Abstract
Over 50% of patients with 22q11.2 deletion syndrome (DS) have a conotruncal or related cardiac defect (CTRD). We hypothesized that similar genetic variants, developmental pathways and biological functions, contribute to disease risk for CTRD in patients without a 22q11.2 deletion (ND-CTRD) and with a 22q11.2 deletion (DS-CTRD). To test this hypothesis, we performed rare CNV (rCNV)-based analyses on 630 ND-CTRD cases and 602 DS-CTRD cases with comparable cardiac lesions separately and jointly. First, we detected a collection of heart development related pathways from Gene Ontology and Mammalian Phenotype Ontology analysis. We then constructed gene regulation networks using unique genes collected from the rCNVs found in the ND-CTRD and DS-CTRD cohorts. These gene networks were clustered and their predicted functions were examined. We further investigated expression patterns of those unique genes using publicly available mouse embryo microarray expression data from single-cell embryos to fully developed hearts. By these bioinformatics approaches, we identified a commonly shared gene expression pattern in both the ND-CTRD and DS-CTRD cohorts. Computational analysis of gene functions characterized with this expression pattern revealed a collection of significantly enriched terms related to cardiovascular development. By our combined analysis of rCNVs in the ND-CTRD and DS-CTRD cohorts, a group of statistically significant shared pathways, biological functions, and gene expression patterns were identified that can be tested in future studies for their biological relevance.
Keywords: 22q11.2 deletion syndrome, conotruncal or related cardiac defect, functional analysis, gene interaction networks, mouse heart gene expression, pathway analysis, rare CNV
1 |. INTRODUCTION
Congenital heart defects (CHDs) are the leading cause of birth defect-related deaths in newborns and are estimated to affect 35,000 live births each year in the United States (Hoffman & Kaplan, 2002; Reller, Strickland, Riehle-Colarusso, Mahle, & Correa, 2008). The spectrum of defects varies from simple to complex heart malformations. They can be diagnosed as an isolated finding or as part of a collection of findings, such as in the 22q11.2 deletion syndrome (22q11.2DS). The etiology of CHDs is complex, resulting from both genetic and epigenetic factors. In addition to known pathogenic CNVs such as the 22q11.2 deletion, numerous studies report putatively disease-related, rare CNVs (rCNVs) in approximately 10% of patients with CHDs (Andersen, Troelsen, & Larsen, 2014; Lalani & Belmont, 2014). While most studies reported a similar prevalence of de novo and/or inherited rCNVs within the CHD population, with rare exception, each study reports enrichment of different putative-disease-related rCNVs (Soemedi et al., 2012; Xie et al., 2017). Given the limited study cohort size in each report and the observed heterogeneity of rCNVs, the mechanisms by which these rCNVs might increase risk of disease is challenging for any single study to identify and not necessarily replicated by another. For example, in our recent studies on cases with CHDs without a recognized genetic cause, we observed a significantly increased burden of rCNVs among patients with CHDs (White et al., 2014; Xie et al., 2017). With rare exception, our particular list of individual CNVs demonstrated little overlap with those in other reports. It is unclear how rCNVs or their gene content influences the etiology of CHDs in spite of extensive analysis. Moreover, it is unclear whether the same CNVs exert an influence on the risk of CHDs in the patient with or without a recognizable genomic alternation.
The 22q11.2DS (velo-cardio-facial syndrome; DiGeorge syndrome, VCFS/DGS; MIM #192430; 188400) is the most common rCNV syndrome. It affects approximately one in 2,000–4,000 newborns each year (McDonald-McGinn et al., 2015). The vast majority of patients with 22q11.2DS carry the typical 3 million base pair (3 Mb) deletion located between low copy repeats A-D in the 22q11.2 region. Phenotypic findings in patients with 22q11.2DS are highly variable including CTRDs, which are reported in 60–75% of patients. We refer to the 22q11.2DS cases with CTRDs as DS-CTRDs. The cardiac phenotypes range from major intracardiac malformations such as tetralogy of Fallot to minor aortic arch anomalies, such as an isolated right-sided aortic arch or abnormal origin of the subclavian arteries. The etiology of CTRD phenotypic variability is currently unknown. It has been suggested that common and rare CNVs influence the risk for CTRDs (Mlynarski et al., 2015, 2016). Our recent studies suggested that a duplication of a commonly occurring CNV harboring the glucose transporter gene, SLC2A3, (solute carrier family 2 member 3), may serve as a genetic modifier in approximately 8–10% of patients with 22q11.2DS (Mlynarski et al., 2015). When we examined rCNVs in the same cohort, we did not identify any genes, networks, or functions significantly enriched within DS-CTRD cases compared with 22q11.2DS patients without CTRD, even though functions such as the WNT pathway were suggested.
Separately, we also previously studied a distinct CTRD patient cohort without 22q11.2DS for rCNVs. We refer to this cohort of cases as ND-CTRD. A burden of rCNVs was detected in ND-CTRD. Functional and pathway analyses revealed enrichment of terms involved in heart development. We were interested to test whether the rCNVs, functional pathways, or gene networks were shared between the CTRDs cases without a 22q11.2 deletion (ND-CTRD) or with a 22q11.2 deletion (DS-CTRD).
In the present study, we used the existing ND-CTRD and DS-CTRD cohorts to test the hypothesis that genes or gene networks associated with the CTRD phenotype were shared. Given their comparable cardiac phenotypes, and the possibility that the same genetic factors could contribute to disease risk for CTRD, we combined the data from the two cohorts in order to enhance our power to detect any likely etiologic disease-associated genes and/or functional networks. We first confirmed the putative pathogenic rCNVs in each of the CTRD cohorts. We then used the combined gene-lists within rCNVs in both cohorts to identify shared gene-based features (pathways, functions, and gene regulation networks) that were statistically enriched.
2 |. METHODS
2.1 |. Study cohorts and array genotyping
The Children’s Hospital of Philadelphia (CHOP) Institutional Review Board approved this study. Subjects were diagnosed with CHDs in a clinical setting in a uniform manner at CHOP’s Cardiac Center (ND-CTRD; cohorts 1 and 2) (Xie et al., 2017). Subjects with clinically defined genetic syndromes were excluded from the ND-CTRD cohort. Subjects with 22q11.2DS were identified within the 22q and You Center at CHOP and other centers (DS-CTRD) (Mlynarski et al., 2016; Xie et al., 2017). Reports from echocardiograms, cardiac catheterizations, cardiac magnetic resonance imaging, and cardiac operative notes were reviewed to record cardiac and aortic arch anomalies. Parents were recruited when available and when they agreed to consent for this study.
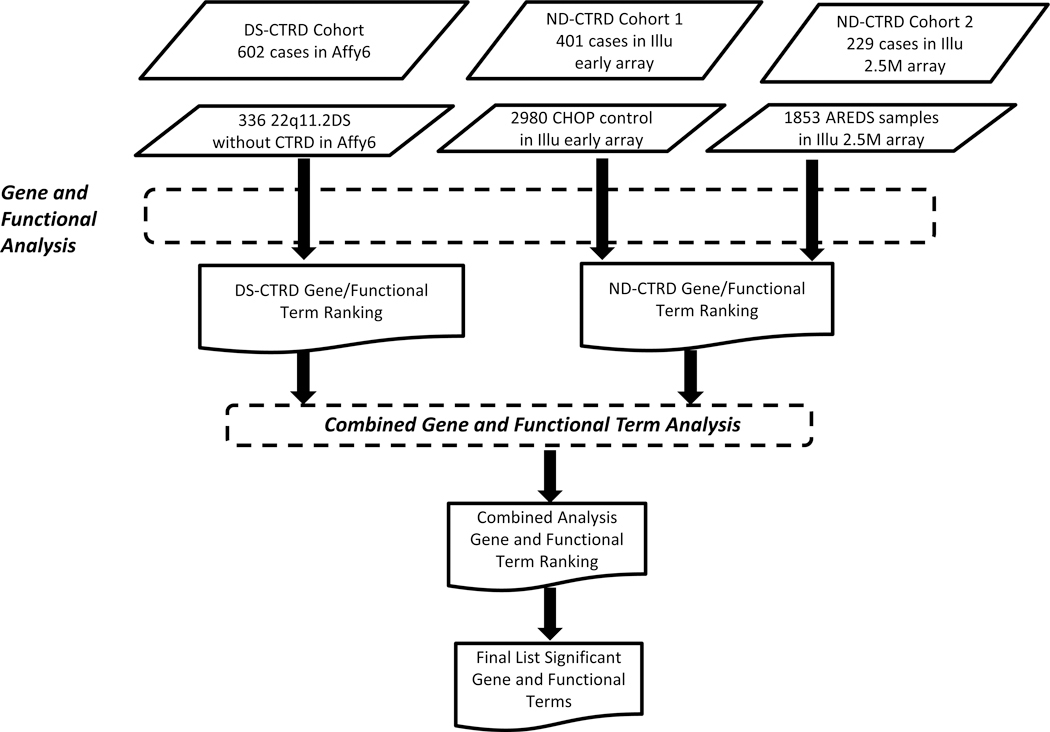
The overall work flow and details about each study cohort are described in Figure 1. A detailed description of the ND-CTRD cohort’s patient cardiac phenotypic information, genotyping procedures, and the quality control procedure and metrics used in vetting the cohort have been described previously (Xie et al., 2017). All cases were screened for a 22q11.2 deletion by fluorescence in situ hybridization, MLPA or microarray analysis. Additionally, we only selected ND-CTRD patients carrying CTRDs typically present in the 22q11.2DS cohort, namely, tetralogy of Fallot, truncus arteriosus, interrupted aortic arch type B, ventricular septal defects (specifically conoventricular, posterior malalignment, and conoseptal hypoplasia types), and isolated aortic arch anomalies that we refer to as CTRDs. We used a previously described healthy population as controls for the ND-CTRD cohort (Xie et al., 2017).
FIGURE 1.
