Abstract
Alternative splicing of pre-messenger RNA is a key feature of transcriptome expansion in eukaryotic cells, yet its regulation is poorly understood. Spliceosome assembly occurs co-transcriptionally, raising the possibility that DNA structure may directly influence alternative splicing. Supporting such an association, recent reports have identified distinct histone methylation patterns, elevated nucleosome occupancy and enriched DNA methylation at exons relative to introns. Moreover, the rate of transcription elongation has been linked to alternative splicing. Here we provide the first evidence that a DNA-binding protein, CCCTC-binding factor (CTCF), can promote inclusion of weak upstream exons by mediating local RNA polymerase II pausing both in a mammalian model system for alternative splicing, CD45, and genome-wide. We further show that CTCF binding to CD45 exon 5 is inhibited by DNA methylation, leading to reciprocal effects on exon 5 inclusion. These findings provide a mechanistic basis for developmental regulation of splicing outcome through heritable epigenetic marks.
It is estimated that greater than 90% of human genes undergo alternative splicing of pre-mRNA1,2 and aberrant splicing has been implicated in a number of human diseases3. Alternative splicing decisions are determined by the ability of weak splice sites to effectively compete with strong splice sites for detection by the spliceosome4. The balance between splice site selection is principally influenced by two variables5: (1) the availability of splicing factors that detect enhancer or silencer sequences encoded within nascent RNA4,6 and (2) the rate of RNA polymerase II (pol II) transcription elongation, wherein a slow rate favours co-transcriptional spliceosome assembly at weak splice sites7,8.
A surprising result of genome-wide chromatin-immunoprecipitation-sequencing (ChIP-seq) studies is the non-random distribution of several epigenetic marks in exons relative to introns. In particular, exons show elevated nucleosome density, DNA methylation of cytosine, and over-representation of certain histone modifications, relative to introns9–15. Differential ‘marking’ of exons on DNA highlighted a possible connection between DNA structure and co-transcriptional RNA processing. Accordingly, several recent studies suggest that exonic histone modification may affect variable inclusion of alternative exons16,17. Collectively, these studies raise the intriguing possibility that epigenetic modifications are maintained on DNA to aid the spliceosome in the process of exon definition9,13, and that differential chromatin assembly may represent a critical aspect of alternative splicing regulation.
Processing of CD45 pre-mRNA (also known as PTPRC) is a well established model system to study the regulatory mechanisms of alternative splicing. CD45 is a trans-membrane protein tyrosine phosphatase that initiates signalling through antigen receptors by dephosphorylating the inhibitory tyrosine on Src family kinases18. Variable exclusion of exons 4–6 (A–C) of CD45 transcripts is tightly correlated with stages of lymphocyte development and expressed splice variants can be distinguished using isoform-specific antibodies and flow cytometry19 (Supplementary Fig. 1). In general, the larger, exon4-containing isoforms (CD45RA) are expressed early in peripheral lymphocyte development, whereas the shortest isoform (CD45RO), which lacks all three variable exons, is expressed in terminally differentiated lymphocytes19. We recently identified heterogeneous ribonucleoprotein L-like (hnRNPLL) as a tissue-specific master regulator of the CD45RA to CD45RO transition in peripheral lymphocytes20. hnRNPLL binds to exons 4 and 6 of CD45 mRNA and blocks the inclusion of both exons in the mature message21. In contrast, hnRNPLL expression does not influence exclusion of exon 5 (refs 20, 22; Supplementary Fig. 2a). In vitro studies aimed at uncovering regulators of exon 5 exclusion have identified several ubiquitously expressed splicing factors22,23. However, peripheral lymphocytes retain exon 5 until the terminal stages of development24 (Supplementary Fig. 2b), portending a yet uncovered layer of regulation.
CTCF regulates exon 5 inclusion in CD45 mRNA
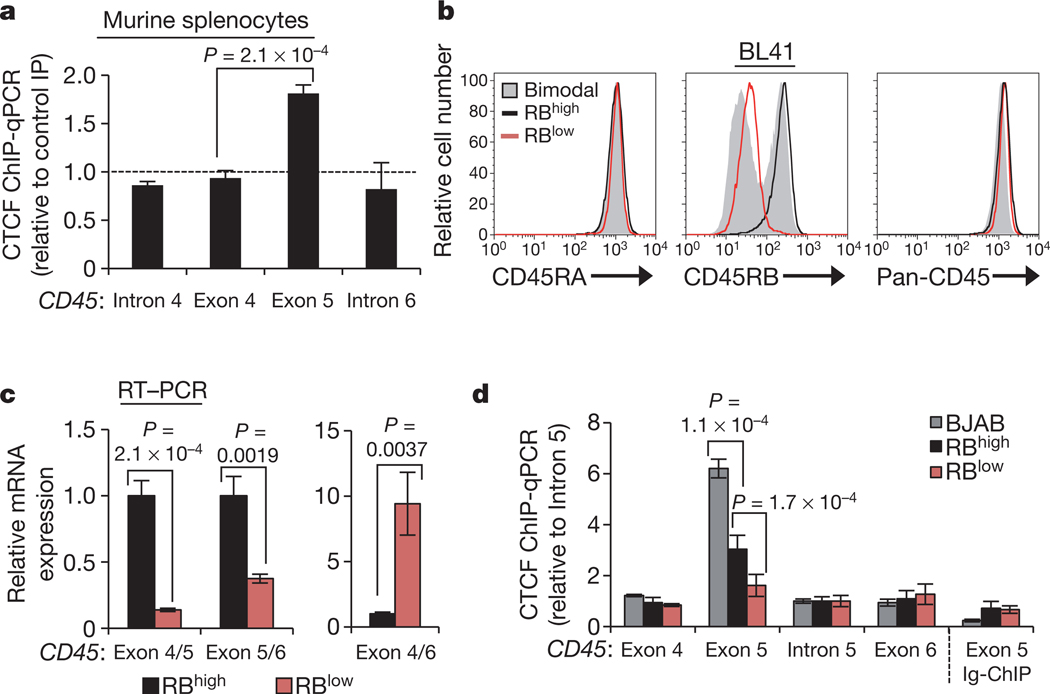
Considering the growing evidence for DNA-mediated regulation of spliceosome assembly, we explored the hypothesis that exon 5 inclusion is mediated by the epigenetic structure of the gene encoding CD45. By analysing published ChIP-seq data within the UCSC genome browser25,26, we identified a strong CTCF peak overlapping with exon 5 across cell types. CTCF is an 11 zinc-finger DNA-binding protein with multiple nuclear functions, largely grouped into two categories: insulating inactive regions of the genome from active regions and promoting long-range interactions between distal regions of the genome27,28. Whereas intergenic CTCF is an effective barrier to transcription27, we found that CTCF binding at exon 5 is maintained in cells that actively transcribe abundant CD45 (ref. 26) and that exon 5 binding is conserved in murine splenocytes (Fig. 1a and Supplementary Fig. 2c), indicating an important, position-dependent ‘non-insulator’ function. We thus explored whether and how CTCF binding at CD45 exon 5 DNA influences processing of CD45 transcripts.
