Abstract
The FANCI-FANCD2 (ID) complex, mutated in the Fanconi Anemia (FA) cancer predisposition syndrome, is required for the repair of interstrand crosslinks (ICL) and related lesions1. The FA pathway is activated when a replication fork stalls at an ICL2, triggering the mono-ubiquitination of the ID complex. ID mono-ubiquitination is essential for ICL repair by excision, translesion synthesis and homologous recombination, but its function was hitherto unknown1,3. Here, the 3.5 Å cryo-EM structure of mono-ubiquitinated ID (IDUb) bound to DNA reveals that it forms a closed ring that encircles the DNA. Compared to the cryo-EM structure of the non-ubiquitinated ID complex bound to ICL DNA, described here as well, mono-ubiquitination triggers a complete re-arrangement of the open, trough-like ID structure through the ubiquitin of one protomer binding to the other protomer in a reciprocal fashion. The structures, in conjunction with biochemical data, indicate the mono-ubiquitinated ID complex looses its preference for ICL and related branched DNA structures, becoming a sliding DNA clamp that can coordinate the subsequent repair reactions. Our findings also reveal how mono-ubiquitination in general can induce an alternate structure with a new function.
FANCI and FANCD2 are paralogs that bind to DNA with preference for branched structures including Holliday junction, overhang and replication fork DNA4–7. The previous crystal structure of the mouse ID complex showed that it forms an open trough-like structure with two basic grooves, one on each paralog7. A 7.8 Å crystallographic map of FANCI bound to splayed Y DNA confirmed that its basic groove is the site of dsDNA binding, and also identified a likely single-stranded DNA binding region7. However, it has not been clear how these DNA binding activities relate to the function of the ID complex in replication and ICL repair. The mouse ID structure also showed the mono-ubiquitination sites are embedded inside the FANCI-FANCD2 interface7, but did not shed light on the function of mono-ubiquitination.
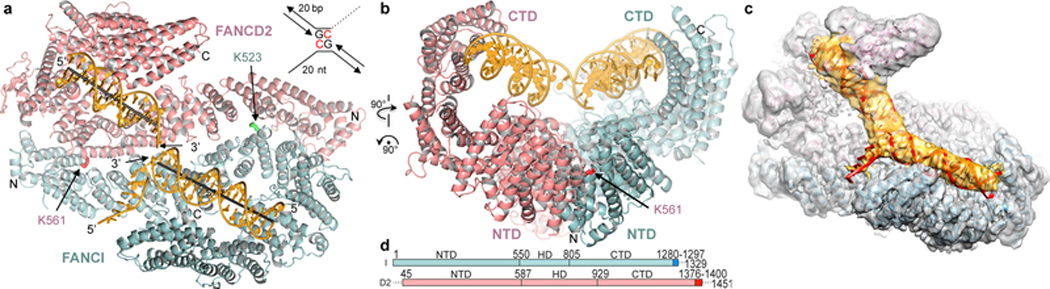
To address these questions, we first collected cryo-EM data on the human, full-length ID complex bound to an ICL-containing DNA constructed by crosslinking two modified oligonucleotides with a triazole moiety8,9 (Fig. 1a). Although it has not been clear if the ID complex recognizes ICLs, the triazole ICL DNA mimics two replication forks converging on an ICL, an event shown to trigger ICL repair in a cell-free Xenopus laevis system2. The initial consensus reconstruction with 231,943 particles extended to 3.4 Å as determined by the gold-standard fourier shell correlation (FSC) procedure10,11 (Extended Data Fig. 1). The map showed an overall structure and FANCI-FANCD2 interface very similar to the mouse ID complex7 (Fig. 1a, Extended Data Fig. 1d). Each paralog consists of N-terminal helical repeats that form a long α−α solenoid (henceforth NTD), followed by a helical domain (HD) that reverses the direction of the solenoid, and a C-terminal helical repeat domain (CTD; Fig 1d, Extended Data Fig. 1g). The consensus reconstruction showed clear density for FANCI and its bound dsDNA with single-stranded DNA (ssDNA) density extending from it. FANCD2 had clear density for its NTD, but poor density for its CTD and its bound dsDNA (Extended Data Fig. 2a). 3D classification indicated that the FANCD2 CTD exhibited substantial conformational flexibility, its relative position moving by up to 23 Å due to rotation within the HD domain (Extended Data Figs 2b and c). FANCI did not exhibit this flexibility, as its NTD contains a helical protrusion that packs with and stabilizes the CTD7 (Extended Data Fig. 1e). Accounting for FANCD2 CTD flexibility with multi-body refinement improved the solvent-corrected resolution of FANCD2 CTD and its associated dsDNA to 3.8 Å, and the remainder of the complex to 3.3 Å (Extended Data Figs 1 and 3).
Figure 1 |. Human ID complex bound to ICL DNA.
a, Overall structure with FANCI colored cyan, FANCD2 salmon, and ICL DNA yellow. The mono-ubiquitination sites are shown as sticks (labeled). The helical axes of the DNA duplexes are shown as gray lines. Inset shows schematic of the ICL DNA, with triazole-linked deoxycytidine residues in red (not built in structure). b, View looking down the left side of the horizontal axis of a. c, Cryo-EM density without temperature-factor sharpening. Orientation similar to a. DNA model is red, and its density yellow. d, Linear representation marking domain boundaries. Darker hues are disordered C-terminal segments that become ordered on ubiquitination; dashed lines disordered in both.
The improved maps showed continuous double-stranded DNA density extending from the FANCI groove to the FANCD2 groove (Fig. 1c). The DNA is sharply kinked near the center where the ICL would be based on the lengths of the flanking duplexes. There was no density that could correspond to the 5’ overhang ssDNA of the FANCD2-bound duplex. We modeled the overall DNA with an 18 base pair (bp) duplex and an 8 nucleotide ssDNA bound to FANCI, and a 15 bp duplex on FANCD2, and refined the model with the composite map option of REFMAC512 (Extended Data Table 1). We have not modeled the ICL and its immediate surroundings due to the overall high temperature factor and limited resolution of the DNA density. In the refined model, the duplexes bound to FANCI and FANCD2 are at a ~33° angle. Their helical axes are non-collinear, dislocated laterally by ~14 Å (Fig. 1a). This non-collinear arrangement of the two duplexes is largely unaffected by the conformational flexibility of the FANCD2 CTD (Extended Data Fig. 2d, e; Supplementary Videos 1 and 2).
FANCI binds to DNA using an extended basic groove consisting of parts of the NTD, HD and CTD domains. A 4 bp portion of dsDNA distal from the ICL is bound by a semi-circular constriction between the HD and CTD domains, while the rest of the duplex is bound by the NTD, and the ssDNA runs across the last two helical repeats of the CTD (Extended Data Fig. 4a). FANCD2’s DNA binding activity is divergent, as the HD portion of its semi-circular groove is acidic and is uninvolved in DNA binding (Extended Data Fig. 4b to e). Rather, FANCD2 binds to DNA using a localized basic patch on its CTD largely non-overlapping with that of FANCI. The density corresponding to the ICL rests against the NTD domains of both FANCI and FANCD2 (Extended Data Fig. 4a and b). DNA binding does not cause any conformational changes, as the 3.4 Å cryo-EM structure of the apo-ID, also reported here, is essentially indistinguishable from the ID-ICL complex (Extended Data Fig. 1b, f and Table 1).
The ID complex can associate with replication forks independent of ICLs13–15, and it is implicated in replication fork recovery after stalling 3,16. We thus also collected cryo-EM data of ID with 5’ flap DNA, a reversed fork-like Holliday junction (HJ), and a replication fork, mimicking DNA structures that can arise during replication. All three substrates contained clear density for a FANCI-bound duplex that extended partway to the FANCD2 NTD (Extended Data Fig. 5). However, there was essentially no density for a duplex bound to the FANCD2 CTD, even in individual 3D classifications. This suggests that engagement of the FANCD2 CTD requires a translocation of the helical axes of the two duplexes, because while these DNA substrates have a discontinuity in the DNA backbone, they remain stacked17,18.
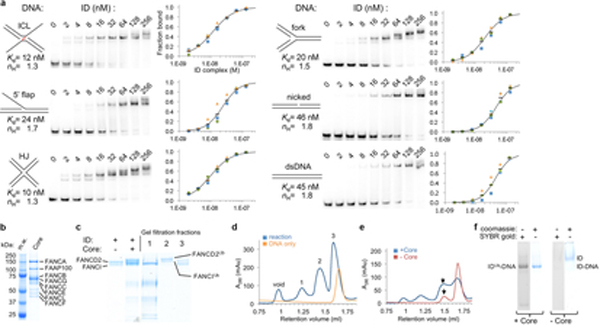
Together, these data indicate that the principal dsDNA binding activity resides with FANCI, with canonical dsDNA sufficing for FANCI binding. This may account for observations that FANCI alone can accumulate at active replication forks before stalling15. It is also consistent with the ID complex exhibiting only modest specificity for branched DNA structures in biochemical assays5,7 (Extended Data Fig. 6a). We presume that once FANCD2 engages in DNA binding at an ICL or a related DNA structure, this would likely prevent the ID complex from laterally diffusing along the DNA, stabilizing it at the lesion.
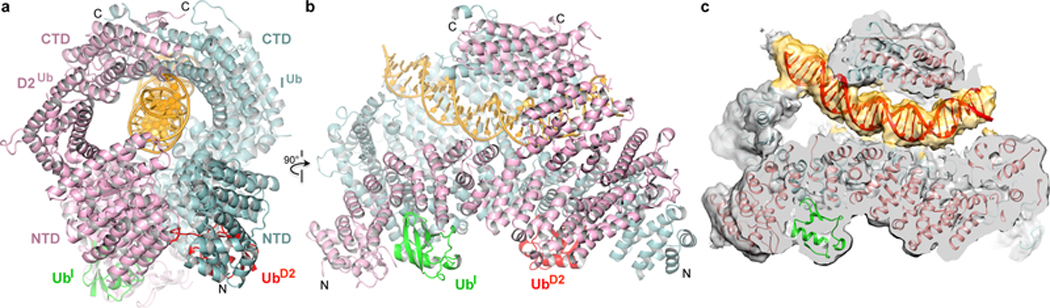
We next investigated the function of mono-ubiquitination by determining the structure of the mono-ubiquitinated ID (henceforth IDUb). For this, we constructed a stably-transfected HEK-293F cell line overexpressing eight subunits of the FA Core complex ubiquitin ligase (Extended Data Fig. 6b). Using the purified FA Core complex with the UBE2T E2 and E1, we ubiquitinated the ID complex in the presence of ICL DNA or a variety of other DNA molecules shown to promote ubiquitination 19–21 (Extended Data Fig. 6c, d). We purified the reaction products by preparative size exclusion chromatography and found that IDUb remained bound to DNA, in contrast to non-ubiquitinated ID (Extended Data Fig. 6e, f). We collected cryo-EM data of IDUb bound to four different DNA molecules: ICL DNA, 5’ flap DNA, nicked DNA and dsDNA. The largest data set was of the nicked DNA complex, and its 3D auto-refinement with 301,058 particles led to a 3.6 Å consensus reconstruction (Extended Data Fig. 7). After focused reconstructions with three masks (3.4, 3.5, and 3.5 Å reconstructions), we refined the model of IDUb bound to a 28 bp DNA duplex at 3.5 Å (Fig. 2, Extended Data Table 1). The reconstructions of the 5’ flap DNA, dsDNA and ICL DNA complexes extend to 3.8 Å, 3.8 Å and 4.4 Å, respectively (Extended Data Fig. 8a–c).
Figure 2 |. Mono-ubiquitinated human IDUb complex bound to nicked DNA.
a, Overall IDUb-DNA structure with FANCD2Ub pink (labeled D2), FANCIUb cyan (I), FANCI ubiquitin (UbI) green, FANCD2 ubiquitin (UbD2) red, and DNA yellow. The N and C termini are labeled (FANCD2 N-terminus obscured in this view). FANCI is oriented as in Fig. 1b. b, View looking down the left side of the horizontal axis of a. c, Cryo-EM density without temperature-factor sharpening. Orientation as in b, but parts of FANCD2 are cropped to reveal the internal DNA (red, density yellow).
