Abstract
Candida auris is an emerging pathogenic yeast of significant clinical concern because of its frequent intrinsic resistance to fluconazole and often other antifungal drugs and the high mortality rates associated with systemic infections. Furthermore, C. auris has a propensity for persistence and transmission in health care environments. The reasons for this efficient transmission are not well understood, and therefore we tested whether enhanced resistance to environmental stresses might contribute to the ability of C. auris to spread in health care environments. We compared C. auris to other pathogenic Candida species with respect to their resistance to individual stresses and combinations of stresses. Stress resistance was examined using in vitro assays on laboratory media and also on hospital linen. In general, the 17 C. auris isolates examined displayed similar degrees of resistance to oxidative, nitrosative, cationic and cell wall stresses as clinical isolates of C. albicans, C. glabrata, C. tropicalis, C. parapsilosis, C. krusei, C. guilliermondii, C. lusitaniae and C. kefyr. All of the C. auris isolates examined were more sensitive to low pH (pH 2, but not pH 4) compared to C. albicans, but were more resistant to high pH (pH 13). C. auris was also sensitive to low pH, when tested on contaminated hospital linen. Most C. auris isolates were relatively thermotolerant, displaying significant growth at 47°C. Furthermore, C. auris was relatively resistant to certain combinations of combinatorial stress (e.g., pH 13 plus 47°C). Significantly, C. auris was sensitive to the stress combinations imposed by hospital laundering protocol (pH > 12 plus heat shock at >80°C), suggesting that current laundering procedures are sufficient to limit the transmission of this fungal pathogen via hospital linen.
Keywords: Candida auris, Candida pathogens, stress resistance, decontamination, hospital laundry
Introduction
A number of Candida species cause infections in humans. These yeasts are estimated to cause about 400,000 of the estimated two million life-threatening fungal infections that occur annually worldwide, generally in individuals undergoing surgery or with severely compromised immune systems.1 Indeed, the Candida genus is the fourth most common cause of hospital acquired bloodstream infections (candidemia) and the third most common cause in intensive care units.2 Historically, the majority of candidemia cases have been attributed to Candida albicans.3 However, the frequency of infections caused by non-Candida albicans Candida (NAC) species is increasing.4,5 The rising proportion of Candida glabrata, Candida krusei and Candida guilliermondii infections is thought to be due to their intrinsic resistance to certain antifungal drugs,6,7 such as fluconazole, which, when used routinely as a prophylaxis, can lead to selection of resistant species.8 Meanwhile, the prevalence of Candida parapsilosis outbreaks in intensive care units and premature neonates is probably due to this yeast's effective adherence to catheters and skin and the lipid rich environment favorable for its growth afforded by the frequent use of total parenteral nutrition (TPN) in this group.9
Candida auris has recently emerged as a global health concern.10–12C. auris was first isolated in South Korea in 1996, but was not identified as such at the time.13 Although present, C. auris infections were rare prior to 2009, when it was isolated from the ear canal of a patient in Japan and described as a new species.14,15 Evidence for C. auris as a cause of systemic infection in a health care setting was not documented until 2011 in South Korea.13 Since then, C. auris has spread around the globe at an alarming rate. March 2012 saw the first recorded outbreak in the Americas, in a hospital in Venezuela,16 this was quickly followed by the first outbreak in the USA in 2013.16 Sporadic cases also appeared in the UK from 2013,17 with the first UK outbreak in 2015.18 The propensity of C. auris to cause outbreaks in health care settings is now a major global health concern. The rapid emergence of C. auris has caused concern for several reasons. First, this species exhibits a propensity for patient-to-patient transmission in care settings, more so than other species of Candida and other pathogenic fungi.19 Amplified fragment length polymorphism (AFLP) analysis and genome sequencing has suggested that outbreak strains are highly clonal within geographical regions and distinct between continents.18,20,21 Based on these distinctions, C. auris isolates are considered to represent four phylogenetic clades,14 but recently a potential fifth clade has been described.22 Second, a significant proportion of C. auris strains display multidrug resistance. Approximately 90% of C. auris isolates are reported to be resistant to fluconazole, about 8% are resistant to amphotericin B, about 2% are resistant to echinocandins, and about 25% are resistant to more than one class of agent,23 with patterns of resistance closely linked to the phylogenetic clades.24 This drug resistance can lead to therapeutic failure. Third, in some settings C. auris is associated with an unusually high mortality rate compared to other Candida species. For example, whilst C. albicans is associated with ∼40% mortality in hospital patients,25 the mortality rate for C. auris in healthcare environments is 30–60%.16,18,26 Fourth, C. auris has proved difficult to identify using standard biochemical methods and consequently was often misidentified as other Candida species, particularly Candida haemulonii. This increased the chance of inappropriate treatment and, as a result, therapeutic failure.26,27
Currently, C. auris is the focus of a considerable research effort. This research has revealed that C. auris is closely related, phylogenetically, to C. krusei, C. haemulonii, and C. lusitaniae, all of which have been shown to exhibit resistance to azoles and/or amphotericin B.14 Genome sequencing combined with phenotypic analysis has revealed that C. auris shares many of the virulence factors and fitness attributes of C. albicans, such as multidrug efflux systems and cell wall remodeling pathways.21,26,28 Some C. auris strains exhibit salt tolerance, thermotolerance up to 42°C, as well as an aggregating phenotype that has been proposed to contribute to its efficient transmission.28,29
The persistence of C. auris in healthcare settings is also thought to have contributed to its emergence as a global health concern. C. auris has been found to particularly colonize the groin and axilla of infected and noninfected people, it was first isolated from aural cavities and has also been found colonizing the nostrils.18,30C. auris can also persist in patient rooms for extended periods of time, even after cleaning,31 on mattresses, chairs, windowsills, and areas around patient's beds.18,30 Quaternary ammonia products, such as Lysol and Virex II 256, have been shown to be relatively ineffective in killing C. auris,32 but evidence suggests that chlorine products are effective,29,33–35 and the Centers for Disease Control and Prevention (CDC) guidelines recommend use of disinfectants approved for use against Clostridioides difficile.30
We reasoned that resistance to certain environmental stresses might contribute to the spread of C. auris in healthcare environments. Hence, in this study we compared the stress phenome of C. auris to those of other pathogenic Candida species (C. albicans, C. glabrata, C. tropicalis, C. parapsilosis, C. krusei, C. guilliermondii, C. lusitaniae, and C. kefyr), revealing sensitivities to specific combinations of environmental stresses in vitro. The focus of most disinfection studies to date has been the removal of C. auris from hard surfaces or the prevention of transfer between individuals.18,29,33–35 Therefore, we extended our analyses to hospital bed linen, demonstrating that the environmental stresses imposed during hospital laundry procedures are effective in killing C. auris, as well as other pathogenic Candida species.
Methods
Candida clinical isolates
Seventeen C. auris isolates from the South Asian, East Asian, and South Africa clades were selected for analysis (Table 1). These isolates displayed different drug resistance profiles and aggregation phenotypes.28 For comparison with other Candida pathogens, we selected three isolates each of C. albicans, C. glabrata, C. tropicalis, C. parapsilosis, C. krusei, C. guilliermondii, C. lusitaniae, and C. kefyr from the collections of Donna MacCallum and Frank Odds (University of Aberdeen, UK) (Table 2). These 17 C. auris isolates and 24 other Candida isolates were arrayed in a microtiter plate format for stress resistance assays (Fig. 1).