Figure 1 |. Binding of CTCF to exon 5 of CD45 DNA is associated with inclusion of exon 5 in CD45 transcripts.
a, CTCF ChIP in murine splenocytes and quantitative PCR (qPCR) relative to rabbit Ig control ChIP (n = 2). b, Cell-surface staining for CD45RA (exon 4-containing) and CD45RB (exon 5-containing) isoforms and total CD45 (pan) in parental BL41 B cells (RBhigh), cell-culture-derived CD45RB bimodal cells, and CD45RB low (RBlow) cells sorted from the bimodal BL41 population. c, GAPDH-normalized qRT–PCR data from RBhigh and RBlow cells using the indicated junction-spanning primers (n = 3). d, CTCF ChIP in BJAB, RBhigh and RBlow cells and qPCR for CD45 exons and introns (n = 3–6). All graphs show mean values ± standard deviation (s.d.). P = two-tailed Student’s t test comparing the indicated samples.
To dissect the impact of CTCF on exon 5 splicing in a cell-based system, we screened several human Burkitt lymphoma B cell lines for differences in expression of the exon 5 containing CD45RB isoform. Whereas lymphocyte cell lines generally express high levels of CD45RB (Supplementary Fig. 2d, e), culturing BL41 cells in non-heat-inactivated fetal bovine serum (FBS) resulted in bimodal CD45RB expression. CD45RB low cells (RBlow) were sorted from the bimodal population and stably maintained (Fig. 1b). Parental BL41 cells (RBhigh) and sorted RBlow cells express equivalent exon 4-containing CD45RA isoforms and total CD45 (Fig. 1b), indicating specific exclusion of exon 5 in the RBlow population. Quantitative RT–PCR with exon junction spanning primers validated exon 5 skipping: RBlow cells showed reduced exon 4/5 and exon 5/6 junctions, but enhanced exon 4/6 junctions relative to RBhigh cells (Fig. 1c). Notably, several histone modifications that that have been previously linked to alternative splicing (H3K36me3, H3K27me3, H3K4me3)16,29 are equivalently detected at exon 5 in RBhigh and RBlow cells (Supplementary Fig. 3a, b). CTCF-ChIP in the newly identified RBhigh and RBlow BL41 cells, and CD45RB-high BJAB cells revealed a strong positive correlation between exon 5 inclusion in CD45 mRNA and CTCF binding at CD45 exon 5 DNA (Fig. 1b–d and Supplementary Fig. 2e), particularly in BJAB cells, which also express elevated CTCF protein (Supplementary Fig. 3c). In agreement with the observation that exon 5 splicing is independent of hnRNPLL, modulation of hnRNPLL expression did not influence CTCF binding to CD45 exon 5 (Supplementary Fig. 3d).
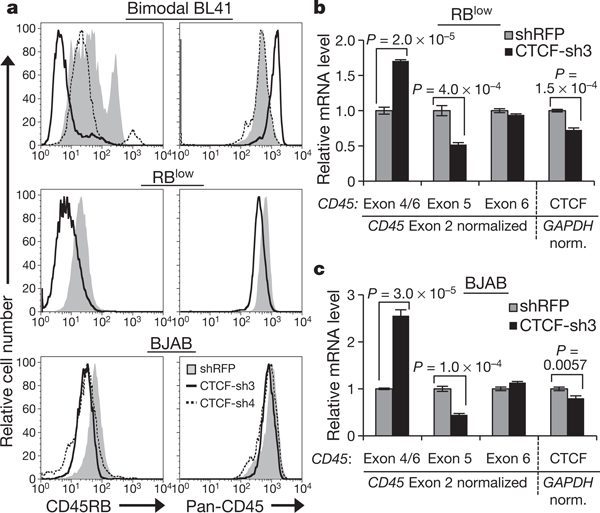
To assess whether the association between CTCF binding and exon 5 inclusion reflects a direct role for CTCF in CD45 alternative splicing, we used RNA interference to deplete CTCF from our B cell lines (Supplementary Fig. 4a). Decreasing CTCF levels in bimodal BL41 cells led to a marked loss of CD45RB expression without reducing overall CD45 levels (Fig. 2a and Supplementary Fig. 4b). Similarly, CTCF depletion in RBlow cells and BJAB cells led to a substantial loss of CD45RB staining with little effect on overall CD45 levels (Fig. 2a). Quantitative RT–PCR of CD45 mRNA in CTCF-depleted RBlow and BJAB cells validated reduced exon 5 expression and increased exon 4/6 junctions (Fig. 2b, c, respectively; Supplementary Fig. 4c, additional transductions), confirming that CTCF mediates exon 5 inclusion.
Figure 2 |. CTCF depletion leads to reduced exon 5 inclusion in CD45 transcripts.
a, Cell-surface CD45RB isoform and total CD45 expression in cells transduced with short hairpin RNA (shRNA)against CTCF (CTCF-sh3 and/or sh-4) or control shRNA against red fluorescent protein (RFP). b, c, qRT–PCR in CTCF-depleted RBlow (b) and BJAB cells (c) from a to detect CD45 (left) and CTCF (right) mRNA levels (n = 3). Graphs show mean values ± s.d. P, two-tailed Student’s t test.
CTCF promotes pol II pausing at CD45 exon 5
We next investigated the mechanism by which CTCF binding to CD45 DNA influences mRNA splicing outcomes. Given that genome-wide ChIP-seq studies have revealed overlapping intragenic CTCF and pol II peaks30, we examined whether CTCF promotes inclusion of exon 5 through interference with pol II elongation. ChIP confirmed significant enrichment of pol II at CD45 exon 5 DNA, but not at adjacent regions in RBhigh cells as compared to RBlow cells (Fig. 3a). Using antibodies specific to pol II phosphorylated on the carboxy-terminal domain (CTD), we further showed that elevated pol II at CD45 exon 5 in RBhigh cells is associated with the elongating form phosphorylated on serine 2 of CTD YSPTSPS heptad repeats31 (Supplementary Fig. 5a, b). Notably, CTCF depletion from RBhigh cells (Supplementary Fig. 5c, d) reduced both CTCF binding (Fig. 3b) and pol II levels at CD45 exon 5 (Fig. 3c).
Figure 3 |. CTCF binding at CD45 exon 5 DNA facilitates exon 5 inclusion in CD45 transcripts through local pol II pausing.
a, RNA pol II ChIP and qPCR relative to mouse Ig control IP (n = 3). b, CTCF ChIP in RBhigh cells transduced with shRNA against CTCF versus shRFP-transduced cells and qPCR relative to rabbit Ig control IP (n = 2). c, RNA pol II ChIP of RBhigh cells from b and qPCR relative to mouse Ig control IP (n = 2). d, In vitro transcription with a DNA oligo incorporating a CTCF binding site at position 26 relative to elongation complex assembly. Recombinant CTCF and TFIIS protein were introduced as indicated, with variable effects on pausing at adenine 21 (A21). e, Representation of CD45 minigenes with wild-type (I3-I7) or mutated exon 5 CTCF binding site (I3-I7*CTCF), used in f–j. f, CTCF-ChIP in NIH3T3 and CHO cells transfected with the CD45 minigenes and qPCR relative to rabbit Ig control IP. Error bars represent standard error of the mean (s.e.m.) (n = 3). g–i, qRT–PCR from minigene-transfected HEK293, NIH3T3 and CHO cells to detect the junctions of exons 4/5 (g), 5/6 (h) and 4/6 (i) relative to exon 6 (n = 3). j, RNA pol II ChIP in CHO cells transfected with the CD45 minigenes and qPCR relative to mouse Ig control IP (n = 3). Unless indicated otherwise, graphs show mean values ± s.d. P, two-tailed Student’s t test.