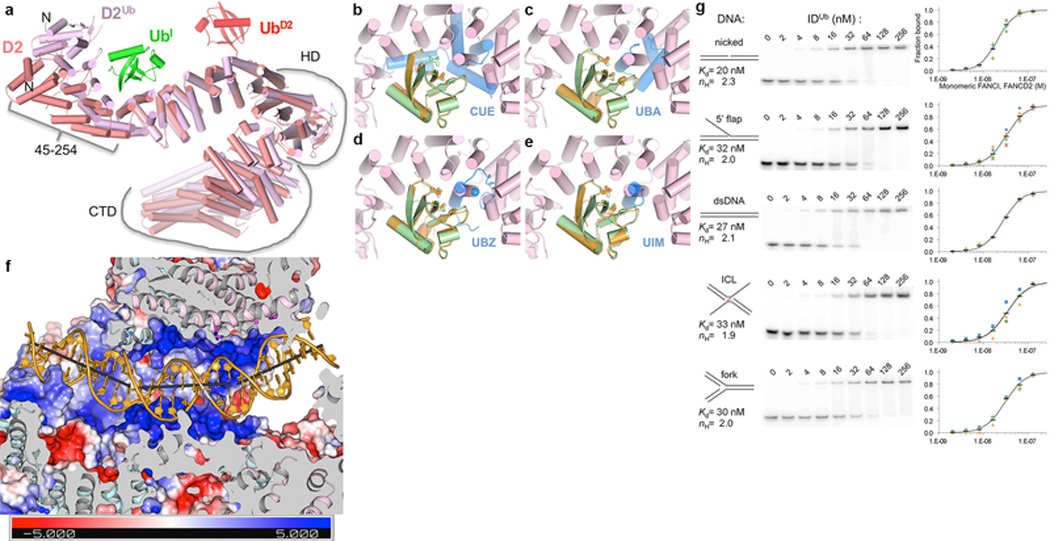
Mono-ubiquitination induces a new mode of FANCI-FANCD2 association that results in the conversion of the open-trough structure to a closed ring with the DNA inside (Fig. 2a). Central to the new mode of heterodimerization are the two ubiquitin molecules, whereby the ubiquitin covalently attached to one paralog binds to the other paralog in a reciprocal fashion.
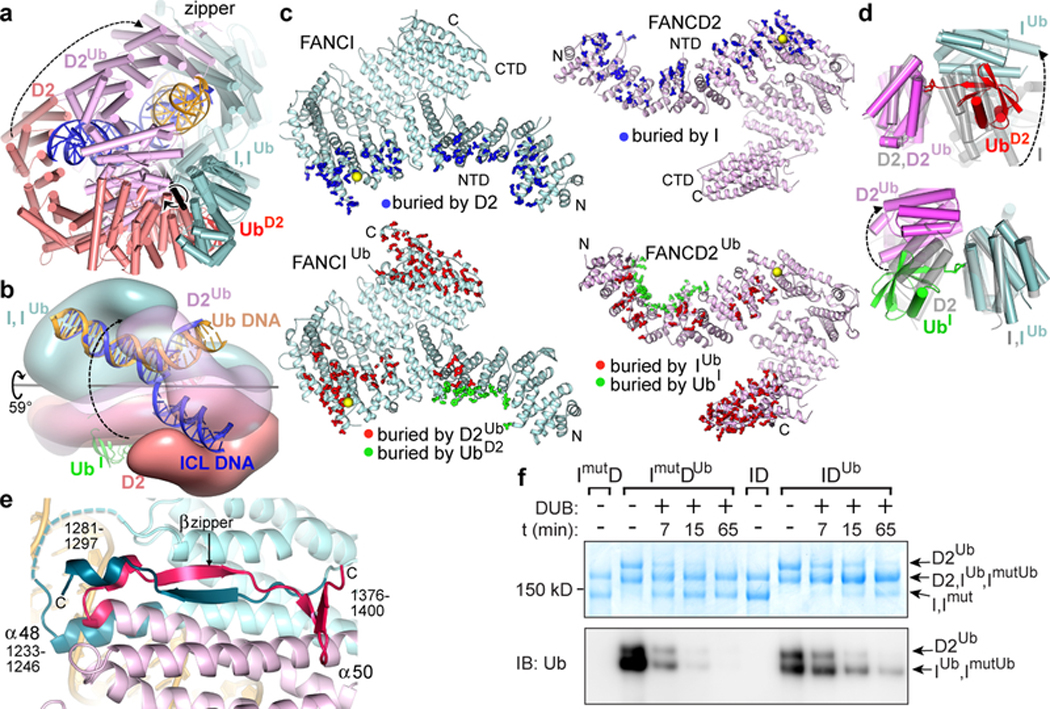
In the non-ubiquitinated ID complex, the two proteins interact along the length of their NTD solenoids in an antiparallel direction, forming an extended interface that buries a total surface area of ~4,950 Å2. The interface is continuous except for two narrow openings wherein the ubiquitination sites (Lys523 and Lys561 of FANCI and FANCD2, respectively) are embedded (Figs 1a and b). After ubiquitination, this interface opens up through a relative rotation of FANCI and FANCD2 by 59° and translation by 15 Å about an axis that runs through the NTD-NTD interface (Fig. 3a and b). None of the intermolecular contacts of the non-ubiquitinated interface are retained (Fig. 3c). Some of these residues are repurposed for interactions with the ubiquitin of the other paralog, with the reciprocal interactions contributing ~3,250 Å2 of buried surface area (Figs 3c and d). Other residues are repurposed for new intermolecular contacts in a smaller NTD-NTD interface of ~2,800 Å2.
Figure 3 |. Conformational changes on ubiquitination.
a, Non-ubiquitinated ID (cyan, salmon and blue for FANCI, FANCD2 and DNA, respectively) superimposed on IDUb (cyan, magenta and yellow for FANCIUb, FANCD2Ub, and DNA, respectively) by aligning FANCI. UbD2 is red (UbI obscured in this view). The rotation axis is shown as a thick black stick with the rotation indicated by a circular arrow. FANCD2 movement is indicated by a dashed arrow linking equivalent structural elements. The CTD-CTD interaction region is labeled “zipper”. Orientation similar to Fig. 2a. b, View looking approximately down the vertical axis of a with the two complexes rendered as semi-transparent surfaces to reveal the DNA inside. c, FANCI (left top, cyan), FANCIUb (left bottom, cyan), FANCD2 (right top, pink), and FANCD2Ub (right bottom, pink) proteins showing residues (thick sticks) with a reduction in solvent accessibility due to interactions between FANCI and FANCD2 (top pair, blue sticks), between FANCIUb and FANCD2Ub (bottom pair, red sticks), and between each protein and the ubiquitin of the other paralog (bottom pair, green sticks). Yellow spheres mark ubiquitination sites. d, Close-up view of the ID and IDUb complexes superimposed on the FANCD2 ubiquitination site (top) or on the FANCI ubiquitination site (bottom). Non-ubiquitinated ID is transparent gray. e, The IDUb CTD zipper looking down the vertical axis of b. Residues unstructured in non-ubiquitinated ID are shown in darker shades and are labeled. Dashed cyan line is a loop unstructured in both complexes. f, SDS-PAGE gel of the de-ubiquitination of wild type IDUb and mutant ImutDUb complexes, both at 940 nM, by USP1-UAF1 (400 nM) at the indicated time points, detected by Coomassie staining (top) or anti-ubiquitin immunoblot (bottom). The positions of the substrates and products are marked. Repeated n=3 times.
The relative rotation of the two proteins also juxtaposes their CTD domains, thereby portions of the previously unstructured C-terminal extensions of FANCI and FANCD2 become structured in a zipper-like intermolecular interaction (Fig. 3e). The zipper involves the FANCIUb α48 helix forming a coiled coil with α50 of the newly juxtaposed FANCD2Ub CTD, as well as the inter-digitation of the two chains to form an intermolecular β sheet, capped by a short helix in FANCIUb and a β hairpin in FANCD2Ub (Fig. 3e). The zipper buries an additional ~4,350 Å2 of surface area, completing the transformation of the open trough to a closed ring. The importance of ring closing is underscored by the Fanconi Anemia R1285Q mutation19,22 in FANCI. Arg1285, which is on the zipper β sheet, forms an intermolecular salt bridge with Glu1365 of FANCD2 in a buried environment, whereas it is unstructured in the non-ubiquitinated ID complex (Extended Data Figs 7f, g). We tested whether R1285Q destabilizes the zipper interface using de-ubiquitination as a surrogate assay, as the lysine-ubiquitin bonds inside the I-D interface should be more accessible in a destabilized heterodimer. As shown in Figure 3f, the de-ubiquitination of R1285Q IDUb by USP1-UAF1 is indeed significantly faster than the wild type IDUb. This mutation also reduces levels of ID ubiquitination in cells14 as well as in vitro (S.W. not shown), suggesting that the transient closure of the zipper, aided by the mobility of the FANCD2 CTD, may be an intermediate in the ubiquitination reaction that allows the UBE2T to access the ubiquitination sites.
The conformation of FANCI does not change significantly in IDUb, but FANCD2 undergoes two conformational changes important in the remodeling of the complex. The N-terminal part of the NTD rotates by 38° towards FANCI, which is now farther away, to better embrace UbI and also to form a new interface with FANCI, while the CTD rotates 20° relative to its NTD to form the zipper interface with the FANCI CTD (Extended Data Fig. 9a).
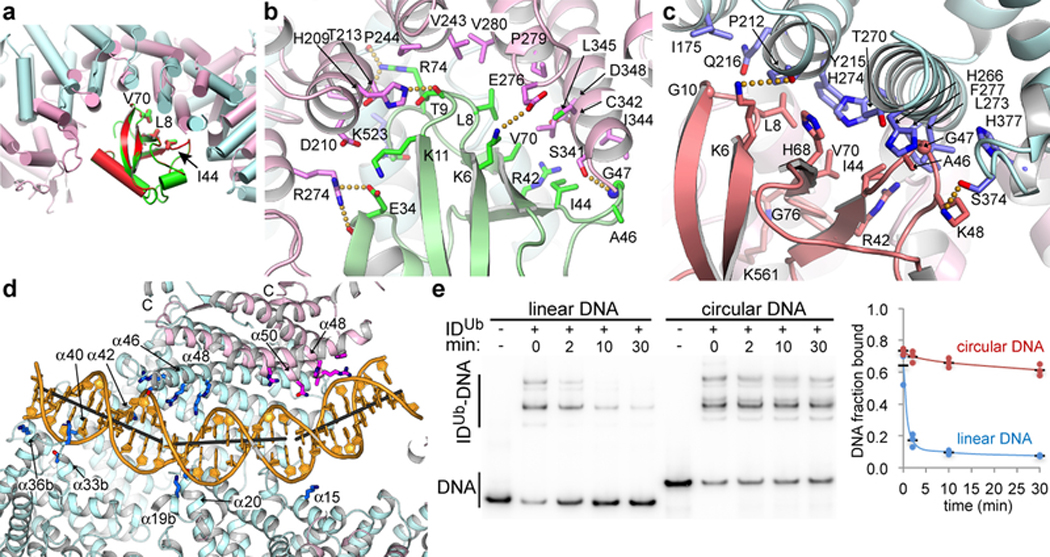
FANCI and FANCD2 use similar concave regions of their NTD solenoids (residues 175 to 377 and 174 to 348, respectively) to bind to ubiquitin (Fig. 4a). Both interfaces completely cover the ubiquitin hydrophobic patch consisting of Leu8, Ile44 and Val70 (Figs 4b and c). Ile44 in various combinations with the other two hydrophobic residues is key to the binding of diverse ubiquitin-binding domains such as UBA, UBZ, UIM and CUE23, which have structures unrelated to the ubiquitin-binding helical repeats of FANCI and FANCD2 (Extended Data Fig. 9b–e). The FANCD2Ub-UbI interactions extend beyond the hydrophobic patch owing to the aforementioned conformational change of the N-terminal portion of FANCD2Ub that embraces more of the ubiquitin compared to FANCIUb (1,820 and 1,450 Å2 buried, respectively; Figs 4a to c).
Figure 4 |. Interactions with ubiquitin and DNA.
a, Superposition of the FANCD2Ub-UbI and FANCIUb-UbD2 interfaces by aligning UbD2 (red) with UbI (green). The paralogs that each ubiquitin is attached to are not shown for clarity. The ubiquitin hydrophobic patch residues (Leu8, Ile44, and Val70) are shown as sticks for both ubiquitin molecules. Colored as in Fig. 2a. b, Close-up view of the non-covalent interface between FANCD2Ub (pink) and UbI (green) showing the residues within interaction distance as sticks (darker colors). Yellow dotted lines indicate potential hydrogen bonds. c, Close-up view of the reciprocal interface between FANCIUb (cyan) and UbD2 (green). d, Cartoon, colored as in Fig. 2a, showing IDUb side chains within contact distance of the DNA as sticks (darker hues). Gray sticks show the helical axes for the three relatively straight segments of the DNA. The majority of FANCD2Ub and parts of FANCIUb are clipped above the plane of the figure to make the DNA visible (Extended Data Fig. 9f legend lists side chains shown). e, Autoradiogram of DNA binding competition time course after 8 μM of unlabeled 67 bp dsDNA was added to the IDUb complex (800 nM) pre-assembled on a 95 bp circular nicked DNA or the corresponding linear nicked DNA, both at 400 nM with only 2 nM of each 32P labeled. 32P DNA-protein complexes contain multiple IDUb proteins due to the length of each DNA. Fraction of DNA bound is quantified in the chart (repetitions marked by circles, and their mean by a black dash; n=3).
