Abstract
Azelaic acid is a dicarboxylic acid that has recently been shown to play a role in plant-bacteria signalling and also occurs naturally in several cereals. Several bacteria have been reported to be able to utilize azelaic acid as a unique source of carbon and energy, including Pseudomonas nitroreducens . In this study, we utilize P. nitroreducens as a model organism to study bacterial degradation of and response to azelaic acid. We report genetic evidence of azelaic acid degradation and the identification of a transcriptional regulator that responds to azelaic acid in P. nitroreducens DSM 9128. Three mutants possessing transposons in genes of an acyl-CoA ligase, an acyl-CoA dehydrogenase and an isocitrate lyase display a deficient ability in growing in azelaic acid. Studies on transcriptional regulation of these genes resulted in the identification of an IclR family repressor that we designated as AzeR, which specifically responds to azelaic acid. A bioinformatics survey reveals that AzeR is confined to a few proteobacterial genera that are likely to be able to degrade and utilize azelaic acid as the sole source of carbon and energy.
Keywords: Bacteria, Gene regulation, azelaic acid, pseudomonas
Introduction
Bacteria of the plant microbiome are exposed to many plant-derived compounds that are likely to play important roles in plant-bacteria communication and facilitate bacterial growth and colonization of plant niches. For example, lipid-derived molecules of both microbes and plants have recently been shown to play a role as signals for plant-bacteria communication [1, 2]. In addition, fatty acids are also the precursors for the synthesis of compounds involved in intracellular plant signalling including jasmonic and azelaic acids, which are part of the cascade that triggers systemic defences [1].
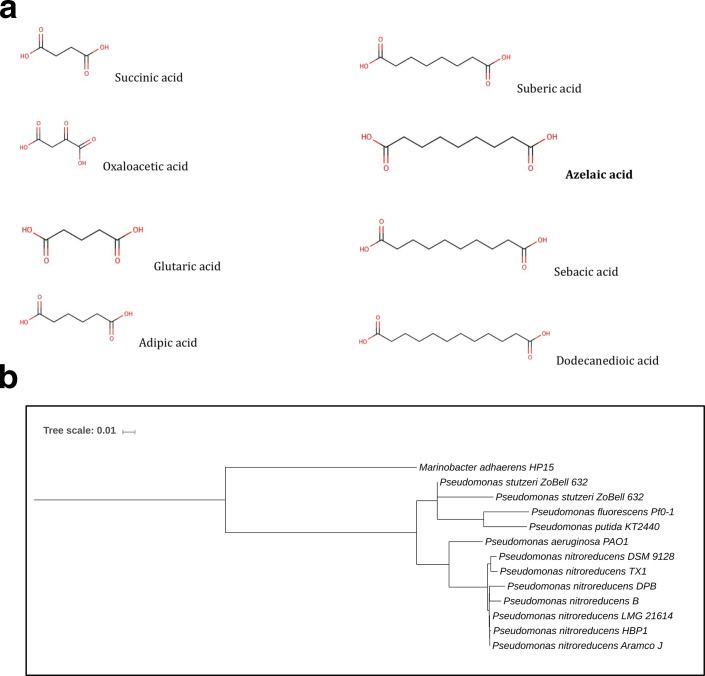
Azelaic acid (Fig. 1a) is a straight chained nine-carbon (C9) saturated dicarboxylic acid that occurs naturally in whole-grain cereals, rye and barley [3]. Azelaic acid in plants is derived from lipids via lipid peroxidation and primes systemic defence in Arabidopsis, suggesting that this mobile molecule can move systemically, most likely through the plant vasculature, and contribute to long-distance signalling in plant defence against pathogens. Azelaic acid specifically promotes disease resistance by priming salicylic acid (SA) signalling as part of systemic plant immunity [4, 5]. Recently, plant-associated Pseudomonas syringae pathovars have been reported to most likely produce azelaic acid, hinting a role in plant-bacteria communication [6]. In soil, azelaic acid along with other short-chain fatty acids are considered the products of microbial enzymatic oxidation of unsaturated lipids and aromatic hydrocarbons [7]. Several reports have shown that bacteria such as Acinetobacter spp., P. fluorescens and P. aeruginosa are capable of degrading dicarboxylic acids and utilizing them as their sole carbon source [8, 9]. Established work reported the isolation and identification of a Pseudomonas strain that can very efficiently degrade azelaic acid and utilize it as the sole source of carbon and energy [10, 11]. The bacterial isolate was classified as Pseudomonas azelaica DSM 9128 and later reclassified as P. nitroreducens [12].
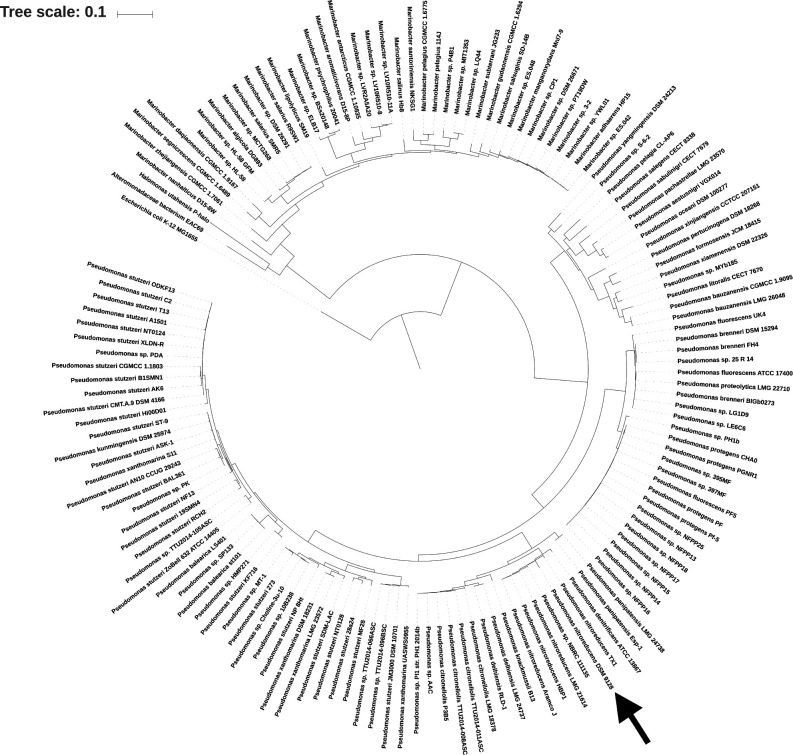
Fig. 1.
