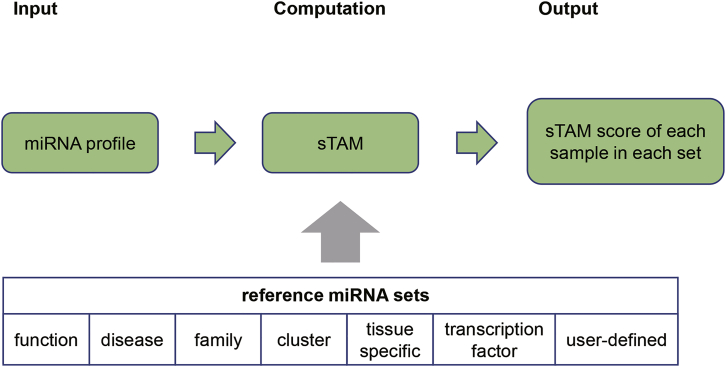
Abstract
MicroRNAs (miRNAs) are an important class of small noncoding RNA molecules that serve as excellent biomarkers of various diseases. However, current miRNA biomarkers, including those comprised of multiple miRNAs, work at a single-miRNA level but not at a miRNA-set level, which is defined as a group of miRNAs sharing common biological characteristics. Given the rapidly accumulating miRNA omics data, we believe that the miRNA-set level analysis could be an important supplement to the single-miRNA level analysis. Therefore, we present sTAM (http://mir.rnanut.net/stam), a computational tool for single-sample miRNA-set enrichment analysis. Moreover, we demonstrate the utility of sTAM scores in discovering miRNA-set level biomarkers through two case studies. We conduct a pan-cancer analysis of the sTAM scores of the “tumor suppressor miRNA set” on 15 types of cancers from The Cancer Genome Atlas (TCGA) and 14 from Gene Expression Omnibus (GEO), results of which indicated a good performance in distinguishing cancers from controls. Moreover, we reveal that the sTAM scores of the “brain development miRNA set” can effectively predict cerebrovascular disorder (CVD). Finally, we believe that sTAM can be used to discover disease-related biomarkers at a miRNA-set level.
Keywords: miRNA set, biomarker, single-sample enrichment analysis, gene expression
Graphical Abstract
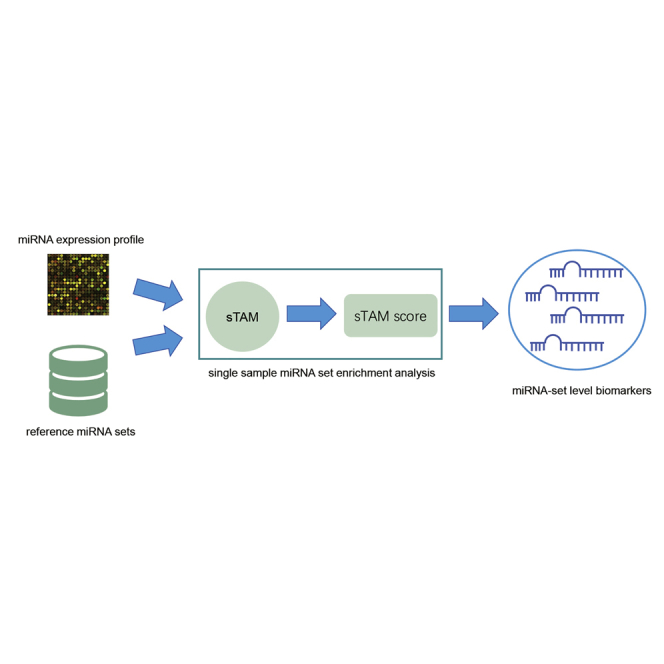
Cui and colleagues presented a computational method for single-sample miRNA-set enrichment analysis and developed the sTAM tool, which can be used to discover disease-related biomarkers at a miRNA-set level, making an important supplement to the single-miRNA level analysis.
Introduction
MicroRNAs (miRNAs) are an important class of small noncoding RNA molecules with a remarkable variety of biological functions.1 In the past decade, miRNAs have been proposed as being useful as diagnostic and prognostic biomarkers for a series of diseases.2 However, most of the current miRNA-based biomarkers work at a single-miRNA level. It is known that no molecules work in isolation but instead, have various connections with others. To address these connections, the concept of a miRNA set was proposed and defined as groups of miRNAs with similar biological characteristics, such as common biological functions, genome locations, or associated diseases.3 Therefore, enrichment analysis becomes an important and popular method to infer the meaningful relationships. In addition, single-sample gene set enrichment analysis (ssGSEA) is a powerful tool to find gene-set level biomarkers for various diseases.4,5 Gene-set level biomarkers represent an important supplement to single-gene level biomarkers. However, methods and tools for single-sample miRNA-set enrichment analysis (ssMSEA) are still not available.
