Abstract
Objective
Due to the global increase in obesity rates and success of bariatric surgery in weight reduction, an increasing number of women now present pregnant with a previous bariatric procedure. This study investigates the extent of bariatric-associated metabolic and gut microbial alterations during pregnancy and their impact on fetal development.
Design
A parallel metabonomic (molecular phenotyping based on proton nuclear magnetic resonance spectroscopy) and gut bacterial (16S ribosomal RNA gene amplicon sequencing) profiling approach was used to determine maternal longitudinal phenotypes associated with malabsorptive/mixed (n=25) or restrictive (n=16) procedures, compared with women with similar early pregnancy body mass index but without bariatric surgery (n=70). Metabolic profiles of offspring at birth were also analysed.
Results
Previous malabsorptive, but not restrictive, procedures induced significant changes in maternal metabolic pathways involving branched-chain and aromatic amino acids with decreased circulation of leucine, isoleucine and isobutyrate, increased excretion of microbial-associated metabolites of protein putrefaction (phenylacetlyglutamine, p-cresol sulfate, indoxyl sulfate and p-hydroxyphenylacetate), and a shift in the gut microbiota. The urinary concentration of phenylacetylglutamine was significantly elevated in malabsorptive patients relative to controls (p=0.001) and was also elevated in urine of neonates born from these mothers (p=0.021). Furthermore, the maternal metabolic changes induced by malabsorptive surgery were associated with reduced maternal insulin resistance and fetal/birth weight.
Conclusion
Metabolism is altered in pregnant women with a previous malabsorptive bariatric surgery. These alterations may be beneficial for maternal outcomes, but the effect of elevated levels of phenolic and indolic compounds on fetal and infant health should be investigated further.
Keywords: enteric bacterial microflora, colonic fermentation, colonic bacteria, gastric surgery, obesity surgery
Significance of this study.
What is already known on this subject?
Bariatric surgery, particularly the malabsorptive type, alters the gut microbiota and host metabolism.
Pregnancy following bariatric surgery, especially a malabsorptive procedure, is associated with a reduced prevalence of maternal gestational diabetes and increased prevalence of small for gestational age babies.
What are the new findings?
We demonstrated that malabsorptive surgery-induced changes in the maternal gut microbiota persist throughout pregnancy compared with pregnancies with similar maternal body mass index but no history of such surgery, and may have a trans-generational impact.
Gut microbial changes in malabsorptive patients are associated with increased excretion of protein putrefaction metabolites during pregnancy.
How might it impact on clinical practice in the foreseeable future?
The impact of maternal malabsorption on fetal health warrants further investigation to determine the most appropriate type of weight loss surgery for women of reproductive age.
Introduction
Currently more than a third of women are classified as overweight or obese1 with obesity reaching epidemic levels globally. Obesity is associated with a number of adverse metabolic effects resulting in cardiovascular disease, metabolic syndrome, diabetes and cancer.2 Bariatric surgery (BS) has proven a successful treatment modality for lasting weight loss and has been shown to reduce obesity-related morbidity and mortality.3 4 Altered bile flow, reduction in gastric capacity, anatomical gut rearrangement and altered nutrient flow, vagal manipulation and modulation of enteric gut hormones, collectively referred to as the BRAVE effects,3 5 have been proposed to contribute to the mechanisms of weight loss and diabetes resolution in bariatric patients. There are two main types of BS: the restrictive (RES) procedures such as gastric banding and sleeve gastrectomy, which reduce the stomach size, and the malabsorptive/mixed (MAL) type, such as bilio-pancreatic diversion and Roux-en-Y-gastric bypass, which aim to decrease caloric absorption.6 BS has been shown to impact metabolic profiles reflecting altered metabolism and changes in the gut microbiome that play a role in improved lipid and glucose metabolism.7–11 The contribution of the gut mictobiota to host metabolism and metabolic control is well-established. Microbes degrade dietary components that are undigested by the host, convert dietary molecules into bioactive metabolites, influence host response to drugs and participate in many homeostatic processes and signalling pathways.12 13 However, the mechanisms by which BS confers metabolic benefit and the potential role of the microbiome in achieving this are poorly understood.
Evidence continues to accumulate that pregnancy post-BS is associated with lower prevalence of gestational diabetes mellitus (GDM) and large for gestational age neonates but higher risk of small for gestational age neonates and late preterm delivery compared with pregnancies in obese women without a previous BS.14 To investigate the impact of previous BS on maternal phenotype during pregnancy, we adopted a longitudinal metabolic (serum and urine) and gut bacterial (faeces) profiling strategy using proton nuclear magnetic resonance (1H NMR) spectroscopy and 16S rRNA gene amplicon sequencing, respectively, to characterise women with previous BS (RES and MAL) throughout the course of their pregnancy. Profiles were compared against women with similar early pregnancy body mass index (BMI) and no history of BS (NBS). For a subset of the cohort, we also characterise neonatal cord serum and urine metabolomes to assess potential trans-generational metabolic effects of BS.
Methods
Study population, sampling and clinical data
The population is part of an ongoing prospective cohort study investigating the impact of maternal BS on perinatal outcomes. All women gave written, informed consent for their data and samples to be used. Pregnant women with (n=47) and without (n=118) previous BS were recruited from May 2015 to April 2017 at Chelsea and Westminster Hospital (London, UK) as previously described.15 Women were seen at five time points during pregnancy (T1: 11+0–14+0, T2: 20+0–24+0, T3: 28+0–30+0, T4: 30+0–33+0 and T5: 35+0–37+6 weeks gestation) and within 72 hours of delivery (T6). Maternal blood (serum) and urine samples were collected at each visit while faecal samples were requested at the T1, T2 and T4 visits (see online supplementary figure S1 and table S1). A full oral glucose tolerance test (2 hour, 75 g) was conducted at T3 and maternal insulin resistance was calculated using the homeostatic model assessment for insulin resistance (HOMA-IR=fasting serum insulin (µU/L) × fasting glucose (mmol/L)/22.5). Estimated fetal weight was calculated by trans-abdominal ultrasound scans at T2, T4 and T5. At T6, birth weight was recorded, percentiles for the gestation were calculated and, where possible, neonatal samples (cord serum and urine) were collected. All samples were stored at −80°C for future analysis. For the study population, women with diagnosis of type 2 diabetes mellitus or GDM (due to the effect of diabetes on the metabolic profile) and those that had a miscarriage were excluded. Only NBS participants with a BMI of 25–50 kg/m2 at T1 were included to match the BMI range of the included bariatric patients at T1.