Structure of dicarboxylic acids and P. nitroreducens DSM 9128 phylogenetic tree. (a) Structure of dicarboxylic acids used in this study. (b) Phylogenetic tree showing the phylogenetic position of P. nitroreducens strain DSM9128 based on evolutionary distance of concatenated COGs listed in Table 3. Included in the tree are Pseudomonas and Marinobacter species, which have a homologue of AzeR (see text).
Initial experiments from Janota–Bassalik and Wright using P. nitroreducens DSM 9128 [10, 11] support the hypothesis that the trait of degrading the azelaic acid is a consequence of β-oxidation steps analogous to those of fatty acid degradation. Due to its possible role in plant-bacteria communication, it was of interest to isolate a bacterial regulator that can respond to the presence of azelaic acid and determine its presence in plant-associated bacteria. In this study, we report genetic evidence of azelaic acid degradation and the identification of a transcriptional regulator that responds to azelaic acid in P. nitroreducens DSM 9128. Three mutants possessing transposon insertions in genes of acyl-CoA ligase, of acyl-CoA dehydrogenase and in an isocitrate lyase display a growth deficiency in the presence of azelaic acid. Studies on transcriptional regulation of these genes resulted in the identification of a IclR family repressor that we designated as AzeR that specifically responds to azelaic acid. AzeR is widespread in Pseudomonas and Marinobacter species.
Methods
Bacterial strains, constructs, media and growth conditions
P. nitroreducens and Escherichia coli strains DH5α [13], CC118 [14] and HB101 [15] were routinely grown at 28 and 37 °C respectively, in Luria–Bertani broth/agar medium [16] or in defined M9 minimal medium [15]. All plasmid constructs used in this study are listed in Table 1. When azelaic acid was used as the carbon source, it was added to the growth medium at a final concentration of 0.1–0.2 % (5–10 mM). The following antibiotic concentrations were used: ampicillin (Amp) 100 µg ml−1, kanamycin (Km) 100 µg ml−1, gentamicin (Gm) 50 µg ml−1, tetracycline (Tc) 40 µg ml−1, nalidixic (Nx) 25 µg ml−1 and nitrofurantoin (Nf) 50 µg ml−1 for P. nitroreducens and Amp100 μg ml−1, Km50 μg ml−1, Gm 15 µg ml−1 and Tc 15 µg ml−1 for E. coli strains.
Table 1.
Plasmids used in this study
|
Plasmids |
Characteristics |
References |
|---|---|---|
|
pGem T-Easy |
Cloning vector; AmpR LacZ reporter |
(Promega) |
|
pBluescript II KS |
Cloning vector; AmpR LacZ reporter, sito polylinker, da pUC19 |
(Stratagene) |
|
pLAFR3 |
Broad-host-range cosmid cloning vector, IncP1; Tetr |
[37] |
|
pMP220 |
Promoter probe vector, IncQ; TetR |
[38] |
|
pKNOCK- Km |
Conjugative suicide vector; KmR |
[14] |
|
pCRS530 |
pAB2001 derivative containing CAS-GNm with flanking region NotI-HindIII-EcoRI-SmaI; Apr, Kmr |
[19] |
|
pRK2013 |
helper triparental conjugation, Kmr |
[18] |
|
pCos1 |
pLAFR3 carrying P. nitroreducens DSM 9128 genomic DNA |
This study |
|
pCos 5 |
pLAFR3 carrying P. nitroreducens DSM 9128 genomic DNA |
This study |
|
pCos G |
pLAFR3 carrying P. nitroreducens DSM 9128 genomic DNA |
This study |
|
pCos M |
pLAFR3 carrying P. nitroreducens DSM 9128 genomic DNA |
This study |
|
pKnReg |
Internal PCR fragment of P. nitroreducens DSM 9128 azeR cloned in Pknock-Km |
This study |
|
pMPReg |
Promoter of transcriptional regulator gene cloned in pMP220 vector |
This study |
|
pMPLig |
Promoter of Acyl-CoA ligase cloned in pMP220 vector |
This study |
|
pMPDeh |
Promoter of Acyl-CoA dehydrogenase cloned in pMP220 vector |
This study |
|
pMPIsocit |
Promoter of Isocitrate lyase cloned in pMP220 vector |
This study |
Recombinant DNA techniques
DNA manipulations, including digestion with restriction enzymes, agarose gel electrophoresis, purification of DNA fragments, ligation with T4 DNA ligase, transformation of E. coli , colony hybridization and radioactive labelling by random priming, were performed as previously described [15]. Southern hybridizations were performed using Hybond-N+membrane (Amersham Pharmacia Biotech). Plasmids were purified using EuroClone columns (EuroClone S.p.A., Italy). Total DNA from P. nitroreducens was isolated with the sarkosyl-pronase lysis method Better et al. [17]. Triparental matings to mobilize DNA from E. coli to P. nitroreducens were carried out with the helper strain E. coli (pRK2013) [18]. PCR amplifications were performed on P. nitroreducens genomic DNA using GoTaq Flexi DNA Polymerase (Promega, Madison, WI, USA) and the automated sequencing was performed by GATC-Biotech services (Kostanz, Germany). The oligonucleotide primers used in this study are listed in Table 2.
Table 2.
