Abstract
Mass spectrometry (MS)-based sequencing approaches have been shown to be useful in direct sequencing RNA without the need for a complementary DNA (cDNA) intermediate. However, such approaches are rarely applied as a de novo RNA sequencing method, but used mainly as a tool that can assist in quality assurance for confirming known sequences of purified single-stranded RNA samples. Recently, we developed a direct RNA sequencing method by integrating a 2-dimensional mass-retention time hydrophobic end-labeling strategy into MS-based sequencing (2D-HELS MS Seq). This method is capable of accurately sequencing single RNA sequences as well as mixtures containing up to 12 distinct RNA sequences. In addition to the four canonical ribonucleotides (A, C, G, and U), the method has the capacity to sequence RNA oligonucleotides containing modified nucleotides. This is possible because the modified nucleobase either has an intrinsically unique mass that can help in its identification and its location in the RNA sequence, or can be converted into a product with a unique mass. In this study, we have used RNA, incorporating two representative modified nucleotides (pseudouridine (ψ) and 5-methylcytosine (m5 C)), to illustrate the application of the method for the de novo sequencing of a single RNA oligonucleotide as well as a mixture of RNA oligonucleotides, each with a different sequence and/or modified nucleotides. The procedures and protocols described here to sequence these model RNAs will be applicable to other short RNA samples (<35 nt) when using a standard high-resolution LC-MS system, and can also be used for sequence verification of modified therapeutic RNA oligonucleotides. In the future, with the development of more robust algorithms and with better instruments, this method could allow sequencing of more complex biological samples.
Introduction
Mass spectrometry (MS)-based sequencing methods, including top-down MS and tandem MS1, 2, 3, 4 , have been developed for direct sequencing of RNA. However, in situ fragmentation techniques for effectively generating high-quality RNA ladders in mass spectrometers currently can not be applied to de novo sequencing5, 6. Furthermore, it is not very trivial to analyze the traditional one-dimensional (1D) MS data for de novo sequencing of even one purified RNA sequence, and it would be even more challenging for MS sequencing of mixed RNA samples7, 8. Therefore, a two-dimensional (2D) liquid chromatography (LC)-MS-based RNA sequencing method has been developed, incorporating production of 2D mass-retention time (tR) ladders to replace 1D mass ladders, making it much easier to identify ladder components needed for de novo sequencing of RNAs8. However, the 2D LC-MS-based RNA sequencing method is mainly limited to purified synthetic short RNA, as it cannot read a complete sequence solely based on one single ladder, but must rely on two co-existing adjacent ladders (5′- and 3′-ladders)8. More specifically, this approach requires bidirectional paired-end reads for reading terminal nucleobases in the low-mass region8. The added complexity of the paired-end reading results in this method being untenable for sequencing of RNA mixtures because confusion is raised on which ladder fragment belongs to which ladder for the unknown samples.
To overcome the abovementioned barriers in MS-based RNA sequencing approaches and to broaden such applications in direct RNA sequencing, two issues must be addressed: 1) how to generate a high-quality mass ladder that can be used to read a complete sequence, from the first nucleotide to the last in an RNA strand, and 2) how to effectively identify each RNA/mass ladder in a complex MS dataset. Together with well-controlled acid degradation, we have developed a new sequencing method by introducing a hydrophobic end labeling strategy (HELS) into the MS-based sequencing technique, and successfully addressed these two issues by adding a hydrophobic tag at either 5′- and/or 3′-end of the RNAs to be sequenced9. This method creates an “ideal” sequence ladder from RNA—each ladder fragment derives from site-specific RNA cleavage exclusively at each phosphodiester bond, and the mass difference between two adjacent ladder fragments is the exact mass of either the nucleotide or nucleotide modification at that position 8, 9, 10. This is possible because we include a highly controlled acidic hydrolysis step, which fragments the RNA, on average, once per molecule, before it is injected into the instrument. As a result, each degradation fragment product is detected on the mass spectrometer and all fragments together form a sequencing ladder8, 9, 10. This new strategy enables complete reading of an RNA sequence from one single ladder of an RNA strand without paired-end reading from the other ladder of the RNA, and additionally allows MS sequencing of RNA mixtures with multiple different strands that contain combinatorial nucleotide modifications9. By adding a tag at the 5′- and/or 3′-end of the RNA, the labeled ladder fragments display a significant delay of tR, which can help to distinguish the two mass ladders from each other and also from the noisy low-mass region. The mass-tR shift caused by adding the hydrophobic tag facilitates mass ladder identification and simplifies data analysis for sequence generation. Furthermore, the addition of the hydrophobic tag can help to identify the terminal base in the strand by preventing its corresponding ladder fragment from being in the noisy low-mass-tR region due to the mass and hydrophobicity increase caused by the tag, thus allowing identification of the complete sequence of an RNA from a single ladder; no paired-end reads are required. As a result, we have previously demonstrated the successful sequencing of a complex mixture of up to 12 RNA distinct strands without the use of any advanced sequencing algorithm9, which opens the door for de novo MS sequencing of RNA containing both canonical and modified nucleotides and makes it more feasible for the sequencing of mixed and more complex RNA samples. In fact, using 2D-HELS MS Seq, we have even successfully sequenced a mixed population of tRNA samples10 and are actively expanding its application to other complex RNA samples.
