Abstract
Background:
Human type-2 innate lymphoid cells (ILC2s) are identified by coupled-detection of CRTH2 and IL7Rα on lineage-negative cells. Type-2 cytokine production by CRTH2-IL7Rα-ILCs is unknown.
Objective:
To identify CRTH2-IL7Rα- type-2 cytokine producing ILCs and their disease relevance.
Methods:
We studied human blood and lung ILCs from asthmatic and control subjects by flow cytometry, ELISA, RNA-seq, qPCR, adoptive transfer to mice and measurement of airway hyperreactivity by Flexivent.
Results:
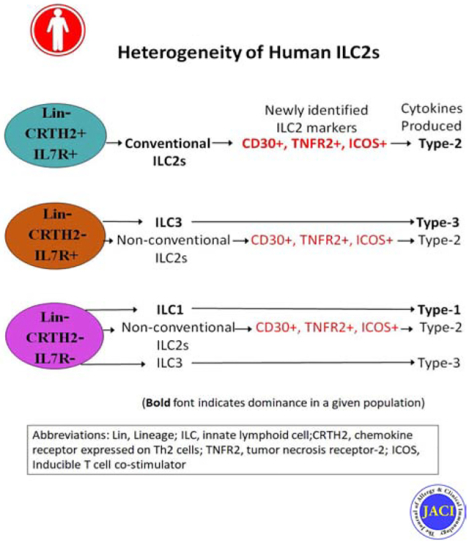
We found that IL5 and IL13 were expressed not only by CRTH2+ but also by CRTH2-IL7Rα+ and CRTH2-IL7Rα- (DN: double negative) human blood and lung cells. All three ILC populations expressed type-2 genes and induced airway hyperreactivity when adoptively transferred to mice. The frequency of type-2 cytokine+ IL7Rα and DN ILCs were similar to that of CRTH2 ILCs in the blood and lung. Their frequency was higher in asthmatic patients compared to disease controls. Transcriptomic analysis of CRTH2, IL7Rα and DN ILCs confirmed the expression of mRNA for type-2 transcription factors in all three populations. Unexpectedly, the mRNA for GATA3 and IL5 correlated better with mRNA for CD30, TNFR2, ICOS, CCR4 and CD200R1 than CRTH2. Using a combination of these surface markers, especially CD30/TNFR2, we identified a previously unrecognized ILC2 population.
Conclusions:
The commonly used surface markers for human ILC2s leave a majority of type-2 cytokine-producing ILC2s unaccounted for. We identified top GATA3-correlated cell-surface expressed genes in human ILCs by RNA-seq. These new surface markers such as CD30 and TNFR2 identified a previously unrecognized human ILC2 population. This ILC2 population is likely to contribute to asthma.
Keywords: Asthma, type 2 innate lymphoid cells, novel ILC2 population, cytokines
Graphical Abstract
Capsule summary
We identified top GATA3-correlated cell-surface expressed genes in human ILCs by RNA-seq. The new surface markers such as CD30 and TNFR2 identified a previously unrecognized human ILC2 population, which is likely to contribute to asthma.
Introduction
Innate lymphoid cells (ILCs) represent a distinct lymphoid lineage that diverges from T and B lymphocytes early in development under the milieu of a high level of the transcription factor ID2 (reviewed in ref1 and2). These cells are a potent source of a variety of cytokines belonging to the type-1, −2, and −3 immune responses. As such, they are classified into 3 types—ILC1, ILC2 and ILC3 to reflect their functional analogy to Th1, Th2 and Th17 cells. ILC2s have been traditionally identified by their expression of cell surface markers that are co-regulated with the expression GATA3 and type-2 cytokines. The most widely used human ILC2 markers are CRTH2/IL7Rα3, 4 and the less frequently used markers include CD1613 and ST25, 6. IL7Rα (CD127)+ ILCs give rise to all subtypes of ILCs. IL7Rα is expressed on mature ILCs4. For this reason it is a commonly used marker for detection of ILCs. CD161 (KLRB1) is expressed by NK cells, all T helper cell subtypes including Th2 cell, Tregs, MAIT cells, follicular dendritic cells and various subpopulations of ILCs, especially ILC17. CD161 is expressed by highly activated cells regardless of the cell type. CRTH2, a receptor for prostaglandin D2 (PTGDR2), is expressed by Th2 cells, type-2 CD8 T cells, eosinophils, basophils and a subset of monocytes8–10. CRTH2 is specific for type-2 polarized cells and its expression is regulated by GATA311. For these reasons CRTH2 is used as a type-2-specific marker for ILC2s. Indeed, numerous publications reported the frequency ILC2s in various diseases using a combination of CRTH2/IL7Rα as ILC2-specific surface markers12–14. CRTH2+ ILC2s express high levels of type-2 cytokines. However, it is unknown whether CRTH2- ILCs are capable of producing type-2 cytokines. Recently, Lin-IL7Rα- cells have been shown to give rise to various ILC populations including ILC2s15, 16. The foregoing studies raise concerns about accuracy of ILC2 frequency based solely upon their expression of CRTH2 and IL7Rα as this may underestimate the true frequency of ILC2s. The objective of this study was to assess the capacity of lin-CRTH2-IL7Rα- cells to express GATA3 and other ILC2 markers including type-2 cytokines. Through transcriptional profiling of CRTH2+, IL7Rα+ and CRTH2-IL7Rα- ILCs we identified cell surface markers that strongly correlated with type-2 signature genes. Using these new cell surface markers we detected a novel CRTH2-IL7Rα- but type-2 cytokine+ ILC2 subpopulation in asthmatic patients and disease control subjects.
Materials and Methods
Human subjects:
We studied blood and bronchoalveolar lavage (BAL) ILC2 from allergic asthmatic patients, disease controls and healthy donors. Asthmatic patients and disease controls were recruited during July, 2015 through December, 2019 from the outpatient clinics at National Jewish Health. Healthy donors were recruited from the blood bank of National Jewish Health. Bronchoscopy and BAL were performed as a part of their clinical work-up for poorly controlled asthma as defined by EPR3 guidelines. We recruited all patients who were eligible, i.e. 1) Met criteria for poorly controlled asthma despite guideline-based therapy; and 2) Qualified for clinical bronchoscopy. None of the disease control patients met the American Thoracic Society diagnostic criteria for asthma. Subjects in the disease control group were selected to match to those in the poorly controlled asthma group regarding age and sex. Demographic and clinical characteristics of asthma and disease control subjects are shown in Supplemental Table-1. Asthma patients, who were recruited for blood studies met the criteria for asthma as per ATS guidelines (presence of reversible airway obstruction and/or a positive methacholine test). The clinical severity of their asthma varied from mild to severe, and well-controlled to poorly-controlled. Asthmatic patients and disease control subjects maintained their controller medications at the time of bronchoscopy and blood draw. The protocols for blood and BAL studies of lymphoid cells from asthmatic patients and disease controls were approved by the institutional IRB. Written informed consent was obtained from each participant. The data using all available samples are shown in each experiment in this manuscript.
Isolation of PBMCs, lin-cells, culture and flow cytometry for ILC2s:
PBMCs were isolated from EDTA-treated blood by density centrifugation with Ficoll-Hypaque. PBMCs were cultured in RPMI 1640 plus 10% FBS in the medium alone or with the cytokine combo (as indicated in the text) as described previously51. The following cytokines were used at 20 ng/ml concentration: IL2, IL7, IL25, and IL33 (all from Peprotech, Rocky Hills, NJ). In select experiments lin- cells were isolated from PBMCs by negative selection using antibody-coated magnetic beads (lineage cell depletion kit) from Miltenyi, Inc. San Diego, CA. PBMCs and lin- cells were cultured for 5 days unless otherwise stated in the text. Monensin (2 μM, Biolegend, Inc., San Diego, CA) was added to all cultures 4 hr before conclusion. Following culture cells were pelleted and then stained for flow cytometry as described previously14, 51.
Processing and culture of BAL cells for ILC2 studies:
BAL cells were pelleted, washed and incubated with Monensin (2 μM) in RPMI 1640 plus 10% FBS for 4 hr and then stained for flow cytometry as described previously51, 52.
Flow cytometric staining of cells:
Staining for flow cytometry was carried out as described previously14, 51. A list of all fluorophore-labeled antibodies, isotype control antibodies and other relevant reagents is given Supplemental Table-2. We added a labeled FcεRI antibody to the human lineage cocktail to eliminate basophils. We first stained cells with the eBioscience fixable viability dye and then stained surface markers on live cells. We fixed cells using 4% paraformaldehyde, permeabilized them with 0.1% saponin and performed staining for intracellular molecules (cytokines, signaling molecules and transcription factors). Flow cytometry was performed on LSRII (BD Biosciences). The data was analyzed by the FlowJo software. Isotype antibody controls and FMO (fluorescence minus one) were used to develop the gating and data analysis strategies.
Isolation of ILCs from the human lung:
Deidentified human lungs were obtained from the Donor Alliance of Colorado and International Institute for the Advancement of Medicine (Edison, NJ). A fragment of the lung tissue was finely minced and then treated with 0.5 mg/ml of collagenase (Worthington # LS004197) in presence of 1% penicillin/streptomycin for 30 min as described previously53. Cell suspensions were agitated at room temperature for 10 minutes in RPMI 1640 with 100U/mL DNAse I and 10% FBS prior to filtration through a 40μm filter and red blood cell lysis. The cells were then washed 3x with PBS. The pelleted single cells were subjected to a density centrifugation on a Ficoll-Hypaque (relative density 1.077) gradient. Live mononuclear cells were collected from above the gradient, washed 3x and cultured overnight in RPMI1640 containing 10% FBS, IL2 (100 units/ml) and 1% penicillin/streptomycin. Cells were stained for FACS for the following surface markers: CD45, Lin, CD25, CRTH2, IL7Rα and the fixable viability dye e-Fluor 780 (e-Bioscience). Single cells were gated for e-Fluor 780-CD45+lin-CD25+cells and then sorted for CRTH2+, IL7Rα+ and DN cells. The sorted cells were cultured for 5 days with IL2 (100 units/ml). The culture supernatant was collected for measurement of IL5. The pelleted cells were used in adoptive transfer experiments.
Adoptive transfer of ILCs to immunodeficient mice and measurement of airway hyperreactivity:
FACS-sorted CRTH2+, IL7Rα+ and DN cells from the human lung were cultured overnight with IL2 (100 units/ml) before adoptive transfer. A total of 105 ILCs (CRTH2+, IL7Rα+ and DN) cells in 1 ml volume per mouse was intravenously injected into Rag2−/−:γc−/− mice. As a control for cell transfer we sorted lung CD45+CD3+CD4+ T cells, cultured them with IL2 as above and transferred to Rag2−/−:γc−/− mice at 106 cells/mouse. The mice were rested for 1 day and then intranasally challenged with the Alternaria allergen (10 μg/mouse) for 3 consecutive days. Airway hyperreactivity in response to inhaled methacholine was measured by Flexivent 1 day later as described previously54. The protocol for mouse studies was approved by the institutional IACUC.
Airway hyperreactivity measurement:
Measurement methodologies have been explained in depth elsewhere54. Briefly, mice were anesthetized with ketamine (180 mg/kg), xylazine (9 mg/kg), and acepromazine (4 mg/kg). After loss of foot-pad pinch reflex, a tracheotomy was performed and the mouse was attached via an 18 gauge cannula to a small-animal ventilator with a computer-controlled piston (Flexivent; Scireq). Mice were ventilated at a frequency of 90 breaths/ min with a tidal volume of 20 mL/kg during nebulization and otherwise with a frequency of 150 breaths/ min with a tidal volume of 20 mL/kg while breathing against an artificial positive end-expiratory pressure of 2.5 to 3 cm H2O. Lungs were inflated to total lung capacity twice to standardize volume history. Resistance measurements were then taken to establish the baseline for the total lung resistance and at each methacholine dose. Group averages were expressed as fold increase over the baseline resistance (mean ± SEM).
ELISA for cytokines:
IL5 was measured in the culture supernatant from CRTH2+, IL7Rα+ and DN cells following their FACS-sorting of CD45+lin-CD25+ cells isolated from the human lung. The IL5 ELISA kit was from Thermo Fisher Scientific, Inc., Waltham, MA (detection threshold 4 pg/ml) ELISA was performed as per manufacturer’s instruction as described previously51, 55.