The above data definitely link CTCF binding, pol II pausing and exon 5 inclusion, but do not exclude additional, context-dependent secondary effects. To query whether CTCF binding to an actively transcribed template is sufficient to promote pol II pausing, we assembled a pol II ternary elongation complex from synthetic DNA and RNA oligonucleotides and highly purified yeast pol II32. A CTCF binding site was incorporated into the template DNA at position 26 relative to the hybridization location of a 9-nucleotide RNA primer (Fig. 3d). CTCF binding to the target sequence was confirmed by electrophoretic mobility shift assay (EMSA) (Supplementary Fig. 6a). Incubation with pol II and increasing amounts of recombinant CTCF resulted in pausing immediately upstream of the CTCF binding site (Fig. 3d). Extended incubation or introduction of the elongation factor TFIIS substantially reduced pausing and led to near complete escape of paused pol II (Fig. 3d and Supplementary Fig. 6b). Thus, CTCF can autonomously promote pol II pausing, but not complete arrest, on a naked DNA template. These data establish CTCF as a direct impediment to transcription that can act in the absence of a particular nucleosome structure or chromatin context. Furthermore, the ability of paused pol II to resume transcription efficiently in the presence of CTCF supports a physiological role for CTCF in favouring exon inclusion through transient, spatiotemporal pol II pausing.
Having demonstrated that CTCF can promote pol II pausing, we explored the relationship between CTCF, pol II and exon inclusion in a tractable, endogenous system. We generated a wild-type minigene extending from intron 3 through intron 7 of human CD45 genomic DNA (I3-I7), as well as a mutant analogue, in which the exon 5 CTCF binding site was disrupted through nucleotide substitution (I3-I7*CTCF) (Fig. 3e and Supplementary Fig. 6c). The 11 zinc fingers of CTCF support multiple contacts to substrate DNA33,34 and a minimum of five substitutions within the core motif35 were required to significantly ablate CTCF binding (EMSA, Supplementary Fig. 6d). To avoid detection of endogenous CD45, which is confined to the haematopoietic lineage18, the minigenes were transfected into several fibroblast cell lines. In addition to human HEK293 cells, murine NIH3T3 and hamster CHO cells were used to specifically amplify human minigene CD45 DNA in ChIP analyses. CTCF ChIP of transfected NIH3T3 and CHO cells confirmed robust binding to exon 5 of the I3-I7 minigene, and complete disruption of binding to the mutated, I3-I7*CTCF minigene (Fig. 3f). Quantitative RT–PCR indicated that both minigenes were comparably expressed and approximated endogenous CD45 levels in immune cells (Supplementary Fig. 6e). Mutation of the CTCF binding site in exon 5 led to a marked decrease in 4/5 and 5/6 junctions, and increase in 4/6 junctions in all three cell types (Fig. 3g, h, i, respectively), resulting in an overall 50–100× decrease in exon 5 inclusion. Notably, ChIP confirmed increased pol II occupancy at exon 5 in the I3-I7 minigene, but not in the mutated, I3-I7*CTCF minigene (Fig. 3j). As the two minigenes are identical in every regard minus the five core nucleotides of the CTCF binding site, these data establish CTCF as a direct regulator of CD45 exon 5 inclusion, which operates through promoting local pol II pausing.
DNA methylation inhibits exon 5 CTCF binding
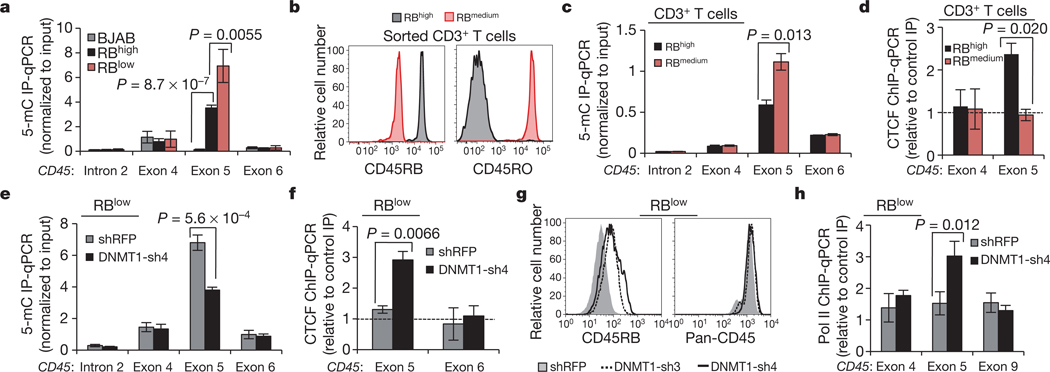
Armed with the knowledge that CTCF binding to exon 5 DNA regulates inclusion, and given that exon 5 is variably excluded during lymphocyte maturation, we asked whether and how CTCF binding is modulated to influence splicing outcome. Whereas CTCF is ubiquitously expressed, binding to DNA is inhibited by methylation on CpG dinucleotides27,34. Several recent studies have shown that DNA methylation is substantially enriched at exons relative to introns14,15,36, suggesting a role in pre-mRNA processing, yet a causal relationship between these processes has not been demonstrated. Methylated DNA immunoprecipitation (MedIP) in our B-cell lines suggested that CTCF binding at CD45 exon 5 and associated exon inclusion are indeed regulated by DNA methylation: we detected a strong inverse correlation between CTCF and 5-methylcytosine at CD45 exon 5, but not at adjacent exons (Figs 1d, 4a). To assess whether DNA methylation of CD45 exon 5 and reciprocal loss of CTCF binding contribute to exon 5 exclusion during the transition from naïve to mature T lymphocytes, CD3+ T cells were isolated from human peripheral blood and sorted into RBhigh (naive) and RBmedium (mature) populations19 (Fig. 4b). MedIP confirmed significant enrichment of CD45 exon 5 methylation (Fig. 4c) and reduced CTCF binding (Fig. 4d) in RBmedium cells compared to RBhigh peripheral T cells. Thus, CTCF binding and CD45 exon 5 inclusion are inversely related to DNA methylation in several transformed cell lines and in primary T cells.
Figure 4 |. 5-methylcytosine levels (5-mC) are inversely related to CTCF binding and exon 5 inclusion.
a, Methylated DNA immunoprecipitation (MedIP) in B cell line genomic DNA and qPCR relative to input (n = 5). b, Representative CD45 isoform expression in primary peripheral human CD3+ T cells sorted on the basis of cell-surface CD45RB and CD45RO. c, MedIP and qPCR relative to input in sorted primary human CD3+ T cells (n = 6, compiled from two donors). d, CTCF-ChIP and qPCR relative to rabbit Ig control IP, in sorted primary CD3+ T cells (n = 2). e, MedIP and qPCR relative to input in BL41 RBlow cells transduced with shRNA against DNMT1 versus shRFP-transduced cells (n = 3). f, CTCF ChIP in cells from e and qPCR relative to rabbit Ig control IP (n = 3). g, Cell-surface CD45RB expression in cells from e. h, RNA pol II ChIP and qPCR in cells from e relative to mouse Ig control IP (n = 3). Unless indicated otherwise, graphs show mean values ± s.d. P, two-tailed Student’s t test.