Table 1.
Candida auris strains.
| Mating | |||||||
|---|---|---|---|---|---|---|---|
| Working name | Collection name | Isolated from | Clade | Aggregating | type | Drug resistance | Source |
| auris 1 | Gow1715, 470026 | blood | South Asian | a | FLZ, CAS | 1 | |
| auris 2 | Gow1716, 470027 | blood | South Asian | a | FLZ, VORI, CAS | 1 | |
| auris 3 | Gow1717, 470028 | blood | South Asian | a | FLZ, CAS | 1 | |
| auris 4 | Gow1718, 470029 | blood | South Asian | a | FLZ, VORI, CAS | 1 | |
| auris 5 | Gow1719, 470030 | blood | South Asian | a | FLZ, VORI, CAS | 1 | |
| auris 6 | Gow1720, 470031 | blood | South Asian | a | FLZ, CAS | 1 | |
| auris 7 | Gow1721, 470032 | blood | South Asian | a | FLZ, CAS | 1 | |
| auris 8 | Gow1722, 470033 | blood | South Asian | a | FLZ | 1 | |
| auris 9 | Gow1723, 470034 | blood | South Asian | a | FLZ, 5FC | 1 | |
| auris 10 | Gow1725, 470036 | blood | South Asian | a | FLZ, CAS, 5FC | 1 | |
| auris 11 | Gow1726, 470037 | blood | South Asian | a | FLZ, CAS, 5FC | 1 | |
| auris 12 | Gow1747, NCPF8980#9 | blood | South African | Y | α | FLZ, CAS | 2 |
| auris 13 | Gow1748, NCPF8984#15 | unknown | East Asian | Y | α | FLZ | 2 |
| auris 14 | Gow1750, NCPF8985#20 | wound | South Asian | a | FLZ, ISAV, POSA, VORI, FLY, ANID, CAS | 2 | |
| auris 15 | Gow1749, NCPF13001#16 | unknown | South Asian | a | FLZ | 2 | |
| auris 16 | Gow1751, NCPF13005#95 | urine | South African | Y | α | FLZ, VORI, ANID, AMB, CAS | 2 |
| auris 17 | VPCI479/P/13 | blood | South Asian | a | FLZ | 3 |
Sources: (1) Chakrabarti A. Incidence, characteristics and outcome of ICU-acquired candidemia in India. Intensive Care Med. 2015; 41: 285–295. doi: 10.1007/s00134-014-3603-2 (2) Dr. L. Johnson, Public Health England, Bristol (3) Sharma C, Kumar N, Meis JF, Pandey R, Chowdhary A. 2015. Draft genome sequence of a fluconazole-resistant Candida auris strain from a candidemia patient in India. Genome Announc 3(4): e00722-15. doi:10.1128/genomeA.00722-15.
Table 2.
Other Candida strains.
| Working name | Species | Collection name | Isolated from | Geographic source | Source | Biotyper results | Biotyper score | Chromogenic ID |
|---|---|---|---|---|---|---|---|---|
| Alb 1 | Candida albicans | SC5314 | blood | 3 | Candida albicans | 2.17 | C. albicans | |
| Alb 2 | Candida albicans | SCSB42457 | blood | UK | 2 | Candida albicans | 2.13 | C. albicans |
| Alb 3 | Candida albicans | AM2002/0089 | catheter tip | UK | 1 | Candida albicans | 2.189 | C. albicans |
| Gla 1 | Candida glabrata | SCS130399L | blood | UK | 2 | Candida glabrata | 2.009 | C. glabrata |
| Gla 2 | Candida glabrata | SCS123636D | blood | UK | 2 | Candida glabrata | 1.994 | C. glabrata |
| Gla 3 | Candida glabrata | AM2004/0091 | central line | UK | 1 | Candida glabrata | 1.955 | C. glabrata |
| Tro 1 | Candida tropicalis | SCS122443V | blood | UK | 2 | Candida tropicalis | 2.102 | C. tropicalis |
| Tro 2 | Candida tropicalis | SCS76638K | blood | UK | 2 | Candida tropicalis | 1.959 | C. tropicalis |
| Tro 3 | Candida tropicalis | AM2007/0111 | IV catheter | UK | 1 | Candida tropicalis | 2.049 | C. tropicalis |
| Par 1 | Candida parapsilosis | SCS5080820 | blood | UK | 2 | Candida glabrata | 1.8 | C. glabrata |
| Candida tropicalis | 1.7 | C. tropicalis | ||||||
| C. albicans | ||||||||
| Par 2 | Candida parapsilosis | SCSXM70052 | blood | UK | 2 | Candida parapsilosis | 2.113 | C. parapsilosis |
| Par 3 | Candida parapsilosis | SCSBB425167 | blood | UK | 2 | Candida parapsilosis | 2.003 | C. parapsilosis |
| Kru 1 | Candida krusei | SCS71987M | blood | UK | 2 | Candida krusei | 2.062 | C. krusei |
| Kru 2 | Candida krusei | SCS73972P | blood | UK | 2 | Candida parapsilosis | 2.084 | C. parapsilosis |
| Kru 3 | Candida krusei | AM2019/001 | femoral line tip | UK | 1 | Candida krusei | 2.14 | C. krusei |
| Gui 1 | Candida guilliermondii | SCSBB418097 | blood | UK | 2 | Candida guilliermondii | 2.104 | C. guilliermondii |
| C. albicans | ||||||||
| Gui 2 | Candida guilliermondii | SCS74937K | blood | UK | 2 | Candida guilliermondii | 2.127 | C. guilliermondii |
| Gui 3 | Candida guilliermondii | SCSMB042615 | blood | UK | 2 | Candida guilliermondii | 2.1 | C. guilliermondii |
| Lus 1 | Candida lusitaniae | SCS211362H | blood | UK | 2 | Candida lusitaniae | 2.275 | C. lusitaniae |
| Lus 2 | Candida lusitaniae | SCS17999 | blood | UK | 2 | not reliable identification | 1.367 | C. lusitaniae |
| C. krusei | ||||||||
| Lus 3 | Candida lusitaniae | SCS401413 | blood | UK | 2 | Candida lusitaniae | 1.743 | C. lusitaniae |
| Kef 1 | Candida kefyr | J981143 | unknown | USA | 1 | Candida tropicalis | 2.262 | C. tropicalis |
| Kef 2 | Candida kefyr | 81/017 | unknown | unknown | 1 | Candida kefyr | 1.765 | C. kefyr |
| Kef 3 | Candida kefyr | Y128 | unknown | unknown | 1 | Candida kefyr | 2.143 | C. kefyr |
Sources: (1) Dr. D. MacCallum, University of Aberdeen (2) Odds, F. C., Hanson, M. F., Davidson, A. D., Jacobsen, M. D., Wright, P., Whyte, J. A., et al. (2007). One year prospective survey of Candida bloodstream infections in Scotland. J. Med. Microbiol. 56, 1066–1075. doi: 10.1099/jmm.0.47239-0 (3) Gillum, A. M., Tsay, E. Y. H. & Kirsch, D. R. (1984). Isolation of the Candida albicans gene for orotidine-5′-phosphate decarboxylase by complementation of S. cerevisiae ura3 and E. coli pyrF mutations. Mol Gen Genet 198, 179–182.