To facilitate 2D-HELS MS Seq to directly sequence a broader range of RNA samples, here we will focus on the technical aspects of this sequencing approach and will cover all of the essential steps needed when applying the technique towards direct sequencing of RNA samples. Specific examples will be used to illustrate the sequencing technique, including synthetic single RNA sequences, mixtures of multiple distinct RNA sequences, and modified RNAs containing both canonical and modified nucleotides such as pseudouridine (ψ) and 5-methylcytosine (m5C). Since RNAs all contain phosphodiester bonds, any type of RNA can be acid-hydrolyzed to generate an ideal sequence ladder for 2D-HELS MS Seq under optimal conditions8, 9. However, detection of all ladder fragments of a given RNA is instrument dependent. On a standard high-resolution LC-MS (40K), the minimal loading amount for sequencing a purified short RNA sample (<35 nt) is 100 pmol per run. However, more material is required (up to 400 pmol per RNA sample) when additional experiments must be conducted (e.g., to distinguish isomeric base modifications that share identical masses). The protocol used in sequencing the model synthetic modified RNAs will also be applicable to sequencing broader RNA samples, including biological RNA samples with unknown base modifications. However, an even larger sample amount, such as 1000 pmol for sequencing tRNA (~76 nt) using a standard LC-MS instrument, is required to sequence the complete tRNA with all the modifications, and an advanced algorithm must be developed for its de novo sequencing10.
Protocol
1. Design RNA oligonucleotides
-
Design synthetic RNA oligonucleotides with different lengths (19 nt, 20 nt and 21 nt), including one (RNA #6) with both canonical and modified nucleotides, ψ is employed as a model for non-mass-altering modifications, which is challenging for MS sequencing because it has an identical mass to U. m5 C is chosen as a model for mass-altering modifications to demonstrate the robustness of the approach.
RNA #1: 5′-HO-CGCAUCUGACUGACCAAAA-OH-3′
RNA #2: 5′-HO-AUAGCCCAGUCAGUCUACGC-OH-3′
RNA #3: 5′-HO-AAACCGUUACCAUUACUGAG-OH-3′
RNA #4: 5′-HO-GCGUACAUCUUCCCCUUUAU-OH-3′
RNA #5: 5′-HO-GCGGAUUUAGCUCAGUUGGGA-OH-3′
RNA #6: 5′-HO-AAACCGUψACCAUUAm5 CUGAG-OH-3′
Dissolve each synthetic RNA in nuclease-free diethyl pyrocarbonate (DEPC)-treated water (expressed as DEPC-treated H2O unless otherwise indicated) to obtain a 100 mM RNA stock solution. Stock solutions are stored long-term at −20 °C.
To avoid possible RNA sample degradation, use RNase-free experimental supplies including DEPC-treated water, microcentrifuge tubes, and pipette tips. Frequently wipe down surfaces of lab supplies using RNase elimination wipes.
2. Label the 3′-end of RNAs with biotin
1. Two-step reaction protocol (adenylation and ligation)
-
Add 1 μL of 10x adenylation reaction buffer containing 50 mM sodium acetate, pH 6.0, 10 mM MgCl2, 5 mM dichlorodiphenyltrichloroethane (DTT), 0.1 mM ethylenediaminetetraacetic acid (EDTA), 1 μL of 1 mM ATP, 1 μL of 100 μM biotinylated cytidine bisphosphate (pCp-biotin), 1 μL of 50 μM Mth RNA ligase, and 6 μL of DEPC-treated H2O (a total volume of 10 μL) into an RNase-free thin-walled 0.2 mL PCR tube.
NOTE: Store the reagents at −20 °C before the two-step reaction. Thaw the reagents at room temperature and mix well by vortexing and centrifuging before adding to the reaction.
Incubate the reaction in a PCR machine at 65 °C for 1 h and inactivate the reaction at 85 °C for 5 min.
-
Conduct the ligation step in an RNase-free, thin walled 0.2 mL PCR tube containing 10 μL of reaction solution from the previous step by adding 3 μL of 10x T4 RNA ligase reaction buffer containing 50 mM tris(hydroxymethyl)aminomethane (Tris)-HCl, pH 7.8, 10 mM MgCl2, 1 mM DTT, 1.5 μL of the 100 mM sample stock of the RNA to be sequenced, 3 μL of anhydrous dimethyl sulfoxide (DMSO) to reach 10% (v/v), 1 μL of T4 RNA ligase (10 units/μL), and 11.5 μL of DEPC-treated H2O (for a total volume of 30 mL). Incubate the reaction overnight at 16 °C in a PCR machine.
NOTE: Combine reaction components at room temperature due to the high freezing point of DMSO (18.45 °C).
Incubate the reaction overnight at 16 °C.
Quench and purify the reaction by column purification to remove enzymes and free pCp-biotin using Oligo Clean & Concentrator (Zymo Research, Irvine, CA, USA). Oligo Binding Buffer, DNA Wash Buffer, spin columns and collection tubes are provided in the kit. Add 20 mL of DEPC-treated H2O to the reaction solution to reach a 50 mL sample volume prior to adding the Binding Buffer.
Add 100 mL of binding buffer to each reaction solution. Add 400 μL of ethanol, mix by pipetting, and transfer the mixture to the column. Centrifuge at 10,000 x g for 30 s. Discard the flow-through.
Add 750 μL of DNA Wash Buffer to the column. Centrifuge at 10,000 x g and maximum speed for 30 s and 1 minute, respectively.