To determine whether dynamic methylation of CD45 exon 5 DNA is a regulatory mechanism contributing to CD45 alternative splicing, we modulated methylation through inhibition of the DNA maintenance methyltransferase, DNMT1. We reasoned that, if elevated exon 5 methylation and consequent reduced CTCF binding were the principal components distinguishing RBlow and RBhigh cells, inhibition of methylation should cause RBlow cells to revert to an RBhigh phenotype. Indeed, DNMT1 depletion in RBlow cells (Supplementary Fig. 7a, b) reduced 5-methylcytosine levels (Fig. 4e) and restored CTCF binding at CD45 exon 5 (Fig. 4f), leading to enhanced exon 5 inclusion in CD45 mRNA, as evidenced by increased exon 4/5 and 5/6 junctions (Supplementary Fig. 7c) and cell-surface CD45RB (Fig. 4g). Notably, increasing CTCF binding in RBlow cells through reduced exon 5 methylation also reinstated local pol II pausing (Fig. 4h). In addition to identifying dynamic DNA methylation as a possible regulatory mechanism governing CD45 alternative splicing in vivo, these data establish CTCF as the first mechanistic link between DNA methylation and alternative pre-mRNA splicing.
Global effects of intragenic CTCF on splicing
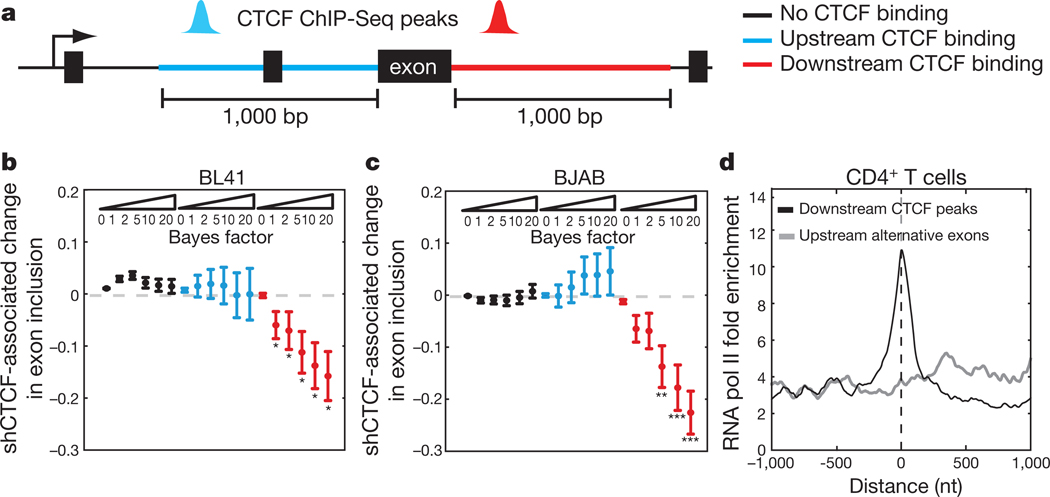
Although studies of CTCF function have been largely restricted to intergenic activities, CTCF ChIP-seq studies found that approximately 40–45% of CTCF binding sites are located intragenically35,37,38. Based on our observations with CD45, we propose that some portion of intragenic CTCF binding sites operate to influence pre-mRNA processing decisions. To globally address the impact of CTCF on alternative splicing, we performed CTCF ChIP-seq in BL41 and BJAB cells to produce cell-type-specific CTCF binding maps, and high-throughput RNA-sequencing (RNA-seq) of total RNA from CTCF-depleted BL41 and BJAB cells and their relevant controls (CTCF-sh3, Fig. 2a). Mapping of overall CTCF binding sites in BL41 and BJAB cells indicated comparable distribution patterns to previous reports (Supplementary Table 2). The mixture of isoforms (MISO) model was applied to RNA-seq data (Supplementary Table 3) to identify exons with a high probability of differential expression in response to CTCF depletion, as assessed by the Bayes factor confidence index39. Exons showing altered inclusion in response to CTCF depletion were further subdivided into three categories based on proximity to a local CTCF binding site: unbound by CTCF, or CTCF-bound within 1 kilobase downstream or upstream of the exon (Fig. 5a and Supplementary Table 4). CTCF is a global regulator of transcription27 and depletion would be expected to result in some level of alternative splicing due to alterations in upstream pathways. Accordingly, MISO identified exons that were differentially included in mRNA in response to CTCF depletion, but were not locally bound by CTCF on the corresponding DNA. Importantly, in BL41 and BJAB cells, alternative exons not bound by CTCF were centred at zero across Bayes factor thresholds, indicating that secondary effects of CTCF depletion showed no preference towards exon inclusion or exclusion (Fig. 5b, c, respectively). Similarly, CTCF binding upstream of the differentially expressed exon did not show a statistically significant bias towards exon inclusion or exclusion (Fig. 5b, c and Supplementary Fig. 8a, b). However, we detected a strong correlation between CTCF depletion and exon exclusion if CTCF is bound downstream of the alternative exon in both BL41 and BJAB cells (Fig. 5b, c and Supplementary Fig. 8a, b). We additionally identified CTCF-bound exons that showed reduced inclusion in BL41 and BJAB cells, as well as unique examples, indicating a degree of cell-type specificity (Supplementary Figs 8c, 9a and Supplementary Table 4).
Figure 5 |. Global identification of CTCF-dependent exons.
a, Alternative exons were classified on the basis of the relative location of an exclusive CTCF peak within 1 kb of the exon. b, Difference in the mean exon inclusion level between bimodal BL41 cells transduced with shRNA against CTCF versus shRFP-transduced cells (from Fig. 2a) for exons with CTCF peak in upstream (blue) or downstream regions (red) but not in the exon body and for exons with no CTCF binding (black). The mean ± s.e.m. for each class of exons is plotted against increasing Bayes factor thresholds. *P < 0.05, **P < 0.01, ***P < 0.001, Wilcoxon rank sum test for differences in exon inclusion at the different thresholds. c, Same as b for BJAB shCTCF compared with wild-type BJAB cells (from Fig. 2a). d, Normalized CD4+ T cell RNA pol II read signal centred on the alternative exon or the corresponding downstream CTCF peak summit.
These genome-level data are consistent with our observations in the CD45 model system, wherein CTCF binding downstream of the weak 3′ splice site flanking exon 5 promoted inclusion of exon 5 in mature message, but had no effect on exon 6 (Figs 1c, d and 2b, c). As we had mechanistically linked CTCF-associated pol II pausing to CD45 exon 5 inclusion, we examined pol II occupancy at the downstream CTCF sites that led to reduced exon inclusion upon CTCF depletion. Inspection of publicly available CTCF ChIP-seq data from CD4+ T cells37 indicated high conservation of these CTCF binding sites26. Analysis of the corresponding pol II ChIP-seq data37 revealed a stronger enrichment of pol II at downstream CTCF binding sites relative to upstream exons (Fig. 5d). Enrichment of pol II occupancy at CTCF binding sites compared to associated upstream alternative exons was confirmed for several genes in BJAB and BL41 cells (Supplementary Fig. 9b). Together with our CD45 data, we conclude that CTCF bound downstream of alternative exons promotes pol II pausing, providing the necessary temporal context for co-transcriptional spliceosome assembly at weak upstream splice sites.