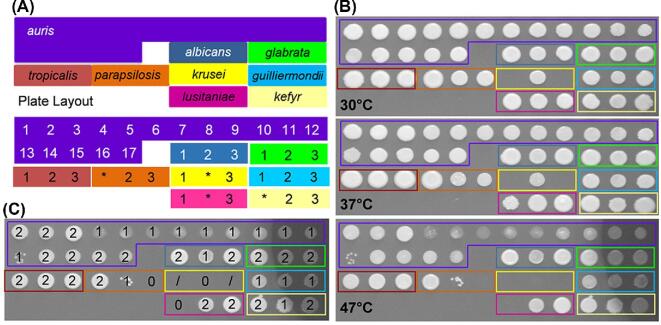
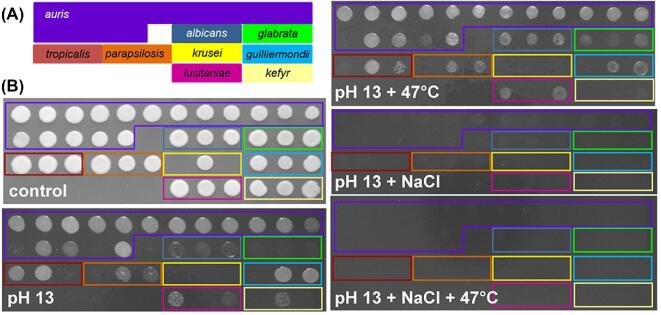
Figure 1.
Comparison of C. auris thermotolerance with other Candida pathogens. (A) The microtiter plate layout of the Candida clinical isolates listed in Tables 1 and 2. Misidentified isolates are highlighted with an asterisk. (B) Growth of Candida isolates for 24 h on YPD agar at the stated temperatures. (C) Illustration of the semi-quantitative analysis of stress resistance, where 0 = no growth, 1 = less growth, 2 = ‘normal growth’ 3 = more growth, in comparison to the same isolate grown on YPD at 30°C, using the 47°C panel from (B) which illustrates one representative of six replicates. Data from the six replicate experiments were used to generate the averages presented in Tables S1 and S2. Note that while ‘C. krusei’ strains were included in the panel (yellow), for technical reasons these were excluded from our data analysis. This Figure is reproduced in color in the online version of Medical Mycology.
The identity of the C. auris isolates had been confirmed by ITS sequence analysis.36,37 To confirm the identity of the other Candida isolates, these were analyzed by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI ToF MS)38 and by growth on chromogenic media (Brilliance™ Candida Agar, Oxoid, Hampshire, UK), which highlighted that some cultures were mixed. The initial species identification was confirmed for most isolates (Table 2). However, in a few cases, initial misidentifications were corrected by MALDI ToF MS: C. krusei 2 was in fact C. parapsilosis; C. kefyr 2 was C. tropicalis; and C. parapsilosis 1 was a mixture of C. glabrata and C. tropicalis (Table 2; Fig. S1). As strain identification was performed after the isolates were arrayed, misidentified Candida isolates were included in the stress panels, below (these are marked with an asterisk in the figures). C. krusei 1 and 3 were excluded from some assays (by washing these wells with 100% ethanol before replication onto test plates) because these isolates spread and overgrew neighboring isolates under certain growth conditions.
Candida strains were grown overnight in yeast-extract peptone dextrose (YPD) (1% yeast extract, 2% mycological peptone, 2% glucose)39 at the specified temperature for each experiment. Similar inocula of each isolate (based on optical density [OD]600), from early stationary phase liquid cultures, were aliquoted into a microtiter plate format (Fig. 1), glycerol was added to a final concentration of 20%, and replicate plates frozen at −80°C. Plates were then thawed at room temperature for 45 minutes before replica plating onto the specified media.
Stress resistance on plates
To test the stress sensitivity of the Candida isolates, isolates were replica plated using an 8 × 12 prong replicator (Sigma-Aldrich, Dorset, UK) onto YPD agar supplemented with one of the following: 1 M NaCl; 0.6 M KCl; 25 mM succinic acid; 25 mM succinic acid plus 5 mM sodium nitrite (NaNO2); 1 mM tert-Butyl hydroperoxide (t-BOOH); 2.5, 5 or 7.5 mM hydrogen peroxide (H2O2); 100 µg/ml Calcofluor White (CFW); 100 or 150 µg/ml Congo Red (CR); or YPD adjusted to pH2, pH4, pH10, pH11, pH12, or pH13. Plates were incubated at 30°C, 37°C or 47°C for 24, 48, or 72 hours, as specified. Every experiment included a control YPD plate incubated at 30°C for 24 hours. Also, clinical isolates were exposed to the following stresses for 10 minutes in liquid suspension40,41 before plating on YPD: 0.5% chlorhexidine (Chlor); 0.5% sodium hypochlorite (NaClO); 2% alcohol ethoxylate (AlcEth). The growth of each isolate was rated qualitatively on a scale of 0 to 3, compared to the growth of the same isolate on YPD at 30°C in the absence of stress (where ‘normal’ growth was assigned 2, no growth was assigned 0, less growth was assigned 1, and more growth was assigned 3) (Fig. 1C). The average from six independent replicate experiments was then calculated to provide a semiquantitative measure of the effect of the stress upon growth.
Stress resistance on hospital linen
To test the stress sensitivities of Candida clinical isolates on hospital linen, bedsheets were obtained from NHS Grampian, one of the regional health boards for the National Health Service Scotland, cut into required sizes and sterilized by autoclaving. The 41 Candida isolates were replica plated onto 12 × 8 cm rectangles of linen (in the pattern shown in Fig. 1) and, after inoculation, these sheets were laid onto YPD plates containing different stressors. After incubation for 48 hours at 30°C, the linen was removed to assess the growth of each isolate. Alternatively, where specified, individual Candida isolates were inoculated onto clean, sterilized 1 × 1 cm squares (approximately 1 × 106 cells per square). To simulate patient soiling, 30 µl of test soil (Browne washer/disinfector test soil, Serris, France), a substance used to test washer efficiency by mimicking contamination by bodily fluids, was added to the squares after inoculation. Inoculated squares were then laid on YPD plates (containing different stressors, as above) and incubated at the relevant temperature for 1–3 days. The squares were removed, and cells were recovered by vigorously vortexing each square in 1 ml sterile water. The cells were harvested by centrifugation and resuspended in 1 ml water, then the OD600 of the cell suspension measured to estimate the amount of Candida recovered after growth on each inoculated linen square. While the degree of growth in the presence of stress varied, generally over 95% of those cells harvested after growth were viable, based on cfu assays.
Results
C. auris isolates generally display similar stress sensitivities to other Candida species
The Candida isolates were exposed to a range of individual stress conditions including growth at different temperatures (ranging from 30 to 47°C) and pH (2 to 13), cationic stresses (NaCl, KCl), oxidative stresses (H2O2, t-BOOH), nitrosative stresses (sodium nitrite), and cell wall stresses (Calcofluor White, Congo Red). The amount of growth on plates was estimated (Fig. 1), and the average from six experiments (two technical replicates in each of three biological repeats) was used to generate a semi-quantitative measure of the effect of a stress upon growth. These data are summarized in Table S1.
