral progestins. Protocol A5316 of the AIDS Clinical Trials Group (ACTG) characterized effects of efavirenz- and atazanavir/ritonavir (atazanavir/r)-containing ART on plasma pharmacokinetics (PK) of etonogestrel and ethinyl estradiol administered by vaginal ring over 21 days.[11] On Day 21, concomitant efavirenz was associated with 79% lower etonogestrel and 59% lower ethinyl estradiol concentrations compared to the control group not on ART, while concomitant atazanavir/r was associated with 71% higher etonogestrel and 38% lower ethinyl estradiol concentrations. There were no statistically significant changes in plasma efavirenz or atazanavir exposure.
There is extensive literature regarding the pharmacogenetics of efavirenz [12]. Frequent CYP2B6 polymorphisms predict increased plasma efavirenz exposure, including CYP2B6 516G→T (rs3745274) [13–17], 983T→C (rs28399499) [17–20], and 15582C→T (rs4803419) [17]. A CYP2A6 polymorphism, −48T→G (rs28399433), may also affect efavirenz pharmacokinetics [21–24] when present with CYP2B6 slow metabolizer genotypes [21, 24]. These polymorphisms explain approximately 10-fold greater plasma efavirenz exposure in slow versus normal metabolizers [17]. CYP2B6 slow metabolizer genotypes (i.e., two copies of decreased-function or no-function alleles) are present in approximately 30% of Asians, 25% of Africans, and 5% of Europeans.
The present study characterized the impact of known functional pharmacogenetic variants on drug-drug interactions between ART and plasma hormone exposure among A5316 participants.
Methods
Protocol A5316 (NCT01903031) was a multisite, international, non-randomized, open-label, three-group parallel PK study among women living with HIV. Primary results are described elsewhere [11]. All study procedures followed the Declaration of Helsinki and were approved by ethics boards at each participating clinical site. Eligible participants were at least 16 years of age, were of reproductive potential, were willing to use a second, non-hormonal form of contraception during the study period, and agreed to avoid concurrent hormonal therapies or interacting medications.
Participants enrolled between December 2014 and September 2016, and were assigned to one of three study groups based on ART use at screening. The control group comprised women who had not yet begun ART with CD4+ T-cell counts of at least 350 cells/mm3. The efavirenz group comprised women receiving efavirenz 600 mg daily plus at least two nucleoside/nucleotide reverse transcriptase inhibitors (NRTI), and the atazanavir/ritonavir (atazanavir/r) group comprised women receiving atazanavir/r 300mg/100mg daily, tenofovir 300mg daily, plus at least one additional NRTI. Women on ART were required to be on stable ART for at least 30 days and have screening plasma HIV-1 RNA ≤400 copies/mL. For all participants, a vaginal ring releasing etonogestrel/ethinyl estradiol 120/15 μg/day was inserted on day 0. Prior to placement of the vaginal ring on day 0, participants in the ART groups underwent intensive 8-hour PK sampling to analyze efavirenz, atazanavir and ritonavir. With an observed ART dose, plasma for antiretroviral assays were collected pre-dose and 1, 3, 4, 5, and 8 hours post-dose. Participants returned on days 7, 14 and 21 for single plasma samples for hormone PK (etonogestrel and ethinyl estradiol). On day 21, 8-hour ART PK sampling was repeated before removing the vaginal ring. Adherence to ART and the vaginal ring were evaluated by self-report.[25] The 8-hour PK sampling was rescheduled if any of the prior three ART doses were missed, or if the vaginal ring was outside of the body during a specified time leading up to the PK sampling. This time (12 hours) was based on the known PK of the hormones, time to appropriate concentrations, and half-life. Hormone and antiretroviral concentrations were analyzed by LC-MS/MS. All PK assays were validated in accordance with guidance from the Food and Drug Administration.[26] Efavirenz, atazanavir, and ritonavir PK parameters were estimated using Phoenix WinNonLin® version 7.0 (Certara USA, Inc., Princeton, NJ). AUC was calculated using non-compartmental methods over the 8-hour intensive PK sampling (AUC0–8h). Resultant PK parameters were log10 transformed to approximate normality.
Characterization of Genetic Polymorphisms
We genotyped polymorphisms that are known to predict plasma efavirenz PK (11–15). For efavirenz, CYP2B6 983T→C and 15582C→T, and CYP2A6 rs28399433 were assayed using MassARRAY® iPLEX Gold (Sequenom Inc., San Diego, California, USA), while CYP2B6 516G→T was genotyped by Taqman. The CYP3A5*3 variant 6986A→G (rs776746) (22) was genotyped by MassARRAY® iPLEX Gold. For atazanavir, UGT1A1 rs887829 which is associated with bilirubin levels was genotyped [27]. No SNPs are known to be associated with atazanavir/r PK [27]. For etonogestrel and ethinyl estradiol SNPs were genotyped that have been genome-wide associated with estradiol phenotypes, including rs1864729 [28] , rs2414095, rs2445762 [29], rs117585797 [30], rs727428 [31]. For CYP3A4 we genotyped rs17277546 [32], rs34670419 [30], rs62471956 [33], rs34670419 [30], and CYP3A4 SNPs associated with changes in in vivo activity, rs28371759, rs35599367 [34]. For CYP3A5 we genotyped rs4646450 [35] and rs776746 [34]. For CYP1A1 we genotyped rs2470893 [36], rs2472297 [37], rs2470893 [38], which have been associated with various traits in genome-wide association studies. For CYP1A2 we genotyped SNPs associated with altered in vivo activity, including rs2069514, rs762551, rs56276455, rs72547516, rs28399424, and rs56107638 [34].
Human DNA was extracted from whole blood. Genotyping was done in the Vanderbilt DNA Resources Core using MassARRAY® iPLEX Gold (Agena Bioscience™, California, USA) and Taqman (ThermoFisher Scientific, Massachusetts, USA). Final Sequenom assay design is available upon request. Laboratory personnel with no knowledge of clinical data performed genotyping. Ample duplicate and blank assays were included to assure validity, and all samples were assayed in duplicate. Genotyping efficiency was 100% for all SNPs. Of 27 SNPs assayed, we excluded 10 with minor allele frequencies less than 5%, leaving 17 for association analyses.