RNA-seq:
PBMCs were isolated from 4 healthy donors by Ficoll-Hypaque density centrifugation. Lin- cells were separated by negative selection using antibody-coated magnetic beads (Lineage Cell Depletion Kit, Miltenyi, Inc). Lin- cells were then surface stained and FACS-sorted for CRTH2+IL7Rα+ (CRTH2), CRTH2-IL7Rα+ (IL7Rα) and DN (CRTH2-IL7Rα-) cells. RNA was isolated from the sorted cells (105 cell per ILC population) using RNeasy Mini Kit (Qiagen, Inc. San Diego, CA) and used for RNA-seq analyses in the NJH Genomic Facility. The isolated total RNA was processed for next-generation sequencing (NGS) library construction as developed in the NJH Genomics Facility for analysis with a Life Technologies (Carlsbad, CA, USA) Ion Proton NGS platform. A modified Kapa Biosystems (Wilmington, MA, USA) KAPA Stranded mRNA-Seq kit for whole transcriptome libraries was used to primarily target all polyA RNA. Briefly, library construction started from isolation of total RNA species, followed by mRNA (poly-A) isolation, 1st and 2nd strand cDNA synthesis, adaptor ligation, amplification, and bead templating. Once validated, the libraries were sequenced as barcoded-pooled samples on a P1 Ion Proton chip, as routinely performed by the NJH Genomics Facility.
Basecalling, barcode demultiplexing and adapter trimming was performed by the Ion Torrent Suite 5.0.4 software. Sequence reads of at least 30 nt length were mapped to the hg19 release of the human genome using STAR (version 2.5.1b) with splice site data from the Ensembl annotation release 75.
Reads mapping unambiguously to each gene in the Ensembl annotation were quantified using the featureCounts program (version 1.5.0-p1) from the subread package (Liao & al.: https://doi.org/10.1093/bioinformatics/btt656). Pairwise statistical comparisons of the gene expression between the cell genotypes were performed with Wald test of the DESeq2 package version 1.8.1 (Loeve & al.: https://doi.org/10.1186/s13059-014-0550-8) for the R statistical software version 3.2.0 (https://www.R-project.org/) while accounting for any donor-specific effects. Resulting p-values were adjusted for multiple testing using the method by Benjamini and Hochberg (1995 [: https://www.jstor.org/stable/2346101]). For plotting, gene expression values were normalized to transcripts per million (TPM). Principal component analysis (PCA) was calculated using the prcomp function in R version 3.5.1 based on unscaled TPM.
Statistical Analyses:
Comparison between the study groups was done by Mann-Whitney U test unless otherwise stated. Comparison among multiple study groups was performed by Kruskal Wallis test. For correlations based on RNA-Seq results, gene expression values in TPM from all samples were used. The CORREL function in Microsoft Excel was used to calculate Pearson correlation coefficients. Statistical analyses were performed and graphs were generated using the GraphPad Prism 6 software (San Diego, CA).
Results
Type-2 cytokines are expressed by CRTH2- and IL7Rα- ILCs:
CRTH2 and IL7Rα are commonly used to detect human ILC2s in the lineage- (the lin antibody cocktail contained antibodies against CD3, CD14, CD16, CD19, CD20, CD56 and FcεRI) cell population of blood or other tissues. We asked whether CRTH2 and IL7Rα captured all type-2 cytokine+ ILCs. To this goal, we examined type-2 cytokine expression by peripheral blood CRTH2+IL7Rα−, CRTH2+IL7Rα+, CRTH2-IL7Rα+ and CRTH2-IL7Rα- ILC populations (Fig. 1A–J) cultured with IL2/IL25. The gating strategy is shown in Figure 1 and is based upon isotype (CRTH2 isotype control antibody: rat IgG2a, IL7Ra isotype control: mouse IgG1; IL5 isotype control: rat IgG1; Fig. S1A) and FMO (fluorescence minus one) controls (Fig. S1B). We observed that the CRTH2+IL7Rα− and CRTH2+IL7Rα+ populations (referred to as CRTH2+ cells) of Lin- cells had the highest frequency of IL5+ cells (Fig. 1G and H). The CRTH2+ population contained a total of 15.7% of all IL5+lin- cells. However, CRTH2-IL7Rα+ (referred to as IL7Rα+, Fig. 1I) and CRTH2-IL7Rα- (referred to as double negative: DN, Fig. 1J) populations of Lin- cells also contained a significant, albeit a lower frequency of IL5+ cells. The IL7Rα+ and the DN populations contained 4.2% and 16.1% of all IL5+lin- cells, respectively. As the DN cells outnumbered the CRTH2+ cells, the absolute number of IL5+ DN cells was higher than that of IL5+ CRTH2+ cells. Data presented in this figure comes from a single asthmatic donor. Cumulative data from18 asthmatic patients and 12 disease controls is shown in a latter figure (Fig. 4A). We checked the contamination of lineage- cells (identified through our gating strategy) with CD4 and CD5 T cells. The lineage- cells contained 0.23% and 0.51% of CD4 and CD5 T cells (Fig. S1C). We examined the frequency of IL5+ cells in the lineage+ (containing CD4 T cells) population. The frequency was 5%. The results suggest that a negligible ~0.001% cells (0.23 × 0.005) within the IL5+Lin- population were CD4 T cells.
Figure 1.
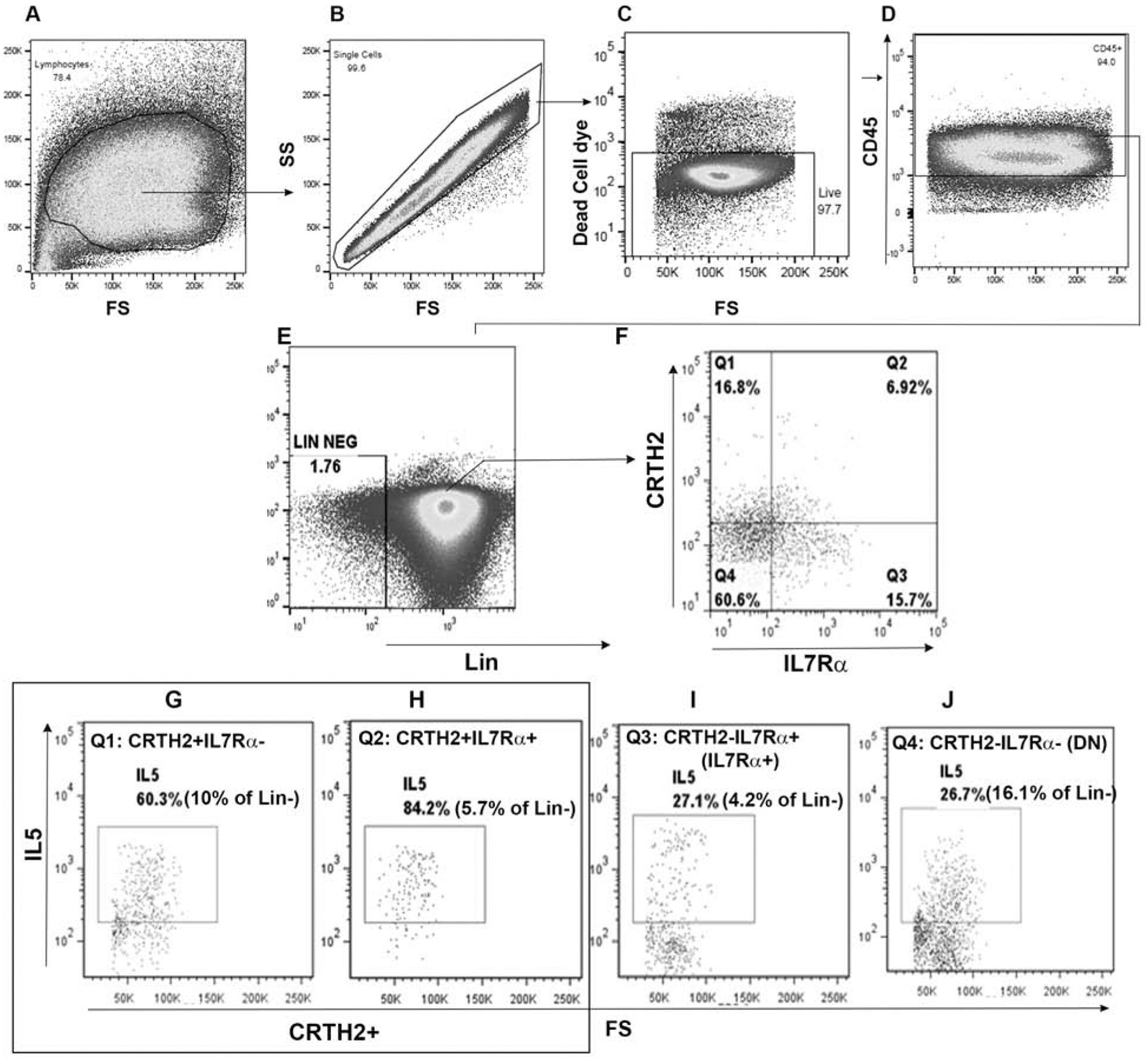
Expression of IL5 by ILC2 subpopulations. PBMCs from an allergic asthmatic subject were cultured with IL2/IL25 for 5 days and analyzed for IL5+ cells by flow cytometry. Mononuclear cells (A) were sequentially gated for single cells (B), CD45+ cells (C) and lineage (lin) – cells (D). Gated lin- cells were analyzed for expression of CRTH2 and IL7Rα (E). Each quadrant (Q1: CRTH2+IL7Rα−[F]; Q2: CRTH2+IL7Rα+ [G]; Q3: CRTH2-IL7Rα+ [H]; and Q4: CRTH2-IL7Rα- ([I]) was then analyzed for expression of IL5. The threshold for positive staining was determined by isotype antibody staining and FMO (fluorescence minus one) (Fig. S1).
Figure 4.
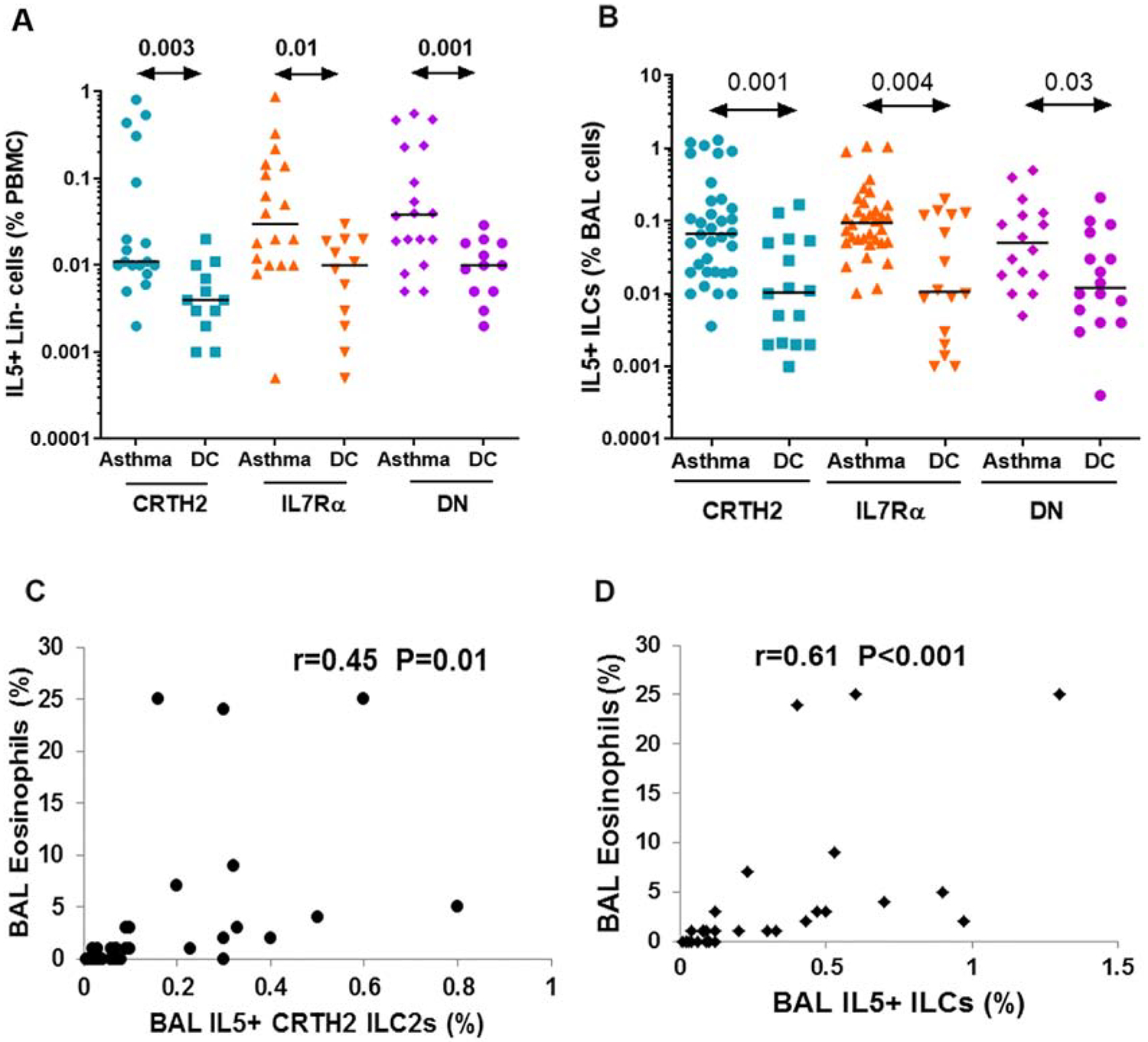
Comparison of blood and bronchoalveolar lavage (BAL) ILC2 populations in asthmatic patients and disease controls (DC). A: PBMCs from 18 asthmatic patients and 12 disease controls were cultured in the medium alone for 5 days and then analyzed for IL5+ ILC2 populations. The frequency of IL5+lin- cells is presented as % PBMCs. ILC B: BAL cells from 34 asthmatic patients and 16 disease controls were stained ex vivo for IL5+ (CD45+Lin-) ILC2 populations by flow cytometry. The frequency of IL5+ ILCs is presented as % BAL cells. Statistical significance (Mann-Whitney U test) is shown at the top of the dot plots. C& D: Correlation (r) between BAL eosinophils and BAL IL5+ CRTH2+ ILC2s (C) and BAL total IL5+ ILC2s (CRTH2+, IL7Rα+ and DN).
