Abstract
Local adaptation directs populations towards environment-specific fitness maxima through acquisition of positively-selected traits. However, rapid environmental changes can identify hidden fitness trade-offs that turn adaptation into maladaptation, resulting in evolutionary traps. Cancer, a disease prone to drug resistance, is in principle susceptible to such traps. We therefore performed pooled CRISPR/Cas9 knockout screens in acute myeloid leukemia (AML) cells treated with various chemotherapies to map the drug-dependent genetic basis of fitness trade-offs, a concept known as antagonistic pleiotropy (AP). We identified a PRC2-NSD2/3-mediated MYC regulatory axis as a drug-induced AP pathway whose ability to confer resistance to bromodomain inhibition and sensitivity to BCL-2 inhibition templates an evolutionary trap. Across diverse AML cell line and PDX models, we find that acquisition of resistance to bromodomain inhibition through this pathway exposes coincident hypersensitivity to BCL-2 inhibition. Thus, drug-induced AP can be leveraged to design evolutionary traps that selectively target drug resistance in cancer.
Introduction
All populations are subject to selective pressures that enforce local adaptation. In certain cases, the evolved traits that confer local adaptation to a given environment may become maladaptive following changes to that environment [1, 2]. These reversals of fortune are known as evolutionary mismatches or evolutionary traps [3, 4]. Provided that the environmental shift occurs abruptly enough to evade adaptation and the mismatch is sufficiently large, evolutionary traps can drive local extinction [5].
This concept should be applicable to the construction of two-body chemotherapeutic regimens for the eradication of cancer cell populations. Specifically, local adaptation to chemotherapy could prime an evolutionary trap, which would be triggered upon administration of a carefully-selected second drug. This approach is potentially well-suited to address the problem of acquired drug resistance in cancer for several reasons. First, acquired drug resistance is often a genetically- or epigenetically-mediated, cell-autonomous process that is selected for in the presence of drug but could in principle be selected against [6-9]. Second, a growing body of evidence suggests that diverse upstream drivers of chemotherapeutic resistance often converge on and activate common effectors, permitting the design of wide-reaching evolutionary traps that exploit recurrent downstream mediators of resistance. [7, 10-16]. Lastly, many chemotherapeutic strategies already require multiple drugs administered in sequence, allowing for the adoption of evolutionary traps by amending or shuffling existing chemotherapy regimens.
Recognizing this opportunity, a handful of studies have identified “collateral sensitivities” in cancer cells, scenarios where acquired resistance to an initial therapy produces heightened sensitivity to a second therapy, providing the first experimental support for evolutionary traps in cancer [7, 13, 17]. However, for evolutionary traps to be rationally applied, we require a framework for connecting the specific genetic events that drive acquired drug resistance to the collateral sensitivities that can be used to exploit this resistance. Here we provide such a framework, producing a genetically-coherent model of chemotherapy-induced evolutionary traps.
This framework is built on, and requires, an understanding of drug-induced antagonistic pleiotropy (AP), the concept that genes can exert diametrically-opposed effects on fitness in different drug contexts [18, 19]. The gold-standard method for testing local adaptation and/or AP is reciprocal transplantation, a laborious process that artificially restricts the number of putative AP interactions that can be validated at once [18, 19]. To overcome this inefficiency, we used a pooled, loss-of-function CRISPR/Cas9 library targeting major growth, proliferative, and oncogenic pathways, to screen nine chemotherapies, effectively performing thousands of reciprocal transplantation experiments in parallel. This approach allowed us to produce the first systematic analysis of drug-induced AP, which we use to design an evolutionary trap that subverts acquired drug resistance. The principles described here should be broadly applicable, both for the phenomenological study of AP in cancer and for the rational construction of chemotherapy-driven evolutionary traps designed to address acquired drug resistance.
Results
Using loss-of-function genetics to identify drug-induced AP
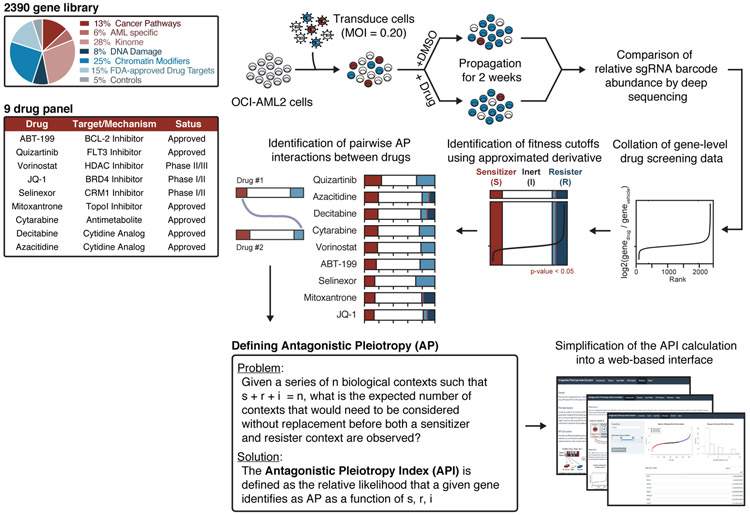
To identify genes that exhibit drug-induced AP, we assembled a CRISPR/Cas9 loss-of-function library composed of 11,200 short guide RNA (sgRNA) constructs targeting 2,240 genes from major cellular and oncogenic signaling pathways, as well as 50 non-targeting constructs and 750 constructs targeting 150 control genes, chosen for their predicted dispensability or essentiality (Supplementary Table 1) [20]. This library was cloned into a published lentiviral system [21] and used to screen a panel of nine chemotherapies in AML cells as previously described [22]. In brief, cells were transduced with library lentivirus, selected with puromycin, treated with vehicle or drug, sampled at zero and two weeks, and deconvoluted with deep sequencing (Fig. 1). The compositional abundance of sgRNAs in each drug-treated population was compared to that of the untreated population. Construct scores were averaged into gene scores. (Methods, Supplementary Table 2,3).
Figure 1: Loss-of-function CRISPR screens identify drug-induced antagonistic pleiotropy.
a, Pooled drug-modifier CRISPR/Cas9 screening and API analysis strategy. CRISPR/Cas9 drug-modifier screens represented as fractions of sensitizer (red), inert (white), and resister (blue) genes. Gene cutoffs controlled by p-value of 0.05. MOI, multiplicity of infection.
We sought to identify the AP genes within these drug-modifier screens. A minimal demonstration of drug-induced AP requires that a gene scores as a sensitizer to at least one drug (gene loss potentiates drug effect; depleted representation) and as a resister to at least one drug (gene loss refutes drug effect; replete representation). Because it is difficult to impose equivalent selection pressures using different drugs, we used the topology of each drug screen to define fitness thresholds. In brief, genes from each drug screen, ranked from fitness-loss to fitness-gain, were trichotomized by imposing two cutoffs, distinguishing fitness loss from inertness and inertness from fitness gain (Methods). These cutoffs were evaluated against the distribution of non-targeting control genes and controlled at p<0.05. The resulting drug-induced, gene-level designations were used to define three types of pairwise gene-gene interactions within the drug-treated datasets: sensitizer-sensitizer (shared detriment), resister-resister (shared benefit), and sensitizer-resister interactions. Sensitizer-resister interactions are particularly notable because they represent examples of drug-induced AP, instances where loss of a gene confers a fitness benefit in the presence of one drug and a fitness penalty in the presence of another. Applying this generous definition of drug-induced AP to our data classified nearly half (1,174 of 2,390 genes) of the genes in our library as AP.
Defining an antagonistic pleiotropy index (API)
To prioritize genes by the strength of their drug-induced AP properties, we formalized an antagonistic pleiotropy index (API), expressed in terms of the total number of contexts (n) and the number of contexts in which a gene is classified as a sensitizer (s), resister (r), or inert (i). An analytical solution can be obtained by solving the following problem: given a series of contexts such that s + r + i = n, what is the expected number of contexts without replacement before both sensitizer and resister contexts are observed, qualifying the gene as AP? A solution to this problem is provided (Supplementary Note) in which the genes most likely to be observed as AP have the lowest API and vice versa.
To validate the basic functions of the API, we reanalyzed two publicly-available datasets. These analyses serve to affirm the basic functions of the API and provide an intuitive understanding for the quantification of AP (Supplementary Note, Supplementary Fig. 1a-k). To facilitate adoption of the API, we have provided a web interface (https://apicalculator.shinyapps.io/api_shiny_directory/), which users can use to analyze their own datasets.
Analyzing drug-treated screens using API
Next, we applied the API to our drug-modifier screens (Supplementary Fig. 2a, Supplementary Table 4). Ontology analysis of the top AP genes, defined as API < 4.5 (348 genes, top 15%; Supplementary Table 4), showed enrichment for transcriptional and epigenetic modifiers (Supplementary Fig. 2b). This coheres with the notion that chromatin modifications can drive both drug-tolerance and chemosensitivity in cancer cells [23-26].