Flow chart outlining process of data analysis
We grouped our ND-CTRD cases and controls into two mutually exclusive cohorts as described in (Xie et al., 2017). ND-CTRD cohort 1 included all cases and controls (Healthy_CHOP) genotyped using the early Illumina array technologies (Illumina Infinium™ II HumanHap550 v1, Illumina Infinium™ II HumanHap550 v3, or Illumina BeadChip 610 array). We corrected differences in SNP probe content among all three SNP array versions used in cohort 1 by limiting our analysis to the subset of SNPs shared by all three genotyping arrays (535, 606 SNPs) as described before (Xie et al., 2017). ND-CTRD cohort 2 included cases (Illumina HumanOmni2.5–8v1) and Healthy_AREDS samples (Illumina HumanOmni2.5–4) genotyped on later arrays. For the 2.5M arrays, the subset of common SNPs (n = 2,332,518) between the two platforms was used to predict CNV regions in genotyped samples as described in Xie et al. (2017). The two cohorts were evaluated separately and then aggregately.
Description of phenotypes in DS-CTRD subjects and the distribution of 22q11.2 deletion sizes were previously reported (Mlynarski et al., 2015). 22q11.2 deletions of DS-CTRD subjects were confirmed by fluorescence in situ hybridization or multiplex ligation-dependent probe amplification (MLPA). The genotyping procedure and quality control criteria of those DS-CTRD subjects were as previously described in Mlynarski et al. (2015, 2016).
2.2 |. CNV detection and quality control
CNV detection and quality control methods have been described previously (Mlynarski et al., 2015; Xie et al., 2017). In brief, both CNV Workshop (Gai et al., 2010) and PennCNV (Wang et al., 2007) were used to define CNV regions. To reduce type I error of CNV detection, based on our previous effort detecting and validating CNVs (Mlynarski et al., 2015; Xie et al., 2017), deletions spanning <5 consecutive SNPs and duplications with <10 consecutive SNPs in the ND-CTRD cohort 1 were excluded. Due to higher probe density, density in the Illumina 2.5M array and Affymetrix 1.8M array, which contain 3–4 times more SNPs across the human genome as compared with earlier Illumina arrays, higher thresholds for ND-CTRD cohort 2 and DS-CTRD were used to remove deletions spanning <10 SNPs and duplications spanning <20 SNPs. In the ND-CTRD cohorts, deletions spanning <10 Kbps and duplications spanning <20 Kbps were removed. DS-CTRD cases with deletions nested within the typical 3 Mbps 22q11.2 deletion were also removed from any further analysis as we only examined CNVs outside the typical 22q11.2DS deletion region in the DS-CTRD cohort as described in Mlynarski et al. (2015, 2016).
Additional exclusion criteria included CNVs with >50% overlap with centromere, telomere, and immunoglobulin variable regions and CNVs with SNP densities <1 SNP/30 Kbps as described in Hasin et al. (2008), Hellemans, Mortier, De Paepe, Speleman, and Vandesompele (2007), and Young et al. (2008). All olfactory receptor genes were removed from further analysis. Samples detected with a total CNV burden ≥3 standard deviations from the cohort mean were removed (Pankratz et al., 2011).
Large CNVs were defined as those whose length fell within the 20th vigintile of CNVs observed in the corresponding control cohorts. Predicted CNVs were annotated using the coordinates of RefSeq genes and their corresponding official gene symbol, as represented in the UCSC Genome/Table Browser (genome.ucsc.edu).
2.3 |. Rare CNV definition
CNVs were considered as equivalent if CNVs were the same type (deletions or duplications) and their genomic regions reciprocally overlapped for greater than 60% of their length as described in Mlynarski et al. (2016), White et al. (2014), and Xie et al. (2017)). To minimize the artifacts introduced by different genotyping technologies, we compared CNVs detected in our cases directly with CNVs detected in matched controls using the same or similar genotyping arrays to estimate case CNV occurrence frequency in controls. We adopted our previous definition of rare CNVs (rCNV) for the ND-CTRD as those CNVs being observed in no more than one healthy control subject (estimated as <0.1% in healthy CHOP controls as well as within Healthy_AREDS), and defined unique CNVs as those not observed in the control cohort (White et al., 2014; Xie et al., 2017). Similarly, we adopted our previous definition of rCNVs for the DS-CTRD cohort as CNVs found in <0.1% of a previously published control population, and defined unique CNVs as those not observed in this previously published control population (dbVar accession nstd100; Coe et al., 2014) as described in Mlynarski et al. (2016).
2.4 |. Gene and functional analysis
The significance and details of analytical approaches to identify genes and their known function in CNVs has been described previously (Elia et al., 2010; Silversides et al., 2012; Xie et al., 2017). As copy number variation of amplification and deletion events may impact phenotypic outcome through different mechanisms, they were considered both separately and aggregately at each locus for global CNV and gene analyses. For the DS-CTRD cohorts, we disregarded the gene content within the hemizygous 22q11.2 deletion from all DS-CTRD cases and controls as they all carry the same deletion. We associated gene content of rCNVs with gene ontology (GO) terms and Mammalian Phenotype Ontology. GO annotations were downloaded from Ensembl. org (huseast.ensembl.org/index.html) using the BioMart data-mining tool. Mammalian Phenotype Ontology (MPO) term annotations were obtained from the Mammalian Genome Informatics resource (MGI) (www.informatics.jax.org). The GO and MPO terms were expanded as described previously (Xie et al., 2017). For each functional term (GO and MPO), we directly compared the frequency of occurrence between cases and controls using Fisher’s exact test (two-tailed). The Benjamini–Hochberg False Discovery Rate (BH-FDR) method was used on all gene or functional terms evaluated to reduce family-wise type I error. We only reported functions if their nominal p values were <.05 in the combined ND-CTRD cohort (i.e., merging the ND-CTRD cohort 1 and ND-CTRD cohort 2) as well as the DS-CTRD when each cohort was considered individually and the False Discovery Rates (FDR) were <0.05 when evaluated in the combined CTRD cohorts (i.e., combined ND-CTRD cohort 1, ND-CTRD cohort 2, and the DS-CTRD cohort).
2.5 |. Gene network construction
We used the ReactomeFIViz application (version 6.1) within Cytoscape (version 3.6) (f1000research.com/articles/3-146/v2) (Wu, Dawson, Duong, Haw, & Stein, 2014) to construct a network among genes uniquely detected in rCNVs in cases from the ND-CTRD cohorts and the DS-CTRD cohort as compared with their respective controls, separately and aggregately, as previously described (Mlynarski et al., 2016; Xie et al., 2017). We applied the ReactomeFIViz “Gene Set / Mutation Analysis” function following the protocol described in the ReactomeFIViz user guide with all default parameters. The resulting gene interaction networks were grouped into different “modules” using ReactomeFIViz’s built-in “cluster FI network” function. Within each module, the “Analyze module functions” tool was used to measure pathway enrichment. Only pathways with FDR <0.05 were reported.
2.6 |. Mouse embryo homolog gene expression analysis
A total of 74 Affymetrix Mouse Genome 430 2.0 expression arrays from nine studies were downloaded through NCBI/GEO (https://www.ncbi.nlm.nih.gov/geo/): C1_EB: single-cell embryo; C2_EB: two-cell embryo (Vassena, Han, Gao, & Latham, 2007); E1.5_EB: E1.5 embryo; E2.5_EB: E2.5 embryo; E3.5_EB: E3.5 embryo (Maekawa, Yamamoto, Kohno, Takeichi, & Nishida, 2007); E9.5_AVC: E9.5 atrioventricular canal (Rivera-Feliciano et al., 2006); E10.5_WH: E10.5 whole heart; E11.5_WH: E11.5 whole heart; E12.5_Vc: E12.5 ventricles; E12.5_ACH: E12.5 atrial chamber of heart; E13.5_Vc: E13.5 ventricles; E13.5_ACH: E13.5 atrial chamber of heart; E14.5_Vc: E14.5 ventricles; E14.5_ACH: E14.5 atrial chamber of heart; E16.5_Vc: E16.5 ventricles; E16.5_ACH: E16.5 atrial chamber of heart; E18.5_Vc: E18.5 ventricles; E18.5_ACH: E18.5 atrial chamber of heart (Schinke, Jay, Brown, & Izumo, 2004); E17.5_Mc: E17.5 myocardium (Trivedi et al., 2007); PN_WH: Post Natal whole heart (Dufour et al., 2007; Muchir et al., 2007; Zhao et al., 2007); PN_Vc: Post Natal ventricle (Bisping et al., 2006). Detailed description for each study is listed in Supporting Information Table S1.
Gene expression data were normalized with respect to expression at the single cell stage (C1_EB) using the Robust Multi-array Average method (RMA, R 3.21, Library Affy_1.55 [Irizarry et al., 2003]). Differentially expressed genes were identified by using ANOVA. Hierarchical clustering on differentially expressed genes was performed as previously described (Eisen et al, 1998). A re-clustering process was then applied to remove weakly associated genes and filter out small clusters. We first calculated the cluster centroid (median expression level of all genes in the cluster) for each cluster, and then computed the correlation coefficient of each gene to every cluster centroid. A gene was assigned to a cluster if its correlation coefficient to the cluster was >0.5 and the correlation coefficient to any other cluster was at least 0.2 lower. This re-clustering procedure was repeated 50 times unless the re-clustering process converged earlier. Finally, any clusters with sizes below twice the standard deviation of the average cluster size were excluded. Gene Set Enrichment Analysis (GESA) (Subramanian et al., 2005) was used to investigate the gene functions and pathways (MSigDB v5.0) (Subramanian et al., 2005) enriched within each cluster. The BH-FDR method was applied to the p values from the GSEA procedure to reduce multiple testing type I error.
2.7 |. Statistical testing
Two-tailed Fisher’s exact tests, as appropriate, were used to test significance in CNV and gene enrichment analyses comparing cases with controls. The BH-FDR method was applied to adjust family-wise multiple testing errors. All statistical analyses were processed using R (version 3.4)
3 |. RESULTS
3.1 |. Study cohort
The total number of subjects in the ND-CTRD and DS-CTRD cohorts is detailed in Figure 1 and Table 1. Briefly, a total of 630 cases were identified with a definitive diagnosis of CTRDs without a diagnosis of a known genetic condition upon review of medical records and without a 22q11.2DS deletion. They comprise the two ND-CTRD cohorts that include 401 cases in ND-CTRD cohort 1 and 229 cases in ND-CTRD cohort 2. In addition, cohort 1 consists of 227 trios, 130 duos (one of the parents and the proband), and 44 probands only. A total of 602 cases with both typical 22q11.2 deletions and CTRD diagnoses were deemed as cases for the DS-CTRD cohort as described in Mlynarski et al. (2016)).