Statistical analysis
The population for analysis of antiretroviral exposure comprised participants for whom intensive ARV sampling was conducted on both days 0 and 21, and who had genetic data available. Genetic associations were assessed using linear regression models. Statistical analyses for associations between PK parameters and composite CYP2B6 genotype levels were performed with STATA version 15.1 (StataCorp, College Station, Texas, USA). Composite CYP2B6 genotype that predicts 12 levels of progressively higher plasma efavirenz exposure was defined based on combinations of four SNPs as follows: normal metabolizer genotype (1: 15582CC-516GG-983TT or 2: 15582CT-516GG-983TT); intermediate metabolizer genotype (3: 15582TT-516GG-983TT; 4: 15582CC-516GT-983TT; 5: 15582CC-516GG-983CT; 6: 15582CT-516GT-983TT; or 7: 15582CT-516GG-983CT); and slow metabolizer genotype (8: 15582CC-516TT-983TT; 9: 15582CC-516GT-983CT; or 10: 15582CC-516GG-983CC) [17]. With slow metabolizer genotypes, two additional composite genotypes were defined by the presence of a CYP2A6 SNP as follows: 11: −48GT; and 12: −48GG. Statistical analyses for associations with SNPs were performed with PLINK version 1.07 [39]. For composite CYP2B6 genotype associations in the efavirenz group we did not correct for multiple comparisons because the primary analysis focused on this genotype and etonogestrel and ethinyl estradiol concentrations in the efavirenz group. this was the primary focus of analyses. For other SNPs we corrected for 17 comparisons, giving a significance cut-off of P = 0.0029. For CYP2B6, directionality of β coefficients considers CYP2B6 normal metabolizer genotype as the reference (i.e., a positive β indicates that CYP2B6 slow metabolizer genotype is associated with a greater PK parameter value). Two-sided tests were used for all analyses. All SNPs were in Hardy-Weinberg equilibrium after correcting for multiple comparisons. Only one SNP, CYP2B6 rs3745274, deviated nominally from Hardy-Weinberg equilibrium (p = 0.041).
Results
Study participants
The total group for analysis comprised 72 participants with both PK and genotype data, and included 25 in the control group, 24 in the efavirenz group, and 23 in the atazanavir/r group. Participants enrolled at ACTG sites in Asia, South America, Sub-Saharan Africa, and the United States. In the total group, median age was 34 years, 35 (49%) self-identified as Black, 26 (36%) reported Hispanic ethnicity, and median baseline weight was 67.5 kg. Characteristic of participants by study group are provided in Table 1. Black participants were overrepresented in the efavirenz group, Asian/Pacific Islander participants overrepresented in the atazanavir/r group, and CYP2B6 slow metabolizer genotypes underrepresented in the atazanavir/r group. Frequencies of non-CYP2B6 SNPs by study group are available in Supplemental Online Material.
Table 1.
Baseline characteristics of participants included in genetic association analyses
| Total (n=72) | Control Group (n=25) | Efavirenz Group (n=24) | Atazanavir/r Group (n=23) | |
|---|---|---|---|---|
| Age in years, median (range) | 34.0 (22 – 55) | 31 (22 – 48) | 36 (24 – 55) | 37 (24 – 48) |
| Sex (female); n (%) | 72 (100) | 25 (100) | 24 (100) | 23 (100) |
| Race/Ethnicity; n (%) | ||||
| White | 3 (4) | 1 (4) | 1 (4) | 1 (4) |
| Black | 35 (49) | 11 (44) | 16 (64) | 9 (39) |
| Asian/ Pacific Islandera | 8 (11) | 3 (12) | 0 (0) | 5 (22) |
| Hispanic | 26 (36) | 10 (40) | 8 (32) | 8 (35) |
| Weight in kg, median (range) | 67.5 (36.9 – 170.6) | 66.8 (36.9 – 112.9) | 68.9 (46.5 – 170.6) | 64 (47.3 – 152.4) |
| CYP2B6 metabolizer genotype; n (%) | ||||
| normal | 22 (31) | 5 (20) | 9 (38) | 8 (35) |
| intermediate | 32 (44) | 12 (48) | 7 (29) | 13 (57) |
| slow | 18 (25) | 8 (32) | 8 (33) | 2 (9) |
defined based on NIH policy on reporting race and ethnicity data.
CYP2B6/CYP2A6 associations with plasma efavirenz PK parameters
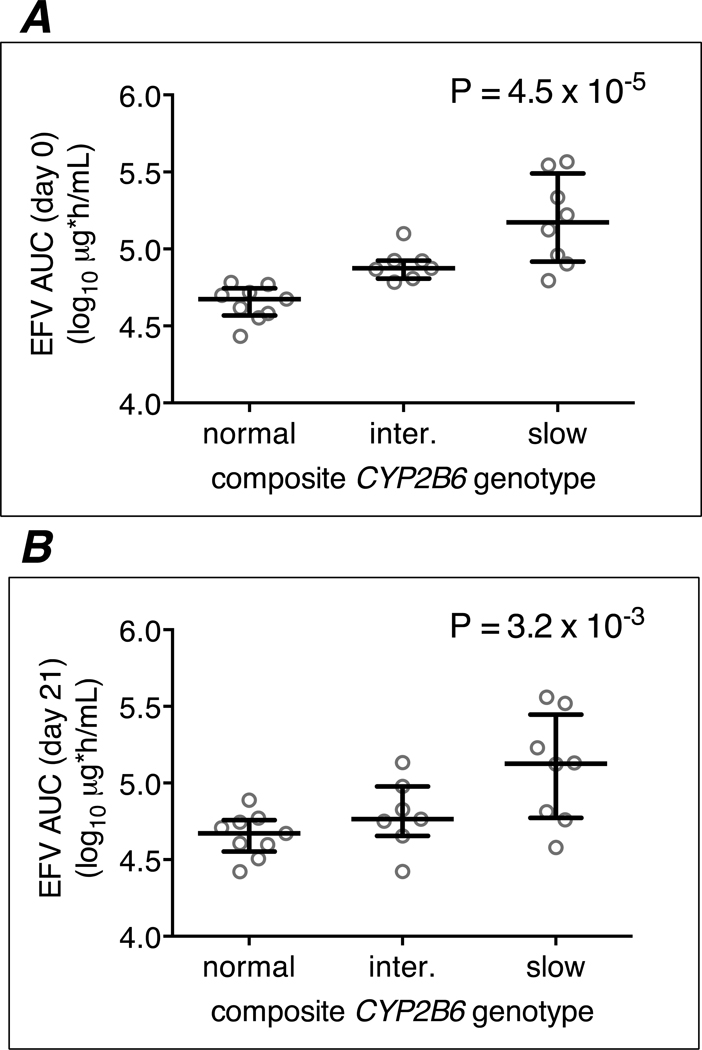
As expected, among the 24 efavirenz group participants, CYP2B6 slow metabolizer genotypes were associated with higher plasma efavirenz log10 AUC0–8h, log10 Cmax and log10 Cmin values at days 0 and 21. For example, for CYP2B6 genotype (stratified into 3 levels) and log10 plasma efavirenz AUC0–8h at day 0: β coefficient 0.27, P = 4.5 × 10−5. Relationships between CYP2B6/CYP2A6 genotype, stratified into 3 genotype levels, and log10 plasma efavirenz AUC0–8h values at days 0 and 21, are presented in Figure 1. These relationships stratified into 12 CYP2B6/CYP2A6 genotype levels are presented in Supplemental On-line Material. Relationships between CYP2B6 genotype and each efavirenz PK parameter are presented in Table 2. These genetic associations persisted after adjusting for weight and/or age (data not shown).
