Summary
Human herpesviruses (HHV) are large, double stranded, DNA viruses with high sero-prevalence across the globe. Clinical manifestation of primary HHV infection resolve shortly, however, this period is prolonged in immunocompromised patients or individuals with suppressed immunity. Examining molecular mechanisms of HHV-encoded virulence factors can provide finer details of HHV-host interaction. A unique genetic feature of most members of HHV is that they encode multiple microRNAs (miR). In this review, I will provide mechanistic insights into the immunomodulatory functions of herpesvirus-encoded viral miR (v-miR) that favor viral persistence and spread by ingenious immune evasion schemes. Similar to host miR, v-miR can simultaneously regulate expression of multiple transcripts including host- and virus-derived. V-miRs, by virtue of their direct interaction with various transcripts, can regulate expression of critical components of host innate and adaptive immune system. V-miRs are also exported through exosomal route and gain entry into various cells even at distant sites, thereby allowing HHV to manipulate cellular and tissue immunity. Targeting v-miR may serve as a novel and promising therapeutic candidate to mitigate HHV-mediated clinical manifestations.
Keywords: human herpesvirus, immune responses, viral microRNA
1 |. INTRODUCTION
Human herpesviruses (HHV) are large, enveloped, linear double-stranded DNA (dsDNA) viruses that have evolved over millions of years to persist lifelong inside host. HHV establish lifelong infection, interchanging between latent (non-productive) and lytic (productive) infections.1–3 The abilty of the virus to survive is dependent on its ability to evade host immunosurveillance.2 The Herpesviridae family is subdivided into three subfamilies: Alphaherpesvirinae, Betaherpesvirinae, and Gammaherpesvirinae. Members of Alphaherpesvirinae family (HSV-1, HSV-2, and VZV) are cytolytic viruses that infect a variety of human cell-types and establish latency in neurons.1,2 Frequent clinical manifestations of HSV-1 infection are gingivostomatitis and pharyngitis, whereas herpes labialis is the most frequent sign of reactivation disease.2 HSV-1 can also cause ocular infections (eg, herpetic keratitis) and serious systemic illnesses such as encephalitis and neonatal disease involving multiple organs. HCMV, HHV-6A, and HHV-6B are members of the Betaherpesvirinae family.1 These viruses maintain latency in cells of myeloid and lymphoid compartments and exhibit clinical manifestation particularly in immunocompromised or immunosuppressed individuals. While latent infection may not have immediate ramifications to the host, latent infection of two gammaherpesviruses, Epstein-Barr Virus (EBV) and Kaposi sarcoma-associated herpesvirus (KSHV), may lead to carcinogenesis. EBV and KSHV are two of seven oncoviruses, otherwise known as human cancer viruses.4 Specifically, EBV is shown to cause Hodgkin lymphoma, Burkitt lymphoma, gastric cancer, and nasopharyngeal carcinoma.5 KSHV is shown to cause Kaposi sarcoma, a cancer seen in Acquired Immunodeficiency Syndrome (AIDS) patients,4 as well as primary effusion lymphoma (PEL)6 and mulicentric Castleman disease.
With relatively large genomes (approximately 120–180 kb), these viruses encode a large repertoire of protein-coding genes to overcome numerous host defense and cellular functions. The discovery of herpesvirus encoded v-miRs by Pfeffer et al7 showed that these viruses have evolved with yet another critical regulatory RNA called miR, previously thought to be restricted to metazoans and plants. Incorporation of tiny, multifunctional, non-protein coding RNAs that are tightly clustered within few viral transcripts, demonstrate a classical example of incessant host-pathogen co-evolution. Most of the HHV are reported to encode a large number of (22–44) v-miRs, indicating unique requirement of these noncoding RNAs in HHV-host interaction with numerous studies experimentally supporting the notion. Lack of v-miR sequence conservation across different HHV further supports these findings. Similar to host miR, v-miR can interact with the 3′ untranslated region (UTR) of the target mRNA to regulate gene expression.8–10 This interaction results in translational inhibition and/or destabilization of cognate transcripts including host- and virus-encoded.11,12
Viruses develop ingenious mechanisms to overcome host defenses. In this review, critical functions of HHV-encoded v-miR in shaping host-pathogen interaction through modulation of host and viral functions are highlighted. Specifically, v-miR-mediated subversion of host immune responses (innate and adaptive) is discussed. HHV v-miRs are demonstrated to utilize host exosomal pathway to safely reach bystander cells and alter their transcriptome. Evidence demonstrating exosome-mediated delivery of functional v-miR and how this alters the function of recipient cells are provided. Figure 1 illustrates an overview of herpesvirus entry into target host cell, v-miR biogenesis and modulation of infected cell function and regulation of viral life cycle, secretion of v-miRs in exosomes and their entry and potential impact on bystander cells.
FIGURE 1.
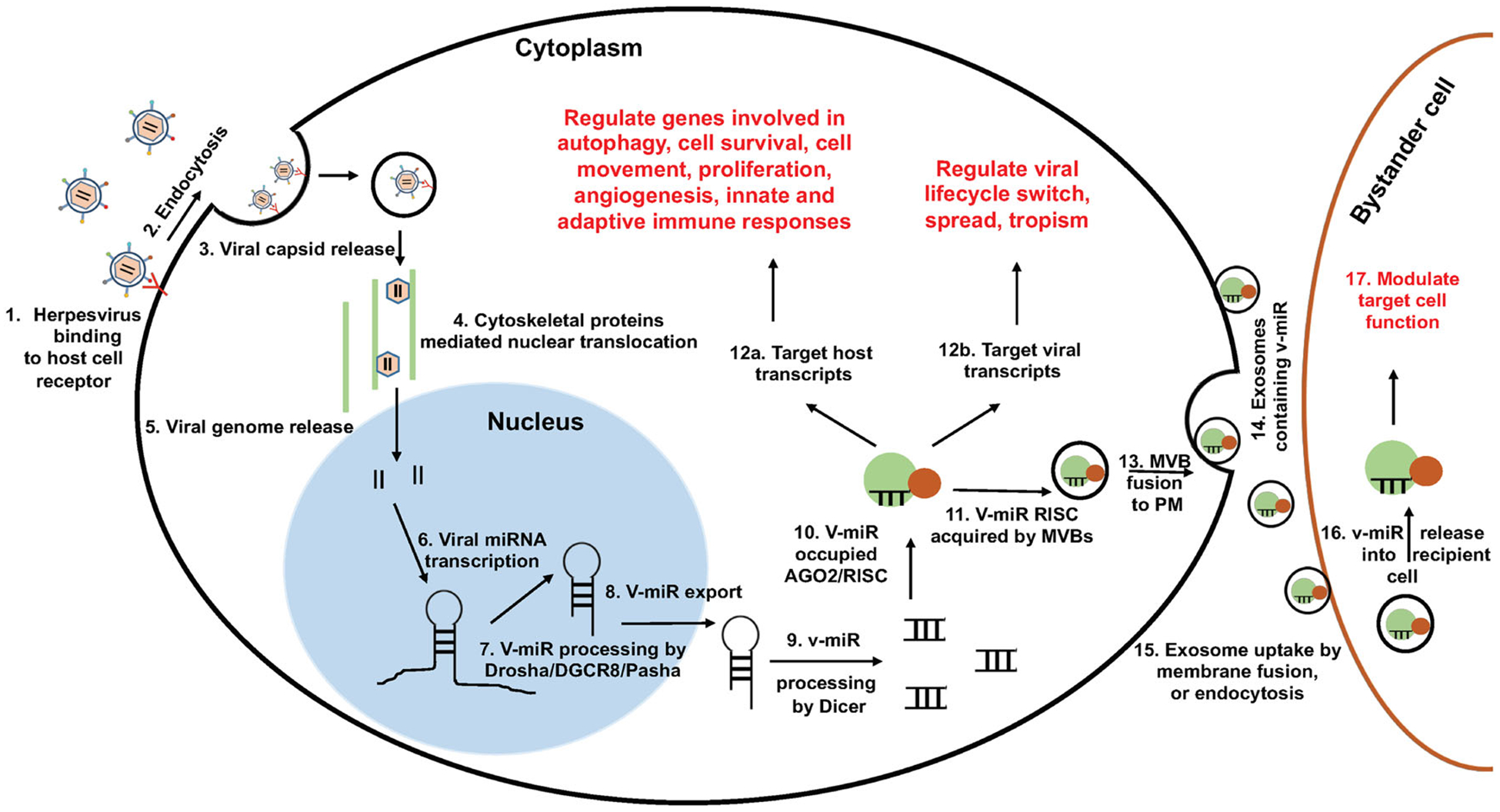
Schematic depiction of viral miR biogenesis and their impact on infected and bystander host cells. A series of events leading to the production of viral miR and modulation of host functions is summarized. Virus bind to cell surface receptors via glycoproteins leading to fusion at the plasma membrane or fusion with an endocytic membrane after endocytosis. Internalized virion sheds envelop and is directed towards nucleus, facilitated by host cytoskeleton proteins. In nucleus, viral transcripts (including v-miR precursors), are generated. Host miR biogenesis machinery (eg, DGCR8 and Dicer) recognize viral miR stem-loop structure and generate mature v-miRs. These molecules acts as autocrine and paracrine modulators. In the infected cells, v-miRs can regulate both host and viral transcripts thereby shifting host transcriptome more favorable towards virus. Additionally, v-miRs can be packaged into exosomes and gain entry into bystander cells. Steady influx of v-miRs in non-infected bystander cells can also modulate their cellular functions, including immune responses and thus contribute to viral persistence and immune evasion
2 |. VIRUS-ENCODED miRNAs
In 2004, the first v-miRs were discovered in human B cells that were latently infected with γ-herpesvirus EBV.7 Subsequently, many other miRs were identified in human and animal herpesviruses.13 To date, more than 250 different miR of viral origin have been identified and the list continues to expand (“miRBase: the microRNA database”). Both host and viral miR are involved in viral tropism, pathogenesis, and latency.14–17 Interestingly, while host miR can function as antiviral defenses, viruses encode suppressors (RNA/proteins) to counteract miR functions and have evolved mechanisms to utilize host miR for their benefit.18–21 According to miRBase (version 22.1), more than 30 different viruses are known to encode miRs. These predominantly include members of herpesviruses representing both human- and animal-infecting viruses. The focus of this review primarily involves HHV derived v-miRs and their functions in host-pathogen interaction. Among human herpesviruses, miR are encoded by herpes simplex virus 1 (HSV-1), herpes simplex virus 2 (HSV-2), human cytomegalovirus (HCMV), human herpesvirus-6B (HHV-6B), Epstein-Barr virus (EBV), and Kaposi sarcoma-associated herpesvirus (KSHV).
Within the alpha subfamily of HHV, miR have been documented in HSV-1 and HSV-2. While the discovery of miR in HSV-1 was first reported in 2006,22 27 miR in HSV-1 have now been documented. Interestingly, over half of the miR are generated from latency-associated transcript (LAT) locus, while others are expressed during productive infection from other regions of the viral genome. It has been proposed that miR work in concert with other viral proteins during different life-cycle stages. More specifically, LAT-encoded miR may repress transcripts that are essential for lytic gene expression, thereby working to establish or maintain latency.23 MiR have been isolated in betaherpesvirus family including HCMV and HHV-6B. While miR genes are seen throughout the viral genome, they are most commonly associated with the lytic stage of the viral lifecycle.24,25 Finally, both gammaherpesviruses viz., KSHV and EBV encode large repertoire of miR. Interestingly, a cluster of 12 pre-miR are highly expressed in latently infected B cells.24,26 These v-miRs may play a role in viral transformation of B cells, as multiple mRNA targets have been documented. In addition to the v-miRs associated with latency, others have been found to be associated with lytic reactivation and viral replication.27 While some HHV express miR during all the phases of the viral life cycle, others may predominantly express them during latency. For instance, specific EBV miR are required in abundance during every stage of virus life cycle indicating their critical role in host-virus interaction.28
HHV miR biogenesis is completely dependent on host miR machinery and evidences clearly suggest that none of the viral protein is required to facilitate this process. Multiple EBV miR precursors were cloned in an expression vector to check if EBV miR are produced in the absence of viral protein. Interestingly, generation of mature EBV miR was observed using various quantitative expression and reporter assays indicating that host miR machinery is sufficient to process v-miR precursors.12,27,29 However, viruses can obstruct miR biogenesis. Pan et al demonstrated impaired precursor-miR/mature miR ratios in HSV-1 infected cells or murine trigeminal ganglia during different life cycle stages.30 Higher levels of precursor-miR were observed in nuclear fraction indicating that miR biogenesis is inhibited at the step of nuclear export. This inhibition was mediated by viral protein ICP27 with a known role in mRNA nuclear export and deletion of ICP27 enhances nuclear export of precursor-miR (both virus and host-derived) with almost no impact on mature miRs. This strategy allows HSV-1 to block nuclear export of host precursor-miR and eventually mature miRs which are antiviral in function. While generation of v-miR requires host machinery, virus-encoded proteins can impair miR biogenesis pathway to facilitate virus survival.