-
Transfer the column to a 1.5 mL microcentrifuge tube. Add 15 μL of DEPC-treated H2O to the column and centrifuge at 10,000 x g for 30 s to elute the RNA product.
NOTE: Samples can be stored at −20 °C at this stage until the next step is performed.
2. One-step reaction protocol
Perform a one-step labeling reaction by combining 2 μL of 150 μM adenosine-5′-5′-diphosphate-{5′-(cytidine-2′-O-methyl-3′-phosphate-TEG}C-biotin (AppCp-biotin), 3 μL of 10x ligase reaction buffer, 1.5 μL of the 100 mM sample stock of the RNA to be sequenced, 3 μL of anhydrous DMSO to reach 10% (v/v), 1 μL of T4 RNA ligase (10 units/μL), and 19.5 μL of DEPC-treated H2O (for a total volume of 30 mL) in a 1.5 mL RNase-free microcentrifuge tube.
Incubate the reaction overnight at 16 °C in a PCR machine.
-
Perform column purification as described above in steps 2.1.5-2.1.8.
NOTE: Prepare a separate/exclusive reaction tube for each RNA sample (150 pmol scale of RNA). Labeling of the 5′-end of the RNA(s) with sulfo-Cyanine3 (Cy3) or Cy3 may be needed (e.g., for bidirectional sequencing verification). The method is different than that of 3′-biotinylation and is described in a previous publication9.
3. Capture biotinylated RNA sample on streptavidin beads
Activate 200 ¼L of streptavidin C1 magnet beads by adding 200 μL of 1x B&W buffer (5 mM Tris-HCl, pH 7.5, 0.5 mM EDTA, 1 M NaCl) in a 1.5 mL RNase-free microcentrifuge tube. Vortex this solution and place it on a magnet stand for 2 min. Then discard the supernatant by carefully pipetting out the solution.
Wash the beads twice with 200 μL of Solution A (DEPC-treated 0.1 M NaOH and DEPC-treated 0.05 M NaCl) and once in 200 μL of Solution B (DEPC-treated 0.1 M NaCl). For each wash step, vortex the solution and place it on a magnet stand for 2 min, followed by discarding of the supernatant. Then add 100 μL of 2x B&W buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 2 M NaCl).
Add 1x B&W buffer to the biotinylated RNA sample until the volume is 100 μL. Then add this solution to the washed beads stored in 100 μL of 2x B&W buffer. Incubate for 30 min at room temperature on a rocking platform shaker at 100 rpm. Place the tube on a magnet stand for 2 min and discard the supernatant.
Wash the coated beads 3 times in 1x B&W buffer and measure the final concentration of supernatant in each wash step by Nanodrop for recovery analysis, to confirm that the target RNA molecules remain on the beads.
-
Incubate the beads in 10 mM EDTA, pH 8.2 with 95% formamide at 65 °C for 5 min in a PCR machine. Keep the tube on the magnet stand for 2 min and collect the supernatant (containing the biotinylated RNAs released from the streptavidin beads) by pipet.
NOTE: This physical separation step prior to acid degradation is only used for sequencing of RNA#1 in Figure 1c, and is not mandatory for the 2D-HELS MS Seq since the hydrophobic biotin label can cause the 3′-labeled ladder fragments to have a significantly delayed tR during LC-MS measurement, which can clearly distinguish the labeled 3′-ladder fragments from the unlabeled 5′-ladder fragments in the 2D mass-tR plot.
Figure 1. 2D-HELS MS Seq of representative RNA samples.
(a) Workflow for 2D-HELS MS Seq. The major steps include 1) hydrophobic tag-labeling of RNA to be sequenced, 2) acid hydrolysis, 3) LC-MS measurement, 4) extraction and analysis of MFE data, and 5) sequence generation via algorithms or manual calculation. (b) Sample preparation protocol including introducing a biotin tag to the 3′-end of RNA for 2D-HELS MS Seq. (c) Separation of the 3′-ladder from the 5′-ladder and other undesired fragments in a 2D mass-retention time (tR) plot based on systematic changes in tRS of 3′-biotin-labeled mass-tR ladder fragments of RNA #1 (19 nt). The sequences are de novo and automatically read out directly by a base-calling algorithm9 . (d) Simultaneous sequencing of 5′-biotin labeled RNA #1 and RNA #2, 19 nt and 20 nt, respectively. Methods for introducing a biotin tag to the 5′-end of RNA are different than that of 3′-biotinylation, and can be found in the previous published protocol9 . The 5′-end of two RNAs (RNA #1 and RNA #2) are biotinylated and their 5′-biotinylated ladders can be easily identified; both 5′-biotinylated ladders are easily separated from their unlabeled 3′-ladders in the 2D mass-tR plot after LC-MS, because the biotinylated ladder components have the larger tR shifts due to the hydrophobicity of the biotin, while unlabeled ladder components are in the lower tR region. Although the 5′-ladders and 3′-ladders co-exist, they do not interfere with the sequence interpretation of two mixed RNA strands. Each sequence of these two RNAs are manually acquired from 5′-biotinylated ladders based on the computational algorithm-processed data8 , 9 . This figure has been modified from Zhang et al.9 .
4. Acid hydrolysis of RNA to generate MS ladders for sequencing
Divide each RNA sample into three equal aliquots. For instance, divide an RNA sample with a volume of 15 μL RNA sample into three aliquots of 5 μL.
