Abstract
Aims
Thalassaemia is one of the most common genetics disorders in the world, especially in southern China. The aim of the present study was to investigate the feasibility of combining the gap-PCR and next-generation sequencing (NGS) for thalassaemia carrier screening in the Chinese population.
Methods
Blood samples were obtained from 944 prepregnancy couples; thalassaemia carrier screening was performed by using a routine haematological method and a combination of gap-PCR and NGS method.
Results
We found that the α thalassaemia carrier rate was 11% (207/1888); the β thalassaemia carrier rate was 3.7% (70/1888); the composite α thalassaemia and β thalassaemia carrier rate was 0.4% (8/1888). We also identified seven novel mutations, including HBA1: c.412A>G, −50 (G>A), HBB: c.*+129T>A, HBB: c.-64G>C, HBB: c.-180G>C, HBB: c.*+5G>A and HBB: c.-113A>G. By comparing the combined gap-PCR and NGS method, the MCV+MCH and HbA2 detection strategy showed a lower sensitivity of 61.05% (105/172) and a higher missed diagnosis ratio of 38.95% (67/172) for α thalassaemia mutations. The sensitivity was improved with the MCV+MCH and HbA2 detection screen when compared with MCV+MCH detection for β thalassaemia (98.51% vs 85.90%).
Conclusions
Our study suggests the combined gap-PCR and NGS method is a cost-effective method for the thalassaemia carrier screening, particularly for the α thalassaemia mutation carriers.
Keywords: diagnostic screening, haematology, genetics
Introduction
Thalassaemia is one of the most common genetic disorders in the world and is caused by α-globin and β-globin gene mutations. Thalassaemia is distributed mainly in coastal areas of the Mediterranean sea, Africa, the Middle East, India and southeastern Asia.1 In China, thalassaemia is mainly distributed in southern China, particularly in Guangxi, Guangdong and Hainan provinces.2–4 Most patients with severe thalassaemia may die in utero or during early childhood if untreated. Thus, effective preconception screening is essential for reducing the incidence of thalassaemia in high-prevalence regions of thalassaemia.
Over 1530 mutations ranging from single-nucleotide variations to large rearrangements are associated with thalassaemia or abnormal haemoglobin variants (HbVar database for human HbVar and thalassaemia mutations). Moreover, several variants at various loci have been identified to modify β thalassaemia phenotypes.5–7 The traditional method uses haematological and biochemical tests and subsequent molecular genetic tests to screen for thalassaemia carriers with phenotypic traits associated with thalassaemia.1 8 Whereas a phenotypic screening approach has a certain weakness in carrier testing, many carriers who have normal or borderline red cell indices and/or HbA2 levels may be missed.8 9
Next-generation sequencing (NGS) allows for multiplex and high-throughput detection of genetic variants10 and is commonly used for diagnosis of genetic disorders.11 12 Two recent studies have shown that NGS largely improved thalassaemia carrier screening and had a higher sensitivity and specificity than traditional methods; however, these results might be limited to certain ethnic population or rely only on the NGS technique.13 14 In the present study, we applied the gap-PCR and NGS techniques to test more than 300 mutations associated with thalassaemia in a cohort of 944 couples pretyped by routine screening methods. Our results suggested that the combined method outperformed traditional methods in the detection of pathogenic or likely pathogenic variants, thus providing an effective platform for application in clinical management of monogenic diseases in a large-scale population.
Materials and methods
Samples and demographic data
A total of 944 prepregnancy couples between August 2017 and August 2018 were screened for thalassaemia mutations in Xiaolan People’s Hospital of Zhongshan. Their age ranged from 23 to 40 years. All individuals provided informed written consent.