Primers used in this study
|
Primer name |
Sequence of primer |
References |
|---|---|---|
|
Tn5ext |
5′-TATTGCTGAAGAGCTTGGC-3′ |
This study |
|
arb1 |
5′-GGCCACGCGTCGACTAGTACNNNNNNNNNN-3′ |
[21] |
|
Tn5int |
5′-GCATCGCCTTCTATCGCCTTC −3’ |
This study |
|
arb2 |
5′-GGCCACGCGTCGACTAGTAC-3′ |
[21] |
|
prMutS1 |
5′-TATCAAAACGACATCAAAGCCGTTGC-3′ |
This study |
|
prMutS2 |
5′-TTAGTGGAACTGGTTCATGGTGTT-3′ |
This study |
|
prMutD1 |
5′-ATGCCCGCTCAACCGACC-3′ |
This study |
|
prMutD2 |
5′-TCACGGGGTCCTTCT-3′ |
This study |
|
prRegBam |
5′-AGGATCCGGAGGGTTGAGCAGACAT-3′ |
This study |
|
prRegEco |
5′-AGAATTCCGCGTGTGCTCGTTGCG-3′ |
This study |
|
prLigBam |
5′-AGGATCCTTGGGGTTGCTGCTGTCT-3′ |
This study |
|
prLigEco |
5′-AGAATTCGGAGGGTTGAGCAGACAT-3′ |
This study |
|
prIsocitBam |
5′-AGGATCCAGAACGGTTTCGCCCG-3′ |
This study |
|
prIsocitEco |
5′-AGAATTCTTTCAGAGCGGCAACGG-3′ |
This study |
|
KnRegXho |
5′-ACTCGAGGAAATCTCGCGGATCACCG-3′ |
This study |
|
KnRegXba |
5′-ATCTAGATCATCGACAGCCAGTCACGG-3′ |
This study |
|
prDehEco |
5′-AGAATTCACCGGTGGGTTGTCGACT-3′ |
This study |
|
prDehBam |
5′-AGGATCCGCCAGCAACTGGTGTT-3′ |
This study |
|
ΔReg mut |
5′-ATCACCGACCAATCGCCTTCG-3′ |
This study |
|
prRegSensBam |
5′-AGGATCCGAGCAGGCGATCCAG-3′ |
This study |
Construction of genomic transposon mutant bank and isolation of mutants unable to use azelaic acid as the carbon source
Plasmid pCRS530 [19] carrying the mTn5-GNm (containing gusA-Ptac- nptII-TtrpA) was harboured in E. coli S17-1 [20] and biparental conjugation was performed in order to conjugate this plasmid into P. nitroreducens DSM 9128. The P. nitroreducens strains harbouring the mTn5-GNm into the chromosome were selected on Luria–Bertani agar plates containing Km, Nf and Nx and incubated for 18 h at 28 °C. Several conjugations were performed in order to obtain approximately 30 000 independent transposon insertions. Among these, 5000 mutants were screened for inability to grow on azelaic acid as the unique source of carbon and energy as follows: purified colonies from the mTn5-GNm genomic mutant bank were individually hand-picked and grown at 28 °C in replicate on minimal M9 plates containing glucose or azelaic acid (0.1%). Six P. nitroreducens DSM 9128 transposon mutants were selected, which were unable or could very poorly grow on azelaic acid as the sole carbon source, but retaining the ability to grow on glucose. Their azelaic acid utilization inability was confirmed by growth-curve studies.
Mapping of transposon insertion sites and complementation of genomic mutants
The DNA sequence-flanking transposon mutants in P. nitroreducens DSM 9128 were determined using an arbitrary PCR procedure, as previously described [21]. In this method, the DNA flanking the transposon insertion site was enriched in two rounds of amplification using primers specific to the ends of the mTn5-GNm element and primers of random sequence that annealed to chromosomal sequences flanking the transposon (for primers see Table 2). For mutants 9128E, 9128I, 9128F and 9128M the arbitrary PCR did not work and in order to identify/clone the chromosomal DNA flanking the transposon, digestion of genomic DNA was performed using restriction enzymes that do not recognize the DNA sequence of the transposon. Sequences obtained were subjected to homology searches using a local NCBI blast and also with the draft genome sequence of P. nitroreducens DSM 9128 using a local blast algorithm confirming the genomic location of the mutants.
The mutants selected were complemented for their defective growth as follows: a genomic bank (cosmid library) of P. nitroreducens DSM 9128 was constructed from a partial EcoRI digested P. nitroreducens DSM 9128 genomic DNA and ligated into EcoRI digested cosmid pLAFR3. The concatameric constructs obtained were then packaged in phage lambda and transfected into E. coli HB101 cells using Gigapack III XL-4 packaging kit as recommended by the supplier (Stratagene-Agilent, Santa Clara, CA, USA). The genomic cosmid bank (7500 clones) was then screened for the gene of interest by hybridization with labelled DNA probes via colony blot hybridization and then verified by Southern analysis. Triparental matings between the E. coli HB1010 harbouring the cosmids selected were then conjugated into P. nitroreducens using E. coli (pRK2013) as the helper strain.
Construction of the azeR mutant in P. nitroreducens
The azeR gene was inactivated in P. nitroreducens by amplifying via PCR an internal fragment of this gene using primers KnRegXho, KnRegXba listed in Table 2, cloning it in pKNOCK-Km [14] and then using it as a suicide plasmid via conjugation into P. nitroreducens and selecting for insertion into the chromosome via homologous recombination. Km-resistant colonies were selected and the mutation of the targeted gene was confirmed by PCR.
Construction of plasmid gene promoter transcriptional fusions and β-galactosidase assays
Transcriptional gene promoter activity studies of four promoters were performed in P. nitroreducens . All constructs were made in the promoter probe vector pMP220 (TcR); this is a plasmid which carries a promoterless lacZ gene and harbours the gene for tetracycline resistance. The four gene promoter regions were amplified from P. nitroreducens DSM 9128 genomic DNA by using the primers listed in Table 2. The PCR amplified fragments were first cloned in pGEM-T Easy vector (Promega, Madison, WI, USA), sequenced and then excised as EcoRI/BamHI fragments and cloned into the BglII/EcoRI sites in pMP220 obtaining pMPLig, pMPDeh, pMPIsocit, pMPReg. The constructs obtained are listed in Table 1.
β-galactosidase activities were determined essentially as described by Miller [16], with the modifications of Stachel et al. [22]. Each experiment was performed in triplicate. Gene promoter activity was determined by monitoring β-galactosidase activity in overnight cultures.
Draft genome sequence of P. nitroreducens DSM 9128.
The genome sequence of P. nitroreducens was determined using a 250 bp paired-end library with the Illumina MiSeq sequencing system (University of Exeter Sequencing Service, Exeter, UK), which generated a total of 1 611 521 pairs of reads. Reads were assembled using SPAdes 3.9.03 [23] and the assembled sequence annotated using the NCBI Prokaryotic Genome Annotation Pipeline (PGAP). The P. nitroreducens DSM 9128 genome was also uploaded to the Integrated Microbial Genomes and Metagenomes (IMG/M) database and was automatically annotated using annotation pipeline IMG Annotation Pipeline v.4.16.6 [24]. The IMG genome ID is 2818991220.