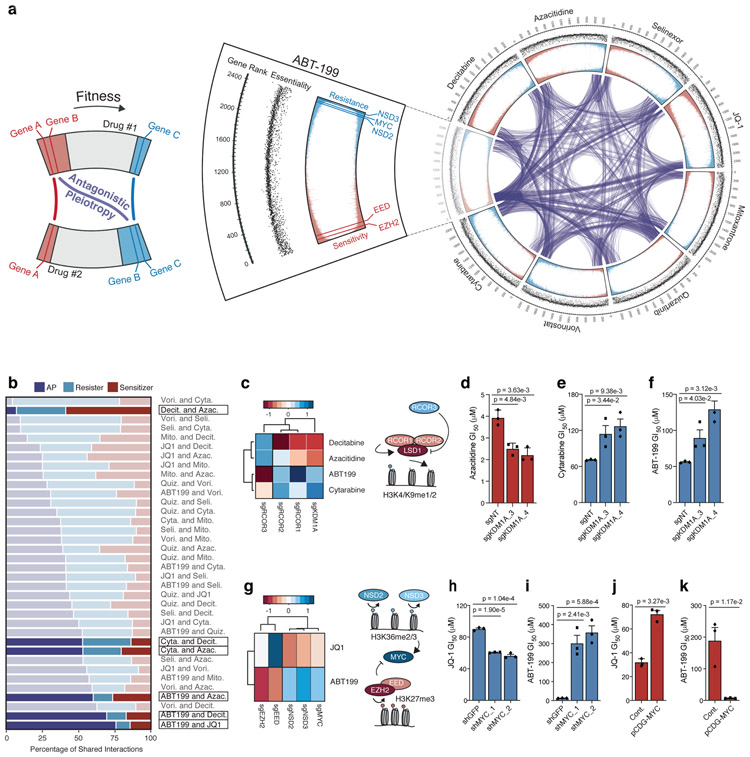
To map the connections between fitness-detrimental and fitness-beneficial genes, we arranged our CRISPR screens circumferentially as segments of a circos plot, the segments connected to each other through links representing shared chemosensitizers or chemoresisters (Fig. 2a and Supplementary Fig. 2c). Within each drug pair, our analysis identified genes that were fitness-detrimental in the presence of one drug and fitness-beneficial for another—AP genes (Fig. 2a). The linkage patterns that connected drug tracts to one another uncovered broad trends. For instance, decitabine, azacitidine, and JQ-1 share many fitness-detrimental and fitness-beneficial genes but few AP genes, consistent with PCA analysis of the screens (Supplementary Fig. 2d).
Figure 2: Analysis of drug-induced antagonistic pleiotropy reveals gene-gene and drug-drug interactions.
a, Circos plot displaying data from drug-modifier CRISPR screens. Screen conducted in presence of ABT-199 shown as cutout with annotations from outermost rim: genes ranked from most sensitizing (rank = 1) to most resisting (rank = 2,390); scatterplot of corresponding gene essentiality score; drug-modifier score (shown in replicate), colored to depict sensitizers (red) and resisters (blue). Gene-gene relationships that exhibit AP between drug screens indicated by purple connections. b, Breakdown of shared sensitizer, shared resister and AP gene interactions between 36 drug pairs. Drug pairs ranked by percentage of AP gene interactions. c, Heatmap representing effect of sgRNAs targeting KDM1A (encoding LSD1) and RCOR1/2/3 (n = 5 sgRNAs per gene) on ABT-199, cytarabine, decitabine and azacitidine; schematic depicts known relationship between genes identified in screen. d-f, GI50 values of azacitidine (d), cytarabine (e), and ABT-199 (f) in OCI-AML2 cells following CRISPR/Cas9 mediated knockout of KDM1A versus non-targeting control. g, Heatmap representing effect of sgRNAs targeting MYC, NSD2/3, EED, EZH2 (n = 5 sgRNAs per gene) on JQ-1 and ABT-199 from CRISPR/Cas9 screens; schematic depicts reciprocal effects of NSD2/3 and EED, EZH2 on their respective methylation marks and, indirectly, MYC. h-i, GI50 values of JQ-1 (h) and ABT-199 (i) following short-hairpin knockdown of MYC versus shGFP. j-k, GI50 values of JQ-1 (j) and ABT-199 (k) following overexpression of pCDH-MYC versus control vector. d-f; h-k, P-values computed by two-sided t-Test for equal means. Data are presented as mean ± SEM for n = 3 biologically independent experiments.
Importantly, screens also resolved differences between drugs. Azacitidine requires activation by uridine-cytidine kinase 2 (encoded by UCK2) [27] while its deoxy-derivative, decitabine, is activated by deoxycytidine kinase (encoded by DCK) [28, 29]. Both genes scored strongly as resisters in their respective drug screens and inertly in the other drug screens (Supplementary Fig. 2e). Accordingly, the screen conducted with cytarabine, a deoxycytidine antimetabolite which shares structural similarities with decitabine, identified DCK as a resister and the equilibrative nucleoside transporter hENT1 (encoded by SLC29A1), required for uptake of deoxycytidines [29], scored as a strong resister in the decitabine and cytarabine screens but not in the azacitidine screen (Supplementary Fig. 2e-j).
The PRC2 complex and NSD2/3 comprise an AP axis
Next, we evaluated each of the 36 possible drug pairs for their shared sensitizer, resister, and AP interactions across the set of top AP genes (API < 4.5) (Fig. 2b). We observed wide variation in the composition of shared interactions across drug pairs. For instance, the hypomethylating agents decitabine and azacitidine harbored few AP interactions along with the highest percentage of shared sensitizer interactions. In contrast, there were many AP interactions between ABT-199 and both decitabine and azacitidine which stood apart from pairwise relationships identified using PCA or correlative analyses (Supplementary Fig. 3a,b). We validated an exemplar AP axis composed of KDM1A (encoding LSD1) and known LSD1 regulators repressor element-1 silencing transcription factor corepressors 1, 2, and 3 (RCOR1/2/3) (Fig. 2c-f). Additional development of KDM1A as an AP gene can be found in the supplement (Supplementary Note, Supplementary Fig. 4a-i).
Among the drug pairs, ABT-199 and JQ-1 recorded the highest percentage of AP interactions, suggesting that the resister and sensitizer landscapes of these drugs were most opposed (Fig. 2b). In particular, we identified four AP genes that were unified on the basis of their reciprocal transcriptional regulation. EZH2 (API = 4.2) and EED (API = 3.9), which encode components of PRC2, a gene silencing complex that catalyzes H3K27 trimethylation [30, 31], scored as sensitizers in the ABT-199 screen; EED also scored as a resister in the JQ-1 screen (Fig. 2g, Supplementary Fig. 5a). The transcriptionally-repressive H3K27me3 marks deposited by PRC2 are opposed by transcriptionally-activating H3K36me1/2 marks [32, 33]. Accordingly, two H3K36 methyltransferases, NSD2 (API = 3.3) and NSD3 (API = 4.7) [34], scored in opposition to the PRC2 members (Fig. 2g, Supplementary Fig. 5a). While these findings were consistent, it was unclear why these epigenetic modifiers should modulate the effect of these two drugs. To find possible effectors, we canvassed our dataset for genes that behaved like EZH2/EED or NSD2/NSD3 in the ABT-199 and JQ-1 screens. Our search identified the transcription factor MYC, which scored alongside NSD2/3 and opposite EZH2/EED in the ABT-199 and JQ-1 screens (Fig. 2g, Supplementary Fig. 5a). These correlations agree with prior studies, which describe the negative transcriptional control of MYC by PRC2 [35], and together delineate an AP axis featuring the reciprocal regulation of MYC by NSD2/3 and EZH2/EED (PRC2) (Fig. 2g).
EZH2/EED counterpoise NSD2/3 at the level of MYC
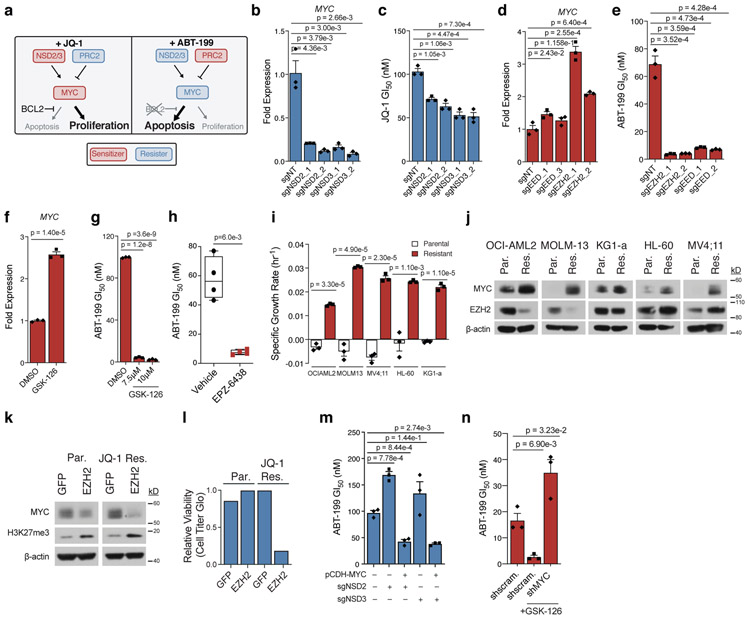
To investigate MYC’s capacity to affect drug sensitivity, we performed knockdown and overexpression studies. Compared to cells expressing scrambled short hairpin RNAs (shRNAs), cells expressing MYC-targeting shRNAs were sensitized to JQ-1 and resistant to ABT-199 (Fig. 2h,i, Supplementary Fig. 5b-e). Reciprocally, overexpression of MYC conferred resistance to JQ-1 and sensitized cells to ABT-199 (Fig. 2j,k, Supplementary Fig. 5f-i). Taken alongside data from the screen, these studies validated the sufficiency of MYC to modulate drug sensitization (Fig. 3a).
Figure 3: MYC and its epigenetic regulators are drug-induced AP genes.
a, Schematic depicting drug-dependent polarity of MYC axis, as defined by drug-modifier screens. b-c, Effect of sgRNAs targeting NSD2 or NSD3 versus non-targeting control in OCI-AML2 cells on MYC transcripts (b) and JQ-1 GI50 (c). d-e, Effect of sgRNAs targeting EED or EZH2 versus non-targeting control in OCI-AML2 cells on MYC transcripts (d) and ABT-199 GI50 (e). f, Fold-change of MYC transcripts following 48-hour treatment with EZH2 inhibitor GSK-126. g, ABT-199 GI50 combined with GSK-126 in OCI-AML2. h, ABT-199 GI50 values from drug naïve and EPZ-6438 relapsed AML PDX cells. hCD45+ blasts from whole bone marrow were subjected to 72-hour ABT-199 incubation. Significance determined by Welch’s two-sample t-Test. Boxplot elements defined in Methods. Data are mean ± SEM of n = 4 biologically independent animals. i, Specific growth rate in the presence of JQ-1 in matched parental and JQ-1 resistant AML cell lines. Representative growth rate of n = 4 independent samplings. j, Immunoblot analysis of MYC and EZH2 across matched parental (Par.) and JQ-1 resistant (Res.) AML cell lines. Representative immunoblot of n = 3 independent experiments. k, Immunoblot analysis of MYC and H3K27me3 following overexpression of GFP control and EZH2 in parental and JQ-1 resistant OCI-AML2 cells. Representative immunoblot of n = 3 independent experiments. l, Relative cell viability of parental and JQ-1 resistant OCI-AML2 cells following overexpression of GFP control or EZH2 for n = 1 biologically independent experiments. m, ABT-199 GI50 following overexpression of pCDH-MYC in NSD2/3 knockout cells. n, ABT-199 GI50 following short-hairpin knockdown of MYC versus scrambled control. b-g; i; m-n, P-values computed by two-sided t-Test for equal means. Data are mean ± SEM for n = 3 biologically independent experiments. Uncropped blots in Source Data.