Comparative analysis of human and herpesviruses miR, genome size and gene/miR ratio highlights how efficiently these viruses have successfully integrated miR-mediated post-transcriptional regulation. The human genome contains approximately 3 billion bases and encodes approximately 73 000 transcripts and approximately 1900 miR precursors (approximately 2300 mature miR).31,32 In humans, each miR precursor will have approximately 1.5 million bases of genomic landscape to encode for its gene and will regulate approximately 38 different transcripts among the whole transcriptome. However, it can be noted that several human miR have not been validated and are not considered as bona fide miR. This will likely further increase the gene/miR ratio.
Depending on the herpesvirus in context, genome size of human herpesvirus range from 140 to 235 kb and encode 74 to 213 genes and protein-coding genes and 8 to 44 mature miR.33–35 For herpesvirus, the genomic space and gene/miR ratio vary remarkably indicating some herpesviruses have evolved more robust miR-mediated post-transcriptional regulation than others. Table 1 highlights the differences between the human and human herpesvirus with respect to integration of miR-encoding genes and their predictive post-transcriptional utility. For instance, the gene/miR ratio in HSV-1, HSV-2, EBV, and KSHV is close to approximately 3 to 6, indicating that viral transcripts are under a robust v-miR control. Other miR encoding HHV, HCMV, and HHV-6B (both beta family members) exhibit a gene/miR ratio of 14 and 28, respectively. Compared with humans, even HCMV and HHV-6B exhibit more efficient integration of miR genes and mature miR control of viral transcriptome. Indeed, cis- and trans-regulation of viral transcripts is a common feature of HHV-encoded v-miRs indicating their critical role in viral life cycle.12–18 This in silico analysis shows human herpevirus have evolved this post-transcriptional mechanism to regulate large proportion of its transcriptome. However, further experimental testing is required to support the findings of bioinformatic analysis.
TABLE 1.
Comparative analysis of human and human herpesviruses highlights integration of miR-mediated post-transcriptional regulation by most herpesviruses. Computed genomic landscape occupancy (per kb per miR) and protein coding gene to miR ratio of human and miR-encoding HHV
| Genome Size (Transcripts Encoded) | Precursor miR Expressed (Mature miR) | Ratio of Genome size/ miR | Ratio of Gene/miR | ||
|---|---|---|---|---|---|
| Human | 3 billion base (~73 000) | 1917 (2300) | 1.5 Mb/miR | ~38 | |
| Human Herpesvirus | Common Clinical Manifestations | ||||
| HSV-1 (α) | gingivostomatitis, herpetic keratitis, dermal whitlows; encephalitis in neonates | 152 kb (90) | 18 (27) | 8.4 kb/miR | 5 |
| HSV-2 (α) | Genital herpes | 155 kb (74) | 18 (24) | 8.6 kb/miR | 4 |
| HCMV (β) | mononucleosis syndrome, systemic diseases observed in congenital infection or immunocompromised individuals | 235 kb (213) | 15 (26) | 15 kb/miR | 14 |
| HHV-6B (β) | exanthem subitum | 161 kb (115) | 4 (8) | 40 kb/miR | 28 |
| EBV(γ) | Infectious mononucleosis, oral hairy leukoplakia, lymphoproliferative syndrome in immunocompromised hosts | 172 kb (85) | 25 (44) | 6.8 kb/miR | 3.4 |
| KSHV (γ) | Kaposi’s sarcoma, Primary effusion lymphomas and Castleman’s disease | 140 kb (87) | 13 (25) | 10.7 kb/miR | 6.6 |
While research in the field has introduced the influence of herpesvirus-encoded miR in the regulation of virus lifecycle regulation, much is left to learn regarding their direct and indirect downstream functional impact on host defense responses. This is particularly significant as herpesvirus have mastered multiple ingenious mechanisms to escape host immune pathways. In the following sections, I discuss the role of HHV-derived miR in targeting various aspects of host innate and adaptive immunity pathways that play critical role in virus survival, spread, and evasion.
3 |. ROLE OF VIRAL MIRS IN MODULATING HOST IMMUNITY
3.1 |. Innate immune responses
All living organisms are subject to attack from disease-causing agents. Metazoans, over millions of years, have evolved to precisely recognize self from non-self. Acquisition of these mechanisms, in particular, have allowed them to fend off plethora of microbes that are ubiquitously present and pose danger to host. Orchestrated response by various cell types (both immune and non-immune cells) confer protection against incessant invasion of microbes (harmful or innocuous) by recognizing and eliciting efficient immune response to contain and clear microbes. Immune mechanisms that participate in the recognition and recruitment of leukocytes acts as first line of defense for host. Innate (non-specific) immune system, which responds very quickly-within minutes to hours after infection, can halt spread of virus. For a successful viral pathogen, like HHV, it is required to subdue innate immune response in order to avoid robust adaptive immune responses. V-miRs play critical role in overcoming host innate immune response by suppressing various genes involved in the pathway.
3.1.1 |. Pathogen recognition receptors
A key aspect of host defense is the ability to recognize and initiate responses to counter invading pathogens and destroy infected cells. Host and pathogen interactions has co-evolved over millennia and the host has evolved diverse mechanisms to elicit adept responses by distinguishing between self and non-self (pathogenic) antigens. This is achieved primarily by precise recognition of patterns displayed by pathogenic biomolecules. This includes proteins, carbohydrates moieties, DNA methylation patterns, and nucleic acid structure and sequence. Proteins present on host cell surface, endosomal membranes, and cytosol act as receptors for various pathogen associated molecular patterns (PAMPs). Upon recognition, these receptors can initiate a signaling cascade that consequently activates proinflammatory cytokine production.36,37 On the basis of the motifs, these recognition molecules are categorized into three major categories viz, (a) toll-like receptors (TLR), (b) Nucleotide oligomerization domain (NOD)-like receptors (NLR), and (c) retinoid acid-inducible gene-I (RIG-I)-like receptors (RLR).38–40 While TLR are localized on cellular and endosomal membranes, NLR and RLR are intracellular cytosolic sensors. Together, these molecules scan both extra and intracellular PAMPs and elicit adept immune response.
Toll-like receptors
TLRs are present on cell and endosomal surfaces and play a key role in innate immune activation. In general, extracellular pathogens are recognized by surface TLRs, while intracellular pathogens and their ligands are recognized by endosomal TLRs. Depending on the ligand binding and location of TLRs, a specific set of transcription factors are activated leading to expression of genes required to potentiate immune response against incumbent pathogens.36,38,39,41
Not surprisingly, HHV has devised mechanisms to thwart innate immune responses. HHV utilizes v-miRs as an important tool to suppress innate immunity. Figure 2 summarizes v-miR–mediated inhibition of selective host innate immune genes. Among the TLRs, TLR2 in conjunction with TLR1 or TLR6 is a key receptor in recognizing HCMV, EBV, HSV-1/2, and HSV-derived ligands.42–44 HCMV encoded miR-UL-112–3p has been demonstrated to silence TLR2 which binds to HCMV glycoproteins B and H.45 This is highly significant for viral persistence as it prevents NFκB-mediated proinflammatory cytokine production. The cytokines in turn will recruit other immune cells of both innate and adaptive arms of immunity that work in concert to clear virus-infected cells. Moreover, viral ligand-mediated internalization of TLR2 complex can also occur. This, unlike NFκB activation, triggers IRF2, IRF3, IRF7 activation by yet unknown signaling cascade, consequently upregulating production of antiviral genes IFNα (IRF2) and IFNβ (IRF3/7).46–50
FIGURE 2.
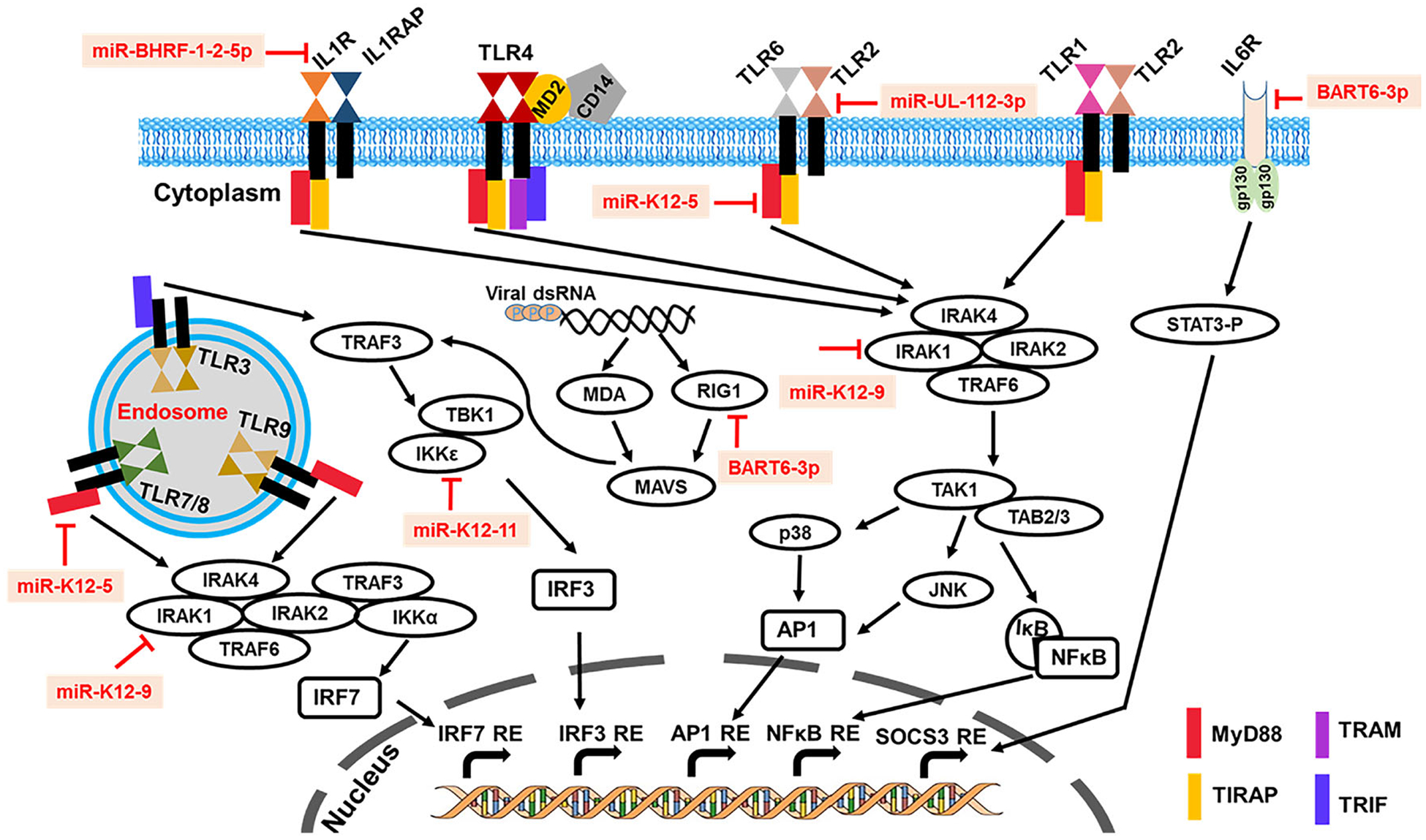
Overview of viral miR-mediated suppression of host innate immune pathways. The illustration summarizes known mechanisms by which individual viral miR interact with host genes involved in virus recognition and activation of innate immune responses. Viral miR promote immune evasion by directly targeting host genes involved in the recognition of virion, virus-derived nucleic acids (DNA/RNA) or proteins to suppress antiviral pathways. Viral miR have evolved to target, essentially, all of the known viral defense mechanisms by targeting one or more critical components of the innate immune pathway. Functions of selected validated herpesvirus-encoded miR and host innate immunity genes are highlighted in the schematic. Viral miR inhibit production of proinflammatory cytokines and interferons that are required for recruitment and activation of adaptive arm of immunity, which can elicit robust antiviral response to contain herpesvirus infection by clearing infected cells
Various components of TLR signaling proteins are also reported as direct targets of HHV miR. Myeloid differentiation primary-response gene (MyD88) and interleukin-1 receptor-associated kinase (IRAK1) are two key components of TLR signaling. Multiple KSHV-encoded v-miRs are demonstrated to specifically interfere with TLR signaling, signifying the importance of innate responses in KSHV infection. KSHV miR-K12–5 and miR-K12–9, respectively, downregulate expression of MyD88 and IRAK1 and hence inhibit proinflammatory cytokine production.51 Activation of NFκB is mediated by two IκB kinases (IKK), namely, IKKα and IKKβ that can phosphorylate IκBα and release the p50 and p65 (RelA) subunits. Two HCMV miR, viz, miR-US5–1 and miR-UL112–3p suppress IKKα and IKKβ levels thereby limiting NFκB activation allowing viral persistence by mitigating cellular inflammation.52 Virus mutated for miR-US5–1 and miR-UL112–3p increased levels of IKKα and IKKβ proteins, impaired NF-κB signaling at late times of lytic infection, and increased proinflammatory cytokine production compared to wild-type virus in HCMV infection in vivo.