TABLE 1.
CTRD cohorts description
| Cohort name | Abbreviation | Number of cases | Number of controls | Genotyping array | References |
|---|---|---|---|---|---|
| Conotruncal and related defects, without a 22q11.2 deletion | ND-CTRD 1 | 407 | 2,980 | Illumina 550, 610 | Xie et al. (2017) |
| ND-CTRD 2 | 229 | 1853 | Illumina 2.5M | ||
| Conotruncal and related defects, with 22q11.2 deletion syndrome | DS-CTRD | 602 | 336 | Affymetrix 6.0 | Mlynarski et al. (2016) |
A total of 4,833 healthy subjects served as controls for the ND-CTRD cohorts including 2,980 healthy controls for the ND-CTRD cohort 1 and 1,853 different healthy controls for the ND-CTRD cohort 2 (Xie et al., 2017). These subjects passed our quality control measures described in section 2. A total of 336 subjects carrying a typical 22q11.2DS deletion without an intracardiac congenital malformation were used as DS-CTRD study controls.
3.2 |. CNV burden in patient cohorts
CNVs detected in the two ND-CTRD cohorts and the DS-CTRD cohort as well as their gene annotations are listed in Supporting Information Table S2 (S2a_ND-CTRD COHORT 1, S2b_ND-CTRD COHORT 2, and S2c_DS-CTRD). For each CNV, frequencies in control populations were also measured and presented (Supporting Information Tables S2a–c). The overview of total CNVs and rCNVs detected in each cohort is detailed in Table 2.
TABLE 2.
Rare CNV burden among different CTRD cohorts
| Rarea |
|||||||
|---|---|---|---|---|---|---|---|
| CNV type | Count | Count | CNV burdenb | Case/control CNV burden odds ratio | Significance (sample count-based) | Significance (CNV count-based) | |
| ND-CTRD cohort 1 | Duplications | 341 | 170 | 0.42 | 1.58 | 2.35E-06 | 4.98E-09 |
| Deletions | 1,392 | 366 | 0.91 | 1.50 | 1.12E-05 | 1.59E-16 | |
| All CNVs | 1,733 | 536 | 1.34 | 1.52 | 1.18E-07 | 2.05E-24 | |
| Large CNVs | |||||||
| Duplications | 61 | 45 | 0.11 | 1.56 | 1.80E-02 | 1.15E-04 | |
| Deletions | 33 | 25 | 0.06 | 2.14 | 2.38E-03 | 7.16E-03 | |
| All CNVs | 94 | 70 | 0.17 | 1.73 | 5.23E-04 | 1.45E-06 | |
| ND-CTRD cohort 2 | Duplications | 391 | 170 | 0.74 | 1.64 | 9.80E-09 | 4.10E-33 |
| Deletions | 1,714 | 415 | 1.81 | 1.96 | 1.12E-07 | 1.43E-27 | |
| All CNVs | 2,105 | 585 | 2.55 | 1.85 | 4.36E-09 | 1.65E-49 | |
| Large CNVs | |||||||
| Duplications | 65 | 35 | 0.15 | 1.64 | 9.50E-03 | 6.55E-05 | |
| Deletions | 33 | 22 | 0.10 | 1.84 | 8.45E-02 | 8.69E-03 | |
| All CNVs | 98 | 57 | 0.25 | 1.71 | 2.29E-03 | 9.91E-07 | |
| DS-CTRD | Duplications | 784 | 311 | 0.52 | 1.11 | 4.60E-01 | 5.76E-01 |
| Deletions | 7,767 | 2082 | 3.46 | 1.00 | 2.73E-01 | 3.78E-01 | |
| All CNVs | 8,551 | 2,393 | 3.97 | 1.01 | 4.09E-01 | 5.43E-01 | |
| Large CNVs | |||||||
| Duplications | 219 | 128 | 0.21 | 1.13 | 3.82E-01 | 7.25E-01 | |
| Deletions | 258 | 74 | 0.12 | 0.92 | 3.26E-01 | 3.57E-01 | |
| All CNVs | 477 | 202 | 0.34 | 1.04 | 9.26E-01 | 7.52E-01 | |
Fisher Exact Test, two-side, bold type indicates significance.
CNV Burden = Number of CNV/Sample.
Structural variant content within the 401 cases in ND-CTRD cohort 1 totaled 1,733 common and rare CNVs combined. They consisted of 341 duplications, 1,341 heterozygous deletions, and 51 homozygous deletions (Table 2 and Supporting Information Table S2a). We detected no significant differences in the overall CNV (including both common and rare) frequency (p > .05, case/control ratio = 0.98), size (p > .05, case/control ratio = 1.14), or gene content (p > .05, case/control ratio = 1.32) between cases and controls.
Of all detected CNVs in ND-CTRD cohort 1, 850 (49.0%) could be definitively identified as inherited (446 maternal, 391 paternal, and 39 present in both parents), while 300 were deemed as de novo events. There was no bias of maternal or paternal inheritance (p value >.05). Of those de novo CNVs, 103 were unique (5.9% of total CNVs, not observed in control cohorts) and identified in 73 subjects (18.2% of subjects). Certain of these de novo detections were potentially due to Type II error (Itsara et al., 2010).
We detected 2,105 CNVs from 229 singletons of ND-CTRD Cohort 2. They included 1,517 heterozygous deletions, 196 homozygous deletions, and 391 duplications (Table 2 and Supporting Information Table S2b). We once again detected no significant differences in the overall CNV frequency (p > .05, case/control ratio = 0.96), size (p > .05, case/control ratio = 1.17), or gene content (p > .05, case/control ratio = 1.55) between cases and controls in Cohort 2. We were unable to determine inheritance status in cohort 2 as no parental data were available.
In the DS-CTRD cohort, we detected 8,551 CNVs (excluding the typical 22q11.2DS deletion) in 602 cases. Among them, 7,767 were deletions and 784 were duplications (Table 2 and Supporting Information Table S2c). There was no significant difference in overall CNV frequency (p > .05, case/control ratio = 1.02), sizes (p > .05, case/control ratio = 1.05), or gene content (p > .05, case/control ratio = 1.17) comparing cases with controls in the DS-CTRD cohort.
ND-CTRD Cohort 1 contained 536 rCNVs (30.9% of the total CNVs detected within this cohort, 170 duplications and 366 heterozygous deletions) and ND-CTRD Cohort 2 contained 585 rCNVs (27.8% of the total CNVs detected within this cohort, 170 duplications and 415 heterozygous deletions). There was a total of 2,393 rCNVs in DS-CTRD cohort (30.8% of the total CNVs being detected within this cohort, 2082 deletions and 311 duplications).
The burden of rCNVs is depicted for each cohort in Table 2. rCNVs were significantly overrepresented in ND-CTRD cases as compared with controls, whether comparing the proportion of subjects with rCNVs or the total number of rCNVs in cases and controls. rCNV burden remained significant for large CNVs in the ND-CTRD cohort as defined in section 2. These results were similar to our previous report studying a conotruncal cohort with a broader phenotype (Xie et al., 2017).
We did not detect any significant difference in rCNV burden comparing cases with CTRD as compared with controls without CTRD in the DS-CTRD cohort (Table 2) regardless of CNV size. These results were consistent with our previous findings (Mlynarski et al., 2016).
3.3 |. Gene analysis in patient cohorts
In ND-CTRD cohort 1, a total of 786 CNVs included one or more genes, collectively representing 1,251 individual genes (Supporting Information Table S3). Of these, 160 genes were fully or partially contained within CNVs in two or more cases, of which 25 genes were not contained within CNVs in controls. In ND-CTRD Cohort 2, 922 CNVs included 1,083 unique genes (Supporting Information Table S3), of which 249 genes were contained within CNVs from two or more individuals, of which 33 genes were not contained within CNVs in controls. Collectively, 28 genes were contained by rCNVs in both CTRD-cohorts at least once but not in any controls (7 genes were in deletions in both cohorts, 5 genes were in duplications in both cohorts, and 16 genes were in different types of CNVs in the two cohorts; Supporting Information Table S3). This number of shared genes is not statistically significant nor more than what would be expected by chance alone.
A total of 3,087 CNVs in the DS-CTRD cohort included 1,080 individual genes (Supporting Information Table S3). A total of 208 of these genes were contained by CNVs in two or more individuals. We found that 125 of these 208 genes were not included in CNVs identified in DS-CTRD controls.
The combination of all three cohorts (ND-CTRD 1 and 2, DS-CTRD) identified 59 genes that were contained by rCNVs in both the combined ND-CTRD cohort and DS-CTRD cohort and deemed as recurrent in CTRD cohorts and not seen in controls (15 genes were in deletions in both cohorts, 22 genes were in duplications in both cohorts, and 38 genes were in different types of CNVs in the two case cohorts; Supporting Information Table S4). This number of shared genes is not statistically significant nor more than what would be expected by chance alone.
We used a gene-based case-control enrichment analysis on genes disrupted by CNVs in the combined ND-CTRD cohorts and the DS-CTRD cohort to determine if any genes were overrepresented in the CTRD cases as compared with their respective controls (Fisher’s Exact Test, two-tailed). The results are listed in Supporting Information Table S3. No genes remained significantly enriched in CTRD cases when all CNVs or only deletions or duplications were evaluated following the procedures and filtering criteria outlined in section 2.