Discussion
In recent years, the link between DNA structure and pre-mRNA processing has been gaining increasing attention. Reports of increased nucleosome occupancy and DNA methylation as well as distinct histone methylation patterns at exons relative to introns have fuelled the hypothesis that exons are differentially marked to aid the spliceosome in the process of exon definition16,17. It has further been shown that pol II occupancy increases in the vicinity of exons, although whether a function of DNA sequence, chromatin structure or the presence of DNA-binding proteins has not been defined. Recently, several studies have linked modification of distinct histone methylation patterns to alternative splicing16,17. However, exonic histone methylation was shown to be equivalent in other models of robust exon inclusion versus exclusion40, suggesting that, although histone methylation patterns may prime splicing decisions, they probably do so in concert with other factors. Consistent with the latter, we observed comparable histone methylation at exon 5 whether or not exon 5 was included in the CD45 message (Supplementary Fig. 3a, b). Rather, we show that mutually exclusive DNA methylation and CTCF binding regulate exon 5 inclusion through influencing pol II elongation dynamics (Supplementary Fig. 10). Given that mapping of CTCF binding sites shows roughly 40–70% conservation between tissues27, it is tempting to speculate that altered DNA methylation patterns during development can lead to variations in intragenic CTCF binding that thereby contribute to tissue-specific alternative splicing patterns. This may be especially relevant in pathological conditions, such as cancer, where widespread changes in DNA methylation, altered CTCF binding, and aberrant alternative pre-mRNA splicing have been reported15,41–43. We predict that our identification of CTCF as a DNA-binding regulator of alternative pre-mRNA splicing represents the tip of the iceberg, and that a long list of location-specific DNA-binding ‘splicing factors’ will follow.
METHODS SUMMARY
Experiments were performed with BJAB and BL41 cells or primary lymphocytes. CD45 isoform analysis was achieved with isoform-specific antibodies or pan-antibody directed against a common region of CD45. Transductions were executed with vesicular stomatitis virus G (VSV-G)-pseudotyped lentivirus, and selected for puromycin resistance. Quantitative RT–PCR was performed on cDNA from total RNA. Protein lysates were prepared with RIPA buffer. ChIP and MedIP were conducted with formaldehyde cross-linked, sonicated material. In vitro transcription elongation was performed with yeast RNA pol II, yeast TFIIS and human CTCF. Minigenes were cloned into the pCI-neo (Promega) construct and transfected with Lipofectamine 2000 (Invitrogen). ChIP-Seq and RNA-Seq were executed with the Illumina platform. For ChIP-Seq, Illumina FastQ files were mapped to the human genome (hg19). Peak calling was run using Rabbit Ig control sequencing data as background. For RNA-Seq, exon inclusion levels were determined using the MISO program39.
METHODS
Cell culture.
BJAB and BL41 cells were maintained at 37 °C, 5% CO2 in RPMI (Invitrogen) supplemented with 10% FBS (Hyclone), and 1% L-glutamine. BJAB and parental RBhigh BL41 cells were cultured in heat-inactivated FBS, whereas RBlow BL41 cells were initially kept in native FBS, but were ultimately transitioned into inactivated serum. JSL1 cells were maintained at 37 °C, 5% CO2 in RPMI (Invitrogen) supplemented with 5% FBS (Hyclone), and 1% L-glutamine. Primary human peripheral blood lymphocytes were purified by spinning through Ficoll Paque (GE Healthcare). Isolated cells were washed twice with PBS and CD3+ T cells were isolated with CD3+ microbeads (Miltenyi Biotech). Primary murine splenocytes were isolated from whole spleen of BL/6 mice. Single-cell suspensions were lysed with ACK lysis buffer (0.15 M NH4CL, 10 mM KHCO3, 0.1 mM Na2EDTA) to remove red blood cells before ChIP assay. JSL1 cells were stimulated at a concentration of 3 × 105 cells per ml. Phorbol 12-myristate 13-acetate (PMA) was added at a final concentration of 20 nM. Flow cytometry was performed 2 days post-stimulation.
Virus production.
Constructs encoding shRNA directed against CTCF and DNMT1 were obtained from Open Biosystems and were transfected (Lipofectamine 2000, Invitrogen) along with VSV-G and gag/pol (courtesy of The RNAi Consortium of the Broad Institute) into 293T cells for viral production. Viral supernatants were concentrated 50× and aliquoted for storage.
Cell line infection.
BJAB and BL41 cells were plated in 96-well round-bottom plates at 100,000 cells per well. Five microlitre of virus and 8 μg ml−1 polybrene were added per well and the plate was spun at 760g for 90 min. The supernatants were removed and fresh media was added. Puromycin was added at a final concentration of 5 μg ml−1 on day 2. Depletion of CTCF from cells resulted in significant cell death after 1 week in culture and depletion of DNMT1 resulted in silencing after 10 days in culture. Cells for downstream analysis were collected 5 (shCTCF) or 7 (shDNMT1) days post-infection. To scale up infections for ChIP and western blotting, infections were performed in individual wells of 96-well plates and pooled before harvesting for RNA, ChIP and western blot. Three plates were pooled for shCTCF experiments and 3.5 plates were pooled for shDNMT1 experiments. Three individual RNA and ChIP samples were taken from each of the bulk cultures.
Target sequences of shRNAs.
DNMT1-sh3, 5′-CGAGAAGAATATCGAACTCTT-3′; DNMT1-sh4, 5′-CGACTACATCAAAGGCAGCAA-3′; CTCF-sh3, 5′-CCTCCTGAGGAATCACCTTAA-3′; CTCF-sh4, 5′-GCGGAAAGTGAACCCATGATA-3′; shRFP, 5′-GAATTAAGAGAGGCTCAGTTA-3′; LL-sh4, 5′-CGACAGGCTCTAGTGGAATTT-3′.
Flow cytometry.
The following antibodies were used for flow cytometry: CD45RO clone UCHL1 (eBioscience, 12–0457-42, batch no. E034572), CD45RA clone MEM-56 (ExBio, 1P-223, batch no. 11827), CD45RB clone MT4 (BD Pharmingen, 555904, batch no. 89956) and pan-CD45 clone HI30 (BD Pharmingen, 555483, batch no. 555483). Staining of CD45 isoforms was performed in separate tubes, to avoid competition for antibody binding. Flow cytometry was performed on either a BD FACSCalibur or BD LSR II cytometer.
Quantitative RT–PCR.
RNA was isolated with the Qiagen RNeasy Mini Kit and reverse transcription was performed with SuperScript II (Invitrogen) according to the manufacturer’s instructions. PCR measurements were performed in triplicate in the presence of SYBR green reagent (Roche) and amplification was performed on a 480 Light Cycler (Roche). The average cycle thresholds for the technical triplicates were calculated to yield one value per primer set for each biological replicate. Normalization was performed to GAPDH, RPS16 or surrounding exon level values using the formula to determine relative expression. Averages and standard deviations of the normalized biological replicate values were plotted in the figures and used in t-test calculations. Figure legends indicate the number of biological replicates (individual RNA preparations) used in each experiment.
Western blots.
Cells were lysed in RIPA buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1% NP40, 0.1% SDS, 0.5% sodium deoxycholate, and 1× Halt protease inhibitor cocktail (Thermo Scientific)). Proteins (35 μg) were loaded per lane on a 4–20% gradient SDS–PAGE gel. Western blot was performed with anti-CTCF clone D31H2(Cell signaling 3418S, batch no. 1), DNMT1 antibody (Abcam ab13537, batch no. GR16960–1), or anti-p65 RelA (BD Bioscience 610869, batch no. 50886) antibodies. Anti-RelA immunoblotting served as a loading control for protein levels.
Chromatin immunoprecipitation (ChIP).