We observed incomplete separation of CRTH2+IL7Ra+ cells in Figure 1. To test if this was related to a poorly performing antibody, we performed additional experiments and stained cells with anti-IL7Ra antibodies from 3 different vendors (BD, Biolegend and R&D). With cells cultured with medium alone (Fig. S2), only the Biolegend antibody showed a small double positive (CRTH2+IL7Ra+) cell population (1.47% vs. 0.59 and 0.1%). In IL2/IL33-stimulated cultures a small cluster of cells was separated and visible with all three antibodies. The BD antibody, which was conjugated with PE (a stronger fluorophore than A488), showed the best separation. The clusters were smaller with the Biolegend and R&D antibodies. However, if one counts all CRTH2+ cells (CRTH2+IL7Ra− and CRTH2+IL7Ra+), the total number of cells was similar with BD and Biologend antibodies. The total number of CRTH2+ cells was 5.9+6.5=12.4% with the BD antibody and 10.3+1.8= 12.1% with the Biolegend antibody. The R&D antibody performed poorly. We do find a high degree of variability and heterogeneity of these measurements in the population. Although the Biolegend antibody detected a smaller and not fully separated cluster of CRTH2+IL7Ra+ cells in donor 1, it did detect a distinct and well separated cluster of CRTH2+IL7Ra+ cells in three other donors (lower panel).
Type-2 cytokine expression by IL7Rα+ and DN cells occurred not only under stimulated but also under non-stimulated conditions (Fig. S3). We cultured PBMCs with various combinations of cytokines—IL2/IL33, IL2/7, IL2/TSLP, IL2/IL7/IL33 and in addition to IL2/IL25. All these cytokine combinations induced a similar profile of type-2 cytokine expression by the ILC populations and there was no significant difference among the culture conditions (not shown). IL5 expression by the three ILC population following stimulation with IL2/7/33 is shown in Fig. S3. We asked if the expression of type-2 cytokines by non-CRTH2 ILCs was due to the mixed cell culture milieu of the PBMCs. To address this question, we isolated lineage- cells, cultured them with and without IL2/IL25 and compared the results with that from the PBMC culture. IL13+ ILC2s were present both in CRTH2+ and CRTH2- ILC subpopulations (Fig. S5). The isotype antibody control for IL13 was rat IgG1 (Fig. S1A) and the FMO for IL13 is shown in Fig. S1B. We studied airway ILC2s obtained through bronchoalveolar lavage (BAL) from asthmatic patients. BAL ILC2s were studied ex vivo without cytokine stimulation. Fig. S4 shows representative flow cytographs of IL5 expression by BAL CRTH2+, IL7Rα+ and DN cells from an asthmatic patient. The isotype and FMO controls were the same as those described for Fig. 1. We observed a similar distribution of IL5+ cells in BAL ILCs. Cumulative data of IL5 expression by BAL CRTH2+, IL7Rα+ and DN cells from 34 asthmatic patients and 16 disease controls is shown in a latter figure (Fig. 4B).
We next asked if the DN ILC population contained precursors for CRTH2+ and IL7Rα+ ILC2s, which could explain the capacity to express type-2 cytokines. ILCs manifest a high degree of plasticity17–19. The ILC transition from one type to another may require a transient state, when they do not express typical surface markers. To address this question, we sorted DN cells (lin-CRTH2-IL7Rα-cells) (Fig. 2A), IL7Rα+ and CRTH2+ cells (the isotype and FMO controls are shown in Fig. S1A–B). The purity of DN cells is shown in Fig. S1D. We cultured them with IL2/IL25 for 7 days. We cultured the cells for a longer period of time in order to allow differentiation. The cultured cells were then examined for expression of CRTH2, IL7Rα and IL13 (Fig. 2B). Of the DN cells 18% converted to CRTH2+ and 37% to IL7Rα+.
Figure 2.
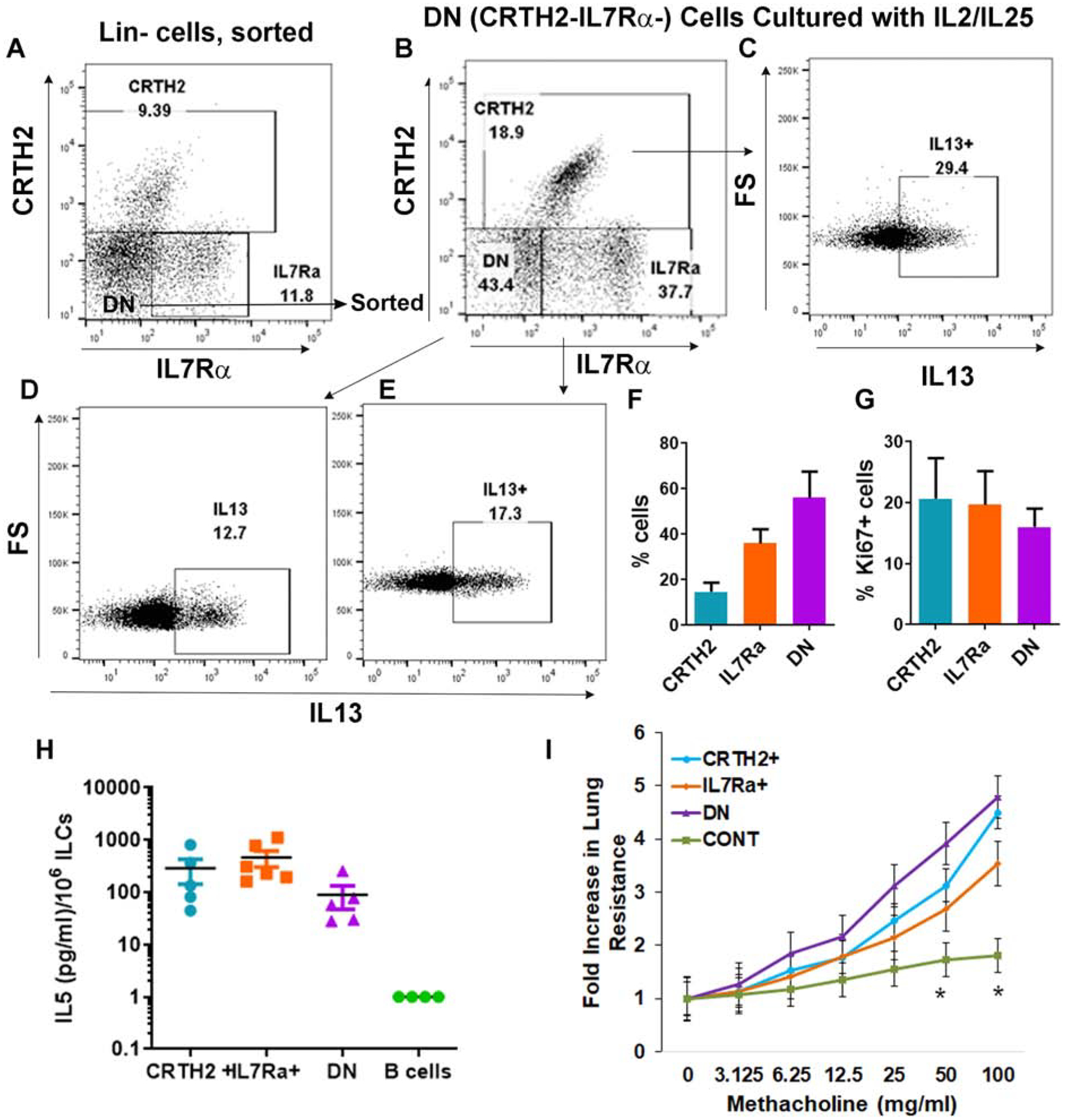
Differentiation, proliferation, cytokine secretion and in vivo activity of DN ILCs. A-E: FACS-sorted DN (lin-CRTH2-IL7Rα-) cells from PBMCs obtained from an asthmatic patient (A) were cultured with IL2/IL25 for 7 days and their expression of CRTH2 and IL7Rα was analyzed (B). The newly emerged CRTH2+ (C), IL7Rα+ (E) and the remaining DN cells (D) were gated for expression of IL13. F: Results of experiments from 3 different asthmatic donors. G: FACS-sorted CRTH2+, IL7Rα+ and DN cells were cultured with IL2/IL25 for 7 days and then analyzed for expression ki67 by flow cytometry (N=3). (H) CD45+Lin- CRTH2+, IL7Ra+ and DN ILCs from the human lung were cultured with IL2/IL25 for 5 days and then supernatant was assayed for IL5 by ELISA. Culture supernatant from IL2/IL25-treated B cells (negatively selected from PBMCs) was used a control. Each symbol represents a single lung donor. (I) Human lung-derived ILC2 subpopulations were cultured with IL2 overnight and adoptively transferred (105 cells/mouse) to Rag2−/−:γc−/− mice. Lung-derived CD45+CD3+CD4 T cells were injected (106 cells/mouse) as a control. One-day latter the mice were challenged with the Alternaria allergen for 3 consecutive days before methacholine challenge. The increase in lung resistance was statistically significant (*=P<0.05, t test) for all ILC2 populations as compared to the control (CONT).
Approximately 29% and 17% of the “new” CRTH2+ and IL7Rα+ cells expressed IL13, respectively (Fig. 2C–F). About 43% of the cells (cumulatively, slightly over 50%, Fig. 2F) remained DN, and 12% of these DN cells expressed IL13. The culture of IL7Rα+ cells showed that the majority (87%) retained their surface expression (Fig. S6A and C). About 3.6% became CRTH2+ and 3.5% became DN. All three populations expressed IL13. Unlike IL7Rα+ cells, about 87% of CRTH2+ cells lost their surface expression during the 7-day culture (Fig. S6B and D). Nonetheless, both the CRTH2− and CRTH2+ cells expressed IL13. These findings in aggregate suggest that the DN ILC population contains a previously unidentified ILC2 population that does not express CRTH2 or IL7Rα. In addition, it contains ILCs that are capable of expressing (or re-expressing) CRTH2 and IL7Rα upon culture with ILC2 growth factors. Finally, the CRTH2 population and to a lower extent, the IL7Rα population manifest dynamic changes in their surface marker expression over time. We compared the proliferation of the ILC populations in IL2/IL25-stimulated cultures by measuring Ki67+ cells. All three subpopulations showed similar levels of Ki67 staining (Fig. 2G) suggesting an equivalent proliferative response to growth factors.