Add an equal volume of formic acid to achieve 50% (v/v) formic acid in the reaction mixture8, 9.
Incubate the reaction at 40 °C in a PCR machine, with one reaction running for 2 min, one for 5 min, and one for 15 min, respectively.
Quench the acid degradation by immediately freezing the sample on dry ice after each reaction finishes.
Use a centrifugal vacuum concentrator to dry the sample. The sample is typically completely dried within 30 min, and formic acid is removed together with H2O during the drying process because formic acid has a boiling point (100.8 °C) similar to that of H2O (100 °C).
-
Suspend and combine a total of three dried samples in 20 μL of DEPC-treated H2O for LC-MS measurement.
NOTE: Samples can be stored at −20 °C at this stage while waiting for LC-MS measurement.
5. Convert ψ to CMC-ψ adduct
-
Add 80 μL of DEPC-treated H2O into a 1.5 mL RNase-free microcentrifuge tube containing 0.0141 g of N-cyclohexyl-N'-(2-morpholinoethyl)-carbodiimide metho-p-toluenesulfonate (CMC) and 0.07 g of urea. Add 10 μL of the 100 μM sample stock of the RNA to be sequenced, 8 μL of 1 M bicine buffer (pH 8.3), and 1.28 μL of 0.5 M EDTA. Add DEPC-treated H2O to reach a total volume of 160 μL. Final concentrations are 0.17 M CMC, 7 M urea, and 4 mM EDTA in 50 mM bicine (pH 8.3)11 .
NOTE: This protocol is applicable to either a single synthetic RNA sequence or RNA mixtures.
-
Divide the 160 μL reaction solution into four equal aliquots in RNase-free, thin walled 0.2 mL PCR tubes and incubate at 37 °C for 20 min in a PCR machine.
NOTE: 50 μL per tube is the maximum reaction volume that can be used in a PCR machine.
Quench each reaction with 10 μL of 1.5 M sodium acetate and 0.5 mM EDTA (pH 5.6).
Perform column purification with four parallel spin columns to remove excessive reactants according to the procedure as described in steps 2.1.5-2.1.8. Dissolve the purified product in 15 μL of DEPC-treated H2O in each 1.5 mL RNase-free microcentrifuge tube.
Transfer the purified product to four RNase-free, thin walled 0.2 mL PCR tubes. Add 20 μL of 0.1 M Na2CO3 buffer (pH 10.4) into each 15 μL of purified product and add DEPC-treated H2O to make a final volume of 40 μL for each reaction tube (in total four tubes). Incubate the reaction at 37 °C for 2 h in a PCR machine.
Quench and purify the reaction by column purification with four parallel spin columns as described in step 2.1.5. Elute the CMC-ψ converted product to a 1.5 mL RNase-free microcentrifuge tube each with 15 μL of DEPC-treated H2O.
Combine the purified CMC-ψ converted sample from four collection tubes into one tube. Perform formic acid degradation 50% (v/v) according to the procedures as described in steps 4.1-4.6 to generate MS ladders for sequencing.
6. LC-MS measurement
Prepare mobile phases for LC-MS measurement. Mobile phase A is 25 mM hexafluoro-2-propanol with 10 mM diisopropylamine in LC-MS grade water; mobile phase B is methanol.
Transfer the sample to LC-MS sample vial for analysis. Each sample injection volume is 20 μL containing 100-400 pmol of RNA.
-
Use the following LC conditions: column temperature of 35 °C, flow rate of 0.3 mL/min; a linear gradient from 2–20% mobile phase B over 15 min followed by a 2 min wash step with 90% mobile phase B.
NOTE: For more hydrophobic end-labels such as Cy3 and sulfo-Cy3 as mentioned in Section 2, a higher percentage of organic solvent may be necessary for sample elution (i.e., a similar gradient can be used but with an increased percentage range of mobile phase B). For instance, 2–38% mobile phase B over 30 min with a 2 min wash step with 90% mobile phase B.
Separate and analyze samples on an Agilent Q-TOF (Quadrupole Time-of-Flight) mass spectrometer coupled to an LC system equipped with an autosampler and an MS HPLC (High Performance Liquid Chromatography) system. The LC column is a 50 mm x 2.1 mm C18 column with a particle size of 1.7 μm. Use the following MS settings: negative ion mode; range, 350 m/z to 3200 m/z; scan rate, 2 spectra/s; drying gas flow, 17 L/min; drying gas temperature, 250 °C; nebulizer pressure, 30 psig; capillary voltage, 3500 V; and fragmentor voltage, 365 V. Please note that these parameters are specific to the type or model of mass spectrometer being used.
-
Acquire data with Agilent MassHunter acquisition software. Use Agilent molecular feature extraction (MFE) workflow to extract compound information including mass, retention time, volume (the MFE abundance for the respective ion species), and quality score, etc. Use the following MFE settings: “centroid data format, small molecules (chromatographic), peak with height ≥ 100, up to a maximum of 1000, quality score ≥ 50”.
NOTE: Optimize MFE settings to extract as many potential compounds as possible, up to a maximum of 1000, with quality scores of ≥ 50.
7. Automate RNA sequence generation by a computational algorithm
NOTE: This procedure is shown only for RNA #1 in Figure 1c.