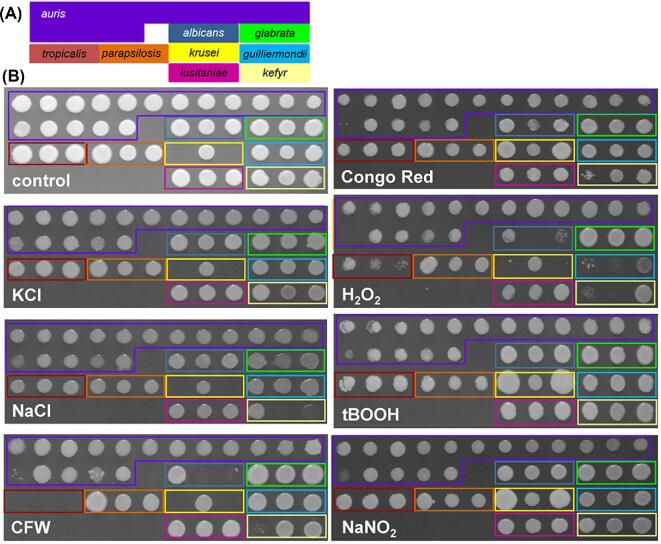
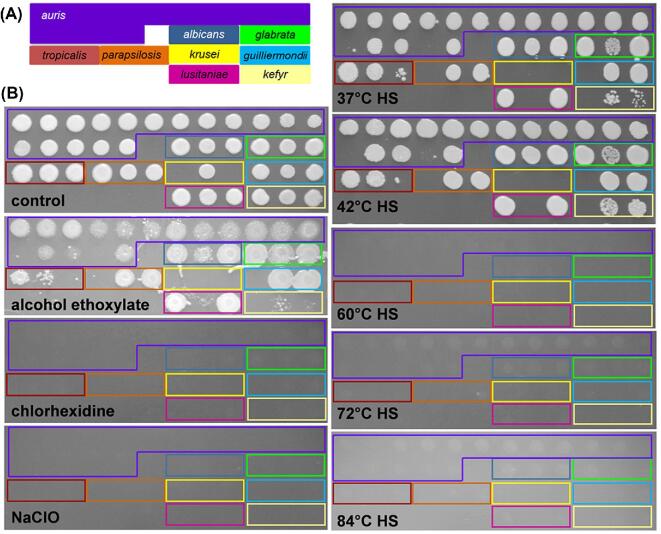
All of the Candida isolates tested grew well at 30 and 37°C, but the growth of a subset of C. auris isolates (4–13) was attenuated at 42°C and 47°C (Fig. 1B, Table S1). The C. auris isolates were generally resistant to the cationic, oxidative, nitrosative and cell wall stresses we examined, with the exception of C. auris 13 and 16 (Fig. 2; Table S1). C. auris 13 and 16 are from the East Asian and South African clades, respectively. Therefore, their sensitivities appear to be isolate-, rather than clade-, related. The stress resistance of most C. auris isolates we examined is consistent with recent data showing that some C. auris isolates are resistant to double the concentration of CFW and Congo Red used in this study and to similar levels of cationic and oxidative stress.42 Furthermore, the C. auris isolates generally displayed similar levels of resistance to those demonstrated by the other Candida isolates examined. However, all three C. tropicalis isolates and two C. albicans isolates (2 and 3) were particularly sensitive to Calcofluor White. Also, C. glabrata was particularly resistant to H2O2 (Fig. 2; Table S1).
Figure 2.
Resistance of C. auris and other Candida pathogens to individual environmental stresses. (A) The layout of the Candida clinical isolates to facilitate interpretation. (B) Growth of these Candida clinical isolates on YPD at 30°C for 24 h in the presence of the following stressors: no stress, control; 0.6 M KCl; 1 M NaCl; 100 µg/ml Calcofluor White, CFW; 150 µg/ml Congo Red; 7.5 mM H2O2; 1 mM t-BOOH); 25 mM succinic acid plus 5 mM NaNO2. Illustrates one representative of six replicates. This Figure is reproduced in color in the online version of Medical Mycology.
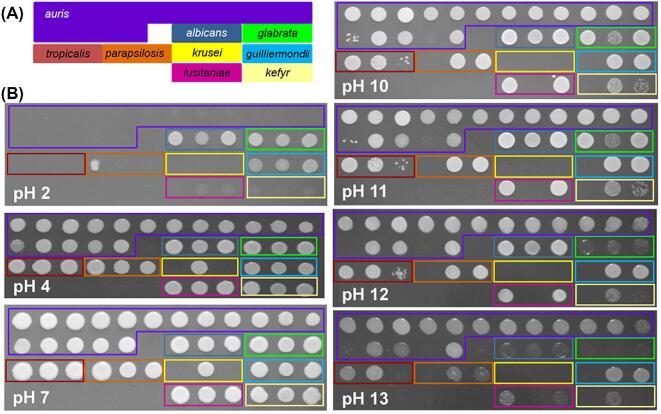
Significant differences in pH tolerance were observed between species (Fig. 3). C. albicans was tolerant to extremely low pH (pH 2) as reported previously,43 with C .glabrata and C. guilliermondii displaying similar degrees of acid tolerance. Interestingly, all C. auris isolates were unable to grow at acidic pH (pH 2) but, with the exception of isolates 13 and 16, were resistant to highly alkaline conditions (pH 13) (Fig. 3). Therefore, like other Candida pathogens, C. auris can tolerate a wide range of pH but, unlike most of the other species tested, it can grow at the high pH associated with detergents used during hospital laundering.44
Figure 3.
Resistance of the C. auris and other Candida isolates to pH stresses. (A) The layout of the Candida clinical isolates. (B) Growth of these Candida clinical isolates on YPD at 30°C for 24 h at the specified pH. Illustrates one representative of six replicates. This Figure is reproduced in color in the online version of Medical Mycology.
C. auris isolates and other Candida isolates are sensitive to certain combinatorial stresses
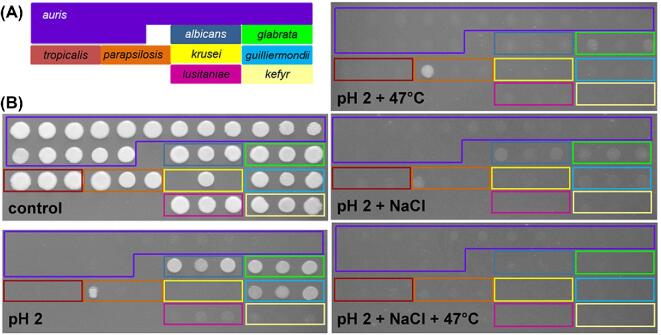
C. albicans and C. glabrata are sensitive to certain combinations of stress, notably salt plus oxidative stress.45 Therefore, we compared the resistance of the Candida isolates to certain types of combinatorial stress, focusing initially on combinations of salt stress with those stresses to which some sensitivity had been observed, as described in the section above. We found that, unlike C. albicans,46C. auris isolates were relatively resistant to the combinatorial salt plus oxidative stress (Table S2). C. parapsilosis and C. guilliermondii were also resistant to this combinatorial stress, whereas C. lusitaniae and C. kefyr were sensitive (Table S2). Interestingly, all of the pathogenic Candida species tested were sensitive to combinations of NaCl with pH extremes, whether at pH 2 or pH 13 (Figs 4, 5), and where residual growth was observed at 30°C, all growth was inhibited when the temperature was raised to 47°C (Table S1). Therefore, combinatorial salt plus pH plus thermal stresses are particularly effective at killing C. auris and other pathogenic Candida species (Fig. 4).
Figure 4.
Resistance of the Candida isolates to combinations of salt, thermal and acid stress. (A) The layout of the Candida clinical isolates. (B) Growth of these Candida clinical isolates for 24 h at 30°C on unbuffered YPD (control) or, where specified, at pH2 with 1 M NaCl, or at 47°C. This Figure is reproduced in color in the online version of Medical Mycology.