Haematological phenotype analysis
We collected 8 mL blood samples from each individual for routine blood examinations. An automated haematology analyser was used to detect mean corpuscular volume (MCV) and mean corpuscular haemoglobin (MCH). Participants with an MCV of <82 fL or an MCH of <27 pg underwent Hemoglobin A2 (HbA2) test by the alkaline agarose gel electrophoresis (Helena Spife 3000). An HbA2 value between 2.5% and 3.5% was considered normal. When HbA2 was ≤2.5%, α thalassaemia was suspected, whereas when HbA2 was ≥3.5%, β thalassaemia was considered.13
Reverse dot blot hybridisation (RDB)
Suspected α thalassaemia and β thalassaemia carriers with positive HbA2 results underwent routine genotyping tests. RDB was performed to detect common α thalassaemia or β thalassaemia mutations, including non-deletion-type thalassemia hemoglobin Constant Spring (CS) (HBA2: c.427T>C), Quong Sze (QS) (HBA2: c.377T>C), Westmead (WS) (HBA2: c.369C>G) and 17 β thalassaemia mutations, Int (HBB: c.2T>G), CD41-42 (HBB: c.124_127delTTCT), CD31(HBB: c.94delC), CD14-15 (HBB: c.45_46insG), CD17 (HBB: c.52A>T), CD71-72 (HBB: c.216_217insA), IVS-I-1 (HBB: c.92+1G>T), CD43 (HBB: c.130G>T), CD27/28 (HBB: c.84_85insC), IVS-II-654 (HBB: c.316–197C>T), −28 (HBB: c.-78A>G), −29 (HBB: c.-79A>G), IVS-1-5 (HBB: c.92+5G>C), βE (HBB: c.79G>A), −32 (HBB: c.-82C>A), −30 (HBB: c.-80T>C) and HBB: c.-50A>C. In the RDB assay, the amplified target DNA is first fixed to a nylon membrane to form a filter-fixed DNA dot; the dot is then hybridised to allele-specific oligonucleotide (ASO) probes whose 5′ terminal was conjugated with either P-labelled deoxynucleoside triphosphates, biotin or horseradish peroxidase. All procedures of RDB were conducted following the kit’s instructions (YanengBio, Shenzhen, China).
Gap-PCR tests and NGS screening
As for suspected α thalassaemia and β thalassaemia carriers, multiplex gap-PCR was used to detect deletion-type α thalassaemia α3.7, -α4.2, --SEA, --FIL and --THAI and β thalassaemia deletions (Chinese Ggamma (Agammadeltabeta) 0, South-East Asia type hereditary persistence of fetal hemoglobin (SEA-HPFH) and Taiwanese). NGS was applied to detect all mutations in HBA1, HBA2 and HBB genes.
Genomic DNA was extracted from 200 µL blood samples using the Kingfisher Flex (Thermo Scientific, Rockford, Illinois, USA) and isolated using the GenMag Nucleic Acid Isolation kit (Magnetic bead method) (GenMagBio, Beijing, China). DNA extracts were arrayed in 96-well plates, and the concentration was quantified by Nanodrop-8000 (Thermo Scientific, Waltham, MA, USA). We restricted our analysis to samples with a DNA concentration >20 µg/mL and an A260:A280 ratio between 1.8 and 2.0.We designed forward primers and specific reverse primers to amplify HBA1, HBA2 and HBB genes. PCR indexes were used to distinguish the amplified products. The primers used in this study came from a published article15 and patent (patent ID CN108796054A). The HBA1 amplicon size was 919 bp; the HBA2 amplicon size was 914 bp; and the HBB amplicon size was 599 and 771 bp. PCR reactions were performed in 96-well plates, with each sample corresponding to one library. Ninety-six kinds of index sequences were designed, corresponding to each well of the plate. Ninety-six samples were pooled into one tube for further library construction. The detailed experimental protocol was described in a previous study.13 15 ABI 9700 Thermal Cycler (Life Technologies, Foster City, California, USA) was used in PCR reactions. We adopted NGS library preparation protocol for library construction, including purified genomic DNA (Qiagen DNA Purification kit, Hilden, Germany), DNA quantification (NanoDrop 8000 UV-Vis Spectrophotometer, Thermo Fisher Scientific, Waltham, MA, USA), DNA fragmentation (excluding copy number variation amplicons), blunt-ended fragmentation (Enzymatics kits, Qiagen, Hilden, Germany), 3′-dA overhang, paired-end adapters ligation, DNA fragment separation (copy number variations not included) and size selection using magnetic beads. Sequencing was performed using the paired-end tag (PE100) protocol with an BGISEQ-500 machine (Beijing Genomics Institute, shenzhen, China).