IL7Rα+ and DN ILCs produce and secrete IL5:
We examined secretion of IL5 by CRTH2+, IL7Rα+ and DN ILCs. To this goal we FACS-sorted lin-CRTH2+IL7Rα+ (CRTH2+), lin-IL7Rα+ and lin-CRTH2-IL7Rα- subpopulations (Fig. S7) from the human lung following collagenase digestion and isolation of mononuclear cells. Note that the gating strategy for the lung ILC populations was slightly different from the one that was used for human blood. This is because 1) we used the cell sorter BD FACSAria Fusion to sort cells; 2) human lung cells showed higher intensity of staining for CRTH2 and IL7Rα. The combination of these two factors resulted in better separation of lung ILC populations than that was observed with blood ILC populations using the cell analyzer LSRFortessa. The sorted cells were cultured with IL2/IL25 for 3 days and the culture supernatant was assayed for IL5 by ELISA. IL5 was detected in the culture supernatant from all three ILC subpopulations but not from a B cell population, which was used as a control (Fig. 2H). IL7Rα+ and CRTH2+ ILCs produced higher levels of IL5 as compared to DN ILCs although the difference did not reach statistical significance likely due to the donor-to-donor variation and low sample size.
ILC2 subpopulations induce airway hyperreactivity following adoptive transfer to Rag2−/−:γc−/− mice:
To test their biological relevance we FACS-sorted lin-IL7Rα+ and lin-DN ILCs along with lin-CRTH2+ ILCs and CD3+CD4+ T cells from the human lung as described above. We transferred the sorted ILC populations adoptively to Rag2−/−:γc−/− mice. The number of CRTH2+, IL7Rα+, DN and CD4 T cells that were transferred to each mouse were 105, 105, 105 and 106 cells, respectively. One day after the adoptive transfer we administered the allergen extract of Alternaria intranasally on 3 consecutive days. We measured airway hyperreactivity to methacholine one day later. All three ILC subpopulations, and expectedly, not control (CONT) CD4 T cells induced a significant level of airway resistance following the adoptive transfer (Fig. 2I). The results suggest that both IL7Rα+ and DN ILCs are equally effective in inducing an asthma-type airway response in mice. The limitation of this experiment is that it utilizes an ectopic (human cells transferred to mice) and non-physiologic approach.
Characterization of ILC2 subpopulations:
We asked if DN cells expressed typical markers of ILC2. All three ILC populations from the blood expressed GATA3, PLZF, and Bcl11b (Fig. 3), three key ILC2 transcription factors. Expectedly, the CRTH2+ population expressed the highest level of these transcription factors. ICOS and CD161 expression was highest in CRTH2+ cells (Fig. S8). IL25R and KLRG1 were expressed mostly on CRTH2+ and IL7Rα+ cells and only minimally on DN cells. The expression of KLRG1 was modest in our study subjects. Low levels of RORγT are known to be expressed by ILC2s20. Accordingly, all three subpopulations expressed low levels of RORγT and there was no difference in their expression level among the subpopulations. TSLPR was primarily expressed by CRTH2+ cells.
Figure 3.
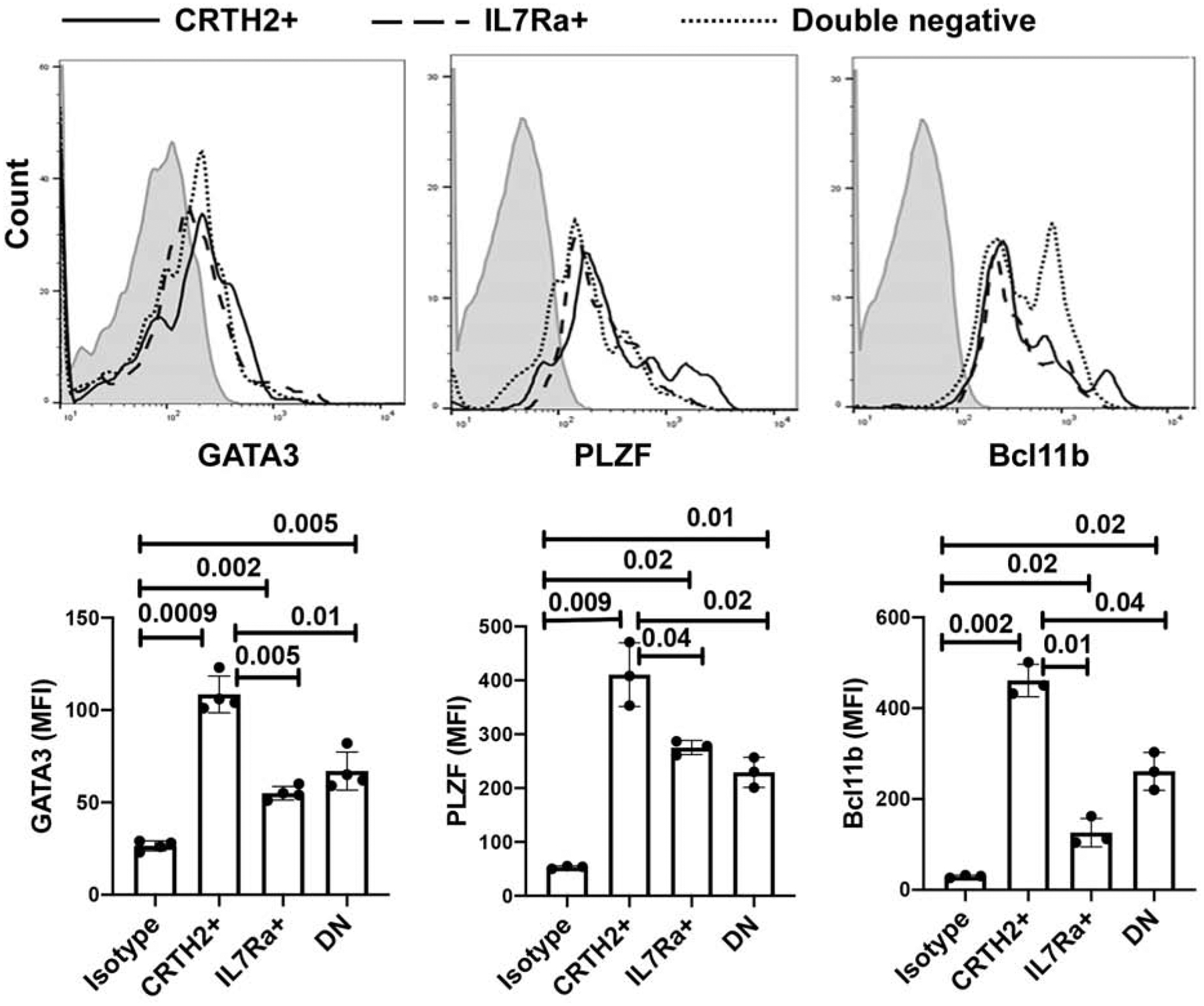
Expression of ILC2-related markers in ILC2 subpopulations. PBMCs from 3–4 donors were cultured with IL2/IL25 for 3 days and then analyzed for expression of GATA3, PLZF, and Bcl11b, in lin- CRTH2+, IL7Rα+ and DN ILCs by flow cytometry. Histograms of mean fluorescence intensity are shown in the upper panel and bar graphs with statistical analysis are shown in the lower panel. Each symbol represents a donor.
Frequency of ILC2 subpopulations in asthma:
To establish the relevance of type-2 cytokine-producing ILC2 subpopulations we studied their frequency in bronchoalveolar lavage (BAL) from 34 asthmatic patients and 16 disease controls. The latter included patients with other allergic and pulmonary conditions without asthma, who required diagnostic bronchoscopy and BAL. These patients had one of the following diseases—chronic cough, bronchiectasis, chronic aspiration, tracheobronchomalacia, COPD, and pulmonary infiltrates of unknown etiology. The patient characteristics and their medications are shown in Supplemental Table-1. All BAL ILCs were studied ex vivo. We also studied type-2 cytokine-producing ILC2s in the blood from 18 of these 34 asthma patients and 12 of the disease controls. Blood ILCs were studied following isolation and culture of PBMCs with medium alone or with cytokines for 5 days. This is based upon our initial optimization experiments where we cultured cells for 3, 5 and 7 days and then studied expression of cytokine+ cells. The culture of PBMC for 5 days produced the highest level of cytokine+ ILCs (data not shown). Since BAL ILCs were studied ex vivo (Figure 4B), we show ILC data from non-stimulated PBMCs that were cultured in the medium alone (Fig. 4A). We recognize that the BAL and PBMC data are not comparable because of the difference in the culture condition. Our primary goal is to show the difference in the frequency of cytokine+ ILCs between asthma and disease controls in these two tissue compartments (blood and BAL). We observed an increased frequency of IL5+CRTH2+, IL5+IL7Rα+ and IL5+DN cells in the blood and BAL from asthma patients as compared to disease controls (Fig. 4A and B). The median frequencies of IL5+ CRTH2+, IL5+IL7Rα+ and IL5+DN cells in the PBMCs were 0.011%, 0.030% and 0.038% in asthmatic patients and 0.004%, 0.010% and 0.010% in disease controls, respectively. The frequency of IL5+ ILC populations was significantly higher in asthma than disease controls in the blood. The median frequency of IL5+IL7Rα+ and IL5+DN cells was slightly higher than that of IL5+CRTH2+ cells in the blood although the difference did not reach statistical significance (P=0.2 and 0.1, respectively). The median frequencies of IL5+ CRTH2+, IL5+IL7Rα+ and IL5+DN cells in the BAL were 0.06%, 0.09% and 0.05% in asthmatic patients and 0.01%, 0.01% and 0.01% in disease controls, respectively. The frequency of IL5+ ILC populations in BAL was largely similar within the study group but was significantly higher in the asthma as compared to the disease control group. The foregoing results suggest that the measurement of IL5+ cells only in the CRTH2 population will miss approximately two-thirds of all IL5+ ILCs. In order to establish the clinical relevance, we studied correlation of BAL IL5+CRTH2+ ILC2s with the BAL eosinophil count and compared it with that obtained using the total number of BAL IL5+ILCs (CRTH2+, IL7Rα+ and DN). BAL IL5+CRTH2+ ILC2s showed a low level correlation (r=0.45) with BAL eosinophils (Figure 4C). This correlation became stronger and reached a coefficient (r) of 0.61 when the total number of IL5+ILCs (CRTH2+, IL7Rα+ and DN) were correlated with eosinophils (Figure 4D). The results demonstrate that the measurement of type-2 cytokine+ CRTH2 cells as effector ILC2s underestimates the true frequency of human type-2 cytokine+ ILCs, and undervalues their contribution to asthma.
Since CRTH2 is commonly used as a marker of human ILC2s, we examined the frequency of CRTH2+ ILCs in the blood from 30 asthmatic patients and 17 disease controls. The frequency of CRTH2+ lin- ILCs was assessed in the blood under 3 different conditions: 1) Ex vivo, 2) Culture in medium alone for 5 days; and 3) Culture in the presence of cytokines (IL2/IL25) for 5 days. We also studied the frequency of CRTH2+ ILCs in BAL from 33 asthmatic patients and 24 disease controls. The frequency of CRTH2+ ILCs (lin-CD45+ cells) in BAL was assessed ex vivo. The frequency of CRTH2+ ILCs in PBMCs was low ex vivo (Fig. S8A). The median frequencies of CRTH2+ ILCs were 0.05% and 0.01% of PBMCs in asthma and disease controls, respectively. These frequencies increased to 0.18% and 0.10%, respectively, when PBMCs were cultured in medium alone for 5 days. They further increased to 0.48% and 0.19% of PBMCs when cultured with IL2/IL33 for 5 days. The increases after cytokine stimulation were statistically (P<0.05) significant. The median frequencies of CRTH2+ ILCs ex vivo in BAL were 0.50% and 0.29% in asthma and disease controls, respectively (Fig. S9B). These frequencies were 10 to 29-fold higher than those in the blood and were similar to those observed upon stimulation of blood ILCs in vitro. The results suggest that the airway milieu favors type-2 ILC differentiation or expansion of differentiated ILC2s.