It has been suggested that one function of ID mono-ubiquitination may be the recruitment of downstream effectors that contain ubiquitin-binding domains1,24. However, the sequestration of the entire ubiquitin hydrophobic patch on both UbI and UbD2 indicates the ubiquitin is unlikely to play this role with effectors containing the aforementioned ubiquitin-binding domains. This is consistent with the UBZ-containing FANCP nuclease scaffold not requiring FANCD2 ubiquitination for recruitment to ICL sites25. IDUb could still have a recruitment function through other elements such as the C-terminal EDGE motifs26 that remain unstructured and accessible. We also cannot rule out other factors associated with the stalled fork inducing a conformational change in IDUb that exposes the ubiquitin hydrophobic patch, or dissociates the complex into monomers.
While FANCD2 mono-ubiquitination is essential for the FA pathway, FANCI mono-ubiquitination can be of minor importance in certain settings14,27, raising the question of whether the singly-monoubiquitinated ID (ID-/Ub) can also assume the closed-ring structure. Indeed, the 4.0 Å refined cryo-EM structure of ID-/Ub has essentially the same structure as IDUb, with only minor shifts localized to the now empty UbI binding site of FANCD2 (Extended Data Fig. 8e–g).
The IDUb conformational changes result in the remodeling of the bound DNA, converting the non-collinear arrangement of the FANCI- and FANCD2-associated DNA duplexes to a continuous but bent duplex (Fig. 4d and Extended Data Fig. 9f). IDUb uses the FANCI groove and the localized FANCD2 CTD patch to bind to opposite ends of the DNA as with the ID-ICL DNA complex. Additional DNA contacts occur near the middle of the DNA, where the extension of FANCI α48 at the CTD-CTD zipper gives rise to a new semicircular basic groove into which dsDNA binds (Fig. 4d). This is associated with one of the two DNA bends. The second bend occurs as the dsDNA is redirected by the binding site at the FANCD2 CTD. The new position of the FANCD2 CTD overlaps with and blocks the FANCI ssDNA-binding site (Fig. 3b).
We have not been able to locate the DNA nick in the map, which looks indistinguishable from the 3.8 Å map of IDUb bound to canonical dsDNA (Extended Data Fig. 8c). While this could be due to inadequate resolution, the IDUb-5’ flap DNA map shows no trace of the ssDNA branch neither (Extended Data Fig. 8b). This suggests that either IDUb binds to these substrates in multiple registers without a preferred location for the nick or flap, or it is just binding to the dsDNA arms. The latter seems to be the case with the ICL DNA whose duplex arms bind to either end of the clamp (Extended Data Fig. 8a). These findings imply that IDUb has lost the specificity the non-ubiquitinated ID has for branched DNA structures (Extended Data Figs 6a and 9g).
This raises the possibility that IDUb functions as a sliding DNA clamp. To address this, we first assembled the IDUb complex on either a circular nicked DNA or the corresponding linear nicked DNA, then added a 20-fold molar excess of unlabeled 67 bp dsDNA and monitored the DNA binding in a time course. At 2 minutes, 73 % of the linear DNA complex had dissociated compared to only 4 % of the circular DNA complex, 84 % of which persisted at 30 minutes (Fig. 4e). This indicates the clamp dissociates from linear DNA by sliding off the end and not by opening up, because in the latter case it would come off the circular DNA as well.
Our data reveal the function of ID mono-ubiquitination is to completely re-model FANCI-FANCD2 association, expanding our understanding of the roles of mono-ubiquitination. The functional significance of this re-modeling is to convert the ID complex to a clamp that could slide away from its initial location at the ICL or related DNA structures. In principle, this would allow downstream nucleases and other factors to act on the ICL, with the IDUb clamp coordinating the repair reactions, serving as a processivity factor, or protecting the DNA.
METHODS
Protein Expression and purification.
Full length human FANCI with a non-cleavable C-terminal His6 tag and full length human FANCD2 with an N-terminal His6 tag followed by a TEV protease cleavage site were co-expressed in Hi5 insect cells (Invitrogen, not authenticated, not tested for mycoplasma contamination) using baculovirus. Cells were lysed in 50 mM Tris-HCl, 200 mM NaCl, 5 % (v/v) glycerol, 0.5 mM TCEP, pH 8.0, and protease inhibitors. After Ni2+ affinity chromatography and overnight cleavage of the FANCD2 His6 tag by TEV protease, the FANCI and FANCD2 proteins were purified by ion exchange (MonoQ) chromatography, which dissociated the two proteins. The FANCI and FANCD2 proteins were then combined at a 1:1 molar ratio, and concentrated by ultrafiltration to ~20 mg/ml in 20 mM Tris-HCl, 150 mM NaCl, 0.5 mM TCEP, pH 8.0. For the FA Core complex, Flag-tagged FANC A, B, C, E, F, G, L, and FAAP100 were cloned into three modified pCDNA3.1 plasmids with different drug resistance genes, and were used to transfect HEK 293F cells (Invitrogen, not authenticated, not tested for mycoplasma contamination). A stably-transfected cell line with the highest expression of all eight subunits was then adapted for growth in suspension. The FA Core complex was purified using anti-Flag M2 agarose beads (Sigma), ion-exchange (MonoQ) and gel-filtration chromatography (Superose 6) in 20 mM Bicine-HCl, 150 mM NaCl, 0.1 mM TCEP, pH 8.0.
Preparation of the IDUb complexes.
Ubiquitination reactions, carried out in 20 mM Tris-HCl, 150 mM NaCl, 1 mM DTT, pH 8.0, contained 40 μM human Flag-tagged ubiquitin (Fisher Scientific U-120), 0.5 μM human His6-tagged Ubiquitin E1 enzyme (Fisher Scientific, E304050), 2.4 μM human His6-UBE2T E2 enzyme (Fisher Scientific, E2–695), 5 mM adenosine triphosphate, 3 μM core complex, 10 μM ID complex and 30 μM 58 bp nicked DNA, or the other DNA substrates. 60 μL reactions were set up on ice and incubated at 28°C for 1–2 hours. The reaction products were separated by gel-filtration chromatography (Superose 6) in 20 mM Tris-HCl, 150 mM NaCl, 0.2 mM TCEP, pH 8.0, and concentrated by ultrafiltration. Samples were analyzed by SDS-PAGE with NuPAGE 3%–8% Tris-Acetate gels (Invitrogen) and the mono-ubiquitinated FANCI and FANCD2 were verified using mass spectrometry. To prepare the ID-/Ub that is mono-ubiquitinated only on FANCD2 we took advantage of the FA Core complex ubiquitinating the ID complex sequentially starting with FANCD2 (S.W. not shown). We run ubiquitination reactions in the presence of nicked DNA for shorter time periods (10–30 min), and separated the product ID complex that was mostly singly-ubiquitinated on FANCD2 away from the FANCI and FANCD2 substrates by gel filtration chromatography as with the IDUb complex.
Deubiquitination of the ImutDUb complex.
The R1285Q mutant of human FANCI (FANCImut) was generated using QuickChange Lightning Site-Directed Mutagenesis Kit (Agilent), it was cloned with a C-terminal non-cleavable His8 tag into pFastBac vector, and it was overexpressed in Hi-5 insect cells. The recombinant protein was purified by Nickel-affinity, anion-exchange and gel-filtration chromatography and concentrated by ultrafiltration (Amicon) to 85 µM in 20 mM Tris-HCl, 150 mM NaCl, 10% (v/v) glycerol, 1mM DTT, pH 8.0. FANCI and FANCImut were mono-ubiquitinated by the FA Core complex in a FANCD2 independent reaction (S.W. not shown). The mono-ubiquitinated FANCImutUb and the FANCIUb control were purified by anion exchange chromatography and concentrated to 26 µM and 8 µM respectively. Mono-ubiquitinated FANCD2Ub was prepared by running the IDUb complex on an anion exchange column (MonoQ), which separates it from FANCIUb, and concentrated to 42 µM. The IDUb and ImutDUb complexes were then prepared by mixing FANCIUb or FANCImutUb with FANCD2Ub at 1 µM, in 20 mM Tris-HCl, 100 mM NaCl, pH 8.0. After 10 min. incubation on ice, 400 nM of the USP1-UAF1 complex (Boston Biochem, E-568) was added. The final concentration of IDUb or ImutDUb in the reactions was 940 nM. 15 µL reactions were started at 28 ˚C. At the indicated time points, a 3 µL aliquote was mixed with 9 µL NuPAGE LDS sample buffer (Invitrogen) and heated at 95˚C for 2 min to stop the reaction. Samples were separated by NuPAGE 3–8% Tris-Acetate SDS-PAGE (Invitrogen) in duplicate sets for either Coomassie blue staining (6.5 µL loading) or western blotting (4.5 µL loading; anti-ubiquitin antibody from SANTA CRUZ, sc-271289).
Cryo-EM sample preparation and data collection.
For the non-ubiquitinated ID complex bound to the indicated DNA substrates, the concentrated ID complex was combined with a 3-fold molar excess of DNA. For ID-ICL DNA, the mixture was diluted to 2 mg/ml ID (6.5 μM) and 0.8 mg/ml ICL DNA (19.5 μM) in 20 mM Tris-HCl, 150 mM NaCl, 0.5 mM TCEP, pH 8.0. For the other DNA substrates, the ID-DNA mixture was diluted to 3 mg/ml ID (10 μM) and 27 μM DNA. The samples (3 μl) were applied to glow discharged UltrAuFoil 300 mesh R1.2/1.3 grids (Quantifoil). Grids were blotted for 1.5 s at 16° C or 22° C and ~100 % humidity and plunge-frozen in liquid ethane using a FEI Vitrobot Mark IV. For all the mono-ubiquitinated IDUb-DNA complexes, the ubiquitination reactions, performed as described above, were fractionated by gel-filtration chromatography (Superose 6), and the peak corresponding to the IDUb-DNA complex was concentrated by ultrafiltration to ~1.5 mg/ml in 20 mM Tris-HCl, 150 mM NaCl, 0.2 mM TCEP, pH 8.0. Grids were prepared as with the non-ubiquitinated complex. All data were collected with a Titan Krios microscope operated at 300 kV and Gatan K2 Summit camera. Most data were collected with a 1.089 Å pixel size and 10.0 electrons per pixel per second at the MSKCC Cryo-EM facility, with additional data collected at the NYSBC Simons Electron Microscopy Center (1.09 Å pixel size), and at the HHMI Cryo-EM facility (1.04 Å pixel size and 8.0 electrons per pixel per second).
Cryo-EM image processing.
The super-resolution videos were initially aligned with MOTIONCOR228, and the contrast transfer function (CTF) parameters were estimated with CTFFIND429. All 2D/3D classifications, 3D refinements and other image processing were carried out with RELION-310. For the data from the ID-ICL DNA complex and all the IDUb-DNA complexes, Bayesian beam induced motion correction, scale and B-factors for radiation-damage weighting, and per particle refinement of CTF parameters were also applied30. All reported map resolutions are from gold-standard refinement procedures with the FSC=0.143 criterion after post-processing by applying a soft mask. For the non-ubiquitinated ID-DNA complexes, the DNA density in the initial 3D reconstructions was weaker than the protein, even with low-passed maps, suggesting partial DNA occupancy. We thus used partial signal subtraction outside the FANCI-bound DNA duplex followed by 3D classification without alignment of the density inside the DNA mask, all with RELION-3 (Extended Data Fig. 1b). The fraction of the data set that contained FANCI-bound DNA ranged from 48 % for the ICL DNA complex to 28 % for the 5’ flap DNA (Extended Data Figs 1b and 5a, f, k). The thus separated particles were then restored and used for the consensus 3D reconstruction of the ID-DNA complexes. For the reconstruction of apo-ID, we used the DNA-free particles from the ID-ICL DNA data set (Extended Data Fig. 1b, right part of flow-chart). The discord between the EMSA-derived low nanomolar Kd values and the partial DNA-occupancy in the cryo-EM maps is likely due, at least in part, to the significant non-specific DNA binding activity the non-ubiquitinated ID complex exhibits in EMSA as described below (the “DNA binding assays” paragraph in METHODS). Because the non-ubiquitinated ID complex exhibits substantial conformational flexibility in the FANCD2 HD and CTD domains, we performed multi-body refinement with two soft masks in RELION-3. The larger-body mask (body1 in Extended Data Fig. 1b, c) covered FANCI, the FANCI-bound dsDNA and ssDNA, and residues 45 to 623 of FANCD2, and the smaller body mask (body2) covered FANCD2 residues 624 to 1376 and the FANCD2-associated dsDNA. Multi-body refinement was carried out with signal subtraction outside the masks. The mono-ubiquitinated IDUb-DNA complexes appeared to have full occupancy of DNA, except for the IDUb-ICL DNA complex that has dsDNA arms shorter than those of the other DNA substrates (Extended Data 8a). IDUb does not exhibit conformational flexibility in the FANCD2 CTD. The three focused 3D refinements of the IDUb-nicked DNA data improved the resolution only marginally, but there was noticeable improvement in side chain density and continuity. Of the three partially overlapping masks, one mask (focus1 in Extended Data Figs 7b, c) covered FANCI residues 288–1297, FANCD2 residues 45–466, UbI, and the FANCI-proximal half of the DNA, corresponding roughly to the left half of Fig. 2b. A second mask (focus2) covered the C-terminal portions of FANCI (residues 598–1298) and FANCD2 (residues 625–1400) and the entire DNA, corresponding roughly to the top half of Fig. 2a. The third mask (focus3) covered FANCI residues 1–376, FANCD2 residues 447–1400, UbD2, and the FANCD2-proximal half of the DNA, and corresponds roughly to the right half of Fig. 2b. For the singly-ubiquitinated ID-/Ub-nicked DNA complex, we removed residual particles containing FANCIUb from the partial ubiquitination reaction using partial signal subtraction outside the UbI softmask, followed by 3D classification without alignment. Subsequent 3D auto-refinement and focused refinements of the particles lacking UbI were performed as with the IDUb complex (Extended Data Fig. 8e, f).