Bioinformatics analyses
We identified α thalassaemia and β thalassaemia deletion genotypes according to the expected amplicon sizes for the deletion junction fragments by agarose gel electrophoresis. For transcripts used to call genetic variants, we required that the sequence depth should be greater than 50×. We developed a bioinformatics pipeline to detect Hb gene point mutations. First, we filtered low-quality reads and classified remaining reads based on adapter information (index primer). Then, we aligned reads to target region reference using BWA software16 and obtained consensus sequence by SAMtools.17 Coverage, depth and length were recorded for each consensus using ReSeqTools.18 Finally, single-nucleotide variation and indel results were filtered based on sequencing quality and depth. Mutation types were annotated according to the HbVar database of human HbVar and thalassaemia mutations.
Validation of NGS methods
We applied two different strategies to validate the gap-PCR and NGS approach for detection of thalassaemia carriers. First, we compared four α thalassaemia mutations and eight β thalassaemia mutations detected by NGS with the Sanger sequencing results. The mutations analysed included α thalassaemia mutations Hemoglobin (Hb) WS, Hb CS, Hb QS and alpha2 codon 30 del GAG and β thalassaemia mutations Hb E, codons 41/42 (-TTCT), IVS-II-654 (C>T), codon 17 (A>T), −28 (A>G), −50 (G>A), codons 71/72 (+A) and codon 43 (G>T). Second, we compared thalassaemia mutations detected by gap-PCR and NGS with routine genotyping tests (the gap-PCR and RDB method) in 184 individuals with positive HbA2 results.
Results
NGS methodological validation
We initially performed the validation experiment by comparing results generated by NGS and Sanger sequencing independently. Sanger sequencing confirmed 4 α thalassaemia mutations and 8 β thalassaemia mutations detected as positive by NGS results from 12 samples were found to completely match the NGS results (online supplementary figures 1 and 2). Then, among the 184 individuals who underwent routine genotyping and gap-PCR and NGS tests, 152 showed positive α thalassaemia mutations and β thalassaemia mutations. Thalassaemia mutations detected by the gap-PCR and NGS were all successfully confirmed by gap-PCR and RDB (online supplementary table S1).
jclinpath-2019-206339supp001.pdf (340KB, pdf)
jclinpath-2019-206339supp002.pdf (157.8KB, pdf)
Combined gap-PCR and NGS screening reveals thalassaemia mutation carriers
We applied the combined gap-PCR and NGS method to 944 couples and found that 15.1% (285/1888) of couples were thalassaemia mutation carriers. Of these, the α thalassaemia carrier rate was 11% (207/1888); the β thalassaemia carrier rate was 3.7% (70/1888); and the composite α thalassaemia and β thalassaemia carrier rate was 0.4% (8/1888). Moreover, 2.9% (27/944) of couples were both estimated as thalassaemia mutation carriers. We identified 20 distinct genotypes of α thalassaemia mutations (online supplementary table S2 and table 1). Among these genotypes, --SEA/αα was the most common genotype and accounted for 44.4% of all cases (92/207). -α3.7/αα was the second most common genotype, with an incidence rate of 25.6% (53/207). In addition, we identified 12 carriers with rare genotypes, such as Hb Hekinan II and Hb Owari. We identified 14 distinct genotypes of β thalassaemia mutations (online supplementary table S3 and table 1). The most common genotypes were codons 41/42 (-TTCT) (34.3%, 24/70) and IVS-II-654 (C>T) (15.7%, 11/70). We also identified 14 β thalassaemia carriers with rare genotypes which were not detected in previous studies.13 19 20 For example, we found that five β thalassaemia carriers had a −50 (G>A) mutation. In addition, we identified eight composite α thalassaemia and β thalassaemia carriers (online supplementary table S4). Of these, two cases showed a composite -α3.7/αα and codons 41/42 (-TTCT) genotype, and remaining cases showed a unique genotype (online supplementary table S4 and table 2).
Table 1.
