Abstract
The association between distress caused by tinnitus and psychological factors such as depression and anxiety has been examined and reported. However, prognostic factors remain poorly understood because there are only a few reports on genetic associations. We theorized there might be an association between the grade of tinnitus distress and the genetic background related to psychological factors which might lead us to identify prognostic markers. We enrolled 138 patients who had suffered from tinnitus for over 3 months. Using Tinnitus Handicap Inventory (THI) scores, we examined the association between tinnitus distress and a genetic background related to depression or anxiety. A significant association between single nucleotide polymorphism rs131702 of the Breakpoint Cluster Region (BCR) gene and the severe THI score was identified. In addition, there was an association with the severity of the State-Trait Anxiety Inventory, an index of state anxiety severity. No association was found with the Self-Rating Depression Scale, an index of depression severity. It is reported that rs131702 of BCR in Japanese patients are related to bipolar II depression characterized by fluctuation between abnormal mood states of mania and depression. Our results indicate that rs131702 of BCR is independent of depression in this study and is, therefore, a prognostic factor unique to tinnitus. We conclude that the severity of tinnitus is associated with genes related to depression.
Subject terms: Medical research, Neurology
Introduction
Tinnitus is a perceived symptom that affects 15% of the population. Although no external sound source can be attributed to tinnitus, in 20% of cases, the patient often experiences severe enough distress, impairing their daily activities1. In addition to hearing impairment, tinnitus is associated with different types of clinical symptoms due to the involvement of various pathologies resulting from environmental, psychological (depression or anxiety disorder), or other factors.
In examining tinnitus, the maximum slope within audiograms is determined to be higher in people with tinnitus than in people with hearing loss without tinnitus, despite the latter having a greater mean hearing loss2. The additional involvement of non-auditory areas of the brain, particularly areas associated with awareness and salience, can explain why some people with hearing loss develop tinnitus3,4. Whether tinnitus is perceived as bothersome or not may be related to the additional involvement of emotion-processing areas5–7. Some models have proposed that tinnitus reflects “an emergent property of multiple parallel dynamically changing and partially overlapping sub-networks”. This suggests that various brain networks associated with memory and emotional processing are involved in tinnitus and that the degree of involvement of the different networks reflects the variable aspects of an individual’s tinnitus3,4,8.
The psychological models, which use the concept of habituation, explain how and why some people experience a negative effects of tinnitus on quality of life9. For example, Jastreboff’s model features classical conditioning mechanisms10 and there are also cognitive-behavioral models11–13. These models underpin the rationale for and development of cognitive-behavioral interventions for reducing the impact of tinnitus on quality of life.
According to the neurophysiological model of Jastreboff14 and the maladaptive neural plasticity model of Shore15, tinnitus has been found to be correlated with the plasticity of the central nervous system.
Tinnitus Retraining Therapy (TRT), consisting of sound therapy and psychotherapy based on the neurophysiological model proposed by Jastreboff in the latter half of the 1980s, demonstrates good therapeutic effect as a standard therapy in Japan16. Additionally, according to the Multidisciplinary European Guideline for tinnitus17 there is evidence for safety but little high-level evidence for the effectiveness of TRT (one RCT and two systematic reviews). However, some tinnitus patients have experienced treatment with difficulties10.
There are several tests in use for assessing the level of severity of tinnitus complaints. These questionnaires including Tinnitus Handicap Inventory (THI), Tinnitus Questionnaire, and Visual Analogue Scale have been used to examine tinnitus distress and related factors.
In recent years, gap detection tests, proposed by Turner in 200618, for patients with tinnitus19 and tinnitus animal models has enabled the investigation of tinnitus pathology. Gene analysis is also identified as an effective method for elucidating disease pathology and developing novel therapeutic approaches. Thus, in the field of otolaryngology, next-generation sequencing (NGS) can be used to identify the causative genes in congenital deafness, elucidate the pathology, and predict the phenotype of hearing loss. In recent years, there have been reports of tinnitus-related gene analysis being used to clarify the mechanisms of tinnitus and discover tinnitus biomarkers.
Sand et al. analyzed the relevance of tinnitus and single nucleotide polymorphisms (SNPs) in brain-derived neurotrophic factor (BDNF) and glial-derived neurotrophic factor (GDNF) genes. Some genotypes predicted tinnitus severity in women (but not in men). There was no significant difference in mean tinnitus severity scores between carriers and non-carriers of the minor alleles (p > 0.19), nor did a positive family history of tinnitus in first-degree relatives predict minor allele carrier status (p = 0.08)20. There are, however, conflicting reports on the association between tinnitus and plasma or serum BDNF. Goto et al. reported that plasma BDNF levels were significantly higher in the group of patients mildly handicapped by tinnitus than in the severely handicapped and control groups (p < 0.01)21. Xiong et al. found plasma BDNF levels were elevated in patients with tinnitus compared with healthy controls22. Coskunoglu et al. found lower serum BDNF levels in tinnitus patients than controls. There was no correlation between BDNF gene polymorphism and tinnitus23.The contribution of BDNF to tinnitus severity is still under debate as mentioned above.
KCTD12 genes are auxiliary subunits of GABAB receptors, and GABAB receptor agonists are known tinnitus suppressors24.
Previous reports proposed that the KCNE3 gene involved in voltage-gated potassium channels correlated to non-syndromic hearing loss. Implication of the KCNE3 gene in tinnitus was investigated; however, no KCNE3 variants was implicated25.