Next, we tested the capacity of CRISPR/Cas9-mediated NSD2/3 and EZH2 knockout to modulate MYC and sensitivity to JQ-1 and ABT-199. NSD2/3 knockout resulted in reduction of MYC mRNA (Fig. 3b, Supplementary Fig. 6a,b) and sensitization to JQ-1 (Fig. 3c, Supplementary Fig. 6c). Short term MYC-suppressive sensitization to JQ-1 is slight because JQ-1 treatment acutely suppresses MYC, leaving little room for additional downregulation. Conversely, knockout of EED or EZH2 increased MYC mRNA (Fig. 3d, Supplementary Fig. 6d,e) and sensitized AML cells to ABT-199 (Fig. 3e, Supplementary Fig. 6f). These effects were phenocopied using the EZH2 inhibitors GSK-126 in vitro (Fig. 3f,g) and EPZ-6438 in NSG-SGM3 (NSGS) mice xenografted with patient-derived AML cells (Fig. 3h).
To determine whether PRC2-NSD2/3-MYC-driven resistance arises from chronic drug treatment, we cultured OCI-AML2 cells in escalating doses of ABT-199 or JQ-1 until they exhibited drug resistance (Fig. 3i, Supplementary Fig. 7a). Western blot analysis showed that, compared to cells cultured in DMSO, ABT-199-resistant OCI-AML2 cells did not exhibit changes in MYC (Supplementary Fig. 7b); rather, prior studies have shown that resistance to ABT-199 often relies on upregulation of BCL-XL and MCL-1 [36, 37]. However, JQ-1-resistant OCI-AML2 cells upregulated MYC at the protein level (Fig. 3j). This prompted us to culture four additional AML cell lines in the presence of JQ-1 until resistance was achieved (Fig. 3i). In each case, the acquisition of JQ-1 resistance was associated with MYC upregulation, suggesting that MYC upregulation is a pervasive feature of acquired JQ-1 resistance in AML (Fig. 3j).
Subsequently, we sought to characterize the means of MYC upregulation. qRT-PCR analysis showed a relative increase in MYC transcripts in all but one model (Supplementary Fig. 7c), consistent with epigenetic regulation and potential involvement of the PRC2-NSD2/3-MYC axis. Protein levels of NSD2 and NSD3 were not increased in any of the resistant lines (Supplementary Fig. 7d). However, two of the JQ-1-resistant lines displayed a reduction in EZH2 protein (Fig. 3j), in line with MYC upregulation and a role for EZH2 loss in JQ-1 resistance. Those lines also exhibited collateral resistance to the BET degrader ARV771 (Supplementary Fig. 7e,f), suggesting that control of MYC expression may have shifted from BRD4 to EZH2 loss in these JQ1-resistant cells. To test this, we overexpressed GFP or EZH2 in parental and JQ-1-resistant OCI-AML2 lines. In parental cells, EZH2 overexpression increased global H3K27me3 without significantly affecting MYC expression or cell viability (Fig. 3k,l). In contrast, EZH2 overexpression in resistant cells led to increased global H3K27me3, loss of MYC protein, and loss of cellular viability (Fig. 3k,l), implying that MYC’s upregulation was now dependent on EZH2 loss. Mechanistically, western blot analysis of JQ-1 resistant OCI-AML2 cells identified an increase in CDK1-mediated phosphorylation of EZH2 at Thr487 (Supplementary Fig. 7g), a ubiquitin-dependent phosphodegron [38]. Immunoprecipitation of EZH2 showed increased bound ubiquitin (Supplementary Fig. 7h). Inhibition of CDK1 or the proteasome was sufficient to rescue EZH2 in JQ-1-resistant OCI-AML2 cells (Supplementary Fig. 7i).
Because we did not observe transcriptional upregulation of MYC in all JQ-1-resistant models, we reasoned that other modes of regulation also play a role. MYC is known to be the object of successive phosphorylation events catalyzed by ERK and GSK3β, the latter of which is controlled by AKT and whose phosphorylation of MYC permits consequent ubiquitination and proteolysis [39]. To explore this paradigm in our models, we performed cycloheximide chase experiments on each parental-resistant pair, finding a relative increase in MYC stability in three out of five resistant lines (Supplementary Fig. 8a,b). Those lines exhibited evidence of AKT and ERK activation by western blot (Supplementary Fig. 8) while the two lines with unchanged MYC stability did not have increased AKT or ERK signaling. Furthermore, MYC was more sensitive to ERK inhibition in AKT/ERK-active resistant models than in their parental counterparts (Supplementary Fig. 8d), consistent with an increased dependence on AKT/ERK signaling. These resistant models were also sensitive to the combination of JQ-1 and ERK inhibition (Supplementary Fig. 8e,f). These data indicate that transcriptional and post-translational regulation of MYC can jointly account for the MYC upregulation observed in our JQ-1-resistant models although additional mechanisms cannot be excluded.
Together, these data demonstrate that MYC upregulation is a common feature of JQ-1-resistant AML cells and that this upregulation can be driven by dysregulation of the PRC2-NSD2/3-MYC regulatory axis nominated through our AP analysis. Finally, to establish that the effects of NSD2/3 loss or EZH2 inhibition on ABT-199 sensitivity were mediated through MYC, we sought to rescue their observed effects by overexpressing or knocking out MYC. In NSD2/3 knockout cells, overexpression of MYC restored sensitivity to ABT-199 (Fig. 3m, Supplementary Fig. 8g-i). Correspondingly, sensitization to ABT-199 mediated by EZH2 inhibition was rescued by MYC knockdown (Fig. 3n). These observations point to MYC upregulation as the basis for synergy between EZH2 and BCL-2 inhibition that we observe here in AML and previously described in non-Hodgkin’s lymphoma [40].
MYC upregulation sensitizes to BH3 mimetics
Up to this point, our data assert that MYC’s pleiotropic effects on cellular growth and death allow it to assume one of two roles: its incumbent oncogenic, pro-proliferative role, observed in most cancer-related contexts, or a pro-apoptotic role, unmasked in the presence of BH3 mimetics or circumstances that otherwise compromise the cancer cell’s anti-apoptotic machinery. This duality should empower MYC, capable of subverting the effects of one drug while abetting another, to act as the driving mechanism for an evolutionary trap. However, in order to realize the trap, it is essential to determine whether MYC-driven resistance gives rise to enhanced sensitivity to BCL2 inhibition.
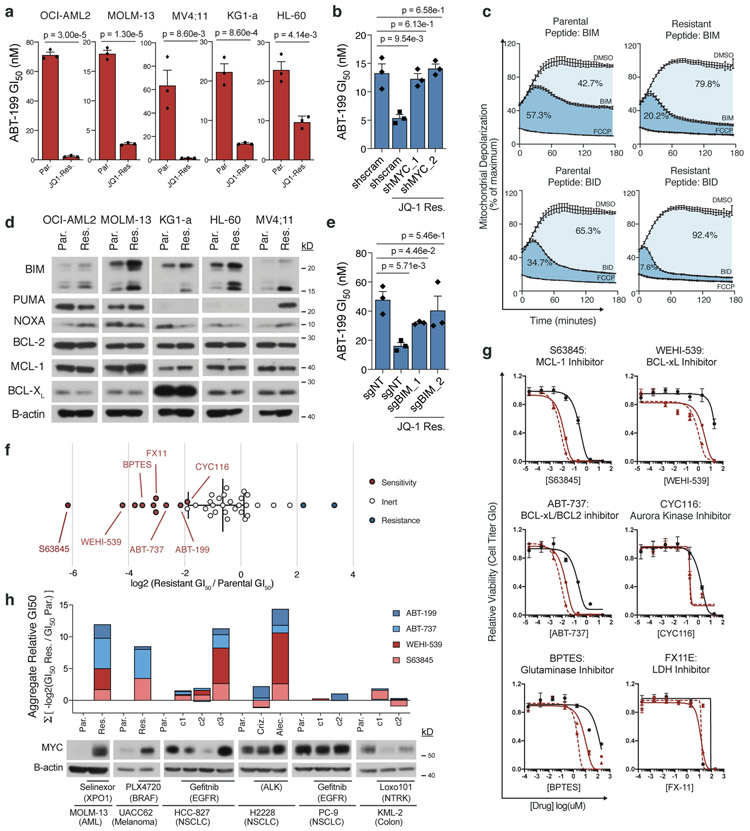
To investigate this, we tested the sensitivity of our JQ-1-resistant AML cell line models to ABT-199. Each JQ-1-resistant model was markedly more sensitive to ABT-199 than its parental counterpart (in contrast, ABT-199-resistant cells were not sensitized to JQ-1) (Fig. 4a, Supplementary Fig. 9a). The collateral sensitivity to ABT-199 was MYC-dependent and rescuable by shRNA-mediated MYC knockdown (Fig. 4b, Supplementary Fig. 9b). Both resistance to JQ-1 and collateral sensitivity to ABT-199 were stable following removal of JQ-1 for ten days (Supplementary Fig. 9c-f). Having demonstrated that parallel evolution converges upon MYC as an adaptive mechanism of resistance to JQ-1, the finding that MYC upregulation is responsible for heightened sensitivity to ABT-199 qualifies it to act as the functional requisite of an evolutionary trap: an adaptive response to an initial selective pressure (JQ-1) that is rendered maladaptive by a successive selective pressure (ABT-199).