Figure 5.
Resistance of the Candida isolates to combinations of salt, thermal and alkaline stress. (A) The layout of the Candida clinical isolates. (B) The growth of these Candida clinical isolates for 24 hours at 30°C on unbuffered YPD (control) or, where specified, at pH 13, with 1 M NaCl, or at 47°C. Illustrates one representative of six replicates. This Figure is reproduced in color in the online version of Medical Mycology.
Stress resistance of Candida clinical isolates on hospital linen
It has been suggested that, in the hospital setting, C. auris might spread via contaminated fomites, such as bed linen.18,30,47 Therefore, we tested the stress sensitivities of the Candida isolates when they were contaminating hospital linen under controlled laboratory conditions. All of the isolates were inoculated onto linen squares, dried for 1 hour, laid on a YPD plate inoculated side up, and incubated at 30°C for 72 hours. The linen was then removed and the plate photographed (Fig. 6A), showing that most isolates remained viable and were able to grow under these conditions. However, despite testing a number of approaches, our attempts to quantify the degree to which each Candida isolate grew on the linen over YPD plates proved fruitless. The challenges were compounded by the aggregation phenotype of some C. auris isolates.28 Therefore, we estimated the growth of individual isolates on linen squares by resuspending the cells in water and measuring the resultant OD600.
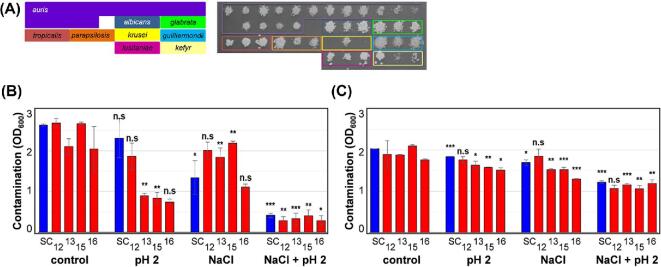
Figure 6.
Resistance of the C. auris to combinations of salt and pH stress when contaminating ‘clean’ or ‘soiled’ hospital linen. (A) The layout of the Candida clinical isolates and their growth on YPD after contamination of clean linen. (B) Growth of selected clinical isolates on clean 1 × 1 cm linen squares in YPD in the presence or absence of stress (pH2 and/or 1 M NaCl) for 24 h at 30°C: C. albicans SC5314, SC; C. auris clinical isolates 12, 13, 15, 16. Growth was estimated by re-suspending the Candida cells in H2O and measuring the OD600/ml. (C) The resistance of these Candida isolates to combinations of acid and salt stress on soiled linen. Means and standard deviations from three independent replicate experiments are shown, and stressed isolates were compared to the relevant unstressed control using the student t-test: ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. This Figure is reproduced in color in the online version of Medical Mycology.
Using this approach, we focused on five representative isolates: C. albicans SC5314 and C. auris isolates 12, 13, 15, and 16. First we examined them on ‘clean’ linen. The growth of three C. auris isolates (13, 15, 16) was attenuated at pH2 and none of these isolates were particularly affected by the salt stress, but the growth of all five isolates was inhibited by the combination of these two stresses (Fig. 6B). These data suggest that, for these Candida isolates, growth on linen does not confer resistance to combinatorial acid plus salt stress.
We then examined the impact of soiling upon the stress sensitivities of the Candida isolates on linen. Soiling enhanced the resistance of most clinical isolates to pH2 and/or salt stress, but particularly to combinatorial salt plus acid stress (Fig. 6C). Clearly this could be significant in the clinical setting.
Candida resistance to clinically relevant disinfectants and laundry conditions
Next we examined the impact of clinically relevant disinfectants and laundry conditions upon the full set of 41 Candida isolates. We tested the susceptibility of these isolates to chlorhexidine and sodium hypochlorite as these are frequently used to clean surfaces in patient rooms.40,41,48,49 The isolates were exposed to 0.5% chlorhexidine or sodium hypochlorite for 10 minutes before plating. These concentrations were based on standard cleaning recommendations and previous studies.29,32,41,50,51 All Candida isolates were sensitive to chlorhexidine and sodium hypochlorite under these conditions (Fig. 7B).
Figure 7.
Resistance of the Candida isolates to hospital laundry-related environmental stresses. (A) The layout of the Candida clinical isolates. (B) The growth of these Candida clinical isolates for 24 h at 30°C on YPD after exposure to 0.5% chlorhexidine, 0.5% sodium hypochlorite (NaClO) or 2% alcohol ethoxylate for 10 min, or to the specified heat shock (HS) for 5 min. Illustrates one representative of six replicates. This Figure is reproduced in color in the online version of Medical Mycology.
We also examined the effects of alcohol ethoxylate on the Candida isolates as this is the primary ingredient in detergents used in National Health Service (NHS) Scotland laundries (Dermasil Plus). The alcohol ethoxylate concentration we used (2%) was based on information provided by the Laundry Facility at the Foresterhill Healthcare Campus, NHS Grampian. Many of the clinical isolates, and most of the C. auris isolates in particular, were resistant to alcohol ethoxylate at 30°C (Fig. 7B). However, hospital laundry conditions also include rapid cycles of washing at high temperatures. For example, during the laundry process at the Foresterhill Healthcare Campus, the temperature is raised rapidly from 38°C to 52°C, and then to 81°C, 84°C, and 80°C. Each load spends at least 4.5 min above 80°C. Therefore, we tested the effects of a 5-min heat shock at different temperatures on the Candida isolates. Heat shock at 60°C or above killed all of the isolates examined (Fig. 7B).
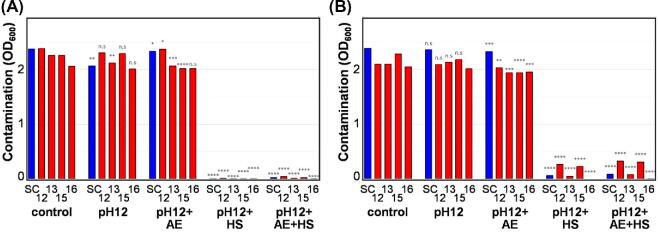
We extended our analyses to hospital linen. We included alkaline pH because hospital laundry cycles can approach pH12. Consistent with our earlier finding (Fig. 5), C. auris isolates were resistant to pH12 when contaminating hospital linen (Fig. 8). However, the combination of alkaline stress with heat shock (84°C) was particularly effective at killing cells on ‘clean’ linen (Fig. 8A). However, some residual cells were observed on ‘soiled’ linen (Fig. 8B). We conclude that, while care might be required for badly soiled linen, hospital laundry conditions effectively kill the pathogenic Candida species we examined, including C. auris.
Figure 8.
Resistance of the C. auris to a combination of laundry-related stresses on ‘clean’ or ‘soiled’ hospital linen. (A) The resistance of selected Candida isolates on clean linen to a combination of alkaline stress (pH12), alcohol ethoxylate (2%) and heat shock (84°C): C. albicans SC5314, SC; C. auris clinical isolates 12, 13, 15, 16. (B) Resistance of the same isolates on soiled linen to a combination of alkaline stress, alcohol ethoxylate and heat shock. Means and standard deviations from three independent replicate experiments are shown, and stressed isolates were compared to the relevant unstressed control using the student t-test: ns, not significant; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001. This Figure is reproduced in color in the online version of Medical Mycology.