Cryo-EM structure refinement.
Model refinement was done with REFMAC5 modified for cryo-EM12 and with PHENIX31. For the ID-ICL DNA and apo-ID complexes, the two multi-body refinement maps were combined with the composite sfcalc option of REFMAC5 to construct a single set of structure factors to 3.3 Å, the resolution of the larger body1, and refinement was carried out at this resolution. As the resolution of the smaller body2 map is lower (3.8 Å), this results in higher temperature factors for the portion of the model within this map. The model was built based on the mouse ID structure. Because the DNA has significantly higher temperature factors than the overall protein, it was modeled as dA-dT base pairs. The model was first refined in reciprocal space using REFMAC5, then in real space with PHENIX, and again in reciprocal space including TLS refinement. REFMAC5 refinement included secondary structure restrains (SSR) generated by ProSMART12. The structure factors for the IDUb-nicked DNA complex were calculated by combining the three focused 3D reconstructions similarly, except refinement was carried out at 3.48 Å, the highest resolution common to all 3 maps. The coordinates were assigned to the three focused maps as follows: FANCI residues 327–959, FANCD2 residues 45–454, and UbI to focus1; FANCI residues 960–1297, FANCD2 residues 1150–1400, and the DNA to focus2; FANCI residues 1–326, FANCD2 residues 455–1145, and UbD2 to focus3. Refinement was carried out with REFMAC5 and PHENIX as with the ID-ICL DNA complex. The ID-/Ub-nicked DNA complex was refined similarly.
The refined ID-ICL DNA model lacks the following unstructured regions: 250–259, 401–408, 551–574, 685–695, 935–948, 1111–1125, 1222–1246, 1281–1328 (C-terminus) of FANCI, and 1–44, 122–129, 313–337, 589–604, 708–725, 852–915, 947–959, 982–1000, 1043–1050, 1146–1149, 1216–1219, 1377–1451 of FANCD2. As discussed in the main text (Fig. 3e), in the refined IDUb-nicked DNA model FANCI residues 1233–1246 (α48 extension) and 1281–1297 (C-terminal residues), and FANCD2 residues 1377–1400 become ordered as part of the zipper interface. The unstructured regions of the non-ubiquitinated ID-ICL DNA complex generally correspond well with the unstructured regions in the mouse ID crystal structure, or the regions deleted from the mouse FANCD2 construct used in those crystallization experiments based on susceptibility to limited proteolysis7.
DNA-binding assays.
Electrophoretic mobility shift assays (EMSA) assays were performed using the 40 to 42 bp (or equivalent) DNA substrates shown in the table below. The non-ubiquitinated ID complex exhibits considerable non-specific DNA binding that seems to correlate with the length and thus total charge of the DNA. To try to mitigate that, we used unlabeled 20 bp dsDNA as nonspecific competitor. However, we cannot rule out the possibility that the Kd value of a substrate like HJ DNA, which differs significantly from the other substrates in the number of nucleotides, having a larger contribution from non-specific DNA binding. Reactions (15 μl) were assembled by mixing the indicated 32P-labeled DNA substrates (0.5 nM) with the unlabeled 20 bp dsDNA (1.4 μM) and adding the ID complex in 20 mM Tris-HCl, 150 mM NaCl, 5 % Glycerol, 0.1 mg/ml BSA, 0.5 mM TCEP, pH 8.0. They were incubated on ice for 30 min, followed by electrophoresis at 4 °C on 4 % (w/v) polyacrymide gels in 0.5x TBE buffer. The dried gels were quantitated using a phosphorimager, and the data were fit to a one-site cooperative binding model by minimizing the sum of square of the differences. For the mono-ubiquitinated IDUb complex, the DNA that was carried over from the ubiquitination reaction and remained bound to the IDUb complex during gel-filtration was removed by anion-exchange chromatography. This also dissociated the FANCIUb and FANCD2Ub proteins, which were then concentrated separately by ultrafiltration. Binding reactions were assembled by first mixing FANCIUb with the indicated 32P-labeled DNA substrate (0.5 nM) and then adding FANCD2Ub at a one molar ratio to FANCIUb. The subsequent steps were performed as with the non-ubiquitinated ID complex. The IDUb reactions did not contain the unlabeled dsDNA competitor used with the non-ubiquitinated ID complex. For the competition experiments of Fig. 4f, DNA-binding reactions were assembled by first adding monomeric FANCD2Ub (800 nM) to the indicated 5’ 32P labeled DNA (400 nM, with only 2 nM labeled), then adding FANCIUb (800 nM) in the same buffer as above. Reactions were incubated on ice for 30 min before adding a 20-fold molar excess (to the substrate DNA) of unlabeled 67 bp dsDNA competitor (8 μM) to start the time course, followed by electrophoresis at 4 °C as above.
DNA substrates.
All of the DNA substrates were prepared by annealing the oligonucleotides listed in Supplementary Table. The 95 nt circular oligonucleotide was synthesized by Bio-Synthesis Inc. Its sequence is based on a 91 nt case study on the company’s webpage, except for the addition of 4 nts to create a restriction enzyme site. The double-stranded circular and its corresponding linear DNA molecules were prepared by annealing complementary oligonucleotides. The ICL DNA was prepared as described9. Briefly, we used the Cu(I)-catalyzed azide-alkyne cycloaddition between an N4-(3-azidopropyl) modified cytosine on a dCdG step of one DNA strand, and an N4-propargyl modified cytosine on the complementary strand, crosslinking the N4 positions of the two cytosines with a triazole moiety. The A3 and A4 oligonucleotides were synthesized by Sigma incorporating N4-(3-azidopropyl) deoxycytidine and N4-propargyl deoxycytidine, respectively, at the positions indicated by the bold “C”. For the A3 oligonucleotide, the synthesis involved first adding an N4-chloropropyl deoxycytidine phosphoramidite to avoid an azide-phosphoramidite side reaction, with the chloropropyl group subsequently converted to azidopropyl on the beads. The oligonucleotides were HPLC purified by Sigma. For the crosslinking, the two oligonucleotides, each at 0.2 mM concentration, were incubated with 2 mM CuSO4, 10 mM Sodium Ascorbate and 1 mM Tris(3-hydroxypropyltriazolylmethyl)amine in 10 mM HEPES-Na, 50 mM NaCl, pH 7.5, for 30 minutes at room temperature. The reaction products were separated by denaturing PAGE (12% polyacrylamide, 8 M urea), and the crosslinked product was isolated from the gel slice by electroelution (Whatman Elutrap). The ICL was confirmed by liquid chromatography coupled electrospray ionization mass spectroscopy, performed by Novatia (Newtown, PA).
Extended Data
Extended Data Figure 1 |. Cryo-EM reconstructions of the non-ubiquitinated ID-ICL DNA and apoID complexes.
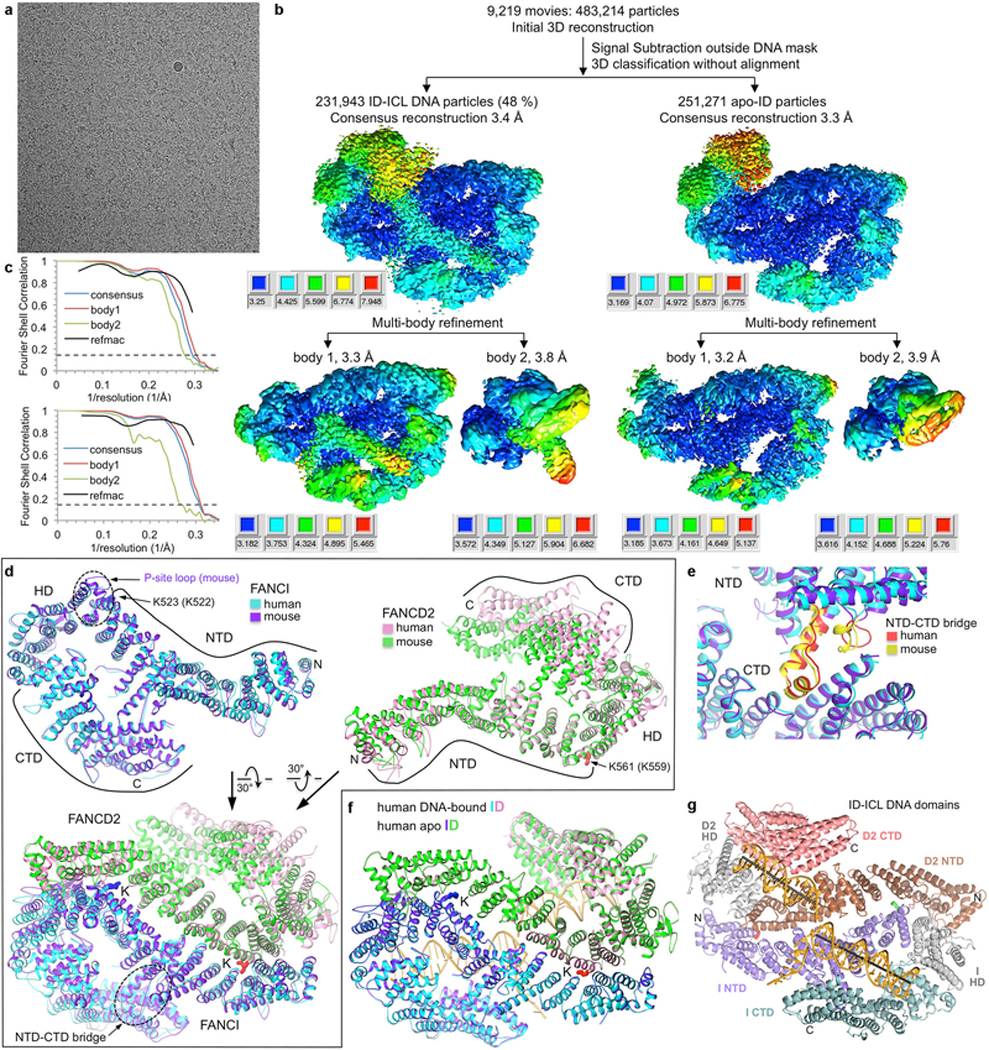
a, Micrograph of non-ubiquitinated ID-ICL DNA particles. The presence of excess DNA at 0.8 mg/ml obscures the protein particles. The particles were collected in five data sets. b, Flow chart of single particle cryo-EM data processing for the ID-ICL DNA and apo-ID complexes. Consensus (top) and focused maps from the RELION3 multi-body refinement, temperature-factor sharpened and masked, are colored by local resolution estimated with the RELION3 postprocess program. The resolution range is mapped to the colors in the inset next to each map. Orientation is similar to Figure 1a. The particle has dimensions of 164 Å, 116 Å, 96 Å. Additional details are in Methods. c, Top graph shows gold-standard FSC plots between two independently refined half-maps for the ID-ICL DNA consensus reconstruction (blue curve), for the RELION3 multi-body refinement of the larger body1 consisting of FANCI, FANCI-bound dsDNA and ssDNA, and FANCD2 residues 43–623 (red curve), and for the smaller body2 consisting of FANCD2 residues 624–1376 and the FANCD2-associated dsDNA (green curve). The FSC curve for the final model versus the composite map combining the cryo-EM maps of the two bodies is shown in black. Dashed line marks the FSC cutoff of 0.143. Bottom graph shows the gold-standard FSC plots between two independently refined half-maps for the consensus reconstruction of the apo-ID complex (blue curve), of the RELION3 multi-body refinement of the two bodies, and of the model refined with REFMAC5 (black). d, Top left panel shows the superposition of the human FANCI structure on mouse FANCI from the crystal structure of the mouse ID complex32 with an r.m.s.d. of 1.8 Å for 926 out of 1,168 common Cα atoms (colored blue and purple for human and mouse, respectively). The majority of the residues that do not superimpose are in the N-terminal 183 amino-acid segment that packs with the FANCD2 HD domain that is mobile in the cryo-EM structure. Other differences include FANCI residues 551–574 that are disordered in human FANCI, while the corresponding mouse FANCI segment (marked by dotted oval and labeled P-site loop) is well ordered at the FANCI-FANCD2 interface, and affects the conformations of flanking helical repeats. The FANCD2 region with which this segment packs in the mouse ID complex is shifted by ~5 Å away from FANCI in the human ID complex. This segment also contains the phosphorylation sites of ATR kinase32,33. Ubiquitination sites, N- and C- termini and individual domains are marked. Top right panel shows the superposition of the human FANCD2 on the mouse structure with an r.m.s.d. of 2.0 Å for 501 out of 1,131 common Cα atoms (colored pink and green for human and mouse, respectively). The low level of overall structural overlap is due to the flexibility of the FANCD2 CTD and HD domains in the cryo-EM structure compared to the mouse ID crystal structure, where the FANCD2 CTD is involved in crystal packing contacts that limit its mobility. Another difference involves the first ~200 residues that cap the FANCD2 NTD solenoid. This segment is displaced by 5 Å in the mouse structure owing to the ordering of the aforementioned FANCI phosphorylation-site loop at the interface. Bottom panel shows the superposition of human ID on the crystal structure of mouse ID with an r.m.s.d. of 2.2 Å for 1,302 out of 2,312 common Cα atoms. The superimposed segments are primarily the FANCI and FANCD2 NTD solenoids, including their ubiquitination sites but excluding their N-termini as discussed above, and most of the FANCI CTD domain. The dashed oval marks the helical protrusion (NTD-CTD bridge) from the FANCI NTD that packs with its CTD and rigidifies the structure. The lack of an NTD-CTD bridge in FANCD2 is associated with the increased mobility of its CTD relative to the NTD. e, Close-up view of the FANCI NTD-CTD bridge of the human and mouse structures colored red and yellow, respectively. f, DNA binding does not cause any substantial conformational differences, except for the FANCD2 CTD that exhibits increased mobility in apo-ID. The human ID-ICL complex (cyan FANCI and pink FANCD2) can be superimposed on the human apo-ID complex (purple FANCI and green FANCD2) in its entirety with a Cα r.m.s.d. of 0.9 Å (2,291 common Cα atoms). g, The human ID-ICL DNA complex colored according to the individual proteins’ domains as indicated, in the same orientation as Figure 1a.