Figure 4: MYC upregulation enables a drug-induced evolutionary trap.
a, ABT-199 GI50 in matched parental and JQ-1 resistant AML cell lines. b, ABT-199 GI50 in parental and JQ-1 resistant OCI-AML2 cells following short-hairpin knockdown of MYC or scrambled control. c, BH3 profiling traces of parental and JQ-1 resistant OCI-AML2 cells stimulated with BIM or BID peptides. Mitochondrial potential was measured every 2 minutes over 3-hours. Percent depolarization relative to maximal depolarization by carbonyl cyanide-4-(trifluoromethoxy)-phenylhydrazone (FCCP). Data are mean ± SEM for n = 3 biologically independent experiments. d, Immunoblot analysis of BIM, PUMA, NOXA, BCL-2, MCL-1, and BCL-XL protein levels across matched parental and JQ-1 resistant AML cell lines. Representative immunoblot of n = 3 independent experiments yielding similar results. e, ABT-199 GI50 in parental and JQ-1 resistant OCI-AML2 cells following CRISPR/Cas9 knockout of BIM or non-targeting control. f, Log2-fold change in GI50 values from 40 compound screen of JQ-1-resistant OCI-AML2 cells relative to parental cells. g, Relative cell viability of matched parental and JQ-1 resistant OCI-AML2 cells following 72-hour incubation with indicated drugs across an 8-point drug dilution series. Data are mean ± SEM for n = 3 biologically independent experiments. h, Aggregate −log2(GI50) of drug-resistant versus parental cell lines, summed across four different BH3 mimetics. Distinct resistant clones marked c1-c3. Data are matched to corresponding MYC immunoblots. Representative immunoblots of n = 3 independent experiments yielding similar results. a-b; e, P-values computed by two-sided t-Test for equal means. Data are presented as mean ± SEM for n = 3 biologically independent experiments. Uncropped blots in Source Data.
To understand the mechanism of acquired sensitivity to ABT-199, we performed BH3 profiling [41] on each parental-resistant pair. JQ-1-resistant cells exhibited increased mitochondrial depolarization when acquainted with BCL-2-like protein 11 (BIM) and/or BH3 interacting-domain death agonist (BID) (Fig. 4c). This suggested that the JQ-1-resistant cells were more primed for intrinsic, mitochondrial apoptosis than their parental counterparts. Such a state signals a shift in the balance of pro-apoptotic versus anti-apoptotic BCL-2 family members. Expression analysis of BCL-2 family members showed an increase in BIM in each JQ-1-resistant derivative compared to its parental pair (Fig. 4d, Supplementary Fig. 9g), consistent with BIM upregulation secondary to MYC overexpression (Supplementary Fig. 9h), and the fact that BIM is an established, direct transcriptional target of MYC [42-44]. CRISPR-mediated knockout of BIM neutralized the sensitivity to ABT-199 observed in JQ-1-resistant cells (Fig. 4e, Supplementary Fig. 9i). Notably, MYC upregulation was not associated with consistent alterations in other BCL-2 family proteins, including NOXA and PUMA, known p53 targets that can be indirectly activated by MYC through p19, and the anti-apoptotic proteins BCL-2, MCL-1, and BCL-XL (Fig. 4d) [45, 46].
Having established that BIM upregulation was the means for the observed collateral sensitivity to ABT-199, we reasoned that upregulation of a pro-apoptotic protein like BIM should sensitize to other BCL-2 family inhibitors and pro-apoptotic chemotherapeutics [47]. We examined this by profiling JQ-1-resistant OCI-AML2 cells and their vehicle-treated counterparts using a panel of 40 chemotherapies (Fig. 4f, Supplementary Fig. 9j). Here, we observed a general shift toward sensitivity in the JQ-1-resistant cells. Among the drugs that elicited a potentiated response in JQ-1-resistant cells were BH3 mimetics: ABT-199, WEHI-539, the selective MCL-1 inhibitor S63845, and the combined BCL2/BCL-XL inhibitor ABT-737 (Fig. 4g). We also noticed hypersensitivities to inhibitors of aurora kinase A (CYC116), glutaminase (BPTES), and lactate dehydrogenase (FX11), each of which has been implicated as a synthetic dependency of oncogenic MYC (Fig. 4f,g, Supplementary Fig. 9j) [48-50]. Thus, many of the collateral sensitivities observed in JQ-1-resistant cells are associated with MYC, either as a requirement for MYC dependence or related to apoptotic priming from MYC-driven BIM upregulation.
Finally, the notion that MYC-driven resistance to JQ-1 lays an evolutionary trap that can be deployed in AML through treatment with BH3 mimetics suggests that a similar approach could be used in other acquired resistance paradigms, particularly given the emerging role of MYC as a widespread mediator of acquired resistance to targeted therapies [7, 51-56]. To test this concept, we evolved a panel of cell lines from distinct lineages and harboring different driver mutations to resistance to their cognate inhibitors (Fig. 4h, Supplementary Fig. 9k-p). Across all models, we observed that drug resistance associated with MYC upregulation was accompanied by collateral sensitivities to BH3 mimetics, while MYC-independent models were indifferent to BH3 mimetic treatment (Fig. 4h, Supplementary Fig. 9k-p). This was true, even when considering distinct drug-resistant clones derived from the same parental population but exhibiting variable MYC expression. Collectively, these data suggested that the principles described and defined here in AML cells are generalizable and robust to tissue-specific variation. Furthermore, these findings imply that solid tumors, which are largely thought to be insensitive to BH3 mimetics, can be primed for apoptosis by MYC upregulation and rendered vulnerable to MYC-dependent evolutionary traps.
Validating the evolutionary trap in a PDX model of AML
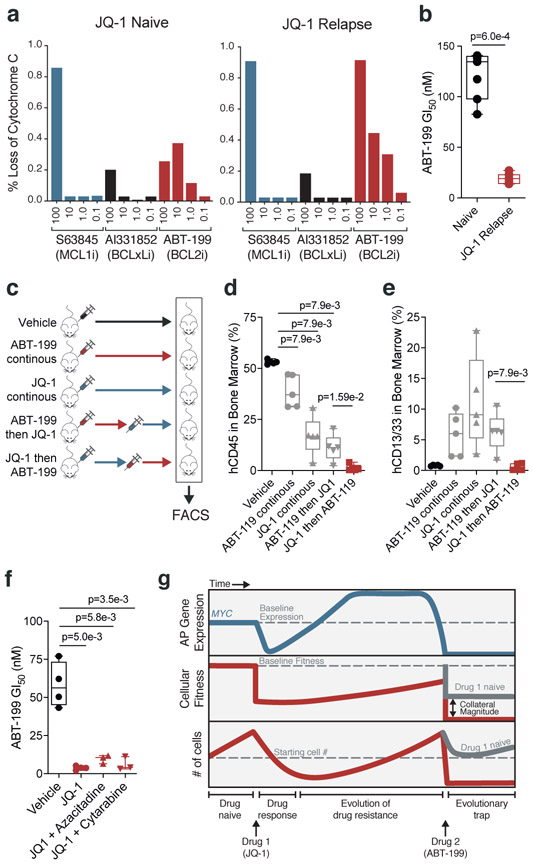
To determine if a MYC-dependent evolutionary trap could be therapeutically exploited, we asked whether JQ-1 treatment of a PDX model of AML could prime cells for apoptosis in a BCL-2-dependent manner. First, we engrafted NSGS mice with an AML PDX and treated with JQ-1 for ten days. Engrafted PDX cells were collected after resurgence of leukemia in the peripheral blood. BH3 profiling showed that the relapsed JQ-1-treated PDX cells exhibited BCL-2-dependent priming, evidenced by the potentiated release of cytochrome C upon treatment with ABT-199 in JQ-1-treated cells compared to JQ-1-naive cells (Fig. 5a). The same relapsed JQ-1-treated PDX cells were also more sensitive to ABT-199 in vitro, corroborating data from cell line models (Fig. 5b). By comparison, treatment with the MCL-1 inhibitor S63845 and the BCL-XL inhibitor A1331852 elicited the same amount of cytochrome C release in JQ-1-treated and JQ-1-naïve cells, suggesting a dominant role for BCL-2 in this model.
Figure 5: JQ-1 treatment primes PDX model of AML for treatment with ABT-199.
a, Release of cytochrome c upon drug stimulation in JQ-1 naïve and JQ-1 relapsed (50mg/kg, intraperitoneal injection) AML PDX cells. b, ABT-199 GI50 values from JQ-1 naïve and JQ-1 relapsed AML PDX cells. Significance determined by two-tailed Mann Whitney test for n = 5 biologically independent animals. c, Schematic depicting drug scheduling study in murine PDX model of AML treated with JQ-1 (50mg/kg, intraperitoneal injection) and/or ABT-199 (100mg/kg, oral gavage) according to indicated drug schedules. d-e, FACS quantification of human CD45+ (d) and human CD13/33+ (e) cells from murine bone marrow aspirates of mice in drug scheduling study. Significance determined by two-tailed Mann Whitney test. Data are presented as mean ± SEM for n = 4 biologically independent animals. f, ABT-199 GI50 values in AML PDX cells from treatment naïve versus mice with relapsed disease following treatment with JQ-1 alone or in combination with cytarabine (5mg/kg, intraperitoneal injection) and azacitidine (150mg/kg, intraperitoneal injection). Significance determined by two-tailed Mann Whitney test for n = 3 biologically independent animals. g, Proposed model for a chemotherapy-induced evolutionary trap. Treatment of AML cells with JQ-1 prompts compensatory upregulation of MYC, an AP gene that harbors pro-apoptotic potential. Subsequent treatment with ABT-199 produces a potentiated apoptotic response. b,d-f; Boxplot elements defined in Methods.