Sort MFE extracted compounds in order of decreasing volume (peak intensity) and tR. Perform data preselection via 1) setting tR from 4 to 10 min to select the RNA fragments labeled by the biotin, since the tRS of the biotin-labeled mass ladder components are shifted to this tR window (4 min to 10 min), and 2) using an order-of-magnitude higher of input compounds than the number of ladder fragments for algorithm computation to reduce data amount based on volume. For instance, for a 20 nt RNA, 20 labeled mass-tR ladder components will be required for sequencing of the 20 nt RNA, and thus, 200 compounds from MFE data file will be selected based on volume. Please note that the tR window may be different when a different type or model of mass spectrometer is used.
Perform data processing and sequence generation of RNA #1 using a revised version of a published algorithm8. The source codes of the revised algorithm are described previously (https://academic.oup.com/nar/article/47/20/e125/5558343#supplementary-data)9.
In addition to automating sequence generation using the algorithm, manually calculate the mass differences between two adjacent ladder components for base calling. All bases in the RNA can be called manually and matched with the theoretical ones in the RNA nucleotide and modification database8; thus, the complete sequence of the RNA strand can be accurately read out manually, which is used to confirm the accuracy of the algorithm-reported sequence read. More structures of RNA modifications can be found in RNA modification databases12 , and their corresponding theoretical masses are obtained by ChemBioDraw. In Tables S1-S2, the ppm (parts-per-million) mass difference is shown when comparing the observed mass to its theoretical mass for a specific ladder component, and a value less than 10 ppm is considered a good match for each base calling.
8. Sequencing RNA mixtures
Label a mixture of five RNA strands (RNA #1 to #5) at their 3′-ends with A(5′)pp(5′)Cp-TEG-biotin using a one-step protocol described in step 2.2. In a total volume of 150 μL reaction solution, add 15 μL of 10x T4 RNA ligase reaction buffer, 1.5 μL of each RNA strand (100 μM stock of RNA #1 to #5, respectively, for a total volume of 7.5 μL), 10 μL of 150 μM A(5′)pp(5′)Cp-TEG-biotin, 15 μL of anhydrous DMSO, 5 μL of T4 RNA ligase (10 units/μL), and 97.5 μL of DEPC-treated H2O. Equally distribute the reaction solution into five aliquots. Each RNase-free microcentrifuge tube contains 30 μL of reaction solution.
Incubate the reaction overnight at 16 °C in a PCR machine.
Perform column purification according to the procedure as described in steps 2.1.5-2.1.8 with five parallel spin columns. Elute a mixture sample of 3′-biotinylated 5 RNA strands (mixture of RNA #1 to #5) to a 1.5 mL RNase-free microcentrifuge tube each with 15 –L of DEPC-treated H2O.
Combine the purified mixture samples from the five collection tubes into one tube. Perform formic acid degradation according to the procedure described in Section 4.
Measure samples by LC-MS as described in Section 6, and analyze the data using the data analysis software with optimized MFE settings to extract data containing mass, tR, and volume as described in step 6.5. The typical processing and base-calling algorithm is not applied due to the significantly increased data complexity resulting from the mixture. All bases in the RNA of the mixed sample are called manually in a method similar to Section 7.3 and match well with the theoretical ones in the RNA nucleotide and modification database8, thus the complete sequences of all five RNA strands in the mixed sample are accurately read out. In Tables S7-S11, all information is listed including observed mass, tR, volume, quality score and ppm mass difference.
Representative Results
Introducing a biotin tag to the 3′-end of RNA to produce easily-identifiable mass-tR ladders.
The workflow of the 2D-HELS MS Seq approach is demonstrated in Figure 1a. The hydrophobic biotin label introduced to the 3′-end of the RNA (see Section 2) increases the masses and tRS of the 3′-labeled ladder components when compared to those of their unlabeled counterparts. Thus, the 3′-ladder curve is shifted to greater y-axis values (due to the increase in the tRS) and shifted to greater x-axis values (due to the increase in masses) in the 2D mass-tR plot. Figure 1b shows the sample preparation protocol including introducing a biotin tag to the 3′-end of RNA for 2D-HELS MS Seq. Figure 1c demonstrates separation of the 3′-ladder from the 5′-ladder and other undesired fragments on a 2D mass-tR plot based on systematic changes in tRS of the 3′-biotin-labeled mass-tR ladder fragments of RNA #1. The 3′-ladder curve alone gives a complete sequence of RNA #1, and the 5′-ladder curve that does not show a tR shift provides the reverse sequence, but it requires end-pairing for reading the terminal base8. With this strategy of 2D-HELS, end-pairing is not required as reported before and the entire RNA sequence can be read out completely from only one labeled ladder curve8. As such, it is possible to sequence mixed samples containing multiple RNAs, e.g., two RNA strands of different lengths (RNA #1 and RNA #2, 19 nt and 20 nt, respectively) with a 5′-biotin label at each RNA (Figure 1d).
Converting ψ to its CMC-ψ adduct for 2D-HELS MS Seq.