Extended Data Figure 2 |. Conformational flexibility of the FANCD2 CTD domain and its associated DNA.
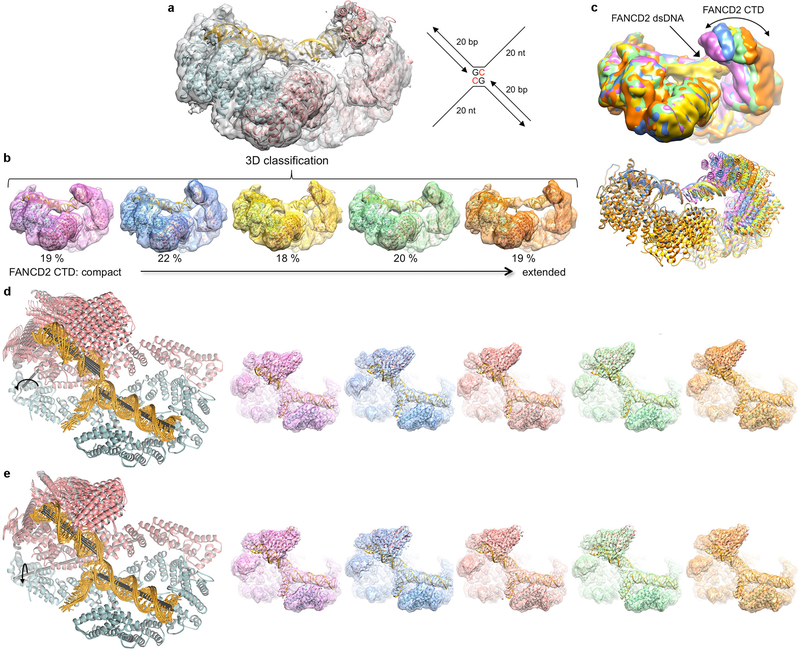
a, The FANCD2 CTD and its associated DNA are evident in the consensus reconstruction prior to temperature-factor sharpening. Because the DNA has higher temperature factors than the protein (Extended Data Table 1), the temperature-factor calculated from the overall map degrades the continuity of the DNA density. The cartoon representation of the refined model is colored cyan for FANCI, pink for FANCD2, and gold for DNA. The schematic of the ICL DNA is shown to the right of the map, with the deoxycytidine bases that are crosslinked by a triazole colored red. The 20 nt ssDNA arms consist of (dT)20 to minimize secondary structure. b, 3D classification of the particles showing the conformational flexibility of FANCD2. The 3D classes are arranged starting with the most compact conformation where the FANCD2 CTD is closer to its NTD. Also shown is the refined consensus model rigid-body fitted into each class and colored as in a. The conformational flexibility starts within the HD domain (starting around residue 645). c, The five 3D classes are superimposed by aligning the FANCI-portion of each map (left), or of each pdb (right) colored according to their map in b. d, Cryo-EM reconstructions using particles from the top component of the principle component analysis (PCA) of the multi-body refinement angles, separated into 5 bins. This component accounts for 21.5 % of the variance in the relative orientation of the FANCD2 CTD (Supplementary Video 1). Left, five ID-ICL DNA models refined in real-space with PHENIX (overall solvent-corrected resolution ranging from 3.7 to 3.9 Å) against maps reconstructed with particles derived from five bins of eigenvalues for the top eigenvector. This PCA component corresponds to a rotation of up to 16° (curved arrow) about an axis running through the HD domain roughly perpendicular to the plane of the figure (gray stick). The helical axes of the individual duplexes are shown as black sticks. Right, the corresponding maps, without temperature-factor sharpening, starting with the conformation (pink map) where the FANCD2 CTD is closest to the FANCI CTD. e, The second component from the PCA analysis accounts for 17% of the variance in the relative orientation of the FANCD2 CTD (Supplementary Video 2). It corresponds to an up to ~10° rotation (curved arrow) about an axis (gray stick) roughly parallel to the plane of the figure. Left are the refined models, and right the maps as in d.
Extended Data Figure 3 |. Cryo-EM density from post-processed maps of non-ubiquitinated ID bound to ICL DNA.
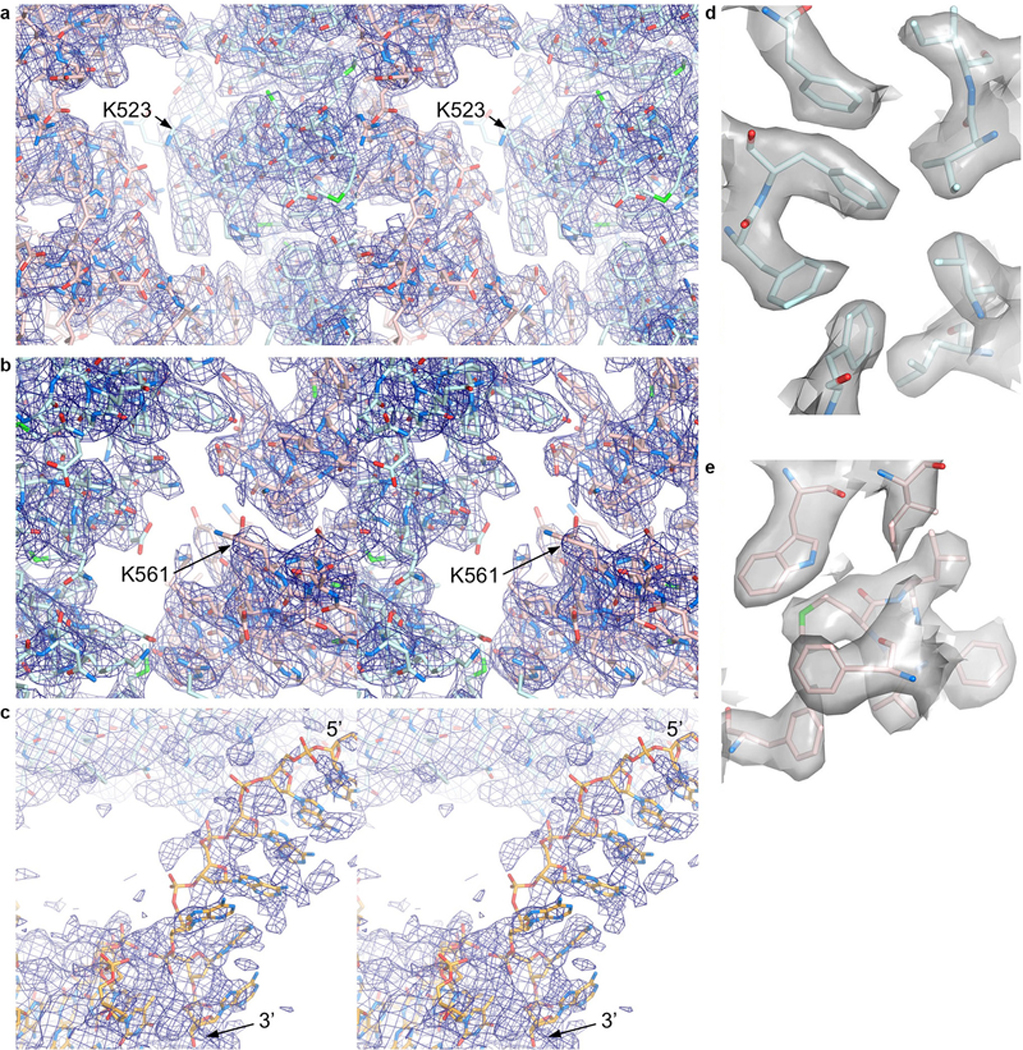
a-b, Stereo view of the 3.3 Å cryo-EM density from the post-processed reconstruction using multi-body refinement. Map shows the vicinity of a, the FANCI ubiquitination site (Lys523 marked) with portions of FANCI residues 475–593 (cyan) and FANCD2 residues 174–287 (pink) shown in stick representation, and b, the FANCD2 ubiquitination site (Lys561 marked), with portions of FANCD2 residues 482–578 and of FANCI residues 123–223. O and N atoms are colored half-bonded red and blue, respectively, for both proteins. c, Stereo view of the map from a focusing on the ssDNA (5’ and 3’ ends marked) at the FANCI CTD, as well as a portion of the FANCI-bound dsDNA. The DNA is in stick representation colored half-bonded yellow, red and blue for C, O, N atoms, respectively. The map is shown at a low contour level because the ssDNA has high temperature factors, and its density is broken up due to the temperature-factor applied in post-processing being calculated from the entire map. ssDNA density before post-processing can be seen in the panel of maps in Extended Data Fig. 2, d and e. d-e, Mono view of the 3.3 Å cryo-EM density depicted as a semi-transparent surface at the hydrophobic core of d, the FANCINTD, showing the residues 202, 214, 217, 236–237, 271–272, and 300, and e, of the FANCD2NTD, showing the residues 347, 364, 368, 380, and 383–386.
Extended Data Figure 4 |. DNA binding by the non-ubiquitinated ID complex.