Hence, we developed sTAM and implemented it onto the web server. Moreover, by applying sTAM to 29 cancer miRNA expression datasets from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO), we show that the sTAM scores of the tumor suppressor miRNA set6 can discriminate tumors from controls (healthy or adjacent tissues). In addition, with the use of sTAM, we identified the brain development miRNA set as an effective biomarker for predicting cerebrovascular disorder (CVD). These results suggest that miRNA sets with specific biological consequences have the potential to be biomarkers for monitoring disease formation and development.
Results
Overview of the sTAM Server
The interface of the sTAM server is shown in Figure 1A. sTAM works according to the following procedures. First, users are required to upload their whole genome-wide miRNA expression profiles. Then, users need to select a certain category of reference miRNA sets (e.g., function category) or input their own annotated miRNA sets, the format of which should be a tab-delimited gene matrix-transposed (GMT) file in which each row represents a miRNA set and is described by a name, description, and miRNA list in the miRNA set. Example files of expression profiles and miRNA sets are provided in the sTAM server. In addition, users can adjust advanced parameters. Parameters, such as the size of miRNA sets, normalization method of expression profiles, and modest weight to the rank, are explained in more details in the “Help” page of the sTAM server. Finally, after running sTAM, users will get the sTAM scores of each inputted sample in each miRNA set (Figure 1B). Also, users can download the results from the web server. In addition to sTAM scores, users can further quantitatively evaluate the discriminating ability of each miRNA set and identify candidate miRNA sets as the potential miRNA-set level biomarkers.
Figure 1.
The sTAM Server Interface
(A and B) The main page (A) and the results page (B) of sTAM are shown.
Along with sTAM, we implemented the MSEA method, which evaluates the enrichment of miRNA sets on miRNA expression profiles from samples belonging to two classes. That is, the MSEA method does class (e.g., disease samples as a class versus normal samples as a class)-level enrichment analysis and does not do single-sample level enrichment analysis.
Case Studies
Case 1: sTAM Score in the Tumor Suppressor miRNA Set Effectively Discriminates Cancers from Controls
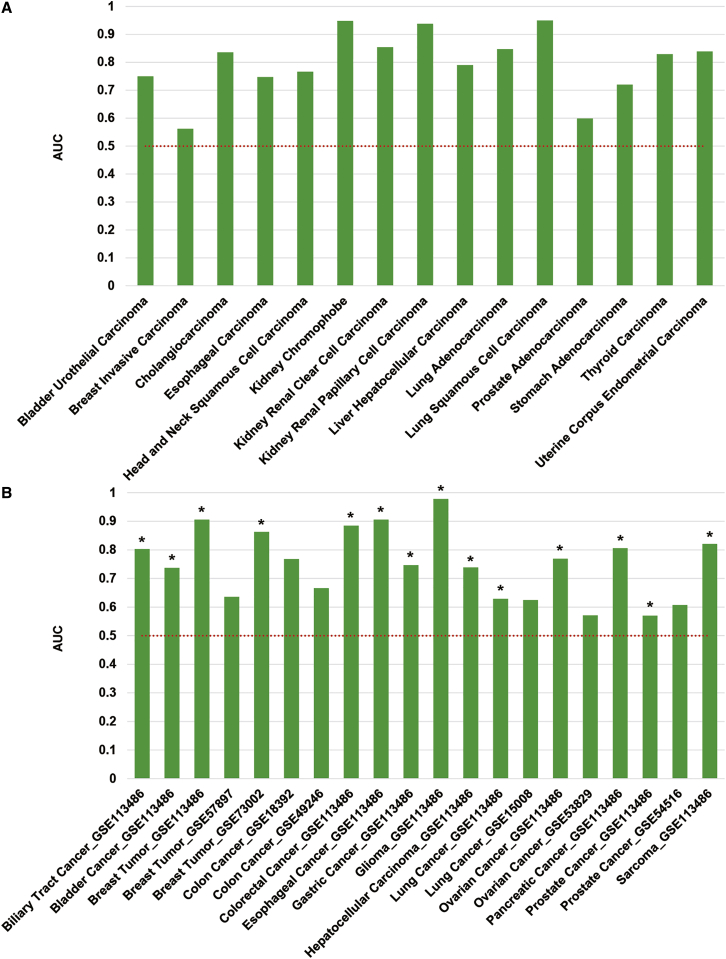
It is well known that some miRNAs are tumor suppressors, which play critical roles in cancer formation and development.7 Thus, we hypothesized that the sTAM scores of the tumor suppressor miRNA set in miRNA expression profiles can discriminate cancers from normal controls. To validate this hypothesis, we applied the sTAM tool to 15 datasets from TCGA database and 14 datasets from GEO. In addition, we drew a receiver operating characteristic (ROC) curve and used the area under the curve (AUC) to estimate the performance of the sTAM scores. As a result, the sTAM scores of the tumor suppressor miRNA set were able to discriminate cancers effectively from normal controls for most of the datasets from TCGA (Figure 2A), as well as most of the datasets from GEO (Figure 2B).