Figure 1. Relationships between CYP2B6/CYP2A6 genotype levels and plasma efavirenz log10 AUC0–8h values among 24 efavirenz group participants.
Panel A: associations at day 0 with CYP2B6/CYP2A6 genotype stratified into 3 levels; Panel B: associations at day 21 with CYP2B6/CYP2A6 genotype stratified into 3 levels. Error bars indicate median and interquartile range. Linear regression model P-values are shown. EFV = efavirenz; AUC = area under the concentration-time curve.
Table 2.
Relationships between CYP2B6 genotype levels, plasma efavirenz pharmacokinetic parameters, and hormone concentrations among the 24 efavirenz group participants.
| Pharmacokinetic parameter | day | CYP2B6 Normal median (IQR) | CYP2B6 Intermediate median (IQR) | CYP2B6 Slow median (IQR) | β coefficient, P-valuea |
|---|---|---|---|---|---|
| efavirenz | |||||
| AUC0–8h (log10 ng*h/mL) | 0 | 4.67 (4.57 – 4.74) | 4.87 (4.81 – 4.92) | 5.17 (4.92 – 5.49) | 0.27, 4.5 ×10−5 |
| 21 | 4.67 (4.55 – 4.76) | 4.76 (4.65 – 4.98) | 5.13 (4.77 – 5.45) | 0.21, 3.2 ×10−3 | |
| Cmin (log10 ng/mL) | 0 | 3.12 (3.00 – 3.22) | 3.39 (3.33 – 3.42) | 3.59 (3.33 – 4.07) | 0.27, 4.3 ×10−4 |
| 21 | 3.09 (2.93 – 3.16) | 3.30 (3.09 – 3.37) | 3.42 (3.29 – 3.91) | 0.32, 3.0 ×10−2 | |
| Cmax (log10 ng/mL) | 0 | 3.49 (3.40 – 3.61) | 3.68 (3.60 – 3.75) | 3.91 (3.71 – 4.19) | 0.21, 7.0 ×10−4 |
| 21 | 3.51 (3.41 – 3.67) | 3.60 (3.54 – 3.78) | 3.89 (3.53 – 4.17) | 0.16, 1.3 ×10−2 | |
| etonogestrel | |||||
| concentration (log10 pg/mL) | 21 | 2.70 (2.64 – 2.85) | 2.64 (2.54 – 2.82) | 2.10 (2.10 – 2.55) | −0.21, 1.7 ×10−3 |
| ethinyl estradiol | |||||
| concentration (log10 pg/mL) | 21 | 1.09 (1.06 – 1.12) | 1.09 (0.99 – 1.16) | 0.72 (0.40 – 0.92) | −0.19, 6.7 ×10−4 |
Abbreviations: AUC, area under the concentration time curve; Cmax, maximum concentration; Cmin, minimum concentration.
P-values and beta coefficients were generated with linear regression models.
CYP2B6/CYP2A6 and etonogestrel and ethinyl estradiol concentrations
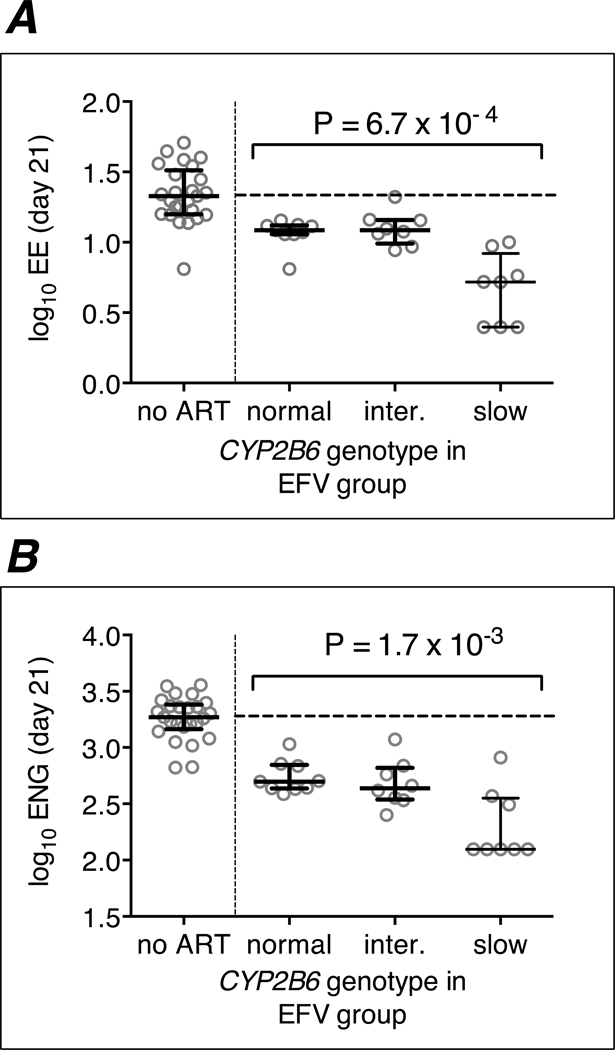
Relationships between CYP2B6/CYP2A6 genotype, stratified into 3 genotype levels, and log10 ethinyl estradiol and log10 ethinyl estradiol at day 21 are presented in Figure 2. These relationships stratified into 12 CYP2B6/CYP2A6 genotype levels are presented in Supplemental On-line Material. Detailed information regarding CYP2B6 genotype, stratified into 3 genotype levels, log10 ethinyl estradiol and log10 ethinyl estradiol are presented in Table 2. Among the 24 efavirenz group participants, CYP2B6 slow metabolizer genotypes were associated with significantly lower plasma concentrations of both etonogestrel and ethinyl estradiol at day 21. For example, considering CYP2B6 genotype (stratified into 3 levels) and log10 plasma etonogestrel concentrations, the β coefficient was −0.21, P = 1.7 × 10−3, and considering log10 plasma ethinyl estradiol concentrations, the β coefficient was −0.19, P = 6.7 × 10−4. These genetic associations persisted after adjusting for weight and/or age (data not shown). Plasma hormone concentrations below limits of quantification on day 21 occurred only among participants with CYP2B6 slow metabolizer genotypes, including 3 participants with plasma ethinyl estradiol concentrations below 5 pg/mL, and 5 participants with plasma etonogestrel concentrations below 25 pg/mL (Figure 2).
Figure 2. Relationships between CYP2B6/CYP2A6 genotype levels and day 21 log10 etonogestrel and ethinyl estradiol concentrations among the 24 efavirenz group participants.
Panel A: associations of CYP2B6/CYP2A6 genotype stratified into 3 levels and log10 ethinyl estradiol concentrations at day 21; Panel B: associations of CYP2B6/CYP2A6 genotype stratified into 3 levels and log10 etonogestrel concentrations at day 21. Error bars indicate median and interquartile range. Linear regression model P-values are shown. ENG = etonogestrel; EE = ethinyl estradiol; EFV = efavirenz. Data from 25 individuals not receiving antiretriviral therapy (ART) are included to the left.