3.4 |. In silico functional and pathway analyses in patient cohorts
We examined several functional and pathway domains to determine whether genes listed in Supporting Information Table S3, sharing particular biological functions were enriched within rCNVs in ND-CTRD cases (combined ND-CTRD cohort 1 and ND-CTRD cohort 2) as well as in DS-CTRD cases, separately first and then in aggregate, as described in section 2. GO analysis was performed on gene content from the full list of rCNVs to examine the annotated biological processes, cellular components, and molecular functions of genes impacted by CNVs in CTRD (combined ND-CTRD and DS-CTRD) cases versus controls. Only those GO terms (GO terms) identified in the ND-CTRD cohort as well as the DS-CTRD cohort with nominal p values <.05 were selected for further analysis as described in section 2. The final GO terms were reported if their FDRs were <0.05 measured after combining all CTRD cases. The final results from the GO analyses are listed in Table 3. Thirty-seven terms were found to be significantly enriched in the combined CTRD cohorts (ND-CTRD and DS-CTRD) that met our criteria as described in section 2. Among them, several GO terms were found to be significantly enriched: “GO:0016477: Cell Migration” (FDR < 4.18E-05), “GO: 0048870: Cell Motility” (FDR < 5.30E-05), and “GO: 0043405: Regulation of MAP Kinase Activity” (FDR < 4.46E-03). Migration of cardiac progenitor cells during embryogene-sis is a process tightly regulated and essential for proper heart development (Buckingham, Meilhac, & Zaffran, 2005). Pathogenic mutations in genes related to cell migration have been previously reported in patients with CTRDs (Buckingham et al., 2005; Di Felice & Zummo, 2009; Silversides et al., 2012). Further, mitogen-activated protein kinase (MAPK) signaling cascades have been shown to play critical roles in the pathogenesis of cardiac and vascular disease (Muslin, 2008).
TABLE 3.
Enriched gene ontology terms
| ND-CTRD cohort |
|||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ND-CTRD cohort 1 |
ND-CTRD cohort 2 |
DS-CTRD cohort |
Total | ||||||||||||
| Term | Description | GO type | CNV type | Case count | Control count | p valuea Cohort1 | Case count | Control count | p valuea Cohort2 | P_TOTAL ND-CTRD | Case count | Control count | p valuea DS-CTRD | P_TOTAL CTRD | FDRb |
| GO:0006629 | Lipid metabolic process | Biological process | Dup | 16 | 73 | 9.363E-02 | 25 | 51 | 1.459E-07 | 1.287E-06 | 40 | 10 | 1.546E-02 | 1.566E-10 | 2.795E-07 |
| GO:0044255 | Cellular lipid metabolic process | Biological process | Dup | 15 | 63 | 5.021E-02 | 23 | 45 | 2.695E-07 | 9.745E-07 | 36 | 9 | 2.519E-02 | 2.641E-10 | 3.536E-07 |
| GO:0032787 | Monocarboxylic acid metabolic process | Biological process | Dup | 11 | 43 | 5.716E-02 | 16 | 24 | 1.541E-06 | 3.989E-06 | 27 | 5 | 1.448E-02 | 7.717E-10 | 5.903E-07 |
| GO:0044765 | Single-organism transport | Biological process | Dup | 34 | 169 | 3.278E-02 | 42 | 115 | 6.730E-09 | 7.676E-08 | 61 | 20 | 2.931E-02 | 8.612E-10 | 6.148E-07 |
| GO:0006082 | Organic acid metabolic process | Biological process | Dup | 19 | 76 | 2.244E-02 | 24 | 47 | 1.469E-07 | 1.727E-07 | 34 | 8 | 2.085E-02 | 1.014E-09 | 6.389E-07 |
| GO:0043436 | Oxoacid metabolic process | Biological process | Dup | 18 | 75 | 3.282E-02 | 24 | 47 | 1.469E-07 | 3.237E-07 | 34 | 8 | 2.085E-02 | 1.552E-09 | 8.480E-07 |
| GO:0019752 | Carboxylic acid metabolic process | Biological process | Dup | 16 | 66 | 3.728E-02 | 19 | 39 | 5.455E-06 | 5.824E-06 | 32 | 7 | 1.683E-02 | 1.140E-08 | 3.928E-06 |
| GO:0051649 | Establishment of localization in cell | Biological process | Dup | 22 | 113 | 1.040E-01 | 31 | 64 | 4.895E-09 | 4.232E-07 | 41 | 12 | 3.973E-02 | 1.174E-08 | 3.928E-06 |
| GO:0006631 | Fatty acid metabolic process | Biological process | Dup | 6 | 23 | 1.458E-01 | 11 | 16 | 5.924E-05 | 1.165E-04 | 21 | 4 | 3.592E-02 | 1.253E-08 | 4.066E-06 |
| GO:0071702 | Organic substance transport | Biological process | Dup | 30 | 126 | 7.167E-03 | 22 | 77 | 8.196E-04 | 3.031E-05 | 47 | 13 | 1.796E-02 | 1.448E-07 | 2.584E-05 |
| GO:0016477 | Cell migration | Biological process | Dup | 8 | 50 | 6.801E-01 | 17 | 37 | 3.159E-05 | 8.780E-04 | 30 | 6 | 1.315E-02 | 2.421E-0 | 4.181E-05 |
| GO:0048870 | Cell motility | Biological process | Dup | 10 | 56 | 4.389E-01 | 17 | 38 | 4.129E-05 | 7.291E-04 | 31 | 7 | 2.412E-02 | 3.464E-07 | 5.299E-05 |
| GO:0045859 | Regulation of protein kinase activity | Biological process | Dup | 11 | 46 | 9.500E-02 | 12 | 24 | 2.648E-04 | 3.944E-04 | 23 | 5 | 4.639E-02 | 1.107E-06 | 1.108E-04 |
| GO:0015980 | Energy derivation by oxidation of organic compounds | Biological process | Dup | 4 | 12 | 1.120E-01 | 6 | 12 | 9.846E-03 | 3.844E-03 | 15 | 2 | 4.086E-02 | 1.432E-06 | 1.381E-04 |
| GO:0051259 | Protein oligomerization | Biological process | Dup | 7 | 34 | 3.254E-01 | 10 | 13 | 6.704E-05 | 1.050E-03 | 18 | 3 | 3.907E-02 | 2.521E-06 | 1.987E-04 |
| GO:0044429 | Mitochondrial part | Cellular component | Dup | 12 | 59 | 1.922E-01 | 13 | 28 | 2.585E-04 | 8.780E-04 | 26 | 6 | 4.048E-02 | 4.763E-06 | 3.249E-04 |
| GO:0033559 | Unsaturated fatty acid metabolic process | Biological process | Dup | 2 | 11 | 6.608E-01 | 7 | 5 | 8.700E-05 | 1.261E-03 | 9 | 0 | 3.060E-02 | 1.105E-05 | 6.612E-04 |
| GO:0071900 | Regulation of protein serine/threonine kinase activity | Biological process | Dup | 5 | 26 | 4.057E-01 | 7 | 13 | 3.916E-03 | 1.326E-02 | 16 | 2 | 2.628E-02 | 3.844E-05 | 1.774E-03 |
| GO:0071705 | Nitrogen compound transport | Biological process | Dup | 11 | 36 | 2.132E-02 | 5 | 21 | 1.968E-01 | 9.101E-03 | 18 | 2 | 1.641E-02 | 8.725E-05 | 3.373E-03 |
| GO:0043405 | Regulation of MAP kinase activity | Biological process | Dup | 4 | 22 | 5.402E-01 | 6 | 7 | 1.462E-03 | 1.097E-02 | 12 | 1 | 3.940E-02 | 1.209E-04 | 4.455E-03 |
| GO:0004672 | Protein kinase activity | Molecular function | Dup | 10 | 55 | 3.365E-01 | 17 | 42 | 1.116E-04 | 9.008E-04 | 21 | 4 | 3.592E-02 | 1.320E-04 | 4.760E-03 |
| GO:0009891 | Positive regulation of biosynthetic process | Biological process | Dup | 16 | 93 | 3.65E-01 | 17 | 62 | 5.12E-03 | 1.40E-02 | 35 | 8 | 1.47E-02 | 1.64E-04 | 5.69E-03 |
| GO:0016301 | Kinase activity | Molecular function | Dup | 15 | 69 | 8.82E-02 | 17 | 51 | 1.04E-03 | 6.70E-04 | 24 | 5 | 4.71E-02 | 1.65E-04 | 5.72E-03 |
| GO:0050867 | Positive regulation of cell activation | Biological process | Dup | 6 | 22 | 1.34E-01 | 5 | 13 | 3.98E-02 | 1.69E-02 | 13 | 1 | 2.40E-02 | 1.91E-04 | 6.41E-03 |
| GO:0030529 | Ribonucleoprotein complex | Cellular component | Dup | 10 | 43 | 1.30E-01 | 7 | 32 | 1.89E-01 | 4.63E-02 | 22 | 3 | 1.06E-02 | 3.16E-04 | 9.18E-03 |
| GO:0002696 | Positive regulation of leukocyte activation | Biological process | Dup | 5 | 19 | 1.95E-01 | 4 | 13 | 1.08E-01 | 4.69E-02 | 13 | 1 | 2.40E-02 | 3.89E-04 | 1.10E-02 |
| GO:0050865 | Regulation of cell activation | Biological process | Dup | 7 | 35 | 3.33E-01 | 9 | 21 | 3.61E-03 | 8.25E-03 | 15 | 2 | 4.09E-02 | 5.70E-04 | 1.48E-02 |
| GO:0006935 | Chemotaxis | Biological process | Dup | 4 | 23 | 5.53E-01 | 8 | 14 | 1.54E-03 | 1.06E-02 | 9 | 0 | 3.06E-02 | 2.21E-03 | 4.00E-02 |
| GO:0042330 | Taxis | Biological process | Dup | 4 | 23 | 5.53E-01 | 8 | 14 | 1.54E-03 | 1.06E-02 | 9 | 0 | 3.06E-02 | 2.21E-03 | 4.00E-02 |
| GO:0050678 | Regulation of epithelial cell proliferation | Biological process | Del | 12 | 15 | 2.16E-05 | 4 | 12 | 8.99E-02 | 1.07E-05 | 12 | 1 | 3.94E-02 | 1.97E-07 | 9.54E-06 |
| GO:0009968 | Negative regulation of signal transduction | Biological process | Del | 14 | 60 | 6.76E-02 | 7 | 30 | 1.77E-01 | 2.28E-02 | 27 | 5 | 1.45E-02 | 3.78E-05 | 9.34E-04 |
| GO:0023057 | Negative regulation of signaling | Biological process | Del | 14 | 62 | 1.03E-01 | 8 | 30 | 6.14E-02 | 1.64E-02 | 27 | 5 | 1.45E-02 | 4.22E-05 | 1.02E-03 |
| GO:0010648 | Negative regulation of cell communication | Biological process | Del | 14 | 63 | 1.05E-01 | 8 | 30 | 6.14E-02 | 1.69E-02 | 27 | 5 | 1.45E-02 | 4.54E-05 | 1.09E-03 |
| GO:0006691 | Leukotriene metabolic process | Biological process | All | 3 | 9 | 1.62E-01 | 6 | 2 | 3.85E-05 | 1.77E-04 | 10 | 0 | 1.70E-02 | 1.39E-07 | 4.90E-06 |
| GO:0030100 | Regulation of endocytosis | Biological process | All | 8 | 31 | 1.27E-01 | 4 | 16 | 2.66E-01 | 4.04E-02 | 25 | 3 | 4.30E-03 | 6.09E-07 | 1.77E-05 |
| GO:0031253 | Cell projection membrane | Cellular component | All | 11 | 35 | 1.89E-02 | 9 | 37 | 8.86E-02 | 4.45E-03 | 28 | 7 | 4.90E-02 | 9.90E-07 | 2.73E-05 |
| GO:0001952 | Regulation of cell-matrix adhesion | Biological process | All | 5 | 17 | 1.72E-01 | 3 | 7 | 8.80E-02 | 2.55E-02 | 16 | 2 | 2.63E-02 | 3.73E-06 | 9.28E-05 |
| GO:0050679 | Positive regulation of epithelial cell proliferation | Biological process | All | 9 | 21 | 6.18E-03 | 4 | 12 | 8.99E-02 | 1.54E-03 | 12 | 1 | 3.94E-02 | 3.87E-05 | 7.36E-04 |
Two sided Fisher Exact test.