Figure 2.
The Performance of the Tumor Suppressor miRNA Set in Discriminating Cancers from the Controls
(A and B) AUCs for the sTAM scores of the tumor suppressor miRNA set from the TCGA (A) and GEO (B) datasets. The height of each bar represents the AUC in the corresponding dataset. The asterisk (∗) represents samples of the corresponding datasets that are blood based.
Case 2: sTAM Score in the Brain Development miRNA Set Effectively Predicts CVD
There exist a large number of miRNAs in body fluids, such as blood, urine, and saliva.8 Thus, circulating miRNAs can be used as potentially valuable disease-related biomarkers. CVD is a common and severe complex disease with limited availability of effective biomarkers. Here, we sought to determine whether sTAM can be used to predict CVD. Given that CVD is highly associated with the process of brain development, we investigated sTAM scores’ ability for predicting CVD. In order to determine whether the sTAM scores of the brain development miRNA set are different between patients with CVD and non-CVD controls and further validate the biomarkers of miRNA-set level associated with CVD, we obtained serum miRNA expression profiles of CVD patients and non-CVD controls from GEO. Thereafter, we performed sTAM analysis on the brain development miRNA set and drew a ROC curve of the sTAM scores. Interestingly, we found significant differences when comparing the sTAM scores of the brain development miRNA set of CVD patients with non-CVD controls (Figure 3A; p = 2.88e−44, Wilcoxon rank-sum test). In addition, the sTAM scores of the brain-development miRNA set achieve an AUC of 0.822 for discriminating CVD patients from non-CVD controls (Figure 3B), suggesting that miRNAs in the brain development set can potentially serve as biomarkers for CVD prediction at a miRNA-set level. Furthermore, we evaluated the discriminating ability of each miRNA set in the whole function category. Results showed that miRNA sets of “angiogenesis,” “hematopoiesis,” “vascular inflammation,” and “aging” perform well in discriminating CVD patients from non-CVD controls (Figure S1; Table S1).
Figure 3.
The Performance of the Brain Development miRNA Set in Discriminating the Cerebrovascular Disorder (CVD) Patients from the Non-CVD Controls
(A and B) Comparison of sTAM score of the brain development miRNA set between CVD patients and healthy controls (p value = 2.88e-44, Wilcoxon rank-sum test) (A) and the AUC in discriminating CVD patients from non-CVD controls (B). The blue line represents the ROC curve, and the red dashed line represents a random classifier (AUC = 0.5).
Discussion
miRNAs are an important class of small noncoding RNAs that are mainly involved in the post-transcriptional regulation of gene expression and play crucial roles in a series of biological processes and various disease development, such as cancer.9 In the past decade, miRNAs have shown their excellent ability as biomarkers for various cancers and other diseases.10,11 Given that several miRNAs have been identified in a number of body fluids, and miRNAs normally show high stability,12,13 it makes them suitable biomarkers for many diseases. However, the current miRNA-based biomarkers work at a single-miRNA level. Even for biomarkers composed of multiple miRNAs, each miRNA represents one component of the biomarker but does not function at the level of a miRNA set.