Deniz et al. reported SLC6A4 as a candidate for the serotonin transporter gene25. Although no SNPs associated with tinnitus have been identified, Deniz et al. identified a significant association between the 5-HTTLPR polymorphism and scores from the Visual Analogue Scale of the patients. The generation of the tinnitus signal is not associated with SLC6A4 polymorphism and possibly with serotonergic mechanisms. However, the genotype variant of the SLC6A4 polymorphic promoter region seems associated with the limbic and autonomic nervous system symptoms of patients with tinnitus. Selective serotonin reuptake inhibitors are also determined to be effective in reducing tinnitus distress, indicating that further elucidation of the relationship between tinnitus and SLC6A4 should be considered25.
COCH is a known causative gene of dominantly inherited non-syndromic hearing loss. An association between tinnitus and COCH gene mutation has been reported in the analysis of a family with the mutation26. The study involved a basic genetic analysis related to tinnitus. They identified a heterozygous 18 base pair deletion on exon 11 of the COCH gene in a large multigenerational family, segregating late-onset progressive bilateral sensorineural hearing impairment and tinnitus. However, hearing loss can explain the association between tinnitus and the COCH variant, judging from the fact that the tinnitus of patients with COCH variants is secondary from hearing loss.
In another pilot study, Gilles et al. performed a genome-wide association study27 of 4,000,000 SNPs in 167 patients experiencing tinnitus lasting > 5 min and 749 non-tinnitus groups. However, no clearly related SNPs were detected.
Despite the existence of several reports on the tinnitus-related genes, their contribution to the development of tinnitus is still under debate due to the variety of tinnitus phenotypes caused by clinical heterogeneity such as anxiety, age, hearing loss, occupational/recreational noise exposure, and distress. Among the phenotype variety, psychological factors in the heterogeneous aspects of tinnitus are reported to be most influential to the THI score. Additionally, the psychological models proposed the involvement of memory and emotional processing area in the central nerve system in the tinnitus. Thus, the hypothesis was that the focus on the gene linked with psychological factors such as anxiety, depression, fear and panic disorder can facilitate the exploration of genetically contributing factors to tinnitus distress.
Therefore, we analyzed the relationship between genetic backgrounds correlated to psychological condition and tinnitus distress levels and searched for prognostic markers. Online Mendelian Inheritance in Man (OMIM) lists the genes contributing to depression/anxiety, fear, and panic disorder (https://www.omim.org/). Among them, we found eight SNPs with a minor allele frequency (MAF) more than or equal to 0.1 according to the HapMap for Japan.
Three SNPs (rs140504, rs131690, and rs131702)28 of BCR in Japanese patients are related to bipolar II depression characterized by fluctuation between abnormal mood states of mania and depression. Bipolar I patients experience full manic episodes, while a person with bipolar II will experience only hypomanic episodes.
The rs1545843 allele (MAF = 0.41 in controls) of Solute Carrier Family6 (Neurotransmitter Transporter), Member 15; SLC6A15 gene on chr12q21.31 showed experiment-wide significance in a recessive mode of inheritance (AA versus AG+GG) on 12q21.31 and was associated with a higher risk for major depression, but only in individuals less than 55 years old and only when homozygous with an odds ratio of 1.4 (p = 2 × 10e-8).
The rs2267735 allele of Adenylate Cyclase-activating polypeptide1 Receptor For; ADCYAP1R1 predicts post-traumatic stress disorder (PTSD) diagnoses and symptoms in females only. This may occur due to the estrogen regulation of ADCYAP1R1. Total PTSD symptoms are differentially associated with rs2267735 genotype (CC is a high-risk variant) in females (p ≤ 0.001).
The SNP of the serotonin 2A receptor; HTR2A, rs7997012, is associated with the outcome of antidepressant treatment29.
The rs10997870 allele of Sirtuin 1; SIRT1 is activates Monoamine oxidase A; MAO-A in the brain to mediate anxiety and exploratory drive30 and the rs1799836 in Monoamine oxidase B; MAOB affect the levels of negative emotions in healthy human volunteers31.
Results
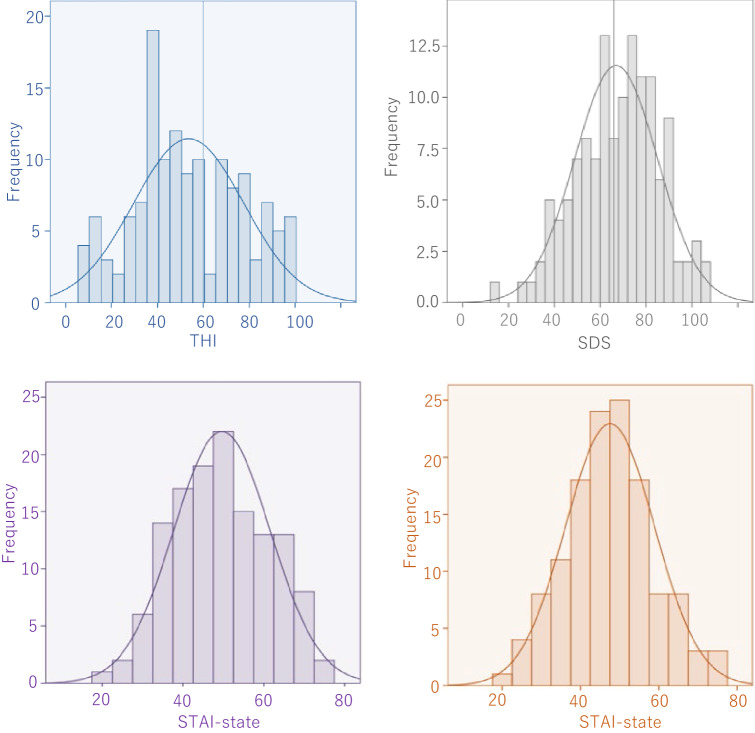
The average THI score was 53.64 (s.d. 24.05). The average Self-Rating Depression Scale (SDS) score was determined to be at 43.48 (s.d. 9.05). The average STAI-state score was 49.61 (s.d. 11.97). The average STAI-trait score was 47.51 (s.d. 11.41). The THI score, the SDS score, the STAI-state score, and the STAI trait score were distributed broadly from mild to severe, although they did not exhibit a normal distribution (Fig. 1, Table 1).