In addition, miR-K12–11 targets IKKε, a non-canonical NFκB activator and inducer of antiviral IRF3/7 signaling.53 By silencing IKKε, which is involved in KSHV reactivation, viral miR suppresses antiviral pathway and also promotes latency maintenance. Similarly, KSHV miR-K12–3 and miR-K12–7 target an important transcriptional regulator C/EBPβ that suppresses IL-6 and IL-10 cytokines and promotes virus reactivation.54 HHV miR not only target PRR but also downstream effector molecules consequently inhibiting inflammatory as well as interferon pathways. These findings clearly indicate that multiple KSHV miR have evolved to target different critical components of the innate signaling cascade. List of validated v-miR and their host targets are summarized in Table 2.
TABLE 2.
List of validated viral miR targets involved in the innate immune subversion
| Viral miR Modulation of Innate Immune Responses | |||
|---|---|---|---|
| Viral miR | Target | Function | References |
| HCMV miR-UL112–3p | TLR2 | Inhibition of proinflammatory responses by HCMV ligand | Landais et al45 |
| KSHV miR-K12–5 | MYD88 | Suppression of TLR-mediated activation of cytokine production | Abend et al51 |
| KSHV miR-K12–9 | IRAK1 | Suppression of TLR-mediated activation of cytokine production | Abend et al51 |
| HCMV miR-US5–1 and miR-US112–3p | IKKA and IKKB | Blocks NFκB activation | Hancock et al52 |
| KSHV miR-K12–11 | IKKɛ | Inhibition of NFκB and antiviral cytokine activating IRF3 | Liang et al53 |
| EBV BART6–3p | RIG-1 | Impairs RLR pathway-mediated activation of antiviral cytokines | Lu et al55 |
| KSHV miR-K12–3, −7 | C/EBPβ | Modulate cytokine secretion by immune cells | Qin et al54 |
| EBV miR-BART-15 | NLRP3 | Suppress inflammasome formation and hence IL-1p/IL-18/ROS production | Haneklaus et al56 |
| EBV miR-BART16 | CREBBP | Inhibits type I interferon signaling | Hooykas et al57 |
| EBV miR-BHRF-1-2-5p | IL1R1 | Dampens proinflammatory IL-1 signaling | Skinner et al58 |
| EBV BART-6–3p | IL6R | Impairs IL-6 signaling | Zhang et al59 |
| HCMV miR-UL148D | ACVR1B | Suppress cytokine production by blocking activing A signaling | Lau et al60 |
| HSV2 miR-H9–5p | SOCS2 | Impaired immune cell activation and differentiation | Wang et al61 |
| KSHV miR-K12–10 | TWEAKR | Reduced induction of IL-6 and IL-10 | Abend et al62 |
| HCMV miR-UL112–1 | IL-32 | Reduced NK cell activity & Mϕ proinflammatory function | Huang et al63 |
| HSV-1 miR-H1 | SORT1/Integrin | Attenuates bacterial phagocytosis and cytokine production | Naqvi et al64 |
| KSHV miR-K12–3 | RAB3B/RAB3D | Attenuates bacterial phagocytosis and altered cytokine production | Naqvi et al64 |
NOD-like receptors and RIG-1–like receptors
As mentionend earlier, NLR and RLR are a family of multiple cytosolic helicases that detect invading pathogens or host-derived damage signals in the cytosol and trigger innate immune response primarily by NFκB and MAPK singaling cascade.37 A key feature of NLR proteins is they oligomerize to form large complexes called inflammasomes. EBV miR have been shown to influence inflammation and chemotaxis and thereby contribute to host immune system evasion. EBV-miR-BART15 binds and controls levels of NLR family, pyrin domain containing 3 (NLRP3). NLRP3 inflammasomes are among the well-characterized NLR-containing inflammasomes. NLRP3 is involved in the production of numerous proinflammatory cytokines such as IL-1β and IL-18, suggesting that downregulation of this protein by EBV miR can suppress innate immune activation.56
RLR are important in detecting viral infection (RNA or replication transcripts) and initiate the production of IFN and induction of an antiviral response.65,66 Thus, silencing the components of this pathway is crucial for viral persistence. Not surprisingly, EBV miR-BART6–3p directly bind to and post-transcriptionally suppresses RIG-1 mRNA. Overexpression of miR-BART6–3p was able to suppress EBV-triggered IFN-β response production.55 Another EBV miR, miR-BART16, also inhibits type I IFN signaling by targeting CREB-binding protein (CREBBP), a transcriptional coactivator required for IFN pathway.57 Overall, these studies clearly support the role of HHV miR in the early phase of virus infection by simultaneously interfering with multiple host defense systems involved in virus sensing and immune activation.
Cytokine receptors
Cytokine-mediated autocrine and paracrine immune signaling is critical in containing virus infection. IL-1 and IL-6 are two major proinflammatory cytokines that are induced during initial herpesvirus infection both in vitro and in vivo implying their role in paracrine antiviral immunity.67,68 Cytokine signaling cascade is triggered upon binding of cytokine to its cognate receptors which in turn activate other cytokine/chemokine production. In order to block the spread of antiviral cytokines, it is critical for HHV to interfere with cytokine signaling. Indeed, different HHV are reported to block the receptors for IL-1 and IL-6 cytokines, namely, IL-1R and IL-6R.
Using a combinatorial approach of miR targetome analysis, v-miR pathway analysis and reporter-based assays, Skinner et al. showed that EBV encoded miR-BHRF-1-2-5p is shown to target interleukin 1 receptor (IL-1R1).58 Ectopic expression of miR-BHRF-1-2-5p downregulated IL-1R1 and hence IL-1 signaling, while inhibition of miRBHRF-1-2-5p augments IL-1 activation in latently infected B cells.
Zhang et al showed that IL-6R transcript was downregulated in EBV-positive BL.59 IL6/IL-6R signaling is critical in acute phase response, immune activation, T cell growth, and NK cell activity. They predicted EBV-BART-6–3p binding site on IL-6R and validated that v-miR directly regulate IL-6R levels by reporter gene assays.59 Inhibtion of IL-6 pathway is also important for latent HCMV infection in myeloid cells, which are important latent HCMV reservoirs. Lau et al showed that miR-UL148D targets cellular receptor ACVR1B, an important component of the activin signaling axis and promotes monocyte-to-dendritic cell differentiation.60 Monocytes challenged with activin A secrete proinflammatory cytokines including IL-1 and IL-6.69 Comparative analysis of activin A-treated monocytes challenged with wild type HCMV and ΔmiR-UL148D HCMV showed higher expression of ACVR1B and concomitantly induced IL-6 production in cell infected with miR-UL148D deleted HCMV. Yet another HCMV miR-UL112–1 also impairs activation of inflammatory signaling by binding to and inhibiting IL-32, a key cytokine that participates in the induction of other proinflammatory cytokines including IL-1β, IL-6, IL-8, and TNF-α through the p38MAPK, NF-kappa B, and AP-1 signaling pathways.63 On the other hand, HSV-2 encoded miR-H9–5p targets suppressors of cytokine signaling 2 (SOCS2), a multifaceted immunoregulatory protein involved in immune cell differentiation, NK cell function, and resolution of inflammatory processes.61
KSHV-encoded miR target genes involved in the secretion of cytokines. More specifically, KSHV miR-K10 was found to target TNF-like weak inducer of apoptosis receptor (TWEAKR), which is the receptor for the proinflammatory cytokine TNF-like weak inducer of apoptosis (TWEAK).62 This leads to a decrease in proinflammatory cytokine expression.70 Another example of v-miR involvement in inflammation and chemotaxis is found in KSHV miR-K6 and miR-K9, which were found to target transcripts involved in the NFκB signaling pathway, and thereby decrease the expression of cytokines involved in inflammation.71 V-miR-mediated suppression of these critical, early proinflammatory and antiviral cytokines are not only helpful in blocking cytokine signaling but also enhances survival of HHV infected cell by dampening inflammatory microenvironment.
3.1.2 |. Phagocytosis and immune activation
Studies from our lab have identifed v-miR in diseased oral tissues including gingiva and dental pulp, which are also sites of herpesvirus infection and reactivation.72–75 Global mRNA and miR profiling of miR-H1 and miR-K12–3 expressing human oral keratinocytes and primary macrophages together reveal that these v-miRs specifically target pathways related to endosomal trafficking, suggesting their role in deregulation of endocytic pathways which are required for phagocytosis and immune activation.64,74,76 Genes downregulated by miR-H1 and miR-K12–3 harbors one or more potential binding sites for genes involved in secretory pathway and some of these sites were validated using dual luciferase assays.64,74 miR-K12–3 regulate RAB3B and RAB3D, while miR-H1 target sortilin 1 (SORT1) transcripts by directly binding to the 3′UTR. Targeting of endosomal/secretory pathway-related genes is also common in other HHV members with various HCMV and KSHV miR known to specifically target cellular genes associated with endosomal pathways.71,77
Macrophages expressing miR-H1 and miR-K12–3 exhibit reduced uptake of florescently labeled Escherichia coli, strongly suggesting impaired phagocytosis in presence of v-miRs. Furthermore, this translates to dysregulated immune response against incumbent pathogen as observed by cytokine profile analysis. Levels of multiple cytokines/ chemokines (including TNF-α, IL-1β, GM-CSF, IL12-p70, and IL-10) were significantly altered in miR-H1 and miR-K12–3 expressing macrophages.64
Overexpression of miR-H1 in HOK showed inhibitory role of v-miR in entry and infection. Given that miR-H1 is expressed in copious amounts during productive infection, our results suggest that this v-miR likely facilitate optimization of virion generation during the productive phase. Two other HSV-1 miRs, viz, miR-H28 and miR-H29, are also reported to regulate HSV-1 infection and spread.78 Similar to miR-H1, both v-miRs are produced during productive infection, are exported into exosomes and prevent spread and infection of HSV-1. Thus, v-miRs act as multifunctional viral factors that regulate viral transcripts, participate in viral life cycle switching, suppress host immune response and facilitate viral spread in a contained manner.
3.2 |. Adaptive immune responses
Adaptive (specific) immune responses are initiated by activation of naïve lymphocytes by cells of the innate immune pathway, in particular dendritic cells and macrophages. Proper induction of this pathway takes a long time—days to weeks—and is critical in acquiring immunity. However, the adaptive immune system specializes in retaining memory of foreign antigens. Adaptive immunity effectively contains herpesviruses in immunocompetent individuals, while high prevalence of herpesvirus and clinical manifestations are observed in immunocompromised individuals. Blocking activation of adaptive immune response is therefore critical to herpesvirus persistence and spread. The contribution of v-miR in evading adaptive immunity is discussed in this section.