Given that in vivo JQ-1 treatment primes AML cells in a BCL-2-dependent manner, increasing in vitro sensitivity to ABT-199, we predicted that murine PDX models treated with JQ-1 would also exhibit a heightened in vivo response to ABT-199. Moreover, because evolutionary traps do not necessarily work in reverse, and according to our in vitro data, we did not expect mice treated with ABT-199 followed by JQ-1 to respond as well; in theory, the order of treatment matters. To test these concepts, we performed a drug-switching study where mice engrafted with the PDX were treated with either JQ-1 or ABT-199 for ten days, after which half of the mice treated with JQ-1 were switched to ABT-199, and vice versa (Fig. 5c). Analysis of bone marrow aspirates showed that mice treated with JQ-1 followed by ABT-199 exhibited lower human CD45+ leukemic infiltration than other groups (Fig. 5d, Supplementary Fig. 9q). Flow cytometry analysis of the myeloid markers CD13 and CD33 indicated that, unlike cells from the other treatment groups, AML cells from mice treated with the progression of JQ-1 to ABT-199 did not exhibit evidence of differentiation, suggesting that the mechanism of blast reduction in these mice was induction of cell death, consistent with hypersensitivity to ABT-199-induced apoptosis (Fig. 5e, Supplementary Fig. 9q).
Lastly, chemotherapies that fail to perform as monotherapies frequently gain approval as part of combination regimens [57-59]. This led us to consider whether combination regimens that include JQ-1 could prime AML cells in the same way that JQ-1 monotherapy does. To test this, we treated xenotransplanted mice with JQ-1 combined with cytarabine or azacitidine, chemotherapies that are routinely used to treat AML. Upon disease relapse, engrafted PDX cells were isolated and tested for sensitivity to ABT-199. We observed that, similar to PDX cells treated with JQ-1 alone, PDX cells treated with combinations that included JQ-1 were sensitized to ABT-199 (Fig. 5f). This result indicates that chemotherapeutic traps can be set with combination therapies, broadening the translatability of this principle.
Discussion
Here, we define a mechanistically-coherent chemotherapeutic paradigm that turns the expected acquisition of resistance to an initial drug treatment into an evolutionary trap, setting up a second, more powerful selection event. We design this trap using genetic screens coupled with the concept of drug-induced AP. This framework enables the rational design of evolutionary traps by connecting the specific mechanisms that drive resistance to an initial drug treatment to their resultant collateral sensitivities (Fig. 5g).
Present circumstances favor the adoption of the principles described here. The majority of modern chemotherapeutic regimens require multiple drugs, often administered sequentially to avoid dose-limiting toxicities. Few of those regimens are molecularly grounded, and they could be restructured to incorporate evolutionary traps, thereby taking advantage of drug-induced AP relationships without drastically upending the status quo. Take for instance ABT-199 (venetoclax), recently approved by the FDA in combination with cytarabine or a hypomethylating agent for the treatment of AML. Our work suggests that an additional role of venetoclax in the management of AML may be as a second chemotherapy, used to spring a MYC-dependent evolutionary trap in a drug-resistant tumor, especially because many drug resistance mechanisms in AML appear to feature upregulation of MYC [51, 52] or downregulation of EZH2 [38]. Furthermore, the development of inhibitors of MCL-1 [37, 60] and BCL-XL [61], which comprise the anti-apoptotic defense in many solid tumors [62], suggest that BH3 mimetics could be used to trigger MYC-dependent traps beyond AML, an important concept given the prevalence of MYC upregulation in drug resistant tumors across lineages. With respect to the specific progression of JQ-1 to ABT-199, several bromodomain inhibitors are in clinical trials in AML [63-65]. Our data indicate that patients who relapse on or are refractory to treatment with bromodomain inhibitors could be candidates for venetoclax treatment. It is worth mentioning that the upfront combination of bromodomain inhibition and BCL-2 inhibition has also been reported [66, 67]. The potency of this combination could be accounted for by our model, through simultaneous eradication of MYC-high and MYC-low cell populations, but the synergy most likely stems from the bromodomain inhibition-induced downregulation of MCL-1 and BCL-XL.
The ideas explored here motivate several key directions for future study. First, this work focuses on validating an exemplar AP interaction as the substrate for an evolutionary trap. However, the results of our screens performed in the presence of diverse chemotherapies identify a rich landscape of interconnected, drug-induced AP relationships. We expect that further analyses, performed on this or similar chemical-genetics datasets, will uncover additional interactions that can be used to design mechanism-based evolutionary traps. Second, the study of AP in evolutionary biology tends to lack formalism. As a result, a majority of AP studies simply identify AP, rather than quantify it. By establishing the API model which quantifies the relative frequency a given gene is observed as AP, we provide a means for prioritizing AP genes that can adapted for other studies. Finally, the study of AP in cancer has been limited to only a small number of genes that display context-dependent oncogenic or tumor suppressive characteristics. A comprehensive, systems-level identification and stratification of AP genes in cancer, driven by the application of the quantitative AP model presented here to existing and emerging structural or functional genomic datasets obtained from diverse tumors and cell lines, should allow us to begin to challenge preconceived notions of gene function and essentiality in cancer.
In summary, our work describes how an understanding of the fitness trade-offs inherent to drug-induced AP relationships, mapped through CRISPR/Cas9 loss-of-function screens, can be used to lay an evolutionary trap for cancer. This trap preys on genetic weaknesses exposed in the acquisition of drug resistance, channeling the very mechanism(s) of treatment failure into a second, more powerful selection event. The concepts explored here are valuable, in part because they directly motivate the design of evolutionarily-rational chemotherapeutic traps, but more broadly because they have the potential to reframe our understanding of how fundamental evolutionary principles relate to chemotherapeutic response.
Methods
Cell lines and reagents
All cell lines were maintained in a humidified incubator at 37 °C with 5% CO2. HCC827, KM-12, PC9, MOLM-13, OCI-AML2, MV4;11, KG-1a, HL-60 and UACC-62 cells were cultured in RPMI-1640 medium with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. OCI-AML2, MOLM-13, and MV4;11 have wild type p53 while HL-60 and KG-1a have mutated p53. SKMEL5 were cultured in DMEM with 10% FBS and 1% penicillin/streptomycin. 293FT cells were cultured in DMEM high glucose medium with 10% FBS, 1% penicillin/streptomycin, 1% sodium pyruvate, 1% non-essential amino acids, and 1% GlutaMax. All cell lines were purchased from American Type Culture Collection (ATCC) or Duke University Cell Culture Facility (CCF). Drugs were purchased from ApexBio (S63485), Cayman Chemical (WEHI-539), ARV-771 (MedChem), and SelleckChem (all other inhibitors).
Evolving drug resistant cell lines
To achieve drug resistance in vitro, AML cells were continuously cultured in increasing concentrations of drug. Cells were first drugged at a dose approximately equal to their GI50 value. The growth rate was monitored with weekly passaging and the concentration of drug was increased once a stable growth rate was achieved. Solid tumor cell lines were evolved to resistance either as described above or by treating cells with a high drug concentration and selecting resistant clones.
The maximally-tolerated dose of JQ-1 corresponding to each resistant cell line is as follows: OCI-AML2 (250nM), MV4;11 (150nM), MOLM-13 (200nM), KG-1a (250nM), HL-60 (200nM). These doses were achieved gradually over the course of two months.
Short-term cell viability assay (GI50)
AML cells were seeded into 96-well plates at a density of 7,500 cells/well. Immediately following plating, AML cells were treated with vehicle (DMSO) or a 10-fold serial dilution of drug. Each treatment condition was conducted in triplicate. Solid tumor cells were plated at a density of 2,000 cells/well and treated after 24 hours. Three days following addition of drug, cell viability was quantified using Cell Titer Glo (Promega). The relative cell viability was determined by normalizing the raw luminescence values for each treatment condition to DMSO-treated wells. For experiments involving two drugs, slight modifications were made. One drug was kept at a constant concentration across all wells and a serial dilution of a second drug was added on top of the background drug. One well was treated with DMSO only and one well was treated with background drug only. The relative cell viability was normalized to the luminescence of the background drug only. Dose-response curves were fit using GraphPad/Prism 7/8 software. GI50 values were interpolated from the resultant graphs as the dose corresponding to 50% cell viability relative to DMSO-treated cells.
Cloning CRISPR Library
Our CRISPR library was cloned according to previously described methods [21]. Each gene in the library is represented by 5 sgRNAs, pulled from a previously published, full-genome CRISPR library [68]. Also included were 50 non-targeting controls. In brief, each unique 20-mer sgRNA sequence was appended by constant prefix and suffix sequences (Supplementary Table 5) and synthesized as an oligo pool by Custom Array Inc. The pooled inserts were diluted 1:10 in molecular-grade water and amplified with NEB Phusion Hotstart Flex enzyme mix using the following primers:
Array_Forward: see Supplementary Table 5
Array_Reverse: see Supplementary Table 5
Protocol: 98C (30s), 18x [98C (10s), 63C (10s), 72C (15s)], 72C (3 min)
Double-stranded inserts were cleaned up with Axygen magnetic PCR bead purification kits (Fisher Scientific) and eluted in molecular-grade water. lentiCRISPRv2 (Addgene plasmid #52961) was digested with FastDigest BsmBI at 37 °C for 2 hours and the cut vector was gel extracted. A 20μL Gibson assembly reaction was performed using 100 ng of cut lentiCRISPRv2 and 40 ng of prepped sgRNA inserts. 1μL of the Gibson reaction product was transformed into electrocompetent cells (E. cloni 10G ELITE, Lucigen #60052-2) and spread onto LB-ampicillin plates. Following overnight incubation at 37 °C, plates were counted to ensure 40x library coverage, colonies were scraped, and plasmid DNA was isolated using a Maxiprep kit (QIAGEN). 50 colonies were individually cultured and sequenced to validate cloning fidelity.
Individual sgRNA oligonucleotides were prepared, cleaned-up, cloned, transformed, isolated, and validated in an analogous manner.
pCDH-puro-cMyc was a gift from Jialiang Wang (Addgene plasmid # 46970).