Figure 1.
Results of patient questionnaires. Although the THI score was generally biased toward moderate symptoms, the results were globally distributed from mild to severe. The scores of STAI-state, STAI-trait, and SDS characteristics were broadly distributed from mild to severe; it was not a normal distribution. THI, Tinnitus Handicap Inventory; SDS, Self-Rating Depression Scale; STAI, State-Trait Anxiety Inventory.
Table 1.
Clinical information of the subjects.
| Total | Male | Female | |||||||
|---|---|---|---|---|---|---|---|---|---|
| N | Mean | SD | N | Mean | SD | N | Mean | SD | |
| Age | 138 | 61.31 | 13.12 | 59 | 60.92 | 13.21 | 79 | 61.61 | 13.13 |
| THI | 138 | 53.64 | 24.05 | 59 | 54.58 | 21.72 | 79 | 52.95 | 25.77 |
| SDS | 131 | 43.48 | 9.05 | 56 | 44.91 | 8.68 | 75 | 42.41 | 9.23 |
| STAI-State | 132 | 49.61 | 11.97 | 55 | 51.82 | 10.66 | 77 | 48.03 | 12.65 |
| STAI-Trait | 131 | 47.51 | 11.4 | 54 | 48.81 | 10.45 | 77 | 46.6 | 12.01 |
The association between SNP rs131702 of the BCR gene and the severity of THI was found to be significant (Tables 2, 3). The results of the Fisher test indicated that the carriers of minor allele G showed a severe THI score with odds ratio of 2.019 (Table 2). The trend test also revealed that the minor allele G was positively associated with the severity of the THI score (Table 3). Furthermore, there was a significant allelic association between SNP rs131702 of the BCR gene and the STAI-state score with the trend test. The severe STAI-state score, as well as the severe THI score was detected in the carriers of the minor allele G (Table 4). On the other hand, SNP rs131702 did not show any association with the STAI-trait score and the SDS (Tables 5, 6). No other SNP showed any relation with the THI, STAI, or SDS score.
Table 2.
Statistical analysis for single SNP; the comparison of allele frequencies between cases with mild-moderate THI and with severe-catstrophic THI.
| Gene | SNP | Allele | All subjects (n = 134) | ||||
|---|---|---|---|---|---|---|---|
| Mild and moderate (%) | Severe and catastrophic (%) | Total | P for Fisher's test | Odds ratio (for considering minor allele as risk allele) | |||
| BCR | rs140504 | A allele | 92(57.5) | 49(45.4) | 141(52.6) | 0.061 | 1.629 (0.996–2.664) |
| G allele | 68(42.5) | 59(54.6) | 127(47.4) | ||||
| Total | 160 | 108 | 268 | ||||
| BCR | rs131690 | A allele | 123(76.9) | 79(73.1) | 202(75.4) | 0.563 | 1.22 (0.695–2.141) |
| G allele | 37(23.1) | 29(26.9) | 66(24.6) | ||||
| Total | 160 | 108 | 268 | ||||
| BCR | rs131702 | G allele | 53(33.1) | 53(50) | 106(39.8) | 0.007 | 2.019 (1.220–3.340) |
| T allele | 107(66.9) | 53(50) | 160(60.2) | ||||
| Total | 160 | 106 | 266 | ||||
| SLC6A15 | rs1545843 | A allele | 53(33.1) | 41(38) | 94(35.1) | 0.436 | 1.235 (0.742–2.056) |
| G allele | 107(66.9) | 67(62) | 174(67.9) | ||||
| Total | 160 | 108 | 268 | ||||
| ADCYAP1R1 | rs2267735 | C allele | 81(50.6) | 48(44.4) | 129(48.1) | 0.383 | 0.78 (0.478–1.274) |
| G allele | 79(49.4) | 60(55.6) | 139(51.9) | ||||
| Total | 160 | 108 | 268 | ||||
| SIRT1 | rs10997870 | G allele | 131(81.9) | 87(80.6) | 218(81.3) | 0.873 | 1.353 (0.659–2.780) |
| T allele | 29(18.1) | 21(19.4) | 50(18.7) | ||||
| Total | 160 | 108 | 268 | ||||
| HTR2A | rs7997012 | A allele | 32(20) | 25(23.1) | 57(21.3) | 0.546 | 1.205 (0.667–2.177) |
| G allele | 128(80) | 83(76.9) | 211(78.7) | ||||
| Total | 160 | 108 | 268 | ||||
| MAOB (male) | rs1799836 | C allele | 4(12.1) | 3(13) | 7(12.5) | 1 | 1.088 (0.219–5.395) |
| T allele | 29(87.9) | 20(87) | 49(87.5) | ||||
| Total | 33 | 23 | 56 | ||||
| MAOB (female) | rs1799836 | C allele | 19(22.1) | 8(13.3) | 27(18.5) | 0.201 | 0.543 (0.220–1.337) |
| T allele | 67(77.9) | 52(86.7) | 119(81.5) | ||||
| Total | 86 | 60 | 146 | ||||
Table 3.