gutjnl-2019-319620supp001.pdf (5.8MB, pdf)
Metabolic profiling of biofluid samples
Serum and urine samples were prepared according to an established protocol.16 1H NMR spectra were acquired on a Bruker 600 MHz spectrometer (Bruker BioSpin) following a published method16 (see online supplementary methods for further detail). Methodology for preprocessing of spectral data is described in online supplementary methods. Multivariate modelling of the spectral data was performed in the software package SIMCA V.14.1 (Sartorius Stedim Biotech). Principal component analysis (PCA) was used to assess variation in metabolic profiles over all time points and to identify extreme outliers to exclude from supervised models for each time point. Orthogonal partial least squares discriminant analysis (OPLS-DA) was used to identify spectral variables (relating to specific chemical compounds) that contributed to discrimination of clinical classes (pairwise comparisons between NBS, MAL and RES groups) at each time point. From the discriminatory NMR peaks, metabolite identities were confirmed using statistical tools,17 18 2D NMR experiments and by spiking in authentic standards (see online supplementary methods). Relative concentrations of each discriminatory metabolite were calculated by integrating a representative peak of that metabolite. Downstream analysis was performed in the R software environment.19 For each discriminatory metabolite identified in the cross-sectional analysis, time series curves were generated through a spline-fitting method implemented in ‘santaR’ (see online supplementary methods) to visualise their behaviour in different groups over the pregnancy time course. Individuals with at least five data points were included. Correlation analysis including partial correlation to adjust for confounders is detailed in the online supplementary methods along with a list of all R packages used. Missing data were excluded from calculations. All p values were adjusted (padj) where necessary to control for the false discovery rate according to the Benjamini-Hochberg method.20 An alpha of 0.05 was used for p and padj values.
gutjnl-2019-319620supp002.pdf (629.9KB, pdf)
Gut bacterial community profiling
Stool samples were randomised for processing and DNA was extracted (see online supplementary methods) using the PowerLyzer PowerSoil DNA Isolation Kit (Mo Bio). 16S rRNA gene amplicon sequencing targeting the V1-V2 regions was performed on the Illumina MiSeq platform as previously described.21 Raw reads were processed in the R software environment19 following a published workflow22 which includes amplicon denoising implemented in ‘DADA2’.23 See online supplementary methods for full details. Functions in the ‘vegan’ R package were used to calculate Shannon Diversity Indices (α-diversity) on data rarefied to the minimum sequencing depth and Bray-Curtis dissimilarity (β-diversity) on log-transformed data (pseudocount of 1 added to each value). Significance of group separation in β-diversity was assessed by permutational multivariate analysis of variance. Changes in relative abundance were tested at each taxonomic rank from phylum to genus using the Mann-Whitney U test while differentially abundant 16S rRNA gene sequences were identified using ‘DESeq2’.24 For ‘DESeq2’ analysis, data were pooled for each individual rather than analysing distinct time points.
Integrative analysis of metabolic and taxonomic data
Relationships between the serum, urine and faecal data sets were modelled using the DIABLO method in ‘mixOmics’.25 This is a multi-block latent variable-based approach which aims to identify concordance between multiple data sets. Metabolites significant throughout the time course (serum: leucine, isoleucine, isobutyrate, d-β-hydroxybutyrate; urine: phenylacetylglutamine (PAG), p-cresol sulfate (PCS), indoxyl sulfate (IS), p-hydroxyphenylacetate (PHPA), unknown, α-ketoisovalerate, creatinine) and a subset of bacterial genera (log-transformed; selected using the least absolute shrinkage and selection operator (LASSO) penalisation method implemented in ‘mixOmics’) were modelled. Sampling points for each individual, where matching microbiome and metabolite data were obtained (T1, T2 and T4), were included in the model.
Results
Characteristics of the study participants
Of the 165 women enrolled in the study, 54 were excluded from longitudinal profiling due to BMI (n=36), diagnosis of diabetes (n=16), miscarriage (n=1) or withdrawal from the study (n=1). The final study population included 111 women (NBS n=70, RES n=16, MAL n=25) who contributed 491 maternal urine, 394 maternal blood, 68 maternal stool (online supplementary figure S1), 54 cord blood and 28 neonatal urine samples (online supplementary table S1). Two women (RES n=1, MAL n=1) contributed only neonatal samples. All women in the MAL group had a previous gastric bypass while the RES group had gastric banding (n=8) or sleeve gastrectomy (n=8). Maternal demographics, pregnancy characteristics and outcomes of the study population (online supplementary table S2) were consistent with our earlier report.15 Compared with the NBS group, women with a previous BS were on average 3.7 years older and, especially those with a MAL procedure, had lower insulin resistance at T3 and delivered smaller babies earlier (online supplementary table S2). Among the BS group, women with a previous MAL procedure had a higher pre-surgery BMI compared with those with a previous RES procedure.
Metabolic differences in maternal and neonatal serum metabolomes associated with bariatric surgery
Clear differences between the serum metabolic profiles of MAL, but not RES, participants and NBS participants were detected in the third trimester of pregnancy (time points T4 and T5; online supplementary table S3) as evidenced by the OPLS-DA models. The serum metabolic profiles associated with BS at the time of labour and in the first and second trimesters could not be differentiated from the control group. Serum samples obtained from MAL patients contained lower concentrations of unsaturated lipids (CH=CH, δ 5.31 (m)) at T5 and the lipids group corresponding to C=CCH2C=C (δ 2.79 (m)) at both T4 and T5 (online supplementary figure S2) compared with the NBS group. Other metabolic differences between the MAL and NBS groups included (online supplementary table S4) N-acetyl glycoprotein (GlycA), leucine, isoleucine, isobutyrate (decreased in MAL) and glutamine, d-β-hydroxybutyrate (increased in MAL). The behaviour of these metabolites throughout the pregnancy period was investigated further using time series analysis to model individual metabolites. Serum leucine, isoleucine and isobutyrate concentrations were significantly lower in MAL (n=12; NBS n=34) throughout the time course (figure 1A), whereas increases in d-β-hydroxybutyrate were associated with the last few weeks of pregnancy and delivery. GlycA and glutamine were not significantly changed in MAL during pregnancy. Significance of the differential serum metabolite concentrations at each time point is shown in online supplementary figure S3A. There was no significant difference between NBS and RES participants at any time point (online supplementary table S5). Although OPLS-DA and time series modelling showed clear evidence that previous BS impacted on the metabolic profiles, PCA indicated that the greatest metabolic variation in plasma composition was due to the changes over time during pregnancy, irrespective of surgery, with samples collected at T1 being markedly different from those obtained at later time points in the pregnancy (online supplementary figure S4). Thus no inherent structure associated with study group, age or BMI was evident in the 1H NMR maternal serum data. No differences in the metabolic profiles of cord serum from neonates in each study group were detected (online supplementary table S3).