We previously defined the concept of miRNA set and found that diseases often associated with miRNAs at a miRNA-set level.3 Therefore, we developed TAM for MSEA using a list of miRNAs, such as those that are deregulated in some cancers.14,15 In addition, two other tools for MSEA, miSEA,16 and miEAA,17 have also been developed with different statistical methods. These tools provide help in discovering valuable clues and predicting novel disease-related miRNAs based on miRNA expression data. However, none of these can perform enrichment analysis at a single-sample level, which is very important for miRNA-set-based biomarker discovery.5
In this study, we presented a computational method for ssMSEA and developed the sTAM software. We integrated 1,236 miRNA sets into sTAM, making it easy for users to select the reference miRNA sets for ssMSEA. In addition, users can input their own defined miRNA sets. Finally, we proposed two case studies that reveal that the sTAM scores of the tumor suppressor miRNA set can effectively discriminate cancers from controls, whereas sTAM scores of the brain development miRNA set are able to discriminate CVDs from controls. It should be noted that this type of biomarker works at the miRNA-set level. Therefore, miRNA profiles from different platforms or experiments may result in different numbers of miRNAs, which can influence analysis results. Hence, users should consider this situation when selecting their candidate miRNA-set level biomarkers. According to our knowledge, sTAM represents the first tool for ssMSEA. With the continuous improvement of miRNA-set annotations and rapid accumulation of miRNA omics data, we believe that sTAM will be useful for the discovery of miRNA-set-based biomarkers in various diseases.
Materials and Methods
Data Sources and Data Preprocessing
A total of 1,236 reference miRNA sets were downloaded from TAM 2.014 and were composed of six categories, including function, disease, family, cluster, tissue specific, and transcription factor. For the case studies, we downloaded 15 cancer miRNA expression datasets from TCGA (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga) and 14 cancer miRNA expression datasets from GEO (datasets with at least 50 samples; https://www.ncbi.nlm.nih.gov/geo/). The “DESeq2” package was used to estimate expression value and “sva” package to remove batch effects on TCGA datasets.18,19 In addition, we obtained the serum miRNA expression profiles of 173 CVD patients and 1,612 non-CVD controls from a GEO dataset (GEO: GSE117064).
Workflow of sTAM
The workflow of sTAM is presented in Figure 4. sTAM allows submission of whole genome-wide miRNA expression profiles. Since the reference miRNA sets are at a pre-miRNA level, sTAM will convert the mature miRNA names to precursor names and automatically average the value of duplicated miRNAs. Then, users need to select a certain category of the reference miRNA set or upload their own defined miRNA sets instead. sTAM will then perform ssMSEA in the background. Finally, the server will deliver the sTAM scores of each sample in each miRNA set. Furthermore, users can download the compressed result file generated by the server and conduct customized analyses.
Figure 4.
The Workflow of sTAM
The main procedures of sTAM are as follows: (1) upload a whole genome-wide miRNA expression profile; (2) select one certain category of the miRNA set, or upload user-defined miRNA sets; (3) perform single-sample miRNA-set enrichment analysis; and (4) sTAM generates the analysis result.
sTAM Algorithm
In order to perform ssMSEA, we adopted the algorithm for ssGSEA.5 Given a miRNA expression profile, sTAM ranks miRNAs by absolute expression values in one sample and then calculates the enrichment score (ES) by going down the ranked list of miRNAs. This leads to an increase in a running-sum statistic when a miRNA is present in the miRNA set and a decrease when it is not. The ES is the maximum deviation from zero encountered when going down the list. In other words, for a given miRNA-set of size and single sample of the dataset of miRNAs, the miRNAs are sorted according to their absolute expression values from high to low: . An enrichment score, , is obtained by a sum of the difference between a weighted empirical cumulative distribution function (ECDF) of the miRNAs in the miRNA-set and the ECDF of the remaining miRNAs :
sTAM repeated this calculation for each miRNA set and each sample in the expression dataset. Note that the exponent is set to 0.25 by default, adding a modest weight to the rank.
Server Construction
The web server was established using the “Linux + Apache + Thinkphp (version 3.2)” framework. The ssMSEA was implemented using GSEAPY (version 0.9.15), a Python wrapper for enrichment analysis. The MSEA analysis was performed by applying the JAR (Java Archive) file, which is based on the GSEA algorithm.20 The sTAM web server is freely accessible at http://mir.rnanut.net/stam/.
Statistical Analysis
ROC curve and AUC were used to evaluate the performance of the sTAM score. Differences in sTAM scores between CVD patients and non-CVD controls were analyzed by Wilcoxon rank-sum test performed by SciPy (version 1.1.0), an open-source scientific computing library for the Python programming language. A p value of less than 0.05 was considered statistically significant.
Author Contributions
Q.C. conceived the study. J.S. developed the web server and performed bioinformatics analysis. Both authors wrote the manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work has been supported by grants from the Natural Science Foundation of China (81970440, 81670462, and 31930056); Peking University Basic Research Program (BMU2020JC001); Peking University Clinical Scientist Program (BMU2019LCKXJ001); and Fundamental Research Funds for the Central Universities.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2020.07.004.