On Day 21, the median log10 etonogestrel concentration was 3.27 pg/mL in the non-ART control group, but was 2.70 pg/mL, 2.64 pg/mL, and at most 2.10 pg/mL in CYP2B6 normal, intermediate, and slow metabolizers, respectively, which represent 73%, 77% and at least 93% reductions in median absolute etonogestrel concentrations.
On Day 21, the median log10 ethinyl estradiol concentration was 1.33 pg/mL in the non-ART control group, but was 1.10 pg/mL, 1.10 pg/mL, and 0.72 pg/mL in CYP2B6 normal, intermediate, and slow metabolizers, respectively, which represent 41%, 41%, and at least 75% reductions in median absolute ethinyl estradiol concentrations.
Within the control group and atazanavir/r group, each group analyzed separately, there were no associations between CYP2B6/CYP2A6 slow metabolizer genotypes and plasma concentrations of either etonogestrel or ethinyl estradiol at day 21 (P > 0.05 for each model).
Plasma efavirenz PK, etonogestrel and ethinyl estradiol concentrations
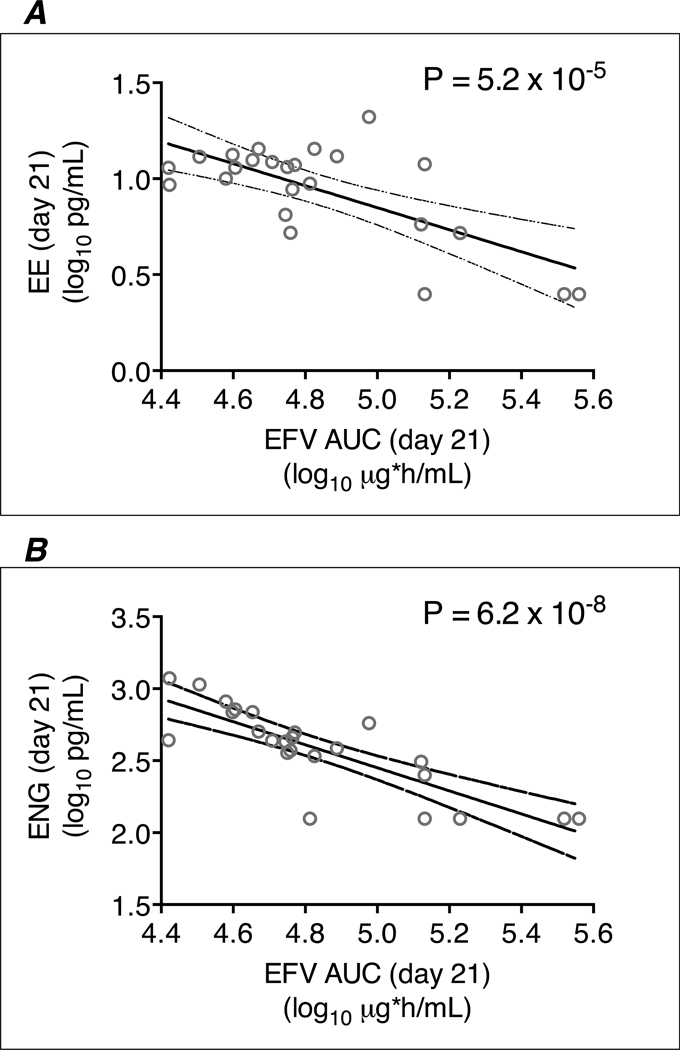
To assess whether the association of CYP2B6/CYP2A6 with etonogestrel and ethinyl estradiol concentrations was mediated by plasma efavirenz exposure, we characterized such relationships (Table 3 and Figure 3). Among the 24 efavirenz group participants, increased plasma efavirenz exposure was significantly associated with decreased plasma concentrations of both etonogestrel and ethinyl estradiol at day 21. Day 21 efavirenz AUC0–8h were significantly associated with day 21 log10 plasma etonogestrel concentrations (β coefficient −0.80, P = 6.2 ×10−8) and day 21 log10 plasma ethinyl estradiol concentrations (β coefficient −0.57, P = 5.2 ×10−5). Participants with plasma etonogestrel and ethinyl estradiol concentration below limits of quantification on day 21 tended to have the highest plasma efavirenz AUC0–8h values (Figure 3). Considering each efavirenz PK parameter, plasma etonogestrel and plasma ethinyl estradiol concentration at day 21 were significantly associated with efavirenz AUC0–8h, Cmin and Cmax at day 0 and/or day 21. For all parameters, P-values for association were lower for day 0 efavirenz PK parameters than for day 21 efavirenz PK parameters. The lowest P-values for association for both etonogestrel and ethinyl estradiol were with efavirenz AUC0–8h at day 0. Relationships between each efavirenz PK parameter, etonogestrel and ethinyl estradiol concentrations are presented in Table 3.
Table 3.
Relationships between efavirenz pharmacokinetic parameters and day 21 etonogestrel and ethinyl estradiol concentrations among the 24 efavirenz group participants.
| Efavirenz pharmacokinetic parameter | daya | log10 etonogestrel β coefficient, P-valuea | log10 ethinyl estradiol β coefficient, P-valueb |
|---|---|---|---|
| AUC0–8h (log10 ng*h/mL) | 0 | −0.80, 1.0 ×10−9 | −0.68, 5.9 ×10−7 |
| 21 | −0.80, 6.2 ×10−8 | −0.57, 5.2 ×10−5 | |
| Cmin (log10 ng/mL) | 0 | −0.63, 3.4 ×10−7 | −0.58, 4.0 ×10−6 |
| 21 | −0.27, 1.1 ×10−1 | −0.20, 1.0 ×10−1 | |
| Cmax (log10 ng/mL) | 0 | −0.88, 5.4 ×10−9 | −.075, 1.2 ×10−5 |
| 21 | −0.86, 4.7 ×10−7 | −0.67, 1.4 ×10−4 | |
Abbreviations: AUC, area under the concentration time curve; Cmax, maximum concentration; Cmin, minimum concentration.
Day 0 or 21 refers to the day on which efavirenz pharmacokinetic parameters were determined.
P-values and beta coefficients were generated with linear regression models.
Figure 3. Relationships between day 21 plasma efavirenz log10 AUC0–8h values and day 21 log10 etonogestrel and ethinyl estradiol concentrations among the 24 efavirenz group participants.
Panel A: associations of plasma efavirenz log10 AUC0–8h values on day 21 and log10 ethinyl estradiol concentrations on day 21; Panel B: associations of plasma efavirenz log10 AUC0–8h values on day 21 and log10 etonogestrel concentrations on day 21; Line of regression and 95% confidence intervals are shown. EFV = efavirenz; AUC = area under the concentration-time curve; ENG = etonogestrel; EE = ethinyl estradiol.