3.2.1 |. Antigen processing and presentation
To persist inside the host, it is imperative for HHV to interfere with antigenic presentation of viral proteins. This is achieved by encoding proteins that have specifically evolved to block the antigen processing pathway. There are two distinct antigen processing and presentation pathways. In general, major histocompatibility complex (MHC) I and MHC II process intracellular and extracellular antigens, respectively.79–81 MHC-II presentation to CD4+ T cells results in polarized cells with specific antiviral immunity.82–85
Several HHV-encoded proteins inhibit both pathways, however, the propensity to interfere with cytotoxic T lymphocyte (CTL) activation is remarkably common. Indeed, EBV-specific CD8+ T cells are frequently detected in asymptomatic individuals and decrease in the EBV-specific T cells is associated with the emergence of EBV-related clinical manifestations in immunocompromised subjects.86 Multiple HHV-encoded proteins target the generation of peptides (including viral antigens) by blocking the activity of host proteins required in the process.87,88 HHV proteins have also been shown to interfere with proteasome activity, transport of viral peptide to MHC I, degradation of MHC I, secretion of mature MHC I-peptide complex into the golgi, directing of MHC I to cytosolic degradation, and redirecting surface-bound MHC I-peptide complex to lysosome for degradation.89,90 For instance, HSV-1 ICP47, HCMV US3 and US6, and EBV BNLF2a all target Tapasin, a protein that monitors the integrity of peptide loading to MHC I.91–93 Tagawa et al showed that two different EBV miRs, viz, BHRF1–3 and BART17, directly target TAP2 mRNA and downregulate its protein levels. Given that these v-miR are expressed in early infection (BHRF1–3) as well as latency (BART17) cleary suggest that v-miR assure suppression of antigenic peptide transport.94
The role of v-miRs in interfering antigen processing/presentation pathway is reported. MHC class I and II pathways and selected genes of the pathways targeted by v-miRs are highlighted in Figure 3. An increasing number of HHV-encoded v-miRs are demonstrated to specifically target genes that participate in antigen presentation and activation of T cells as well as NK cells. Different HHV encode miR that target the same gene, while multiple v-miRs from the same HHV exhibit sequence complementarity for a single gene. MHC class I polypeptide-related sequence A/B (MICA/MICB), a stress-induced or virus infection-induced glycosylated protein is targeted by v-miRs encoded by HCMV (miR-UL112), EBV (BART-2–5p), and KSHV (miR-K12–1).95,96 MICB is a ligand for C-type lectin receptor NKG2D expressed on NK cells and γδ T cells. Signals from NKG2D induces cytolytic activity of NK or T cells. HCMV utilizes another ingenious strategy to downmodulate MICA by miR-US25–2–3p targeting of TIMP3 transcript, leading to enhanced shedding of MICA.97 Silencing MICA/ MICB by v-miR prevents clearance of virus-infected cells.
FIGURE 3.
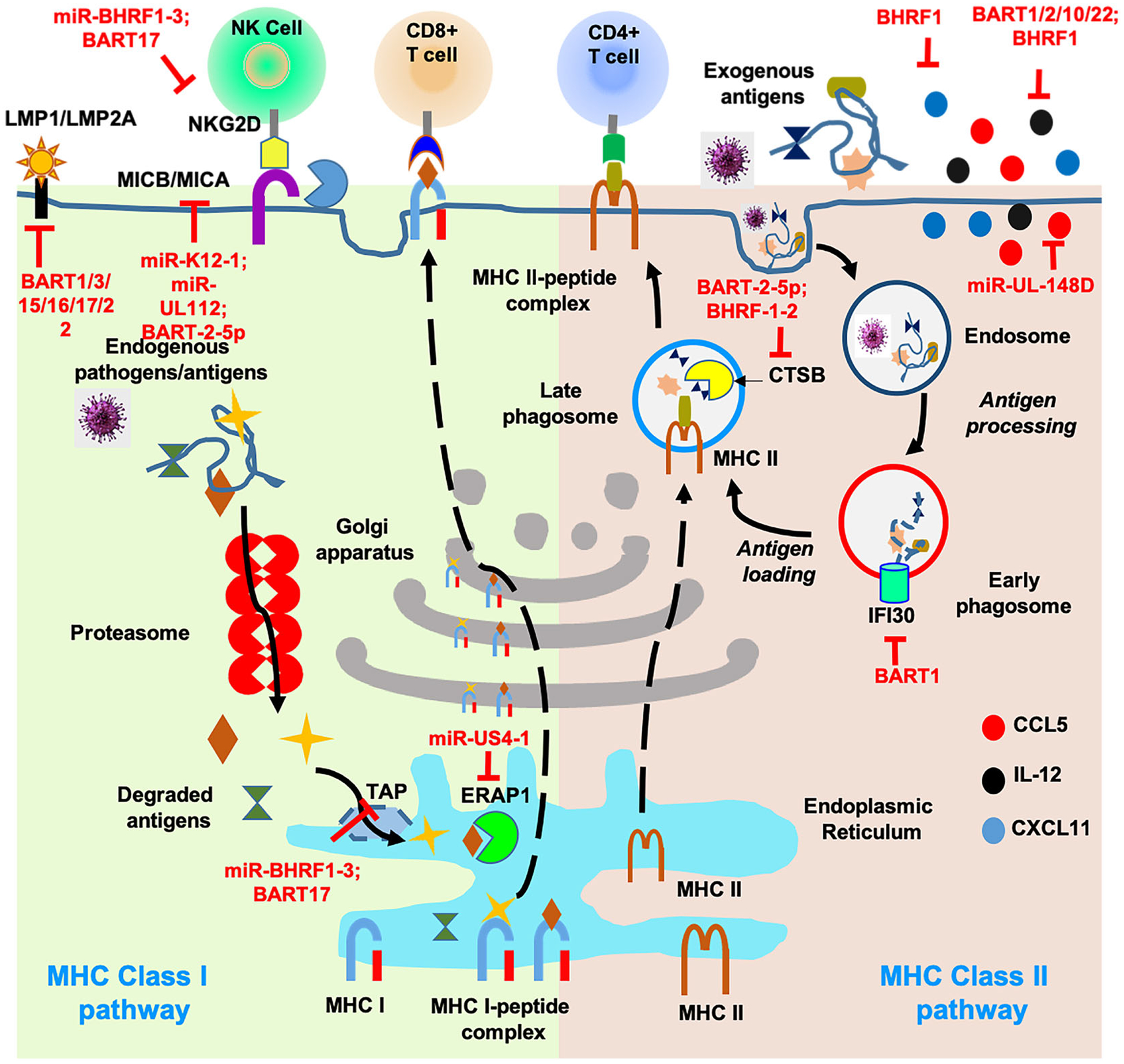
Viral miR-mediated evasion of adaptive immune responses. Antigens are presented by two major pathways, MHC class I and MHC class II. In MHC class I pathway, endogenous viral antigens are degraded via proteasome machinery, translocated to endoplasmic reticulum through TAP proteins and are processed by proteases like ERAP. Processed peptides are assembled with MHC-I complex to be presented predominantly to CD8+ T cells. MHC class II pathway involves exogenous antigen internalization by several pathways, including phagocytosis, macropinocytosis, and endocytosis. Antigens are trafficked to a mature or late endosomal compartment where they are processed and loaded onto MHC-II molecules that present antigens to CD4+ T cells. Numerous viral miR are known to impair both MHC class I and II pathways by targeting one or more components of the same pathways. In addition, some critical genes (eg, MICB) are targeted by multiple herpesvirus miR indicating selective evolution of miR sequences that can interfere with antigen processing and presentation pathway. Inhibition by viral miR is shown as blocked line highlighted in red. By virtue of their direct interaction with host and viral transcripts, v-miRs can impair components of MHC class I (antigenic peptide transporter TAP, and protease ERAP1) and class II (cathepsin B); inhibit release of proinflammatory and antiviral cytokines; protect infected cell from CD4+/CD8+ T cell response and NK cell-mediated cytolysis; reduce highly antigenic viral antigens (LMP1/ LMP2A)
HCMV miR-US4–1 binds to and suppresses endoplasmic reticulum aminopeptidase 1 (ERAP1) expression.98 This gene is responsible for trimming peptides to be properly loaded onto MHC I. In this regard, ERAP1 is important in activating cytolysis of virus-infected cells through activation of CD8+ T cells. Tagawa et al showed multiple genes involved in antigen processing directly regulated by EBV miR.94 Three critical endosomal genes involved in antigen processing, namely, interferon gamma-inducible protein 30 (IFI30), cathepsin B (CTSB), and asparagine endopeptidase (LGMN), were targeted by multiple EBV miR to restrict viral antigen processing to MHC pathway.94 Expression of CTSB, a lysosomal cysteine protease, is directly targeted by two different miRs encoded by EBV, viz, BART-2 and BHRF-1–2. CTSB is an important protein involved in antigen processing of intracellular antigens. Downregulation of IFI30 and CTSB by miR-BART1 and miR-BART2 assures reduced antigen presentation of exogenously loaded protein in presence of v-miR as observed by impaired recognition of virus-infected B cells by EBV-specific CD4+ T cells.94 V-miR are therefore capable of restricting the antigen porcessing pathway at multiple levels including lysosomal protein degradation, HLA class II, and co-stimulatory molecule expression. By targeting multiple proteins of the same pathway, v-miR facilitate successful immune evasion by virus. These examples and others of v-miR targeting of antigen processing/presentation pathways reveal that they exhibit a propensity toward the same targets as shown for viral proteins. For example, MICB is targeted by KSHV kK3 and kK5, VZV ORF66, HCMV Il18, and also by KSHV BART2–5p and miR-K12–7. These observations indicate that viruses have evolved multi-prong approaches to thwart antigen processing and presentation to T/NK cells, suggesting the importance of this pathway in HHV persistence.
LY75, a C-type lectin receptor is a key molecule involved in antigen presentation on MHC. PAR-CLIP–based EBV miR target identification yielded two potential EBV miR with putative binding sites on LY75 3′UTR. Using mutation analysis, miR-BART1–5p was confirmed as a bonafide regulator of LY75 transcript.12 Overexpression of miR-BART1–5p can evade recognition of EBV-infected cells by CD4+ or CD8+ T cells. Interestingly, bioinformatic analysis also predicted binding sites of miR from other HHV including KSHV, and HCMV indicating LY75 (like many other host targets) are selectively targeted by HHV encoded miR to evade immune responses. Studies from our lab have reported increased accumulation and prevalence of various HHV-encoded miR in healthy and diseased gingiva, suggesting their role in the pathogenesis of oral inflammatory diseases. miR-K12–3 identified in our analysis was assessed for its potential role in antigen processing and uptake. We observed that ectopic expression of miRK12–3 in macrophages and DC interferes with the processing of antigen as observed by reduced processing of ovalbumin, conjugated with a quencher dye (Naqvi et al, Unpublished data).
Interestingly, viral miR not only suppress host genes involved in antigen processing/presentation, but also actively degrade viral transcripts that generate highly immunogenic proteins. For instance, EBV-encoded latency-associated membrane proteins LMP1 and LMP2A are post-transcriptionally silenced by v-miR. LMP1 is regulated by miR-BART16, miR-BART17–5p, and miR-BART1–5p, while LMP2A expression levels are controlled by miR-BART22.99,100 Down-regulation of LMP protein levels by EBV miRs allow evasion of host antigen responses against latently infected cells. List of validated v-miR and their host targets involved in shaping adaptive immunity are summarized in Table 3.
TABLE 3.