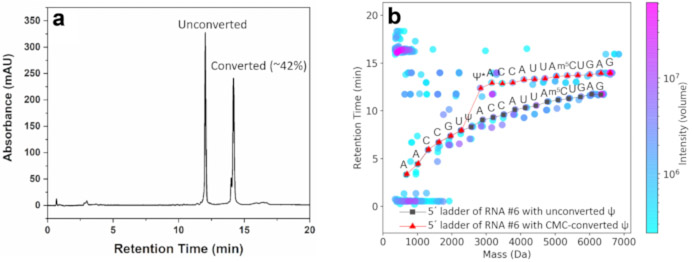
ψ is a difficult nucleotide modification for MS-based sequencing because it has the same mass as uridine (U). To differentiate these two bases from each other, we treat the RNA with CMC, which converts a ψ to a CMC-ψ adduct (see Section 5). The adduct has a different mass than U and can be differentiated in the 2D-HELS MS Seq. Figure 2a shows the HPLC profile of the crude product of the reaction converting ψ to its CMC-adduct in RNA #6. By integrating their UV peaks, we calculated the percent conversion and 42% ψ is converted to its CMC-ψ adduct after the process illustrated in Section 5. After acid degradation and LC-MS measurement, we manually acquired the sequence based on both non-CMC-converted ladders and CMC-converted ladders identified from the algorithm-processed data8 ,9. A red curve branches up off of the grey curve starting from ψ at position 8 in RNA #6 (Figure 2b) due to partial conversion of ψ to the CMC-ψ adduct. Because of the mass and hydrophobicity of the CMC, this conversion results in a 252.2076 Dalton increase in mass and a significant increase in tR for each CMC-ψ adduct-containing ladder component when compared to its unconverted counterpart. Thus, a dramatic shift starting at position 8 in RNA #6 can be observed in the 2D mass-tR plot, indicating that position 8 is indeed a ψ in RNA #6.
Figure 2. Converting pseudouridine (ψ) to its adduct for 2D-HELS MS Seq.
(a) HPLC profile of the crude product of the reaction converting ψ to its CMC adduct in a 20 nt RNA (RNA #6) that contains one ψ. (b) Sequencing of a ψ-containing RNA #6. The conversion of the ψ to the CMC-ψ adducts (ψ*) results in a 252.2076 Dalton increase in mass and a significant increase in tR because of its mass and hydrophobicity of the CMC. Thus, a dramatic shift starting at the position of 8 can be observed in the mass-tR plot, indicating that this is a ψ at the position of 8 in the RNA sequence. The sequences are manually acquired based on the computational algorithm-processed data8 , 9 . This figure has been modified from Zhang et al.9 .
Sequencing RNA mixtures.
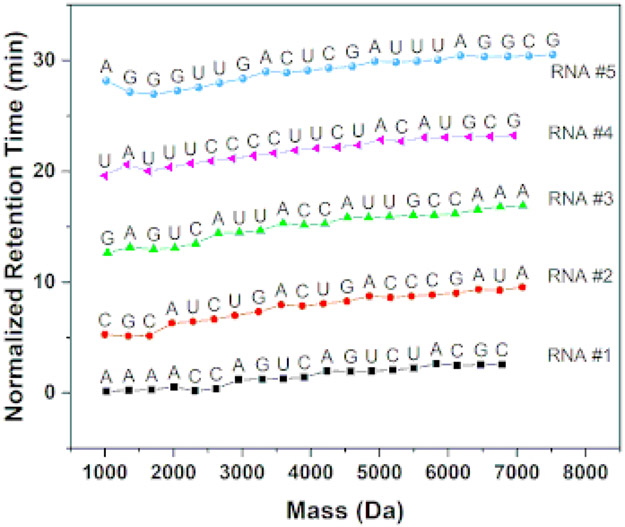
A mixture of five different RNA strands is sequenced by the 2D-HELS MS Seq approach with 3′-end labeling (see Section 8). The concern for sequencing mixed RNAs is that multiple ladder curves in the 2D mass-tR plot may overlap with each other when they all share the same starting points (the hydrophobic tag in the 2D mass-tR plot). However, base calling is made one by one, each based on a mass difference between two adjacent ladder fragments in the MFE data. The correct base-call can be made as long as each mass difference matches well (a PPM MS difference < 10) with one of the theoretical masses of canonical or modified nucleotides in the data pool8 , 9. In the analysis of the multiplexed RNA samples, the typical processing and base-calling algorithm used in Figure 1 and 2 is not used mainly due to the significantly increased data complexity resulting from the mixture. These sequences are base-called manually via calculating the mass difference between two adjacent mass ladder fragments, and comparing it to the theoretical mass of the nucleotide in the data pool9. Any matched base with a mass PPM <10 is chosen as the base identity at this position. With this base-by-base manual calculation for base-calling, all sequences in the mixture are accurately sequenced. OriginLab software is used to re-construct a 2D mass-tR plot, in which the starting tR for each sequence is normalized systematically for better visualizing five different RNA sequences (Figure 3). Without such normalization, the letter codes (i.e., A, C, G, and U) for the sequences of all five RNA would be crowded together on the plot (Figure S1), resulting in less ease of visualization compared to that reported in Figure 3. The sequencing results demonstrate that 2D-HELS MS Seq approach is not just limited to sequencing of purified single-stranded RNAs, but also, more importantly, RNA mixtures with multiple RNA strands. Algorithms are currently under development to automate the process of base-calling and sequence generation.
Figure 3. Sequencing RNA mixtures containing five distinct RNAs.