Statistical analysis for single SNP to evaluate the correlation between genotype and THI score.
| Gene | SNP | Genotype | All Subjects (n =134) | |||
|---|---|---|---|---|---|---|
| N | Mean | SD | P for trend | |||
| BCR | rs140504 | AA | 35 | 47.26 | 22.57 | 0.103 |
| AG | 71 | 55 | 23.75 | |||
| GG | 28 | 56.86 | 26.22 | |||
| BCR | rs131690 | AA | 76 | 52.72 | 24.8 | 0.567 |
| AG | 50 | 53.36 | 22.51 | |||
| GG | 8 | 59.5 | 28.96 | |||
| BCR | rs131702 | GG | 18 | 66.67 | 24.49 | 0.008 |
| GT | 70 | 53.3 | 24.04 | |||
| TT | 45 | 47.82 | 22.53 | |||
| SLC6A15 | rs1545843 | AA | 17 | 50 | 13.95 | 0.514 |
| AG | 60 | 57.33 | 25.8 | |||
| GG | 57 | 50.19 | 24.31 | |||
| ADCYAP1R1 | rs2267735 | CC | 29 | 53.55 | 25.12 | 0.763 |
| CG | 71 | 52.39 | 24.21 | |||
| GG | 34 | 55.24 | 23.55 | |||
| SIRT1 | rs10997870 | GG | 90 | 53.78 | 25.78 | 0.772 |
| GT | 38 | 52.63 | 20.46 | |||
| TT | 6 | 51.83 | 22.26 | |||
| HTR2A | rs7997012 | AA | 9 | 60 | 30.74 | 0.308 |
| AG | 39 | 54.82 | 23.44 | |||
| GG | 86 | 52.01 | 23.79 | |||
| MAOB (male) | rs1799836 | C | 7 | 65.43 | 22.11 | 0.195 |
| T | 49 | 53.96 | 21.59 | |||
| MAOB (female) | rs1799836 | CC | 2 | 51 | 60.81 | 0.343 |
| CT | 23 | 47.04 | 23.64 | |||
| TT | 48 | 54.19 | 25.86 | |||
Table 4.
Statistical analysis for single SNP to evaluate the correlation between the genotypes and STAI-state score.
| Gene | SNP | Genotype | All Subjects (n = 128) | |||
|---|---|---|---|---|---|---|
| N | Mean | SD | P for trend | |||
| BCR | rs140504 | AA | 35 | 46.89 | 11.85 | 0.101 |
| AG | 65 | 50.2 | 11 | |||
| GG | 28 | 51.75 | 13.79 | |||
| BCR | rs131690 | AA | 73 | 48.68 | 11.46 | 0.193 |
| AG | 47 | 50.28 | 12.63 | |||
| GG | 8 | 54.5 | 11.92 | |||
| BCR | rs131702 | GG | 18 | 54.56 | 13.31 | 0.016 |
| GT | 66 | 50.15 | 11.62 | |||
| TT | 43 | 46.67 | 11.37 | |||
| SLC6A15 | rs1545843 | AA | 15 | 45.4 | 8.89 | 0.908 |
| AG | 58 | 52 | 12.27 | |||
| GG | 55 | 48.29 | 11.92 | |||
| ADCYAP1R1 | rs2267735 | CC | 28 | 47.57 | 13.2 | 0.146 |
| CG | 70 | 49.39 | 11.96 | |||
| GG | 30 | 52.13 | 0.45 | |||
| SIRT1 | rs10997870 | GG | 85 | 50.22 | 12.33 | 0.897 |
| GT | 37 | 47.19 | 10.74 | |||
| TT | 6 | 56.33 | 11.15 | |||
| HTR2A | rs7997012 | AA | 9 | 44.78 | 12.68 | 0.895 |
| AG | 39 | 51.59 | 13.03 | |||
| GG | 80 | 49.23 | 11.22 | |||
| MAOB (male) | rs1799836 | C | 6 | 55.33 | 13.87 | 0.478 |
| T | 46 | 52 | 10.33 | |||
| MAOB (female) | rs1799836 | CC | 2 | 60 | 19.8 | 0.35 |
| CT | 23 | 47.7 | 12.94 | |||
| TT | 46 | 46.91 | 12.04 | |||
Table 5.