List of validated v-miR targets involved in the regualtion of antigen processing and presentation and T cell activation and polarization
| Viral miR Modulation of Adaptive Immune Responses | |||
|---|---|---|---|
| Antigen processing and presentation | |||
| Viral miR | Target | Functional impact | References |
| HCMV miR-UL112; EBV miR-BART-2–5p; KSHV miR-K12–1 | MICB | NK cell/CD8+ T cell/γδ T cell cytolytic evasion by reducing stimulatory signals through NKG2D receptor | Nachmani et al95; Stern-Ginossar et al96 |
| HCMV miR-US25-2-3p | TIMP3 | Degradation of MICA and hence reduced NKG2D activation | Esteso et al97 |
| BHRF1–3; BART17 | TAP2 | Blocks peptide transport | Tagawa et al94 |
| EBV miR-BART2 BHRF1–2 | CTSB | Interferes with MHC class I antigen processing | Tagawa et al94 |
| HCMV miR-US4–1 | ERAP1 | Reduced antigen degradation and presentation to CD8 + T cells | Kim et al98 |
| EBV miR-BART1–5p; BART3; BART15; BART16; BART17–5p | LMP1 (viral) | Downregulation of highly immunogenic viral latency membrane protein 1 | Lo et al99 |
| EBV miR-BART22 | LMP2A | Downregulation of highly immunogenic viral latency membrane protein 2A | Lung et al100 |
| EBV miR-BART2 | LGMN | Reduced processing of MHC II bound pathogenic antigens | Tagawa et al94 |
| EBV miR-BART1 | IFI30 | Inhibition of MHC class II-restricted antigen processing | Tagawa et al94 |
| EBV miR-BART1–5p | LY75 | Impaired antigen presentation on MHC | Skalsky et al12 |
| T cell activation and polarization | |||
| EBV BART1; BART2; BART10; BART22; BHRF1 | IL12B | Prevents polarization of Th cells to antiviral Th1 subtype | Tagawa et al94 |
| HCMV-UL-148D | CCL5 | Reduced NK cell activation and T cell recruitment | Kimet al101 |
| EBV BHRF1–3 | CXCL11 | Inhibition of chemotaxis of activated T cells | Xia et al102 |
3.2.2 |. T cell activation and polarization
Antigen-presenting cells (APCs) display viral epitopes that are recognized by T cells leading to their activation. This is mediated by a set of stimulatory and costimulatory molecules present on APC and T cells. MHCI/II (APCs) and TCR (T cells) are primary stimulatory molecules, while multiple factors present on APCs (CD86/CD80/ICAM1/CD40/PD-L1/2) and T cells (CD28/CD40L/CTLA/PD-1) act as costimulatory molecules.103,104
In order to survive inside the host, it is imperative for HHV to interfere with cytokine-mediated feed-forward activity. Recently, Tagawa et al compared the impact of EBV miR on B cell function, an important APC.94 Using an EBV wild-type strain that expresses 13 v-miRs and a deletion strain lacking any miRs, they demonstrated that v-miRs can modulate various pathways including apoptosis, cell cycle, cytokine signaling, and antigen processing relevant to viral persistence. Several proinflammatory cytokines (TNF-α, IL-6, IL-8, IL-12, IL-23) but not the anti-inflammatory cytokine IL-10, were also observed to be downregulated in wild-type EBV strain compared with the v-miR deleted strain. Five different EBV encoded miRs, viz, BART-1, −2, −10, −22, and BHRF1 were shown to directly target IL12B by binding to the 3′UTR. Reduced IL-12 production by B cells interferes with polarization of Th cells to the Th1 subtype. Antibody against IL-12 relieved Th1 suppression caused by v-miRs indicating a specific role of this cytokine in T cell polarization. These findings indicate that targeting of IL-12–mediated activation of Th1 cells is critical for EBV survival. IL-12 also activates expression of IFN responsive genes required for antiviral immunity. Furthermore, IL-12 not only derives Th1 differentiation but also suppresses Th2 polarization by opposing IL-4 activity.94
Further examples of the role of v-miR in immune evasion via altered chemotaxis can be found in HCMV. It has been demonstrated that HCMV-miR-UL148D may contribute to evasion of the immune response via targeting of the gene encoding the chemokine (C-C Motif) ligand 5 (CCL5) transcript.101 This protein acts as an inducer of cell activation as well as proliferation in NK cells.3,105 EBV miR-BHRF1–3 is yet another example of v-miR regulating chemotaxis. This v-miR targets CXCL11, a T cell attracting chemokine.102 By interfering with chemotaxis, HHV can simultaneously minimize the intensity of the innate response and block efficient activation of adaptive responses.
4 |. IMPACT OF VIRAL miRNA MUTANTS ON HERPESVIRUS PATHOGENESIS
Deletion or mutation of HHV miRs has shown contrasting results in vitro and in vivo. Phenotypic effects of v-miR knockout differed on the basis of the infection model used, viral strain used, number of v-miRs targeted, and experimental conditions. In HSV-1, impact of mutation or deletion of three highly expressed and RISC-associated v-miRs, viz, miR-H2, miR-H3, and miR-H4 on viral replication was examined in different cell types.106 Mutation/deletion of these miR-H2 and miR-H4 also impacted expression of other v-miRs. Surprisingly, it was observed that disruption of miR-H3 and miR-H4 significantly enhanced viral replication in Neuro2A cells but not in NIH 3 T3 fibroblasts. On the other hand, mutational inactivation of miR-H2 reduced viral replication only in Neuro2A. These findings indicate that v-miR have a critical role to play in viral replication, but the effects may be specific to cell type or lifecycle associated. The impact of mutated miR-H2 HSV-1 was evaluated for viral replication and reactivation in vivo by the same research group. Contrary to the previous in vitro findings, no significant changes in viral replication, virulence, or reactivation was observed in the mice trigeminal ganglia acutely or latently infected with wild type or miR-H2 mutant virus.107 Furthermore, expression of ICP0, a well-characterized miR-H2 target in vitro, were not significantly changed in TG infected with mutant virus, suggesting it is a weak regulator of ICP0 in vivo. On the contrary, Jiang et al showed miR-H2 mutant virus exhibits increased ICP0 during productive infection and higher virulence and rate of reactivation, implying that targeting of v-miR could be a promising antiviral strategy.108,109 The differences observed could likely be attributed to the redundant functions of v-miRs. Simultaneous deletion of multiple miR might be required to observe more consistent phenotypes. Different viral strains, mutation impact on viral miRs or transcripts, and HHV-specific differences in humans and mice could contribute to disparities observed in the v-miR mutation analysis.
Mutational analysis of EBV miRs strongly support their oncogenic role. The BHRF region encodes three v-miR, viz, BHRF1–1, BHRF1–2, and BHRF1–3. To evaluate thier role in EBV infection and oncogenic potential, Feederle et al deleted the BHRF1 miR cluster from the B95.8 genome. B cells infected with mutant showed reduced transformation compared with wild type or revertant virus, suggesting that BHRF1 miR cluster markedly increases the efficiency of transformation.110 Cell cycle analysis revealed that in the absence of these genetic elements, infected B cells grow markedly more slowly. Using a humanized mouse model, Wahl et al demonstrated a role of v-miR in EBV infection.111 Compared with the wild type virus, systemic EBV infection is delayed in mice infected with mutant virus lacking BHRF miR cluster. However, v-miR mutant virus had no effect on the generation of tumors.
Employing CRISPR/Cas9 to directly mutate v-miR has shown promising results indicating their role as critical viral genetic elements required for both productive and latent herpesvirus infection.112–115 Guide RNAs (gRNA) targeting v-miRs BART5, BART6, and BART15 in latently infected gastric carcinoma cell line NSU-719-generated viruses with mutant v-miR genes. Using a florescent reporter-based assay, direct editing of v-miR on the EBV genome impaired their function by preventing v-miR binding to their target transcripts.112 Comparing different mutation techniques for assessing v-miR function both in vivo and in vitro will provide better and novel insights of their biological roles.
KSHV miRs are highly expressed in latency and KS tumors implicating their role in oncogenesis. The oncogenic role of KSHV miR cluster was assessed by deleting 10 pre-miRs. Cells infected with mutated virus showed similar anchorage-dependence and contact-inhibition, and thus were not transformed.116 Mutant virus infected cells fail to develop tumors in nude mice. Genetic complementation by stably expressing the miR cluster rescues KSHV-induced cellular transformation. Mechanistically, miRs in this cluster promote cell cycle progression and inhibit apoptosis.
Mutating the viral gene targets of v-miRs further highlight their role in viral persistence. In HCMV, immediate early 72 (IE72) is a target of miR-UL112–1. Mutating miR-binding site on IE72 remarkably enhanced expression of IE72 and allowed productive virus infection.117 Monocytes latently infected with mutant virus are recognized specifically by IE-specific CD8+ T cells suggesting an immunosuppressive role of v-miR by inhibiting recognition of latently infected cells by the host’s CTL response. Overall, v-miR mutation/deletion studies have emphasized the role of these regulatory RNAs in pathogenesis, viral spread and persistence but the observations are not consistent in different models. Development of tractable and consistent disease models to measure in vivo impact of v-miR in disease pathogenesis, oncogenesis, and herpesvirus reactivation will provide conclusive information on their biological functions and mechanisms through which they contribute to these pathways.
5 |. VIRAL miRNA AND EXOSOME PATHWAY
Similar to cellular miR, v-miRs can be packaged into exosomes and delivered to various cell types. Exosomes are membrane bound nanovesicles (20–100 nm in diameter) secreted ubiquitously by various cell types. These vesicles contain RNA, miR, proteins, and DNA and can reach other cell types through membrane fusion.118 To exert systemic modulation of cellular functions, viruses have hijacked the host exosomal pathway to disseminate their modulatory biomolecules, including proteins and v-miR.119–121 This allows viruses to shut down key host cell functions. In terms of immune responses, this may allow viruses to persist. Furthermore, unlike proteins, these v-miRs are nonimmunogenic thus providing viruses an advantage. Studies from our lab and others have shown the presence of v-miRs in host cell– derived exosomes in vivo and in vitro, further signifying the functional relevance of v-miRs in viral immunoevasion.64,122–124 EBV miR-BART-15 is secreted in exosomes of EBV-infected B cells as well as macrophages ectopically expressing v-miR. This v-miR share the same seed region as that of cellular miR-223 for its target NLRP3.56 Such interactions provide an evolutionary benefit as these sequences are less likely to undergo mutation to avoid v-miR regulation. B cell–derived exosome mediated delivery of BART-15 into recipient macrophages suppresses inflammasome formation by silencing NLRP3. Considering that exosomal v-miRs can theoretically be delivered to any cell type, these can be considered as broad spectrum virulence factors. While copious amounts of v-miRs are produced in both the latency as well as lytic phase of viral infection, the functional relevance of v-miRs as virulence factors is plausible in latency where they have ample time to downmodulate target genes in a wide range of host cells, particularly immune cells. Overall, non-immunogenic v-miRs are employed as tools by HHV to modulate functions of a wide range of non-permissive cells.
In a recent study, HCMV virions are shown to carry different RNA molecules, both of viral and cellular origin.125 Purified virions and dense bodies (DB) isolated from HCMV infected lung fibroblast cells MRC-5 were profiled for RNA contents. Interestingly, various small RNAs including cellular and viral miR, protein coding, and long non-coding RNA were identified in virions and DB. Among the miRs, 14 HCMV-encoded miR (more than half of the HCMV repertoire) and two cellular miR, viz, miR-218–5p and miR-21–5p, were detected. This indicates that v-miRs are preferentially sorted into the virions; however, the underlying mechanisms remain obscure. Using dual luciferase reporter assays, it was shown that virion contained miR were functionally delivered to the recipient cells. Virion mediated delivery of miR-US25-1-5p into recipient cells expressing luciferase-tagged CCNE2 5′UTR, a known target of candidate v-miR,126 was functionally assessed by silencing of reporter gene expression. Overall, both exosomes and virions are key sources for virus to modulate host cell functions, primarily by sorting large repertoire of v-miRs.
6 |. CONCLUSIONS AND FUTURE PERSPECTIVE
Numerous mechanisms of HHV protein mediated immune evasion and modulation of host cell function are well-known. However, these viruses maintain latent life cycle during which there is little or no viral protein production except LAT transcripts that also encode for miR. Incessant production of v-miRs over a long period might even alter cellular functions. Ability of v-miRs to reach distant sites through exosome pathway can further cause a ripple effect in impairing functions of various cells at tissue or even systemic level. Altered genetic and immune function of host can further facilitate this long-term take-over. Dissecting molecular mechanisms underlying each v-miR will be critical in our understanding of their contribution in viral tropism, persistence and disease manifestation. While the discovery of v-miRs in most of the human viruses have become stagnant, a more challenging path forward is to annotate biological functions to these enigmatic molecules deployed by virus to evade host defenses.