Supplemental Information
References
- 1.Kim D., Sung Y.M., Park J., Kim S., Kim J., Park J., Ha H., Bae J.Y., Kim S., Baek D. General rules for functional microRNA targeting. Nat. Genet. 2016;48:1517–1526. doi: 10.1038/ng.3694. [DOI] [PubMed] [Google Scholar]
- 2.Wang J., Chen J., Sen S. MicroRNA as Biomarkers and Diagnostics. J. Cell. Physiol. 2016;231:25–30. doi: 10.1002/jcp.25056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu M., Zhang Q., Deng M., Miao J., Guo Y., Gao W., Cui Q. An analysis of human microRNA and disease associations. PLoS ONE. 2008;3:e3420. doi: 10.1371/journal.pone.0003420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Abazeed M.E., Adams D.J., Hurov K.E., Tamayo P., Creighton C.J., Sonkin D., Giacomelli A.O., Du C., Fries D.F., Wong K.K. Integrative radiogenomic profiling of squamous cell lung cancer. Cancer Res. 2013;73:6289–6298. doi: 10.1158/0008-5472.CAN-13-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barbie D.A., Tamayo P., Boehm J.S., Kim S.Y., Moody S.E., Dunn I.F., Schinzel A.C., Sandy P., Meylan E., Scholl C. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang D., Qiu C., Zhang H., Wang J., Cui Q., Yin Y. Human microRNA oncogenes and tumor suppressors show significantly different biological patterns: from functions to targets. PLoS ONE. 2010;5:e13067. doi: 10.1371/journal.pone.0013067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Svoronos A.A., Engelman D.M., Slack F.J. OncomiR or Tumor Suppressor? The Duplicity of MicroRNAs in Cancer. Cancer Res. 2016;76:3666–3670. doi: 10.1158/0008-5472.CAN-16-0359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuzaki J., Ochiya T. Circulating microRNAs and extracellular vesicles as potential cancer biomarkers: a systematic review. Int. J. Clin. Oncol. 2017;22:413–420. doi: 10.1007/s10147-017-1104-3. [DOI] [PubMed] [Google Scholar]
- 9.Rupaimoole R., Slack F.J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 2017;16:203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 10.Eslamizadeh S., Akbari A. Serum or plasma; which is a more competent molecular source for investigating the blood-based tumor-specific miRNA biomarkers? J. Cell. Physiol. 2020;235:5858–5859. doi: 10.1002/jcp.29598. [DOI] [PubMed] [Google Scholar]
- 11.Condrat C.E., Thompson D.C., Barbu M.G., Bugnar O.L., Boboc A., Cretoiu D., Suciu N., Cretoiu S.M., Voinea S.C. miRNAs as Biomarkers in Disease: Latest Findings Regarding Their Role in Diagnosis and Prognosis. Cells. 2020;9:276. doi: 10.3390/cells9020276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Srinivasan S., Yeri A., Cheah P.S., Chung A., Danielson K., De Hoff P., Filant J., Laurent C.D., Laurent L.D., Magee R. Small RNA Sequencing across Diverse Biofluids Identifies Optimal Methods for exRNA Isolation. Cell. 2019;177:446–462.e16. doi: 10.1016/j.cell.2019.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balzano F., Deiana M., Dei Giudici S., Oggiano A., Baralla A., Pasella S., Mannu A., Pescatori M., Porcu B., Fanciulli G. miRNA Stability in Frozen Plasma Samples. Molecules. 2015;20:19030–19040. doi: 10.3390/molecules201019030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li J., Han X., Wan Y., Zhang S., Zhao Y., Fan R., Cui Q., Zhou Y. TAM 2.0: tool for MicroRNA set analysis. Nucleic Acids Res. 2018;46(W1):W180–W185. doi: 10.1093/nar/gky509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu M., Shi B., Wang J., Cao Q., Cui Q. TAM: a method for enrichment and depletion analysis of a microRNA category in a list of microRNAs. BMC Bioinformatics. 2010;11:419. doi: 10.1186/1471-2105-11-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Çorapçıoğlu M.E., Oğul H. miSEA: microRNA set enrichment analysis. Biosystems. 2015;134:37–42. doi: 10.1016/j.biosystems.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 17.Backes C., Khaleeq Q.T., Meese E., Keller A. miEAA: microRNA enrichment analysis and annotation. Nucleic Acids Res. 2016;44 doi: 10.1093/nar/gkw345. W110–W116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leek J.T., Johnson W.E., Parker H.S., Jaffe A.E., Storey J.D. The sva package for removing batch effects and other unwanted variation in high-throughput experiments. Bioinformatics. 2012;28:882–883. doi: 10.1093/bioinformatics/bts034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., Mesirov J.P. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.