α-Globin gene mutation spectrum
| Mutation type | HGVS name | Number | Ratio (%) | Class |
| --SEA | NG_000006.1:g.26264_45564del19301 | 96 | 42.1 | Common |
| -α3.7 | NG_000006.1:g.34164_37967del3804 | 61 | 26.8 | Common |
| Hb Westmead | HBA2: c.369C>G | 24 | 10.5 | Common |
| -α4.2 | NA | 20 | 8.8 | Common |
| Hb Constant Spring | HBA2: c.427T>C | 9 | 3.9 | Common |
| Hb Quong Sze | HBA2: c.377T>C | 4 | 1.8 | Common |
| Hb G-Honolulu | HBA2: c.91G>C | 4 | 1.8 | Rare |
| Hb Hekinan II | HBA1: c.84G>T | 3 | 1.3 | Rare |
| HBA2: c.46G>A | HBA2: c.46G>A | 2 | 0.9 | Rare |
| Hb Owari | HBA1: c.364G>A | 2 | 0.9 | Rare |
| HBA2: c.190G>A | HBA2: c.190G>A | 1 | 0.4 | Rare |
| HBA1: c.412A>G | HBA1: c.412A>G | 1 | 0.4 | Novel |
| Alpha2 codon 30 del GAG | HBA2: c.91_93delGAG | 1 | 0.4 | Rare |
| Total | 228 | 100 |
Table 2.
β-Globin gene mutation spectrum
| Mutation type | HGVS name | Number | Ratio (%) | Class |
| Codons 41/42 (-TTCT) | HBB: c.124_127delTTCT | 29 | 35.8 | Common |
| IVS-II-654 (C>T) | HBB: c.316–197C>T | 14 | 17.3 | Common |
| Codon 17 (A>T) | HBB: c.52A>T | 8 | 9.9 | Common |
| −28 (A>G) | HBB: c.-78A>G | 7 | 8.6 | Common |
| −50 (G>A) | HBB: c.-100G>A | 5 | 6.2 | Novel |
| Hb E | HBB: c.79G>A | 4 | 4.9 | Common |
| HBB: c.*+129T>A | HBB: c.*+129T>A | 3 | 3.7 | Novel |
| Codons 71/72 (+A) | HBB: c.216_217insA | 3 | 3.7 | Common |
| Codon 43 (G>T) | HBB: c.130G>T | 2 | 2.5 | Common |
| HBB: c.-64G>C | HBB: c.-64G>C | 2 | 2.5 | Novel |
| HBB: c.-180G>C | HBB: c.-180G>C | 1 | 1.2 | Novel |
| HBB: c.*+5G>A | HBB: c.*+5G>A | 1 | 1.2 | Novel |
| Hb New York | HBB: c.341T>A | 1 | 1.2 | Rare |
| HBB: c.-113A>G | HBB: c.-113A>G | 1 | 1.2 | Novel |
| Total | 81 | 100 |
Mutation spectrum of α-globin and β-globin genes
We identified 13 distinct mutations in α-globin genes (table 1). Most recurrent mutations were consistent with the report of Guangdong province.20 The most recurrent two mutations were --SEA and -α3.7, accounting for approximately 68.9% of all mutations (157/228). Unexpectedly, we noted that the mutation rate of Hb Westmead was higher than -α4.2 in Zhongshan city, which was discordant with that of Guangdong province.20 We additionally identified seven rare mutations, such as Hb Hekinan II, Hb G-Honolulu and Hb Owari, accounting for 6.1% (14/228) of all mutations. Of these, the HBA1: c.412A>G variant was first identified as a novel α-globin gene mutation.
We identified 14 distinct mutations in β-globin genes (table 2). Among these mutations, codons 41/42 (-TTCT) and IVS-II-654 (C>T) were most prevalent, accounting for 53.1% (43/81) of all mutations. This was consistent with previous reports.19 We additionally identified seven rare mutations, such as −50 (G>A) and Hb New York, accounting for 17.3% (14/81) of all mutations. Of these, −50 (G>A), HBB: c.*+129T>A, HBB: c.-64G>C, HBB: c.-180G>C, HBB: c.*+5G>A and HBB: c.-113A>G were identified as β-globin gene mutations.