Discussion
The pathogenic yeasts, C. albicans and C. glabrata, display relatively high levels of resistance to certain environmental stresses compared to less-pathogenic yeasts, such as C. lusitaniae, and this is thought to enhance their pathogenicity.52,53 We reasoned therefore that the ability of C. auris to persist in hospital environments and on individuals might be attributed, at least in part, to a relatively high resistance to environmental stresses compared with other Candida pathogens. With a view to obtaining a general, rather than an isolate-specific, view of their stress phenomes, 17 different C. auris isolates from different geographical locations were compared with at least two clinical isolates each of C. albicans, C. glabrata, C. tropicalis, C. parapsilosis, C. guilliermondii, C. lusitaniae, and C. kefyr.
Initially, we examined resistance to a range of individual environmental stresses. Few significant differences were observed between C. auris and the other Candida pathogens with respect to their thermotolerance or their resistance to salt, oxidative, nitrosative or cell wall stresses. This is consistent with a recent study that describes the importance of the stress activated protein kinase, Hog1, in mediating the resistance of C. auris to such stresses.42 Similar to other Candida isolates examined, C. auris isolates were tolerant over a broad pH range. However, C. auris was more resistant under alkaline conditions relative to C. albicans and C. glabrata (pH13). This is significant because hospital bleach is typically only pH12, and the activity range for quaternary ammonia products commonly used in health care environments is between pH3 and pH10.54 It has been reported that C. auris is less susceptible than C. albicans to 2% and 4% chlorhexidine55 and that C. parapsilosis, which has shown similar persistence levels on skin and environmental surfaces,56 is not killed by chlorine based cleaners after standard exposure times. In our hands, none of the Candida isolates grew after exposure to bleach or chlorhexidine in vitro. This confirmed and extended the findings of a recent report showing that C. auris, C. albicans, and C. glabrata are sensitive to sodium hypochlorite.34,35,55,57,58 However, it should be noted that, even after repeated application of chlorhexidine (a common skin disinfectant also used to disinfect surgical instruments), C. auris can still persist on patient skin.18,34,59 It has also been shown that C. auris can persist on hospital linen for several days.60 This suggests that sensitivity of C. auris to disinfection depends on the physical and physiological context.
We also examined combinatorial stresses because C. albicans is particularly sensitive to combinations of salt plus oxidative stress.46 We found that the C. auris isolates displayed similar sensitivity to combinations of salt, alkaline and thermal stress as the other Candida isolates we examined. Therefore, in general, our analyses of individual and combinatorial stresses did not support our initial working hypothesis that persistence of C. auris is attributable to high levels of environmental stress resistance compared to other Candida pathogens. The possible exception was the resistance of C. auris to combinatorial salt plus oxidative stress which could, in principle, compromise the ability of innate immune cells to kill this fungus.46 However, we reasoned that this would be unlikely to directly affect the transmission of C. auris in hospital settings.
Having examined C. auris and other Candida pathogens in vitro, we then tested the stress resistance of this pathogen when contaminating hospital linen. In general, the results were similar to those observed in vitro, where most C. auris isolates remained sensitive to combinatorial salt plus acid stress and the combination of alkali, alcohol ethoxylate and heat shock. However, the soiling of linen appeared to enhance resistance to these stresses. This observation, which is highly relevant to the clinical setting, strengthens the suggestion that the stress sensitivities of C. auris depends on the physiological context (above). For example, it is entirely conceivable that C. auris stress resistance might be enhanced during growth in mixed-species populations or biofilms.29 This would certainly be consistent with the fact that the growth conditions significantly affect the responses of C. albicans to environmental stresses.61–63
We also examined environmental stresses of direct relevance to hospital laundering, alcohol ethoxylate, thermal and alkaline stresses. Our data suggest that the heat shocks imposed during the laundry process promote the killing of the Candida pathogens we tested. However, it should be noted that our data do not exclude the possibility that nongrowing persister cells may survive these treatments.59 Furthermore, particular care might need to be taken with the washing process for heavily soiled laundry, as there appears to be an increased level of cells present on soiled linen after exposure to laundry conditions, compared to that of clean linen.
In conclusion, the profiles of Candida stress resistance observed in this study do not support our original working hypothesis that the nosocomial transmission of C. auris isolates is enhanced by their relatively high resistance to environmental stresses. Nevertheless, it would be prudent to test whether the stress resistance of Candida species, and C. auris in particular, is affected by other fabrics found in health care environments, such as healthcare professionals’ uniforms, some of which are washed by these professionals at home. This information, together with more accurate replication of the laundry process in vitro, would help to inform hospital laundering protocols in general.
Supplementary Material
Acknowledgments
We are grateful to Dr. David Stead, Evelyn Argo, and Craig Pattison (Aberdeen Proteomics Core Facility) for their expert identification of Candida isolates by MALDI ToF MS, and to Dr. Jill King and our colleagues in the Aberdeen Fungal Group for their helpful advice. AJPB and NARG were supported by a programme grant from the Medical Research Council [www.mrc.ac.uk] (grant number MR/M026663/1) and by the Medical Research Council Centre for Medical Mycology and the University of Aberdeen (grant number MR/N006364/1). A.J.P.B. was also supported by the UK Biotechnology and Biological Research Council [www.bbsrc.ac.uk] (grant numbers BB/F00513X/1, BB/P020119/1), and A.W.W. by the Scottish Government's Rural and Environment Science and Analytical Services (RESAS) division. N.A.R.G. was also supported by grants from the Wellcome Trust [www.wellcome.ac.uk] (grant numbers 075470, 086827, 093378, 097377, 099197, 101873, 102705, 200208). D.M.M. was supported by National Centre for the Replacement, Refinement and Reduction of Animals in Research (NC3Rs) [www.nc3rs.org.uk] (grant numbers NC/S001557/1 and NC/N002482/1) and the UK Biotechnology and Biological Research Council [www.bbsrc.ac.uk] (grant number BB/P02050X/1). H.H. was supported by the Dorothy Campbell Scholarship from the University of Aberdeen's Development Trust. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the paper.
Contributor Information
Helen Heaney, Aberdeen Fungal Group, Institute of Medical Sciences, University of Aberdeen, Aberdeen, UK.
Juliette Laing, NHS Grampian Central Decontamination Unit, Foresterhill Health Campus, Aberdeen, UK.
Linda Paterson, NHS Grampian Central Decontamination Unit, Foresterhill Health Campus, Aberdeen, UK.
Alan W Walker, Rowett Institute, University of Aberdeen, Aberdeen, UK.
Neil A R Gow, Aberdeen Fungal Group, Institute of Medical Sciences, University of Aberdeen, Aberdeen, UK; MRC Centre for Medical Mycology, University of Exeter, School of Biosciences, Exeter, UK.
Elizabeth M Johnson, Mycology Reference Laboratory, PHE South West Laboratory, Southmead Hospital, Bristol, UK.
Donna M MacCallum, Aberdeen Fungal Group, Institute of Medical Sciences, University of Aberdeen, Aberdeen, UK.
Alistair J P Brown, Aberdeen Fungal Group, Institute of Medical Sciences, University of Aberdeen, Aberdeen, UK; MRC Centre for Medical Mycology, University of Exeter, School of Biosciences, Exeter, UK.
Declaration of interest
The authors report no conflicts of interest. The authors alone are responsible for the content and the writing of the paper.