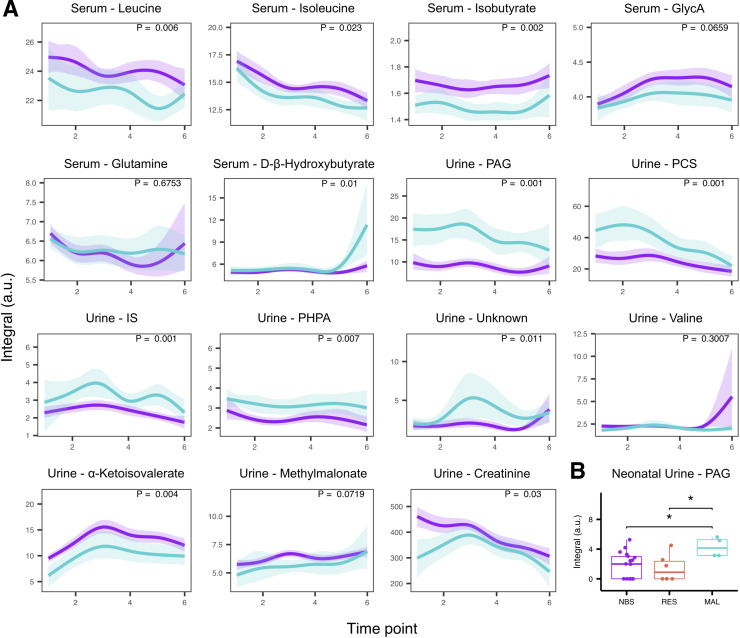
Figure 1.
Modelling of metabolite concentrations. (A) Time series analysis of discriminatory metabolites with lines representing group mean curves while shaded areas denote 95% confidence bands. Malabsorptive (MAL, blue) patients relative to No bariatric surgery (NBS, purple) controls. (B) Relative concentration of phenylacetylglutamine (PAG) is increased in neonatal urine from the MAL group compared with the NBS or restrictive (RES) groups. NMR peak integrals are reported in arbitrary units (a.u.). *P<0.05 (Mann-Whitney U test); GlycA, N-acetyl glycoprotein; IS, indoxyl sulfate; PCS, p-cresol sulfate; PHPA, p-hydroxyphenylacetate.
Surgery-associated metabolic differences in maternal and neonatal urine metabolomes
Similar to the serum samples, PCA models indicated that the metabolic variation driven by temporal changes throughout pregnancy was stronger than that associated with study group, age or BMI (online supplementary figure S4). Urine samples collected at delivery (T6) clearly differed from those taken at any other time point during pregnancy. In addition to endogenous changes, drug metabolites of paracetamol as well as mannitol, a common ingredient in drug tablet coating, were detected in urine obtained at T6. Outliers identified from PCA models at each time point were samples containing high concentrations of glucose or drug metabolites and were excluded from subsequent supervised models (online supplementary table S3). Based on the OPLS-DA models, no significant differences were identified between NBS and RES groups or between RES and MAL groups at any of the six time points; however, urine metabolic profiles of MAL patients were significantly altered compared with the NBS group as early as T1 and this difference persisted throughout pregnancy (online supplementary table S3). The strongest discriminatory metabolites (online supplementary table S4 and online supplementary figure S5) contributing to the differences between MAL and NBS groups were host-gut bacterial co-metabolites PAG, PCS, IS, PHPA, as well as an unidentified aromatic metabolite (unknown) with structural similarities to PCS (based on both 1H and 13C shifts; online supplementary figure S6). These metabolites were significantly elevated in MAL (n=14; NBS n=50) patients during pregnancy (figure 1A). Creatinine and α-ketoisovalerate were present in lower concentrations in MAL patients; methylmalonate and valine were not significantly associated with the MAL group over the time course (figure 1A) but were significantly lower in MAL patients at specific time points (online supplementary figure S3B). None of these metabolites were significantly changed in the RES group after correcting for multiple hypothesis tests (online supplementary table S5). Interestingly, when the RES group was divided into gastric banding (purely mechanical) and sleeve gastrectomy (removal of part of the stomach with metabolic effects), patients with a sleeve tended to have higher PAG and PCS than patients with a band (online supplementary figure S7). Maternal urine samples collected at T6 could not be discriminated for any pairwise comparison by OPLS-DA (online supplementary table S3). However PAG, the metabolite with the largest effect size in maternal samples, retained significance in the offspring’s urine at T6 (figure 1B). The relative concentration of PAG was significantly increased in babies born from MAL mothers (n=4) compared with babies born from RES (n=6, p=0.04) and NBS (n=18, p=0.021) mothers, mirroring the difference observed between the MAL and NBS maternal samples.
Taxonomic changes in the maternal gut microbiota of MAL patients
To support the finding of altered microbiota-associated metabolites in MAL patients detected by 1H NMR spectroscopy, faecal microbiome compositions from a subset of the cohort were analysed. A total of 68 stool samples representing 39 mothers (NBS n=25, MAL n=14) were studied. After excluding one maternal NBS sample due to low sequencing depth (2181 reads), the mean number of high-quality, paired-end 16S rRNA gene amplicon sequences per sample was 22 405 (±5535 SD) with a minimum sequencing depth of 12 967.