A biotin is used to label each RNA at their 3′-end before 2D-HELS MS Seq. For each sequence, the starting tR values are normalized systematically to start at 7 min intervals for ease of visualization. The absolute differences between the starting tR value and subsequent tRS remain unchanged for each of the five RNAs, and thus it is easier to visualize each of them in the same plot. All bases are identified by manually calculating the mass differences of two adjacent ladder components and matching them with the theoretical mass differences in the RNA nucleotide and modification database8 ; plots for Figure 3 are re-constructed using OriginLab based on manual base-calling and sequencing data (see Section of Sequencing RNA mixtures in Representative Results). The 2D mass-tR figure of the five mixed RNAs without tR normalization is shown in Figure S1.
Discussion
Unlike tandem-based MS fragmentation, highly controlled acidic hydrolysis is used in the 2D-HELS MS Seq approach to fragment the RNA before analysis with a mass spectrometer9 , 10. As a result, each acid-degraded fragment can be detected by the instrument, forming the equivalent of a sequencing ladder. Under optimal conditions, this method creates an “ideal” sequence ladder from RNA via, on average, one-per-molecule site-specific RNA cleavage exclusively at a phosphodiester bond8 , 9 , 10 . After each degraded fragment is measured by the mass spectrometer in a single run, the mass difference between two adjacent ladder fragments corresponds to the exact mass of the RNA nucleotide or modification at that position. Each RNA modification either has an intrinsic unique mass that can help to identify and locate it in the RNA, or can be converted to one with a unique mass. Thus, in theory, this method can report the identity and location of both canonical and modified nucleotides for de novo and direct sequencing of any RNA. However, different sequence ladders may overlap with each other, complicating MS data analysis and making it difficult for RNA sequencing by MS in practice.
One of the benefits of the 3′-hydrophobic tag is that it overcomes a major challenge in any fragmentation method, i.e., that every RNA molecule must be cleaved into exactly two fragments (and ideally no more): one fragment containing the original 5′-end, and the other containing the original 3′-end of the RNA. Therefore, each cleavage event produces two fragments, producing two ladders—one measured from the 5′-end, and the other from the 3′-end. There is always ambiguity in determining which MS peak belongs to which ladder. This becomes more problematic in a mixture of several different RNAs, due to generation of a large number of overlapping sequence ladders. However, since all ladder fragments from the 3′-ends are labeled with a hydrophobic tag, they exhibit much longer tRS (Figure 1a). As a result, we can obtain clear and unambiguous ladders in the 2D mass-tR data exclusively derived from just the 3′-labeled RNA. Notably, we are optimizing approaches to selectively tag either the 5′- or 3′-end of any RNA using different chemical conjugation methods9 . We can also perform bidirectional sequencing, which is not used to determine the terminal base(s) here, but is used to provide identical sequence information twice when reading from both 5′- and 3′-directions (i.e., bidirectional sequencing verification), and thus further improving the accuracy of sequencing.
For de novo sequencing of unknown RNA samples, especially for complex biological samples, a general and robust algorithm is required to process a large amount of LC-MS data for sequence generation in an accurate and efficient manner, which has recently become available via other published work10 . Although these algorithms have been used for sequencing of more complicated samples10 , in this study, we performed manual base calling for sequence generation unless indicated otherwise. We aim to cover all key steps in the 2D-HELS MS Seq, and would like to illustrate the process during which even without using additional sequencing algorithms, we can still manually read out sequences of the RNA to be sequenced. For ease of visualization and to more quickly identify ladder fragments needed for sequencing in the 2D mass-tR plot, the MFE files of each LC-MS run are processed by a revised version of a published algorithm8 before reading their sequences, unless indicated otherwise. The published algorithm cannot be used directly to read out the sequences from the LC-MS data, but part of its function can still be used to process the data—hierarchically clustering mass adducts through this algorithm will augment the intensity of each ladder component, which in turn reduces the data complexity, especially in the crucial region where sequence reads are generated8 , 9 .
One of the crucial steps during sample preparation for 2D-HELS MS Seq results in the improvement of RNA hydrophobic tag end-labeling efficiency. A high labeling efficiency can help to reduce the amount of RNA sample needed for generating MS signals that sequence data rely on. In order to increase the labeling efficiency, we employ new labeling strategies, including using activated AppCp-biotin to avoid the adenylation step when labeling the 3′-end of the RNA. The yield of the reaction for labeling the 3′-end of a 19 nt RNA with biotin (see step 2.2) can be improved from 60% to ~95%9 using this one-step method. With the efficient labeling, we are able to sequence a mixed sample containing up to 12 distinct RNAs as previously described9 . In this study, we use a mixture of five RNAs as a representative example to illustrate the sequencing process. We also detect all ladder fragments needed for accurate sequencing and read out the complete sequences of each of the five RNA sequences in the mixture. Higher labeling efficiency not only assists in minimizing the sample loading amount, but it also assists in significant reduction of data complexity during downstream data analysis for sequence generation. Novel reactions are currently under development to achieve quantitative yield in labeling RNAs on both 5′- and 3′-ends.
When sequencing RNA #1 as shown in Figure 1c, streptavidin capture and release steps are used to physically separate biotinylated RNA #1 prior to acid degradation (see Section 3). This removes a small portion of unlabeled RNA, and subsequently results in greater ease of visual identification of the labeled mass ladders in the 2D mass-tR plot. However, the physical separation step is not mandatory because the biotinylated RNA ladder fragments have delayed/longer tRS due to the hydrophobicity from the biotin tag when compared to their unlabeled counterparts. In addition, base calling does not rely on physical separation, but relies on the mass differences of adjacent mass ladder components, thus, the correct base call can be achieved as long as the mass differences of two adjacent ladder components match well with the corresponding masses of a particular nucleotide or modification in the RNA nucleotide and modification datebase8 . A computational algorithm is currently under development to automate base-calling and sequence generation.