Statistical analysis for single SNP to evaluate the correlation between the genotypes and STAI-trait score.
| Gene | SNP | Genotype | All subjects (n = 127) | |||
|---|---|---|---|---|---|---|
| N | Mean | SD | P for trend | |||
| BCR | rs140504 | AA | 35 | 45.91 | 10.6 | 0.331 |
| AG | 65 | 47.77 | 10.76 | |||
| GG | 27 | 48.67 | 13.34 | |||
| BCR | rs131690 | AA | 73 | 46.75 | 10.66 | 0.127 |
| AG | 46 | 47.09 | 11.83 | |||
| GG | 8 | 55.88 | 11.31 | |||
| BCR | rs131702 | GG | 18 | 50.72 | 11.85 | 0.076 |
| GT | 65 | 47.78 | 11.61 | |||
| TT | 43 | 45.28 | 10.24 | |||
| SLC6A15 | rs1545843 | AA | 14 | 45.07 | 10.87 | 0.31 |
| AG | 58 | 50.05 | 11.31 | |||
| GG | 55 | 45.31 | 10.91 | |||
| ADCYAP1R1 | rs2267735 | CC | 28 | 46.71 | 11.67 | 0.211 |
| CG | 70 | 46.51 | 11.61 | |||
| GG | 29 | 50.41 | 9.78 | |||
| SIRT1 | rs10997870 | GG | 84 | 47.29 | 12.01 | 0.342 |
| GT | 37 | 46.32 | 9.28 | |||
| TT | 6 | 56.67 | 8.5 | |||
| HTR2A | rs7997012 | AA | 9 | 47.33 | 11.74 | 0.536 |
| AG | 39 | 48.77 | 13.23 | |||
| GG | 79 | 46.81 | 10.21 | |||
| MAOB (male) | rs1799836 | C | 6 | 54.17 | 16.27 | 0.231 |
| T | 45 | 48.62 | 9.65 | |||
| MAOB (female) | rs1799836 | CC | 2 | 50.5 | 13.44 | 0.521 |
| CT | 23 | 46.74 | 10.7 | |||
| TT | 46 | 45.46 | 12.26 | |||
Table 6.
Statistical analysis for single SNP to evaluate the correlation between the genotypes and SDS score.
| Gene | SNP | Genotype | All subjects (n = 127) | |||
|---|---|---|---|---|---|---|
| N | Mean | SD | P for trend | |||
| BCR | rs140504 | AA | 35 | 41.63 | 8.71 | 0.218 |
| AG | 64 | 43.8 | 8.36 | |||
| GG | 28 | 44.36 | 10.7 | |||
| BCR | rs131690 | AA | 73 | 43.4 | 8.7 | 0.899 |
| AG | 46 | 42.91 | 9.54 | |||
| GG | 8 | 45 | 9.55 | |||
| BCR | rs131702 | GG | 18 | 45.11 | 9.71 | 0.169 |
| GT | 64 | 43.7 | 9.24 | |||
| TT | 44 | 41.89 | 8.41 | |||
| SLC6A15 | rs1545843 | AA | 15 | 42.53 | 9.4 | 0.381 |
| AG | 58 | 44.76 | 8.98 | |||
| GG | 54 | 42 | 8.86 | |||
| ADCYAP1R1 | rs2267735 | CC | 27 | 42.59 | 9.06 | 0.229 |
| CG | 70 | 42.71 | 8.9 | |||
| GG | 30 | 45.4 | 9.18 | |||
| SIRT1 | rs10997870 | GG | 85 | 43.34 | 9.28 | 0.537 |
| GT | 37 | 42.3 | 8.44 | |||
| TT | 5 | 50.6 | 5.32 | |||
| HTR2A | rs7997012 | AA | 9 | 39.44 | 9.29 | 0.666 |
| AG | 39 | 44.41 | 10.2 | |||
| GG | 79 | 43.23 | 8.31 | |||
| MAOB (male) | rs1799836 | C | 7 | 47.71 | 9.86 | 0.453 |
| T | 46 | 45.07 | 8.46 | |||
| MAOB (female) | rs1799836 | CC | 2 | 47.5 | 9.19 | 0.203 |
| CT | 23 | 43.13 | 8.29 | |||
| TT | 44 | 40.86 | 9.52 | |||
Multiple regression analysis was performed to assess whether the STAI-state score was a confounder between the THI score and SNP rs131702 of the BCR gene (Table 7). The THI score is coded as 1 = 56 and under and 2 = 58 and over. STAI-state is coded as 1 = 44 or under for male, 40 or under for female and 2 = 45 and over for male, 41 and over for female. The genotype of SNP rs131702 of BCR gene is coded as 0 = TT, 1 = GT, 2 = GG. The THI score was identified as the dependent variable and STAI-state and SNP rs131702 as independent variables. The regression coefficient associated with the rs131702 of BCR was 0.175 in the simple linear regression analysis. The association was statistically significant (p = 0.008). The association between THI score and SNP rs131702 of BCR was 0.147 after adjustment for the STAI-state score. SNP rs131702 of BCR retains a statistically significant association with THI score (p = 0.02), but the value of the regression coefficient decreased by 16.7%.
Table 7.
(a) The simple linear regression analysis for THI score and (b) the multiple regression analysis for THI score.
| Regression coefficient | P value | |
|---|---|---|
| (a) Independent variables | ||
| SNP rs131702 of BCR gene | 0.175566826 | 0.008047161 |
| STAI-state | 0.317073171 | 0.000621627 |
| (b) Independent variables | ||
| SNP rs131702 of BCR gene | 0.147354015 | 0.021741808 |
| STAI-state | 0.288321168 | 0.001639804 |
Discussion
We theorized there might be an association between the genetic background related to psychological factors and the grade of tinnitus distress, which might lead us to identify prognostic markers.