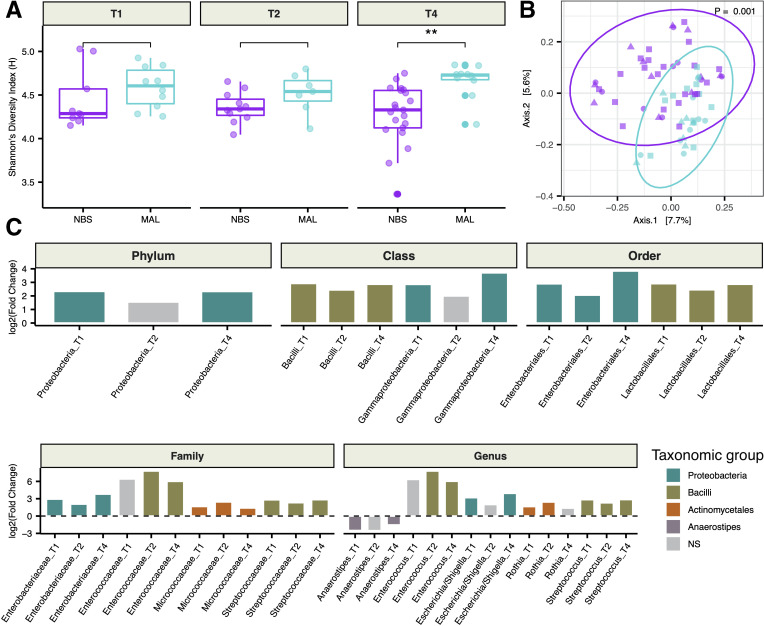
Women with a previous MAL surgery exhibited greater α-diversity, or a higher number of distinct taxa with more even distributions, compared with the NBS group but this was only statistically significant in the third trimester of pregnancy (figure 2A). β-diversity analysis comparing pairwise dissimilarities between samples revealed that MAL patients had a distinct (p=0.001) gut microbiota from the NBS group throughout pregnancy (figure 2B). MAL patients could be distinguished by an increase in relative abundance of Escherichia/Shigella, Streptococcus and Enterococcus genera and these changes manifested at all higher taxonomic ranks (figure 2C). Relative increases in abundances of Rothia and its family Micrococcaceae were also observed. Anaerostipes was the only genus found in significantly lower relative abundance in the MAL group at multiple time points. Only taxa that exhibited differential abundance in the MAL group at more than one time point were included in figure 2C, but taxa displaying differences between groups at a single time point during pregnancy are detailed in online supplementary table S6. Differential abundance at the amplicon sequence level was investigated for sequences assigned to the genera Streptococcus, Enterococcus, Escherichia/Shigella and Rothia. Species assignments were compared against SILVA26 and RDB27 databases. From the 25 16S rRNA gene sequences (Escherichia/Shigella n=11, Streptococcus n=12, Enterococcus n=1 and Rothia n=1) found in significantly higher abundance in MAL patients (online supplementary table S7), the following species were identified: Escherichia coli, Streptococcus salivarius, Streptococcus vestibularis, Streptococcus mutans and Streptococcus parasanguinis.
Figure 2.
Gut bacterial alterations in malabsorptive patients. (A) Comparison of Shannon diversity between malabsorptive (MAL) and no bariatric surgery (NBS) groups at 11–14 weeks (T1), 20–24 weeks (T2) and 30–33 weeks (T4) gestation. (B) Principal coordinates analysis of Bray-Curtis dissimilarities between MAL (blue) and NBS (purple) during pregnancy (T1: circle, T2: triangle, T4: square). P value obtained from permutational multivariate analysis of variance test. (C) Taxa that exhibited differential abundance (Mann-Whitney U test; padj<0.05) at more than one time point are plotted. Values are reported in online supplementary table S6. **P<0.01 (Mann-Whitney U test); NS, not significant.
Integrative analysis of phenotypes
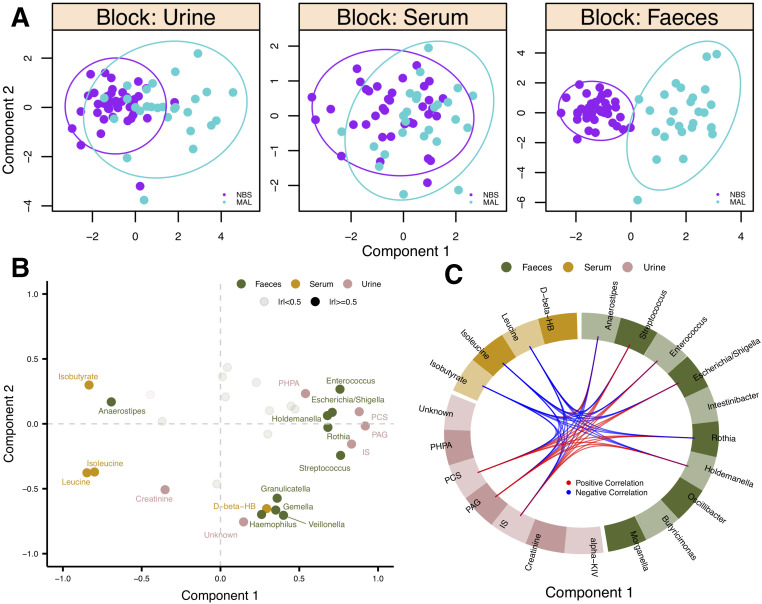
To determine if the observed MAL signatures were complementary across the data sets, an integrative approach was used to model the bacterial (n=68) and metabolic (serum n=60; urine n=66) data together. Clinical classes could be discriminated on the first component of the model for both urine and faecal data sets while this difference was not apparent in the serum data (figure 3A). Urine PAG, PCS, and IS concentrations were highly correlated and had the highest contribution to the model followed by leucine, isoleucine (also highly correlated) and isobutyrate (figure 3B). PAG, PCS and IS were most correlated with Streptococcus, Enterococcus, Escherichia/Shigella, Rothia and Holdemanella (figure 3C). Variables in this cluster also had a strong negative correlation with serum leucine, isoleucine and isobutyrate. Anaerostipes, the only genus that decreased in abundance in the MAL group, was negatively correlated with PAG, PCS and IS but positively correlated with leucine, isoleucine and isobutyrate (figure 3C).
Figure 3.
Integration of serum, urine and faecal profiles. (A) Scores plots showing samples from each data set projected in latent space. Clinical classes are discriminated along component 1. (B) Variables are plotted (coloured by biofluid) where each point represents the Pearson correlation between that variable and each component. High correlations with either component (|r|≥0.5) are BOLD and labelled. (C) Pearson correlations (|r|≥0.5) between variables in different data sets along component 1 are plotted. NBS, no bariatric surgery; MAL, malabsorptive; alpha-KIV, α-ketoisovalerate; D-beta-HB, d-β-hydroxybutyrate; IS, indoxyl sulfate; PAG, phenylacetylglutamine; PCS, p-cresol sulfate; PHPA, p-hydroxyphenylacetate.