Ten million cells were cross-linked for 10 min in 1% formaldehyde (Sigma) at room temperature, and quenched by adding glycine to a final concentration of 0.125 M for 5 min at room temperature. Cells were washed twice in chilled PBS, resuspended in buffer containing 50 mM HEPES-KOH (pH 7.5), 140 mM NaCl, 1 mM EDTA, 10% glycerol, 0.5% NP-40, 0.25% Triton X-100 and protease inhibitors (Thermo Scientific) and kept on ice for 10 min. Nuclei were pelleted at 800g for 5 min at 4 °C and resuspended in buffer containing 10 mM Tris-HCl pH 8.0, 200 mM NaCl, 1 mM EDTA, 0.5 mM EGTA and protease inhibitors (Thermo Scientific) followed by a 10-min incubation on ice. Nuclei were collected and resuspended in sonication buffer containing 10 mM Tri-HCl pH 8.0, 100 mM NaCl, 1 mM EDTA, 0.5% EGTA, 0.1% Nadeoxycholate, 0.5% N-lauryl sarcosine and protease inhibitors (Thermo Scientific). Sonication of DNA was performed in an ultra sonicator water bath (Bioruptor) using two ten cycle runs of 30 s ‘on’ and 30 s ‘off’ to achieve an average fragment length of 200–400 bp. After addition of 1% Triton X-100, samples were centrifuged at 16000g for 10 min at 4 °C. An aliquot of sonicated DNA was reverse-crosslinked and run on a 1% agarose gel to confirm fragment size during each ChIP procedure. Chromatin (25 μg) was immunoprecipitated by adding the antibody of interest followed by overnight incubation at 4 °C. The following antibodies were used for ChIP: anti-CTCF (Millipore 07–729, batch no. DAM1772428), anti-RNA polymerase II clone 4H8 (Millipore 05–623, batch no. DAM1731474), anti-Ser2P RNA polymerase II clone H5 (Covance MMS129R, batch no. E10017AF), anti-Ser5P RNA polymerase II clone H14 (Covance MMS134R, batch no. E10142DF), anti-H3K36Me3 (Abcam ab9050, batch no.947467), anti-H3K27Me3 (Abcam ab6002, batch no. 934602), anti-H3K4Me3 clone MC315 (Millipore 04–745, batch no. NG1717145), Normal Rabbit IgG (Cell signaling Technology 2729, batch no. 4), and normal mouse IgG (Millipore 12–371, batch no. 1718089). After overnight incubation, 30 μl of Dynal Protein A/G beads (Invitrogen) or Protein L magnetic beads (Biovision) (for phosphorylated RNA Pol II antibodies) were added and incubated for 1 h at 4 °C. Beads were washed sequentially for 3 min each in low salt (20 mM Tris-HCl pH 8.0, 150 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), high salt (20 mM Tris-HCl pH 8.0, 500 mM NaCl, 2 mM EDTA, 0.1% SDS, 1% Triton X-100), LiCl buffer (10 mM Tris-HCl pH 8.0, 0.25 M LiCl, 1% NP40, 1% Nadeoxycholate) and TE buffer. Beads were eluted in 150 μl elution buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% SDS, 50 mM NaHCO3) and treated with 1 μl RNase A (1 mg ml−1 Ambion) at 37 °C for 30 min. Cross-linking was reversed and proteins were degraded by addition of 1 μl proteinase K (20 mg ml−1 Ambion) and incubation at 65 °C for 4 h. Eluted DNA was purified with QIAquick PCR purification (Qiagen), according to the manufacturer instructions.
Immunoprecipitated DNA and 5% input DNA were analysed by SYBR-Green real-time quantitative PCR. PCR measurements were performed in duplicate. The average cycle thresholds for the technical replicates were calculated to yield one value per primer set for each biological replicate and normalized to input using the formula . These values were further normalized relative to the rabbit or mouse Ig control IP values for the primer set. Averages and standard deviations of the normalized biological replicate values were plotted in the figures and used in t-test calculations. Figure legends indicate the number of biological replicates (individual IPs) used in each experiment.
Methylated DNA immunoprecipitation (MedIP).
MedIP was performed essentially according to the protocol described in ref. 44. Genomic DNA was purified from approximately 25 million cells using Zymo research Quick gDNA Midiprep kit (D3100), according to the manufacturer’s instructions. For primary cells, CD3+ T cells were isolated from peripheral blood using CD3 microbeads (Miltenyi Biotech). CD3+ T cells were sorted into CD45RB high and CD45RB medium populations based on surface receptor staining of CD45RB and CD45RO. Purified genomic DNA was diluted into a total of 300 μl TE buffer and sonicated with a Bioruptor (10 cycles at low power, of 30 s ‘on’ and 30 s ‘off’) to an average size of 300–500 bp. An aliquot of sonicated DNA was run on 1% agarose gel to confirm fragment size during each MedIP procedure. Sonicated DNA (4 μg; 3 μg for primary cells) was denatured by incubation at 95 °C for 10 min and was immediately transferred to ice for 10 min. Immunoprecipitation buffer containing 10 mM sodium phosphate, 140 mM NaCl and 0.05% Triton X-100 was added to a final volume of 500 μl. For each IP reaction, 10 μg (8 μg for primary cells) of 5-methyl cytidine antibody clone b (Diagenode MAb-006–100, batch no. DA-0018) was added and incubated overnight at 4 °C with shaking. Five percent of DNA was kept as input.
After incubation, 30 μl of Dynal Protein G beads were added and further incubated for 1 h at 4 °C. Beads were washed thrice with 500 μl of IP buffer. Elution buffer (150 μl) containing 50 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% SDS, 50 mM NaHCO3 and 20 μg proteinase K was added and incubated at 55 °C for 3 h. Tubes were applied to a magnetic rack and eluted DNA and input DNA were purified with the Qiaquick PCR purification kit (Qiagen) followed by SYBR-Green real-time quantitative PCR to identify methylated regions. PCR measurements were performed in duplicate. The average cycle thresholds for the technical replicates were calculated to yield one value per primer set for each biological replicate and normalized to input using the formula . Averages and standard deviations of the normalized biological replicate values were plotted in the figures and used in t-test calculations. Figure legends indicate the number of biological replicates (individual IPs) used in each experiment.
In vitro transcription elongation assay.
RNA pol II from yeast Saccharomyces cerevisiae containing a histidine-tagged Rpb3 subunit was purified as described previously45. Histidine-tagged TFIIS expression plasmid46 was a gift from C. Kane. Recombinant TFIIS was purified according to ref. 46, with an additional purification on a Mono-S column (GE Helthcare). Human CTCF recombinant protein was obtained from Abnova (catalogue no. H00010664-P01, batch no. 0991020–2).
Elongation complex incorporating a 9-nt RNA was assembled as described previously47, purified with Amicon Ultra-0.5 ml centrifugal filter (Millipore), and diluted with transcription buffer (TB; 20 mM Tris-HCl pH 7.9, 5 mM MgCl2, 10 mM 2-mercaptoehanol, 40 mM KCl, 0.1 mg ml−1 BSA). The reaction was initiated by mixing 5 μl of TEC +/− XμM CTCF with 5 μl of 0.1–0.5 mM NTP (GE Healthcare) +/− 1 μM TFIIS in TB and was terminated with gelloading buffer (5 M urea, 25 mM EDTA at final concentration). RNA products were resolved in 20% denaturing polyacrylamide gels and visualized with a Typhoon 8600 phosphoimager (GE Helthcare).
Oligonucleotides used for elongation complex.
Sequences of RNA and DNA oligonucleotides are as follows. RNA, 5′-AUCGAGAGG-3′; DNA with CTCF binding site, non-template strand, 5′-GGTATAGGATACTTACAGCCATCGAGAGGGACAAGGCGAAAGCATCCACCAGGGGGCGCCAGCTAAT-3′; template strand, 5′-ATTAGCTGGCGCCCCCTGGTGGATGCTTTCGCCTTGTCCCTCTCGATGGCTGTAAGTATCCTATACC-3′.