Association of HHV in diverse inflammation-related diseases and/or disorders is increasingly being recognized. For instance, increased prevalence of various members of HHV have been demonstated in Alzheimer’s disease, cancers, multiple sclerosis, periodontitis, peri-apical periodontitis, etc, indicating their contribution in manifesting chronic inflammation, a common feature of most of these diseases.127–131 This is not surprising given that most of these viruses are acquired during childhood, possess multiple immune subversion strategies and persist lifelong. Therefore, v-miRs may serve as a reliable diagnostic marker for HHV infection. A thorough examination of HHV genome detection and its correlation with v-miR may further shed light on how expression levels of these promising viral biomolecules correlate in health and disease.72–74,132–134 This will facilitate future research avenues on employing v-miRs as therapeutic targets, not only for HHV-mediated infections but likely for other diseases as well. How v-miRs can alter the life cycle of HHV in host is another key question that remains unanswered. If exogenous supply of disease-associated v-miRs can support HHV infection and/or persistence, how this changes the microenvironment for other diseases (eg, oral cancers) and pathogens (bacteria and fungi) will be critical questions to address. Studies focusing on chronic inflammatory diseases with HHV association will unravel new etiological role of these elusive viruses. Future studies examining HHV association with other known etiological factors in chronic diseases like periodontitis, Alzheimer’s, multiple sclerosis, and cancers could provide a novel perspective of dynamic players in the etiopathogenesis of multifactorial diseases.
ACKNOWLEDGMENTS
I thank Dr. Salvador Nares, Dr. Alexandra Seal, and Dr. Jennifer Shango for their help in discussion and critical reading of the manuscript. This work was funded by the NIH/NIDCR grants R21 DE026259-01A1, R01 DE027980 and R03DE027147 to ARN.
Funding information
National Institute of Dental and Craniofacial Research, Grant/Award Numbers: R01 DE027980, R03 DE027147, R21 DE026259; NIH/NIDCR, Grant/Award Number: R01 DE027980, R21 DE026259-01A1 and R03 DE027147
Abbreviations used
- AIDS
Acquired Immunodeficiency Syndrome
- APC
antigen presenting cells
- BART
BamHI fragment A rightward transcript
- BHRF1
BamHI fragment H rightward reading frame 1
- C/EBP
CCAAT-enhancer-binding proteins
- CCL5
chemokine (C–C motif) ligand 5
- CREBBP
CREB-binding protein
- CTL
cytotoxic T lymphocyte
- EBV
Epstein-Barr virus
- ERAP1
endoplasmic reticulum aminopeptidase 1
- HCMV
human cytomegalovirus
- HHV 6B
human herpesvirus 6B
- HHV
human herpesviruses
- HSV-1
herpes simplex virus 1
- HSV-2
herpes simplex virus 2
- IFI30
interferon gamma-inducible protein 30
- IFN
interferon
- IKK
inhibitor of kappa B kinase
- IL
interleukin
- IRAK1
interleukin-1 receptor–associated Kinase
- IRF
interferon regulatory factor
- KSHV
Kaposi sarcoma-associated herpesvirus
- LAT
latency-associated transcript
- LMP
latency-associated membrane proteins
- MHC
major histocompatibility complex
- MICA/MICB
MHC class I polypeptide-related sequence A/B
- MyD88
myeloid differentiation primary-response gene
- NFκB
nuclear factor kappa-light-chain-enhancer of activated B cells
- NLR
nucleotide oligomerization domain (NOD)-like receptors
- NLRP3
NLR family, pyrin domain containing 3
- PAMP
pathogen associated molecular patterns
- PEL
primary effusion lymphoma
- RLR
retinoid acid-inducible gene-I (RIG-I)-like receptors
- SOCS2
suppressors of cytokine signaling 2
- Th
T helper
- TIMP
tissue inhibitor of metalloproteinases
- TLR
toll-like receptors
- TNF
tumor necrosis factor
- TWEAKR
TNF-like weak inducer of apoptosis receptor
- UTR
untranslated region
Footnotes
CONFLICT OF INTEREST
The authors have stated explicitly that there are no conflicts of interest in connection with this article.
REFERENCES
- 1.Pellet P, Roizman B. The Family Herpesviridae: A Brief Introduction In: Knipe DM, Howley PM, eds. Fields Virology. Philadelphia, PA: Lippincott Williams, Wilkins; 2007:2479–2499. [Google Scholar]
- 2.Roizman B, Sears AE. Herpes simplex viruses and their replication In: The Human Herpesviruses. New York: Raven Press; 1993. [Google Scholar]
- 3.Piedade D, Azevedo-Pereira JM. The role of microRNAs in the pathogenesis of herpesvirus infection. Viruses. 2016;8(6). 10.3390/v8060156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boshoff C, Weiss R. AIDS-related malignancies. Nat Rev Cancer. 2002;2(5):373–382. 10.1038/nrc797 [DOI] [PubMed] [Google Scholar]
- 5.Maeda E, Akahane M, Kiryu S, et al. Spectrum of Epstein-Barr virus-related diseases: a pictorial review. Jpn J Radiol. 2009;27(1):4–19. 10.1007/s11604-008-0291-2 [DOI] [PubMed] [Google Scholar]
- 6.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332(18):1186–1191. 10.1056/nejm199505043321802 [DOI] [PubMed] [Google Scholar]
- 7.Pfeffer S, Zavolan M, Grässer FA, et al. Identification of virus-encoded microRNAs. Science. 2004;304(5671):734–736. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. [DOI] [PubMed] [Google Scholar]
- 9.Almeida MI, Reis RM, Calin GA. MicroRNA history: discovery, recent applications, and next frontiers. Mutat Res. 2011;717(1–2):1–8. 10.1016/j.mrfmmm.2011.03.009 [DOI] [PubMed] [Google Scholar]
- 10.Naqvi AR, Islam MN, Choudhury NR, Haq QM. The fascinating world of RNA interference. Int J Biol Sci. 2009;5(2):97–117. 10.7150/ijbs.5.97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Skalsky RL, Cullen BR. Viruses, microRNAs, and host interactions. Annu Rev Microbiol. 2010;64(1):123–141. 10.1146/annurev.micro.112408.134243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skalsky RL, Corcoran DL, Gottwein E, et al. The viral and cellular microRNA targetome in lymphoblastoid cell lines. PLoS Pathog. 2012;8(1):e1002484 10.1371/journal.ppat.1002484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grundhoff A, Sullivan CS. Virus-encoded microRNAs. Virology. 2011; 411(2):325–343. 10.1016/j.virol.2011.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR. An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34.5, a viral neurovirulence factor. Proc Natl Acad Sci U S A. 2008;105(31):10931–10936. 10.1073/pnas.0801845105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454(7205):780–783. 10.1038/nature07103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lei X, Bai Z, Ye F, et al. Regulation of NF-kappaB inhibitor IkappaBalpha and viral replication by a KSHV microRNA. Nat Cell Biol. 2010;12(2):193–199. 10.1038/ncb2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan D, Flores O, Umbach JL, et al. A neuron-specific host microRNA targets herpes simplex virus-1 ICP0 expression and promotes latency. Cell Host Microbe. 2014;15(4):446–456. 10.1016/j.chom.2014.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barth S, Pfuhl T, Mamiani A, et al. Epstein-Barr virus-encoded micro-RNA miR-BART2 down-regulates the viral DNA polymerase BALF5. Nucleic Acids Res. 2008;36(2):666–675. 10.1093/nar/gkm1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haasnoot J, Berkhout B. RNAi and cellular miR in infections by mammalian viruses. Methods Mol Biol. 2011;721:23–41. 10.1007/978-1-61779-037-9_2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Seto E, Moosmann A, Grömminger S, Walz N, Grundhoff A, Hammerschmidt W. Micro RNAs of Epstein-Barr virus promote cell cycle progression and prevent apoptosis of primary human B cells. PLoS Pathog. 2010;6(8):e1001063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Umbach JL, Cullen BR. The role of RNAi and microRNAs in animal virus replication and antiviral immunity. Genes Dev. 2009;23(10): 1151–1164. 10.1101/gad.1793309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cui C, Griffiths A, Li G, et al. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol. 2006;80(11):5499–5508. 10.1128/JVI.00200-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kramer MF, Jurak I, Pesola JM, Boissel S, Knipe DM, Coen DM. Herpes simplex virus 1 microRNAs expressed abundantly during latent infection are not essential for latency in mouse trigeminal ganglia. Virology. 2011;417(2):239–247. 10.1016/j.virol.2011.06.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfeffer S, Sewer A, Lagos-Quintana M, et al. Identification of micro-RNAs of the herpesvirus family. Nat Methods. 2005;2(4):269–276. 10.1038/nmeth746 [DOI] [PubMed] [Google Scholar]
- 25.Stark TJ, Arnold JD, Spector DH, Yeo GW. High-resolution profiling and analysis of viral and host small RNAs during human cytomegalovirus infection. J Virol. 2012;86(1):226–235. 10.1128/JVI.05903-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cai X, Lu S, Zhang Z, Gonzalez CM, Damania B, Cullen BR. Kaposi’s sarcoma-associated herpesvirus expresses an array of viral micro-RNAs in latently infected cells. Proc Natl Acad Sci U S A. 2005;102 (15):5570–5575. 10.1073/pnas.040819210227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lin YT, Sullivan CS. Expanding the role of Drosha to the regulation of viral gene expression. Proc Natl Acad Sci U S A. 2011;108(27): 11229–11234. 10.1073/pnas.1105799108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Qiu J, Cosmopoulos K, Pegtel M, et al. A novel persistence associated EBV miR expression profile is disrupted in neoplasia. PLoS Pathog. 2011;7(8):e1002193 10.1371/journal.ppat.1002193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Umbach JL, Cullen BR. In-depth analysis of Kaposi’s sarcoma-associated herpesvirus microRNA expression provides insights into the mammalian microRNA-processing machinery. J Virol. 2010;84(2): 695–703. 10.1128/JVI.02013-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pan D, Li G, Morris-Love J, et al. Herpes simplex virus 1 lytic infection blocks microRNA (miRNA) biogenesis at the stage of nuclear export of pre-miRNAs. MBio. 2019;10(1). pii:e02856–18. 10.1128/mBio.02856-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abecasis GR, Auton A, Brooks LD, De Pristo MA, Durbin RM, Handsaker RE. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. 10.1038/nature11233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davison AJ. Herpesvirus genes. Rev Med Virol. 1993;3(4):237–244. [Google Scholar]
- 34.Davison AJ. Herpesvirus systematics. Vet Microbiol. 2010;143(1):52–69. 10.1016/j.vetmic.2010.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McGeoch DJ, Rixon FJ, Davison AJ. Topics in herpesvirus genomics and evolution. Virus Res. 2006;117(1):90–104. 10.1016/j.virusres.2006.01.002 [DOI] [PubMed] [Google Scholar]
- 36.Barton GM, Medzhitov R. Toll-like receptor signaling pathways. Science. 2003;300(5625):1524–1525. 10.1126/science.1085536 [DOI] [PubMed] [Google Scholar]
- 37.Takeda K, Akira S. Toll-like receptors. Curr Protoc Immunol. 2015; 109:14.12.1–14.12.10. 10.1002/0471142735.im1412s109 [DOI] [PubMed] [Google Scholar]
- 38.Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol. 2009;21(4):317–337. 10.1093/intimm/dxp017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kumar H, Kawai T, Akira S. Pathogen recognition by the innate immune system. Int Rev Immunol. 2011;30(1):16–34. 10.3109/08830185.2010.529976 [DOI] [PubMed] [Google Scholar]
- 40.Ozinsky A, Underhill DM, Fontenot JD, et al. The repertoire for pattern recognition of pathogens by the innate immune system is defined by cooperation between toll-like receptors. Proc Natl Acad Sci U S A. 2000;7(25):13766–13771. 10.1073/pnas.250476497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kawasaki T, Kawai T. Toll-like receptor signaling pathways. Front Immunol. 2014;5:461 10.3389/fimmu.2014.00461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boehme KW, Guerrero M, Compton T. Human cytomegalovirus envelope glycoproteins B and H are necessary for TLR2 activation in permissive cells. J Immunol. 2006;177(10):7094–7102. 10.4049/jimmunol.177.10.7094 [DOI] [PubMed] [Google Scholar]