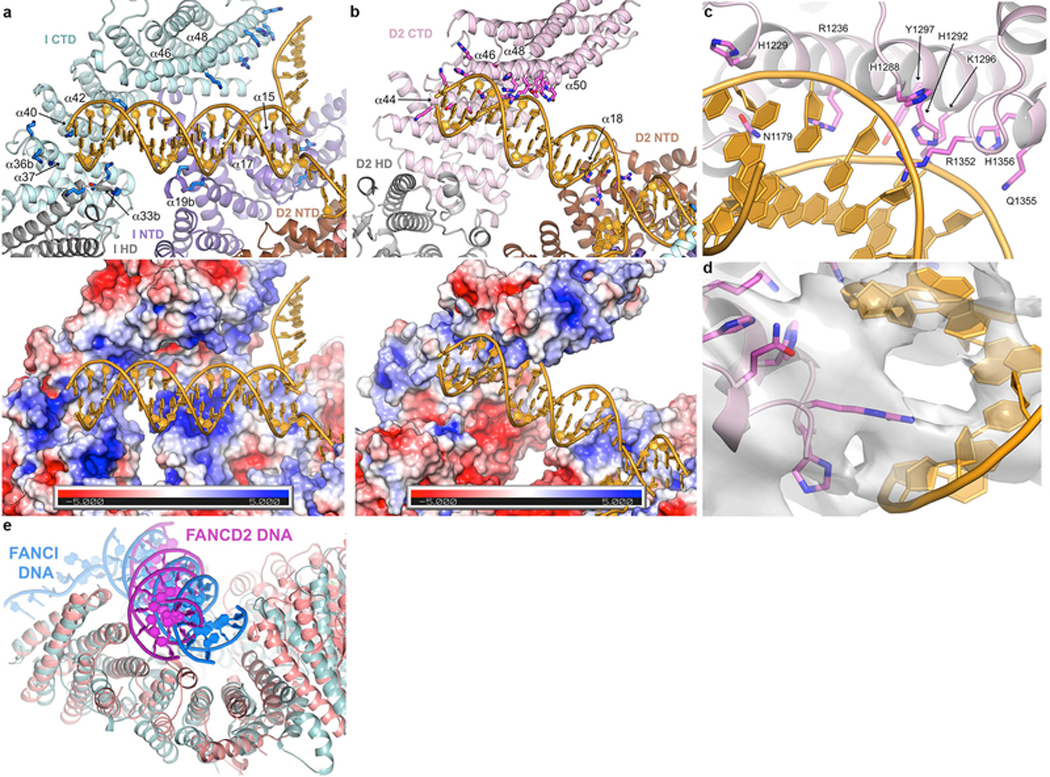
a, DNA binds to an extended basic surface of FANCI. Cartoon representation showing FANCI side chains within potential contact distance of the DNA (top), and molecular surface colored according to the electrostatic potential calculated with PYMOL (bottom, colored −5 to +5 kT blue to red). The proteins are colored according to their domains, in purple, gray and cyan for the FANCI NTD, HD and CTD domains, respectively, and brown, gray, pink for the FANCD2 NTD, HD and CTD domains, respectively. The end of the dsDNA binds to a semi-circular groove consisting of helices α33b, α36b, α37, α40 and α42 (secondary structure elements numbered as in the mouse ID structure32, with insertions denoted by letters after the helix number). This is analogous to the 7.8 Å crystallographic map of mouse FANCI bound to Y DNA, with the N-termini of helices and inter-helix loops providing multiple basic residues. The ICL-proximal portion of the duplex, which is absent from the shorter DNA used in the mouse FANCI crystals, is positioned against basic residues emanating from helices α15, α17 and α19b. The ssDNA rests against the sides of the α48 and α49 helices. The overall DNA density is of lower resolution than the surrounding protein, and in the refined model the DNA has high temperature factors suggesting it is significantly more mobile than the surrounding protein elements. Figure shows side chains for Arg287 on α15, Arg321, Lys336 and Lys339 on α17, Lys396 and Lys397 on α19b, Lys791, Lys793, Thr794 and Lys795 on α33b, Lys897, Lys898 and Lys902 on α36b, Lys980 on α40, Lys1026 on α42, Arg1178 on α46, and Lys1262, His1266, Lys1269 and Lys1270 on α48. b, FANCD2-DNA contacts are localized to the last four helical repeats of the CTD and a patch of basic residues on the NTD. Top figure shows the residues within contact distance of the DNA, colored as in a. The CTD residues involve the N-terminal portions of the inner helices of the helical repeats: Lys1172, Lys1174, Ser1175, Ser1178, Asn1179 and His1183 on α44, Arg1128, His1229 and Arg1236 on α46, Ser1287, His1288, His1292, Lys1296 and Tyr1297 on α48, and Thr1351, Arg1352, Gln1355 and His1356 on α50. NTD residues on α50 are Arg401, Arg404, Asn405 and Arg408. Bottom figure shows the corresponding molecular surface colored according to the electrostatic potential calculated with PYMOL (bottom, colored −5 to +5 kT blue to red). Note the absence of a basic patch at the HD-portion of the semi-circular groove (lower left portion of figure) compared to that of FANCI in a. c, Close-up view of the FANCD2 Arg1352 side chain in the DNA minor groove, and the residues that are in the vicinity of the flanking phosphodiester backbone.
d, Close-up view of c, showing the 3.8 Å cryo-EM density centered on Arg1352, shown in stick representation. Only a subset of the side chains shown in c are visible in this view (His1288, His1292, Lys1236, Lys1296, Gln1355 and His1356). e, Superposition of the DNA-binding region of FANCD2 (pink) CTD on the corresponding region of the FANCI paralog (cyan) showing the different orientations of the FANCI dsDNA (blue) and FANCD2 dsDNA (magenta) in the semi-circular grooves of the respective proteins. Residues 905–1269 of FANCD2 were aligned on residues 1058–1376 of FANCI with a 2.2 Å r.m.s.d. in the positions of 185 Cα atoms.
Extended Data Figure 5 |. Cryo-EM reconstructions of the non-ubiquitinated ID complex bound to 5’ flap DNA, Holliday junction DNA, and replication fork DNA.
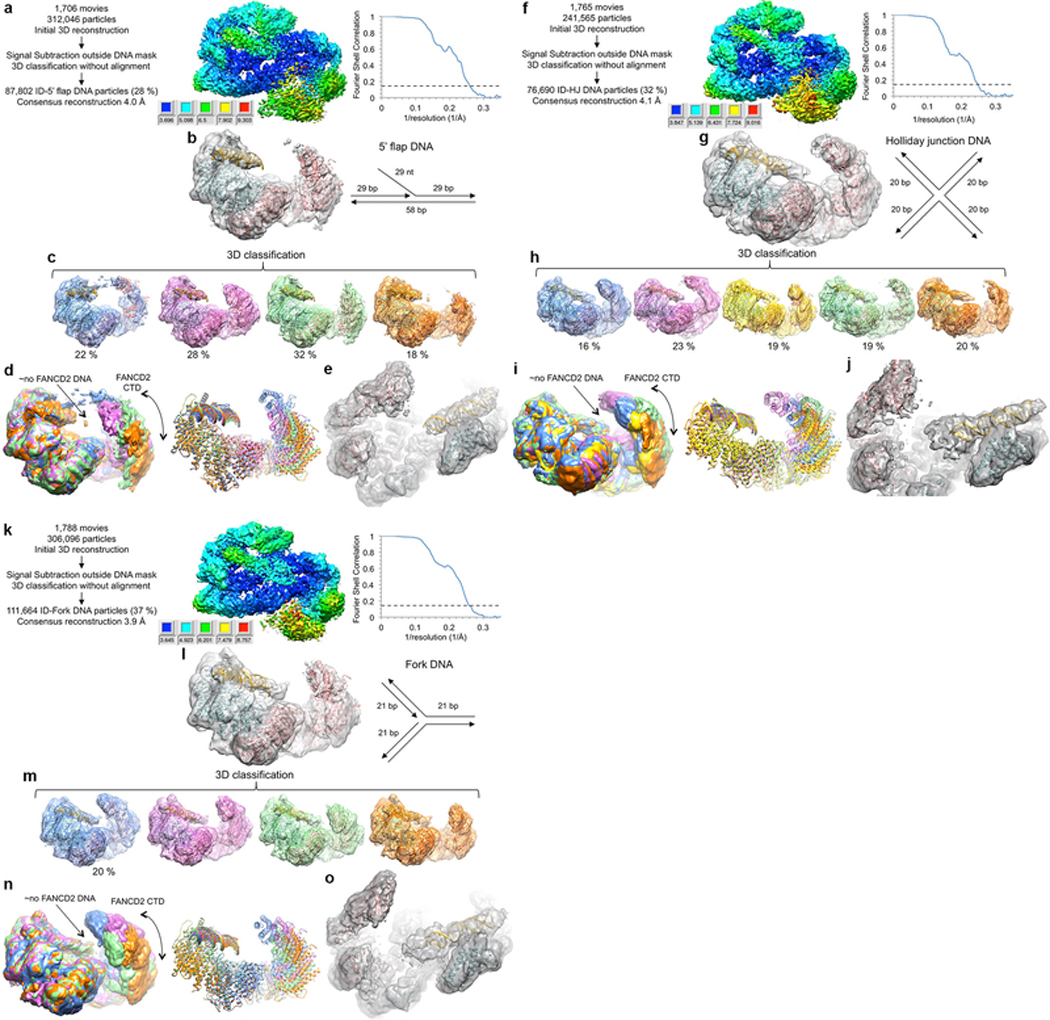
a, Flow chart of single particle cryo-EM data processing. The consensus reconstruction map, temperature-factor sharpened and masked, is colored by local resolution estimated with the RELION3 postprocess program. Graph on the right shows gold-standard FSC plot between two independently refined half-maps for the consensus reconstruction with 87,802 particles. Dashed line marks the FSC cutoff of 0.143 that the FSC curve intersects at 4.0 Å. b, Cryo-EM map from the consensus reconstruction without temperature-factor sharpening. Also shown are cartoon representations of the FANCI (cyan), FANCD2 (pink) and FANCI-bound dsDNA (gold) from the ID-ICL DNA complex rigid-body fitted as multiple domains into the map. Schematic of the 5’ flap DNA is shown to the right of the map. c, 3D classification of the particles showing the conformational flexibility of FANCD2. Maps shown are after the particles from each 3D class were further refined in RELION to 4.7, 4.3, 4.3 and 4.9 Å, respectively. The maps are without temperature-factor sharpening to make the DNA easier to see. The ID complex and dsDNA, rigid body fitted into each map are also shown. d, The five 3D classes are superimposed by aligning the FANCI-portion of each map (left), or of each pdb (right) colored as in c. The lack of FANCD2-bound dsDNA is indicated by the label “ño FANCD2 DNA”. The density of the FANCD2 CTD is significantly weaker and flatter than the similarly calculated maps of the of the ID-ICL DNA complex. The curved arrow indicates the motion suggested by the 3D classification. In the most compact class (blue map), there is density extending from the FANCD2 CTD to the FANCI CTD. While we could not improve this density due to the limited number of particles, it suggests that the flexibility of the FANCD2 CTD may be important for the closing of the structure on ubiquitination. e, Close-up view of the best 3D class (pink in c) after 3D refinement of the particles prior to post processing. Orientation is similar to Figure 1c in the main text. Neither this map nor those of the other 3D classes have any evidence for a localized fork junction or for the 5’ ssDNA flap, suggesting the 5’ flap DNA binds to FANCI in multiple registers with no specificity for the junction. f, Flow chart of cryo-EM data processing for the ID complex bound to Holliday junction DNA. The consensus reconstruction map, temperature-factor sharpened and masked, is colored by local resolution estimated with the RELION3 postprocess program. Right graph shows the gold-standard FSC plot between two independently refined half-maps for the consensus reconstruction with 76,690 particles, with an FSC value of 0.143 (dashed line) at 4.1 Å. g, Cryo-EM map from the consensus reconstruction prior to post processing. Also shown are cartoon representations of the FANCI (cyan), FANCD2 (pink) and FANCI-bound dsDNA (gold) from the ID-ICL DNA complex rigid-body fitted into the map. Schematic of the Holliday junction DNA used is shown to the right of the map. h, 3D classification of the particles showing the conformational flexibility of FANCD2. Maps shown are after the particles from each 3D class were further refined in RELION to 6.9, 4.7, 6.6, 4.7 and 6.5 Å, respectively. The maps are without temperature-factor sharpening. The ID complex and dsDNA, rigid body fitted into each map are also shown. i, The five 3D classes are superimposed by aligning the FANCI-portion of each map (left), or of each pdb (right) colored as in h. j, Close-up view of the best 3D class (green in h) after 3D refinement of the particles prior to post processing. Unlike the map with the 5’ flap DNA, this map shows some bifurcation at one end of the duplex suggestive of the presence of the Holliday junction at a preferred location. This may be due to its shorter duplexes of 20 bp being just long enough to fill the FANCI groove. k, Cryo-EM data processing flow chart of ID bound to replication fork DNA. The consensus reconstruction map, temperature-factor sharpened and masked, is colored by local resolution estimated with the RELION3 postprocess program. The graph on the right shows the gold-standard FSC plot between two independently refined half-maps for the consensus reconstruction with 111,664 particles, with an FSC value of 0.143 (dashed line) at 3.9 Å. l, Cryo-EM map from the consensus reconstruction prior to post processing. Also shown are cartoon representations of the FANCI (cyan), FANCD2 (pink) and FANCI-bound dsDNA (gold) from the ID-ICL DNA complex rigid-body fitted into the map. Schematic of the replication fork DNA used is shown to the right of the map. m, 3D classification of the particles showing the conformational flexibility of FANCD2. Maps shown are after the particles from each 3D class were further refined in RELION to 4.8, 4.7, 4.6 and 4.4 Å, respectively. The maps are without temperature-factor sharpening. The ID complex and dsDNA, rigid body fitted into each map are also shown. n, The five 3D classes are superimposed by aligning the FANCI-portion of each map (left), or of each pdb (right) colored as in m. o, Close-up view of the best 3D class (orange in m) after 3D refinement of the particles prior to post processing.