Electrophoretic mobility shift assay (EMSA).
The CTCF-binding oligonucleotides used for EMSA correspond to either the template used for in vitro transcription (Supplementary Fig. 6a) or the CTCF binding sites in the wild-type and mutated I3-I7 minigenes (Supplementary Fig. 6d). The two strands of DNA were annealed, 5′ end-labelled with [γ−32P] ATP and purified with a G-50 Micro column (GE healthcare). DNA probe (3 pM) equalling approximately 70,000 c.p.m. was mixed with glutathione S-transferase (GST)-tagged CTCF in binding buffer containing PBS and 5 mM MgCl2, 0.1 mM ZnSO4, 1 mM DTT, 0.1% NP40 and 10% glycerol. EMSA reaction mixtures (20 μl final volume) were incubated for 20min at room temperature followed by electrophoresis on 5% native polyacrylamide gels and visualized as described above for in vitro transcription.
EMSA DNA probe sequences.
In vitro transcription probe, 5′-CATCCACCAGGGGGCGCCAGCTAAT-3′ and 5′-ATTAGCTGGCGCCCCCTGGTGGATG-3′; wild-type exon 5 probe, 5′-TCAGTTCCAGCAGAGGGCGTCTGCG-3′ and 5′-CGCAGACGCCCTCTGCTGGAACTGA-3′; mutated exon 5 probe, 5′-TCAGTTAAAGCTGAGTACGTCTGCG-3′ and 5′-CGCAGACGTACTCAGCTTTAACTGA-3′.
ChIP-Seq analyses.
Illumina FastQ files were mapped to the human genome (hg19) using Bowtie48 requiring a unique match (using the ‘-m 1’ flag). The aligned reads in SAM format were converted to BED format before running the MACS peak caller49. MACS peak calling was run using the rabbit Ig control sequencing data as background files for BJAB and BL41 CTCF ChIP-Seq data, respectively. The number of peaks identified per ChIP-Seq sample and sequenced reads are listed in Supplementary Table 2.
To investigate the effects of CTCF binding upon pre-mRNA splicing, we compared CTCF ChIP-Seq peaks with a set of alternative exons1 requiring that the CTCF peak summit was located within 1,000 bp of the alternative exon boundaries (see Fig. 5a). Based on the presence of local CTCF peaks in BL41, BJAB and CD4 we classified each alternative exon into exons that were unbound by CTCF, and exons with either downstream or upstream CTCF binding. Classified unbound exons lacked CTCF peak summits in both the alternative exon body and within 1,000 bp on either side of the alternative exon. Exons with downstream CTCF binding had one or more CTCF peak summits within the region spanning from the alternative exon 5′ splice site and 1,000 bp downstream in one or more of the CTCF data in BJAB, BL41 or CD4. Any alternative exons with a downstream CTCF peak but additional peak summits in the upstream region or within the alternative exon were not considered. The reciprocal procedure was used to classify exons with upstream CTCF binding. Alternative exons classified by local CTCF binding together with exon inclusion levels are provided in Supplementary Table 4.
RNA-Seq analyses.
Illumina FastQ files were mapped to the human genome (hg19) and a collection of junctions using Tophat version 1.1.4 (ref. 50), using the paired-end mode and requiring a unique match. The resulting SAM file with uniquely mapped reads was converted to BAM format using samtools51. Mapping statistics for the RNA-Seq data are provided in Supplementary Table 3. We estimated exon inclusion levels of a collection of 42,557 alternative exons (approximately the same as in ref. 1) using the MISO program39 with the default parameters using the ‘compute-genes-psi’ function. The estimated exon inclusion levels from different RNA-Seq experiments were compared using MISO function ‘compare-samples’ to obtain exon inclusion level differences and Bayes factors. Statistically significant differences in exon inclusion levels between CTCF-bound and -unbound exons at different thresholds were evaluated using the Wilcoxon rank sum test. The overall difference in gene expression across RNA-Seq samples was evaluated using singular value decomposition. First we computed the expression level of each Refseq transcript as reads per kilobase and million mappable reads using the rpkmforgenes program52. The full gene expression matrix was normalized to unit length per transcript and subsequently used as input for singular value decomposition using the svdman program53. The result in Supplementary Fig. 8c was obtained by projecting each sample onto the first two ‘eigenarrays’54.
RNA polymerase II ChIP-Seq analysis.
We generated normalized RNA polymerase II fold enrichment signals over a set of regions by dividing the observed read sum at each position with the expected read sum computed as: (total_reads * read_length * number_of_regions) / genome_length. The normalized fold enrichment at each position was smoothened by window averaging using a window size of 100 nucleotides. We analysed all alternative exons with a downstream CTCF peak summit conserved in CD4+ T cells (Supplementary Table 4) after removing exons with a CTCF peak within 1,000 bp of an annotated transcript start site or poly A site in Ensembl (to remove effects from strong RNA pol II signals at transcript start and end locations). This procedure rendered 408 exons from which we computed both the CTCF peak summit position and exon middle coordinate. These two sets of 408 genomic coordinates each were used as the centre for the analysis in Fig. 5d. The same procedure was used to generate CTCF peak summits and middle exon positions for alternative exons with a conserved CTCF peak in the upstream region for Supplementary Fig. 8e (number of exons identified was 416).
Minigenes and transfection.
CD45 minigenes were cloned into the pC1-neo mammalian expression vector (Promega). The wild-type CD45 minigene consists of 9.7 kb of CD45 genomic DNA sequence extending from 2.4 kb of intron 3 through 588 bp of intron 7. The I3-I7*CTCF minigene was made by mutating the CTCF binding site of exon 5 using site-directed mutagenesis with the primers indicated in Supplementary Table 1. The minigenes were transfected (Lipofectamine 2000, Invitrogen) along with pC1-neo vector control into HEK293 cells, CHO cells and NIH-3T3 cells. Cells were collected 48h after transfection for RNA isolation (RNeasy, Qiagen) and chromatin immunoprecipitation. Transfection was performed in triplicate for HEK293 and CHO cells, with an individual RNA preparation (HEK293 and CHO) and duplicate ChIPs (CHO) derived from each of the three dishes. Transfection was performed in a single dish for NIH3T3 cells with three individual RNA preparations and triplicate ChIPs derived from the one dish.