Transcriptomic profiling by RNA-seq confirms heterogeneity of human ILCs:
To determine if other cell surface markers would correlate better with type-2 cytokines and thus, provide a superior means to capture type-2 cytokine+ ILCs, we analyzed the transcriptomic profile of CRTH2, IL7Rα and DN ILCs. We FACS-sorted lin- CRTH2+, IL7Rα+ and DN cells from the PBMC obtained from 4 healthy subjects, isolated RNA and then performed RNA-seq at the NJH Genomics Core. A representative sorting strategy is shown in Fig. S10. We first analyzed the expression of mRNA for CRTH2 (PTGDR2) and IL7Rα to determine the purity of the sorted cells. The mRNA for CRTH2 was expectedly present in the CRTH2 population but not in other two populations (Fig. 5A). This result verified that the IL7Rα and DN populations did not contain CRTH2+ cells. The mRNA for IL7Rα was highest in the IL7Rα population. The CRTH2 population consists of CRTH2+IL7Rα− and CRTH2+IL7Rα+ cells. Expectedly, the CRTH2 population expressed a lower level of mRNA for IL7Rα. The lowest level of mRNA for IL7Rα was present in the DN population. This is also expected as the distribution of cells among these two populations (IL7Rα+ and IL7Rα-) is a continuum (Fig. S10). Next, we analyzed the RNA-seq data for the presence of core ILC genes (CXCR6, TMEM176B, TMEM176A and TRGV3) as identified in a recent paper21. All three ILC populations expressed the core ILC genes (Fig. 5A). They did not express the NK cell-associated genes CD56 and NCR1. Principal component analysis (PCA) showed that the CRTH2 populations clustered together tightly as did the IL7Rα populations, which were very close to the CRTH2 cluster (Fig. S11). Three of four DN samples were either embedded or very close to the CRTH2 and IL7Rα clusters. The results suggest that despite the heterogeneity of surface marker expression, ILC gene expression profiles are quite similar.
Figure 5.
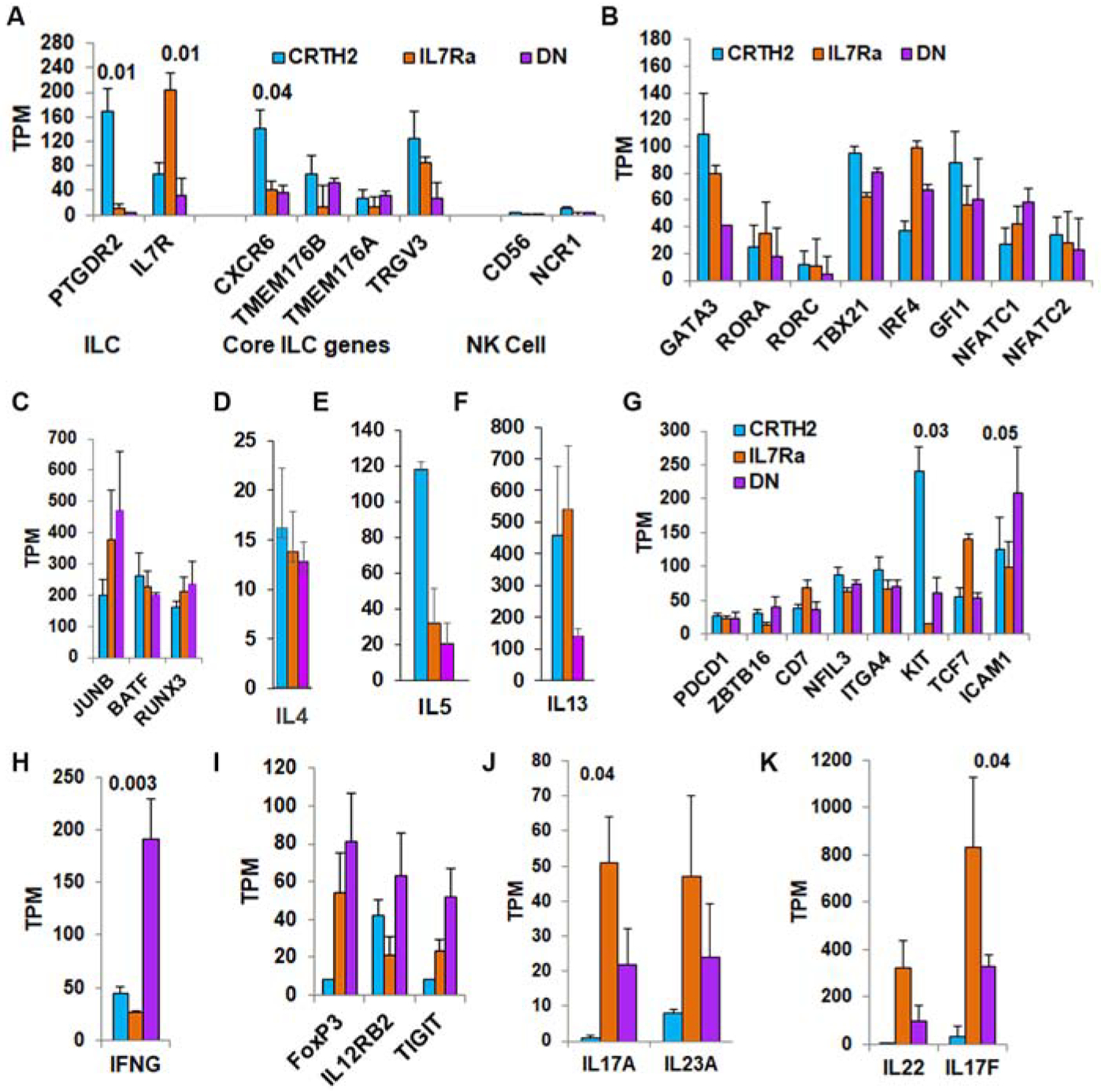
Expression profile of ILC-related genes in ILC2 subpopulations by RNA-seq. FACS-sorted CRTH2+IL7Rα+ (CRTH2), CRTH2-IL7Rα+ (IL7Ra) and DN lin- cells from the peripheral blood of 4 healthy donors were used to isolate RNA and perform RNA-seq. A: Expression of genes used for sorting of ILCs (PTGDR2, IL7R), and core ILC- and NK cell-related genes (TPM: transcripts per million). B and C: Expression of genes related to ILC1 (TBX21), ILC2 (, GATA3, RORA, IRF4, GFI1, NFATC1, NFATC2, JUNB, BATF), and ILC3 (RORC and RUNX3). D-F: Expression of type-2 cytokine (IL4, IL5, and IL13) genes. G. Expression of ILC2 precursor-related genes. H and I: Top immune response genes expressed by DN cells. J and K: Top cytokine genes expressed by IL7Rα+ cells. Gene expression values are normalized to transcripts per million (TPM). Statistical significance, when present, is shown on the top of the bars.
The expression of mRNA for GATA3 was highest in CRTH2 and lowest in DN cells (Fig. 5B), which is in agreement with the flow data. The expression of mRNA for RORA was similar in all three populations. The expression of RORC was low. The mRNA for TBX21 was present at a similar level in all three populations. To validate this finding we examined IFNγ expression in CRTH2+ ILCs. Blood ILCs from most asthma patients did not express IFNγ but expressed IL5 (Fig. S12A–C). However, in some asthma subjects all ILC populations including the CRTH2 population (Fig. S12D and E) expressed IFNγ in addition to IL5. The foregoing cytokines co-localized in CRTH2+ cells (Fig. S11F). The frequency of dual IFNγ+ IL5+ CRTH2+ ILCs is quite low and represented about 11% (N=18).
Next we examined genes known to regulate ILC2s and type-2 cytokines (Fig. 5B and C). These genes included IRF4, GFI1, NFATC1, NFATC2, JUNB, and BATF. GFI1, NFATC1 & 2, and BATF were expressed at a similar level by all three ILC populations. IRF4 tended to be higher in CRTH2- cells and JUNB tended to be higher in DN cells but the differences did not reach significance due to donor variability. Similar expression of these genes in addition to the expression of GATA3 provides a mechanistic basis for expression of type-2 cytokines by IL7Rα and DN populations. RUNX3, which controls the expression of RORC22 was present at a higher level in all three populations. All three subpopulations expressed mRNA for IL5 and IL13 and negligibly, for IL4 (Fig. 5D–F). The expression of mRNA for IL5 and IL13 was lowest in the DN population.
We asked if mRNA for ILC precursors was selectively increased in the IL7Rα and especially, in the DN population. Previous studies identified a series of genes that marked precursor ILCs in humans and mice23–29. We examined the expression of top 8 ILC precursor genes—PDCD1, ZBTB16, CD7, NFIL3, ITGA4, KIT, TCF7 and ICAM1. The expression of these genes was similar in the DN population, except for KIT, which was significantly lower in the IL7Rα population, especially when compared to the CRTH2 population (Fig. 5G). The expression of ICAM1 mRNA tended to be higher in DN cells. We analyzed immune response-associated genes that were expressed at a high level in the DN population. IFNG expression was significantly higher in DN cells (Fig. 5H) and did not correlate with TBX21 expression. The expression of FoxP3, IL12RB2 and TIGIT showed a tendency for higher expression in DN cells but the difference was not significant (Fig. 5I). The IL7Rα population was characterized by dominant expression of ILC3-related genes—IL17A & IL17F (Fig. 5J and K). The expression of IL22 and IL23A showed a tendency for higher expression. The foregoing results suggest that IFNγ-producing cells (ILC1?) and type-3 cytokine-producing ILC3s were dominant in DN and IL7Rα populations, respectively. Nonetheless, both populations were heterogeneous and contained poorly distinguishable ILCs and GATA3+ ILC2s.
Correlates of GATA3 and type-2 cytokines:
We analyzed the RNA-seq data for correlation with GATA3, IL5 and IL13. We observed a strong correlation of GATA3 with IL5 (r=0.83) and IL4 (r=0.80) but not with IL13 (r=0.13). The result suggests that IL13 is regulated differently than GATA3 and IL5 in ILCs. We examined the top genes that correlated with GATA3, IL5 and IL13 (Fig. 6A and B). In this analysis we primarily (but not exclusively) focused on genes that are expressed on the cell surface with the goal to use them for ILC2 detection. The top genes that correlated with GATA3 and IL5 were CD30, CD200R1, TMEM156, TNFR2, DPP4, IL17RB, ICOS and CCR4. As anticipated, CRTH2 and CD161 showed a much lower correlation with GATA3/IL5. The top genes that correlated with IL13 were TMEM173 (STING), IL1RL1, RBPJ, JUNB, NFKB1, CD74, CD58, INSIG2, TREML1, and OX40. A higher degree of correlation of these genes with GATA3, IL5 and IL13 suggests that they might be superior to CRTH2 and IL7Rα in detecting type-2 ILCs.
Figure 6.