In A5316, geometric mean efavirenz Cmin concentrations were 36% lower on day 21 (with hormones) than on day 0 (without hormones). In the present analysis, the proportion change in median log10 efavirenz Cmin was similar regardless of CYP2B6 genotype level, and was −0.24 log10 (42.5% reduction), −0.2 log10 (36.9% reduction), and −0.15 log10 (29.9% reduction) with CYP2B6 normal, intermediate, and slow metabolizer genotypes, respectively.
Other polymorphisms, etonogestrel and ethinyl estradiol concentrations
Among the 24 efavirenz group participants, and after adjusting for CYP2B6/CYP2A6 composite genotype, and correcting for multiple comparisons, there were no statistically significant associations between any of the 17 SNPs and concentrations of either etonogestrel or ethinyl estradiol at day 21. For log10 etonogestrel, the lowest P-values was for rs727428 which is intronic between SHBG and ATP1B2 (P =0.16), and for log10 ethinyl estradiol was for UGT1A1 rs887829 (P = 0.011).
Among the 25 control group participants, without adjusting for CYP2B6/CYP2A6 composite genotype but correcting for multiple comparisons, there were no statistically significant associations between any of the 17 SNPs and concentrations of either etonogestrel or ethinyl estradiol at day 21. For log10 etonogestrel the lowest P-values was for CYP3A5 rs776746 (P = 0.078), and for log10 ethinyl estradiol was also CYP3A5 rs776746 (P = 0.026).
Among the 23 atazanavir/r group participants, without adjusting for CYP2B6/CYP2A6 composite genotype but correcting for multiple comparisons, there were no statistically significant associations between any of the 17 SNPs and concentrations of either etonogestrel or ethinyl estradiol at day 21. For etonogestrel the lowest P-values was for CYP19A1 rs2445762 (P = 0.018), and for ethinyl estradiol was rs2472297 which is intronic between CYP1A1 and CYP1A2 (P = 0.16).
Discussion
Primary analyses from study A5316 showed that, compared to women with HIV but not yet receiving ART, efavirenz-based ART was associated with significantly reduced plasma exposure of both etonogestrel and ethinyl estradiol released from a vaginal ring. [11] The present study extended that observation by showing that, in the efavirenz arm, plasma etonogestrel and ethinyl estradiol concentrations were reduced to a significantly greater extent among CYP2B6 slow metabolizers than among CYP2B6 intermediate and normal metabolizers. Efavirenz is known to induce hepatic CYP3A4 and other CYP isoforms. We hypothesize that higher plasma efavirenz concentrations, which result from reduced CYP2B6 activity, cause greater induction of CYP3A4 and therefore more rapid plasma clearance of both etonogestrel and ethinyl estradiol. This mechanism is supported by the highly significant inverse correlation between day 21 plasma efavirenz exposure and plasma concentrations of both etonogestrel and ethinyl estradiol. These findings support another pharmacogenetic evaluation of a similar progestin, levonorgestrel, which shares the same metabolic pathway as etonogestrel. When efavirenz-based ART was combined with a levonorgestrel-releasing contraceptive implant, CYP2B6 516G→T was associated with lower levonorgestrel Cmax and AUC. [40]
The present study suggests that the detrimental interaction between efavirenz and both etonogestrel and ethinyl estradiol administered by vaginal ring could be reduced by individualizing efavirenz dosing based on CYP2B6 genotype. However, even with CYP2B6 normal metabolizer genotypes the magnitude of effect is substantial (73% lower etonogestrel and 41% lower ethinyl estradiol concentrations), suggesting that individualizing efavirenz dosing based on CYP2B6 genotype would not be sufficient to mitigate this effect. We suspect that drug-drug interactions between efavirenz and other concomitant medications could be similarly reduced by individualizing efavirenz dosing based on CYP2B6 genotype. Physiologically-based pharmacokinetic modeling of levonorgestrel, a progestin with a similar metabolism pathway as etonogestrel, predicted that dose reduction of efavirenz to 400 mg daily would not significantly influence the clinical significance of the drug-drug interaction between efavirenz and the progestin (53% lower levonorgestrel with efavirenz 600 mg versus 45% lower levonorgestrel with efavirenz 400 mg).[41] However, the present study suggests that influence of efavirenz dose reduction on the extent of the drug-interaction may vary depending on CYP2B6 genotype.
The effect of efavirenz-based ART on vaginally administered ethinyl estradiol in A5316 was greater (59% lower exposure) than previously described between efavirenz and oral ethinyl estradiol, which only observed a 10% lower exposure of oral ethinyl estradiol. [4] The former study was an HIV-negative, healthy volunteer, cross-over, PK intraindividual comparison of ethinyl estradiol from a combined oral contraceptive pill administered with or without efavirenz. That trial was entirely enrolled in the United States, 68% of participants were White and 21% were Black, and pharmacogenetics were not analyzed. In A5316, all participants were living with HIV, the efavirenz group was receiving fully suppressive ART, and enrolled from diverse international clinical trials sites, reflected in a high proportion of Black participants in both the control and ART groups (44% and 64%, respectively). The potential mechanism for differences is the extent of the drug-drug interaction between efavirenz and ethinyl estradiol given orally or via a vaginal ring is unclear, but it is possible that more patients with CYP2B6 slow metabolizer genotypes were included in A5316. Results of the present study are consistent with a recent report describing the pharmacogenetic interaction between efavirenz and etonogestrel delivered by subdermal implant. In that study, among 19 women receiving efavirenz-based antiretroviral therapy, CYP2B6 516G→T was associated with 43% lower etonogestrel Cmin and 34% lower AUC0–24weeks.[42]
Study A5316 previously showed that, compared to women with HIV but not yet receiving ART, atazanavir/r was associated with 71% higher plasma etonogestrel concentrations and 38% lower ethinyl estradiol concentrations. The present study showed that, in the atazanavir/r arm, there were no significant associations between 17 candidate SNPs and concentrations of either etonogestrel or ethinyl estradiol at day 21 after controlling for multiple comparisons. Similarly, in the control groups there were no significant associations between 17 candidate SNPs and concentrations of either etonogestrel or ethinyl estradiol at day 21 after controlling for multiple comparisons.
Efavirenz 600 mg daily is being phased out in many low- and middle-income settings in favor of efavirenz 400mg daily.[43] This dose reduction is reported to be associated with approximately 25% lower efavirenz exposure across CYP2B6 normal-, intermediate- and slow-metabolizer genotype groups.[44] Study A5316 observed modestly lower efavirenz exposure during vaginal ring use. The efavirenz minimum concentration was 36% lower during hormone use, but remained above the concentration proposed to be associated with efavirenz effectiveness.[45] In the present study, none of the genotypes studied were associated with the magnitude of effect of vaginal ring use on plasma efavirenz exposure. Therefore, the risk of drug-drug interaction between vaginal ring hormones and ART will not be mitigated by genotype, and patients in the normal metabolizer genotypes will remain at greatest risk of sub-therapeutic concentrations, particularly in the context of suboptimal adherence.