References
- 1. Brown GD, Denning DW, Gow NA, Levitz SM, Netea MG, White TC. Hidden killers: human fungal infections. Sci Transl Med. 2012; 4: 165rv13. [DOI] [PubMed] [Google Scholar]
- 2. Orsini J, Mainardi C, Muzylo E, Karki N, Cohen N, Sakoulas G. Microbiological profile of organisms causing bloodstream infection in critically ill patients. J Clin Med Res. 2012; 4: 371–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kett DH, Azoulay E, Echeverria PM, Vincent JL. Extended Prevalence of Infection in ICU Study (EPIC II) Group of Investigators . Candida bloodstream infections in intensive care units: analysis of the extended prevalence of infection in intensive care unit study. Crit Care Med. 2011; 39: 665–670. [DOI] [PubMed] [Google Scholar]
- 4. Lamoth F, Lockhart SR, Berkow EL, Calandra T. Changes in the epidemiological landscape of invasive candidiasis. J Antimicrob Chemother. 2018; 73: i4–i13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Quindos G, Marcos-Arias C, San-Millan R, Mateo E, Eraso E. The continuous changes in the aetiology and epidemiology of invasive candidiasis: from familiar Candida albicans to multiresistant Candida auris. Int Microbiol. 2018; 21: 107–119. [DOI] [PubMed] [Google Scholar]
- 6. Arendrup MC. Update on antifungal resistance in Aspergillus and Candida. Clin Microbiol Infect. 2014; 20: 42–48. [DOI] [PubMed] [Google Scholar]
- 7. Perlin DS, Shor E, Zhao Y. Update on antifungal drug resistance. Curr Clin Microbiol Rep. 2015; 2: 84–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zilberberg M, Yu HT, Chaudhari P, Emons MF, Khandelwal N, Shorr AF. Relationship of fluconazole prophylaxis with fungal microbiology in hospitalized intra-abdominal surgery patients: a descriptive cohort study. Crit Care. 2014; 18: 590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pammi M, Holland L, Butler G, Gacser A, Bliss JM. Candida parapsilosis is a significant neonatal pathogen: a systematic review and meta-analysis. Pediatr Infect Dis J. 2013; 32: e206–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cortegiani A, Misseri G, Fasciana T, Giammanco A, Giarratano A, Chowdhary A. Epidemiology, clinical characteristics, resistance, and treatment of infections by Candida auris. J Intensive Care. 2018; 6: 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lone SA, Ahmad A. Candida auris: the growing menace to global health. Mycoses. 2019; 62: 620–637. [DOI] [PubMed] [Google Scholar]
- 12. Fisher MC, Hawkins NJ, Sanglard D, Gurr SJ. Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science. 2018; 360: 739–742. [DOI] [PubMed] [Google Scholar]
- 13. Lee WG, Shin JH, Uh Y et al.. First three reported cases of nosocomial fungemia caused by Candida auris. J Clin Microbiol. 2011; 49: 3139–3142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lockhart SR, Etienne KA, Vallabhaneni S et al.. Simultaneous emergence of multidrug-resistant candida auris on 3 continents confirmed by whole-genome sequencing and epidemiological analyses. Clin Infect Dis. 2017; 64: 134–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Satoh K, Makimura K, Hasumi Y, Uchida K, Yamaguchi H. Candida auris sp. nov., a novel ascomycetous yeast isolated from the external ear canal of an inpatient in a Japanese hospital. Microbiol Immunol. 2009; 1: 41–44. [DOI] [PubMed] [Google Scholar]
- 16. Calvo B, Melo AS, Perozo-Mena A et al.. First report of Candida auris in America: clinical and microbiological aspects of 18 episodes of candidemia. J Infect. 2016; 73: 369–374. [DOI] [PubMed] [Google Scholar]
- 17. Public Health England Candida auris: The characteristics, diagnosis and management of Candida auris. 2017, Available from: https://www.gov.uk/government/collections/candida-auris, Accessed July 31, 2019. [Google Scholar]
- 18. Schelenz S, Hagen F, Rhodes JL et al.. First hospital outbreak of the globally emerging Candida auris in a European hospital. Antimicrob Resist Infect Control. 2016; 5: 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eyre DW, Sheppard AE, Madder H et al.. A Candida auris outbreak and its control in an intensive care setting. N Engl J Med. 2018; 379: 1322–1331. [DOI] [PubMed] [Google Scholar]
- 20. Sarma S, Upadhyay S. Current perspective on emergence, diagnosis and drug resistance in Candida auris. Infect Drug Resist. 2017; 10: 155–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rhodes J, Abdolrasouli A, Farrer RA et al.. Genomic epidemiology of the UK outbreak of the emerging human fungal pathogen Candida auris. Emerg Microbes Infect. 2018; 7: 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Chow NA, de Groot T, Badali H, Abastabar M, Chiller TM, Meis JF. Potential fifth clade of Candida auris, Iran, 2018. Emerg Infect Dis. 2019; 25: 1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chowdhary A, Prakash A, Sharma C et al.. A multicentre study of antifungal susceptibility patterns among 350 Candida auris isolates (2009–17) in India: role of the ERG11 and FKS1 genes in azole and echinocandin resistance. J Antimicrob Chemother. 2018; 73: 891–899. [DOI] [PubMed] [Google Scholar]
- 24. Welsh RM, Sexton DJ, Forsberg K, Vallabhaneni S, Litvintseva A. Insights into the unique nature of the east Asian clade of the emerging pathogenic yeast Candida auris. J Clin Microbiol. 2019; 57: e00007––19.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moran C, Grussemeyer CA, Spalding JR, Benjamin DK Jr, Reed SD. Candida albicans and non-albicans bloodstream infections in adult and pediatric patients: comparison of mortality and costs. Pediatr Infect Dis J. 2009; 28: 433–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chatterjee S, Alampalli SV, Nageshan RK, Chettiar ST, Joshi S, Tatu US. Draft genome of a commonly misdiagnosed multidrug resistant pathogen Candida auris. BMC Genomics. 2015; 16: 686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ben-Ami R, Berman J, Novikov A et al.. Multidrug-resistant Candida haemulonii and C. auris, Tel Aviv, Israel. Emerg Infect Dis. 2017; 23: 195–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Borman AM, Szekely A, Johnson EM. Comparative pathogenicity of United Kingdom isolates of the emerging pathogen Candida auris and other key pathogenic Candida species. mSphere. 2016; 1 doi:4:10.1128/mSphere.00189-16. eCollection 2016 Jul-Aug, doi:10.1128/mSphere.00189-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sherry L, Ramage G, Kean R et al.. biofilm-forming capability of highly virulent, multidrug-resistant Candida auris. Emerg Infect Dis. 2017; 23: 328–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vallabhaneni S, Kallen A, Tsay S et al.. Investigation of the first seven reported cases of Candida auris, a globally emerging invasive, multidrug-resistant fungus-United States, May 2013–August 2016. Am J Transplant. 2017; 17: 296–299. [DOI] [PubMed] [Google Scholar]
- 31. Piedrahita CT, Cadnum JL, Jencson AL, Shaikh AA, Ghannoum MA, Donskey CJ. Environmental surfaces in healthcare facilities are a potential source for transmission of Candida auris and other Candida species. Infect Control Hosp Epidemiol. 2017; 38: 1107–1109. [DOI] [PubMed] [Google Scholar]