Comparison analysis of traditional methods and the combined method
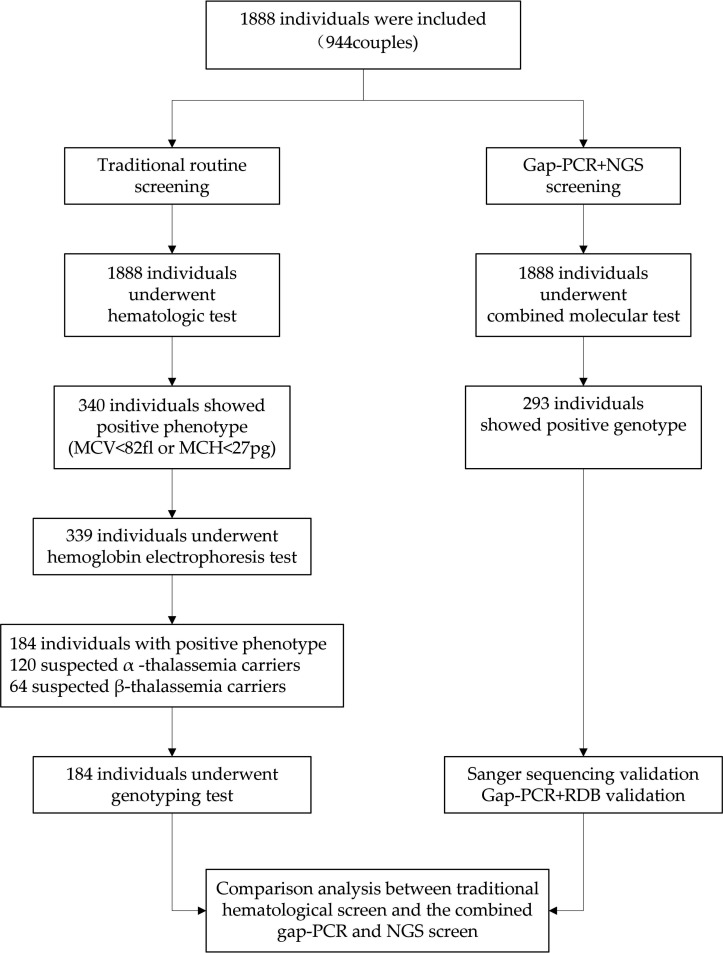
Based on MCV and MCH (MM) metrics, 18% (340/1888) of the people showed an abnormal haematological phenotype. Of these, 339 individuals further had a haemoglobin electrophoresis test based on data availability. As a result, 120 and 64 individuals were suspected of being the α thalassaemia and β thalassaemia carriers, respectively (figure 1). We estimated the detection rates of α thalassaemia mutations to be 73.49% by MM (158/215) and
Figure 1.
Diagram of work flow and screening results. MCH, mean corpuscular haemoglobin; MCV, mean corpuscular volume; NGS, next-generation sequencing; RDB, reverse dot blot hybridisation.
65.12% (112/172) by haemoglobin electrophoresis. The detection rates of β thalassaemia mutations by MM were 85.90% (67/78) and 95.65% (66/69) by haemoglobin electrophoresis (online supplementary table S5). Compared with the combined method results, the MCV+MCH and HbA2 detection strategy resulted in a lower sensitivity of 61.05% (105/172) and a high missed diagnosis ratio of 38.95% (67/172) for α thalassaemia. The sensitivity improved with the MM and HbA2 detection screen when compared with MM detection for β thalassaemia (98.51% vs 85.90%; table 3 and online supplementary tables S5 and S6). Notably, we identified six high-risk couples who have same and abnormal α thalassaemia genotype via NGS. Of these, only three couples were discovered through traditional methods. This demonstrated that the combined approach largely increased the sensitivity and outperformed conventional haematological methods in screening for thalassaemia mutation carriers.
Table 3.