This study had several limitations. Given the small sample size we could only evaluate relatively frequent genetic polymorphisms. With relatively few individuals representing any given genotype, this study should be considered exploratory. Hormone and ART adherence was assessed by self-report and not by direct measures of adherence. In addition, the study was not designed to evaluate the pharmacodynamic relationship between hormone concentrations and ovulation suppression or other measures of contraceptive effectiveness.
In summary, CYP2B6 slow metabolizer genotype worsens the detrimental drug-drug interaction between hormones administered by vaginal ring (93% versus 73% lower etonogestrel concentrations, and 75% versus 41% lower ethinyl estradiol with CYP2B6 slow versus normal metabolizer genotypes, respectively). These findings suggest that this detrimental interaction could be reduced, but not eliminated, by individualizing the efavirenz dose based on CYP2B6 genotype. It is likely that drug-drug interactions between efavirenz and other concomitant medications may be similarly reduced by individualized efavirenz dosing based on CYP2B6 genotype.
Supplementary Material
Acknowledgements
Research reported in this publication was supported by the AIDS Clinical Trials Group (ACTG) through the National Institutes of Health (NIH), National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award numbers UMI AI068636; UMI AI068634; UMI AI106701; U01AI69471. This work was also supported by the International Maternal Pediatric Adolescent AIDS Clinical Trials (IMPAACT) Network through the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award numbers UM1AI068632; UM1AI068616; UM1AI106716 with cofounding from the Eunice Kennedy Shriver National Institute of Child Health and Human Development and the National Institute of Mental Health under award number HHSN275201300003C. This work was also supported by the Eunice Kennedy Shriver National Institute of Child Health and Human Development under award number 1R01HD085887 (PI Scarsi). This work was also supported by P30AI110527, R01AI077505, and UL1TR002243 (David Haas). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. We gratefully acknowledge the support of Merck Sharp & Dohme Corp., a subsidiary of Merck & Co., Inc., Kenilworth, NJ, USA, for providing NuvaRing® for use in this study.
We gratefully acknowledge the patients who participated in this research, and site personnel who contributed to this work, including: Dr. Ruth K. Friedman, Dr. Angela Cristina Andrade, and Dr. Vânia Stiepanowez Regina de Oliveira Rocha, Instituto de Pesquisa Clínica Evandro Chagas (IPEC) – Fiocruz (Site 12101; 5UM1AI069476); Dr. Mariam Aziz and Maureen McNichols, Rush University Medical Center/Ruth M Rothstein CORE Center (Site 5083; HHSN275201800001I); Dr. Victor Akelo and Johnson Ondiek, KISUMU CRS (Site 31460; UM1AI069418); Dr. Shobha Swaminathan, Nila Dharan, and Christie Lyn Costanza, Rutgers New Jersey Medical School (Site 31786; AI069419–12); Dr. Carmel Ganoza and Maria Esther Guevara, Barranco CRS (Site 11301; 1U01AI069438–01); San Miguel CRS (Site 11302; 1U01AI069438–01); Gaborone CRS (Site 12701; UM1 AI069456–08); Dr. Karen Tashima and Deborah Perez, The Miriam Hospital (Site 2951; 2UM1A1069412–08); Professor Kiat Ruxrungtham, The Thai Red Cross AIDS Research Centre (TRC-ARC) CRS (Site 31802; 5UM1AI069399); Suwat Chariyalertsak and Daralak Tavornprasit, Chiang Mai University HIV Treatment CRS (Site 31784; 5UM1AI069399–10); USC LA NICHD CRS (Site 5048); Dr. Michael S. Saag, Alabama CRS (Site 31788; UM1 AI069452–08); University of Colorado CRS (Site 5052); Dr. Dr. Jaime G. Deville and Dr. Carla Janzen, David Geffen School of Medicine at UCLA CRS (Site 5112; NICHD Contract HHSN275201300003C, Westat ID-6101-S059, UL1TR001881); Dr. Mhleli Masango and Dr. Lee Fairlie, Wits RHI Shandukani Research Centre (Site 8051; 5UM1A1068632); Columbia Physicians & Surgeons (P & S) CRS (Site 30329; UM1 AI069470–08); Raphaelle Auguste and Marlene Burey, Jacobi Medical Center (Site 5013; HHSN275201300003C); San Juan City Hospital (Site 5031); Dr. Mobeen H Rathore and Saniyyah Mahmoudi, University of Florida Center for HIV/AIDS Research, Education and Service (UF CARES) (Site 5051; HHSN275201300003C); Dr. Jorge L Santana and Marielly Lopez-Rivera, Puerto Rico AIDS Clinical Trails Unit (PR-AIDS) CRS (Site 5401; 5 UM1 AI069415–13); Emily Barr and Adriana Weinberg, Westat ID-6101-S035 The University of Puerto Rico Pediatric HIV/AIDS Research Program CRS (Site 6601; UM1 AI069415–09).
We thank the other team members affiliated with A5316, including Liz Barr, Christina Blanchard-Horan, Elizabeth Connick, Mary Allegra Cermak, Nahida Chakhtoura, Cecilia Chang-Ching, Andee Fox, Alan Landay, Mey Leon, Jeong-Gun Park, Kristine Patterson, Thucuma Sise, Greg Spear, David Shugarts, Pamela Tshandu, and Charles Wira.
Footnotes
Declaration of Interests:
Susan E. Cohn is a scientific advisor to Merck.
None: David W. Haas, Paxton Baker; Yoninah S. Cramer; Catherine Godfrey; Susan L. Rosenkranz, Francesca Aweeka; Baiba Berzins; Robert Coombs; Kristine Coughlin; Laura E. Moran; David Gingrich; Carmen D. Zorrilla; Kimberly K. Scarsi
Contributor Information
David W. Haas, Vanderbilt University School of Medicine, Nashville, TN; Meharry Medical College, Nashville, TN, USA
Yoninah S. Cramer, Harvard TH Chan School of Public Health, Boston MA, USA; Frontier Science Foundation, Brookline, MA, USA.
Catherine Godfrey, Division of AIDS, National Institutions of Allergy and Infectious Disease, National Institutes of Health, Bethesda, MD, USA.
Susan L. Rosenkranz, Harvard TH Chan School of Public Health, Boston MA, USA; Frontier Science Foundation, Brookline, MA, USA.
Francesca Aweeka, Department of Clinical Pharmacy, School of Pharmacy, University of California San Francisco, San Francisco, CA, USA.
Baiba Berzins, Division of Infectious Diseases, Northwestern University, Chicago, IL, USA.
Robert Coombs, Departments of Medicine and Laboratory Medicine, University of Washington, Seattle, WA, USA.