Lentivirus production
Lentivirus production was conducted as previously described [22], with slight modification. Briefly, 293 FT cells grown to 60-70% confluency and transfected using Fugene6 (Promega), 2.80 μg of psPAX2, 0.280 μg of pVSVg, and 2.80 μg of plasmid. After 30 minutes of incubation at room temperature, the transfection mixture was added dropwise to 293 FT. After 24 hours, media was aspirated and replaced with harvest media (DMEM with 30% FBS, 1% penicillin/streptomycin). After 48 hours, the virus was harvested, cleared through 5-minute centrifugation at 1200rpm, and passed through a 0.45-μm filter to remove cell particles. Viral titers and transductions were performed as previously described [22].
Pooled CRISPR Screening
OCI-AML2 cells were seeded into six-well plates at a density of 3E6 cells per well and transduced at an MOI of 0.2. A total of 144E6 cells were transduced in 48 wells. 24 hours after viral transduction, cells were replated into puromycin-containing media. A sample was collected at 48 hours of puromycin exposure to confirm library coverage in the transduced population. Transduced cells were expanded in puromycin for a total of ten days prior to drug introduction, at which point the transduced cell population was split into vehicle (DMSO) and drug treatment conditions and maintained for two weeks. All conditions were performed in replicate. Drugs were used at doses sufficient to achieve 20-30% loss of viability as follows: 100nM ABT-199, 100nM Selinexor, 50nM JQ-1, 1μM Quizartinib, 600nM Vorinostat, 60nM Cytarabine, 2nM Mitoxantrone, 100nM Decitabine, 1μM Azacitidine. Cells were counted, replated, and drug replenished every 3-4 days. At any given point during the screen, each replicate was represented by a minimum of 12E6 cells, sufficient to provide 1000x coverage of the library (~1000 cells per unique sgRNA). Samples of 25E6 cells were collected upon screen initiation, termination, and at weekly intervals. Following completion of the screens, DNA was extracted (DNeasy Blood & Tissue Kit, QIAGEN) and prepared for sequencing as previously described [22].
CRISPR Screen Analysis
Deep sequencing was performed on an Illumina Nextseq platform (75 bp, single-ended) to identify differences in library composition. All sequencing was performed by Hudson Alpha Institute for Biotechnology. Barcoded reads were matched and binned into guide-level counts. Determinations of genetic essentiality and drug sensitization/resistance were made by evaluating differential guide compositions between the initial population and subsequent drug-treated and vehicle-treated cells populations. Briefly, the fractional representation (FR) for a guide within a sample was normalized to the sum of all guides attributed to that sample. A direct comparison between two samples entailed the quotient of the respective FRs, which we term the depletion metric (DM). The five guide-level DMs for each gene were then collapsed to gene-level scores by taking the average. Guides which totaled fewer than 200 counts for a given sample were excluded from analysis. Genetic essentiality was calculated by considering the depletion/enrichment of the vehicle-treated population over time (DMSOfinal / initial). Drug sensitization/resistance was calculated by considering the depletion/enrichment of the drug-treated population relative to the vehicle-treated population (Drugfinal / DMSOfinal). All depletion/enrichment effects are reported as log2 ratios. All described manipulations were performed in R.
Identification of Antagonistically Pleiotropic (AP) Genes
To establish a framework for the systematic identification of AP genes, we accepted the following definition of an AP gene: a gene whose null allele (here achieved through CRISPR-Cas9-mediated genetic ablation) confers relative fitness under one condition and relative loss of fitness under another. This definition permits flexibility at two points: the (number and type of) conditions observed, and the relative threshold for defining gain or loss of fitness. The identified AP genes in a given analysis are a function of these variables. As the number of conditions approaches infinity, the number of AP genes identified approaches the total number of genes monitored. Our examination of a nine-chemotherapy panel does not exhaust but approaches saturation of our 2,390-gene library. Second, the definition of an observed effect on relative fitness is necessarily measured against a fitness threshold. As the fitness threshold approaches zero (null effect), the number of AP genes identified approaches the total number of genes monitored. The stringency of this threshold dictates both the quantity and the ‘quality’ of the AP genes identified. Because the absolute depletion/enrichment engendered by drug treatment on top of gene knockout is a function of the dose and duration of drug exposure, an absolute depletion/enrichment cutoff was avoided in favor of a relative cutoff defined by the topology of the curve. In short, a five-gene, moving average of the slope was calculated across the ranked, gene-level scores corresponding to each drug-treated screen. The cutoffs that distinguished fitness loss from inertness from fitness gain were defined as the points where the moving average of the slope first equaled the slope of the middle 50% of the curve (Supplementary Fig. 2a). Each cutoff was evaluated against the distribution of non-targeting control genes (outliers excluded by Tukey’s rule) and controlled at p < 0.05. Complete sets of fitness-beneficial and fitness-detrimental genes were compiled by identifying the genes that score above (fitness-beneficial) and below (fitness-detrimental) both topological and statistical cutoffs in at least one condition. The intersection of those two gene sets yielded the full set of AP genes.
Calculation of Antagonistic Pleiotropy Index (API)
Circos Plot
Circos plot shown in Fig. 2a and Supplementary Fig. 2c was assembled using Circos (http://circos.ca/).
Clustered Heatmaps
Heatmaps produced in R with the gplots package. Unsupervised clustering performed from Euclidean distance matrix according to Ward’s clustering criterion.
Correlogram
Correlograms shown in Supplementary Fig. 2f, 3a, 3j were assembled by first calculating correlations between relevant AP genes across all conditions screened. Correlograms were produced in R with the corrplot package.
Gene Ontology Analysis
Gene Ontology of extrinsic AP genes performed using GeneOntology biological process platform (http://geneontology.org). Default GO terms included in analysis.
qRT-PCR
RNA was isolated from whole cells with QIAshredder Homogenizers and the RNEasy Mini kit (Qiagen). cDNA was reverse transcribed from total RNA samples using iScript cDNA Synthesis Kit with 1 μg of RNA template. qRT-PCR was carried out using iQ SYBR Green Supermix and a CFX384 Touch Real-Time PCR Detection System, according to manufacturer instructions. Fold expression was determined by normalizing cycle threshold (Cq) values to ACTB reference gene and normalizing samples to control sample, in accordance with the ΔΔCq method.
For primers, see Supplementary Table 5.
Western blot
Immunoblotting:
Immunoblotting was performed as previously described [22], with slight modification. Protein lysates were prepared with RIPA lysis buffer supplemented with 1X protease inhibitor cocktail. Crude lysates were cleared using QIAshredder Homogenizers (Qiagen) and centrifuged at 13,000 rpm for 2 minutes at 4 °C. Membranes were probed with primary antibodies B-actin (CST #4970 CST #5453 diluted 1:3000 in 5% BSA), Myc [Y69] (Abcam ab32072), Ezh2 (D2C9) (CST #5246 diluted 1:1000 in 5% BSA), Acetyl-Histone H3 (Lys27) (CST #8173 CST #5453 diluted 1:1000 in 5% BSA), Bim (CST #2933 diluted 1:500 in 5% BSA), Puma (CST #12450 diluted 1:500 in 5% BSA), Noxa (CST #14766 diluted 1:500 in 5% BSA), Bcl-2 (CST #4223 CST #5453 diluted 1:1000 in 5% BSA), Mcl-1 (CST #5453 diluted 1:1000 in 5% BSA), Bcl-xL (CST #2764), Nsd2 (Abcam ab75359 diluted 1:500 in 5% BSA), Nsd3 (Abcam ab4514 diluted 1:500 in 5% BSA), LSD1 (CST #2139 diluted 1:1000 in 5% BSA), H3K4me1 (Abcam ab8895 diluted 1:2000 in 5% BSA) and H3K9me2 (CST #4658 diluted 1:1000 in 5% BSA) overnight (16 hours). Following incubation with HRP-conjugated secondary antibody, blots were developed with SuperSignal West Pico PLUS Chemiluminescent Substrate (ThermoFisher) or ECL Western Blotting Substrate (ThermoFisher). All uncropped images have been provided as source data.
Immunoprecipitation:
For coimmunoprecipitation cells were resuspended in lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 20 mM EDTA, 50mM NaF, 0.5% NP-40, 1 mM dithiothreitol with EDTA-free protease inhibitor and PhosSTOP phosphatase inhibitor tablets), rotated for 1 hour at 4 °C, and cleared by centrifugation at 14,000 rpm for 20 minutes at 4 °C. Lysates then equilibrated to 500ug of protein in 1mL lysis buffer and acquainted with 50uL protein G-Sepharose (Invitrogen) pre-incubated with 1:100 dilution of anti-EZH2 primary antibody (CST #5246) and rotated overnight at 4 °C. Samples washed 5 times with lysis buffer, boiled for 5 minutes, loaded into sample buffer and run on gel.
Cycloheximide chase:
Parental and resistant AML cells were treated with cycloheximide (CST #2112 20ug/mL in DMSO) for indicated times. Cells were collected and subjected to western blot analysis to assess MYC stability following inhibition of protein synthesis. Immunoblots quantified using ImageJ 1.51S software.
Flow cytometry
Live cells (1.5E6 cells/sample) were washed with PBS then with FACS buffer (PBS with 2% FBS, 2mM EDTA and 0.9% sodium azide) prior to staining with CD11b-PE antibody (BioLegend #101207 diluted 1:500 in FACS buffer) for 30 minutes at 4 °C and subjected to cytometric analysis with BD FACSCanto II.
BH3 profiling
BH3 profiling was performed as described previously [69]. In short, parental and JQ-1-resistant OCI-AML2 cells were collected, washed with PBS, and resuspended in Newmeyer buffer followed by permeabilization with digitonin. Samples were exposed to BH3 peptides and monitored for mitochondrial transmembrane potential loss using JC-1. Measurements were taken at 590nM at 30C every 5 minutes for 3 hours.