False Discovery Rate Adjustment following Benjamini-Hochberg procedure.
Mammalian phenotype analysis was performed as a complementary case-control study to investigate phenotypes associated with genes impacted by rCNVs in ND-CTRD and DS-CTRD cases versus controls, separately and then in aggregate, as described in section 2. MGI-derived MPO terms were associated with gene orthologs in the CTRD cases as compared with their controls. The results from the MP analyses are listed in Table 4. Thirty-eight mammalian phenotype terms were identified as significantly enriched in the combined CTRD cohorts (ND-CTRD and DS-CTRD) that met our criteria as described in section 2. Relevant developmental phenotypes included: “Abnormal muscle physiology” (FDR < 1.98E-03), “Abnormal Heart Development” (FDR < 2.56E-04), “Abnormal Cell Differentiation” (FDR < 6.18E-04), and “Abnormal Cardiovascular System Morphology” (FDR < 1.25E-03).
TABLE 4.
Enriched mammalian phenotype
| ND-CTRD cohort |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ND-CTRD cohort 1 |
ND-CTRD cohort 2 |
DS-CTRD cohort |
Total |
|||||||||||
| Term | Description | CNV type | Case count | Control count | p valuea Cohort1 | Case count | Control count | p valuea Cohort2 | P_TOTAL ND-CTRD | Case count | Control count | p valuea DS-CTRD | P_TOTAL CTRD | FDRb |
| MP:0005621 | Abnormal cell physiology | Dup | 35 | 160 | 1.148E-02 | 34 | 123 | 5.016E-05 | 5.739E-06 | 60 | 20 | 3.805E-02 | 4.272E-08 | 2.770E-05 |
| MP:0002078 | Abnormal glucose homeostasis | Dup | 9 | 54 | 5.532E-01 | 14 | 44 | 4.097E-03 | 1.376E-02 | 38 | 9 | 1.822E-02 | 1.475E-07 | 6.667E-05 |
| MP:0000188 | Abnormal circulating glucose level | Dup | 7 | 34 | 3.254E-01 | 8 | 26 | 2.720E-02 | 2.794E-02 | 29 | 5 | 9.726E-03 | 2.512E-07 | 1.018E-04 |
| MP:0005076 | Abnormal cell differentiation | Dup | 10 | 45 | 1.421E-01 | 14 | 33 | 3.128E-04 | 4.755E-04 | 23 | 4 | 2.370E-02 | 4.099E-06 | 6.182E-04 |
| MP:0002163 | Abnormal gland morphology | Dup | 20 | 110 | 2.122E-01 | 25 | 78 | 7.133E-05 | 4.726E-04 | 40 | 11 | 3.466E-02 | 9.754E-06 | 8.908E-04 |
| MP:0004937 | Dilated heart | Dup | 5 | 15 | 7.886E-02 | 8 | 16 | 2.908E-03 | 9.758E-04 | 13 | 1 | 2.402E-02 | 1.113E-05 | 9.890E-04 |
| MP:0002019 | Abnormal tumor incidence | Dup | 13 | 59 | 1.367E-01 | 12 | 49 | 3.656E-02 | 1.258E-02 | 32 | 8 | 4.183E-02 | 1.368E-05 | 1.095E-03 |
| MP:0002127 | Abnormal cardiovascular system morphology | Dup | 16 | 110 | 7.784E-01 | 22 | 86 | 3.796E-03 | 2.742E-02 | 50 | 15 | 3.125E-02 | 1.598E-05 | 1.249E-03 |
| MP:0002166 | Altered tumor susceptibility | Dup | 13 | 59 | 1.367E-01 | 12 | 50 | 3.962E-02 | 1.315E-02 | 32 | 8 | 4.183E-02 | 2.164E-0 | 1.478E-03 |
| MP:0005369 | Muscle phenotype | Dup | 15 | 89 | 4.394E-01 | 21 | 72 | 9.589E-04 | 4.283E-03 | 39 | 11 | 4.762E-02 | 2.282E-05 | 1.526E-03 |
| MP:0002459 | Abnormal B cell physiology | Dup | 9 | 44 | 2.795E-01 | 9 | 28 | 1.581E-02 | 1.829E-02 | 24 | 4 | 1.566E-02 | 2.655E-05 | 1.688E-03 |
| MP:0005418 | Abnormal circulating hormone level | Dup | 9 | 59 | 7.041E-01 | 16 | 39 | 1.578E-04 | 3.936E-03 | 27 | 6 | 4.044E-02 | 3.030E-05 | 1.794E-03 |
| MP:0000163 | Abnormal cartilage morphology | Dup | 4 | 21 | 5.288E-01 | 8 | 9 | 1.911E-04 | 2.089E-03 | 12 | 1 | 3.940E-02 | 3.569E-05 | 1.978E-03 |
| MP:0000738 | Impaired muscle contractility | Dup | 5 | 24 | 3.804E-01 | 8 | 23 | 1.579E-02 | 2.268E-02 | 18 | 1 | 3.141E-03 | 3.539E-05 | 1.978E-03 |
| MP:0002106 | Abnormal muscle physiology | Dup | 12 | 62 | 2.720E-01 | 13 | 56 | 4.745E-02 | 3.260E-02 | 33 | 6 | 5.904E-03 | 3.530E-05 | 1.978E-03 |
| MP:0003953 | Abnormal hormone level | Dup | 11 | 70 | 6.017E-01 | 16 | 47 | 1.436E-03 | 1.115E-02 | 30 | 6 | 1.315E-02 | 4.975E-05 | 2.520E-03 |
| MP:0002498 | Abnormal acute inflammation | Dup | 4 | 22 | 5.402E-01 | 10 | 20 | 8.740E-04 | 4.652E-03 | 14 | 1 | 1.486E-02 | 9.731E-05 | 4.123E-03 |
| MP:0005620 | Abnormal muscle contractility | Dup | 5 | 29 | 5.913E-01 | 9 | 28 | 1.581E-02 | 3.829E-02 | 19 | 1 | 3.341E-03 | 1.310E-04 | 5.088E-03 |
| MP:0004087 | Abnormal muscle fiber morphology | Dup | 6 | 34 | 4.656E-01 | 9 | 24 | 7.251E-03 | 2.456E-02 | 15 | 2 | 4.086E-02 | 1.673E-03 | 3.218E-02 |
| MP:0002164 | Abnormal gland physiology | Del | 17 | 43 | 3.923E-04 | 6 | 24 | 1.329E-01 | 1.676E-04 | 23 | 4 | 2.370E-02 | 3.503E-07 | 2.287E-05 |
| MP:0005667 | Abnormal circulating leptin level | Del | 7 | 10 | 2.104E-03 | 2 | 8 | 3.027E-01 | 2.340E-03 | 12 | 1 | 3.940E-02 | 2.433E-06 | 1.128E-04 |
| MP:0000274 | Enlarged heart | All | 18 | 72 | 2.051E-02 | 11 | 47 | 5.591E-02 | 3.728E-03 | 46 | 9 | 1.364E-03 | 1.964E-09 | 1.125E-07 |
| MP:0000920 | Abnormal myelination | All | 8 | 29 | 7.301E-02 | 6 | 24 | 1.329E-01 | 3.114E-02 | 29 | 5 | 9.726E-03 | 5.941E-08 | 2.263E-06 |
| MP:0003856 | Abnormal hindlimb stylopod morphology | All | 13 | 27 | 4.461E-04 | 3 | 13 | 4.080E-01 | 3.988E-04 | 19 | 1 | 3.341E-03 | 8.467E-08 | 3.174E-06 |
| MP:0000559 | Abnormal femur morphology | All | 13 | 25 | 2.510E-04 | 2 | 13 | 6.767E-01 | 6.723E-04 | 19 | 1 | 3.341E-03 | 9.189E-08 | 3.409E-06 |
| MP:0005438 | Abnormal glycogen homeostasis | All | 7 | 19 | 2.768E-02 | 4 | 13 | 1.078E-01 | 1.297E-02 | 21 | 3 | 1.643E-02 | 1.054E-07 | 3.871E-06 |
| MP:0005437 | Abnormal glycogen level | All | 7 | 17 | 1.793E-02 | 3 | 13 | 4.080E-01 | 2.046E-02 | 21 | 3 | 1.643E-02 | 1.112E-07 | 4.040E-06 |
| MP:0005560 | Decreased circulating glucose level | All | 10 | 38 | 6.803E-02 | 6 | 24 | 1.329E-01 | 1.895E-02 | 30 | 7 | 3.460E-02 | 2.331E-07 | 7.722E-06 |
| MP:0008772 | Increased heart ventricle size | All | 11 | 35 | 1.892E-02 | 8 | 34 | 1.272E-01 | 6.182E-03 | 27 | 5 | 1.448E-02 | 9.078E-07 | 2.597E-05 |
| MP:0000558 | Abnormal tibia morphology | All | 8 | 17 | 6.336E-03 | 4 | 15 | 1.480E-01 | 3.210E-03 | 16 | 1 | 8.754E-03 | 1.812E-06 | 4.817E-05 |
| MP:0003109 | Short femu | All | 9 | 18 | 2.801E-03 | 2 | 12 | 6.609E-01 | 5.397E-03 | 16 | 1 | 8.754E-03 | 1.972E-06 | 5.183E-05 |
| MP:0003857 | Abnormal hindlimb zeugopod morphology | All | 8 | 20 | 1.324E-02 | 5 | 16 | 7.209E-02 | 2.865E-03 | 16 | 1 | 8.754E-03 | 4.252E-06 | 9.707E-05 |
| MP:0002764 | Short tibia | All | 5 | 12 | 4.226E-02 | 4 | 11 | 7.348E-02 | 8.269E-03 | 13 | 1 | 2.402E-02 | 1.021E-05 | 2.034E-04 |
| MP:0000267 | Abnormal heart development | All | 16 | 46 | 2.119E-03 | 8 | 30 | 6.137E-02 | 3.724E-04 | 20 | 3 | 2.563E-02 | 1.371E-05 | 2.557E-04 |
| MP:0001176 | Abnormal lung development | All | 7 | 21 | 4.051E-02 | 4 | 14 | 1.273E-01 | 1.685E-02 | 16 | 1 | 8.754E-03 | 1.486E-05 | 2.722E-04 |
| MP:0004982 | Abnormal osteoclast morphology | All | 6 | 21 | 1.242E-01 | 3 | 8 | 1.115E-01 | 3.503E-02 | 15 | 2 | 4.086E-0 | 3.569E-05 | 5.726E-04 |
| MP:0003055 | Abnormal long bone epiphyseal plate morphology | All | 7 | 25 | 9.363E-02 | 4 | 11 | 7.348E-02 | 1.877E-02 | 13 | 1 | 2.402E-02 | 2.374E-04 | 2.689E-03 |
| MP:0002182 | Abnormal astrocyte morphology | All | 9 | 23 | 9.776E-03 | 3 | 17 | 4.764E-01 | 1.495E-02 | 12 | 1 | 3.940E-02 | 7.410E-04 | 6.689E-03 |
Two-sided Fisher Exact Test.