Supplementary Material
Acknowledgements
We thank A. Rao, C. Burge and K. Lynch for critical reading of this manuscript. We also thank A. Rao for reagents and K. Nyswaner and M. Prigge for technical assistance. This work is supported by the Intramural Research Program of NIH, the National Cancer Institute, The Center for Cancer Research (S.O., P.O., M.K.), and the Swedish Research Council Foundation and the Foundation for Strategic Research (R.S.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
References
- 1.Wang ET et al. Alternative isoform regulation in human tissue transcriptomes. Nature 456, 470–476 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pan Q, Shai O, Lee LJ, Frey BJ & Blencowe BJ Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nature Genet. 40, 1413–1415 (2008). [DOI] [PubMed] [Google Scholar]
- 3.Tazi J, Bakkour N. & Stamm S. Alternative splicing and disease. Biochim. Biophys. Acta 1792, 14–26 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Matlin AJ, Clark F. & Smith CW Understanding alternative splicing: towards a cellular code. Nature Rev. Mol. Cell Biol 6, 386–398 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Han J, Xiong J, Wang D. & Fu XD Pre-mRNA splicing: where and when in the nucleus. Trends Cell Biol. 21, 336–343 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh R. & Valcarcel J. Building specificity with nonspecific RNA-binding proteins. Nature Struct. Mol. Biol 12, 645–653 (2005). [DOI] [PubMed] [Google Scholar]
- 7.Kornblihtt AR Coupling transcription and alternative splicing. Adv. Exp. Med. Biol 623, 175–189 (2007). [DOI] [PubMed] [Google Scholar]
- 8.de la Mata M. et al. A slow RNA polymerase II affects alternative splicing in vivo. Mol. Cell 12, 525–532 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Tilgner H. et al. Nucleosome positioning as a determinant of exon recognition. Nature Struct. Mol. Biol 16, 996–1001 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Spies N, Nielsen CB, Padgett RA & Burge CB Biased chromatin signatures around polyadenylation sites and exons. Mol. Cell 36, 245–254 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andersson R, Enroth S, Rada-Iglesias A, Wadelius C. & Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 19, 1732–1741 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolasinska-Zwierz P. et al. Differential chromatin marking of introns and expressed exons by H3K36me3. Nature Genet. 41, 376–381 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartz S, Meshorer E. & Ast G. Chromatin organization marks exon-intron structure. Nature Struct. Mol. Biol 16, 990–995 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Chodavarapu RK et al. Relationship between nucleosome positioning and DNA methylation. Nature 466, 388–392 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hodges E. et al. High definition profiling of mammalian DNA methylation by array capture and single molecule bisulfite sequencing. Genome Res. 19, 1593–1605 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luco RF, Allo M, Schor IE, Kornblihtt AR& Misteli T. Epigenetics in alternative pre-mRNA splicing. Cell 144, 16–26 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alló M. et al. Chromatin and alternative splicing. Cold Spring Harb. Symp. Quant. Biol 75, 103–111 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Trowbridge IS & Thomas ML CD45: an emerging role as a protein tyrosine phosphatase required for lymphocyte activation and development. Annu. Rev. Immunol 12, 85–116 (1994). [DOI] [PubMed] [Google Scholar]
- 19.Hermiston ML, Xu Z. & Weiss A. CD45: a critical regulator of signaling thresholds in immune cells. Annu. Rev. Immunol 21, 107–137 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Oberdoerffer S. et al. Regulation of CD45 alternative splicing by heterogeneous ribonucleoprotein, hnRNPLL. Science 321, 686–691 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Topp JD, Jackson J, Melton AA & Lynch KW A cell-based screen for splicing regulators identifies hnRNP LL as a distinct signal-induced repressor of CD45 variable exon 4. RNA 14, 2038–2049 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Motta-Mena LB, Heyd F. & Lynch KW Context-dependent regulatory mechanism of the splicing factor hnRNP L. Mol. Cell 37, 223–234 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tong A, Nguyen J. & Lynch KW Differential expression of CD45 isoforms is controlled by the combined activity of basal and inducible splicing-regulatory elements in each of the variable exons. J. Biol. Chem 280, 38297–38304 (2005). [DOI] [PubMed] [Google Scholar]
- 24.Horgan KJ et al. CD45RB expression defines two interconvertible subsets of human CD4+ T cells with memory function. Eur. J. Immunol 24, 1240–1243 (1994). [DOI] [PubMed] [Google Scholar]
- 25.Birney E. et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447, 799–816 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.UCSC Genome Browser, GRC37/hg19, ENCODE Histone Modification Tracks. [Google Scholar]
- 27.Phillips JE & Corces VG CTCF: master weaver of the genome. Cell 137, 1194–1211 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohlsson R, Bartkuhn M. & Renkawitz R. CTCF shapes chromatin by multiple mechanisms: the impact of 20 years of CTCF research on understanding the workings of chromatin. Chromosoma 119, 351–360 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Luco RF et al. Regulation of alternative splicing by histone modifications. Science 327, 996–1000 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wada Y. et al. A wave of nascent transcription on activated human genes. Proc. Natl Acad. Sci. USA 106, 18357–18361 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol. Cell 36, 541–546 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Komissarova N, Kireeva ML, Becker J, Sidorenkov I. & Kashlev M. Engineering of elongation complexes of bacterial and yeast RNA polymerases. Methods Enzymol. 371, 233–251 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Filippova GN et al. An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol 16, 2802–2813 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Renda M. et al. Critical DNA binding interactions of the insulator protein CTCF: a small number of zinc fingers mediate strong binding, and a single finger-DNA interaction controls binding at imprinted loci. J. Biol. Chem 282, 33336–33345 (2007). [DOI] [PubMed] [Google Scholar]
- 35.Kim TH et al. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128, 1231–1245 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lyko F. et al. The honey bee epigenomes: differential methylation of brain DNA in queens and workers. PLoS Biol. 8, e1000506 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barski A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007). [DOI] [PubMed] [Google Scholar]
- 38.Jothi R, Cuddapah S, Barski A, Cui K.& Zhao K. Genome-wide identification of in vivo protein-DNA binding sites from ChIP-Seq data. Nucleic Acids Res. 36, 5221–5231 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Katz Y, Wang ET, Airoldi EM & Burge CB Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nature Methods 7, 1009–1015 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Huff JT, Plocik AM, Guthrie C. & Yamamoto KR Reciprocal intronic and exonic histone modification regions in humans. Nature Struct. Mol. Biol 17, 1495–1499 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Berdasco M. & Esteller M. Aberrant epigenetic landscape in cancer: how cellular identity goes awry. Dev. Cell 19, 698–711 (2010). [DOI] [PubMed] [Google Scholar]
- 42.David CJ & Manley JL Alternative pre-mRNA splicing regulation in cancer: pathways and programs unhinged. Genes Dev. 24, 2343–2364 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jelinic P. & Shaw P. Loss of imprinting and cancer. J. Pathol 211, 261–268 (2007). [DOI] [PubMed] [Google Scholar]
- 44.Mohn F, Weber M, Schubeler D. & Roloff TC Methylated DNA immunoprecipitation (MeDIP). Methods Mol. Biol 507, 55–64 (2009). [DOI] [PubMed] [Google Scholar]
- 45.Kireeva ML et al. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol. Cell 30, 557–566 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Awrey DE et al. Transcription elongation through DNA arrest sites. A multistep process involving both RNA polymerase II subunit RPB9 and TFIIS. J. Biol. Chem 272, 14747–14754 (1997). [DOI] [PubMed] [Google Scholar]
- 47.Kireeva ML, Lubkowska L, Komissarova N. & Kashlev M. Assays and affinity purification of biotinylated and nonbiotinylated forms of double-tagged core RNA polymerase II from Saccharomyces cerevisiae. Methods Enzymol. 370, 138–155 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Langmead B, Trapnell C, Pop M. & Salzberg SL Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang Y. et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trapnell C, Pachter L. & Salzberg SL TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramsköld D, Wang ET, Burge CB & Sandberg R. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput. Biol 5, e1000598 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wall ME, Dyck PA & Brettin TS SVDMAN–singular value decomposition analysis of microarray data. Bioinformatics 17, 566–568 (2001). [DOI] [PubMed] [Google Scholar]
- 54.Wall ME, Rechsteiner A. & Rocha LM in A Practical Approach to Microarray Data Analysis (eds Berrar DP, Dubitzky W. & Granzow M.) pp. 91–109 (Springer, 2003). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.