Metabolic profile and clinical outcomes
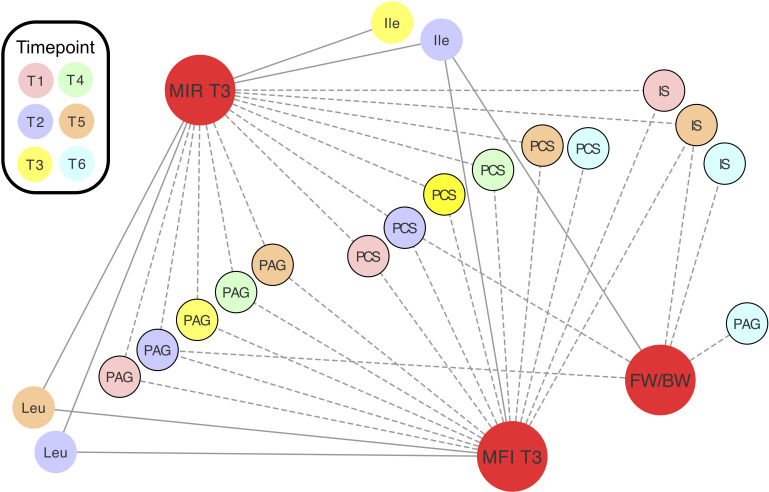
Spearman’s correlation coefficients (ρ) were calculated to assess the relationships between relative concentrations of maternal metabolites and clinical measurements (maternal: fasting insulin, fasting glucose and HOMA-IR measured at T3; neonatal: estimated fetal weight (measured at T2, T4 and T5) or birth weight percentiles, and gestational age at delivery). Throughout pregnancy, urinary host-microbial co-metabolites PAG and PCS were negatively associated with maternal HOMA-IR and fasting insulin while the branched-chain amino acids (mainly at T2) had a positive correlation (figure 4). PAG and PCS relative concentrations at T2, IS and unknown at T5, and IS and PAG at T6 were also negatively correlated with the weight percentiles of the babies at the corresponding time points. Partial correlations controlling for the effect of maternal age and BMI on maternal measurements as well as the effect of maternal age, BMI and HOMA-IR on birthweight percentile are reported in online supplementary table S8, but these confounders did not alter the overall results. Importantly, the concentrations of these metabolites did not correlate (online supplementary figure S8) with percentage of weight lost or time interval between BS and conception (which ranged from 12 months to 11 years in our cohort) in MAL patients, suggesting that the phenotype is persistent.
Figure 4.
Relative concentrations of metabolites correlate with maternal insulin resistance and fetal/birth weight. Spearman’s correlations between clinical measurements (red) and metabolite relative concentrations. Correlations plotted are significant (padj<0.05). Values are reported in online supplementary table S8. Negative correlations are denoted by dashed lines. Urine metabolites are denoted by black borders. FW/BW, estimated fetal weight or birth weight; Ile, isoleucine; IS, indoxyl sulfate; Leu, leucine; MFI, maternal fasting insulin; MIR, maternal HOMA-IR; PAG, phenylacetylglutamine; PCS, p-cresol sulfate.
Discussion
Our results demonstrate that pregnancies following a MAL bariatric procedure are characterised by altered maternal and neonatal metabolic profiles compared with pregnancies without such surgery. In particular, women with previous MAL surgery had lower serum concentrations of branched-chain amino acids (leucine and isoleucine) and branched-chain fatty acids (isobutyrate) and excreted higher concentrations of urinary host-microbial co-metabolites of protein putrefaction (PAG, PCS, IS and PHPA). This signature was correlated with a shift in the gut microbiota which included increases in relative abundance of bacterial genera Enterococcus, Streptococcus, Escherichia/Shigella and Rothia as well as a decrease in Anaerostipes. The observed MAL-associated changes in metabolism were also inversely associated with maternal insulin resistance and the offspring’s weight, suggesting that a previous MAL surgery may have both beneficial and detrimental effects on pregnancy. With regard to the RES group, we posit these patients, especially those with a previous sleeve gastrectomy, represent an intermediate phenotype due to the lower risk of malabsorption imposed by these procedures and less extreme changes to the gastrointestinal environment which would likely result in lesser alterations in the gut microbiome.28
Our results assessing the influence of BS on the maternal faecal microbiome composition are consistent with previous findings in non-pregnant individuals where MAL, but not RES, surgery resulted in increased relative abundances of Bacilli and Gammaproteobacteria (including genera Enterococcus, Streptococcus and Escherichia) as well as associated faecal metabolites28; abundances of both classes positively correlated with products of protein fermentation. Studies consistently show a gut bacterial signature of facultative anaerobes and oral-associated bacteria in BS (mainly MAL) patients with an increase in relative abundance of enterobacteria being the most consistent bacterial marker across multiple studies in both humans7 8 10 28–30 and animals.11 31 This shift in gut microbial community structure is believed to be, at least partially, a result of the increased oxygen and pH in the digestive tract following the anatomical rearrangement of a MAL surgery.30 32 Enterobacteria are Gram-negative and generally present in very low densities (<<108 CFU/g) in the normal gut, but increased abundances of these bacteria, some of which are known pathogens, have been associated with both inflammatory bowel disease and colon cancer.33 This underscores the need for understanding the mechanisms by which the altered bariatric microbiome can impact on maternal and neonatal metabolism and downstream health.