Phylogenomics and AzeR homology analysis
The amino acid sequence of AzeR of P. nitroreducens DSM 9128 was queried by a blast search against all genomes in the US Department of Energy IGM/M database. All hits with greater than 60 % amino acid sequence identity were considered matches, and the species associated with each protein were taken for further comparison (n=192). Overall, our methodology compared sequence variation of 30 COGs (Clusters of Orthologous Genes) that are considered universal single-copy genes [25] to calculate the evolutionary distance between the species. Of the 192 species matches to AzeR, poor-quality genomes that were missing universal COGs as well as those with more than one copy of COG genes were removed from the analysis, with a final list of 143 genomes continuing in the pipeline. The COGs from each species were aligned using Clustal Omega, with FASTA-formatted output. Each alignment was trimmed using TrimAl v1.3 [26] using the ‘automated1’ flag. The COGs from each species were concatenated, all in the same order. These 143 sequences were inputted into fasttree 2.1.10 [27] using standard settings. The outputted tree was visualized using Interactive Tree of Life [28]. E. coli K12 MG1655 served as an outgroup. The same analysis was performed for comparison of relatives of P. nitroreducens , using the marked COGs indicated in Table 3.
Table 3.
COGs used for P. nitroreducens phylogenomics and AzeR phylogenetic tree
|
COG0012* |
|
COG0016 |
|
COG0018 |
|
COG0049* |
|
COG0052* |
|
COG0080* |
|
COG0081 |
|
COG0087* |
|
COG0088* |
|
COG0090* |
|
COG0092* |
|
COG0093* |
|
COG0094* |
|
COG0096* |
|
COG0097* |
|
COG0098* |
|
COG0103* |
|
COG0124 |
|
COG0172 |
|
COG0185* |
|
COG0186* |
|
COG0197* |
|
COG0200* |
|
COG0201* |
|
COG0202* |
|
COG0215 |
|
COG0256* |
|
COG0495* |
|
COG0522* |
|
COG0525* |
Those with a star *were used for Fig. 1. All COGs in this table were used for Fig. 6.
Nucleotide sequence accession numbers
This whole-genome shotgun project of the P. nitroreducens genome has been deposited at DDBJ/EMBL/GenBank under the accession number VASG00000000. Sequence reads are available in the Sequence Read Archive under accession number SRR9041312.
Results
Draft genome sequence of P. nitroreducens DSM 9128
In order to begin to study the possible regulatory response in bacteria to azelaic acid, the genome of P. nitroreducens DSM 9128 was sequenced, since this strain was isolated and reported as being able to efficiently degrade and utilize azelaic acid as the sole carbon and energy source [10, 11]. We obtained a total of 1 611 521 pairs of reads, which we assembled into 15 contigs with a total length of 6 707 527 bp and an N50 length of 782 887. The G+C content was 65.5 %, similar to that of other sequenced Pseudomonas genomes. Automated annotation of the P. nitroreducens draft genome sequence assigned a total of 6055 candidate protein-coding genes. A total of 6 rRNA and 60 tRNA genes were also identified. The phylogenetic analysis (Fig. 1b) showed that the strain of P. nitroreducens DSM 9128 is most closely related to other strains of P. nitroreducens spp. such as P. nitroreducens strain TX1, which was isolated from a rice field's recycle drainage, and has the ability to metabolize industrial surfactants that pollute aquatic environments [29].
Isolation and identification of genomic transposon mutants affected in azelaic acid degradation in P. nitroreducens DSM 9128
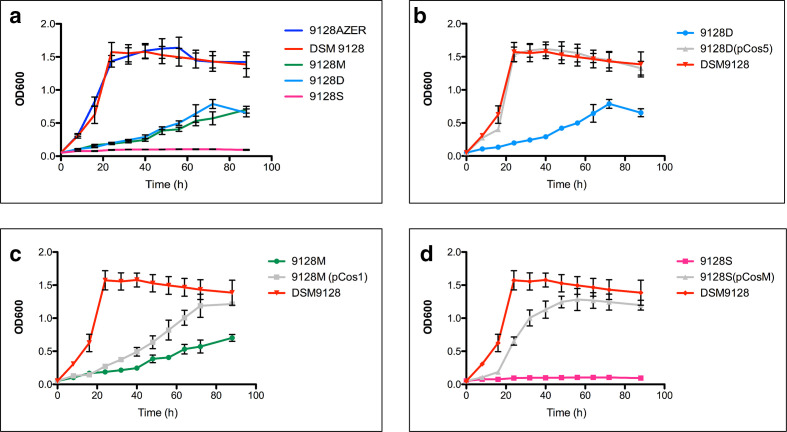
We confirmed that P. nitroreducens strain DSM 9128 can utilize azelaic acid as the sole carbon source since it could grow well in M9 minimal medium supplemented with 0.2 % w/v of azelaic acid as the unique source of carbon and energy. In order to identify the regulatory mechanism(s) in response to azelaic acid a P. nitroreducens transposon mutant bank was constructed and screened for inability to grow on azelaic acid. In total, 5000 transposon mutants of P. nitroreducens DSM 9128 were screened on solid media for their ability to grow on azelaic acid. Six mutants, designated 9128D, 9128E, 9128F, 9128I, 9128M, 9128S, which could not grow on minimal media with azelaic acid but grew on glucose as the sole carbon source were isolated. Mutants 9128S, 9128E, 9128I, and 9128F displayed a complete lack of growth phenotype whereas 9128D and 9128M were characterized by very slow growth compared to the wild-type strain. Growth curves were determined for mutants 9128M, 9128D and 9128S, which represent the three different classes along with the wild-type strain and the results show significant differences in growth (Fig. 2a). It was therefore concluded that the identified mutants most likely had transposon insertions in loci, which were involved in the utilization of azelaic acid as the unique source for carbon in P. nitroreducens 9128.
Fig. 2.
Growth rates of P. nitroreducens DSM 9128 and mutant derivatives. (a) Growth rates of P. nitroreducens DSM 9128 and mutant derivatives 9128M, 9128D, 9128S and 9128AZER grown in M9 minimal media with azelaic acid 0.2 % as the unique source of carbon and energy. (b) Growth rates of P. nitroreducens DSM 9128 and mutant derivative 9128D and 9128D (pCos5) grown in M9 minimal media with azelaic acid 0.2 % as the unique source of carbon and energy. (c) Growth rates of P. nitroreducens DSM 9128 and mutant derivative 9128M and 9128M (pCos1) grown in M9 minimal media with azelaic acid 0.2 % as the unique source of carbon and energy. (d) Growth rates of P. nitroreducens DSM 9128 and mutant derivative 9128S and 9128S (pCosM) grown in M9 minimal media with azelaic acid 0.2 % as the unique source of carbon and energy.