The BCR SNP rs131702 is found on chromosome 22. The BCR gene encodes the Rho family low molecular weight G-protein, which is abundantly expressed in the central nervous system and considered crucial for neurogenesis.
The results could support the suggestion that the BCR gene is implicated in the process of making tinnitus distress severe. The SDS score and the STAI-state score are known to contribute significantly to the THI score14,32. Therefore, in addition to the THI score, we analyzed the association between these two scores and each SNPs.
In 171 Japanese patients, bipolar depression and related disorders (odds ratio minor allele (G) 1.49, CI: 1.15–1.93, p = 0.0026) have been reported to be associated with rs131702 (https://www.snpedia.com/index.php/Rs131702).
In this study, no significant association between BCR rs131702 and the SDS score was detected, because the patients with mental illness were excluded.
In this study, a significant association between rs131702 of BCR and the severe THI score was recognized. rs131702 was independent of the SDS score and the STAI-trait score. But a significant association between rs131702 and the severe STAI-state score was identified. It could be considered that the STAI-state score affected the result of the correlation between the severe THI score and rs131702 as a confounding factor, as the STAI-state score has a significant association with both the THI score and rs131702. Therefore, multiple regression analysis was then performed to assess whether the STAI-state score is a confounder. Although the association between the severe THI score and rs131702 was significant even after adjustment for the STAI-state score, the value of the coefficient after adjustment decreased by about 16%. On the other hand, the coefficient of association between THI score and STAI-state score was almost the same even after adjustment for rs131702. It may indicate that the part of the association between the severe THI score and rs131702 of BCR can be explained by the STAI-state score, because the change by more than 10% of the regression coefficient in the multiple linear regression model indicates the influence of cofounders.
Although the STAI-state score was identified as cofounder between THI score and SNP rs131702 of the BCR gene, it could mean that the patients with SNP rs131702 of BCR gene have potential to suffer from more severe tinnitus. Therefore, it is considered a prognostic factor specific to tinnitus.
The study had several limitations. Because of the small sample size, our ability to evaluate other patient groups to confirm reproducibility was limited. Further, we evaluated only eight candidate genes. The likelihood of tinnitus being caused by multiple genes (susceptibility genes) is high. Therefore, future studies should examine more candidate genes. Finally, we limited our study to those with a minor allele frequency ≥ 0.1, but this could have eliminated a strongly associated SNP or a rare variant. Therefore, the sample size and number of genes evaluated would need to be increased in future studies.
Subjects and methods
This study was approved by the Ethics Committee of Keio University (UMIN000017306). The research was performed in accordance with clinical research guidelines in Japan, and informed consent was obtained from all patients.
Subjects
We enrolled patients who presented to the Department of Otolaryngology, Keio University Hospital, Japan, from January 2013 until April 2015, who complained of tinnitus continuing for over 3 months (the average duration was 56.2 ± 82.0 months) which is defined as chronic tinnitus in Japan16. After excluding patients with mental illness, a final cohort of 138 patients remained. These patients answered the validated Japanese version of THI33,34, SDS35,36, and STAI questionnaires37,38, and blood samples were drawn to investigate SNPs.
Genotyping
Genomic DNA samples were extracted from the peripheral blood of patients using a DNA extraction kit (Genomix, Biologica, Japan).
Genotyping of the eight SNPs (rs14050428, rs13169028, rs13170228, rs154584339, rs226773540, rs1099787030, rs799701229, and rs179983631) was performed by allelic discrimination analysis (Light cycler 480 system,Roche, Basel, Switzerland), using TaqMan SNP genotyping assay probes (Thermo Fisher Scientific, Waltham, USA. The genotyping call rate was 100%.
The correlation between a genetic background related to depression/anxiety and tinnitus distress levels was evaluated. The SNPs targeted in our study were determined to be correlated with depression, anxiety disorder, and obsessive–compulsive disorder, according to Online Mendelian Inheritance in Man (https://www.omim.org/). We found eight SNPs with a MAF more than or equal to 0.1 according to the HapMap for Japan (Table 8).
Table 8.
Candidate gene polymorphism.
| Gene | Chromosome | Polymorphism | Genotypes | Reported minor allele | Reported minor allele frequency (JPT) |
|---|---|---|---|---|---|
| BCR | 22 | rs140504 | GG/GA/AA | G | 0.500* |
| BCR | 22 | rs131690 | GG/GA/AA | G | 0.235* |
| BCR | 22 | rs131702 | GG/GA/AA | G | 0.407* |
| SLC6A15 | 12 | rs1545843 | AA/AG/GG | A | 0.367* |
| ADCYAP1R1 | 7 | rs2267735 | GG/GC/CC | C | 0.479* |
| SIRT1 | 10 | rs10997870 | TT/TG/GG | T | 0.149* |
| HTR2A | 13 | rs7997012 | AA/AG/GG | A | 0.173+ |
| MAOB (male) | X | rs1799836 | C/T | C | 0.147# |
| MAOB (female) | rs1799836 | CC/CT/TT |
*Integrative Japanese Genome Variation Database, +1000 Genomes: phase_3 (JPT), #Hapmap_JPT.
The target SNPs were analyzed using the TaqMan method.