Extended Data Figure 6 |. Biochemical characterization of ID DNA binding and reconstitution of ID mono-ubiquitination.
a, Electrophoretic mobility shift assay (EMSA) of the ID complex binding to the indicated 32P-labeled DNA substrates (0.5 nM) in the presence of 1.4 μM unlabeled, 20 bp dsDNA as nonspecific competitor. The plots with a logarithmic X-axis show fraction bound in three repetitions of each experiment (blue, green, orange markers), and their mean value (black dash). Each binding isotherm fits a Hill slope model significantly better than a non-competitive binding model, even after excluding the highest protein concentration reactions where multiple shifted bands are apparent, and also in the absence of nonspecific competitor DNA (not shown). A binding curve (black line) simulated with the indicated Kd and Hill coefficient (ηH) values is shown on each plot. b, SDS-PAGE gel of the purified FA core complex. M.w.: molecular weight markers with their mass labeled; Core: FA Core complex with the constituent proteins labeled. The Core prep was performed at least 3 times, and an additional time with a different isolate of a stably transfected cell line with very similar results. c, SDS-PAGE of the ubiquitination reaction of the ID complex in the presence of a 58 bp nicked-DNA molecule and of three peaks from the fractionation of the reaction products on a Superose 6 gel-filtration column shown in d. The gel and gel filtration run of d are typical of the preparative reaction and purification performed at least 3 times with nicked DNA and once each for the DNA substrates of Extended Data 8a–c with similar results. d, Gel-filtration chromatography of the ubiquitination reaction products (blue plot) and of the DNA-only control (orange plot). The fraction marked 1 contains the core complex, fraction 2 the complex of uniquitinated FANCI and ubiquitinated FANCD2, and fraction 3 contains monomeric non-ubiquitinated FANCI and FANCD2, as well as the overlapping peak of excess DNA. e, Comparison of the gel-filtration chromatography profiles of the ID ubiquitination reaction product (blue plot) and of non-ubiquitinated ID (red plot), both at 8 μM, in the presence of 16 μM ICL-DNA and 100 mM NaCl. The chromatography and SDS-PAGE of f were repeated twice with similar results. f, 4 % TBE gels of the gel-filtration fractions marked with an arrow in e stained for protein with Coomassie Blue and for DNA with SYBR Gold. Compared to ubiquitinated ID, only a residual amount of non-ubiquitinated ID remains bound to DNA, and its DNA and ID-DNA bands are faint. Because gel filtration chromatography is more sensitive to the off rate of a complex than the EMSA assay, this suggests that the IDUb complex has a slower off rate consistent with its closed ring structure compared to the open-trough structure prior to ubiquitination. Complexes of IDUb with nicked DNA and with the other DNA substrates of Extended Data Fig. 8 behave similarly to the IDUb-ICL complex under these conditions (not shown).
Extended Data Figure 7 |. Cryo-EM reconstruction of the IDUb complex bound to nicked DNA.
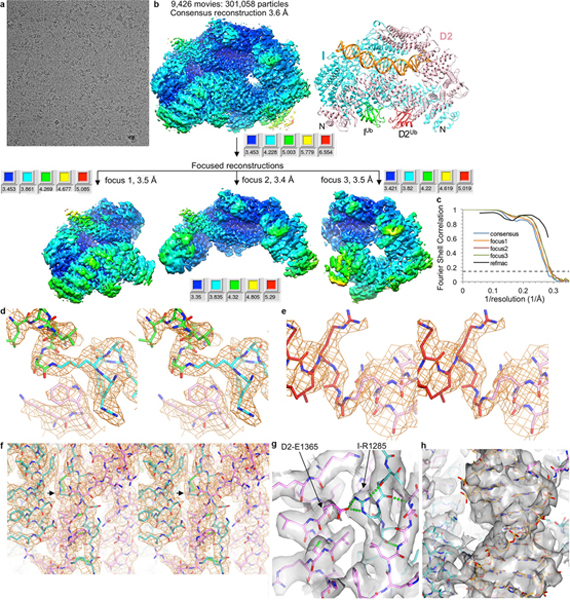
a, Micrograph of IDUb-nicked DNA particles. The particles were collected in three data sets. b, Flow chart of single particle cryo-EM data processing. Consensus (top) and focused maps from RELION3, temperature-factor sharpened and masked, are colored by local resolution estimated with the RELION3 postprocess program. The focused maps below roughly correspond to the left, top, and right portions of the consensus map. The maps are all oriented as the structure on the right (colored as in Fig. 2a). The resolution range is mapped to the colors in the inset next to each map. The particle has dimensions of 155 Å, 115 Å, 101 Å. Additional details are in Methods. c, Graph shows gold-standard FSC plots between two independently refined half-maps for the consensus reconstruction of the ID-ICL DNA (blue curve) and the three focused maps. The FSC curve for the final model versus the composite map combining the focused maps in REFMAC5 is shown in black. Dashed line marks the FSC cutoff of 0.143. d, Stereo view of the 3.5 Å cryo-EM density of the isopeptide bond between the ubiquitin Gly76 C atom and the Nζ atom of Lys523 of FANCI from the post-processed reconstruction of the focused refinement. UbI is green, FANCI cyan. O and N atoms are colored half-bonded red and blue, respectively, for both proteins. Also shown is FANCD2 Trp182 (pink) that packs with Lys523. e, Stereo view of the 3.5 Å cryo-EM density of the isopeptide bond between the ubiquitin Gly76 C atom and the Nζ atom of Lys561 of FANCD2 from the post-processed reconstruction. UbD2 is dark red, FANCD2 pink. f, Stereo view of the 3.4 Å cryo-EM density of the zipper β sheet of the IDUb complex. FANCI is cyan and FANCD2 pink. The arrow points to FANCI Arg1285 that is mutated to glutamine in Fanconi Anemia. Select hydrogen bonds (made by the β sheet and by Arg1285) are shown as green dotted lines. g, Mono view of the density in f, rendered semi-transparent, focusing on the vicinity of FANCI Arg1285. Arg1285 and the FANCD2 Glu1365 with which it forms a salt bridge are both in a buried environment as can be seen in e (the structural elements and density above the plane of the figure are not shown for clarity). Because there are no other basic residues near Glu1365 to neutralize its charge, the loss of the arginine charge in the FA R1285Q mutant would leave a net charge in a buried environment and destabilize the zipper. h, Mono view of the DNA density inside the ring, abutted by the zipper on the left side.
Extended Data Figure 8 |. Cryo-EM reconstructions of the IDUb complex bound to ICL DNA, 5’ flap DNA and dsDNA, and of the ID-/Ub complex ubiquitinated only on FANCD2 bound to nicked DNA.
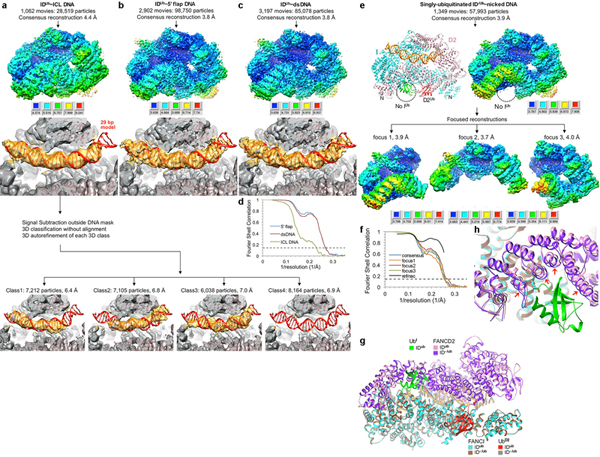
a, Cryo-EM reconstruction of IDUb bound to ICL DNA (same DNA as Extended Data Fig. 2a). The consensus map (top), temperature-factor sharpened and masked, is colored by local resolution estimated with the RELION3 postprocess program. The resolution range is mapped to the colors in the inset. The map is oriented as in Extended Data Fig. 7b. To make the DNA easier to see, the consensus map is also shown prior to temperature-factor sharpening with the FANCD2 portion above the plane of the figure clipped (middle, gray map) in the same orientation as Fig. 2c. The model shown is the IDUb-nicked DNA complex structure that was fit as a single body into the map with CHIMERA, without any further refinement of the coordinates. Because the DNA density in the consensus map appeared longer than the 20 bp dsDNA arms of the ICL DNA, we used 3D classification with partial signal subtraction followed by the reconstruction of the non-signal subtracted particles from four 3D classes (bottom row of four maps). This revealed that a 20 bp duplex arm goes in from either end of the clamp resulting in the consensus DNA density looking longer. In the 1st class, the DNA duplex reaches into the clamp from the FANCI side, and in the 2nd class from the FANCD2 side. The 3rd class appears to have mixed DNA registers with some bulbous density at the FANCD2 side and flat density at the FANCI side (the 4th class is devoid of DNA). This suggests that the ICL DNA has no preferred orientation in binding to IDUb. b, Cryo-EM reconstruction of IDUb bound to 5-flap DNA containing two 29 bp duplexes flanking the flap (same DNA as the non-ubiquitinated complex of Extended Data Fig. 5b). The top and middle density are the consensus maps with and without temperature-factor sharpening, respectively, colored as in a. The model shown is the IDUb-nicked DNA structure that was fit as a single body into the map. We did not refine the coordinates for this reconstruction, yet the DNA density overlaps well with the nicked DNA model. Unlike the ICL DNA complex, 3D classification failed to identify unique DNA conformations, and there was no evidence for any ssDNA branch (not shown). This suggests that either the flap is past the 29 bp duplex ends, or it can be accommodated within the clamp but not in a specific register that can be identified by 3D classification. c, Cryo-EM reconstruction of IDUb bound to 58 bp dsDNA, shown as in b. d, Gold-standard FSC plots for the consensus reconstructions of IDUb bound to the other DNA molecules discussed in text. The blue curve is the 3.8 Å reconstruction from 98,750 particles of IDUb bound to 5’ flap DNA, the red curve is the 3.8 Å reconstruction from 85,078 particles of IDUb bound to dsDNA, and the green curve is the 4.4 Å reconstruction from 28,519 particles of IDUb bound to ICL-DNA. e, Flow chart of single particle cryo-EM data processing for the ID-/Ub complex ubiquitinated only on FANCD2 bound to nicked DNA. Consensus (top) and focused maps, temperature-factor sharpened and masked, are colored by local resolution estimated with the RELION3 postprocess program. The resolution range is mapped to the colors in the inset next to each map. f, Graph shows gold-standard FSC plots between two independently refined half-maps for the consensus reconstruction (blue curve) and the three focused maps. The FSC curve for the final model versus the composite map is shown in black. Dashed line marks the FSC cutoff of 0.143. g-h, The singly mono-ubiquitinated ID-/Ub structure is overall very similar to IDUb, and the two superimpose with a 0.57 Å r.m.s.d. in the positions of 2,429 common Cα atoms. There is only a small shift in the FANCD2 segment that interacts with the FANCI ubiquitin (UbI) as shown in the close-up view of h, where the red arrows indicate the ~1.5 Å local motion the FANCD2 helical repeats undergo when FANCI is also ubiquitinated.
Extended Data Figure 9 |. Ubiquitination induces FANCD2 conformational changes associated with alternative FANCI-FANCD2 contacts; DNA binding activity of IDUb.
a, After ubiquitination, FANCI does not change significantly (entire molecule can be superimposed with a 1.6 Å r.m.s.d in 1132 Cα positions), but FANCD2 undergoes two changes. FANCD2 from the ID complex (salmon) is superimposed on that of IDUb (pink) by aligning the 2nd half of their NTDs (residues 255–587; 1.8 Å r.m.s.d. for 308 Cα positions), a segment that changes little on ubiquitination. UbD2 that is covalently attached to FANCD2 is in red, and the UbI with which it packs in green. The N-terminal portion of the NTD, which rotates by 38° towards FANCI as a mostly rigid body (residues 45 to 254, 0.68 Å r.m.s.d. for 202 Cα) is approximately marked by a bracket. This rotation at the center of the UbI binding site allows FANCD2 to better embrace UbI, and also to interact with FANCI (NTD-HD junction, residues 529 to 593), the latter involving similar residues on FANCD2 but mostly different ones on FANCI. Similarly marked is the HD domain (residues 604–928) that rotates relative to the NTD by 15°. Additional tilting of helices within the HD domain results in the CTD that follows (residues 929 to C-termini also marked) being rotated by 20° degrees relative to the invariant portion of the NTD. b-e, The FANCI and FANCD2 ubiquitin binding structural elements are distinct from commonly occurring ubiquitin-binding domains. Superposition of the UbI (green) bound to FANCD2Ub (pink) on the ubiquitin (orange) bound to the dimeric Vps29 CUE domain (blue) from PDBID 1P3Q in b, to the Cbl-b UBA domain (PDBID 2OOP) in c, to the UBZ domain of Faap20 (PDBID 3WWQ) in d, and to the UIM domain of Vps27 (PDBID 1Q0W) in e. The ubiquitin hydrophobic patch residues (Leu8, Ile44, and Val70) are shown in stick representation for both ubiquitin molecules in each figure. The blue sphere in d is the Zn atom of the UBZ domain. Orientation similar to that of Figure 4a. It has been suggested that FANCD2 shares sequence homology with the CUE domain34. While the 47-residue region of proposed homology (residues 191–237) partially overlaps the Ub-binding site, its structure is unrelated to the CUE domain, and Ub binding by FANCD2 is distinct. f, Molecular surface colored according to electrostatic potential calculated with PYMOL in the absence of DNA (colored −5 to +5 kT blue to red), in the same view as Figure 4d and with structural elements and surfaces above the DNA similarly clipped to reveal the DNA. As with the ID-ICL DNA complex, the FANCIUb groove partially encircles the DNA through basic and polar residues from α33b and α36b on one side of the groove and α40, α42 on the other. Near the middle of the DNA, however, the extension of α48 that results from coiling with FANCD2Ub α50 (Fig. 3e) gives rise to a new semicircular basic groove, between α46 and α48 on one side and the NTD α19b and α20 on the other, into which dsDNA binds (Fig. 4d). This is associated with one of the two DNA bends, which redirects the duplex away from clashing with the new position of FANCD2. Thereafter, the second bend occurs as the dsDNA is redirected by the FANCD2 CTD, which uses the localized patch of α48 and α50 to bind to the duplex analogously to the ID-ICL DNA complex. The side chains near the DNA, shown in Figure 4d as sticks, are from FANCIUb residues Lys291 (α15), Lys397 (α19b), Ser411 (α20), Lys793, Thr794 on α33b, Lys898 (α36b), Lys980 (α40), Lys1026 (α42), Lys1164 (α46), and Thr1238, Arg1242, Arg1245 and Lys1248 on the extended α48; and from FANCD2Ub residues His1288, His1292, and Arg1299 on α48, and Thr1351, Arg1352 and Gln1355 on α50. The sites of DNA bending, of 26° and 31°, are centered on the 11th and 21st base pairs from the FANCI end, respectively, with large roll values over three base-pair steps. g, The IDUb Kd values for dsDNA, nicked, 5’ flap, ICL and fork DNA vary by less than a factor of two, with dsDNA and nicked DNA exhibiting slightly tighter binding. EMSA of the equimolar mixture of the mono-ubiquitinated FANCIUb and FANCD2Ub, each at the indicated concentrations, binding to the 32P-labeled DNA substrates (0.5 nM) shown schematically. The plots with a logarithmic X-axis show fraction bound in at least three repetitions of each experiment (different color and shape markers) and their mean value (black dash). As with the non-ubiquitinated complex, the binding isotherms fit a Hill slope model best. A binding curve (black line) simulated with the indicated Kd and Hill coefficient (ηH) values is shown on each plot.