Characterization of the six transposon genomic mutants of P. nitroreducens DSM 9128 affected in azelaic acid utilization
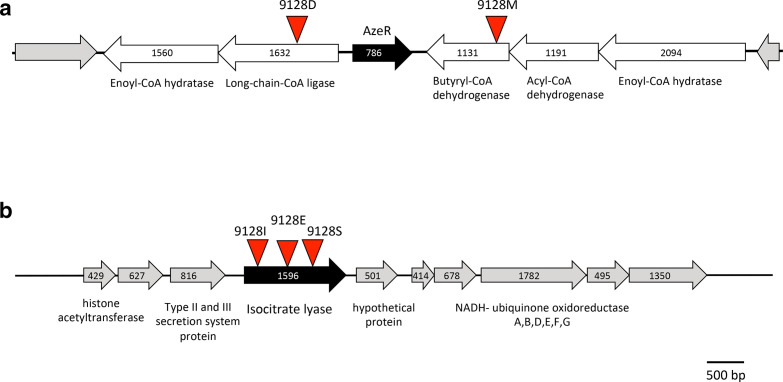
In order to identify the genes in the transposon insertion mutants that were defective in azelaic acid utilization, the location of the transposon in the mutants was mapped. Mutant 9128D had the transposon inserted in a gene encoding for an acyl-CoA ligase for long-chain fatty acids whereas mutant 9128S had the insertion in the isocitrate lyase gene (Fig. 3). Mutants 9128E, 9128F, 9128I and 9128S carried the transposon in an isocitrate lyase gene but in different positions within its nucleotide sequence thus representing independent insertion mutants and strengthening the association of this gene with azelaic acid utilization. The transposon in the mutant 9128M was located in a locus encoding an acyl-CoA dehydrogenase, which was genetically adjacent to the acyl-CoA ligase, which had the transposon insertion of mutant 9128D (Fig. 3b). The acyl-CoA dehydrogenase is part of an operon of three genes and the acyl-CoA ligase is part of an operon with the enoyl-CoA hydratase. The two operons are separated by a transcriptional regulator that belongs to the IclR family (Fig. 3, designated as AzeR).
Fig. 3.
Map of genetic loci harbouring the Tn5 mutations of the identified mutants. (a) Genetic locus harbouring the Tn5 mutants 9128D and 9128M, which were affected in growth in azelaic acid as the unique source of carbon. The position of the Tn5 is shown by a red triangle, the positions of the ORFs are shown and the encoding gene product is described below the open arrows. The AzeR IclR-family transcriptional regulator is indicated by a black-filled arrow. (b) Genetic locus harbouring the Tn5 mutants 9128E, 9128I and 9128S, which were affected in growth in azelaic acid as the unique source of carbon. The position of the Tn5 is shown by a red triangle, the positions of the ORFs are shown and the encoding gene product is described below the open arrows.
Next, we tried to complement the mutations. A pLAFR3 cosmid genomic library of P. nitroreducens DSM9128 was constructed as described in Methods. The cosmid colonies harboured in E. coli were then hybridized with a labelled DNA probe of the iclR regulatory gene located in between the two operons and four cosmids were identified and designated pCos1, pCos5, pCosM and pCosG, which contained the genomic loci harbouring the genes of interest.
It was then determined that pCos5 complemented the defect of growth phenotype on azelaic acid displayed by the acyl-CoA ligase 9128D mutant (Fig. 2b); pCos1 and pCos5 could significantly complement (P<0.001) the acyl-CoA dehydrogenase 9128M (Fig. 2c) mutant and pCosM complemented the isocitrate lyase mutant 9128S (Fig. 2d). It was therefore concluded that the phenotypes observed by the transposon mutants were due to the loci in which the insertion occurred.
AzeR is an IclR family repressor that responds to azelaic acid
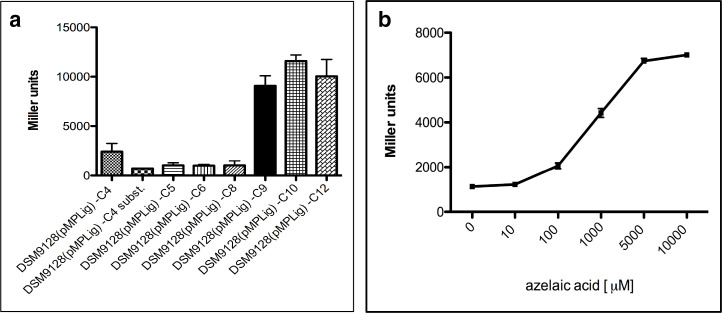
The main aim of this work was to study the regulation of genes in response to azelaic acid. The gene promoters regulating the transcription of the operons harbouring the acyl-CoA ligase and acyl-CoA dehydrogenase and the gene promoter of the isocitrate lyase were cloned in a promoter probe vector, which harboured a promoterless β-galactosidase (lacZ) gene. P. nitroreducens DSM9128 harbouring the plasmids constructs were assayed for promoter activity and it was established that the presence of 0.1 % w/v of azelaic acid dramatically induced the transcription from the promoters controlling the expression of the operons containing the acyl-CoA dehydrogenase and of the acyl-CoA ligase (Fig. 4a and b). If other very closely related dicarboxylic acids were used instead, like suberic, sebacic and adipic acid (Fig. 1a), no activation of the two promoters was observed indicating that the response was specific for azelaic acid (Fig. 5a). Intergenically located between the operons, which harbour two loci identified in this study, is a locus encoding a putative iclR family transcriptional regulator (Fig. 3a); it was therefore postulated that this regulator, which we designated as AzeR, might play a role in their regulation. A genomic knock-out mutant of this regulator was constructed, which we designated as strain 9128AZER. The gene promoter assays were then performed in the wild-type strain and the P. nitroreducens 9128AZER establishing that the two promoters displayed significantly high constitutive expression even in the absence of azelaic acid (Fig. 4a and b). It was therefore concluded that AzeR behaved as a negative transcriptional regulator, which dramatically de-repressed transcription of the two operons in the presence of azelaic acid. We also determined whether AzeR was autoregulating its own expression; gene promoter studies showed that it was negatively autoregulating its own transcription in response to azelaic acid (Fig. 4c). The isocitrate lyase promoter on the other hand did not respond to azelaic acid and it was active and constitutive in all the conditions tested (Fig. 4d). With the aim to understand at which concentration of azelaic acid the AzeR de-repressed the operon promoters, a calibration curve in response to different concentrations of azelaic acid was determined. The results obtained revealed that the genes involved in the azelaic acid degradation pathway were strongly regulated by the azelaic acid, starting from a concentration higher than 10 µM (Fig. 5b).