- 32. Cadnum JL, Shaikh AA, Piedrahita CT et al.. Effectiveness of disinfectants against Candida auris and other Candida species. Infect Control Hosp Epidemiol. 2017; 38: 1240–1243. [DOI] [PubMed] [Google Scholar]
- 33. Tsay S, Kallen A, Jackson BR, Chiller TM, Vallabhaneni S. Approach to the investigation and management of patients with Candida auris, an emerging multidrug-resistant yeast. Clin Infect Dis. 2018; 66: 306–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ku TSN, Walraven CJ, Lee SA. Candida auris: disinfectants and implications for infection control. Front Microbiol. 2018; 9: 726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kean R, Sherry L, Townsend E et al.. Surface disinfection challenges for Candida auris: an in vitro study. J Hosp Infect. 2018; 98: 433–436. [DOI] [PubMed] [Google Scholar]
- 36. Chakrabarti A, Sood P, Rudramurthy S et al.. Incidence, characteristics and outcome of ICU-acquired candidemia in India. J Intensive Care. 2015; 41: 285–295. [DOI] [PubMed] [Google Scholar]
- 37. Sharma C, Kumar N, Meis JF, Pandey R, Chowdhary A. Draft genome sequence of a fluconazole-resistant Candida auris strain from a candidemia patient in India. Genome Announc. 2015; 3 doi:10.1128/genomeA.00722-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Schmitt BH, Cunningham SA, Dailey AL, Gustafson DR, Patel R. Identification of anaerobic bacteria by Bruker Biotyper matrix-assisted laser desorption ionization-time of flight mass spectrometry with on-plate formic acid preparation. J Clin Microbiol. 2013; 51: 782–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sherman F. Getting started with yeast. Methods Enzymol. 1991; 194: 3–21. [DOI] [PubMed] [Google Scholar]
- 40. Abreu AC, Tavares RR, Borges A, Mergulhao F, Simoes M. Current and emergent strategies for disinfection of hospital environments. J Antimicrob Chemother. 2013; 68: 2718–2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rutala WA, Weber DJ. Infection control: the role of disinfection and sterilization. J Hosp Infect. 1999; 43Suppl: S43–55. [DOI] [PubMed] [Google Scholar]
- 42. Day AM, McNiff MM, da Silva Dantas A, Gow NAR, Quinn J. Hog1 regulates stress tolerance and virulence in the emerging fungal pathogen Candida auris. mSphere. 2018; 3:5. doi:10.1128/mSphere.00506-18, e00506-18 [pii]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vylkova S, Carman AJ, Danhof HA, Collette JR, Zhou H, Lorenz MC. The fungal pathogen Candida albicans autoinduces hyphal morphogenesis by raising extracellular pH. mBio. 2011; 2: e00055–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Linen Services Advisory group National Guidance for Sage Management of Linen in NHS Scotland. Health Protection Scotland, 2018. Available from: https://www.hps.scot.nhs.uk/web-resources-container/national-guidance-for-safe-management-of-linen-in-nhsscotland-health-and-care-environments-for-laundry-servicesdistribution/, Accessed July 15, 2019. [Google Scholar]
- 45. Kaloriti D, Tillmann A, Cook E et al.. Combinatorial stresses kill pathogenic Candida species. Med Mycol. 2012; 50: 699–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kaloriti D, Jacobsen M, Yin Z et al.. Mechanisms underlying the exquisite sensitivity of Candida albicans to combinatorial cationic and oxidative stress that enhances the potent fungicidal activity of phagocytes. MBio. 2014; 5: e01334–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Centers for Disease Control and Prevention Clinical Alert to U.S. Healthcare Facilities: Global Emergence of Invasive Infections Caused by the Multidrug-Resistant Yeast Candida Auris. 2016. Available from: http://www.cdc.gov/fungal/diseases/candidiasis/candida-auris-alert.html, Accessed February 24, 2017. [Google Scholar]
- 48. Infection Control Team Transmission Based Precautions Literature Review: Management of Care Equipment and Environmental Decontamination. Health Protection Scotland; 2017. Available from: http://www.nipcm.hps.scot.nhs.uk/documents/tbp-management-of-care-equipment-and-environmental-decontamination/, Accessed July 31, 2019. [Google Scholar]
- 49. Loveday HP, Wilson JA, Pratt RJ et al.. epic3: national evidence-based guidelines for preventing healthcare-associated infections in NHS hospitals in England. J Hosp Infect. 2014; 86: S1–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. McDonnell G, Russell AD. Antiseptics and disinfectants: activity, action, and resistance. Clin Microbiol Rev. 1999; 12: 147–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lowbury EJ, Lilly HA. Use of 4 per cent chlorhexidine detergent solution (Hibiscrub) and other methods of skin disinfection. Br Med J. 1973; 1: 510–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Jamieson DJ, Stephen DW, Terriere EC. Analysis of the adaptive oxidative stress response of Candida albicans. FEMS Microbiol Lett. 1996; 138: 83–88. [DOI] [PubMed] [Google Scholar]
- 53. Nikolaou E, Agrafioti I, Stumpf M, Quinn J, Stansfield I, Brown AJ. Phylogenetic diversity of stress signalling pathways in fungi. BMC Evol Biol. 2009; 9:44-2148-9-44, doi:10.1186/1471-2148-9-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Soike K, Miller D, Elliker P. Effect of pH of solution on germicidal activity of quaternary a mmonium compounds. J Dairy Sci. 1952; 35: 764–771. [Google Scholar]
- 55. Rutala WA, Kanamori H, Gergen M, Sickbert-Bennett E, Weber DJ. A preliminary study of germicidal efficacy against Candida auris. Open Forum Infect Dis. 2017; 4: S184. [Google Scholar]
- 56. Trofa D, Gacser A, Nosanchuk JD. Candida parapsilosis, an emerging fungal pathogen. Clin Microbiol Rev. 2008; 21: 606–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Abdolrasouli A, Armstrong-James D, Ryan L, Schelenz S. In vitro efficacy of disinfectants utilised for skin decolonisation and environmental decontamination during a hospital outbreak with Candida auris. Mycoses. 2017; 60: 11. [DOI] [PubMed] [Google Scholar]
- 58. Moore G, Schelenz S, Borman AM, Johnson EM, Brown CS. Yeasticidal activity of chemical disinfectants and antiseptics against Candida auris. J Hosp Infect. 2017; 97: 371–375. [DOI] [PubMed] [Google Scholar]
- 59. Welsh RM, Bentz ML, Shams A et al.. Survival, persistence, and isolation of the emerging multidrug-resistant pathogenic yeast Candida auris on a plastic health care surface. J Clin Microbiol. 2017; 55: 2996–3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Biswal M, Rudramurthy SM, Jain N et al.. Controlling a possible outbreak of Candida auris infection: lessons learnt from multiple interventions. J Hosp Infect. 2017; 97: 363–370. [DOI] [PubMed] [Google Scholar]
- 61. Ene IV, Adya AK, Wehmeier S et al.. Host carbon sources modulate cell wall architecture, drug resistance and virulence in a fungal pathogen. Cell Microbiol. 2012; 14: 1319–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ene IV, Walker LA, Schiavone M et al.. Cell wall remodeling enzymes modulate fungal cell wall elasticity and osmotic stress resistance. MBio. 2015; 6: e00986–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kastora SL, Herrero-de-Dios C, Avelar GM, Munro CA, Brown AJP. Sfp1 and Rtg3 reciprocally modulate carbon source-conditional stress adaptation in the pathogenic yeast Candida albicans. Mol Microbiol. 2017; 105: 620–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.