Cytochrome C loss assay
Cytochrome C loss assay was performed as described previously [70]. 5.0×106 cells were washed in PBS, then stained with Zombie Aqua Fixable Viability dye (Bio Legend) and anti CD45 APC H7 (BD Biosciences, clone 2D1). Cells were resuspended in DTEB Buffer and permeabilized with digitonin. 200×103 cells were incubated with different concentrations (100μM, 10μM, 1μM and 0,1μM) of S63845, AI331852 and ABT-199 for 1 hour. Cells were then fixated with formaldehyde 8%. After fixation termination with N2 buffer, cells were labeled with anti-human cytochrome C (CytC) Alexa Fluor 488 (BD Biosciences, clone 6H2.B4). All experiments included two DMSO 2% sample (without BH3 mimetics peptide), one labeled with anti-Cytochrome C, and one unlabeled as positive and negative controls for mitochondrial Cytochrome C content, respectively. Flow Cytometry analysis was then performed on a BD Fortessa analyzer (BD Biosciences).
In vivo transplantation
The French National Committee on Animal Care reviewed and approved all mouse experiments described in this study. Primary patient AML blasts were collected from bone marrow aspirates after obtaining patient informed consent under a St Louis Hospital Internal Review Board-approved protocol. Mononuclear cells were isolated using Ficoll-Paque Plus (Amersham Biosciences) and red blood cells were lysed before flow cytometry analysis. These cells were maintained in StemSpan SFEM (#09650, StemCell Technologies Inc.) medium supplemented with 20 ng/ml IL-3 (#200-03, Peprotech), 20 ng/ml IL-6 (#200-06, Peprotech), 20 ng/ml GM-CSF (#300-03, Peprotech), 100 ng/ml FLT3-Ligand (#300-19, Peprotech) and 100 ng/ml SCF (#300-07, Peprotech) prior to injection into NOD.Cg-Prkdcscid Il2rgtm1Sug Tg(SV40/HTLV-IL3,CSF2)10-7Jic/JicTac (huNOG-EXL) mice purchased from Taconic. Sample size was chosen in light of the fact that these in vivo models were historically highly penetrant and consistent. Animals were excluded from the study if any signs of distress were observed without clinical signs of leukemia: absence of leukemic blasts in bone marrow, spleen, and blood. None of our animals were excluded based on these criteria. Blinded observers visually inspected mice for obvious signs of distress, such as loss of appetite, hunched posture, and lethargy. Approximately 0.65 × 106 cells were tail vein injected as a secondary transplant into sublethally irradiated (125 cGy) 6-8-week-old male huNOG-EXL mice.
Drug scheduling experiments:
Twelve days after injection, mice were randomized and treated daily either by oral gavage with 100mg/kg ABT-199 (60% Phosal 50 +30 % PEG400 + 10% ethanol), by intraperitoneal injection with 50mg/kg JQ-1 (10% DMSO + 90% G5W), or with these two drugs used consecutively as indicated in the figure. Bone marrow biopsies were performed on anesthetized animals 28 days after cell injection, and biopsies were washed once in PBS and resuspended in 0.5% BSA, 2mM EDTA PBS prior to staining with either APC-conjugated anti-human CD45 (BioLegend, 368512, 3:100) or a cocktail of PE-Cy7-conjugated anti-human CD13/33 (eBioscience, 25-0138-42 & 25-0338-42, 3:100) antibodies and flow cytometry analysis.
Drug relapsed experiments:
Twelve days after injection, mice were randomized into 5 groups of four mice each and treated daily either by oral gavage 15 mg/kg EPZ-6438 (20% Captisol, 7 days), intraperitoneal injection with 50mg/kg JQ-1 (10% DMSO + 90% G5W, 7 days), JQ-1 + intraperitoneal injection 5mg/kg azacitidine (HBSS, 5 days), JQ-1 + intraperitoneal injection 150mg/kg cytarabine (HBSS, 5 days) or vehicle. Upon disease relapse, mice were sacrificed, the whole bone marrow was collected and stained with an anti-hCD45 antibody prior to flow cytometry-based sorting of hCD45-positive AML blasts. ABT-199 GI50 values obtained by plating sorted blasts in 384-well plates in presence of increasing concentrations of ABT-199 for 3 days (top concentration of 5 μM).
Statistical analyses
All results are shown as mean ± SEM. Unless specified, p-values determined using unpaired, two-tailed Student’s t-tests. P-values < 0.05 were considered significant. P-values provided as exact values whenever significant. Unless otherwise noted, all measurements were taken from distinct samples. Boxplot elements defined as: box extends from 25th to 75th percentile; whiskers extend from the minimum to maximum value; median indicated by traversing line.
Data availability
The data supporting the findings of this study are found within the paper and supplementary files.
Code availability
Scripts for analyzing CRISPR/Cas9 screens and calculating API are available on Github (https://github.com/linkvein/).
Supplementary Material
Acknowledgements:
We would like to thank the members of the K.C.W. laboratory and Alexandre Puissant laboratory for helpful discussions and scientific input. We also thank Kevin Wood (University of Michigan), Gerard Blobe and Scott Floyd (Duke Pharmacology & Cancer Biology) for providing helpful feedback. This work was supported by Duke University School of Medicine start-up funds and support from the Duke Cancer Institute (K.C.W.), NIH awards (R01CA207083 to K.C.W., F30CA206348 to K.H.L., F31CA195967 to P.S.W.), National Science Foundation Graduate Research Fellowship awards (DGE-1106401 to G.R.A. and DGF-1106401 to L.C.), the Duke Medical Scientist Training Program (T32 GM007171 to K.H.L.), the Duke Undergraduate Research Support Office (to J.C.R. and A.X.), the ATIP/AVENIR French research program (to A.P.), and the EHA research grant for Non-Clinical Advanced Fellow (to A.P.). A.P. is a recipient of support from the ERC Starting program (758848) and supported by the St Louis Association for leukemia research. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors(s) and do not necessarily reflect the views of the National Science Foundation or the NIH. Finally, we dedicate this work to the memory of our friend, Kimberly Brigati Wang, and her courageous fight against AML.
Footnotes
Declaration of interests:
J.W.L. serves on the scientific advisory board and owns equity in Nanocare Technologies and Raphael Pharmaceuticals. R.I. has received previous funding from Oncoethix SA for work on the bromodomain inhibitor OTX015. The remaining authors have no competing interests to declare.
References
- 1.Kawecki TJ and Ebert D, Conceptual issues in local adaptation. Ecology Letters, 2004. 7(12): p. 1225–1241. [Google Scholar]
- 2.Singer MC and Parmesan C, Lethal trap created by adaptive evolutionary response to an exotic resource. Nature, 2018. 557(7704): p. 238–241. [DOI] [PubMed] [Google Scholar]
- 3.Schlaepfer MA, Runge MC, and Sherman PW, Ecological and evolutionary traps. Trends in Ecology & Evolution, 2002. 17(10): p. 474–480. [Google Scholar]
- 4.Robertson BA, Rehage JS, and Sih A, Ecological novelty and the emergence of evolutionary traps. Trends Ecol Evol, 2013. 28(9): p. 552–60. [DOI] [PubMed] [Google Scholar]
- 5.Walther V, et al. , Can oncology recapitulate paleontology? Lessons from species extinctions. Nat Rev Clin Oncol, 2015. 12(5): p. 273–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Allen EM, et al. , The genetic landscape of clinical resistance to RAF inhibition in metastatic melanoma. Cancer Discov, 2014. 4(1): p. 94–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singleton KR, et al. , Melanoma Therapeutic Strategies that Select against Resistance by Exploiting MYC-Driven Evolutionary Convergence. Cell Rep, 2017. 21(10): p. 2796–2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holohan C, et al. , Cancer drug resistance: an evolving paradigm. Nat Rev Cancer, 2013. 13(10): p. 714–26. [DOI] [PubMed] [Google Scholar]
- 9.Gatenby R and Brown J, The Evolution and Ecology of Resistance in Cancer Therapy. Cold Spring Harb Perspect Med, 2018. 8(3). [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Konieczkowski DJ, Johannessen CM, and Garraway LA, A Convergence-Based Framework for Cancer Drug Resistance. Cancer Cell, 2018. 33(5): p. 801–815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gundem G, et al. , The evolutionary history of lethal metastatic prostate cancer. Nature, 2015. 520(7547): p. 353–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Juric D, et al. , Convergent loss of PTEN leads to clinical resistance to a PI(3)Kalpha inhibitor. Nature, 2015. 518(7538): p. 240–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang L, et al. , An Acquired Vulnerability of Drug-Resistant Melanoma with Therapeutic Potential. Cell, 2018. 173(6): p. 1413–1425 e14. [DOI] [PubMed] [Google Scholar]
- 14.Chen G, et al. , Targeting the adaptability of heterogeneous aneuploids. Cell, 2015. 160(4): p. 771–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Imamovic L, et al. , Drug-Driven Phenotypic Convergence Supports Rational Treatment Strategies of Chronic Infections. Cell, 2018. 172(1–2): p. 121–134 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amirouchene-Angelozzi N, Swanton C, and Bardelli A, Tumor Evolution as a Therapeutic Target. Cancer Discov, 2017. [DOI] [PubMed] [Google Scholar]
- 17.Zhao B, et al. , Exploiting Temporal Collateral Sensitivity in Tumor Clonal Evolution. Cell, 2016. 165(1): p. 234–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Savolainen O, Lascoux M, and Merila J, Ecological genomics of local adaptation. Nat Rev Genet, 2013. 14(11): p. 807–20. [DOI] [PubMed] [Google Scholar]
- 19.Tiffin P and Ross-Ibarra J, Advances and limits of using population genetics to understand local adaptation. Trends Ecol Evol, 2014. 29(12): p. 673–80. [DOI] [PubMed] [Google Scholar]