Kristine Coughlin, Frontier Science & Technology Research Foundation, Inc, Amherst, NY, USA.
Laura E. Moran, Social & Scientific Systems, Silver Spring, MD, USA.
David Gingrich, Department of Clinical Pharmacy, School of Pharmacy, University of California San Francisco, San Francisco, CA, USA.
Carmen D. Zorrilla, University of Puerto Rico School of Medicine, San Juan, Puerto Rico
Paxton Baker, Vanderbilt University Medical Center, Nashville, TN, USA.
Susan E. Cohn, Division of Infectious Diseases, Northwestern University, Chicago, IL, USA.
Kimberly K. Scarsi, Department of Pharmacy Practice and Science, College of Pharmacy; University of Nebraska Medical Center, Omaha, NE, USA.
REFERENCES
- 1.Carten ML, Kiser JJ, Kwara A, Mawhinney S, Cu-Uvin S. Pharmacokinetic interactions between the hormonal emergency contraception, levonorgestrel (Plan B), and Efavirenz. Infect Dis Obstet Gynecol. 2012;2012:137192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chappell CA, Lamorde M, Nakalema S, Chen BA, Mackline H, Riddler SA, et al. Efavirenz decreases etonogestrel exposure: a pharmacokinetic evaluation of implantable contraception with antiretroviral therapy. AIDS. 2017;31(14):1965–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scarsi KK, Darin KM, Nakalema S, Back DJ, Byakika-Kibwika P, Else LJ, et al. Unintended Pregnancies Observed With Combined Use of the Levonorgestrel Contraceptive Implant and Efavirenz-based Antiretroviral Therapy: A Three-Arm Pharmacokinetic Evaluation Over 48 Weeks. Clin Infect Dis. 2016;62(6):675–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sevinsky H, Eley T, Persson A, Garner D, Yones C, Nettles R, et al. The effect of efavirenz on the pharmacokinetics of an oral contraceptive containing ethinyl estradiol and norgestimate in healthy HIV-negative women. Antivir Ther. 2011;16(2):149–56. [DOI] [PubMed] [Google Scholar]
- 5.Vieira CS, Bahamondes MV, de Souza RM, Brito MB, Rocha Prandini TR, Amaral E, et al. Effect of antiretroviral therapy including lopinavir/ritonavir or efavirenz on etonogestrel-releasing implant pharmacokinetics in HIV-positive women. J Acquir Immune Defic Syndr. 2014;66(4):378–85. [DOI] [PubMed] [Google Scholar]
- 6.Shrader SP, Ragucci KR. Contraception In: DiPiro JT, Talbert RL, Yee GC, Matzke GR, Wells BG, Posey LM, eds. Pharmacotherapy: A Pathophysiologic Approach Ninth Edition: McGaw Hill Education; 2014. p. 1271–86. [Google Scholar]
- 7.Nanda K, Stuart GS, Robinson J, Gray AL, Tepper NK, Gaffield ME. Drug interactions between hormonal contraceptives and antiretrovirals. AIDS. 2017;31(7):917–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scarsi KK, Darin KM, Chappell CA, Nitz SM, Lamorde M. Drug-Drug Interactions, Effectiveness, and Safety of Hormonal Contraceptives in Women Living with HIV. Drug Saf. 2016;39(11):1053–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Back DJ, Houlgrave R, Tjia JF, Ward S, Orme ML. Effect of the progestogens, gestodene, 3-keto desogestrel, levonorgestrel, norethisterone and norgestimate on the oxidation of ethinyloestradiol and other substrates by human liver microsomes. J Steroid Biochem Mol Biol. 1991;38(2):219–25. [DOI] [PubMed] [Google Scholar]
- 10.Palovaara S, Pelkonen O, Uusitalo J, Lundgren S, Laine K. Inhibition of cytochrome P450 2B6 activity by hormone replacement therapy and oral contraceptive as measured by bupropion hydroxylation. ClinPharmacolTher. 2003;74(4):326–33. [DOI] [PubMed] [Google Scholar]
- 11.Scarsi KK, Cramer YS, Rosenkranz SL, Aweeka F, Berzins B, Coombs RW, et al. Antiretroviral therapy and vaginally administered contraceptive hormones: a three-arm, pharmacokinetic study. Lancet HIV. 2019;6(9):e601–e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Desta Z, Gammal RS, Gong L, Whirl-Carrillo M, Gaur AH, Sukasem C, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2B6 and Efavirenz-Containing Antiretroviral Therapy. Clin Pharmacol Ther. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Haas DW, Ribaudo HJ, Kim RB, Tierney C, Wilkinson GR, Gulick RM, et al. Pharmacogenetics of efavirenz and central nervous system side effects: an Adult AIDS Clinical Trials Group study. AIDS. 2004;18(18):2391–400. [PubMed] [Google Scholar]
- 14.Tsuchiya K, Gatanaga H, Tachikawa N, Teruya K, Kikuchi Y, Yoshino M, et al. Homozygous CYP2B6 *6 (Q172H and K262R) correlates with high plasma efavirenz concentrations in HIV-1 patients treated with standard efavirenz-containing regimens. Biochem Biophys Res Commun. 2004;319(4):1322–6. [DOI] [PubMed] [Google Scholar]
- 15.Rotger M, Colombo S, Furrer H, Bleiber G, Buclin T, Lee BL, et al. Influence of CYP2B6 polymorphism on plasma and intracellular concentrations and toxicity of efavirenz and nevirapine in HIV-infected patients. Pharmacogenet Genom. 2005;15(1):1–5. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez-Novoa S, Barreiro P, Rendon A, Jimenez-Nacher I, Gonzalez-Lahoz J, Soriano V. Influence of 516G>T polymorphisms at the gene encoding the CYP450–2B6 isoenzyme on efavirenz plasma concentrations in HIV-infected subjects. Clin Infect Dis. 2005;40(9):1358–61. [DOI] [PubMed] [Google Scholar]
- 17.Holzinger ER, Grady B, Ritchie MD, Ribaudo HJ, Acosta EP, Morse GD, et al. Genome-wide association study of plasma efavirenz pharmacokinetics in AIDS Clinical Trials Group protocols implicates several CYP2B6 variants. Pharmacogenet Genom. 2012;22(12):858–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wyen C, Hendra H, Vogel M, Hoffmann C, Knechten H, Brockmeyer NH, et al. Impact of CYP2B6 983T>C polymorphism on non-nucleoside reverse transcriptase inhibitor plasma concentrations in HIV-infected patients. J Antimicrob Chemo. 2008;61(4):914–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J, Sonnerborg A, Rane A, Josephson F, Lundgren S, Stahle L, et al. Identification of a novel specific CYP2B6 allele in Africans causing impaired metabolism of the HIV drug efavirenz. Pharmacogenet Genom. 2006;16(3):191–8. [DOI] [PubMed] [Google Scholar]