Increased concentrations of urinary microbial-associated metabolites following MAL BS have been reported in animals11 34 and in one human study with only two bariatric patients.9 Carbohydrates are the preferred source of energy for microbes inhabiting the large intestine.35 However, we hypothesise that the MAL condition not only changes the gastrointestinal environment resulting in an altered microbial community but also results in more undigested protein reaching the large intestine36 and both of these factors likely contribute to the shift from carbohydrate to protein metabolism. Fermentation of protein by gut bacteria, or putrefaction, in the colon is generally considered detrimental to health.37 Gut bacterial metabolism of aromatic amino acids (phenylalanine, tyrosine and tryptophan) results in the production of phenols and indoles,38 which are inflammatory and potentially toxic compounds.39 40 Bacterial enzymes are necessary for metabolism of phenylalanine and tyrosine to phenolic compounds phenylacetate and p-cresol, respectively, and for the metabolism of tryptophan to indole. Indole and p-cresol are sulfated to IS and PCS, respectively, prior to excretion while phenylacetate is converted to PAG by host metabolism. PHPA is an intermediate in the tyrosine degradation pathway but is also formed from phenylacetate.38 Enterococcus spp. contain enzymes necessary for both phenylalanine and tyrosine metabolism; E. coli is involved in tyrosine metabolism.41 Both species are also known as indole producers.42 Although Streptococcus may not be directly involved in the formation of these metabolites, it does have proteolytic activity43 and could contribute to the observed phenotype by making amino acids available from dietary protein. To our knowledge, this is the first report of upregulation of host-microbe co-metabolism of aromatic amino acids post-MAL surgery in humans. Previous reports have shown that the gut microbiota shifts to protein putrefaction after a MAL BS10 28 29 but, having focused on faecal samples, the host component of these pathways was overlooked. Lacking dietary data, we cannot exclude the possibility that the MAL metabolic phenotype results from increased protein intake. However, reports of protein malnutrition and protein intolerance post-BS44 45 suggest that high protein intake is unlikely in these patients. An inverse association between PAG/PCS and BMI has been reported although the relationship between adipiosity and the gut microbiota is still under debate.46 Other reports, in agreement with our data, have identified decreased concentrations of branched-chain amino acids in non-pregnant BS patients which supports our hypothesis that protein metabolism is altered in these individuals. Branched-chain amino acids have previously been linked to insulin resistance associated with obesity.47–50
Our data suggest that PAG and PCS, or their respective metabolic pathways, are also associated with reduction in maternal insulin resistance following BS. We are aware of recent discussion surrounding the suitability of oral glucose tolerance testing in bariatric patients and have since revised our methods. In spite of this caveat, it is conceivable that the gut microbiota plays a role in insulin resistance.49 51 Our finding of an inverse correlation between maternal urinary host-microbial co-metabolites and fetal/birth weight indicates that the maternal metabolic changes may also be related to the risk of these women delivering small for gestational age neonates. Although malnutrition resulting from maternal malabsorption likely contributes to reduced fetal growth, the downstream metabolic consequences of malabsorption also warrant further investigation given the potential toxicity of the final metabolic products. It has also been shown that higher urinary concentrations of PCS are indicative of reduced sulfation capacity in the host, which presents significant competition for similar reactions, namely acetaminophen detoxification, which also require sulfation prior to excretion.52 Pharmacokinetics in these patients should be investigated, especially during pregnancy, given that MAL patients have higher urinary concentrations of two sulfated compounds (PCS and IS).
Pregnancy has been shown to induce substantial time-dependent alterations in physiology and metabolism53 54 under the constantly changing physiological demand as the mother responds to the needs of the growing fetus. Nevertheless, the systematic effect of BS on the metabolic profiles is superimposed on the changing metabolic landscape and can be clearly observed in the metabolic phenotypes. In particular, GlycA, a metabolite associated with systemic inflammation, increased during the first two trimesters but remained lower in MAL patients during the third trimester indicating that these women have a lower grade of pregnancy-related inflammation compared with obese women without BS.50 55 Drastic metabolic changes could also be seen at the time of delivery in all groups which is likely due to the metabolic consequences of the labour process and masked surgery-related differences.56 Intriguingly, despite prophylactic administration of antibiotics (given to 59% of the women at delivery), PAG was detected in the neonatal urine soon after birth and in significantly higher concentration in those born from women with previous MAL surgery compared with the offspring of the NBS women. Although this result should be treated with caution due to the low number of newborns enrolled in the study, it suggests transfer of the modified maternal profile to the offspring. Further independent studies are required to validate the generalisability of this observation with longer-term follow-up of the infants to determine whether this neonatal metabolic phenotype is maintained and how it impacts their future risk of obesity and diabetes. BS is not an appropriate intervention at population scale but identification of beneficially altered physiology may allow for development of targeted interventions aimed at specifically modulating key pathways.
Acknowledgments
The authors would like to thank Dr Ivan Serrano Contreras for assistance with carrying out 2D NMR spectroscopy. We would also like to acknowledge that KAW is supported by the STRATiGRAD doctoral training program and JVL is supported by Medical Research Council New Investigator Grant (P62104) and European Research Council Starting Grant (P61481). EH is supported by the Western Australian Government through the Premier’s Science Fellowship Program. Infrastructure support was provided by the National Institute for Health Research Imperial Biomedical Research Centre aligned with the gut health theme.
Footnotes
Twitter: @kiana_a_west
KAW and CK contributed equally.
Contributors: EH, MDS, JVL and MRJ designed the study. CK and TM recruited patients and collected biospecimens. KAW, CK and JAKM performed the experiments. KAW analysed the data. All authors contributed to the interpretation of results. KAW, MDS and EH wrote the manuscript. All authors read and approved the final manuscript.
Funding: The study was supported partially by BORNE (UK Registered Charity number: 1167073).
Disclaimer: The funding body did not contribute to the design of the study, collection, analysis, interpretation of data or writing the manuscript.
Competing interests: None declared.
Patient consent for publication: Not required.
Ethics approval: The study was approved by the West London Research Ethics Committee (No: 14/LO/0592).
Provenance and peer review: Not commissioned; externally peer reviewed.
Data availability statement: Data are available upon reasonable request. Raw sequencing data are available in a public, open access repository: Sequence Read Archive under BioProject PRJNA493625. Raw NMR data are available upon reasonable request. All pre-processed data and code to reproduce the results are available in a public, open access repository: GitHub (https://github.com/ka-west/PBS_manuscript). The R script is also included as a pdf file created with ‘knitr’ (see online supplementary methods).