- 20.Hart T, et al. , Measuring error rates in genomic perturbation screens: gold standards for human functional genomics. Mol Syst Biol, 2014. 10: p. 733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shalem O, et al. , Genome-scale CRISPR-Cas9 knockout screening in human cells. Science, 2014. 343(6166): p. 84–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin KH, et al. , Systematic Dissection of the Metabolic-Apoptotic Interface in AML Reveals Heme Biosynthesis to Be a Regulator of Drug Sensitivity. Cell Metab, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fiskus W, et al. , Highly effective combination of LSD1 (KDM1A) antagonist and pan-histone deacetylase inhibitor against human AML cells. Leukemia, 2014. 28(11): p. 2155–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fiskus W, et al. , Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells. Blood, 2009. 114(13): p. 2733–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beaumont KA, et al. , Cell Cycle Phase-Specific Drug Resistance as an Escape Mechanism of Melanoma Cells. J Invest Dermatol, 2016. 136(7): p. 1479–1489. [DOI] [PubMed] [Google Scholar]
- 26.Knutson SK, et al. , Synergistic Anti-Tumor Activity of EZH2 Inhibitors and Glucocorticoid Receptor Agonists in Models of Germinal Center Non-Hodgkin Lymphomas. PLoS One, 2014. 9(12): p. e111840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee T, Karon M, and Momparler RL, Kinetic studies on phosphorylation of 5-azacytidine with the purified uridine-cytidine kinase from calf thymus. Cancer Res, 1974. 34(10): p. 2482–8. [PubMed] [Google Scholar]
- 28.Liliemark JO, Plunkett W, and Dixon DO, Relationship of 1-beta-D-arabinofuranosylcytosine in plasma to 1-beta-D-arabinofuranosylcytosine 5’-triphosphate levels in leukemic cells during treatment with high-dose 1-beta-D-arabinofuranosylcytosine. Cancer Res, 1985. 45(11 Pt 2): p. 5952–7. [PubMed] [Google Scholar]
- 29.Cai J, et al. , Two distinct molecular mechanisms underlying cytarabine resistance in human leukemic cells. Cancer Res, 2008. 68(7): p. 2349–57. [DOI] [PubMed] [Google Scholar]
- 30.Margueron R and Reinberg D, The Polycomb complex PRC2 and its mark in life. Nature, 2011. 469(7330): p. 343–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holoch D and Margueron R, Mechanisms Regulating PRC2 Recruitment and Enzymatic Activity. Trends Biochem Sci, 2017. 42(7): p. 531–542. [DOI] [PubMed] [Google Scholar]
- 32.Schmitges FW, et al. , Histone methylation by PRC2 is inhibited by active chromatin marks. Mol Cell, 2011. 42(3): p. 330–41. [DOI] [PubMed] [Google Scholar]
- 33.Zheng Y, et al. , Total kinetic analysis reveals how combinatorial methylation patterns are established on lysines 27 and 36 of histone H3. Proc Natl Acad Sci U S A, 2012. 109(34): p. 13549–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bennett RL, et al. , The Role of Nuclear Receptor-Binding SET Domain Family Histone Lysine Methyltransferases in Cancer. Cold Spring Harb Perspect Med, 2017. 7(6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kaur M and Cole MD, MYC acts via the PTEN tumor suppressor to elicit autoregulation and genome-wide gene repression by activation of the Ezh2 methyltransferase. Cancer Res, 2013. 73(2): p. 695–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lin KH, et al. , Targeting MCL-1/BCL-XL Forestalls the Acquisition of Resistance to ABT-199 in Acute Myeloid Leukemia. Sci Rep, 2016. 6: p. 27696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ramsey HE, et al. , A Novel MCL1 Inhibitor Combined with Venetoclax Rescues Venetoclax-Resistant Acute Myelogenous Leukemia. Cancer Discov, 2018. 8(12): p. 1566–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gollner S, et al. , Loss of the histone methyltransferase EZH2 induces resistance to multiple drugs in acute myeloid leukemia. Nat Med, 2017. 23(1): p. 69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farrell AS and Sears RC, MYC degradation. Cold Spring Harb Perspect Med, 2014. 4(3). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bradley WD, et al. , EZH2 inhibitor efficacy in non-Hodgkin’s lymphoma does not require suppression of H3K27 monomethylation. Chem Biol, 2014. 21(11): p. 1463–75. [DOI] [PubMed] [Google Scholar]
- 41.Ryan J and Letai A, BH3 profiling in whole cells by fluorimeter or FACS. Methods, 2013. 61(2): p. 156–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campone M, et al. , c-Myc dependent expression of pro-apoptotic Bim renders HER2-overexpressing breast cancer cells dependent on anti-apoptotic Mcl-1. Mol Cancer, 2011. 10: p. 110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee YY, et al. , CREB-binding protein (CBP) regulates beta-adrenoceptor (beta-AR)-mediated apoptosis. Cell Death Differ, 2013. 20(7): p. 941–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Muthalagu N, et al. , BIM is the primary mediator of MYC-induced apoptosis in multiple solid tissues. Cell Rep, 2014. 8(5): p. 1347–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Villunger A, et al. , p53- and drug-induced apoptotic responses mediated by BH3-only proteins puma and noxa. Science, 2003. 302(5647): p. 1036–8. [DOI] [PubMed] [Google Scholar]
- 46.Nakano K and Vousden KH, PUMA, a novel proapoptotic gene, is induced by p53. Mol Cell, 2001. 7(3): p. 683–94. [DOI] [PubMed] [Google Scholar]
- 47.Ni Chonghaile T, et al. , Pretreatment mitochondrial priming correlates with clinical response to cytotoxic chemotherapy. Science, 2011. 334(6059): p. 1129–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dauch D, et al. , A MYC-aurora kinase A protein complex represents an actionable drug target in p53-altered liver cancer. Nat Med, 2016. 22(7): p. 744–53. [DOI] [PubMed] [Google Scholar]
- 49.Stine ZE, et al. , MYC, Metabolism, and Cancer. Cancer Discov, 2015. 5(10): p. 1024–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.den Hollander J, et al. , Aurora kinases A and B are up-regulated by Myc and are essential for maintenance of the malignant state. Blood, 2010. 116(9): p. 1498–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fong CY, et al. , BET inhibitor resistance emerges from leukaemia stem cells. Nature, 2015. 525(7570): p. 538–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rathert P, et al. , Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature, 2015. 525(7570): p. 543–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shu S, et al. , Response and resistance to BET bromodomain inhibitors in triple-negative breast cancer. Nature, 2016. 529(7586): p. 413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xia B, et al. , c-Myc plays part in drug resistance mediated by bone marrow stromal cells in acute myeloid leukemia. Leuk Res, 2015. 39(1): p. 92–9. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y, et al. , Sp1 and c-Myc modulate drug resistance of leukemia stem cells by regulating survivin expression through the ERK-MSK MAPK signaling pathway. Mol Cancer, 2015. 14: p. 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pan XN, et al. , Inhibition of c-Myc overcomes cytotoxic drug resistance in acute myeloid leukemia cells by promoting differentiation. PLoS One, 2014. 9(8): p. e105381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cortes JE, et al. , Glasdegib plus intensive/nonintensive chemotherapy in untreated acute myeloid leukemia: BRIGHT AML 1019 Phase III trials. Future Oncol, 2019. 15(31): p. 3531–3545. [DOI] [PubMed] [Google Scholar]
- 58.Cortes JE, et al. , Randomized comparison of low dose cytarabine with or without glasdegib in patients with newly diagnosed acute myeloid leukemia or high-risk myelodysplastic syndrome. Leukemia, 2019. 33(2): p. 379–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.DiNardo CD, et al. , Venetoclax combined with decitabine or azacitidine in treatment-naive, elderly patients with acute myeloid leukemia. Blood, 2019. 133(1): p. 7–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kotschy A, et al. , The MCL1 inhibitor S63845 is tolerable and effective in diverse cancer models. Nature, 2016. 538(7626): p. 477–482. [DOI] [PubMed] [Google Scholar]
- 61.Leverson JD, et al. , Exploiting selective BCL-2 family inhibitors to dissect cell survival dependencies and define improved strategies for cancer therapy. Sci Transl Med, 2015. 7(279): p. 279ra40. [DOI] [PubMed] [Google Scholar]
- 62.Soderquist RS, et al. , Systematic mapping of BCL-2 gene dependencies in cancer reveals molecular determinants of BH3 mimetic sensitivity. Nat Commun, 2018. 9(1): p. 3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Xu Y and Vakoc CR, Targeting Cancer Cells with BET Bromodomain Inhibitors. Cold Spring Harb Perspect Med, 2017. 7(7). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stathis A and Bertoni F, BET Proteins as Targets for Anticancer Treatment. Cancer Discov, 2018. 8(1): p. 24–36. [DOI] [PubMed] [Google Scholar]
- 65.Berthon C, et al. , Bromodomain inhibitor OTX015 in patients with acute leukaemia: a dose-escalation, phase 1 study. Lancet Haematol, 2016. 3(4): p. e186–95. [DOI] [PubMed] [Google Scholar]
- 66.Fiskus W, et al. , Superior efficacy of cotreatment with BET protein inhibitor and BCL2 or MCL1 inhibitor against AML blast progenitor cells. Blood Cancer J, 2019. 9(2): p. 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Esteve-Arenys A, et al. , The BET bromodomain inhibitor CPI203 overcomes resistance to ABT-199 (venetoclax) by downregulation of BFL-1/A1 in in vitro and in vivo models of MYC+/BCL2+ double hit lymphoma. Oncogene, 2018. 37(14): p. 1830–1844. [DOI] [PubMed] [Google Scholar]
Methods-only references
- 68.Wang T, et al. , Genetic screens in human cells using the CRISPR-Cas9 system. Science, 2014. 343(6166): p. 80–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sarosiek KA, et al. , BID preferentially activates BAK while BIM preferentially activates BAX, affecting chemotherapy response. Mol Cell, 2013. 51(6): p. 751–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vo TT, et al. , Relative mitochondrial priming of myeloblasts and normal HSCs determines chemotherapeutic success in AML. Cell, 2012. 151(2): p. 344–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are found within the paper and supplementary files.