- 43.Kurt-Jones EA, Chan M, Zhou S, et al. Herpes simplex virus 1 interaction with toll-like receptor 2 contributes to lethal encephalitis. Proc Natl Acad Sci U S A. 2004;101(5):1315–1320. 10.1073/pnas.0308057100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.West JA, Gregory SM, Damania B. Toll-like receptor sensing of human herpesvirus infection. Front Cell Infect Microbiol. 2012;2:122 10.3389/fcimb.2012.00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Landais I, Pelton C, Streblow D, DeFilippis V, McWeeney S, Nelson JA. Human cytomegalovirus miR-UL112–3p targets TLR2 and modulates the TLR2/IRAK1/NFkappaB signaling pathway. PLoS Pathog. 2015;11(5):e1004881 10.1371/journalppat.1004881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barbalat R, Lau L, Locksley RM, Barton GM. Toll-like receptor 2 on inflammatory monocytes induces type I interferon in response to viral but not bacterial ligands. Nat Immunol. 2009;10(11):1200–1207. 10.1038/ni.1792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dietrich N, Lienenklaus S, Weiss S, Gekara NO. Murine toll-like receptor 2 activation induces type I interferon responses from endolysosomal compartments. PLoS ONE. 2010;5(4):e10250 10.1371/journal.pone.0010250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dunne A, Carpenter S, Brikos C, et al. IRAK1 and IRAK4 promote phosphorylation, ubiquitination, and degradation of MyD88 adaptor-like (Mal). J Biol Chem. 2010;285(24):18276–18282. 10.1074/jbc.M109.098137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liljeroos M, Vuolteenaho R, Rounioja S, Henriques-Normark B, Hallman M, Ojaniemi M. Bacterial ligand of TLR2 signals Stat activation via induction of IRF1/2 and interferon-alpha production. Cell Signal. 2008;20(10):1873–1881. 10.1016/j.cellsig.2008.06.017 [DOI] [PubMed] [Google Scholar]
- 50.Watters TM, Kenny EF, O’Neill LA. Structure, function and regulation of the Toll/IL-1 receptor adaptor proteins. Immunol Cell Biol. 2007;85(6):411–419. 10.1038/sj.icb.7100095 [DOI] [PubMed] [Google Scholar]
- 51.Abend JR, Ramalingam D, Kieffer-Kwon P, Uldrick TS, Yarchoan R, Ziegelbauer JM. Kaposi’s sarcoma-associated herpesvirus micro-RNAs target IRAK1 and MYD88, two components of the toll-like receptor/interleukin-1R signaling cascade, to reduce inflammatory-cytokine expression. J Virol. 2012;86(21):11663–11674. 10.1128/JVI.01147-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hancock MH, Hook LM, Mitchell J, Nelson JA. Human cytomegalovirus microRNAs miR-US5–1 and miR-UL112–3p block proinflammatory cytokine production in response to NF-κB-activating factors through direct downregulation of IKKα and IKKβ. MBio. 2017;8(2). pii: e00109–17. 10.1128/mBio.00109-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liang D, Gao Y, Lin X, et al. A human herpesvirus miR attenuates interferon signaling and contributes to maintenance of viral latency by targeting IKKepsilon. Cell Res. 2011;21(5):793–806. 10.1038/cr.2011.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qin Z, Kearney P, Plaisance K, Parsons CH. Kaposi’s sarcoma-associated herpesvirus (KSHV)-encoded microRNA specifically induce IL-6 and IL-10 secretion by macrophages and monocytes. J Leukoc Biol. 2010;87(1):25–34. 10.1189/jlb.0409251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lu Y, Qin Z, Wang J, et al. Epstein-Barr virus miR-BART6–3p inhibits the RIG-I pathway. J Innate Immun. 2017;9(6):574–586. 10.1159/000479749 [DOI] [PubMed] [Google Scholar]
- 56.Haneklaus M, Gerlic M, Kurowska-Stolarska M, et al. miR-223 and EBV miR-BART15 regulate the NLRP3 inflammasome and IL-1beta production. J Immunol. 2012;189(8):3795–3799. 10.4049/jimmunol.1200312 [DOI] [PubMed] [Google Scholar]
- 57.Hooykaas MJG, van Gent M, Soppe JA, et al. EBV MicroRNA BART16 suppresses type I IFN signaling. J Immunol. 2017;198(10): 4062–4073. 10.4049/jimmunol.1501605 [DOI] [PubMed] [Google Scholar]
- 58.Skinner CM, Ivanov NS, Barr SA, Chen Y, Skalsky RL. An Epstein-Barr virus MicroRNA blocks interleukin-1 (IL-1) signaling by targeting IL-1 receptor 1. J Virol. 2017;91(21). pii: e00530–17. 10.1128/JVI.00530-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang YM, Yu Y, Zhao HP. EBV-BART-6–3p and cellular microRNA-197 compromise the immune defense of host cells in EBV-positive Burkitt lymphoma. Mol Med Rep. 2017;15(4):1877–1883. 10.3892/mmr.2017.6173 [DOI] [PubMed] [Google Scholar]
- 60.Lau B, Poole E, Krishna B, et al. The expression of human cytomegalovirus microRNA MiR-UL148D during latent infection in primary myeloid cells inhibits activin A-triggered secretion of IL-6. Sci Rep. 2016;6(1):31205 10.1038/srep31205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X, Liu S, Zhou Z, Yan H, Xiao J. A herpes simplex virus type 2-encoded microRNA promotes tumor cell metastasis by targeting suppressor of cytokine signaling 2 in lung cancer. Tumour Biol. 2017; 39(5):1010428317701633 10.1177/1010428317701633 [DOI] [PubMed] [Google Scholar]
- 62.Abend JR, Uldrick T, Ziegelbauer JM. Regulation of tumor necrosis factor-like weak inducer of apoptosis receptor protein (TWEAKR) expression by Kaposi’s sarcoma-associated herpesvirus microRNA prevents TWEAK-induced apoptosis and inflammatory cytokine expression. J Virol. 2010;84(23):12139–12151. 10.1128/jvi.00884-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang Y, Qi Y, Ma Y, et al. The expression of interleukin-32 is activated by human cytomegalovirus infection and down regulated by hcmv-miR-UL112–1. Virol J. 2013;10(1):51 10.1186/1743-422x-10-51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Naqvi AR, Shango J, Seal A, Shukla D, Nares S. Viral miR alter host cell miR profiles and modulate innate immune responses. Front Immunol. 2018;9:433 10.3389/fimmu.2018.00433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Franchi L, Warner N, Viani K, Nuñez G. Function of nod-like receptors in microbial recognition and host defense. Immunol Rev. 2009; 227(1):106–128. 10.1111/j.1600-065X.2008.00734.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xu XX, Wan H, Nie L, Shao T, Xiang LX, Shao JZ. RIG-I: a multifunctional protein beyond a pattern recognition receptor. Protein Cell. 2018;9(3):246–253. 10.1007/s13238-017-0431-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Devergne O, Peuchmaur M, Humbert M, et al. In vivo expression of IL-1 beta and IL-6 genes during viral infections in human. Eur Cytokine Netw. 1991;2(3):183–194. [PubMed] [Google Scholar]
- 68.Li H, Zhang J, Kumar A, Zheng M, Atherton SS, Yu FS. Herpes simplex virus 1 infection induces the expression of proinflammatory cytokines, interferons and TLR7 in human corneal epithelial cells. Immunology. 2006;117(2):167–176. 10.1111/j.1365-2567.2005.02275.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ohguchi M, Yamato K, Ishihara Y, et al. Activin A regulates the production of mature interleukin-1beta and interleukin-1 receptor antagonist in human monocytic cells. J Interferon Cytokine Res. 1998; 18(7):491–498. [DOI] [PubMed] [Google Scholar]
- 70.Wiley SR, Winkles JA. TWEAK, a member of the TNF superfamily, is a multifunctional cytokine that binds the TweakR/Fn14 receptor. Cytokine Growth Factor Rev. 2003;14(3–4):241–249. 10.1016/S1359-6101(03)00019-4 [DOI] [PubMed] [Google Scholar]
- 71.Ramalingam D, Kieffer-Kwon P, Ziegelbauer JM. Emerging themes from EBV and KSHV microRNA targets. Viruses. 2012;4(9):1687–1710. 10.3390/v4091687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhong S, Naqvi A, Bair E, Nares S, Khan AA. Viral microRNAs identified in human dental pulp. J Endod. 2017;43(1):84–89. 10.1016/j.joen.2016.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Naqvi AR, Brambila MF, Martínez G, Chapa G, Nares S. Dys-regulation of human miR and increased prevalence of HHV miR in obese periodontitis subjects. J Clin Periodontol. 2019;46(1):51–61. 10.1111/jcpe.13040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Naqvi AR, Seal A, Shango J, et al. Herpesvirus-encoded microRNAs detected in human gingiva alter host cell transcriptome and regulate viral infection. Biochim Biophys Acta Gene Regul Mech. 2018;1861(5): 497–508. 10.1016/j.bbagrm.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Naqvi AR, Shango J, Seal A, Shukla D, Nares S. Herpesviruses and MicroRNAs: new pathogenesis factors in oral infection and disease? Front Immunol. 2018;9:2099 10.3389/fimmu.2018.02099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Naqvi AR, Seal A, Shango J, Shukla D, Nares S. In silico prediction of cellular gene targets of herpesvirus encoded microRNAs. Data Brief. 2018;19:249–255. 10.1016/j.dib.2018.05.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hook L, Hancock M, Landais I, Grabski R, Britt W, Nelson JA. Cytomegalovirus microRNAs. Curr Opin Virol. 2014;7:40–46. 10.1016/j.coviro.2014.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Han Z, Liu X, Chen X, et al. miR-H28 and miR-H29 expressed late in productive infection are exported and restrict HSV-1 replication and spread in recipient cells. Proc Natl Acad Sci U S A. 2016;113(7):E894–E901. 10.1073/pnas.1525674113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Blum JS, Wearsch PA, Cresswell P. Pathways of antigen processing. Annu Rev Immunol. 2013;31(1):443–473. 10.1146/annurev-immunol-032712-095910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Neefjes J, Jongsma ML, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol. 2011;11(12):823–836. 10.1038/nri3084 [DOI] [PubMed] [Google Scholar]
- 81.Roche PA, Furuta K. The ins and outs of MHC class II-mediated antigen processing and presentation. Nat Rev Immunol. 2015;15(4):203–216. 10.1038/nri3818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Raghavan M, Del Cid N, Rizvi SM, Peters LR. MHC class I assembly: out and about. Trends Immunol. 2008;29(9):436–443. 10.1016/j.it.2008.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.van de Weijer ML, Luteijn RD, Wiertz EJ. Viral immune evasion: lessons in MHC class I antigen presentation. Semin Immunol. 2015;27 (2):125–137. 10.1016/j.smim.2015.03.010 [DOI] [PubMed] [Google Scholar]
- 84.Kidd P Th1/Th2 balance: the hypothesis, its limitations, and implications for health and disease. Altern Med Rev. 2003;8(3):223–246. [PubMed] [Google Scholar]
- 85.Romagnani S T-cell subsets (Th1 versus Th2). Ann Allergy Asthma Immunol. 2000;85(1):9–21. 10.1016/s1081-1206(10)62426-x [DOI] [PubMed] [Google Scholar]
- 86.van Baarle D, Hovenkamp E, Callan MF, et al. Dysfunctional Epstein-Barr virus (EBV)-specific CD8(+) T lymphocytes and increased EBV load in HIV-1 infected individuals progressing to AIDS-related non-Hodgkin lymphoma. Blood. 2001;98(1):146–155. 10.1182/blood.V98.1.146 [DOI] [PubMed] [Google Scholar]
- 87.DuPage M, Bluestone JA. Harnessing the plasticity of CD4(+) T cells to treat immune-mediated disease. Nat Rev Immunol. 2016;16(3): 149–163. 10.1038/nri.2015.18 [DOI] [PubMed] [Google Scholar]