- 20.Ribaudo HJ, Liu H, Schwab M, Schaeffeler E, Eichelbaum M, Motsinger-Reif AA, et al. Effect of CYP2B6, ABCB1, and CYP3A5 polymorphisms on efavirenz pharmacokinetics and treatment response: an AIDS Clinical Trials Group study. J Infect Dis. 2010;202(5):717–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.di Iulio J, Fayet A, Arab-Alameddine M, Rotger M, Lubomirov R, Cavassini M, et al. In vivo analysis of efavirenz metabolism in individuals with impaired CYP2A6 function. Pharmacogenet Genom. 2009;19(4):300–9. [DOI] [PubMed] [Google Scholar]
- 22.Kwara A, Lartey M, Sagoe KW, Rzek NL, Court MH. CYP2B6 (c.516G-->T) and CYP2A6 (*9B and/or *17) polymorphisms are independent predictors of efavirenz plasma concentrations in HIV-infected patients. Br J Clin Pharmacol. 2009;67(4):427–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwara A, Lartey M, Sagoe KW, Kenu E, Court MH. CYP2B6, CYP2A6 and UGT2B7 genetic polymorphisms are predictors of efavirenz mid-dose concentration in HIV-infected patients. AIDS. 2009;23(16):2101–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haas DW, Kwara A, Richardson DM, Baker P, Papageorgiou I, Acosta EP, et al. Secondary metabolism pathway polymorphisms and plasma efavirenz concentrations in HIV-infected adults with CYP2B6 slow metabolizer genotypes. J Antimicrob Chemother. 2014;69(8):2175–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reynolds NR, Sun J, Nagaraja HN, Gifford AL, Wu AW, Chesney MA. Optimizing measurement of self-reported adherence with the ACTG Adherence Questionnaire: a cross-protocol analysis. J AcquirImmuneDeficSyndr. 2007;46(4):402–9. [DOI] [PubMed] [Google Scholar]
- 26.Agostini HT, Ryschkewitsch CF, Mory R, Singer EJ, Stoner GL. JC virus (JCV) genotypes in brain tissue from patients with progressive multifocal leukoencephalopathy (PML) and in urine from controls without PML: increased frequency of JCV type 2 in PML. JournalofInfectiousDiseases. 1997;176(1):1–8. [DOI] [PubMed] [Google Scholar]
- 27.Johnson DH, Venuto C, Ritchie MD, Morse GD, Daar ES, McLaren PJ, et al. Genomewide association study of atazanavir pharmacokinetics and hyperbilirubinemia in AIDS Clinical Trials Group protocol A5202. Pharmacogenet Genom. 2014;24(4):195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu M, Ingle JN, Fridley BL, Buzdar AU, Robson ME, Kubo M, et al. TSPYL5 SNPs: association with plasma estradiol concentrations and aromatase expression. Mol Endocrinol. 2013;27(4):657–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Z, Tao S, Gao Y, Zhang J, Hu Y, Mo L, et al. Genome-wide association study of sex hormones, gonadotropins and sex hormone-binding protein in Chinese men. J Med Genet. 2013;50(12):794–801. [DOI] [PubMed] [Google Scholar]
- 30.Ruth KS, Campbell PJ, Chew S, Lim EM, Hadlow N, Stuckey BG, et al. Genome-wide association study with 1000 genomes imputation identifies signals for nine sex hormone-related phenotypes. Eur J Hum Genet. 2016;24(2):284–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prescott J, Thompson DJ, Kraft P, Chanock SJ, Audley T, Brown J, et al. Genome-wide association study of circulating estradiol, testosterone, and sex hormone-binding globulin in postmenopausal women. PLoS One. 2012;7(6):e37815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Suhre K, Shin SY, Petersen AK, Mohney RP, Meredith D, Wagele B, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477(7362):54–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Varenhorst C, Eriksson N, Johansson A, Barratt BJ, Hagstrom E, Akerblom A, et al. Effect of genetic variations on ticagrelor plasma levels and clinical outcomes. Eur Heart J. 2015;36(29):1901–12. [DOI] [PubMed] [Google Scholar]
- 34.Pharmacogene Variation (PharmVar) Consortium. Available at: https://www.pharmvar.org/gene/CYP3A4. Accessed August 24, 2018.
- 35.Korostishevsky M, Steves CJ, Malkin I, Spector T, Williams FM, Livshits G. Genomics and metabolomics of muscular mass in a community-based sample of UK females. Eur J Hum Genet. 2016;24(2):277–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amin N, Byrne E, Johnson J, Chenevix-Trench G, Walter S, Nolte IM, et al. Genome-wide association analysis of coffee drinking suggests association with CYP1A1/CYP1A2 and NRCAM. Mol Psychiatry. 2012;17(11):1116–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sulem P, Gudbjartsson DF, Geller F, Prokopenko I, Feenstra B, Aben KK, et al. Sequence variants at CYP1A1-CYP1A2 and AHR associate with coffee consumption. Hum Mol Genet. 2011;20(10):2071–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cornelis MC, Monda KL, Yu K, Paynter N, Azzato EM, Bennett SN, et al. Genome-wide meta-analysis identifies regions on 7p21 (AHR) and 15q24 (CYP1A2) as determinants of habitual caffeine consumption. PLoS Genet. 2011;7(4):e1002033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neary M, Lamorde M, Olagunju A, Darin KM, Merry C, Byakika-Kibwika P, et al. The Effect of Gene Variants on Levonorgestrel Pharmacokinetics When Combined With Antiretroviral Therapy Containing Efavirenz or Nevirapine. Clin Pharmacol Ther. 2017;102(3):529–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roberts O, Rajoli RKR, Back DJ, Owen A, Darin KM, Fletcher CV, et al. Physiologically based pharmacokinetic modelling prediction of the effects of dose adjustment in drug-drug interactions between levonorgestrel contraceptive implants and efavirenz-based ART. J Antimicrob Chemother. 2018;73(4):1004–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neary M, Chappell CA, Scarsi KK, Nakalema S, Matovu J, Achilles SL, et al. Effect of patient genetics on etonogestrel pharmacokinetics when combined with efavirenz or nevirapine ART. J Antimicrob Chemother. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.PEPFAR. PEPFAR 2019 Country Operational Plan Guidance for all PEPFAR Countries. https://wwwpepfargov/documents/organization/288731pdf. 2019. [Google Scholar]
- 44.Dickinson L, Amin J, Else L, Boffito M, Egan D, Owen A, et al. Pharmacokinetic and Pharmacodynamic Comparison of Once-Daily Efavirenz (400 mg vs. 600 mg) in Treatment-Naive HIV-Infected Patients: Results of the ENCORE1 Study. Clin Pharmacol Ther. 2015;98(4):406–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Punyawudho B, Singkham N, Thammajaruk N, Dalodom T, Kerr SJ, Burger DM, et al. Therapeutic drug monitoring of antiretroviral drugs in HIV-infected patients. Expert Rev Clin Pharmacol. 2016;9(12):1583–95. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.