Fig. 4.
Gene-promoter studies in P. nitroreducens . (a) β-galactosidase assays in P. nitroreducens DSM 9128, 9128AZER and 9128AZER(pAzeR) harbouring the gene promoter regulating the transcription of the acyl-CoA ligase (inactivated by transposon 9128D) fused to a promoterless lacZ in plasmid construct pMPLig. All strains were grown in minimal M9 media contain either 0.2 % glucose (M9 GLU) or azelaic acid (M9 AZ) as the unique source of carbon and energy. (b) β-galactosidase assays in P. nitroreducens DSM 9128, 9128AZER and 9128AZER(pAzeR) harbouring the gene promoter regulating the transcription of the acyl-CoA dehydrogenase (inactivated by transposon 9128M) fused to a reportless lacZ in plasmid construct pMPDeh. All strains were grown in minimal M9 media containing either 0.2 % glucose (M9 GLU) or azelaic acid (M9 AZ) as the unique source of carbon and energy. (c) β-galactosidase assays in P. nitroreducens DSM 9128, 9128AZER and 9128AZER(pAzeR) harbouring the gene promoter regulating the transcription of the azeR regulator gene fused to a reportless lacZ in plasmid construct pMPazeR. All strains were grown in minimal M9 media containing either 0.2 % glucose (M9 GLU) or azelaic acid (M9 AZ) as the unique source of carbon and energy. (d) β-galactosidase assays in P. nitroreducens DSM 9128, 9128AZER and 9128AZER(pAzeR) harbouring the gene promoter regulating the transcription of the isocitrate lyase fused to a reportless lacZ in plasmid construct pMPIsocit. All strains were grown in minimal M9 media containing either 0.2 % glucose (M9 GLU) or azelaic acid (M9 AZ) as the unique source of carbon and energy. The data are means of three repeats, error bars indicate standard deviation and and t-test was performed (*P<0.05, **P<0.01, ***P<0.001), using Prism 7 (GraphPad Software).
Fig. 5.
Gene promoter studies in P. nitroreducens grown in the presence of different dicarboxylic acids. (a) β-galactosidase assays in P. nitroreducens DSM 9128 harbouring the gene promoter regulating the transcription of the acyl-CoA ligase (fused to a reportless lacZ in plasmid construct pMPLig) grown in minimal M9 medium in the presence of different dicarboxylic acids at a concentration of 0.2 %; C4 is succinic acid, C4 subst. is oxaloacetic acid, C5 is glutaric acid, C6 is adipic acid, C8 is suberic acid, C9 is azelaic acid, C10 is sebacic acid and C12 is dodecanedioic acid. (b) β-galactosidase assays in P. nitroreducens DSM 9128 harboring the gene promoter regulating the transcription of the acyl-CoA ligase (inactivated by transposon 9128D) fused to a reportless lacZ in a plasmid construct pMPLig in M9 minimal medium with azelaic acid as the unique source of carbon and energy in different concentrations.
AzeR is a novel bacterial regulator
AzeR belongs to the IclR family of bacterial transcriptional regulators, which consists of both activators and repressors [30]. These regulators are approximately 250 amino acids long and possess two domains: a DNA-binding helix-turn-helix domain at their N-terminus and an effector/inducer domain at their C-terminus. IclR-family regulators have been implicated in the regulation of many different loci, which are involved in degradation of aromatic compounds, in the glyoxylate cycle, in multidrug resistance and in virulence [30]. In order to establish the presence of orthologues and homologues of AzeR in bacteria, the amino acid sequence was compared to available sequences in the databases. Fig. 6 depicts the highly similar proteins (>60 %) in bacteria showing that although AzeR-like proteins occur almost exclusively in Pseudomonas species, they are also found in Marinobacter species. To our knowledge, none of these proteins has yet been studied or their biological role reported making AzeR a new IclR-family regulator in bacteria. The Pseudomonas species that harbour an AzeR-like regulator are known to be able to degrade many compounds for their energy and carbon source and many are plant-associated. Marinobacter are gamma-Proteobacteria that live in seawater and have been isolated and studied for their ability to degrade hydrocarbons [31]. Moreover, azelaic acid is enriched in marine aerosols of high biological activity in the ocean vs. regions of low biological activity (Bikkina et al. [32]). Marinobacter in these aquatic regions may be responding to azelaic acid thorough its AzeR regulon. AzeR therefore is confined to a few proteobacterial genera that are likely to be able to degrade and utilize azelaic acid as the sole source of carbon and energy.
Fig. 6.
AzeR homologies in other bacteria. Phylogenetic tree showing organisms with a strong (>60 % amino acid sequence identity) AzeR homologue. The evolutionary distance between the organisms was determined using concatenated COGs listed in Table 3.
Discussion
The bacterial strain P. nitroreducens DSM 9128 was used as a model for the study of genes necessary for the catabolism of azelaic acid. In bacteria, which utilize fatty acids as a carbon source, azelaic acid is relatively less preferred over other carbon sources [7]. A genetic screen using a genomic transposon library resulted in the identification of a set of mutants, which were unable, or significantly altered, in the ability to utilize azelaic acid as a unique source of carbon and energy. Mutants 9128D and 9128M (in an acyl-CoA ligase, an acyl-CoA dehydrogenase genes, respectively) that showed slow growth phenotype can possibly have mutated a functionally redundant gene for the degradation of azelaic acid. The total absence of growth on azelaic acid observed in mutants 9128S, 9128E, 9128I and 9128F, which are null mutants in an isocitrate lyase gene, indicated that the locus of the transposon insertion is absolutely necessary for this catabolic pathway.
The acyl-CoA ligase catalyses the pre-step reaction of β-oxidation by joining the acyl-CoA to the fatty acids that activates the breakdown of the molecule [33]. The acyl-CoA dehydrogenase enzyme on the other hand, catalyses the second step of β-oxidation of fatty acids and in particular transfers electrons from the substrate to the cofactor FAD in order to form a molecule of enoyl-CoA acting as a substrate for subsequent reactions [34]. Inactivation of these two genes in P. nitroreducens DSM 9128 results in the bacterium being significantly less efficient in the catabolism of azelaic acid (C9 dicarboxylic acid), as well as suberic acid (C8) and sebacic acid (C10). From in silico analysis it was observed that genes encoding for the acyl-CoA ligase and for the acyl-CoA dehydrogenase are part of a cluster of genes encoding for other enzymes in the pathway of β-oxidation (Fig. 3a). Specifically, these genes are organized into two operons: one is composed of three genes, an enoyl-CoA hydratase, an acyl-CoA dehydrogenase and a long-chain fatty acid acyl-CoA dehydrogenase (this last locus identified and studied in this work) and the second one is composed of two genes, the long-chain fatty acid acyl-CoA ligase identified in this study and an enoyl-CoA hydratase. These two operons are separated by a nucleotide sequence encoding for a transcriptional regulator belonging to the IclR family.