False Discovery Rate Adjustment following Benjamini-Hochberg procedure.
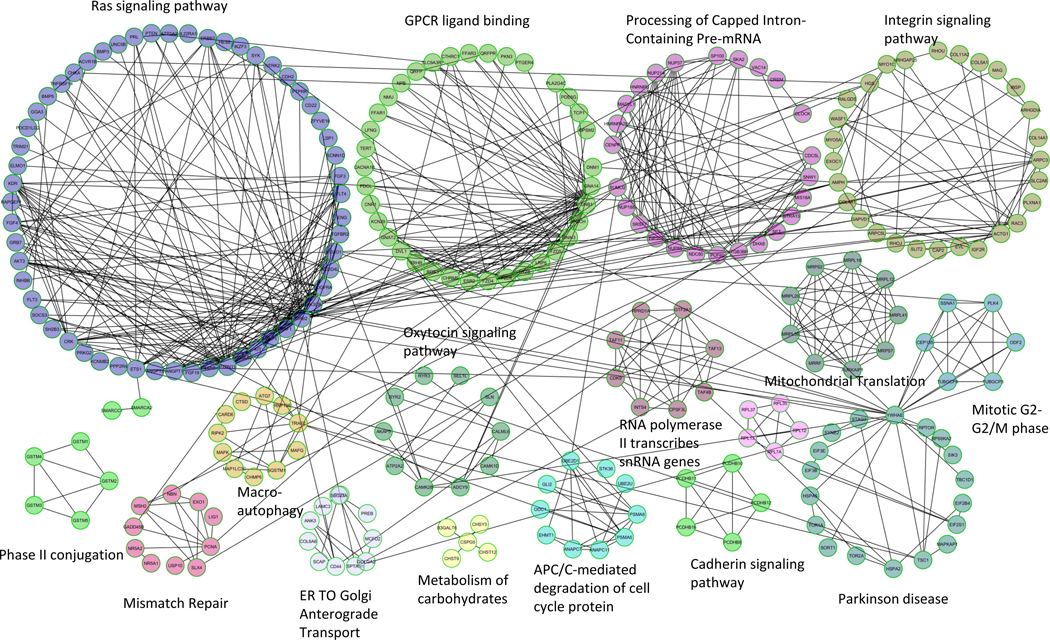
Gene interaction network analyses were performed using unique genes within rCNVs in the CTRD cohorts (both ND-CRTD and DS-CTRD cohorts) that were not found in any of the healthy controls or DS-CTRD controls. We collected 950 unique genes from the combined ND-CTRD cohort and 896 genes from the DS-CTRD cohort. Gene sets from each collection were imported separately, then in aggregate into the ReactomeFIViz component (https://f1000research.com/articles/3-146/v2) (Wu et al., 2014) of gene interaction networks visualization software CytoScape (http://www.cytoscape.org) (v3.6) (Shannon et al., 2003). After importing the gene list into ReactomeFIViz software, we first built gene interaction and regulatory networks using the knowledge base of Reactome. The resulting gene networks were further grouped into modules based on their properties of connectivity using ReactomeFIViz’s internal build-in application. Functional enrichment rankings were provided for each module as well. Those modules were filtered by size and pathway enrichment; only statistically significant modules (FDR < 0.05) that contained five or more genes were retained. The gene network results from the merged unique genes identified from ND-CTRD and DS-CTRD cohorts are shown in Figure 2. We also characterized ND-CTRD and DS-CTRD gene interaction networks separately (Supporting Information Figure S1a, b, respectively). Several modules with functions and pathways that were significantly enriched in cases pertain to functions that are related to cell signaling such as the “Ras signaling pathway”, “G protein-coupled receptor (GPCR) signaling”, “Integrin Signaling Pathway”, and mRNA processing network (Figure 2, Supporting Information Figure S1,b). It is known that RNA binding proteins are tightly associated with regulation of heart development in general (Blech-Hermoni & Ladd, 2013). The combined unique gene interaction network identified a module with functional enrichment of “GPCR ligand binding”. G protein-coupled receptors (GPCRs) collect a large group of membrane receptors and regulate various physiological processes. Some of the GPCR genes have been deemed therapeutic targets in cardiovascular diseases due to their relevance to cardiogenesis and are essential for proper heart development (Belmonte & Blaxall, 2011).
FIGURE 2.
CTRD unique gene interaction network using ReactomeFIViz. Top function within each cluster is highlighted at the top of each cluster. To simplify figure presentation, we annotated each module using its top enriched function or more abundant functional categories to illustrate each module’s functional characterization. Different connecting lines represent different biological events as illustrated in the legend in the figure
3.5 |. Mouse embryo homolog gene expression analysis
To further characterize the involvement of unique genes revealed by CNVs in the shared CTRD cohorts in heart development, we utilized a systems biology approach and publicly available mouse expression data to investigate the expression patterns of CTRD unique genes in different tissues and stages throughout heart development.
In the ND-CTRD cases, 855 of 950 unique genes within CNVs with mouse homologs were identified; 600 of these genes were significantly differentially expressed at p values <.01 level along the time course of embryonic and heart development. Those 600 genes were clustered using the expression level data, which produced eight clusters with distinct expression patterns (Supporting Information Figure S2). Gene set enrichment analyses were performed among genes within each cluster. Cluster #5 was the only cluster that contained functions identified as statistically significantly enriched within the cluster after FDR adjustment (FDR < 0.05). Many of the significantly enriched functions in this cluster are directly related to cardiac development (the top 20 identified functions are shown in Supporting Information Table S5): “GO:0072358: Cardiovascular System Development” (FDR < 0.05), “GO:0072359: Circulatory System Development”, and “GO:0003013: Circulatory System Process” (FDR < 0.05).
The same process was repeated using the 848 unique genes with mouse homologs identified in the DS-CTRD cohorts; 576 genes were significantly differentially expressed at a nominal p value <.01 level. A total of 10 clusters were identified (Supporting Information Figure S2b). Interestingly, the gene expression profile of DS-CTRD cluster #8 was remarkably similar to cluster #5 in the ND-CTRD analysis. More interestingly, similar heart development related functions enriched in ND-CTRD cluster #5 were also listed as most significantly enriched in DS-CTRD cluster #8 (Supporting Information Table S6). Again, this cluster was the only cluster that contained functions identified as statistically significantly enriched within the cluster after FDR adjustment (FDR < 0.05).