- 88.Hirahara K, Poholek A, Vahedi G, et al. Mechanisms underlying helper T-cell plasticity: implications for immune-mediated disease. J Allergy Clin Immunol. 2013;131(5):1276–1287. 10.1016/j.jaci.2013.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Roder G, Geironson L, Bressendorff I, Paulsson K. Viral proteins interfering with antigen presentation target the major histocompatibility complex class I peptide-loading complex. J Virol. 2008;82(17): 8246–8252. 10.1128/jvi.00207-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Yewdell JW, Bennink JR. Mechanisms of viral interference with MHC class I antigen processing and presentation. Annu Rev Cell Dev Biol. 1999;15(1):579–606. 10.1146/annurev.cellbio.15.1.579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Horst D, van Leeuwen D, Croft NP, et al. Specific targeting of the EBV lytic phase protein BNLF2a to the transporter associated with antigen processing results in impairment of HLA class I-restricted antigen presentation. J Immunol. 2009;182(4):2313–2324. 10.4049/jimmunol.0803218 [DOI] [PubMed] [Google Scholar]
- 92.Oosten LE, Koppers-Lalic D, Blokland E, et al. TAP-inhibiting proteins US6, ICP47 and UL49.5 differentially affect minor and major histocompatibility antigen-specific recognition by cytotoxic T lymphocytes. Int Immunol. 2007;19(9):1115–1122. 10.1093/intimm/dxm082 [DOI] [PubMed] [Google Scholar]
- 93.Verweij MC, Horst D, Griffin BD, et al. Viral inhibition of the transporter associated with antigen processing (TAP): a striking example of functional convergent evolution. PLoS Pathog. 2015;11(4):e1004743 10.1371/journal.ppat.1004743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Tagawa T, Albanese M, Bouvet M, et al. Epstein-Barr viral miR inhibit antiviral CD4+ T cell responses targeting IL-12 and peptide processing. J Exp Med. 2016;213(10):2065–2080. 10.1084/jem.20160248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nachmani D, Stern-Ginossar N, Sarid R, Mandelboim O. Diverse herpesvirus microRNAs target the stress-induced immune ligand MICB to escape recognition by natural killer cells. Cell Host Microbe. 2009;5(4):376–385. 10.1016/j.chom.2009.03.003 [DOI] [PubMed] [Google Scholar]
- 96.Stern-Ginossar N, Elefant N, Zimmermann A, et al. Host immune system gene targeting by a viral miR. Science. 2007;317(5836):376–381. 10.1126/science.1140956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Esteso G, Luzon E, Sarmiento E, et al. Altered microRNA expression after infection with human cytomegalovirus leads to TIMP3 down-regulation and increased shedding of metalloprotease substrates, including MICA. J Immunol. 2014;193(3):1344–1352. 10.4049/jimmunol.1303441 [DOI] [PubMed] [Google Scholar]
- 98.Kim S, Lee S, Shin J, et al. Human cytomegalovirus microRNA miR-US4–1 inhibits CD8(+) T cell responses by targeting the aminopeptidase ERAP1. Nat Immunol. 2011;12(10):984–991. 10.1038/ni.2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lo AK, To KF, Lo KW, et al. Modulation of LMP1 protein expression by EBV-encoded microRNAs. Proc Natl Acad Sci U S A. 2007;104 (41):16164–16169. 10.1073/pnas.0702896104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lung RW, Tong JH, Sung YM, et al. Modulation of LMP2A expression by a newly identified Epstein-Barr virus-encoded microRNA miR-BART22. Neoplasia. 2009;11(11):1174–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kim Y, Lee S, Kim S, Kim D, Ahn JH, Ahn K. Human cytomegalovirus clinical strain-specific microRNA miR-UL148D targets the human chemokine RANTES during infection. PLoS Pathog. 2012;8(3): e1002577 10.1371/journal.ppat.1002577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Xia T, O’Hara A, Araujo I, et al. EBV microRNAs in primary lymphomas and targeting of CXCL-11 by ebv-mir-BHRF1–3. Cancer Res. 2008;68(5):1436–1442. 10.1158/0008-5472.can-07-5126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Keir ME, Butte MJ, Freeman GJ, Sharpe AH. PD-1 and its ligands in tolerance and immunity. Annu Rev Immunol. 2008;26(1):677–704. 10.1146/annurev.immunol.26.021607.090331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Tomescu C, Law WK, Kedes DH. Surface downregulation of major histocompatibility complex class I, PE-CAM, and ICAM-1 following de novo infection of endothelial cells with Kaposi’s sarcoma-associated herpesvirus. J Virol. 2003;77(17):9669–9684. 10.1128/jvi.77.17.9669-9684.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Maghazachi AA, Al-Aoukaty A, Schall TJ. CC chemokines induce the generation of killer cells from CD56+ cells. Eur J Immunol. 1996;26 (2):315–319. 10.1002/eji.1830260207 [DOI] [PubMed] [Google Scholar]
- 106.Flores O, Nakayama S, Whisnant AW, Javanbakht H, Cullen BR, Bloom DC. Mutational inactivation of herpes simplex virus 1 micro-RNAs identifies viral mRNA targets and reveals phenotypic effects in culture. J Virol. 2013;87(12):6589–6603. 10.1128/JVI.00504-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Pan D, Pesola JM, Li G, McCarron S, Coen DM. Mutations inactivating herpes simplex virus 1 microRNA miR-H2 do not detectably increase ICP0 gene expression in infected cultured cells or mouse trigeminal ganglia. J Virol. 2017;91(2). 10.1128/JVI.02001-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jiang X, Brown D, Osorio N, Hsiang C, BenMohamed L, Wechsler SL. Increased neurovirulence and reactivation of the herpes simplex virus type 1 latency-associated transcript (LAT)-negative mutant dLAT2903 with a disrupted LAT miR-H2. J Neurovirol. 2016;22(1):38–49. 10.1007/s13365-015-0362-y109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jiang X, Brown D, Osorio N, et al. A herpes simplex virus type 1 mutant disrupted for microRNA H2 with increased neurovirulence and rate of reactivation. J Neurovirol. 2015;21(2):199–209. 10.1007/s13365-015-0319-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wahl A, Linnstaedt SD, Esoda C, et al. A cluster of virus-encoded microRNAs accelerates acute systemic Epstein-Barr virus infection but does not significantly enhance virus-induced oncogenesis in vivo. J Virol. 2013;87(10):5437–5446. 10.1128/JVI.00281-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Feederle R, Linnstaedt SD, Bannert H, et al. A viral microRNA cluster strongly potentiates the transforming properties of a human herpesvirus. PLoS Pathog. 2011;7(2):e1001294 10.1371/journal.ppat.1001294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.van Diemen FR, Kruse EM, Hooykaas MJ, et al. CRISPR/Cas9-mediated genome editing of herpesviruses limits productive and latent infections. PLoS Pathog. 2016;12(6):e1005701 10.1371/journal.ppat.1005701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lin C, Li H, Hao M, et al. Increasing the efficiency of CRISPR/Cas9-mediated precise genome editing of HSV-1 virus in human cells. Sci Rep. 2016;6(1):34531 10.1038/srep34531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Wang J, Quake SR. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proc Natl Acad Sci U S A. 2014;111(36):13157–13162. 10.1073/pnas.1410785111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Koujah L, Shukla D, Naqvi AR. CRISPR-Cas based targeting of host and viral genes as an antiviral strategy. Semin Cell Dev Biol. 2019:pii: S1084–9521(18)30108–3. 10.1016/j.semcdb.2019.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Moody R, Zhu Y, Huang Y, et al. KSHV microRNAs mediate cellular transformation and tumorigenesis by redundantly targeting cell growth and survival pathways. PLoS Pathog. 2013;9(12):e1003857 10.1371/journal.ppat.1003857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Lau B, Poole E, Van Damme E, et al. Human cytomegalovirus miR-UL112–1 promotes the down-regulation of viral immediate early-gene expression during latency to prevent T-cell recognition of latently infected cells. J Gen Virol. 2016;97(9):2387–2398. 10.1099/jgv.0.000546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Colombo M, Raposo G, Théry C. Biogenesis, secretion, and inter-cellular interactions of exosomes and other extracellular vesicles. Annu Rev Cell Dev Biol. 2014;30(1):255–289. 10.1146/annurev-cellbio-101512-122326 [DOI] [PubMed] [Google Scholar]
- 119.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9 (6):654–659. 10.1038/ncb1596 [DOI] [PubMed] [Google Scholar]
- 120.Sadeghipour S, Mathias RA. Herpesviruses hijack host exosomes for viral pathogenesis. Semin Cell Dev Biol. 2017;67:91–100. 10.1016/j.semcdb.2017.03.005 [DOI] [PubMed] [Google Scholar]
- 121.Aqil M, Naqvi AR, Bano AS, Jameel S. The HIV-1 Nef protein binds argonaute-2 and functions as a viral suppressor of RNA interference. PLoS ONE. 2013;8(9):e74472 10.1371/journal.pone.0074472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Aqil M, Naqvi AR, Mallik S, Bandyopadhyay S, Maulik U, Jameel S. The HIV Nef protein modulates cellular and exosomal miR profiles in human monocytic cells. J Extracell Vesicles. 2014;3(1):23129 10.3402/jev.v3.23129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, et al. Functional delivery of viral miR via exosomes. Proc Natl Acad Sci U S A. 2010; 107(14):6328–6333. 10.1073/pnas.0914843107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Chugh PE, Sin SH, Ozgur S, et al. Systemically circulating viral and tumor-derived microRNAs in KSHV-associated malignancies. PLoS Pathog. 2013;9(7):e1003484 10.1371/journal.ppat.1003484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Mohammad AA, Costa H, Landázuri N, et al. Human cytomegalovirus microRNAs are carried by virions and dense bodies and are delivered to target cells. J Gen Virol. 2017;98(5):1058–1072. 10.1099/jgv.0.000736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Grey F, Tirabassi R, Meyers H, et al. A viral microRNA down-regulates multiple cell cycle genes through mRNA 5’UTRs. PLoS Pathog. 2010;6(6):e1000967 10.1371/journal.ppat.1000967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Lin WR, Wozniak MA, Cooper RJ, Wilcock GK, Itzhaki RF. Herpesviruses in brain and Alzheimer’s disease. J Pathol. 2002;197(3):395–402. 10.1002/path.1127 [DOI] [PubMed] [Google Scholar]
- 128.Alibek K, Baiken Y, Kakpenova A, et al. Implication of human herpesviruses in oncogenesis through immune evasion and supression. Infect Agent Cancer. 2014;9(1):3 10.1186/1750-9378-9-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Meyding-Lamadé U, Strank C. Herpesvirus infections of the central nervous system in immunocompromised patients. Ther Adv Neurol Disord. 2012;5(5):279–296. 10.1177/1756285612456234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Jakovljevic A, Andric M. Human cytomegalovirus and Epstein-Barr virus in etiopathogenesis of apical periodontitis: a systematic review. J Endod. 2014;40(1):6–15. 10.1016/j.joen.2013.10.001 [DOI] [PubMed] [Google Scholar]
- 131.Jankovic S, Aleksic Z, Dimitrijevic B, Lekovic V, Camargo P, Kenney B. Prevalence of human cytomegalovirus and Epstein-Barr virus in subgingival plaque at peri-implantitis, mucositis and healthy sites. A pilot study. Int J Oral Maxillofac Surg. 2011;40(3):271–276. 10.1016/j.ijom.2010.11.004 [DOI] [PubMed] [Google Scholar]
- 132.Wang YF, He DD, Liang HW, et al. The identification of up-regulated ebv-miR-BHRF1-2-5p targeting MALT1 and ebv-miR-BHRF1–3 in the circulation of patients with multiple sclerosis. Clin Exp Immunol. 2017;189(1):120–126. 10.1111/cei.12954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kang BW, Choi Y, Kwon OK, et al. High level of viral microRNA-BART20–5p expression is associated with worse survival of patients with Epstein-Barr virus-associated gastric cancer. Oncotarget. 2017; 8(9):14988–14994. 10.18632/oncotarget.14744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Yun SJ, Jeong P, Kang HW, et al. Increased expression of herpes virus-encoded hsv1-mir-h18 and hsv2-mir-h9–5p in cancer-containing prostate tissue compared to that in benign prostate hyper-plasia tissue. Int Neurourol J. 2016;20(2):122–130. 10.5213/inj.1632552.276 [DOI] [PMC free article] [PubMed] [Google Scholar]