Previous studies made in E. coli and in Rhodopseudomonas palustris suggest the presence in the genomes of these bacteria of a number of enzymes involved in β-fatty acid oxidation; some are organized in an operon called fad operon in E. coli and the pim operon in R. palustris [8, 35] both encoding for five enzymes involved in long and medium fatty acid degradation, reminiscent of the ones found in P. nitroreducens DSM 9128 reported here. The organization of the loci and the sequence of individual genes do not show however a particularly high homology.
An iclR family regulator is located adjacent to the two operons involved in azelaic acid degradation (see above). We established that this regulator in P. nitroreducens, which we designated as AzeR, is involved in the stringent negative regulation of the two adjacent operons in response to azelaic acid. As far as we know, this is the first transcriptional regulator responding to azelaic acid, an important molecule involved in plant–microbe interactions. The fact that it responds very poorly to other closely related dicarboxylic acids (Fig. 5) indicates that P. nitroreducens has evolved the ability to specifically respond to the presence of azelaic acid and via AzeR de-repress the transcription of the two operons (Fig. 5). AzeR also negatively auto-regulates its own transcription in response to azelaic acid (Fig. 5); auto-regulation of transcription factors is a common trait in bacteria since it allows a more efficient response to the stimulus affecting the activity of the regulator. Interestingly AzeR homologes are only found in Pseudomonas (homology from 70–99 %) and Marinobacter sp. (homology from 60–70 %); it remains to be established if these species are able to utilize azelaic acid as the carbon source and if AzeR is the regulator which is responsible for the regulation of the genetic loci. The ability to respond to azelaic acid by these two genera indicates that they encounter it in the environment that they colonize. As AzeR is an IclR-family regulator, which specifically responds to azelaic acid and de-represses the transcriptional regulation, it is likely that it binds azelaic at its C-terminus and this results in AzeR being unable to bind to DNA through its helix-turn-helix N-terminus DNA-binding domain, thereby allowing transcription of the target genes [30]. Biochemical and structural studies are needed to confirm this likely model of regulation by AzeR.
The isocitrate lyase mutant is characterized by the total lack of growth of P. nitroreducens with azelaic acid as the unique source of carbon and energy. This is an enzyme of the glyoxylate cycle, which acts within the TCA cycle allowing it to skip the last steps of the TCA cycle [36]. The growth on fatty acids requires the induction of this glyoxalate bypass since it prevents the loss of carbon as CO2. The absolute requirement of isocitrate lyase indicates that azelaic degradation also occurs via the glyoxalate shunt. The isocitrate lyase is not regulated by AzeR or induced by the presence of azelaic acid and displayed constitutive expression during all the conditions tested in this study.
In summary, this study has revealed the presence of an IclR family repressor in bacteria, designated as AzeR, which responds to azelaic acid and de-represses transcription of two operons involved in the degradation of azelaic acid. AzeR is widespread only in Pseudomonas spp. and in Marinebacter indicating that these two genera have evolved the ability to sense the presence and respond to azelaic acid and regulate gene expression and this might be an important train in plant–microbe interaction.
Funding information
This work received no specific grant from any funding agency.
Acknowledgements
C.B. and S.G.J. are beneficiary of an ICGEB fellowship. We thank ICGEB for supporting this work. The bacterial P. nitroreducens strain DSM9128 sequenced in this work was obtained from the DSMZ collection and was isolated by Janot-Bassalik and Wright as cited in this work.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: AZ, azelaic acid; GLU, glucose.
Edited by: D. Grainger and D. Lee
References
- 1.Shine MB, Xiao X, Kachroo P, Kachroo A. Signaling mechanisms underlying systemic acquired resistance to microbial pathogens. Plant Sci. 2019;279:81–86. doi: 10.1016/j.plantsci.2018.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Siebers M, Brands M, Wewer V, Duan Y, Hölzl G, et al. Lipids in plant-microbe interactions. Biochim Biophys Acta. 2016;1861:1379–1395. doi: 10.1016/j.bbalip.2016.02.021. [DOI] [PubMed] [Google Scholar]
- 3.Khakimov B, Jespersen BM, Engelsen SB. Comprehensive and comparative metabolomic profiling of wheat, barley, oat and rye using gas chromatography-mass spectrometry and advanced chemometrics. Foods. 2014;3:569–585. doi: 10.3390/foods3040569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cecchini NM, Roychoudhry S, Speed DJ, Steffes K, Tambe A, et al. Underground Azelaic Acid–Conferred Resistance to Pseudomonas syringae in Arabidopsis . Mol Plant Microbe Interact. 2019;32:86–94. doi: 10.1094/MPMI-07-18-0185-R. [DOI] [PubMed] [Google Scholar]
- 5.Jung HW, Tschaplinski TJ, Wang L, Glazebrook J, Greenberg JT. Priming in systemic plant immunity. Science. 2009;324:89–91. doi: 10.1126/science.1170025. [DOI] [PubMed] [Google Scholar]
- 6.Javvadi SG, Cescutti P, Rizzo R, Lonzarich V, Navarini L, et al. The spent culture supernatant of Pseudomonas syringae contains azelaic acid. BMC Microbiol. 2018;18:199. doi: 10.1186/s12866-018-1352-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chapman PJ, Duggleby RG. Dicarboxylic acid catabolism by bacteria. Biochem J. 1967;103:7C–9.:7contd-9c. doi: 10.1042/bj1030007C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harrison FH, Harwood CS. The pimFABCDE operon from Rhodopseudomonas palustris mediates dicarboxylic acid degradation and participates in anaerobic benzoate degradation. Microbiology. 2005;151:727–736. doi: 10.1099/mic.0.27731-0. [DOI] [PubMed] [Google Scholar]