Extended Data Table 1 |.
Cryo-EM data collection, refinement and validation statistics.
| ID–ICL DNA (EMDB-21134) (PDB-6VAA) | IDUb–DNA (EMD-21138) (PDB-6VAE) | ID−Ub–DNA (EMD-21139) (PDB-6VAF) | apoID (EMDB-21137) (PDB-6VAD) | |
|---|---|---|---|---|
| Data collection and processing | ||||
| Magnification | 45,914 | 45,914 | 45,914 | 45,914 |
| Voltage (kV) | 300 | 300 | 300 | 300 |
| Electron exposure (e−/Å2) | 65.6 | 65.6 | 65.6 | 65.6 |
| Defocus range (urn) | 1.0–3.2 | 1.0–3.2 | 1.0–3.2 | 1.0–3.2 |
| Pixel size (Å) | 1.090 | 1.089 | 1.089 | 1.090 |
| Symmetry imposed | C1 | C1 | C1 | C1 |
| Initial partiC1e images (no.) | 483,214 | 301,058 | 57,993 | 483,214 |
| Final partiC1e images (no.) | 231,943 | 301,058 | 57,993 | 251,271 |
| Map resolution (Å) | ||||
| Consensus reconstruction | 3.4 | 3.6 | 3.9 | 3.3 |
| Focus 1 reconstruction | 3.3 | 3.5 | 3.9 | 3.2 |
| Focus 2 reconstruction | 3.8 | 3.4 | 3.7 | 3.9 |
| Focus 3 reconstruction | - | 3.5 | 4.0 | - |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | ||||
| Consensus reconstruction | 3.3–8.0 | 3.5–6.6 | 3.8–7.9 | 3.2–6.8 |
| Focus 1 reconstruction | 3.2–5.5 | 3.5–5.1 | 3.8–7.4 | 3.2–5.1 |
| Focus 2 reconstruction | 3.6–6.7 | 3.4–5.3 | 3.7–6.8 | 3.6–5.8 |
| Focus 3 reconstruction | - | 3.4–5.0 | 3.8–6.6 | - |
| Refinement | ||||
| Initial model used (PDB code) | 3S4W | - | - | - |
| Model resolution (Å) | 3.4 | 3.5 | 3.9 | 3.4 |
| FSC threshold | 0.59 | 0.62 | 0.63 | 0.69 |
| Model resolution range (Å) | 173.3–3.4 | 30.0–3.5 | 30.0–3.9 | 40.0–3.4 |
| Map sharpening B factor (Å2) | 101–150 | 103–114 | 84–113 | 95–229 |
| Model composition | ||||
| Non-hydrogen atoms | 20,055 | 21,167 | 20,566 | 18,517 |
| Protein residues | 2,321 | 2,505 | 2,429 | 2,321 |
| DNA residues | 75 | 58 | 58 | - |
| B factors (Å2) | ||||
| Protein | 169.4 | 149.6 | 195.3 | 196.20 |
| DNA | 292.3 | 226.2 | 290.6 | - |
| R.m.s. deviations | ||||
| Bond lengths (Å) | 0.011 | 0.010 | 0.009 | 0.012 |
| Bond angles (°) | 1.42 | 1.42 | 1.66 | 1.61 |
| B factors main chain (Å2) | 2.9 | 2.6 | 3.2 | 4.4 |
| B factors side chain (Å2) | 0.8 | 3.6 | 3.1 | 3.6 |
| Validation | ||||
| MolProbity score | 1.62 | 1.51 | 1.71 | 1.70 |
| Clashscore | 4.35 | 3.79 | 3.90 | 4.69 |
| Poor rotamers (%) | 0.75 | 0.87 | 0.54 | 1.13 |
| Ramachandran plot | ||||
| Favored (%) | 93.97 | 95.08 | 90.73 | 93.57 |
| Allowed (%) | 5.99 | 4.84 | 8.89 | 6.21 |
| Disallowed (%) | 0.04 | 0.08 | 0.38 | 0.22 |
| Rwork (%) | 34.3 | 30.2 | 32.8 | 30.3 |
| Average FSC | 0.84 | 0.85 | 0.83 | 0.87 |
Supplementary Material
Acknowledgments
We thank the staff of the MSKCC Cryo-EM facility, the NYSBC Simons Electron Microscopy Center, and the HHMI Cryo-EM facility for help with data collection. Supported by HHMI and National Institutes of Health grant CA008748.
Footnotes
Author Information The authors declare no competing financial or non-financial interests.
Data availability. The ID-ICL DNA, IDUb-DNA, ID-/Ub-DNA and apoID coordinates, corresponding cryo-EM maps, including the focused reconstructions and the composite map used in refinement, have been deposited with the Protein Data Bank and the Electron Microscopy Data Bank under accession codes PDB-6VAA and EMDB-21134, PDB-6VAE and EMDB-21138, PDB-6VAF and EMDB-21139, PDB-6VAD and EMDB-21137, respectively.
References
- 1.Ceccaldi R, Sarangi P & D’Andrea AD The Fanconi anaemia pathway: new players and new functions. Nature reviews. Molecular cell biology 17, 337–349, doi: 10.1038/nrm.2016.48 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Zhang J et al. DNA interstrand cross-link repair requires replication-fork convergence. Nature structural & molecular biology 22, 242–247, doi: 10.1038/nsmb.2956 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boisvert RA & Howlett NG The Fanconi anemia ID2 complex: dueling saxes at the crossroads. Cell cycle 13, 2999–3015, doi: 10.4161/15384101.2014.956475 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park WH et al. Direct DNA binding activity of the Fanconi anemia D2 protein. J Biol Chem 280, 23593–23598, doi:M503730200 [pii] 0.1074/jbc.M503730200 (2005). [DOI] [PubMed] [Google Scholar]
- 5.Yuan F, El Hokayem J, Zhou W & Zhang Y FANCI protein binds to DNA and interacts with FANCD2 to recognize branched structures. J Biol Chem 284, 24443–24452, doi:M109.016006 [pii] 10.1074/jbc.M109.016006 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Longerich S, San Filippo J, Liu D & Sung P FANCI binds branched DNA and is monoubiquitinated by UBE2T-FANCL. J Biol Chem 284, 23182–23186, doi:C109.038075 [pii] 10.1074/jbc.C109.038075 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joo W et al. Structure of the FANCI-FANCD2 complex: insights into the Fanconi anemia DNA repair pathway. Science 333, 312–316, doi: 10.1126/science.1205805 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Raschle M et al. Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134, 969–980, doi:S0092–8674(08)01075–1 [pii] 10.1016/j.cell.2008.08.030 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang R et al. DNA repair. Mechanism of DNA interstrand cross-link processing by repair nuclease FAN1. Science 346, 1127–1130, doi: 10.1126/science.1258973 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zivanov J et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, doi: 10.7554/eLife.42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakane T, Kimanius D, Lindahl E & Scheres SH Characterisation of molecular motions in cryo-EM single-particle data by multi-body refinement in RELION. eLife 7, doi: 10.7554/eLife.36861 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brown A et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr 71, 136–153, doi: 10.1107/S1399004714021683 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Howlett NG, Taniguchi T, Durkin SG, D’Andrea AD & Glover TW The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Human molecular genetics 14, 693–701, doi: 10.1093/hmg/ddi065 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Smogorzewska A et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129, 289–301, doi:S0092–8674(07)00320–0 [pii] 10.1016/j.cell.2007.03.009 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sirbu BM et al. Identification of proteins at active, stalled, and collapsed replication forks using isolation of proteins on nascent DNA (iPOND) coupled with mass spectrometry. J Biol Chem 288, 31458–31467, doi: 10.1074/jbc.M113.511337 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yates M & Marechal A Ubiquitylation at the Fork: Making and Breaking Chains to Complete DNA Replication. International journal of molecular sciences 19, doi: 10.3390/ijms19102909 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aymami J et al. Molecular structure of nicked DNA: a substrate for DNA repair enzymes. Proceedings of the National Academy of Sciences of the United States of America 87, 2526–2530, doi: 10.1073/pnas.87.7.2526 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ho PS Structure of the Holliday junction: applications beyond recombination. Biochemical Society transactions 45, 1149–1158, doi: 10.1042/BST20170048 (2017). [DOI] [PubMed] [Google Scholar]
- 19.Sato K, Toda K, Ishiai M, Takata M & Kurumizaka H DNA robustly stimulates FANCD2 monoubiquitylation in the complex with FANCI. Nucleic Acids Res 40, 4553–4561, doi: 10.1093/nar/gks053 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Longerich S et al. Regulation of FANCD2 and FANCI monoubiquitination by their interaction and by DNA. Nucleic Acids Res 42, 5657–5670, doi: 10.1093/nar/gku198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rajendra E et al. The genetic and biochemical basis of FANCD2 monoubiquitination. Molecular cell 54, 858–869, doi: 10.1016/j.molcel.2014.05.001 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colnaghi L et al. Patient-derived C-terminal mutation of FANCI causes protein mislocalization and reveals putative EDGE motif function in DNA repair. Blood 117, 2247–2256, doi: 10.1182/blood-2010-07-295758 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Husnjak K & Dikic I Ubiquitin-binding proteins: decoders of ubiquitin-mediated cellular functions. Annual review of biochemistry 81, 291–322, doi: 10.1146/annurev-biochem-051810-094654 (2012). [DOI] [PubMed] [Google Scholar]
- 24.Yamamoto KN et al. Involvement of SLX4 in interstrand cross-link repair is regulated by the Fanconi anemia pathway. Proceedings of the National Academy of Sciences of the United States of America 108, 6492–6496, doi:1018487108 [pii] 10.1073/pnas.1018487108 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lachaud C et al. Distinct functional roles for the two SLX4 ubiquitin-binding UBZ domains mutated in Fanconi anemia. Journal of cell science 127, 2811–2817, doi: 10.1242/jcs.146167 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montes de Oca R et al. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood 105, 1003–1009, doi: 10.1182/blood-2003-11-3997 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Ishiai M et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nature structural & molecular biology 15, 1138–1146, doi: 10.1038/nsmb.1504 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zheng SQ et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature methods 14, 331–332, doi: 10.1038/nmeth.4193 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rohou A & Grigorieff N CTFFIND4: Fast and accurate defocus estimation from electron micrographs. Journal of structural biology 192, 216–221, doi: 10.1016/j.jsb.2015.08.008 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zivanov J, Nakane T & Scheres SHW A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ 6, 5–17, doi: 10.1107/S205225251801463X (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Afonine PV et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr 68, 352–367, doi: 10.1107/S0907444912001308 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Extended Data References
- 32.Joo W et al. Structure of the FANCI-FANCD2 complex: insights into the Fanconi anemia DNA repair pathway. Science 333, 312–316, doi: 10.1126/science.1205805 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ishiai M et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nature structural & molecular biology 15, 1138–1146, doi: 10.1038/nsmb.1504 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rego MA, Kolling F. W. t., Vuono EA, Mauro M & Howlett NG Regulation of the Fanconi anemia pathway by a CUE ubiquitin-binding domain in the FANCD2 protein. Blood 120, 2109–2117, doi: 10.1182/blood-2012-02-410472 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.