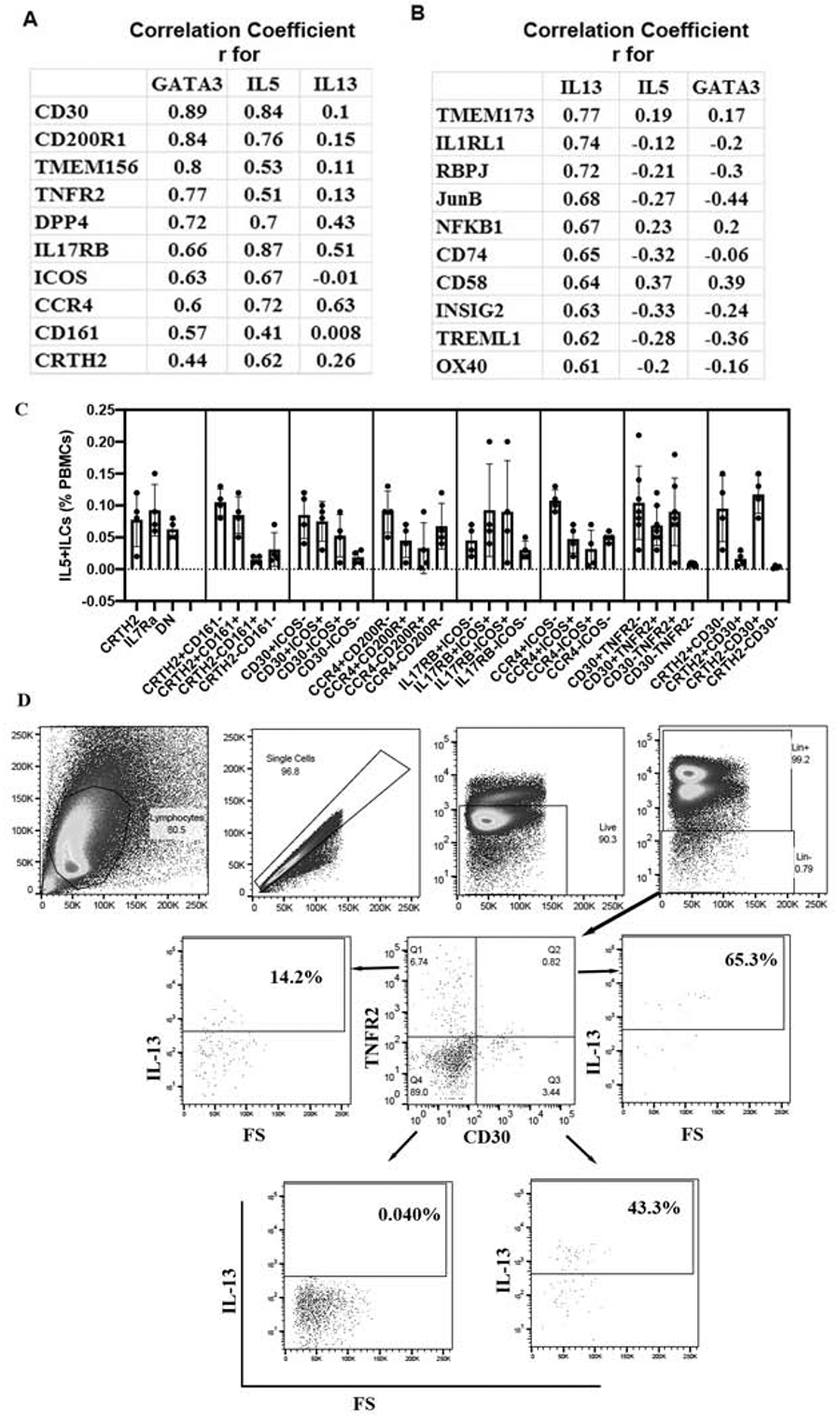
A list of the top genes that correlated with GATA3 and type-2 cytokines, and their surface expression on ILC2 populations. RNA-seq data was analyzed for Pearson correlation (r) with type-2 genes, and the top genes that correlated with GATA3/IL5 and IL13 are shown in panels A & B, respectively. C: PBMCs from asthmatic patients were cultured with IL2/IL25 for 5 days, stained for flow cytometry and then analyzed for expression of IL5 by CD45+lin- cells expressing various combination of surface markers (N=4 for CCR4/CD200R, CCR4/ICOS, IL17RB/ICOS, CD30/ICOS and CD30/CRTH2; N=7 for CD30/TNFR2). D: Representative flow cytometry plots from a single donor showing the gating strategy for CD30 and TNFR2 expressing ILCs and their expression of IL13.
The CD30/TNFR2 surface marker combination correlates better with GATA3 and type-2 cytokine+ ILC2s:
Next, we examined combinations of the top six GATA3-correlated genes to detect GATA3/type-2 cytokine+ ILC2s by flow cytometry and compared them to CRTH2/IL7Rα, and CRTH2/CD161 combinations. These gene combinations included CD30/ICOS, CCR4/CD200R, IL17RB/ICOS, CCR4/ICOS, CD30/TNFR2 and CRTH2/CD30 (Fig. 6C)). The primary objective is to identify a combination of surface markers that captures nearly all type-2 cytokine+ ILCs. The CCR4-CD200R-, CCR4-ICOS-, IL17RB-ICOS- and CD30-ICOS- populations contained relatively high number of IL5+ ILCs (>0.02% of PBMCs). The CD30-TNFR2- and CRTH2-CD30- populations contained the least number of IL5+ ILCs (<0.02% of PBMCs) suggesting that the combinations of CD30/TNFR2 and CRTH2/CD30 captured all type-2 cytokine+ ILCs. Representative flow cytometry plots for IL13+ cells identified by the CD30/TNFR2 gating strategy are shown in Fig. 6D. We compared GATA3 expression among CD30/TNFR2 populations. Unlike CRTH2-IL7Rα- (DN) cells, which expressed GATA3, CD30-TNFR2- cells did not express GATA3 (Fig. 7A). The results suggest that the CD30/TNFR2 combination is more sensitive than CRTH2/IL7Rα in capturing GATA3 and IL5+ ILCs.
Figure 7.
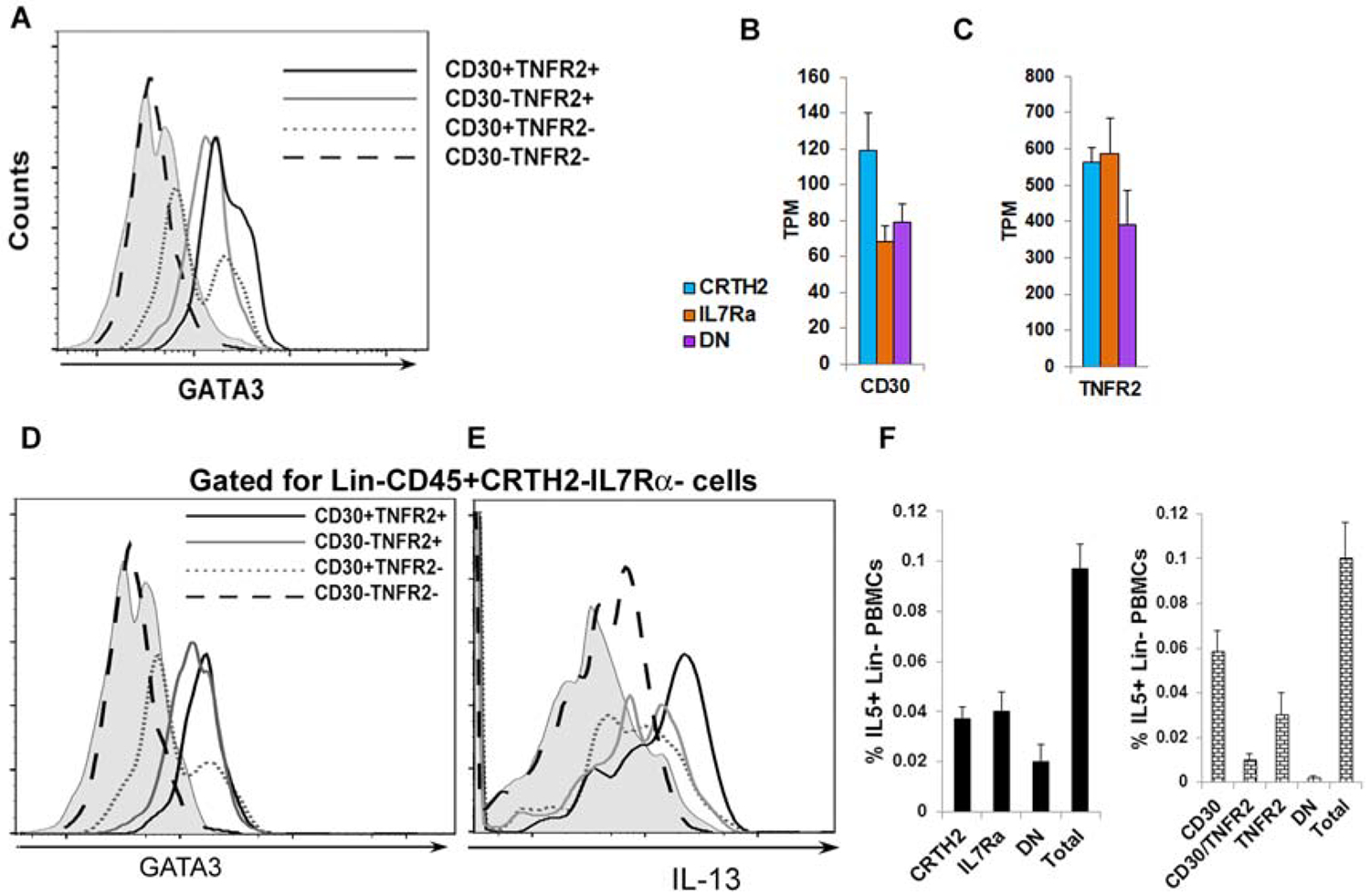
CD30/TNFR2 as ILC2 surface markers. A: A histogram of expression of GATA3 in various populations of lin-CD30/TNFR2 +/− cells. PBMCs were cultured with IL2/IL25 for 5 days, stained for flow cytometry and then gated sequentially for live mononuclear cells, single cells, lin- cells, CD30+TNFR2+, CD30+TNFR2−, CD30-TNFR2+ and CD30-TNFR2- cells. The expression of GATA3 was analyzed in the latter 4 ILC populations (N=4). B & C: Expression of mRNA for CD30 and TNFR2 in ILC populations as measured by RNA-seq. D & E: Expression of GATA3 and IL13 in DN, CD30/TNFR2+/− cells. PBMCs were cultured with IL2/IL25 for 5 days, stained for flow cytometry and then gated sequentially for live mononuclear cells, single cells, CD45+, lin- cells, CRTH2-IL7Ra-, CD30+TNFR2+, CD30+TNFR2−, CD30-TNFR2+ and CD30-TNFR2- cells. The expression of GATA3 and IL13 in the latter 4 ILC populations is shown (N=3). F: Comparison of flow cytometric enumeration of IL5+ ILCs in PBMCs using the CRTH2/IL7Rα and CD30/TNFR2 antibody-based detection approaches. IL5+ cells were first analyzed in lin- cells (Total). The lin- cells were then gated for CRTH2/IL7Rα (left) or CD30/TNFR2 (right) and IL5 expression was analyzed in the indicated cell populations (N=4). CD30: CD30+TNFR2−; TNFR2: CD30-TNFR2+; DN: CD30-TNFR2−.
Next, we interrogated the expression of mRNA for CD30 and TNFR2 in CRTH2, IL7Rα and DN populations. The mRNA for CD30 and TNFR2 was expressed not only by CRTH2+ ILC2s but also by IL7Rα+ and DN ILCs (Fig. 7B and C). We examined the expression of type-2 genes (GATA3 and IL13) within the DN ILC population using CD30 and TNFR2 as surface markers. GATA3 was detected in CD30+, TNFR2+, CD30+TNFR2+ cells but not in CD30-TNFR2- cells within this DN ILC population (Fig. 7D). Likewise, IL13 was not detected in CD30-TNFR2- cells (Fig. 7E). Finally, we compared the total number of IL5+ ILCs in PBMCs as detected by the CRTH2/IL7Rα strategy (the sum of IL5+ cells in CRTH2+, IL7Rα+ and DN populations) and CD30/TNFR2 strategy (the sum of IL5+ cells in the CD30/TNFR2+ populations). Both strategies yielded similar results (Fig. 7F) indicating that all type-2 cytokine+ ILCs were accounted for by the CD30/TNFR2 approach. The results in aggregate suggest that the CD30/TNFR2 approach detected a previously unrecognized CRTH2-IL7Rα-(DN) ILC2 population. The CD30/TNFR2+ population captured nearly all GATA3 and IL5+ ILCs. Finally, we analyzed the flow cytometry data first by gating for type-2 cytokine+ cells in the lin- population and then examining the expression of surface markers. We tested the combination of CRTH2/TNFR2 and compared it with the conventional markers CRTH2/IL7Ra. We found that 11.5% of the DN population from the CRTH2/IL7Rα combination expressed IL13 (Fig. S13). In contrast, only 3.3% of DN cells from the new combination of CRTH2/TNFR2 expressed IL13. Thus, the combination of CD30/TNFR2 and CRTH2/TNFR2 represented superior markers for quantification of human ILC2s.