In this study, a THI score of less than 56 was considered light/moderate, and a score greater than or equal to 58 was considered severe33. Both trait anxiety (STAI-trait) and state anxiety (STAI-state) are evaluated in the STAI. High trait anxiety implies a remarkable tendency to respond with anxiety to perceived threats in the environment, and high state anxiety implies a transitory high symptom of anxiety. The depression level is measured by the SDS. Higher score of the SDS indicates a severer depressive condition. Statistical analysis was performed using Fisher’s exact test (software PLINK v1.07 https://zzz.bwh.harvard.edu/plink/index.shtml).
Furthermore, the association between SNPs and the severity of THI, SDS, and STAI was evaluated using the Jonckheere trend test. In each of the above tests, the significance level was set at 5%.
Acknowledgements
This study was supported by a Grant from the Ministry of Health, Labour, and Welfare (Japan).
Author contributions
T.W.: planning, data collection, analyzing, writing, S.K.: planning, data collection, analyzing, writing, editing, T.M.: planning, DNA extraction, N.S. and M.M.: planning, genotyping and analyzing, K.O.: planning, data collection.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Tunkel DE, et al. Clinical practice guideline: Tinnitus executive summary. Otolaryngol. Head Neck Surg. 2014;151:533–541. doi: 10.1177/0194599814547475. [DOI] [PubMed] [Google Scholar]
- 2.Konig O, Schaette R, Kempter R, Gross M. Course of hearing loss and occurrence of tinnitus. Hear. Res. 2006;221:59–64. doi: 10.1016/j.heares.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 3.De Ridder D, Elgoyhen AB, Romo R, Langguth B. Phantom percepts: Tinnitus and pain as persisting aversive memory networks. Proc. Natl. Acad. Sci. USA. 2011;108:8075–8080. doi: 10.1073/pnas.1018466108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Ridder D, et al. An integrative model of auditory phantom perception: Tinnitus as a unified percept of interacting separable subnetworks. Neurosci. Biobehav. Rev. 2014;44:16–32. doi: 10.1016/j.neubiorev.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 5.Rauschecker JP, Leaver AM, Muhlau M. Tuning out the noise: Limbic–auditory interactions in tinnitus. Neuron. 2010;66:819–826. doi: 10.1016/j.neuron.2010.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schecklmann M, et al. Neural correlates of tinnitus duration and distress: A positron emission tomography study. Hum. Brain Mapp. 2013;34:233–240. doi: 10.1002/hbm.21426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanneste S, Joos K, De Ridder D. Prefrontal cortex based sex differences in tinnitus perception: Same tinnitus intensity, same tinnitus distress, different mood. PLoS One. 2012;7:e31182. doi: 10.1371/journal.pone.0031182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elgoyhen AB, Langguth B, De Ridder D, Vanneste S. Tinnitus: Perspectives from human neuroimaging. Nat. Rev. Neurosci. 2015;16:632–642. doi: 10.1038/nrn4003. [DOI] [PubMed] [Google Scholar]
- 9.Hallam, R. Psychological aspects of tinnitus. In Contributions to Medical Psychology. Vol. 84 (Oxford, 1984).
- 10.Jastreboff PJ. Phantom auditory perception (tinnitus): Mechanisms of generation and perception. Neurosci. Res. 1990;8:221–254. doi: 10.1016/0168-0102(90)90031-9. [DOI] [PubMed] [Google Scholar]
- 11.McKenna L, Handscomb L, Hoare DJ, Hall DA. A scientific cognitive-behavioral model of tinnitus: Novel conceptualizations of tinnitus distress. Front. Neurol. 2014;5:196. doi: 10.3389/fneur.2014.00196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cima RF, Crombez G, Vlaeyen JW. Catastrophizing and fear of tinnitus predict quality of life in patients with chronic tinnitus. Ear Hear. 2011;32:634–641. doi: 10.1097/AUD.0b013e31821106dd. [DOI] [PubMed] [Google Scholar]
- 13.Kleinstauber M, et al. The role of fear-avoidance cognitions and behaviors in patients with chronic tinnitus. Cogn. Behav. Ther. 2013;42:84–99. doi: 10.1080/16506073.2012.717301. [DOI] [PubMed] [Google Scholar]
- 14.Jastreboff PJ, Hazell JW. A neurophysiological approach to tinnitus: Clinical implications. Br. J. Audiol. 1993;27:7–17. doi: 10.3109/03005369309077884. [DOI] [PubMed] [Google Scholar]
- 15.Shore SE, Roberts LE, Langguth B. Maladaptive plasticity in tinnitus–triggers, mechanisms and treatment. Nat. Rev. Neurol. 2016;12:150–160. doi: 10.1038/nrneurol.2016.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ogawa K, et al. Clinical practice guidelines for diagnosis and treatment of chronic tinnitus in Japan. Auris Nasus Larynx. 2019 doi: 10.1016/j.anl.2019.09.007. [DOI] [PubMed] [Google Scholar]
- 17.Cima RFF, et al. A multidisciplinary European guideline for tinnitus: Diagnostics, assessment, and treatment. Hno. 2019;67:10–42. doi: 10.1007/s00106-019-0633-7. [DOI] [PubMed] [Google Scholar]