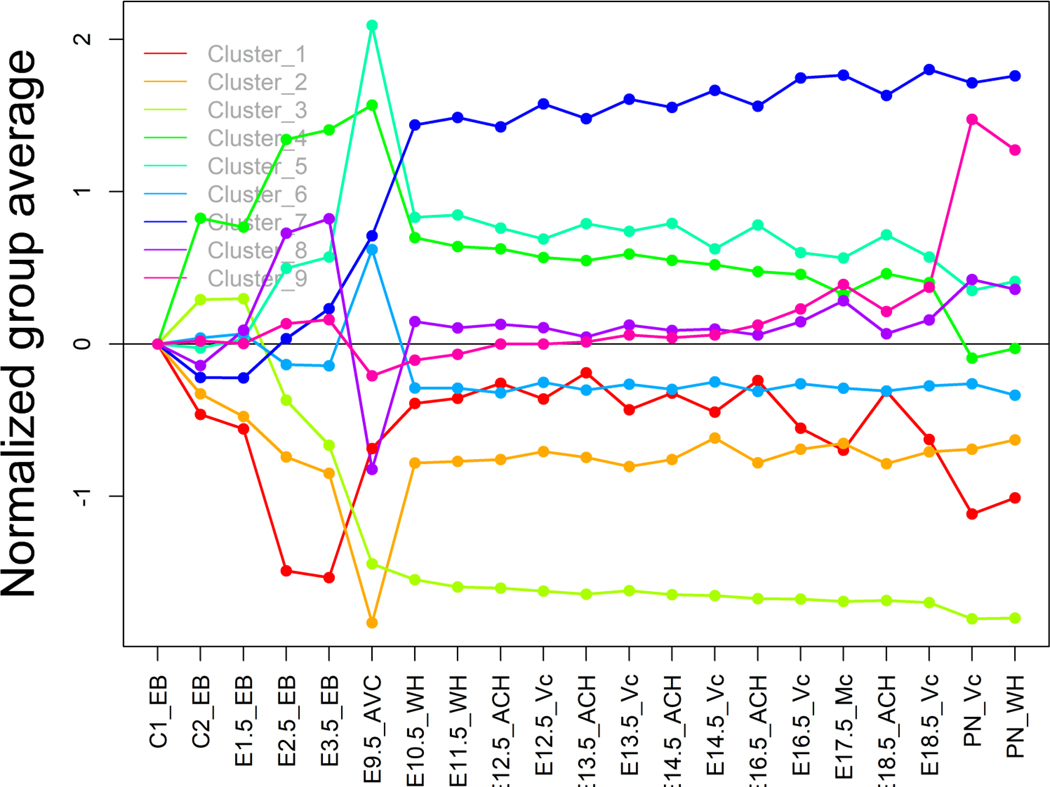
We summed all unique genes identified from both the ND-CTRD and DS-CTRD cohorts and repeated the process. A total of 894 genes were selected and nine clusters were identified. As shown in Figure 3, the gene expression levels after E9.5 in cluster #7 in this merged analysis, as well as those in cluster #5 from the ND-CTRD cohort and those in cluster #8 from the DS-CTRD cohort remained consistently higher than other clusters throughout the rest of embryonic development. The functionality of cluster #7 using all CTRD unique genes is provided in Supporting Information Table S8. Many heart development terms were consistently enriched, with high significance, as detected in the DS-CTRD cohort specific cluster #8 and the ND-CTRD cohort specific cluster #5 as well. Among them, “GO:0072358: Cardiovascular System Development” (FDR < 1.80E-05), “GO:0072359: Circulatory System Development” (FDR < 1.80E-05), and “GO:0001944: Vasculature Development”(FDR < 7.80E-05) were ranked as the top three most significantly enriched functions within cluster #7 (Supporting Information Table S7).
FIGURE 3.
CTRD unique gene expression in mouse heart development. Genes are clustered into groups based on their gene expression level at each embryonic developmental stage. Each line represents average gene expression levels among genes within a given cluster
The cardiac outflow tract greatly enlarges from embryonic day (E) 8.5–10.5 in mouse development, while mesodermal precursor cells from the second heart field are added (Review by Kelly, PMID: 22449840; Meilhac & Buckingham, 2018, PMID: 30266935). One interesting observation is that the expression level of all genes under interrogation remained constant after mouse E9.5 and throughout the rest of development, whereas expression levels were dynamic and highly variable prior to E9.5. This trend was consistent across all clusters and all CTRD data sets whether we studied ND-CTRD and DS-CTRD genes separately or aggregately.
4 |. DISCUSSION
We studied rCNVs in the largest cohorts with and without 22q11.2 deletion who carry the same cardiac malformations to identify shared genes, developmental pathways or biological functions that could explain disease risk for CTRD. Even though ND-CTRD and DS-CTRD subjects carry comparable cardiac phenotypes, the genetic architecture of the two cohorts are considerably different given the 1.5–3 million base-pair 22q11.2 deletion in the DS-CTRD subjects. Nonetheless, our combined analyses allowed us to identify several statistically significant functional networks comprised of gene pathways associated with the cardiac phenotypes that were only suggested by our previous independent studies of the two cohorts (Mlynarski et al., 2015, 2016; Xie et al., 2017). Thus, we demonstrated that rCNVs collected from the ND-CTRD cohorts and DS-CTRD cohort did not share the same individual genes, but impacted genes sharing certain common pathways and gene networks with known functions. Many of the pathways and functions were previously reported as being associated with cardiac phenotypes, such as “GO:0016477: Cell Migration” that was identified from both the DS-CTRD and ND-CTRD cohorts. Likewise, it was very intriguing that cardiac developmental and morphology relevant terms from mammalian phenotype, such as “Abnormal Heart Development” and “Abnormal Cardiovascular System Morphology”, were significantly enriched in combined CTRD cohort cases as compared with their respective controls.
Constructing a regulation network using unique genes derived from the rCNV in cases generated meaningful insights into the etiology of CTRDs. Common pathways, such as pathways involving mRNA processing, were identified in both ND-CTRD and DS-CTRD cases independently, even though each cohort harbored different genes. Those results provided additional support to the hypothesis that ND-CTRD and DS-CTRD cases likely develop CTRD through common mechanisms.
We used publicly available mouse embryo microarray data to further investigate the expression patterns of unique genes from rCNVs in the CTRD cohorts. A gene expression pattern identified from ND-CTRD unique genes was also observed when using DS-CTRD unique genes, and also in the combined ND-CTRD and DS-CTRD unique gene list. Functional analysis of this particular expression profile revealed that genes recruited in this cluster were significantly enriched in various cardiac development terms.
CHD is characterized by significant genetic heterogeneity (Pierpont et al., 2007). rCNVs are considered important in the etiology of CHD and, as such, have been studied in detail in recent years. With rare exception (e.g., CNVs at 1q21 or 8p23), the majority of studies report mostly novel or rCNVs. Likewise, we did not identify any single rCNV or gene significantly enriched in the CTRD cohorts. Instead, function, pathway, and gene regulation network analyses appeared to be more informative. Even so, the functions, pathways, and gene regulation networks identified in this study do not overlap with those defined by other early, large-scale cardiac rCNV studies (Glessner et al., 2014; Soemedi et al., 2012). This variability could potentially be due to the incomplete nature and evolving nature of the functional annotation of human genes. Limitations for different CHD rCNVs centered studies, as well as ours, such as genotyping platform limitations, CNV detection algorithm inefficiency, heterozygous cardiac phenotypic presentations and small study cohort sizes (incomplete sampling of the full CTRD population) could also be important factors underlying variable results.
As before, we observed an increased burden of rCNVs in the ND-CTRD cases, even though the ND-CTRD cases in this study represent only a subset of those previously reported (Xie et al., 2017). One might say that the rCNVs in ND-CTRD cases are more likely to represent the pathogenic driving events while the rCNVs in DS-CTRDs cases are more likely to modify the risk of CTRD given a 22q11.2 deletion (Mlynarski et al., 2015), but they appear to contribute to disease risk for CTRD in both cohorts. Although our rCNVs events are less likely to be recurrent within or between different CTRD cohorts, it was considered likely that they were linked to each other via shared developmental pathways. Our data support this hypothesis.
We also thoroughly inspected pathways known to be involved in heart development, such as the Notch pathway and chromatin modification genes. We did not detect any statistically significant difference comparing CTRD cases with controls. Given the known disease association of these pathways with CHDs, either we were underpowered to detect the association or the genetic architecture (i.e., rCNVs as opposed to single nucleotide variants) may be of importance. A much larger cohort and inclusion of various genotyping technologies is required to better understand the genetic etiology of these complex traits.
In summary, the combined gene content in rCNVs identified in two distinct CTRD cohorts, those with and without a 22q11.2 deletion, appears to contribute to pathways and functions critical to cardiovascular development not previously identified or determined to be significantly enriched. Studied separately and in combination, we observed that the rCNVs in these two genetically distinct CTRD patient cohorts (ND-CTRD and DS-CTRD cohorts) share a collection of functions, pathways, and gene regulation networks directly relevant to cardiac phenotypes despite the fact that the explicit gene content varied between the two cohorts. Gene expression pattern analyses also support the hypothesis that these two CTRD groups share genetic and developmental mechanisms underlying their cardiac phenotype. Increasing the size of the patient cohorts, with analyses emphasizing gene sets and incorporating gene expression data will likely enhance our understanding of the etiology of cardiac defects in the future.
Supplementary Material
ACKNOWLEDGMENTS
We thank all of the families for their participation and the members of the Cardiac Center for their support of case ascertainment. We thank Jennifer Garbarini and Stacy Woyciechowski for case ascertainment, Sharon Edman for data management for ND-CTRD cohort, and Oanh Tran and Andrea Jin for their technical assistance for the DS-CTRD cohort. This work was funded by U.S. National Institutes of Health funding HL84410 (B.M., B.E.), P01-HD070454 (B.M., E.G., B.E.), HD026979 (B.M., B.E.), and P50-HL074731 (E.G.), National Center for Research Resources funding UL1RR024134 / National Center for Advancing Translational Sciences funding UL1TR000003 (E.G.), and the National Eye Institute funding R01 EY020483 (D.S.).
Funding information
National Eye Institute, Grant/Award Number: R01 EY020483; National Center for Advancing Translational Sciences, Grant/Award Number: UL1TR000003; National Center for Research Resources, Grant/Award Number: UL1RR024134; National Institutes of Health, Grant/Award Numbers: HD026979, HL84410, P01-HD070454, P50-HL074731
Footnotes
CONFLICT OF INTEREST
The content is the sole responsibility of the authors and does not necessarily represent the official views of the NIH.
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of this article.