Discussion
In this paper we showed that the commonly used ILC2 surface marker CRTH2 did not identify all type-2 cytokine+ ILCs. The CRTH2-IL7Rα+ and DN (CRTH2-IL7Rα-) subpopulations also contained type-2 cytokine+ ILC2s and expressed the type-2 transcription factors. The CRTH2+ ILC population had the highest frequency of type-2 cytokine+ ILC2s when compared to IL7Rα+ and DN populations. However, the CRTH2+ population was much smaller in size than the other two populations. Consequently, the absolute number of type-2 cytokine+ ILCs in IL7Rα+ and DN populations was similar to or higher than that in the CRTH2+ population. The enumeration of lin-CRTH2+ cells as ILC2s underestimated the total type-2 cytokine+ ILCs in the blood and lung. To address this shortcoming, we examined the gene expression profile of CRTH2+, IL7Rα+ and DN cells by RNA-seq and then correlated them with the type-2 signature genes. This approach identified multiple surface markers that correlated with GATA3 better than CRTH2. The new surface markers (CD30, ICOS, TNFR2 and CCR4) captured more than 95% of type-2 cytokine+ ILCs. Of these new markers the combinations—CD30/TNFR2, CD30/ICOS and CD30/CRTH2 showed the highest level of sensitivity. The application of the foregoing markers helped detect a previously unrecognized CRTH2-IL7Rα- subpopulation of ILC2s. This subpopulation expressed GATA3 and type-2 cytokines. Thus, the use of CD30/TNFR2, CD30/ICOS and CD30/CRTH2 represents additional strategies to capture type-2 cytokine+ human ILC2s.
ILCs have previously shown to be very plastic18. ILC2s become ILC1s upon exposure to IL1 and IL12 and begin to express T-bet and IFNγ while losing their ability to produce type-2 cytokines19. We observed that about 56% of DN cells converted to CRTH2+ and IL7Rα+ ILCs upon culture with type-2 inducing cytokines. This might suggest that DN cells represent progenitors. However, the progenitor status cannot explain their expression of GATA3, RORA and, especially, the type-2 cytokines. Our RNA-seq data did not show any preferential expression of known progenitor-associated molecules in DN cells (Fig. 5C). In their single cell RNA-seq study of tonsil cells, Bjorklund et al4 reported that about 20% of ILC1s, which were sorted as cells lacking specific surface markers—CRTH2 and CD117, clustered with ILC2s or ILC3s due to their genomic profile. In our PCA analysis of RNA-seq data DN cells from 2 of 4 subjects clustered with the CRTH2 and IL7Rα populations. We believe that the DN cells represent a mixture of progenitors and mature ILCs. The mature ILCs temporarily and intermittently downregulate the expression of surface markers such as CRTH2 and IL7Rα (Fig. S5) but retain the maturation-associated function of cytokine expression. Our results also suggest that some ILC2 -associated surface markers such as CD30, TNFR2 and ICOS are less plastic.
All three ILC populations expressed GATA3, RORA and other type-2 transcription factors. On the other hand, the CRTH2 population from a small group of asthmatic patients expressed IFNγ. The expression of IFNγ by CRTH2+ ILC2s is not a surprise. Lim et al.17 previously reported that a significant fraction of ILC2 clones co-express GATA3 with T-bet, and IL13 with IFNγ. Beuraud et al.30 reported that human and mouse CCR10+ ILC2s, especially, those obtained from the lungs, express IFNγ. The results suggest that a subtype of ILCs are inherently versatile and pluripotent. Some transcriptional master regulators (e.g. TBX21) are epigenetically marked by H3K4me3 and H3K27me331. Gene loci that are simultaneously marked by H3K4me3 and H3K27me3 are considered bivalent and poised32. It is possible that bivalence of TBX21 is particularly prominent in select ILCs, which explains their versatility.
One important aspect of our study is that we measured cytokines without PMA/ionomycin stimulation. This approach distinguishes this study from many previous studies where PMA and ionomycin were added to maximally stimulate cytokine production before flow cytometric analysis3, 4, 18, 33, 34. In our opinion the foregoing approach detects the potential of the cell to produce cytokines but does not detect their ongoing production of cytokines. The frequency of cytokine positive cells measured under ex vivo conditions is relatively low when compared to PMA/ionomycin stimulated cells. Nonetheless, this low level of cytokine positive cells is clinically relevant.
CRTH2 was originally identified as a cell surface receptor that was highly expressed on Th2 cells8, 9. CRTH2 had a high predictive value for type-2 cytokine expression in Th2 cells. However, the negative predictive value of CRTH2- CD4 T cells for production of type-2 cytokines was low in early papers. In the original CRTH2 paper Nagata et al.8 reported that 89% and 95% of CRTH2+ CD4 T cells expressed IL4 and IL13, respectively, while 19% and 28% of CRTH2- CD4 T cells expressed IL4 and IL13, respectively. The result suggested that CRTH2 expression did not capture all type-2 cytokine-producing cells. Similarly, Di Fanis et al.11 demonstrated that CRTH2- T cells produced ¼ the amount of IL4 secreted by CRTH2+ T cells. Our results with ILC2s are in agreement with the foregoing Th2 cell studies.
We identified CD30, TNFR2, ICOS, and CCR4 as additional ILC2 markers. These molecules are expressed by many T cell subtypes, and other lymphoid and myeloid cells35–38. CD30 knockout mice are unable to mount an eosinophilic inflammation in an acute model of asthma and in allergic rhinitis39, 40. TNFR2 knockout mice did not have any effect on allergen-induced airway hyperreactivity or eosinophilic inflammation but they had decreased airway hyperreactivity and neutrophilic inflammation in an ozone-induced airway inflammation model41. An antibody-mediated inhibition of TNFR2 blocked allergic inflammation elicited by a combination of an allergen and the viral mimic poly-IC42. ICOS plays a key role in ILC2-mediated allergic inflammation43–45. CCR4 knockout mice have impaired allergic inflammation46. The high degree of correlation of the foregoing molecules with GATA3 and IL5 suggests that they are co-regulated. Our results suggest that many of these surface markers, especially CD30, TNFR2 and ICOS are useful for identification of cytokine-producing ILC2s.
Comparison studies showed that BAL had 3 to 8-fold higher levels of IL5+ ILC2s than blood in asthmatic subjects. This applies to all subtypes of ILC2s—CRTH2, IL7Rα and DN ILC2s. The frequency of CRTH2+ ILC2s is 10-fold higher in BAL than in blood. The results suggest that the airway milieu favors differentiation and/or expansion of CRTH2+ ILC2s as well as type-2 cytokine+ ILCs. This is consistent with the notion that tissue signals imprint ILC2 identity47. For example, lung ILC2s express higher levels of ST2 as compared to gut ILC2s. One of the caveats of BAL studies is that we compared ILCs between asthma patients and disease controls. The latter included patients with other respiratory illnesses. Some respiratory illnesses (e.g. COPD) are associated with abnormal ILC numbers48, which might have affected our comparison analysis.
Two recent publications clearly demonstrated expression of type-2 transcription factors in CRTH2- ILCs from the peripheral blood. Lim et al.17 performed RNA-seq on peripheral blood lin-CRTH2-CD117+ cells, which are considered precursors of ILCs. The foregoing cells expressed GATA3 and RORA, two known ILC2 transcription factors. However, these cells did not produce type-2 or other cytokines upon stimulation with PMA and ionomycin, thus, supporting their precursor nature. It is unclear if they had capacity to produce cytokines upon stimulation with ILC2-promoting cytokines such as IL2, IL33 or IL25. Nagasawa et al.49 demonstrated the expression of type-2 transcription factor such as GATA3 in CRTH2-KLRG1+ ILCs. Finally, Cavagnero et al.50 reported that Alternaria induced non-conventional ILC2s in the mouse lung that did not express the signature ILC2 markers—ST2 and IL7Rα. The foregoing studies indirectly suggested the existence of ILC2s in CRTH2- and IL7Rα- ILC populations. As mentioned previously, Bjorklund et al.4 reported the transcriptome of human tonsil ILCs that were examined by single cell RNA-seq. They reported the expression of mRNA for GATA3, RORA and ZBTB16 in all three ILC populations and that for Bcl11b in ILC1s and ILC2s. Simoni et al.20 conducted a thorough transcriptomic and mass cytometric analyses of ILCs from cord blood and various human organs. Similar to our findings they concluded that GATA3 and RORγT cannot be used to distinguish various human ILC populations.
In summary, a significant fraction of type-2 cytokine+ cells are not detected by the commonly used ILC2 markers—CRTH2 and IL7Rα. As a consequence, the true frequency of ILC2s is underestimated by the currently applied flow cytometric approaches. We have identified a number of cell surface receptors that capture the highest number of type-2 cytokine+ cells from the ILC population. These receptors include CD30 and TNFR2. A combination of these receptors outperforms the currently used ILC2 markers—CRTH2 and IL7Rα and represents an additional approach for enumeration of type-2 cytokine+ ILCs. We believe that most surface markers are likely to under- or over-estimate the true frequency of ILC2s because of highly plastic nature of ILCs. As such, the most meaningful strategy is to enumerate the frequency of type-2 cytokine+ ILCs as a measure of active ILC2s.
Supplementary Material
Clinical Implications.
The measurement of CRTH2- and IL7Rα- ILC2 populations through the use of novel markers such as CD30 and TNFR2 will more accurately assess the contribution of ILC2s to human pathophysiology.
Acknowledgements:
The work was supported by NIH grants AI102943, AI137970 and HL126895; and grants from the Cohen Family Foundation, NBL Fellowship Foundation and Colorado Technology and Department of Medicine of NJH. We are indebted to Allen Stevens and Christina Kolakowski for the outstanding work with patient recruitment.
Abbreviations:
- BAL
bronchoalveolar lavage
- CRTH2
chemoattractant receptor homologous molecule expressed on T helper-2 cells
- DN
double negative
- ILC
innate lymphoid cell
- lin
lineage
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no competing financial interests.