- 18.Turner JG, et al. Gap detection deficits in rats with tinnitus: A potential novel screening tool. Behav. Neurosci. 2006;120:188–195. doi: 10.1037/0735-7044.120.1.188. [DOI] [PubMed] [Google Scholar]
- 19.Boyen K, Baskent D, van Dijk P. The gap detection test: Can it be used to diagnose tinnitus? Ear Hear. 2015;36:e138–145. doi: 10.1097/AUD.0000000000000156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sand PG, Langguth PG, Schecklmann M, Kleinjung T. GDNF and BDNF gene interplay in chronic tinnitus. Int. J. Mol. Epidemiol. Genet. 2012;3:245–251. [PMC free article] [PubMed] [Google Scholar]
- 21.Goto F, et al. Various levels of plasma brain-derived neurotrophic factor in patients with tinnitus. Neurosci. Lett. 2012;510:73–77. doi: 10.1016/j.neulet.2012.01.001. [DOI] [PubMed] [Google Scholar]
- 22.Xiong H, et al. Plasma brain-derived neurotrophic factor levels are increased in patients with tinnitus and correlated with therapeutic effects. Neurosci. Lett. 2016;622:15–18. doi: 10.1016/j.neulet.2016.04.032. [DOI] [PubMed] [Google Scholar]
- 23.Coskunoglu A, et al. Evidence of associations between brain-derived neurotrophic factor (BDNF) serum levels and gene polymorphisms with tinnitus. Noise Health. 2017;19:140–148. doi: 10.4103/nah.NAH_74_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sand PG, et al. Resequencing of the auxiliary GABA(B) receptor subunit gene KCTD12 in chronic tinnitus. Front. Syst. Neurosci. 2012;6:41. doi: 10.3389/fnsys.2012.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deniz M, et al. Significance of serotonin transporter gene polymorphism in tinnitus. Otol. Neurotol. 2010;31:19–24. doi: 10.1097/MAO.0b013e3181c2dcbc. [DOI] [PubMed] [Google Scholar]
- 26.Gallant E, et al. Novel COCH mutation in a family with autosomal dominant late onset sensorineural hearing impairment and tinnitus. Am. J. Otolaryngol. 2013;34:230–235. doi: 10.1016/j.amjoto.2012.11.002. [DOI] [PubMed] [Google Scholar]
- 27.Gilles A, Van Camp G, Van de Heyning P, Fransen E. A pilot genome-wide association study identifies potential metabolic pathways involved in tinnitus. Front. Neurosci. 2017;11:71. doi: 10.3389/fnins.2017.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hashimoto R, et al. The breakpoint cluster region gene on chromosome 22q11 is associated with bipolar disorder. Biol. Psychiatry. 2005;57:1097–1102. doi: 10.1016/j.biopsych.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 29.McMahon FJ, et al. Variation in the gene encoding the serotonin 2A receptor is associated with outcome of antidepressant treatment. Am. J. Hum. Genet. 2006;78:804–814. doi: 10.1086/503820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Libert S, et al. SIRT1 activates MAO-A in the brain to mediate anxiety and exploratory drive. Cell. 2011;147:1459–1472. doi: 10.1016/j.cell.2011.10.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dlugos AM, Palmer AA, de Wit H. Negative emotionality: Monoamine oxidase B gene variants modulate personality traits in healthy humans. J. Neural. Transm. (Vienna) 2009;116:1323–1334. doi: 10.1007/s00702-009-0281-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oishi N, et al. Influence of depressive symptoms, state anxiety, and pure-tone thresholds on the tinnitus handicap inventory in Japan. Int. J. Audiol. 2011;50:491–495. doi: 10.3109/14992027.2011.560904. [DOI] [PubMed] [Google Scholar]
- 33.Newman CW, Jacobson GP, Spitzer JB. Development of the Tinnitus Handicap Inventory. Arch. Otolaryngol. Head Neck Surg. 1996;122:143–148. doi: 10.1001/archotol.1996.01890140029007. [DOI] [PubMed] [Google Scholar]
- 34.Shinden S, Ogawa K, Inoue Y, Tazoe M, Asano K. Methods for assessing the level of distress and difficulty in activities of daily living due to tinnitus. Audiol. Jpn. 2002;45:685–691. doi: 10.4295/audiology.45.685. [DOI] [Google Scholar]
- 35.Zung W. A self-rating depression scale. Arch. Gen. Psychiatry. 1965;12:63–70. doi: 10.1001/archpsyc.1965.01720310065008. [DOI] [PubMed] [Google Scholar]
- 36.Zung, W., Fukuda, K., Kobayashi, S. & Shiyoutebiki, SDS. (Sankyobo, Kyoto, 1983).
- 37.Spielberger C. State-Trait Anxiety Inventory: A Comprehensive Bibliography. California: Consulting Psychologists Press; 1984. [Google Scholar]
- 38.Hidano T, Fukuhara M, Iwawaki M, Soga S, Spielberger C. State-Trait Anxiety Inventory-JYZ. Tokyo: Jitsumukyouiku; 2000. [Google Scholar]
- 39.Kohli MA, et al. The neuronal transporter gene SLC6A15 confers risk to major depression. Neuron. 2011;70:252–265. doi: 10.1016/j.neuron.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ressler KJ, et al. Post-traumatic stress disorder is associated with PACAP and the PAC1 receptor. Nature. 2011;470:492–497. doi: 10.1038/nature09856. [DOI] [PMC free article] [PubMed] [Google Scholar]