Sensitivity and specificity of MCV+MCH and HbA2 levels for thalassaemia carriers screened by NGS
| MM and HbA2 | Sensitivity (95% CI) |
Specificity (95% CI) |
PPV (95% CI) |
NPV (95% CI) |
True positive | False positive | True negative | False negative |
| α thalassaemia | 61.05% (53.33% to 68.38%) |
87.65 % (84.05% to 90.70%) |
67.74% (61.21% to 73.64%) |
84.12 % (81.41% to 86.51%) |
105 | 50 | 355 | 67 |
| αα/--SEA | 86.81% (78.10% to 93.00%) |
84.36% (80.82% to 87.48%) |
50.97% (45.44% to 56.47%) |
97.16 % (95.27% to 98.30%) |
79 | 76 | 410 | 12 |
| αα/-α3.7 | 34.21% (19.63% to 51.35%) |
73.65% (69.72% to 77.33%) |
8.39% (5.45% to 12.70%) |
94.08 % (92.62% to 95.26%) |
13 | 142 | 397 | 25 |
| αα/-α4.2 | 37.50% (15.20% to 64.57%) |
73.44% (69.58% to 77.05%) |
3.87% (2.06% to 7.14%) |
97.63 % (96.56% to 98.37%) |
6 | 149 | 412 | 10 |
| β thalassaemia | 98.51% (91.96% to 99.96%) |
99.61 % (98.59% to 99.95%) |
97.06% (89.22% to 99.25%) |
99.8 % (98.64% to 99.97%) |
66 | 2 | 508 | 1 |
| Codons 41/42 (-TTCT) | 96.55% (82.24% to 99.91%) |
92.7 % (90.19% to 94.73%) |
41.18% (34.01% to 48.74%) |
99.8% (98.67% to 99.97%) |
28 | 40 | 508 | 1 |
| IVS-II-654 (C>T) | 100.00% (76.84% to 100.00%) |
90.41 % (87.67% to 92.71%) |
20.59% (16.75% to 25.04%) |
100.00 % | 14 | 54 | 509 | 0 |
| −28 (A>G) | 100.00% (59.04% to 100.00%) |
89.3 % (86.47% to 91.71%) |
10.29% (8.30% to 12.70%) |
100.00 % | 7 | 61 | 509 | 0 |
| Codon 17 (A>T) | 100.00% (63.06% to 100.00%) |
89.46 % (86.64% to 91.86%) |
11.76% (9.50% to 14.48%) |
100.00% | 8 | 60 | 509 | 0 |
CI, Confidence interval; MCH, mean corpuscular haemoglobin; MCV, mean corpuscular volume; MM, MCV and MCH; NGS, next-generation sequencing; NPV, negative predictive value; PPV, positive predictive value.
Discussion
In the present study, we revealed that the α thalassaemia carrier rate was 11% and the β thalassaemia carrier rate was 3.7%, and the composite α thalassaemia and β thalassaemia carrier rate was 0.4% in the Chinese population. The most common α thalassaemia genotype is --SEA/αα, followed by other less common α thalassaemia genotypes, including -α3.7/αα, Hb Westmead and -α4.2/αα. The prevalent genotypes of β thalassaemia are codons 41/42 (-TTCT), IVS-II-654 (C>T) and codon 17 (A>T). Among β-globin mutants, codons 41/42 show a higher carrier frequency than codon 17. All these findings are in line with previous studies.13 14 Additionally, we detected some rare α-globin and β-globin gene mutations unpublished by previous studies,5 6 representing 6.1% and 17.3% of all mutations, respectively. For example, we identified four cases with an Hb G-Honolulu mutation. This mutation was first identified in a Chinese woman in Singapore and was subsequently observed in southern China.21 We also identified seven novel mutations, including HBA1: c.412A>G, −50 (G>A), HBB: c.*+129T>A, HBB: c.-64G>C, HBB: c.-180G>C, HBB: c.*+5G>A and HBB: c.-113A>G. Patients with these novel variants showed normal values in haematological indexes. Therefore, it remains to be further elucidated in future studies if these variants are pathogenic
Through comparison analysis, we demonstrated that conventional haematological methods have lower sensitivity, especially for α thalassaemia detection, and a large fraction of thalassaemia variants with clinical significance like --SEA, -α3.7 and -α4.2 were not detected because the patients usually have normal values in MCV, MCH and HbA2 indexes. The sensitivity of MM combined with HbA2 improved when compared with MM in the detection for β thalassaemia mutations (92.75% vs 85.90%). These findings are consistent with a previous study.13 Although there are some similar studies on the application of NGS to the detection of thalassaemia mutations in China, He et al’s13 and Yao et al’s3 studies focused on the spectrum of thalassaemia mutations and evaluated the performance of NGS in screening for thalassaemia mutations in Chinese ethnic minorities. Zhang et al’s study22 lacked quantitative measures of the performance of NGS in the general Chinese population. For the first time, our study provided an in-depth comparison between traditional haematological methods and combined gap-PCR and NGS, which further demonstrated that the combined method outperformed the traditional haematological method in screening for thalassaemia mutations, particularly for α thalassaemia mutation carriers, in the general Chinese population.