The MFE settings during original LC-MS data export (in the file type of .d) into spreadsheet files are highly crucial to the data processing and subsequent sequence generation (see Section 6.5). For instance, we tested the MFE setting “peak with height” in a range from 100 to 1000 and noticed that setting of 100 can provide us with 2-fold more compounds than those of setting 1000. In order to avoid missing any ladder components, we can adjust the MFE setting during the sequencing workflow. This setting is likely dependent on instrument mass resolution, the amount of mass ladder fragments, and data complexity. In addition, it is important to use the centroid dataset and chromatographic type setting for small molecules. The quality score can be varied from 50 % to 100% based on the data quality.
The LC-MS instrument we use in the study has an upper mass resolution of ~40K, limiting the method to only sequencing RNA less than 35 bases long. However, the exact read length of this method is instrument-dependent; more advanced instruments with higher resolving power may lead to longer read length. Similarly, the throughput, i.e., how many RNA sequences can be simultaneously sequenced in a single LC-MS run, remains to be explored, although we manually sequenced a mixture of RNA sample up to 12 distinct RNA strands even without use of any algorithm9 . With the current workflow, ~100 pmol short RNA (<35 nt) is required for each LC-MS run. The loading amount increases when additional experiments are needed: for differentiating isomeric nucleotide modifications, typically up to 400 pmol of RNA is required. For sequencing specific tRNA like tRNAPhe , ~1000 pmol of sample may be needed for sequencing and modification analysis. However, we expect required sample loading amounts will be decreased on LC-MS instruments with greater sensitivity. With improvements in sample labeling efficiency, sequencing algorithm, and instrument sensitivity and resolution, we expect our method to be applicable to a wider range of RNA samples, especially those with various RNA modifications.
Supplementary Material
Acknowledgments
The authors acknowledge the R21 grant from National Institutes of Health (1R21HG009576) to S. Z. and W. L. and New York Institute of Technology (NYIT) Institutional Support for Research and Creativity grants to S. Z., which supported this work. The authors would like to thank PhD student Xuanting Wang (Columbia University) for assisting in figure-making, and thank Prof. Michael Hadjiargyrou (NYIT), Prof. Jingyue Ju (Columbia University), Drs. James Russo, Shiv Kumar, Xiaoxu Li, Steffen Jockusch, and other members of the Ju lab (Columbia University), Dr. Yongdong Wang (Cerno Bioscience), Meina Aziz (NYIT), and Wenhao Ni (NYIT) for helpful discussions and suggestions for our manuscript.
Footnotes
Disclosures
The authors have filed a provisional patent related to the technology discussed in this manuscript.
References
- 1.Addepalli B, Venus S, Thakur P, Limbach PA Novel ribonuclease activity of cusativin from Cucumis sativus for mapping nucleoside modifications in RNA. Analytical and Bioanalytical Chemistry. 409 (24), 5645–5654 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gao H, Liu Y, Rumley M, Yuan H, Mao B Sequence confirmation of chemically modified RNAs using exonuclease digestion and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Communications in Mass Spectrometry. 23 (21), 3423–3430 (2009). [DOI] [PubMed] [Google Scholar]
- 3.McLuckey SA, Van Berkel GJ, Glish GL Tandem mass spectrometry of small, multiply charged oligonucleotides. Journal of The American Society for Mass Spectrometry. 3 (1), 60–70 (1992). [DOI] [PubMed] [Google Scholar]
- 4.Fountain KJ, Gilar M, Gebler JC Analysis of native and chemically modified oligonucleotides by tandem ion-pair reversed-phase high-performance liquid chromatography/electrospray ionization mass spectrometry. Rapid Communications in Mass Spectrometry. 17 (7), 646–653 (2003). [DOI] [PubMed] [Google Scholar]
- 5.Taucher M, Breuker K Characterization of modified RNA by top-down mass spectrometry. Angewandte Chemie International Edition in English. 51 (45), 11289–11292 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kellner S, Burhenne J, Helm M Detection of RNA modifications. RNA Biology. 7 (2), 237–247 (2010). [DOI] [PubMed] [Google Scholar]
- 7.Thomas B, Akoulitchev AV Mass spectrometry of RNA. Trends in Biochemical Sciences. 31 (3), 173–181 (2006). [DOI] [PubMed] [Google Scholar]
- 8.Bjorkbom A et al. Bidirectional direct sequencing of noncanonical RNA by two-dimensional analysis of mass chromatograms. Journal of the American Chemical Society. 137 (45), 14430–14438 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang N et al. A general LC-MS-based RNA sequencing method for direct analysis of multiple-base modifications in RNA mixtures. Nucleic Acids Research. 47 (20), e125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang N et al. Direct sequencing of tRNA by 2D-HELS-AA MS Seq reveals its different isoforms and dynamic base modifications. ACS Chemical Biology. 15 (6), 1464–1472 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bakin A, & Ofengand J Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyltransferase center: analysis by the application of a new sequencing technique. Biochemistry. 32 (37), 9754–9762 (1993). [DOI] [PubMed] [Google Scholar]
- 12.Cantara WA et al. The RNA Modification Database, RNAMDB: 2011 update. Nucleic Acids Research. 39 (Database issue), D195–201 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.