References
- 1. World Health Organization Obesity and overweight, 2017. [Google Scholar]
- 2. Abdelaal M, le Roux CW, Docherty NG. Morbidity and mortality associated with obesity. Ann Transl Med 2017;5 10.21037/atm.2017.03.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ashrafian H, Ahmed K, Rowland SP, et al. Metabolic surgery and cancer. Cancer 2011;117:1788–99. 10.1002/cncr.25738 [DOI] [PubMed] [Google Scholar]
- 4. Noria SF, Grantcharov T. Biological effects of bariatric surgery on obesity-related comorbidities. Can J Surg 2013;56:47–57. 10.1503/cjs.036111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ashrafian H, Athanasiou T, Li JV, et al. Diabetes resolution and hyperinsulinaemia after metabolic Roux-en-Y gastric bypass. Obes Rev 2011;12:e257–72. 10.1111/j.1467-789X.2010.00802.x [DOI] [PubMed] [Google Scholar]
- 6. Elder KA, Wolfe BM. Bariatric surgery: a review of procedures and outcomes. Gastroenterology 2007;132:2253–71. 10.1053/j.gastro.2007.03.057 [DOI] [PubMed] [Google Scholar]
- 7. Graessler J, Qin Y, Zhong H, et al. Metagenomic sequencing of the human gut microbiome before and after bariatric surgery in obese patients with type 2 diabetes: correlation with inflammatory and metabolic parameters. Pharmacogenomics J 2013;13:514–22. 10.1038/tpj.2012.43 [DOI] [PubMed] [Google Scholar]
- 8. Kong L-C, Tap J, Aron-Wisnewsky J, et al. Gut microbiota after gastric bypass in human obesity: increased richness and associations of bacterial genera with adipose tissue genes. Am J Clin Nutr 2013;98:16–24. 10.3945/ajcn.113.058743 [DOI] [PubMed] [Google Scholar]
- 9. Calvani R, Miccheli A, Capuani G, et al. Gut microbiome-derived metabolites characterize a peculiar obese urinary metabotype. Int J Obes 2010;34:1095–8. 10.1038/ijo.2010.44 [DOI] [PubMed] [Google Scholar]
- 10. Tremaroli V, Karlsson F, Werling M, et al. Roux-En-Y gastric bypass and vertical banded gastroplasty induce long-term changes on the human gut microbiome contributing to fat mass regulation. Cell Metab 2015;22:228–38. 10.1016/j.cmet.2015.07.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Li JV, Ashrafian H, Bueter M, et al. Metabolic surgery profoundly influences gut microbial-host metabolic cross-talk. Gut 2011;60:1214–23. 10.1136/gut.2010.234708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nicholson JK, Holmes E, Kinross J, et al. Host-gut microbiota metabolic interactions. Science 2012;336:1262–7. 10.1126/science.1223813 [DOI] [PubMed] [Google Scholar]
- 13. Holmes E, Li JV, Athanasiou T, et al. Understanding the role of gut microbiome-host metabolic signal disruption in health and disease. Trends Microbiol 2011;19:349–59. 10.1016/j.tim.2011.05.006 [DOI] [PubMed] [Google Scholar]
- 14. Kwong W, Tomlinson G, Feig DS. Maternal and neonatal outcomes after bariatric surgery; a systematic review and meta-analysis: do the benefits outweigh the risks? Am J Obstet Gynecol 2018;218:573–80. 10.1016/j.ajog.2018.02.003 [DOI] [PubMed] [Google Scholar]
- 15. Maric T, Kanu C, Johnson MR, et al. Maternal, neonatal insulin resistance and neonatal anthropometrics in pregnancies following bariatric surgery. Metabolism 2019;97:25–31. 10.1016/j.metabol.2019.04.005 [DOI] [PubMed] [Google Scholar]
- 16. Dona AC, Jiménez B, Schäfer H, et al. Precision high-throughput proton NMR spectroscopy of human urine, serum, and plasma for large-scale metabolic phenotyping. Anal Chem 2014;86:9887–94. 10.1021/ac5025039 [DOI] [PubMed] [Google Scholar]
- 17. Cloarec O, Dumas M-E, Craig A, et al. Statistical total correlation spectroscopy: an exploratory approach for latent biomarker identification from metabolic 1H NMR data sets. Anal Chem 2005;77:1282–9. 10.1021/ac048630x [DOI] [PubMed] [Google Scholar]
- 18. Posma JM, Garcia-Perez I, De Iorio M, et al. Subset optimization by reference matching (storm): an optimized statistical approach for recovery of metabolic biomarker structural information from 1H NMR spectra of biofluids. Anal Chem 2012;84:10694–701. 10.1021/ac302360v [DOI] [PubMed] [Google Scholar]
- 19. R Core Team R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing, 2017. https://www.R-project.org/ [Google Scholar]
- 20. Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Royal Stat Soc 1995;57B:289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 21. Mullish BH, Pechlivanis A, Barker GF, et al. Functional microbiomics: evaluation of gut microbiota-bile acid metabolism interactions in health and disease. Methods 2018;149:49–58. 10.1016/j.ymeth.2018.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Callahan BJ, Sankaran K, Fukuyama JA, et al. Bioconductor workflow for microbiome data analysis: from raw reads to community analyses. Version 2. F1000Res 2016;5:1492 10.12688/f1000research.8986.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: high-resolution sample inference from illumina amplicon data. Nat Methods 2016;13:581–3. 10.1038/nmeth.3869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-Seq data with DESeq2. Genome Biol 2014;15:550 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rohart F, Gautier B, Singh A, et al. mixOmics: An R package for 'omics feature selection and multiple data integration. PLoS Comput Biol 2017;13:e1005752 10.1371/journal.pcbi.1005752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Quast C, Pruesse E, Yilmaz P, et al. The Silva ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 2013;41:D590–6. 10.1093/nar/gks1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cole JR, Wang Q, Fish JA, et al. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 2014;42:D633–42. 10.1093/nar/gkt1244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ilhan ZE, DiBaise JK, Isern NG, et al. Distinctive microbiomes and metabolites linked with weight loss after gastric bypass, but not gastric banding. ISME J 2017;11:2047–58. 10.1038/ismej.2017.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Palleja A, Kashani A, Allin KH, et al. Roux-En-Y gastric bypass surgery of morbidly obese patients induces swift and persistent changes of the individual gut microbiota. Genome Med 2016;8:67 10.1186/s13073-016-0312-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhang H, DiBaise JK, Zuccolo A, et al. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci U S A 2009;106:2365–70. 10.1073/pnas.0812600106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liou AP, Paziuk M, Luevano J-M, et al. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci Transl Med 2013;5:178ra41 10.1126/scitranslmed.3005687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Aron-Wisnewsky J, Doré J, Clement K. The importance of the gut microbiota after bariatric surgery. Nat Rev Gastroenterol Hepatol 2012;9:590–8. 10.1038/nrgastro.2012.161 [DOI] [PubMed] [Google Scholar]