References
- 1.Klose CS, Artis D. Innate lymphoid cells as regulators of immunity, inflammation and tissue homeostasis. Nat Immunol 2016; 17:765–74. [DOI] [PubMed] [Google Scholar]
- 2.Vivier E, Artis D, Colonna M, Diefenbach A, Di Santo JP, Eberl G, et al. Innate Lymphoid Cells: 10 Years On. Cell 2018; 174:1054–66. [DOI] [PubMed] [Google Scholar]
- 3.Mjosberg JM, Trifari S, Crellin NK, Peters CP, van Drunen CM, Piet B, et al. Human IL-25- and IL-33-responsive type 2 innate lymphoid cells are defined by expression of CRTH2 and CD161. Nat Immunol 2011; 12:1055–62. [DOI] [PubMed] [Google Scholar]
- 4.Bjorklund AK, Forkel M, Picelli S, Konya V, Theorell J, Friberg D, et al. The heterogeneity of human CD127(+) innate lymphoid cells revealed by single-cell RNA sequencing. Nat Immunol 2016; 17:451–60. [DOI] [PubMed] [Google Scholar]
- 5.Barlow JL, Peel S, Fox J, Panova V, Hardman CS, Camelo A, et al. IL-33 is more potent than IL-25 in provoking IL-13-producing nuocytes (type 2 innate lymphoid cells) and airway contraction. J Allergy Clin Immunol 2013; 132:933–41. [DOI] [PubMed] [Google Scholar]
- 6.Salimi M, Barlow JL, Saunders SP, Xue L, Gutowska-Owsiak D, Wang X, et al. A role for IL-25 and IL-33-driven type-2 innate lymphoid cells in atopic dermatitis. J Exp Med 2013; 210:2939–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirkham CL, Carlyle JR. Complexity and Diversity of the NKR-P1:Clr (Klrb1:Clec2) Recognition Systems. Front Immunol 2014; 5:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagata K, Hirai H, Tanaka K, Ogawa K, Aso T, Sugamura K, et al. CRTH2, an orphan receptor of T-helper-2-cells, is expressed on basophils and eosinophils and responds to mast cell-derived factor(s). FEBS Lett 1999; 459:195–9. [DOI] [PubMed] [Google Scholar]
- 9.Nagata K, Tanaka K, Ogawa K, Kemmotsu K, Imai T, Yoshie O, et al. Selective expression of a novel surface molecule by human Th2 cells in vivo. J Immunol 1999; 162:1278–86. [PubMed] [Google Scholar]
- 10.Cosmi L, Annunziato F, Galli MIG, Maggi RME, Nagata K, Romagnani S. CRTH2 is the most reliable marker for the detection of circulating human type 2 Th and type 2 T cytotoxic cells in health and disease. Eur J Immunol 2000; 30:2972–9. [DOI] [PubMed] [Google Scholar]
- 11.De Fanis U, Mori F, Kurnat RJ, Lee WK, Bova M, Adkinson NF, et al. GATA3 up-regulation associated with surface expression of CD294/CRTH2: a unique feature of human Th cells. Blood 2007; 109:4343–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bartemes KR, Kephart GM, Fox SJ, Kita H. Enhanced innate type 2 immune response in peripheral blood from patients with asthma. J Allergy Clin Immunol 2014; 134:671–8 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson DJ, Makrinioti H, Rana BM, Shamji BW, Trujillo-Torralbo MB, Footitt J, et al. IL-33-dependent type 2 inflammation during rhinovirus-induced asthma exacerbations in vivo. Am J Respir Crit Care Med 2014; 190:1373–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Christianson CA, Goplen NP, Zafar I, Irvin C, Good JT Jr., Rollins DR, et al. Persistence of asthma requires multiple feedback circuits involving type 2 innate lymphoid cells and IL-33. J Allergy Clin Immunol 2015; 136:59–68 e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robinette ML, Bando JK, Song W, Ulland TK, Gilfillan S, Colonna M. IL-15 sustains IL-7R-independent ILC2 and ILC3 development. Nat Commun 2017; 8:14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mora-Velandia LM, Castro-Escamilla O, Mendez AG, Aguilar-Flores C, Velazquez-Avila M, Tussie-Luna MI, et al. A Human Lin(−) CD123(+) CD127(low) Population Endowed with ILC Features and Migratory Capabilities Contributes to Immunopathological Hallmarks of Psoriasis. Front Immunol 2017; 8:176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lim AI, Menegatti S, Bustamante J, Le Bourhis L, Allez M, Rogge L, et al. IL-12 drives functional plasticity of human group 2 innate lymphoid cells. J Exp Med 2016; 213:569–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ohne Y, Silver JS, Thompson-Snipes L, Collet MA, Blanck JP, Cantarel BL, et al. IL-1 is a critical regulator of group 2 innate lymphoid cell function and plasticity. Nat Immunol 2016; 17:646–55. [DOI] [PubMed] [Google Scholar]
- 19.Bal SM, Bernink JH, Nagasawa M, Groot J, Shikhagaie MM, Golebski K, et al. IL-1beta, IL-4 and IL-12 control the fate of group 2 innate lymphoid cells in human airway inflammation in the lungs. Nat Immunol 2016; 17:636–45. [DOI] [PubMed] [Google Scholar]
- 20.Simoni Y, Fehlings M, Kloverpris HN, McGovern N, Koo SL, Loh CY, et al. Human Innate Lymphoid Cell Subsets Possess Tissue-Type Based Heterogeneity in Phenotype and Frequency. Immunity 2018; 48:1060. [DOI] [PubMed] [Google Scholar]
- 21.Robinette ML, Fuchs A, Cortez VS, Lee JS, Wang Y, Durum SK, et al. Transcriptional programs define molecular characteristics of innate lymphoid cell classes and subsets. Nat Immunol 2015; 16:306–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ebihara T, Song C, Ryu SH, Plougastel-Douglas B, Yang L, Levanon D, et al. Runx3 specifies lineage commitment of innate lymphoid cells. Nat Immunol 2015; 16:1124–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Constantinides MG, McDonald BD, Verhoef PA, Bendelac A. A committed precursor to innate lymphoid cells. Nature 2014; 508:397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geiger TL, Abt MC, Gasteiger G, Firth MA, O’Connor MH, Geary CD, et al. Nfil3 is crucial for development of innate lymphoid cells and host protection against intestinal pathogens. J Exp Med 2014; 211:1723–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lei AH, Xiao Q, Liu GY, Shi K, Yang Q, Li X, et al. ICAM-1 controls development and function of ILC2. J Exp Med 2018; 215:2157–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li N, van Unen V, Hollt T, Thompson A, van Bergen J, Pezzotti N, et al. Mass cytometry reveals innate lymphoid cell differentiation pathways in the human fetal intestine. J Exp Med 2018; 215:1383–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mielke LA, Groom JR, Rankin LC, Seillet C, Masson F, Putoczki T, et al. TCF-1 controls ILC2 and NKp46+RORgammat+ innate lymphocyte differentiation and protection in intestinal inflammation. J Immunol 2013; 191:4383–91. [DOI] [PubMed] [Google Scholar]
- 28.Seillet C, Rankin LC, Groom JR, Mielke LA, Tellier J, Chopin M, et al. Nfil3 is required for the development of all innate lymphoid cell subsets. J Exp Med 2014; 211:1733–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu Y, Tsang JC, Wang C, Clare S, Wang J, Chen X, et al. Single-cell RNA-seq identifies a PD-1(hi) ILC progenitor and defines its development pathway. Nature 2016; 539:102–6. [DOI] [PubMed] [Google Scholar]
- 30.Beuraud C, Lombardi V, Luce S, Horiot S, Naline E, Neukirch C, et al. CCR10(+) ILC2s with ILC1-like properties exhibit a protective function in severe allergic asthma. Allergy 2018. [DOI] [PubMed] [Google Scholar]
- 31.Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity 2009; 30:155–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fang D, Zhu J. Dynamic balance between master transcription factors determines the fates and functions of CD4 T cell and innate lymphoid cell subsets. J Exp Med 2017; 214:1861–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Halim TY, Krauss RH, Sun AC, Takei F. Lung natural helper cells are a critical source of Th2 cell-type cytokines in protease allergen-induced airway inflammation. Immunity 2012; 36:451–63. [DOI] [PubMed] [Google Scholar]
- 34.Barnig C, Cernadas M, Dutile S, Liu X, Perrella MA, Kazani S, et al. Lipoxin A4 regulates natural killer cell and type 2 innate lymphoid cell activation in asthma. Sci Transl Med 2013; 5:174ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sun X, Yamada H, Shibata K, Muta H, Tani K, Podack ER, et al. CD30 ligand/CD30 plays a critical role in Th17 differentiation in mice. J Immunol 2010; 185:2222–30. [DOI] [PubMed] [Google Scholar]
- 36.Tang C, Yamada H, Shibata K, Muta H, Wajjwalku W, Podack ER, et al. A novel role of CD30L/CD30 signaling by T-T cell interaction in Th1 response against mycobacterial infection. J Immunol 2008; 181:6316–27. [DOI] [PubMed] [Google Scholar]
- 37.Rudmann DG, Moore MW, Tepper JS, Aldrich MC, Pfeiffer JW, Hogenesch H, et al. Modulation of allergic inflammation in mice deficient in TNF receptors. Am J Physiol Lung Cell Mol Physiol 2000; 279:L1047–57. [DOI] [PubMed] [Google Scholar]
- 38.Faustino L, da Fonseca DM, Takenaka MC, Mirotti L, Florsheim EB, Guereschi MG, et al. Regulatory T cells migrate to airways via CCR4 and attenuate the severity of airway allergic inflammation. J Immunol 2013; 190:2614–21. [DOI] [PubMed] [Google Scholar]
- 39.Fuchiwaki T, Sun X, Fujimura K, Yamada H, Shibata K, Muta H, et al. The central role of CD30L/CD30 interactions in allergic rhinitis pathogenesis in mice. Eur J Immunol 2011; 41:2947–54. [DOI] [PubMed] [Google Scholar]
- 40.Polte T, Behrendt AK, Hansen G. Direct evidence for a critical role of CD30 in the development of allergic asthma. J Allergy Clin Immunol 2006; 118:942–8. [DOI] [PubMed] [Google Scholar]
- 41.Cho HY, Zhang LY, Kleeberger SR. Ozone-induced lung inflammation and hyperreactivity are mediated via tumor necrosis factor-alpha receptors. Am J Physiol Lung Cell Mol Physiol 2001; 280:L537–46. [DOI] [PubMed] [Google Scholar]
- 42.Choi JP, Kim YM, Choi HI, Choi SJ, Park HT, Lee WH, et al. An important role of tumor necrosis factor receptor-2 on natural killer T cells on the development of dsRNA-enhanced Th2 cell response to inhaled allergens. Allergy 2014; 69:186–98. [DOI] [PubMed] [Google Scholar]
- 43.Wikenheiser DJ, Stumhofer JS. ICOS Co-Stimulation: Friend or Foe? Front Immunol 2016; 7:304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maazi H, Patel N, Sankaranarayanan I, Suzuki Y, Rigas D, Soroosh P, et al. ICOS:ICOS-ligand interaction is required for type 2 innate lymphoid cell function, homeostasis, and induction of airway hyperreactivity. Immunity 2015; 42:538–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Paclik D, Stehle C, Lahmann A, Hutloff A, Romagnani C. ICOS regulates the pool of group 2 innate lymphoid cells under homeostatic and inflammatory conditions in mice. Eur J Immunol 2015; 45:2766–72. [DOI] [PubMed] [Google Scholar]
- 46.Schuh JM, Power C, Proudfoot AE, Kunkel SL, Lukacs NW, Hogaboam CM. Airway hyperresponsiveness, but not airway remodeling, is attenuated during chronic pulmonary allergic responses to Aspergillus in CCR4−/− mice. FASEB J 2002; 16:1313–5. [DOI] [PubMed] [Google Scholar]
- 47.Ricardo-Gonzalez RR, Van Dyken SJ, Schneider C, Lee J, Nussbaum JC, Liang HE, et al. Tissue signals imprint ILC2 identity with anticipatory function. Nat Immunol 2018; 19:1093–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Silver JS, Kearley J, Copenhaver AM, Sanden C, Mori M, Yu L, Pritchard GH, Berlin AA, Hunter CA, Bowler R, Erjefalt JS, Kolbeck R, Humbles AA. Inflammatory triggers associated with exacerbations of COPD orchestrate plasticity of group 2 innate lymphoid cells in the lungs. Nat Immunol. 2016;17:626–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nagasawa M, Heesters BA, Kradolfer CMA, Krabbendam L, Martinez-Gonzalez I, de Bruijn MJW, Golebski K, Hendriks RW, Stadhouders R, Spits H, Bal SM. KLRG1 and NKp46 discriminate subpopulations of human CD117(+)CRTH2(−) ILCs biased toward ILC2 or ILC3. J Exp Med. 2019;216:1762–1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cavagnero KJ, Badrani JH, Najii LH, Amadeo MB, Shah VS, Gasparian S, Pham A, Wang AW, Seumois G, Croft M, Broide D, Doherty TA. Unconventional ST2- and CD127-negative lung ILC2 populations are induced by the fungal allergen Alternaria alternate. JACI (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu S, Verma M, Michalec L, Liu W, Sripada A, Rollins D, et al. Steroid resistance of airway type 2 innate lymphoid cells from patients with severe asthma: The role of thymic stromal lymphopoietin. J Allergy Clin Immunol 2018; 141:257–68 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Irvin C, Zafar I, Good J, Rollins D, Christianson C, Gorska MM, et al. Increased frequency of dual-positive TH2/TH17 cells in bronchoalveolar lavage fluid characterizes a population of patients with severe asthma. J Allergy Clin Immunol 2014; 134:1175–86 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Quatromoni JG, Singhal S, Bhojnagarwala P, Hancock WW, Albelda SM, Eruslanov E. An optimized disaggregation method for human lung tumors that preserves the phenotype and function of the immune cells. J Leukoc Biol 2015; 97:201–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Goplen N, Karim MZ, Liang Q, Gorska MM, Rozario S, Guo L, et al. Combined sensitization of mice to extracts of dust mite, ragweed, and Aspergillus species breaks through tolerance and establishes chronic features of asthma. J Allergy Clin Immunol 2009; 123:925–32 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Verma M, Liu S, Michalec L, Sripada A, Gorska MM, Alam R. Experimental asthma persists in IL-33 receptor knockout mice because of the emergence of thymic stromal lymphopoietin-driven IL-9(+) and IL-13(+) type 2 innate lymphoid cell subpopulations. J Allergy Clin Immunol 2018; 142:793–803 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