The gap-PCR and NGS combined method has various advantages over the traditional haematological screening method. The major factors affecting the application of NGS are the high cost of sequencer, reagent and additional bioinformatics technicians. Whereas the NGS strategy shows a substantial advantage in time and cost in a large-scale population screening, in this study, the NGS had a throughput of 3000 samples per run, and the combined gap-PCR and NGS costs approximately US$25 for each sample, which is similar to that of the traditional method combined with gap-PCR or PCR-RBD ($25). Therefore, NGS is cost-efficient to screen for thalassaemia mutations in a large-scale population. Third, the combined method detects a wider spectrum of mutations, including common, rare, annotated and novel variants; therefore, it remarkably increases the detection rates of carrier status and therefore improves the detection rate of at-risk couples at a relatively low cost. Given the strengths of the combined gap-PCR and NGS method, we believe it can completely replace the haemoglobin electrophoresis screening step. In the prepregnancy or pregnancy screening, if the routine blood tests suggest small cell low-pigment anaemia, the gap-PCR and NGS combined detection should be carried out for the patients. Based on the results of gap-PCR and NGS combined detection, prenatal counselling can be more accurate so as to avoid abortion of fetuses with severe thalassaemia at late pregnancy or before pregnancy.
Though our study demonstrated the combined gap-PCR and NGS method outperformed routine haematological method in the detection of the thalassaemia mutations, it might have certain limitations. First of all, the study population was relatively small and confined to the Zhongshan city from Guangdong province. The results might have regional bias, therefore, further studies are required to validate the findings in a larger sample population from multiple regions of South China. Secondly, the pathogenicity of novel thalassaemia mutations remained unknown; their phenotypic effect should be validated in future studies. In summary, we systematically characterised thalassaemia carrier rate and mutation spectrum of α-globin and β-globin genes using gap-PCR and NGS among Chinese people. Our study has great value for preventing major thalassaemia in China while laying the groundwork for clinical application of the gap-PCR and NGS combined method to thalassaemia carrier screening.
Take home messages.
The first comprehensive survey of thalassaemia gene mutations was performed in the Zhongshan region.
The α thalassaemia carrier rate was 11% (207/1888); the β thalassaemia carrier rate was 3.7% (70/1888); and the composite α thalassaemia and β thalassaemia carrier rate was 0.4% (8/1888). Seven novel mutations were identified, including HBA1: c.412A>G, −50 (G>A), HBB: c.*+129T>A, HBB: c.-64G>C, HBB: c.-180G>C, HBB: c.*+5G>A and HBB: c.-113A>G.
The combined gap-PCR and NGS method outperformed the MCV+MCH and HbA2 method and MCV+MCH method in the detection of α thalassaemia and β thalassaemia mutations.
Acknowledgments
The authors thank all the staff of their department for their kind help and Dr Shiping Chen
for technical support.
Footnotes
Handling editor: Mary Frances McMullin.
Contributors: YY designed the study and wrote the article; JZ and QL performed routine thalassemia analysis; QL performed the specific gap-PCR amplification and the DNA sequencing; JL wrote the manuscript and provided technical support. All authors approved the final manuscript.
Funding: This research was financially supported by Science and Technology Planning Project of Zhongshan (grant/award number 2017B1007).
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: This study was approved by the medical ethics committee of Xiaolan People's Hospital of Zhongshan (XLLL-2017-KY-001).
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available upon reasonable request. All data relevant to the study are included in the article or uploaded as supplementary information.