REFERENCES
- Andersen TA, Troelsen K.d. L. L ., & Larsen LA. (2014). Of mice and men: Molecular genetics of congenital heart disease. Cellular and Molecular Life Sciences, 71(8), 1327–1352. 10.1007/s00018-013-1430-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belmonte SL, & Blaxall BC (2011). G protein coupled receptor kinases as therapeutic targets in cardiovascular disease. Circulation Research, 109(3), 309–319. 10.1161/CIRCRESAHA.110.231233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisping E, Ikeda S, Kong SW, Tarnavski O, Bodyak N, McMullen JR, … Pu WT (2006). Gata4 is required for maintenance of postnatal cardiac function and protection from pressure overload-induced heart failure. Proceedings of the National Academy of Sciences of the United States of America, 103(39), 14471–14476. 10.1073/pnas.0602543103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blech-Hermoni Y, & Ladd AN (2013). RNA binding proteins in the regulation of heart development. The International Journal of Biochemistry & Cell Biology, 45(11), 2467–2478. 10.1016/j.biocel.2013.08.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M, Meilhac S, & Zaffran S. (2005). Building the mammalian heart from two sources of myocardial cells. Nature Reviews Genetics, 6(11), 826–835. 10.1038/nrg1710 [DOI] [PubMed] [Google Scholar]
- Coe BP, Witherspoon K, Rosenfeld JA, van Bon BWM, Vulto-van Silfhout AT, Bosco P, … Eichler EE (2014). Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nature Genetics, 46 (10), 1063–1071. 10.1038/ng.3092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Felice V, & Zummo G. (2009). Tetralogy of Fallot as a model to study cardiac progenitor cell migration and differentiation during heart development. Trends in Cardiovascular Medicine, 19(4), 130–135. 10.1016/j.tcm.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Dufour CR, Wilson BJ, Huss JM, Kelly DP, Alaynick WA, Downes M, … Giguère V. (2007). Genome-wide orchestration of cardiac functions by the orphan nuclear receptors ERRα and γ. Cell Metabolism, 5(5), 345–356. 10.1016/j.cmet.2007.03.007 [DOI] [PubMed] [Google Scholar]
- Eisen M, Spellman P, Brown P. and Botstein D. (1998). Cluster analysis and display of genome-wide expression patterns. PNAS ,95 (25) 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elia J, Gai X, Xie HM, Perin JC, Geiger E, Glessner JT, … White PS (2010). Rare structural variants found in attention-deficit hyperactivity disorder are preferentially associated with neurodevelopmental genes. Molecular Psychiatry, 15(6), 637–646. 10.1038/mp.2009.57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gai X, Perin JC, Murphy K, O’Hara R, D’arcy M, Wenocur A, … White PS (2010). CNV workshop: An integrated platform for high-throughput copy number variation discovery and clinical diagnostics. BMC Bioinformatics, 11(1), 74 10.1186/1471-2105-11-74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner JT, Bick AG, Ito K, Homsy J, Rodriguez-Murillo L, Fromer M, … Chung WK (2014). Increased frequency of de novo copy number variants in congenital heart disease by integrative analysis of single nucleotide polymorphism array and exome sequence data. Circulation Research, 115(10), 884–896. 10.1161/CIRCRESAHA.115.304458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasin Y, Olender T, Khen M, Gonzaga-Jauregui C, Kim PM, Urban AE, … Korbel JO (2008). High-resolution copy-number variation map reflects human olfactory receptor diversity and evolution. PLoS Genetics, 4(11), e1000249 10.1371/journal.pgen.1000249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellemans J, Mortier G, De Paepe A, Speleman F, & Vandesompele J. (2007). qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology, 8(2), R19 10.1186/gb-2007-8-2-r19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman JIE, & Kaplan S. (2002). The incidence of congenital heart disease. Journal of the American College of Cardiology, 39 (12), 1890–1900 Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/12084585 [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, & Speed TP (2003). Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics, 4(2), 249–264. 10.1093/biostatistics/4.2.249 [DOI] [PubMed] [Google Scholar]
- Itsara A, Wu H, Smith JD, Nickerson DA, Romieu I, London SJ, & Eichler EE (2010). De novo rates and selection of large copy number variation. Genome Research, 20(11), 1469–1481. 10.1101/gr.107680.110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalani SR, & Belmont JW (2014). Genetic basis of congenital cardiovascular malformations. European Journal of Medical Genetics, 57(8), 402–413. 10.1016/j.ejmg.2014.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maekawa M, Yamamoto T, Kohno M, Takeichi M, & Nishida E. (2007). Requirement for ERK MAP kinase in mouse preimplantation development. Development, 134(15), 2751–2759. 10.1242/dev.003756 [DOI] [PubMed] [Google Scholar]
- McDonald-McGinn DM, Sullivan KE, Marino B, Philip N, Swillen A, Vorstman JAS, … Bassett AS (2015). 22q11.2 deletion syndrome. Nature Reviews Disease Primers, 1(15071), 15071 10.1038/nrdp.2015.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynarski EE, Sheridan MB, Xie M, Guo T, Racedo SE, McDonald-McGinn DM, … International Chromosome 22q11.2 Consortium. (2015). Copy-number variation of the glucose transporter gene SLC2A3 and congenital heart defects in the 22q11.2 deletion syndrome. The American Journal of Human Genetics, 96 (5), 753–764. 10.1016/j.ajhg.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mlynarski EE, Xie M, Taylor D, Sheridan MB, Guo T, Racedo SE, … International Chromosome 22q11.2 Consortium. (2016). Rare copy number variants and congenital heart defects in the 22q11.2 deletion syndrome. Human Genetics, 135(3), 273–285. 10.1007/s00439-015-1623-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchir A, Pavlidis P, Decostre V, Herron AJ, Arimura T, Bonne G, & Worman HJ (2007). Activation of MAPK pathways links LMNA mutations to cardiomyopathy in Emery-Dreifuss muscular dystrophy. Journal of Clinical Investigation, 117(5), 1282–1293. 10.1172/JCI29042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muslin AJ (2008). MAPK signalling in cardiovascular health and disease: Molecular mechanisms and therapeutic targets. Clinical Science (London, England : 1979), 115(7), 203–218. 10.1042/CS20070430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankratz N, Dumitriu A, Hetrick KN, Sun M, Latourelle JC, Wilk JB, … PSG-PROGENI and GenePD Investigators, Coordinators and Molecular Genetic Laboratories. (2011). Copy number variation in familial Parkinson disease. PLoS ONE, 6(8), e20988 10.1371/journal.pone.0020988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierpont ME, Basson CT, Benson DW, Gelb BD, Giglia TM, Goldmuntz E, … American Heart Association Congenital Cardiac Defects Committee, Council on Cardiovascular Disease in the Young. (2007). Genetic basis for congenital heart defects: Current knowledge: A scientific statement from the American Heart Association congenital cardiac defects committee, council on cardiovascular disease in the Young: Endorsed by the American Academy of Pediatrics. Circulation, 115(23), 3015–3038. 10.1161/CIRCULATIONAHA.106.183056 [DOI] [PubMed] [Google Scholar]
- Reller MD, Strickland MJ, Riehle-Colarusso T, Mahle WT, & Correa A. (2008). Prevalence of congenital heart defects in metropolitan Atlanta, 1998–2005. The Journal of Pediatrics, 153(6), 807–813. 10.1016/j.jpeds.2008.05.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Feliciano J, Lee K-H, Kong SW, Rajagopal S, Ma Q, Springer Z, … Pu WT (2006). Development of heart valves requires Gata4 expression in endothelial-derived cells. Development, 133(18), 3607–3618. 10.1242/dev.02519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schinke M, Jay P, Brown J, & Izumo S. (2004). C57BL/6 Benchmark set for early cardiac development. Series GSE1479. Retrieved from http://www.cardiogenomics.org
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, … Ideker T. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Research, 13(11), 2498–2504. 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silversides CK, Lionel AC, Costain G, Merico D, Migita O, Liu B, … Bassett AS (2012). Rare copy number variations in adults with tetralogy of Fallot implicate novel risk gene pathways. PLoS Genetics, 8(8), e1002843 10.1371/journal.pgen.1002843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soemedi R, Wilson IJ, Bentham J, Darlay R, Töpf A, Zelenika D, … Keavney BD (2012). Contribution of global rare copy-number variants to the risk of sporadic congenital heart disease. American Journal of Human Genetics, 91(3), 489–501. 10.1016/j.ajhg.2012.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, … Mesirov JP (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America, 102(43), 15545–15550. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi CM, Luo Y, Yin Z, Zhang M, Zhu W, Wang T, … Epstein JA (2007). Hdac2 regulates the cardiac hypertrophic response by modulating Gsk3β activity. Nature Medicine, 13(3), 324–331. 10.1038/nm1552 [DOI] [PubMed] [Google Scholar]
- Vassena R, Han Z, Gao S, & Latham KE (2007). Deficiency in recapitulation of stage-specific embryonic gene transcription in two-cell stage cloned mouse embryos. Molecular Reproduction and Development, 74(12), 1548–1556. 10.1002/mrd.20723 [DOI] [PubMed] [Google Scholar]
- Wang K, Li M, Hadley D, Liu R, Glessner J, Grant SFA, … Bucan M. (2007). PennCNV: An integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Research, 17(11), 1665–1674. 10.1101/gr.6861907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White PS, Xie HM, Werner P, Glessner J, Latney B, Hakonarson H, & Goldmuntz E. (2014). Analysis of chromosomal structural variation in patients with congenital left-sided cardiac lesions. Birth Defects Research Part A: Clinical and Molecular Teratology, 100(12), 951–964. 10.1002/bdra.23279 [DOI] [PubMed] [Google Scholar]
- Wu G, Dawson E, Duong A, Haw R, & Stein L. (2014). ReactomeFIViz: A Cytoscape app for pathway and network-based data analysis. F1000Research, 3, 146 10.12688/f1000research.4431.2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie HM, Werner P, Stambolian D, Bailey-Wilson JE, Hakonarson H, White PS, … Goldmuntz E. (2017). Rare copy number variants in patients with congenital conotruncal heart defects. Birth Defects Research, 109(4), 271–295. 10.1002/bdra.23609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JM, Endicott RM, Parghi SS, Walker M, Kidd JM, & Trask BJ (2008). Extensive copy-number variation of the human olfactory receptor gene family. The American Journal of Human Genetics, 83(2), 228–242. 10.1016/j.ajhg.2008.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, … Srivastava D. (2007). Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1–2. Cell, 129(2), 303–317. 10.1016/j.cell.2007.03.030 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.