- 9.Parke D, Garcia MA, Ornston LN. Cloning and genetic characterization of dca genes required for beta-oxidation of straight-chain dicarboxylic acids in Acinetobacter sp. strain ADP1. Appl Environ Microbiol. 2001;67:4817–4827. doi: 10.1128/AEM.67.10.4817-4827.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janota-Bassalik L, Wright LD. Pimelic Acid as a by-Product of Azelaic Acid Degradation by Pseudomonas sp. Nature. 1964;204:501–502. doi: 10.1038/204501a0. [DOI] [PubMed] [Google Scholar]
- 11.Janota-Bassalik L, Wright LD. Azelaic acid utilization by a Pseudomonas . J Gen Microbiol. 1964;36:405–414. doi: 10.1099/00221287-36-3-405. [DOI] [PubMed] [Google Scholar]
- 12.Lang E, Griese B, Spröer C, Schumann P, Steffen M, et al. Characterization of 'Pseudomonas azelaica' DSM 9128, leading to emended descriptions of Pseudomonas citronellolis Seubert 1960 (Approved Lists 1980) and Pseudomonas nitroreducens Iizuka and Komagata 1964 (Approved Lists 1980), including Pseudomonas multiresinivorans as its later heterotypic synonym. Int J Syst Evol Microbiol. 2007;57:878–882. doi: 10.1099/ijs.0.64849-0. [DOI] [PubMed] [Google Scholar]
- 13.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 14.Alexeyev MF. The pKNOCK series of broad-host-range mobilizable suicide vectors for gene knockout and targeted DNA insertion into the chromosome of gram-negative bacteria. Biotechniques. 1999;26:828. doi: 10.2144/99265bm05. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: a Laboratory Manual. 2nd ed. N.Y: Cold Spring Harbor; 1989. [Google Scholar]
- 16.Miller JH. Experiments in Molecular Genetics. N.Y: Cold Spring Harbor; 1972. [Google Scholar]
- 17.Better M, Lewis B, Corbin D, Ditta G, Helinski DR. Structural relationships among Rhizobium meliloti symbiotic promoters. Cell. 1983;35:479–485. doi: 10.1016/0092-8674(83)90181-2. [DOI] [PubMed] [Google Scholar]
- 18.Figurski DH, Helinski DR. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans . Proc Natl Acad Sci U S A. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reeve WG, Tiwari RP, Worsley PS, Dilworth MJ, Glenn AR, et al. Constructs for insertional mutagenesis, transcriptional signal localization and gene regulation studies in root nodule and other bacteria. Microbiology. 1999;145:1307–1316. doi: 10.1099/13500872-145-6-1307. [DOI] [PubMed] [Google Scholar]
- 20.Simon R, Priefer U, Pühler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in Gram negative bacteria. Biotechnology. 1983;1:784–791. doi: 10.1038/nbt1183-784. [DOI] [Google Scholar]
- 21.O'Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 22.Stachel SE, An G, Flores C, Nester EW. A Tn3 lacZ transposon for the random generation of beta-galactosidase gene fusions: application to the analysis of gene expression in Agrobacterium . EMBO J. 1985;4:891–898. doi: 10.1002/j.1460-2075.1985.tb03715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen I-MA, Chu K, Palaniappan K, Pillay M, Ratner A, et al. IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Res. 2019;47:D666–D677. doi: 10.1093/nar/gky901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Creevey CJ, Doerks T, Fitzpatrick DA, Raes J, Bork P. Universally distributed single-copy genes indicate a constant rate of horizontal transfer. PLoS One. 2011;6:e22099. doi: 10.1371/journal.pone.0022099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price MN, Dehal PS, Arkin AP. FastTree 2-approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Letunic I, Bork P. Interactive tree of life (iTOL) V3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen H-J, Guo G-L, Tseng D-H, Cheng C-L, Huang S-L. Growth factors, kinetics and biodegradation mechanism associated with Pseudomonas nitroreducens TX1 grown on octylphenol polyethoxylates. J Environ Manage. 2006;80:279–286. doi: 10.1016/j.jenvman.2005.09.009. [DOI] [PubMed] [Google Scholar]
- 30.Molina-Henares AJ, Krell T, Eugenia Guazzaroni M, Segura A, Ramos JL. Members of the IclR family of bacterial transcriptional regulators function as activators and/or repressors. FEMS Microbiol Rev. 2006;30:157–186. doi: 10.1111/j.1574-6976.2005.00008.x. [DOI] [PubMed] [Google Scholar]
- 31.Fathepure BZ. Recent studies in microbial degradation of petroleum hydrocarbons in hypersaline environments. Front Microbiol. 2014;5:173. doi: 10.3389/fmicb.2014.00173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bikkina S, Kawamura K, Miyazaki Y, Fu P. High abundances of oxalic, azelaic, and glyoxylic acids and methylglyoxal in the open Ocean with high biological activity: implication for secondary oa formation from isoprene. Geophys Res Lett. 2014;41:3649–3657. doi: 10.1002/2014GL059913. [DOI] [Google Scholar]
- 33.Weimar JD, DiRusso CC, Delio R, Black PN. Functional role of fatty acyl-coenzyme A synthetase in the transmembrane movement and activation of exogenous long-chain fatty acids. Amino acid residues within the ATP/AMP signature motif of Escherichia coli FadD are required for enzyme activity and fatty acid transport. J Biol Chem. 2002;277:29369–29376. doi: 10.1074/jbc.M107022200. [DOI] [PubMed] [Google Scholar]
- 34.Klein K, Steinberg R, Fiethen B, Overath P. Fatty acid degradation in Escherichia coli. An inducible system for the uptake of fatty acids and further characterization of old mutants. Eur J Biochem. 1971;19:442–450. doi: 10.1111/j.1432-1033.1971.tb01334.x. [DOI] [PubMed] [Google Scholar]
- 35.My L, Rekoske B, Lemke JJ, Viala JP, Gourse RL, et al. Transcription of the Escherichia coli fatty acid synthesis operon fabHDG is directly activated by FadR and inhibited by ppGpp. J Bacteriol. 2013;195:3784–3795. doi: 10.1128/JB.00384-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dolan SK, Welch M. The glyoxylate shunt, 60 years on. Annu Rev Microbiol. 2018;72:309–330. doi: 10.1146/annurev-micro-090817-062257. [DOI] [PubMed] [Google Scholar]
- 37.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spaink HP, Okker RJ, Wijffelman CA, Pees E, Lugtenberg BJ. Promoters in the nodulation region of the Rhizobium leguminosarum Sym plasmid pRL1JI. Plant Mol Biol. 1987;9:27–39. doi: 10.1007/BF00017984. [DOI] [PubMed] [Google Scholar]