References
- 1. Cao A, Kan YW. The prevention of thalassemia. Cold Spring Harb Perspect Med 2013;3:11775 10.1101/cshperspect.a011775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Xiong F, Sun M, Zhang X, et al. Molecular epidemiological survey of haemoglobinopathies in the Guangxi Zhuang autonomous region of southern China. Clin Genet 2010;78:139–48. 10.1111/j.1399-0004.2010.01430.x [DOI] [PubMed] [Google Scholar]
- 3. Yao H, Chen X, Lin L, et al. The spectrum of α- and β-thalassemia mutations of the Li people in Hainan Province of China. Blood Cells, Mol Dis 2014;53:16–20. 10.1016/j.bcmd.2014.01.003 [DOI] [PubMed] [Google Scholar]
- 4. XM X, Zhou YQ, Luo GX, et al. The prevalence and spectrum of α and β thalassaemia in Guangdong Province: implications for the future health burden and population screening. J Clin Pathol 2004;57:517–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Giardine B, Borg J, Higgs DR, et al. Systematic documentation and analysis of human genetic variation in hemoglobinopathies using the microattribution approach. Nat Genet 2011;43:295–301. 10.1038/ng.785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sankaran VG, Weiss MJ. Anemia: progress in molecular mechanisms and therapies. Nat Med 2015;21:221–30. 10.1038/nm.3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Thein SL. Genetic association studies in β-Hemoglobinopathies. Hematology 2013;2013:354–61. 10.1182/asheducation-2013.1.354 [DOI] [PubMed] [Google Scholar]
- 8. Traeger-Synodinos J, Harteveld CL, Old JM, et al. EMQN best practice guidelines for molecular and haematology methods for carrier identification and prenatal diagnosis of the haemoglobinopathies. Eur J Hum Genet 2015;23:426–37. 10.1038/ejhg.2014.131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Piel FB, Weatherall DJ. The α-thalassemias. N Engl J Med 2014;371:1908–16. 10.1056/NEJMra1404415 [DOI] [PubMed] [Google Scholar]
- 10. Korf BR, Rehm HL. New approaches to molecular DiagnosisNew approaches to molecular diagnosis. JAMA 2013;309:1511–21. [DOI] [PubMed] [Google Scholar]
- 11. Stark Z, Tan TY, Chong B, et al. A prospective evaluation of whole-exome sequencing as a first-tier molecular test in infants with suspected monogenic disorders. Genet Med 2016;18:1090–6. 10.1038/gim.2016.1 [DOI] [PubMed] [Google Scholar]
- 12. Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of Mendelian disorders. N Engl J Med 2013;369:1502–11. 10.1056/NEJMoa1306555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. He J, Song W, Yang J, et al. Next-Generation sequencing improves thalassemia carrier screening among premarital adults in a high prevalence population: the DAI nationality, China. Genet Med 2017;19:1022–31. 10.1038/gim.2016.218 [DOI] [PubMed] [Google Scholar]
- 14. Shang X, Peng Z, Ye Y, et al. Rapid targeted next-generation sequencing platform for molecular screening and clinical genotyping in subjects with hemoglobinopathies. EBioMedicine 2017;23:150–9. 10.1016/j.ebiom.2017.08.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tan AS, Quah TC, Low PS, et al. A rapid and reliable 7-deletion multiplex polymerase chain reaction assay for alpha-thalassemia. Blood 2001;98:250–1. 10.1182/blood.v98.1.250 [DOI] [PubMed] [Google Scholar]
- 16. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li H, Handsaker B, Wysoker A, et al. The sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–9. 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. He W, Zhao S, Liu X, et al. ReSeqTools: an integrated toolkit for large-scale next-generation sequencing based resequencing analysis. Genet Mol Res 2013;12:6275–83. 10.4238/2013.December.4.15 [DOI] [PubMed] [Google Scholar]
- 19. Zhang C-M, Wang Y, Gao L-S, et al. Molecular epidemiology investigation of β-thalassemia in Zhongshan City, Guangdong Province, people's Republic of China. Hemoglobin 2010;34:55–60. 10.3109/03630260903547724 [DOI] [PubMed] [Google Scholar]
- 20. Yin A, Li B, Luo M, et al. The prevalence and molecular spectrum of α- and β-globin gene mutations in 14,332 families of Guangdong Province, China. PLoS One 2014;9:e89855 10.1371/journal.pone.0089855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wang PP, Lin M, JR W, et al. Wang PP1, Lin M, Wu JR, Wang XY yl. three cases of the hemoglobin G-Chinese variant detected in patients of southern Chinese origin. Mol Med Rep 2010;3:459–61. [DOI] [PubMed] [Google Scholar]
- 22. Zhang H, Li C, Li J, et al. Next‐generation sequencing improves molecular epidemiological characterization of thalassemia in Chenzhou region, P.R. China. J Clin Lab Anal 2019;33:1–9. 10.1002/jcla.22845 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
jclinpath-2019-206339supp001.pdf (340KB, pdf)
jclinpath-2019-206339supp002.pdf (157.8KB, pdf)