- 33. Liu Z, Cao AT, Cong Y. Microbiota regulation of inflammatory bowel disease and colorectal cancer. Semin Cancer Biol 2013;23:543–52. 10.1016/j.semcancer.2013.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Seyfried F, Li JV, Miras AD, et al. Urinary phenotyping indicates weight loss-independent metabolic effects of Roux-en-Y gastric bypass in mice. J Proteome Res 2013;12:1245–53. 10.1021/pr300909v [DOI] [PubMed] [Google Scholar]
- 35. Conlon M, Bird A. The impact of diet and lifestyle on gut microbiota and human health. Nutrients 2015;7:17–44. 10.3390/nu7010017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ponsky TA, Brody F, Pucci E. Alterations in gastrointestinal physiology after Roux-en-Y gastric bypass. J Am Coll Surg 2005;201:125–31. 10.1016/j.jamcollsurg.2005.03.021 [DOI] [PubMed] [Google Scholar]
- 37. Windey K, De Preter V, Verbeke K. Relevance of protein fermentation to gut health. Mol Nutr Food Res 2012;56:184–96. 10.1002/mnfr.201100542 [DOI] [PubMed] [Google Scholar]
- 38. Smith EA, Macfarlane GT. Formation of phenolic and indolic compounds by anaerobic bacteria in the human large intestine. Microb Ecol 1997;33:180–8. 10.1007/s002489900020 [DOI] [PubMed] [Google Scholar]
- 39. Meyer TW, Hostetter TH. Uremic solutes from colon microbes. Kidney Int 2012;81:949–54. 10.1038/ki.2011.504 [DOI] [PubMed] [Google Scholar]
- 40. Sun C-Y, Hsu H-H, Wu M-S. P-Cresol sulfate and indoxyl sulfate induce similar cellular inflammatory gene expressions in cultured proximal renal tubular cells. Nephrol Dial Transplant 2013;28:70–8. 10.1093/ndt/gfs133 [DOI] [PubMed] [Google Scholar]
- 41. Gryp T, Vanholder R, Vaneechoutte M, et al. p-Cresyl sulfate. Toxins (Basel) 2017;9:52 10.3390/toxins9020052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Lee J-H, Lee J. Indole as an intercellular signal in microbial communities. FEMS Microbiol Rev 2010;34:426–44. 10.1111/j.1574-6976.2009.00204.x [DOI] [PubMed] [Google Scholar]
- 43. Macfarlane GT, Cummings JH, Allison C. Protein degradation by human intestinal bacteria. Microbiology 1986;132:1647–56. 10.1099/00221287-132-6-1647 [DOI] [PubMed] [Google Scholar]
- 44. Steenackers N, Gesquiere I, Matthys C. The relevance of dietary protein after bariatric surgery: what do we know? Curr Opin Clin Nutr Metab Care 2018;21:58–63. 10.1097/MCO.0000000000000437 [DOI] [PubMed] [Google Scholar]
- 45. Lupoli R, Lembo E, Saldalamacchia G, et al. Bariatric surgery and long-term nutritional issues. World J Diabetes 2017;8:464–74. 10.4239/wjd.v8.i11.464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Elliott P, Posma JM, Chan Q, et al. Urinary metabolic signatures of human adiposity. Sci Transl Med 2015;7:285ra62-ra62 10.1126/scitranslmed.aaa5680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Newgard CB, An J, Bain JR, et al. A branched-chain amino acid-related metabolic signature that differentiates obese and lean humans and contributes to insulin resistance. Cell Metab 2009;9:311–26. 10.1016/j.cmet.2009.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tai ES, Tan MLS, Stevens RD, et al. Insulin resistance is associated with a metabolic profile of altered protein metabolism in Chinese and Asian-Indian men. Diabetologia 2010;53:757–67. 10.1007/s00125-009-1637-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pedersen HK, Gudmundsdottir V, Nielsen HB, et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016;535:376–81. 10.1038/nature18646 [DOI] [PubMed] [Google Scholar]
- 50. Houttu N, Mokkala K, Laitinen K. Overweight and obesity status in pregnant women are related to intestinal microbiota and serum metabolic and inflammatory profiles. Clin Nutr 2018;37:1955–66. 10.1016/j.clnu.2017.12.013 [DOI] [PubMed] [Google Scholar]
- 51. Kootte RS, Levin E, Salojärvi J, et al. Improvement of insulin sensitivity after lean donor feces in metabolic syndrome is driven by baseline intestinal microbiota composition. Cell Metab 2017;26:611–9. 10.1016/j.cmet.2017.09.008 [DOI] [PubMed] [Google Scholar]
- 52. Clayton TA, Baker D, Lindon JC, et al. Pharmacometabonomic identification of a significant Host-microbiome metabolic interaction affecting human drug metabolism. Proc Natl Acad Sci U S A 2009;106:14728–33. 10.1073/pnas.0904489106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu X, Wang X, Sun H, et al. Urinary metabolic variation analysis during pregnancy and application in gestational diabetes mellitus and spontaneous abortion biomarker discovery. Sci Rep 2019;9:2605 10.1038/s41598-019-39259-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wang M, Xia W, Li H, et al. Normal pregnancy induced glucose metabolic stress in a longitudinal cohort of healthy women: novel insights generated from a urine metabolomics study. Medicine 2018;97:e12417 10.1097/MD.0000000000012417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Otvos JD, Shalaurova I, Wolak-Dinsmore J, et al. GlycA: a composite nuclear magnetic resonance biomarker of systemic inflammation. Clin Chem 2015;61:714–23. 10.1373/clinchem.2014.232918 [DOI] [PubMed] [Google Scholar]
- 56. Caboni P, Meloni A, Lussu M, et al. Urinary metabolomics of pregnant women at term: a combined GC/MS and NMR approach. J Matern Fetal Neonatal Med 2014;27(Suppl 2):4–12. 10.3109/14767058.2014.956403 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
gutjnl-2019-319620supp001.pdf (5.8MB, pdf)
gutjnl-2019-319620supp002.pdf (629.9KB, pdf)